Opsin evolution: trichromatic ancestral mammal
See also: Curated Sequences | Opsin Origins | Ancestral Introns | Informative Indels | Update Blog
So-so recovery of trichomatic vision in mammals
Textbooks often portray evolution as unidirectional progress chugging steadily along to the pinnacle of human innovative perfection, even as more primitive creatures drop by the wayside for lack of a forward-looking vision of manifest destiny. However newly available comparative genomics of opsins -- the primary photon receptors underlying vision -- clearly refute those notions and show instead serious backsliding in mammalian vision.
The fairy tale recovery of trichromaticity (old world primate duplication of LWS) is hardly an unmitigated success story. Humans have lost 10 of the 14 ciliary and rhabdomeric opsins present from lamprey to amniote, not to mention oil droplets that refine color vision. The advent of the MWS gene duplicate represents a minor partial recovery of color vision capabilities, though not nearly to that still enjoyed by birds and lizards. However this highly unstable locus experiences chronic non-homologous recombination mishaps, gene conversion erasing critical spectral differences, chimeric single genes, bizarre tandem arrays, leading to dichromatic color vision in perhaps 15% of the population.
The locus cannot stabilize itself by translocating away the gene duplication because such an event would not bring along the essential control region, which is tethered upstream to the primary LWS gene. The initial duplication and continuing instability may be driven by flanking Alu units. Recent selective forces acting on the region are difficult to disambiguate from gene conversion homogenization.
Howler monkeys independently duplicated LWS and importantly also its LCR control region, perhaps avoiding problems that have plagued anthropoid primates for 35 million years. New world monkeys exhibit a different form of partial recovery (in females) utilizing spectrally significant coding polymorphisms of a single X-linked LWS locus and random X inactivation.
This latter mechanism may illustrate Piatiogorsky's evolutionary principle: genes, exemplified by crystallins, develop their multi-functionality prior to the gene duplication event, thus coming online already possessing the selective umbrella of neo- or sub-functionalization, rather than racing against time to acquire useful mutations before inactivation. Here, a single-copy LWS gene developed spectrally useful polymorphisms in the diploid copy that had selective advantage (perhaps in food foraging) to females. In howler monkeys, their gene duplication could have captured these polymorphisms, providing in effect instant selectively protected gene divergence. Old World monkeys utilize different spectral tuning changes but the same mechanism could have been operative.
Lemurs have been the subject a fairy tale of their own: the strepsirrhines with polymorphic trichromacy (ie, females) are the diurnal species. That is in fact applicable to Propithecus coquereli, Varecia variegata and Varecia rubra. The corollary that should follow -- but doesn't -- is cathemeral lemurs (active sporadically day or night acquiring food) will lack the M/L opsin gene polymorphism that is the basis for trichromacy. However Eulemur macaco flavifrons has the identical change in the critical amino acid (position 285 in exon 5) as the diurnal lemurs.
Although this could reflect convergent evolution (same mutation, multiple occurrences), more likely this was a balanced polymorphism A <-> T (or the reduced alphabet choices) even though the mutation is generally a CpG hotspot when alanine, many tuning options exist with similar outcomes given 20 amino acids and 6-7 tuning residues and no observational support is at hand for separate selective advantages in diurnal and cathemeral species.
An Australian marsupial, the fat-tailed dunnart Sminthopsis, appears to utilize a fourth mechanism: expression of rod rhodopsin additionally in cone cells (conversely salamanders can express SWS2 in rod cells). None of these developments remotely approach full recovery of oil-droplet supplemented tetrachromatic color enjoyed by the ancestor at 310 myr and its descendants in extant non-mammalian tetrapods.
Evolution only lives in the selective present -- genes are retained on a "use or lose it" basis. A gene cannot be kept on unless it continuously retains function -- there's no long-term warehousing mechanism within the genome that can anticipate re-use later down the road. The blind mole rat, once in fully subterranean lifestyle, had no further selective pressure on its imaging opsin genes to weed out deleterious random mutations. Today those opsins are all rapidly deteriorating pseudogenes. The mole rat is now committed; there's no turning back.
Platypus color vision and implications for ancestral mammal
Gordon Walls proposed in 1942 that mammals -- but not turtles, birds and lizards -- experienced a prolonged bottleneck of strictly nocturnal lifestyle during which certain vision genes were not used and so lost irrevocably to mutation. Upon returning to sunlight, mammals had to go forward with distinctly inferior color vision plus loss of color oil droplet spectral enhancement. This epoch should not be conflated with episodes of nocturnality affecting various primate clades.
While some marsupials and placentals (including human) have regained trichromatic vision by various mechanisms, despite 100 million years of playing catchup, no theran mammal has ever regained the superior sharp tetrachromatic color vision enjoyed by the amniote common ancestor and contemporary turtles, birds and lizards. In other words, 250 million years of evolutionary "progress" from lamprey to amniote was lost in mammals. It's possible to have much better color vision than human -- and many earlier diverging vertebrate species do.
The advent of the genomic era has vastly improved our prospects for working out a detailed evolutionary history of photoreception -- ciliary opsin genes can be extracted from the 57 deuterostome genomes available (as of November 2007) and visual capabilities of last common ancestors reliably deduced. The newly sequenced platypus genome especially informs the history of color vision.
