CDH23 SNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 172: | Line 172: | ||
=== Normal function of CDH23 === | === Normal function of CDH23 === | ||
(to be continued shortly) | |||
=== CDH23 allele assessment by PolyPhen === | === CDH23 allele assessment by PolyPhen === | ||
Revision as of 17:33, 10 August 2009
CDH23 SNPs
CDH23 (cadherin 23) on 10q22.1 is one of the better understood genes of the Usher disease complex. These genes generally encode structural proteins utilized in both hearing and visual systems -- and so at the mutational level by effects on both. Stop codons within CDH23 cause both deafness and blindness (USH1D) whereas missense alleles can affect hearing only (DFNB12). Both conditions are autosomal recessive. However one bad copy of CDH23 in conjunction with one bad allele of PCDH15 (protocadherin 15) on 10q21.1 (17 million bp over, not tandem) can give rise to the digenic disease USH1H. That has a simple physical explanation in defective heteroligomeric binding of the two terminal domains where the respective cSNPs occur.
Many Usher genes function both transiently during development of cochlea and retina and permenantly in adult structures. These functions may localize to multiple sites within each organ, for example ribbon synapses and stereocilia. CDH23 like many of these proteins has different binding partners to its cytoplasimic and extracellular domains as well as a transmembrane region. Other unrelated cell types elsewhere in the body may use these gene products though mutant alleles to date first manifest in hearing and vision. The role of CDH23 in hair tip links has recently been disentangled from its transient but critical role in hair cell development.
However some coding variants of CDH23 are simply near-normal (or even adaptive) polymorphic variants not giving rise to problems during the carrier's lifespan, though subtle subclinical effects on age related (or noise-induced) hearing loss or night vision acuity might still occur. In the past, such variations would be occasionally be detected within geneologies of affected indiviuals but not track with their disease; today, coding SNPs are far more likely to emerge -- and in far greater numbers -- simply in the course of genomic screening. That trend will only accerate with the advent of rapid screening platforms such as Nimblegen that can affordably screen the entire human proteome.
Note these myriad new cSNPs needing interpretation will come with accurate population frequencies further stratified by ethnic group distribution. That can be viewed as 'close-up' comparative genomics that complements the longer view of reduced alphabet afforded currently by CDH23 orthologs in 50-odd vertebrate genome phylogenetic tree. These considerations, along with accurate 3D models of both the cadherin module affected and protein binding partner, greatly help in interpreting disease implications of particular observed SNPs (for example E737V), yet uncertainty will remain in many instances.
Here a newly observed cSNP in a Kalahari Bushmen, heterozygous L1122V in exon 26, lies fall just before the boundary of the 11th of 27 cadherin ectodomains of the 3354 residue, 67 exon protein. This would appear unremarkable except for the observation that valine is ancestral mammalian value here which is conserved over vast phylogenetic time.
Comparative genomics
Orthologs of CDH32 are available from 42 vertebrates in the exon containing L1122V. The following exon is quite short so difficult to obtain broadly; transcripts are uncommon so deep in a gene. This does not affect modeling because residue 1122 lies in an interdomain region anyway.
Observe that while leucine is sometimes found at this position in other species, that occurence is concentrated strictly in early diverging vertebrates. In all 33 species of tetrapods (where sound is conducted primarily through air), the value here is exclusively valine. Note in particular the four other species of great apes have valine with no indication of heterozygosity.
From this perspective, L1122V may reflect retention of the ancestral value in one allele, rather than result from de novo back mutation from a L1122L homozygote. In other words, L1122 could be viewed as a mutation apparently fixed for better or worse in all other human populatons -- at a position conserved over billions of years of branch length in phylogenetically related species.
<----------cad10----------><------interdomain------><---cad11--->
................................................^.
