Marsupial phyloSNPs: Difference between revisions
From genomewiki
Jump to navigationJump to search
Tomemerald (talk | contribs) (New page: == Introduction to Marsupial phyloSNPs == In this project, new genomic data from the Tasmanian devil (Sarcophilus harrisii), Tasmanian tiger (Thylacinus cynocephalus), and echidna (Tachyg...) |
Tomemerald (talk | contribs) |
||
| Line 1: | Line 1: | ||
== Introduction to Marsupial phyloSNPs == | == Introduction to Marsupial phyloSNPs == | ||
In this project, new genomic data from the Tasmanian devil (Sarcophilus harrisii), Tasmanian tiger (Thylacinus cynocephalus), and echidna (Tachyglossus aculeatus) are analyzed for significant changes at the protein coding level. The goal is to find single amino acid changes in one of these species at a highly invariant residue in a well-conserved exon in a gene with known or predictable tertiary structure. Such changes are thought to enrich for genetic changes with significant, adaptive biochemical or phenotypic consequences ([[Opsin_evolution:_RGR_phyloSNPs#PhyloSNPs_in_RGR|1],[[Opsin_evolution:_RGR_phyloSNPs#PhyloSNPs_in_RGR|2][[Opsin_evolution:_RGR_phyloSNPs#PhyloSNPs_in_RGR|3],[[Opsin_evolution:_LWS_PhyloSNPs#PhyloSNPs_in_LWS_opsins:_alignment_of_94_LWS_vertebrate_opsins|4]), in contrast to ordinary SNPs at positions of low conservation. | In this project, new genomic data from the Tasmanian devil (Sarcophilus harrisii), Tasmanian tiger (Thylacinus cynocephalus), and echidna (Tachyglossus aculeatus) are analyzed for significant changes at the protein coding level. The goal is to find single amino acid changes in one of these species at a highly invariant residue in a well-conserved exon in a gene with known or predictable tertiary structure. Such changes are thought to enrich for genetic changes with significant, adaptive biochemical or phenotypic consequences ([[Opsin_evolution:_RGR_phyloSNPs#PhyloSNPs_in_RGR|1]],[[Opsin_evolution:_RGR_phyloSNPs#PhyloSNPs_in_RGR|2]][[Opsin_evolution:_RGR_phyloSNPs#PhyloSNPs_in_RGR|3]],[[Opsin_evolution:_LWS_PhyloSNPs#PhyloSNPs_in_LWS_opsins:_alignment_of_94_LWS_vertebrate_opsins|4]]), in contrast to ordinary SNPs at positions of low conservation. | ||
Revision as of 15:40, 9 February 2009
Introduction to Marsupial phyloSNPs
In this project, new genomic data from the Tasmanian devil (Sarcophilus harrisii), Tasmanian tiger (Thylacinus cynocephalus), and echidna (Tachyglossus aculeatus) are analyzed for significant changes at the protein coding level. The goal is to find single amino acid changes in one of these species at a highly invariant residue in a well-conserved exon in a gene with known or predictable tertiary structure. Such changes are thought to enrich for genetic changes with significant, adaptive biochemical or phenotypic consequences (1,23,4), in contrast to ordinary SNPs at positions of low conservation.
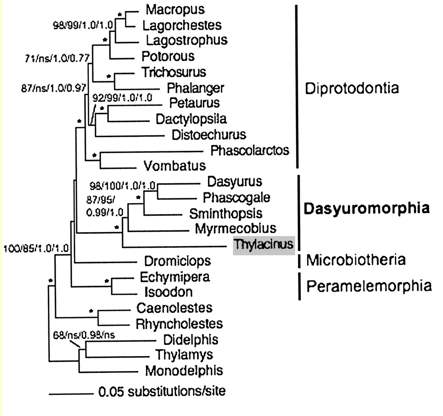
Assumed vertebrate phylogenetic tree
Marsupial relationships taken from 2009 paper establishing the mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus):
Newick tree that generates vertebrate phylogenetic tree used in the analysis here: ((((((((((((((((((homSap,panTro),gorGor),ponPyg),macMul),calJac),tarSyr),(micMur,otoGar)),tupBel), (((((musMus,ratNor),dipOrd),cavPor),speTri),(oryCun,ochPri))), (((((vicPac,susScr),turTru),bosTau),((equCab,(felCat,canFam)),(myoLuc,pteVam))),(eriEur,sorAra))), (((loxAfr,proCap),echTel),(dasNov,choHof))), (monDom,((macEug,triVul),(sarHar,thyCyn)))), (ornAna,tacAcu)), ((galGal,taeGut),anoCar)), xenTro), (((tetNig,takRub),(gasAcu,oryLap)),danRer)), calMil), petMar);
