Opsin evolution: Cytoplasmic face
Comparative genomics of the cytoplasmic face of GPCR proteins
The cytoplasmic 'face' of opsin (or any GPCR) is presumably responsible for all interactions with downstream signal relaying partners because these latter are cytoplasmic proteins having no access to extracellular loops or transmembrane segments. Here it must be noted that ligand photoisomerization and release from Schiff base deep within the transmembrane region must drive a significant change in conformation in the cytoplasmic face that differentiates inactive from active states.
The cytoplasmic face is comprised of three loops and carboxy terminus. For bioinformatic purposes, it is convenient to 'reorganize' each linear protein sequence into its intracellular, membrane and outer regions for separate consideration. This is done below for the cytoplasmic face of 500 curated opsins from each of the 18 vertebrate opsin orthology classes using multiple representatives for each phylogenetic node and intense bracketing at eras of change (eg between DRY and GRY opsins of RGR class). A range of non-opsin GPCR are included to define properties common to all members of this large gene family (not specific to opsins).
The two critical goals in GPCR research are to determine the natural ligands (which largely concerns the extracellular and transmembrane regions) notably for orphan receptors and to determine their specific Galpha signaling partner among the 16 such paralogs in the vertebrate genome. For the 18 orthology classes of vertebrate opsins, the ligand is already known (11-cis retinal or related) but the signaling partner is generally not. As an example, does RGR opsin signal, to what purpose, and what is the meaning of the abrupt shift in the DRY motif to GRY in boreoeutheres?
Cytoplasmic loop C2 at 18 opsin genetic loci
DRY loop motif transmemb L 7 9 signaling
ENCEPH_hom ERYIRVVHARVINFSW AWRAITYIW 16 V A G?
RGR_homSap GRYHHYCTRSQLAWNS AVSLVLFVW 16 C R G?
RGR2_gasAc DRYHQYCTRQKLFWST TLTMSAIIW 16 C R G?
RHO1_homSa ERYVVVCKPMSNFRFGENH AIMGVAFTW 19 C P GNAT1
RHO2_galGa ERYIVVCKPMGNFRFSATH AMMGIAFTW 19 C P GNAT2
SWS2_ornAn ERFLVICKPLGNLSFRGTH AIFGCAATW 19 C P GNAT2
PIN_galGal ERYVVVCRPLGDFQFQRRH AVSGCAFTW 19 C P G?
SWS1_homSa ERYIVICKPFGNFRFSSKH ALTVVLATW 19 C P GNAT2
LWS_homSap ERWMVVCKPFGNVRFDAKL AIVGIAFSW 19 C P GNAT2
VAOP_galGa ERYIVICRPVGNMRLRGKH AAQGIAFVW 19 C P Gt
PARIE_utaS ERYNVVCQPLGTLQMSTKR GYQLLGFIW 19 C P Gd+Go
PPIN_xenTr DRVFVVCKPMGTLTFTPKQ ALAGIAASW 19 C P Gt
PER_homSap DRYLTICLPDVGRRMTTNT YIGLILGAW 19 C P Go
NEUR1_homS DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L G?
NEUR2_galG VCCLKICFPAYGNRFRRKH GQILIACAW 19 C P G?
TMT_monDom ERYRTL-TLCPGQGADYQK ALLAVAGSW 19 - L G?