The platypus genome does not encode a single ancient, ancestral, antiquated, archaic, basal, dead-end, failed experiment, fossil, frozen, immature, obsolete, outmoded, primitive, primordial, relic, retro, stationary, sub-human or vestigial gene. On the contrary, the platypus is a fully modern organism successfully adapted to its current preferred habitat. Indeed by any objective measures of evolutionary distance from the last common ancestor with human (such as lower percent identity of the average selected coding gene), the platypus is more highly evolved from the ancestral condition.
Further, the platypus retains hundreds of genes (shared in conserved syntentic position with bird and lizard) that are lost in the human, including non-imaging opsins, and may well have more genes than human. These genes represent functionality conserved in ancestral vertebrates for hundreds of millions of years that subsequently degenerated in placentals. Living fossils, strictly defined, cannot exist since no species is immune to mutational processes (followed by drift and selection that fix them).
Reasoning from the platypus genome, new echidna data and other extant species, the ancestral amniote and its immediate descendants were tetrachromatic (4 cone opsins plus rod opsin) -- indeed all those genes had originated not later than the Ordovician (in the common stem ancestor with jawless fish). At the platypus node, the common ancestor had RHO1, no RHO2, but SWS2, SWS1, and LWS. This implies RHO2 was lost at some point on the stem. Prior to that loss, the mammalian ancestor had a full complement of imaging opsins (ie RHO1, RHO2, SWS2, SWS1, and LWS).
Thus the 'first mammal' was certainly trichromatic and even tetrachromatic for a time, depending on how that term is defined along the stem relative to RHO2 loss (via fossils or molecular reconstruction). This can never be settled because no datable pseudogene remnants of RHO2 remain even with sensitive searching of the syntenic sweet spot in reconstructed ancestral genomes. The last hope, that echidna (the only other surviving monotreme) relative) retained RHO2 or at least some datable fragment of it, now appears ruled out by PCR and 454 transcripts.
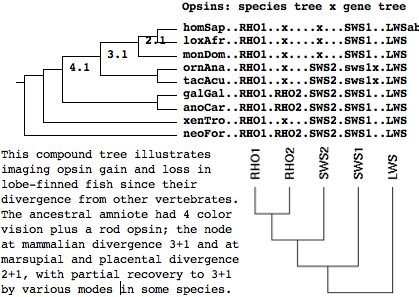
The labeled gene tree was drawn by the online tool Phylodendron using the following Newick-format tree:
((((((homSap..RHO1..x....x...SWS1..LWSab,loxAfr..RHO1..x....x...SWS1..LWS),monDom..RHO1..x....x...SWS1..LWS)[2.1], (ornAna..RHO1..x...SWS2.sws1x.LWS,tacAcu..RHO1..x...SWS2.sws1x.LWS))[3.1], (galGal..RHO1.RHO2.SWS2.SWS1..LWS,anoCar..RHO1.RHO2.SWS2.SWS1..LWS))[4.1],xenTro..RHO1..x...SWS2.SWS1..LWS), neoFor..RHO1.RHO2.SWS2.SWS1..LWS);
The proof that ancestral mammal at monotreme divergence had trichromatic (rather than tetrachomatic) vision involves a significant technical issue: showing that the first 4 exons of SWS1 are truly deleted rather than simply missing from the assembly: vertebrate genomes being incomplete, absence of evidence in an assembly (or trace archives) does not constitute evidence of absence in the genome but more commonly lack of coverage, even for a species such as platypus with 33,353,710 reads.
Opsins don't lend themselves to simplistic reconstructions based on retina spectroscopy and processed transcripts because of potential evolutionary convergence of spectral properties and multiple lineage-specific expansions and contractions of genes. What's needed here are complete genomes and syntenic validation of orthology, lacking hereto a certain degree here in key species (eg 3 of 6 known chicken opsins are missing from its current assembly).
However using the platypus genome, it is possible to reliably reconstruct the cone opsin situation at the ancestral mammal node, building on ref 2. This is a highly technical argument because platypus assembly initially exhibits only exon 5 of an opsin gene clustering to the short wavelength sensitive opsin SWS1 (as observed in ref 2). This last coding exon has the phylogenetically expected position and phase of its intron (ie is not a processed retrogene). It must be shown that the rest of platypus SWS1 has been deleted from the genome, rather than reflect a coverage gap in the platypus assembly:
Close synteny analysis of the gapless platypus assembly contig 45.4 using a conserved non-coding phastCons marker and adjacent CALU gene shows that exons 1-4 do not lie in adjacent assembly gaps but were lost in a multi-kbp deletion. Exon 5 exhibits a small number of disabling non-synonymous changes despite retaining overall high (63%) sequence identity to human, consistent with millions of years of normal divergence followed by rapid neutral evolution after the deletion of exons 1-4 pseudogenized residual exon 5. The deletion can be heuristically dated to perhaps 40 mya, an estimate that echidna SWS1 sequence would greatly improve. No opsin data currently exists for echidna; they may retain functional SWS1.
It follows that ancestral monotreme (respectively ancestral mammal) had 3-color vision (one step down from ancestral amniote) even though the platypus itself today has 2-color vision (similar to marsupials and most placentals). (This assumes intact SWS1 was not exapted to some unrelated function; this is testable from its spectral reconstruction.)
