Bison: mitochondrial genomics: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 327: | Line 327: | ||
1 L yak | 1 L yak | ||
1 N lemur | 1 N lemur | ||
<font color=red> | <font color=red>I118T</font>: At position 118, the reduced alphabet consists predominantly of ILV with some A and M, a very common occurence proteomewide. TSF are all deleterious mutations in domestic yak. | ||
I118T | I118T | ||
2597 I yak has most common amino acid at position 118 | 2597 I yak has most common amino acid at position 118 | ||
Revision as of 18:09, 3 December 2010
Introduction to bison conservation genomics
(to be continued)
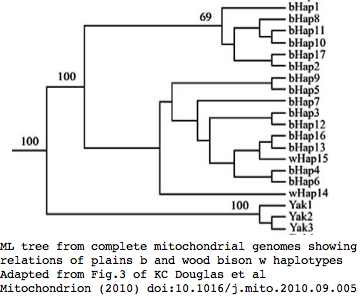
Phylogeny: bison and yak are sister groups
(to be continued)
Interpreting bison CYTB variation
Bison mitochondrial genomes became well-represented at GenBank with the 1 Dec 10 release by the Derr group of 31 complete genomes from 6 herds including two woods bison (Bison bison athabascae) from the non-admixed Elk Island herd (along with various cow-bison hybrid and cow breed genomes). The cow-bison hybrids represent crossing of a bison male with a domestic cow (or rather a continuous line of female descent from such a cross) and so have strictly cow mitochondrial dna, not relevent to this section. The haplotype of all hybrids studied (from an unnamed private ranch in Montana, presumably Turner's Flying D) cluster with cow haplotype cHap32.
Bison accession numbers: GU946976 GU946977 GU946978 GU946979 GU946980 GU946981 GU946982 GU946983 GU946984 GU946985 GU946986 GU946987 GU946988 GU946989 GU946990 GU946991 GU946992 GU946993 GU946994 GU946995 GU946996 GU946997 GU946998 GU946999 GU947000 GU947001 GU947002 GU947003 GU947004 GU947005 GU947006
The CYTB sequences retrieved from these genomic entries (they are not yet in the database used by blastp) show haplotype notation. The 15 previously existing bison sequences at GenBank (some just fragments are also provided. Older fragmentary sequences are demonstrably error-prone and will be used here only as support -- never as sole source -- of a polymorphism. Redundancy introduced via non-standard SwissProt (UniProt) entries also has to be manually removed -- the Swiss did no sequencing on their own, simply deriving protein sequences from existing GenBank entries. This leaves 5 older complete sequences for Bison bison and 4 fragments, 2 attributed to Bison bonasus and 1 fossil dna sequence from Bos primigenius to serve as outgroup (rather than an inbred domestic cow).
Here it is necessary to pick a terminology. This must accommodate NCBI taxonomy -- irregardless of its correctness -- because otherwise blastp searches cannot be restricted by taxon. Note although bison are definitely sistered with yak to the exclusion of all other extant species, that creates problems because yak has been put in the genus Bos. Many relic wild cattle have no english language common name but rather that of a local language. Terminology table must show synonyms to allow PubMed and google searches -- especially important in a fast-moving field to locate preprints and conference proceedings. The table below does not attempt to implicitly resolve any scientific issue; it simply states preferred terminology at this site along with synonyms in common use.
(editing to be continued) Bison bison plains bison Bison athabascae woods bison Bison bonasus euro bison Bison priscus steppe bison Bos primigenius auroch (extinct except for Korean and Italian cattle with auroch mitochondrial genomes) Bos grunniens yak Bos indicus zebu kourey Bos taurus common cow gaur wisent Leptobos last common ancestor to cows and bison
Sequences are color clustered according to the phylogenetic tree above. bHap1 is not shown. Note the woods bison cannot be resolved from the plains bison even though the Elk Island woods bison are a relic herd that did not mix with 7,000 plains bison imported from the Flathead Reservation in Montana up to Canada's Wood Buffalo National Park in the 1920's.
