Bison: mitochondrial genomics
Update blog
The notes below describe significant fixes and additions in reverse chronological order and link to their sub-section. This makes it easier to find recent changes within the long document. Comments and suggestions for improving the article are welcome and can be emailed here.
16 Jan 11: trimmed down article to 5-6 pages for journal publication later this month 14 Jan 11: compiled bibliography of 163 articles, pdf, reports, master theses, and scanned older texts 13 Jan 11: re-analyzed unpublished Gardipee YNP and GTNP bison RFLP and sequence data, finding horrific levels of implied mitochondrial disease 12 Jan 12: worked on all-vs-all yak vs bison whole genome comparisons and yak-vs-stepper bison control region comparisons 11 Jan 11: added all-vs-all yak whole genome blastn comparison; spellchecked document; updated management recommendations 10 Jan 11: added all-vs-all whole genome blast comparisons of micro-haplotypes 9 Jan 11: included details on two fossil bison with possible mitochondrial disease 8 Jan 11: improved table content and ordering 7 Jan 11: fixed treatment of control region data 6 Jan 11: raised disease status prediction to 130 animals 5 Jan 11: characterized steppe bison sequencing error types 4 Jan 11: reconstructed ancestral bison control loop sequence using steppe bison and yak as outgroups 3 Jan 11: wrapped up mutational analysis of the 22 mitochondrial tRNAs for both yak and bison 2 Jan 11: simplified table of tRNA stem region changes and added explanation of how it was made 1 Jan 11: added cybrid genetic engineering section to steppe bison
Introduction to bison and yak conservation genomics
Bison and wild yak are but two of many genomically endangered species impacted by past and present human activities: historic population bottlenecks (from overhunting and take of habitat) and unnatural selection from uninformed culls, loss of best bulls to trophy hunting, gender imbalance practices, interference with predator selection, competition for forage, breeding opportunities, and disease resistance, selection for docility, and introgression from inbred domestic animals unfit for the wild.
Entire mitochondrial genomes on a population level first became available in December 2010, with several dozen sequences now available for both bison and its sister species yak. A nuclear genome for cattle is available now and one is underway in China for yak, with a genome expected for bison by 2016. In the meantime, a very extensive SNP bead chip allows querying of the nuclear genome on a herd scale. Thus for the first time, it has become possible to consider the genetic status of the herd and make rational conservation management decisions. It is the genome that must be conserved -- humps and shaggy appearance will follow.
The expected genetic impacts of a population expanding out from a severe bottleneck include undesirably high frequencies -- or even total fixation -- of maladaptive amino acid alleles originally present as rare mutations in the founder population (eg all wolves on Isle Royale descend from two that crossed a rare ice bridge in 1949, the mitochondria descending from a wolf-coyote hybrid; their idiosyncratic genomes providing the new allele frequencies). Deleterious mutations can be reliably identified and distinguished from adaptive change or desirable miscellaneous genetic diversity by the comparative genomics techniques described below, provided sufficient sampling data is available.
Mitochondria encode 13 distinct proteins central to energy metabolism. Well-studied human and canine mitochondrial diseases associated with specific mutations in these proteins give rise to clinical conditions, typically exercise intolerance for cytochrome b. A polymorphism at any site in this gene can currently be compared across 1300 mammalian species with individual animal multiplicities bringing the total feasible comparison to over 12,600 sequences.
With such an incredible data set and a fully resolved mammalian phylogenetic tree, the admissible amino acid spectrum (reduced alphabet) is defined to very high sensitivity. Although the function of each residue is seldom entirely known, this reduced alphabet has already been thoroughly vetted by a hundred million years of placental evolution and suffices to evaluate variation in moderately conserved genes. It is not currently possible to attain cutting edge sensitivity for nuclear encoded proteins because data from only 55 vertebrates might typically be available.
However mitochondrial inheritance has nine complexities that strongly affect conservation genomics:
- mitochondrial dna is maternally inherited, meaning that any and all mutations in bull mitochondrial dna are lost in their descendants but any hybrid resulting from cow introgression retain strictly cattle mitochondrial dna that persists indefinitely without any prospects for dilution by back-breeding within a bison herd.
- although mitochondrial dna is present in very high copy number in bovine oocytes, the strands are effectively non-recombining, meaning no prospects exist for compiling good variations from the multiple haplotypes present in an individual animal (heteroplasmy).
- mitochondrial dna can be erratically replicated (not proportionally to haplotype abundance), allowing copy number of mutation-bearing mitochondria dna to surge (or fall) unpredictably relative to residual wildtype haplotypes both in oocytes and somatic cell lineages.
- stochastic segregation of mitochondria at cell division both in terms of haplotypes and number of mitochondria inherited by daughter cells, a bottleneck effect.
- mitochondrial dna sequences at GenBank do not describe the germline oocyte haplotype proportions but rather are taken from leucocytes, skin or muscle whose polymorphisms are not necessarily applicable to germline inheritance. It is only when the same mutation surfaces in multiple animals in a phylogenetically coherent clade that sporadic mutations (possibly somatic) can be distinguished from stably heritable mutational haplotypes (where a given mutational haplotype has expanded to become the only haplotype present).
- in the heteroplasmic case, only the predominant haplotype in the tissue sampled will get reported, even though the dire nature of some mutations and the essentiality of the cytochrome b imply internal compensation by unreported wildtype haplotypes must be occurring at some level.
- functional compensation could conceivably occur within a single mitochondrion carrying multiple haplotypes, one of them wildtype or for that matter between an allele of an imported nuclear gene serving as part of the mitochondrial oxidative phosphorylation complex. The bc1 complex involves 11 gene products, with all but cytochrome nuclear encoded.
- though the mutation rate in mitochondria is high and hotspots may exist, actual homoplasy (recurrent mutation) is rare. That is, it is fairly uncommon for the same amino substitution to occur in the oocyte, much less surface from low heteroplasmy to full heritability. This can be seen either from the low occurrence of the same mutation across tens of thousands of sequences and also from human mitochondrial disease statistics. However it is not unusual for multiple haplotypes to wax and wane across a species divergence, with sampling artifacts than picking one of these out in preference to others, giving the appearance of a fixed substitution.
- lineage sorting of haplotypes at the time of speciation is quite different from nuclear genes because of heteroplasmic persistence, making the determination, indeed definition, of ancestral state quite difficult, though that remains important in establishing the fully functional amino acid alphabet at a given position.
The complexities of heteroplasmy make sequence data difficult to interpret and inheritance of mitochondrial polymorphisms problematic to predict, much less affect by management. Cattle have been specifically studied, with those results probably transferable to bison and yak. However mitochondrial disease has proven exceedingly difficult to understand even in human and cannot be treated.
"Heteroplasmy is the presence of a mixture of more than one type of an organellar genome within a cell or individual. It is a factor for the severity of mitochondrial diseases, since every eukaryotic cell contains many hundreds of mitochondria with hundreds of copies of mtDNA, it is possible and indeed very frequent for mutations to affect only some of the copies, while the remaining ones are unaffected."
GS Michaels 1982: "Restriction endonuclease analysis and direct nucleotide sequencing of bovine mitochondrial DNA have revealed a high apparent rate of sequence divergence between maternally related individuals. Oocytes had 260,000 dna genomic copies per cell, whereas primary bovine tissue culture cells contained only 2,600 copies. These experiments ... are consistent with models which generate mitochondrial DNA polymorphisms by unequal amplification of mitochondrial genomes within an animal/"
"Mitochondrial diseases arise frequently: 1 in 4000 individuals is at risk of developing a mitochondrial disease sometime in their lifetime. Half of those affected are children who show symptoms before age five, and approximately 80% of them will die before age 20. The mortality rate is roughly that of cancer... The mutation rate of the mitochondrial genome is 10–20 times greater than of nuclear DNA, and mtDNA is more prone to oxidative damage than is nuclear DNA. Mutations in human mtDNA cause premature aging, severe neuromuscular pathologies and maternally inherited metabolic diseases, and influence apoptosis."
An alarming situation has arisen in North American bison at position 98 of cytochrome b. A majority of animals sampled to date (17 of 33, none hybrids, one from Yellowstone) have alanine at this position, a seemingly innocuous but -- as shown below -- clearly a deleterious change from wildtype valine, which is otherwise invariant here throughout mammals and indeed vertebrates (ie unchanged over a hundred billion years of observed branch length). Note canine spongiform leukoencephalomyelopathy arises from the very similar V98M.
The single basepair change resulting in V98A suffices to define the same two major clades of bison established by whole genome comparison of 16,322 bp. The A98 mutation evidently arose in a single female bison, expanded over time to become the sole haplotype in female descendants who then provided -- through the bottleneck effect -- all mitochondria of the vast dispersed herd corresponding to the A98 clade. The rise of a maladaptive allele is not surprising in view of human interference with natural selection.
The V98A mutation is considered at greater detail below, along with 5 mostly deleterious sporadic haplotypes of lower frequency (lesser concern) and 10 other sites where all bison sequences differ from the ancestral amino acid at the time of divergence from yak. These latter probably reflect heteroplasmic lineage sorting of haplotype frequencies though two substitution I316M and M353L are of some concern.
While here only one of 13 mitochondrial proteins is considered below, disturbing findings have been reported for bison mitochondrial tRNAs. This raises the question of the current severity of genetic burden of all endangered wildlife species, not just bison. It may prove very difficult to recover these species to their previous adaptive genetics.
Management options for mitochondrial dna genetics begin with hybrids. Here bison or yak with cattle mitochondria will also have cattle nuclear gene introgression (whose dilutional state today depends on subsequent backcrossing history); this follows without testing for all progeny. Bison residing in fenced preserves are not commonly limited in population size by predation, disease or winter starvation. Since vegetation productivity will only sustainably support a certain population, removal of surplus animals could emphasize female hybrids, as any desirable authentic nuclear genome diversity can be carried forward by bulls (provided the herd is not gender-imbalanced). Because of cheap and reliable testing, cattle mitochondrial introgression may soon be a thing of the past for confined herds under conservation-minded management.
Given the complexities of mitochondrial inheritance however, even in pure bison no selective breeding strategy may exist should multiple mitochondrial genes have widespread adverse polymorphisms, perhaps leaving all surviving haplotypes adversely affected one way or another. Needless to say, alleles of the 20,000 nuclear genes have to be considered at the same time.
Phylogeny: bison and yak are sister species
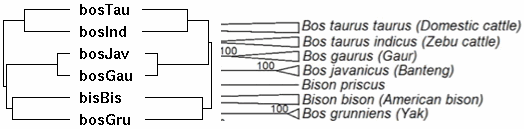
Bison genomics is best considered within its phylogenetic context. This means first of all parallel consideration of its sister species (nearest living relative) the yak. Although not tropical, both species were dramatically affected by closing of the Darien gap in Panama at 2.5 million years and ensuing unstable climatic change. This led to Pleistocene ice ages: episodic glacial barriers isolating regional herds yet promoting repeated dispersion across Beringia as sea levels fell. Those events manifest today as deep bifurcations of the mitochondrial phylogenetic tree of both species.
However a broader phylogenetic perspective is also essential to provide the outgroup sequences that influence ancestral sequence reconstruction. Here the evolutionary history of cetartiodactyls has taken decades to sort out: the position of whales, once controversial, has been settled (sister, together with hippopotamus, of Ruminantia), as has the non-intuitive branching order of pigs and lamas (Camelidae are basal).
Within pecoran ruminants, difficulty arises not so much from conflicts between fossils morphology and molecular trees but rather rapid radiation of species (hard polytomy), only recently resolved (we hope and assume below) with the bovine SNP bead chip. This samples nuclear genes vastly better than homoplasy-prone microsatellites and sidesteps limitations of mitochondrial inheritance.
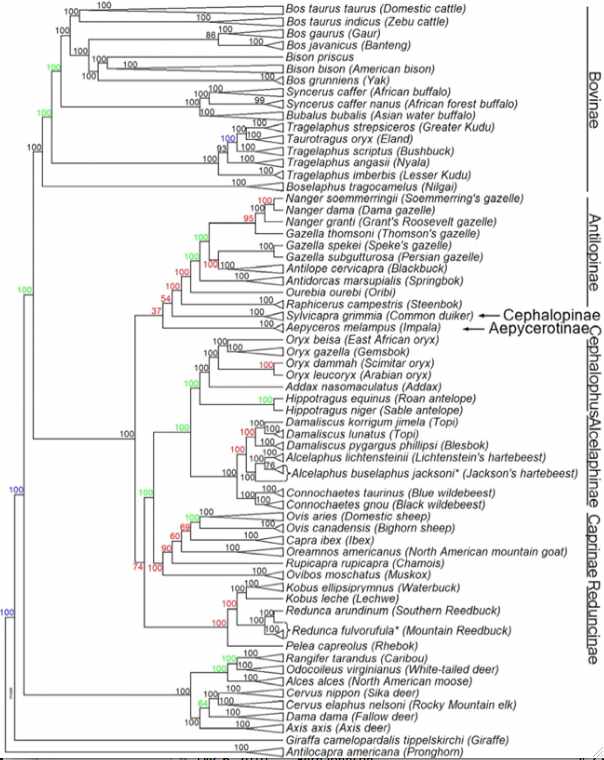
In the figure at left, JE Decker et al evaluated 52,356 sites across the nuclear genome not only of cows but throughout ruminants. The resulting tree (antelopes,(giraffes,(deer,((gazelles,sheep),bovinae)))) is critical to understanding the evolution of mitochondrial proteins and evaluating amino acid substitutions -- which are of grave concern for conservation of bottlenecked species such as bison and yak.
Notice that Linnaean taxonomy requires substantial revision according to the tree below -- genera such as Bos, Tragelaphus and Gazella are inconsistent with it. This could be remedied for bison by either placing them in Bos or putting yak, gaur and banteng in the genus Bison. Here the position of gaur and banteng has less bootstrap support than other nodes and has long been contentious. It is shown below that the mitochondrial proteome of Bos javanicus associates with cattle at all informative sites so this species cannot be sister to bison and yak as shown in the figure. However nuclear and mitochondrial genes can give different species trees. The position of kouprey and mithun (gayal), Bos sauveli and Bos frontalis, have not been analyzed with the bead cheap and little mitochondrial proteome data is available.
There may not be any simplistic nomenclatural resolution because of male introgression as illustrated in european bison (wisent) and zebu cattle. The speciation process is far messier than suggested by bifurcating tree nodes. For example, subsequent to some measure of genomic divergence, wandering bulls from one population can join another or mixed herds of wild taurine cows form. While this does not affect mitochondrial lineages, it does result in periodic introgressions into the nuclear genomes. Since Holocene domestication, cattle have hybridized with aurochs, yak and bison, indicating full speciation barriers still do not exist. Polymorphic alleles represented in an ancestral population at various frequencies may sort out differently in descendant lineages, though this plays out quite differently for nuclear and mitochondrial genomes.
The data situation is otherwise very favorable with over 214 mammalian species having sequenced mitochondrial genomes, with high multiplicities for some individual species such as eland, cow, bison and yak. Individual genes such as CYTB may have extensive additional data from targeted studies. However all data, especially fragmentary older GenBank entries, must be carefully screened for errors and implausible sequence anomalies.
The table below makes no nomenclature proposals whatsoever but simply describes the heuristic terminology adopted here -- driven by that used, right or wrong, at GenBank Taxonomy -- because only that can be used to restrict the blast searches necessary for comparative genomics. To open all 72 article abstracts, click here. Free full text is available for 27.
Acronm Species Common Mito CYTB NucG PubMed bosSau Bos sauveli (kouprey) 0 5 0 15522811 16439342 17848372 bosFro Bos frontalis (mithun gayal) 0 16 0 20331596 20433524 18244904 17560527 bosGau Bos gaurus (gaur) 0 17 0 19436739 19777782 19367625 17986322 bosJav Bos javanicus (banteng) 2 39 0 18937038 18937038 17614913 16922247 12522420 bosTau Bos taurus (cattle) 168 500 1 19603063 19484124 19393053 19393048 19393045 20347826 bosPri Bos primigenius (auroch) 1 17 0 18199470 19456314 20346116 bosInd Bos indicus (zebu) 3 387 0 12648092 19436739 19770222 20597883 18467841 12399392 bosGru Bos grunniens (yak) 72 53 * 19917041 17257194 18439980 16942892 12137333 bisBis Bison bison (plains bison) 33 7 0 20870040 20637048 19414501 bisAth Bison athabascae (woods bison) 2 3 0 20808568 18191321 bisBon Bison bonasus (wisent eurobison) 4 9 0 14739241 19623210 17177698 15125253 14703870 bisPri Bison priscus (steppe bison) 0 0 0 15567864 20409351 20212118 18653730 18199470 bisAnt Bison antiquus (ancient bison) 0 0 0 17256570 9826742 17686730 bubBub Bubalus bubalis (water buffalo) 4 342 0 17459014 15621663 11212504 19140976 19462514 19207933 synCaf Syncerus caffer (cape buffalo) 0 10 0 10603253 9126673 9987926 17313588 17459014 14715223 traScr Tragelaphus scriptus (eland) 0 172 0 10222159 7723053 17520013 traSpp Tragelaphus others (7 spp eland) 0 7 0 10380679 bseTra Boselaphus tragocamelus (nilgai) 0 3 0 10603253 17158073
In the table, sequence availability counts do not include poor quality fragments or inadvertent hybrid data, eg 13 nominal Bos frontalis entries are instead introgressions from Bos indicus and misplaced at GenBank.
Yak nuclear genome sequencing is in progress at Beijing Genomics Institute. Other cetartiodactyl genomes in progress include Camelus bactrianus and Ovis aries with Camelus dromedarius and Pantholops hodgsonii completed but not released. Other relevant genomes said to be underway include Bubalus bubalis, Addax nasomaculatus, Muntiacus muntjak, Hippopotamus amphibius, and Balaena mysticetus. Cow, pig, sheep, and vicuna genomes have long been available for blast search.
These additional genomes would allow fossil nuclear numts to contribute to understanding of mitochondrial gene evolution, making the mitochondrial proteome of ancestral species such as Leptobos (last common ancestor to cattle, bison and yak) easy to work out. Note too that the mitochondrial genome, although not targeted, gets sequenced to very high multiplicity as a byproduct. To date, such projects have produced single mitochondrial genomes. This however is surely wrong in view of the prevalence of heteroplasmy: most species host a population of significantly different mitochondrial genomes. Thus these genome projects are a golden opportunity to characterize mitochondrial genome diversity within single species.
GenBank sequences are often retrieved blindly and run through extensive software pipelines to provide some conclusion. However it is imperative to manually curate accessions prior to analysis because a certain percentage of legacy entries are completely inappropriate. This ranges from attribution to the wrong species, gross and subtle sequence errors, reduced reliability at sequence termini, redundant entries, unpublishable submissions from third-world countries, mixups of mitochondrial and nuclear dna, lab dna contamination, text processing mishaps during the submission process, to outright data fraud. Below, bison and yak and their contextual species are considered individually.
- Both complete and fragmentary aurochs (Bos primigenius) accessions condense to two sequences sufficient to represent all GenBank aurochs data on 8 Dec 10 namely ACE76876 ADE05539 which differ as I4F T23A V372I (latter two changes are sporadic for ACE76876). Aurochsen became extinct in 1627 due to overhunting and the loss of habitat. Their mitochondrial genome still persists in a few Italian and Korean cows.
- The nine GenBank sequences for european bison (wisent) condense to a single representative sequence, for example ADF29596. Here it must be noted that ADQ12704 has a terrible sequencing error introducing ETTAEF for VNYGWI -- unfortunately this sequence has been used uncritically in published analyses. CAA75238 is also defective distally, a poor quality sequence from 2005 that was never published. Blast shows beyond any doubt that the known wisent sequences are not remotely affiliated with bison but instead are Bos taurus (not even aurochs Bos primigenius). It has long been suspected that wisent originated from a bison bull naturally crossed with a taurine cow. It follows wisent mitochondrial genomes will not be terribly informative for bison or yak.
- The extinct steppe bison, Bison priscus, has no protein sequences among its 298 GenBank entries, only control regions. Complete mitochondrial genomes from this species would be very informative -- evidently the dna is readily collected.
- The kouprey Bos sauveli has five CYTB sequences but only one full length, AAV51239. Two fragmentary entries are polymorphic relative to this at T248I, namely ABB88561 ABN73101. Here care must be taken as kouprey bull x banteng cow hybrids are known, causing confusion as to kouprey status as distinct wild cow species.
- Domestic cattle have a vast amount of sequence data, much breed-specific. The detail anomalies of inbred animals are not especially informative to wild bison or yak. Since however many of the observed cattle substitutions are radical chemical changes at highly conserved sites in a vital enzyme, the question arises as to how these animals survived to adulthood. The answer is probably heteroplasmy, with late onset, that is compensation via wildtype mitochondrial dna that persists in some mitochondria to some extent. Exercise intolerance -- a common outcome of human cytochrome b deficiency -- would hardly be noticed in a cow prior to the animal's arrival at a slaughterhouse. Two CYTB sequences have very high multiplicity, represented by AAM12814 AAW78524 at 208 and 71 copies. The latter differs at V356I I372V. The single-site polymorphisms shown below arise in AAV88174 BAC54760 AAZ16727 AAW83829 AAT80776 AAV88122 AAS93073 AAZ16896 AAT80776 AAZ95368 AAZ95339 AAS93061 AAM08329 AAZ95354 AAZ16545 AAZ95379 AAZ95338 AAZ95338 ABV70594 AAW78531 AAZ95334 AAZ95385 AAZ95378 ABV70763 ACQ73865 ACQ73761 AAZ95359 AAZ17091 BAA07016 AAV88161 ABV70555 AAZ95331 AAV88135 AAZ95405 AAZ95389 AAV88187 AAZ95386 AAZ16688 AAZ95339 AAQ06605 BAC20256 ACQ73813 AAW78527 respectively.
MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAILRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMMFRPLSQCLFWALVADLLTLTWIGGQPVEHPYITIGQLASVLYFLLILVLMPTAGTIENKLLKW ................T..........................................................D...W...................................................L.........................................................K.........................................................N..............................................V.....S....................................................................S......... .................L...........................................................M............................................................................................N..................T...............................................................N...............................................T....................................................................AV....... ......................T......................................................V...................................................................................................................T....................................................................................................T.........................T...S....T.........................I...............V....... ..............................S..............................................V....................................................................................................................I.................................................................................................................................S....T.........................I....................... ................................................................................C.............................................................................................................................I.......................................................................................................................T.................................................... .........................................................................................V....................................................................................................................I....L.....................................................................................................................................S................................. ...................................................................................................G..................................................................................................................A....................................................................................................................................T............................... ......................................................................................................N...............................................................................................................A......................................................................................................................................F............................. ...............................................................................................................A......................................................................................................M............................................................................................................................................I....................... .....................................................................................................................L.................................................................................................N.........F......................................................................................................................................................... .......................................................................................................................................................................................................................................T...................................................................................................................................................
- The gayal (or mithun) Bos frontalis has 28 full length CYTB sequences. These fall into two very distinct groups, suggesting introgression of female mitochondria from another species. According to blastp, this species is Bos taurus or Bos indicus, a conclusion also reached for Yunnan gayal. Here ABO07421 most parsimoniously represents the first group should that be desired, with ACF17717 BAJ05325 identical, ABO07426 differing by a sporadic L376V, and ACN12147 differing by F296L and K375N. A derived subgroup has I356V and I372V and sporadic A291V, namely ABO07428 ABO07427 ABO07425 ABO07422 ACF17716 ABO07419.
- The second group of 16 near-identical gayal sequences can be represented by ABO07423. This set contains four sporadic mutations N3S D252N F276L I298L and two sites of shared polymorphism with the first group, T232A K375N. It differs consistently from the first group at 6 sites, I39V V215A A232T A302I A327T L357M and so bears much closer relationships -- given the strong conservation of this protein -- to Bos gaurus and secondarily Bos javanicus (2 and 4 differences respectively) than to Bos taurus or Bos indicus (6 differences at best). Only this second group is usefully included as an outgroup to yak and bison.
- Thus the first choice for Bos frontalis conservation genomics -- based solely on CYTB -- involves animals represented by the second group ABO07424 ACF17720 BAJ05320 ABO07423 ABO07420 ACF17718 ABS18292 AAV51237 BAJ05321 BAJ05322 ABS18291 with possible inclusion of ABO07418 ACF17719 for diversity but not sporadics BAJ05323 BAJ05324 ACM24710 unless other considerations warrant it. Based on skimpy GenBank entries, these animals are called Dulong cattle in China but mithun in Myanmar and Bhutan. This is apparently corroborated by a 2010 study utilizing 16S mitochondrial rRNA. Nuclear genes also are very important to consider.
- Bos gaurus has 17 entries including 3 where a nucleotide was submitted but not a translation (causing protein queries to miss them). After observing that the fragmentary sequences where not flawed are merely supportive, the set can be pruned to six. However two of these (ABF20228 ABF20227) are actually maternal Bos indicus/taurus sequences. The remaining four are practically identical to Bos javanicus/frontalis but differ from each other at 6 sporadic sites V39I A62V Y95H T108P L105P T190M N206I. This species will not prove useful to bison/yak comparative genomics but one sequence ADB80894 is retained below.
- The banteng Bos javanicus has an excellent set of complete sequences among its 35 entries for cytochrome b. After noting sporadic variation and checking for hybrids, a set of three sequences ABS18295 ABW82495 ABW82494 suffices to represent population diversity. Banteng do appear quite diverse, with several substantial variants supported by sequences from multiple individuals. Some clearly deleterious mutations are also evident, such as R80W in ADC53249. Sequences such as ABW82495 are peculiar in having 8 substitutions, suggesting a hybrid, yet with what is unclear: possibly a remote ancestor of Bos taurus or some extinct lineage not otherwise represented today. This sequence is supported by AAV51238 BAA11625 BAA07017 and so cannot be sequencing error; their disparate GenBank entries do not provide locational information.
- For the zebu, Bos indicus, 20 full length sequences are available (in addition to hundreds of fragments not considered further). These however are all identical with the exception of a sporadic variation T67I in ABS18290. Thus ABO07435 can serve to represent this species. It differs from the most abundant Bos taurus allele (208 entries) at only two distal positions I356V and V372I.
- Bison, yak and cattle have buffaloes as outgroup. Here Syncerus caffer (cape buffalo) has 10 CYTB sequences, only 2 of which are informative, AF036275 BAA11624. The latter differs at H3N T56S I295V.
- An extraordinary amount of data exists for water buffalo (Bubalus bubalis) -- some 165 CYTB sequences (after dropping defective entries ABO20788, ABO26586, BAJ05824 and discarding boundary variation of fragmentary sequences) of which 44 are essentially full length. However very little polymorphism occurs. In the first half of the molecule, 8 sites exhibit variation but only in unique individuals, making it impossible to distinguish sequencing error from authentic one-off events(which themselves could be non-heritable heteroplasmy. This is remarkably low (0.02%) in an alignment with 165 x 190 aa = 31,350 residues.
- The second half of Bubalus cytochrome b exhibits higher variation. Three individuals carry A191G, 28 have T246A, five are I365V and seven I372V, in addition to eight scattered sporadic variations. All the I372V individuals -- chinese water buffaloes -- are also T246A. The remaining 21 T246A animals apparently originated in China, Japan and Thailand but details remain unpublished. Non-sporadic variation in water buffalo is satisfactorily represented by GenBank accessions ACF17726 ABR08397.
- Syncerus is surprisingly diverged from Bubalus (12 positions): L102M T122A N159S I195V S246T I290V I293L L320F D331N M357T T371M. Only two of these positions are polymorphic in cape buffalo H3N and I295V; water buffalo are all 3N and 295V making those ancestral, with no indication of lineage sorting. This species is satisfactorily represented by AF036275 BAA11624.
- Can there be too much data? GenBank carries 172 CYTB sequences for Tragelaphus scriptus and its 30 subspecies (sylvaticus, uellensis, signatus, scriptus, simplex, sassae, roualeyni, punctatus, powelli, pictus, phaleratus, ornatus, meruensis, meridionalis, meneliki, massaicus, locorinae, knutsoni, johannae, heterochrous, haywoodi, fasciatus, dodingae, dianae, delameri, decula, dama, cottoni, bor, barkeri). However only two of these are full length, AF036277 AAD13501 (and differ at 7 sites) with the rest older and running from residue 138 to 232. Despite dropping poor quality sequences, considerable variation remains, both of sporadic and sub-clade type. To track this without sequences proliferating too much, a third quasi-sequence consisting of AF036277 substituted in silico with all major non-sporadic alleles -- which cannot represent sequencing error -- was made below, called CYTB_traScr3.
- Seven other species of Tragelaphus also have full length sequences available -- T. eurycerus, strepsiceros, imberbis, oryx, angasii, spekii, and derbianus. These sequences are moderately diverged from each other. They are fairly old in terms of sequencing technology used - 1999. Nonetheless, AAD51427 AAD51431 AAD13498 AAD13491 AAD42706 CAA10935 AAD13496 have been added to the sequence base below to represent this diversity. Tragelaphus is a large and important outgroup for bison/yak/cattle.
- Five of seven posted sequences for Boselaphus tragocamelus (nilgai) are poor quality fragments, illustrating a pitfall for blast searches. However the two full length sequences are in complete agreement. Here CAA10934 will be taken as reference sequence.
The goals here are to reduce the clutter from redundant sequences allowing an informative final alignment without discarding significant allele data or losing track of species multiplicities. This information can be retained within the alignment by a carefully designed fasta header. (Some web tools cut off the header at 10 characters but others allow any length.)
Interpreting bison CYTB variation
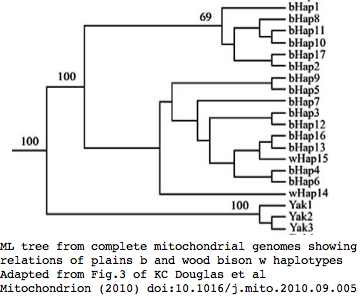
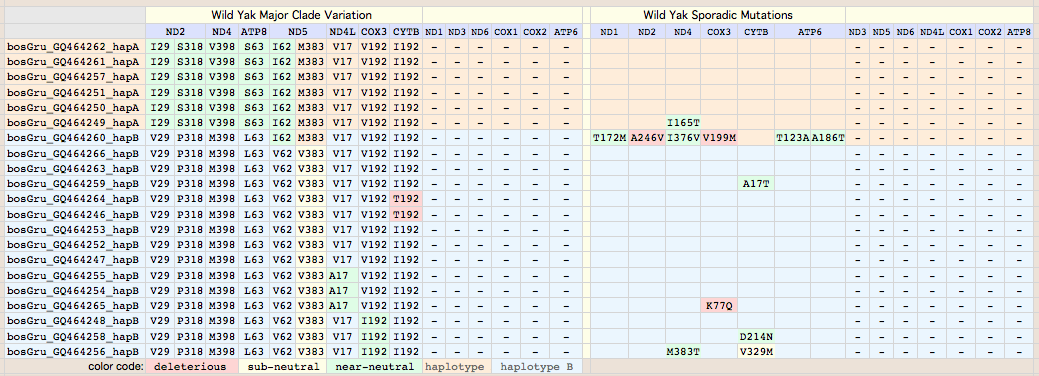
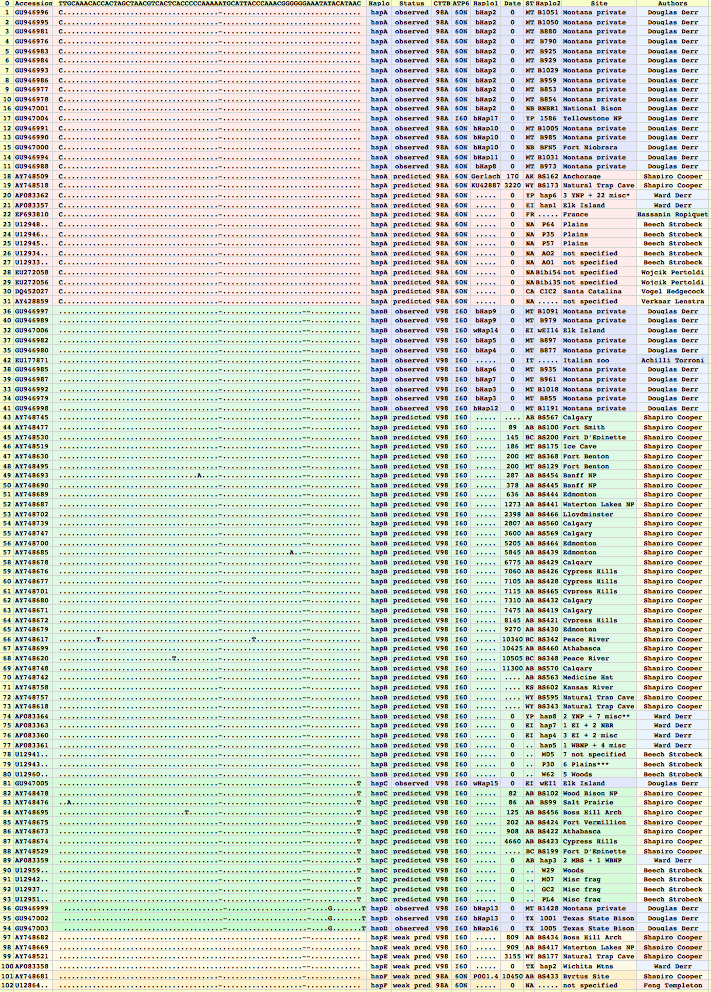
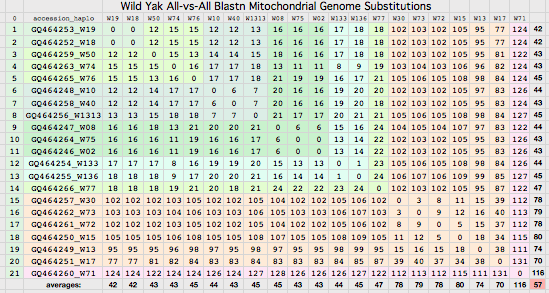
Bison mitochondrial genomes are well-represented at GenBank because of a Dec 2010 release by the JN Derr group of 31 complete genomes (along with various cow-bison hybrids and cow breeds) from 6 herds including two woods bison (sometimes denoted Bison athabascae) from a wood bison herd in Elk Island, Canada that was not historically admixed with plains bison. Their mitochondrial genomes did not however form a separate clade expected of a distinct taxon.
Cattle-bison hybrids represent crossing a bison male with domestic cow (or rather a continuous line of female descent from such a cross) and so have strictly cow mitochondrial dna, not relevant here because wild yak and aurochs provide more appropriate outgroups than a domesticated animal. Note however the haplotype of all bison hybrids studied cluster with cow haplotype cHap32 which may shed light on the historic cow lineage involved in late nineteenth century cattalo experiments. (The Derr group also posted a complete mitochondrial genome HQ223450 from European bison on 15 Nov 10 that -- like all to date -- is a taurine hybrid.)