Queries by known SWS1 genes -- but not other opsins -- return a strong match to platypus chr10:3,960,795-3,960,908, which translates to FHACIMEMMRGKLMVDDSESSSQETKTSTVSSRQVGPS* with conserved intron position and phase zero characteristic of SWS1 exon 5. The 11,205 bp contig 45.4 also contains terminal exons of long-conserved syntenic gene CALU in the expected opposite strand orientation. Total coding gene span seems short enough (3,646 bp in opossum, 3,248 bp in human, 11,959 bp in anole) that the contig might be expected to contain some or all of exons 1-4 as well, even adjusting contig length to reflect observed lineage-specific retroposons of platypus. However exons 1-4 are missing from the contig and indeed the assembly. Furthermore missing exons cannot be found among GenBank transcripts now raw trace reads which include singleton reads and reads from other centers not used in the assembly.
This raises the question whether platypus lacks full length SWS1 or whether exons 1-4 merely lie in nearby gaps of the current 6x assembly. Possibly earlier introns have expanded so much that exons 1-4 have been pushed off the contig into these gaps. However contig 45.4 contains the phylogenetically conserved non-coding phastCons element lod109 (ref 3) at its extreme 3' end chr10:3,964,439-3,964,588 (the contig ends at 3,964,658!).
The UCSC genome browser provides putative orthologs of lod109 in marsupial and placental. These blat as standalone queries into full syntenic position with matching strand orientation with respect to marsupial and mouse where they are named lod15+lod45 and lod78 respectively. Here phastCons strand convention, which is inherently arbitrary, can be taken as positive for platypus and carried forward to mouse and opossum. CALU is positive but SWS1 exon 5 is negative in all three species.
Thus exons 1-4 of platypus are required to lie between SWS1 exon 5 and lod109, a region spanned by a single gapless contig, but they do not. This requires either a deletion or translocation elsewhere (to a trace read and assembly gap). A local inversion might move exons 1-4 into an assembly gap but that would disrupt reading frame. Implausibly, the missing exons could be translocated into an adjacent bridged gap of estimated modest size preceding the right-flanking gene NAG6 while retaining strand; this is incompatible with all four exons remaining recalcitrant to repeated attempts at genomic amplification (ref 2) and still missing in 6x coverage.
In summary, SWS1 is non-functional in platypus because the highly conserved exons 1-4 critical to opsin structure and function have been deleted. Conceivably this could be a copy-number polymorphism specific to the individual female platypus chosen for the genome project -- a seemingly viable concern given such polymorphisms are common for the tandem chrX opsin array in human. (Note the current human assembly shows 3 tandem opsins on chrX!) However platypus SWS1 does not lie in tandem array that might foster frequent inhomogeneous recombination.
Further close analysis of exon 5 shows the deletion event lies between ancient and recent:
Alignment of orthologous exons from 19 species phylogenetically bracketing platypus establish an arginine anomalously replacing conserved cysteine and a one residue deletion in platypus but otherwise very respectable percent identity that is not compatible with gene loss 166 mya ago (date for divergence taken in platypus genome paper). Non-selected pseudogenized coding dna does not leave detectable remnants at this age -- indeed no remnants can be detected of amniote RHO2 in mammals nor SWS2 in therans even focusing the search on syntenic dna, ref 2 and here.
Tachyglossus (echidna) transcripts became available from a massive 454 program at WUGSC in April 2008. Opsin sequences are not well represented there (compared to other gene families such as selenoproteins), apparently because library construction did not involve retinal tissue. However, a fragmentary exon 5 for echidna was locatable (which does not address presence of other exons) as well as two exons from SWS2.
Full length echidna SWS2 and LWS genes were provided in June 2008 by a study focused on monotreme opsins. SWS1 could not be found in echidna by plausibly primed PCR, suggesting the deletion of upstream exons occurred gene prior to echidna-platypus divergence, which the authors put at 21 myr. That date is compatible with the high 96% and 93% identity of the SWS1 and LWS genes of echidna to platypus but less so with a much earlier date of 120 myr reasoned from fossils, given what we know about rates of overall evolution in these opsins within vertebrates and overall rapid rate of protein evolution in monotremes.
Both echidna and platypus retain the ancestral syntenic tandem ordering of LWS and SWS2 seen in bird, lizard, frog and fish with that linkage probably perpetuated by a shared upstream locus control region. This association also implies SWS2 arose as a duplication of LWS back in lamprey; the linkage may be retained in species such as Geotria.
Note SWS1 is closer than SWS2 to LWS by various independent criteria, suggesting this gene copy was thrown off earlier possibly as a segmental duplication bringing with it a copy of the control region. However RHO2 and RHO1 appear to be descended from the tandem SWS2. These events seemingly all took place between amphioxus and lamprey divergence, making them difficult to investigate for lack of intermediate extant species.