>CYTB_bisBis.GU946988 bHap8 plains bison b973 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946994 bHap11 plains bison b1031 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946990 bHap10 plains bison b985 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU947000 bHap10 plains bison bFN5 Niobrara MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946991 bHap10 plains bison b1005 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU947004 bHap17 plains bison bYNP1586 Yellowstone NP MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946976 bHap2 plains bison b790 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946977 bHap2 plains bison b853 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946978 bHap2 plains bison b854 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946981 bHap2 plains bison b880 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946983 bHap2 plains bison b925 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946984 bHap2 plains bison b929 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946993 bHap2 plains bison b1029 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946995 bHap2 plains bison b1050 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946996 bHap2 plains bison b1051 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU947001 bHap2 plains bison bNBR1 National Bison Range MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946986 bHap2 plains bison b959 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946989 bHap9 plains bison b979 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946997 bHap9 plains bison b1091 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946982 bHap5 plains bison b897 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946982 bHap5 plains bison b897 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946987 bHap7 plains bison b961 Montana MTSLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946979 bHap3 plains bison b855 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946992 bHap3 plains bison b1018 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946992 bHap3 plains bison b1018 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946998 bHap12 plains bison b1191 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU947003 bHap16 plains bison bTSBH1005 Texas State Bison Herd MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946999 bHap13 plains bison b1428 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU947002 bHap13 plains bison bTSBH1001 Texas State Bison Herd MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisAth.GU947005 wHap15 woods bison wEI1 Elk Island MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946980 bHap4 plains bison b877 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.GU946985 bHap6 plains bison b935 Montana MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisAth.GU947006 wHap14 woods bison wEI14 Elk Island MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTMMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKWDEFINITION
(editing to be continued) YP_002791041 Bison bison Q9T9C1 Bison bison YP_003587278 Bison bonasus ACE76876 Bos primigenius YP_003541096 Bos primigenius O20998 Bison bonasus ADQ12704 Bison bonasus 61 ......T.....ETTAEF...................V...................... 120 AAL85955 Bison bison AAL85956 Bison bonasus ADM87433 Bison bison AAW28803 Bison bison AAW28804 Bison bison AAW28802 Bison bonasus AAN28295 Bison bison CAA76013 Bison bonasus
Interpreting CYTB variation in yak
Yaks are the closest living sister species to bison. Although 15,000 wild yaks still persist, they have been subject to very similar pressures to those experienced by bison: bottlenecks, population fragmentation, introgression from long domesticated yaks and hybridization with cattle. Adaptations specific to mitochondria may exist as yak live at altitudes exceeeding 3500 meters where average annual temperatures in rearing areas are –8°C, surviving winter temperatures of –40°C.
Because yaks provide the immediate outgroup for bison genetics (and vice versa), their parallel mitochondrial proteomics are investigated in depth here. This further enables reconstruction of their last common ancestor and correct placement of Pleistocene dna sequences.
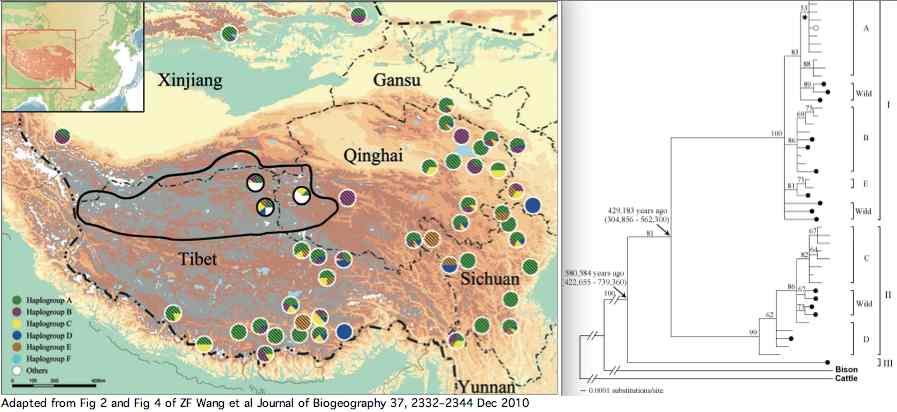
Data availability for yaks was greatly improved by a Dec 2010 paper investigating yak phylogeographical structure and demographic history on the Qinghai-Tibetan Plateau. Complete mitochondrial genomes were determined for 48 domesticated and 21 wild yaks. The three lineages article supplemental established diverged at 420 kyr and 580 kyr in accordance with allopatric migration barriers created by two large plateau glaciations.
