C7orf10: Difference between revisions
m (fix regardless) |
(Updated genome-test.cse links to .gi, one link broken) |
||
| Line 5: | Line 5: | ||
By way of background, mutations in the preceding step (genes undetermined) give rise to [http://www.wjgnet.com/1007-9327/6/766.asp alpha-ketoadipic aciduria (KA)] and mutations in the following step (GCDH: Glutaryl-CoA dehydrogenase 19p13.2, mitochondrial homotetramer) cause glutaric aciduria type 1 (GA1). Glutaric aciduria type 2 (GA2) affects this and other acyl-CoA dehydrogenases via mutations in the accessory flavoprotein cofactor genes (ETFA, ETFB, ETFDH). | By way of background, mutations in the preceding step (genes undetermined) give rise to [http://www.wjgnet.com/1007-9327/6/766.asp alpha-ketoadipic aciduria (KA)] and mutations in the following step (GCDH: Glutaryl-CoA dehydrogenase 19p13.2, mitochondrial homotetramer) cause glutaric aciduria type 1 (GA1). Glutaric aciduria type 2 (GA2) affects this and other acyl-CoA dehydrogenases via mutations in the accessory flavoprotein cofactor genes (ETFA, ETFB, ETFDH). | ||
It turns out that quite a bit of comparative genomics is [http://genome-test. | It turns out that quite a bit of comparative genomics is [http://genome-test.gi.ucsc.edu/cgi-bin/hgGene?hgsid=157956 already available] for C7orf10. Most intrigingly, it has a weak [http://genome-test.gi.ucsc.edu/cgi-bin/hgGene?hgsid=1579568 full length paralog AMACR] on chr 5, evidently an old gene duplication as it lacks any flanking synteny. That gene has been [http://www.jbc.org/cgi/content/full/280/13/12611 previously characterized] as an alpha-methylacyl-CoA racemase, an enzyme that converts alpha-branched acyl CoA substrates to their S-stereoisomer, the form can be degraded via peroxisomal beta-oxidation. However no comparable substituent is immediately at hand for C7orf10 as its presumed substrate lacks stereoisomeric carbon centers. | ||
Both C7orf10 and AMACR classify as type 3 CoA-transferases, that being their only detectable Pfam domain (residues 45-381 and 113-302 respectively). These are the only two such proteins in the human proteome. This class of enzyme, which bears no sequence or structural resemblance to type 1 or type 2 CoA-transferases, catalyses the reversible transfer of coenzyme A from a CoA-thioester to a free carboxylic acid in a highly substrate- and stereo-specific manner. Type 3 CoA-transferases do not utilize free CoA as a substrate. | Both C7orf10 and AMACR classify as type 3 CoA-transferases, that being their only detectable Pfam domain (residues 45-381 and 113-302 respectively). These are the only two such proteins in the human proteome. This class of enzyme, which bears no sequence or structural resemblance to type 1 or type 2 CoA-transferases, catalyses the reversible transfer of coenzyme A from a CoA-thioester to a free carboxylic acid in a highly substrate- and stereo-specific manner. Type 3 CoA-transferases do not utilize free CoA as a substrate. | ||
Latest revision as of 20:49, 24 September 2018
According to Swiss Prot, as of November 2008, over half of the human genome encoded proteins (that is, 10,000) have never been seen by mass spectrometry nor been the subject of a focused publication. Accurate models of the encoding genes may or may not lie nested in the debris of pipeline algorithms and transcript collections; homologies, domains, tertiary structure, and comparative genomics may already be preposed. However no one has assessed this machine output so the biological functions of half of human genes remain undetermined. Consequently when a new human disease is mapped via SNP associations to one of these gene products, medically-oriented researchers hit an interpretive brick wall.
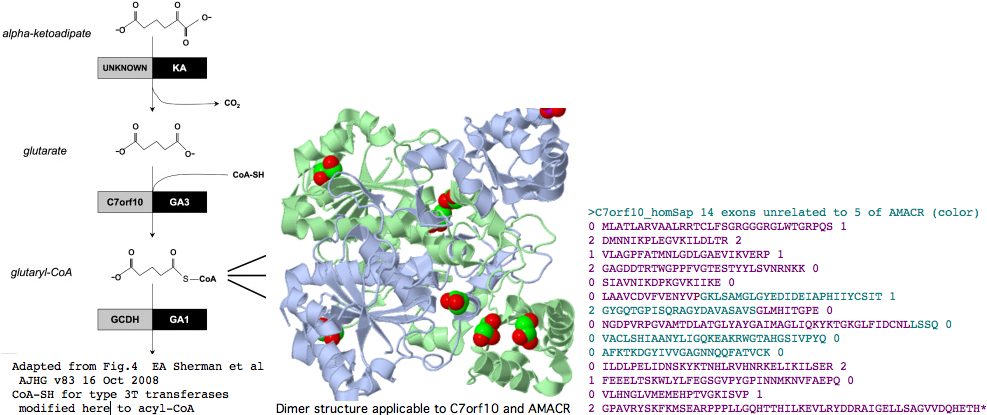
C7orf10 is an example of such a gene. As the name suggests, it lies on chromosome 7 and originated as a pipeline prediction of open reading frame. The gene recently surfaced in Old Order Amish as the cause of type 3 glutaric aciduria (GA3). However the authors write "there is no existing functional data on the C7orf10 protein" noting intracellular location (mitochondria not peroxisome) is key to understanding accruing metabolite toxicity. The authors note further that meagre existing annotation suggests a role linking CoA to the dicarboxylic acid glutarate in the mitochondrial lysine--tryptophan degradation pathway. (It should have noted that hydroxylysine -- from collagen -- is also catabolized by that same shared pathway.)
