Marsupial phyloSNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) m (→Csae of SPON1) |
||
| (24 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
=== Assumed vertebrate phylogenetic tree === | === Assumed vertebrate phylogenetic tree === | ||
Marsupial relationships are taken from a [http://genome.cshlp.org/content/19/2/213.full 2009 paper] establishing the mitochondrial genome sequences of the Tasmanian tiger (Thylacinus cynocephalus) and numbat (Myrmecobius fasciatus). | Marsupial relationships are taken from a [http://genome.cshlp.org/content/19/2/213.full 2009 paper] establishing the mitochondrial genome sequences of the Tasmanian tiger (Thylacinus cynocephalus) and numbat (Myrmecobius fasciatus). A slightly different topology was found using transposons in an excellent July 2010 [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000436 PLOS paper] (right). | ||
[[Image:MarsupTree.jpg]][[Image:MarsupPhylo.jpg]] | [[Image:MarsupTree.jpg]][[Image:MarsupPhylo.jpg]][[Image:marsupRetro.jpg]] | ||
<pre>Newick tree that generates a marsupial-centric vertebrate phylogenetic tree: | <pre>Newick tree that generates a marsupial-centric vertebrate phylogenetic tree: | ||
| Line 1,255: | Line 1,255: | ||
WDFY4_gasAcu1 DMMEPEVLPHPFD-R-LRFQCSS M LVPGQWHHLAVVLSKDVKKSCIASAYFNGKAVGTGK | WDFY4_gasAcu1 DMMEPEVLPHPFD-R-LRFQCSS M LVPGQWHHLAVVLSKDVKKSCIASAYFNGKAVGTGK | ||
=== Case of XYLT1 === | === Case of XYLT1 === | ||
| Line 1,266: | Line 1,266: | ||
This non-conservative change A-->D is backed by three Sarcophilus reads. However all three are fairly near the end of a minus strand read so none cover the whole exon (raising mild concerns over read quality, given the unusual c-->a base transversion), yet none are long enough to span the next intron to reach the short following exon (leaving some mild pseudogene and paralog issues). Although blastn of extended opossum dna shows that the expected downstream phase 2 splice donor is present, that would also be expected in a close paralog or segmental duplication. | This non-conservative change A-->D is backed by three Sarcophilus reads. However all three are fairly near the end of a minus strand read so none cover the whole exon (raising mild concerns over read quality, given the unusual c-->a base transversion), yet none are long enough to span the next intron to reach the short following exon (leaving some mild pseudogene and paralog issues). Although blastn of extended opossum dna shows that the expected downstream phase 2 splice donor is present, that would also be expected in a close paralog or segmental duplication. | ||
'''Pseudogene issues:''' None observed in any mammal using tblastn at wgs database. The detection technique here is a multi-exon query. Because the target database is genomic, recent processed pseudogenes actually give stronger matches because of longer contiguous matches, whereas ortholog matches are weakened by the attempt by blast to extend them. Hence processed pseudogenes surface at the top of match list. | |||
Only a fragment of the gene can be recovered from current Sarcophilus reads, about 8 of 12 exons. However it cannot be determined without genomic assembly which exons 'belong' to the D containing exon, nor can the risk of including matches from the paralog be excluded. This gene has so-so conservation between human and opossum (270 myr roundtrip), 78% identity. which is somewhat puzzling in view of its enzymatic importance. However within marsupials conservation of most exons is in the mid-90's. | |||
'''Paralog issues:''' XYLT2 (xylosyltransferase II) gives a moderate match but is not an issue in terms of accurately scoring tasmanian devil populations for the A-->D change. It does create problems in conserved exons in recovering full length genes in species where reads span only single exons. Note XYLT2 also has a conserved A at this position in all 34 available species back to lamprey, proving it an important invariant. Adjacent residues however are only moderately conserved. | |||
XYLT1_homSap RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGASLLSTYLQSMRDLLEMTDWPWDFFINLS<font color="blue">A</font>ADYPIRTNDQLVAFLSRYRDMNFLKSHGRDNAR | |||
RS+YLHR+V+++++ Y NVRVTPWRM TIWGGASLL+ YL+SMRDLLE+ W WDFFINLS<font color="blue">A</font> DYP RTN++LVAFLS+ RD NFLKSHGRDN+R | |||
XYLT2_homSap RSDYLHREVVELAQGYDNVRVTPWRMVTIWGGASLLTMYLRSMRDLLEVPGWAWDFFINLS<font color="blue">A</font>TDYPTRTNEELVAFLSKNRDKNFLKSHGRDNSR | |||
'''Homoplasy (recurrent mutation) issues:''' The sole homolog in Drosophila (CG17771, 41% identity) has been [http://www.jbc.org/cgi/content/full/277/24/21207?ijkey=f35fe326b1b8b46e1fd369a6d575bea2bbded521 previously studied]. Here the 424 residue is large and charged E in a motif SESD conserved within arthropods but not lophotrochozoa nor cnidarians such as Hydra magnipapillata (where the corresponding fragment has 63% identity) or Nematostella where A424 are A and G respectively. This is not the drosophila DxD motif however -- this occurs much later in the protein. A further very remote crystallographic paralog MGAT1 also has D here as discussed later. | |||
XYLT1_homSap WRMATIWGGASLLSTYLQSMRDLLEMTDWPWDFFINLS<font color="blue">A</font>ADYPIRTNDQLVAFLSRYRDM | |||
R +TIWGGASLL+ LQ M DLL+ ++W WDF INLS +D+P++T D+LV FLS | |||
OXT_droMel KRFSTIWGGASLLTMLLQCMEDLLQ-SNWHWDFVINLS<font color="red">E</font>SDFPVKTLDKLVDFLSANPGR | |||
XYLT_hydMag WRMATIWGGASLLSMLLQMMEDTLKIKEWKWDFFINLSASDYPVQ | |||
XYLT_nemVec WSMATIWGGATLLQMLLKSMEDLIARKEWKWDFFINLSGNDFPIKVNT | |||
'''Known variations:''' Not a known disease gene. Natural human polymorphisms in XYLT1 have been observed, P325R, P766A, V8391 and R892Q but these do not include changes near the locus under consideration here A424. | |||
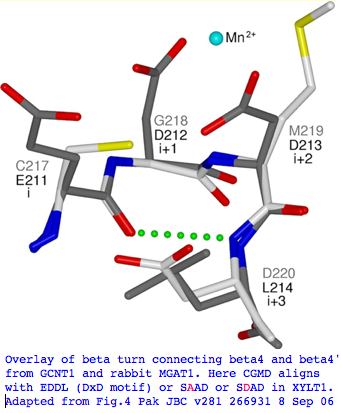
'''Structural significance:''' The region enveloping the key residue has a weak 30% match encompassing residues 328-535 (thus including the A-->D residue at 424) that nonetheless is adequate for structural modeling to PDB structure [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=protein&dopt=GenPept&RID=UAEKAVWX016&log%24=protalign&blast_rank=1&list_uids=112490721 2GAK] -- our residue is part of a short type I beta turn connecting strands 4 and 4' of the donor Rossmann domain. The [http://www.jbc.org/cgi/pmidlookup?view=long&pmid=16829524 determined structure] match to residues 86-289 is a somewhat similar enzyme, 6-N-Acetylglucosaminyltransferase, a product of the GCNT1 gene. A glycine has replaced the alanine, showing the latter is not a deep invariant critical to this class of enzyme. | |||
[[Image:XYLT1align.jpg]] | |||
This region has been compared in the structural overlay below to yet another glycosylating enzyme, rabbit MGAT1 beta-1,2-N-acetylglucosaminyltransferase. In this enzyme, this short beta turn carries the critical DxD motif that provides bound Mn++ for the UDP of incoming substrate. Comparing GCNT1 and XYLT1 aligns CGMD to SAAD (SDAD in Sarcophilus) in XYLT1. These residues are EDDL (DxD motif) in MGAT1. In other words, A424D of Sarcophilus is in fact physically realizable by D in functional MGAT1. This middle D is invariant throughout vertebrate MGAT1 even as the 'x' residue. However XYLT1 and MGAT1 have no significant alignment at the amino acid level and A-->D (or any other residue) is never observed in XYLT1. | |||
[[Image:XYLT1 struct.jpg|left]] | |||
The size of XYLT1 presents an [http://www.jbc.org/cgi/content/full/279/41/42566 unresolved mystery] requiring a crystallographic determination. A simply glycosylation reaction could be accomplished in a bacterium with perhaps 250 residues, yet here the enzyme is 959 residues long, almost 4x the minimum even allowing for targeting peptides and a transmembrane segment. | |||
A second puzzling aspect of glycosylases generally is their lack of homology -- 91 families exist of which only 29 have determined representatives (as tracked at the [http://www.cazy.org/fam/acc_GT.html CAZy database]. XYLT1 and XYLT2 are typical in belonging to a small isolated glycosyltransferase family 14 sharing no real sequence homology with other glycosylases (other than the DxD divalent cation coordination motif which could have arisen convergently). Structurally, known glycosylase folds are [http://www.jbc.org/cgi/pmidlookup?view=long&pmid=15148316 classified] as GT-A (DxD plus single Rossmann-like UDP-binding fold) or GT-B (double). | |||
Note the immediately preceding residues NLS constitute a potential glycosylation site, plausibly realized given the localization of the enzyme (Golgi or extracellular matrix) yet completely consistency with the beta role is required. NLS is invariantly conserved in both XYLT1 (even in drosophila and cnidaria) and XYLT2. While adjacent residues are not normally considered relevant to the NxT/S motif, potentially the substitution of D could interfer with this post-translational modification, were it to occur. This would require the glycosylated serine would be at the surface of the protein, contrary to the best PDB fit. Clearly a large attached carbohydrate would block interactions of immediately adjacent residues. | |||
<br clear="all"> | |||
'''Functional significance:''' The protein has been the subject of about a [http://www.ncbi.nlm.nih.gov/pubmed/10383739,11087729,11099377,11814476,15294915,17189266,17635914,17980567,18294457,19014925,15461586 dozen publications]. Xylosyltransferases I and II are the chain-initiating enzymes in the biosynthesis of glycosaminoglycans. XYLT1 is the initial and rate-limiting enzyme, transfering UDP-xylose to specific serine residues of a target protein. It is localized to the endoplasmic reticulum and Golgi apparatus as a single-pass membrane protein, but with some fraction also secreted to the extracellular space. The domain match is pfam02485, defined as 'core-2/I-branching' reflecting the branch the added carbohydrate introduces to the growing chain in chondroitin and heparan sulfate and post-translational proteoglycan production. The precise function of XYLT2 has not been established. | |||
Some 19 residues have been subject to [http://www.uniprot.org/uniprot/Q86Y38 experimental mutation] though none of the glycosylation sites. Only 8 of the 19 induced mutations affected enzymatic activity (yet without lowering UDP=xylose binding), even though the comparative genomics at bottom shows all 19 sites are equally invariant back to lamprey. Thus residues can be under tremendous selection for a variety of reasons other than substrate binding or direct or indirect role in catalysis. | |||
It is known that formation of abdominal aortic aneurysms can be caused by a destructive remodeling of the extracellular matrix in the vascular wall -- A115S enhances this risk. This bears no apparent relation to the A424D allele (human numbering) in tasmanian devil. The [http://www.jbc.org/cgi/pmidlookup?view=long&pmid=15294915 745DWD747 motif] has been shown essential to catalytic activity but again lacks immediate relevence. Reduced XYLT1 activity is a [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=search&term=11814476 known contributor] to male sterility. XYLT1 is elevated in [http://www.clinchem.org/cgi/content/full/52/12/2243 connective tissue diseases] such as systemic sclerosis, osteoarthritis, and pseudoxanthoma elasticum. | |||
The connection to tumors or cancers is tenuous. GCNT1 expression is highly correlated with tumor progression in a number of cancers. It is overexpressed in colorectal, lung, and prostate cancer. It is a very weak paralog. Similarly the proteoglycans produced by XYLT1 are important regulators in extracellular matrix deposition, cell membrane signal transfer, morphogenesis, cell migration, normal and tumor cell growth. Mouse [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17517600 knockouts of XYLT2] produce polycystic liver and kidney disease. | |||
In summary, this putative change in tasmanian devil could use additional sequences validation. While not likely linked to facial tumors, the A-->D allele is very undesirable in an inbreed population in view of its role in aortic aneurisms and male sterility. The several billion years of branch length invariance of the alanine argues for no tolerance for variation at this position. | |||
exon 5 ^ exon 6 | exon 5 ^ exon 6 | ||
homSap RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLS<font color="blue">A</font>ADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR | |||
panTro2 RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR | panTro2 RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR | ||
gorGor1 RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSXGRDNA- | gorGor1 RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSXGRDNA- | ||
| Line 1,295: | Line 1,340: | ||
echTel1 RSNYLHRQVLQFTGQYDNVRVTPWRMATIWGGA SLLTTYLQSMRDLLEMADWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR | echTel1 RSNYLHRQVLQFTGQYDNVRVTPWRMATIWGGA SLLTTYLQSMRDLLEMADWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR | ||
dasNov2 RSNYLHRQVLQFARQYANVRITPWRMATIWGGA SLLSTYLQSMRDLLEMSDWPWDFFINLSAADYPIR --------------------------- | dasNov2 RSNYLHRQVLQFARQYANVRITPWRMATIWGGA SLLSTYLQSMRDLLEMSDWPWDFFINLSAADYPIR --------------------------- | ||
monDom4 RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGA | monDom4 RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGA SLLSTYLQSMRDLMEMTDWPWDFFINLS<font color="blue">A</font>ADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR | ||
macEug RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGA | macEug RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGA SLLSTYLQSMRDLMEMTDWPWDFFINLS<font color="blue">A</font>ADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR | ||
sacHar1 rSNYLHRQVLQFAGQYQNVRVTSWRMATIWGGA | sacHar1 rSNYLHRQVLQFAGQYQNVRVTSWRMATIWGGA SLLSTYLQSMRDLMEMTDWPWDFFINLS<font color="blue">A</font>ADYPIr TNDQLVAFLSRYRDMNFLKSHGRDNAR | ||
sacHar2 | sacHar2 SLLSTYLQSMRDLMEMTDWPWDFFINLS<font color="red">D</font>ADYPIr TNDQLVAFLSRYRDMNFLKSHGRDNAR | ||
ornAna1 RSNYLYRQVLQFAGQYPNVRVTSWRMATIWGGA SLLTTYLQSMRDLMEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYREMNFLKSHGRDNAR | ornAna1 RSNYLYRQVLQFAGQYPNVRVTSWRMATIWGGA SLLTTYLQSMRDLMEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYREMNFLKSHGRDNAR | ||
galGal3 RSNYLHRQVLQFANQYPNVRVTSWRMATIWGGA SLLSTYLQSMRDLMEMNDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR | galGal3 RSNYLHRQVLQFANQYPNVRVTSWRMATIWGGA SLLSTYLQSMRDLMEMNDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR | ||
| Line 1,312: | Line 1,357: | ||
petMar1 RSNYLQRQVLQVAERYPNVRVTPWRMATIWGGA SLLTMYLRTMKDLLDMADWAWDFFINLSATDYPIR TNDQLVAFLTKYRDKNFLKSHGRDNNR | petMar1 RSNYLQRQVLQVAERYPNVRVTPWRMATIWGGA SLLTMYLRTMKDLLDMADWAWDFFINLSATDYPIR TNDQLVAFLTKYRDKNFLKSHGRDNNR | ||
The A is also conserved in the paralog XYLT2: | |||
^ | |||
XYLT2_hg18_4_ RSDYLHREVVELAQGYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_gorGor1 RSNYLHREVVELAQGYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_ponAbe2 RSNYLHREVVELAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_rheMac2 RSDYLHREVVELAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_calJac1 RSNYLHREVAELAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_tupBel1 RSNYLHREVVELAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_mm9_4_1 RSNYLYREVVELAQHYENVRVTPWRMVTIWGGASLLRMYLRSMKDLLEIPGWTWDFFINLSATDYPTR | |||
XYLT2_rn4_4_1 RSNYLYREVVELAQHYDNVRVTPWRMVTIWGGASLLRMYLRSMKDLLEIPGWTWDFFINLSATDYPTR | |||
XYLT2_dipOrd1 RSDYLHREVVELAKQYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_cavPor3 RSNYLHREVVALAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_speTri1 RSNYLHREVVELAQRYENVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_ochPri2 ---YLHREVVELAQQYENVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWTWDFFINLSATDYPTR | |||
XYLT2_turTru1 RSNYLHREVVELARQYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPDWAWDFFINLSATDYPTR | |||
XYLT2_bosTau4 RSNYLHREVVELARQYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_equCab2 RSNYLHREVVELARQYDNVQVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_felCat3 RSNYLHREVVELARRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_canFam2 RSNYLHREVVELARQYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_myoLuc1 RSNYLHREVVELARQYDNIRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_pteVam1 RSNYLHREVVELARQYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_eriEur1 RSNYLHREVVELARHYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_proCap1 RSNYLHREVVELARQYDNMRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR | |||
XYLT2_monDom4 RSNYLHREVVALAQHYANVRVTPWRMGTIWGGASLLKMYLRSMQDLLEAPGWTWDFFINLSATDYPTR | |||
XYLT2_macEug RSNYLHREVVALAQHYANVRVTPWRMGTIWGGASLLKMYLRSMQDLLEAPGWTWDFFINLSATDYPTR | |||
XYLT2_sarHar RSNYLHREVVALAQHYANVRVTPWRMGTIWGGASLLKMYLRSMQDLLEAPGWTWDFFINLSATDYPTR | |||
XYLT2_galGal3 RSNYLHREAVELAQHYPNIRVTPWRMVTIWGGASLLKMYLRSMKDLLELTEWPWDFFINLSATDYPTR | |||
XYLT2_taeGut1 RSSYLHREAVELARHYPNIRVTPWRMVTIWGGASLLKMYLRSMKDLLELSEWPWDFFINLSATDYPTR | |||
XYLT2_anoCar1 RSTYLHREVVEMAQHYPNIRVTPWRMVTIWGGASLLKMYLHSMKDLLEMTDWTWDYYINLSATDYPTR | |||
XYLT2_xenTro2 RSNYLHREVVRLAQSYENMRVTPWRMVTIWGGASLLTMYLRSMKDLLEVPDWPWDFFINLSATDYPTR | |||
XYLT2_tetNig1 RSGYMHREVLQVAQQYPNIRATPWRMVTIWGGASLLKAYLHSMQDLLSMLDWKWDFFINLSATDFPTR | |||
XYLT2_fr2_4_1 RSNYLHRQVQALAALYPNVRVTPWRMATIWGGASLLTMYLRSMADLLAMRDWSWDFFINLSAADYPIR | |||
XYLT2_gasAcu1 RSNYLHRQVLSLAAQYSNVRATPWRMATIWGGASLLTMYLRSMADLLAMRDWSWDFFINLSAADYPIR | |||
XYLT2_oryLat2 RCSYMHREVLQMAKHYPNIRATPWRMVTIWGGASLLKAYLRSMQDLLSMAEWKWDFFINLSATDFPTR | |||
XYLT2_danRer5 RSNYLHRQMVALAHQYPNVRVTSWRMSTIWGGASLLTMYLQSMKDLLAMRDWSWDFFINLSAADYPIR | |||
XYLT2_petMar1 RSNYLQRQVLQVAERYPNVRVTPWRMATIWGGASLLTMYLRTMKDLLDMADWAWDFFINLSATDYPIR | |||
The comparative genomics of the 19 XYLT1_homSap residues replaced by experimental mutagenesis. The key residue columns have been sliced out of intact protein accompanied by a few residues of flanking context and then concatenated to make a compact display (dots used if identical to human). | |||
chr4_18550 ATP4A 6 16 C=4(130) R=3(74) | C257A none C542A none D745G enz- | ||
C276A enz- C561A enz- D745E none | |||
C285A none C563A none W746DNG enz- | |||
C301A none C572A enz- D747GE enz- | |||
D314G none C574A enz- C920A none | |||
D316G none C675A none C927A none | |||
C471A enz- C933A none | |||
* * * * * * * * * *** * * * | |||
homSap CDISGKEAISALSRAKSKHCRQEIGETYCRHKLGLLMPEKVTRFCPLEDEDECDCDTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSSCRVGTDWDAKERDICATGPTACPVMQTCSQ | |||
panTro2 ....................................................................................................................... | |||
gorGor1 .....................................................................X................................................. | |||
ponAbe2 ....................................................................................................................... | |||
rheMac2 ...............................................................................................................R...I... | |||
calJac1 ....................................................................................................................... | |||
tarSyr1 ................................H...................................I.................................V.....S.......... | |||
micMur1 ........V.................................P....D....................................................................... | |||
otoGar1 ........................................................................................................T.............. | |||
tupBel1 .E............S........................................H................................................T.............. | |||
musMus1 ............T...........A................A..............................................I.V.............T.............. | |||
ratNor ............T...........A................A............................................................V................ | |||
dipOrd1 ........................A.........Q..................................................................EV.SV.LS...T.PA... | |||
cavPor3 ..............S..............................................................................................S......... | |||
speTri1 ..............S..R...........Q.........RL..L..........................................................V................ | |||
oryCun1 .E............S.......Q................R..............................................................V.S..........S..H | |||
ochPri2 .E.....................................R................................................................T...S.......... | |||
turTru1 ..............S.............................................................................................S.......... | |||
bosTau4 ..............S............................L.................................................................V......... | |||
equCab2 ..............S.................................................................................E...........S.......... | |||
felCat3 ......................................K................................................................................ | |||
canFam2 .........................D........M...K......S..........................................................T.............. | |||
myoLuc1 ..............S......................................................................................................TH | |||
pteVam1 ..............S...........-............................................................................................ | |||
proCap1 ..............S.........V................A................................................L...........-GGA.AR...MLPAG.. | |||
echTel1 ..............S.........A.A..S....R......A.L............................................................S..........A... | |||
dasNov2 ..............S.........A..................L...........G...................................I..E.......V.T........L.-... | |||
monDom4 ..............S...Q.....A.I..Q..V.K.....................S...............................R..I..E.........T..........A... | |||
ornAna1 ..............S...Q.....A....Q..Y.K.....................S..................................I..E........................ | |||
galGal3 .EVT.......M......P.....ADV..Q..H.K..........T.............................................I..E..........I............. | |||
taeGut1 .EVT.......M.....QQ.....ADV..Q....K....Q.....A.........ES...............................T.....E..........A...S.....A... | |||
anoCar1 .E......L.........P.....A......RQ.K....Q...L...Q.D.....ES..................................I.AE.........S..........A.T. | |||
xenTro2 .E.T..............Q.....A.V..Q..Q.K........L...........ES..................................I.A........V.SV.......L.G.A. | |||
tetNig1 .E..........A.....E...Q.A.V.....E.Q....R...Y...........ES..................................I..E..P......T...SS.....S.A. | |||
fr2_3_1 .E..........A.....E...Q.A.VF....E.Q........Y...........ES.....................................E..........V.........A.PK | |||
gasAcu1 .E............V...E...Q.A.V.....E.Q....T...Y......H....GSL.................................I.............V.........A.PK | |||
oryLat2 .E................D...Q.A.V....RE.R........Y....E......GSL..............................A..IS....P....V..V.........A.PK | |||
danRer5 .E..............T.E...Q.V.V.....EHQ........Y..V...V....GSL..............................A..IS....P....V..V...S.....A.AK | |||
petMar1 .E.A....L......R.AQ.K...ADVV.L.QE.K....SLP....I...V....GSL..............................A..IS....P....V.S...SG.......RE | |||
<pre> | |||
>XYLT1_homSap | |||
MVAAPCARRLARRSHSALLAALTVLLLQTLVVWNFSSLDSGAGERRGGAAVGGGEQPPPAPAPRRERRDLPAEPAAARGGGGGGGGGGGGRGPQARARGGGPGEPRGQQPASRGALPARAL | |||
DPHPSPLITLETQ | |||
DGYFSHRPKEKVRTDSNNENSVPKDFENVDNSNFAPRTQKQKHQPELAKKPPSRQKELLKRKLEQQEKGKGHTFPGKGPGEVLPPGDRAAANSSHGKDVSRPPHARKTGGSSPETKYDQPPKCDISGKEAISALSRAKSKHCRQEIGETYCRHKLGLLMPEKVTRFCPLE | |||
GKANKNVQWDEDSVEYMPANPVRIAFVLVVHGRASRQLQRMFKAIYHKDHFYYIHVDK | |||
RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGASLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR | |||
TNDQLVAFLSRYRDMNFLKSHGRDNAR | |||
FIRKQGLDRLFLECDAHMWRLGDRRIPEGIAVDGGSDWFLLNRRFVEYVTFSTDDLVTKMKQFYSYTLLPAE | |||
SFFHTVLENSPHCDTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSPNDFKPQDFHRFQ | |||
QTARPTFFARKFEAVVNQEIIGQLDYYLYGNYPAGTPGLRSYWENVYDEPDGIHSLSDVTLTLYHSFARLGLRRAETSLHTDGENSCR | |||
YYPMGHPASVHLYFLADRFQGFLIKHHATNLAVSKLETLETWVMPKKVFKIASPPSDFGRLQFSE | |||
VGTDWDAKERLFRNFGGLLGPMDEPVGMQKWGKGPNVTVTVIWVDPVNVIAATYDILIESTAEFTHYKPPLNLPLRPGVWTVKILHHWVPVAETKFLVAPLTFSNRQPIKP | |||
EEALKLHNGPLRNAYMEQSFQSLNPVLSLPINPAQVEQARRNAASTGTALEGWLDSLVGGMWTAMDICATGPTACPVMQTCSQTAWSSFSPDPKSELGAVKPDGRLR* | |||
>XYLT1_monDom | |||
MVAALCARRLARRSHSALIAALTVLLLQTLIVWNFSSLDSGAGDHRGGAAAGGPPPAPRRERRDLPLEPAAAGEGERGPAGGQLLRERGGGHGEHRAQHPPRRGGLPGRAL | |||
EPPPSPFTSLETQ DGYFSHRPKEKMRTDSNNENSVPKDFENIDNSNFAPRTQRQKHQPDLGKKPLSKQKEHLKKKLEQDEKVKENSLLGKGSNEALQYSNQAAQNSSQGKKSSRLPHSRKNGAGSPELKYDQPPRCDISGKEAISALSRSKSKQCRQEIAEIYCQHKVGKLMPEKVTRFCPLE | |||
GKANNNVRWDEDSVEYMPANPVRIVFVLVVHGRASRQLQRMFKAIYHKDHFYYIHVDK | |||
RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGASLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPIR | |||
TNDQLVAFLSRYRDMNFLKSHGRDNAR | |||
FIRKQGLDRLFLECDTHMWRLGDRKIPEGITVDGGSDWFLLNRKFVEYVTFSNDDLVTKMKQFYSYTLLPAE | |||
SFFHTVLENSPHCGTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSPNDFKPADFHRFQ | |||
QTARPTFFARKFEAVVNQEIIGQLDYYLYGNYPSGTPGLRSYWENVYDEPDGIHSISDVVLTMYHSFTRLGLRRAETSLHTDGENSCR | |||
YYPMGHPVSVHLYFLADHFQGFLIKHHATNLAVSKLETLETWVMPKKVFKIANPPSDFGRLQFSE | |||
IGTEWDAKERIFRNFGGLLGPMDEPVGMQKWGKGPNVTVTVIWVDPVNIIAATYDILIESSAEFTHYKPPLNLPLRPGVWTVKILHHWVPVAETKFLVTPLTFSNKQPIKP | |||
DESLKLHNGPPRNAYMEQSFQGLNPVLNIPINLAHVEQARRNAATTGAKLESWVDSLVGGIWSAVDICAIGPTACPVMQTCSQTSWSSLSPDPKSELGAIKPDGRLR* | |||
>XYLT1_macEug fragment | |||
MVAALCARRLARRSHSALIAALTVLLLQTLIVWNFSSLDSGAGDHRGGEQHAGGEPPPAPRRERRDLAPESRAAAGEEGGGGGRGPQPRGYKLPLERGGGGGGGHREHRPQQTPRRGGPAAGAAQLPGQAL | |||
... | |||
DGYFSHRPKEKMRTDSNNENSVPKDFENIDNSNFAPRTQRQKHQPDLGKKSLSKQKEQLKKKLEQEEKAKENSLLGKSSNEAMQYSNQAAQNSSAAKASPKSSKQPHTRKNGAGSPELKYDQLPRCDISGKEAISALSRSKSKQCRQEIAEIYCQHKVGKLMPEKVTRFCSLE | |||
GKANNNVRWDEDSVEYMPANPVRIAFVLVVHGRASRQLQRMFKAIYHKDHFYYIHVDK | |||
RSNYLHRQVLQFAGQYQNVRVTSWRMATIWGGASLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPIR | |||
... | |||
FIRKQGLDRLFLECDTHMWRLGDRKIPEGITVDGGSDWFLLNRKFVEYVTFSNDDLVTKMK... | |||
SFFHTVLENSPHCDTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSPNDFKPADFHRFQ | |||
... | |||
YYPMGHPVSVHLYFLADRFQGFLIKHHATNLAVSKLETLETWVMPKKVFKIANPPSDFGRLQFSE | |||
IGTDWDAKERIFRNFGGLLGPKDEPVGMQKWGKGP... | |||
DESLKLHGGPPHNAYMEQSFQGLNPVLNIPINLAHVEQARRNAATTGPKLESWVDSLVGGVWSAMDICAIGPTACPVMQTCSQTSWSSLSPDPKSELGAVKPDGRLR* | |||
>XYLT1_sarHar fragment missing 5-6 exons | |||
... | |||
... | |||
DGYFSHRPKEKMRTDSNNENSVPKDFENIDNSNFAPRTQRQKHQPDLG...PHVRKNGVGSPELKYDQPPRCDISGKEAISALSRSKSKQCRQEIAEIYCQHKVGKLMPEKVTRFCPL. | |||
rSNYLHRQVLQFAGQYQNVRVTSWRMATIWGGASLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPIR | |||
TNDQLVAFLSRYRDMNFLKSHGRDNAR | |||
FIKKQGLDRLFHECDSHMWRLGERQIPEGIVVDGGSDWFALTRSFVEYVVYTDDPLVAQLRQFYTYTLLPAE | |||
SFFHTVLENSPHCDTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSPNDFKPADFHRFQ | |||
... | |||
YYPMGHPVSVHLYFLADRFQGFLIKHHATNLAVS... | |||
IGTDWDAKERIFRNFGGLLGPMDEPVGMQKWGKGPNVTVTVIWVDPVNVIAATYDILIESSAEFTHYKPPLNLPLRPGVWTVKILHHWVPVAETKFLVTPLTFSNRQPIKP | |||
DESLKLHNGPPRNAYMEQSFQGLNPVLNIPINLAHVEQARRNAAITGPKLENWVDSLVGGIWSAVDICAIGPTACPVMQTCSQTSWSSLSPDPKSELGAIKPDGRLR* | |||
</pre> | |||
=== Case of ATP4A === | |||
chr4_18550 ATP4A 6 16 C=4(130) R=3(74) | |||
>contig00001 length=906 numreads=10 | >contig00001 length=906 numreads=10 | ||
TAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT | TAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT | ||
| Line 1,320: | Line 1,510: | ||
^ | ^ | ||
This is a common non-conservative substitution resulting from the CpG hotspot effect. The gene involved, ATP4A, is a member of an extensive well-studied family of hydrogen-potassium membrane pumps coupled to ATP hydrolysis, with this one responsible for acid secretion into the stomach from electroneutral exchange of cytoplasmic hydrogen ion with external potassium ions. The enzyme resides in gastric parietal cells, localized in cytoplasmic vesicles and apical plasma membranes of the secretory canaliculus. It is comprised of alpha chains such as this as well as beta and gamma chains. The protein is large at 1,035 residues. The R280C variant occurs in exon 7 of the 22 coding exons. | |||
'''Pseudogene issues:''' Opossum has a processed pseudogene covering the critical residue at chr2:88378354-88379057. However the parent gene here is ATP12A rather than ATP4A. It may be lineage-specific because a counterpart could not be found in Sarcophilus (at this stage of assembly). | |||
'''Paralog issues:''' ATP4A is part of a sizeable gene family with a half-dozen paralogs showing good percent identity over this exon. ATP4A may be a relatively new gene because it cannot be located in sauropsids or platypus -- its telltale location on human chromosome 19, lack of good syntenic conservation, and tandem location of its best counterpart with respect to ATP12A in species such as lizard. With so many paralogs, loss with compensation may have occured in some species. | |||
Although the history of this gene family will prove complex, to a certain extent it is irrelevent because the R of R280C is found in homologous position in all members of the family. There is no reduced alphabet flexibility at this residue. That is illustrated for marsupials below. One cnidarian sequence is included from Nematostella to show this R is quite ancient. | |||
* chr strand pos | |||
monDom1 GTAQGLVVNTGDRTIIG<font color="red">R</font>IASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT 4 - 373500709 | |||
monDom2 GTATGIVINTGDRTIIG<font color="red">R</font>IASLASSVGQEKTPIAIEIEHFVHIVAGVAVSIGIVFFIIAICMKYRVLDAVVFLIGIILANVPEGLVAAVT 4 + 278084055 | |||
monDom5 GTATGMVINTGDRTIIG<font color="red">R</font>IASLASGVGNEKTPIAIEIEHFVHMVAGVAVSIGVIFFIIAVSMKYPVLESIIFLIGIIVANVPEGLLAAVT 4 + 277916123 | |||
monDom3 GTARGIVIATGDRTVMG<font color="red">R</font>IATLASGLEVGRTPIAMEIEHFIQLITGVAVFLGVSFFVLSLILGYSWLEAVIFLIGIIVANVPEGLLATVT 2 - 165703122 | |||
