Marsupial phyloSNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 227: | Line 227: | ||
'''Side issues:''' The column marked with an asterisk in the difference alignment below indicates a non-conservative phyloSNP K-->I that occured in the theran mammal stem after platypus divergence. All three marsupial sequences including tasmanian devil have isoleucine in this position as do all 30 of the available placental mammal sequences, suggesting that both the lysine and the isoleucine continue to be under strong selection. No comparable shift occured in the theran stem for MGAT5B where the residue is arginine in all species, a basic residue similar to lysine. | '''Side issues:''' The column marked with an asterisk in the difference alignment below indicates a non-conservative phyloSNP K-->I that occured in the theran mammal stem after platypus divergence. All three marsupial sequences including tasmanian devil have isoleucine in this position as do all 30 of the available placental mammal sequences, suggesting that both the lysine and the isoleucine continue to be under strong selection. No comparable shift occured in the theran stem for MGAT5B where the residue is arginine in all species, a basic residue similar to lysine. | ||
'''Structural significance:''' The MGAT5 gene encodes a conventional enzyme, mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase | '''Structural significance:''' The MGAT5 gene supposedly encodes a conventional enzyme, mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase involved in the synthesis of protein-bound and lipid-bound oligosaccharides. Yet surprisingly, no determined 3D structure exists at PDB relevent to the configuration of this exon -- nor indeed the large 741 residue protein. This is very peculiar because glycosyl transerfases are a well-studied group of enzymes (nearly 100 loci in human) and might be expected to bind UDP-GlcNAc (like MGAT4A or MGAT3). | ||
[http://www.expasy.org/uniprot/MGT5A_HUMAN SwissProt] does not annotate the cysteine at position 532 as part of a disulfide or active site; the predicted location (Golgi) can have [http://www.jbc.org/cgi/content/abstract/271/13/7758 homodimer disulfides] of similar enzymes, though this is a [http://www.pnas.org/content/104/26/10813.full complex topic]. Highest MGAT5 expression occurs in brain, heart, kidney, and placenta. No domains other than a signal peptide and 6 of its own glycosylation target sites are found by online tools such as [http://smart.embl-heidelberg.de/ SMART]. | [[Image:MGAT5.jpg|left]] | ||
Only a small region of the protein have a [http://salilab.org/modbase/search?databaseID=Q09328 prediction] at ModBase using 2f9fA, a remote mannosyltransferasee from Archaeoglobus fulgidus. Luckily the model covers the cysteine at issue, showing two helices and a beta sheet. | |||
[http://www.expasy.org/uniprot/MGT5A_HUMAN SwissProt] does not annotate the cysteine at position 532 as part of a disulfide or active site; the predicted location (Golgi) can have [http://www.jbc.org/cgi/content/abstract/271/13/7758 homodimer disulfides] of similar enzymes, though this is a [http://www.pnas.org/content/104/26/10813.full complex topic]. Although all 20 cysteines in this protein are conserved human to opossum, this could be a consequence of the overall sequence identity of 90%. | |||
Highest MGAT5 expression occurs in brain, heart, kidney, and placenta. No domains other than a signal peptide and 6 of its own glycosylation target sites are found by online tools such as [http://smart.embl-heidelberg.de/ SMART]. | |||
Although the bulky tyrosine substitution is conservative in the sense of polar nature and perhaps hydrogen-bonding capacity, it cannot replace these specialized functions of cysteine. Considering the extreme conservation of this cysteine, this substitution must have a substantial-- <font color="blue">perhaps even disabling</font> -- impact on enzymatic function. | Although the bulky tyrosine substitution is conservative in the sense of polar nature and perhaps hydrogen-bonding capacity, it cannot replace these specialized functions of cysteine. Considering the extreme conservation of this cysteine, this substitution must have a substantial-- <font color="blue">perhaps even disabling</font> -- impact on enzymatic function. | ||
Revision as of 11:33, 13 February 2009
Introduction to Marsupial phyloSNPs
In this project, new genomic data from the Tasmanian devil (Sarcophilus harrisii), Tasmanian tiger (Thylacinus cynocephalus), and echidna (Tachyglossus aculeatus) are analyzed for significant changes at the protein coding level. The goal is to find single amino acid changes in one of these species at a highly invariant residue in a well-conserved exon in a gene with known or predictable tertiary structure. Such changes are thought to enrich for genetic changes with significant, adaptive biochemical or phenotypic consequences (1,2,3,4), in contrast to ordinary SNPs at positions of low conservation. Thus phyloSNPs are informative to the distinctive biology of the species carrying them and suggest a focus for subsequent experiment.
It is also of particular interest to determine the levels of variation within the Tasmanian devil population as a whole because the number of individuals have become low and possibly inbreed with adverse sequelae. For this it will be necessary to first determine sites of variation and then to genotype them across a large number of individuals.
Marsupial genomic and cDNA data to date has been quite limited compared to placental mammal. Yet as outgroup, metatheran animals provide important context to placentals and represent important context in understanding human protein evolution. The monotheres are inevitably limited by the paucity of extant species (basically platypus and echidna) and dim prospects for fossil DNA. Consequently echidna provides an important adjunct to the existing but incomplete platypus assembly. While extant birds and reptiles -- the preceding divergence node -- are abundant it must be remembered that a very considerable time elapsed (from 310 mry to 175 mry) prior to divergence of mammals with living representatives. This gap of 135 myr is comparable to the whole evolutionary record of theran mammals.
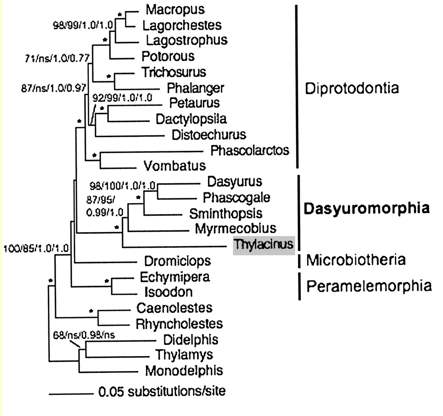
Assumed vertebrate phylogenetic tree
Marsupial relationships taken from 2009 paper establishing the mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus):
Newick tree that generates vertebrate phylogenetic tree used in the analysis here: ((((((((((((((((((homSap,panTro),gorGor),ponPyg),macMul),calJac),tarSyr),(micMur,otoGar)),tupBel), (((((musMus,ratNor),dipOrd),cavPor),speTri),(oryCun,ochPri))), (((((vicPac,susScr),turTru),bosTau),((equCab,(felCat,canFam)),(myoLuc,pteVam))),(eriEur,sorAra))), (((loxAfr,proCap),echTel),(dasNov,choHof))), (monDom,((macEug,triVul),(sarHar,thyCyn)))), (ornAna,tacAcu)), ((galGal,taeGut),anoCar)), xenTro), (((tetNig,takRub),(gasAcu,oryLap)),danRer)), calMil), petMar);
Phylo-sorting data
This tab-delimited table enables four different sort orders. These are needed because data can be missing from species in a manner that varies by gene, making data alignment difficult. Some alignment tools also lose input order, so that needs to be recovered. The ordering here flattens the phylogenetic tree by taking human (arbitrarily) at the top and resolving ambiguous situations (eg mouse, rat) by putting species with the best assemblies first.
The first two columns provide sort order number for the 44 species alignment at UCSC as phylogenetic and alphabetic order respectively. The third and fifth columns do this for a larger set of species for which data is commonly available. The fourth column provides a fasta line indicator. The sixth column is a dummy gene name to be replaced as needed. The next column has stripped out the syntax from Newick tree format. This column and column six together will correctly draw the vertebrate phylogenetic tree in all online software without further editing. The final columns provide genus, species, and common name.
