Cryptochrome evolution: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| (163 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
See also: [[Cryptochrome_refSeqs|Curated reference sequences for cryptochromes and photolyases]] | |||
'''Updates''': fixes and additions become difficult to locate within a long article | |||
so these are provided below in reverse chronological order linked to their approximate location. | |||
08 Jun 12: significant additions to [[#4Fe-4S_photolyases_and_their_relation_to_primases|iron-sulfur photolyases and primases]] | |||
21 May 12: determined DASH's [[#DASH:_spotty_phylogenetic_distribution_and_unexplained_carboxy_terminal_extension|phylogenetic distribution and terminal motif]] using a greatly improved [[Cryptochrome_refSeqs#DASH|sequence set]]. | |||
== Introduction to Cryptochromes == | == Introduction to Cryptochromes == | ||
Cryptochromes are large flavoproteins with a curiously complex evolutionary history, beginning billions of years ago as repair enzymes | Cryptochromes are large flavoproteins with a curiously complex evolutionary history, beginning billions of years ago as dna repair enzymes (or even earlier as replication primase). An old gene duplication followed by specializing divergence gave rise to two paralogs repairing distinct types of dna damage (cyclobutane pyrimidine dimers and 6-4 pyrimidine-pyrimidone pairs). These photolyases initially used FAD activated by visible blue light to undo the damage done by UV and other processes. | ||
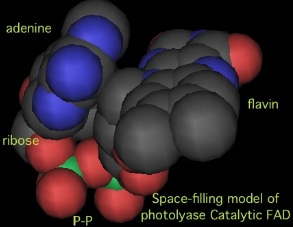
Since FAD has relatively low adsorbance, photolyases evolved a second site for an antenna chromophore with better light harvesting capabilities that could transfer its excitation to the FAD at the active site. This elusive antenna molecule may be FMN, a folate, lumazine, or a 5-deazariboflavin called Fo once thought restrict to methanogenic archaea. In the case of the much-studied Drosophila, [http://www.ncbi.nlm.nih.gov/pubmed/22175817 both the photolyases] utilize Fo, making it a [http://www.ncbi.nlm.nih.gov/pubmed/19570997 new vitamin] for this species since the biosynthetic genes are absent. Cryptochrome so far lack antenna molecules but retain the binding domain and substrate pocket. | |||
The next round of gene duplication of the 6-4 photolyase gave rise to a cryptochrome which retained the conformational change induced by FAD binding of blue light but lost dna repair capacity, instead specializing in entraining the day/night circadian rhythm cycle. However the distinction between signalling (non-enzymatic) and catalytic gene family members is muddled. Later rounds of gene duplication gave rise to yet more orthology classes to be followed -- sometimes hundreds of millions of years later -- by gene loss in some large lineages. | |||
The seven main classes were retained in various combinations in different clades during the subsequent course of evolution, causing endless comparative nomenclatural confusion (when in doubt, look at the [[Cryptochrome_refSeqs|amino acid sequences]]). For example, Drosophila did not retain CRY1A unlike other insects while placental mammals lost all three photolyases though marsupials retained one and monotremes two. Gallinaceous birds also lost a photolyase. Rayfinned fish had a series of further duplications within the gene family. Despite this, the primary sequence, exon structure, fold and FAD, antenna and dna binding sites have largely been conserved -- along with key regulatory binding sites to other proteins -- even as antenna molecules and dna repair capacity might be dispensed with. | |||
=== A new vertebrate cryptochrome CRY7 with a ubiquitin binding domain UIM === | |||
Even ten years into the whole genome era, the comparative genomics of cryptochromes and photolyases has never been considered, perhaps because of a narrow experimental focus on 'model' organisms such as mouse and fruit fly that, as it turns out, have rather restricted and unrepresentative gene family complements. Since most annotation effort goes into human (which are very deficient in their repertoire), the lack of a suitable homology probe there lets novel photolyases and cryptochromes in other species go undiscovered. | |||
This section describes a new cryptochrome orthology class (designated CRY7 here) with an extensive but not universal phylogenetic distribution. It apparently arose in the pre-Cambrian as a segmental gene duplication of CRY64 (or vice versa) based on its independent intronation pattern. Most remarkably, CRY7 possesses an amino terminal ubiquitin binding domain. The new protein is evolving overall rather rapidly for a cryptochrome and has been lost from many clades but it still retains the two core domains. Although the antenna molecule cannot be predicted, the FAD cofactor is likely present, based on structurally modelling with 1U3C and 3CVW (from CRY64_droMel, 34% identity and CRY1A_araTha, 29% identity). | |||
CRY7 is absent from mammals and indeed all amniotes but still present in amphibians, lobe-finned, ray-finned fish including basal gar, and two molluscs. These genes form a single new orthology class with distinct syntenic location, intronation pattern, and domain structure. The unusual phylogenetic distribution cannot be plausibly explained by prokaryotic endosymbiont, DNA contamination by xenobiotics (in filter-feeders), nor horizontal gene transfer. There is also affinity to two placozoan cryptochrome but these lack the ubiquitin binding domain. | |||
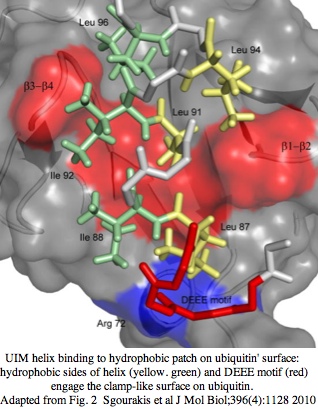
CRY7 in frog has 20 overlapping transcripts at GenBank dating back to 2003 that cover all but the middle of the gene. Expression has been reported from egg (BX771555, AL893008), neurulation embryo (BX699228, AL662439), whole embryo (CX470086, CX470087), tailbud head (CR562794, CR562774), adult testes (CX928370 and 7 others), and adult ovary (DR850985 and 3 others). These sites of expression do not distinguish between a DNA repair role and photosignalling. However the presence of the N-terminal UIM domain strongly suggests the latter because protein turnover is a well-established component of the cryptochrome circadian system. | |||
In non-mammalian species, circadian regulation of other genes can take place directly at cellular sites indpendently of the central nervous system, often in species with [http://rstb.royalsocietypublishing.org/content/364/1531/2849.full.pdf extra-retinal opsin expression]. Frog expresses melanopsin in [http://www.xenbase.org/literature/article.do?method=display&articleId=2463 skin melanophores]; fish also express an opsin in lateral line iridophores which exhibit [http://www.ncbi.nlm.nih.gov/pubmed/17043404 circadian color changes]; and squid utilize an [http://rsbl.royalsocietypublishing.org/content/6/5/600.full external opsin] to manage camouflage. Ultra-structural coexpression studies of CRY7 and the respective opsins might establish an association. | |||
[[Image:CRY7uim.jpg|left]] | |||
(to be | The ubiquitin interacting motif (UIM) consists of 20 amino acid residues first described in the 26S proteasome subunit that recognises ubiquitin. Ubiquitin binds UIM so the motif triggers a cascade of downstream signalling events. The UIM forms a short alpha-helix that can [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3080845/?tool=pubmed fits into the ubiquitin pocket] via hydrophobic and electrostatic interactions. The UIM motif of a frog CRY7 gene model was predicted by subsequent automatic procedures at [http://www.genome.jp/dbget-bin/www_bget?xtr:100486741 KEGG] but the short UIM motif was neither homologously confirmed in other species nor shown actually part of the cryptochrome gene (rather than belonging to an upstream adjacent gene with a missed stop codon). UIM domains are widespread but not necessarily homologous (ie mobile chimeric domains) because short motif can evolve in situ. | ||
However here the amino terminus begins with about 70 semi-conserved residues, followed by the UIM domain beginning a new exon. This extended motif has no Blast counterpart in other known proteins even using a consensus sequence probe. It is followed by a long spacer region of about 140 amino acids that is evolving chaotically in both length and composition. This pattern suggests a fusion with a UIM donor protein with the spacer region in the process of being discarded. Conservation begins again as the antenna domain is reached and continues through the FAD domain all the way to the carboxy terminus (which extends nearly 100 amino acids beyond any homology with the CRY64 FAD domain). A crystallographic structure for CRY7 might reveal more distant relationships for the conserved N- and C-terminal extensions. | |||
<br clear=all> | |||
Species UIM motif UIM conservation Genus species (common) | |||
CRY7_xenTro GYETDLELAIALSLQEHNQL GYETD....I....Q.HNQL Xenopus tropicalis (frog) | |||
CRY7_lepOcu VEEEEVEVALALSLQELGVS SV.EE.V.V......Q.LGV Lepisosteus oculatus (gar) | |||
CRY7_danRer DESEELELALTLSLYETKQI D.SE......T...Y.T.QI Danio rerio (zebrafish) | |||
CRY7_salSal DEDDELAVALALSLLEVKRQ D.....AV........V.R. Salmo salar (salmon) | |||
CRY7_gadMor DEEDELEVALALSLLDVKPQ ...............D..GH Haplochromis burtoni (chichlid) | |||
CRY7_hapBur TEDDELELALALSLLDMKGH .Q........S...V..D.H Gasterosteus aculeatus (stickleback) | |||
CRY7_oreNil TEDDELELALALSLLDMKGQ ....D.........M..E.. Oryzias latipes (medaka) | |||
CRY7_xipMac MEDDELELALALSLLDMKDQ ...............D..G. Oreochromis niloticus (tilapia) | |||
CRY7_gasAcu TQDDELELALSLSLVEMDDH ...ED.........V....C Tetraodon nigroviridis (fugu) | |||
CRY7_takRub TEDDELELALALSLVETKDY ..............V.T..Y Takifugu rubripes (fugu) | |||
CRY7_oryLat TEDDDLELALALSLMEMEDQ D.E....V.......DV.P. Gadus morhua (cod) | |||
CRY7_tetNig TEDEDLELALALSLVEMKDC M..............D.... Xiphophorus maculatus (platyfish) | |||
consensus TEDDELELALALSLLEMKDQ TEDDELELALALSLLEMKDQ UIM motif PFAM: PF02809 | |||
Synteny is not helpful in the CRY7 situation with only Oryzias latipes (medaka) sharing a neighboring gene with frog. CRY7 does not represent a segmental duplication of CRY64 because its intronation pattern is totally different, plus the percent identity is very low for a pair of cryptochromes. Vertebrates do lose and gain introns but that process is extremely slow. More likely the gene duplication took place in single-celled eukaryotes prior to the principal era of intronation, with the two ortholog classes then acquiring introns independently at essentially random positions. CRY7 is a misclassified paralog cross-over in Genomicus and not represented in the UCSC 46-way whole genome alignment because human lacks the gene. | |||
CRY7 has a completely unique intronation pattern lacking any relationship to CRY64 (its best blast match within the gene family) or any other cryptochrome or photolyase. Since this pattern is strongly conserved in the CRY7 ortholog set, it is likely ancient. If so, this protein represents a very old branch of the gene family but one that is unrecognizable or lost from most lineages. CRY7 is not an evolutionary novelty, having persisted for 450,000,000 years in vertebrates; nearly half of the [http://en.wikipedia.org/wiki/Vertebrates 58,000 living species of vertebrates] retain it, though not any amniotes studied to date. The position and phase of CRY64 intron breaks are positioned by homology into frog CRY7 below and contrasted with a comparison of CRY64 to human CRY1 (which share 5 identical intron sites). <font color =blue>Blue</font> indicates phase 00, <font color =orange>orange</font> phase 12, <font color =red>red</font> phase 21, <font color =magenta>magenta </font>perfect match of position and phase: | |||
CRY7_xenTro Xenopus tropicalis (frog) introns relative to CRY64 | |||
0 MDLEPFERAQIDDVLQQLESGSVQADEFLCLVLSILGSSRTYSQFPAILQSLSRKEPAMYRELMDLHAEYFRK 0 | |||
0 EPADLETLGYETDLELAIALSLQEHNQLTDTASFASEVDPAPKISFADAAKLSHFSHKHNKKNSSSKTEITKLKDNVAAMNLYQERKRYHINGQEKTCISN | |||
CYNGQPEPEDCVLKSEDGEDVFHVETSRPRESKAKHSRRSRKKKKSAPSRG<font color =red>L^V</font>AMKPVLVWFRRDLRLHDNPALISALEHGVPVIPVFLWCINEETGQNFTLATGGAT | |||
KYWLHHALLKLNQSLIQRFGS<font color =red>H^I</font>IFRVARSCEEELVSLVHETGADTIIINAVYEPWLKERDDLISETLRRHGVELKKHHSYCLYEPD<font color =orange>S^V</font>STEGVGLR 1 | |||
2 GIGSVSHFMSCCKRNNSAPIGMPLDAPRCLPAP<font color =blue>C^N</font>WPESDHLDTLELGKMPHRKDGTL 0 | |||
0 IDWAVTIRESWDFSEDGAYTCLANFL<font color =blue>Q^D</font>1 | |||
2 GVKHYEKESGRADKPYTSHISPYLHFGQISPRTVLHEAYFTKKN<font color =blue>V^P</font>KFLRKLAWRDLAYWLLILFPDMPSEPVRPAYK 0 | |||
0 SQRWSSDLNHLRAWQ<font color =blue>K^G</font>LTGYPLVDAAMRELWLTGWMCNYSRHVVASFLVAYLHIHWVHGY<font color =blue>R^W</font>FQ 0 | |||
0 DTLLDADVAINAMMWQNGGMSGLDHWNFVMHPVDSALTCDPYGSYV<font color =red>R^K</font>WCPELAGLPDEYIHKPWKCAPSQLRRA 1 | |||
2 GVILGRNYPHRIVLDLEERREQSLKDVVEVRKKHLEYLDEVSGCDMVQIPDQLLA<font color =lightblue>L^T</font>LGHTSGEDEVVRNRTGSFLLPVITRKEFKYKTLQPDTKDNPYNTVLKGYV | |||
SRKRDETIAYMNERHFTASTINEGAQRHERIERTNRLMEGLPAPSDAKNKSRRTPKKDPFSIIPPSYLHLAN* 0 | |||
>CRY1_homSap Homo sapiens (human) introns relative to CRY64 | |||
0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 | |||
1 FLLQCLEDLDANLRKLNS<font color =red>R^L</font>FVIRGQPADVFPRLFK 0 | |||
0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLD<font color =magenta>K 2 | |||
1 I</font>IELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVI<font color =orange>E^K</font>CTTPLSDDHDEKYGVPSLEEL 1 | |||
2 GFDTDGLSSAVWPGGETEALTRLERHLER<font color =magenta>K 0 | |||
0 A</font>WVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYK<font color =magenta>K 0 | |||
0 V</font>KKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWA<font color =blue>E^G</font>RTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGM<font color =magenta>K 0 | |||
0 V</font>FEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYI<font color =magenta>R 2 | |||
1 R</font>YLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 | |||
2 GLLASVPSNPN<font color =orange>G^N</font>GGFMGYSAENIPGCSSSG 1 | |||
2 SCSQGSGILHYAHGDSQQTHLLKQ 1 | |||
2 GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN* 0 | |||
Below the frog protein CRY7 is marked up for its various domains and motifs according to Pfam, Blast and PDB searches. <font color =blue>Blue</font> shows the antenna domain with predicted α/β secondary structure, <font color =purple>purple</font> the possibly catalytic FAD domain with predicted all α secondary structure, <font color =magenta>magenta</font> the UIM ubiquitin motif, <font color =purple>purple</font> two compositionally simple regions rich is basic residues predicted not to have definite fold, <font color =darkred>dark red</font> the conserved region of unknown function upstream of the UIM ubiquitin motif, and <font color =darkblue>dark blue</font> the conserved carboxy terminal motif of unknown function. | |||
>CRY7_xenTro Xenopus tropicalis (frog) | |||
0 MDLEPFERAQIDDVLQ<font color =darkred>QLESGSVQADEFLCLVLSILGSSRTYSQFPAILQSLSRKEPAMYRELMDLHAEYFRK</font> 0 | |||
0 EPADLETL<font color =magenta>GYETDLELAIALSLQEHNQL</font>TDTASFASEVDPAPKISFADAA<font color =purple>KLSHFSHKHNKKNSSSK</font>TEITKLKDNVAAMNLYQERKRYHINGQEKTCISN | |||
CYNGQPEPEDCVLKSEDGEDVFHVETSRP<font color =purple>RESKAKHSRRSRKKKKSAPSR</font>GLVAMK<font color =blue>PVLVWFRRDLRLHDNPALISALEHGVPVIPVFLWCINEETGQNFTLATGGAT | |||
KYWLHHALLKLNQSLIQRFGSHIIFRVARSCEEELVSLVHETGADTIIINAVYEPWLKERDDLISETLRRHGVELKKHHSYCLYEPDSVST</font>EGVGLR 1 | |||
2 GIGSVSHFMSCCKRNNSAPIGMPLDAPRCLPAPCNWPESDHLDTLELGKMPHRKDGTL 0 | |||
0 IDWAVTIRESWDFS<font color =purple>EDGAYTCLANFLQD 1 | |||
2 GVKHYEKESGRADKPYTSHISPYLHFGQISPRTVLHEAYFTKKNVPKFLRKLAWRDLAYWLLILFPDMPSEPVRPAYK 0 | |||
0 SQRWSSDLNHLRAWQKGLTGYPLVDAAMRELWLTGWMCNYSRHVVASFLVAYLHIHWVHGYRWFQ 0 | |||
0 DTLLDADVAINAMMWQNGGMSGLDHWNFVMHPVDSALTCDPYGSYVRKWCPELAGLPDEYIHKPWKCAPSQLRRA 1 | |||
2 GVILGRNYPHRIVLDLEERREQSLKDVVEVRKK</font><font color =darkblue>HLEYLDEVSGCDMVQIPDQLLALTLGHTSGEDEVVRNRTGSFLLPVITRKEFKYKTLQPDTKDNPYNTVLKGYV | |||
SRKRDETIAYMNERHFTASTINEGAQRHERIERTNRLMEGLPAPSDAKNKSRRTPKKDPFSIIPPSY</font>LHLAN* 0 | |||
[[Image:CRY7pdb.jpg|left]] | |||
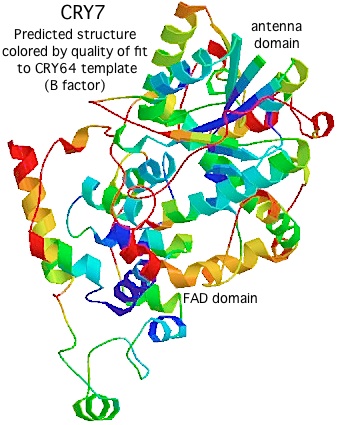
Using [http://swissmodel.expasy.org/ Swissmodel] with CRY64 from Drosophila as template (PDB:1UC3. 31% identity), the tertiary structure of CRY7 can be successfully modeled from over the region PVLLWF...ALVRRR of salmon CRY7 (corresponding to residues 13-497 of the experimentally determined structure). The predicted two domain structure very much resembles that of any cryptochrome or photolyase and allows preliminary identification of beta strands and helices. | |||
The quality of the model varies by position, as shown by the B factor score coloring in the figure on the left. The overall Z-Score quality of fit is -3.74, not too shabby for a large protein but no substitute for an actual experimental structure determination. (The other template option, an Arabidopsis cryptochrome, gives an unsatisfactory Z-Score.) Note the amino terminal conserved domain, the UIM motif, and the C-terminal conserved domain cannot be modeled at all without a template. | |||
The FAD binding site exhibits moderate steric interference but that molecule could be docked if a few residues were re-positioned slightly. The antenna site is more problematic: while present in the 3D structure, the nature of the antenna molecule (if any) cannot really be predicted. The early divergence of CRY7 from CRY64 and the lack of evolutionary persistence (consistency) of antenna molecules makes it very uncertain whether the Drosophila antenna molecule in CRY64 and CPD -- [http://www.ncbi.nlm.nih.gov/pubmed/22175817 recently determined] to be 5-deazariboflavin -- is actually the antenna molecule for CRY7. | |||
Although 5-deazariboflavin is the best historic option, CRY7 has been lost from Drosophila and 5-deazariboflavin is not known to occur in vertebrates or molluscs (the phylogenetic setting for CRY7 today). Since they cannot synthesize it, it would amount to a new vitamin in these species. | |||
Alternatively, the antenna molecule could be 6,7-dimethyl-8-ribityl-lumazine, folate, FMN, FAD or related molecules (as seen in other members of the gene family. No antenna molecule might be appropriate in view of the UIM domain and the implied signaling role, yet that does not account for the observed conservation of the antenna domain. | |||
<br clear=all> | |||
Predicted alpha helices (h) and beta strands (s) of CRY7: | |||
CRY7 PVLLWFRRDL RLHDNPAVIG SLEAGGPVIP VFIWCPEEEE GPGVTVAMGG ACKFWLHQAL SCLSSALEHI GSHLVFLRPD EEREGIGSSL RALRSLVRET | |||
CRY7 csivwfrrdl rvednpalaa avrag-pvia lfvwapeeeg hyhpg----r vsrwwlknsl aqldsslrsl gtclitkrs- ------tdsv aslldvvkst | |||
CRY7 sssss hhhhh hhh ss ssss hh hhhhhhhhhh hhhhhhhhhh sssss h hhhhhhh | |||
CRY7 sssss hhhhh hhhh ssss ssss hh h hhhhhhhhhh hhhhhhhhhh sssss h hhhhhhh | |||
CRY7 GAQTVLASAL YEPWLRERDQ VVVSALQKDR VEVNMVHSYC LRDPYTVTTE GVGLRGIGSV SHFMSCCQMN PGPGLGVPLD PPISLPSPSV WPRGCPLEGL | |||
CRY7 gasqiffnhl ydplslvrdh rakdvltaqg iavrsfnadl lyepwevtde lgrpfsm-fa afwerclsmp ydpesp--ll ppkkiisgdv sk--cvadpl | |||
CRY7 sssssss hhhhhhhh hhhhhhh sssss hhhhhhh | |||
CRY7 sssssss hhhhhhhh hhhhhhh sssss hh hhhhhhh hh | |||
CRY7 GLARMPCRKD GTTIDWAANI RSSWDFSEEG AQSRLEAFLN DGVYRYEKES GRADAPNTSC LSPYLHFGQL SARWLLWDTK GA-------- ----RCRPPK | |||
CRY7 v------fed dsekgsnall arawspgwsn gdkalttfin gplleysknr rkadsattsf lsphlhfgev svrkvfhlvr ikqvawaneg neageesvnl | |||
CRY7 hhhhhhhhhh h hhh hhhhhhhh hh hh hhhh hhhhhhh hhh | |||
CRY7 hhhhhhhhhh hhh hhh hhhhhhhhhh hhhh hhhhhhhhh hhhhhhhh hhhhhhhhh | |||
CRY7 FIRKLAWRDL AYWQLTLFPD LPWESLRPPY KALRWSNERG HLKAWQKGRT GYPLVDAAMR QLWLTGWMNN YMRHVVASFL IAYLHLPWQE GYRWFQDTLV | |||
CRY7 flksiglrey sryisfnhpy sherpllghl kffpwavden yfkawrqgrt gyplvdagmr elwatgwlhd rirvvvssff vkvlqlpwrw gmkyfwdtll | |||
CRY7 hhhhhhhhhh hhhhhhh hh hhhhhhh hhhhhhhh hhhh h hhhhhhhhhh hhh hh hhhhhhh | |||
CRY7 hhhhhhhhhh hhhhhhh hh hhhhhhh hhhhhhhh hhhh h hhhhhhhhhh hhh hh hhhhhhh | |||
CRY7 DADVAIDAMM WQNGGMCGLD H--WNFVMHP VDAAMTCDPY GNYVRKWCTE LAVLPDDLIH KPWKCPASML RRAGVVLGQS YPERVVTDLE ERRSQSLQDV | |||
CRY7 dadlesdalg wqyitgtlpd srefdridnp qfegykfdpn geyvrrwlpe lsrlptdwih hpwnapesvl qaagielgsn yplpiv-gld eakarlheal | |||
CRY7 hhhhhhh hhhhh h hhhhhhh hhhhh h h hhhh hhh h hhhhhhhhhh | |||
CRY7 hhhhhhh hhhhh h hhhhhhh hhhhh h h hhhh hhh hh hhhhhhhhhh | |||
CRY7 LAVLPDDLIH KPWKCPASML RRAGVVLGQS YPERVVTDLE ERRSQSLQDV ALVRRR | |||
CRY7 lsrlptdwih hpwnapesvl qaagielgsn yplpiv-gld eakarlheal sqmwql | |||
CRY7 h hhhh hhh h hhhhhhhhhh hhhhhh | |||
CRY7 h hhhh hhh hh hhhhhhhhhh hhhhhh | |||
=== Standard lab mouse C57BL/6J has a mutated CRY1 cryptochrome gene === | === Standard lab mouse C57BL/6J has a mutated CRY1 cryptochrome gene === | ||
| Line 35: | Line 167: | ||
"genetic screening assay for mutant circadian clock proteins that is based on real-time circadian rhythm monitoring in cultured fibroblasts. By using this assay, we identified a domain in the extreme C terminus of BMAL1 that plays an essential role in the rhythmic control of E-box-mediated circadian transcription. Remarkably, the last 43 aa of BMAL1 are required for transcriptional activation, as well as for association with the circadian transcriptional repressor CRY1"</blockquote> | "genetic screening assay for mutant circadian clock proteins that is based on real-time circadian rhythm monitoring in cultured fibroblasts. By using this assay, we identified a domain in the extreme C terminus of BMAL1 that plays an essential role in the rhythmic control of E-box-mediated circadian transcription. Remarkably, the last 43 aa of BMAL1 are required for transcriptional activation, as well as for association with the circadian transcriptional repressor CRY1"</blockquote> | ||
507 517 527 537 547 557 567 577 587 597 | |||
| | | | | | | | | | | |||
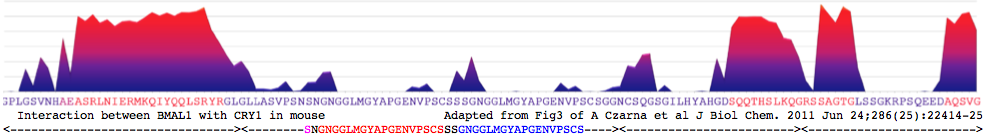
CRY1_musMus GLLASVPSN<font color=magenta>S</font>N<font color = red>GNGGLMGYAPGENVPSCS</font>SS<font color = blue>GNGGLMGYAPGENVPSCS</font>GG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* | CRY1_musMus NHAEAS<font color=purple>RL</font><font color=blue>NIERMKQ</font><font color=green>IYQQLSR</font><font color=purple>YR</font>GL GLLASVPSN<font color=magenta>S</font>N<font color = red>GNGGLMGYAPGENVPSCS</font>SS<font color = blue>GNGGLMGYAPGENVPSCS</font>GG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* | ||
CRY1_ratNor GLLASVPSNPNGNGGLMGYAPGENVPSGGSGG------------------G NCSQGSGILHYAHGDSQQTNPLKQ GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* | CRY1_ratNor NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYAPGENVPSGGSGG------------------G NCSQGSGILHYAHGDSQQTNPLKQ GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* | ||
CRY1_criGri GLLASVPSNPNGNGGLMGYTTGENLPSCSGGG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN* | CRY1_criGri NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYTTGENLPSCSGGG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN* | ||
CRY1_spaJud GLLASVPSNPNGNGGLMGYTPGENIPNCSSSG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN* | CRY1_spaJud NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYTPGENIPNCSSSG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN* | ||
CRY1_dipOrd GLLASVPSNPNGNGGLMGYAAGDNLPGSSSSG------------------- SCSQGSGILHYAHGDSQQMHLLKQ GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN* | CRY1_dipOrd NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYAAGDNLPGSSSSG------------------- SCSQGSGILHYAHGDSQQMHLLKQ GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN* | ||
CRY1_hetGla GLLASVPSNPNGNGGLMGYAPGESIPGSSGSG------------------- SCAHGSGILPCAHTDGQQAHLLKP GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD* | CRY1_hetGla NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYAPGESIPGSSGSG------------------- SCAHGSGILPCAHTDGQQAHLLKP GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD* | ||
CRY1_cavPor GLLASVPSNPNGNGGLLGYAPGESTPGSGGG-------------------- SCVPGSSSAGVSHCAQGEAPQAPP GRDPAGPGLGGGKRPSQEEDAQSTGHKIQRQSPD* | CRY1_cavPor HHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLLGYAPGESTPGSGGG-------------------- SCVPGSSSAGVSHCAQGEAPQAPP GRDPAGPGLGGGKRPSQEEDAQSTGHKIQRQSPD* | ||
CRY1_speTri GLLASVPSNPNGNGGLMAYAPGENIPGCSSSG------------------- SCTQGSSILHNAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* | CRY1_speTri NHEASL NIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMAYAPGENIPGCSSSG------------------- SCTQGSSILHNAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* | ||
CRY1_oryCun GLLASVPSNPNGNGGLMGYSPGENIPGCSSSG------------------- SCSQGSGILHYAQGDTQQTQLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* | CRY1_oryCun NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYSPGENIPGCSSSG------------------- SCSQGSGILHYAQGDTQQTQLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* | ||
CRY1_musMus GLLASVPSN<font color=magenta>S</font>N<font color = red>GNGGLMGYAPGENVPSCS</font>SS<font color = blue>GNGGLMGYAPGENVPSCS</font>GG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* | CRY1_musMus NHAEAS<font color=purple>RL</font><font color=blue>NIERMKQ</font><font color=green>IYQQLSR</font><font color=purple>YR</font>GL GLLASVPSN<font color=magenta>S</font>N<font color = red>GNGGLMGYAPGENVPSCS</font>SS<font color = blue>GNGGLMGYAPGENVPSCS</font>GG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* | ||
CRY1_ratNor .........P.................GG.G.------------------. ...................NP... ....M............................. | CRY1_ratNor .......................... .........P.................GG.G.------------------. ...................NP... ....M............................. | ||
CRY1_criGri .........P.........TT...L....GG.------------------- S.................A.L... ....M..S...........ETR..D......... | CRY1_criGri .......................... .........P.........TT...L....GG.------------------- S.................A.L... ....M..S...........ETR..D......... | ||
CRY1_spaJud .........P.........T....I.N.....------------------- S.................A.L... .S..M.H...N.........T..I........T. | CRY1_spaJud .......................... .........P.........T....I.N.....------------------- S.................A.L... .S..M.H...N.........T..I........T. | ||
CRY1_dipOrd .........P..........A.D.L.GS....------------------- S.................M.L.... ...M...............S..I........T. | CRY1_dipOrd .......................... .........P..........A.D.L.GS....------------------- S.................M.L.... ...M...............S..I........T. | ||
CRY1_hetGla .........P.............SI.GS.G..------------------- S.AH.....PC..T.G..A.L..P. .NCV.PV...............I...L....TD | CRY1_hetGla .......................... .........P.............SI.GS.G..------------------- S.AH.....PC..T.G..A.L..P. .NCV.PV...............I...L....TD | ||
CRY1_cavPor .........P......L......ST.GSGGG-------------------- S.VP..SSAGVS.CAQGEAPQAPP. .DP..P..GG............T.H.I....PD | CRY1_cavPor H......................... .........P......L......ST.GSGGG-------------------- S.VP..SSAGVS.CAQGEAPQAPP. .DP..P..GG............T.H.I....PD | ||
CRY1_speTri .........P.......A......I.G.....------------------- S.T...S...N.........L.... ...M...............T..I........T. | CRY1_speTri ......--.................. .........P.......A......I.G.....------------------- S.T...S...N.........L.... ...M...............T..I........T. | ||
CRY1_oryCun .........P.........S....I.G.....------------------- S...........Q..T...QL.... ...M...............T..I........T. | CRY1_oryCun .......................... .........P.........S....I.G.....------------------- S...........Q..T...QL.... ...M...............T..I........T. | ||
[http://www.ch.embnet.org/software/COILS_form.html | [http://www.ch.embnet.org/software/COILS_form.html Coiled coil]: <font color=purple>RL</font><font color=blue>NIERMKQ</font><font color=green>IYQQLSR</font><font color=purple>YR</font> for CRY1_musMus 480-493 | ||
<font color=purple>478 R e 0.644 | <font color=purple>478 R e 0.644 | ||
479 L f 0.644</font> | 479 L f 0.644</font> | ||
| Line 77: | Line 209: | ||
495 R e 0.375</font> | 495 R e 0.375</font> | ||
Full length CRY1 sequences are available for 10 Glires in the [[Cryptochrome_refSeqs|cryptochrome refSeq collection]]: | |||
CRY1_musMus Mus musculus (mouse) NM_007771 CRY1_ratNor Rattus norvegicus (rat) NM_198750 | |||
CRY1_criGri Cricetulus griseus (hamster) XM_003505292 CRY1_spaJud Spalax judaei (blind_mole_rat) AJ606298 | |||
CRY1_dipOrd Dipodomys ordii (kangaroo_rat) ABRO01202522 CRY1_hetGla Heterocephalus glaber (blind_mole-rat) | |||
CRY1_cavPor Cavia porcellus (guinea_pig) CRY1_speTri Spermophilus tridecemlineatus (squirrel) | |||
CRY1_oryCun Oryctolagus cuniculus (rabbit) CRY1_ochPri Ochotona princeps (pika) | |||
=== Lost distal exon in placental cryptochrome CRY1 === | === Lost distal exon in placental cryptochrome CRY1 === | ||
Although cryptochromes are highly conserved in their two main domains, the C-terminal region in CRY1 has a reputation for variability. This is attributable in part to | Although cryptochromes are highly conserved in their two main domains, the C-terminal region in CRY1 has a reputation for variability. This is attributable in part to loss of an ancient exon encoding 32 amino acids in placental mammals. However this exon persists in contemporary marsupials, monotremes, birds, alligators, turtles, lizards, snakes and frogs, so its conservation implies a continuing functional role maintained by selective pressure for several hundred million years of tetrapod evolution. | ||
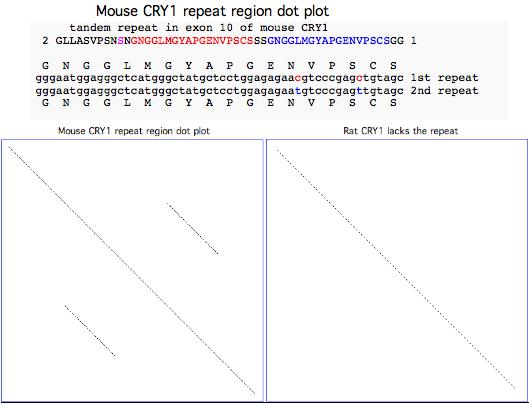
In addition, some distal motifs in CRY1 are compositionally simple, predisposing not only to the replication slippage event described above for mouse but also to smaller indels in the repetitive regions, notably the 2 aa deletional synapomorphy in placentals in GLLASVPSNPNGN--GGFM (the conserved methionine is at position 514 in human) and possibly the loss of proline (P518) in post-tarsier divergence primates. | |||
The loss may have preceded in stages, | The exon loss may have preceded in stages, beginning with alternative splicing that skipped it (this conserves reading frame as the ancestral gene ends with three consecutive phase 12 exons). Later, the exon came not to be used at all and thereafter rapidly degenerated to the point it cannot be detected today by blastx of the relevant region in any placental mammal. The exon does not plausibly contribute to the core fold (photolyase and FAD domains) though it could form a better defined structure upon interacting with other proteins. | ||
The loss of this exon raises | The functional consequences of exon loss are unknown; the timing matches that of overall collapse of the photolyase family in placentals. (Note the first half of placental evolution -- about 90 myr -- lacks any living representative, so events can pile up there by coincidence.) Possibly when CYT4, Cyt64, DASH and CPD were lost, the remaining two cryptochromes, especially CRY1, compensated for that loss (without however taking up catalytic roles in dna repair), with exon loss somehow contributing adaptively to that adjustment. | ||
The loss of this exon raises certain questions about the use of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3156801/ marsupial model systems] to understand CRY1 functionality in mouse (in turn a model system for human). For example, CRY1 of the marsupial Potorous tridactylus would still retain the exon but to date it has not been placed in a CRY1-- mouse. It would also be feasible to insert just the missing exon into an otherwise intact, ectopically expressed rat CRY1 gene, after first disentangling the effects of the mouse expansion in this same region (shown as ^^ below) as well as proline P518 removal. Note the lab mouse expansion somewhat restores length relative to marsupials, but in the wrong place. | |||
<font color=blue>CRY1_homSap MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCSSSG <-- lost exon in placentals --> SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN | <font color=blue>CRY1_homSap MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCSSSG <-- lost exon in placentals --> SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN | ||
| Line 165: | Line 249: | ||
CRY1_susScr MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCPQGSGILHYAHGESQQNHLLKQ GRSSTGSGLSSAKRPSQEEDTQSIIGPKVQRQSTN | CRY1_susScr MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCPQGSGILHYAHGESQQNHLLKQ GRSSTGSGLSSAKRPSQEEDTQSIIGPKVQRQSTN | ||
CRY1_ailMel MKQIYQQLSRYRGL GLLASVPANPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGSGLSSGKRPSEEEDTQSIGPKVQRQSTN | CRY1_ailMel MKQIYQQLSRYRGL GLLASVPANPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGSGLSSGKRPSEEEDTQSIGPKVQRQSTN | ||
CRY1_turTru MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGYSSSG SCTPGSGILHYAYGDSQQTHLLKQ | CRY1_turTru MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGYSSSG SCTPGSGILHYAYGDSQQTHLLKQ GRSSTCTGLSSGKRPSQEEDTQSIGPKVQRQSTN | ||
CRY1_equCab MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ | CRY1_equCab MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSLGPGLSSGKRPGPEEDTQGIGPKVQRQSTT | ||
CRY1_canFam MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ | CRY1_canFam MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSEEEDTQTISPKVQRQSTN | ||
CRY1_myoLuc MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SYAQGSGILHYALGDSQQTHLLKQ | CRY1_myoLuc MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SYAQGSGILHYALGDSQQTHLLKQ GRSSVGTGLSSGKRPSQEEDTQSIGRKVQRQSTN | ||
CRY1_pteVam MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGSLHYAHGDCQQTHLLKQ | CRY1_pteVam MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGSLHYAHGDCQQTHLLKQ GRSSMGTGLSSGKRPSQEEDMQSIGPKVQRQSTN | ||
CRY1_loxAfr MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENTPGCNSSG SCSQGSGILHYVHGDS....LLKQ GRSPTGTGVSSGKRPSQDEETQTLGPKVQRQSTN | CRY1_loxAfr MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENTPGCNSSG SCSQGSGILHYVHGDS....LLKQ GRSPTGTGVSSGKRPSQDEETQTLGPKVQRQSTN | ||
CRY1_triMan MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSNG SCPQGNGILHYAHRDSQQAHLLKQ GRSPTGTGVSSGKRPSQEEETQSIGPKVQRQSAN | CRY1_triMan MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSNG SCPQGNGILHYAHRDSQQAHLLKQ GRSPTGTGVSSGKRPSQEEETQSIGPKVQRQSAN | ||
| Line 187: | Line 271: | ||
CRY1_sylBor MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGAGDGHSV.VQSCALGDSHTGTSGVQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN | CRY1_sylBor MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGAGDGHSV.VQSCALGDSHTGTSGVQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN | ||
CRY1_taeGut MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGTGDGHSV.VQSCALGDSHTGTSGIQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN | CRY1_taeGut MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGTGDGHSV.VQSCALGDSHTGTSGIQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN | ||
CRY1_parWeb MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGTGDGHSV.VQSCALGDSHTGTSGIQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN | |||
CRY1_allMis MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGENVSGCGSTG GAQMGSSDGHTVSVQPCALGESHGGSNGIQQQ GYFQASSILHFPHGDDQQSHLLQQ GRTSLSSGISAGKRPNPEEETQSIGPKVQRQSTN | CRY1_allMis MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGENVSGCGSTG GAQMGSSDGHTVSVQPCALGESHGGSNGIQQQ GYFQASSILHFPHGDDQQSHLLQQ GRTSLSSGISAGKRPNPEEETQSIGPKVQRQSTN | ||
CRY1_anoCar MKQMYQQLSRYRGL GLLASVPSNGNGNGNGGLMGYSTGENIPGCTNTN GSQMGMNEGHIGNVQACTMGESHTGTSGIQQQ GYSQGSGILLYSHGDNQKTHSAQK GRISLGTGVCTGKRPSPEVETQSVGPKVQRQSSN | CRY1_anoCar MKQMYQQLSRYRGL GLLASVPSNGNGNGNGGLMGYSTGENIPGCTNTN GSQMGMNEGHIGNVQACTMGESHTGTSGIQQQ GYSQGSGILLYSHGDNQKTHSAQK GRISLGTGVCTGKRPSPEVETQSVGPKVQRQSSN | ||
| Line 194: | Line 279: | ||
CRY1_xenTro MKQIYQQLSRYRGL GLLASVPSNPNGNGNGGLMSYSPGESMSGCSNNG GGQMGVNEGSSASNPNANKGEVHPGTSGLQ.. GYWQGSSILHYSHSDSQQSY LMQ ARNPLHSVVSSGKRPNPEEETQSIGPKVQRQSSH | CRY1_xenTro MKQIYQQLSRYRGL GLLASVPSNPNGNGNGGLMSYSPGESMSGCSNNG GGQMGVNEGSSASNPNANKGEVHPGTSGLQ.. GYWQGSSILHYSHSDSQQSY LMQ ARNPLHSVVSSGKRPNPEEETQSIGPKVQRQSSH | ||
CRY1_xenLae MKQIYQQLSRYRGL GLLASVPSNPNG..NGGLMSYSPGESMPGCSNNG GGQMGAIEGSSASNPNPNQGEVLPGTSGLQ.. GYWQGSSILHYSHSDNQQSY LMQ ARNPLHSVVSSGKRPNPEEETQSVGPKVQRQSTH | CRY1_xenLae MKQIYQQLSRYRGL GLLASVPSNPNG..NGGLMSYSPGESMPGCSNNG GGQMGAIEGSSASNPNPNQGEVLPGTSGLQ.. GYWQGSSILHYSHSDNQQSY LMQ ARNPLHSVVSSGKRPNPEEETQSVGPKVQRQSTH | ||
CRY1_latCha MKQIYQQLSRYRGM GLLASVPSNPNGNGGLGCSLAENIPVCNSAA GAQMGGDDGHKVSVLAYTQGDSRAGEIEMQQQ | |||
CRY1_danRer MKQIYQQLSCYRGL GLLAMVPSNPNGNGENSTSLMGFQTGDMTKEVTTPS GYQMPPTSQGEWHGRTMVYSQGDQQTSSIMTSQ GFGNNGSTMCYRQDAQQIT GRGLHSSIIQTSGKRHSEESGPTTVSKVQRQCSS | |||
When the terminal four exons of CRY1 are compared to those of its nearest homolog class CRY2, no similarity can be detected beyond the first 8 residues of the tenth exon of CRY1 (2 GLLASVPS) vs the tenth and penultimate exon of CRY2 (2 CLLASVPS). This raises the question of what the last common ancestor had for terminal exons and -- given no counterpart in CRY4, CRY64, DASH, or CPD -- where they originated. Note that last two exons of CRY2 are strongly conserved in their own right, proving a separate conserved functionality from that of CRY1. Since the tenth exons begin homologously and end after a similar length with a phase 1 splice donor, these exons could possibly be homologous their entire length, just diverged distally. The eleventh exon of CRY2 could then correspond (allowing for total sequence divergence) to any of exons 11-13 in CRY1. | |||
CRY2_homSap CLLASVPSCVEDLSHPVAEPSSSQAGSMSSA GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDA | |||
CRY2_panTro ............................... .............................................. | |||
CRY2_gorGor ...........................V... .............................................. | |||
CRY2_ponAbe ...........................V... .............................................. | |||
CRY2_rheMac ...........................VN.. ...............................K.............. | |||
CRY2_papHam ...........................VN.. ...............................K.............. | |||
CRY2_calJac ............................... .............................................V | |||
CRY2_micMur ..............................T .................................T............ | |||
CRY2_musMus ....................G......I.NT ...A.S.....................T.....T.M..Q.PA...S | |||
CRY2_ratNor ....................G......I.NT .....S...........................T.M.AQ.P....S | |||
CRY2_criGri ...........................I.NT .S...S...........................T.M.AQ.PQT... | |||
CRY2_spaJud ........................P..ITNT .....ST..........................T...A..PA.... | |||
CRY2_cavPor .....................L.....ST.T ......G.................................P..... | |||
CRY2_hetGla ....................TL.....S..T ...S..D..............................A..PT.... | |||
CRY2_speTri ....................G......I..T .....S..Q..................................... | |||
CRY2_oryCun ...........................V.G. A..................................V........AV | |||
CRY2_turTru .........M....N...........G.... ................G.................G..PS..L...V | |||
CRY2_bosTau ..............N.......I....S..V ......G.................G..........SLPS....RGV | |||
CRY2_susScr ..............N............V.A. .....................................PT...GR.V | |||
CRY2_canFam ..............N.........T...... ..........................................CR.V | |||
CRY2_ailMel ..............N.........T...... .....................................A..P..R.V | |||
CRY2_myoLuc .........M....N......L..T...... ..K..................................AT....R.V | |||
CRY2_pteVam .............NN.........T...NN. .....................................A.....R.V | |||
CRY2_loxAfr ..............S............SN...........T........................K..G.......V | |||
CRY2_proCap ..............N........P..H.....L................................K..G.....T.. | |||
CRY2_choHof ..............N....................V............................T...........V | |||
CRY2_macEug .........M....S.M..T.MG....V..T..K...CS..........T..ASR..H.....M.A..V...A.--- | |||
CRY2_monDom .........L....S.MV.A.LG...AV.GP.LK...CS..........T..A....H.......R..GS..AG..V | |||
CRY2_ornAna ..............SAA..SGLG....NI.TA...-.P.............GL.....C..PK..GR.G..P.GE.. | |||
CRY2_galGal ..............G..TDSAPG.-..ST.TAV.LPQ.DQ......H.G...LCT...Y...K.TG..A..I.G.SS | |||
CRY2_taeGut ............I.G..PDSA.G.-.CST.TAV.LSQAEQ......H.G....CS...Y...K.TG...S.ISG.SL | |||
CRY2_allMis G........A....G..TD.A.V.-.CST.TALK.SQ..Q......H.GI..MCT.D.Y...K.TG.HG..I...SL | |||
CRY2_anoCar .........M....N...DT...H-.NCIGTAS.QTHC.QT.....HDVVQ.YK-...Y...K.VASQFA.N.RQEL | |||
CRY2_xenTro .I.......M...GG.M.DS.QNISEAGKM.P.SHTSGESVLAAQYTAGI--------------------------- | |||
CRY2_ranCat .I......S.....G.M.D.A...Q..SD---.A.RLCAVD.....H.DLD----G..C.K..LQCVQEM.RAA..F | |||
=== A distal alternative splice in avian cryptochrome CRY1 not used for magnetosensing === | |||
Bird CRY1 presents a further curious situation with respect to the terminal extentional exons of CRY1: an alternative splice in exon 11, more accurately a failure to consistently recognize its splice donor (or the following acceptor) leading to translational read-out of the mRNA to the first stop codon following. The vast majority of such events are misinterpreted artifacts -- the transcript simply terminated too soon, providing no splice acceptor and consequently no way for the intervening intron to be removed. | |||
However here two types of transcripts were found in both Erithacus rubecula (Euro robin) and Sylvia borin (warbler) in targeted experiments by separate research groups. The long form, called there CRY1A, has the usual four terminal exons of vertebrates; the short form, CRY1B, provides 25 new amino acids before a stop codon. | |||
Comparative genomics is capable of resolving artifact, coincidence, and functionality. First note that GenBank chicken transcripts contain a supportive entry (BU143111) that surfaced in a large transcript program not focused on particular genes. Secondly, the read-out of exon 11 in species without transcripts is implied by highly conserved amino acid sequence. While a certain amount of nucleotide conservation might be expected because splice sites are larger than just GT-AG, the intron could contain enhancers or other conserved non-coding elements (of this or an adjacent gene), and conservation can persist for a time via coldspots and failure of a mutation to fix in a population, the conservation here at the protein level significantly exceeds what these factors could contribute. Gray shows species lacking conservation; blue conserved amino acids within birds. | |||
This conservation was in fact already established in the early diverging lineage of duck + chicken but deteriorated as shown by early stop codons and distal sequence restored by a shared frameshift (lower case below) in gallinaceous birds. However nothing resembling the bird read-out sequence is found in alligator, turtles, snakes, lizard or frog in any reading frame. Thus, the simplest scenario is it arose early in bird evolution and so is restricted to them. (Here we await an ostrich genome to see if the event took place already in Paleognathae.) | |||
If the selective pressure truly operates on the level of amino acids here and if the region is not a mutational cold spot, then relatively higher levels of variation should be observed at redundant codon positions within the DNA, eg 3rd position in 4-codon amino acids. However, by collecting the DNA sequences, it emerges that synonymous changes do not noticably predominate (after minimizing events needed by branching of the avian phylogenetic tree and ignoring the breakdown of this region in duck, chicken, turkey) nor do non-synonymous changes conserve amino acid properties. This argues strongly that the region has not been conserved by selection on amino acid sequence but rather selection on the underlying DNA. | |||
[[File:AvianCRY1.jpg]] | |||
> | Exon 11 read-out of CRY1 genSpp transcript support of read-out (or wgs accession) | ||
<font color=gray> | |||
GISKNTF* monDom Monodelphis domestica (opossum) | |||
GISDNTFLTLTQSRGSLGIPHQS..* macEug Macropus eugenii (wallaby) | |||
GISQNTFESVRLS* sarHar Sarcophilus harrisii (tasmanian_devil) | |||
GISKLFSFIFKNTFN* ornAna Ornithorhynchus anatinus (platypus) | |||
GRSSLTPGLSGGKRHCQEEESQN..* tacAcu Tachyglossus aculeatus (echidna)</font> | |||
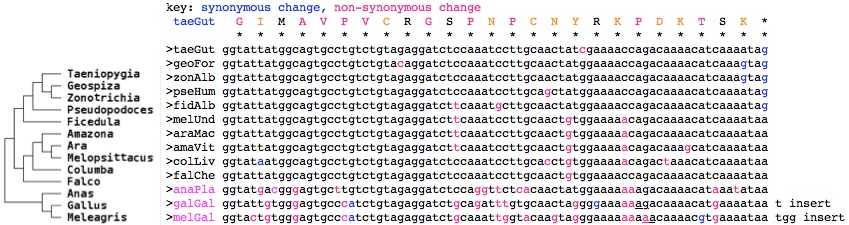
<font color=blue>GIMAVPVCRGSPNPCNYRKPDKTSK* taeGut Taeniopygia guttata (finch) | |||
GIMAVPVCRGSPNACNYGKPDKTSK* eriRub Erithacus rubecula (robin) AY585717 | |||
GIVAVAVCRGSPNPCNYGKPDKTSE* sylBor Sylvia borin (warbler) DQ838738 | |||
GIMAVPVCRGSSNPCNCGKTDKTSK* melUnd Melopsittacus undulatus (parakeet) | |||
GIMAVPVCRGSPNPCNYGKPDKTSK* zonAlb Zonotrichia albicollis (sparrow) (ARWJ01011250) | |||
GIMAVPVCRGSPNPCSYGKPDKTSK* pseHum Pseudopodoces humilis (ground-tit) (ANZD01003613) | |||
GIMAVPVCRGSPNPCNCGKPDKTSK* falChe Falco cherrug (falcon) (AKMU01039249) | |||
GIMAVPVCRGSSNPCNCGKTDKTSK* araMac Ara macao (scarlet macaw) (AMXX01097310) | |||
GIMAVPVCRGSPNPCTCGKTD*TSK* colLiv Columba livia (rock pigeon) (AKCR01045195) | |||
GMTGVLVCRGSPGSHNYGKKDKT*K* anaPla Anas platyrhynchos (duck) | |||
GIVGVPICRGSADLCN*GKKdkt*k* galGal Gallus gallus (chicken) BU143111 | |||
GTVGVPICRGSANWYK*GKKdkt*k* melGal Meleagris gallopavo (turkey)</font><font color=gray> | |||
KCLQRICKFL*LKFSKY.. . allMis Alligator mississippiensis (alligator) | |||
KNVFKEVLAILEIVKIP... pelSin Pelodiscus sinensis (turtle) | |||
II*QIKCVQRHFSRFLK... chrPic Chrysemys picta (turtle) | |||
IIQQIKCVQRGSRYS*NC*... apaSpi Apalone spinifera (turtle) | |||
YCQGNSGILHYAHGD.. . croHor Crotalus horridus (snake) | |||
KTL*KSLI*YSS*NTACVHG... anoCar Anolis carolinensis (lizard) | |||
GKLAAPLISVSSIIGVFHTHEPQ... xenTro Xenopus tropicalis (frog)</font> | |||
The data thus support the notion of birds having evolved a distinct function for the read-out option at exon 11 -- with nothing comparable in the immediate outgroups (crocodile, turtle) or mammals. While more bird genomes are expected in 2014, these don't include basal Paleognathae such as ostrich and other non-passerine species needed to check read-out conservation patterns conform to the [http://ldl.genomics.cn/page/pa-bird.jsp avian phylogenetic tree]. However the more common CRY1 form retaining the usual extra exons is also conserved in birds (as seen in the [[Cryptochrome_evolution#Lost_distal_exon_in_placental_cryptochrome_CRY1|earlier alignment]] of this region). | |||
[[Image:CRY1retina.jpg|left]] | |||
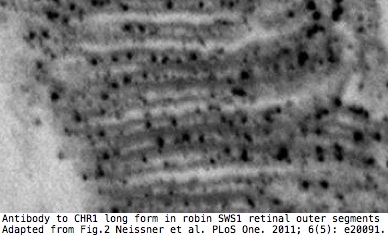
It has been [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3102070/ reported] that only the long form is expressed in [[Opsin_evolution:_update_blog|SWS1 opsin]] cones of retinas of migrating passerine birds where it detects the earth's magnetic field via electron spin pairing in [http://www.pnas.org/content/early/2012/03/08/1118959109.full.pdf+html?with-ds=yes tryptophan and FAD]. The short form is apparently expressed in the [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC521149 ganglion cell layer] where it may represent an adaptive synapomorphy for a large part of the avian tree. | |||
Note the vertebrate ciliary opsin SWS1 has [[Opsin_evolution:_key_critters_(ecdysozoa)#Hexapoda:_Drosophila_melanogaster_.28fruitfly.29_.._7_opsins|no counterpart]] in fruit flies. Since invertebrate cryptochromes correspond poorly too, Drosophila is completely unsuitable here as model species. However dipterans do have two rhabdomeric opsins with peak sensitivity in the ultraviolet, RH5 and RH7, with characteristic lysine at position 90 and a short third cytoplasmic loop. RH5 is located in the larval Bolwig organ; RH7 has not been assigned an anatomical site but may be located in antenna. Conceivably analogous co-expression with a different cryptochrome could couple these photosensing systems too. | |||
Human CRY2, also strongly expressed in retina but not so specifically in cone cell outer segment membranes, can [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3128388 reportedly replace] the invertebrate cryptochrome CRY1B in the drosophila magnetic field detection system (as can insect CRY1A). The final exon of human CRY2 bears no clear relationship to the terminal exons of CRY1 nor to the read-out exon 13 of birds and is only secondarily related homologically to invertebrate CRY1B cryptochromes. | |||
