Gene Set Summary Statistics: Difference between revisions
From genomewiki
Jump to navigationJump to search
(Updated genome-test or hgwdev links from .cse/.soe to .gi, most links validated) |
|||
| (32 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
* hg17 - knownGenes version 2 | * hg17 - knownGenes version 2 | ||
* hg18 - knownGenes version 3 | * hg18 - knownGenes version 3 | ||
* hg19 - knownGenes version 7 | |||
* mm8 - knownGenes version 2 | * mm8 - knownGenes version 2 | ||
* mm9 - knownGenes version 3 | * mm9 - knownGenes version 3 | ||
* mm10 - knownGenes version 6 | |||
The min, max and mean measurements are ''per gene'' | The min, max and mean measurements are ''per gene'' | ||
| Line 12: | Line 14: | ||
<TR><TH>hg17</TH><TD ALIGN=RIGHT>39368</TD><TD ALIGN=RIGHT>405720</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>149</TD><TD ALIGN=RIGHT>10</TD></TR> | <TR><TH>hg17</TH><TD ALIGN=RIGHT>39368</TD><TD ALIGN=RIGHT>405720</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>149</TD><TD ALIGN=RIGHT>10</TD></TR> | ||
<TR><TH>hg18</TH><TD ALIGN=RIGHT>56722</TD><TD ALIGN=RIGHT>519308</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>2899</TD><TD ALIGN=RIGHT>9</TD></TR> | <TR><TH>hg18</TH><TD ALIGN=RIGHT>56722</TD><TD ALIGN=RIGHT>519308</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>2899</TD><TD ALIGN=RIGHT>9</TD></TR> | ||
<TR><TH>hg19</TH><TD ALIGN=RIGHT>82960</TD><TD ALIGN=RIGHT>742493</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>5065</TD><TD ALIGN=RIGHT>9</TD></TR> | |||
<TR><TH>mm8</TH><TD ALIGN=RIGHT>31863</TD><TD ALIGN=RIGHT>314628</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>313</TD><TD ALIGN=RIGHT>10</TD></TR> | <TR><TH>mm8</TH><TD ALIGN=RIGHT>31863</TD><TD ALIGN=RIGHT>314628</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>313</TD><TD ALIGN=RIGHT>10</TD></TR> | ||
<TR><TH>mm9</TH><TD ALIGN=RIGHT>49220</TD><TD ALIGN=RIGHT>417114</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>610</TD><TD ALIGN=RIGHT>8</TD></TR> | <TR><TH>mm9</TH><TD ALIGN=RIGHT>49220</TD><TD ALIGN=RIGHT>417114</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>610</TD><TD ALIGN=RIGHT>8</TD></TR> | ||
<TR><TH>mm10</TH><TD ALIGN=RIGHT>59121</TD><TD ALIGN=RIGHT>486425</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>813</TD><TD ALIGN=RIGHT>8</TD></TR> | |||
</TABLE> | </TABLE> | ||
| Line 21: | Line 25: | ||
<TR><TH>hg17</TH><TD ALIGN=RIGHT>106839720</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>18172</TD><TD ALIGN=RIGHT>263</TD></TR> | <TR><TH>hg17</TH><TD ALIGN=RIGHT>106839720</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>18172</TD><TD ALIGN=RIGHT>263</TD></TR> | ||
<TR><TH>hg18</TH><TD ALIGN=RIGHT>146371091</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>36861</TD><TD ALIGN=RIGHT>282</TD></TR> | <TR><TH>hg18</TH><TD ALIGN=RIGHT>146371091</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>36861</TD><TD ALIGN=RIGHT>282</TD></TR> | ||
<TR><TH>hg19</TH><TD ALIGN=RIGHT>221924089</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>205012</TD><TD ALIGN=RIGHT>299</TD></TR> | |||
<TR><TH>mm8</TH><TD ALIGN=RIGHT>83159087</TD><TD ALIGN=RIGHT>4</TD><TD ALIGN=RIGHT>17497</TD><TD ALIGN=RIGHT>264</TD></TR> | <TR><TH>mm8</TH><TD ALIGN=RIGHT>83159087</TD><TD ALIGN=RIGHT>4</TD><TD ALIGN=RIGHT>17497</TD><TD ALIGN=RIGHT>264</TD></TR> | ||
