Pegasoferae?
Introduction to Pegasoferae
Pegasoferae is a novel proposal for the phylogenetic ordering within Laurasiatheres, grouping bats, perissodactyls, and carnivores to the exclusion of the other hoofed mammalian group artiodactyls. Bats have been placed in many previous locations, notably in the Euarchonta wing (with primates). While that particular idea is clearly refuted by many lines of evidence, the proper placement of bats remains under discussion.
Rare genomic events may be more useful for this than maximal likelihood because the orders of Laurasiatheres may have diverged relatively rapidly. Retroposon events are so numerous per million years however that they may be able to resolve branching at these tight nodes. However they suffer from homoplasy in that separate insertion events from a given parental element can look very similar and because deletions over time (no selection for their retention) can cause their disappearance and so confusion with lineages that never had the insertion.
Analysis of L1MA9 retroposon INT189
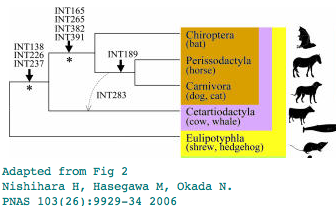
The phlogenetic distribution of the L1MA9 retroposon INT189 has been taken as evidence for bats being the immediate outgroup of horse + dog. That interpretation can be revisited using newly available genomes. Yet only two sequences representing perissodactyl and carnivore are at GenBank as cat assembly has a gap in the critical region. But other new data in 3 bats and 4 cetartiodactyls and 2 shrew/hedgehog confirm the lack of L1MA9 near the distal exon.
The trouble is a second L1MA9 element lies upstream of the MER58A middle marker. This is lacking in both carnivores. Evidently it was deleted in stem carnivore -- otherwise it would be providing evidence for carnivores being outgroup to cows + bats + horse. In short this single intron is providing 'support' for two contradictory topologies.
The sizes of many bat genomes have been experimentally determined: the 30-genus average of 2.6 gbp is about 500,000,000 bp less than human. Since bats in essence have the same 20,000 coding genes as other mammals, that discrepancy has to arise from less intronic and intergenic dna. Possibly bats had fewer active retroposing elements. Far more likely, bats they have an average number and the discrepancy arises from a faster rate of deletions than insertions.
Thus for taxonomically informative (ancestral laurasiathere) retroposons, many millions of deletion events have occured. Since the L1MA9 elements here are only 100bp or so, it would come as no surprise if a high percentage of the older relevent ones have experienced partial (or full) deletions making them unrecognizable with RepeatMasker.
Thus presence of a retroposon in a given orthologous position bat can be informative but absence is not so informative. INT189 is an absence. That one event isn't insufficient anyway to establish branching order. So bat/horse/carnivore tree topology remains unresolved. If horse is the outgroup to carnivore + bat -- and cow outgroup to all of these -- then hoofed animals are parsimoniously ancestral (rather than arising twice by convergent evolution) and bat and carnivore lost hooves (a bit unreasonable as dog and bats retain the ancestral 5 digits).
Summary of the phylogenetic distribution of the L1MA9 retroposon INT189: >PGM2_canFam Canis familiaris (dog) abseny -MER58 182-265 23% -L1MA9 6069-6302 27% >PGM2_felCat Felis catus (cat) genomic del absent -MER58 no data >PGM2_equCab Equus caballus (horse) -L1MA9 6172-6264 26% -MER58A 1-145 23% -L1MA9 6050-6302 23% >PGM2_myoLuc Myotis lucifugus (microbat) -L1MA9 6174-6264 20% -MER58A 38-157 26% >PGM2_pteVam Pteropus vampyrus (macrobat) -L1MA9 6161-6291 25% -MER58A 35-145 29% >PGM2_pipAbr Pipistrellus abramus (microbat) -L1MA9 6180-6301 252% -MER58A 38-157 24% >PGM2_bosTau Bos taurus (cow) -L1MA9 6155-6263 28% -MER58A 7-157 21% >PGM2_turTru Tursiops truncatus (dolphin) -L1MA9 6155-6265 29% -MER58A 37-148 21% >PGM2_susScr Sus scrofa (pig) cdna + tiled -L1MA9 6159-6264 27% -MER58 212-271 28% >PGM2_vicVic Vicugna vicugna (vicugna) tiled -L1MA9 6162-6310 24% -MER58A 35-157 20% >PGM2_ateAlb Atelerix albiventris (hedgehog) ... ... >PGM2_sorAra Sorex araneus (shrew) ... ...