monDom4 GTARGIVVYTGDRTVMG<font color="red">R</font>IATLASGLEGGQTPIAAEIEHFIHLITGVAVFLGVTFFILSLILEYTWLEAVIFLIGIIVANVPEGLLATVT 2 + 487887988 | |||
monDom6 GTATGIVINMGDHTIIG<font color="red">R</font>IASLDSSVGHEKTPIAIEIEPFVHIVAGVAVSFGIGFFIIAIFMKYWVLDVVIFLIGIILANVPEGLVAAVT 2 + 88378354 | |||
sarHar1 GTAQGLVVNTGDRTIIG<font color="red">R</font>IASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT | |||
sarHar2 GTARGVVVATGDRTVMG<font color="red">R</font>IATLASGLEVGKTPIAIEIEHFIQLITGVAVFLGVSFFILSLILGYTWLEAVIFLIGIIVANVPEGLLATVT | |||
sarHar3 GTATGMVINTGDRTVIG<font color="red">R</font>IASLASSVGHEKTPIAIEIEHFVHIVAGVAVSIGIVFFIIAICMKYRVLDAVIFLIGIILANVPEGLVAAVT | |||
sarHar4 GTARGIVIATGDHTVMG<font color="red">R</font>IASLTSVLEAGKTPIAIEIEHFIHIITGVAVFLGVTFFILSLLLGYGWLHAVIFLIGIIVANVPEGLLATVT | |||
macEug1 gTATGMVINTGDRTIIG<font color="red">R</font>IASLASGVGNEKTPIAIEIEHFVHIVAGVAVSLGVIFFIIAVSMKYPVLESIIFLIGIIVANVPEGLLAAVT | |||
macEug2 gTARGVVVATGDRTVMG<font color="red">R</font>IATLASGLEVGKTPIAIEIEHFIQLITGVAVFLGVSFFILSLILGYTWLEAVIFLIGIIVANVPEGLLATVT | |||
macEug3 gTARGIVVYTGDRTVMG<font color="red">R</font>IATLASGLEGGQTPIAAEIEHFIHLITGVAVFLGVTFFILSLILEYTWLEAVIFLIGIIVANVPEGLLATVT | |||
macEug4 gTAQGIVIATGDNTVMG<font color="red">R</font>IASLTSVLEAGQTPIAIEIEHFIHLITAVAVFLGVSFFILSLVLGYGWLQAVIFLIGIIVANVPEGLLATVT | |||
macEug5 gTATGVVINTGDQTIIG<font color="red">R</font>IALLTSSVGHEKTPSAIEIEHFVHIVAEVAVSLGMVFFTIAICTKYQVLDAVIFLIGIILGSVPESLVAAVT | |||
nemVec1 GNATGVVVQTGDNTVMG<font color="red">R</font>IANLASGLGSGKTPIAVEIEHFIHIITGVAVFLGVTFFIIAFILKYKWLEAVIFLIGIIVANVPEGLLATVT XP_001632743 | |||
Closest paralogs of ATP4A within human genome: | |||
ATP4A ATPase, H+/K+ transporting, nongastric, alpha | |||
ATP1A3 Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9). | |||
ATP1A1 Na+/K+ -ATPase alpha 1 subunit isoform a | |||
ATP1A2 Na+/K+ -ATPase alpha 2 subunit proprotein | |||
ATP1A4 Na+/K+ -ATPase alpha 4 subunit isoform 1 | |||
ATP2A3 sarco/endoplasmic reticulum Ca2+ -ATPase isoform | |||
ATP2A2 ATPase, Ca++ transporting, cardiac muscle, slow | |||
ATP2A1 ATPase, Ca++ transporting, fast twitch 1 isoform | |||
ATP2C1 calcium-transporting ATPase 2C1 isoform 1d | |||
ATP2C2 calcium-transporting ATPase 2C2 | |||
ATP2B4 plasma membrane calcium ATPase 4 isoform 4b | |||
ATP2B3 plasma membrane calcium ATPase 3 isoform 3b | |||
ATP2B1 plasma membrane calcium ATPase 1 isoform 1b | |||
ATP2B2 plasma membrane calcium ATPase 2 isoform 1 | |||
'''Homoplasy (recurrent mutation) issues:''' None, as discussed above. The CpG at the start of this arginine codon occurs in all vertebrates back to lamprey for which sequence is available, meaning the CpG hotspot is ancient. Yet R140C is never observed in other species, even as an allele, even though it is likely to have been generated many times in various populations. That would imply negative selection against this substitution. | |||
'''Known variations:''' Not a known disease gene at OMIM. Natural human polymorphisms have been observed, notably the T-->V substitution at position 3 of the exon. | |||
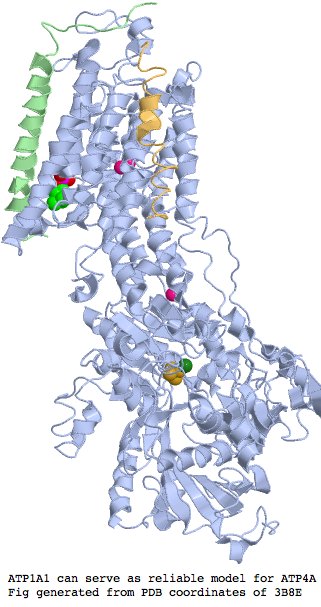
'''Structural significance:''' The region enveloping the key residue, according to an excellent 72% blastp match at PDB (3B8E) to the ATP1A1 paralog in pig using three exons about the critical residue. This suffices for an accurate model of both Sarcophilus ATP4A wildtype as well as R280C, though it must be kept in mind that the pig crystal was [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=pubmed&list_uids=18075585 only determined to 3.5 angstroms] due to its large size and integral membrane aspects. R280C lies in the sixth alpha helix of this structure which lies in the cytoplasm (rather than lumen) some 20 residues before the next transmembrane helix enters the membrane | |||
=== | Alignment of human ATP4A to pig ATP1A1 about <font color="red">R280C</font> showing <font color="blue">strand 11</font>, <font color="green">helix 5</font>, <font color="brown">helix 6</font>, and active site <font color="magenta">D</font>: | ||
ATP4A 1 QATVIRDGDKFQINADQLVVGDLVEMKGGDRVPADIRILAAQGCKVDNSSLTGESEPQTR 60 | |||
QA VIR+G+K INA+++VVGDLVE+KGGDR+PAD+RI++A GCKVDNSSLTGESEPQTR | |||
ATP1A1 143 QALVIRNGEKMSINAEEVVVGDLVEVKGGDRIPADLRIISANGCKVDNSSLTGESEPQTR 202 | |||
ATP4A 61 SPECTHESPLETRNIAFFSTMCL<font color="blue">EGTVQ</font>GLVVNTGDRT<font color="green">IIG<font color="red">R</font>IAS</font>LASGVENEKT<font color="brown">PIAIE 120</font> | |||
SP+ T+E+PLETRNIAFFST C+<font color="blue">EGT +</font>G+VV TGDRT<font color="green">++G<font color="red">R</font>IA+</font>LASG+E +T<font color="brown">PIA E</font> | |||
ATP1A1 203 SPDFTNENPLETRNIAFFSTNCV<font color="blue">EGTAR</font>GIVVYTGDRT<font color="green">VMG<font color="red">R</font>IAT</font>LASGLEGGQT<font color="brown">PIAAE 262</font> | |||
ATP4A 121 <font color="brown">IEHFVDIIAGLA</font>ILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVTVCLSLT 180 | |||
<font color="brown">IEHF+ II G+A</font>+ G +FFI+++ + YT+L A++F + I+VA VPEGLLATVTVCL+LT | |||
ATP1A1 263 <font color="brown">IEHFIHIITGVA</font>VFLGVSFFILSLILEYTWLEAVIFLIGIIVANVPEGLLATVTVCLTLT 322 | |||
ATP4A 181 AKRLASKNCVVKNLEAVETLGSTSVICS<font color="magenta">D</font>KTGTLTQNRMTVSHLWFDNHIHTADTTEDQS 240 | |||
AKR+A KNC+VKNLEAVETLGSTS ICS<font color="magenta">D</font>KTGTLTQNRMTV+H+W DN IH ADTTE+QS | |||
ATP1A1 323 AKRMARKNCLVKNLEAVETLGSTSTICS<font color="magenta">D</font>KTGTLTQNRMTVAHMWSDNQIHEADTTENQS 382 | |||
[[Image:ATP4Astruc.jpg]] | |||
<br clear="all"> | |||
'''Functional significance:''' Clearly it would be disadvantageous to lose function in a key enzyme in the gastric digestive process. It is unlikely to be an adaptation to carnivory because all other mammals with such a diet retain the arginine. It remains conceivable that amino acid change elsewhere in this molecule or its hetero-oligomer partners could compensate. However R240C may not induce loss but rather suboptimal functioning in this otherwise extremely conserved regin of the protein. As such it likely spread from an inbreeding artefact attributable to low population levels. It is not plausibly associated with facial tumors but still would be a high priority to breed out. | |||
>ATP4A_homSap 263-352 chr 19 flanking exons 20 phase tandem to anoCar: -FFAR3 +ATP4A +ATP12A -TMEM147 -GAPDHS | |||
>contig00001 length= | QATVIRDGDKFQINADQLVVGDLVEMKGGDRVPADIRILAAQGCKVDNSSLTGESEPQTRSPECTHESPLETRNIAFFSTMCLE | ||
GTVQGLVVNTGDRTIIG<font color="red">R</font>IASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT | |||
................................. | VCLSLTAKRLASKNCVVKNLEAVETLGSTSVICSDKTGTLTQNRMTVSHLWFDNHIHTADTTEDQS | ||
>ATP4A_monDom (note smaller introns relative to human) | |||
QATVIREGDKFQINADQLVVGDLVEIKGGDRVPADIRILAAQGCKVDNSSLTGESEPQTRSPECTHESPLETRNIAFFSTMCLE | |||
GTAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT | |||
VCLSLTAKRLARKNCVVKNLEAVETLGSTSVICSDKTGTLTQNRMTVSHLWFDNHVHTADTTEDQS | |||
>ATP4A_sarHar (other exons provisional: lack of assembly, paralogs) | |||
QATVIREGDKFQINADQLVVGDLVEIKGGDRVPADIRVLAAQGCKVDNSSLTGESEPQTRSPECTHDSPLETRNIAFFSTMCLE | |||
GTAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT | |||
VCLSLTAKRLARKNCVVKNLEAVETLGSTSVICSDKTGTLTQNRMTVSHLWFDNHVHTADTTEDQS | |||
=== Case of VPS72 === | |||
chr2_30280 VPS72 5 15 R=3(59) K=2(51) | |||
>contig00001 length=591 numreads=6 | |||
NYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLAEPGPKEENVDVE | |||
...............R..................T............ | |||
^ | |||
The K-->R substitution K204 in exon 5 of the six exon VPS72 (vacuolar protein sorting-associated protein 72) would be innocuous if the role of the residue were simply to provide a positively charged side chain. However here the lysine is invariant back to cnidaria with no arginine accepted into the reduced alphabet. | |||
'''Pseudogene issues:''' No recent pseudogenes occur in opossum or human genomes at the sensitivity of Blat. The Sarcophilus exon variant has normal splice junctions and its extension lacks amino acids of flanking exons, so it itself is not part of a processed pseudogene. A full length gene is readily recovered; other exons are quite close in sequence to opossum and do not support the notion of gene loss. | |||
'''Paralog issues:''' This gene has only weak partial paralogs in mammal, ATAD2 and MYO9B at 1e-05, that could not cause confusion. | |||
'''Homoplasy (recurrent mutation) issues:''' None. No variation is seen at position K204 in other species back to cnidaria: | |||
nemVec: LTQEELLAEARITEEENTASLLAYQRHEADK<font color="red">K</font>KTKIQKVTHKGPIIRFCSLSMPV XP_001632443 | |||
hydMag: LTQQELLAEAKITAEKNLASLAQFLKLEEEK<font color="red">K</font>HIKISKVRYQGPIIRYQSVRMPL 207 XP_002165194 | |||
LTQ+ELLEAKIT E NL SL + +LE +K<font color="red">K</font> K + GPII Y SV +PL | |||
homSap: LTQEELLREAKITEELNLRSLETYERLEADK<font color="red">K</font>KQVHKKRKCPGPIITYHSVTVPL 221 | |||
* | |||
homSap ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDIEG | |||
gorGor1 eTYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDIEG | |||
ponAbe2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDIEG | |||
rheMac2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENIDIEG | |||
calJac1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGLKEENVDIEG | |||
tarSyr1 ETYERLEADKKKQVHKKRKCPGPIITFHSVTVPLVGEPGPKEENVDVEg | |||
micMur1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGELGPKEETVDIEG | |||
otoGar1 ETYERLEADKKKQVHKKRKCPGPIITYHSMAVPLVGELGPK-ETVDVEG | |||
tupBel1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
mm9_5_6 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
rn4_5_6 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
cavPor3 ETYERLEADKKKQVHKKRKCPGPIITYHSMTVPLVGEPGPKEENVDVEG | |||
speTri1 ETYERLEADKKKPVHKETECPGPIITYHSMTVPLIGELGPKEENVDVEG | |||
ochPri2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLIGELGPKEENVDVEG | |||
turTru1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
bosTau4 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
equCab2 ETYERLEADKKKQVHKKRKCP-PIITYHSVTVPLVGEPGPKEENVDVEG | |||
felCat3 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
canFam2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
pteVam1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGKPGPREETVDVEG | |||
eriEur1 ETYERLEADKKKQVHKKRKCPGPIITYHSLTVPLIGELGPKEENVDVEG | |||
sorAra1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGELGPKEENVDVEG | |||
loxAfr2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
proCap1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
echTel1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG | |||
choHof1 eRRALLKADKRKQVHKKRKCPGPIITYHSVSVPLVR-PGPKEENVDAEg | |||
monDom4 ENYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLAEPGPKEENVDVEG | |||
macEug ENYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLTEPGPKEENVDVEG | |||
sarHar ENYERLEADK<font color="red">R</font>KQVHKRRKCPGPVITYHSMTVPLLTEPGPKEENVDVEG | |||
ornAna1 ------------------------ISFHSLTVPLLADPGAREENVDVEG | |||
galGal3 ENYERLEADKKKQVQKKRKCVGPVIRYWSVTMPLITELG-KEENVDVEG | |||
melGal ENYERLEADKKKQVQKKRKCVGPVIRYWSVTMPLITELG-KEENVDVEG | |||
anoCar1 ETYERLEADKKRQVQKKRKCVGPTIRYYSGTMPLITDLGCKEETVDVEG | |||
xenTro2 ENYERLEADRKKQVHKKRRCVGPTIRHHSLVMPLITELNVKEENVDVEG | |||
tetNig1 ENYERLEADKKKQVQKKRRFDGPTIRYHSVLMPVVSHSVLKEENVDVEG | |||
takRub ENYERLEADKKKQVQKKRRFDGPTVRYHSVLMPIVSHSVLKEENVDVEG | |||
gasAcu1 ENYERLEADKKKQVHKKRRFEGPTIRYHSVLMPLVSHSVLKEENVDVEG | |||
oryLat2 ENYERLEADKKKQVHKKRRFEGPTIRYHSLLMPIVSHSVLKEENVDVEg | |||
danRer5 ENYERLEADKKRQVHMKRQCVGSVIRYHSVLMPLVSDVTLKEENVDVEg | |||
petMar1 ENYERLEADKKKQVLKKHHYTGPVIRYHSLTMPLITELPIKEENVDVEg | |||
* | |||
'''Known variations:''' A breast cancer sample identified I318V as a somatic mutation in this gene; the significance of this is unclear. An [http://www.ncbi.nlm.nih.gov/pubmed/7664828? early report] associates it with repression of transformed cells. These links do not provide a specific connection to the Sarcophilus facial tumor situation. | |||
'''Structural significance:''' No structural matches exist at PDB using blastp. Modbase predicts helical fragments of the 3D structure. Pfam domains are circular references to YL1 (the name of the encoded protein). SwissProt [http://www.expasy.org/uniprot/Q15906 notes] various compositional biases (DE- and P-rich regions) and a phosphoserine at residue 168. | |||
'''Functional significance:''' The specific function is not well understood. VPS72 is generally described as a dna-binding transcriptional regulator possibly involved in chromatin modification and remodeling as a subunit of the NuA4 histone acetyltransferase complex. whose metazoan counterpart is called the [http://www.jbc.org/cgi/content/full/280/14/13665 TRRAP/TIP60 HAT complex]. It is also a subunit of the SNF2-related helicase SRCAP complex. Thus it is localized in the nucleus. | |||
In summary this substitution, if confirmed, could have significant but probably not disabling impacts on the functionality of this gene in view of the extreme intolerance for any kind of substitution at the lysine. However it would be difficult to pursue the impact further given the lack of available structure and complexitities of the VPS72 protein complex and its role in histone modification. | |||
<pre> | |||
>VPS72_homSap | |||
MSLAGGRAPRKTAGNRLSGLLEAEEEDEFYQTTYGGFTE | |||
ESGDDEYQGDQSDTEDEVDSDFDIDEGDEPSSDGEAEEPRRKRRVVTKAYK | |||
EPLKSLRPRKVNTPAGSSQKAREEKALLPLELQDDGSD | |||
SRKSMRQSTAEHTRQTFLRVQERQGQSRRRKGPHCERPLTQEELLREAKITEELNLRSL | |||
ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDIEG | |||
LDPAPSVSALTPHAGTGPVNPPARCSRTFITFSDDATFEEWFPQGRPPKVPVREVCPVTHRPALYRDPVTDIPYATARAFKIIREAYKKYITAHGLPPTASALGPGPPPPEPLPGSGPRALRQKIVIK* | |||
>VPS72_monDom4 | |||
MSLAGGRAPRKTAGNRLSGLLEAEEEDEFYQTTYGGFTE | |||
ESGDDEYQGDQSDTEDEVDSDFDIDEGDEPASDGDGDEPRRKRRVVTKAYK | |||
EPIKSLRPRKVSTPAGSSQKTREEKTLLPLELQDDGLD | |||
SRKSMRQSTAEHTRQTFLRVQERQGQSRRRKGPHCERPLTQEELLREAKITEELNLRSL | |||
ENYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLAEPGPKEENVDVEG | |||
LEPTPVVSAVAPHSGAGPVLPPARCSRTFITFSDDATFEECFPRGKPPKIPVREVCPVTHRPALYRDPVTDIPYATARAFKIIREAYKKYITAHGLPPAASALGPGPPPPEPLPGPGPRALRQKIIIK* | |||
>VPS72_macEug Macropus eugenii cDNA EX201397 | |||
MSLAGGRAPRKTAGNRLSGLLEAEEEDEFYQTTYGGFTE | |||
ESGDDEYQGDQSDTEDEVDSDFDIDEGDEPASDGDGDEPRRKRRVVTKAYK | |||
EPIKSLRPRKVSTPAGSSQKAREEKTLLPLELQDDGVD | |||
SRKSMRQSTAEHTRQTFLRVQERQGQSRRRKGPHCERPLTQEELLREAKITEELNLRSL | |||
ENYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLTEPGPKEENVDVEG | |||
LEPPTLVSTVAPHSGTGPLIPPARCSRTFITFSDDAFEECFPRGKPPKIPVREVCPVTHRPALYRDPVTDIPYATARAFKIIREAYKKYITAHGLSPAASALGPGPPPPEHLPGPGPRALRQKIVIK* | |||
>VPS72_sarHar | |||
MSLAGGRAPRKTAGNRLSGLLEAEEEDEFYQTTYGGFTE | |||
ESGDDEYQGDQSDTEDEVDSDFDIDEGDEPASDGEGDEPRRKRRVVTKAYK | |||
ePIKSLRPRKVSTPAGSSQKAREEKTLLPLELQDDGLD | |||
sRKSMRQSTAEHTRQTFLRVQERQGQSRRRKGPHCERPLTQEELLREAKITEELNLRSL | |||
ENYERLEADKKKQVHKRRKCPGPVITYHSMTVPLLTEPGPKEENVDVEG | |||
LEPIPAVPTAAPHSATGPVIPPARCSRTFITFSDDATFEECFPRGKPPKIPVREVCPVTHRPALYRDPVTDIPYATARAFKIIREAYKKYITAHGLPRLPRPWGPGPPPPEPLPGPGPRALRQKIIIK* | |||
</pre> | |||
=== Case of ABCC1 === | |||
chr6_5144 ABCC1 23 4 Q=2(69) P=2(80) looks like a frame-shift problem in monDom5 | |||
>contig00001 length=802 numreads=10 | |||
HLCFPRLHLDLLHNVLRSPMSFFERTPSGNLVNRFSKEMDTVDSMIPQIIKMFMGSLFNVIGACIIILLATPIAAIIIPPLGLIYFFVQ | |||
....Q.................................................................................... | |||
^ | |||
== Discarded candidates == | |||
Below are three initial candidates that had to be discarded without detailed followup. One arose from repeated frameshifts in the critical region, another exhibited homoplasy with marsupials, and the third too extensive of an accepted reduced alphabet at the site. Thus while these three genes do not meeet the search criteria, they are nonetheless instructive in illustrating those criteria and making clear these are quite restrictive. | |||
=== Case of ACOT12 === | |||
chr3_5872 ACOT12 14 14 I=3(95) V=3(110) 'wobbly' | |||
>contig00001 length=472 numreads=6 | |||
NTYVVAVKSVTLASIPPSPQYNRSEITCAGFLIRAVDSNSCT | |||
.................................Q....S... | |||
^ | |||
Here an I-->V change is seen in some tasmanian devils reads relative to opossum and wallaby. Here V is more typical of a theran mammal. Note I is also seen in armadillo, a placental, and A occurs in platypus and various other mammals. ACOT12, a acyl-CoA thioesterase, does not track back well in earlier diverging species. Because of the observed homoplasy, this locus is an unsuitable example of a significant amino acid change in Sarcophilus. However it illuminates the nature of suitable candidates and so is retained here. | |||
^ | |||
ACOT12_hg18_14 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSNSCI | |||
ACOT12_panTro2 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSNSCI | |||
ACOT12_gorGor1 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSNSCI | |||
ACOT12_ponAbe2 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSNSCI | |||
ACOT12_rheMac2 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSSSCI | |||
ACOT12_calJac1 NTYTVAVKSVMLPS V PPSPQYIRSEIICAGFLIHAIDSNSCI | |||
ACOT12_micMur1 NTYTVAVKSVILPS V PPSPQHVRSEIICAGFLIHAADSNSCT | |||
ACOT12_otoGar1 NTYMVAAKSVILPS V PPSPQYIRSEIICAGFLIHTIDSTSCT | |||
ACOT12_tupBel1 NTYTVAVKSVTLPS V PPSPQYIRSDIICAGFLIRPVDSSSCT | |||
ACOT12_mm9_14_ NTYTVALRSVVLPS V PSSPQYIRSEVICAGFLIQAVDSNSCT | |||
ACOT12_rn4_14_ NTYIVALMSVVLPS V PPSPQYIRSQVICAGFLIQPVDSSSCT | |||
ACOT12_dipOrd1 NTYVVATKSVILPS V PPSPAYIRSEAVCSGFLIKAVDSSSCT | |||
ACOT12_cavPor3 DTYLVAVKSVVLPA V PPSPGYTRSEVALAGFLIQPTDHSSCT | |||
ACOT12_oryCun1 HAYTVAAKSVMLPS A PPSPDHTRSEIICAGFLIHAIDSHSCT | |||
ACOT12_ochPri2 HAYVVAVKSVVLPS A PPSPEYIRGEIVCAGFLIHAIDSHACT | |||
ACOT12_vicPac1 NTYTVAVKSVILPS V PPSPQYVRSEITCAGFLIHAIDNSSCT | |||
ACOT12_turTru1 HTYTVAVRSVILAS V PPSPQYSRSEIISAGFLIRAIDSSSCT | |||
ACOT12_bosTau4 HTYVVAVRSVILPS V PPSPQYVRSEIECAGFLIHATDSSSCT | |||
ACOT12_equCab2 KTFSVAAKSVILPS V PPSPQYMRSEIRCAGFLICAIDNSSCT | |||
ACOT12_felCat3 STYTVAVKSVLLPS V PPCPHYIRSEIICAGFLIRAIDSSSCT | |||
ACOT12_canFam2 NTYTVAVKSVTLPS V PPSPQYSRSEILCAGFLIHAIDSSSCT | |||
ACOT12_myoLuc1 NTYTVAVKSVILPS V PPSPQYVRSEIICAGFLIHAIDSSSCT | |||
ACOT12_pteVam1 NTYTVAVKSVILPS V PPSPZYVRSEIVCAGFLIHAIDGSLCI | |||
ACOT12_eriEur1 STFTVAMKSVLLAS V PSSPQYIRSEITCAGFVIHAVSSNSCI | |||
ACOT12_sorAra1 NAFTVAVKSVILPS V PPSPQYMRSEIICAGFLIHATDSNSCI | |||
ACOT12_loxAfr2 D--TVAVKSVLLPS V PPCPQYIRSEIIRAGFLIHTIDSNSCT | |||
ACOT12_echTel1 TTYTVALRSVLLPS V PSSPNYVRGEIICAGFLVHPIDSSACT | |||
ACOT12_dasNov2 NTYTVAVKSVVLPS I PPSPQYIRSEIICAGFLIHAIDSSSCT | |||
ACOT12_choHof1 NSYTVAAKSVVLPS V PPSPQYIRSETICAGFLINAIDSSSCT | |||
ACOT12_monDom4 NTYVVAVKSVTLAS I PPSPQYNRSEITCAGFLIRAVDSNSCT | |||
ACOT12_macEug NTYVVAMKSVTLAS I PPSPQYNRSEITSAGFLIQAVDSNSCT | |||
ACOT12_sacHar1 NTYVVAVKSVTLAS I PPSPQYNRSEITCAGFLIQAVDSSSCT | |||
ACOT12_sacHar2 NTYVVAVKSVTLAS V PPSPQYNRSEITCAGFLIQAVDSSSCT | |||
ACOT12_ornAna1 DSYLVAVKSVILAS A PPSHQYIRSEIPCAGFLVEALDSSSCK | |||
=== Case of FLI1 === | |||
chr4_11174 FLI1 3 32 N=2(63) K=3(47) | |||
>contig00001 length=575 numreads=9 | |||
ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEKNGPPPNMTTNERRVIVPA | |||
>contig00001 length= | .................................................. | ||
^ | |||
................................... | Here the N-->K change is a non-conservative substitution in the sense asperagine is merely polar whereas lysine is bulkier and negatively charged. The N is highly invariant at this position back to teleost fish. FLI1 is a transcription factor associated with a leukemia virus integration site and Ewing sarcoma. | ||
Here | |||
This would be a promising candidate except for the fact that the three reads establishing K clearly are plagued by frameshifts at the critical region. Possibly anomalous base composition is responsible here (ggatgagaagaacggcccccctcc) -- which is no doubt giving rising to transcriptional slippage generating homoplasic deletions of polyP -- or perhaps low coverage. This change is unlikely to be validated upon additional bulk or targeted sequencing because these lack motivating evidence. | |||
>FP1JAYN01BA7O5 and FP5M7SR01ERAQP Frame = +1 Frame = +2 | |||
Query: 1 ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEKNGPP 36 KNGPPPNMTTNERRVIVPA 50 | |||
ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEK P KNGPPPNMTTNERRVIVPA | |||
Sbjct: 37 ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEKKRSP 144 KNGPPPNMTTNERRVIVPA 187 | |||
>FP1JAYN01DX0A1 length=254 | |||
Query: 1 ESPVDCSVNKCSKLVGGNESNPMN-YNTYMDEKNGPPPNMTTNERRVIVPA 50 | |||
ESPVDCSVNKCSKLVGGNESNPMN + + EK PPPNMTTNERRVIVPA | |||
Sbjct: 37 ESPVDCSVNKCSKLVGGNESNPMNLQHLHG*EKTVPPPNMTTNERRVIVPA 189 | |||
N P M N Y N T Y M D E K N G P P P N M T | |||
| | | | | | | | | | | | | | | | | | | | | |||
aatcctatgaattacaatacctacatggatgagaagaacggcccccctcctaacatgacc | |||
FLI1_hg18_3_ ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_panTro2 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_gorGor1 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_ponAbe2 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_rheMac2 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_calJac1 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_tarSyr1 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_micMur1 ESPVDCSVSKCGKLIGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_otoGar1 ESPVDCSVSKCSKLIGGGEANPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_mm9_3_9 ESPVDCSVSKCNKLVGGGEANPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_rn4_3_9 ESPVDCSVSKCNKLVGGGEANPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_dipOrd1 ESPVDCSVSKCSKLVGGGESNPMNYNSYIDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_cavPor3 ESPVDCSVSKCSKLVGTGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_speTri1 ---VDCSVSKCSKLVFGGESNPMNYNSYLDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_oryCun1 ESPVDCSISKCGKLVGGGEANAMSYNNYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_vicPac1 ESPVDCSVSKCGKLVGGGESNTMSYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_turTru1 ESPVDCSVSKCGKLVGGGESNAMSYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_bosTau4 ESPVDCSVSKCGKLVGGGESNTMSYTSYVDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_equCab2 ESPVDCSVSKCSKLVGGGESNAMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_canFam2 ESPVDCSVSKCSKLVGGSESNAMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_myoLuc1 ESPVDCSVSKCSKLVGGGESNAMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_pteVam1 ESPVDCSVSKCSKLVGGGESNAMNYNSYIDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_eriEur1 ESPVDCSVSKCSKLVGGGESNAMNYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_proCap1 ESPVDCSVSKCSKLAGGGESNPMNYNTYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_dasNov2 ESPVDCSVSKYSKLVGGGESNPMTYSTYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_choHof1 ESPVDCSVSKCSKLVGGGEATPMTYNTYMDEK N GPP-PNMTTNERRVIVPA | |||
FLI1_monDom4 ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEK N GPP-PNMTTNERRVIVPA | |||
FLI1_macEug ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEK N GPP-PNMTTNERRVIVPA | |||
FLI1_sarHar1 ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEK N GPP-PNMTTNERRVIVPA | |||
FLI1_ornAna1 ESPVDCSVSKCGKLVGSGESNPMNYNSYMEEK N GPPPPNMTTNERRVIVPA | |||
FLI1_galGal3 ESPVDCSVNKCSKLVGAGESNPMSYSTYMDEK N GPP-PNMTTNERRVIVPA | |||
FLI1_taeGut1 ESPVDCSMNKCGKLVGAGESNPMSYSTYMDEK N GPP-PNMTTNERRVIVPA | |||
FLI1_anoCar1 ESPVDCSVSKCNKLVPAGESNSLNYGTYMDEK N GPP-PNMTTNERRVIVPA | |||
FLI1_xenTro2 ESPVDCSISKCSKLIGGSENNAVTYNSYMDEK N GPPPPNMTTNERRVIVPA | |||
FLI1_tetNig1 ESPVDCSVGKCNKLVGGNDVSQMSYGSYMDEK N APP-PNMTTNERRVIVPA | |||
FLI1_fr2_3_9 ESPVDCSVGKCNKLVGGNDVSQMNYGSYMDEK N APP-PNMTTNERRVIVPA | |||
FLI1_gasAcu1 ESPVDCSVGKCNKLVGSNDTSQMNYGNYMDEK N APP-PNMTTNERRVIVPA | |||
FLI1_oryLat2 ESPVDCSVGKCNKLVGGNDTSQMTYGNYMDEK S APP-PNMTTNERRVIVPA | |||
FLI1_danRer5 ESPVDCSVGKCNKMVGGTEASQMNYTGYMDEK C APP-PNMTTNERRVIVPA | |||
== | === Case of SPON1 === | ||
chr5_8347 SPON1 11 20 V=3(65) I=2(66) wobbly | |||
>contig00001 length=433 numreads=5 | |||
GSTCTMSEWITWSPCSISCGVGMRSRERYVKQFPEDGSVCTVPTEETEKCTVNEEC | |||
......................................I.N............... | |||
^ | |||
Here two Sarcophilus reads show V-->I following residue 20 while three are V like opossum. It quickly emerges that wallaby also has I. Thus the change in tasmanian devil is within the normal reduced alphabet of this residue position. Various placentals show that T and M and even P are also accepted substituents here. Note too these are used clade-incoherently (eg primates alone are variable). Consequently this site is not under strong selection for V to begin with so SPON1 does not meet the selection criteria being used here. | |||
^ | |||
SPON1_hg18_13 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_panTro2 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_gorGor1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_ponAbe2 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_rheMac2 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_calJac1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_tarSyr1 -GSTCTMSEWITWSPCCLSCV P GMRSREYYLK-FFEDGSVCSLTPKKTQNRTV-EZC | |||
SPON1_micMur1 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_otoGar1 DGSTCTMSEWITW-PCSISCG T GMRSRERYVKQFPEDVSVCTLPTEETEKCTVNEEC | |||
SPON1_tupBel1 EGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_mm9_13_ DGSTCTMSEWITWSPCSVSCG M GMRSRERYVKQFPEDGSVCMLPTEETEKCTVNEEC | |||
SPON1_rn4_13_ DGSTCTMSEWITWSPCSVSCG M GMRSRERYVKQFPEDGSVCMLPTEETEKCTVNEEC | |||
SPON1_dipOrd1 -GSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_cavPor3 DGSTCTMSEWIIWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_speTri1 EHSTCTMSEWITWSPCCISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_oryCun1 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_ochPri2 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_turTru1 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPT-ETEKCTVNEEC | |||
SPON1_bosTau4 DGSTCTMSEWITWSPCSISCG T GTRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_equCab2 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_canFam2 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_myoLuc1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_pteVam1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_eriEur1 DGSACTMSEWITWSPCSLSCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_sorAra1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_proCap1 -GSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_echTel1 ----CPMSEWITWSPRSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_dasNov2 -GSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC | |||
SPON1_monDom4 DGSTCTMSEWITWSPCSISCG V GMRSRERYVKQFPEDGSVCTVPTEETEKCTVNEEC | |||
SPON1_macEug GSTCTMSEWMTWSPCSISCG I GMRSRERYVKQFPEDGSVCTVPTEETEK | |||
SPON1_sacHar1 GSTCTMSEWITWSPCSISCG V GMRSRERYVKQFPEDGSICNVPTEETEKCTVNEEC | |||
SPON1_sacHar2 GSTCTMSEWITWSPCSISCG I GMRSRERYVKQFPEDGSICNVPTEETEKCTVNEEC | |||
SPON1_ornAna1 DGSTCTMSEWITWSPCSVSCG M GMRSRERYVKQFPDDGSMCKVPTEETEKCVVNEDC | |||
SPON1_anoCar1 DGSTCMMSEWITWSPCSVSCG M GMRSRERYVKQFPDDGSMCKVPTEETEKCIVNEEC | |||
SPON1_xenTro2 EASTCMMSEWITWSPCSASCG M GMRSRERYVKQFPEDGSMCKVPTEETEKCIVNEEC | |||
SPON1_tetNig1 DASTCMMSEWITWSPCSASCG M GSRSRERYVKQFPDDGSICTLPTEETEDCVVNEEC | |||
SPON1_fr2_13_ DASTCMMSEWITWSPCSASCG M GSRSRERYVKQFPDDGSICTLPTEETEDCVVNEEC | |||
SPON1_gasAcu1 DASTCMLSEWITWSPCSLSCG M GTRSRERYVKQFPDDGSLCSLPTEETDNCVVNEEC | |||
SPON1_oryLat2 DGSTCMMSEWITWSPCSMSCG A GIRSRERYVKQFPDDGSICTLPTEETENCVVNEEC | |||
SPON1_danRer5 DSSTCMMSEWITWSPCSVSCG S GLRSRERYVKQFPDDGFACTHPTEETEPCTVNEEC | |||
== | == Marsupial data availability == | ||
Scattered data is available for other marsupials and monotremes from 454 reads, Sanger trace data and transcripts: | |||
Didelphis virginiana 88,207 traces 248 nuc | |||
Trichosurus vulpecula 169,115 traces 321 nuc 147,199 ests | |||
Sminthopsis crassicaudata 59 nuc 1,669 ests | |||
Sminthopsis macroura 3,411 traces 89 nuc | |||
Isoodon macrourus 6,144 traces, 70 nuc 1,319 ests | |||
Tachyglossus aculeatus 93,653 traces 243 nuc | |||
SRX000015 Baylor 454 sequencing of Monodelphis domestica genomic fragment library | |||
SRX000086 WUGSC 454 sequencing of Macropus eugenii genomic fragment library | |||
SRX000186 WUGSC 454 sequencing of Ornithorhynchus anatinus transcript | |||
SRX000122 WUGSC 454 sequencing of Tachyglossus aculeatus transcript | |||
SRX000121 WUGSC 454 sequencing of Tachyglossus aculeatus transcript | |||
The | The running estimate of coverage of Sarcophilus genome combining all runs for 11 expected genes on different chromosomes: | ||
59 of 68 exons found (87%) | |||
3883 of 4339 amino acids available (89%) | |||
Newbler has a bad tendency to create non-existent frameshifts as seen in these three reads for the same query gene: | |||
Query: 82 ggtctctacggcagtgtcattgtcactggagggaacacactcttgcaagg | |||
|||||||||||||||||||||| |||||||||||||||||| |||||||| | |||
Sbjct: 167 ggtctctacggcagtgtcattg-cactggagggaacacactgttgcaagg FP1I63R01APY7E | |||
Query: 82 ggtctctacggcagtgt-cattgtcactggagggaacacactcttgcaagg | |||
||||||||||||||||| |||||||||||||||||||||||| |||||||| | |||
Sbjct: 268 ggtctctacggcagtgttcattgtcactggagggaacacactgttgcaagg FKUJDAX01AWWZ3 | |||
Query: 82 ggtctctacggcagtgtcattgtcactggagggaac-acactcttgcaagg | |||
|||||||||||||||||||||||||||||||||||| ||||| |||||||| | |||
Sbjct: 268 ggtctctacggcagtgtcattgtcactggagggaacgacactgttgcaagg FKUJDAX01DZSZO | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 11:01, 27 January 2011
Introduction to Marsupial phyloSNPs
In this project, new genomic data from the Tasmanian devil (Sarcophilus harrisii), Tasmanian tiger (Thylacinus cynocephalus), and echidna (Tachyglossus aculeatus) are analyzed for significant changes at the protein coding level. The goal is to find single amino acid changes in one of these species at a highly invariant residue in a well-conserved exon in a gene with known or predictable tertiary structure. Such changes are thought to enrich for genetic changes with significant, adaptive biochemical or phenotypic consequences (1,2,3,4), in contrast to ordinary SNPs at positions of low conservation. Thus phyloSNPs are informative to the distinctive biology of the species carrying them and suggest a focus for subsequent experiment.
