IMPDH duplication and CBS domain: Difference between revisions
Tomemerald (talk | contribs) No edit summary |
Tomemerald (talk | contribs) No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== | == Introduction to the IMPDH Gene Family == | ||
Inosine 5' monophosphate dehydrogenase (IMPDH) is a highly conserved ubiquitous enzyme that catalyzes the first step unique to GTP synthesis, playing a key role in de novo synthesis of nucleic acids, maintaining intracellular balance of A and G nucleotides, hence regulating cell cycle pauses, cell growth, differentiation, apoptosis, and signalling (via say cGMP). | |||
Vertebrates in general have two very close paralogs, constitutively expressed IMPDH1 and inducible IMPDH2, otherwise indistinguishable in catalytic activity, substrate affinities, tetrameric structure, and interaction with inhibitors. IMPDH2 is greatly up-regulated in proliferating cells, notably activated leukocytes and tumor cells. These two genes lie on different autosomal chromosomes but share 12 identically placed and phased introns, suggesting segmental duplication. That duplication took place after lamprey divergence but prior chondrichtyhian; the deuterostome chain of events should not be conflated with independent duplications in other species (eg yeast and cnidarian) that may not offer valid structural, expressional, or functional parallels. | |||
IMPDH genes are unusually conserved, in the top 95th percentile of all human genes. Remarkably, there is no drift of amino or carboxy termini nor any indel of any length tolerated in any species over many billions of years of branch length. That conservation extends to both members of the IMPDH duplication; for example human IMPDH1 is still 84% identical and 91% similar to IMPDH2 some 500 million years after the duplication (without maintainence from gene conversion). More typically, one gene copy subfunctionalizes or neofunctionalizes after a duplication event and so diverges considerably more rapidly to optimize to that changed role. Here divergence may have been concentrated in upstream regulatory regions relevent to constitutive vs inducible expression. | |||
Since structural features are maintained to 20% identity and less, very high correspondence can be expected between and across IMPDH proteins, even comparing remote clades. Thus small molecule affectors of activity will in general not discriminate well between paralogs in a given species. If IMPDH2 was targeted but IMPDH1 was also impacted, that could result in chemotherapeutic toxicity. However some degree of discrimination is possible since mycophenolic acid (MPA) has higher affinity for IMPDH2. | |||
IMPDH | Curiously, IMPDH has two further remote paralogs in GMPR and GMPR2. These 9-exon guanosine monophosphate reductases catalyze the preceding reaction in the pathway, one of very few instances of support for the 1945 Horowitz retrograde theory of the origin of metabolic pathways. Their intronation pattern could illuminate the relative timing of the gene duplications involved -- if totally unrelated, then the gene duplication took place in prokaryotes or very early unicellular eukaryotes prior to the main intronation era. | ||
IMPDH1 in human has generated 11 quite recent processed pseudogenes, indicating significant germ line expression. These have integrated into the genome at seemingly random locations, most recently in a gene desert in chrX p11.4. None of these appear to have retained functionality, though they still translate to percent identity in the high 80's, either indicative of a burst at some point in the phylogenetic tree (ie a shift in expression after a divergence) or simply better detectability of less decayed events. It emerges that rampant steady pseudogene production is restricted to old world primates (based on two dozen available genomes). | |||
IMPDH2 has not generated a single pseudogene. Consequently specific targeting of IMPDH2 would not be expected to affect germ line nucleotide metabolism. | |||
=== CBS domain === | === CBS domain === | ||
CBS is an ancient non-catalytic paired | The CBS domain is an ancient non-catalytic paired region found in a wide range of [http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=pfam0057 otherwise non-homologous] genes. These fusions are all very old; the CBS domain is not a mobile domain today. The interface between the two CBS domains forms a cleft that is a potential ligand binding site. In IMPDH, the domain comprises part of exons 5,6,7 but does not correspond cleanly to exon boundaries, which fits the domain being appended to the catalytic domain in a common ancestor with prokaryotes prior to establishment of introns. GMPR and GMPR2 lack the CBS domain. | ||
The CBS region can be deleted in its entirety without affecting catalytic properties or the ability to form homotetramers. However R224P and D226N mutations at the end of the CBS domain affect deeply invariant residues and result in retinitis pigmentosa (through a complex chain of events not understood). IMPDH1 may control the rate-limiting regulated step in regeneration of cyclic GMP needed intensively in GPCR photoreceptor signalling -- the bulk of GTP within photoreceptors is generated by IMPDH1 (found predominately in retinal inner segment and synaptic termini). However this role would be relevent solely to ciliary opsins, meaning rhabodomeric photoreceptors in Drosophila (and other arthropods) could not serve as model systems for RP10 retinitis pigmentosa. | |||
[[Image:IMPDH_CBS.png|left|]] | |||
A [http://ajpcell.physiology.org/cgi/content/full/289/6/C1369 comparative review] of CBS domains in otherwise non-homologous proteins (IMPDH, cystathionine beta synthase, AMP kinase gamma2 subunit PRKAG2, chloride channels) suggests a common function: ATP (or related) binding as cooperative allosteric effector of catalytic activity, possibly in part through indirect regulation of oligomericity. The rate-limiting step in a pathway is a logical place for such a sensor of conditions. Mutations in the CBS domain of these proteins also can initiate disease: homocystinuria (I435T, D444N, and S466L abolish AdoMet activation), familial hypertrophic cardiomyopathy (H142R causes insensitivity to energy depletion signals -- high AMP, low ATP), and 5 CLC chloride channel paralogous conditions. | |||
By cross-aligning mutations in these other genes onto the CBS domain of IMPDH1, residues important to CBS ligand specificities (and avoidance of toxic CBS cross-reactivity) can perhaps be identified. For example, H142R of AMP kinase transmaps to a exceedingly conserved glycine in the SKKGKLPIV region of the CBS domain of human IMPDH1. Because the CBS domains are so anciently diverged, sequence-based mappings are barely possible and need to be affirmed by structural alignment. (The cystathionine beta synthase mutation numbering is offset with respect to RefSeq.) | |||
The ability of IMPDH to bind single stranded nucleic acid (not including cognate mRNA) is likely just a byproduct of the ability of the CBS domain to bind mononucleotides. This additional capability could still have utility in various assays of observed amino acid polymorphisms. | |||
IMPDH1 knockouts in mice exhibit only slow retinal degeneration; the rapid course of RP10 in CBS mutations in humans has been [http://hmg.oxfordjournals.org/cgi/content/abstract/13/6/641 attributed] to IMPDH1 misfolding and aggregation [http://www.jbc.org/cgi/content/abstract/281/1/206 also seen with MPA] (and alleviated by elevated GTP). Toxic gain of dysfunction would account for autosomal dominance perhaps better than partial loss of function in a heterozygote. In this scenario, suppression of transcript translation from the bad allele could have value in RP10 in conjunction with elevated guanosine. | |||
Five different IMPDH1 variants, Thr116Met, Asp226Asn, Val268Ile, Gly324Asp, and His 372Pro, were identified in eight autosomal dominant RP families. Two additional IMPDH1 variants, Arg105Trp and Asn198Lys, were found in two patients with isolated LCA. None of the novel IMPDH1 mutants identified in this study altered the enzymatic activity of the corresponding proteins. In contrast, the affinity and/or the specificity of single-stranded nucleic acid binding were altered for each IMPDH1 mutant except the Gly324Asp variant. 2% of families with adRP, and de novo IMPDH1 mutations are also rare causes of isolated Leber congenital amaurosis. | |||
The alignment below of CBS domains reflects the evolutionary history before and after the gene duplication. Stable differences arose between CBS-IMPDH1 and CBS-IMPDH2 despite outstanding conservation in constrained regions that form the binding cleft, determine sensor specificity, and provide ability to communicate nucleotide cell status to the catalytic core. The degree of conservation, if visualized on the 3D structural coordinates of human IMPDH1 bound to 6-Cl-IMP (IJCN at PDB) or human IMPDH2 complexed with MPA (1JR1) using [http://www.bioinformatics.org/combosa3d/index_b.html Combosa], would likely be most informative. | |||