The wild yaks are found in all three branches of the tree (solid circles at left). Their entries at GenBank are apparently distinguished by a W (for wild) prefix, eg isolate W77 GQ464266. There is potential for confusion here because NCBI taxonomy uses Bos grunniens mutus subspecies notation for wild yak (a concept contradicted by the mixed distribution of wild and domestic yaks in the tree). Related concepts such as Bos mutus (Przewalski, 1883), Bos mutus grunniens, and Poephagus mutus also don't fit the facts.
Protein polymorphisms in wild yak cytochrome b sequences are the primary focus here as domestic yak may exhibit inbreeding issues and evolutionary artefacts. Consequently it is important to track which GenBank entries reference wild yaks.
Bos grunniens mutus has two GenBank entries relevent to CYTB: AAX53006 containing V195A, I348F otherwise lacking support and CAA76015, an older fragmentary sequence of no allelic interest. The Myanmar/Bhutan mithun sequence BAJ05329 attributed to Bos grunniens at GenBank has 12 differances but is 100% identical to 94 Bos indicus entries, ie the mitochondrial genome of this hybrid originated there.
The 21 new genome accessions of wild yak are GQ464266, GQ464265, GQ464264, GQ464263, GQ464262, GQ464261, GQ464260, GQ464259, GQ464258, GQ464257, GQ464256, GQ464255, GQ464254, GQ464253, GQ464252, GQ464251, GQ464250, GQ464249, GQ464248, GQ464247, GQ464246.
In terms of protein accessions (which will be shown at NCBI blastp output), these are ACU81659, ACU81646, ACU81633, ACU81620, ACU81607, ACU81594, ACU81581, ACU81568, ACU81555, ACU81542, ACU81529, ACU81516, ACU81503, ACU81490, ACU81477, ACU81464, ACU81451, ACU81438, ACU81425, ACU81412, ACU81399.
Of these, 16 fall in the main reference sequence group but 5 wild Tibetan plateau yaks exhibit polymorphisms that cannot be attributed to domestication. Two additional wild yaks from extreme NW China have additional double alleles but no associated PubMed publication. There is no overlap between wild yak polymorphism sites and the five of domestic yak. Alleles occurring in full length sequences are analyzed further below.
The summary table of yak CYTB amino acid polymorphisms below arises from alignment of 5000 full-length mammalian cytochrome b orthologs. Red indicates deleterious mutation, green a possibly acceptable change but of restricted distribution and fitness, and blue a near-neutral substitution. Gray is reserved for probable sequencing error. It can be seen that the smallish yak population sampled (21 wild, 48 domestic added in Aug 10 to 3-4 previously available) already contains 5 deleterious alleles in CYTB which represents only 10% of the mitochondrial proteome.
In summary, out of 70 individual yaks, 10 are carrying deleterious mutations at five sites. That seems like an extraordinary number for a central enzyme in energy metabolism for which it is difficult to envision compensation by another gene. Restricting to the 21 wild yaks, 3 have deleterious polymorphism and 1 has a marginal change. Overall 1 in 7 animals is affected just in this one gene. However CYTB is but one of 13 encoded by the mitochondrial genome -- what sort of genetic burden are yaks carrying overall?
