Opsin evolution: RPE65: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 32: | Line 32: | ||
[[Image:RPE65 cycle.jpg]] | [[Image:RPE65 cycle.jpg]] | ||
<br clear="all"> | |||
=== BCO2 role in chicken yellow skin === | |||
It turns out that Darwin was wrong about domestic chicken: it is a [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=18454198#pgen.1000010-Sunde1 hybrid] of two south Asian species of junglefowl, red junglefowl (Gallus gallus) and closely related grey junglefowl (Gallus sonneratii). This was proposed in 1949 by Hirt but only recently confirmed by extensive mapping of the recessive allele for yellow skin and yellow legs. | |||
Most likely, this can be attributed to a cis-regulatory region just upstream of BCO2 that apparently reduces gene expression in skin, resulting in dietary carotenoids accumulating there. While the exact causal mutation cannot yet be identified within the minimum 23.8 kb haplotype chr24:6,264,083-6,287,900 May 06 assembly coordinates, it is very unlikely to be the only non-synonymous SNP in this region because as shown below it occurs at a highly variable position in poorly conserved exon 8 of a rapidly evolving overall gene (67% identity between chicken and human orthologs). | |||
Thus BCDO2 regulation (rather than coding sequence per se) could be selectively important (for mating displays) by controlling skin, scale, and feather carotenoid pigmentation in birds, fish and other vertebrates. If so, it would be an important example of evolution at the regulatory level rather than at protein sequence. | |||
In terms of domestication, early farmers may have wanted egg yolks with extensive carotenoids coloring and may have taken bright yellow skin as a proxy for healthy birds, thus imposing selection on their flocks, though QTL studies today do not support such an association. | |||
Two chicken ESTs (CN221314 and DR424873) indicate that the promoter region and its CpG island occur upstream of 6282641, suggesting that the SNP should be located in the 5,260 bp region between there and 6287900. However no striking conserved non-coding region exists in chicken; human regulatory tracks at the UCSC also do not illuminate how or where expression is regulated here. | |||
The K416N SNP can be N, G, D, E, H, L, K, and R in vertebrate orthologs indicating no catalytic significance: | |||
* | |||
>BCO2_homSap VHNSAAKSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT | |||
>BCO2_panTro VHNSAAKSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT | |||
>BCO2_ponPyg VYNSAAKSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT | |||
>BCO2_macMul VYNSAGRSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT | |||
>BCO2_calJac VYNSAVRSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGK | |||
>BCO2_tarSyr VYDSIARSCPRRFVLPLNVSLNAPEEENLSPLSYSSATAVKKTDGK | |||
>BCO2_otoGar VYNSAARSFPRRFVLPLNVGLNAPEGENLSPLSYSSASAVKQADGK | |||
>BCO2_tupBel VYNSVARSFPRRFVLPLGVSSNAPEGENLSPLSYSSASAVKLSDGK | |||
>BCO2_musMus VYELKAKSFPRRFVLPLDVSVDAAEGKNLSPLSYSSASAVKQGDGE | |||
>BCO2_ratNor VYKAKAKSFPRRFVLPLDISVGAPEGENLRPLPYSSASVVKQGDRE | |||
>BCO2_cavPor VYRSVARSFPRRFVLPLDVTVNTPEGETLSPLSYSSASAVKQADGK | |||
>BCO2_speTri VYNSAALSFPRRFVLPLHISENDPAGENLSPLSYSSASAVKQSDGK | |||
>BCO2_oryCun VYNSVARSFPRRFVLPLNVSINAPEGKNLSPLTYSSASAVKQADGK | |||
>BCO2_vicPac VYNSVARSFPRRFVLPLHVSLNAPERENLSPLSYSSASAVKQADGK | |||
>BCO2_turTru VYNSVARSFPRRFVLPLHVGLNAPEGKNLSPLTYSSASAVKQADGK | |||
>BCO2_bosTau VYNLIARNSPRRFVLPLLGNLNAPEGENLSPLTYSSASAVKQADGK | |||
>BCO2_equCab VYNSAAKSFPRRFVLPLHVSLDAPEGKNLSPLSYSSASAVKQADGK | |||
>BCO2_felCat VYNSVGRSFPRRFVLPLHVSLNDPEGQNLSPLSYSSASAVKQADGK | |||
>BCO2_canFam VYNSVGRSFPRRFVLPLHVSLNDPEGENLSPLSYSSASAVKHADGK | |||
>BCO2_myoLuc VYNSVATSFPRRFVLPLRVSLNAPEGENLSLLSYPSASAVKQADGK | |||
>BCO2_pteVam VYNSVARSFPRRFVLPLHINLNAPEGENLSPLPYSSASAVKQADGK | |||
>BCO2_eriEur VYNSVARSFPRRFVLPLNVNLDTPEEENLSPLSYSSASAVKQADGT | |||
>BCO2_sorAra VYNSLARSFPRRFVLPLNISPNAPQGENLSSLSYSSASAVKQADGK | |||
>BCO2_loxAfr VZISVARSFPQRFVVPLNVSLNAPEGQTLGLLSRSLASVVEQADGK | |||
>BCO2_dasNov VYNSVARSFPRRFVLPLNVSLNAPEGENLSPLSYSSASAVK-ADG- | |||
>BCO2_choHof VYNSVARSFPRRFVLPLDVSLNAPEGENLSSLSYSSASAVKQADGK | |||
>BCO2_monDom VYNSVARAFPRRFVLPLDISTKASVGQNLSPLTYSSASAVKQADGT | |||
>BCO2_ornAna VYNSAARTFPRRFVLPLDAFLDAPTGENLSPLTYSTAKAMRGEDGK | |||
>BCO2_galGal IFGSVARTFPRRFVLPLKVNSDTPVGKNLNPLSYTSAKAVKDSDGK | |||
>BCO2_galLaf IFGSVARTFPRRFVLPLNVNSDTPVGKNLNPLSYTSAKAVKDSDGK Gallus lafayetii | |||
>BCO2_taeGut ---------------------------------------------- finch trace coverage unavailable | |||
>BCO2_anoCar TYNSIATPYPRRFVLPLNIDDKKPVGENLSPLSYTSATAVKEADGK | |||
>BCO2_tetNig VYNSTGRAFPRRFVLPLHVTSQTAAGHNLNTRPZSKETCVKTDKDT | |||
>BCO2_takRub VYNSTGRAFPRRFVLPLHVTSEMATGQNLNTRASSQATCVKTGKDT | |||
>BCO2_gasAcu VYNTLCRVFPRRFVLPLNVDQETPYGRNLN--PKGSATSVRFAKNK | |||
>BCO2_oryLap VYNTTERSFPRRFVLPLNLTSETPTNQNLNTRPFSKASCTKISPDK | |||
>BCO2_danRer FFNSLCTNLPRRYVLPLEVKEDEPNDQNLINLPYTTASAVKT-QTG | |||
* | |||
[[Image:BCO2junglefowl.jpg]] | |||
<br clear="all"> | <br clear="all"> | ||
Revision as of 16:52, 8 January 2009
Introduction to the RPE65 gene family
This gene family is critical to the metabolism of provitamin A dietary carotenoids and thus to retinoic acid signaling and the recycling of all-trans retinal needed to replenish vertebrate ciliary opsins after exposure to light. RPE65, as its illegimate but irrevocably established gene name suggests, is a protein of 61 kdaltons found in the retinal pigment epithelium adjacent to photoreceptor layers. It isomerizes all-trans-retinyl palmitate extracted from phospholipid membranes to 11-cis-retinol.
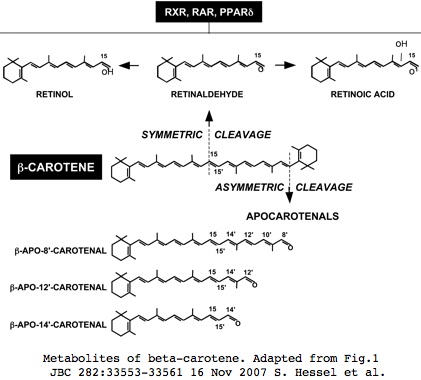
In vertebrates including human, RPE65 has two full-length paralogs on different chromosomes, BCMO1 (ß-carotene-15,15'-oxygenase cleaving symmetricaly to yield two retinals) and BCO2 (ß-carotene-9',10'-oxygenase cleaving asymmetricaly to yield a retinoic acid precursor and also cleaving lycopene), as recently reviewed. Insects have but a single gene NinaB but as shown below, this is attributable to gene loss rather than being the ancestral bilateran state.
The nomenclature of these three genes violates the spirit and letter of international agreement for naming human gene families, yet the responsible committee has approved the names! While an improvement over monstrosities such as B-DIOX-II and BCDO2 beta-carotene dioxygenase (for a mono-oxygenase!), it should be noted that RPE65 is 61 kd and expressed in a great many other tissues (such as skin) beyond retinal pigment epithelium (where BCMO1 also occurs). A more appropriate nomenclature capturing the paralogous relationships and key features might be BCO1S, BCO2A, and BCO3R, where the letters denote symmetric, asymmetric, and retinal.