[[Image:IMPDH.png|left|]] | [[Image:IMPDH.png|left|]] | ||
| Line 30: | Line 46: | ||
The cartilaginous fish, Callorhinchus milii, is also in a state of incomplete assembly. Here however distinct multi-exon fragments can be recovered, with duplicate coverage at five of the 13 exons. These cluster with IMPDH1 and IMPDH2 respectively. However, the contigs are too short to incorporate neighboring genes and syntenic correlation with other vertebrates is not yet possible. (The paralogs in elephantfish should have the same neighbors, unless subsequent chromosomal rearrangements have irrevocably scrambled gene order.) | The cartilaginous fish, Callorhinchus milii, is also in a state of incomplete assembly. Here however distinct multi-exon fragments can be recovered, with duplicate coverage at five of the 13 exons. These cluster with IMPDH1 and IMPDH2 respectively. However, the contigs are too short to incorporate neighboring genes and syntenic correlation with other vertebrates is not yet possible. (The paralogs in elephantfish should have the same neighbors, unless subsequent chromosomal rearrangements have irrevocably scrambled gene order.) | ||
Transcripts for skate, Leucoraja erinacea, support this view: EE991359 clusters with blastx with IMPDH1 and DT726645 with IMPDH2. The situation is the same dogfish, Squalus acanthias, for EG027286 and CV720525 respectively. No genome projects are underway in these species so again a duplication specific to chondrichtythes is difficult to rule out. | Transcripts for skate, Leucoraja erinacea, support this view: EE991359 clusters with blastx with IMPDH1 and DT726645 with IMPDH2. The situation is the same dogfish, Squalus acanthias, for EG027286 and CV720525 respectively. No genome projects are underway in these species so again a duplication specific to chondrichtythes is difficult to rule out. These species confirm that distinctions between the respective CBS domains were established shortly after divergence and conserved ever since. | ||
Five teleost fish genomes are available but these experienced a whole genome duplication with complex retention patterns here. making them unsuitable for evolutionary issues. Frog has the expected paralog pair, as does the amniote, Anolis. Chicken presents an odd situation with a clear IMPDH2 ortholog but an assembly gap where IMPDH1 should be. At the trace archives, a diverged and gappy second gene can be located that resembles IMPDH1. It is likely a pseudogene despite the lack of internal stop codons. The second bird genome, zebrafinch, also lacks IMPDH1. Thus in Aves it appears from the available data that IMPDH1 has been lost. | Five teleost fish genomes are available but these experienced a whole genome duplication with complex retention patterns here. making them unsuitable for evolutionary issues. Frog has the expected paralog pair, as does the amniote, Anolis. Chicken presents an odd situation with a clear IMPDH2 ortholog but an assembly gap where IMPDH1 should be. At the trace archives, a diverged and gappy second gene can be located that resembles IMPDH1. It is likely a pseudogene despite the lack of internal stop codons. The second bird genome, zebrafinch, also lacks IMPDH1. Thus in Aves it appears from the available data that IMPDH1 has been lost. | ||
Synteny is important in this particular gene family for building an accurately labelled set of reference sequences. That is, in a fast evolving clade, IMPDH1 and IMPDH2 might be confused if neighboring gene information was not available to establish orthology. However, the useful lifespan of synteny for IMPDH1 especially is quite short. For example, Xenopus gene order (+SND1 -IMPDH1 -OPN1SW +CALU -SAPS2) already differs from human either by inversion or | Synteny is important in this particular gene family for building an accurately labelled set of reference sequences. That is, in a fast evolving clade, IMPDH1 and IMPDH2 might be confused if neighboring gene information was not available to establish orthology. However, the useful lifespan of synteny for IMPDH1 especially is quite short. For example, Xenopus gene order (+SND1 -IMPDH1 -OPN1SW +CALU -SAPS2) already differs from human either by local inversion, misassembly, gene indels, or growing ortholog unrecognizability. Where synteny would be really helpful, say Branchiostoma or Petromyzon of Callorhinchus, it is either entirely lost or not yet available at the current state of assembly. | ||
<pre>human gene order | <pre>human gene order about IMPDH | ||
+SND1 PH4 | +SND1 PH4 | ||
+LEP DALRD3 | +LEP DALRD3 | ||
| Line 47: | Line 63: | ||
</pre> | </pre> | ||
== IMPDH Sequence Resources == | |||
<pre> Phylogenetically representative IMPDH inosine monophosphate dehdrogenase sequences from eumetazoans. Both genes -- and their regulatory regions -- could be sampled much more densely within mammals. | |||
>IMPDH1_homSap Homo sapiens (human) chr7 127,828,554 | |||
0 MADYLISGGTGYVPEDGLTAQQLFASADGLTYN 2 | |||
1 DFLILPGFIDFIADEV 0 | |||
0 DLTSALTRKITLKTPLISSPMDTVTEADMAIAMA 0 | |||
0 LMGGIGFIHHNCTPEFQANEVRKVK 0 | |||
0 KFEQGFITDPVVLSPSHTVGDVLEAKMRHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHTTLLSE 0 | |||
0 VMTPRIELVVAPAGVTLKEANEILQRSKK 1 | |||
2 GKLPIVNDCDELVAIIARTDLKKNRDYPLASKDSQKQLLCGAAVGTREDDKYRLDLLTQAGVDVIVL 0 | |||
0 DSSQGNSVYQIAMVHYIKQKYPHLQVIGGN 2 | |||
1 VVTAAQAKNLIDAGVDGLRVGMGCGSICITQE 1 | |||
2 VMACGRPQGTAVYKVAEYARRFGVPIIADGGIQTVGHVVKALALGAST 1 | |||
2 VMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGSLDAMEKSSSSQKRYFS 2 | |||
1 EGDKVKIAQGVSGSIQDKGSIQKFVPYLIAGIQHGCQDIGARSLSVLR 2 | |||
1 SMMYSGELKFEKRTMSAQIEGGVHGLHS 2 | |||
1 YEKRLY* 0 | |||
>IMPDH2_homSap Homo sapiens (human) paired CDS domain exons 5-7 chr3 49,039,322 | |||
0 MADYLISGGTSYVPDDGLTAQQLFNCGDGLTYN 2 | |||
1 DFLILPGYIDFTADQV 0 | |||
0 DLTSALTKKITLKTPLVSSPMDTVTEAGMAIAMA 0 | |||
0 LTGGIGFIHHNCTPEFQANEVRKVK 0 | |||
0 KYEQGFITDPVVLSPKDRVRDVFEAKARHGFCGIPITDTGRMGSRLVGIISSRDIDFLKEEEHDCFLEE 0 | |||
0 IMTKREDLVVAPAGITLKEANEILQRSKK 1 | |||
2 GKLPIVNEDDELVAIIARTDLKKNRDYPLASKDAKKQLLCGAAIGTHEDDKYRLDLLAQAGVDVVVL 0 | |||
0 DSSQGNSIFQINMIKYIKDKYPNLQVIGGN 1 | |||
2 VVTAAQAKNLIDAGVDALRVGMGSGSICITQE 1 | |||
2 VLACGRPQATAVYKVSEYARRFGVPVIADGGIQNVGHIAKALALGAST 1 | |||
2 VMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAMDKHLSSQNRYFS 2 | |||
1 EADKIKVAQGVSGAVQDKGSIHKFVPYLIAGIQHSCQDIGAKSLTQVR 2 | |||
1 AMMYSGELKFEKRTSSAQVEGGVHSLHS 2 | |||
1 YEKRLF* | |||
>IMPDH1_musMus Mus musculus (mouse) | |||
MADYLISGGTGYVPEDGLTAQQLFANADGLTYNDFLILPGFIDFIADEVDLTSALTRKITLKTPLISSPM | |||
DTVTEADMAIAMALMGGIGFIHHNCTPEFQANEVRKVKKFEQGFITDPVVLSPSHTVGDVLEAKIQHGFS | |||
GIPITATGTMGSKLVGIVTSRDIDFLAEKDHTTLLSEVMTPRVELVVAPAGVTLKEANEILQRSKKGKLP | |||
IVNDQDELVAIIARTDLKKNRDYPLASKDSHKQLLCGAAVGTREDDKYRLDLLTQAGADVIVLDSSQGNS | |||
VYQIAMVHYIKQKYPHLQVIGGNVVTAAQAKNLIDAGVDGLRVGMGCGSICITQEVMACGRPQGTAVYKV | |||
AEYARRFGVPVIADGGIQTVGHVVKALALGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGSLDAM | |||
EKSSSSQKRYFSEGDKVKIAQGVSGSIQDKGSIQKFVPYLIAGIQHGCQDIGAQSLSVLRSMMYSGELKF | |||
EKRTMSAQIEGGVHGLHSYEKRLY* | |||
>IMPDH2_musMus Mus musculus (mouse) | |||
MADYLISGGTSYVPDDGLTAQQLFNCGDGLTYNDFLILPGYIDFTADQVDLTSALTKKITLKTPLVSSPM | |||
DTVTEAGMAIAMALTGGIGFIHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPKDRVRDVFEAKARHGFC | |||
GIPITDTGRMGSRLVGIISSRDIDFLKEEEHDRFLEEIMTKREDLVVAPAGVTLKEANEILQRSKKGKLP | |||
IVNENDELVAIIARTDLKKNRDYPLASKDAKKQLLCGAAIGTHEDDKYRLDLLALAGVDVVVLDSSQGNS | |||
IFQINMIKYIKEKYPSLQVIGGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKV | |||
SEYARRFGVPVIADGGIQNVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAM | |||
DKHLSSQNRYFSEADKIKVAQGVSGAVQDKGSIHKFVPYLIAGIQHSCQDIGAKSLTQVRAMMYSGELKF | |||
EKRTSSAQVEGGVHSLHSYEKRLF* | |||
>IMPDH1_bosTau Bos taurus (cow) | |||
MEEPRAPPAGSAPFPVSLQGGGTADVPEPGARQHPGHETAAQRY | |||
SARLLQAGYEPESMADYLISGGTGYVPEDGLTAQQLFANADGLTYNDFLILPGFIDFT | |||
ADEVDLTSALTRKITLKTPLISSPMDTVTEADMAIAMALMGGIGFIHHNCTPEFQANE | |||
VRKVKKFEQGFITDPVVLSPSHTVGDVLEAKIRHGFSGIPITETGTMGSKLVGIVTSR | |||
DIDFLAEKDHTTLLSEVMTPRNELVVAPAGVTLKEANEILQRSKKGKLPIVNDRDELV | |||
AIIARTDLKKNRDYPLASKDSHKQLLCGAAVGTREDDKYRLDLLTQAGADVIVLDSSQ | |||
GNSVYQIAMVHYIKQKYPHLQVIGGNVVTAAQAKNLIDAGVDGLRVGMGCGSICITQE | |||
VMACGRPQGTAVYKVAEYARRFGVPVIADGGIQTVGHVVKALALGASTVMMGSLLAAT | |||
TEAPGEYFFSDGVRLKKYRGMGSLDAMEKSSSSQKRYFSEGDKVKIAQGVSGSIQDKG | |||
SIQKFVPYLIAGIQHGCQDIGARSLSVLRSMMYSGELKFEKRTMSAQIEGGVHGLHSY | |||
EKRLY* | |||
>IMPDH2_bosTau Bos taurus (cow) | |||
MADYLISGGTSYVPDDGLTAQQLFNCGDGLTYNDFLILPGYIDFTADQVDLTSALTKKITLKTPLVSSPM | |||
DTVTEAGMAIAMALTGGIGFIHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPRDRVRDVFEAKARHGFC | |||