1 ACU81568 A017T wild yak isolate W50 GQ464259 2 ACU81399 I192T wild yak isolate W02 GQ464246 ACU81633 I192T wild yak isolate W75 GQ464264 3 ACU81555 D214N wild yak isolate W40 GQ464258 4 AAX53006 V195A I348F mutus isolate Xinjiang01 unpublished Liu,Q Wu,M Li,Y AAX53007 V195A I348F mutus isolate Xinjiang02 unpublished Liu,Q Wu,M Li,Y 5 ACU81529 V329M wild yak isolate W1313 GQ464256 6 ABI15999 V039I A067T domestic yak fragment PUBMED:17257194 Poephagus 7 ABI16000 V039I A067T domestic yak fragment PUBMED:17257194 Poephagus ACU82153 A084T domestic yak isolate HY5 8 ACU82101 V098L domestic yak isolate HY1 9 AAU89116 I118T domestic yak =SP:Q5Y4Q0 PUBMED:16942892 ACU81711 I118T domestic yak isolate HZ3 ACU81737 I118T domestic yak isolate MQ1 AAS93096 I118T domestic yak fragment PUBMED:17257194 AAS93099 I118T domestic yak fragment PUBMED:17257194
Although the mitochondria encodes the usual 20 amino acids, only a subset of physio-chemically similar residues (the reduced alphabet) ever appear at a given position in a given protein. This subset describes the acceptable substitutions that do not significantly disrupt protein functionality. Discovery of this reduced alphabet can be achieved with greater sensitivity when the number of available species and their individual sequences multiplicities are high. For mitochondrial proteins, that sensitivity is 1 in 10,000 (0.01% occurrence frequency) for a given amino acid.
Interpretive certainty is never attained without experimentation but improves (up to a point) with more sequence data. Here it is important to check whether certain less common substitutions have persisted over evolutionary time in a phylogenetically coherent manner (ie a sub-clade) or are novel adaptations perhaps in conjunction with a co-evolving residue at another site (or another protein, perhaps even nuclear-encoded). After these considerations, the remaining rare changes are either deleterious or sequencing error. Polymorphism significance can be pursued at the xray structural level for only 3 of the 13 mitochondrial proteins (CYTB, COX2, COX1) and even this is complicated in the case of CYTB by its oligomeric association with 3 nuclear encoded proteins.
Aligning CTYB from the 70 complete yak mitochondrial genomes available on 1 Dec 10 shows variation at just 9 sites along the protein (ie 9 nsSNPs). These are quickly found when the web alignment tool retains input sequence order, displays residues identical to the top sequence as dots, gaps fragmentary data correctly, and allows a wide display permitting effective cross-species comparisons.
Yak and bison -- despite being sister species -- share variation only at one site, position 98. Here yak is exclusively valine with the exception of a single deleterious occurrence (see below) of leucine, whereas bison have a mix of valine and alanine (which otherwise is very rare at this position in mammals), ie the ancestral residue was valine. Thus no lineage sorting occurred at any amino acid position in CYTB at the time these species diverged. Lineage sorting however may be important in the overall evolution of the Bovini: 53 ancient polymorphisms (at the dna level) are said to have persisted since Bos and Bison diverged from Bubalus 5–8 million years ago.
The changes can also be displayed in context by coloring the appropriate residues in a reference sequence relative to a composite sequence consolidating all the polymorphisms from distinct animals (no one animal has more than two of the 9; V195A + I348F occurs in two animals). This latter sequence is quite useful in comparing polymorphism sites across species as explained in the annotation tricks section.