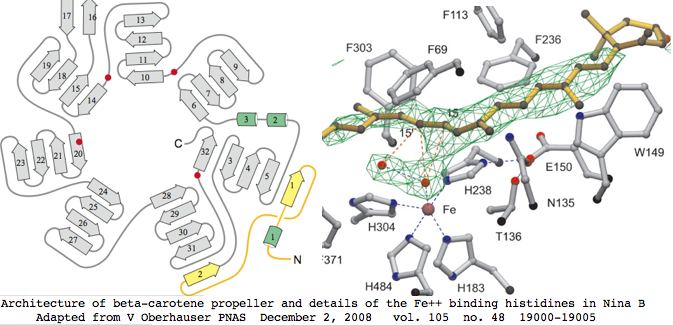
This gene family is readily tracked back to bacteria where a 3D structure has been determined that suffices to model the entire family in all species. Four invariant histidines that hold the catalytic ferrous iron lie on the axis of a seven-bladed beta-propeller fold. The Fe2+ is accessible to carotenoids via a long nonpolar tunnel capable of promoting cis-trans double bond conversions. RPE65 shares all these features even though as a mere isomerase.
Since this complex structure consists of a single coherent domain fold, alternative coding splices can be dismissed out of hand as transcriptional noise not leading to stable functional enzyme. The domain structure implies all homologs from all species must be full length or around 550 amino acids, which is validated below over tens of billions of years of evolutionary branch length. That conclusion must be qualified to the extent that N- and C-terminal extensions could be trimmed post-translationally and small indels not affecting the propellers might be tolerated.
RPE65 is a well-known disease gene for retinitis pigmentosa and type II Leber congenital amaurosis. Its two paralogs are not currently associated with human disease but have various consequences in knockout mice consistent with enzymatic expectations. Retinol dehydrogenase RDH5 is associated with fundus albipunctatus; LRAT lecithin retinol acyltransferase with severe, early-onset retinal dystrophy.
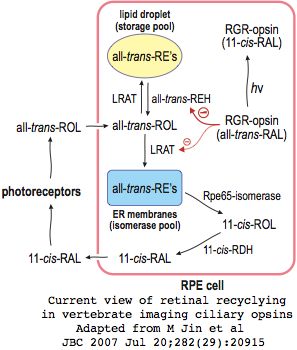
Mouse knockouts have been studied for RPE65, BCMO1, LRAT, and curiously RGR opsin which mediates all-trans retinyl ester mobilization from lipid droplet storage to the endoplasmic reticulum explaining the intrinsic light sensitivity of the retinal pigment epithelium.
Vitamin A (all-trans-retinol) is a commercial dietary substitute for natural dietary carotenoids that itself is a waste product of the ciliary opsin photoreception but scavengible to 11-cis-retinal (and related opsin chromophores such as 11-cis-3-hydroxy-retinal) and retinoic acid (all-trans and 9-cis). Less characterized asymmetric BCO2 cleavage products may have unsuspected functionality. Retinoic acid is a ligand regulating gene transcription via heterodimeric nuclear hormone receptors (RA receptor and retinoid X receptor RXR) as recently reviewed. CMO1 is transcriptionally regulated by various heterodimers of RXR including PPARs (peroxisome proliferator-activated receptors).
The confused history of RPE65
RPE65 has a history of serious experimental and interpretive error assigning its isomerase activity to LRAT (ecithin:retinol acyl transferase), assigning palmitolyation to various RPE65 cysteine residues that did not occur, and attributing its (non-transiting) membrane association to these lipids when in fact that is intrinsic to a surface hydrophobic patch. Still earlier confusion over enzyme mechanism introduced dioxygenases for BCO2 (which is a mono-oxygenase like BCMO1).
While this June 2004 paper conflicted from the get-go with full-length alignment of RPE65 to well-studied carotenoid isomeroxygenases, the brief debacle exposed a multi-year delay in update cycling at primary hub databases such as RefSeq, SwissProt, and OMIM. None of these have not been corrected 55 months later. It's not clear when these pivotal descriptions will ever be revisited. Meanwhile hundreds of derived databases continue to perpetuate serious misunderstandings about this important but straighforward disease gene and the visual pigment cycle in vertebrates. How many thousands of other genes lie outside an update cycle?
The current view of retinal cycling in vertebrate eyes envisions the primary photosensitive event as isomerization of opsin-bound 11-cis retinal to all-trans retinol. After importation to the adjacent retinal pigment epithelia, that is esterified by LRAT palmitolyation and subsequently isomerized by RPE65 to 11-cis retinol, which must then be oxidized to 11-cis retinal by RDH5 before it can be re-exported back to the retina and re-form its Schiff base in a newly recharged opsin molecule. This system seems quite bizarre in comparison to self-recharging insect melanopsins.
BCO2 role in chicken yellow skin
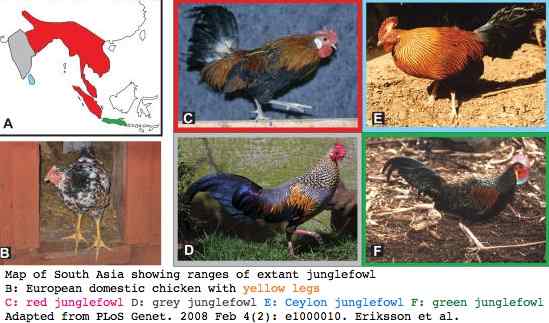
It turns out that Darwin was wrong about domestic chicken: it is a hybrid of two south Asian species of junglefowl, red junglefowl (Gallus gallus) and closely related grey junglefowl (Gallus sonneratii). This was proposed in 1949 by Hirt but only recently confirmed by extensive mapping of the recessive allele for yellow skin and yellow legs.
Most likely, this can be attributed to a cis-regulatory region just upstream of BCO2 that apparently reduces gene expression in skin, resulting in dietary carotenoids accumulating there. While the exact causal mutation cannot yet be identified within the minimum 23.8 kb haplotype chr24:6,264,083-6,287,900 May 06 assembly coordinates, it is very unlikely to be the only non-synonymous SNP in this region because as shown below it occurs at a highly variable position in poorly conserved exon 8 of a rapidly evolving overall gene (67% identity between chicken and human orthologs).
Thus BCDO2 regulation (rather than coding sequence per se) could be selectively important (for mating displays) by controlling skin, scale, and feather carotenoid pigmentation in birds, fish and other vertebrates. If so, it would be an important example of evolution at the regulatory level rather than at protein sequence.
In terms of domestication, early farmers may have wanted egg yolks with extensive carotenoids coloring and may have taken bright yellow skin as a proxy for healthy birds, thus imposing selection on their flocks, though QTL studies today do not support such an association.
Two chicken ESTs (CN221314 and DR424873) indicate that the promoter region and its CpG island occur upstream of 6282641, suggesting that the SNP should be located in the 5,260 bp region between there and 6287900. However no striking conserved non-coding region exists in chicken; human regulatory tracks at the UCSC also do not illuminate how or where expression is regulated here.
