Opsin evolution: RBP3 (IRBP)
RPB3 (IRBP): introduction
Interstitial retinol-binding protein, oddly named by IGNC as RBP3 despite a lack of paralogs RBP1 or RBP2, confusion with ribosomal binding protein genes and widespread prior use of protein name IRBP, is a 4 exon 1247 residue glycoprotein thought to shuttle retinoids interstitally between the photoreceptor cells and the retinal pigment epithelium. This role would only make sense for ciliary opsin systems that are unable to regenerate cis-retinal without an auxillary pathway in an anatomically separate tissue (here RPE). Consequently -- since nearly all protein folds are extremely ancient -- RPB3 must have been co-opted from some other role.
The protein's size results from four ancient internal tandem duplictions that became established prior to intronation (that is, the gene structure does not reflect the repeat structure; the repeats happened first, introns were inserted randomly later within the fourth repeat). Any given repeat module clusters markedly better to the same-numbered repeat in other species than to any of the internal repeats, establishing that repeats had arisen and diverged already prior to speciation rather than arising independently in descendent lineages (like RHO1 and RHO2 in lamprey).
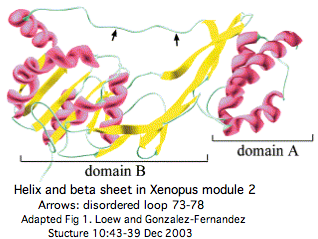
It was initially expected that IRBP as a self-contained homotetramer would load four molecules of trans-retinol to accomplish its passive shuttling efficiently. However experiments decisively show this to be false -- the subunits are quite inequivalent and only one molecule is transported. The structure of module 2 of frog was [determined in 2002 (PDB: 1J7X); it consists of two sub-domains and a cleft. The fold matches obscure proteases and hydrolases found in little-studied bacteria; this might represent convergent evolution as seen in TIM beta barrels rather than provide valid clues to ancestral function. A structural determination of the entire molecule is reportedly underway.
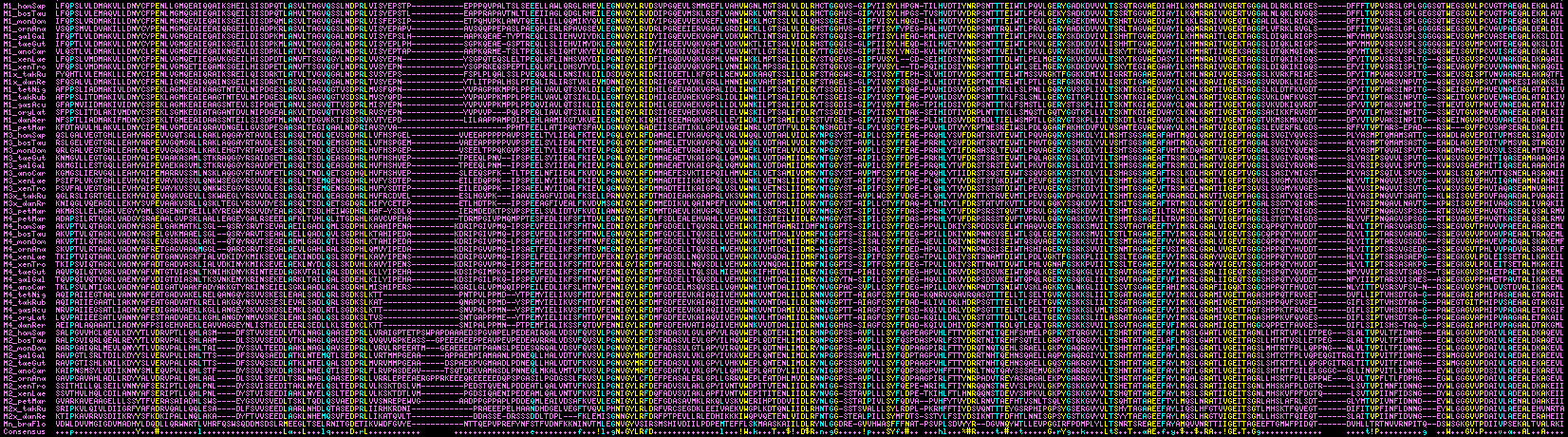
Fragments of the protein have been sequenced from an immense number of species for phylogenetic purposes. That's because it evolves fast enough to provide a large number of seemingly informative sites and these can be obtained conveniently exploiting the large size of the first reading frame. While there is minimal risk of accidentally cross-matching different modules, the internal repeat structure implies that residues might not be evolving independently. While the modules might seem too diverged for cross-module gene conversion to still be operative, they retain patches of near identity.
Some very odd aspects of marsupial RBP3 are investigated below because of the implications in a gene so widely used in taxonomy. Teleost fish further raise the question of inhomogeneous recombination events arising from module mix-up. In fish, the ancestral four-module gene has given rise to a two-module gene (M1 and M4) accompanied by an intronless upstream tandem fragment duplicate of M1-M3 without any indication of M4 in the residual 10 residues. Both genes are transcribed and apparently functional since the establishment event occured prior to zebrafish and fugu divergence, though the upstream gene has been lost in many lineages including tetraodon, medaka, and stickleback but not cichlids.
The upstream fragment suggests a truncated version of the first large exon. The lack of intron does not suggest retroprocessing in view of their lack in the parental gene and the locational adjacency but rather standard recombinational or flanking retroposon-driven tandem duplicating. The loss of M2 and M3 in the downstream gene but retention of standard introns again suggests recombinant loss, either as part of the initial tandem event or subsequent to it as a consequence of the new potential for exact misalignment. Note otherwise the four modules would have been quite diverged from each other prior to the emergence of rayfinned fish.
Conceivably whole genome or segmental duplication in fish set the stage for inhomogeneous recombination (as modelled by Nickerson), though simple local tandem duplication event mediated by retroposons during replication is another option. A great many tandem loci exist in genomes such as human for which wholesale duplication is inapplicable. This could possibly be resolved by sequencing basal teleost fish that diverged prior to the putative genome duplication to determine if the odd arrangement of genes was already established.
IRBP module alignment, conservation and structural aspects
The image below aligns the four module types from human to lamprey and amphioxus, with yellow showing exceptionally conserved residues and cyan moderately conserved. The number of species shown could be greatly expanded especially for module 1 but for clarity only a phylogenetically representative set is displayed. Module boundaries vary slightly according to the bioinformatics tool used to define the domain (eg cystallographic homology transfer, SuperFamily, Pfam, SwissProt, blastp of reference domains) but can be standardized by counting back a fixed number of residues N-terminally from the universally conserved tyrosine and forward from the C-terminal alanines because conserved residues must occupy near-identical positions within the tertiary structure.
It can be seen that M2 has two variable length insertional regions, no doubt relatively disordered loops. Amphioxus additionally has inserted residues in three regions that are suppressed in the figure -- observe that although its overall identity is low, identity is high in conserved residues. The modules each have distinctive class indels and characteristic residues that allow them to be readily distinguished from each other. These features were most likely established prior to lamprey divergence rather than later by convergence. Only two conserved patches are observed, GNvGLYRvD and RAivVGErT in human; other conserved residues are dispersed (at least within the linear structure).
The sole structural determination to date, 1J7X_A the single module M2 of Xenopus laevis to 1.8 Å, describes two domains separated by a hydrophobic binding cleft (based on analogy to very similar protease and hydrolase folds). The smaller domain A is an an amino terminal three helix bundle that extends 8 residues past the M2 insertion region (to PGDSIQAEN in xenLae) . M2 was an unfortunate choice because it is most diverged and bares the least resemblance to early-diverging or ancestral sequence.
The observed fold similarities could not plausibly have arisen by chance. Crotonase has a binding site for hydrophobic chains of fatty acids and isomerizes them, with possible relevence in RBP3 to cis and trans retinoid stabilization and transport; however IRBP does not appear to be an enzyme or isomerase itself and RPE65 carries out the regenerative isomerization reaction.
M2 may have two distinct binding sites, one in the cleft between domains A and B and the other solely inside domain B. The assignment of conserved tryptophans to these sites and other conserved residues to the surface (suggesting a protein binding partner, possibly intra-module binding) has to be revisited in view of the much deeper phylogenetic array of sequences now available.
A peculiar feature of full four-module protein (after discounting a couple dozen residues for signal peptide and carboxy terminal wander) is the cheek-to-jowl adjacency of consecutive modules -- in human at most 3 amino acids separate the modules from each other.
This is far too short a bridge to allow many arrangement options for module-module quasi-homodimerization (or allosteric regulation of the binding subunit). Instead the modules must be arranged more as beads on a string. Linear donor-receptor docking in a M1-M2-M3-M4 arrangement risks endless polymerization unless the donor in M4 docks to the receptor in M1. However the bridge sequences are too short to permit closure into a tetrameric cycle.
Despite subunits quite diverged in sequence, a more likely arrangement is an anti-parallel dimer of M1-M2-M3-M4 to M4-M3-M2-M1 (as seen in actinin spectrin repeats). This arrangement allows each module to dimerize with the quasi dihedral symmetry, leaving no docking sites vacant. Because the binding patches are in paralogous modules, this explains why observed surface residue conservation in frog M2 carries over to the other modules.
If true, this implies co-evolution of amino acids in these binding patches on top of what already occurs internally to an individual module. All phylogenetic software assumes -- ridiculously -- independent evolution of sites. However like morphological characters, molecular characters can have dependencies that amount to couplings and coordination between individual reduced alphabets. Here the much studied M1 module has surface residues co-evoving with counterparts on M4.
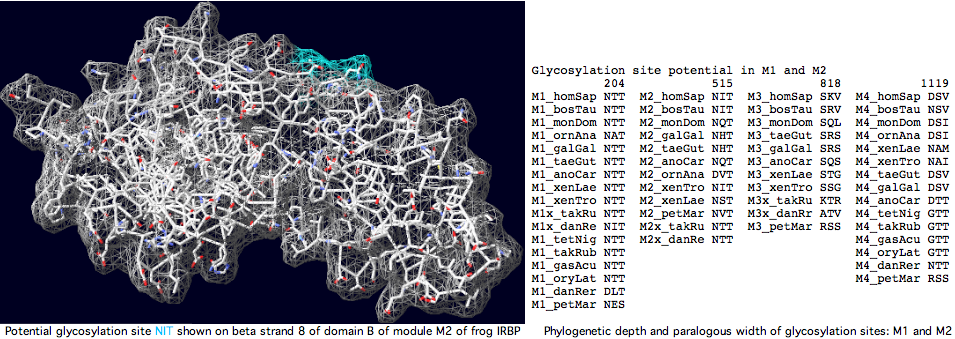
IRBP has only a single site with glycosylation potential (in beta strand 8 of the hinge region of domain B at residues 204 and 515 which are paralogous under module alignment). Although the NxT motif occurs with excellent phylogenetic conservation depth (human to lamprey, also in fish fragmentary genes), only modules M1 and M2 exhibit it. Given the interstitial location and experimental support for glycoprotein nature, IRBP is very likely glycosylated. Bulky carbohydrate chains need to be out of the way of the putative dimerization patches.
The situation with cysteines is not so clear. Here M1 has a cysteine at position 19 that is conserved back to lamprey (also in fish fragmentary genes) but A/T/S in other modules. A second conserved cysteine is found only in modules M2 and M3 at position 163 (human M1 numbering, residue is I). M4 has no conserved cysteines. A few other cysteines occur elsewhere but only sporadicaly in the comparative genomics sense. The cysteines at M2 and M3 coud potentially form a disulfide in the antiparallel dimer model discussed above. Inter-modular disulfides are not feasibly if modules are arranged linearly. No explanation is currently available for the conserved cysteine in M1.
An initial study of knockout mice disasterously used a strain carrying a severe mutation at L450 in RPE65, the enxyme catalyzing the rate-limiting isomerization step in the visual regeneration cycle. Lack of IRBP alone delays transfer of both bleached from photoreceptor cells and newly regenerated chromophore from RPE to photoreceptors.
A single known disease mutation D1080N causes a very rare recessive retinitis pigmentosa. This aspartate may disrupt an internal salt bridge according to the Xenopus M2 structure (and homology transfer to other modules]. It lies in M4 near the end of the second exon. This residue is conserved back to lamprey and amphioxus, indeed to crotonase and other weakly aligning homologs.
* AIILDLRYNLGGDREGVVHWASFFF RBP3_braFlo A+I+D+R+N+GG + S+FF AMIIDMRFNIGGPTSSIPILCSYFF RBP3_homSap +IID+R+N+GG ++SIPILCSYFF ILIIDLRYNMGGYSTSIPILCSYFF RBP3_petMar
Evolutionary origin of RBP3 (IRBP)
Lamprey sequence was recovered by a French group in 2008 from unassembled genome project contigs but not explicitly provided in the article or posted at GenBank. They reported four modules despite uncertainty in whether these all resided in the same gene. Below, a full length lamprey gene is independently recovered from the initial assembly and parsed into its modules using Superfamily HMM. It indeed has four modules classifying to the expected types and ordering, proving the gene was fully established prior to the common ancestor of mammals and lamprey. As the lamprey ancestor already had fully modern ciliary color vision (so need for retinol shuttling), the role of RBP3 (IRBP) in extant species may have already been established 500 myr ago.
Unexpectedly, lamprey lacks the final intron: exons 3/4 are fused. While no full length chondrichthyes sequence is available, close study of elephantshark contigs establishes that exon 3 has acquired an intron with the same phase 0 and position seen in later-diverging tetrapods. While intron loss in lamprey is possible, the simpler explanation is intron gain in the stem for reasons provided below.
Callorhinchus milii contig AAVX01012059 establishes that exon 3 has standard flanking phase 1 introns on both sides: VIASTSSLIVDLRYNIGGPTSSIPILCSYFFDDDKTVLLDTVYSRPTDTISEMKAIPQVAGNGSTESSVHSYIGERYGSKKSMVIL +I T+SL D RYNIGGPTSSIPILCSYFFDDDKTVLLDTVYSRPTDTISEMKAIPQVAGNGSTESSVHSYI E K +I+ LIIETNSL-RDHRYNIGGPTSSIPILCSYFFDDDKTVLLDTVYSRPTDTISEMKAIPQVAGNGSTESSVHSYICEDLHHCKYGLII
Previous efforts to trace the gene back to earlier deuterostomes (or metazoa) proved futile. That remains the case in March 2009 for tunicate (with a third assembly and massive transcript set) and much-studied sea urchin, establishing that in all likelihood homologs have been lost. However it is possible but implausible that sequences have diverged to the point of unrecognizability. It is equally implausible that a complex fold matching known bacterial folds arose de novo in deuterostomes. This creates the unresolved dilemma of ghost gene retention over an immense time frame in ancestral eyeless species yet loss in almost all extant clades.
With the advent of the second assembly of the cephalochordate Branchiostoma floridae (which has far fewer polymorphism-related assembly stutters than the first release), it is straightforward to recover a homolog in this species (after rubbishing an unmotivated fusion to the adjacent sulfotransferase in the JGI gene model). The considerably smaller gene here consists of a single module that clusters best with M3 and M4. This suggests that the module number expansion -- like so many key events in vertebrate gene evolution -- took place between cephalochordate and agnathan divergences.