>CYTB_bosGruR Bos grunniens cytochrome b ref seq taken as gi|147744503
MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF
WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIITAIAMVHLLFLHETGSNNPTGISSDADKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI
LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQLASIMYFLLILVLMPTAGTIENKLLKW
>CYTB_bosGruP Bos grunniens composite polymorphisms: A017T A084T V098L I188T I192T V195A D214N V329M I348F
MTNIRKSHPLMKIVNNTFIDLPAPSNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHTNGASMFFICLYMHLGRGLYYGSYTFLETWNIGVTLLLTVMATAFMGYVLPWGQMSF
WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIITATAMAHLLFLHETGSNNPTGISSNADKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI
LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLMADLLTLTWIGGQPVEHPYFIIGQLASIMYFLLILVLMPTAGTIENKLLKW
A017T A084T V098L I118T I192T V195A D214N V329M I348F
927 A 4,994 A 4522 V 4309 I 94 I 4528 V 4429 D 4610 V 4232 I
4018 S 3 T 430 I 667 S 4353 L 427 I 512 N 188 T 651 V
46 T 1 P 34 M 14 I 505 M 25 T 43 E 133 A 63 T
3 L 1 V 11 A 1 T 31 T 4 G 8 S 44 I 45 M
3 M 1 L 3 F 4 M 2 Y 22 M 4 N
1 F 1 N 2 V 1 A 1 H 2 G 2 F
1 P 1 A 1 E 1 A
1 S
A017T: At position 98, the mammalian reduced alphabet consists primarily of serine with yak alanine also well represented at 18%. Threonine occurs in 46 sequences so cannot be sequence error or serious mutation. Bulk seems to be the main criterion at this site rather than polarity -- threonine though polar is bulkier residue than serine or alanine. To determine whether it has arisen multiple times or just in one clade, the phylogenetic distribution of the 46 occurrences needs consideration.
It can be seen from the graphic at left that A017T has arisen multiple times with no common denominator (such as high elevation lifestyle) but -- with the exception of monotremes -- never in a deep stem ancestor. That is, A017T occurs here and there but only in recently speciated clades. This suggests that while not lethal, over time it gets replaced by more adaptive serine or alanine.
A017T
927 A
4018 S
46 T
3 L
3 M
1 F
1 P
A084T: At position 84, alanine is strictly invariant. Thus threonine is an umistakable deleterious mutation in domestic yak.
A084T
4994 A
3 T
1 P
1 V
V098L: At position 98, the reduced alphabet consists of valine 90% of the time regardless of mammalian clade with the similar (branched chain aliphatic) isoleucine having substantial dispersed representation at nearly 9%. The 430 species in which it occurs are scattered incoherently within mammal clades, meaning that it has arisen independently many times. V098I may be slightly suboptimal as there is an evident bias (at some level) against equal occurence. It likely co-exists with valine in most non-bottlenecked populations of mammals, observed if enough individuals of a given species are sequenced.
However leucine, the seemingly similar third aliphatic residue, occurs one once despite being but a single base change transition away from the dominant residue. Were leucine a near-neutral substitution, its incidence would be vastly higher. Thus the change V098L reported for yak represents either a deleterious mutation or an unprecedented adaptation (eg to high altitude) or sequencing error in GenBank entry ACU82101. The same can be said for the more overtly radical change V098N in lemur AAS00156. The 34 methionines occur sporadically in the phylogenetic tree suggesting they are sub-adaptive and blink out over time. Indeed, canine spongiform leukoencephalomyelopathy is attributed to V98M. Dog CYTB is 89% identical to that of yak and numbering corresponds.
V098L 4522 V yak has most common amino acid at position 98 of CYTB 430 I 34 M 11 A bison 1 L yak 1 N lemur
I118T: At position 118, the reduced alphabet consists predominantly of ILV with some A and M, a very common occurence proteomewide. TSF are all deleterious mutations in domestic yak.
I118T
2597 I yak has most common amino acid at position 118
1843 L
404 V
87 A
61 M
6 T (all yak)
1 S
1 F
I192T: At position 192 of wild yak, the dominant residue is leucine instead of the yak ancestral value isoleucine, which is disfavored relative to methionine, ie isoleucine is a mild polymorphism in its own right but the associated taxonomy shows it narrowly restricted to 83 sequences in Bos, Bison, and separately in 5 Kobus (waterbucks), too persistent to be disfunctional and indeed a candidate for adaptive. However change to polar threonine is seen in 31 nominal species but after removal of redundancy, only in two species of pocket mice. Thus the yak change is deleterious.