CDH23_homSap DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSILQ
CDH23_panTro DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_gorGor dNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_ponAbe DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASIPEDIPEGHSIVQ
CDH23_nomLeu DNGPVGKRHTGTATVFITVLDVNDNRPIFLQSSYEASIPEDIPEGHSIVQ
CDH23_rheMac DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_calJac DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_tarSyr DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_micMur DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_musMus DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_ratNor DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_cavPor DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_speTri DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_oryCun DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_ochPri DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIVEGHSIVQ
CDH23_bosTau DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVSEDIPEGHSIVQ
CDH23_canFam DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_felCat DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_pteVam DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_turTru DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_susScr DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_equCab DNGPVGKRRTGTATVFITVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_eriEur dNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_loxAfr DNGPVGKRRTGTTTVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_proCap DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASIPEDIPEGHSIVQ
CDH23_echTel dNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_choHof dNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGRSIVQ
CDH23_monDom DNGPVGKRRTGTATIYVTVLDVNDNRPIFLQSSYEASVPEDIPEGSSIVQ
CDH23_macEug DNGPVGKRRTGTATVYVTVLDVNDNRPIFLHSSYEASISEDIPEGSSIVQ
CDH23_ornAna DNGPSGKRRTGTATVYVTVLDVNDNRPIFLQSSYEASVPEDIPEASSIVQ
CDH23_galGal DNGPTGNRRTGTATVYVTVLDVNDNRPIFLQSSYEASVPEDIPAASSIVQ
CDH23_taeGut DNGPSGNRRTGTATVYVTVLDVNDNRPIFLQSSYEVSVPEDIPAASSIVQ
CDH23_anoCar DNGPTGKRRTGTATVHVTVLDVNDNRPYFLQSSYEATVPEDIPDYSSIVQ
CDH23_xenTro DNGPAGNRKTGTATVSVTVLDINDNKPIFLKSSYEASVPENVPFSSSIVQ
CDH23_oryLat DNGPAGSRRTGTATVFVEVLDVNDNRPIFLQNSYETSVLETVPQGTSILQ
CDH23_takRub DNGPAGSRRTGTATVFVEVQDVNDNRPIFLQNSYETGILESVPQGTSILQ
CDH23_danRer DNGPAGGRRTGTATVYVEVLDVNDNRPIFLQNSYETSVLENIPRGTSILQ
CDH23_gasAcu DNGPAGSRRTGTATVFVEVQDVNDNRPIFLQNSYETSILESVPQRTSILK
CDH23_tetNig DNGPAGSRRTGTATVFVEVQDVNDNRPIFLQNSYETSVLESVPQGTSILQ
CDH23_ictPun DNGPAGDRKTGTATVYVEVLDVNDNRPIFLQNSYETTVLENVPRGSSVLQ
CDH23_calMil DNGPAGSRRTGTATVYIRVLDVNDNRPIFLQNTYEASVPENITMSTSILQ
CDH23_petMar DHGPAGSRRTGTTTLDVLVLDVNDNRPLFLEGSYZVSVPDNVTRGAIFLQ
................................................^.
CDH23_homSap DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSILQ
CDH23_panTro ................................................V.
CDH23_gorGor ................................................V.
CDH23_rheMac ................................................V.
CDH23_calJac ................................................V.
CDH23_pteVam ................................................V.
CDH23_ponAbe .....................................I..........V.
CDH23_nomLeu ................I....................I..........V.
CDH23_tarSyr ........R.......................................V.
CDH23_micMur ........R.......................................V.
CDH23_musMus ........R.......................................V.
CDH23_ratNor ........R.......................................V.
CDH23_cavPor ........R.......................................V.
CDH23_speTri ........R.......................................V.
CDH23_oryCun ........R.......................................V.
CDH23_canFam ........R.......................................V.
CDH23_felCat ........R.......................................V.
CDH23_turTru ........R.......................................V.
CDH23_susScr ........R.......................................V.
CDH23_echTel ........R.......................................V.
CDH23_eriEur ........R.......................................V.
CDH23_proCap ........R............................I..........V.
CDH23_equCab ........R.......I...............................V.
CDH23_loxAfr ........R...T...................................V.
CDH23_choHof ........R....................................R..V.
CDH23_bosTau ........R.............................S.........V.
CDH23_ochPri ..........................................V.....V.
CDH23_monDom ........R.....IY.............................S..V.
CDH23_macEug ........R......Y..............H......IS......S..V.
CDH23_ornAna ....S...R......Y............................AS..V.
CDH23_galGal ....T.N.R......Y...........................AAS..V.
CDH23_taeGut ....S.N.R......Y...................V.......AAS..V.
CDH23_anoCar ....T...R......H...........Y........T......DYS..V.