Bison CYTB protein accessions: wood bison ADF48936 ADF48949 ADF48962 ADF48975 ADF48988 ADF49001 ADF49014 ADF49027 ADF49040 ADF49053 ADF49066 ADF49079 ADF49092 ADF49105 ADF49118 ADF49131 ADF49144 ADF49157 ADF49170 ADF49183 ADF49196 ADF49209 ADF49222 ADF49235 ADF49248 ADF49261 ADF49274 ADF49287 ADF49300 ADF49313 ADF49326 Earlier bison protein accessions: ABV70945 AAD51424 (ABV70945 complete genome: YP_002791041 derived from it; AAD51424 complete gene only) AAW28804 AAW28803 AAL85955 (fragmentary) ADM87433 (uninformative fragment) AAN28295 (taurine hybrid poor quality) Non-redundant protein set (with multiplicities): pick one from each row 18 98A: ADF49092 ADF49170 ADF49118 ADF49248 ADF49131 ADF49300 ADF48936 ADF48949 ADF48962 ADF49001 ADF49027 ADF49040 ADF49157 ADF49183 ADF49196 ADF49261 ADF49066 AAW28803 (frag) 1 98A V132D: AAD51424 1 98A Q322R: AAL85955 (frag) 13 V98: ADF49105 ADF49209 ADF49014 ADF48975 ADF49144 ADF49222 ADF49287 ADF49235 ADF49274 ADF49313 ADF48988 ADF49053 AAW28804 (frag) 1 V98 N3S: ADF49079 1 V98 I42T: ABV70945 1 V98 V123M: ADF49326
Thus for comparative genomics purposes, all available authentic bison cytochrome b data on 11 Dec 10 can be represented by just three sequences (one a constructed composite of all polymorphisms). This facilitates comparison of amino acid variation with yak and other species. The fasta headers are designed to display informatively after alignment. Apart from V98A, the other 5 variations are sporadic (observed in only one animal to date). They are analyzed in great detail below to determine which are deleterious mutations.
CYTB_bisBis_V98 wild type MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis_98A major variant MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis_all N3S I42T V98A V123M V132D Q322R all-allele composite MTSLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLTLQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTMMATAFMGYDLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSRCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW
The alignment below shows bison CYTB aligned against its nearest living relatives within Bovinae. Data from nearly a thousand individual animals are compressed without significant loss of information into the 28 lines of the alignment. The order of species corresponds to the topology of the phylogenetic tree, facilitating interpretation of individual sites in bison (or its sister species yak). If a residue is invariant in the preceding 4-5 levels of outgroup but changes to another amino acid in bison, that change needs detailed evaluation. The main possibilities are:
- near-neutral wander within the acceptable reduced alphabet for that site (blue) with modestly increased sampling likely to reveal the outgroup value within bison and lineage sorting likely as the site is persistently polymorphic
- deleterious mutations (red) that nonetheless persist in bison due to population bottleneck expansions or drift to unnaturally high frequencies under non-adaptive management. This includes private polymorphisms affecting one known animal, semi-population level changes such as V98A and changes fixed since divergence from yak.
- synapomorphic change in cytochrome b (green) at the bison/yak divergence node, possibly adaptive (improving fitness relative to environment) but more likely just sites affected by lineage sorting of a reduced alphabet present in the ancestral population.
These substitutions are then discussed individually below using an advanced nsSNP evaluation protocol that considers the physical-chemical nature of amino acid change (Grantham value and later refinements such as PolyPhen2), site-specific phylogenetic tree-aware comparative genomics (along the lines of TreeSAAP), and clade pattern analysis (ie random dispersement or sub-clade persistent of synapomorphic or phyloSNP type) of homoplasic occurrences of the change elsewhere in mammals.
While nsSNP interpretation can never be perfect, here the analysis will be extraordinarily reliable for two reasons: the truly massive data set that exists for this particular protein (12,603 sequences in 1,637 mammals utilized below) and the relatively slow evolution of CYTB (still 83% identity between bison and platypus/echidna proteins) that allows the data set to retain applicability.
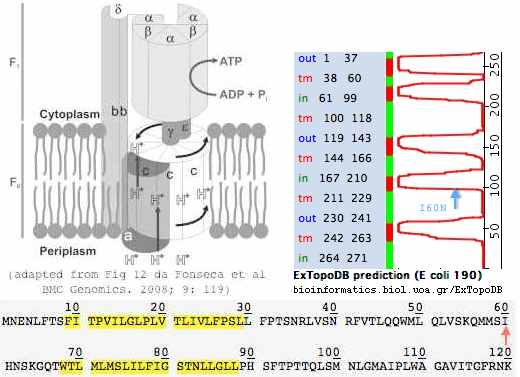
10 20 30 40 50 60 70 80 b562 90 b566 100 110 120 130 140 150 160 170 180 b562 190
| | | <------- tmA ----|--> | | <---|--*- tmB |------*> | | <-------|- tmC ---|----> <-|----- tmD|-------> | | <-|-* tmE --|
CYTB_bisBis_V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIM
CYTB_bisBis_all ..S......................................T.......................................................A........................M........D..........................................................
CYTB_bosPriW ...F..................................I...........................T...........................................................................................................................
CYTB_bosPriM ...I..................T...............I...........................T...........................................................................................................................
CYTB_bosSau ...I....................P.............V...........................T.....................................................I....................................................................A
CYTB_bosfroI ...I..................................I...........................T...........................................................................................................................
CYTB_bosFroW ...I..................................V...........................T..........................................................................................................................T
CYTB_bosGau1 ...I..................................V...........................T..........................................................................................................................T
CYTB_bosJav1 ...I....................P.............V...........................T.....................................................I....................................................................T
CYTB_bosJav2 ...I....................P.............V.................P.........T...........................................................................................................................
CYTB_bosJav3 ...I............T.......P.............V...........................T.....................................................I....................................................................T
CYTB_bosInd ...I..................................I...........................T...........................................................................................................................
CYTB_bisBon ...I..................................V...........................T...........................................................................................................................
CYTB_bosTau1 ...I..................................I...........................T...........................................................................................................................
CYTB_bosTau2 ...I..................................I...........................T...........................................................................................................................
CYTB_synCafW ..HI.........L........................I.................................................................................F....................................................................A
CYTB_synCafP ...I.........L........................I................S................................................................F....................................................................A
CYTB_bubBubW ...I.........L........................I..............................................................M..................FA....................................S..............................A
CYTB_bubBubP ...I.........L........................I..............................................................M..................FA....................................S..............................A
CYTB_traScr1 ...I..................................I....................M......T.......H..........................M..................F.....................................S..............................A
CYTB_traEur .I.I..................................I...........................T..................................M..................F.......T.............................S..............................T
CYTB_traStr ...I..................................I...........................T............................V.....M..................F....................................................................A
CYTB_traImb .I.I..................T.P.............I..V.................M......T.....................................................F....................................................................A
CYTB_traOry ...I..................T...............I..T........................TD.................................M..................F.....................................S..............................A
CYTB_traAng ...I..................................V....................M......T...............................................V.....FM...................................................................T
CYTB_traSpi ...I..................................I...........................T..................................M..................F.....................................S.........................F....A
CYTB_traDer ...I..................................I...........................T..................................M..................F.....................................S..............................A
CYTB_bseTra ...I..................................I....................M...A..T.....................................................F....................................................................A
b566 200 210 220 230 240 250 260 270 280 290 300 310 320 330 340 350 360 370
- tmE*-> | | | <---- tmF |---------> | | | <-|--- tmG -|-------> | | <------|tmH -----|--> <|--------|-tmI----->|
CYTB_bisBis_V98 AIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAILRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW
CYTB_bisBis_all ...................................................................................................................................R.........................................................
CYTB_bosPriW ........................V..............................A.....................................................................M..........A.....................T...L..VL......................
CYTB_bosPriM ........................V..............................A.....................................................................M..........A.....................T...L..VL..............V.......
CYTB_bosSau ....................V...V.............T................A.......................................................I.............M...............................TT...L..................V.......
CYTB_bosfroI ........................V..............................A.....................................................................M..........A.....................T...L...L..............V.......
CYTB_bosFroW ........................A................T.............A.......................................................I.............M................................T...L..................V.......
CYTB_bosGau1 ........................A................T.............A.......................................................I.............M................................T...L..................V.......
CYTB_bosJav1 ....................V...A..............................A.......................................................I.............M................................T...L..................V.......
CYTB_bosJav2 ........................V..............................A.....................................................................M..........I.M...................T...L..................V.......
CYTB_bosJav3 ....................V...A..............................A.......................................................I.............M................................T...L...T..............V.......
CYTB_bosInd ........................V..............................A.....................................................................M..........A.....................T...L...L..............V.......
CYTB_bisBon ........................T......................T.......A.......................................................I.............M..........A.....................T...L..........................
CYTB_bosTau1 ........................V..............................A.....................................................................M..........A.....................T...L..VL......................
CYTB_bosTau2 ........................V..............................A.....................................................................M..........A.....................T...L...L..............V.......
CYTB_synCafW .L..I...................T..............................S................................................IL.....IIM...........M..........I.........................L................S....N....
CYTB_synCafP .L..I...................T..............................S................................................VL.....IIM...........M..........I.........................L................S....N....
CYTB_bubBubW .L......................T...............................................................................VL.....I.M...........M...F......I...N.....................L...T............SM...N....
CYTB_bubBubP .L......................T..............................A................................................VL.....I.M...........M...F......I...N.....................L...T............SMV..N....
CYTB_traScr1 .L...................P.........................I.......A................................................V......I.M...........M..........I.A.......................L.......I......ATSM...SF...
CYTB_traEur .L.....................N.......................T.......A................................................VL.....I.M....M......M..........I.A.......................L.......I......VTSM...NF...
CYTB_traStr .L...........................................V..........................................................VL.....IFL...........M..........I....................M....L..............VTSM...NF...
CYTB_traImb .L.L....................T..............................A................................................ILT..MPI.M....A......M..........I.........................L..............M..S...N....
CYTB_traOry .L......................T........H.............T.......A................................................VL.....I.M...........M..........V.A.......................L..............V.SM...NF.--
CYTB_traAng .LV...............................................V.....................................................VL.....I.M....M......M..........L.........................L...I..........VIS....N....
CYTB_traSpi .L.......................................V.....T.......A................I...............................VL.....I.M....V......M..........I.A.......................L.......I......ATSM...NF...
CYTB_traDer .L.I...................................................A................S...................L...........VL...V.M.M...........M.......F..I.A............L..........L..............V.SM...N....
CYTB_bseTra .L..I...................A.........................M....A................S............................M..VL.....I.M...........M.......M..I...N.....................L................SM...N....
Site by site analysis of bison sporadic and sub-population variation in CYTB:
N3S I42T V98A V123M V132D Q322R
4345 N 2403 I 4522 V 4409 V 4981 V 4993 Q
132 I 719 A 430 I 483 T 9 I 2 R
70 H 645 G 34 M 73 I 5 L 2 P
14 Y 640 M 11 A 18 L 3 C 2 K
9 K 359 V 1 L 7 A 2 D 1 D
5 S 167 T 1 N 5 M
2 T 52 L 2 G
10 S
N3S: MT plains bison GU946987. Deleterious despite physical-chemical similarities of asparagine and serine. Based on 12,603 CYTB sequences from 1,637 species of mammals, this substitution has never gained traction in any clade despite being a simple 1 bp transition (codon AAC to AGC) that must have arisen a great many times in one species or another.
I42T: IT plains bison EU177871. Considerable amino acid flexibility exists at this site for cytochrome b. While threonine might be sub-optimal, it is not plausible that 167 species all have mitochondrial disease because of it. Nor could a substantial fraction of these other occurrences reflect sequencing error. Since the species are phylogenetically quite dispersed, the codon change here ATC to ACC arose and gained predominance many times. The Italian sequencers have not disclosed any details about the source of this bison dna.
V123M: EI woods bison GU947006. Threonine appears to be a fully functional alternative to valine but methionine is not. The 5 accessions having methionine do not comprise a coherent taxonomic clade but rather occur sporadically. As in this woods bison, the mutation in the other species may simply reflect heteroplasmy in the tissue sample used as dna source rather than a germline condition that would give rise to full-blown mitochondrial disease.
V132D: FR plains bison AF036273. The bison here resided at a Paris zoo. The peculiar change reported, AT --> TA, might be viewed as a 2 bp inversion rather than a double point mutation. However it is far likelier, given that the sequencing was done with 1999 technology, that an error occurred in the process of making the GenBank submission. The substitution is so radical at such a conserved site that lethal mitochondrial disease would likely have resulted if not a minor heteroplasmic haplotype.
Q322R: found only in a fragmentary plains bison sequence, AAL85955. This is again a radical change at an invariant site so either sequence error, minor heteroplasmy, or causative for mitochondrial disease. As with other sporadic mutations, there are no management implications unless broader sampling uncovers more individuals with this haplotype.
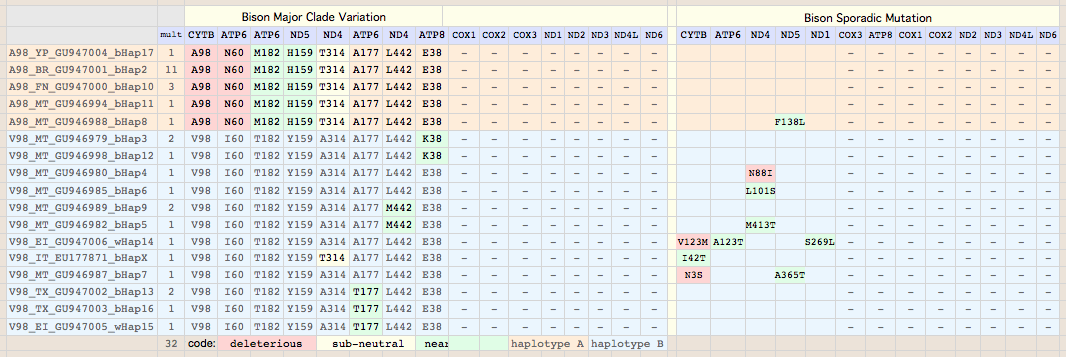
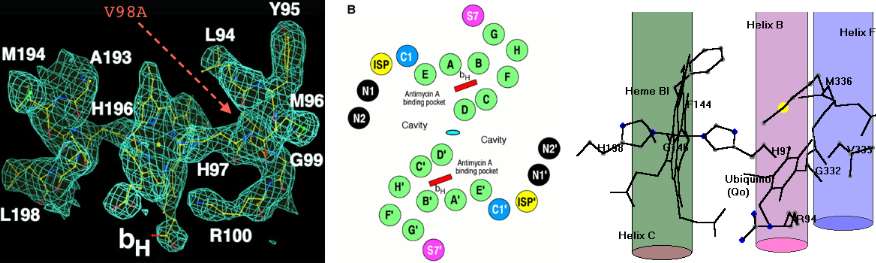
V98A: 18 plains bison. This site, located at the very end of transmembrane helix 2 just past the axial iron histidine ligand, is very important to bison conservation genomics. Of the 33 bison evaluated at this position, ancestrally valine), 18 animals are V98A but not either wood bison nor outgroups yak, aurochs or other Bovinae. Two of the sporadic variations occur among the 15 bison comprising the V98 clade, namely N3S in a MT private herd bison and V123M (in outlier wood bison wHap14). V123M may not remain sporadic as more Elk Island animals are sequenced; if more common, it becomes of management concern as it too is deleterious.
The single change V98A corresponds perfectly to the two major clades, with 98A shared by all individuals in the upper half of the tree ending in bHap2. In the overall mammalian context, A98 is a non-adaptive derived condition (synapomorphy) of the upper clade of bison. Note V98L is a domestic yak mutation described below. Although the vast majority of mammals are V98, isoleucine is also common with methionine fairly rare among the 12,603 CYTB sequences in 1,637 mammals considered. The statistics: 4522 V, 430I, 34M, 11A, 1L, 1N.
The other occurrences of alanine are scattered and shallow in the phylogenetic sense, ie alanine has never become established in another mammalian subclade despite tens of billions of (geologic) branch time accessible to study. These other species with V98A are Castor fiber (4 subspecies of beaver), Anomalurus (rodent), Eptesicus hottentotus (bat), Herpestes naso (mongoose), Genetta johnstoni (carnivore), Hyaena hyaena (hyena), and Macroscelides proboscideus (elephant shrew).
No internal compensation by a co-evolving residue elsewhere in CYTB can occur since V98A is the sole residue change. Conceivably a change in one of the ten nuclear encoded proteins targeted to mitochondria Complex III (of which oligomeric partners cytochrome c1 and Rieske iron sulfur proteins are the likeliest candidates). The concept of balanced polymorphism (along the lines of E6V of sickle cell hemoglobin) also seems inapplicable.
Looking now at 500 non-mammalian cytochrome b -- ie at species predating the bison divergence from birds at 310 myr, not a single alanine occurs. Valine no longer dominates at 186 species (37%) but instead the closely related branched chain aliphatic isoleucine at 314 occurrences. Cytochrome b has been very particular about position 98 for a very long time.
The next level of consideration, beyond the private and sub-population variation considered above, are sites the same in all bison but different from a conserved ancestral value at yak divergence. These can be seen in the dot alignment above as residue columns identical for the progressive outgroup (ie yak, wild cattle, water buffaloes, elands, nilgai) but another amino acid at bison.
These 9 additional sites need to be carefully evaluated because they might be deleterious alleles that have spread to all bison (rather than just a sub-population in the manner of V98A). Alternatively, they might simply reflect neutral drift within the acceptable reduced alphabet for the respective sites, an innocent haplotype that become predominant in the stem lineage. Another less common possibility is adaptive change, part of what makes a bison a bison rather than a yak. Note heteroplasmy is inapplicable here because the change in so many bison is clearly being inherited.
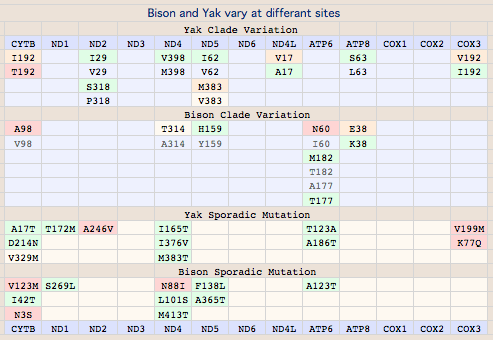
Evaluating these as before (phylogeny-aware frequencies), most of the changes are fully consistent with near-neutral drift within the reduced alphabet. Here the notation is slightly different: in I4L etc, I is the outgroup consensus, 4 the numbering within CYTB, and L is bison variant. The only causes for concern here are I39M, I316M and L353M because in these the observed frequency of the bison amino acid is quite low. L353M may be significantly sub-adaptive given the invariance of leucine and rarity of methionine and similarly for I39M and M3126I. The same holds for M316I. A246T is a bit peculiar in that both bison and outgroups have less common residues. V215M does not have that decisive an outgroup value and can be dismissed as neutral.
I4L I39M T67A V215M A246T M316I T349I L353M V372I
3757 I 2399 I 4513 T 1682 S 3096 S 3857 I 3701 I 3913 L 2805 I
319 L 1640 V 456 A 1522 A 765 F 650 T 761 T 413 T 616 M
287 M 688 L 14 S 741 M 611 T 230 M 199 V 395 V 526 V
244 T 140 M 7 M 528 T 455 A 127 A 165 L 146 M 415 L
16 F 101 A 6 V 354 P 26 M 95 S 85 M 69 I 363 F
10 V 17 T 2 I 91 V 16 V 24 L 75 A 44 A 202 A
3 Y 13 F 34 L 12 L 15 V 3 S 13 F 40 T
3 P 2 M 32 C 7 Y 3 F 5 S
2 S 6 I 5 N 2 H
2 L 5 Y 2 I
2 C 2 C
1 Y
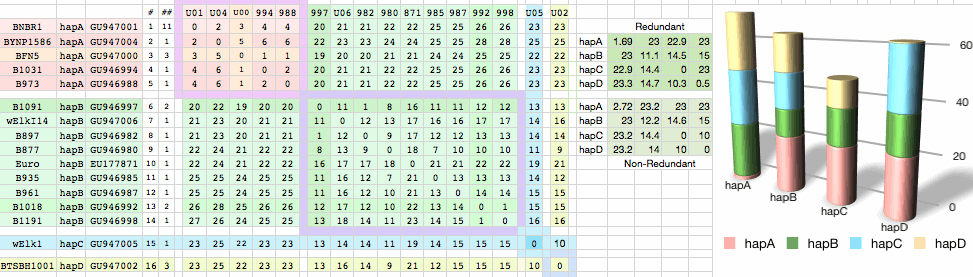
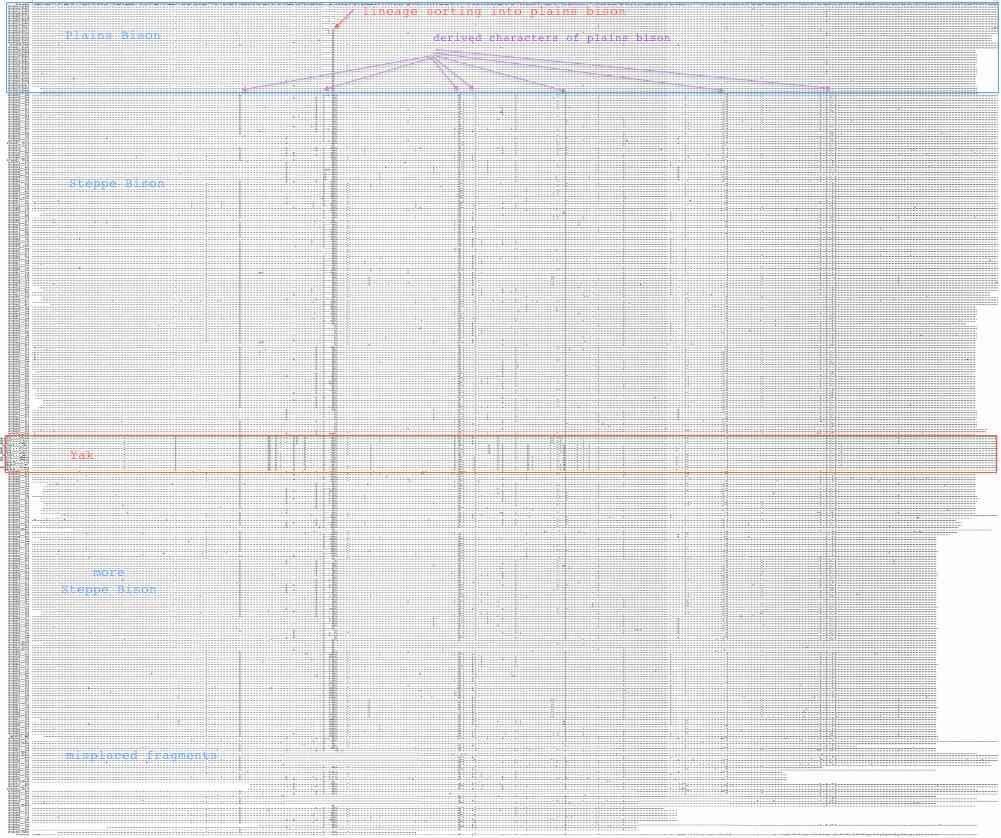
Sequences are color clustered according to the haplotype tree. bHap1 is not shown. Note the woods bison cannot be resolved from the plains bison even though the Elk Island woods bison are a relic herd that did not mix with 7,000 plains bison imported from the Flathead Reservation in Montana up to Canada's Wood Buffalo National Park in the 1920's. (The same conclusion was reached earlier after sequencing D-loops from 3 museum species predating the 1925 introductions.) Clearly these animals are a mixture of the second major clade of bison with an earlier diverged lineage represented by wHap14 surviving (at least in mitochondrial dna) from the founder herd. This could represent allopatric separation during a glaciation epoch with subsequent reunification. However the prevalence of wHap14 needs to be established along with uniqueness of its nuclear dna.
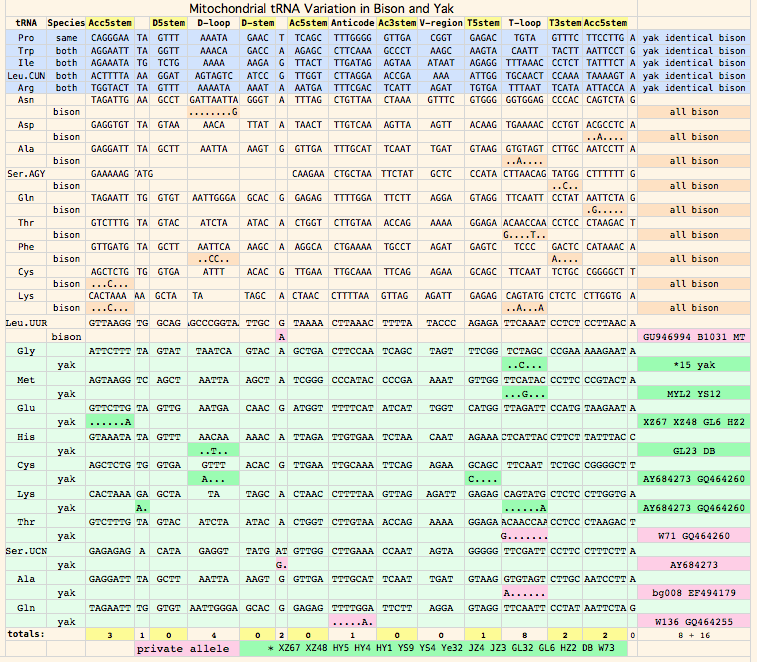
NucAcc ProAcc PubMed ST Locale TYP MUT BP Change Isolate Haplo Source Herd GU946976 ADF48936 20870040 MT plains A98 V.98A GTA to GCA B790 bHap2 Montana private herd GU946977 ADF48949 20870040 MT plains A98 V.98A GTA to GCA B853 bHap2 Montana private herd GU946978 ADF48962 20870040 MT plains A98 V.98A GTA to GCA B854 bHap2 Montana private herd GU946981 ADF49001 20870040 MT plains A98 V.98A GTA to GCA B880 bHap2 Montana private herd GU946983 ADF49027 20870040 MT plains A98 V.98A GTA to GCA B925 bHap2 Montana private herd GU946984 ADF49040 20870040 MT plains A98 V.98A GTA to GCA B929 bHap2 Montana private herd GU946986 ADF49066 20870040 MT plains A98 V.98A GTA to GCA B959 bHap2 Montana private herd GU946993 ADF49157 20870040 MT plains A98 V.98A GTA to GCA B1029 bHap2 Montana private herd GU946995 ADF49183 20870040 MT plains A98 V.98A GTA to GCA B1050 bHap2 Montana private herd GU946996 ADF49196 20870040 MT plains A98 V.98A GTA to GCA B1051 bHap2 Montana private herd GU947001 ADF49261 20870040 NB plains A98 V.98A GTA to GCA BNBR1 bHap2 National Bison Refuge GU947004 ADF49300 20870040 YP plains A98 V.98A GTA to GCA BYNP1586 bHap17 Yellowstone Natl Park GU946990 ADF49118 20870040 MT plains A98 V.98A GTA to GCA B985 bHap10 Montana private herd GU946991 ADF49131 20870040 MT plains A98 V.98A GTA to GCA B1005 bHap10 Montana private herd GU947000 ADF49248 20870040 NB plains A98 V.98A GTA to GCA BFN5 bHap10 Fort Niobrara GU946994 ADF49170 20870040 MT plains A98 V.98A GTA to GCA B1031 bHap11 Montana private herd GU946988 ADF49092 20870040 MT plains A98 V.98A GTA to GCA B973 bHap8 Montana private herd AF036273 AAD51424 10603253 FR plains A98 V132D AT to TA ..... ..... Vincennes Zoo 1999 GU946979 ADF48975 20870040 MT plains V98 ..... .......... B855 bHap3 Montana private herd GU946992 ADF49144 20870040 MT plains V98 ..... .......... B1018 bHap3 Montana private herd GU946998 ADF49222 20870040 MT plains V98 ..... .......... B1191 bHap12 Montana private herd GU946980 ADF48988 20870040 MT plains V98 ..... .......... B877 bHap4 Montana private herd GU946985 ADF49053 20870040 MT plains V98 ..... .......... B935 bHap6 Montana private herd GU946989 ADF49105 20870040 MT plains V98 ..... .......... B979 bHap9 Montana private herd GU946997 ADF49209 20870040 MT plains V98 ..... .......... B1091 bHap9 Montana private herd GU946982 ADF49014 20870040 MT plains V98 ..... .......... B897 bHap5 Montana private herd GU947006 ADF49326 20870040 EI woodsB V98 V123M ATA to GTA wEI14 wHap14 Elk Island EU177871 ABV70945 18302915 IT plains V98 I.42T ATC to ACC ..... ..... unknown Italy GU946987 ADF49079 20870040 MT plains V98 N..3S AAC to AGC B961 bHap7 Montana private herd GU946999 ADF49235 20870040 MT plains V98 ..... .......... B1428 bHap13 Montana private herd GU947002 ADF49274 20870040 TX plains V98 ..... .......... BTSBH1001 bHap13 Texas Sate Bison Herd GU947003 ADF49287 20870040 TX plains V98 ..... .......... BTSBH1005 bHap16 Texas Sate Bison Herd GU947005 ADF49313 20870040 EI woodsB V98 ..... .......... wEI1 wHap15 Elk Island
Variation in all 13 bison and yak mitochondrial proteins
The variation observed in the entire mitochondrial proteome can be readily interpreted along the lines of CYTB above. First note COX1 COX2 COX3 ND2 ND3 ND4L ND6 are completely conserved in all bison complete genome data. ND1 is also conserved with the exception of a sporadic near-neutral substitution S269L in a woods bison (ND1: EI_GU947006_wHap14). This degree of conservation makes it unlikely that the founder population harbored a high frequency deleterious allele of hyper-mutating nuclear genes such as POLG.
Next note that 9 of 10 overall sporadic substitutions (F138L in ND5: A98_MT_GU946988_bHap8 being the exception), are concentrated in the CYTB-determined V98 clade even though it represents fewer animals (the multiplicity column shows 17 bison in the A98 clade and 15 in V98). This is consistent with the A98 haplotype being of much less diverse (more recent origin) and indeed this is borne out by whole genome blastn comparisons.
Recall that sporadic substitutions -- even when clearly deleterious like N88I in ND4:V98_MT_GU946980_bHap4 -- may not be fully heritable but rather simply reflect heteroplasmic amplification of an uncommon germline haplotype in the tissue used for dna sequencing. Alternatively they could simply represent somatic mutation and not be represented at all in germline dna. Consequently even deleterious sporadic mutations may not have major phenotypic effects. However if the same mutation shows up again as more bison are sampled, the interpretation shifts towards heritability (since the same mutation would not arise independently in a still-small sample) and, for N88I, significant effect on fitness.
Semi-systemic (clade-level) variations affecting multiple animals are definitely maternally heritable to an extent determined by germline haplotype prevalence. Bison have 8 such substitutions in 5 of their 13 mitochondrial proteins, namely CYTB:V98, ND5:Y159H, ND4:A314T and ND4:L442M, ATP6:T182M and ATP6:A177T, ATP8:E38K.
All of these fall along the lines already established by CYTB:V98A with the exception of ATP6:A177T, ND4:L442M and ATP8:E38K which are restricted to (and define) subclades of the major CYTB:V98 subgroup. All three of these substitutions classify as somewhat sub-normal but not outright deleterious. That is, the same substitutions are observed in too many other species to be outright mutations (consistent with fairly benign amino acid attribute change), yet are not so common as to be on an equal fitness footing with the major components of the reduced alphabet. Bison carrying ATP6:A177T can be predicted to be least affected in view of threonine being the second most frequent residue at this position. Since we don't fully understand the significance of these changes, they represent genetic variation that should be protected by conservation genomic management as adaptive or adaptive in combination with other alleles in other mitochondrial or nuclear genes, now or under later environmental circumstances.
ATP6:A177T ND4:L442M ATP8:E38K
998 A 221 L 215 E
88 T 46 I 34 K
4 S 19 M 17 S
2 V 8 T 15 M
1 P 2 V 5 T
1 A 1 F 5 G
3 V
2 A
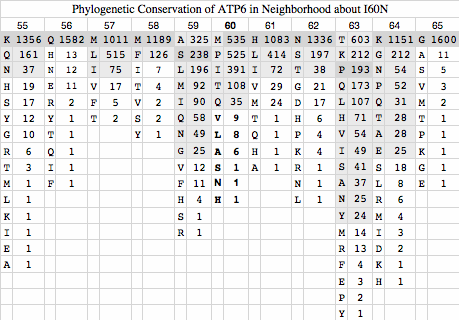
The remaining five major sites are distributed precisely along the clade lines established by V98A of CYTB. These are likely fully heteroplasmically penetrant in both clades and inherited in all descendants. ATP6:I60N can immediately be seen to be a second deleterious change in the A98 clade as asparagine never occurs here in thousands of other species and its polar nature is a substantial change from branched chain aliphatic isoleucine.
ND4:A314T is borderline deleterious -- while a very rare substitution within mammals, it has become established in all Camelidae with available sequence and so is unlikely to be harmful there. However bison and camel ND4 differ at many other sites -- 73 of 459 -- so the status of ND4:T314 in bison is likely sub-neutral but mild phenotypically, additionally as alanine and threonine are not too dissimilar. The peculiar appearance of T314 in an Italian zoo bison of the V98 I60 clade, if not sequence error, suggests lineage sorting and possible heteroplasmic persistence in some A314 bison.
ATP6:I60N ATP6:T182M ND5:Y159H ND4:A314T ATP6:A177T ND4:L442M ATP8:E38K
531 M 553 S 225 Y 281 A 998 A 221 L 215 E
392 I 286 M 73 H 8 T 88 T 46 I 34 K
106 T 98 T 5 I 4 S 19 M 17 S
37 V 92 L 3 V 2 V 8 T 15 M
6 A 57 I 1 P 2 V 5 T
5 N 10 A 1 A 1 F 5 G
4 L 4 V 3 V
2 P 2 F 2 A
S 1 1 M
N 1 1 C
The species sharing the errant variation with bison: note A314 forms a clade within Camelidae
ATP6:I60N ND4:T314A
Panthera tigris carnivore Camelus ferus artiodactyl
Erinaceus europaeus insectivore Camelus bactrianus artiodactyl
Cebus capucinus primate Lama glama artiodactyl
Cebus albifrons primate Lama guanicoe artiodactyl
Callicebus donacophilus primate Lama pacos artiodactyl
Callicebus donacophilus primate Vicugna vicugna artiodactyl
Pontoporia blainvillei cetacean
Physeter catodon cetacean
ND5:Y159H ND4:L442M ATP8:E38K
11 carnivores 8 carnivores 20 cetaceans
11 artiodactyls 3 bats 4 ruminants
2 cetaceans 3 carnivores
1 ruminant 1 rodent
The third class of site variation is defined by synapomorphies, eg the same substitution seen in all bison relative to a residue invariant in yak, cattle, water buffalo and other close-in Bovidae. Changes here affect all bison. They may either be adaptive, part of what makes a bison a bison (not a yak or cow), or sub-normal variations that rose to high frequency because of severe historical bottlenecking of the bison population, or simply neutral fixation of a particular amino acid within the normal reduced alphabet at a given site. These residues are located using methods described in the following section but are provided here for analysis. The notation shows gene, ancestral value, protein position, and variation found in all bison (respectively yak) studied to date.
