Gene Set Summary Statistics: Difference between revisions
From genomewiki
Jump to navigationJump to search
Matt Speir (talk | contribs) |
(Updated genome-test or hgwdev links from .cse/.soe to .gi, most links validated) |
||
| (8 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
* hg17 - knownGenes version 2 | * hg17 - knownGenes version 2 | ||
* hg18 - knownGenes version 3 | * hg18 - knownGenes version 3 | ||
* hg19 - knownGenes version 7 | |||
* mm8 - knownGenes version 2 | * mm8 - knownGenes version 2 | ||
* mm9 - knownGenes version 3 | * mm9 - knownGenes version 3 | ||
* mm10 - knownGenes version 6 | |||
The min, max and mean measurements are ''per gene'' | The min, max and mean measurements are ''per gene'' | ||
| Line 15: | Line 17: | ||
<TR><TH>mm8</TH><TD ALIGN=RIGHT>31863</TD><TD ALIGN=RIGHT>314628</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>313</TD><TD ALIGN=RIGHT>10</TD></TR> | <TR><TH>mm8</TH><TD ALIGN=RIGHT>31863</TD><TD ALIGN=RIGHT>314628</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>313</TD><TD ALIGN=RIGHT>10</TD></TR> | ||
<TR><TH>mm9</TH><TD ALIGN=RIGHT>49220</TD><TD ALIGN=RIGHT>417114</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>610</TD><TD ALIGN=RIGHT>8</TD></TR> | <TR><TH>mm9</TH><TD ALIGN=RIGHT>49220</TD><TD ALIGN=RIGHT>417114</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>610</TD><TD ALIGN=RIGHT>8</TD></TR> | ||
<TR><TH>mm10</TH><TD ALIGN=RIGHT>59121</TD><TD ALIGN=RIGHT>486425</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>813</TD><TD ALIGN=RIGHT>8</TD></TR> | |||
</TABLE> | </TABLE> | ||
| Line 25: | Line 28: | ||
<TR><TH>mm8</TH><TD ALIGN=RIGHT>83159087</TD><TD ALIGN=RIGHT>4</TD><TD ALIGN=RIGHT>17497</TD><TD ALIGN=RIGHT>264</TD></TR> | <TR><TH>mm8</TH><TD ALIGN=RIGHT>83159087</TD><TD ALIGN=RIGHT>4</TD><TD ALIGN=RIGHT>17497</TD><TD ALIGN=RIGHT>264</TD></TR> | ||
<TR><TH>mm9</TH><TD ALIGN=RIGHT>117671086</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>29698</TD><TD ALIGN=RIGHT>282</TD></TR> | <TR><TH>mm9</TH><TD ALIGN=RIGHT>117671086</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>29698</TD><TD ALIGN=RIGHT>282</TD></TR> | ||
<TR><TH>mm10</TH><TD ALIGN=RIGHT>143087341</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>83437</TD><TD ALIGN=RIGHT>294</TD></TR> | |||
</TABLE> | </TABLE> | ||
| Line 35: | Line 39: | ||
<TR><TH>mm8</TH><TD ALIGN=RIGHT>1476081990</TD><TD ALIGN=RIGHT>9</TD><TD ALIGN=RIGHT>1347550</TD><TD ALIGN=RIGHT>5220</TD></TR> | <TR><TH>mm8</TH><TD ALIGN=RIGHT>1476081990</TD><TD ALIGN=RIGHT>9</TD><TD ALIGN=RIGHT>1347550</TD><TD ALIGN=RIGHT>5220</TD></TR> | ||
<TR><TH>mm9</TH><TD ALIGN=RIGHT>2055504784</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>1253430</TD><TD ALIGN=RIGHT>5589</TD></TR> | <TR><TH>mm9</TH><TD ALIGN=RIGHT>2055504784</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>1253430</TD><TD ALIGN=RIGHT>5589</TD></TR> | ||
<TR><TH>mm10</TH><TD ALIGN=RIGHT>2420504661</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>3003780</TD><TD ALIGN=RIGHT>5665</TD></TR> | |||
</TABLE> | </TABLE> | ||
| Line 53: | Line 58: | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:88937989-89411302 uc002stk.1] (217)</TD> | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:88937989-89411302 uc002stk.1] (217)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:179098964-179380395 uc002umt.1] (194)</TD> | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:179098964-179380395 uc002umt.1] (194)</TD> | ||
</TR> | |||
<TR><TH>hg19</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr14:105994256-107283085 uc031qqx.1] (5065)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr22:22385572-23265082 uc031rxg.1] (462)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr2:179390717-179672150 uc031rqc.1] (363)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr2:179390717-179672150 uc031rqd.1] (313)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr2:179390717-179672150 uc021vsy.2] (312)</TD> | |||
</TR> | </TR> | ||
<TR><TH>mm8</TH> | <TR><TH>mm8</TH> | ||