GIPITDTGRMGSHLVGIISSRDIDFLKEEEHDRLLGEIMTKREDLVVAPAGITLKEANEILQRSKKGKLP | |||
IVNENDELVAIIARTDLKKNRDYPLASKDAKKQLLCGAAIGTHEDDKYRLDLLSQAGVDVVVLDSSQGNS | |||
IFQINMIKYIKEKYPSIQVIGGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKV | |||
SEYARRFGVPVIADGGIQNVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAM | |||
DKHLSSQNRYFSEADKIKVAQGVSGAVQDKGSIHKFVPYLIAGIQHSCQDIGAKSLTQVRAMMYSGELKF | |||
EKRTSSAQVEGGVHSLHSYEKRLF* | |||
>IMPDH1_ornAna Ornithorhynchus anatinus frag | |||
MEGTPLISGGTGYVPLDGLTAQQLFAIADGLTYNDFLILPGFIDFTADEVDLTSALTRKITLK | |||
TPLISSPMDTVTEADMAIAMALMGGIGFIHHNCTPEFQANEVRKVKKFEQGFITDPVV | |||
LSPSHTVGDVLEAKARHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHATYLSEV | |||
MTRRTELVVAPAGVTLKEANEILQRSKKGKLPIVNDSDELVAIIARTDLKKNRDYPLA | |||
SKDAHKQLLCGAAVGTREDDKYRLDLLTQAGTDVIVLDSSQGNSVYQIAMVHYIKQKY | |||
PQLQVIGGNVVTAAQAKNLIDAGVDGLRVGMGCGSICITQEVMACGRPQGTAVYKVAE | |||
YARRFGVPVIADGGIQTVGHVVKALALGAST EGDKVKVAQGVSGSIQDKGSIQKFVP | |||
YLIAGIQHGCQDIGARSLSVLRSMMYSGELKFEKRTMSAQIEGGVHGLHSYEKRLY* | |||
>IMPDH2_ornAna Ornithorhynchus anatinus frag | |||
DFLILPGYIDFTADQVDLTSALTKKITLKTPLIS | |||
SPMDTVTEAGMAIAMALTGGIGFIHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPKD | |||
RVRDVFEAKARHGFCGIPITDNGKMGSRLMGIISSRDIDFLKEEEHDLYLGEIMTKWE | |||
DLVVAPAGVTLKEANEILQRSKKGKLPIVNEDNELVAIIARTDLKKNRDYPLASKDAK | |||
KQLLCGAAIGTHEDDKYRLDLLAQAGVDVVVLDSSQGNSIFQINMIKYIKEKYHNLQV | |||
IGGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKVSEYARRF | |||
GVPVIADGGIQNVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMG | |||
SLDAMDKNLGSQNRYFSEADKIKVAQGVSGAVQDKGSIHKFVPYLIAGIQHSCQDIGA | |||
KSLTLVRAMMYSGELKFEKRTSSAQVEGGVHGLHSYEKRLF* | |||
>IMPDH1_galGal Gallus gallus (chicken) 60% indels eye (hatched) DR427747 tr 251351904 inconsistentTaeniopygia guttata missing too | |||
AIPATDGESGRLQMVAEAGADVVLLDSSQGNSACQISMIRYIKERYPDMQVVGGCVVTAA | |||
QCRRLIEAGADALRVGMGGGADCGDSDVPWGRPPGTAVYRVADYARRFGVPVIADGGIRA | |||
VGHIVKAAAVGASTVMLGSLLWGTTEAPGVLFSDGAVLKKFRGGAALEVTETGGGTQQHC | |||
YSDVAEGQMAQGVPAALQDKGSVQKFLPYLSAGIQRGCQHVGARSLSALRSMAYSGELKF | |||
ERRTLAAQLEGLQGLRACEKRLY* | |||
>IMPDH1_anoCar Anolis carolinensis (lizard) misassembly frag | |||
0 2 | |||
1 0 | |||
0 0 | |||
0 LMGGIGIIHHNCTPEFQANEVRKVK 0 | |||
0 VQKFEQGFITDPVVLSPSHSVGDVFEAKVRHGFSGIPVTEAGKMGSTLVGIVTSRDIDFLSEKDYDTPLSE 0 | |||
0 VMTKRSDLVVAPAGVTLKEANEILQRSKK 1 | |||
2 GKLPIVNDADELVAIIARTDLKKNRDYPLASKDPRKQLLCGAAIGTREDDKYRLDLLTQAGVDVVVL 0 | |||
0 DSSQGNSVYQISMIHYIKHKYPELQVIGGN 2 | |||
1 VVTAAQAKNLIDAGVDALRVGMGCGSICITQE 1 | |||
2 VMACGRPQGTAVYKVAEYARRFGVPVIADGGIQTVGHVVKALSLGAST 1 | |||
2 VMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGSLDAMEKNSSSQKRYFs 2 | |||
1 EGDKVKVAQGVSGSIQDKGSIQKFVPYLIAGIQHGCQDIGAKSLSILR 2 | |||
1 2 | |||
1* 0 | |||
>IMPDH2_galGal Gallus gallus (chicken) fully syntenic. no IMPDH1 | |||
MADYLISGGTGYVPDDGLTAQQLFSCGDGLTYNDFLILPGYIDFTADQVD | |||
LTSALTKRITLKTPLVSSPMDTVTEAGMAIAMALTGGIGFIHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPNDRVRDVFEAKARHGFCGIPITDNGKM | |||
GGKLVGIISSRDIDFLKESEHDLPLGEIMTKREDLVVAPSGVMLKEANEILQRSKKGKLPIVNEDDELVAIIARTDLKKNRDYPLASKDSKKQLLCGAAI | |||
GTHEDDKYRLDLLVQAGVDAVVLDSSQGNSIFQINMIKYIKEKYPNLQVIGGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKV | |||
SEYARRFGVPVIADGGIQTVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAMDKNLGSQNRYFSETDKIKVAQGVSGAVQDK | |||
GSIHKFIPYLIAGIQHSCQDIGAKSLTQVRAMMYSGELKFEKRTTSAQVEGGVHGLHSYEKRLF* | |||
>IMPDH1_xenTro Xenopus tropicalis (frog) SND1 ... OPN1SW CALU SAPS2 not syntenic | |||
MADYLISGGTGYVPEDGLTAQHLFANSDGLTYNDFLILPGFIDFTADEVDLTSALTRKITLKTPLISSPM | |||
DTVTESDMAIAMALMGGIGIIHHNCTPEFQANEVRKVKKFEQGFITDPVVMSLNHTVGDVFEAKNRHGFS | |||
GIPVTETGKMGSKLVGIVTSRDIDFLTEKDYSTYLSEVMTKRDELVVAPAGVTLKEANEILQRSKKGKLP | |||
IVNDSDELVAIIARTDLKKNRDYPLASKDCRKQLLCGAAIGTREDDKYRLDLLTQAGVDVVVLDSSQGNS | |||
VYQINMIHYIKQKYPELQVVGGNVVTAAQAKNLIDAGVDALRVGMGCGSICITQEVMACGRPQGTAVYKV | |||
AEYARRFGVPVIADGGIQTVGHVVKALALGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGSLDAM | |||
EKNTSSQKRYFSEGDKVKVAQGVSGSIQDKGSIHKFVPYLIAGIQHGCQDIGAKSLSILRSMMYSGELKL | |||
EKRTMSAQVEGGVHGLHSYEKRLY* | |||
>IMPDH2_xenTro Xenopus tropicalis (frog) fully syntenic | |||
MADYLISGGTSYVPDDGLTAQQLFGAGDGLTYNDFLILPGYIDFTADQVD | |||
LTSALTKKITLKTPMVSSPMDTVTEASMAIAMALTGGIGIMHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPKHRVRDVFEAKARHGFCGIPITENGKM | |||
GSKLAGIISSRDIDFLKSEEHDLALSEIMTRREDLVVAPAGVTLKEANEILQRSKKGKLPIVNGNDELVAIIARTDLKKNRDYPLASKDAKKQLLCGAAI | |||
GTHEDDKYRLDLLVQAGVDAVVLDSSQGNSIFQINMIKFIKEKYQDLQVIAGNVVTAAQAKNLIDAGADALRVGMGSGSICITQEVLACGRPQATAVYKV | |||
SEYARRFGVPVIADGGIQTVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAMDKNVSSQKRYFSEADKIKVAQGVSGAVQDK | |||
GSIHKFIPYLIAGIQHSCQDIGAKSLTQVRAMMYSGELKFEKRTMSAQVEGGVHGLHSYEKRLF* | |||
>IMPDH1_calMil Callorhinchus milii (elephantshark) fragment | |||
0 2 | |||
1 DFLILPGFIDFSAEEV 0 | |||
0 0 | |||
0 0 | |||
0 0 | |||
0 1 | |||
2 GKLPIVNDADELIAIIARTDLKKNRDYPLASKDSRKQLLCGAAIGTREDDKYRLDLLVQAGVDVIVL 0 | |||
0 DSSQGNSVFQINMIHCIKQRYPELQVVGGN 2 | |||
2 1 | |||
2 1 | |||
2 2 | |||
1 EGDKVKVAQGVSGSVQDKGSIHKFAPYLITGIQHGCQDIGAKSLSILR 2 | |||
1 SMMYSGELKFEKRTISAQVEGGVHGLHS 2 | |||
1 * 0 | |||
>IMPDH2_calMil Callorhinchus milii (elephantshark) fragment | |||
0 MADYLISGGTGYVPDDGQSAQQLLGAGDGLTYn 2 | |||
1 DFLILPGYIDFTSDQV 0 | |||
0 DLTSALTKKITLKTPLVSSPMDTVTEANLAIAMA 0 | |||
0 LMGGIGIIHHNCTPEFQANEVRKVK 0 | |||
0 KYEQGFITDPVVMSPSEKVRDVFEAKARHGFSGIPITDNGKMGGKLKGIISSRDIDFLTEKEHDLLLSE 0 | |||
0 VMTRREDLVVAPAAVTLKEANEILQRSKK 1 | |||
2 GKLPIVNDNDELVAIIARTDLKKNRDYPLASKDSKKQLLCGAAIGTHEDDKYRLDQLFQAGVDVVVL 0 | |||
0 2 | |||
2 VVTAAQAKNLIDAGVDALRVGMGCGSICITQE 1 | |||
2 VMACGRPQGTAVYKVAEYARRFGVPVIADGGIQTVGHVVKALALGAST 1 | |||
2 2 | |||
1 ESDKIKVAQGVSGAVQDKGSIQKFIPYLIAGIQHGCQDIGAKSLTQLR 2 | |||
1 AMMYSGELKFEKRTLCAQVEGGVHGLHS 2 | |||
1 * 0 | |||
>IMPDH_petMar_frag2 Petromyzon marinus (lamprey) 854345740 854350173 | |||
0 MADFLISGGTGYVPDDGMTAQQLFAGADGLTYn 2 | |||
1 0 | |||
0 DLASALTKQITLKTPLVSSPMDTVTESSMAIAMA 0 | |||
0 LMGGVGVIHHNCTPEFQANEVRKVk 0 | |||
>IMPDH_petMar_frag3 Petromyzon marinus (lamprey) 565582434 565582613 | |||
0 MADYLISGGAGYVPEDGLTATQLFAGTDGLTYn 2 | |||
1 DFLILPGFIDFTVEEV 0 | |||
0 0 | |||
0 0 | |||
0 KYEQGFITDPVVLSPAHTVGDVLEAKARHGFSGIPITETGRMGGRLAGVITSRDIDFLTE 0 | |||
>IMPDH_petMar_frag1 Petromyzon marinus (lamprey) 700532459 700537638 | |||
2 GKLPIVNEAGELVAIMARTDLKKNRDFPLASKDAHKQLLCGAAVGTRPDDQHRLELLVKAGVDVVIL 0 | |||
0 DSSQGNSVFQINMIRYIKQTYPELQVIGGN 1 | |||
2 VVTAAQAKNLIDAGVDALRVGMGAGSICITQE 1 | |||
2 1 | |||
2 VMMGSLLATTEAPGEYFFSDGVRLKKYRGMGSLDAMEKNQGSQSRYFS 2 | |||
1 ENDKMKVAQGVSGSVQRQG 2 | |||
1 SMMYTGELKFEKRTMSAQVEGGVHGLHS 2 | |||
1 * 0 | |||
>IMPDH1_petMar Petromyzon marinus (lamprey) composite | |||
0 MADYLISGGAGYVPEDGLTATQLFAGTDGLTYn 2 | |||
1 DFLILPGFIDFTVEEV 0 | |||
0 DLASALTKQITLKTPLVSSPMDTVTESSMAIAMA 0 | |||
0 LMGGVGVIHHNCTPEFQANEVRKVk 0 | |||
0 KYEQGFITDPVVLSPAHTVGDVLEAKARHGFSGIPITETGRMGGRLAGVITSRDIDFLTEKDHGIPLHQ 0 | |||
0 VMTKREDLVVAPSGVTLTEANELLQRSKK 1 | |||
2 GKLPIVNEAGELVAIMARTDLKKNRDFPLASKDAHKQLLCGAAVGTRPDDQHRLELLVKAGVDVVIL 0 | |||
0 DSSQGNSVFQINMIRYIKQTYPELQVIGGN 1 | |||
2 VVTAAQAKNLIDAGVDALRVGMGAGSICITQE 1 | |||
2 VMAVGRPQGTAVYRVAEYARRFGVPVMADGGIQTVGHITKALALGAST 1 | |||
2 VMMGSLLATTEAPGEYFFSDGVRLKKYRGMGSLDAMEKNQGSQSRYFS 2 | |||
1 ENDKMKVAQGVSGSVQRQG 2 | |||
1 SMMYTGELKFEKRTMSAQVEGGVHGLHS 2 | |||
1 * 0 | |||
>IMPDH_braFlo Branchiostoma floridae (lancelet) one copy, severe misassembly | |||
0 MADYLISGGTGYVPDDGMTASQLFGTGEGLTYn 2 | |||
1 DFLILPGFIDFTADEV 0 | |||
0 DLTSAMTKKIQLKAPLVSSPMDTVSESDMAIAMA 0 | |||
0 LTGGIGIIHSNCTPEFQANEVRKVK 0 | |||
0 KYEQGFIMDPIVLSPEHTVGDVCEMKRKHGFSGIPITENGKLGGKLLGIVTSRDIDFMNSDHHHIKLRD 0 | |||
0 VMTPFEELVVGHAGVSLKEANETLQRSKK 1 | |||
2 GKLPIVNENDELVSLIARTDLKKNRDYPLASKDSKKQLLCGAAIGTREEDKYRVELLVQAGVDLVVL 0 | |||
0 DSSQGNSIYQINMIRYLKQKYSELQVIGGN 2 | |||
1 VVTAAQAKNLIDAGVDGLRVGMGSGSICITQE 1 | |||
2 VMAVGRPQGTAVYKVAEYARRFGVPVIADGGISTVGHITKALALGAST 1 | |||
2 VMMGSLLAGTSEAPGEYFFQDGVRLKKYRGMGSLEAMEKGKASQNRYFR 2 | |||
1 ESDKLKVAQGVTGSIQDKGSVHKFVPYLIAGIQHGCQDIGAKSLSSLR 2 | |||
1 SMMYSGELRFETRTVSAQVEGGVHGLHS 2 | |||
1 * 0 | |||
>IMPDH_cioInt Ciona intestinalis (tunicate) mRNA AK114441 | |||
MAEFLISGETSYVPEDGLTAAQLLNTGDGLTYNDFLILPGFIDFTASEVDLTSALTKKISL | |||
KTPLLSSPMDTVTESDMAIGMALMGGMGFIHYNCTPEFQAAEVRRVKKYEQGFIQNPVTL | |||
GPKATVRDVTDVKAMYGFSGIPVTDDGTPTGKLIGLVSSRDFDFLKPEESNTPLEQVMTG | |||
RDKLITADTSVTLQEANHILSQSKKGKLPIVDADDRLVSLIARTDLKKNREFPLASKDER | |||
KQLLCGAAISTREEDKHRLELLVEAGVDAVILDSSQGNSIYQINSIRYIRHKYPHLQVIA | |||
GNVVTAAQAKNLIDAGADALRVGMGSGSICITQEVMAVGRPQATAVYKVSEYARRFNVPV | |||
IADGGIQNVGHVTKALALGASTVMMGSLLAATTESPGEYFYSDGIRLKKYRGMGSVDAME | |||
SCKSSQSRYFSEKDKIRVAQGVSGAVQDKGSVHTFLPYLIAGIQHGCQDIGSRSMPMLRS | |||
MMYSGELKFERRSTSAQVEGGVHGLHSFEKPHL* | |||
>IMPDH_strPur Strongylocentrotus purpuratus (sea urchin) single gene 2 intronation changes | |||