<br clear=all> | |||
The alignment below shows very limited distal homology between tetrapod CRY2 and invertebrate CRY1B. The primary sequence correspondence does not even extend to the <font color=green>coiled coil</font> region of vertebrate CRY2 which is not always evident in invertebrate CRY1B, much less to distal exons of CRY2 (indicated by spacing). On the flip side, just distal to the its missing coiled coil, invertebrate CRY1B has a conserved 16 residue motif [http://www.ncbi.nlm.nih.gov/pubmed/22080955 known] to imitate a damaged DNA base with a tryptophan; vertebrate CRY2 is itself conserved here but not relative to the CRY1B spoof motif and contains no counterpart to the key aromatic residue. | |||
Amino acids are shown only when 50% or more conserved within the total alignment column: | |||
> | CRY2_homSap RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.S<font color=green>RLNIERMKQIYQQLSRYR</font>GL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | ||
CRY2_rheMac RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_calJac RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_micMur RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_musMus RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_cavPor RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_oryCun RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_bosTau RYLP.LK.FPSRYIYEPWNAPES.QKAAKC.IGVDYP.PIVNHAE.SRLNIERMKQ.YQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_ailMel RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_pteVam RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDL..P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_loxAfr RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | |||
2 | CRY2_choHof RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E...... | ||
CRY2_monDom RYLP.LK.FP.RYIYEPWNAPE.VQKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSC.EDLS.P.......Q.G............ ........SPKRK.E........E..KRA.V......E...... | |||
CRY2_ornAna RYLP.LK.FPSRYIYEPWNAPESVQKAAKC.IGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.........Q.G............ ........SPKRK.E........EL.KR..V......E...... | |||
CRY2_galGal RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.P.VNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q-G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_taeGut RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.P.VNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVED.S.P.......Q-G............ ........SPKRK.E........EL.KRA.V......E...... | |||
CRY2_allMis RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.P.VNHAE.SRLNIERMKQIYQQLSRYRGL .LLASVPSC.EDLS.P.......Q-G............ ........SPKRK.E.........L.KRA.V......E...... | |||
CRY2_anoCar RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.P.VNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSC.EDLS.P........-G............ ........SPKRK.......-..EL.KRA.V......E...... | |||
CRY2_ranCat RYLP.LK..PSRYIYEPWNAPESVQK.AKCI.GVDYP.P.VNHAE.S<font color=green>RLNIERMKQ.YQQLSRYR</font>GL C.LASVPS.VEDLS.P.......Q.G...---...... ........SPKRK.E....----EL.K.A........E...... | |||
<font color=blue>PPHCRPSNEEEVRQFM<font color=red>W</font>LP</font>: helix conserved within CR!B whose [[Cryptochrome_evolution#Invertebrate_cryptochromes:_distal_CRY1B_spoofs_a_damaged_DNA_base|tryptophan spoofs damaged DNA base]] | |||
CRY1B_strPur RYLP.LK..P.RY..EPW.AP..VQ..AKCI.G.DYP.P.V.H...S..N.E.M......L.... ......S....V....... | |||
CRY1B_lytVar RYLP.LK..P.RY..EPW.AP..VQ..AKCI.G.DYP.PIV.H...S..N.E.M......L.... ......S....V....... | |||
CRY1B_parLiv RYLP.LK..P.RY..EPW.AP..VQ..AKCI.G.DYP.PIV.H...S..N.E.M......L.... ......S....V....... | |||
CRY1B_aplCal RY.P.LK..P..Y..EPW.AP...Q....CIIG.DYP.P.V.H...S......M..I.--..... ...........V..L.... | |||
CRY1B_octVul .Y.P.LK..P..Y...PW.AP...Q..A.CIIG.DYP.PIV.H...S..N...M......L.... ...........V....... | |||
CRY1B_craGig RYLP.LK..P.RY..EPW.AP..VQ..AKCI.--DYP.P.V.H...S...I..MK.....L.... ......S........S... | |||
CRY1B_acyPis RY.P.LK..P....YEPW..PESVQK...CIIG.DYP..IV.H...S..N...M........... ......S....V....... | |||
CRY1B_dapPul RY.P.L..F...YI.EPW.AP...Q..A.CIIG.DYP...V.H.E....N.E.MK...Q..-... ......S..S.V....... | |||
CRY1B_diaNig RY.P.LK..P..Y.YEPW.AP..VQ..A.CI.G.DYP..I..H...S..N...M..I.-...... ......S............ | |||
CRY1B_danPle RY.P.L...P..YIYEPW.AP..VQ.AA.C.IG.DYP.P.V.H......N...M....-.L.... ......S....V....... * | |||
CRY1B_mamBra RY.P.L...P..YIYEPW.AP...Q..A.CIIG.DYP.P.VNH......N...MK...-...... ......S............ * | |||
CRY1B_helArm RY.P.L...P..YIYEPW.AP..VQ..A.C.IG.DYP.P.VNH......N...MK...-...... ......S............ * | |||
CRY1B_bomMor RY.P.L...P..YIYEPW.AP..VQ..A.CIIG.DYP.P.VNH......N...M....-.L.... ......S............ | |||
CRY1B_droMel .Y.P.L...P.....EPW......Q....C.IGV.YP..I.........N...MK.....L.... .....S....V....... | |||
CRY1B_anoGam RYLP.L...P.....EPW.A....Q....C.IG..YP.P.V..A..S..N...M......L.... ......S............ | |||
CRY1B_neoBul .Y.P.L...P..YI.EPW..P...Q....C.IG..YP............N...M......L.... ......S............ | |||
CRY1B_bacCuc .Y.P.L...P..YI.EPW..P...Q....C.IGV.YP..IV..A..S..N...M....Q.L.... ......S....V....... | |||
[[Image:CRY1Bcoils.png|center]] | |||
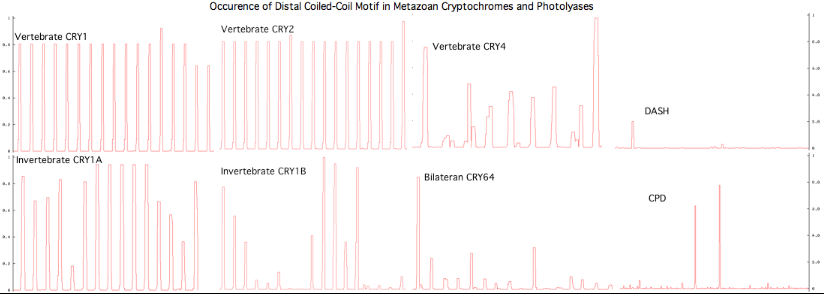
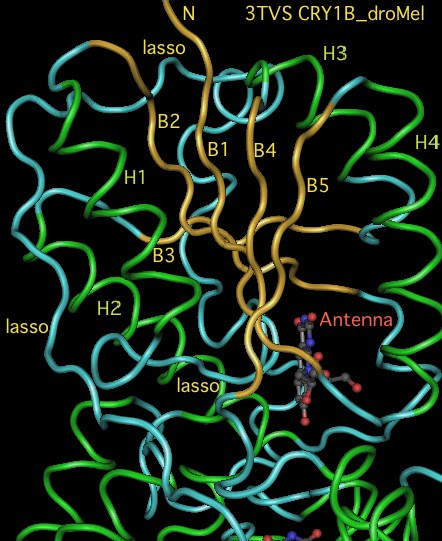
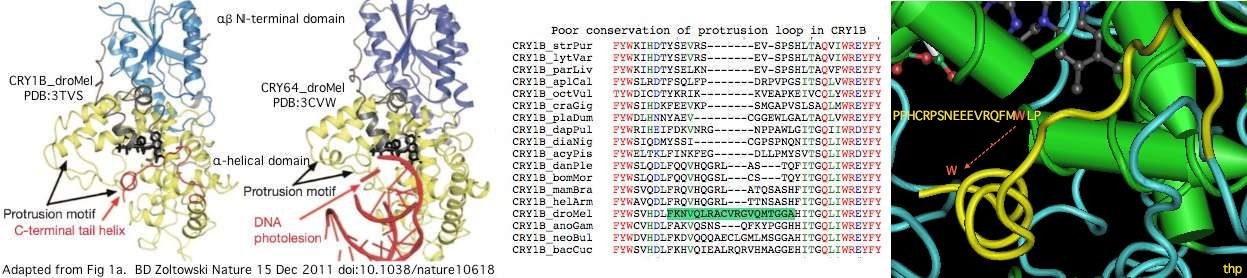
The graphic above shows separate predictions for distal coiled coil [http://www.ch.embnet.org/software/COILS_form.html prediction] for each of 17-20 concatenated vertebrate distal sequences for each of the eight cryptochromes and photolyases that occur in bilaterans. The species are presented in phylogenetic order left to right (ie as listed in [[Cryptochrome_refSeqs|refSeq collection]]). Invertebrate CRY1B clearly does not have the domain not consistently present. The three largest CRY1B peaks (indicated by asterisks in the alignment) are all lepidoptera; the Drosophila protein does not contain this structural motif motif. Given the duplications of the gene tree, the coiled coil domain probably arose once in an early ancetral cryptochrome but was been lostin some species groups such as dipteran flies. The new crystallographic structure PDB:3TVS confirms the lack of coiled-coil motif in CRY1B. | |||
C-terminal deletions of the Drosophila cryptochrome have been [http://www.pnas.org/content/108/2/516.full extensively studied]. While informative, the poor distal correspondence to mammalian cryptochromes makes carry-over of such results -- annotation transfer -- to mammalian cryptochromes a dubious proposition since key sequence motifs used in signalling are not present in the C-terminus of this model species (and vice versa!). | |||
=== Evolutionary origin of the α/β photolyase fold === | |||
Comparative genomics (lots of phylogenetically structured primary sequences) synergizes strongly with three-dimensional structural determinations, the former providing the conserved so presumably functional regions and the latter their structural interpretation. In the case of cryptochrome and photolyase structures, it is quite important that full length proteins be considered because N- and C-terminal extensions can provide the very properties that distinguish an orthology class from its paralogs. | |||
However the N-terminus can also be evolving haphazardly from compositionally simple sequence, be quickly trimmed from newly synthesized protein by cellular proteases, lack assignable structure in a crystal, and be functionally irrelevant. Similarly, an extended C-terminus can represent meaningless run-out through junk DNA to the first stop codon encountered. In these situations, sequence conservation will not extend beyond the genus level (a few million years). | |||
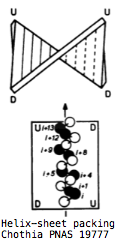
The overall fold of all cryptochromes and photolyases is basically the same: two distinct globular domains held together in part by a long lasso thrown out by the second domain. The amino terminal domain lies at the far end of the protein from the DNA binding site. It consists of a 5-stranded parallel beta sheet sandwiched between 4 alpha helices whose axes are anti-parallel to the sheet. The strands are ordered 32145 with the helices alternating in position. The first two helices form the top of a sandwich, the second two the bottom with strand 3 transitioning. The binding site for the antenna molecule is at the edge of the sandwich between the two domains; it is not intimately associated with the helices or central strands themselves but rather with helix-strand turns. | |||
[[Image:CRYHS.png|left]] | |||
Surprisingly, the βαβαβαβαβ pattern of alternating helix and sheet with the outer layer of helices packing against the central core in 32145 order is not necessarily indicative of evolutionary relatedness but instead a [http://swissmodel.expasy.org/course/text/chapter4.htm default supersecondary structure] for cytoplasmic proteins. Its inevitability was [http://www.mrc-lmb.cam.ac.uk/tcb/pdf/chc/31_pnas_74_4130_77.pdf first explained] by C. Chothia et al in 1977 as complementarity between the right handed twist of a beta sheet and the rotating i+4 ridge of helix side chains (due to its 3.4 residues per turn) -- close packing of side chains in the hydrophobic core is entropically favorable and so the same basic fold commonly arises regardless of evolutionary relatedness. | |||
In terms of evolutionary characters, the fold is homoplasic, having arisen many times independently rather than having descended from a single ancestral fold. (The same is true for the more complex [http://en.wikipedia.org/wiki/TIM_barrel TIM beta barrel], an eightfold repeat of the βαβ pattern found in 15 gene families with no bona fide sequence homology.) | |||
With photolyases, coincidence extends to antenna molecules, some of which are similar to the NAD of the Rossmann fold homology group. However the binding site location is different. Photolyases do not have a stand-alone pocket in the α/β amino terminal domain but utilize portions of the following fold (not to mention a the composite route of excitation transfer). In fact, it's not clear that the antenna binding site is fixed in all homologs. Further, there is no conservation of key residues nor any convergence of ancestral sequences to homology. | |||
In summary, the photolyase fold is '''not homologous''' to the classic nucleotide binding fold. Searching PDB with a given protein to find related fold structures thus requires careful overall evaluation of candidates to ensure actual evolutionary relatedness. While the α/β domain draws a blank, the resemblance found by Dali in the catalytic domain of primases and 4Fe-4S photolyases to previously studied photolyases/cryptochromes is beyond coincidence. | |||
[[Image:CRY1B3TVS.jpg|left]] | |||
Many large eukaryotic proteins are chimeric, having arisen from genetic fusions of mobile domains. Alternatively, certain common folds have arisen independently in situ in different gene families rather than been shuffled in. Initially, modular proteins fold as their constituent pieces, with less substantive interaction in the final product than an ordinary non-covalent heterodimer might have, but over time more intimate structural codependencies evolve. Photolyase may once have been a heterodimer of a small redox protein that passes antenna excitations to a larger catalytic subunit, becoming later a genetically fused modular protein, but today the α/β amino terminal domain is quite integrated with the all-alpha domain -- the long lasso holding them together is preceded by the essential protrusion loop that binds DNA in the second domain. (This connector region was reported attached by a [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC514401/ reported disulfide] but the cysteines are not conserved and cytoplasmic proteins generally lack disulfides in vivo.) | |||
Separating the two domains with limited trypsin digestion (or better, genetic methods) has not yet been attempted and might not be feasible with retention of functionality if the domains are structurally interdependent. This could explain why cryptochromes that lack antenna molecules have not lost the α/β domain under the evolutionary principle of [http://www.ncbi.nlm.nih.gov/pubmed/18085818 'use it or lose it']. That is, if no selective pressure persisted in this region, what weeds out structurally deleterious mutations or keeps a large N-terminal deletion from being fixed? Not only has CRY1B of drosophila retained the antenna pocket, but it also exhibits very high levels of conservation of individual amino acids and small motifs beyond what is needed for folding and stability. | |||
The main alternatives to structural integration are (1) evolution has not caught up yet with very recent loss of antenna molecule in CRY1B and other cryptochromes, (2) an unsuspected, undetected new antenna molecule is present and important in vivo which maintains selective pressure and (3) a signalling or magnetosensing role for the α/β domain, either from direct participation in a conformational shift or through homodimeric or heterologous binding to other proteins. The first possiblity can be rejected because seemingly antenna-less cryptochromes fall into different groups, each of long standing. The second seems inconsistent with careful experimentation, yet reconstitution experiments are no better than the antenna molecules included, with the very recent discovery of lumazine casting further doubt on the completeness of that set. The third is a distinct possibility yet does not seem sufficent to provide the level of conservation observed. | |||
Taking phylogenetically distributed representatives from each cryptochrome/photolyase class (excluding 4Fe-4S photolyases and primases), the alignment below shows the regions of conservation within the α/β domain. While it is easy and informative to align all 250 sequences, to avoid excess display only 4 of each orthology class are shown. However the full set of sequences was separately aligned to determine conservation at the 70% level, again with key species (experimental models and those with PDB structures) shown. It can be seen immediately that universally conserved residues do not correlate particularly with secondary structure (even though that is strongly conserved). | |||
<br clear=all> | |||
10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 | |||
| | | | | | | | | | | | | | | | | | |||
bbbbbb aaaaaaaaa bbbbbbbbb aaaaaaaaaaaaaaaaaaa bbbbbb aaaaaaaaaa bbbbbbbb aaaaaaaaaaaa bbbbbbb | |||
<font color=blue>CRY1_homSap MGVNAVHWFRKGLRLHDNPALKECIQGAD-TIRCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFKEWN-ITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDKIIELNGGQPPLTYKRFQTLISKMEPLEIP | |||
CRY1_musMus MGVNAVHWFRKGLRLHDNPALKECIQGAD-TIRCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFKEWN-ITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDKIIELNGGQPPLTYKRFQTLVSKMEPLEMP | |||
CRY1_galGal MGVNAVHWFRKGLRLHDNPALRECIRGAD-TVRCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFKEWS-IAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDKIIELNGGQPPLTYKRFQTLISRMEPLEMP | |||
CRY1_xenTro MGVNAVHWFRKGLRLHDNPALRECIQGAD-TVRCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFKEWK-ITKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDKIIELNGGQPPLTYKRFQTLISKMDPLEIP</font> | |||
<font color=darkblue>CRY2_homSap DSASSVHWFRKGLRLHDNPALLAAVRGAR-CVRCVYILDP------WFAASSSVGINRWRFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFKEWG-VTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDRIIELNGQKPPLTYKRFQAIISRMELPKKP | |||
CRY2_musMus DGASSVHWFRKGLRLHDNPALLAAVRGAR-CVRCVYILDP------WFAASSSVGINRWRFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFKEWG-VTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDRIIELNGQKPPLTYKRFQALISRMELPKKP | |||
CRY2_galGal GFCRSVHWFRRGLRLHDNPALQAALRGAA-SLRCIYILDP------WFAASSAVGINRWRFLLQSLEDLDNSLRKLNSRLFVVRGQPTDVFPRLFKEWG-VTRLTFEYDSEPFGKERDAAIIKLAKEAGVEVVIENSHTLYDLDRIIELNGNKPPLTYKRFQAIISRMELPKKP | |||
CRY2_xenTro PSVSSVHWFRKGLRLHDNPALLSALRGAN-SVRCVYILDP------WFAASSSGGVNRWRFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFKEWG-VSRLTFEYDSEPFGKERDAVIMKLAKEAGVEVVVENSHTLYDLDRVIELNGHSPPLTYKRFQAIISRMELPRRP</font> | |||
<font color=green>CRY1A_triCas QDKHMVHWFRRGLRLHDNPSLREGLKGAR-TFRCVFVLDP------WFAGSSNVGINKWRFLLQCLEDLDRSLRKL-SRLFVIRGQPADALPKLFKEWG-TTALTFEEDPEPFGGVRDHNLTTLCQELGISVVQKVSHTLYHLQDIIDRNGGRAPLTYHQFLAIIACMGPPPQP | |||
CRY1A_bomImp MGKHTVHWFRKGLRLHDNPSLREGLTGAT-TFRCVFVLDP------WFAGSTNVGINKWRFLLQCLEDLDCSLRKLNSRLFVIRGQPADALPKLFKEWG-TTNLTFEEDPEPFGRVRDHNISALCKELGISVVQKVSHTLYKLDEIIERNGGKPPLTYHQFQNVVASMDPPEPS | |||
CRY1A_nasVit MKKHTVHWFRKGLRLHDNPSLREGLAGAS-TFRCVFVLDP------WFAGSANVSINKWRFLLQCLEDLDRSLHQLNSRLFVIRGQPADALPKLFREWG-TTSLTFEEDPEPYGRVRDENITTLCKELGITVVQRVSHTLYKLDEIIEKNGGKPPLTYHQFQNVIARMDPPEYP | |||
CRY1A_anoGam RDKHTVHWFRKGLRLHDNPALREGLRGAR-TFRCVFIIDP------WFAGSSNVGINKWRFLLQCLDDLDRNLRKLNSRLFVIRGQPADALPKLFKEWG-TTCLTFEEDPEPFGRVRDHNISEMCKELGIEVISAASHTLYNLERIIEKNGGRAPLTYHQFQAIIASMDAPPQP</font> | |||
<font color=darkgreen>CRY1B_strPur PGGACIHWFRHGLRLHDNPALLEGMTLGK-EFYPVFIFDN------EVAGTKTSGYNRWRFLHDCLVDLDEQLKAAGGRLFVFHGDPCLIFKEMFLEWG-VRYLTFESDPEPIWTERDRRVKALCKEMKVECIERVSHTLWNPDIIIEKNGGTPPITYSMFMECVTEIGHPPRP | |||
CRY1B_octVul KQKIAVHWFRHGQRLHDNPALLDALKDCD-EFYPVFIFDG------EVAGTKLCGFNRWRFLLENLKDLDESFSEYGGRLYTFQGKPVEVFANLQNEWG-ITHITAEIDPEPIWQERDDAVKEFCQKSGIKCDFFNSHTLWDPKRLLKKNGGTPPLTFELFQLVTSSLGPPPRP | |||
CRY1B_danPle MLGGNVIWFRHGLRLHDNPSLHSALEDASSPFFPIFIFDG------ETAGTKMVGYNRMRYLLEALNDLDQQFRKYGGKLLMIKGRPDLIFRRLWEEFG-IRTLCFEQDCEPIWRPRDASVRALCRDIGVSCREHVAHTLWNPDTVIKANGGIPPLTYQMFLHTVEIIGNPPRP | |||
CRY1B_droMel TRGANVIWFRHGLRLHDNPALLAALKDQGIALIPVFIFDG------ESAGTKNVGYNRMRFLLDSLQDIDDQLQDGRGRLLVFEGEPAYIFRRLHEQVR-LHRICIEQDCEPIWNERDESIRSLCRELNIDFVEKVSHTLWDPQLVIETNGGIPPLTYQMFLHTVQIIGLPPRP 3TVS</font> | |||
<font color=brown>CRY4_galGal MRHRTIHLFRKGLRLHDNPALLAALQSSE-VVYPVYILDR------AFTSSMHIGALRWHFLLQSLEDLRSSLRQLGSCLLVIQGEYESVVRDHVQKWN-ITQVTLDAEMEPFYKEMEANIRGLGEELGFQVLSLMGHSLYNTQRILELNGGTPPLTYKRFLRILSLLGDPEVP | |||
CRY4_xenTro MPHRTIHIFRKGLRLHDNPTLVTALETSD-VVYPVYILDR------NFTSSSVIGSKRWNFFLQSIEDLHCNLQKLNSCLFVIQGDYERVLREHVEKWN-ITQVTFDLEIEPYYKGLDERIRAMGQELGFEVVSMVAHTLYDIKKILALNCGKPPLTYKNFLRVLSMLGNPDKP | |||
CRY4_latCha MTHRTIHIFRKGLRLHDNPILLAALEFSR-VVYPVYILDR------KLESGVIIGALRWRFILQSLEDLHRNLVKLNSRLFVIQGDYEQILREYVQKWT-ITQVTFDTEIEPFYKEMDKKVRLMGKEMGFTVLFSVAHALYDVARIVENNGGQPPLTYKKFLHVLSKLGDPERP | |||
CRY4_danRer MSHRTIHLFRKGLRLHDNPSLLGALASSS-ALYPVYVLDR------VFQGAMHMGALRWRFLLQSLEDLDTRLQAIGSRLFVLCGSTANILRELVAQWG-ITQISYDTEVEPYYTRMDKDIQTVAQENGLQTYTCVSHTLYDVKRIVKANGGSPPLTYKKFLHVLSVLGEPEKP</font> | |||
<font color=darkbrown>CRY64_xenTro KHNSTIHWFRKGLRLHDNPALLAAMKDCA-ELYPIFILDP------WFPRNMKVSVNRWRFLIEALKDLDENLKKINSRLFVVRGKPTEVFPLLFKKWK-VTRLTFEVDTEPYSRQRDADVEKLAAEHNVQVIQKVSNTLYAIDRIIAENNGKPPLTYVRFQTVLALLGPPKRP | |||
CRY64_danRer SHNTTIHWFRKGLRLHDNPALIAALKDCR-HIYPLFLLDP------WFPKNTRIGINRWRFLIEALKDLDSSLKKLNSRLFVVRGSPTEVLPKLFKQWK-ITRLTFEVDTEPYSQSRDKEVMKLAKEYGVEVTPKISHTLYNIDRIIDENNGKTPMTYIRLQSVVKAMGHPKKP | |||
CRY64_droMel QRSTLVHWFRKGLRLHDNPALSHIFTGKY-FVRPIFILDP------GILDWMQVGANRWRFLQQTLEDLDNQLRKLNSRLFVVRGKPAEVFPRIFKSWR-VEMLTFETDIEPYSVTRDAAVQKLAKAEGVRVETHCSHTIYNPELVIAKNLGKAPITYQKFLGIVEQLKVPKKV 3CVU | |||
CRY64_danPle KVASVIHWFRLDLRLHDNLALRNAINRKQ-ILRPIYVIDP------DIKNWMRVGCNRLRFLFQSLKNLDTSLRKINTRLYVIKGKAIECLPKLFDEWH-VKFLTLQVDIDADLVKQDEVIEEFCEANNIFVVKRMQHTVYDFNSVVKKNNGSIPLTYQKFLSLVSDVQVKDKI</font> | |||
CRY1C_araTha TGSGSLIWFRKGLRVHDNPALEYASKGSE-FMYPVFVIDP------HYPGSSRAGVNRIRFLLESLKDLDSSLKKLGSRLLVFKGEPGEVLVRCLQEWK-VKRLCFEYDTDPYYQALDVKVKDYASSTGVEVFSPVSHTLFNPAHIIEKNGGKPPLSYQSFLKVAGEPSCAKSE 3FY4 | |||
CRY1A_araTha SGGCSIVWFRRDLRVEDNPALAAAVRAGR-PVIALFVWAP------EEEGHYHPGRVSRWWLKNSLAQLDSSLRSLGTCLITKRSDSVASLLDVVKSTG-ASQIFFNHLYDPLSLVRDHRAKDVLTAQGIAVRSFNADLLYEPWEVTDELGRPFSMFAAFWERCLSMPYDPESP 1U3C | |||
<font color=purple>DASH_taeGut MAGTAICLLRCDLRAHDNQQVLHWAQHNADFVIPLYCFDPRHYLGTHCYRLPKTGPHRLRFLLESVKDLRETLKKKGSTLVVRKGKPEDVVCDLITQLGSVTAVVFHEEATQEELDVEKGLCQVCRQHGVKIQTFWGSTLYHRDDLPFRPIDRLPDVYTHFPKGLESGAKVRPT | |||
DASH_xenTro RARVIICLLRNDLRLHDNEVLHHWAHRNADQIVPLYCFDPRHYGGTHYFNFPKTGPHRLKFLLESVQDLRNTLKERGSNLLLRRGKPEEIIAGLVKQLGNVSAVTLHEEATKEETDVESAVRRVCTQLGVRYQTFWGSTLYHREDLPFRHISSLPDVYTQFRKAAETQGKVRST | |||
DASH_danRer ASRTVICLLRNDLRLHDNEVFHHWAQRNAEHIIPLYCFDPRHYQGTYHYNFPKTGPFRLRFLLDSVKDLRALLKKHGSTLLVRQGKPEDVVCELIKQLGSVSTVAFHEEVASEEKSVEEKLKEICCQNKVRVQTFWGSTLYHRDDLPFSHIGGLPDVYTQFRKAVEAQGRVRPV | |||
DASH2_araTha GKGVTILWFRNDLRVLDNDALYK-AWSSSDTILPVYCLDPRLFHTTHFFNFPKTGALRGGFLMECLVDLRKNLMKRGLNLLIRSGKPEEILPSLAKDFGA-RTVFAHKETCSEEVDVERLVNQGLKRVGTKLELIWGSTMYHKDDLPFD-VFDLPDVYTQFRKSVEAKCSIRSS 2VTB</font> | |||
<font color=#0066CC>CPD_galGal GAECILYWMCRDQRVQDNWAFLYAQRLALKQELPLRVCFC------LVPAFLDATIRHYGFMLRGLREVAKECAELDIPFHVLLGCPKDVLPSFVVEHGVGGLVTDFCPLRVPRQWVEEVKERLPED--VPFAQVDAHNIVPCWVASPKQEYSARTIRAKIHSQLPEFLTEFPP | |||
CPD_xenTro DAQGIVYWMSRDQRVQDNWAFLYAQRLALKQKLPLHVTFC------LVPKFLDATIRHYGFMVKGLQEVAEECKELNIPFHLLIGYAKDILPNFVKKHAIGGVVTDFSPLRVPLQWVEDVSKRLPKD--VPLVQVDAHNIVPCWVASNKQEYGARTIRKKIHDQLSQFLTEFPP | |||
CPD_droMel SSLGVVYWMSRDGRVQDNWALLFAQRLALKLELPLTVVFC------LVPKFLNATIRHYKFMMGGLQEVEQQCRALDIPFHLLMGSAVEKLPQFVKSKDIGAVVCDFAPLRLPRQWVEDVGKALPKS--VPLVQVDAHNVVPLWVASDKQEYAARTIRNKINSKLGEYLSEFPP | |||
CPD_orySat PGGPVVYWMLRDQRLADNWALLHAAGLAAASASPLAVAFA------LFPRLLSARRRQLGFLLRGLRRLAADAAARHLPFFLFTGGPAE-IPALVQRLGASTLVADFSPLRPVREALDAVVGDLRRG--VAVHQVDAHNVVPVWTASAKMEYSAKTFRGKVSKVMDEYLVEFPE 3UMV</font> | |||
bbbbbb aaaaaaaaa bbbbbbbbb aaaaaaaaaaaaaaaaaaa bbbbbb aaaaaaaaaa bbbbbbbb aaaaaaaaaaaa bbbbbbb | |||
<font color=blue>CRY1_homSap M..N..HWFRKGLRLHDNP.L.....G..-..RCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDA.LRKLNSRLFVIRGQP.DVFPRLFKEW.-I..LS.EYDSEPFGKERDAAIKKLA.EAGVEVI.R.SHTLY.LD.IIELNGGQ.PLTYKRFQ.L.S.M.P...P 95% conservation | |||
CRY1_musMus M..N..HWFRKGLRLHDNP.L.....G..-..RCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDA.LRKLNSRLFVIRGQP.DVFPRLFKEW.-I..LS.EYDSEPFGKERDAAIKKLA.EAGVEVI.R.SHTLY.LD.IIELNGGQ.PLTYKRFQ.L.S.M.P...P | |||
CRY1_galGal M..N..HWFRKGLRLHDNP.L.....G..-..RCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDA.LRKLNSRLFVIRGQP.DVFPRLFKEW.-I..LS.EYDSEPFGKERDAAIKKLA.EAGVEVI.R.SHTLY.LD.IIELNGGQ.PLTYKRFQ.L.S.M.P...P | |||
CRY1_xenTro M..N..HWFRKGLRLHDNP.L.....G..-..RCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDA.LRKLNSRLFVIRGQP.DVFPRLFKEW.-I..LS.EYDSEPFGKERDAAIKKLA.EAGVEVI.R.SHTLY.LD.IIELNGGQ.PLTYKRFQ.L.S.M.P...P</font> | |||
<font color=darkblue>CRY2_homSap ....SVHWFR.GLRLHDNPAL..A.....-..RC.YILDP------WFA....VG.NRWRFLL.SLEDLD.SLRKLNSRLFVVRGQP.DVFPRLFKEW.-VTRLTFEYDSEP.GKERDAAI.K.A.E.GVE....NSHTLY.LDRIIE.N...PPLT.KRFQ.I.SR..LP..P 95% conservation | |||
CRY2_musMus .....VHWFR.GLR.HDNPAL..A.....-..RC.YILDP------.FA.....G.NRWRFLL..LEDLD.SL.KL.SRLFVVRGQP.DVFPRLFKEW.-V..LTFEYD.EP.GKERD..I.K.A.E.GVE......HTLY.....IE.N...PPLT.KRFQ....R..LP..P | |||
CRY2_galGal .....VHWFR.GLR.HDNPAL..A.....-..RC.YILDP------.FA.....G.NRWRFLL..LEDLD.SL.KL.SRLFVVRGQP.DVFPRLFKEW.-V..LTFEYD.EP.GKERD..I.K.A.E.GVE......HTLY.....IE.N...PPLT.KRFQ....R..LP..P | |||
CRY2_xenTro .....VHWFR.GLR.HDNPAL..A.....-..RC.YILDP------.FA.....G.NRWRFLL..LEDLD.SL.KL.SRLFVVRGQP.DVFPRLFKEW.-V..LTFEYD.EP.GKERD..I.K.A.E.GVE......HTLY.....IE.N...PPLT.KRFQ....R..LP..P</font> | |||
<font color=green>CRY1A_triCas ..K..VHWFR.GLR.HDNP.L..G.....-T.R..F..DP------WFA...N..INKWRFLL..L.DLD..L..L-.RLFV..GQPA..LP.L...W.-TT..TFE.DPEP.G.VRD.N.........I.V.....HTLY....II..N....PLTY..F.........P... 95% conservation | |||
CRY1A_bomImp ..K..VHWFR.GLR.HDNP.L..G.....-T.R..F..DP------WFA...N..INKWRFLL..L.DLD..L..L..RLFV..GQPA..LP.L...W.-TT..TFE.DPEP.G.VRD.N.........I.V.....HTLY....II..N....PLTY..F.........P... | |||
CRY1A_nasVit ..K..VHWFR.GLR.HDNP.L..G.....-T.R..F..DP------WFA...N..INKWRFLL..L.DLD..L..L..RLFV..GQPA..LP.L...W.-TT..TFE.DPEP.G.VRD.N.........I.V.....HTLY....II..N....PLTY..F.........P... | |||
CRY1A_anoGam ..K..VHWFR.GLR.HDNP.L..G.....-T.R..F..DP------WFA...N..INKWRFLL..L.DLD..L..L..RLFV..GQPA..LP.L...W.-TT..TFE.DPEP.G.VRD.N.........I.V.....HTLY....II..N....PLTY..F.........P...</font> | |||
<font color=darkgreeen>CRY1B_strPur .......WFRHGLRLHDNP.L........-.F.P.FIFD.------E.AGT...GYNR..FL...L.DLD......GGRL....G.P...F.....E.G-.....FE.D.EP.W..RD..VK..C......C.E.VSHTLW.P...I..NGG.PP.TY.MF......IG.PPRP 70% conservation | |||
CRY1B_octVul .....V.WFRHG.RLHDNP.L........-.F.P.FIFD.------E.AGT...G.NR..FLL..L.DLD......GGRL....G.P...F.....E.G-......E.D.EP.W..RD..VK..C......C....SHTLW.P......NGG.PPLT...F.......G.PPRP | |||
CRY1B_danPle .....V.WFRHGLRLHDNP.L........-.F.P.FIFD.------E.AGT...GYNR...LL..L.DLD......GG.L....G.P...F.....E.G-.....FE.D.EP.W..RD..V...C......C.E.V.HTLW.P...I..NGG.PPLTY.MF......IG.PPRP | |||
CRY1B_droMel .....V.WFRHGLRLHDNP.L........-...P.FIFD.------E.AGT...GYNR..FLL..L.D.D.......GRL....G.P...F........-......E.D.EP.W..RD......C........E.VSHTLW.P...I..NGG.PPLTY.MF......IG.PPRP</font> | |||
<font color=brown>CRY4_galGal M.HRTIH.FRKGLRLHDNP.LL.AL..S.-..YPVYILDR------.F......GALRW.F.LQSLEDL...L...GS.L.V..G......R..V.KW.I-TQ.T.D.E.EP.Y..M...I.....E.G..V.....H.LY...RI...NGG.PPLTYK.FL..LS.LG.PE.P 70% conservation | |||
CRY4_xenTro M.HRTIH.FRKGLRLHDNP.L..AL..S.-..YPVYILDR------.F......G..RW.F.LQS.EDL...L....S.L.V..G......R..V.KW.I-TQ.T.D.E.EP.Y......I.....E.G..V...V.H.LY....I...N.G.PPLTYK.FL..LS.LG.P..P | |||
CRY4_latCha M.HRTIH.FRKGLRLHDNP.LL.AL..S.-..YPVYILDR------........GALRW.F.LQSLEDL...L....S.L.V..G......R..V.KW.I-TQ.T.D.E.EP.Y..M.........E.G..V...V.H.LY...RI...NGG.PPLTYK.FL..LS.LG.PE.P | |||
CRY4_danRer M.HRTIH.FRKGLRLHDNP.LL.AL..S.-..YPVY.LDR------.F......GALRW.F.LQSLEDL...L...GS.L.V..G......R..V..W.I-TQ...D.E.EP.Y..M...I.....E.G......V.H.LY...RI...NGG.PPLTYK.FL..LS.LG.PE.P</font> | |||
<font color=darkbrown>CRY64_xenTro .....IHWFRKGLRLHDNPAL..A.....-...PIF.LDP------.......V..NRWRFL...L.DLD..L.K.N.RLFV.RG.P.E..P.LF..W.V-..LT.EVDTEPY...RD..V...A....V.V...VS.T.Y........N.G..PLTY............P..P 70% conservation | |||
CRY64_danRer .....IHWFRKGLRLHDNPAL..A.....-...P.F.LDP------..........NRWRFL...L.DLD..L.KLN.RLFV.RG.P.E..P.LF..W..-..LT.EVDTEPY...RD..V...A....V.V....SHT.Y........N.G..P.TY............P..P | |||
CRY64_droMel ......HWFRKGLRLHDNPAL........-...PIF.LDP------.......V..NRWRFL...L.DLD..L.KLN.RLFV.RG.P.E..P..F..W.V-..LT.E.D.EPY...RD..V...A....V.V....SHT.Y........N.G..P.TY............P... | |||
CRY64_danPle .....IHWFR..LRLHDN.AL..A.....-...PI...DP------.......V..NR.RFL...L..LD..L.K.N.RL.V..G...E..P.LF..W.V-..LT..VD........D.............V.....HT.YD.......N.G..PLTY................</font> | |||
<font color=purple>DASH_taeGut .....ICLLR.DLR.HDN....HWA...A....PLYCFDPRHY.GT.....PKTGP.RL.FLLES..DLR..L...GS.L..R.GKPE.V...L..QLG.V..V....E.T.EE.DVE......C....V...T.WGSTLYHR.DLPF..I..LPDVYT.F.K..E....VR.. 70% conservation | |||
DASH_xenTro .....ICLLRNDLR.HDNE...HWA...A....PLYCFDPRHY.GT....FPKTGP.RL.FLLES..DLR..L...GS.L..R.GKPE.....L..QLG.V..V....E.T.EE.DVE......C....V...T.WGSTLYHR.DLPF.HI..LPDVYT.FRK..E....VR.. | |||
DASH_danRer .....ICLLRNDLR.HDNE...HWA...A....PLYCFDPRHY.GT....FPKTGP.RL.FLL.S..DLR..L...GS.L..R.GKPE.V...L..QLG.V..V....E...EE..VE......C....V...T.WGSTLYHR.DLPF.HI..LPDVYT.FRK.VE....VR.. | |||
DASH2_araTha .....I...RNDLR..DN.....-A........P.YC.DPR....T....FPKTG..R..FL.E...DLR..L...G..L..R.GKPE.....L....G.-..V....E...EE.DVE.................WGST.YH..DLPF.-...LPDVYT.FRK.VE.....R..</font> | |||
<font color=#0066CC>CPD_galGal ......YWM.RDQRVQDNWA.L.AQ.LALK...PL.VCF------CL.P.FL.AT.R...F.L.GL.EV..EC..L.I.FH.L.G.....LP.FV.....G..V.DF.PLR.P..W...V...LP..--VP..QVDAHNIVPCW.AS.K.EY.ARTIR.KI...L..FLTEFPP 70% conservation | |||
CPD_xenTro ......YWM.RDQRVQDNWA.L.AQ.LALK...PL.V.F------CL.P.FL.AT.R...F...GL.EV..EC..L.I.FHLL.G.....LP.FV.....G..V.DF.PLR.P..W...V...LP..--VP..QVDAHNIVPCW.AS.K.EY.ARTIR.KI...L..FLTEFPP | |||
CPD_orySat ......YWM.RDQR..DNWA.L.A..LA.....PL.V.F------.L.P..L.A..R...F.L.GL.............F.L..G....-.P..V........V.DF.PLR........V...L...--V...QVDAHN.VP.W.AS.K.EY.A.T.R.K........L.EFP. | |||
CPD_metMaz ......YWM.RDQR..DNWA.L.....A.....P..V.F------CL...FL.A..R...F.L.GL.E.........I....L.G........FV.....G..V.DF.PLR....W...V....--.--.P...VDAHN.VPCW.AS.K.EY.A.T.R.K....L..FL.EFP.</font> | |||
bbbbbb aaaaaaaaa bbbbbbbbb aaaaaaaaaaaaaaaaaaa bbbbbb aaaaaaaaaa bbbbbbbb aaaaaaaaaaaa bbbbbbb | |||
Three major indels occur. Using CPD and 4Fe-4S photolyases as outgroup, these can be resolved as either insertions or deletions (ie as derived traits or synapomorphies). The first indel is a 6 residue insertion found only in the DASH group; the second is a 1 residue deletion in stem post-DASH divergence proteins; and the third is a 2 residue insertion that occurred shortly after divergence from CPD. Even though 3D coverage of cryptochromes is inadequate, enough exists that each of these indels can be localized in an existing stucture and so visualized by [http://www.ncbi.nlm.nih.gov/Structure/VAST/vast.shtml precomputed structural co-alignments]. | |||
Indels can work as a standalone classifier. However this third of the protein provides discriminants only for CPD and DASH which are more easily identified just by a [http://www.proweb.org/proweb/Tools/WU-blast.html blast classifier] using the reference sequences. Note too at position 30, ecdysozoans (minus orthopterans and crustaceans) show homoplasy, re-inserting a residue at a site where it had long been deleted. | |||
[[Image:CRYFAD.jpg|left]] | |||
A deeper history of photolyase structural history must find a place for the 4Fe-4S cluster family (likelier as ancestral condition rathner than a development off to the side) and an explanation for the same cluster and structural homology to primase which is extensive butdoes not extend to the α/β domain. Here the unusual but conserved U-shaped conformation of the catalytic FAD may be a key piece of the history. | |||
Here the rings of FAD's adenine and flavin each lie in a plane but these planes while not quite parallel are alignable by a rotation, and ring long axes are almost perpendicular. This configuration may allow them to bind two primer pyrmimidines much as the damaged DNA thymine dimer is bound. Indeed the 4Fe-4S cluster may create a transient cyclobutane bond in the primer. As usual, divalent magnesium binds the diphosphate and offsets its charge. | |||
<br clear=all> | |||
The table below list the current structural determinations for eukaryotic cryptochromes, photolyases and related folds available in March 2012. Archaeal and bacterial structures are included in the table when their eukaryotic alignment coverage or blast score warrants it -- they are surprisingly well conserved relative to metazoan, probably because of a 'floor' of essential core residues that prevents further percent divergence. (Opsins and the huge GPCR gene family have a [[Opsin_evolution|similar floor]] but much lower, about 24%.) Overall, coverage is not ideal because not all orthology classes are represented yet -- while their core fold is easily predictable given high percent identities (eg 65% human to plant), critical functional nuances provided by actual extensions are not. | |||
Date PDB Class PubMed Species BestBlastP Accession Cofactors Alternates | |||
Nov 2011 3TVS CRY1B 22080955 Drosophila melanogaster (fruit_fly) musMus 40% AB019389 FAD no antenna CRY1 cryptochrome 19722240 | |||
Dec 2008 1U3C CRY1A 15299148 Arabidopsis thaliana (cress) homSap 29% NM_116961 FAD MTHF CRY1-PHR | |||
Oct 2009 3CVU CRY64 18956392 Drosophila melanogaster (fruitfly) xenTro 57% NM_165334 FAD [deazaflavin Fo] phr 6-4 | |||
Apr 2009 3FY4 CRY1C 19359474 Arabidopsis thaliana (cress) musMus 51% NM_001035626 FAD UVR3 CRY3 | |||
Dec 2008 2VTB DASH2 19074258 Arabidopsis thaliana (cress) xenTro 50% NM_122394 FAD CRY3 | |||
Dec 2011 3UMV CPD 22170053 Oryza sativa (rice) galGal 53% B096003 FAD PhrII Class II | |||
Sep 2011 2XRY CPD 21892138 Methanosarcina mazei (euryarchaeota) xenTro 49% AE008384 FAD Class II | |||
Mar 2012 3ZXS PFES 22290493 Rhodobacter sphaeroides (bacteria) .......... CP000144 FAD 4Fe-4S lumazine CryPro | |||
Aug 2010 3L9Q PRIM2 21346410 Homo sapiens (human) .......... NM_000947 FAD 4Fe-4S primase large subunit 3Q36 PMC3204975 | |||
Apr 2010 3LGB PRIM2 20404922 Saccharomyces cerevisiae (yeast) .......... NM_001179611 FAD 4Fe-4S primase large subunit PriL PRI2_YEAST | |||
To what extent can the structures available now be used to model the vertebrate ones that so far are missing? That can be done at the primary and secondary structure level simply by [http://www.proweb.org/proweb/Tools/WU-blast.html aligning a batch of orthologs] under a structurally determined sequence and transfering its features to non-gappy regions having signficant conservation. Multiple target sequences are essential to purge one-off accidental matches and to assess phylogenetic depth (ancestral persistance vs recent convergent origin). | |||
When percent identity exceeds 25%, reasonably accurate 3D coordinates can be obtained by fitting the unstudied primary sequence to a PDB entry using [http://swissmodel.expasy.org/ SwissModel]. A third approach uses [http://www.ebi.ac.uk/Tools/dalilite/ DaliLite] for pairwise comparisons of proteins with PDB coordinates, either real or modelled. NCBI's VAST allows any number of structures to be simultaneously aligned. The antenna pocket has also been examined by docking candidate receptor molecules. | |||
For example, human CRY1 is not available. According to [http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&BLAST_PROGRAMS=blastp Blastp] of PDB on 25 Mar 2012, the structure with the highest percent identity 53% and most extensive coverage (positions 6-522 out of 587 amino acids) is 3CVU from CRY64_droMel. Setting aside concerns about what the extra 65 amino acids at the end of the human protein might contribute to structure, that template request quickly yields PDB coordinates (with local error estimates) from SwissModel. And those in turn can be uploaded at DaliLite to structurally align human CRY1 with CPD and 4Fe-4S photolyases and primases which are too distant to align by primary sequence. This shows where the 4Fe-4S would sit in human cryptochrome had it been retained and also identifies the ancient structural core in the all-alpha domain that cryptochromes share with primases. These tools also rotate sequences so that all are in the same orientation. | |||
In all three approaches, what emerges is not fact but prediction. Only the first provides homological (genetic) alignment; structural alignments provide the best geometrical fit but do not necessarily recapitulate evolutionary relatedness of residues near gaps. Because folds are far more deeply conserved than sequence, structural alignment of greatly diverged sequences can uncover very faint but real relationships. However the N- and C-terminal extensions are not modelable yet their strong conservation in some instances argues for important function and possibly fixed structure. | |||
=== Syntentic relationships in vertebrate cryptochromes === | |||
Synteny -- the conservation of flanking gene relationships -- is critical (along with indels and intron structure) in establishing orthology and so to transferal of experimental information from a model species to another because primary sequence analysis alone can be give misleading results: | |||
After gene duplication, both members of a retained pair may diverge rapidly in primary sequence if they subfunctionalize, whereas if one gene -- not necessarily the parent -- retains the original function and the other neofunctionalizes, only the latter may diverge rapidly. This behavior can lead to long branch attraction artefacts and major misclassification of relationships. | |||
Cryptochromes and photolyases have experienced numerous duplications over evolutionary time. Those within multi-cellular organisms have all have been segmental duplications of limited extent. The alternative, retroprocessing, removes all introns. The sole known retroprocessed cryptochromes are CRY1 pseudogenes in naked mole rat, marmoset and sloth (AHKG01086374 ACFV01087645 ABVD01272190). These are easily recognized -- even far into pseudogenization -- as the top hits at genomic blast because as long contiguous matches they outscore multi-exonic ones. The existence of such features implies transcription of the parental gene in germline cells, typically testis. | |||
Syntenic relationships can persist for billions of years of branch length but more commonly dissipate over a few hundred million years because of local inversions and other chromosomal rearrangements that shuffle gene order. The rate of dissipation varies greatly by clade, with vertebrates much slower than arthropods. | |||
In vertebrates, synteny is typically well-retained back to the human-coelocanth divergence, with less certain correspondences extending to ray-finned fish and sometimes to chondrichthyes. Although complicated by poor quality assemblies, little synteny appears to persist back to lamprey, tunicate, amphioxus, sea urchin, and hemichordate. In contrast, synteny for a Drosophila gene rarely extends through dipteran flies, much less Insecta. | |||
The primary method for determining synteny at the phylogenetic level is a Blast search against multiple assemblies. This can be done very efficiently by concatenating conserved and diagnostic regions of 4-5 adjacent human proteins centered on the target gene. As the percent identity falls off, the human probe can be replaced with an orthologous concatenate from chicken or frog using the UCSC 46-way to collect orthologs. If probes aren't known, blastx of the contig containing the cryptochrome will reveal its neighbors. | |||
[[Image:CRY1INVERT.gif|left]] | |||
However the outcome also has been precomputed on a massive scale by [http://www.dyogen.ens.fr/genomicus-66.01/cgi-bin/search.pl Genomicus], the complication here being that only two cryptochromes persist into humans (meaning no HUGO gene names are available to enter Genomicus). To procede with CPD, DASH, CRY64, CRY4 in the tetrapods that have them, it is necessary to blat into a UCSC assembly that carries an Ensembl gene name track. | |||
The figure (taken from the [http://www.dyogen.ens.fr/genomicus-66.01/cgi-bin/search.pl Genomicus] synteny tool) shows that CRY1 experienced a small local inversion in amniotes subsequent to mammalian divergence. This may have carried all upstream regulatory regions along with it or left some behind, perhaps with significant effects altering gene expression. Since the event occurred some 350 myr ago, the boundaries of the inversion cannot be precisely determined today. | |||
Genomicus works surprisingly well given that almost all the Ensembl gene models it uses are wrong, the explanation being a few missed exons, erroneous termini and retained introns don't signficantly affect best reciprocal blast. But Ensembl models are often absent altogether in non-mammalian tetrapods, for example missing out entirely for DASH in frog and lizard which have full length conserved genes in their assembly. Unlike the UCSC 46-way, Genomicus does not begin with a whole genome alignment. Consequently it can stub in an erroneous paralogs when a gene is missing (eg CRY4_latCha in place of DASH_latCha). | |||
In some cases a gene appears absent but pseudogene debris in the expected syntenic position is still detectable. That says gene loss was fairly recent (last five million years), for example DASH pseudogenes in gallinaceous birds (chicken and turkey) but not duck (the immediate outgroup) or songbirds. Only very recent pseudogenes would be represented in either Genomicus or at the UCSC 46-way. Genes can also seem absent in spotty assemblies but individual exons can be recovered from raw trace reads (eg platypus CPD). Long processing lags prevent certain strategic assemblies from being represented in the 46-way or Genomicus (eg alligator, turtle, python, spotted gar) so they were considered separately here. | |||
These sites also provide no resources for invertebrate synteny. It isn't currently possible to study CRY1A (which is missing from fruit fly) even though orthologs extend phylogenetically from honey bee (ecdysozoa) to molluscs (lophotrochozoa) to sea urchin (echinoderms) so method of concatenated queries must be employed. Convenient queries for the other ortholog classes are provided below: | |||
<br clear=all> | |||
Gene Species Genomicus entry <--------------------------------- Synteny ----------------------------------> Phylogenetic Depth | |||
CRY1 homSap CRY1 CMKLR1 ASCL4 <font color=red>PRDM4</font> PWP1 BTBD11 <font color=blue>CRY1</font> MTERFD3 C12orf23 RIC8B RFX4 to coelocanth, ray-finned fish | |||
CRY2 homSap CRY2 <font color=red>PRDM11</font> SYT13 CHST1 CTD SLC35C1 <font color=blue>CRY2</font> MAPK8IP1 C11orf94 PEX16 GYLTL1B barely to ray-finned fish | |||
CRY4 anoCar ENSACAG00000004583 GPR37L1 ARL8A PTPN7 LGR6 UBE2T <font color=blue>CRY4</font> LRIF1 CEPT1 ADORA1 MYOG to coelocanth | |||
CRY64 xenTro ENSXETG00000003913 UBASH3B STS1 RPL27A <font color=blue>CRY64</font> FOXRED1 SRPR FAM118B FAM55A lizard to frog | |||
CPD monDom ENSMODG00000018409 PCYT1A ZDHHC19 TFRC TNK2 IGFBP5 <font color=blue>CPD</font> KIAA0226 FYTTD1 LRCH3 IQCG barely to ray-finned fish | |||
DASH danRer ENSDARG00000002396 CTDSPLA VILL PLCD1A DLEC1 ACAA1 <font color=blue>DASH</font> MYD66/88 OXSR1 SLC22A13 CSRNP1B birds to ray-finned fish | |||
CRY1B droMel FBgn0025680 SQZ CG14282 CG5555 CG31475 <font color=blue>CRY1B</font> VIB CG11703 CG5250 CG3773 ... | |||
CRY1A apiMel concatenated blast XM_395048 XM_393681 <font color=blue>CRY1A</font> Amel_5586 XM_391835 ... | |||
Synteny can be used to disentangle paralogs, especially important in zebrafish which has been [http://www.ncbi.nlm.nih.gov/pubmed/21232144,20830285,19663499,19652538,19346448,18494763,17785416,16176122,16000406,12832412,12560489,12354100,12015122 studied to a certain extent] -- despite the poor correspondence of its oversize cryptochrome repertoire to mammals -- because individual cell lines are cryptochrome-entrainable. Here zebrafish have four cryptochromes on four different chromosomes related to mammalian CRY1, a single copy of a cryptochrome clustering with mammalian CRY2, and single copies of CRY4, CRY64, DASH and CPD. | |||
Because chondrichthyes, lobe-finned fish (coelocanth), and basal ray-finned fish (gars) already have two separate genes classifying as CRY1 (ie distinct from CRY2 and other photolyases), a gene duplication occurred in early vertebrates and persisted almost to amphibians. Since it is [http://www.ncbi.nlm.nih.gov/pubmed/15486693 generally believed] that a latter whole genome duplication took place in ray-finned fish following the [http://www.ncbi.nlm.nih.gov/pubmed/22445447,20931168 divergence of gar], then the four CRY1 genes in zebrafish may represent retention of all copies whereas presumptive second copies of CRY4, CRY64, DASH and CPD were lost. | |||
The new spotted gar assembly (Lepisosteus oculatus) has the photolyase and cryptochrome repertoire (ie two CRY1 and one CRY2) expected from this scenario but the contigs are too small to contain flanking genes. The same can be said for the new coelocanth assembly (here Genomicus works with scaffolds of unordered unoriented contigs, which really pushes the limits for synteny). The shark and skate assemblies consist mostly of kilobase-size contigs that cover at most 60% of the coding exons; both have CRY1A and CRY1B but not CRY2 (and skate CPD has pseudogenized). | |||
The gar genome assembly as of April 2012 consists of 45,199 contigs (eg AHAT01025403) organized into 185 scaffolds (eg JH591278) organized further into 15 superscaffolds (eg CM001411) comprising 29 lingage groups (eg LG8) in 1012 pieces separated by gaps. Blast at NCBI can only access contigs. The entries for these contigs do not indicate their scaffold, superscaffold or linkage group. However those can be ferreted out with Entrez queries such as 'AHAT01025403 AND JH591*'. The cryptochrome contigs are too small to have any syntentic information, but the presence of a second gene within the same scaffold implies synteny. | |||
Here CRY1B and CRY4 both lie in the JH591232 gar scaffold, as they do in coelocanth and zebrafish scaffolds, establishing this as an ancestral synteny. Shark and skate, cartilaginous vertebrates, have CRY1A and CRY1B but the assemblies are too poor to even consider syntenic questions. Lamprey assembly is a non-starter, tunicates are too diverged, and amphioxus is uninformative (the flanking genes NAV2 GIT2 TUBB5 CRY1 TBC1D17 do not correspond at all to vertebrate gene order). | |||
Since frog and amniotes still have CRY4 in syntenic position with 7 other zebrafish genes but no cryptochrome, it follows that CRY1B was lost in the tetrapod stem. Thus it is CRY1A of fish that has continued in land animals under the name CRY1, as driven by human nomenclature convention (which is oblivious to other species). | |||
The near-adjacency of CRY1B and CRY4 could either be coincidence or indicative of an earlier tandem duplication relationship. CRY4 has a limited phylogenetic distribution in fish but continues on to frog, lizard and birds. In one scenario, CRY4 is not particularly related to CRY64 but instead arose from CRY1B in early ray-finned fish. The tandem pair persisted to extant fish but only CRY4 continued on in amniotes, with both CRY4 and CRY1B absent from mammals. | |||
Old RefSeq Chr S Start End N-term Pub Accession New RefSeq #Syn Comment | |||
CRY1A_danRer 4 + 11078260 11088888 MVVNTVH cry1a ENSDARG00000045768 CRY1P2_danRer 2 whole genome duplicate of retained CRY1 duplicate | |||
CRY1A2_danRer 18 + 14692957 14714934 MVVNTVH cry1b ENSDARG00000011583 CRY1P1_danRer 7 old CRY1 duplicate retained as tetrapod CRY1 | |||
1 | CRY1C_danRer 22 + 748902 787935 MSVNSVH cry2b ENSDARG00000091131 CRY1Q1_danRer 3 old CRY1 duplicate lost in tetrapods CRY1 C12ORF23 CRY4 in latCha too | ||
CRY1B_danRer 8 + 21767736 21788261 MAPNSIH cry2a ENSDARG00000069074 CRY1Q2_danRer 1 whole genome duplicate of lost CRY1 duplicate | |||
1 | CRY2_danRer 25 - 4289163 4311451 MVVNSVH cry3 ENSDARG00000024049 CRY2_danRer 3 synteny retained far better in coelocanth | ||
2 | CRY4_danRer 22 - 800173 811759 MSHRTIH cry4 ENSDARG00000011890 CRY4_danRer 7 adjacency to lost CRY1 suggests relationship | ||
CRY64_danRer 10 - 40633074 40645329 MSHNTIH cry5 ENSDARG00000019498 CRY64_danRer 2 poor synteny within fish, adjacent FOXRED1 in frog too | |||
DASH_danRer 24 - 20802832 20816799 MSASRTV cry-dash ENSDARG00000002396 DASH_danRer 1 strong synteny within fish, none to tetrapods | |||
CPD_danRer 2 + 13740732 13773635 MSANKNN cry-phr ENSDARG00000054999 CPD_danRer 3 mediocre preservation of synteny | |||
1 | |||
CRY1A_lepOcu JH591278 . LG8 CM001411 MVVNTVH AHAT01025403 CRY1P_lepOcu 1 old CRY1 duplicate retained as tetrapod CRY1 | |||
CRY1B_lepOcu JH591232 . LG3 CM001406 MGPNSIH AHAT01016727 CRY1Q1_lepOcu 1 old CRY1 duplicate lost in tetrapods | |||
CRY4_lepOcu JH591232 . LG3 CM001406 MTHRTIH AHAT01016726 CRY1Q2_lepOcu 1 adjacency to lost CRY1 suggests relationship | |||
CRY2_lepOcu JH591436 . UNK23 ........ MVVNSVH AHAT01038797 CRY2_lepOcu 1 isolated small contig | |||
CRY64_lepOcu JH591390 . LG26 CM001429 MMHRSIH AHAT01024141 CRY64_lepOcu 1 retained | |||
DASH_lepOcu JH591300 . LG9 CM001412 MSTIRTI AHAT01010414 DASH_lepOcu 1 retained | |||
CPD_lepOcu JH591341 . LG14 CM001417 MSGRSPP AHAT01034265 CPD_lepOcu 1 retained | |||
CRY1A_latCha JH126600 - 326804 392530 MGVNAIH ENSLACG00000008174 CRY1P_latCha 5 old CRY1 duplicate retained as tetrapod CRY1 | |||
CRY1B_latCha JH126576 + 512532 551664 MVVNSVH ENSLACG00000010538 CRY1Q1_latCha 2 old CRY1 duplicate lost in tetrapods | |||
CRY4_latCha JH126576 - 707590 727148 MTHRTIH ENSLACG00000012369 CRY1Q2_latCha 9 adjacency to lost CRY1 suggests relationship | |||
CRY2_latCha JH126568 + 3409780 3427400 MVVNSVH ENSLACG00000018488 CRY2_latCha 25 remarkably conserved synteny | |||
CRY64_latCha ........ . ....... ....... ....... .................. CRY64_latCha . apparently lost recently | |||
DASH_latCha ........ . ....... ....... ....... .................. DASH_latCha . apparently lost | |||
CPD_latCha ........ . ....... ....... ....... .................. CPD_latCha . apparently lost | |||