The K416N SNP can be N, G, D, E, H, L, K, and R in vertebrate orthologs indicating no catalytic significance:
*
>BCO2_homSap VHNSAAKSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT
>BCO2_panTro VHNSAAKSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT
>BCO2_ponPyg VYNSAAKSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT
>BCO2_macMul VYNSAGRSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT
>BCO2_calJac VYNSAVRSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGK
>BCO2_tarSyr VYDSIARSCPRRFVLPLNVSLNAPEEENLSPLSYSSATAVKKTDGK
>BCO2_otoGar VYNSAARSFPRRFVLPLNVGLNAPEGENLSPLSYSSASAVKQADGK
>BCO2_tupBel VYNSVARSFPRRFVLPLGVSSNAPEGENLSPLSYSSASAVKLSDGK
>BCO2_musMus VYELKAKSFPRRFVLPLDVSVDAAEGKNLSPLSYSSASAVKQGDGE
>BCO2_ratNor VYKAKAKSFPRRFVLPLDISVGAPEGENLRPLPYSSASVVKQGDRE
>BCO2_cavPor VYRSVARSFPRRFVLPLDVTVNTPEGETLSPLSYSSASAVKQADGK
>BCO2_speTri VYNSAALSFPRRFVLPLHISENDPAGENLSPLSYSSASAVKQSDGK
>BCO2_oryCun VYNSVARSFPRRFVLPLNVSINAPEGKNLSPLTYSSASAVKQADGK
>BCO2_vicPac VYNSVARSFPRRFVLPLHVSLNAPERENLSPLSYSSASAVKQADGK
>BCO2_turTru VYNSVARSFPRRFVLPLHVGLNAPEGKNLSPLTYSSASAVKQADGK
>BCO2_bosTau VYNLIARNSPRRFVLPLLGNLNAPEGENLSPLTYSSASAVKQADGK
>BCO2_equCab VYNSAAKSFPRRFVLPLHVSLDAPEGKNLSPLSYSSASAVKQADGK
>BCO2_felCat VYNSVGRSFPRRFVLPLHVSLNDPEGQNLSPLSYSSASAVKQADGK
>BCO2_canFam VYNSVGRSFPRRFVLPLHVSLNDPEGENLSPLSYSSASAVKHADGK
>BCO2_myoLuc VYNSVATSFPRRFVLPLRVSLNAPEGENLSLLSYPSASAVKQADGK
>BCO2_pteVam VYNSVARSFPRRFVLPLHINLNAPEGENLSPLPYSSASAVKQADGK
>BCO2_eriEur VYNSVARSFPRRFVLPLNVNLDTPEEENLSPLSYSSASAVKQADGT
>BCO2_sorAra VYNSLARSFPRRFVLPLNISPNAPQGENLSSLSYSSASAVKQADGK
>BCO2_loxAfr VZISVARSFPQRFVVPLNVSLNAPEGQTLGLLSRSLASVVEQADGK
>BCO2_dasNov VYNSVARSFPRRFVLPLNVSLNAPEGENLSPLSYSSASAVK-ADG-
>BCO2_choHof VYNSVARSFPRRFVLPLDVSLNAPEGENLSSLSYSSASAVKQADGK
>BCO2_monDom VYNSVARAFPRRFVLPLDISTKASVGQNLSPLTYSSASAVKQADGT
>BCO2_ornAna VYNSAARTFPRRFVLPLDAFLDAPTGENLSPLTYSTAKAMRGEDGK
>BCO2_galGal IFGSVARTFPRRFVLPLKVNSDTPVGKNLNPLSYTSAKAVKDSDGK
>BCO2_galLaf IFGSVARTFPRRFVLPLNVNSDTPVGKNLNPLSYTSAKAVKDSDGK Gallus lafayetii
>BCO2_taeGut ---------------------------------------------- finch trace coverage unavailable
>BCO2_anoCar TYNSIATPYPRRFVLPLNIDDKKPVGENLSPLSYTSATAVKEADGK
>BCO2_tetNig VYNSTGRAFPRRFVLPLHVTSQTAAGHNLNTRPZSKETCVKTDKDT
>BCO2_takRub VYNSTGRAFPRRFVLPLHVTSEMATGQNLNTRASSQATCVKTGKDT
>BCO2_gasAcu VYNTLCRVFPRRFVLPLNVDQETPYGRNLN--PKGSATSVRFAKNK
>BCO2_oryLap VYNTTERSFPRRFVLPLNLTSETPTNQNLNTRPFSKASCTKISPDK
>BCO2_danRer FFNSLCTNLPRRYVLPLEVKEDEPNDQNLINLPYTTASAVKT-QTG
*
Reference gene collection: non-teleosts
Here the primary focus is obtaining reliable full length genes for the three members of this gene family from early diverging species (rather than standard teleost sequences more closely related to human), the idea being to possibly correlate gene expansions of the RPE65 family with the origin of imaging vision in deuterostomes. However partial sequences are also of valuable for purposes of establishing paralog numbers and presence.
Here it must be immediately noted that many Gnomon predicted sequences at GenBank are so deeply flawed that their uncritical use would hopelessly taint any comparative genomics effort. Without transcripts and given the rates of divergence, it is quite difficult to recover full length genes by blast into incomplete and sometimes garbled assemblies. Of the early deuterostome invertebrates, only Ciona has an adequate (tileable) set of transcripts vis-a-vis this gene family. Yet tunicates have lost one family member altogether, BCO2.
This gene family is quite unusual in that new members are quite difficult to classify by blast clustering relative to a reference collection. The match qualities tend to be quite similar, possibly attributable to gap issues and regions of uncertain and unpersuasive alignment. Here it is best to classify by individual exons because each paralog is intronated distinctively.
Because intronation patterns are strongly conserved from human back to pre-bilaterans (eg cnidarian and placamorpha), the fact that the three paralogs are intronated quite differently implies gene duplication and divergence occured very early, prior to the main era of intron establishment in early pre-metazoan eukaryotes. This is consistent with full length counterparts with conserved 3D structure already existing in bacteria.
This in turn implies that loss of one or more family members has occured in many lineages. For example, arthropods (notably many insect species) contain but a single gene copy (denoted NinaB), lophotrochozoan genomes have none (despite ciliary opsins), and tunicates have two. Gene expansions have also occured in species such as Nematostella yet no expansion occured in deuterostomes despite supposed 1R and 2R whole genome expansions.
>RPE65_homSap length=547 14 exons 0 MSIQ 21 VEHPAGGYKKLFETVEELSSPLTAHVT 1 2 GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR 2 1 FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR 2 1 FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ 0 0 VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA 1 2 DKEDPISKSEIVVQFPCSDRFKPSYVHS 21 FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG 0 0 VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG 21 FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK 0 0 ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ 1 2 AFEFPQINYQKYCGKPYTYAYGLGLNHFVPDR 0 0 LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD 1 2 GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* 0 >BCMO1_homSap length=547 11 exons 51,666 bp 0 MDIIFGRNRKEQLEPVRAKVT 1 2 GKIPAWLQGTLLRNGPGMHTVGESRYNHWFDGLALLHSFTIRD 1 2 GEVYYRSKYLRSDTYNTNIEANRIVVSEFGTMAYPDPCKNIFSK 2 1 AFSYLSHTIPDFTDNCLINIMKCGEDFYATSETNYIRKINPQTLETLEK 0 0 VDYRKYVAVNLATSHPHYDEAGNVLNMGTSIVEKGKTKYVIFKIPATVP 1 2 EGKKQGKSPWKHTEVFCSIPSRSLLSPSYYHSFGVTENYVIFLEQPFRLDILKMATAYIRRMSWASCLAFHREEK 0 0 TYIHIIDQRTRQPVQTKFYTDAMVVFHHVNAYEEDGCIVFDVIAYEDNSLYQLFYLANLNQDFKENSRLTSVPTLRRFAVPLHVDK 0 0 NAEVGTNLIKVASTTATALKEEDGQVYCQPEFLYE 1 2 GLELPRVNYAHNGKQYRYVFATGVQWSPIPTK 0 0 IIKYDILTKSSLKWREDDCWPAEPLFVPAPGAKDEDD 1 2 GVILSAIVSTDPQKLPFLLILDAKSFTELARASVDVDMHMDLHGLFITDMDWDTKKQAASEEQRDRASDCHGAPLT* 0 >BCO2_homSap length=579 12 exons alt leader peptide not shown 0 MGNTPQKKAVFGQCRGLPCVAPLLTTVEEAPRGISARVWGHFPKWLNGSLLRIGPGKFEFGKDK 2 1 YNHWFDGMALLHQFRMAKGTVTYRSKFLQSDTYKANSAKNRIVISEFGTLALPDPCKNVFERFMSRFELPGKAA 1 2 AMTDNTNVNYVRYKGDYYLCTETNFMNKVDIETLEKTEK 0 0 VDWSKFIAVNGATAHPHYDLDGTAYNMGNSFGPY 1 2 GFSYKVIRVPPEKVDLGETIHGVQVICSIASTEKGKPSYYHSF 1 2 GMTRNYIIFIEQPLKMNLWKIATSKIRGKAFSDGISWEPQCNTRFHVVEKRTGQ 0 0 LLPGRYYSKPFVTFHQINAFEDQGCVIIDLCCQDNGRTLEVYQLQNLRKAGEGLDQ 0 0 VHNSAAKSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT 0 0 IWCSHENLHQEDLEKEGGIEFPQIYYDRFSGKKYHFFYGCGFRHLVGDSLIKVDVVNKTLK 0 0 VWREDGFYPSEPVFVPAPGTNEEDGGVILSVVITPNQ 0 0 NESNFILVLDAKNFEELGRAEVPVQMPYGFHGTFIPI* 0 >BCDO2_galGal 67% identical homSap, FVLPLK yellow SNP, RefSeq XM_417929 in error 0 MQFVPGLKNSLPSPLEEQFTLPTLQCISPLLQTVEETPEPIPAKIKGHIPGWINGNLLRNGPGKFEFGEEK 2 1 YNHWFDGMALLHQFQLRNGTVTYQSKFLQSNSYLINNQHNRIVVSEFGTLAMPDPCKSVFARFMSRFDPP 1 2 PSDNANVSYVVYKGDYYVTGENNCMYKVDPETLEMKEK 0 0 VDWTKFVAVNGATAHPHYAPDGTAYNMGNSYGKF 1 2 GTTYNIIEVPPQKSNCNETLEGAKVLCSIAPTDNMKPSYYHSF 1 2 GMSENYIIFIEQPIKLNLLRIITSKFRGKPISEGINWEPQYNTRFHVVDKRTGK 0 0 VLPGQWYTKPFVTFHQINAFEDRGCVVLDLCCQDDGKTLAVYKLQNMRKSGADLDQ 0 0 IFGSVARTFPRRFVLPLKVNSDTPVGKNLNPLSYTSAKAVKDSDGK 0 0 VWCTHENLHPDGFENFGGLEFPQINYSQYSGRKYRYFYGCGFRHFIGDSLMKVDVETKNFK 0 0 IWQEDGSYPSEPVFVPVPNATAEDSGVILSVVISPDE 0 0 NRSAFLLVLDAETFRELGRAEVPVQMPYGFHGIFSSR* 0 >RPE65_cioInt 40% but about equally similar, slightly different intronation 0 MFAIQRRPFFAVFRNFNKMSAPSKTKSYVKLLQKAEERANAECVVT 1 2 GCIPEWLNGDVLRNGPAEFDIGPDTFKHWFDGHALLHK 2 1 FSMFEGKVTYSSKFLRSGTYKTNHENSRIIIGEFGTASRPDPCKNMFSR 2 1 FFTNFVEIAPRSDNANVSVAQLGEAYYAITDGPTAYGFDPETLETKNLITDCGPANMTVTAAHPHY 1 2 DRNGDYLNLGTTFGRTPHYHVIKVPAAKMTSPDPMNELEVFMKFPSTTSNASYHHS 2 1 FGLSENWIIFHEQPFSFSTPKLLIGLKLWNPILSSFYEDKQTIS 0 0 FHIINKTTGEKIATKYEARGMFCFHHINAYETKENDGKRFIVVDMCGSDRSLVWLL 2 1 GLDTLLDEEAHDKVVSNLDEKYLTRPRRIVIPLDISSDTPN 1 2 DTNLVTIPGCKATAMLNKSGVVSLTYELLVPDDFPNTELGIELPRINYDGYNGREYK 2 1 FIYAISSEYILPSHLVKINVETKEIKYWKEK 0 0 DKYTSEPIFVPRPGSQDEDDGVVLSTVISPTDDKTFLLILDGQSFKEIARAE 0 0 IETKMSYPLHGLFSK* 0 >BCMO1_cioInt 525 aa tiled transcripts agree with genomic 0 MDFPVSAFPHLTALATTKNIEYAEAVQGKVQ 1 2 GEVPSWLNGSWYRNGPGVVHFREESVKHWFDGMALARK 2 1 FCIEDGKVSYMSRLVDGESLQKNTAAGRVVVAEFGTTTHSEGFLGR 2 1 VKSALTMPEFTDNCLINFMNLGDHLFAITESNFIRQIDPVTLDTKDK 0 0 VDLAKHLPINIMSSHPLVDGEGNVYTFSSSIFNMGRTKYNLLKFPAAAP 1 2 GTPLETILSQSESICSIDSSWRVSPSYHHSFAMSEKYAVFVEMPLKIDIPKMAVAHLRHMCYSDCIEVLEDTK 0 0 TRIYLVNKETGKQHPITFLCDPLIVYHHVNAYDDGDHVVLDLSCYKKNSFYDKFTMSNLEKTPQEFSKLFDSDEQAVKAMRIVLPLANDS 0 0 KTTGNLVSVANTSCTAEFQGNNIFCTSEMLSVGTECAVINNKYIGKKYKYFYSPGGLKLPPGEM 0 0 LTKIDVETKQRVQTWQEKGCWASQPVFVAKPGATQEDE 1 2 GILMSSVVNENGNPFLLMLDAKSFTEVARIHFDANIPPDVHGVFVPKA* 0
Regularized reference gene collection: teleost RPE65