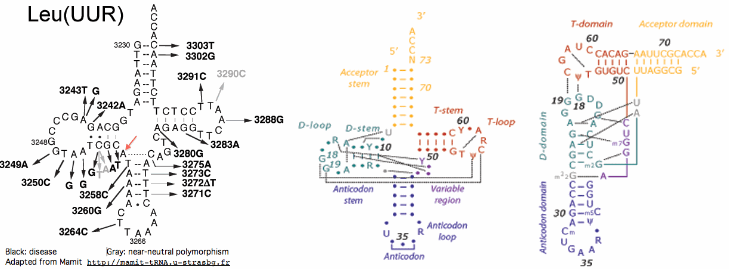
Bison Synap Yak Synap Status Conservation Significance (to be continued) ATP6 I195V ND3 M16L ATP6 I201V ATP6 A186T ATP8 Y34H ATP6 T123A COX2 S94F CYTB T190M CYTB A215M ND1 H92Y CYTB I4L ND1 T171M CYTB L99M ND1 T67A CYTB V39M ND2 T209I ND2 M320T ND2 T7I ND2 M92V ND3 I24V ND4 Y421H ND3 T28A ND5 A519T ND4 M185V ND5 L449F ND5 H272Y ND5 M87V ND5 T21M ND6 F101S ND5 Y56H
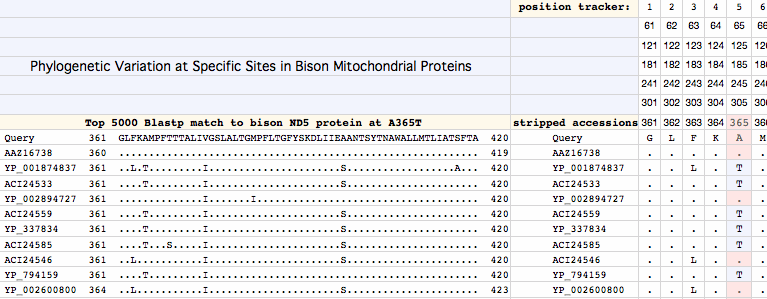
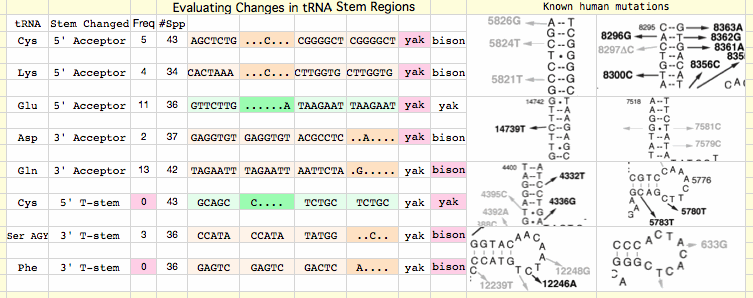
Methods here are important to understand because a vast amount of empirical data is being compressed to a small but important bit of management information -- the healthy haplotypes. The screenshot below illustrates the simple desktop method used for extracting variation at a given site from 5000 Blastp matches to a given protein. In the example, A365T has been previously identified as a site of variation within the alignment of all available bison NADH dehydrogenase subunit 5 (ND5) proteins. Using a bison sequence with A365 as query, output formatting of blastp output is set to "query-anchored with dots for identity".
Pasting the relevant section of higher quality sequences (which varies from protein to protein depending on indels and sequence divergence) into the spreadsheet causes its text-extracting formulas (provided below in the methods section) to separate the match into individual columns for accession number and each amino acid. Columns of interest are then processed to obtain frequencies of each of the 20 amino acids at the site under study (here only A and T occur).
Blast output order is by similarity, so it corresponds approximately to phylogenetic distance from bison. However the set of protein accessions corresponding to a particular variant (here A or T) can more precisely processed back at NCBI Entrez for phylogenetic position (indeed tree) using the NCBI Taxonomy extractor. This eliminates over-counting of frequencies attributable to high multiplicities of sequenced individuals of some species. A computer algorithm here is ill-advised since NCBI can and does change formatting practices without notice.
The frequency tables and phylogenetic distribution pattern then determine the interpretation of the amino acid variation. For A365T, a mild physical-chemical change (according to Dayhoff, Blosum, Kyte-Doolittle or Grantham value), both A and T occur in widely dispersed clades of mammals, as do proline, valine and isoleucine, so it follows that the substitution is near-neutral for bison. It may not have arisen by recent mutation but instead may reflect lineage sorting during bison cladogenesis or speciation, simply rising to prominence in the bison studied (V98_MT_GU946987_bHap7) via heteroplasmic amplification in the leucocytes sampled. Consequently it is part of normal natural variation at this site and its elimination should not be a conservation genomics management objective.
Synapomorphies in bison, yak and cattle mitochondrial proteomes
What makes a bison a bison? Synapomorphies (derived characters) are those amino acids invariant within bison but differing from the ancestral value as determined by yak, cow, water buffalo and other pecoran ruminant outgroups. To collect these across the entire mitochondrial proteome (13 proteins, 3790 amino acids in bison), 80 complete mitochondrial genomes were aligned at the protein level, with 102 sites of interest then extracted (below) and grouped by type.
It is important here to include all available bison, yak, cattle and water buffalo data so that bona fide synapomorphies -- rather than sub-clade features or one-off mutations -- are properly defined. (Here 15 cattle genomes and 11 cattle-bison hybrids were chosen from the many available.)
Yak and cattle synapomorphies provide important context to bison so they too are displayed along with synapomorphies applicable to the yak-bison ancestral divergence node. Note cattle have more synapomorphic sites than bison and yak put together, possibly a byproduct of breeding for certain features during their long history of domestication. Sub-clade features of bison and yak, considered above, are also shown along with a couple of bison + yak + cattle synapomorphies relative to water buffalo (which can be determined with less precision because only 4 genomes are available).
These changes may represent adaptive variations that swept across the entire population because of their selective value. Alternatively, they might represent maladaptive alleles that nonetheless rose to high frequency because historic bottlenecks accidentally favored a small subpopulation of animals carrying a mutation (much as seen above for V98a and N10I in bison sub-clades). Another option is simply neutral drift that fixed a particular amino acid from the normal reduced alphabet at that position. Only the maladaptive alleles have relevance to conservation genomics management.
One striking feature of the table is the apparent over-representation by amino acids from the first two columns of the genetic code, especially methionine. Many variations are known for the mitochondrial genetic code, so this raises concerns that bison, yak or cattle might use a slightly different translation table (making the synapomorphies into artifacts), perhaps only at certain sites along certain proteins. This scenario could be consistent with the oddities in bison mitochondrial tRNAs reported by the Derr group. Both mitochondrial mRNA and tRNA editing have been documented -- these could hypothetically result in a normal protein encoded by a mutant gene. Recall here protein sequences are not routinely determined experimentally but rather inferred from dna sequences, the exceptions being bovine cytochrome b and cytochrome oxidases used in xray crystallography which agree with standard mitochondrial genetic code translation and show no sign of mRNA editing.
Sequence error must always be considered, even for data originating from experienced laboratories using modern sequencing technologies at high multiplicities. For example, the widely used Cambridge Reference Sequence for human contained 11 errors in addition to 7 very rare valid polymorphisms. For bison and yak, this seems highly implausible at sub-clade and synapomorphic sites where the same error would have to occur in processing distinct individuals. While somatic heteroplasmy could result in less represented wildtype being discarded as a minor allele, that again is unlikely for a stably inherited character. Laboratory contamination with dna from another species -- a great concern in sequencing extinct species -- makes no sense here: contamination by cattle dna (which is too remote) could not give the observed results in bison or yak.
Errors could not arise systematically from either abnormal base composition or local tertiary structures because these mitochondrial genomes are nearly identical overall. Sporadic mutations are a different matter and for nuclear genes are normally validated by independent methods (eg Sanger sequencing). GenBank in fact contains many erroneous sequences, notably affecting fragmentary mitochondrial proteins sequenced in the 1990's. Here, since non-synonymous change in yak and bison was not the original focus, it would be worthwhile to revisit the raw data to confirm the reported mutations and any observable heteroplasmy. Short-read technologies trade off high error rates against high coverage, making authentic heteroplasmy difficult to distinguish from read error.
The first five columns of table below is sorted by over-representation of synapomorphies. For example, ATP8 has five of these in a very short sequence of 66 residues. While ND5 has more synapomorphies (25), it is a much longer protein at 606 residues and so has a lower density of synapomorphies than ATP8 (1.5 vs 2.8). The next three columns show the composition of the bison mitochondrial proteome, sorted according to decreasing occurrence. Note the very low abundances of charged amino acids attributable to the many transmembrane domains which utilize apolar amino acids. Overall, the abundances of amino acids does not resemble that of nuclear-encoded cytoplasmic proteins. The last three columns show abundances taken from the large synapomorphy table below. The top line of that was provided by the Yellowstone bison sequence and dots indicate identity relative to it.
Bison Synapomorphy Table Gene #AAs Syn %syn %AA Over AA Freq % 3790 102 100 100 3790 100 . 3683 ATP8 66 5 4.9 1.7 2.8 L 593 15.7 T 589 CYTB 379 14 13.7 6.9 2 I 328 8.7 I 487 ATP6 226 11 10.8 6 1.8 T 308 8.1 V 478 ND3 115 5 4.9 3 1.6 S 274 7.2 M 414 ND5 606 25 24.5 16 1.5 M 265 7 L 292 ND2 347 10 9.8 9.2 1.1 A 250 6.6 A 277 ND6 175 5 4.9 4.6 1.1 F 244 6.4 Y 260 ND4 459 12 11.8 12.1 1 G 219 5.8 S 172 ND4L 98 2 2 2.6 0.8 P 191 5 P 89 COX3 260 4 3.9 6 0.7 V 185 4.9 F 81 ND1 318 5 4.9 8.4 0.6 N 164 4.3 H 73 COX2 227 3 2.9 13.6 0.2 Y 132 3.5 N 35 COX1 514 1 1 10 0.1 W 104 2.7 H 98 2.6 K 99 2.6 E 95 2.5 Q 87 2.3 D 69 1.8 R 63 1.7 C 22 0.6
(The table will paste properly into common desktop spreadsheets if all spaces are replaced with tabs:) .gene.. A A C N N N N A N C N C A A C C N N C N N N A N N N N N N A N N N N N C N A N N N C N C C N N N N N N N N N A N N A N N N A N C N N A N N C N N C C N C N N N N C N A C A C C N N A A C C N N N N N N N N N C N .gene.. T T Y D D D D T D O D Y T T Y Y D D Y D D D T D D D D D D T D D D D D Y D T D D D Y D O O D D D D D D D D D T D D T D D D T D Y D D T D D Y D D Y O D O D D D D O D T O T Y Y D D T T O Y D D D D D D D D D Y D .gene.. P P T 4 5 2 2 P 4 X 5 T P P T T 5 5 T 6 2 2 P 4 5 5 3 4 3 P 1 2 2 5 3 T 1 P 5 1 5 T 5 X X 5 5 6 5 5 5 4 4 5 P 4 3 P 2 6 5 P 2 T 4 5 P 5 2 T 2 4 T X 2 X 1 5 6 3 X 4 P X P T T 4 1 P P X T 4 5 4 6 4 5 5 5 5 T 2 .gene.. 6 6 B . . . . 8 . 2 . B 6 6 B B . . B . . . 8 . . . . . . 6 . . . . . B . 6 . . . B . 2 1 . . . . . . . . . 8 L . 8 . . . 6 . B L . 6 . . B . . B 3 . 3 . . . . 2 . 6 3 6 B B . . 6 8 3 B . . . . . . . . . B . . ..pos.. 6 1 9 3 1 9 3 3 4 9 4 9 1 2 4 3 8 5 2 1 6 2 6 3 6 3 1 1 2 1 6 7 2 2 2 1 1 1 5 9 2 6 1 9 4 1 3 1 4 4 4 1 3 7 4 5 1 3 2 9 3 3 3 2 1 1 6 2 3 1 6 5 7 3 1 4 7 2 1 8 1 1 7 4 1 6 9 4 1 1 2 6 1 2 4 1 1 1 9 2 1 5 1 1 . ..pos.. 0 9 8 1 5 2 2 4 2 4 4 9 9 0 . 9 7 1 1 0 3 8 3 9 2 8 6 8 4 2 7 . 0 1 8 9 7 8 6 2 7 2 2 9 5 7 4 6 3 9 7 6 8 . 1 . 9 5 4 3 9 9 0 4 3 7 3 9 2 0 . 4 3 2 5 0 6 2 4 1 8 0 . 1 9 7 5 4 6 3 8 1 0 3 4 9 1 1 0 4 0 1 9 6 . ..pos.. . 2 . 4 9 . 0 . 1 . 9 . 5 1 . . . 9 5 1 . . . 8 . 3 . 5 . 3 . . 9 . . 0 1 6 . . 2 . 9 . 3 2 6 1 8 9 7 9 2 . . . . . 1 . . . 1 6 . . . . 5 3 . . . . 8 . . . . . 7 0 . . 4 . . 8 6 . . . 2 3 1 2 4 6 . . 9 3 2 2 . bisBisY N M A T H V T H H F F M V V L M V T M S P V L V I M L V V A A I I M A M M T H Y Y I M A V V M F S T V M M F S H G Y I I T N A T A V P S T M T M T A I T L T I T T L A A I A I S I M I I I I I I I F V L H F I L ATP6_GU947004_bHap17_A98_YP_Bison bisBisA . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU947001_bHap2_A98_BR_Bison bisBisA . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU947000_bHap10_A98_FN_Bison bisBisA . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU946994_bHap11_A98_MT_Bison bisBisV . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU946988_bHap8_A98_MT_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU946979_bHap3_V98_MT_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU946998_bHap12_V98_MT_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU946980_bHap4_V98_MT_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . S . . . . . . . . . . . . . . . . . . . . . . ATP6_GU946985_bHap6_V98_MT_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU946989_bHap9_V98_MT_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU946982_bHap5_V98_MT_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU947006_wHap14_V98_EI_Bison bisBisV I T V . Y . . . . . . . . . . . . . . . . . . . . . . . . T . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_EU177871_bHapX_V98_IT_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU946987_bHap7_V98_MT_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU947002_bHap13_V98_TX_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU947003_bHap16_V98_TX_Bison bisBisV I T V A Y . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GU947005_wHap15_V98_EI_Bison bosGruA I T V A Y M M Y Y S L L I I I V M A A F S I S . . . M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464262_hapA_yak bosGruA I T V A Y M M Y Y S L L I I I V M A A F S I S . . . M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464261_hapA_yak bosGruA I T V A Y M M Y Y S L L I I I V M A A F S I S . . . M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464257_hapA_yak bosGruA I T V A Y M M Y Y S L L I I I V M A A F S I S . . . M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464251_hapA_yak bosGruA I T V A Y M M Y Y S L L I I I V M A A F S I S . . . M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464250_hapA_yak bosGruA I T V A Y M M Y Y S L L I I I V M A A F S I S . . . M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464249_hapA_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M . . M M I . T T T T T T . . Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464260_hapA_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464266_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464265_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . T . ATP6_GQ464264_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464263_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464259_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464258_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464256_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464255_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464254_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464253_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464252_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464248_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ATP6_GQ464247_hapB_yak bosGruB I T V A Y M M Y Y S L L I I I V M A A F . . . M V V M M I T T T T T T T T A Y H H . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . T . . . . . . ATP6_GQ464246_hapB_yak bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947021_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A . T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947020_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947019_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_HM045018_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . F M L T I . I L P I I T V L P Y N H V V A S T A T M S P M L I V A T V M M M M . S S . T T T T T . V . V . V V V V L T . . Y . . ATP6_AF492351_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . F M L T I . I L P I I T V L P Y N H V V A S T A T M S P M L I V A T V M M M M . S S . T T T T T . V . V . V V V V L T . . Y . . ATP6_EU177870_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A . T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L T F . Y . . ATP6_EU177868_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_EU177866_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_EU177864_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_EU177862_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . V . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_EU177860_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_EU177858_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_EU177856_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_EU177854_BosTau bosTauD I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V . V V V V L I F Y Y . . ATP6_EU177852_BosTau bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947018_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947017_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947016_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947015_BosHyb bosTauH I T V A Y M . Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947014_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947013_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947012_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947011_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947010_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947009_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947008_BosHyb bosTauH I T V A Y M M Y Y S L L I I I I M A V F . . . . . . . . . . . . . . . . . . . . . M L T I I I L P I I T V L P Y N H V V A S T A T M S P M L I L A T V M M M M A S S T T T T T T T V V V V V V V V L I F Y Y . . ATP6_GU947007_BosHyb bubBub. I T V A . . . . . S L L I I I I M A T L S I S A . . . M I . . . . . . A T M . . F M L T I I I L P I I T V L P Y N H V V A S T A . L T N L T F V I I M . . . . . . . . . . . . . . . . . . . . . . . . . . . L M ATP6_NC_006295_Bubalus bubBub. I T V A . . . . . S L L I I I I M A T L S I S A . . . M I . . . . . . A T M . . F M L T I I I L P I I T V L P Y N H V V A S T A . L T N L T F V I I M . . . . . . . . . . . . . . . . . . . . . . . . . . . L M ATP6_AY702618_Bubalus bubBub. I T V A . . . Y . . L L I I I I M A T L S I S A . . . M I . . . . . T A T M . . L M L T . I I L P I I T V L P Y N H V V A S T A T M T N L T F V I I . . . . . . . . . . . . . . . . . . . . . . . . . . . . L M ATP6_AY488491_Bubalus bubBub. I T V A . T . . . S L L I I I I M A T L S I S A . . . M I . . . . . T A T M . . L M L T I I I L P I I T V L P Y N H V V A S T . T M T N L T F V I I M . . . . . . . . . . . . . . . . . . . . . . . . . . . L M ATP6_AF547270_Bubalus antCer. I T V A Y T . Y Y S . L I I T I M . A L S I S L V . . M I . I L . . T A V . . . L M L T I I I L P I . T V . P Y S H V V . . . S T I . T L . I I V I . . . . V . . . . T . T . . . . . V V . . V . . . S . . L M ATP6_NC_012098_Antilope ammLer. I T V A Y T M Y . S L L I I I I . M A F S I S L V . . M I . I . . T L T V . . . F M L T I I I L P . I T V L A Y N . . V . . T . . A T Y L . I I I I . M . I M . . . T T . . . . T . . V . . . V . L . . . . L M ATP6_NC_009510_Ammotragus budTax. T T V A Y M . Y . S . L I I I T T M T F S I S L . . . M I . I . . T L V V . . . L M L T I I I L P I I T V L T Y N . . . . . T . M . T . L . I T L I . . . . M . . . T . T T . . . V . V . . . T . L I . . . L M ATP6_NC_013069_Budorcas hydIne. I T V A Y . . Y . S . L I . M I M I A V S . . L . . . L I . I L . I T A V . . . F M L T I I I L P I I I V . . Y S F V . . S . S T I T T L L I L I I . I . . L D . . . I . T . . . . V . . . V V . . . S . . L M ATP6_NC_011821_Hydropotes elaCeo. I T V A Y T . . . S . L I . I I M I S M . . . L . . . L I . I . . T T A V . . . F M L T I I I L P I I I V . L Y . F V . . . . A T I T T L L I . I I . L . . L D . . . I . T . . . . . . V V . V . . . S . . L M ATP6_NC_008749_Elaphodus panHod. I T V A Y T . Y . S L L I I I I M I A F S I S L V . . M I . I . . . L A . . . . F M L T I I I L P I I T V . L Y . . V . . . . S . M I T L . I T I I . M . . . . . . T I . T . . . . . . . . . T . . . F . . L M ATP6_NC_007441_Pantholops capCri. I T V A Y M . Y . S L L I I I I M . T F S I S L V . . M I T I . . T L T V . . . F M L T I I I L P I . T . . T Y N . . . . . . . . T V . L . I T I T . M . . L . . . T I . T . . . . . V . . . T . . . . . . L M ATP6_NC_012096_Capricornis mosBer. I T V A Y . . F . S . L I I I I M . T V S I S . . I . M I . I L . . T A V . . . F M L T I I I L P I . T V . L Y N H A . . . T . T M T T L . I V . I . . . . . . . . . T . T . . T . . . . . . V V L . F . . L M ATP6_NC_012694_Moschus cerUni. I T V A Y M M . . S . L I I I I M I A M S . . . V . . L I . I . . T T A V . . . F M L T I I I L P I I I V . L Y . F . V . . . A M I A T L L I V I I . M . . L N . . . I . T . . . . . . . . . V . . . S . . L M ATP6_NC_008414_Cervus ranTar. I T V A Y T . F . S . L I . I I M I S V S I . L . . . L I . I . . T T A V . . . F M L T I I I . L I I I V . . Y . F V M . . . A M I A T L L I V I I . . . . L N . . . I . T T . . V V . . . T V V . . S . . L M ATP6_NC_007703_Rangifer girCam. I T M A . H . Y . S T L . I I I M S . L . I S . V . . . I . T . . . . A T A . H L T L T I I I . P I I . . L P . N H . . A . T . . I T T L . . V I T . . . . . N . . . I A T . . . T . V . V . . . L I S . . L M ATP6_NC_012100_Giraffa
Predicting Protein Mutations from Control Region Data
It appears that the status of cytochrome b and ATP6 proteins can be reliably predicted from a small region in the control region (ie these micro-haplotypes can serve as proxy for CYTB and ATP6 amino acid alleles). Because many more bison have been sequenced just in the control region (rather than whole mitochondrial genome), this considerably expands the statistics and locale distribution of bison with 'known' V98A and I60N status. (Haplotype definition here does not consider sporadic variations occurring in individual animals.)
Assuming the prediction method is reliable, the status of 165 plains bison (119 contemporary and 46 fossil) is known (32) or inferable (133). Of these, 42% of contemporary and 2 of 48 fossil bison may have double mitochondrial disease V98A I60N (exercise intolerance, lactic acidemia, ragged red muscle fiber). Bison-cattle hybrids never carry the mutations since the bison bull is always on the paternal side and does not contribute to mitochondrial dna in progeny. Since haplotype, 98A, 60N arose in an unknown order over time, intermediate stages may eventually be found (eg haplotype, V98A, 60N), though because of the bottleneck this is more likely in geographically remote fossil dna.
The graphic shows a screenshot of a database whose plain text version is furnished below. The minimal diagnostic region (micro-haplotypes A-E) became apparent during the massive alignment of bison, steppe bison and yak control regions described in the steppe bison section. It corresponds to bases 15894-15965 in the Yellowstone NP bison numbering scheme of GU947004, namely TTGCAAACACCACTAGCT AACGTCACTCACCCCCAA AAATGCATTACCCAAACG GGGGGAAATATACATAAC.
This region begins with a T to C transition characteristic of mutational status, continues to two variable homopolymer runs (microsatellites), AAAAA and GGGGGG in the last common ancestor of plains and steppe bison, includes two indel sites, and terminates with a C to T transition characteristic of the Elk Island woods bison (GU947005) cohorts. While repeat and revertant mutations can muddy the interpretive waters even for rare genomic events, these sites do not vary in the 247 available yak control regions. Wild cattle and water buffalo do not work well as earlier outgroups because too much divergence has accumulated.
Gardipee determined haplotypes of 151 YNP and 28 GTNP bison for a 2007 MS thesis at UM. While this never resulted in a journal publication or GenBank entries, the level of scientific detail and genetics qualifications of the thesis review committee warrant inclusion of these additional statistics here, especially since they represent almost all of the Yellowstone bison data set -- all but 7 of 158 total individuals.
Two methodological limitations of the 2006 study by 2011 standards-- using cheaper, obsolete RFLP fragment proxies in place of actual dna sequencing (ie reliance on restriction endonuclease Ssp1 fragment lengths of 372 and 98 bp to identify hap8 defined by Ward/Derr) and incomplete sequencing of even the control region -- of the 470 bp PCR, only the first 408 bp relates to hap6 and hap8. These were validated to a certain extent by controls.
RFLP was an inappropriate technology then or now -- it misses all the subclades and private mutations outside the restriction enzyme site. And the short PCR reads -- Sanger technology routinely gave 1100 bp reads during the 2001 human genome project. One has to wonder what the senior thesis advisors were thinking -- RFLP baffles molecular biologists the same way the bizarre era of starch gel electrophoresis in population genetics baffled the protein biochemistry community.
It's necessary to revisit the RFLP specificity of endonuclease SspI and sensitivity to nucleotide modification and degraded dna in view of the much broader plains and steppe bison haplotypes known to exist today. SspI makes a blunt cut on palindromic AATATT in the positive strand notation of GenBank entries. This short pattern occurs up to 18 times in typical bison whole mitochondrial genome, for example GU946997 (MT) or GU947004 (YNP) out of 16,323 bp so has serious potential for ambiguity. Thus with the Anchorage, Alaska bison AY748509, SspI would result in a classification error because this has a private mutation to uncleavable AATACT.
Applying SspI in silico to hap8 AF083364 does not give fragments of length 372 and 98 (= 470 bp) but instead fragments of length 38 and 615 (= 653) bp. This is the only GenBank sequence of Ward/Derr labeled hap8. The Gardipee sequences apparently began 62 bp upstream of the 5' start of AF083364. Subtracting 62 bp to get 36 and 310 reconciles the issue (the 310 premature terminus is irrelevant) if Gardipee took the Ssp1 motif start position rather than distance to the actual central cleavage site.
More puzzling is the lack of private mutations reported by Gardipee, especially in view of those observed in whole-genome defined haplotypes. That is, nothing posted to GenBank implies no variants were discovered for hap6 and hap 8 in the 120 animals for which actual sequencing was performed. Expectations can readily be tested by blastn against all GenBank Bison bison of AF083364 shortened to 408 bp but lengthened on the 5' side by an additional 62 bp borrowed from GU946997 (respectively GU947004 for hap6) to recreate the Gardipee region.
That exercise shows some 29 private mutations and 83 micro-clade substitutions over the 470 sites might be expected from 120 animals, that is 29 + 83 = 111 variations out of 470*120 = 56,400 sites (ignoring haplotype correlation) if YNP bison were representative of bison genetic diversity at GenBank. However GenBank represents the diversity of all herds ever sampled, including Canadian and Texan bison and animals in European zoos not immediately related to current YNP stock. Further, the YNP bison appear to be quite homogeneous, representing only 2 of the main 5 micro-haplotypes. (Positions 102-170 of the sequence cover the diagnostic region used to define micro-haplotypes hapA-hapE.) Nonetheless, zero departures from the reference sequence suggests some variants were missed, discarded as sequence error, or deemed tangential to project purpose.
The bottom line here is that the Gardipee study holds up quite well. RFLP and partly sequenced control regions together provide -- after fixing Table 3-1 -- an additional 145 bisons with mitochondrial disease and 34 without, again under the assumptions that truncated micro-haplotypes still provide a valid window on protein coding genes and that the bison sampled are representative of the YNP herds. If so, this is very bad news indeed since it implies the (super-bottlenecked historically) Grand Teton herd is entirely composed of animals with mitochondrial disease as are 78% (117 of 151) of the Hayden, Lamar Valley and Mirror Plateau bison.
hap6 hap8
Hayden Valley 88 6
Lamar Valley 19 22
Mirror Plateau 10 6
Tetons Antelope Flats 20 0 [note bad typo of 50]
Tetons Wolf Creek 8 0
totals: 145 + 34 = 179
Note the table already maps the 1999 Ward/Derr haplotypes (hap notation) into 2010 whole genome haplotypes (bhap notation) defined by Douglas/Derr in Dec 2010 and the diagnostic micro-haplotypes defined here. From this, it appears that the old control-region based hap6 splits into whole-genome based deleterious bHap2, bHap17, bHap10, bHap11 and confusingly bHap8 but not bHap6. The old hap8 could be any of non-deleterious bHap3, bHap4, bHap5, confusingly bHap6 but not bHap8, bHap7, bHap9, bHap12, wHap14.
Blastn of the defining GenBank entry for the 653 bp hap8 of Ward/Derr, namely AF083364, shows complete identity only with 4 complete genomes (bHap4,bHap5, bHap9) from Montana private (GU946980, GU946982, GU946997, GU946989), 11 fossil bison sequences of Shapiro/Cooper, and 6 never-described bison Beech/Strobeck 1995 unpublished. None of these bison would carry the mitochondrial disease alleles.
For the 655 bp hap6 of Ward/Derr defined by GenBank entry AF083362, blastn against all bison entries gives 100% identity to:
GU946988 B973 bHap8 GU946993 B1029 bHap2 GU946995 B1050 bHap2 GU946996 B1051 bHap2 GU946986 B959 bHap2 GU946984 B929 bHap2 GU946983 B925 bHap2 GU946981 B880 bHap2 GU946978 B854 bHap2 GU946977 B853 bHap2 GU946976 B790 bHap2 GU947000 BFN5 bHap10 GU946991 B1005 bHap10 GU946990 B985 bHap10 GU946994 B1031 bHap11 GU947004 BNBR1 bHap17 EU272056 Bibi35 EF693810 U12934 A02 U12933 A01 DQ452027 CIC2 but not quite (1 bp difference): U12945 U12946 U12948 EU272058 AY428859 AF083357 AY748518 AY748509
One shortcoming of the statistics: some bison sequences could be duplicates if the same dna samples were passed around, given different haplotype acronyms by different groups or sequenced more fully at later dates. Few groups ever explain their acronyms and here the Beech Strobeck sequences of 1995 still don't have an associated publication -- their unsatisfactory GenBank entries provide no clues as to herd sourcing.
The two fossil bison were found in Anchorage, Alaska and Natural Cave Trap, Wyoming. These date from 170 and 3,220 years ago (AY748509 BS162 and AY748518 BS173). Blastn and haplotype signatures establishes definitively that these are Bison bison (plains bison), not Bison priscus (steppe bison), as classified in Shapiro's GenBank entries. It would be of great interest to sequence more of the mitochondrial genome than just the control region.
The Anchorage find in 1969 by Guthrie came as a surprise because Alaskan bison were not thought to range outside the Yukon basin or thrive in such a wet habitat. The male skull, found partly exposed in a streambank of Chester Creek, was questioned by Heuer but reinvestigation by Gerlach in 2003 has confirmed the original interpretation and dating -- 'nearby' recent bones cite by Heuer proved to be irrelevant small mammals. The skull could not plausibly been brought from interior Alaska for trade purposes in 1800, then discarded upstream and fortuitously buried by sediment in low gradient streamside habitat bison favor today in the far North. Since a minuscule fraction of living animals end up as discovered fossils, this bison represented a population, not a wandering solo Yukon bull. Neither hoax nor fraud from a distinguished academic paleobiologist merit serious consideration. Bison can escape notice -- the last herd of Canadian wild bison was not discovered until 1957 near the Nyarling river in Wood Buffalo National Park.
Although called a woods bison, the mitochondrial control region haplotype is typical of southern plains bison, again no support here for woods bison as a valid taxon. Other recent bison fossils in the far North include a 370-year-old bison molar from Yukon, Canada and a 420-year-old skull from Northwest Territories, so regional bison populations persisting to modern times can no longer be doubted. Part of the bias arises from misunderstanding of habitat requirements: bison are often said obligate grass grazers whereas carefully monitoring of feces of Nahanni bison proves these bison winter almost exclusively on willow and rose browse, plus horsetail and sedges. Equisetum and Carex do not remotely qualify as Poaceae. Their high silicate content relative to grasses causes very rapid wear of bison molars.
The other fossil plains bison potentially with mitochondrial disease comes from a 3,220 year old metapodial bone in Natural Trap Cave, a karst sinkhole in Wyoming. Natural Trap Cave may not be ideal for either stratigraphic or C14 dating. Animals perish after a long fall, landing on 250,000 years of previously accumulated debris. They are only slowly buried though bones are not scattered by mammalian scavengers. Rainwater (high C14) falls directly on the bones, yet water percolating through ancient limestone (low-C14) could give rise to a spurious older date. Purified collagen should have been used here for contaminant-free dating. One wonders what became of the skull and teeth.
Sequencing error cannot explain the perfect match to disease haplotype -- the Alaskan specimen has a very recent date and minimal thermal history. As noted elsewhere, fossil bison sequences of Shapiro are significantly affected by cytosine deamination as claimed and here retention of initial C is a hallmark of the micro haplotype hapA here. On the contrary, the predominant error type is on homopolymer runs as can be seen from the implausible proliferation of distinct steppe bison haplotypes but this affects accuracy of modern dna as well for Sanger and Illumina technologies but especially Roche 454. The 2004 study used Sanger sequencing (ABI 377, 310 and 3700 instruments).
It does not follow that either of the two fossil specimens carries either or both CYTB and ATP6 mutations. Overall haplotype establishment likely preceded the sequential addition of mutations. More data might locate 'intermediate' stages with just one (earlier) mutation. If the mutations are not new and affected bison ranged from Wyoming to Alaska, how could this be reconciled with natural selection? Possibly through heteroplasmy: V98A and I60N could have arisen thousands of years ago yet co-existed for long periods with compensatory residual wildtype mitochondrial dna. Sequencing at high enough coverage could establish the degree of heteroplasmy in these fossils. However damaged dna produces enough error that variation is commonly discarded as mere sequencing error. Somatic cell data also does not speak directly to the germline situation. In humans, mitochondrial diseases often arise late in life in restricted somatic cell lineages. In relatively short-lived animals, natural selection would not come into play.
The Yellowstone bison are represented by two micro-haplotypes, hapA and hapB. One animal is known affected from whole mitogenomic sequencing but for three others, known herd founder history predicts maladaptive CYTB and ATP6 genes. The remaining two animals likely have normal mitochondrial proteins. One hopes that the Ward-Derr group has retained the dna for these latter 5 animals. It seems the bad haplotype rose to high frequency in the severely bottlenecked population of the 1890's, having been neutralized selectively by prevailing unnatural selection. It appears the disease haplotype is now being maternally inherited in a simple manner, ie the compensatory wildtype haplotype is no longer prevalent and quite possibly has disappeared.
The Elk Island bison are represented by one individual predicted to have the bad haplotype, two known and four predicted to have a good haplotype, almost the opposite statistics from the YNP bison (sampling is too limited to establish significance). Since no gene flow has occurred in historic times, the data as it stands is consistent with an unfortunate enrichment of the bad haplotype during the YNP bottleneck and its subsequent export to many other American herds (National Bison Refuge, Fort Niobrara and Montana private proven and a dozen others predicted).
The 32 animals with complete mitochondrial genomes fall into four micro-haplotypes hapA-D (represented by 17, 11, 1, 3 sequences respectively), but this obscures major differences in overall sequence clustering. Upon all-vs-all whole genome blastn, HapA emerges as much more narrowly defined, with ten bison completely identical over 16322 bp. The hapB group has much greater internal divergence suggesting that it is much older. All the numbers are very low compared to differences between human population groups consistent with more recent bottlenecks in bison evolution.
A set of 16 non-redundant sequences suffices to represent the whole set of 32 individual genomes. The table shows the number of nucleotide differences between any pair of complete genomes. The outcome is similar whether gaps or substitutions are used, or redundant or non-redundant sequence sets.
One Elk Island bison (U12960 hapC Hap15 wEI1) is of special interest. If there is to be a woods bison mitochondrion, this is the best candidate so far based on locations of fossil bison and the nine contemporary bison (which are not all well-described however in their GenBank entries). It is quite remote from hapA but not very close to hapB or hapD either.
The Texas State Bison Herd has an interesting history of founding bison. One sequenced individual there (GU947003 hapD bHap16 TSBH1005) represents significant mitochondrial diversity. These bison may have been the source for the Montana private herd, which otherwise has poor diversity and high representation of V98A and I60N.
The set of 21 complete genomes from wild yak differ from each other to a much greater extent than intra-bison comparisons, even within major subgroups. Thus the average number of substitutions between two randomly chosen yak mitochondrial genomes is 57 whereas that number is 14 in bison (a quarter of yak diversity). These numbers hardly change when both are reduced to non-redundant sets (average substitutions yak 61 to bison 16) to avoid sampling artifacts.