Phylo-sorting data
46 10 52 > 10 gene anoCar Anolis carolinensis (lizard) anoCar 29 11 30 > 11 gene bosTau Bos taurus (cow) bosTau 15 12 15 > 12 gene calJac Callithrix jacchus (marmoset) calJac 60 54 59 > 13 gene calMil Callorhinchus milii (elephantfish) 32 13 33 > 14 gene canFam Canis familiaris (dog) canFam 23 14 23 > 15 gene cavPor Cavia porcellus (guinea_pig) cavPor 41 15 42 > 16 gene choHof Choloepus hoffmanni (sloth) choHof 52 16 58 > 17 gene danRer Danio rerio (zebrafish) danRer 40 17 41 > 18 gene dasNov Dasypus novemcinctus (armadillo) dasNov 22 18 22 > 19 gene dipOrd Dipodomys ordii (kangaroo_rat) dipOrd 39 19 40 > 20 gene echTel Echinops telfairi (tenrec) echTel 30 20 31 > 21 gene equCab Equus caballus (horse) equCab 35 21 36 > 22 gene eriEur Erinaceus europaeus (hedgehog) eriEur 31 22 32 > 23 gene felCat Felis catus (cat) felCat 44 23 50 > 24 gene galGal Gallus gallus (chicken) galGal 50 24 56 > 25 gene gasAcu Gasterosteus aculeatus (stickleback) gasAcu 12 25 12 > 26 gene gorGor Gorilla gorilla (gorilla) gorGor 10 26 10 > 27 gene homSap Homo sapiens (human) hg181 37 27 38 > 28 gene loxAfr Loxodonta africana (elephant) loxAfr 55 55 44 > 29 gene macEug Macropus eugenii (wallaby) 14 28 14 > 30 gene macMul Macaca mulatta (rhesus) rheMac 17 29 17 > 31 gene micMur Microcebus murinus (mouse_lemur) micMur 42 30 43 > 32 gene monDom Monodelphis domestica (opossum) monDom 20 31 20 > 33 gene musMus Mus musculus (mouse) mm91 33 32 34 > 34 gene myoLuc Myotis lucifugus (microbat) myoLuc 26 33 26 > 35 gene ochPri Ochotona princeps (pika) ochPri 43 34 48 > 36 gene ornAna Ornithorhynchus anatinus (platypus) ornAna 25 35 25 > 37 gene oryCun Oryctolagus cuniculus (rabbit) oryCun 51 36 57 > 38 gene oryLap Oryzias latipes (medaka) oryLat 18 37 18 > 39 gene otoGar Otolemur garnettii (bushbaby) otoGar 11 38 11 > 40 gene panTro Pan troglodytes (chimp) panTro 53 39 60 > 41 gene petMar Petromyzon marinus (lamprey) petMar 13 40 13 > 42 gene ponPyg Pongo pygmaeus (orang) ponAbe 38 41 39 > 43 gene proCap Procavia capensis (hyrax) proCap 34 42 35 > 44 gene pteVam Pteropus vampyrus (macrobat) pteVam 21 43 21 > 45 gene ratNor Rattus norvegicus (rat) rn41 56 56 45 > 46 gene sarHar Sarcophilus harrisii (tasmanian_devil) 36 44 37 > 47 gene sorAra Sorex araneus (shrew) sorAra 24 45 24 > 48 gene speTri Spermophilus tridecemlineatus (squirrel) speTri 54 57 28 > 49 gene susScr Sus scrofa (pig) 59 58 49 > 50 gene tacAcu Tachyglossus aculeatus (echidna) 45 46 51 > 51 gene taeGut Taeniopygia guttata (finch) taeGut 49 47 55 > 52 gene takRub Takifugu rubripes (fugu) fr21 16 48 16 > 53 gene tarSyr Tarsius syrichta (tarsier) tarSyr 48 49 54 > 54 gene tetNig Tetraodon nigroviridis (pufferfish) tetNig 58 59 47 > 55 gene thyCyn Thylacinus cynocephalus (tasmanian_tiger) 57 60 46 > 56 gene triVul Trichosurus vulpecula (bushytail_possum) 19 50 19 > 57 gene tupBel Tupaia belangeri (tree_shrew) tupBel 28 51 29 > 58 gene turTru Tursiops truncatus (dolphin) turTru 27 52 27 > 59 gene vicPac Vicugna pacos (lama) vicPac 47 53 53 > 60 gene xenTro Xenopus tropicalis (frog) xenTro 44 44 51 f 51 gene fasta genus species common ucsc phy alp phy alp