Gene Set Summary Statistics: Difference between revisions
From genomewiki
Jump to navigationJump to search
| Line 38: | Line 38: | ||
<TR><TH>db</TH><TH COLSPAN=5>gene name (exon count)</TH></TR> | <TR><TH>db</TH><TH COLSPAN=5>gene name (exon count)</TH></TR> | ||
<TR><TH>hg17</TH> | <TR><TH>hg17</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr2:152167363-152416454 NM_004543] (149)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6:152534937-153050100 AF535142] (146)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6:152534937-153050100 AF535142] (146)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6:152534937-153050100 NM_033071] (146)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6:152535029-153050648 AF495910] (146)</TD> | ||
</TR> | </TR> | ||
<TR><TH>hg18</TH> | <TR><TH>hg18</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr14: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr14:105065301-106352275 uc001yrq.1] (2899)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr22: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr22:20715572-21595078 uc002zvw.1] (322)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:179098964-179380395 uc002umr.1] (313)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:88937989-89411302 uc002stk.1] (217)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:179098964-179380395 uc002umt.1] (194)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm8</TH> | <TR><TH>mm8</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2:76504823-76783386 NM_011652] (313)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2:76504823-76783386 NM_028004] (192)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr9: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr9:108810682-108841812 NM_007738] (118)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr1: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr1:33956371-34252174 