Yaks too experienced glacial bottlenecks and recent habitat loss but nothing temporally comparable to bison (nor their post-bottleneck expansion of the disease haplotype hapA). Overall the wild yak population carries much greater genetic diversity than bison. No yak-bison hybrids have been located -- even the extremely isolated yak GQ464260 W71 nests convincingly within that taxon.
In terms of haplotype coalescence, the maximal divergence of yak haplotypes is 131 bp versus 28 bp for bison. Since they are sister species sharing a common set of ancestral haplotypes at 2.5 million years, bison have lost about 80% of their mitochondrial genome diversity.
All-vs-all whole genome comparisons of non-redundant sets of yak and bison haplotypes gives a curious result: no matter which pair is compared, the result is always the same: a difference of about 442 bp (with an incredibly small standard deviation of 3 bp overall). Assuming a goodly diversity of haplotypes at the time of bison/yak divergence -- which was allopatric as bison are Beringian -- and each population then went its own way in terms of where mutations were fixed in the descendant haplotypes, then substructure should be observable in the all-vs-all bison vs yak table (according to matching or cross-matching). Yet bison and yak haplotype signatures are completely uncorrelated today (even though any two pair of mitochondrial genomes are 97% identical overall).
Even if bison haplotypes are impoverished today because of the 19th century bottleneck, this is not applicable to yak (all of whose haplotypes are equally distant from bison). Thus complete lineage sorting around the time of population disjunction may have occurred (possibly via a homogeneous founder population that took a whole haplotype away with it). If so, the current sets would coalesce earlier back in the stem common ancester.
From this ancestral haplotype, assuming equal rates of divergence, each lineage accrued 221 substitutions over the minimum 2.5 million years, for a maximum rate of 1 substitution per 11,300 years. Since this is approximately the length of the Holocene, bison haplotypes became established considerably earlier in the Pleistocene, the main coalescences dating to roughly 30,700 years for the hapA group and 262,000 for divergence of hapA from the others. In yak the haplogroups are more internally diverged, so the dates are older, corresponding all likelihood to specific glaciation events on the Tibetan plateau as note by Zhaofeng Wang et al.
Here are the summary statistics from the spreadsheet. Data from the Gardipee study, 145 YNP bisons with mitochondrial disease and 34 without, represents a large survey of a single herd and so would skew the overall estimate of prevalence in North American conservation herds. This too is only proxy data -- a portion of the mitochondrial control region being used to predict distant nucleotides in two coding genes.
17 bison from 4 herds known to have mitochondrial disease hapA 36 bison from 15 herds predicted to have mitochondrial disease hapA 2 fossils from 0 sites predicted to have mitochondrial disease hapA 11 bison from 3 herds known not to have mitochondrial disease hapB 33 bison from 13 herds predicted not to have mitochondrial disease hapB 38 fossils from 14 sites predicted not to have mitochondrial disease hapB 1 bison from 1 herd known not to have mitochondrial disease hapC 12 bison from 9 herds known not to have mitochondrial disease hapC 7 fossils from 7 sites predicted not to have mitochondrial disease hapC 3 bison from 2 herds known not to have mitochondrial disease hapD 1 bison from 1 herd seems not to have mitochondrial disease hapE 1 fossil from 1 site seems not to have mitochondrial disease hapE 53 unhealthy/77 healthy = 42% living bison have mitochondrial disease
Here is a hand-gapped alignment of the micro-haplotype region between yak, cattle and bison: steppe bison situation is too complicated to include:
TTACAAACACCACTAGCTAACAACACACATCC----------CCAAAAATGCATTATCCA---AACGGGGGAATACGTACATAAc GQ464236 yak TTACAAACACCACTAGCTAACATAACACGCCCATACACAGACCACAGAATGAATTACCCAGGCAA-GAGGTAAT--GTACATAAC GU256940 indicus TTGCAAACACCACTAGCTAACGTCACTCACCC-----------CCAAAATGCATTACCCA---AACGGGGGAAT--ATACATAAC GU947006 bison
Here are haplotype frequencies in outgroup species and ancestral micro-haplotypes: (to be continued)
BosGru ..A..................AA...A..T................T...........AATACG--...... 1 BosGru ..A..................AA...A..T...............GT............ATACG--...... 2 BosGru ..A..................AA...A..T...............GT...........AATA.G--...... 9 BosGru ..A..................AA...A..T...............GT...........AATACG--.....t 235 BisBis ..................................AAAA-..............GGGGG-A............ 2 BisBis ..................................AAAA-..............GGGGG-A...G.......T 3 BisBis ..................................AAAA-..............GGGGG--...........T 13 BisBis ..................................AAAA-..............GGGGG--............ 49 BisBis C.................................AAAA-..............GGGGGGA............ 31 BisBis ..................................AAAAA..............GGGGG--............ 4 ..A...........AGCTAACGTCACTCACCCCCAAAAATGCATTACCCAAACGGGGGGAAACGTACATAAC LCA bison/yak TTGCAAACACCACTAGCTAACGTCACTCACCCCCAAAAATGCATTACCCAAACGGGGGGAAATATACATAAC LCA plains/steppe TTGCAAACACCACTAGCTAACGTCACTCACCCCCAAAAATGCATTACCCAAACGGGGGGAAATATACATAAC BisPri ..A...................................-..............A.....-..CG........ 41 BisPri .....................................................A.....-..CG........ 5 BisPri ...................-...................A.....-..CG........ 3 BisPri ........................-..............A.....-...G........ 2 BisPri ...................-...................A.....--.CG........ 2 BisPri ...................A...................A.....-G.CG........ 6 BisPri ...................-...................A.....-G.CG........ 6 BisPri ....................-..................A.....-G.CG........ 3 BisPri ........................-..............A.....G..CG........ 4 BisPri ...................-...................A.....GG.CG........ 3 BisPri ...................A...................A.....GG.CG........ 2 BisPri ...................-.........................-..CG........ 2 BisPri ........................-....................-..CG........ 9 BisPri ...................--........................-..CG........ 8 BisPri ........................-...........G........-..CG........ 7 BisPri ...................--...........................CG........ 5 BisPri ........................-...................--............ 3 BisPri ........................-...................--..CG........ 8 BisPri ...................-........................--..CG........ 2 BisPri ........................-..............-......G.CG........ 12 BisPri ........................-...................A.G.CG........ 6 BisPri ...................--........................G..CG........ 3
Here is the text version of the spreadsheet. Minor adjustments are made frequently.
Accession TTGCAAACACCACTAGCTAACGTCACTCACCCCCAAAAATGCATTACCCAAACGGGGGGAAATATACATAAC Haplo Status CYTB ATP6 Haplo1 Date ST Haplo2 Site Authors GU946996 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B1051 Montana private Douglas Derr GU946995 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B1050 Montana private Douglas Derr GU946981 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B880 Montana private Douglas Derr GU946976 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B790 Montana private Douglas Derr GU946983 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B925 Montana private Douglas Derr GU946984 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B929 Montana private Douglas Derr GU946993 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B1029 Montana private Douglas Derr GU946986 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B959 Montana private Douglas Derr GU946977 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B853 Montana private Douglas Derr GU946978 C.....................................-................................. hapA observed 98A 60N bHap2 0 MT B854 Montana private Douglas Derr GU946988 C.....................................-................................. hapA observed 98A 60N bHap8 0 MT B973 Montana private Douglas Derr GU946991 C.....................................-................................. hapA observed 98A 60N bHap10 0 MT B1005 Montana private Douglas Derr GU946990 C.....................................-................................. hapA observed 98A 60N bHap10 0 MT B985 Montana private Douglas Derr GU946994 C.....................................-................................. hapA observed 98A 60N bHap11 0 MT B1031 Montana private Douglas Derr GU947000 C.....................................-................................. hapA observed 98A 60N bHap10 0 NB BFN5 Fort Niobrara Douglas Derr GU947001 C.....................................-................................. hapA observed 98A 60N bHap2 0 NB BNBR1 National Bison Douglas Derr GU947004 C.....................................-................................. hapA observed 98A I60 bHap17 0 YP 1586 Yellowstone NP Douglas Derr AY748509 C.....................................-................................. hapA predicted 98A 60N Gerlach 170 AK BS162 Anchorage Shapiro Cooper AY748518 C.....................................-................................. hapA predicted 98A 60N KU42887 3220 WY BS173 Natural Trap Cave Shapiro Cooper AF083362 C.....................................-................................. hapA predicted 98A 60N ..... 0 YP hap6 3 YNP + 22 misc* Ward Derr AF083357 C.....................................-................................. hapA predicted 98A 60N ..... 0 EI hap1 Elk Island Ward Derr EF693810 C.....................................-................................. hapA predicted 98A 60N ..... 0 FR ..... France Hassanin Ropiquet U12948.. C.....................................-................................. hapA predicted 98A 60N ..... 0 NA P64 Plains Beech Strobeck U12946.. C.....................................-................................. hapA predicted 98A 60N ..... 0 NA P35 Plains Beech Strobeck U12945.. C.....................................-................................. hapA predicted 98A 60N ..... 0 NA P57 Plains Beech Strobeck U12934.. C.....................................-................................. hapA predicted 98A 60N ..... 0 NA AO2 not specified Beech Strobeck U12933.. C.....................................-................................. hapA predicted 98A 60N ..... 0 NA AO1 not specified Beech Strobeck EU272058 C.....................................-................................. hapA predicted 98A 60N ..... 0 NA Bibi54 not specified Wojcik Pertoldi EU272056 C.....................................-................................. hapA predicted 98A 60N ..... 0 NA Bibi35 not specified Wojcik Pertoldi DQ452027 C.....................................-................................. hapA predicted 98A 60N ..... 0 CA CIC2 Santa Catalina Vogel Hedgecock AY428859 C.....................................-................................. hapA predicted 98A 60N ..... 0 NA ..... not specified Verkaar Lenstra GU947006 ......................................-...................--............ hapB observed V98 I60 wHap14 0 EI wEI14 Elk Island Douglas Derr GU946992 ......................................-...................--............ hapB observed V98 I60 bHap3 0 MT B1018 Montana private Douglas Derr GU946979 ......................................-...................--............ hapB observed V98 I60 bHap3 0 MT B855 Montana private Douglas Derr GU946980 ......................................-...................--............ hapB observed V98 I60 bHap4 0 MT B877 Montana private Douglas Derr GU946997 ......................................-...................--............ hapB observed V98 I60 bHap9 0 MT B1091 Montana private Douglas Derr GU946982 ......................................-...................--............ hapB observed V98 I60 bHap5 0 MT B897 Montana private Douglas Derr GU946985 ......................................-...................--............ hapB observed V98 I60 bHap6 0 MT B935 Montana private Douglas Derr GU946987 ......................................-...................--............ hapB observed V98 I60 bHap7 0 MT B961 Montana private Douglas Derr GU946989 ......................................-...................--............ hapB observed V98 I60 bHap9 0 MT B979 Montana private Douglas Derr GU946998 ......................................-...................--............ hapB observed V98 I60 bHap12 0 MT B1191 Montana private Douglas Derr EU177871 ......................................-...................--............ hapB observed V98 I60 ..... 0 IT ..... Italian zoo Achilli Torroni AY748745 ......................................-...................--............ hapB predicted V98 I60 ..... .... AB BS567 Calgary Shapiro Cooper AY748477 ......................................-...................--............ hapB predicted V98 I60 ..... 89 AB BS100 Fort Smith Shapiro Cooper AY748530 ......................................-...................--............ hapB predicted V98 I60 ..... 145 BC BS200 Fort D’Epinette Shapiro Cooper AY748519 ......................................-...................--............ hapB predicted V98 I60 ..... 186 MT BS175 Ice Cave Shapiro Cooper AY748630 ......................................-...................--............ hapB predicted V98 I60 ..... 200 MT BS368 Fort Benton Shapiro Cooper AY748495 ......................................-...................--............ hapB predicted V98 I60 ..... 200 MT BS129 Fort Benton Shapiro Cooper AY748693 .................................A....-...................--............ hapB predicted V98 I60 ..... 287 AB BS454 Banff NP Shapiro Cooper AY748690 ......................................-...................--............ hapB predicted V98 I60 ..... 378 AB BS445 Banff NP Shapiro Cooper AY748689 ......................................-...................--............ hapB predicted V98 I60 ..... 636 AB BS444 Edmonton Shapiro Cooper AY748687 ......................................-...................--............ hapB predicted V98 I60 ..... 1273 AB BS441 Waterton Lakes NP Shapiro Cooper AY748702 ......................................-...................--............ hapB predicted V98 I60 ..... 2398 AB BS466 Lloydminster Shapiro Cooper AY748739 ......................................-...................--............ hapB predicted V98 I60 ..... 2807 AB BS560 Calgary Shapiro Cooper AY748747 ......................................-...................--............ hapB predicted V98 I60 ..... 3600 AB BS569 Calgary Shapiro Cooper AY748700 ......................................-...................--............ hapB predicted V98 I60 ..... 5205 AB BS464 Edmonton Shapiro Cooper AY748685 ......................................-................A..--............ hapB predicted V98 I60 ..... 5845 AB BS439 Edmonton Shapiro Cooper AY748678 ......................................-...................--............ hapB predicted V98 I60 ..... 6775 AB BS429 Calgary Shapiro Cooper AY748676 ......................................-...................--............ hapB predicted V98 I60 ..... 7060 AB BS426 Cypress Hills Shapiro Cooper AY748677 ......................................-...................--............ hapB predicted V98 I60 ..... 7105 AB BS428 Cypress Hills Shapiro Cooper AY748701 ......................................-...................--............ hapB predicted V98 I60 ..... 7115 AB BS465 Cypress Hills Shapiro Cooper AY748680 ......................................-...................--............ hapB predicted V98 I60 ..... 7310 AB BS432 Calgary Shapiro Cooper AY748671 ......................................-...................--............ hapB predicted V98 I60 ..... 7475 AB BS419 Calgary Shapiro Cooper AY748672 ......................................-...................--............ hapB predicted V98 I60 ..... 8145 AB BS421 Cypress Hills Shapiro Cooper AY748679 ......................................-...................--............ hapB predicted V98 I60 ..... 9270 AB BS430 Edmonton Shapiro Cooper AY748617 .........T............................-.......T...........--............ hapB predicted V98 I60 ..... 10340 BC BS342 Peace River Shapiro Cooper AY748699 ......................................-...................--............ hapB predicted V98 I60 ..... 10425 AB BS460 Athabasca Shapiro Cooper AY748620 ...........................T..........-...................--............ hapB predicted V98 I60 ..... 10505 BC BS348 Peace River Shapiro Cooper AY748748 ......................................-...................--............ hapB predicted V98 I60 ..... 11300 AB BS570 Calgary Shapiro Cooper AY748742 ......................................-...................--............ hapB predicted V98 I60 ..... .... AB BS563 Medicine Hat Shapiro Cooper AY748758 ......................................-...................--............ hapB predicted V98 I60 ..... .... KS BS602 Kansas River Shapiro Cooper AY748757 ......................................-...................--............ hapB predicted V98 I60 ..... .... WY BS595 Natural Trap Cave Shapiro Cooper AY748618 ......................................-...................--............ hapB predicted V98 I60 ..... .... WY BS343 Natural Trap Cave Shapiro Cooper AF083364 ......................................-...................--............ hapB predicted V98 I60 ..... 0 YP hap8 2 YNP + 7 misc** Ward Derr AF083363 ......................................-...................--............ hapB predicted V98 I60 ..... 0 EI hap7 1 EI + 2 NBR Ward Derr AF083360 ......................................-...................--............ hapB predicted V98 I60 ..... 0 EI hap4 3 EI + 2 misc Ward Derr AF083361 ......................................-...................--............ hapB predicted V98 I60 ..... 0 .. hap5 1 WBNP + 4 misc Ward Derr U12941.. ......................................-...................--............ hapB predicted V98 I60 ..... 0 .. M05 7 not specified Beech Strobeck U12943.. ......................................-...................--............ hapB predicted V98 I60 ..... 0 .. P30 6 Plains*** Beech Strobeck U12960.. ......................................-...................--............ hapB predicted V98 I60 ..... 0 .. W62 5 Woods Beech Strobeck GU947005 ......................................-...................--...........T hapC observed V98 I60 wHap15 0 EI wEI1 Elk Island Douglas Derr AY748478 ......................................-...................--...........T hapC predicted V98 I60 ..... 82 AB BS102 Wood Bison NP Shapiro Cooper AY748476 ..A...................................-...................--...........T hapC predicted V98 I60 ..... 86 AB BS99 Salt Prairie Shapiro Cooper AY748695 ..............................T.......-...................--...........T hapC predicted V98 I60 ..... 125 AB BS456 Boss Hill Arch Shapiro Cooper AY748675 ......................................-...................--...........T hapC predicted V98 I60 ..... 202 AB BS424 Fort Vermillion Shapiro Cooper AY748673 ......................................-...................--...........T hapC predicted V98 I60 ..... 908 AB BS422 Athabasca Shapiro Cooper AY748674 ......................................-...................--...........T hapC predicted V98 I60 ..... 4660 AB BS423 Cypress Hills Shapiro Cooper AY748529 ......................................-...................--...........T hapC predicted V98 I60 ..... .... BC BS199 Fort D’Epinette Shapiro Cooper AF083359 ......................................-...................--...........T hapC predicted V98 I60 ..... 0 AB hap3 2 MBS + 1 WBNP Ward Derr U12959.. ......................................-...................--...........T hapC predicted V98 I60 ..... 0 .. W29 Woods Beech Strobeck U12942.. ......................................-...................--...........T hapC predicted V98 I60 ..... 0 .. M07 Misc frag Beech Strobeck U12937.. ......................................-...................--...........T hapC predicted V98 I60 ..... 0 .. GC2 Misc frag Beech Strobeck U12951.. ......................................-...................--...........T hapC predicted V98 I60 ..... 0 .. PL4 Misc frag Beech Strobeck GU947003 ......................................-...................-....G.......T hapD observed V98 I60 bHap16 0 TX 1005 Texas State Bison Douglas Derr GU947002 ......................................-...................-....G.......T hapD observed V98 I60 bHap13 0 TX 1001 Texas State Bison Douglas Derr GU946999 ......................................-...................-....G.......T hapD observed V98 I60 bHap13 0 MT B1428 Montana private Douglas Derr AY748682 ..........................................................--............ hapE weak pred V98 I60 ..... 809 AB BS434 Boss Hill Arch Shapiro Cooper AY748669 ..........................................................--............ hapE weak pred V98 I60 ..... 909 AB BS417 Waterton Lakes NP Shapiro Cooper AY748521 ..........................................................--............ hapE weak pred V98 I60 ..... 3155 WY BS177 Natural Trap Cave Shapiro Cooper AF083358 ..........................................................--............ hapE weak pred V98 I60 ..... 0 TX hap2 Wichita Mtns Ward Derr AY748681 ......................................-...................-............. hapF weak pred 98A 60N P001.4 10450 AB BS433 Byrtus Site Shapiro Cooper U12864.. ......................................-...................-............. hapF weak pred 98A 60N ..... 0 NA ..... not specified Feng Templeton 17 bison from 4 herds known to have mitochondrial disease hapA ** 2 Mackenzie Sanctuary * 1 Custer State Park 36 bison from 15 herds predicted to have mitochondrial disease hapA 1 Custer State Park 2 Elk Island 2 fossils from 2 sites predicted to have mitochondrial disease hapA 2 Natl Bison Refuge 5 Fort Niobrara NWR 11 bison from 3 herds known not to have mitochondrial disease hapB *** U12947 P63 5 Henry Mountains 33 bison from 13 herds predicted not to have mitochondrial disease hapB U12944 P33 1 National Bison Range 38 fossils from 14 sites predicted not to have mitochondrial disease hapB U12947 P63 5 Wind Cave National Park 1 bison from 1 herd known not to have mitochondrial disease hapC U12944 P33 3 Wichita Mountains NWR 12 bison from 9 herds known not to have mitochondrial disease hapC U12950 PL4 U12940 LB3 7 fossils from 7 sites predicted not to have mitochondrial disease hapC U12958 W26 U12938 LB3 3 bison from 2 herds known not to have mitochondrial disease hapD U12957 W03 U12936 C03 1 bison from 1 herd seems not to have mitochondrial disease hapE U12956 W01 U12935 C02 1 fossil from 1 site seems not to have mitochondrial disease hapE U12955 W05 U12952 SW04 53 diseased/77 healthy = 42% living bison have mitochondrial disease .... U12939 GC2 U12949 SW03
Structural and functional consequences of bison mitochondrial mutations
Structural studies of amino acid substitutions in mitochondrial proteins can supplement analysis by comparative genomics and physical-chemical change in properties. This requires a pre-existing xray structural determination of the protein in question, a relevant motif established in a paralogous protein (eg Rossmann fold for NADH binding) or a reliable ab initio structural prediction amenable to molecular dynamics modeling.
Most structural studies of mammalian protein use proteins purified from a bovine source -- not exactly bison but close enough to reliably thread bison sequence onto them and then introduce substitutions, optimize rotamers and evaluate the overall effect. However oxidative phosphorylation is a very complex process involving some 125 proteins (mostly nuclear), so a full understanding would need to encompass hetero-oligomeric partners both stable and transitory.
While cytochrome b and the three cytochrome oxidases have available high resolution models of their full complexes, little such data is available for the other 9 mitochondrially encoded proteins. For example, a structure for ATP6 has been determined but only for a very distant E. coli homolog and the alignable region does not cover the bison mutation of interest. Even if it did, the evolutionary distance is too great to allow annotation transfer.
The structure of bison ATP6 (and others) cannot be usefully predicted ab initio to the extent it consists of loops connecting transmembrane regions. The loops, too short for a hydrophobic interior, cannot follow the folding rules of cytoplasmic proteins; the distinct angles of transmembrane helices relative to membrane perpendicular and inter-helical associations are unpredictable, just as with GPCR even given the carefully refined structure of rhodopsin.
In terms of membrane topology, it can sometimes be established whether a given loop lies on the matrix or interstitial side of the mitochondrial inner membrane. That can roughly halve the number of potential interacting partners but cannot be expected to illuminate the consequences of particular amino acid changes in a loop. Compensatory mutations in such proteins are sometimes found as yeast suppressors but here too it is problematic whether that information would carry over to bison (where nuclear-encoded gene sequences are not yet available for any bison, much less the individual mitochondrial haplotypes).
Yeast however is a surprisingly relevant model system. Most work focus on putting mutant alleles of human genes into the nucleus with mitochondrial targeting signals (and suitably altered to use the nuclear gene translation table), while suppressing mitochondrial dna -- and its competing protein -- with ethidium bromide. This results in a heterologous oxidative phosphorylation system called allotopic expression of one human protein interacting with 12 yeast mitochondrial proteins and all the other yeast nuclear encoded ones.
This appears to reliably characterize structural and functional attributes of numerous mutations -- surprising perhaps yet what has changed in oxidative phosphorylation over the last billion years -- nothing. Allotopic expression has also been achieved in a hamster cell line which addresses some issues arising from extreme sequence divergence. The yeast or hamster system could be adapted to specifically study the effect of variations on wildtype bison protein for both CYTB and ATP6.
Cybrids -- cells which combines the nuclear genome from one source with the mitochondrial genome from another may provide a route to genetic re-engineering of bison mitochondrial dna back to that of steppe bison, should none of the surviving bison today have a wildtype mitochondrial genome. It is otherwise quite difficult to get at this dna because of its compartmentalization and lack of recombination.
A final method considered here for interpreting unusual bison alleles relies on clinically analyzed human variation known for the same site. Conservation genomics of bison will never attain the funding level or investigative intensity of human mitochondrial disease (which often affects children). Consequently, when such information is available, it should be considered even though correspondences can only suggest specific avenues to experimentally pursue in bison (eg lactic acid buildup, encephalomyopathies, exercise intolerance). As noted above V98A in cytochrome b does have an approximate canine counterpart in V98M disease. That will not prove the case for I60N of ATP6. However, even there we shall see that all 21 known mutations in the human gene have similar outcomes, suggesting something similar for bison.
Structural impact of I60N on ATP synthase F0 subunit 6
The bison substitution I60N follows the same sub-clade pattern as V98A. It too is a mutation. Fortunately both reside on the same haplotype, a very small 19th century cohort (in terms of little overall mitochondrial dna divergence) that is greatly expanded in the bison herds of today. These bison, though genetically pure in the sense of no introgression of cattle genes, very likely are overtly affected by mitochondrial disease given the double mutation (which cannot be mutually compensatory since they occur in different oxidative phosphorylation complexes).
According to SwissProt secondary structure analysis of ATP6 of 97% identical yak and cattle, I60N is located in a loop region connecting the first two transmembrane helices, as shown above. For tertiary structure, it is currently necessary to use the E. coli PDB model 1C17|M for bison ATP6, even though the alignment is quite weak at 27% identity and does not really extend to the I60N site:
Bison: KQMMSNHNPKGQTWTLMLMLMSLILFIGSTNLLGLLPHSFTPTTQLSMNLGMAIPLWAGAVITGFRNKTKASLAHFLPQGTPTPL--
L+ + L+ +I ++LGL P+ +++ L MA+ ++ +I + K K + F + T P
Ecoli: PLALTIFVWVFLMNLMDLLPIDLLPYIAE-HVLGLPALRVVPSADVNVTLSMALGVFI--LILFYSIKMKG-IGGFTKELTLQPFNH
Bison: ---IPMLVIIETISLFIQPMALAVRLTANITAGHLLIHLIGGATLALMSISTTTALIMFIVLILLTVLEFAVAMIQAYVFTLLVSLYL
IP+ +I+E +SL +P++L +RL N+ AG L+ LI G L S +I+ + + + +QA++F +L +YL
Ecoli: WAFIPVNLILEGVSLLSKPVSLGLRLFGNMYAGELIFILIAGL-LPWWS--------QWILNVPWAIFHILIITLQAFIFMVLTIVYL
The global topology of the entire E. coli membrane proteome was established in 2005, enabling the localization of this region to cytoplasm or periplasmic space and so by homology transfer, perhaps reliable localization of the loop containing I60N to either mitochondrial matrix or inter-membrane region. ExTopDB, a web database carrying this information for all protein for which it has been experimentally determined, shows position 108 of the E. coli protein to lie within the second membrane helix tm2:100-118 which is headed inwards at this point. This conflicts with the assignment of the comparable bison region to a loop, showing the limitations of the weak alignment.
The conservation of residue 60 indicates its overall importance. Asparagine, which arises here from from wildtype valine via a T --> A transversion, is clearly highly anomalous, not being seen in 1635 other mammalian ATP6 sequences. This substitution is radical in terms of properties (branched aliphatic hydrophobic to polar) and seldom seen in large scale compilations of fixed amino acid substitutions. Consequently it is beyond doubt a serious mutation.
The figure at left also shows conservation within a small patch of 5 residues on each side. This has relevance if I60N occurs within a structural or functional local motif. However motifs in the three dimensional structure do not necessarily involve contiguous residues in the primary structure. Should the loop containing I60N form an alpha helix, it would not be unusual for one face to be conserved and the rest variable. This takes the form of periodic conservation (every 3-4 residues as 3.4 amino acids form a full turn of helix). That's not observed in this instance.
Human disease alleles in ATP6 do not include any changes at position 60. Note that the many clinical descriptors are similar to those of any mitochondrial disease and include here lack of exercise endurance, lactic acid blood acidemia, muscle weakness, and heart ventricular problems. thus the bison phenotype for I60N is more or less predictable. The columns indicate wildtype value, position of the mutation, the amino acid substitution, and whether seen homoplasmically, heteroplasmically or both in the range of patients studied.
wt pos mut hom het L 72 R - + Leigh syndrome: Maternal inheritance protective factor H 90 Y + - Exercise Endurance H 90 R - + Leigh syndrome: Maternal inheritance protective factor M 104 V + - Leber Hereditary Optic Neuropathy W 109 R + + Bilateral striatal necrosis P 136 S + - Prostate Cancer V 142 I + - Leber hereditary optic neuropathy and dystonia L 156 P - + Neurogenic muscle weakness, Ataxia, and Retinitis Pigmentosa/Leigh syndrome L 156 R - + Neurogenic muscle weakness, Ataxia, and Retinitis Pigmentosa/Leigh syndrome I 164 V - + Leber Hereditary Optic Neuropathy L 170 P + - Progressive ataxia A 177 T + - PD protective factor T 178 A + - Left ventricular noncompaction-associated I 191 T + - Predisposition to anti-retroviral mito disease I 192 T + - Leber Hereditary Optic Neuropathy A 205 T + - Leber Hereditary Optic Neuropathy L 217 P + + Familial Bilateral Striatal Necrosis/Leigh Disease L 217 R - + Leigh Disease L 220 P + + Leigh Disease/Neurogenic muscle weakness, Ataxia, and Retinitis Pigmentosa L 222 P - + Leigh Disease/Seizures/Lacticacidemia
Structural impact of V98A on cytochrome b
This amino acid variation in bison cytochrome b is straightforward to understand structurally. It has long been known that the preceding residue, H97, is one of two critical axial histidine ligands to the high potential heme. Two residues later, arginine R100 provides an essential salt bridge to a buried proprionic side chain of this same heme. It is scarcely necessary to perform a blastp comparative genomics study of these two residues -- they will prove strictly invariant over trillions of years of branch length (up to GenBank error and outright mutations in other species) because none of the other amino acids have the required properties.
An alignment of 5000 cytochrome b sequences establishes that either valine and isoleucine (and less commonly methionine) occur here in 99.6% of mammalian species, indeed vertebrates. This cannot be because opportunity for change is rare -- we shall see shortly that 18 of the 20 possible amino acids occur at position 62 of ATP6. The mutation rate of mitochondrial dna is so high that ample time has elapsed for position 98 in cytochrome b to equilibrate across the genetic code -- were not strong selective pressure enforcing the restriction to isoleucine and valine here.
The location of V98 is well-known from high resolution xray crystallography. It occurs at the end of transmembrane helix B, sandwiched between strongly conserved residues L94 Y95 M96 H97 G99 and R100. Any structural perturbation of axial liganding to the heme could change its redox potential which was optimized long ago; any disruption of the proprionate salt bridge would affect thermodynamic stability and folding or perhaps production of reactive oxygen species. Thus it is no surprise that position 98 is under strong selection -- but why isoleucine and valine and no others?
The explanation must lie in the post-beta carbon side chain. It cannot involve the alpha carbon or peptide bond -- any amino acid would provide these. A beta carbon alone cannot suffice -- in that case glycine and proline might be excluded from the reduced alphabet but nothing else. Alanine, the bison mutation under consideration, terminates its side chain with a methylene group here. That's evidently not good enough because alanine has never gained a toehold at this site despite being 'next door' to valine in the mitochondrial genetic code (ie is a simple T --> C transition away).
The complete absence of leucine at position 98 is also quite informative because it, like isoleucine and valine, is a branched chain aliphatic amino acid. However the branching comes later, at the gamma carbon. Thus rules out a role as simple insertion in the hydrophobic membrane milieu because leucine could do that equally well. Similarly phenylalanine, tryptophan and tyrosine do not occur here, again establishing the hydrophobicity, while necessary, is not sufficient. Consequently only isoleucine and valine work because the constraint must be a packing issue in a hydrophobic environment. It follows then that alanine is unacceptable because it leaves a packing void that is energetically unfavorable to protein folding, stability or functionality at the axial or proprionate ligands.
(to be continued shortly)
Mitchondrial tRNA variation in bison and yak
Mammalian mitochondrial dna encodes 22 tRNAs as well as 13 proteins. A great variety of mutations in these tRNAs cause human mitochondrial disease, likely through inefficient or incorrect production of mitochondrial proteins as the end clinical symptoms are very similar to mitochondrial disease of the proteins.
The Derr group raised concerns about the fitness of certain bison tRNAs. Here all tRNAs of both bison and yak were compared to each other and to other Bovidae. This can be done quickly using the remarkable Strasbourg tRNA database Mamit.
Five tRNAs are completely identical in the 69 yak and 32 bison mitochondrial genomes, namely Pro, Trp, Ile, Leu (CUN), and Arg. It is very unlikely adverse substitutions could persist because bison and yak have evolved under natural selection for all but the last fraction of the 2.5 million years since their divergence.
Bison does differ from yak at the species level at 11 sites in 9 tRNAs. That is, all bison have a substitution relative to all yak at these sites. In most cases, it is possible to unambiguously determine which species carries the ancestral nucleotide value. Other tRNAs are evolving so rapidly and erratically (indels) in the vicinity of the change that the ancestral state is unclear, with lineage sorting possibly at work as some changes are also seen in wild cattle. These changes are again unlikely to be deleterious mutations, especially in yak which has not experienced quite the same bottleneck as bison.
Bison have no tRNA changes that track with sub-clades. However one bison has a private allele in a leucine tRNA recognizing UUR codons, an animal from the Montana herd (already in the V98A I60N cull group). This occurs at the junction of D-stem with anti-codon stem so its functional interpretation is not clear. However in generic tRNA the G here is post-translationally modified (di-methylated) and has a role in formation of correct tertiary structure through an association with another purine at the start of the T-loop. Even a sub-optimal phenotype here does not have sweeping management implications unless the haplotype proves more abundant.
Yak present a quite different situation from bison. There are six tRNAs that have the same substitution in multiple animals (2-15), presumably tracking with recognized subclades, and four private substitutions occur in single animals. This greater mitochondrial diversity in yak tRNAs may reflect a larger wild population of 15,000 that may not have been as historically bottlenecked as bison.
Substitutions in tRNAs can be evaluated both by comparative genomics and by reference to known human mitochondrial diseases. However human tRNAs differ at a fair number of other sites so annotation may not reliably transfer over to bison. MitoMap maintains a current list of human tRNA alleles and their associated diseases, even carrying pre-publication data.
The status of the homologous leucine tRNA in humans is shown at left; the red arrow locates the site of substitution in bison. Change at this site has not been observed in humans to date. In the tertiary tRNA fold, this nucleotide forms an association with the first nucleotide of the variable region preceding the T-stem. However in a curated alignment of 127 mammals that includes yak and 41 other cetartiodactyls, A and G are used indiscriminately at the first position without any evident correlation with the second site. Consequently this variation is probably innocuous. The same can be said for the serine UCN change at the same site.
Comparative genomics takes a few twists here. Because tRNAs are short and indel rich, nucleotide alignment programs will not reliably place gaps, in part because they do not recognize -- and anchor the alignment to -- reverse complementary stem base pairs and other non-local constraints. Even if they could, homological (evolutionary descent) alignment may differ from structural alignment (to the standard folded-over cloverleaf). This latter alignment is the one relevant to dysfunction analysis.
Here Mamit-tRNA provides convenient structural alignments for 127 phylogenetically ordered mammalian species for each of the 22 tRNAs. The sequences have been parsed by tRNA experts into functional columns such as stems and loops. This allows such tricks as reverse-complementing one column of a stem pair (causing it to agree with the other column if hydrogen-bonded as A-T G-C or wobble pairs) which permits, after concatenation, a single difference alignment to display all exceptions to stem base pairing and so to evaluate the functional significance of departures. The outcome of that exercise is summarized at left. If the same change commonly appears in other cetartiodactyls, it is probably innocuous.
This applies to 8 of the 24 tabulated tRNA variations. Since these are all in different rows, none are compensated by a second mutation in the same stem that could restore complementarity. However many tRNAs have imperfect base pairing in their stems. Mutations in the T-stem may be of greater concern than those in the acceptor stem. In particular, the bison phenylalanine tRNA change lacks phylogenetic precedent. However it affects all known bison so has no management implications. Here it would be of interest to sequence this tRNA gene from steppe bison.