- - - - - - - (((((((((((((((((( - - - - 10 26 10 > 27 gene homSap , Homo sapiens (human) hg181 11 38 11 > 40 gene panTro ), Pan troglodytes (chimp) panTro 12 25 12 > 26 gene gorGor ), Gorilla gorilla (gorilla) gorGor 13 40 13 > 42 gene ponPyg ), Pongo pygmaeus (orang) ponAbe 14 28 14 > 30 gene macMul ), Macaca mulatta (rhesus) rheMac 15 12 15 > 12 gene calJac ), Callithrix jacchus (marmoset) calJac 16 48 16 > 53 gene tarSyr ),( Tarsius syrichta (tarsier) tarSyr 17 29 17 > 31 gene micMur , Microcebus murinus (mouse_lemur) micMur 18 37 18 > 39 gene otoGar )), Otolemur garnettii (bushbaby) otoGar 19 50 19 > 57 gene tupBel ),((((( Tupaia belangeri (tree_shrew) tupBel 20 31 20 > 33 gene musMus , Mus musculus (mouse) mm91 21 43 21 > 45 gene ratNor ), Rattus norvegicus (rat) rn41 22 18 22 > 19 gene dipOrd ), Dipodomys ordii (kangaroo_rat) dipOrd 23 14 23 > 15 gene cavPor ), Cavia porcellus (guinea_pig) cavPor 24 45 24 > 48 gene speTri ),( Spermophilus tridecemlineatus (squirrel) speTri 25 35 25 > 37 gene oryCun , Oryctolagus cuniculus (rabbit) oryCun 26 33 26 > 35 gene ochPri ))),((((( Ochotona princeps (pika) ochPri 27 52 27 > 59 gene vicPac , Vicugna pacos (lama) vicPac 54 57 28 > 49 gene susScr ), Sus scrofa (pig) 28 51 29 > 58 gene turTru ), Tursiops truncatus (dolphin) turTru 29 11 30 > 11 gene bosTau ),(( Bos taurus (cow) bosTau 30 20 31 > 21 gene equCab ,( Equus caballus (horse) equCab 31 22 32 > 23 gene felCat , Felis catus (cat) felCat 32 13 33 > 14 gene canFam )),( Canis familiaris (dog) canFam 33 32 34 > 34 gene myoLuc , Myotis lucifugus (microbat) myoLuc 34 42 35 > 44 gene pteVam ))),( Pteropus vampyrus (macrobat) pteVam 35 21 36 > 22 gene eriEur , Erinaceus europaeus (hedgehog) eriEur 36 44 37 > 47 gene sorAra ))),((( Sorex araneus (shrew) sorAra 37 27 38 > 28 gene loxAfr , Loxodonta africana (elephant) loxAfr 38 41 39 > 43 gene proCap ), Procavia capensis (hyrax) proCap 39 19 40 > 20 gene echTel ),( Echinops telfairi (tenrec) echTel 40 17 41 > 18 gene dasNov , Dasypus novemcinctus (armadillo) dasNov 41 15 42 > 16 gene choHof ))),( Choloepus hoffmanni (sloth) choHof 42 30 43 > 32 gene monDom ,(( Monodelphis domestica (opossum) monDom 55 55 44 > 29 gene macEug , Macropus eugenii (wallaby) 56 56 45 > 46 gene sarHar ),( Sarcophilus harrisii (tasmanian_devil) 57 60 46 > 56 gene triVul , Trichosurus vulpecula (bushytail_possum) 58 59 47 > 55 gene thyCyn )))),( Thylacinus cynocephalus (tasmanian_tiger) 43 34 48 > 36 gene ornAna , Ornithorhynchus anatinus (platypus) ornAna 59 58 49 > 50 gene tacAcu )),(( Tachyglossus aculeatus (echidna) 44 23 50 > 24 gene galGal , Gallus gallus (chicken) galGal 45 46 51 > 51 gene taeGut ), Taeniopygia guttata (finch) taeGut 46 10 52 > 10 gene anoCar )), Anolis carolinensis (lizard) anoCar 47 53 53 > 60 gene xenTro ),((( Xenopus tropicalis (frog) xenTro 48 49 54 > 54 gene tetNig , Tetraodon nigroviridis (pufferfish) tetNig 49 47 55 > 52 gene takRub ),( Takifugu rubripes (fugu) fr21 50 24 56 > 25 gene gasAcu , Gasterosteus aculeatus (stickleback) gasAcu 51 36 57 > 38 gene oryLap )), Oryzias latipes (medaka) oryLat 52 16 58 > 17 gene danRer )), Danio rerio (zebrafish) danRer 60 54 59 > 13 gene calMil ), Callorhinchus milii (elephantfish) 53 39 60 > 41 gene petMar ) Petromyzon marinus (lamprey) petMar 44 44 51 f 51 gene fasta tree_syntax genus species common ucsc phy alp phy alp
Candidate analysis
The first issue is error within the reads themselves; the second is whether the default 454 Newbler assembler correctly identified overelapping reads and put them together properly to give exon-spanning reads. Those issues are discussed elsewhere -- here it is assumed the data is correct, so the entire focus is on subsequent bioinformatics.
(methods explained more shortly)
Case of ERN2
chr6_5971 ERN2 4
contig00001 length=355 numreads=5
KLPFTIPELVHASPCRSSDGVLYT
.....................F..
^
15 R=3(75) H=2(50
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by L->F), then differences between the two thylacines
(here one individual has R at position 15, the other has H), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues: ERN2 has not generated potentially confusing recent processed pseudogenes in mammals (lack of human, opossum or platypus genome Blat matches to ERN2 query). The variation observed here between individual tasmanian devils is implausibly an early stage in the loss of parent gene because of ERN2 functional essentiality; the exon cannot come from a decaying segmental duplication because coverage is high enough to also detect the main gene.
Paralog issues: The GeneSorter tool at UCSC shows a single significant full-length paralog in human, ERN1, also with 22 coding exons. The genes reside on different chromosomes but in regions with local homology of synteny. However this particular exon is a good match (3 differences out of 23), so there is potential for experimental difficulties in distinguishing them in short reads (including the following exon readily resolves them bioinformatically). In any event, at positions 15 and 20, ERN1 is identical at the amino acid level to ERN2. The gene duplication appears to have occured subsequent to amphioxus divergence earlier diverging metazoans are single-copy.
Homoplasy (recurrent mutation) issues: This exon is very conserved and does not exhibit repetitive sequence, compositional simplicity, or indels in any species in either paralog that could foster experimental error or alignment ambiguity. At position 15, the ancestral value is arginine in both paralogs. The G--> A transition to histidine in one individual is conservative under most circumstances (still basic) and arises from an arginine codon CpG hotspot conserved back to lamprey in 30 of 32 species with available data, yet histidine is not observed part of a reduced alphabet (ie R/H) at this position over many billions of years of branch length. Consequently R-->H is a significant change in this individual tasmanian devil.
Known variations: No human disease variants have been reported for either ERN2 or ERN1, probably attributable to essentiality. Site-specific mutation close to the exon here have been generated for K121P, D123P, W125A, and Q105E but only for ERN1. Naturally occuring coding SNPs in the human population relevent to the ERN2 exon are not known but low frequency alleles could emerge from the 1000 Genomes Project.
Side issues: a very ancient conserved leucine at position 21 appears to be transitioning to phenylalanine at marsupial node but has not been fixed, so settles out as L or F depending on lineage-sorting on each terminal marsupial leaf whereas placentals are all changed to phenylalanine (a phyloSNP caught in mid-air). While L and F might seem about the 'same' as amino acids, the branch length conservation totals say both are important but for different reasons: this is not a waffle codon nor reduced alphabet situation. This raises the question -- given the extreme conservation of this exon otherwise -- of whether the L-->F change at position 21 in both individuals has 'enabled' (made neutral or adaptive) an otherwise unfavorable R-->H change at position 15 in one individual.
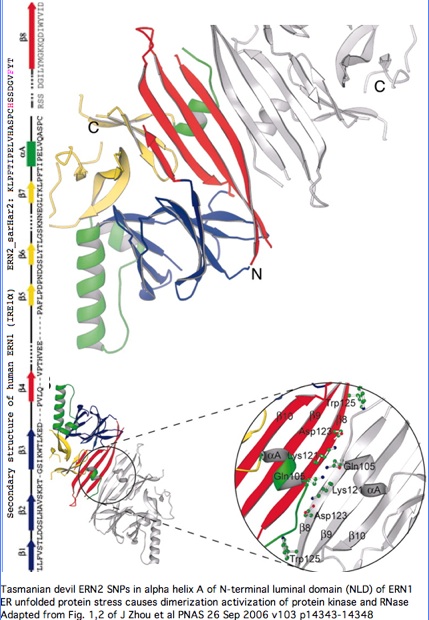
Structural significance: By good fortune, the crystal structure of ERN1 (alternately called IRE1) has been published. The PDB 2HZ6 structure has good coverage of this particular exon. Consequently the marsupial ERN2 could be very accurately modelled and the structural effects of L-->F with or without R-->H computed by submission to online SwissProt modelling service.
Monodelphis ERN2 (key exon: sarHar2) aligned to human ERN1 luminal domain
Expect = 5.8e-65 Identities = 109/180 (60%), Positives = 141/180 (78%)
ERN2_monDom 1 PESLLFISTLDGSLHAVSKKTGDIQWTLKDDPIIQGPVYATEPAFLPDPSDGSLYILGEE 60
PE+LLF+STLDGSLHAVSK+TG I+WTLK+DP++Q P + EPAFLPDP+DGSLY LG +
ERN1_homSap 8 PETLLFVSTLDGSLHAVSKRTGSIKWTLKEDPVLQVPTHVEEPAFLPDPNDGSLYTLGSK 67
ERN2_monDom 61 SKQGLMKLPFTIPELVHASPCHSSDGVFYTGRKQDTWFMVDPKSGKKQTMLSTETWDGLY 120
+ +GL KLPFTIPELV ASPCRSSDG+LY G+KQD W+++D +G+KQ LS+ D L
ERN1_homSap 68 NNEGLTKLPFTIPELVQASPCRSSDGILYMGKKQDIWYVIDLLTGEKQQTLSSAFADSLC 127
ERN2_monDom 121 PSAPLLYIGRTQYTVTMYDPRSQALRWNTTYRGYSAPLLDHLPGYQVGHFTCSGEGLVVT 180
PS LLY+GRT+YT+TMYD +++ LRWN TY Y+A L + Y++ HF +G+GLVVT
ERN1_homSap 128 PSTSLLYLGRTEYTITMYDTKTRELRWNATYFDYAASLPEDDVDYKMSHFVSNGDGLVVT 187
Functional significance: A considerable amount is known about the paralog ERN1. Annotation transfer is likely applicable to ERN2. The two gene products differ primarily in expression -- ERN1 ubiquitious but ERN2 restricted to intestinal epithelial cells:
"The unfolded protein response (UPR) is an evolutionarily conserved mechanism by which all eukaryotic cells adapt to the accumulation of unfolded proteins in the endoplasmic reticulum (ER). Inositol-requiring kinase 1 (IRE1 or ERN1) and PKR-related ER kinase (PERK) are two type I transmembrane ER-localized protein kinase receptors that signal the UPR through a process that involves homodimerization and autophosphorylation... The monomer of the luminal domain comprises a unique fold of a triangular assembly of beta-sheet clusters. Structural analysis identified an extensive dimerization interface stabilized by hydrogen bonds and hydrophobic interactions... Mutations that disrupt the dimerization interface produced ERN1 protein that failed to either dimerize or activate the UPR upon ER stress."