<TR><TH>mm9</TH><TD ALIGN=RIGHT>117671086</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>29698</TD><TD ALIGN=RIGHT>282</TD></TR> | <TR><TH>mm9</TH><TD ALIGN=RIGHT>117671086</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>29698</TD><TD ALIGN=RIGHT>282</TD></TR> | ||
<TR><TH>mm10</TH><TD ALIGN=RIGHT>143087341</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>83437</TD><TD ALIGN=RIGHT>294</TD></TR> | |||
</TABLE> | </TABLE> | ||
| Line 30: | Line 36: | ||
<TR><TH>hg17</TH><TD ALIGN=RIGHT>2223224397</TD><TD ALIGN=RIGHT>6</TD><TD ALIGN=RIGHT>1096450</TD><TD ALIGN=RIGHT>6069</TD></TR> | <TR><TH>hg17</TH><TD ALIGN=RIGHT>2223224397</TD><TD ALIGN=RIGHT>6</TD><TD ALIGN=RIGHT>1096450</TD><TD ALIGN=RIGHT>6069</TD></TR> | ||
<TR><TH>hg18</TH><TD ALIGN=RIGHT>2784923600</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>1047320</TD><TD ALIGN=RIGHT>6023</TD></TR> | <TR><TH>hg18</TH><TD ALIGN=RIGHT>2784923600</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>1047320</TD><TD ALIGN=RIGHT>6023</TD></TR> | ||
<TR><TH>hg19</TH><TD ALIGN=RIGHT>4127555916</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>1160410</TD><TD ALIGN=RIGHT>6260</TD></TR> | |||
<TR><TH>mm8</TH><TD ALIGN=RIGHT>1476081990</TD><TD ALIGN=RIGHT>9</TD><TD ALIGN=RIGHT>1347550</TD><TD ALIGN=RIGHT>5220</TD></TR> | <TR><TH>mm8</TH><TD ALIGN=RIGHT>1476081990</TD><TD ALIGN=RIGHT>9</TD><TD ALIGN=RIGHT>1347550</TD><TD ALIGN=RIGHT>5220</TD></TR> | ||
<TR><TH>mm9</TH><TD ALIGN=RIGHT>2055504784</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>1253430</TD><TD ALIGN=RIGHT>5589</TD></TR> | <TR><TH>mm9</TH><TD ALIGN=RIGHT>2055504784</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>1253430</TD><TD ALIGN=RIGHT>5589</TD></TR> | ||
<TR><TH>mm10</TH><TD ALIGN=RIGHT>2420504661</TD><TD ALIGN=RIGHT>1</TD><TD ALIGN=RIGHT>3003780</TD><TD ALIGN=RIGHT>5665</TD></TR> | |||
</TABLE> | </TABLE> | ||
| Line 38: | Line 46: | ||
<TR><TH>db</TH><TH COLSPAN=5>gene name (exon count)</TH></TR> | <TR><TH>db</TH><TH COLSPAN=5>gene name (exon count)</TH></TR> | ||
<TR><TH>hg17</TH> | <TR><TH>hg17</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr2:152167363-152416454 NM_004543] (149)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6:152534937-153050100 AF535142] (146)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6:152534937-153050100 AF535142] (146)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6:152534937-153050100 NM_033071] (146)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr6:152535029-153050648 AF495910] (146)</TD> | ||
</TR> | </TR> | ||
<TR><TH>hg18</TH> | <TR><TH>hg18</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr14: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr14:105065301-106352275 uc001yrq.1] (2899)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr22: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr22:20715572-21595078 uc002zvw.1] (322)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:179098964-179380395 uc002umr.1] (313)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:88937989-89411302 uc002stk.1] (217)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr2:179098964-179380395 uc002umt.1] (194)</TD> | ||