CDH23_xenTro ....A.N.K......S.....I...K....K.........NV.FSS..V.
CDH23_oryLat ....A.S.R........E.............N...T..L.TV.Q.T....
CDH23_takRub ....A.S.R........E.Q...........N...TGIL.SV.Q.T....
CDH23_tetNig ....A.S.R........E.Q...........N...T..L.SV.Q.T....
CDH23_gasAcu ....A.S.R........E.Q...........N...T.IL.SV.QRT...K
CDH23_danRer ....A.G.R......Y.E.............N...T..L.N..R.T....
CDH23_ictPun ....A.D.K......Y.E.............N...TT.L.NV.R.S.V..
CDH23_calMil ....A.S.R......YIR.............NT.......N.TMST....
CDH23_petMar .H..A.S.R...T.LD.L.........L..EG..ZV...DNVTR.AIF..
Consensus .n..a...r...a.v..t.l.......i..qs..#as!p#.!p.g.siv.
................................................^.
Comparative anatomy
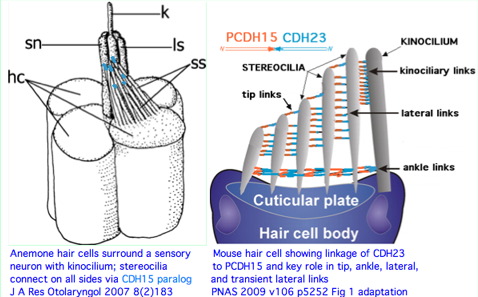
The remarkable auditory hair cell linker provided by CDH23 and PCDH15 is not a vertebrate innovation. Instead it must date back to pre-bilateran because contemporary cnidarians such as Nematostella have a very similar overall structure that incorporates an apparent ortholog to CDH23. This cannot be plausibly attributed to convergent evolution given the extent of structural agreement.
The Nematostella protein most resembling CDH23 has 6,074 residues, three transmembrane helices and 44 contiguous cadherin ectodomains with 4x-periodicity. Thus the correspondence at the protein level is imperfect. However antibodies show it is distributed on stereocilia of anemone hair bundles and required for tenticle sensitivity to vibration (prey detection). It provides both lateral and tip linkages. Nematocyst discharge is sensitive to calcium levels and streptomycin (like vertebrate mechanotransduction channels) but is insensitive to the MET channel blocker amiloride.
Nematostella also has long but weak matches to PCDH15, namely XM_001638202 and EU289217. However upon back-blast to human, these do not quite pull up PCDH15 as best match but rather its closest paralog FAT4, perhaps because of lineage-specific expansion. Possibly CDH23 had not yet undergone duplication and divergence to protocadherins and it alone may play a doubled linker role in anemone stereocilia.
Prey capture can result in severe trauma to anemone tenacle hair bundles but this [can be repaired using a protein again with similarities to a vertebrate stereocilia repair protein ARL5B which acts on the extracellular face of the plasma membrane along stereocilia n the vicinity of tip links.
It's important to note that 'stereocilia' is an anatomical misnomer. These are instead actin-based protrusions. It is the kinocilium that is a true cilium in both anemone and mouse. If parallels to ciliary photoreceptors are sought, these should be with the kinocilium rather than stereocilia. Since no known counterpart to the PCDH15-CDH23 linker occurs in vision, the commonality (Usher syndrome) may reside primarily in ribbon synapses of auditory and photoreceptive neurons.
Pseudogene and paralog issues
No potential exists here for mis-determining orthologous exons, even in remote species such as lamprey with poor assemblies. Exceedingly long genes such as this are not well-represented as retrogenes (which begin 3' and truncate early). Position 1122 is too remote from the 3' terminus. Relevent pseudogenes are not observed by Blat of human, macaque, and dog. Indeed, this exon gives a unique match at this level of sensitivity, even though cadherins are very widespread in the proteome.
The top matches to CDH23 within the human genome are shown as provided by GeneSorter at UCSC. Observe the established binding partner PCDH15 is by no means the best match; the e-value is high but only because of many weak alignments of tandem cadherin domains.
PCDH15 has 11 cadherin domains and a transmembrane region C-terminally. It's problematic to call it a paralog of CDH23 even though all cadherin domains must ultimately coalesce because PCDH15 tandem domains, which cannot be put in 1:1 correspondence with the 27 cadherin domains of CDH23, might have had quite a different history of assembly. The history of these gene families might instead best be worked out using interdomain spacer regions. Particular regions can be very conserved in themselves while not display much conservation between spacers.