The other 16 anomalies are not constrained by stem base pairing. Two yak sequences (GQ464255 AY684273) are not considered further because excessive anomalies (including anti-codon change) indicate sequence error. The rest have no immediate opportunities for functional impact analysis other than conservation within cetartiodactyls and known homologous human disease alleles.
Some tRNA mutations seem inexplicable, such as the double base change in yak phenylalanine tRNA following a deletion relative to other Pecora (which is seemingly recurrent in Neotragus). One might suppose a inefficient but functional tRNA would not affect rates of translation for a rare amino acid (and this makes some sense for cysteine with just 22 sites) but phenylalanine occurs 244 times (6.4%)in the bison mitochondrial proteome. Mitochondrial tRNAs make surprisingly weak characters overall compared to amino acids.
GTTGATGTAGCTTAAT TCAAAGCAAGGCACTGAAAATGCCTAGATGAGT Bison grunniens tRNA Phe GTTGATGTAGCTTAAC CCAAAGCAAGGCACTGAAAATGCCTAGATGAGT Bison bison GU947006 GTTGATGTAGCTTAAC CCAAAGCAAGGCACTGAAAATGCCTAGATGAGT Bison bonasus HQ437666 GTTGATGTAGCTTAAC CCAAAGCAAGGCACTGAAAATGCCTAGATGAGT Bos primigenius GU985279 GTTGATGTAGCTTAAC CCAAAGCAAGGCACTGAAAATGCCTAGATGAGT Bos gaurus GU324988 GTTGATGTAGCTTAAC CCAAAGCAAGGCACTGAAAATGCCTAGATGAGT Bos javanicus FJ997262 GTTGATGTAGCTTAAC CCAAAGCAAGGCACTGAAAATGCCTAGATGAGT Bos indicus AF492350 GTTAATGTAGCTTAAAACCAAAGCAAGGCACTGAAAATGCCTAGATGAGT Bubalus bubalis AY488491 GTTAATGTAGCTTAAATTTAAAGCAAGGCACTGAAAATGCCTAGATGAGT Budorcas taxicolor FJ207524 GTTGATGTAGCTTAAACTTAAAGCAAGGCACTGAAAATGCCTAGATGAGT Capra hircus GU295658 GTTAATGTAGCTTAAACTTAAAGCAAGGCACTGAAAATGCCTAGATGAGT Ovis ammon HM236188 GTTGATGTAGCTTAAA TTAAAGCAAGGCACTGAAAATGCCTAGATGAGT Neotragus moschatus AJ235323
Steppe bison could be helpful here but only the last 33 bp of tRNA Pro are available (eg AY748559). Since the entire tRNA pro is identical in all bison and yak, the observed 100% identity of steppe bison here comes as no surprise. (There is additional sporadic variation but this probably arises from dna damage that affects sequencing accuracy.)
The phylogenetic relationships of artiodactyls was very much improved in a recent PNAS paper that assayed nuclear genes with a large SNP array. The resulting phylogenetic tree associated gaur and banteng with bison and yak rather than cattle. However the bootstrap value was low at 86. Since countless studies reporting 100% bootstrap confidence levels have subsequently been repudiated, the low bootstrap value here simply adds to the long-running controversy over placement of these two species of wild cattle.
However, restricting the analysis to informative mitochondrial amino acid sites greatly improves the ratio of phylogenetic signal to homoplasic noise. It makes no sense to include sites where the residue is simply rattling around within its reduced alphabet. The best sites to use are cattle synapomorphies. These sites are derived relative to ancestral and disagree with the comparable conserved ancestral value in bison and yak. It quickly emerges that banteng (Bos javanicus) has overwhelming affinities to the cattle lineage and disagrees everywhere with bison and yak. The divergence node is thus not remotely polytomic. There is less data available for gaur (Bos gaurus) but that also favors the same tree topology (right side).
Thus it suffices to correct the genus of wild yak from Bos grunniens to Bison grunniens. There is no particular justification for replacing Bison bison with Bos bison as no recognized criteria exists for genus level divergence.
Note this analysis greatly benefits from the availability of multiple complete genomes for cattle, yak and bison because these serve to suppress private mutations, sub-clade confusion and tissue heteroplasmy artifacts that result when only a single individual is sequenced and only the dominant haplotype reported.
Nuclear gene trees not uncommonly differ from mitochondrial trees. Here it could be conjectured that wild female cattle joined herds of bison or wandering bison bulls displaced wild cattle bulls, leading to nuclear/mitochondrial hybrids. There is ample precedent for this within Bovidae. The situation is not very different within humans, notably in Neanderthal/modern and Denisovan/Melanesian data, with claims also for chimp/human hybridizations continuing a million years after speciation.
However here the nuclear data is ambiguous (low bootstrap) and the mitochondrial data overwhelming. Nothing resembling a bison or yak mitochondria persists in banteng or gaur. Yet few individuals have been sampled and the precedent of aurochs mitochondria persisting in Korean and Italian cattle emerged only after extensive surveys. Still, the data available at this time do not support anything other than a straightforward speciation of bison/yak and taurine lineages.
Bos indicus Bos primigenius Bos javanicus Bos gaurus Bos frontalis Bos sauveli
cow bison cow bison cow bison cow bison cow bison cow bison
CYTB 4 0 4 0 4 0 4 0 4 0 - -
ATP8 1 1 2 0 2 0 - - - - - -
ND5 14 0 14 0 14 0 - - - - - -
ND6 4 0 - - 4 0 - - - - - -
ND4L 2 0 - - 2 0 - - - - - -
ND3 0 0 0 0 0 0 - - - - - -
COX1 - - 1 0 1 0 - - - - - -
COX3 5 0 5 0 5 0 - - - - - -
COX2 2 0 2 0 2 0 2 0 - - - -
ND2 5 0 5 0 5 0 - - - - - -
ND1 2 0 2 0 2 0 - - - - - -
ATP6 3 1 4 0 4 0 - - - - - -
ND4 8 0 8 0 8 0 - - - - - -
totals 50 1 47 0 47 0 6 0 4 0 0 0
The phylogenetic trees (upper left) were generated with the Newick trees below; distances shown are heuristic. The tree at right is adapted from Decker.
(((bosTau:15,bosInd:15):7,(bosJav:4,bosGau:4):17):1,(bisBis:19,bosGru:19):3); (((bosTau:15,bosInd:15):7,(bosJav:4,bosGau:4):17):1,(bisBis:19,bosGru:19):3);
Steppe bison to the rescue?
It is not any great technical feat today to sequence an entire nuclear genome or provide population-scale mitochondrial genomes of surviving late Pleistocene dna. That would be particularly useful in the case of the steppe bison, Bison priscus, because of its implications for conservation genomics management of contemporary bison herds. Although a particular frozen carcass may lay off to the side (not literally be a grandparent) of living bison, it still provides a much more immediate outgroup than wild yak or aurochs, thus allowing more reliable reconstruction of the last common ancestor of steppe and plains bison. The european bison (wisent) cannot serve this purpose because all examined to date are hybrids carrying cattle mitochondria.
However an individual steppe bison mitochondrial genome might not be representative of the population at the time. Indeed mitochondrial mutations were as common then as now. An especially favorable situation would be samples from multiple herds (because of matrilineal clustering) dating to the era and geographic region of divergence of steppe and plains bison that were each sequenced to deep coverage to uncover heteroplasmy. These sequences would be very close to inferred ancestral node sequences and indeed clarify ambiguous sites (where yak does not agree with either steppe or plains bison). Other than lineage sorting, ambiguous sites are rare because transitions are two-valued and transversions are uncommon.
Note an individual chosen for nuclear genome sequencing generates (as byproduct) colossal coverage of mitochondrial dna because the latter is present in great copy number excess. Consequently the haplotype heteroplasmy (differing mitochondrial genomes within the same cell) could be determined. This still does not get at heritable heteroplasmy unless multiple oocytes are dissected out from frozen carcasses and used as separate (non-pooled) sources of dna.
Considerable steppe bison dna has indeed been sequenced already. Unfortunately these studies sequenced only the control region which has little relevance to either fitness or conservation proteomics. Worse, degradation of dna -- primarily in the form of deaminated cytosine -- may have have lead to numerous artifactual C-->T transitions in the reported sequences (1,2). Yet the data make clear that steppe bison population genetics history must be considered if steppe bison are to be used in infer the plains bison ancestral situation.
A few radiocarbon-dated plains bison sequences were also determined. Provided these predate the advent of non-natural selection, they are more directly relevant than steppe bison to defining wildtype values at ambiguous sites in mitochondrial proteins and tRNA. Yet it is the synapomorphies of plains bison relative to steppe bison that really define plains bison as a distinct species.
As of 16 Dec 10, none of the 298 steppe bison GenBank submissions relate to mitochondrial proteins. Only a single entry AY748559 provides any steppe bison tRNA -- the last 33 bp of tRNA Pro but here the entire tRNA is identical to all sampled bison and yak and so steppe bison is not informative. These coverage overall ranges from 313 to 761 bp in length, averages 599 bp and so captures together only 5% of the mitochondrial genome. A single short 23S rRNA fragment from another extinct species Bison antiquus (ancient bison) is available; its genetic relationship to steppe bison and Bison latifrons is obscure.
Number Type PubMed Date Authors Title 5 A5630 D-loop -------- 14-JUL-2010 Chen K Llamas B Cooper A --- 1 voucher NWT 984.80 in press 11-JUN-2009 Zazula GD MacKay G Late Pleistocene steppe bison partial carcass from Tsiigehtchic pdf 10 BP100 control region msthesis 04-DEC-2006 Douglas KC Baker LE Comparing Genetic Diversity of Late Pleistocene Bison with Modern Bison 7 IB73 control region 15567864 14-MAR-2008 Shapiro B Cooper A Rise and fall of the Beringian steppe bison 274 BS163 control region 15567864 14-MAR-2008 Shapiro B Cooper A Rise and fall of the Beringian steppe bison
Steppe bison D-loop sequences could be used -- in conjunction with yak and cattle -- to determine the ancestral D-loop at the time of plains bison/steppe bison divergence. Bison changes relative to that could be either adaptive synapomorphies, deleterious old or new bottleneck alleles, or just near-neutral drift.
However this quest is complicated by GenBank entries filed under the wrong species name. This disrupts taxon-delimited blast searches: for example, AB177774 is designated Bos taurus despite obviously originating from a yak maternal hybrid (ditto AB177775 AB177776 EU281351 AB177769 AB065127). These should be included to obtain the full measure of yak outgroup D-loop diversity yet actual Bos taurus sequences should not. It's not clear with fossil sequences that Bison priscus have been properly discriminated from Bison bison at GenBank, vastly confusing that issue.
A second complication at GenBank arises from new bison entries with D-loops split by a bizarre choice of non-homologous numbering. For example the YNP bison GU947004 has its first 530 bp at the end (15794-16323) and its last 362 bp at the beginning (1-362). Earlier bison D-loop entries (with the exception of EU177871) follow the standard numbering system, as do most yak, cattle, and water buffalo. Thus full length D-loop bison entry U12959 and 18 others use standard numbering. This causes splitting of blast matches to bison with YNP numbering into two segments which results in lower scores and thus inappropriate placement lower in output. However this is corrected to some extent in certain blast formatting options.
Fragmentary sequences cause a third and fourth complication: uneven coverage and uneven quality. While numbered conventionally, partial sequences are from an older sequencing era and so higher in base miscalls, undetermined nucleotide N's and sequencing gaps. Nonetheless, they often score higher than split bison entries. To omit these loses considerable sampling diversity; to include increases the need for problematic line item curation.
These complications come together in fragmentary sequences with reduced basis for unambiguous species determination (eg DQ45202 from Santa Catalina Island, 45% hybrid bison). Normally back-blast would correctly pick out the species (Bos taurus for DQ45202) but bison split numbering and sequence quality issues could disrupt the best hit order.
Then there is the issue of the 2004 steppe bison data set being possibly riddled with systematic sequencing error arising from degraded dna. Expected primarily to enrich for non-existent C to T transitions, these would not necessarily pose a problem for ancestral sequence reconstruction provided they mostly gave rise to sporadic errors (not consistently affecting clades defined by transversions). However, local structural features of dna could hypothetically give rise to hotspots of deaminated cytosines and thus pervasive artifacts. Large-scale reliable sequencing of yak then place them among genuine steppe bison synapomorphies.
However the extent of degraded dna C to T transitions is easily measured by examining steppe bison at C sites invariant in modern yak and plains bison, the expectation being steppe bison will exhibit T's sprinkled erratically in these dozens of alignment columns. If the alignment order is by thermal or moisture history (for which date serves as rough proxy), these T's might occur less haphazardly. No such effect is observed. Whatever low level of artifactual T's that occurs is completely swamped out by accurate sequence, meaning no impact whatsoever on determination of ancestral sequence.
Three aurochsen genomes have survived into contemporary cattle. Could something comparable have happened with steppe bison and plains bison? This would require separation and contemporaneous existence of two separate lineages, followed by introgression into the lineage that descended to today's bison while the other went extinct. The first condition might have been repeatedly satisfied by glacial barriers and Beringian passages and for the second, steppe bison are assuredly extinct. If this amounts to two steppe bison lineages, one of which we decided to call bison at some point in time, then the question amounts to whether the second lineage is still represented.
This amounts to observing a seriously outlying sequence of bison control region that clusters in the joint phylogenetic tree with steppe bison. That would be very surprising given the tremendous bottleneck experienced in nineteenth century and more plausibly encountered if more diverse plains bison museum specimens were added. Indeed, the tree below -- which could be refined by pruning sequences to the same length and dropping poor quality entries -- does not show any bison sequence stepping 'out of line'. The indels alone strongly cluster them to the exclusion of steppe bison. The latter has distinct haplotype groups that may reflect introgression of another lineage (not necessarily) after a long separation or simply a long separation of two populations or data from different ages of steppe bison. These issues were addressed in the 2004 paper that collected the bulk of steppe bison data.
Shapiro et al make a puzzling claim about possible survival of Beringian bison to the near present (which would greatly improve the changes for observing hybrids). The value quoted of 50±75 ka BP means anything from 125,000 years ago to the present. Here ka must be taken as an unfortunate typo (as it conflicts with dates in supplementary table S2). Yet a date of 1881 for steppe bison surviving along a transcontinental railroad line by Banff National Park seems a stretch -- where are the living hybrid plains bison with steppe bison mitochondria?
"One of the specimens belonging to clade 4b (Fig. S2) appears to be much younger than the others, and suggests the survival of Beringian bison haplotypes to the near present. This sample (BS469) has been associated with two independent radiocarbon dates placing it at 50±75 ka BP (BGS-2054) and 305±24 ka BP (OxA-11988), and has been extracted and cloned twice, each time resulting in identical mitochondrial DNA sequences. This specimen originates from Banff National Park in Alberta, not far from the location of all other specimens in clade 4b, however was found ex-situ. The radiocarbon ages assigned to this specimen may be the result of contamination by modern bone material, and require further analysis. If the dates are correct, however, this specimen is the only evidence of a Beringian steppe bison mitochondrial haplotype surviving to the near present."
Other early dates in this article also imply steppe bison surviving well into the modern era, for example BS289 from Yukon Flats dated 2172±37, BS198 from Yukon Territories dated 2460±40 and BS177 from Natural Cave Trap, WY dated 3155±36. The GenBank accession of BS469 is AY748705. It differs from the next nearest steppe bison AY748551 at 3 sites, all of the type enriched for artifacts (C to T transitions) and from the nearest plains bison (AY748476) at 21 sites.
The Wyoming site would bring both species of bison together at the same place and time. Natural Cave Trap is a karst sinkhole from which 30,000 bones from 21 mammals have been collected, including mammoth, cheetah, bear, equids, camels and scavenging carnivores. However it reportedly contains no modern bison. Only two publications have ever been written on it: one an offline article from 1978 [Martin LD, Gilbert BM. Excavation at Natural Trap Cave. Trans Nebraska Acad Sci 1978; 6: 107-16] and the other an investigation of erosive arthritis which affected a high proportion of the bovids including steppe bison. A reporter who worked at the 1975 excavation describes barely funded excavators gluing teeth back into skulls at the site (glue derived from bovine collagen?). The steppe bison bones with arthritis derived from the 17,000 to 20,000 year level. This seems to conflict with the reported 3155 BP date but there is no discussion of it.
The alignment of all 331 steppe bison sequences at left, compressed beyond the point of readability to keep file size small but showing the entire alignment (250,000 letters), easily differentiates plains bison (top group of 38) from steppe bison. A few sequences mis-align due to data gaps. The vast majority of sites are invariant up to noise from private alleles and putative sequencing error. The dozen or so remaining synapomorphic sites suffice to define all major clades and subclades (and these are quickly extracted as text using the magic blast stripper), with the lineage carrying the derived state readily determined by adding a dozen yak sequences to the alignment -- essentially all of them are in plains bison because steppe bison and outgroup yak agree (ie have the ancestral value). Finally, sequences at GenBank assigned to the wrong taxon can be corrected the informative sites as classifying tool.
The informative sites extracted from the graphic above are shown in the following table:
2 3 1 2 5 2 2 4 2 1 1 2 2 1 9 1 2 5 1 6 1 1 3 3 3 3 3 3 3 3 1 1 2 1 4 4 5 3 4 4 1 3 3 5 4 2 3 2 3 4 4 3 3 3 3 2 2 4 2
8 5 3 9 3 0 0 6 8 5 6 8 0 7 6 9 0 2 7 2 5 6 4 6 1 5 6 7 3 5 8 8 1 9 2 3 2 9 6 6 1 7 2 3 8 8 5 9 1 3 3 2 4 0 4 2 2 3 9
5 5 9 5 5 2 3 4 6 9 3 2 0 5 5 1 9 0 8 6 5 7 0 4 3 1 0 2 2 3 1 0 6 6 5 5 1 3 7 7 2 2 7 7 0 3 3 1 7 9 7 3 6 5 6 0 4
PlainsBRef T G G C C T A C C T T T A - - - - G C C G C T T A G C T G T A C C G A T C T A C C T T T C C C G G G C A T A C A A A A
AY748700_B . . . . . . . . . . . . . - - - - . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748739_B . . . . . . . . . . . . . - - - - . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748529_B . . . . . . . . . . . . . - - - - . . . . . . . . . . . . . . . T . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748702_B . . . . . . . . . . . . . - - - - . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748706_B . . . . . - - . . . . . - - - - - . - . . . . . . . . . . . - - - - . . . . . . . . . . . . . . . . . . . . . - - . .
AY748683_B . . . . . - - . . . . . - - - - - . - . . . . . . . . . . . - - - - . . . . . . . . . . . T . . . . . . . . . - - . .
AY748477_B . . . T . . . . . . . . . - - - - . . . . . . . . . . . . . . . . . . . . . . . . C . . T . . . . . . . . . . . . . .
AY748745_B . . . . . . . . . . . . . - - - - . . . . . . . . . . . . . . . . . . . . . . T . C . . . . . . . . . . . . . . . . .
AY748701_B . . . . . . . . . . . . . - - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748699_B . . . . . . . . . . . . . - - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748495_B . . . . . . . . . . . . . - - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748681_B . . . . . . . . . . . . . - - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748509_B . . . . . . . . . . . . . - - - A A . . . . . C . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748748_B . . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748617_B . . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . T . . . . . . . . . C . . . . . . . . . . . . . . . . .
AY748672_B . . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748747_B . . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748674_B . . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . Y . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748680_B . . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748620_B . . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748691_B . . . . . . . . . . A A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748758_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748689_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748742_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748676_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748671_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748757_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . T . . . . . . . . . . . . . .
AY748693_B . . . . . . . . . . . . . - - - A . A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748678_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748677_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748518_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . T . . T . . . . . . . . . . . . . . . . . . .
AY748519_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . T . . T . . . . . . . . . . . . . . . . . . .
AY748630_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748685_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748687_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748675_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . T . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748673_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . T . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748476_B . . A . . . . . . . . . . - - - A . . . . . . . . . . . . . . . T . . . . C . . T . . . . . . . . A . . . . . . . . .
AY748478_B . . . . . . . . . . . . . - - - A . . . . . . . . . . . . . . . T . . . . C . . . . . . . . . . . . . . . . . . . . .
AY748695_B . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . . T . . . . . . . . . . . . . . . . . . . . . . - - . .
AY748679_B . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748690_B . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748618_B . . . . . . . . . . . . - - - A A . . . . . . C . . . . . . . . . . . . . . . . . . . . . . T . . . . . . . . . . . .
AY748530_B . . . . . . . . . . . . - - - A A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
AY748521_B . . . . . . . . . . . . - A - A A . . . . . . . . . . . . . . . . . . . . C . . . C . . . . . . . . . . . . . . . . .
AY748682_B . . . . . . . . . . . . . A - - A . . . . . . . . . . . . . . . . . . . . . . T . . . . . T . A . . . . . . . . . . .
AY748669_B . . . . . . . . . . . . . A - - A . . . . . . C . . . . . . . . . . . . . . . . . . . . . . T . . . . . . . . . . . .
DQ139154_Y A A A T T C G T . A A A T A A A A A . T A . C C G A T C A C G T T . . C . C . . . C C C . T T . A . . . . . . . G G .
DQ139214_Y A A A T T C G T T A A A T A A A A A . T A T C C G A T C A C G T T . . C . . . . . . C C . T T . A . . . . . . . G . .
DQ138999_Y A A A T T C G T . A A A T A A A A A . T A T C C G A T C A C G T T . . C . C . . . C C C . T T . A . . . . . . . G . .
GQ464236_Y A A A T T C G T T A A A T A A A A A . T A T C C G A T C A C . T T . . C . C . . . C C C . T T . A . T . . . . . G . G
GQ464191_Y A A A T T C G T T A A A T A A A A A . T A T C C G A T C A C G T T . . C . . . . . C C C . T T . A . T . . . . . G G G
GQ464223_Y A A A T T . G T T A A A T A A A A A . T A T C C G A T C A C G T . . . C . C . . . C . C . T T . A . T . . . . . G G G
DQ139104_Y A A A T T C G T T A A A T A A A A A . T A T C C G A T C A . G T . . . C . C . . . C C C . T T . A . T . . . . . G G G
DQ139103_Y A A A T T C G T T A A A T A A A A A . T A T C C G A T C A . G T . . . C . C . . . C C C . T T . A . T . . . . . G G G
GQ464172_Y A A A T T C G T T A A A T A A A A A . T A T C C G A T C A . G T . . . C . C . . . C C C . T T . A . T . . . . . G G G
DQ139208_Y A A A T T C G T T A A A T A A A A A . T A T C C G A T C A . G T T . . C . C . . . C C C . . T . A . T . . . . . G . .
DQ139193_Y A A A T T C G T . A A A T A A A A A . T A T C C G A T C A C G T T . . C . C . . . C C C . . T . A . . . . . . . G G .
GQ464150_Y A A A T T C G T T A A A T A A A A A T T A T C C G . . . . C G T T . . C . . . . . C C C . . T A . . T . C . T . . . .
DQ139162_Y A A A T T C G T T A A A T A A A A A T T A T C C G . . . . C G T T . . C . . . . . C C C . . T A . . T . C . T . . . .
DQ139146_Y A A A T T C G T T A A A T A A A A A T T A T C C G . . . . C G T T . . C . . . . . C C C . . T A . . T . C . T . . . .
DQ139010_Y A A A T T C G T T A A A T A A A A A T T A T C C G . . . . C G T T . . C . . . . . C C C . . T A . . T . C . T . . . .
AY748639_S A A A T T - - - T . . . - - - - - A - . . . . C . . . . . . - - - - - - T C - - T . . C . . T A . - - . C . . - - - .
AY748640_S G A A T T - - - T . . . - - - - - A - . . . . . . . . . . . - - - - - - . C - - T . C . . T T . . - - . . . . - - - .
AY748703_S A A A T T - - - T . . . - - - - - A - . . . . . . . . . . . - - - - - - . . - - T C . . . . . . . - - G . . . - - - .
AY748629_S A A . T T - - - . . . . - - - - - A - . . . . . G . . . . . - - - - . - T C - - . . C C . . . . . - - . . . . - - . .
AY748716_S A A A T T - - T T . . . - - - - - A - . . . . . . . . . . . - - - - G . T C . . T . . . . T . A . . . . . . . - - . .
AY748552_S A A R T T - - T T . . . - - - - - A - . . . . . . . . . . . - - - - . . T C G T T . C C . T . . . A . . . . T - - . G
AY748631_S A A A . T - - T T . . . - - - - - A - . . . . . . . . . . . - - - - . C T C . T T . . . . T . . . . . . . . . - - . .
AY748625_S A A A T . - - T T . . . - - - - - . - . . . . . . . . . . . - - - - . C . C . T T . . . . Y . R . . T . . . . - - . .
AY748721_S A A A T - - T . . . - - - - - - . . . . . . . . . . . - - - - C T C . . . . . G . . . - - .
AY748636_S A A A T T - - T T . . . - - - - - A - . . . . . . . . . . . - - - - . C T . G . . C C C . T T . A . . . . . . - - . .
AY748483_S A A . T T C G T . . . . . - - - - A A . . . . . . . . . . . . . . . . . T C G T . C C C . T . . . A . . . G . . . . .
AY748474_S A A . T T C G T . . . . . - - - A A A . . . . . . . . . . . . . . . . . T C G T . C C C . T T . . A . . . . . . . . .
AY748749_S A A A T T . G T T . . . - - - - A A A . . . . . . . . . . . . . . A . C T . . T . C C C T . . . . . T . . . . . . . .
AY748471_S A A . T T . . . . . . . . - - - A A . . . . . . . . . . . . . . . . . C T C G T . . C C . T T . . . . . . . . . . . .
AY748705_S A A A T T C G T T . . C - - - - A A . . . . . . . . . . . . . . T . G C T C . . T . C . . . . . . . . . . . . . . . .
AY748551_S A A A T T C G T T . . C - - - - A A . . . . . . . . . . . . . . T . G C T C . . T . . . . . . . . . . . . . . . . . .
AY748532_S A A A T T C G T T . . . - - - - A A . . . . . . . . . . . . . . T . G C T C . . T . . . . . . . . . . . . . . . . . .
AY748746_S . . A Y T C G T . . . . . - - - A A . . . . . . . . . . . . . . T . . C T C . T T . . . . . . . . . . . . . . . . . .
AY748485_S G A A T T C G T T . . C G - - - A A . . . . . . . . . . . . . . T . . C T . . T T . . . . T . . . . . . . . . . . . .
AY748500_S A A A T T C G T . . . . G - - - A A . . . . . . . . . . . . . . T . . C . C G T T C . . . T T . . . . G . . . G . . .
AY748730_S A A A T T C G T T . . . G A - - A A . . . . . . . . . . . . . . T . . C . C . T T . . . . T T . . . T G . . . . G . .
AY748722_S A A A T T C G T T . . . . - - G - A . . . . . . . . . . . . . . T . G C T C . . T C . . . . . A . . . . . . . . . . .
AY748665_S A A A T T C G T T . . . . - - G - A . . . . . . . . . . . . . . T . G C T C . . T . . . . . . A . . . . . . . . . . .
AY748488_S A A A T T C G T T . . . . - - G - A . . . . . . . . . . . . . . T . G C T C . . T . . . . . . A . . . . . . . . . . .
AY748645_S A A A T T C G T T . . . . - - G - A . . . . . . . . . . . . . . T . G C T C G . T . C . . . . A . . . . . . . . . . .
AY748650_S A A A T T C G T T . . C . - - G - A . . . . . . . . . . . . . . T . G C T C . . T . . . . . . A . . . . . . . . . . .
AY748642_S A A A T T C G T T . . . . - - G - A . . . . . C . . . . . . . . T . . . T C . . T . . . . . . . . . . . . . . . . . .
AY748482_S A A A T T C G T . . . . . - - G - A . . . . . . . . . . . . . . T . . C T C . . T . . . . T . A . . . . . . . . . . .
AY748688_S A A . T T C G T . . . . . - - G - A . . . . . . . . . . . . . . T . . . . C . . . . . C . . . . A . . G . . . G . . .
AY748606_S A A . T T C G T . . . . . - - G - A A . . . . . . . . . . . . . . . . . T C G T . . C C . . . . . A . . . . . . . . .
AY748740_S A A . T T C G T . . . . . - - G - A A . . . . . . . . . . . . . . . . . T C . T . C C C . T . . . A . . . G . . . . .
AY748667_S A A . T T C G T . . . . . - - G - A A . . . . . . . . . . . . . . . . . T C . T . C C C . T T . . A . . . G . . . . .
AY748658_S A A A T T C G T T . . . - - - G - A A . . . . . . . . . . . . . . A . C T . . T . C C C T . T . . . . . . . . . . . .
AY748593_S A A A T T C G T T . . . . - - G - A . . . . . . . . . . . . . . . . . . . . G T . C . C . . . . A . . . . . . . . . .
AY748533_S A A A T T C G T . . . . . - - G - A . . . . . . . . . . . . . . . A . C T C G T . C . C T T . . . . T . . . . . . . .
AY748666_S A A A T T . G T T . . C . - - G - A . . . . . . . . . . . . . . . A . . T . G T . C C C . . . . A . . . . . . . . . .
AY748646_S A A A T T C G . T . . . . - - G - A . . . . . . . . . . . . . . . . . . . C G T . . . C . . . A A . . . . . . . . . .
AY748643_S A A A T T C G T T . . . . - - G - A . . . . . . . . . . . . . . . . . C . Y G T . . . C . . . . . . . . . . . . . . .
AY748710_S A A A T T C G T T . . . . A - G - A . . . . . . . . . . . . . . T . G C T C . . T C . . . . . A A . . . . . . . . . .
AY748586_S A A . T T C G T . . . . . A - A - A . . . . . . . . . . . . . . . . . . . . . T . C . C . . . . . . . . . . . . . . .
AY748550_S A A . T T C G T . . . . . A - G - A - . . . . . . . . . . . . . Y . . . T C . T . . C C . . . . A A . . . . . . . . .
AY748594_S A A A T T C G T . . . . . A - A - A - . . . . . . . . . . . . . . . . . T C G T . . C C . . . . . A . . . . . . . . .
AY748576_S A A . T T C G T T . . . . A - A - A A . . . . . . . . . . . . . . . . . T . G T . C C C . T . . . A . . . . . . . . .
AY748619_S A A A T T C G T T . . . - A - G A A A . . . . . . . . . . . . . . A . C T . G . . C C C . . T . A . . . . . . . . . .
AY748603_S A A A T T C G T T . . . - A - G A A A . . . . . . . . . . . . . . A . C T . G T . C C C . T T . A . . . . . . . . . .
AY748596_S A A A T T C G T T . . . - A - G A A A . . . . . . . . . . . . . . A . C . . G T . C C C T T T . . . . . C . . . . . .
AY748608_S A A A T T C G T T . . . - A - G A A . . A . . . . . . . . . . . . A . C T . G T . . C C T . T . . . . . . . . . . . .
AY748743_S A A A T T C G T T . . . - A - G A A - . . . . . . . . . . . . . . A . C T . . T . C C C T . T . . . . . C . . . . . .
AY748769_S A A A T T C G T T . . C - - - G A A . . . . . . . . . . . . . . . A . C . C G T . C C . . T T . A . T . . . . . . . .
AY748793_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T C G T . C C . . . . . A . T . . . . . . . .
AY748475_S A A A T T C G . T . . . - A - G A A . . . . . . . . . . . . . . T . . C T . G T T . . C . . . A . . . . . . . . . . .
AY748633_S A A A T T C G T T . . . - A - G A A . . . . . C . . . . . . . . T . G C T C . . T . . . . . . A . . T . . . . . . . .
AY748655_S A A A T T C G T T . . C - - - G A A . . . . . . . . . . . . . . T . . C T C . T T . . . . . . . . . . . . . . . . . .
AY748508_S A A A T T C G T T . . C - - - G A A . . . . . . . . . . . . . . T . . C . C . T T . . . . . . A . . . . . . . . . . .
AY748503_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . . C T . . . T . . . . . . A . . . . . . . . . . .
AY748526_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . . C T C . . T . . . T . . A . . . . . . . . . . .
AY748696_S A A . T T . . T T . . . - - - A A A . . . . . . . . . . . . . . T . . C T C . T Y C . . . T . . . . . G . . . . . . .
AY748648_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . . C . C . T T . C C . T . . . . . . . . . . . . G
AY748724_S A A A T T C G T T . . C - - - G A A . . . . . C . . . . . . . . T . G C T C . . T . . . . . . A . . . . . . . . . . .
AY748752_S A A A T T C G T T . . . - - - G A A . . . . . . . A . . . . . . T . G C T C . . T . . . . . . A . . . . . . . . . . .
AY748741_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . G C T C . . T . . . . . . . A . . . . . . . . . .
AY748663_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . G C T C . . T . . . T . . A . . . . . . . . . . .
AY748522_S A A A T T C G T T . . . - - - A A A . . . . . . . . . . . . . . T . G C T C . . T . C . . . . A . . . . . . . . . . .
AY748547_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . G C T C . . T . . . . . T A . . . . C . . . . . .
AY748511_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . G C T C . . T . . . . . . A . . . . . . . . . . .
AY748707_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . G . T C . . T . . . T . . A . . . . C . T G . . .
AY748504_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . G . T C . . T C . . T . . A . . . . C . T G . . .
AY748595_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . G . T C . . T . . . T . . A . . . . C . T G . . .
AY748510_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . G . T C . T T . . . T . . A . . . . C . T G . . .
AY748531_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T . . . T . . A . . . . C . T G . . .
AY748513_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T . . . T . . A . . . . C . T G . . .
AY748501_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T C . . T . . A . . . . C . T G . . .
AY748515_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T . . . T . . A . . . . C . T G . . .
AY748570_S . A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T . . . T . . A . . . . C . T G . . .
AY748568_S . A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T . . . T . . A . . . . C . T G . . .
AY748767_S A A A T T C G T . . . . - - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . T . . . . . . . . . . . .
AY748772_S A A A T T C G T T . . . - - - G A A . . . . . C . . . . . . . . . A . . . . . . . C . C T T . . . . . G . G . . . . .
AY748791_S A A A T T C G T T . . . - - - A A A . . . . . . . . . . . . . . . A . C T . G . . C C C T . T . A . . . . . . . . . .
AY748572_S A A A T T C G T . . . . - - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . T . . . . . . . . . . . .
AY748502_S A A A T T C G T T . . . - - - G A A . . . . . . G . . . . . . . . A . C T C G T . C C C T . . . . . T . . . . . . . .
AY748492_S A A . T T C G T . . . . - - - G A A . T . . . C . . . . . . . . . . . . T . G . . . C C . . . . . . . . . . . . . . .
AY748493_S A A A T . C G . . . . . - - - G A A . . . . . . . . . . . . . . T . . . . C . . T . . . T . . . . . . . . . . . . . .
AY748604_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T . C . . . . A . . . . . . . . . . .
AY748496_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T . C . . . . A . . . . . . . . . . .
AY748516_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T . C . . T . A . . . . . . . . . . .
AY748612_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G C T C . . T . . . . . . A . . . . . . . . . . .
AY748582_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G C T C . . T . . . . . . A . . . . . . . . . . .
AY748609_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G C T C . . T . . . . . . A . . . . . . . . . . .
AY748641_S A A T T C G - T . . . - - G A A . . . . . . . . . . . . . T . - - T C - - C . C . . . A . - - . . . . . . - .
AY748719_S A A A T C G T T . . . - - - G A . . . . . . . . . . . . . . T . . C T C . T T . . T . . . . . . G . . . . . . .
AY748660_S A A A T C G T . . . - - - G A . . . . . . . . . . . . . . T . C T . C T . . . G . . . . . G
AY641550_T A C G . . - - - G A . . . . . . T . T . .