It is also of particular interest to determine the levels of variation within the Tasmanian devil population as a whole because the number of individuals have become low and possibly inbreed with adverse sequelae. For this it will be necessary to first determine sites of variation and then to genotype them across a large number of individuals.
Marsupial genomic and cDNA data to date has been quite limited compared to placental mammal. Yet as outgroup, metatheran animals provide important context to placentals and represent important context in understanding human protein evolution. The monotheres are inevitably limited by the paucity of extant species (basically platypus and echidna) and dim prospects for fossil DNA. Consequently echidna provides an important adjunct to the existing but incomplete platypus assembly. While extant birds and reptiles -- the preceding divergence node -- are abundant it must be remembered that a very considerable time elapsed (from 310 mry to 175 mry) prior to divergence of mammals with living representatives. This gap of 135 myr is comparable to the whole evolutionary record of theran mammals.
Assumed vertebrate phylogenetic tree
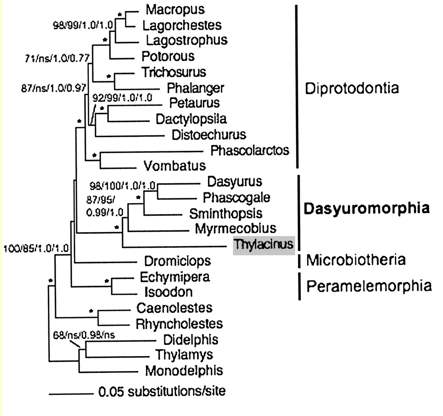
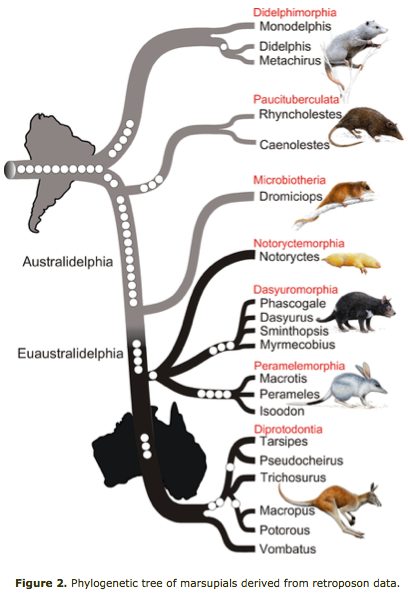
Marsupial relationships are taken from a 2009 paper establishing the mitochondrial genome sequences of the Tasmanian tiger (Thylacinus cynocephalus) and numbat (Myrmecobius fasciatus). A slightly different topology was found using transposons in an excellent July 2010 PLOS paper (right).
Newick tree that generates a marsupial-centric vertebrate phylogenetic tree: ((((((((((((sarHar,smiCra),myrFas),thyCyn),(macEug,triVul)),monDom), ((((loxAfr,proCap),echTel),(dasNov,choHof)), ((((((bosTau,turTru),susScr),vicPac),((equCab,(felCat,canFam)),(myoLuc,pteVam))),(eriEur,sorAra)), (((((((((homSap,panTro),gorGor),ponPyg),macMul),calJac),tarSyr),(micMur,otoGar)),tupBel), (((((musMus,ratNor),dipOrd),cavPor),speTri),(oryCun,ochPri)))))), (ornAna,tacAcu)), ((galGal,taeGut),anoCar)), xenTro), (((tetNig,takRub),(gasAcu,oryLap)),danRer)), calMil), petMar); Newick tree that generates the homo-centric vertebrate phylogenetic tree: ((((((((((((((((((homSap,panTro),gorGor),ponPyg),macMul),calJac),tarSyr),(micMur,otoGar)),tupBel), (((((musMus,ratNor),dipOrd),cavPor),speTri),(oryCun,ochPri))), (((((vicPac,susScr),turTru),bosTau),((equCab,(felCat,canFam)),(myoLuc,pteVam))),(eriEur,sorAra))), (((loxAfr,proCap),echTel),(dasNov,choHof))), (monDom,((macEug,triVul),(sarHar,thyCyn)))), (ornAna,tacAcu)), ((galGal,taeGut),anoCar)), xenTro), (((tetNig,takRub),(gasAcu,oryLap)),danRer)), calMil), petMar);
Phylo-sorting data
This tab-delimited table enables four different sort orders. These are needed because data can be missing from species in a manner that varies by gene, making data alignment difficult. Some alignment tools also lose input order, so that needs to be recovered. The ordering here flattens the phylogenetic tree by taking human (arbitrarily) at the top and resolving ambiguous situations (eg mouse, rat) by putting species with the best assemblies first.
The first two columns provide sort order number for the 44 species alignment at UCSC as phylogenetic and alphabetic order respectively. The third and fourth columns do this for a larger set of 53 species for which data is commonly available (notably in marsupials). The fifth column supplies the genSpp acronym and the sixth the Newick tree format syntax. These two columns by themselves will correctly draw the vertebrate phylogenetic tree in all online software without further editing. The final columns provide genus, species, and common name.
.. .. .. .. ...... (((((((((((( 46 10 54 10 anoCar )), Anolis carolinensis (lizard) 29 11 22 11 bosTau , Bos taurus (cow) 15 12 38 12 calJac ), Callithrix jacchus (marmoset) 62 54 61 13 calMil ), Callorhinchus milii (elephantfish) 32 13 28 14 canFam )),( Canis familiaris (dog) 23 14 46 15 cavPor ), Cavia porcellus (guinea_pig) 41 15 21 16 choHof )),(((((( Choloepus hoffmanni (sloth) 52 16 60 17 danRer )), Danio rerio (zebrafish) 40 17 20 18 dasNov , Dasypus novemcinctus (armadillo) 22 18 45 19 dipOrd ), Dipodomys ordii (kangaroo_rat) 39 19 19 20 echTel ),( Echinops telfairi (tenrec) 30 20 26 21 equCab ,( Equus caballus (horse) 35 21 31 22 eriEur , Erinaceus europaeus (hedgehog) 31 22 27 23 felCat , Felis catus (cat) 44 23 52 24 galGal , Gallus gallus (chicken) 50 24 58 25 gasAcu , Gasterosteus aculeatus (stickleback) 12 25 35 26 gorGor ), Gorilla gorilla (gorilla) 10 26 33 27 homSap , Homo sapiens (human) 37 27 17 28 loxAfr , Loxodonta africana (elephant) 58 56 14 29 macEug , Macropus eugenii (wallaby) 14 28 37 30 macMul ), Macaca mulatta (rhesus) 17 29 40 31 micMur , Microcebus murinus (mouse_lemur) 42 30 16 32 monDom ),(((( Monodelphis domestica (opossum) 20 31 43 33 musMus , Mus musculus (mouse) 33 32 29 34 myoLuc , Myotis lucifugus (microbat) 56 57 12 35 myrFas ), Myrmecobius fasciatus (numbat) 26 33 49 36 ochPri )))))),( Ochotona princeps (pika) 43 34 50 37 ornAna , Ornithorhynchus anatinus (platypus) 25 35 48 38 oryCun , Oryctolagus cuniculus (rabbit) 51 36 59 39 oryLap )), Oryzias latipes (medaka) 18 37 41 40 otoGar )), Otolemur garnettii (bushbaby) 11 38 34 41 panTro ), Pan troglodytes (chimp) 53 39 62 42 petMar ) Petromyzon marinus (lamprey) 13 40 36 43 ponPyg ), Pongo pygmaeus (orang) 38 41 18 44 proCap ), Procavia capensis (hyrax) 34 42 30 45 pteVam ))),( Pteropus vampyrus (macrobat) 21 43 44 46 ratNor ), Rattus norvegicus (rat) 54 58 10 47 sarHar , Sarcophilus harrisii (tasmanian_devil) 55 59 11 48 smiCra ), Sminthopsis crassicaudata (dunnart) 36 44 32 49 sorAra )),((((((((( Sorex araneus (shrew) 24 45 47 50 speTri ),( Spermophilus tridecemlineatus (squirrel) 60 60 24 51 susScr ), Sus scrofa (pig) 61 61 51 52 tacAcu )),(( Tachyglossus aculeatus (echidna) 45 46 53 53 taeGut ), Taeniopygia guttata (finch) 49 47 57 54 takRub ),( Takifugu rubripes (fugu) 16 48 39 55 tarSyr ),( Tarsius syrichta (tarsier) 48 49 56 56 tetNig , Tetraodon nigroviridis (pufferfish) 57 62 13 57 thyCyn ),( Thylacinus cynocephalus (tasmanian_tiger) 59 63 15 58 triVul )), Trichosurus vulpecula (bushytail_possum) 19 50 42 59 tupBel ),((((( Tupaia belangeri (tree_shrew) 28 51 23 60 turTru ), Tursiops truncatus (dolphin) 27 52 25 61 vicPac ),(( Vicugna pacos (lama) 47 53 55 62 xenTro ),((( Xenopus tropicalis (frog) 44 44 53 53 genSpp tree_syntax genus species common ph al ph al
Candidate analysis
The first issue is error within the primary reads themselves; the second is whether the default 454 Newbler assembler correctly identified overelapping reads and put them together properly to give exon-spanning reads. Those issues are discussed elsewhere -- here it is assumed the reads at the PSU blast site are correct, so the entire focus is on subsequent bioinformatics. In some cases that results in retrospective identification and correction of errors, notably introduced frameshifts that are far too common.
After thorough evaluation, candidates are given a final heuristic score based on awarding 0,1,or 2 points for the following 13 critera:
- the change is real: multiple reads support each of the two amino acid values
- quality coverage: the entire exon can be recovered from multiple reads without manual frameshift correction
- processed pseudogenes can be recognized by reads long enough at flanks to identify neighoring exons now adjacent (resp GT-AG splice donors)
- non-processed pseudogenes can be distinguished by recovery of additional exons of the gene with expected levels of conservation
- paralogs and internal repeats are readily distinguishable from the exon under stead
- phylogenetic depth: multiple marsupials, monotremes, all placental branches, fish, chondrichthyes, possibly lamprey available
- homoplasy: the reduced alphabet consists of a single amino acid with the exception of Sarcophilus
- appropriate character of the change in amino acid properties
- amenability to accurate rapid scoring in many individual animals
- interpretability of structural significance of change within 3D structure or characterized domain
- interpretability of functional role of overall gene and of region containing the amino acid change
- previous relevant publications, animal kockout models, known human ortholog disease SNPs
- plausible relevancy of the change to cancer or facial tumor
When scoring is finished, the dummy table below will be filled in with real data and genes will become sorted by highest overall score (or by preferred columns appropriate to specialized purposes).
..... valid cover psgen paral depth alpha AAcha popul struc funct pubmd tumor ERN2 1 1 1 1 1 1 1 1 1 1 1 1 12 MGAT5 1 1 1 1 1 1 1 1 1 1 1 1 12 ACTL6B 1 1 1 1 1 1 1 1 1 1 1 1 12 IPO7 1 1 1 1 1 1 1 1 1 1 1 1 12 PPFIA3 1 1 1 1 1 1 1 1 1 1 1 1 12 WDFY3 1 1 1 1 1 1 1 1 1 1 1 1 12 XYLT1 1 1 1 1 1 1 1 1 1 1 1 1 12 ATP4A 1 1 1 1 1 1 1 1 1 1 1 1 12 VPS72 1 1 1 1 1 1 1 1 1 1 1 1 12 ABCC1 1 1 1 1 1 1 1 1 1 1 1 1 12 ACOT12 1 1 1 1 1 1 1 1 1 1 1 1 12 FLI1 1 1 1 1 1 1 1 1 1 1 1 1 12 SPON1 1 1 1 1 1 1 1 1 1 1 1 1 12 ..... 13 13 13 13 13 13 13 13 13 13 13 13 .....
Case of ERN2
chr6_5971 ERN2 4
contig00001 length=355 numreads=5
KLPFTIPELVHASPCRSSDGVLYT
.....................F..
^
15 R=3(75) H=2(50
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by L->F), then differences between the two tasmanian devils
(here one individual has R at position 15, the other has H), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues: ERN2 has not generated potentially confusing recent processed pseudogenes in mammals (lack of human, opossum or platypus genome Blat matches to ERN2 query). The variation observed here between individual tasmanian devils is implausibly an early stage in the loss of parent gene because of ERN2 functional essentiality; the exon cannot come from a decaying segmental duplication because coverage is high enough to also detect the main gene.
Paralog issues: The GeneSorter tool at UCSC shows a single significant full-length paralog in human, ERN1, also with 22 coding exons. The genes reside on different chromosomes but in regions with local homology of synteny. However this particular exon is a good match (3 differences out of 23), so there is potential for experimental difficulties in distinguishing them in short reads (including the following exon readily resolves them bioinformatically). In any event, at positions 15 and 20, ERN1 is identical at the amino acid level to ERN2. The gene duplication appears to have occured subsequent to amphioxus divergence earlier diverging metazoans are single-copy.
Homoplasy (recurrent mutation) issues: This exon is very conserved and does not exhibit repetitive sequence, compositional simplicity, or indels in any species in either paralog that could foster experimental error or alignment ambiguity. At position 15, the ancestral value is arginine in both paralogs. The G--> A transition to histidine in one individual is conservative under most circumstances (still basic) and arises from an arginine codon CpG hotspot conserved back to lamprey in 30 of 32 species with available data, yet histidine is not observed part of a reduced alphabet (ie R/H) at this position over many billions of years of branch length. Consequently R-->H is a significant change in this individual tasmanian devil.
Known variations: No human disease variants have been reported for either ERN2 or ERN1, probably attributable to essentiality. Site-specific mutation close to the exon here have been generated for K121P, D123P, W125A, and Q105E but only for ERN1. Naturally occuring coding SNPs in the human population relevent to the ERN2 exon are not known but low frequency alleles could emerge from the 1000 Genomes Project.
Side issues: a very ancient conserved leucine at position 21 appears to be transitioning to phenylalanine at marsupial node but has not been fixed, so settles out as L or F depending on lineage-sorting on each terminal marsupial leaf whereas placentals are all changed to phenylalanine (a phyloSNP caught in mid-air). While L and F might seem about the 'same' as amino acids, the branch length conservation totals say both are important but for different reasons: this is not a waffle codon nor reduced alphabet situation. This raises the question -- given the extreme conservation of this exon otherwise -- of whether the L-->F change at position 21 in both individuals has 'enabled' (made neutral or adaptive) an otherwise unfavorable R-->H change at position 15 in one individual.
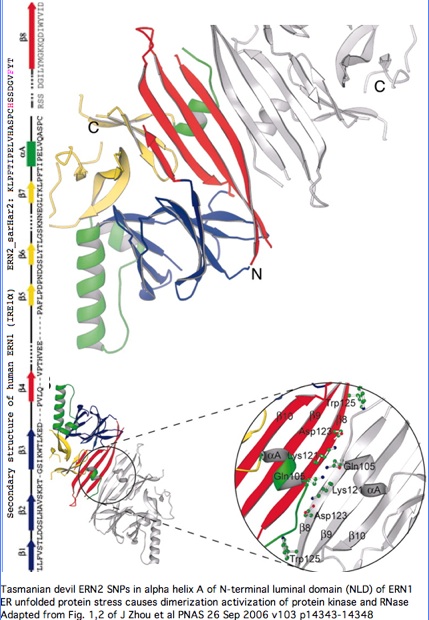
Structural significance: By good fortune, the crystal structure of ERN1 (alternately called IRE1) has been published. The PDB 2HZ6 structure has good coverage of this particular exon. Consequently the marsupial ERN2 could be very accurately modelled and the structural effects of L-->F with or without R-->H computed by submission to online SwissProt modelling service.
Monodelphis ERN2 (key exon: sarHar2) aligned to human ERN1 luminal domain
Expect = 5.8e-65 Identities = 109/180 (60%), Positives = 141/180 (78%)
ERN2_monDom 1 PESLLFISTLDGSLHAVSKKTGDIQWTLKDDPIIQGPVYATEPAFLPDPSDGSLYILGEE 60
PE+LLF+STLDGSLHAVSK+TG I+WTLK+DP++Q P + EPAFLPDP+DGSLY LG +
ERN1_homSap 8 PETLLFVSTLDGSLHAVSKRTGSIKWTLKEDPVLQVPTHVEEPAFLPDPNDGSLYTLGSK 67
ERN2_monDom 61 SKQGLMKLPFTIPELVHASPCHSSDGVFYTGRKQDTWFMVDPKSGKKQTMLSTETWDGLY 120
+ +GL KLPFTIPELV ASPCRSSDG+LY G+KQD W+++D +G+KQ LS+ D L
ERN1_homSap 68 NNEGLTKLPFTIPELVQASPCRSSDGILYMGKKQDIWYVIDLLTGEKQQTLSSAFADSLC 127
ERN2_monDom 121 PSAPLLYIGRTQYTVTMYDPRSQALRWNTTYRGYSAPLLDHLPGYQVGHFTCSGEGLVVT 180
PS LLY+GRT+YT+TMYD +++ LRWN TY Y+A L + Y++ HF +G+GLVVT
ERN1_homSap 128 PSTSLLYLGRTEYTITMYDTKTRELRWNATYFDYAASLPEDDVDYKMSHFVSNGDGLVVT 187
Functional significance: A considerable amount is known about the paralog ERN1. Annotation transfer is likely applicable to ERN2. The two gene products differ primarily in expression -- ERN1 ubiquitious but ERN2 restricted to intestinal epithelial cells:
"The unfolded protein response (UPR) is an evolutionarily conserved mechanism by which all eukaryotic cells adapt to the accumulation of unfolded proteins in the endoplasmic reticulum (ER). Inositol-requiring kinase 1 (IRE1 or ERN1) and PKR-related ER kinase (PERK) are two type I transmembrane ER-localized protein kinase receptors that signal the UPR through a process that involves homodimerization and autophosphorylation... The monomer of the luminal domain comprises a unique fold of a triangular assembly of beta-sheet clusters. Structural analysis identified an extensive dimerization interface stabilized by hydrogen bonds and hydrophobic interactions... Mutations that disrupt the dimerization interface produced ERN1 protein that failed to either dimerize or activate the UPR upon ER stress."
"ERN1 is a type I transmembrane protein kinase receptor that also has a site-specific RNase activity that, upon activation, initiates a site-specific unconventional splicing reaction. The substrate for IRE1 RNase in metazoans is Xbp1 mRNA, which encodes a basic leucine zipper transcription factor of the ATF/CREB family. XBP1 controls expression of genes containing an X-box element or a UPR element in their promoter regions. The IRE1-mediated splicing reaction introduces into XBP1 an alternative C terminus, thereby generating an XBP1 molecule that is a more potent transcriptional activator. Therefore, activation of IRE1 and its RNase increases the transcription of genes encoding ER chaperones and folding catalysts... the ERN1 N-terminal luminal domain (NLD) functions as an ER stress sensor... under normal conditions IRE1 is maintained in a monomeric state through interaction of the NLD with the ER resident chaperone BiP. Upon ER stress, Grp78 binds to unfolded proteins as they accumulate, permitting the released NLD to form homodimers. Dimerization of the NLD in turn leads to the activation of the protein kinase and RNase activities in the cytosolic domain of ERN1."
ENR2 is readily distinguished from its ERN1 paralog at tBlastn by including the two following exons which bring percent identity to 62%:
ERN2_monDom KLPFTIPELVHASPCRSSDGVLYTGRKQDTWFMVDPKSGKKQTMLSTETWDGLYPSAPLLYIGRTQYTVTMYDPRSQALRWNTTYRGYSA
KLPFTIPELV ASPCRSSDG+LY G+KQD W++VD +G+KQ LS+ + L PS LLY+GRT+YT+TM+D +S+ LRWN TY Y+A
ERN1_monDom KLPFTIPELVQASPCRSSDGILYMGKKQDIWYVVDLMTGEKQQTLSSAFAESLCPSTSLLYLGRTEYTITMFDTKSRELRWNATYFDYAA
The first alignment shows ERN2 orthologs in vertebrates, the second as difference relative to opossum, the third ERN1 orthologs.
The ancestral nature of the CpG hotspot is shown in nucleotides in the final columns.
^ * ^ * ^ *
ERN2_homSap KLPFTIPELVHASPCRSSDGVFYT ERN2_homSa .....................F.. ERN1_homSap KLPFTIPELVQASPCRSSDGILYM CG Human
ERN2_panTro KLPFTIPELVHASPCRSSDGVFYT ERN2_panTr .....................F.. ERN1_panTro KLPFTIPELVQASPCRSSDGILYM CG Chimp
ERN2_ponAbe KLPFTIPELVHASPCRSSDGVFYT ERN2_ponAb .....................F.. ERN1_ponAbe KLPFTIPELVQASPCRSSDGILYM -- Gorilla
ERN2_rheMac KLPFTIPELVHASPCRSSDGVFYT ERN2_rheMa .....................F.. ERN1_rheMac KLPFTIPELVQASPCRSSDGILYM CG Orangutan
ERN2_calJac KLPFTIPELVHASPCRSSDGVFYT ERN2_calJa .....................F.. ERN1_calJac KLPFTIPELVQASPCRSSDGILYM CG Rhesus
ERN2_tarSyr KLPFTIPELVHASPCRSSDGVFYT ERN2_tarSy .....................F.. ERN1_tarSyr KLPFTIPELVQASPCRSSDGILYM CG Marmoset
ERN2_micMur KLPFTIPELVHASPCRSSDGVFYT ERN2_micMu .....................F.. ERN1_micMur KLPFTIPELVQASPCRSTDGILYM CG Tarsier
ERN2_tupBel KLPFTIPELVHASPCRSSDGVFYT ERN2_tupBe .....................F.. ERN1_otoGar KLPFTIPELVQASPCRSSDGILYM CG Mouse_lemur
ERN2_musMus KLPFTIPELVHASPCRSSDGVFYT ERN2_musMu .....................F.. ERN1_tupBel KLPFTIPELVQASPCRSSDGILYM -- Bushbaby
ERN2_ratNor KLPFTIPELVHASPCRSSDGVFYT ERN2_ratNo .....................F.. ERN1_musMus KLPFTIPELVQASPCRSSDGILYM CG TreeShrew
ERN2_cavPor KLPFTIPELVHTSPCRSSDGVFYT ERN2_cavPo ...........T.........F.. ERN1_ratNor KLPFTIPELVQASPCRSSDGILYM CG Mouse
ERN2_speTri KLPFTIPELVHASPCRSSDGVFYT ERN2_speTr .....................F.. ERN1_dipOrd KLPFTIPELVQASPCRSSDGILYM CG Rat
ERN2_oryCun KLPFTIPELVHASPCRSSDGVFYT ERN2_oryCu .....................F.. ERN1_cavPor KLPFTIPELVQASPCRSSDGILYM -- Kangaroo_rat
ERN2_ochPri KLPFSIPELVHASPCRSSDGVFYT ERN2_ochPr ....S................F.. ERN1_speTri KLPFTIPELVQASPCRSSDGILYM CG Guinea_pig
ERN2_turTru RLPFTIPELVHASPCRSSDGVFYT ERN2_turTr R....................F.. ERN1_oryCun KLPFTIPELVQASPCRSSDGILYM CG Squirrel
ERN2_bosTau RLPFTIPELVHASPCRSSDGVFYT ERN2_bosTa R....................F.. ERN1_vicPac KLPFTIPELVQASPCRSSDGILYM CG Rabbit
ERN2_equCab KLPFTIPELVHASPCRSSDGVFYT ERN2_equCa .....................F.. ERN1_turTru KLPFTIPELVQASPCRSSDGILYM CG Pika
ERN2_felCat RLPFTIPELVHASPCRSSDGVFYT ERN2_felCa R....................F.. ERN1_bosTau KLPFTIPELVQASPCRSSDGILYM -- Alpaca
ERN2_canFam KLPFTIPELVHASPCRSSDGVFYT ERN2_canFa .....................F.. ERN1_equCab KLPFTIPELVQASPCRSSDGILYM CG Dolphin
ERN2_myoLuc KLPFTIPELVHASPCRSSDGVFYT ERN2_myoLu .....................F.. ERN1_canFam KLPFTIPELVQASPCRSSDGILYM CG Cow
ERN2_eriEur KLPFTVPELVHTSPCRSSDGVFYT ERN2_eriEu .....V.....T.........F.. ERN1_myoLuc KLPFTIPELVQASPCRSSDGILYM CG Horse
ERN2_sorAra KLPFTIPELVHASPCRSSDGVFYT ERN2_sorAr .....................F.. ERN1_pteVam KLPFTIPELVQASPCRSSDGILYM CG Cat
ERN2_loxAfr KLPFTIPELVHASPCRSSDGVFYT ERN2_loxAf .....................F.. ERN1_eriEur KLPFTIPELVQASPCRSSDGILYM CG Dog
ERN2_echTel KLPFTIPELVLASPCRSSDGVFYT ERN2_echTe ..........L..........F.. ERN1_sorAra KLPFTIPELVQASPCRSSDGILYM CG Microbat
ERN2_dasNov KLPFTIPELVHTSPCRSSDGIFYT ERN2_dasNo ...........T........IF.. ERN1_loxAfr KLPFTIPELVQASPCRSSDGILYM -- Megabat
ERN2_monDom KLPFTIPELVHASPCRSSDGVLYT ERN2_monDo KLPFTIPELVHASPCRSSDGVLYT ERN1_proCap KLPFTIPELVQASPCRSSDGILYM CG Hedgehog
ERN2_macEug KLPFTIPELVHASPCRSSDGVFYT ERN2_macEu .....................F.. ERN1_echTel KLPFTIPELVQASPCRSSDGILYM CG Shrew
ERN2_sarHar1 KLPFTIPELVQASPCRSSDGIFYM ERN2_sarHa ..........Q.........IF.M ERN1_dasNov KLPFTIPELVQASPCRSSDGILYM -- Elephant
ERN2_sarHar2 KLPFTIPELVQASPCHSSDGIFYM ERN2_sarHa ..........Q....H....IF.M ERN1_choHof KLPFTIPELVQASPCRSSDGILYM -- Rock_hyrax
ERN2_ornAna KLPFTIPELVQSSPCRSSDGILYT ERN2_ornAn ..........QS........I... ERN1_monDom KLPFTIPELVQASPCRSSDGILYM CG Tenrec
ERN2_anoCar KLPFTIPELVQSSPCRSSDGIIYT ERN2_anoCa ..........QS........II.. ERN1_ornAna KLPFTIPELVHASPCRSSDGILYM CG Armadillo
ERN2_taeGut KLPFTIPELVQSSPCRSSDGVLYT ERN2_taeGu ..........QS............ ERN1_galGal KLPFTIPELVQASPCRSSDGILYM CG Opossum
ERN2_galGal KLPFTIPELVQASPCRSSDGILYM ERN2_galGa ..........Q.........I..M ERN1_taeGut KLPFTIPELVQASPCRSSDGILYM CG Platypus
ERN2_xenTro KLPFTIPELVQSSPCRSSDGILYT ERN2_xenTr ..........QS........I... ERN1_anoCar KLPFTIPELVQASPCRSSDGILYM CG Lizard
ERN2_xenLae KLPFTIPELVQSSPCRSSDGILYT ERN2_xenLa ..........QS........I... ERN1_xenTro KLPFTIPELVQSSPCRSSDGILYT CG Tetraodon
ERN2_tetNig KLPFTIPELVQASPCRSSDGVLYM ERN2_tetNi ..........Q............M ERN1_tetNig KLPFTIPELVQASPCRSSDGVLYM CG Fugu
ERN2_takRub KLPFTIPELVQASPCRSSDGVLYM ERN2_takRu ..........Q............M ERN1_takRub KLPFTIPELVQASPCRSSDGVLYM CT Stickleback
ERN2_gasAcu KLPFTIPDLVQSAPCRSSDGILYT ERN2_gasAc .......D..QSA.......I... ERN1_gasAcu KLPFTIPELVQASPCRSSDGVLYM CT Medaka
ERN2_oryLat KLPFTIPELVQSAPCRSSDGILYT ERN2_oryLa ..........QSA.......I... ERN1_oryLat KLPFTIPELVQASPCRSSDGVLYM CG Lamprey
ERN2_calMil KLPFTIPELVQSSPCRSSDGILYT ERN2_calMi ..........QS........I... ERN1_danRer KLPFTIPELVQASPCRSSDGILYM
ERN2_petMar KLPFTIPELVHASPCRTSDGVLYT ERN2_petMa ................T.......