"ERN1 is a type I transmembrane protein kinase receptor that also has a site-specific RNase activity that, upon activation, initiates a site-specific unconventional splicing reaction. The substrate for IRE1 RNase in metazoans is Xbp1 mRNA, which encodes a basic leucine zipper transcription factor of the ATF/CREB family. XBP1 controls expression of genes containing an X-box element or a UPR element in their promoter regions. The IRE1-mediated splicing reaction introduces into XBP1 an alternative C terminus, thereby generating an XBP1 molecule that is a more potent transcriptional activator. Therefore, activation of IRE1 and its RNase increases the transcription of genes encoding ER chaperones and folding catalysts... the ERN1 N-terminal luminal domain (NLD) functions as an ER stress sensor... under normal conditions IRE1 is maintained in a monomeric state through interaction of the NLD with the ER resident chaperone BiP. Upon ER stress, Grp78 binds to unfolded proteins as they accumulate, permitting the released NLD to form homodimers. Dimerization of the NLD in turn leads to the activation of the protein kinase and RNase activities in the cytosolic domain of ERN1."
ENR2 is readily distinguished from its ERN1 paralog at tBlastn by including the two following exons which bring percent identity to 62%:
ERN2_monDom KLPFTIPELVHASPCRSSDGVLYTGRKQDTWFMVDPKSGKKQTMLSTETWDGLYPSAPLLYIGRTQYTVTMYDPRSQALRWNTTYRGYSA
KLPFTIPELV ASPCRSSDG+LY G+KQD W++VD +G+KQ LS+ + L PS LLY+GRT+YT+TM+D +S+ LRWN TY Y+A
ERN1_monDom KLPFTIPELVQASPCRSSDGILYMGKKQDIWYVVDLMTGEKQQTLSSAFAESLCPSTSLLYLGRTEYTITMFDTKSRELRWNATYFDYAA
The first alignment shows ERN2 orthologs in vertebrates, the second as difference relative to opossum, the third ERN1 orthologs.
The ancestral nature of the CpG hotspot is shown in nucleotides in the final columns.
^ * ^ * ^ *
ERN2_homSap KLPFTIPELVHASPCRSSDGVFYT ERN2_homSa .....................F.. ERN1_homSap KLPFTIPELVQASPCRSSDGILYM CG Human
ERN2_panTro KLPFTIPELVHASPCRSSDGVFYT ERN2_panTr .....................F.. ERN1_panTro KLPFTIPELVQASPCRSSDGILYM CG Chimp
ERN2_ponAbe KLPFTIPELVHASPCRSSDGVFYT ERN2_ponAb .....................F.. ERN1_ponAbe KLPFTIPELVQASPCRSSDGILYM -- Gorilla
ERN2_rheMac KLPFTIPELVHASPCRSSDGVFYT ERN2_rheMa .....................F.. ERN1_rheMac KLPFTIPELVQASPCRSSDGILYM CG Orangutan
ERN2_calJac KLPFTIPELVHASPCRSSDGVFYT ERN2_calJa .....................F.. ERN1_calJac KLPFTIPELVQASPCRSSDGILYM CG Rhesus
ERN2_tarSyr KLPFTIPELVHASPCRSSDGVFYT ERN2_tarSy .....................F.. ERN1_tarSyr KLPFTIPELVQASPCRSSDGILYM CG Marmoset
ERN2_micMur KLPFTIPELVHASPCRSSDGVFYT ERN2_micMu .....................F.. ERN1_micMur KLPFTIPELVQASPCRSTDGILYM CG Tarsier
ERN2_tupBel KLPFTIPELVHASPCRSSDGVFYT ERN2_tupBe .....................F.. ERN1_otoGar KLPFTIPELVQASPCRSSDGILYM CG Mouse_lemur
ERN2_musMus KLPFTIPELVHASPCRSSDGVFYT ERN2_musMu .....................F.. ERN1_tupBel KLPFTIPELVQASPCRSSDGILYM -- Bushbaby
ERN2_ratNor KLPFTIPELVHASPCRSSDGVFYT ERN2_ratNo .....................F.. ERN1_musMus KLPFTIPELVQASPCRSSDGILYM CG TreeShrew
ERN2_cavPor KLPFTIPELVHTSPCRSSDGVFYT ERN2_cavPo ...........T.........F.. ERN1_ratNor KLPFTIPELVQASPCRSSDGILYM CG Mouse
ERN2_speTri KLPFTIPELVHASPCRSSDGVFYT ERN2_speTr .....................F.. ERN1_dipOrd KLPFTIPELVQASPCRSSDGILYM CG Rat
ERN2_oryCun KLPFTIPELVHASPCRSSDGVFYT ERN2_oryCu .....................F.. ERN1_cavPor KLPFTIPELVQASPCRSSDGILYM -- Kangaroo_rat
ERN2_ochPri KLPFSIPELVHASPCRSSDGVFYT ERN2_ochPr ....S................F.. ERN1_speTri KLPFTIPELVQASPCRSSDGILYM CG Guinea_pig
ERN2_turTru RLPFTIPELVHASPCRSSDGVFYT ERN2_turTr R....................F.. ERN1_oryCun KLPFTIPELVQASPCRSSDGILYM CG Squirrel
ERN2_bosTau RLPFTIPELVHASPCRSSDGVFYT ERN2_bosTa R....................F.. ERN1_vicPac KLPFTIPELVQASPCRSSDGILYM CG Rabbit
ERN2_equCab KLPFTIPELVHASPCRSSDGVFYT ERN2_equCa .....................F.. ERN1_turTru KLPFTIPELVQASPCRSSDGILYM CG Pika
ERN2_felCat RLPFTIPELVHASPCRSSDGVFYT ERN2_felCa R....................F.. ERN1_bosTau KLPFTIPELVQASPCRSSDGILYM -- Alpaca
ERN2_canFam KLPFTIPELVHASPCRSSDGVFYT ERN2_canFa .....................F.. ERN1_equCab KLPFTIPELVQASPCRSSDGILYM CG Dolphin
ERN2_myoLuc KLPFTIPELVHASPCRSSDGVFYT ERN2_myoLu .....................F.. ERN1_canFam KLPFTIPELVQASPCRSSDGILYM CG Cow
ERN2_eriEur KLPFTVPELVHTSPCRSSDGVFYT ERN2_eriEu .....V.....T.........F.. ERN1_myoLuc KLPFTIPELVQASPCRSSDGILYM CG Horse
ERN2_sorAra KLPFTIPELVHASPCRSSDGVFYT ERN2_sorAr .....................F.. ERN1_pteVam KLPFTIPELVQASPCRSSDGILYM CG Cat
ERN2_loxAfr KLPFTIPELVHASPCRSSDGVFYT ERN2_loxAf .....................F.. ERN1_eriEur KLPFTIPELVQASPCRSSDGILYM CG Dog
ERN2_echTel KLPFTIPELVLASPCRSSDGVFYT ERN2_echTe ..........L..........F.. ERN1_sorAra KLPFTIPELVQASPCRSSDGILYM CG Microbat
ERN2_dasNov KLPFTIPELVHTSPCRSSDGIFYT ERN2_dasNo ...........T........IF.. ERN1_loxAfr KLPFTIPELVQASPCRSSDGILYM -- Megabat
ERN2_monDom KLPFTIPELVHASPCRSSDGVLYT ERN2_monDo KLPFTIPELVHASPCRSSDGVLYT ERN1_proCap KLPFTIPELVQASPCRSSDGILYM CG Hedgehog
ERN2_macEug KLPFTIPELVHASPCRSSDGVFYT ERN2_macEu .....................F.. ERN1_echTel KLPFTIPELVQASPCRSSDGILYM CG Shrew
ERN2_sarHar1 KLPFTIPELVQASPCRSSDGIFYM ERN2_sarHa ..........Q.........IF.M ERN1_dasNov KLPFTIPELVQASPCRSSDGILYM -- Elephant
ERN2_sarHar2 KLPFTIPELVQASPCHSSDGIFYM ERN2_sarHa ..........Q....H....IF.M ERN1_choHof KLPFTIPELVQASPCRSSDGILYM -- Rock_hyrax
ERN2_ornAna KLPFTIPELVQSSPCRSSDGILYT ERN2_ornAn ..........QS........I... ERN1_monDom KLPFTIPELVQASPCRSSDGILYM CG Tenrec
ERN2_anoCar KLPFTIPELVQSSPCRSSDGIIYT ERN2_anoCa ..........QS........II.. ERN1_ornAna KLPFTIPELVHASPCRSSDGILYM CG Armadillo
ERN2_taeGut KLPFTIPELVQSSPCRSSDGVLYT ERN2_taeGu ..........QS............ ERN1_galGal KLPFTIPELVQASPCRSSDGILYM CG Opossum
ERN2_galGal KLPFTIPELVQASPCRSSDGILYM ERN2_galGa ..........Q.........I..M ERN1_taeGut KLPFTIPELVQASPCRSSDGILYM CG Platypus
ERN2_xenTro KLPFTIPELVQSSPCRSSDGILYT ERN2_xenTr ..........QS........I... ERN1_anoCar KLPFTIPELVQASPCRSSDGILYM CG Lizard
ERN2_xenLae KLPFTIPELVQSSPCRSSDGILYT ERN2_xenLa ..........QS........I... ERN1_xenTro KLPFTIPELVQSSPCRSSDGILYT CG Tetraodon
ERN2_tetNig KLPFTIPELVQASPCRSSDGVLYM ERN2_tetNi ..........Q............M ERN1_tetNig KLPFTIPELVQASPCRSSDGVLYM CG Fugu
ERN2_takRub KLPFTIPELVQASPCRSSDGVLYM ERN2_takRu ..........Q............M ERN1_takRub KLPFTIPELVQASPCRSSDGVLYM CT Stickleback
ERN2_gasAcu KLPFTIPDLVQSAPCRSSDGILYT ERN2_gasAc .......D..QSA.......I... ERN1_gasAcu KLPFTIPELVQASPCRSSDGVLYM CT Medaka
ERN2_oryLat KLPFTIPELVQSAPCRSSDGILYT ERN2_oryLa ..........QSA.......I... ERN1_oryLat KLPFTIPELVQASPCRSSDGVLYM CG Lamprey
ERN2_calMil KLPFTIPELVQSSPCRSSDGILYT ERN2_calMi ..........QS........I... ERN1_danRer KLPFTIPELVQASPCRSSDGILYM
ERN2_petMar KLPFTIPELVHASPCRTSDGVLYT ERN2_petMa ................T.......