NM_134448] (99)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr4: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr4:122852023-123186663 DQ067088] (99)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm9</TH> | <TR><TH>mm9</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12: | <TD>[http://genome-test.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12:114498400-117248164 uc007pgj.1] (610)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome-test.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:76542041-76820604 uc008kfn.1] (313)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome-test.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:76542041-76820604 uc008kfo.1] (192)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome-test.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:51992167-52194318 uc008jqv.1] (157)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr9: | <TD>[http://genome-test.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr9:108855790-108886920 uc009rrh.1] (118)</TD> | ||
</TR> | </TR> | ||
</TABLE> | </TABLE> | ||
==Top five largest CDS extent genes== | ==Top five largest CDS extent genes== | ||
Revision as of 17:38, 19 September 2007
gene sets measured
- hg17 - knownGenes version 2
- hg18 - knownGenes version 3
- mm8 - knownGenes version 2
- mm9 - knownGenes version 3
The min, max and mean measurements are per gene
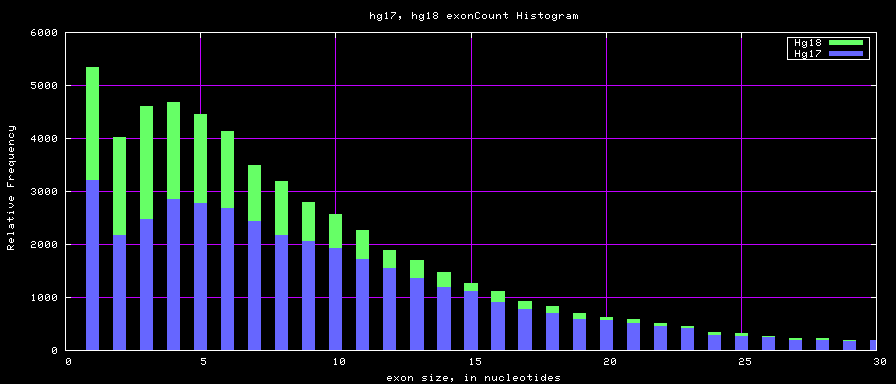
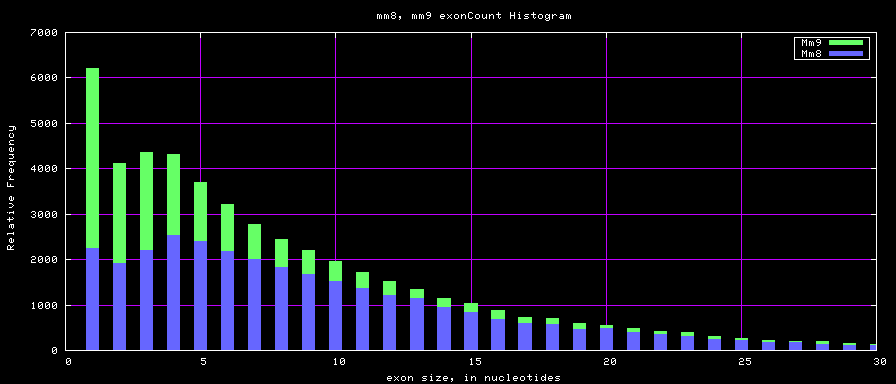
summary of gene and exon counts
| db | gene count | total exon count | min exon count | max exon count | mean exon count |
|---|---|---|---|---|---|
| hg17 | 39368 | 405720 | 1 | 149 | 10 |
| hg18 | 56722 | 519308 | 1 | 2899 | 9 |
| mm8 | 31863 | 314628 | 1 | 313 | 10 |
| mm9 | 49220 | 417114 | 1 | 610 | 8 |
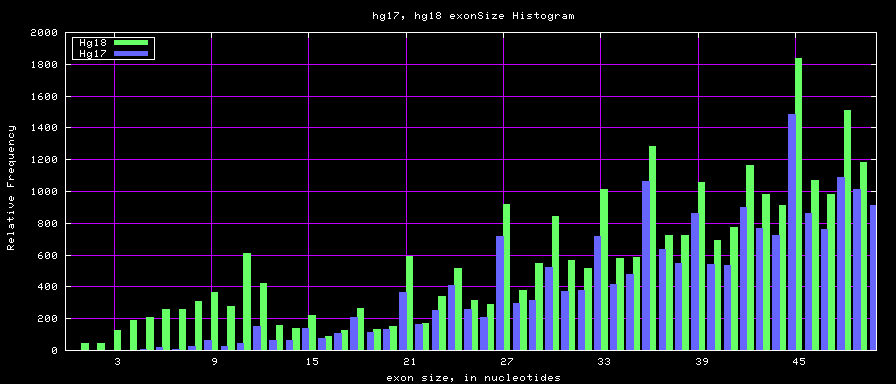
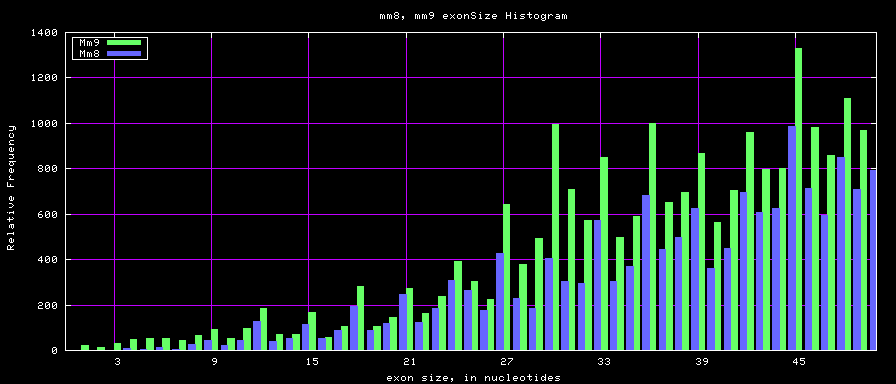
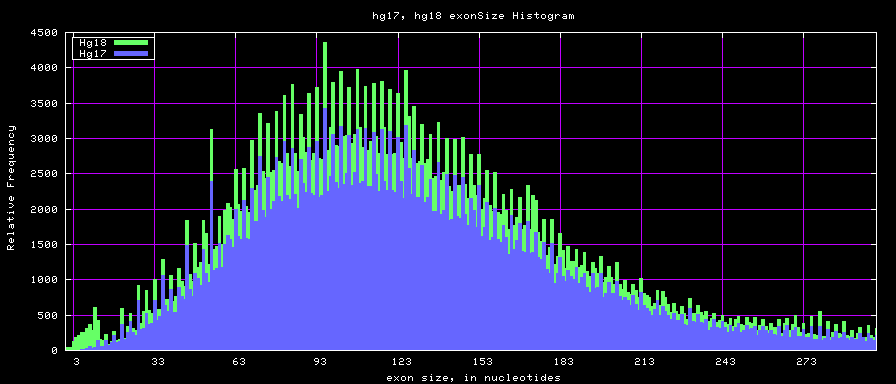
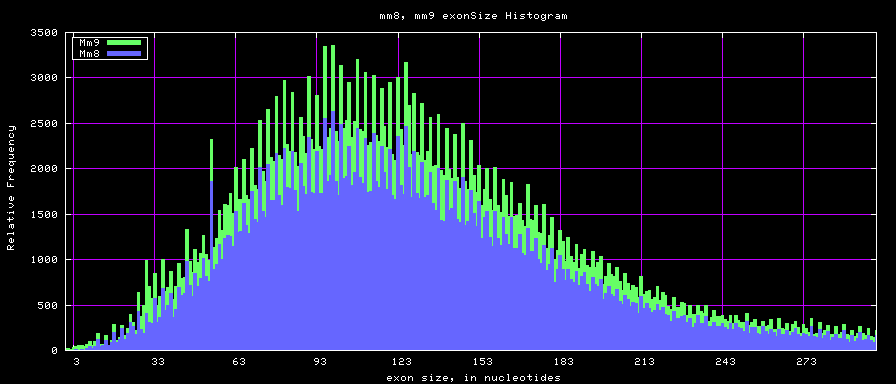