MEL1_homSa DRYLVITRPLATFGVASKRR AAFVLLGVW 20 T P Gq
MEL2_anoCa DRYCVITKPLQSIKRTSKKR TCIIIVFVW 20 T P Gq
While it might seem straightforward to thread any opsin onto its best fit among the five newly available crystallographic structures, that does not work for distantly related paralogs beyond the universal 7-transmembrane feature because loop regions can be of quite different length and so lack discernable alignability, having diverged greatly in amino acid sequence (even though they are all ultimately homologous). While these structures entail various compromises (to enable stable crystallization), they are hugely important to annotation transfer of sequence/function relationships via comparative genomics. Yet most of the 18 vertebrate opsin orthology classes have only remote models to date:
Gene PDB Protein PubMed Best human opsin Next Best Signaling RHO1_bosTau 1JFP 3C9M 2J4Y bovine rod rhodopsin 17825322 RHO1_homSap 93% SWS1_homSap 45% Gt GNAT1 raises cGMP MEL1_todPac 2Z73 2ZIY squid melanopsin 18480818 MEL1_homSap 43% PER1_homSap 30% Gq GNAQ? inositol trisphosphate ADORA2A_homSap 3EML adenosine receptor 2A 18832607 MEL1_homSap 27% ENCEPH_homSap 27% Gs GNAT3 raises cAMP ADRB1_melGal 2VT4 beta 1 adrenergic receptor 18594507 MEL1_homSap 29% ENCEPH_homSap 25% Gs GNAT3 raises cAMP ADRB2_homSap 2R4R beta 2 adrenergic receptor 17962520 MEL1_homSap 28% PER1_homSap 29% Gs GNAT3 raises cAMP
It has not proven feasible to predict loop conformations ab initio or from peptide libraries; it is folly to consider individual loop structure in isolation (rather than the cytoplasmic face in its entirety) or fail to specify the activation state being computed. Any predicted structure and special roles for individual residues be consistent with the comparative genomics of close and even distant orthologs because binding relationships to Galpha and other proteins do not change rapidly in evolutionary time (as seen from heterologous substitution experiments). Even when a cytoplasmic loop seems to lack a definable structure, individual residues can be conserved over vast branch length times. That conservation must ultimately be explained.
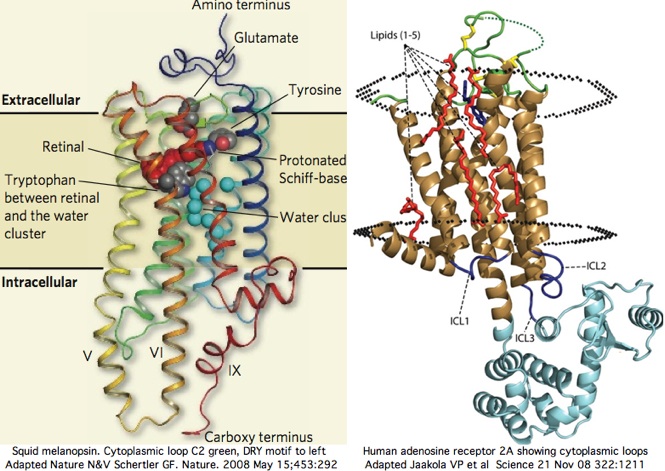
Two new high resolution structures of squid melanopsin establish that the cytoplasmic face is not structurally homologous as a whole across paralogous opsin classes. We knew this already from comparative genomics alone but not specifically why. The xray structure exhibits unprecedented rigid extensions of transmembrane helices 5 and 6 of order 25 angstroms out into the cytoplasm, greatly constraining the intermediate residues of cytoplasmic loop C3. The proximal carboxy terminus also contributes importantly to the overall structure here.
The squid melanopsin structure, used at SwissModel, could readily predict the structure of the cytoplasmic face of all opsins of melanopsin class, of which 48 vertebrate sequences, 9 lophotrochozoan, 43 arthropod, and 1 cnidarian sequences are available here. The Gq signalling partner will be used throughout these melanopsins, yet what features the Galpha protein specifically recognizes in the cytoplasmic face remains obscure. It cannot really be the helical extensions per se because the Gq protein is structurally still homologous to its 15 paralogs (in vertebrates) of different signaling types.
The second cytoplasmic loop
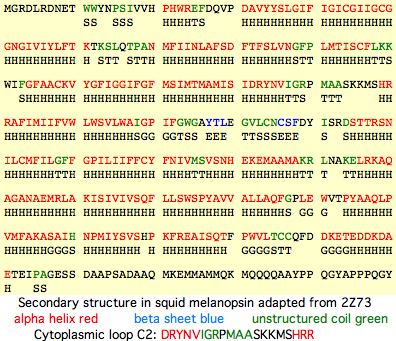
In squid melanopsin, first six residues of cytoplasmic loop C2 also form an extensional helix in squid melanopsin beginning with the DRY motif and surprisingly terminating three residues before the deeply conserved proline (normally a helix breaker as in adrenergic receptors). This proline alone cannot define the two states through its cis and trans configurations because glycine or leucine can also characterize whole opsin orthology classes at this position. The last 3 residues of basic character HRR of loop C2 also preface a transmembrane helix as RAR do in turkey receptor.
