Gbib auto updates: Difference between revisions
No edit summary |
(Rewording sentence to emphasize that hgdownload just has the data for GBiB) |
||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
Jorge's Sunday update of hgdownload is run by /root/mirror-mysql-apache-gbdb. At the end, it runs the command "touch /mirrordata/gbib/lastUpdate". | Jorge's Sunday update of hgdownload is run by /root/mirror-mysql-apache-gbdb. At the end, it runs the command "touch /mirrordata/gbib/lastUpdate". | ||
By default the Gbib update script /root/updateBrowser.sh checks every few minutes if | By default the Gbib update script /root/updateBrowser.sh checks every few minutes if the flag file on the hgDownload server http://hgdownload.soe.ucsc.edu/gbib/lastUpdate is more recent than the date of the last update of the Gbib. This check is randomly spread over 30 seconds (crontab + /root/cronUpdate.sh). | ||
Auto updates can be deactivated on a box with autoUpdateOff and reactivated with autoUpdateOn. | Auto updates can be deactivated on a box with autoUpdateOff and reactivated with autoUpdateOn. | ||
| Line 20: | Line 20: | ||
Note that if you have downloaded other binaries than the release ones, then the box will loose them by auto-updating to release state again on the next Sunday, when hgdownload is updated. If you do not want to have your special CGIs/htdocs overwritten, deactivate auto updates on your own gbib. Auto updates can be deactivated on a box with autoUpdateOff and reactivated with autoUpdateOn. You can also run '''VBoxManage guestproperty set browserbox gbibAutoUpdateOff yes''' more [http://genomewiki.ucsc.edu/genecats/index.php/GBiB_Testing#Other here.] | Note that if you have downloaded other binaries than the release ones, then the box will loose them by auto-updating to release state again on the next Sunday, when hgdownload is updated. If you do not want to have your special CGIs/htdocs overwritten, deactivate auto updates on your own gbib. Auto updates can be deactivated on a box with autoUpdateOff and reactivated with autoUpdateOn. You can also run '''VBoxManage guestproperty set browserbox gbibAutoUpdateOff yes''' more [http://genomewiki.ucsc.edu/genecats/index.php/GBiB_Testing#Other here.] | ||
See this [http://genomewiki.ucsc.edu/genecats/index.php/Gbib_updates#updateBrowser_hgwdev_USER_RELEASE lastUpdate file] note about how if you have a beta candidate, after the beta week, the Sunday touch of hgdownload files will update the date of lastUpdate and trigger your beta to update to the version on hgdownload. | |||
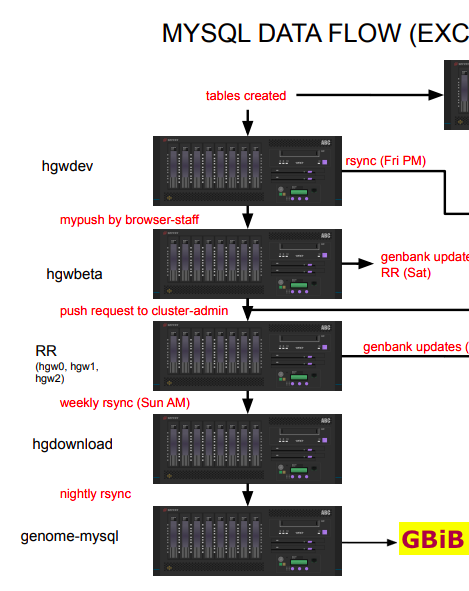
Here is also a picture of the MySQL data flow. | |||
[[File:MySQL_GBiB.png|alt=GBiB_MySQL_flow]] | |||
The flow of other GBiB data (/gbdb /cgi-bin /htdocs) is similar, where the data ends up on hgdownload. The following rsync command reflects how the data exists at hgdownload: | |||
<pre> | |||
rsync rsync://hgdownload.soe.ucsc.edu | |||
genome UCSC Human Genome Downloads | |||
sars UCSC Human Genome SARS Downloads | |||
htdocs UCSC Human Genome Web Site Htdocs | |||
goldenPath UCSC Human Golden Path Downloads | |||
cgi-bin UCSC Human Genome Web Site CGI Binaries x86_64 | |||
cgi-bin-i386 UCSC Human Genome Web Site CGI Binaries i386 | |||
