Bison: nuclear genomics
Bison conservation genomics: introduction
The main nuclear genome of bison, like the mitochondrial genome, will have significant conservation management issues because the consequences of nineteenth and twentieth century bottlenecks (and consequent inbreeding) are still with us today.
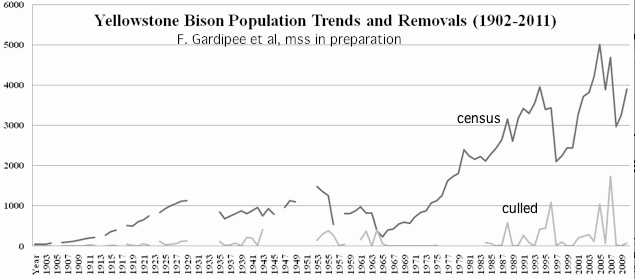
Most conservation herds are derived from a tiny founding populations and maintained for many decades at far too low a population level, with surplus animals removed episodically without the slightest consideration of population genetic impacts. Other management practices such elimination of predators, winter feeding, gender imbalance, culling of unruly bulls, and trophy hunts also interfere with natural selection (survival of the fittest).
The founding individuals of a given herd -- even previously wild animals experiencing millenia of natural selection -- still have a substantial genetic load . Autosomal recessives form an important component of that load and are the primary focus here. These are gene mutations found in one of the two copies of non-sex chromosomes that are more or less masked by compensation by the properly functioning copy.
When the founding population is small, the gene frequency of an autosomal recessive mutation is necessarily high. As inbreeding is unavoidable, offspring can inherit two bad copies of the gene. In this homozygous state, no compensation can occur and the disease associated with the mutation is fully manifested. Note populations often harbor mutations at different sites in the same gene. Here the affected offspring can be a compound heterozygote -- two bad copies but at different sites in the same protein.
Extensive whole genome sequencing in humans has established that each individual human carries 275 loss-of-function variants and 75 variants previously implicated in inherited disease (both classes typically heterozygous and differing from person to person), additionally varying from the reference human proteome of 9,000,000 amino acids at 10,500 other sites (0.12%). The deleterious alleles include 200 in-frame indels, 90 premature stop codons, 45 splice-site-disrupting variants and 235 deletions shifting reading frame.
In bison the overall genetic load will surely be worse in view of extreme bottlenecks, small herd size history and unavoidable inbreeding. Offspring will deleterious nuclear genes in the homozygous state will be more abundant than in humans who are inbred too but nearly to the same extent.