Marsupial phyloSNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 824: | Line 824: | ||
equCab EALQDYYTLVDRVPALLHHLASMDFSSVVSEDDLVAKLNAGLQAVSEDPRLLVWVVRSK | equCab EALQDYYTLVDRVPALLHHLASMDFSSVVSEDDLVAKLNAGLQAVSEDPRLLVWVVRSK | ||
=== Rod rhodopsin RHO1 (4 marsupials) === | === Rod rhodopsin RHO1 (4+ marsupials) === | ||
The optimal wavelength for scotopic (dim light) vision of Sarcophilus is easily predictable provided key tuning residues are covered by the assembly. The 97% match to Sminthopsis and agreement at tuning residues suggests this aspect of vision will be nearly identical between the two species. | The optimal wavelength for scotopic (dim light) vision of Sarcophilus is easily predictable provided key tuning residues are covered by the assembly. The 97% match to Sminthopsis and agreement at tuning residues suggests this aspect of vision will be nearly identical between the two species. | ||
Revision as of 09:07, 20 February 2009
Introduction to Marsupial phyloSNPs
In this project, new genomic data from the Tasmanian devil (Sarcophilus harrisii), Tasmanian tiger (Thylacinus cynocephalus), and echidna (Tachyglossus aculeatus) are analyzed for significant changes at the protein coding level. The goal is to find single amino acid changes in one of these species at a highly invariant residue in a well-conserved exon in a gene with known or predictable tertiary structure. Such changes are thought to enrich for genetic changes with significant, adaptive biochemical or phenotypic consequences (1,2,3,4), in contrast to ordinary SNPs at positions of low conservation. Thus phyloSNPs are informative to the distinctive biology of the species carrying them and suggest a focus for subsequent experiment.
It is also of particular interest to determine the levels of variation within the Tasmanian devil population as a whole because the number of individuals have become low and possibly inbreed with adverse sequelae. For this it will be necessary to first determine sites of variation and then to genotype them across a large number of individuals.
Marsupial genomic and cDNA data to date has been quite limited compared to placental mammal. Yet as outgroup, metatheran animals provide important context to placentals and represent important context in understanding human protein evolution. The monotheres are inevitably limited by the paucity of extant species (basically platypus and echidna) and dim prospects for fossil DNA. Consequently echidna provides an important adjunct to the existing but incomplete platypus assembly. While extant birds and reptiles -- the preceding divergence node -- are abundant it must be remembered that a very considerable time elapsed (from 310 mry to 175 mry) prior to divergence of mammals with living representatives. This gap of 135 myr is comparable to the whole evolutionary record of theran mammals.
Assumed vertebrate phylogenetic tree
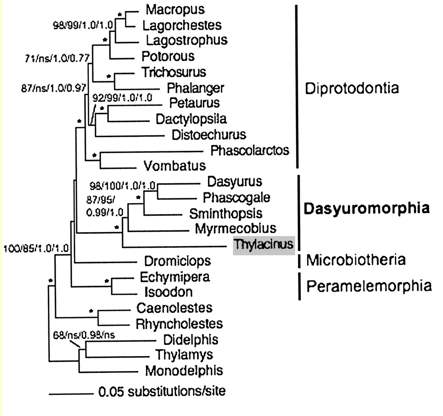
Marsupial relationships are taken from a 2009 paper establishing the mitochondrial genome sequences of the Tasmanian tiger (Thylacinus cynocephalus) and numbat (Myrmecobius fasciatus).
Newick tree that generates a marsupial-centric vertebrate phylogenetic tree: ((((((((((((sarHar,smiCra),myrFas),thyCyn),(macEug,triVul)),monDom), ((((loxAfr,proCap),echTel),(dasNov,choHof)), ((((((bosTau,turTru),susScr),vicPac),((equCab,(felCat,canFam)),(myoLuc,pteVam))),(eriEur,sorAra)), (((((((((homSap,panTro),gorGor),ponPyg),macMul),calJac),tarSyr),(micMur,otoGar)),tupBel), (((((musMus,ratNor),dipOrd),cavPor),speTri),(oryCun,ochPri)))))), (ornAna,tacAcu)), ((galGal,taeGut),anoCar)), xenTro), (((tetNig,takRub),(gasAcu,oryLap)),danRer)), calMil), petMar); Newick tree that generates the homo-centric vertebrate phylogenetic tree: ((((((((((((((((((homSap,panTro),gorGor),ponPyg),macMul),calJac),tarSyr),(micMur,otoGar)),tupBel), (((((musMus,ratNor),dipOrd),cavPor),speTri),(oryCun,ochPri))), (((((vicPac,susScr),turTru),bosTau),((equCab,(felCat,canFam)),(myoLuc,pteVam))),(eriEur,sorAra))), (((loxAfr,proCap),echTel),(dasNov,choHof))), (monDom,((macEug,triVul),(sarHar,thyCyn)))), (ornAna,tacAcu)), ((galGal,taeGut),anoCar)), xenTro), (((tetNig,takRub),(gasAcu,oryLap)),danRer)), calMil), petMar);
Phylo-sorting data
This tab-delimited table enables four different sort orders. These are needed because data can be missing from species in a manner that varies by gene, making data alignment difficult. Some alignment tools also lose input order, so that needs to be recovered. The ordering here flattens the phylogenetic tree by taking human (arbitrarily) at the top and resolving ambiguous situations (eg mouse, rat) by putting species with the best assemblies first.
The first two columns provide sort order number for the 44 species alignment at UCSC as phylogenetic and alphabetic order respectively. The third and fourth columns do this for a larger set of 53 species for which data is commonly available (notably in marsupials). The fifth column supplies the genSpp acronym and the sixth the Newick tree format syntax. These two columns by themselves will correctly draw the vertebrate phylogenetic tree in all online software without further editing. The final columns provide genus, species, and common name.
.. .. .. .. ...... (((((((((((( 46 10 54 10 anoCar )), Anolis carolinensis (lizard) 29 11 22 11 bosTau , Bos taurus (cow) 15 12 38 12 calJac ), Callithrix jacchus (marmoset) 62 54 61 13 calMil ), Callorhinchus milii (elephantfish) 32 13 28 14 canFam )),( Canis familiaris (dog) 23 14 46 15 cavPor ), Cavia porcellus (guinea_pig) 41 15 21 16 choHof )),(((((( Choloepus hoffmanni (sloth) 52 16 60 17 danRer )), Danio rerio (zebrafish) 40 17 20 18 dasNov , Dasypus novemcinctus (armadillo) 22 18 45 19 dipOrd ), Dipodomys ordii (kangaroo_rat) 39 19 19 20 echTel ),( Echinops telfairi (tenrec) 30 20 26 21 equCab ,( Equus caballus (horse) 35 21 31 22 eriEur , Erinaceus europaeus (hedgehog) 31 22 27 23 felCat , Felis catus (cat) 44 23 52 24 galGal , Gallus gallus (chicken) 50 24 58 25 gasAcu , Gasterosteus aculeatus (stickleback) 12 25 35 26 gorGor ), Gorilla gorilla (gorilla) 10 26 33 27 homSap , Homo sapiens (human) 37 27 17 28 loxAfr , Loxodonta africana (elephant) 58 56 14 29 macEug , Macropus eugenii (wallaby) 14 28 37 30 macMul ), Macaca mulatta (rhesus) 17 29 40 31 micMur , Microcebus murinus (mouse_lemur) 42 30 16 32 monDom ),(((( Monodelphis domestica (opossum) 20 31 43 33 musMus , Mus musculus (mouse) 33 32 29 34 myoLuc , Myotis lucifugus (microbat) 56 57 12 35 myrFas ), Myrmecobius fasciatus (numbat) 26 33 49 36 ochPri )))))),( Ochotona princeps (pika) 43 34 50 37 ornAna , Ornithorhynchus anatinus (platypus) 25 35 48 38 oryCun , Oryctolagus cuniculus (rabbit) 51 36 59 39 oryLap )), Oryzias latipes (medaka) 18 37 41 40 otoGar )), Otolemur garnettii (bushbaby) 11 38 34 41 panTro ), Pan troglodytes (chimp) 53 39 62 42 petMar ) Petromyzon marinus (lamprey) 13 40 36 43 ponPyg ), Pongo pygmaeus (orang) 38 41 18 44 proCap ), Procavia capensis (hyrax) 34 42 30 45 pteVam ))),( Pteropus vampyrus (macrobat) 21 43 44 46 ratNor ), Rattus norvegicus (rat) 54 58 10 47 sarHar , Sarcophilus harrisii (tasmanian_devil) 55 59 11 48 smiCra ), Sminthopsis crassicaudata (dunnart) 36 44 32 49 sorAra )),((((((((( Sorex araneus (shrew) 24 45 47 50 speTri ),( Spermophilus tridecemlineatus (squirrel) 60 60 24 51 susScr ), Sus scrofa (pig) 61 61 51 52 tacAcu )),(( Tachyglossus aculeatus (echidna) 45 46 53 53 taeGut ), Taeniopygia guttata (finch) 49 47 57 54 takRub ),( Takifugu rubripes (fugu) 16 48 39 55 tarSyr ),( Tarsius syrichta (tarsier) 48 49 56 56 tetNig , Tetraodon nigroviridis (pufferfish) 57 62 13 57 thyCyn ),( Thylacinus cynocephalus (tasmanian_tiger) 59 63 15 58 triVul )), Trichosurus vulpecula (bushytail_possum) 19 50 42 59 tupBel ),((((( Tupaia belangeri (tree_shrew) 28 51 23 60 turTru ), Tursiops truncatus (dolphin) 27 52 25 61 vicPac ),(( Vicugna pacos (lama) 47 53 55 62 xenTro ),((( Xenopus tropicalis (frog) 44 44 53 53 genSpp tree_syntax genus species common ph al ph al
Candidate analysis
The first issue is error within the reads themselves; the second is whether the default 454 Newbler assembler correctly identified overelapping reads and put them together properly to give exon-spanning reads. Those issues are discussed elsewhere -- here it is assumed the data is correct, so the entire focus is on subsequent bioinformatics.
(methods explained more shortly)
Case of ERN2
chr6_5971 ERN2 4
contig00001 length=355 numreads=5
KLPFTIPELVHASPCRSSDGVLYT
.....................F..
^
15 R=3(75) H=2(50
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by L->F), then differences between the two thylacines
(here one individual has R at position 15, the other has H), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues: ERN2 has not generated potentially confusing recent processed pseudogenes in mammals (lack of human, opossum or platypus genome Blat matches to ERN2 query). The variation observed here between individual tasmanian devils is implausibly an early stage in the loss of parent gene because of ERN2 functional essentiality; the exon cannot come from a decaying segmental duplication because coverage is high enough to also detect the main gene.
Paralog issues: The GeneSorter tool at UCSC shows a single significant full-length paralog in human, ERN1, also with 22 coding exons. The genes reside on different chromosomes but in regions with local homology of synteny. However this particular exon is a good match (3 differences out of 23), so there is potential for experimental difficulties in distinguishing them in short reads (including the following exon readily resolves them bioinformatically). In any event, at positions 15 and 20, ERN1 is identical at the amino acid level to ERN2. The gene duplication appears to have occured subsequent to amphioxus divergence earlier diverging metazoans are single-copy.
Homoplasy (recurrent mutation) issues: This exon is very conserved and does not exhibit repetitive sequence, compositional simplicity, or indels in any species in either paralog that could foster experimental error or alignment ambiguity. At position 15, the ancestral value is arginine in both paralogs. The G--> A transition to histidine in one individual is conservative under most circumstances (still basic) and arises from an arginine codon CpG hotspot conserved back to lamprey in 30 of 32 species with available data, yet histidine is not observed part of a reduced alphabet (ie R/H) at this position over many billions of years of branch length. Consequently R-->H is a significant change in this individual tasmanian devil.
Known variations: No human disease variants have been reported for either ERN2 or ERN1, probably attributable to essentiality. Site-specific mutation close to the exon here have been generated for K121P, D123P, W125A, and Q105E but only for ERN1. Naturally occuring coding SNPs in the human population relevent to the ERN2 exon are not known but low frequency alleles could emerge from the 1000 Genomes Project.
Side issues: a very ancient conserved leucine at position 21 appears to be transitioning to phenylalanine at marsupial node but has not been fixed, so settles out as L or F depending on lineage-sorting on each terminal marsupial leaf whereas placentals are all changed to phenylalanine (a phyloSNP caught in mid-air). While L and F might seem about the 'same' as amino acids, the branch length conservation totals say both are important but for different reasons: this is not a waffle codon nor reduced alphabet situation. This raises the question -- given the extreme conservation of this exon otherwise -- of whether the L-->F change at position 21 in both individuals has 'enabled' (made neutral or adaptive) an otherwise unfavorable R-->H change at position 15 in one individual.
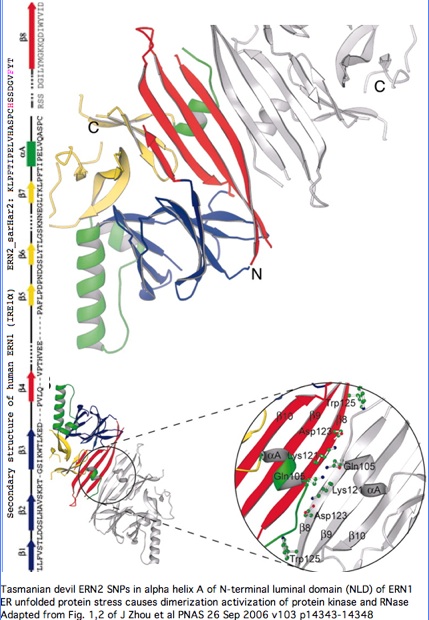
Structural significance: By good fortune, the crystal structure of ERN1 (alternately called IRE1) has been published. The PDB 2HZ6 structure has good coverage of this particular exon. Consequently the marsupial ERN2 could be very accurately modelled and the structural effects of L-->F with or without R-->H computed by submission to online SwissProt modelling service.
Monodelphis ERN2 (key exon: sarHar2) aligned to human ERN1 luminal domain
Expect = 5.8e-65 Identities = 109/180 (60%), Positives = 141/180 (78%)
ERN2_monDom 1 PESLLFISTLDGSLHAVSKKTGDIQWTLKDDPIIQGPVYATEPAFLPDPSDGSLYILGEE 60
PE+LLF+STLDGSLHAVSK+TG I+WTLK+DP++Q P + EPAFLPDP+DGSLY LG +
ERN1_homSap 8 PETLLFVSTLDGSLHAVSKRTGSIKWTLKEDPVLQVPTHVEEPAFLPDPNDGSLYTLGSK 67
ERN2_monDom 61 SKQGLMKLPFTIPELVHASPCHSSDGVFYTGRKQDTWFMVDPKSGKKQTMLSTETWDGLY 120
+ +GL KLPFTIPELV ASPCRSSDG+LY G+KQD W+++D +G+KQ LS+ D L
ERN1_homSap 68 NNEGLTKLPFTIPELVQASPCRSSDGILYMGKKQDIWYVIDLLTGEKQQTLSSAFADSLC 127
ERN2_monDom 121 PSAPLLYIGRTQYTVTMYDPRSQALRWNTTYRGYSAPLLDHLPGYQVGHFTCSGEGLVVT 180
PS LLY+GRT+YT+TMYD +++ LRWN TY Y+A L + Y++ HF +G+GLVVT
ERN1_homSap 128 PSTSLLYLGRTEYTITMYDTKTRELRWNATYFDYAASLPEDDVDYKMSHFVSNGDGLVVT 187
Functional significance: A considerable amount is known about the paralog ERN1. Annotation transfer is likely applicable to ERN2. The two gene products differ primarily in expression -- ERN1 ubiquitious but ERN2 restricted to intestinal epithelial cells:
"The unfolded protein response (UPR) is an evolutionarily conserved mechanism by which all eukaryotic cells adapt to the accumulation of unfolded proteins in the endoplasmic reticulum (ER). Inositol-requiring kinase 1 (IRE1 or ERN1) and PKR-related ER kinase (PERK) are two type I transmembrane ER-localized protein kinase receptors that signal the UPR through a process that involves homodimerization and autophosphorylation... The monomer of the luminal domain comprises a unique fold of a triangular assembly of beta-sheet clusters. Structural analysis identified an extensive dimerization interface stabilized by hydrogen bonds and hydrophobic interactions... Mutations that disrupt the dimerization interface produced ERN1 protein that failed to either dimerize or activate the UPR upon ER stress."