summary of exon size statistics
| db | sum exon sizes | min exon size | max exon size | mean exon size |
|---|---|---|---|---|
| hg17 | 106839720 | 1 | 18172 | 263 |
| hg18 | 146371091 | 1 | 36861 | 282 |
| mm8 | 83159087 | 4 | 17497 | 264 |
| mm9 | 117671086 | 1 | 29698 | 282 |
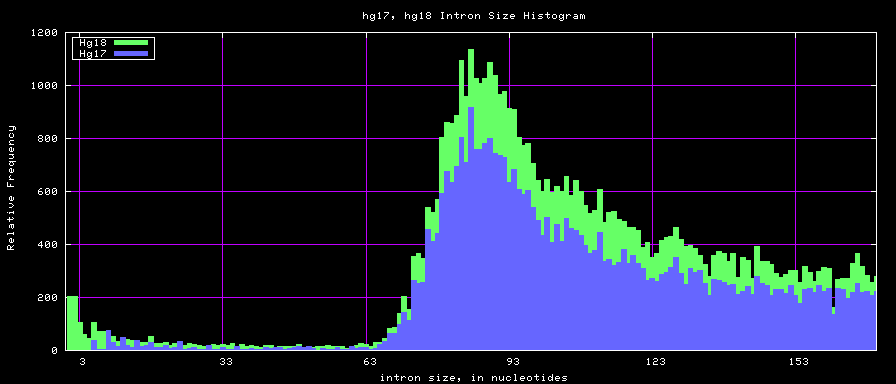
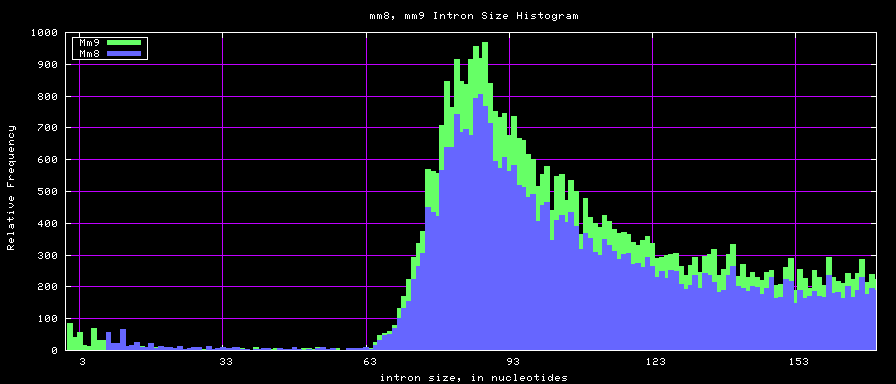
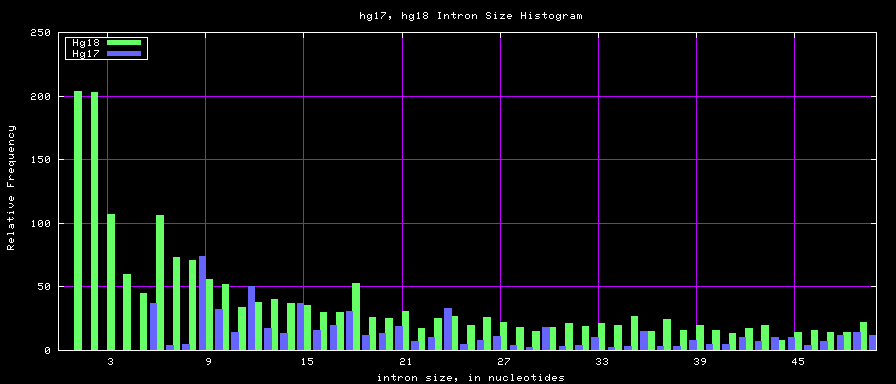
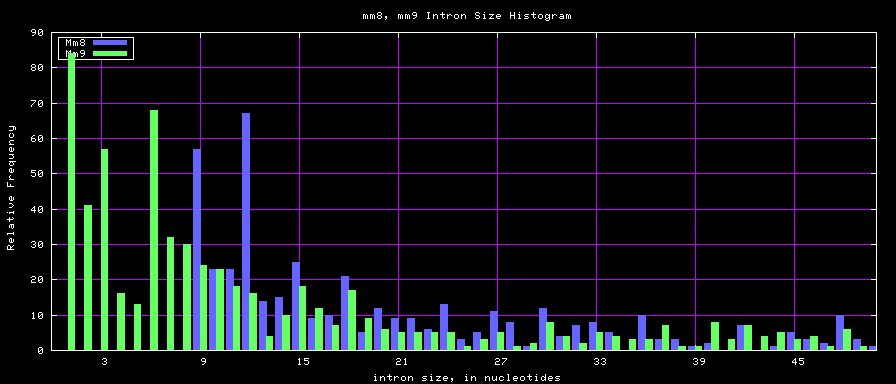
summary of intron size statistics
| db | sum intron sizes | min intron size | max intron size | mean intron size |
|---|---|---|---|---|
| hg17 | 2223224397 | 6 | 1096450 | 6069 |
| hg18 | 2784923600 | 1 | 1047320 | 6023 |
| mm8 | 1476081990 | 9 | 1347550 | 5220 |
| mm9 | 2055504784 | 1 | 1253430 | 5589 |
Top five exon count genes
| db | gene name (exon count) | ||||
|---|---|---|---|---|---|
| hg17 | NM_004543 (149) | AF535142 (146) | AF535142 (146) | NM_033071 (146) | AF495910 (146) |
| hg18 | uc001yrq.1 (2899) | uc002zvw.1 (322) | uc002umr.1 (313) | uc002stk.1 (217) | uc002umt.1 (194) |
| mm8 | NM_011652 (313) | NM_028004 (192) | NM_007738 (118) | NM_134448 (99) | DQ067088 (99) |
| mm9 | uc007pgj.1 (610) | uc008kfn.1 (313) | uc008kfo.1 (192) | uc008jqv.1 (157) | uc009rrh.1 (118) |
Top five largest CDS extent genes
| db | gene name (CDS extent size: thickEnd-thickStart) | ||||
|---|---|---|---|---|---|
| hg17 | NM_014141 (2298740) | NM_000109 (2217347) | CR749820 (2138880) | NM_004006 (2089394) | X14298 (2089394) |
| hg18 | uc003weu.1 (2298740) | uc004ddb.1 (2217347) | uc001pak.1 (2138880) | uc004dda.1 (2089394) | uc003wqd.1 (2055833) |
| mm8 | NM_007868 (2253366) | NM_001004357 (2238304) | NM_053011 (2055883) | AK134694 (1988713) | NM_053171 (1639258) |
| mm9 | uc009tri.1 (2253366) | uc009bst.1 (2238325) | uc007zfr.1 (2189582) | uc008jon.1 (2055883) | uc008mpv.1 (1988713) |
Top five smallest transcript genes
| db | gene name (transcript size: txEnd-txStart) | ||||
|---|---|---|---|---|---|