</TR> | |||
<TR><TH>hg19</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr14:105994256-107283085 uc031qqx.1] (5065)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr22:22385572-23265082 uc031rxg.1] (462)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr2:179390717-179672150 uc031rqc.1] (363)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr2:179390717-179672150 uc031rqd.1] (313)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr2:179390717-179672150 uc021vsy.2] (312)</TD> | |||
</TR> | </TR> | ||
<TR><TH>mm8</TH> | <TR><TH>mm8</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2:76504823-76783386 NM_011652] (313)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2:76504823-76783386 NM_028004] (192)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr9: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr9:108810682-108841812 NM_007738] (118)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr1: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr1:33956371-34252174 NM_134448] (99)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr4: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr4:122852023-123186663 DQ067088] (99)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm9</TH> | <TR><TH>mm9</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12:114498400-117248164 uc007pgj.1] (610)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:76542041-76820604 uc008kfn.1] (313)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:76542041-76820604 uc008kfo.1] (192)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:51992167-52194318 uc008jqv.1] (157)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr9: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr9:108855790-108886920 uc009rrh.1] (118)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm10</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr12:113260189-116009879 uc029ryi.1] (813)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:76703984-76982547 uc008kfn.1] (313)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:76703984-76982547 uc008kfo.1] (192)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:52136647-52338798 uc008jqv.1] (157)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr9:108953745-108984875 uc009rrh.1] (118)</TD> | |||
</TABLE> | </TABLE> | ||
==Top five largest CDS extent genes== | ==Top five largest CDS extent genes== | ||
| Line 72: | Line 92: | ||
<TR><TH>db</TH><TH COLSPAN=5>gene name (CDS extent size: thickEnd-thickStart)</TH></TR> | <TR><TH>db</TH><TH COLSPAN=5>gene name (CDS extent size: thickEnd-thickStart)</TH></TR> | ||
<TR><TH>hg17</TH> | <TR><TH>hg17</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7:145251477-147555734 NM_014141] (2298740)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX:30897002-33117383 NM_000109] (2217347)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr11: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr11:82846625-85015962 CR749820] (2138880)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX:30897002-32989330 NM_004006] (2089394)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX:30898403-32989184 X14298] (2089394)</TD> | ||
</TR> | </TR> | ||