>PGM2_canFam Canis familiaris (dog) -MER58 182-265 23% -L1MA9 6069-6302 27% VISAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICHDQGTIKKLFENLRNY GTCATCAGCGCCGAGTTGGCTAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAGGCCATTTATGTTGAGTACGTTTCTATTAACTCTG TTTAATTGAAATAATACTTTTTAAAAGTTTTATTATGTTTTTATGTGTGACACTAATATTCTAACCCTCTTACTTTGGGTGAGGGTTCTTCTGAAAACTA AAGGATCACTTTTTCTTTTAATGCTTAACTATTCAATACTAATTATCACTTATGACTGTGTTAATCCTTAACAAATGAGAACATCAGTTGCAGAAATAGC TAATTGAGGAGGGTGATTCCCTGATGTCAGAAAGGACAAAGGTTTTCGTGAAACATCTATTACGTGTTTAGAgccactagtcaagtctgcctttgtagtg caaaagcagctgatggcaagacgtacaggaatgggtgtggtgtggctgcaatgaaaTGAAACTTTCACCTCCCAAGATAGGCCGAAGGCCAGGCAGCAGT TTGGCAATACCTGGGGTCAATAGTTATACCTCTTTTTTATGCTAAATTATTCCTTTGAAGCTAGTCATTGTTATCGTTTCATTTAGCTTAAAATATACTG ATTGCTACATGTTCTGTATACACCACGTGAGATTATTTGTTCCTCATTTTGCATATTTGTACTTTTtttattgagatgtaattgacattaatgtcaggta taataacataatgattcgatatttatatattattacaaagtgatcaccatagtaagtcgagttaacatccacaccacatataatcacaaatattcattct tgtgatgatagcttttatgatctgtggtcttagcaactttcaaatatacagtacaatactagtagatacagtcaccaagttatatatATATAATTTTATT TCTTTTGATAGATATGGCTACCATATTACCAAAGCTTCCTATTTTATCTGCCATGATCAAGGCACCATTAAAAAATTGTTTGAAAACCTTAGAAACTAC >PGM2_felCat Felis catus (cat) genomic del incomplete coverage -MER58 ASFLATKNLsLSQQLKAIYGE YGYRITKASYFICHDQGTIKQLFENLRNY GCTAGCTTTCTAGCAACCAAGAATTTGTTTGTCTCAGCAGCTAAAGGCCATCTACGGCGAGTAAGTGTCTTCTAACCTGGTAAAGAAGTAATAG TGTTAAATATTTTCTTATGGTTCTACGTGTGAGATATTAATATTCTTTCTAATGCTCTTTGGTTGTGAATTCTATTTCTTTTTCTTTTTTTAATGTTTAT TTATTTTTGAGAGAGAGAGAGAGAGAGATGGAGTATGAGCAGGGGAGGGGCAGAGAGAGAGGGAGATACAGAATCCAAAGCAGGCTCCAGGCTCTGAGCT GTCAGCACAGAGCTCCACACGGGGCTTAAACTCACAAACCATGAGATCATGACCTGAGCTGAAGTCAGACACTCAACCGTTTGAGCCACCCACGTGCCCC ATGAATTCTATTTCTTATGAAACTAAATAATCATCTTTTCTTTTGATACTTAACCATGTAATGGTAATTATCATTCACGATTGCACGAATCCTTAACAAA TGAGGGCATCAGTTGCAGAAATAGCTAATTGAAGAATGTGATTTTAAGTGTGTGATGTCAAAAAAGATTAAAGGTGTTCATGAAATCTCTATTAAGTTTT TAGAGCAATGACCCAGGTCTGCCTTTATAAAGTGCAAAAGCAGCCCGTGGCAACACGTTGCAGTAAGACTCTTACTTACAAATACAGGCTAAAGGCCAGG