It is easy to compile large sets of bony vertebrate exons for these three genes using the 44-species genomic alignments at UCSC. To do this, look up the human gene by name at GeneSorter (or blat in a reference sequence) and click on "protein fasta" on the gene details page. Among the various output configurations provided, select all species checking the options as below. Note these alignments, being exon-aware, are intrinsically homological unlike blast alignments in gappy regions which lack this local constraint.
For some species, no data is available (shown as dashes). For others, some exons or parts of exons are misssing. This can be due to incompleteness of the respective genome or technical difficulties at exon edges due to split codons. To facilitate uniform comparison of paralogs, the output can be 'regularized' by filling in missing data using the nearest species in the taxonomic sense, for example using chicken data if finch is missing an exon. Most human genes can be completely regularized back to teleost fish. In some cases, notably frog, it may be necessary to stub in an orthologous region from salamander cDNA.
This introduces artefacts that have the effect of understating comparative genomics variability at the locus but that error is offset by the benefits of having 'complete' copies of the three genes in the same set of 44 species. As long as regularization is limited (topologically) to species diverging off the same node (eg lizard could regularize finch and chicken but not frog or platypus), the impact on gene history at the ancestral nodes leading to human will be very minimal. The regularized set works quite well in a blast classificatory tool.
The sequences for RPE65, BCMO1, and BCO2 have been completely regularized in this manner below. This allows comparison of variation by various techniques such as percent identity at nodes (ie averaging human matches to all species coming off that node if ancestral sequences are not computed) and residue-by-residue conservation by Multalin and similar tools.
Regularization can be done by hand in a spreadsheet using fill-down and similar commands. Begin by creating a template whose first column consists of the 44 species. Color rows by subclade as a visual assist to filling in using tree topology as constraint. Now copy the exons, replace all Z by *, replace double carriage returns by a placeholder, replace remaining carriage returns by tabs, replace placeholder with single carriage return, place into spreadsheet, and reverse rows and columns. Now each column is a separate exon and guided regularization can begin.
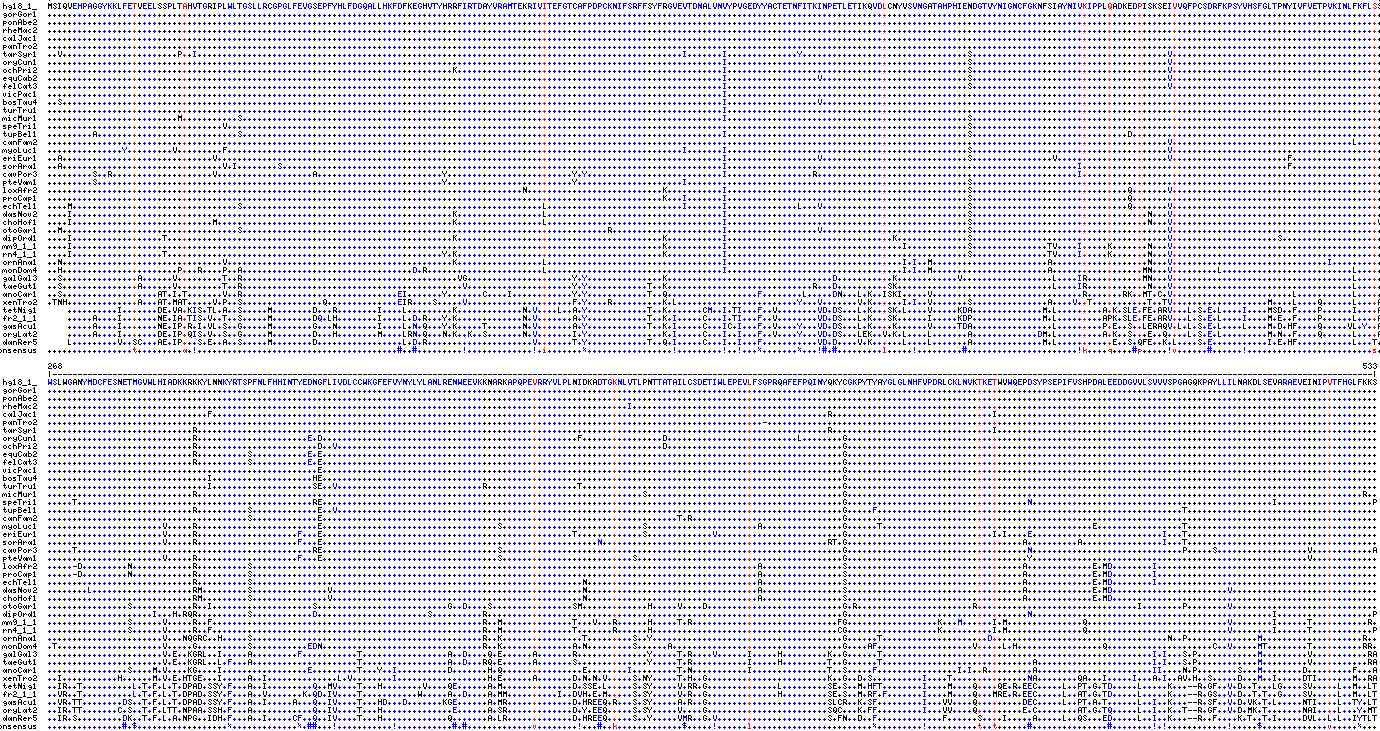
Differance alignment of RPE65 relative to homSap: hg18_1_ MSIQVEHPAGGYKKLFETVEELSSPLTAHVTGRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRRFIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSRFFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQVDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQADKEDPISKSEIVVQFPCSDRFKPSYVHSFGLTPNYIVFVETPVKINLFKFLSS gorGor1 ........................................................................................................................................................................................................................................................................... ponAbe2 ........................................................................................................................................................................................................................................................................... rheMac2 ........................................................................................................................................................................................................................................................................... calJac1 ........................................................................................................................................................................................................................................................................... panTro2 ........................................................................................................................................................................................................................................................................... tarSyr1 ..V.......................P..I.................................................................................................I.......I..............Y.................................S.......................................V.......................................... oryCun1 .......................................................................................................................................I................................................S.......................................V.......................................... ochPri2 .................................................................................K.....................................................I................................................S.......................................V.......................................... equCab2 .......................................................................................................................................I..................V.............................S.......................................V.......................................... felCat3 .......................................................................................................................................I................................................S.......................................V.......................................... vicPac1 .......................................................................................................................................I................................................................................................................................... bosTau4 ..S....................................................................................................................................I..................V................................................................................................................ turTru1 .......................................................................................................................................I................................................................................................................................... micMur1 ..........................M...........S................................................................................................I................................................S.................................................................................. speTri1 ...................................V....................................................................................................................................................S.................................................................................. tupBel1 .........A............................S................................................................................................I................................................S...............................D.................................................. canFam2 ................................................................................................................................................................................................................................V....................................L..... myoLuc1 ...............Y.........V.........F...........................................................................................I.......I................................................S.......................................V.......................................... eriEur1 ..A..............................V.....................................................................................................I..................V.............................S................V......................V.......................F.................. sorAra1 ..A................................V.I........S........................................................................................I......................................................................I.........................................F.................. cavPor3 .........S..R....................V...................A.........................Y.........................Y.Y...........................I.................R....................................................I............................................................ pteVam1 .........S.....................................................................Y.........................Y.....................I........................................................S.................................................................................. loxAfr2 ...............................................................................................N...........................K...........I................................................S...............................Q.......V.......................................... proCap1 ...........................................................................................................................K...I.......I................................................S...............................Q.................................................. echTel1 ....M.................................S............................................................L...........................I.......I..............L...V.............................S...............................Q.......V.......................................... dasNov2 ....I............................................................................K.................L...................................I................................................S...................................N...V.......................................... choHof1 ....I............................M...............................................K.................L...................................I................................................S...................................N...V.........................................V otoGar1 ..M...................................S.........................................................................R......................I..................V.............................S.......................................V.......................................... dipOrd1 ....I..................T.........................................................K.........................................K...........I.................................K..............S.......................................V.....................S.................... mm9_1_1 ....I......................................................................................................................K...........I...................................I............S...............TV....I.....K.......N...V.......................................... rn4_1_1 ....I..................T.......................................................Y.K.........................................K...........I................................................S...............TV....I.....K.......N...V.......................................... ornAna1 ..N................................V.............................................K.................L...................................I...................................I.I..M.......................A...................N...V...............................I.......... monDom4 ..H.......................P...R....P..A..................................D.R.......................L...................................I...................................I.I..M.......................A..................MN...V.........L..........................L..... galGal3 ..S...............A......V.........T..R...........................................VG.....................Y.Y...............K.................................D...........K............V..................L....IR...........MN...V....................................L..... taeGut1 ..S...............A......V.........T..R............................................V.....................Y.Y............T..K.............................R...D..........SK...............................L....IR............N...V....................................L..... anoCar1 ..S...................AT.I.T.......V..R...............................EI.......Y.......C....I............Y.Y............T..Q..................F........L.....DN...L.K..ISKI.....V..............................R.......RK..MT.C.V........................L...........L..... xenTro2 .TNH..............A...AT.MAT.....V.P..S................Q..............EIR....S.....V.....................F.Y....R..........K.L........................Y...V.......V.K......I.I..V.......S...............A....V..T...........T..TV...................M....L....Q......L....A tetNig1 ..........I.......DE.VA.KIS.TL.A..S.....M........D...R........I....L.........K.............N.V....L..A.Y............T..K.......CM..I.TI...F..V........VD.DS...L.K...SK.L....V.....KDA..............M.L.........A.K.SLE.FE.ARV...L.S.E.L....I....MSD..F....P......I....A fr2_1_1 .....A....I.......NE.IA.TIS.V..T..S.....M........DQ.LH........I....L.D.R...Y.K.............N.V.......A.Y............T..K.I.....C...I.TI...F..V........VD.DS...L.K...SK......I.....KDP..............M.L.........APK.SLE.FE.ARV...L.S.E.L....I....M.E..F....P......L....A gasAcu1 .....A....I.......NE.IP.R.I.VL.S..G.....M........G...N........I....L.N.Q...Y.K.....T.......N.V.......A.Y............T..K.I.....C...IC.I...F..V....Y...VD.DS...L.K....K......V.....TDA................L.........P.E..S..LERAQV.L.L.S.E.L....L....M.D.HF....Q.....VL.Y..A oryLat2 .....A....I.......DE.IP.QIS.V..S..G.....M........D...H........M....LRN.Q...N.K..K..S.......N.V.......I.Y...............K.I.....C...I..I...F..V....Y...VD.DS...LEK..V.K.L....L.......A.............DM.L.........A.K..S...E..Q....L.S.E.L.........M.N..F....Q......L....A danRer5 L.........V..SC...AE.IP...S.E..A..S.....M........D.................L.D.R.....................V.......T.Y............T..Q.I.....C...I..I...F...........VD.D....V.K......L....L.......A..............M.L...........E..S.QFE..K.L....S.E...........I.E.HF...........L...T. hg18_1_ WSLWGANYMDCFESNETMGVWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKGFEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDKADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQAFEFPQINYQKYCGKPYTYAYGLGLNHFVPDRLCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDDGVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS gorGor1 .......................................................................................................................................................................................................................................................................... ponAbe2 .......................................................................................................................................................................................................................................................................... rheMac2 ....................................................................................................................I..................................................................................................................................................... calJac1 ................................F...........................................................................................................................R................................I............................................................................ panTro2 ...............................................................................................................................................-.......................................................................................................................... tarSyr1 .............................R..............................................................................................................................R................................I............................................................................ oryCun1 .............................R......................E.D...................................................F................D..........................L........G.......................................................................................................... ochPri2 .............................R........................D..V.................................................................D...................................G.......................................................................................................... equCab2 .............................R..........S...........E.E........................................................................................................G.......................................................................................................... felCat3 .............................R..........S...........E.E........................................................................................................G.......................................................................................................... vicPac1 ......................................................E........................................................................................................G.......................................................................................................... bosTau4 ................................I....................HE........................................................................................................G.......................................................................................................... turTru1 ................................I....................SE..V.............................R..................T....................................................G.......................................................................................................... micMur1 .............................R.........................................................................................S.......................................G.......................................................................................................R.. speTri1 .....T...............................................RE........................................................................................................G....................................N................................................I...................P tupBel1 .............................R..........S.............E..V.....................................................................................................G.....F.............................................................T...................................... canFam2 ........................................S.............E.......................................................................T.R..............................G.......................................................................................................... myoLuc1 .......................V.....R........................E...................................S............................S......................A................G......T..........................................E.................T...................................... eriEur1 .......................V.....R....................F...E..................................................T.............S.............V.........................G.............................I.I....E..........................I...T...................................... sorAra1 .......................V.....R....................F...E.......................................................N.............................................RT.G...................................A..........A................I...T...................................... cavPor3 .....T...............................................RE...................................S....................................................................G....................................N..............................P.....S..................V............A pteVam1 .......................V.....R....................F...E...................................S..........................S.........................................G....................................Y.......................................................V............. loxAfr2 .....-D.........N............R..........S.....................................................................................................A................S...................................A.............E.MD........I............................................ proCap1 .....-D.........N............R..........S.....................................................................................................A................S...................................A.............E.MD........I............................................ echTel1 .............................R..........S..................................................................N..................................A................S...................................A.............E.MD........I............................................ dasNov2 ........L....................RM.........S...............V..................................................N..................................A................S...................................A.............E.MD.......................V............................. choHof1 .............................RM.........S...............V..................................................N..................................A................S...................................A.............E.MD..................................................... otoGar1 ................S............R..I....................S..........................G..D....S................SM.............H.....V....D...........................G.R.......................R................................I........P........V............................. dipOrd1 .....................I...H.RQR..........S............D......................S..........R.......................................................................G....................................N................................................I...................P mm9_1_1 ................S......V.....R..F......................................................R..M..............T...V...R......H.....T.R.............................FG..................K...M......I.M...............Q............................V........I......T...........R. rn4_1_1 ................S......V.....R..F......................................................R..M..............T.......R......H.....................................CG..................K..........I.M...............Q............................V........I......T............P ornAna1 .......................V...NQGRC..H.....S..............................................R..K................T.........S..Y......................................G......T.....................D...................................N..P..............M....................R.. monDom4 .T.....................V.....G..........S...........EDN................................R..K..............A.N.........S......A..................................G....AF...................V.....L.H.................Q.....................C..V.....MT........T..........RR. galGal3 .......................V.E..KGRL..I.....A.........F...........T.................A..D....Q.E......A.......R..............Y.....T.R....V......I.....H.........K..G......T......................................................I.I...S.P............M.........V...........RA taeGut1 .......................V.E..KGRL..L.F...A.....................T.................A..D...RQ.E......A.......S..............Y.....T.R...........I.....H.........T..G......T..............................A.......................I.I...S.P............M.........V...........RA anoCar1 ................S....M.V....KG....I.................E.........T...Y..I..........D.......H.Q................E........I.........T.N...........I...............T..S.....F................I.I..R.........A.......................II....N.P........S...M........D.........F...A xenTro2 ..I...........H......M.V.E.HTGE...I.....A..I..............L.V........I..................H.E......A.......D.N.N.V.....S.NY.....T.H..G........................K..G..D.S..........I....T...............NA........QA...I.....I...A.I..AV.H..S.....D...M..I.....DTI.......M..RA tetNig1 ..IR..T..........L.T.F.L.T.DPAD.SSY.F...A..I.........Q..MV....T...H........V....QE......A.M..............D.SSE.L.....S.SY.....V.RR.G..............L.........SE.S..M.HFT........I....M....Q....QE.R.EEC.....L..PT.G.TD.....L.....K...--R.GF..V.D..T...LG....SV.....L..M..LT fr2_1_1 ..VR..T..........L.T.F.L.T.DPAD.SSY.F...A..V.......K.QD.IV....T...........W.....A..D....A.MM..........I..DVH.E.M.....S.SY.....V.R..G..............H.........SE.S..M.RF.F.......I....M....Q...MRE.R.EEC.....L..AT.G.TD.....L.....K...--R.GS..V.D..T.T..G....SV.....L..M..LT gasAcu1 ..VR.TT........DS..T.F.L.T.DPAD.SSY.F...A..I.....A...Q..IV....T...HD...D.......KGE......A.MR.............D.HREEQ.R...S.SY.....A.R..G........................SLCR..K.SF.........I...VV....Q.........DEC.....L..PT.A.T......L..I..K...--R.SF..V.D.VK.T.L.....NTI....L..TY.LT oryLat2 ..IR.TT.......C.S..T.F.LTT.NPAA.SSH.F...A............Q..IV....T...H..............E......A.SR.............D.Y.EEQ.....S.SY.....V....G........................SQC...K.FF.........I...VV....Q.........E.C........AT.G.TQ.....L..I..K..T--R.GF..V.D.MK.T.......NAI....L...Y.MT danRer5 ..IR.S.........DK..T.F.L.A.NPG..IDH.F...A..I.....CF..Q..IV....T...H.............Q.......A.LR.............D.HREEQ.....S..Y.....VMR..G.V......................S.FN..D..F.............F......S....I.....A.....L..QS...ED.....L..I..K...--R..F....K.T..T.I.....DVL..L.L..IYTLT