0 MSKKVKLMNGAMEPQVDDGLSGQQLFGSGDGLTYn 2 | |||
1 DFLILPGYIDFTSDQV 0 | |||
0 DLQSQLTKDITLKAPLVSSPMDTVTESSMAIAMA 0 | |||
0 LCGGIGIIHHNCSPEFQANEVRKVK 0 | |||
0 KYEQGFIMDPVVLGPNDTVGDVFGSKAKHGFS 1 | |||
2 GIPITDTGRLGGKLLGIVTARDIDFLKPESYVKPLST 00 AMTCREDLVVAPANVTLKQANDLLQKAKK 1 | |||
2 GKLPIVNEKDELVSLISRTDLKKHREFPLASKDPRKQLLCGAAIGTREEDKHRLDLLVQAGVDVVIL 0 | |||
0 DSSQGNSSFQVSMIKCTKAKYPELQVVAGN 1 | |||
2 VVTVAQAKNLIQAGADALRVGMGSGSICITQE 1 | |||
2 VMAVGRPQGTAVYRVAQYARSCGVPIIADGGITTVGHITKALSLGASS 1 | |||
2 VMMGSLLAGTTEAPGEYFFSDGVRLKKYRGMGSLDAMEKNQSSAKRYFR 2 | |||
1 ERDKLKVAQGVSGSIVDKGSIHKFVPYLIAGIQHGCQDIGALSLTVLR 2 | |||
1 EKMYSGEVRFERRSPSAQVEGGVHSLHS 2 | |||
1 *0 | |||
>IMPDH_sacKow Saccoglossus kowalevskii (acornworm) frag one copy 82% homSap | |||
0 2 | |||
1 0 | |||
0 0 | |||
0 0 | |||
0 KYEQGFIMDALVMSANTTIKEVFAAKSQHGFSGIPITDNGKLGGRLLGIVTARDIDFVEPEFNDKPLEQFMTKREDLVVAPANVTLKEANDILQKSKK 1 | |||
2 GKLPIVNENDELVSLISRTDLKKHREFPLASKDSKKQLLCGAAIGTHESDKNRLDLLVQAGVDVIIL 0 | |||
0 1 | |||
2 VVTAAQAKNLIDAGVDALRVGMGSGSICITQE 1 | |||
2 VMAVGRPQGTSVYKVAEYARRFGVPVIADGGIGTVGHITKALALGAST 1 | |||
2 VMMGSLLAGTSEAPGEYYFSDGVRLKKYRGMGSLDAMEiNQSDRYFS 2 | |||
1 ESDKLKVAQGVSGSIIDKGSIHKFIPYLIAGIQHGCQDIGAKSMSMLR 2 | |||
1 2 | |||
1 *0 | |||
>IMPDH_helRob Helobdella robusta (leech) frag cDNA | |||
QAGVDFVVLDSSQGNSIYQIKLIKYIKEKYPNLQVIGGNVVTAAQAKNLIDAGVDALRVG | |||
MGSGSICITQEVMAVGRPQGTAVYKVAEYARRFGVPIIADGGIENVGHIVKALALGASTV | |||
MMGSLLAGTTEAPGEYYFADGVRLKKYRGMGSLDAMEQHKASQSRYFSDSDKVKVAQGVS | |||
GAVVDKGSIHKFLPYLISGVQHGCQDLGAKSLSCLRSMMYQGELKFEKRTTSSQIEGGVH | |||
GLHSYEKRLY* | |||
>IMPDH_schMan Schistosoma japonicum (flatworm) fragment | |||
FSLKVPFASSPMDTVTEAKMAIAMS | |||
LCGSIGFVHNNCSVEAQANEVKKVK | |||
KYNQGFILSPVVVSPRQPIYDIIEIKKKYGFG | |||
GIPVTEDGYMGSRLVGLVTLRDVDFLDPNDFNTPVEKVMTPFDDLVTAFSCVTLSEANDLLRKSKK | |||
GKLPIINENRELVALIARTDLQKNRDYPLASRDDENQLIVGAAISTHEGDFARVKALINSGVDIIVI | |||
DSSQGNSIYQLDMIKRIKSSFPDLQIIGGN | |||
IVTCAQAKNLIDAGVDGLRVGMGSGSICITQE | |||
VTAIGRSQAKAVYSVSEYAHKYDIPVIADGGIQNTGHIVKALSFGASS | |||
VMMGGLLAGTTESAGEYIFSDGVKLKKYRGMGSIEAMSQHTESQARYFSESDRIKVAQGVSGTIVDR | |||
GSVHQLVPYLVAGVKHGLQQIGARNITELHNMSRSGKLRFELRSPSAQLEGGVHSLYS | |||
YDKSMF* | |||
>IMPDH_schMed Schmidtea mediterranea (planaria) frag NZ_AAWT01010891 more | |||
GFSASELFNKKTGLTYNDFILLPGYIDFRSEEVDITTRITKNMYLKTPLVSSPMDTVTES | |||
NMAIAMALSGGIGILHHNCTIDTQANEIRRVKKYEQGFIVDPVVMSIDNTVGEVMNIKIK | |||
NGFTGIPITDNGKLGGKLVGLVTLRDIDFLPKSNWDLKVSEVMTPFENLITAPYGVALHE | |||
ANGILQRSKKGKLPIINENRELVALISRTDLKKNREYPLSSKDDR | |||
>IMPDH_ixoSca Ixodes scapularis | |||
MSDKKTTFSEDGLTAQHLIGSGDGLTYNDFLILPGFIDFNADDVD | |||
LTSKLTKNITLQAPLVSSPMDTVTESEMAIAMALCGGIGIIHHNCTPEHQANEVHKVKKYKHGFIHDPVVLSPNNC | |||
VADVFEVKRKHGFAGVPITENGKLGGKLVGMVTSRDIDFIPIEDHNRLLSEVMTSLK | |||
DLTVASSKVTLSEANSLLQKSKKGKLPLVNEGGELVSLIARTDLKKSRSYPLASKDENKQLIVGAAIGTREADKPRL | |||
ELLVQAGVDVVVLDSSQGNSVYQIEMVKYIKSKYPGLQVIGGNVVTTAQAKNLIEAGVDG | |||
LRVGMGSGSICITQEVMACGRPQATAVYKVAEYARRFGVPCVADGGVSSVGHIIKALALG | |||
ASTVMMGSMLAGTTESPGEYFFSNGVRLKKYRGMGSLDAMQSTEGGGSLNRYYQSDQDKV | |||
RVAQGVSGTIVDKGSIHRYVPYLITGIKYGCQDIGARSLDVLKANMYSGDIKFEKRSVSA | |||
QIEGGVHGLHSYEKRLY* | |||
>IMPDH_nemVec Nematostella vectenisis (anemone) 78% XM_001641825 | |||
0 MMADFLIAGGTSYVPEDGMTAGQLFQSDGLTYS 2 | |||
1 DFIILPGFIDFPATDV 0 | |||
0 DLTSPLTRRITIKTPLVSSPMDTVTESALATAMA 0 | |||
0 LNGGIGIIHHNCSIEFQANEIRKVKKFEQGFIMAPLVLSATNTVADVIDAKQRHGFSGIPIT 1 | |||
2 ENGQLGGILQGIVTSRDIDFLHGVENHKQLGE 0 | |||
0 VMTRLEDLVVAKAGITLNEANKILQMSKK 1 | |||
2 GKLPIVNEKGELVSLIARTDLKKNRDYPLASKDENKQLL 1 | |||
2 VGAAIGTREDDKARLHALVEAGVDVVVIDSSQGNSIYQLSLIS 2 | |||
1 HIKENYPNLQIVGGNVVTASQAKNLIDAGVDALRVGMGSGSICITQEVMAVGRPQGTAVYKVAEYARRFGVPVLADGGIQNVGHITKALSLGAST 1 | |||
2 VMMGSLLAGTSEAPGEYFFADGVRLKKYR 1 | |||
2 GMGSLSAMEKNSSSASRYFS 2 | |||
1 ENDKVKVAQGVSGSVVDKGSIHKFVPYLTAGIQHGCQDLGAKSLTSLR 2 | |||
1 SMMYSGELKFERRTTSSQIEGGVHGLHS 2 | |||
1 YEKRLF* 0 | |||
</pre> | |||
== CBS Domain Sequence Resources == | |||
It is difficult to recover full length IMPDH sequences for a gene with 13 exons because of incomplete assemblies, confusion between IMPDH1 and IMPDH2 fragments, and the tendency for cDNA projects to begin at one end and not proceed quite to full length. Sequences more commonly are complete enough to have full length CBS domains. Thus to the extent that this domain regulates enzymatic activity through allosteric effects and that small molecule effectors should discriminate between the two CBS domains, it is informative to determine where conserved residues in the respective domains differ, ie determine a subtractive profiles. | |||
<pre> | |||
>IMPDH1_homSap | |||
ITDPVVLSPSHTVGDVLEAKMRHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHTTLLSEVMTPRIELVVAPAGVTLKEANEILQRSKKGKLPIVNDCDELVAIIARTDLKKN | |||
>IMPDH1_bosTau | |||
ITDPVVLSPSHTVGDVLEAKIRHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHTTLLSEVMTPRNELVVAPAGVTLKEANEILQRSKKGKLPIVNDRDELVAIIARTDLKKN | |||
>IMPDH1_musMus | |||
ITDPVVLSPSHTVGDVLEAKIQHGFSGIPITATGTMGSKLVGIVTSRDIDFLAEKDHTTLLSEVMTPRVELVVAPAGVTLKEANEILQRSKKGKLPIVNDQDELVAIIARTDLKKN | |||
>IMPDH1_ornAna | |||
ITDPVVLSPSHTVGDVLEAKARHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHATYLSEVMTRRTELVVAPAGVTLKEANEILQRSKKGKLPIVNDSDELVAIIARTDLKKN | |||
>IMPDH1_anoCar | |||
ITDPVVLSPSHSVGDVFEAKVRHGFSGIPVTEAGKMGSTLVGIVTSRDIDFLSEKDYDTPLSEVMTKRSDLVVAPAGVTLKEANEILQRSKKGKLPIVNDADELVAIIARTDLKKN | |||
>IMPDH1_xenTro | |||
ITDPVVMSLNHTVGDVFEAKNRHGFSGIPVTETGKMGSKLVGIVTSRDIDFLTEKDYSTYLSEVMTKRDELVVAPAGVTLKEANEILQRSKKGKLPIVNDSDELVAIIARTDLKKN | |||
>IMPDH1_danRer | |||
ITDPVVMSPRHTVGDVFEAKVRHGFSGIPVTETGKMGSKLVGIVTSRDIDFLSEKDYDRPLEESMTKREDLVVAPAGVTLKEANDILQRSKKGKLPIVNDSDELVAIIARTDLKKN | |||
>IMPDH1_takRer | |||
ITDPVVMSPRHTVGDVFEAKIRHGFSGIPVTETGKMGSKLVGIVTSRDIDFLSEKDHDRPLEEAMTKREDLVVAPAGVTLKEANDILQRSKKGKLPIVNDSDELVAIIARTDLKKN | |||
>IMPDH1_leuRaj | |||
ITDPVVMSLHHTVGDVFEAKSRHGFSGIPITETGKMGSKLMGIVTSRDIDFLSEKDYDTALNEVMTKREDLVVAPAGVTLKEANGILQRSKKRKVPIVNDMDELISIIARTDLkkn | |||
>IMPDH1_petMar | |||
ITDPVVLSPAHTVGDVLEAKARHGFSGIPITETGRMGGRLAGVITSRDIDFLTEKDHGIPLHQVMTKREDLVVAPSGVTLTEANELLQRSKKGKLPIVNEAGELVAIMARTDLKKN | |||
>IMPDH2_homSap | |||
ITDPVVLSPKDRVRDVFEAKARHGFCGIPITDTGRMGSRLVGIISSRDIDFLKEEEHDCFLEEIMTKREDLVVAPAGITLKEANEILQRSKKGKLPIVNEDDELVAIIARTDLKKN | |||
>IMPDH2_musMus | |||
ITDPVVLSPKDRVRDVFEAKARHGFCGIPITDTGRMGSRLVGIISSRDIDFLKEEEHDRFLEEIMTKREDLVVAPAGVTLKEANEILQRSKKGKLPIVNENDELVAIIARTDLKKN | |||
>IMPDH2_bosTau | |||
ITDPVVLSPRDRVRDVFEAKARHGFCGIPITDTGRMGSHLVGIISSRDIDFLKEEEHDRLLGEIMTKREDLVVAPAGITLKEANEILQRSKKGKLPIVNENDELVAIIARTDLKKN | |||
>IMPDH2_galGal | |||
ITDPVVLSPNDRVRDVFEAKARHGFCGIPITDNGKMGGKLVGIISSRDIDFLKESEHDLPLGEIMTKREDLVVAPSGVMLKEANEILQRSKKGKLPIVNEDDELVAIIARTDLKKN | |||
>IMPDH2_ornAna | |||
ITDPVVLSPKDRVRDVFEAKARHGFCGIPITDNGKMGSRLMGIISSRDIDFLKEEEHDLYLGEIMTKWEDLVVAPAGVTLKEANEILQRSKKGKLPIVNEDNELVAIIARTDLKKN | |||
>IMPDH2_xenTro | |||
ITDPVVLSPKHRVRDVFEAKARHGFCGIPITENGKMGSKLAGIISSRDIDFLKSEEHDLALSEIMTRREDLVVAPAGVTLKEANEILQRSKKGKLPIVNGNDELVAIIARTDLKKN | |||
>IMPDH2_calMil | |||
ITDPVVMSPSEKVRDVFEAKARHGFSGIPITDNGKMGGKLKGIISSRDIDFLTEKEHDLLLSEVMTRREDLVVAPAAVTLKEANEILQRSKKGKLPIVNDNDELVAIIARTDLKKN | |||
>IMPDH2_leuRaj | |||
ITDPVVLSPRHRVRDVFEAKARHGFCGIPITDTGTLGGRLAGIISSRDIDFLEESEQELPLEQVMTRREELVVAPAGVRLKEANEILQRSKKGKLPIVNEQDQLVAIISRTDLkkn | |||
>IMPDH_braFlo | |||
IMDPIVLSPEHTVGDVCEMKRKHGFSGIPITENGKLGGKLLGIVTSRDIDFMNSDHHHIKLRDVMTPFEELVVGHAGVSLKEANETLQRSKKGKLPIVNENDELVSLIARTDLKKN | |||
>IMPDH_cioInt | |||
IQNPVTLGPKATVRDVTDVKAMYGFSGIPVTDDGTPTGKLIGLVSSRDFDFLKPEESNTPLEQVMTGRDKLITADTSVTLQEANHILSQSKKGKLPIVDADDRLVSLIARTDLKKN | |||
>IMPDH_strPur | |||
IMDPVVLGPNDTVGDVFGSKAKHGFSGIPITDTGRLGGKLLGIVTARDIDFLKPESYVKPLSTAMTCREDLVVAPANVTLKQANDLLQKAKKGKLPIVNEKDELVSLISRTDLKKH | |||
>IMPDH_sacKow | |||
IMDALVMSANTTIKEVFAAKSQHGFSGIPITDNGKLGGRLLGIVTARDIDFVEPEFNDKPLEQFMTKREDLVVAPANVTLKEANDILQKSKKGKLPIVNENDELVSLISRTDLKKH | |||
>IMPDH_schMed | |||
IVDPVVMSIDNTVGEVMNIKIKNGFTGIPITDNGKLGGKLVGLVTLRDIDFLPKSNWDLKVSEVMTPFENLITAPYGVALHEANGILQRSKKGKLPIINENRELVALISRTDLKKN | |||
>IMPDH_schMan | |||
ILSPVVVSPRQPIYDIIEIKKKYGFGGIPVTEDGYMGSRLVGLVTLRDVDFLDPNDFNTPVEKVMTPFDDLVTAFSCVTLSEANDLLRKSKKGKLPIINENRELVALIARTDLQKN | |||
>IMPDH_ixoSca | |||
IHDPVVLSPNNCVADVFEVKRKHGFAGVPITENGKLGGKLVGMVTSRDIDFIPIEDHNRLLSEVMTSLKDLTVASSKVTLSEANSLLQKSKKGKLPLVNEGGELVSLIARTDLKKS | |||
>IMPDH_nemVec | |||
IMAPLVLSATNTVADVIDAKQRHGFSGIPITENGQLGGILQGIVTSRDIDFLHGVENHKQLGEVMTRLEDLVVAKAGITLNEANKILQMSKKGKLPIVNEKGELVSLIARTDLKKN | |||
</pre> | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 17:27, 28 January 2008
Introduction to the IMPDH Gene Family
Inosine 5' monophosphate dehydrogenase (IMPDH) is a highly conserved ubiquitous enzyme that catalyzes the first step unique to GTP synthesis, playing a key role in de novo synthesis of nucleic acids, maintaining intracellular balance of A and G nucleotides, hence regulating cell cycle pauses, cell growth, differentiation, apoptosis, and signalling (via say cGMP).