CAGCAGTTTGGCAATCCCCAGGGTTAATTGTTGTACCTCTTTTTTATGCTAAATTATTCCTTTGAAGGTACTCATGGCTATTTGTTTCATTTGGTTTAAA ATATACTGGTTGACAAATGTACACTGTGTGGAATTATGTGTTCCTCATTTTGCATATTTGTATTTCCTTAACTGAGATATAACTGACATTAGTTTCAGGT ATGCGATACAGTGTTTCAATATCTGTATATATTACAAAATGATCATCACAGTACATCTAGTAACAGTCGCACCACACTTAATACAAAAGT TCCaTATGGCTACCGTATTACCAAAGCTTCATATTTTATTTGCCATGATCAAGGCACCATTAAACAATTATTTGAAAACCTTAGAAACTAT >PGM2_equCab Equus caballus (horse) -L1MA9 6172-6264 24% -MER58A 1-145 23% -L1MA9 6172-6264 26%-L1MA9 6050-6302 23% VISAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICYDQDTIKKLFENLRNY GTCATAAGCGCAGAGTTGGCTAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAGGCCATCTATGTTGAGTAAGTTTCTATTAACTCTC TTTAACTGAGGTAATTTTTTTTATTAGtttcaaatgtacaacataatgattcaatgtatgtatatattttgaaatgatcgccacaataagtctggctaac ctgtatcaccgacatagGGCTCTTTTTAAATGTTTTATGTTCTTTTGCATGAAACAGTAATATTCTTTTGAATGCTCTTACTTTAGCTATGAATTGTTCC TTATGAAAACTAAGTAAGAGATCACTTTTTCCTTTCGATACTTAACCACTTAGTAGTATTACCCTTTGTGATTGCATTAATCCTTAACAAATGAGAACAT TAGTCACGGAAATGGTGAAGTGAAGAATGTAATTTTCAGTGTCTGAGGTCAAAAAAGATTAAATGTGTTCATGAAACATCTATTTAGTCTTTAACTTCat tgctcagctctgcctttgtagtgcagaaacagccggggacaatacataatgtaatgggtgtggggtggctgtgttccagtagatcttttacttaaaaata caggccgaaggccaggcagcagtttggcaatccctgGGGGAGATTATTGTACCTTTTTTTAATGTTAAATTATCCCTTTGAAGTTAGTCATGGTTATTTC ATTTAGTTTAGAATATAATGGTTAATACATAGTGTATGTACACCATGTGGAATTATTTTTTCCCATTTTGCATTTCTTCTtttgttgagatataattaac atagaacattatattagcttcaggtgtacagtgtaattatttgataattgtatatattgcagattgatcaccaccataagactagttaacatccatcacc acacatagttataaatttttttcttgtgatgagaacttttaaggtctattctcttagcaaccttcaaatatacaatacagtattattaattctagtcacc gtgctgtgtattatatcctcatgacccattTTATTATTTTGTTTCGAAAGGTATGGCTACCATATTACCAAAGCTTCATATTTTATCTGCTATGATCAAG ACACCATTAAAAAATTGTTTGAAAACCTTAGAAACTAC >PGM2_myoLuc Myotis lucifugus (microbat) -L1MA9 6174-6264 20% -MER58A 38-157 26% VISAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICHDQGTIKKLFENLRNY AAPE01636299 GTCATAAGCGCAGAGCTGGCTAGCTTTCTTGCAACCAAAAATTTGTCTCTGTCTCAGCAGCTAAAGGCCATCTACGTTGAGTAAGTTTCTATTGATTATTG AATTGAAGTAATATAGTTTGATTAGTTTCATGTGTACAATGTAATGATTCAATATGTGTATATATTGGGACATGGTTGCCACAATAAGTCGTTAACATAC