AY748565_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T - G . T C . . T . . . T . . A . . . . C . T G . . .
AY748751_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . T . . . . C . T T . . . T . . . . . . G . . . . . . .
AY748567_S A A A T T - G T T . . . - - - G - A A . . . . C . . . . . . . . - A . . T C . . . . C C T . T . . . . . . . . - - . .
AY748790_S G A A T T C G T T . . . - - - G A A A . . . . . . . . . . . . . T . . . T . . T . C C C T . T . A . . . . . . . . . .
AY748787_S A A A T T C G T T . . . . - - G A A A . . . . . G . . . . . . . . A . C T . G T . C C C . T T . . . . . . . . . . . .
AY748786_S A A A T T C G T T . . . - - - G A A A . . . . . G . . . . . . . . A . C T . G . . C C C . . T . A . . . . . . . . . .
AY748651_S A A A T T C G T T . . . - - - G A A A . . . . . . . . . . . . . . A . C T . G . . C C C . . T . A . . . . . . . . . .
AY748768_S A A A T T C G T . . . . - - - G A A A . . . . . . . . . . . . . . A . C T C . T . C C C T . . . . . . . . . . . . . .
AY748759_S A A . T T . G T T . . . - - - A A A A . . . . . . . . . . . . . . . . . . C G . . C . C . . . . . . . . . . . . . . .
AY748670_S A A A T T C G T T . . . - - - G A A A . . . . . . . . . . . . . . A . . T . G T . . C C . . T . A . . . . . . . . . .
AY748599_S A A A T T C G T T . . . - - - G A A A . . . . . . . . . . . . . . A . C T . . T . C C C T . T . A . . . C . . . . . .
AY748584_S A A A T T C G T T . . . - - - G A A A . . . . . . . . . . . . . . A . C T . G T . C C C T . . . . . . . . . . . . . .
AY748578_S A A A T T C G T T . . . - - - G A A A . . . . . . . . . . . . . . A . C T . G T . C C C . T T . . . . . . . . . . . .
AY748540_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T . G T . . C C T . T . . . . . . . . . . . .
AY748536_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T . . T . C C C T . T . A . . . . . . . . . .
AY748534_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . T . A . . . . G . . . . .
AY748543_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T . . T . C C C T T . . . . . G . . . . G . .
AY748574_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . . . C . . G T . C C C . . . . A . . . . . . . . . .
AY748588_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T . G T . . C C T T . . . . . . . G . . . . .
AY748590_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T C . T . C C C T . . . . . T . . . . . . . .
AY748591_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . T . . . . . . . . . . . .
AY748597_S A A A T T C G T T . . C - - - G A A . . . . . . G . . . . . . . . A . C . C G T . . C . . T . . . . T . . . . . . . .
AY748601_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C . . . T . . . . . . . . . . . G
AY748605_S A A A T T C G T T . . . - - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T T T . . . . . . G . . . . .
AY748610_S A A A T . C G T T . . C - - - G A A . . . . . . G . . . . . . . . A T . . C T T . . C . . T . . . . . . . . . . . . .
AY748613_S A A A T T C G T T . . . - - - G A A . . . . . . . A . . . . . . . A . C T . G T . C C C T . T . . . . . . . . . . . .
AY748627_S A A A T T C G T . . . . - - - G A A . . . . . . . . . . . . T . . A . C T C G T . . C . T . . . . . . . . . . . . . .
AY748600_S A A A T T C G T T . . . G - - G A A A . . . . . . . . . . . . . . A . C T . G T . . C C . . T . A . . . . . . . . . .
AY748733_S A A A T T C G T T . . . G - - G A A A . . . . . G . . . . . . . T A . C T . G . . C C C . . T . A . . . . . . . . . .
AY748545_S A A . T T C G T . . . . . - - G A A A . . . . . . . . . . . . . . . . . T C . T . C C C . T . . . A . . . G . . . . .
AY748542_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T C C . . T T . . . . . C . . . . . .
AY748734_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . G . T . . . T . C . . . . A A . . . . . . . . . .
AY748537_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . T . . . . . . . . . . . .
AY748698_S A A A T T C G T T . . A . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C . T T . . . . . . . G . . . . .
AY748535_S G A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . . T . G T . C C C T . T . A . . . . . . . . . .
AY748731_S A A A T T . G T . . . . . - - G A A . . . . . . . . . C . . . . . A . C T C . T . C . . T T . . A . . . . . . . . . .
AY748539_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C . T T . . A . . . . . . . . . .
AY748546_S A A A T T C G T . . . . . - - G A A . . . . . . . . . . . . . . . A . C T C G T . C C . T . . . . . . . . . . . . . .
AY748538_S A A A T T C G T . . . . . - - G A A . . . . . . . . . . . . . . . A . . T . . . . C C C . T T A . . . . . . . . . . .
AY748736_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . . T . G T . C C C T . T . . . . . . . . . . . .
AY748623_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T T . . A . . . . . . . . . .
AY748662_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . T . . . . . . . . . . . .
AY748735_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C . . G T . C C C . T . . A . . . . . . . . . .
AY748738_S - - A - T C G T - . . - . - - G A A . . . . - - - - - - - - . . . A . C T - G T . - - C T - - - - . T - - - - . . . -
AY748544_S A A A T T C G T T . . . . A - G A A . . . . . C . . . . . . . . T . . C . C G T T . . . . T . . . . . R . . . . . . .
AY748541_S A A A T T C G T T . . . . A - G A A . . . . . . . . . . . . . . . A . . T . G T . C C C T . T . . . . . . . . . . . .
AY748659_S A A A T T C G T T . . . . A - G A A . . . . . C . . . . . . . . . A . C T C G T . . C C . . . . . . . . . . . . . . .
AY748737_S - - A - T . G T - . . - . A - G A A . . . . - - - - - - - - . . . A . C T - . T . - - . T - - - - . . - - - - . . . -
AY748470_S A A T T C G T T . . . . A G A A . . . . . . . . . . . . . . A . C T C G T C C . . T . . A . T G . . . . . . .
AY748781_S A A A T T C G T T . . . . A - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . T . . . . . . . . . . . .
AY748773_S A A A T T C G T T . . . . A - G A A . . . . . . . . . . . . . . . A . C . . G T . C C C T T . . . . T . . . . . . . .
AY748776_S A A A T T C G T T . . . . A - G A A . . . . . . . . . . . . . . . A . . T . G T . C C C T . T . . . . G . . . . . . .
AY748473_S A A T T C G T T . . . . A G A A . . . . . . . . . . . . . T . G C T C . . . . . . . . A A . - . . . . . . . .
AY748558_S A A A T T C G T T . . . G A - G A A . . . . . C . . . . . . . . T - G C T C . . T . . C . . T A . . T . C . . . . . .
AY748715_S A A T T C G T T . . . G A G A A . . . . . . . . . . . . . T . . C T C . . . . . . . . A . . - . . . . . . . .
AY748498_S A A A T T C G T T . . . G A - - A A . . . . . . . . . . . . . . T . . . . C . T T . C . T T . . A . . . . . . . . . .
AY748486_S A A A T T C G T T . . . G A - A A A . . . . . C . . . . . . . . T . . C T . . . T . . . . . . . A . . G . . . . . . .
AY748723_S A A A T T C G T T . . . G A - G A A A . . . . . . . . . . . . . . - . C T . G . . C C C . . T . A . . . . . . . . . .
AY748553_S A A A T T C G T T . . . G A - G A A A . . . . . . . . . . . . . . A . C T C G . . C C C T . T . . . . . . . . . . . .
AY748497_S A A A T T C G T . . . . G A - G A A A . . . . . . . . . . . . . . A . . T . G . . . C C . T T . . . T . . . . . . . .
AY748777_S G A A T T C G T T . . . G A - G A A A . . . . . . . . . . . . . . A . C . . G T . C C C . . T . A . T . . . . . . . .
AY748712_S A A A T T C G T T . . . G A - G A A A . . . . . . . . . . . . . . A . C T . G . . C C C . . T . A . . . . . . . . . .
AY748560_S A A A T C G T . . . G A - G A A . . . . . . . . . . . . . . A . C . . . . . A . . C . T . . . .
AY748499_S A A A T - G T T . . . . A - G C A . . . . . . . . . . . . . . A . C . G . . C C . . T . A . . . . . . . . . .
AY748469_S A A A T T C G T T . . . G - - A A A A . . . . . . . . . . . . . T . . . . C . T T C . . . T . . . . . G . . . . . . .
AY748718_S A A A T T C G T T . . . G - - A A A A . . . . . . . . . . . . . T . . . T Y . T T . C . . T . . A . . . . . . . . . .
AY748765_S A A A T T C G T T . . . G - - G A A A . . . . . G . . . . . . . . A . C T . G . . C C C . . . . A . . . . . . . . . .
AY748783_S A A A T T C G T T . . . G - - G A A A . . . . . . . . . . . . . T A . . T . . T . C C C T . T . . . . . . G . . . . .
AY748755_S A A A T T C G T T . . . G - - G A A A . . . . C . . . . . . . . . A . C T C G T . . . C . T . . A . . . . . . . . . .
AY748686_S A A . T T C G T . . . . G - - A A A A . . . . . . . . . . . . . . . . . . C G . . . . C . T T . . . . . . . . . . . .
AY748668_S A A A T T C G T T . . . G - - G A A A . . . . . . . . . . . . . . A . C T . G . . C C C . . T . A . . . . . . . . . .
AY748744_S A A . T T C G T . . . . . - - G A A A . . . . . . . . . . . . . . . . . T . G T . . C C . T . . . A . . . . . . . . .
AY748634_S A A . T T C G T . . . . . - - G A A A . . . . . . . . . . . . . . A . . T C G T . C C C . T . . A A . . . G . . . . .
AY748561_S A A A T - G T T . . . . - - A C A . . . . . . . . . . . . . T . . . C . T T . C . . . A A . . . . . . . . . .
AY748637_S A A . T C G T . . . . - - G A A . . . . . . . . . . . . . . . . . . . T . A . . . . . . . .
AY748635_S A A . T T C G T . . . C . - - G A A A . . . . . . . . . . . . . . . . . T C G T . . C C . . . . . A . . . . . . . . .
AY748479_S A A A T T C G T T . . . . - - G A A A . . . . . . . . . . . . . . A . C T . G . . C C C . . T . A . . . . . . . . . .
AY748711_S A A . T T C G T . . . . . - - G A A A . . . . . . . . . . . . . . . G C T C . . . C C . . T . . . . . . . G . . . . .
AY748520_S A A . T T C G T . . . . . - - G A A A . . . . . . . . . . . . . . . G . T C . . . C C . . T T . . . . . . G . . . . .
AY748704_S A A . T T C G T . . . . . - - G A A A . . . . . . . . . . . . . . . . . T C G T . C C C . T T . . A . . . G . . . . .
AY748697_S A A . T T C G T . . . . . - - G A A A . . . . . . . . . . . . . . . . . T C . T . C C C . T . . . A . . . G . . . . .
AY748638_S A A . T . C G T Y . . . . - - G A . A . . . . . . . . . . . . . . . . . . C G T . . C . . . . . . A . . . . . . . . .
AY748554_S A A . T T C G T . . . . . - - G A A A . . . . . . . . C . . . . . . . . T C G T . . C C . . . . . A . . . . . . . . .
AY748512_S A A . T T C G T . . . . . - - G A A A . . . . . . . . C . . . . . . . . T C G T . . C C . . . . . A . . . . . . . . .
AY748556_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T - G . T C . . T . . . T . . A . . . . C . T G . . .
AY748549_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T - G C T . . . T . . . . . . A . . . . . . . . . . .
AY748626_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T - G C T C . . T . . . . . . A . . . . . . . . . . .
AY748614_S A A A T T C G T T . . C G - - G A A . . . . . . . . . . . . . . T - G C T C . . T . . . . . . . . . . . . . . . . . .
AY748563_S A A A T T C G T T . . C G - - G A A . . . . . . . . . . . . . . T - G C T C . . T . . . . . . . . . . . . . . . . . .
AY748611_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T - G C T C . . T . . . . . . A . . . . . . . . . . .
AY748602_S A A A T T C G T T . . . G - - G A A . . . . . . G A . . . . . . T - G C T . . . T . C . . . . A . . T . . . . . . . .
AY748555_S A A A T T C G T T . . . G - - G A A . . . . . . . A . . . . . . T - G C T . . . T . C . . . . A . . T . . . . . . . .
AY748571_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T - G . T C . . T . . . . . . A . . . . . . . . . . .
AY748763_S A A A T T C G T T . . . . - - G A A - . . . . . . . . . . . . . . . . C T . . T . C C C T . T . . . . . . . . . . . .
AY748762_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T . . T . C C C T . . . . . . . . . . . . . .
AY748652_S A A A T T C G T . . . C . - - G A A . . . . . . G . . . . . . . . A . C . C G T . . C . T T . . . . . . . . . . . . .
AY748548_S A A A T T C G T . . . . . - - G A A . . . . . . . . . . . . . . . A . C T C G T . C C . T . . . . . . . . . . . . . .
AY748720_S A A A T T C G T T . . C . - - G A A . . . . . . . . . . . . . . . . . C . C G T . . . C . . . . . . . . . . . . . . .
AY748527_S A A . T T C G T . . . . . - - G A A . . . . . . . . . . . . . . . . . . . C G T . . . C . . T . . . . . . . . . . . .
AY748480_S A A . T T C G T . . . . . - - G A A . . . . . C . . . . . . . . . . . . T C G . . . . C . . . . . . . . . . . . . . .
AY748507_S A A A T T C G T . . . . . - - G A A . . . . . . . . . . . . . . T . . . . C . T T C . . . . T . . . . G . . . . . . .
AY748750_S A A A T T C G T T . . . . - - G A A . . . . . C . . . . . . . . T . G C T C . . T . . C . . T A . . . . C . . . . . .
AY748490_S A A . T T . . T T . . . . - - - A A . . . . . C . . . . . . . . . . G C T C . . . . . C . . T A . . . . C . . . . . .
AY748528_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . T . G . T C . . T . C . . . . A A . . . . . . . . . .
AY748489_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . T . G . T C . . T . C . . . . A A . . . . . . . . . .
AY748472_S G A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . T . G . T C . . T . . . . . . A . . . . . . . . . . .
AY748729_S A A A T T C G T T . . C . - - G A A . . . . . . . . . . . . . . T . . . T C . T T . . . . . . . . . . . . . . . . . .
AY748728_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . T . G . T C . . T . C . . . . A A . . . . . . . . . .
AY748717_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . T . G . . C . . T . C . . . . A A . . . . . . . . . .
AY748647_S A A A T T C G T T . . . . - - G A A . . . . . C . . . . . . . . T . . C . C . . T . . . T . T . . . . G . . . . . . .
AY748487_S G A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . T . . C . C . T T . . C . T . . . . . . . . . . . . .
AY748789_S A A A T T C G T T . . . . - - G A A . . . . . C . . . . . . . . . A . C T . . T . C C . T . T . . . . . . . . . . . .
AY748788_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . . . . . . . . . . . . . .
AY748725_S A A A T T C G T T . . C . - - G A A . . . . . . . . . . . . . . T . G C T C . . T . . . . . . A . . . . . . . . . . .
AY748760_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . . . . . . . . . . . . . .
AY748753_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . . . . . . . . . . . . . .
AY748785_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T C G T . C C C T . T . A . . . . . . . . . .
AY748778_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T C G T . C C C T . . . A . . . . . . . . . .
AY748775_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T C G T . C C . . . . . A . T . . . . . . . .
AY748766_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T C G T . C C . . . . . A . T . . . . . . . .
AY748784_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . . . C . . G T . C C C . . . . A . . . . . . . . . .
AY748782_S A A A T T C G T T . . C . - - G A A . . . . . . . . . . . . . . . A . C . . G . . C C C T T . . . . . . . G . . . . .
AY748792_S A A A T T C G T . . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T T T . . . . . . . . . . . .
AY748764_S A A A T T . G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T C G T . C C C T T . . A . . . . . . . . . .
AY748771_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . . . C . C G T . . . C . . . . . . . . . . . . . . .
AY748770_S A A A T T C G T . . . . . - - R A A . . . . . . . . . . . . . . . A . C T . G T . . . C T T . . A . . . . . . . . . .
AY748774_S A A A T T C G T . . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . T . A . . . . . . . . . .
AY748754_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T . T . . . . . . . . . . . .
AY748780_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C Y . R T . C C C T T T . A . . . . G . . . . .
AY748598_S A A . T T C G T . . . . . - - G A A - . . . . . . . . . . . . . . . . . T C G T . C C C . T . . A A . . . G . . . . .
AY748517_S A A . T T C G T . . . . . - - G A A - . . . . . . . . . . . . . . . . . T C G T . C C C . T . . . A . . . G . . . . .
AY748562_S A A . T T C G T . . . . . - - G A A - . . . . . . . . . . . . . . . . . T C . T . C C C . T . . . A . . . G . . . . .
AY748569_S A A . T T C G T T . . . . - - G A A - . . . . . G . . . . . . . . . . . T C G T . C C C . . . . . A . . . . . . . . .
AY748506_S A A . T T . . T . . . . . - - G A A . . . . . . . . . . . . . . . . . . T . G T . C C C . T . . . A . . . . . . . . .
AY748581_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . T - G . T C . . T . C . . . . A A . . . . . . . . . .
AY748622_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . A . C T C . . T . . . . . . . . . . . . . . . . . .
AY748621_S A A A T T C G T T . . C . - - G A A . . . . . . . . . . . . . . T - G C T C . . T . . . . . . A . . . . . . . . . . .
AY748575_S A A A T T C G T T . . C . - - G A A . . . . . . . . . . . . . . T - G C T C . . T . . . . . . A . . . . . . . . . . .
AY748592_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . . . . - G C T C . . T . . . . . . . . . . . . . . . . . .
AY748628_S A A A T T C G T T . . . . - - G A A . . . . . . . . . . . . T . T . . C T C . . T . . . . . . A . . . . . . . . . . .
AY748684_S A A . T . G T . . . . . - - G A . . . . . . . . . . . . . . . . . . C G T . C C . . . . . A . . . . . . . . .
AY748580_S A A A T - G T T . . . . - - G C . . . . . . . . . . . . . . T . G . C . . T . C . . . A . . . . . . . . . . .
AY748795_S A . A T C G . . . . . - - G A . . . . . . . . . . . . . . . A C . C C T . . . G . . . . . .
AY748632_S A A A T C G T . . . . - - G A . . A . . . . . . . . . . . T - . C T . C . . A . . . . . . . . .
AY748491_S A A A T T C G T T . . C G - - G A A . . . . . . . . . . . . . . T . . C T C G T T C . . T . . A . . . . . . . . . . .
AY748692_S A A A T Y C G T T . . . G - - A A A . . . T . . . . . . . . . . T . . C T C . T T C . . . T . . . - T G . . . . . . .
AY748761_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . . . T T C . . . T . . . . . . . . . . . . .
AY748525_S A A A T T C G T . . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T C . . . T T . . . . . . . . . G . .
AY748654_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . . . . T . . . . T . . . . . G . . . . . . .
AY748657_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T . . . . T T . . . . G . . . . . . .
AY748587_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T C . . . T T . . . . . C . . . . . .
AY748505_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T C C . . T T . . . . . C . . . . . .
AY748523_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T . C C . T . . . . . . . . . . . . G
AY748559_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . . C T C . T T . . . T . . . . . . G . . . . . . .
AY748732_S A A A T T C G T T . . C G - - G A A . . . . . . . . . . . . . . T . . C T C . T T . . . . . . . . . . . . . . . . . .
AY748714_S A A A T T C G T T . . C G - - G A A . . . . . . . . . . . . . . T . . C T C . . T C . . . . . A . . . . . . . . . . .
AY748713_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . G C T C . . T C . . T . . A . . . . . . . . . . .
AY748653_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . . C T . . T T . . . . . . . . . . . . . . . . . .
AY748514_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . . C T C . T T . . . . . . . . . . . . . . . . . .
AY748524_S A A . T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . G C T C . . . . . C . . . A . . . . . . . . . . .
AY748644_S A A A T T C G T T . . C G - - G A A . . . . . . . . . . . . . . T . . C T C . T T . . . . . . . . . . . . . . . . . .
AY748727_S A A A T T C G T . . . . G - - A A A . . . . . C . . . . . . . . T . . C . C . T T . C . . T . . . . . . . . . . . . .
AY748557_S A A A T T C G T . . . . G - - A A A . . . . . . . . . . . . . . T . G C T C . . T . . . . T T . . . . . . . . . G . .
AY748481_S A A A T T C G T . . . . G - - G A A . . . . . . . . . . . . . . T . . . T C . . T . . . . . . A . . T . . . . . . . .
AY748726_S A A A T T C G T T . . C G - - A A A . . . . . . . . . . . . . . T . . Y . C . T T . C . . T T . A . . . . . . . . . G
AY748661_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T . . . . T . . . . . G . . . . . . .
AY748796_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . . C . C . T T . . . . T . . . . . G . . . . . . .
AY748649_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . . C . C . T T C . . T . . . . . . G . . . . . . .
AY748794_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . . A . . T . . T . C C C . . T . . . . . C . . . . . .
AY748756_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . . A . C T . G T . C C C T T T . A . . . . G . . . . .
AY748779_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . . A . . . . G T . C C . T . T . . . . . . . . . . . .
AY748709_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . G C T C . . T . . . . T . A . . . . . . . . . . .
AY748708_S A A A T T C G T T . . C G - - A A A . . . . . . . . . . . . . T T . . C . C . T T . . . . . . . . A T G . . . . . . .
AY748694_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . . . T T . . . . . . . . A T . . . . . . . .
AY748616_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T C . . . T . . . . . . . . . . . . .
AY748585_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T C . . . . . . . . . . . . . . . . .
AY748583_S A A A T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T C . . . T . . . . . G . . . . . . .
AY748579_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T . . C . C . T T . . . . T . . . . T G . . . . . . .
AY748573_S A A . T T C G T T . . . G - - A A A . . . . . . . . . . . . . . T . . C . C . T T . . . . T . . . . . G . . . . . . .
AY748607_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T - . C T C . T T . . . . . . . . . . . . . . . . . .
AY748577_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T - . C T . . T T . . . . . . . . . . . . . . . . . .
AY748566_S A A A T T C G T T . . . G - - G A A . . . . . C . . . . . . . . T - . C T . . T T . . . . . . . . . T . . . . . . . .
AY748589_S A A A T T C G T T . . . G - - G A A - . . . . . . . . . . . . . T - . C T C . . T . C . . . . . . . . . . . . . . . .
GQ258703_S A A A T T C G T T . . . G - - G A A . . . . . C . . . . . . . . T - G C T C . . T . . C . . T A . . . . C . . . . . .
AY748664_S - A A - T C G T - . . - G - - A A A . . . . . . - . . . . . . . T . . C . C . T T . - . . - . - - . . . . - . . . . -
AY748615_S A - A T T C G T T . . . G - - A A A . . . . - - . - - - . - . . T . . C . . G T T - . . . T - . . . . G - . - . . . .
AY748564_S A A A T C G T . . . G - - G A . . . . . . . . . . . . . . T - . . T C C T . . . G . . . . . . .
AY748484_S A A A T C G T . . . G - - G A . . . . . . . . . . . . . . T - . C T . C T . . . . . . . . . . G
AY748494_S A A A T C G . . . . G - - G A . . . . . . . . . . . . . . T - . . T C . T T . . . . . . . G .
AY748656_S A A A T T C G T T . . C G - - G A A . . . . . . . . . . . . . T T - G C T C . . T . . . . T . A . . . . . . . . . . .
AY748624_S A A A T T C G T T . . . G - - G A A . . . . . . . . . . . . . . T - G . T C . . T . C . . . . A . . . . . . . . . . .
For the nuclear genome, only a single 49 aa of osteocalcin fragment dating from a 55.6 kyr permafrost steppe bison is available corresponding to gene BGLAP. The protein was sequenced directly by mass spectroscopy so the dna sequence remains unknown. This gene has not been sequenced in either bison or yak. It is only moderately conserved within laurasiatheres. Given that BGLAP has numerous but variably implemented post-translational modifications mediated by vitamin K, it is not a good choice for MS comparative genomics. The sequence does appear accurate however -- cow, not steppe bison, has the odd tryptophan variation.
Steppe bison YLDHGLGAPAPYPDPLEPKREVCELNPDCDELADHIGFQEAYRRFYGPV Bison priscus NP_776674 ....W............................................ Bos taurus NP_001035098 ...P..............R.............................. Ovis aries NP_001157476 ..................R............................ Sus scrofa XP_002927002 ...S.......................N...........D......... Ailuropoda melanoleuca XP_547536 ...S.....V.................N..............Q...... Canis familiaris Bison bone collagen isotopic values track climate fluctuations and vegetation change in Late Pleistocene and Early Holocene Northern Eurasia and North America Richards M, Shapiro B, Ditchfield P, Cooper A Geological Journal (in press 2011)
In summary, steppe bison mitochondrial protein sequences -- though not available as yet -- would be very helpful in establishing wildtype in bison. In particular they could help time the origin of V98A and address the significance of the other sites where contemporary bison differ from the conserved value seen in yak and other Bovidae cytochrome b, namely I4L I39M T67A V215M A246T M316I T349I L353M V372I. The predictions here for steppe bison are I39M, M316 and L353. The rest are lineage-sorted alleles that require population-level sampling.
Among tRNA genes, phenylalanine raises the most concerns. The exercise phenotype of such bison may be suboptimal. If this is a bottleneck expansion of a rare bad allele, steppe bison will have GACTC in its 3' T-loop stem (instead of AACTC).
The prospects for using steppe bison in genetic re-engineering of bison mitochondrial dna seem far-fetched at this time: a bison cell line is cured of mitochondrial (but not nuclear) dna with ethidium bromide and fused with implausibly viable frozen steppe bison fibroblasts from which the nuclear (but not mitochondrial) dna has been removed by centrifugation or electrophoresis. (Alternatively, intact steppe bison mitochondria might hypothetically be recoverable and somehow injectable). The resulting cybrid has bison nuclear dna and steppe bison mitochondria. It is then cloned in cattle. Examples to date (extinct ibex, post-mortem wolf) focus on inter-species somatic cell nuclear transfer which addresses nuclear gene issues, not mitochondrial.
Interpreting yak CYTB variation
Yaks are the closest living sister species to bison. Although 15,000 wild yaks still persist, they have been subject to very similar pressures to those experienced by bison: bottlenecks, population fragmentation, introgression from long domesticated yaks and hybridization with cattle. Adaptations specific to mitochondria may exist as yak live at altitudes exceeding 4000 meters with average annual temperatures in rearing areas –8°C, with animals surviving winter temperatures of –40°C.
Because yaks provide the immediate outgroup for bison genetics (and vice versa), their parallel mitochondrial proteomics are investigated in depth here. This further enables reconstruction of mitochondrial proteins of their last common ancestor (after consideration of lineage sorting) and correct placement of Pleistocene genomic sequences.
Data availability for yaks was greatly improved by a Dec 2010 paper by Zhaofeng Wang et al. that investigated yak phylogeographical structure and demographic history on the Qinghai-Tibetan Plateau. Complete mitochondrial genomes were determined for 48 domesticated and 21 wild yaks. The three lineages shown in article supplemental established diverged at 420 kyr and 580 kyr in accordance with extended but temporary allopatric migration barriers created by two large plateau glaciations.
The wild yaks are found in all three branches of the tree (solid circles in figure). Their entries at GenBank are distinguished by a W (for wild) prefix, eg isolate W77 GQ464266. There is potential for confusion here because NCBI taxonomy uses Bos grunniens mutus for wild yak, yet the subspecies concept is contradicted by the mixed distribution of wild and domestic yaks in the mitochondrial tree. Related taxa such as Bos mutus (Przewalski, 1883), Bos mutus grunniens, and Poephagus mutus also conflict with the facts. Yak and bison -- diverging at 2.5 million years -- need to reside in the same genus.
The primary focus here are protein polymorphisms in wild yak because domesticated animals may exhibit inbreeding issues and other evolutionary artifacts due to their estrangement from darwinian selection. Consequently it is important to track which GenBank entries reference wild yaks.
Bos grunniens mutus has three GenBank entries relevant to cytochrome b: proteins AAX53006 and AY955226 both containing unique V195A, I348F mutations in an otherwise wildtype background and CAA76015, an older fragmentary wildtype sequence not considered further here. The first two animals add samples to the large, remote Xinjiang province but remain unpublished (Liu,Q Wu,M Li,Y) despite the 27 Mar 2005 submission date at GenBank. (A number of D-loop sequences submitted for this taxon on 19 Jan 2009 by 27-MAR-2005 by Ma,ZJ also remain unpublished.)
The Myanmar/Bhutan mithun sequence BAJ05329 attributed to Bos grunniens at GenBank has 12 differences to wild yak but is 100% identical to 94 Bos indicus entries, ie it is a hybrid and its mitochondrial genome is irrelevant here. Such GenBank errors are all but impossible to correct.
The 21 new genome accessions of wild yak are GQ464266, GQ464265, GQ464264, GQ464263, GQ464262, GQ464261, GQ464260, GQ464259, GQ464258, GQ464257, GQ464256, GQ464255, GQ464254, GQ464253, GQ464252, GQ464251, GQ464250, GQ464249, GQ464248, GQ464247, GQ464246. These were not labelled on the published tree.
In terms of protein accessions (which will be shown at NCBI blastp output), these are ACU81659, ACU81646, ACU81633, ACU81620, ACU81607, ACU81594, ACU81581, ACU81568, ACU81555, ACU81542, ACU81529, ACU81516, ACU81503, ACU81490, ACU81477, ACU81464, ACU81451, ACU81438, ACU81425, ACU81412, ACU81399 to which AAX53006 and AY955226 can be added.
Of these, 16 fall in the main reference sequence group (wildtype) but 5 wild plateau yaks exhibit polymorphisms that cannot be attributed to domestication. As noted, two additional wild yaks from extreme NW China have additional double mutations but no associated PubMed publication nor tissue source indicated. As either change alone would inactivate an essential enzyme, these represent either heteroplasmic oddities or sequence error (to be pursued as other proteins are considered). The remaining sequences were derived from muscle and skin dna.
There is no overlap between wild yak polymorphism sites and the five of domestic yak. Alleles occurring in full length sequences are analyzed further below.
The summary table of yak CYTB amino acid polymorphisms below arises from alignment of 5000 full-length mammalian cytochrome b orthologs. Magenta indicates a deleterious change at an invariant position, red a deleterious mutation at a naturally polymorphic site, green a possibly acceptable change but of restricted distribution and fitness, and blue a near-neutral substitution. Gray is reserved for probable sequencing error. It can be seen that the smallish yak population sampled (72 animals) already contains 5 deleterious alleles in CYTB which represents only 10% of the amino acids of the mitochondrial proteome.
In summary, out of 70 individual yaks, 10 are carrying deleterious mutations at five sites. That seems like an extraordinary number for a central enzyme in energy metabolism for which it is difficult to envision compensation by another gene. Restricting to the 21 wild yaks, 3 have deleterious polymorphism and 1 has a marginal change. Overall 1 in 7 animals is affected just in this one gene. However CYTB is but one of 13 encoded by the mitochondrial genome -- what sort of genetic burden are yaks carrying overall?
1 ACU81568 A017T wild yak isolate W50 GQ464259 2 ACU81399 I192T wild yak isolate W02 GQ464246 ACU81633 I192T wild yak isolate W75 GQ464264 3 ACU81555 D214N wild yak isolate W40 GQ464258 4 AAX53006 V195A I348F mutus isolate Xinjiang01 unpublished Liu,Q Wu,M Li,Y AAX53007 V195A I348F mutus isolate Xinjiang02 unpublished Liu,Q Wu,M Li,Y 5 ACU81529 V329M wild yak isolate W1313 GQ464256 6 ABI15999 V039I A067T domestic yak fragment PUBMED:17257194 Poephagus 7 ABI16000 V039I A067T domestic yak fragment PUBMED:17257194 Poephagus ACU82153 A084T domestic yak isolate HY5 8 ACU82101 V098L domestic yak isolate HY1 9 AAU89116 I118T domestic yak =SP:Q5Y4Q0 PUBMED:16942892 ACU81711 I118T domestic yak isolate HZ3 ACU81737 I118T domestic yak isolate MQ1 AAS93096 I118T domestic yak fragment PUBMED:17257194 AAS93099 I118T domestic yak fragment PUBMED:17257194
Although the mitochondria encodes the usual 20 amino acids, only a subset of chemically similar residues ever appear at a given position in a given protein -- its reduced alphabet. This subset describes the evolutionarily acceptable substitutions that do not significantly disrupt protein functionality. Discovery of this reduced alphabet can be achieved with greater precision the higher the number of available species and individual sequences multiplicities. For mitochondrial proteins, that sensitivity is 1 in 10,000 (0.01% occurrence frequency) for a given amino acid, much better than even the much-studied human nuclear genome.
Interpretive certainty is never attained without experimentation (yeast is a surprisingly informative model system) but improves up to a point with more sequence data. Here it is important to check whether less common substitutions have persisted over evolutionary time in a phylogenetically coherent manner (ie a sub-clade) or are novel adaptations perhaps in conjunction with a co-evolving residue at another site (or another protein, perhaps nuclear-encoded). After these considerations, the remaining rare changes are mostly deleterious (or sequencing error) but rarely adaptive. Polymorphism significance can be pursued at the xray structural level for only 3 of the 13 mitochondrial proteins (CYTB, COX2, COX1) and even this is complicated in the case of CYTB by its oligomeric association with 3 nuclear encoded proteins.
Aligning CTYB from the 72 complete yak mitochondrial genomes available on 1 Dec 10 shows variation at just 9 sites along the protein (ie 9 nsSNPs). These are quickly found when the web alignment tool retains input sequence order, displays residues identical to the top sequence as dots, gaps fragmentary data correctly, and allows a wide display permitting effective cross-species comparisons.
Yak and bison -- despite being sister species -- share variation only at one site, position 98. Here yak is exclusively valine with the exception of a single deleterious occurrence (see below) of leucine, whereas bison have a mix of valine and alanine (which otherwise is very rare at this position in mammals), ie the ancestral residue was valine. Thus no lineage sorting occurred at any amino acid position in CYTB at the time these two species diverged at 2.5 myr. Lineage sorting however may be important in the overall evolution of the Bovini: 53 ancient polymorphisms (at the dna level) are said to have persisted since Bos and Bison diverged from Bubalus 5–8 million years ago.
The changes can also be displayed in context by coloring the appropriate residues in a reference sequence relative to a composite sequence consolidating all the polymorphisms from distinct animals (no one animal has more than two of the 9; V195A + I348F occurs in two animals). The composite sequence is quite useful in comparing polymorphism sites across species as explained in the annotation tricks section.
