VariantAnnotationTool: Difference between revisions
(→Input) |
(→High-level interfaces: annoGrator.h: Updating with more details about core objects.) |
||
| (37 intermediate revisions by 3 users not shown) | |||
| Line 19: | Line 19: | ||
== Novel variant calls from sequencing experiment == | == Novel variant calls from sequencing experiment == | ||
The user gets a bunch of short reads and uses a commonly available NGS pipeline to align short reads to the genome and report discovered variants. The pipeline produces a big file of genomic positions and observed variant alleles. | The user gets a bunch of short reads and uses a commonly available NGS pipeline to align short reads to the genome and report discovered variants. The pipeline produces a big VCF file of genomic positions and observed variant alleles. | ||
Now they're wondering which variants are the interesting ones. They want to upload the file that their pipeline spit out, and get back some clue about which variants might have a functional effect. | Now they're wondering which variants are the interesting ones. They want to upload the file that their pipeline spit out, and get back some clue about which variants might have a functional effect. | ||
| Line 26: | Line 26: | ||
The user reads a paper that lists ~20 SNPs associated with some trait, and wants to know more about them: coding? conserved? etc. | The user reads a paper that lists ~20 SNPs associated with some trait, and wants to know more about them: coding? conserved? etc. | ||
== User gets BAM file from seq facility == | |||
The user is a PI in a lab and the seq data come to her in a BAM file. Would likle to see the read depth as a custom track and also the amino acid diffs for the non-reference alleles in a Custom Track. It could be we simply show any DNA base that does not meatch (even it only one of ten reads), and not try tomake a judgement on how likely it is a real diff. | |||
== Command line mode == | == Command line mode == | ||
Standalone binary takes | Standalone binary takes a config file in the .ra format on the command line. The config.ra file contains stanzas describing inputs (file names, database tables etc.; filters; fields to include in output) and outputs (format, filename, options). | ||
= Implementation plan = | |||
Most of the code will be in library modules. We will develop a command-line tool first, with web UI design in parallel, and then will implement the web UI. This allows more time for web UI design, and a command-line interface makes it easier to develop automated tests for the lib modules. Also, power-users will probably ask for a command-line tool. | |||
== Shared git branch == | |||
Initial prototyping is underway in a shared branch in hgwdev's central repository. To check out your own local branch that tracks the shared branch, do this: | |||
<pre> | |||
# Update your local git data -- git pull does this too: | |||
git fetch | |||
# Make your own local annoGrator branch: | |||
git checkout --track -b annoGrator origin/annoGrator | |||