>SWS1_xenTro FRGCIMETVCGRPMTDDSSVSSTSQKTEVSTVSSSQVSPA* amphibian >SWS1_anoCar FRACILETVCGKPMSDESDVSSSAQKTEVSSVSSSQVSPS* lizard >SWS1_pheMad FRGCIMEMVCGKPMSDDSEASTS-QKTEVSSVSSSQVSPS* lizard >SWS1_gekGek FRGCILEMVCGKTMAEESEVSSASQKTEVSSVSSSQVGPS* lizard >SWS1_utaSta FRACIMETVCGKPMTDESDVSSSAQKTEVSSVSSSQVSPS* lizard >SWS1_galGal FRACIMETVCGKPLTDDSDASTSAQRTEVSSVSSSQVGPT* bird >SWS1_taeGut FRACIMETVCGRPMTDDSEVSSSAQRTEVSSVSSSQVGPS* bird >SWS1_phaCar FRACIMETVCGKPMADDSEASSSAQRTEVSSVSSSQVSPS* bird >SWS1_ancMam FHACIMEMVCGKPMTDDSdVSSS-QKTEVSTVSSSQVGPS* ancestral mammal >SWS1_ornAna FHACIMEMMRGKLMVDDSE-SSS-QETKTSTVSSRQVGPS* monotreme >SWS1_monDom FHACIMEMVCRKPMTDDSDVSSS-QKTEVSAVSSSQVGPT* marsupial opossum Didelphimorphia >SWS1_macEug FHACIMEMVCRKPMTDDSEASSS-QKTEVSTVSSSQVGPS* marsupial wallaby Diprotodontia >SWS1_smiCra FHACIMEMICKKPMTDDSETTSS-QKTEVSTVSSSQVGPS* marsupial dunnart Dasyuromorphia >SWS1_setBra FHACIMEMVCRKPMTDDSEASSS-QKTEVSTVSSSQVGPS* marsupial quokka Diprotodontia >SWS1_isoObe FHACIMEMICRKPMTDDSETSSS-QKTEVSTVSSSQVSPS* marsupial bandicoots Peramelemorphia >SWS1_canFam FRACIMEMVCGKSMTEDSEMSSS-QKTEVSTVSPSQVGPN* placental >SWS1_susScr FRACIMEMVCGKPMTDESDMSSS-QKTEVSTVSSTQVGPN* placental >SWS1_homSap FQACIMKMVCGKAMTDESDTCSS-QKTEVSTVSSTQVGPN* placental >SWS1_sciBol FRACIMEMVCGKAMTDESDISSS-QKTEVSTVSSSQVGPN* placental >SWS1_cavPor FRACIMELVCRKPMADESDMSTS-QKTEVSAVSSSKVGPN* placental >SWS1_musMus FRACILEMVCRKPMADESDVSGS-QKTEVSTVSSSKVGPH* placental Consensus FraCImEmvcgkpMtD#S#.sss.QkTevStVSSsqVgP.*
The anomalous arginine can be validated in raw trace data, necessary because the trace assembly process resolves discrepancies by quality scores weighting that might disadvantage a read more accurate locally at the cysteine position. Viewing original data requires blastn against separate platypus WGS and OTHER databases at the NCBI trace archive repository. Some 752,000 additional traces from TIGR and NISC are found there in addition to 3,595,283 cDNA WUGSC traces. These numbers are a significant addition to the 27,607,516 conventional WGS traces used (but not used up) in the current platypus assembly.
The usual error types occur in these traces but none would result in R --> C and similarly for the 3bp indel. This cysteine is deeply conserved in homologous position in other opsins such as RHO1, SWS2, and LWS though there is 6 residues of spacing in the latter two families instead of 5 here.Therefore arginine, manifestly unsuited for a disulfide bond, is not acceptable structurally in this position.
The SWS1 exon 5 platypus residues are a bit unusual at four other sites. Some of these changes could be incapacitating but others tolerated or even advantageous (ie cannot be reliably assigned relative to the deletion event without echidna). While other species have their anomalies as well, platypus is a real outlier in this regard which can be seen for example by MultiAlign tree construction.
The alignment contains a heuristic prediction of exon 5 in the last common ancestor of mammals that is useful in counting changes in descendant lineages. Overall platypus exon 5 has evolved anomalously in that its variations are not concentrated at commonly variable positions but disproportionately at conserved ones, a pattern characteristic of a pseudogene. Opsin rate change are difficult to assess because to be retained over long timeframes, SWS1 and SWS2 in the same species may push each other apart spectrally (subfunctionalize) rather than just drift at soft positions.
Platypus exon 5 can only have become nonfunctional fairly recently -- retaining 63% identity to human given 332 mya of round-trip evolutionary time is quite respectable and implies long periods of conserved functionality. (Chicken/human have 65% identity.) This degree of conservation is incompatible with ancestral monotreme already having pseudogenized SWS1 from consideration of neutral rates.
Thus a platypus ancestor prior to echidna divergence at 25 mya (ref 4) still retained functional SWS1 and so likely had trichromatic cone vision. It cannot be proven that intact SWS1 functioned as a cone cell color vision receptor in ancestral platypus (because opsins are occasionally exapted to other purposes); echidna sequence might have allowed appropriateness of its adsorption spectrum to be adduced. However the key exons are deleted in echidna too. Exon 5 alone does not have a sufficient set of modifying residues to allow spectral prediction.
That trichromaticity must also hold further back at the ancestral mammalian node because platypus and echidna also retain SWS2 and LWS, still in amniote-syntenic tandem position at Ultra401:228519-263967 (blat genome or see ref 2). It appears marsupials + placentals went dichromatic earlier than monotremes, losing SWS2 without a trace (sooner than SWS1 loss in monotremes).
Indeed, RHO2 and tetrachomatic vision was present at the ancestral node with chicken/lizard. The subsequent (complete) loss of RHO2 in the mammalian stem cannot be dated. However all mammals experienced sequential degradation of color vision from four to three to two. This same conclusion has been reached by Australian researchers in ref 5.