<TR><TH>hg18</TH> | <TR><TH>hg18</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr7: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr7:145444386-147749019 uc003weu.1] (2298740)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chrX:31047266-33267647 uc004ddb.1] (2217347)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr11: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr11:82843701-85015962 uc001pak.1] (2138880)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chrX:31047266-33139594 uc004dda.1] (2089394)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr8: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr8:2782789-4839736 uc003wqd.1] (2055833)</TD> | ||
</TR> | |||
<TR><TH>hg19</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr1:144146811-146467744 uc021ott.2] (2307732)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr7:145813453-148118088 uc003weu.2] (2298740)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chrX:31137345-33357726 uc004ddb.1] (2217347)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr11:83166056-85338314 uc001pak.2] (2138880)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chrX:31137345-33229673 uc004dda.1] (2089394)</TD> | |||
</TR> | </TR> | ||
<TR><TH>mm8</TH> | <TR><TH>mm8</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chrX:79201622-81457760 NM_007868] (2253366)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr6:44989695-47230955 NM_001004357] (2238304)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2:40418782-42475607 NM_053011] (2055883)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2:140086871-142081491 AK134694] (1988713)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr8: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr8:15895480-17535258 NM_053171] (1639258)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm9</TH> | <TR><TH>mm9</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chrX: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chrX:80194242-82450380 uc009tri.1] (2253366)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr6: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr6:45010087-47251368 uc009bst.1] (2238325)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr16: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr16:39984484-42176405 uc007zfr.1] (2189582)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:40452293-42509118 uc008jon.1] (2055883)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:140221166-142215786 uc008mpv.1] (1988713)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm10</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chrX:82948870-85205050 uc009tri.2] (2253366)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr6:45060061-47301371 uc009bst.2] (2238325)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr16:39984371-42176292 uc007zfr.1](2189582)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:40596773-42653598 uc008jon.1] (2055883)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr2:140395430-142390050 uc008mpv.1] (1988713)</TD> | |||
</TABLE> | </TABLE> | ||
| Line 106: | Line 139: | ||
txEnd-txStart)</TH></TR> | txEnd-txStart)</TH></TR> | ||