ERN_braFlo KLPFTIPELVNASPCKSSDGILYT ERN_braFlo ..........N....K....I...
Case of MGAT5
chr4_4859 MGAT5 12
>contig00001 length=538 numreads=5 21 C=2(61) Y=2(56)
LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE
.................................................
^
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in two
tasmanian devil (here one is identical and the other differs from Monodelphis by C->Y) and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler).
Pseudogene issues: No processed pseudogenes relevent to this exon are seen by Blat of human and opossum sequence. Some questionable sequence occurs in tarsier and sloth but may be due to low coverage read or assembly error. These fragmentary sequences also have cysteine at the position in question.
Paralog issues: This gene has a moderately similar paralog, MGAT5B, with a similar enzymatic role (beta1,6-N-acetylglucosaminyltransferase). The opossum MGAT5B protein differs at 12 positions out of 49 from opossum MGAT5, whereas human and marsupial MGAT5A differ at one residue. Consequently the two paralogs are readily distinguished within vertebrates. This is moot because 33 of 33 available MGAT5B also have cysteine at the position in question (data not shown).
Homoplasy (recurrent mutation) issues: The alignments below show tyrosine has never replaced cysteine in any other species. This cysteine is extremely invariant in both paralogs, tracing back to lophotrochozoa and cnidaria.
Known variations: No human disease alleles have been mapped to either paralog. None of 9 SNP tracks at the UCSC browser show human variation in this exon.
Side issues: The column marked with an asterisk in the difference alignment below indicates a non-conservative phyloSNP K-->I that occured in the theran mammal stem after platypus divergence. All three marsupial sequences including tasmanian devil have isoleucine in this position as do all 30 of the available placental mammal sequences, suggesting that both the lysine and the isoleucine continue to be under strong selection. No comparable shift occured in the theran stem for MGAT5B where the residue is arginine in all species, a basic residue similar to lysine.
Structural significance: The MGAT5 gene supposedly encodes a conventional enzyme, mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase involved in the synthesis of protein-bound and lipid-bound oligosaccharides. Yet surprisingly, no determined 3D structure exists at PDB relevent to the configuration of this exon -- nor indeed the large 741 residue protein. This is very peculiar because glycosyl transerfases are a well-studied group of enzymes (nearly 100 loci in human) and might be expected to bind UDP-GlcNAc (like MGAT4A or MGAT3).
Only a small region of the protein have a prediction at ModBase using 2f9fA, a remote mannosyltransferasee from Archaeoglobus fulgidus. Luckily the model covers the cysteine at issue, showing two helices and a beta sheet.
SwissProt does not annotate the cysteine at position 532 as part of a disulfide or active site; the predicted location (Golgi) can have homodimer disulfides of similar enzymes, though this is a complex topic. Although all 20 cysteines in this protein are conserved human to opossum, this could be a consequence of the overall sequence identity of 90%. Twelve of the cysteines, not including the Sarcophilus variant, are found in the last 140 residues, perhaps forming a disulfide knot. All but 1 of these cysteines is conserved in the pre-Bilateran anemone Nematostella (which enriches relative to overall percent identity of 43%).
Highest MGAT5 expression occurs in brain, heart, kidney, and placenta. No domains other than a signal peptide and 6 of its own glycosylation target sites are found by online tools such as SMART.
Although the bulky tyrosine substitution is conservative in the sense of polar nature and perhaps hydrogen-bonding capacity, it cannot replace these specialized functions of cysteine. Considering the extreme conservation of this cysteine, this substitution must have a substantial-- perhaps even disabling -- impact on enzymatic function.
Functional significance: In view of the facial tumor situation in tasmanian devils, OMIM's account of prior research in mouse on this gene is quite interesting. Less is known about MGAT5B though it also functions in the synthesis of complex cell surface N-glycans.
" Malignant transformation is accompanied by increased beta-1,6-GlcNAc branching of N-glycans attached to Asn-X-Ser/Thr sequences in mature glycoproteins... The amount of MGAT5 products correlates with disease progression... Mgat5-deficient mice, which are born healthy but develop various abnormalities as adults...Mgat5-deficient mice showed kidney autoimmune disease, enhanced delayed-type hypersensitivity, and increased susceptibility to experimental autoimmune encephalomyelitis...The Golgi enzyme beta1,6 N-acetylglucosaminyltransferase V (Mgat5) is up-regulated in carcinomas and promotes the substitution of N-glycan with poly N-acetyllactosamine, the preferred ligand for galectin-3 (Gal-3)...inhibitors of MGAT5 might be useful in the treatment of malignancies by targeting their dependency on focal adhesion signaling for growth and metastasis."
^ ^ * MGAT5_homSap LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE homSap MGAT5_panTro LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. panTro MGAT5_gorGor LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. gorGor MGAT5_ponAbe LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. ponAbe MGAT5_rheMac LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. rheMac MGAT5_calJac LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. calJac MGAT5_micMur LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... micMur MGAT5_otoGar LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. otoGar MGAT5_tupBel LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. tupBel MGAT5_musMus LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. musMus MGAT5_ratNor LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. ratNor MGAT5_criGri LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... criGri MGAT5_dipOrd LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. dipOrd MGAT5_cavPor LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. cavPor MGAT5_speTri LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. speTri MGAT5_oryCun LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. oryCun MGAT5_ochPri LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. ochPri MGAT5_vicPac LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. vicPac MGAT5_susScr LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. susScr MGAT5_turTru LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. turTru MGAT5_bosTau LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. bosTau MGAT5_equCab LFAGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ..A.............................................. equCab MGAT5_felCat lfvgLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... felCat MGAT5_canFam LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... canFam MGAT5_myoLuc LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. myoLuc MGAT5_eriEur LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. eriEur MGAT5_sorAra LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... sorAra MGAT5_loxAfr LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. loxAfr MGAT5_proCap LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. proCap MGAT5_echTel LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. echTel MGAT5_monDom LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... monDom MGAT5_macEug LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... macEug MGAT5_sarHar1 LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... sarHar1 MGAT5_sarHar2 LFVGLGFPYEGPAPLEAIANGYAFLNVKFNPPKSSKNTDFFIGKPTLRE .....................Y....V...................... sarHar2 MGAT5_ornAna LFVGLGFPYEGPAPLEAIANGCAFLNLKFNPPKSSKNTDFFKGKPTLRE ..........................L..............K....... ornAna MGAT5_galGal LFVGLGFPYEGPAPLEAIANGCAFLNLRFNPPKSSKNTEFFKGKPTLRE ..........................LR..........E..K....... galGal MGAT5_taeGut LFVGLGFPYEGPAPLEAIANGCAFLNLRFNPPKSSKNTDFFKGKPTLRE ..........................LR.............K....... taeGut MGAT5_anoCar LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFKGKPTLRE .........................................K....... anoCar MGAT5_xenTro LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSRNTDFFKGKPTLRE ...................................R.....K....... xenTro MGAT5_tetNig VFVGLSFPYEGPAPLEALANGCIFLNPRLKPPQSSLNSEFFKEKPNIRE V....S...........L....I....RLK..Q..L.SE..KE..NI.. tetNig MGAT5_takRub LFVGLSFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFKGKPTLRE .....S...................................K....... takRub MGAT5_gasAcu LFVGLSFPYEGPAPLEAIANGCAFLNPKFSPAKSSKNTDFFKGKPTLRE .....S.......................S.A.........K....... gasAcu MGAT5_oryLat LFVGLSFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFKGKPTLRE .....S...................................K....... oryLat MGAT5_danRer LFVGLSFPYEGPAPLEAIANGCAFLNPRFDPAKSSKNTDFFKGKPTLRE .....S.....................R.D.A.........K....... danRer MGAT5_oncMyk LFVGLSFPYEGPAPLEAIANGCAFLNPKFTPPKSSKNTDFFKGKPTLRE .....S.......................T...........K....... oncMyk MGAT5_pimPro LFVGLSFPYEGPAPLEAIANGCAFLNPRFDPSKSSKNTDFFKGKPTLRE .....S.....................R.D.S.........K....... pimPro MGAT5_calMil LFVGLGFPYEGPAPLEAIANGCAFLNPRFNPPKSSKNTEFFKGKPTLRE ...........................R..........E..K....... calMil MGAT5_petMar LFVGLGFPYEGPAPLEAIANGCVFLNPRFRPPKSSKNTDFFKGKPTLRE ......................V....R.R...........K....... petMar MGAT5_braFlo LFVGLGFPYEGPAPLEAIASGCVFLNPKFTQPKSRLNTKFFEGKPTFRE ...................S..V......TQ...RL..K..E....F.. braFlo MGAT5_strPur LFIGLGFPYEGPAPLEAVANGCVFLNPKFNPPKNYQNTKFFQGKPTSR. MGAT5_helRob LFIGLGFPYEGPAPLEAIAAGCVFINPKFNPPHSSLNTKFFKGKPTARE MGAT5_nemVec VFIGLGFPYEGPAPLEAIQSGCVFLNAKFDPPHDRVNTPFFKNKPTLRK
Note: the species with unfamiliar genSpp acronyms are Cricetulus griseus, Oncorhynchus mykiss, Pimephales promelas , Callorhinchus milii, Branchiostoma floridae, Strongylocentrotus purpuratus, Helobdella robusta, Nematostella vectensis, and Acropora millepora.
Here the opossum protein is broken into its 16 coding exons with phases (base overhangs at split codons) shown:
>MGAT5_monDom length=743
0 MAFFAPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQHESSSMLREQILDLSKRYIKALAEENRNVVDGPYVGVMTAY 1
2 DLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTTTTAAPSSIAAFEKISVA 1
2 DIINGAQEKCELPPMDGFPHCEGKIK 0
0 WMKDMWRTDPCYANYGVDGSTCSFFIYLSE 0
0 VENWCPHLPWRAKNPYEEPDQNSM 0
0 AEIRTDFNLLYGMMKRHEEFRWMILRIRRMADAWIEAIKSLAEKQNLEKRKRKK 0
0 ILVHLGLLTKESGFKIAENAFSGGPLGELVQWSDLITSLYLLGHDIRISASLAELKE 2
1 IMKRVVGNRSGCPTVGDRIVELIYIDIVGLAQFKKTLGPSWVHYQ 2
1 CMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMF 1
2 PHTPDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDSFWK 0
0 NKKEYLDIIHTYMEVHATVYGSSTNHMPSYVKNHGILSGRDLQFLLRETK 0
0 LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE 0
0 LTSQHPYAEVFIGRPHVWTVNPTDHREVENAVKAILNQK 0
0 IEPYMPYEFTCEGMLQRMNAFIEKQ 0
0 DFCHGQVMWPPLNALQVKLSEPGKSCKQVCQENQLICEPSFFQHLNKDKDVLK 2
1 YEVICHTTELANDILVPSYDDKKKHCVFQGDLLLFSCAGAHPKHKRICPCRDYIKGQVALCQDCL* 0
>MGAT5_sacHar Sarcophilus harrisii (tasmanian_devil) one match to exon 1: FPUIIJ301C96S1
0 MAFFAPWKLSSQN*GFSWLTFGFIWGMMLLHFTIQQRTQHESSSMLREQILDLSKRYIKALAEENRNVVDGPYVGVMTAY 1
2 DLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTTTTAVPSSIAAFEKIsVA 1
2 DIINGAQEKCELPPMDGFPHCEGKIK 0
0 0
0 0
0 AEIRTDFHLLYGMMKRHEEFRWMILRIRRMADAWIEAIKSLAEKQNLEKRKRKK 0
0 ILVHLGLLTKESGFKIAENAFSGGPLGELVQWSDLITSLYLLGHDIRISASLAELKE 2
1 IMKRVVGNRSGCPTVGDRIVELIYIDIVGLAQFKKTLGPSWVHYQ 2
1 CMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMF 1
2 AHTPDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDNFWK 0
0 NKKEYLDIIHTYMEVHATVYGSSTNHMPSYVKNHGILSGRDLQFLLRETK 0
0 LFVGLGFPYEGPAPLEAIANGYAFLNVKFNPPKSSKNTDFFIGKPTLRE 0
0 0
0 IEPYMPYEFTCEGMLQRMNAFIEKQ 0
0 2
1 YEVVCHTTELANDILVPSYDDRKKHCVFQGDLLLFSCAGAHPKHKRICPCRDYIKGQVALCQDCL* 0
The premature stop codon in the first exon is likely read error (1 bp dropped, 1 bp later added):
atggctttctttgctccatggaaattatcctctcagaaactagggtttttcctggtgact
M A F F A P W K L S S Q K L G F F L V T correct monDom frame
W L S L L H G N Y P L R N - G F S W - L 6 residue observed frameshifts in sarHar N*GFSWL
G F L C S M E I I L S E T R V F P G D F irrelevent 3rd reading frame
MGAT5 has 16 exons. The key one here is 12. Alignment of MGAT5_sarHar to opossum shows only 5 differences in 589 residues available for comparison.
Alignment of Monodelphis to human establishes that MGAT5 is better conserved than the average gene:
Identities = 673/744 (90%), Positives = 708/744 (95%), Gaps = 2/744 (0%)
monDo 1 MAFFAPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQHESSSMLREQILDLSKRYIKA 60
MA F PWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQ ESSSMLREQILDLSKRYIKA
homSa 146 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKA 325
monDo 61 LAEENRNVVDGPYVGVMTAYDLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTT 120
LAEENRNVVDGPY GVMTAYDLKKTLAVLLDNILQRIGKLESKVDNLV+ G + +T
homSa 326 LAEENRNVVDGPYAGVMTAYDLKKTLAVLLDNILQRIGKLESKVDNLVV--NGTGTNSTN 499
monDo 121 TTAAPSSIAAFEKISVADIINGAQEKCELPPMDGFPHCEGKIKWMKDMWRTDPCYANYGV 180
+T A S+ A EKI+VADIINGAQEKC LPPMDG+PHCEGKIKWMKDMWR+DPCYA+YGV
homSa 500 STTAVPSLVALEKINVADIINGAQEKCVLPPMDGYPHCEGKIKWMKDMWRSDPCYADYGV 679
monDo 181 DGSTCSFFIYLSEVENWCPHLPWRAKNPYEEPDQNSMAEIRTDFNLLYGMMKRHEEFRWM 240
DGSTCSFFIYLSEVENWCPHLPWRAKNPYEE D NS+AEIRTDFN+LY MMK+HEEFRWM
homSa 680 DGSTCSFFIYLSEVENWCPHLPWRAKNPYEEADHNSLAEIRTDFNILYSMMKKHEEFRWM 859
monDo 241 ILRIRRMADAWIEAIKSLAEKQNLEKRKRKKILVHLGLLTKESGFKIAENAFSGGPLGEL 300
LRIRRMADAWI+AIKSLAEKQNLEKRKRKK+LVHLGLLTKESGFKIAE AFSGGPLGEL
homSa 860 RLRIRRMADAWIQAIKSLAEKQNLEKRKRKKVLVHLGLLTKESGFKIAETAFSGGPLGEL 1039
monDo 301 VQWSDLITSLYLLGHDIRISASLAELKEIMKRVVGNRSGCPTVGDRIVELIYIDIVGLAQ 360
VQWSDLITSLYLLGHDIRISASLAELKEIMK+VVGNRSGCPTVGDRIVELIYIDIVGLAQ
homSa 1040 VQWSDLITSLYLLGHDIRISASLAELKEIMKKVVGNRSGCPTVGDRIVELIYIDIVGLAQ 1219
monDo 361 FKKTLGPSWVHYQCMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMFPHT 420
FKKTLGPSWVHYQCMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMFPHT
homSa 1220 FKKTLGPSWVHYQCMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMFPHT 1399
monDo 421 PDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDSFWKNKKEYLDIIHTYMEVHAT 480
PDNSFLGFVVEQHLNSSDI HINEIKRQNQSLVYGKVDSFWKNKK YLDIIHTYMEVHAT
homSa 1400 PDNSFLGFVVEQHLNSSDIHHINEIKRQNQSLVYGKVDSFWKNKKIYLDIIHTYMEVHAT 1579
monDo 481 VYGSSTNHMPSYVKNHGILSGRDLQFLLRETKLFVGLGFPYEGPAPLEAIANGCAFLNVK 540
VYGSST ++PSYVKNHGILSGRDLQFLLRETKLFVGLGFPYEGPAPLEAIANGCAFLN K
homSa 1580 VYGSSTKNIPSYVKNHGILSGRDLQFLLRETKLFVGLGFPYEGPAPLEAIANGCAFLNPK 1759
monDo 541 FNPPKSSKNTDFFIGKPTLRELTSQHPYAEVFIGRPHVWTVNPTDHREVENAVKAILNQK 600
FNPPKSSKNTDFFIGKPTLRELTSQHPYAEVFIGRPHVWTV+ + EVE+AVKAILNQK
homSa 1760 FNPPKSSKNTDFFIGKPTLRELTSQHPYAEVFIGRPHVWTVDLNNQEEVEDAVKAILNQK 1939
monDo 601 IEPYMPYEFTCEGMLQRMNAFIEKQDFCHGQVMWPPLNALQVKLSEPGKSCKQVCQENQL 660
IEPYMPYEFTCEGMLQR+NAFIEKQDFCHGQVMWPPL+ALQVKL+EPG+SCKQVCQE+QL
homSa 1940 IEPYMPYEFTCEGMLQRINAFIEKQDFCHGQVMWPPLSALQVKLAEPGQSCKQVCQESQL 2119
monDo 661 ICEPSFFQHLNKDKDVLKYEVICHTTELANDILVPSYDDKKKHCVFQGDLLLFSCAGAHP 720
ICEPSFFQHLNKDKD+LKY+V C ++ELA DILVPS+D K KHCVFQGDLLLFSCAGAHP
homSa 2120 ICEPSFFQHLNKDKDMLKYKVTCQSSELAKDILVPSFDPKNKHCVFQGDLLLFSCAGAHP 2299
monDo 721 KHKRICPCRDYIKGQVALCQDCL* 744
+H+R+CPCRD+IKGQVALC+DCL
homSa 2300 RHQRVCPCRDFIKGQVALCKDCL* 2371
Full length genes appear available from GenBank and genome projects for mouse, rat (NM_001107068), dog (wgs exons), horse (XM_001489091), wallaby (wgs exons), and platypus (XM_001520380). Because this gene is 90% conserved at marsupial, placental mammals will not be informative -- indeed it is necessary to go to greater phylogenetic depth than lamprey to define the ultra-conserved residues in this protein:
>MGAT5_macEug nearly identical to monDom; 3 exons are missing, 2 partial exons, exon 4 has frameshifts 0 MAFFAPWKLSSQKLGFFL 1 2 DLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTTTTAVPSSIAAFEKISVA 1 2 DIINGAQEKCELPPMDGFPHCEGKIK 0 0 WMKDiWRTDPCYANYGVDGSTCSFFIYLSE 0 0 VENWCPHLPWRAKNPYEEPDQNSM 0 0 0 0 ILVHLGLLTKESGFKIAENAFSGGPLGELVQWSDLITSLYLLGHDIRISASLAELKE 2 1 2 1 GTEPEFNHANYAQSKGHKTP 1 2 aHTPDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDSFWK 0 0 NKKEYLDIIHTYMEVHATVYGSSTNHMPSYVKNHGILSGRDLQFLLRETK 0 0 LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE 0 0 LTSQHPYAEVFIGRPHVWTVNPTDHREVENAVKAILNQK 0 0 IEPYMPYEFTCEGMLQRMNAFIEKQ 0 0 DFCHGQVMWPPLNALQVKLSEPGKSCKQVCQENQLICEPSFFQHLNKDKDVLK 2 1 YEVICHTTELANDILVPSYDDKKKHCVFQGDLLLFSCAGAHPKHKRICPCRDYIKGQVALCQDCL* 0 >MGAT5_galGal 87% identical to opossum MAFPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQQTQHESSSVLREQILDLSKRYIKALAEENKNVVDGPYVGTVTAY DLKKTLAVLLDNILQRIGKLESKVENLVLNGTGANSTNTTTPAPSLGAVEKLNVA DLINGAQEQCELPPMDGFPHCEGKIK WMKDMWRSDPCYASYGVDGSTCSFFIYLSE VENWCPRLPWRAKNPNEETDQKTV AEIRINFDPLYKMMSRHEEFRWMTLRIRRMADTWIEAIKSLAEKQNLENRKRKK ILVHLGLLTKESGFKIAENAFSGGPLGELVQWSDLITSLYLLGHDIRISASLAELKE IMKKVVGNRSGCPTQGDKVVELIYIDIVGLTQFKKTLGPSWVHYQ CMLRVLDSFGTEPEFNHAHYAQSKGHKTPWGKWNLNPQQFYTMF PHTPDNSFLGFVVEQHLNSSDIKHINDIKRQNQSLVYGKVDNFWK DKKAYLDVIHTYMEVHGTVHGTSTIYIPGYVKNHGILSGRDLQFLLRETK LFVGLGFPYEGPAPLEAIANGCAFLNLRFNPPKSSKNTEFFKGKPTLRE LTSQHPYAEVYIGKPHVWTVDINNLSEVEKAVKSILNQK IDPYLPYEFTCEGMLQRMNAFIERQ DFCHGQVMWPPLSALQVKLAEPGKSCKQVCQESQLICEPSFFQHLNKDKALLK HNIECLTTESANDILVPSFDGRRKHCVFQGDLLLFSCAGSHPTHRRICPCRDYIKGQVALCKDCL* >MGAT5_nemVec Nematostella vectensis (sea anemone) XM_001641404 43% identical to opossum 19 of 20 cysteines conserved MIATKGRPTFKLSAHRIGIVFIIISFIWGLYLIKIQLDERNSQPDYLKGRIIHLSKEYIRALAREKGVYGIDGQPSTQQGVGDLKKATAVLLQSMLERIHVL EKQVEGVIVNSTLEFEILASQIKSLNTTFSLHLSNHSYVSANSCVIPDDPSYPECRQKVMWMRNFWKTHECYAKDHGVNGTICSFLVYLSEVENWCPKFPGRMKPTSRATTEGADL HRSDVQGLLGLLNDQDPIKFKWIKNRINQMWPQWLSALEDLKKKRDLKKIKQKKILVHIGLLANERALHFAANADKGGPLGELVQWSDLIASLYLLGHDVTVTADIPRLQGIFGKL RGPAKKPCPTTIKNDYDLIYLDYYGVKQMQTKVGQFTQSFKCKFRIVDSFGTEAQFNYAGFTEKVPGGSMALWGRHNLNLKQFMTMFPHSPDNSFLGFVVGEEPTPDPHPKKKKAR ALVYGKHYYMWKDLKQRSFLDVINKYMEIHATVGGGIKKWVPSYVINHGVLPSLEVQKLLQDSMIFVGLGFPYEGPAPLEAIAHGCFFLNTKYHPPRNRINTPFFKDKPTLRQITS QHPYAEDYIGQPYVYTVDINDLNKIEAVMKEIMMAEPVSPYLPYEFTHKGMLERLHVFIENQNFCGQNLWPPLNALQARKGAMGSSCKETCHSLGLVCEPQYFPAINTKERMTRSG FPCNTTRVEDMPSLVAPGYRDDPPVCLRQAQNLLFSCTANSPTTKRLCPCRDFKKGQVALCSKC*
Case of ACTL6B
chr2_18546 ACTL6B 11 >contig00001 length=502 numreads=11 GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ^ 3 G=4(94) R=7(213) Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5 and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in tasmanian devil (in this case, one individual differs from Monodelphis by G->R), then differences between the two devils, and finally the number of experimental reads that confirm the nucleotide difference and the sum of the quality scores. The sequences were assembled by Newbler.
The change from small non-polar glycine to bulky positively charged lysine is highly non-conservative, especially at a highly conserved residue such as this. Again the change in Sarcophilus is at a CpG hotspot, this time with a mildly unusual transversion of the C to the purine G.
The well-studied protein here is a member of a family of actin-related proteins (ARPs) which have significant homology to conventional actins, in particular sharing the actin fold (an ATP-binding cleft) as common feature. ACTL6B and its 83% identical paralog ACTL6A are involved in diverse cellular processes such as vesicular transport, spindle orientation, nuclear migration and chromatin remodeling. Both have 14 coding exons. The entire exon containing the G-->R is highly conserved including the glycine.
Pseudogene issues: Blat of full length sequence to human shows no recent processed or segmental pseudogenes. However more sensitive methods show a half dozen processed pseudogenes on different chromosomes plus one for ACTL6A. And opossum assembly, which has all 14 exons, also contains a fairly recent processed pseudogene with 91.5% identity. This locus has internal stop codons and ELSD in place of GLSG for the key glycine. This pseudogene arose from ACTL6A, not ACTL6B.
Retroposed Genes, Including Pseudogenes (retroMrnaInfo UCSC track): ACTL6B at chrX:53188763-53189824 ACTL6B at chr9:110656744-110657692 ACTL6A at chr14:49217726-49219292 ACTL6B at chr7:5533936-5535808 ACTL6B at chr6:46280879-46281761 ACTL6B at chr17:77092347-77093972 ACTL6B at chr1:227633849-227635482
Sarcophilus also has one or more processed pseudogenes which considerably complicates the interpretation of tblastn output. However reads FP1I63R01ARR6N etc show two consecutive exons, the first of which is the G-->R version of the exon and the second identical to the following exon from opossum. The spacing between the two exons is 132 bp, more than adequate for a mammalian intron (whose lower limit is about 78 bp). Other reads span two exon for the normal version of the exon such as FKUJDAX01DZSZO etc again with same intron spacing. (Processed pseudogenes may later acquire pseudo-introns in the form of retroposons so RepeatMaskers needs to be run on the intervening sequence.)
>FP1JAYN01EIJD3 length=493 xy=1734_1049 region=1 run=R_2009_01_29_12_22_00_
monDo: 37 VKGLSGNTMLGVGHVVTTSIGMCDIDIRP 65
++GLS NTMLGVGHVVTTSIGMCDIDIRP
sacHa: 386 LQGLSRNTMLGVGHVVTTSIGMCDIDIRP 300
monDo: 66 GLYGSVIVTGGNTLLQGFTDRLNRELSQKTPP 97
GLYGSVIVTGGNTLLQGFTDRLNRELSQKTPP
sacHa: 168 GLYGSVIVTGGNTLLQGFTDRLNRELSQKTPP 73
Newbler has a bad tendency to create faux frameshifts:
Query: 82 ggtctctacggcagtgtcattgtcactggagggaacacactcttgcaaggctttactgac 141
|||||||||||||||||||||| |||||||||||||||||| |||||||| |||||||||
Sbjct: 167 ggtctctacggcagtgtcattg-cactggagggaacacactgttgcaaggttttactgac 109 FP1I63R01APY7E
Query: 82 ggtctctacggcagtgt-cattgtcactggagggaacacactcttgcaaggctttactga 140
||||||||||||||||| |||||||||||||||||||||||| |||||||| ||||||||
Sbjct: 268 ggtctctacggcagtgttcattgtcactggagggaacacactgttgcaaggttttactga 327 FKUJDAX01AWWZ3
Query: 82 ggtctctacggcagtgtcattgtcactggagggaac-acactcttgcaagg 131
|||||||||||||||||||||||||||||||||||| ||||| ||||||||
Sbjct: 268 ggtctctacggcagtgtcattgtcactggagggaacgacactgttgcaagg 318 FKUJDAX01DZSZO
Paralog issues: There is potential for confusion with the paralog ACTL6A. This wouldn't normally matter because all species in this gene too have glycine at the arginine-substituted site. However its pseudogene could present problems because its decay may have taken a different path in Sarcophilus than in Monodelphis giving the R (instead of D), assuming the pseudogene was formed prior to divergence of these species. Indeed, Macropus eugenii appears to have two processed pseudogenes; one of this has R in place of a glycine 4 residues earlier. It will prove necessary to consider adjacent regions in Sarcophilus reads to determine whether the feature is a pseudogene.
To summarize, this appears to be a valid coding SNP but the situation with paralogs, pseudogenes, and errors intrinsic to the 454 platform makes it unfavorable for rapid screening. It would be necessary to require matches of flanking intronic regions on both sides to be sure that the right locus is being investigated.
