Cryptochrome evolution: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 1,026: | Line 1,026: | ||
It appears that the last exon in fish has lost all homology (and so functionality), in some cases simply running out into junk dna until a stop codon is encountered. Exon seven is broken up in some fish with an extra intron that might have some use in fish taxonomy as a derived characteristic. | It appears that the last exon in fish has lost all homology (and so functionality), in some cases simply running out into junk dna until a stop codon is encountered. Exon seven is broken up in some fish with an extra intron that might have some use in fish taxonomy as a derived characteristic. | ||
No evidence for CRY2 currently exists in cartilaginous fish or earlier deuterostomes suggesting that ancestral CRY1 duplicated in stem bony vertebrates, giving rise to CRY2. It appears that all insect CRY2 entries at GenBank mislabelled and actually represent an independent gene duplication of CRY1 in arthropods (that was lost in drosophilids). | No evidence for CRY2 currently exists in cartilaginous fish or earlier deuterostomes suggesting that ancestral CRY1 duplicated in stem bony vertebrates, giving rise to CRY2. It appears that all insect CRY2 entries at GenBank are grievously mislabelled and actually represent an independent gene duplication of CRY1 in arthropods (that was lost in drosophilids). If so, these 'CRY2' sequences cannot serve as valid model systems for vertebrate CRY2. | ||
>CRY2_homSap Homo sapiens (human) 11 exons | >CRY2_homSap Homo sapiens (human) 11 exons | ||
Revision as of 15:51, 12 March 2012
Introduction to Cryptochromes
Cryptochromes are large flavoproteins with a curiously complex evolutionary history, beginning billions of years ago as repair enzymes for dna damaged by ultraviolet light. An old gene duplication followed by specializing divergence gave rise to two paralogs repairing distinct types of dna damage (cyclobutane pyrimidine dimers and 6-4 pyrimidine-pyrimidone pairs). These photolyases initially used FAD activated by visible blue light to undo the damage done by UV.
Since FAD has relatively low adsorbance, photolyases evolved a second site for an antenna chromophore with better light harvesting capabilities that could transfer its excitation to the FAD at the active site. This elusive second molecule may be FMN, folate, or a 5-deazariboflavin called Fo (once thought restrict to methanogenic archaea). In the case of the much-studied Drosophila, both the photolyases utilize Fo, making it a new vitamin for this species since the Fo biosynthetic genes are absent.
A second round of gene duplication of the 6-4 photolyase gave rise to a cryptochrome which retained the conformational change induced by FAD binding of blue light but lost dna repair capacity, instead specializing in entraining the day/night circadian rhythm cycle. A third round of gene duplication gave rise to two cryptochromes CRY1 and CRY2.
These five genes were retained in various combinations in different clades during the subsequent course of evolution, causing endless comparative nomenclatural confusion. For example, Drosophila did not retain CRY2 unlike other insects while placental mammals lost all three photolyases though marsupials retained one and monotremes two. Gallinaceous birds also lost a photolyase. Rayfinned fish had a series of further duplications within the gene family. Despite this, the primary sequence, exon structure, fold and FAD, antenna and dna binding sites have largely been conserved -- along with key regulatory binding sites to other proteins -- even as antenna molecules and dna repair capacity was dispensed with.
Standard lab mouse C57BL/6J has a mutated CRY1 cryptochrome gene
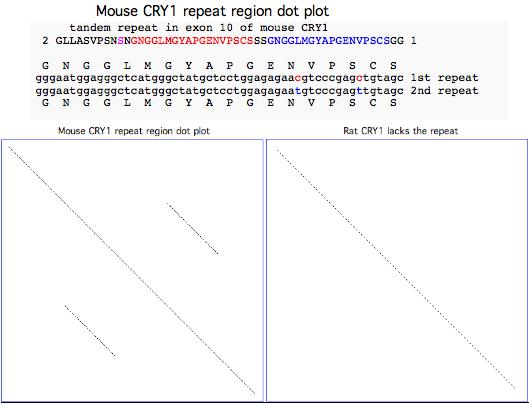
Lab mouse has an odd mutation in its 10th exon where a century of inbreeding may have inadvertently fixed a very serious 54 bp tandem stutter mutation resulting in 18 additional amino acids (the NGGLMGYAPGENVPSCSGG red and blue repeats in NM_007771 reference sequence) that would very likely disrupt the C-terminal region of the protein. The repeat is preceded by the substitution of a serine (shown in magenta in the alignment below) for a strictly invariant proline (back to chondrichthyes).
Although this region lies beyond the two main domains and has a complex evolutionary history, phylogenetic comparison to the eight available rodent and lagomorph sequences implies that this change in lab mouse will have serious functional consequences. A mutation in this critical pacemaker gene could plausibly affect lifespan, metabolic disorder and tumor progression; such a change is completely unprecedented in rodents including rat and indeed in vertebrates.
All 14 available transcripts exhibit the same anomaly -- this is not limited to one strain of mouse, not a somatic mutation, not an unfortunate heterozygous allele. The affected ESTs came from C57BL/6J, C57BL/6, C57BL/6J x DBA/2J, 129 FVB/N and embryo, eye, ventricle, thymus, mammary tumor; the affected GenBank NR entries add a keratinocyte cell line Pam. The mouse genome project used C57BL/6J, the most widely used inbred strain according to the Jackson Laboratory:
"Although C57BL/6J is refractory to many tumors, it is a permissive background for maximal expression of most mutations. C57BL/6J mice are resistant to audiogenic seizures, have a relatively low bone density, and develop age related hearing loss. They are also susceptible to diet-induced obesity, type 2 diabetes, and atherosclerosis. C57BL/6J mice are used in a wide variety of research areas including cardiovascular biology, developmental biology, diabetes and obesity, genetics, immunology, neurobiology, and sensorineural research. C57BL/6J mice are also commonly used in the production of transgenic mice. Overall, C57BL/6 mice breed well, are long-lived, and have a low susceptibility to tumors. Primitive hematopoietic stem cells from C57BL/6J mice show greatly delayed senescence relative to BALB/c and DBA/2J. This is a dominant trait. Other characteristics include: 1) a high susceptibility to diet-induced obesity, type 2 diabetes, and atherosclerosis; 2) a high incidence of microphthalmia and other associated eye abnormalities; 3) resistance to audiogenic seizures; 4) low bone density; 5) hereditary hydrocephalus (early reports indicate 1 - 4 %); 6) hairloss associated with overgrooming, 7) a preference for alcohol and morphine; 8) late-onset hearing loss; and 9) increased incidence of hydrocephalus and malocclusion."
Although this distal region is not modelled in any PDB structure as of March 2012, it has been specifically addressed in 4 of the 195 articles on mouse CRY1 or CRY2.
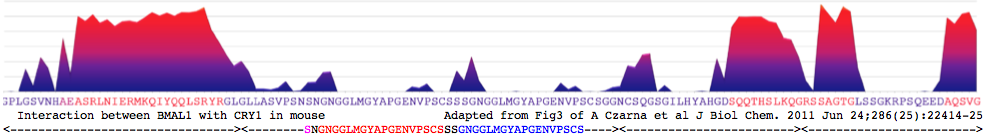
"purified mCRY1/2CCtail proteins form stable heterodimeric complexes with two C-terminal mBMAL1 fragments. The longer mBMAL1 fragment (BMAL490) includes Lys-537, which is rhythmically acetylated by mCLOCK in vivo. mCRY1 (but not mCRY2) has a lower affinity to BMAL490 than to the shorter mBMAL1 fragment (BMAL577) and a K537Q mutant version of BMAL490. Using peptide scan analysis we identify two mBMAL1 binding epitopes within the coiled coil RLNIERMKQIYQQLSRYR and tail regions of mCRY1/2 and document the importance of positively charged mCRY1 residues for mBMAL1 binding."
"mammalian CRY1 and CRY2 are integral components of the circadian oscillator. However, the function of their C terminus remains to be resolved. Here, we show that the C-terminal extension of mCRY1 harbors a nuclear localization signal and a putative coiled-coil domain that drive nuclear localization via two independent mechanisms and shift the equilibrium of shuttling mammalian CRY1 (mCRY1)/mammalian PER2 (mPER2) complexes towards the nucleus. Importantly, deletion of the complete C terminus prevents mCRY1 from repressing CLOCK/BMAL1-mediated transcription, whereas a plant photolyase gains this key clock function upon fusion to the last 100 amino acids of the mCRY1 core and its C terminus. Thus, the acquirement of different (species-specific) C termini during evolution not only functionally separated cryptochromes from photolyase but also caused diversity within the cryptochrome family."
"The mCRY1 and mCRY2 genes are located on chromosome 10C and 2E, respectively, and are expressed in all mouse organs examined. We raised antibodies specific against each gene product using its C-terminal sequence, which differs completely between the genes. Immunofluorescent staining of cultured mouse cells revealed that mCRY1 is localized in mitochondria whereas mCRY2 was found mainly in the nucleus. The subcellular distribution of CRY proteins was confirmed by immunoblot analysis of fractionated mouse liver cell extracts. Using green fluorescent protein fused peptides we showed that the C-terminal region of the mouse CRY2 protein contains a unique nuclear localization signal, which is absent in the CRY1 protein. The N-terminal region of CRY1 was shown to contain the mitochondrial transport signal. Recombinant as well as native CRY1 proteins from mouse and human cells showed a tight binding activity to DNA Sepharose, while CRY2 protein did not"
"genetic screening assay for mutant circadian clock proteins that is based on real-time circadian rhythm monitoring in cultured fibroblasts. By using this assay, we identified a domain in the extreme C terminus of BMAL1 that plays an essential role in the rhythmic control of E-box-mediated circadian transcription. Remarkably, the last 43 aa of BMAL1 are required for transcriptional activation, as well as for association with the circadian transcriptional repressor CRY1"
507 517 527 537 547 557 567 577 587 597
| | | | | | | | | |
CRY1_musMus GLLASVPSNSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN*
CRY1_ratNor GLLASVPSNPNGNGGLMGYAPGENVPSGGSGG------------------G NCSQGSGILHYAHGDSQQTNPLKQ GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN*
CRY1_criGri GLLASVPSNPNGNGGLMGYTTGENLPSCSGGG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN*
CRY1_spaJud GLLASVPSNPNGNGGLMGYTPGENIPNCSSSG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN*
CRY1_dipOrd GLLASVPSNPNGNGGLMGYAAGDNLPGSSSSG------------------- SCSQGSGILHYAHGDSQQMHLLKQ GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN*
CRY1_hetGla GLLASVPSNPNGNGGLMGYAPGESIPGSSGSG------------------- SCAHGSGILPCAHTDGQQAHLLKP GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD*
CRY1_cavPor GLLASVPSNPNGNGGLLGYAPGESTPGSGGG-------------------- SCVPGSSSAGVSHCAQGEAPQAPP GRDPAGPGLGGGKRPSQEEDAQSTGHKIQRQSPD*
CRY1_speTri GLLASVPSNPNGNGGLMAYAPGENIPGCSSSG------------------- SCTQGSSILHNAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN*
CRY1_oryCun GLLASVPSNPNGNGGLMGYSPGENIPGCSSSG------------------- SCSQGSGILHYAQGDTQQTQLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN*
CRY1_musMus GLLASVPSNSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN*
CRY1_ratNor .........P.................GG.G.------------------. ...................NP... ....M.............................
CRY1_criGri .........P.........TT...L....GG.------------------- S.................A.L... ....M..S...........ETR..D.........
CRY1_spaJud .........P.........T....I.N.....------------------- S.................A.L... .S..M.H...N.........T..I........T.
CRY1_dipOrd .........P..........A.D.L.GS....------------------- S.................M.L.... ...M...............S..I........T.
CRY1_hetGla .........P.............SI.GS.G..------------------- S.AH.....PC..T.G..A.L..P. .NCV.PV...............I...L....TD
CRY1_cavPor .........P......L......ST.GSGGG-------------------- S.VP..SSAGVS.CAQGEAPQAPP. .DP..P..GG............T.H.I....PD
CRY1_speTri .........P.......A......I.G.....------------------- S.T...S...N.........L.... ...M...............T..I........T.
CRY1_oryCun .........P.........S....I.G.....------------------- S...........Q..T...QL.... ...M...............T..I........T.
COILs coiled coil prediction for mouse CRY1 480-493 RLNIERMKQIYQQLSRYR
478 R e 0.644
479 L f 0.644
480 N g 0.806
481 I a 0.806
482 E b 0.806
483 R c 0.806
484 M d 0.806
485 K e 0.806
486 Q f 0.806
487 I g 0.806
488 Y a 0.806
489 Q b 0.806
490 Q c 0.806
491 L d 0.806
492 S e 0.806
493 R f 0.806
494 Y d 0.375
495 R e 0.375
>CRY1_musMus Mus musculus (mouse) NM_007771 all transcripts support longer exon 10 lost splice donor
1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRYRGL 1
2 GLLASVPSSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG 1
2 NCSQGSGILHYAHGDSQQTHSLKQ 1
2 GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* 0
>CRY1_ratNor Rattus norvegicus (rat) NM_198750
1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1
2 GLLASVPSNPNGNGGLMGYAPGENVPSGGSGGG 1
2 NCSQGSGILHYAHGDSQQTNPLKQ 1
2 GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* 0
>CRY1_criGri Cricetulus griseus (hamster) XM_003505292
1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1
2 GLLASVPSNPNGNGGLMGYTTGENLPSCSGGG 1
2 SCSQGSGILHYAHGDSQQAHLLKQ 1
2 GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN* 0
>CRY1_spaJud Spalax judaei (blind_mole_rat) AJ606298
1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1
2 GLLASVPSNPNGNGGLMGYTPGENIPNCSSSG 1
2 SCSQGSGILHYAHGDSQQAHLLKQ 1
2 GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN* 0
>CRY1_dipOrd Dipodomys ordii (kangaroo_rat) ABRO01202522 ABRO01202521
1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1
2 GLLASVPSNPNGNGGLMGYAAGDNLPGSSSSG 1
2 SCSQGSGILHYAHGDSQQMHLLKQ 1
2 GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN* 0
>CRY1_hetGla Heterocephalus glaber (blind_mole-rat)
1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1
2 GLLASVPSNPNGNGGLMGYAPGESIPGSSGSG 1
2 SCAHGSGILPCAHTDGQQAHLLKP 1
2 GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD* 0
>CRY1_cavPor Cavia porcellus (guinea_pig) last two exons diverged
1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVHHAEASRLNIERMKQIYQQLSRYRGL 1
2 GLLASVPSNPNGNGGLLGYAPGESTPGSGGG 1
2 SCVPGSSSAGVSHCAQGEAPQAPP 1
2 GRDPAGPGLGGGKRPSQEEDAQSTGHKIQRQSPD* 0
>CRY1_speTri Spermophilus tridecemlineatus (squirrel) Ictidomys
1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHEASLNIERMKQIYQQLSRYRGL 1
2 GLLASVPSNPNGNGGLMAyAPGENIPGCSSSG 1
2 SCTQGSSILHNAHGDSQQTHLLKQ 1
2 GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* 0
>CRY1_oryCun Oryctolagus cuniculus (rabbit)
1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGINYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1
2 GLLASVPSNPNGNGGLMGYSPGENIPGCSSSG 1
2 SCSQGSGILHYAQGDTQQTQLLKQ 1
2 GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* 0
>CRY1_ochPri Ochotona princeps (pika)
1 1
2 1
2 GCPQGSGILHCGHGDSQQTHLLKQ 1
2 GRGSPDTGISSGKRPSQEEDTQSIGPKVQRQSTN* 0
Lost distal exon in placental cryptochrome CRY1
Although cryptochromes are highly conserved in their two main domains, the C-terminal region in CRY1 has a reputation for variability. This is attributable in part to loss of an ancient exon encoding 32 amino acids in placental mammals. However this exon persists in contemporary marsupials, monotremes, birds, alligators, turtles, lizards, snakes and frogs, so its conservation implies a continuing functional role maintained by selective pressure for several hundred million years of tetrapod evolution.
In addition, some distal motifs in CRY1 are compositionally simple, predisposing not only to the replication slippage event described above for mouse but also to smaller indels in the repetitive regions, notably the 2 aa deletional synapomorphy in placentals in GLLASVPSNPNGN--GGFM (the conserved methionine is at position 514 in human) and possibly the loss of proline (P518) in post-tarsier divergence primates.
The exon loss may have preceded in stages, beginning with alternative splicing that skipped it (this conserves reading frame as the ancestral gene ends with three consecutive phase 12 exons). Later, the exon came not to be used at all and thereafter rapidly degenerated to the point it cannot be detected today by blastx of the relevant region in any placental mammal. The exon does not plausibly contribute to the core fold (photolyase and FAD domains) though it could form a better defined structure upon interacting with other proteins.
The functional consequences of exon loss are unknown; the timing matches that of overall collapse of the photolyase family in placentals. (Note the first half of placental evolution -- about 90 myr -- lacks any living representative, so events can pile up there by coincidence.) Possibly when CYT4, Cyt64, DASH and CPD were lost, the remaining two cryptochromes, especially CRY1, compensated for that loss (without however taking up catalytic roles in dna repair), with exon loss somehow contributing adaptively to that adjustment.
The loss of this exon raises certain questions about the use of marsupial model systems to understand CRY1 functionality in mouse (in turn a model system for human). For example, CRY1 of the marsupial Potorous tridactylus would still retain the exon but to date it has not been placed in a CRY1-- mouse. It would also be feasible to insert just the missing exon into an otherwise intact, ectopically expressed rat CRY1 gene, after first disentangling the effects of the mouse expansion in this same region (shown as ^^ below) as well as proline P518 removal. Note the lab mouse expansion somewhat restores length relative to marsupials, but in the wrong place.
CRY1_homSap MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCSSSG <-- lost exon in placentals --> SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN CRY1_ponAbe MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENVPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSGGKRASQEEDTQSIGPKVQRQSTN CRY1_nomLeu MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN CRY1_macMul MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS TENIPGCSSSG SCSQGSGILHYTHGDSQQTHLLKQ GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN CRY1_calJac MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCTSSG SCSQGSGILHCAHGDSQQTHLLKQ GRSSMSTGISGGKRPSQEEDTQSIGPKVQRQSTN CRY1_saiBol MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYS AENIPGCTSSG SCSQGSGILHCAHGDSQQTHLLKQ GRSSMSTGLGGGKRPSQEEDTQSIGPKVQRQSTN CRY1_tarSyr MKQIYQQLSRYRGL GLLASVPSNPNGN GGFMGYSPAENTPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSVGTGLSGGKRPSQEEDPQSIGPKVQRQSTN CRY1_otoGar MKQIYQQLSRYRGL GLLASVPSNPNGN GSFMEYSPPENIPGCSSSG NCSQGSGILHYAPGDGQQPHLLKQ GRSSMGTGLSGGKRPSQEEDMQSVGPKVQRQSTN CRY1_musMus MKQIYQQLSRYRGL GLLASVPSNSNGN^^GGLMGYAPGENVPSCSSSG NGGLGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN CRY1_ratNor MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAPGENVPSGGSGG GNCSQGGILHYAHGDSQQTNPLKQ GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN CRY1_criGri MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYTTGENLPSCSGGG SCSQGSGILHYAHGDSQQAHLLKQ GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN CRY1_spaJud MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYTPGENIPNCSSSG SCSQGSGILHYAHGDSQQAHLLKQ GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN CRY1_dipOrd MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAAGDNLPGSSSSG SCSQGSGILHYAHGDSQQMHLLKQ GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN CRY1_hetGla MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAPGESIPGSSGSG SCAHGSGILPCAHTDGQQAHLLKP GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD CRY1_speTri MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMAYAPGENIPGCSSSG SCTQGSSILHNAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN CRY1_oryCun MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAQGDTQQTQLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN CRY1_oviAri MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSA SCTQGSGILHYAHGDSQQTHLLKQ GRSSTAAGLGSGKRPSQEEDTQSVGPKVQRQSTN CRY1_bosTau MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSNA SCTQGSGILHYAHGDSQQTHLLKQ GRSSTGAGLGSGKRPSQEEDTQSIGPKVQRQSTN CRY1_susScr MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCPQGSGILHYAHGESQQNHLLKQ GRSSTGSGLSSAKRPSQEEDTQSIIGPKVQRQSTN CRY1_ailMel MKQIYQQLSRYRGL GLLASVPANPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGSGLSSGKRPSEEEDTQSIGPKVQRQSTN CRY1_turTru MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGYSSSG SCTPGSGILHYAYGDSQQTHLLKQ GRSSTCTGLSSGKRPSQEEDTQSIGPKVQRQSTN CRY1_equCab MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSLGPGLSSGKRPGPEEDTQGIGPKVQRQSTT CRY1_canFam MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGILHYAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSEEEDTQTISPKVQRQSTN CRY1_myoLuc MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SYAQGSGILHYALGDSQQTHLLKQ GRSSVGTGLSSGKRPSQEEDTQSIGRKVQRQSTN CRY1_pteVam MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG SCSQGSGSLHYAHGDCQQTHLLKQ GRSSMGTGLSSGKRPSQEEDMQSIGPKVQRQSTN CRY1_loxAfr MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENTPGCNSSG SCSQGSGILHYVHGDS....LLKQ GRSPTGTGVSSGKRPSQDEETQTLGPKVQRQSTN CRY1_triMan MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSNG SCPQGNGILHYAHRDSQQAHLLKQ GRSPTGTGVSSGKRPSQEEETQSIGPKVQRQSAN CRY1_proCap MKQIYQQLSRYRGL GLLASVPSNPNGN GGLIGYSPGESIPGCSNSG SCSQGSGILHYAHGDSQQAHLLKP GRSPMGTGISSGKRPSQEEETQTVGRKVQRQSTN CRY1_echTel MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENTTGCSSGG GCPPGNGILHYAHGDSQQAALLKQ GRSPLGTGLSSGKRPSQEEDTQSVGPKVQRQSSN CRY1_dasNov MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAPGENILGCSSSG SCAQGSSILHYAHGDNQQTHLLKQ GRSSMGTVLSSGKRPSQEEETQSIGPKVQRQSTN CRY1_choHof MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYSPGENIPGCSSSG sCSQGSGILHYAHGDSQQTHLLKQ GRSSMGIGLSSGKRPSQEEETQGIGPKVQRQSTN CRY1_monDom MKQIYQQLSRYRGL GLLASVPSNPNGN GSLMAYTPGENIPGCSSGG GAPVGASDGQIL..QACVLPEPPTGTSGVQQP GYSQGSGISHYSHEDSQQAYMLKQ GRSSL..GVGGGKRPRQEEETQSINPKVQRQSTN CRY1_macEug MKQIYQQLSRYRGL GLLASVPSNPNGN GSLMGYTTGENIPTCSSSGG GAPAGASDGQIL..QACVLPEPPTGTSGVQQP GGYSQGGISHYSHEDSQQAYVLKQ GRNSL....GGGKRHRQEEETQSIGSKMQRQSVN CRY1_sarHar MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYTSGENGPACNSGG GAPVGASDGQIL..QSCALPEPPAGASCIQQS GYSQGSGISHYSHEDSQQAYILKQ GRSSL....SGGKRPRQEEETQSVGPKVQRQSVN CRY1_triVul MKQIYQQLSRYRGL GLLASVPSNPNGN GGLMGYAPGENIPACSSSGG GAPAGVGDGQIL..QACALPEPPTGASGVQQP GYSQGSGISHYAHEDSQQAYMLKQ GRSSL...SGGGKRHRQEEEAQSIGPKMQRQSVN CRY1_ornAna MKQIYQQLSRYRGL GLLASVPSNPNANGSGGLMAYSPGENIPGCSSGGG GVQMGASESHLL..QTCVLGESHLGPSGIQQQ GYCQGSGVLYYANGE....SHLTQ GRSSLTPGLSGGKRPCQEEESQSIGPKVQRQSTD CRY1_tacAcu MKQIYQQLSRYRGL GLLASVPSNPNANGSGGLMAYSPGENIPGCSSGG GAQIGASESHLL..QTCVLGESHLGPSGIQQQ GRSSLTPGLSGGKRHCQEEESQSIGPKVQRQSTD CRY1_galGal MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMSFSPGESISGCSSAG GAQLGTGDGQTVGVQTCALADSHTGGSGVQQQ GYCQASSILRYAHGDNQQSHLMQP GRASLGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_melGal MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMSFSPGESISGCSSAG GAQLGTGDGQTVGVQSCALGDSHTGGNGVQQQ GYCQASSILRYAHGDNQQPHLMQP GRASLGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_eriRub MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGTGDGHTV.VQSCTLGDSHSGTSGIQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_sylBor MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGAGDGHSV.VQSCALGDSHTGTSGVQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_taeGut MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG GAQLGTGDGHSV.VQSCALGDSHTGTSGIQQQ GYCQASSILHYAHGDNQQSHLLQA GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN CRY1_allMis MKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGENVSGCGSTG GAQMGSSDGHTVSVQPCALGESHGGSNGIQQQ GYFQASSILHFPHGDDQQSHLLQQ GRTSLSSGISAGKRPNPEEETQSIGPKVQRQSTN CRY1_anoCar MKQMYQQLSRYRGL GLLASVPSNGNGNGNGGLMGYSTGENIPGCTNTN GSQMGMNEGHIGNVQACTMGESHTGTSGIQQQ GYSQGSGILLYSHGDNQKTHSAQK GRISLGTGVCTGKRPSPEVETQSVGPKVQRQSSN CRY1_podSic MKQIYQQLSRYRGL GLLASVPLNGNGNGNGGLMGYSTGENIPGCTNTN GSQMGTNEAHTGSVQTCTLGESHTGTSGIQQQ GYPQGSDILHYAHGEGQKTHLIQQ GRASLVAGVCTGKRPNPEEETQSIGPKVQRQSSK CRY1_pytMol MKQIYQQLSRYRGL GAQMGTSEGHTGNVQACTLGETHTGTSGIQQQ GYSQGNSGILHYAHGDSQKTLLMQ GRTSLSVGVCTGKRPNPEEGIQSIGPKVQRQSSN CRY1_chrPic MKQIYQQLSRYRGL GLLATVPSNPNG..NGGLMGYSPGENISGCSSAS GAQMGSNDGHTVGVQTCSLEDSHAGSSGIQQH GYSQGNSIVHYAQGDHQQSHLLQQG GRTVST GISTGKRPNPEKETQSIGPKVQRQSTN CRY1_xenTro MKQIYQQLSRYRGL GLLASVPSNPNGNGNGGLMSYSPGESMSGCSNNG GGQMGVNEGSSASNPNANKGEVHPGTSGLQ.. GYWQGSSILHYSHSDSQQSY LMQ ARNPLHSVVSSGKRPNPEEETQSIGPKVQRQSSH CRY1_xenLae MKQIYQQLSRYRGL GLLASVPSNPNG..NGGLMSYSPGESMPGCSNNG GGQMGAIEGSSASNPNPNQGEVLPGTSGLQ.. GYWQGSSILHYSHSDNQQSY LMQ ARNPLHSVVSSGKRPNPEEETQSVGPKVQRQSTH CRY1_latCha MKQIYQQLSRYRGM GLLASVPSNPNGNGGLGCSLAENIPVCNSAA GAQMGGDDGHKVSVLAYTQGDSRAGEIEMQQQ CRY1_danRer MKQIYQQLSCYRGL GLLAMVPSNPNGNGENSTSLMGFQTGDMTKEVTTPS GYQMPPTSQGEWHGRTMVYSQGDQQTSSIMTSQ GFGNNGSTMCYRQDAQQIT GRGLHSSIIQTSGKRHSEESGPTTVSKVQRQCSS
When the terminal four exons of CRY1 are compared to those of its nearest homolog class CRY2, no similarity can be detected beyond the first 8 residues of the tenth exon of CRY1 (2 GLLASVPS) vs the tenth and penultimate exon of CRY2 (2 CLLASVPS). This raises the question of what the last common ancestor had for terminal exons and -- given no counterpart in CRY4, CRY64, DASH, or CPD -- where they originated. Note that last two exons of CRY2 are strongly conserved in their own right, proving a separate conserved functionality from that of CRY1. Since the tenth exons begin homologously and end after a similar length with a phase 1 splice donor, these exons could possibly be homologous their entire length, just diverged distally. The eleventh exon of CRY2 could then correspond (allowing for total sequence divergence) to any of exons 11-13 in CRY1.