<TR><TH>hg17</TH> | <TR><TH>hg17</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr4: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr4:69806230-69806397 AF241539] (168)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7:129369319-129369494 AF277175] (176)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX:9353026-9353265 AY459291] (240)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr1: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr1:35869683-35869925 AY605064] (243)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7:128643905-128644162 AF503918] (258)</TD> | ||
</TR> | </TR> | ||
<TR><TH>hg18</TH> | <TR><TH>hg18</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr9: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr9:130062323-130062342 uc004buj.1] (20)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr1: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr1:65874524-65874545 uc001dcm.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr12: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr12:52671813-52671834 uc001seo.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr12: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr12:56504708-56504729 uc001sqn.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr20: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr20:15523127-15523148 uc002wpa.1] (22)</TD> | ||
</TR> | |||
<TR><TH>hg19</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr10:122806840-122806858 uc031pxj.1] (19)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr12:58034966-58034984 uc021qzo.1] (19)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr1:179170963-179170982 uc021pfi.1] (20)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr1:68649302-68649321 uc021oot.1] (20)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr10:134958378-134958397 uc021qbd.1] (20)</TD> | |||
</TR> | </TR> | ||
<TR><TH>mm8</TH> | <TR><TH>mm8</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chrX:12185020-12185236 AJ319753] (217)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr3: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr3:92585666-92585896 BC107019] (231)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr11: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr11:70546334-70546619 BC016221] (286)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr16: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr16:88757957-88758259 NM_130876] (303)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr16: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr16:88773645-88773948 NM_130873] (304)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm9</TH> | <TR><TH>mm9</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr1: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr1:74440898-74440919 uc007bma.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr10: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr10:85230424-85230445 uc007gmr.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr11: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr11:77886549-77886570 uc007khz.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr12:110831098-110831119 uc007pay.1] (22)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr13: | <TD>[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr13:54944553-54944574 uc007qpn.1] (22)</TD> | ||