This raises the question of the ancestral origin of mammalian CRY2. It has it earliest representatives in early diverging teleost fish. It may have arisen from CRY1 after its duplication to CRY1P and CRY1Q and then diverged rather rapidly. Alternatively, it might be an older duplication and simply be lost from the chrondrichthyes studied to date. The CRY2 region exhibits remarkable conservation of gene order which may help resolve this issue once better early assemblies become available. Note that gene order has also been quite stable around CRY1 for several hundred million years. | |||
CRY2 must have arisen from a segmental duplication of the older CRY1 because their identical pattern of intronation could not have arisen independently. The size of the region duplicated in this event (ranging from one to several gene, or to a chromosome or even whole genome) could still be reflected by the extent of paralogous gene pairs. However subsequent inversions, gene losses, and rapid divergence might render these relationships opaque today. | |||
[[Image:Cry12syn.png|left]] | |||
Seeking homologous pairs (using gene names as a proxy for homology) in 25 genes flanking each side of CRY1 and CRY2 in human turns up 5 candidate pairs, not particularly supportive of a large segmental duplication given the many intervening non-homologous genes. The most intriguing pair, [[PRDM9:_meiosis_and_recombination|PRDM4]] and [[PRDM11:_giant_missing_exon|PRDM11]], are closely related and documented to have arisen in the same time frame. Since these are nearly adjacent to CRY1 and CRY2 respectively, a small duplication of 2-3 genes is the best fit to the data. | |||
One or even two rounds of whole genome duplication supposedly took place prior to vertebrate origins. Little supporting data for that scenario actually exists -- contrary to dozens of papers (all citing the same meagre investigations). The critical genome assemblies (amphioxus, tunicate and lamprey) are poor quality yet appear to have very similar numbers of protein coding genes. After a [http://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi?REQUEST=SHOW_STATISTICS decade of manual curation], no more than 18,500 coding genes can be documented in human. That's a very long ways from the 30,000 expected from 1R (or 60,000 from 2R) relative to the 15,000 genes of cephalochordate. | |||
The cryptochromes and photolyase gene family conflicts with both 1R and 2R hypotheses. If DASH, CPD, CRY64 and CRY1 were duplicated in such an event, then all duplicates were lost in all surviving lineages. The same is true for [[Opsin_evolution:_orgins_of_opsins|all three classes of opsins]] and many other gene families. If almost all duplicates from a whole genome duplication are lost, the outcome is effectively indistinguishable from the always-ongoing process of small segmental duplications with retention (the default hypothesis for paralogous gene origin). | |||
However CRY1 did experience three separate duplications-with-retention at various points in vertebrate evolution, implying functionality for all paralogous copies. One of these did arise from a whole genome duplication in a sub-lineage of ray-finned fish; the other two did not. Continued retention has been uneven in land animals (as with DASH, CPD, and CRY64) and the process of loss continues to the present day (still-recognizable pseudogenes cannot be old). In the same time frame, vertebrates also experienced a great expansion of the other main photoreceptor family, the opsins. Tunicates and amphioxus have no imaging vision but lenses, four cone opsins and rhodopsin were firmly established at the time of lamprey divergence. | |||
These quasi-simultaneous expansions may be correlated: cryptochromes are co-expressed in retinal, pineal and other light sensing cells and can function coordinately with opsins, as in the SWS1 cone cells of bird. This association may have a very long history as sponges, which have numerous GPCR genes but none with K296 retinol binding, may [http://www.ncbi.nlm.nih.gov/pubmed/14499587,20121950,20830749,22442365 use cryptochromes alone] as their larval photosensing system. In this scenario, they paired with a GPCR gene to signal; later that protein adapted retinoic acid signalling to retinal and took over the primary photosensing role in ctenophores and cnidarians which do have conventional opsins and neurons. The reference sequence collection contains [[Cryptochrome_refSeqs#Cryptochromes_from_cnidarians.2C_trichoplax.2C_sponges.2C_choanoflagellate.2C_plant_and_diatoms|5 cryptochromes]] from 4 sponge species, Amphimedon, Suberites, Crateromorpha and Aphrocallistes. | |||
=== Inconsequential N-terminal extension in vertebrate CRY2 === | |||
CRY1 duplicated in early vertebrates giving rise to CRY2 which evidently carved out a distinct functional role as it has persisted in all species since including committed subterranean and cave species. The status of the gene in lamprey, hagfish and chondrichthyes cannot be resolved without better assemblies but there is no indication of a duplication there or in urochordates or cephalochordates. | |||
Confusingly, Cry2 is also used for non-orthologous insect cryptochromes entries at GenBank. These represent a different duplication of a different parent gene called CRY1A here (which itself was subsequently lost in dipterans). The Drosophila 'CRY2' sequence, called CRY1B here, is not a valid model system for vertebrate CRY2 and indeed is equally unsuited as a invertebrate CRY1 proxy because properties can be expected to pull apart in species retaining both copies. | |||
2 | Vertebrate CRY2 has an extended amino terminus that arose in amniotes. Prior to that time, it was similar in length to vertebrate CRY1 (and still is from fish to frog). A few residues of this extension are conserved but overall the sequences displays compositional simplicity, to the point that RepeatMasker finds a variable length simple repeat (CCG)n within coding and in some species also upstream. Base composition alone then leads to inevitable amino acid conservation which however does not imply selective pressure or functionality. | ||
While no applicable crystallographic structure has been determined, homology modeling reliably locates these extra residues outside the closed globular alternating beta strand/alpha helix structure of this domain. They are likely proteolytically trimmed off in mature protein leaving only 3-4 extra residues. | |||
One scenario here posits that the initial methionine in stem amniotes was lost to mutation, leading to an upstream random in-phase ATG stepping in, followed by evolution of variable length as the new ATG lay in the (CCG)n repeat which was subject to expansions and contractions. While this had no adverse consequences, it did not lead to functional innovation in this region either -- a short amino terminal extension is necessarily remote from excitation transfer pathways, and antenna, FAD and substrate binding sites. | |||
<font color=gray>CRY1_homSap ...............................MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR CRY1_homSap ...............................MGVNA................KECIQ..DTI...........G..N.......</font> | |||
<font color=blue>CRY2_homSap ............MAATVATAAAVAPAPAPGTDSASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_homSap ...........MAATV.ATAAAVAPAPAPGTDSASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR | |||
CRY2_panTro ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_panTro ................................G................................................... | |||
CRY2_gorGor ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_gorGor ................................G................................................... | |||
CRY2_ponAbe ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_ponAbe ................................G................................................... | |||
CRY2_rheMac ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_rheMac ................................G................................................... | |||
CRY2_papHam ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_papHam ................................G................................................... | |||
CRY2_calJac ............MAATVATAAAAVPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_calJac ......................AV........G................................................... | |||
CRY2_micMur .............MATAVATAAAAPTPASSTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_micMur ............M..A.VAT..A..T..SS..G...................................................</font> | |||
<font color=darkblue>CRY2_musMus .............MAAAAVVAATVPAQSMGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_musMus ........MAAA.VVA......TV..QSM.A.G................................................... | |||
CRY2_ratNor .............MAAAAVVAATVPAQSMGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_ratNor ........MAAA.VVA......TV..QSM.A.G................................................... | |||
CRY2_criGri ........MAAAAVVAGAPRGARVPALTMGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_criGri ........MAAA.VVAG.PRG.RV..LTM.A.G................................................... | |||
CRY2_spaJud .............MAAASVVVATSAAPAMAVDGGSSVHWFRKGLRLHDNPSLLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_spaJud ........MAAASVV.......TSA...MAV.GG................S................................. | |||
CRY2_dipOrd ............MAAAMVTAAVAVPAPPSGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_dipOrd ..............AM.V...VAV...PS.A.G................................................... | |||
CRY2_cavPor ............MAAAVGTGTAAAPTPVTGAEGACSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_cavPor ..............A..G.GT.A..T.VT.AEG.C................................................. | |||
CRY2_hetGla ............MAAAVGTGTGAAPTPATGAEGACSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_hetGla ..............A..G.GTGA..T..T.AEG.C................................................. | |||
CRY2_speTri ...............MSASVVTTSATLLTPTSADVSSVHWFRKGLRLDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_speTri ............S.S..V.TS.TLLT.TSA..DV.................................................. | |||
CRY2_oryCun ............MAAAAAAAAAAVPAPAASANGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_oryCun ..............AA..A...AV....ASANG...................................................</font> | |||
<font color=brown>CRY2_turTru ............MAAAVATSAVAAPAPAARAEGASSVHWFRKGLRLHDNPALQAAVRGAHCVRCVYILDPWFAASSSVGINRWR CRY2_turTru ..............A....S.VA.....ARAEG...................Q......H........................ | |||
CRY2_bosTau ...............MAAAAAAATQAPAARGDGASSVHWFRKGLRLHDNPALLAAVRGAHCVRCVYILDPWFAASSSVGINRWR CRY2_bosTau ........MAAA..........ATQ...ARG.G..........................H........................ | |||
CRY2_oviAri .........MAAAAAATASAAAAAQAPAPRGDGASSVHWFRKGLRLHDNPALLAAVRGAHCVRCVYILDPWFAASSSVGINRWR CRY2_oviAri ........MAAA..AT..S...A.Q....RG.G..........................H........................ | |||
CRY2_susScr ............MAAAVATAAASSPAPAAGAEGASSVHWFRKGLRLHDNPALLAAVRGAHCVRCVYILDPWFAASSSVGINRWR CRY2_susScr ..............A.......SS....A.AEG..........................H........................ | |||
CRY2_equCab MKKAAAPVRFIATSEAPAASAAAAATAAAGADGDSSVHWFRKGLRLHVNPALLAAVRFLRSVLCVYKNDPWFVASSSVGINRWR CRY2_equCab MKKAAAPVRFIATSEAP.AS..A.ATA.A.A.GD.............V.........FL.S.L...KN....V........... | |||
CRY2_canFam ............MAAAVVAAAAAAPVPTAGVDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_canFam ..............A..VA...A..V.TA.V.G................................................... | |||
CRY2_ailMel ........................PAPAAGVDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_ailMel ............................A.V.G................................................... | |||
CRY2_myoLuc ............MAANAVTAAAAAPAPAAGTDGASSVYWFRKGLRLHDNPALLAAVRGARCVLCVYILDPWFAASSSVGINRWR CRY2_myoLuc ..............NA.V....A.....A...G....Y........................L..................... | |||
CRY2_pteVam ............MAATVGTAAAAASAPAAGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_pteVam .................G....A.S...A...G...................................................</font> | |||
<font color=green>CRY2_loxAfr ............MAAAVVTAGAAALVPIPSMDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_loxAfr ..............A..V..G.A.LV.I.SM.G................................................... | |||
CRY2_triMan ............MAATVVTAAAAALAPAPSIDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_triMan .................V....A.L....SI.G................................................... | |||
CRY2_choHof .............MAATAVMAGSAAPAPASGTEGASSVHWFRKGLLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_choHof ...............A.VM.GSA.....S..EG...................................................</font> | |||
<font color=#00CC66>CRY2_monDom ............MAAAVVTMTAAAPAPAPSPEGASSVHWFRKGLRLHDNPALQAALRGARCVRCVYILDPWFAASSSVGINRWR CRY2_monDom ..............A..V.MT.A......SPEG...................Q..L............................ | |||
CRY2_macEug ..........MAATTAVTVTVPAAAPAPAPEEGASSVHWFRMGLRLHYNPVLYSALRGARCVRCVFFLYSWFAASSSVVFFLWL CRY2_macEug .........MAATTA.TV.VP.A......E.EG........M.....Y..V.YS.L.........FF.YS........VFFL.L</font> | |||
<font color=#990099>CRY2_galGal ......................MAAAASPPRGFCRSVHWFRRGLRLHDNPALQAALRGAASLRCIYILDPWFAASSAVGINRWR CRY2_galGal ......................M.A.AS.PRGFCR......R..........Q..L...ASL..I...........A....... | |||
CRY2_allMis ................MAASRSFPSSVPARAGPCRAVHWFRRGLRMHDNPALQAALRDAASVRCIYILDPWFAASSAVGINRWR CRY2_allMis ................M.ASRSFPSSVPARAGPCRA.....R...M......Q..L.D.AS...I...........A....... | |||
CRY2_anoCar ........................MAALPGPLGRCSVHWFRRGLRLHDNPALQAAIRDGGPVRCIYILDPWFAASSSVGINRWR CRY2_anoCar ........................M.AL..PLGRC......R..........Q..I.DGGP...I...................</font> | |||
<font color=#0066CC>CRY2_xenTro ...........................MEGKPSVSSVHWFRKGLRLHDNPALLSALRGANSVRCVYILDPWFAASSSGGVNRWR CRY2_xenTro ...........................ME.KP.V...................S.L...NS................G.V.... | |||
CRY2_ranCat ............................MEGPAVSSVHWFRKGLRLHDNPALLAALRGARCVRCVYILDPWFAASSSGGVNRWR CRY2_ranCat ...........................ME..PAV.....................L.....................G.V....</font> | |||
<font color=#990000>CRY2_lepOcu ...............................MVVNSVHWFRKGLRLHDNPALQEALNISDTVRCVYILDPWFAASANVGINRWR CRY2_lepOcu ...............................MVVN.................QE.LNISDT..............AN....... | |||
CRY2_danRer ...............................MVVNSVHWFRKGLRLHDNPALQEALNGADTVRCVYILDPWFAGSANVGVNRWR CRY2_danRer ...............................MVVN.................QE.LN..DT............G.AN..V.... | |||
CRY2_oreNil ...............................MVVNSVHWFRKGLRLHDNPALQEALNGADAVRCVYILDPFFAGAANVGINRWR CRY2_oreNil ...............................MVVN.................QE.LN..DA.........F..GAAN....... | |||
CRY2_tetNig ...............................MVVNSVHWFRKGLRLHDNPALQEALSGADSLRCVYVLDPWFAGAANVGINRWR CRY2_tetNig ...............................MVVN.................QE.LS..DSL....V......GAAN....... | |||
CRY2_takRub ...............................MVVNSVHWFRKGLRLHDNPALQEALSGADSLRCVYVLDPWFAGAANVGINRWR CRY2_takRub ...............................MVVN.................QE.LS..DSL....V......GAAN.......</font> | |||
<font color=gray>CRY1_homSap ...............................MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR CRY1_homSap ...............................MGVNA................KECIQ..DTI...........G..N.......</font> | |||
CRY2_homSap (CGG)n 46 bp atggcggcgactgtggcgacggcggcagctgtggccccggcgccagcg | |||
<font color=blue> M A A T V A T A A A V A P A P A </font> | |||
CRY2_rheMac (CCG)n 46 bp atggcggcgactgtggcgacggcggcagctgtggccccggcgccagcg | |||
<font color=blue> M A A T V A T A A A V A P A P A </font> | |||
CRY2_musMus (CCG)n 38 bp ggcggcgatggcggcggctgctgtggtggcagcgacgg | |||
A A <font color=blue> M A A A A V V A A T </font> | |||
CRY2_oryCun (CGG)n 116 bp ggcggggctcgcggcgccgccgggggcggagcggcggtggctccggcagtctgagctgtgatggcggcggcggcggcagtgggtcctgcggcggcggcggtccccgcgccggcggc | |||
G G A R G A A G G G A A V A P A V - A V <font color=blue> M A A A A A V G P A A A A V P A P A </font> | |||
CRY2_canFam (CCG)n 101 bp gcggcgccggcgggggcggagcggcggagcggcggagcggcggaggcctgagcagtcggagcggtgatggcggcggctgtggtggcggcggcagcggcggc | |||
A A P A G A E R R S G G A A E A - A V G A V <font color=blue> M A A A V V A A A A A </font> | |||
CRY2_bosTau (CGG)n 31 bp ggcggtgatggcggcggcggcggcggcggcg | |||
A V <font color=blue> M A A A A A A A </font> | |||
CRY2_myoLuc CCG)n 57 bp ggcggcgatggcggcgaatgcggtgacggcagcagcggcggccccagcgccagcggc | |||
A A <font color=blue> M A A N A V T A A A A A P A P A </font> | |||
CRY2_loxAfr (CCG)n 98 bp ggaggtggggcccgcggcgctgtcgggggcggagcgccgccggccagagcagtctaggcggtgatggcggcggcagtggtgacggcgggagcggcggc | |||
G G G A R G A V G G G A P P A R A V - A V <font color=blue> M A A A V V T A G A A </font> | |||
The full set of 43 vertebrate CRY2 sequences is [[Cryptochrome_refSeqs|here]]. These are mostly pre-curated provisional sequences taken from the UCSC 46-way genomic alignment relative to human except where the fasta header displays an accession number. This source misses small insertions and indeed whole exons when very diverged or lie in isolated small contigs or unassembled traces. | |||
The final exons, being quite variable especially in fish, are best determined from transcripts when available and then extended by blastx homology to species within the same clade that lack transcripts. After these corrections, the sequences are [http://npsa-pbil.ibcp.fr/cgi-bin/align_multalin.pl aligned] and additional anomalies are confirmed or discarded on a case-by-base basis. | |||
It has been [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC147960/pdf previously reported] that the PKRK motif in the last exon of mouse CRY2 represents a nuclear localization signal. This motif is indeed conserved in all tetrapod sequences from frog to human. While neither coelocanth nor gar sequence extends to this region, it is definitely absent from other ray-finned fish. Thus nuclear localization may be an innovation of land vertebrates. | |||
It appears that the last exon in fish has lost all homology (and so functionality), in some cases simply running out into junk dna until a stop codon is encountered. Exon seven is broken up in some fish with an extra intron that might have some use in fish taxonomy as a derived characteristic. | |||
=== Invertebrate cryptochromes: distal CRY1B spoofs a damaged DNA base === | |||
Nomenclature of cryptochromes is both a historic and continuing source of confusion, with experimentalists oblivious to anything outside a personal narrow clade and seemingly befuddled by simple concepts such as the timing of gene duplications within eukaryotic phylogeny and orthologouse classification. Another bizarre aspect is functional taxonomy -- like lumping penguins and fur seals together because both eat fish -- whereas enzyme vs circadian signaling vs magnetosensing, single-stranded vs duplex DNA, 6-4 lesions vs thimine dimer have every prospect of multiple origins, gains and losses, cross-overs and reversibility. For example, Arabidopsis UVR3/CRY3 may very well repair 6-4 lesions but it certainly has nothing to do with the CRY64 ortholog group, classifying as it does with CRY1. | |||
With the number of complete genomes available today, it is clear that early bilateran ancestor contained two distinct cryptochromes (in addition to three photolyases), all of which persisted into early deuterostomes (sea urchin). These cannot be denoted CRY1 and CRY2 because those gene names have been assigned to human cryptochromes [http://www.genenames.org/ by international agreement], excluding their re-use in invertebrates or plants for weakly related homologs. The two invertebrate cryptochromes are denoted CRY1A and CRY1B here to distinguish them from the later gene duplication of CRY1A in vertebrates giving rise to tetrapod CRY1 and CRY2. CRY1B is the most commonly studied gene in Drosophila. It has no orthologous counterpart in vertebrates because the definition of orthology requires descent from a single genetic locus in the last common ancestor. | |||
[[Image:CryAcryB.png|left]] | |||
Some lophotrochozoa, notably molluscs, retained both cryptochromes (and indeed a third in a few species not clustering with either CR1A OR CRY1B). Within arthropods, generally one cryptochrome was retained but not always the same one. However some dipterans, hemipterans and lepidopterans retain both. It appears that the CRY1A/CRY1B gene duplication itself took place after divergence from cnidarians. | |||
The two cryptochromes are intronated quite characteristically, with the first class (called CRY1A here) most similiar to that of vertebrate CRY1/2, in agreement with closer blastp clustering. The second class (called CRY1B below) bears less relevance to the cryptochromes retained in mammals but unfortunately is the only one retained in Drosophila and [http://www.ncbi.nlm.nih.gov/pubmed/22080955 most studied]. Annotation transfer from study of CRY1B proteins to mammals is thus exceedingly problematic given that the CRY1A family retains far more sequence similarity and is not descended from CRY1B. Clade-delimited gene duplications can lead to accelerated divergence as the two copies must subfunctionalize to garner selective pressure support. | |||
<br clear=all> | |||
A remarkable recent [http://www.ncbi.nlm.nih.gov/pubmed/22080955 crystallographic result] establishes an important role for tryptophan W536 near the end of the variable region of drosophila CRY1B. This aromatic residue and its associated helix arch back to occupy the site normally occupied by a damaged dna nucleotide, spoofing the presence of a damaged dna residue for conformational change purposes. | |||
[[Image:Cry1Bspoof.jpg]] | |||
The tryptophan is part of a larger motif <font color=blue>PPHCRPSNEEEVRQFM<font color=red>W</font>LP</font> conserved in the CRY1B orthologs from insects, crustaceans, molluscs and surprisingly three echinoderms. It is erroneously denoted the FFW motif in the fruit fly cryptochrome literature. Since 3.4 residues are needed for a full alpha helix turn, the 16 residues of the motif are enough for 4.7 turns (more than the 3 actually observed). Note the full motif is quite well conserved in amino acid sequence whereas the protrusion motif is not conserved either in residue or length. | |||
[[Image:CRY1BlogoMotif.png|left]] | |||
However two substitutions should be noted, cysteine and tyrosine in daphnia and aphid respectively, suggesting that the overall motif is more critical than just a tryptophan. Further, no comparable residue or motif exists in invertebrate CRY1A proteins, vertebrate cryptochromes or other photolyase homologs. The observed phylogenetic distribution (which is unlikely to reflect convergent evolution) implies the spoofing mechanism arose in an early bilateran after to the gene duplication giving rise to CRY1A/CRY1B but before the protostome/deuterostome split. | |||
A threonine at position 518 is reported phosphorylated in CRY1B_droMel but has no real phylogenetic support even within drosophilids, also lying outside the motif detected by [http://weblogo.berkeley.edu/logo.cgi WebLogo]. This post-translational modification could nonetheless have regulatory signficance in the limited spectrum of species that could have it but more likely it represents an aberrant event. | |||
For a full set of 38 invertebrate CRY1A and CRY1B sequence available in Mar 2012, see the [[Cryptochrome_refSeqs|curated reference sequences]]. | |||
<br clear=all> | |||
<font color=blue>PPHCRPSNEEEVRQFM<font color=red>W</font>LP</font> | |||
CRY1B_strPur KVVNKLRDTGIVHCAPSTQREVREFV<font color=red>W</font>LPEKMAGGGSCRADQNCEGILGL echinoderm | |||
CRY1B_lytVar KVINRLRDSGIVHCAPSTQKEVREFV<font color=red>W</font>LPEKMAGGGSCRADQNCEGILGL echinoderm | |||
CRY1B_parLiv KVINRLRDSGIVHCAPSTQKEVREFV<font color=red>W</font>LPEKMAGGGSCRASQNCEGRTGS echinoderm | |||
> | CRY1B_aplCal MEAIKKVSKDVPHIAPANEEEVLTLM<font color=red>W</font>SGKQTRSELMDA----------- mollusc | ||
CRY1B_craGig AVKDALIGKEIPHCAPSEEIEARRFS<font color=red>W</font>LP--------------------- mollusc | |||
CRY1B_octVul KVKEHLLHQDVPHCGPTNETEVWKFA<font color=red>W</font>LPPIEHHDLAHNI---------- mollusc | |||
CRY1B_rudPhi KNKLVQQGKDLEHCRPTNVEEVRMFV<font color=red>W</font>MPGAHKGACGQEVPLDDKELCDG mollusc | |||
CRY1B_plaOce MDRIKNLCKGIPHVAPTNENEVLSYM<font color=red>W</font>LDKSNSEAMEESLFEACSHLSSV mollusc | |||
> | CRY1B_dapPul KEFRQKFKETPAHCQPSSNSEVYKFF<font color=magenta>C</font>LPDDSLPF--------------- crustacean | ||
CRY1B_diaNig DEIRNRLMNPPPHCRPSSEKETRQFM<font color=red>W</font>FPDDCSEHSSQ------------ orthoptera | |||
CRY1B_acyPis LRVSMTNENRVPHCCPSDREEVQKFM<font color=magenta>Y</font>LPDECMQQLLPLENQDSKAYDIY hemiptera | |||
CRY1B_danPle QELRRLLEKAPPHCCPSSEDEVRQFM<font color=red>W</font>LGDDSQPELTTT----------- lepidoptera | |||
CRY1B_bomMor EELRMLLEKAPPHCCPSSEDEIRQFM<font color=red>W</font>LNE-------------------- lepidoptera | |||
CRY1B_mamBra GELRHFLQKAPPHCCPSSEDEIRQFM<font color=red>W</font>LNE-------------------- lepidoptera | |||
CRY1B_helArm KELRHMLQKAPPHCCPSSEDEIRQFM<font color=red>W</font>LNE-------------------- lepidoptera | |||
CRY1B_droMel KSLRNSLITPPPHCRPSNEEEVRQFF<font color=red>W</font>LADVVV----------------- diptera | |||
CRY1B_anoGam REKLVDGGSTPPHCRPSDIEEIRQFF<font color=red>W</font>LADDAATEA-------------- diptera | |||
CRY1B_neoBul LIAEGAPDNGPPHCRPSNEEEIRNFF<font color=red>W</font>LAD-------------------- diptera | |||
CRY1B_bacCuc LIAGGAPDEGPPHCRPSNEEEVHQFF<font color=red>W</font>LVE-------------------- diptera | |||
CRY1B: C-terminal conservation in drosophilids * * | |||
Drosophila melanogaster YECLIGVHYPERIIDLSMAVKRNMLAMKSLRNSLI T PPPHCRPSNEEEVRQFFWLAD | |||
Drosophila simulans ................................... . ..................... | |||
Drosophila sechellia ................................... . ..................... | |||
Drosophila yakuba ............................T...... . ..................... | |||
Drosophila erecta ................T...Q.......A...... . ..................... | |||
Drosophila rhopaloa ............................A.....M . ..................... | |||
Drosophila elegans ............................A.....M . ..................... | |||
Drosophila takahashii ..................Y.........A.....M . ..................... | |||
Drosophila ficusphila .................L..........A...... . ..................... | |||
Drosophila eugracilis ............M....L..........A...... . ..................... | |||
Drosophila biarmipes .................VY.....M...A...... . ..................... | |||
Drosophila kikkawai .................K.........TA...... . ..................... | |||
Drosophila mojavensis ..........D......L.S........A...... E ..................... | |||
Drosophila persimilis ................K......M..TA...... . ....................N | |||
Drosophila pseudoobscur .................K......M..TA...... . ....................N | |||
Drosophila bipectinata ..........D.L...TK...G.........D... . ..............T...... | |||
Drosophila ananassae ..........D.L....K...G......T..D... . ..............T...... | |||
Drosophila willistoni ...........P.....L.L...T...TN...... . ....................E | |||
Drosophila grimshawi ................L.S....A...A......E . T.................DE. | |||
Drosophila virilis ....L.F...Q......L.S...TM...A...... E ...................TN | |||
=== Cryptochrome CRY4 evolutionary origin === | |||
This orthology class has been [http://www.ncbi.nlm.nih.gov/pubmed/10971654,16687285,19663499,21909239,20174658 studied to a limited extent] in fish, frog and birds, often without full knowledge of the overall cryptochrome repertoire in the given species. Literature search is confused by erratic nomenclature practises in publications and gross mislabelling of GenBank reference sequences such as NM_001095521, which is CRY4 of Xenopus laevis rather than CRY1 as stated. | |||
CRY4 has 19 reported transcripts originating from frog testes, oocytes, ovary and whole embryo whereas orthologous zebrafish transcripts have come from retina, eye, brain, heart, liver, paraxial mesoderm, caudal fin, tail bud, and embryo. Chicken has transcripts from brain, heart, kidney, limb, muscle, ovary; sparrow from brain; finch from embryonic brain. If these are representative -- rather than merely reflecting experimental focus -- this does not suggest continuity of function. | |||
Cry4 has been lost in multiple clades and is missing from echinoderms,chondrichthyes, perciform fish, crocodillians, turtles and snakes, and all mammals. It is diverging quite rapidly in amphibians, with Xenopus laevis only 89% identical to Xenopus tropicalis. However according to Blast classification, it is present in tunicate and amphioxus. | |||
The evolutionary orign of CRY4 has some puzzling aspects, though being restricted to deuterostomes it has limited parental gene options, basically CRY64 or CRY1. The 8th exon -- the far end of the FAD domain -- is split by a phase 21 intron. This is a derived condition since it is absent at homologous position in bilateran CRY64, CRY1, CRY2, CRY1A and CRY1B. Intron gain is quite rare in vertebrates but not in some earlier diverging bilaterans. | |||
Split exon 8 cryptochromes (imputed* for processed transcripts in non-genomic species): | |||
CRY4_galGal Gallus gallus (chicken) | |||
CRY4_melGal Meleagris gallopavo (turkey) | |||
CRY4_anaPla Anas platyrhynchos (duck) | |||
CRY4_pasDom Passer domesticus (sparrow)* | |||
CRY4_taeGut Taeniopygia guttata (finch) | |||
CRY4_anoCar Anolis carolinensis (lizard) | |||
CRY4_xenTro Xenopus tropicalis (frog) | |||
CRY4_xenLae Xenopus laevis (frog) | |||
CRY4_latCha Latimeria chalumnae (coelocanth) | |||
CRY4_lepOcu Lepisosteus oculatus (spotted_gar) | |||
CRY4_danRer Danio rerio (zebrafish) | |||
CRY4_molTec Molgula tectiformis (tunicate)* | |||
CRY4_braFlo Branchiostoma floridae (amphioxus) | |||
> | CRY1_braFlo Branchiostoma floridae (amphioxus) | ||
CRY1A_strPur Strongylocentrotus purpuratus (urchin) | |||
However the same intron is present in two amphioxus and sea urchin cryptochromes which classify as CRY1 and CRY1A rather than CRY4. These early deuterostome cryptochromes may be misclassified -- divergence is fairly high and synteny is gone, leaving match quality, introns and indels as the remaining diagnostic criteria. However blast classifier data shows high confidence classification of both, with the top 43 matches of the 278 reference gene set all being CRY1-class cryptochromes, with the best match of CRY64 and CRY4 from any species far more distantly related: | |||
<font color=blue>CRY1_braFlo Branchiostoma floridae (amphioxus) XM_00260... 2982 3.9e-314</font> | |||
<font color=purple>CRY1_melUnd Melopsittacus undulatus (parakeet) AGAI0106... 2303 3.5e-242 | |||
CRY2_anoCar Anolis carolinensis (lizard) XM_003214641 2141 5.2e-225 | |||
1 | CRY1A_aedAeg Aedes aegypti (mosquito) XM_001655728 dipte... 2062 1.2e-216 | ||
CRY64_xenTro Xenopus tropicalis (frog) synteny: STS1 RPL... 1595 3.7e-167 | |||
CRY4_xenTro Xenopus tropicalis (frog) NP_001123706 1428 1.9e-149 | |||
CRY1B_octVul Octopus vulgaris (octopus) JR450373 transcr... 1166 1.1e-121 | |||
CRY7_hapBur Haplochromis burtoni (chichlid) AFNZ01022319 380 7.0e-36</font> | |||
> | <font color=blue>CRY1A_strPur Strongylocentrotus purpuratus (urchin) XM_0... 3045 7.7e-321</font> | ||
<font color=purple>CRY1_xenTro Xenopus tropicalis (frog) NM_001087660 1153... 1966 1.8e-206 | |||
CRY2_anoCar Anolis carolinensis (lizard) XM_003214641 1919 1.7e-201 | |||
CRY1A_apiMel Apis mellifera (bee) NM_001083630 AADG06001... 1900 1.8e-199 | |||
CRY64_anoCar Anolis carolinensis (lizard) XM_003225714 6... 1562 1.2e-163 | |||
CRY4_lepOcu Lepisosteus oculatus (spotted_gar) AHAT0101... 1429 1.5e-149 | |||
CRY1B_parLiv Paracentrotus lividus (sea_urchin) AM599080... 1292 4.8e-135 | |||
CRY7_tetNig Tetraodon nigroviridis (fugu) 369 1.8e-34</font> | |||
In a gene with 530aa x 3bp/aa x 3 phases, there are 4770 ways of creating a new intron, so it is wholly implausible that the event was fixed twice. Thus the origin of CRY4 must be entangled with these other cryptochromes. | |||
1 | |||
1 | |||
1 | |||
1 | |||
1 | |||
1 | |||
The amphioxus CRY4 sequence also has a fusion of exon 2 and exon 3. This also occurs in amphioxus CRY64 but nowhere else. Echinoderms lack CRY4, either having lost it or never having had one. A third peculiar feature of amphioxus CRY4 is the phase 12 intron between exon 4 and exon 5, representing a shift from ancestral phase 00. Amphioxus CRY64 also shares this phase shift but again earlier and later diverging orthologs do not. | |||
The anomalous CRY1 sequences of amphioxus and sea urchin share another odd feature, a new phase 00 intron internal to exon 7. This intron does not occur in any other CRY1, CRY4 or CRY64 sequence and so is derived. Echinoderms and cephalochordates arose from separate divergence nodes; the event could not have taken place in a common ancestral stem. Regretably, no cryptochromes have been retained in the hemichordates, sister group to echinoderms, based on the Saccoglossus kowalevskii genome project. | |||
Recall CRY4 is nearly adjacent to CRY1B of fish, the earlier CRY1 gene duplication preceding fish whole genome duplication lost in tetrapods. That could reflect tandem duplication or improbable but possible accidental juxtapositioning. | |||
It takes a fairly complicated scenario to reconcile these observations. Suppose the earliest deuterostomes acquired the intron gain between exon 8-9 in CRY64. Assume further that this engaged in heterologous recombination with CRY1, leading to a polymorphic state that was never really fixed but persisted across the divergence of echinoderms and cephalochordates, with lineage sorting leaving CRY1 and CRY1A in those species with the extra intron but not those genes vertebrates. | |||
After amphioxus diverged, its CRY64 acquired the fusion of exon 2-3 and phase shift between exon 4-5. It then duplicated, giving rise to amphioxus CRY4, but later lost the intron between exon 8-9 (though amphioxus CRY4 retained it). This would cause best-blast of amphioxus CRY4 to be amphioxus CRY64 rather than lie with other CRY4, as observed. In this case, the two amphioxus genes should be renamed CRY64A and CRY64B (replacing CRY4). | |||
Gene_genSpp Exon 2-3 fusion Exon 4-5 phase shift Exon7 new 00 intron Exon 8 new 21 intron CRY1B sytney | |||
CRY1_other no no no no no | |||
CRY1_braFlo no no <font color=blue>yes</font> <font color=blue>yes</font> no | |||
CRY1A_strPur no no <font color=blue>yes</font> <font color=blue>yes</font> no | |||
> | CRY4_other no no no <font color=blue>yes</font> <font color=blue>yes</font> | ||
CRY4_braFlo <font color=blue>yes</font> <font color=blue>yes</font> no <font color=blue>yes</font> no | |||
CRY4_strPur --- --- --- --- --- | |||
CRY64_other no no no no no | |||
CRY64_braFlo <font color=blue>yes</font> <font color=blue>yes</font> no no no | |||
CRY64_strPur no no no no no | |||
> | |||
=== Cryptochrome 6-4 photolyases === | === Cryptochrome 6-4 photolyases === | ||
>CRY64_anoCar Anolis carolinensis (lizard) XM_003225714 6-4 photolyase synteny: DCPS TIRAP CRY64 SRPR FOXRED1 | CRY64 is a mainstream catalytic photolyase that reverses UV-induced (6-4) photoproducts in DNA using blue light. CRY64 gave rise to many cryptochromes over evolutionary time via sequential gene duplications. It [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3645588/ originated in prokaryotes] and persists into many invertebrates and amniotes. However it has been lost both in birds and mammals (which given the phylogenetic tree, requires two distinct loss events at a minimum). The reasons for loss, the adequacy of residual compensatory repair processes, and the consequences to final mutational rate are not well understood. | ||
[[IMAGE:2WQ7limits.jpg|left]] | |||
The carboxy terminus of CRY64 is a bit curious. The last two exons are provided below. Note the sequences become unalignable, despite all members retaining the same phase 12 splice donor. However clamping to the conserved exon break does not put the sequences into register for long -- even restricting to ray-finned fishes for which data is unfortunately overweighted. However the deuterostome sequences all retain a high content of basic residues at the end as shown below. (Other invertebrates have lost their introns and so lack this device for re-registration of the alignment.) | |||
The available 3D structures (from Drosophila and Agrobacterium) only partly clarify the role of the carboxy terminal basic residues in vertebrates. The last determinable residues of CRY64 form a long terminal alpha helix highlighted in yellow in the adjacent image (and as hhh in the alignment below). However this extends (magenta) beyond the limit of blast alignability to vertebrates (blue) and the remained residues studied (gray dots) do not form a sufficiently stable conformation to display as fixed electron density. However this region clearly is positioned near the substrate binding site. | |||
Possibly the terminal residues provide positively charged residues that offset negative backbone phosphates in the DNA strands. In this scenario, the primary sequence itself is not so important as long as it provides a sufficient number of flexibly positionable lysines and arginines. In this scenario, there is selection for amino acid composition rather than conventional residue by residue conservation. This could readily be tested by small terminal deletions. | |||
<br clear=all> | |||
<font color=blue>hhhhhhhhhhhhhhhh</font><font color=magenta>hhhhhhhhh</font><font color=gray>.............</font> | |||
CRY64_droMel 2WQ7 HEVVH<font color=red>K</font>ENI<font color=red>K</font><font color=red>R</font>MGAAY<font color=red>K</font>VN<font color=red>R</font>EV<font color=red>R</font>TG<font color=red>K</font>EEESSFEE<font color=red>K</font>SETSTSG<font color=red>K</font><font color=red>R</font><font color=red>K</font>V<font color=red>R</font><font color=red>R</font>ATGSAP<font color=red>K</font><font color=red>R</font><font color=red>K</font><font color=red>R</font> | |||
CRY64_anoCar <font color=red>K</font>YLPFL<font color=red>RK</font>FSNDYIYEPW<font color=red>K</font>AP<font color=red>R</font>SLQE<font color=red>R</font>AGCIIGQDYP<font color=red>K</font>PIVEHE<font color=red>K</font>VY<font color=red>KR</font>NLE<font color=red>R</font>M<font color=red>K</font>AAYA<font color=red>RR</font>SPNLVIQA<font color=red>K</font>D<font color=red>K</font>VSQ<font color=red>KK</font> GVN<font color=red>RK</font><font color=red>R</font>PEAPT<font color=red>K</font>A<font color=red>K</font>VQA<font color=red>KK</font>V | |||
CRY64_chrPic <font color=red>K</font>YLPFL<font color=red>RK</font>FPAEYIYEPW<font color=red>K</font>AP<font color=red>R</font>SMQEQAGCVIG<font color=red>R</font>DYP<font color=red>K</font>PIVVHEVVS<font color=red>KR</font>NVE<font color=red>R</font>M<font color=red>K</font>AAYA<font color=red>RR</font>SSSTTAQLEGGGG<font color=red>KK</font>GI GA<font color=red>K</font><font color=red>RR</font>TPAGPSVAELLT<font color=red>KK</font>P | |||
CRY64_allMis <font color=red>K</font>YLPIL<font color=red>RK</font>FPAEYIYEPW<font color=red>K</font>AP<font color=red>R</font>SMQEQAGCIIG<font color=red>R</font>DYP<font color=red>K</font>PIVEHEALS<font color=red>KR</font>NIM<font color=red>R</font>M<font color=red>K</font>AAYAQ<font color=red>R</font>SHS<font color=red>K</font>AAQVE<font color=red>K</font>EST<font color=red>KK</font>GN GG<font color=red>K</font><font color=red>RK</font>LPAGPSVVELLT<font color=red>KK</font>P | |||
CRY64_croPor <font color=red>K</font>YLPIL<font color=red>RK</font>FPAEYIYEPW<font color=red>K</font>AP<font color=red>R</font>SMQEQAGCIIG<font color=red>R</font>DYP<font color=red>R</font>PIVEHEAVS<font color=red>KR</font>NIM<font color=red>R</font>M<font color=red>K</font>AAYAQ<font color=red>R</font>SHS<font color=red>K</font>SAQVE<font color=red>K</font>EGT<font color=red>KK</font>GN GG<font color=red>K</font><font color=red>RK</font>LPAGPSVVELLT<font color=red>KK</font>P | |||
CRY64_xenTro <font color=red>K</font>YLPIL<font color=red>KK</font>FPAEYIYEPW<font color=red>K</font>AP<font color=red>R</font>SLQE<font color=red>R</font>AGCIIG<font color=red>K</font>DYP<font color=red>K</font>PIVEHDVAS<font color=red>K</font>QNIQ<font color=red>R</font>M<font color=red>K</font>AAYA<font color=red>RR</font>SGSTAEVD<font color=red>K</font>DSGQSN<font color=red>K</font>N GA<font color=red>K</font><font color=red>RK</font>VAGGPSVAELF<font color=red>KK</font>N<font color=red>K</font> | |||
CRY64_lepOcu <font color=red>K</font>YLPVL<font color=red>KK</font>FPSAYIYEPW<font color=red>K</font>AP<font color=red>R</font>SVQEQAGCIVG<font color=red>K</font>DYP<font color=red>R</font>PIVDHDVVS<font color=red>KK</font>NIQ<font color=red>R</font>M<font color=red>K</font>LAYA<font color=red>RR</font>AQLGGEQEGTG<font color=red>K</font> GM<font color=red>K</font><font color=red>RK</font>GQSVADLLT<font color=red>KK</font>Q<font color=red>KR</font>N | |||
CRY64_danRer <font color=red>K</font>YLPVL<font color=red>KK</font>FSTEYIYEPW<font color=red>K</font>AP<font color=red>R</font>SVQE<font color=red>R</font>AGCIVG<font color=red>K</font>DYP<font color=red>R</font>PIVDHEVVH<font color=red>KK</font>NIL<font color=red>R</font>M<font color=red>K</font>AAYA<font color=red>KR</font>SPED<font color=red>K</font>TIN<font color=red>K</font> GE<font color=red>K</font><font color=red>RK</font>ASPSI<font color=red>K</font>EMFQ<font color=red>KK</font>A<font color=red>KR</font> | |||
CRY64_salSal <font color=red>K</font>YLPHL<font color=red>KK</font>YPAQYIYEPW<font color=red>K</font>AP<font color=red>R</font>SVQEAAGCIVG<font color=red>K</font>DYP<font color=red>R</font>PIVEHEVIS<font color=red>KK</font>NIQ<font color=red>R</font>M<font color=red>K</font>AAYA<font color=red>KR</font>SPHSSEESP G<font color=red>KK</font>E<font color=red>K</font>G<font color=red>RK</font>H<font color=red>K</font>APSVVDMLM<font color=red>K</font> | |||
CRY64_gadMor <font color=red>K</font>YLPVL<font color=red>KK</font>FPVEYIYEPW<font color=red>K</font>APLSVQ<font color=red>K</font>AAGCIVG<font color=red>K</font>DYPSPIVEHEVIS<font color=red>K</font>QNIQ<font color=red>R</font>M<font color=red>K</font>TSYG<font color=red>KR</font>SQGVSESPQPM<font color=red>K</font>AE<font color=red>K</font><font color=red>RK</font> GPSVLDMM<font color=red>K</font>N<font color=red>KK</font><font color=red>KK</font> | |||
CRY64_takRub <font color=red>K</font>FLPHL<font color=red>KK</font>FPAEYIFEPW<font color=red>K</font>APQSVQQAAGCIVG<font color=red>K</font>DYPHPIVQHEVVS<font color=red>KK</font>NIQ<font color=red>R</font>M<font color=red>K</font>AAYA<font color=red>KR</font>SANTA<font color=red>K</font>SLS<font color=red>K</font>IQ GL<font color=red>K</font><font color=red>RK</font>PSSSVDML<font color=red>KK</font><font color=red>KK</font><font color=red>K</font>NN | |||
CRY64_tetNig <font color=red>K</font>YLPHL<font color=red>KK</font>FPAQYIYEPW<font color=red>K</font>APQSIQ<font color=red>K</font>AAGCIIG<font color=red>K</font>DYPHPIV<font color=red>K</font>HEEVS<font color=red>KK</font>NIQ<font color=red>R</font>M<font color=red>K</font>LAYA<font color=red>RR</font>STSNAASP<font color=red>KK</font>T GV<font color=red>K</font><font color=red>RK</font>GPSVVDLL<font color=red>KK</font><font color=red>K</font><font color=red>RK</font><font color=red>K</font>I | |||
CRY64_gasAcu <font color=red>K</font>YLPLL<font color=red>KK</font>FPAEYIYEPW<font color=red>K</font>AP<font color=red>R</font>SVQQAAGCIVG<font color=red>K</font>DYPQPIA<font color=red>K</font>HEVIS<font color=red>KK</font>NIQ<font color=red>R</font>M<font color=red>K</font>LAYA<font color=red>KR</font>SGDSAESAN<font color=red>K</font>SPV<font color=red>KR</font>Q GT<font color=red>K</font><font color=red>RK</font>APSVVDML<font color=red>KK</font><font color=red>K</font>D<font color=red>RR</font><font color=red>K</font> | |||
CRY64_oryLat <font color=red>K</font>YLPIL<font color=red>KK</font>FPPQYIYEPW<font color=red>K</font>AP<font color=red>R</font>SVQQAAGCIVG<font color=red>K</font>DYP<font color=red>K</font>PIIEHEVIS<font color=red>KK</font>NIQ<font color=red>R</font>M<font color=red>K</font>QAYA<font color=red>RR</font>TSGSTESPT<font color=red>KK</font>Q GV<font color=red>K</font><font color=red>RK</font>APTVVDLIQ<font color=red>KK</font>Q<font color=red>KR</font>S | |||
CRY64_oreNil <font color=red>K</font>YLPLL<font color=red>KK</font>FPAEYIYEPW<font color=red>K</font>AP<font color=red>R</font>SIQQAAGCIVG<font color=red>K</font>DYPHPIVQHEVIS<font color=red>KK</font>NIQ<font color=red>R</font>M<font color=red>K</font>LAYA<font color=red>KR</font>SPDTTESPS<font color=red>K</font>S<font color=red>K</font> GV<font color=red>K</font><font color=red>RK</font>APSIIEMI<font color=red>KK</font><font color=red>K</font>A<font color=red>K</font>V<font color=red>K</font> | |||
CRY64_braFlo HYLPVL<font color=red>K</font>NFP<font color=red>K</font>EYIYEPW<font color=red>K</font>AP<font color=red>R</font>NVQE<font color=red>K</font>AGCIVG<font color=red>K</font>DYP<font color=red>R</font>PIVDH<font color=red>K</font>EASQ<font color=red>R</font>NLDIM<font color=red>R</font>DV<font color=red>RK</font>DQ<font color=red>K</font>ETAAVTL GYG<font color=red>K</font> | |||
CRY64_strPur <font color=red>K</font>YIPALN<font color=red>K</font>LPAEYIYEPWTAP<font color=red>R</font>SVQEAAGCIIG<font color=red>R</font>DYP<font color=red>R</font>PIVDHSIVS<font color=red>KR</font>NIG<font color=red>R</font>M<font color=red>K</font>DA<font color=red>R</font>ACQPG<font color=red>KK</font>A E<font color=red>KR</font>PAEPS<font color=red>K</font>QDNNG<font color=red>KK</font>V<font color=red>RK</font>ITSML<font color=red>KK</font><font color=red>K</font> | |||
CRY64_lytVar <font color=red>K</font>YIPIME<font color=red>R</font>FPAQYIYEPWTAP<font color=red>R</font>SVQEAAGCIIG<font color=red>R</font>DYP<font color=red>R</font>PIVDHSVVS<font color=red>KR</font>NIG<font color=red>R</font>M<font color=red>K</font>DA<font color=red>R</font>ACQPG<font color=red>K</font>SA E<font color=red>KR</font>PTDASN<font color=red>K</font>NSNG<font color=red>K</font>V<font color=red>RK</font>ITSML<font color=red>KK</font><font color=red>K</font> | |||
For the full set of 22 metazoan CRY64 sequences, see the [[Cryptochrome_refSeqs|curated reference sequence section]]. A single representative sequence is shown below. | |||
>CRY64_anoCar Anolis carolinensis (lizard) XM_003225714 6-4 photolyase synteny: DCPS TIRAP CRY64 SRPR FOXRED1 | |||
0 MAHVSIHWFRKGLRLHDNPALLAAMKNSAEIYPIFILDPWFPKNMQVSINRWRFLIESLKDLDESLKKLNSR 2 | 0 MAHVSIHWFRKGLRLHDNPALLAAMKNSAEIYPIFILDPWFPKNMQVSINRWRFLIESLKDLDESLKKLNSR 2 | ||
1 LFVVRGRPAEVFPELFTKWKVTRLAFEVDTEPYARRDAEVVRLAAEHGVQVIQKVSHTLYDTER 2 | 1 LFVVRGRPAEVFPELFTKWKVTRLAFEVDTEPYARRDAEVVRLAAEHGVQVIQKVSHTLYDTER 2 | ||
| Line 1,559: | Line 916: | ||
0 SWVCNFKKPETHPNSLTPSTTVLSPYVKFGCLSVRMFWWKLAEVYQG 0 | 0 SWVCNFKKPETHPNSLTPSTTVLSPYVKFGCLSVRMFWWKLAEVYQG 0 | ||
0 RKHSDPPVSLHGQLLWREFFYTAGAGIPNFDRMENNPVCVQVDWDNNQEYLRAWRE 0 | 0 RKHSDPPVSLHGQLLWREFFYTAGAGIPNFDRMENNPVCVQVDWDNNQEYLRAWRE 0 | ||
0 | 0 GQTGYPFIDAIMTQLRTEGWIHHLARHAVACFLTRGDLWISWEEGQK 0 | ||
0 VFEELLLDADWSLNAANWQWLSASAFFHQFFRVYSPVTFGKKTDKNGEYIK 2 | 0 VFEELLLDADWSLNAANWQWLSASAFFHQFFRVYSPVTFGKKTDKNGEYIK 2 | ||
1 KYLPFLRKFSNDYIYEPWKAPRSLQERAGCIIGQDYPKPIVEHEKVYKRNLERMKAAYARRSPNLVIQAKDKVSQKKGV 1 | 1 KYLPFLRKFSNDYIYEPWKAPRSLQERAGCIIGQDYPKPIVEHEKVYKRNLERMKAAYARRSPNLVIQAKDKVSQKKGV 1 | ||
2 NRKRPEAPTKAKVQAKKVKTKSS* 0 | 2 NRKRPEAPTKAKVQAKKVKTKSS* 0 | ||
=== DASH | === Cryptochrome CRY7 photolyases === | ||
For the full set of 14 bilateran CRY7 sequences, see the [[Cryptochrome_refSeqs|curated reference sequence section]]. | |||
Below the frog protein CRY7 is marked up for its various domains and motifs according to Pfam, Blast and PDB searches. <font color =blue>Blue</font> shows the antenna domain with predicted α/β secondary structure, <font color =purple>purple</font> the possibly catalytic FAD domain with predicted all α secondary structure, <font color =magenta>magenta</font> the UIM ubiquitin motif, <font color =green>green</font> two compositionally simple regions rich is basic residues predicted not to have definite fold, <font color =red>red</font> the conserved region of unknown function upstream of the UIM ubiquitin motif, and <font color =lightblue>light blue</font> the conserved carboxy terminal motif of unknown function. | |||