CRY2_homSap CLLASVPSCVEDLSHPVAEPSSSQAGSMSSA GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDA CRY2_panTro ............................... .............................................. CRY2_gorGor ...........................V... .............................................. CRY2_ponAbe ...........................V... .............................................. CRY2_rheMac ...........................VN.. ...............................K.............. CRY2_papHam ...........................VN.. ...............................K.............. CRY2_calJac ............................... .............................................V CRY2_micMur ..............................T .................................T............ CRY2_musMus ....................G......I.NT ...A.S.....................T.....T.M..Q.PA...S CRY2_ratNor ....................G......I.NT .....S...........................T.M.AQ.P....S CRY2_criGri ...........................I.NT .S...S...........................T.M.AQ.PQT... CRY2_spaJud ........................P..ITNT .....ST..........................T...A..PA.... CRY2_cavPor .....................L.....ST.T ......G.................................P..... CRY2_hetGla ....................TL.....S..T ...S..D..............................A..PT.... CRY2_speTri ....................G......I..T .....S..Q..................................... CRY2_oryCun ...........................V.G. A..................................V........AV CRY2_turTru .........M....N...........G.... ................G.................G..PS..L...V CRY2_bosTau ..............N.......I....S..V ......G.................G..........SLPS....RGV CRY2_susScr ..............N............V.A. .....................................PT...GR.V CRY2_canFam ..............N.........T...... ..........................................CR.V CRY2_ailMel ..............N.........T...... .....................................A..P..R.V CRY2_myoLuc .........M....N......L..T...... ..K..................................AT....R.V CRY2_pteVam .............NN.........T...NN. .....................................A.....R.V CRY2_loxAfr ..............S............SN...........T........................K..G.......V CRY2_proCap ..............N........P..H.....L................................K..G.....T.. CRY2_choHof ..............N....................V............................T...........V CRY2_macEug .........M....S.M..T.MG....V..T..K...CS..........T..ASR..H.....M.A..V...A.--- CRY2_monDom .........L....S.MV.A.LG...AV.GP.LK...CS..........T..A....H.......R..GS..AG..V CRY2_ornAna ..............SAA..SGLG....NI.TA...-.P.............GL.....C..PK..GR.G..P.GE.. CRY2_galGal ..............G..TDSAPG.-..ST.TAV.LPQ.DQ......H.G...LCT...Y...K.TG..A..I.G.SS CRY2_taeGut ............I.G..PDSA.G.-.CST.TAV.LSQAEQ......H.G....CS...Y...K.TG...S.ISG.SL CRY2_allMis G........A....G..TD.A.V.-.CST.TALK.SQ..Q......H.GI..MCT.D.Y...K.TG.HG..I...SL CRY2_anoCar .........M....N...DT...H-.NCIGTAS.QTHC.QT.....HDVVQ.YK-...Y...K.VASQFA.N.RQEL CRY2_xenTro .I.......M...GG.M.DS.QNISEAGKM.P.SHTSGESVLAAQYTAGI--------------------------- CRY2_ranCat .I......S.....G.M.D.A...Q..SD---.A.RLCAVD.....H.DLD----G..C.K..LQCVQEM.RAA..F
Vertebrate CRY1 reference sequences
Here it is quite important to straighten out misnamed homologs, especially in zebrafish which has been studied extensively. Because chondrichthyes, lobe-finned fish, and basal ray-finned fish (gars) have two separate genes classifying as CRY1 (with CRY2 ruled out), a gene duplication must occurred in early vertebrates and persisted almost to land animals.
Both syntentic sites can be explored in Xenopus and amniotes but only one location hosts a cryptochrome today. These two syntenic regions are not more broadly paralogous, so not supportive of a large scale duplication. The figure (taken from the Genomicus synteny tool) suggests that CRY1 experienced an inversion in amniotes subsequent to mammalian divergence. This may have carried all upstream regulatory regions along with it or left some portion orphaned in a new downstream position with unknown functional consequences. Since the event occurred some 300 myr ago, the boundaries of the inversion cannot be precisely determined
Zebrafish, which has four distinct CRY1 paralogs in addition to a CRY2, may have had a doubling of this ancestral pair through whole genome duplication but the sequences don't quite cluster in this way. All four CRY1 genes are actively transcribed.
>CRY1_homSap Homo sapiens (human) 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGFMGYSAENIPGCSSSG 1 2 SCSQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_panTro Pan troglodytes (chimpanzee) XM_509339 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGFMGYSAENIPGCSSSG 1 2 SCSQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSVGTGLSGGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_ponAbe Pongo abelii (orangutan) XM_002823690 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDASLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGFMGYSAENVPGCSSSG 1 2 SCSQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSMGTGLSGGKRASQEEDTQSIGPKVQRQSTN* 0 >CRY1_nomLeu Nomascus leucogenys (gibbon) XM_003269977 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGFMGYSAENIPGCSSSG 1 2 SCSQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_macMul Macaca mulatta (rhesus) NM_001194159 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGINYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGFMGYSTENIPGCSSSGS 1 2 CSQGSGILHYTHGDSQQTHLLKQ 1 2 GRSSMGTGLSGGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_calJac Callithrix jacchus (marmoset) XM_002752946 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGFMGYSAENIPGCTSSG 1 2 SCSQGSGILHCAHGDSQQTHLLKQ 1 2 GRSSMSTGISGGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_saiBol Saimiri boliviensis (squirrel_monkey) nearly identical to marmoset 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGFMGYSAENIPGCTSSG 1 2 SCSQGSGILHCAHGDSQQTHLLKQ 1 2 GRSSMSTGLGGGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_tarSyr Tarsius syrichta (tarsier) ABRT010205577 unsure if exon 2 is CRY1 or CRY2 0 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWNIAKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 0 0 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGFMGYSPAENTPGCSSSG 1 2 SCSQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSVGTGLSGGKRPSQEEDPQSIGPKVQRQSTN* 0 >CRY1_micMur Microcebus murinus (mouse_lemur) 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 IIELNGGQPPLTYKRFQTLISK SEVIEKCTTPLSDD 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKSSPPLSLYGRYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGFMEYSPAENIPGCSSSG 1 2 NCSQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSMGTGFSGGKRPSQEEDTQSIGSKVQRQSTN* 0 >CRY1_otoGar Otolemur garnettii (bushbaby) AAQR03016495 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITRLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 0 0 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEIIEKCTTPLFDDHDEKYGVPSLEEL 1 2 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPASFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGINYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGSFMEYSPPENIPGCSSSG 1 2 NCSQGSGILHYAPGDGQQPHLLKQ 1 2 GRSSMGTGLSGGKRPSQEEDMQSVGPKVQRQSTN* 0 >CRY1_tupBel Tupaia belangeri (treeshrew) 0 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 0 0 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKFGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRVNSNSLLASPTGL 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLSSSGSCSQGSGILHYAHGDSQQTHLLKQGRSSMGP 1 2 1 2 GCSQGSGILHYAHGDSQQTHLLKQ 1 2 GLSSGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_musMus Mus musculus (mouse) NM_007771 all transcripts support longer exon 10 lost splice donor 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLVSKMEPLEMPADTITSDVIGKCMTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLER 0 0 KAWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG 1 2 NCSQGSGILHYAHGDSQQTHSLKQ 1 2 GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* 0 >CRY1_ratNor Rattus norvegicus (rat) NM_198750 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLVSKMEPLEMPADTITSDVIGKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYAPGENVPSGGSGGG 1 2 NCSQGSGILHYAHGDSQQTNPLKQ 1 2 GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* 0 >CRY1_criGri Cricetulus griseus (hamster) XM_003505292 0 MGVNAVHWFRKglrlhDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDSNLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLVSKMEPLEMPAETITSDVIGKCMTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLER 0 0 KAWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYTTGENLPSCSGGG 1 2 SCSQGSGILHYAHGDSQQAHLLKQ 1 2 GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN* 0 >CRY1_spaJud Spalax judaei (blind_mole_rat) AJ606298 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRW 2 1 RFLLQCLEDLDANLRKLNSRLLVIRGQPADVFPRLFKEWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERR 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTGSDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYTPGENIPNCSSSG 1 2 SCSQGSGILHYAHGDSQQAHLLKQ 1 2 GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_dipOrd Dipodomys ordii (kangaroo_rat) ABRO01202522 ABRO01202521 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRW 2 1 RFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCSVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYAAGDNLPGSSSSG 1 2 SCSQGSGILHYAHGDSQQMHLLKQ 1 2 GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN* 0 >CRY1_hetGla Heterocephalus glaber (mole-rat) stop codon in place of conserved W8, last two exons very diverged 0 MAMNAVH*FRKGLRLHDNPALKECTRGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCVTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYAPGESIPGSSGSG 1 2 SCAHGSGILPCAHTDGQQAHLLKP 1 2 GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD* 0 >CRY1_cavPor Cavia porcellus (guinea pig) last two exons diverged 69 bp separation 0 MGVNAVHWFRKGLRLHDNPALKECIRGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVVEKCVTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVHHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLLGYAPGESTPGSGGG 1 2 SCVPGSSSAGVSHCAQGEAPQAPP 1 2 GRDPAGPGLGGGKRPSQEEDAQSTGHKIQRQSPD* 0 >CRY1_speTri Spermophilus tridecemlineatus (squirrel) Ictidomys 0 2 1 0 RDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHEASLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMAyAPGENIPGCSSSG 1 2 SCTQGSSILHNAHGDSQQTHLLKQ 1 2 GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_oryCun Oryctolagus cuniculus (rabbit) 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRW 2 1 RFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITRLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSDVIEKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGQTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGINYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYSPGENIPGCSSSG 1 2 SCSQGSGILHYAQGDTQQTQLLKQ 1 2 GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_oviAri Ovis aries (sheep) NM_001129735 19341811 19150926 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIIRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVMEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYSPGENIPGCSSSA 1 2 SCTQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSTAAGLGSGKRPSQEEDTQSVGPKVQRQSTN* 0 >CRY1_bosTau Bos taurus (cow) NM_001105415 XM_616063 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYSPGENIPGCSSNA 1 2 SCTQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSTGAGLGSGKRPSQEEDTQSIGPKVQRQSTN* 0 + >CRY1_susScr Sus scrofa (pig) XM_003126079 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYSPGENIPGCSSSG 1 2 SCPQGSGILHYAHGESQQNHLLKQ 1 2 GRSSTGSGLSSAKRPSQEEDTQSIIGPKVQRQSTN* 0 >CRY1_ailMel Ailuropoda melanoleuca (panda) XM_002927658 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPANPNGNGGLMGYSPGENIPGCSSSG 1 2 SCSQGSGILHYAHGDSQQTHLLKQ 1 2 GRSSMGSGLSSGKRPSEEEDTQSIGPKVQRQSTN* 0 >CRY1_loxAfr Loxodonta africana (elephant) XM_003405313 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWDITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIGKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYSPGENTPGCNSSG 1 2 SCSQGSGILHYVHGDSLLKQ 1 2 GRSPTGTGVSSGKRPSQDEETQTLGPKVQRQSTN* 0 >CRY1_triMan Trichechus manatus (manatee) AHIN01036366 AHIN01036362 very similar to elephant 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWDITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVMEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCSIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYSPGENIPGCSSNG 1 2 SCPQGNGILHYAHRDSQQAHLLKQ 1 2 GRSPTGTGVSSGKRPSQEEETQSIGPKVQRQSAN* 0 >CRY1_monDom Monodelphis domestica (opossum) XM_003341966 0 MGVNAGHWFRKRLRLPHNPALKGCIQGADTVCCVYIRDPWFGGSSNFGANEWK 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNIAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 0 0 IIELNGGQPPLTYKRFQTLISKMEPLATPVETITPEVMHKCVTPLSDEHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGSLMAYTPGENIPGCSSGG 1 2 GAPVGASDGQILQACVLPEPPTGTSGVQQP 1 2 GYSQGSGISHYSHEDSQQAYMLKQ 1 2 GRSSLGVGGGKRPRQEEETQSINPKVQRQSTN* 0 >CRY1_macEug Macropus eugenii (wallaby) assembly frameshift 0 MGVNAVHWFRKGLRLHDNPALKECIEGADTVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPLGRNAMQLLKNWASEAGVEVIVRISHTLYDLDK 2 1 1 2 gFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGVQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGSLMGYTTGENIPTCSSSGG 1 2 GAPAGASDGQILQACVLPEPPTGTSGVQQP 1 2 GYSQGSGISHYSHEDSQQAYVLKQ 1 2 GRNSLGGGKRHRQEEETQSIGSKMQRQSVN* 0 >CRY1_sarHar Sarcophilus harrisii (tasmanian_devil) nearly identical to oppossum 0 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNIAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 0 0 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYTSGENGPACNSGG 1 2 GAPVGASDGQILQSCALPEPPAGASCIQQS 1 2 GYSQGSGISHYSHEDSQQAYILKQ 1 2 GRSSLSGGKRPRQEEETQSVGPKVQRQSVN* 0 >CRY1_ornAna Ornithorhynchus anatinus (platypus) XM_001508563 = rubbish, genomic frameshift, continuing exon 12 0 MGVNAVHWFRKGLRLHDNPALRDCVRGADTVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNIAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYELDK 2 1 IIELNGGQPPLTYKRFQTLISRMDPLAMPVETITAEVMGKCMTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATSNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNANGSGGLMAYSPGENIPGCSSGGG 1 2 GVQMGASESH LLQTCVLGESHLGPSGIQQQ 1 2 GYCQGSGVLYYANGESHLTQ 1 2 GRSSLTPGLSGGKRPCQEEESQSIGPKVQRQSTD* 0 >CRY1_tacAcu Tachyglossus aculeatus (echidna) SRR000649.130490 short read transcripts corrected for frameshifts, skipped penultimate exon 0 2 1 0 0 2 1 ETITAEVMGKCVTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGEAEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLTPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATSNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAImtqlrqeGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNANGSGGLMAYSPGENIPGCSSGG 1 2 GAQIGASESHLLQTCVLGESHLGPSGIQQQ 1 2 1 2 GRSSLTPGLSGGKRHCQEEESQSIGPKVQRQSTD* 0 >CRY1_galGal Gallus gallus (chicken) 11684328 17324421 15459395 0 MGVNAVHWFRKGLRLHDNPALRECIRGADTVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWSIAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISRMEPLEMPVETITPEVMQKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERH 0 0 LERKAWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPESVQKAAKCVIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLATVPSNPNGNGNGGLMSFSPGESISGCSSAG 1 2 GAQLGTGDGQTVGVQTCALADSHTGGSGVQQQ 1 2 GYCQASSILRYAHGDNQQSHLMQP 1 2 GRASLGTGISAGKRPNPEEETQSVGPKVQRQSTN* 0 >CRY1_melGal Meleagris gallopavo (turkey) XM_003202363 0 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWSIAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISRMEPLEMPVETITPEVMQKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERH 0 0 LERKAWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPESVQKAAKCVIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLATVPSNPNGNGNGGLMSFSPGESISGCSSAG 1 2 GAQLGTGDGQTVGVQSCALGDSHTGGNGVQQQ 1 2 GYCQASSILRYAHGDNQQPHLMQP 1 2 GRASLGTGISAGKRPNPEEETQSVGPKVQRQSTN* 0 >CRY1_eriRub Erithacus rubecula (robin) AY585716 0 MGVNAVHWFRKGLRLHDNPALRECIRGADTVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNIAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISRMEPLEMPVETITPEVMKKCTTPVFDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 ASVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPESIQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG 1 2 GAQLGTGDGHTVVQSCTLGDSHSGTSGIQQQ 1 2 GYCQASSILHYAHGDNQQSHLLQA 1 2 GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN* 0 >CRY1_sylBor Sylvia borin (warbler) AJ632120 15381765 0 MGVNAVHWFRKGLRLHDNPALRECIQGADTVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNIAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISRMEPLEMPVETITPEVMKKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPESIQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG 1 2 GAQLGAGDGHSVVQSCALGDSHTGTSGVQQQ 1 3 GYCQASSILHYAHGDNQQSHLLQA 1 2 GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN* 0 >CRY1_taeGut Taeniopygia guttata (finch) XM_002196518 0 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNIAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 0 0 IIELNGGQPPLTYKRFQTLISRMEPLEMPVETITPEVMKKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPESIQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLATVPSNPNGNGNGGLMGYSPGESISGCGSTG 1 2 GAQLGTGDGHSVVQSCALGDSHTGTSGIQQQ 1 2 GYCQASSILHYAHGDNQQSHLLQA 1 2 GRTALGTGISAGKRPNPEEETQSVGPKVQRQSTN* 0 >CRY1_melUnd Melopsittacus undulatus (budgerigar) AGAI01062111 1 RYLPVLRGFPAKYIYDPWNAPESIQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL GLLATVPSNPNGNGNGGLMGYSPGESISGCSSAGG GAQLGTGDGHTVVQPCALGDSHTAGASGIQQQ GYCQASSILHYAHGDNQQSHLLQAG VGTGISAGKRPNPEEETQSVGPKVQRQSTN* 0 >CRY1_allMis Alligator mississippiensis (alligator) genome/blat 0 MGVNAVHWFRKGLRLHDNPALRECIQGADTVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISRMEPLEMPVETITAEVMKKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGVQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLATVPSNPNGNGNGGLMGYSPGENVSGCGSTG 1 2 GAQMGSSDGHTVSVQPCALGESHGGSNGIQQQ 1 3 GYFQASSILHFPHGDDQQSHLLQQ 1 2 GRTSLSSGISAGKRPNPEEETQSIGPKVQRQSTN* >CRY1_anoCar Anolis carolinensis (lizard) XM_003220923 0 MGVNAVHWFRKGLRLHDNPALREVIQGADTVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYELDK 2 1 IIELNGGQPPLTYKRFQTLISRMEPLEIPVETITAEVMSKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPDGVQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQMYQQLSRYRGL 1 2 GLLASVPSNGNGNGNGGLMGYSTGENIPGCTNTN 1 2 GSQMGMNEGHIGNVQACTMGESHTGTSGIQQQ 1 2 GYSQGSGILLYSHGDNQKTHSAQKGRISLGT 1 2 GVCTGKRPSPEVETQSVGPKVQRQSSN* 0 >CRY1_podSic Podarcis siculus (wall_lizard) DQ376040 16809482 0 MGVNAVHWVRQGLRLPDNRALREVIQGADTARCVYILDPSFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISRMEPLEMPVETITAEVMSKCTTPVSDDHDEKCGVPSLEEL 1 2 GFDTGGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARRAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGVQKVAKCVIGVNYPKPMVNHTEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPLMEWNVIVLMGYHQGTLLVYHAS 1 2 GSQMGTNEAHTGSVQTCTLGESHTGTSGIQQQ 1 2 GYPQGSDILHYAHGEGQKTHLIQQGRASLVA 1 2 GVCTGKRPNPEEETQSIGPKVQRQSSK* 0 >CRY1_pytMol Python molurus (python) AEQU010547455 0 MGVNSVHWFRKGLRLHDNPALREVIQGADTVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFk 0 0 EWNISKLSIEYDSEPFGKERDAAIKKLATEAGLEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISRMEPLEMPVETITAEVMSKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLER 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 2 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPILRGFPAKYIYDPWNAPEGVQKAAKCIIGINYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 2 1 2 GAQMGTSEGHTGNVQACTLGETHTGTSGIQQQ 1 2 GYSQGNSGILHYAHGDSQKTLLMQ 1 2 GRTSLSVGVCTGKRPNPEEGIQSIGPKVQRQSSN* 0 >CRY1_chrPic Chrysemys picta (turtle) AHGY01469963 AHGY01469969 0 MGVNAVHWFRKGLRLHDNPALRECIQGADSVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNIAKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISRMEPLEMPVETITAEVMEKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNRSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVSFGRRTDPNGDYIR 2 1 RYLPILRGFPAKYIYDPWNAPESIQKAAKCVIGVHYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLATVPSNP NGNGGLMGYSPGENISGCSSAS 1 2 GAQMGSNDGHTVGVQTCSLEDSHAGSSGIQQH 1 2 GYSQGNSIVHYAQGDHQQSHLLQQG 1 2 GRTVSTGISTGKRPNPEKETQSIGPKVQRQSTN* 0 >CRY1_xenTro Xenopus tropicalis (frog) NM_001087660 11533577 final four exons confirmed by many ESTs 0 MGVNAVHWFRKGLRLHDNPALRECIQGADTVRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWKITKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMDPLEIPVETITAEVMEKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTEGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASTTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMDGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGKRTDPNGDYIR 2 1 RYLPILKGFPPKYIYDPWNAPETVQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGNGGLMSYSPGESMSGCSNNG 1 2 GGQMGVNEGSSASNPNANKGEVHPGTSGLQ 1 2 GYWQGSSILHYSHSDSQQSYLMQ 1 2 ARNPLHSVVSSGKRPNPEEETQSIGPKVQRQSSH* 0 >CRY1A_latCha Latimeria chalumnae (coelocanth) 0 MGVNAIHWFRKGLRLHDNPALREAIQGGDCIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNISRLSFEYDSEPFGKERDAAIKKLASEAGVEVIVRITHTLYDLDR 2 1 IIELNGGQPPLTYKRFQTLISRMEPLDLPVETINSEVMGNCSTPVSDDHDEKYGVPSLEEL 1 2 GFDTEGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGVQKAAKCIIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGM 1 2 GLLASVPSNPNGNGGLGCSLAENIPVCNSAA 1 2 GAQMGGDDGHKVSVLAYTQGDSRAGEIEMQQQ* 1 2 1 2 * 0 >CRY1B_latCha Latimeria chalumnae (coelocanth) 0 MVVNSVHWFRKGLRLHDNPALQEALKGADTVRCVYMLDPRFAGSSNLGINRWR 2 1 FLLQCLEDLDASLRKLNSRLFVIRGQPANVFPRLFK 0 0 QEWKITRLTIEYDSEPFGKERDAAIKKLASEAGVEVIVKISHTLYDLDK 2 1 IIELNGGHPPLTYKRFQTLVSRMEHPEMPVEAITASFMGKCITPVSEDHHEKYGVPSLEEL 1 2 GFDTEGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFEKPRMNANSLLASPTGLSPYLRFGCLSCHLFYFKLTDLYK 0 0 VKKNGSPPLSLYGQLLWREFFYTAATNNPQFDKMEGNPICVRIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEDLLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWSAPESVQTAAKCIIGVHYPKPVVNHAEASRLNIERMKQIYQQLSRYRGM 1 2 GLLASVPSNHNGNGNGGMMVYSPVEPTLGTNAAH 1 2 1 2 1 2 * 0 >CRY1A_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01025403 0 MVVNTVHWFRKGLRLHDSPALRESLKGAASLRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDASLRKLNSRLFVIRGQPADVFPRLFK 0 EWNINRLSFEYDSEPFGKERDAAIKKLASEADVEVIVRVSHTLYDLDK 2 0 IIELNGGQPPLTYKRFQTLISRMDPLETPAEPITAEVMGKCTTPVSDDHDEKFGVPSLEEL 1 2 GFDTEGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRHEGWIHHLARHAVACFLTRGDLWVSWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPDSVQKAAKCVIGVHYPKPMVHHAEASRLNIERMKQIYQQLSCYRGL 1 2 GLLASVPSNPNGSGGAGMTGFSSGEGLHSSNAA 1 2 GYQIPSGRSSLPSYSSGDSQAGSNLLPQ 1 2 GYTRGSTVVHYTQGDSQQTS 1 2 GRGHLPSVLTTVKRPSPEESTQGIVPKVQRQSSN* 0 >CRY1B_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01016727 AHAT01016728 0 MGPNSIHWFRKGLRLHDNPALKEAIRGASTVRCVYFLDPWFAGSSNLGVNRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPANVFPRLFK 0 EWKISRLTFEYDSEPFGKERDAAIKKLAKEAGVEVIVKISHTLYDLDR 2 0 IIELNGGQPPLTYKRFQTLISRMDPPELPVDPLSEVFMGKCVTPVSDDHGDKYGVPSLEEL 1 2 GFDTEGLPSAVWPGGETEALTRIERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYR 0 0 VKKNSSPPLSLYGQLLWREFFYTAATTNPRFDKMEGNPICVRIPWDRNSEALAKWAEAKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPDSVQNAAKCVIGVHYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNHNGNGNGGMMAYSPGEQQPAHGTTY 1 2 GPGSSVASGNGSSSILLNFDGEEQPGPSGVHH 1 2 1 2 GRWAHGGAGGGSSAPGKRERETERDAAGDEEARSSA* 0 >CRY1A_danRer Danio rerio (zebrafish) NM_001077297 0 MVVNTVHWFRKGLRLHDNPSLRDSILGAHSVRCVYILDPWFAGSSNVGISRWR 2 1 FLLQCLEDLDASLRKLNSRLFVIRGQPTDVFPRLFK 0 EWNINRLSYEYDSEPFGKERDAAIKKLANEAGVEVIVRISHTLYDLDK 2 0 IIELNGGQSPLTYKRFQTLISRMEAVETPAETITAEVMGPCTTPLSDDHDEKFGVPSLEEL 1 2 GFDTEGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYRK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVSFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPESVQKAAKCIIGVHYPMPMVHHAEASRLNIERMKQIYQQLSCYRGL 1 2 GLLAMVPSNPNGNGENSTSLMGFQTGDMTKEVTTPS 1 2 GYQMPPTSQGEWHGRTMVYSQGDQQTSSIMTSQ 1 2 GFGNNGSTMCYRQDAQQIT 1 2 GRGLHSSIIQTSGKRHSEESGPTTVSKVQRQCSS* 0 >CRY1A2_danRer Danio rerio (zebrafish) BC044558 AW184635 olfactory 0 MVVNTVHWFRKGLRLHDNPSLRDSIKGADNLRCVYILDPWFAGSSNVGISRWR 2 1 FLLQCLEDLDASLRKLNSRLFVIRGQPTDVFPRLFK 0 0 EWKISRLSYEYDSEPFGKDRDAAIRKLATEAGVEVFVRISHTLYDLDK 2 1 IIEFNGGQSPLTYKRFQTLISRMDPVEMPAETITAEIMGKCSTPVSDDHDDKFGVPSLEEL 1 2 GFETEGLSTAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYRK 0 0 VKKNSTPSLSLYGQLLWREFFYTAATNNPHFDKMEFNPICVQIPWDRNPEALAKWAEGQTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPILRGFPAKFIYDPWNAPESVQKVAKCIIGVHYPKPMVNHAEASRINIERMKQIYQQLSCYRGL 1 2 GLLATIPSNTANNEDSNGAGPSSGDTQESTHIT 1 2 VHTLPASSQSEFPAFAVSCGQQLVNSVQPQP 1 2 GYSGAGMVGQAHKQTQR 1 2 VRGMTAFKRHGEDRTTDFPSKIQRNDCN* 0 >CRY1B_danRer Danio rerio (zebrafish) BC095305 EB921055 aka CRY2A 0 MAPNSIHWFRKGLRLHDNPALQEAVRGADTVRCVYFLDPWFAGSSNLGVNRWR 2 1 FLLQCLDDLDSNLRKLNSRLFVVRGQPANVFPRLFK 0 0 EWKISRLTFEYDSEPFGKERDAAIKKLAMEAGVEVIVKTSHTLYNLDK 2 1 IIELNGGQPPLTYKRFQTLISRMDPPEMPVETLSNSIMGCCVTPVSEDHGDKYGVPSLEEL 1 2 GFDIEGLPSAVWPGGETEALTRIERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYRK 0 0 VKKTSTPPLSLYGQLLWREFFYTAATTNPRFDKMEGNPICVRIPWDKNPEALAKWAEAKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDFIR 2 1 RYLPILRGFPAKYIYDPWNAPDSVQAAAKCIIGVHYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSTHNGNGNGMAYSPGEQQSGTNTP 1 2 APAVSSGSVASGNRSGSILLNFDSEEHQGPSGIQQQHQQQQQQQQQQQL 1 2 GYHHMPDSGHNSRFYKSNVSHDMA 1 2 GHLLHKGGSVTGKRERESERDLDGEDESLSTSHKLQRQIAEVTSVYASSGNQSSMVNITLLNL* 0 >CRY1C_danRer Danio rerio (zebrafish) BC164795 EE210836 aka CRY2B 0 MSVNSVHWFRKGLRLHDNPALLEALNGADTLRCVYFLDPWFAGASNLGVNRWR 2 1 FLLQSLEDLDASLRKLNSCLFVIRGQPADIFPRLFK 0 0 EWKVSRLTFEFDSEPFGKERDAAIKKLACEAGVEVIVKISHTLYDLDR 2 1 IIELNGGQSPLTYKRFQTLVSSMEPPDPPLSSPDRGMMGKCVTPISENHRDKYGVPLLEEL 1 2 GFDTEGLAPvVWPGGESEALKRMERHLdrt 0 0 AWQENFERPKMNASPLMASPLGLSPYLRFGCLSCRLFYCKLTQLYKK 0 0 VKKNMNPSISLYDKILWREFFYTAATNNPRFDRMEGNPICIRIPWDRNAEALAKWAEAKTGFPWIDAIMMQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWLCHSCSSFFQQFFHCYCPVGFGRRIDPNGDFIR 2 1 RYLPVLRDFPAKYIYDPWNAPHDVQLAAKCVIGVDYPKPMVNHAEASRLNIERMRQIYQQLSRYRGL 1 2 GLLATVPSNHIEDAAANVNRGAMAPPTRRKTRNDASSY 1 2 RVRREQPGPSGAKHRHRHSLS 1 2 HRPSPYATQSELTDHNHEFAVPQQP 1 2 GRVSWPNVGGTGKKEGEAEHNCCRGDNGPSSSALKAQQQDNEVCRK* 0 >CRY1_petMar Petromyzon marinus (lamprey) Contig24766 0 MVLNSIHWFRKGLRLHDNPALREALQGADTVRCVYILDPWFAGASNVGINRWR 2 1 FLLQSLEDLDASLHKLNSRLFVIRGQPADVFPRLFK 0 0 EWNISRLTFEYDSEPFGKERDAAIKKLASEAGVEVLVRISHTLYNLDR 2 1 IIELNEGQAPLTYKRFQALVSRMEPPERPVGTITSEVMGPCRTPLWEDHDERYGVPSLEEL 1 2 GFDTDGLTTAVWQGGESEALTRLDRHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFHYKLTELYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPKFDRMEGNPICVQIPWDRNPEALAKWAEGRTGYPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYIPLLRAFPAKYIYDPWNAPESVQKTARCIIGVDYPKPMANHAEASRLNIERMKQIYQHLSRYRGL 1 2 1 2 1 2 1 2 * 0 >CRY1_braFlo Branchiostoma floridae (amphioxus) XM_002609455 end uncertain 0 MKSTIHWFRKGLRLHDNPSLRDALQMLQPGDVWRCVYVLDPWFAGSSNVGVNRWR 2 1 FLLQCLEDLDASLRKLNSRLYVIRGQPTDVFPRLFK 0 0 EWKVSCLSFEEDSEPFGRERDMAVMKLAKEAGVKVILRTSHTLYKLQD 2 1 IIDVNGGQPPLTYKRFQAILSKMDPPDQPVDSITASTVDNLTTPIRDDHDEKFGVPSLEEL 1 2 GFDTDNLNPVVWPGGETEALTRLERHLERK 0 0 AWVASFERPKMTASSLLASPTGLSPYLRFGCLSPRLFYKKLTELYKK 0 0 VKRSNHPPLSLYGQLLWREFFYTAATNNPNFDKMVNNTICVQIPWDKNPEALAKWAE 00 GQTGFPWIDAIMAQLQQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSSFFQQFFHVYCPVNFGRRTDPNGDFIR 2 1 KYLPKLRGFPAKYIYDPWNSPESVQKAAKCII 12 GKDYPLPMVNHAETSRINVERMKQVYQQLSRYRGA 1 2 GLLATIPYPDGGNGNPTGHKKHAIVQGMERQHAERREHGQPAHVSRQHI * 0 >CRY1_strPur Strongylocentrotus purpuratus (urchin) XM_001194752 same split exons as braFlo, end of gene uncertain, partial duplicate elsewhere 0 MPKRKHRSSRRDREEKPGCLVHWFRKGLRLHDNPALKEGLKSASGFRCIYILDPWFAGSCSKGVNRWR 2 1 FLLECLEDLDSSLRKLNSRLFLIRGQPADVLPRLFK 0 0 EWKVTQLSFEEDSEPFGRTRDKAISTLAQEAGVKVISKVSHTLYDPQE 2 1 ILALNNNEPPLTYKRFQDIISLMGIPIYPAEALEAEDVEGLDTFIDPNHEDKYGIPTLEEL 1 2 GFDPEDVPPPMWIGGETEAKQRLDRHLERK 0 0 AWVANFERPRMSPASLMASPAGLSPYLRFGCLSPRTFYWKLTELYQK 0 0 VRKTTNTPLSLHGQLLWREFFFTVACNNRQFDHMVDNPICIQIPWDKNTALLNKWAN 00 GETGYPWIDAIMTQLRLEGWIHPLARHAVACFLTRGDLWISWEEGMK 0 0 VFDEYLLDADWSVNAGNWIWLSCSSFYQQFFHCYCPVKFGRRTDPNGDYVR 2 1 KYLPFLKNFPSKYIFEPWTAPIEVQEKAKCII 12 GKDYPLPIVDHAEASHRNIERMRKVYNNLSRRGGP 1 2 GILGSVPSSQAASAPPQLGKFNMSSKHGQLLCITVYYCVLLCITV* 0 >CRY1_aphVas Aphrocallistes vastus (sponge) AJ437143 PubMed:14499587 MESLSITEFSPSAPSPISSSSSPDEETDRDIVAKDIEQDVEPQGKVLLHIFNNRHLRLKDNTALYQAMAQNPDKFYAVYIFDGFDSKPVAPVRWQFLIDCLE DLKEQLNGFGLELYCFRGETIDVLATLVQAWKVKLLSINMDPDVNFTFFNEKIVKMCTINAVQLYNDMDSHRLLYLPPKYKSAIPMSKFRVLLAEAITAKQNNLESEAKIQDITPP LNPEQLSDLGNKPRLDSPLPSEIPKLNALFTEEEIAKLNFIFQGGERRTEDYLNEYREARLRDVSGDEDASPIAAKAMGISPHLRFGCITPRHLFNFLVKTIKDANYSRIKINKVL AGIMARDFALQVSQLQTIPERIISLNKICLPIPWDKNNNEIVEKLTDAQTGFPFFDAAITQLKTEGYVINEVSEALATFVTNSLLWVSWEEGQNFFSQHLICFDLAMSTHSWLEAS GSTMVTGRQKSYQDPLLFVSKKLDPNGEYIKRYLPKFINFPIEFIHKPGNASLEAQQAANCVIDIDYPKPLFEYECRNGICCKRLRVFMEVVDSAAKATKLPHVIENCSGKFP* 0
Vertebrate CRY2 reference sequences
These are pre-curated provisional sequences, taken from the UCSC 46-way genomic alignment relative to human except where confirmed by an accession number. This can in omission of insertions in other species and missing exons in the situation where they are too diverged or lie in isolated small contigs. The final exons, being quite variable especially in fish, are best determined from transcripts when available and then extended by blastx homology to species within the same clade lacking transcripts. After these corrections, the sequences are aligned and further anomalies are confirmed or discarded on a case-by-base basis.
It appears that the last exon in fish has lost all homology (and so functionality), in some cases simply running out into junk dna until a stop codon is encountered. Exon seven is broken up in some fish with an extra intron that might have some use in fish taxonomy as a derived characteristic.
No evidence for CRY2 currently exists in cartilaginous fish or earlier deuterostomes suggesting that ancestral CRY1 duplicated in stem bony vertebrates, giving rise to CRY2. It appears that all insect CRY2 entries at GenBank are grievously mislabelled and actually represent an independent gene duplication of CRY1 in arthropods (that was lost in drosophilids). If so, these 'CRY2' sequences cannot serve as valid model systems for vertebrate CRY2.
>CRY2_homSap Homo sapiens (human) 11 exons 0 MAATVATAAAVAPAPAPGTDSASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGLVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 1 RYLPKLKAFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSMSSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDA* 0 >CRY2_panTro Pan troglodytes (chimp) 0 MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKAFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSMSSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDA* 0 >CRY2_gorGor Gorilla gorilla (gorilla) 0 MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKAFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSVSSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDA* 0 >CRY2_ponAbe Pongo pygmaeus (orangutan) 0 MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKAFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSVSSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDA* 0 >CRY2_rheMac Macaca mulatta (rhesus) CJ488220 testis 0 MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLER 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSVNSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRAKVAELPTPELPSKDA* 0 >CRY2_papHam Papio hamadryas (baboon) 0 MAATVATAAAVAPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLER 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSVNSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRAKVAELPTPELPSKDA* 0 >CRY2_calJac Callithrix jacchus (marmoset) 0 MAATVATAAAAVPAPAPGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSMSSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDV* 0 >CRY2_micMur Microcebus murinus (mouse_lemur) 0 MATAVA-TAAAAPTPASSTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRW- 2 2 ------------------------------------ 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNSGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSMSST 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVTELPTPELPSKDA* 0 >CRY2_musMus Mus musculus (mouse) CF898022 0 MAAAAV-VAATVPAQSMGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQALISRMELPKKPAVAVSSQQMESCRAEIQENHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPGSSQAGSISNT 1 2 GPRALSSGPASPKRKLEAAEEPPGEELTKRARVTEMPTQEPASKDS* 0 >CRY2_ratNor Rattus norvegicus (rat) DN948283 prostate 0 MAAAAVVAATVPAQSMGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQALISRMELPKKPVGAVSSQHMENCRAEIQENHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYRK 1 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPGSSQAGSISNT 1 2 GPRPLSSGPASPKRKLEAAEEPPGEELSKRARVTEMPAQEPPSKDS* 0 >CRY2_criGri Cricetulus griseus (hamster) XR_135830 0 MAAAAVVAGAPRGARVPALTMGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGAVTSQQMENCRAEIQENHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 1 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSISNT 1 2 GSRPLSSGPASPKRKLEAAEEPPGEELSKRARVTEMPAQEPQTKDA* 0 >CRY2_spaJud Spalax judaei (blind_mole_rat) AJ606300 0 MAAASVVVATSAAPAMAVDGGSSVHWFRKGLRLHDNPSLLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIDLNGQKPPLTYKRFQAIISRMELPKKPVVAVTSQQMESCRAEIQDNHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 1 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFPTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAARCIIGVDYPRPMVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQPGSITNT 1 2 GPRPLSTGPASPKRKLEAAEEPPGEELSKRARVTELPAPEPASKDA* 0 >CRY2_dipOrd Dipodomys ordii (kangaroo_rat) 0 MAAAMVTAAVAVPAPPSGADGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQL-QEGWIHHLARHAVTCFLTRGDLWVSW----- 0 --------------------------------------------------- 2 2 RYLPRLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSISST 1 2 