Vertebrates in general have two very close paralogs, constitutively expressed IMPDH1 and inducible IMPDH2, otherwise indistinguishable in catalytic activity, substrate affinities, tetrameric structure, and interaction with inhibitors. IMPDH2 is greatly up-regulated in proliferating cells, notably activated leukocytes and tumor cells. These two genes lie on different autosomal chromosomes but share 12 identically placed and phased introns, suggesting segmental duplication. That duplication took place after lamprey divergence but prior chondrichtyhian; the deuterostome chain of events should not be conflated with independent duplications in other species (eg yeast and cnidarian) that may not offer valid structural, expressional, or functional parallels.
IMPDH genes are unusually conserved, in the top 95th percentile of all human genes. Remarkably, there is no drift of amino or carboxy termini nor any indel of any length tolerated in any species over many billions of years of branch length. That conservation extends to both members of the IMPDH duplication; for example human IMPDH1 is still 84% identical and 91% similar to IMPDH2 some 500 million years after the duplication (without maintainence from gene conversion). More typically, one gene copy subfunctionalizes or neofunctionalizes after a duplication event and so diverges considerably more rapidly to optimize to that changed role. Here divergence may have been concentrated in upstream regulatory regions relevent to constitutive vs inducible expression.
Since structural features are maintained to 20% identity and less, very high correspondence can be expected between and across IMPDH proteins, even comparing remote clades. Thus small molecule affectors of activity will in general not discriminate well between paralogs in a given species. If IMPDH2 was targeted but IMPDH1 was also impacted, that could result in chemotherapeutic toxicity. However some degree of discrimination is possible since mycophenolic acid (MPA) has higher affinity for IMPDH2.
Curiously, IMPDH has two further remote paralogs in GMPR and GMPR2. These 9-exon guanosine monophosphate reductases catalyze the preceding reaction in the pathway, one of very few instances of support for the 1945 Horowitz retrograde theory of the origin of metabolic pathways. Their intronation pattern could illuminate the relative timing of the gene duplications involved -- if totally unrelated, then the gene duplication took place in prokaryotes or very early unicellular eukaryotes prior to the main intronation era.
IMPDH1 in human has generated 11 quite recent processed pseudogenes, indicating significant germ line expression. These have integrated into the genome at seemingly random locations, most recently in a gene desert in chrX p11.4. None of these appear to have retained functionality, though they still translate to percent identity in the high 80's, either indicative of a burst at some point in the phylogenetic tree (ie a shift in expression after a divergence) or simply better detectability of less decayed events. It emerges that rampant steady pseudogene production is restricted to old world primates (based on two dozen available genomes).
IMPDH2 has not generated a single pseudogene. Consequently specific targeting of IMPDH2 would not be expected to affect germ line nucleotide metabolism.
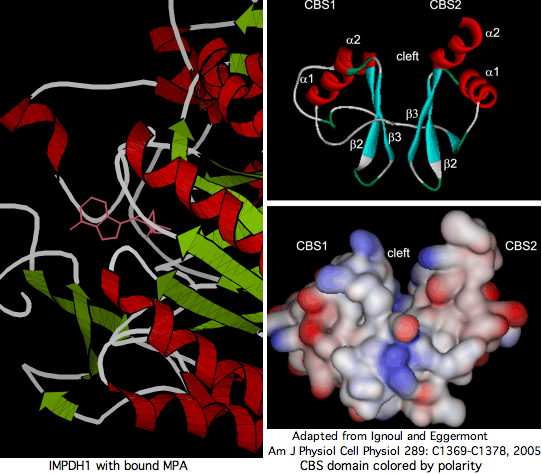
CBS domain
The CBS domain is an ancient non-catalytic paired region found in a wide range of otherwise non-homologous genes. These fusions are all very old; the CBS domain is not a mobile domain today. The interface between the two CBS domains forms a cleft that is a potential ligand binding site. In IMPDH, the domain comprises part of exons 5,6,7 but does not correspond cleanly to exon boundaries, which fits the domain being appended to the catalytic domain in a common ancestor with prokaryotes prior to establishment of introns. GMPR and GMPR2 lack the CBS domain.
The CBS region can be deleted in its entirety without affecting catalytic properties or the ability to form homotetramers. However R224P and D226N mutations at the end of the CBS domain affect deeply invariant residues and result in retinitis pigmentosa (through a complex chain of events not understood). IMPDH1 may control the rate-limiting regulated step in regeneration of cyclic GMP needed intensively in GPCR photoreceptor signalling -- the bulk of GTP within photoreceptors is generated by IMPDH1 (found predominately in retinal inner segment and synaptic termini). However this role would be relevent solely to ciliary opsins, meaning rhabodomeric photoreceptors in Drosophila (and other arthropods) could not serve as model systems for RP10 retinitis pigmentosa.
A comparative review of CBS domains in otherwise non-homologous proteins (IMPDH, cystathionine beta synthase, AMP kinase gamma2 subunit PRKAG2, chloride channels) suggests a common function: ATP (or related) binding as cooperative allosteric effector of catalytic activity, possibly in part through indirect regulation of oligomericity. The rate-limiting step in a pathway is a logical place for such a sensor of conditions. Mutations in the CBS domain of these proteins also can initiate disease: homocystinuria (I435T, D444N, and S466L abolish AdoMet activation), familial hypertrophic cardiomyopathy (H142R causes insensitivity to energy depletion signals -- high AMP, low ATP), and 5 CLC chloride channel paralogous conditions.
By cross-aligning mutations in these other genes onto the CBS domain of IMPDH1, residues important to CBS ligand specificities (and avoidance of toxic CBS cross-reactivity) can perhaps be identified. For example, H142R of AMP kinase transmaps to a exceedingly conserved glycine in the SKKGKLPIV region of the CBS domain of human IMPDH1. Because the CBS domains are so anciently diverged, sequence-based mappings are barely possible and need to be affirmed by structural alignment. (The cystathionine beta synthase mutation numbering is offset with respect to RefSeq.)
The ability of IMPDH to bind single stranded nucleic acid (not including cognate mRNA) is likely just a byproduct of the ability of the CBS domain to bind mononucleotides. This additional capability could still have utility in various assays of observed amino acid polymorphisms.
IMPDH1 knockouts in mice exhibit only slow retinal degeneration; the rapid course of RP10 in CBS mutations in humans has been attributed to IMPDH1 misfolding and aggregation also seen with MPA (and alleviated by elevated GTP). Toxic gain of dysfunction would account for autosomal dominance perhaps better than partial loss of function in a heterozygote. In this scenario, suppression of transcript translation from the bad allele could have value in RP10 in conjunction with elevated guanosine.
Five different IMPDH1 variants, Thr116Met, Asp226Asn, Val268Ile, Gly324Asp, and His 372Pro, were identified in eight autosomal dominant RP families. Two additional IMPDH1 variants, Arg105Trp and Asn198Lys, were found in two patients with isolated LCA. None of the novel IMPDH1 mutants identified in this study altered the enzymatic activity of the corresponding proteins. In contrast, the affinity and/or the specificity of single-stranded nucleic acid binding were altered for each IMPDH1 mutant except the Gly324Asp variant. 2% of families with adRP, and de novo IMPDH1 mutations are also rare causes of isolated Leber congenital amaurosis.
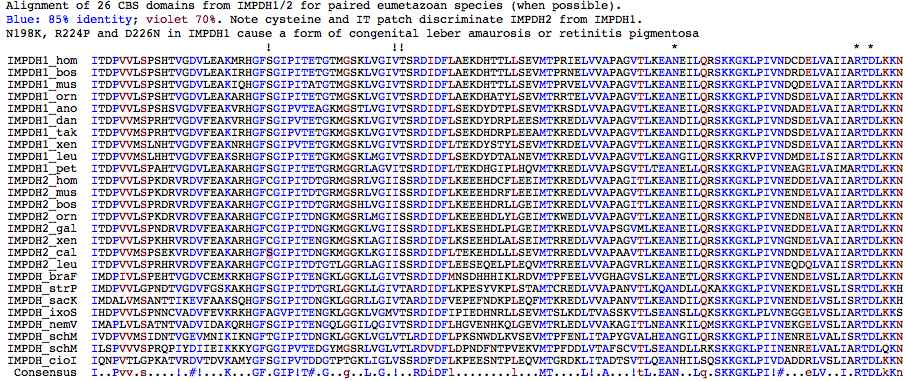
The alignment below of CBS domains reflects the evolutionary history before and after the gene duplication. Stable differences arose between CBS-IMPDH1 and CBS-IMPDH2 despite outstanding conservation in constrained regions that form the binding cleft, determine sensor specificity, and provide ability to communicate nucleotide cell status to the catalytic core. The degree of conservation, if visualized on the 3D structural coordinates of human IMPDH1 bound to 6-Cl-IMP (IJCN at PDB) or human IMPDH2 complexed with MPA (1JR1) using Combosa, would likely be most informative.
Timing the IMPDH duplication
Using released genomes, wgs contigs, trace archives, cDNA, and ad hoc GenBank sequences, the evolutionary history of IMPDH can be traced. It emerges that echinoderms, hemichordates, cephalochordates, and urochordates have but a single copy of the gene. Since it is implausible that a second copy was lost repeatedly in separate clades, by parsimony early deuterostomes did not experience an IMPDH duplication. Consequently, earlier species with two copies, such as the cnidarian Nematostella and yeast, experienced independent duplications not necessarily to the same purpose, making them irrelevent as model species to human.
The situation in lamprey is slightly ambiguous because, while numerous exons or short blocks of contiguous exons can be located, these are hard to assign unambiguously to IMPDH1 or IMPDH2 due to the latter's high percent identity. However it appears that lamprey contains a single gene of IMPDH1 character -- trace coverage averages 3-4 per exon and these alway of the same amino acid sequence (up to inherent trace sequence errors).
The cartilaginous fish, Callorhinchus milii, is also in a state of incomplete assembly. Here however distinct multi-exon fragments can be recovered, with duplicate coverage at five of the 13 exons. These cluster with IMPDH1 and IMPDH2 respectively. However, the contigs are too short to incorporate neighboring genes and syntenic correlation with other vertebrates is not yet possible. (The paralogs in elephantfish should have the same neighbors, unless subsequent chromosomal rearrangements have irrevocably scrambled gene order.)
Transcripts for skate, Leucoraja erinacea, support this view: EE991359 clusters with blastx with IMPDH1 and DT726645 with IMPDH2. The situation is the same dogfish, Squalus acanthias, for EG027286 and CV720525 respectively. No genome projects are underway in these species so again a duplication specific to chondrichtythes is difficult to rule out. These species confirm that distinctions between the respective CBS domains were established shortly after divergence and conserved ever since.
