Bison: nuclear genomics: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 42: | Line 42: | ||
[[Image:IncestSnp.jpg|left]] | [[Image:IncestSnp.jpg|left]] | ||
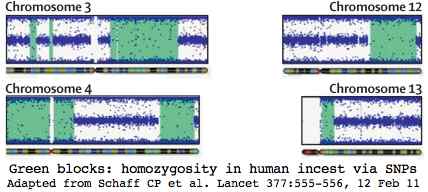
The green blocks show 668 million base pairs of DNA homozygosity out of the 716 Mbp expected for parent-child incest (coefficient of inbreeding 1/4, human genome size 3.000 Mbp). This represents a quarter of the genes, so approximately 5,000 of which 62 would be expected to have carried deleterious mutations of which 31 on average would now be homozygous deleterious in the child | The green blocks show 668 million base pairs of DNA homozygosity out of the 716 Mbp expected for parent-child incest (coefficient of inbreeding 1/4, human genome size 3.000 Mbp). This represents a quarter of the genes, so approximately 5,000 of which 62 would be expected to have carried deleterious mutations of which 31 on average would now be homozygous deleterious in the child. | ||
[[Image:Cousins.gif|left]] | |||
Incest is a [http://en.wikipedia.org/wiki/Incest_laws crime] in nearly all human societies but management-driven incest in bison is not. The SNP chip here had 620,901 markers, representing 12x the resolution available for the comparable cattle chip applied to bison. Thus the bison chip would give clear results but not the sharp resolution because the median marker spacing would slip to 32.4 kbp and the average spacing to 56.4 kbp. For matings between bison related at the second degree (uncle-niece, double first cousins), the inbreeding coefficient is 1/8 and expected level of homozygosity 358 Mb. Here the calf would carry roughly 15 deleterious mutations. | |||
Bison are routinely corralled and tested for previous exposure to brucellosis. The blood samples taken also serve for DNA sampling, where a tiny volume placed on special filter paper would be stable for years at room temperature. These [http://www.biomedcentral.com/1756-0500/2/107 FTA cards] provide DNA suitable for readout on the widely used bovine SNP beadchip Illumina. Thus it is fast and cheap to determine the extent of inbreeding at Yellowstone National Park even though cattle introgression (the main use of the chip in bison) is not the issue there. Inbreeding and long-ago introgression are the same thing, just opposite extremes. | |||
Actual opportunistic measurement of corralled, radio-collared, or naturally expired animals is vastly preferable to academic exercises in theoretic population modelling. There is no real interest in neutral alleles measured by microsatellites or junk DNA SNPs but rather in consequential frequencies of deleterious alleles in specific genes that are the legacy of the initial bottleneck. The genes of interest are yet to be determined. | |||
Real bison herds are impossibly complex. The femals are strongly matrilocal. Distinct herds may persist for decades because of physical barriers between valleys but bulls may wander between them. The herd sizes and composition fluctuate dramatically from year to year depending primarily on the severity of culls, its targeting to extended family groups, snowfall in winter, and susceptibility to predation and disease. | |||
One wonders what conservation genomics management purpose the obscure theoretical constructs of population ecology can serve, given real bison herds have constantly shifting and essentially unmeasurable hereditary allele parameters in 20,000 genes, with two weakly related bison differing at more than 10,500 amino acid sites. | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Revision as of 14:28, 16 February 2011
Bison conservation genomics: introduction
The main nuclear genome of bison, like the mitochondrial genome, will have significant conservation management issues because the consequences of nineteenth and twentieth century bottlenecks (and consequent inbreeding) are still with us today.
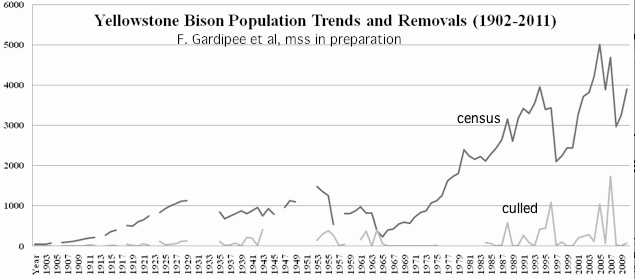
Most conservation herds are derived from a tiny founding populations and maintained for many decades at far too low a population level, with surplus animals removed episodically without the slightest consideration of population genetic impacts. Other management practices such elimination of predators, winter feeding, gender imbalance, culling of unruly bulls, and trophy hunts also interfere with natural selection (survival of the fittest).
The founding individuals of a given herd -- even previously wild animals experiencing millenia of natural selection -- still have a substantial genetic load . Autosomal recessives form an important component of that load and are the primary focus here. These are gene mutations found in one of the two copies of non-sex chromosomes that are more or less masked by compensation by the properly functioning copy.
When the founding population is small, the gene frequency of an autosomal recessive mutation is necessarily high. As inbreeding is unavoidable, offspring can inherit two bad copies of the gene. In this homozygous state, no compensation can occur and the disease associated with the mutation is fully manifested. Note populations often harbor mutations at different sites in the same gene. Here the affected offspring can be a compound heterozygote -- two bad copies but at different sites in the same protein.
Looking just at same-site autosomal recessives, the two variables are the frequency q of the bad allele in the population and the coefficient of inbreeding f. The latter simply tallies the percentage of identical alleles by inherited descent (autozygosity). There is an assumption here that would not be valid for the YNP bison nineteenth century bottleneck, namely that the surviving parental animals were not already inbred.