"ERN1 is a type I transmembrane protein kinase receptor that also has a site-specific RNase activity that, upon activation, initiates a site-specific unconventional splicing reaction. The substrate for IRE1 RNase in metazoans is Xbp1 mRNA, which encodes a basic leucine zipper transcription factor of the ATF/CREB family. XBP1 controls expression of genes containing an X-box element or a UPR element in their promoter regions. The IRE1-mediated splicing reaction introduces into XBP1 an alternative C terminus, thereby generating an XBP1 molecule that is a more potent transcriptional activator. Therefore, activation of IRE1 and its RNase increases the transcription of genes encoding ER chaperones and folding catalysts... the ERN1 N-terminal luminal domain (NLD) functions as an ER stress sensor... under normal conditions IRE1 is maintained in a monomeric state through interaction of the NLD with the ER resident chaperone BiP. Upon ER stress, Grp78 binds to unfolded proteins as they accumulate, permitting the released NLD to form homodimers. Dimerization of the NLD in turn leads to the activation of the protein kinase and RNase activities in the cytosolic domain of ERN1."
ENR2 is readily distinguished from its ERN1 paralog at tBlastn by including the two following exons which bring percent identity to 62%:
ERN2_monDom KLPFTIPELVHASPCRSSDGVLYTGRKQDTWFMVDPKSGKKQTMLSTETWDGLYPSAPLLYIGRTQYTVTMYDPRSQALRWNTTYRGYSA
KLPFTIPELV ASPCRSSDG+LY G+KQD W++VD +G+KQ LS+ + L PS LLY+GRT+YT+TM+D +S+ LRWN TY Y+A
ERN1_monDom KLPFTIPELVQASPCRSSDGILYMGKKQDIWYVVDLMTGEKQQTLSSAFAESLCPSTSLLYLGRTEYTITMFDTKSRELRWNATYFDYAA
The first alignment shows ERN2 orthologs in vertebrates, the second as difference relative to opossum, the third ERN1 orthologs.
The ancestral nature of the CpG hotspot is shown in nucleotides in the final columns.
^ * ^ * ^ *
ERN2_homSap KLPFTIPELVHASPCRSSDGVFYT ERN2_homSa .....................F.. ERN1_homSap KLPFTIPELVQASPCRSSDGILYM CG Human
ERN2_panTro KLPFTIPELVHASPCRSSDGVFYT ERN2_panTr .....................F.. ERN1_panTro KLPFTIPELVQASPCRSSDGILYM CG Chimp
ERN2_ponAbe KLPFTIPELVHASPCRSSDGVFYT ERN2_ponAb .....................F.. ERN1_ponAbe KLPFTIPELVQASPCRSSDGILYM -- Gorilla
ERN2_rheMac KLPFTIPELVHASPCRSSDGVFYT ERN2_rheMa .....................F.. ERN1_rheMac KLPFTIPELVQASPCRSSDGILYM CG Orangutan
ERN2_calJac KLPFTIPELVHASPCRSSDGVFYT ERN2_calJa .....................F.. ERN1_calJac KLPFTIPELVQASPCRSSDGILYM CG Rhesus
ERN2_tarSyr KLPFTIPELVHASPCRSSDGVFYT ERN2_tarSy .....................F.. ERN1_tarSyr KLPFTIPELVQASPCRSSDGILYM CG Marmoset
ERN2_micMur KLPFTIPELVHASPCRSSDGVFYT ERN2_micMu .....................F.. ERN1_micMur KLPFTIPELVQASPCRSTDGILYM CG Tarsier
ERN2_tupBel KLPFTIPELVHASPCRSSDGVFYT ERN2_tupBe .....................F.. ERN1_otoGar KLPFTIPELVQASPCRSSDGILYM CG Mouse_lemur
ERN2_musMus KLPFTIPELVHASPCRSSDGVFYT ERN2_musMu .....................F.. ERN1_tupBel KLPFTIPELVQASPCRSSDGILYM -- Bushbaby
ERN2_ratNor KLPFTIPELVHASPCRSSDGVFYT ERN2_ratNo .....................F.. ERN1_musMus KLPFTIPELVQASPCRSSDGILYM CG TreeShrew
ERN2_cavPor KLPFTIPELVHTSPCRSSDGVFYT ERN2_cavPo ...........T.........F.. ERN1_ratNor KLPFTIPELVQASPCRSSDGILYM CG Mouse
ERN2_speTri KLPFTIPELVHASPCRSSDGVFYT ERN2_speTr .....................F.. ERN1_dipOrd KLPFTIPELVQASPCRSSDGILYM CG Rat
ERN2_oryCun KLPFTIPELVHASPCRSSDGVFYT ERN2_oryCu .....................F.. ERN1_cavPor KLPFTIPELVQASPCRSSDGILYM -- Kangaroo_rat
ERN2_ochPri KLPFSIPELVHASPCRSSDGVFYT ERN2_ochPr ....S................F.. ERN1_speTri KLPFTIPELVQASPCRSSDGILYM CG Guinea_pig
ERN2_turTru RLPFTIPELVHASPCRSSDGVFYT ERN2_turTr R....................F.. ERN1_oryCun KLPFTIPELVQASPCRSSDGILYM CG Squirrel
ERN2_bosTau RLPFTIPELVHASPCRSSDGVFYT ERN2_bosTa R....................F.. ERN1_vicPac KLPFTIPELVQASPCRSSDGILYM CG Rabbit
ERN2_equCab KLPFTIPELVHASPCRSSDGVFYT ERN2_equCa .....................F.. ERN1_turTru KLPFTIPELVQASPCRSSDGILYM CG Pika
ERN2_felCat RLPFTIPELVHASPCRSSDGVFYT ERN2_felCa R....................F.. ERN1_bosTau KLPFTIPELVQASPCRSSDGILYM -- Alpaca
ERN2_canFam KLPFTIPELVHASPCRSSDGVFYT ERN2_canFa .....................F.. ERN1_equCab KLPFTIPELVQASPCRSSDGILYM CG Dolphin
ERN2_myoLuc KLPFTIPELVHASPCRSSDGVFYT ERN2_myoLu .....................F.. ERN1_canFam KLPFTIPELVQASPCRSSDGILYM CG Cow
ERN2_eriEur KLPFTVPELVHTSPCRSSDGVFYT ERN2_eriEu .....V.....T.........F.. ERN1_myoLuc KLPFTIPELVQASPCRSSDGILYM CG Horse
ERN2_sorAra KLPFTIPELVHASPCRSSDGVFYT ERN2_sorAr .....................F.. ERN1_pteVam KLPFTIPELVQASPCRSSDGILYM CG Cat
ERN2_loxAfr KLPFTIPELVHASPCRSSDGVFYT ERN2_loxAf .....................F.. ERN1_eriEur KLPFTIPELVQASPCRSSDGILYM CG Dog
ERN2_echTel KLPFTIPELVLASPCRSSDGVFYT ERN2_echTe ..........L..........F.. ERN1_sorAra KLPFTIPELVQASPCRSSDGILYM CG Microbat
ERN2_dasNov KLPFTIPELVHTSPCRSSDGIFYT ERN2_dasNo ...........T........IF.. ERN1_loxAfr KLPFTIPELVQASPCRSSDGILYM -- Megabat
ERN2_monDom KLPFTIPELVHASPCRSSDGVLYT ERN2_monDo KLPFTIPELVHASPCRSSDGVLYT ERN1_proCap KLPFTIPELVQASPCRSSDGILYM CG Hedgehog
ERN2_macEug KLPFTIPELVHASPCRSSDGVFYT ERN2_macEu .....................F.. ERN1_echTel KLPFTIPELVQASPCRSSDGILYM CG Shrew
ERN2_sarHar1 KLPFTIPELVQASPCRSSDGIFYM ERN2_sarHa ..........Q.........IF.M ERN1_dasNov KLPFTIPELVQASPCRSSDGILYM -- Elephant
ERN2_sarHar2 KLPFTIPELVQASPCHSSDGIFYM ERN2_sarHa ..........Q....H....IF.M ERN1_choHof KLPFTIPELVQASPCRSSDGILYM -- Rock_hyrax
ERN2_ornAna KLPFTIPELVQSSPCRSSDGILYT ERN2_ornAn ..........QS........I... ERN1_monDom KLPFTIPELVQASPCRSSDGILYM CG Tenrec
ERN2_anoCar KLPFTIPELVQSSPCRSSDGIIYT ERN2_anoCa ..........QS........II.. ERN1_ornAna KLPFTIPELVHASPCRSSDGILYM CG Armadillo
ERN2_taeGut KLPFTIPELVQSSPCRSSDGVLYT ERN2_taeGu ..........QS............ ERN1_galGal KLPFTIPELVQASPCRSSDGILYM CG Opossum
ERN2_galGal KLPFTIPELVQASPCRSSDGILYM ERN2_galGa ..........Q.........I..M ERN1_taeGut KLPFTIPELVQASPCRSSDGILYM CG Platypus
ERN2_xenTro KLPFTIPELVQSSPCRSSDGILYT ERN2_xenTr ..........QS........I... ERN1_anoCar KLPFTIPELVQASPCRSSDGILYM CG Lizard
ERN2_xenLae KLPFTIPELVQSSPCRSSDGILYT ERN2_xenLa ..........QS........I... ERN1_xenTro KLPFTIPELVQSSPCRSSDGILYT CG Tetraodon
ERN2_tetNig KLPFTIPELVQASPCRSSDGVLYM ERN2_tetNi ..........Q............M ERN1_tetNig KLPFTIPELVQASPCRSSDGVLYM CG Fugu
ERN2_takRub KLPFTIPELVQASPCRSSDGVLYM ERN2_takRu ..........Q............M ERN1_takRub KLPFTIPELVQASPCRSSDGVLYM CT Stickleback
ERN2_gasAcu KLPFTIPDLVQSAPCRSSDGILYT ERN2_gasAc .......D..QSA.......I... ERN1_gasAcu KLPFTIPELVQASPCRSSDGVLYM CT Medaka
ERN2_oryLat KLPFTIPELVQSAPCRSSDGILYT ERN2_oryLa ..........QSA.......I... ERN1_oryLat KLPFTIPELVQASPCRSSDGVLYM CG Lamprey
ERN2_calMil KLPFTIPELVQSSPCRSSDGILYT ERN2_calMi ..........QS........I... ERN1_danRer KLPFTIPELVQASPCRSSDGILYM
ERN2_petMar KLPFTIPELVHASPCRTSDGVLYT ERN2_petMa ................T.......
ERN_braFlo KLPFTIPELVNASPCKSSDGILYT ERN_braFlo ..........N....K....I...
Case of MGAT5
chr4_4859 MGAT5 12
>contig00001 length=538 numreads=5 21 C=2(61) Y=2(56)
LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE
.................................................
^
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in two
tasmanian devil (here one is identical and the other differs from Monodelphis by C->Y) and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler).
Pseudogene issues: No processed pseudogenes relevent to this exon are seen by Blat of human and opossum sequence. Some questionable sequence occurs in tarsier and sloth but may be due to low coverage read or assembly error. These fragmentary sequences also have cysteine at the position in question.
Paralog issues: This gene has a moderately similar paralog, MGAT5B, with a similar enzymatic role (beta1,6-N-acetylglucosaminyltransferase). The opossum MGAT5B protein differs at 12 positions out of 49 from opossum MGAT5, whereas human and marsupial MGAT5A differ at one residue. Consequently the two paralogs are readily distinguished within vertebrates. This is moot because 33 of 33 available MGAT5B also have cysteine at the position in question (data not shown).
Homoplasy (recurrent mutation) issues: The alignments below show tyrosine has never replaced cysteine in any other species. This cysteine is extremely invariant in both paralogs, tracing back to lophotrochozoa and cnidaria.
Known variations: No human disease alleles have been mapped to either paralog. None of 9 SNP tracks at the UCSC browser show human variation in this exon.
Side issues: The column marked with an asterisk in the difference alignment below indicates a non-conservative phyloSNP K-->I that occured in the theran mammal stem after platypus divergence. All three marsupial sequences including tasmanian devil have isoleucine in this position as do all 30 of the available placental mammal sequences, suggesting that both the lysine and the isoleucine continue to be under strong selection. No comparable shift occured in the theran stem for MGAT5B where the residue is arginine in all species, a basic residue similar to lysine.
Structural significance: The MGAT5 gene supposedly encodes a conventional enzyme, mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase involved in the synthesis of protein-bound and lipid-bound oligosaccharides. Yet surprisingly, no determined 3D structure exists at PDB relevent to the configuration of this exon -- nor indeed the large 741 residue protein. This is very peculiar because glycosyl transerfases are a well-studied group of enzymes (nearly 100 loci in human) and might be expected to bind UDP-GlcNAc (like MGAT4A or MGAT3).
Only a small region of the protein have a prediction at ModBase using 2f9fA, a remote mannosyltransferasee from Archaeoglobus fulgidus. Luckily the model covers the cysteine at issue, showing two helices and a beta sheet.
SwissProt does not annotate the cysteine at position 532 as part of a disulfide or active site; the predicted location (Golgi) can have homodimer disulfides of similar enzymes, though this is a complex topic. Although all 20 cysteines in this protein are conserved human to opossum, this could be a consequence of the overall sequence identity of 90%. Twelve of the cysteines, not including the Sarcophilus variant, are found in the last 140 residues, perhaps forming a disulfide knot. All but 1 of these cysteines is conserved in the pre-Bilateran anemone Nematostella (which enriches relative to overall percent identity of 43%).
Highest MGAT5 expression occurs in brain, heart, kidney, and placenta. No domains other than a signal peptide and 6 of its own glycosylation target sites are found by online tools such as SMART.
Although the bulky tyrosine substitution is conservative in the sense of polar nature and perhaps hydrogen-bonding capacity, it cannot replace these specialized functions of cysteine. Considering the extreme conservation of this cysteine, this substitution must have a substantial-- perhaps even disabling -- impact on enzymatic function.
Functional significance: In view of the facial tumor situation in tasmanian devils, OMIM's account of prior research in mouse on this gene is quite interesting. Less is known about MGAT5B though it also functions in the synthesis of complex cell surface N-glycans.