Five teleost fish genomes are available but these experienced a whole genome duplication with complex retention patterns here. making them unsuitable for evolutionary issues. Frog has the expected paralog pair, as does the amniote, Anolis. Chicken presents an odd situation with a clear IMPDH2 ortholog but an assembly gap where IMPDH1 should be. At the trace archives, a diverged and gappy second gene can be located that resembles IMPDH1. It is likely a pseudogene despite the lack of internal stop codons. The second bird genome, zebrafinch, also lacks IMPDH1. Thus in Aves it appears from the available data that IMPDH1 has been lost.
Synteny is important in this particular gene family for building an accurately labelled set of reference sequences. That is, in a fast evolving clade, IMPDH1 and IMPDH2 might be confused if neighboring gene information was not available to establish orthology. However, the useful lifespan of synteny for IMPDH1 especially is quite short. For example, Xenopus gene order (+SND1 -IMPDH1 -OPN1SW +CALU -SAPS2) already differs from human either by local inversion, misassembly, gene indels, or growing ortholog unrecognizability. Where synteny would be really helpful, say Branchiostoma or Petromyzon of Callorhinchus, it is either entirely lost or not yet available at the current state of assembly.
human gene order about IMPDH +SND1 PH4 +LEP DALRD3 -RBM28 C3orf60 -IMPDH1 IMPDH2 +HIG2 QARS +METTL2B USP19 +CALU -OPN1SW
IMPDH Sequence Resources
Phylogenetically representative IMPDH inosine monophosphate dehdrogenase sequences from eumetazoans. Both genes -- and their regulatory regions -- could be sampled much more densely within mammals. >IMPDH1_homSap Homo sapiens (human) chr7 127,828,554 0 MADYLISGGTGYVPEDGLTAQQLFASADGLTYN 2 1 DFLILPGFIDFIADEV 0 0 DLTSALTRKITLKTPLISSPMDTVTEADMAIAMA 0 0 LMGGIGFIHHNCTPEFQANEVRKVK 0 0 KFEQGFITDPVVLSPSHTVGDVLEAKMRHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHTTLLSE 0 0 VMTPRIELVVAPAGVTLKEANEILQRSKK 1 2 GKLPIVNDCDELVAIIARTDLKKNRDYPLASKDSQKQLLCGAAVGTREDDKYRLDLLTQAGVDVIVL 0 0 DSSQGNSVYQIAMVHYIKQKYPHLQVIGGN 2 1 VVTAAQAKNLIDAGVDGLRVGMGCGSICITQE 1 2 VMACGRPQGTAVYKVAEYARRFGVPIIADGGIQTVGHVVKALALGAST 1 2 VMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGSLDAMEKSSSSQKRYFS 2 1 EGDKVKIAQGVSGSIQDKGSIQKFVPYLIAGIQHGCQDIGARSLSVLR 2 1 SMMYSGELKFEKRTMSAQIEGGVHGLHS 2 1 YEKRLY* 0 >IMPDH2_homSap Homo sapiens (human) paired CDS domain exons 5-7 chr3 49,039,322 0 MADYLISGGTSYVPDDGLTAQQLFNCGDGLTYN 2 1 DFLILPGYIDFTADQV 0 0 DLTSALTKKITLKTPLVSSPMDTVTEAGMAIAMA 0 0 LTGGIGFIHHNCTPEFQANEVRKVK 0 0 KYEQGFITDPVVLSPKDRVRDVFEAKARHGFCGIPITDTGRMGSRLVGIISSRDIDFLKEEEHDCFLEE 0 0 IMTKREDLVVAPAGITLKEANEILQRSKK 1 2 GKLPIVNEDDELVAIIARTDLKKNRDYPLASKDAKKQLLCGAAIGTHEDDKYRLDLLAQAGVDVVVL 0 0 DSSQGNSIFQINMIKYIKDKYPNLQVIGGN 1 2 VVTAAQAKNLIDAGVDALRVGMGSGSICITQE 1 2 VLACGRPQATAVYKVSEYARRFGVPVIADGGIQNVGHIAKALALGAST 1 2 VMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAMDKHLSSQNRYFS 2 1 EADKIKVAQGVSGAVQDKGSIHKFVPYLIAGIQHSCQDIGAKSLTQVR 2 1 AMMYSGELKFEKRTSSAQVEGGVHSLHS 2 1 YEKRLF* >IMPDH1_musMus Mus musculus (mouse) MADYLISGGTGYVPEDGLTAQQLFANADGLTYNDFLILPGFIDFIADEVDLTSALTRKITLKTPLISSPM DTVTEADMAIAMALMGGIGFIHHNCTPEFQANEVRKVKKFEQGFITDPVVLSPSHTVGDVLEAKIQHGFS GIPITATGTMGSKLVGIVTSRDIDFLAEKDHTTLLSEVMTPRVELVVAPAGVTLKEANEILQRSKKGKLP IVNDQDELVAIIARTDLKKNRDYPLASKDSHKQLLCGAAVGTREDDKYRLDLLTQAGADVIVLDSSQGNS VYQIAMVHYIKQKYPHLQVIGGNVVTAAQAKNLIDAGVDGLRVGMGCGSICITQEVMACGRPQGTAVYKV AEYARRFGVPVIADGGIQTVGHVVKALALGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGSLDAM EKSSSSQKRYFSEGDKVKIAQGVSGSIQDKGSIQKFVPYLIAGIQHGCQDIGAQSLSVLRSMMYSGELKF EKRTMSAQIEGGVHGLHSYEKRLY* >IMPDH2_musMus Mus musculus (mouse) MADYLISGGTSYVPDDGLTAQQLFNCGDGLTYNDFLILPGYIDFTADQVDLTSALTKKITLKTPLVSSPM DTVTEAGMAIAMALTGGIGFIHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPKDRVRDVFEAKARHGFC GIPITDTGRMGSRLVGIISSRDIDFLKEEEHDRFLEEIMTKREDLVVAPAGVTLKEANEILQRSKKGKLP IVNENDELVAIIARTDLKKNRDYPLASKDAKKQLLCGAAIGTHEDDKYRLDLLALAGVDVVVLDSSQGNS IFQINMIKYIKEKYPSLQVIGGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKV SEYARRFGVPVIADGGIQNVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAM DKHLSSQNRYFSEADKIKVAQGVSGAVQDKGSIHKFVPYLIAGIQHSCQDIGAKSLTQVRAMMYSGELKF EKRTSSAQVEGGVHSLHSYEKRLF* >IMPDH1_bosTau Bos taurus (cow) MEEPRAPPAGSAPFPVSLQGGGTADVPEPGARQHPGHETAAQRY SARLLQAGYEPESMADYLISGGTGYVPEDGLTAQQLFANADGLTYNDFLILPGFIDFT ADEVDLTSALTRKITLKTPLISSPMDTVTEADMAIAMALMGGIGFIHHNCTPEFQANE VRKVKKFEQGFITDPVVLSPSHTVGDVLEAKIRHGFSGIPITETGTMGSKLVGIVTSR DIDFLAEKDHTTLLSEVMTPRNELVVAPAGVTLKEANEILQRSKKGKLPIVNDRDELV AIIARTDLKKNRDYPLASKDSHKQLLCGAAVGTREDDKYRLDLLTQAGADVIVLDSSQ GNSVYQIAMVHYIKQKYPHLQVIGGNVVTAAQAKNLIDAGVDGLRVGMGCGSICITQE VMACGRPQGTAVYKVAEYARRFGVPVIADGGIQTVGHVVKALALGASTVMMGSLLAAT TEAPGEYFFSDGVRLKKYRGMGSLDAMEKSSSSQKRYFSEGDKVKIAQGVSGSIQDKG SIQKFVPYLIAGIQHGCQDIGARSLSVLRSMMYSGELKFEKRTMSAQIEGGVHGLHSY EKRLY* >IMPDH2_bosTau Bos taurus (cow) MADYLISGGTSYVPDDGLTAQQLFNCGDGLTYNDFLILPGYIDFTADQVDLTSALTKKITLKTPLVSSPM DTVTEAGMAIAMALTGGIGFIHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPRDRVRDVFEAKARHGFC GIPITDTGRMGSHLVGIISSRDIDFLKEEEHDRLLGEIMTKREDLVVAPAGITLKEANEILQRSKKGKLP IVNENDELVAIIARTDLKKNRDYPLASKDAKKQLLCGAAIGTHEDDKYRLDLLSQAGVDVVVLDSSQGNS IFQINMIKYIKEKYPSIQVIGGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKV SEYARRFGVPVIADGGIQNVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAM DKHLSSQNRYFSEADKIKVAQGVSGAVQDKGSIHKFVPYLIAGIQHSCQDIGAKSLTQVRAMMYSGELKF EKRTSSAQVEGGVHSLHSYEKRLF* >IMPDH1_ornAna Ornithorhynchus anatinus frag MEGTPLISGGTGYVPLDGLTAQQLFAIADGLTYNDFLILPGFIDFTADEVDLTSALTRKITLK TPLISSPMDTVTEADMAIAMALMGGIGFIHHNCTPEFQANEVRKVKKFEQGFITDPVV LSPSHTVGDVLEAKARHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHATYLSEV MTRRTELVVAPAGVTLKEANEILQRSKKGKLPIVNDSDELVAIIARTDLKKNRDYPLA SKDAHKQLLCGAAVGTREDDKYRLDLLTQAGTDVIVLDSSQGNSVYQIAMVHYIKQKY PQLQVIGGNVVTAAQAKNLIDAGVDGLRVGMGCGSICITQEVMACGRPQGTAVYKVAE YARRFGVPVIADGGIQTVGHVVKALALGAST EGDKVKVAQGVSGSIQDKGSIQKFVP YLIAGIQHGCQDIGARSLSVLRSMMYSGELKFEKRTMSAQIEGGVHGLHSYEKRLY* >IMPDH2_ornAna Ornithorhynchus anatinus frag DFLILPGYIDFTADQVDLTSALTKKITLKTPLIS SPMDTVTEAGMAIAMALTGGIGFIHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPKD RVRDVFEAKARHGFCGIPITDNGKMGSRLMGIISSRDIDFLKEEEHDLYLGEIMTKWE DLVVAPAGVTLKEANEILQRSKKGKLPIVNEDNELVAIIARTDLKKNRDYPLASKDAK KQLLCGAAIGTHEDDKYRLDLLAQAGVDVVVLDSSQGNSIFQINMIKYIKEKYHNLQV IGGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKVSEYARRF GVPVIADGGIQNVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMG SLDAMDKNLGSQNRYFSEADKIKVAQGVSGAVQDKGSIHKFVPYLIAGIQHSCQDIGA KSLTLVRAMMYSGELKFEKRTSSAQVEGGVHGLHSYEKRLF* >IMPDH1_galGal Gallus gallus (chicken) 60% indels eye (hatched) DR427747 tr 251351904 inconsistentTaeniopygia guttata missing too AIPATDGESGRLQMVAEAGADVVLLDSSQGNSACQISMIRYIKERYPDMQVVGGCVVTAA QCRRLIEAGADALRVGMGGGADCGDSDVPWGRPPGTAVYRVADYARRFGVPVIADGGIRA