| Line 62: | Line 74: | ||
</TR> | </TR> | ||
<TR><TH>mm9</TH> | <TR><TH>mm9</TH> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12:114498400-117248164 uc007pgj.1] (610)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:76542041-76820604 uc008kfn.1] (313)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:76542041-76820604 uc008kfo.1] (192)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:51992167-52194318 uc008jqv.1] (157)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr9:108855790-108886920 uc009rrh.1] (118)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm10</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr12:113260189-116009879 uc029ryi.1] (813)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:76703984-76982547 uc008kfn.1] (313)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:76703984-76982547 uc008kfo.1] (192)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:52136647-52338798 uc008jqv.1] (157)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr9:108953745-108984875 uc009rrh.1] (118)</TD> | |||
</TABLE> | </TABLE> | ||
| Line 102: | Line 120: | ||
</TR> | </TR> | ||
<TR><TH>mm9</TH> | <TR><TH>mm9</TH> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chrX:80194242-82450380 uc009tri.1] (2253366)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr6:45010087-47251368 uc009bst.1] (2238325)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr16:39984484-42176405 uc007zfr.1] (2189582)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:40452293-42509118 uc008jon.1] (2055883)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:140221166-142215786 uc008mpv.1] (1988713)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm10</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chrX:82948870-85205050 uc009tri.2] (2253366)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr6:45060061-47301371 uc009bst.2] (2238325)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr16:39984371-42176292 uc007zfr.1](2189582)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:40596773-42653598 uc008jon.1] (2055883)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:140395430-142390050 uc008mpv.1] (1988713)</TD> | |||
</TABLE> | </TABLE> | ||
| Line 143: | Line 167: | ||
</TR> | </TR> | ||
<TR><TH>mm9</TH> | <TR><TH>mm9</TH> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr1:74440898-74440919 uc007bma.1] (22)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr10:85230424-85230445 uc007gmr.1] (22)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr11:77886549-77886570 uc007khz.1] (22)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12:110831098-110831119 uc007pay.1] (22)</TD> | ||
<TD>[http://genome-test. | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr13:54944553-54944574 uc007qpn.1] (22)</TD> | ||
</TR> | |||
<TR><TH>mm10</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr17:15192492-15192509 uc029tak.1] (18)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr1:137031387-137031406 uc029qtn.1] (20)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr1:187624235-187624254 uc029qvd.1] (20)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr1:82585127-82585146 uc029qpl.1] (20)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr11:46266296-46266315 uc029rlk.1] (20)</TD> | |||
</TR> | </TR> | ||
</TABLE> | </TABLE> | ||
| Line 155: | Line 186: | ||
Custom track: | Custom track: | ||
[http://genome-test. | [http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=hg18&hgt.customText=http://genomewiki.ucsc.edu/images/3/37/Hg18KnownGenesSmallIntronsExonsCt.txt.gz Hg18 small exons and introns] on the UCSC Genes track | ||
These are exons of size less than 22 bases, and introns of size less than 12 bases. The score column contains the size and thus you can filter smaller subsets via the score column in the table browser. | These are exons of size less than 22 bases, and introns of size less than 12 bases. The score column contains the size and thus you can filter smaller subsets via the score column in the table browser. | ||