GPRPLSSGPASPKRKLEAAEEPPGEELSKRARVTEMPAQEPPSKDS* 0 >CRY2_cavPor Cavia porcellus (guinea_pig) 0 MAAAVGTGTAAAPTPVTGAEGACSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGAVSSQQMESCRAEIQENHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSLSQAGSSTST 1 2 GPRPLPGGPASPKRKLEAAEEPPGEELSKRARVAELPTPEPPSKDA* 0 >CRY2_hetGla Heterocephalus glaber (blind_mole_rat) EHA99865 0 MAAAVGTGTGAAPTPATGAEGACSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRW 2 1 RFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGAVSSQQMKSCRAEIQENHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 1 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 2 CLLASVPSCVEDLSHPVAEPTLSQAGSSSSTG 1 2 PRSLPDGPASPKRKLEAAEEPPGEELSKRARVAELPAPEPTSKDA* 0 >CRY2_speTri Spermophilus tridecemlineatus (squirrel) 0 MSASVVTTSATLLTP-TSADV-SSVHWFRKGLRL-DNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHT------ 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGAVSSQQMESCKAEIQEDHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKKNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPGSSQAGSISST 1 2 GPRPLSSGQASPKRKLEAAEEPPGEELSKRARVAELPTPELPSKDA* 0 >CRY2_oryCun Oryctolagus cuniculus (rabbit) 0 MAAAAAAAAAAVPAPAASANGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPAGSVTSQQLEGCRAEIQDNHDETYGVPSLEEL 1 2 GFPTEGLSPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYVR 2 2 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSHPVAEPSSSQAGSVSGA 1 2 APRPLPSGPASPKRKLEAAEEPPGEELSKRARVAEVPTPELPSKAV* 0 >CRY2_turTru Tursiops truncatus (dolphin) 0 MAAAVATSAVAAPAPAARAEGASSVHWFRKGLRLHDNPALQAAVRGAHCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDK 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMEGCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRSSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCMEDLSNPVAEPSSSQAGGMSSA 1 2 GPRPLPSGPASPKRKLGAAEEPPGEELSKRARVAGLPPSELLSKDV* 0 >CRY2_bosTau Bos taurus (cow) EG706191 lens 0 MAAAAAAATQAPAARGDGASSVHWFRKGLRLHDNPALLAAVRGAHCVRCVYILDPWFAASSSVGINRWR 2 2 FLLQSLEDLDRSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDTAIMKMAKEAGVEVVTENSHTLYDLDK 2 1 IIELNGQKPPLTYKRFQAIISRMELPRKPVGSVTSQQMEGCRAEIQESHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCVIGVDYPRPIVNHAEASRLNIERMKQVYQQLSRYRGL 1 2 CLLASVPSCVEDLSNPVAEPSSIQAGSSSSV 1 2 GPRPLPGGPASPKRKLEAAEEPPGGELSKRARVAESLPSELPSRGV* 0 >CRY2_oveAri Ovis aries (sheep) NM_001129736 PubMed:19341811 0 MAAAAAATASAAAAAQAPAPRGDGASSVHWFRKGLRLHDNPALLAAVRGAHCVRCVYILDPWFAASSSVGINRWR 2 2 FLLQSLEDLDRSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDK 2 1 IIELNGQKPPLTYKRFQAIISRMELPRKPVGSVTSQQMEGCQAEIQESHDETYGVPSLEEL 1 2 GFPTEGLGPAVWRGGETEALARLDKHLERK 0 0 AWVASYERPRMNASSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYRK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMAQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAEASRLNIERMKQVYQQLSRYRGL 1 2 CLLASVPSCVEDLSTPVAEPSSSQAGSSSSA 1 2 GPRPLPGGPASPKRKLEAAEEPPGGELSKRARVAESLPSELPSRGV* 0 >CRY2_susScr Sus scrofa (pig) XM_003122835 0 MAAAVATAAASSPAPAAGAEGASSVHWFRKGLRLHDNPALLAAVRGAHCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPALFQ 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDK 2 1 IIELNGQKPPLTYKRFQALISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSNPVAEPSSSQAGSVSAA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPPTELPGRDV* 0 >CRY2_equCab Equus caballus (horse) 0 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPRKPVGSVTSQQMESCRADIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNAASLLASPTGLSPYLRFGCLSCRLFYYRLWDLYRK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEAKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKAFPSRYIYEPWNAPEAVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSNPVAEPSSSQAGSVSSA 1 2 GPRPPPSGPASPKRKLEAAEEPPGEELSKRARVAEPPS* 0 >CRY2_canFam Canis familiaris (dog) XM_540761 0 MAAAVVAAAAAAPVPTAGVDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDK 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRADIQDNHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPILKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSNPVAEPSSSQTGSMSSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPTPELPCRDV* 0 >CRY2_ailMel Ailuropoda melanoleuca (panda) XM_002922310 iMet lost to assembly gap 0 PAPAAGVDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDK 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRADIQESHDDTYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPILKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSNPVAEPSSSQTGSMSSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPAPEPPSRDV* 0 >CRY2_myoLuc Myotis lucifugus (microbat) 0 MAANAVTAAAAAPAPAAGTDGASSVYWFRKGLRLHDNPALLAAVRGARCVLCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVASVTRHQMESCPAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSSPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGFR 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCMEDLSNPVAEPSLSQTGSMSSA 1 2 GPKPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPATELPSRDV* 0 >CRY2_pteVam Pteropus vampyrus (macrobat) 0 MAATVGTAAAAASAPAAGTDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQHMESCTAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLNNPVAEPSSSQTGSMNNA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAELPAPELPSRDV* 0 >CRY2_loxAfr Loxodonta africana (elephant) 0 MAAAVVTAGAAALVPIPSMDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWELYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSSPVAEPSSSQAGSSNSA 1 2 GPRPLPSGPTSPKRKLEAAEEPPGEELSKRARVAKLPGPELPSKDV* 0 >CRY2_triMan Trichechus manatus (manatee) AHIN01126950 AHIN01126951 .gt 0 MAATVVTAAAAALAPAPSIDGASSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDRSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVPSLEEL 1 2 GFPTEGLGPAVWQGGETEAMARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCAEDLSNPVAEPSLSQAGSMSSA 1 2 GPRPLPSGPASPKRKLEAAEEPPGEELSKRARVAKLPSPELPSKDV* 0 >CRY2_choHof Choloepus hoffmanni (sloth) 0 MAATAVMAGSAAPAPASGTEGASSVHWFRKGL*LHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVGSVTSQQMESCRAEIQENHDETYGVP----- 1 1 -FPTEGLGPAVWQGGEAEALARLDKHLERK 0 0 AWVANFERPQMNANSLLASPTGLSPYLQFGCLSCQLFYFKLTDLYKK 0 0 VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNSGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYLPKLKGFPSRYIYEPWNAPESIQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSNPVAEPSSSQAGSMSSA 1 2 GPRPVPSGPASPKRKLEAAEEPPGEELSKRARVTELPTPELPSKDV* 0 >CRY2_macEug Macropus eugenii (wallaby) FY652314 testis 0 ------------------------------------------------------------------------ 2 1 FLLQSLEDLDISLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIVKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVSCVTSQQMEQCQAEIQENHDDTYGVPSLEEL 1 2 GFPTDGLGPAVWQGGEMEALARLDKHLERK 0 0 AWVANYERPRMNATSLLASPTGLSPYLRFGCLSCRLFYYRLWELYKK 0 0 VKRNNTPPLSLYGQLLWREFFYTAATNNPKFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYVR 2 2 RYLPQLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPKPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCMEDLSSPMAETSMGQAGSVSST 1 2 GPKPLPCSPASPKRK LEATEEASREEHSKRARMAALPVPELAS----* 0 >CRY2_monDom Monodelphis domestica (opossum) 0 MAAAVVTMTAAAPAPAPSPEGASSVHWFRKGLRLHDNPALQAALRGARCVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDISLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIVKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPRKPVNSVTSQQMERCQAEIQENHDDAYGVPSLEEL 1 2 GFPTDGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNAASLLASPTGLSPYLRFGCLSCRLFYYRLWELYKK 0 0 VKRNNTPPLSLYGQLLWREFFYTAATNNPKFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNSGSWMWLSCSAFFQQFFHCYCP-GFGRRTDPSGDYVR 2 2 RYLPQLKGFPARYIYEPWNAPEPVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCLEDLSSPMVEASLGQAGAVSGP 1 2 GLKPLPCSPASPKRKLEATEEAPGEEHSKRARVARLPGSELAGKDV* 0 >CRY2_ornAna Ornithorhynchus anatinus (platypus) 0 ---------------------------------LHDNPALQAALRGARSVRCVYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIVKMAKEAGVEVVTENSHTLYDLDR 2 1 IIELNGQKPPLTYKRFQAIISRMELPKKPVSCVTSQQMEGCKAEIQDNHDDTYGVPSLEEL 1 2 GFPTDGLGPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK 0 0 VKRNSTPPLSLYGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDFIR 2 2 RYLPKLKAFPSRYIYEPWNAPESVQKAAKCVIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSSAAAESGLGQASGSNIST 1 2 APRPPPPPYGPASPKRKLEAAEGLPGEELCKRPKVAGRPGPEPPGEDA* 0 >CRY2_galGal Gallus gallus (chicken) AJ396745 bursa 19456395 15459395 0 MAAAASPPRGFCRSVHWFRRGLRLHDNPALQAALRGAASLRCIYILDPWFAASSAVGINRWR 2 1 FLLQSLEDLDNSLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIIKLAKEAGVEVVIENSHTLYDLDR 2 1 IIELNGNKPPLTYKRFQAIISRMELPKKPVSSIVSQQMETCKVDIQENHDDVYGVPSLEEL 1 2 GFPTDGLAPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWELYKK 0 0 VKRNSTPPLSLYGQLLWREFFYTAATNNPKFDRMEGNPICIQIPWDKNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYVK 2 2 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPKPMVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDLSGPVTDSAPGQGSSTST 1 2 AVRLPQSDQASPKRKHEGAEELCTEELYKRAKVTGLPAPEIPGKSS* 0 >CRY2_taeGut Taeniopygia guttata (finch) FE716439 brain 0 --------------------------------------------IYILDPWFAATSAVGINRWR 2 1 FLLQSLEDLDNSLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIVKLAKEAGVEVVIENSHTLYDLDR 2 1 IIELNGHKPPLTYKRFQAIISRMELPKKPVSTVISQQMETCKVDIQENHDDVYGVPSLEEL 1 2 GFPTDGLAPAVWQGGETEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWELYKK 0 0 VKRNSTPPLSLYGQLLWREFFYTAATNNPKFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWESGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYVK 2 2 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPKPMVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCVEDISGPVPDSASGQGCSTST 1 2 AVRLSQAEQASPKRKHEGAEEPCSEELYKRAKVTGLPTSEISGKSL* 0 >CRY2_allMis Alligator mississippiensis (alligator) genome/blat 0 MAASRSFPSSVPARAGPCRAVHWFRRGLRMHDNPALQAALRDAASVRCIYILDPWFAASSAVGINRWR 2 1 FLLQSLEDLDNSLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWGVTRLTFEYDSEPFGKERDAAIIKLAKEAGVEVVIENSHTLYDLDR 2 1 IIELNGNKPPLTYKRFQAIISRMELPKKPVSSITSQQMEKCKAEIQENHDDVYGVPSLEEL 1 2 GFPMDGLAAAVWQGGEMEALARLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWELYKK 0 0 VKRNSTPPLSLYGQLLWREFFYTAATNNPKFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWECGVR 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYVK 2 2 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPKPMVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSCAEDLSGPVTDPASVQGCSTST 1 2 ALKPSQSGQASPKRKHEGIEEMCTEDLYKRAKVTGLHGPEIPSKSL* 0 >CRY2_anoCar Anolis carolinensis (lizard) XM_003214641 0 MAALPGPLGRCSVHWFRRGLRLHDNPALQAAIRDGGPVRCIYILDPWFAASSSVGINRWR 2 1 FLLQSLEDLDNSLRKLGSRLFVVRGQPTDVFPRLFK 0 0 EWRVTRLTFEYDSEPFGKERDAAIVKLAKEAGVEVITENSHTLYDLDR 2 1 IIELNGHKPPLTYKRFQTIISRMDLPKKPVASITHQQMEMCKTEIQDNHDETYGVPSLEEL 1 2 GFPTESLAPAVWLGGETEALTRLDKHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWELYKK 0 0 VKRNSTPPLSLYGQLLWREFFYTAATNNPKFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWESGVR 0 0 VFDELLLDADFSVNSGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYVR 2 2 RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPKPMVNHAETSRLNIERMKQIYQQLSRYRGL 1 2 CLLASVPSCMEDLSNPVADTSSSHGNCIGT 1 2 ASRQTHCGQTSPKRKHDVVQEYKEELYKRAKVVASQFAENPRQELVHHEMGN* 0 >CRY2_xenTro Xenopus tropicalis (frog) NM_001088670 AY049035 CX389867 11533577 discrepancies 0 MEGKPSVSSVHWFRKGLRLHDNPALLSALRGANSVRCVYILDPWFAASSSGGVNRWR 2 1 FLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWGVSRLTFEYDSEPFGKERDAVIMKLAKEaGVEVVVENSHTLYDLDR 2 2 VIELNGHSPPLTYKRFQAIISRMELPRRPAPSVTRQQMEACRAEIKRNHDETYGVPSLEEL 1 1 GFHsEiKGpsIWPGGETEALARLDRHLERK 0 0 AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLKELYKK 0 0 VKKNSPPPLSLYGQLLWREFFYTAATNNPKFDQMEGNPICVQIPWDKNPKALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWNSWECGVK 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYVR 2 2 RYLPVLKAFPSRYIYEPWSAPESVQKEAKCIIGIDYPKPIVNHAEASRMNIERMKQTYQQLSRYRGL 1 2 CILASVPSCMEDLGGPMADSSQNISE 1 2 AGKMAPPSHTSGESVLAAQYTAGI* 0 >CRY2_ranCat Rana catesbeiana (bullfrog) GO458565 AY256684 0 MEGPAVSSVHWFRKGLRLHDNPALLAALRGARCVRCVYILDPWFAASSSSSGGVNRWR 2 1 FLLQSLEDLDSSLRKLGSRLFVGRGQPADVFPRLFK 0 0 EWGVTRLTFQYYSEPFGKERDAAIMKLAKEAGVEVIVESSHTLYDLDK 2 2 IIELNGNSPPLTYKRFQAIVSRMELPRRPVPSITRQQMEKCRAEIKSTHDDTYGVPSLEEL 1 1 GFPRDNPGAAVWPGGETEALARLDRHLERK 0 0 AWVAHYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLRELYQK 0 0 VKKNSPPPLSLFGQLLWREFFYTAATNNPKFDQMEGNPICVQIPWDKNPEALAKWTEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWNSWECGVK 0 0 VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYVR 2 2 RYLPVLKGYPSRYIYEPWNAPESVQKEAKCIVGVDYPKPVVNHAEASRLNIERMKQTYQQLSRYRGL 1 2 CILASVPSSVEDLSGPMADPASSQQGSS 1 2 DPAPRLCAVDSPKRKHEDLDGELCKKARLQCVQEMERAAKDFL* 0 >CRY2_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01038797 0 MVVNSVHWFRKGLRLHDNPALQEALNISDTVRCVYILDPWFAASANVGINRWR 2 1 FLLESLEDLDASLRKLNSRLFVVRGQPADVFPRLFK 0 0 EWNVTHLTFEYDSEPFGKERDAAIMKLARESGVETIVQESHTLYSLDR 2 2 IIELNNNSPPLTFKRFQAIVSRLGLPKKPLGTVTPQQMERCHTPISENHDECYRVPTLEEL 1 1 GFKTHGLSPSIWKGGETEAVRRLNKHLERK 0 0 AWVASFERPKINVYSLIASPTGLSPYLRFGCVSCRLFYYSLLELYKK 0 0 VRKSSSPPLSLFGQLLWREFFYTAATNNPKFDQMEGNPICVQIPWDRNPEALAKWAEGRTGYPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGMR 0 0 VFEELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYIPKLKEYPNRYIYEPWNAPEIVQKAANCVIGVDYPKPMINHAQASRLNIERMKQIYQQLSHYKGL 1 2 GLLASVPSVPDDARSPVTDDPHGHSS 1 2 * 0 >CRY2_danRer Danio rerio (zebrafish) aka CRY3 NM_131786 0 MVVNSVHWFRKGLRLHDNPALQEALNGADTVRCVYILDPWFAGSANVGVNRWR 2 1 FLLESLEDLDTSLRKLNSRLFVVRGQPTDVFPRLFK 0 0 EWNVTRLTFEYDSEPYGKERDAAIIKMAQEYGVETVVRNTHTLYNPDR 2 2 IIEMNNHSPPLTFKRFQAIVNRLELPRKPLPTITQEQMARCRTQISDNHDEHYGVPSLEEL 1 1 GFRTQGDSLHVWKGGETEALERLNKHLDRK 0 0 AWVANFERPRISGQSLFPSPTGLSPYLRFGCLSCRVFYYNLRDLFMK 0 0 LRRRSSPPLSLFGQLLWREFFYTAGTNNPNFDHMEGNPICVQIPWDHNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWESGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR 2 2 RYIPKLKDYPNRYIYEPWNAPESVQKAANCIVGVDYPKPMINHAESSRLNIERMKQVYQQLSHYRGL 1 2 SLLASVPTIQEEAEPPMSDDSQASSSST 1 2 GQASSPPHLSITAPSTPPLSESSSPNSSPTASTSVPHTQRKRVRPSETPSKQKTKVKHTSQARGLELKMDDKQ >CRY2_oreNil Oreochromis niloticus (tilapia) XM_003449249 split exon 7 also in gasAcu, oryLat, tetNig not danRef or lepOcu 0 MVVNSVHWFRKGLRLHDNPALQEALNGADAVRCVYILDPFFAGAANVGINRWR 2 1 FLLEALEDLDSSLKKLNSRLFVVRGQPTDVFPRLFK 0 0 EWNVTRLTFEYDPEPYGKERDGAIIKMAQEFGVETIVRNSHTLYNLDR 2 2 IIEMNNNSPPLTFKRFQTIVSRLELPRRPLPPITQQQMDKCHTKIGDNHDQLYSIPSLEEL 1 1 GFRTAGLPPAVWRGGESQALDRLSKHLDKK 0 0 VWVTSLEHPRVNTCSLYASPTGLSPYLRFGCLSCRVLYYNLRELYMK 0 0 VRKRCSPPLSLFGQLLWREFFYTAATNNPNFDRMDGNPICVQ 00 IPWDQNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWESGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPTGDYIR 2 2 HYIPILKDYPNRYIYEPWNAPESLQKAANCVVGVDYPKPMINHAESSRLNIERMKQVYQQLSHYRGL 1 2 SLLASVPTIQEEAEPPMTDESQTSSGP 1 2 DSPQRVPANTEGAGCYEAPDSSTVCPSSTSAPHPDQEHQAPCISSGCHTDPS >CRY2_tetNig Tetraodon nigroviridis (fugu) CAAE01010345 0 MVVNSVHWFRKGLRLHDNPALQEALSGADSLRCVYVLDPWFAGAANVGINRWR 2 1 FLLEALEDLDCSLKKLNSRLFVVRGQPTDVFPRLLK 0 0 EWKVTRLTFEYDPEPYGKERDGAIIKMAQQFGVETIVRNSHTLYNLDR 2 2 IVEMNNNSPPLTFKRFQTIVSRLELPRRPLPSVTQQQMDKCGTPVADNHDQLYSIPSLEEL 1 1 GFRTGGLAPAVWRGGESEALERLRKHLEKK 0 0 VWVSHLEHSRANTCSLYASPAGLSPYLRFGCLSCRVFYYNLREHYIR 0 0 LCKGCSPPLSLFGQLLWREFFYTAATNNPNFDRMAGNPICVQ 00 IPWDQNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHQARRASACFLTRGDLWISWECGMK 0 0 VFEELLLDADWSVNAGSWMWLSCSAFFQQFFKCYCPVGFGRRTDPSGDYIR 2 2 RYIPILKDYPNRYIYEPWNAPEAVQKAANCVVGVDYPRPMINHAEGSRLNIERMKQVYQQLSHYRGL 1 2 SLLASVPTIQEEADTP 1 2 * 0 >CRY2_takRub Takifugu rubripes (fugu) HE592015 0 MVVNSVHWFRKGLRLHDNPALQEALSGADSLRCVYVLDPWFAGAANVGINRWR 2 1 FLLEALEDLDCSLRKLSSRLLVVRGQPTDVFPRLLK 0 0 DWKVTRLTFEFDPEPYGKERDGAIIKLAQQFGVETIVRNSHTLYNLDR 2 2 IIEVNNNSPPLTFKRFQTIVSRLELPRRPLPTVTQHQIHKCGAKMADSQEQLYSIPSLEEL 1 1 GFRTEGLPPAVWRGGESEALERLHKHLDKK 0 0 VWVANLEHSRVSTCSLYASPAGLSPYLRFGCLSCRVLYYNLRELYVK 0 0 LRKGCSPPPSLFGQLLWREFFYTAATNNPNFDRMEGNPICVQ 00 IPWDQNPEALAKWAEGHTGFPWIDAIMTQLRQEGWIHHQARRAVACFLTRGDLWISWE 0 0 VFEELLLDADWSVNAGSWMWLSCSAFFQQFFKCYCPVGFGRRTDPSGDYIR 2 2 RYIPILKDYPNRYIYEPWNAPEAVQKAANCVVGVDYPRPMINHAEGSRLNIERMKQVYQQLSHYRGL 1 2 SLLASVPTIQEEAEPPMTDEFQNSSGP 1 2 * 0
Cryptochrome CRY4 sequences
This subfamily has its 8th and 9th exons split identically in position and phase to CRY1 in sea urchin and amphioxus whereas these exons are fused from lamprey to human. This suggests the split state is ancestral and that CRY4 arose as a gene duplication in stem vertebrates. Although CRY4 has persisted in most tetrapods to the present day, it has been lost in all mammals and also in crocodileans.