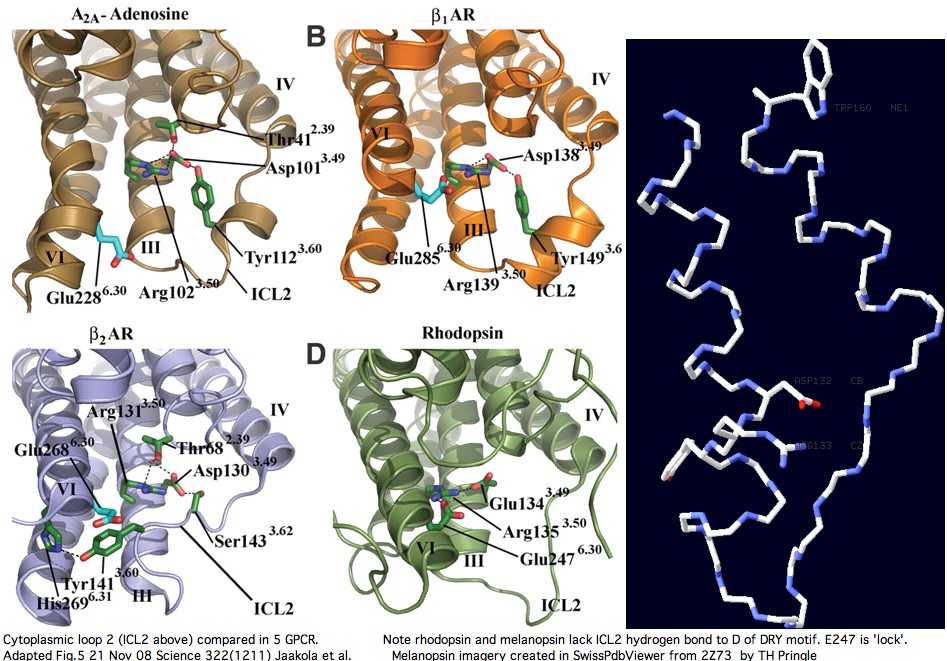
Cytoplasmic loop C2 has conserved length of 16-20 in all opsins with much more rigid constraint within individual opsin classes (eg all vertebrate imaging opsins have length 19. The structure of the C2 loop of over 100 melanopsins can readily be modelled based on its closest match among the determined structures. Because adrenergic loop C2 is a structural outgroup, yet has a very similar fold, means all opsin C2 loops have a very similar structure.
The adenosine and adrenergic receptor structures -- however useful they might be for annotation transfer to the other 350 non-oderant human GPCR -- ultimately will not prove helpful to modeling the second cytoplasmic loop of opsins (squid melanopsin does that better already). Note C2 in these three structures is consistently stablized by a mid-loop hydrogen bond to the DRY residues. This constraint is not observed in squid melanopsins; indeed it is not feasible because no hydrogen bond-capable residue occurs there (in the comparative genomics sense of conserved residue).
