Marsupial phyloSNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) No edit summary |
||
| Line 87: | Line 87: | ||
</pre> | </pre> | ||
== Candidate analysis == | |||
(methods explained here shortly) | |||
=== Case of ERN2 === | |||
Here a very ancient L appears to be transitioning to F at marsupial node but has not settled down so ends up L or F depending on lineage-sorting on each terminal marsupial leaf whereas placentals have settled on the phenylalanine (a phyloSNP caught in mid-air). While L and F might seem about the 'same' as amino acids, the branch length conservation totals say both are important but for different reasons: this is not a waffle codon or reduced alphabet situation. | |||
<pre> | |||
ERN2 endoplasmic reticulum to nucleus signalling 2 | |||
remote hoverall homology to ERN1 yet for this exon, only 3 diffs so some possibilities for confusion | |||
ERN1_monDom KLPFTIPELVQASPCRSSDGILYM all ERN1s are leucine | |||
uc002dma.2_hg18_5 KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_panTro KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_ponAbe KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_rheMac KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_calJac KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_tarSyr KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_micMur KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_tupBel KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_mm9_5_ KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_rn4_5_ KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_cavPor KLPFTIPELVHTSPCRSSDGVFYT | |||
uc002dma.2_speTri KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_oryCun KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_ochPri KLPFSIPELVHASPCRSSDGVFYT | |||
uc002dma.2_turTru RLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_bosTau RLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_equCab KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_felCat RLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_canFam KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_myoLuc KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_eriEur KLPFTVPELVHTSPCRSSDGVFYT | |||
uc002dma.2_sorAra KLPFTIPELVHASPCRSSDGVFYT | |||
uc002dma.2_loxAfr KLPFTIPELVHAS----------- | |||
uc002dma.2_proCap ---------------------FYT | |||
uc002dma.2_echTel KLPFTIPELVLASPCRSSDGVFYT | |||
uc002dma.2_dasNov KLPFTIPELVHTSPCRSSDGIFYT | |||
uc002dma.2_monDom KLPFTIPELVHASPCRSSDGVLYT | |||
macropus eugeneii KLPFTIPELVQASPCRSSDGILYM | |||
uc002dma.2_ornAna KLPFTIPELVQSSPCRSSDGILYT | |||
uc002dma.2_anoCar KLPFTIPELVQSSPCRSSDGIIYT | |||
finch KLPFTIPELVQSSPCRSSDGVLYT | |||
chicken KLPFTIPELVQASPCRSSDGILYM | |||
XenopusT KLPFTIPELVQSSPCRSSDGILYT | |||
XenopusL KLPFTIPELVQSSPCRSSDGILYT | |||
uc002dma.2_tetNig KLPFTIPELVQASPCRSSDGVLYM | |||
uc002dma.2_fr2_5_ KLPFTIPELVQASPCRSSDGVLYM | |||
uc002dma.2_gasAcu KLPFTIPDLVQSAPCRSSDGILYT | |||
uc002dma.2_oryLat KLPFTIPELVQSAPCRSSDGILYT | |||
uc002dma.2_petMar KLPFTIPELVHASPCRTSDGVLYT | |||
ERN1 are all L | |||
uc002jdz.2_hg18_5 KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_panTro KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_ponAbe KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_rheMac KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_calJac KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_tarSyr KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_micMur KLPFTIPELVQASPCRSTDGILYM | |||