By way of background, mutations in the preceding step (genes undetermined) give rise to alpha-ketoadipic aciduria (KA) and mutations in the following step (GCDH: Glutaryl-CoA dehydrogenase 19p13.2, mitochondrial homotetramer) cause glutaric aciduria type 1 (GA1). Glutaric aciduria type 2 (GA2) affects this and other acyl-CoA dehydrogenases via mutations in the accessory flavoprotein cofactor genes (ETFA, ETFB, ETFDH).
It turns out that quite a bit of comparative genomics is already available for C7orf10. Most intrigingly, it has a weak full length paralog AMACR on chr 5, evidently an old gene duplication as it lacks any flanking synteny. That gene has been previously characterized as an alpha-methylacyl-CoA racemase, an enzyme that converts alpha-branched acyl CoA substrates to their S-stereoisomer, the form can be degraded via peroxisomal beta-oxidation. However no comparable substituent is immediately at hand for C7orf10 as its presumed substrate lacks stereoisomeric carbon centers.
Both C7orf10 and AMACR classify as type 3 CoA-transferases, that being their only detectable Pfam domain (residues 45-381 and 113-302 respectively). These are the only two such proteins in the human proteome. This class of enzyme, which bears no sequence or structural resemblance to type 1 or type 2 CoA-transferases, catalyses the reversible transfer of coenzyme A from a CoA-thioester to a free carboxylic acid in a highly substrate- and stereo-specific manner. Type 3 CoA-transferases do not utilize free CoA as a substrate.
It's been argued persuasively on structural and mechanistic grounds that two subcategories of type III CoA-transferases should be distinguished, which we call here 3T and 3R for transferase and racemase respectively. AMACR is type 3R and it will turn out that C7orf10 is type 3T.
The initial crystal structure of the E. coli enzyme CaiB, which as type 3T transfers the CoA of butyrobetaine-CoA to carnitine forming carnityl-CoA and gammabutyrobetaine, shows an unprecedented homodimer of interlocked rings each consisting of a large domain with a acyl-CoA binding Rossmann fold and a small secondary domain. While very diverged in primary sequence from the two human paralogs, even this structure will suffice to model their 3D structure quite accurately. Structures that came later provide even better PDB matches; 1X74 and 1X74, can be viewed interactively. The former is a type 3T CoA-transferase with 45% identity to the internal core C7orf10. The latter, type 3R from Mycobacterium tuberculosis, bears an astonishing 46% sequence identity with human AMACR and provides an overwhelmingly strong template for the latter's structure.
C7orf10, like AMACR and other ancient metabolic pathway enzymes (eg homogentisate 1,2-dioxygenase of tyrosine-phenylalanine catabolism) is a slowly evolving protein with 58% conservation in pre-Bilateran Cnidaria, a 53% identity homolog in fungi, and a 45% match in bacteria. This -- and lack of additional gene copies -- suggests retained consistency of function over many billions of years of branch length. That function seems never to have been specificaly studied by mutational effects in common model organisms. It would not be difficult to knock out orthologs in various model species, observe phenotype, and check for complementation by the human gene.
The explanation for the percent identity plateau is likely that about half the residues are important for fixed structural or catalytic roles. They are either invariant or admit cycling only within a small reduced alphabet of 2-3 residues. These on average contribute a match about half the time regardless of phylogenetic depth. Other parts of the protein are unconstrained relatively speaking and contribute very little to match percentage outside a narrow clade. The net effect is the possiblities for divergence become saturated, here at about 55% whether human is compared to a basal animal, a fungus, or a bacterium. Another way of putting this: this enzyme stopped evolving adaptively two billion years ago as by then its structure and function were already perfected.
Pairs of paralogs such as C7orf10 and AMACR are sometimes cited as supporting evidence for 1R whole genome duplication in chordates, with the divergence also claimed indicative of that. This is complete rubbish. This pair of genes duplicated billions of years earlier and, like many enzymes of intermediary metabolism, did not give rise to additional paralogs. Indeed, they provide evidence *against* 2R which predicts 8 members in vertebrates instead of the observed 2.