# See what has been done since this branch was started: | |||
git log --stat 010ad06e.. | |||
# Return to your main branch: | |||
git checkout master | |||
</pre> | |||
Since your local branch was created with "--track", git pull from within your local branch will use the shared branch origin/annoGrator instead of the main shared branch origin/master. But watch out for git push! That still wants to push both your local annoGrator branch and your master branch! So to be safe, do a git push like this: | |||
<pre> | |||
git push origin annoGrator | |||
</pre> | |||
== High-level interfaces: anno*.h == | |||
Several new library modules, anno*.[ch], will perform the core functions: | |||
=== annoRow === | |||
The basic unit of data interchange between modules: a genomic position plus an array of strings that correspond to columns defined in the data source's autoSql description. | |||
=== annoColumn === | |||
For communication with UI: a column's autoSql definition, and a boolean flag for whether this column should appear in the output (e.g. as a column in tab-separated output, or an attribute in VCF output). | |||
=== annoFilter === | |||
For communication with UI: a specification of a filter on a column's data values: the column's autoSql definition, filtering operation, and current values (threshold, range or wildcard pattern). Also has a flag for "right join" (a la SQL) behavior: i.e. if this filter fails on a secondary table, do we filter out the primary table's variant too, or simply ignore this secondary table row? | |||
=== annoStreamer === | |||
Interface to data source: get autoSql description, get/set filters and columns, get next row of data. Subclasses of this handle details such as whether the data come from a db table, file, etc. | |||
=== annoGrator === | |||
Integrates each row of data from the primary source with data from an internal source, returning zero or more rows of data that overlap the primary source's row. Same external interface as annoStreamer, except nextRow() is replaced by integrate(). Subclasses of this can add integrated data columns, for example a module that integrates variants and genePreds also outputs predicted functional effects. | |||
=== annoFormatterOption === | |||
This is how configuration parameters are passed to annoFormatter: it is an optionSpec plus a value. | |||
=== annoFormatter === | |||
Subclasses of this write output, as tab-separated text, bigBed, custom track, HTML summary etc. | |||
=== annoGratorQuery === | |||
A complete description of a query, constructed from a primary source (annoStreamer), 0 or more annoGrators, and 1 or more annoFormatters. Call annoGratorQueryNew(), annoGratorQuerySetRegion(), annoGratorQueryExecute() and annoGratorQueryFree(). | |||
== Input modules == | |||
annoStreamer subclasses, in rough order of importance/implementation: | |||
* annoStreamDb | |||
* annoStreamTabFile | |||
* annoStreamTabix | |||
* annoStreamBigBed | |||
* annoStreamWig (including bedGraph, bigWig) | |||