Comparison of gene to pseudogene in opossum:
000000889 E R L R I P E G L F D P S N V K G L S G 000000948
<<<<<<<<< | X | K | | | | | | | | | | | | E | | D <<<<<<<<<
250390825 gagtgactcaagattcctgaagggttatttgacccatctaatgtgaaggaattgtcagac 250390766
000000949 N T M L G V G H V V T T S I G M C D I D 000001008
<<<<<<<<< | | | | | | S | | | | | | F | | | | | | <<<<<<<<<
250390765 aacacaatgttgggagtcagtcatgttgttaccacaagctttgggatgtgtgacattgac 250390706
000001009 I R P G L Y G S V I V T G G N T L 000001059
<<<<<<<<< F | | | | | D N M L G A | | | I | <<<<<<<<<
250390705 tttagaccgggactttatgacaatatgttaggggcgggaggaaacattctg 250390655
Comparison of ACTL6A_homSap gene to pseudogenes in wallaby:
macEu: 1063 FPVGYNCNFGVEQLKITERLFDPSNVKRLSGNPMLGVSHVVTTRIGMCDIDIRPGLYGTV 1242
FP GYNC+FG E+LKI E LFDPSNVK LSGN MLGVSHVVTT +GMCDIDIRPGLYG+V
homSa: 289 FPNGYNCDFGAERLKIPEGLFDPSNVKGLSGNTMLGVSHVVTTSVGMCDIDIRPGLYGSV 348
macEu: 48 PNVYKCGFGAEHFKIPEGLFDRSNMKGLSGNTMLGISHVVTKSTGMCDIDIRPGFYISVI 227
PN Y C FGAE KIPEGLFD SN+KGLSGNTMLG+SHVVT S GMCDIDIRPG Y SVI
homSa: 290 PNGYNCDFGAERLKIPEGLFDPSNVKGLSGNTMLGVSHVVTTSVGMCDIDIRPGLYGSVI 349
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
Structural significance:
Functional significance:
* * *
ACTL6B_homSap GLSGNTMLGVGHVVTTSIGMCDIDIRP GLSGNTMLGVGHVVTTSIGMCDIDIRP ACTL6B_homSap GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_panTro GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_panTro GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_gorGor GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_gorGor GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_ponAbe GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_ponAbe GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_rheMac GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_rheMac GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_calJac GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_calJac GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_tarSyr GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_tarSyr GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_micMur GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_micMur GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_otoGar GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_otoGar GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_tupBel GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_tupBel GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_musMus GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_musMus GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_ratNor GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_ratNor GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_dipOrd GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_dipOrd GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_cavPor GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_cavPor GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_ochPri GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_ochPri GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_turTru GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_turTru GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_bosTau GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_bosTau GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_equCab GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_equCab GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_felCat GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_felCat GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_canFam GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_canFam GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_myoLuc GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_myoLuc GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_pteVam GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_pteVam GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_eriEur GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_eriEur GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_loxAfr GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_loxAfr GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_proCap GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_proCap GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_echTel GLSGNTMLGVGHVVTTSIGMCDNDIRP ......................N.... ACTL6B_echTel GLSGNTMLGVGHVVTTSIGMCDNDIRP
ACTL6B_monDom GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_monDom GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_ornAna GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_ornAna GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_galGal GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_galGal GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_taeGut GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_taeGut GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_anoCar GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_anoCar GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_xenTro GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_xenTro GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_tetNig GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_tetNig GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_takRub GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_takRub GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_gasAcu GLSGNTMLGVGHVVTTSVGMCDIDIRP .................V......... ACTL6B_gasAcu GLSGNTMLGVGHVVTTSVGMCDIDIRP
ACTL6B_oryLat GLSGNTMLGVGHVVTTSVGMCDIDIRP .................V......... ACTL6B_oryLat GLSGNTMLGVGHVVTTSVGMCDIDIRP
ACTL6B_danRer GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_danRer GLSGNTMLGVGHVVTTSIGMCDIDIRP
* * *
Consensus gLsGnTMlgvgHVVTts!g$CDi.Ir. gLsGnTMlgvgHVVTts!g$CDi.Ir.
>ACTL6B_homSap
MSGGVYGG
DEVGALVFDIGSFSVRAGYAGEDCPK
ADFPTTVGLLAAEEGGGLELEGDKEKKGKIFHIDTNALHVPRDGAEVMSPLKNGM
IEDWECFRAILDHTYSKHVKSEPNLHPVLMSEAP
WNTRAKREKLTELMFEQYNIPAFFLCKTAVLTA
FANGRSTGLVLDSGATHTTAIPVHDGYVLQQ
GIVKSPLAGDFISMQCRELFQEMAIDIIPPYMIAAK
EPVREGAPPNWKKKEKLPQVSKSWHNYMCN
EVIQDFQASVLQVSDSPYDEQ
VAAQMPTVHYEMPNGYNTDYGAERLRIPEGLFDPSNVK
GLSGNTMLGVGHVVTTSIGMCDIDIRP
GLYGSVIVTGGNTLLQGFTDRLNRELSQKTPP
SMRLKLIASNSTMERKFSPWIGGSILASL
GTFQQMWISKQEYEEGGKQCVERKCP*
>ACTL6B_monDom
MSGGVYGG
DEVGALVFDIGSFSVRAGYAGEDCPK
ADFPTTVGLLTLEEGGGLELDGEKEKKGKTFHIDTNALHVPRDGAEVMSPLKNGM
IEDWECFRAILDHTYSKHVKSEPNLHPVLMSEAP
WNTRAKREKLTELMFEQYNIPAFFLCKTAVLTA
FANGRSTGLVLDSGATHTTAIPVHDGYVLQQ
GIVKSPLAGDFISMQCRELFQEMAIDIIPPYMIAAK
EPVREGAPPNWKKKEKLPQVSKSWHNYMCN
EVIQDFQASVLQVSDSPYDEQ
VAAQMPTVHYEMPNGYNTDYGAERLRIPEGLFDPSNVK
GLSGNTMLGVGHVVTTSIGMCDIDIRP
GLYGSVIVTGGNTLLQGFTDRLNRELSQKTPP
SMRLKLIASNSTMERKFSPWIGGSILASL
GTFQQMWISKQEYEEGGKQCVERKCP*
Case of IPO7
chr5_9037 IPO7 23
>contig00001 length=680 numreads=8
SSQVEKHSCSLTEELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYSTIIDDEENPVDEYQIFKAIFQ
....*N.....................................................F.....................
^
59 F=2(72) S=3(53)
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by -> ), then differences between the two devils
(here one individual has S at position 59, the other has F), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores.
Here the Ensembl-predicted sequence for opossum IPO7 is wrong. The exon begins with EELGSD... and the preceding residues are rubbish. The stop codon and N are thus extraneous.
Pseudogene issues: Human has 4 processed pseudogenes originating at various dates. However opossum lacks any detectable by Blat.
Retroposed Genes, Including Pseudogenes (from pseudoGeneLink and retroMrnaInfo UCSC tracks) IPO7 at chr1:209097616-209101414 IPO7 at chr13:23593176-23594670 IPO7 at chr20:25520871-25521227 IPO7 at chrX:51680122-51682234
Paralog issues: IPO8 is somewhat similar but not sufficiently in this exon to engender confusion.
monDom7 EEIPSDEEDTNEARQALHE---RGGGEDEEEDDDDWDEEVLEETALEGFSTPLDLDDG-VDEYQFFT---QALLSRS
EE+ SDE+D +E Q E + GED DD++W+E+ EETALEG+ST +D ++ VDEYQ F QA+ SR+
monDom8 EELGSDEDDIDEDGQEYLEILAKQAGEDG--DDEEWEEDDAEETALEGYSTIIDDEENPVDEYQIFKAIFQAIQSRN
sacHar7 EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYSTIIDDEENPVDEYQIF
EE+ SDE+D +E Q E ++ +D++W+ED EETALEG+ST +D E++ VDEYQ F
sacHar8 EEIPSDEEDTNETSQTMHENNGGGDEDEEEDDDWDEDVLEETALEGFSTPLDLEDS-VDEYQFF
Homoplasy (recurrent mutation) issues: It can be seen from the 44-species alignment that the serine here is quite invariant, being conserved in all amniotes. In frog and all earlier diverging species, threonine is utilized. However serine is used in all vertebrates at the comparable position in the paralog IPO8 except for tetraodon which again uses threonine, as do weaker homologs in the protostomes Tribolium and Ixodes and cnidarians Nematostella and Acropora. This could be described as a reduced alphabet situation where the residue is strongly restricted to a small residue with hydroxyl side chain. Phenylalanine here, as in Sarcophilus, is thus an immensely non-conservative change as it is bulkly, unable to hydrogen bond, and unsuitable for the protein surface.
Query 2 ELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYFTIIDDEENPVDEYQIFKAIFQ IPO7_sarHar
EL SDED+I+ED +Y+E LA +A E DD++ E EETALE + T +D EE +DE+ F+ Q
Sbjct 1788 ELASDEDEINEDDVQYIESLALKAAEHLDDDDVCE---EETALENFTTSVDTEE--IDEFIAFRTSLQ Acropora millepora
Query 3 LGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYFTIIDDEENPVDEYQIFK IPO7_sarHar
L SDED+ +ED EY+E LAK+A D D+E ++DD EET LE Y T ID E +DEY FK
Sbjct 2611 LASDEDEFNEDDVEYIENLAKKAA-DHFDDEDDDDDDEETPLEEYTTSIDGEN--MDEYIAFK Nematostella vectensis
Known variations: No disease variants are known according to OMIM for either IPO7 or IPO8. No relevent structure at PDB has been determined for the central or distal region of the protein. The protein is quite large and thus it will be very difficult to predict the environment of the serine, much less the impact of phenylalanine substitution.
Side issues: Importin IPO7 has a broad and extremely important function in nuclear protein import, either autonomously as nuclear transport receptor or as an adapter in association with KPNB1. Havilng a receptor for nuclear localization signals, it can promote translocation of import substrates through the nuclear pore complex (NPC) by the energy requiring RAN-dependent mechanism. It mediates autonomously the nuclear import of ribosomal proteins RPL23A, RPS7 and RPL5, but in association with KPNB1 the import of five histones. The role of the paralog IPO8 is similar.
The question here is to what extent could IPO8 compensate for the S-->F change observed in Sarcophilus. It seems implausible given the divergence of the two proteins and the great conservation of IPO7 in the enveloping exon -- what selective force could maintain this if an auxillary gene is available to take on the nuclear import role?
^ ^
IPO7_homSap EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ
IPO7_panTro EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_ponAbe EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_rheMac EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_calJac EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_tarSyr EELGSDEDDIDEDGQEYLEILAKQAGEDGDEEEWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ ................................E.....................E..............
IPO7_micMur EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_tupBel EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_musMus EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_ratNor EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_dipOrd EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_cavPor EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_speTri EELGSDEDDIDEDGQEYLEILAKQAGEDGDDDDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ ...............................D.....................................
IPO7_oryCun EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_ochPri EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_vicPac EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_turTru EELGSDEDDIDVDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ ...........V.........................................................
IPO7_bosTau EELGSDEDDIDEDGQEYLEILAKQAGEDGDEEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ ..............................E......................................
IPO7_equCab EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_canFam EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_myoLuc EELGSDEDDIDEDGQEYLEILAKQAGEDGDDDEWEENDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ ...............................DE...N................................
IPO7_pteVam EELGSDEDDIDEDGQEYLEILAKQA-EDGDDEDWR-DDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .........................-........R-.................................
IPO7_eriEur EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_sorAra EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_loxAfr EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_proCap EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_echTel EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_dasNov EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_choHof EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_monDom EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYSTIIDDEENPVDEYQIFKAIFQ ................................E.....................E..............
IPO7_sarHar1 EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYSTIIDDEENPVDEYQIFKAIFQ ................................E.....................E..............
IPO7_sarHar2 EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYFTIIDDEENPVDEYQIFKAIFQ ................................E..............F......E..............
IPO7_ornAna EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ .....................................................................
IPO7_galGal EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKTIFQ .................................................................T...
IPO7_taeGut EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPIDEYQIFKTIFQ .........................................................I.......T...
IPO7_anoCar EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPIDEYQIFKAIFQ .........................................................I...........
IPO7_xenTro AELGSDEDDIDEEGQEYLEILAKQAGEDGDDEDWEDDDAEETALEGYTTLIDDEDTPIDEYQIFKAIFQ A...........E......................D...........T.L.....T.I...........
IPO7_tetNig AELGSDEDDIDEEGQEYLEMLAKQAGEDGDDEDWEEDDAEETALEGYTTTVDDEDNFVDEYQIFKAILQ A...........E......M...........................T.TV.....F..........L.
IPO7_takRub AELGSDEDDIDEEGQEYLEMLAKQAGEDGDDEDWEDDDAEETALEGYTTNIDDEDNFVDEYQIFKAILQ A...........E......M...............D...........T.N......F..........L.
IPO7_gasAcu AELGSDEDDIDEEGQEYLEMLAKQAGEDGDDEDWEEDDAEETALEGYTTAVDDEDNLVDEYQIFKAILQ A...........E......M...........................T.AV.....L..........L.
IPO7_oryLat AELGSDEDDIDEEGQEYLEMLAKQAGEDGDDDDWEEDDAEETALEGYTTAIDDEDNFVDEYQIFKAVLQ A...........E......M...........D...............T.A......F.........VL.
IPO7_danRer AELGSDEDDIDDEGQEYLEMLAKQAGEDGDDEDWEEDDAEETALEGYTTLVDDEDNLVDEYQIFKAIMQ A..........DE......M...........................T.LV.....L..........M.
^ ^
^
IPO8_hg18_23 EEISSDEEETNVTAQAMQSNNGRGEDEEEEDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALI
IPO8_panTro2 EEISSDEEETNVTAQAMQSNNGRGEDEEEEDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALI
IPO8_gorGor1 -EISSDEEETNVTAQAMQSNNGRGEDEEEEDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALI
IPO8_ponAbe2 EEISSDEEETNVTAQAMQSNNGRGEDEEEEDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALI
IPO8_rheMac2 EEISSDEEETNVTAQAMQSNNGRGEDEEEEDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALI
IPO8_calJac1 EEISSDEEETNVTAQAMQSNNGRGEDEEEDDEDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALI
IPO8_tarSyr1 EEISSDEEETTVTAQAMQSNNGRGEDEEEDDDDWDEEVLEETALEGFSTPIDLDHSVDEYQFFTQALL
IPO8_micMur1 -EIASDEEEMNVNAQAMQSSNGRGEDEEEDDDDWDDEVVEETALEGFSTPLDLDSSVDEYQFFTQALL
IPO8_otoGar1 KEISSDEEESNVKAQAMQSNNGRGDDEEEEEDDWDEEVLEETALEGFSTPLDLDSSVDEYQFFTQALL
IPO8_dipOrd1 EEISSDEEEKSVSVQAMQSVNRRGADEEDEDEDWEEEILEETALEGFSTPLDLDNSVDEYQFFTQALL
IPO8_cavPor3 EEISSDEEETNANAQAMQSNTRKG--EEEEDDDWDEEVLEETALEGFSTPLDLDDSVDEYQFFTQALL
IPO8_speTri1 EEISSDEEDTNITAQAMQANNGRSGDEEEEQDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALL
IPO8_oryCun1 EEISSDEEETNVASQAVQSSSGRGEDEEEDDDDWADEVLEETALEGFSTPLDLDNSVDEYQFFTQALL
IPO8_ochPri2 -EISSDEEETNPSTQAMQSSTGRGEDEDEEEEEWDDEVLEETALESFSTP----ECVDEYQFFTQALL
IPO8_vicPac1 EEISSDEEETNVTAQAMQSNNGRGEDEEEDDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALL
IPO8_turTru1 EEISSDEEETNVTAQAMQSNNGRGEDEEEDDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALL
IPO8_bosTau4 EEISSDEEETNVTAQAMQSNNGRGEDEEEDDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALL
IPO8_equCab2 EEISSDEEETNVTAQAMQSNNGRGEDEEEDDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALL
IPO8_felCat3 EEISSDEEETNVTAQAMQSNNGRGEDEEEEEDDWDEEVLEETALEGFSTPLDLDNSVDEYQIFTQALL
IPO8_canFam2 EEISSDEEETNVTAQAMQSNNGRGEDEEEEDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALL
IPO8_myoLuc1 EEISSDEEEANITAQAMQSKNGRGEEEEEDDDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFTQALL
IPO8_pteVam1 E-ISSDEE-ANVTAQAMQPNNGRGEDEEEDDDDWDEEVLEETALEGFSTPLDLDNSVDEYLFFTQALL
IPO8_eriEur1 EEISSDEEETTVGVQAKQPSNGRVEAEEDDDDDWEEELLEETTLEGFSTPLDLDGSVDEYQFFTQALL
IPO8_loxAfr2 -EISSDEEETNVTAQAMQSNNGRGEDEEEDDDDWDEEVLEETALEGFSTPLDLDSSVDEYQFFTQALL
IPO8_proCap1 EEISSDEEETNVTAQAMQSNNGRGEDEEEDDDDWDEEVLEETALEGFSTPLDLDSSVDEYQFFTQALL
IPO8_echTel1 EEISSDEEETNVTAQAMQSTNGRGDNEEEEEDDWDEEVLEETALEGFSTPLDLDNSVDEYQFFAQALL
IPO8_choHof1 EEISSDEEETSVTAQAMQSNNGRGEDEEEDDDDWDEEVLEETALEGFSTPLDLDSNVDEYQFFTQALL
IPO8_monDom4 EEIPSDEEDTNEARQAL--S-GGGEDEEEDDDDWDEEVLEETALEGFSTPLDLDDGVDEYQFFTQALL
IPO8_ornAna1 EEIPSDEEETNETGQLMQENLGGDEEEDDEDDDWDEDVLEETALEGFSTPLDLENSVDEYQFFTQALL
IPO8_galGal3 EEIPSDEEETNEVSQAMQENHGEEEDDDDDDDDWDEDALEETALEGFSTPLDLENGVDEYQFFTQALL
IPO8_taeGut1 EEIPSDEDETNEVSQAMQENHGEEEDEDDDDDDWDEDALEETALEGFSTPLDLENGVDEYQFFTQALL
IPO8_anoCar1 EEIPSDEEEANEVTQEMQENHVGDEDDDDDDDDWDDDALEETALEGFSTPIDLEDAVDEYQFFTQALI
IPO8_xenTro2 EEIASDEEEAN---QAMQQN---GEDAEEEDEDWDDEVLEETALEGFSTPLDCEDALDEYQFFTNALL
IPO8_tetNig1 QEIPSDEDEVNENH-A-QQASRNGAEDEEEDDYWEDDCFEGTALEEYTTPLDFDNGEDEYLFFTSTLL
IPO8_fr2_23_ QEIPSDEDEVSENHSA-PLPNMSGEDDEEEDDYWDDDGFEGTPLEEYSTPLDFENGEDEFHFFTSTLL
IPO8_gasAcu1 QEIPSDEDEVTENRKAVQHANR-EEEEEDDEDDWDNDCFEGTPLEEYSTPLDYDNGEDEYQFFASALL
IPO8_oryLat2 EEIPSDEDEVNENREAVQHHSR-EDDDDDEEDYWEEDGFEGTPLEEYSTSLDYDNGEDEYEFFTCALL
IPO8_danRer5 EEIPSDEDEVGEKGVAIRRSHREDDDDEDDDEYWDDEGLEGTPLEEYSTPLDCDNGEDEYQFFTASLL
^ ^
>IPO7_homSap
MDPNTIIEALRGTMDPALREAAERQLNE
AHKSLNFVSTLLQITMSEQLDLPVRQA
GVIYLKNMITQYWPDRETAPGDISPYTIPEEDRHCIRENIVEAIIHSPELIR
VQLTTCIHHIIKHDYPSRWTAIVDKIGFYLQSDNSACWLGILLCLYQLVKNYE
YKKPEERSPLVAAMQHFLPVLKDRFIQLLSDQSDQSVLIQKQIFKIFYALVQ
YTLPLELINQQNLTEWIEILKTVVNRDVPN
ETLQVEEDDRPELPWWKCKKWALHILARLFER
YGSPGNVSKEYNEFAEVFLKAFAVGVQQ
VLLKVLYQYKEKQYMAPRVLQQTLNYINQGVSHALTWKNLKPHIQ
GIIQDVIFPLMCYTDADEELWQEDPYEYIRMKF
DVFEDFISPTTAAQTLLFTACSKRKE
VLQKTMGFCYQILTEPNADPRKKDGALHMIGSLAEILLK
KKIYKDQMEYMLQNHVFPLFSSELGYMRAR
ACWVLHYFCEVKFKSDQNLQTALELTRRCLIDDREMPVKVEAAIALQVLISNQEK
AKEYITPFIRPVMQALLHIIRETENDDLTNVIQKMICEYSEEVTPIAVEMTQHL
AMTFNQVIQTGPDEEGSDDKAVTAMGILNTIDTLLSVVEDHKE
ITQQLEGICLQVIGTVLQQHVL
EFYEEIFSLAHSLTCQQVSPQMWQLLPLVFEVFQQDGFDYFT
DMMPLLHNYVTVDTDTLLSDTKYLEMIYSMCKK
VLTGVAGEDAECHAAKLLEVIILQCKGRGIDQ
CIPLFVEAALERLTREVKTSELRTMCLQVAIAALYYNPHLLLNTLENLRFPNNVEPVTNHFITQWLNDVDCFLG
1 LHDRKMCVLGLCALIDMEQIPQVLNQVSGQILPAFILLFNGLKRAYACHAEHENDSDDDDEAEDDDET 1
2 EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEDWEEDDAEETALEGYSTIIDDEDNPVDEYQIFKAIFQ 1
2 TIQNRNPVWYQALTHGLNEEQRKQLQDIATLADQRRAAH 1
ESKMIEKHGGYKFSAPVVPSSFNFGGPAPGMN*
>IPO7_monDom
MDPNTIIEALRGTMDPALREAAERQLNE
AHKSVNFVSTLLQITMSEQLDLPVRQA
GVIYLKNMITQYWPDRETTPGEIPPYTIPEEDRHCIRENIVEAIIHSPELIR
VQLTTCIHHIIKHDYPSRWTAVVDKIGFYLQSENSACWLGILLCLYQLVKNYE
YKKPEERSPLVAAMQHFLPVLKDRFIQLLPDQSDQSVLIQKQIFKIFYALVQ
YTLPLELINQANLTEWIEILKTVVNRDVPP
ETLQVEEDDRPELPWWKCKKWALHILARLFER
YGSPGNVSKEYNEFAEVFLKAFAVGVQQ
VLLKVLYQYKEKQYMAPRVLQQTLNYINQGVSHAVTWKNLKPHIQ
GIIQDVIFPLMCYTDADEELWQEDPYEYIRMKF
DVFEDFISPTTAAQTLLFTACSKRKE
VLQKTMGFCYQILTEPNADPRKKDGALHMIGSLAEILLK
KKIYKDQMEYMLQNHVFPLFSSDLGYMRAR
ACWVLHYFCEVKFKSDQNLQTALELTRRCLIDDREMPVKVEAAIALQVLISNQEK
AKEYITPFIRPVMQALLHIIRETENDDLTNVIQKMICEYSEEVTPIAVEMTQHL
AMTFNQVIQTGPDEEGSDDKAVTAMGILNTIDTLLSVVEDHKE
ITQQLEGICLQVIGTVLQQHVL
EFYEEIFSLAHSLTCQQVSPQMWQLLPLVFEVFQQDGFDYFT
DMMPLLHNYVTVDTDTLLSDTKYLEMIYSMCKK
VLTGVAGEDAECHAAKLLEVIILQCKGRGIDQ
CIPLFVEAALERLTREVKTSELRTMCLQVAIAALYYNPHLLLNTLENLRFPNNVEPVTNHFITQWLNDVDCFLG
LHDRKMCVLGLCALIDLEQIPQVLNQVSGQILPAFILLFNGLKRAYACHAEHENDSDEDDEADDDEET
EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYSTIIDDEENPVDEYQIFKAIFQ
AIQSRNPVWYQALTHGLNEEQRKQLQDIATLADQRRAAH
ESKMIEKHGGYKFNAPVVPSSFNFGGPAPGMN*
>IPO7_sarHar
LHDRKMCVLGLCALIDLEQIPQVLNQVSGQILPAFILLFNGLKRAYACHAEHENDSDEDDEADDDEET
EELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYSTIIDDEENPVDEYQIFKAIFQ
AIQSRNPVWYQALTHGLNEEQRKQLQDIATLADQRRAAH
>IPO8_sarHar
EEIPSDEEDTNETSQTMHENNGGGDEDEEEDDDWDEDVLEETALEGFSTPLDLEDS-VDEYQFF
Case of PPFIA3
chr4_22002 PPFIA3 15 'anomalous mapping from monDom5 to human'
>contig00001 length=298 numreads=4
LIQEEKETTEQRAEELESRVSGSGLDSLGRYRASCSLPPSLTTSTLASPSPPSSGHSTPRPAPPSPAREAPANSTSNTAEKP
........................................................F..................G.V.
^
56 F=2(43) S=2(37)
Here both individuals differed from Monodelphis by S->F at position 56 of PPFIA3 with a confusing end to the exon.
Pseudogene issues: Not applicable.
Paralog issues: PPFIA3 (liprin) has 3 paralogs with considerable (but readily differentiable) sequence identity in this exon. These latter genes are more similar to each other than to PPFIA3, yet all 4 have S at the position occupied by F in tasmanian devil. The ancestral gene duplications must be quite old because lamprey has at least two copies and PPFIA3 itself is readily traced to shark.
PPFIA3 is missing in chicken and finch (proving it is not an essential gene in vertebrates) though present in lizard and frog. These latter species have a one residue mid-exon insert relative to mammals compensated for well past the key residue with a one residue deletion. All three species of marsupials with available data have an 8 residue insert three residues from the end of the exon (which still ends in phase 0 like all other orthologs). These indels have seriously affected the UCSC 44-species alignment quality. The batch of sequences immediately below are hand-curated directly from trace reads but otherwise are provided 'as is.'
The S-->F changed observed in tasmanian devil is likely very significant to protein function given the immense conservation of this residue and its flanking environment. However given the numerous independent indels still within this exon -- especially the 8 residue insert in the marsupial stem -- it would be difficult to argue that S-->F could not somehow be compensated with material impact on function. The complete loss of the gene in two birds (together these species have overwhelming trace coverage and many transcripts) establishes either that PPFIA3 lost its importance important or that one of its three paralogs can assume its function.
No structural data relevent to this exon exists at PDB. The entry at SwissProt shows two predicted phosphoserines within the exon but not at the serine here. Predicted domains and secondary structure coils are not applicable to this exon either. The function is somewhat understood: it may regulate the disassembly of focal adhesions, localize receptor-like tyrosine phosphatases type 2A at specific sites on the plasma membrane and forms homodimers and heterodimers with other liprins.