" Malignant transformation is accompanied by increased beta-1,6-GlcNAc branching of N-glycans attached to Asn-X-Ser/Thr sequences in mature glycoproteins... The amount of MGAT5 products correlates with disease progression... Mgat5-deficient mice, which are born healthy but develop various abnormalities as adults...Mgat5-deficient mice showed kidney autoimmune disease, enhanced delayed-type hypersensitivity, and increased susceptibility to experimental autoimmune encephalomyelitis...The Golgi enzyme beta1,6 N-acetylglucosaminyltransferase V (Mgat5) is up-regulated in carcinomas and promotes the substitution of N-glycan with poly N-acetyllactosamine, the preferred ligand for galectin-3 (Gal-3)...inhibitors of MGAT5 might be useful in the treatment of malignancies by targeting their dependency on focal adhesion signaling for growth and metastasis."
^ ^ * MGAT5_homSap LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE homSap MGAT5_panTro LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. panTro MGAT5_gorGor LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. gorGor MGAT5_ponAbe LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. ponAbe MGAT5_rheMac LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. rheMac MGAT5_calJac LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. calJac MGAT5_micMur LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... micMur MGAT5_otoGar LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. otoGar MGAT5_tupBel LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. tupBel MGAT5_musMus LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. musMus MGAT5_ratNor LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. ratNor MGAT5_criGri LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... criGri MGAT5_dipOrd LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. dipOrd MGAT5_cavPor LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. cavPor MGAT5_speTri LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. speTri MGAT5_oryCun LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. oryCun MGAT5_ochPri LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. ochPri MGAT5_vicPac LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. vicPac MGAT5_susScr LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. susScr MGAT5_turTru LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. turTru MGAT5_bosTau LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. bosTau MGAT5_equCab LFAGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ..A.............................................. equCab MGAT5_felCat lfvgLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... felCat MGAT5_canFam LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... canFam MGAT5_myoLuc LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. myoLuc MGAT5_eriEur LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. eriEur MGAT5_sorAra LFVGLGFPYEGPAPLEAIANGCAFLNPKFSPPKSSKNTDFFIGKPTLRE .............................S................... sorAra MGAT5_loxAfr LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. loxAfr MGAT5_proCap LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. proCap MGAT5_echTel LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFIGKPTLRE ................................................. echTel MGAT5_monDom LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... monDom MGAT5_macEug LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... macEug MGAT5_sarHar1 LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE ..........................V...................... sarHar1 MGAT5_sarHar2 LFVGLGFPYEGPAPLEAIANGYAFLNVKFNPPKSSKNTDFFIGKPTLRE .....................Y....V...................... sarHar2 MGAT5_ornAna LFVGLGFPYEGPAPLEAIANGCAFLNLKFNPPKSSKNTDFFKGKPTLRE ..........................L..............K....... ornAna MGAT5_galGal LFVGLGFPYEGPAPLEAIANGCAFLNLRFNPPKSSKNTEFFKGKPTLRE ..........................LR..........E..K....... galGal MGAT5_taeGut LFVGLGFPYEGPAPLEAIANGCAFLNLRFNPPKSSKNTDFFKGKPTLRE ..........................LR.............K....... taeGut MGAT5_anoCar LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFKGKPTLRE .........................................K....... anoCar MGAT5_xenTro LFVGLGFPYEGPAPLEAIANGCAFLNPKFNPPKSSRNTDFFKGKPTLRE ...................................R.....K....... xenTro MGAT5_tetNig VFVGLSFPYEGPAPLEALANGCIFLNPRLKPPQSSLNSEFFKEKPNIRE V....S...........L....I....RLK..Q..L.SE..KE..NI.. tetNig MGAT5_takRub LFVGLSFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFKGKPTLRE .....S...................................K....... takRub MGAT5_gasAcu LFVGLSFPYEGPAPLEAIANGCAFLNPKFSPAKSSKNTDFFKGKPTLRE .....S.......................S.A.........K....... gasAcu MGAT5_oryLat LFVGLSFPYEGPAPLEAIANGCAFLNPKFNPPKSSKNTDFFKGKPTLRE .....S...................................K....... oryLat MGAT5_danRer LFVGLSFPYEGPAPLEAIANGCAFLNPRFDPAKSSKNTDFFKGKPTLRE .....S.....................R.D.A.........K....... danRer MGAT5_oncMyk LFVGLSFPYEGPAPLEAIANGCAFLNPKFTPPKSSKNTDFFKGKPTLRE .....S.......................T...........K....... oncMyk MGAT5_pimPro LFVGLSFPYEGPAPLEAIANGCAFLNPRFDPSKSSKNTDFFKGKPTLRE .....S.....................R.D.S.........K....... pimPro MGAT5_calMil LFVGLGFPYEGPAPLEAIANGCAFLNPRFNPPKSSKNTEFFKGKPTLRE ...........................R..........E..K....... calMil MGAT5_petMar LFVGLGFPYEGPAPLEAIANGCVFLNPRFRPPKSSKNTDFFKGKPTLRE ......................V....R.R...........K....... petMar MGAT5_braFlo LFVGLGFPYEGPAPLEAIASGCVFLNPKFTQPKSRLNTKFFEGKPTFRE ...................S..V......TQ...RL..K..E....F.. braFlo MGAT5_strPur LFIGLGFPYEGPAPLEAVANGCVFLNPKFNPPKNYQNTKFFQGKPTSR. MGAT5_helRob LFIGLGFPYEGPAPLEAIAAGCVFINPKFNPPHSSLNTKFFKGKPTARE MGAT5_nemVec VFIGLGFPYEGPAPLEAIQSGCVFLNAKFDPPHDRVNTPFFKNKPTLRK
Note: the species with unfamiliar genSpp acronyms are Cricetulus griseus, Oncorhynchus mykiss, Pimephales promelas , Callorhinchus milii, Branchiostoma floridae, Strongylocentrotus purpuratus, Helobdella robusta, Nematostella vectensis, and Acropora millepora.
Here the opossum protein is broken into its 16 coding exons with phases (base overhangs at split codons) shown:
>MGAT5_monDom length=743
0 MAFFAPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQHESSSMLREQILDLSKRYIKALAEENRNVVDGPYVGVMTAY 1
2 DLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTTTTAAPSSIAAFEKISVA 1
2 DIINGAQEKCELPPMDGFPHCEGKIK 0
0 WMKDMWRTDPCYANYGVDGSTCSFFIYLSE 0
0 VENWCPHLPWRAKNPYEEPDQNSM 0
0 AEIRTDFNLLYGMMKRHEEFRWMILRIRRMADAWIEAIKSLAEKQNLEKRKRKK 0
0 ILVHLGLLTKESGFKIAENAFSGGPLGELVQWSDLITSLYLLGHDIRISASLAELKE 2
1 IMKRVVGNRSGCPTVGDRIVELIYIDIVGLAQFKKTLGPSWVHYQ 2
1 CMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMF 1
2 PHTPDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDSFWK 0
0 NKKEYLDIIHTYMEVHATVYGSSTNHMPSYVKNHGILSGRDLQFLLRETK 0
0 LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE 0
0 LTSQHPYAEVFIGRPHVWTVNPTDHREVENAVKAILNQK 0
0 IEPYMPYEFTCEGMLQRMNAFIEKQ 0
0 DFCHGQVMWPPLNALQVKLSEPGKSCKQVCQENQLICEPSFFQHLNKDKDVLK 2
1 YEVICHTTELANDILVPSYDDKKKHCVFQGDLLLFSCAGAHPKHKRICPCRDYIKGQVALCQDCL* 0
>MGAT5_sacHar one match to exon 1: FPUIIJ301C96S1
0 MAFFAPWKLSSQN*GFSWLTFGFIWGMMLLHFTIQQRTQHESSSMLREQILDLSKRYIKALAEENRNVVDGPYVGVMTAY 1
2 DLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTTTTAVPSSIAAFEKIsVA 1
2 DIINGAQEKCELPPMDGFPHCEGKIK 0
0 0
0 0
0 AEIRTDFHLLYGMMKRHEEFRWMILRIRRMADAWIEAIKSLAEKQNLEKRKRKK 0
0 ILVHLGLLTKESGFKIAENAFSGGPLGELVQWSDLITSLYLLGHDIRISASLAELKE 2
1 IMKRVVGNRSGCPTVGDRIVELIYIDIVGLAQFKKTLGPSWVHYQ 2
1 CMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMF 1
2 AHTPDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDNFWK 0
0 NKKEYLDIIHTYMEVHATVYGSSTNHMPSYVKNHGILSGRDLQFLLRETK 0
0 LFVGLGFPYEGPAPLEAIANGYAFLNVKFNPPKSSKNTDFFIGKPTLRE 0
0 0
0 IEPYMPYEFTCEGMLQRMNAFIEKQ 0
0 2
1 YEVVCHTTELANDILVPSYDDRKKHCVFQGDLLLFSCAGAHPKHKRICPCRDYIKGQVALCQDCL* 0
The premature stop codon in the first exon is likely read error (1 bp dropped, 1 bp later added):
atggctttctttgctccatggaaattatcctctcagaaactagggtttttcctggtgact
M A F F A P W K L S S Q K L G F F L V T correct monDom frame
W L S L L H G N Y P L R N - G F S W - L 6 residue observed frameshifts in sarHar N*GFSWL
G F L C S M E I I L S E T R V F P G D F irrelevent 3rd reading frame
MGAT5 has 16 exons. The key one here is 12. Alignment of MGAT5_sarHar to opossum shows only 5 differences in 589 residues available for comparison.
Alignment of Monodelphis to human establishes that MGAT5 is better conserved than the average gene:
Identities = 673/744 (90%), Positives = 708/744 (95%), Gaps = 2/744 (0%)
monDo 1 MAFFAPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQHESSSMLREQILDLSKRYIKA 60
MA F PWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQ ESSSMLREQILDLSKRYIKA
homSa 146 MALFTPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQRTQPESSSMLREQILDLSKRYIKA 325
monDo 61 LAEENRNVVDGPYVGVMTAYDLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTT 120
LAEENRNVVDGPY GVMTAYDLKKTLAVLLDNILQRIGKLESKVDNLV+ G + +T
homSa 326 LAEENRNVVDGPYAGVMTAYDLKKTLAVLLDNILQRIGKLESKVDNLVV--NGTGTNSTN 499
monDo 121 TTAAPSSIAAFEKISVADIINGAQEKCELPPMDGFPHCEGKIKWMKDMWRTDPCYANYGV 180
+T A S+ A EKI+VADIINGAQEKC LPPMDG+PHCEGKIKWMKDMWR+DPCYA+YGV
homSa 500 STTAVPSLVALEKINVADIINGAQEKCVLPPMDGYPHCEGKIKWMKDMWRSDPCYADYGV 679
monDo 181 DGSTCSFFIYLSEVENWCPHLPWRAKNPYEEPDQNSMAEIRTDFNLLYGMMKRHEEFRWM 240
DGSTCSFFIYLSEVENWCPHLPWRAKNPYEE D NS+AEIRTDFN+LY MMK+HEEFRWM
homSa 680 DGSTCSFFIYLSEVENWCPHLPWRAKNPYEEADHNSLAEIRTDFNILYSMMKKHEEFRWM 859
monDo 241 ILRIRRMADAWIEAIKSLAEKQNLEKRKRKKILVHLGLLTKESGFKIAENAFSGGPLGEL 300
LRIRRMADAWI+AIKSLAEKQNLEKRKRKK+LVHLGLLTKESGFKIAE AFSGGPLGEL
homSa 860 RLRIRRMADAWIQAIKSLAEKQNLEKRKRKKVLVHLGLLTKESGFKIAETAFSGGPLGEL 1039
monDo 301 VQWSDLITSLYLLGHDIRISASLAELKEIMKRVVGNRSGCPTVGDRIVELIYIDIVGLAQ 360
VQWSDLITSLYLLGHDIRISASLAELKEIMK+VVGNRSGCPTVGDRIVELIYIDIVGLAQ
homSa 1040 VQWSDLITSLYLLGHDIRISASLAELKEIMKKVVGNRSGCPTVGDRIVELIYIDIVGLAQ 1219
monDo 361 FKKTLGPSWVHYQCMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMFPHT 420
FKKTLGPSWVHYQCMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMFPHT
homSa 1220 FKKTLGPSWVHYQCMLRVLDSFGTEPEFNHANYAQSKGHKTPWGKWNLNPQQFYTMFPHT 1399
monDo 421 PDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDSFWKNKKEYLDIIHTYMEVHAT 480
PDNSFLGFVVEQHLNSSDI HINEIKRQNQSLVYGKVDSFWKNKK YLDIIHTYMEVHAT
homSa 1400 PDNSFLGFVVEQHLNSSDIHHINEIKRQNQSLVYGKVDSFWKNKKIYLDIIHTYMEVHAT 1579
monDo 481 VYGSSTNHMPSYVKNHGILSGRDLQFLLRETKLFVGLGFPYEGPAPLEAIANGCAFLNVK 540
VYGSST ++PSYVKNHGILSGRDLQFLLRETKLFVGLGFPYEGPAPLEAIANGCAFLN K
homSa 1580 VYGSSTKNIPSYVKNHGILSGRDLQFLLRETKLFVGLGFPYEGPAPLEAIANGCAFLNPK 1759
monDo 541 FNPPKSSKNTDFFIGKPTLRELTSQHPYAEVFIGRPHVWTVNPTDHREVENAVKAILNQK 600
FNPPKSSKNTDFFIGKPTLRELTSQHPYAEVFIGRPHVWTV+ + EVE+AVKAILNQK
homSa 1760 FNPPKSSKNTDFFIGKPTLRELTSQHPYAEVFIGRPHVWTVDLNNQEEVEDAVKAILNQK 1939
monDo 601 IEPYMPYEFTCEGMLQRMNAFIEKQDFCHGQVMWPPLNALQVKLSEPGKSCKQVCQENQL 660
IEPYMPYEFTCEGMLQR+NAFIEKQDFCHGQVMWPPL+ALQVKL+EPG+SCKQVCQE+QL
homSa 1940 IEPYMPYEFTCEGMLQRINAFIEKQDFCHGQVMWPPLSALQVKLAEPGQSCKQVCQESQL 2119
monDo 661 ICEPSFFQHLNKDKDVLKYEVICHTTELANDILVPSYDDKKKHCVFQGDLLLFSCAGAHP 720
ICEPSFFQHLNKDKD+LKY+V C ++ELA DILVPS+D K KHCVFQGDLLLFSCAGAHP
homSa 2120 ICEPSFFQHLNKDKDMLKYKVTCQSSELAKDILVPSFDPKNKHCVFQGDLLLFSCAGAHP 2299
monDo 721 KHKRICPCRDYIKGQVALCQDCL* 744
+H+R+CPCRD+IKGQVALC+DCL
homSa 2300 RHQRVCPCRDFIKGQVALCKDCL* 2371
Full length genes appear available from GenBank and genome projects for mouse, rat (NM_001107068), dog (wgs exons), horse (XM_001489091), wallaby (wgs exons), and platypus (XM_001520380). Because this gene is 90% conserved at marsupial, placental mammals will not be informative -- indeed it is necessary to go to greater phylogenetic depth than lamprey to define the ultra-conserved residues in this protein:
>MGAT5_macEug nearly identical to monDom; 3 exons are missing, 2 partial exons, exon 4 has frameshifts 0 MAFFAPWKLSSQKLGFFL 1 2 DLKKTLAVLLDNILQRIGKLESKVDNLVINGTGANSTNTTTTAVPSSIAAFEKISVA 1 2 DIINGAQEKCELPPMDGFPHCEGKIK 0 0 WMKDiWRTDPCYANYGVDGSTCSFFIYLSE 0 0 VENWCPHLPWRAKNPYEEPDQNSM 0 0 0 0 ILVHLGLLTKESGFKIAENAFSGGPLGELVQWSDLITSLYLLGHDIRISASLAELKE 2 1 2 1 GTEPEFNHANYAQSKGHKTP 1 2 aHTPDNSFLGFVVEQHLNSSDIKHINEIKRQNQSLVYGKVDSFWK 0 0 NKKEYLDIIHTYMEVHATVYGSSTNHMPSYVKNHGILSGRDLQFLLRETK 0 0 LFVGLGFPYEGPAPLEAIANGCAFLNVKFNPPKSSKNTDFFIGKPTLRE 0 0 LTSQHPYAEVFIGRPHVWTVNPTDHREVENAVKAILNQK 0 0 IEPYMPYEFTCEGMLQRMNAFIEKQ 0 0 DFCHGQVMWPPLNALQVKLSEPGKSCKQVCQENQLICEPSFFQHLNKDKDVLK 2 1 YEVICHTTELANDILVPSYDDKKKHCVFQGDLLLFSCAGAHPKHKRICPCRDYIKGQVALCQDCL* 0 >MGAT5_galGal 87% identical to opossum MAFPWKLSSQKLGFFLVTFGFIWGMMLLHFTIQQQTQHESSSVLREQILDLSKRYIKALAEENKNVVDGPYVGTVTAY DLKKTLAVLLDNILQRIGKLESKVENLVLNGTGANSTNTTTPAPSLGAVEKLNVA DLINGAQEQCELPPMDGFPHCEGKIK WMKDMWRSDPCYASYGVDGSTCSFFIYLSE VENWCPRLPWRAKNPNEETDQKTV AEIRINFDPLYKMMSRHEEFRWMTLRIRRMADTWIEAIKSLAEKQNLENRKRKK ILVHLGLLTKESGFKIAENAFSGGPLGELVQWSDLITSLYLLGHDIRISASLAELKE IMKKVVGNRSGCPTQGDKVVELIYIDIVGLTQFKKTLGPSWVHYQ CMLRVLDSFGTEPEFNHAHYAQSKGHKTPWGKWNLNPQQFYTMF PHTPDNSFLGFVVEQHLNSSDIKHINDIKRQNQSLVYGKVDNFWK DKKAYLDVIHTYMEVHGTVHGTSTIYIPGYVKNHGILSGRDLQFLLRETK LFVGLGFPYEGPAPLEAIANGCAFLNLRFNPPKSSKNTEFFKGKPTLRE LTSQHPYAEVYIGKPHVWTVDINNLSEVEKAVKSILNQK IDPYLPYEFTCEGMLQRMNAFIERQ DFCHGQVMWPPLSALQVKLAEPGKSCKQVCQESQLICEPSFFQHLNKDKALLK HNIECLTTESANDILVPSFDGRRKHCVFQGDLLLFSCAGSHPTHRRICPCRDYIKGQVALCKDCL* >MGAT5_nemVec Nematostella vectensis (sea anemone) XM_001641404 43% identical to opossum 19 of 20 cysteines conserved MIATKGRPTFKLSAHRIGIVFIIISFIWGLYLIKIQLDERNSQPDYLKGRIIHLSKEYIRALAREKGVYGIDGQPSTQQGVGDLKKATAVLLQSMLERIHVL EKQVEGVIVNSTLEFEILASQIKSLNTTFSLHLSNHSYVSANSCVIPDDPSYPECRQKVMWMRNFWKTHECYAKDHGVNGTICSFLVYLSEVENWCPKFPGRMKPTSRATTEGADL HRSDVQGLLGLLNDQDPIKFKWIKNRINQMWPQWLSALEDLKKKRDLKKIKQKKILVHIGLLANERALHFAANADKGGPLGELVQWSDLIASLYLLGHDVTVTADIPRLQGIFGKL RGPAKKPCPTTIKNDYDLIYLDYYGVKQMQTKVGQFTQSFKCKFRIVDSFGTEAQFNYAGFTEKVPGGSMALWGRHNLNLKQFMTMFPHSPDNSFLGFVVGEEPTPDPHPKKKKAR ALVYGKHYYMWKDLKQRSFLDVINKYMEIHATVGGGIKKWVPSYVINHGVLPSLEVQKLLQDSMIFVGLGFPYEGPAPLEAIAHGCFFLNTKYHPPRNRINTPFFKDKPTLRQITS QHPYAEDYIGQPYVYTVDINDLNKIEAVMKEIMMAEPVSPYLPYEFTHKGMLERLHVFIENQNFCGQNLWPPLNALQARKGAMGSSCKETCHSLGLVCEPQYFPAINTKERMTRSG FPCNTTRVEDMPSLVAPGYRDDPPVCLRQAQNLLFSCTANSPTTKRLCPCRDFKKGQVALCSKC*