Latest revision as of 22:01, 31 August 2018
gene sets measured
- hg17 - knownGenes version 2
- hg18 - knownGenes version 3
- hg19 - knownGenes version 7
- mm8 - knownGenes version 2
- mm9 - knownGenes version 3
- mm10 - knownGenes version 6
The min, max and mean measurements are per gene
summary of gene and exon counts
| db | gene count | total exon count | min exon count | max exon count | mean exon count |
|---|---|---|---|---|---|
| hg17 | 39368 | 405720 | 1 | 149 | 10 |
| hg18 | 56722 | 519308 | 1 | 2899 | 9 |
| hg19 | 82960 | 742493 | 1 | 5065 | 9 |
| mm8 | 31863 | 314628 | 1 | 313 | 10 |
| mm9 | 49220 | 417114 | 1 | 610 | 8 |
| mm10 | 59121 | 486425 | 1 | 813 | 8 |
summary of exon size statistics
| db | sum exon sizes | min exon size | max exon size | mean exon size |
|---|---|---|---|---|
| hg17 | 106839720 | 1 | 18172 | 263 |
| hg18 | 146371091 | 1 | 36861 | 282 |
| hg19 | 221924089 | 1 | 205012 | 299 |
| mm8 | 83159087 | 4 | 17497 | 264 |
| mm9 | 117671086 | 1 | 29698 | 282 |
| mm10 | 143087341 | 1 | 83437 | 294 |
summary of intron size statistics
| db | sum intron sizes | min intron size | max intron size | mean intron size |
|---|---|---|---|---|
| hg17 | 2223224397 | 6 | 1096450 | 6069 |
| hg18 | 2784923600 | 1 | 1047320 | 6023 |
| hg19 | 4127555916 | 1 | 1160410 | 6260 |
| mm8 | 1476081990 | 9 | 1347550 | 5220 |
| mm9 | 2055504784 | 1 | 1253430 | 5589 |
| mm10 | 2420504661 | 1 | 3003780 | 5665 |
Top five exon count genes
| db | gene name (exon count) | ||||
|---|---|---|---|---|---|
| hg17 | NM_004543 (149) | AF535142 (146) | AF535142 (146) | NM_033071 (146) | AF495910 (146) |
| hg18 | uc001yrq.1 (2899) | uc002zvw.1 (322) | uc002umr.1 (313) | uc002stk.1 (217) | uc002umt.1 (194) |
| hg19 | uc031qqx.1 (5065) | uc031rxg.1 (462) | uc031rqc.1 (363) | uc031rqd.1 (313) | uc021vsy.2 (312) |
| mm8 | NM_011652 (313) | NM_028004 (192) | NM_007738 (118) | NM_134448 (99) | DQ067088 (99) |
| mm9 | uc007pgj.1 (610) | uc008kfn.1 (313) | uc008kfo.1 (192) | uc008jqv.1 (157) | uc009rrh.1 (118) |
| mm10 | uc029ryi.1 (813) | uc008kfn.1 (313) | uc008kfo.1 (192) | uc008jqv.1 (157) | uc009rrh.1 (118) |
Top five largest CDS extent genes
| db | gene name (CDS extent size: thickEnd-thickStart) | ||||
|---|---|---|---|---|---|
| hg17 | NM_014141 (2298740) | NM_000109 (2217347) | CR749820 (2138880) | NM_004006 (2089394) | X14298 (2089394) |
| hg18 | uc003weu.1 (2298740) | uc004ddb.1 (2217347) | uc001pak.1 (2138880) | uc004dda.1 (2089394) | uc003wqd.1 (2055833) |
| hg19 | uc021ott.2 (2307732) | uc003weu.2 (2298740) | uc004ddb.1 (2217347) | uc001pak.2 (2138880) | uc004dda.1 (2089394) |
| mm8 | NM_007868 (2253366) | NM_001004357 (2238304) | NM_053011 (2055883) | AK134694 (1988713) | NM_053171 (1639258) |
| mm9 | uc009tri.1 (2253366) | uc009bst.1 (2238325) | uc007zfr.1 (2189582) | uc008jon.1 (2055883) | uc008mpv.1 (1988713) |
| mm10 | uc009tri.2 (2253366) | uc009bst.2 (2238325) | uc007zfr.1(2189582) | uc008jon.1 (2055883) | uc008mpv.1 (1988713) |
Top five smallest transcript genes
| db | gene name (transcript size: txEnd-txStart) | ||||
|---|---|---|---|---|---|
| hg17 | AF241539 (168) | AF277175 (176) | AY459291 (240) | AY605064 (243) | AF503918 (258) |
| hg18 | uc004buj.1 (20) | uc001dcm.1 (22) | uc001seo.1 (22) | uc001sqn.1 (22) | uc002wpa.1 (22) |
| hg19 | uc031pxj.1 (19) | uc021qzo.1 (19) | uc021pfi.1 (20) | uc021oot.1 (20) | uc021qbd.1 (20) |
| mm8 | AJ319753 (217) | BC107019 (231) | BC016221 (286) | NM_130876 (303) | NM_130873 (304) |
| mm9 | uc007bma.1 (22) | uc007gmr.1 (22) | uc007khz.1 (22) | uc007pay.1 (22) | uc007qpn.1 (22) |
| mm10 | uc029tak.1 (18) | uc029qtn.1 (20) | uc029qvd.1 (20) | uc029qpl.1 (20) | uc029rlk.1 (20) |
Custom Track of Small Exons and Introns
Custom track: Hg18 small exons and introns on the UCSC Genes track
These are exons of size less than 22 bases, and introns of size less than 12 bases. The score column contains the size and thus you can filter smaller subsets via the score column in the table browser.