I192T
94 I
4353 L
505 M
31 T
3 F
2 V
1 A
1 S
V195A: This allele occurs together with I348F in two wild yaks from a remote region in NW China. Despite sequence submission, no article has appeared in the three subsequent years. In can be seen from the reduced alphabet frequencies that this is a severe mutation but more likely sequence error, as is I348F.
V195A
4528 V
427 I
25 T
4 G
4 M
1 A
D214N: This polymorphism of wild yak is seen quite widely, in some 10% of mammals. The 223 taxa with D214N are mostly confined to laurasiatheres and glires but not a hallmark of these clades. Nor do the species with asparagine have any common lifestyle denominator. This is an acceptable variation at this site if perhaps not optimal.
D214N
4429 D
512 N
43 E
8 S
4 X
2 Y
1 H
V329M: This allele occurs in wild yak. Methionine is not a radical substitution in terms of chemical properties and various other similar amino acids appear at low levels, even though valine is in a huge majority of species. Methionine occurs sporadically in 17 other species include Bos javanicus, Ovis, Budorcas, Naemorhedus, Mus, Rattus, bats and sloth. Thus it is likely suboptimal but not significantly deleterious.
V329M
4610 V
188 T
133 A
44 I
22 M
2 G
1 E
I348F: This allele occurs together with V195A in two wild yaks from a remote region in NW China. Despite sequence submission, no article has appeared in the three subsequent years. In can be seen from the reduced alphabet frequencies that this is a severe mutation but more likely sequence error, as is V195A.
I348F
4232 I
651 V
63 T
45 M
4 N
2 F
1 A
Known human CYTB alleles
Polymorphisms for human CYTB have been very helpfully compiled by mtDB -- the Human Mitochondrial Genome Database. The numbering system works without change for bison and yak since no indels occur in this gene within mammals.
A number of these sites overlap, so any information about human disease associated with the human polymorphism raises issues for the fitness of identical or similar changes in these species. This neglects the possibly compensatory effect of changes elsewhere in this gene (or nuclear genes that interact with it).
Here color indicates human sites that have an allele of concern wild yak, domestic yak, or questionable yak sequences. In two significant cases -- both in domestic yak the initial and final residue of human are identical to that of yak, A084T* and I118T*. Here annotation can be transferred more reliably from what is known about these alleles in human to their expected effect in yak.
human yak H214Y D214N A329T V329M A084T A084T* I098V V098L I118V I118T I118T I118T* T2A S56A I117V D171N I211T G251S M316T A354T T2I S56L I118V D171G T212A E251D Y325H V356M M4V T61A I118T S172N T212I Y256H A329T V356A M4T T70A L121F P173S H214Y T257I A330T T360A R5G Y75C A122T T174A T219A L258P A330V T360M I7T I78V T123A F181L T219I A259T I334V T368A N8S I78T A125T I184V I226V N260D T336A T368I N15S L82F E136D L185S A229T V284I I338V I369V H16R A84T F140L I189V L230F V291A P342S I369T F18L G86S L149M A190T L233V S297P V343M I372V I19M C93Y I153T A190V F235L I300T V343A M376V A29T I98V Y155H A191T L236I I304T S344G A380T A39T G101S I156V A191D S238P I306V S344N A380V A39V Y109H I156T A193T S238F I306T Y345F ---- I42V E111K T158A T194A T241A M309V T348I ---- I42T T112A D159N T194V T241M M309T I349V ---- F50L W113R I164V F199L T243A S310P I349T ---- F50L I115T G167S I211V F245L M316V V353M ----
Kilo-sequence alignment tricks
New sequencing technologies have greatly affected the amount of mammalian mitochondrial genomic data available at GenBank. Five years ago, it was acceptable to publish population-level D loop sequences accompanied by a few fragmentary coding reads; today, a publication might offer 60-70 entire mitochondrial genomes. This favors evolutionary study of mitochondrial proteins over comparative genomics of nuclear genome products because the latter is still restricted to around 50 species (Dec 2010) almost all incompletely sequenced.