Case of ACTL6B
chr2_18546 ACTL6B 11 >contig00001 length=502 numreads=11 GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ^ 3 G=4(94) R=7(213) Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5 and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in tasmanian devil (in this case, one individual differs from Monodelphis by G->R), then differences between the two thylacines, and finally the number of experimental reads that confirm the nucleotide difference and the sum of the quality scores. The sequences were assembled by Newbler.
The change from small non-polar glycine to bulky positively charged lysine is highly non-conservative, especially at a highly conserved residue such as this. Again the change in Sarcophilus is at a CpG hotspot, this time with a mildly unusual transversion of the C to the purine G.
The well-studied protein here is a member of a family of actin-related proteins (ARPs) which have significant homology to conventional actins, in particular sharing the actin fold (an ATP-binding cleft) as common feature. ACTL6B and its 83 identical paralog ACTL6A are involved in diverse cellular processes such as vesicular transport, spindle orientation, nuclear migration and chromatin remodeling. Both have 14 coding exons. The entire exon containing the G-->R is highly conserved including the glycine.
Pseudogene issues: Blat of full length sequence to human shows no recent processed or segmental pseudogenes. However opossum assembly, which has all 14 exons, also contains a fairly recent processed pseudogene with 91.5% identity. This locus has internal stop codons and ELSD in place of GLSG for the key glycine. This arose from ACTL6A, not ACTL6B.
Paralog issues: There is potential for confusion with the paralog ACTL6A. This wouldn't normally matter because all species in this gene too have glycine at the arginine-substituted site. However its pseudogene could present problems because its decay may have taken a different path in Sarcophilus than in Monodelphis giving the R (instead of D), assuming the pseudogene was formed prior to divergence of these species. Indeed, Macropus eugenii appears to have two processed pseudogenes; one of this has R in place of a glycine 4 residues earlier. It will prove necessary to consider adjacent regions in Sarcophilus reads to determine whether the feature is a pseudogene.
Comparison of gene to pseudogene in opossum:
000000889 E R L R I P E G L F D P S N V K G L S G 000000948
<<<<<<<<< | X | K | | | | | | | | | | | | E | | D <<<<<<<<<
250390825 gagtgactcaagattcctgaagggttatttgacccatctaatgtgaaggaattgtcagac 250390766
000000949 N T M L G V G H V V T T S I G M C D I D 000001008
<<<<<<<<< | | | | | | S | | | | | | F | | | | | | <<<<<<<<<
250390765 aacacaatgttgggagtcagtcatgttgttaccacaagctttgggatgtgtgacattgac 250390706
000001009 I R P G L Y G S V I V T G G N T L 000001059
<<<<<<<<< F | | | | | D N M L G A | | | I | <<<<<<<<<
250390705 tttagaccgggactttatgacaatatgttaggggcgggaggaaacattctg 250390655
Comparison of ACTL6A_homSap gene to pseudogenes in wallaby:
macEu: 1063 FPVGYNCNFGVEQLKITERLFDPSNVKRLSGNPMLGVSHVVTTRIGMCDIDIRPGLYGTV 1242
FP GYNC+FG E+LKI E LFDPSNVK LSGN MLGVSHVVTT +GMCDIDIRPGLYG+V
homSa: 289 FPNGYNCDFGAERLKIPEGLFDPSNVKGLSGNTMLGVSHVVTTSVGMCDIDIRPGLYGSV 348
macEu: 48 PNVYKCGFGAEHFKIPEGLFDRSNMKGLSGNTMLGISHVVTKSTGMCDIDIRPGFYISVI 227
PN Y C FGAE KIPEGLFD SN+KGLSGNTMLG+SHVVT S GMCDIDIRPG Y SVI
homSa: 290 PNGYNCDFGAERLKIPEGLFDPSNVKGLSGNTMLGVSHVVTTSVGMCDIDIRPGLYGSVI 349
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
Structural significance:
Functional significance:
* * *
ACTL6B_homSap GLSGNTMLGVGHVVTTSIGMCDIDIRP GLSGNTMLGVGHVVTTSIGMCDIDIRP ACTL6B_homSap GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_panTro GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_panTro GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_gorGor GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_gorGor GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_ponAbe GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_ponAbe GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_rheMac GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_rheMac GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_calJac GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_calJac GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_tarSyr GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_tarSyr GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_micMur GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_micMur GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_otoGar GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_otoGar GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_tupBel GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_tupBel GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_musMus GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_musMus GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_ratNor GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_ratNor GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_dipOrd GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_dipOrd GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_cavPor GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_cavPor GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_ochPri GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_ochPri GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_turTru GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_turTru GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_bosTau GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_bosTau GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_equCab GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_equCab GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_felCat GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_felCat GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_canFam GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_canFam GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_myoLuc GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_myoLuc GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_pteVam GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_pteVam GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_eriEur GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_eriEur GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_loxAfr GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_loxAfr GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_proCap GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_proCap GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_echTel GLSGNTMLGVGHVVTTSIGMCDNDIRP ......................N.... ACTL6B_echTel GLSGNTMLGVGHVVTTSIGMCDNDIRP
ACTL6B_monDom GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_monDom GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_ornAna GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_ornAna GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_galGal GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_galGal GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_taeGut GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_taeGut GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_anoCar GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_anoCar GLSGNTMLGVGHVVTTSIGMCDIDIRP
ACTL6B_xenTro GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_xenTro GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_tetNig GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_tetNig GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_takRub GLSGNTMLGVSHVVTTSVGMCDIDIRP ..........S......V......... ACTL6B_takRub GLSGNTMLGVSHVVTTSVGMCDIDIRP
ACTL6B_gasAcu GLSGNTMLGVGHVVTTSVGMCDIDIRP .................V......... ACTL6B_gasAcu GLSGNTMLGVGHVVTTSVGMCDIDIRP
ACTL6B_oryLat GLSGNTMLGVGHVVTTSVGMCDIDIRP .................V......... ACTL6B_oryLat GLSGNTMLGVGHVVTTSVGMCDIDIRP
ACTL6B_danRer GLSGNTMLGVGHVVTTSIGMCDIDIRP ........................... ACTL6B_danRer GLSGNTMLGVGHVVTTSIGMCDIDIRP
* * *
Consensus gLsGnTMlgvgHVVTts!g$CDi.Ir. gLsGnTMlgvgHVVTts!g$CDi.Ir.
(more shortly)
Case of IPO7
chr5_9037 IPO7 23
>contig00001 length=680 numreads=8
SSQVEKHSCSLTEELGSDEDDIDEDGQEYLEILAKQAGEDGDDEEWEEDDAEETALEGYSTIIDDEENPVDEYQIFKAIFQ
....*N.....................................................F.....................
^
59 F=2(72) S=3(53)
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by -> ), then differences between the two thylacines
(here one individual has at position , the other has ), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues:
Paralog issues:
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
Case of WDFY3
chr5_2532 WDFY3 19
>contig00001 length=482 numreads=8
DDFSEESSFYEILPCCARFRCGDLIVEGQWHHLVLVMSKGMLKNSTAALYIDGQLVSTVK
................T..............................T..L.....N...
^
16 T=3(117) A=5(138)
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by -> ), then differences between the two thylacines
(here one individual has at position , the other has ), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues:
Paralog issues:
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
Structural significance:
Functional significance:
(more shortly)
Case of PPFIA3
chr4_22002 PPFIA3 15 incorrectly mapped from monDom5 to human
>contig00001 length=298 numreads=4
LIQEEKETTEQRAEELESRVSGSGLDSLGRYRASCSLPPSLTTSTLASPSPPSSGHSTPRPAPPSPAREAPANSTSNTAEKP
........................................................F..................G.V.
^
56 F=2(43) S=2(37)
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by -> ), then differences between the two thylacines
(here one individual has at position , the other has ), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues:
Paralog issues:
Homoplasy (recurrent mutation) issues:
Known variations:
Side issues:
Structural significance:
Functional significance:
(more shortly)
Structural significance:
Functional significance:
(more shortly)
Other cases to be considered
chr6_2360 XYLT1 5 61 D=3(110) A=5(107)
>contig00001 length=488 numreads=10
RSNYMHRQVLQFAGQYQNVRVTSWRMATIWGGASLLSTYLQSMRDLMEMTDWPWDFFINLSAADYPI
...L........................................................D.....
^
chr4_18550 ATP4A 6 16 C=4(130) R=3(74)
>contig00001 length=906 numreads=10
TAQGLVVNTGDRTIIGRIASLASGVENEKTPIAIEIEHFVDIIAGLAILFGATFFIVAMCIGYTFLRAMVFFMAIVVAYVPEGLLATVT
................C........................................................................
^
chr4_11174 FLI1 3 32 N=2(63) K=3(47)
>contig00001 length=575 numreads=9
ESPVDCSVNKCSKLVGGNESNPMNYNTYMDEKNGPPPNMTTNERRVIVPA
..................................................
^
chr2_30280 VPS72 5 15 R=3(59) K=2(51)
>contig00001 length=591 numreads=6
NYERLEADKKKQVHKKRKCPGPVITYHSMTVPLLAEPGPKEENVDVE
...............R..................T............
^
chr6_5144 ABCC1 23 4 Q=2(69) P=2(80) looks like a frame-shift problem in monDom5
>contig00001 length=802 numreads=10
HLCFPRLHLDLLHNVLRSPMSFFERTPSGNLVNRFSKEMDTVDSMIPQIIKMFMGSLFNVIGACIIILLATPIAAIIIPPLGLIYFFVQ
....Q....................................................................................
^
chr5_8347 SPON1 11 20 V=3(65) I=2(66) wobbly
>contig00001 length=433 numreads=5
GSTCTMSEWITWSPCSISCGVGMRSRERYVKQFPEDGSVCTVPTEETEKCTVNEEC
......................................I.N...............
^
chr3_5872 ACOT12 14 14 I=3(95) V=3(110) wobbly
>contig00001 length=472 numreads=6
NTYVVAVKSVTLASIPPSPQYNRSEITCAGFLIRAVDSNSCT
.................................Q....S...
^
Other marsupial genes of interest
The collections below contain well-understood genes with very extensive comparative genomics. They can serve as a test bed for Sarcophilus assembly quality, a place where genuine anomalies or distinct adaptive features might surface (perhaps as phyloSNPs) and where marsupial phylogeny might be refined using rare genomic events in nuclear genes.