These small exons and introns are used to maintain frame coding boundaries as found in mRNAs compared to the reference genome coordinates.
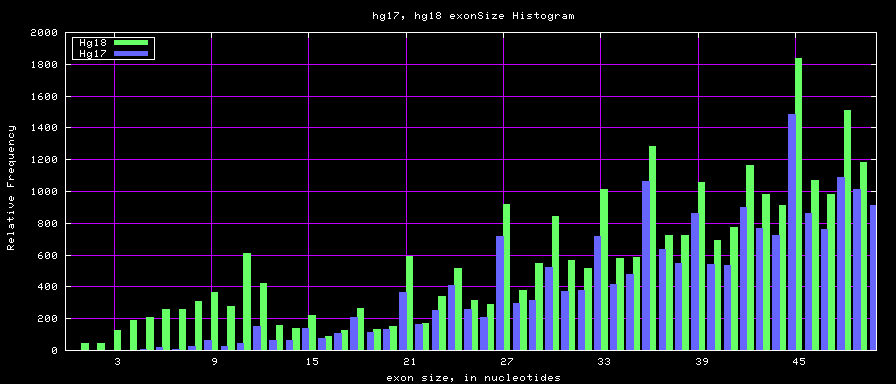
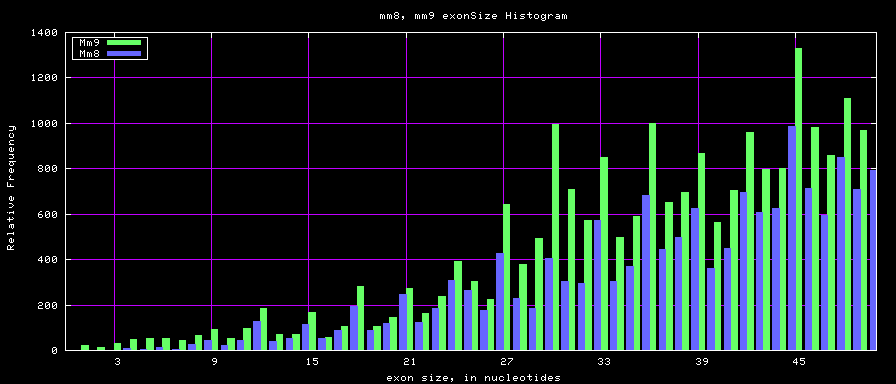
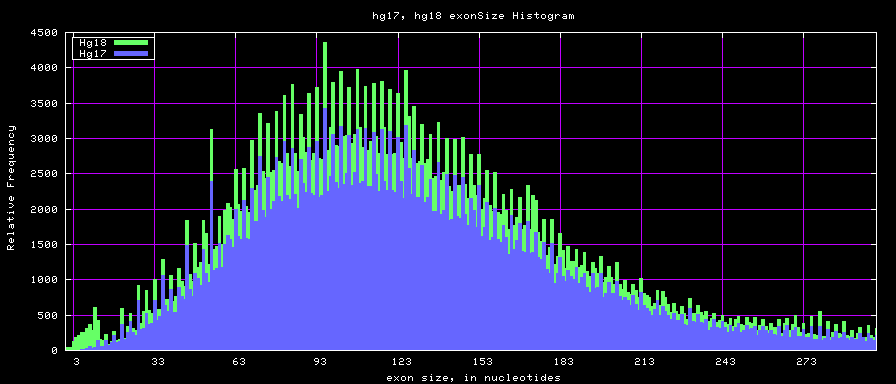
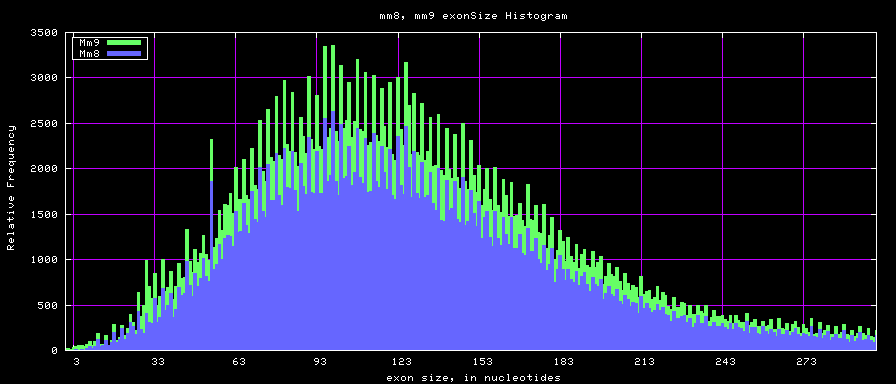
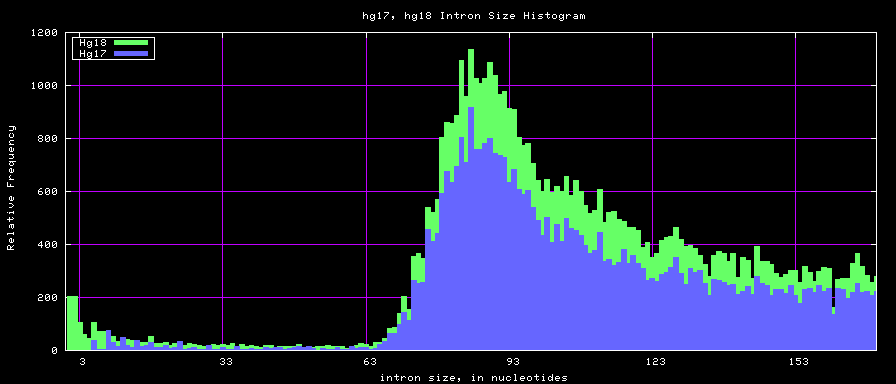
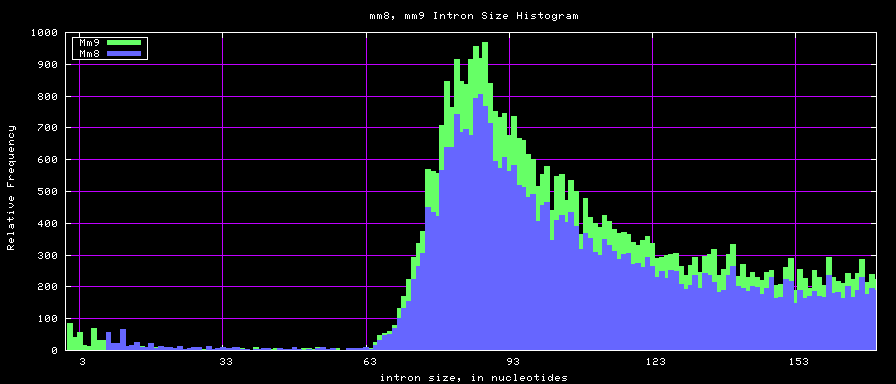
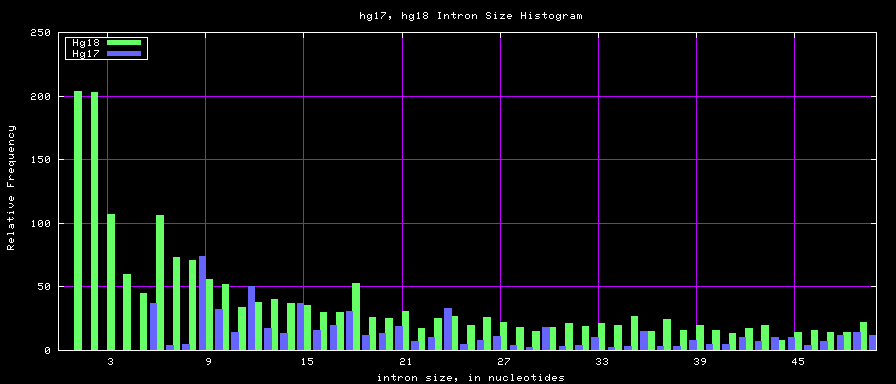
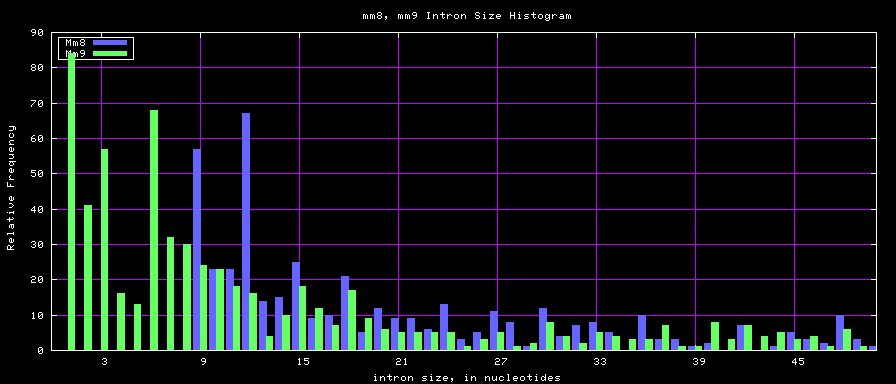
Histogram graphs
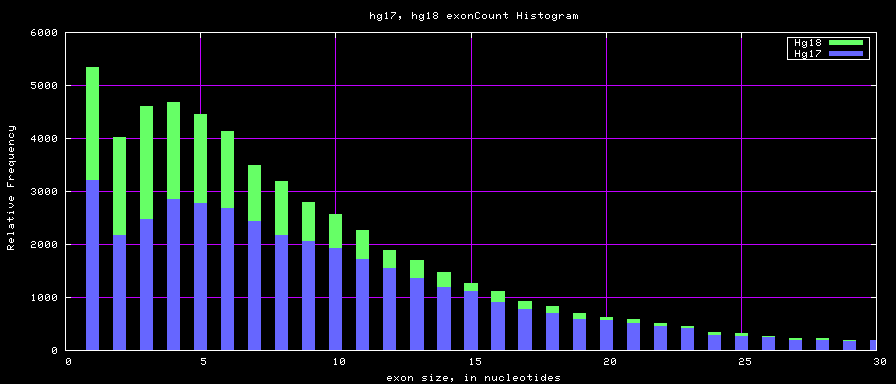 The caption on the graph above is incorrect. The X axis is Exon Count per Gene
The caption on the graph above is incorrect. The X axis is Exon Count per Gene