</TR> | |||
<TR><TH>mm10</TH> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr17:15192492-15192509 uc029tak.1] (18)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr1:137031387-137031406 uc029qtn.1] (20)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr1:187624235-187624254 uc029qvd.1] (20)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr1:82585127-82585146 uc029qpl.1] (20)</TD> | |||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&position=chr11:46266296-46266315 uc029rlk.1] (20)</TD> | |||
</TR> | </TR> | ||
</TABLE> | </TABLE> | ||
==Custom Track of Small Exons and Introns== | |||
Custom track: | |||
[http://genome-test.gi.ucsc.edu/cgi-bin/hgTracks?db=hg18&hgt.customText=http://genomewiki.ucsc.edu/images/3/37/Hg18KnownGenesSmallIntronsExonsCt.txt.gz Hg18 small exons and introns] on the UCSC Genes track | |||
These are exons of size less than 22 bases, and introns of size less than 12 bases. The score column contains the size and thus you can filter smaller subsets via the score column in the table browser. | |||
These small exons and introns are used to maintain frame coding boundaries | |||
as found in mRNAs compared to the reference genome coordinates. | |||
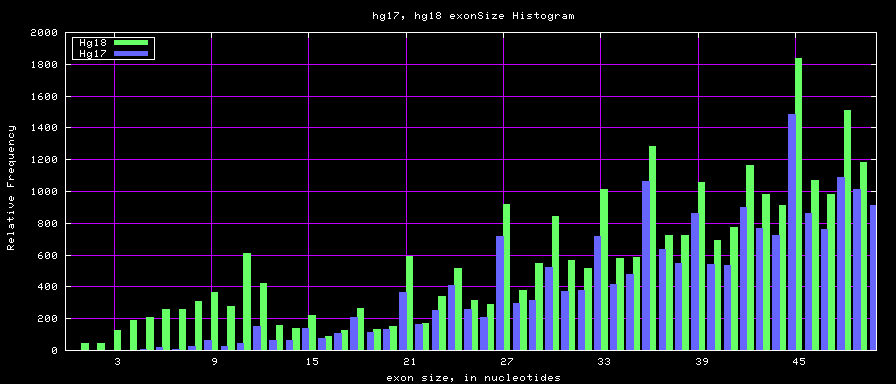
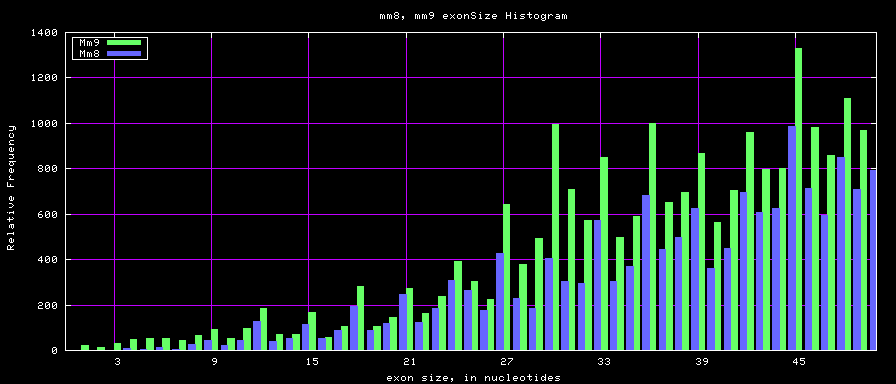
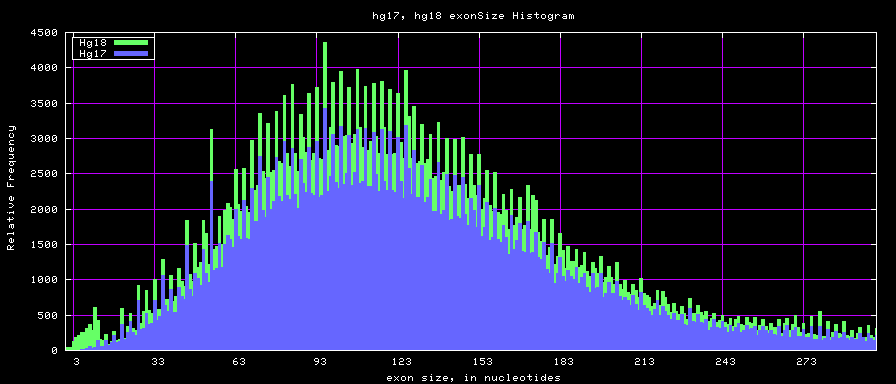
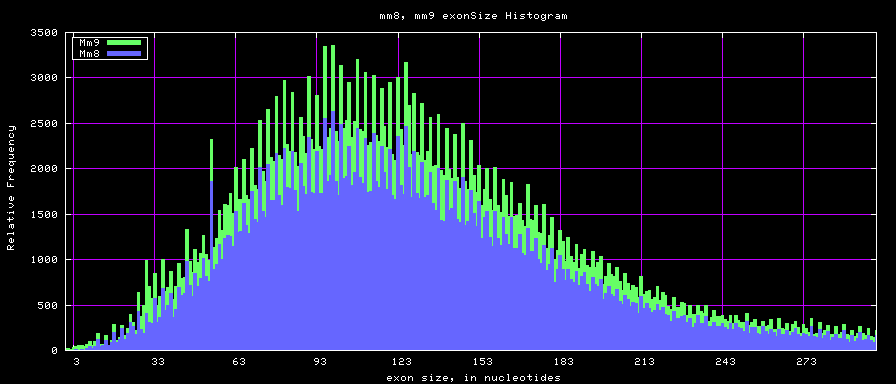
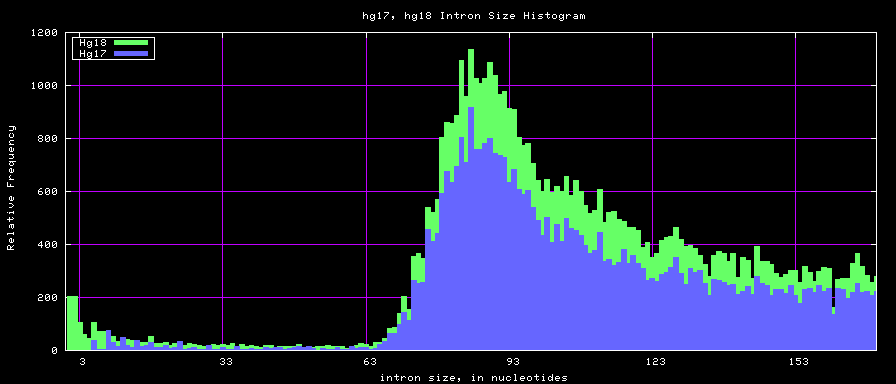
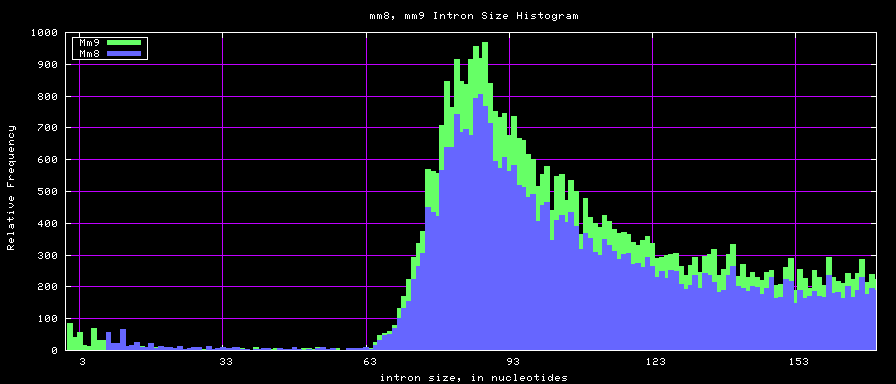
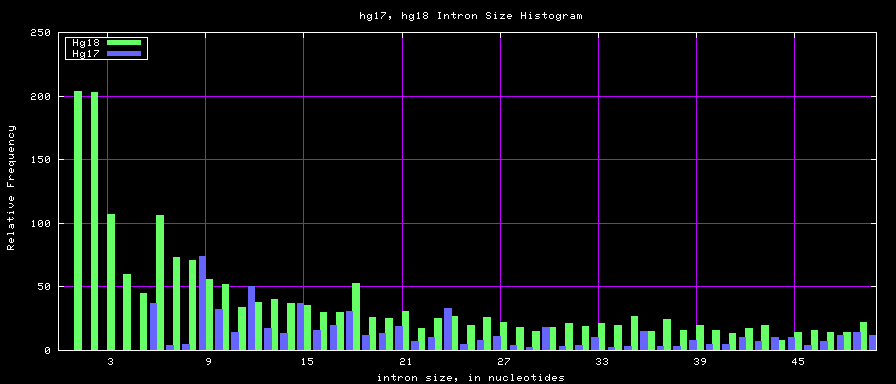
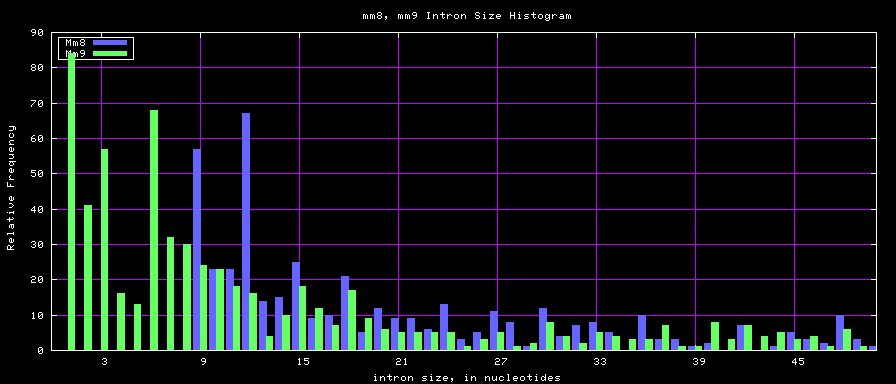
==Histogram graphs== | ==Histogram graphs== | ||
[[Image: | [[Image:Hg17_hg18.exonCount.png]] | ||
The caption on the graph above is incorrect. The X axis is Exon Count per Gene | |||
[[Image:Mm8_mm9.exonCount.png]] | |||
The caption on the graph above is incorrect. The X axis is Exon Count per Gene | |||
<HR><HR><HR> | |||
[[Image:Hg17_hg18.exonSize.png]] | |||
[[Image:Mm8_mm9.exonSize.png]] | |||
[[Image:Hg17_hg18_exonsTo300.png]] | |||
[[Image:Mm8_mm9_exonsTo300.png]] | [[Image:Mm8_mm9_exonsTo300.png]] | ||
[[Image:Hg17_hg18.intronsTo170.png]] | |||
[[Image:Mm8_mm9.intronsTo170.png]] | |||
[[Image:Hg17_hg18.intronSize.png]] | |||
[[Image:Mm8_mm9.intronSize.png]] | |||
==Methods== | ==Methods== | ||
| Line 183: | Line 258: | ||
}' wholeGene.bed | sort -k5n | head -5 | }' wholeGene.bed | sort -k5n | head -5 | ||
</pre> | </pre> | ||
[[Category:Gene Set Summary Statistics]] | |||
Latest revision as of 22:01, 31 August 2018
gene sets measured
- hg17 - knownGenes version 2
- hg18 - knownGenes version 3
- hg19 - knownGenes version 7
- mm8 - knownGenes version 2
- mm9 - knownGenes version 3
- mm10 - knownGenes version 6
The min, max and mean measurements are per gene
summary of gene and exon counts
| db | gene count | total exon count | min exon count | max exon count | mean exon count |
|---|---|---|---|---|---|
| hg17 | 39368 | 405720 | 1 | 149 | 10 |
| hg18 | 56722 | 519308 | 1 | 2899 | 9 |
| hg19 | 82960 | 742493 | 1 | 5065 | 9 |
| mm8 | 31863 | 314628 | 1 | 313 | 10 |
| mm9 | 49220 | 417114 | 1 | 610 | 8 |
| mm10 | 59121 | 486425 | 1 | 813 | 8 |
summary of exon size statistics
| db | sum exon sizes | min exon size | max exon size | mean exon size |