>CRY7_xenTro Xenopus tropicalis (frog) | |||
0 MDLEPFERAQIDDVLQ <font color =red>QLESGSVQADEFLCLVLSILGSSRTYSQFPAILQSLSRKEPAMYRELMDLHAEYFRK</font> 0 | |||
0 EPADLETL<font color =magenta>GYETDLELAIALSLQEHNQL</font>TDTASFASEVDPAPKISFADAA<font color =green>KLSHFSHKHNKKNSSSK</font>TEITKLKDNVAAMNLYQERKRYHINGQEKTCISN | |||
CYNGQPEPEDCVLKSEDGEDVFHVETSRP<font color =green>RESKAKHSRRSRKKKKSAPSR</font>GLVAMKP<font color =blue>VLVWFRRDLRLHDNPALISALEHGVPVIPVFLWCINEETGQNFTLATGGAT | |||
KYWLHHALLKLNQSLIQRFGSHIIFRVARSCEEELVSLVHETGADTIIINAVYEPWLKERDDLISETLRRHGVELKKHHSYCLYEPDSVST</font>EGVGLR 1 | |||
2 GIGSVSHFMSCCKRNNSAPIGMPLDAPRCLPAPCNWPESDHLDTLELGKMPHRKDGTL 0 | |||
0 IDWAVTIRESWDFS<font color =purple>EDGAYTCLANFLQD 1 | |||
2 GVKHYEKESGRADKPYTSHISPYLHFGQISPRTVLHEAYFTKKNVPKFLRKLAWRDLAYWLLILFPDMPSEPVRPAYK 0 | |||
0 SQRWSSDLNHLRAWQKGLTGYPLVDAAMRELWLTGWMCNYSRHVVASFLVAYLHIHWVHGYRWFQ 0 | |||
0 DTLLDADVAINAMMWQNGGMSGLDHWNFVMHPVDSALTCDPYGSYVRKWCPELAGLPDEYIHKPWKCAPSQLRRA 1 | |||
2 GVILGRNYPHRIVLDLEERREQSLKDVVEVR</font><font color =lightblue>KKHLEYLDEVSGCDMVQIPDQLLALTLGHTSGEDEVVRNRTGSFLLPVITRKEFKYKTLQPDTKDNPYNTVLKGYV | |||
SRKRDETIAYMNERHFTASTINEGAQRHERIERTNRLMEGLPAPSDAKNKSRRTPKKDPFSIIPPSY</font>LHLAN* 0 | |||
=== DASH: spotty phylogenetic distribution and unexplained carboxy terminal extension === | |||
DASH is yet another member of the cryptochrome and photolyase family. It was identified [http://www.ncbi.nlm.nih.gov/pubmed/15147276,19228922,20684534,19074258,20174658,21128641,20828134,19945437,19888752,19477164,19345672 only recently] as active only on ssDNA repair, reportedly because of a barrier to flipping the damaged cyclobutane pyrimidine dimer dinucleotide out of dsDNA into | DASH is yet another member of the cryptochrome and photolyase family. It was identified [http://www.ncbi.nlm.nih.gov/pubmed/15147276,19228922,20684534,19074258,20174658,21128641,20828134,19945437,19888752,19477164,19345672 only recently] as active only on ssDNA repair, reportedly because of a barrier to flipping the damaged cyclobutane pyrimidine dimer dinucleotide out of dsDNA into the active repair site unless the damaged base lies in a loop. In species investigated to date, this enzyme uses folate (MTHF) as antenna and FAD activated by blue light. It is a fairly remote outgroup to cytochromes, with only CPD further diverged. | ||
Its name is a [http://www.sciencedirect.com/science/article/pii/S109727650300008X peculiar] | Its name is a [http://www.sciencedirect.com/science/article/pii/S109727650300008X peculiar acronym] of Drosophila, Arabidopsis, Synechocystis and Homo -- yet the gene was never present in Drosophila or placentals. In Arabidopsis, the principal copy is called CRY3, again in contravention of photolyase naming conventions. The numerous genome projects available today allow a quick determination of its rather unusual phylogenetic distribution. | ||
[[IMAGE:CyrDASH.jpg|left]] | [[IMAGE:CyrDASH.jpg|left]] | ||
Although originally studied in plants and cyanobacteria, the DASH photolyase surprisingly extends into fish, frogs, salamanders, turtle, lizard, and birds | Although originally studied in plants and cyanobacteria, the DASH photolyase surprisingly extends into fish, frogs, salamanders, turtle, lizard, and birds -- duck, finch and budgerigar (chicken and turkey have pseudogenes) -- but not any mammal. It is not known if the DNA repair function has been retained in all these taxa or has drifted in new roles like CRY1 and CRY2 in land vertebrates. | ||
Blastx on the syntenic region in gallinaceous birds (chicken and turkey) establishes rather degenerate multi-exonic pseudogenes at the expected location and strand orientation. Here duck, which has an intact gene, is the [http://ldl.genomics.cn/page/pa-bird.jsp immediate outgroup], diverging at 80 myr. It is not currently possible to date this more precisely nor determine whether pseudogenization occurred in a common ancestor or independently, perhaps on account of separate domestications. Platypus lacks pseudogene debris at the expected location but the assembly is currently unsatisfactory here. Marsupials and placentals would never have had this enzyme assuming lost shortly after divergence with the last common ancestor with birds. | |||
DASH is also | The phylogenetic loss pattern of DASH in mammals is reminiscent the [[Opsin_evolution:_trichromatic_ancestral_mammal|massive loss of opsins]] that also occurred early in mammalian evolution -- which GT Walls in 1942 [http://www.crsltd.com/research-topics/walls/index.html attributed] to mammals experiencing a sustained period of deep nocturnality where these systems did not need to function (no UV damage) and indeed could not function (insufficient blue light even with antenna) and so were lost, implying they were not sustained by a [http://en.wikipedia.org/wiki/Protein_moonlighting Piatigorskyian secondary functionality] such as circadian rhythm, lunar calendaring, or magnetosensing. | ||
DASH is also missing from alligator and crocodile assemblies, deep-water lobe-finned fish (coelocanth) have a pseudogene, and cartilaginous fish to date lack it completely. These probably reflect multiple independent gene losses rather than inadequate assemblies. DASH is restricted within invertebrates to crustaceans and mollusks, a pattern which could have arisen from stem losses in insects etc. The sole insect DASH at GenBank (whitefly EZ942653) appears to be a fungal contaminant. | |||
The | The [http://en.wikipedia.org/wiki/Great_Oxygenation_Event great oxygenation event] gave rise to a stratospheric ozone protective layer at 2.4 gyr but reached an even higher lever during the early Cambrian (based indirectly on oxygen levels). If more ozone meant less DNA damage from UV, this may have favored independent but simultaneous gene loss events in various clades. However the persistence of DASH in ray-finned fish for 450 myrs raises the question of whether UV light penetration of sea water is the sole or even principle cause of DASH-repairable DNA damage -- if indeed DASH is still a repair enzyme in benthic species. However the first land plants and animals were plausibly exposed to greatly increased levels of UV damage that may correlate with DASH retention. | ||
[[Image:DashDistal.png|left]] | |||
Multi-cellular animals from cnidarian to amniote all have a short C-terminal extensional exon whose distal region contains a conserved motif of unknown function. This is positioned to cap the binding site in the manner of CRY64_droMel but there is no evidence that it does -- while positively charged arginines and lysines that might offset negative DNA phosphates are among the conserved residues, so are negatively charged glutamates, polar, neutral and aromatic residues. If this domain does prove to be a structural cap, it represents convergent evolution with respect to the CRY64 cap domain because the two orthology classes diverged long before the caps evolved. | |||
Overall sequence conservation of DASH is less stringent than other photolyases and cytochromes, suggesting loosened constraints or a measure of functional redundancy with respect to other repair enzymes. However it is difficult to understand how antenna domain -- though less conserved than the FAD domain -- could be conserved over vast spans of branch length in the absence of function (antenna molecule binding and/or something else). | |||
Amniote DASH proteins can be modeled structurally using nearly 50% matches in Arabidopsis (cryptochrome 3: 2IJG) or equally suitable cyanobacterium Synechocystis (1NP7). However these structures do not provide any information on the C-terminal extension. The 14 exons share only one match with vertebrate CRY1 and CRY2 -- and that is more likely coincidental than indicative of a shared ancestral protein subsequent to the main era of eukaryotic intronation. | |||
For the full set of 30 metazoan DASH sequences and conservation alignments, see: [[Cryptochrome_refSeqs|Curated reference sequences for cryptochromes and photolyases]] | |||
<br clear=all> | <br clear=all> | ||
>DASH_taeGut Taeniopygia guttata (finch) | >DASH_taeGut Taeniopygia guttata (finch) <font color=purple>antenna</font> catalytic</font> <font color=red>C-terminal motif</font> | ||
0 | 0 MSGTAGT<font color=purple>AICLLRCDLRAHDNQ 0 | ||
0 QVLHWAQHNADFVIPLYCFDPRHYLGTHCYRLPKTGPHRLRFLLESVKDLRETLKKKGS 2 | 0 QVLHWAQHNADFVIPLYCFDPRHYLGTHCYRLPKTGPHRLRFLLESVKDLRETLKKKGS 2 | ||
1 TLVVRKGKPEDVVCDLITQLGSVTAVVFHEE 0 | 1 TLVVRKGKPEDVVCDLITQLGSVTAVVFHEE 0 | ||
0 ATQEELDVEKGLCQVCRQHGVKIQTFWGSTLYHRDDLPFRPIDR 2 | 0 ATQEELDVEKGLCQVCRQHGVKIQTFWGSTLYHRDDLPFRPIDR 2 | ||
1 | 1 LPDVYTHF</font>PKGLESGAKVRPTLRMADQLKPLAPGLEEGSIPTMEDFGQK 1 | ||
2 | 2 DPVADPRTAFPCS<font color=blue>GGETQALMRLQYYFWDT 0 | ||
0 NLVASYKETRNGLVGMDYSTKFAPW 2 | 0 NLVASYKETRNGLVGMDYSTKFAPW 2 | ||
1 LALGCISPRYIYEQIQKYERERTANESTYW 2 | 1 LALGCISPRYIYEQIQKYERERTANESTYW 2 | ||
| Line 1,700: | Line 982: | ||
0 EGKTGVPFVDANMRELSATGFMSNRGRQNVASFLTKDLGLDWRMGAEWFEYLL 0 | 0 EGKTGVPFVDANMRELSATGFMSNRGRQNVASFLTKDLGLDWRMGAEWFEYLL 0 | ||
0 VDYDVCSNYGNWLYSAGIGNDPRDNRKFNMIKQGLDYDGN 0 | 0 VDYDVCSNYGNWLYSAGIGNDPRDNRKFNMIKQGLDYDGN 0 | ||
0 | 0 GDYVRLWVPELQGIKGADIHTPWALSSAALSQAGVTLGETYPQPVV</font>TAPEWSRHIHRRP 0 | ||
0 GGSPHP<font color=red>RGRRGPAQRKDRGIDFYFSRKKD</font>AC* 0 | |||
0 | |||
=== Cryptochrome CPD photolyases === | === Cryptochrome CPD photolyases === | ||
This dna repair enzyme was [http://www.ncbi.nlm.nih.gov/pubmed/7937136,7813451 studied in marsupials] during the pre-genomic era (1994), with two groups concluding even that that no ortholog existed in placentals. | This dna repair enzyme (cyclobutane pyrimidine dimers for CPD) was [http://www.ncbi.nlm.nih.gov/pubmed/7937136,7813451 studied in marsupials] during the pre-genomic era (1994), with two groups concluding even that that no ortholog existed in placentals. Today we are certain of that because the gene is not present in any complete placental mammal genome; no pseudogene debris exists in the partly conserved syntenic location in any species. This strongly suggests that the gene was lost once in stem placental rather than many times in later subclades (as happened with encephalopsin). The gene remains very strongly conserved in species such as opossum with no indication of impending loss. | ||
The loss in placentals is somewhat peculiar given that CPD is a very ancient (pre-eukaryotal) member of the photolyase family, with highly conserved orthologs readily recoverable in other commonly studied marsupials, monotremes, birds, alligators, turtle, lizard, snakes, frog, fish, agnathan, amphioxus, sea urchin, many invertebrates, cnidarians, plants and so forth. However it also appears to be lost in tunicate -- indeed Ciona has lost all its photolyases leaving it a bit mysterious how it repairs these types of dna damage. Hemichordates have also lost all members of this gene family including CPD. | |||
It is very unlikely that placentals displaced CPD with something better. More likely, CPD was lost during a dark phase of placental evolution when UV damage to dna was a non-issue and its photo-repair infeasible. Genes cannot be retained without selection (use it or lose it). Coming back out into the light millions of years later (having also lost DASH, CRY64 and [[Opsin_evolution:_update_blog|13 of 21 opsin genes]), they evidently made do with a less efficient excision repair that overlaps repair photolyase functionality. | |||
The CPD gene product is very diverged from other photolyases though still retains the photolyase and FAD binding domain folds. The antenna moiety is usually reported as MTHF (folate). The best available structures are from rice (3UMV: 53% identity to marsupial) and an archaeal methanogen (Methanosarcina mazei 2XRZ) which likely uses 5-deazariboflavin Fo as antenna (which it can synthesize de novo). The latter enzyme [http://www.ncbi.nlm.nih.gov/pubmed/21892138 repairs cyclobutane pyrimidine dimers] in duplex DNA using blue or near-UV light. | |||
Despite great divergence in primary sequence from other members of the gene family, fold conservation may explain in part the unexpected [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3156801/?tool=pubmed circadian compensatory capacity] of marsupial CPD expressed in double CRY1/2 knockout mouse, seemingly driven by interaction of CPD with CLOCK of the CLOCK/BMAL1 system. CPD lacks any counterpart to the distal exons of placental CRY1. | |||
It | CPD presents no special problems in classification as it clearly originated early in the history of prokaryotes and today serves as the outgroup to the overall metazoan photolyase gene family (though not as usefully as the less diverged DASH). It has never undergone gene duplication and divergence, at least none that stuck, and has been retained as single copy in the vast majority of species from choanflagellate to mammal. There are no noteworthy C-terminal expansions or supplemental exons within metazoan -- CPD is the exception among photolyases and cryptochromes for its lack of overt innovation. However as the knock-in experiment in mouse shows, CPD has unexpected properties. | ||
The | The N-terminus has various extensions -- indeed the initial methionine is problematic -- but these are poorly conserved even within closely related taxa. Conservation sets in some 38 residues upstream of the first conserved methionine. While these 114 bp could represent conserved 5'UTR nucleotides rather than conserved amino acids, the two relevent crystallographic structures include this region (Methanosarcina 2XRY and rice 3UMV) as do many transcripts. Two in Xenopus (ES684787 BX851972) seem to rule out a cryptic short first exon splicing into the conserved region. | ||
[[Image:CPDragged.png]] | |||
Some 32 curated CPD sequences spanning the whole of metazoan evolution are provided at the [[Cryptochrome_refSeqs|reference sequences]]. Many more could be extracted from GenBank should some research issue warrant more intensive surveying. | |||
=== 4Fe-4S photolyases and their relation to primases === | |||
An intriguing new subfamily of photolyases ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3204975/ 1],[http://www.ncbi.nlm.nih.gov/pubmed/22290493 2]) contains a 4Fe-4S cluster in the catalytic domain in addition to an FAD binding site. This makes sense given the [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0010083 equally surprising finding] of unmistakable fold homology between photolyases and the large subunit of archaeal-eukaryotic primase (eg the PRIM2 gene product of human). | |||
This [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2846230/ ancient enzyme] is critical to de novo synthesis of the short RNA primers essential to DNA replication. Primase also contains a 4Fe-4S cluster as do numerous non-homologous DNA repair enzymes such as [http://nar.oxfordjournals.org/content/early/2012/01/28/nar.gks039.full helicases and endonucleases]. Such clusters have a redox role elsewhere in the cell but it is not immediately evident that's applicable here. | |||
The photolyase antenna molecule is Rhodobacter is new but not entirely novel: the final intermediate in riboflavin biosynthesis, 6,7-dimethyl-8-ribityl-lumazine (which serves a similar role in bioluminescence). This illustrates again the plasticity of the antenna site -- the antenna molecule is unpredictable from primary sequence (indeed tertiary structure). | |||
Since the list of possible antenna molecules is still growing, reconstitution experiments that don't find a suitable antenna molecule may simply have tested an insufficient range of molecules -- they have to be repeated as new ones emerge. Similarly, in silico docking can only fit what is on the list. Here we cannot be sure that other members of this new subfamily of photolyases will use this (or indeed any) antenna molecule. | |||
The new class of photolyase conflicts with the notion of a universal tryptophan triad chain in photolyases, agreeing instead with [http://www.ncbi.nlm.nih.gov/pubmed/22139370 reports] in other photolyases suggesting that the whole concept -- or at least invariance part -- was limited in applicability. | |||
Most gene families members in this class of proteins have more than the three ultra-conserved tryptophans. Simply knocking in a tyrosine at a site that has never tolerated a substitution for a hundred billion years of branch length evolution does not for test electron flow specifically any substitution at any invariant residue necessarily has major adverse effects: how else could it have been conserved for such a huge multiple of the neutral substitution rate? | |||
Three inappropriate gene names for this new photolyase class -- PhrB is already in use at GenBank for a different photolyase class, CRYB suggests non-repair cryptochrome, FeS-BCP has an erroneous phylogenetic distribution and disallowed hyphen -- won't be used here but rather a provisional name PFES (photolyase iron sulfide). Reference sequences are provided below for two bacteria and two archaeal FeS photolyases, as well as yeast and human FeS primases; these suffice as GenBank blast probes. | |||
Some confusion surrounds the human primase sequence because the NCBI reference genome (Build 37.1) carries only a [http://www.ncbi.nlm.nih.gov/pubmed/22437878 pseudogene] -- a copy number variant bordering the centromere of chromosome 6, with the actual gene is still missing from the June 2012 reference genome, causing transcripts to mis-align with genome at 11 of 509 amino acids. Bizarrely, these discrepancies -- including an internal stop codon in exon 11 -- were [http://www.ncbi.nlm.nih.gov/nuccore/40675621 noted by NCBI] in accession BC064931 but never resolved because the chimpanzee assembly was also wrong in the same way. It is inconceivable that project DNA donors lacked a working copy of this very essential gene. | |||
Note human primase has two components. The smaller catalytic unit is encoded by PRIM1 on chr 12. This gene has a very distant homolog CCDC111 (unofficially renamed [http://www.ncbi.nlm.nih.gov/pubmed/24207056,24267451,24240614,24682820 PrimPol]) on chr 4 containing an extra C-terminal zinc finger domain. Neither of these contains a 4Fe-4S cluster. The larger auxillary subunit is encoded by PRIM2 on chr 6. It contains the 4Fe-4S cluster discussed here. PRIM2 has no additional homologs in human (or other multi-cellular organisms). | |||
Using blastp and the 4 conserved cysteines as guide to presence of the iron sulfur cluster , bacterial representatives of the new photolyase class are readily located in 150 genera, largely alphaproteobacter) but are more narrowly distributed in Archaea (8 of 49 genera of Euryarchaeota but no Thaumarchaeota, Aigarchaeota, Korarchaeota, Crenarchaeota in 33 genomes tested) suggesting horizontal gene transfer to (or from) Euryarchaeota or stem gene loss in the TACT group. | |||
Since the eukaryotes acquired mitochondria from a [http://rspb.royalsocietypublishing.org/content/278/1708/1009.short relatively late endosymbiosis] with an alphaproteobacter, a gene copy might initially have been present. | |||
No eukaryotic photolyase to date (18 April 2014) has retained or independently re-developed a 4Fe-4S domain (ignoring blast matches such as XM_002537565 in castor bean that represents Agrobacterium contamination). The Agrobacterium photolyase itself is [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3645588/ surprisingly] a 6-4 photolyase with a well-established Fe-S cluster and 6,7-dimethyl-8-ribityllumazine antenna molecule. As noted by Zhang et al, its structural homology to primases places the 6-4 photolyases at the root of the whole cryptochrome/photolyase family. | |||
Curiously, the four cysteines coordinating iron are not deeply conserved. For example, yeast primase cysteines 336/417/434/474 cannot be fully homologous to Agrobacterium cysteines where the linear order is different 350/454/438/441. During evolution, other cysteines must have arisen and as the fold diverged, replaced some of the originals. (A similar phenomenon occurs with disulfides, for example in sulfatases.) However the four cysteines and their exact spacing (up to a one residue indel) are conserved between yeast and human primases, representing conservation over a billion years since their divergence. | |||
The 4Fe-4S cluster of primase is surely an ancient feature of primase and thus of the whole fold family descended from it, suggesting that FeS-photolyases are a relic of an old gene duplication, retaining a feature lost in subsequent duplications giving rise first to CPD and then to the overall photolyase/cryptochrome gene family. | |||
The alternative scenario, that the 4Fe-4S cluster represents convergent evolution in photolyases (later independent acquisition) at first seems implausible given the requirements of cubane geometry, the complexity of the auxiliary enzymes and scaffolding proteins [http://www.ncbi.nlm.nih.gov/pubmed/22609301 involved in 4Fe-4S assembly], and the lack of functioning intermediate states. However the eukaryotic proteome overall contains a large and heterogeneous set of iron-sulfur proteins; there is no support for a 4Fe-4S cluster as a mobile duplicated domain. | |||
It is not clear how many distinct homology classes exist for 4Fe-4S domains even restricting to DNA proteins -- primary sequence is not immediately helpful given deep divergences of these ancient proteins, cysteines anchors of an alignment might only represent convergent evolution, as could short fold similarities recognized by Dali given the geometrical constraints. If one supposes a late-stage cluster assembly protein [http://www.sciencemag.org/content/early/2012/06/06/science.1219664 such as MMS19] provides 4Fe-4S cluster to structurally dissimilar fold classes localized to the nucleus, then what is the common ground biochemically for recognition of apoprotein? | |||
It has not proved feasibly to date to develop a bioinformatic screen that catches the full repertoire of 4Fe-4S clusters in DNA proteins in the yeast/human proteomes because the conserved cysteine pattern can be confused with bona fide zinc binding sites (eg zinc ribbons) that themselves lack distinctive signatures. Proof of that can be seen from the large number of 4Fe-4S clusters only recognized in 2011-2013 -- in enzymes studied intensively for decades. | |||
A 4Fe-4S cluster has a clear enough spectroscopic signature, the problem arises from the lability of clusters when the protein is purified in the presence of oxygen. When the cluster is lost, its binding domain loses its rigidity, becoming structurally indeterminable in crystallographic studies. Alternatively, a zinc ion occupies the site, causing the structural determination to proceed to an erroneous conclusion. Zinc in 4Fe-4S cluster proteins could represent artifact, placeholder, protection, idle cycle, or even functionally viable alternative. | |||
While zinc ions are ubiquitous throughout the cell as more or less harmless (in contrast to iron) atoms that spontaneously find to their target sites by diffusion (like magnesium ions), 4Fe-4S clusters are not free-floating constituents of the mitochondria, cytoplasm or nucleoplasm. Instead, they are built and held on scaffolding proteins, then passed along a complex chain of chaperones and assembly proteins for insertion into apoprotein, with no aspect of the process left to chance chemistry. | |||
Although in most of biochemistry, 4Fe-4S clusters serve a clear redox function, such a role has not been established for primases, helicases, other DNA repair enzymes, much less PFES photolyases. Conceivably the redox state of the 4Fe-4S cluster can sense the status of a DNA helix and facilitate rapid scanning for the odd damaged base among billions of normal ones. The photolyases present an interesting situation because only one of many orthology classes utilizes an iron sulfur cluster, whereas it would make sense given the newly recognized ubiquity for all of them to have it. Thus the novelty is turned around -- how can other photolyases work without an iron sulfur cluster? | |||
Primase may be among the very oldest of enzymes since it is essential for DNA replication (ie, perhaps for exiting the hypothetical earlier RNA world). However UV damage is also a very old issue, especially for the billion years of life preceding oxygenation of the atmosphere (which led to the ozone shield of today). Priming is not needed for RNA replication or transcription nor in DNA replication in mitochondria; bacteria use a non-homologous system based on the DNAG protein. | |||
One intriguing idea starts with the observation that FAD mimics two free RNA bases with its flavin and adenine rings which are are stacked like bases (U-folded) in all studied photolyases. In primase -- which has no FAD -- two purine ribonucleotides at the FAD site may recognize two bases of template DNA by conventional hydrogen bonding that perhaps resemble the flipped out cyclobutane pair needing repair by a photolyase. | |||
Indeed, the template dinucleotide could even be stabilized temporarily as a cyclobutane pair, reversing the normal sense of the reaction, borrowing reductive units from the 4Fe-4S cluster (UV/blue light is not a known primase requirement). This would explain primase preference for a pyrimidine template. Photolyases then arose by replacing the two mononucleotides with FAD and adding a Rossmann-like domain for the antenna, with the utilization of light displacing the need for the 4Fe-4S cluster except in the PFES class of photolyases. | |||
Human primase also undergoes a profound conformational change from a three-helix binding site for DNA to a helix-sheet site as it counts primer size and passes it along to the catalytic subunit and other protein partners. That's not so clear for not-so-large subunits archaeal primases which seem to lack an internal domain duplication. A large conformational change -- not just internal changes in FAD redox status -- is also needed in cryptochrome signaling, possibly this same one. | |||
>PFES_agrTum Agrobacterium tumefaciens (bacteria) NP_355900 aka: PhrB pdb:4DJA | |||
MSQLVLILGDQLSPSIAALDGVDKKQDTIVLCEVMAEASYVGHHKKKIAFIFSAMRHFAEELRGEGYRVRYTRIDDADNAGSFTGEVKRAIDDLTPSRIC | |||
VTEPGEWRVRSEMDGFAGAFGIQVDIRSDRRFLSSHGEFRNWAAGRKSLTMEYFYREMRRKTGLLMNGEQPVGGRWNFDAENRQPARPDLLRPKHPVFAP | |||
DKITKEVIDTVERLFPDNFGKLENFGFAVTRTDAERALSAFIDDFLCNFGATQDAMLQDDPNLNHSLLSFYINCGLLDALDVCKAAERAYHEGGAPLNAV | |||
EGFIRQIIGWREYMRGIYWLAGPDYVDSNFFENDRSLPVFYWTGKTHMN<font color=magenta>C</font>MAKVITETIENAYAHHIQRLMITGNFALLAGIDPKAVHRWYLEVYADAYE | |||
WVELPNVIGMSQFADGGFLGTKPYAASGNYINRMSDY<font color=magenta>C</font>DT<font color=magenta>C</font>RYDPKERLGDNA<font color=magenta>C</font>PFNALYWDFLARNREKLKSNHRLAQPYATWARMSEDVRHDLRAKAAAFLRKLD* | |||
> | >PFES_rhoSph Rhodobacter sphaeroides (bacteria) CP000144 Alphaproteobacteria <font color=blue>PDB|3ZXS</font> PMID:22290493 6,7-dimethyl-8-ribityl-lumazine antenna aka CryPro 4Fe-4S photolyase | ||
<font color=blue>MRGSHHHHHHGIRMLTRLILVLGDQLSDDLPALRAADPAADLVVMAEVMEEGTYVPHHPQKIALILAAMRKFARRLQERGFRVAYSRLDDPDTGPSIGAE | |||
LLRRAAETGAREAVATRPGDWRLIEALEAMPLPVRFLPDDRFLCPADEFARWTEGRKQLRMEWFYREMRRRTGLLMEGDEPAGGKWNFDTENRKPAAPDL | |||
LRPRPLRFEPDAEVRAVLDLVEARFPRHFGRLRPFHWATDRAEALRALDHFIRESLPRFGDEQDAMLADDPFLSHALLSSSMNLGLLGPMEVCRRAETEW | |||
REGRAPLNAVEGFIRQILGWREYVRGIWTLSGPDYIRSNGLGHSAALPPLYWGKPTRMA<font color=magenta>C</font>LSAAVAQTRDLAYAHHIQRLMVTGNFALLAGVDPAEVHEW | |||
YLSVYIDALEWVEAPNTIGMSQFADHGLLGSKPYVSSGAYIDRMSDY<font color=magenta>C</font>RG<font color=magenta>C</font>AYAVKDRTGPRA<font color=magenta>C</font>PFNLLYWHFLNRHRARFERNPRMVQMYRTWDRMEET | |||
HRARVLTEAEAFLGRLHAGEPV*</font> | |||
> | >PFES_metMah Methanohalophilus mahii (Euryarchaeota) CP001994 4Fe-4S photolyase | ||
MRHYAEKLRNRGADITYIKTAELEKSLSRWIKKKGIDELNIAEPANITLKEYLGKLNIDCKIVFVDNKQFIWSIPEFNTWASSRKNLIMEDFYRTGRKNSEI | |||
LLEKDGKPSGGKWNLDRENRKLPPKNGFQKKPPQHIKFSPDKITKEIIAEVERSEYPTYGKGKDFNLAVTHEDAQKALDFFIEEKLSNFGPYQDIMLTGDNVLWHSILSPYLNLGL | |||
LHPLNVIKKAELAYYQKNLPLNSIEGFIRQILGWREYMHCIYKYTGDKYLKSNWFDHERELPDIYWYPERTSMN<font color=magenta>C</font>MASVIEEVLNTGYAHHIQRLMILSNFALLAEVNPAKVKNWF | |||
HAAFIDAYDWVMQPNVIGMGQFADGGILATKPYISSANYINKMSDY<font color=magenta>C</font>QN<font color=magenta>C</font>TYNHNHRTGEDA<font color=magenta>C</font>PFNYLYWAFLHKNNEKLRDIGRMKLILKNLDRINKKELKQIMTHADDFLKSLK* | |||
> | >PFES_natPha Natronomonas pharaonis (Euryarchaeota) CR936257 4Fe-4S photolyase | ||
MTVLVLGDCLTEFGPLASDARSTDERVLCIEARAFARRKPYHPHKLTLVFSAMRHFRDRLREAGYTVDYRRVETFAEGLDAHFAAHPEDHIVTVRRTAHGAT | |||
DRLQRLVANRGGTVEFVADPRFHCSREEFDAWADGDPPYRHESFYRHMRRETGYLMDGDEPVGGEWNFDDENREFPGPEYVPPEPPQFEPDETTREVREWVDATFGEDGYDDAPYG | |||
GAWADPEPFSWPVTREGALQALEAFIEERLPTFGPYQDAMLGDEWAMNHALLSSSLNLGLLSPSEVIEAALAAFEEGSVSIASVEGFLRQVLGWREFVRHAYRRTPGMAAANQLGA | |||
AEPLPEFFWTGDTDMA<font color=magenta>C</font>VADAVDGVRTRGYAHHIERLMVLSNFATLYGVEPSRLNEWFHAAFVDAYHWVTTPNVVGMGTFGTDTLSTKPYVASANYIDRMSDH<font color=magenta>C</font>SG<font color=magenta>C</font>PYYKTKTTG | |||
DGA<font color=magenta>C</font>PFNALYWDFLGRNESQLRSNHRMGLVYSHYDDKSDGEREAIADRAETLRQRARNGTL* | |||
> | >PRIM2_homSap Homo sapiens (human) NM_000947 primase large subunit 4Fe-4S <font color=blue>pdb|3L9Q,3Q36</font> | ||
0 | 0 MEFSGRKWRKLRLAGDQRNASYPHCLQFYLQPPSENISLIEFENLAIDRVK 1 | ||
2 | 2 LLKSVENLGVSYVKGTEQYQSKLESELR 0 | ||
0 | 0 KLKFSYRENLEDEYEPRRRDHISHFILRLAYCQS 2 | ||
0 | 1 EELRRWFIQQEMDLLRFRFSILPKDKIQDFLKDSQLQFEA 0 | ||
0 | 0 ISDEEKTLREQEIVASSPSLSGLKLGFESIYK 0 | ||
2 | 0 IPFADALDLFRGRKVYLEDGFAYVPLKDIVAIILNEFRAKLSKALA 0 | ||
0 LTARSLPAVQSDERLQPLLNHLS 2 | |||
0 | 1 HSYTGQDYSTQGNVGKI<font color=blue>SLDQIDL 0 | ||
2 | 0 LSTKSFPP<font color=red>C</font>MRQLHKALRENHHLRHGGRMQYGLFLKGIGLTLEQALQFWKQEFIKGKMDPDK 0 | ||
0 FDKGYSYNIRHSFGKEGKRTDYTPFS<font color=red>C</font>LKIILSNPPSQGDYH 1 | |||
2 G<font color=red>C</font>PFRHSDPELLKQKLQSYKISPGGISQ 0 | |||
0 ILDLVKGTHYQVA<font color=red>C</font>QKYFEMIHN 0 | |||
0 VDDCGFSLNHPNQFFCESQRILNGGKDIKKE</font>PIQPETPQPKPSVQKTKDASSALASLNSSLEMDMEGLEDYFSEDS* | |||
> | >PRIM2_sacCer Saccharomyces cerevisiae (yeast) P20457 aka: PRI2_YEAST primase large subunit <font color=blue>PDB|3LGB</font> | ||
MFRQSKRRIASRKNFSSYDDIVKSELDVGNTNAANQIILSSSSSEEEKKLYARLYESKLSFYDLPPQGEITLEQFEIWAIDRLKILLEIESCLSRNKSIK | |||
EIETIIKPQFQKLLPFNTESLEDRKKDYYSHFILRLCFCRSKELREKFVRAETFLFKIRFNMLTSTDQTKFVQSLDLPLLQFISNEEKAELSHQLYQTVS | |||
ASLQFQLNLNEEHQRKQYFQQEKFIKLPFENVIELVGNRLVFLKDGYAYLPQFQQLNLLSNEFASKLNQELIKTYQYLPRLNEDDRLLPILNHLSSGYTI | |||
ADFNQQKANQFSENVD<font color=blue>DEINAQSVWSEEISSNYPL<font color=red>C</font>IKNLMEGLKKNHHLRYYGRQQLSLFLKGIGLSADEALKFWSEAFTRNGNMTMEKFNKEYRYSFR | |||
HNYGLEGNRINYKPWD<font color=red>C</font>HTILSKPRPGRGDYHG<font color=red>C</font>PFRDWSHERLSAELRSMKLTQAQIISVLDSCQKGEYTIA<font color=red>C</font>TKVFEMTHNSASADLEIGEQTHIAHP | |||
NLYFERSRQLQK</font>KQQKLEKEKLFNNGNH* | |||
=== 278 curated refSeqs for metazoan cryptochromes and photolyases === | |||
The full length sequences have been moved to a [[Cryptochrome_refSeqs|separate page]]; only headers are shown below. The sequences use augmented fasta format transparent to web tools: primary sequence broken into exons, codon phase (bp overhang) shown, marked up for features with color, grouped into orthologous clusters, and presented in phylogenetic order relative to human evolutionary history, with subtree order determined by assembly quality. | |||
The fasta headers themselves are little databases showing gene name (following HUGO symbol rules), genus, species, common name, genomic and transcript accession number when not a routine NCBI blast match, PubMed id if specifically studied in a journal article, followed by an unstructured comment field. Both headers and sequences fall readily into desktop databases, allowing different sort orders for other investigative priorities. | |||
The availability of some orthology classes is inherently limited due to recent origin, restricted phylogenetic retention and the uneven focus of sequencing effort across the phylogenetic tree of metazoans. Genomic sequencing of 10,000 vertebrates will not greatly benefit cryptochrome research because the vast majority will be mammals, birds and perch-like fish which are excessively represented already. What is needed are more and better assemblies for a handful of keystone species such as lamprey, hagfish, sharks and rays, bichir, lungfish, and especially amphibians and herptiles. | |||
For species with good assemblies, the entire repertoire of cryptochromes and photolyases has been deduced. It is foolish to compare a gene in isolation across two species with different overall gene family complements because multiple roles and functional complementation may have evolved. | |||
For a large gene with numerous exons, absence from the assembly usually means genuine absence from the genome. Even when only an exon or two gene fragment is available, the classifier can almost always assign the correct orthology class to it. However it is risky to assemble an entire gene from many unlinked single-exon contigs and that was not done here; however certain important clades such as cartilaginous fish lack coherent assemblies and adequate transcripts so provisional gene assemblies are provided. | |||
A remarkable amount of the data has surfaced at GenBank only in the last six months, implying much weaker results had the project been done in 2011 but also that much better phylogenetic coverage will surface this year. For the full set of fasta sequences available in April 2012, see the [[Cryptochrome_refSeqs|reference sequence repository]]. Manually curated sequences -- which use all available data and internal orthology class consistency checks -- should not be equated with provisional unsupervised computerized efforts at GenBank (XM_ gnomon entries), the UCSC 46-way or Ensembl. | |||
CRY1_homSap Homo sapiens (human) | |||
CRY1_panTro Pan troglodytes (chimpanzee) XM_509339 | |||
CRY1_ponAbe Pongo abelii (orangutan) XM_002823690 | |||
CRY1_nomLeu Nomascus leucogenys (gibbon) XM_003269977 | |||
CRY1_macMul Macaca mulatta (rhesus) NM_001194159 | |||
CRY1_calJac Callithrix jacchus (marmoset) XM_002752946 | |||
CRY1_saiBol Saimiri boliviensis (squirrel_monkey) nearly identical to marmoset | |||
CRY1_tarSyr Tarsius syrichta (tarsier) ABRT010205577 unsure if exon 2 is CRY1 or CRY2 | |||
CRY1_micMur Microcebus murinus (mouse_lemur) | |||
CRY1_otoGar Otolemur garnettii (bushbaby) AAQR03016495 | |||
CRY1_tupBel Tupaia belangeri (treeshrew) | |||
CRY1_musMus Mus musculus (mouse) NM_007771 all transcripts support longer exon 10 lost splice donor | |||
CRY1_ratNor Rattus norvegicus (rat) NM_198750 | |||
CRY1_criGri Cricetulus griseus (hamster) XM_003505292 | |||
CRY1_spaJud Spalax judaei (blind_mole_rat) AJ606298 | |||
CRY1_dipOrd Dipodomys ordii (kangaroo_rat) ABRO01202522 ABRO01202521 | |||
CRY1_hetGla Heterocephalus glaber (mole-rat) stop codon in place of conserved W8, last two exons very diverged | |||
CRY1_cavPor Cavia porcellus (guinea pig) last two exons diverged 69 bp separation | |||
CRY1_speTri Spermophilus tridecemlineatus (squirrel) Ictidomys | |||
CRY1_oryCun Oryctolagus cuniculus (rabbit) | |||
CRY1_oviAri Ovis aries (sheep) NM_001129735 19341811 19150926 | |||
CRY1_bosTau Bos taurus (cow) NM_001105415 XM_616063 | |||
CRY1_susScr Sus scrofa (pig) XM_003126079 | |||
CRY1_ailMel Ailuropoda melanoleuca (panda) XM_002927658 | |||
CRY1_loxAfr Loxodonta africana (elephant) XM_003405313 | |||
CRY1_triMan Trichechus manatus (manatee) AHIN01036366 AHIN01036362 very similar to elephant | |||
CRY1_monDom Monodelphis domestica (opossum) XM_003341966 | |||
CRY1_macEug Macropus eugenii (wallaby) assembly frameshift | |||
CRY1_sarHar Sarcophilus harrisii (tasmanian_devil) nearly identical to oppossum | |||
CRY1_triVul Trichosurus vulpecula (possum) EC362500 terminal transcript | |||
CRY1_ornAna Ornithorhynchus anatinus (platypus) XM_001508563 = rubbish, genomic frameshift, continuing exon 12 | |||
CRY1_tacAcu Tachyglossus aculeatus (echidna) SRR000649.130490 short read transcripts corrected for frameshifts, penultimate exon | |||
CRY1_galGal Gallus gallus (chicken) PMID: 11684328,17324421,15459395 altSplExon11: GIVGVPICRGSADLCN* BU143111 | |||
CRY1_melGal Meleagris gallopavo (turkey) XM_003202363 altSplExon11: GTVGVPICRGSANWYK* | |||
CRY1_anaPla Anas platyrhynchos (duck) scaffold157 altSplExon11: GMTGVLVCRGSPGSHNYGKKDKT* | |||
CRY1_eriRub Erithacus rubecula (robin) AY585716 aka: CRY1A altSplExon11: GIMAVPVCRGSPNACNYGKPDKTSK* CRY1B | |||
CRY1_sylBor Sylvia borin (warbler) AJ632120 aka: CRY1A PMID:15381765 altSplExon11: GIVAVAVCRGSPNPCNYGKPDKTSE* sylBor DQ838738 CRY1B | |||
CRY1_taeGut Taeniopygia guttata (finch) XM_002196518 altSplExon11: GIMAVPVCRGSPNPCNYRKPDKTSK* | |||
CRY1_melUnd Melopsittacus undulatus (parakeet) AGAI01062111 altSplExon11: GIMAVPVCRGSSNPCNCGKTDKTSK* | |||
CRY1_parWeb Paradoxornis webbianus (parrotbill) JR867166 TSA transcript | |||
CRY1_allMis Alligator mississippiensis (alligator) genome/blat | |||
CRY1_anoCar Anolis carolinensis (lizard) XM_003220923 AAWZ02014443 | |||
CRY1_podSic Podarcis siculus (wall_lizard) DQ376040 16809482 | |||
CRY1_pytMol Python molurus (python) AEQU010547455 | |||
CRY1_chrPic Chrysemys picta (turtle) AHGY01469963 AHGY01469969 | |||
CRY1_xenTro Xenopus tropicalis (frog) NM_001087660 11533577 final four exons confirmed by many ESTs | |||
CRY1A_latCha Latimeria chalumnae (coelocanth) AFYH01018055 AFYH01018053 AFYH01018050 | |||
CRY1B_latCha Latimeria chalumnae (coelocanth) last exons uncertain | |||
CRY1A_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01025403 | |||
CRY1B_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01016727 AHAT01016728 | |||
CRY1A_danRer Danio rerio (zebrafish) NM_001077297 whole genome duplicate of retained CRY1 duplicate | |||
CRY1A2_danRer Danio rerio (zebrafish) BC044558 AW184635 olfactory old teleost CRY1 duplicate syntenically retained as tetrapod CRY1 | |||
CRY1B_danRer Danio rerio (zebrafish) BC095305 EB921055 aka CRY2A whole genome duplicate of lost CRY1 duplicate | |||
CRY1C_danRer Danio rerio (zebrafish) BC164795 EE210836 aka CRY2B old CRY1 duplicate lost in tetrapods CRY1 C12ORF23 CRY4 | |||
CRY1A_leuEri Leucoraja erinacea (skate) AESE010236716 AESE011153531 AESE010038968 AESE010673288 AESE012524396 | |||
CRY1B_leuEri Leucoraja erinacea (skate) AESE011669465 AESE012563587 AESE010604630 AESE011547252 | |||
CRY1A_calMil Callorhinchus milii (shark) AAVX01551101 AAVX01266331 AAVX01354908 AAVX01055947 | |||
CRY1B_calMil Callorhinchus milii (shark) AAVX01090452 AAVX01101328 AAVX01636526 AAVX01201905 | |||
CRY1_petMar Petromyzon marinus (lamprey) Contig24766 | |||
CRY1_braFlo Branchiostoma floridae (amphioxus) XM_002609455 end uncertain | |||
CRY1A_strPur Strongylocentrotus purpuratus (urchin) XM_001194752 same split exons as braFlo, end of gene uncertain, partially duplicated | |||
CRY2_homSap Homo sapiens (human) 11 exons | |||
CRY2_panTro Pan troglodytes (chimp) | |||
CRY2_gorGor Gorilla gorilla (gorilla) | |||
CRY2_ponAbe Pongo pygmaeus (orangutan) | |||
CRY2_rheMac Macaca mulatta (rhesus) CJ488220 testis | |||
CRY2_papHam Papio hamadryas (baboon) | |||
CRY2_calJac Callithrix jacchus (marmoset) | |||
CRY2_micMur Microcebus murinus (mouse_lemur) | |||
CRY2_musMus Mus musculus (mouse) CF898022 | |||
CRY2_ratNor Rattus norvegicus (rat) DN948283 prostate | |||
CRY2_criGri Cricetulus griseus (hamster) XR_135830 | |||
CRY2_spaJud Spalax judaei (blind_mole_rat) AJ606300 | |||
CRY2_dipOrd Dipodomys ordii (kangaroo_rat) | |||
CRY2_cavPor Cavia porcellus (guinea_pig) | |||
CRY2_hetGla Heterocephalus glaber (blind_mole_rat) EHA99865 | |||
CRY2_speTri Spermophilus tridecemlineatus (squirrel) | |||
CRY2_oryCun Oryctolagus cuniculus (rabbit) | |||
CRY2_turTru Tursiops truncatus (dolphin) | |||
CRY2_bosTau Bos taurus (cow) EG706191 lens | |||
CRY2_oviAri Ovis aries (sheep) NM_001129736 PubMed:19341811 | |||
CRY2_susScr Sus scrofa (pig) XM_003122835 | |||
CRY2_equCab Equus caballus (horse) | |||
CRY2_canFam Canis familiaris (dog) XM_540761 | |||
CRY2_ailMel Ailuropoda melanoleuca (panda) XM_002922310 iMet lost to assembly gap | |||
CRY2_myoLuc Myotis lucifugus (microbat) | |||
CRY2_pteVam Pteropus vampyrus (macrobat) | |||
CRY2_loxAfr Loxodonta africana (elephant) | |||
CRY2_triMan Trichechus manatus (manatee) AHIN01126950 AHIN01126951 | |||
CRY2_choHof Choloepus hoffmanni (sloth) | |||
CRY2_macEug Macropus eugenii (wallaby) FY652314 testis | |||
CRY2_monDom Monodelphis domestica (opossum) | |||
CRY2_ornAna Ornithorhynchus anatinus (platypus) | |||
CRY2_galGal Gallus gallus (chicken) AJ396745 bursa 19456395 15459395 | |||
CRY2_taeGut Taeniopygia guttata (finch) FE716439 brain | |||
CRY2_allMis Alligator mississippiensis (alligator) genome/blat | |||
CRY2_anoCar Anolis carolinensis (lizard) XM_003214641 | |||
CRY2_xenTro Xenopus tropicalis (frog) NM_001088670 AY049035 CX389867 11533577 discrepancies | |||
CRY2_ranCat Rana catesbeiana (bullfrog) GO458565 AY256684 extra SS removed | |||
CRY2_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01038797 | |||
CRY2_latCha Latimeria chalumnae (coelocanth) AFYH01005158 AFYH01005161 AFYH01005164 | |||
CRY2_danRer Danio rerio (zebrafish) aka CRY3 NM_131786 | |||
CRY2_oreNil Oreochromis niloticus (tilapia) XM_003449249 split exon 7 also in gasAcu, oryLat, tetNig not danRef or lepOcu | |||
CRY2_sigGut Siganus guttatus (spinefoot) AB643456 full length? imputed introns Percomorpha PUBMED 22163321 lunar phase-recognition | |||
CRY2_tetNig Tetraodon nigroviridis (fugu) CAAE01010345 | |||
CRY2_takRub Takifugu rubripes (fugu) HE592015 | |||
CRY1B_strPur Strongylocentrotus purpuratus (sea_urchin) XM_001183029 echinoderm lacks final 2 exons | |||
CRY1B_lytVar Lytechinus variegatus (sea_urchin) AGCV01081039 echinoderm many small contigs | |||
CRY1B_parLiv Paracentrotus lividus (sea_urchin) AM599080 echinoderm many transcripts | |||
CRY1B_aplCal Aplysia californica (sea_hare) FF067636 AASC02010117 scaffold_151 mollusc | |||
CRY1B_octVul Octopus vulgaris (octopus) JR450373 transcript assembly mollusc | |||
CRY1B_craGig Crassostrea gigas (oyster) GQ415324 HS189569 mollusc | |||
CRY1B_rudPhi Ruditapes philippinarum (clam) JO113369 mollusc | |||
CRY1B_vilLie Villosa lienosa (mussel) JR510441 transcript assembly mollusc fragment | |||
CRY1B_lymSta Lymnaea stagnalis (snail) ES576734 mollusc | |||
CRY1B_plaDum Platynereis dumerilii (clam_worm) GU322429 annelid mRNA fragment | |||
CRY1B_dapPul Daphnia pulex (water_flea) ACJG01002273 FE370447 FE356368 crustacean | |||
CRY1B_diaNig Dianemobius nigrofasciatus (cricket) AB291231 orthoptera | |||
CRY1B_acyPis Acyrthosiphon pisum (aphid) NM_001171061 ABLF02032292 HP303737 hemiptera | |||
CRY1B_danPle Danaus plexippus (butterfly) AY860425 AGBW01012954 lepidoptera | |||
CRY1B_bomMor Bombyx mori (silkworm) NM_001195699 wrong BABH01015108 moth lepidoptera | |||
CRY1B_mamBra Mamestra brassicae (moth) AY947639 Glossata lepidoptera | |||
CRY1B_helArm Helicoverpa armigera (cotton_bollworm) JN997418 moth lepidoptera | |||
CRY1B_droMel Drosophila melanogaster (fruit_fly) AB019389 diptera PubMed:22080955 PDB:3TVS | |||
CRY1B_anoGam Anopheles gambiae (mosquito) DQ219482 diptera PubMed:16332522 | |||
CRY1B_neoBul Neobellieria bullata (fleshfly) FJ373353 diptera | |||
CRY1B_bacCuc Bactrocera cucurbitae (melon_fly) AB517608 diptera | |||
CRY1A_dapPul Daphnia pulex (water_flea) FE418063 FE356487 ACJG01001137 crustacean | |||
CRY1A_eupSup Euphausia superba (krill) FM200054 contig crustacean | |||
2 | CRY1A_pedHum Pediculus humanus (louse) XM_002430500=wrong AAZO01005932 phthiraptera very similar intron pattern to vertebrate but lacks last 4 exons | ||
CRY1A_acyPis Acyrthosiphon pisum (aphid) NM_001171102 ABLF02035823 hemiptera cry2-2 PubMed:20482645 end uncertain | |||
CRY1A_ripPed Riptortus pedestris (bean_bug) AB379863 hemiptera PubMed:18547745 | |||
CRY1A_triCas Tribolium castaneum (flour_beetle) AAJJ01000096 coleopetera | |||
CRY1A_bomImp Bombus impatiens (bumble_bee) EF110521 AEQM02008194 hymenoptera PubMed:17244599 | |||
CRY1A_apiMel Apis mellifera (bee) NM_001083630 AADG06001305 hymenoptera | |||
CRY1A_attCep Atta cephalotes (ant) ADTU01021771 hymenoptera | |||
CRY1A_exoRob Exoneura robusta (bee) HP928681 hymenoptera fragment | |||
CRY1A_nylPub Nylanderia pubens (crazy_ant) JP792144 hymenoptera fragment | |||
CRY1A_nasVit Nasonia vitripennis (wasp) XM_001606355 AAZX01001169 hymenoptera N-term shortened | |||
CRY1A_antPer Antheraea pernyi (silkmoth) EF117812 lepidoptera PubMed:17244599 dropped long C-terminus | |||
CRY1A_anoGam Anopheles gambiae (mosquito) DQ219483 diptera dropped long C-terminus | |||
CRY1A_aedAeg Aedes aegypti (mosquito) XM_001655728 diptera dropped long C-terminus | |||
CRY1_vilLie Villosa lienosa (mussel) JR505030 mollusc transcript assembly mollusc | |||
CRY1_tetUrt Tetranychus urticae (spider-mite) CAEY01002034 chelicerate N-terminus uncertain | |||
CRY1_aplCal Aplysia californica (sea_hare) scaffold_2275 mollusc small fragment | |||
CRY4_galGal Gallus gallus (chicken) NP_001034685 CRY4 PubMed:19663499 synteny: ADIPOR1 UBE2T CRY4 LRIF1 DRAM2 CEPT1 | |||
CRY4_melGal Meleagris gallopavo (turkey) XM_003212851 | |||
CRY4_anaPla Anas platyrhynchos (duck) scaffold1663 | |||
CRY4_taeGut Taeniopygia guttata (finch) XM_002198497 | |||
CRY4_pasDom Passer domesticus (sparrow) AY494987 16687285 fragment | |||
CRY4_anoCar Anolis carolinensis (lizard) FG650345 synteny: UBE2T CRY4 LRIF1 DRAM2 verified indel exon 3 | |||
CRY4_xenTro Xenopus tropicalis (frog) NP_001123706 | |||
CRY4_xenLae Xenopus laevis (frog) BC167313 only 89% identical to CRY4_xentro | |||
CRY4_latCha Latimeria chalumnae (coelocanth) AFYH01009222 | |||
CRY4_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01016726 | |||
CRY4_danRer Danio rerio (zebrafish) BC164413 adjacency to lost CRY1 suggests relationship | |||
CRY4_molTec Molgula tectiformis (tunicate) CJ347377 CJ411442 CJ358785 fragment imputed introns | |||
CRY4_braFlo Branchiostoma floridae (amphioxus) Un:610812841 XM_002609457 exon 4,7,8 wrong | |||
CRY64_anoCar Anolis carolinensis (lizard) XM_003225714 6-4 photolyase synteny: DCPS TIRAP CRY64 SRPR FOXRED1 | |||
CRY64_chrPic Chrysemys picta (turtle) AHGY01135270 AHGY01135271 no synteny | |||
2 | CRY64_allMis Alligator mississippiensis (alligator) blat | ||
CRY64_croPor Crocodylus porosus (crocodile) blat/genome | |||
CRY64_xenTro Xenopus tropicalis (frog) synteny: STS1 RPL27A CRY64 FOXRED1 SRPR PubMed:19715341 19345672 9016626 | |||
CRY64_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01024141 | |||
CRY64_danRer Danio rerio (zebrafish) BC044204 6-4 photolyase aka CRY5 synteny: FOXRED1 | |||
CRY64_salSal Salmo salar (salmon) BT058852 | |||
CRY64_oreNil Oreochromis niloticus (tilapia) XM_003437598 AERX01000034 | |||
CRY64_braFlo Branchiostoma floridae (amphioxus) BW780666 FE555184 XM_002595028 fused exons 2-3 fusion exons 2-3 odd splice phases exon 5-6, no split 8-9 short final exon | |||
CRY64_strPur Strongylocentrotus purpuratus (urchin) XM_001189626 extra 1st exon unwarranted MCGAPRSYVEIRDSEEHSRRHVARLQFQFQSDLP 12 K | |||
CRY64_eucTri Eucidaris tribuloides (pencil_urchin) JI324408 fragment imputed introns | |||
CRY64_aplCal Aplysia californica (sea_hare) scaffold_427 | |||
CRY64_vilLie Villosa lienosa JR505030 transcript assembly mollusc | |||
CRY64_droMel Drosophila melanogaster (fruitfly) 6-4 photolyase PDB:3CVW CG2488 uses 5-deazariboflavin | |||
CRY64_danPle Danaus plexippus (butterfly) EF117813 PubMed:17244599 two novel exons | |||
CRY64_acyPis Acyrthosiphon pisum (aphid) XM_001945977 single exon | |||
CRY64_anoGam Anopheles gambiae (mosquito) XM_314748 | |||
CRY64_bomMor Bombyx mori (silkworm) AK381942 frameshift | |||
CRY64_craMey Crateromorpha meyeri (sponge) PubMed:20121950 | |||
CRY64A_triAdh Trichoplax adhaerens (placozoa) XM_002108524 ABGP01000049 no UIM domain affinity to CRY class | |||
CRY64B_triAdh Trichoplax adhaerens (placozoa) XM_002107723 ABGP01000051 anti-parallel tandem no UIM domain | |||
CRY7_xenTro Xenopus tropicalis (frog) XP_002938187 AAMC01077621 AAMC01077620 many transcripts CDK10+ CRYM+ GCSH- PDK1L2- BCMO1+ GL172982 1U3C 34% 3CVW 29% | |||
CRY7_xenLae Xenopus laevis (frog) transcripts DC068968 EG576829 BU901325 | |||
CRY7_latCha Latimeria chalumnae (coelocanth) AFYH01265207 pseudogene | |||
CRY7_lepOcu Lepisosteus oculatus (gar) AHAT01010533 AHAT01010534 | |||