ERN_braFlo KLPFTIPELVNASPCKSSDGILYT ERN_braFlo ..........N....K....I...
Case of MGAT5
chr4_4859 MGAT5 12
>contig00001 length=538 numreads=5 21 C=2(61) Y=2(56)
LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE
.................................................
^
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in two
tasmanian devil (here one is identical and the other differs from Monodelphis by C->Y) and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler).
Pseudogene issues: No processed pseudogenes relevent to this exon are seen by Blat of human and opossum sequence. Some questionable sequence occurs in tarsier and sloth but may be due to low coverage read or assembly error. These fragmentary sequences also have cysteine at the position in question.
Paralog issues: This gene has a moderately similar paralgog, MGAT5B, with a similar enzymatic role (beta1,6-N-acetylglucosaminyltransferase). The opossum MGAT5B protein differs at 12 positions out of 49 from opossum MGAT5, whereas human and marsupial MGAT5A differ at one residue. Consequently the two paralogs are readily distinguished within vertebrates. This is moot because 33 of 33 available MGAT5B also have cysteine at the position in question (data not shown).
Homoplasy (recurrent mutation) issues: The alignments below show tyrosine has never replaced cysteine in any other species. This cysteine is extremely invariant in both paralogs, tracing back to lophotrochozoa and cnidaria.
Known variations: No human disease alleles have been mapped to either paralog. None of 9 SNP tracks at the UCSC browser show human variatin in this exon.
Side issues: The column marked with an asterisk in the difference alignment below indicates a non-conservative phyloSNP K-->I that occured in the theran mammal stem after platypus divergence. All three marsupial sequences including tasmanian devil have isoleucine in this position as do all 30 of the available placental mammal sequences, suggesting that both the lysine and the isoleucine continue to be under strong selection. No comparable shift occured in the theran stem for MGAT5B where the residue is arginine in all species, a basic residue similar to lysine.
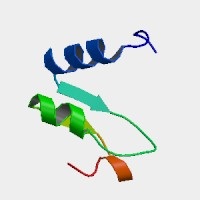
Structural significance: The MGAT5 gene supposedly encodes a conventional enzyme, mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase involved in the synthesis of protein-bound and lipid-bound oligosaccharides. Yet surprisingly, no determined 3D structure exists at PDB relevent to the configuration of this exon -- nor indeed the large 741 residue protein. This is very peculiar because glycosyl transerfases are a well-studied group of enzymes (nearly 100 loci in human) and might be expected to bind UDP-GlcNAc (like MGAT4A or MGAT3).
Only a small region of the protein have a prediction at ModBase using 2f9fA, a remote mannosyltransferasee from Archaeoglobus fulgidus. Luckily the model covers the cysteine at issue, showing two helices and a beta sheet.
SwissProt does not annotate the cysteine at position 532 as part of a disulfide or active site; the predicted location (Golgi) can have homodimer disulfides of similar enzymes, though this is a complex topic. Although all 20 cysteines in this protein are conserved human to opossum, this could be a consequence of the overall sequence identity of 90%.
Highest MGAT5 expression occurs in brain, heart, kidney, and placenta. No domains other than a signal peptide and 6 of its own glycosylation target sites are found by online tools such as SMART.
Although the bulky tyrosine substitution is conservative in the sense of polar nature and perhaps hydrogen-bonding capacity, it cannot replace these specialized functions of cysteine. Considering the extreme conservation of this cysteine, this substitution must have a substantial-- perhaps even disabling -- impact on enzymatic function.
Functional significance: In view of the facial tumor situation in tasmanian devils, OMIM's account of prior research in mouse on this gene is quite interesting. Less is known about MGAT5B though it also functions in the synthesis of complex cell surface N-glycans.
" Malignant transformation is accompanied by increased beta-1,6-GlcNAc branching of N-glycans attached to Asn-X-Ser/Thr sequences in mature glycoproteins... The amount of MGAT5 products correlates with disease progression... Mgat5-deficient mice, which are born healthy but develop various abnormalities as adults...Mgat5-deficient mice showed kidney autoimmune disease, enhanced delayed-type hypersensitivity, and increased susceptibility to experimental autoimmune encephalomyelitis...The Golgi enzyme beta1,6 N-acetylglucosaminyltransferase V (Mgat5) is up-regulated in carcinomas and promotes the substitution of N-glycan with poly N-acetyllactosamine, the preferred ligand for galectin-3 (Gal-3)...inhibitors of MGAT5 might be useful in the treatment of malignancies by targeting their dependency on focal adhesion signaling for growth and metastasis."
^ ^ * MGAT5_homSap LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE homSap MGAT5_panTro LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. panTro MGAT5_gorGor LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. gorGor MGAT5_ponAbe LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. ponAbe MGAT5_rheMac LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. rheMac MGAT5_calJac LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. calJac MGAT5_micMur LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... micMur MGAT5_otoGar LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. otoGar MGAT5_tupBel LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. tupBel MGAT5_musMus LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. musMus MGAT5_ratNor LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. ratNor MGAT5_criGri LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... criGri MGAT5_dipOrd LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. dipOrd MGAT5_cavPor LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. cavPor MGAT5_speTri LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. speTri MGAT5_oryCun LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. oryCun MGAT5_ochPri LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. ochPri MGAT5_vicPac LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. vicPac MGAT5_susScr LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. susScr MGAT5_turTru LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. turTru MGAT5_bosTau LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. bosTau MGAT5_equCab LFAGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ..A.............................................. equCab MGAT5_felCat lfvgLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... felCat MGAT5_canFam LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... canFam MGAT5_myoLuc LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. myoLuc MGAT5_eriEur LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. eriEur MGAT5_sorAra LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... sorAra MGAT5_loxAfr LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. loxAfr MGAT5_proCap LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. proCap MGAT5_echTel LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. echTel MGAT5_monDom LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... monDom MGAT5_macEug LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... macEug MGAT5_sarHar1 LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... sarHar1 MGAT5_sarHar2 LFVGLGFPYEGPAPLEAIANGYAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... sarHar2 MGAT5_ornAna LFVGLGFPYEGPAPLEAIANGCAFLNLKFNPPKSSKNTDFFKGKPTLRE ..........................L..............K....... ornAna MGAT5_galGal LFVGLGFPYEGPAPLEAIANGCAFLNLRFNPPKSSKNTEFFKGKPTLRE ..........................LR..........E..K....... galGal MGAT5_taeGut LFVGLGFPYEGPAPLEAIANGCAFLNLRFNPPKSSKNTDFFKGKPTLRE ..........................LR.............K....... taeGut MGAT5_anoCar LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFKGKPTLRE .........................................K....... anoCar MGAT5_xenTro LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSRNTDFFKGKPTLRE ...................................R.....K....... xenTro MGAT5_tetNig VFVGLSFPYEGPAPLEALANGCIFLNPRLKPPQSSLNSEFFKEKPNIRE V....S...........L....I....RLK..Q..L.SE..KE..NI.. tetNig MGAT5_takRub LFVGLSFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFKGKPTLRE .....S...................................K....... takRub MGAT5_gasAcu LFVGLSFPYEGPAPLEAIANGCAFLNPKFSPAKSSKNTDFFKGKPTLRE .....S.......................S.A.........K....... gasAcu MGAT5_oryLat LFVGLSFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFKGKPTLRE .....S...................................K....... oryLat MGAT5_danRer LFVGLSFPYEGPAPLEAIANGCAFLNPRFDPAKSSKNTDFFKGKPTLRE .....S.....................R.D.A.........K....... danRer MGAT5_oncMyk LFVGLSFPYEGPAPLEAIANGCAFLNPKFTPPKSSKNTDFFKGKPTLRE .....S.......................T...........K....... oncMyk MGAT5_pimPro LFVGLSFPYEGPAPLEAIANGCAFLNPRFDPSKSSKNTDFFKGKPTLRE .....S.....................R.D.S.........K....... pimPro MGAT5_calMil LFVGLGFPYEGPAPLEAIANGCAFLNPRFNPPKSSKNTEFFKGKPTLRE ...........................R..........E..K....... calMil MGAT5_petMar LFVGLGFPYEGPAPLEAIANGCVFLNPRFRPPKSSKNTDFFKGKPTLRE ......................V....R.R...........K....... petMar MGAT5_braFlo LFVGLGFPYEGPAPLEAIASGCVFLNPKFTQPKSRLNTKFFEGKPTFRE ...................S..V......TQ...RL..K..E....F.. braFlo MGAT5_strPur LFIGLGFPYEGPAPLEAVANGCVFLNPKFNPPKNYQNTKFFQGKPTSR. MGAT5_helRob LFIGLGFPYEGPAPLEAIAAGCVFINPKFNPPHSSLNTKFFKGKPTARE MGAT5_nemVec VFIGLGFPYEGPAPLEAIQSGCVFLNAKFDPPHDRVNTPFFKNKPTLRK MGAT5_acrMil VFIGLGFPYEGPAPLEVISQGCVFLNPKFEPPLSSSNSDFF........
Note: the species with unfamiliar genSpp acronyms are Cricetulus griseus, Oncorhynchus mykiss, Pimephales promelas , Callorhinchus milii, Branchiostoma floridae, Strongylocentrotus purpuratus, Helobdella robusta, Nematostella vectensis, and Acropora millepora.