| hg17 | AF241539 (168) | AF277175 (176) | AY459291 (240) | AY605064 (243) | AF503918 (258) |
| hg18 | uc004buj.1 (20) | uc001dcm.1 (22) | uc001seo.1 (22) | uc001sqn.1 (22) | uc002wpa.1 (22) |
| mm8 | AJ319753 (217) | BC107019 (231) | BC016221 (286) | NM_130876 (303) | NM_130873 (304) |
| mm9 | uc007bma.1 (22) | uc007gmr.1 (22) | uc007khz.1 (22) | uc007pay.1 (22) | uc007qpn.1 (22) |
Custom Track of Small Exons and Introns
Custom track: Hg18 small exons and introns on the UCSC Genes track
These are exons of size less than 22 bases, and introns of size less than 12 bases. The score column contains the size and thus you can filter smaller subsets via the score column in the table browser.
These small exons and introns are used to maintain frame coding boundaries as found in mRNAs compared to the reference genome coordinates.
Histogram graphs
Methods
- From the table browser, request three different bed files for the knownGenes track:
- whole gene
- exons only
- introns only
- From those bed files, stats can be extracted
- gene count from: 'wc -l wholeGene.bed'
- exon count stats from:
STATS=`ave -col=10 wholeGene.bed -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
COUNT=`echo $STATS | cut -d' ' -f8 | awk '{printf "%d", $1}'`
- for exon or intron size stats:
STATS=`awk '{print $3-$2}' {introns,exons}.bed \
| ave -col=1 stdin -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5 | awk '{printf "%d", $1}'`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
SUM_SIZE=`awk '{sum += $3-$2} END{printf "%d", sum}' {introns,exons}.bed`
- top five exon count genes
sort -k10nr wholeGene.bed | head -5
- top five CDS size genes
awk '{cdsSize=$8-$7
if (cdsSize > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,cdsSize}
}' wholeGene.bed | sort -k5nr | head -5
- top five smallest transcript genes
awk '{size=$3-$2
if (size > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,size}
}' wholeGene.bed | sort -k5n | head -5