p * p homSap LIQEEKETTEQRAEELESRVSSSGLD-SLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGT--------DKA sarHar LIQEEKETTEQRAEELESRVSGSGLD-SLGRYRASCSLPPSLTTSTLASPSPPSSGHFTPRPAPPSPAREAPANSTGNVADKP monDom LIQEEKETTEQRAEELESRVSGSGLD-SLGRYRASCSLPPSLTTSTLASPSPPSSGHSTPRPAPPSPAREAPANSTSNTAEKP macEug LIQEEKETTEQRAEELESRVSGSGLD-SLGRYRASCSLPPSLTTSTLASPSPPSSGHSTPRPAPPSPAREAPANSTSNAADKP ornAna LIQEEKETTEQRAEELESRVSGSGLD-SLGRYRGGSALPASLTSSTLASPSPPSSGHSTPRLAPPSPAREGS--------EKT anoCar LIQEEKESTEQRAEEIESRVTSASLDGSLGRYRSGASIPPSVTSSTLASPSPPSSGHSTPRLAPHSPARDG---------EKM xenTro LIQEEKETTELRAEEIESRVTSGTLDGSLGRYRSASSIPTSVTTSTLASPSPPSSGHSTPRITPHSPAREG---------DKF PPFIA3 monDom LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGIMTL PPFIA2 monDom LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSLSSLPPHPSSCLSG--SSPPGSGRSTPRRHPHSPAREVDRLGIMTL PPFIA1 monDom MIQEEKESTELRAEEIETRVTSGSMEALNLQLRKRSSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL PPFIA4
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
*
>PPFIA3_sarHar FHGHM9L01BYK1T length=435 xy=0686_3455 region=1
aggcttatccaggaggaaaaggagaccacggagcagcgggccgaggaactggagagccgc
L I Q E E K E T T E Q R A E E L E S R
gtgtccggctctggcctggactccctgggacgctaccgggccagctgctccctcccgcct
V S G S G L D S L G R Y R A S C S L P P
tccctgaccacgtccaccctggctagcccttccccccccagctctgggcactccacgccc
S L T T S T L A S P S P P S S G H S T P
cgccctgctccccccagtcccgcccgggaagccccggccaacagcaccggcaacgtggca
R P A P P S P A R E A P A N S T G N V A
gataagcccgtgagt
D K P
>PPFIA3_monDom phase 0
ggccactccacccctcgccctgccccgcccagccctgctcgggaagctccagccaacagcactagcaacactgcagaaaagcctgtgagt
G H S T P R P A P P S P A R E A P A N S T S N T A E K P V S
>PPFIA3_macEug Macropus eugenii phase 0, assembly has early frameshift due to extra G
aggctcatccaggaggagaaggagacgacggaacagcgggcagaggagctggagagccgg
R L I Q E E K E T T E Q R A E E L E S R
gtgtctggctctggcctggactccttgggacgctaccgggccagctgctcccttccacct
V S G S G L D S L G R Y R A S C S L P P
tccctgactacatccaccctggccagcccttcaccccccagctctggtcactccacaccc
S L T T S T L A S P S P P S S G H S T P
cgccctgccccacccagccctgcccgagaagccccagccaacagcactagcaacgctgca
R P A P P S P A R E A P A N S T S N A A
gataagcctgtgagt
D K P V S
>PPFIA3_xenTro
aggttaatccaagaggaaaaggagacaacagagttgcgggctgaagaaatagagagtcga
L I Q E E K E T T E L R A E E I E S R
gtgaccagcggcactctggacggatcactgggacgctaccgttctgccagttccatcccc
V T S G T L D G S L G R Y R S A S S I P
acctccgtcaccacatcaactctagccagtccctcaccacccagcagtgggcattccacc
T S V T T S T L A S P S P P S S G H S T
ccgcgcatcacgccacacagccctgccagagaaggagacaaatttgtaagttcctttcaa
P R I T P H S P A R E G D K F V S
>PPFIA3_ornAna platypus phase 0
ctgatccaggaggaaaaggagacgacagagcagcgggccgaggagctggagagccgggtg
L I Q E E K E T T E Q R A E E L E S R V
tccggctcggggttggactccctgggccggtaccggggcggcagtgccctgcccgcctcc
S G S G L D S L G R Y R G G S A L P A S
ctcacctcctccaccctggccagcccctctccccccagcagcggccactccaccccccgc
L T S S T L A S P S P P S S G H S T P R
ctggcgccccccagccccgcccgcgaggggtccgaaaaaaccgtaagtggaaaaggccgc
L A P P S P A R E G S E K T
>PPFIA3_anoCar
aggttgatccaggaggaaaaagaatccacagaacaacgggcagaggaaatcgagagccga
L I Q E E K E S T E Q R A E E I E S R
gtgactagtgccagcttggacggttccctcggccgctaccgctcaggcgcttccatccct
V T S A S L D G S L G R Y R S G A S I P
ccctccgtcaccagctccaccctggccagcccttctccccccagcagtggccactccacc
P S V T S S T L A S P S P P S S G H S T
ccccgcttggcgccccatagccctgctcgcgatggggaaaaaatggtatgtcatgactgt
P R L A P H S P A R D G E K M
>PPFIA3_homSap phase 0
aggctgatccaagaggagaaggagacaacagaacagagggcagaggagctggagagtcgg
R L I Q E E K E T T E Q R A E E L E S R
gtgtccagctctggcttggactcgttgggccgctaccgcagcagctgctccctgcccccc
V S S S G L D S L G R Y R S S C S L P P
tccctcaccacctctacccttgccagcccctcccctcccagctctggccactcaacaccc
S L T T S T L A S P S P P S S G H S T P
cgcctggcaccccctagccctgcccgtgagggcaccgacaaggctgtgagtgctctgaag
R L A P P S P A R E G T D K A V S A L K
tctccccagcctagt
S P Q P S
>PPFIA3_calMil Callorhinchus milii
MIQEEKETNELRAEEIESRVGSGTLEGPQGGGYRSAASLSHSVTASTLASPSPPNSGHSPRMAPHSPAREGDRVGIGNTVS
atggcacctcacagcccagccagggagggggacagggtcggcatcggcaacacagtgagt
M A P H S P A R E G D R V G I G N T V S
PPFIA3_homSap LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_panTro LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_rheMac LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_calJac LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_tarSyr LIQEEKDTTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKT
PPFIA3_micMur LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_mm9_15_ LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKT
PPFIA3_rn4_15_ LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPARE-TDKT del verified
PPFIA3_dipOrd LIQEEKETTEQRAEELESRVSSSGLDSLSRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_cavPor LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_ochPri LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_turTru LIQEEKETTEQRAEELESRVSGSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_bosTau LIQEEKETTEQRAEELESRVSGSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_equCab LIQEEKETTEQRAEELESRVSGSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_canFam LIQEEKETTEQRAEELESRVSGSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_sorAra LIQEEKETTEQRAEELESRVSGSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKT
PPFIA3_loxAfr LIQEEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKA
PPFIA3_proCap LIQZEKETTEQRAEELESRVSSSGLDSLGRYRSSCSLPPSLTTSTLASPSPPSSGHSTPRLAPPSPAREGTDKT
PPFIA3_monDom LIQEEKETTEQRAEELESRVSGSGLDSLGRYRASCSLPPSLTTSTLASPSPPSSGHSTPRPAPPSPAREAPANSTSNTAEKP
PPFIA3_ornAna LIQEEKETTEQRAEELESRVSGSGLDSLGRYRGGSALPASLTSSTLASPSPPSSGHSTPRLAPPSPAREGSEKT
PPFIA3_anoCar LIQEEKESTEQRAEEIESRVTSASLGSLGRYRSGASIPPSVTSSTLASPSPPSSGHSTPRLAPHSPARDGEKM
PPFIA3_galGal missing
PPFIA3_taeGut missing
PPFIA3_xenTro LIQEEKETTELRAEEIESRVTSGTLGSLGRYRSASSIPTSVTTSTLASPSPPSSGHSTPRITPHSPARE--DKF
PPFIA3_tetNig LIQEEKENTELRAEEIENR--SVALATLGRDAAGRFLPSSITSSTLASPSPPSSGHSTPRL-PHSPAREPSDR-
PPFIA3_fr2_15 LIQEEKESTELRAGEIESRVSSVALASLGRDSIGRYMTPSITSSTLASPSPPSSGHSTPRL-PHSPARETTDR-
PPFIA3_gasAcu LIQEEKENTELRAEEIESRVSSVALASLGGDSVGRYMTPSITSSTLASPSPPSSGHSTPRL-PHSPARETTDR-
PPFIA3_oryLat LIQEEKENTELRAEEIESR--SVALASLGRDSAGRFIPSSITSSTLASPSPPSSGTSTPRL-PHSPAREMTDR-
PPFIA3_danRer LIQEEKESTELRAEEIESRVSSVALASLGRDSTGRFIPPSLTSSTLASPSPPSSGHSTPRL-PHSPARETTDR-
PPFIA2_homSap LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_panTro LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_ponAbe LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_rheMac LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_calJac LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_tarSyr LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_micMur LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_otoGar LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_tupBel LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_mm9_16 LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_rn4_16 LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_cavPor LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_speTri LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_oryCun LIQEEKESTELRAEEIENRVASVSLEGLNLARVHQGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_vicPac LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_turTru LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_bosTau LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_equCab LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_felCat LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_canFam LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTTITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_pteVam LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_eriEur LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_sorAra LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_proCap LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_echTel LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_dasNov LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_choHof LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_monDom LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGIMTL
PPFIA2_ornAna LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_galGal LIQEEKESTELRAEEIENRVASVSLEGLNLARVHQGTSITGSVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_taeGut LIQEEKESTELRAEEIENRVASVSLEGLNLARVHQGTSITGSVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_anoCar LIQEEKESTELRAEEIENRVASVSLEGLNLARMHPGTSITASITASSLASSSPPSGHSTPKLTPRSPAREMDRMGIMTL
PPFIA2_xenTro LIQEEKESTELRAEEIENRVASVSLEGLNLARMHPGTSITASVTASSLASSSPPSGHSTPKLTPRSPAREMDRMGVMTL
PPFIA2_tetNig LIQEEKESTELRAEEIEHRVASVSLEGLNLARIHHGASITASATASSLASSSPPSGHSTPKLDPRSPARDMERMGVMTL
PPFIA2_fr2_16 LIQEEKESTELRAEEIENRVASVSLEGLNLARIHHGASITASATASSLASSSPPSGHSTPKLDPRSPARDMERMGVMTL
PPFIA2_gasAcu LIQEEKESTELRAEEIENRVASVSLEGLNLARIHHGASITASATASSLASSSPPSGHSTPKLDPRSPARDMERMGVMTL
PPFIA2_oryLat LIQEEKESTELRAEEIENRVASVSLEGLNLARIHHGVSMTASATASSLASSSPPSGHSTPKLDPRSPARDMERMGVMTL
PPFIA2_danRer LIQEEKESTELRAEEIENRVASVSLEGLNLARVHPGTSITASATASSLASSSPPSGHSTPKLTPRSPARDMERMGVMTL
PPFIA1_homSap LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLASSSPPGSGRSTPRRIPHSPAREVDRLGVMTL
PPFIA1_panTro LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLASSSPPGSGRSTPRRIPHSPAREVDRLGVMTL
PPFIA1_gorGor LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLASSSPPGSGRSTPRRIPHSPAREVDRLGVMTL
PPFIA1_ponAbe LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLASSSPPGSGRSTPRRIPHSPAREVDRLGVMTL
PPFIA1_rheMac LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLASSSPPGSGRSTPRRIPHSPAREVDRLGVMTL
PPFIA1_calJac LIQEEKENTEQRAEEIENRVGSGSLDNLGRFRSMSSIPPYPASSLASSSPPSSGRSTPRRIPHSPAREVDRLGVMTL
PPFIA1_tarSyr LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRPVSSIPPCPASSLAGSSPPGSGRSTPRRIPHSPAREVDRLGIMTL
PPFIA1_micMur LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLGGSSPPGSGRSTPRRIPHSPAREVDRLGIMTL
PPFIA1_otoGar LIQEEKENTEQRAEEIESRVGSGSLDSLGRFRSMSSIPPYPASSLAGSSPPGSGRSTPRRIPHSPAREVDRLGIMTL
PPFIA1_tupBel LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLAGSSPPGSGRSTPRRMPHSPAREVDRLGIMTL
PPFIA1_mm9_15_ LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLAGSSPPGSGRSTPRRVPHSPAREVDRLGVMTL
PPFIA1_rn4_15_ LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLAGSSPPGSGRSTPRRVPHSPAREVDRLGVMTL
PPFIA1_dipOrd LIQEEKESTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLASSSPPGSGRSTPRRVPHSPAREVDRLGVMTL
PPFIA1_cavPor LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLAGSSPPGSGRSTPRMVPHSPAREVDRLGVMTL
PPFIA1_speTri -------------EEIESRVGSGSLDNLGRFRSMSSLPPYPASSLAGSSPPGSGRSTPRRVPHSPAREVDRLGVMTL
PPFIA1_turTru LIQEEKENTEQRAEEIESRVGSGSLDSLGRFRSVSSIPPYPASSRASSSPPSSGRPTPRRAPHSPAREVDRLGVMTL
PPFIA1_bosTau LIQEEKENTEQRAEEIESRVGSGSLDSLGRFRSMSSIPPYPASSLAGSSPPSSGRSTPRRMPHSPAREVDRLGIMTL
PPFIA1_equCab LIQEEKENTEQRAEEIESRVGSGSFGNLR-FRSVSSIPLYPASSLAGSSPPNSGRSTPRRIPHSPAREVDRLGIMTL
PPFIA1_felCat LIQEEKESAEQRAEEIESRVGSVFLDSPGRFRPAGSGAPHPASPLAGPSPPHSGRSTPRRGPHSPAREVDRLGVMTL
PPFIA1_canFam LIQEEKESTEQRAEEIESRVGSGSLDSPGRFRSLGDAPPHPTSVLTGPSPPHSGRSTPRRGPHSPAREVDRLGVMTL
PPFIA1_myoLuc LIQEEKESTEQRAEEIESRVGSGSLDNLGRFRSMSS--PYPGSSLAGSSPPNSGRSTPRRIPHSPAREVDRLGIMTL
PPFIA1_pteVam LIQEEKENTEQRAEEIESRVGSGSLDSLGRFRSMSAIPPYPASSLAGSSPPNSGRSTPRRIPHSPAREVDRLGIMTL
PPFIA1_sorAra LIQEEKESTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPASSLAGSSPPNSGRSTPRRIPHSPAREVDRLGIMTL
PPFIA1_loxAfr LIQEEKESAEQRAEEIESRVGSGSLDNLDRFRSMSSIPPYPAPSLAGSSPPGSGRSTPRRIPQSPAREVDRLGIMTL
PPFIA1_proCap LIQEEKESAEQRAEEIESRVGSGSLDNLDRFRSVSSIPPYPASSLAGSSPPGSGRSTPRRIPQSPAREVDRLGIMTL
PPFIA1_echTel LIQEEKENAEQRAEEIESRVGSGSLSDLGHFRPLGSVPPHPSSALAGSSPPGSGRSTPRRIPQSPSREVDQLGIMTL
PPFIA1_dasNov LIQEEKENTEQRAEEIESRVGSGTLDNLGRFRSLSSIPPYPASSLAGSSPPGSGRSTPRRIPHSPAREVDRLGIMTL
PPFIA1_choHof LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSLSAIPPYPASSLASSSPPGSGRSTPRRMPHSPAREVDRLGVMTL
PPFIA1_monDom LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSLSSLPPHPSSCLSGSSPPGSGRSTPRRHPHSPAREVDRLGIMTL
PPFIA1_ornAna LIQEEKENTEQRAEEIESRVGSGSLDNLGRFRSMSSIPPYPGSSLAGSSPPGSGRSTPRRIPHSPAREVDRLGIMTL
PPFIA1_galGal LIQEEKENTEQRAEEIESRVGSGSLDAHGRFRSMSSIPPPYGGSLAGSSPPGSGRSTPRRIPHSPTREVDRLGIMTL
PPFIA1_taeGut LIQEEKENTEQRAEEIESRVGSGSLEAHGRFRSLGSIAPALGGALAGSSPPGSGRSTPRRIPHSPAREVDKLGIMTL
PPFIA1_anoCar LIQEEKENTEQRAEEIESRVGSGSLENLGRFRSMSSLPAPFRGSLSGTSPPGSGRSTPRRMPHSPAREVDRLGIMTL
PPFIA1_xenTro LIQEEKETTEQRAEEIESRVGSGSLDNLGRFRSITSIPPFTGTSLAGSSPPGSGRSTPRRIPHSPAREVDRLGVMTL
PPFIA1_tetNig MIQEEKESTAIRAEEIECRVGSEGLG--GRFRSMSSIPPCMGSSLGG-SPPGSGHSTPRRIPCSPNRELDRMGVMTL
PPFIA1_fr2_15 MIQEEKESTAIRAEEIECRVGSDGLG--GRFRSMSSIPPCMGSSVGG-SPPGSGHSTPRRIPRSPNRELDRMGVMTL
PPFIA1_gasAcu MIQEEKENTVIRAEEIECRVGSDSLG--GRFRSMGSIPPCPGSSLGG-SPPGSGHSTPRRVPRSPNRELDRMGVMTL
PPFIA1_oryLat MIQEEKESTAIRAEEIECRVGSDSIG--GRFRSLSSIPPCAGSSLGG-SPPSSGHSTPRRIPRSPNRELDRMGVMTL
PPFIA1_danRer LIQEEKESTELRAEEIENRVASVSLE--GRIWHESTIPPSTASSLAS-SSPPSGHSTPKLTPRSPARDMERMGVMTL
PPFIA1_petMar LIQEEKESTEQLAEEIEIRVGGSSGGGGGRLRSARSIPGSATATLATNSAPVSGYATPKRLTHSPAHDPDRHGAMTL
PPFIA4_homSap MIQEEKESTELRAEEIETRVTSGSMEALNLKQLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_gorGor MIQEEKESTELRAEEIETRVTSGSMEALNLKQLRKRGSIPTSLTALSLASASPPLSGRSTAKLTSRSAAQDLDRMGVMTL
PPFIA4_ponAbe MIQEEKESTELRAEEIETRVTSGSMEALNLKQLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_rheMac MIQEEKESTELRAEEIETRVTSGSMEALNLKQLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_calJac MIQEEKESTELRAEEIETRVTSGSMEALNLKQLRKRGSIPTSLTALSLASTSPPLSGRSTPKLTSRSAAQGLDRMGVM--
PPFIA4_micMur MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRSSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_tupBel MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRSSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_musMus MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_ratNor MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_dipOrd MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRGSIPTSLTALSLASASPSLSGRSTPKLTSRSPAQDLDRMGVMTL
PPFIA4_cavPor MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGIMTL
PPFIA4_ochPri MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSTAQDLDRMGVMTL
PPFIA4_vicPac M-QEEKESTELRA-EIDTEVTSGSLEVLKLXLKLQCGGI------------SPPLSGRSAPKLTSRSAAQDLDRMGVMTL
PPFIA4_turTru MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRGSIPTSLTALSLASASPPFSGRSTPKLTSRSATQDLDRMGVMTL
PPFIA4_bosTau MIQEEKESTELRAEELETRVTSGSMEALDLTQLHKRGSIPTSLTALSLASASPPLSGRATPKLTSRSAAQDLDRMGVMTL
PPFIA4_equCab MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_felCat MIQEEKESTELRAEEIETRVTSGSMEALNLTQLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_canFam MIQEEKESTELRAEEIETRVSSGSVEALNLTQLRKRGSIPTSLTALSLASASPPLSGRSTPKLASRSAAQDLDRMGVMTL
PPFIA4_myoLuc MIQEEKESTELRAEEIETRVTSGSMEALNLTQPHRRGPIPTSLTALSLASGSPAFSGRSTAKCASRSAVQDLDRMGVMTL
PPFIA4_eriEur MIQEEKESTELRAEENETRVTSGSMEALNLSQRRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_sorAra MILEEKEATELRAEEIETRMNSASIE-LDSSQLRKRASITTPZMPLSLARASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_loxAfr MIQEEKESTELRAEEIETQVTSGSMEALNL-QLRKRASIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_proCap MIQEEKESTELRAEEIETRVTSGSMEALNL-QLRKRASIPTSLTALSLASTSPQLSGRSTPKLTSRSTAQDLDRMGVMTL
PPFIA4_echTel MIQEEKESAELRAEEIETRVTSGSMEALNL-QLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_dasNov MIQEEKESTELRAEEIETRVTSGSMEALNL-QLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_choHof MIQEEKESTELRAEEIETRVTSGSMEALNL-QLRKRGSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_monDom MIQEEKESTELRAEEIETRVTSGSMEALNL-QLRKRSSIPTSLTALSLASASPPLSGRSTPKLTSRSAAQDLDRMGVMTL
PPFIA4_ornAna LIQEEKESTELRAEEIENRVASVSLEGLNL-RVHPGTSITASVTASSLASSSP--SGHSTPKLTPRSPAREMDRMGVMTL
PPFIA4_galGal MIQEEKESTELRAEELETRVTSGSMEGLNL-QLCKRASIPTSLTALSLASSSPPLSGRSTPKLTSRSAAQDLDRMGIMTL
PPFIA4_taeGut MIQEEKESTELRAEELETRVTSGSMEGLNL-QLCKRASIPTSLTALSLASSSPPLSGRSTPKLSSRSAAQDLDRMGIMTL
PPFIA4_anoCar MIQEEKESTELRAEQLESRVTSGSMEALNL-QLRKRASIPTSLTALSLASSSPPISGRSTPKLSSRSAAQDLDRICSMTL
PPFIA4_xenTro LIQEEKETTEQRAEEIESRVGSGSLDNLG---FQVHHFNSPFZVV-SLAGSSPPGSGRSTPRRIPHSPAREVDRLGVMTL
PPFIA4_tetNig LIQEEKESTELRAEEIEHRVASVSLEGLNL--PPPRR--PASATASSLASSSP--SGHSTPKLDPRSPARDMERMGVMTL
PPFIA4_takRub MIQVERESADLRSDEIESRVNSGSMDGLNV--LRPRA--PTSATAQSLASSCSPHSGHSTPKHHSRNAGHH---LGIMTL
PPFIA4_gasAcu MIQVERESADLRSDEIESRVNSGSMDGLNV--LRPRA--PTSATAQSLASSSSPPSGHSTPKHHSRNASHH---LGIMTL
PPFIA4_oryLat MIQVERESADLRSGDIESRVNSGSMDGLNV--LRPRA--PTSATAQSLASSSSPHSGHSTPKHHGRNASHH---LGIMTL
PPFIA4_danRer MIQVERESAELRADEIESRVNSGSMDGLNV--LRPRSSIPTSVTALSLASSSP--SGRSTPKLTSGSTAHE---LGIMTL
PPFIA4_petMar LIQ-EKESTEQRAEEIESRVGSGSLDSLSL-QQRDGGSLPVSLTGSSLASSSPPVSGRSTPKFTPRSPARDADRAGA---
Case of WDFY3
chr5_2532 WDFY3 19
>contig00001 length=482 numreads=8
DDFSEESSFYEILPCCARFRCGDLIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVSTVK
................T..............................T..L.....N...
^
16 T=3(117) A=5(138)
Tasmanian devil differs from Monodelphis by A->T at position 16. The variation observed later in the exon
is largely in opossum rather than other marsupials and occurs at positions with tight reduced alphabets.
Pseudogene issues: None.
Paralog issues: Some weak paralogs exist in human. These are poorly conserved in the region in question even about the key residue. While this weakens the overall invariance of the key residue, it also eliminates any possibility of cross-alignment to inappropriate homologs.
WDFY3 0 WD repeat and FYVE domain containing 3 isoform WDFY4 0 WDFY family member 4 LYST 3e-114 lysosomal trafficking regulator NBEAL1 6e-109 neurobeachin-like 1 isoform 1 NBEAL2 7e-109 neurobeachin-like 2 LRBA 5e-100 LPS-responsive vesicle trafficking, beach and NBEA 3e-98 eurobeachin NSMAF 1e-78 neutral sphingomyelinase (N-SMase) activation
Homoplasy (recurrent mutation) issues: None.
Known variations: Not a known disease gene; no relevent human variants known.
Side issues: None.
Structural significance: WDFY3 encodes a very large peripheral membrane protein of 3526 aa and 65 codinbg exons containing two leucine-rich repeats, a BEACH doman, five WD domains, a FYVE-domain,3 phosphotyrosines, 2 phosphoserine, and 1 phosphothreonine. However none of these are immediately relevent to the three exons centered on the SNP-containing exon. SuperFamily identifies the key exon significant matches (4e-09) as a concanavalin A-like lectin/glucanase domain. It co-localizes with autophagic structures in starved cells. The few transcripts that cover this region arise from testes (Xenopus), heart (chicken), early embryo (pig), and colon and hypothalamus (human), not informative as to function.
Functional significance: The substitution of threonine for alanine in proteins in general has quite mild effects. Alanine is the most generic amino acid and never catalytically active; threonine is polar but not charged and only somewhat bulkier. However the comparative genomics of this alanine in WDFY3 says this alanine is very different -- it is completely invariant over immense branch length back to chondrichthyes with the sole exception of Sarcophilus.
The embedding exon has more nearby variability than some of the other candidates. Its rather diverged paralog WDFY4 has leucine in place of alanine; this leucine is quite well conserved but has some exceptions. Note the alanine is not one of the better conserved residue patches in the overall region. Thus it appears that the substitution A-->T will have a significant effect on function but not a catastrophic one on core properties.
WDFY3 VSTKEELLQNYVDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVKLHYVHSTPG
VST+E+ Q +D E C + RCG+L+ GQWHHL +V++K M ++ T + +DGQ++ + K+ Y+ + PG
WDFY4 VSTEEKEFQP-LDVMEPEDDSEPSAGCQLQVRCGQLLACGQWHHLAVVVTKEMKRHCTVSTCLDGQVIGSAKMLYIQALPG
* *
homSap VDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK VDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK homSap
panTro VDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ............................................................. panTro
gorGor VDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ............................................................. gorGor
ponPyg VDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ............................................................. ponPyg
macMul VDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ............................................................. macMul
calJac VDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ............................................................. calJac
otoGar VDDFSEESSFYEILPCCARFRCGDLIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK .......................D..V.................................. otoGar
musMus VDDFSEESSFYEILPCCARFRCGELVVEGQWHHLALLMSRGMLKNSTAALYLDGQLVSTVK .........................VV.......A.L..R...........L.....S... musMus
ratNor VDDFSEESSFYEILPCCARFRCGELVVEGQWHHLALLMSRGMLKNSTAALYIDGQLVSTVK .........................VV.......A.L..R.................S... ratNor
dipOrd VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLVLVMSKGMLKNSTAALYLDGQLVSTVK ..........................V........................L.....S... dipOrd
cavPor VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLALVMSKGMLKNSTATLYIDGQLVSTVK ................................................T........S... vicPac
speTri VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ..........................V.................................. speTri
ochPri VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVSTVK ..........................V..............................S... ochPri
vicPac VDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSKGMLKNSTATLYIDGQLVSTVK ..........................V.....................T........S... bosTau
turTru VDDFSEESSFYEILPCCARFRCGELIIEGQWHHLVLVMSRGMLKNSTAALYIDGQLVSTVK .......................................R.................S... turTru
bosTau VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLVLVMSKGMLKNSTATLYIDGQLVSTVK .......................D..V........................L......I.. taeGut
equCab VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ..........................V.................................. equCab
felCat VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ..........................V.................................. felCat
canFam VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ..........................V.................................. canFam
echTel VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVNTVK ..........................V.................................. echTel
dasNov VDDFSEESSFYEILPCCARFRCGELIVEGQWHHLVLVMSKGMLKNSTAALYIDGQPVTTVK ..........................V............................P.T... dasNov
choHof VDDFSEESSFYEILPCCAHFRCGELIVEGQWHHLVLVMSRGMLKNSTAALYIDGQLVNTVK ..........................V.......A.............T........S... cavPor
monDom VDDFSEESSFYEILPCCARFRCGDLIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVSTVK .......................D..V..............................S... monDom
macEug VDDFSEESSFYEILPCCARFRCGDLIVEGQWHHLVLVMSKGMLKNSTAALYLDGQLVNTVK .......................D..V........................L......... macEug
sarHa1 VDDFSEESSFYEILPCCARFRCGDLIVEGQWHHLVLVMSKGMLKNSTATLYLDGQLVNTVK .......................D..V.....................T..L......... sarHar
sarHa2 VDDFSEESSFYEILPCCTRFRCGDLIVEGQWHHLVLVMSKGMLKNSTATLYLDGQLVNTVK .................T.....D..V.....................T..L......... sarHar
ornAna ADDFSEESSFYELLPCCAHFRCGDLIAEGQWHHLVLVMSKGMLKNSTATLYIDGQLVNTVK A...........L.....H....D..A.....................T............ ornAna
galGal VDDFSEESSFYEILPCCARFRCGELIAEGQWHHLVLVMSKGMLKNSTAALYLDGQLVNTVK ..........................A........................L......... galGal
taeGut VDDFSEESSFYEILPCCARFRCGDLIVEGQWHHLVLVMSKGMLKNSTAALYLDGQLVNIVK ..................H.......V............R..................... choHof
anoCar VDDFGEESSCYEILPCCARFRCGDHIVEGQWHHMVLVMSKGMLKNSTAALYIDGQLINTVK ....G....C.............DH.V......M......................I.... anoCar
xenTro VDDFSEEASFYEILPCCARFRCSDLIMEGQWHHLVLVMSKGMLKNSTAALYIDGQLVSTVK .......A..............SD..M..............................S... xenTro
tetNig SDESSEEASFYEILPCCARFRCGEAIAEGQWHHLVLVMSKGMLKNSMATLYIDGQLINTVK S.ES...A................A.A...................M.T.......I.... tetNig
takRub SDESSEEASFYEILPCCARFRCGEVIAEGQWHHLVLVMSKGMLKNSMATLYLDGQLINTVK S.ES...A................V.A...................M.T..L....I.... takRub
gasAcu SDDSREDSFFYEILPCCARFRCGELIAEGQWQHLVLVMSKGMLKNSMATLYLDGQLVNTVK S..SR.D.F.................A....Q..............M.T..L......... gasAcu
oryLap SDESSEEASFYEILPCCARFRCADLIAEGQWHHLVLVMSKGMLKNSMATLYIDGQLVNTVK S.ES...A..............AD..A...................M.T............ oryLap
danRer VDDFSEESSFYEILPCCARFRCADLITEGQWHHLLLVMSKGMLKNSMATLYIDGQMVSTVK ......................AD..T.......L...........M.T......M.S... danRer
calMil VDDFSEESSFYEILPCCARFRCTDLINEGQWHHLVLVMSKGMLKNSTATLYVDGQHVNTVK ......................TD..N.....................T..V...H..... calMil
* *
Less conseervation of this position in paralog WDFY4:
WDFY4_hg18_18 DVMEPEDDSEPSAGCQLQVRCGQ L LACGQWHHLAVVVTKEMKRHCTVSTCLDGQVIGSAK
WDFY4_panTro2 DVMEPEDDSEPSAGCQLQVRCGQ L LACGQWHHLAVVVTKEMKRHCTVSTCLDGQVIGSAK
WDFY4_gorGor1 DVMEPEDDSEPSAGCQLQVRCGQ L LACGQWHHLAVVVTKEMKRHCTVSTCLDGQVIGSAK
WDFY4_ponAbe2 DVMEPEDDSEPSAGCQLQVRCGQ L LTCGQWHHLAVVVTKEMKRHCTVSTCLDGQVIGSAK
WDFY4_rheMac2 DVMEPEDDSEPSAGRQLQVRCGQ L LACGQWHHLAVVVTKEMKRHCTVSTCLDGQVIGSAK
WDFY4_calJac1 DVMEPEDDSEPSAGCQLQVRCGQ L LACGQWHHLAVVVTKEMKRHCTVSTCLNGQVIGSAK
WDFY4_micMur1 DVMEPEDDSEPSGGRQLLVRWSQ L LTWGQGHHLGGVVTKEMKRHCTISTYLDGQGIGSAK
WDFY4_otoGar1 -IMEPEDDSEPSAGCQLQVRCGQ L LACGQWHHLAVVVTKEMKRNCTVSTYLDGQAIGSAK
WDFY4_tupBel1 -VMEPEDDAEPSAGRQLQVRCGQ L LACGQWHHLAVVVTKEVKRSCTVSTYLDGQGIGSAK
WDFY4_mm9_18_ DAMEPEDEAEPSAGRQLQVRCSQ L LTCGQWYHLAVVVSKEMKRNCSVTTYLDGQAIGSAK
WDFY4_rn4_18_ DIMEPEDEAEPSAGRQLQVRCSQ L LACGQWYHLAVVVSKEMKRNCTVTMYLDGQAIGSAK
WDFY4_dipOrd1 DIMEPEDEGEPSAGRQLQVRCGQ H LACGQWHHLAVVVTKEMKRNCTVSTYLDGQAIGLAK
WDFY4_cavPor3 DFMEPEDTIEPSAGRQLQVRCGQ L LACGQWHHLAVVVTKEMKRNCTVSTYLDGQAIGSAK
WDFY4_speTri1 DIMEPEDESEPSAGCQLQVRCGQ L LACGQWHHLAVVVTKEMKRNCIISTYLDGQVIGSAK
WDFY4_oryCun1 DVMEPEDDAEPSAGRQLQVRCGQ L LACGQWHHLAVVVTKEMKRNCTVSTYLDGQLTGSAK
WDFY4_ochPri2 DVMEPEDDAEPSAGRQLQVRCGQ L LACGQWHHLAVVVTKEMKRNCTVSTYLDGQLTGSAK
WDFY4_turTru1 DVMEPEGDPEPSAGRQLRVRCGQ M LACAQWHHLAVVVTKEMKRNCTVSTYLDGQVVGSAK
WDFY4_bosTau4 DVMELEDDPEPSAGRQLRVRCGQ L LACGQWHHLAVVVTKEMKRNCTVFTYLDGQVIGSAK
WDFY4_equCab2 DIMEPEDDPEPSAGRQLRVRCGQ L LACGQWHHLAVVVTKEMKRNCTVSTYLDGQAVGSAK
WDFY4_felCat3 DIMEPEDDPEPSAGRQLRVRCGQ L LACGQWHHLAVVVTKEMKRNCTVSTYLDGQIVGSAK
WDFY4_canFam2 DVMEPEDDPEPSAGRQLRVRCGQ L LACGQWHHLAVVVTKEMKRNCTVSTYLDGQITGSAK
WDFY4_myoLuc1 DVMEPEDNAEPSAGRQLQVRCGQ L LACGQWHHLAVVVTKEMKRNCTVSTYLDGQAIGSAK
WDFY4_pteVam1 DVMEPEDDSEPSAGRQLRVRCGQ L LACGQWHHLAVVVTKEMKKNCTVSTYLDGQVIGSAK
WDFY4_sorAra1 DVMEPEEDFEPSAGRQLRVRCGQ L LTCGQWHHLTVVVTKEMKRNCTISAYLDGQVIGSAK
WDFY4_loxAfr2 -AMEPEDVAEPSAGRQLQIRCGQ L LACGQWHHLAVVVTKEMKRNCTVSTYLDGQIIGSAK
WDFY4_proCap1 DTMEPEDVAEPSAGCQLQVRCGQ L LACGQWHHLAVVVNKEMKRNCTVSTYLDGQIIGSAK
WDFY4_echTel1 DAMEPEGDAEPSAGCQLQVKCGQ L LACGQWHHLAVVITKEMKRNCIVSTYLDGQIIGSAK
WDFY4_dasNov2 -AMEPEDAAEPSAGCQLQVRCGQ Q LTCGKWYHLVVVVTKEMKRNCTVSTYLDGQIIGSAK
WDFY4_choHof1 DVMEPEDDTEPSAGRQLQVRCGQ L LACGQWYHLVVVVTKEMKRNC-ISTYLDGQLIGSAK
WDFY4_monDom4 DVMEPEDIHEPSAGSRLQFHCGN L LSSGQWHHLAVVVSKEMKRNCAVSTYINGQLIGSAK
WDFY4_ornAna1 DIMEPEETSEPPAGSRVQFKCVK L ITTGQWHHLAIVVAKEMKRTCVVRAFIDGQLVGSAK
WDFY4_galGal3 DIMEPEGEVQPFPE-QVQFGCGK L LVTGQWHHLTVTVAKEAKKNCTVSAFINGQMLGSAK
WDFY4_taeGut1 DIMEPEGEVLPFPG-QVKFGCGK L LVTGQWHHLTVTVAKEAKKSCIVAAYINGQMLGSAK
WDFY4_tetNig1 DIMEAEVYSDITA-R-LRFRCSS M LIPGQWHHLVVVMTKDVKKSCVTSVYFNGKAFGSGK
WDFY4_fr2_18_ NIMEPEVHSYITP-R-LRFRCSN M LVPGQWHHLAVVMSKDVKKSCVTSVYFNGKAFGSRK
WDFY4_gasAcu1 DMMEPEVLPHPFD-R-LRFQCSS M LVPGQWHHLAVVLSKDVKKSCIASAYFNGKAVGTGK
Case of XYLT1
chr6_2360 XYLT1 5 61 D=3(110) A=5(107)
>contig00001 length=488 numreads=10
RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGASLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPI
....L........................................................D.....