* annoStreamBam | |||
== Integrators == | |||
annoGrator and its subclasses; each contains a streamer plus integration method. | |||
* annoGrator (base class: just intersect by position and keep fields intact) | |||
* annoGrateGenePredVariant (predict functional changes!) | |||
== Output formatters == | |||
= | * annoOutTab | ||
* annoOutVcf | |||
* annoOutBigBed | |||
* annoOutCustomTrack | |||
* annoOutSummary | |||
* annoOutRanking | |||
== Who's going to do what? == | |||
Angie and Brian will divvy up the work. For starters, Angie is working on basic annoStreamers, annoFormatters and the generic (data-agnostic position-joiner) annoGrator. Brian is working on functional annotation given a variant {genomic position, observed alleles}, genePred and reference transcript sequence. Remaining modules TBD. | |||
= Features = | = Features = | ||
| Line 42: | Line 138: | ||
== Input == | == Input == | ||
The primary input will be variant calls: fundamentally, genomic position plus observed alleles. | |||
* pgSnp | * pgSnp | ||
* VCF | * VCF | ||
* Other formats, e.g. outputs of popular NGS pipelines? | * Other formats, e.g. outputs of popular NGS pipelines? | ||
** 23andMe? :) | ** 23andMe? :) | ||
** maybe eventually BAM and pileup; but there are established, sophisticated variant callers for BAM, better to take variant calls from those tools. | |||
Other inputs will be annotations to relate to the variant calls. These annotations may be stored in database tables, bigData files, flat files; they might be found in trackDb, custom tracks, or hubs. For TCGA and the cancer group, we need to support the Generic Annotation Format (GAF). | |||
== Types of Variation == | |||
The architecture must support all known forms of variation with respect to a reference assembly. Initially, the implementation will support single nucleotide variants only. We will work our way up through multi-nucleotide variants, small indels, and ultimately large-scale rearrangements. | |||
== Output == | == Output == | ||
* tab-separated file | * tab-separated file with all/selected fields (with BED+ as an option) | ||
* bigBed with embedded autoSql to define extra columns | |||
* custom track in GB/TB | * VCF with added INFO column tags | ||
* intermediate level to summarize and filter findings | * custom track in GB/TB | ||
** display should show any AA diff from ref. | |||
** probably should show two alleles if input in heterozygous SNPs - can be simply two BED boxes. | |||
** coloring of diffs in amino-acid space, anyway, can use "different codons" as we do with mRNAs now. Should show amino acids downstream of a frameshift in yellow all the way to any in-frame stop. If user is looking at a window on the gene that does not include the actual variant, she is going to want to know that the protein is messed up. | |||
* intermediate level to summarize, sort / rank, and filter findings | |||
* ?highlight in multiple alignment? | |||
* ?ancestral polarization? | |||
* ?binding motif disruption? | |||
