Cell Browser data flow and architecture: Difference between revisions
| Line 18: | Line 18: | ||
</pre> | </pre> | ||
2. The output files from <code>cbBuild</code> are placed inside <code>/usr/local/apache/htdocs-cells</code>. Note that the original configuration files and expression matrices inside the dataset directory are converted into either JSON or binary files (BIN). These files are used by the Cell Browser to . | 2. The output files from <code>cbBuild</code> are placed inside <code>/usr/local/apache/htdocs-cells</code>. Note that the original configuration files and expression matrices inside the dataset directory are converted into either JSON or binary files (BIN). These files are used by the Cell Browser website to display the visualization. The original files are human readable; whereas, the ones used by the browser are for faster access. | ||
3. Once the dataset is on [https://cells-test.gi.ucsc.edu/ cells-test], the next destination is [https://cells-beta.gi.ucsc.edu/ cells-beta]. You will push the directory and files from <code>htdocs-cells</code> onto <code>/usr/local/apache/htdocs-cells-beta</code> using the command: | 3. Once the dataset is on [https://cells-test.gi.ucsc.edu/ cells-test], the next destination is [https://cells-beta.gi.ucsc.edu/ cells-beta]. You will push the directory and files from <code>htdocs-cells</code> onto <code>/usr/local/apache/htdocs-cells-beta</code> using the command: | ||
<pre> | <pre> | ||
cbPush dir-name | cbPush dir-name | ||
</pre> | </pre> | ||
Note that cbPush requires you to input a directory name | Note that cbPush requires you to input a directory name. | ||
A good alias to have in your .bashrc that pushes the current directory you are in onto beta: | A good alias to have in your .bashrc that pushes the current directory you are in onto beta: | ||
| Line 33: | Line 33: | ||
alias cbPushDir='cbPush "${PWD##*/}"' | alias cbPushDir='cbPush "${PWD##*/}"' | ||
</pre> | </pre> | ||
You could name this alias whatever you prefer. | |||
4. Once your dataset is on beta, you are almost there! Once the dataset is checked over for potential bugs, you will use the command: | 4. Once your dataset is on beta, you are almost there! Once the dataset is checked over for potential bugs, you will use the command: | ||
Revision as of 17:49, 26 August 2022
How does data flow between the different machines?
How does building a cell browser work?
What files are copied over? Which ones are transformed into another format?
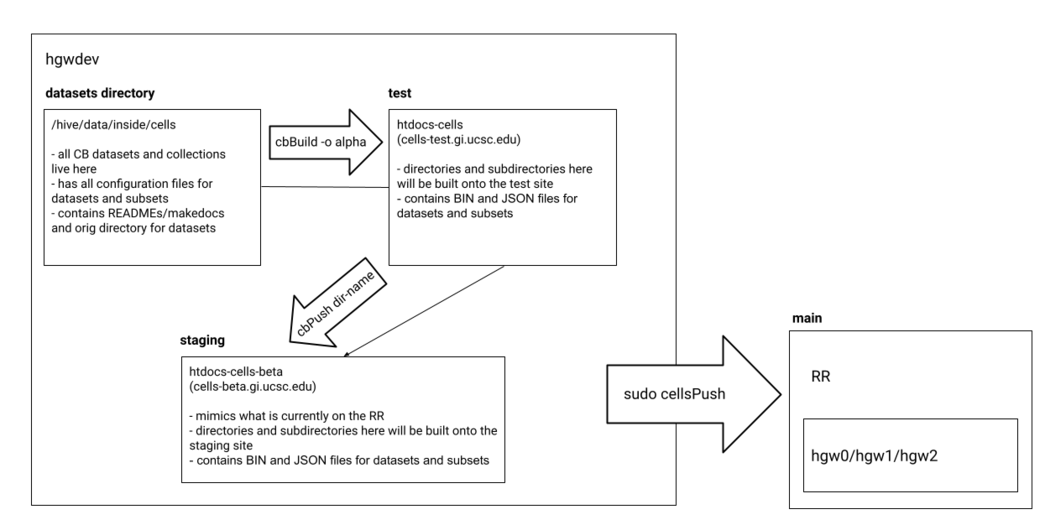
1. Data is first deposited in a dataset directory inside /hive/data/inside/cells on hgwdev and then gets built onto cells-test using the command:
# For datasets with no additional subsets cbBuild -o alpha # For dataset collections you will use the recursive option "-r" cbBuild -r -o alpha
2. The output files from cbBuild are placed inside /usr/local/apache/htdocs-cells. Note that the original configuration files and expression matrices inside the dataset directory are converted into either JSON or binary files (BIN). These files are used by the Cell Browser website to display the visualization. The original files are human readable; whereas, the ones used by the browser are for faster access.
3. Once the dataset is on cells-test, the next destination is cells-beta. You will push the directory and files from htdocs-cells onto /usr/local/apache/htdocs-cells-beta using the command:
cbPush dir-name
Note that cbPush requires you to input a directory name.
A good alias to have in your .bashrc that pushes the current directory you are in onto beta:
alias cbPushDir='cbPush "${PWD##*/}"'
You could name this alias whatever you prefer.
4. Once your dataset is on beta, you are almost there! Once the dataset is checked over for potential bugs, you will use the command:
sudo cellsPush
You will be prompted to type in a password, use your hgwdev password. Once you do that, the datasets will be built onto the hgw0, hgw1, and hgw2 machines! Voila!