The shocking aspect of the Branchiostoma RBP3 (IRBP) gene is its 9 exons. These range in size from 30 to 66 amino acids, quite typical of the average vertebrate gene. The anomaly here is really in the immense size of the first exon of the later four-module gene which extends for 1018 residues in human. That can be placed in prospective using the UCSC Table Browser to determine the overall size distribution of the 190,000-odd human coding exons. The average protein has 450 residues and 8-9 exons.
The second anomaly is that the placement and phasing of the Branchiostoma introns do not correspond at all via blastp alignment to those of module 4, the only vertebrate module with internal exons (3 of them). A great majority of introns are immensely conserved -- from human to cnidarian -- so the notion of massive erasure followed by de novo intronation in either the Branchiostoma or vertebrate gene can be discarded. However some explanation is required because the genes are strongly homologous (chance expectation e-30) though at best 31% identical and gappy in alignment.
The Branchiostoma gene can be assumed orthologous for lack of a better candidate, but nearby genes in the assembly (EARS2 DNAJC19 UGT2B4 CD79B) bear no obvious synteny to those flanking the human RBP3 gene (RBP3 ZNF488 GDF2 GDF10 ANXA8L1) and only broken chromosomal correspondence is seen in whole genome alignment to human (net track).
It follows that searches for earlier diverging species that still carry a homolog should be carried out with a single-domain protein as query. The amphioxus protein is the only one currently available and may be highly diverged from its ancestral form. In any event, it does not expose any cryptic homologs in tunicate or echinoderm, much less lophotrochozoa, arthropods or cnidaria established to have ciliary opsin systems.
Under the twin assumptions of orthology and approximately ancestral intronation pattern in Branchiostoma, how do we get from a one module gene with 8 introns to a four module gene with 2 unrelated introns in lamprey? This in some ways may have been the critical step in the evolution of imaging eyes with their massive requirement for retinol recycling that vastly exceeded that of the ancestral cephalochordate. Thus the gene co-evolved its interstitial location and shuttling function with the newly developed supporting role of the retina pigmented epithelium, which together were essential for effective vertebrate imaging vision.
One hypothetical scenario is formation first of a retroprocessed intronless gene. This may have displaced the nine-exon parental gene because of selection for more efficient translation. The protein may have been initially a tetramer of discrete subunits. The gene dosage then doubled allowing the protein to become a dimer of dimers, perhaps through a transcript that read through two close tandem copies. Next the intervening untranslated region experienced deletions fusing the previously independent modules. The gene complex then doubled again perhaps through recombinational mismatch to meet the newly evolving need for rapid retinol recycling -- at this point all four modules bound retinol. After two introns were acquired in the terminal module and lamprey diverged, a third and final intron was gained. Finally a need for allosteric regulation or interaction with other proteins allowed subfunctionalization of other domains to non-retinol shuttling roles despite the apparent loss of previously selected efficiency.
While this scenario is speculation and non-unique, elements of it are not uncommon in other genes, even in opsins. For example, a retroprocessed fish RHO1 completely displaced the parental 5 exon gene and LWS repeated spawned secondary cone opsin genes that were initially tandem. A great many human proteins have internally repeated domains, some like titin quite large numbers of them. There may not exist sufficient surviving members of the cephalochordate, hagfish and lamprey clades to specifically illuminate what happened here. The highest priority would be to study localization and function of RBP3_braFlo within that organisms visual systems.
>RBP3_braFlo Branchiostoma floridae Region: 9 exons; 1 domain: 83-381 0 MTRPSKVDIVFPIKPFTIPTAHEQVKGEGPVDINKNALCKSADEGHTHP 1 2 VSIAMAPTAYIVFVALVPTVLSVDWLDVVMGIGDVMADHYLDQDLRALNDQSLLQRWNRTLVHRFQ 0 0 SWSQDDMSDSLRMEEGLTSELRNITGDETIK 0 0 VWDFGVYENTTQEPVPREFYNFSTFVDNFK 2 1 KNREKHINVTMLEGNVGYVSIRSMSHIVDIILPDPEMTEFFLSKMAALNESK 0 0 AIILDLRYNLGGDREGVVHWASFFFNATPSVPLSDVYYRDGVNQYWTLLE 0 0 VPGGIRFPDMPLYLLTSNRTSREAEEFAYAMQVVNRTTIIGETT 1 2 AGEEFTGMWFPIDQTDVHLLTRTNVVRNPITQDSWSGK 1 2 GVTPDIIVPSEKALTVALRKIQGSEDTKMAASSGNIEPPRWTVYLVFICTSIAILTYPTFM*
RBP3 (IRBP) use in marsupial phylogeny
The first three homology domains and part of the fourth are all encoded by the first large exon of 1090 amino acids. Fragments of this exon has been much used in marsupial phylogeny (along with the first intron of transthyretin). Indeed the 96 marsupial species in 51 genera having determined IRBP sequences at GenBank include a Dec 2008 partial sequence for Thylacinus cynocephalus, as well as for Sarcophilus harrisii.
The closest matches to the thylacine IRBP are shown in the difference alignment of the first 60 residues below. These species all lie with the Dasyuromorphia. The indicated E-->K may be one of several phyloSNPs breaking this group into blue and green subclades.
The numbat Myrmecobius fits implausibly (its amino terminal sequence EF028750 needs verification) -- its affinities seem to lie with the Didelphimorphia. Thylacinus is not basal within Dasyuromorphia relative to Myrmecobius using IRBP. However this may be a case of mis-comparison of genes.
* * * STSKAPQHDSKFTNATQEELLALFQQIIKYQVLEGNVGYLRVDYIPGREMIEEVGEFLVN EU091365 0 Thylacinus cynocephalus .........P..A..................I............................ AY532676 3 Myoictis wallacei ........NP..A............................................... AY532687 3 Neophascogale lorentzii ........NP..A........T...................................... AY532686 4 Phascolosorex dorsalis .........P..V............................................... AY532670 2 Parantechinus apicalis ....V....P..A..................I.....................L...... AY532675 5 Myoictis melas .........P..A...................................D........... AY532679 3 Dasyurus hallucatus ...E.....P..A............K........D.............D........... AY532685 6 Sarcophilus harrisii ...E.......RA..........L............................Q..K.... EF028748 6 Sminthopsis crassicaudata .......R.P.LA.........SL.......................Q....Q....... EF028749 8 Planigale ingrami ..A......P.LA.V.....................................K....... EF028736 6 Antechinus stuartii ..A......P.L..V.....................................K....... EF028743 5 Micromurexia habbema ..A......P.LA.V.....................................K....... EF028744 6 Murexchinus melanurus ..A......P.L..V....V................................K....... EF028746 6 Paramurexia rothschildi ..A......P.LA.V.....................................K....... EF028747 6 Phascogale calura ..A......P.LA.V.....................................K....... EF028745 6 Phascomurexia naso .SA......P.LA.V.....................................K....... AY532667 7 Murexia longicaudata ......K..PNLA........T.L..R....................Q.VV.K....... EF028750 12 Myrmecobius fasciatus ..PET...VP..A.V........L..M....................Q.VV.K....... AY233765 13 Caluromys philander ..PET...VP.LA.V.......QL..M....................Q.VV.K....... AF257675 15 Caluromysiops irrupta ..PET...VP.LA.V......T.L..M....................Q.VV.K....... AF257688 15 Glironia venusta .IPET...VP..A.V.R....T.L..M....................Q.VV.K....... AF257683 16 Didelphis albiventris .IPE....VP.LA.I......T.L..M....................Q.VV.K....... AF257686 15 Gracilinanus microtarsus .IPET...VP..A.V......T.L..M....................Q.VV.K....... AF257676 15 Marmosops noctivagus .IPET...VP.LA.V........L..M....................Q.VV.K....... AY233788 15 Philander opossum .IPET...VP.LA.I......T.L..M....................Q.VV.K....... AF257689 16 Thylamys pallidior
Using Sarcophilus as probe in a different region, 721-900, we find this peculiar outcome: what appears to be a second very odd gene, XY difference, pseudogene, weird balanced polymorphism, nonhomologous recombination, sequence submission error, frameshifts, or systemic experimental error (eg Dasyurus maculatus AY532680 is identical to AY243439 outside the 15 amino acid block). However the genomic reads from individual Sarcophilus used in this project show no sign of this gene despite excellent coverage of the second type of gene.
Macropus and Monodelphis genomes only contain the second type of gene. All Didelphimorphia and Diprotodontia are of this type, as are platypus and all placentals. With the Sarcophilus genome, this can be resolved as it should have both and be the such first genome. Perhaps the alignment above is a mixture of type 1 and type 2 genes (resp. alleles). The Myrmecobius anomaly makes it more likely two distinct genes are present.
A definite pecularity seen in blast searches is the occurence earlier in the sequence of a very homologous segment for this very block, likely the homologous part of another of the internal tandem repeats. It is seen in both types of genes. Possibly internal non-homologus recombination or gene conversion has inserted first repeat sequence again in this distal block in place of what was relatively diverged sequence. Internal gene conversion would make IRBP extremely difficult to use in alignment-based phylogeny. As rare genomic event, it unites the species that have it but species that don't have it would have to be re-examined to exclude the possiblity that only the type 2 gene happened to be sequenced.
It emerges from direct tblastn that the Sacrophilus individual sequenced was female. That is, ATRX is well represented but not ATRY (though the situation is somewhat confused due to additional paralogs). Marsupial XY are quite different from placentals:
"Many or most genes on the mammal Y chromosome evolved a testis-specific function after diverging from an X-borne copy with a general function in both sexes. In marsupial but not eutherian mammals, a testis-specific orthologue (ATRY) of the widely expressed X-borne ATRX gene lies on the Y chromosome. Since mutations in human ATRX cause sex reversal, it is possible that one function of ATRY in marsupials is testicular differentiation. We report here the isolation and sequencing of the tammar wallaby (Macropus eugenii) ATRY cDNA, and comparison of its sequence with that of tammar ATRX. The evolution of a testis-specific function for the ATRY protein distinct from the general role of ATRX in both sexes has been accompanied by sequence changes in many protein domains that would alter protein binding partners. A large open reading frame encodes a 1771 amino acid ATRY protein that has diverged extensively from ATRX. The conservation and loss of particular motifs identify those required for testicular function (ATRY) and function in other tissues (ATRX)."
AY532685 MEILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE Sarcophilus harrisii
AY532684 ....E................................S....................P. Dasyurus geoffroii
AY532681 ....E................................S....................P. Dasyurus albopunctatus
AY532683 ....E................................S....................P. Dasyurus viverrinus
AY532682 ....E........................P.......SE...................P. Dasyurus spartacus
AY532680 ....E..............R.................SR...................P. Dasyurus maculatus
AY532678 ..V..................................S....................P. Dasycercus cristicauda
AY532669 ..V..................................S....................P. Dasykaluta rosamondae
AY532676 ..V..................S...............S....................P. Myoictis wallacei
AY532675 ..V..................S...............S....................P. Myoictis melas
AY532687 ..V........N.L.......................S....................P. Neophascogale lorentzii
AY532671 ..V..................................S....................P. Parantechinus bilarni
AY532670 ..V.................................TS.........RG.........P. Parantechinus apicalis
AY532686 ..V..................................S........P...........p. Phascolosorex dorsalis
AY532674 ..V.......................................................P. Pseudantechinus ningbing
AY532672 ..V..................................S....................P. Pseudantechinus woolleyae
AY532673 ..V........N..R......................S...................SP. Pseudantechinus roryi
454 read MEILQKYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAMLQAVSEDP Sarcophilus harrisii
EF028739 ............................V.TEEDLAAKLNAMLQA.............P. Antechinus minimus
AY243439 ....E..............R........V.TEEDLAAKLNAMLQA.............P. Dasyurus maculatus
EF028750 ....K................KT.....I.TEEDLAAKLNAILQA.............P. Myrmecobius fasciatus
EF028737 ..V.........................V.TEEDLAAKINAMLQA.............P. Antechinus flavipes
EF028748 ..V.........................V.TEEDLAAKLNA.LQA.............P. Sminthopsis crassicaudata
AY243438 ..V.........................V.TEEDLAAKLNA.LQA.............P. Planigale sp.
EF028749 ..V.........................V.TEEDLAAKLNA.LQA.............P. Planigale ingrami
AY532679 ..V.........................V.TEEDLAAKLNAMLQA............... Dasyurus hallucatus
AF025382 ..V.........................V.TEEDLAAKLNAMLQA.............P. Phascogale tapoatafa
EF028741 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus godmani
AY532666 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus swainsonii
EF028736 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus stuartii
EF028742 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus agilis
EF028738 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus bellus
EF028740 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus leo
EF028747 ..V.........................V.TEEDLAAKLNAMLQA.............P. Phascogale calura
EF028744 ..V.........................V.TEEDLAAKLNAMLQA.............P. Murexchinus melanurus
EF028743 ..V.........................V.TEEDLAAKLNAMLQA.............P. Micromurexia habbema
EU086688 ..V.........................V.TEEDLAAKLNAMLQA.............P. Pseudantechinus macdonnellensis
EU086689 ..V.........................V.TEEDLAAKLNAMLQA.............P. Pseudantechinus roryi
EU086686 ..V.........................V.TEEDLAAKLNAMLQA............SP. Pseudantechinus macdonnellensis
EU086687 ..V.........................V.TEEDLAAKLNAMLQA..........G..P. Pseudantechinus mimulus
AY532667 ..V.........................V.TEEDLAAKLNAMLQA.............P. Murexia longicaudata
EF028746 ..V.........................V.TEEDLAAKLNAMLQA.............P. Paramurexia rothschildi
AY532677 ..V.........................V.TEEDLAAKLNAMLQA.............P. Dasyuroides byrnei
EF028745 ..V..........I..............V.TEEDLAAKLNAMLQA.............P. Phascomurexia naso
Macropus eugenii assembly
sacHar MEILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE
ME+LQ YYTLVDRVPALLHHLTAIDYSS L + ++ VSEDPRLLVRVLR E
macEug MEVLQNYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAGLQAVSEDPRLLVRVLRPE
Monodelphis domestica assembly TSSLVLDLQHSSGGEISG
sacHar MEILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE
ME+LQ YYTLVDRVPALLHHLTAIDYSS L + ++ VSEDPRLLVRVLR E
monDom MEVLQNYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAGLQAVSEDPRLLVRVLRPE
Ornithorhynchus anatinus assembly
sacHar EILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE
++L+ YY LVDRVPALL HL A+D SS L + SR SEDPRLLVR L E
ornAna DLLRDYYALVDRVPALLRHLAALDLSSVLSEEDLTSRLNAGLQAASEDPRLLVRRLEPE
Equus caballus assembly
sacHar EILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE
E LQ YYTLVDRVPALLHHL ++D+SS + D ++ VSEDPRLLV V+RS+
equCab EALQDYYTLVDRVPALLHHLASMDFSSVVSEDDLVAKLNAGLQAVSEDPRLLVWVVRSK
Reference sequences
These are organized both parsed into functional modules (signal peptide and spacer residues are dropped) as determined by global multiple alignment consistency and as intronated genes showing position and phase of intron breaks. Both sets are in phylogenetic order with respect to the canonical deuterostome tree.