^
This non-conservative change A-->D is backed by three Sarcophilus reads. However all three are fairly near the end of a minus strand read so none cover the whole exon (raising mild concerns over read quality, given the unusual c-->a base transversion), yet none are long enough to span the next intron to reach the short following exon (leaving some mild pseudogene and paralog issues). Although blastn of extended opossum dna shows that the expected downstream phase 2 splice donor is present, that would also be expected in a close paralog or segmental duplication.
Pseudogene issues: None observed in any mammal using tblastn at wgs database. The detection technique here is a multi-exon query. Because the target database is genomic, recent processed pseudogenes actually give stronger matches because of longer contiguous matches, whereas ortholog matches are weakened by the attempt by blast to extend them. Hence processed pseudogenes surface at the top of match list.
Only a fragment of the gene can be recovered from current Sarcophilus reads, about 8 of 12 exons. However it cannot be determined without genomic assembly which exons 'belong' to the D containing exon, nor can the risk of including matches from the paralog be excluded. This gene has so-so conservation between human and opossum (270 myr roundtrip), 78% identity. which is somewhat puzzling in view of its enzymatic importance. However within marsupials conservation of most exons is in the mid-90's.
Paralog issues: XYLT2 (xylosyltransferase II) gives a moderate match but is not an issue in terms of accurately scoring tasmanian devil populations for the A-->D change. It does create problems in conserved exons in recovering full length genes in species where reads span only single exons. Note XYLT2 also has a conserved A at this position in all 34 available species back to lamprey, proving it an important invariant. Adjacent residues however are only moderately conserved.
XYLT1_homSap RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGASLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIRTNDQLVAFLSRYRDMNFLKSHGRDNAR
RS+YLHR+V+++++ Y NVRVTPWRM TIWGGASLL+ YL+SMRDLLE+ W WDFFINLSA DYP RTN++LVAFLS+ RD NFLKSHGRDN+R
XYLT2_homSap RSDYLHREVVELAQGYDNVRVTPWRMVTIWGGASLLTMYLRSMRDLLEVPGWAWDFFINLSATDYPTRTNEELVAFLSKNRDKNFLKSHGRDNSR
Homoplasy (recurrent mutation) issues: The sole homolog in Drosophila (CG17771, 41% identity) has been previously studied. Here the 424 residue is large and charged E in a motif SESD conserved within arthropods but not lophotrochozoa nor cnidarians such as Hydra magnipapillata (where the corresponding fragment has 63% identity) or Nematostella where A424 are A and G respectively. This is not the drosophila DxD motif however -- this occurs much later in the protein. A further very remote crystallographic paralog MGAT1 also has D here as discussed later.
XYLT1_homSap WRMATIWGGASLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIRTNDQLVAFLSRYRDM
R +TIWGGASLL+ LQ M DLL+ ++W WDF INLS +D+P++T D+LV FLS
OXT_droMel KRFSTIWGGASLLTMLLQCMEDLLQ-SNWHWDFVINLSESDFPVKTLDKLVDFLSANPGR
XYLT_hydMag WRMATIWGGASLLSMLLQMMEDTLKIKEWKWDFFINLSASDYPVQ
XYLT_nemVec WSMATIWGGATLLQMLLKSMEDLIARKEWKWDFFINLSGNDFPIKVNT
Known variations: Not a known disease gene. Natural human polymorphisms in XYLT1 have been observed, P325R, P766A, V8391 and R892Q but these do not include changes near the locus under consideration here A424.
Structural significance: The region enveloping the key residue has a weak 30% match encompassing residues 328-535 (thus including the A-->D residue at 424) that nonetheless is adequate for structural modeling to PDB structure 2GAK -- our residue is part of a short type I beta turn connecting strands 4 and 4' of the donor Rossmann domain. The determined structure match to residues 86-289 is a somewhat similar enzyme, 6-N-Acetylglucosaminyltransferase, a product of the GCNT1 gene. A glycine has replaced the alanine, showing the latter is not a deep invariant critical to this class of enzyme.
This region has been compared in the structural overlay below to yet another glycosylating enzyme, rabbit MGAT1 beta-1,2-N-acetylglucosaminyltransferase. In this enzyme, this short beta turn carries the critical DxD motif that provides bound Mn++ for the UDP of incoming substrate. Comparing GCNT1 and XYLT1 aligns CGMD to SAAD (SDAD in Sarcophilus) in XYLT1. These residues are EDDL (DxD motif) in MGAT1. In other words, A424D of Sarcophilus is in fact physically realizable by D in functional MGAT1. This middle D is invariant throughout vertebrate MGAT1 even as the 'x' residue. However XYLT1 and MGAT1 have no significant alignment at the amino acid level and A-->D (or any other residue) is never observed in XYLT1.
The size of XYLT1 presents an unresolved mystery requiring a crystallographic determination. A simply glycosylation reaction could be accomplished in a bacterium with perhaps 250 residues, yet here the enzyme is 959 residues long, almost 4x the minimum even allowing for targeting peptides and a transmembrane segment.
A second puzzling aspect of glycosylases generally is their lack of homology -- 91 families exist of which only 29 have determined representatives (as tracked at the CAZy database. XYLT1 and XYLT2 are typical in belonging to a small isolated glycosyltransferase family 14 sharing no real sequence homology with other glycosylases (other than the DxD divalent cation coordination motif which could have arisen convergently). Structurally, known glycosylase folds are classified as GT-A (DxD plus single Rossmann-like UDP-binding fold) or GT-B (double).
Note the immediately preceding residues NLS constitute a potential glycosylation site, plausibly realized given the localization of the enzyme (Golgi or extracellular matrix) yet completely consistency with the beta role is required. NLS is invariantly conserved in both XYLT1 (even in drosophila and cnidaria) and XYLT2. While adjacent residues are not normally considered relevant to the NxT/S motif, potentially the substitution of D could interfer with this post-translational modification, were it to occur. This would require the glycosylated serine would be at the surface of the protein, contrary to the best PDB fit. Clearly a large attached carbohydrate would block interactions of immediately adjacent residues.
Functional significance: The protein has been the subject of about a dozen publications. Xylosyltransferases I and II are the chain-initiating enzymes in the biosynthesis of glycosaminoglycans. XYLT1 is the initial and rate-limiting enzyme, transfering UDP-xylose to specific serine residues of a target protein. It is localized to the endoplasmic reticulum and Golgi apparatus as a single-pass membrane protein, but with some fraction also secreted to the extracellular space. The domain match is pfam02485, defined as 'core-2/I-branching' reflecting the branch the added carbohydrate introduces to the growing chain in chondroitin and heparan sulfate and post-translational proteoglycan production. The precise function of XYLT2 has not been established.
Some 19 residues have been subject to experimental mutation though none of the glycosylation sites. Only 8 of the 19 induced mutations affected enzymatic activity (yet without lowering UDP=xylose binding), even though the comparative genomics at bottom shows all 19 sites are equally invariant back to lamprey. Thus residues can be under tremendous selection for a variety of reasons other than substrate binding or direct or indirect role in catalysis.
It is known that formation of abdominal aortic aneurysms can be caused by a destructive remodeling of the extracellular matrix in the vascular wall -- A115S enhances this risk. This bears no apparent relation to the A424D allele (human numbering) in tasmanian devil. The 745DWD747 motif has been shown essential to catalytic activity but again lacks immediate relevence. Reduced XYLT1 activity is a known contributor to male sterility. XYLT1 is elevated in connective tissue diseases such as systemic sclerosis, osteoarthritis, and pseudoxanthoma elasticum.
The connection to tumors or cancers is tenuous. GCNT1 expression is highly correlated with tumor progression in a number of cancers. It is overexpressed in colorectal, lung, and prostate cancer. It is a very weak paralog. Similarly the proteoglycans produced by XYLT1 are important regulators in extracellular matrix deposition, cell membrane signal transfer, morphogenesis, cell migration, normal and tumor cell growth. Mouse knockouts of XYLT2 produce polycystic liver and kidney disease.
In summary, this putative change in tasmanian devil could use additional sequences validation. While not likely linked to facial tumors, the A-->D allele is very undesirable in an inbreed population in view of its role in aortic aneurisms and male sterility. The several billion years of branch length invariance of the alanine argues for no tolerance for variation at this position.
exon 5 ^ exon 6 homSap RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR panTro2 RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR gorGor1 RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSXGRDNA- ponAbe2 RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR rheMac2 RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR calJac1 RSNYLHRQVLQFSRQYGNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR tarSyr1 RSNYLHRQVLQFARQYDNIRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNA- otoGar1 RSNYLHRQVLQFARQYGNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR --------------------------- tupBel1 RSNYLHRQVLQFARQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR mm9_5_1 RSNYLHRQVLQFSRQYDNVRVTSWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR rn4_5_1 RSNYLHRQVLQFSRQYDNVRVTSWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR dipOrd1 RSNYLHRQVLQFATQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLQMPDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR cavPor3 RSNYLHRQVLQFARQYSNVRVTPWRMATIWGGA SLLSTYLQSMQDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR speTri1 RSNYLHRQVLQFAGQYGNVRVTPWRMATIWGGA SLLATYLQSMRDLLEMTDWPWDFFINLSAADYPIR --------------------------- ochPri2 RSNYLHRQVLQMARQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMPDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR vicPac1 rSDYLHRQVLQFARQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR --------------------------- bosTau4 rSNYLHRQVLQFARQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR equCab2 RSNYLHRQVLQFARQYSNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR felCat3 RSNYLHRQVLQFARQYDNVRVTPWRMATIWGGA SLLSTYLQGMRDLLEMTDWPWDFFINLSAADYPIR --------------------------- canFam2 RSNYLHRQVLQFARQYGNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR myoLuc1 RSNYLHRQVLQFARQYSNVRVTPWRMATIWGGA SLLATYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR pteVam1 RSNYLHRQVVQVARQYDNVRVTPWRRATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR --------------------------- loxAfr2 RSNYLHRQVLZFARQYANVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR proCap1 RSNYLHRQVLQLARQYPNVRVTPWRMATIWGGA SLLSTYLQSMRDLLEMTSWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR echTel1 RSNYLHRQVLQFTGQYDNVRVTPWRMATIWGGA SLLTTYLQSMRDLLEMADWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR dasNov2 RSNYLHRQVLQFARQYANVRITPWRMATIWGGA SLLSTYLQSMRDLLEMSDWPWDFFINLSAADYPIR --------------------------- monDom4 RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGA SLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR macEug RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGA SLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR sacHar1 rSNYLHRQVLQFAGQYQNVRVTSWRMATIWGGA SLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPIr TNDQLVAFLSRYRDMNFLKSHGRDNAR sacHar2 SLLSTYLQSMRDLMEMTDWPWDFFINLSDADYPIr TNDQLVAFLSRYRDMNFLKSHGRDNAR ornAna1 RSNYLYRQVLQFAGQYPNVRVTSWRMATIWGGA SLLTTYLQSMRDLMEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYREMNFLKSHGRDNAR galGal3 RSNYLHRQVLQFANQYPNVRVTSWRMATIWGGA SLLSTYLQSMRDLMEMNDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR taeGut1 RSNYLHRQVLQFASQYPNVRVTSWRMATIWGGA SLLTTYLQTMKDLMEMSDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR xenTro2 RSHYLHRQVLQFASQYPNVRVTSWRMSTIWGGA SLLSTYLQSMRDLLEMSDWSWDFFINLSAADYPVR --------------------------- tetNig1 RSNYLHRQVQALAALYPNVRVTPWRMATIWGGA SLLTMYLRSMADLLAMRDWSWDFFINLSAADYPIR --------------------------- fr2_5_1 RSNYLHRQVQALAALYPNVRVTPWRMATIWGGA SLLTMYLRSMADLLAMRDWSWDFFINLSAADYPIR TNDQLVAFLSKYRNMNFIKSHGRDNAR gasAcu1 RSNYLHRQVLSLAAQYSNVRATPWRMATIWGGA SLLTMYLRSMADLLAMRDWSWDFFINLSAADYPIR --------------------------- oryLat2 -SNYLHRQVQIMAMKYPNVRVTPWRMATIWGGA SLLTMYLRSMADLLAMRDWSWDFFINLSAADYPIR --------------------------- danRer5 RSNYLHRQMVALAHQYPNVRVTSWRMSTIWGGA SLLTMYLQSMKDLLAMRDWSWDFFINLSAADYPIR --------------------------- squAca1 RSNYLHREAMQLAQRYSNIRITPWRMVTIWGGA SLLKMYLHCMKDLLEMTDWQWDYFINLSATDYPTR TNDELMGFLSKYRGKNFLKSHGRDNAR leuEri1 RSNYLHREVMQLAQQYPNVRVTPWRMVTIWGGA SLLKMYLNCMKDLLEMTDWHWDYFINLSATDYPTR TNDELVGFLSRYREKNFLKISR----- petMar1 RSNYLQRQVLQVAERYPNVRVTPWRMATIWGGA SLLTMYLRTMKDLLDMADWAWDFFINLSATDYPIR TNDQLVAFLTKYRDKNFLKSHGRDNNR
The A is also conserved in the paralog XYLT2:
^
XYLT2_hg18_4_ RSDYLHREVVELAQGYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_gorGor1 RSNYLHREVVELAQGYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_ponAbe2 RSNYLHREVVELAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_rheMac2 RSDYLHREVVELAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_calJac1 RSNYLHREVAELAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_tupBel1 RSNYLHREVVELAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_mm9_4_1 RSNYLYREVVELAQHYENVRVTPWRMVTIWGGASLLRMYLRSMKDLLEIPGWTWDFFINLSATDYPTR
XYLT2_rn4_4_1 RSNYLYREVVELAQHYDNVRVTPWRMVTIWGGASLLRMYLRSMKDLLEIPGWTWDFFINLSATDYPTR
XYLT2_dipOrd1 RSDYLHREVVELAKQYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR
XYLT2_cavPor3 RSNYLHREVVALAQRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_speTri1 RSNYLHREVVELAQRYENVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_ochPri2 ---YLHREVVELAQQYENVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWTWDFFINLSATDYPTR
XYLT2_turTru1 RSNYLHREVVELARQYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPDWAWDFFINLSATDYPTR
XYLT2_bosTau4 RSNYLHREVVELARQYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR
XYLT2_equCab2 RSNYLHREVVELARQYDNVQVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR
XYLT2_felCat3 RSNYLHREVVELARRYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_canFam2 RSNYLHREVVELARQYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR
XYLT2_myoLuc1 RSNYLHREVVELARQYDNIRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR
XYLT2_pteVam1 RSNYLHREVVELARQYDNVRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_eriEur1 RSNYLHREVVELARHYDNVRVTPWRMVTIWGGASLLRMYLRSMQDLLEVPGWAWDFFINLSATDYPTR
XYLT2_proCap1 RSNYLHREVVELARQYDNMRVTPWRMVTIWGGASLLRMYLRSMRDLLEVPGWAWDFFINLSATDYPTR
XYLT2_monDom4 RSNYLHREVVALAQHYANVRVTPWRMGTIWGGASLLKMYLRSMQDLLEAPGWTWDFFINLSATDYPTR
XYLT2_macEug RSNYLHREVVALAQHYANVRVTPWRMGTIWGGASLLKMYLRSMQDLLEAPGWTWDFFINLSATDYPTR
XYLT2_sarHar RSNYLHREVVALAQHYANVRVTPWRMGTIWGGASLLKMYLRSMQDLLEAPGWTWDFFINLSATDYPTR
XYLT2_galGal3 RSNYLHREAVELAQHYPNIRVTPWRMVTIWGGASLLKMYLRSMKDLLELTEWPWDFFINLSATDYPTR
XYLT2_taeGut1 RSSYLHREAVELARHYPNIRVTPWRMVTIWGGASLLKMYLRSMKDLLELSEWPWDFFINLSATDYPTR
XYLT2_anoCar1 RSTYLHREVVEMAQHYPNIRVTPWRMVTIWGGASLLKMYLHSMKDLLEMTDWTWDYYINLSATDYPTR
XYLT2_xenTro2 RSNYLHREVVRLAQSYENMRVTPWRMVTIWGGASLLTMYLRSMKDLLEVPDWPWDFFINLSATDYPTR
XYLT2_tetNig1 RSGYMHREVLQVAQQYPNIRATPWRMVTIWGGASLLKAYLHSMQDLLSMLDWKWDFFINLSATDFPTR
XYLT2_fr2_4_1 RSNYLHRQVQALAALYPNVRVTPWRMATIWGGASLLTMYLRSMADLLAMRDWSWDFFINLSAADYPIR
XYLT2_gasAcu1 RSNYLHRQVLSLAAQYSNVRATPWRMATIWGGASLLTMYLRSMADLLAMRDWSWDFFINLSAADYPIR
XYLT2_oryLat2 RCSYMHREVLQMAKHYPNIRATPWRMVTIWGGASLLKAYLRSMQDLLSMAEWKWDFFINLSATDFPTR
XYLT2_danRer5 RSNYLHRQMVALAHQYPNVRVTSWRMSTIWGGASLLTMYLQSMKDLLAMRDWSWDFFINLSAADYPIR
XYLT2_petMar1 RSNYLQRQVLQVAERYPNVRVTPWRMATIWGGASLLTMYLRTMKDLLDMADWAWDFFINLSATDYPIR
The comparative genomics of the 19 XYLT1_homSap residues replaced by experimental mutagenesis. The key residue columns have been sliced out of intact protein accompanied by a few residues of flanking context and then concatenated to make a compact display (dots used if identical to human).
C257A none C542A none D745G enz-
C276A enz- C561A enz- D745E none
C285A none C563A none W746DNG enz-
C301A none C572A enz- D747GE enz-
D314G none C574A enz- C920A none
D316G none C675A none C927A none
C471A enz- C933A none
* * * * * * * * * *** * * *
homSap CDISGKEAISALSRAKSKHCRQEIGETYCRHKLGLLMPEKVTRFCPLEDEDECDCDTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSSCRVGTDWDAKERDICATGPTACPVMQTCSQ
panTro2 .......................................................................................................................
gorGor1 .....................................................................X.................................................
ponAbe2 .......................................................................................................................
rheMac2 ...............................................................................................................R...I...
calJac1 .......................................................................................................................
tarSyr1 ................................H...................................I.................................V.....S..........
micMur1 ........V.................................P....D.......................................................................
otoGar1 ........................................................................................................T..............
tupBel1 .E............S........................................H................................................T..............
musMus1 ............T...........A................A..............................................I.V.............T..............
ratNor ............T...........A................A............................................................V................
dipOrd1 ........................A.........Q..................................................................EV.SV.LS...T.PA...
cavPor3 ..............S..............................................................................................S.........
speTri1 ..............S..R...........Q.........RL..L..........................................................V................
oryCun1 .E............S.......Q................R..............................................................V.S..........S..H
ochPri2 .E.....................................R................................................................T...S..........
turTru1 ..............S.............................................................................................S..........
bosTau4 ..............S............................L.................................................................V.........
equCab2 ..............S.................................................................................E...........S..........
felCat3 ......................................K................................................................................
canFam2 .........................D........M...K......S..........................................................T..............
myoLuc1 ..............S......................................................................................................TH
pteVam1 ..............S...........-............................................................................................
proCap1 ..............S.........V................A................................................L...........-GGA.AR...MLPAG..
echTel1 ..............S.........A.A..S....R......A.L............................................................S..........A...
dasNov2 ..............S.........A..................L...........G...................................I..E.......V.T........L.-...
monDom4 ..............S...Q.....A.I..Q..V.K.....................S...............................R..I..E.........T..........A...
ornAna1 ..............S...Q.....A....Q..Y.K.....................S..................................I..E........................
galGal3 .EVT.......M......P.....ADV..Q..H.K..........T.............................................I..E..........I.............
taeGut1 .EVT.......M.....QQ.....ADV..Q....K....Q.....A.........ES...............................T.....E..........A...S.....A...
anoCar1 .E......L.........P.....A......RQ.K....Q...L...Q.D.....ES..................................I.AE.........S..........A.T.
xenTro2 .E.T..............Q.....A.V..Q..Q.K........L...........ES..................................I.A........V.SV.......L.G.A.
tetNig1 .E..........A.....E...Q.A.V.....E.Q....R...Y...........ES..................................I..E..P......T...SS.....S.A.
fr2_3_1 .E..........A.....E...Q.A.VF....E.Q........Y...........ES.....................................E..........V.........A.PK
gasAcu1 .E............V...E...Q.A.V.....E.Q....T...Y......H....GSL.................................I.............V.........A.PK
oryLat2 .E................D...Q.A.V....RE.R........Y....E......GSL..............................A..IS....P....V..V.........A.PK
danRer5 .E..............T.E...Q.V.V.....EHQ........Y..V...V....GSL..............................A..IS....P....V..V...S.....A.AK
petMar1 .E.A....L......R.AQ.K...ADVV.L.QE.K....SLP....I...V....GSL..............................A..IS....P....V.S...SG.......RE
>XYLT1_homSap MVAAPCARRLARRSHSALLAALTVLLLQTLVVWNFSSLDSGAGERRGGAAVGGGEQPPPAPAPRRERRDLPAEPAAARGGGGGGGGGGGGRGPQARARGGGPGEPRGQQPASRGALPARAL DPHPSPLITLETQ DGYFSHRPKEKVRTDSNNENSVPKDFENVDNSNFAPRTQKQKHQPELAKKPPSRQKELLKRKLEQQEKGKGHTFPGKGPGEVLPPGDRAAANSSHGKDVSRPPHARKTGGSSPETKYDQPPKCDISGKEAISALSRAKSKHCRQEIGETYCRHKLGLLMPEKVTRFCPLE GKANKNVQWDEDSVEYMPANPVRIAFVLVVHGRASRQLQRMFKAIYHKDHFYYIHVDK RSNYLHRQVLQVSRQYSNVRVTPWRMATIWGGASLLSTYLQSMRDLLEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR FIRKQGLDRLFLECDAHMWRLGDRRIPEGIAVDGGSDWFLLNRRFVEYVTFSTDDLVTKMKQFYSYTLLPAE SFFHTVLENSPHCDTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSPNDFKPQDFHRFQ QTARPTFFARKFEAVVNQEIIGQLDYYLYGNYPAGTPGLRSYWENVYDEPDGIHSLSDVTLTLYHSFARLGLRRAETSLHTDGENSCR YYPMGHPASVHLYFLADRFQGFLIKHHATNLAVSKLETLETWVMPKKVFKIASPPSDFGRLQFSE VGTDWDAKERLFRNFGGLLGPMDEPVGMQKWGKGPNVTVTVIWVDPVNVIAATYDILIESTAEFTHYKPPLNLPLRPGVWTVKILHHWVPVAETKFLVAPLTFSNRQPIKP EEALKLHNGPLRNAYMEQSFQSLNPVLSLPINPAQVEQARRNAASTGTALEGWLDSLVGGMWTAMDICATGPTACPVMQTCSQTAWSSFSPDPKSELGAVKPDGRLR* >XYLT1_monDom MVAALCARRLARRSHSALIAALTVLLLQTLIVWNFSSLDSGAGDHRGGAAAGGPPPAPRRERRDLPLEPAAAGEGERGPAGGQLLRERGGGHGEHRAQHPPRRGGLPGRAL EPPPSPFTSLETQ DGYFSHRPKEKMRTDSNNENSVPKDFENIDNSNFAPRTQRQKHQPDLGKKPLSKQKEHLKKKLEQDEKVKENSLLGKGSNEALQYSNQAAQNSSQGKKSSRLPHSRKNGAGSPELKYDQPPRCDISGKEAISALSRSKSKQCRQEIAEIYCQHKVGKLMPEKVTRFCPLE GKANNNVRWDEDSVEYMPANPVRIVFVLVVHGRASRQLQRMFKAIYHKDHFYYIHVDK RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGASLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR FIRKQGLDRLFLECDTHMWRLGDRKIPEGITVDGGSDWFLLNRKFVEYVTFSNDDLVTKMKQFYSYTLLPAE SFFHTVLENSPHCGTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSPNDFKPADFHRFQ QTARPTFFARKFEAVVNQEIIGQLDYYLYGNYPSGTPGLRSYWENVYDEPDGIHSISDVVLTMYHSFTRLGLRRAETSLHTDGENSCR YYPMGHPVSVHLYFLADHFQGFLIKHHATNLAVSKLETLETWVMPKKVFKIANPPSDFGRLQFSE IGTEWDAKERIFRNFGGLLGPMDEPVGMQKWGKGPNVTVTVIWVDPVNIIAATYDILIESSAEFTHYKPPLNLPLRPGVWTVKILHHWVPVAETKFLVTPLTFSNKQPIKP DESLKLHNGPPRNAYMEQSFQGLNPVLNIPINLAHVEQARRNAATTGAKLESWVDSLVGGIWSAVDICAIGPTACPVMQTCSQTSWSSLSPDPKSELGAIKPDGRLR* >XYLT1_macEug fragment MVAALCARRLARRSHSALIAALTVLLLQTLIVWNFSSLDSGAGDHRGGEQHAGGEPPPAPRRERRDLAPESRAAAGEEGGGGGRGPQPRGYKLPLERGGGGGGGHREHRPQQTPRRGGPAAGAAQLPGQAL ... DGYFSHRPKEKMRTDSNNENSVPKDFENIDNSNFAPRTQRQKHQPDLGKKSLSKQKEQLKKKLEQEEKAKENSLLGKSSNEAMQYSNQAAQNSSAAKASPKSSKQPHTRKNGAGSPELKYDQLPRCDISGKEAISALSRSKSKQCRQEIAEIYCQHKVGKLMPEKVTRFCSLE GKANNNVRWDEDSVEYMPANPVRIAFVLVVHGRASRQLQRMFKAIYHKDHFYYIHVDK RSNYLHRQVLQFAGQYQNVRVTSWRMATIWGGASLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPIR ... FIRKQGLDRLFLECDTHMWRLGDRKIPEGITVDGGSDWFLLNRKFVEYVTFSNDDLVTKMK... SFFHTVLENSPHCDTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSPNDFKPADFHRFQ ... YYPMGHPVSVHLYFLADRFQGFLIKHHATNLAVSKLETLETWVMPKKVFKIANPPSDFGRLQFSE IGTDWDAKERIFRNFGGLLGPKDEPVGMQKWGKGP... DESLKLHGGPPHNAYMEQSFQGLNPVLNIPINLAHVEQARRNAATTGPKLESWVDSLVGGVWSAMDICAIGPTACPVMQTCSQTSWSSLSPDPKSELGAVKPDGRLR* >XYLT1_sarHar fragment missing 5-6 exons ... ... DGYFSHRPKEKMRTDSNNENSVPKDFENIDNSNFAPRTQRQKHQPDLG...PHVRKNGVGSPELKYDQPPRCDISGKEAISALSRSKSKQCRQEIAEIYCQHKVGKLMPEKVTRFCPL. rSNYLHRQVLQFAGQYQNVRVTSWRMATIWGGASLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPIR TNDQLVAFLSRYRDMNFLKSHGRDNAR FIKKQGLDRLFHECDSHMWRLGERQIPEGIVVDGGSDWFALTRSFVEYVVYTDDPLVAQLRQFYTYTLLPAE SFFHTVLENSPHCDTMVDNNLRITNWNRKLGCKCQYKHIVDWCGCSPNDFKPADFHRFQ ... YYPMGHPVSVHLYFLADRFQGFLIKHHATNLAVS... IGTDWDAKERIFRNFGGLLGPMDEPVGMQKWGKGPNVTVTVIWVDPVNVIAATYDILIESSAEFTHYKPPLNLPLRPGVWTVKILHHWVPVAETKFLVTPLTFSNRQPIKP DESLKLHNGPPRNAYMEQSFQGLNPVLNIPINLAHVEQARRNAAITGPKLENWVDSLVGGIWSAVDICAIGPTACPVMQTCSQTSWSSLSPDPKSELGAIKPDGRLR*
Case of ATP4A
chr4_18550 ATP4A 6 16 C=4(130) R=3(74)
>contig00001 length=906 numreads=10
TAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT
................C........................................................................
^
This is a common non-conservative substitution resulting from the CpG hotspot effect. The gene involved, ATP4A, is a member of an extensive well-studied family of hydrogen-potassium membrane pumps coupled to ATP hydrolysis, with this one responsible for acid secretion into the stomach from electroneutral exchange of cytoplasmic hydrogen ion with external potassium ions. The enzyme resides in gastric parietal cells, localized in cytoplasmic vesicles and apical plasma membranes of the secretory canaliculus. It is comprised of alpha chains such as this as well as beta and gamma chains. The protein is large at 1,035 residues. The R280C variant occurs in exon 7 of the 22 coding exons.
Pseudogene issues: Opossum has a processed pseudogene covering the critical residue at chr2:88378354-88379057. However the parent gene here is ATP12A rather than ATP4A. It may be lineage-specific because a counterpart could not be found in Sarcophilus (at this stage of assembly).
Paralog issues: ATP4A is part of a sizeable gene family with a half-dozen paralogs showing good percent identity over this exon. ATP4A may be a relatively new gene because it cannot be located in sauropsids or platypus -- its telltale location on human chromosome 19, lack of good syntenic conservation, and tandem location of its best counterpart with respect to ATP12A in species such as lizard. With so many paralogs, loss with compensation may have occured in some species.
Although the history of this gene family will prove complex, to a certain extent it is irrelevent because the R of R280C is found in homologous position in all members of the family. There is no reduced alphabet flexibility at this residue. That is illustrated for marsupials below. One cnidarian sequence is included from Nematostella to show this R is quite ancient.
* chr strand pos monDom1 GTAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT 4 - 373500709 monDom2 GTATGIVINTGDRTIIGRIASLASSVGQEKTPIAIEIEHFVHIVAGVAVSIGIVFFIIAICMKYRVLDAVVFLIGIILANVPEGLVAAVT 4 + 278084055 monDom5 GTATGMVINTGDRTIIGRIASLASGVGNEKTPIAIEIEHFVHMVAGVAVSIGVIFFIIAVSMKYPVLESIIFLIGIIVANVPEGLLAAVT 4 + 277916123 monDom3 GTARGIVIATGDRTVMGRIATLASGLEVGRTPIAMEIEHFIQLITGVAVFLGVSFFVLSLILGYSWLEAVIFLIGIIVANVPEGLLATVT 2 - 165703122 monDom4 GTARGIVVYTGDRTVMGRIATLASGLEGGQTPIAAEIEHFIHLITGVAVFLGVTFFILSLILEYTWLEAVIFLIGIIVANVPEGLLATVT 2 + 487887988 monDom6 GTATGIVINMGDHTIIGRIASLDSSVGHEKTPIAIEIEPFVHIVAGVAVSFGIGFFIIAIFMKYWVLDVVIFLIGIILANVPEGLVAAVT 2 + 88378354 sarHar1 GTAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT sarHar2 GTARGVVVATGDRTVMGRIATLASGLEVGKTPIAIEIEHFIQLITGVAVFLGVSFFILSLILGYTWLEAVIFLIGIIVANVPEGLLATVT sarHar3 GTATGMVINTGDRTVIGRIASLASSVGHEKTPIAIEIEHFVHIVAGVAVSIGIVFFIIAICMKYRVLDAVIFLIGIILANVPEGLVAAVT sarHar4 GTARGIVIATGDHTVMGRIASLTSVLEAGKTPIAIEIEHFIHIITGVAVFLGVTFFILSLLLGYGWLHAVIFLIGIIVANVPEGLLATVT macEug1 gTATGMVINTGDRTIIGRIASLASGVGNEKTPIAIEIEHFVHIVAGVAVSLGVIFFIIAVSMKYPVLESIIFLIGIIVANVPEGLLAAVT macEug2 gTARGVVVATGDRTVMGRIATLASGLEVGKTPIAIEIEHFIQLITGVAVFLGVSFFILSLILGYTWLEAVIFLIGIIVANVPEGLLATVT macEug3 gTARGIVVYTGDRTVMGRIATLASGLEGGQTPIAAEIEHFIHLITGVAVFLGVTFFILSLILEYTWLEAVIFLIGIIVANVPEGLLATVT macEug4 gTAQGIVIATGDNTVMGRIASLTSVLEAGQTPIAIEIEHFIHLITAVAVFLGVSFFILSLVLGYGWLQAVIFLIGIIVANVPEGLLATVT macEug5 gTATGVVINTGDQTIIGRIALLTSSVGHEKTPSAIEIEHFVHIVAEVAVSLGMVFFTIAICTKYQVLDAVIFLIGIILGSVPESLVAAVT nemVec1 GNATGVVVQTGDNTVMGRIANLASGLGSGKTPIAVEIEHFIHIITGVAVFLGVTFFIIAFILKYKWLEAVIFLIGIIVANVPEGLLATVT XP_001632743 Closest paralogs of ATP4A within human genome: ATP4A ATPase, H+/K+ transporting, nongastric, alpha ATP1A3 Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9). ATP1A1 Na+/K+ -ATPase alpha 1 subunit isoform a ATP1A2 Na+/K+ -ATPase alpha 2 subunit proprotein ATP1A4 Na+/K+ -ATPase alpha 4 subunit isoform 1 ATP2A3 sarco/endoplasmic reticulum Ca2+ -ATPase isoform ATP2A2 ATPase, Ca++ transporting, cardiac muscle, slow ATP2A1 ATPase, Ca++ transporting, fast twitch 1 isoform ATP2C1 calcium-transporting ATPase 2C1 isoform 1d ATP2C2 calcium-transporting ATPase 2C2 ATP2B4 plasma membrane calcium ATPase 4 isoform 4b ATP2B3 plasma membrane calcium ATPase 3 isoform 3b ATP2B1 plasma membrane calcium ATPase 1 isoform 1b ATP2B2 plasma membrane calcium ATPase 2 isoform 1
Homoplasy (recurrent mutation) issues: None, as discussed above. The CpG at the start of this arginine codon occurs in all vertebrates back to lamprey for which sequence is available, meaning the CpG hotspot is ancient. Yet R140C is never observed in other species, even as an allele, even though it is likely to have been generated many times in various populations. That would imply negative selection against this substitution.