>RPE65_homSap Homo sapiens (human) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYCGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_panTro Pan troglodytes (chimp) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFS-PRQ AFEFPQINYQKYCGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_gorGor Gorilla gorilla (gorilla) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYCGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD gVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_ponPyg Pongo pygmaeus (orang_sumatran) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYCGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_macMul Macaca mulatta (rhesus) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLITLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYCGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_calJac Callithrix jacchus (marmoset) MSIQ vEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYFNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYRKYCGKPYTYAYGLGLNHFVPDR LCKLNVKTKEIWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_tarSyr Tarsius syrichta (tarsier) MSVQ VEHPAGGYKKLFETVEELSSPLPAHIT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEITDNALVNIYPVGEDYYACTETNYITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRRKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYRKYCGKPYTYAYGLGLNHFVPDR LCKLNVKTKEIWVWQEPDSYPSEPIFVSHPDALEEDD gVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_micMur Microcebus murinus (mouse_lemur) MSIQ VEHPAGGYKKLFETVEELSSPLMAHVT GRIPLWLSGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA dKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRRKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKg FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLSNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFRKS* >RPE65_otoGar Otolemur garnettii (bushbaby) MSMQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLSGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCRNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKVNPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNESMG VWLHIADKKRRKYINNKYRTSPFNLFHHINTYEDSGFLIVDLCCWKG FEFVYNYLYLANLRGNWDEVKKSARKAPQPEVRRYVLPLSMDK ADTGKNLVTLPHTTATAVLCSDDTIWLEPEVLFSGPRQ AFEFPQINYQKYGGRPYTYAYGLGLNHFVPDR LCKLNVRTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVILSVVVSPGPGQKPAYLLVLNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_tupBel Tupaia belangeri (tree_shrew) MSIQ VEHPAAGYKKLFETVEELSSPLTAHVT GRIPLWLSGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA dKDDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRRKYLNNKYRTSSFNLFHHINTYEDNEFLVVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTFAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGTGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_musMus Mus musculus (mouse) MSIQ IEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFKGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYISVNGATAHPHIESDGTVYNIGNCFGKNFTVAYNIIKIPPLKA DKEDPINKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNESMG VWLHVADKKRRKYFNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKRNAMKAPQPEVRRYVLPLTIDK VDTGRNLVTLPHTTATATLRSDETIWLEPEVLFSGPRQ AFEFPQINYQKFGGKPYTYAYGLGLNHFVPDK LCKMNVKTKEIWMWQEPDSYPSEPIFVSQPDALEEDD GVVLSVVVSPGAGQKPAYLLVLNAKDLSEIARAEVETNIPVTFHGLFKRS* >RPE65_ratNor Rattus norvegicus (rat) MSIQ IEHPAGGYKKLFETVEELSTPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYYRK FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFKGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFTVAYNIIKIPPLKA DKEDPINKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNESMG VWLHVADKKRRKYFNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKRNAMKAPQPEVRRYVLPLTIDK ADTGRNLVTLPHTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKCGGKPYTYAYGLGLNHFVPDK LCKLNVKTKEIWMWQEPDSYPSEPIFVSQPDALEEDD GVVLSVVVSPGAGQKPAYLLVLNAKDLSEIARAEVETNIPVTFHGLFKKP* >RPE65_dipOrd Dipodomys ordii (kangaroo_rat) MSIQ IEHPAGGYKKLFETVEELSTPLTAHVT gRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRK FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFKGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCKYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTSNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWIHIAHKRQRKYLNNKYRTSSFNLFHHINTYEDDGFLIVDLCCWKG FEFVYNYLYLSNLRENWEEVKRNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPNSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEIARAEVEINIPVTFHGLFKKP* >RPE65_cavPor Cavia porcellus (guinea_pig) MSIQ VEHPASGYRKLFETVEELSSPLTAHVT GRVPLWLTGSLLRCGPGLFEVGAEPFYHLFDGQALLHKFDFKEGHVTYYRR FIRTDAYVRAMTEKRIVITEFGTYAYPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITRINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIIKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGTNYMDCFESNETMG VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDREFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNASKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPNSYPSEPIFVSHPDALEEDD gVVLSVVVSPGPGQKPASLLILNAKDLSEVARAEVEVNIPVTFHGLFKKA* >RPE65_speTri Spermophilus tridecemlineatus (squirrel) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPVWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGTNYMDCFESNETMG VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDREFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPNSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEIARAEVEINIPVTFHGLFKKP* >RPE65_oryCun Oryctolagus cuniculus (rabbit) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT gRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRRKYLNNKYRTSPFNLFHHINTYEENDFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNFDK ADTGKNLVTLPNTTDTAILCSDETIWLEPEVLFSGPRQ aFELPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_ochPri Ochotona princeps (pika) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRK FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRRKYLNNKYRTSPFNLFHHINTYEDNDFLVVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTDTAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_vicPac Vicugna pacos (lama) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDNEFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_turTru Tursiops truncatus (dolphin) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYINNKYRTSPFNLFHHINTYEDSEFLVVDLCCWKG FEFVYNYLYLANLRENWEEVKRNARKAPQPEVRRYVLPLNTDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_bosTau Bos taurus (cow) MSSQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKVNPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYINNKYRTSPFNLFHHINTYEDHEFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_equCab Equus caballus (horse) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKVNPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRRKYLNNKYRTSSFNLFHHINTYEENEFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD gVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_felCat Felis catus (cat) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT gRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA dKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRRKYLNNKYRTSSFNLFHHINTYEENEFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD gVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_canFam Canis familiaris (dog) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLLKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRKKYLNNKYRTSSFNLFHHINTYEDNEFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATATLRSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_myoLuc Myotis lucifugus (microbat) MSIQ VEHPAGGYKKLYETVEELSSPVTAHVT GRIPFWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHkFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEITDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHVADKKRRKYLNNKYRTSPFNLFHHINTYEDNEFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNASKAPQPEVRRYVLPLNIDK ADTGKNLVTLSNTTATAILCSDETIWLEPEVLFAGPRQ AFEFPQINYQKYGGKPYTYTYGLGLNHFVPDR LCKLNVKTKETWVWQEPDSYPSEPIFVSHPEALEEDD GVVLSVVVSPGTGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_pteVam Pteropus vampyrus (macrobat) MSIQ VEHPASGYKKLFETVEELSSPLTAHVT GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYYRR FIRTDAYVRAMTEKRIVITEFGTYAFPDPCKNIFSR FFSYFRGVEITDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA dKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHVADKKRRKYLNNKYRTSPFNLFHHINTFEDNEFLIVDLCCWKg FEFVYNYLYLANLRENWEEVKKNASKAPQPEVRRYVLPLNIDK ADTGKNLVSLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEPYSYPSEPIFVSHPDALEEDD GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEVNIPVTFHGLFKKS- >RPE65_eriEur Erinaceus europaeus (hedgehog) MSAQ VEHPAGGYKKLFETVEELSSPLTAHVT GRVPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKVNPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSVAYNIVKIPPLQA DKEDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNFIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHVADKKRRKYLNNKYRTSPFNLFHHINTFEDNEFLIVDLCCWKg FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLTIDK ADTGKNLVTLSNTTATAILCSDETVWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKEIWIWQEPESYPSEPIFVSHPDALEEDD GVVLSVVISPGTGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_sorAra Sorex araneus (shrew) MSAQ VEHPAGGYKKLFETVEELSSPLTAHVT gRIPVWITGSLLRCGSGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIIKIPPLQA DKEDPISKSEIVVQFPCSDRFKPSYVHS FGLTPNFIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHVADKKRRKYLNNKYRTSPFNLFHHINTFEDNEFLIVDLCCWKg FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ANTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYRTYGGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEADSYPSEPIFVAHPDALEEDD GVVLSVVISPGTGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_loxAfr Loxodonta africana (elephant) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT gRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTENRIVITEFGTCAFPDPCKNIFSR FFSYFKGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKQDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWG-DYMDCFESNENMG VWLHIADKKRRKYLNNKYRTSSFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFAGPRQ AFEFPQINYQKYSGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEADSYPSEPIFVSHPEAMDEDD gVVLSIVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_proCap Procavia capensis (hyrax) MSIQ VEHPAGGYKKLFETVEELSSPLTAHVT gRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR FFSYFKGVEITDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA dKQDPISKSEIVVQFPCSDRFKPSYVHs FGLTPNYIVFVETPVKINLFKFLSSWSLWG-DYMDCFESNENMG VWLHIADKKRRKYLNNKYRTSSFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFAGPRQ AFEFPQINYQKYSGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEADSYPSEPIFVSHPEAMDEDD GVVLSIVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_echTel Echinops telfairi (tenrec) MSIQ MEHPAGGYKKLFETVEELSSPLTAHVT GRIPLWLSGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FIRTDAYVRAMTEKRIVLTEFGTCAFPDPCKNIFSR FFSYFRGVEITDNALVNIYPVGEDYYACTETNLITKVNPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKQDPISKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG VWLHIADKKRRKYLNNKYRTSSFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNINK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFAGPRQ AFEFPQINYQKYSGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEADSYPSEPIFVSHPEAMDEDD GVVLSIVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_dasNov Dasypus novemcinctus (armadillo) MSIQ IEHPAGGYKKLFETVEELSSPLTAHVT gRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRK FIRTDAYVRAMTEKRIVLTEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPINKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYLDCFESNETMG VWLHIADKKRRMYLNNKYRTSSFNLFHHINTYEDNGFVIVDLCCWKg FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNINK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFAGPRQ AFEFPQINYQKYSGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEADSYPSEPIFVSHPEAMDEDD gVVLSVVVSPGAGQKPAYLLVLNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_choHof Choloepus hoffmanni (sloth) MSIQ IEHPAGGYKKLFETVEELSSPLTAHVT GRMPLWLTGSLLRCGPGLFEvGSEPFYHLFDGQALLHKFDFKEGHVTYHRK FIRTDAYVRAMTEKRIVLTEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYVSVNGATAHPHIESDGTVYNIGNCFGKNFSIAYNIVKIPPLQA DKEDPINKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLFKFLSVWSLWGANYMDCFESNETMG VWLHIADKKRRMYLNNKYRTSSFNLFHHINTYEDNGFVIVDLCCWKg FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNINK ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFAGPRQ AFEFPQINYQKYSGKPYTYAYGLGLNHFVPDR LCKLNVKTKETWVWQEADSYPSEPIFVSHPEAMDEDD gVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* >RPE65_monDom Monodelphis domestica (opossum) MSHQ VEHPAGGYKKLFETVEELSSPLPAHVR GRIPPWLAGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKDGRVTYHRR FIRTDAYVRAMTEKRIVLTEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYISINGMTAHPHIENDGTVYNIGNCFGKNFAIAYNIVKIPPLQA dKEDPMNKSEVVVQFPCSDRLKPSYVHS FGLTPNYIVFVETPVKINLLKFLSSWTLWGANYMDCFESNETMG VWLHVADKKRGKYLNNKYRTSSFNLFHHINTYEEDNFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKRNAKKAPQPEVRRYVLPLAINK ADTGKNLVSLPNTTAAAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYAFAYGLGLNHFVPDR LCKLNVVTKETWLWHEPDSYPSEPIFVSHPDAQEEDD GVVLSVVVSPGAGQKPACLLVLNAKDMTEVARAEVETNIPVTFHGLFRRS* >RPE65_ornAna Ornithorhynchus anatinus (platypus) MSNQ VEHPAGGYKKLFETVEELSSPLTAHVT GRIPVWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRK FIRTDAYVRAMTEKRIVLTEFGTCAFPDPCKNIFSR FFSYFRGVEVTDNALVNIYPVGEDYYACTETNFITKINPETLETIKQ VDLCNYISINGMTAHPHIENDGTVYNIGNCFGKNFAIAYNIVKIPPLQA DKEDPINKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPIKINLFKFLSSWSLWGANYMDCFESNETMG VWLHVADKNQGRCLNHKYRTSSFNLFHHINTYEDNGFLIVDLCCWKG FEFVYNYLYLANLRENWEEVKRNAKKAPQPEVRRYVLPLNITK ADTGKNLVSLPYTTATAILCSDETIWLEPEVLFSGPRQ AFEFPQINYQKYGGKPYTYTYGLGLNHFVPDR LCKLNVKTKDTWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSVVVNPGPGQKPAYLLILNAKDMSEVARAEVEINIPVTFHGLFRKS* >RPE65_galGal Gallus gallus (chicken) MSSQ VEHPAGGYKKLFETAEELSSPVTAHVT GRIPTWLRGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR VGRTDAYVRAMTEKRIVITEFGTYAYPDPCKNIFSR FFSYFKGVEVTDNALVNVYPVGEDYYACTETNFITKINPDTLETIKQ VDLCKYVSVNGATAHPHVENDGTVYNIGNCFGKNFSLAYNIIRIPPLQA DKEDPMNKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLLKFLSSWSLWGANYMDCFESNETMG VWLHVAEKKKGRLLNIKYRTSAFNLFHHINTFEDNGFLIVDLCTWKG FEFVYNYLYLANLRANWDEVKKQAEKAPQPEARRYVLPLRIDK ADTGKNLVTLPYTTATATLRSDETVWLEPEVIFSGPRH AFEFPQINYKKYGGKPYTYTYGLGLNHFVPDR lCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD GVVLSIVISPGSGPKPAYLLILNAKDMSEVARAEVEVNIPVTFHGLFKRA* >RPE65_taeGut Taeniopygia guttata (finch) MSSQ VEHPAGGYKKLFETAEELSSPVTAHVT GRIPTWLRGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR FVRTDAYVRAMTEKRIVITEFGTYAYPDPCKNIFSR FFTYFKGVEVTDNALVNVYPVGEDYYACTETNFITRINPDTLETIKQ VDLSKYVSVNGATAHPHIENDGTVYNIGNCFGKNFSLAYNIIRIPPLQA DKEDPINKSEVVVQFPCSDRFKPSYVHS FGLTPNYIVFVETPVKINLLKFLSSWSLWGANYMDCFESNETMG VWLHVAEKKKGRLLNLKFRTSAFNLFHHINTYEDNGFLIVDLCTWKG FEFVYNYLYLANLRANWDEVKRQAEKAPQPEARRYVLPLSIDK ADTGKNLVTLPYTTATATLRSDETIWLEPEVIFSGPRH AFEFPQINYTKYGGKPYTYTYGLGLNHFVPDR lCKLNVKTKETWVWQEPDAYPSEPIFVSHPDALEEDD GVVLSIVISPGSGPKPAYLLILNAKDMSEVARAEVEVNIPVTFHGLFKRA* >RPE65_anoCar Anolis carolinensis (lizard) MSSQ VEHPAGGYKKLFETVEELATPITTHVT GRIPVWLRGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFEIKEGHVTYYRR FIRTDCYVRAITEKRIVITEFGTYAYPDPCKNIFSR FFTYFQGVEVTDNALVNVYPVGEDFYACTETNFLTKINPDNLETLKK VDISKIVSVNGVTAHPHIENDGTVYNIGNCFGKNFSIAYNIVRIPPLQA DRKDPMTKCEVVVQFPCSDRFKPSYVHS FGLTPNYLVFVETPVKINLLKFLSSWSLWGANYMDCFESNESMG VWMHVADKKKGKYLNIKYRTSPFNLFHHINTYEENGFLIVDLCTWKG YEFIYNYLYLANLRDNWEEVKKHAQKAPQPEVRRYVLPLNIEK ADTGKNLITLPNTTATATLNSDETIWLEPEVIFSGPRQ AFEFPQINYTKYSGKPYTFAYGLGLNHFVPDR lCKINIKTRETWVWQEPDAYPSEPIFVSHPDALEEDD GVVLSIIVSPGNGPKPAYLLILSAKDMSEVARAEVDINIPVTFHGFFKKA* >RPE65_xenTro Xenopus tropicalis (frog) MTNH VEHPAGGYKKLFETAEELATPMATHVT GRVPPWLSGSLLRCGPGLFEVGSEQFYHLFDGQALLHKFEIREGHVSYHRR FVRTDAYVRAMTEKRIVITEFGTFAYPDPCRNIFSR FFSYFKGLEVTDNALVNVYPVGEDYYACTETNYITKVNPETLETVKK VDLCNYISINGVTAHPHIESDGTVYNIGNCFGKNFAIAYNVVKTPPLQA DKEDPITKSTVVVQFPCSDRFKPSYVHS FGMTPNYLVFVEQPVKINLLKFLSAWSIWGANYMDCFESHETMG VWMHVAEKHTGEYLNIKYRTSAFNIFHHINTYEDNGFLILDVCCWKG FEFIYNYLYLANLRENWEEVKKHAEKAPQPEARRYVLPLDINK NDVGKNLVSLNYTTATATLHSDGTIWLEPEVLFSGPRQ AFEFPQINYKKYGGKDYSYAYGLGLNHFIPDR lTKLNVKTKETWVWQEPNAYPSEPIFVQAPDAIEEDD GIVLSAVISPAVGHKPSYLLILDAKDMSEIARAEVDTIIPVTFHGMFKRA* >RPE65_tetNig Tetraodon nigroviridis (pufferfish) ---- VEHPAGGYKKIFETVEELDEPVAAKIS GTLPAWLSGSLLRMGPGLFEVGDEPFRHLFDGQALIHKFDLKEGHVTYHRK FIRTDAYVRAMTENRVVITELGTAAYPDPCKNIFSR FFTYFKGVEVTDNCMVNIYTIGEDFYAVTETNFITKVDPDSLETLKK VDLSKYLSVNGVTAHPHKDADGTVYNIGNCFGKNMSLAYNIVKIPPAQK DSLEPFEKARVVVQLPSSERLKPSYIHS FGMSDNYFVFVEPPVKINLIKFLSAWSIRGATYMDCFESNETLG TWFHLATKDPADYSSYKFRTSAFNIFHHINTYEDQGFMVVDLCTWKG HEFVYNYLYVANLRQEWEEVKKAAMKAPQPEVRRYVLPLDISS EDLGKNLVSLSYTTATAVLRRDGTIWLEPEVLFSGPRL AFEFPQINYSEYSGKMYHFTYGLGLNHFIPDR LMKLNVQTKETQEWREEECYPSEPLFVPTPGATDEDD GVLLSVVVKPGA--RPGFLLVLDAKTLSELGRAEVSVNIPVTLHGMFKLT* >RPE65_takRub Takifugu rubripes (fugu) ---- VEHPAAGYKKIFETVEELNEPIAATIS GVIPTWLSGSLLRMGPGLFEVGDQPLHHLFDGQALIHKFDLKDGRVTYYRK FIRTDAYVRAMTENRVVITEFGTAAYPDPCKNIFSR FFTYFKGIEVTDNCLVNIYTIGEDFYAVTETNFITKVDPDSLETLKK VDLSKYVSVNGITAHPHKDPDGTVYNIGNCFGKNMSLAYNIVKIPPAPK DSLEPFEKARVVVQLPSSERLKPSYIHS FGMTENYFVFVEPPVKINLLKFLSAWSVRGATYMDCFESNETLG TWFHLATKDPADYSSYKFRTSAFNVFHHINTYKDQDFIVVDLCTWKG FEFVYNYLWLANLRANWDEVKKAAMMAPQPEVRRYVIPLDVHK EDMGKNLVSLSYTTATAVLRSDGTIWLEPEVLFSGPRH AFEFPQINYSEYSGKMYRFAFGLGLNHFIPDR lMKLNVQTKEMREWREEECYPSEPLFVATPGATDEDD GVLLSVVVKPGA--RPGSLLVLDAKTLTEVGRAEVSVNIPVTLHGMFKLT* >RPE65_gasAcu Gasterosteus aculeatus (stickleback) ---- VEHPAAGYKKIFETVEELNEPIPARVI GVLPSWLGGSLLRMGPGLFEVGGEPFNHLFDGQALIHKFDLKNGQVTYYRK FIRTDTYVRAMTENRVVITEFGTAAYPDPCKNIFSR FFTYFKGIEVTDNCLVNICPIGEDFYAVTETNYITKVDPDSLETLKK vDLCKYVSVNGVTAHPHTDADGTVYNIGNCFGKNFSLAYNIVKIPPPQE DKSDPLERAQVVLQLPSSERLKPSYLHS FGMTDNHFVFVEQPVKINVLKYLSAWSVRGTTYMDCFESNDSMG TWFHLATKDPADYSSYKFRTSAFNIFHHINAYEDQGFIVVDLCTWKG HDFVYDYLYLANLKGEWEEVKKAAMRAPQPEVRRYVLPLDIHR EEQGRNLVSLSYTTATAALRSDGTIWLEPEVLFSGPRQ AFEFPQINYSLCRGKKYSFAYGLGLNHFIPDR VVKLNVQTKETWVWQEDECYPSEPLFVPTPAATEEDD GVLLSIVVKPGA--RPSFLLVLDAVKLTELARAEVNTIIPVTLHGTYKlt* >RPE65_oryLap Oryzias latipes (medaka) ---- VEHPAAGYKKIFETVEELDEPIPAQIS GVIPSWLGGSLLRMGPGLFEVGDEPFHHLFDGQALMHKFDLRNGQVTYNRK FIKTDSYVRAMTENRVVITEFGTIAYPDPCKNIFSR FFSYFKGIEVTDNCLVNIYPIGEDFYAVTETNYITKVDPDSLETLEK VDVCKYLSVNGLTAHPHIEADGTVYNIGNCFGKDMSLAYNIVKIPPAQK DKSDPIEKSQIVVQLPSSERLKPSYVHS FGMTNNYFVFVEQPVKINLLKFLSAWSIRGTTYMDCFESCESMG TWFHLTTKNPAAYSSHKFRTSAFNLFHHINTYEDQGFIVVDLCTWKG HEFVYNYLYLANLREEWEEVKKAASRAPQPEVRRYVLPLDIYK EEQGKNLVSLSYTTATAVLCSDGTIWLEPEVLFSGPRQ AFEFPQINYSQCCGKKYFFAYGLGLNHFIPDR VVKLNVQTKETWVWQEEDCYPSEPIFVATPGATQEDD GVLLSIVVKPGT--RPGFLLVLDAMKLTEVARAEVNAIIPVTLHGLYKMT* >RPE65_danRer Danio rerio (zebrafish) ---- LEHPAGGYKKVFESCEELAEPIPAHVS GEIPAWLSGSLLRMGPGLFEVGDEPFYHLFDGQALLHKFDLKDGRVTYHRR FIRTDAYVRAMTEKRVVITEFGTTAYPDPCKNIFSR FFTYFQGIEVTDNCLVNIYPIGEDFYACTETNFITKVDPDTLETVKK VDLCNYLSVNGLTAHPHIEADGTVYNIGNCFGKNMSLAYNIVKIPPLQE DKSDQFEKSKILVQFPSSERFKPSYVHS FGITENHFVFVETPVKINLLKFLTSWSIRGSNYMDCFESNDKMG TWFHLAAKNPGKYIDHKFRTSAFNIFHHINCFEDQGFIVVDLCTWKG HEFVYNYLYLANLRQNWEEVKKAALRAPQPEVRRYVLPLDIHR EEQGKNLVSLPYTTATAVMRSDGTVWLEPEVLFSGPRQ AFEFPQINYSKFNGKDYTFAYGLGLNHFVPDR FCKLNVKSKETWIWQEPDAYPSEPLFVQSPDAEDEDD GVLLSIVVKPGA--RPAFLLILKATDLTEIARAEVDVLIPLTLHGIYTLt*