Here the protein is broken into its 16 coding exons with phases (base overhangs at split codons) shown:
>MGAT5_monDom length=743
0 MAFFAPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQHESSSMLREQILDLSKRYIKALAEENRNVVDGPYVGVMTAY 1
2 DLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTTTTAAPSSIAAFEKISVA 1
2 DIINGAQEKCELPPMDGFPHCEGKIK 0
0 WMKDMWRTDPCYANYGVDGSTCSFFIYLSE 0
0 VENWCPHLPWRAKNPYEEPDQNSM 0
0 AEIRTDFNLLYGMMKRHEEFRWMILRIRRMADAWIEAIKSLAEKQNLEKRKRKK 0
0 ILVHLGLLTKESGFKIAENAFSGGPLGELVQWSDLITSLYLLGHDIRISASLAELKE 2
1 IMKRVVGNRSGCPTVGDRIVELIYIDIVGLAQFKKTLGPSWVHYQ 2
1 CMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMF 1
2 PHTPDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDSFWK 0
0 NKKEYLDIIHTYMEVHATVYGSSTNHMPSYVKNHGILSGRDLQFLLRETK 0
0 LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE 0
0 LTSQHPYAEVFIGRPHVWTVNPTDHREVENAVKAILNQK 0
0 IEPYMPYEFTCEGMLQRMNAFIEKQ 0
0 DFCHGQVMWPPLNALQVKLSEPGKSCKQVCQENQLICEPSFFQHLNKDKDVLK 2
1 YEVICHTTELANDILVPSYDDKKKHCVFQGDLLLFSCAGAHPKHKRICPCRDYIKGQVALCQDCL* 0
MGAT5 has 16 exons, here numbered from 0:
>monDom4_MGAT5_monDom_1 range=chr4:131201988-131202228
ATGGCTTTCTTTGCTCCATGGAAATTATCCTCTCAGAAACTAGGGTTTTT
CCTGGTGACTTTTGGCTTCATTTGGGGTATGATGCTTTTGCATTTTACCA
TCCAGCAACGAACTCAACATGAAAGCAGCTCAATGCTTCGTGAACAAATA
TTGGACTTAAGCAAAAGGTATATCAAAGCTCTTGCAGAAGAAAATAGGAA
TGTGGTGGATGGCCCCTATGTTGGTGTGATGACAGCCTATG
>monDom4_MGAT5_monDom_2 range=chr4:131219922-131220092
ATCTGAAGAAAACTCTTGCTGTCTTATTGGACAATATTTTACAACGTATT
GGCAAGTTGGAATCGAAGGTGGACAATCTTGTAATTAATGGCACTGGAGC
AAATTCAACTAATACCACCACAACTGCTGCTCCTAGTAGCATAGCAGCAT
TTGAGAAAATTAGTGTAGCAG
>monDom4_MGAT5_monDom_3 range=chr4:131294347-131294423
ATATCATTAATGGAGCTCAAGAAAAATGTGAATTGCCTCCTATGGATGGC
TTCCCGCACTGCGAAGGGAAAATCAAG
>monDom4_MGAT5_monDom_4 range=chr4:131295527-131295616
TGGATGAAAGATATGTGGAGAACAGATCCCTGCTACGCAAATTATGGTGT
GGATGGATCCACGTGCTCTTTTTTTATTTACCTCAGTGAG
>monDom4_MGAT5_monDom_5 range=chr4:131308004-131308075
GTTGAAAATTGGTGTCCTCATTTACCTTGGAGAGCCAAAAATCCCTATGA
AGAACCAGATCAGAATTCAATG
>monDom4_MGAT5_monDom_6 range=chr4:131310550-131310711
GCGGAAATTCGTACGGATTTTAATCTACTCTATGGAATGATGAAACGGCA
TGAAGAATTCCGATGGATGATACTAAGGATCCGGCGGATGGCTGATGCAT
GGATTGAAGCAATTAAGTCACTGGCAGAGAAGCAAAACCTTGAAAAGAGA
AAACGGAAAAAA
>monDom4_MGAT5_monDom_7 range=chr4:131313210-131313379
ATTCTCGTCCACCTGGGACTTCTGACCAAGGAATCTGGCTTCAAGATTGC
AGAGAATGCATTTAGTGGAGGCCCACTTGGTGAATTAGTTCAGTGGAGTG
ATTTAATTACATCTCTCTACTTACTGGGCCATGACATTAGGATTTCAGCT
TCACTGGCTGAGCTCAAGGA
>monDom4_MGAT5_monDom_8 range=chr4:131314690-131314824
GATAATGAAGAGGGTTGTAGGTAATAGATCTGGCTGCCCAACTGTAGGCG
ATAGGATTGTGGAGCTCATTTATATTGATATCGTGGGACTTGCTCAATTT
AAGAAAACTCTTGGCCCTTCCTGGGTTCATTACCA
>monDom4_MGAT5_monDom_9 range=chr4:131320626-131320759
GTGCATGCTCCGAGTGCTGGATTCATTTGGTACTGAACCAGAATTTAATC
ATGCTAATTATGCCCAGTCTAAAGGCCACAAGACCCCATGGGGAAAATGG
AACCTTAATCCTCAACAATTTTATACAATGTTCC
>monDom4_MGAT5_monDom_10 range=chr4:131341860-131341993
CCCACACTCCAGACAACAGCTTTTTAGGATTTGTGGTAGAACAACACCTG
AATTCAAGTGATATCAAGCACATTAATGAAATTAAAAGGCAAAATCAATC
CCTTGTGTATGGCAAAGTTGATAGCTTCTGGAAG
>monDom4_MGAT5_monDom_11 range=chr4:131384716-131384865
AATAAGAAAGAGTACTTGGACATCATTCACACTTACATGGAAGTACATGC
AACTGTCTATGGGTCTAGCACTAACCACATGCCCAGCTATGTGAAGAATC
ATGGAATCCTCAGTGGACGAGATTTACAATTTCTTCTCAGAGAAACTAAG
>monDom4_MGAT5_monDom_12 range=chr4:131400553-131400699
TTGTTTGTTGGACTGGGGTTCCCATATGAAGGTCCTGCTCCCCTTGAGGC
TATTGCTAATGGATGCGCATTCCTGAATGTCAAGTTTAATCCACCCAAAA
GCAGCAAAAATACGGACTTTTTTATAGGCAAACCAACCCTGAGAGAG
>monDom4_MGAT5_monDom_13 range=chr4:131416458-131416574
TTAACTTCTCAGCATCCTTACGCTGAAGTTTTCATTGGGCGACCACATGT
CTGGACCGTAAACCCAACTGATCATCGCGAAGTAGAGAATGCAGTGAAAG
CAATTTTAAATCAAAAG
>monDom4_MGAT5_monDom_14 range=chr4:131420486-131420560
ATTGAACCTTATATGCCATATGAGTTCACATGTGAAGGGATGCTACAAAG
GATGAATGCTTTCATTGAGAAACAG
>monDom4_MGAT5_monDom_15 range=chr4:131427641-131427798
GATTTCTGTCATGGGCAAGTTATGTGGCCACCACTTAATGCTCTTCAGGT
GAAGCTTTCTGAACCTGGGAAATCATGCAAGCAAGTCTGCCAAGAAAACC
AACTCATCTGTGAGCCATCTTTTTTCCAACACCTCAACAAGGACAAGGAT
GTGCTGAA
>monDom4_MGAT5_monDom_16 range=chr4:131441114-131441309
GTATGAGGTGATATGCCACACCACAGAACTGGCCAATGACATCCTGGTGC
CCTCATATGATGACAAGAAAAAACACTGTGTGTTTCAAGGTGATCTTTTG
CTGTTCAGCTGTGCAGGAGCCCACCCCAAGCACAAGAGGATATGCCCTTG
CAGGGACTATATCAAGGGCCAAGTCGCTCTCTGTCAAGACTGCCTA
full length alignment of Monodelphis gene to human gene
Identities = 673/744 (90%), Positives = 708/744 (95%), Gaps = 2/744 (0%)
monDo 1 MAFFAPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQHESSSMLREQILDLSKRYIKA 60
MA F PWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQ ESSSMLREQILDLSKRYIKA
homSa 146 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKA 325
monDo 61 LAEENRNVVDGPYVGVMTAYDLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTT 120
LAEENRNVVDGPY GVMTAYDLKKTLAVLLDNILQRIGKLESKVDNLV+ G + +T
homSa 326 LAEENRNVVDGPYAGVMTAYDLKKTLAVLLDNILQRIGKLESKVDNLVV--NGTGTNSTN 499
monDo 121 TTAAPSSIAAFEKISVADIINGAQEKCELPPMDGFPHCEGKIKWMKDMWRTDPCYANYGV 180
+T A S+ A EKI+VADIINGAQEKC LPPMDG+PHCEGKIKWMKDMWR+DPCYA+YGV
homSa 500 STTAVPSLVALEKINVADIINGAQEKCVLPPMDGYPHCEGKIKWMKDMWRSDPCYADYGV 679
monDo 181 DGSTCSFFIYLSEVENWCPHLPWRAKNPYEEPDQNSMAEIRTDFNLLYGMMKRHEEFRWM 240
DGSTCSFFIYLSEVENWCPHLPWRAKNPYEE D NS+AEIRTDFN+LY MMK+HEEFRWM
homSa 680 DGSTCSFFIYLSEVENWCPHLPWRAKNPYEEADHNSLAEIRTDFNILYSMMKKHEEFRWM 859
monDo 241 ILRIRRMADAWIEAIKSLAEKQNLEKRKRKKILVHLGLLTKESGFKIAENAFSGGPLGEL 300
LRIRRMADAWI+AIKSLAEKQNLEKRKRKK+LVHLGLLTKESGFKIAE AFSGGPLGEL
homSa 860 RLRIRRMADAWIQAIKSLAEKQNLEKRKRKKVLVHLGLLTKESGFKIAETAFSGGPLGEL 1039
monDo 301 VQWSDLITSLYLLGHDIRISASLAELKEIMKRVVGNRSGCPTVGDRIVELIYIDIVGLAQ 360
VQWSDLITSLYLLGHDIRISASLAELKEIMK+VVGNRSGCPTVGDRIVELIYIDIVGLAQ
homSa 1040 VQWSDLITSLYLLGHDIRISASLAELKEIMKKVVGNRSGCPTVGDRIVELIYIDIVGLAQ 1219
monDo 361 FKKTLGPSWVHYQCMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMFPHT 420
FKKTLGPSWVHYQCMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMFPHT
homSa 1220 FKKTLGPSWVHYQCMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMFPHT 1399
monDo 421 PDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDSFWKNKKEYLDIIHTYMEVHAT 480
PDNSFLGFVVEQHLNSSDI HINEIKRQNQSLVYGKVDSFWKNKK YLDIIHTYMEVHAT
homSa 1400 PDNSFLGFVVEQHLNSSDIHHINEIKRQNQSLVYGKVDSFWKNKKIYLDIIHTYMEVHAT 1579
monDo 481 VYGSSTNHMPSYVKNHGILSGRDLQFLLRETKLFVGLGFPYEGPAPLEAIANGCAFLNVK 540
VYGSST ++PSYVKNHGILSGRDLQFLLRETKLFVGLGFPYEGPAPLEAIANGCAFLN K
homSa 1580 VYGSSTKNIPSYVKNHGILSGRDLQFLLRETKLFVGLGFPYEGPAPLEAIANGCAFLNPK 1759
monDo 541 FNPPKSSKNTDFFIGKPTLRELTSQHPYAEVFIGRPHVWTVNPTDHREVENAVKAILNQK 600
FNPPKSSKNTDFFIGKPTLRELTSQHPYAEVFIGRPHVWTV+ + EVE+AVKAILNQK
homSa 1760 FNPPKSSKNTDFFIGKPTLRELTSQHPYAEVFIGRPHVWTVDLNNQEEVEDAVKAILNQK 1939
monDo 601 IEPYMPYEFTCEGMLQRMNAFIEKQDFCHGQVMWPPLNALQVKLSEPGKSCKQVCQENQL 660
IEPYMPYEFTCEGMLQR+NAFIEKQDFCHGQVMWPPL+ALQVKL+EPG+SCKQVCQE+QL
homSa 1940 IEPYMPYEFTCEGMLQRINAFIEKQDFCHGQVMWPPLSALQVKLAEPGQSCKQVCQESQL 2119
monDo 661 ICEPSFFQHLNKDKDVLKYEVICHTTELANDILVPSYDDKKKHCVFQGDLLLFSCAGAHP 720
ICEPSFFQHLNKDKD+LKY+V C ++ELA DILVPS+D K KHCVFQGDLLLFSCAGAHP
homSa 2120 ICEPSFFQHLNKDKDMLKYKVTCQSSELAKDILVPSFDPKNKHCVFQGDLLLFSCAGAHP 2299
monDo 721 KHKRICPCRDYIKGQVALCQDCL* 744
+H+R+CPCRD+IKGQVALC+DCL
homSa 2300 RHQRVCPCRDFIKGQVALCKDCL* 2371
Case of ACTL6B