RBP3 (IRBP) sequences parsed into structural modules
>M1_homSap LFQPSLVLDMAKVLLDNYCFPENLLGMQEAIQQAIKSHEILSISDPQTLASVLTAGVQSSLNDPRLVISYEPSTPEPPPQVPALTSLSEEELLAWLQRGLRHEVLEGNVGYLRVDSVPGQEVLSMMGEFLVAHVWGNLMGTSALVLDLRHCTGGQVSGIPYIISYLHPGNTILHVDTIYNRPSNTTTEIWTLPQVLGERYGADKDVVVLTSSQTRGVAEDIAHILKQMRRAIVVGERTGGGALDLRKLRIGESDFFFTVPVSRSLGPLGGGSQTWEGSGVLPCVGTPAEQALEKALAIL >M1_bosTau LFQPSLVLEMAQVLLDNYCFPENLMGMQGAIEQAIKSQEILSISDPQTLAHVLTAGVQSSLNDPRLVISYEPSTLEAPPRAPAVTNLTLEEIIAGLQDGLRHEILEGNVGYLRVDDIPGQEVMSKLRSFLVANVWRKLVNTSALVLDLRHCTGGHVSGIPYVISYLHPGSTVSHVDTVYDRPSNTTTEIWTLPEALGEKYSADKDVVVLTSSRTGGVAEDIAYILKQMRRAIVVGERTVGGALNLQKLRVGQSDFFLTVPVSRSLGPLGEGSQTWEGSGVLPCVGTPAEQALEKALAVL >M1_monDom IFQPSLVRDMAKILLDNYCFPENLMGMQEVIEQAIKSGEILDISDPQMLASVLTAGVQGALNDPRLVISFEPSIPETPQHVPKLANVTQEELLILLQQMIKYQVLEGNVGYLRVDYIPGQEVVEKVGEFLVNNIWKKLMGTSSLVLDLQHSSGGEISGIPFVISYLHQGDILLHVDTVYDRPSNTTTEIWTLPQVLGERYGGEKDMVVLTSHRTVGVAEDIAYILKKLRRAIVVGEQTLGGALDLRKLRIGQSDFFITVPVSRSLSPLGGGSQTWEGSGVLPCVGIPAEQALGKALAIL >M1_macEug IFQPSLVLDMAKILLDNYCFPENLMGMQEAIEQAIKSGEILDISDPQMLASVLTAGVQGSLNDPRLVISYEPSPAEAPQQSPKLTSLTQEELLTLLQQMIKYQVLDGNVGYLRVDYIPGQEVVEKVGEFLVNDIWKKLMGTSSLVLDLQHSTEGEVSGIPFVISYLHEGDILLHVDTVYDRPSNTTTEIWTLPQVLGERYSGEKDLVILTSHRTVGVAEDIAYILKKMRRAIVVGEQTLGGALDLRKLRIGQSDFFITVPVSRSLSPLGGGSQTWEGSGVLPCVGIPAEQALEKALAIL >M1_sacHar IFQPTLVLDMAKILLDNYCFPENLMGMQEAIEQAIKSGEILDISDPQMLANVLTAGVQGSLNDPRLVISYEPSTSEAPQHDPKFANATQEELLALFQKIIKYQVLEDNVGYLRVDYIPGRDMIEEVGEFLVNDIWKKVMETSSLVLDLQHSRGGEVSGIPFVISYLHQGDILLHVDTIYDRPSNTTTEIWTLPQVLGERYSGEKDIVVLTSHHTVGVAEDIAYILKKMRRAIVVGEQTQGGSLDLRKLRIGQSDFFITVPVARSLSPLGGGSQTWEGSGVVPCVGIPAEQALEKALAIL >M1_ornAna VSQPSMVLDVAKILLDNYCYPENLMGMQEAIEEAIQRGEILDIADPKRLASVLTAGVQGSLNDPRLVISYEPAPVAVSQQPPEPASLPAEQPLERLRPAVGSEVLEGNVGYLRVDRLPGREEIERVGAVLGRDIWEKLLGTSALVLDLRHSTGGHVSGIPFFISYFYPEGPALHVDTVYDRPSNATRQLWTLPRVLGARYAADKDVVVLTSRLTAGVAEDVAYILQQMRRAIVVGERTAGGPLVFRKLRVGLSDFFITVPVACSLGPLGGGGRSWEGSGVLPCVAVPADRALDEALDIL >M1_galGal IFQPTLVLDMAKVLLDNYCYPENLVGMQEAIEQAIKSGEILDISDPKMLANVLTAGVQGALNDPRLVISYEPSLHAAPKQEAETYPTREQLLSLIEHVVIYDKLEGNVGYLRIDYIIGQEVVEKVGAFLVDKVWKTLINTSALVIDLRYSTGGQISGIPFIISYLHEADKMLHVETVYNRPSNTTTEIWTLPKVLGERYSKDKDVIVLISHHTTGVAEDVAYILKHMNRAITLGEKTAGGSLDIQKLRIGPSNFYMMVPVSRSVSPLSGGGQSWEVSGVMPCVASEAEQALKKSLDIL >M1_taeGut IFQPTLVLDMAKVLLDNYCYPENLVGMQEAIEQAIKSGEILDISDPKMLANVLTAGVQGALNDPRLVISYEPLPHSGPKQEAEGSPTREQLLSLIEHVIMYDKLEGNVGYLRIDYIIGEEVVQKVGAFLVDKVWKTLIETSALVIDLRHSTGGQISGLPFIISYLHEQDKILHVETVYNRPSNTTTEIWTLPKVLGERYSKDKDVIVLISHHTTGVAEDVAYILKHMNRAITVGEKTAGGSLDIQKLRIGPSNFYMMVPVSRSVSPLSGGGQSWEVSGVMPCVATEAEQALQKSLDIL >M1_anoCar VLQSTLVLDMAKLLLDNYCLPENLVGMREAIEQAIKNGEVLDISDPKLLATVLTAGVQGALNDPRLVISYEPTAPAAPKQRMETSLTPEQLLSLIQHTVKYEVLDDNVGYLRIDYIMGQDIVQKIGSFLVEKVWKTLLGTSALILDLRYTTGGDVSGIPFIISYLYNGDKVLHVDTVYNRPSNTTVEILTLPKVLGVRYSKDKDVILLISKYTTGVAENVAYILKHMHRTIIVGEKSAGGSLDTQKMQIGNSQFYMTVPLSCSVSPLSGSGQSWEISGVTPCVVISAEQALDKALAIL >M1_xenLae LFQPSLVMDMAKVLLDNYCFPENLVGMQETIEQAVKGGEILHISDPDTLANVFTSGVQGYLNDPRLVVSYEPNYSGPQTEQSLELTPEQLKFLINHSVKYDILPGNIGYLRIDFIIGQDVVQKVGPHLVNNIWKKLMPTSALILDLRYSTQGEVSGIPFVVSYLCDSEIHIDSIYNRPSNTTTDLWTLPELMGERYGKVKDVVVLTSKYTKGVAEDASYILKHMNRAIVVGEKTAGGSLDTQKIKIGQSDFYITVPVSRSLSPLTGQSWEVSGVSPCVVVNAKDALDKAQAIL >M1_xenTro VFQPSLVMDMAKVLLDNYCFPENLVGMQETIEQAMKSGEILHISDPETLANVFTSGVQGFLNDPRLVVSYEPNYSGPRKEQSPEPTLEQLKFLLDHSVTYDLLPGNIGYLRIDFIIGQDVVQKVGPLLVNNIWKKLMPSSALILDLRYSTQGKVSGIPFVVSYLTDPQIHIDSIYNRPSNTTTDLWTLSELMGERYGKDKDVVVLTSKYTEGIAEGAAYILKHMSRAIVVGEKTAGGSLDIQKIKIGQSEFYITVPVSRSISPLTGQSWEVAGVFPCVVVNANNALNKAQGIL >M1_tetNig AFPPSLIADMAKIVLDNYCSPEKLAGMKEAIKAAGTNTEVLNIPDGESLARVLSAGVQGTVSDPRLMVSFQPNYVPAGPHKMPPLPPEHLVAVLQTSVKLDILEGNTGYLRIDHILGEEVADKVGPALIDLIWNKILPTSALIFDLRYTSSGDISGIPYIVSYFTQAEPVVHIDSVYDRPSNTTTKLLSLPNLLGQRYGVSKPLIVLTSKNTKGIAEDVAYCLKNLKRATIVGEKTAGGSLKLDTFKVGDTDFYITVPTAKSINPITGSSWEIRGVTPHVEVNAEDALATAIKIV >M1_takRub AFPPSLITDMAKIVLDNYCSPEKLAGMKEAIEAAGTNTEVLNIPDGESLARVLSAGVQGTVSDSRLMVSYQPDYVPAVPPKMPPLPPEHLVAVLQTSIKLDLLEGNTGYLRIDHIIGEDVAEKVGPSLIDLIWNKILPTSALIFDLRYTSSGEISGIPYIVSYFTQAEPVVHIDSVYDRPSNTTTKLFSLSNLLGERYGITKPLIILTSKNTKGIAEDVAYCLKNLKRATIVGERTAGGSVKLDNFKVGSTDFYITVPTAKSINPVTGSSWEITGVKPDVEVNAEDALATAIKIV >M1_gasAcu GFAPNVIIDMAKIVIDNYCSPEKLAGMKEAIEAAGSNTEVLSIPDAETLANVLSAGVQTTVSDPRLMISYEPNYVPVVPPKMPPLPPDQVIAVLQTSIKLDILEGNIGYLRIDHILGEDVAEKVGPLLLDLVWNKILPTSALIFDLRYTSSGDISGIPYIVSYFTEAGTPIHIDSIYDRPSNTTTKLFSMSTLLGERYSTSKPLIILTSKNTKGIAEDVAYCLQNLKRATIVGEKTAGGSVKVDKIQVRDTGFYVTVPTAKSVNPITGSTWEVTGVTPNVEVNAEDALATAIKIV >M1_oryLat SFPPSLITDLAKIVMDNYCSPEKLSGMKEDIATAGANTDVLNIPDGEALAKVLTDGVQTTVSDPRLRVSYEPNYVPVVPPQLPPEQLIAVLQTSIKLDILEGNIGYLRIDSIIGEEVAEKVGPLLLELVWSKILPTSALIFDLRYTSSGDITGIPYIISYLTDAKSEIHIDTIYDRPLNTTTKLLSMQSTLGQTYGGTKPLLVLTSKNTKDIAEDVAYCLKNLKRATIVGEKTAGGSAKIKKFRVGDTDFYVTLPTAKSINPITGSSWEVTGVKPNVEVNAEEALATALKII >M1_danRer NFSPTLIADMAKIFMDNYCSPEKLTGMEEAIDAASSNTEILSISDPTMLANVLTDGVKKTISDSRVKVTYEPDLILAAPPAMPDIPLEHLAAMIKGTVKVEILEGNIGYLKIQHIIGEEMAQKVGPLLLEYIWDKILPTSAMILDFRSTVTGELSGIPYIVSYFTDPEPLIHIDSVYDRTADLTIELWSMPTLLGKRYGTSKPLIILTSKDTLGIAEDVAYCLKNLKRATIVGENTAGGTVKMSKMKVGDTDFYVTVPVAKSINPITGKSWEINGVAPDVDVAAEDALDAAIAII >M1_petMar KFDTAVVLHLAKVLLDNYCIPENLVGMDEAIQRAVDNGELLGVSDPESAASALTEGIQAALNDPRIAVSYVAPPHTFEELLATIPQKTSFAVLDGNVGYLRADEIISEATIKKLGPVIVQRIWNRLVDTDTFVLDLRYNSHGDITGLPYLVSCFCEPRPVVHLDTVYYRPTNESKEIWSLPDLQGARFAKHKDVFVLVSANTEGVAENVAYVLKHLHRATVIGEQTAGGSLEVERFRLGDSRFFVTVPTARSEPADRSWGVFPCVSAPSERALDKALEIL >M1x_takRu FYQHTLVLEMAKLLLENYCIPENLVGMQEAIQRAIKSREILQISDRKTLATVLTVGVQGALNDPRLSVSYEPSFSPLPLQALSSLPVEQQLRLLRNSIKLDILDSDVGYLRIDRIIDEETLLKFGPLLRENVWDKAAQTSSLILDLRFSTAGGWSGIPSIVSYFTEPHSLVHIDTVYDRPSNTTTELWTMSSVRGKTFGGKKDMIVLIGRRTAGAAEAVAYTLKHLNRAIVVGERSAGGSLKVRKFRIAESDFYITMPVARSVSPITGKSWEVSGISPTVNVAAREALAKAQTFL >M1x_danRe SFQSALVLDMAKILLDNYCFPENLIGMQEAIQQAINSGEILHISDRKTLASVLTAGVQGALNDPRLTVSYEPNYTLITPPALHSLPTEQLIRLIRSTVKLEVMDNNIGYLRIDRIIGQETVVKLGRLLHNNIWKKVAHTSAMIFDLRFSTAGELSGLPYIVSYFSDSDPLLHIDTIYERPTNITRELWTLPTLLGERFGKRKDLIVLISKRTIGAAEGVAYILKHLKRAVIIGERSAGGSVRVDKLKIGDSGFYITVPVARSVNPVTGQSWEVSGVAPSVTVNPKESIAKAKSLI >M2_homSap SALPGVVHCLQEVLKDYYTLVDRVPTLLQHLASMDFSTVVSEEDLVTKLNAGLQAASEDPRLLVRAIGPTETPSWPAPDAAAEDSPGVAPELPEDEAIRQALVDSVFQVSVLPGNVGYLRFDSFADASVLGVLAPYVLRQVWEPLQDTEHLIMDLRHNPGGPSSAVPLLLSYFQGPEAGPVHLFTTYDRRTNITQEHFSHMELPGPRYSTQRGVYLLTSHRTATAAEEFAFLMQSLGWATLVGEITAGNLLHTRTVPLLDTPEGSLALTVPVLTFIDNHGEAWLGGGVVPDAIVLAEEALDKAQEVL >M2_bosTau RALPGVIQRLQEALREYYTLVDRVPALLSHLAAMDLSSVVSEDDLVTKLNAGLQAVSEDPRLQVQVVRPKEASSGPEEEAEEPPEAVPEVPEDEAVRRALVDSVFQVSVLPGNVGYLRFDSFADASVLEVLGPYILHQVWEPLQDTEHLIMDLRQNPGGPSSAVPLLLSYFQSPDASPVRLFSTYDRRTNITREHFSQTELLGRPYGTQRGVYLLTSHRTATAAEELAFLMQSLGWATLVGEITAGSLLHTHTVSLLETPEGGLALTVPVLTFIDNHGECWLGGGVVPDAIVLAEEALDRAQEVL >M2_monDom RARPGAIQRLMEVLQNYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAGLQAVSEDPRLLVRVLRPEEATMGEAEEEDATPAANSLPEDESQRQALVDSVFQVSVLPGNVGYLRFDEFADSSVLGTLAPYVIRQVWEPLQDTNHLIMDLRYNPGGPSSAVPLLLSYFQDPAAGPIRLFTTYDRQTNQTQEHLSRAELLGKPYGAQRGVYLLTSHHTATAAEEFAFLMQSLGRATLVGEITAGSLMHTRTFPLLQPPNGNLVLTVPILTFIDNNGECWLGGGVVPDAIVLAEEALDKAKEVL >M2_macEug RARPAAIQRLMEVLQNYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAGLQAVSEDPRLLVRVLRPEEATAGAESREEAATAAPVPLPDGESQRQALVNSVFQVSVLPGNVGYLRFDEFADSSVLGALAPYVLQQVWEPLQDTDHLIMDLRYNPGGPSSAVPLLLSYFQDPAAGPVRLFATYDRQTNQTREYRSQAELLGKPYGAQRGVYLLTSHHTATAAEEFAFLMQSLGRATLVGEITAGNLMHTRTFSLLQPPDGSLVLTVPILTFIDNHGECWLGGGVVPDAIVLAEEALDKAKEVI >M2_sacHar RARPGAIQRLMEILQKYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAMLQAVSEDPRLLVRVLRPEEATVEAEPGEESATPASVSLPESDAERQALIDSVFQVSVLPGNVGYLRFDEFADNSVLGTLAPYVLRQVWEPLQDTDHLIMDLRYNPGGPSSAVPLLLSYFQDPSAGPVRLFATYDRQTNQTQEYRSRAELLGKPYGAERGVYLLTSYHTATAAEEFAFLMQSLGRATLVGEITAGSLMHTRTFPLLQPPNGSLVLTVPTLTFIDNHGECWLGGGVVPDAIVLAEEALDKAKEVL >M2_ornAna GAVPGAVAHLADLLRDYYALVDRVPALLRHLAALDLSSVLSEEDLTSRLNAGLQAASEDPRLLVRRLEPEEAERGPPRKEEEQKEEEEEDQPSPGASILPGDGSSLFRVSVLPGNVGYLCFDEFPEASALERLGPLLGRRVWEPLEATDHLMVDLRNNPGGPSSAVPLLLSYFQDPAAGPIRLFTTYNRPADVTREYASRAGALEKPYGARRGVYLLTSHRTATAAEEFAYLMQALGRATLVGEITAGRLLHSRTFPLLRPPWEGLVLTVPFLTLFDPHGEGWLGGGVVPDAIVLAEEALEKAGEVL >M2_galGal RAVPGTLSRLTDILKDYYSLVERVPVLLRHLTTSDFSSVQSAEDLATKLNTEMQTLSEDPRLLVRTMMPGEAAAPPAEMPIAMAANLPDNEQLLHALVDTVFKVSVLPGNVGYMRFDEFADASVLVKLGPYIVKKVWEPLQNTENLIMDLRYNPGGPSSSAVPMLISYFQDPTAGPVHLFTTYDRRTNHTQEHNSQAELLAQPYGAQRGIYVLTSRHTATAAEEFAYLMQSLGRATLIGEITAGSLSHTCTFPLVQPEQGITRGLTITVPVITFIDNHGESWMGGGVVPDAIVLAEDALEKAEEVL >M2_taeGut RAVPGTISHLKNILKDYYSLVERVPALLRRLTTSDFSSVQSSEDLATKLNTELQALSDDPRLMVRVMMPGEAADSPAEKPVGMAADLPDNEQLLHALVDTVFKVSVLPGNVGYMRFDEFADASVLVKLGPYLVHKVWEPLQNTENLIMDLRYNLGGPSSSAVPVLLSYFQDPAAGPVHLFTTYDRRTNHTQEHNSQAELLGQSYGAKRGVYLLTSHHTATAAEEFAYLMQSLGRATLIGEITAGSLSHTRTFPLLQPGPGITRGLTITVPVITFIDNHGESWMGGGVVPDAIVLAEDALEKAEEVL >M2_anoCar KAIPNSMSYLVDIIKNNYSMLEQVPVLLQHLSTFDYSSVLSVKDLASKLNAELQTISEDPRLFLRVPASDEAVTSQTDEKVAMASDLPNNEQLMKALVMTVFKVSVLPGNVGYMRFDEFGDATVLVKLGPYLLQHVWEPLQATDYLIIDLRYNIGGPSSSAVPVLLSYFQDPSAGPVHFFTTYNRLTNQTQAYSSSAEMVGKPYGARRGVYLLTSHNTATAAEEFAYLMQTLGRATLVGEITAGSLSHTHTFCILELGGGCGLLINVPVITLIDNHGEYWLGGGVVPDSIVLADEALEKAREVL >M2_xenTro SSITHILLQLSEILVNNYAFSERIPTLLQHLPNLDYSSVISEEDITAKLNYELQSLTEDPRLVLKSKTDSLVMPEDSTQVENLPDDEATLQALVNTVFKVSILPGNIGYLRFDEFADVSVLAKLGPYIVNTVWDPITVTENLIIDLRYNIGGSSTSIPLLLSYFQEPENRIHLFTIYNRQQNSTNEVYSLPKVLGKPYGSKKGVYVLTSHETATAAEEFAYLMQSLSRATIIGEITSGNLMHSKAFPLDGTRLSVTVPIMNFIDNNGDYWLGGGVVPDAIVLADEALDKAKEII >M2_xenLae SSVTHVLHQLCDILANNYAFSERIPTLLQHLPNLDYSTVISEEDIAAKLNYELQSLTEDPRLVLKSKTDTLVMPGDSIQAENIPEDEAMLQALVNTVFKVSILPGNIGYLRFDQFADVSVIAKLAPFIVNTVWEPITITENLIIDLRYNVGGSSTAVPLLLSYFLDPETKIHLFTLHNRQQNSTDEVYSHPKVLGKPYGSKKGVYVLTSHQTATAAEEFAYLMQSLSRATIIGEITSGNLMHSKVFPFDGTQLSVTVPIINFIDSNGDYWLGGGVVPDAIVLADEALDKAKEII >M2_petMar GVARKAVEAAGELLLSSYTFVERASAIADHLSWSEYGSVVSVEDLTSKLTQDLQSVAEDPRLVVSNREPEWVGAADPPGPPAPLPDDEQMLEAIVDSAFKVEVLEGNIGYLRFDEFGDASAVMKLRKQLVSKVWERIHPTDDVIIDLRYNLGGSSTAIPIVLSYFQDVAPVHFYTVYDRLRNVTAEFHTVSNLTSQLYGSKKGVYLLTSQHTATAAEEFTYLMQSLNRATIVGEITSGRLAHSLAFRLSDTGLYMTVPIVNFIDNNDEYWLGGGVVPDAIVLAENALDAAKEII >M2x_takRu SRIPKVLQIVLDIIGRFYAFADRVQALLQQLESADLFSVVSEEDLAARLNHDLQTASEDPRLIIRHKRDNIPRAEEEPELHAANDHDGELVEGFTVQVLPHNTGYLRLDRFVRCSEGDKLEEIVAEKVWGPLKDTQNLIIDLRHNTGGSSTSVALLLSYLRDPLPKRHFFTIYDSVQNTTTEYGSRPHIPGPSYGSERGVYVLTSHYTAGAAEEFAYLIQSLHFGTVVGEITSGTLMHSKTFQVEGTDIFITVPFINFLDNNGEYWLGGGVVPDAIVLAEEALEHVNRTA >M2x_danRe KTIPKAVRRVSDIIKRYYSFKDKIPALLNQLAKADYFTVVSEEDLAGKLNHEMQSVFEDPRLLIKATQVLTDDASSEDRSSSDDLTDPLFKLEMISGNNGYLRFDRFPTPEVLLRLEDHIKKKIWQPVQETENLVIDLRFNTGGSTEALPILLSYMFDTSSSTYLFSIYDSIKNTTFDFHTLNNISGPSYGSTKGVYVLTSYYTAEAGEEFAYLMQSLHRGTVIGEITSGMLLHSKTFQIEQTSLAITVPIINFIDVNGECWLGGGVVPDAIVLAEEALERAHEII >M3_homSap QSLGALVEGTGHLLEAHYARPEVVGQTSALLRAKLAQGAYRTAVDLESLASQLTADLQEVSGDHRLLVFHSPGELVVEEAPPPPPAVPSPEELTYLIEALFKTEVLPGQLGYLRFDAMAELETVKAVGPQLVRLVWQQLVDTAALVIDLRYNPGSYSTAIPLLCSYFFEAEPRQHLYSVFDRATSKVTEVWTLPQVAGQRYGSHKDLYILMSHTSGSAAEAFAHTMQDLQRATVIGEPTAGGALSVGIYQVGSSPLYASMPTQMAMSATTGKAWDLAGVEPDITVPMSEALSIAQDIV >M3_bosTau RSLGELVEGTGRLLEAHYARPEVVGQMGALLRAKLAQGAYRTAVDLESLASQLTADLQEMSGDHRLLVFHSPGEMVAEEAPPPPPVVPSPEELSYLIEALFKTEVLPGQLGYLRFDAMAELETVKAVGPQLVQLVWQKLVDTAALVVDLRYNPGSYSTAVPLLCSYFFEAEPRRHLYSVFDRATSRVTEVWTLPHVTGQRYGSHKDLYVLVSHTSGSAAEAFAHTMQDLQRATIIGEPTAGGALSVGIYQVGSSALYASMPTQMAMSASTGEAWDLAGVEPDITVPMSVALSTARDIV >M3_monDom QRLGALVEGTGHLLEAHYALPEVVGQASALLKAKLEHGTYRTAVDFESLASQLTSDLQEVSGDHRLHVFHSPGEPVSEELTPPQKGVPSPEELTYLIEALFKTEVLPGQLGYLRFDMMAEAETVRAIAPQLVELVWEKLVHTEALVVDLRYNPGGYSTAVPLLCSYFFEAEPRRHLYTIFDRAASQLTEVWTLPQVAGERYGSQKDLYILISHTSGSAAEAFVHTMKDQHRATVIGEPTGGGALSVGIYQVENSPLYASMPTQVAISPVTGKAWDMAGVEPDVSVLSSEALMTTQGIV >M3_macEug QRLGALVEDAGHLLEAHYALPEVVGQASALLRARLVHGTYRTAVDFESLASQLTSDLQEVSGDHRVHVFHSPGELIPEELSPPQNVVPSPEELTYLIEALFKTEVLPGQLGYLRFDMMAEAETVRAIGPQLIELVWEKLVNTEALVVDLRYNPGSYSTSVPLLCSYFFEAEPRKHLYTIFDRAASQATEVWTLPQVAGERYGSQKDLYILISHTSGSAAEAFAHAMKDLRRATVIGEPTAGGALSVGIYQVSNSPLYASMPTQVAISPVTGKAWDIAGVEPDVS >M3_sacHar QRLGALVEGTGHLVEAHYALPEVVGQASAFLRATLAHGTYRTAVDFESLASQLTSDLQEVSGDHRLHVFHSPGEPVPEESSPPHKGVPSPEELTYLIEALFKTDVLPQLGYLRFDMMAEVETVRAIGPQLVELVWEKLVNTEALVVDLRYNPGSYSTTVPLLCSYFFEAEPRKHLYTVFDRATSQFTEVWTLPQVTGERYGSQKDLYILISHTSGSAAEAFAHIMKDLQRATVIGEPTAGGALSVGIYQVGDSPLYVSMPTQVALSPVTGKAWDMAGVEPDVS >M3_taeGut KNMGVLLEGTGQLLEDHYAIPEVAAKASAMLSTKRAQGGYRSAIDSETLASQLTSDLQEASGDHRLHVFHSHVEPTPEEQLPNVIPSPEELSYIIEALFKIEVLPGNLGYLRFDMMAEAETVKAIGPQLLQMVWNKLVDTDAMIIDMRYNTGGYSTAIPILCSYFFDPEPRKHLYTVFDRSTSRSTEVWTLPQLAGKRYGSLKDIYILTSHMSGSAAEAFTRSMKDLHRATVVGEPTVGGSLSVGIYRVGNSSLYASIPSQVVLSPVTGKVWSVSGVEPHITIQASEAMAAAQHIA >M3_galGal RKMGILLESTGQLLEAHYAIPEVAEKASVMLSTKRVQGGYRSAVDFETLASQLTSDLQEASGDHRLHVFHSHVEPTPEEQLPNMIPSPEELSYIIEALFKIEVLPGNLGYLRFDMMAEAETVKAIGPQLVQMVWNKLVDTDAMIIDMRYNTGGYSTAVPILCSYFFEPEPRQHLYTVFDRSTSRSTEVWTLPKVTGKRYGSLKDIYILTSHMSGSAAEAFTRSMKDLHRATVIGEPTVGGSLSVGIYRVGNSSLYRSIPSQVVLSPVTGKVWSVSGAEPHITIQASEALAAAKHIA >M3_anoCar KGMGSLIERVGQLLEAHYAIPEMARRVSSMLNSKLAQGGYRTAVDFETLASQLTNDLQETSGDHQLHVFHSHVEPSLEEQSPFKTLTPEELNFIIEALFKVDVLPGNVGYLRFDMMAEFESVKTIEPQILHMVWEKLVETSAMIVDMRYNTGSYSTAVPMFCSYFFDAEPQQHLYTIIDRSTSQSTEVWTSSQVSGKRYGSTKDLYILISHASGSAAEAFTRSLKDLHRATVIGEPTVGGSLSASIYNIGSTPLYASIPSQIVLSPVSGKVWSLSGIQPHVTTQSNEALASAQNII >M3_xenTro PSVFALVEGTGHLLEVHYAIPEVAYKVSSVLQNKWSEGGYRSVVDLESLASQLTSEMQENSGDHRLHVFYSDTEPEILEDQPPKIPSAEELNYIIDALFKIEVLQGNVGYLRFDMMADTEIIKAIGPQLVSLVWNKLVETNSLIIDMRYNTGGYSTAIPIFCSYFFDPEPLQHLYTVYDRSTSSGTDIWTLPEVVGERYGSTKDIYILTSHMTGSAAEVFTRSMKELNRATIIGEPTSGVSLSVGMYKVGESNLYVSIPNQVVISSVTGKVWSVSGVEPHVIAQASEAMNVAHHII >M3_xenLae PSIFPLVKGTGHLLEVHYAIPEVAYKVSSVLQNKWSEGGYRSVVDLESLASLLTSEMQENSGDHRLHVFYSDTEPEILEDQPPKIPSPEELNYIIDALFKIEVLPGNVGYLRFDMMADTEIIKAIGPQLVSLVWNKLVETNSLIIDMRYNTGGYSTAIPIFCSYFFDPEPLQHLYTVYDRSTSTGKDIWTLPEVFGERYGSTKDIYILTSHMTGSAAEVFTRSLKDLNRATLIGEPTSGVSLSVGMYKVGDSNLYVTIPNQVVISSVTGKVWSVSGVEPHVIIQANEAMNIAHRII >M3_petMar AKMASLLELAGALVEGYYAMLSDGENATAEILLKYREGWYRSVVDYEALASQLTSDLHEIWGDHRLHAFYSDLQIERMDEDKTPSVPSPEELSVLIDTVFKVDILANNVGYLRFDMMTDAEVLKHVGPQLVEKVWNKISSTRSLVIDVRYNMGGYSTSIPILCSYFFDASPPRHLYTVFDRPSRSSTQVFTVPRVLGQRYGASKDVYILTSHMTGSAGEILTRVMSDLKRATVIGEPTAGGSLSTGTYRIGDSRLYVFIPNQAGVSPSGGRTWSVAGVEPHVQTKASEALQSALRMV >M3x_takRu