ATTACCACATGTGGCAATGTATTTTAAGTGTATTATGTTCTTGCGTATGAGATGCTAATGTTCTTTCCAAAGCTCTGACTTTAGTTATGAATTCTATTTC TTAAGAAAACGAAACGAGATTATCTTTTCCTTTTGATACTTACCATTTGTGATAGCACTAATCTTTACTAAATGAGAACATGACACAGAATGTGATTTTA AGTGTCTGATGCCAAAAAAGATTAAATGTGTTCATGAAACGTCTATTTAGTCTTTATAGCAGTTTCTCAACTCTTGCCTTTCTGATGCAAAAGGAGCCAG ACACAGTACATAATGCAATGGGCGTGGTATGGCTGTTCCAGTATAATTTTACTTACAAGTATAGGCTGAAGGCAAGGTAGCAGCTTGGTGAGCCCTCGGG TAAATTGTTGCACCTCCTTTTAATGCTAAATGATTGCTTTGAAGCTAGTCATGGTCATTTGTCTCATTACGTATTTGAGAATGTGCTGGTTGGTGCCCGT TCTGTATATGCTATGCATAATTATTTGTTCCTCATTTTGCATGTATTTGTATTTGTTTTGATAGGTATGGCTACCATATTACCAAAGCTTCATATTTTAT CTGCCATGATCAAGGAACCATTAAGAAATTATTTGAGAACCTTAGAAACTAT >PGM2_pteVam Pteropus vampyrus (macrobat) -L1MA9 6161-6291 25% -MER58A 35-145 29% VISAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICHDQGTIKKLFENLRNY GTCATAAGCGCGGAGTTGGCTAGCTTTTTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAGGCCATCTATGTTGAGTAAGTTTCTATTGACTCTA CATAACTGAAATAATATTTTTTATTAGTTTCAGGTGTACAGCACAGTGATTCGGTATATGTATATATTATGACATGATTGCTATAAGTCTATTGCATGCA TCAGTCTATTACTACATGCATCACCACACGTAGTAATATTTTTAAATGTATTATGTACTTGTGCACAAGATACTAATATTCTTTCCAATGCTCTTACTTT AGTTATGAATTCTATTTCTTATAAAAACCAAATAAGAAATTACCTTTTCCCTTTGATACTTAGCCATTTAATAGTAATTACCATTTGTGATGACAGTAAC CTTTACCAGATGAGACATTAGCCACAGAAACAGCTAAAGAATATGATTTTAAGTGTCCGATGTCAAAAGATTAAATGTGTTTATGAAACATCCTATTTAG TCTTTTTATAGCATTATTCAGCTGTGCCTTTGTAGTACAAAAGCAGCCAGACCCGATGCATATGTAATGGGTGCAGCGTGGCTACATTTCTGTAAAATTT TTACTTACAAATATAGGCTGAAGGCCAGGCAACAGTTTGGTGATCCCCTGAGTAAATTGTTATACTTCTTTCTTAATGCTGAACTATTCCTTTGAAGCTA GTCATGGTCATTTGTTTCATTAAGCGTTTTAGAATGTACTGGTTGATACATGTTCTGTGTACACTATGCAGAATGATTTGTTCCTTATTTTGCATGTGTT TGTATTTATTTTGATAGGTATGGCTACCATATTACCAAAGCTTCATATTTCATCTGCCATGATCAAGGCACCATCAAAAAATTATTTGAAAACCTTAGAAACTAT >PGM2_pipAbr Pipistrellus abramus (microbat) -L1MA9 6180-6301 25% -MER58A 38-157 24% AB258957 AIYVE YGYHITKASYFICHDQGTIKKLFENLRNY GGCCATCTATGTCGAGTAAGTTTCTATTGATTATTGAATTAAAGTAATATAATTTGATTAGATTCATGCGTACAGTGTAATGATTCAATACATGTATATA