== Interface == | == Interface == | ||
=== | === Web UI === | ||
This is still wide open -- everyone's input would be very much appreciated! | |||
==== Main page ==== | ==== Main page ==== | ||
| Line 81: | Line 191: | ||
** ...? | ** ...? | ||
=== command line === | === Command line === | ||
Since this tool is so highly configurable, instead of cramming so many options into command line arguments, the tool will read a configuration file (possibly stdin) in .ra format. It might look something like this: | |||
<pre> | |||
primarySource ct_myVars | |||
sourceType customTrack | |||
dataType pgSnp | |||
filterSpecs filter1,filter2 | |||
outFields allButBin | |||
filter filter1 | |||
alleleCount == 2 | |||
filter filter2 | |||
alleleFreq noMatch 0,0 | |||
source snp135Common | |||
sourceType dbTrack | |||
dataType bed 6 + | |||
outFields chrom,chromStart,chromEnd,name,strand,observed,exceptions,alleleFreqCount,alleles,alleleNs | |||
source pubMatches | |||
sourceType dbTrack | |||
dataType bed12 | |||
outFields name | |||
outputFormat tabSep | |||
fileName ./myAnnotatedVars.txt.gz | |||
</pre> | |||
Then the tool might be invoked like this: | |||
<pre> | |||
annoGrate hg19 config.ra | |||
</pre> | |||
= Name = | |||
Not variant Effect Predictor or other names in use. | |||
* Variantizor? | |||
* Predictorator? | |||
* Diffmeister? | |||
* Differizerator? | |||
* Variant Annotator | |||
* Varannosaurus Rex | |||
* Varannozilla | |||
* Global Variant Annotator | |||
* Integrated Variant Annotator (except rings of Broad IGV) | |||
* Variant Annotation Integrator | |||
* Variant Annotatoon Tool ( except VAT is not good acronym) | |||
* VarAnnoGrator | |||
= Links to Similar Tools = | |||
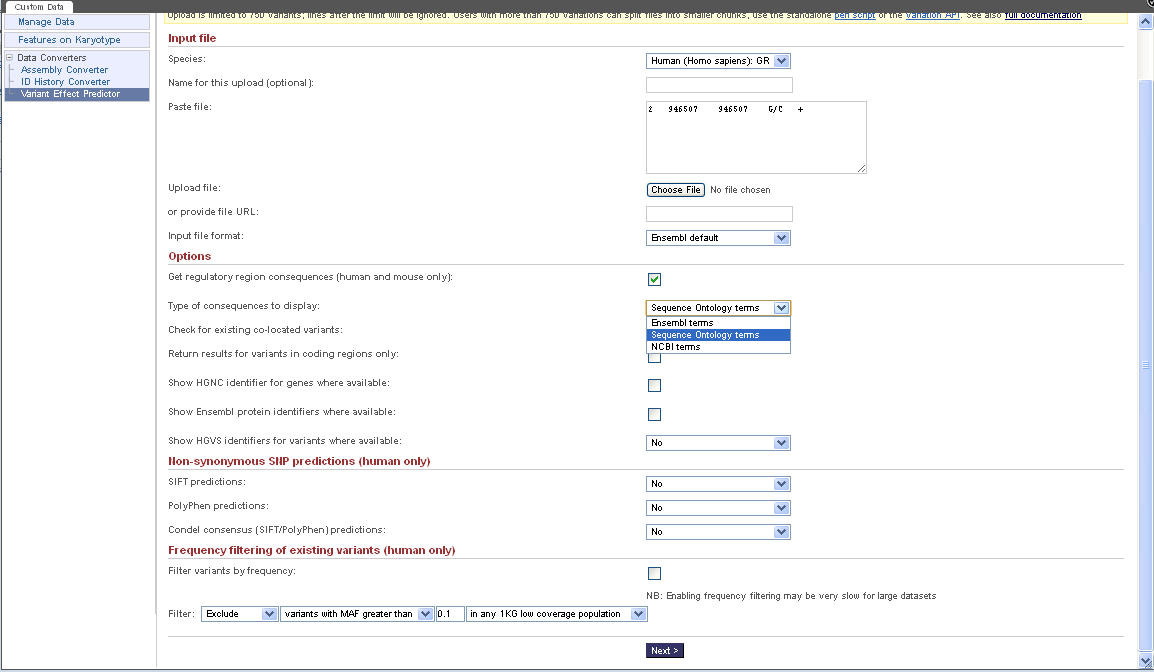
http://uswest.ensembl.org/info/docs/variation/vep/index.html | |||
http://snpeff.sourceforge.net/faq.html | |||
http://www.ncbi.nlm.nih.gov/variation/tools/reporter | |||
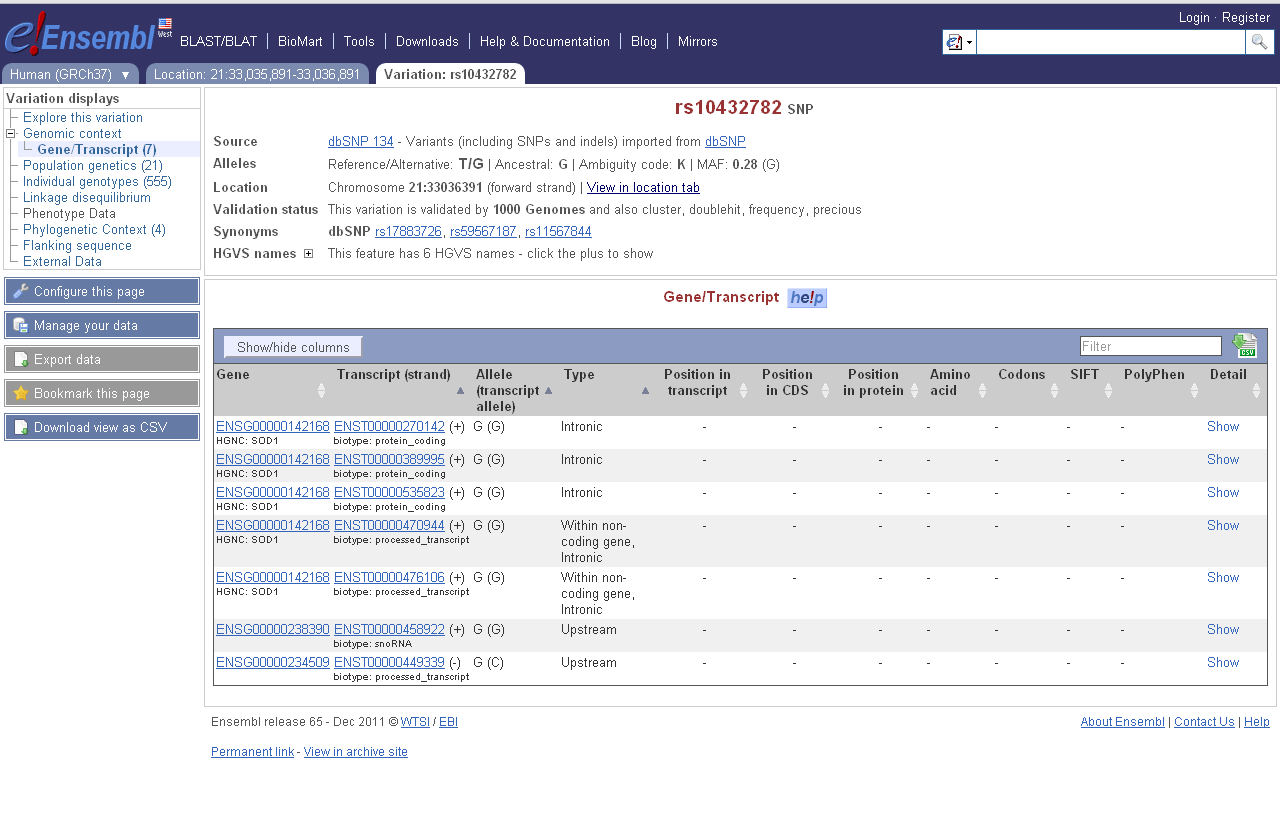
= Screen Shots of Other Tools = | |||
[[File:EnsemblTranscriptSnp.png]] | |||
[[File:EnsemblVariantPredictorFron.png]] | |||
Latest revision as of 17:35, 12 March 2012
Introduction
This is a design document for a proposed variant annotation tool, Feature #6152 in redmine.
Numerous existing tools combine variant calls and gene annotations into predicted functional effects of SNPs. For example, an A/G variant at hg19 chr1:15687059-15687059 (rs4661330) is coding-non-synonymous: having an A makes the 919th codon of canonical FHAD1 code for E (GAA), while having a G makes it code for peptide G (GGA). For alt-splice variants, the same variant falls in the 172nd, 107th or 66th codon. Another variant might be intergenic, another may fall in the UTR or splice site of a gene, another variant may be coding but synonymous, etc.
Our tool will of course produce protein-coding effect predictions such as those, but since we have such a rich annotation database, we can relate a variant to many types of data beyond protein-coding gene annotations.