uc002jdz.2_otoGar KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_tupBel KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_mm9_5_ KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_rn4_5_ KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_dipOrd KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_cavPor KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_speTri KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_oryCun KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_vicPac KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_turTru KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_bosTau KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_equCab KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_canFam KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_myoLuc KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_pteVam KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_eriEur KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_sorAra KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_loxAfr KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_proCap KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_echTel KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_dasNov KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_choHof KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_monDom KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_ornAna KLPFTIPELVHASPCRSSDGILYM | |||
uc002jdz.2_galGal KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_taeGut KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_anoCar KLPFTIPELVQASPCRSSDGILYM | |||
uc002jdz.2_xenTro KLPFTIPELVQSSPCRSSDGILYT | |||
uc002jdz.2_tetNig KLPFTIPELVQASPCRSSDGVLYM | |||
uc002jdz.2_fr2_5_ KLPFTIPELVQASPCRSSDGVLYM | |||
uc002jdz.2_gasAcu KLPFTIPELVQASPCRSSDGVLYM | |||
uc002jdz.2_oryLat KLPFTIPELVQASPCRSSDGVLYM | |||
uc002jdz.2_danRer KLPFTIPELVQASPCRSSDGILYM | |||
</pre> | |||
=== Case of XXXX === | |||
(more shortly) | |||
=== Case of YYYY === | |||
(more shortly) | |||
=== Case of ZZZZ === | |||
(more shortly) | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Revision as of 16:03, 9 February 2009
Introduction to Marsupial phyloSNPs
In this project, new genomic data from the Tasmanian devil (Sarcophilus harrisii), Tasmanian tiger (Thylacinus cynocephalus), and echidna (Tachyglossus aculeatus) are analyzed for significant changes at the protein coding level. The goal is to find single amino acid changes in one of these species at a highly invariant residue in a well-conserved exon in a gene with known or predictable tertiary structure. Such changes are thought to enrich for genetic changes with significant, adaptive biochemical or phenotypic consequences (1,2,3,4), in contrast to ordinary SNPs at positions of low conservation. Thus phyloSNPs are informative to the distinctive biology of the species carrying them and suggest a focus for subsequent experiment.
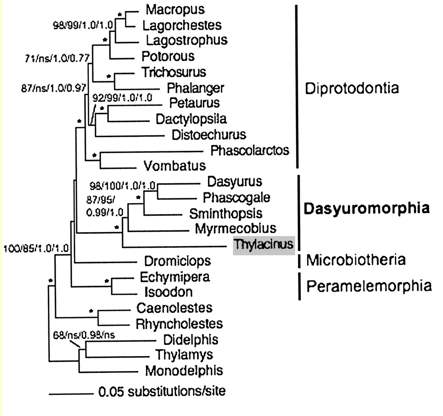
Marsupial genomic and cDNA data to date has been quite limited compared to placental mammal. Yet as outgroup, metatheran animals provide important context to placentals and represent important context in understanding human protein evolution. The monotheres are inevitably limited by the paucity of extant species (basically platypus and echidna) and dim prospects for fossil DNA. Consequently echidna provides an important adjunct to the existing but incomplete platypus assembly. While extant birds and reptiles -- the preceding divergence node -- are abundant it must be remembered that a very considerable time elapsed (from 310 mry to 175 mry) prior to divergence of mammals with living representatives. This gap of 135 myr is comparable to the whole evolutionary record of theran mammals.
Assumed vertebrate phylogenetic tree
Marsupial relationships taken from 2009 paper establishing the mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus):
Newick tree that generates vertebrate phylogenetic tree used in the analysis here: ((((((((((((((((((homSap,panTro),gorGor),ponPyg),macMul),calJac),tarSyr),(micMur,otoGar)),tupBel), (((((musMus,ratNor),dipOrd),cavPor),speTri),(oryCun,ochPri))), (((((vicPac,susScr),turTru),bosTau),((equCab,(felCat,canFam)),(myoLuc,pteVam))),(eriEur,sorAra))), (((loxAfr,proCap),echTel),(dasNov,choHof))), (monDom,((macEug,triVul),(sarHar,thyCyn)))), (ornAna,tacAcu)), ((galGal,taeGut),anoCar)), xenTro), (((tetNig,takRub),(gasAcu,oryLap)),danRer)), calMil), petMar);