QGLRSLIGRTGELLEKHYAIQEVAQKVGEVLLSKWAEGLYRSVVDLESLASQLTADLQEASGDHRLHVFRCDVELESLHGVPKIAAVEEAGFVIDALFKSELLPRNVGYLRFDTMADIEAAKGAAPRLVKSVWNKLVDTDSLIIDMRYNAGGSSTAVPLWCSYFVDGEPLQHLYTVYDRTTKTRVEVMTLPEVSGQRYDPGKDVYILTSHMTGSAAEAFVRAMRDLNRVTIVGEPTAGGSLSSATYQIGESVLYASIPNQVVTSAATGKLWSISGVEPDVFAQARDALPVAQRII >M3x_danRr KNIQGLVQEAGDLLEKHYSVPEVAAKVSRLLQSKLTEGLYRSVVDYESLASQLTSDLQETSGDQRLHIFYCETEPETLHDTPKIPSPEEAGFIVEALFKVDVMSGNIGYLRFDMMEDIKVLQAINPEFLKVVWNKLVNTDMLIIDVRYNTGGYSTAIPLLCTYFFDAQPLTHIYTLFDRSTATVTKVTTLPDVLGQKYSSQKDVYILTSHITGSAAEAFTRTMKDLKRATVIGEPTIGGALSSGTYQIGNSILYASIPNQAVLNAVTGKPWSISGVEPHIVAQASDALIVAQKII >M4_homSap AKVPTVLQTAGKLVADNYASAELGAKMATKLSGLQSRYSRVTSEVALAEILGADLQMLSGDPHLKAAHIPENAKDRIPGIVPMQIPSPEVFEELIKFSFHTNVLEDNIGYLRFDMFGDGELLTQVSRLLVEHIWKKIMHTDAMRIIDMFNIGGPTSSIPILCSYFFDEGPPVLLDKIYSRPDDSVSELWTHAQVVGERYGSKKSMVILTSSVTAGTAEEFTYIMKRLGRALVIGEVTSGGCQPPQTYHVDDTNLYLTIPTARSVGASDGSSWEGVGVTPHVVVPAEEALARAKEML >M4_bosTau AKVPTVLQTAGKLVADNYASPELGVKMAAELSGLQSRYARVTSEAALAELLQADLQVLSGDPHLKTAHIPEDAKDRIPGIVPMQIPSPEVFEDLIKFSFHTNVLEGNVGYLRFDMFGDCELLTQVSELLVEHVWKKIVHTDALIVDMRFNIGGPTSSISALCSYFFDEGPPILLDKIYNRPNNSVSELWTLSQLEGERYGSKKSMVILTSTLTAGAAEEFTYIMKRLGRALVIGEVTSGGCQPPQTYHVDDTDLYLTIPTARSVGAADGSSWEGVGVVPDVAVPAEAALTRAQEML >M4_monDom AKVPTILQTAGKLVADNYASLEVGSRVASKLAKLQTQYRQVTSEGELADMLGADLQTLSGDRHLKTAHIPEDAKDRIPGIVPMQLPSPEAFEDLIKFSFHTNVFEGNIGYLRFDMFGDCELLTQVSDLLVEHVWKKVVHTDGMIIDMRFNIGGPTSSISALCSYFFDEGQEVLLDQIYNRPNDSISEIWTQSQVAGERYGSKKSVIILTSSMTAGAAEEFVYVMQRLGRALVIGEVTSGGCQPPQTYHVDDTDLYITIPTARSVGSGDKPSWEGVGVAPHVEVPADQALSKAKEMF >M4_macEug lower case: sacHar fix SKVPTILQTAGKLVADNYASPEVGSRVAAKLARLQTQYRQVTSEGELADMLGADLQTLSGDSHLKTAHIPEDSKDRIPGIVPMQlpspeafedlikfsFHTNVFEGNIGYLRFDMFGDCELLTQVSDLLVEHVWKKVVHTDGMIIDMRFNIGGPTSSISALCSYFFDEGQKVLLDRIYNRPNDSIVEIWTQPHVTGERYGSKKSVIILTSSTTAGAAEEFVYIMQGLGRALVIGEVTSGGCQPPQTYHVDDTDLYITIPTAQSVGSGDRPSWEGIGVTPHVEVPADQALSKAKEMFIHHLQR >M4_sacHar AKVPTILQTAGKLVADNYASPEVGSRVAAKLASLQIQYGKVTSEGELADMLGADLQTLSGDRHLKTAHIPEDAKDRIPGIVPMQLPSPEAFEDLIKFSFHTNVFEGNIGYLRFDMFGDCELLIQVSDLLVEHVWKKVMHTDGMIIDMRFNIGGPTSSISAMCSYFFDEGQGVLLDRIYNRPNDSISEIWTQPQVIGERYGSKKTVVILTSSMTAGAAEEFVYIMQRLGRALVIGEVTSGGCQPPQTYHVDDTDLYITIPTTRSVVSGDKSSWEGVGVVPHMEVPADQALSKAKEMFTHQLQKTK >M4_ornAna SKVPTVLRTAAKLVADNYAFRETGAGVAAQMGGLQARCGRVTSEGALAEVLGAHLRALSGDPHLQMVYIPEDAKDRIPGVVPMQIPSAETFEDLIKFSFHTSVMEGNIGYLRFDMFGDCELLTQVSELMVEHVWKKIVHTDGLIIDMRNIGGPTSSISALCSYFFDEDHPVLLDKIYNRPNDSISEIWTHSHIAGERYGSRKSVVILTSNMTAGAAEEFVSIMKRLGRALVVGEVTGGGCHPPQTYHVDDTHLYITIPTSRSVGSEDGSSWEGVGVTPHLVVPADVALSRAKDLF >M4_taeGut AQVPQILQTVGKLVADNYAFVNTGTVIASNLTKNIHKDNYKRINTEEDLAGKVTAILQALSDDKHLKLLYIPEHAKDSIPGIMPKQIPPPEVFEDLIKFSFHTNVFENNIGYLRFDMFGDSELLTQLSDLMIEHVWKKIFHTDALIIDLRYNIGGSTTPIAILCSYFFDEGHPVLLDRVYDRPSDSVKEIWTQPQLKGERYGSQKGLVILTSAVTAGAAEEFVYIMKRLSRALIIGEQTSGGCHSPQTYQVDETNFYVVIPTSRSVTSADSTSWEGKGVSPHIETPAETALIKAKEML >M4_galGal TQVPQIVQTVGKLVAENYAFVDIGTDIASNLTKSVNKENYKRINSEKELARKLTAILQALSDDEHLKILYIPEHAKDSIPGILPKQIPSPEVFEDLIKFSFHTNVFENNIGYLRFDMFGDCELLTQVSDLLVEHVWKKIVHTDALIIDMRYNIGGYTNSIPILCSYFFDEGHQVLLDKVYDRPSDSVKEIWTQPQLRGERYGSQKGLIILTSAVTAGAAEEFVFIMKRLGRALIIGEQTSGGSHSPQTYQVDDTNFYIIIPTARSVISAESASWEGKGVPPHMETPAVTALIKAKEVL >M4_anoCar TKLPSVLNTIGKLVADNYAFADIGATVAAKFADYAKKGTYRKINSEIELSGKLAADLKALSGDRHLMISHIPERSKGRILGLVPMQQIPPPEILEDLIKFSLHTNVFENNIGYLRFDMFGDCELMSQVSELLVQHVWNKIVNTDALIIDMRYNVGGPACSVPLLCSYFFDEGHPILLDKVYNRPNDTTSNIWTVSKLAGKRYGLNKGLIILTSSVTSGAAEEFAHIMKRLGRAFIIGQKTSGGCHPPQTFHVDGTNLYITTPVSRSVFSVNDSWEGVGVSPHLDVSTDVALIKAKEML >M4_xenLae TKIPTVIQTAAKLVADNYAFADTGANVASKFIALVDKIDYKMIKSEVELAEKINDDLQSLSKDFHLKAVYIPENSKDRIPGVVPMQIPSPELFEELIKFSFHTDVFEKNIGYIRFDMFADSDLLNQVSDLLVEHVWKKVVDQDALIIDMRFNIGGPTSSIPIFCSYFFDEGTPVLLDKIYSRTSNAMTDIWTLPDLVGKTFGSKKPLIILTSSLTEGAAEEFVYIMKRLGRAYVVGEVTSGGCHPPQTYHVDDTHLYLTIPTSRSASAEPGESWEGKGVLPDLEISSETALLKAKEIL >M4_xenTro TKIPSVIQTAGKLVADNYAFADTGADVASKLIALVDKINYKMIKSEVELAEKLNYDLQSLSKDVHLKAVYIPENSKDRIPGVVPMQIPSPEMFEDLIKFSFHTDVFEKNLGYIRFDMFADSDLLNQVSDLLVEHVWKKVVNQDALIIDMRFNIGGPTSSIPTFCSYFFDEGTPVLLDKIYSRTTNAITDVWTLPHLVGNAFGSKKPVIILTSSLTEGAAEEFVYIMKRLGRAYVIGEVTSGGCHPPQTYHVDDTHLYLTIPTSRSASAKPGESWEGKGVLPDLEITSETALMKAKEIL >M4_tetNig AQIPAIIEGTAALVANNYAFEATGADVAKELRELQANGQYSSVVSKESLEAALSADLQRLSGDKSLKTTPNTPVLPPMDYTPEMYIELIKVSFHTDVFENNIGYLRFDMFGDFEEVKAIAQIIVEHVWNKVVNTDALILDLRNNVGGPTTAIAGFCSYFFDADKQNRVGQAVRQASGTTTELLTLSELTGVRYGSKKSLIILTSGATAGAAEEFVYIMKKLGRAMIVGETTAGASHPPQTFRVGETDVFLLIPTVHSDTGAGPAWEGAGIAPHIPASAEAALGTARAIL >M4_takRub AQIPAIIEGAATLIAKNYAFEATGADVATKLRELLAKGQYNSVVSSESLEVALSADLQRLSGDKSLKATQNAPVLPPMDYSPEMYIELIKVSFHTDVFENNIGYLRFDMFGDFEEVKAIAQIIVEHVWNKVVNTDALILDLRNNVGGPTTAIAGFCSYFFDADKLIVLDKLHDRPSGTTTELLTLPELTGVRYGSKKSLIILTSGATAGAAEEFVYIMKKLGRAMIVGETTAGASHPPQVFSVGEIGIFLSIPTVHSDTAAGPAWEGTGITPHIPVSAEAALGTAKGIL >M4_gasAcu NRVPAIIEGSATLIADNYAFEDIGAAVAEKLKGLLANGEYSKVVSKDSLEMKLSADLRTLSGDKSLKTTSNVPALPPMNYSPEMYIELIKVSFHTDVFEDNIGYLRFDMFGDFEEVKAIAQIIVEHVWNKVVNTDAMIVDLRNNIGGPTTAIAGFCSYFFDSDKQIVLDRLYDRPSGTTTELRTLPELTGTRYGSKKSLVMLTSRATAGAAEEFVYIMKKLGRAMIVGETTAGTSHPPKTFRVGETDIFLSIPTVHSDTAAGPAWEGAGVAPHIPVPADAALETAKGIF >M4_oryLat LQVPAIIEESATLVANNYAFESTAADVAEKLKGHLANGDYNMVVSKESLEAKLSADLQSLSGDKSLTVSSNTGAPPPMEYTPEMYIELIKISFHTDVFENNIGYLRFDMFGDFEEVKAIAQVIVEHVWNKVLHTDAMIIDLRNNVGGPTTAIAGFCSYFFDGDKQILLDKLYDRSTGTTTDLLTLGELTGERYGSKKSLIILASRATAGAAEEFVYIMKRLGRAMIVGETTAGASHPPKVFQVGESDIFLSIPTVHSDTSAGPGWEGAGVAPHIPVAAGAALETAKAIL >M4_danRer AEIPALAQAAATLIADNYAFPSIGEHVAEKLEAVVAGGEYNLISTKEDLEERLSEDLLKLSEDKCLKTTSNIPALPPMNPTPEMFIALIKSSFQTDVFENNIGYLRFDMFGDFEHVATIAQIIVEHVWNKVVDTDALIIDLRNNIGGHASSIAGFCSYFFDADKQIVLDHIYDRPSNTTRDLQTLEQLTGRRYGSKKSVVILTSGVTAGAAEEFVFIMKRLGRAMIIGETTHGGCQPPETFAVGESDIFLSIPISHSTAQGPSWEGAGIAPHIPVPAGAALDTAKGML >M4_petMar ADAPSILRTVGKLVADGYSRAEAALGVPSKLAALLEAGEYGALRSEEELAFKLTVHLQLITGDRHLKAVCVPEHATDRMPGIVPMQMPPTESFEDLIKFSFITDVLEGNIGYLRFDLFSDLEALEHVAHLLVEHVWKKICDTEILIIDLRYNMGGYSTSIPILCSYFFDASPPRHLYTVFDRPSRSSTQVFTVPRVLGQRYGASKDVYILTSHMTGSAGEILTRVMSDLKRATVIGEPTAGGSLSTGTYRIGDSRLYVFIPNQAGVSPSGGRTWSVAGVEPHVQTKASEALQSALRMV >Mn_braFlo RALNDQSL SKAIILD LNEL insertions omitted VDWLDVVMGIGDVMADHYLDQDLLQRWNRTLVHRFQSWSQDDMSDSLRMEEGLTSELRNITGDETIKVWDFGVYENTTQEPVPREFYNFSTFVDNFKKNINVTMLEGNVGYVSIRSMSHIVDIILPDPEMTEFFLSKMAASKAIILDLRYNLGGDREGVVHWASFFFNATPSVPLSDVYYRDGVNQYWTLLEVPGGIRFPDMPLYLLTSNRTSREAEEFAYAMQVVNRTTIIGETTAGEEFTGMWFPIDQTDVHLLTRTNVVRNPITQDSWSGKGVTPDIIVPSEKALTVALRKI
RBP3 (IRBP) sequences from human to amphioxus