VGHIVKAAAVGASTVMLGSLLWGTTEAPGVLFSDGAVLKKFRGGAALEVTETGGGTQQHC YSDVAEGQMAQGVPAALQDKGSVQKFLPYLSAGIQRGCQHVGARSLSALRSMAYSGELKF ERRTLAAQLEGLQGLRACEKRLY* >IMPDH1_anoCar Anolis carolinensis (lizard) misassembly frag 0 2 1 0 0 0 0 LMGGIGIIHHNCTPEFQANEVRKVK 0 0 VQKFEQGFITDPVVLSPSHSVGDVFEAKVRHGFSGIPVTEAGKMGSTLVGIVTSRDIDFLSEKDYDTPLSE 0 0 VMTKRSDLVVAPAGVTLKEANEILQRSKK 1 2 GKLPIVNDADELVAIIARTDLKKNRDYPLASKDPRKQLLCGAAIGTREDDKYRLDLLTQAGVDVVVL 0 0 DSSQGNSVYQISMIHYIKHKYPELQVIGGN 2 1 VVTAAQAKNLIDAGVDALRVGMGCGSICITQE 1 2 VMACGRPQGTAVYKVAEYARRFGVPVIADGGIQTVGHVVKALSLGAST 1 2 VMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGSLDAMEKNSSSQKRYFs 2 1 EGDKVKVAQGVSGSIQDKGSIQKFVPYLIAGIQHGCQDIGAKSLSILR 2 1 2 1* 0 >IMPDH2_galGal Gallus gallus (chicken) fully syntenic. no IMPDH1 MADYLISGGTGYVPDDGLTAQQLFSCGDGLTYNDFLILPGYIDFTADQVD LTSALTKRITLKTPLVSSPMDTVTEAGMAIAMALTGGIGFIHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPNDRVRDVFEAKARHGFCGIPITDNGKM GGKLVGIISSRDIDFLKESEHDLPLGEIMTKREDLVVAPSGVMLKEANEILQRSKKGKLPIVNEDDELVAIIARTDLKKNRDYPLASKDSKKQLLCGAAI GTHEDDKYRLDLLVQAGVDAVVLDSSQGNSIFQINMIKYIKEKYPNLQVIGGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKV SEYARRFGVPVIADGGIQTVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAMDKNLGSQNRYFSETDKIKVAQGVSGAVQDK GSIHKFIPYLIAGIQHSCQDIGAKSLTQVRAMMYSGELKFEKRTTSAQVEGGVHGLHSYEKRLF* >IMPDH1_xenTro Xenopus tropicalis (frog) SND1 ... OPN1SW CALU SAPS2 not syntenic MADYLISGGTGYVPEDGLTAQHLFANSDGLTYNDFLILPGFIDFTADEVDLTSALTRKITLKTPLISSPM DTVTESDMAIAMALMGGIGIIHHNCTPEFQANEVRKVKKFEQGFITDPVVMSLNHTVGDVFEAKNRHGFS GIPVTETGKMGSKLVGIVTSRDIDFLTEKDYSTYLSEVMTKRDELVVAPAGVTLKEANEILQRSKKGKLP IVNDSDELVAIIARTDLKKNRDYPLASKDCRKQLLCGAAIGTREDDKYRLDLLTQAGVDVVVLDSSQGNS VYQINMIHYIKQKYPELQVVGGNVVTAAQAKNLIDAGVDALRVGMGCGSICITQEVMACGRPQGTAVYKV AEYARRFGVPVIADGGIQTVGHVVKALALGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGSLDAM EKNTSSQKRYFSEGDKVKVAQGVSGSIQDKGSIHKFVPYLIAGIQHGCQDIGAKSLSILRSMMYSGELKL EKRTMSAQVEGGVHGLHSYEKRLY* >IMPDH2_xenTro Xenopus tropicalis (frog) fully syntenic MADYLISGGTSYVPDDGLTAQQLFGAGDGLTYNDFLILPGYIDFTADQVD LTSALTKKITLKTPMVSSPMDTVTEASMAIAMALTGGIGIMHHNCTPEFQANEVRKVKKYEQGFITDPVVLSPKHRVRDVFEAKARHGFCGIPITENGKM GSKLAGIISSRDIDFLKSEEHDLALSEIMTRREDLVVAPAGVTLKEANEILQRSKKGKLPIVNGNDELVAIIARTDLKKNRDYPLASKDAKKQLLCGAAI GTHEDDKYRLDLLVQAGVDAVVLDSSQGNSIFQINMIKFIKEKYQDLQVIAGNVVTAAQAKNLIDAGADALRVGMGSGSICITQEVLACGRPQATAVYKV SEYARRFGVPVIADGGIQTVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGSLDAMDKNVSSQKRYFSEADKIKVAQGVSGAVQDK GSIHKFIPYLIAGIQHSCQDIGAKSLTQVRAMMYSGELKFEKRTMSAQVEGGVHGLHSYEKRLF* >IMPDH1_calMil Callorhinchus milii (elephantshark) fragment 0 2 1 DFLILPGFIDFSAEEV 0 0 0 0 0 0 0 0 1 2 GKLPIVNDADELIAIIARTDLKKNRDYPLASKDSRKQLLCGAAIGTREDDKYRLDLLVQAGVDVIVL 0 0 DSSQGNSVFQINMIHCIKQRYPELQVVGGN 2 2 1 2 1 2 2 1 EGDKVKVAQGVSGSVQDKGSIHKFAPYLITGIQHGCQDIGAKSLSILR 2 1 SMMYSGELKFEKRTISAQVEGGVHGLHS 2 1 * 0 >IMPDH2_calMil Callorhinchus milii (elephantshark) fragment 0 MADYLISGGTGYVPDDGQSAQQLLGAGDGLTYn 2 1 DFLILPGYIDFTSDQV 0 0 DLTSALTKKITLKTPLVSSPMDTVTEANLAIAMA 0 0 LMGGIGIIHHNCTPEFQANEVRKVK 0 0 KYEQGFITDPVVMSPSEKVRDVFEAKARHGFSGIPITDNGKMGGKLKGIISSRDIDFLTEKEHDLLLSE 0 0 VMTRREDLVVAPAAVTLKEANEILQRSKK 1 2 GKLPIVNDNDELVAIIARTDLKKNRDYPLASKDSKKQLLCGAAIGTHEDDKYRLDQLFQAGVDVVVL 0 0 2 2 VVTAAQAKNLIDAGVDALRVGMGCGSICITQE 1 2 VMACGRPQGTAVYKVAEYARRFGVPVIADGGIQTVGHVVKALALGAST 1 2 2 1 ESDKIKVAQGVSGAVQDKGSIQKFIPYLIAGIQHGCQDIGAKSLTQLR 2 1 AMMYSGELKFEKRTLCAQVEGGVHGLHS 2 1 * 0 >IMPDH_petMar_frag2 Petromyzon marinus (lamprey) 854345740 854350173 0 MADFLISGGTGYVPDDGMTAQQLFAGADGLTYn 2 1 0 0 DLASALTKQITLKTPLVSSPMDTVTESSMAIAMA 0 0 LMGGVGVIHHNCTPEFQANEVRKVk 0 >IMPDH_petMar_frag3 Petromyzon marinus (lamprey) 565582434 565582613 0 MADYLISGGAGYVPEDGLTATQLFAGTDGLTYn 2 1 DFLILPGFIDFTVEEV 0 0 0 0 0 0 KYEQGFITDPVVLSPAHTVGDVLEAKARHGFSGIPITETGRMGGRLAGVITSRDIDFLTE 0 >IMPDH_petMar_frag1 Petromyzon marinus (lamprey) 700532459 700537638 2 GKLPIVNEAGELVAIMARTDLKKNRDFPLASKDAHKQLLCGAAVGTRPDDQHRLELLVKAGVDVVIL 0 0 DSSQGNSVFQINMIRYIKQTYPELQVIGGN 1 2 VVTAAQAKNLIDAGVDALRVGMGAGSICITQE 1 2 1 2 VMMGSLLATTEAPGEYFFSDGVRLKKYRGMGSLDAMEKNQGSQSRYFS 2 1 ENDKMKVAQGVSGSVQRQG 2 1 SMMYTGELKFEKRTMSAQVEGGVHGLHS 2 1 * 0 >IMPDH1_petMar Petromyzon marinus (lamprey) composite 0 MADYLISGGAGYVPEDGLTATQLFAGTDGLTYn 2 1 DFLILPGFIDFTVEEV 0 0 DLASALTKQITLKTPLVSSPMDTVTESSMAIAMA 0 0 LMGGVGVIHHNCTPEFQANEVRKVk 0 0 KYEQGFITDPVVLSPAHTVGDVLEAKARHGFSGIPITETGRMGGRLAGVITSRDIDFLTEKDHGIPLHQ 0 0 VMTKREDLVVAPSGVTLTEANELLQRSKK 1 2 GKLPIVNEAGELVAIMARTDLKKNRDFPLASKDAHKQLLCGAAVGTRPDDQHRLELLVKAGVDVVIL 0 0 DSSQGNSVFQINMIRYIKQTYPELQVIGGN 1 2 VVTAAQAKNLIDAGVDALRVGMGAGSICITQE 1 2 VMAVGRPQGTAVYRVAEYARRFGVPVMADGGIQTVGHITKALALGAST 1 2 VMMGSLLATTEAPGEYFFSDGVRLKKYRGMGSLDAMEKNQGSQSRYFS 2 1 ENDKMKVAQGVSGSVQRQG 2 1 SMMYTGELKFEKRTMSAQVEGGVHGLHS 2 1 * 0 >IMPDH_braFlo Branchiostoma floridae (lancelet) one copy, severe misassembly 0 MADYLISGGTGYVPDDGMTASQLFGTGEGLTYn 2 1 DFLILPGFIDFTADEV 0 0 DLTSAMTKKIQLKAPLVSSPMDTVSESDMAIAMA 0 0 LTGGIGIIHSNCTPEFQANEVRKVK 0 0 KYEQGFIMDPIVLSPEHTVGDVCEMKRKHGFSGIPITENGKLGGKLLGIVTSRDIDFMNSDHHHIKLRD 0 0 VMTPFEELVVGHAGVSLKEANETLQRSKK 1 2 GKLPIVNENDELVSLIARTDLKKNRDYPLASKDSKKQLLCGAAIGTREEDKYRVELLVQAGVDLVVL 0 0 DSSQGNSIYQINMIRYLKQKYSELQVIGGN 2 1 VVTAAQAKNLIDAGVDGLRVGMGSGSICITQE 1 2 VMAVGRPQGTAVYKVAEYARRFGVPVIADGGISTVGHITKALALGAST 1 2 VMMGSLLAGTSEAPGEYFFQDGVRLKKYRGMGSLEAMEKGKASQNRYFR 2 1 ESDKLKVAQGVTGSIQDKGSVHKFVPYLIAGIQHGCQDIGAKSLSSLR 2 1 SMMYSGELRFETRTVSAQVEGGVHGLHS 2 1 * 0 >IMPDH_cioInt Ciona intestinalis (tunicate) mRNA AK114441 MAEFLISGETSYVPEDGLTAAQLLNTGDGLTYNDFLILPGFIDFTASEVDLTSALTKKISL KTPLLSSPMDTVTESDMAIGMALMGGMGFIHYNCTPEFQAAEVRRVKKYEQGFIQNPVTL GPKATVRDVTDVKAMYGFSGIPVTDDGTPTGKLIGLVSSRDFDFLKPEESNTPLEQVMTG RDKLITADTSVTLQEANHILSQSKKGKLPIVDADDRLVSLIARTDLKKNREFPLASKDER KQLLCGAAISTREEDKHRLELLVEAGVDAVILDSSQGNSIYQINSIRYIRHKYPHLQVIA GNVVTAAQAKNLIDAGADALRVGMGSGSICITQEVMAVGRPQATAVYKVSEYARRFNVPV IADGGIQNVGHVTKALALGASTVMMGSLLAATTESPGEYFYSDGIRLKKYRGMGSVDAME SCKSSQSRYFSEKDKIRVAQGVSGAVQDKGSVHTFLPYLIAGIQHGCQDIGSRSMPMLRS MMYSGELKFERRSTSAQVEGGVHGLHSFEKPHL* >IMPDH_strPur Strongylocentrotus purpuratus (sea urchin) single gene 2 intronation changes 0 MSKKVKLMNGAMEPQVDDGLSGQQLFGSGDGLTYn 2 1 DFLILPGYIDFTSDQV 0 0 DLQSQLTKDITLKAPLVSSPMDTVTESSMAIAMA 0 0 LCGGIGIIHHNCSPEFQANEVRKVK 0 0 KYEQGFIMDPVVLGPNDTVGDVFGSKAKHGFS 1 2 GIPITDTGRLGGKLLGIVTARDIDFLKPESYVKPLST 00 AMTCREDLVVAPANVTLKQANDLLQKAKK 1 2 GKLPIVNEKDELVSLISRTDLKKHREFPLASKDPRKQLLCGAAIGTREEDKHRLDLLVQAGVDVVIL 0 0 DSSQGNSSFQVSMIKCTKAKYPELQVVAGN 1 2 VVTVAQAKNLIQAGADALRVGMGSGSICITQE 1 2 VMAVGRPQGTAVYRVAQYARSCGVPIIADGGITTVGHITKALSLGASS 1 2 VMMGSLLAGTTEAPGEYFFSDGVRLKKYRGMGSLDAMEKNQSSAKRYFR 2 1 ERDKLKVAQGVSGSIVDKGSIHKFVPYLIAGIQHGCQDIGALSLTVLR 2 1 EKMYSGEVRFERRSPSAQVEGGVHSLHS 2 1 *0 >IMPDH_sacKow Saccoglossus kowalevskii (acornworm) frag one copy 82% homSap 0 2 1 0 0 0 0 0 0 KYEQGFIMDALVMSANTTIKEVFAAKSQHGFSGIPITDNGKLGGRLLGIVTARDIDFVEPEFNDKPLEQFMTKREDLVVAPANVTLKEANDILQKSKK 