The second cytoplasmic loop in melanopsin
Cytoplasmic loop C2 from 101 Melanopsins species helix bridge area hel transmemb Le 7 9 MEL1_homSa DRYLV ITRPLATFGVAS KRR AAFVLLGVW 20 T P MEL1_panTr DRYLV ITRPLATFGVAS KRR AAFVLLGVW 20 T P MEL1_gorGo DRYLV ITRPLATFGVAS KRR AAFVLLGVW 20 T P MEL1_ponAb DRYLV ITRPLATIGVAS KRR AAFVLLGVW 20 T P MEL1_rheMa DRYLV ITRPLATIGVAS KRR AAFVLLGVW 20 T P MEL1_calJa DRYLV ITRPLATIGVAS TKR AAFVLLGVW 20 T P MEL1_micMu DRYLV ITRPLASVGTAS KRR AGLVLLGVW 20 T P MEL1_otoGa DRYLV ITRPLTTVGVAS KRR AALVLLGVW 20 T P MEL1_musMu DRYLV ITRPLATIGRGS KRR TALVLLGVW 20 T P MEL1_ratNo DRYLV ITRPLATIGMRS KRR TALVLLGVW 20 T P MEL1_nanEh DRYLV ITRPLATIGVAS KRR TALVLLGVW 20 T P MEL1_phoSu DRYLV ITRPLATIGMGS KRR TALVLLGIW 20 T P MEL1_dipOr DRYLV ITRPLATIGVTS KRR TAFVLLGVW 20 T P MEL1_cavPo DRYLV ITRPLATIGVAS KRQ AALVLLGVW 20 T P MEL1_speTr DRYLV ITRPLATIGMAS KKR AAFFLLGVW 20 T P MEL1_oryCu DRYLV ITRPLAAVGMVS KKR AGLVLLGVW 20 T P MEL1_ochPr DRYLV ITRPLAAVGMVS KRR TGLVLLGVW 20 T P MEL1_bosTa DRYLV ITRPLATVGMVS KRR AALVLLGVW 20 T P MEL1_turTr DRYLV ITRPLATVGMVS KRR AALVLLGVW 20 T P MEL1_susSc DRYLV ITHPLATVGMVS KRR AALVLLGVW 20 T P MEL1_equCa DRYLV ITRPLATVGVVS KRW AALVLLGIW 20 T P MEL1_felCa DRYLV ITHPLATIGVVS KRR AALVLLGVW 20 T P MEL1_canFa DRYLV ITHPLAAVGVVS KRR AALVLLGVW 20 T P MEL1_myoLu DRYLV ITRPLA-IGVVS KRR AALVLLGVW 19 T P MEL1_pteVa DRYLV ITRPLAAIGVVS KRR AALVLLGVW 20 T P MEL1_eriEu DRYLV ITRPLATIGVVS KRR VALVLLGVW 20 T P MEL1_loxAf DRYLV ITRPLATIGVVS KRR AALVLLGIW 20 T P MEL1_proCa DRYLV ITRPLATIGVVS KRR TALVLLGTW 20 T P MEL1_echTe DRYLV ITRPLATIGVVS KRR AALVLLVIW 20 T P MEL1_smiCr DRYFV ITRPLASIGMIS KKK TGLILLGVW 20 T P MEL1_monDo DRYFV ITRPLASIGVIS KKK TGFILLGVW 20 T P MEL1_ornAn DRYFV ITRPLASIGVIS KKR ALLILTGVW 20 T P MEL1_anoCa DRYFV ITRPLASIGAMS TKK ALLILSGVW 20 T P MEL1_taeGu DRYFV ITKPLASVGVTS KKK ALIILVGVW 20 T P MEL1_galGa DRYFV ITKPLASVRVMS KKK ALIILVGVW 20 T P MEL1_xenTr DRYFV ITRPLTSIGVMS KKR AVLILSGVW 20 T P MEL1_danRe DRYFV ITRPLASIGVLS QKR ALLILLVAW 20 T P MEL1_danRe DRYFV ITRPLASIGVMS RKR ALLILSAAW 20 T P MEL1_takRu DRYFV ITRPLTSIGVLS RKR AFVILMTVW 20 T P MEL1_gasAc DRYFV ITRPLTSIGMMS RRR ALLILMGAW 20 T P MEL1_oryLa DRYFV ITRPLTSIGVLS RKR ALLILSAAW 20 T P MEL1_calMi DRYFV ITRPLASIGVLS HRR AGLIILSLW 20 T P MEL1_petMa DRYLV LTRPLASIGAMS KRR AMYITAAVW 20 T P MEL2_galGa DRYLV ITKPLRSIQWTS KKR TIQIIAAVW 20 T P MEL2_anoCa DRYCV ITKPLQSIKRTS KKR TCIIIVFVW 20 T P MEL2_xenLa NRYIV ITKPLQSIQWSS KKR