>CYTB_bosGruR Bos grunniens cytochrome b ref seq taken as gi|147744503
MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF
WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIITAIAMVHLLFLHETGSNNPTGISSDADKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI
LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQLASIMYFLLILVLMPTAGTIENKLLKW
>CYTB_bosGruP Bos grunniens composite polymorphisms: A017T A084T V098L I188T I192T V195A D214N V329M I348F
MTNIRKSHPLMKIVNNTFIDLPAPSNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHTNGASMFFICLYMHLGRGLYYGSYTFLETWNIGVTLLLTVMATAFMGYVLPWGQMSF
WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIITATAMAHLLFLHETGSNNPTGISSNADKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI
LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLMADLLTLTWIGGQPVEHPYFIIGQLASIMYFLLILVLMPTAGTIENKLLKW
wild dom dom dom wild dom wild wild dom
A017T A084T V098L I118T I192T V195A D214N V329M I348F
4018S 4994A 4522V 4309I 4353L 4528V 4429D 4610V 4232I
927A* 3T 430I 667S 505M 427I 512N 188T 651V
46T 1P 34M 14I 94I* 25T 43E 133A 63T
3L 1V 11A 1T 31T 4G 8S 44I 45M
3M 1L 3F 4M 2Y 22M 4N
1F 1N 2V 1A 1H 2G 2F
1P 1A 1E 1A
1S
A017T: At position 98, the mammalian reduced alphabet consists primarily of serine but with yak alanine also well represented at 18%. Threonine occurs in 46 sequences so cannot be sequence error or serious mutation. Bulk seems to be the main criterion at this site rather than polarity -- threonine though polar is bulkier residue than serine or alanine. To determine whether it has arisen multiple times or just in one clade, the phylogenetic distribution of the 46 occurrences needs consideration.
It can be seen from the graphic at left that A017T has arisen multiple times with no common denominator (such as high elevation lifestyle) but -- with the exception of monotremes -- never in a deep stem ancestor. That is, A017T occurs here and there but only in recently speciated clades. This suggests that while not lethal, over time it gets replaced by more adaptive serine or alanine.
A017T
4018 S
927 A yak do not have the most common amino acid at position 17
46 T
3 L
3 M
1 F
1 P
A084T: At position 84, alanine is strictly invariant. Thus threonine is an unmistakable deleterious mutation in domestic yak.
A084T
4994 A
3 T
1 P
1 V
V098L: At position 98, the reduced alphabet consists of valine 90% of the time regardless of mammalian clade with the similar (branched chain aliphatic) isoleucine having substantial dispersed representation at nearly 9%. The 430 species in which it occurs are scattered incoherently within mammal clades, meaning that it has arisen independently many times. V098I may be slightly suboptimal as there is an evident bias (at some level) against equal occurrence. It likely co-exists with valine in most non-bottlenecked populations of mammals, observed if enough individuals of a given species are sequenced.
However leucine, the seemingly similar third aliphatic residue, occurs one once despite being but a single base change transition away from the dominant residue. Were leucine a near-neutral substitution, its incidence would be vastly higher. Thus the change V098L reported for yak represents either a deleterious mutation or an unprecedented adaptation (eg to high altitude) or sequencing error in GenBank entry ACU82101. The same can be said for the more overtly radical change V098N in lemur AAS00156. The 34 methionines occur sporadically in the phylogenetic tree suggesting they are sub-adaptive and blink out over time. Indeed, canine spongiform leukoencephalomyelopathy is attributed to V98M. Dog CYTB is 89% identical to that of yak and numbering corresponds.
V098L 4522 V 430 I 34 M 11 A bison 1 L yak 1 N lemur
I118T: At position 118, the reduced alphabet consists predominantly of ILV with some A and M, a very common occurrence proteomewide. TSF are all deleterious mutations in domestic yak.
I118T
2597 I
1843 L
404 V
87 A
61 M
6 T (all yak)
1 S
1 F
I192T: At position 192 of wild yak, the dominant residue is leucine instead of the yak ancestral value isoleucine, which is disfavored relative to methionine, ie isoleucine is a mild polymorphism in its own right but the associated taxonomy shows it narrowly restricted to 83 sequences in Bos, Bison, and separately in 5 Kobus (waterbucks), too persistent to be dysfunctional and indeed a candidate for adaptive. However change to polar threonine is seen in 31 nominal species but after removal of redundancy, only in two species of pocket mice. Thus the yak change is deleterious.
I192T
4353 L
505 M
94 I
31 T
3 F
2 V
1 A
1 S
V195A: This allele occurs together with I348F in two wild yaks from a remote region in NW China. Despite sequence submission, no article has appeared in the three subsequent years. It can be seen from the reduced alphabet frequencies that this is a severe mutation (as is I348F) so taken together likely sequence just error. No further analysis will be done here until such time as the polymorphisms are confirmed.
V195A
4528 V
427 I
25 T
4 G
4 M
1 A
D214N: This polymorphism of wild yak is seen quite widely, in some 10% of mammals. The 223 taxa with D214N are mostly confined to laurasiatheres and glires but are not a hallmark of these clades. Nor do the species with asparagine have any common lifestyle denominator. Asparagine is an acceptable variation for aspartate at this site if perhaps not optimal.
D214N
4429 D
512 N
43 E
8 S
4 X
2 Y
1 H
V329M: This allele occurs in wild yak. Methionine is not a radical substitution in terms of physical/chemical properties and similar additional amino acids appear at low levels, even though valine occurs in a huge majority of species. Methionine occurs in 17 other species phylogenetically scattered species include Bos javanicus, Ovis, Budorcas, Naemorhedus, Mus, Rattus, bats and sloth. Thus it is likely suboptimal but not significantly deleterious.
V329M
4610 V
188 T
133 A
44 I
22 M
2 G
1 E
I348F: This allele occurs together with V195A in two wild yaks from a remote region in NW China. Despite sequence submission, no article has appeared in the three subsequent years. In can be seen from the reduced alphabet frequencies that this is a severe mutation but more likely sequence error, as is V195A.
I348F
4232 I348F
651 V
63 T
45 M
4 N
2 I348F
1 A
Human CYTB polymorphism and disease
Polymorphisms and pathogenic mutations disease for human CYTB have been very helpfully compiled by mtDB and MitoMap, with other mammals at OMIA. Fortunately, numbering systems carry over without change to bison and yak since no indels occur in this gene within mammals.
A poor tradition in mitochondrial research allows amino acid changes to be described just by a single nucleotide coordinate relative to the Cambridge Reference Sequence, NC_012920. That requires the user to have a numbered translation via the mitochondrial genetic code showing in-frame amino acids; however the change from the protein perspective (eg V98T) is often conveniently displayed at Uniprot. Coordinates for all mitochondrial features are tabulated here; CYTB extends from position 14747-15887.
Given over 7000 complete human mitochondrial genomes and a high mutation rate, some human polymorphic sites will inevitably overlap with yak and bison alleles. Thus any information about associated human disease at the 16 known disease sites might be transferable. However many rare and obviously dysfunctional alleles were collected for population haplotype mapping and no disease information was collected.
Annotation transfer is vastly complicated by heteroplasmy, experimentalist inability to establish the heritability of the allele, and differences in tissues used to obtain dna for sequencing and so neglects the possibly compensatory effect of changes elsewhere in this or another gene (eg S152P and G291D are suppressed by [compensatory hinge region substitutions in nuclear-encoded rieske protein), a substantial issue in a protein like cytochrome b where 10% of the residues between bovids and human are non-identical and so many proteins participate in Complex III. At such sites (eg H214Y human, D214N yak), transfer of phenotypic information is dubious.
brown in the human allele table below indicates human polymorphisms corresponding to an allele of concern in yak or bison. In two significant cases -- both in domestic yak -- the initial and final residue of human are identical to that of yak, namely A084T* and I118T*. Both are predicted to be deleterious in both human and yak. Unfortunately no clinical information was collected on the human side and the health status of the yaks is unknown (eg level of exercise intolerance).
However even A084T (a strongly invariant site in all mammals) was evidently not early-lethal for its adult human carrier (dna samples are collected from adult volunteers whose health status is not recorded). Here the vast and still unsettled complexities of mitochondrial genomics may come into play:
- a single mitochondrion may up to 10 replicated copies of its genome which need not be identical
- cells can carry thousands of mitochondria inherited erratically during embryogenesis and later stem cells
- dna samples, not being collected from germline cells, may represent non-heritable somatic mutations in restricted descendent cells of the tissue sampled
- disease onset is often in late adulthood due to the nature of mitochondrial replication and dispersal to daughter cells and so may not be applicable to shorter-lived species
Symptoms of severe heteroplasmic mitochondrial disorders frequently do not appear until adulthood because many cell divisions and much time is required for a cell to receive enough mitochondria containing the mutant alleles to cause symptoms. An example of this phenomenon is Leber optic atrophy (LHON). Affected individuals may not experience vision difficulties until they have reached adulthood. Another example is MERRF syndrome (Myoclonic Epilepsy with Ragged Red Fibers). Heteroplasmy here explains the variation in severity of the disease among siblings. The incidence of heteroplasmy in human mtDNA is unknown, as the number of individuals who have been subjected to mtDNA testing for reasons other than the diagnosis of mitochondrial disorders is small."
The oft-observed disease Leber Hereditary Optic Neuropathy (LHON) is genetically heterogeneous, arising from mutations in other mitochondrial genes (R340H in ND4, A52T in ND1 and M64V in ND6, subunits of complex I of the oxidative phosphorylation chain in mitochondria) as well as from CYTB variants A29T and secondarily D171N and V356M.
tRNA alterations in bison were analyzed by Douglas et al. Here it is known the human disease MERRF disrupts mitochondrial tRNA-Lys in 80% of cases and so biosynthesis of mitochondrial proteins essential for oxidative phosphorylation. It too is genetically heterogeneous as tRNAs for leucine, histidine, serine and phenylalanine can be affected in other individuals. In all 126 distinct mitochondrial tRNA disease mutations have been reported.
human yak A084T A084T* seen twice in Japanese population I098V V098L I118V I118T I118T I118T* seen once in Japan and once in India H214Y D214N A329T V329M T2A S56A I117V D171N I211T G251S M316T A354T T2I S56L I118V D171G T212A E251D Y325H V356M M4V T61A I118T S172N T212I Y256H A329T V356A M4T T70A L121F P173S H214Y T257I A330T T360A R5G Y75C A122T T174A T219A L258P A330V T360M I7T I78V T123A F181L T219I A259T I334V T368A N8S I78T A125T I184V I226V N260D T336A T368I N15S L82F E136D L185S A229T V284I I338V I369V H16R A84T F140L I189V L230F V291A P342S I369T F18L G86S L149M A190T L233V S297P V343M I372V I19M C93Y I153T A190V F235L I300T V343A M376V A29T I98V Y155H A191T L236I I304T S344G A380T A39T G101S I156V A191D S238P I306V S344N A380V A39V Y109H I156T A193T S238F I306T Y345F ---- I42V E111K T158A T194A T241A M309V T348I ---- I42T T112A D159N T194V T241M M309T I349V ---- F50L W113R I164V F199L T243A S310P I349T ---- F50L I115T G167S I211V F245L M316V V353M ---- Of known disease mutations, only V98M corresponds to a bison allele. Disease alleles in blue have been thoroughly studied in yeast. A29T LHON Leber hereditary optic neuropathy G34S mitochondrial myopathy; sporadic S35P exercice intolerance V98M dog leukoencephalomyelopathy V98L human polymorphism with unknown consequences S151P exercise intolerance G166E hyperthrophic cardiomyopathy D171N secondary LHON G231D 16026996 mouse G251D CMIH G251S obesity N255H cardiomyopathy Y278C multisystem disorder G290D exercise intolerance S297P neonatal polyvisceral failure G339E mitochondrial myopathy V356M secondary LHON
- See abstracts for all 16 disease sites
- Adaptive rates of evolution in all 13 genes from an alignment of 214 mammalian mitochondrial genomes
- Adaptive evolution of the mammalian mitochondrial genome
- A series of 12 papers on mitochondrial dna inheritance issues
Cytochrome b mutations in Leber hereditary optic neuropathy CYTB:D171N CYTB:V356M ND5:A458T New mutations were discovered in the apocytochrome b gene in Leber hereditary optic neuropathy probands who did not harbor either of the two known Complex I mutations (positions 3,460 and 11,778). A mutation at position 15,257 was found in eight independent probands which changed a highly conserved D to N, was not found in controls, and appears to be pathogenetically significant. The 15,257 mutation occurred in association with a known synergistic mutation at position 13,708 in 7/8 probands (ie ND5 A458T) and in association with a new apocytochrome b mutation at position 15,812 (ie V356M) in 4/8 probands. Mutations in Complex III genes may be involved in Leber hereditary optic neuropathy and multiple, simultaneous mutations occur frequently.
Mazunin IO (2010) Mitochondrial genome and human mitochondrial diseases. Molecular Biology 44(5) Today there are described more than 400 point mutations and more than hundred of structural rearrangements of mitochondrial DNA associated with characteristic neuromuscular and other mitochondrial syndromes, from lethal in the neonatal period of life to the disease with late onset. The defects of oxidative phosphorylation are the main reasons of mitochondrial disease development. Phenotypic diversity and phenomenon of heteroplasmy are the hallmark of mitochondrial human diseases. It is necessary to assess the amount of mutant mtDNA accurately, since the level of heteroplasmy largely determines the phenotypic manifestation. In spite of tremendous progress in mitochondrial biology since the cause-and-effect relations between mtDNA mutation and the human diseases was established over 20 years ago, there is still no cure for mitochondrial diseases.
Pathogenic mitochondrial DNA mutations in protein-coding genes
Lee-Jun C. Wong PhD Muscle Nerve, 2007
More than 200 disease-related mitochondrial DNA (mtDNA) point mutations have been reported in the Mitomap database. These mutations can be divided into two groups: mutations affecting mitochondrial protein synthesis, including mutations in tRNA and rRNA genes; and mutations in protein-encoding genes (mRNAs). This review focuses on mutations in mitochondrial genes that encode proteins. These mutations are involved in a broad spectrum of human diseases, including a variety of multisystem disorders as well as more tissue-specific diseases such as isolated myopathy and Leber hereditary optic neuropathy (LHON). Because the mitochondrial genome contains a large number of apparently neutral polymorphisms that have little pathogenic significance, along with secondary homoplasmic mutations that do not have primary disease-causing effect, the pathogenic role of all newly discovered mutations must be rigorously established. A scoring system has been applied to evaluate the pathogenicity of the mutations in mtDNA protein-encoding genes and to review the predominant clinical features and the molecular characteristics of mutations in each mtDNA-encoded respiratory chain complex.
S297P homoplasmic in all tissues tested, undetectable in mother PMID: 19563916
Eur J Hum Genet. 2004 Mar;12(3):220-4.
The deleterious G15498A mutation in mitochondrial DNA-encoded cytochrome b may remain clinically silent in homoplasmic carriers.
We report on a patient with severe growth retardation and IgF1 deficiency, in which a mitochondrial abnormality was suspected. An isolated mitochondrial respiratory chain complex III deficiency was found in blood lymphocytes and skin fibroblasts. Sequence analysis of the cytochrome b, which is the only mitochondrial DNA-encoded subunit of complex III, revealed a homoplasmic G15498A mutation, resulting in the substitution of a highly conserved amino acid (glycine 251 into an aspartic acid). The mutation was found to be homoplasmic in all tissues examined from the mother and her brother (lymphocytes, fibroblasts, hair roots and buccal cells). Complex III deficiency was also demonstrated in these cells. Nevertheless, the mother and the brother were asymptomatic. This mutation had been considered as a cardiomyopathy-generating mutation in a previously reported case, and its pathogenicity has been demonstrated recently in yeast. However, it seems not to fulfil the classical criteria for pathogenicity of a mitochondrial DNA mutation, especially the heteroplasmic status, and to be clinically silent, albeit present, in nonaffected relatives. We suggest that other factors are contributing to the clinical variability expression of the G15498A mtDNA mutation.
Mitochondrial DNA mutations cause disease in >1 in 5000 of the population and approximately 1 in 200 of the population are asymptomatic carriers of a pathogenic mtDNA mutation. Many patients with these pathogenic mtDNA mutations present with a progressive, disabling neurological syndrome that leads to major disability and premature death. There is currently no effective treatment for mitochondrial disorders, placing great emphasis on preventing the transmission of these diseases. An empiric approach can be used to guide genetic counseling for common mtDNA mutations, but many families transmit rare or unique molecular defects. There is therefore a pressing need to develop techniques to prevent transmission based on a solid understanding of the biological mechanisms. Several recent studies have cast new light on the genetics and cell biology of mtDNA inheritance, but these studies have also raised new controversies.
Nuclear proteins that raise mitochondrial mutation rates
The genetic stability of mtDNA in every mammal (indeed every eukaryote) depends critically on the accuracy of dna replication. The consequences of any mutation in this machinery would be greatly amplified (like the broomsticks in the Sorcerer's Apprentice) by subsequent somatic errors created in replicating mitochondrial genomes. It is essential to consider these genes given the apparent elevated rate of mitochondrial polymorphism reported for bison and yak.
The nuclear encoded, mitochondrially functioning dna polymerase POLG on chr 15, the catalytic subunit The catalytic subunit (dna polymerase itself, 3’-5’ exonuclease for proofreading, 5’deoxyribosephosphate lyase for base excision repair), deserves special mention in regards to the extraordinary observed rates of yak and bison coding polymorphisms. Some 90 distinct [human disease alleles are known along the 1239 residue protein, causing progressive external ophthalmoplegia, sensory and ataxic neuropathy, Alpers syndrome, and male infertility (see PEOA1, SANDO, AHS, MNGIE at OMIM). POLG also contains a polyglutamine tract near its N-terminus of length 13 in human that may be subject to polymorphic replication slippage.
POLG is accompanied by an accessory dimer of POLG2. Now receiving considerable attention, two mitochondrial disease alleles have been found, G416A and G451E (causing adPEO). A helicase (PEO1 or twinkle) causing an adult-onset progressive external ophthalmoplegia PEO and topoisomerase TOP1MT are other nuclear encoded proteins critical to mitochondrial dna replication. The latter binds a specific site in the D loop control region. These too have been implicated in rare mitochondrial diseases.
These enzymes, especially POLG, needs extensive sequencing in bison and yak (indeed every once-bottlenecked endangered species). That might done economically on a population scale with whole-exome chips rather than sequencing whole genomes. The POLG gene itself is difficult to study in isolation, being comprised of 23 exons spread out over 18490 bp.
No sequencing of yak or bison POLG has been done yet but that of cow, sheep and pig etc are readily retrieved from their respective genome projects. The Bos taurus POLG protein is 90% identical to human; it has not been specifically studied.
Numts: excluding mitochondrial pseudogenes
Mitochondrial research has been plagued by numt pseudogene alleles mistakenly obtained from the nuclear genome by primer cross-over. Here rna transcribed from mitochondrial genes somehow exits the mitochondria, enters the cell nucleus, gets reverse-transcribed into dna, and then gets heritably integrated into the nuclear junk genome (where it generally is not transcribed and rapidly accrues the mutation pattern of a pseudogene), sometimes becoming fixed across the entire population and even diagnostic of it.
This seemingly implausible sequence of events is surprisingly common. Counts for any species with assembled genome can quickly be conducted by Blat at the UCSC genome browser, though very old events would be missed. Querying cow genome for CYTB nuclear pseudogenes shows 19 nuclear genome matches to cytochrome b, ranging from quite strong to barely significant.
The best match occurs on cow chr28:34924178-34924995. Not quite full length 3', it contains 7 internal stop codons, 52 addition missense mutations (that characteristically do not follow site conservation patterns), and various indels and frameshifts. Not particularly recent, its date of formation could be bracketed by examining sheep and pig genomes for an orthologous numt at syntenic location (+psCYTB +SFTPD (or CGN1, bovine conglutinin).
It's not clear pig contains the orthologous pseudogene at chr14:34281258-34281963 because this feature is not immediately syntenic to porcine SFTPD at chr14:85511174-85522800. If so, the most recent CYTB pseudogene in cow predates the divergence of cow and pig. It then will be found in both yak and bison genomes unless lost through large-scale deletion. Note pig has a much more recent CYTB at chr2:104178282-104179415.
The sheep genome is not currently in a satisfactory state of assembly. This is far more likely than pig to contain a demonstrably syntenic CYTB pseudogene. No sheep pseudogenes are posted at GenBank nr nor locatable by tblastn against wgs or hgts databases. Although 31 CYTB pseudogenes from 11 pecoran species are available, these species all lack genome projects. However upon blastn of the cow chr28 feature, Kobus kob (AF052940) and Capra hircus (GU120393) have very strong matches.
Recent numt pseudogenes can capture ancestral values that prevailed in the mitochondria at the time of formation. Unlike bone fossils, this dna has steadily accrued changes up to the present, but the benefit is nuclear pseudogenes evolve up to 12 times slower than the mitochondrial parental gene. Thus it might represent an atypical heteroplasmic allele existing at that time, be affected by lineage sorting, or reflect a parallel nuclear mutation and so not really settle the issue of ancestral value. A joint tree (1, 2) containing both mitochondrial CYTBs and nuclear pseudogenes (as outgroups) considered in chamois (Rupicapra) has many complexities because genes evolve so differently in the two compartments (for example the pseudogene might have arisen from a heteroplasmic variant that existed at the time).
The yak study specifically considered whether numts could explain divergent, low-frequency mtDNA haplotypes, but ruled out all but the very most recent on the basis of the separate confirmatory D-loop haplotype phylogeny and great similarity to other haplotypes without unusual sequence features.
Conservation genomics
(to be continued shortly with a brief survey of recent conservation informed by modern genetics)
Recommendations for bison conservation genomics
The deleterious mutations V98A and I60N in the mitochondrial genes CYTB and ATP6 have reached high frequencies (42% of the 32 known or 133 of inferable genomes sampled) in North American bison herds. Both occur within a single haplotype -- bad news in that the two together assuredly cause mitochondrial disease, good news in that many bison have healthy mitochondrial dna. Because tRNAs are not functionally affected (contrary to a initial evaluation), there is no indication of an elevated mitochondrial mutation rate attributable to a imported nuclear-encoded protein encoded by a defective nuclear gene.
Affected bison can be predicted to experience significant impairment of oxidative phosphorylation and exhibit significant exercise intolerance and lactic acidemia relative to healthy bison, based on known phenotypes in dogs and human with similar substitutions. The alanine and asparagine double mutation is likely maladaptive with respect to predators, cold winters, and competition with other bison for forage and breeding.
Given the predicted scope of the maladaptive haplotype, further validation of the proxy region inference method by direct sequencing of affected coding genes is needed. More important still is direct experimental verification of bison mitochondrial dysfunction is essential prior to considering a management practise based on genomic analysis alone. Such experiments have been conducted thousands of times already in humans suspected of mitochondrial disease so established protocols simply need repeating on bison.
Note these mutations have nothing to do with whether the herd history includes hybridization with domestic cattle as mitochondrial dna is maternally inherited. Ironically, hybrids (always involving a bison bull) and their descendants would be spared because cattle do not carry these mutations. The haplotype has spread heritably to many animals so cannot be wished away by invoking heteroplasmic compensation or somatic cell manifestation or origin.
How is it possible that a bad polymorphism has spread so widely? This is not at all surprising given a severely bottlenecked nineteenth century bison population expanding from a small number of individuals, in conjunction with a greatly diminished role for natural selection ever since (ie disruptive management practices such as random culls, gender imbalance and trophy hunting).
Even if experimental data establishes significant dysfunction, the bison are evidently viable. Herds such as those at Grand Teton National Park -- which appear 100% affected and have no prospects for correction without outside re-introductions -- persist through long winters, fend off predators and expand their population each year. Yet genomic management is still important (as argued for many years by Derr) because these bison do not accurately represent the genetics of wild nineteenth century bison and consequently not their capabilities, behaviors or even ecosystem interactions.
Because of its peculiar inheritance and lack of recombination, the mitochondrial genome can be managed in isolation -- but should it? Its 13 encoded proteins represent less than 0.1% of the 20,000 total. Although little is currently known about the distribution of advantageous and deleterious nuclear alleles, that situation is changing very rapidly as new genome projects are completed for yak, bison and steppe bison. (The cattle SNP chip is an interim survey technique that does not satisfactorily inventory or assess amino acid changes genomewide.)
If bison nuclear gene variation approximates that of human, we can expect a bison genome to have 275 coding genes with a dysfunctional allele (but heterozygously compensated by normal allele except for chr X) and another 75 proteins with troubling amino acid substitutions. While this is perhaps a natural level of genetic burden, certain deleterious changes may again have attained unnatural frequencies relative to ancestral bison populations and thus be targets for reduction.
1000 Genomes Project Oct 2010: On average, each individual nuclear genome carries 275 loss-of-function variants in annotated genes and 75 variants previously implicated in inherited disease [both classes typically heterozygous]. We estimated that an individual typically differs from the reference human genome sequence at 10,488 non-synonymous sites [out of 9,000,000 proteomewide, O.12%]. Each individual has 200 in-frame indels, 90 premature stop codons, 45 splice-site-disrupting variants and 235 deletions that shift reading frame."[small edits made]
No North American bison herd is currently able to freely expand its population subject only to the natural constraints of habitat availability, predators and disease. If management culling is to occur, better that it be genetically informed than uninformed. This has already occurred in terms of eliminating cattle mitochondrial hybrids in various conservation herds (such as the TNC Ordway herd). That reduces but does not eliminate introgressed cattle nuclear genes and could inadvertently enhance deleterious bison mitochondrial haplotypes and lower important bison nuclear alleles.
The next priority for mitochondrial genomic management are V98A-I60N bison. Over time, resumption of natural selection would presumably accomplish this without human intervention, provided each herd had at least some V98-I60 component. Nuclear genetic diversity of 98A-60N bison could still carried forward by bulls, recombining into nuclear genomes of V98-I60 cows. Current culling practices at Yellowstone National Park do not move the genetics situation forward.
Bison also carry various deleterious private mitochondrial alleles. Here it is not possible without sequencing many more mitochondrial genomes whether these simply reflect heteroplasmy in tissues sampled (typically leucocytes, skin or muscle) and are not currently heritable via oocytes. While these mutations adversely affect individual animals, the haplotype frequency appears low. These alleles can be tolerated (keeping anomalies in view, eg overtly deleterious human hemoglobin genes protecting against malaria) but with higher frequencies attained (or not) only via natural selections.
In free-ranging bison herds such as Yellowstone, it is currently impractical to track individual animal genotypes. Here natural selection is the best option; current uninformed culling is very dangerous. The Yellowstone herd size of 3900 may well suffice to maintain genetic diversity over time -- the problem is, we don't want to maintain that genetic diversity as it now stands. Certain haplotypes are deleterious and far too common. Even genetically pure bison have been adversely affected by past human actions.
Random culls will never winnow out V98A-I60N or other deleterious phenotypes in any herd, at best holding haplotype frequencies constant while interfering with beneficial losses that would have come from natural selection or genomically informed management. In effect, no management is the best management -- just as beaver taught salmon how to jump, wolves and winter will keep bison aerobically fit.
Recommendations for yak conservation genomics
When yak and bison mitochondrial genomes are sequenced and a polymorphism is reported to GenBank, what does that mean? Presumably it reflects an overwhelmingly dominant value of whatever heteroplasmy existed in the tissue sample used to sequence the dna.
The key bison study used white blood cells as dna source, rather than muscle/skin of yak data. One might imagine this fraction of whole blood is quite heterogeneous in terms of stem cell origin -- five different, diverse leukocyte types exist -- but in fact these all derive from a single hematopoietic stem cell type of bone marrow. Consequently, no other cell types were sampled. Thus we do not know whether the observed polymorphisms are heritable, apart from those observed in multiple animals.
For yak, the observed polymorphisms are again not necessarily heritable even for female individuals (male mitochondria are not passed on). However in the case of yak polymorphisms I118T (domestic) and I192T (wild), multiple individuals (5, 2 respectively) sampled carried the same rare change, strongly implying (unless these are mutational hotspots) that these are entrenched in the germline and so inherited. Oocyte heteroplasmy however is also heritable so wildtype may still persist. The other polymorphisms may be mere somatic mutations that attained abundance in the sampled tissue but are still complemented by residual wildtype. This would have to be pursued in additional tissues or more definitively by sequencing offspring, perhaps not feasible in wild yak.
In summary, even deleterious polymorphisms may have limited effects, depending on stem cell origin and compensation by the wildtype component of heteroplasmy. On the other hand, should a bad alleles exert a negative dominant effect even as the minority allele in the mitochondria in which it resides (eg tainting oligomeric proteins), it would still have a deleterious phenotype even though it never comes to 100% frequency in any particular cell type. Somatic mutations in bison and yak may have limited impacts if onset of disease is delayed to late adulthood as in human. For conservation genomics, we are primarily concerned with heritable mitochondrial mutations, though enhanced levels of somatic mutations (due say to a faulty POLG dna polymerase) are also a concern.
In domestic yak, animals bearing I118T should not be encouraged to reproduce. To be on the safe side, higher frequencies of the other deleterious alleles are also undesirable, even though not quite proven to be heritable.
In wild yak, I192T is the primary cause of concern. It should be avoided if captive breeding comes into play. A017T, D214N, and V329M are not deleterious mutations but rather natural and possibly adaptive parts of yak diversity whose continuation should be encouraged.
These preliminary recommendations are based solely on CYTB. Since only rare recombination occurs in mitochondria (that could bring good alleles on different genes together) and no paternal contribution can dilute out undesirable heteroplasmy, it is unclear how these recommendations can be implemented, much less reconciled from those emerging from independent considerations of the other 12 mitochondrial genes.
Bioinformatic methods, curated sequences, free full text sources
These sections describe analytic methods used, provide various curated data sets, and compile sites hosting free full text of bison and yak research genomics articles. The latter is especially important in terms of leveling the informational playing field and enhancing public understanding of management issues involving bison conservation herds.
The analytic methods used above are all fully accessible to anyone with an internet connection and do not require any programming skills or installation of software, but assume a basic understanding of molecular biology and biochemistry, familiarity with widely used databases such GenBank and PubMed, and the use of paste-and-submit web tools. While many advanced modeling programs exist, they inevitably involve a raft of arbitrary parameters and hidden assumptions seldom justified by data quality, paleo-uncertainties, and the issue at hand. It is far better to keep the data in view (even if that entails hand-editing of screen-scrapes) and produce a jargon-free product that anyone can understand and replicate.
Curated sequences are provided because GenBank can be quite tedious to winnow and re-format. It is not unusual for hybrid dna sequences to be filed under the wrong taxon. Control region sequences are quite variable in region covered, have submission age dependent quality, and contain inadequate description of bison dna sources that must be supplemented from their underlying article (if one exists). The main innovation here is to treat fasta headers as small flatfile databases. This causes them to display informatively (unlike accession number gibberish) within the small number of characters allotted in the margin by alignment programs. This is especially important when many hundreds of sequences are aligned.
Methods: bioinformatic tips and tricks
New sequencing technologies have greatly affected the amount of mammalian mitochondrial genomic data available at GenBank. Five years ago, it was acceptable to publish population-level D loop sequences accompanied by a few fragmentary coding reads; today, a publication might offer 60-70 entire mitochondrial genomes. This favors evolutionary study of mitochondrial proteins over comparative genomics of nuclear genome products because the latter is still restricted to around 50 species (Dec 2010) almost all incompletely sequenced.
Many long-standing issues such as introgression, historic bottlenecks, population mixing, accrual of deleterious coding variants, hard polytomies, and lineage sorting during speciation can now be approached and resolved, especially with the increasing sequencing of end-Pleistocene frozen dna. This may allow more enlightened management of endangered species such as bison where populations reached rock bottom -- recovering numbers is not enough if genomic integrity is still at risk.
However, the flood of data raises significant issues in extraction of significant information: it is not instructive to align the tens of thousands of sequences available for each of 13 mitochondrial proteins -- that give a an intractable array of 3789 amino acids by 12500 sequences, enough to fill 20 x 100 = 2000 screens on the largest possible computer monitor. That data must be distilled down somehow to take-away information.
This section explains a practical desktop protocol for extracting the 'reduced phylogenetic alphabet' at each residue of the mitochondrial proteome. The method depends heavily on current capabilities of Blastp at NCBI and so may not be completely stable to changes made there over time.
First note that tBlastn cannot be used against the nr or wgs nucleotide databases at NCBI (or with Blat at UCSC) since the significantly different genetic code of mammalian mitochondria is no longer supported as a parameter option. Other oddities involve missing terminal nucleotides that are added before translation. However mitochondrial dna is usually translated sensibly at GenBank protein entries.
The vertebrate mitochondrial code:
TTT F Phe TCT S Ser TAT Y Tyr TGT C Cys
TTC F Phe TCC S Ser TAC Y Tyr TGC C Cys
TTA L Leu TCA S Ser TAA * Ter TGA W Trp
TTG L Leu TCG S Ser TAG * Ter TGG W Trp
CTT L Leu CCT P Pro CAT H His CGT R Arg
CTC L Leu CCC P Pro CAC H His CGC R Arg
CTA L Leu CCA P Pro CAA Q Gln CGA R Arg
CTG L Leu CCG P Pro CAG Q Gln CGG R Arg
ATT I Ile ACT T Thr AAT N Asn AGT S Ser
ATC I Ile i ACC T Thr AAC N Asn AGC S Ser
ATA M Met i ACA T Thr AAA K Lys AGA * Ter Bos can use ATA as initiation codon
ATG M Met i ACG T Thr AAG K Lys AGG * Ter
GTT V Val GCT A Ala GAT D Asp GGT G Gly
GTC V Val GCC A Ala GAC D Asp GGC G Gly
GTA V Val GCA A Ala GAA E Glu GGA G Gly
GTG V Val i GCG A Ala GAG E Glu GGG G Gly
AAs = FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIMMTTTTNNKKSS**VVVVAAAADDEEGGGG
Start = --------------------------------MMMM---------------M------------
Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG
Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG
Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
Blastp output at NCBI now has a very useful feature: clustering of identical individual sequences into single alignments, display of multiplicities, with all the accessions visible with an extra click. The only exception involves double-counting of UniProt entries which, since that institution conducts no sequencing, always arise from another entry. These entries are poorly formatted and inconsistent in many ways with GenBank protocols but very well done at UniProt itself. This does not affect overall analysis because individual alleles are scrutinized at the end of the procedure.
However the multiplicities are not retained in the output format needed, query-based with dots for identities. This results in a single representative accession for multiplexed matches. However upon pasting accession containing a given allele into GenBank taxonomy, species redundancy is discarded, yield a count of the number of distinct species with that allele as well as a measure of overall multiplicity.
After collecting high resolution amino acid frequencies at a given site, it is necessary to determine the phylogenetic distribution of each variant (in practice just those of moderate occurrence). That is now very convenient to do provided the associated accessions have been saved:
Simply paste the blastp match list of protein accessions having the chosen amino acid variant into the Entrez text query box. Never mind if it only returns 20 out of your 157 input sequences -- it hasn't forgotten. It doesn't matter if the list has redundant entries (typically SwissProt and the protein giving rise to the SwissProt entry). After retrieval, set the "Find Related Data" to "Taxonomy" and wait for the options to load, then click "Find Items".
Miraculously, this returns a page that can be set to display a text phylogenetic tree your input sequences, the full set entered with all redundancy removed. That text tree has labelled higher taxonomic nodes and individual species deeper down. Final edits can be made quickly that capture the phylogenetic spread of the variant allele for interpretive purposes.
The two most common outcomes:
- all the species carrying the variant comprise a monophyletic clade. If the origin of the clade is fairly ancient, then the variation is a derived informative adaptive change relative to ancestral (synapomorphy). If the site is invariant in all members of the co-clade (meaning the ancestral state has persisted to all other extant species), then the site is a phyloSNP (definition and examples: 1 2 3 4).
- species carrying the variation are scattered incoherently across the mammalian phylogenetic tree. This means that the variation has arisen multiple times (all fairly recently) but has not persisted when it arose earlier, ie it is not a preferred allele for this protein at this site and gets replaced.