This can be translated into millions of DNA base pairs lacking heterozygosity assuming a bison nuclear genome size of three billion. Then for f at 1/4, there 716,000,000 base pairs of inbreeding derived homozygosity, enough for 5,000 protein coding genes. This DNA will be somewhat broken up into blocks by recombination. For f at 1/8, these numbers are 358,000,000 bp and 2,500 genes. Other coefficients of inbreeding are quickly computed:
- Father/daughter, mother/son or brother/sister → 25%
- Grandfather/granddaughter or grandmother/grandson → 12.5%
- Half-brother/half-sister → 12.5%
- Uncle/niece or aunt/nephew → 12.5%
- Great-grandfather/great-granddaughter or great-grandmother/great-grandson → 6.25%
- Half-uncle/niece or half-aunt/nephew → 6.25%
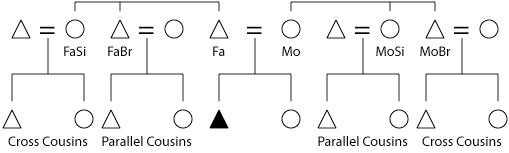
- First cousins → 6.25%
- First cousins once removed or half-first cousins → 3.1%
- Second cousins or first cousins twice removed → 1.6%
- Second cousins once removed or half-second cousins → 0.78%
The frequency of an autosomal recessive disorder in the offspring of a consanguineous mating is then qf + qq (1-f). Inserting various coefficients of inbreeding and realistic values of deleterious alleles, it quickly emerges that almost all autosomal recessive disease in bison arises from inbreeding. Very rarely does it arise in the offspring of remotely related animals.
Example: suppose a bison bull is dominant for a few years. If it breeds with a calf it previously sired, f is 1/4. The odds that recessive disease did not result from inbreeding only exceed 50-50 when q exceeds 1/3. However q = 0.1 is the largest q known in human disease(hemochromatosis). Cystic fibrosis is another extreme case but there q is only 0.02. For a bison disease allele at that frequency, with a disease observed in the offspring, the odds are overwhelming (94%) that it came from inbreeding, not mating of unrelated bison representative of the whole populations. The odds are still high for the grandparent and first cousin situations (88% and 77%) and are higher still for lower q that are more typical. In summary, autosomal recessive disease in bison can be brought back to natural levels simply by avoidance of inbreeding.
Extensive whole genome sequencing in humans has established that each individual human carries 275 loss-of-function variants and 75 variants previously implicated in inherited disease (both classes typically heterozygous and differing from person to person), additionally varying from the reference human proteome of 9,000,000 amino acids at 10,500 other sites (0.12%). The deleterious alleles include 200 in-frame indels, 90 premature stop codons, 45 splice-site-disrupting variants and 235 deletions shifting reading frame.
In bison the overall genetic load will surely be worse in view of extreme bottlenecks, small herd size history and unavoidable inbreeding. Offspring will deleterious nuclear genes in the homozygous state will be more abundant than in humans who are inbred too but nearly to the same extent.
Measuring incest in bison
Little bison nuclear genome data is currently available but that situation is changing rapidly with ongoing whole genome sequencing projects not only for bison but also of closely related species such as yak, water buffalo, domestic cow and fossil steppe and plains bison that can help establish a baseline of normality for current conservation herd bison.
Humans however are already intensively studied. Here incest studies in human have transferable implications to bison herds with limited a number of bulls or a single bull maintaining breeding dominance across generations. The graphic at left shows how a human SNP chip detected incest in a 3-year-old boy with multiple medical issues without access to parental dna.
The green blocks show 668 million base pairs of DNA homozygosity out of the 716 Mbp expected for parent-child incest (coefficient of inbreeding 1/4, human genome size 3.000 Mbp). This represents a quarter of the genes, so approximately 5,000 of which 62 would be expected to have carried deleterious mutations of which 31 on average would now be homozygous deleterious in the child.
Incest is a crime in nearly all human societies but management-driven incest in bison is not. The SNP chip here had 620,901 markers, representing 12x the resolution available for the comparable cattle chip applied to bison. Thus the bison chip would give clear results but not the sharp resolution because the median marker spacing would slip to 32.4 kbp and the average spacing to 56.4 kbp. For matings between bison related at the second degree (uncle-niece, double first cousins), the inbreeding coefficient is 1/8 and expected level of homozygosity 358 Mb. Here the calf would carry roughly 15 deleterious mutations.
Bison are routinely corralled and tested for previous exposure to brucellosis. The blood samples taken also serve for DNA sampling, where a tiny volume placed on special filter paper would be stable for years at room temperature. These FTA cards provide DNA suitable for readout on the widely used bovine SNP beadchip Illumina. Thus it is fast and cheap to determine the extent of inbreeding at Yellowstone National Park even though cattle introgression (the main use of the chip in bison) is not the issue there. Inbreeding and long-ago introgression are the same thing, just opposite extremes.
Actual opportunistic measurement of corralled, radio-collared, or naturally expired animals is vastly preferable to academic exercises in theoretic population modelling. There is no real interest in neutral alleles measured by microsatellites or junk DNA SNPs but rather in consequential frequencies of deleterious alleles in specific genes that are the legacy of the initial bottleneck. The genes of interest are yet to be determined.
Real bison herds are impossibly complex. The femals are strongly matrilocal. Distinct herds may persist for decades because of physical barriers between valleys but bulls may wander between them. The herd sizes and composition fluctuate dramatically from year to year depending primarily on the severity of culls, its targeting to extended family groups, snowfall in winter, and susceptibility to predation and disease.
One wonders what conservation genomics management purpose the obscure theoretical constructs of population ecology can serve, given real bison herds have constantly shifting and essentially unmeasurable hereditary allele parameters in 20,000 genes, with two weakly related bison differing at more than 10,500 amino acid sites.