TSQIIVLVW 20 T P MEL2_danRe DRYLV ITKPLQTIQWNS KRR TGLAILCIW 20 T P MEL2_tetNi DRYVV ITKPLQTIRRSS KRR TALAILMVW 20 T P MEL2_gasAc DRYLV ITKPLQAIHWGS KRR TTLAILLVW 20 T P MEL1_plaDu DRFYV ITNPLGAAQTMT KKR AFIILTIIW 20 T P MEL1_capCa DRYMV IAKPFYAMKHVS HKR SLIQIILAW 20 A P MEL1_helRo DRYLV VGQPLAMLNQSH FRR SFYHVLIIW 20 G P MEL1_todPa DRYNV IGRPMAASKKMS HRR AFIMIIFVW 20 G P MEL1_schMe DRYFV IAQPFQTMKSLT IKR AIIMLVFVW 20 A P MEL2_schMa DRYLV IATPFESVFQTT PRR TLLLMLFLW 20 A P MEL1_lotGi DRYLV ITSPFTAMRNMT HKR AFLMIVGVW 20 T P MEL1_sepOf DRYNV IGRPMAASKKMS HRR AFLMIIFVW 20 G P MEL1_entDo DRYNV IGRPMAASKKMS HRR AFLMIIFVW 20 G P UVV_camAb DRYST IARPLDGKLS RGQ VLLLIMLIW 18 A P UVV_catBo DRYST IARPLDGKLS RGQ VILLIALIW 18 A P UVV_apiMe DRYST IARPLDGKLS RGQ VILFIVLIW 18 A P BLU_apiMe DRYRT ISCPIDGRLN SKQ AAVIIAFTW 18 S P BLU_ DRoMe DRYKT ISNPIDGRLS YGQ IVLLILFTW 18 S P BLU_manSe DRYKT ISSPLDGRIN TVQ AGLLIAFTW 18 S P UVV1_droMe DRYNV ITKPMNRNMT FTK AVIMNIIIW 18 T P UVV1_pedHu DRCET ITNPL-QKSG KKK AFLLAAFTW 18 T P UVV_manSe DRHST ITRPLDGRLS EGK VLLMVAFVW 18 T P UVV_papXu DRHST ITRPLDGRLS RGK VLLMMVCVW 18 T P UVV2_droMe DRFNV ITRPMEGKMT HGK AIAMIIFIY 18 T P UVV2_pedHu DRYQV IVHPLER-KT KAA VYFQILLIW 18 V P LWS_nemVe DRYIV IVHPMKKIMT RKK AALMIVGVW 18 V P LWS_pedHu DRYNV IVKGLSAKPMT IKM ALLNILFVW 19 V G LWS_vanCa DRYNV IVKGIAAKPLT ING AMLRVLGIW 19 V G LWS_papXu DRYNV IVKGIAAKPMT ING ALLRILGIW 19 V G LWS_helSa DRYNV IVKGIAAKPMT ING ALLRVFGIW 19 V G LWS_pieRa DRYNV IVKGIAAKPMT INS ALLRILGVW 19 V G LWS_manSe DRYNV IVKGIAAKPMT SNG ALLRILGIW 19 V G MWS2_droMe DRYNV IVKGINGTPMT IKT SIMKILFIW 19 V G LWS_rhoPr DRYNV IVKGISAKPMT NKT AMLRILLVW 19 V G LWS_meoOe DRYNV IVKGISGTPLS QKN TTLQVLFVW 19 V G LWS_catBo DRYNV IVKGLSAKPMT ING ALLRILGIW 19 V G LWS_schGr DRYNV IVKGLSAKPMT NKT AMLRILFIW 19 V G LWS_triCa DRYNV IVKGLSAQPLT KKG AMLRILIIW 19 V G LWS2_apiMe DRYNV IVKGLSGKPLS ING ALIRIIAIW 19 V G LWS_bomTe DRYNV IVKGLSGKPLT ING ALLRILGIW 19 V G MWS_calEr DRYNV IVKGMAGQPMT IKL AIMKIALIW 19 V G MWS1_droMe DRYQV IVKGMAGRPMT IPL ALGKIAYIW 19 V G LWS_droMe DRYCV IVKGMARKPLT ATA AVLRLMVVW 19 V G LWS_arcGr DRYNV IVKGVAAEPLT SKG ASIRILFVW 19 V G LWS_eupSu DRYNV IVKGVAATPLT NKG AFARNIFSW 19 V G LWS_camLu DRYNV IVKGVAGEPLS TKK ASLWILTVW 19 V G LWS_proMi DRYNV IVKGVAGEPLS TKK ASLWILIVW 19 V G LWS_holCo DRYNV IVKGVSAEPLT SGG AMMRIAGTW 19 V G LWS_homGa DRYNV IVKGVSATPLT TNG AMLRNLFSW 19 V G LWS_neoAm DRYNV IVKGVSGEPLT NSG AMTRIAGTW 19 V G LWS_neoOe DRYNV