chr2_18546 ACTL6B 11 >contig00001 length=502 numreads=11 GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ^ 3 G=4(94) R=7(213) Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5 and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in tasmanian devil (in this case, both individuals differ from Monodelphis by -> ), then differences between the two thylacines (here one individual has at position , the other has ), and finally the number of experimental reads that confirm the nucleotide difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses lower-case letters for less confident calls.
Pseudogene issues:
Paralog issues:
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
Structural significance:
Functional significance:
(more shortly)
Case of IPO7
chr5_9037 IPO7 23
>contig00001 length=680 numreads=8
SSQVEKHSCSLTEELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYSTIIDDEENPVDEYQIFKAIFQ
....*N.....................................................F.....................
^
59 F=2(72) S=3(53)
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by -> ), then differences between the two thylacines
(here one individual has at position , the other has ), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues:
Paralog issues:
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
Case of WDFY3
chr5_2532 WDFY3 19
>contig00001 length=482 numreads=8
DDFSEESSFYEILPCCARFRCGDLIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVSTVK
................T..............................T..L.....N...
^
16 T=3(117) A=5(138)
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by -> ), then differences between the two thylacines
(here one individual has at position , the other has ), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues:
Paralog issues:
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
Structural significance:
Functional significance:
(more shortly)
Case of PPFIA3
chr4_22002 PPFIA3 15 incorrectly mapped from monDom5 to human
>contig00001 length=298 numreads=4
LIQEEKETTEQRAEELESRVSGSGLDSLGRYRASCSLPPSLTTSTLASPSPPSSGHSTPRPAPPSPAREAPANSTSNTAEKP
........................................................F..................G.V.
^
56 F=2(43) S=2(37)
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by -> ), then differences between the two thylacines
(here one individual has at position , the other has ), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues:
Paralog issues:
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
Structural significance:
Functional significance:
(more shortly)
Structural significance:
Functional significance:
(more shortly)
Other cases to be considered
chr6_2360 XYLT1 5 61 D=3(110) A=5(107)
>contig00001 length=488 numreads=10
RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGASLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPI
...L........................................................D.....
^
chr4_18550 ATP4A 6 16 C=4(130) R=3(74)
>contig00001 length=906 numreads=10
TAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT
................C........................................................................
^
chr4_11174 FLI1 3 32 N=2(63) K=3(47)
>contig00001 length=575 numreads=9
ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEKNGPPPNMTTNERRVIVPA
..................................................
^
chr2_30280 VPS72 5 15 R=3(59) K=2(51)
>contig00001 length=591 numreads=6
NYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLAEPGPKEENVDVE
...............R..................T............
^
chr6_5144 ABCC1 23 4 Q=2(69) P=2(80) looks like a frame-shift problem in monDom5
>contig00001 length=802 numreads=10
HLCFPRLHLDLLHNVLRSPMSFFERTPSGNLVNRFSKEMDTVDSMIPQIIKMFMGSLFNVIGACIIILLATPIAAIIIPPLGLIYFFVQ
....Q....................................................................................
^
chr5_8347 SPON1 11 20 V=3(65) I=2(66) wobbly
>contig00001 length=433 numreads=5
GSTCTMSEWITWSPCSISCGVGMRSRERYVKQFPEDGSVCTVPTEETEKCTVNEEC
......................................I.N...............
^
chr3_5872 ACOT12 14 14 I=3(95) V=3(110) wobbly
>contig00001 length=472 numreads=6
NTYVVAVKSVTLASIPPSPQYNRSEITCAGFLIRAVDSNSCT
.................................Q....S...
^
Other marsupial genes of interest
The collections below contain well-understood genes with very extensive comparative genomics. They can serve as a test bed for Sarcophilus assembly quality, a place where genuine anomalies or distinct adaptive features might surface (perhaps as phyloSNPs) and where marsupial phylogeny might be refined using rare genomic events in nuclear genes.
The gene sets contain all available marsupial orthologs plus for context one flanking gene each from placentals and monotremes. These genes are available in much broader hand-curated sets elsewhere on this site.
Rod rhodopsin RHO1 (4 marsupials)
The optimal wavelength for scotopic (dim light) vision of Sarcophilus is easily predictable provided key tuning residues are covered by the assembly.
>RHO1_homSap Homo sapiens (human) 0 MNGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSR 2 1 YIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQ 0 0 FRNCMLTTICCGKNPLGDDEASATVSKTETSQVAPA* 0 >RHO1_monDom Monodelphis domesticus (opossum) Didelphimorphia 0 MNGTEGPNFYVPFSNKTGTVRSPFEEPQYYLADPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAIADLFMVFGGFTMTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIIGVAFTWVMALACAFPPLIGWSR 2 1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSNFGPIFMTIPAFFAKSSSVYNPVIYIMMNKQ 0 0 FRTCMITTLCCGKNPLGDDEASATASKTETSQVAPA* 0 >RHO1_macEug Macropus eugenii (wallaby) Diprotodontia frag, traces not yet consulted 0 MNGTEGPNFYVPFSNKTGVVRSPFEEPQYYLAEPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLADADLFMDFGGFT 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACSTPPLLGWSR 2 1 0 0 ESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTLPAFFAKTSAVYNPVIYIMMNKQ 0 0 FRNCMITTLCCGKNPLGDDEASATTSKTETSQVAPA* 0 >RHO1_smiCra Sminthopsis crassicaudata (fat-tailed dunnart) Dasyuromorphia MNGTEGPNFYVPYSNKSGVVRSPYEEPQYYLAEPWMFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAVADLFMVICGFTTTLVTSLNGY FVFGTTGCLVEGFFATTGGEVALWALVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACSVPPIFGWSRYIPEGMQCSCGIDYYTLNPEFNNESFVIYMFVVHFIIPLTV IFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSDFGPIFMTLPAFFAKSSSIYNPVIYIMMNKQFRNCMITTLCCGKNPLGDDEASTTASKTETSQVAPA* >RHO1_calPhi Caluromys philander (woolly opossum) Didelphimorphia abstract:14659889 MNGTEGPNFYVPFSNKTGVVRSPFEEPQYYLAEPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAIADLFMVFGGFTTTLYTSLHGY FVFGPTGCDLEGFFATLGGEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMVV IFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSNFGPILMTLPAFFAKTSAVYNPVIYIMLNKQFRTCMLTTLCCGKIPLGDDEASATASKTETSQVAPA* >RHO1_ornAna Ornithorhynchus anatinus (platypus) 0 MNGTEGQDFYIPMSNKTGVVRSPFEYPQYYLAEPWQYSVLAAYMFMLIMLGFPINFLTLYVTIQHKKLRTPLNYILLNLAFANHFMVLGGFTTTLYTSLHGYFVFGPTGCNIEGFFATLG 1 2 GEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACALPPLVGWSR 2 1 YIPEGMQCSCGIDYYTLRPEVNNESFVIYMFVVHFTIPMTIIFFCYGRLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTVPAFFAKSSAIYNPVIYIMMNKQ 0 0 FRNCMLTTICCGKNPLGDDEASATASKTEQSSVSTSQVSPA* 0
Cone rhodopsin SWS2 (9 marsupials)
Cone rhodopsin RHO2 has been lost in all mammals and no debris from this gene is expected in Sacrophilus). The short wavelength cone opsin SWS2, while still present in platypus, has also been lost in all theran opsion too long ago to leave detectable remnants in syntenic position. Cone opsin SWS1 has this turned around, being present in theran mammals but only as debris in platypus.