>CRY4_galGal Gallus gallus (chicken) NP_001034685 CRY4 PumMed:19663499 synteny: ADIPOR1 UBE2T CRY4 LRIF1 DRAM2 CEPT1 0 MRHRTIHLFRKGLRLHDNPALLAALQSSEVVYPVYILDRAFMTSSMHIGALRWHFLLQSLEDLRSSLRQLGSCLLVIQGEYESVVRDHVQKWNITQVTLDAEMEPFYKEMEANIRGLGEELGFQVLSLMGHSLYNTQR 2 1 ILELNGGTPPLTYKRFLRILSLLGDPEVPVRNPTAEDFQ 2 1 RCSPPELGLAECYGVPLPTDLKIPPESISPWRGGESEGLQRLEQHLADQ 0 0 GWVASFTKPKTVPNSLLPSTTGLSPYFSTGCLSVRSFFYRLSNIYAQ 0 0 AKHHSLPPVSLQGQLLWREFFYTVASATPNFTKMAGNPICLQIRWYEDAERLHKWKT 0 0 AQTGFPWIDAIMTQLRQEGWIHHLARHAAACFLTRGDLWISWEEGMK 0 0 VFEELLLDADYSINAGNWMWLSASAFFHHYTRIFCPVRFGRRTDPEGQYIR 2 1 KYLPILKNFPSKYIYEPWTASEEEQKQAGCII 12 GRDYPFPMVDHKEASDHNLQLMKQAREEQHRIAQLTR 1 2 DDADDPMEMKLKRDHSEESFTKTKAARMTEQT* 0 >CRY4_taeGut Taeniopygia guttata (finch) XM_002198497 0 MLHRTIHLFRKELRLHDNPVLLAALESSEALYPVYILDRAFLTSSMHIGALRWNFLLQSLEDLHKNLGQLGSCLLVIQGEYEIVLRDHIQKWNITQVTLDAEMEPFYKEMEANIQRLGAELGFEVLSLVSHSLYNTQR 2 1 ILDLNGGSPPLTYKRFLHILSLLGDPELPVRNLTAEDFQ 2 1 RCRAPEPGLAECYRVPLPVDLKISPESLSPWRGGETEGLRRLEQHLIDQ 0 0 GWVTSFAKPRTSPNSLLPSTTGLSPYFSMGCLSVRTFFYRLSNIYAQ 0 0 AKHHSLPPVSLQGQLLWREFFYTVASATPNFTQMAGNPICLQISWYKDAERLHKWKT 0 0 AKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADYSINAGNWMWLSASAFFHQYTRIFCPVRFGKRTDPQGNYIR 2 1 KYLPILKNFPSKYIYEPWTASEEEQKLAGCII 12 GQDYPFPMVNHKEASDHNLQLMKQVREEQHRTVQLTR 1 2 DDADDPMEIRVKRDHSEENISKGKVARTTE* 0 >CRY4_melGal Meleagris gallopavo (turkey) XM_003212851 0 MRHRTIHLFRKGLRLHDNPALLAALQSSEVVYPVYILDRAFMTSSMHIGALRWHFLLQSLEDLRSSLCQLGSCLLVIQGEYDAVVRDHVQKWNITQVTLDAEMEPFYKEMEANIRALGEELGFEVLSLMGHSLYNTQR 2 1 ILELNGGTPPLTYKRFLHILSLLGDPEMPIRNLTAEDFQ 2 1 RCSPPELCLAECYRVPLPTDLKIPPESISPWRGGESEGLQRLEQHLADR 0 0 GWVASFTKPKTVPNSLLPSTTGLSPYFSMGCLSVRSFFYRLSNIYAQ 0 0 AKHHSLPPVSLQGQLLWREFFYTVASATPNFTKMAGNPICLQIRWYEDAERLHRWKT 0 0 AQTGFPWIDAIMTQLRQEGWIHHLARHAAACFLTRGDLWISWEEGMK 0 0 VFEELLLDADYSINAGNWMWLSASAFFHHYTRIFCPVRFGRRTDPEGQYIR 2 1 KYLPILKNFPSKYIYEPWTASEEEQKQAGCII 12 GRDYPFPMVDHKEASDHNLQLMKQVREEQYRTAQLTR 1 2 DDADDPMEMKLKRDHSEENLTKTKAARMTKQT* 0 >CRY4_pasDom Passer domesticus (sparrow) AY494987 16687285 fragment 0 KGLRLHDNPVLLAALESSEALYPVYILDRAFLTSSMHIGALRWNFLLQSLEDLHKNLGQLGSCLLVIQGEYEIVLRDHIQKWNITQVTLDAEMEPFYKEMEANIQRLGVELGFEVFSLVSHSLYNTQR 2 1 ILDLNGGSPPLTYKRFLHILSLLGDPEVPVRNVTAEDFQ 2 1 RCRAPDPGLAECYRVPLPVDLKISPESLSPWRGGETEGLQRLERHLTDQ 0 0 GWVTSFTKPRTVPNSLLPSTTGLSPYFSMGCLSVRTFFYRLSNIYAQ 0 0 AKHHSLPPVSLQGQLLWREFFYTVASATPNFTQMAGNPICLQICWYKDAERLHKWKT 0 0 AQTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLW 0 0 2 1 12 1 2 * 0 >CRY4_anoCar Anolis carolinensis (lizard) synteny: UBE2T CRY4 LRIF1 DRAM2 0 2 1 IVELNRGTPPLTYKRFLQILTTLGDPEMPVEDLTAENSQ 2 1 KLCPLPEEDLDECYRVPLPEDLGISKEHLSPWRGGETTGLQRLEEHMKNQ 0 0 GWIAKFTKPRTIPNSLLPSTTGLSPYFSLGCLSVRMFFYRLSKIYAQ 0 0 SKNHSLPPVSLQGQLLWREFFYTVASATPNFTKMVGNPICLQIEWYEDAEKLHKWKT 0 AQTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDSDYSINAGNWMWLSASAFFHQYSRIFCPVRFGKRTDPEGHYIR 2 1 KYLPVLKNFPSKYIYEPWTAPAEAQREAGCII 12 GKEYPFPMVNHKEVSDNNLQLMKQVREEQYKTAQLTR 1 2 DDVDDPMEMGLKHRHSEEVTPKGKHARKAEEL* 0 >CRY4_xenTro Xenopus tropicalis (frog) NP_001123706 0 MPHRTIHIFRKGLRLHDNPTLVTALETSDVVYPVYILDRNFMTSSSVIGSKRWNFFLQSIEDLHCNLQKLNSCLFVIQGDYERVLREHVEKWNITQVTFDLEIEPYYKGLDERIRAMGQELGFEVVSMVAHTLYDIKK 1 ILALNCGKPPLTYKNFLRVLSMLGNPDKPARQITSEDFIK 2 1 CITPTKLAAEEYYRIPKPEDLGISKDCPTNWIGGESEALSRLEQHLEKQ 0 GWVANFKKPQTIPNSLLPSTTGLSPYFSLGCLSVRVFFHRLSNIYAQ 0 0 SKNHSLPPVSLQGQLLWREFFYTVASSTPNFTHMVGNPICLQIDWYKNEEQLQKWKE 0 0 AKTGFPWIDAIMTQLHNEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEEFLLDADYCINAGNWMWLSASAFFHHYTRIFCPVRFGKRSDPEGNYIR 2 1 KYLPVLKNFPAKYIYEPWTAPEEIQKQAGCLI 12 GKDYPLPMVDHKTASEHNLQLMRQVREEQQKTAQLTK 1 2 DIADDPMELNLKHPFKNDKECGLENNKVKRRCLEEMTAEDGTRQSDLVQSLDPCEVKVC* 0 >CRY4_latCha Latimeria chalumnae (coelocanth) AFYH01009222 0 MTHRTIHIFRKGLRLHDNPILLAALEFSRVVYPVYILDRKLLESGVIIGALRWRFILQSLEDLHRNLVKLNSRLFVIQGDYEQILREYVQKWTITQVTFDTEIEPFYKEMDKKKVRLMGKEMGFTVLFSVAHALYDVAR 2 1 IVENNGGQPPLTYKKFLHVLSKLGDPERPVRDITVDDFQ 2 1 KCMPPNPGLKELFRVPLPEELGLQSEHNASWIGGESQGLQRLEQHLENQ 0 0 GWIAHFTKPRTIPNSLLPSTTGLSPYFTLGCLSVRNNFYRLSHIYAQ 0 0 SKSHSLPPVSLQGQLLWREFFYTVASATPNFTKMVGNPICLQIGWYEDPEGLRKWRT 0 0 AETGFPWIDAIMTQLRQEGWVHHLARHAVACFLTRGDLWISWEGGMK 0 0 VFEELLLDADYSMNAGNWMWLSASAFFHQYTRILCPVRFGKRTDPEGHYIR 2 1 KYLPVLKNFPSKYIYEPWTAPEEVQQQAGCVI 12 GKDYPFPMVNHKEVSKNSLHLMKQVREEQHKVTQLTR 1 2 DVADDPMEMRMKRKFPDDFKNKGIFSETAERSSTT* 0 >CRY4_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01016726 0 MTHRTIHLFRKGLRLHDNPTLLGALESSAVLYPVFILDRAFMEEAMARGVLRWRFILQSLEDLDTRLQAIGSRLFVLCGSTANILRELVAQWGITQISYDTEVEPYYTRMDKDIQTVAQENGLQTYTCVSHTLYDVKR 2 1 IIKANGGEAPLTYKKFLHVLALLGAPETPARRITREDFR 2 1 CRTPTEEESEEKYRVPSLEDLGIVVESEALWVGGETEGLQRLEQHMQNQ 0 0 GWIENFTKPRTIPNSLLPSTTGLSPYFSLGCLSVRMFYHRLSNIYAQ 0 0 SKNHSLPPVSLQGQVLWREFFYTVAAATPNFTRMAGNPICLQIDWYKDQEALDRWKT 0 0 ARTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEEHLLDADYSVNAGNWMWLSASAFFHQYTRIFCPVRFGRRTDPEGHYLR 2 1 KYLPVLKNFPSQYIYEPWTAPQQVQLEAGCII 12 GKDYPLPMVNHREVSESNLALMREVRREQEKTAQLTR 1 2 DTADDPMEVGKKRACQREAETDRSDAFLEKPERAKRFSHAEGEAKALACSWPSETLRLPALGREVM* 0 >CRY4_danRer Danio rerio (zebrafish) BC164413 0 MSHRTIHLFRKGLRLHDNPSLLGALASSSALYPVYVLDRVFLQGAMHMGALRWRFLLQSLEDLDTRLQAIGSRLFVLCGSTANILRELVAQWGITQISYDTEVEPYYTRMDKDIQTVAQENGLQTYTCVSHTLYDVKR 2 1 IVKANGGSPPLTYKKFLHVLSVLGEPEKPARDVSIEDFQ 2 1 RCVTPVDVDRVYAVPSLADLGLQVEAEVLWPGGESHALQRLEKHFQSQ 0 0 GWVANFSKPRTIPNSLLPSTTGLSPYLSLGCLSVRTFYHRLNSIYAQ 0 0 SKNHSLPPVSLQGQVLWREFFYTVASATPNFTKMEGNSICLQIDWYHDPERLEKWRT 0 0 AQTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWITWEEGMK 0 0 VFEEFLLDADYSVNAGNWMWLSASAFFHKYTRIFCPVRFGRRTDPQGEYLR 2 1 KYLPVLKNFPSQYIYEPWKAPEDVQLSAGCII 12 GKDYPRPIVSHIEASQRNLALMRQVRTEQQTTAELTR 1 2 DVADDPMEAGLKRELREEEGLLEEAESQCTSKRFSGSSDHKSRPCSWTPETLQLSELSGEVM* 0 >CRY4_braFlo Branchiostoma floridae (amphioxus) XM_002595028 fused exons 2-3 BW780666 odd splice phases exon 5-6, no split 8-9, no last exon 0 MANKVQHSTIHWFRKGLRFHDNPSLLHALRTSRHVYPVYVVDQNWMKEHDIRYGANWWRFVIQCLEELDTRLRKYGLRLFVVRGSAEDFFKEHFRKWKVTQLTHDVDTEPFYRIRDVAVRKIASDMGVEVVTHVAHTLYDIDR 2 1 TVERNGGTPPLTYRKFLKVYNEMGKPPKEKETATTEHFA NCTLPPEALQDKKFDMVTLEELGMRCDFPAKFVGGETEALRRLEKSLEDK 0 0 DWVLKFEKPKTSPNSLLPSTTVLSPYITVGCLSARLFYHQLDKIYSK 0 0 AKNHSQPPVSLHGQLIWREFFYTAAAHTPNFNQMEGNRVCLQIPWNKNDEHLTRWRN 1 2 AQTGFPWIDAAMTQLRKEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSLNAGNWMWLSASAFFHQYFRVYSPIVFGKKTDPKGTFIR 2 1 HYLPVLKNFPKEYIYEPWKAPRNVQEKAGCIVGKDYPRPIVDHKEASQRNLDIMRDVRKDQKETAAVTLGYGK* 0 >CRY4_nemVec Nematostella vectensis (sea_anemone) XP_001636303 ABAV01006592 last exon uncertain 0 MASKKTHQTVHWFRNGLRLHDNPALKEAFETSQTVRPLYVLDPDVLKNGNIGVVRWRFILESLADLDNNLKKLNSR 2 1 LFVVRGRPSEVFPKLFKEWKISKLTFEVDTTEPARKQDAEVLKIANKLGVDIEQRVSHTLYDLDR 2 1 IVNKNGGTAPLTYKKFQSIVSSLGPPAAPLPAIDKKMVAGCNTPTTANHDKIYGVPTLE EIGQEVPEESREVLYPGGETEALERLEVYMKKE 0 0 DWVCKFEKPNTAPNSIEPSTTVLSPYMTYGCLSARLFYTRLAEIYAK 0 0 KKKHSQPPVSLHGQLLWREFFTTAAYKTPNFNKMVGNSLCLQVPWDQNDEYLAAWAE 0 0 ARTGYPYIDAIMTQLRREGWVHHLARHSVACFLTRGDLWISWEEGLK 0 0 VFERLLIDHDWNLNAGNWMWLSASSFFHAYFRVYSPVAFGKKTDPNGDYIR 2 1 KYIPKLSRYPPKYIYEPWNAPLAVQEKAGCVIGRDYPCPIVDHNKVVTRNMSRMKEARSLKYGKTDMGVDKRE 1 2 EASTPSKRKSKTTTDKPAKKTKKITDFLADSSE* 0
Cryptochrome 6-4 photolyases
>CRY64_anoCar Anolis carolinensis (lizard) XM_003225714 6-4 photolyase synteny: DCPS TIRAP CRY64 SRPR FOXRED1 0 MAHVSIHWFRKGLRLHDNPALLAAMKNSAEIYPIFILDPWFPKNMQVSINRWRFLIESLKDLDESLKKLNSR 2 1 LFVVRGRPAEVFPELFTKWKVTRLAFEVDTEPYARRDAEVVRLAAEHGVQVIQKVSHTLYDTER 2 1 IIVENSGKAPLTYTRLQTLVASLGPPKQPVPAPKLEDMK 1 2 DCCTPVKEDHDLEYGTPSYEELGQDPKTAGPHLYPGGETEALARLDLHMKRT 0 0 SWVCNFKKPETHPNSLTPSTTVLSPYVKFGCLSVRMFWWKLAEVYQG 0 0 RKHSDPPVSLHGQLLWREFFYTAGAGIPNFDRMENNPVCVQVDWDNNQEYLRAWRE 0 0 GQTGYPFIDAIMTQLRTEGWIHHLARHAVACFLTRGDLWISWEEGQ 0 0 VFEELLLDADWSLNAANWQWLSASAFFHQFFRVYSPVTFGKKTDKNGEYIK 2 1 KYLPFLRKFSNDYIYEPWKAPRSLQERAGCIIGQDYPKPIVEHEKVYKRNLERMKAAYARRSPNLVIQAKDKVSQKKGV 1 2 NRKRPEAPTKAKVQAKKVKTKSS* 0 >CRY64_chrPic Chrysemys picta (turtle) AHGY01135270 AHGY01135271 no synteny 0 MKHVSIHWFRKGLRLHDNPALLAATTDCRKLYPIFVLDPWFPKNMRVSVNRWRFLVESLRDLDESLQKLNSR 2 1 LFVVRGRPTEIFPVLFKEWKVTRLTFEVDTEPYSRQRDSEVASLAAEHGVQVIQKVSNTLYDTDR 2 1 VIAENNGKVPLTYVRLQTLLATLGPPKRPVPAPTLENLK 1 2 DCCTPCKDNHDAEYGIPTLEELGQDPEQAGLRLYPGGESEALCRLDLHMNRT 0 0 AWVCNFQKPQTEPNSLNPSTTVLSPYIKFGCLSVRTFWWRLAEVYQG 0 0 KKHSNPPVSLHGQLLWREFFYTAGAGIPNFNKMEGNPVCVQVDWDNNPEHLRAWSE 0 0 GRTGYPFIDAIMAQLRTEGWIHHLARHAVACFLTRGDLWISWEQGQ 0 0 VFEELLLDADWSLNAGNWQWLSASAFFHQFFRVYSPIAFGKKTDKSGEYIK 2 1 KYLPFLRKFPAEYIYEPWKAPRSMQEQAGCVIGRDYPKPIVVHEVVSKRNVERMKAAYARRSSSTTAQLEGGGGKKGIN 1 2 GAKRRTPAGPSVAELLTKKPKT * 0 >CRY64_allMis Alligator mississippiensis (alligator) 0 MKHSCIHWFRKGLRLHDNPALLAAMKDCSELYPIFILDPWFPKNMQVSVNRWRFLVESLKDLDESLKKLNSR 2 1 LFVVRGHPAEVFPGLFKAWKVTRLTFEVDTEPYSKQRDAEVVRLAAAHGVQVIQKVSHTLYDTDR 2 1 VIAENNGKVPLTYRQLQAVLAGLGPPKQPVLAPTLETLK 1 2 DCCTPGRDNRDPKYEIPTLEELGQDPKEAGPCLYPGGESEALSRLAFHMKRM 0 0 TWVCNFKKPDTEPNTLSPSTTVLSPYVKFGCLSVRTFWWKLAEIYRG 0 0 KKHSSPPVSLHGQLLWREFFYTAGAGIPNFNRMEGNPICVQVDWDDNPEYVKAWKE 0 0 GRTGYPFIDAIMTQLRTEGWIHHLARHAVACFLTRGDLWVSWEEGQ 0 0 VFEELLLDADWSLNAGNWQWLSASAFFHQFFRVYSPVAFGKKTDKSGAYIK 2 1 KYLPILRKFPAEYIYEPWKAPRSMQEQAGCIIGRDYPKPIVEHEALSKRNIMRMKAAYAQRSHSKAAQVEKESTKKGN 1 2 GGKRKLPAGPSVVELLTKKPKAKSS* 0 >CRY64_croPor Crocodylus porosus (crocodile) blat/genome 0 MKHSCIHWFRKGLRLHDNPALLAAMKDCSDLYPIFILDPWFPKNMQVSVNRWRFLIESLKDLDESLKKLNSR 2 1 LFVVRGHPAEVFPGLFKAWKVTRLTFEVDTEPYSKLRDAEVVRLAAAHGVQVIQKVSHTLYDTDR 2 1 IIAENNGKVPLTYRQLQAVLAGLGSPKQPMLAPTLETLK 1 2 DCCTPARDNHDPKYEIPTLEELGQDPKEAGPRLYPGGESEALSRLDFHMKRM 0 0 TWVCNFKKPDTEPNTLSPSTTVLSPYIKFGCLSVRTFWWKLAEIYRG 0 0 KKHSSPPVSLHGQLLWREFFYTAGAGIPNFNRMEGNPICVQVDWDDNLEYVKAWKE 0 0 GRTGYPFIDAIMTQLRTEGWIHHLARHAVACFLTRGDLWVSWEEGQ 0 0 VFEELLLDADWSLNAGNWQWLSASAFFHQFFRVYSPVAFGKKTDKSGAYIK 2 1 KYLPILRKFPAEYIYEPWKAPRSMQEQAGCIIGRDYPRPIVEHEAVSKRNIMRMKAAYAQRSRSHSKSAQVEKEGTKKGN 1 2 GGKRKLPAGPSVVELLTKKPKAKSS* 0 >CRY64_xenTro Xenopus tropicalis (frog) synteny: STS1 RPL27A CRY64 FOXRED1 SRPR 0 MKHNSIHWFRKGLRLHDNPALLAAMKDCAELYPIFILDPWFPRNMKVSVNRWRFLIEALKDLDENLKKINSR 2 1 LFVVRGKPTEVFPLLFKKWKVTRLTFEVDTEPYSRQRDADVEKLAAEHNVQVIQKVSNTLYAIDR 2 1 IIAENNGKPPLTYVRFQTVLALLGPPKRPVQVPTQENMK 1 2 DCCTLWKSSYNEKYGVPTLEELGQDSLKLGPRLYPGGESEALSRLELHMKRT 0 0 TWVCNFKKPETEPNSLTPSTTVLSPYVKFGCLSARTFWWRIADIYQG 0 0 KKHSDPPVSLHGQLLWREFFYTAGVGIPNFNKMEGNTVCVQVDWGNNKEHLQAWSE 0 0 GRTGYPFIDAIMTQLRTEGWIHHLARHAVACFLTRGDLWISWEEGQK 0 0 VFEELLLDADWSLNAGNWQWLSASTFFHQFFRVYSPVAFGKKTDKNGDYIK 2 1 KYLPILKKFPAEYIYEPWKAPRSLQERAGCIIGKDYPKPIVEHDVASKQNIQRMKAAYARRSGSTAEVDKDSGQSNKN 1 2 GAKRKVAGGPSVAELFKKNKSKKD* 0 >CRY64_xenLae Xenopus laevis (frog) PubMed:19715341 19345672 9016626 0 MRHNSIHWFRKGLRLHDNPALLAAMKDCAELHPIFILDPWFPKNMQVSVNRWRFLIDALKDLDENLKKINSR 2 1 LFVVRGKPAEVFPLLFKKWKVTRLTFEVDIEPYSRQRDAEVEKLAAEHDVQVIQKVSNTLYDIDR 2 1 IIAENNGKPPLTYVRFQTVLAPLGPPKRPIKAPTLENMK 1 2 DCHTPWKSSYDEKYGVPTLEELGQDPMKLGPHLYPGGESEALSRLDLHMKRT 0 0 SWVCNFKKPETEPNSLTPSTTVLSPYVKFGCLSARTFWWKIADIYQG 0 0 KKHSDPPVSLHGQLLWREFYYTTGAGIPNFNKMEGNPVCVQVDWDNNKEHLEAWSE 0 0 GRTGYPFIDAIMTQLRTEGWIHHLARHAVACFLTRGDLWISWEEGQK 0 0 VFEELLLDADWSLNAGNWLWLSASAFFHQFFRVYSPVAFGKKTDKNGDYIK 2 1 KYLPILKKFPAEYIYEPWKSPRSLQERAGCIIGKDYPKPIVEHNVVSKQNIQRMKAAYARRSGSTEGVDKDSGQNNK 1 2 KGvKRKVAAGpSVAELFKKK* 0 >CRY64_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01024141 0 MMHRSIHWFRKGLRLHDNPALLASLRDCAELWPVFLLDPWFPKNARVSVNRWRFLLRALQDLDGNLRKLGSR 2 1 LFVVRGSPAEVFPRLFEQWKVTRLTFEVDTEPYARQRDAQVGKIAEEHGVEVIQKVSHTLYDTER 2 1 ILVENNGKAPLTYNRLQALLKTLGAPKRPVPPPTAEDMK 1 2 GVCTPCSERHDEEFGVPTLEELGQDPRTAGPELYPGGETEALSRLDRHMQRT 0 0 AWVCGFQKPNTEPNALSPSTTVLSPYLKFGCLSARTFWWRLTDVYRG 0 0 0 0 GCTGFPFIDAIMTQLRSEGWVHHLARHAVACFLTRGDLWISWQEGQK 0 0 VFEELLLDADWALNAGNWQWLSASAFFHQYYRVYSPIAFGKKTDKNGDYIR 2 1 KYLPVLKKFPSAYIYEPWKAPRSVQEQAGCIVGKDYPRPIVDHDVVSKKNIQRMKLAYARRAQLGGEQEGTGK 1 2 * 0 >CRY64_danRer Danio rerio (zebrafish) BC044204 6-4 photolyase aka CRY5 0 MSHNTIHWFRKGLRLHDNPALIAALKDCRHIYPLFLLDPWFPKNTRIGINRWRFLIEALKDLDSSLKKLNSR 2 1 LFVVRGSPTEVLPKLFKQWKITRLTFEVDTEPYSQSRDKEVMKLAKEYGVEVTPKISHTLYNIDR 2 1 IIDENNGKTPMTYIRLQSVVKAMGHPKKPIPAPTNEDMR 1 2 GVSTPLSDDHEEKFGIPTLEDLGLDTSSLGPHLFPGGEQEALRRLDEHMERT 0 0 NWVCKFEKPKTSPNSLIPSTTVLSPYVRFGCLSARIFWWRLADVYRG 0 0 KTHSDPPVSLHGQLLWREFFYTTAVGIPNFNKMEGNSACVQVDWDNNPEHLAAWRE 0 0 ARTGFPFIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGQK 0 0 VFEELLLDSDWSLNAGNWQWLSASTFFHQYFRVYSPIAFGKKTDKHGDYIK 2 1 KYLPVLKKFSTEYIYEPWKAPRSVQERAGCIVGKDYPRPIVDHEVVHKKNILRMKAAYAKRSPEDKTINK 1 2 GEKRKASPSIKEMFQKKAKR* 0 >CRY64_strPur Strongylocentrotus purpuratus (urchin) XM_001189626 extra 1st exon unwarranted MCGAPRSYVEIRDSEEHSRRHVARLQFQFQSDLP 12 K 0 MPHNSTIHWFRKGLRIHDNPALLTAIQGTKVFRPIFILDPHFIESEKVGINRWRFLLETLQDLDYSFRALGTR 2 1 LFVVRGNPTTVFPEIFKKWNVTRLTFEVDTEPYARRRDQEVIELAKKNDVEVITKVSHTLYDTER 2 1 TIKANKYKPPMTYQRMVGLLSEIGAPAIPELPPLMANFT 1 2 GVATPVKPDHDNEYGVPSLEDLGLDLEGLGPRLYPGGETEGLQRMDLHLARK 0 0 SWVCGFEKPKTSPNSLEPSTTVLSPYLKWGCLSPRKFYYAIKEVYAQQ 0 0 TNCTKPPVSLMGQLIWREFFYTVAAGTPNFHQMEENPICIQVPWDENPEFLAAWKE 0 0 GRTGYPYIDAIMTQLRTEGWIHHLARHSAACFLTRGDLWQSWVKGQE 0 0 VFDEWLLDADYSLNAANWMWLSSSAFFHQYYRVYSPVVFGKKTDKTGEYIR 2 1 KYIPALNKLPAEYIYEPWTAPRSVQEAAGCIIGRDYPRPIVDHSIVSKRNIGRMKDARACQPGKKA 1 2 EKRPAEPSKQDNNGKKVRKITSMLKKK* 0 >CRY64_droMel Drosophila melanogaster (fruitfly) 6-4 photolyase 3CVW CG2488 uses 5-deazariboflavin 0 MDSQRSTLVHWFRKGLRLHDNPALSHIFTAANAAPGKYFVRPIFILDPGILDWMQVGANRWRFLQQTLEDLDNQLRKLNSRLFVVRGKPAEVFPRIFKSWRVEMLTFETDIEPYSVTRDA AVQKLAKAEGVRVETHCSHTIYNPELVIAKNLGKAPITYQKFLGIVEQLKVPKVLGVPEKLKNMPTPPKDEVEQKDSAAYDCPTMKQLVKRPEELGPNKFPG 1 2 GETEALRRMEESLKDEIW VARFEKPNTAPNSLEPSTTVLSPYLKFGCLSARLFNQKLKEIIKRQPKHSQPPVSLIGQLMWREFYYTVAAAEPNFDRMLGNVYCMQIPWQEHPDHLEAWTHGRTGYPFIDAIMRQLRQE GWIHHLARHAVACFLTRGDLWISWEEGQRVFEQLLLDQDWALNAGNWMWLSASAFFHQYFRVYSPVAFGKKTDPQGHYIRKYVPELSKYPAGCIYEPWKASLVDQRAYGCVLGTDYPHRI VKHEVVHKENIKRMGAAYKVNREVRTGKEEESSFEEKSETSTSGKRKVRRATGSAPKRKR* 0 >CRY64_danPle Danaus plexippus (butterfly) EF117813 PubMed:17244599 MTKVASVIHWFRLDLRLHDNLALRNAINEAENRKQILRPIYVIDPDIKNRVGCNRLRFLFQSLKNLDTSLRKINTRLYVIKGKAIECLPKLFDEWHVKFLTL QVDIDADLVKQDEVIEEFCEANNIFVVKRMQHTVYDFNSVVKKNNGSIPLTYQKFLSLVSDVQVKDIIQISKGVSDECKASDYDSQGYDIPSLEEFGVNESELSECKYPGGESEGL KRLDVYMAKKQWVCNFEKPKSSPNSIEPSTTVLSPYISHGCLSAKLFYHKLKQVENGSKHTLPPVSLMGQLMWREFYYTAGSGTENFDKMVGNSVCTQIPWKKNDAHLKAWAEGKT GYPFVDAIMRQLKQEGWIHHLARHMVACFLTRGDLWISWEEGAKVFEDFLLDYDWSLNAGNWMWLSASAFFYKYYRVYSPVAFGKKTDKDGLYIRKYVPELKKYPSEFIYEPWKAP KGVQKTAGCIIGEGYPNRIVDHDKVHKDNIQKMNAAYKVNKEKKAMKRPRQ* 0
DASH cryptochrome CRY3 sequences
DASH is yet another member of the cryptochrome and photolyase family. It was identified only recently as active only on ssDNA repair, reportedly because of a barrier to flipping the damaged cyclobutane pyrimidine dimer dinucleotide out of dsDNA into its active repair site unless it is in a loop. In the species investigated to date, this enzyme uses folate (MTHF) and FAD, activated by blue light.
Its name is a peculiar acronym of Drosophila, Arabidopsis, Synechocystis and Homo. The gene has been lost in Drosophila or Homo. In Arabidopsis, it is denoted CRY3. The many genome projects available today allow a quick determination of its very unusual phylogenetic distribution.
Although originally studied in plants and cyanobacteria, the DASH photolyase surprisingly extends into fish, frogs, salamanders, turtle, lizard, and birds (duck, finch and budgerigar but not chicken or turkey). It's not clear however if the protein function has stayed the same in all these taxa or drifted in new roles like CRY1 and CRY2 or has acquired multiple functions in single species.
Using blastx on the syntenic region in gallinaceous birds (chicken and turkey) establishes that they have a fairly degenerate multi-exonic pseudogene at the expected location and orientation. It is not currently possible to date this more precisely nor determine whether pseudogenization occurred in a common ancestor or independently on account of domestication.
DASH is also missing from alligator and crocodile assemblies, the lobe-finned coelocanth assembly, and chondrichthyes genomes and transcripts. These probably represent additional, independent gene losses rather than just poor assemblies -- the overall conservation of DASH is much less stringent suggesting loosened constraints or even a measure of functional redundancy.
Platypus does not have any pseudogene debris at the expected location but the assembly has a break here. Marsupials and placentals again appear never to have had this enzyme as it had been lost in the earliest mammals.
The phylogenetic loss pattern of DASH parallels the [Opsin_evolution:_trichromatic_ancestral_mammal|massive loss of opsins] that also occured early in mammalian evolution -- which GT Walls in 1942 [attributed] to mammals experiencing a sustained period of deep nocturnality where these systems did not need to function (no UV damage) and indeed could not function (insufficient blue light even with antenna) and so were lost, implying they were not sustained by a Piatigorskyian secondary functionality such as circadian rhythm or magnetosensing.
The great oxygenation event gave rise to a stratospheric ozone protective layer at 2.4 gyr but reached an even higher lever during the early Cambrian (based indirectly on oxygen levels). This may have favored independent but simultaneous gene loss events in various clades.
However the first land plants and animals risked greatly increased levels of UV damage that may correlate with DASH retention. Here it must be clarified whether repairable damage can arise from direct oxidative damage in addition to ultraviolet light which could not plausibly be an issue for benthic marine organisms that still retain this enzyme.
The amniote proteins can easily be modeled structurally using nearly 50% matches in Arabidopsis (cryptochrome 3: 2IJG) and or equally suitable cyanobacterium Synechocystis (1NP7). The 14 exons share only one match with vertebrate CRY1 and CRY2 which is more likely coincidental than indicative of a common ancestor subsequent to the era of eukaryotic intronation onset.