ATGGGACATGGTTGCCACAATAAGTCGTTAACATACATCACCACCTGTGGCAATATATTTTAGGTGTATTATGTTCTTTAGTATGAGACACTAGTACTAA TATTCTTTCCAAGGCTCTGACTTTAGTTATGAATTCTATTTCTTAAGAAAATGAAACGAGATTATCTTTTCCTTTGGATACTTACCATTTGTGATTGCAC TAATCTTGATTAAACGAGAACATTACACAGAATGTGATTTTAAGTGTCTGATGCCAAAAAAGATTACATGTGTTCATGAAACATCTATTTAGTCTTTATA GCAATTTCTCAACTCTTGCCTTTCTGGTGCAAAAGCAGCCTGACACAATACATAATGTAATCGGCGAGGGATGGCTGGTCCAATAAAACTGTACTTACCA ATGTAGGCTGAAGGCAAGGTAGCAGCGTGGTGTTCCCTCAGAATTATTTGTTCCTCATTTTGCACGTATTATTTGTTTTGATAGGTATGGCTACCATATT ACCAAAGCTTCATATTTTATCTGCCATGATCAAGGCACCATTAAGAAATTATTTGAAAACCTAAGAAACTACGATGGGAAGAATAATTAT >PGM2_bosTau Bos taurus (cow) -L1MA9 6155-6263 28% -MER58A 7-157 21% VITAELASFLATKNLSLSQQLKAIYVE YGYHITRASYFICHDQETIKQLFENLRNY AB258958 [L1_Carn7] GTCATAACTGCAGAGTTGGCCAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAAGCCATCTATGTTGAGTAAGTTTCTATTGACTATT TAATTGAAGTAATTTTTTTTTATCAGttcaggtatacaacacagtgattcagtgtatgtctatattgtgaaatgatcacagtggatacaattaacatgca tccccacacaggaatattttttaatgtTTTACTCTCTTCTTGTGCACCCGATACTCATATTCTTTCTGATGCTCTTGCTTTAGTTATGAATTCTATTTCG TATGAAAACTAAATAAGAGATCACCTTTTCCTTTTGCTACTTAAGCAGTTAATAGTAATTACCATTCATGATGACGTTAATCCTTAATAAATGAGAACGT TAGCTGCAGAAATGGCTAAGGGAAGAATGTGATTTTTTAAATGTCCAGTGTTGAAAAAGACTAAATGTGTTCATTAAACATCTATTTAgtctttgtagca attacttatttctgcctttctagtgcaaaagcaaccagacacaaggtaatgggcatgacgtggctgtattccaatgataaaacttttacttacaaacaga gactgagggccACACAGCAGGGCAGTGATTCCTGGTGTAGATTGTTGGACCTCTTTATTTAATGCTGAATTACTCCTTTGAAATTAGTCATGGTTGTTTG TTTTAGAATATACTGTTTGATAGATACATGTTCAGTGTACACTGTGCCCAATTATTTGTCCCTCATTTGCATGTAACCATGTTTGTATTGATAGGTATGG CTACCATATCACCAGAGCTTCGTATTTTATCTGCCATGATCAAGAAACTATTAAACAATTATTTGAAAACCTTAGAAACTAT >PGM2_turTru Tursiops truncatus (dolphin) -L1MA9 6155-6265 29% -MER58A 37-148 21% FISAEVGSFLAQNCLVSAAKAIYV YGYHITKASYFICHDQGTIKKLFENLRNY TTCATAAGTGCAGAGGTTGGCAGCTTTCTAGCACAGAATTGTCTTGTTTCAGCAGCTAAAGCCATCTATGTTGAGTAAGTTCTTCTATGACTGTTAAATG AGTAATGTTTTTTTTCATTTCAGTTGTGCAACACAATGATTCAATGTATATCTATTATTGTGAAATGATTGCAACAAATACAGTTTACATGTATCCCCAC ATGTAGTAATATTTTTTAATGTTTTACTCCGTTCTTATGCATGAGATACTAATATTCTTTCTGATGTCCTTACTTTGGCTATGAATTCTATTGCCTATAA