Phylo-sorting data
46 10 52 > 10 gene anoCar Anolis carolinensis (lizard) anoCar 29 11 30 > 11 gene bosTau Bos taurus (cow) bosTau 15 12 15 > 12 gene calJac Callithrix jacchus (marmoset) calJac 60 54 59 > 13 gene calMil Callorhinchus milii (elephantfish) 32 13 33 > 14 gene canFam Canis familiaris (dog) canFam 23 14 23 > 15 gene cavPor Cavia porcellus (guinea_pig) cavPor 41 15 42 > 16 gene choHof Choloepus hoffmanni (sloth) choHof 52 16 58 > 17 gene danRer Danio rerio (zebrafish) danRer 40 17 41 > 18 gene dasNov Dasypus novemcinctus (armadillo) dasNov 22 18 22 > 19 gene dipOrd Dipodomys ordii (kangaroo_rat) dipOrd 39 19 40 > 20 gene echTel Echinops telfairi (tenrec) echTel 30 20 31 > 21 gene equCab Equus caballus (horse) equCab 35 21 36 > 22 gene eriEur Erinaceus europaeus (hedgehog) eriEur 31 22 32 > 23 gene felCat Felis catus (cat) felCat 44 23 50 > 24 gene galGal Gallus gallus (chicken) galGal 50 24 56 > 25 gene gasAcu Gasterosteus aculeatus (stickleback) gasAcu 12 25 12 > 26 gene gorGor Gorilla gorilla (gorilla) gorGor 10 26 10 > 27 gene homSap Homo sapiens (human) hg181 37 27 38 > 28 gene loxAfr Loxodonta africana (elephant) loxAfr 55 55 44 > 29 gene macEug Macropus eugenii (wallaby) 14 28 14 > 30 gene macMul Macaca mulatta (rhesus) rheMac 17 29 17 > 31 gene micMur Microcebus murinus (mouse_lemur) micMur 42 30 43 > 32 gene monDom Monodelphis domestica (opossum) monDom 20 31 20 > 33 gene musMus Mus musculus (mouse) mm91 33 32 34 > 34 gene myoLuc Myotis lucifugus (microbat) myoLuc 26 33 26 > 35 gene ochPri Ochotona princeps (pika) ochPri 43 34 48 > 36 gene ornAna Ornithorhynchus anatinus (platypus) ornAna 25 35 25 > 37 gene oryCun Oryctolagus cuniculus (rabbit) oryCun 51 36 57 > 38 gene oryLap Oryzias latipes (medaka) oryLat 18 37 18 > 39 gene otoGar Otolemur garnettii (bushbaby) otoGar 11 38 11 > 40 gene panTro Pan troglodytes (chimp) panTro 53 39 60 > 41 gene petMar Petromyzon marinus (lamprey) petMar 13 40 13 > 42 gene ponPyg Pongo pygmaeus (orang) ponAbe 38 41 39 > 43 gene proCap Procavia capensis (hyrax) proCap 34 42 35 > 44 gene pteVam Pteropus vampyrus (macrobat) pteVam 21 43 21 > 45 gene ratNor Rattus norvegicus (rat) rn41 56 56 45 > 46 gene sarHar Sarcophilus harrisii (tasmanian_devil) 36 44 37 > 47 gene sorAra Sorex araneus (shrew) sorAra 24 45 24 > 48 gene speTri Spermophilus tridecemlineatus (squirrel) speTri 54 57 28 > 49 gene susScr Sus scrofa (pig) 59 58 49 > 50 gene tacAcu Tachyglossus aculeatus (echidna) 45 46 51 > 51 gene taeGut Taeniopygia guttata (finch) taeGut 49 47 55 > 52 gene takRub Takifugu rubripes (fugu) fr21 16 48 16 > 53 gene tarSyr Tarsius syrichta (tarsier) tarSyr 48 49 54 > 54 gene tetNig Tetraodon nigroviridis (pufferfish) tetNig 58 59 47 > 55 gene thyCyn Thylacinus cynocephalus (tasmanian_tiger) 57 60 46 > 56 gene triVul Trichosurus vulpecula (bushytail_possum) 19 50 19 > 57 gene tupBel Tupaia belangeri (tree_shrew) tupBel 28 51 29 > 58 gene turTru Tursiops truncatus (dolphin) turTru 27 52 27 > 59 gene vicPac Vicugna pacos (lama) vicPac 47 53 53 > 60 gene xenTro Xenopus tropicalis (frog) xenTro 44 44 51 f 51 gene fasta genus species common ucsc phy alp phy alp
Candidate analysis
(methods explained here shortly)
Case of ERN2
Here a very ancient L appears to be transitioning to F at marsupial node but has not settled down so ends up L or F depending on lineage-sorting on each terminal marsupial leaf whereas placentals have settled on the phenylalanine (a phyloSNP caught in mid-air). While L and F might seem about the 'same' as amino acids, the branch length conservation totals say both are important but for different reasons: this is not a waffle codon or reduced alphabet situation.