>DASH_taeGut Taeniopygia guttata (finch) ABQF01044665 ABQF01044669 ABQF01044671 synteny: ACAA1 DASH MYD66 OXSR1 0 MSGTAGTAICLLRCDLRAHDNQ 0 0 QVLHWAQHNADFVIPLYCFDPRHYLGTHCYRLPKTGPHRLRFLLESVKDLRETLKKKGS 2 1 TLVVRKGKPEDVVCDLITQLGSVTAVVFHEE 0 0 ATQEELDVEKGLCQVCRQHGVKIQTFWGSTLYHRDDLPFRPIDR 2 1 LPDVYTHFPKGLESGAKVRPTLRMADQLKPLAPGLEEGSIPTMEDFGQK 1 2 DPVADPRTAFPCSGGETQALMRLQYYFWDT 0 0 NLVASYKETRNGLVGMDYSTKFAPW 2 1 LALGCISPRYIYEQIQKYERERTANESTYW 2 1 VLFELLWRDYFRFVALKYGRRIFSLR 1 2 GLQSKDIPWKKDLQLFSCWQ 0 0 EGKTGVPFVDANMRELSATGFMSNRGRQNVASFLTKDLGLDWRMGAEWFEYLL 0 0 VDYDVCSNYGNWLYSAGIGNDPRDNRKFNMIKQGLDYDGN 0 0 GDYVRLWVPELQGIKGADIHTPWALSSAALSQAGVTLGETYPQPVVTAPEWSRHIHRRP 0 0 GGSPHPRGRRGPAQRKDRGIDFYFSRKKDAC* 0 >DASH_anaPla Anas platyrhynchos (duck) scaffold1769 0 0 0 QVLHWAASNADYLIPLYCFDPRHYAGTHCYGFPKTGPHRLRFLLESVKDLRETLKKKGS 2 1 TLVVRKGKPEDVVRDLITQLGCVGVVAFHEE 0 0 ATQEELDVEKELCQVCEQHGVKVQPFWAATLYHRDDLPFGPIAR 2 1 LPDVYTQFRKAVEAEAKVRPALQLPDQLKPLAPGVEEGCIPTMEDLGQK 1 2 DPGTDPRTAFPCCGGETQALARLQYYFWDT 0 0 NLVASYKETRNGLIGMDYSTKFAPW 2 1 LALGCISPRYIYEQLRKYEKERGANDSTYW 2 1 VLFELLWRDYFRFVALKHGRRIFSVR 1 2 GLQSKEVPWKKDLQLFDCWK 0 0 EGKTGVPFVDANMRELAATGFMSNRGRQNVASFLTKDLGLDWRMGAEWFEHLL 0 0 VDYDVCSNYGNWLYSAGVGNDPRDKRKFNVIKQGLDYDGN 0 0 GDFVRLWLPELQGIAGADAHAPWALSSTALRQAGIILGQTYPQPLVRAPEWSRHINQRL 0 0 GRSPHPRGRRGPADAPAQRKDRGIDFYFSRKKDE* 0 >DASH_melUnd Melopsittacus undulatus (budgerigar) AGAI01061648 0 msgtSARTAICLLRCDLRAHDNQ 0 0 QVLHWAQSNAEFVVPLYCFDPRHYLRTHWYGFPKTGPYRLRFLLESVKDLRETLKKKGS 2 1 TLVVRKGKPEDVVRDLITQLGSVSAVVFHEE 0 0 ATQEELDVENELCQVCSQHGVKVYTFWASTLYHRDDLPFRPIAR 2 1 LPDVYTHFRKAVESEAKVRPTLRMADQLQPLAPGVEEGCIPTMEDFGQK 1 2 DPDTDPRTAFPGSGGETQALMRLQYIYFWDT 0 0 NLVASYKEVRNGLVGMDYSTKFAPW 2 1 LALGCISPRYIYEQIKKYERERTANQSTYW 2 1 VLFELLWRDYFRFVALKYGRRIFSLR 1 2 GLQSKEVPWKKDLQLFNCWK 0 0 EGKTGVPFVDANMRELAATGFMSNRGRQNVASFLTKDLGLDWRMGAEWFEYLL 0 0 VDYDVCSNYGNWLYSAGVGNDPRDNRKFNMVKQGLDYDGN 0 0 GDYVRLWVPELQGIKGADIHTPWALSSAALSQAGVTLGETYPQPIVTAVEWSRHISQRP 0 0 GRSLHPRGSRRPAHSPVQHKDRGIDFYFSRKKDA* 0 >DASH_galGal Gallus gallus (chicken) syntentic pseudogene, numerous indels, frameshifts, internal stops 0 0 0 2 1 MVVRKGKLEDMVHDVITSLGCVGVVAFHEE 0 0 2 1 IQPDVYTNF*KAVESEVKTWPALQRADQLKPLAGG&^EEGCIPTTEDLGQK 1 2 DPGTDPQTAF-LGFNETAAL 0 0 LLFASYNETWSGLIGVD*STKFAPW 2 1 LVLGCLSP*YIFSQIGKYVKERAANHST*W 2 1 AVFELLWWDCF*FVALKYGRRILSLK 1 2 0 0 LGLDWRMGAEWFEYLL 0 0 VDYNVFSN*STWLYSAGIGNHPKDNRKFNVIKQGPDYDGN 0 0 GDCIQLRVTELQGMKEADIHSPWALNSATLSQVGVTLGKTYPQVQSDSP^EWSRQIRQRA 0 0 GRSPHPKGRRGPAHSPLQHKERGTDCFSLKK* 0 >DASH_melGal Meleagris gallopavo (turkey) ADDD01036185 syntenic pseudogene 0 0 0 2 1 0 0 2 1 DVYTNL*KAVESEVKVWPALQRADQLKPLA-GSGRKMYPYNKGFG 1 2 0 0 LFASYKEIWSGLTGADNSTKFAPW 2 1 LVLDCLSP*CIFSQVQKYIKERAANQSTFW 2 1 VFELLWWDCF*FVPLKYGRRIFSL 1 2 0 0 LKVDYNVPNN*VT*LYSADIGNHPKENRKFNMIKEGPNDDG 0 0 VPELQGIKGPDIHSPWALNSASVFQVGGTLVETYPQPAV 0 0 * 0 >DASH_anoCar Anolis carolinensis (lizard) XM_003221869 14 exons 0 MASSLPRTALCLLRNDLRCHDNE 0 0 VLHWAQSHADRIVPLYCFDPRHYAQTYHYNFPKTGPFRLRFLLESVKDLRETLKKKGS 2 1 NLVVRKGKPEDVVRDLIIQLGSVASVSFHEE 0 0 ATKEELDVEKALIRVCTEHGVEVQTFWGSTLYHRQDLPFKHISQ 2 1 LPDVYTQFRKAVESQARVRPVLKMEGQMKSLPPEIEEGNIPSLEDFGMT 1 2 EPLRDPRSAFLCSGGETQALMRLQHYFWET 0 0 NLVASYKETRNGLIGLDYSTKFAPW 2 1 LALGCISPRYIYDQIQKYEKERTANQSTYW 2 1 VLFELAWRDYFRFVALKYGRKIFFPR 1 2 GLQDKDISWKKDPKLFDAWK 0 0 EGRTGVPFVDANMRELATTGFMSNRGRQNVASFLTKDLGLDWRMGAEWFESLL 0 0 VDYDVCSNYGNWLYSAGIGNDPRENRKFNMIKQALDYDGN 0 0 GDYARLWVPELQCIRGGDVHTPWALSNGALSQAGVSLGSSYPVPIIKAPEWGRHINQKPVSM 0 0 SNQGPKGRKGPSHTPKQHKNRGVDFYFSHKKDLS* 0 >DASH_chrPic Chrysemys picta (turtle) AHGY01416294 first exon off contig 0 0 0 VLRWAQNNADYVIPLYCFDPRQYLRTYYYGFPKTGPHRLKFLLESVKDLRQTLKKKGS 2 1 NLVVRKGNPEDVVHDLITQLGSVTAVAFHEE 0 0 ATKEELDVEKALQRVCVQHGVKIQTFWASTLYHRDDLPFRHISR 2 1 LPDVYTQFRKAVEAQAKVRPTVQMPDQLKPLAPGVEDGCIPVPEEFGQK 1 2 DPLTDPRTAFPCCGGETQALMRLQHYFWDT 0 0 NLVASYKETRNGLIGIDYSTKFAPW 2 1 LALGCISPRYIYEQIRKYEKERTANQSTYW 2 1 VIFELLWRDYFRFVALKYGTRIFLLK 1 2 GLQDKEVPWKKDPKLFDAWK 0 0 EGRTGVPFVDANMRELAATGFMSNRGRQNVASFLTKDLGLDWRMGAEWFEYLL 0 0 VDYDVCSNYGNWLYSAGIGNDPRENRKFNMIKQGLDYDSN 0 0 GDYVRLWVPEIQCVKGGEIHTLWALSNAALSQAGIALGETYPLPVVLAREWSRHINQKP 0 0 RDHPPRGRKDPSDTPRQHKDRGMDFYFSRNKDLS* 0 >DASH_xenTro Xenopus tropicalis (frog) XM_002938001 PubMed:15147276 synteny: ACAA1 DASH MYD66 transcripts AL790297 CR419606 etc 0 MAARARVIICLLRNDLRLHDNE 0 0 VLHWAHRNADQIVPLYCFDPRHYGGTHYFNFPKTGPHRLKFLLESVQDLRNTLKERGS 2 1 NLLLRRGKPEEIIAGLVKQLGNVSAVTLHEE 0 0 ATKEETDVESAVRRVCTQLGVRYQTFWGSTLYHREDLPFRHISS 2 1 LPDVYTQFRKAAETQGKVRSTFQMPDRLKPLPSGLEEGSVPTHQDFDQQ 1 2 DPLTDPRSAFPCCGGETQALQRLHHYFWET 0 0 NLVASYKDTRNGLIGIDYSTKFAPW 2 1 LALGCISPRYIYEQIRKYEKERTANQSTYW 2 1 VIFELLWRDYFRFVALKYGRRIFFLR 1 2 GLQDKDVPWKKDPKLFDAWK 0 0 EGRTGVPFVDANMRELAMTGFMSNRGRQNVASFLTKDLXIDWRLGAEWFEYLL 0 0 VDYDVCSNYGNWLYSAGIGNDPRENRKFNMIKQGLDYDAG 0 0 GDYIRLWVPELQQIKGGDAHTPWALSTASLAHSNVSLGETYPYPIVMAPEWSRHINQKPSGS 0 0 WETSARRGKGPSHTPKQHRNRGIDFYFSRNKNV* 0 >DASH_hymCut Hymenochirus curtipes (frog) 1 QMPKLKPLPDGLEEGKVPTYEEFDQE 1 2 DPLTDPRTAFPCSGGETQALQRLHHYFWDT 0 0 NLVPPYKDTRNGLIGLDYSTKLAPW 2 1 LALGCVSPRFIYEQIKKYEQERTANQSTYW 0 0 VIFELLWRDYFRFVALKYGRRIFFLR 1 2 GLAHTNV >DASH_ambMex Ambystoma mexicanum (axolotl) CO785483 0 MSVQARTIICLLRNDLRFHDNE 0 0 VLLWALKNAERIVPLYCFDPRHYVGTHNYNFPKTGPHRLKFLLESVKDLRETLKKKGS 2 1 NLLVRKGKPEDVIQELLKQLGSVSAVAFHNE 0 0 VTKEELDVEAAVKQVCvqhgGKIQTFWGSTLYHRDDLPFRHISR 2 1 LPDVYTQFRKGVESQGTSAATLQMPQKVPPLPSGLE >DASH_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01010414 0 MSTIRTIICLLRNDLRFHDNE 0 0 vfhHWAQGNAEQIVPLYCFDPRHYLGTHCYNFPKTGPFRLRFLLESVKDLRDTLKKNGS 2 1 NLLVKRGKPENVVSDLIKQLGSVTAVAFHEE 0 0 VTKEEQDVETEVTRVCAQFKVRVHTCWGSTLYHREDLPFNHIAR 2 1 LPDVYTQFRKAVETQSQVRPTIRMPEQLKPFPHNLEEGPIPTPEDLGQP 1 2 DAVSDPRTAFPCIGGETQALARLKHYFWDT 0 0 NLVASYKETRNGLIGMDYSTKFAPW 2 1 LALGCISPRYIYEQIKQYEKERTANQSTYW 2 1 VIFELLWRDYFRFVAVKYGNRIFTVK 1 2 GLQQKSVSWKKDVKLFDAWK 0 0 EGKTGVPFVDANMRELALTGFMSNRGRQNVASFLTKDLGIDWRMGAEWFEFLL 0 0 VDYDVCSNYGNWLYSAGIGNDPRENRKFNMIKQGLDYDNN 0 0 GDYVRLWVPELRGLKGGDVHAPWVLSSASLADAEVSLNQTYPVPMVVAPEWSRHTTSKA 0 0 SGGGLSQKGKRGPSHTPKQHRDRGIDFYFSRNNKKLQ* 0 >DASH_danRer Danio rerio (zebrafish) NM_205686 0 MSASRTVICLLRNDLRLHDNE 0 0 VFHWAQRNAEHIIPLYCFDPRHYQGTYHYNFPKTGPFRLRFLLDSVKDLRALLKKHGS 2 1 TLLVRQGKPEDVVCELIKQLGSVSTVAFHEE 0 0 VASEEKSVEEKLKEICCQNKVRVQTFWGSTLYHRDDLPFSHIGG 2 1 LPDVYTQFRKAVEAQGRVRPVLSTPEQVKSPPSGLEEGPIPTFDSLGQT 1 2 EPLDDCRSAFPCRGGETEALARLKHYFWDT 0 0 NAVATYKETRNGMIGVDFSTKFSPW 2 1 LALGCISPRYIYEQIKKYEVERTANQSTYW 2 1 VIFELLWRDYFKFVALKYGNRIFYMN 1 2 GLQDKHVPWKTDMKMFDAWK 0 0 EGRTGVPFVDANMRELALTGFMSNRGRQNVASFLTKDLGLDWRLGAEWFEYLL 0 0 VDHDVCSNYGNWLYSAGIGNDPRENRKFNMIKQGLDYDNN 0 0 GDYVRQWVPELRGIKGGDVHTPWTLSNSALSHAQVSLNQTYPCPIITAPEWSRHVNNKS 0 0 SGPSSSKGRKGSSYTARQHKDRGIDFYFSKNKHF* 0 >DASH_oreNil Oreochromis niloticus (tilapa) XM_003439198 0 MSSSRTVICLLRNDLRLHDNE 0 0 LFHWAQRNAEHIVPLYCFDPTHYVGTYNYSLPKTGPFRLRFLLEGIRDLRNTLINKGS 2 1 NLVVRRGKPEEVVADLIRQLGSVSSVAFHEE 0 0 VTSEELNVEKRVKDVCAQMKVKVHTCWGSTLYHRDDLPFPHMSR 2 1 LPDVYTQFRKAVESEGRVRPVFSTPEKLNPLPPGLEEGAIPTAEDLQQT 1 2 EPVTDPRSAFPCGGGESQALARLKHYFWDT 0 0 DAVATYKETRNGLIGVDYSTKFSPW 2 1 LAMGCISPRYIYHQIKQYEKERTANQSTYW 2 1 VIFELLWRDYFRFVSVKYGNRIFQVK 1 2 GLQDKSVPWKKDMKLFDAWK 0 0 EGRTGVPFVDANMRELAATGFMSNRGRQNVASFLTKDLGLDWRMGAEWFESLL 0 0 IDHDVCSNYGNWLYSAGIGNDPRENRKFNMIKQGLDYDNN 0 0 GDYVRGWVPELQNIRGGDVHTPWALSTAVLAHAHVTLGDTYPAPIVTAPEWSRHVNKKAGGT 0 0 GPSPRGKKGPSHTPKQHRDRGIDFYFSRSKNL* 0 >DASH_patPec Patiria pectinifera (starfish) HP101597 0 MAGRMKIIICLLRNDLRYRDNE 0 0 VLHWAHQNADILPLYCLDPRHYHGTWHFSFPKTGPHRLKFLLESLRDLKETLRQVGS 2 1 NLVISRGKPETVIQDLIRQLGQDNVKAVALQQE 0 0 ATREELDVEAAIEKSCQVPVHKFWGSTLYHKDDIPFRIRD 2 1 LPNVFTEFRKKTENQSDVRPTLPMPSKLKPLPPGIDPGHAPSVEEMGMK 1 2 DPASDERTAFPYSGGEKQALARIQHYFWDY 0 0 DYVSSYKETRNGLIGGDYSTKFSSW 2 1 LALGCLSPRKIYEEIRKYEKERRANQSTYW 2 1 VVFELLWRDYFKFVALKFGDRLFYLS 1 2 GLLGNHREWRENKALFDAWR 0 0 EGRTGVPFVDANMRELAATGFMSNRGRQNVASFLTKDLKLDWRLGAEWFEHLL 0 0 IDHDVCSNYGNWLYCAGVGNDPRQDRKFNMIKQGLDYDAQ 0 0 GDYVRLWVPELQGIKGGAVHTPWILSSSMLIQANVRLGHTYPLPVVKAPEWNKHMRP 0 0 QERGKSGKGPGQRQQTRGIDFYFSSGKQGKFKK* 0 >DASH_strPur Strongylocentrotus purpuratus (urchin) 0 MAGRMKIIICLLRNDLRYRDNE 0 0 VLFWAHKNATNVIPLYCFDPRHYKGTHQFGFPKTGPHRLKFLLESVRDLRTTLQSVGS 2 1 GLVVRSGNPEDVVPQLIQQFGEGEVAAVALQEE 0 0 ATREELDVEAGLHQSCSKLGVNVKKFWGSTLYHREDVPFSPG 2 1 VPNVYTEFRKKVENRSHVRPPINMPKPLKGLPEGVEEGDIPVFSSFGLK 1 2 DPGSDSRTAFPFQGGETTGLARIEDYFWKS 0 0 DCIAKYKETRNGLIGSEYSTKFAAW 2 1 LAHGCVSPRQIHAEVKRYEKERTENQSTYW 2 1 VIFELIWRDYFKFVALKFGDKMFYLS 1 2 GLLGVHKPWNKNKQWFDAWC 0 0 KGQTGVPFVDANIREMAATGFMSNRGRQNVASFLTKDLVLDWRLGAEWFEQML 0 0 VDHDVTSNYGNWLYSAGVGNDPRQDRKFNMIKQGLDYDPN 0 0 GDYIRLWVPELSGIKGGSIHMPWTLSSAQLNQAGVSLGETYPSPIVTAPEWSRHSKRP 0 0 qsgrggaggsqgggsqgGGRNQSGKGQRRNQGPPRGQQRGLDFYFSNPSQK* 0 >DASH_nemVec Nematostella vectensis (sea_anemone) XP_001623243 ABAV01026885 0 MATSKTQKTVICLLRNDLRYHDNEALLWAHKNADFVLPLYCFDPDHYKTTWRFSLPKTAQYRAKFLLESVTDLRSTLQIHGSNLIIRQCQPLEAVTKLTE LLKPVAPVTSIVFQQEVTYEELNVEKALVEFCKKSGIHMHTIWGSTLFHKDDIPYKAKTVPDTYTQFRKGVENQSTVRNLIDMPKNLKPLPPVKGELGTI PDLKSLLNDSEIKEVDQRSAFPFMGGEQEALSRLGSYLWGTDSVAKYKETRNGLLGENYSTKLSPWLANGSLSPRMVYHRIKQYEEERVANHSTYWVLFE LIWRDYFKFVCLKYGDRVFYRSGIMGKSLPWKHDKMTFKLWCEGKTGVPFVDANMRELKETGWMSNRGRQNVASFLIK 0 0 DLGLDWRYGAEWFESLLLDHDVCSNYGNWNYAAGIGNDPREGRKFNMVKQGLDYDPDGDYIKTWVPELAKIPGAKVHVPWTLKPGELR 2 1 SSGVELGVTYPNPIVIAPEWARHTSKTISGKKPSNFNPRQKGVDFYFKSANNGPSGSR* 0 >DASH_hydMag Hydra magnipapillata (cnidarian) XM_002166508 single exon ABRM01055505 MAGFSSRKTIIYWIRNDLRLHDNECLNFACRDGSHVLPLYCFDPDHYKLTHHFGFLKTGIHRAKLILDAVSGLRNSLQKKGSNLVISKHRPKEAIEKLIL ECEQTAPVTSIVFQKEITKEETDVEASVFKVCDSKGIKVLSFWGSTLYHIEDIPFQINQIPNTYTLFRKGVEKCPVRKLNPIPDKISSLPNLKNFPLGEV PLLNELGFDKNEYEIDVRSVFPFNGSEVDAIKRLNEYLWGNNAILTYKETRNGLLGKDYSTKFSPWLALGSLSPRMIYHKIKDFEEQKKIKENDSTYWVV FELIWRDYFKFICSKHGNKVFFKGGLKGGKGPWTWKQDLNLFMKWANGDTGIPFVDANMKELLATGWMSNRGRQNVASFLVKDLNLDWRMGAEWFESLLI DSDVCSNYGNWNYIAGIGNDPRSNRKFNVIKQAADYDEEGDYVRTWLPVLKKLGICHTPWRLSASELNAGGVDLGKNYPHPIIVAPEWAKHALKMNSKNC FHEHRGKQNGQKRSIDFYFKSGKTGE* 0 >DASH_monBre Monosiga brevicollis (choanoflagellate) XP_001745157 ABFJ01000402 0 MAKAGNPRPVVVWFRNDLRVHDNEVLLQAAK 0 0 ASHNHVVPVYCFDIRQ 1 2 YSLVITHRSRRCGQFPKCGRPRARFLIESVDDLRTRLQELGSGLVVRTGLPEEEVARVAAQVGATQVFAHQEVCSEEVAAEHRLKRQLEVPLSLHWGAVTLCHLDDLDFGPRCKHLPSVFTQFRKRVEADMHVRPVVAAPARLAPLPSDLELGSIPTVEDLCPGQH 0 0 EPDERAVLPFKGGETAARARLQYYLWESNLLAS 2 1 YKDTRNGLVGGDYSSKFSPWLAHGNLTARWIYHE 0 0 VKRYEQERTENTSTYWLIFELLWRDYFRFVALQHGTAIFKP 1 2 GGVQHKDVPWRHSPADFEAWQNGQTGFPFIDANMRELAKTGFMSNRGRQNVASFLTK 0 0 DLQIDWRLGAEWFETLLLDHDPCSNY 1 2 GNWNYAAGVGNDPRQGRHFNVIKQ 1 2 AKTYDPTAEYVHLWVPELRGLDAPQAHVPFK 0 0 LSSEELAQANIQLGSTYPRPVVDRLDGGRLPLKVRDETAYGPHVQLPQQGLIKMILFDTLFTGLSW* 0
Cryptochrome CPD photolyases
This dna repair enzyme was studied in marsupials during the pre-genomic era (1994), with two groups concluding even that that no ortholog existed in placentals. That is still the case today despite several dozen complete placental mammal genomes. Further, no pseudogene debris exist in the partly conserved syntenic location. This strongly suggests that the gene was lost once in stem placental rather than many times in later subclades (as happened with encephalopsin). The gene remains very strongly conserved in species such as opossum in which it is still present today.
This loss in placentals is somewhat peculiar given that CPD is a very ancient (pre-eukaryotal) member of the photolyase family, with highly conserved orthologs readily recoverable in other commonly studied marsupials, monotremes, birds, alligators, turtle, lizard, snakes, frog, fish, agnathan, amphioxus and sea urchin. However it also appears to be lost in tunicates -- indeed Ciona has lost all its photolyases leaving it a bit mysterious how it repairs these types of dna damage.
It is very unlikely that placentals evolved something better. More likely, they lost CPD along with most other photoproteins during a dark phase of their evolution when UV damage to their dna was a non-issue. Coming back out into the light millions of years later, they evidently make do with a less efficient excision repair that overlaps repair photolyase functionality.
The CPD gene product is very diverged from other photolyases though still retaining weak homology along its photolyase and FAD bindng domains. The antenna moiety is usually reported as MTHF folate. The best available structres are from rice (3UMV: 53% identity) and an archael methanogen (Methanosarcina mazei 2XRZ) which likely uses 5-deazariboflavin Fo as antenna (which it can synthesize de novo). The latter enzyme repairs cyclobutane pyrimidine dimers in duplex DNA using blue or near-UV light.
Despite great divergence in primary sequence, the fold itself is largely conserved. That may explain in part the unexpected circadian compensatory capacity of marsupial CPD expressed in double CRY knockout mouse, seemingly driven by interaction of CPD with CLOCK of the CLOCK/BMAL1 system. CPD lacks any counterpart to the distal exons of CRY1.