AAACTAAATAAGGGATCACCTTTTCCTTTCGATATTTAACTACTTAATAGTAGTTACCCCTTCATGATGACATTGATTCTTAACAAATGAGAACATTAGT TGCAGAAATGGCTAAGGGAAGAATGTGATTTTTAAGTGTCCAATGTCAAAAAAGACACATGTGTTCACAAAACATGTTTAGCCTTTAAAGCAATTATTCA CCAGTGTCTTTGTAGTGCAAAAGCAGCCAGACACAATACATAAGGTAATGGGCATGGCATGGCTACGTTCCAATAGAGAAACTTTTACTTAGAAATACAG GCTGAGGGCCACAGAGCAGTTCAGCGATCCCTGGGGTAGATTGTTGGACCTCTTTTATAAAATTGGACCTCTTTTTTTTTTTTTTTTTTTTTGGCGGGGG GTACGTGGACCTCTCACTGTTGTGGCCTCTCCCGTTGCAGAGCACAGGCTCCAGACGCGCAGGCTCAGTGGCCATGGCTCGCGGGCCCAGCCGCTCCACG GCATGTGGGATCTTCCCAGACCGGGGCACGAACCCGTGTCCCCTGCGTCGGCAGGCGGACTCTCAACCACTGCGCCACCAGGGAAGCCCTGAACCTCTTT TTTAATGCTGAATTATTCCTTTGAAATTAGTCGCGGTTATTTGTTTTAGAATATACTGGTTGATACATGTTCAGTGTACACTGTGCAGAATTATTTGTTC CTCGTTTTGCATGTAATTGTGTTTGTATTGATAG GTATGGCTACCATATCACCAAGGCTTCGTATTTTATCTGCCACGATCAAGGCACTATTAAAAAATTATTTGAAAACCTTAGAAACTAC >PGM2_susScr Sus scrofa (pig) cdna + tiled -L1MA9 6159-6264 27% MER58 212-271 28% VISAELASFLATKNLSLSQQLNAIYVE YGYHVTKGTYFICHDQGNVKKLFENLRNY GTCATAAGCGCAGAGTTGGCCAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAATGCCATCTATGTTGA GTAAGTTTCTATTGACTGCATTTAATTGAAGTAATTTTTTTAATCAGTTTCGGGTGTACGACATAATGATTCAGTGTATATGTATTGTGAAATGATCCCAA TGAGTACAGCTAACATGCATCCCACACGTAATAATATTTTTTTTTCTTTCTTTTTCTTTTTTTAGGGCTACTCCTGTGGCATATGGAAGTTCCCAGGCTA AGGGTCGAATAGGATCCATAGCCGCTAGCCTAAGCCACAGCCACAGCAGCACGGAATTCGAGCCACATCTTTGACCTCCGCTACAGCTCATGGCAATGCC AGATCCTTAACCCACTGAGCAAAGCCAGGGATCAAACCCAACATCTCATGGATCCTAGTCGGGTTTGTTAACCCTTGAGCTGCAAAGGGAACTCCCATAA TAATCCTTTTAAATGTTTTACTCTGTTCTGATGCATGAGACTAATATTCTTTCTGATACTCTCATTTTAGCTATAAAGTTGATTTCTTATGAAAACTCAG TAAGAGATCACTCTTTCCTTTTGATATTTAACCCCTTAATAGTAATTACCATTCATGATGACATTAATCCATAACAGATGAGAACAGTAGTTGCAGAAATGGGTAAT GGAAGAATGTGATTTCAACTAAATGTCCAATATCAAAAAAGACTAAGTGTGTTCATGAAACATCTATTTACTATTTATAGCAGTTATTCAGCTCTGCCTT TGTAGTGGTAAAGTGGTCAGACACAATACTTAAGGTAAAAGTTTCCAGTTATGAAACTTTTACTTACAAATATGGGCTGAGACTGGGCAATAGTTCAGTG ATTCCTTGGGGTAGATTCTTGGACCTCTTTTTTTAAATGTTGGACCTCTTTTTTAATGCTAAGTTATTCCTTTGAAATTAGTCTTGCTTATTTGTGTCAT TTGTATTGAAGTATACTGGTGAATTACATGTTCTGTGTATGCTGTGTGGAATTATTTGTTCCTCATTTTGCATGTAATTGTATTTGTATTGATAGG