ERN2 endoplasmic reticulum to nucleus signalling 2 remote hoverall homology to ERN1 yet for this exon, only 3 diffs so some possibilities for confusion ERN1_monDom KLPFTIPELVQASPCRSSDGILYM all ERN1s are leucine uc002dma.2_hg18_5 KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_panTro KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_ponAbe KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_rheMac KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_calJac KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_tarSyr KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_micMur KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_tupBel KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_mm9_5_ KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_rn4_5_ KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_cavPor KLPFTIPELVHTSPCRSSDGVFYT uc002dma.2_speTri KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_oryCun KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_ochPri KLPFSIPELVHASPCRSSDGVFYT uc002dma.2_turTru RLPFTIPELVHASPCRSSDGVFYT uc002dma.2_bosTau RLPFTIPELVHASPCRSSDGVFYT uc002dma.2_equCab KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_felCat RLPFTIPELVHASPCRSSDGVFYT uc002dma.2_canFam KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_myoLuc KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_eriEur KLPFTVPELVHTSPCRSSDGVFYT uc002dma.2_sorAra KLPFTIPELVHASPCRSSDGVFYT uc002dma.2_loxAfr KLPFTIPELVHAS----------- uc002dma.2_proCap ---------------------FYT uc002dma.2_echTel KLPFTIPELVLASPCRSSDGVFYT uc002dma.2_dasNov KLPFTIPELVHTSPCRSSDGIFYT uc002dma.2_monDom KLPFTIPELVHASPCRSSDGVLYT macropus eugeneii KLPFTIPELVQASPCRSSDGILYM uc002dma.2_ornAna KLPFTIPELVQSSPCRSSDGILYT uc002dma.2_anoCar KLPFTIPELVQSSPCRSSDGIIYT finch KLPFTIPELVQSSPCRSSDGVLYT chicken KLPFTIPELVQASPCRSSDGILYM XenopusT KLPFTIPELVQSSPCRSSDGILYT XenopusL KLPFTIPELVQSSPCRSSDGILYT uc002dma.2_tetNig KLPFTIPELVQASPCRSSDGVLYM uc002dma.2_fr2_5_ KLPFTIPELVQASPCRSSDGVLYM uc002dma.2_gasAcu KLPFTIPDLVQSAPCRSSDGILYT uc002dma.2_oryLat KLPFTIPELVQSAPCRSSDGILYT uc002dma.2_petMar KLPFTIPELVHASPCRTSDGVLYT ERN1 are all L uc002jdz.2_hg18_5 KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_panTro KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_ponAbe KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_rheMac KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_calJac KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_tarSyr KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_micMur KLPFTIPELVQASPCRSTDGILYM uc002jdz.2_otoGar KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_tupBel KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_mm9_5_ KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_rn4_5_ KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_dipOrd KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_cavPor KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_speTri KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_oryCun KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_vicPac KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_turTru KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_bosTau KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_equCab KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_canFam KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_myoLuc KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_pteVam KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_eriEur KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_sorAra KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_loxAfr KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_proCap KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_echTel KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_dasNov KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_choHof KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_monDom KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_ornAna KLPFTIPELVHASPCRSSDGILYM uc002jdz.2_galGal KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_taeGut KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_anoCar KLPFTIPELVQASPCRSSDGILYM uc002jdz.2_xenTro KLPFTIPELVQSSPCRSSDGILYT uc002jdz.2_tetNig KLPFTIPELVQASPCRSSDGVLYM uc002jdz.2_fr2_5_ KLPFTIPELVQASPCRSSDGVLYM uc002jdz.2_gasAcu KLPFTIPELVQASPCRSSDGVLYM uc002jdz.2_oryLat KLPFTIPELVQASPCRSSDGVLYM uc002jdz.2_danRer KLPFTIPELVQASPCRSSDGILYM
Case of XXXX
(more shortly)
Case of YYYY
(more shortly)
Case of ZZZZ
(more shortly)