Personal genomics: ACTN3
Introduction to ACTN3 comparative genomics
The alpha actinin gene ACTN3 is a coding gene on human chromosome 11, quite interesting in its own right but best known as ground zero in the debate over frivolity and unexpected consequences of personal genomics. This gene first of all needs careful and exhaustive re-annotation before considering this controversy because its existing peer-reviewed scientific literature (some 22 papers) is a mixture of pre-genomic era obsolescence and gross factual errors such as expression said specific to skeletal muscle.
Some unfortunate historic terminology needs to be explained. Actinins were erroneously thought similar to actin in early studies of myofibrillar components; instead they are homologically unrelated proteins that happen to bind actin. These 'actinins' were then improperly divided into 3 classes (alpha, beta, gamma) before it became known that their respective gene families were wholly unrelated (not homologous). For example, 'beta' actinins refer to heterodimers functioning as actin barbed-end capping proteins in skeletal muscle; they are comprised of the distinct gene families CAPZA and CAPZB themselves non-homologous and thus further misnamed. 'Gamma' actinins refer to yet other unrelated genes.
In this article, actinin shall mean alpha actinin, ie a protein encoded by one of the four paralogous genes ACTN1-ACTN4. Gene names are used for both gene or gene product (as this is always clear from context). Genus and species are indicated with standard 6-letter code (eg ACTN3_homSap). Care still must be taken with published articles and genBank entries that may fail to specify the alpha actinin under consideration despite a 2003 article calling for adherence to HGNC international nomenclature standards followed here.
The comparative genomics situation is further confused by high sequence conservation within the ACTN gene family, by paralog loss in some clades, by possible independent duplication events, and by pre-duplication parental genes only in early deuterostomes. It is not easy to assign transcripts or genomic fragments to correct orthology class by methods such as best reciprocal Blast, especially when the query itself is a fragment (eg third spectrin domain of ACTN3). Many genBank entries are unlabelled, mislabelled, or ambiguously labelled as to correct ortholog family.
However a reliable actinin classifier can be built by requiring flanking gene synteny, diagnostic signature residues and indels in building the reference sequence seed collection that focus on signature regions in which ortholog classes differ significantly from each other. For example ACTN2/3 share a five-residue deletion in exon 19 relative to (ancestral-length) ACTN1/4.
The human ortholog of ACTN3 is unusual (but not unique) in having a fairly abundant null allele, R577x, meaning the arginine at position 577 of the 901 residue protein has been replaced by a stop codon. Worldwide 18% of the human population is homozygous 577x 577x. It is very unlikely that a functional truncated protein can be produced because even if the promoter region is still functional, the mRNA would be degraded by nonsense-mediated decay (with no possibility of selenocysteine substitution) and the stable quaternary dimer necessary for function cannot form (as explained below).
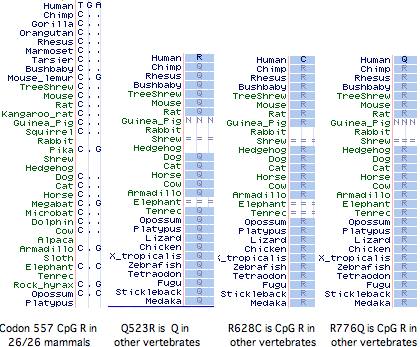
It has not been established whether the 577x was the initial inactivating mutation, as 3 additional amino acid changes (Q523R, R628C, R776Q) have also accrued at otherwise invariant sites in this allele (ie, in the dna donor to the public human genome relative to genBank reference sequence NM_001104). The latter two substitutions are also CpG mutational hotspots (the entire mRNA has 131 such sites). It is not known whether these other changes became widespread before or after R577x nor whether they affect ACTN3 function. However it is not easy to inactivate a large structural protein comprised of independent modules by single substitutions.
Curiously Q523R, R577x, and R628C all occur in the third spectrin repeat despite this region constituting only 11% of the gene, yet not a single nonsynonymous base change has occured in the 2706 bp coding region. With the advent of HapMap and similar projects, the phenotypic associations of these changes, possible co-occurence with wildtype R577, and the date(s) of 577x founder mutations could be resolved.
All mammals with assembled genomes encode a CpG hotspot at codon 577. This has transitioned to TpG in the human 577x allele but is not a polymorphic site in any other known mammal, though the search has been restricted to the individual animals used in genome projects (since transcripts rarely extend this far), plus 36 unrelated baboons and 33 chimpanzees all genotyping to ‘wild-type’ 577R. Thus there is no support for 577x as balanced polymorphism in any mammal other than human even though Z-line skeletal muscle structures may be very similar.