TATGGCTACCATGTTACCAAAGGTACATATTTTATCTGCCATGATCAAGGCAATGTTAAAAAATTATTTGAAAACCTTAGAAACTACGATGGGAAGAATAATTAT >PGM2_vicVic Vicugna vicugna (vicugna) tiled -L1MA9 6162-6310 24% -MER58A 35-157 20% VITAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICHDQGTVKKLFENLRNY GTCATAACTGCAGAGTTGGCTAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAGGCCATTTACGTTGAGTAAGTTTCTATTAATGCTG TTTAATTGGAGTAAGCTTTTTATCCATTTCAGATGTACCACATTATGACTCAGTATACGTCTACATTGTGAAATGATCACAATTAGTAAAGTTAACGTGT ATCATCACACATAGTAATATTTTATAATGCTTTACTCTGTTCTTGTGCATGGGACACTAATGTTCTTTCTGATGCTCTTTCTTTAGTTATGAATTCTGTT TCTTATGAGAACTAGATAAGAGATCATCTTTTCCTTTTGATACCTAATCACTTAATAGTAATTACCATTCATGATGACATTAATCCTTACAAATGAGAAA ATTAGTTGCAGAAATGGCTAATGGAAGAATGCGATTTTAAGTGTCTAATGTCAAAAAAGACTAAATGTGTTCATGAAACATCTGTTTAGTCTTTATAGCA ATTACTCAACTCTACCTTTGTAGTGCAGAAGCAGCCAGACTCAACACATAAGGTAATGATGTGGCTGTTCCACTAATAAAACTTTTACTCAAAAACACTG GCTGAGGGCCAGGCAACAGTTCAGCAATCCCTGGGGTAGATAGTTGGACCTCTTTTTTTTAATTCTAAATTATTCCTTTGAAACTCATCATGGTTATTTG TGTCATTTATTTTAGAGTATACTGGTTGATGACATGTTCAGTGTACACTGTGCAGAATTCTTTGTTCCTTGTTTGCATGTAATTGTATTTGTATTGATAG GTATGGCTACCATATTACCAAAGCTTCATATTTCATCTGTCACGATCAAGGCACTGTTAAAAAATTATTTGAAAACCTTAGAAACTAC >PGM2_ateAlb Atelerix albiventris (African hedgehog) No repetitive sequences [VISAELASFLATKNLSLSQQLKAIYVE] YGYHITKASYFICHDQVTIKKLFENLRNY AB258952 TTAATGTGTTTGTTAAACATCTATTTATTCTTTACAGCTATCACTCAACTCTGACTTTGTAATACAAAATAGCCACACTTAGTCCATGAGGTCATGGACC TGATGTGACTGCCCCAATAAAACTTATACCTACAGATATAATCAAAATAAGATAAAATGGATGCTATCAATACTTAAGAATATTGGCTAAGTAAAAACAA AGAACTAGTTTAGAAACCTACAGGGGGTTATTGTTCTTCCTTTTTTTCATGCTATATTATTCCTTTGAAGCCAGTCATAGTTATTAGTCTCATTAACTTT ATAATATACTGGTTATATATGTTCTGTGTATACTAGGTAAAGTTATTTCTACCTAATTTTGCATACGTTTTATTTGTTTGCTAGGTATGGCTACCATATA ACCAAAGCTTCATATTTTATCTGCCATGATCAAGTCACCATTAAAAAATTATTTGAAAACCTTAGAAATTAT >PGM2_sorAra Sorex araneus (shrew) +SOR1_SINE +SOR1_SINE VISAELASFLATRNLSLSQQLKAIYVE YGYHITKASYFICHDQSIIKKLFENLRNY AALT01183695 AALT01470682 GTCATTAGCGCGGAGCTGGCCAGCTTTCTCGCCACCAGGAACCTGAGTTTGTCCCAGCAGCTAAAGGCCATCTATGTGGAGTAAGTTCCCTACTGACTGT