>SWS1_homSap Homo sapiens (human) Gt 0.2.2.1.0.0 indel -FAM137A -CALU -NAG6 -FLNC 348 aa 000 nm 1385866 NP_990769 cone short 0 MRKMSEEEFYLFKNISSVGPWDGPQYHIAPVWAFYLQAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVA 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQ 0 0 FQACIMKMVCGKAMTDESDTCSSQKTEVSTVSSTQVGPN* 0 >SWS1_monDom Monodelphis domesticus (opossum) Didelphimorphia 0 MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHFQTVFMGFVFCAGTPLNAVVLVATLRYKKLRQPLNYILVNVSLCGFIFCIFAVFTVFISSSQGYFIFGRHVCAMEAFLGSVA 1 2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWVIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIMPLFLICFSYSQLLRALRA 0 0 VAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNQNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FHACIMEMVCRKPMTDDSDVSSSQKTEVSAVSSSQVGPT* 0 >SWS1_thyEle Thylamys elegans (fat-tailed opossum) Didelphimorphia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHLQTVFMGFVFC AGTPLNAVVLVATLRYKKLRQPLNYILVNVSFSGFIFCIFAVFTVFISSSQGYFIFGH HVCAMEAFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWVIGI GVSIPPFFGWSRFIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLFLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNRNH GLDLRLVTIPAFFSKSACVYNPIIYCFMNKQFHACIMEMVCRKPMTDDSDVSSSQKTE VSAVSSSQVGPS >SWS1_didAur Didelphis aurita (big-eared opossum) Didelphimorphia MSGDEEFYLFKNISSVGPWDGPQHHIAPAWAFHFQTVFMGFVFC AGTPLNAVVLVATLRYKKLRQPLNYILVNVSLSGFIFCIFAVFTVFISSSRGYFVFGR HVCAMEAFLGSVAGLVMGWSLAFLAFERFVVICKPFGNFRFNAKHAMMVVLATWVIGI GVSIPPFFGWSRFIPEGLQCSCGPDWYTVGTKYRSEYYAWFLFLSCFIGPLFLICFSY AQLLGALRAVAAQQQESTTTQKAEREVSRMVVMMVGSFCLCYVPYAALGMYMINNRNH GLDLRLVTIPAFFSKSACVYNPIIYCFMNKQFHACIMEMVCRKPMADDSDITSSQKTE VSTVSSSQVGPS >SWS1_macEug Macropus eugenii (wallaby) Diprotodontia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHCQTVFMGFVFFAGTPLNAVVLIATFRYKKLRQPLNYILVNISLAGFIYCIFSVFTVFISSSQGYFIFGR HVCAMEGFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFNSKHSMMVVLATWVIGIGVSIPPFFGWSRYIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFILCFIMPLSLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNRNHGIDLRLVTIPAFFSKSSCVYNPIIYCFMNKQFHACIMEMVCRKPMTDDSEASSSQKTEVSTVSSSQVGPS* >SWS1_setBra Setonix brachyurus (quokka) Diprotodontia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHCQTVFMGFVFF AGTPLNAVVLIATFRYKKLRQPLNYILVNISLAGFIYCIISVFTVFISSSQGYFIFGR HVCAMEGFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFNSKHSMMVVLATWVIGI GVSIPPFFGWSRYIPEGLQCSCGPDWYTVGTKYHSEYYTWFLFILCFIMPLSLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNRNH GIDLRLVTIPAFFSKSACVYNPIIYCFMNKQFHACIMEMVCRKPMTDDSEASSSQKTE VSTVSSSQVGPS >SWS1_tarRos Tarsipes rostratus (honey possum) Diprotodontia MSGDEEFYLFKDISSVGPWDGPQYHIAPAWAFHFQTTFMGFVFF AGTPLNAVVLIATLRYKKLRQPLNYILVNISLAGFIFCVISVFTVFISSSQGYFIFGR HVCAMEAFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFSSKHAMMVVLATWVIGI GVSIPPFFGWSRYIPEGLQCSCGPDWYTVGTKYHSEYYTGFLFIFCFIVPLSLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNH GLDLRLVTIPAFFSKSACVYNPIVYWFMNKQFHACIMEMVCRKPMTDDSEISSSQKTE VSTVSSSQVGPS >SWS1_cerCon Cercartetus concinnus (pygmy possum) Diprotodontia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHFQTAFMGFVFF VGTPLNAVVLVATLCYKKLRQPLNYILVNVSLAGFIFCIISVFTVFISSSQGYFIFGR HVCAMEAFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFSSKHAMMVVLATWVIGI GVSIPPFFGWSRYIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNH GLDLRLVTIPACFSK >SWS1_smiCra Sminthopsis crassicaudata (dunnart) Dasyuromorphia 0 MSGDEEFYLFKNISLVGPWDGPQYHLAPAWAFHFQTAFMGFVFFAGTSLNGVVLIATLRYKKLRQPLNYILVNISLAGFIFCVFSVFTVFVSSSQGYFVFGRHVCAMEGFLGSVA 1 2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWIIGIGVSIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLICFSYSQLLGALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAAMAMYMVNNRNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FHACIMEMICKKPMTDDSETTSSQKTEVSTVSSSQVGPS* 0 >SWS1_isoObe Isoodon obesulus (bandicoot) Peramelemorphia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHCQTVFMGFVFF AGTPLNAVVLIATLRYKKLRQPLNYILVNISLAGFIFCIFSVFTVFISSSQGYFIFGR HVCAMEAFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFHSKHAMMVVLATWVIGI GVSIPPFFGWSRFIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIIPLSLICFSY SQLLRALRTVAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNH GLDLRLVTIPAFFSKSACVYNPIIYCFMNKQFHACIMEMICRKPMTDDSETSSSQKTE VSTVSSSQVSPS >SWS1_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel x x x x 348 aa 000 nm no_ref genome cone short1 violet 0 MSSDDDFYLFTNGSVPGPWDGPQYHIAPPWAFYLQTAFMGIVFAVGTPLNAVVLWVTVRYKRLRQPLNYILVNISASGFVSCVLSVFVVFVASARGYFVFGKRVCELEAFVGTHG 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSRHALLVVVATWLIGVGVGLPPFFGWSR 2 1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FRACIMETVCGKPLTDDSDASTSAQRTEVSSVSSSQVGPT* 0
Cone rhodopsin LWS (9 marsupials)
This basal imaging opsin is available from 97 vertebrates and has already been analyzed for phyloSNPs and rare genomic events. The Didelphimorphia experienced a 3-4 residue insert in exon 1 that separates them from all other marsupials. Note this region has quite a complicated indel history. The extra residues have repeat character DVNE DDND suggesting replication slippage.
LWS_loxAfr MAQQWGPHRLTGARLQDASE---DSTQASIFVYTNTNT elephant LWS_echTel MAQRWGAHRLTGGQLQDTYE---GSTRTSIFVYTNSTS tenrec LWS_monDom MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNN Didelphimorphia LWS_didAur MTQAWDPVGFLARRRDENEDDHDDTTRASLFVYTNSNN Didelphimorphia LWS_tarRos MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia LWS_macEug MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia LWS_smiCra MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Dasyuromorphia LWS_setBra MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia LWS_cerCon MTQAWDPAGFLAWQEDENE----ETTRASLFVYTNSNN Diprotodontia LWS_myrFas MTQAWDPAGFLAWRREENE----ETTRASLFTYTNSNN Dasyuromorphia LWS_isoObe MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Peramelemorphia LWS_ornAna MTPAWNSGVYAARRRFEDEE---DTTRTSVFVYTNSNN platupus LWS_tacAcu MTQAWDPAGFLAWRRDENEE---TTRASLFVYTNSNNT echidna
>LWS_homSap Homo sapiens (human) 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_monDom Monodelphis domesticus (opossum) Didelphimorphia 4aa insert 0 MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYVQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYSFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSKTEGSSVSSVAPA* 0 >LWS_didAur Didelphis aurita (big-eared opossum) Didelphimorphia 4aa insert 0 MTQAWDPVGFLARRRDENEDDHDDTTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDLGVQSYMIVLMATCCIFPLSIILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA* 0 >LWS_tarRos Tarsipes rostratus (honey possum) Diprotodontia ENED insert 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAIWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGIQSYMIVLMSTCCILPLSIILLCYVQVWRAIRA 2 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_macEug Macropus eugenii (wallaby) Diprotodontia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGVQSYMIVLMSTCCILPLSVIFLCYIQVWLAIRS 2 0 VAKQQKESESTQKAEKEVSRMVVVMILAFCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_setBra Setonix brachyurus (quokka) Diprotodonti 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETMIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGVQSYMIVLMSTCCILPLSVILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVPA* 0 >LWS_cerCon Cercartetus concinnus (pygmy possum) Diprotodontia 0 MTQAWDPAGFLAWQEDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAIWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGIQSYMIVLMSTCCILPLSIILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTFFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* >LWS_smiCra Sminthopsis crassicaudata (dunnart) Dasyuromorphia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIILCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVMVMILAFCFCWGPYALFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_myrFas Myrmecobius fasciatus (numbat) Dasyuromorphia 0 MTQAWDPAGFLAWRREENEETTRASLFTYTNSNNTK 1 2 GPFEGPNYHIAPRWVYNLTSFWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSVILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_isoObe Isoodon obesulus (bandicoot) Peramelemorphia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSFWMFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMTTCCILPLSIILLCYVQVWLAIRA 0 0 VAKQQKDSESTQKAEKEVSRMVVVMIRAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSGTSRTEVSSVSSAPA* 0 >LWS_ornAna Ornithorhynchus anatinus (platypus) 0 MTPAWNSGVYAARRRFEDEEDTTRTSVFVYTNSNNTR 1 2 DPFEGPNYHIAPRWAYNVTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA* 0 >LWS_tacAcu Tachyglossus aculeatus (echidna) 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAVWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA* 0
Encephalopsin (2 marsupials)
Encephalopsin has a very peculiar history of gene loss in tetrapods, requiring some seven independent events. While this limits the phylogenetic utility of gene loss within marsupials, it is still of interest to determine the status of the gene within Sarcophilus.