Use cases
MLQ requests
MLQ #3582 "Ideally I'd like to submit a list of variations (in BED format), the reference genome (hg18/19) and get type of effect back"
MLQ #5242 note 17: user has (chr, coord, strand); wants ref. base, known SNP data if any, refseq codon if any
MLQ #6294: "I have a list of genomic positions for putative mutations that I would like to convert into mutant mRNA and peptide sequences. Could you recommend an automated way to do this?"
Novel variant calls from sequencing experiment
The user gets a bunch of short reads and uses a commonly available NGS pipeline to align short reads to the genome and report discovered variants. The pipeline produces a big VCF file of genomic positions and observed variant alleles.
Now they're wondering which variants are the interesting ones. They want to upload the file that their pipeline spit out, and get back some clue about which variants might have a functional effect.
List of rs IDs
The user reads a paper that lists ~20 SNPs associated with some trait, and wants to know more about them: coding? conserved? etc.
User gets BAM file from seq facility
The user is a PI in a lab and the seq data come to her in a BAM file. Would likle to see the read depth as a custom track and also the amino acid diffs for the non-reference alleles in a Custom Track. It could be we simply show any DNA base that does not meatch (even it only one of ten reads), and not try tomake a judgement on how likely it is a real diff.
Command line mode
Standalone binary takes a config file in the .ra format on the command line. The config.ra file contains stanzas describing inputs (file names, database tables etc.; filters; fields to include in output) and outputs (format, filename, options).
Implementation plan
Most of the code will be in library modules. We will develop a command-line tool first, with web UI design in parallel, and then will implement the web UI. This allows more time for web UI design, and a command-line interface makes it easier to develop automated tests for the lib modules. Also, power-users will probably ask for a command-line tool.
Initial prototyping is underway in a shared branch in hgwdev's central repository. To check out your own local branch that tracks the shared branch, do this:
# Update your local git data -- git pull does this too: git fetch # Make your own local annoGrator branch: git checkout --track -b annoGrator origin/annoGrator # See what has been done since this branch was started: git log --stat 010ad06e.. # Return to your main branch: git checkout master
Since your local branch was created with "--track", git pull from within your local branch will use the shared branch origin/annoGrator instead of the main shared branch origin/master. But watch out for git push! That still wants to push both your local annoGrator branch and your master branch! So to be safe, do a git push like this:
git push origin annoGrator
High-level interfaces: anno*.h
Several new library modules, anno*.[ch], will perform the core functions:
annoRow
The basic unit of data interchange between modules: a genomic position plus an array of strings that correspond to columns defined in the data source's autoSql description.
annoColumn
For communication with UI: a column's autoSql definition, and a boolean flag for whether this column should appear in the output (e.g. as a column in tab-separated output, or an attribute in VCF output).
annoFilter
For communication with UI: a specification of a filter on a column's data values: the column's autoSql definition, filtering operation, and current values (threshold, range or wildcard pattern). Also has a flag for "right join" (a la SQL) behavior: i.e. if this filter fails on a secondary table, do we filter out the primary table's variant too, or simply ignore this secondary table row?
annoStreamer
Interface to data source: get autoSql description, get/set filters and columns, get next row of data. Subclasses of this handle details such as whether the data come from a db table, file, etc.
annoGrator
Integrates each row of data from the primary source with data from an internal source, returning zero or more rows of data that overlap the primary source's row. Same external interface as annoStreamer, except nextRow() is replaced by integrate(). Subclasses of this can add integrated data columns, for example a module that integrates variants and genePreds also outputs predicted functional effects.
annoFormatterOption
This is how configuration parameters are passed to annoFormatter: it is an optionSpec plus a value.
annoFormatter
Subclasses of this write output, as tab-separated text, bigBed, custom track, HTML summary etc.
annoGratorQuery
A complete description of a query, constructed from a primary source (annoStreamer), 0 or more annoGrators, and 1 or more annoFormatters. Call annoGratorQueryNew(), annoGratorQuerySetRegion(), annoGratorQueryExecute() and annoGratorQueryFree().
Input modules
annoStreamer subclasses, in rough order of importance/implementation:
- annoStreamDb
- annoStreamTabFile
- annoStreamTabix
- annoStreamBigBed
- annoStreamWig (including bedGraph, bigWig)
- annoStreamBam
Integrators
annoGrator and its subclasses; each contains a streamer plus integration method.
- annoGrator (base class: just intersect by position and keep fields intact)
- annoGrateGenePredVariant (predict functional changes!)