CRY7_danRer Danio rerio (zebrafish) ENSDART00000125725 no synteny to frog | |||
CRY7_salSal Salmo salar (salmon) AGKD01006863 | |||
CRY7_hapBur Haplochromis burtoni (chichlid) AFNZ01022319 | |||
CRY7_gasAcu Gasterosteus aculeatus (stickleback) DN725444 | |||
CRY7_oryLat Oryzias latipes (medaka) CRYM+ GCSH- two transcripts, very small introns | |||
CRY7_oreNil Oreochromis niloticus (tilapia) | |||
CRY7_tetNig Tetraodon nigroviridis (fugu) | |||
CRY7_takRub Takifugu rubripes (fugu) | |||
CRY7_gadMor Gadus morhua (cod) CAEA01536921 | |||
CRY7_xipMac Xiphophorus maculatus (platyfish) AGAJ01012112 | |||
CRY7_rudPhi Ruditapes philippinarum (clam) JO112203 gonad transcript missing first half of antenna domain note filter feeder | |||
CRY7_craGig Crassostrea gigas (oyster) HS138673 | |||
DASH_taeGut Taeniopygia guttata (finch) ABQF01044665 ABQF01044669 ABQF01044671 synteny: ACAA1 DASH MYD66 OXSR1 | |||
DASH_anaPla Anas platyrhynchos (duck) scaffold1769 | |||
DASH_melUnd Melopsittacus undulatus (budgerigar) AGAI01061648 | |||
DASH_galGal Gallus gallus (chicken) syntentic pseudogene, numerous indels, frameshifts, internal stops | |||
DASH_melGal Meleagris gallopavo (turkey) ADDD01036185 syntenic pseudogene | |||
DASH_anoCar Anolis carolinensis (lizard) XM_003221869 14 exons | |||
DASH_chrPic Chrysemys picta (turtle) AHGY01416294 first exon off contig | |||
DASH_xenTro Xenopus tropicalis (frog) XM_002938001 PubMed:15147276 synteny: ACAA1 DASH MYD66 transcripts AL790297 CR419606 etc | |||
DASH_hymCut Hymenochirus curtipes (frog) fragment | |||
DASH_ambMex Ambystoma mexicanum (axolotl) CO785483 fragment | |||
DASH_latCha Latimeria chalumnae (coelocanth) AFYH01055296 AFYH01281932 probable pseudogene | |||
DASH_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01010414 | |||
DASH_danRer Danio rerio (zebrafish) NM_205686 | |||
DASH_oreNil Oreochromis niloticus (tilapa) XM_003439198 | |||
DASH_patPec Patiria pectinifera (starfish) HP101597 | |||
DASH_strPur Strongylocentrotus purpuratus (urchin) | |||
DASH_aplCal Aplysia californica (sea_hare) scaffold_151:75,790-145,485 | |||
DASH_vilLie Villosa lienosa (mussel) JR504188 transcript assembly mollusc | |||
DASH_nemVec Nematostella vectensis (sea_anemone) XP_001623243 ABAV01026885 | |||
DASH_hydMag Hydra magnipapillata (cnidarian) XM_002166508 single exon ABRM01055505 | |||
DASH_monBre Monosiga brevicollis (choanoflagellate) XP_001745157 ABFJ01000402 | |||
DASH1_araTha Arabidopsis thaliana (cress) PHR2 NM_130327 AFNA01010806 | |||
DASH2_araTha Arabidopsis thaliana (cress) NM_122394 AFMZ01019177 aka:CRY3 PDB:2VTB | |||
DASH_phaTri Phaeodactylum tricornutum (diatom) XM_002178853 CPF2 | |||
DASH_thaPse Thalassiosira pseudonana (diatom) XM_002291289 | |||
CPD_monDom Monodelphis domestica (opossum) NP_001028149:wrong OPC1 PubMed:7937136 synteny: TNK1 MUC4 CPD KIAA0226 FYTTD1 | |||
CPD_sarHar Sarcophilus harrisii (tasmanian_devil) AEFK01107967 | |||
CPD_potTri Potorous tridactylus (rat_kangaroo) D26020 PubMed:7813451 | |||
CPD_ornAna Ornithorhynchus anatinus (platypus) | |||
CPD_taeGut Taeniopygia guttata (finch) XM_002190577 | |||
CPD_melUnd Melopsittacus undulatus (budgerigar) AGAI01046895 | |||
CPD_galGal Gallus gallus (chicken) XM_422729 | |||
CPD_melGal Meleagris gallopavo (turkey) XM_003209143 | |||
CPD_allMis Alligator mississippiensis (alligator) genome/blat | |||
CPD_chrPic Chrysemys picta (turtle) AHGY01112360 incomplete | |||
CPD_anoCar Anolis carolinensis (lizard) XM_003226963 | |||
CPD_pytMol Python molurus (python) | |||
CPD_xenTro Xenopus tropicalis (frog) NP_001135721 | |||
CPD_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01034265 | |||
CPD_danRer Danio rerio (zebrafish) NM_201064 | |||
CPD_petMar Petromyzon marinus (lamprey) rough revised sequence | |||
CPD_braFlo Branchiostoma floridae (amphioxus) XP_002586934 FE570347 fixed frameshift exon 4 | |||
CPD_strPur Strongylocentrotus purpuratus (urchin) JT122393 JT102939 FJ812411 | |||
CPD_aplCal Aplysia californica (sea_hare) scaffold_446:238,174 | |||
CPD_vilLie Villosa lienosa (mussel) JR505029 transcript assembly mollusc | |||
CPD_droMel Drosophila melanogaster (fruitfly) thymidine dimer photolyase CG11205 uses 5-deazariboflavin | |||
CPD_nasVit Nasonia vitripennis (wasp) XM_001603235 trimmed N-terminal | |||
CPD_bomImp Bombus impatiens (bumble_bee) XM_003488984 | |||
CPD_apiMel Apis mellifera (bee) XM_003250426 | |||
CPD_anoGam Anopheles gambiae (mosquito) XM_313925 trimmed N-terminal | |||
CPD_aedAeg Aedes aegypti (mosquito) XM_001653905 trimmed N-terminal | |||
CPD_acyPis Acyrthosiphon pisum (aphid) XM_001949116 trimmed N-terminal | |||
CPD_nemVec Nematostella vectensis (anemone) ABAV01006764 XM_001636204 bad BACK01030119 | |||
CPD_acrDig Acropora digitifera (coral) BACK01030119 cnidarian one intron missing | |||
CPD_ampQue Amphimedon queenslandica (sponge) ACUQ01006132 XM_003388698 bad | |||
CPD_monBre Monosiga brevicollis (choanflagellate) ABFJ01000652 related intronation but numerous differences | |||
CPD_salSpp Salpingoeca species (choanflagellate) ACSY01000967 different intronation still | |||
CPD_araTha Arabidopsis thaliana (cress) PHR1 NM_179320 AFMZ01000529 GC-AG splice exon 6-7 | |||
CPD_orySat Oryza sativa (rice) B096003 BACJ01049170 aka:PhrII,Class II PMID:22170053 PDB:3UMV | |||
CRY1A_acrMil Acropora millepora (coral) EF202589 | |||
CRY1B_acrMil Acropora millepora (coral) EF202590 | |||
CRY1A_nemVec Nematostella vectensis (anemone) XM_001623096 | |||
CRY1B_nemVec Nematostella vectensis (anemone) XM_001623096 | |||
CRY1C_nemVec Nematostella vectensis (anemone) XM_001630979 | |||
CRY1D_nemVec Nematostella vectensis (anemone) XM_001632799 | |||
CRY1E_nemVec Nematostella vectensis (anemone) XM_001632800 | |||
CRY64_nemVec Nematostella vectensis (anemone) XP_001636303 ABAV01006592 last exon uncertain | |||
CRY2_ampQue Amphimedon queenslandica (sponge) XM_003386521 | |||
CRY_ampQue Amphimedon queenslandica (sponge) XM_003386534 | |||
CRY_subDom Suberites domuncula (sponge) FN421335 | |||
CRY_aphVas Aphrocallistes vastus (sponge) PubMed:14499587 | |||
CRY1A_araTha Arabidopsis thaliana (cress) NM_116961 AFNC01018176 aka:CRY1,HY4 PDB:2VTB | |||
CRY1B_araTha Arabidopsis thaliana (cress) CRY2 PHH1 NM_100320 AFNB01000167 no antennal chromophore | |||
CRY1C_araTha Arabidopsis thaliana (cress) NM_001035626 AFNC01013058 aka:UVR3,CRY3 PDB:3FY4 | |||
CRY_phaTri Phaeodactylum tricornutum (diatom) XM_002180059 PMID:19424294 | |||
CRY_thaPse Thalassiosira pseudonana (diatom) XM_002291108 | |||
PFES_agrTum Agrobacterium tumefaciens (bacteria) NP_355900 aka: PhrB | |||
PFES_rhoSph Rhodobacter sphaeroides (bacteria) CP000144 PDB|3ZXS PMID:22290493 6,7-dimethyl-8-ribityl-lumazine antenna aka CryPro 4Fe-4S photolyase | |||
PFES_metMah Methanohalophilus mahii (Euryarchaeota) CP001994 4Fe-4S photolyase | |||
PFES_natPha Natronomonas pharaonis (Euryarchaeota) CR936257 4Fe-4S photolyase | |||
PRIM2_homSap Homo sapiens (human) primase large subunit 4Fe-4S pdb|3L9Q,3Q36 | |||
PRIM2_sacCer Saccharomyces cerevisiae (yeast) P20457 aka: PRI2_YEAST primase large subunit PDB|3LGB | |||
== | == Article authorship and data usage policy == | ||
[[Image:Author.jpg|left]] | |||
I researched this article in its entirety in the winter of 2012, not paying attention initially to previous studies which are excellent on reaction mechanisms and regulatory cycles but completely clueless on comparative genomics (10 years into the genomic era!). Cryptochromes are a moderately difficult topic as metazoan genes go because the timing of gene duplications largely falls between the cracks of phylogenetic coverage and because extenive gene losses in unrepresentative model organisms have distorted the overall evolutionary picture. I plan to greatly expand the treatment of 3D structural implications of comparative genomics during the summer of 2012. | |||
My interests are primarily in the long range evolutionary acquisition and divergence of function-enabling structure, starting from primases and 4Fe-4S cluster photolyases and ending with circadian, magnetosensing and couplings with opsins. However comparative genomics has major applications to rapid hypothesis-testing in all aspects of cryptochrome and photolyase research, the main point being the strong coupling between sequence conservation (ie selective pressure) and functional importance. This means conservation never lasts very long without a reason, and conversely non-conserved features are not important. | |||
Although copyrighted, all the information here is in the public domain and can be used by anyone without additional permissions if properly sourced; however if data, figures or original observations are taken wholesale for a peer-reviewed scientific publication, it might be appropriate (after consultation early on) to include me among secondary co-authors. | |||
Rather than make article edits yourself, please contact me by email with clarifications, corrections or additions to the content so I can make edits while maintaining a consistent approach. For broader disagreements or different interests, a better option is to simply register at the UCSC genomeWiki site and create your own page within the comparative genomics category. | |||
This is just a scientific research article on an old gene family, not an advisory resource for personal genomics issues, melatonin dietary supplementation or medical advice on jet lag and insomnia -- thanks in advance for not sending inappropriate email. Technical terms from genetics and molecular biology are not explained in the article when keywords have a satisfactory treatment at wikipedia or in undergraduate genetics texts; because of good keywords, the scientific literature is easily searched at PubMed so not duplicated here. | |||
My last dozen published research papers in PNAS, Nature, Science etc can be found [http://www.ncbi.nlm.nih.gov/pubmed/21709235,20164927,19020620,18464734,18266766,17984227,18085818,17975064,17322288,15608236,12045153,22012981 here]. Watch for 4 additional comparative genomics paper to appear in 2012. I've also written over a [[:Category:Comparative_Genomics|thousand pages of comparative genomics]] for other human genes, authored the original user manual to the UCSC human genome browser and in 1999 an advanced tutorial on metazoan genome annotation still [http://www.mad-cow.org/00/annotation_tutorial.html widely available online]. I thank the UCSC Genomics Group (Hiram Clawson, Brian Raney, Maximilian Haeussler) for software, manuscript and literature resources, Evim Foundation for logistical support, and the Sperling Foundation for financial support under project grant 2012.GNTCS.006. | |||
<br clear=all> | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 08:38, 18 April 2014
See also: Curated reference sequences for cryptochromes and photolyases
Updates: fixes and additions become difficult to locate within a long article so these are provided below in reverse chronological order linked to their approximate location. 08 Jun 12: significant additions to iron-sulfur photolyases and primases 21 May 12: determined DASH's phylogenetic distribution and terminal motif using a greatly improved sequence set.
Introduction to Cryptochromes
Cryptochromes are large flavoproteins with a curiously complex evolutionary history, beginning billions of years ago as dna repair enzymes (or even earlier as replication primase). An old gene duplication followed by specializing divergence gave rise to two paralogs repairing distinct types of dna damage (cyclobutane pyrimidine dimers and 6-4 pyrimidine-pyrimidone pairs). These photolyases initially used FAD activated by visible blue light to undo the damage done by UV and other processes.
Since FAD has relatively low adsorbance, photolyases evolved a second site for an antenna chromophore with better light harvesting capabilities that could transfer its excitation to the FAD at the active site. This elusive antenna molecule may be FMN, a folate, lumazine, or a 5-deazariboflavin called Fo once thought restrict to methanogenic archaea. In the case of the much-studied Drosophila, both the photolyases utilize Fo, making it a new vitamin for this species since the biosynthetic genes are absent. Cryptochrome so far lack antenna molecules but retain the binding domain and substrate pocket.
The next round of gene duplication of the 6-4 photolyase gave rise to a cryptochrome which retained the conformational change induced by FAD binding of blue light but lost dna repair capacity, instead specializing in entraining the day/night circadian rhythm cycle. However the distinction between signalling (non-enzymatic) and catalytic gene family members is muddled. Later rounds of gene duplication gave rise to yet more orthology classes to be followed -- sometimes hundreds of millions of years later -- by gene loss in some large lineages.
The seven main classes were retained in various combinations in different clades during the subsequent course of evolution, causing endless comparative nomenclatural confusion (when in doubt, look at the amino acid sequences). For example, Drosophila did not retain CRY1A unlike other insects while placental mammals lost all three photolyases though marsupials retained one and monotremes two. Gallinaceous birds also lost a photolyase. Rayfinned fish had a series of further duplications within the gene family. Despite this, the primary sequence, exon structure, fold and FAD, antenna and dna binding sites have largely been conserved -- along with key regulatory binding sites to other proteins -- even as antenna molecules and dna repair capacity might be dispensed with.
A new vertebrate cryptochrome CRY7 with a ubiquitin binding domain UIM
Even ten years into the whole genome era, the comparative genomics of cryptochromes and photolyases has never been considered, perhaps because of a narrow experimental focus on 'model' organisms such as mouse and fruit fly that, as it turns out, have rather restricted and unrepresentative gene family complements. Since most annotation effort goes into human (which are very deficient in their repertoire), the lack of a suitable homology probe there lets novel photolyases and cryptochromes in other species go undiscovered.
This section describes a new cryptochrome orthology class (designated CRY7 here) with an extensive but not universal phylogenetic distribution. It apparently arose in the pre-Cambrian as a segmental gene duplication of CRY64 (or vice versa) based on its independent intronation pattern. Most remarkably, CRY7 possesses an amino terminal ubiquitin binding domain. The new protein is evolving overall rather rapidly for a cryptochrome and has been lost from many clades but it still retains the two core domains. Although the antenna molecule cannot be predicted, the FAD cofactor is likely present, based on structurally modelling with 1U3C and 3CVW (from CRY64_droMel, 34% identity and CRY1A_araTha, 29% identity).
CRY7 is absent from mammals and indeed all amniotes but still present in amphibians, lobe-finned, ray-finned fish including basal gar, and two molluscs. These genes form a single new orthology class with distinct syntenic location, intronation pattern, and domain structure. The unusual phylogenetic distribution cannot be plausibly explained by prokaryotic endosymbiont, DNA contamination by xenobiotics (in filter-feeders), nor horizontal gene transfer. There is also affinity to two placozoan cryptochrome but these lack the ubiquitin binding domain.
CRY7 in frog has 20 overlapping transcripts at GenBank dating back to 2003 that cover all but the middle of the gene. Expression has been reported from egg (BX771555, AL893008), neurulation embryo (BX699228, AL662439), whole embryo (CX470086, CX470087), tailbud head (CR562794, CR562774), adult testes (CX928370 and 7 others), and adult ovary (DR850985 and 3 others). These sites of expression do not distinguish between a DNA repair role and photosignalling. However the presence of the N-terminal UIM domain strongly suggests the latter because protein turnover is a well-established component of the cryptochrome circadian system.
In non-mammalian species, circadian regulation of other genes can take place directly at cellular sites indpendently of the central nervous system, often in species with extra-retinal opsin expression. Frog expresses melanopsin in skin melanophores; fish also express an opsin in lateral line iridophores which exhibit circadian color changes; and squid utilize an external opsin to manage camouflage. Ultra-structural coexpression studies of CRY7 and the respective opsins might establish an association.
The ubiquitin interacting motif (UIM) consists of 20 amino acid residues first described in the 26S proteasome subunit that recognises ubiquitin. Ubiquitin binds UIM so the motif triggers a cascade of downstream signalling events. The UIM forms a short alpha-helix that can fits into the ubiquitin pocket via hydrophobic and electrostatic interactions. The UIM motif of a frog CRY7 gene model was predicted by subsequent automatic procedures at KEGG but the short UIM motif was neither homologously confirmed in other species nor shown actually part of the cryptochrome gene (rather than belonging to an upstream adjacent gene with a missed stop codon). UIM domains are widespread but not necessarily homologous (ie mobile chimeric domains) because short motif can evolve in situ.
However here the amino terminus begins with about 70 semi-conserved residues, followed by the UIM domain beginning a new exon. This extended motif has no Blast counterpart in other known proteins even using a consensus sequence probe. It is followed by a long spacer region of about 140 amino acids that is evolving chaotically in both length and composition. This pattern suggests a fusion with a UIM donor protein with the spacer region in the process of being discarded. Conservation begins again as the antenna domain is reached and continues through the FAD domain all the way to the carboxy terminus (which extends nearly 100 amino acids beyond any homology with the CRY64 FAD domain). A crystallographic structure for CRY7 might reveal more distant relationships for the conserved N- and C-terminal extensions.
Species UIM motif UIM conservation Genus species (common) CRY7_xenTro GYETDLELAIALSLQEHNQL GYETD....I....Q.HNQL Xenopus tropicalis (frog) CRY7_lepOcu VEEEEVEVALALSLQELGVS SV.EE.V.V......Q.LGV Lepisosteus oculatus (gar) CRY7_danRer DESEELELALTLSLYETKQI D.SE......T...Y.T.QI Danio rerio (zebrafish) CRY7_salSal DEDDELAVALALSLLEVKRQ D.....AV........V.R. Salmo salar (salmon) CRY7_gadMor DEEDELEVALALSLLDVKPQ ...............D..GH Haplochromis burtoni (chichlid) CRY7_hapBur TEDDELELALALSLLDMKGH .Q........S...V..D.H Gasterosteus aculeatus (stickleback) CRY7_oreNil TEDDELELALALSLLDMKGQ ....D.........M..E.. Oryzias latipes (medaka) CRY7_xipMac MEDDELELALALSLLDMKDQ ...............D..G. Oreochromis niloticus (tilapia) CRY7_gasAcu TQDDELELALSLSLVEMDDH ...ED.........V....C Tetraodon nigroviridis (fugu) CRY7_takRub TEDDELELALALSLVETKDY ..............V.T..Y Takifugu rubripes (fugu) CRY7_oryLat TEDDDLELALALSLMEMEDQ D.E....V.......DV.P. Gadus morhua (cod) CRY7_tetNig TEDEDLELALALSLVEMKDC M..............D.... Xiphophorus maculatus (platyfish) consensus TEDDELELALALSLLEMKDQ TEDDELELALALSLLEMKDQ UIM motif PFAM: PF02809
Synteny is not helpful in the CRY7 situation with only Oryzias latipes (medaka) sharing a neighboring gene with frog. CRY7 does not represent a segmental duplication of CRY64 because its intronation pattern is totally different, plus the percent identity is very low for a pair of cryptochromes. Vertebrates do lose and gain introns but that process is extremely slow. More likely the gene duplication took place in single-celled eukaryotes prior to the principal era of intronation, with the two ortholog classes then acquiring introns independently at essentially random positions. CRY7 is a misclassified paralog cross-over in Genomicus and not represented in the UCSC 46-way whole genome alignment because human lacks the gene.
CRY7 has a completely unique intronation pattern lacking any relationship to CRY64 (its best blast match within the gene family) or any other cryptochrome or photolyase. Since this pattern is strongly conserved in the CRY7 ortholog set, it is likely ancient. If so, this protein represents a very old branch of the gene family but one that is unrecognizable or lost from most lineages. CRY7 is not an evolutionary novelty, having persisted for 450,000,000 years in vertebrates; nearly half of the 58,000 living species of vertebrates retain it, though not any amniotes studied to date. The position and phase of CRY64 intron breaks are positioned by homology into frog CRY7 below and contrasted with a comparison of CRY64 to human CRY1 (which share 5 identical intron sites). Blue indicates phase 00, orange phase 12, red phase 21, magenta perfect match of position and phase:
CRY7_xenTro Xenopus tropicalis (frog) introns relative to CRY64 0 MDLEPFERAQIDDVLQQLESGSVQADEFLCLVLSILGSSRTYSQFPAILQSLSRKEPAMYRELMDLHAEYFRK 0 0 EPADLETLGYETDLELAIALSLQEHNQLTDTASFASEVDPAPKISFADAAKLSHFSHKHNKKNSSSKTEITKLKDNVAAMNLYQERKRYHINGQEKTCISN CYNGQPEPEDCVLKSEDGEDVFHVETSRPRESKAKHSRRSRKKKKSAPSRGL^VAMKPVLVWFRRDLRLHDNPALISALEHGVPVIPVFLWCINEETGQNFTLATGGAT KYWLHHALLKLNQSLIQRFGSH^IIFRVARSCEEELVSLVHETGADTIIINAVYEPWLKERDDLISETLRRHGVELKKHHSYCLYEPDS^VSTEGVGLR 1 2 GIGSVSHFMSCCKRNNSAPIGMPLDAPRCLPAPC^NWPESDHLDTLELGKMPHRKDGTL 0 0 IDWAVTIRESWDFSEDGAYTCLANFLQ^D1 2 GVKHYEKESGRADKPYTSHISPYLHFGQISPRTVLHEAYFTKKNV^PKFLRKLAWRDLAYWLLILFPDMPSEPVRPAYK 0 0 SQRWSSDLNHLRAWQK^GLTGYPLVDAAMRELWLTGWMCNYSRHVVASFLVAYLHIHWVHGYR^WFQ 0 0 DTLLDADVAINAMMWQNGGMSGLDHWNFVMHPVDSALTCDPYGSYVR^KWCPELAGLPDEYIHKPWKCAPSQLRRA 1 2 GVILGRNYPHRIVLDLEERREQSLKDVVEVRKKHLEYLDEVSGCDMVQIPDQLLAL^TLGHTSGEDEVVRNRTGSFLLPVITRKEFKYKTLQPDTKDNPYNTVLKGYV SRKRDETIAYMNERHFTASTINEGAQRHERIERTNRLMEGLPAPSDAKNKSRRTPKKDPFSIIPPSYLHLAN* 0 >CRY1_homSap Homo sapiens (human) introns relative to CRY64 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSR^LFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIE^KCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAE^GRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNG^NGGFMGYSAENIPGCSSSG 1 2 SCSQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN* 0
Below the frog protein CRY7 is marked up for its various domains and motifs according to Pfam, Blast and PDB searches. Blue shows the antenna domain with predicted α/β secondary structure, purple the possibly catalytic FAD domain with predicted all α secondary structure, magenta the UIM ubiquitin motif, purple two compositionally simple regions rich is basic residues predicted not to have definite fold, dark red the conserved region of unknown function upstream of the UIM ubiquitin motif, and dark blue the conserved carboxy terminal motif of unknown function.
>CRY7_xenTro Xenopus tropicalis (frog) 0 MDLEPFERAQIDDVLQQLESGSVQADEFLCLVLSILGSSRTYSQFPAILQSLSRKEPAMYRELMDLHAEYFRK 0 0 EPADLETLGYETDLELAIALSLQEHNQLTDTASFASEVDPAPKISFADAAKLSHFSHKHNKKNSSSKTEITKLKDNVAAMNLYQERKRYHINGQEKTCISN CYNGQPEPEDCVLKSEDGEDVFHVETSRPRESKAKHSRRSRKKKKSAPSRGLVAMKPVLVWFRRDLRLHDNPALISALEHGVPVIPVFLWCINEETGQNFTLATGGAT KYWLHHALLKLNQSLIQRFGSHIIFRVARSCEEELVSLVHETGADTIIINAVYEPWLKERDDLISETLRRHGVELKKHHSYCLYEPDSVSTEGVGLR 1 2 GIGSVSHFMSCCKRNNSAPIGMPLDAPRCLPAPCNWPESDHLDTLELGKMPHRKDGTL 0 0 IDWAVTIRESWDFSEDGAYTCLANFLQD 1 2 GVKHYEKESGRADKPYTSHISPYLHFGQISPRTVLHEAYFTKKNVPKFLRKLAWRDLAYWLLILFPDMPSEPVRPAYK 0 0 SQRWSSDLNHLRAWQKGLTGYPLVDAAMRELWLTGWMCNYSRHVVASFLVAYLHIHWVHGYRWFQ 0 0 DTLLDADVAINAMMWQNGGMSGLDHWNFVMHPVDSALTCDPYGSYVRKWCPELAGLPDEYIHKPWKCAPSQLRRA 1 2 GVILGRNYPHRIVLDLEERREQSLKDVVEVRKKHLEYLDEVSGCDMVQIPDQLLALTLGHTSGEDEVVRNRTGSFLLPVITRKEFKYKTLQPDTKDNPYNTVLKGYV SRKRDETIAYMNERHFTASTINEGAQRHERIERTNRLMEGLPAPSDAKNKSRRTPKKDPFSIIPPSYLHLAN* 0
Using Swissmodel with CRY64 from Drosophila as template (PDB:1UC3. 31% identity), the tertiary structure of CRY7 can be successfully modeled from over the region PVLLWF...ALVRRR of salmon CRY7 (corresponding to residues 13-497 of the experimentally determined structure). The predicted two domain structure very much resembles that of any cryptochrome or photolyase and allows preliminary identification of beta strands and helices.
The quality of the model varies by position, as shown by the B factor score coloring in the figure on the left. The overall Z-Score quality of fit is -3.74, not too shabby for a large protein but no substitute for an actual experimental structure determination. (The other template option, an Arabidopsis cryptochrome, gives an unsatisfactory Z-Score.) Note the amino terminal conserved domain, the UIM motif, and the C-terminal conserved domain cannot be modeled at all without a template.
The FAD binding site exhibits moderate steric interference but that molecule could be docked if a few residues were re-positioned slightly. The antenna site is more problematic: while present in the 3D structure, the nature of the antenna molecule (if any) cannot really be predicted. The early divergence of CRY7 from CRY64 and the lack of evolutionary persistence (consistency) of antenna molecules makes it very uncertain whether the Drosophila antenna molecule in CRY64 and CPD -- recently determined to be 5-deazariboflavin -- is actually the antenna molecule for CRY7.
Although 5-deazariboflavin is the best historic option, CRY7 has been lost from Drosophila and 5-deazariboflavin is not known to occur in vertebrates or molluscs (the phylogenetic setting for CRY7 today). Since they cannot synthesize it, it would amount to a new vitamin in these species.
Alternatively, the antenna molecule could be 6,7-dimethyl-8-ribityl-lumazine, folate, FMN, FAD or related molecules (as seen in other members of the gene family. No antenna molecule might be appropriate in view of the UIM domain and the implied signaling role, yet that does not account for the observed conservation of the antenna domain.
Predicted alpha helices (h) and beta strands (s) of CRY7: CRY7 PVLLWFRRDL RLHDNPAVIG SLEAGGPVIP VFIWCPEEEE GPGVTVAMGG ACKFWLHQAL SCLSSALEHI GSHLVFLRPD EEREGIGSSL RALRSLVRET CRY7 csivwfrrdl rvednpalaa avrag-pvia lfvwapeeeg hyhpg----r vsrwwlknsl aqldsslrsl gtclitkrs- ------tdsv aslldvvkst CRY7 sssss hhhhh hhh ss ssss hh hhhhhhhhhh hhhhhhhhhh sssss h hhhhhhh CRY7 sssss hhhhh hhhh ssss ssss hh h hhhhhhhhhh hhhhhhhhhh sssss h hhhhhhh CRY7 GAQTVLASAL YEPWLRERDQ VVVSALQKDR VEVNMVHSYC LRDPYTVTTE GVGLRGIGSV SHFMSCCQMN PGPGLGVPLD PPISLPSPSV WPRGCPLEGL CRY7 gasqiffnhl ydplslvrdh rakdvltaqg iavrsfnadl lyepwevtde lgrpfsm-fa afwerclsmp ydpesp--ll ppkkiisgdv sk--cvadpl CRY7 sssssss hhhhhhhh hhhhhhh sssss hhhhhhh CRY7 sssssss hhhhhhhh hhhhhhh sssss hh hhhhhhh hh CRY7 GLARMPCRKD GTTIDWAANI RSSWDFSEEG AQSRLEAFLN DGVYRYEKES GRADAPNTSC LSPYLHFGQL SARWLLWDTK GA-------- ----RCRPPK CRY7 v------fed dsekgsnall arawspgwsn gdkalttfin gplleysknr rkadsattsf lsphlhfgev svrkvfhlvr ikqvawaneg neageesvnl CRY7 hhhhhhhhhh h hhh hhhhhhhh hh hh hhhh hhhhhhh hhh CRY7 hhhhhhhhhh hhh hhh hhhhhhhhhh hhhh hhhhhhhhh hhhhhhhh hhhhhhhhh CRY7 FIRKLAWRDL AYWQLTLFPD LPWESLRPPY KALRWSNERG HLKAWQKGRT GYPLVDAAMR QLWLTGWMNN YMRHVVASFL IAYLHLPWQE GYRWFQDTLV CRY7 flksiglrey sryisfnhpy sherpllghl kffpwavden yfkawrqgrt gyplvdagmr elwatgwlhd rirvvvssff vkvlqlpwrw gmkyfwdtll CRY7 hhhhhhhhhh hhhhhhh hh hhhhhhh hhhhhhhh hhhh h hhhhhhhhhh hhh hh hhhhhhh CRY7 hhhhhhhhhh hhhhhhh hh hhhhhhh hhhhhhhh hhhh h hhhhhhhhhh hhh hh hhhhhhh CRY7 DADVAIDAMM WQNGGMCGLD H--WNFVMHP VDAAMTCDPY GNYVRKWCTE LAVLPDDLIH KPWKCPASML RRAGVVLGQS YPERVVTDLE ERRSQSLQDV CRY7 dadlesdalg wqyitgtlpd srefdridnp qfegykfdpn geyvrrwlpe lsrlptdwih hpwnapesvl qaagielgsn yplpiv-gld eakarlheal CRY7 hhhhhhh hhhhh h hhhhhhh hhhhh h h hhhh hhh h hhhhhhhhhh CRY7 hhhhhhh hhhhh h hhhhhhh hhhhh h h hhhh hhh hh hhhhhhhhhh CRY7 LAVLPDDLIH KPWKCPASML RRAGVVLGQS YPERVVTDLE ERRSQSLQDV ALVRRR CRY7 lsrlptdwih hpwnapesvl qaagielgsn yplpiv-gld eakarlheal sqmwql CRY7 h hhhh hhh h hhhhhhhhhh hhhhhh CRY7 h hhhh hhh hh hhhhhhhhhh hhhhhh
Standard lab mouse C57BL/6J has a mutated CRY1 cryptochrome gene
Lab mouse has an odd mutation in its 10th exon where a century of inbreeding may have inadvertently fixed a very serious 54 bp tandem stutter mutation resulting in 18 additional amino acids (the NGGLMGYAPGENVPSCSGG red and blue repeats in NM_007771 reference sequence) that would very likely disrupt the C-terminal region of the protein. The repeat is preceded by the substitution of a serine (shown in magenta in the alignment below) for a strictly invariant proline (back to chondrichthyes).
Although this region lies beyond the two main domains and has a complex evolutionary history, phylogenetic comparison to the eight available rodent and lagomorph sequences implies that this change in lab mouse will have serious functional consequences. A mutation in this critical pacemaker gene could plausibly affect lifespan, metabolic disorder and tumor progression; such a change is completely unprecedented in rodents including rat and indeed in vertebrates.
All 14 available transcripts exhibit the same anomaly -- this is not limited to one strain of mouse, not a somatic mutation, not an unfortunate heterozygous allele. The affected ESTs came from C57BL/6J, C57BL/6, C57BL/6J x DBA/2J, 129 FVB/N and embryo, eye, ventricle, thymus, mammary tumor; the affected GenBank NR entries add a keratinocyte cell line Pam. The mouse genome project used C57BL/6J, the most widely used inbred strain according to the Jackson Laboratory:
"Although C57BL/6J is refractory to many tumors, it is a permissive background for maximal expression of most mutations. C57BL/6J mice are resistant to audiogenic seizures, have a relatively low bone density, and develop age related hearing loss. They are also susceptible to diet-induced obesity, type 2 diabetes, and atherosclerosis. C57BL/6J mice are used in a wide variety of research areas including cardiovascular biology, developmental biology, diabetes and obesity, genetics, immunology, neurobiology, and sensorineural research. C57BL/6J mice are also commonly used in the production of transgenic mice. Overall, C57BL/6 mice breed well, are long-lived, and have a low susceptibility to tumors. Primitive hematopoietic stem cells from C57BL/6J mice show greatly delayed senescence relative to BALB/c and DBA/2J. This is a dominant trait. Other characteristics include: 1) a high susceptibility to diet-induced obesity, type 2 diabetes, and atherosclerosis; 2) a high incidence of microphthalmia and other associated eye abnormalities; 3) resistance to audiogenic seizures; 4) low bone density; 5) hereditary hydrocephalus (early reports indicate 1 - 4 %); 6) hairloss associated with overgrooming, 7) a preference for alcohol and morphine; 8) late-onset hearing loss; and 9) increased incidence of hydrocephalus and malocclusion."
Although this distal region is not modelled in any PDB structure as of March 2012, it has been specifically addressed in 4 of the 195 articles on mouse CRY1 or CRY2.
"purified mCRY1/2CCtail proteins form stable heterodimeric complexes with two C-terminal mBMAL1 fragments. The longer mBMAL1 fragment (BMAL490) includes Lys-537, which is rhythmically acetylated by mCLOCK in vivo. mCRY1 (but not mCRY2) has a lower affinity to BMAL490 than to the shorter mBMAL1 fragment (BMAL577) and a K537Q mutant version of BMAL490. Using peptide scan analysis we identify two mBMAL1 binding epitopes within the coiled coil RLNIERMKQIYQQLSRYR and tail regions of mCRY1/2 and document the importance of positively charged mCRY1 residues for mBMAL1 binding."
"mammalian CRY1 and CRY2 are integral components of the circadian oscillator. However, the function of their C terminus remains to be resolved. Here, we show that the C-terminal extension of mCRY1 harbors a nuclear localization signal and a putative coiled-coil domain that drive nuclear localization via two independent mechanisms and shift the equilibrium of shuttling mammalian CRY1 (mCRY1)/mammalian PER2 (mPER2) complexes towards the nucleus. Importantly, deletion of the complete C terminus prevents mCRY1 from repressing CLOCK/BMAL1-mediated transcription, whereas a plant photolyase gains this key clock function upon fusion to the last 100 amino acids of the mCRY1 core and its C terminus. Thus, the acquirement of different (species-specific) C termini during evolution not only functionally separated cryptochromes from photolyase but also caused diversity within the cryptochrome family."
"The mCRY1 and mCRY2 genes are located on chromosome 10C and 2E, respectively, and are expressed in all mouse organs examined. We raised antibodies specific against each gene product using its C-terminal sequence, which differs completely between the genes. Immunofluorescent staining of cultured mouse cells revealed that mCRY1 is localized in mitochondria whereas mCRY2 was found mainly in the nucleus. The subcellular distribution of CRY proteins was confirmed by immunoblot analysis of fractionated mouse liver cell extracts. Using green fluorescent protein fused peptides we showed that the C-terminal region of the mouse CRY2 protein contains a unique nuclear localization signal, which is absent in the CRY1 protein. The N-terminal region of CRY1 was shown to contain the mitochondrial transport signal. Recombinant as well as native CRY1 proteins from mouse and human cells showed a tight binding activity to DNA Sepharose, while CRY2 protein did not"
"genetic screening assay for mutant circadian clock proteins that is based on real-time circadian rhythm monitoring in cultured fibroblasts. By using this assay, we identified a domain in the extreme C terminus of BMAL1 that plays an essential role in the rhythmic control of E-box-mediated circadian transcription. Remarkably, the last 43 aa of BMAL1 are required for transcriptional activation, as well as for association with the circadian transcriptional repressor CRY1"
507 517 527 537 547 557 567 577 587 597
| | | | | | | | | |
CRY1_musMus NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN*
CRY1_ratNor NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYAPGENVPSGGSGG------------------G NCSQGSGILHYAHGDSQQTNPLKQ GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN*
CRY1_criGri NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYTTGENLPSCSGGG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN*
CRY1_spaJud NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYTPGENIPNCSSSG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN*
CRY1_dipOrd NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYAAGDNLPGSSSSG------------------- SCSQGSGILHYAHGDSQQMHLLKQ GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN*
CRY1_hetGla NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYAPGESIPGSSGSG------------------- SCAHGSGILPCAHTDGQQAHLLKP GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD*
CRY1_cavPor HHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLLGYAPGESTPGSGGG-------------------- SCVPGSSSAGVSHCAQGEAPQAPP GRDPAGPGLGGGKRPSQEEDAQSTGHKIQRQSPD*
CRY1_speTri NHEASL NIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMAYAPGENIPGCSSSG------------------- SCTQGSSILHNAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN*
CRY1_oryCun NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNPNGNGGLMGYSPGENIPGCSSSG------------------- SCSQGSGILHYAQGDTQQTQLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN*
CRY1_musMus NHAEASRLNIERMKQIYQQLSRYRGL GLLASVPSNSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN*
CRY1_ratNor .......................... .........P.................GG.G.------------------. ...................NP... ....M.............................
CRY1_criGri .......................... .........P.........TT...L....GG.------------------- S.................A.L... ....M..S...........ETR..D.........
CRY1_spaJud .......................... .........P.........T....I.N.....------------------- S.................A.L... .S..M.H...N.........T..I........T.
CRY1_dipOrd .......................... .........P..........A.D.L.GS....------------------- S.................M.L.... ...M...............S..I........T.
CRY1_hetGla .......................... .........P.............SI.GS.G..------------------- S.AH.....PC..T.G..A.L..P. .NCV.PV...............I...L....TD
CRY1_cavPor H......................... .........P......L......ST.GSGGG-------------------- S.VP..SSAGVS.CAQGEAPQAPP. .DP..P..GG............T.H.I....PD
CRY1_speTri ......--.................. .........P.......A......I.G.....------------------- S.T...S...N.........L.... ...M...............T..I........T.
CRY1_oryCun .......................... .........P.........S....I.G.....------------------- S...........Q..T...QL.... ...M...............T..I........T.
Coiled coil: RLNIERMKQIYQQLSRYR for CRY1_musMus 480-493
478 R e 0.644
479 L f 0.644
480 N g 0.806
481 I a 0.806
482 E b 0.806
483 R c 0.806
484 M d 0.806
485 K e 0.806
486 Q f 0.806
487 I g 0.806
488 Y a 0.806
489 Q b 0.806
490 Q c 0.806
491 L d 0.806
492 S e 0.806
493 R f 0.806
494 Y d 0.375
495 R e 0.375
Full length CRY1 sequences are available for 10 Glires in the cryptochrome refSeq collection:
CRY1_musMus Mus musculus (mouse) NM_007771 CRY1_ratNor Rattus norvegicus (rat) NM_198750
CRY1_criGri Cricetulus griseus (hamster) XM_003505292 CRY1_spaJud Spalax judaei (blind_mole_rat) AJ606298
CRY1_dipOrd Dipodomys ordii (kangaroo_rat) ABRO01202522 CRY1_hetGla Heterocephalus glaber (blind_mole-rat)
CRY1_cavPor Cavia porcellus (guinea_pig) CRY1_speTri Spermophilus tridecemlineatus (squirrel)
CRY1_oryCun Oryctolagus cuniculus (rabbit) CRY1_ochPri Ochotona princeps (pika)
Lost distal exon in placental cryptochrome CRY1
Although cryptochromes are highly conserved in their two main domains, the C-terminal region in CRY1 has a reputation for variability. This is attributable in part to loss of an ancient exon encoding 32 amino acids in placental mammals. However this exon persists in contemporary marsupials, monotremes, birds, alligators, turtles, lizards, snakes and frogs, so its conservation implies a continuing functional role maintained by selective pressure for several hundred million years of tetrapod evolution.
In addition, some distal motifs in CRY1 are compositionally simple, predisposing not only to the replication slippage event described above for mouse but also to smaller indels in the repetitive regions, notably the 2 aa deletional synapomorphy in placentals in GLLASVPSNPNGN--GGFM (the conserved methionine is at position 514 in human) and possibly the loss of proline (P518) in post-tarsier divergence primates.
The exon loss may have preceded in stages, beginning with alternative splicing that skipped it (this conserves reading frame as the ancestral gene ends with three consecutive phase 12 exons). Later, the exon came not to be used at all and thereafter rapidly degenerated to the point it cannot be detected today by blastx of the relevant region in any placental mammal. The exon does not plausibly contribute to the core fold (photolyase and FAD domains) though it could form a better defined structure upon interacting with other proteins.
The functional consequences of exon loss are unknown; the timing matches that of overall collapse of the photolyase family in placentals. (Note the first half of placental evolution -- about 90 myr -- lacks any living representative, so events can pile up there by coincidence.) Possibly when CYT4, Cyt64, DASH and CPD were lost, the remaining two cryptochromes, especially CRY1, compensated for that loss (without however taking up catalytic roles in dna repair), with exon loss somehow contributing adaptively to that adjustment.
The loss of this exon raises certain questions about the use of marsupial model systems to understand CRY1 functionality in mouse (in turn a model system for human). For example, CRY1 of the marsupial Potorous tridactylus would still retain the exon but to date it has not been placed in a CRY1-- mouse. It would also be feasible to insert just the missing exon into an otherwise intact, ectopically expressed rat CRY1 gene, after first disentangling the effects of the mouse expansion in this same region (shown as ^^ below) as well as proline P518 removal. Note the lab mouse expansion somewhat restores length relative to marsupials, but in the wrong place.
CRY1_homSap MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCSSSG <-- lost exon in placentals --> SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN CRY1_ponAbe MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENVPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSGGKRASQEEDTQSIGPKVQRQSTN CRY1_nomLeu MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN CRY1_macMul MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS TENIPGCSSSG SCSQGSGILHYTHGDSQQTHLLKQ GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN CRY1_calJac MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCTSSG SCSQGSGILHCAHGDSQQTHLLKQ GRSSMSTGISGGKRPSQEEDTQSIGPKVQRQSTN CRY1_saiBol MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCTSSG SCSQGSGILHCAHGDSQQTHLLKQ GRSSMSTGLGGGKRPSQEEDTQSIGPKVQRQSTN CRY1_tarSyr MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYSPAENTPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSVGTGLSGGKRPSQEEDPQSIGPKVQRQSTN CRY1_otoGar MKQIYQQLSRYRGL GLLASVPSNPNGN GSFMEYSPPENIPGCSSSG NCSQGSGILHYAPGDGQQPHLLKQ GRSSMGTGLSGGKRPSQEEDMQSVGPKVQRQSTN CRY1_musMus MKQIYQQLSRYRGL GLLASVPSNSNGN^^GGLMGYAPGENVPSCSSSG NGGLGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN CRY1_ratNor MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAPGENVPSGGSGG GNCSQGGILHYAHGDSQQTNPLKQ GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN CRY1_criGri MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYTTGENLPSCSGGG SCSQGSGILHYAHGDSQQAHLLKQ GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN CRY1_spaJud MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYTPGENIPNCSSSG SCSQGSGILHYAHGDSQQAHLLKQ GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN CRY1_dipOrd MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAAGDNLPGSSSSG SCSQGSGILHYAHGDSQQMHLLKQ GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN CRY1_hetGla MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAPGESIPGSSGSG SCAHGSGILPCAHTDGQQAHLLKP GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD CRY1_speTri MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMAYAPGENIPGCSSSG SCTQGSSILHNAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN CRY1_oryCun MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAQGDTQQTQLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN CRY1_oviAri MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSA SCTQGSGILHYAHGDSQQTHLLKQ GRSSTAAGLGSGKRPSQEEDTQSVGPKVQRQSTN CRY1_bosTau MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSNA SCTQGSGILHYAHGDSQQTHLLKQ GRSSTGAGLGSGKRPSQEEDTQSIGPKVQRQSTN CRY1_susScr MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCPQGSGILHYAHGESQQNHLLKQ GRSSTGSGLSSAKRPSQEEDTQSIIGPKVQRQSTN CRY1_ailMel MKQIYQQLSRYRGL GLLASVPANPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGSGLSSGKRPSEEEDTQSIGPKVQRQSTN CRY1_turTru MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGYSSSG SCTPGSGILHYAYGDSQQTHLLKQ GRSSTCTGLSSGKRPSQEEDTQSIGPKVQRQSTN CRY1_equCab MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSLGPGLSSGKRPGPEEDTQGIGPKVQRQSTT CRY1_canFam MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSEEEDTQTISPKVQRQSTN CRY1_myoLuc MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SYAQGSGILHYALGDSQQTHLLKQ GRSSVGTGLSSGKRPSQEEDTQSIGRKVQRQSTN CRY1_pteVam MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGSLHYAHGDCQQTHLLKQ GRSSMGTGLSSGKRPSQEEDMQSIGPKVQRQSTN CRY1_loxAfr MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENTPGCNSSG SCSQGSGILHYVHGDS....LLKQ GRSPTGTGVSSGKRPSQDEETQTLGPKVQRQSTN CRY1_triMan MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSNG SCPQGNGILHYAHRDSQQAHLLKQ GRSPTGTGVSSGKRPSQEEETQSIGPKVQRQSAN CRY1_proCap MKQIYQQLSRYRGL GLLASVPSNPNGN GGLIGYSPGESIPGCSNSG SCSQGSGILHYAHGDSQQAHLLKP GRSPMGTGISSGKRPSQEEETQTVGRKVQRQSTN CRY1_echTel MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENTTGCSSGG GCPPGNGILHYAHGDSQQAALLKQ GRSPLGTGLSSGKRPSQEEDTQSVGPKVQRQSSN CRY1_dasNov MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAPGENILGCSSSG SCAQGSSILHYAHGDNQQTHLLKQ GRSSMGTVLSSGKRPSQEEETQSIGPKVQRQSTN CRY1_choHof MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG sCSQGSGILHYAHGDSQQTHLLKQ GRSSMGIGLSSGKRPSQEEETQGIGPKVQRQSTN CRY1_monDom MKQIYQQLSRYRGL GLLASVPSNPNGN GSLMAYTPGENIPGCSSGG GAPVGASDGQIL..QACVLPEPPTGTSGVQQP GYSQGSGISHYSHEDSQQAYMLKQ GRSSL..GVGGGKRPRQEEETQSINPKVQRQSTN CRY1_macEug MKQIYQQLSRYRGL GLLASVPSNPNGN GSLMGYTTGENIPTCSSSGG GAPAGASDGQIL..QACVLPEPPTGTSGVQQP GGYSQGGISHYSHEDSQQAYVLKQ GRNSL....GGGKRHRQEEETQSIGSKMQRQSVN CRY1_sarHar MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYTSGENGPACNSGG GAPVGASDGQIL..QSCALPEPPAGASCIQQS GYSQGSGISHYSHEDSQQAYILKQ GRSSL....SGGKRPRQEEETQSVGPKVQRQSVN CRY1_triVul MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAPGENIPACSSSGG GAPAGVGDGQIL..QACALPEPPTGASGVQQP GYSQGSGISHYAHEDSQQAYMLKQ GRSSL...SGGGKRHRQEEEAQSIGPKMQRQSVN CRY1_ornAna MKQIYQQLSRYRGL GLLASVPSNPNANGSGGLMAYSPGENIPGCSSGGG GVQMGASESHLL..QTCVLGESHLGPSGIQQQ GYCQGSGVLYYANGE....SHLTQ GRSSLTPGLSGGKRPCQEEESQSIGPKVQRQSTD CRY1_tacAcu MKQIYQQLSRYRGL GLLASVPSNPNANGSGGLMAYSPGENIPGCSSGG GAQIGASESHLL..QTCVLGESHLGPSGIQQQ GRSSLTPGLSGGKRHCQEEESQSIGPKVQRQSTD CRY1_galGal MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMSFSPGESISGCSSAG GAQLGTGDGQTVGVQTCALADSHTGGSGVQQQ GYCQASSILRYAHGDNQQSHLMQP GRASLGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_melGal MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMSFSPGESISGCSSAG GAQLGTGDGQTVGVQSCALGDSHTGGNGVQQQ GYCQASSILRYAHGDNQQPHLMQP GRASLGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_eriRub MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGTGDGHTV.VQSCTLGDSHSGTSGIQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_sylBor MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGAGDGHSV.VQSCALGDSHTGTSGVQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_taeGut MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGTGDGHSV.VQSCALGDSHTGTSGIQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_parWeb MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGTGDGHSV.VQSCALGDSHTGTSGIQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_allMis MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGENVSGCGSTG GAQMGSSDGHTVSVQPCALGESHGGSNGIQQQ GYFQASSILHFPHGDDQQSHLLQQ GRTSLSSGISAGKRPNPEEETQSIGPKVQRQSTN CRY1_anoCar MKQMYQQLSRYRGL GLLASVPSNGNGNGNGGLMGYSTGENIPGCTNTN GSQMGMNEGHIGNVQACTMGESHTGTSGIQQQ GYSQGSGILLYSHGDNQKTHSAQK GRISLGTGVCTGKRPSPEVETQSVGPKVQRQSSN CRY1_podSic MKQIYQQLSRYRGL GLLASVPLNGNGNGNGGLMGYSTGENIPGCTNTN GSQMGTNEAHTGSVQTCTLGESHTGTSGIQQQ GYPQGSDILHYAHGEGQKTHLIQQ GRASLVAGVCTGKRPNPEEETQSIGPKVQRQSSK CRY1_pytMol MKQIYQQLSRYRGL GAQMGTSEGHTGNVQACTLGETHTGTSGIQQQ GYSQGNSGILHYAHGDSQKTLLMQ GRTSLSVGVCTGKRPNPEEGIQSIGPKVQRQSSN CRY1_chrPic MKQIYQQLSRYRGL GLLATVPSNPNG..NGGLMGYSPGENISGCSSAS GAQMGSNDGHTVGVQTCSLEDSHAGSSGIQQH GYSQGNSIVHYAQGDHQQSHLLQQG GRTVST GISTGKRPNPEKETQSIGPKVQRQSTN CRY1_xenTro MKQIYQQLSRYRGL GLLASVPSNPNGNGNGGLMSYSPGESMSGCSNNG GGQMGVNEGSSASNPNANKGEVHPGTSGLQ.. GYWQGSSILHYSHSDSQQSY LMQ ARNPLHSVVSSGKRPNPEEETQSIGPKVQRQSSH CRY1_xenLae MKQIYQQLSRYRGL GLLASVPSNPNG..NGGLMSYSPGESMPGCSNNG GGQMGAIEGSSASNPNPNQGEVLPGTSGLQ.. GYWQGSSILHYSHSDNQQSY LMQ ARNPLHSVVSSGKRPNPEEETQSVGPKVQRQSTH CRY1_latCha MKQIYQQLSRYRGM GLLASVPSNPNGNGGLGCSLAENIPVCNSAA GAQMGGDDGHKVSVLAYTQGDSRAGEIEMQQQ CRY1_danRer MKQIYQQLSCYRGL GLLAMVPSNPNGNGENSTSLMGFQTGDMTKEVTTPS GYQMPPTSQGEWHGRTMVYSQGDQQTSSIMTSQ GFGNNGSTMCYRQDAQQIT GRGLHSSIIQTSGKRHSEESGPTTVSKVQRQCSS
When the terminal four exons of CRY1 are compared to those of its nearest homolog class CRY2, no similarity can be detected beyond the first 8 residues of the tenth exon of CRY1 (2 GLLASVPS) vs the tenth and penultimate exon of CRY2 (2 CLLASVPS). This raises the question of what the last common ancestor had for terminal exons and -- given no counterpart in CRY4, CRY64, DASH, or CPD -- where they originated. Note that last two exons of CRY2 are strongly conserved in their own right, proving a separate conserved functionality from that of CRY1. Since the tenth exons begin homologously and end after a similar length with a phase 1 splice donor, these exons could possibly be homologous their entire length, just diverged distally. The eleventh exon of CRY2 could then correspond (allowing for total sequence divergence) to any of exons 11-13 in CRY1.