Here the spacer region of CDH23 containing L1122 has best match within the human proteome to PCDHB14 (protocadherin beta 14), far down on the list of overall best matches. The interdomain region is shown in blue below. Note the best matches internally to other spacers are quite weak and neither L nor V is conserved in them.
PCDH15 S-TLTLAIKVLDIDDNSPVFTNSTYTVLVEENLPAGTTILQIEAKDVDLG---ANVSYRIRS
+ | |+ + |||++|| |+| |+| | |++| | +|||++| | | | | |||
CDH23 TGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSILQILKATDADEGEFGRVWYRILH
+GT V + VLD+NDN P F QS YE VPED P G I + A D D G +G++ Y H
PCDHB14 SGTTLVLIKVLDINDNAPEFPQSLYEVQVPEDRPLGSWIATIISAKDLDAGNYGKISYTFFH
IFLQSSYEASVPEDIPEGHSILQLK 1122
TFQNLPFVAEVLEGIPAGVSIYQVV 928 CDH23 internal spacer
IFSQPLYNISLYENVTVGTSVLTVL 497 CDH23 internal spacer
TFFPAVYNVSVSEDVPREFRVVWLN 1034 CDH23 internal spacer
TFHNQPYSVRIPENTPVGTPIFIVN 169 CDH23 internal spacer
TWKDAPYYINLVEMTPPDSDVTTVV 809 CDH23 internal spacer
CDH23 0 chr10:72826710-73245710 cadherin-like 23
FAT4 0 chr4:126457017-126633537 FAT tumor suppressor homolog 4
DCHS1 0 chr11:6599134-6633650 dachsous 1
FAT3 0 chr11:91724910-92269283 FAT tumor suppressor homolog 3
FAT1 0 chr4:187745931-187881981 FAT tumor suppressor 1
FAT2 0 chr5:150863846-150928698 FAT tumor suppressor 2
DCHS2 0 chr4:155375138-155531899 dachsous 2 isoform 1
CELSR2 1e-115 chr1:109594164-109619901 cadherin EGF LAG seven-pass G-type receptor 2
CELSR1 5e-113 chr22:45135395-45311731 cadherin EGF LAG seven-pass G-type receptor 1
CELSR3 5e-109 chr3:48637835-48684985 cadherin EGF LAG seven-pass G-type receptor 3
PCDH24 3e-87 chr5:175908971-175955375 protocadherin LKC
...
PCDHB14 5e-53 chr5 140,584,653 protocadherin beta 14
Structural significance
Blastp at PDB of the region around residue L1122 establishes that the best match to an already determined structure is a 39% identity match to mouse cadherin CDH8, 2A62. Within the 25 residue interdomain region, the percent identity is somewhat higher at nearly 50%. While not ideal, this should still allow accurate modeling of the adjacent cadherin domains and the critical spacer region, although the structural effects of the L1122V may be fairly subtle.
Since CDH23 is known to form a homodimer in tip links -- and such binding patches can involve hydrophobic residues that otherwise would be buried -- the quaternary structure here is the main unknown. Crystallographic adjacency in the unit cell does not always reflect oligomeric solution structure. Consequently, it may not be possible to fully evaluate L1122V despite the favorable match at PDB.
CDH23 1 GTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSILQLKATDADEGEFGRVWYRILHGNHGNNFRI
GT T+ VT+ DVNDN P F QS Y SVPED+ G +I ++KA D D GE + Y I+ G+ F I
CDH8 174 GTTTLTVTLTDVNDNPPKFAQSLYHFSVPEDVVLGTAIGRVKANDQDIGENAQSSYDIIDGDGTALFEI
Normal function of CDH23
(to be continued shortly)
CDH23 allele assessment by PolyPhen
Here L1122V will test out as benign because it is a common conservative substitution and comparative genomics support in fish. PolyPhen and SIFT have comparative genomics limited to sequences at SwissProt and do not incorporate phylogenetic relations; consequently they miss the L-->V transition at the level of the tetrapod clade. Consequently such tools do not utilize a significant part of the available information and are not informative here.