gbdb UCSC Human Genome Browser Gbdb Config Files | |||
archives UCSC Human Genome Browser Archived Config Files | |||
mysql UCSC Human Genome Raw Mysql Tables | |||
gbib UCSC Genome Browser in a Box | |||
hubs UCSC Genome Browser Public Hubs | |||
</pre> | |||
[[Category:GBiB]] | [[Category:GBiB]] | ||
[[Category:Browser QA]] | [[Category:Browser QA]] | ||
[[Category:Browser Development]] | [[Category:Browser Development]] | ||
Latest revision as of 21:14, 9 December 2020
Jorge's Sunday update of hgdownload is run by /root/mirror-mysql-apache-gbdb. At the end, it runs the command "touch /mirrordata/gbib/lastUpdate".
By default the Gbib update script /root/updateBrowser.sh checks every few minutes if the flag file on the hgDownload server http://hgdownload.soe.ucsc.edu/gbib/lastUpdate is more recent than the date of the last update of the Gbib. This check is randomly spread over 30 seconds (crontab + /root/cronUpdate.sh).
Auto updates can be deactivated on a box with autoUpdateOff and reactivated with autoUpdateOn.
By supplying the parameter "notSelf" to updateBrowser, the box will skip this check and update without checking the flag file.
If the file on hgDownload is newer than the flagfile, it will start the update (/root/updateBrowser.sh):
- Update the update script itself from hgdownload.soe.ucsc.edu/gbib
- Update all existing /gbdb files
- Update all mysql files
- Update all hgcentral files + patch the blat server table
- Patch the menu to remove hgNear and add hgMirror
- Place the symlink to /media from htdocs/folders
- Run hgMirror to hide some tracks on hg19 and remove some searches from hg19
updateBrowser.sh also accepts an option: "hgwdev <hgwdevUsername> <cgiSuffix>". These pull the CGIs from hgwdev into the current gbib. "updateBrowser hgwdev" shows some more help and examples.
Note that if you have downloaded other binaries than the release ones, then the box will loose them by auto-updating to release state again on the next Sunday, when hgdownload is updated. If you do not want to have your special CGIs/htdocs overwritten, deactivate auto updates on your own gbib. Auto updates can be deactivated on a box with autoUpdateOff and reactivated with autoUpdateOn. You can also run VBoxManage guestproperty set browserbox gbibAutoUpdateOff yes more here.
See this lastUpdate file note about how if you have a beta candidate, after the beta week, the Sunday touch of hgdownload files will update the date of lastUpdate and trigger your beta to update to the version on hgdownload.
Here is also a picture of the MySQL data flow.
The flow of other GBiB data (/gbdb /cgi-bin /htdocs) is similar, where the data ends up on hgdownload. The following rsync command reflects how the data exists at hgdownload:
rsync rsync://hgdownload.soe.ucsc.edu genome UCSC Human Genome Downloads sars UCSC Human Genome SARS Downloads htdocs UCSC Human Genome Web Site Htdocs goldenPath UCSC Human Golden Path Downloads cgi-bin UCSC Human Genome Web Site CGI Binaries x86_64 cgi-bin-i386 UCSC Human Genome Web Site CGI Binaries i386 gbdb UCSC Human Genome Browser Gbdb Config Files archives UCSC Human Genome Browser Archived Config Files mysql UCSC Human Genome Raw Mysql Tables gbib UCSC Genome Browser in a Box hubs UCSC Genome Browser Public Hubs