Since types 3T and 3R arose already in bacteria prior to emergence of eukaryotes and gene intronation, this predicts that the human paralogs C7orf10 and AMACR reflect a very ancient gene duplication within bacteria and consequently the number, location, and phase of their introns (being from a much later era) should be entirely different.
Indeed, C7orf10 has 14 coding exons (after discarding alternate splice artefacts lacking phylogenetic support) whereas AMACR has but 5. The positions and phases of exon breaks in C7orf10 are shown below. The sequence has been colored according to exons of AMACR using blastp alignment to determine placement. It can be seen that the intronation patterns -- normally conserved to immense phylogenetic depth -- are completely uncorrelated in these two paralogs, as expected from a very early gene duplication. This again weakens the case for C7orf10 as a racemase based on paralogy to AMACR.
AMACR carries both N- and C-terminal targeting sequences for input into mitochondria and peroxisomes, respectively. C7orf10 on the other hand has only mitochondrial targeting (according to 3 separate tools at SwissProt). Consequently, acyl-CoA compounds abundant in the mitochondia are appropriate candidates for the CoA donor. These include Krebs cycle compounds such as acetyl-CoA and succinyl-CoA. Note that shortly after crotonyl-CoA, the catabolism pathway here itself enters the Krebs cycle.
In summary, despite the initially intriguing paralogous relationship to AMACR, C7orf10 is not a type 3R racemase but belongs instead with type 3T transferases. This could be strengthened by carefully compiling classifying sets of phylogenetically diverse validated 3T and 3R sequences and verifying that C7orf10 clusters with the former and lacks the defining specializations of the latter, as well as by considering non-conservation of specialized racemization protic residues.
C7orf10 is a simple nuclear-encoded mitochondrial enzyme that exchanges an already-charged CoA (of succinyl-CoA or similar) with glutarate to form glutaryl-CoA and succinate. No ATP, NAD, or protein partners are needed. It may or may not have other substrates and functions.
This leaves 9,999 human protein left undescribed. A fair number of these could be, like this one, obscure enzymes of intermediary metabolism. Notice that without the observed human phenotype, C7orf10 could have been annotated but only up to a point.
In 1967, Linus Pauling proposed widespread screening of anomalous compounds accumulating in urine of seemingly healthy individuals as part of routine preventative health, much as (less affordable) whole genome sequencing for everyone is proposed today. While that screening would be a highly inefficient way of annotating that portion of the human genome compared to screening focused on inbred populations, the combination of metabolite accrual with modern genomic mapping could be quite effective identifying new gene functions.
>C7orf10_homSap (uc003thn) span 725,456 bp 14 exons unrelated to those of AMACR (color) 0 MLATLARVAALRRTCLFSGRGGGRGLWTGRPQS 1 2 DMNNIKPLEGVKILDLTR 2 1 VLAGPFATMNLGDLGAEVIKVERP 1 2 GAGDDTRTWGPPFVGTESTYYLSVNRNKK 0 0 SIAVNIKDPKGVKIIKE 0 0 LAAVCDVFVENYVPGKLSAMGLGYEDIDEIAPHIIYCSIT 1 2 GYGQTGPISQRAGYDAVASAVSGLMHITGPE 0 0 NGDPVRPGVAMTDLATGLYAYGAIMAGLIQKYKTGKGLFIDCNLLSSQ 0 0 VACLSHIAANYLIGQKEAKRWGTAHGSIVPYQ 0 0 AFKTKDGYIVVGAGNNQQFATVCK 0 0 ILDLPELIDNSKYKTNHLRVHNRKELIKILSER 2 1 FEEELTSKWLYLFEGSGVPYGPINNMKNVFAEPQ 0 0 VLHNGLVMEMEHPTVGKISVP 1 2 GPAVRYSKFKMSEARPPPLLGQHTTHILKEVLRYDDRAIGELLSAGVVDQHETH* 0 >AMACR_homSap (uc003jig) 5 exons 0 MALQGISVVELSGLAPGPFCAMVLADFGARVVRVDRPGSRYDVSRLGRGKRSLVLDLKQPRGAAVLRRLCKRSDVLLEPFRR 1 2 GVMEKLQLGPEILQRENPRLIYARLSGFGQSGSFCRLAGHDINYLALS 1 2 GVLSKIGRSGENPYAPLNLLADFAGGGLMCALGIIMALFDRTRTGKGQVIDANM 0 0 VEGTAYLSSFLWKTQKLSLWEAPRGQNMLDGGAPFYTTYRTADGEFMAVGAIEPQFYELLIK 1 2 GLGLKSDELPNQMSMDDWPEMKKKFADVFAEKTKAEWCQIFDGTDACVTPVLTFEEVVHHDHNKERGSFITSEEQDVSPRPA PLLLNTPAIPSFKRDPFIGEHTEEILEEFGFSREEIYQLNSDKIIESNKVKASL* 0