Below is the magic spreadsheet formula that correctly strips NCBI blastp output into individual columns: use exactly as tabbed in excel, iworks, apple numbers etc. with blast output occupying the first column beginning at row 8:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 =mid(a8,1,14) =mid(a8,20,1) =mid(a8,21,1) =mid(a8,22,1) =mid(a8,23,1) =mid(a8,24,1) =mid(a8,25,1) =mid(a8,26,1) =mid(a8,27,1) =mid(a8,28,1) =mid(a8,29,1) =mid(a8,30,1) =mid(a8,31,1) =mid(a8,32,1) =mid(a8,33,1) =mid(a8,34,1) =mid(a8,35,1) =mid(a8,36,1) =mid(a8,37,1) =mid(a8,38,1) =mid(a8,39,1) =mid(a8,40,1) =mid(a8,41,1) =mid(a8,42,1) =mid(a8,43,1) =mid(a8,44,1) =mid(a8,45,1) =mid(a8,46,1) =mid(a8,47,1) =mid(a8,48,1) =mid(a8,49,1) =mid(a8,50,1) =mid(a8,51,1) =mid(a8,52,1) =mid(a8,53,1) =mid(a8,54,1) =mid(a8,55,1) =mid(a8,56,1) =mid(a8,57,1) =mid(a8,58,1) =mid(a8,59,1) =mid(a8,60,1) =mid(a8,61,1) =mid(a8,62,1) =mid(a8,63,1) =mid(a8,64,1) =mid(a8,65,1) =mid(a8,66,1) =mid(a8,67,1) =mid(a8,68,1) =mid(a8,69,1) =mid(a8,70,1) =mid(a8,71,1) =mid(a8,72,1) =mid(a8,73,1) =mid(a8,74,1) =mid(a8,75,1) =mid(a8,76,1) =mid(a8,77,1) =mid(a8,78,1) =mid(a8,79,1) =mid(a8,80,1)
Literature sources
Conservation genomics is a fast-moving field, particularly for bison and yak. It would serve no purpose here to duplicate what PubMed, Google and alerting services already offer in terms of abstracts, text search and breaking developments.
However locating free full text for older articles can be tedious, yet few people will read the scientific literature at the $35 per document charged by journals. The very most recent articles already provide comprehensive bibliographies and links to the later publications that cite them. The links used in the main article above are collected here, along with 167 PubMed links, articles with free full text online, and pdf files for older items, theses and reports. Run-on titles have been truncated at 120 characters. The list is sorted by topic (eg bis for bison,dis for mitochondrial disease, ple for Pleistocene etc) and reverse chronological date.
Open all 129 abstracts in reverse chronological order. Or open just 23 bison and yak genetics abstracts.
abs bis 2011 Complete mitochondrial DNA sequence analysis of Bison bison and bison-cattle hybrids: function and phylogeny htm bis 2011 Buffalo conservation bibliography abs bis 2010 Cattle ancestry in bison: explanations for higher mtDNA than autosomal ancestry pdf bis 2010 American Bison: Status Survey and Conservation Guidelines 2010 htm bis 2010 The IUCN Red List of Threatened Species v2010.4 abs bis 2009 Wood bison population recovery and forage availability in northwestern Canada htm bis 2009 Conservation: The genome of the American West htm bis 2009 Conservation genetics and North American bison (Bison bison) htm bis 2009 Effectiveness of microsatellite and SNP markers for parentage and identity analysis in species with low genetic diversit htm bis 2009 Depauperate genetic variability detected in the American and European bison using genomic techniques pdf bis 2009 A late Pleistocene steppe bison (Bison priscus) partial carcass from Tsiigehtchic, Northwest Territories, Canada pdf bis 2009 Population viability targets abs bis 2008 The ecological future of the North American bison: conceiving long-term, large-scale conservation of wildlife abs bis 2008 Cloning endangered gray wolves (Canis lupus) from somatic cells collected postmortem pdf bis 2008 Patterns of genetic variation in US federal bison herds htm bis 2007 Detection of mitochondrial DNA from domestic cattle in bison on Santa Catalina Island abs bis 2007 Historic distribution and challenges to bison recovery in the northern Chihuahuan desert pdf bis 2007 History and current status of the Nahanni wood bison population pdf bis 2007 Second Chance for the Plains Bison pdf bis 2007 Development of fecal dna sampling methods to assess genetic population structure of Greater Yellowstone bison pdf bis 2007 A Comprehensive Evaluation of Cattle Introgression into US Federal Bison Herds pdf bis 2007 Ecological future of North American bison pdf bis 2006 Marker Genotypes Association With Body Weight, Height and Relative Body Mass in United States Federal Bison Herds abs bis 2006 Application of bovine microsatellite markers for genetic diversity analysis of European bison (Bison bonasus) htm bis 2006 Comparing the genetic diversity of late Pleistocene Bison with modern Bison bison using ancient DNA techniques pdf bis 2006 Brief Reviw of Status of Plains Bison in North America pdf bis 2005 Conservation genomics: disequilibrium mapping of domestic cattle chromosomal segments in North American bison populations pdf bis 2005 Genetic and demographic consequences of importing animals into a small population: a simulation model of the Texas State Bison Herd pdf bis 2005 The Ecology of bison movements in and beyond Yellowstone pdf bis 2005 Effect of Population Control on Retention of Genetics of NPS bison pdf bis 2005 Spatial Population Structure of Yellowstone Bison abs bis 2004 Genetic characteristics and prognosis of the existence of the European bison free-living population created in the Orlov pdf bis 2004 Conservation genetic analysis of the Texas State Bison Herd pdf bis 2004 Supporting Online Material: Rise and fall of the Beringian Steppe bison pdf bis 2003 Review of Wood Bison in Alaska and adjacent Canada with reference to Yukon Flats pdf bis 2003 Conservation of North American Bison: Status and Recommendations (Boyd MS) pdf bis 2003 Utilization of genetic markers to resolve management issues in historic bison populations pdf bis 2001 Differential introgression of uniparentally inherited markers in bison populations with hybrid ancestries pdf bis 2001 Contribution of prehistoric bison studies to modern bison management pdf bis 2001 Late pre-Historic Game Sink in Northwestern US pdf bis 2000 Destruction of bison: 1750-1920 pdf bis 2000 Validation of 15 microsatellites for parentage testing in North American bison pdf bis 2000 Spatial Aspects of Bison Density Dependence pdf bis 1999 Genetic variation within and relatedness among wood and plains bison populations pdf bis 1999 Keystone Role of Bison in North American Tallgrass Prairie pdf bis 1999 Identification of domestic cattle hybrids in wild cattle and bison species: a general approach using mtDNA markers and t pdf bis 1997 Analysis of a Late Holocene Bison Skull from Fawn Creek, Lemhi County, Idaho pdf bis 1995 Bovine mtDNA Discovered in North American Bison Populations pdf bis 1994 On the Origin of Brucellosis in Bison of Yellowstone National Park: A Review pdf bis 1991 Bison Ecology and Bison Diplomacy: The Southern Plains from 1800 to 1850 pdf bis 1991 Phantom subspecies: the wood bison Bison bison athabascae Rhoads 1897 is not a valid taxon but an ecotype pdf bis 1973 Meagher: the bison of YNP 1973 htm bis 1889 Hornaday WT 1889: The Extermination of the American Bison pdf bis 1885 1885: Present Condition of Yellowstone National Park (Cope) pdf bis ---- Bison conservation genetics and disease pdf bis ---- Greater Yellowstone Bison Distribution and Abundance in the Early Historical Period pdf bis ---- Genetic variation of mitochondrial DNA within domestic yak populations abs bos 2010 Genetic diversity and structure in Bos taurus and Bos indicus populations analyzed by SNP markers abs bos 2010 Genetic diversity and origin of Gayal and cattle in Yunnan revealed by mtDNA control region and SRY gene sequence variat abs bos 2010 A new insight into cattle's maternal origin in six Asian countries abs bos 2010 Diversity and phylogeny of mitochondrial DNA isolated from mithun Bos frontalis located in Bhutan htm bos 2010 Diversity and phylogeny of mitochondrial DNA isolated from mithun Bos frontalis located in Bhutan htm bos 2010 Population dynamic of the extinct European aurochs: genetic evidence of a north-south differentiation pattern and no evi pdf bos 2010 Zebu cattle are an exclusive legacy of the South Asia neolithic abs bos 2009 Karyotype of Malayan Gaur (Bos gaurus hubbacki), Sahiwal-Friesian cattle and Gaur x cattle hybrid backcrosses abs bos 2009 Cytochrome b sequences of ancient cattle and wild ox support phylogenetic complexity in the ancient and modern bovine po abs bos 2009 Species identification, molecular sexing and genotyping using non-invasive approaches in two wild bovids species: Bos ga abs bos 2009 Conservation value of non-native banteng in northern Australia htm bos 2009 Has the kouprey (Bos sauveli Urbain, 1937) been domesticated in Cambodia? htm bos 2009 Maternal and paternal genealogy of Eurasian taurine cattle (Bos taurus) htm bos 2009 The multifaceted origin of taurine cattle reflected by the mitochondrial genome htm bos 2009 On the origin of Indonesian cattle htm bos 2009 An examination of positive selection and changing effective population size in Angus and Holstein cattle populations (Bo htm bos 2009 Molecular evolution of the Bovini tribe (Bovidae, Bovinae): is there evidence of rapid evolution or reduced selective co abs bos 2008 Phylogenetic analysis and comparison between cow and buffalo (including Egyptian buffaloes) mitochondrial displacement-l abs bos 2008 Chromosome evolution in the subtribe Bovina (Mammalia, Bovidae): the karyotype of the Cambodian banteng (Bos javanicus b abs bos 2008 Complete mitochondrial genomes of Bos taurus and Bos indicus provide new insights into intra-species variation, taxonomy abs bos 2008 [Molecular phylogeny of the gayal inferred from the analysis of cytochrome b gene entire sequences] abs bos 2007 Low genetic diversity in the bottlenecked population of endangered non-native banteng in northern Australia abs bos 2007 Phylogenetic relationships and status quo of colonies for gayal based on analysis of cytochrome B gene partial sequences htm bos 2007 Genomic conservation of cattle microsatellite loci in wild gaur (Bos gaurus) and current genetic status of this species htm bos 2007 Resolving a zoological mystery: the kouprey is a real species abs bos 2004 Molecular phylogeny of the tribe Bovini (Bovidae, Bovinae) and the taxonomic status of the Kouprey, Bos sauveli Urbain 1 htm bos 2004 Maternal and paternal lineages in cross-breeding bovine species Has wisent a hybrid origin? abs bos 2003 Phylogenies using mtDNA and SRY provide evidence for male-mediated introgression in Asian domestic cattle htm bos 2003 Hybridization of banteng (Bos javanicus) and zebu (Bos indicus) revealed by mitochondrial DNA, satellite DNA, AFLP and m htm bos 2002 An outbreak of bovine tuberculosis in an intensively managed conservation herd of wild bison in the Northwest Territories abs bos 2001 Evidence from DNA that the mysterious 'linh duong' (Pseudonovibos spiralis) is not a new bovid abs bub 2010 Mitochondrial DNA analyses of Indian water buffalo support a distinct genetic origin of river and swamp buffalo abs bub 2010 Independent maternal origin of Chinese swamp buffalo (Bubalus bubalis) abs bub 2009 Riverine status and genetic structure of Chilika buffalo of eastern India as inferred from cytogenetic and molecular mar abs bub 2008 Mid-Holocene decline in African buffalos inferred from Bayesian coalescent-based analyses of microsatellites and mitocho abs bub 2004 Water buffalo (Bubalus bubalis): complete nucleotide mitochondrial genome sequence abs bub 2004 Analysis of mitochondrial D-loop region casts new light on domestic water buffalo (Bubalus bubalis) phylogeny abs bub 1996 Phylogenetic relationship among all living species of the genus Bubalus based on DNA sequences of the cytochrome b gene pdf cyt 2010 Dindel: Accurate indel calls from short-read data htm cyt 2009 Cytochrome b Phylogeography of Chamois (Rupicapra spp) Population Contractions, Expansions and Hybridizations Governed htm cyt 2008 The adaptive evolution of the mammalian mitochondrial genome abs cyt 2007 Molecular phylogeny of musk deer: a genomic view with mitochondrial 16S rRNA and cytochrome b gene htm cyt 2007 Cytochrome b Pseudogene Originated from a Highly Divergent Mitochondrial Lineage in Genus Rupicapra abs cyt 2005 Global topology analysis of the Escherichia coli inner membrane proteome abs cyt 1999 The tribal radiation of the family Bovidae (Artiodactyla) and the evolution of the mitochondrial cytochrome b gene abs cyt 1999 Cytochrome b phylogeny of the family bovidae: resolution within the alcelaphini, antilopini, neotragini, and tragelaphin htm cyt 1999 Cytochrome b gene haplotypes characterize chromosomal lineages of anoa, the Sulawesi dwarf buffalo (Bovidae: Bubalus sp abs cyt 1995 Mammalian mitochondrial DNA evolution: a comparison of the cytochrome b and cytochrome c oxidase II genes htm dis 2011 Identification of rare DNA variants in mitochondrial disorders with improved array-based sequencing htm dis 2010 An outbreak of bovine tuberculosis in an intensively managed conservation herd of wild bison in the Northwest Territories abs dis 2010 Mitochondrial dysfunction in autism htm dis 2010 Connecting Variability in Global Transcription Rate to Mitochondrial Variability htm dis 2010 Previous Estimates of Mitochondrial DNA Mutation Level Variance Did Not Account for Sampling Error: Comparing the mtDNA abs dis 2009 A neonatal polyvisceral failure linked to a de novo homoplasmic mutation in the mitochondrially encoded cytochrome b gen htm dis 2009 The inheritance of pathogenic mitochondrial DNA mutations htm dis 2009 DNA polymerase gamma and mitochondrial disease: understanding the consequence of POLG mutations abs dis 2009 Biochemical consequences in yeast of the human mitochondrial DNA 8993T>C mutation in the ATPase6 gene found in NARP/MILS abs dis 2009 First birth of an animal from an extinct subspecies (Capra pyrenaica pyrenaica) by cloning abs dis 2008 A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes htm dis 2008 Pathogenic mitochondrial DNA mutations are common in the general population htm dis 2008 Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A-->G mutation in bl htm dis 2008 Mitochondrial Topoisomerase I Sites in the Regulatory D-Loop Region of Mitochondrial DNA pdf dis 2008 The distribution of mitochondrial DNA heteroplasmy due to random genetic drift htm dis 2007 Finding coevolving amino acid residues using row and column weighting of mutual information and multi-dimensional amino htm dis 2007 Depletion of mitochondrial DNA in leucocytes harbouring the 3243A->G mtDNA mutation htm dis 2007 Normal levels of wild-type mitochondrial DNA maintain cytochrome c oxidase activity for two pathogenic mitochondrial DNA htm dis 2006 Holocene coccidioidomycosis: Valley Fever in early Holocene bison (Bison antiquus) abs dis 2006 Canine spongiform leukoencephalomyelopathy is associated with a missense mutation in cytochrome b abs dis 2006 Is selection required for the accumulation of somatic mitochondrial DNA mutations in post-mitotic cells? htm dis 2005 Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice htm dis 2005 A mitochondrial cytochrome b mutation causing severe respiratory chain enzyme deficiency in humans and yeast abs dis 2005 Mitochondrial DNA copy number threshold in mtDNA depletion myopathy abs dis 2005 Stable transformation of CHO Cells and human NARP cybrids confers oligomycin resistance abs dis 2004 Two direct repeats cause most human mtDNA deletions htm dis 2004 Human Disease-related Mutations in Cytochrome b Studied in Yeast abs dis 2003 Association of the mitochondrial DNA 15497G/A polymorphism with obesity in a middle-aged and elderly Japanese population abs dis 2002 Sequence analysis of the complete mitochondrial genome in patients with Leber's hereditary optic neuropathy abs dis 2002 Septo-optic dysplasia associated with a new mitochondrial cytochrome b mutation abs dis 2001 Multisystem disorder associated with a missense mutation in the mitochondrial cytochrome b gene htm dis 2001 Identification of rare DNA variants in mitochondrial disorders with improved array-based sequencing abs dis 2000 A missense mutation in the mitochondrial cytochrome b gene in a revisited case with histiocytoid cardiomyopathy abs dis 1999 A mitochondrial cytochrome b mutation but no mutations of nuclearly encoded subunits htm dis 1999 Exercise intolerance due to mutations in the cytochrome b gene of mitochondrial DNA abs dis 1998 Missense mutation in the mtDNA cytochrome b gene in a patient with myopathy abs dis 1996 A novel gly290asp mitochondrial cytochrome b mutation linked to a complex III deficiency in progressive exercise intoler abs dis 1996 A point mutation in the cytb gene of cardiac mtDNA associated with complex III deficiency in ischemic cardiomyopathy htm dis 1992 Mitochondrial DNA complex I and III mutations associated with Leber's hereditary optic neuropathy abs dis 1991 Cytochrome b mutations in Leber hereditary optic neuropathy abs dis 1982 Mitochondrial DNA copy number in bovine oocytes and somatic cells abs dis 1974 Amino acid difference formula to help explain protein evolution htm phy 2010 Unraveling bovin phylogeny: accomplishments and challenges htm phy 2009 Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics htm phy 2009 Phylogenetic reconstruction and the identification of ancient polymorphism in the Bovini tribe htm phy 2007 28-way vertebrate alignment and conservation track in the UCSC Genome Browser htm phy 2007 Molecular and genomic data identify the closest living relative of primates htm phy 2007 Using genomic data to unravel the root of the placental mammal phylogeny htm ple 2010 Ancient DNA analyses exclude humans as the driving force behind late Pleistocene musk ox (Ovibos moschatus) population d htm ple 2010 Population dynamic of the extinct European aurochs: genetic evidence of a north-south differentiation pattern and no evi abs ple 2010 Genetic history of an archaic hominin group from Denisova Cave in Siberia htm ple 2010 Diversity lost: are all Holarctic large mammal species just relict populations? abs ple 2009 Phylogeography of lions reveals three distinct taxa and a late Pleistocene reduction in genetic diversity htm ple 2009 Accommodating the Effect of Ancient DNA Damage on Inferences of Demographic Histories abs ple 2008 Human influence on distribution and extinctions of the late Pleistocene Eurasian megafauna pdf ple 2007 Stable isotopes, ecological integration and environmental change: wolves record atmospheric carbon isotope trend better htm ple 2004 Rise and fall of the Beringian steppe bison htm ple 1998 Paleoindian large mammal hunters on the plains of North America htm yak 2010 Assessment of cattle genetic introgression into domestic yak populations using mitochondrial and microsatellite DNA mark htm yak 2010 Phylogeographical analyses of domestic and wild yaks based on mitochondrial DNA: new data and reappraisal htm yak 2009 Schaller: Bos grunniens and Bos mutus abs yak 2008 Analysis of genetic diversity and population structure of Chinese yak breeds (Bos grunniens) using microsatellite marker abs yak 2007 Mitochondrial DNA sequence diversity and origin of Chinese domestic yak abs yak 2007 Complete sequence of the yak (Bos grunniens) mitochondrial genome and its evolutionary relationship with other ruminants
Authorship
I researched and wrote this article in its entirety between 1 Dec 2010 and 15 Jan 2011 and take full responsibility for its accuracy. A much shortened citable journal version will appear at Nature Proceedings on 25 Jan 2011 in pdf format. After review by the scientific community, an amended form of article will appear in a conventional peer-reviewed open source journal sometime in 2011.
Although copyrighted, all the information above is in the public domain and can be used by anyone if properly sourced and linked to this url. Please contact me by email if you would like clarifications or additions to the content -- I will make edits as appropriate but may not otherwise respond. Words and concepts not understood are usually explained at the main wikipedia or in undergraduate genetics text.
This is a scientific research article on bison and yak mitochondrial conservation genomics, not a wildlife blog, animal rights forum, or government policy comment site -- thanks in advance for not sending inappropriate email.
My last ten published genomics research papers can be found at PubMed. Watch for 6 additional papers on conservation genomics to appear in 2011. I've previously written over a thousand pages of comparative genomics evaluating variation in hundreds of other genes. I wrote the original user guide to the human genome browser and in 1999 a widely used tutorial from on human genome annotation -- applicable to any mammalian genome -- still available online. I thank Hiram Clawson and the UCSC Genomics Group for software support, Evim Foundation for logistical support and Sperling Foundation for financial support.
Curated reference sequences
The CYTB sequences retrieved from these genomic entries show haplotype notation. The 15 previously existing bison sequences at GenBank (some just fragments) are also provided. Older fragmentary sequences are demonstrably error-prone and will be used here only as support -- never as sole source -- of a polymorphism. Redundancy introduced via non-standard SwissProt (UniProt) entries also has to be manually removed -- the Swiss did no sequencing on their own, simply deriving protein sequences from existing GenBank entries. This leaves 5 older complete sequences for Bison bison and 4 fragments, 2 attributed to Bison bonasus and 1 fossil dna sequence from Bos primigenius to serve as outgroup (rather than an inbred domestic cow).
Here it is necessary to pick a terminology. This must accommodate NCBI taxonomy -- regardless of its correctness -- because otherwise blastp searches cannot be restricted by taxon. Note although bison are definitely sistered with yak to the exclusion of all other extant species, that creates problems because yak has been put in the genus Bos. Many relic wild cattle have no english language common name but rather that of a local language. Terminology table must show synonyms to allow PubMed and google searches -- especially important in a fast-moving field to locate preprints and conference proceedings. The table below does not attempt to implicitly resolve any scientific issue; it simply states preferred terminology at this site along with synonyms in common use.
>CYTB_bisBis.ADF49092 bHap8 plains bison b973 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49170 bHap11 plains bison b1031 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49118 bHap10 plains bison b985 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49248 bHap10 plains bison bFN5 Niobrara A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49131 bHap10 plains bison b1005 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49300 bHap17 plains bison bYNP1586 Yellowstone A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF48936 bHap2 plains bison b790 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF48949 bHap2 plains bison b853 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF48962 bHap2 plains bison b854 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49001 bHap2 plains bison b880 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49027 bHap2 plains bison b925 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49040 bHap2 plains bison b929 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49157 bHap2 plains bison b1029 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49183 bHap2 plains bison b1050 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49196 bHap2 plains bison b1051 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49261 bHap2 plains bison bNBR1 National Bison Range A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49066 bHap2 plains bison b959 Montana A98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49105 bHap9 plains bison b979 Montana V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49209 bHap9 plains bison b1091 Montana V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49014 bHap5 plains bison b897 Montana V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49079 bHap7 plains bison b961 Montana N3S V98 MTSLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF48975 bHap3 plains bison b855 Montana V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49144 bHap3 plains bison b1018 Montana V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49222 bHap12 plains bison b1191 Montana V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49287 bHap16 plains bison bTSBH1005 Texas State Bison Herd V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49235 bHap13 plains bison b1428 Montana V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49274 bHap13 plains bison bTSBH1001 Texas State Bison Herd V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisAth.ADF49313 wHap15 woods bison wEI1 Elk Island V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF48988 bHap4 plains bison b877 Montana V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis.ADF49053 bHap6 plains bison b935 Montana V98 MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisAth.ADF49326 wHap14 woods bison wEI14 Elk Island V98 V123M MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTMMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis_V98_I42T ABV70945 V98 I42T Bison bison MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLTLQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis_98A_V132D AAD51424 Bison bison MTNLRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGMCLILXILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYDLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYIIIGQMASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bisBis_V98 AAW28804 Bison bison NFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSFW >CYTB_bisBis_98A AAW28803 Bison bison NFGSLLGMCLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSFW >CYTB_bisBis_98A_Q322R AAL85955 Bison bison ILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHAGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFI LPFIIMAIAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAILRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMIFRPLSRCLFWTLVADLLTL >CYTB_bosPriW Bos primigenius gi|190360872|gb|ACE76876 MTNFRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMMFRPLSQCLFWALVADLLTLTWIGGQPVEHPYITIGQLASVLYFLLILVLMPTAGTIENKLLKW >CYTB_bosPriM Bos primigenius gi|291463835|gb|ADE05539 alleles F004I A023T I372V MTNIRKSHPLMKIVNNAFIDLPTPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMMFRPLSQCLFWALVADLLTLTWIGGQPVEHPYITIGQLASVLYFLLILVLMPTAGTVENKLLKW >CYTB_bosSau Bos sauveli AAV51239 MTNIRKSHPLMKIVNNAFIDLPAPPNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLITVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIAAIAMVHLLFLHETGSNNPTGVSSDVDKIPFHPYYTIKDTLGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILILIPLLHTSKQRSMMFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYTTIGQLASIMYFLLILVLMPTAGTVENKLLKW >CYTB_bosfroI Bos frontalis ABO07421 (maternal Bos indicus) MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMMFRPLSQCLFWALVADLLTLTWIGGQPVEHPYITIGQLASILYFLLILVLMPTAGTVENKLLKW >CYTB_bosFroW Bos frontalis ABO07423 I39V V215A A232T A302I A327T L357M non-hybrid MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIITAIAMVHLLFLHETGSNNPTGISSDADKIPFHPYYTIKDILGTLLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILILIPLLHTSKQRSMMFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYITIGQLASIMYFLLILVLMPTAGTVENKLLKW >CYTB_bosGau1 Bos gaurus ADB80894 V39I A62V Y95H T108P L105P T190M N206I ADB80893 ADB80892 EU878387 MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIITAIAMVHLLFLHETGSNNPTGISSDADKIPFHPYYTIKDILGTLLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILILIPLLHTSKQRSMMFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYITIGQLASIMYFLLILVLMPTAGTVENKLLKW >CYTB_bosJav Bos javanicus ABS18295 S29A R80W E110K I121F K375N MTNIRKSHPLMKIVNNAFIDLPAPPNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLITVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIITAIAMVHLLFLHETGSNNPTGVSSDADKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILILIPLLHTSKQRSMMFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYITIGQLASIMYFLLILVLMPTAGTVENKLLKW >CYTB_bosJav Bos javanicus ABW82495 MTNIRKSHPLMKIVNNAFIDLPAPPNISSWWNFGSLLGVCLILQILTGLFLAMHYTPDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMMFRPLSQCLFWILMADLLTLTWIGGQPVEHPYITIGQLASIMYFLLILVLMPTAGTVENKLLKW >CYTB_bosJav Bos javanicus ABW82494 MTNIRKSHPLMKIVNNTFIDLPAPPNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLITVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIITAIAMVHLLFLHETGSNNPTGVSSDADKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILILIPLLHTSKQRSMMFRPLSQCLFWTLVADLLTLTWIGGQPVEHPYITIGQLASITYFLLILVLMPTAGTVENKLLKW >CYTB_bosInd Bos indicus ABO07435 T67I sporadic, differs from Bos taurus at I356V and V372I MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILALIPLLHTSKQRSMMFRPLSQCLFWALVADLLTLTWIGGQPVEHPYITIGQLASILYFLLILVLMPTAGTVENKLLKW >CYTB_bisBon Bison bonasus 295065508 YP_003587278 MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIMAIAMVHLLFLHETGSNNPTG ISSDTDKIPFHPYYTIKDILGALLLILTLMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALAFSILILILIPLLHTSKQRSMMFRPLSQCLFWALVADLLTLTWIGGQPVEHPYITIGQLASIMYFLLILVLMPTAGTIENKLLKW >CYTB_bosTau1 Bos taurus AAM12814 208 instances MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICL YMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFI IMAIAMVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAILR SIPNKLGGVLALAFSILILALIPLLHTSKQRSMMFRPLSQCLFWALVADLLTLTWIGGQPVEHPYITIGQLASVLYFLLILVLMPTAGTIENKLLKW >CYTB_bosTau2 Bos taurus AAW78524 72 instances V356I I372V MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICL YMHVGRGLYYGSYTFLETWNIGVILLLTVMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFI IMAIAMVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAILR SIPNKLGGVLALAFSILILALIPLLHTSKQRSMMFRPLSQCLFWALVADLLTLTWIGGQPVEHPYITIGQLASILYFLLILVLMPTAGTVENKLLKW >CYTB_synCafW Syncerus caffer 5777912 AAD51426 AF036275 MTHIRKSHPLMKILNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVAHICrDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAMIHLLFLHETGSNNPTGISSDTDKIPFHPYYTIKDILGALLLILALMLLVLFSPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALILSILILIIMPLLHTSKQRSMMFRPLSQCLFWILVADLLTLTWIGGQPVEHPYIIIGQLASIMYFLLILVLMPTASTIENNLLKW >CYTB_synCafP Syncerus caffer 1813355 BAA11624 H3N T56S I295V MTNIRKSHPLMKILNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYSSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAMIHLLFLHETGSNNPTGISSDTDKIPFHPYYTIKDILGALLLILALMLLVLFSPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVLSILILIIMPLLHTSKQRSMMFRPLSQCLFWILVADLLTLTWIGGQPVEHPYIIIGQLASIMYFLLILVLMPTASTIENNLLKW >CYTB_bubBubW Bubalus bubalis ACF17726 MTNIRKSHPLMKILNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGMYYGSYTFLETWNIGVILLFAVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTSLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAMVHLLFLHETGSNNPTGISSDTDKIPFHPYYTIKDILGALLLILALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVLSILILILMPLLHTSKQRSMMFRPFSQCLFWILVANLLTLTWIGGQPVEHPYIIIGQLASITYFLLILVLMPTASMIENNLLKW >CYTB_bubBubP Bubalus bubalis ABR08397 MTNIRKSHPLMKILNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVAHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGMYYGSYTFLETWNIGVILLFAVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTSLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAMVHLLFLHETGSNNPTGISSDTDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVLSILILILMPLLHTSKQRSMMFRPFSQCLFWILVANLLTLTWIGGQPVEHPYIIIGQLASITYFLLILVLMPTASMVENNLLKW >CYTB_traScr1 Tragelaphus scriptus AF036277 MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTMTAFSSVTHICRDVNHGWIIRYMHANGASMFFICLYMHVGRGMYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTSLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAMVHLLFLHETGSNNPTGIPSDMDKIPFHPYYTIKDILGALLLILILMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVFSILILILMPLLHTSKQRSMMFRPLSQCLFWILAADLLTLTWIGGQPVEHPYIIIGQLASIMYFLIILVLMPATSMIENSFLKW >CYTB_traScr2 Tragelaphus scriptus AAD13501 MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTWDTMTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGMYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTSLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAMVHLLFLHETGSNNPTGIPSDMDKIPFHPYYTIKDILGALLLILILMLLVLFAPDLLGDPDNYAPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVLSILILILMPLLHTSKQRSMMFRPLSQCLFWILAADLLTLTWIGGQPVEHPYIIIGQLASIMYFLIILVLMPAVSMIENNLLKW >CYTB_traScr3 Tragelaphus scriptus non-sporadic alleles S159N A190T M205T A232V L234M I238T V243T F296L I302V lower case MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTMTAFSSVTHICRDVNHGWIIRYMHANGASMFFICLYMHVGRGMYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTnLVEWIWGGFSVDKATLTRFFAFHFILPFIItALAMVHLLFLHETGSNNPTGIPSDtDKIPFHPYYTIKDILGvLLLILtLMLLtLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVlSILILvLMPLLHTSKQRSMMFRPLSQCLFWILAADLLTLTWIGGQPVEHPYIIIGQLASIMYFLIILVLMPATSMIENSFLKW >CYTB_traEur Tragelaphus eurycerus (bongo) AAD51427 MINIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGMYYGSYTFLETWNIGVILLFTVMATAFTGYVLPWGQMSF WGATVITNLLSAIPYIGTSLVEWIWGGFSVDKATLTRFFAFHFILPFIITALAMVHLLFLHETGSNNPTGISSNMDKIPFHPYYTIKDILGALLLILTLMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVLSILILILMPLLHMSKQRSMMFRPLSQCLFWILAADLLTLTWIGGQPVEHPYIIIGQLASIMYFLIILVLMPVTSMIENNFLKW >CYTB_traStr Tragelaphus strepsiceros (greater kudu) AAD51431 MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYVHVGRGMYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLVLALMLLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVLSILILIFLPLLHTSKQRSMMFRPLSQCLFWILVADLLTLTWIGGQPVEHPYMIIGQLASIMYfLLILVLMPVTSMIENNFLKW >CYTB_traImb Tragelaphus imberbis (lesser kudu) AAD13498 MINIRKSHPLMKIVNNAFIDLPTPPNISSWWNFGSLLGICLVLQILTGLFLAMHYTSDTMTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALALVHLLFLHETGSNNPTGISSDTDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALILTILMPILMPLLHASKQRSMMFRPLSQCLFWILVADLLTLTWIGGQPVEHPYIIIGQLASIMYFLLILVLMPMAGSIENNLLKW >CYTB_traOry Tragelaphus oryx (eland) AAD13491 MTNIRKSHPLMKIVNNAFIDLPTPSNISSWWNFGSLLGICLTLQILTGLFLAMHYTSDTTTAFSSVTDICRDVNYGWIIRYMHANGASMFFICLYMHVGRGMYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTSLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAMVHLLFLHETGSNNPTGISSDTDKIPFHPYHTIKDILGALLLILTLMLLVLFAPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVLSILILILMPLLHTSKQRSMMFRPLSQCLFWVLAADLLTLTWIGGQPVEHPYIIIGQLASIMYFLLILVLMPVASMIENNFL >CYTB_traAng Tragelaphus angasii (nyala) AAD42706 MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGVCLILQILTGLFLAMHYTSDTMTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNVGVILLFMVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIITALVMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMVLVLFTPDLLGDPDNYTPANPLNTPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVLSILILILMPLLHMSKQRSMMFRPLSQCLFWLLVADLLTLTWIGGQPVEHPYIIIGQLASIIYFLLILVLMPVISTIENNLLKW >CYTB_traSpi Tragelaphus spekii (sitatunga) CAA10935 MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGMYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTSLVEWIWGGFSVDKATLTRFFAFHFIFPFIIAALAMVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGVLLLILTLMLLVLFAPDLLGDPDNYTPANPLITPPHIKPEWYFLFAYAI LRSIPNKLGGVLALVLSILILILMPLLHVSKQRSMMFRPLSQCLFWILAADLLTLTWIGGQPVEHPYIIIGQLASIMYFLIILVLMPATSMIENNFLKW >CYTB_traDer Taurotragus derbianus (giant eland) AAD13496 MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGMYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTSLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAIVHLLFLHETGSNNPTGISSDMDKIPFHPYYTIKDILGALLLILALMLLVLFAPDLLGDPDNYTPANPLSTPPHIKPEWYFLFAYAI LRLIPNKLGGVLALVLSILVLMLMPLLHTSKQRSMMFRPLSQCFFWILAADLLTLTWIGGQLVEHPYIIIGQLASIMYFLLILVLMPVASMIENNLLKW >CYTB_bseTra Boselaphus tragocamelus CAA10934 MTNIRKSHPLMKIVNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTMTAFASVTHICRDVNYGWIIRYMHANGASMFFICLYMHVGRGLYYGSYTFLETWNIGVILLFTVMATAFMGYVLPWGQMSF WGATVITNLLSAIPYIGTNLVEWIWGGFSVDKATLTRFFAFHFILPFIIAALAMIHLLFLHETGSNNPTGISSDADKIPFHPYYTIKDILGALLLILALMMLVLFAPDLLGDPDNYTPANPLSTPPHIKPEWYFLFAYAI LRSIPNKLGGVMALVLSILILILMPLLHTSKQRSMMFRPLSQCMFWILVANLLTLTWIGGQPVEHPYIIIGQLASIMYFLLILVLMPTASMIENNLLKW