In summary, the preferred scenario is platypus/echidna common ancestor possessing a functional SWS1 gene evolving at a moderate rate until the fairly recent deletion of exons 1-4. After the deletion the remaining exon 5 had no constraints and so evolved at the faster neutral rate of pseudogenic dna.
How recent was this deletion? That would require partitioning changes in platypus exon 5 since divergence from ancestral mammal between drift and selective change during the functional era from subsequent neutral change in the pseudogene era, based on anomalous character and what is known generally about opsin evolution and neutral rates in general. That calculation would greatly benefit from determining the sequence and status of exon 5 in echidna. If echidna still had functional SWS1 and the divergence time was 25 mya, the deletion event in platypus must post-date that. On the other hand, if echidna also contained the indel, the departure of its exon 5 sequence from platypus would significantly inform event partitioning and so improve dating of the indel.
ref 1. Walls, GL The Vertebrate Eye and its Adaptive Radiation Cranbrook Institute 1942
ref 2. Davies WL, Carvalho LS, Cowing JA, Beazley LD, Hunt DM, Arrese CA Visual pigments of the platypus Curr Biol. 2007 Mar 6;17(5):R161-3.
ref 3. Siepel A, Bejerano G, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes Genome Res. 2005 Aug;15(8):1034-50.
ref 4. Pettigrew JD. Electroreception in monotremes J Exp Biol. 1999 May;202(Pt 10):1447-54.
ref 5. Wakefield MJ, Anderson M, Chang E, Wei KJ, Kaul R, Graves JA, Grützner F, Deeb SS. Cone visual pigments of monotremes: filling the phylogenetic gap. Vis Neurosci. 2008 May-Jun;25(3):257-64.
Platypus TMT pseudogene barely lingers on
The ancient ciliary ur-opsin represented by TMTa still has the deep ancestral N D P C region in the second transmembrane helix. However the gene, which can be tracked back to early deuterostomes such as sea urchin, only continues intact through fish to early tetrapods (fish and lizard). Pseudogenes can be detected by blast focused on the syntenic region. Chicken, finch, and platypus have significant but barely detectable exonic debris from the lost TMTa gene here.
This situation is reminiscent of SWS1: platypus has lost the TMT gene as well perhaps 100 myr after its divergence. Other mammals apparently lost it early because no debris can be located. It is possible that the gene continues on in echidna.
Opsin data for monotremes
The critical conserved non-coding markers used to prove a deletion event in platypus SWS1:
>lod109_ornAna+ chr10:3964439-3964588 tgctgctgatgtggcaaagaagaccacatctgtccagctacttctatcaccggctcgttggtctaggggtatgattctcgcttagggtgcgagaggtcccgggttcaaatcccggacgagccctgtttttcccacctggttttatttggg >lod109_monDom+ chr8:184,539,020-184,560,157 from platypus alignment tgctgatgatgtggcaaaaccacatctggacaactattttcaacactggctcgttggtctaggggtatgattctcgcttagggtgcgagaggtcccgggttcaaatcccggacgagcccaattttac >lod78_musMus+ chr6:29338830-29338906 from platypus alignment tgttgatgatgtggtagag-aggatacatctggccaggagtaaccgccagtggctcgttggtctaggggtatgattctcgcttagggtgcgagaggtcccgggttcaaatcccggacgagccctgcttatc
The status of the five imaging opsins in platypus genome and echidna PCR:
>RHO1_ornAna Ornithorhynchus anatinus (platypus) rod rhodopsin full 0 MNGTEGQDFYIPMSNKTGVVRSPFEYPQYYLAEPWQYSVLAAYMFMLIMLGFPINFLTLYVTIQHKKLRTPLNYILLNLAFANHFMVLGGFTTTLYTSLHGYFVFGPTGCNIEGFFATLG 1 2 GEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACALPPLVGWSR 2 1 YIPEGMQCSCGIDYYTLRPEVNNESFVIYMFVVHFTIPMTIIFFCYGRLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTVPAFFAKSSAIYNPVIYIMMNKQ 0 0 FRNCMLTTICCGKNPLGDDEASATASKTEQSSVSTSQVSPA* 0 >RHO2_ornAna Ornithorhynchus anatinus (platypus) gene completely absent from genome and by PCR >SWS2_ornAna Ornithorhynchus anatinus (platypus) ancestral tandem position with LWS 0 MHKTHRNLQNELPEDFFIPLPLDTDNITSLSPFLVPQTHLGGSGIFMSLAAFMFLLITLGFPINLLTVICTIKYKKLRSHLNYILVNLAVSNMLVVCVGSATAFYSFAHMYFVLGPTACKIEGFAATLG 1 2 GMVSLWSLAVIAFERFLVICKPLGNLSFRGTHAIFGCAATWVFGLAASLPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFSFCFGVPLSIIIFSYGRLLLTLRA 0 0 VAKQQEQSATTQKAEREVTKMVIVMVLGFLVCWLPYASFSLWVVTNRGQVFDLRMASIPSVFSKASTIYNPIIYVFMNKQ 0 0 FRSCMLKLVFCGKSPFGDEDEISGSSQATQVSSVSSSQVSPA* 0 >SWS1_ornAna Ornithorhynchus anatinus (platypus): terminal exon 5 surviving deletion event chr10:3,935,884-3,985,883 FHACIMEMMRGKLMVDDSESSSQETKTSTVSSRQVGPS* >LWS_ornAna Ornithorhynchus anatinus (platypus) cone long LWS green full 0 MTPAWNSGVYAARRRFEDEEDTTRTSVFVYTNSNNTR 1 2 DPFEGPNYHIAPRWAYNVTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA* 0 >RHO1_tacAcu Tachyglossus aculeatus (echidna) AY894355 frag covering parts of exons 3-4 identical to platypus NESFVIYMFVVHFTIPMTIIFFCYGRLVFTVKE 0 0 AAAQQQESATTQKA >RHO2_tacAcu Tachyglossus aculeatus (echidna) assumed absent from genome >SWS2_tacAcu Tachyglossus aculeatus (echidna) EU636012 ancestral tandem position with LWS 0 MHKTHQNLQNEPPEDFFIPLPLDTDNITSLSPFLVPQTHLGGAGIFLSLAAFMFLLVTLGFPINLLTVICTVRYKKLRSHLNYILVNLAVSNMLVVCVGSATAFYSFAHMYFVLGPTACKIEGFAATLG 1 2 GMVSLWSLAVIAFERFLVICKPLGNLSFRGTHAIFGCAATWVFGLAASLPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFSFCFGVPLSIIIFSYGRLLLTLRS 0 0 VAKQQEQSATTQKAEREVTKMVIVMVLGFLVCWLPYASFSLWVVTNRGQAFDLRMASIPSVFSKASTVYNPVIYVFMNKQ 0 0 FRSCMLKLVFCGKSPFGDEDEMSGSSQATQVSSVTSSQVSPA* 0 >SWS1_tacAcu Tachyglossus aculeatus (echidna) exon 5 but other exons deleted >LWS_tacAcu Tachyglossus aculeatus (echidna) EU636011 recurrent del at asp 21 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNVTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWSAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFTGSSDPGVQSYMIVLMCTCCILPLSIIVLCYLQVWMAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAAHPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 VRNCLMQLFGKKVDDVSELSSTSRTEVSSVSSVSPA* 0 [Note: position 21 lies in a compositionally simple region of repeat acid amino acids with poor conservation, experiencing numerous homoplasic events over evolutionary time: LWS_homSap MAQQWSLQRLAGRHPQDSYEDSTQSSIFTY LWS_loxAfr MAQQWGPHRLTGARLQDASEDSTQASIFVY LWS_monDom MTQAWDPAGFLARRRDDDNDETTRSSLFVY LWS_myrFas MTQAWDPAGFLAWRREEN-EETTRASLFTY LWS_macEug MTQAWDPAGFLAWRRDENE-ETTRASLFVY LWS_setBra MTQAWDPAGFLAWR-RDEN-EETTRASLFVY LWS_cerCon MTQAWDPAGFLAWQEDEN-EETTRASLFVY LWS_tarRos MTQAWDPAGFLAWRRDEN-EETTRASLFVY LWS_tacAcu MTQAWDPAGFLAWRRDEN-EETTRASLFVY LWS_isoObe MTQAWDPAGFLAWRRDEN-EETTRASLFVY LWS_ornAna MTPAWNSGVYAARRRFEDEEDTTRTSVFVY LWS_galGal MAAWEAA-FAARRRHEE-EDTTRDSVFTY LWS_taeGut MATWDGAVFAARRRHDD-EDTTRDSIFTY LWS_colLiv MDGFAAARRRHED-EDTTRDSVFTY LWS_anoCar MAEAWDVAVFAARRRNDE-DDTTRDSLFTY LWS_ambTig MAHSWNSGAYAARRRYDD-EDTTRSSIFTY LWS_cynPyr MAYSWNSGAYAARRRYDD-EDTTRSSVFVY LWS_xenTro MASHWNEAVFAARRRNDD-DDTTRSSVFTY LWS_xenLae MASQLNEAIFAARRRNDD-DDTTRSSVFTY LWS_petMar MTASWQGAMFAARRRQDD-EDTTMESLFRY LWS_letJap MTASWHGAVFAARRRNDD-EDTTKDSIFRY LWS_geoAus MAQSWERAMFAARRRQD--EDTTKGDLFRY LWS_neoFor MAEPWD-AVLAARRRHQD-EETTRSTIFVY LWS_gecGec MTEAWNVAVFAARRSRDD-DDTTRGSVFTY LWS_pheMad MTEAWNVAVFAARRSRDDDDDTTRGSVFTY LWS_danRer MAEHWGDAIYAARRKG---DETTREAMFTY LWS_cypCar MAEQWGDAIFAARRRG---DETTRETMFVY LWS_pleAlt MTDEWGNAVFAARRRN---EDTTRESSFTY LWS_takRub MAEEWGKQSFAARRYH---EDTTRGSAFVY LWS_oryLat MAEQWGKQVFAARRQN---EDTTRGSAFTY LWS_gasAcu MAEEWGKQAFAARRYN---EDTTRGSMFVY
An old platypus pseudogene recovered by blast restricted to syntentic position. Chicken and finch also have a pseudogene here but nothing is detectable in marsupials or placentals (ie the gene was lost far earlier).