The gene sets contain all available marsupial orthologs plus for context one flanking gene each from placentals and monotremes. These genes are available in much broader hand-curated sets elsewhere on this site.
IRBP (96 marsupials)
Interphotoreceptor retinol-binding protein, poorly named by IGNC as RBP3 despite its complete lack of paralogs, is a 4 exon 1247 residue glycoprotein that shuttles retinoids between the photoreceptor cells and the retinal pigment epithelium. The protein's size results from four ancient internal tandem duplictions that became established prior to the intronation era.
The first three homology domains and part of the fourth are all encoded by the first large exon of 1090 amino acids. This exon has been much used in marsupial phylogeny (along with the first intron of transthyretin). Indeed the 96 marsupial species in 51 genera having determined IRBP sequences at GenBank include a Dec 2008 partial sequence for Thylacinus cynocephalus, as well as for Sarcophilus harrisii.
The closest matches to the thylacine IRBP are shown in the difference alignment of the first 60 residues below. These species all lie with the Dasyuromorphia. The indicated E-->K may be one of several phyloSNPs breaking this group into blue and green subclades.
The numbat Myrmecobius fits implausibly (its amino terminal sequence EF028750 needs verification) -- its affinities seem to lie with the Didelphimorphia. Thylacinus is not basal within Dasyuromorphia relative to Myrmecobius using IRBP. However this may be a case of mis-comparison of genes.
* * * STSKAPQHDSKFTNATQEELLALFQQIIKYQVLEGNVGYLRVDYIPGREMIEEVGEFLVN EU091365 0 Thylacinus cynocephalus .........P..A..................I............................ AY532676 3 Myoictis wallacei ........NP..A............................................... AY532687 3 Neophascogale lorentzii ........NP..A........T...................................... AY532686 4 Phascolosorex dorsalis .........P..V............................................... AY532670 2 Parantechinus apicalis ....V....P..A..................I.....................L...... AY532675 5 Myoictis melas .........P..A...................................D........... AY532679 3 Dasyurus hallucatus ...E.....P..A............K........D.............D........... AY532685 6 Sarcophilus harrisii ...E.......RA..........L............................Q..K.... EF028748 6 Sminthopsis crassicaudata .......R.P.LA.........SL.......................Q....Q....... EF028749 8 Planigale ingrami ..A......P.LA.V.....................................K....... EF028736 6 Antechinus stuartii ..A......P.L..V.....................................K....... EF028743 5 Micromurexia habbema ..A......P.LA.V.....................................K....... EF028744 6 Murexchinus melanurus ..A......P.L..V....V................................K....... EF028746 6 Paramurexia rothschildi ..A......P.LA.V.....................................K....... EF028747 6 Phascogale calura ..A......P.LA.V.....................................K....... EF028745 6 Phascomurexia naso .SA......P.LA.V.....................................K....... AY532667 7 Murexia longicaudata ......K..PNLA........T.L..R....................Q.VV.K....... EF028750 12 Myrmecobius fasciatus ..PET...VP..A.V........L..M....................Q.VV.K....... AY233765 13 Caluromys philander ..PET...VP.LA.V.......QL..M....................Q.VV.K....... AF257675 15 Caluromysiops irrupta ..PET...VP.LA.V......T.L..M....................Q.VV.K....... AF257688 15 Glironia venusta .IPET...VP..A.V.R....T.L..M....................Q.VV.K....... AF257683 16 Didelphis albiventris .IPE....VP.LA.I......T.L..M....................Q.VV.K....... AF257686 15 Gracilinanus microtarsus .IPET...VP..A.V......T.L..M....................Q.VV.K....... AF257676 15 Marmosops noctivagus .IPET...VP.LA.V........L..M....................Q.VV.K....... AY233788 15 Philander opossum .IPET...VP.LA.I......T.L..M....................Q.VV.K....... AF257689 16 Thylamys pallidior
Using Sarcophilus as probe in a different region, 721-900, we find this peculiar outcome: what appears to be a second very odd gene, XY difference, pseudogene, weird balanced polymorphism, nonhomologous recombination, sequence submission error, frameshifts, or systemic experimental error (eg Dasyurus maculatus AY532680 is identical to AY243439 outside the 15 amino acid block). However the genomic reads from individual Sarcophilus used in this project show no sign of this gene despite excellent coverage of the second type of gene.
Macropus and Monodelphis genomes only contain the second type of gene. All Didelphimorphia and Diprotodontia are of this type, as are platypus and all placentals. With the Sarcophilus genome, this can be resolved as it should have both and be the such first genome. Perhaps the alignment above is a mixture of type 1 and type 2 genes (resp. alleles). The Myrmecobius anomaly makes it more likely two distinct genes are present.
A definite pecularity seen in blast searches is the occurence earlier in the sequence of a very homologous segment for this very block, likely the homologous part of another of the internal tandem repeats. It is seen in both types of genes. Possibly internal non-homologus recombination or gene conversion has inserted first repeat sequence again in this distal block in place of what was relatively diverged sequence. Internal gene conversion would make IRBP extremely difficult to use in alignment-based phylogeny. As rare genomic event, it unites the species that have it but species that don't have it would have to be re-examined to exclude the possiblity that only the type 2 gene happened to be sequenced.
It emerges from direct tblastn that the Sacrophilus individual sequenced was female. That is, ATRX is well represented but not ATRY (though the situation is somewhat confused due to additional paralogs). Marsupial XY are quite different from placentals:
"Many or most genes on the mammal Y chromosome evolved a testis-specific function after diverging from an X-borne copy with a general function in both sexes. In marsupial but not eutherian mammals, a testis-specific orthologue (ATRY) of the widely expressed X-borne ATRX gene lies on the Y chromosome. Since mutations in human ATRX cause sex reversal, it is possible that one function of ATRY in marsupials is testicular differentiation. We report here the isolation and sequencing of the tammar wallaby (Macropus eugenii) ATRY cDNA, and comparison of its sequence with that of tammar ATRX. The evolution of a testis-specific function for the ATRY protein distinct from the general role of ATRX in both sexes has been accompanied by sequence changes in many protein domains that would alter protein binding partners. A large open reading frame encodes a 1771 amino acid ATRY protein that has diverged extensively from ATRX. The conservation and loss of particular motifs identify those required for testicular function (ATRY) and function in other tissues (ATRX)."
AY532685 MEILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE Sarcophilus harrisii
AY532684 ....E................................S....................P. Dasyurus geoffroii
AY532681 ....E................................S....................P. Dasyurus albopunctatus
AY532683 ....E................................S....................P. Dasyurus viverrinus
AY532682 ....E........................P.......SE...................P. Dasyurus spartacus
AY532680 ....E..............R.................SR...................P. Dasyurus maculatus
AY532678 ..V..................................S....................P. Dasycercus cristicauda
AY532669 ..V..................................S....................P. Dasykaluta rosamondae
AY532676 ..V..................S...............S....................P. Myoictis wallacei
AY532675 ..V..................S...............S....................P. Myoictis melas
AY532687 ..V........N.L.......................S....................P. Neophascogale lorentzii
AY532671 ..V..................................S....................P. Parantechinus bilarni
AY532670 ..V.................................TS.........RG.........P. Parantechinus apicalis
AY532686 ..V..................................S........P...........p. Phascolosorex dorsalis
AY532674 ..V.......................................................P. Pseudantechinus ningbing
AY532672 ..V..................................S....................P. Pseudantechinus woolleyae
AY532673 ..V........N..R......................S...................SP. Pseudantechinus roryi
454 read MEILQKYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAMLQAVSEDP Sarcophilus harrisii
EF028739 ............................V.TEEDLAAKLNAMLQA.............P. Antechinus minimus
AY243439 ....E..............R........V.TEEDLAAKLNAMLQA.............P. Dasyurus maculatus
EF028750 ....K................KT.....I.TEEDLAAKLNAILQA.............P. Myrmecobius fasciatus
EF028737 ..V.........................V.TEEDLAAKINAMLQA.............P. Antechinus flavipes
EF028748 ..V.........................V.TEEDLAAKLNA.LQA.............P. Sminthopsis crassicaudata
AY243438 ..V.........................V.TEEDLAAKLNA.LQA.............P. Planigale sp.
EF028749 ..V.........................V.TEEDLAAKLNA.LQA.............P. Planigale ingrami
AY532679 ..V.........................V.TEEDLAAKLNAMLQA............... Dasyurus hallucatus
AF025382 ..V.........................V.TEEDLAAKLNAMLQA.............P. Phascogale tapoatafa
EF028741 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus godmani
AY532666 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus swainsonii
EF028736 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus stuartii
EF028742 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus agilis
EF028738 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus bellus
EF028740 ..V.........................V.TEEDLAAKLNAMLQA.............P. Antechinus leo
EF028747 ..V.........................V.TEEDLAAKLNAMLQA.............P. Phascogale calura
EF028744 ..V.........................V.TEEDLAAKLNAMLQA.............P. Murexchinus melanurus
EF028743 ..V.........................V.TEEDLAAKLNAMLQA.............P. Micromurexia habbema
EU086688 ..V.........................V.TEEDLAAKLNAMLQA.............P. Pseudantechinus macdonnellensis
EU086689 ..V.........................V.TEEDLAAKLNAMLQA.............P. Pseudantechinus roryi
EU086686 ..V.........................V.TEEDLAAKLNAMLQA............SP. Pseudantechinus macdonnellensis
EU086687 ..V.........................V.TEEDLAAKLNAMLQA..........G..P. Pseudantechinus mimulus
AY532667 ..V.........................V.TEEDLAAKLNAMLQA.............P. Murexia longicaudata
EF028746 ..V.........................V.TEEDLAAKLNAMLQA.............P. Paramurexia rothschildi
AY532677 ..V.........................V.TEEDLAAKLNAMLQA.............P. Dasyuroides byrnei
EF028745 ..V..........I..............V.TEEDLAAKLNAMLQA.............P. Phascomurexia naso
Macropus eugenii assembly
sacHar MEILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE
ME+LQ YYTLVDRVPALLHHLTAIDYSS L + ++ VSEDPRLLVRVLR E
macEug MEVLQNYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAGLQAVSEDPRLLVRVLRPE
Monodelphis domestica assembly TSSLVLDLQHSSGGEISG
sacHar MEILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE
ME+LQ YYTLVDRVPALLHHLTAIDYSS L + ++ VSEDPRLLVRVLR E
monDom MEVLQNYYTLVDRVPALLHHLTAIDYSSVLTEEDLAAKLNAGLQAVSEDPRLLVRVLRPE
Ornithorhynchus anatinus assembly
sacHar EILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE
++L+ YY LVDRVPALL HL A+D SS L + SR SEDPRLLVR L E
ornAna DLLRDYYALVDRVPALLRHLAALDLSSVLSEEDLTSRLNAGLQAASEDPRLLVRRLEPE
Equus caballus assembly
sacHar EILQKYYTLVDRVPALLHHLTAIDYSSSLVLDLQHSRGGEVSGTVSEDPRLLVRVLRSE
E LQ YYTLVDRVPALLHHL ++D+SS + D ++ VSEDPRLLV V+RS+
equCab EALQDYYTLVDRVPALLHHLASMDFSSVVSEDDLVAKLNAGLQAVSEDPRLLVWVVRSK
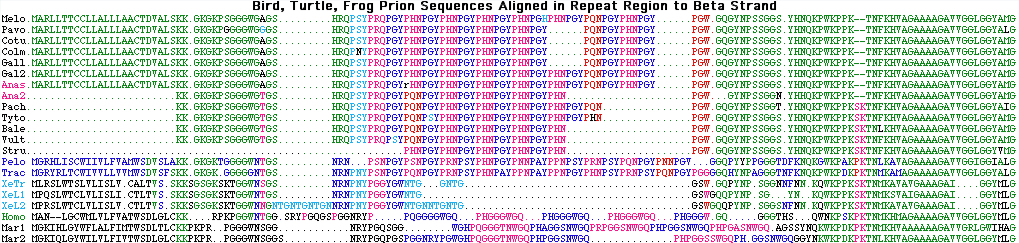
Rod rhodopsin RHO1 (4+ marsupials)
The optimal wavelength for scotopic (dim light) vision of Sarcophilus is easily predictable provided key tuning residues are covered by the assembly. The 97% match to Sminthopsis and agreement at tuning residues suggests this aspect of vision will be nearly identical between the two species.
>RHO1_homSap Homo sapiens (human) 0 MNGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSR 2 1 YIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQ 0 0 FRNCMLTTICCGKNPLGDDEASATVSKTETSQVAPA* 0 >RHO1_monDom Monodelphis domesticus (opossum) Didelphimorphia 0 MNGTEGPNFYVPFSNKTGTVRSPFEEPQYYLADPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAIADLFMVFGGFTMTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIIGVAFTWVMALACAFPPLIGWSR 2 1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSNFGPIFMTIPAFFAKSSSVYNPVIYIMMNKQ 0 0 FRTCMITTLCCGKNPLGDDEASATASKTETSQVAPA* 0 >RHO1_macEug Macropus eugenii (wallaby) Diprotodontia frag, traces not yet consulted 0 MNGTEGPNFYVPFSNKTGVVRSPFEEPQYYLAEPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLADADLFMDFGGFT 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACSTPPLLGWSR 2 1 0 0 ESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTLPAFFAKTSAVYNPVIYIMMNKQ 0 0 FRNCMITTLCCGKNPLGDDEASATTSKTETSQVAPA* 0 >RHO1_smiCra Sminthopsis crassicaudata (fat-tailed dunnart) Dasyuromorphia 0 MNGTEGPNFYVPYSNKSGVVRSPYEEPQYYLAEPWMFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAVADLFMVICGFTTTLVTSLNGYFVFGTTGCLVEGFFATTG 1 2 GEVALWALVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACSVPPIFGWSR 2 1 YIPEGMQCSCGIDYYTLNPEFNNESFVIYMFVVHFIIPLTVIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSDFGPIFMTLPAFFAKSSSIYNPVIYIMMNKQ 0 0 FRNCMITTLCCGKNPLGDDEASTTASKTETSQVAPA* 0 >RHO1_sacHar 9 Sarcophilus harrisii (tasmanian_devil) 97% identity Sminthopsis crassicaudata 0 MNGTEGPNFYVPHSNKTGVVRSPYEEPQYYLAEPWMFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAVADLFMVICGFTTTLVTSLNGYFVFGTTGCQIEGFFATTG 1 2 GEVALWALVVLAIERYIVVCKPMSNFRFGENHAIMGVVFTWIMALACSVPPLFGWSR 2 1 YIPEGMQCSCGIDYYTLNPEFNNESFVIYMFVVHFTIPLTVIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSDFGPIFMTLPAFFAKSSSIYNPVIYIMMNKQ 0 0 FRTCMITTLCCGKNPLGDDEASATVSKTETSQVAPA* 0 >RHO1_calPhi Caluromys philander (woolly opossum) Didelphimorphia abstract:14659889 0 MNGTEGPNFYVPFSNKTGVVRSPFEEPQYYLAEPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAIADLFMVFGGFTTTLYTSLHGYFVFGPTGCDLEGFFATLG 1 2 GEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSR 2 1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMVVIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSNFGPILMTLPAFFAKTSAVYNPVIYIMLNKQ 0 0 FRTCMLTTLCCGKIPLGDDEASATASKTETSQVAPA* >RHO1_ornAna Ornithorhynchus anatinus (platypus) 0 MNGTEGQDFYIPMSNKTGVVRSPFEYPQYYLAEPWQYSVLAAYMFMLIMLGFPINFLTLYVTIQHKKLRTPLNYILLNLAFANHFMVLGGFTTTLYTSLHGYFVFGPTGCNIEGFFATLG 1 2 GEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACALPPLVGWSR 2 1 YIPEGMQCSCGIDYYTLRPEVNNESFVIYMFVVHFTIPMTIIFFCYGRLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTVPAFFAKSSAIYNPVIYIMMNKQ 0 0 FRNCMLTTICCGKNPLGDDEASATASKTEQSSVSTSQVSPA* 0
Cone rhodopsin SWS2 (9+ marsupials)
Cone rhodopsin RHO2 has been lost in all mammals and no debris from this gene is expected in Sacrophilus). The short wavelength cone opsin SWS2, while still present in platypus, has also been lost in all theran opsin too long ago to leave detectable remnants in syntenic position. Cone opsin SWS1 has this turned around, being present in theran mammals but only as debris in platypus. A nearly full length gene, most simiilar to Sminthopsis, can be recovered from Sarcophilus read coverage.