IVKGVSGKPLS QKN ATLQVLFVW 19 V G LWS_mysDi ERYNV IVKGVSSKPLS VKG AITRIVLTW 19 V G LWS1_apiMe DRYNV IVKGMSGTPLT IKR AMLQILGIW 19 V G LWS_limPo DRYNV IVRGMAAAPLT HKK ATLLLLFVW 19 V G LWS_limPo DRYNV IVRGMAAAPLT HKK ATLLLLFVW 19 V G LWS_ixoSc DRYNV IVRGVAAAPLT HKR AALMIFFVW 19 V G ADRB2_homS DRYFA ITSPFKYQSLLT KNK ARVIILMVW 20 T P ADRA2A_hom DRYWS ITQAIEYNLKRT PRR IKAIIITVW 20 T A ADRA2C_hom DRYWS VTQAVEYNLKRT PRR VKATIVAVW 20 T A HTR1A_homS DRYWA ITDPIDYVNKRT PRR AAALISLTW 20 T P CHRM1_homS DRYFS VTRPLSYRAKRT PRR AALMIGLAW 20 T P DRD2_homSa DRYTA VAMPMLYNTRYS KRR VTVMISIVW 21 A P TAAR9_homS DRYIA VTDPLTYPTKFT VSV SGICIVLSW 20 T P ADRA2B_hom DRYWA VSRALEYNSKRT PRR IKCIILTVW 20 S A
Reference collection of structurally determined GPCR
>RHO1_bosTau cow rod rhodopsin MNGTEGPNFYVPFSNKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAI ERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWL PYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA* >MEL1_todPac Todarodes pacificus (squid) Gq X70498 480 11106382 Mollusca 'squid rhodopsin' 3D: May 2008 Cys 337 palmitoyled MGRDLRDNETWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISI DRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGANAEMRLAKI SIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAETEIPAGESSDAAPSADAAQMKEMMAMMQKMQQQQAAYPPQGY APPPQGYPPQGYPPQGYPPQGYPPQGYPPPPQGAPPQGAPPAAPPQGVDNQAYQA* >ADRB1_melGal turkey Beta 1 adrenergic receptor with stabilising mutations And bound cyanopindolol MGAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAI DRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREA KEQIRKIDRASKRKRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCRSPDFRKAFKRLLAFPRKADRRLHHHHHH* >ADRB2_homSap beta 2 adrenergic receptor 365 aa MGQPGNGSAFLLAPNRSHAPDHDVTQQRDEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAV DRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYANETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRFHVQNLSQVEQDGRTGHGL RRSSKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLCLRRSSLKAYGNGYSSNGNTGEQSG* >ADORA2A_homSap adenosine adrenergic receptor 2A MPIMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAI DRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKNHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRI FLAARRQLKQMESQPLPGERARSTLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFR KIIRSHVLRQQEPFKAAGTSARVLAAHGSDGEQVSLRLNGHPPGVWANGSAPHPERRPNGYALGLVSGGSAQESQGNTGLPDVELLSHELKGVCPEPPGLDDPLAQDGAGVS*