|---|---|---|---|---|
| hg17 | 106839720 | 1 | 18172 | 263 |
| hg18 | 146371091 | 1 | 36861 | 282 |
| hg19 | 221924089 | 1 | 205012 | 299 |
| mm8 | 83159087 | 4 | 17497 | 264 |
| mm9 | 117671086 | 1 | 29698 | 282 |
| mm10 | 143087341 | 1 | 83437 | 294 |
summary of intron size statistics
| db | sum intron sizes | min intron size | max intron size | mean intron size |
|---|---|---|---|---|
| hg17 | 2223224397 | 6 | 1096450 | 6069 |
| hg18 | 2784923600 | 1 | 1047320 | 6023 |
| hg19 | 4127555916 | 1 | 1160410 | 6260 |
| mm8 | 1476081990 | 9 | 1347550 | 5220 |
| mm9 | 2055504784 | 1 | 1253430 | 5589 |
| mm10 | 2420504661 | 1 | 3003780 | 5665 |
Top five exon count genes
| db | gene name (exon count) | ||||
|---|---|---|---|---|---|
| hg17 | NM_004543 (149) | AF535142 (146) | AF535142 (146) | NM_033071 (146) | AF495910 (146) |
| hg18 | uc001yrq.1 (2899) | uc002zvw.1 (322) | uc002umr.1 (313) | uc002stk.1 (217) | uc002umt.1 (194) |
| hg19 | uc031qqx.1 (5065) | uc031rxg.1 (462) | uc031rqc.1 (363) | uc031rqd.1 (313) | uc021vsy.2 (312) |
| mm8 | NM_011652 (313) | NM_028004 (192) | NM_007738 (118) | NM_134448 (99) | DQ067088 (99) |
| mm9 | uc007pgj.1 (610) | uc008kfn.1 (313) | uc008kfo.1 (192) | uc008jqv.1 (157) | uc009rrh.1 (118) |
| mm10 | uc029ryi.1 (813) | uc008kfn.1 (313) | uc008kfo.1 (192) | uc008jqv.1 (157) | uc009rrh.1 (118) |
Top five largest CDS extent genes
| db | gene name (CDS extent size: thickEnd-thickStart) | ||||
|---|---|---|---|---|---|
| hg17 | NM_014141 (2298740) | NM_000109 (2217347) | CR749820 (2138880) | NM_004006 (2089394) | X14298 (2089394) |
| hg18 | uc003weu.1 (2298740) | uc004ddb.1 (2217347) | uc001pak.1 (2138880) | uc004dda.1 (2089394) | uc003wqd.1 (2055833) |
| hg19 | uc021ott.2 (2307732) | uc003weu.2 (2298740) | uc004ddb.1 (2217347) | uc001pak.2 (2138880) | uc004dda.1 (2089394) |
| mm8 | NM_007868 (2253366) | NM_001004357 (2238304) | NM_053011 (2055883) | AK134694 (1988713) | NM_053171 (1639258) |
| mm9 | uc009tri.1 (2253366) | uc009bst.1 (2238325) | uc007zfr.1 (2189582) | uc008jon.1 (2055883) | uc008mpv.1 (1988713) |
| mm10 | uc009tri.2 (2253366) | uc009bst.2 (2238325) | uc007zfr.1(2189582) | uc008jon.1 (2055883) | uc008mpv.1 (1988713) |
Top five smallest transcript genes
| db | gene name (transcript size: txEnd-txStart) | ||||
|---|---|---|---|---|---|
| hg17 | AF241539 (168) | AF277175 (176) | AY459291 (240) | AY605064 (243) | AF503918 (258) |
| hg18 | uc004buj.1 (20) | uc001dcm.1 (22) | uc001seo.1 (22) | uc001sqn.1 (22) | uc002wpa.1 (22) |
| hg19 | uc031pxj.1 (19) | uc021qzo.1 (19) | uc021pfi.1 (20) | uc021oot.1 (20) | uc021qbd.1 (20) |
| mm8 | AJ319753 (217) | BC107019 (231) | BC016221 (286) | NM_130876 (303) | NM_130873 (304) |
| mm9 | uc007bma.1 (22) | uc007gmr.1 (22) | uc007khz.1 (22) | uc007pay.1 (22) | uc007qpn.1 (22) |
| mm10 | uc029tak.1 (18) | uc029qtn.1 (20) | uc029qvd.1 (20) | uc029qpl.1 (20) | uc029rlk.1 (20) |
Custom Track of Small Exons and Introns
Custom track: Hg18 small exons and introns on the UCSC Genes track
These are exons of size less than 22 bases, and introns of size less than 12 bases. The score column contains the size and thus you can filter smaller subsets via the score column in the table browser.