>TMTa_ornAnaPS Ornithorhynchus anatinus (platypus) pseudogene frags adjacent to syntenic GPR35, weak assembly from ST6GAL1 side 2 WAYASFWATMPLVGLGNYAPEPFGTSCTLDWWLAQASVAGQAFILNILFFCLLLPTAVIV 0 0 SKGVSKGMEKIGEQ*VQLTVFVVVICFLFCWLPYGTMASISTCGKPGLITPT SSVFPLVLG KNSTVLNPVIYGFLNvk 0 0 FYRCFHALMSF KDFTSSISEVSPIPFDFSCVTPRIQNNH-FPSASEGRP
Opsin 454 transcripts of salamander Ambystoma tigrinum
Amphibians are the last stop before the complications introduced by teleost fish (large-scale expansions of certain loci, overall whole genome duplication, and inherently rapid divergence). Currently the frog Xenopus is the only amphibian with a genome project and that has lain dormant for 5 years. Consequently it is quite important to collect other amphibian opsin sequences so that the corresponding divergence node can be better understood (an issue heightened by loss of several ancestral loci in Xenopus).
A large scale Ambystoma transcriptome project at the University of Kentucky provides a public blast server for assembled 454 reads. A vertebrate with this phylogenetic position should have 21 distinct opsin loci. Four complete ones were already available at GenBank nr from previous ad hoc projects. Using tBlastn of each plausible query in turn, fragmental genes for 8 more opsin loci are currently recoverable, yielding a total of 12 Ambystoma loci. Here the dna of the blast-match contigs are retrievable under 'cDNA V3.0' using 'ContigName'.
This is really an excellent outcome considering that transcripts at some opsin loci are very rare to date in other species. While partial genes are unsatisfactory in many respects, they do at least establish that the gene is present, help establish ancestral values in areas of coverage and provide a handle for targeted experimental retrieval of it. A great many more runs with the same material could yield full length versions of these genes.
A few other opsin genes are present at GenBank for other species of amphibians, for example SWS2 and a nearly complete RGR1 sequence for Cynops pyrrhogaster (newt). These (non-Xenopus) sequences are included below. Outside of rhodopsin, amphibian opsin coverage remains quite poor as of April 2010.
>RHO1_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud ATU36574 8759361 full Gt 0 MNGTEGPNFYVPFSNKSGVVRSPFEYPQYYLAEPWQYSVLAAYMFLLILLGFPVNFLTLYVTIQHKKLRTPLNYILLNLAFANHFMVFGGFPVTMYSSMHGYFVFGQTGCYIEGFFATMG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVMMTWIMALACAAPPLFGWSR 2 1 YIPEGMQCSCGVDYYTLKPEVNNESFVIYMFLVHFTIPLMIIFFCYGRLVCTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVVAFLICWVPYASVAFYIFSNQGTDFGPIFMTVPAFFAKSSAIYNPVIYIVLNKQ 0 0 FRNCMITTICCGKNPFGDDETTSAATSKTEASSVSSSQVSPA* 0 >SWS1_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud AF038948 full Gt 0 MLEEEEFYLYKNISKVGPWDGPQYHIAPAWTFYFQTAFMGFVFFVGTPLNAIVLIVTVKYKKLRQPLNYILVNVSLAGFTFCIFSVFTVFVSSSQGYFIFGKTICELEAFLGSVS 1 2 GLVTGWSLAFLAIERYIVICKPMGSFRFSSKHATMVVLATWAIGFSVSIGPLVGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYNSETYTWFLFIFCFIIPLSLICFCYAQLLGALRA 0 0 VAAQQQESATTQKAEREVTRMVIVMVVSFCLCYVPYAAMAMYMVNNRNHGLDLRLVTIPAFFSKSACVYNPIIYSFMNKQ 0 0 FRACIMETVCGTPMTDESDISSSSNKTEVSSVSSSQVSPS* 0 >SWS2_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud AF038946 9675215 full Gt 0 MYKGKQEMMAELSDDFYIPVPMETTNISALSPFLVPQTHLGSPAVFMSLAAFMFLKVVFGFPINLLTVICTIKYKKLRSHLNYILVNLAIANLLVVSVGSTVAFYSNAQMYFALGPLACKVEGFTATLG 1 2 GMVGLWSLAVVAFERFLVICKPLGSFTFRESHAIMGCAFTWIVGLAAATPPLLGWSR 2 1 YIPEGLHCSCGPDWYTVNNKWNNESYVLFLFCFCFGVPLTTIIFSYGRLLITLRA 0 0 VAKQQEQSATTQKAEREVTRMVIFMVAGFLVCWLPYASFALWATTHRGELFDLRMASIPSVFSKASTVYNPVIYIFMNRQ 