>CPD_monDom Monodelphis domestica (opossum) NP_001028149:wrong OPC1 PubMed:7937136 synteny: TNK1 MUC4 CPD KIAA0226 FYTTD1 0 MAPKKRSLGDGDEQEKTETQENKTKRKPLQKHQFSKSNVVQKEEENKREGEKKGGAEGLQEVVMQSRLQTASSVLEFRFNKQRVRLISQDCHLQDHSQAFVYWMSRDQRVQ 1 2 DNWAFLYAQRLALKQKLPLHVCFCLAPCFLGATIRHYDFMLRGLEEVAE 0 0 ECEKLHIPFHLLLGLPKDVLPAFVQAHSIGGIVTDFSPLLHHTQWVKDVQDGLPKQVPFVQ 0 0 VDAHNIVPCWIASDKQEYGARTIRHKIHDRLPHFLTEFPPVICHPYPSNIQAE 0 0 PVDWNACRAGLQVDRSVKEVSWAKPGTASGLTMLQSFISQRLPYFGSDRNNPNKDALSNLSPWFHF 1 2 GQVSVQRAILEVQKHRSRYPDSVANFVEEAVVRRELADNFCFYNKNYDKLE 1 2 GAYDWAQTTLRLHAKDKRPHLYSLEQLESGKTHDPLWNAAQ 0 0 MQMVQEGKMHGFLRMYWAKKILEWTRSPEEALEFAIYLNDRFQLDGRDPNGYV 1 2 GCMWSICGIHDQGWAEREIFGKIRYMNYAGCKRKFDVAEFERKYSPAD* 0 >CPD_sarHar Sarcophilus harrisii (tasmanian_devil) AEFK01107967 0 MAPKKRSHGTGSEPEKTDSQESKAKRKPLQKHPSSKNNVVQPEREDKKEGQKRAGAEGLQEAVRQARLRTAPSVLEFRFNKQRVRLISQDCHLQDHSQAFVYWMARDQRVQ 1 2 DNWAFLYAQRLALKQKLPLHVCFCLAPCFLGATLRHYDFMLRGLEEVAE 0 0 ECEKLCIPFHLLLGLPKDVLPPFVQKHSIGGIVTDFSPLVHHTQWVKDVQDALPRQVPFVQ 0 0 VDAHNIVPCWVASDKQEYGARTIRHKIHDRLPHFLTEFPPIICHPYPSSIQAE 0 0 PVDWNACRAGLQVDRSVKEVSWAKPGTAFGLTMLQSFIAERLPYFGSDRNNPNKDVLSNLSPWFHF 1 2 GQVSVQRAILEVQKHRGRYPDSVANFVEEAVVRRELADNFCFYNKNYDKLE 1 2 GAYDWAQTTLRLHAKDKRPHLYSLEQLESGKTHDPLWNAAQ 0 0 MQMVKEGKMHGFLRMYWAKKILEWTHSPEEALEFAIYLNDRFQLDGRDPNGYVG 1 2 GCMWSICGIHDQGWAEREIFGKIRYMNYAGCKRKFDVAEFERKYSPVD* 0 >CPD_potTri Potorous tridactylus (rat kangaroo) D26020 PubMed:7813451 0 MDSKKRSHSTGGEAENMESQESKAKRKPLQKHQFSKSNVVQKEEKDKTEGEEKGAEGLQEVVRQSRLRTAPSVLEFRFNKQRVRLISQDCHLQDQSQAFVYWMSRDQRVQ 1 2 DNWAFLYAQRLALKQKLPLHVCFCLAPCFLGATIRHYDFMLRGLEEVAE 0 0 ECEKLCIPFHLLLGLPKDVLPAFVQTHGIGGIVTDFSPLLHHTQWVKDVQDALPRQVPFVQ 0 0 VDAHNIVPCWVASDKQEYGARTIRHKIHDRLPHFLTEFPPVICHPYTSNVQAE 0 0 PVDWNGCRAGLQVDRSVKEVSWAKPGTASGLTMLQSFIAERLPYFGSDRNNPNKDALSNLSPWFHF 1 2 GQVSVQRAILEVQKHRSRYPDSVTNFVEEAVVRRELADNFCFYNKNYDKLE 1 2 GAYDWAQTTLRLHAKDKRPHLYSLEQLESGKTHDPLWNAAQ 0 0 MQTVKEGKMHGFLRMYWAKKILEWTRSPEEALEFAIYLNDRFQLDGWDPNGYV 1 2 GCMWSICGIHDQGWAEREIFGKIRYMNYAGCKRKFDVAEFERKISPAD* 0 >CPD_ornAna Ornithorhynchus anatinus (platypus) 0 1 2 0 0 ECEKLCIPFHLLLGLPKDVLPAFVQTHGIGGIVTDFSPLLHHTQWVKDVQDALPRQVPFVQ 0 0 VADNWAFLYAQRLALQRKLPLHVCFCLVPSFLGATIRHFGFMLGGLKEVAK 0 0 1 2 GQVSVQRAILEVRKQRHRYPDSVASFVEEAMVRRELADNFCFYNENYDKVE 1 2 DTLWNAAQ 0 0 LQMVQEGKMHGFLRMYWAKKILEWTRSPEEALEFAIHLNDRFQLDGRDPNGYV 1 2 GCMWSICGIHDQGWAEREVFGKIRYMNYAGCKRKFDVAQFEHKYSPRE* 0 >CPD_taeGut Taeniopygia guttata (finch) XM_002190577 0 1 2 DNWAFLYAQRLALKQELPLHVCFCLVPKFLGATIRHYRFMLRGLQEVAE 0 0 ECAELNIPFHLLLGYAKDVLPTFVVEHGVGGLVTDFSPLRLPRQWVEDVKERLPEDVPFAQ 0 0 VDAHNIVPCWVASPKQEYSARTIRGKIHAQLPEFLTEFPPVVPHPHLPSCPAE 0 0 PIAWEACYSSLQVDHTVKEVEWATPGTAAGMAVLKSFIAERLKSFSTHRNDPNKAALSNLSPWLHF 1 2 GQVSTQRAILEVQKQRRNYKDSVDAFVEEAVVRRELAENFCYYNENYDSVQ 1 2 GAYDWAQTTLKLHAKDKRPYLYSLQELEQGTTHDPLWNAAQ 0 0 LQMVQEGKMHGFLRMYWAKKILEWTRSPEEALQFAIYLNDRYELDGRDPNGYV 1 2 GCLWSICGIHDQGWAERPIFGKIRYMNYAGCKRKFDVDQFERRYTPTHSQ* 0 >CPD_melUnd Melopsittacus undulatus (budgerigar) AGAI01046895 0 MLRRQRKRKASAPEEIPRVSRKRTEQEEELHEARRRAAPSVREFKFNKKRVRLISQASDLKDGGRCILYWMSRDQRLQ 1 2 DNWAFLYAQRLALKQELPLCVCFCLVPKFLEATIRHYGFMLRGLQEVAE 0 0 ECAELNIAFHLLRGYAKDVLPAFVEEHGVGGLVTDFSPLRLPWQWVQDVREWLPEDVPFAQ 0 0 VDAHNIVPCWVASPKQEYSAYTIRRKIQAQLPEFLTEFPPVVRHPYSPSCPAE 0 0 PIAWEACYSSLEVDRTVKEVEWATPGTAAGLAVLQSFIRERLKYFGSHRNDPNKAALSNLSPWFHF 1 2 GQVSTQRAILEVQKHQHKYKESVDMFVEEAIVRRELAENFCYYNENYDSVQ 1 2 GAYPWAQTTLKLHAKDERPFLYKLQELEQGTTHDPLWNAAQ 0 0 LQMVREGKMHGFLRMYWAKKILEWTHSPEEALQFAIYLNDRYELDGMDPNGYV 1 2 GCLWSICGIHDHGWTERAVFGKIRYMNYAGCKRKFDVRQFERRYAPCTLSQ* 0 >CPD_galGal Gallus gallus (chicken) XM_422729 0 MPRGNGKGRKERDAGREEEEAVGTLEAAVREARRRTAPSVRDFRYNKQRARLVSRGSELKEGAECILYWMCRDQRVQ 1 2 DNWAFLYAQRLALKQELPLRVCFCLVPAFLDATIRHYGFMLRGLREVAK 0 0 ECAELDIPFHVLLGCPKDVLPSFVVEHGVGGLVTDFCPLRVPRQWVEEVKERLPEDVPFAQ 0 0 VDAHNIVPCWVASPKQEYSARTIRAKIHSQLPEFLTEFPPVIRHPHPPPNPPE 0 0 PIAWDACYSSLQVDRTVTEVAWATPGTAAGLAMLQSFITERLKSFGSQRNDPNKAALSNLSPWFHF 1 2 GQVSTQRAILEVQKHRRVYKESVDAFVEEAVVRRELAENFCYYNENYDSVR 1 2 GAYDWAQSTLKLHAKDKRPFLYKLPQLEQATTHDPLWNAAQ 0 0 LQMVREGKMHGFLRMYWAKKILEWTRSPEEALQFAIYLNDRYELDGMDPNGYV 1 2 GCLWSICGIHDQGWKERDVFGKIRYMNYAGCKRKFDVDQFERRYAHCK* 0 >CPD_melGal Meleagris gallopavo (turkey) XM_003209143 0 MPRGSGKGRKERDAGREKEEAEAGAVGSLEAAVREARSRAAPSVRDFRYNKQRARLVSRGSELKEGAECILYWMCRDQRVQ 1 2 DNWAFLYAQRLALKQELPLRVCFCLVPTFLDATIRHYGFMLRGLREVAE 0 0 ECAELDIPFHLLLGCPKDVLPSFVVEHSVGGLVTDFCPLRVPRQWVEEVKERLPEDVPFAQ 0 0 VDAHNIVPCWVTSPKQEYSARTIRAKIHSKLPEFLTEFPPVIRHPHPPPNPPE 0 0 PIAWDACYSSLQVDHTVTEVAWATPGTAAGLAMLQSFITKRLKSFGSQRNDPNKAALSNLSPWFHF 1 2 GQVSTQRAILEVQKHRCAYKESVDAFVEEAVVRRELAENFCYYNENYDSVR 1 2 GAYDWAQSTLKLHAKDKRPFLYELPQLEQATTHDPLWNAAQ 0 0 LQMVQEGKMHGFLRMYWAKKILEWTRSPEEALQFAIYLNDRYELDGMDPNGYV 1 2 GCLWSICGIHDQGWKERDVFGKIRYMNYAGCKRKFDVDQFESRYTHCK* 0 >CPD_allMis Alligator mississippiensis (alligator) genome/blat 0 MFPKKQRQAAGADTEPSGSRKRGRQDAEVVPGAESLAEAVKQARRRAALSVREFKFNKKRVRLLSQESHLQDGALGILYWMSRDQRVQ 1 2 DNWAFLYAQRLALKQQLPLHVCFCLVPKFLDATIRHFGFLLKGLQEVAE 0 0 ECGELGIPFHLLQGYAKDVLPAFVTRHGIGGVVTDFSPLRVPLQWVEDVKERLPQGVPLVQ 0 0 VDAHNIVPCWVASNKQEYGARTIRPKIHAQLPEFLTEFPPIIQHPFPAAAPAQ 0 0 PIDWDACYAGLEVDRTVPEVKWATPGTAAGLAVLQAFVTRRLPGYSEYRNNPNKAALSNLSPWFHF 1 2 GQVSVQRAVLEVQKSRDRHRESVNSFVEEAVIRRELADNFCFYNKSYDQLE 1 2 GAHDWARDTLILHARDKREYLYDLHQLEQGKTHDQLWNAAQ 0 0 IQMVREGKMHGFLRMYWAKKILEWTRSPEDALRFAIYLNDRFELDGRDPNGYV 1 2 GCMWSICGTHDQGWAERNVFGKIRYMNYAGCKRKFNVGQFEHKYRP* 0 >CPD_chrPic Chrysemys picta (turtle) AHGY01112360 incomplete 0 MSQKSRNRQARGGEEISGDAARPGAAAAPRGQLKGKRKVLAEPGGAEVLTVEKRRKEGEAAEGGGAGSLGEAVRQARLQVAPSVQDFKYNKKRVRLVSQGSDLKDAAQGVVYWMSRDQRVQ 1 2 DNWAFLYAQRLALKQQLPLHVCFCLVPKFLEATIRHFGFMLKGLQEVAE 0 0 ECQELDIPFHLLIGFAKDVLPAFVTGRGLGGVVTDFSPLRVPMQWVEDVKERLPGDVPFVQ 0 0 VDAHNIVPCWVASDKQEYGARTIRRKIHDQLSEFLTEFPPLIKHPYPSTAPAE 0 0 PIDWNACRASLQVDCSVKEVTWATPGTAAGLAVLESFIGERLKSFGTDRNNPNR------------ 1 2 ---------------------SVDGFIEEAVIRRELADNFCYYNRNYDKVEG 1 2 GAYDWAKTSLKLHAQDKRSHLYELKQLEEGKTHDPLWNAAQ 0 0 LQMVHEGKMHGFLRMYWAKKILEWTRSPEEALEFTIYLNDRYELDGRDPSGYV 1 2 GCMWSICGIHDQGWAERAVFGKIRYMNYAGCKRKFDVGQFERRY-------* 0 >CPD_anoCar Anolis carolinensis (lizard) XM_003226963 0 MPQKRKKRKEPASGDCENQVEGDGGTSFGSESGDKKTEGKPEEKKPKKTAAVAKGPPLTEAVKASRLKMAPSIAEFHFNKKRVRMISENSDLKEGAQGILYWMSRDQRVQ 1 2 DNWAFLYAQRLALKQNLPLHVAFCLVPKFLEATIRQFGFMLKGLKEVAE 0 0 ECQELSIPFHLLIGFAKDTLPPFVAKHNIAGVVTDFAPLRVPMQWVQEVQERLPPDVPFVQ 0 0 VDAHNVVPCWVASEKQEYGARTIRRKIHDRLCEFLTGFPPVIQHPHQAASQAE 0 0 PIDWDACSASLEVDRSVPEVEWAKPGSAAGLLVLGEFVQERLKFFSSDRNNPNRAALSNLSPWFHF 1 2 GQVSVQRAILEVRKHRTRCRESVEAFIEEALIRRELADNFCFYNPNYDKVE 1 2 GAYEWAKTTLKLHATDKRPHLYTLKQLEEGKTHDQLWNAAQ 0 0 LQMVHEGKMHGFLRMYWAKKILEWTNSPEEALKFSIYLNDRYELDGRDPNGYV 1 2 GCMWSVCGIHDQGWAERAVFGKIRYMNYAGCTKKFDVGQFERKYNPRKIAY* 0 >CPD_pytMol Python molurus (python) 0 YNKKRVRLVSQAPELKEGAQGILYWMSRDQRVQ 1 2 DNWAFLYAQRLALKQRLPLHVCFCLVPKFLDATIRHFGFMLRGLEEVAE 0 0 ECRTLDIPFHLLIGYAKDTLPPFVAQRGIGGVVTDFSPLRLPMKWIQEVLEQLPSDVPFAQ 0 0 VDAHNIVPCWVASDKQEYGARTIRRKIHDQLAEFLTEFPPVIKHPFPVGLQEE 0 0 PISWDACYSSLQVDCSVKEVDWAKPGAASGLKVLEEFVKERLKHFASGRNNPNQAALSNLSPWFRF 1 2 GQVAVQRAILEVYKHRTRYKESVEGFIEEAVVRRELADNFCFYNPNYDKIE 1 2 GAYDWAKTTLKIHAKDKRPHLYTLKQLEEGKTHDPLWNAAQ 0 0 LQMVQDGKMHGFLRMYWAKKILEWTRSPEEALQFSIYLNDRYELDGRDPNGYV 1 2 GCMWSICGIHDQGWAERAVFGKIRYMNYAGCKRKFDVGQFERKYGAHSFTQ* 0 >CPD_xenTro Xenopus tropicalis (frog) 0 MSHKGKVSKSKDEESGSEKSDGTLTKGMRMDTKAKSKTITKSPGKRKTPGKVLSETSSGESSDVDAKKKKKMVEQTEEKGVGGAVKISRVEAAPSVSEFKFNKKRVRLvSTEADLKDDAQGIVYWMSRDQRVQ 1 2 DNWAFLYAQRLALKQKLPLHVTFCLVPKFLDATIRHYGFMVKGLQEVAE 0 0 ECKELNIPFHLLIGYAKDILPNFVKKHAIGGVVTDFSPLRVPLQWVEDVSKRLPKDVPLVQ 0 0 VDAHNIVPCWVASNKQEYGARTIRKKIHDQLSQFLTEFPPVIKHPYEPKLEAE 0 0 PVDWDNCYNSLEVDRTIGEVEWAKPGTNAGFNVLQSFISERLRHFNSDRNNPNQNALSNLSPWFHF 1 2 GQLSVQRAILEVQKYRSKCKDSVDSFVEEAVVRRELADNFCFYNKNYDKIE 1 2 GAYDWAKNTLKDHAKDKRTHLYTLEKLEAGKTHDPLWNAAQ 0 0 LQMVHEGKMHGFLRMYWAKKILEWTSSPEEALHFSLYLNDRYELDGRDPNGYV 1 2 GCMWSICGIHDQGWAERAVFGKIRYMNYQGCKRKFDVAQFERRYHPKKFSQ* 0 >CPD_lepOcu Lepisosteus oculatus (spotted_gar) AHAT01034265 0 MSGRSPPSPQGGAAGKGAAKRKLQGGEGTGEKKAKKTAPEQGAGWLAEAVAQSRRQAGLSVQGFGFDKKRMRVLSKASAMKGGSEGVLYWMSRDQRVQ 1 2 DNWALLYAQQLALEEQLPLHVCFCLVPCFLDATIRQFGFLLRGLEEVAK 0 0 ECSALGLEFHLLRGSAGEVLPQFVQKQGLGAVVTDFAPLRLPLQWVEEVKERLPQDIPLIQ 0 0 VDAHNVVPCWVASDKQEYSAKTIRKKITSKLPDFLKEFPLVAEHPHRASKPAK 0 0 IIDWEQARSSLAVDRSVKEVSWARPGAAAGLEVLESFIDHRLKDFATQRNNPNGEALSNLSPWLHF 1 2 GQVSAQRVVLEVRRHGRRWPQSVDVFVEEVVVRRELADNFCFYNKKYDRVD 1 2 GAYEWAQKTLREHAKDRRPYLYSRGELEASRTHDKLWNAAQ 0 0 FQMVSEGKMHGFLRMYWAKKILEWTASPEEALATAIYLNDRYELDGRDPNGYV 1 2 GCMWSICGIHDQGWKEREIFGKVRYMNYAGCTRKFNVPKFEIKYNPKRCDDEDGNRKRGGD* 0 >CPD_danRer Danio rerio (zebrafish) NM_201064 0 MSANKNNLKRQMKSTISAGGKQPKLTGEKGKESGWLLKEVAELRRAAQGCEFNKKRLRYLSDTQKVKQSSQGVLYWMSRDQRIQ 1 2 DNWALIYSQQLALAEKLPLHICFCLVPKFLDATYRQYAFMLKGLQEVVK 0 0 ECKSLDIEFHLLSGEPVHNLPAFVKSWNIGAVVTDFNPLRISLQWIDTVKKHLPSDIPFIQ 0 0 VDAHNVVPCWEASTKLEYGARTIRGKITKHLQEFLTDMPLVDTHPYCASRAAK 0 0 IIDWEEVLSSLEVDHNVCEVEWAQPGTTGGMFMLESFIDQRLHIFATHRNNPNSDAVSHLSPWIHA 1 2 GQLSAQRVVMQVKRKKNASESVASFIEEIVVRRELADNFCFYNQNYDNIS 1 2 GAYDWAKKTLQEHAKDRRQYLYTKKELESAETHDQLWNAAQ 0 0 RQLLLEGKMHGFLRMYWAKKILEWTASPEEALSIAIYLNDRLSLDGCDPNGYV 1 2 GCMWSICGIHDQGWAERPIFGKIRYMNYAGCKRKFDVEQFERKYAAIKKNPNLNAKISV* 0 >CPD_petMar Petromyzon marinus (lamprey) rough but revised sequence 0 MCLSRLTIVFKLSSWLVNVVGSGNFEEQISKTRAALASSVLEFRFNKKRVRAVSNADDMAANASGILYWMSRDQRVQ 1 2 DNWALLYAQRLALKQRLPLYVCFCLVPKFMDATIRHFGFMLRGLQEVEK 0 0 EFRSLDISFHLLKGYAKDVLPPFVNEKNIGAVVTDFLPLRVPRQWVKDLKKALPKNVPLAQ 0 0 VDAHNVVPCWVTSDKLEYGARTIRRKIHDKLPQFFTEFPPAIKHTYPAKSLPE 0 0 ETDWEAAWQSLEVDRSVGEVAWAQPGTSAGLEMLNSFINERLKLFSTDRNNPTAMHSATSSPVIHF 1 2 1 2 GAYDWAKRSLDAHTQDNREYIYSLQQLEEAKTHDPLWNAAQ 0 0 LQMVVEGKMHGFMRMYWAKKILEWTSSPEEALRIAIYLNDRYQLDGSDPNGYV 1 2 GCMWSICGTHDQGWAERAVFGKIRYMNYAGCKRKFDVGAYESRYSSK* 0 >CPD_braFlo Branchiostoma floridae (amphioxus) XP_002586934 FE570347 0 MSSGDKRKSSGSGDSPAKKAKVETGRSSAQDGAGSEDFLDQIAERRQNVCESVAGFKFNKKRCRQMSDVGDLKQNCGGIMYWMSRDQRVQ 1 2 DNWALLFAQRLAMKHKVPLYVCFCLVPKFLDANIRHYGFMLKGLEEVEK 0 0 FPSPDWLCCGFLPGFVDEKNIGGVVTDFSPLRTPLKWVHDLAGKLPSDVPFVQ 0 0 VDAHNIVPCWEASPKQEYGARTIRGKIHNQLSSYLTEFPPVSQHPHPPKDKPE 0 0 PTDWAAARASLKVDMTVAEVTWATPGTTGGLRMLESFCKQRLKNFGTDRNNPNKRALSNLSPWIHF 1 2 GQISVQRCILTVKKYRSKYKESVDAYIEEAIIRRELSDNFCFYNHDKYDSIK 1 2 GASAWAQKTLKDHAGDKREYLYSREQLEQAKTHDDLWNAAQ 0 0 DKYSLDGRDPNGYV 1 2 GCMWSICGIHDQGWAERPVFGKIRFMNYNGCKRKFDVAQFVTKYKAKGYLKK* 0 >CPD_strPur Strongylocentrotus purpuratus (urchin) JT122393 JT102939 FJ812411 0 MITFSVQGLKLGHHCQCWKVVFLSRFISSRSYSSIMGKDGQTAKRSSEGQDNGETSAAKKVKKEEDGAEVQDLQTKVASLRNASGSSILNFKFNKKRVKILSDTMDIAEDNRGIVYWMSRDQRVQ 1 2 DNWALLFAQRLAMKQEVPLHVCFCLVPKFLEGTIRHFNFVLEGLKEVSQ 0 0 ECKELNIPFHLLIGYAKDILPNFVKKHAIGGVVTDFSPLRVPLQWVEDVSKRLPKDVPLVQ 0 0 VDAHNVVPCWEASNKLEYGARTIRPKITKQLSTYLTEFPPVICHPQKAKAKAE 0 0 PIDWEGAYASLEVDQTVKPVDWAKPGTSEGMKMLDSFVKERLRYFSSARNDPTKSVCSNLSPWIHF 1 2 GQLSSQRAALIVRLYRSRFSESVAAFIEESIIRRELSDNFCFYNDKYDSIE 1 2 GTNDWAKKTLKDHAKDKRDYVYSRETLERAKTYDQLWNSAQ 0 0 KQMVREGKMHGFLRMYWAKKILEWTTSPEEALEIAIYLNDRYSLDGRDPNGYV 1 2 GCMWSICGIHDQGWGERPVFGKIRFMNYKGCKRKFDVDTFVNRYKKYNL* 0 >CPD_droMel Drosophila melanogaster (fruitfly) thymidine dimer photolyase CG11205 uses 5-deazariboflavin 0 MFTLASYWRESFKI 0 0 VLPLQAMKRTKAQKAGPSKKAAKNEKASSEPKSDQESSDEEASTSKALLVSKPDYQNFEQFLTHLEHQRVCTAANIQEFSFRKKRVRVLSKTEDVKESSL GGVVYWMSRDGRVQDNWALLFAQRLALKLELPLTVVFCLVPKFLNATIRHYKFMMGGLQEVEQQCRALDIPFHLLMGSAVEKLPQFVKSKDIGAVVCDFA PLRLPRQWVEDVGKALPKSVPLVQVDAHNVVPLWVASDKQEYAARTIRNKINSKLGEYLSEFPPVVRHPHGTGCKNVNTVDWSAAYASLQCDMEVDEVQW AKPGYKAACQQLYEFCSRRLRHFNDKRNDPTADALSGLSPWLHFGHISAQRCALEVQRFRGQHKASADAFCEEAIVRRELADNFCFYNEHYDSLKGLSSWAYQTLDAHRKDKRDPCYSLEELEK 0 0 SLTYDDLWNSAQLQLVREGKMHGFLRMYWAKKILEWTATPEHALEYAILLNDKYSLDGRDPNGYVGCMWSIGGVHDMGWKERAIFGKVRYMNYQGCRRKFDVNAFVMRYGGKVHKKK* 0 >CPD_nasVit Nasonia vitripennis (wasp) XM_001603235 trimmed N-terminal LVKQIEEQRNQTADSVMTFKFNKKRVRILTDSDEVSSESKGIVYW MFRDARVHDNWAMLFAQKIALKNKVPLHVCFCILPKFLDATIRHYKFLLEALEEVEKDCKELNINFHLLHGEPNTAIINFVEKYKMGAVIADFFPLRLPLFWLEDIKKKLPKKIPL CQVDAHNIVPCWVASEKLEYAARTIRNKINSKLDEFLTEFPPVIKHPHTSDQKFDKINWDTALDDVLVDKSVDKITWAKAGYKGGIAELDKFLKIRLRIYDEKRNNPIFNALSNLS PWFHFGMISVQRCILEAKKYKSQYNKSVEAFMEEAIVRRELSDNFCFYNEHYDSLKGAYDWARETLNQHRNDKRDYIYTLNELENGLTHDDLWNAAEIQLVKEGKIHGFLRMYWAK KILEWTETPDDALKWSIYLNDKYSMDGRDPNGYVGCMWSICGVHDQGWRERPVFGKIRYMNYKGCERKFDVQAFVMKYGAKVQLTKDENKRKGKKK* 0 >CPD_bomImp Bombus impatiens (bumble_bee) XM_003488984 MDELNPSKRKKVSESIMTFNFNKQRIKHLSELNDVKENCKGILYWMFRDIRTQDNWALLFAQKIAVKRNVPLHICFCIMPSFLNASMRYYKFLLKGLMEIEE ECKQLNLNFHLLHGEPNESILKFVKTYNMGAVIVDFYPLKLPMLWIDNVQKNLPEDIPIYQIDAHNIVPCWYASSKQEFAAKTIRNKINTKLQEFLTEFPPVIKHPYLTKEKFENN NWDITLQDVEASKPTTEITWAKPGYRNGIKELENFIQNHLQKYGDERNNPLSNVISNLSPWFHFGMISVQRCILEIQEYKGLYKKSVESFMEEAIIRRELSDNFCFYNEKYDLVEG AYPWAIKTLNKHRKDTRKYVYSLSQLENSKTHDDLWNACQNQMIITGKMHGFLRMYWAKKLLEWTEIPEIALEWANYLNNKYSIDGCDPNGYVGCMWSICGVHDHGWPERDIFGKI RYMNYNGCKRKFNVAEFVMKWGKKKASELI* 0 >CPD_apiMel Apis mellifera (bee) XM_003250426 MEELNPFKRRKTSESIMTFKFNKKRIKRLNNLNDIKENCNGILYWMFRDIRIQDNWALLFAQKAALKNNVPLHICFCLISNFLNASIRYYKFLLKGLEEIEK ECKKLNINFHLLHGEPNINILKFIKIYNMGAIITDFYPLKLPMLWIDNVKKNLPKDIPICQVDAHNIVPCWYASSKQEFAAKTIRNKINTKLEEFLTEFPPVIKHPYTTKEKFEKN NWKIALQNVEVDKSVKEITWAKPGYENGIKELENFLQNRLKKYGDERNNPLSNAISNLSPWFHFGMISVQRCILEIKEYKKLYKKSVESFMEEAIIRKELSDNFCFYNEKYDLIEG AYPWAIETLNKHRKDKRKYIYFLNHLENSETHDDLWNACQNQMVTIGKMHGFLRMYWAKKILEWTETPEIALKWANYLNNKYSIDGCDPNGYVGCMWSICGVHDHGWPERDIFGKI RYMNYEGCKRKFNIAEFVMKWGKKKTNELI* 0 >CPD_anoGam Anopheles gambiae (mosquito) XM_313925 trimmed N-terminal YVAMLKADRKATAKSILDFDFNKKRVRILSDA KTIEDGKQGVLYWMSRDARVQDNWAFLFAQKLALKNDLPLHVCFNLVPKFLDATIRHFKFMLKGLEEVAEECRRLNIQFHLLRGSAGENVPAFVKKHKIGGVVCDFSPLRVPMKWV DEVRKALPMEIPLCQVDAHNIVPVWVTSDKLEYAARTIRTKVNKNLPTYLTPFPPLVKHPFTANFEADPINWTQVLDTLQVDRTVEAVEWATPGYAGGVKTLQTFVEKRLGKFNDK RNDPTENALSNLSPWFHFGQIAVQRAVLTVKKHGKRYSESVASFCEEAIVRRELSDNFCFHNKNYDNLQGAYDWARKTLDDHRKDKRVYCYSREELETAKTHDDLWNSAQLQMVKE GKMHGFLRMYWAKKILEWTKTPEEALETAIYLNDRYSLDGRDPNGYVGCMWSIAGIHDQGWKEREIFGKIRYMNYDGCKRKFNVNAFVVRYGGKVHRRK* 0 >CPD_aedAeg Aedes aegypti (mosquito) XM_001653905 trimmed N-terminal FVEKFKTIRKETAKSILDFDFKKKRVRILSDAKEVEENKKGVIYWMSRDARVQDNWAFLFAQKLALKNELPLHV CFSLVPKFLDATIRHYKFMLKGLEEVAKECESLNINFHMLTGMAKDTIPKFVKTHNIGAVVCDFSPLRVPMKWVEDVRKSLPAEVPLCQVDAHNIVPLWVTSEKQEYAARTIRNKV NNNLNTYLTQFPPVIKHPHKASFKADSIDWVKLLDTIEVDRTVDEVDWAVPGYTGGIGVLQSFVEKRLRKFNAKRNDPTDDALSNLSPWFHFGQISVQRCVLAVKKYGKGYSEGVA AFCEESIVRRELSDNFCYYNKNYDNLKGAYDWAQKTLDDHRKDKRTYVYSRDQLEQARTHDDLWNSAQIQMVKEGKMHGFLRMYWAKKILEWTKTPEEALETAIYLNDRYQLDGRD PNGYVGCMWSIAGIHDQGWREREVFGKIRYMNYEGCKRKFDVAAFVARYGGKVYKK* 0 >CPD_acyPis Acyrthosiphon pisum (aphid) XM_001949116 trimmed N-terminal FLNDVAAERNKTASSIMEFKFNKKRVRVLSEQKEVPEWAEGVIYWTFRDERIHDNWALLYAQKLAIKNKVSLHITFC RLKQFLDCSLRHYKHIFQGLEELETECKSLDIQFHFLIGCAADILPDFVKKHKLGAIIVDFMPVREHISWTKQLADRIGSEVPVIQVDAHNIVPCWVASDKQEYSARTIRNKINNK LPEFLTEFPPVIKHPFRSTFKAKPTNWDEADKTLEVDRSVVSVPGLKAGFKAGMSELEQFLKKRLPKYSTDRNNPVKDGLSKLSPWLHFGQISAQRCILEVSKLSKQYPDSVAAYR EEAIVRRELSDNFCFYNPKYDKIEGAPNWAQTSLNEHRKDKRMFVYTREELESSRTHDDLWNSAQIQLVKEGKMHGFLRMYWAKKILEWTDTPDRALADAIYLNDKYSMDGRDPSG FVGCMWSICGIHDQGWKEREIFGKIRYMNYAGCKRKFDINAFIARYGGMVHKYTKK* 0 >CPD_nemVec Nematostella vectensis (anemone) ABAV01006764 XM_001636204 bad 0 MASDEPAAKRRKQEVTGPNKSELDQLDDIVKTYKEKRMSVCNSVTDFKFNKKRVRMLSKEGSISENQCGGVVYWMWRDQRVQ 1 2 DNWALLYAQRLALKQQAPLHVCFCLPSNFLDATLRQYNFMIKGLQEVEK 0 0 ELKELEISFHLLLGDPGKVLPEFVKSAGIGGIVVDFCPLRLPTQWVNDVVKAVPKDVPVCQ 0 0 VDAHNIVPCWHASPKLEYGARTIRPKIHKVLTEFLTEFPPVIKHSHVSGEKTK 0 0 TTDWDAVDTFIEVDRSVGEVDWAKPGTAEGLFMLESFCKDRLKYFHSSRNDPTKRALSNLSPWFHT 1 2 GQISPQRAILRVRDFRSKFRESVESFIEECIIRRELSDNFCYYNNEKYDSI 1 2 EGTNEWARKTLNDHAKDKREYLYARGKLEKAQTHDDLWNAAQ 0 0 RQLVREGKLHGFLRMYWAKKILEWTDSPETALSDAIYFNDKYALDGIDPNGYV 1 2 GCMWSVCGIHDQGWAERPVFGKIRYMNYKGCQRKFAVAEFVKRYKP* 0 >CPD_ampQue Amphimedon queenslandica (sponge) ACUQ01006132 XM_003388698 bad 0 MQCKQKNKAWSSKLTATPILCKKIKMAAGSQAAGAARGAALQESLLIAGSSFNMKRCRLITKPTAGKSSIVKGPVLYWMSRDQRVQ 1 2 DNWGLVYSQELANKHGVPLLVAFTLVPKFLDATWRQYSFMMSGLQEVEK 0 0 ELLKLKIPFHLLLGKAQSCLPSFIAKESVSVVVCDFSPLRVPLGWVKETGAELDKIKVPLVQ 0 0 VDAHNIVPVWLASDKQEYAARTLRNKIHKFLPEFLTEFPLVTLHTHNSKLTMK 0 0 STNWIKARESLEVDMTVSEVSWATPGTNAGLKVLDDFCTKRLKFFAAQRNDPNKDSLSNLSPWYHF 1 2 GQVGVQRAILKVKSYSSKHSESVSAYIEEAVVRRELADNFCYYNPHYDSI 1 2 YHYCDLFFFFSRAHKKDKREYIYTQEQFESSSTHDPLWNAAQ 0 0 LQMVQEGKMHGFLRMYWAKKILEWTSSPEEGLRIAIYLNDKYEIDGRDPNGYV 1 2 GCMWSICGIHDQGWAERSVFGKIRYMNYQGCKRKFDVGQFERKYNKKRKLGEQ * 0 >CPD_acrDig Acropora digitifera (coral) BACK01030119 one intron missing 0 MASQPPAKRRKEESSENGDDILASCMAKRAQVCASVADFKFNKKRVRLLSKSTDISDDCKAIAYWMWRDQRVQ 1 2 DNWALLYAQRMALKQEVPLVVCFCLPSKYLHSTFRQYSFMIKGLQQVEK 0 0 ELASLN-INFHMLLGEPNIVLPDFVKSENIGGIIADFTPLRKPMKWLNDVMDKLPKNVPVCQ 0 0 VDSHNIIPCWQASPKLEYGARTIRPKIHNQMREFLTEFPPVIKHPHAGKASIK 0 0 TDWKAADEFIEVDRNVQEVNWAEPGTKAGLEMLESFCKDRLKFFATDRNNPTKEALSDLSPWFHA 1 2 GQISVQRAILRVREFRGSSKDSVETFIEEAVVRRELADNFCFYNQEHYDSI KGAKDWAQKTLNDHAADKREYLYTREQLEKGQSHDDLWNAAQ 0 0 LQLVNEGKMHGFLRMYWAKKILEWTESAEEGLEISLYLNDKYSLDGTDPNGYV 1 2 GCMWSVCGIHDQGWAERPVFGKIRYMNYNGCKRKFDVGEFVRKYGATTSKEADGPAKKRKKAK* 0 >CPD_monBre Monosiga brevicollis (choanflagellate) ABFJ01000652 related intronation but numerous differences 0 0 0 LYWMSRDQRVQ DNWAFLYAQRLALKQRLPLHVCFCLVPKFLDATIRHFGFMLRGLEQ 0 0 VVETHLRKKHIPFHLLTGYAKDVLPKFAEEQEACAVVCDMSPLRVPMAWVKETGSKLKDMNVPLYQ 0 0 VDAHNIVPVWHASPKQEYAARTIRNKIHQKLDTCLQPFPELESNSNSVQLPDT VDWKKARESLEINWDVKEVDWLKPGYEGGMKMLEEFINERLHRYADDRNDPNLDALSNLSPYYHFGQ 0 0 ISVQRVVLELRSKQRGKYAEGVKAYIEEAVVRRELSDNFCFYNHRYDSVE GASAWAQETLDVHSKDKR 2 1 EHLYTRKQLENAETADDLWNASQLQLVQEGKMHGFLRMYWAKKILEWTESPEKALEDAIYLNDKYELDGRDPN 1 2 GYVGCMWSICGIHDQGWGERPVFGKIR 21 YMNYKGCKRKFDIAAFVKRYPPAAKNAARGKDGR* 0 >CPD_salSpp Salpingoeca species (choanflagellate) ACSY01000967 different intronation still 0 VYWMSRDQRAK DNWALLYARSLARSARVPLVVVFSLVPKFLDATIRHYGFMLRGLHQ TAKHLHEKLVPFHLLQGSAATTVPAFAAQHEAAAVICDMSPLRVPLRWVKDVGQALEAQNVPLLQ VDAHNIVPVWVTSQKQEYAARTIRPKIHKHLDTYLQPFPELDANDKDTLGDMELPPVFDLEAQFDMLEVDTSVKE 0 0 VDWIEPGYEQGMAAAEAFGRDRAKKFDELRNNPNEDVCSNLSPYFHFGQ ISAAAVVLLLKSKYSKKAAKGVQTFIEEAVVRRE 0 0 LSDNFCFYNRRYDSIDGAAVWARDTLDTHRHDKR 2 1 EYVYTREQLEQGKTHDDLWNAAQLQ 0 0 MVERGKMHGFLRMYWAKKILEWTATPEDALQTALFLNDRYELDGRDPN 1 2 GYVGCMWSIAGIHDQGWAERAVFGKIR 21 RYMNYKGCKRKFDVARFVSRFPQAVANAA* 0
Cryptochromes from cnidarians, trichoplax, sponge and choanoflagellate
These cryptochromes and photolyases are important as metazoan outgroup sequences to Bilatera. These do not classify clearly as yet and may represent clade-specific gene family expansion. These sequences are simply crude GenBank predictions; after validation, they will be moved to their corresponding orthologous class above.