These small exons and introns are used to maintain frame coding boundaries as found in mRNAs compared to the reference genome coordinates.
Histogram graphs
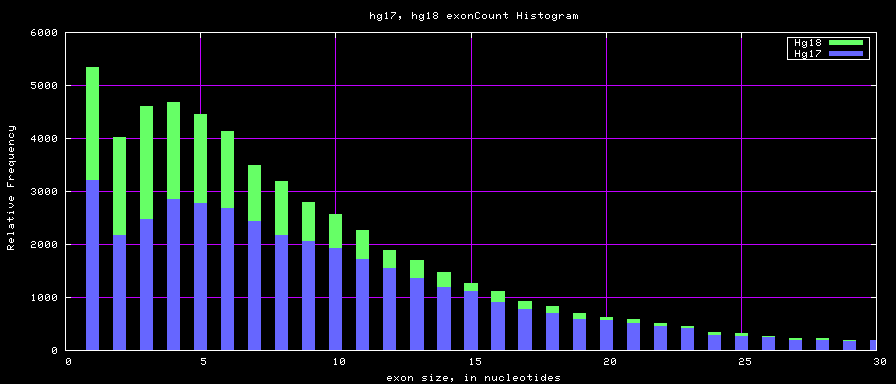 The caption on the graph above is incorrect. The X axis is Exon Count per Gene
The caption on the graph above is incorrect. The X axis is Exon Count per Gene
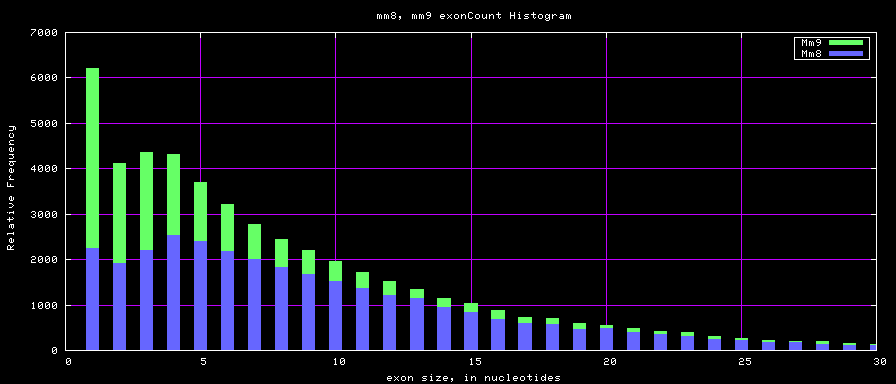 The caption on the graph above is incorrect. The X axis is Exon Count per Gene
The caption on the graph above is incorrect. The X axis is Exon Count per Gene
Methods
- From the table browser, request three different bed files for the knownGenes track:
- whole gene
- exons only
- introns only
- From those bed files, stats can be extracted
- gene count from: 'wc -l wholeGene.bed'
- exon count stats from:
STATS=`ave -col=10 wholeGene.bed -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
COUNT=`echo $STATS | cut -d' ' -f8 | awk '{printf "%d", $1}'`
- for exon or intron size stats:
STATS=`awk '{print $3-$2}' {introns,exons}.bed \
| ave -col=1 stdin -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5 | awk '{printf "%d", $1}'`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
SUM_SIZE=`awk '{sum += $3-$2} END{printf "%d", sum}' {introns,exons}.bed`
- top five exon count genes
sort -k10nr wholeGene.bed | head -5
- top five CDS size genes
awk '{cdsSize=$8-$7
if (cdsSize > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,cdsSize}
}' wholeGene.bed | sort -k5nr | head -5
- top five smallest transcript genes
awk '{size=$3-$2
if (size > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,size}
}' wholeGene.bed | sort -k5n | head -5