>ENCEPH_homSap Homo sapiens (human) OPN3 0 MYSGNRSGGHGYWDGGGAAGAEGPAPAGTLSPAPLFSPGTYERLALLLGSIGLLGVGNNLLVLVLYYKFQRLRTPTHLLLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSGSLF 1 2 GIVSIATLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPLGVIAHCYGHILYSIRM 0 0 LRCVEDLQTIQVIKILKYEKKLAKMCFLMIFTFLVCWMPYIVICFLVVNGHGHLVTPTISIVSYLFAKSNTVYNPVIYVFMIRK 0 0 FRRSLLQLLCLRLLRCQRPAKDLPAAGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDKTNGSKVDVIQVRPL* 0 >ENCEPH_monDom Monodelphis domestica (opossum) 0 MYSDNSSDDGGGGYWGSGRAGGASGTGVTGEPGPEGSPRQAPLFSPGTYELLALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLFLVNISFNDLLVSLFGVTFTFVSCLRSGWVWDSVGCAWDGFSNTLF 1 2 GIVSIMTLTVLAYERYNRIVHAKVINFSWAWRAITYIWLYSLVWTGAPLLGWNRYTLEIHGLGCSVDWKSKDPNDSSFVIFLFFGCLMLPVGVMAYCYGHILYAIRM 0 LRCVEELQTIQVIKILRYEKKVAKMCFLMIAIFLFCWMPYAVICLLVANGYGSLVTPTVAIIASLFAKSSTAYNPIIYIFMSRK 0 0 FRRCLLQLLCFRLLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDETQMIDENDKNSGTKVNVIQVRPL* 0 >ENCEPH_macEug Macropus eugenii frag 0 GALGCREPGQREPSSSAPFSPGTYELLALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLLLVNISFSDLLVSLFGVTFTFVSCLRSGWVWHTVGCAWDGFSNSLF 1 2 GIVSIMTLTVLAYERYHRIVHAKVINFSWTWRAITYIWLYSLVWTGAPLLGWNRYTLEIHGLGCSVDWKSKDPNDSSFVLFLFLGCLVLPVGVMAYCYGHILYAIRM 0 0 0 0 FRRCLLQLLCFRQLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDETQMIDDNDKNNGTKVNVIQVRPL* 0 >ENCEPH_ornAna Ornithorhynchus anatinus pseudo 0 MVPWNGS-GRHLGAVR---GPE--SLPATPGAARPSRPGAGDGRL--LGLF-P-GVGGNLLVLLL--ALPGPPTTTDLYLASVAVSDLL--LL---LPFVYRLWRSRPWVFVCRLLGE-GGSLA 1 2 GIVSLISLAVLSYERYTLTLHPKQSNYQKAVLAVGASWIYSLIWTIPPLLGWSSYGTEGAGTSCSVHWSSKSVC-SYIVCLFI--CLVIPVLVMIYCYGRLLYAVKQ 0 0 LHCVKELQNIQVIGSLRYER*VTEMYFFTIAQFLVCQSPSALVSYPAAH-----VSPVVAKISPVFANSSFVYNPVISIFVRRK 0 0 KASR*KVNVIQVQPPS* 0 >ENCEPH_galGal Gallus gallus (chicken) 71%=homSap encephalopsin OPN3 full 0 MHSGNGTGATSRPQLAAAGHEVPGERPLFSAGTYELLALLIATIGTLGVCNNLLVLVLYYKFKRLRTPTNLFLVNISLSDLLVSVCGVSLTFMSCLRSRWVWDAAGCVWDGFSNSLF 1 2 GIVSIMTLTVLAYERYIRVVHAKVIDFSWSWRAITYIWLYSLAWTGAPLLGWNRYTLEIHGLGCSMDWKSKDPNDTSFVLLFFLGCLVAPVVIMAYCYGHILYAVRM 0 0 LRCVEDFQTSQVIKLLKYEKKVAKMCFLMISTFLICWMPYAVVSLLVTYGYSNLVTPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0 0 FRQCLLQLLCFRLMRFQRIMKEPSGAGNVKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIIASDDTQQIDDNSKHNGTKVNVIQVKPL* 0
TMT opsin (2 marsupials)
TMT is an ancient locus that is present in monotremes and marsupials but lost in all placentals.
>TMT_monDom Monodelphis domestica shortened final exon 0 MSNNLTTNLSLEALLSASEDKQRNGLSRTGHTIVAVFLGIILIFGSISNFIVLVLFCKFKVLRNPVNMLLLNISISDMLVCLSGTTLSFASSIQGRWIGGKHGCRWYGFANSCF 1 2 GIVSLISLAILSYERYRTLTLCPGQGADYQKALLAVAGSWLYSLVWTVPPLIGWSSYGTEGAGTSCSVHWTSKSVESVSYIMCLFIFCLVIPILVMVYFYGRLLYAVKQ 0 0 VGKIRKTAARKREYHVLFMVVTAVICYLICWVPYGMIALLATFGPPGVVSPVANVVPSILAKSSTVCNPIIYVLMNKQ 0 0 FYKCFLILFHCQPAQSGPDVSLCPSNVTVIQLGQRKNKDAPGSI* >TMT_macEug Macropus eugenii frag 0 MSINLTANLSFGTLLPDSEEKQRSGLSRTGHTVTAVFLGLILILGVINNFIVLVLFCKFKVLRNPVNMLLLNISISDMLVCLTGTTLSFASSIRGRWIAGYHGCRWYGFANSCF 1 2 GIVSLISLAVLSYERYRTLTLCPRQGTDYHKALLAVAGSWLYSLIWTVPPLIGWSSYGTEGAGTSCSVHWTSKSVESVSYIMCLFIFCLVIPILFMVYFYGRLLYTVKQ 0 0 VGKIRKSAARKREYHVLFMVVTAVICYLICWVPYGMIALLATFGPPGVVSPVANVVPSILAKSSTVCNPIIYILMNKQ 0 0 FYKCFLILFHCQPASSASDASLCPSKMTVIQLGQRKDKEVPCAIQDLPEVSKKQLCLLSPESNVAPSSGHPQEKMEEKPLSE* 0 >TMT_ornAna Ornithorhynchus anatinus frag 0 GLSRTGHTMVAVFLGIILVFGFMNNLIVLILFCKFKALRNPVNMIMLNISASDMLVCVSGTTLSFASNISGRWIGGDPGCRWYGFVNSCL 1 2 GIVSLISLAVLSYERYRTLTLHPKQSTDYQKAVLAVGASWIYSLIWTIPPLLGWSSYGTEGAGTSCSVHWSSKSPVSVSYIVCLFIFCLVIPVLVMIYCYGRLLYAVKQ 0 0 IGKARKTAARKREYHVLFMVITTVICYLVCWMPYGVTALLATFGQPGTVSPEASVIPSILAKSSTVCNPIIYILMNKQ 0 0 FYKCFLILFHCQPPRAADAPSTYPSQVMVIQLNQRRSRETAGAPQVLLEMKHQTLHLLGPQLHETPSWERSTPVHPE* 0 >TMT_galGal Gallus gallus 0 MNHTWTYNLSFGAPTDPVEPRAGLSRNGHTVVAVFLGFILFFGFLNNLIVLILFCKFKTLRNPVNMLLLNIhttp://genomewiki.ucsc.edu/index.php?title=Opsin_evolution:_Encephalopsin_gene_loss&action=editSISDMLVCISGTTLSFASNIHGKWIGGEHGCRWYGFVNSCF 1 2 GIVSLISLAVLSYERYSTLTLCNKRSDDYRKALLAVGGSWVYSLLWTVPPLLGWSSYGIEGAGTSCSVRWSSETAESTSYIICLFIFCLVIPVMVMMYCYGRLLYAVKQ 0 0 VGKIHKNTARKREYHVLFMVITTVICYLVCWIPYGVIALLATFGKPGVVTPVASIIPSILAKSSTVCNPIIYILMNKQ 0 0 FYKCFRQLFHCQPPSSTDGEPTCHSKVTVIQLNQKTDGGKLCNNKPRPETDNKVTSLLHPEPGLEPAAKTVPPM* 0