Many long-standing issues such as introgression, historic bottlenecks, population mixing, accrual of deleterious coding variants, hard polytomies, and lineage sorting during speciation can now be approached and resolved, especially with the increasing sequencing of end-Pleistocene frozen dna. This may allow more enlightened management of endangered species such as bison where populations reached rock bottom -- recovering numbers is not enough if genomic integrity is still at risk.
However, the flood of data raises significant issues in extraction of significant information: it is not instructive to align the tens of thousands of sequences available for each of 13 mitochondrial proteins -- that give a an intractable array of 3789 amino acids by 12500 sequences, enough to fill 20 x 100 = 2000 screens on the largest possible computer monitor. That data must be distilled down somehow to take-away information.
This section explains a practical desktop protocol for extracting the 'reduced phylogenetic alphabet' at each residue of the mitochondrial proteome. The method depends heavily on current capabilities of Blastp at NCBI and so may not be completely stable to changes made there over time.
First note that tBlastn cannot be used against the nr or wgs nucleotide databases at NCBI (or with Blat at UCSC) since the significantly different genetic code of mammalian mitochondria is no longer supported as a parameter option. Other oddities involve missing terminal nucleotides that are added before translation. However mitochondrial dna is usually translated sensibly at GenBank protein entries.
The vertebrate mitochondrial code:
TTT F Phe TCT S Ser TAT Y Tyr TGT C Cys
TTC F Phe TCC S Ser TAC Y Tyr TGC C Cys
TTA L Leu TCA S Ser TAA * Ter TGA W Trp
TTG L Leu TCG S Ser TAG * Ter TGG W Trp
CTT L Leu CCT P Pro CAT H His CGT R Arg
CTC L Leu CCC P Pro CAC H His CGC R Arg
CTA L Leu CCA P Pro CAA Q Gln CGA R Arg
CTG L Leu CCG P Pro CAG Q Gln CGG R Arg
ATT I Ile ACT T Thr AAT N Asn AGT S Ser
ATC I Ile i ACC T Thr AAC N Asn AGC S Ser
ATA M Met i ACA T Thr AAA K Lys AGA * Ter Bos can use ATA as initiation codon
ATG M Met i ACG T Thr AAG K Lys AGG * Ter
GTT V Val GCT A Ala GAT D Asp GGT G Gly
GTC V Val GCC A Ala GAC D Asp GGC G Gly
GTA V Val GCA A Ala GAA E Glu GGA G Gly
GTG V Val i GCG A Ala GAG E Glu GGG G Gly
AAs = FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIMMTTTTNNKKSS**VVVVAAAADDEEGGGG
Start = --------------------------------MMMM---------------M------------
Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG
Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG
Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
After collecting high resolution amino acid frequencies at a given site, it is necessary to determine the phylogenetic distribution of each variant (in practice just those of moderate occurrence). That is now very convenient to do provided the associated accessions have been saved:
Simply paste the blastp match list of protein accessions having the chosen amino acid variant into the Entrez text query box. Never mind if it only returns 20 out of your 157 input sequences -- it hasn't forgotten. It doesn't matter if the list has redundant entries (typically SwissProt and the protein giving rise to the SwissProt entry). After retrieval, set the "Find Related Data" to "Taxonomy" and wait for the options to load, then click "Find Items".
Miraculously, this returns a page that can be set to display a text phylogenetic tree your input sequences, the full set entered with all redundancy removed. That text tree has labelled higher taxonomic nodes and individual species deeper down. Final edits can be made quickly that capture the phylogenetic spread of the variant allele for interpretive purposes.
The two most common outcomes:
- all the species carrying the variant comprise a monophyletic clade. If the origin of the clade is fairly ancient, then the variation is a derived informative adaptive change relative to ancestral (synapomorphy). If the site is invariant in all members of the co-clade (meaning the ancestral state has persisted to all other extant species), then the site is a phyloSNP (definition and examples: 1 2 3 4).
- species carrying the variation are scattered incoherently across the mammalian phylogenetic tree. This means that the variation has arisen multiple times (all fairly recently) but has not persisted when it arose earlier, ie it is not a preferred allele for this protein at this site and gets replaced.