0 0 FRSCMMKLIFCGKNPFGEDEDTSVSAQSTQVSSVSSSQVAPA* 0 >SWS2_cynPyr Cynops pyrrhogaster (newt) Deut.Amph.Caud AB040148 full 0 MYKVKRVMDAEMSDDFYIPLPLDTTNITALSPFLVPQTHLGSPTIFRSIAVFMFFILLLGIPINFLTILCTFLNKKLRTHFNYILVNMAVANLLVIFIGPTLSFYSNSQMYFALGPLACKIEGFAATLG 1 2 GIVGLWCLAVVAFERYLVICKPVGGFTFRESHAIMGCIFTWIAGFTAAGPPLFGWSR 2 1 YIPEGLQCSCGPDWYTVNNKWNNESYVLFLFCFSFGVPLFIIIFSYGRLLITLRA 0 0 VAKQQEQSATTQKAEREVTKMVIVMVLGYLICWSPYAIFALWATSHRGEIFEPWMASIPAIFSKSSTVYNPVIYVFMNRQ 0 0 FRSSMMKLVFCGKSPFGDDDDSSASGQSTQVSSVSSSQVAPA* 0 >LWS_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud AF038947 9675215 full Gt 0 MAHSWNSGAYAARRRYDDEDTTRSSIFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTTLWMIFVVFASVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETVIASTISVINQMFGYFILGHPLCVIEGYTVSVC 1 2 GISALWSLTIIAWERWFVVCKPFGNIKFDGKLAAAGIIFSWVWSAGWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMMVLMITCCILPLSIIIICYIQVWWAIRQ 0 0 VAMQQKESESTQKAEREVSRMVVVMIIAYIFCWGPYTFFACFAAANPGYAFHPLAASLPAFFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQLFGKKVDDGSEMSSASRTEVSSVSNSSVSPA* 0 >PIN_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud contig29789 frag KHAVMGCAFTWLWSLLWTAPPLFGWSSYVPE 1 2 GLRTSCGPNWYTGGSNNNSYIMVLFITCFIMPLSTIAFSYASLLMALRA 0 0 VAAQQKESETTQRAEKEVTRMVVAMVMAFLICWLPYATFAIVVATNKDIVIEPALASMPSYFSKTATVYNPVIYVFM >PPIN_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud contig02489 frag 2 aSYGYLMWTLRQ 0 0 IAKVGVAESGTTNKAESQVSRMVLLMIIAFLICWLPYALFAMTVVANPGIHIDPIMATVPMYLSKTSTVYNPIIYIFMNRQ 0 0 FRDCIIPLLLCGKNNLTSEMRTSSVTVTSTTSSPSRHGKVVPI* >VAOP_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud EPJ1Z2K02FZXY8 frag 2 RFVVICRPLGNLRLRGKHSALGIAFVWIFSFVWTVPPTMGWSSYTTSKIGTTCEPNW 2 >PER1_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud contig06013 frag 0 HLITA 1 2 GVVSLLSNIVVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGNAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAIDRYLTICRPDV 1 2 GRRMTACNYAALIIASWINAFFWALMPIVGWSSYAPDPTGATCTINWRKNDA 2 1 SLVSYTMSVVAVNFLGPLAVMFYCYYKVSKALKKYTTNGSSLQSLNMDWSDQLDVTK 0 0 VSVVMIVMFLVAWSPYSIVCLWS >MEL1_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud FUQAVB301ESJ4M frag poor quality 1 YSSYPSTRRSTVTS VSDSES 0 0 GWTDTEADTSSVASRPASRQVSYEMSKDTSETTDSISKSKLKSHDSGIFEK 0 0 TSMDVDDVSMVDVSTVERTPPVT 0 0 AVKSLNGIGPRKGGSLRRLPA >RGR1_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud contig42480 contig42478 frag 2 ALLGLILNGLTIVSFYKIRALRTPHNFLIVSLALADSGVCLNAFIASFSSFLR 2 1 YWPYGSEGCQIHGFQGFLTALTSISSCGVIAWDRYNQYCT 1 2 rSKLQWSTALSLVSFVWAFSAFWSVMPLIGWGQYDYEPLRTCCTLDYTKGDK 2 1 NFISYLFPLAFFEFLIPLFIMLTAYQSIEQKFKKTGQHK 0 0 FNTSLPVKTLVMCWGPYSLLCFYATVENATAISPKIRM 0 0 LPAILAKTSPAINAFVYGLGNECYRGGIWQFLTGQKIEKVEVDNKAK* 0 >RGR1_cynPyr Cynops pyrrhogaster (newt) Deut.Amph.Caud FS290827 20090923 frag G? 0 GFTEIEVFGLGT 1 2 ALLIEALLGFILNGLTLLSFYKIRSLRTPHNFLIVSLALADTGVCINAFIAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFVTALSSISSCGVIAWDRYNQYCT 1 2 RTKLQWGTAISLVSFVWAFSAFWSVMPLLGWGQYDYEPLRTCCTLDYTKGDK 2 1 NFISYLFPLAFFEFVIPLFIMLTAYQSVEQKFKKTGQHK 0 0 FNTGLPVKTLVMCWGPYSLLCFYATIENATTISPKIRM 0 0 LPAILAKTAPAINAFLYGMGNESYRGGIWQFlTGQKIEKAEVDNKTK* >NEUR1_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud contig18098 frag 2 LITMTAVSLDRYLKICHLSY 1 2 GTWLKRRHAFLCLTIIWSYASFWATLPLVGVGNYAPEPFGTSCTLDWWLAQASKSGQAFVLCMQVFCLL >NEUR2_ambTig Ambystoma tigrinum (salamander) Deut.Amph.Caud FUQAVB301CB8BY frag 0 DDVILGAVYSLL 1 2 GFLSLCGNSVLLFIAYRKRAMLKPAEYFIVNLSVSDMGMTVTLFPLAIPSLFAHR 2 1 WLFDKIVCKYYAFCGVLFGLSSLTNLTVLSSVCCLKVCYP
See also: Curated Sequences | Opsin Origins | Ancestral Introns | Informative Indels | Update Blog