>CRY1A_acrMil Acropora millepora (sponge) EF202589 MSLNLKSVEDNNSAVSAEKSQGKLKAKHAIHWVRKDLRLHDNPSLLEAVKGSDTVRIIYVLDTKVDHATGIGLNLWRFLLQSLEDVDDSLRKLNSRLFVV RGQPADVFPRLFREWKTSFLTFEEDSEPFGREKDAAIRLLAQESGVEVAVGRSHTLYDPQLIIKHNSGTAPLTYKKFLAIVRSLGNPQHPCATLDVHLLG GCSTPVSEDHEEKFGVPSLKELGLDVAKLSTEIWHGGETEALIRLDRHLERKAWIASFEKPKVTPNSLFPSPTGLSPYLRFGCLSPRLFYHRLSELYRKV KCKDPPISLYGQLLWREFFFTVAANNPNFDQMDENPVCLQIPWTANPEWLKKWEQGQTGFPWIDAIMIQLKQEGXIHHLARHAVGCFLTRGDLWISWEEG MKVFERWLLDAEWSLNAGNWMWLSCSAFFQQFFNCICPVGFGRKLDPNGDYVRKYLPVLKGFPAKYIHAPWTAPENVQRAARCIIGKDYPRPIVDHHKVS TANLEKMRNVFKALLRYKESTVVATSEKSDGSKDKQAKLKQQMLNLEDKENR* 0 >CRY1B_acrMil Acropora millepora (sponge) EF202590 ICVQRLKDLCFSLDILGHTSLHWFRKDLRLHDNPSLRECLRNSKVFYGVYFLPPSEAKQGSVSLNRWGFLLESLRDLDTSLVECGSRLFVIRGNPVEMLP NLFKKWNINQMSFEVDSEPYSNSRDLVISHLAKENGIEVISRVSHTLYDPRILRGLSSGIVPLLFDEFKESVLQKRQPEKPVPKVGRKLFGACVTPVGSD HQQCYGVPRLDEIGGKFSKSGSVCSELYKGGESEALKRMETALQWMVKNDFTEPEITIHSLLXSATHLSPYLRCGCLSPRLLHQRITEEYIKSKGVSPPS ELFVKLLWREFFFVVGGQIPDAHEMINNPISLEIPWEDSVEYLERWKKGTTGFPWIDAIMRQLRTEGWIHNIARRAVASFLTRGCLWVNWEEGFKVFDEF QLDAERSLNVGNWLWVSTSTFVKGPVPWFCPVGVGKKIDPTGEYVRKYVPEIRNLPTEYIFEPWLTPRDLQRSYGCVIGRDYPAPIVNHVKQRAICVQRM QELAFKLASKIRWRQVLNVI* 0 >CRY1A_nemVec Nematostella vectensis (anemone) XM_001623096 MSGMDKFTTDSCNLVKAKKTSHSIHWLRKGLRIHDNPALRDAVLNWGTFRVVYILDTKSVASSNIGLNLWRFLLQALEDLDDSLRKLNSRLFVIRGQPAD VFPRLFREWGITRLTFEEDSEPFGKERDSAICMLAREAGVEVASHRSHTLYHLQGIIDRNGGTPPLTYKKFLSVIEGIAPPDPPVPHIDPSAQELGHTPL SDNHDELYGVPTMEELGLETSKLFVEVWHGGETEALKRLDRHLERKAWIASFGKPKVTSDSLMASPSGVSPYLRFGCLSPRLFYYRLMDLYRKVKGGPPP TSLYGQLLWREFFFVVSTNNPNFDKMESNPICLYIKWRTDKQVASDLQKWTDAQTGFPWIDAIMSQLKQEGWIHYLARVAIASFLTRGCLWISWEAGMKV FAEHLLDADWSSNAGNWMWWSYSAFSQQFFNPPCPVEFSKSLDASGDYIRKYLPVLKGFPARYIHEPWKAPLNVQKMAKCIIGKSYPSPMVDHEKAVSKN MEEMTSVYKSVIQYRISVLSSEEKQIEETKSQFQMPETSMH* 0 >CRY1B_nemVec Nematostella vectensis (anemone) XM_001623096 MSGMDKFTTDSCNLVKAKKTSHSIHWLRKGLRIHDNPALRDAVLNWGTFRVVYILDTKSVASSNIGLNLWRFLLQALEDLDDSLRKLNSRLFVIRGQPAD VFPRLFREWGITRLTFEEDSEPFGKERDSAICMLAREAGVEVASHRSHTLYHLQGIIDRNGGTPPLTYKKFLSVIEGIAPPDPPVPHIDPSAQELGHTPL SDNHDELYGVPTMEELGLETSKLFVEVWHGGETEALKRLDRHLERKAWIASFGKPKVTSDSLMASPSGVSPYLRFGCLSPRLFYYRLMDLYRKVKGGPPP TSLYGQLLWREFFFVVSTNNPNFDKMESNPICLYIKWRTDKQVASDLQKWTDAQTGFPWIDAIMSQLKQEGWIHYLARVAIASFLTRGCLWISWEAGMKV FAEHLLDADWSSNAGNWMWWSYSAFSQQFFNPPCPVEFSKSLDASGDYIRKYLPVLKGFPARYIHEPWKAPLNVQKMAKCIIGKSYPSPMVDHEKAVSKN MEEMTSVYKSVIQYRISVLSSEEKQIEETKSQFQMPETSMH* 0 >CRY1C_nemVec Nematostella vectensis (anemone) XM_001630979 MVNSVHWFRKGLRLHDNEPLRRAIEGSDTFRGIFFLDKAAVKNAKVSPNGWRFLIESLRDLDQSLQKYNSRLFIIQGQPIDVFPKLIKQWNISKLTFEYD SEPFPRQRDLAVKRIAEKAGVDVIVCSSHTLYDIECIEQILEMNEGKPPVLFKDFEAIVRKLGPPENPVESITRKSFMNCICPITSTHSEKFGVPSLAEL GIKPADVTCGDMWVGGEREALNRLGILEDKILGSNSSKNCDGSSVLFPSRTQLSPYLRFGCLSPKLYYKKLTGAYVKMQFSPPPMSLYRQLIWREFFFTL ASKNPNMDQVNDNPLCIQISWEENKDHFLCWKQGNTGFPWIDAIMRQLQKEGWIHHLARLSVGCFLTRGCLWISWEEGLRAFEELQLDAEWSMNSGSWLW LSCSAFFHGQIPWFCPVEVGKKLDPSGSYIRKYVPELRSMPLKYLYEPWTAPDDIQRAAMCVIGEDYPHPIVDHIEQRKACLQRLKTVCRHIQTTSFA* 0 >CRY1D_nemVec Nematostella vectensis (anemone) XM_001632799 MKERILFSSDCPSLGISSMHWFRKDLRLHDNPALLESFKNCQAFYGVYFLDPASVQRSNLSPNRWWFLLESLRDLDYNLRSLGSRLLVVRGQPVQEMPKL LDQWNIKRLTLEYDSEPPAKQRDAVVTHLAKNLGVEVIQRVSHTLYDVETVLETNDGKLPMTFDEMAKTAEQLGPPCPPCQTVDKTVFGACLTPVGPDHA DKYGVPLLSEFGMKELKEATAKKYWTGGEPEALRRLSAALKKCAENDFEERGWTIDEMFSNDAHLSPYMRFGCLSPRLYYQQLALTYMKEKKSIPPATLF TGLVRRELFLHVASHNADLDKMLDNPLSVQFPWEENKEGLERWKEGKTGFPWIDAIMRQLREEGWIHHLARQAVGCFLTRGCLWVSWEEGFKAFDELQLD AEWSLNASNWLWLSCSSYVHGAVPWYCPVEVGKKVDPTGDYIKRYVPEVRGLPSEYVCEPWNAPLSVQKACRCVVGEDYPSPIVDHMEQRMICVQRMQQLSIDLGAKGERVTS* 0 >CRY1E_nemVec Nematostella vectensis (anemone) XM_001632800 GCTSVHWFRKDLRLHDNPSLLASLDNCSTFFPIYVLDMESARASKISANRWNFLCECLEALDRQLRVLGSRLFVIRGRAIDVLPRLFHEWSVNRLTFERE SEPAGRQRDTVIQMLAENANVQLLQHNAHLLYDTDEVLETNDGKLPMTFDEMAKTAEQLGPPCPPCQTVDKTVFGACITPVGLDHAEKYGVPQLSDFGGK ELGRATAKLFWKGGELEAMRRLNLALQKKKFIKPPLSADTLLASDRALSPYMRFGCLSPCYILDRLTTEYQRTMGSKPPETLYTNLLWREFFFATASTNP DHHRMMGNPLALQIPWEQHPEALSLWKQGKTGFPWIDAIMRQLREEGWIHHLARQAVGCFLTRGCLWVSWEEGFKAFDELQLDAEWSLNAGNWLWLSCSS YVHGAVPWYCPVEVGKQVDPTGEYIRTFVPELRRLPTKFLHEPWKAPSSVQREAGCIVGKDYPQPMVNHLERRVVCVQKMQNFTQTLALLNG* 0 >CRY2_ampQue Amphimedon queenslandica (sponge) XM_003386521 MDSYSNGFYEEEERTLTVFNNVDGASEEEQDSFFDSYSASASSSPSSLCSSNWHSERLPVSLHWFRSFSLRLHDNSALMASLLRPQTQFRAVFVLDPWFT ETDKKFGANRLKFLLECLHDLSNQLEALGLKLYVAQGQTTAVLAGLCQEWGVTHLSYQKSQEPRSKVEERTISEMASMMNIEVEEFHNHTLYSPTELIKL NEGKPILTFKDFRTLLSKLKPPAVPISSPSTKQKYNEDNIPMFKVQTFQIPSLASLGCYDVELGTGPWVGGETEALKRLDNYCTVRSKPFDNFLDCLFDK TSLSPYVRFGCLSVRHFWHYVRQLASTDKSKMTLVQEVSSKLLQREFYFLVSSQVPNFDTDTQNPICIQLPWEFDADGLRRWRSGMTGYPWIDAAIRQML KEGWIHNLLRESIATFLTRGDLWISWLKGAESFEMYMLDYEPSVSCGCWMKSSSTAFITGTIEHYNPISYGQQLDPNGDYIRHFVPEVKNLPKEYIFCPW MAPIEVQREAGCIVGTDYPHPVVDHMTAGVICMERLKSFMQSLSSHQTNP* 0 >CRY_ampQue Amphimedon queenslandica (sponge) XM_003386534 METLQGSYDGGGSGFFFPPAGSPRSAMSRHQYPAPEKGHLPSQPPTVLHWFRLDSLRLSDNPAFHHAVSSGKRLKAVVILDPWFNSNNKSGPSANVWRFL LESLHDLDSRLQKRPYNTRLNIYLGQPTVVLSALFHKWNVSELTFQASQTSLESKKHDELIKFAADSQNVKTTSFYSHTLYNPEDILRANNGRFPHSYKE LRRLLPLVGRPREPFPEPDPVLVLLRQNSPEDQDLEEPENRIPSLQDLGFGLEETLYTNSWVGGETEGLSRLSNFCSRRSTQPNEPITWLISKDSLGPYI RFGCLSVRQLFSQLRQYASTSSKGQVLFAELTKNLLMREFAYMVGLTVPKFDTMKDNPLCIQLPWEENERFFYAWKEGRTGYPWIDAAMRQIVRDGWSHY STRQSIAVFLTRGYLWISWEKGLEFFQEHMLDFELPVSTVCWMQSSCSGFFVDFIESYDPCYIGKQMDSDGHFIKLYVPELQDFPSDYIHTPWLAPLHVQ QQANCIIGKDYPKPLVDLCSQGELCCQRVRSILTALRELYGDNP* 0 >CRY_subDom Suberites domuncula (sponge) FN421335 MHCNYPLATQTTFQGQSIPDTTVHWFRLDALRLHDNPAFVDAVKTDGNFKAVFIIDPWFNANYNNGGPQVNVWRFLLEALHDLDSRLQKKPYCARLNVLY GQPTMILPELYKKWNVKKITFQASQVSSESMKHDGIIKILSEQQNVQAVSYFSHTLYDPANVIALNNGRVPLSYKEFRRLMPLMGKPASPIPEPHPMSLC MKAPPSELVPEPEGKIPKLQDLGLSDEFALYTNSWVGGETEALSRLSSFCSRRAAIPNEPVHWLMSKDTLSPYIKFGCLSVRQLFSQLLQFASTSSKGQE LFELLTKNLLLREFAFLVGSSSPKFDVMEGNSLCIQLPWESNNVFIQAFRNGQTGYPWIDAIIRQIRQDGWAHFLARQSIAVFLTRGYLWISWVLGKEFF QEFMIDFELPVSSVCWMQSSCSGFFCTQIESYDPCLVGKQIDTDGHYIKTYVPELKDFPSEYVHQPWKCSSLHTEASWLCDREQYPKPIIDVCKQGELCCKRVQSIMKALADVYGVE* 0 >CRY4_craMey Crateromorpha meyeri (sponge) PUBMED:20121950 MTYNTLHIFTDDALRINDNNSLLSNLENTKNLYTVFYYGHIKKINHIPPHRVIFLLESLKNLKERLDEYGIPLYFIDEPMFQSLRMLISKWNINRVTTEP VVSIVGKRDQLSLRNFLSAFGVMLRTYNSSTLYDTVKVPVNITRSEFFRIITQMEPELSLPEELTEFLSKFSSFPDPFINKKIPSLSEFGITNETMSSID TKFRGGETAAILQLEKLIANRCEPNNLPKVAQLNQYDAISPAIKFGCISVRTIYNRVSKLEPKYNEVKNQIYDGLRNRDYCILVGGNCPNIDNQGSIYTY ILPWDVKQDASTRFQTGRTGYPFIDAAIAQLKREGFIHNSVKNILVRTLTCDLMWIGWHEGVRMFYKWSLDYNAAICALSWMHGSKSTWLLEEISISQIN PIEEAKEIDKDGDYIRKYLPELKDYPSEYIHTPWLAPLDQQIESECVIGQDYPYPNYCDVEERVQQCRKRLQIFYNIMPIAKKRRTLSLLKKRTRININNNDSVINIRAQLECKATKLSQ* 0 >CRY_aphVas Aphrocallistes vastus (sponge) PUBMED:14499587 MESLSITEFSPSAPSPISSSSSPDEETDRDIVAKDIEQDVEPQGKVLLHIFNNRHLRLKDNTALYQAMAQNPDKFYAVYIFDGFDSKPVAPVRWQFLIDC LEDLKEQLNGFGLELYCFRGETIDVLATLVQAWKVKLLSINMDPDVNFTFFNEKIVKMCTINAVQLYNDMDSHRLLYLPPKYKSAIPMSKFRVLLAEAIT AKQNNLESEAKIQDITPPLNPEQLSDLGNKPRLDSPLPSEIPKLNALFTEEEIAKLNFIFQGGERRTEDYLNEYREARLRDVSGDEDASPIAAKAMGISP HLRFGCITPRHLFNFLVKTIKDANYSRIKINKVLAGIMARDFALQVSQLQTIPERIISLNKICLPIPWDKNNNEIVEKLTDAQTGFPFFDAAITQLKTEG YVINEVSEALATFVTNSLLWVSWEEGQNFFSQHLICFDLAMSTHSWLEASGSTMVTGRQKSYQDPLLFVSKKLDPNGEYIKRYLPKFINFPIEFIHKPGN ASLEAQQAANCVIDIDYPKPLFEYECRNGICCKRLRVFMEVVDSAAKATKLPHVIENCSGKFP* 0 >CRY64A_triAdh Trichoplax adhaerens (placozoa) XM_002108524 MVCTVVWLRYDLRLIDNPALFHAAKRGHVVIIYILDQETPGKWKLGKASLWWLHHSLKSLQNDLLKFNIPLILRRGADPLTILRQVLLQSRADAVYWNRC YEPYAVKRDQSIEEALKVDDVTVGSFQAGLLYEPWAVKSAKDEPFKLYIMYWNRCLSSDQPRKPYDKPRFNNTLDIKVQTDTLDSWHLVSAKKDWKDELK IVIGCPGEDGAIRKLKEFIKYKLVDYRKGKETVWPSKTSQLSTHLHFGEISPFVVWHTAKESVHLRSVPSDSSQKFLTELGWRDFCFHLLHYYPDFPEKS YKLNFDETIWKVDKEKLKLWQQGKTGYPIVDAGMRELLNTGVISYRVRTIVASFLTKHLLIPWQNGAEWFWDTLVDADLALNSCSWQRITGCGADISSYF YIINPVTQGERFDSDGNYVRKWIPEIAKLSNDYIQQPWEAPAAVLKKAGIALGKTYPKCIVNHKKARDVALTKFAPLKRQEPMDNLKKLPNKKKMKK* 0 >CRY64B_triAdh Trichoplax adhaerens (placozoa) XM_002107723 MSSSKIPCTLVWLRQDLRLIDNPALFHAAKRGQLITVYIFDEDCSGKRRTGQACLWWLYHSLKSLRNQLSQWNIPLIVKSGKDAFTILKELLDSYNADAI YWNRCYEPFEYNRDCSIEEKLKMENVTVETYQDRVLFEPWTIKTAKGGPVQIYVHFWNQCLSLPQPRKPYDKPKFKNQTGCQSKCDEPAVWQRLAKLSQD MPMKLQDFWHPGEEQAISKLKSFIKNSLNAYAEKSQCLRLNTTSNLSPYLHFGEISPFTVWHSVRHASSSAEPPAVKSESLKQYLRSLGWREYTCHLLFH YPDLPQKCFRSSFEQLKLQVDPVKLQSWQQGNTGYPVVDAGMRQLSVAGIMPNRLRMIVASFLIKHLLVPWQKGEEWFWSKLIDADLAQNAFNWQWAAGS GPNACPYFRVFNPITQGEKFDADGYYVRKWIPELAALPNAYIHKPWEAPSTVLKKANIVLGKTYPRPIVQHKAARELALATFGQTKRKQEPPTSNSNKRVKEE* 0