>SWS1_homSap Homo sapiens (human) Gt -FAM137A -CALU -NAG6 -FLNC 1385866 NP_990769 cone short 0 MRKMSEEEFYLFKNISSVGPWDGPQYHIAPVWAFYLQAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVA 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQ 0 0 FQACIMKMVCGKAMTDESDTCSSQKTEVSTVSSTQVGPN* 0 >SWS1_monDom Monodelphis domesticus (opossum) Didelphimorphia 0 MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHFQTVFMGFVFCAGTPLNAVVLVATLRYKKLRQPLNYILVNVSLCGFIFCIFAVFTVFISSSQGYFIFGRHVCAMEAFLGSVA 1 2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWVIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIMPLFLICFSYSQLLRALRA 0 0 VAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNQNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FHACIMEMVCRKPMTDDSDVSSSQKTEVSAVSSSQVGPT* 0 >SWS1_thyEle Thylamys elegans (fat-tailed opossum) Didelphimorphia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHLQTVFMGFVFC AGTPLNAVVLVATLRYKKLRQPLNYILVNVSFSGFIFCIFAVFTVFISSSQGYFIFGH HVCAMEAFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWVIGI GVSIPPFFGWSRFIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLFLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNRNH GLDLRLVTIPAFFSKSACVYNPIIYCFMNKQFHACIMEMVCRKPMTDDSDVSSSQKTE VSAVSSSQVGPS >SWS1_didAur Didelphis aurita (big-eared opossum) Didelphimorphia MSGDEEFYLFKNISSVGPWDGPQHHIAPAWAFHFQTVFMGFVFC AGTPLNAVVLVATLRYKKLRQPLNYILVNVSLSGFIFCIFAVFTVFISSSRGYFVFGR HVCAMEAFLGSVAGLVMGWSLAFLAFERFVVICKPFGNFRFNAKHAMMVVLATWVIGI GVSIPPFFGWSRFIPEGLQCSCGPDWYTVGTKYRSEYYAWFLFLSCFIGPLFLICFSY AQLLGALRAVAAQQQESTTTQKAEREVSRMVVMMVGSFCLCYVPYAALGMYMINNRNH GLDLRLVTIPAFFSKSACVYNPIIYCFMNKQFHACIMEMVCRKPMADDSDITSSQKTE VSTVSSSQVGPS >SWS1_macEug Macropus eugenii (wallaby) Diprotodontia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHCQTVFMGFVFFAGTPLNAVVLIATFRYKKLRQPLNYILVNISLAGFIYCIFSVFTVFISSSQGYFIFGR HVCAMEGFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFNSKHSMMVVLATWVIGIGVSIPPFFGWSRYIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFILCFIMPLSLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNRNHGIDLRLVTIPAFFSKSSCVYNPIIYCFMNKQFHACIMEMVCRKPMTDDSEASSSQKTEVSTVSSSQVGPS* >SWS1_setBra Setonix brachyurus (quokka) Diprotodontia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHCQTVFMGFVFF AGTPLNAVVLIATFRYKKLRQPLNYILVNISLAGFIYCIISVFTVFISSSQGYFIFGR HVCAMEGFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFNSKHSMMVVLATWVIGI GVSIPPFFGWSRYIPEGLQCSCGPDWYTVGTKYHSEYYTWFLFILCFIMPLSLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNRNH GIDLRLVTIPAFFSKSACVYNPIIYCFMNKQFHACIMEMVCRKPMTDDSEASSSQKTE VSTVSSSQVGPS >SWS1_tarRos Tarsipes rostratus (honey possum) Diprotodontia MSGDEEFYLFKDISSVGPWDGPQYHIAPAWAFHFQTTFMGFVFF AGTPLNAVVLIATLRYKKLRQPLNYILVNISLAGFIFCVISVFTVFISSSQGYFIFGR HVCAMEAFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFSSKHAMMVVLATWVIGI GVSIPPFFGWSRYIPEGLQCSCGPDWYTVGTKYHSEYYTGFLFIFCFIVPLSLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNH GLDLRLVTIPAFFSKSACVYNPIVYWFMNKQFHACIMEMVCRKPMTDDSEISSSQKTE VSTVSSSQVGPS >SWS1_cerCon Cercartetus concinnus (pygmy possum) Diprotodontia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHFQTAFMGFVFF VGTPLNAVVLVATLCYKKLRQPLNYILVNVSLAGFIFCIISVFTVFISSSQGYFIFGR HVCAMEAFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFSSKHAMMVVLATWVIGI GVSIPPFFGWSRYIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLICFSY SQLLGALRAVAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNH GLDLRLVTIPACFSK >SWS1_smiCra Sminthopsis crassicaudata (dunnart) Dasyuromorphia 0 MSGDEEFYLFKNISLVGPWDGPQYHLAPAWAFHFQTAFMGFVFFAGTSLNGVVLIATLRYKKLRQPLNYILVNISLAGFIFCVFSVFTVFVSSSQGYFVFGRHVCAMEGFLGSVA 1 2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWIIGIGVSIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLICFSYSQLLGALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAAMAMYMVNNRNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FHACIMEMICKKPMTDDSETTSSQKTEVSTVSSSQVGPS* 0 >SWS1_sacHar Sarcophilus harrisii (tasmanian_devil) part of last exon missing 96% identity Sminthopsis crassicaudata 0 MSGDEEFYLFKNISPVGPWDGPQYHIAPAWAFHLQTAFMGFVFFAGTPLNGVVLIATLRYKKLRQPLNYILVNISLAGFIFCVFSVFTVFVSSSQGYFVFGRHVCAMEGFLGSVA 1 2 SGLVTGWSLAFLAFERFIVICKPFGNFRFHSKHATMVVLATWVIGIGVSIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLICFSYSQLLGALRAVS 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNQ 0 0 KPMTDDSETTSSQKTEVSTVSSSQVGPS* 0 >SWS1_isoObe Isoodon obesulus (bandicoot) Peramelemorphia MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHCQTVFMGFVFF AGTPLNAVVLIATLRYKKLRQPLNYILVNISLAGFIFCIFSVFTVFISSSQGYFIFGR HVCAMEAFLGSVAGLVTGWSLAFLAFERFIVICKPFGNFRFHSKHAMMVVLATWVIGI GVSIPPFFGWSRFIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIIPLSLICFSY SQLLRALRTVAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNH GLDLRLVTIPAFFSKSACVYNPIIYCFMNKQFHACIMEMICRKPMTDDSETSSSQKTE VSTVSSSQVSPS >SWS1_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel x x x x 348 aa 000 nm no_ref genome cone short1 violet 0 MSSDDDFYLFTNGSVPGPWDGPQYHIAPPWAFYLQTAFMGIVFAVGTPLNAVVLWVTVRYKRLRQPLNYILVNISASGFVSCVLSVFVVFVASARGYFVFGKRVCELEAFVGTHG 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSRHALLVVVATWLIGVGVGLPPFFGWSR 2 1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FRACIMETVCGKPLTDDSDASTSAQRTEVSSVSSSQVGPT* 0
Cone rhodopsin LWS (9+ marsupials)
This basal long wavelength imaging opsin is available from 97 vertebrates and has already been analyzed for phyloSNPs and rare genomic events. The Didelphimorphia experienced a 3-4 residue insert in exon 1 that separates them from all other marsupials. Note this region has quite a complicated indel history. The extra residues have repeat character DVNE DDND suggesting replication slippage. The gene is present and intact in Sarcophilus though two exons are not currently available. LWS in tasmanian devil is identical to the Sminthopsis ortholog.
LWS_loxAfr MAQQWGPHRLTGARLQDASE---DSTQASIFVYTNTNT elephant LWS_echTel MAQRWGAHRLTGGQLQDTYE---GSTRTSIFVYTNSTS tenrec LWS_monDom MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNN Didelphimorphia LWS_didAur MTQAWDPVGFLARRRDENEDDHDDTTRASLFVYTNSNN Didelphimorphia LWS_tarRos MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia LWS_macEug MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia LWS_smiCra MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Dasyuromorphia LWS_sacHar MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Dasyuromorphia LWS_setBra MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Diprotodontia LWS_cerCon MTQAWDPAGFLAWQEDENE----ETTRASLFVYTNSNN Diprotodontia LWS_myrFas MTQAWDPAGFLAWRREENE----ETTRASLFTYTNSNN Dasyuromorphia LWS_isoObe MTQAWDPAGFLAWRRDENE----ETTRASLFVYTNSNN Peramelemorphia LWS_ornAna MTPAWNSGVYAARRRFEDEE---DTTRTSVFVYTNSNN platupus LWS_tacAcu MTQAWDPAGFLAWRRDENEE---TTRASLFVYTNSNNT echidna
>LWS_homSap Homo sapiens (human) 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_monDom Monodelphis domesticus (opossum) Didelphimorphia 4aa insert 0 MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYVQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYSFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSKTEGSSVSSVAPA* 0 >LWS_didAur Didelphis aurita (big-eared opossum) Didelphimorphia 4aa insert 0 MTQAWDPVGFLARRRDENEDDHDDTTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDLGVQSYMIVLMATCCIFPLSIILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA* 0 >LWS_tarRos Tarsipes rostratus (honey possum) Diprotodontia ENED insert 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAIWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGIQSYMIVLMSTCCILPLSIILLCYVQVWRAIRA 2 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_macEug Macropus eugenii (wallaby) Diprotodontia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGVQSYMIVLMSTCCILPLSVIFLCYIQVWLAIRS 2 0 VAKQQKESESTQKAEKEVSRMVVVMILAFCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_setBra Setonix brachyurus (quokka) Diprotodonti 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETMIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGVQSYMIVLMSTCCILPLSVILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVPA* 0 >LWS_cerCon Cercartetus concinnus (pygmy possum) Diprotodontia 0 MTQAWDPAGFLAWQEDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAIWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGNSDPGIQSYMIVLMSTCCILPLSIILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTFFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* >LWS_smiCra Sminthopsis crassicaudata (dunnart) Dasyuromorphia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIILCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVMVMILAFCFCWGPYALFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_sacHar half of exon 2, all of exon 4 missing frag 100% identical to Sminthopsis 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 2 FKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIILCYIQVWLAIRA 0 0 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_myrFas Myrmecobius fasciatus (numbat) Dasyuromorphia 0 MTQAWDPAGFLAWRREENEETTRASLFTYTNSNNTK 1 2 GPFEGPNYHIAPRWVYNLTSFWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSVILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_isoObe Isoodon obesulus (bandicoot) Peramelemorphia 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSFWMFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMTTCCILPLSIILLCYVQVWLAIRA 0 0 VAKQQKDSESTQKAEKEVSRMVVVMIRAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSGTSRTEVSSVSSAPA* 0 >LWS_ornAna Ornithorhynchus anatinus (platypus) 0 MTPAWNSGVYAARRRFEDEEDTTRTSVFVYTNSNNTR 1 2 DPFEGPNYHIAPRWAYNVTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA* 0 >LWS_tacAcu Tachyglossus aculeatus (echidna) 0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVFNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAVWTSPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYIQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA* 0
Encephalopsin (2+ marsupials)
Pinopsin, parapinopsin, parietopsinand VA opsin all terminate in sauropods and are missing in all mammals. Encephalopsin has a very peculiar history of gene loss in tetrapods, requiring some seven independent and asynchronous events including platypus. While this limits the phylogenetic utility of any gene loss within marsupials, the status of the gene within Sarcophilus is still informative. A full length gene can be recovered with 94% identity to opossum, strongly indicating that encephalopsin is fully functional within Sarcophilus.