CRY2_homSap CLLASVPSCVEDLSHPVAEPSSSQAGSMSSA GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDA CRY2_panTro ............................... .............................................. CRY2_gorGor ...........................V... .............................................. CRY2_ponAbe ...........................V... .............................................. CRY2_rheMac ...........................VN.. ...............................K.............. CRY2_papHam ...........................VN.. ...............................K.............. CRY2_calJac ............................... .............................................V CRY2_micMur ..............................T .................................T............ CRY2_musMus ....................G......I.NT ...A.S.....................T.....T.M..Q.PA...S CRY2_ratNor ....................G......I.NT .....S...........................T.M.AQ.P....S CRY2_criGri ...........................I.NT .S...S...........................T.M.AQ.PQT... CRY2_spaJud ........................P..ITNT .....ST..........................T...A..PA.... CRY2_cavPor .....................L.....ST.T ......G.................................P..... CRY2_hetGla ....................TL.....S..T ...S..D..............................A..PT.... CRY2_speTri ....................G......I..T .....S..Q..................................... CRY2_oryCun ...........................V.G. A..................................V........AV CRY2_turTru .........M....N...........G.... ................G.................G..PS..L...V CRY2_bosTau ..............N.......I....S..V ......G.................G..........SLPS....RGV CRY2_susScr ..............N............V.A. .....................................PT...GR.V CRY2_canFam ..............N.........T...... ..........................................CR.V CRY2_ailMel ..............N.........T...... .....................................A..P..R.V CRY2_myoLuc .........M....N......L..T...... ..K..................................AT....R.V CRY2_pteVam .............NN.........T...NN. .....................................A.....R.V CRY2_loxAfr ..............S............SN...........T........................K..G.......V CRY2_proCap ..............N........P..H.....L................................K..G.....T.. CRY2_choHof ..............N....................V............................T...........V CRY2_macEug .........M....S.M..T.MG....V..T..K...CS..........T..ASR..H.....M.A..V...A.--- CRY2_monDom .........L....S.MV.A.LG...AV.GP.LK...CS..........T..A....H.......R..GS..AG..V CRY2_ornAna ..............SAA..SGLG....NI.TA...-.P.............GL.....C..PK..GR.G..P.GE.. CRY2_galGal ..............G..TDSAPG.-..ST.TAV.LPQ.DQ......H.G...LCT...Y...K.TG..A..I.G.SS CRY2_taeGut ............I.G..PDSA.G.-.CST.TAV.LSQAEQ......H.G....CS...Y...K.TG...S.ISG.SL CRY2_allMis G........A....G..TD.A.V.-.CST.TALK.SQ..Q......H.GI..MCT.D.Y...K.TG.HG..I...SL CRY2_anoCar .........M....N...DT...H-.NCIGTAS.QTHC.QT.....HDVVQ.YK-...Y...K.VASQFA.N.RQEL CRY2_xenTro .I.......M...GG.M.DS.QNISEAGKM.P.SHTSGESVLAAQYTAGI--------------------------- CRY2_ranCat .I......S.....G.M.D.A...Q..SD---.A.RLCAVD.....H.DLD----G..C.K..LQCVQEM.RAA..F
A distal alternative splice in avian cryptochrome CRY1 not used for magnetosensing
Bird CRY1 presents a further curious situation with respect to the terminal extentional exons of CRY1: an alternative splice in exon 11, more accurately a failure to consistently recognize its splice donor (or the following acceptor) leading to translational read-out of the mRNA to the first stop codon following. The vast majority of such events are misinterpreted artifacts -- the transcript simply terminated too soon, providing no splice acceptor and consequently no way for the intervening intron to be removed.
However here two types of transcripts were found in both Erithacus rubecula (Euro robin) and Sylvia borin (warbler) in targeted experiments by separate research groups. The long form, called there CRY1A, has the usual four terminal exons of vertebrates; the short form, CRY1B, provides 25 new amino acids before a stop codon.
Comparative genomics is capable of resolving artifact, coincidence, and functionality. First note that GenBank chicken transcripts contain a supportive entry (BU143111) that surfaced in a large transcript program not focused on particular genes. Secondly, the read-out of exon 11 in species without transcripts is implied by highly conserved amino acid sequence. While a certain amount of nucleotide conservation might be expected because splice sites are larger than just GT-AG, the intron could contain enhancers or other conserved non-coding elements (of this or an adjacent gene), and conservation can persist for a time via coldspots and failure of a mutation to fix in a population, the conservation here at the protein level significantly exceeds what these factors could contribute. Gray shows species lacking conservation; blue conserved amino acids within birds.
This conservation was in fact already established in the early diverging lineage of duck + chicken but deteriorated as shown by early stop codons and distal sequence restored by a shared frameshift (lower case below) in gallinaceous birds. However nothing resembling the bird read-out sequence is found in alligator, turtles, snakes, lizard or frog in any reading frame. Thus, the simplest scenario is it arose early in bird evolution and so is restricted to them. (Here we await an ostrich genome to see if the event took place already in Paleognathae.)
If the selective pressure truly operates on the level of amino acids here and if the region is not a mutational cold spot, then relatively higher levels of variation should be observed at redundant codon positions within the DNA, eg 3rd position in 4-codon amino acids. However, by collecting the DNA sequences, it emerges that synonymous changes do not noticably predominate (after minimizing events needed by branching of the avian phylogenetic tree and ignoring the breakdown of this region in duck, chicken, turkey) nor do non-synonymous changes conserve amino acid properties. This argues strongly that the region has not been conserved by selection on amino acid sequence but rather selection on the underlying DNA.
Exon 11 read-out of CRY1 genSpp transcript support of read-out (or wgs accession) GISKNTF* monDom Monodelphis domestica (opossum) GISDNTFLTLTQSRGSLGIPHQS..* macEug Macropus eugenii (wallaby) GISQNTFESVRLS* sarHar Sarcophilus harrisii (tasmanian_devil) GISKLFSFIFKNTFN* ornAna Ornithorhynchus anatinus (platypus) GRSSLTPGLSGGKRHCQEEESQN..* tacAcu Tachyglossus aculeatus (echidna) GIMAVPVCRGSPNPCNYRKPDKTSK* taeGut Taeniopygia guttata (finch) GIMAVPVCRGSPNACNYGKPDKTSK* eriRub Erithacus rubecula (robin) AY585717 GIVAVAVCRGSPNPCNYGKPDKTSE* sylBor Sylvia borin (warbler) DQ838738 GIMAVPVCRGSSNPCNCGKTDKTSK* melUnd Melopsittacus undulatus (parakeet) GIMAVPVCRGSPNPCNYGKPDKTSK* zonAlb Zonotrichia albicollis (sparrow) (ARWJ01011250) GIMAVPVCRGSPNPCSYGKPDKTSK* pseHum Pseudopodoces humilis (ground-tit) (ANZD01003613) GIMAVPVCRGSPNPCNCGKPDKTSK* falChe Falco cherrug (falcon) (AKMU01039249) GIMAVPVCRGSSNPCNCGKTDKTSK* araMac Ara macao (scarlet macaw) (AMXX01097310) GIMAVPVCRGSPNPCTCGKTD*TSK* colLiv Columba livia (rock pigeon) (AKCR01045195) GMTGVLVCRGSPGSHNYGKKDKT*K* anaPla Anas platyrhynchos (duck) GIVGVPICRGSADLCN*GKKdkt*k* galGal Gallus gallus (chicken) BU143111 GTVGVPICRGSANWYK*GKKdkt*k* melGal Meleagris gallopavo (turkey) KCLQRICKFL*LKFSKY.. . allMis Alligator mississippiensis (alligator) KNVFKEVLAILEIVKIP... pelSin Pelodiscus sinensis (turtle) II*QIKCVQRHFSRFLK... chrPic Chrysemys picta (turtle) IIQQIKCVQRGSRYS*NC*... apaSpi Apalone spinifera (turtle) YCQGNSGILHYAHGD.. . croHor Crotalus horridus (snake) KTL*KSLI*YSS*NTACVHG... anoCar Anolis carolinensis (lizard) GKLAAPLISVSSIIGVFHTHEPQ... xenTro Xenopus tropicalis (frog)
The data thus support the notion of birds having evolved a distinct function for the read-out option at exon 11 -- with nothing comparable in the immediate outgroups (crocodile, turtle) or mammals. While more bird genomes are expected in 2014, these don't include basal Paleognathae such as ostrich and other non-passerine species needed to check read-out conservation patterns conform to the avian phylogenetic tree. However the more common CRY1 form retaining the usual extra exons is also conserved in birds (as seen in the earlier alignment of this region).
It has been reported that only the long form is expressed in SWS1 opsin cones of retinas of migrating passerine birds where it detects the earth's magnetic field via electron spin pairing in tryptophan and FAD. The short form is apparently expressed in the ganglion cell layer where it may represent an adaptive synapomorphy for a large part of the avian tree.
Note the vertebrate ciliary opsin SWS1 has no counterpart in fruit flies. Since invertebrate cryptochromes correspond poorly too, Drosophila is completely unsuitable here as model species. However dipterans do have two rhabdomeric opsins with peak sensitivity in the ultraviolet, RH5 and RH7, with characteristic lysine at position 90 and a short third cytoplasmic loop. RH5 is located in the larval Bolwig organ; RH7 has not been assigned an anatomical site but may be located in antenna. Conceivably analogous co-expression with a different cryptochrome could couple these photosensing systems too.
Human CRY2, also strongly expressed in retina but not so specifically in cone cell outer segment membranes, can reportedly replace the invertebrate cryptochrome CRY1B in the drosophila magnetic field detection system (as can insect CRY1A). The final exon of human CRY2 bears no clear relationship to the terminal exons of CRY1 nor to the read-out exon 13 of birds and is only secondarily related homologically to invertebrate CRY1B cryptochromes.
The alignment below shows very limited distal homology between tetrapod CRY2 and invertebrate CRY1B. The primary sequence correspondence does not even extend to the coiled coil region of vertebrate CRY2 which is not always evident in invertebrate CRY1B, much less to distal exons of CRY2 (indicated by spacing). On the flip side, just distal to the its missing coiled coil, invertebrate CRY1B has a conserved 16 residue motif known to imitate a damaged DNA base with a tryptophan; vertebrate CRY2 is itself conserved here but not relative to the CRY1B spoof motif and contains no counterpart to the key aromatic residue.
Amino acids are shown only when 50% or more conserved within the total alignment column:
CRY2_homSap RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_rheMac RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_calJac RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_micMur RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_musMus RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_cavPor RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_oryCun RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_bosTau RYLP.LK.FPSRYIYEPWNAPES.QKAAKC.IGVDYP.PIVNHAE.SRLNIERMKQ.YQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_ailMel RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_pteVam RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDL..P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_loxAfr RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_choHof RYLP.LK.FPSRYIYEPWNAPES.QKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q.G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_monDom RYLP.LK.FP.RYIYEPWNAPE.VQKAAKCIIGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSC.EDLS.P.......Q.G............ ........SPKRK.E........E..KRA.V......E......
CRY2_ornAna RYLP.LK.FPSRYIYEPWNAPESVQKAAKC.IGVDYP.PIVNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.........Q.G............ ........SPKRK.E........EL.KR..V......E......
CRY2_galGal RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.P.VNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLS.P.......Q-G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_taeGut RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.P.VNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSCVED.S.P.......Q-G............ ........SPKRK.E........EL.KRA.V......E......
CRY2_allMis RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.P.VNHAE.SRLNIERMKQIYQQLSRYRGL .LLASVPSC.EDLS.P.......Q-G............ ........SPKRK.E.........L.KRA.V......E......
CRY2_anoCar RYLP.LK.FPSRYIYEPWNAPESVQKAAKCIIGVDYP.P.VNHAE.SRLNIERMKQIYQQLSRYRGL CLLASVPSC.EDLS.P........-G............ ........SPKRK.......-..EL.KRA.V......E......
CRY2_ranCat RYLP.LK..PSRYIYEPWNAPESVQK.AKCI.GVDYP.P.VNHAE.SRLNIERMKQ.YQQLSRYRGL C.LASVPS.VEDLS.P.......Q.G...---...... ........SPKRK.E....----EL.K.A........E......
PPHCRPSNEEEVRQFMWLP: helix conserved within CR!B whose tryptophan spoofs damaged DNA base
CRY1B_strPur RYLP.LK..P.RY..EPW.AP..VQ..AKCI.G.DYP.P.V.H...S..N.E.M......L.... ......S....V.......
CRY1B_lytVar RYLP.LK..P.RY..EPW.AP..VQ..AKCI.G.DYP.PIV.H...S..N.E.M......L.... ......S....V.......
CRY1B_parLiv RYLP.LK..P.RY..EPW.AP..VQ..AKCI.G.DYP.PIV.H...S..N.E.M......L.... ......S....V.......
CRY1B_aplCal RY.P.LK..P..Y..EPW.AP...Q....CIIG.DYP.P.V.H...S......M..I.--..... ...........V..L....
CRY1B_octVul .Y.P.LK..P..Y...PW.AP...Q..A.CIIG.DYP.PIV.H...S..N...M......L.... ...........V.......
CRY1B_craGig RYLP.LK..P.RY..EPW.AP..VQ..AKCI.--DYP.P.V.H...S...I..MK.....L.... ......S........S...
CRY1B_acyPis RY.P.LK..P....YEPW..PESVQK...CIIG.DYP..IV.H...S..N...M........... ......S....V.......
CRY1B_dapPul RY.P.L..F...YI.EPW.AP...Q..A.CIIG.DYP...V.H.E....N.E.MK...Q..-... ......S..S.V.......
CRY1B_diaNig RY.P.LK..P..Y.YEPW.AP..VQ..A.CI.G.DYP..I..H...S..N...M..I.-...... ......S............
CRY1B_danPle RY.P.L...P..YIYEPW.AP..VQ.AA.C.IG.DYP.P.V.H......N...M....-.L.... ......S....V....... *
CRY1B_mamBra RY.P.L...P..YIYEPW.AP...Q..A.CIIG.DYP.P.VNH......N...MK...-...... ......S............ *
CRY1B_helArm RY.P.L...P..YIYEPW.AP..VQ..A.C.IG.DYP.P.VNH......N...MK...-...... ......S............ *
CRY1B_bomMor RY.P.L...P..YIYEPW.AP..VQ..A.CIIG.DYP.P.VNH......N...M....-.L.... ......S............
CRY1B_droMel .Y.P.L...P.....EPW......Q....C.IGV.YP..I.........N...MK.....L.... .....S....V.......
CRY1B_anoGam RYLP.L...P.....EPW.A....Q....C.IG..YP.P.V..A..S..N...M......L.... ......S............
CRY1B_neoBul .Y.P.L...P..YI.EPW..P...Q....C.IG..YP............N...M......L.... ......S............
CRY1B_bacCuc .Y.P.L...P..YI.EPW..P...Q....C.IGV.YP..IV..A..S..N...M....Q.L.... ......S....V.......
The graphic above shows separate predictions for distal coiled coil prediction for each of 17-20 concatenated vertebrate distal sequences for each of the eight cryptochromes and photolyases that occur in bilaterans. The species are presented in phylogenetic order left to right (ie as listed in refSeq collection). Invertebrate CRY1B clearly does not have the domain not consistently present. The three largest CRY1B peaks (indicated by asterisks in the alignment) are all lepidoptera; the Drosophila protein does not contain this structural motif motif. Given the duplications of the gene tree, the coiled coil domain probably arose once in an early ancetral cryptochrome but was been lostin some species groups such as dipteran flies. The new crystallographic structure PDB:3TVS confirms the lack of coiled-coil motif in CRY1B.
C-terminal deletions of the Drosophila cryptochrome have been extensively studied. While informative, the poor distal correspondence to mammalian cryptochromes makes carry-over of such results -- annotation transfer -- to mammalian cryptochromes a dubious proposition since key sequence motifs used in signalling are not present in the C-terminus of this model species (and vice versa!).
Evolutionary origin of the α/β photolyase fold
Comparative genomics (lots of phylogenetically structured primary sequences) synergizes strongly with three-dimensional structural determinations, the former providing the conserved so presumably functional regions and the latter their structural interpretation. In the case of cryptochrome and photolyase structures, it is quite important that full length proteins be considered because N- and C-terminal extensions can provide the very properties that distinguish an orthology class from its paralogs.
However the N-terminus can also be evolving haphazardly from compositionally simple sequence, be quickly trimmed from newly synthesized protein by cellular proteases, lack assignable structure in a crystal, and be functionally irrelevant. Similarly, an extended C-terminus can represent meaningless run-out through junk DNA to the first stop codon encountered. In these situations, sequence conservation will not extend beyond the genus level (a few million years).
The overall fold of all cryptochromes and photolyases is basically the same: two distinct globular domains held together in part by a long lasso thrown out by the second domain. The amino terminal domain lies at the far end of the protein from the DNA binding site. It consists of a 5-stranded parallel beta sheet sandwiched between 4 alpha helices whose axes are anti-parallel to the sheet. The strands are ordered 32145 with the helices alternating in position. The first two helices form the top of a sandwich, the second two the bottom with strand 3 transitioning. The binding site for the antenna molecule is at the edge of the sandwich between the two domains; it is not intimately associated with the helices or central strands themselves but rather with helix-strand turns.
Surprisingly, the βαβαβαβαβ pattern of alternating helix and sheet with the outer layer of helices packing against the central core in 32145 order is not necessarily indicative of evolutionary relatedness but instead a default supersecondary structure for cytoplasmic proteins. Its inevitability was first explained by C. Chothia et al in 1977 as complementarity between the right handed twist of a beta sheet and the rotating i+4 ridge of helix side chains (due to its 3.4 residues per turn) -- close packing of side chains in the hydrophobic core is entropically favorable and so the same basic fold commonly arises regardless of evolutionary relatedness.
In terms of evolutionary characters, the fold is homoplasic, having arisen many times independently rather than having descended from a single ancestral fold. (The same is true for the more complex TIM beta barrel, an eightfold repeat of the βαβ pattern found in 15 gene families with no bona fide sequence homology.)
With photolyases, coincidence extends to antenna molecules, some of which are similar to the NAD of the Rossmann fold homology group. However the binding site location is different. Photolyases do not have a stand-alone pocket in the α/β amino terminal domain but utilize portions of the following fold (not to mention a the composite route of excitation transfer). In fact, it's not clear that the antenna binding site is fixed in all homologs. Further, there is no conservation of key residues nor any convergence of ancestral sequences to homology.
In summary, the photolyase fold is not homologous to the classic nucleotide binding fold. Searching PDB with a given protein to find related fold structures thus requires careful overall evaluation of candidates to ensure actual evolutionary relatedness. While the α/β domain draws a blank, the resemblance found by Dali in the catalytic domain of primases and 4Fe-4S photolyases to previously studied photolyases/cryptochromes is beyond coincidence.
Many large eukaryotic proteins are chimeric, having arisen from genetic fusions of mobile domains. Alternatively, certain common folds have arisen independently in situ in different gene families rather than been shuffled in. Initially, modular proteins fold as their constituent pieces, with less substantive interaction in the final product than an ordinary non-covalent heterodimer might have, but over time more intimate structural codependencies evolve. Photolyase may once have been a heterodimer of a small redox protein that passes antenna excitations to a larger catalytic subunit, becoming later a genetically fused modular protein, but today the α/β amino terminal domain is quite integrated with the all-alpha domain -- the long lasso holding them together is preceded by the essential protrusion loop that binds DNA in the second domain. (This connector region was reported attached by a reported disulfide but the cysteines are not conserved and cytoplasmic proteins generally lack disulfides in vivo.)
Separating the two domains with limited trypsin digestion (or better, genetic methods) has not yet been attempted and might not be feasible with retention of functionality if the domains are structurally interdependent. This could explain why cryptochromes that lack antenna molecules have not lost the α/β domain under the evolutionary principle of 'use it or lose it'. That is, if no selective pressure persisted in this region, what weeds out structurally deleterious mutations or keeps a large N-terminal deletion from being fixed? Not only has CRY1B of drosophila retained the antenna pocket, but it also exhibits very high levels of conservation of individual amino acids and small motifs beyond what is needed for folding and stability.
The main alternatives to structural integration are (1) evolution has not caught up yet with very recent loss of antenna molecule in CRY1B and other cryptochromes, (2) an unsuspected, undetected new antenna molecule is present and important in vivo which maintains selective pressure and (3) a signalling or magnetosensing role for the α/β domain, either from direct participation in a conformational shift or through homodimeric or heterologous binding to other proteins. The first possiblity can be rejected because seemingly antenna-less cryptochromes fall into different groups, each of long standing. The second seems inconsistent with careful experimentation, yet reconstitution experiments are no better than the antenna molecules included, with the very recent discovery of lumazine casting further doubt on the completeness of that set. The third is a distinct possibility yet does not seem sufficent to provide the level of conservation observed.
Taking phylogenetically distributed representatives from each cryptochrome/photolyase class (excluding 4Fe-4S photolyases and primases), the alignment below shows the regions of conservation within the α/β domain. While it is easy and informative to align all 250 sequences, to avoid excess display only 4 of each orthology class are shown. However the full set of sequences was separately aligned to determine conservation at the 70% level, again with key species (experimental models and those with PDB structures) shown. It can be seen immediately that universally conserved residues do not correlate particularly with secondary structure (even though that is strongly conserved).
10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170
| | | | | | | | | | | | | | | | |
bbbbbb aaaaaaaaa bbbbbbbbb aaaaaaaaaaaaaaaaaaa bbbbbb aaaaaaaaaa bbbbbbbb aaaaaaaaaaaa bbbbbbb
CRY1_homSap MGVNAVHWFRKGLRLHDNPALKECIQGAD-TIRCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFKEWN-ITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDKIIELNGGQPPLTYKRFQTLISKMEPLEIP
CRY1_musMus MGVNAVHWFRKGLRLHDNPALKECIQGAD-TIRCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFKEWN-ITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDKIIELNGGQPPLTYKRFQTLVSKMEPLEMP
CRY1_galGal MGVNAVHWFRKGLRLHDNPALRECIRGAD-TVRCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFKEWS-IAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDKIIELNGGQPPLTYKRFQTLISRMEPLEMP
CRY1_xenTro MGVNAVHWFRKGLRLHDNPALRECIQGAD-TVRCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFKEWK-ITKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDKIIELNGGQPPLTYKRFQTLISKMDPLEIP
CRY2_homSap DSASSVHWFRKGLRLHDNPALLAAVRGAR-CVRCVYILDP------WFAASSSVGINRWRFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFKEWG-VTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDRIIELNGQKPPLTYKRFQAIISRMELPKKP
CRY2_musMus DGASSVHWFRKGLRLHDNPALLAAVRGAR-CVRCVYILDP------WFAASSSVGINRWRFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFKEWG-VTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDRIIELNGQKPPLTYKRFQALISRMELPKKP
CRY2_galGal GFCRSVHWFRRGLRLHDNPALQAALRGAA-SLRCIYILDP------WFAASSAVGINRWRFLLQSLEDLDNSLRKLNSRLFVVRGQPTDVFPRLFKEWG-VTRLTFEYDSEPFGKERDAAIIKLAKEAGVEVVIENSHTLYDLDRIIELNGNKPPLTYKRFQAIISRMELPKKP
CRY2_xenTro PSVSSVHWFRKGLRLHDNPALLSALRGAN-SVRCVYILDP------WFAASSSGGVNRWRFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFKEWG-VSRLTFEYDSEPFGKERDAVIMKLAKEAGVEVVVENSHTLYDLDRVIELNGHSPPLTYKRFQAIISRMELPRRP
CRY1A_triCas QDKHMVHWFRRGLRLHDNPSLREGLKGAR-TFRCVFVLDP------WFAGSSNVGINKWRFLLQCLEDLDRSLRKL-SRLFVIRGQPADALPKLFKEWG-TTALTFEEDPEPFGGVRDHNLTTLCQELGISVVQKVSHTLYHLQDIIDRNGGRAPLTYHQFLAIIACMGPPPQP
CRY1A_bomImp MGKHTVHWFRKGLRLHDNPSLREGLTGAT-TFRCVFVLDP------WFAGSTNVGINKWRFLLQCLEDLDCSLRKLNSRLFVIRGQPADALPKLFKEWG-TTNLTFEEDPEPFGRVRDHNISALCKELGISVVQKVSHTLYKLDEIIERNGGKPPLTYHQFQNVVASMDPPEPS
CRY1A_nasVit MKKHTVHWFRKGLRLHDNPSLREGLAGAS-TFRCVFVLDP------WFAGSANVSINKWRFLLQCLEDLDRSLHQLNSRLFVIRGQPADALPKLFREWG-TTSLTFEEDPEPYGRVRDENITTLCKELGITVVQRVSHTLYKLDEIIEKNGGKPPLTYHQFQNVIARMDPPEYP
CRY1A_anoGam RDKHTVHWFRKGLRLHDNPALREGLRGAR-TFRCVFIIDP------WFAGSSNVGINKWRFLLQCLDDLDRNLRKLNSRLFVIRGQPADALPKLFKEWG-TTCLTFEEDPEPFGRVRDHNISEMCKELGIEVISAASHTLYNLERIIEKNGGRAPLTYHQFQAIIASMDAPPQP
CRY1B_strPur PGGACIHWFRHGLRLHDNPALLEGMTLGK-EFYPVFIFDN------EVAGTKTSGYNRWRFLHDCLVDLDEQLKAAGGRLFVFHGDPCLIFKEMFLEWG-VRYLTFESDPEPIWTERDRRVKALCKEMKVECIERVSHTLWNPDIIIEKNGGTPPITYSMFMECVTEIGHPPRP
CRY1B_octVul KQKIAVHWFRHGQRLHDNPALLDALKDCD-EFYPVFIFDG------EVAGTKLCGFNRWRFLLENLKDLDESFSEYGGRLYTFQGKPVEVFANLQNEWG-ITHITAEIDPEPIWQERDDAVKEFCQKSGIKCDFFNSHTLWDPKRLLKKNGGTPPLTFELFQLVTSSLGPPPRP
CRY1B_danPle MLGGNVIWFRHGLRLHDNPSLHSALEDASSPFFPIFIFDG------ETAGTKMVGYNRMRYLLEALNDLDQQFRKYGGKLLMIKGRPDLIFRRLWEEFG-IRTLCFEQDCEPIWRPRDASVRALCRDIGVSCREHVAHTLWNPDTVIKANGGIPPLTYQMFLHTVEIIGNPPRP
CRY1B_droMel TRGANVIWFRHGLRLHDNPALLAALKDQGIALIPVFIFDG------ESAGTKNVGYNRMRFLLDSLQDIDDQLQDGRGRLLVFEGEPAYIFRRLHEQVR-LHRICIEQDCEPIWNERDESIRSLCRELNIDFVEKVSHTLWDPQLVIETNGGIPPLTYQMFLHTVQIIGLPPRP 3TVS
CRY4_galGal MRHRTIHLFRKGLRLHDNPALLAALQSSE-VVYPVYILDR------AFTSSMHIGALRWHFLLQSLEDLRSSLRQLGSCLLVIQGEYESVVRDHVQKWN-ITQVTLDAEMEPFYKEMEANIRGLGEELGFQVLSLMGHSLYNTQRILELNGGTPPLTYKRFLRILSLLGDPEVP
CRY4_xenTro MPHRTIHIFRKGLRLHDNPTLVTALETSD-VVYPVYILDR------NFTSSSVIGSKRWNFFLQSIEDLHCNLQKLNSCLFVIQGDYERVLREHVEKWN-ITQVTFDLEIEPYYKGLDERIRAMGQELGFEVVSMVAHTLYDIKKILALNCGKPPLTYKNFLRVLSMLGNPDKP
CRY4_latCha MTHRTIHIFRKGLRLHDNPILLAALEFSR-VVYPVYILDR------KLESGVIIGALRWRFILQSLEDLHRNLVKLNSRLFVIQGDYEQILREYVQKWT-ITQVTFDTEIEPFYKEMDKKVRLMGKEMGFTVLFSVAHALYDVARIVENNGGQPPLTYKKFLHVLSKLGDPERP
CRY4_danRer MSHRTIHLFRKGLRLHDNPSLLGALASSS-ALYPVYVLDR------VFQGAMHMGALRWRFLLQSLEDLDTRLQAIGSRLFVLCGSTANILRELVAQWG-ITQISYDTEVEPYYTRMDKDIQTVAQENGLQTYTCVSHTLYDVKRIVKANGGSPPLTYKKFLHVLSVLGEPEKP
CRY64_xenTro KHNSTIHWFRKGLRLHDNPALLAAMKDCA-ELYPIFILDP------WFPRNMKVSVNRWRFLIEALKDLDENLKKINSRLFVVRGKPTEVFPLLFKKWK-VTRLTFEVDTEPYSRQRDADVEKLAAEHNVQVIQKVSNTLYAIDRIIAENNGKPPLTYVRFQTVLALLGPPKRP
CRY64_danRer SHNTTIHWFRKGLRLHDNPALIAALKDCR-HIYPLFLLDP------WFPKNTRIGINRWRFLIEALKDLDSSLKKLNSRLFVVRGSPTEVLPKLFKQWK-ITRLTFEVDTEPYSQSRDKEVMKLAKEYGVEVTPKISHTLYNIDRIIDENNGKTPMTYIRLQSVVKAMGHPKKP
CRY64_droMel QRSTLVHWFRKGLRLHDNPALSHIFTGKY-FVRPIFILDP------GILDWMQVGANRWRFLQQTLEDLDNQLRKLNSRLFVVRGKPAEVFPRIFKSWR-VEMLTFETDIEPYSVTRDAAVQKLAKAEGVRVETHCSHTIYNPELVIAKNLGKAPITYQKFLGIVEQLKVPKKV 3CVU
CRY64_danPle KVASVIHWFRLDLRLHDNLALRNAINRKQ-ILRPIYVIDP------DIKNWMRVGCNRLRFLFQSLKNLDTSLRKINTRLYVIKGKAIECLPKLFDEWH-VKFLTLQVDIDADLVKQDEVIEEFCEANNIFVVKRMQHTVYDFNSVVKKNNGSIPLTYQKFLSLVSDVQVKDKI
CRY1C_araTha TGSGSLIWFRKGLRVHDNPALEYASKGSE-FMYPVFVIDP------HYPGSSRAGVNRIRFLLESLKDLDSSLKKLGSRLLVFKGEPGEVLVRCLQEWK-VKRLCFEYDTDPYYQALDVKVKDYASSTGVEVFSPVSHTLFNPAHIIEKNGGKPPLSYQSFLKVAGEPSCAKSE 3FY4
CRY1A_araTha SGGCSIVWFRRDLRVEDNPALAAAVRAGR-PVIALFVWAP------EEEGHYHPGRVSRWWLKNSLAQLDSSLRSLGTCLITKRSDSVASLLDVVKSTG-ASQIFFNHLYDPLSLVRDHRAKDVLTAQGIAVRSFNADLLYEPWEVTDELGRPFSMFAAFWERCLSMPYDPESP 1U3C
DASH_taeGut MAGTAICLLRCDLRAHDNQQVLHWAQHNADFVIPLYCFDPRHYLGTHCYRLPKTGPHRLRFLLESVKDLRETLKKKGSTLVVRKGKPEDVVCDLITQLGSVTAVVFHEEATQEELDVEKGLCQVCRQHGVKIQTFWGSTLYHRDDLPFRPIDRLPDVYTHFPKGLESGAKVRPT
DASH_xenTro RARVIICLLRNDLRLHDNEVLHHWAHRNADQIVPLYCFDPRHYGGTHYFNFPKTGPHRLKFLLESVQDLRNTLKERGSNLLLRRGKPEEIIAGLVKQLGNVSAVTLHEEATKEETDVESAVRRVCTQLGVRYQTFWGSTLYHREDLPFRHISSLPDVYTQFRKAAETQGKVRST
DASH_danRer ASRTVICLLRNDLRLHDNEVFHHWAQRNAEHIIPLYCFDPRHYQGTYHYNFPKTGPFRLRFLLDSVKDLRALLKKHGSTLLVRQGKPEDVVCELIKQLGSVSTVAFHEEVASEEKSVEEKLKEICCQNKVRVQTFWGSTLYHRDDLPFSHIGGLPDVYTQFRKAVEAQGRVRPV
DASH2_araTha GKGVTILWFRNDLRVLDNDALYK-AWSSSDTILPVYCLDPRLFHTTHFFNFPKTGALRGGFLMECLVDLRKNLMKRGLNLLIRSGKPEEILPSLAKDFGA-RTVFAHKETCSEEVDVERLVNQGLKRVGTKLELIWGSTMYHKDDLPFD-VFDLPDVYTQFRKSVEAKCSIRSS 2VTB
CPD_galGal GAECILYWMCRDQRVQDNWAFLYAQRLALKQELPLRVCFC------LVPAFLDATIRHYGFMLRGLREVAKECAELDIPFHVLLGCPKDVLPSFVVEHGVGGLVTDFCPLRVPRQWVEEVKERLPED--VPFAQVDAHNIVPCWVASPKQEYSARTIRAKIHSQLPEFLTEFPP
CPD_xenTro DAQGIVYWMSRDQRVQDNWAFLYAQRLALKQKLPLHVTFC------LVPKFLDATIRHYGFMVKGLQEVAEECKELNIPFHLLIGYAKDILPNFVKKHAIGGVVTDFSPLRVPLQWVEDVSKRLPKD--VPLVQVDAHNIVPCWVASNKQEYGARTIRKKIHDQLSQFLTEFPP
CPD_droMel SSLGVVYWMSRDGRVQDNWALLFAQRLALKLELPLTVVFC------LVPKFLNATIRHYKFMMGGLQEVEQQCRALDIPFHLLMGSAVEKLPQFVKSKDIGAVVCDFAPLRLPRQWVEDVGKALPKS--VPLVQVDAHNVVPLWVASDKQEYAARTIRNKINSKLGEYLSEFPP
CPD_orySat PGGPVVYWMLRDQRLADNWALLHAAGLAAASASPLAVAFA------LFPRLLSARRRQLGFLLRGLRRLAADAAARHLPFFLFTGGPAE-IPALVQRLGASTLVADFSPLRPVREALDAVVGDLRRG--VAVHQVDAHNVVPVWTASAKMEYSAKTFRGKVSKVMDEYLVEFPE 3UMV
bbbbbb aaaaaaaaa bbbbbbbbb aaaaaaaaaaaaaaaaaaa bbbbbb aaaaaaaaaa bbbbbbbb aaaaaaaaaaaa bbbbbbb
CRY1_homSap M..N..HWFRKGLRLHDNP.L.....G..-..RCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDA.LRKLNSRLFVIRGQP.DVFPRLFKEW.-I..LS.EYDSEPFGKERDAAIKKLA.EAGVEVI.R.SHTLY.LD.IIELNGGQ.PLTYKRFQ.L.S.M.P...P 95% conservation
CRY1_musMus M..N..HWFRKGLRLHDNP.L.....G..-..RCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDA.LRKLNSRLFVIRGQP.DVFPRLFKEW.-I..LS.EYDSEPFGKERDAAIKKLA.EAGVEVI.R.SHTLY.LD.IIELNGGQ.PLTYKRFQ.L.S.M.P...P
CRY1_galGal M..N..HWFRKGLRLHDNP.L.....G..-..RCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDA.LRKLNSRLFVIRGQP.DVFPRLFKEW.-I..LS.EYDSEPFGKERDAAIKKLA.EAGVEVI.R.SHTLY.LD.IIELNGGQ.PLTYKRFQ.L.S.M.P...P
CRY1_xenTro M..N..HWFRKGLRLHDNP.L.....G..-..RCVYILDP------WFAGSSNVGINRWRFLLQCLEDLDA.LRKLNSRLFVIRGQP.DVFPRLFKEW.-I..LS.EYDSEPFGKERDAAIKKLA.EAGVEVI.R.SHTLY.LD.IIELNGGQ.PLTYKRFQ.L.S.M.P...P
CRY2_homSap ....SVHWFR.GLRLHDNPAL..A.....-..RC.YILDP------WFA....VG.NRWRFLL.SLEDLD.SLRKLNSRLFVVRGQP.DVFPRLFKEW.-VTRLTFEYDSEP.GKERDAAI.K.A.E.GVE....NSHTLY.LDRIIE.N...PPLT.KRFQ.I.SR..LP..P 95% conservation
CRY2_musMus .....VHWFR.GLR.HDNPAL..A.....-..RC.YILDP------.FA.....G.NRWRFLL..LEDLD.SL.KL.SRLFVVRGQP.DVFPRLFKEW.-V..LTFEYD.EP.GKERD..I.K.A.E.GVE......HTLY.....IE.N...PPLT.KRFQ....R..LP..P
CRY2_galGal .....VHWFR.GLR.HDNPAL..A.....-..RC.YILDP------.FA.....G.NRWRFLL..LEDLD.SL.KL.SRLFVVRGQP.DVFPRLFKEW.-V..LTFEYD.EP.GKERD..I.K.A.E.GVE......HTLY.....IE.N...PPLT.KRFQ....R..LP..P
CRY2_xenTro .....VHWFR.GLR.HDNPAL..A.....-..RC.YILDP------.FA.....G.NRWRFLL..LEDLD.SL.KL.SRLFVVRGQP.DVFPRLFKEW.-V..LTFEYD.EP.GKERD..I.K.A.E.GVE......HTLY.....IE.N...PPLT.KRFQ....R..LP..P
CRY1A_triCas ..K..VHWFR.GLR.HDNP.L..G.....-T.R..F..DP------WFA...N..INKWRFLL..L.DLD..L..L-.RLFV..GQPA..LP.L...W.-TT..TFE.DPEP.G.VRD.N.........I.V.....HTLY....II..N....PLTY..F.........P... 95% conservation
CRY1A_bomImp ..K..VHWFR.GLR.HDNP.L..G.....-T.R..F..DP------WFA...N..INKWRFLL..L.DLD..L..L..RLFV..GQPA..LP.L...W.-TT..TFE.DPEP.G.VRD.N.........I.V.....HTLY....II..N....PLTY..F.........P...
CRY1A_nasVit ..K..VHWFR.GLR.HDNP.L..G.....-T.R..F..DP------WFA...N..INKWRFLL..L.DLD..L..L..RLFV..GQPA..LP.L...W.-TT..TFE.DPEP.G.VRD.N.........I.V.....HTLY....II..N....PLTY..F.........P...
CRY1A_anoGam ..K..VHWFR.GLR.HDNP.L..G.....-T.R..F..DP------WFA...N..INKWRFLL..L.DLD..L..L..RLFV..GQPA..LP.L...W.-TT..TFE.DPEP.G.VRD.N.........I.V.....HTLY....II..N....PLTY..F.........P...
CRY1B_strPur .......WFRHGLRLHDNP.L........-.F.P.FIFD.------E.AGT...GYNR..FL...L.DLD......GGRL....G.P...F.....E.G-.....FE.D.EP.W..RD..VK..C......C.E.VSHTLW.P...I..NGG.PP.TY.MF......IG.PPRP 70% conservation
CRY1B_octVul .....V.WFRHG.RLHDNP.L........-.F.P.FIFD.------E.AGT...G.NR..FLL..L.DLD......GGRL....G.P...F.....E.G-......E.D.EP.W..RD..VK..C......C....SHTLW.P......NGG.PPLT...F.......G.PPRP
CRY1B_danPle .....V.WFRHGLRLHDNP.L........-.F.P.FIFD.------E.AGT...GYNR...LL..L.DLD......GG.L....G.P...F.....E.G-.....FE.D.EP.W..RD..V...C......C.E.V.HTLW.P...I..NGG.PPLTY.MF......IG.PPRP
CRY1B_droMel .....V.WFRHGLRLHDNP.L........-...P.FIFD.------E.AGT...GYNR..FLL..L.D.D.......GRL....G.P...F........-......E.D.EP.W..RD......C........E.VSHTLW.P...I..NGG.PPLTY.MF......IG.PPRP
CRY4_galGal M.HRTIH.FRKGLRLHDNP.LL.AL..S.-..YPVYILDR------.F......GALRW.F.LQSLEDL...L...GS.L.V..G......R..V.KW.I-TQ.T.D.E.EP.Y..M...I.....E.G..V.....H.LY...RI...NGG.PPLTYK.FL..LS.LG.PE.P 70% conservation
CRY4_xenTro M.HRTIH.FRKGLRLHDNP.L..AL..S.-..YPVYILDR------.F......G..RW.F.LQS.EDL...L....S.L.V..G......R..V.KW.I-TQ.T.D.E.EP.Y......I.....E.G..V...V.H.LY....I...N.G.PPLTYK.FL..LS.LG.P..P
CRY4_latCha M.HRTIH.FRKGLRLHDNP.LL.AL..S.-..YPVYILDR------........GALRW.F.LQSLEDL...L....S.L.V..G......R..V.KW.I-TQ.T.D.E.EP.Y..M.........E.G..V...V.H.LY...RI...NGG.PPLTYK.FL..LS.LG.PE.P
CRY4_danRer M.HRTIH.FRKGLRLHDNP.LL.AL..S.-..YPVY.LDR------.F......GALRW.F.LQSLEDL...L...GS.L.V..G......R..V..W.I-TQ...D.E.EP.Y..M...I.....E.G......V.H.LY...RI...NGG.PPLTYK.FL..LS.LG.PE.P
CRY64_xenTro .....IHWFRKGLRLHDNPAL..A.....-...PIF.LDP------.......V..NRWRFL...L.DLD..L.K.N.RLFV.RG.P.E..P.LF..W.V-..LT.EVDTEPY...RD..V...A....V.V...VS.T.Y........N.G..PLTY............P..P 70% conservation
CRY64_danRer .....IHWFRKGLRLHDNPAL..A.....-...P.F.LDP------..........NRWRFL...L.DLD..L.KLN.RLFV.RG.P.E..P.LF..W..-..LT.EVDTEPY...RD..V...A....V.V....SHT.Y........N.G..P.TY............P..P
CRY64_droMel ......HWFRKGLRLHDNPAL........-...PIF.LDP------.......V..NRWRFL...L.DLD..L.KLN.RLFV.RG.P.E..P..F..W.V-..LT.E.D.EPY...RD..V...A....V.V....SHT.Y........N.G..P.TY............P...
CRY64_danPle .....IHWFR..LRLHDN.AL..A.....-...PI...DP------.......V..NR.RFL...L..LD..L.K.N.RL.V..G...E..P.LF..W.V-..LT..VD........D.............V.....HT.YD.......N.G..PLTY................
DASH_taeGut .....ICLLR.DLR.HDN....HWA...A....PLYCFDPRHY.GT.....PKTGP.RL.FLLES..DLR..L...GS.L..R.GKPE.V...L..QLG.V..V....E.T.EE.DVE......C....V...T.WGSTLYHR.DLPF..I..LPDVYT.F.K..E....VR.. 70% conservation
DASH_xenTro .....ICLLRNDLR.HDNE...HWA...A....PLYCFDPRHY.GT....FPKTGP.RL.FLLES..DLR..L...GS.L..R.GKPE.....L..QLG.V..V....E.T.EE.DVE......C....V...T.WGSTLYHR.DLPF.HI..LPDVYT.FRK..E....VR..
DASH_danRer .....ICLLRNDLR.HDNE...HWA...A....PLYCFDPRHY.GT....FPKTGP.RL.FLL.S..DLR..L...GS.L..R.GKPE.V...L..QLG.V..V....E...EE..VE......C....V...T.WGSTLYHR.DLPF.HI..LPDVYT.FRK.VE....VR..
DASH2_araTha .....I...RNDLR..DN.....-A........P.YC.DPR....T....FPKTG..R..FL.E...DLR..L...G..L..R.GKPE.....L....G.-..V....E...EE.DVE.................WGST.YH..DLPF.-...LPDVYT.FRK.VE.....R..
CPD_galGal ......YWM.RDQRVQDNWA.L.AQ.LALK...PL.VCF------CL.P.FL.AT.R...F.L.GL.EV..EC..L.I.FH.L.G.....LP.FV.....G..V.DF.PLR.P..W...V...LP..--VP..QVDAHNIVPCW.AS.K.EY.ARTIR.KI...L..FLTEFPP 70% conservation
CPD_xenTro ......YWM.RDQRVQDNWA.L.AQ.LALK...PL.V.F------CL.P.FL.AT.R...F...GL.EV..EC..L.I.FHLL.G.....LP.FV.....G..V.DF.PLR.P..W...V...LP..--VP..QVDAHNIVPCW.AS.K.EY.ARTIR.KI...L..FLTEFPP
CPD_orySat ......YWM.RDQR..DNWA.L.A..LA.....PL.V.F------.L.P..L.A..R...F.L.GL.............F.L..G....-.P..V........V.DF.PLR........V...L...--V...QVDAHN.VP.W.AS.K.EY.A.T.R.K........L.EFP.
CPD_metMaz ......YWM.RDQR..DNWA.L.....A.....P..V.F------CL...FL.A..R...F.L.GL.E.........I....L.G........FV.....G..V.DF.PLR....W...V....--.--.P...VDAHN.VPCW.AS.K.EY.A.T.R.K....L..FL.EFP.
bbbbbb aaaaaaaaa bbbbbbbbb aaaaaaaaaaaaaaaaaaa bbbbbb aaaaaaaaaa bbbbbbbb aaaaaaaaaaaa bbbbbbb
Three major indels occur. Using CPD and 4Fe-4S photolyases as outgroup, these can be resolved as either insertions or deletions (ie as derived traits or synapomorphies). The first indel is a 6 residue insertion found only in the DASH group; the second is a 1 residue deletion in stem post-DASH divergence proteins; and the third is a 2 residue insertion that occurred shortly after divergence from CPD. Even though 3D coverage of cryptochromes is inadequate, enough exists that each of these indels can be localized in an existing stucture and so visualized by precomputed structural co-alignments.
Indels can work as a standalone classifier. However this third of the protein provides discriminants only for CPD and DASH which are more easily identified just by a blast classifier using the reference sequences. Note too at position 30, ecdysozoans (minus orthopterans and crustaceans) show homoplasy, re-inserting a residue at a site where it had long been deleted.
A deeper history of photolyase structural history must find a place for the 4Fe-4S cluster family (likelier as ancestral condition rathner than a development off to the side) and an explanation for the same cluster and structural homology to primase which is extensive butdoes not extend to the α/β domain. Here the unusual but conserved U-shaped conformation of the catalytic FAD may be a key piece of the history.
Here the rings of FAD's adenine and flavin each lie in a plane but these planes while not quite parallel are alignable by a rotation, and ring long axes are almost perpendicular. This configuration may allow them to bind two primer pyrmimidines much as the damaged DNA thymine dimer is bound. Indeed the 4Fe-4S cluster may create a transient cyclobutane bond in the primer. As usual, divalent magnesium binds the diphosphate and offsets its charge.
The table below list the current structural determinations for eukaryotic cryptochromes, photolyases and related folds available in March 2012. Archaeal and bacterial structures are included in the table when their eukaryotic alignment coverage or blast score warrants it -- they are surprisingly well conserved relative to metazoan, probably because of a 'floor' of essential core residues that prevents further percent divergence. (Opsins and the huge GPCR gene family have a similar floor but much lower, about 24%.) Overall, coverage is not ideal because not all orthology classes are represented yet -- while their core fold is easily predictable given high percent identities (eg 65% human to plant), critical functional nuances provided by actual extensions are not.
Date PDB Class PubMed Species BestBlastP Accession Cofactors Alternates Nov 2011 3TVS CRY1B 22080955 Drosophila melanogaster (fruit_fly) musMus 40% AB019389 FAD no antenna CRY1 cryptochrome 19722240 Dec 2008 1U3C CRY1A 15299148 Arabidopsis thaliana (cress) homSap 29% NM_116961 FAD MTHF CRY1-PHR Oct 2009 3CVU CRY64 18956392 Drosophila melanogaster (fruitfly) xenTro 57% NM_165334 FAD [deazaflavin Fo] phr 6-4 Apr 2009 3FY4 CRY1C 19359474 Arabidopsis thaliana (cress) musMus 51% NM_001035626 FAD UVR3 CRY3 Dec 2008 2VTB DASH2 19074258 Arabidopsis thaliana (cress) xenTro 50% NM_122394 FAD CRY3 Dec 2011 3UMV CPD 22170053 Oryza sativa (rice) galGal 53% B096003 FAD PhrII Class II Sep 2011 2XRY CPD 21892138 Methanosarcina mazei (euryarchaeota) xenTro 49% AE008384 FAD Class II Mar 2012 3ZXS PFES 22290493 Rhodobacter sphaeroides (bacteria) .......... CP000144 FAD 4Fe-4S lumazine CryPro Aug 2010 3L9Q PRIM2 21346410 Homo sapiens (human) .......... NM_000947 FAD 4Fe-4S primase large subunit 3Q36 PMC3204975 Apr 2010 3LGB PRIM2 20404922 Saccharomyces cerevisiae (yeast) .......... NM_001179611 FAD 4Fe-4S primase large subunit PriL PRI2_YEAST
To what extent can the structures available now be used to model the vertebrate ones that so far are missing? That can be done at the primary and secondary structure level simply by aligning a batch of orthologs under a structurally determined sequence and transfering its features to non-gappy regions having signficant conservation. Multiple target sequences are essential to purge one-off accidental matches and to assess phylogenetic depth (ancestral persistance vs recent convergent origin).
When percent identity exceeds 25%, reasonably accurate 3D coordinates can be obtained by fitting the unstudied primary sequence to a PDB entry using SwissModel. A third approach uses DaliLite for pairwise comparisons of proteins with PDB coordinates, either real or modelled. NCBI's VAST allows any number of structures to be simultaneously aligned. The antenna pocket has also been examined by docking candidate receptor molecules.
For example, human CRY1 is not available. According to Blastp of PDB on 25 Mar 2012, the structure with the highest percent identity 53% and most extensive coverage (positions 6-522 out of 587 amino acids) is 3CVU from CRY64_droMel. Setting aside concerns about what the extra 65 amino acids at the end of the human protein might contribute to structure, that template request quickly yields PDB coordinates (with local error estimates) from SwissModel. And those in turn can be uploaded at DaliLite to structurally align human CRY1 with CPD and 4Fe-4S photolyases and primases which are too distant to align by primary sequence. This shows where the 4Fe-4S would sit in human cryptochrome had it been retained and also identifies the ancient structural core in the all-alpha domain that cryptochromes share with primases. These tools also rotate sequences so that all are in the same orientation.
In all three approaches, what emerges is not fact but prediction. Only the first provides homological (genetic) alignment; structural alignments provide the best geometrical fit but do not necessarily recapitulate evolutionary relatedness of residues near gaps. Because folds are far more deeply conserved than sequence, structural alignment of greatly diverged sequences can uncover very faint but real relationships. However the N- and C-terminal extensions are not modelable yet their strong conservation in some instances argues for important function and possibly fixed structure.
Syntentic relationships in vertebrate cryptochromes
Synteny -- the conservation of flanking gene relationships -- is critical (along with indels and intron structure) in establishing orthology and so to transferal of experimental information from a model species to another because primary sequence analysis alone can be give misleading results:
After gene duplication, both members of a retained pair may diverge rapidly in primary sequence if they subfunctionalize, whereas if one gene -- not necessarily the parent -- retains the original function and the other neofunctionalizes, only the latter may diverge rapidly. This behavior can lead to long branch attraction artefacts and major misclassification of relationships.
Cryptochromes and photolyases have experienced numerous duplications over evolutionary time. Those within multi-cellular organisms have all have been segmental duplications of limited extent. The alternative, retroprocessing, removes all introns. The sole known retroprocessed cryptochromes are CRY1 pseudogenes in naked mole rat, marmoset and sloth (AHKG01086374 ACFV01087645 ABVD01272190). These are easily recognized -- even far into pseudogenization -- as the top hits at genomic blast because as long contiguous matches they outscore multi-exonic ones. The existence of such features implies transcription of the parental gene in germline cells, typically testis.
Syntenic relationships can persist for billions of years of branch length but more commonly dissipate over a few hundred million years because of local inversions and other chromosomal rearrangements that shuffle gene order. The rate of dissipation varies greatly by clade, with vertebrates much slower than arthropods.