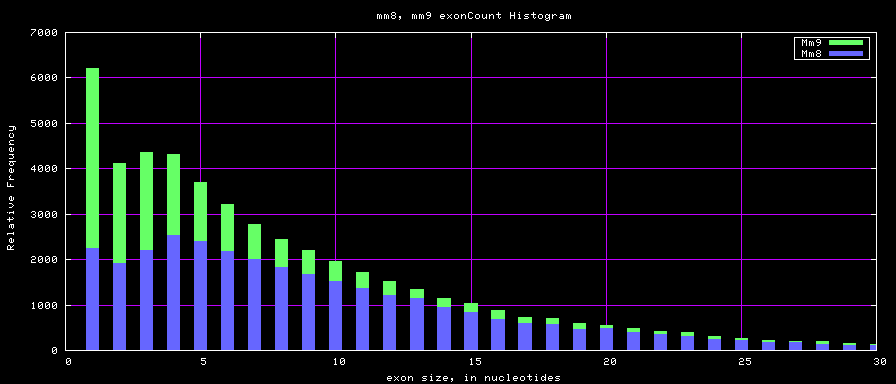 The caption on the graph above is incorrect. The X axis is Exon Count per Gene
The caption on the graph above is incorrect. The X axis is Exon Count per Gene
Methods
- From the table browser, request three different bed files for the knownGenes track:
- whole gene
- exons only
- introns only
- From those bed files, stats can be extracted
- gene count from: 'wc -l wholeGene.bed'
- exon count stats from:
STATS=`ave -col=10 wholeGene.bed -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
COUNT=`echo $STATS | cut -d' ' -f8 | awk '{printf "%d", $1}'`
- for exon or intron size stats:
STATS=`awk '{print $3-$2}' {introns,exons}.bed \
| ave -col=1 stdin -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5 | awk '{printf "%d", $1}'`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
SUM_SIZE=`awk '{sum += $3-$2} END{printf "%d", sum}' {introns,exons}.bed`
- top five exon count genes
sort -k10nr wholeGene.bed | head -5
- top five CDS size genes
awk '{cdsSize=$8-$7
if (cdsSize > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,cdsSize}
}' wholeGene.bed | sort -k5nr | head -5
- top five smallest transcript genes
awk '{size=$3-$2
if (size > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,size}
}' wholeGene.bed | sort -k5n | head -5