>ENCEPH_homSap Homo sapiens (human) OPN3 0 MYSGNRSGGHGYWDGGGAAGAEGPAPAGTLSPAPLFSPGTYERLALLLGSIGLLGVGNNLLVLVLYYKFQRLRTPTHLLLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSGSLF 1 2 GIVSIATLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPLGVIAHCYGHILYSIRM 0 0 LRCVEDLQTIQVIKILKYEKKLAKMCFLMIFTFLVCWMPYIVICFLVVNGHGHLVTPTISIVSYLFAKSNTVYNPVIYVFMIRK 0 0 FRRSLLQLLCLRLLRCQRPAKDLPAAGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDKTNGSKVDVIQVRPL* 0 >ENCEPH_monDom Monodelphis domestica (opossum) 0 MYSDNSSDDGGGGYWGSGRAGGASGTGVTGEPGPEGSPRQAPLFSPGTYELLALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLFLVNISFNDLLVSLFGVTFTFVSCLRSGWVWDSVGCAWDGFSNTLF 1 2 GIVSIMTLTVLAYERYNRIVHAKVINFSWAWRAITYIWLYSLVWTGAPLLGWNRYTLEIHGLGCSVDWKSKDPNDSSFVIFLFFGCLMLPVGVMAYCYGHILYAIRM 0 LRCVEELQTIQVIKILRYEKKVAKMCFLMIAIFLFCWMPYAVICLLVANGYGSLVTPTVAIIASLFAKSSTAYNPIIYIFMSRK 0 0 FRRCLLQLLCFRLLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDETQMIDENDKNSGTKVNVIQVRPL* 0 >ENCEPH_sacHar Sarcophilus harrisii (tasmanian_devil) 94% identity monDom 0 MYSGNSSDDAGGGYWGSGGTGGAGGTGVAGEPAPEGSPRPAPLFSPGTYELLALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLFLVNISFSDLLVSLFGVTFTFVSCLRSGWVWDSVGCAWDGFSNTLF 1 2 GIVSIMTLTVLAYERYNRIVHAKVINFSWAWRAITYIWLYSLIWTGAPLLGWNRYTLEIHGLGCSVDWKSKDPNDSSFVLFLFLGCLVLPVGVMAYCYGHILYAIRM 0 0 FRCVEELQTLQVIKILRYEKKVAKMCFLMIATFLFCWMPHAVICFLVANGYGSLVTPTVAIIPSLFAKSSTAYNPIIYIFMSRK 0 0 FRRCLLQLLcFRQLKFQQPKKDRAIIRTEKQIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDETQMIDDNDKNSETKVNVIQVRPL* 0 >ENCEPH_macEug Macropus eugenii frag 0 GALGCREPGQREPSSSAPFSPGTYELLALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLLLVNISFSDLLVSLFGVTFTFVSCLRSGWVWHTVGCAWDGFSNSLF 1 2 GIVSIMTLTVLAYERYHRIVHAKVINFSWTWRAITYIWLYSLVWTGAPLLGWNRYTLEIHGLGCSVDWKSKDPNDSSFVLFLFLGCLVLPVGVMAYCYGHILYAIRM 0 0 0 0 FRRCLLQLLCFRQLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDETQMIDDNDKNNGTKVNVIQVRPL* 0 >ENCEPH_ornAna Ornithorhynchus anatinus pseudo 0 MVPWNGS-GRHLGAVR---GPE--SLPATPGAARPSRPGAGDGRL--LGLF-P-GVGGNLLVLLL--ALPGPPTTTDLYLASVAVSDLL--LL---LPFVYRLWRSRPWVFVCRLLGE-GGSLA 1 2 GIVSLISLAVLSYERYTLTLHPKQSNYQKAVLAVGASWIYSLIWTIPPLLGWSSYGTEGAGTSCSVHWSSKSVC-SYIVCLFI--CLVIPVLVMIYCYGRLLYAVKQ 0 0 LHCVKELQNIQVIGSLRYER*VTEMYFFTIAQFLVCQSPSALVSYPAAH-----VSPVVAKISPVFANSSFVYNPVISIFVRRK 0 0 KASR*KVNVIQVQPPS* 0 >ENCEPH_galGal Gallus gallus (chicken) 71%=homSap encephalopsin OPN3 full 0 MHSGNGTGATSRPQLAAAGHEVPGERPLFSAGTYELLALLIATIGTLGVCNNLLVLVLYYKFKRLRTPTNLFLVNISLSDLLVSVCGVSLTFMSCLRSRWVWDAAGCVWDGFSNSLF 1 2 GIVSIMTLTVLAYERYIRVVHAKVIDFSWSWRAITYIWLYSLAWTGAPLLGWNRYTLEIHGLGCSMDWKSKDPNDTSFVLLFFLGCLVAPVVIMAYCYGHILYAVRM 0 0 LRCVEDFQTSQVIKLLKYEKKVAKMCFLMISTFLICWMPYAVVSLLVTYGYSNLVTPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0 0 FRQCLLQLLCFRLMRFQRIMKEPSGAGNVKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIIASDDTQQIDDNSKHNGTKVNVIQVKPL* 0
TMT opsin (2+ marsupials)
TMT is an ancient locus that is present in monotremes and marsupials but lost in all placentals.
>TMT_monDom Monodelphis domestica shortened final exon 0 MSNNLTTNLSLEALLSASEDKQRNGLSRTGHTIVAVFLGIILIFGSISNFIVLVLFCKFKVLRNPVNMLLLNISISDMLVCLSGTTLSFASSIQGRWIGGKHGCRWYGFANSCF 1 2 GIVSLISLAILSYERYRTLTLCPGQGADYQKALLAVAGSWLYSLVWTVPPLIGWSSYGTEGAGTSCSVHWTSKSVESVSYIMCLFIFCLVIPILVMVYFYGRLLYAVKQ 0 0 VGKIRKTAARKREYHVLFMVVTAVICYLICWVPYGMIALLATFGPPGVVSPVANVVPSILAKSSTVCNPIIYVLMNKQ 0 0 FYKCFLILFHCQPAQSGPDVSLCPSNVTVIQLGQRKNKDAPGSI* >TMT_macEug Macropus eugenii frag 0 MSINLTANLSFGTLLPDSEEKQRSGLSRTGHTVTAVFLGLILILGVINNFIVLVLFCKFKVLRNPVNMLLLNISISDMLVCLTGTTLSFASSIRGRWIAGYHGCRWYGFANSCF 1 2 GIVSLISLAVLSYERYRTLTLCPRQGTDYHKALLAVAGSWLYSLIWTVPPLIGWSSYGTEGAGTSCSVHWTSKSVESVSYIMCLFIFCLVIPILFMVYFYGRLLYTVKQ 0 0 VGKIRKSAARKREYHVLFMVVTAVICYLICWVPYGMIALLATFGPPGVVSPVANVVPSILAKSSTVCNPIIYILMNKQ 0 0 FYKCFLILFHCQPASSASDASLCPSKMTVIQLGQRKDKEVPCAIQDLPEVSKKQLCLLSPESNVAPSSGHPQEKMEEKPLSE* 0 >TMT_sacHar FP5MBH101BETOZ needed to finish 0 MSINLTTNLSFGPLLIDSEEKPRSGLSRTGHTVVAVFLGIILILGFINNFIVLILFCKFKVLRNPVNMLLLNISISDMLVCLSGTTLSFASSIRGRWIGGYHGCRWYGFANSCF 1 2 GIVSLISLAILSYERYRTLTLCPRRGADYQKALLAVAGSWLYSLIWTVPPLIGWSSYGTEGAGTSCSVHWTSKSVQSVSYIMCLFIFCLVIPILIMIYFYGRLLYTVKQ 0 0 VGKIRKTAARKREYHVLFMVVTAVICYLICWVPYGLIALVATFGPPGVVSPVANIVPSILAKSSTVCNPIIYILMNKQ 0 0 FYKCFLILFHCQPASSAPDASLCPSKVTVIQLGQR * 0 >TMT_ornAna Ornithorhynchus anatinus frag 0 GLSRTGHTMVAVFLGIILVFGFMNNLIVLILFCKFKALRNPVNMIMLNISASDMLVCVSGTTLSFASNISGRWIGGDPGCRWYGFVNSCL 1 2 GIVSLISLAVLSYERYRTLTLHPKQSTDYQKAVLAVGASWIYSLIWTIPPLLGWSSYGTEGAGTSCSVHWSSKSPVSVSYIVCLFIFCLVIPVLVMIYCYGRLLYAVKQ 0 0 IGKARKTAARKREYHVLFMVITTVICYLVCWMPYGVTALLATFGQPGTVSPEASVIPSILAKSSTVCNPIIYILMNKQ 0 0 FYKCFLILFHCQPPRAADAPSTYPSQVMVIQLNQRRSRETAGAPQVLLEMKHQTLHLLGPQLHETPSWERSTPVHPE* 0 >TMT_galGal Gallus gallus 0 MNHTWTYNLSFGAPTDPVEPRAGLSRNGHTVVAVFLGFILFFGFLNNLIVLILFCKFKTLRNPVNMLLLNIhttp://genomewiki.ucsc.edu/index.php?title=Opsin_evolution:_Encephalopsin_gene_loss&action=editSISDMLVCISGTTLSFASNIHGKWIGGEHGCRWYGFVNSCF 1 2 GIVSLISLAVLSYERYSTLTLCNKRSDDYRKALLAVGGSWVYSLLWTVPPLLGWSSYGIEGAGTSCSVRWSSETAESTSYIICLFIFCLVIPVMVMMYCYGRLLYAVKQ 0 0 VGKIHKNTARKREYHVLFMVITTVICYLVCWIPYGVIALLATFGKPGVVTPVASIIPSILAKSSTVCNPIIYILMNKQ 0 0 FYKCFRQLFHCQPPSSTDGEPTCHSKVTVIQLNQKTDGGKLCNNKPRPETDNKVTSLLHPEPGLEPAAKTVPPM* 0 >TMT_taeGut Taeniopygia guttata 0 MNHTWMYNLSFGAPAHPVEPRAGLSRSGHTVVAVFLGLILFFGFLNNLIVLILFCKFKTLRNPVNMLLLNISVSDMLVCISGTTLSFASNIRGKWIGGDHACRWYGFVNSCF 1 2 GVVSLISLAVLSYERYNTLTLCHKRSDDFRKALLAVAGSWIYSLVWTVPPLLGWSSYGVEGAGTSCSVRWSSESAESTSYIICLFVFCLVVPVMVMMYCYGRLLYAVKQ 0 0 VGKIHKNAARKREYHVLFMVIPTVICYLVCWIPYGVIALLATFGKPGAVTPITSIIPSILAKSSTVCNPIIYILMNKQ 0 0 FYKCFRQLFHCQPPSSTDGEPTCHSKVTVIQLDQRADGGNMCNNEPHPETDSKMTSLLCPETTSKATPPTS* 0 >TMT_anoCar full +TMT -ST6GAL2 (overlap) +SLC5A7 0 MSELSSNLTFNMSTSIEEPGSGLSRMGHNIVAVFLGLILVFGFLNNLVVLILFCKFKTLRNPVNMLLLNISASDMLVCISGTTLSFVSNIYGRWIGGEHGCRWYGFVNSCF 1 2 GIVSLISLAILSYERYSTLTQTNKRGSDYQKALLGVGGSWLYSLIWTVPPLIGWSSYGLEGAGTSCSVRWTSETLESVTYIICLFIFCLAIPVLVMIYCYARLFYAVKQ 0 0 VGKLRKTSARKREFHVLFMIITTIICYLICWMPYGVIALLATFGRPGLVSPVASVIPSILAKSSTVFNPIIYILMNKQ 0 0 FYKCFLMLLHCQPSSVADGETICQSKVMAIHQNQKAQGGVILKSQVVPQMDEKAICLLSPESSLDPVLESTPQLSKENSFL* 0 >TMT_xenTro full -UXS1 +TMT -ST6GAL2 (overlap) +SLC5A7 0 MSTIKNWTTNISVENSMSYIENDLSLPTEAVLSRTGHTVVAIFLGFILIFGFLNNFVVLILFCKFKTLRTPVNMMLLNISASDMLVCVSGTTLSFTSSIKGKWIGGEYGCQWYGFVNSCF 1 2 GIVSLISLAILSYERYSTLTLYNKGGPNFKKALLAVASSWLYSLVWTVPPLLGWSSYGREGAGTSCSVRWTSESVESVSYIICLFIFCLALPVFVMLYCYGRLLYAVKQ 0 0 VGKIRKIAARKREYHVLFMVITTVICYLLCWLPYGVVALLATFGRPGVISPVASVVPSILAKSSTVFNPIIYILMNKQ 0 0 FYKCFLILFHCHPTSSADGKSICQSNYTVIQLNQKLNNIVAIPGQTQIPESVDKMPCIHRQNNESPSDQMPQSTTEHLISGT* 0
RGR opsin (0 marsupials)
This gene has apparently been lost specifically in the marsupial clade, though support for that is only provided by the Monodelphis and Macropu genome projects. It would be of considerable interest to find the gene or a fragment thereof in syntenic position in Sarcophilus.
>RGR1_homSap Homo sapiens (human) +PCDH21 -LRIT1 -GRID1 -WAPAL NM_001012720 retinal epithelium Mueller 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSQLAWNSAVSLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKREKDRTK* 0 >RGR_dasNov Dasypus novemcinctus (armadillo) 0 MAGSGVLPPGFGELEVLAVGTVLLVE 1 2 ALSGLVLNGLAIISFCKTPELRSPSRLLVLSLALADSGVSLNALVAATSSLLR 2 1 RWPYGSGGCQAHGFQGFVTALASISSSAAIAWERCHRHCI 1 2 GRRLAWSTAGCLVLCLWMAAAFWAALPLLGWGLYDYEPLGTCCTLDYSRGDR 2 1 NFISFLVTLALFNFFLPLLIMLTSYRLMAQKLKRSGHVQ 0 0 VSTALPGRLLLLGWGPYALLYLYAAVADATSLSPRLQM 0 0 VPALIAKTMPTVNALYYALGRESVHRNA* 0 >RGR_loxAfr Loxodonta africana (elephant) 0 MAEPGHLPAGFQELEVLTVGTVLLLE 1 2 ALSGLSLNGLTILSFCKIPELRTPGHLLVLSLALADSGISLNALVAAMSSLRR 2 1 RWPYGSDGCQAHGFQGFVTALASICSCAAIAWERYHHYCT 1 2 RSRLAWSSASALVLFVWLSSAFWAALPLLGWGRYNYEPLGTCCTLDYSRGDR 2 1 NSTSFLLTMAFFNFLLPLFITLTSYRLMEQKLKKKGPLQ 0 0 VNTTLPARTLLLGWGPYALLYLCAAATDMTSISPRLQM 0 0 VPALVAKAVPVINACHYALGSEVVRGGIWQYLSRQRGESPLRARDRTH* 0 >RGR1_ornAna Ornithorhynchus anatinus (platypus) missing exon 1 DRY motif, afros ERY, other placentals GRY 0 1 2 ALLGLCLNGLTIASFRKIKELRTPSNLLVVSLALADSGICLNALMAALSSFLR 2 1 HWPYGAEGCRLHGFQGFATALASISLSAAIGWDRYLRHCS 1 2 RSKPQWGTAVSTVLFAWGFSAFWSMMPILGWGQYDYEPLRTCCTLDYSKGDR 2 1 NFTTYLFAVAFFNFVIPLFIMLTSYQSIEQRFKKSGLFK 0 0 LNTRLPTRTLLFCWGPYALLCFYATVENVTFISPKLRM 0 0 IPALIAKTVPVIDAFTYALRNEDYRGGIWQFLTGQKIERVEVENKIK* 0 >RGR1_galGal Gallus gallus (chicken) +PCDH21 -LRIT1 +CHAT -PARG 14985289 NM_001031216 0 MVTSHPLPEGFTEIEVFAIGTALLVE 1 2 ALLGFCLNGLTIISFRKIKELRTPSNLLVLSIALADCGICINAFIAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFLTALASISSSAAVAWDRYHHYCT 1 2 RSKLQWSTAISMMVFAWLFAAFWATMPLLGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYITFLFALSIFNFMIPGFIMMTAYQSIHQKFKKSGHYK 0 0 FNTGLPLKTLVICWGPYCLLSFYAAIENVMFISPKYRM 0 0 IPAIIAKTVPTVDSFVYALGNENYRGGIWQFLTGQKIEKAEVDSKTK* 0 >RGR1_xenTro Xenopus tropicalis (frog) ?? 0.2.1.2.1.0.0 indel +PCDH21 -LRIT1 +CHAT -PARG 296 BC135113 0 MVTSYPLPEGFTETEVFAIGTTLLVE 0 0 ALLGLLLNGLTLLSFYKIRELRTPSNLFIISLAVADTGLCLNAFVAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1 2 RSKLHWSTAVSVVFFIWGFSAFWSAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYISYLFTMAFFEFLVPLFILMTAYQSIYQKMKKSGQIR 0 0 FNTSMPVKSLVFCWGPYCLLCFYAVIQDATILSPKLRM 0 0 IPALLAKTSPAVNAYVYGLGNENYRGGIWQYLTGQKLEKAETDNKTK* 0
Peropsin (2+ marsupials)
Sarophillus can be expected to have this gene. Further, the protein sequence should substantiate the 4 previously defined phyloSNPs characteristic of the marsupial/placental transition.