In vertebrates, synteny is typically well-retained back to the human-coelocanth divergence, with less certain correspondences extending to ray-finned fish and sometimes to chondrichthyes. Although complicated by poor quality assemblies, little synteny appears to persist back to lamprey, tunicate, amphioxus, sea urchin, and hemichordate. In contrast, synteny for a Drosophila gene rarely extends through dipteran flies, much less Insecta.
The primary method for determining synteny at the phylogenetic level is a Blast search against multiple assemblies. This can be done very efficiently by concatenating conserved and diagnostic regions of 4-5 adjacent human proteins centered on the target gene. As the percent identity falls off, the human probe can be replaced with an orthologous concatenate from chicken or frog using the UCSC 46-way to collect orthologs. If probes aren't known, blastx of the contig containing the cryptochrome will reveal its neighbors.
However the outcome also has been precomputed on a massive scale by Genomicus, the complication here being that only two cryptochromes persist into humans (meaning no HUGO gene names are available to enter Genomicus). To procede with CPD, DASH, CRY64, CRY4 in the tetrapods that have them, it is necessary to blat into a UCSC assembly that carries an Ensembl gene name track.
The figure (taken from the Genomicus synteny tool) shows that CRY1 experienced a small local inversion in amniotes subsequent to mammalian divergence. This may have carried all upstream regulatory regions along with it or left some behind, perhaps with significant effects altering gene expression. Since the event occurred some 350 myr ago, the boundaries of the inversion cannot be precisely determined today.
Genomicus works surprisingly well given that almost all the Ensembl gene models it uses are wrong, the explanation being a few missed exons, erroneous termini and retained introns don't signficantly affect best reciprocal blast. But Ensembl models are often absent altogether in non-mammalian tetrapods, for example missing out entirely for DASH in frog and lizard which have full length conserved genes in their assembly. Unlike the UCSC 46-way, Genomicus does not begin with a whole genome alignment. Consequently it can stub in an erroneous paralogs when a gene is missing (eg CRY4_latCha in place of DASH_latCha).
In some cases a gene appears absent but pseudogene debris in the expected syntenic position is still detectable. That says gene loss was fairly recent (last five million years), for example DASH pseudogenes in gallinaceous birds (chicken and turkey) but not duck (the immediate outgroup) or songbirds. Only very recent pseudogenes would be represented in either Genomicus or at the UCSC 46-way. Genes can also seem absent in spotty assemblies but individual exons can be recovered from raw trace reads (eg platypus CPD). Long processing lags prevent certain strategic assemblies from being represented in the 46-way or Genomicus (eg alligator, turtle, python, spotted gar) so they were considered separately here.
These sites also provide no resources for invertebrate synteny. It isn't currently possible to study CRY1A (which is missing from fruit fly) even though orthologs extend phylogenetically from honey bee (ecdysozoa) to molluscs (lophotrochozoa) to sea urchin (echinoderms) so method of concatenated queries must be employed. Convenient queries for the other ortholog classes are provided below:
Gene Species Genomicus entry <--------------------------------- Synteny ----------------------------------> Phylogenetic Depth CRY1 homSap CRY1 CMKLR1 ASCL4 PRDM4 PWP1 BTBD11 CRY1 MTERFD3 C12orf23 RIC8B RFX4 to coelocanth, ray-finned fish CRY2 homSap CRY2 PRDM11 SYT13 CHST1 CTD SLC35C1 CRY2 MAPK8IP1 C11orf94 PEX16 GYLTL1B barely to ray-finned fish CRY4 anoCar ENSACAG00000004583 GPR37L1 ARL8A PTPN7 LGR6 UBE2T CRY4 LRIF1 CEPT1 ADORA1 MYOG to coelocanth CRY64 xenTro ENSXETG00000003913 UBASH3B STS1 RPL27A CRY64 FOXRED1 SRPR FAM118B FAM55A lizard to frog CPD monDom ENSMODG00000018409 PCYT1A ZDHHC19 TFRC TNK2 IGFBP5 CPD KIAA0226 FYTTD1 LRCH3 IQCG barely to ray-finned fish DASH danRer ENSDARG00000002396 CTDSPLA VILL PLCD1A DLEC1 ACAA1 DASH MYD66/88 OXSR1 SLC22A13 CSRNP1B birds to ray-finned fish CRY1B droMel FBgn0025680 SQZ CG14282 CG5555 CG31475 CRY1B VIB CG11703 CG5250 CG3773 ... CRY1A apiMel concatenated blast XM_395048 XM_393681 CRY1A Amel_5586 XM_391835 ...
Synteny can be used to disentangle paralogs, especially important in zebrafish which has been studied to a certain extent -- despite the poor correspondence of its oversize cryptochrome repertoire to mammals -- because individual cell lines are cryptochrome-entrainable. Here zebrafish have four cryptochromes on four different chromosomes related to mammalian CRY1, a single copy of a cryptochrome clustering with mammalian CRY2, and single copies of CRY4, CRY64, DASH and CPD.
Because chondrichthyes, lobe-finned fish (coelocanth), and basal ray-finned fish (gars) already have two separate genes classifying as CRY1 (ie distinct from CRY2 and other photolyases), a gene duplication occurred in early vertebrates and persisted almost to amphibians. Since it is generally believed that a latter whole genome duplication took place in ray-finned fish following the divergence of gar, then the four CRY1 genes in zebrafish may represent retention of all copies whereas presumptive second copies of CRY4, CRY64, DASH and CPD were lost.
The new spotted gar assembly (Lepisosteus oculatus) has the photolyase and cryptochrome repertoire (ie two CRY1 and one CRY2) expected from this scenario but the contigs are too small to contain flanking genes. The same can be said for the new coelocanth assembly (here Genomicus works with scaffolds of unordered unoriented contigs, which really pushes the limits for synteny). The shark and skate assemblies consist mostly of kilobase-size contigs that cover at most 60% of the coding exons; both have CRY1A and CRY1B but not CRY2 (and skate CPD has pseudogenized).
The gar genome assembly as of April 2012 consists of 45,199 contigs (eg AHAT01025403) organized into 185 scaffolds (eg JH591278) organized further into 15 superscaffolds (eg CM001411) comprising 29 lingage groups (eg LG8) in 1012 pieces separated by gaps. Blast at NCBI can only access contigs. The entries for these contigs do not indicate their scaffold, superscaffold or linkage group. However those can be ferreted out with Entrez queries such as 'AHAT01025403 AND JH591*'. The cryptochrome contigs are too small to have any syntentic information, but the presence of a second gene within the same scaffold implies synteny.
Here CRY1B and CRY4 both lie in the JH591232 gar scaffold, as they do in coelocanth and zebrafish scaffolds, establishing this as an ancestral synteny. Shark and skate, cartilaginous vertebrates, have CRY1A and CRY1B but the assemblies are too poor to even consider syntenic questions. Lamprey assembly is a non-starter, tunicates are too diverged, and amphioxus is uninformative (the flanking genes NAV2 GIT2 TUBB5 CRY1 TBC1D17 do not correspond at all to vertebrate gene order).
Since frog and amniotes still have CRY4 in syntenic position with 7 other zebrafish genes but no cryptochrome, it follows that CRY1B was lost in the tetrapod stem. Thus it is CRY1A of fish that has continued in land animals under the name CRY1, as driven by human nomenclature convention (which is oblivious to other species).
The near-adjacency of CRY1B and CRY4 could either be coincidence or indicative of an earlier tandem duplication relationship. CRY4 has a limited phylogenetic distribution in fish but continues on to frog, lizard and birds. In one scenario, CRY4 is not particularly related to CRY64 but instead arose from CRY1B in early ray-finned fish. The tandem pair persisted to extant fish but only CRY4 continued on in amniotes, with both CRY4 and CRY1B absent from mammals.
Old RefSeq Chr S Start End N-term Pub Accession New RefSeq #Syn Comment CRY1A_danRer 4 + 11078260 11088888 MVVNTVH cry1a ENSDARG00000045768 CRY1P2_danRer 2 whole genome duplicate of retained CRY1 duplicate CRY1A2_danRer 18 + 14692957 14714934 MVVNTVH cry1b ENSDARG00000011583 CRY1P1_danRer 7 old CRY1 duplicate retained as tetrapod CRY1 CRY1C_danRer 22 + 748902 787935 MSVNSVH cry2b ENSDARG00000091131 CRY1Q1_danRer 3 old CRY1 duplicate lost in tetrapods CRY1 C12ORF23 CRY4 in latCha too CRY1B_danRer 8 + 21767736 21788261 MAPNSIH cry2a ENSDARG00000069074 CRY1Q2_danRer 1 whole genome duplicate of lost CRY1 duplicate CRY2_danRer 25 - 4289163 4311451 MVVNSVH cry3 ENSDARG00000024049 CRY2_danRer 3 synteny retained far better in coelocanth CRY4_danRer 22 - 800173 811759 MSHRTIH cry4 ENSDARG00000011890 CRY4_danRer 7 adjacency to lost CRY1 suggests relationship CRY64_danRer 10 - 40633074 40645329 MSHNTIH cry5 ENSDARG00000019498 CRY64_danRer 2 poor synteny within fish, adjacent FOXRED1 in frog too DASH_danRer 24 - 20802832 20816799 MSASRTV cry-dash ENSDARG00000002396 DASH_danRer 1 strong synteny within fish, none to tetrapods CPD_danRer 2 + 13740732 13773635 MSANKNN cry-phr ENSDARG00000054999 CPD_danRer 3 mediocre preservation of synteny CRY1A_lepOcu JH591278 . LG8 CM001411 MVVNTVH AHAT01025403 CRY1P_lepOcu 1 old CRY1 duplicate retained as tetrapod CRY1 CRY1B_lepOcu JH591232 . LG3 CM001406 MGPNSIH AHAT01016727 CRY1Q1_lepOcu 1 old CRY1 duplicate lost in tetrapods CRY4_lepOcu JH591232 . LG3 CM001406 MTHRTIH AHAT01016726 CRY1Q2_lepOcu 1 adjacency to lost CRY1 suggests relationship CRY2_lepOcu JH591436 . UNK23 ........ MVVNSVH AHAT01038797 CRY2_lepOcu 1 isolated small contig CRY64_lepOcu JH591390 . LG26 CM001429 MMHRSIH AHAT01024141 CRY64_lepOcu 1 retained DASH_lepOcu JH591300 . LG9 CM001412 MSTIRTI AHAT01010414 DASH_lepOcu 1 retained CPD_lepOcu JH591341 . LG14 CM001417 MSGRSPP AHAT01034265 CPD_lepOcu 1 retained CRY1A_latCha JH126600 - 326804 392530 MGVNAIH ENSLACG00000008174 CRY1P_latCha 5 old CRY1 duplicate retained as tetrapod CRY1 CRY1B_latCha JH126576 + 512532 551664 MVVNSVH ENSLACG00000010538 CRY1Q1_latCha 2 old CRY1 duplicate lost in tetrapods CRY4_latCha JH126576 - 707590 727148 MTHRTIH ENSLACG00000012369 CRY1Q2_latCha 9 adjacency to lost CRY1 suggests relationship CRY2_latCha JH126568 + 3409780 3427400 MVVNSVH ENSLACG00000018488 CRY2_latCha 25 remarkably conserved synteny CRY64_latCha ........ . ....... ....... ....... .................. CRY64_latCha . apparently lost recently DASH_latCha ........ . ....... ....... ....... .................. DASH_latCha . apparently lost CPD_latCha ........ . ....... ....... ....... .................. CPD_latCha . apparently lost
This raises the question of the ancestral origin of mammalian CRY2. It has it earliest representatives in early diverging teleost fish. It may have arisen from CRY1 after its duplication to CRY1P and CRY1Q and then diverged rather rapidly. Alternatively, it might be an older duplication and simply be lost from the chrondrichthyes studied to date. The CRY2 region exhibits remarkable conservation of gene order which may help resolve this issue once better early assemblies become available. Note that gene order has also been quite stable around CRY1 for several hundred million years.
CRY2 must have arisen from a segmental duplication of the older CRY1 because their identical pattern of intronation could not have arisen independently. The size of the region duplicated in this event (ranging from one to several gene, or to a chromosome or even whole genome) could still be reflected by the extent of paralogous gene pairs. However subsequent inversions, gene losses, and rapid divergence might render these relationships opaque today.
Seeking homologous pairs (using gene names as a proxy for homology) in 25 genes flanking each side of CRY1 and CRY2 in human turns up 5 candidate pairs, not particularly supportive of a large segmental duplication given the many intervening non-homologous genes. The most intriguing pair, PRDM4 and PRDM11, are closely related and documented to have arisen in the same time frame. Since these are nearly adjacent to CRY1 and CRY2 respectively, a small duplication of 2-3 genes is the best fit to the data.
One or even two rounds of whole genome duplication supposedly took place prior to vertebrate origins. Little supporting data for that scenario actually exists -- contrary to dozens of papers (all citing the same meagre investigations). The critical genome assemblies (amphioxus, tunicate and lamprey) are poor quality yet appear to have very similar numbers of protein coding genes. After a decade of manual curation, no more than 18,500 coding genes can be documented in human. That's a very long ways from the 30,000 expected from 1R (or 60,000 from 2R) relative to the 15,000 genes of cephalochordate.
The cryptochromes and photolyase gene family conflicts with both 1R and 2R hypotheses. If DASH, CPD, CRY64 and CRY1 were duplicated in such an event, then all duplicates were lost in all surviving lineages. The same is true for all three classes of opsins and many other gene families. If almost all duplicates from a whole genome duplication are lost, the outcome is effectively indistinguishable from the always-ongoing process of small segmental duplications with retention (the default hypothesis for paralogous gene origin).
However CRY1 did experience three separate duplications-with-retention at various points in vertebrate evolution, implying functionality for all paralogous copies. One of these did arise from a whole genome duplication in a sub-lineage of ray-finned fish; the other two did not. Continued retention has been uneven in land animals (as with DASH, CPD, and CRY64) and the process of loss continues to the present day (still-recognizable pseudogenes cannot be old). In the same time frame, vertebrates also experienced a great expansion of the other main photoreceptor family, the opsins. Tunicates and amphioxus have no imaging vision but lenses, four cone opsins and rhodopsin were firmly established at the time of lamprey divergence.
These quasi-simultaneous expansions may be correlated: cryptochromes are co-expressed in retinal, pineal and other light sensing cells and can function coordinately with opsins, as in the SWS1 cone cells of bird. This association may have a very long history as sponges, which have numerous GPCR genes but none with K296 retinol binding, may use cryptochromes alone as their larval photosensing system. In this scenario, they paired with a GPCR gene to signal; later that protein adapted retinoic acid signalling to retinal and took over the primary photosensing role in ctenophores and cnidarians which do have conventional opsins and neurons. The reference sequence collection contains 5 cryptochromes from 4 sponge species, Amphimedon, Suberites, Crateromorpha and Aphrocallistes.
Inconsequential N-terminal extension in vertebrate CRY2
CRY1 duplicated in early vertebrates giving rise to CRY2 which evidently carved out a distinct functional role as it has persisted in all species since including committed subterranean and cave species. The status of the gene in lamprey, hagfish and chondrichthyes cannot be resolved without better assemblies but there is no indication of a duplication there or in urochordates or cephalochordates.
Confusingly, Cry2 is also used for non-orthologous insect cryptochromes entries at GenBank. These represent a different duplication of a different parent gene called CRY1A here (which itself was subsequently lost in dipterans). The Drosophila 'CRY2' sequence, called CRY1B here, is not a valid model system for vertebrate CRY2 and indeed is equally unsuited as a invertebrate CRY1 proxy because properties can be expected to pull apart in species retaining both copies.
Vertebrate CRY2 has an extended amino terminus that arose in amniotes. Prior to that time, it was similar in length to vertebrate CRY1 (and still is from fish to frog). A few residues of this extension are conserved but overall the sequences displays compositional simplicity, to the point that RepeatMasker finds a variable length simple repeat (CCG)n within coding and in some species also upstream. Base composition alone then leads to inevitable amino acid conservation which however does not imply selective pressure or functionality.
While no applicable crystallographic structure has been determined, homology modeling reliably locates these extra residues outside the closed globular alternating beta strand/alpha helix structure of this domain. They are likely proteolytically trimmed off in mature protein leaving only 3-4 extra residues.
One scenario here posits that the initial methionine in stem amniotes was lost to mutation, leading to an upstream random in-phase ATG stepping in, followed by evolution of variable length as the new ATG lay in the (CCG)n repeat which was subject to expansions and contractions. While this had no adverse consequences, it did not lead to functional innovation in this region either -- a short amino terminal extension is necessarily remote from excitation transfer pathways, and antenna, FAD and substrate binding sites.
CRY1_homSap ...............................MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR CRY1_homSap ...............................MGVNA................KECIQ..DTI...........G..N....... CRY2_homSap ............MAATVATAAAVAPAPAPGTDSASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_homSap ...........MAATV.ATAAAVAPAPAPGTDSASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_panTro ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_panTro ................................G................................................... CRY2_gorGor ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_gorGor ................................G................................................... CRY2_ponAbe ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_ponAbe ................................G................................................... CRY2_rheMac ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_rheMac ................................G................................................... CRY2_papHam ............MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_papHam ................................G................................................... CRY2_calJac ............MAATVATAAAAVPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_calJac ......................AV........G................................................... CRY2_micMur .............MATAVATAAAAPTPASSTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_micMur ............M..A.VAT..A..T..SS..G................................................... CRY2_musMus .............MAAAAVVAATVPAQSMGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_musMus ........MAAA.VVA......TV..QSM.A.G................................................... CRY2_ratNor .............MAAAAVVAATVPAQSMGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_ratNor ........MAAA.VVA......TV..QSM.A.G................................................... CRY2_criGri ........MAAAAVVAGAPRGARVPALTMGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_criGri ........MAAA.VVAG.PRG.RV..LTM.A.G................................................... CRY2_spaJud .............MAAASVVVATSAAPAMAVDGGSSVHWFRKGLRLHDNPSLLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_spaJud ........MAAASVV.......TSA...MAV.GG................S................................. CRY2_dipOrd ............MAAAMVTAAVAVPAPPSGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_dipOrd ..............AM.V...VAV...PS.A.G................................................... CRY2_cavPor ............MAAAVGTGTAAAPTPVTGAEGACSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_cavPor ..............A..G.GT.A..T.VT.AEG.C................................................. CRY2_hetGla ............MAAAVGTGTGAAPTPATGAEGACSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_hetGla ..............A..G.GTGA..T..T.AEG.C................................................. CRY2_speTri ...............MSASVVTTSATLLTPTSADVSSVHWFRKGLRLDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_speTri ............S.S..V.TS.TLLT.TSA..DV.................................................. CRY2_oryCun ............MAAAAAAAAAAVPAPAASANGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_oryCun ..............AA..A...AV....ASANG................................................... CRY2_turTru ............MAAAVATSAVAAPAPAARAEGASSVHWFRKGLRLHDNPALQAAVRGAHCVRCVYILDPWFAASSSVGINRWR CRY2_turTru ..............A....S.VA.....ARAEG...................Q......H........................ CRY2_bosTau ...............MAAAAAAATQAPAARGDGASSVHWFRKGLRLHDNPALLAAVRGAHCVRCVYILDPWFAASSSVGINRWR CRY2_bosTau ........MAAA..........ATQ...ARG.G..........................H........................ CRY2_oviAri .........MAAAAAATASAAAAAQAPAPRGDGASSVHWFRKGLRLHDNPALLAAVRGAHCVRCVYILDPWFAASSSVGINRWR CRY2_oviAri ........MAAA..AT..S...A.Q....RG.G..........................H........................ CRY2_susScr ............MAAAVATAAASSPAPAAGAEGASSVHWFRKGLRLHDNPALLAAVRGAHCVRCVYILDPWFAASSSVGINRWR CRY2_susScr ..............A.......SS....A.AEG..........................H........................ CRY2_equCab MKKAAAPVRFIATSEAPAASAAAAATAAAGADGDSSVHWFRKGLRLHVNPALLAAVRFLRSVLCVYKNDPWFVASSSVGINRWR CRY2_equCab MKKAAAPVRFIATSEAP.AS..A.ATA.A.A.GD.............V.........FL.S.L...KN....V........... CRY2_canFam ............MAAAVVAAAAAAPVPTAGVDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_canFam ..............A..VA...A..V.TA.V.G................................................... CRY2_ailMel ........................PAPAAGVDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_ailMel ............................A.V.G................................................... CRY2_myoLuc ............MAANAVTAAAAAPAPAAGTDGASSVYWFRKGLRLHDNPALLAAVRGARCVLCVYILDPWFAASSSVGINRWR CRY2_myoLuc ..............NA.V....A.....A...G....Y........................L..................... CRY2_pteVam ............MAATVGTAAAAASAPAAGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_pteVam .................G....A.S...A...G................................................... CRY2_loxAfr ............MAAAVVTAGAAALVPIPSMDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_loxAfr ..............A..V..G.A.LV.I.SM.G................................................... CRY2_triMan ............MAATVVTAAAAALAPAPSIDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_triMan .................V....A.L....SI.G................................................... CRY2_choHof .............MAATAVMAGSAAPAPASGTEGASSVHWFRKGLLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR CRY2_choHof ...............A.VM.GSA.....S..EG................................................... CRY2_monDom ............MAAAVVTMTAAAPAPAPSPEGASSVHWFRKGLRLHDNPALQAALRGARCVRCVYILDPWFAASSSVGINRWR CRY2_monDom ..............A..V.MT.A......SPEG...................Q..L............................ CRY2_macEug ..........MAATTAVTVTVPAAAPAPAPEEGASSVHWFRMGLRLHYNPVLYSALRGARCVRCVFFLYSWFAASSSVVFFLWL CRY2_macEug .........MAATTA.TV.VP.A......E.EG........M.....Y..V.YS.L.........FF.YS........VFFL.L CRY2_galGal ......................MAAAASPPRGFCRSVHWFRRGLRLHDNPALQAALRGAASLRCIYILDPWFAASSAVGINRWR CRY2_galGal ......................M.A.AS.PRGFCR......R..........Q..L...ASL..I...........A....... CRY2_allMis ................MAASRSFPSSVPARAGPCRAVHWFRRGLRMHDNPALQAALRDAASVRCIYILDPWFAASSAVGINRWR CRY2_allMis ................M.ASRSFPSSVPARAGPCRA.....R...M......Q..L.D.AS...I...........A....... CRY2_anoCar ........................MAALPGPLGRCSVHWFRRGLRLHDNPALQAAIRDGGPVRCIYILDPWFAASSSVGINRWR CRY2_anoCar ........................M.AL..PLGRC......R..........Q..I.DGGP...I................... CRY2_xenTro ...........................MEGKPSVSSVHWFRKGLRLHDNPALLSALRGANSVRCVYILDPWFAASSSGGVNRWR CRY2_xenTro ...........................ME.KP.V...................S.L...NS................G.V.... CRY2_ranCat ............................MEGPAVSSVHWFRKGLRLHDNPALLAALRGARCVRCVYILDPWFAASSSGGVNRWR CRY2_ranCat ...........................ME..PAV.....................L.....................G.V.... CRY2_lepOcu ...............................MVVNSVHWFRKGLRLHDNPALQEALNISDTVRCVYILDPWFAASANVGINRWR CRY2_lepOcu ...............................MVVN.................QE.LNISDT..............AN....... CRY2_danRer ...............................MVVNSVHWFRKGLRLHDNPALQEALNGADTVRCVYILDPWFAGSANVGVNRWR CRY2_danRer ...............................MVVN.................QE.LN..DT............G.AN..V.... CRY2_oreNil ...............................MVVNSVHWFRKGLRLHDNPALQEALNGADAVRCVYILDPFFAGAANVGINRWR CRY2_oreNil ...............................MVVN.................QE.LN..DA.........F..GAAN....... CRY2_tetNig ...............................MVVNSVHWFRKGLRLHDNPALQEALSGADSLRCVYVLDPWFAGAANVGINRWR CRY2_tetNig ...............................MVVN.................QE.LS..DSL....V......GAAN....... CRY2_takRub ...............................MVVNSVHWFRKGLRLHDNPALQEALSGADSLRCVYVLDPWFAGAANVGINRWR CRY2_takRub ...............................MVVN.................QE.LS..DSL....V......GAAN....... CRY1_homSap ...............................MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR CRY1_homSap ...............................MGVNA................KECIQ..DTI...........G..N.......
CRY2_homSap (CGG)n 46 bp atggcggcgactgtggcgacggcggcagctgtggccccggcgccagcg
M A A T V A T A A A V A P A P A
CRY2_rheMac (CCG)n 46 bp atggcggcgactgtggcgacggcggcagctgtggccccggcgccagcg
M A A T V A T A A A V A P A P A
CRY2_musMus (CCG)n 38 bp ggcggcgatggcggcggctgctgtggtggcagcgacgg
A A M A A A A V V A A T
CRY2_oryCun (CGG)n 116 bp ggcggggctcgcggcgccgccgggggcggagcggcggtggctccggcagtctgagctgtgatggcggcggcggcggcagtgggtcctgcggcggcggcggtccccgcgccggcggc
G G A R G A A G G G A A V A P A V - A V M A A A A A V G P A A A A V P A P A
CRY2_canFam (CCG)n 101 bp gcggcgccggcgggggcggagcggcggagcggcggagcggcggaggcctgagcagtcggagcggtgatggcggcggctgtggtggcggcggcagcggcggc
A A P A G A E R R S G G A A E A - A V G A V M A A A V V A A A A A
CRY2_bosTau (CGG)n 31 bp ggcggtgatggcggcggcggcggcggcggcg
A V M A A A A A A A
CRY2_myoLuc CCG)n 57 bp ggcggcgatggcggcgaatgcggtgacggcagcagcggcggccccagcgccagcggc
A A M A A N A V T A A A A A P A P A
CRY2_loxAfr (CCG)n 98 bp ggaggtggggcccgcggcgctgtcgggggcggagcgccgccggccagagcagtctaggcggtgatggcggcggcagtggtgacggcgggagcggcggc
G G G A R G A V G G G A P P A R A V - A V M A A A V V T A G A A
The full set of 43 vertebrate CRY2 sequences is here. These are mostly pre-curated provisional sequences taken from the UCSC 46-way genomic alignment relative to human except where the fasta header displays an accession number. This source misses small insertions and indeed whole exons when very diverged or lie in isolated small contigs or unassembled traces.
The final exons, being quite variable especially in fish, are best determined from transcripts when available and then extended by blastx homology to species within the same clade that lack transcripts. After these corrections, the sequences are aligned and additional anomalies are confirmed or discarded on a case-by-base basis.
It has been previously reported that the PKRK motif in the last exon of mouse CRY2 represents a nuclear localization signal. This motif is indeed conserved in all tetrapod sequences from frog to human. While neither coelocanth nor gar sequence extends to this region, it is definitely absent from other ray-finned fish. Thus nuclear localization may be an innovation of land vertebrates.
It appears that the last exon in fish has lost all homology (and so functionality), in some cases simply running out into junk dna until a stop codon is encountered. Exon seven is broken up in some fish with an extra intron that might have some use in fish taxonomy as a derived characteristic.
Invertebrate cryptochromes: distal CRY1B spoofs a damaged DNA base
Nomenclature of cryptochromes is both a historic and continuing source of confusion, with experimentalists oblivious to anything outside a personal narrow clade and seemingly befuddled by simple concepts such as the timing of gene duplications within eukaryotic phylogeny and orthologouse classification. Another bizarre aspect is functional taxonomy -- like lumping penguins and fur seals together because both eat fish -- whereas enzyme vs circadian signaling vs magnetosensing, single-stranded vs duplex DNA, 6-4 lesions vs thimine dimer have every prospect of multiple origins, gains and losses, cross-overs and reversibility. For example, Arabidopsis UVR3/CRY3 may very well repair 6-4 lesions but it certainly has nothing to do with the CRY64 ortholog group, classifying as it does with CRY1.
With the number of complete genomes available today, it is clear that early bilateran ancestor contained two distinct cryptochromes (in addition to three photolyases), all of which persisted into early deuterostomes (sea urchin). These cannot be denoted CRY1 and CRY2 because those gene names have been assigned to human cryptochromes by international agreement, excluding their re-use in invertebrates or plants for weakly related homologs. The two invertebrate cryptochromes are denoted CRY1A and CRY1B here to distinguish them from the later gene duplication of CRY1A in vertebrates giving rise to tetrapod CRY1 and CRY2. CRY1B is the most commonly studied gene in Drosophila. It has no orthologous counterpart in vertebrates because the definition of orthology requires descent from a single genetic locus in the last common ancestor.
Some lophotrochozoa, notably molluscs, retained both cryptochromes (and indeed a third in a few species not clustering with either CR1A OR CRY1B). Within arthropods, generally one cryptochrome was retained but not always the same one. However some dipterans, hemipterans and lepidopterans retain both. It appears that the CRY1A/CRY1B gene duplication itself took place after divergence from cnidarians.
The two cryptochromes are intronated quite characteristically, with the first class (called CRY1A here) most similiar to that of vertebrate CRY1/2, in agreement with closer blastp clustering. The second class (called CRY1B below) bears less relevance to the cryptochromes retained in mammals but unfortunately is the only one retained in Drosophila and most studied. Annotation transfer from study of CRY1B proteins to mammals is thus exceedingly problematic given that the CRY1A family retains far more sequence similarity and is not descended from CRY1B. Clade-delimited gene duplications can lead to accelerated divergence as the two copies must subfunctionalize to garner selective pressure support.
A remarkable recent crystallographic result establishes an important role for tryptophan W536 near the end of the variable region of drosophila CRY1B. This aromatic residue and its associated helix arch back to occupy the site normally occupied by a damaged dna nucleotide, spoofing the presence of a damaged dna residue for conformational change purposes.
The tryptophan is part of a larger motif PPHCRPSNEEEVRQFMWLP conserved in the CRY1B orthologs from insects, crustaceans, molluscs and surprisingly three echinoderms. It is erroneously denoted the FFW motif in the fruit fly cryptochrome literature. Since 3.4 residues are needed for a full alpha helix turn, the 16 residues of the motif are enough for 4.7 turns (more than the 3 actually observed). Note the full motif is quite well conserved in amino acid sequence whereas the protrusion motif is not conserved either in residue or length.
However two substitutions should be noted, cysteine and tyrosine in daphnia and aphid respectively, suggesting that the overall motif is more critical than just a tryptophan. Further, no comparable residue or motif exists in invertebrate CRY1A proteins, vertebrate cryptochromes or other photolyase homologs. The observed phylogenetic distribution (which is unlikely to reflect convergent evolution) implies the spoofing mechanism arose in an early bilateran after to the gene duplication giving rise to CRY1A/CRY1B but before the protostome/deuterostome split.
A threonine at position 518 is reported phosphorylated in CRY1B_droMel but has no real phylogenetic support even within drosophilids, also lying outside the motif detected by WebLogo. This post-translational modification could nonetheless have regulatory signficance in the limited spectrum of species that could have it but more likely it represents an aberrant event.
For a full set of 38 invertebrate CRY1A and CRY1B sequence available in Mar 2012, see the curated reference sequences.
PPHCRPSNEEEVRQFMWLP
CRY1B_strPur KVVNKLRDTGIVHCAPSTQREVREFVWLPEKMAGGGSCRADQNCEGILGL echinoderm
CRY1B_lytVar KVINRLRDSGIVHCAPSTQKEVREFVWLPEKMAGGGSCRADQNCEGILGL echinoderm
CRY1B_parLiv KVINRLRDSGIVHCAPSTQKEVREFVWLPEKMAGGGSCRASQNCEGRTGS echinoderm
CRY1B_aplCal MEAIKKVSKDVPHIAPANEEEVLTLMWSGKQTRSELMDA----------- mollusc
CRY1B_craGig AVKDALIGKEIPHCAPSEEIEARRFSWLP--------------------- mollusc
CRY1B_octVul KVKEHLLHQDVPHCGPTNETEVWKFAWLPPIEHHDLAHNI---------- mollusc
CRY1B_rudPhi KNKLVQQGKDLEHCRPTNVEEVRMFVWMPGAHKGACGQEVPLDDKELCDG mollusc
CRY1B_plaOce MDRIKNLCKGIPHVAPTNENEVLSYMWLDKSNSEAMEESLFEACSHLSSV mollusc
CRY1B_dapPul KEFRQKFKETPAHCQPSSNSEVYKFFCLPDDSLPF--------------- crustacean
CRY1B_diaNig DEIRNRLMNPPPHCRPSSEKETRQFMWFPDDCSEHSSQ------------ orthoptera
CRY1B_acyPis LRVSMTNENRVPHCCPSDREEVQKFMYLPDECMQQLLPLENQDSKAYDIY hemiptera
CRY1B_danPle QELRRLLEKAPPHCCPSSEDEVRQFMWLGDDSQPELTTT----------- lepidoptera
CRY1B_bomMor EELRMLLEKAPPHCCPSSEDEIRQFMWLNE-------------------- lepidoptera
CRY1B_mamBra GELRHFLQKAPPHCCPSSEDEIRQFMWLNE-------------------- lepidoptera
CRY1B_helArm KELRHMLQKAPPHCCPSSEDEIRQFMWLNE-------------------- lepidoptera
CRY1B_droMel KSLRNSLITPPPHCRPSNEEEVRQFFWLADVVV----------------- diptera
CRY1B_anoGam REKLVDGGSTPPHCRPSDIEEIRQFFWLADDAATEA-------------- diptera
CRY1B_neoBul LIAEGAPDNGPPHCRPSNEEEIRNFFWLAD-------------------- diptera
CRY1B_bacCuc LIAGGAPDEGPPHCRPSNEEEVHQFFWLVE-------------------- diptera
CRY1B: C-terminal conservation in drosophilids * *
Drosophila melanogaster YECLIGVHYPERIIDLSMAVKRNMLAMKSLRNSLI T PPPHCRPSNEEEVRQFFWLAD
Drosophila simulans ................................... . .....................
Drosophila sechellia ................................... . .....................
Drosophila yakuba ............................T...... . .....................
Drosophila erecta ................T...Q.......A...... . .....................
Drosophila rhopaloa ............................A.....M . .....................
Drosophila elegans ............................A.....M . .....................
Drosophila takahashii ..................Y.........A.....M . .....................
Drosophila ficusphila .................L..........A...... . .....................
Drosophila eugracilis ............M....L..........A...... . .....................
Drosophila biarmipes .................VY.....M...A...... . .....................
Drosophila kikkawai .................K.........TA...... . .....................
Drosophila mojavensis ..........D......L.S........A...... E .....................
Drosophila persimilis ................K......M..TA...... . ....................N
Drosophila pseudoobscur .................K......M..TA...... . ....................N
Drosophila bipectinata ..........D.L...TK...G.........D... . ..............T......
Drosophila ananassae ..........D.L....K...G......T..D... . ..............T......
Drosophila willistoni ...........P.....L.L...T...TN...... . ....................E
Drosophila grimshawi ................L.S....A...A......E . T.................DE.
Drosophila virilis ....L.F...Q......L.S...TM...A...... E ...................TN
Cryptochrome CRY4 evolutionary origin
This orthology class has been studied to a limited extent in fish, frog and birds, often without full knowledge of the overall cryptochrome repertoire in the given species. Literature search is confused by erratic nomenclature practises in publications and gross mislabelling of GenBank reference sequences such as NM_001095521, which is CRY4 of Xenopus laevis rather than CRY1 as stated.
CRY4 has 19 reported transcripts originating from frog testes, oocytes, ovary and whole embryo whereas orthologous zebrafish transcripts have come from retina, eye, brain, heart, liver, paraxial mesoderm, caudal fin, tail bud, and embryo. Chicken has transcripts from brain, heart, kidney, limb, muscle, ovary; sparrow from brain; finch from embryonic brain. If these are representative -- rather than merely reflecting experimental focus -- this does not suggest continuity of function.
Cry4 has been lost in multiple clades and is missing from echinoderms,chondrichthyes, perciform fish, crocodillians, turtles and snakes, and all mammals. It is diverging quite rapidly in amphibians, with Xenopus laevis only 89% identical to Xenopus tropicalis. However according to Blast classification, it is present in tunicate and amphioxus.
The evolutionary orign of CRY4 has some puzzling aspects, though being restricted to deuterostomes it has limited parental gene options, basically CRY64 or CRY1. The 8th exon -- the far end of the FAD domain -- is split by a phase 21 intron. This is a derived condition since it is absent at homologous position in bilateran CRY64, CRY1, CRY2, CRY1A and CRY1B. Intron gain is quite rare in vertebrates but not in some earlier diverging bilaterans.
Split exon 8 cryptochromes (imputed* for processed transcripts in non-genomic species): CRY4_galGal Gallus gallus (chicken) CRY4_melGal Meleagris gallopavo (turkey) CRY4_anaPla Anas platyrhynchos (duck) CRY4_pasDom Passer domesticus (sparrow)* CRY4_taeGut Taeniopygia guttata (finch) CRY4_anoCar Anolis carolinensis (lizard) CRY4_xenTro Xenopus tropicalis (frog) CRY4_xenLae Xenopus laevis (frog) CRY4_latCha Latimeria chalumnae (coelocanth) CRY4_lepOcu Lepisosteus oculatus (spotted_gar) CRY4_danRer Danio rerio (zebrafish) CRY4_molTec Molgula tectiformis (tunicate)* CRY4_braFlo Branchiostoma floridae (amphioxus) CRY1_braFlo Branchiostoma floridae (amphioxus) CRY1A_strPur Strongylocentrotus purpuratus (urchin)
However the same intron is present in two amphioxus and sea urchin cryptochromes which classify as CRY1 and CRY1A rather than CRY4. These early deuterostome cryptochromes may be misclassified -- divergence is fairly high and synteny is gone, leaving match quality, introns and indels as the remaining diagnostic criteria. However blast classifier data shows high confidence classification of both, with the top 43 matches of the 278 reference gene set all being CRY1-class cryptochromes, with the best match of CRY64 and CRY4 from any species far more distantly related:
CRY1_braFlo Branchiostoma floridae (amphioxus) XM_00260... 2982 3.9e-314 CRY1_melUnd Melopsittacus undulatus (parakeet) AGAI0106... 2303 3.5e-242 CRY2_anoCar Anolis carolinensis (lizard) XM_003214641 2141 5.2e-225 CRY1A_aedAeg Aedes aegypti (mosquito) XM_001655728 dipte... 2062 1.2e-216 CRY64_xenTro Xenopus tropicalis (frog) synteny: STS1 RPL... 1595 3.7e-167 CRY4_xenTro Xenopus tropicalis (frog) NP_001123706 1428 1.9e-149 CRY1B_octVul Octopus vulgaris (octopus) JR450373 transcr... 1166 1.1e-121 CRY7_hapBur Haplochromis burtoni (chichlid) AFNZ01022319 380 7.0e-36 CRY1A_strPur Strongylocentrotus purpuratus (urchin) XM_0... 3045 7.7e-321 CRY1_xenTro Xenopus tropicalis (frog) NM_001087660 1153... 1966 1.8e-206 CRY2_anoCar Anolis carolinensis (lizard) XM_003214641 1919 1.7e-201 CRY1A_apiMel Apis mellifera (bee) NM_001083630 AADG06001... 1900 1.8e-199 CRY64_anoCar Anolis carolinensis (lizard) XM_003225714 6... 1562 1.2e-163 CRY4_lepOcu Lepisosteus oculatus (spotted_gar) AHAT0101... 1429 1.5e-149 CRY1B_parLiv Paracentrotus lividus (sea_urchin) AM599080... 1292 4.8e-135 CRY7_tetNig Tetraodon nigroviridis (fugu) 369 1.8e-34
In a gene with 530aa x 3bp/aa x 3 phases, there are 4770 ways of creating a new intron, so it is wholly implausible that the event was fixed twice. Thus the origin of CRY4 must be entangled with these other cryptochromes.
The amphioxus CRY4 sequence also has a fusion of exon 2 and exon 3. This also occurs in amphioxus CRY64 but nowhere else. Echinoderms lack CRY4, either having lost it or never having had one. A third peculiar feature of amphioxus CRY4 is the phase 12 intron between exon 4 and exon 5, representing a shift from ancestral phase 00. Amphioxus CRY64 also shares this phase shift but again earlier and later diverging orthologs do not.
The anomalous CRY1 sequences of amphioxus and sea urchin share another odd feature, a new phase 00 intron internal to exon 7. This intron does not occur in any other CRY1, CRY4 or CRY64 sequence and so is derived. Echinoderms and cephalochordates arose from separate divergence nodes; the event could not have taken place in a common ancestral stem. Regretably, no cryptochromes have been retained in the hemichordates, sister group to echinoderms, based on the Saccoglossus kowalevskii genome project.
Recall CRY4 is nearly adjacent to CRY1B of fish, the earlier CRY1 gene duplication preceding fish whole genome duplication lost in tetrapods. That could reflect tandem duplication or improbable but possible accidental juxtapositioning.
It takes a fairly complicated scenario to reconcile these observations. Suppose the earliest deuterostomes acquired the intron gain between exon 8-9 in CRY64. Assume further that this engaged in heterologous recombination with CRY1, leading to a polymorphic state that was never really fixed but persisted across the divergence of echinoderms and cephalochordates, with lineage sorting leaving CRY1 and CRY1A in those species with the extra intron but not those genes vertebrates.
After amphioxus diverged, its CRY64 acquired the fusion of exon 2-3 and phase shift between exon 4-5. It then duplicated, giving rise to amphioxus CRY4, but later lost the intron between exon 8-9 (though amphioxus CRY4 retained it). This would cause best-blast of amphioxus CRY4 to be amphioxus CRY64 rather than lie with other CRY4, as observed. In this case, the two amphioxus genes should be renamed CRY64A and CRY64B (replacing CRY4).
Gene_genSpp Exon 2-3 fusion Exon 4-5 phase shift Exon7 new 00 intron Exon 8 new 21 intron CRY1B sytney CRY1_other no no no no no CRY1_braFlo no no yes yes no CRY1A_strPur no no yes yes no CRY4_other no no no yes yes CRY4_braFlo yes yes no yes no CRY4_strPur --- --- --- --- --- CRY64_other no no no no no CRY64_braFlo yes yes no no no CRY64_strPur no no no no no
Cryptochrome 6-4 photolyases
CRY64 is a mainstream catalytic photolyase that reverses UV-induced (6-4) photoproducts in DNA using blue light. CRY64 gave rise to many cryptochromes over evolutionary time via sequential gene duplications. It originated in prokaryotes and persists into many invertebrates and amniotes. However it has been lost both in birds and mammals (which given the phylogenetic tree, requires two distinct loss events at a minimum). The reasons for loss, the adequacy of residual compensatory repair processes, and the consequences to final mutational rate are not well understood.
The carboxy terminus of CRY64 is a bit curious. The last two exons are provided below. Note the sequences become unalignable, despite all members retaining the same phase 12 splice donor. However clamping to the conserved exon break does not put the sequences into register for long -- even restricting to ray-finned fishes for which data is unfortunately overweighted. However the deuterostome sequences all retain a high content of basic residues at the end as shown below. (Other invertebrates have lost their introns and so lack this device for re-registration of the alignment.)
The available 3D structures (from Drosophila and Agrobacterium) only partly clarify the role of the carboxy terminal basic residues in vertebrates. The last determinable residues of CRY64 form a long terminal alpha helix highlighted in yellow in the adjacent image (and as hhh in the alignment below). However this extends (magenta) beyond the limit of blast alignability to vertebrates (blue) and the remained residues studied (gray dots) do not form a sufficiently stable conformation to display as fixed electron density. However this region clearly is positioned near the substrate binding site.
Possibly the terminal residues provide positively charged residues that offset negative backbone phosphates in the DNA strands. In this scenario, the primary sequence itself is not so important as long as it provides a sufficient number of flexibly positionable lysines and arginines. In this scenario, there is selection for amino acid composition rather than conventional residue by residue conservation. This could readily be tested by small terminal deletions.
hhhhhhhhhhhhhhhhhhhhhhhhh............. CRY64_droMel 2WQ7 HEVVHKENIKRMGAAYKVNREVRTGKEEESSFEEKSETSTSGKRKVRRATGSAPKRKR CRY64_anoCar KYLPFLRKFSNDYIYEPWKAPRSLQERAGCIIGQDYPKPIVEHEKVYKRNLERMKAAYARRSPNLVIQAKDKVSQKK GVNRKRPEAPTKAKVQAKKV CRY64_chrPic KYLPFLRKFPAEYIYEPWKAPRSMQEQAGCVIGRDYPKPIVVHEVVSKRNVERMKAAYARRSSSTTAQLEGGGGKKGI GAKRRTPAGPSVAELLTKKP CRY64_allMis KYLPILRKFPAEYIYEPWKAPRSMQEQAGCIIGRDYPKPIVEHEALSKRNIMRMKAAYAQRSHSKAAQVEKESTKKGN GGKRKLPAGPSVVELLTKKP CRY64_croPor KYLPILRKFPAEYIYEPWKAPRSMQEQAGCIIGRDYPRPIVEHEAVSKRNIMRMKAAYAQRSHSKSAQVEKEGTKKGN GGKRKLPAGPSVVELLTKKP CRY64_xenTro KYLPILKKFPAEYIYEPWKAPRSLQERAGCIIGKDYPKPIVEHDVASKQNIQRMKAAYARRSGSTAEVDKDSGQSNKN GAKRKVAGGPSVAELFKKNK CRY64_lepOcu KYLPVLKKFPSAYIYEPWKAPRSVQEQAGCIVGKDYPRPIVDHDVVSKKNIQRMKLAYARRAQLGGEQEGTGK GMKRKGQSVADLLTKKQKRN CRY64_danRer KYLPVLKKFSTEYIYEPWKAPRSVQERAGCIVGKDYPRPIVDHEVVHKKNILRMKAAYAKRSPEDKTINK GEKRKASPSIKEMFQKKAKR CRY64_salSal KYLPHLKKYPAQYIYEPWKAPRSVQEAAGCIVGKDYPRPIVEHEVISKKNIQRMKAAYAKRSPHSSEESP GKKEKGRKHKAPSVVDMLMK CRY64_gadMor KYLPVLKKFPVEYIYEPWKAPLSVQKAAGCIVGKDYPSPIVEHEVISKQNIQRMKTSYGKRSQGVSESPQPMKAEKRK GPSVLDMMKNKKKK CRY64_takRub KFLPHLKKFPAEYIFEPWKAPQSVQQAAGCIVGKDYPHPIVQHEVVSKKNIQRMKAAYAKRSANTAKSLSKIQ GLKRKPSSSVDMLKKKKKNN CRY64_tetNig KYLPHLKKFPAQYIYEPWKAPQSIQKAAGCIIGKDYPHPIVKHEEVSKKNIQRMKLAYARRSTSNAASPKKT GVKRKGPSVVDLLKKKRKKI CRY64_gasAcu KYLPLLKKFPAEYIYEPWKAPRSVQQAAGCIVGKDYPQPIAKHEVISKKNIQRMKLAYAKRSGDSAESANKSPVKRQ GTKRKAPSVVDMLKKKDRRK CRY64_oryLat KYLPILKKFPPQYIYEPWKAPRSVQQAAGCIVGKDYPKPIIEHEVISKKNIQRMKQAYARRTSGSTESPTKKQ GVKRKAPTVVDLIQKKQKRS CRY64_oreNil KYLPLLKKFPAEYIYEPWKAPRSIQQAAGCIVGKDYPHPIVQHEVISKKNIQRMKLAYAKRSPDTTESPSKSK GVKRKAPSIIEMIKKKAKVK CRY64_braFlo HYLPVLKNFPKEYIYEPWKAPRNVQEKAGCIVGKDYPRPIVDHKEASQRNLDIMRDVRKDQKETAAVTL GYGK CRY64_strPur KYIPALNKLPAEYIYEPWTAPRSVQEAAGCIIGRDYPRPIVDHSIVSKRNIGRMKDARACQPGKKA EKRPAEPSKQDNNGKKVRKITSMLKKK CRY64_lytVar KYIPIMERFPAQYIYEPWTAPRSVQEAAGCIIGRDYPRPIVDHSVVSKRNIGRMKDARACQPGKSA EKRPTDASNKNSNGKVRKITSMLKKK
For the full set of 22 metazoan CRY64 sequences, see the curated reference sequence section. A single representative sequence is shown below.
>CRY64_anoCar Anolis carolinensis (lizard) XM_003225714 6-4 photolyase synteny: DCPS TIRAP CRY64 SRPR FOXRED1 0 MAHVSIHWFRKGLRLHDNPALLAAMKNSAEIYPIFILDPWFPKNMQVSINRWRFLIESLKDLDESLKKLNSR 2 1 LFVVRGRPAEVFPELFTKWKVTRLAFEVDTEPYARRDAEVVRLAAEHGVQVIQKVSHTLYDTER 2 1 IIVENSGKAPLTYTRLQTLVASLGPPKQPVPAPKLEDMK 1 2 DCCTPVKEDHDLEYGTPSYEELGQDPKTAGPHLYPGGETEALARLDLHMKRT 0 0 SWVCNFKKPETHPNSLTPSTTVLSPYVKFGCLSVRMFWWKLAEVYQG 0 0 RKHSDPPVSLHGQLLWREFFYTAGAGIPNFDRMENNPVCVQVDWDNNQEYLRAWRE 0 0 GQTGYPFIDAIMTQLRTEGWIHHLARHAVACFLTRGDLWISWEEGQK 0 0 VFEELLLDADWSLNAANWQWLSASAFFHQFFRVYSPVTFGKKTDKNGEYIK 2 1 KYLPFLRKFSNDYIYEPWKAPRSLQERAGCIIGQDYPKPIVEHEKVYKRNLERMKAAYARRSPNLVIQAKDKVSQKKGV 1 2 NRKRPEAPTKAKVQAKKVKTKSS* 0
Cryptochrome CRY7 photolyases
For the full set of 14 bilateran CRY7 sequences, see the curated reference sequence section.
Below the frog protein CRY7 is marked up for its various domains and motifs according to Pfam, Blast and PDB searches. Blue shows the antenna domain with predicted α/β secondary structure, purple the possibly catalytic FAD domain with predicted all α secondary structure, magenta the UIM ubiquitin motif, green two compositionally simple regions rich is basic residues predicted not to have definite fold, red the conserved region of unknown function upstream of the UIM ubiquitin motif, and light blue the conserved carboxy terminal motif of unknown function.
>CRY7_xenTro Xenopus tropicalis (frog) 0 MDLEPFERAQIDDVLQ QLESGSVQADEFLCLVLSILGSSRTYSQFPAILQSLSRKEPAMYRELMDLHAEYFRK 0 0 EPADLETLGYETDLELAIALSLQEHNQLTDTASFASEVDPAPKISFADAAKLSHFSHKHNKKNSSSKTEITKLKDNVAAMNLYQERKRYHINGQEKTCISN CYNGQPEPEDCVLKSEDGEDVFHVETSRPRESKAKHSRRSRKKKKSAPSRGLVAMKPVLVWFRRDLRLHDNPALISALEHGVPVIPVFLWCINEETGQNFTLATGGAT KYWLHHALLKLNQSLIQRFGSHIIFRVARSCEEELVSLVHETGADTIIINAVYEPWLKERDDLISETLRRHGVELKKHHSYCLYEPDSVSTEGVGLR 1 2 GIGSVSHFMSCCKRNNSAPIGMPLDAPRCLPAPCNWPESDHLDTLELGKMPHRKDGTL 0 0 IDWAVTIRESWDFSEDGAYTCLANFLQD 1 2 GVKHYEKESGRADKPYTSHISPYLHFGQISPRTVLHEAYFTKKNVPKFLRKLAWRDLAYWLLILFPDMPSEPVRPAYK 0 0 SQRWSSDLNHLRAWQKGLTGYPLVDAAMRELWLTGWMCNYSRHVVASFLVAYLHIHWVHGYRWFQ 0 0 DTLLDADVAINAMMWQNGGMSGLDHWNFVMHPVDSALTCDPYGSYVRKWCPELAGLPDEYIHKPWKCAPSQLRRA 1 2 GVILGRNYPHRIVLDLEERREQSLKDVVEVRKKHLEYLDEVSGCDMVQIPDQLLALTLGHTSGEDEVVRNRTGSFLLPVITRKEFKYKTLQPDTKDNPYNTVLKGYV SRKRDETIAYMNERHFTASTINEGAQRHERIERTNRLMEGLPAPSDAKNKSRRTPKKDPFSIIPPSYLHLAN* 0
DASH: spotty phylogenetic distribution and unexplained carboxy terminal extension
DASH is yet another member of the cryptochrome and photolyase family. It was identified only recently as active only on ssDNA repair, reportedly because of a barrier to flipping the damaged cyclobutane pyrimidine dimer dinucleotide out of dsDNA into the active repair site unless the damaged base lies in a loop. In species investigated to date, this enzyme uses folate (MTHF) as antenna and FAD activated by blue light. It is a fairly remote outgroup to cytochromes, with only CPD further diverged.
Its name is a peculiar acronym of Drosophila, Arabidopsis, Synechocystis and Homo -- yet the gene was never present in Drosophila or placentals. In Arabidopsis, the principal copy is called CRY3, again in contravention of photolyase naming conventions. The numerous genome projects available today allow a quick determination of its rather unusual phylogenetic distribution.
Although originally studied in plants and cyanobacteria, the DASH photolyase surprisingly extends into fish, frogs, salamanders, turtle, lizard, and birds -- duck, finch and budgerigar (chicken and turkey have pseudogenes) -- but not any mammal. It is not known if the DNA repair function has been retained in all these taxa or has drifted in new roles like CRY1 and CRY2 in land vertebrates.
Blastx on the syntenic region in gallinaceous birds (chicken and turkey) establishes rather degenerate multi-exonic pseudogenes at the expected location and strand orientation. Here duck, which has an intact gene, is the immediate outgroup, diverging at 80 myr. It is not currently possible to date this more precisely nor determine whether pseudogenization occurred in a common ancestor or independently, perhaps on account of separate domestications. Platypus lacks pseudogene debris at the expected location but the assembly is currently unsatisfactory here. Marsupials and placentals would never have had this enzyme assuming lost shortly after divergence with the last common ancestor with birds.
The phylogenetic loss pattern of DASH in mammals is reminiscent the massive loss of opsins that also occurred early in mammalian evolution -- which GT Walls in 1942 attributed to mammals experiencing a sustained period of deep nocturnality where these systems did not need to function (no UV damage) and indeed could not function (insufficient blue light even with antenna) and so were lost, implying they were not sustained by a Piatigorskyian secondary functionality such as circadian rhythm, lunar calendaring, or magnetosensing.
DASH is also missing from alligator and crocodile assemblies, deep-water lobe-finned fish (coelocanth) have a pseudogene, and cartilaginous fish to date lack it completely. These probably reflect multiple independent gene losses rather than inadequate assemblies. DASH is restricted within invertebrates to crustaceans and mollusks, a pattern which could have arisen from stem losses in insects etc. The sole insect DASH at GenBank (whitefly EZ942653) appears to be a fungal contaminant.