>RPB3_homSap human 0 MMREWVLLMSVLLCGLAGPTHLFQPSLVLDMAKVLLDNYCFPENLLGMQEAIQQAIKSHEILSISDPQTLASVLTAGVQSSLNDPRLVISYEPSTPEPPPQV PALTSLSEEELLAWLQRGLRHEVLEGNVGYLRVDSVPGQEVLSMMGEFLVAHVWGNLMGTSALVLDLRHCTGGQVSGIPYIISYLHPGNTILHVDTIYNRPSNTTTEIWTLPQVLG ERYGADKDVVVLTSSQTRGVAEDIAHILKQMRRAIVVGERTGGGALDLRKLRIGESDFFFTVPVSRSLGPLGGGSQTWEGSGVLPCVGTPAEQALEKALAILTLRSALPGVVHCLQ EVLKDYYTLVDRVPTLLQHLASMDFSTVVSEEDLVTKLNAGLQAASEDPRLLVRAIGPTETPSWPAPDAAAEDSPGVAPELPEDEAIRQALVDSVFQVSVLPGNVGYLRFDSFADA SVLGVLAPYVLRQVWEPLQDTEHLIMDLRHNPGGPSSAVPLLLSYFQGPEAGPVHLFTTYDRRTNITQEHFSHMELPGPRYSTQRGVYLLTSHRTATAAEEFAFLMQSLGWATLVG EITAGNLLHTRTVPLLDTPEGSLALTVPVLTFIDNHGEAWLGGGVVPDAIVLAEEALDKAQEVLEFHQSLGALVEGTGHLLEAHYARPEVVGQTSALLRAKLAQGAYRTAVDLESL ASQLTADLQEVSGDHRLLVFHSPGELVVEEAPPPPPAVPSPEELTYLIEALFKTEVLPGQLGYLRFDAMAELETVKAVGPQLVRLVWQQLVDTAALVIDLRYNPGSYSTAIPLLCS YFFEAEPRQHLYSVFDRATSKVTEVWTLPQVAGQRYGSHKDLYILMSHTSGSAAEAFAHTMQDLQRATVIGEPTAGGALSVGIYQVGSSPLYASMPTQMAMSATTGKAWDLAGVEP DITVPMSEALSIAQDIVALRAKVPTVLQTAGKLVADNYASAELGAKMATKLSGLQSRYSRVTSEVALAEILGADLQMLSGDPHLKAAHIPENAKDRIPGIVPMQ 0 0 IPSPEVFEELIKFSFHTNVLEDNIGYLRFDMFGDGELLTQVSRLLVEHIWKKIMHTDAMIIDMR 2 1 FNIGGPTSSIPILCSYFFDEGPPVLLDKIYSRPDDSVSELWTHAQVV 1 2 GERYGSKKSMVILTSSVTAGTAEEFTYIMKRLGRALVIGEVTSGGCQPPQTYHVDDTNLYLTIPTARSVGASDGSSWEGVGVTPHVVVPAEEALARAKEMLQHNQLRVKRSPGLQDHL* 0 >RBP3_bosTau cow run-on terminal exon 0 MVRKWALLLPMLLCGLTGPAHLFQPSLVLEMAQVLLDNYCFPENLMGMQGAIEQAIKSQEILSISDPQTLAHVLTAGVQSSLNDPRLVISYEPSTLEAPP RAPAVTNLTLEEIIAGLQDGLRHEILEGNVGYLRVDDIPGQEVMSKLRSFLVANVWRKLVNTSALVLDLRHCTGGHVSGIPYVISYLHPGSTVSHVDTVY DRPSNTTTEIWTLPEALGEKYSADKDVVVLTSSRTGGVAEDIAYILKQMRRAIVVGERTVGGALNLQKLRVGQSDFFLTVPVSRSLGPLGEGSQTWEGSG VLPCVGTPAEQALEKALAVLMLRRALPGVIQRLQEALREYYTLVDRVPALLSHLAAMDLSSVVSEDDLVTKLNAGLQAVSEDPRLQVQVVRPKEASSGPE EEAEEPPEAVPEVPEDEAVRRALVDSVFQVSVLPGNVGYLRFDSFADASVLEVLGPYILHQVWEPLQDTEHLIMDLRQNPGGPSSAVPLLLSYFQSPDAS PVRLFSTYDRRTNITREHFSQTELLGRPYGTQRGVYLLTSHRTATAAEELAFLMQSLGWATLVGEITAGSLLHTHTVSLLETPEGGLALTVPVLTFIDNH GECWLGGGVVPDAIVLAEEALDRAQEVLEFHRSLGELVEGTGRLLEAHYARPEVVGQMGALLRAKLAQGAYRTAVDLESLASQLTADLQEMSGDHRLLVF HSPGEMVAEEAPPPPPVVPSPEELSYLIEALFKTEVLPGQLGYLRFDAMAELETVKAVGPQLVQLVWQKLVDTAALVVDLRYNPGSYSTAVPLLCSYFFE AEPRRHLYSVFDRATSRVTEVWTLPHVTGQRYGSHKDLYVLVSHTSGSAAEAFAHTMQDLQRATIIGEPTAGGALSVGIYQVGSSALYASMPTQMAMSAS TGEAWDLAGVEPDITVPMSVALSTARDIVTLRAKVPTVLQTAGKLVADNYASPELGVKMAAELSGLQSRYARVTSEAALAELLQADLQVLSGDPHLKTAH IPEDAKDRIPGIVPMQ 0 0 IPSPEVFEDLIKFSFHTNVLEGNVGYLRFDMFGDCELLTQVSELLVEHVWKKIVHTDALIVDMR 2 1 FNIGGPTSSISALCSYFFDEGPPILLDKIYNRPNDSVSELWTLSQLE 1 2 GERYGSKKSMVILTSTLTAGAAEEFTYIMKRLGRALVIGEVTSGGCQPPQTYHVDDTDLYLTIPTARSVGAADGSSWEGVGVVPDVAVPAEAALTRAQEMLQHTPLRARRSPRLHGRRKGHHRQSQGRAGSLGRNQGVgRPEVLTEAPSGQKRGLLQCG* 0 >RBP3_monDom opossum 0 MTSQCLLLFSALLFSLAHAEQIFQPSLVRDMAKILLDNYCFPENLMGMQEVIEQAIKSGEILDISDPQMLASVLTAGVQGALNDPRLVISFEPSIPETPQ HVPKLANVTQEELLILLQQMIKYQVLEGNVGYLRVDYIPGQEVVEKVGEFLVNNIWKKLMGTSSLVLDLQHSSGGEISGIPFVISYLHQGDILLHVDTVY DRPSNTTTEIWTLPQVLGERYGGEKDMVVLTSHRTVGVAEDIAYILKKLRRAIVVGEQTLGGALDLRKLRIGQSDFFITVPVSRSLSPLGGGSQTWEGSG VLPCVGIPAEQALGKALAILTLRRARPGAIQRLMEVLQNYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAGLQAVSEDPRLLVRVLRPEEATMGSEASEEEDATPAANS LPEDESQRQALVDSVFQVSVLPGNVGYLRFDEFADSSVLGTLAPYVIRQVWEPLQDTNHLIMDLRYNPGGPSSAVPLLLSYFQDPAAGP IRLFTTYDRQTNQTQEHLSRAELLGKPYGAQRGVYLLTSHHTATAAEEFAFLMQSLGRATLVGEITAGSLMHTRTFPLLQPPNGNLVLTVPILTFIDNNG ECWLGGGVVPDAIVLAEEALDKAKEVLEFHQRLGALVEGTGHLLEAHYALPEVVGQASALLKAKLEHGTYRTAVDFESLASQLTSDLQEVSGDHRLHVFH SPGEPVSEELTPPQKGVPSPEELTYLIEALFKTEVLPGQLGYLRFDMMAEAETVRAIAPQLVELVWEKLVHTEALVVDLRYNPGGYSTAVPLLCSYFFEA EPRRHLYTIFDRAASQLTEVWTLPQVAGERYGSQKDLYILISHTSGSAAEAFVHTMKDQHRATVIGEPTGGGALSVGIYQVENSPLYASMPTQVAISPVT GKAWDMAGVEPDVSVLSSEALMTTQGIVALRAKVPTILQTAGKLVADNYASLEVGSRVASKLAKLQTQYRQVTSEGELADMLGADLQTLSGDRHLKTAHI PEDAKDRIPGIVPMQ 0 0 LPSPEAFEDLIKFSFHTNVFEGNIGYLRFDMFGDCELLTQVSDLLVEHVWKKVVHTDGMIIDMR 2 1 FNIGGPTSSISALCSYFFDEGQEVLLDQIYNRPNDSISEIWTQSQVA 1 2 GERYGSKKSVIILTSSMTAGAAEEFVYVMQRLGRALVIGEVTSGGCQPPQTYHVDDTDLYITIPTARSVGSGDKPSWEGVGVAPHVEVPADQALSKAKEMFNHHLQRAK* 0 >RBP3_sacHar Sarcophilus harrisii 0 MTSQCLLLFSALLFSLAHGEEIFQPTLVLDMAKILLDNYCFPENLMGMQEAIEQAIKSGEILDISDPQMLANVLTAGVQGSLNDPRLVISYEPSTSEAPQ HDPKFANATQEELLALFQKIIKYQVLEDNVGYLRVDYIPGRDMIEEVGEFLVNDIWKKVMETSSLVLDLQHSR GGEVSGIPFVISYLHQGDILLHVDTIYDRPSNTTTEIWTLPQVLGERYSGEKDIVVLTSHHTVGVAEDIAYILKKMRRAIVVGEQTQGGSLDLRKLRIGQSDFFITVPVARSLSPLGGGSQTWEGSG VVPCVGIPAEQALEKALAILTLRRARPGAIQRLMEILQKYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAMLQAVSEDPRLLVRVLRPEEATVEAEPGEESATPASVSLPESDAERQALI DSVFQVSVLPGNVGYLRFDEFADNSVLGTLAPYVLRQVWEPLQDTDHLIMDLRYNPGGPSSAVPLLLSYFQDPSAGPVRLFATYDRQTNQ TQEYRSRAELLGKPYGAERGVYLLTSYHTATAAEEFAFLMQSLGRATLVGEITAGSLMHTRTFPLLQPPNGSLVLTVPTLTFIDNHGECWLGGGVVPDAIVLAEEALDKAKEVLEFHQRL GALVEGTGHLVEAHYALPEVVGQASAFLRATLAHGTYRTAVDFESLASQLTSDLQEVSGDHRLHVFHSPGEPVPEESSPPHKGVPSPEELTYLIEALFKTDVLP QLGYLRFDMMAEVETVRAIGPQLVELVWEKLVNTEALVVDLRYNPGSYSTTVPLLCSYFFEAEPRKHLYTVFDRATSQFTEVWTLPQVTGERYGSQKDLYILISHTSGSA AEAFAHIMKDLQRATVIGEPTAGGALSVGIYQVGDSPLYVSMPTQVALSPVTGKAWDMAGVEPDVSVLANEALITAQGIVALRAKVPTILQTAGKLVADNYA SPEVGSRVAAKLASLQIQYGKVTSEGELADMLGADLQTLSGDRHLKTAHIPEDAKDRIPGIVPMQ 0 0 LPSPEAFEDLIKFSFHTNVFEGNIGYLRFDMFGDCELLIQVSDLLVEHVWKKVMHTDGMIIDMR 2 1 FNIGGPTSSISAMCSYFFDEGQGVLLDRIYNRPNDSISEIWTQPQVI 1 2 GERYGSKKTVVILTSSMTAGAAEEFVYIMQRLGRALVIGEVTSGGCQPPQTYHVDDTDLYITIPTTRSVVSGDKSSWEGVGVVPHMEVPADQALSKAKEMFTHQLQKTK* 0 >RBP3_ornAna platypus genome rife with frameshifts, dels, misassembly frag 0 MGVCLPLLLVAQFSLTGHVEPVSQPSMVLDVAKILLDNYCYPENLMGMQEAIEEAIQRGEILDIADPKRLASVLTAGVQGSLNDPRLVISYEPAPVAVSQ QPPEPASLPAEQPLERLRPAVGSEVLEGNVGYLRVDRLPGREEIERVGAVLGRDIWEKLLGTSALVLDLRHSTGGHVSGIPFFISYFYPEGPALHVDTVY DRPSNATRQLWTLPRVLGARYAADKDVVVLTSRLTAGVAEDVAYILQQMRRAIVVGERTAGGPLVFRKLRVGLSDFFITVPVACSLGPLGGGGRSWEGSG VLPCVAVPADRALDEALDILALRGAVPGAVAHLADLLRDYYALVDRVPALLRHLAALDLSSVLSEEDLTSRLNAGLQAASEDPRLLVRRLEPEEAERGPP RKEEEQKEEEEEDQPSPGASILPGDGSSLFRVSVLPGNVGYLCFDEFPEASALERLGPLLGRRVWEPLEATDHLMVDLRNNPGGPSSAVPLLLSYF QDPAAGPIRLFTTYNRPADVTREYASRAGALEKPYGARRGVYLLTSHRTATAAEEFAYLMQALGRATLVGEITAGRLLHSRTFPLLRPPWEGLVLTVPFL TLFDPHGEGWLGGGVVPDAIVLAEEALEKAGEVLAFHQTLEALVETTGHLLEAHYCFPAGARRAGAQPWPVAGVEPDVMAQAAEALAVAQGIAALRSKVP TVLRTAAKLVADNYAFRETGAGVAAQMGGLQARCGRVTSEGALAEVLGAHLRALSGDPHLQMVYIPEDAKDRIPGVVPMQ 0 0 IPSAETFEDLIKFSFHTSVMEGNIGYLRFDMFGDCELLTQVSELMVEHVWKKIVHTDGLIIDMR 2 1 NIGGPTSSISALCSYFFDEDHPVLLDKIYNRPNDSISEIWTHSHIA 1 2 GERYGSRKSVVILTSNMTAGAAEEFVSIMKRLGRALVVGEVTGGGCHPPQTYHVDDTHLYITIPTSRSVGSEDGSSWEGVGVTPHLVVPADVALSRAKDLFRAHLEHRD* 0 >RBP3_taeGut Taeniopygia guttata 0 MIRTHFLLLSALIMCSIPAEEIFQPTLVLDMAKVLLDNYCYPENLVGMQEAIEQAIKSGEILDISDPKMLANVLTAGVQGALNDPRLVISYEPLPHSGPK QEAEGSPTREQLLSLIEHVIMYDKLEGNVGYLRIDYIIGEEVVQKVGAFLVDKVWKTLIETSALVIDLRHSTGGQISGLPFIISYLHEQDKILHVETVYN RPSNTTTEIWTLPKVLGERYSKDKDVIVLISHHTTGVAEDVAYILKHMNRAITVGEKTAGGSLDIQKLRIGPSNFYMMVPVSRSVSPLSGGGQSWEVSGV MPCVATEAEQALQKSLDILAVRRAVPGTISHLKNILKDYYSLVERVPALLRRLTTSDFSSVQSSEDLATKLNTELQALSDDPRLMVRVMMPGEAADSPAE KPVGMAADLPDNEQLLHALVDTVFKVSVLPGNVGYMRFDEFADASVLVKLGPYLVHKVWEPLQNTENLIMDLRYNLGGPSSSAVPVLLSYFQDPAAGPVH LFTTYDRRTNHTQEHNSQAELLGQSYGAKRGVYLLTSHHTATAAEEFAYLMQSLGRATLIGEITAGSLSHTRTFPLLQPGPGITRGLTITVPVITFIDNH GESWMGGGVVPDAIVLAEDALEKAEEVLAFHKNMGVLLEGTGQLLEDHYAIPEVAAKASAMLSTKRAQGGYRSAIDSETLASQLTSDLQEASGDHRLHVF HSHVEPTPEEQLPNVIPSPEELSYIIEALFKIEVLPGNLGYLRFDMMAEAETVKAIGPQLLQMVWNKLVDTDAMIIDMRYNTGGYSTAIPILCSYFFDPE PRKHLYTVFDRSTSRSTEVWTLPQLAGKRYGSLKDIYILTSHMSGSAAEAFTRSMKDLHRATVVGEPTVGGSLSVGIYRVGNSSLYASIPSQVVLSPVTG KVWSVSGVEPHITIQASEAMAAAQHIANLRAQVPQILQTVGKLVADNYAFVNTGTVIASNLTKNIHKDNYKRINTEEDLAGKVTAILQALSDDKHLKLLY IPEHAKDSIPGIMPK 0 0 QIPPPEVFEDLIKFSFHTNVFENNIGYLRFDMFGDSELLTQLSDLMIEHVWKKIFHTDALIIDLR 2 1 YNIGGSTTPIAILCSYFFDEGHPVLLDRVYDRPSDSVKEIWTQPQLK 1 2 GERYGSQKGLVILTSAVTAGAAEEFVYIMKRLSRALIIGEQTSGGCHSPQTYQVDETNFYVVIPTSRSVTSADSTSWEGKGVSPHIETPAETALIKAKEMLNAHLHSSR* 0 >RBP3_galGal Gallus gallus 