1 2 GKLPIVNENDELVSLISRTDLKKHREFPLASKDSKKQLLCGAAIGTHESDKNRLDLLVQAGVDVIIL 0 0 1 2 VVTAAQAKNLIDAGVDALRVGMGSGSICITQE 1 2 VMAVGRPQGTSVYKVAEYARRFGVPVIADGGIGTVGHITKALALGAST 1 2 VMMGSLLAGTSEAPGEYYFSDGVRLKKYRGMGSLDAMEiNQSDRYFS 2 1 ESDKLKVAQGVSGSIIDKGSIHKFIPYLIAGIQHGCQDIGAKSMSMLR 2 1 2 1 *0 >IMPDH_helRob Helobdella robusta (leech) frag cDNA QAGVDFVVLDSSQGNSIYQIKLIKYIKEKYPNLQVIGGNVVTAAQAKNLIDAGVDALRVG MGSGSICITQEVMAVGRPQGTAVYKVAEYARRFGVPIIADGGIENVGHIVKALALGASTV MMGSLLAGTTEAPGEYYFADGVRLKKYRGMGSLDAMEQHKASQSRYFSDSDKVKVAQGVS GAVVDKGSIHKFLPYLISGVQHGCQDLGAKSLSCLRSMMYQGELKFEKRTTSSQIEGGVH GLHSYEKRLY* >IMPDH_schMan Schistosoma japonicum (flatworm) fragment FSLKVPFASSPMDTVTEAKMAIAMS LCGSIGFVHNNCSVEAQANEVKKVK KYNQGFILSPVVVSPRQPIYDIIEIKKKYGFG GIPVTEDGYMGSRLVGLVTLRDVDFLDPNDFNTPVEKVMTPFDDLVTAFSCVTLSEANDLLRKSKK GKLPIINENRELVALIARTDLQKNRDYPLASRDDENQLIVGAAISTHEGDFARVKALINSGVDIIVI DSSQGNSIYQLDMIKRIKSSFPDLQIIGGN IVTCAQAKNLIDAGVDGLRVGMGSGSICITQE VTAIGRSQAKAVYSVSEYAHKYDIPVIADGGIQNTGHIVKALSFGASS VMMGGLLAGTTESAGEYIFSDGVKLKKYRGMGSIEAMSQHTESQARYFSESDRIKVAQGVSGTIVDR GSVHQLVPYLVAGVKHGLQQIGARNITELHNMSRSGKLRFELRSPSAQLEGGVHSLYS YDKSMF* >IMPDH_schMed Schmidtea mediterranea (planaria) frag NZ_AAWT01010891 more GFSASELFNKKTGLTYNDFILLPGYIDFRSEEVDITTRITKNMYLKTPLVSSPMDTVTES NMAIAMALSGGIGILHHNCTIDTQANEIRRVKKYEQGFIVDPVVMSIDNTVGEVMNIKIK NGFTGIPITDNGKLGGKLVGLVTLRDIDFLPKSNWDLKVSEVMTPFENLITAPYGVALHE ANGILQRSKKGKLPIINENRELVALISRTDLKKNREYPLSSKDDR >IMPDH_ixoSca Ixodes scapularis MSDKKTTFSEDGLTAQHLIGSGDGLTYNDFLILPGFIDFNADDVD LTSKLTKNITLQAPLVSSPMDTVTESEMAIAMALCGGIGIIHHNCTPEHQANEVHKVKKYKHGFIHDPVVLSPNNC VADVFEVKRKHGFAGVPITENGKLGGKLVGMVTSRDIDFIPIEDHNRLLSEVMTSLK DLTVASSKVTLSEANSLLQKSKKGKLPLVNEGGELVSLIARTDLKKSRSYPLASKDENKQLIVGAAIGTREADKPRL ELLVQAGVDVVVLDSSQGNSVYQIEMVKYIKSKYPGLQVIGGNVVTTAQAKNLIEAGVDG LRVGMGSGSICITQEVMACGRPQATAVYKVAEYARRFGVPCVADGGVSSVGHIIKALALG ASTVMMGSMLAGTTESPGEYFFSNGVRLKKYRGMGSLDAMQSTEGGGSLNRYYQSDQDKV RVAQGVSGTIVDKGSIHRYVPYLITGIKYGCQDIGARSLDVLKANMYSGDIKFEKRSVSA QIEGGVHGLHSYEKRLY* >IMPDH_nemVec Nematostella vectenisis (anemone) 78% XM_001641825 0 MMADFLIAGGTSYVPEDGMTAGQLFQSDGLTYS 2 1 DFIILPGFIDFPATDV 0 0 DLTSPLTRRITIKTPLVSSPMDTVTESALATAMA 0 0 LNGGIGIIHHNCSIEFQANEIRKVKKFEQGFIMAPLVLSATNTVADVIDAKQRHGFSGIPIT 1 2 ENGQLGGILQGIVTSRDIDFLHGVENHKQLGE 0 0 VMTRLEDLVVAKAGITLNEANKILQMSKK 1 2 GKLPIVNEKGELVSLIARTDLKKNRDYPLASKDENKQLL 1 2 VGAAIGTREDDKARLHALVEAGVDVVVIDSSQGNSIYQLSLIS 2 1 HIKENYPNLQIVGGNVVTASQAKNLIDAGVDALRVGMGSGSICITQEVMAVGRPQGTAVYKVAEYARRFGVPVLADGGIQNVGHITKALSLGAST 1 2 VMMGSLLAGTSEAPGEYFFADGVRLKKYR 1 2 GMGSLSAMEKNSSSASRYFS 2 1 ENDKVKVAQGVSGSVVDKGSIHKFVPYLTAGIQHGCQDLGAKSLTSLR 2 1 SMMYSGELKFERRTTSSQIEGGVHGLHS 2 1 YEKRLF* 0
CBS Domain Sequence Resources
It is difficult to recover full length IMPDH sequences for a gene with 13 exons because of incomplete assemblies, confusion between IMPDH1 and IMPDH2 fragments, and the tendency for cDNA projects to begin at one end and not proceed quite to full length. Sequences more commonly are complete enough to have full length CBS domains. Thus to the extent that this domain regulates enzymatic activity through allosteric effects and that small molecule effectors should discriminate between the two CBS domains, it is informative to determine where conserved residues in the respective domains differ, ie determine a subtractive profiles.
>IMPDH1_homSap ITDPVVLSPSHTVGDVLEAKMRHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHTTLLSEVMTPRIELVVAPAGVTLKEANEILQRSKKGKLPIVNDCDELVAIIARTDLKKN >IMPDH1_bosTau ITDPVVLSPSHTVGDVLEAKIRHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHTTLLSEVMTPRNELVVAPAGVTLKEANEILQRSKKGKLPIVNDRDELVAIIARTDLKKN >IMPDH1_musMus ITDPVVLSPSHTVGDVLEAKIQHGFSGIPITATGTMGSKLVGIVTSRDIDFLAEKDHTTLLSEVMTPRVELVVAPAGVTLKEANEILQRSKKGKLPIVNDQDELVAIIARTDLKKN >IMPDH1_ornAna ITDPVVLSPSHTVGDVLEAKARHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHATYLSEVMTRRTELVVAPAGVTLKEANEILQRSKKGKLPIVNDSDELVAIIARTDLKKN >IMPDH1_anoCar ITDPVVLSPSHSVGDVFEAKVRHGFSGIPVTEAGKMGSTLVGIVTSRDIDFLSEKDYDTPLSEVMTKRSDLVVAPAGVTLKEANEILQRSKKGKLPIVNDADELVAIIARTDLKKN >IMPDH1_xenTro ITDPVVMSLNHTVGDVFEAKNRHGFSGIPVTETGKMGSKLVGIVTSRDIDFLTEKDYSTYLSEVMTKRDELVVAPAGVTLKEANEILQRSKKGKLPIVNDSDELVAIIARTDLKKN >IMPDH1_danRer ITDPVVMSPRHTVGDVFEAKVRHGFSGIPVTETGKMGSKLVGIVTSRDIDFLSEKDYDRPLEESMTKREDLVVAPAGVTLKEANDILQRSKKGKLPIVNDSDELVAIIARTDLKKN >IMPDH1_takRer ITDPVVMSPRHTVGDVFEAKIRHGFSGIPVTETGKMGSKLVGIVTSRDIDFLSEKDHDRPLEEAMTKREDLVVAPAGVTLKEANDILQRSKKGKLPIVNDSDELVAIIARTDLKKN >IMPDH1_leuRaj ITDPVVMSLHHTVGDVFEAKSRHGFSGIPITETGKMGSKLMGIVTSRDIDFLSEKDYDTALNEVMTKREDLVVAPAGVTLKEANGILQRSKKRKVPIVNDMDELISIIARTDLkkn >IMPDH1_petMar ITDPVVLSPAHTVGDVLEAKARHGFSGIPITETGRMGGRLAGVITSRDIDFLTEKDHGIPLHQVMTKREDLVVAPSGVTLTEANELLQRSKKGKLPIVNEAGELVAIMARTDLKKN >IMPDH2_homSap ITDPVVLSPKDRVRDVFEAKARHGFCGIPITDTGRMGSRLVGIISSRDIDFLKEEEHDCFLEEIMTKREDLVVAPAGITLKEANEILQRSKKGKLPIVNEDDELVAIIARTDLKKN >IMPDH2_musMus ITDPVVLSPKDRVRDVFEAKARHGFCGIPITDTGRMGSRLVGIISSRDIDFLKEEEHDRFLEEIMTKREDLVVAPAGVTLKEANEILQRSKKGKLPIVNENDELVAIIARTDLKKN >IMPDH2_bosTau ITDPVVLSPRDRVRDVFEAKARHGFCGIPITDTGRMGSHLVGIISSRDIDFLKEEEHDRLLGEIMTKREDLVVAPAGITLKEANEILQRSKKGKLPIVNENDELVAIIARTDLKKN >IMPDH2_galGal ITDPVVLSPNDRVRDVFEAKARHGFCGIPITDNGKMGGKLVGIISSRDIDFLKESEHDLPLGEIMTKREDLVVAPSGVMLKEANEILQRSKKGKLPIVNEDDELVAIIARTDLKKN >IMPDH2_ornAna ITDPVVLSPKDRVRDVFEAKARHGFCGIPITDNGKMGSRLMGIISSRDIDFLKEEEHDLYLGEIMTKWEDLVVAPAGVTLKEANEILQRSKKGKLPIVNEDNELVAIIARTDLKKN >IMPDH2_xenTro ITDPVVLSPKHRVRDVFEAKARHGFCGIPITENGKMGSKLAGIISSRDIDFLKSEEHDLALSEIMTRREDLVVAPAGVTLKEANEILQRSKKGKLPIVNGNDELVAIIARTDLKKN >IMPDH2_calMil ITDPVVMSPSEKVRDVFEAKARHGFSGIPITDNGKMGGKLKGIISSRDIDFLTEKEHDLLLSEVMTRREDLVVAPAAVTLKEANEILQRSKKGKLPIVNDNDELVAIIARTDLKKN >IMPDH2_leuRaj ITDPVVLSPRHRVRDVFEAKARHGFCGIPITDTGTLGGRLAGIISSRDIDFLEESEQELPLEQVMTRREELVVAPAGVRLKEANEILQRSKKGKLPIVNEQDQLVAIISRTDLkkn >IMPDH_braFlo IMDPIVLSPEHTVGDVCEMKRKHGFSGIPITENGKLGGKLLGIVTSRDIDFMNSDHHHIKLRDVMTPFEELVVGHAGVSLKEANETLQRSKKGKLPIVNENDELVSLIARTDLKKN >IMPDH_cioInt IQNPVTLGPKATVRDVTDVKAMYGFSGIPVTDDGTPTGKLIGLVSSRDFDFLKPEESNTPLEQVMTGRDKLITADTSVTLQEANHILSQSKKGKLPIVDADDRLVSLIARTDLKKN >IMPDH_strPur IMDPVVLGPNDTVGDVFGSKAKHGFSGIPITDTGRLGGKLLGIVTARDIDFLKPESYVKPLSTAMTCREDLVVAPANVTLKQANDLLQKAKKGKLPIVNEKDELVSLISRTDLKKH >IMPDH_sacKow IMDALVMSANTTIKEVFAAKSQHGFSGIPITDNGKLGGRLLGIVTARDIDFVEPEFNDKPLEQFMTKREDLVVAPANVTLKEANDILQKSKKGKLPIVNENDELVSLISRTDLKKH >IMPDH_schMed IVDPVVMSIDNTVGEVMNIKIKNGFTGIPITDNGKLGGKLVGLVTLRDIDFLPKSNWDLKVSEVMTPFENLITAPYGVALHEANGILQRSKKGKLPIINENRELVALISRTDLKKN >IMPDH_schMan ILSPVVVSPRQPIYDIIEIKKKYGFGGIPVTEDGYMGSRLVGLVTLRDVDFLDPNDFNTPVEKVMTPFDDLVTAFSCVTLSEANDLLRKSKKGKLPIINENRELVALIARTDLQKN >IMPDH_ixoSca IHDPVVLSPNNCVADVFEVKRKHGFAGVPITENGKLGGKLVGMVTSRDIDFIPIEDHNRLLSEVMTSLKDLTVASSKVTLSEANSLLQKSKKGKLPLVNEGGELVSLIARTDLKKS >IMPDH_nemVec IMAPLVLSATNTVADVIDAKQRHGFSGIPITENGQLGGILQGIVTSRDIDFLHGVENHKQLGEVMTRLEDLVVAKAGITLNEANKILQMSKKGKLPIVNEKGELVSLIARTDLKKN