The great oxygenation event gave rise to a stratospheric ozone protective layer at 2.4 gyr but reached an even higher lever during the early Cambrian (based indirectly on oxygen levels). If more ozone meant less DNA damage from UV, this may have favored independent but simultaneous gene loss events in various clades. However the persistence of DASH in ray-finned fish for 450 myrs raises the question of whether UV light penetration of sea water is the sole or even principle cause of DASH-repairable DNA damage -- if indeed DASH is still a repair enzyme in benthic species. However the first land plants and animals were plausibly exposed to greatly increased levels of UV damage that may correlate with DASH retention.
Multi-cellular animals from cnidarian to amniote all have a short C-terminal extensional exon whose distal region contains a conserved motif of unknown function. This is positioned to cap the binding site in the manner of CRY64_droMel but there is no evidence that it does -- while positively charged arginines and lysines that might offset negative DNA phosphates are among the conserved residues, so are negatively charged glutamates, polar, neutral and aromatic residues. If this domain does prove to be a structural cap, it represents convergent evolution with respect to the CRY64 cap domain because the two orthology classes diverged long before the caps evolved.
Overall sequence conservation of DASH is less stringent than other photolyases and cytochromes, suggesting loosened constraints or a measure of functional redundancy with respect to other repair enzymes. However it is difficult to understand how antenna domain -- though less conserved than the FAD domain -- could be conserved over vast spans of branch length in the absence of function (antenna molecule binding and/or something else).
Amniote DASH proteins can be modeled structurally using nearly 50% matches in Arabidopsis (cryptochrome 3: 2IJG) or equally suitable cyanobacterium Synechocystis (1NP7). However these structures do not provide any information on the C-terminal extension. The 14 exons share only one match with vertebrate CRY1 and CRY2 -- and that is more likely coincidental than indicative of a shared ancestral protein subsequent to the main era of eukaryotic intronation.
For the full set of 30 metazoan DASH sequences and conservation alignments, see: Curated reference sequences for cryptochromes and photolyases
>DASH_taeGut Taeniopygia guttata (finch) antenna catalytic C-terminal motif 0 MSGTAGTAICLLRCDLRAHDNQ 0 0 QVLHWAQHNADFVIPLYCFDPRHYLGTHCYRLPKTGPHRLRFLLESVKDLRETLKKKGS 2 1 TLVVRKGKPEDVVCDLITQLGSVTAVVFHEE 0 0 ATQEELDVEKGLCQVCRQHGVKIQTFWGSTLYHRDDLPFRPIDR 2 1 LPDVYTHFPKGLESGAKVRPTLRMADQLKPLAPGLEEGSIPTMEDFGQK 1 2 DPVADPRTAFPCSGGETQALMRLQYYFWDT 0 0 NLVASYKETRNGLVGMDYSTKFAPW 2 1 LALGCISPRYIYEQIQKYERERTANESTYW 2 1 VLFELLWRDYFRFVALKYGRRIFSLR 1 2 GLQSKDIPWKKDLQLFSCWQ 0 0 EGKTGVPFVDANMRELSATGFMSNRGRQNVASFLTKDLGLDWRMGAEWFEYLL 0 0 VDYDVCSNYGNWLYSAGIGNDPRDNRKFNMIKQGLDYDGN 0 0 GDYVRLWVPELQGIKGADIHTPWALSSAALSQAGVTLGETYPQPVVTAPEWSRHIHRRP 0 0 GGSPHPRGRRGPAQRKDRGIDFYFSRKKDAC* 0
Cryptochrome CPD photolyases
This dna repair enzyme (cyclobutane pyrimidine dimers for CPD) was studied in marsupials during the pre-genomic era (1994), with two groups concluding even that that no ortholog existed in placentals. Today we are certain of that because the gene is not present in any complete placental mammal genome; no pseudogene debris exists in the partly conserved syntenic location in any species. This strongly suggests that the gene was lost once in stem placental rather than many times in later subclades (as happened with encephalopsin). The gene remains very strongly conserved in species such as opossum with no indication of impending loss.
The loss in placentals is somewhat peculiar given that CPD is a very ancient (pre-eukaryotal) member of the photolyase family, with highly conserved orthologs readily recoverable in other commonly studied marsupials, monotremes, birds, alligators, turtle, lizard, snakes, frog, fish, agnathan, amphioxus, sea urchin, many invertebrates, cnidarians, plants and so forth. However it also appears to be lost in tunicate -- indeed Ciona has lost all its photolyases leaving it a bit mysterious how it repairs these types of dna damage. Hemichordates have also lost all members of this gene family including CPD.
It is very unlikely that placentals displaced CPD with something better. More likely, CPD was lost during a dark phase of placental evolution when UV damage to dna was a non-issue and its photo-repair infeasible. Genes cannot be retained without selection (use it or lose it). Coming back out into the light millions of years later (having also lost DASH, CRY64 and [[Opsin_evolution:_update_blog|13 of 21 opsin genes]), they evidently made do with a less efficient excision repair that overlaps repair photolyase functionality.
The CPD gene product is very diverged from other photolyases though still retains the photolyase and FAD binding domain folds. The antenna moiety is usually reported as MTHF (folate). The best available structures are from rice (3UMV: 53% identity to marsupial) and an archaeal methanogen (Methanosarcina mazei 2XRZ) which likely uses 5-deazariboflavin Fo as antenna (which it can synthesize de novo). The latter enzyme repairs cyclobutane pyrimidine dimers in duplex DNA using blue or near-UV light.
Despite great divergence in primary sequence from other members of the gene family, fold conservation may explain in part the unexpected circadian compensatory capacity of marsupial CPD expressed in double CRY1/2 knockout mouse, seemingly driven by interaction of CPD with CLOCK of the CLOCK/BMAL1 system. CPD lacks any counterpart to the distal exons of placental CRY1.
CPD presents no special problems in classification as it clearly originated early in the history of prokaryotes and today serves as the outgroup to the overall metazoan photolyase gene family (though not as usefully as the less diverged DASH). It has never undergone gene duplication and divergence, at least none that stuck, and has been retained as single copy in the vast majority of species from choanflagellate to mammal. There are no noteworthy C-terminal expansions or supplemental exons within metazoan -- CPD is the exception among photolyases and cryptochromes for its lack of overt innovation. However as the knock-in experiment in mouse shows, CPD has unexpected properties.
The N-terminus has various extensions -- indeed the initial methionine is problematic -- but these are poorly conserved even within closely related taxa. Conservation sets in some 38 residues upstream of the first conserved methionine. While these 114 bp could represent conserved 5'UTR nucleotides rather than conserved amino acids, the two relevent crystallographic structures include this region (Methanosarcina 2XRY and rice 3UMV) as do many transcripts. Two in Xenopus (ES684787 BX851972) seem to rule out a cryptic short first exon splicing into the conserved region.
Some 32 curated CPD sequences spanning the whole of metazoan evolution are provided at the reference sequences. Many more could be extracted from GenBank should some research issue warrant more intensive surveying.
4Fe-4S photolyases and their relation to primases
An intriguing new subfamily of photolyases (1,2) contains a 4Fe-4S cluster in the catalytic domain in addition to an FAD binding site. This makes sense given the equally surprising finding of unmistakable fold homology between photolyases and the large subunit of archaeal-eukaryotic primase (eg the PRIM2 gene product of human).
This ancient enzyme is critical to de novo synthesis of the short RNA primers essential to DNA replication. Primase also contains a 4Fe-4S cluster as do numerous non-homologous DNA repair enzymes such as helicases and endonucleases. Such clusters have a redox role elsewhere in the cell but it is not immediately evident that's applicable here.
The photolyase antenna molecule is Rhodobacter is new but not entirely novel: the final intermediate in riboflavin biosynthesis, 6,7-dimethyl-8-ribityl-lumazine (which serves a similar role in bioluminescence). This illustrates again the plasticity of the antenna site -- the antenna molecule is unpredictable from primary sequence (indeed tertiary structure).
Since the list of possible antenna molecules is still growing, reconstitution experiments that don't find a suitable antenna molecule may simply have tested an insufficient range of molecules -- they have to be repeated as new ones emerge. Similarly, in silico docking can only fit what is on the list. Here we cannot be sure that other members of this new subfamily of photolyases will use this (or indeed any) antenna molecule.
The new class of photolyase conflicts with the notion of a universal tryptophan triad chain in photolyases, agreeing instead with reports in other photolyases suggesting that the whole concept -- or at least invariance part -- was limited in applicability.
Most gene families members in this class of proteins have more than the three ultra-conserved tryptophans. Simply knocking in a tyrosine at a site that has never tolerated a substitution for a hundred billion years of branch length evolution does not for test electron flow specifically any substitution at any invariant residue necessarily has major adverse effects: how else could it have been conserved for such a huge multiple of the neutral substitution rate?
Three inappropriate gene names for this new photolyase class -- PhrB is already in use at GenBank for a different photolyase class, CRYB suggests non-repair cryptochrome, FeS-BCP has an erroneous phylogenetic distribution and disallowed hyphen -- won't be used here but rather a provisional name PFES (photolyase iron sulfide). Reference sequences are provided below for two bacteria and two archaeal FeS photolyases, as well as yeast and human FeS primases; these suffice as GenBank blast probes.
Some confusion surrounds the human primase sequence because the NCBI reference genome (Build 37.1) carries only a pseudogene -- a copy number variant bordering the centromere of chromosome 6, with the actual gene is still missing from the June 2012 reference genome, causing transcripts to mis-align with genome at 11 of 509 amino acids. Bizarrely, these discrepancies -- including an internal stop codon in exon 11 -- were noted by NCBI in accession BC064931 but never resolved because the chimpanzee assembly was also wrong in the same way. It is inconceivable that project DNA donors lacked a working copy of this very essential gene.
Note human primase has two components. The smaller catalytic unit is encoded by PRIM1 on chr 12. This gene has a very distant homolog CCDC111 (unofficially renamed PrimPol) on chr 4 containing an extra C-terminal zinc finger domain. Neither of these contains a 4Fe-4S cluster. The larger auxillary subunit is encoded by PRIM2 on chr 6. It contains the 4Fe-4S cluster discussed here. PRIM2 has no additional homologs in human (or other multi-cellular organisms).
Using blastp and the 4 conserved cysteines as guide to presence of the iron sulfur cluster , bacterial representatives of the new photolyase class are readily located in 150 genera, largely alphaproteobacter) but are more narrowly distributed in Archaea (8 of 49 genera of Euryarchaeota but no Thaumarchaeota, Aigarchaeota, Korarchaeota, Crenarchaeota in 33 genomes tested) suggesting horizontal gene transfer to (or from) Euryarchaeota or stem gene loss in the TACT group.
Since the eukaryotes acquired mitochondria from a relatively late endosymbiosis with an alphaproteobacter, a gene copy might initially have been present.
No eukaryotic photolyase to date (18 April 2014) has retained or independently re-developed a 4Fe-4S domain (ignoring blast matches such as XM_002537565 in castor bean that represents Agrobacterium contamination). The Agrobacterium photolyase itself is surprisingly a 6-4 photolyase with a well-established Fe-S cluster and 6,7-dimethyl-8-ribityllumazine antenna molecule. As noted by Zhang et al, its structural homology to primases places the 6-4 photolyases at the root of the whole cryptochrome/photolyase family.
Curiously, the four cysteines coordinating iron are not deeply conserved. For example, yeast primase cysteines 336/417/434/474 cannot be fully homologous to Agrobacterium cysteines where the linear order is different 350/454/438/441. During evolution, other cysteines must have arisen and as the fold diverged, replaced some of the originals. (A similar phenomenon occurs with disulfides, for example in sulfatases.) However the four cysteines and their exact spacing (up to a one residue indel) are conserved between yeast and human primases, representing conservation over a billion years since their divergence.
The 4Fe-4S cluster of primase is surely an ancient feature of primase and thus of the whole fold family descended from it, suggesting that FeS-photolyases are a relic of an old gene duplication, retaining a feature lost in subsequent duplications giving rise first to CPD and then to the overall photolyase/cryptochrome gene family.
The alternative scenario, that the 4Fe-4S cluster represents convergent evolution in photolyases (later independent acquisition) at first seems implausible given the requirements of cubane geometry, the complexity of the auxiliary enzymes and scaffolding proteins involved in 4Fe-4S assembly, and the lack of functioning intermediate states. However the eukaryotic proteome overall contains a large and heterogeneous set of iron-sulfur proteins; there is no support for a 4Fe-4S cluster as a mobile duplicated domain.
It is not clear how many distinct homology classes exist for 4Fe-4S domains even restricting to DNA proteins -- primary sequence is not immediately helpful given deep divergences of these ancient proteins, cysteines anchors of an alignment might only represent convergent evolution, as could short fold similarities recognized by Dali given the geometrical constraints. If one supposes a late-stage cluster assembly protein such as MMS19 provides 4Fe-4S cluster to structurally dissimilar fold classes localized to the nucleus, then what is the common ground biochemically for recognition of apoprotein?
It has not proved feasibly to date to develop a bioinformatic screen that catches the full repertoire of 4Fe-4S clusters in DNA proteins in the yeast/human proteomes because the conserved cysteine pattern can be confused with bona fide zinc binding sites (eg zinc ribbons) that themselves lack distinctive signatures. Proof of that can be seen from the large number of 4Fe-4S clusters only recognized in 2011-2013 -- in enzymes studied intensively for decades.
A 4Fe-4S cluster has a clear enough spectroscopic signature, the problem arises from the lability of clusters when the protein is purified in the presence of oxygen. When the cluster is lost, its binding domain loses its rigidity, becoming structurally indeterminable in crystallographic studies. Alternatively, a zinc ion occupies the site, causing the structural determination to proceed to an erroneous conclusion. Zinc in 4Fe-4S cluster proteins could represent artifact, placeholder, protection, idle cycle, or even functionally viable alternative.
While zinc ions are ubiquitous throughout the cell as more or less harmless (in contrast to iron) atoms that spontaneously find to their target sites by diffusion (like magnesium ions), 4Fe-4S clusters are not free-floating constituents of the mitochondria, cytoplasm or nucleoplasm. Instead, they are built and held on scaffolding proteins, then passed along a complex chain of chaperones and assembly proteins for insertion into apoprotein, with no aspect of the process left to chance chemistry.
Although in most of biochemistry, 4Fe-4S clusters serve a clear redox function, such a role has not been established for primases, helicases, other DNA repair enzymes, much less PFES photolyases. Conceivably the redox state of the 4Fe-4S cluster can sense the status of a DNA helix and facilitate rapid scanning for the odd damaged base among billions of normal ones. The photolyases present an interesting situation because only one of many orthology classes utilizes an iron sulfur cluster, whereas it would make sense given the newly recognized ubiquity for all of them to have it. Thus the novelty is turned around -- how can other photolyases work without an iron sulfur cluster?
Primase may be among the very oldest of enzymes since it is essential for DNA replication (ie, perhaps for exiting the hypothetical earlier RNA world). However UV damage is also a very old issue, especially for the billion years of life preceding oxygenation of the atmosphere (which led to the ozone shield of today). Priming is not needed for RNA replication or transcription nor in DNA replication in mitochondria; bacteria use a non-homologous system based on the DNAG protein.
One intriguing idea starts with the observation that FAD mimics two free RNA bases with its flavin and adenine rings which are are stacked like bases (U-folded) in all studied photolyases. In primase -- which has no FAD -- two purine ribonucleotides at the FAD site may recognize two bases of template DNA by conventional hydrogen bonding that perhaps resemble the flipped out cyclobutane pair needing repair by a photolyase.
Indeed, the template dinucleotide could even be stabilized temporarily as a cyclobutane pair, reversing the normal sense of the reaction, borrowing reductive units from the 4Fe-4S cluster (UV/blue light is not a known primase requirement). This would explain primase preference for a pyrimidine template. Photolyases then arose by replacing the two mononucleotides with FAD and adding a Rossmann-like domain for the antenna, with the utilization of light displacing the need for the 4Fe-4S cluster except in the PFES class of photolyases.
Human primase also undergoes a profound conformational change from a three-helix binding site for DNA to a helix-sheet site as it counts primer size and passes it along to the catalytic subunit and other protein partners. That's not so clear for not-so-large subunits archaeal primases which seem to lack an internal domain duplication. A large conformational change -- not just internal changes in FAD redox status -- is also needed in cryptochrome signaling, possibly this same one.
>PFES_agrTum Agrobacterium tumefaciens (bacteria) NP_355900 aka: PhrB pdb:4DJA MSQLVLILGDQLSPSIAALDGVDKKQDTIVLCEVMAEASYVGHHKKKIAFIFSAMRHFAEELRGEGYRVRYTRIDDADNAGSFTGEVKRAIDDLTPSRIC VTEPGEWRVRSEMDGFAGAFGIQVDIRSDRRFLSSHGEFRNWAAGRKSLTMEYFYREMRRKTGLLMNGEQPVGGRWNFDAENRQPARPDLLRPKHPVFAP DKITKEVIDTVERLFPDNFGKLENFGFAVTRTDAERALSAFIDDFLCNFGATQDAMLQDDPNLNHSLLSFYINCGLLDALDVCKAAERAYHEGGAPLNAV EGFIRQIIGWREYMRGIYWLAGPDYVDSNFFENDRSLPVFYWTGKTHMNCMAKVITETIENAYAHHIQRLMITGNFALLAGIDPKAVHRWYLEVYADAYE WVELPNVIGMSQFADGGFLGTKPYAASGNYINRMSDYCDTCRYDPKERLGDNACPFNALYWDFLARNREKLKSNHRLAQPYATWARMSEDVRHDLRAKAAAFLRKLD* >PFES_rhoSph Rhodobacter sphaeroides (bacteria) CP000144 Alphaproteobacteria PDB|3ZXS PMID:22290493 6,7-dimethyl-8-ribityl-lumazine antenna aka CryPro 4Fe-4S photolyase MRGSHHHHHHGIRMLTRLILVLGDQLSDDLPALRAADPAADLVVMAEVMEEGTYVPHHPQKIALILAAMRKFARRLQERGFRVAYSRLDDPDTGPSIGAE LLRRAAETGAREAVATRPGDWRLIEALEAMPLPVRFLPDDRFLCPADEFARWTEGRKQLRMEWFYREMRRRTGLLMEGDEPAGGKWNFDTENRKPAAPDL LRPRPLRFEPDAEVRAVLDLVEARFPRHFGRLRPFHWATDRAEALRALDHFIRESLPRFGDEQDAMLADDPFLSHALLSSSMNLGLLGPMEVCRRAETEW REGRAPLNAVEGFIRQILGWREYVRGIWTLSGPDYIRSNGLGHSAALPPLYWGKPTRMACLSAAVAQTRDLAYAHHIQRLMVTGNFALLAGVDPAEVHEW YLSVYIDALEWVEAPNTIGMSQFADHGLLGSKPYVSSGAYIDRMSDYCRGCAYAVKDRTGPRACPFNLLYWHFLNRHRARFERNPRMVQMYRTWDRMEET HRARVLTEAEAFLGRLHAGEPV* >PFES_metMah Methanohalophilus mahii (Euryarchaeota) CP001994 4Fe-4S photolyase MRHYAEKLRNRGADITYIKTAELEKSLSRWIKKKGIDELNIAEPANITLKEYLGKLNIDCKIVFVDNKQFIWSIPEFNTWASSRKNLIMEDFYRTGRKNSEI LLEKDGKPSGGKWNLDRENRKLPPKNGFQKKPPQHIKFSPDKITKEIIAEVERSEYPTYGKGKDFNLAVTHEDAQKALDFFIEEKLSNFGPYQDIMLTGDNVLWHSILSPYLNLGL LHPLNVIKKAELAYYQKNLPLNSIEGFIRQILGWREYMHCIYKYTGDKYLKSNWFDHERELPDIYWYPERTSMNCMASVIEEVLNTGYAHHIQRLMILSNFALLAEVNPAKVKNWF HAAFIDAYDWVMQPNVIGMGQFADGGILATKPYISSANYINKMSDYCQNCTYNHNHRTGEDACPFNYLYWAFLHKNNEKLRDIGRMKLILKNLDRINKKELKQIMTHADDFLKSLK* >PFES_natPha Natronomonas pharaonis (Euryarchaeota) CR936257 4Fe-4S photolyase MTVLVLGDCLTEFGPLASDARSTDERVLCIEARAFARRKPYHPHKLTLVFSAMRHFRDRLREAGYTVDYRRVETFAEGLDAHFAAHPEDHIVTVRRTAHGAT DRLQRLVANRGGTVEFVADPRFHCSREEFDAWADGDPPYRHESFYRHMRRETGYLMDGDEPVGGEWNFDDENREFPGPEYVPPEPPQFEPDETTREVREWVDATFGEDGYDDAPYG GAWADPEPFSWPVTREGALQALEAFIEERLPTFGPYQDAMLGDEWAMNHALLSSSLNLGLLSPSEVIEAALAAFEEGSVSIASVEGFLRQVLGWREFVRHAYRRTPGMAAANQLGA AEPLPEFFWTGDTDMACVADAVDGVRTRGYAHHIERLMVLSNFATLYGVEPSRLNEWFHAAFVDAYHWVTTPNVVGMGTFGTDTLSTKPYVASANYIDRMSDHCSGCPYYKTKTTG DGACPFNALYWDFLGRNESQLRSNHRMGLVYSHYDDKSDGEREAIADRAETLRQRARNGTL* >PRIM2_homSap Homo sapiens (human) NM_000947 primase large subunit 4Fe-4S pdb|3L9Q,3Q36 0 MEFSGRKWRKLRLAGDQRNASYPHCLQFYLQPPSENISLIEFENLAIDRVK 1 2 LLKSVENLGVSYVKGTEQYQSKLESELR 0 0 KLKFSYRENLEDEYEPRRRDHISHFILRLAYCQS 2 1 EELRRWFIQQEMDLLRFRFSILPKDKIQDFLKDSQLQFEA 0 0 ISDEEKTLREQEIVASSPSLSGLKLGFESIYK 0 0 IPFADALDLFRGRKVYLEDGFAYVPLKDIVAIILNEFRAKLSKALA 0 0 LTARSLPAVQSDERLQPLLNHLS 2 1 HSYTGQDYSTQGNVGKISLDQIDL 0 0 LSTKSFPPCMRQLHKALRENHHLRHGGRMQYGLFLKGIGLTLEQALQFWKQEFIKGKMDPDK 0 0 FDKGYSYNIRHSFGKEGKRTDYTPFSCLKIILSNPPSQGDYH 1 2 GCPFRHSDPELLKQKLQSYKISPGGISQ 0 0 ILDLVKGTHYQVACQKYFEMIHN 0 0 VDDCGFSLNHPNQFFCESQRILNGGKDIKKEPIQPETPQPKPSVQKTKDASSALASLNSSLEMDMEGLEDYFSEDS* >PRIM2_sacCer Saccharomyces cerevisiae (yeast) P20457 aka: PRI2_YEAST primase large subunit PDB|3LGB MFRQSKRRIASRKNFSSYDDIVKSELDVGNTNAANQIILSSSSSEEEKKLYARLYESKLSFYDLPPQGEITLEQFEIWAIDRLKILLEIESCLSRNKSIK EIETIIKPQFQKLLPFNTESLEDRKKDYYSHFILRLCFCRSKELREKFVRAETFLFKIRFNMLTSTDQTKFVQSLDLPLLQFISNEEKAELSHQLYQTVS ASLQFQLNLNEEHQRKQYFQQEKFIKLPFENVIELVGNRLVFLKDGYAYLPQFQQLNLLSNEFASKLNQELIKTYQYLPRLNEDDRLLPILNHLSSGYTI ADFNQQKANQFSENVDDEINAQSVWSEEISSNYPLCIKNLMEGLKKNHHLRYYGRQQLSLFLKGIGLSADEALKFWSEAFTRNGNMTMEKFNKEYRYSFR HNYGLEGNRINYKPWDCHTILSKPRPGRGDYHGCPFRDWSHERLSAELRSMKLTQAQIISVLDSCQKGEYTIACTKVFEMTHNSASADLEIGEQTHIAHP NLYFERSRQLQKKQQKLEKEKLFNNGNH*
278 curated refSeqs for metazoan cryptochromes and photolyases
The full length sequences have been moved to a separate page; only headers are shown below. The sequences use augmented fasta format transparent to web tools: primary sequence broken into exons, codon phase (bp overhang) shown, marked up for features with color, grouped into orthologous clusters, and presented in phylogenetic order relative to human evolutionary history, with subtree order determined by assembly quality.
The fasta headers themselves are little databases showing gene name (following HUGO symbol rules), genus, species, common name, genomic and transcript accession number when not a routine NCBI blast match, PubMed id if specifically studied in a journal article, followed by an unstructured comment field. Both headers and sequences fall readily into desktop databases, allowing different sort orders for other investigative priorities.
The availability of some orthology classes is inherently limited due to recent origin, restricted phylogenetic retention and the uneven focus of sequencing effort across the phylogenetic tree of metazoans. Genomic sequencing of 10,000 vertebrates will not greatly benefit cryptochrome research because the vast majority will be mammals, birds and perch-like fish which are excessively represented already. What is needed are more and better assemblies for a handful of keystone species such as lamprey, hagfish, sharks and rays, bichir, lungfish, and especially amphibians and herptiles.
For species with good assemblies, the entire repertoire of cryptochromes and photolyases has been deduced. It is foolish to compare a gene in isolation across two species with different overall gene family complements because multiple roles and functional complementation may have evolved.
For a large gene with numerous exons, absence from the assembly usually means genuine absence from the genome. Even when only an exon or two gene fragment is available, the classifier can almost always assign the correct orthology class to it. However it is risky to assemble an entire gene from many unlinked single-exon contigs and that was not done here; however certain important clades such as cartilaginous fish lack coherent assemblies and adequate transcripts so provisional gene assemblies are provided.
A remarkable amount of the data has surfaced at GenBank only in the last six months, implying much weaker results had the project been done in 2011 but also that much better phylogenetic coverage will surface this year. For the full set of fasta sequences available in April 2012, see the reference sequence repository. Manually curated sequences -- which use all available data and internal orthology class consistency checks -- should not be equated with provisional unsupervised computerized efforts at GenBank (XM_ gnomon entries), the UCSC 46-way or Ensembl.
CRY1_homSap Homo sapiens (human) CRY1_panTro Pan troglodytes (chimpanzee) XM_509339 CRY1_ponAbe Pongo abelii (orangutan) XM_002823690 CRY1_nomLeu Nomascus leucogenys (gibbon) XM_003269977 CRY1_macMul Macaca mulatta (rhesus) NM_001194159 CRY1_calJac Callithrix jacchus (marmoset) XM_002752946 CRY1_saiBol Saimiri boliviensis (squirrel_monkey) nearly identical to marmoset CRY1_tarSyr Tarsius syrichta (tarsier) ABRT010205577 unsure if exon 2 is CRY1 or CRY2 CRY1_micMur Microcebus murinus (mouse_lemur) CRY1_otoGar Otolemur garnettii (bushbaby) AAQR03016495 CRY1_tupBel Tupaia belangeri (treeshrew) CRY1_musMus Mus musculus (mouse) NM_007771 all transcripts support longer exon 10 lost splice donor CRY1_ratNor Rattus norvegicus (rat) NM_198750 CRY1_criGri Cricetulus griseus (hamster) XM_003505292 CRY1_spaJud Spalax judaei (blind_mole_rat) AJ606298 CRY1_dipOrd Dipodomys ordii (kangaroo_rat) ABRO01202522 ABRO01202521 CRY1_hetGla Heterocephalus glaber (mole-rat) stop codon in place of conserved W8, last two exons very diverged CRY1_cavPor Cavia porcellus (guinea pig) last two exons diverged 69 bp separation CRY1_speTri Spermophilus tridecemlineatus (squirrel) Ictidomys CRY1_oryCun Oryctolagus cuniculus (rabbit) CRY1_oviAri Ovis aries (sheep) NM_001129735 19341811 19150926 CRY1_bosTau Bos taurus (cow) NM_001105415 XM_616063 CRY1_susScr Sus scrofa (pig) XM_003126079 CRY1_ailMel Ailuropoda melanoleuca (panda) XM_002927658 CRY1_loxAfr Loxodonta africana (elephant) XM_003405313 CRY1_triMan Trichechus manatus (manatee) AHIN01036366 AHIN01036362 very similar to elephant CRY1_monDom Monodelphis domestica (opossum) XM_003341966 CRY1_macEug Macropus eugenii (wallaby) assembly frameshift CRY1_sarHar Sarcophilus harrisii (tasmanian_devil) nearly identical to oppossum CRY1_triVul Trichosurus vulpecula (possum) EC362500 terminal transcript CRY1_ornAna Ornithorhynchus anatinus (platypus) XM_001508563 = rubbish, genomic frameshift, continuing exon 12 CRY1_tacAcu Tachyglossus aculeatus (echidna) SRR000649.130490 short read transcripts corrected for frameshifts, penultimate exon CRY1_galGal Gallus gallus (chicken) PMID: 11684328,17324421,15459395 altSplExon11: GIVGVPICRGSADLCN* BU143111 CRY1_melGal Meleagris gallopavo (turkey) XM_003202363 altSplExon11: GTVGVPICRGSANWYK* CRY1_anaPla Anas platyrhynchos (duck) scaffold157 altSplExon11: GMTGVLVCRGSPGSHNYGKKDKT* CRY1_eriRub Erithacus rubecula (robin) AY585716 aka: CRY1A altSplExon11: GIMAVPVCRGSPNACNYGKPDKTSK* CRY1B CRY1_sylBor Sylvia borin (warbler) AJ632120 aka: CRY1A PMID:15381765 altSplExon11: GIVAVAVCRGSPNPCNYGKPDKTSE* sylBor DQ838738 CRY1B CRY1_taeGut Taeniopygia guttata (finch) XM_002196518 altSplExon11: GIMAVPVCRGSPNPCNYRKPDKTSK* CRY1_melUnd Melopsittacus undulatus (parakeet) AGAI01062111 altSplExon11: GIMAVPVCRGSSNPCNCGKTDKTSK* CRY1_parWeb Paradoxornis webbianus (parrotbill) JR867166 TSA transcript CRY1_allMis Alligator mississippiensis (alligator) genome/blat CRY1_anoCar Anolis carolinensis (lizard) XM_003220923 AAWZ02014443 CRY1_podSic Podarcis siculus (wall_lizard) DQ376040 16809482 CRY1_pytMol Python molurus (python) AEQU010547455 CRY1_chrPic Chrysemys picta (turtle) AHGY01469963 AHGY01469969 CRY1_xenTro Xenopus tropicalis (frog) NM_001087660 11533577 final four exons confirmed by many ESTs CRY1A_latCha Latimeria chalumnae (coelocanth) AFYH01018055 AFYH01018053 AFYH01018050 CRY1B_latCha Latimeria chalumnae (coelocanth) last exons uncertain CRY1A_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01025403 CRY1B_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01016727 AHAT01016728 CRY1A_danRer Danio rerio (zebrafish) NM_001077297 whole genome duplicate of retained CRY1 duplicate CRY1A2_danRer Danio rerio (zebrafish) BC044558 AW184635 olfactory old teleost CRY1 duplicate syntenically retained as tetrapod CRY1 CRY1B_danRer Danio rerio (zebrafish) BC095305 EB921055 aka CRY2A whole genome duplicate of lost CRY1 duplicate CRY1C_danRer Danio rerio (zebrafish) BC164795 EE210836 aka CRY2B old CRY1 duplicate lost in tetrapods CRY1 C12ORF23 CRY4 CRY1A_leuEri Leucoraja erinacea (skate) AESE010236716 AESE011153531 AESE010038968 AESE010673288 AESE012524396 CRY1B_leuEri Leucoraja erinacea (skate) AESE011669465 AESE012563587 AESE010604630 AESE011547252 CRY1A_calMil Callorhinchus milii (shark) AAVX01551101 AAVX01266331 AAVX01354908 AAVX01055947 CRY1B_calMil Callorhinchus milii (shark) AAVX01090452 AAVX01101328 AAVX01636526 AAVX01201905 CRY1_petMar Petromyzon marinus (lamprey) Contig24766 CRY1_braFlo Branchiostoma floridae (amphioxus) XM_002609455 end uncertain CRY1A_strPur Strongylocentrotus purpuratus (urchin) XM_001194752 same split exons as braFlo, end of gene uncertain, partially duplicated CRY2_homSap Homo sapiens (human) 11 exons CRY2_panTro Pan troglodytes (chimp) CRY2_gorGor Gorilla gorilla (gorilla) CRY2_ponAbe Pongo pygmaeus (orangutan) CRY2_rheMac Macaca mulatta (rhesus) CJ488220 testis CRY2_papHam Papio hamadryas (baboon) CRY2_calJac Callithrix jacchus (marmoset) CRY2_micMur Microcebus murinus (mouse_lemur) CRY2_musMus Mus musculus (mouse) CF898022 CRY2_ratNor Rattus norvegicus (rat) DN948283 prostate CRY2_criGri Cricetulus griseus (hamster) XR_135830 CRY2_spaJud Spalax judaei (blind_mole_rat) AJ606300 CRY2_dipOrd Dipodomys ordii (kangaroo_rat) CRY2_cavPor Cavia porcellus (guinea_pig) CRY2_hetGla Heterocephalus glaber (blind_mole_rat) EHA99865 CRY2_speTri Spermophilus tridecemlineatus (squirrel) CRY2_oryCun Oryctolagus cuniculus (rabbit) CRY2_turTru Tursiops truncatus (dolphin) CRY2_bosTau Bos taurus (cow) EG706191 lens CRY2_oviAri Ovis aries (sheep) NM_001129736 PubMed:19341811 CRY2_susScr Sus scrofa (pig) XM_003122835 CRY2_equCab Equus caballus (horse) CRY2_canFam Canis familiaris (dog) XM_540761 CRY2_ailMel Ailuropoda melanoleuca (panda) XM_002922310 iMet lost to assembly gap CRY2_myoLuc Myotis lucifugus (microbat) CRY2_pteVam Pteropus vampyrus (macrobat) CRY2_loxAfr Loxodonta africana (elephant) CRY2_triMan Trichechus manatus (manatee) AHIN01126950 AHIN01126951 CRY2_choHof Choloepus hoffmanni (sloth) CRY2_macEug Macropus eugenii (wallaby) FY652314 testis CRY2_monDom Monodelphis domestica (opossum) CRY2_ornAna Ornithorhynchus anatinus (platypus) CRY2_galGal Gallus gallus (chicken) AJ396745 bursa 19456395 15459395 CRY2_taeGut Taeniopygia guttata (finch) FE716439 brain CRY2_allMis Alligator mississippiensis (alligator) genome/blat CRY2_anoCar Anolis carolinensis (lizard) XM_003214641 CRY2_xenTro Xenopus tropicalis (frog) NM_001088670 AY049035 CX389867 11533577 discrepancies CRY2_ranCat Rana catesbeiana (bullfrog) GO458565 AY256684 extra SS removed CRY2_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01038797 CRY2_latCha Latimeria chalumnae (coelocanth) AFYH01005158 AFYH01005161 AFYH01005164 CRY2_danRer Danio rerio (zebrafish) aka CRY3 NM_131786 CRY2_oreNil Oreochromis niloticus (tilapia) XM_003449249 split exon 7 also in gasAcu, oryLat, tetNig not danRef or lepOcu CRY2_sigGut Siganus guttatus (spinefoot) AB643456 full length? imputed introns Percomorpha PUBMED 22163321 lunar phase-recognition CRY2_tetNig Tetraodon nigroviridis (fugu) CAAE01010345 CRY2_takRub Takifugu rubripes (fugu) HE592015 CRY1B_strPur Strongylocentrotus purpuratus (sea_urchin) XM_001183029 echinoderm lacks final 2 exons CRY1B_lytVar Lytechinus variegatus (sea_urchin) AGCV01081039 echinoderm many small contigs CRY1B_parLiv Paracentrotus lividus (sea_urchin) AM599080 echinoderm many transcripts CRY1B_aplCal Aplysia californica (sea_hare) FF067636 AASC02010117 scaffold_151 mollusc CRY1B_octVul Octopus vulgaris (octopus) JR450373 transcript assembly mollusc CRY1B_craGig Crassostrea gigas (oyster) GQ415324 HS189569 mollusc CRY1B_rudPhi Ruditapes philippinarum (clam) JO113369 mollusc CRY1B_vilLie Villosa lienosa (mussel) JR510441 transcript assembly mollusc fragment CRY1B_lymSta Lymnaea stagnalis (snail) ES576734 mollusc CRY1B_plaDum Platynereis dumerilii (clam_worm) GU322429 annelid mRNA fragment CRY1B_dapPul Daphnia pulex (water_flea) ACJG01002273 FE370447 FE356368 crustacean CRY1B_diaNig Dianemobius nigrofasciatus (cricket) AB291231 orthoptera CRY1B_acyPis Acyrthosiphon pisum (aphid) NM_001171061 ABLF02032292 HP303737 hemiptera CRY1B_danPle Danaus plexippus (butterfly) AY860425 AGBW01012954 lepidoptera CRY1B_bomMor Bombyx mori (silkworm) NM_001195699 wrong BABH01015108 moth lepidoptera CRY1B_mamBra Mamestra brassicae (moth) AY947639 Glossata lepidoptera CRY1B_helArm Helicoverpa armigera (cotton_bollworm) JN997418 moth lepidoptera CRY1B_droMel Drosophila melanogaster (fruit_fly) AB019389 diptera PubMed:22080955 PDB:3TVS CRY1B_anoGam Anopheles gambiae (mosquito) DQ219482 diptera PubMed:16332522 CRY1B_neoBul Neobellieria bullata (fleshfly) FJ373353 diptera CRY1B_bacCuc Bactrocera cucurbitae (melon_fly) AB517608 diptera CRY1A_dapPul Daphnia pulex (water_flea) FE418063 FE356487 ACJG01001137 crustacean CRY1A_eupSup Euphausia superba (krill) FM200054 contig crustacean CRY1A_pedHum Pediculus humanus (louse) XM_002430500=wrong AAZO01005932 phthiraptera very similar intron pattern to vertebrate but lacks last 4 exons CRY1A_acyPis Acyrthosiphon pisum (aphid) NM_001171102 ABLF02035823 hemiptera cry2-2 PubMed:20482645 end uncertain CRY1A_ripPed Riptortus pedestris (bean_bug) AB379863 hemiptera PubMed:18547745 CRY1A_triCas Tribolium castaneum (flour_beetle) AAJJ01000096 coleopetera CRY1A_bomImp Bombus impatiens (bumble_bee) EF110521 AEQM02008194 hymenoptera PubMed:17244599 CRY1A_apiMel Apis mellifera (bee) NM_001083630 AADG06001305 hymenoptera CRY1A_attCep Atta cephalotes (ant) ADTU01021771 hymenoptera CRY1A_exoRob Exoneura robusta (bee) HP928681 hymenoptera fragment CRY1A_nylPub Nylanderia pubens (crazy_ant) JP792144 hymenoptera fragment CRY1A_nasVit Nasonia vitripennis (wasp) XM_001606355 AAZX01001169 hymenoptera N-term shortened CRY1A_antPer Antheraea pernyi (silkmoth) EF117812 lepidoptera PubMed:17244599 dropped long C-terminus CRY1A_anoGam Anopheles gambiae (mosquito) DQ219483 diptera dropped long C-terminus CRY1A_aedAeg Aedes aegypti (mosquito) XM_001655728 diptera dropped long C-terminus CRY1_vilLie Villosa lienosa (mussel) JR505030 mollusc transcript assembly mollusc CRY1_tetUrt Tetranychus urticae (spider-mite) CAEY01002034 chelicerate N-terminus uncertain CRY1_aplCal Aplysia californica (sea_hare) scaffold_2275 mollusc small fragment CRY4_galGal Gallus gallus (chicken) NP_001034685 CRY4 PubMed:19663499 synteny: ADIPOR1 UBE2T CRY4 LRIF1 DRAM2 CEPT1 CRY4_melGal Meleagris gallopavo (turkey) XM_003212851 CRY4_anaPla Anas platyrhynchos (duck) scaffold1663 CRY4_taeGut Taeniopygia guttata (finch) XM_002198497 CRY4_pasDom Passer domesticus (sparrow) AY494987 16687285 fragment CRY4_anoCar Anolis carolinensis (lizard) FG650345 synteny: UBE2T CRY4 LRIF1 DRAM2 verified indel exon 3 CRY4_xenTro Xenopus tropicalis (frog) NP_001123706 CRY4_xenLae Xenopus laevis (frog) BC167313 only 89% identical to CRY4_xentro CRY4_latCha Latimeria chalumnae (coelocanth) AFYH01009222 CRY4_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01016726 CRY4_danRer Danio rerio (zebrafish) BC164413 adjacency to lost CRY1 suggests relationship CRY4_molTec Molgula tectiformis (tunicate) CJ347377 CJ411442 CJ358785 fragment imputed introns CRY4_braFlo Branchiostoma floridae (amphioxus) Un:610812841 XM_002609457 exon 4,7,8 wrong CRY64_anoCar Anolis carolinensis (lizard) XM_003225714 6-4 photolyase synteny: DCPS TIRAP CRY64 SRPR FOXRED1 CRY64_chrPic Chrysemys picta (turtle) AHGY01135270 AHGY01135271 no synteny CRY64_allMis Alligator mississippiensis (alligator) blat CRY64_croPor Crocodylus porosus (crocodile) blat/genome CRY64_xenTro Xenopus tropicalis (frog) synteny: STS1 RPL27A CRY64 FOXRED1 SRPR PubMed:19715341 19345672 9016626 CRY64_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01024141 CRY64_danRer Danio rerio (zebrafish) BC044204 6-4 photolyase aka CRY5 synteny: FOXRED1 CRY64_salSal Salmo salar (salmon) BT058852 CRY64_oreNil Oreochromis niloticus (tilapia) XM_003437598 AERX01000034 CRY64_braFlo Branchiostoma floridae (amphioxus) BW780666 FE555184 XM_002595028 fused exons 2-3 fusion exons 2-3 odd splice phases exon 5-6, no split 8-9 short final exon CRY64_strPur Strongylocentrotus purpuratus (urchin) XM_001189626 extra 1st exon unwarranted MCGAPRSYVEIRDSEEHSRRHVARLQFQFQSDLP 12 K CRY64_eucTri Eucidaris tribuloides (pencil_urchin) JI324408 fragment imputed introns CRY64_aplCal Aplysia californica (sea_hare) scaffold_427 CRY64_vilLie Villosa lienosa JR505030 transcript assembly mollusc CRY64_droMel Drosophila melanogaster (fruitfly) 6-4 photolyase PDB:3CVW CG2488 uses 5-deazariboflavin CRY64_danPle Danaus plexippus (butterfly) EF117813 PubMed:17244599 two novel exons CRY64_acyPis Acyrthosiphon pisum (aphid) XM_001945977 single exon CRY64_anoGam Anopheles gambiae (mosquito) XM_314748 CRY64_bomMor Bombyx mori (silkworm) AK381942 frameshift CRY64_craMey Crateromorpha meyeri (sponge) PubMed:20121950 CRY64A_triAdh Trichoplax adhaerens (placozoa) XM_002108524 ABGP01000049 no UIM domain affinity to CRY class CRY64B_triAdh Trichoplax adhaerens (placozoa) XM_002107723 ABGP01000051 anti-parallel tandem no UIM domain CRY7_xenTro Xenopus tropicalis (frog) XP_002938187 AAMC01077621 AAMC01077620 many transcripts CDK10+ CRYM+ GCSH- PDK1L2- BCMO1+ GL172982 1U3C 34% 3CVW 29% CRY7_xenLae Xenopus laevis (frog) transcripts DC068968 EG576829 BU901325 CRY7_latCha Latimeria chalumnae (coelocanth) AFYH01265207 pseudogene CRY7_lepOcu Lepisosteus oculatus (gar) AHAT01010533 AHAT01010534 CRY7_danRer Danio rerio (zebrafish) ENSDART00000125725 no synteny to frog CRY7_salSal Salmo salar (salmon) AGKD01006863 CRY7_hapBur Haplochromis burtoni (chichlid) AFNZ01022319 CRY7_gasAcu Gasterosteus aculeatus (stickleback) DN725444 CRY7_oryLat Oryzias latipes (medaka) CRYM+ GCSH- two transcripts, very small introns CRY7_oreNil Oreochromis niloticus (tilapia) CRY7_tetNig Tetraodon nigroviridis (fugu) CRY7_takRub Takifugu rubripes (fugu) CRY7_gadMor Gadus morhua (cod) CAEA01536921 CRY7_xipMac Xiphophorus maculatus (platyfish) AGAJ01012112 CRY7_rudPhi Ruditapes philippinarum (clam) JO112203 gonad transcript missing first half of antenna domain note filter feeder CRY7_craGig Crassostrea gigas (oyster) HS138673 DASH_taeGut Taeniopygia guttata (finch) ABQF01044665 ABQF01044669 ABQF01044671 synteny: ACAA1 DASH MYD66 OXSR1 DASH_anaPla Anas platyrhynchos (duck) scaffold1769 DASH_melUnd Melopsittacus undulatus (budgerigar) AGAI01061648 DASH_galGal Gallus gallus (chicken) syntentic pseudogene, numerous indels, frameshifts, internal stops DASH_melGal Meleagris gallopavo (turkey) ADDD01036185 syntenic pseudogene DASH_anoCar Anolis carolinensis (lizard) XM_003221869 14 exons DASH_chrPic Chrysemys picta (turtle) AHGY01416294 first exon off contig DASH_xenTro Xenopus tropicalis (frog) XM_002938001 PubMed:15147276 synteny: ACAA1 DASH MYD66 transcripts AL790297 CR419606 etc DASH_hymCut Hymenochirus curtipes (frog) fragment DASH_ambMex Ambystoma mexicanum (axolotl) CO785483 fragment DASH_latCha Latimeria chalumnae (coelocanth) AFYH01055296 AFYH01281932 probable pseudogene DASH_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01010414 DASH_danRer Danio rerio (zebrafish) NM_205686 DASH_oreNil Oreochromis niloticus (tilapa) XM_003439198 DASH_patPec Patiria pectinifera (starfish) HP101597 DASH_strPur Strongylocentrotus purpuratus (urchin) DASH_aplCal Aplysia californica (sea_hare) scaffold_151:75,790-145,485 DASH_vilLie Villosa lienosa (mussel) JR504188 transcript assembly mollusc DASH_nemVec Nematostella vectensis (sea_anemone) XP_001623243 ABAV01026885 DASH_hydMag Hydra magnipapillata (cnidarian) XM_002166508 single exon ABRM01055505 DASH_monBre Monosiga brevicollis (choanoflagellate) XP_001745157 ABFJ01000402 DASH1_araTha Arabidopsis thaliana (cress) PHR2 NM_130327 AFNA01010806 DASH2_araTha Arabidopsis thaliana (cress) NM_122394 AFMZ01019177 aka:CRY3 PDB:2VTB DASH_phaTri Phaeodactylum tricornutum (diatom) XM_002178853 CPF2 DASH_thaPse Thalassiosira pseudonana (diatom) XM_002291289 CPD_monDom Monodelphis domestica (opossum) NP_001028149:wrong OPC1 PubMed:7937136 synteny: TNK1 MUC4 CPD KIAA0226 FYTTD1 CPD_sarHar Sarcophilus harrisii (tasmanian_devil) AEFK01107967 CPD_potTri Potorous tridactylus (rat_kangaroo) D26020 PubMed:7813451 CPD_ornAna Ornithorhynchus anatinus (platypus) CPD_taeGut Taeniopygia guttata (finch) XM_002190577 CPD_melUnd Melopsittacus undulatus (budgerigar) AGAI01046895 CPD_galGal Gallus gallus (chicken) XM_422729 CPD_melGal Meleagris gallopavo (turkey) XM_003209143 CPD_allMis Alligator mississippiensis (alligator) genome/blat CPD_chrPic Chrysemys picta (turtle) AHGY01112360 incomplete CPD_anoCar Anolis carolinensis (lizard) XM_003226963 CPD_pytMol Python molurus (python) CPD_xenTro Xenopus tropicalis (frog) NP_001135721 CPD_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01034265 CPD_danRer Danio rerio (zebrafish) NM_201064 CPD_petMar Petromyzon marinus (lamprey) rough revised sequence CPD_braFlo Branchiostoma floridae (amphioxus) XP_002586934 FE570347 fixed frameshift exon 4 CPD_strPur Strongylocentrotus purpuratus (urchin) JT122393 JT102939 FJ812411 CPD_aplCal Aplysia californica (sea_hare) scaffold_446:238,174 CPD_vilLie Villosa lienosa (mussel) JR505029 transcript assembly mollusc CPD_droMel Drosophila melanogaster (fruitfly) thymidine dimer photolyase CG11205 uses 5-deazariboflavin CPD_nasVit Nasonia vitripennis (wasp) XM_001603235 trimmed N-terminal CPD_bomImp Bombus impatiens (bumble_bee) XM_003488984 CPD_apiMel Apis mellifera (bee) XM_003250426 CPD_anoGam Anopheles gambiae (mosquito) XM_313925 trimmed N-terminal CPD_aedAeg Aedes aegypti (mosquito) XM_001653905 trimmed N-terminal CPD_acyPis Acyrthosiphon pisum (aphid) XM_001949116 trimmed N-terminal CPD_nemVec Nematostella vectensis (anemone) ABAV01006764 XM_001636204 bad BACK01030119 CPD_acrDig Acropora digitifera (coral) BACK01030119 cnidarian one intron missing CPD_ampQue Amphimedon queenslandica (sponge) ACUQ01006132 XM_003388698 bad CPD_monBre Monosiga brevicollis (choanflagellate) ABFJ01000652 related intronation but numerous differences CPD_salSpp Salpingoeca species (choanflagellate) ACSY01000967 different intronation still CPD_araTha Arabidopsis thaliana (cress) PHR1 NM_179320 AFMZ01000529 GC-AG splice exon 6-7 CPD_orySat Oryza sativa (rice) B096003 BACJ01049170 aka:PhrII,Class II PMID:22170053 PDB:3UMV CRY1A_acrMil Acropora millepora (coral) EF202589 CRY1B_acrMil Acropora millepora (coral) EF202590 CRY1A_nemVec Nematostella vectensis (anemone) XM_001623096 CRY1B_nemVec Nematostella vectensis (anemone) XM_001623096 CRY1C_nemVec Nematostella vectensis (anemone) XM_001630979 CRY1D_nemVec Nematostella vectensis (anemone) XM_001632799 CRY1E_nemVec Nematostella vectensis (anemone) XM_001632800 CRY64_nemVec Nematostella vectensis (anemone) XP_001636303 ABAV01006592 last exon uncertain CRY2_ampQue Amphimedon queenslandica (sponge) XM_003386521 CRY_ampQue Amphimedon queenslandica (sponge) XM_003386534 CRY_subDom Suberites domuncula (sponge) FN421335 CRY_aphVas Aphrocallistes vastus (sponge) PubMed:14499587 CRY1A_araTha Arabidopsis thaliana (cress) NM_116961 AFNC01018176 aka:CRY1,HY4 PDB:2VTB CRY1B_araTha Arabidopsis thaliana (cress) CRY2 PHH1 NM_100320 AFNB01000167 no antennal chromophore CRY1C_araTha Arabidopsis thaliana (cress) NM_001035626 AFNC01013058 aka:UVR3,CRY3 PDB:3FY4 CRY_phaTri Phaeodactylum tricornutum (diatom) XM_002180059 PMID:19424294 CRY_thaPse Thalassiosira pseudonana (diatom) XM_002291108 PFES_agrTum Agrobacterium tumefaciens (bacteria) NP_355900 aka: PhrB PFES_rhoSph Rhodobacter sphaeroides (bacteria) CP000144 PDB|3ZXS PMID:22290493 6,7-dimethyl-8-ribityl-lumazine antenna aka CryPro 4Fe-4S photolyase PFES_metMah Methanohalophilus mahii (Euryarchaeota) CP001994 4Fe-4S photolyase PFES_natPha Natronomonas pharaonis (Euryarchaeota) CR936257 4Fe-4S photolyase PRIM2_homSap Homo sapiens (human) primase large subunit 4Fe-4S pdb|3L9Q,3Q36 PRIM2_sacCer Saccharomyces cerevisiae (yeast) P20457 aka: PRI2_YEAST primase large subunit PDB|3LGB
Article authorship and data usage policy
I researched this article in its entirety in the winter of 2012, not paying attention initially to previous studies which are excellent on reaction mechanisms and regulatory cycles but completely clueless on comparative genomics (10 years into the genomic era!). Cryptochromes are a moderately difficult topic as metazoan genes go because the timing of gene duplications largely falls between the cracks of phylogenetic coverage and because extenive gene losses in unrepresentative model organisms have distorted the overall evolutionary picture. I plan to greatly expand the treatment of 3D structural implications of comparative genomics during the summer of 2012.
My interests are primarily in the long range evolutionary acquisition and divergence of function-enabling structure, starting from primases and 4Fe-4S cluster photolyases and ending with circadian, magnetosensing and couplings with opsins. However comparative genomics has major applications to rapid hypothesis-testing in all aspects of cryptochrome and photolyase research, the main point being the strong coupling between sequence conservation (ie selective pressure) and functional importance. This means conservation never lasts very long without a reason, and conversely non-conserved features are not important.
Although copyrighted, all the information here is in the public domain and can be used by anyone without additional permissions if properly sourced; however if data, figures or original observations are taken wholesale for a peer-reviewed scientific publication, it might be appropriate (after consultation early on) to include me among secondary co-authors.
Rather than make article edits yourself, please contact me by email with clarifications, corrections or additions to the content so I can make edits while maintaining a consistent approach. For broader disagreements or different interests, a better option is to simply register at the UCSC genomeWiki site and create your own page within the comparative genomics category.
This is just a scientific research article on an old gene family, not an advisory resource for personal genomics issues, melatonin dietary supplementation or medical advice on jet lag and insomnia -- thanks in advance for not sending inappropriate email. Technical terms from genetics and molecular biology are not explained in the article when keywords have a satisfactory treatment at wikipedia or in undergraduate genetics texts; because of good keywords, the scientific literature is easily searched at PubMed so not duplicated here.
My last dozen published research papers in PNAS, Nature, Science etc can be found here. Watch for 4 additional comparative genomics paper to appear in 2012. I've also written over a thousand pages of comparative genomics for other human genes, authored the original user manual to the UCSC human genome browser and in 1999 an advanced tutorial on metazoan genome annotation still widely available online. I thank the UCSC Genomics Group (Hiram Clawson, Brian Raney, Maximilian Haeussler) for software, manuscript and literature resources, Evim Foundation for logistical support, and the Sperling Foundation for financial support under project grant 2012.GNTCS.006.