1236 aa N-terminal 21 aa signal peptide 5 glyc (3 unique) two W per repeat 0 MRTYFFLFSVLIVCSISAEEIFQPTLVLDMAKVLLDNYCYPENLVGMQEAIEQAIKSGEILDISDPKMLANVLTAGVQGALNDPRLVISYEPSLHAAPKQ EAETYPTREQLLSLIEHVVIYDKLEGNVGYLRIDYIIGQEVVEKVGAFLVDKVWKTLINTSALVIDLRYSTGGQISGIPFIISYLHEADKMLHVETVYNR PSNTTTEIWTLPKVLGERYSKDKDVIVLISHHTTGVAEDVAYILKHMNRAITLGEKTAGGSLDIQKLRIGPSNFYMMVPVSRSVSPLSGGGQSWEVSGVM PCVASEAEQALKKSLDILAVRRAVPGTLSRLTDILKDYYSLVERVPVLLRHLTTSDFSSVQSAEDLATKLNTEMQTLSEDPRLLVRTMMPGEAAAPPAEM PIAMAANLPDNEQLLHALVDTVFKVSVLPGNVGYMRFDEFADASVLVKLGPYIVKKVWEPLQNTENLIMDLRYNPGGPSSSAVPMLISYFQDPTAGPVHL FTTYDRRTNHTQEHNSQAELLAQPYGAQRGIYVLTSRHTATAAEEFAYLMQSLGRATLIGEITAGSLSHTCTFPLVQPEQGITRGLTITVPVITFIDNHG ESWMGGGVVPDAIVLAEDALEKAEEVLTFHRKMGILLESTGQLLEAHYAIPEVAEKASVMLSTKRVQGGYRSAVDFETLASQLTSDLQEASGDHRLHVFH SHVEPTPEEQLPNMIPSPEELSYIIEALFKIEVLPGNLGYLRFDMMAEAETVKAIGPQLVQMVWNKLVDTDAMIIDMRYNTGGYSTAVPILCSYFFEPEP RQHLYTVFDRSTSRSTEVWTLPKVTGKRYGSLKDIYILTSHMSGSAAEAFTRSMKDLHRATVIGEPTVGGSLSVGIYRVGNSSLYRSIPSQVVLSPVTGK VWSVSGAEPHITIQASEALAAAKHIASLRTQVPQIVQTVGKLVAENYAFVDIGTDIASNLTKSVNKENYKRINSEKELARKLTAILQALSDDEHLKILYI PEHAKDSIPGILPK 0 0 QIPSPEVFEDLIKFSFHTNVFENNIGYLRFDMFGDCELLTQVSDLLVEHVWKKIVHTDALIIDMR 2 1 YNIGGYTNSIPILCSYFFDEGHQVLLDKVYDRPSDSVKEIWTQPQLR 1 2 GERYGSQKGLIILTSAVTAGAAEEFVFIMKRLGRALIIGEQTSGGSHSPQTYQVDDTNFYIIIPTARSVISAESASWEGKGVPPHMETPAVTALIKAKEVLSAHLHSSR* 0 >RBP3_anoCar lizard 0 MLRKCLWLSIVLVCCSSYADSVLQSTLVLDMAKLLLDNYCLPENLVGMREAIEQAIKNGEVLDISDPKLLATVLTAGVQGALNDPRLVISYEPTAPAAPK QRMETSLTPEQLLSLIQHTVKYEVLDDNVGYLRIDYIMGQDIVQKIGSFLVEKVWKTLLGTSALILDLRYTTGGDVSGIPFIISYLYNGDKVLHVDTVYN RPSNTTVEILTLPKVLGVRYSKDKDVILLISKYTTGVAENVAYILKHMHRTIIVGEKSAGGSLDTQKMQIGNSQFYMTVPLSCSVSPLSGSGQSWEISGV TPCVVISAEQALDKALAILSLRKAIPNSMSYLVDIIKNNYSMLEQVPVLLQHLSTFDYSSVLSVKDLASKLNAELQTISEDPRLFLRVPASDEAVTSQTD EKVAMASDLPNNEQLMKALVMTVFKVSVLPGNVGYMRFDEFGDATVLVKLGPYLLQHVWEPLQATDYLIIDLRYNIGGPSSSAVPVLLSYFQDPSAGPVH FFTTYNRLTNQTQAYSSSAEMVGKPYGARRGVYLLTSHNTATAAEEFAYLMQTLGRATLVGEITAGSLSHTHTFCILELGGGCGLLINVPVITLIDNHGE YWLGGGVVPDSIVLADEALEKAREVLEFHKGMGSLIERVGQLLEAHYAIPEMARRVSSMLNSKLAQGGYRTAVDFETLASQLTNDLQETSGDHQLHVFHS HVEPSLEEQSPFKTLTPEELNFIIEALFKVDVLPGNVGYLRFDMMAEFESVKTIEPQILHMVWEKLVETSAMIVDMRYNTGSYSTAVPMFCSYFFDAEPQ QHLYTIIDRSTSQSTEVWTSSQVSGKRYGSTKDLYILISHASGSAAEAFTRSLKDLHRATVIGEPTVGGSLSASIYNIGSTPLYASIPSQIVLSPVSGKV WSLSGIQPHVTTQSNEALASAQNIILFRTKLPSVLNTIGKLVADNYAFADIGATVAAKFADYAKKGTYRKINSEIELSGKLAADLKALSGDRHLMISHIP ERSKGRILGLVPM 0 0 QIPPPEILEDLIKFSLHTNVFENNIGYLRFDMFGDCELMSQVSELLVQHVWNKIVNTDALIIDMR 2 1 YNVGGPACSVPLLCSYFFDEGHPILLDKVYNRPNDTTSNIWTVSKLA 1 2 GKRYGLNKGLIILTSSVTSGAAEEFAHIMKRLGRAFIIGQKTSGGCHPPQTFHVDGTNLYITTPVSRSVFSVNDSWEGVGVSPHLDVSTDVALIKAKEMLKAHLH* 0 >RBP3_xenLae Xenopus laevis 0 MPPLFQALTTALFFCGIASNPLFQPSLVMDMAKVLLDNYCFPENLVGMQETIEQAVKGGEILHISDPDTLANVFTSGVQGYLNDPRLVVSYEPNYSGPQT EQSLELTPEQLKFLINHSVKYDILPGNIGYLRIDFIIGQDVVQKVGPHLVNNIWKKLMPTSALILDLRYSTQGEVSGIPFVVSYLCDSEIHIDSIYNRPS NTTTDLWTLPELMGERYGKVKDVVVLTSKYTKGVAEDASYILKHMNRAIVVGEKTAGGSLDTQKIKIGQSDFYITVPVSRSLSPLTGQSWEVSGVSPCVV VNAKDALDKAQAILAVRSSVTHVLHQLCDILANNYAFSERIPTLLQHLPNLDYSTVISEEDIAAKLNYELQSLTEDPRLVLKSKTDTLVMPGDSIQAENI PEDEAMLQALVNTVFKVSILPGNIGYLRFDQFADVSVIAKLAPFIVNTVWEPITITENLIIDLRYNVGGSSTAVPLLLSYFLDPETKIHLFTLHNRQQNS TDEVYSHPKVLGKPYGSKKGVYVLTSHQTATAAEEFAYLMQSLSRATIIGEITSGNLMHSKVFPFDGTQLSVTVPIINFIDSNGDYWLGGGVVPDAIVLA DEALDKAKEIIAFHPSIFPLVKGTGHLLEVHYAIPEVAYKVSSVLQNKWSEGGYRSVVDLESLASLLTSEMQENSGDHRLHVFYSDTEPEILEDQPPKIP SPEELNYIIDALFKIEVLPGNVGYLRFDMMADTEIIKAIGPQLVSLVWNKLVETNSLIIDMRYNTGGYSTAIPIFCSYFFDPEPLQHLYTVYDRSTSTGK DIWTLPEVFGERYGSTKDIYILTSHMTGSAAEVFTRSLKDLNRATLIGEPTSGVSLSVGMYKVGDSNLYVTIPNQVVISSVTGKVWSVSGVEPHVIIQAN EAMNIAHRIIKLRTKIPTVIQTAAKLVADNYAFADTGANVASKFIALVDKIDYKMIKSEVELAEKINDDLQSLSKDFHLKAVYIPENSKDRIPGVVPM 0 0 QIPSPELFEELIKFSFHTDVFEKNIGYIRFDMFADSDLLNQVSDLLVEHVWKKVVDQDALIIDMR 2 1 FNIGGPTSSIPIFCSYFFDEGTPVLLDKIYSRTSNAMTDIWTLPDLV 1 2 GKTFGSKKPLIILTSSLTEGAAEEFVYIMKRLGRAYVVGEVTSGGCHPPQTYHVDDTHLYLTIPTSRSASAEPGESWEGKGVLPDLEISSETALLKAKEILESQLEGRR* 0 >RBP3_xenTro Xenopus tropicalis 89% xenLae 0 MSPLFKALTTVLFFCIVASNPVFQPSLVMDMAKVLLDNYCFPENLVGMQETIEQAMKSGEILHISDPETLANVFTSGVQGFLNDPRLVVSYEPNYSGPRK EQSPEPTLEQLKFLLDHSVTYDLLPGNIGYLRIDFIIGQDVVQKVGPLLVNNIWKKLMPSSALILDLRYSTQGKVSGIPFVVSYLTDPQIHIDSIYNRPS NTTTDLWTLSELMGERYGKDKDVVVLTSKYTEGIAEGAAYILKHMSRAIVVGEKTAGGSLDIQKIKIGQSEFYITVPVSRSISPLTGQSWEVAGVFPCVV VNANNALNKAQGILAVRSSITHILLQLSEILVNNYAFSERIPTLLQHLPNLDYSSVISEEDITAKLNYELQSLTEDPRLVLKSKTDSLVMPEDSTQVENL PDDEATLQALVNTVFKVSILPGNIGYLRFDEFADVSVLAKLGPYIVNTVWDPITVTENLIIDLRYNIGGSSTSIPLLLSYFQEPENRIHLFTIYNRQQNS TNEVYSLPKVLGKPYGSKKGVYVLTSHETATAAEEFAYLMQSLSRATIIGEITSGNLMHSKAFPLDGTRLSVTVPIMNFIDNNGDYWLGGGVVPDAIVLA DEALDKAKEIIAFHPSVFALVEGTGHLLEVHYAIPEVAYKVSSVLQNKWSEGGYRSVVDLESLASQLTSEMQENSGDHRLHVFYSDTEPEILEDQPPKIP SAEELNYIIDALFKIEVLQGNVGYLRFDMMADTEIIKAIGPQLVSLVWNKLVETNSLIIDMRYNTGGYSTAIPIFCSYFFDPEPLQHLYTVYDRSTSSGT DIWTLPEVVGERYGSTKDIYILTSHMTGSAAEVFTRSMKELNRATIIGEPTSGVSLSVGMYKVGESNLYVSIPNQVVISSVTGKVWSVSGVEPHVIAQAS EAMNVAHHIIKLRTKIPSVIQTAGKLVADNYAFADTGADVASKLIALVDKINYKMIKSEVELAEKLNYDLQSLSKDVHLKAVYIPENSKDRIPGVVPMQ 0 0 IPSPEMFEDLIKFSFHTDVFEKNLGYIRFDMFADSDLLNQVSDLLVEHVWKKVVNQDALIIDMr 2 1 FNIGGPTSSIPTFCSYFFDEGTPVLLDKIYSRTTNAITDVWTLPHLV 1 2 GNAFGSKKPVIILTSSLTEGAAEEFVYIMKRLGRAYVIGEVTSGGCHPPQTYHVDDTHLYLTIPTSRSASAKPGESWEGKGVLPDLEITSETALMKAKEILVSQLEGR* 0 >RBP3_tetNig frameshifts in genome two domains: 23-324,326-612 no upstream dup 0 MAKALFTVASLLLLANGFFVGAAFPPSLIADMAKIVLDNYCSPEKLAGMKEAIKAAGTNTEVLNIPDGESLARVLSAGVQGTVSDPRLMVSFQPNYVPAG PHKMPPLPPEHLVAVLQTSVKLDILEGNTGYLRIDHILGEEVADKVGPALIDLIWNKILPTSALIFDLRYTSSGDISGIPYIVSYFTQAEPVVHIDSVYD RPSNTTTKLLSLPNLLGQRYGVSKPLIVLTSKNTKGIAEDVAYCLKNLKRATIVGEKTAGGSLKLDTFKVGDTDFYITVPTAKSINPITGSSWEIRGVTP HVEVNAEDALATAIKIVNLRAQIPAIIEGTAALVANNYAFEATGADVAKELRELQANGQYSSVVSKESLEAALSADLQRLSGDKSLKTTPNTPVLPPM 0 0 DYTPEMYIELIKVSFHTDVFENNIGYLRFDMFGDFEEVKAIAQIIVEHVWNKVVNTDALILDLr 2 1 NNVGGPTTAIAGFCSYFFDADKQNRVGQAVRQASGTTTELLTLSELT 1 2 GVRYGSKKSLIILTSGATAGAAEEFVYIMKKLGRAMIVGETTAGASHPPQTFRVGETDVFLLIPTVHSDTGAGPAWEGAGIAPHIPASAEAALGTARAILNKHFAGQK* 0 >RBP3_takRub fugu two domains: 23-324,326-612 plus upstream dup 0 MAKALFLVASLLLLANDVLVRAAFPPSLITDMAKIVLDNYCSPEKLAGMKEAIEAAGTNTEVLNIPDGESLARVLSAGVQGTVSDSRLMVSYQPDYVPAV PPKMPPLPPEHLVAVLQTSIKLDLLEGNTGYLRIDHIIGEDVAEKVGPSLIDLIWNKILPTSALIFDLRYTSSGEISGIPYIVSYFTQAEPVVHIDSVYD RPSNTTTKLFSLSNLLGERYGITKPLIILTSKNTKGIAEDVAYCLKNLKRATIVGERTAGGSVKLDNFKVGSTDFYITVPTAKSINPVTGSSWEITGVKP DVEVNAEDALATAIKIVSLRAQIPAIIEGAATLIAKNYAFEATGADVATKLRELLAKGQYNSVVSSESLEVALSADLQRLSGDKSLKATQNAPVLPPM 0 0 DYSPEMYIELIKVSFHTDVFENNIGYLRFDMFGDFEEVKAIAQIIVEHVWNKVVNTDALILDLR 2 1 NNVGGPTTAIAGFCSYFFDADKLIVLDKLHDRPSGTTTELLTLPELT 1 2 GVRYGSKKSLIILTSGATAGAAEEFVYIMKKLGRAMIVGETTAGASHPPQVFSVGEIGIFLSIPTVHSDTAAGPAWEGTGITPHIPVSAEAALGTAKGILNKHFGGQK* 0 >RBP3_gasAcu sticklebck two domains: 27-317,323-612 no upstream dup 0 MAKLIFLVAPLLVLGNIAFIHAGFAPNVIIDMAKIVIDNYCSPEKLAGMKEAIEAAGSNTEVLSIPDAETLANVLSAGVQTTVSDPRLMISYEPNYVPVV PPKMPPLPPDQVIAVLQTSIKLDILEGNIGYLRIDHILGEDVAEKVGPLLLDLVWNKILPTSALIFDLRYTSSGDISGIPYIVSYFTEAGTPIHIDSIYD