Known variations: Not a known disease gene at OMIM. Natural human polymorphisms have been observed, notably the T-->V substitution at position 3 of the exon.
Structural significance: The region enveloping the key residue, according to an excellent 72% blastp match at PDB (3B8E) to the ATP1A1 paralog in pig using three exons about the critical residue. This suffices for an accurate model of both Sarcophilus ATP4A wildtype as well as R280C, though it must be kept in mind that the pig crystal was only determined to 3.5 angstroms due to its large size and integral membrane aspects. R280C lies in the sixth alpha helix of this structure which lies in the cytoplasm (rather than lumen) some 20 residues before the next transmembrane helix enters the membrane
Alignment of human ATP4A to pig ATP1A1 about R280C showing strand 11, helix 5, helix 6, and active site D:
ATP4A 1 QATVIRDGDKFQINADQLVVGDLVEMKGGDRVPADIRILAAQGCKVDNSSLTGESEPQTR 60
QA VIR+G+K INA+++VVGDLVE+KGGDR+PAD+RI++A GCKVDNSSLTGESEPQTR
ATP1A1 143 QALVIRNGEKMSINAEEVVVGDLVEVKGGDRIPADLRIISANGCKVDNSSLTGESEPQTR 202
ATP4A 61 SPECTHESPLETRNIAFFSTMCLEGTVQGLVVNTGDRTIIGRIASLASGVENEKTPIAIE 120
SP+ T+E+PLETRNIAFFST C+EGT +G+VV TGDRT++GRIA+LASG+E +TPIA E
ATP1A1 203 SPDFTNENPLETRNIAFFSTNCVEGTARGIVVYTGDRTVMGRIATLASGLEGGQTPIAAE 262
ATP4A 121 IEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVTVCLSLT 180
IEHF+ II G+A+ G +FFI+++ + YT+L A++F + I+VA VPEGLLATVTVCL+LT
ATP1A1 263 IEHFIHIITGVAVFLGVSFFILSLILEYTWLEAVIFLIGIIVANVPEGLLATVTVCLTLT 322
ATP4A 181 AKRLASKNCVVKNLEAVETLGSTSVICSDKTGTLTQNRMTVSHLWFDNHIHTADTTEDQS 240
AKR+A KNC+VKNLEAVETLGSTS ICSDKTGTLTQNRMTV+H+W DN IH ADTTE+QS
ATP1A1 323 AKRMARKNCLVKNLEAVETLGSTSTICSDKTGTLTQNRMTVAHMWSDNQIHEADTTENQS 382
Functional significance: Clearly it would be disadvantageous to lose function in a key enzyme in the gastric digestive process. It is unlikely to be an adaptation to carnivory because all other mammals with such a diet retain the arginine. It remains conceivable that amino acid change elsewhere in this molecule or its hetero-oligomer partners could compensate. However R240C may not induce loss but rather suboptimal functioning in this otherwise extremely conserved regin of the protein. As such it likely spread from an inbreeding artefact attributable to low population levels. It is not plausibly associated with facial tumors but still would be a high priority to breed out.
>ATP4A_homSap 263-352 chr 19 flanking exons 20 phase tandem to anoCar: -FFAR3 +ATP4A +ATP12A -TMEM147 -GAPDHS QATVIRDGDKFQINADQLVVGDLVEMKGGDRVPADIRILAAQGCKVDNSSLTGESEPQTRSPECTHESPLETRNIAFFSTMCLE GTVQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT VCLSLTAKRLASKNCVVKNLEAVETLGSTSVICSDKTGTLTQNRMTVSHLWFDNHIHTADTTEDQS
>ATP4A_monDom (note smaller introns relative to human) QATVIREGDKFQINADQLVVGDLVEIKGGDRVPADIRILAAQGCKVDNSSLTGESEPQTRSPECTHESPLETRNIAFFSTMCLE GTAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT VCLSLTAKRLARKNCVVKNLEAVETLGSTSVICSDKTGTLTQNRMTVSHLWFDNHVHTADTTEDQS
>ATP4A_sarHar (other exons provisional: lack of assembly, paralogs) QATVIREGDKFQINADQLVVGDLVEIKGGDRVPADIRVLAAQGCKVDNSSLTGESEPQTRSPECTHDSPLETRNIAFFSTMCLE GTAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT VCLSLTAKRLARKNCVVKNLEAVETLGSTSVICSDKTGTLTQNRMTVSHLWFDNHVHTADTTEDQS
Case of VPS72
chr2_30280 VPS72 5 15 R=3(59) K=2(51)
>contig00001 length=591 numreads=6
NYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLAEPGPKEENVDVE
...............R..................T............
^
The K-->R substitution K204 in exon 5 of the six exon VPS72 (vacuolar protein sorting-associated protein 72) would be innocuous if the role of the residue were simply to provide a positively charged side chain. However here the lysine is invariant back to cnidaria with no arginine accepted into the reduced alphabet.
Pseudogene issues: No recent pseudogenes occur in opossum or human genomes at the sensitivity of Blat. The Sarcophilus exon variant has normal splice junctions and its extension lacks amino acids of flanking exons, so it itself is not part of a processed pseudogene. A full length gene is readily recovered; other exons are quite close in sequence to opossum and do not support the notion of gene loss.
Paralog issues: This gene has only weak partial paralogs in mammal, ATAD2 and MYO9B at 1e-05, that could not cause confusion.
Homoplasy (recurrent mutation) issues: None. No variation is seen at position K204 in other species back to cnidaria:
nemVec: LTQEELLAEARITEEENTASLLAYQRHEADKKKTKIQKVTHKGPIIRFCSLSMPV XP_001632443
hydMag: LTQQELLAEAKITAEKNLASLAQFLKLEEEKKHIKISKVRYQGPIIRYQSVRMPL 207 XP_002165194
LTQ+ELLEAKIT E NL SL + +LE +KK K + GPII Y SV +PL
homSap: LTQEELLREAKITEELNLRSLETYERLEADKKKQVHKKRKCPGPIITYHSVTVPL 221
*
homSap ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDIEG
gorGor1 eTYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDIEG
ponAbe2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDIEG
rheMac2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENIDIEG
calJac1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGLKEENVDIEG
tarSyr1 ETYERLEADKKKQVHKKRKCPGPIITFHSVTVPLVGEPGPKEENVDVEg
micMur1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGELGPKEETVDIEG
otoGar1 ETYERLEADKKKQVHKKRKCPGPIITYHSMAVPLVGELGPK-ETVDVEG
tupBel1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
mm9_5_6 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
rn4_5_6 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
cavPor3 ETYERLEADKKKQVHKKRKCPGPIITYHSMTVPLVGEPGPKEENVDVEG
speTri1 ETYERLEADKKKPVHKETECPGPIITYHSMTVPLIGELGPKEENVDVEG
ochPri2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLIGELGPKEENVDVEG
turTru1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
bosTau4 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
equCab2 ETYERLEADKKKQVHKKRKCP-PIITYHSVTVPLVGEPGPKEENVDVEG
felCat3 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
canFam2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
pteVam1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGKPGPREETVDVEG
eriEur1 ETYERLEADKKKQVHKKRKCPGPIITYHSLTVPLIGELGPKEENVDVEG
sorAra1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGELGPKEENVDVEG
loxAfr2 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
proCap1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
echTel1 ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDVEG
choHof1 eRRALLKADKRKQVHKKRKCPGPIITYHSVSVPLVR-PGPKEENVDAEg
monDom4 ENYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLAEPGPKEENVDVEG
macEug ENYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLTEPGPKEENVDVEG
sarHar ENYERLEADKRKQVHKRRKCPGPVITYHSMTVPLLTEPGPKEENVDVEG
ornAna1 ------------------------ISFHSLTVPLLADPGAREENVDVEG
galGal3 ENYERLEADKKKQVQKKRKCVGPVIRYWSVTMPLITELG-KEENVDVEG
melGal ENYERLEADKKKQVQKKRKCVGPVIRYWSVTMPLITELG-KEENVDVEG
anoCar1 ETYERLEADKKRQVQKKRKCVGPTIRYYSGTMPLITDLGCKEETVDVEG
xenTro2 ENYERLEADRKKQVHKKRRCVGPTIRHHSLVMPLITELNVKEENVDVEG
tetNig1 ENYERLEADKKKQVQKKRRFDGPTIRYHSVLMPVVSHSVLKEENVDVEG
takRub ENYERLEADKKKQVQKKRRFDGPTVRYHSVLMPIVSHSVLKEENVDVEG
gasAcu1 ENYERLEADKKKQVHKKRRFEGPTIRYHSVLMPLVSHSVLKEENVDVEG
oryLat2 ENYERLEADKKKQVHKKRRFEGPTIRYHSLLMPIVSHSVLKEENVDVEg
danRer5 ENYERLEADKKRQVHMKRQCVGSVIRYHSVLMPLVSDVTLKEENVDVEg
petMar1 ENYERLEADKKKQVLKKHHYTGPVIRYHSLTMPLITELPIKEENVDVEg
*
Known variations: A breast cancer sample identified I318V as a somatic mutation in this gene; the significance of this is unclear. An early report associates it with repression of transformed cells. These links do not provide a specific connection to the Sarcophilus facial tumor situation.
Structural significance: No structural matches exist at PDB using blastp. Modbase predicts helical fragments of the 3D structure. Pfam domains are circular references to YL1 (the name of the encoded protein). SwissProt notes various compositional biases (DE- and P-rich regions) and a phosphoserine at residue 168.
Functional significance: The specific function is not well understood. VPS72 is generally described as a dna-binding transcriptional regulator possibly involved in chromatin modification and remodeling as a subunit of the NuA4 histone acetyltransferase complex. whose metazoan counterpart is called the TRRAP/TIP60 HAT complex. It is also a subunit of the SNF2-related helicase SRCAP complex. Thus it is localized in the nucleus.
In summary this substitution, if confirmed, could have significant but probably not disabling impacts on the functionality of this gene in view of the extreme intolerance for any kind of substitution at the lysine. However it would be difficult to pursue the impact further given the lack of available structure and complexitities of the VPS72 protein complex and its role in histone modification.
>VPS72_homSap MSLAGGRAPRKTAGNRLSGLLEAEEEDEFYQTTYGGFTE ESGDDEYQGDQSDTEDEVDSDFDIDEGDEPSSDGEAEEPRRKRRVVTKAYK EPLKSLRPRKVNTPAGSSQKAREEKALLPLELQDDGSD SRKSMRQSTAEHTRQTFLRVQERQGQSRRRKGPHCERPLTQEELLREAKITEELNLRSL ETYERLEADKKKQVHKKRKCPGPIITYHSVTVPLVGEPGPKEENVDIEG LDPAPSVSALTPHAGTGPVNPPARCSRTFITFSDDATFEEWFPQGRPPKVPVREVCPVTHRPALYRDPVTDIPYATARAFKIIREAYKKYITAHGLPPTASALGPGPPPPEPLPGSGPRALRQKIVIK* >VPS72_monDom4 MSLAGGRAPRKTAGNRLSGLLEAEEEDEFYQTTYGGFTE ESGDDEYQGDQSDTEDEVDSDFDIDEGDEPASDGDGDEPRRKRRVVTKAYK EPIKSLRPRKVSTPAGSSQKTREEKTLLPLELQDDGLD SRKSMRQSTAEHTRQTFLRVQERQGQSRRRKGPHCERPLTQEELLREAKITEELNLRSL ENYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLAEPGPKEENVDVEG LEPTPVVSAVAPHSGAGPVLPPARCSRTFITFSDDATFEECFPRGKPPKIPVREVCPVTHRPALYRDPVTDIPYATARAFKIIREAYKKYITAHGLPPAASALGPGPPPPEPLPGPGPRALRQKIIIK* >VPS72_macEug Macropus eugenii cDNA EX201397 MSLAGGRAPRKTAGNRLSGLLEAEEEDEFYQTTYGGFTE ESGDDEYQGDQSDTEDEVDSDFDIDEGDEPASDGDGDEPRRKRRVVTKAYK EPIKSLRPRKVSTPAGSSQKAREEKTLLPLELQDDGVD SRKSMRQSTAEHTRQTFLRVQERQGQSRRRKGPHCERPLTQEELLREAKITEELNLRSL ENYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLTEPGPKEENVDVEG LEPPTLVSTVAPHSGTGPLIPPARCSRTFITFSDDAFEECFPRGKPPKIPVREVCPVTHRPALYRDPVTDIPYATARAFKIIREAYKKYITAHGLSPAASALGPGPPPPEHLPGPGPRALRQKIVIK* >VPS72_sarHar MSLAGGRAPRKTAGNRLSGLLEAEEEDEFYQTTYGGFTE ESGDDEYQGDQSDTEDEVDSDFDIDEGDEPASDGEGDEPRRKRRVVTKAYK ePIKSLRPRKVSTPAGSSQKAREEKTLLPLELQDDGLD sRKSMRQSTAEHTRQTFLRVQERQGQSRRRKGPHCERPLTQEELLREAKITEELNLRSL ENYERLEADKKKQVHKRRKCPGPVITYHSMTVPLLTEPGPKEENVDVEG LEPIPAVPTAAPHSATGPVIPPARCSRTFITFSDDATFEECFPRGKPPKIPVREVCPVTHRPALYRDPVTDIPYATARAFKIIREAYKKYITAHGLPRLPRPWGPGPPPPEPLPGPGPRALRQKIIIK*
Case of ABCC1
chr6_5144 ABCC1 23 4 Q=2(69) P=2(80) looks like a frame-shift problem in monDom5
>contig00001 length=802 numreads=10
HLCFPRLHLDLLHNVLRSPMSFFERTPSGNLVNRFSKEMDTVDSMIPQIIKMFMGSLFNVIGACIIILLATPIAAIIIPPLGLIYFFVQ
....Q....................................................................................
^
Discarded candidates
Below are three initial candidates that had to be discarded without detailed followup. One arose from repeated frameshifts in the critical region, another exhibited homoplasy with marsupials, and the third too extensive of an accepted reduced alphabet at the site. Thus while these three genes do not meeet the search criteria, they are nonetheless instructive in illustrating those criteria and making clear these are quite restrictive.
Case of ACOT12
chr3_5872 ACOT12 14 14 I=3(95) V=3(110) 'wobbly'
>contig00001 length=472 numreads=6
NTYVVAVKSVTLASIPPSPQYNRSEITCAGFLIRAVDSNSCT
.................................Q....S...
^
Here an I-->V change is seen in some tasmanian devils reads relative to opossum and wallaby. Here V is more typical of a theran mammal. Note I is also seen in armadillo, a placental, and A occurs in platypus and various other mammals. ACOT12, a acyl-CoA thioesterase, does not track back well in earlier diverging species. Because of the observed homoplasy, this locus is an unsuitable example of a significant amino acid change in Sarcophilus. However it illuminates the nature of suitable candidates and so is retained here.
^ ACOT12_hg18_14 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSNSCI ACOT12_panTro2 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSNSCI ACOT12_gorGor1 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSNSCI ACOT12_ponAbe2 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSNSCI ACOT12_rheMac2 NTYTVAVKSVILPS V PPSPQYIRSEIICAGFLIHAIDSSSCI ACOT12_calJac1 NTYTVAVKSVMLPS V PPSPQYIRSEIICAGFLIHAIDSNSCI ACOT12_micMur1 NTYTVAVKSVILPS V PPSPQHVRSEIICAGFLIHAADSNSCT ACOT12_otoGar1 NTYMVAAKSVILPS V PPSPQYIRSEIICAGFLIHTIDSTSCT ACOT12_tupBel1 NTYTVAVKSVTLPS V PPSPQYIRSDIICAGFLIRPVDSSSCT ACOT12_mm9_14_ NTYTVALRSVVLPS V PSSPQYIRSEVICAGFLIQAVDSNSCT ACOT12_rn4_14_ NTYIVALMSVVLPS V PPSPQYIRSQVICAGFLIQPVDSSSCT ACOT12_dipOrd1 NTYVVATKSVILPS V PPSPAYIRSEAVCSGFLIKAVDSSSCT ACOT12_cavPor3 DTYLVAVKSVVLPA V PPSPGYTRSEVALAGFLIQPTDHSSCT ACOT12_oryCun1 HAYTVAAKSVMLPS A PPSPDHTRSEIICAGFLIHAIDSHSCT ACOT12_ochPri2 HAYVVAVKSVVLPS A PPSPEYIRGEIVCAGFLIHAIDSHACT ACOT12_vicPac1 NTYTVAVKSVILPS V PPSPQYVRSEITCAGFLIHAIDNSSCT ACOT12_turTru1 HTYTVAVRSVILAS V PPSPQYSRSEIISAGFLIRAIDSSSCT ACOT12_bosTau4 HTYVVAVRSVILPS V PPSPQYVRSEIECAGFLIHATDSSSCT ACOT12_equCab2 KTFSVAAKSVILPS V PPSPQYMRSEIRCAGFLICAIDNSSCT ACOT12_felCat3 STYTVAVKSVLLPS V PPCPHYIRSEIICAGFLIRAIDSSSCT ACOT12_canFam2 NTYTVAVKSVTLPS V PPSPQYSRSEILCAGFLIHAIDSSSCT ACOT12_myoLuc1 NTYTVAVKSVILPS V PPSPQYVRSEIICAGFLIHAIDSSSCT ACOT12_pteVam1 NTYTVAVKSVILPS V PPSPZYVRSEIVCAGFLIHAIDGSLCI ACOT12_eriEur1 STFTVAMKSVLLAS V PSSPQYIRSEITCAGFVIHAVSSNSCI ACOT12_sorAra1 NAFTVAVKSVILPS V PPSPQYMRSEIICAGFLIHATDSNSCI ACOT12_loxAfr2 D--TVAVKSVLLPS V PPCPQYIRSEIIRAGFLIHTIDSNSCT ACOT12_echTel1 TTYTVALRSVLLPS V PSSPNYVRGEIICAGFLVHPIDSSACT ACOT12_dasNov2 NTYTVAVKSVVLPS I PPSPQYIRSEIICAGFLIHAIDSSSCT ACOT12_choHof1 NSYTVAAKSVVLPS V PPSPQYIRSETICAGFLINAIDSSSCT ACOT12_monDom4 NTYVVAVKSVTLAS I PPSPQYNRSEITCAGFLIRAVDSNSCT ACOT12_macEug NTYVVAMKSVTLAS I PPSPQYNRSEITSAGFLIQAVDSNSCT ACOT12_sacHar1 NTYVVAVKSVTLAS I PPSPQYNRSEITCAGFLIQAVDSSSCT ACOT12_sacHar2 NTYVVAVKSVTLAS V PPSPQYNRSEITCAGFLIQAVDSSSCT ACOT12_ornAna1 DSYLVAVKSVILAS A PPSHQYIRSEIPCAGFLVEALDSSSCK
Case of FLI1
chr4_11174 FLI1 3 32 N=2(63) K=3(47)
>contig00001 length=575 numreads=9
ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEKNGPPPNMTTNERRVIVPA
..................................................
^
Here the N-->K change is a non-conservative substitution in the sense asperagine is merely polar whereas lysine is bulkier and negatively charged. The N is highly invariant at this position back to teleost fish. FLI1 is a transcription factor associated with a leukemia virus integration site and Ewing sarcoma.
This would be a promising candidate except for the fact that the three reads establishing K clearly are plagued by frameshifts at the critical region. Possibly anomalous base composition is responsible here (ggatgagaagaacggcccccctcc) -- which is no doubt giving rising to transcriptional slippage generating homoplasic deletions of polyP -- or perhaps low coverage. This change is unlikely to be validated upon additional bulk or targeted sequencing because these lack motivating evidence.
>FP1JAYN01BA7O5 and FP5M7SR01ERAQP Frame = +1 Frame = +2
Query: 1 ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEKNGPP 36 KNGPPPNMTTNERRVIVPA 50
ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEK P KNGPPPNMTTNERRVIVPA
Sbjct: 37 ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEKKRSP 144 KNGPPPNMTTNERRVIVPA 187
>FP1JAYN01DX0A1 length=254
Query: 1 ESPVDCSVNKCSKLVGGNESNPMN-YNTYMDEKNGPPPNMTTNERRVIVPA 50
ESPVDCSVNKCSKLVGGNESNPMN + + EK PPPNMTTNERRVIVPA
Sbjct: 37 ESPVDCSVNKCSKLVGGNESNPMNLQHLHG*EKTVPPPNMTTNERRVIVPA 189
N P M N Y N T Y M D E K N G P P P N M T
| | | | | | | | | | | | | | | | | | | |
aatcctatgaattacaatacctacatggatgagaagaacggcccccctcctaacatgacc
FLI1_hg18_3_ ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_panTro2 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_gorGor1 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_ponAbe2 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_rheMac2 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_calJac1 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_tarSyr1 ESPVDCSVSKCSKLVGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_micMur1 ESPVDCSVSKCGKLIGGGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_otoGar1 ESPVDCSVSKCSKLIGGGEANPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_mm9_3_9 ESPVDCSVSKCNKLVGGGEANPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_rn4_3_9 ESPVDCSVSKCNKLVGGGEANPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_dipOrd1 ESPVDCSVSKCSKLVGGGESNPMNYNSYIDEK N GPPPPNMTTNERRVIVPA
FLI1_cavPor3 ESPVDCSVSKCSKLVGTGESNPMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_speTri1 ---VDCSVSKCSKLVFGGESNPMNYNSYLDEK N GPPPPNMTTNERRVIVPA
FLI1_oryCun1 ESPVDCSISKCGKLVGGGEANAMSYNNYMDEK N GPPPPNMTTNERRVIVPA
FLI1_vicPac1 ESPVDCSVSKCGKLVGGGESNTMSYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_turTru1 ESPVDCSVSKCGKLVGGGESNAMSYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_bosTau4 ESPVDCSVSKCGKLVGGGESNTMSYTSYVDEK N GPPPPNMTTNERRVIVPA
FLI1_equCab2 ESPVDCSVSKCSKLVGGGESNAMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_canFam2 ESPVDCSVSKCSKLVGGSESNAMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_myoLuc1 ESPVDCSVSKCSKLVGGGESNAMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_pteVam1 ESPVDCSVSKCSKLVGGGESNAMNYNSYIDEK N GPPPPNMTTNERRVIVPA
FLI1_eriEur1 ESPVDCSVSKCSKLVGGGESNAMNYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_proCap1 ESPVDCSVSKCSKLAGGGESNPMNYNTYMDEK N GPPPPNMTTNERRVIVPA
FLI1_dasNov2 ESPVDCSVSKYSKLVGGGESNPMTYSTYMDEK N GPPPPNMTTNERRVIVPA
FLI1_choHof1 ESPVDCSVSKCSKLVGGGEATPMTYNTYMDEK N GPP-PNMTTNERRVIVPA
FLI1_monDom4 ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEK N GPP-PNMTTNERRVIVPA
FLI1_macEug ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEK N GPP-PNMTTNERRVIVPA
FLI1_sarHar1 ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEK N GPP-PNMTTNERRVIVPA
FLI1_ornAna1 ESPVDCSVSKCGKLVGSGESNPMNYNSYMEEK N GPPPPNMTTNERRVIVPA
FLI1_galGal3 ESPVDCSVNKCSKLVGAGESNPMSYSTYMDEK N GPP-PNMTTNERRVIVPA
FLI1_taeGut1 ESPVDCSMNKCGKLVGAGESNPMSYSTYMDEK N GPP-PNMTTNERRVIVPA
FLI1_anoCar1 ESPVDCSVSKCNKLVPAGESNSLNYGTYMDEK N GPP-PNMTTNERRVIVPA
FLI1_xenTro2 ESPVDCSISKCSKLIGGSENNAVTYNSYMDEK N GPPPPNMTTNERRVIVPA
FLI1_tetNig1 ESPVDCSVGKCNKLVGGNDVSQMSYGSYMDEK N APP-PNMTTNERRVIVPA
FLI1_fr2_3_9 ESPVDCSVGKCNKLVGGNDVSQMNYGSYMDEK N APP-PNMTTNERRVIVPA
FLI1_gasAcu1 ESPVDCSVGKCNKLVGSNDTSQMNYGNYMDEK N APP-PNMTTNERRVIVPA
FLI1_oryLat2 ESPVDCSVGKCNKLVGGNDTSQMTYGNYMDEK S APP-PNMTTNERRVIVPA
FLI1_danRer5 ESPVDCSVGKCNKMVGGTEASQMNYTGYMDEK C APP-PNMTTNERRVIVPA
Case of SPON1
chr5_8347 SPON1 11 20 V=3(65) I=2(66) wobbly
>contig00001 length=433 numreads=5
GSTCTMSEWITWSPCSISCGVGMRSRERYVKQFPEDGSVCTVPTEETEKCTVNEEC
......................................I.N...............
^
Here two Sarcophilus reads show V-->I following residue 20 while three are V like opossum. It quickly emerges that wallaby also has I. Thus the change in tasmanian devil is within the normal reduced alphabet of this residue position. Various placentals show that T and M and even P are also accepted substituents here. Note too these are used clade-incoherently (eg primates alone are variable). Consequently this site is not under strong selection for V to begin with so SPON1 does not meet the selection criteria being used here.
^ SPON1_hg18_13 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_panTro2 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_gorGor1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_ponAbe2 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_rheMac2 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_calJac1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_tarSyr1 -GSTCTMSEWITWSPCCLSCV P GMRSREYYLK-FFEDGSVCSLTPKKTQNRTV-EZC SPON1_micMur1 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_otoGar1 DGSTCTMSEWITW-PCSISCG T GMRSRERYVKQFPEDVSVCTLPTEETEKCTVNEEC SPON1_tupBel1 EGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_mm9_13_ DGSTCTMSEWITWSPCSVSCG M GMRSRERYVKQFPEDGSVCMLPTEETEKCTVNEEC SPON1_rn4_13_ DGSTCTMSEWITWSPCSVSCG M GMRSRERYVKQFPEDGSVCMLPTEETEKCTVNEEC SPON1_dipOrd1 -GSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_cavPor3 DGSTCTMSEWIIWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_speTri1 EHSTCTMSEWITWSPCCISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_oryCun1 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_ochPri2 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_turTru1 DGSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPT-ETEKCTVNEEC SPON1_bosTau4 DGSTCTMSEWITWSPCSISCG T GTRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_equCab2 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_canFam2 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_myoLuc1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_pteVam1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_eriEur1 DGSACTMSEWITWSPCSLSCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_sorAra1 DGSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_proCap1 -GSTCTMSEWITWSPCSISCG T GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_echTel1 ----CPMSEWITWSPRSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_dasNov2 -GSTCTMSEWITWSPCSISCG M GMRSRERYVKQFPEDGSVCTLPTEETEKCTVNEEC SPON1_monDom4 DGSTCTMSEWITWSPCSISCG V GMRSRERYVKQFPEDGSVCTVPTEETEKCTVNEEC SPON1_macEug GSTCTMSEWMTWSPCSISCG I GMRSRERYVKQFPEDGSVCTVPTEETEK SPON1_sacHar1 GSTCTMSEWITWSPCSISCG V GMRSRERYVKQFPEDGSICNVPTEETEKCTVNEEC SPON1_sacHar2 GSTCTMSEWITWSPCSISCG I GMRSRERYVKQFPEDGSICNVPTEETEKCTVNEEC SPON1_ornAna1 DGSTCTMSEWITWSPCSVSCG M GMRSRERYVKQFPDDGSMCKVPTEETEKCVVNEDC SPON1_anoCar1 DGSTCMMSEWITWSPCSVSCG M GMRSRERYVKQFPDDGSMCKVPTEETEKCIVNEEC SPON1_xenTro2 EASTCMMSEWITWSPCSASCG M GMRSRERYVKQFPEDGSMCKVPTEETEKCIVNEEC SPON1_tetNig1 DASTCMMSEWITWSPCSASCG M GSRSRERYVKQFPDDGSICTLPTEETEDCVVNEEC SPON1_fr2_13_ DASTCMMSEWITWSPCSASCG M GSRSRERYVKQFPDDGSICTLPTEETEDCVVNEEC SPON1_gasAcu1 DASTCMLSEWITWSPCSLSCG M GTRSRERYVKQFPDDGSLCSLPTEETDNCVVNEEC SPON1_oryLat2 DGSTCMMSEWITWSPCSMSCG A GIRSRERYVKQFPDDGSICTLPTEETENCVVNEEC SPON1_danRer5 DSSTCMMSEWITWSPCSVSCG S GLRSRERYVKQFPDDGFACTHPTEETEPCTVNEEC
Marsupial data availability
Scattered data is available for other marsupials and monotremes from 454 reads, Sanger trace data and transcripts:
Didelphis virginiana 88,207 traces 248 nuc Trichosurus vulpecula 169,115 traces 321 nuc 147,199 ests Sminthopsis crassicaudata 59 nuc 1,669 ests Sminthopsis macroura 3,411 traces 89 nuc Isoodon macrourus 6,144 traces, 70 nuc 1,319 ests Tachyglossus aculeatus 93,653 traces 243 nuc SRX000015 Baylor 454 sequencing of Monodelphis domestica genomic fragment library SRX000086 WUGSC 454 sequencing of Macropus eugenii genomic fragment library SRX000186 WUGSC 454 sequencing of Ornithorhynchus anatinus transcript SRX000122 WUGSC 454 sequencing of Tachyglossus aculeatus transcript SRX000121 WUGSC 454 sequencing of Tachyglossus aculeatus transcript
The running estimate of coverage of Sarcophilus genome combining all runs for 11 expected genes on different chromosomes:
59 of 68 exons found (87%) 3883 of 4339 amino acids available (89%)
Newbler has a bad tendency to create non-existent frameshifts as seen in these three reads for the same query gene:
Query: 82 ggtctctacggcagtgtcattgtcactggagggaacacactcttgcaagg
|||||||||||||||||||||| |||||||||||||||||| ||||||||
Sbjct: 167 ggtctctacggcagtgtcattg-cactggagggaacacactgttgcaagg FP1I63R01APY7E
Query: 82 ggtctctacggcagtgt-cattgtcactggagggaacacactcttgcaagg
||||||||||||||||| |||||||||||||||||||||||| ||||||||
Sbjct: 268 ggtctctacggcagtgttcattgtcactggagggaacacactgttgcaagg FKUJDAX01AWWZ3
Query: 82 ggtctctacggcagtgtcattgtcactggagggaac-acactcttgcaagg
|||||||||||||||||||||||||||||||||||| ||||| ||||||||
Sbjct: 268 ggtctctacggcagtgtcattgtcactggagggaacgacactgttgcaagg FKUJDAX01DZSZO