GCTTAATCAAAATAACCCGTATTTTTGGATCCATTTTTAACGGTTTATTATGCTCTTGTGTGTGTGATACTGATAGTCTCTCTAATGTCCTCACTTCAGT TATAAATCCTATTTCTTAAAAACATGAAGTTAAGGGGCTGAACCGATAGCACAGCGATAGCAAGGTTTGCCTTGCATGTGACCGATCTGGGTTCGATTCC CAGCATCCCATTTGGTCCCCTGAGCACTGCCAGGAGTAATTCCTGAGTGCATGAGTCAGGAGTAATCCCTGTGCATCGCTGGGTGTCACCAAAAAAAAAA AAACCATGAAGTTAAAAAATCACCTTTTGGGGGGGGTCGGAGAGATAGTGCAGCAGTGGGTAGGGAGCTTGAGTCATTCATGGGTCACCCAGCTTCAATC CCTGGCACGCCCTGTGGCCTCCCAAGTCCCGCCAGGAGTGATCCCTGAGCTCAGAACCATAAGCAAGCCCTGAGCACCATTGGTGTGGCCCCAGAATAAA TAAATTAGAGATAGAAATCACTTTTTCATGCTTAACTACTTAATAATACTTATGATTGCCATACTCCCTAATGAATGAGATCTAATCGCAGAACTAGTTA TTAGTTAAAAGTGTGAATTTAAATGTGTAGTGTCAAAAAAATG ACCAAGATAACCAGCTTATAACTTAGACTTATAAATGACTGCTTATCAATATATCT AAGGCCAGACAGCAGTTTACTTTAGCAGTTCCTAGGATAGGTTATTGTTCCTCTTTTTTTTTTCCCTTTATTTTTTGCCCCCTAGAGATCTACCTTTTAA AAAAATATTTTTTTAATTGAATCACAATGAGATACACAGTTACAAATTGTTTCTGATTTGATTTCAGTCAGACAATGTTCAAATATCTGTCCCTTTCACA GTGTACATTTCCCACCACCAGTGTCCCCACTTTCCTTCCTTGTTCCTCTTTTTTCATGCTCAGTGATTCCTTTGAAGCGAATTATGGTCATTCGCTTCAC TTGCTTAAAAGCAAATGAATCAGCGGCCGATTGATGTCCTGTGTTCGAAAGACAGAATTCTTTGTTCCTTATTTTGCGTGTATTTGTATTGATAGGTATG GCTACCATATAACCAAAGCTTCGTATTTTATCTGTCACGATCAAAGCATCATTAAAAAGTTGTTTGAAAACCTTAGAAATTAT
Analysis of L1MA9 retroposon INT391
Summary of the phylogenetic distribution of the L1MA9 retroposon INT391: >INT391_ACSL5_Peg_canFam +MER91 85-140 23%9 -L1MA9 6082-6298 24% span 762bp >INT391_ACSL5_Peg_felCat -MER91C 97-140 27% -L1MA9 6082-6301 22% >INT391_ACSL5_Peg_equCab -MER91B 62-128 26% -L1MA9 6082-6302 21% >INT391_ACSL5_Peg_myoLuc no MER -L1MA9 6079-6277 23% >INT391_ACSL5_Peg_pteVam -MER91B 8-62 24% -L1MA9 6060-6302 25%
>INT391_ACSL5_Peg_bosTau -MER91C 55 -85 28% No L1MA9 >INT391_ACSL5_Peg_turTru -MER91 261-311 22% No L1MA9 >INT391_ACSL5_Peg_susScr -MER91 257-306 8% No L1MA9 >INT391_ACSL5_Peg_vicVic -MER91 284-337 24% No L1MA9 >INT391_ACSL5_Peg_sorAra no MER91 No L1MA9
Phylogenetically informative coding insertions and deletions
introduction
Analysis of indel 1
etcetc
Analysis of indel 2
etcetc