RPSNTTTKLFSMSTLLGERYSTSKPLIILTSKNTKGIAEDVAYCLQNLKRATIVGEKTAGGSVKVDKIQVRDTGFYVTVPTAKSVNPITGSTWEVTGVTP NVEVNAEDALATAIKIVTLLNRVPAIIEGSATLIADNYAFEDIGAAVAEKLKGLLANGEYSKVVSKDSLEMKLSADLRTLSGDKSLKTTSNVPALPPM 0 0 NYSPEMYIELIKVSFHTDVFEDNIGYLRFDMFGDFEEVKAIAQIIVEHVWNKVVNTDAMIVDLR 2 1 NNIGGPTTAIAGFCSYFFDSDKQIVLDRLYDRPSGTTTELRTLPELT 1 2 GTRYGSKKSLVMLTSRATAGAAEEFVYIMKKLGRAMIVGETTAGTSHPPKTFRVGETDIFLSIPTVHSDTAAGPAWEGAGVAPHIPVPADAALETAKGIFKKHFAGQK* 0 >RBP3_oryLat medaka two domains: 28-314,320-605 no upstream dup 0 MAKTLFLVASLLVLGNVVFLHASFPPSLITDLAKIVMDNYCSPEKLSGMKEDIATAGANTDVLNIPDGEALAKVLTDGVQTTVSDPRLRVSYEPNYVPVV PPQLPPEQLIAVLQTSIKLDILEGNIGYLRIDSIIGEEVAEKVGPLLLELVWSKILPTSALIFDLRYTSSGDITGIPYIISYLTDAKSEIHIDTIYDRPL NTTTKLLSMQSTLGQTYGGTKPLLVLTSKNTKDIAEDVAYCLKNLKRATIVGEKTAGGSAKIKKFRVGDTDFYVTLPTAKSINPITGSSWEVTGVKPNVE VNAEEALATALKIINLRLQVPAIIEESATLVANNYAFESTAADVAEKLKGHLANGDYNMVVSKESLEAKLSADLQSLSGDKSLTVSSNTGAPPPM 0 0 EYTPEMYIELIKISFHTDVFENNIGYLRFDMFGDFEEVKAIAQVIVEHVWNKVLHTDAMIIDLR 2 1 NNVGGPTTAIAGFCSYFFDGDKQILLDKLYDRSTGTTTDLLTLGELT 1 2 GERYGSKKSLIILASRATAGAAEEFVYIMKRLGRAMIVGETTAGASHPPKVFQVGESDIFLSIPTVHSDTSAGPGWEGAGVAPHIPVAAGAALETAKAILNKHIGGQQHAAS* 0 >RBP3_danRer zebrafish upstream frag as well two domains: 22-322,324-609 0 MAQALVLLVSLLFFSNVAHCNFSPTLIADMAKIFMDNYCSPEKLTGMEEAIDAASSNTEILSISDPTMLANVLTDGVKKTISDSRVKVTYEPDLILAAPP AMPDIPLEHLAAMIKGTVKVEILEGNIGYLKIQHIIGEEMAQKVGPLLLEYIWDKILPTSAMILDFRSTVTGELSGIPYIVSYFTDPEPLIHIDSVYDRT ADLTIELWSMPTLLGKRYGTSKPLIILTSKDTLGIAEDVAYCLKNLKRATIVGENTAGGTVKMSKMKVGDTDFYVTVPVAKSINPITGKSWEINGVAPDV DVAAEDALDAAIAIIKLRAEIPALAQAAATLIADNYAFPSIGEHVAEKLEAVVAGGEYNLISTKEDLEERLSEDLLKLSEDKCLKTTSNIPALPPM 0 0 NPTPEMFIALIKSSFQTDVFENNIGYLRFDMFGDFEHVATIAQIIVEHVWNKVVDTDALIIDLr 2 1 NNIGGHASSIAGFCSYFFDADKQIVLDHIYDRPSNTTRDLQTLEQLT 1 2 GRRYGSKKSVVILTSGVTAGAAEEFVFIMKRLGRAMIIGETTHGGCQPPETFAVGESDIFLSIPISHSTAQGPSWEGAGIAPHIPVPAGAALDTAKGMLNKHFSGQK* 0 >RBP3x_takRub fugu single upstream exon 42% frameshift no transcripts three domains: 23-323,325-615,618-907 MAPRTPVLLLVLLFCALPVRSFYQHTLVLEMAKLLLENYCIPENLVGMQEAIQRAIKSREILQISDRKTLATVLTVGVQGALNDPRLSVSYEPSFSPLPLQALSSLPVEQQLRLLRN SIKLDILDSDVGYLRIDRIIDEETLLKFGPLLRENVWDKAAQTSSLILDLRFSTAGGWSGIPSIVSYFTEPHSLVHIDTVYDRPSNTTTELWTMSSVRGK TFGGKKDMIVLIGRRTAGAAEAVAYTLKHLNRAIVVGERSAGGSLKVRKFRIAESDFYITMPVARSVSPITGKSWEVSGISPTVNVAAREALAKAQTFLA VRSRIPKVLQIVLDIIGRFYAFADRVQALLQQLESADLFSVVSEEDLAARLNHDLQTASEDPRLIIRHKRDNIPRAEEEPELHAANDHDGELVEGFTVQV LPHNTGYLRLDRFVRCSEGDKLEEIVAEKVWGPLKDTQNLIIDLRHNTGGSSTSVALLLSYLRDPLPKRHFFTIYDSVQNTTTEYGSRPHIPGPSYGSER GVYVLTSHYTAGAAEEFAYLIQSLHFGTVVGEITSGTLMHSKTFQVEGTDIFITVPFINFLDNNGEYWLGGGVVPDAIVLAEEALEHVNRTATFHQGLRSLIGRTGELLEKHYAIQEVAQKVGEV LLSKWAEGLYRSVVDLESLASQLTADLQEASGDHRLHVFRCDVELESLHGVPKIAAVEEAGFVIDALFKSELLPRNVGYLRFDTMADIEAAKGAAPRLVKSVWNKLVDTDSLIIDMRYNA GGSSTAVPLWCSYFVDGEPLQHLYTVYDRTTKTRVEVMTLPEVSGQRYDPGKDVYILTSHMTGSAAEAFVRAMRDLNRVTIVGEPTAGGSLSSATYQIGESVLYASIPNQVVTSAATGKL WSISGVEPDVFAQARDALPVAQRIISARLLKREKGR* 0 >RBP3x_danRer zebrafish single upstream exon 55%/41% transcript DN857398 3 domains: 21-321,324-609,612-901 expressed: inner nuclear layer and ganglion cell layer MAGVFVFILVTYRVLLVNASFQSALVLDMAKILLDNYCFPENLIGMQEAIQQAINSGEILHISDRKTLASVLTAGVQGALNDPRLTVSYEPNYTLITPPA LHSLPTEQLIRLIRSTVKLEVMDNNIGYLRIDRIIGQETVVKLGRLLHNNIWKKVAHTSAMIFDLRFSTAGELSGLPYIVSYFSDSDPLLHIDTIYERPT NITRELWTLPTLLGERFGKRKDLIVLISKRTIGAAEGVAYILKHLKRAVIIGERSAGGSVRVDKLKIGDSGFYITVPVARSVNPVTGQSWEVSGVAPSVT VNPKESIAKAKSLISVRKTIPKAVRRVSDIIKRYYSFKDKIPALLNQLAKADYFTVVSEEDLAGKLNHEMQSVFEDPRLLIKATQVLTDDASSEDRSSSD DLTDPLFKLEMISGNNGYLRFDRFPTPEVLLRLEDHIKKKIWQPVQETENLVIDLRFNTGGSTEALPILLSYMFDTSSSTYLFSIYDSIKNTTFDFHTLN NISGPSYGSTKGVYVLTSYYTAEAGEEFAYLMQSLHRGTVIGEITSGMLLHSKTFQIEQTSLAITVPIINFIDVNGECWLGGGVVPDAIVLAEEALERAH EIIAFHKNIQGLVQEAGDLLEKHYSVPEVAAKVSRLLQSKLTEGLYRSVVDYESLASQLTSDLQETSGDQRLHIFYCETEPETLHDTPKIPSPEEAGFIV EALFKVDVMSGNIGYLRFDMMEDIKVLQAINPEFLKVVWNKLVNTDMLIIDVRYNTGGYSTAIPLLCTYFFDAQPLTHIYTLFDRSTATVTKVTTLPDVL GQKYSSQKDVYILTSHITGSAAEAFTRTMKDLKRATVIGEPTIGGALSSGTYQIGNSILYASIPNQAVLNAVTGKPWSISGVEPHIVAQASDALIVAQKI IATKQQKKNSGK* 0 >RBP3x_salSal Salmo salar transcript frag DY725143 EETAAKLGPLLRENIWTKVTHASSLIFDLRYSTAGELSGVPFIISYFSDPEPLIHIDTVFDRPSNTTKELWTMSSIMGERYGKRKDLIVLTSKRTMGAAEAIAYTLKHLNRAIIVGERSA GGSVKVQKIRIGDSGFYITVPVARSVNPITGQSWEVSGVSPSVNINAKEAVANAKNLLAVRSAIPNAVQSVSDIIRQYYSFTDRVPALLQHLESTDFFSVISEEDLANKFNNELQSVSEDPRLMIKL >RBP3_calMil elephantfish frag 2 domains 6-243,334-531 PPVTRESSPTSDKLPEDPTFLQALVDTVFKVSVLPDNTGYFRFDEFPEISVMSKLVQYIIEKVWLPVKDTDRLIVDLRHNVGGHSSVVPLLLSYFYDPEP PVGLFTVYNRLTNTTSHTTLPGVGQHVYGSRKDIYVLTSHRTATAAEELAYLLQSLNRATIVGEITSGSLLHSRSFQIPSTHLVITIPFINFMDNHGECW LGGGVVPDSIVLAEDTLERTKEIIGFHAQVAELVESTGKLLAVHYAIPEVAAEVSAVLSAKLTQGLYRSVVDWESLASRLTVDLQETSVWSVSGAEPHVI VQANEAMTVALGIINLRAKIPSIFQAAGKLVADNYAFAQTGAGVAETIADLIEGTGYGMINTEGKLAEVLSDTLQQLSGDKHLKAVHIPGDSKHQTPGIAMIQ 0 0 QMPPPEILEDLVKFSYQTKVLENNVGYLRFDMFGDNEMITQVSELMAKHVWNVIASTSSLIVDLR 2 1 YNIGGPTSSIPILCSYFFDDDKTVLLDTVYSRPTDTISEMKAIPQVAGNGSTESSVHSYI 1 2 * 0 >RBP3_petMar lamprey exon3/4 fused, exon4 run-on, fixed genomic frameshift; four domains: 34-312,327-615,625-914,916-1217 0 MAGSREQRTAFSTRLLLLLLLPLATCPSQAPYKFDTAVVLHLAKVLLDNYCIPENLVGMDEAIQRAVDNGELLGVSDPESAASALTEGIQAALNDPRIAV SYVAPPHTFEELLATIPQKTSFAVLDGNVGYLRADEIISEATIKKLGPVIVQRIWNRLVDTDTFVLDLRYNSHGDITGLPYLVSCFCEPRPVVHLDTVYY RPTNESKEIWSLPDLQGARFAKHKDVFVLVSANTEGVAENVAYVLKHLHRATVIGEQTAGGSLEVERFRLGDSRFFVTVPTARSEPADRSWGVFPCVSAP SERALDKALEILNARGVARKAVEAAGELLLSSYTFVERASAIADHLSWSEYGSVVSVEDLTSKLTQDLQSVAEDPRLVVSNREPEWVGAADPPGPPAPLP DDEQMLEAIVDSAFKVEVLEGNIGYLRFDEFGDASAVMKLRKQLVSKVWERIHPTDDVIIDLRYNLGGSSTAIPIVLSYFQDVAPVHFYTVYDRLRNVTA EFHTVSNLTSQLYGSKKGVYLLTSQHTATAAEEFTYLMQSLNRATIVGEITSGRLAHSLAFRLSDTGLYMTVPIVNFIDNNDEYWLGGGVVPDAIVLAENALDAAKEIIEFHAKMASL LELAGALVEGYYAMLSDGENATAEILLKYREGWYRSVVDYEALASQLTSDLHEIWGDHRLHAFYSDLQIERMDEDKTPSVPSPEELSVLIDTVFKVDILANNVGYLRFDMMTDAEVLKHV GPQLVEKVWNKISSTRSLVIDVRYNMGGYSTSIPILCSYFFDASPPRHLYTVFDRPSRSSTQVFTVPRVLGQRYGASKDVYILTSHMTGSAGEILTRVMSDLKRATVIGEPTAGGSLSTG TYRIGDSRLYVFIPNQAGVSPSGGRTWSVAGVEPHVQTKASEALQSALRMVALRADAPSILRTVGKLVADGYSRAEAALGVPSKLAALLEAGEYGALRSEEELAFKLTVHLQLITGDRHL KAVCVPEHATDRMPGIVPMQ 0 0 MPPTESFEDLIKFSFITDVLEGNIGYLRFDLFSDLEALEHVAHLLVEHVWKKICDTEILIIDLR 2 1 YNMGGYSTSIPILCSYFFDASPPRHLYTVFDRPSRSSTQVFTVPRVL 1^2 GQRYGASKDVYILTSHMTGSAGEILTRVMSDLKRATVIGEPTAGGSLSTGTYRIGDSRLYVFIPNQAGVSPSGGRTWSVAGVEPHVQTKASEALQSALRMVALRADAPSILRTVGKLVADGYSRAEAALGVPSKLAALLEAGEYGALRSEEELAFKLTVHLQLITGDRHLKAVCVPEHATDRMPGIVPMQVNVVRTRI* 0 >RBP3_braFlo Branchiostoma floridae Region: 9 exons 1 domain: 83-381 ClpP/crotonase e-38 419-630; misfused to PAPS sulfotransferase 0 MTRPSKVDIVFPIKPFTIPTAHEQVKGEGPVDINKNALCKSADEGHTHP 1 2 VSIAMAPTAYIVFVALVPTVLSVDWLDVVMGIGDVMADHYLDQDLRALNDQSLLQRWNRTLVHRFQ 0 0 SWSQDDMSDSLRMEEGLTSELRNITGDETIK 0 0 VWDFGVYENTTQEPVPREFYNFSTFVDNFK 2 1 KNREKHINVTMLEGNVGYVSIRSMSHIVDIILPDPEMTEFFLSKMAALNESK 0 0 AIILDLRYNLGGDREGVVHWASFFFNATPSVPLSDVYYRDGVNQYWTLLE 0 0 VPGGIRFPDMPLYLLTSNRTSREAEEFAYAMQVVNRTTIIGETT 1 2 AGEEFTGMWFPIDQTDVHLLTRTNVVRNPITQDSWSGK 1 2 GVTPDIIVPSEKALTVALRKIQGSEDTKMAASSGNIEPPRWTVYLVFICTSIAILTYPTFM* 0