Output formatters
- annoOutTab
- annoOutVcf
- annoOutBigBed
- annoOutCustomTrack
- annoOutSummary
- annoOutRanking
Who's going to do what?
Angie and Brian will divvy up the work. For starters, Angie is working on basic annoStreamers, annoFormatters and the generic (data-agnostic position-joiner) annoGrator. Brian is working on functional annotation given a variant {genomic position, observed alleles}, genePred and reference transcript sequence. Remaining modules TBD.
Features
Like many existing tools, we will report the variants' effects on genes (splice-3, coding-non-synon etc.).
UCSC's major enhancements will be
- the incorporation of the many types of data in our database
- presentation of the results: not just loads of data, but also links to browser views
Input
The primary input will be variant calls: fundamentally, genomic position plus observed alleles.
- pgSnp
- VCF
- Other formats, e.g. outputs of popular NGS pipelines?
- 23andMe? :)
- maybe eventually BAM and pileup; but there are established, sophisticated variant callers for BAM, better to take variant calls from those tools.
Other inputs will be annotations to relate to the variant calls. These annotations may be stored in database tables, bigData files, flat files; they might be found in trackDb, custom tracks, or hubs. For TCGA and the cancer group, we need to support the Generic Annotation Format (GAF).
Types of Variation
The architecture must support all known forms of variation with respect to a reference assembly. Initially, the implementation will support single nucleotide variants only. We will work our way up through multi-nucleotide variants, small indels, and ultimately large-scale rearrangements.
Output
- tab-separated file with all/selected fields (with BED+ as an option)
- bigBed with embedded autoSql to define extra columns
- VCF with added INFO column tags
- custom track in GB/TB
- display should show any AA diff from ref.
- probably should show two alleles if input in heterozygous SNPs - can be simply two BED boxes.
- coloring of diffs in amino-acid space, anyway, can use "different codons" as we do with mRNAs now. Should show amino acids downstream of a frameshift in yellow all the way to any in-frame stop. If user is looking at a window on the gene that does not include the actual variant, she is going to want to know that the protein is messed up.
- intermediate level to summarize, sort / rank, and filter findings
- ?highlight in multiple alignment?
- ?ancestral polarization?
- ?binding motif disruption?
Interface
Web UI
This is still wide open -- everyone's input would be very much appreciated!
Main page
Form:
- paste/upload variants
- select annotation sources
- any track including custom tracks?
- select output format/presentation
- custom track in [Genome Browser | Table Browser]
- summary with filters
- mutant sequence (genomic, mRNA, protein)
- go!
followed by brief how-to and link to more detailed doc.
Summary/Filters
Stats: #variants, #variants intersecting each annotation source (further broken down for protein-coding genes)
Form:
- Select annotation source
- Filters
- For protein-coding gene annotations: coding-non-synon, etc.
- For wiggle tracks: min/max threshold
- ...?
Command line
Since this tool is so highly configurable, instead of cramming so many options into command line arguments, the tool will read a configuration file (possibly stdin) in .ra format. It might look something like this:
primarySource ct_myVars sourceType customTrack dataType pgSnp filterSpecs filter1,filter2 outFields allButBin filter filter1 alleleCount == 2 filter filter2 alleleFreq noMatch 0,0 source snp135Common sourceType dbTrack dataType bed 6 + outFields chrom,chromStart,chromEnd,name,strand,observed,exceptions,alleleFreqCount,alleles,alleleNs source pubMatches sourceType dbTrack dataType bed12 outFields name outputFormat tabSep fileName ./myAnnotatedVars.txt.gz
Then the tool might be invoked like this:
annoGrate hg19 config.ra
Name
Not variant Effect Predictor or other names in use.
- Variantizor?
- Predictorator?
- Diffmeister?
- Differizerator?
- Variant Annotator
- Varannosaurus Rex
- Varannozilla
- Global Variant Annotator
- Integrated Variant Annotator (except rings of Broad IGV)
- Variant Annotation Integrator
- Variant Annotatoon Tool ( except VAT is not good acronym)
- VarAnnoGrator
Links to Similar Tools
http://uswest.ensembl.org/info/docs/variation/vep/index.html
http://snpeff.sourceforge.net/faq.html
http://www.ncbi.nlm.nih.gov/variation/tools/reporter