>PER_homSap Homo sapiens (human) 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDV 1 2 GRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 >PER_loxAfr Loxodonta africana (elephant) 0 MLRNSLDNSSDSKNEDASVFSQTEHNIVATYLIMA 1 2 GMISILSNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLHGRWKFGYTGCQ 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTICHPHI 1 2 GRRMTSNTYVSMILGAWINGLLWALLPITGWASYAPDPTGATCTINWRKNDA 2 1 SFVSYTMTVIVINFVVPLAVMFYCYYHVTRSIKRHTASNCAEYLNRDWSDQLDVTK 0 0 MSVIMILMFLVAWSPYSIVCLWASFGDSKKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMFAMFKCQTHQAEPVTCILPMNVSQNPLAAGRI* 0 >PER_monDom Monodelphis domestica (opossum) 0 MFKNNSVKTLAPEKEGPSVFSPIEHKIVAAYLITA 1 2 GVISIVSNVIVLGIFVKYKALRTATNTIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYDGCQ 0 0 IYAGLNIFFGMASIGLLTAVAIDRYLTICQPDL 1 2 GGRMTSYNYTLMILTAWVNGFFWALMPIVGWAGYAPDPTGATCTINWRKNDV 2 1 SFVSYTMTVITINFAMPLGVMFYCYYNVSQKMKQYSPSNCPDHINRDWSNQVAVTK 0 0 MSVVMILMFLLAWSPYSIVCLWASFGDPKEIPPAMAIVAPLFAKSSTFYNPCIYVAANKK 2 1 FRRAISAMIRCQTHQSMPISNALPMN* 0 >PER_macEug Macropus eugenii (wallaby) 0 MFQNDSLEPEKESYSVFSPTEHNIVAAYLITA 1 2 GVISIPSNIIVLGIFVKYKELRTATNTIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTAVAIDRYLTICQPDL 1 2 2 1 SFVSYTMTVIAINFVMPLVVMFYCYYNVSLKMKQYTRSSCPEHINRDWSNQVDVTK 0 0 MSVIMILMFLLAWSPYSVVCLWASFGDPKEIPPAMAIIAPLFAKSSTFYNPCIYVAANKK 2 1 FRRAISAMMRCETHQSMPVSNALPLNLT* 0 >PER_sarHar Sarcophilus harrisii 5.5 of 7 exons 0 MFKNDSFRSLEPEKEGHSVFSPAEHNIVAAYLITA 1 2 SILSNVIVLGIFVKFKELRTATNAIIINLA 0 0 1 2 GRRMTSFNYTIMILTAWVNGFFWALMPIVGWASYAPDPTGA 2 1 SFVSYTVTVIAINFVMPLVVMIYCYYNVSQKIKQYTPSNCPEYINRDWSNEVAVTK 0 0 MSVIMILMFLLAWSPYSVVCLWASFGDPKEIPPAMAIIAPLFAKSSTFYNPCIYVAANKK 2 1 FRrAISAMIQCQTHQSMSVSKALPMN* 0 >PER_ornAna Ornithorhynchus anatinus (platypus) 0 MRRNDSANLLESEHHDRSAFSQTDHNIVAAYLITA 1 2 GIMSIVSNVIVLGIFVKFEELRTATNAIIINLAVTDIGVSGIGYPMSAASDLHGSWKFGHAGCQ 0 0 IYAGLNIFFGMSSIGLLTVVAVDRYLTICRPAI 1 2 GRKMTRSNYTAMILAAWMNGFFWASMPLLGWASYASDPTGATCTINWRKNDA 2 1 SFISYTMTVIAVNFAVPLIVMFYCYYNVSKAMRQYPASRVLENLNIDWSEQVDVTK 0 0 MSVVMILMFLMAWSPYSIVCLWSSFGDPKKISPAVAIMAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLSMVQCQTHREITITDVLPMNRSRSPLTL* 0 >PER_galGal Gallus gallus (chicken) 0 MHWNDSANSSESDAEAHSVFTQTEHNIVAAYLITA 1 2 GVISIFSNIVVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYTGCQ 0 0 IYAALNIFFGMASIGLLTVVAVDRYLTICRPDI 1 2 GRRMTTRNYAALILAAWINAVFWASMPTVGWAGYASDPTGATCTANWRKNDV 2 1 SFVSYTMSVIAVNFVVPLTVMFYCYYNVSRTMKQYTSSNCLESINMDWSDQVDVTK 0 0 MSVVMIVMFLVAWSPYSIVCLWSSFGDPKKISPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILAMVRCQTRQEITISNALPMTVSLSALTS* 0 >PER_taeGut Taeniopygia guttata (finch) 0 MHWNDSSNSSESDDEAHSAFTQTEHNIVAAYLITA 1 2 GVISIFSNIVVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYTGCQ 0 0 IYAALNIFFGMASIGLLTVVAVDRYLTICRPDI 1 2 GRRMTTRSYATLILAAWINAVFWSSMPTAGWASYAPDPTGATCTVNWRKNDA 2 1 SFISYTMSVIAVNFVVPLTVMFYCYYNVSRTMKQYASSNCLESINIDWSDQVDVTK 0 0 MSVVMIIMFLVAWSPYSIVCLWSSFGDPKKISPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILAMVRCQTRQEITINNALPMSVSQSALTSQNSSHLPA* 0 >PER_anoCar Anolis carolinensis (lizard) 0 MFLNDSANSSESDDEPHSAFSQAEHNIVAAYLITA 1 2 GVISLLSNIVVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYTGCQ 0 0 IYAALNIFFGMASIGLLTVVAIDRYLTICKPHI 1 2 GSRLTATNYTTLILAAWINALFWASMPVVGWASYAPDPTGATCTVNWRKNDT 2 1 SFVSYTMSVIAVNFVIPLSVMFYCYYNVSKTMKYYMRNSCLENINIDWSDQVDVTK 0 0 MSVVMIIMFLLAWSPYSIVCLWSSFGDPKKISPAMAIVAPLFAKSSTFYNPCIYVIANKR 2 1 FRRAILAMIRCQTRQEITINNVLPMSVSQSTIA* 0 >PER_xenTro Xenopus tropicalis (frog) 0 METLAEVSTLLPAGTGTVNISDASSEVHSVFSQSEHNIVAAYLITA 1 2 GVISILSNIIVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYVGCQ 0 0 IYAGLNIFFGMASIGLLTVVAIDRYLTICRPDI 1 2 GRRISGRHYTAMILAAWINAVFWSVMPVVGWSSYAPDPTGATCTINWRKNDV 2 1 SFVSYTMSVVAVNFVVPLMVMFYCYYNVSRTMKGYGSRSSLGGINADWSDQTDVTK 0 0 MSMVMIVMFLVAWSPYSIVCLWSSFGDPRKIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILSMVQCKSRQEVTLDNHFPMNVSQSTLTT* 0
Neuropsin (2+ marsupials)
Here Sarcophilus can be predicted to contain only NEUR1 because the ancient vertebrate genes NEUR2 and NEUR3 appear to terminate in sauropods and NEUR4 in platypus.
>NEUR1_homSap Homo sapiens (human) OPN5 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2 1 LHTVTTVRKSSAVLEIHEEV* 0 >NEUR1_dasNov 0 MALNHTALPQDDRLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 gILSTFGNGYVLYMSSKRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLRATKKKSLEDFR 2 1 LHTVTTVRESSAVLEVHQEV* 0 >NEUR1_monDom 0 MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIISCFSHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPVQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCQSGGQKAAKKESLRTYR 2 1 LHTVTTVRRSSAVLEIHQEv* 0 >NEUR1_macEug 0 1 2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSy 1 2 GTWLKRHHAYICLVIIWAYATFWATMPLAGLGNYAPEPFGTSCTLDWWLAQASVTGQTFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQSSHVLEMKLTK 0 0 2 1 RHTVSTIRKSSSVSETYQEV* 0 >NEUR1_sarHar Sarcophilus harrisii 4 of 6 exons 0 1 2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIISCFSHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAYICLVIIWAYATFWATMPLAGLGNYAPEPFGTSCTLDWWLAQASVTGQTFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQNSHSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPVQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCQSGGQKAAKKESLRDYR 2 1 * 0 >NEUR1_ornAna 0 MTNYSAPQLGDYLPHYLREGDPFVSKLSWEADLVAGVYLVII 1 2 GVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAYICLAIIWAYASFWATMPLVGLGNYAPEPFGTSCTLDWWLAQASVAGQAFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQNSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPIQFSVVPTLLAKSAAMYNPIIYQVIDCRISCCRLGGPKTGKKESLKNSR 2 1 SHSMSTIRKPSAVSGPHQEV* 0 >NEUR1_galGal 0 MASDCNSSSQEEYLPHYMQQEDPFASKLSREADIIAGFYLTVI 1 2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGPKTLQKKSSLKESR 2 1 MYTISSHRDSAALSGTQLEV* 0 >NEUR2_galGal Gallus gallus GenBank 5'UTR mistranslated as coding -B4GALT6 -NEUR2_galGal -KIAA1012 0 MDPSFANSTFQSKITEAADIVVGTCYMVF 1 2 GICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHR 2 1 WLYGKHICLFYAFCGLFFGICSLSTLTLLSVVCCLKICFPAY 1 2 GNRFRRKHGQILIACAWTYAAIFACSPLAHWGEYGEEPYGTACCIDWQSTNVDVMSMSYTVVLFVLCFILPCGVIVTSYSLILVTVKESRKAVEQHVSGPTRINNVQTITAK 0 0 LSIAVCIGFFAAWSPYAIIAMWAAFGSIDKIPPLAFAIPAVFAKSSTLYNPIIHLLLKPNFRSNIAKDFTVIQQLCVRCCFCVKELQTYRSTFNTGLRTFKGKNESSCNALPIMEG CSYFPSEKGSHTFECFKSYPNCFQERLSTMGCHLQDCESLENDLQVEVTQGSRNSMKVVEQEEKSTELDNLEITLEAVPVSCTFTDL* 0 >NEUR3_galGal Gallus gallus cOpn5L2 mRMA for Opsin 5-like 2 AB368183 chr3 XM_420056 CN231992 testis exon 2^3 rel NEUR1/2 0 MEEQYISKLHPVVDYGAGVFLLII 1 2 AILTILGNSAVLATAVKRSSLLKSPELLTVNLAVADIGMAISMYPLAIASAWNHAWLGGDASCIYYALMGFLFGVCSMMTLCAMAVIRFLVTNSSKSN 1 2 SNKISKNTVHILITFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHSSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNINRIPHAAKLEKKLTL 0 0 MAVLISVGFLSAWTPYAAASFWSIFNSSDSLQPIVTLLPCLFAKSSTAYNPFIYYIFSKTFRHEIKQLQCCWGWRVHFFSADNSAENSVSMMWSGRDNIRLSPTAKVESQGAARH* >NEUR4_ornAna Ornithorhynchus anatinus (platypus) XM_001508128 0 MSLSHSLQVPWRNNLTFLNKEAQVSEQGETIIGIYLLAL 1 2 GWMSWFGNSMVIFILHRQRGILNPTDYLTFNLAVSDASVSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 0 VDGFLTLLFGLASINTLAMISVTRYIKGCHPHR 1 2 GHFINTANISVALILIWVSALFWSAGPVLGWGSYT 1 2 DRMYGTCEIDWAEANFSSICKSYIISIFFCCFFLPVSIMFFSYVSIIKMVKSSHTLAGADDPTDRQRRLDRDVTR 0 0 VSVVICTAFIVAWSPYAVISMWSAFGHSVPNLTSVLASLFAKSASFYNPIIYFGMNSKFRKDILVLLPCAKESKEPVKLKKFKNLRQKQGFTLQKPEKAHVLQVPDSGPMSLINTPPLGNRNSFDLACDNSDFECVRL* 0
Melanopsin (3 marsupials)
Here Sarcophilus can be expected to have the main melanopsin but not the paralog MEL2 which terminates in sauropods
>MEL1_homSap Homo sapiens (human) Gq -GRID1 -WAPAL +LDB3 +BMPR1A 483 aa NM_033282 melanopsin OPN4 0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0 0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2 1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2 1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLTSHTSNLSWISIRRRQESLGSESEV 0 0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGK 0 0 TKGLIPSQDPRM* 0 >MEL1_proCap 0 MNPPWGPRVPSRPAQEPSCMSTPASAGRWDSSQATASSLAELPPSSPT 0 0 EARTQTADWVPFPTVDVPDYAHYTLGTVILLVGLTGVLGNLMVIYIFFR 2 1 SRGLRTPANMFIINLAISDFLMSLTQAPVFFASSLYKRWLFGEA 1 2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRTALVLLGTWLYALAWSLPPFFGW 1 2 SAYVPDGLLTSCSWDYKSFMPSARTYTMLLCCFVFFLPLLVIIYCYVFIFKAIRETGR 2 1 ALQTFGACEGASETPRQWQRLQSEWKMAKIALLAILLYVLSWAPYSTVALVGFAG 2 1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 MAIAQHLPCLGVLLGVSDQHTRPYTSYRSTHHSTLSSQASDISWISGRRRQASLGSESEV 0 0 GWTDTEAAAAWEGAQQVSGRASCSQVLESMEANTPPRPQGWGPETPRK 0 0 VKGLPLLDPRA* 0 >MEL1_smiCra Sminthopsis crassicaudata (dunnart) DQ383281 0 MNPSPMLRHLSCPAQDSNCTKIMASISEWNNTEVDAYHLVDLPPITPT 0 0 AVVLPPYSQKVFPTADVPDYAHYTIGATILVVGFTGVLGNLLVIYTFCR 2 1 SRSLRTPANMFIINLAISDFFMSFTQAPVFFASSLYERWIFGEK 2 1 GCEFYAFCGALFGITSMITLMVIALDRYFVITRPLASIGMISKKKTGLILLGVWLYSLAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYTTFTPSVRAYTILLFCFVFFIPLTVIIYCYIFIFRAIKDTNK 2 1 AVQNIGSSEHTPSLRHFQRMKNEWKMAKIALVVILLFVLSWAPYSTVALVAFAG 2 1 YSHVLTPYMNSVPAIIAKASAIHNPIIYAISHPKYR 2 1 MAIAQNFPCLRAVLGIRHPRTQSFSSYRFTHRSTTASQASDISWQSRGRRQLSLGSESEA 0 0 GWNNIETGLTLRSLEGSCGMDEETMDTRELSASTKAKGQSWETLAKTLEE 0 0 MDDLSLLEAGTLLSSLDLQI* 0 >MEL1_macEug Macropus eugenii frag 0 AVVLPPHSRNIFPTADVPDHAHYTVGAIILVVGFTGVLGNLLVIYTFCR 2 1 SRSLRTPANMFIINLAISDFFMSFTQAPVFFANSLYKRWIFGEK 2 2 GCEFYAFCGALFGITSMITLMVIALDRYFVITRPLASIGVVSKKKTGLILLGVWLYSLAWSLPPFFGW 1 2 AYVPEGLLTSCSWDYTTFTPSVRAYTMLLFCFVFFIPLIVIIYCYIFIFKAIQDTNK 2 1 ALQNIRSSESTASPRHFQRMKSEWKMAKIALVVILLFVLSWAPYSTVALVAFAG 2 1 SHILTPYMNSVPAIIAKASAIHNPIIYAISHPKYR 2 >MEL1_monDom Monodelphis domestica (opossum) Gq -GRID1 -WAPAL +LDB3 +BMPR1A 0 MNPSPMLRGLSCPAQDTNCTKIMASMSEWNNTEEDAYHLVDLPSIAPT 0 0 AVVLPPSSQNIFPTVDVPDHAHYTIGAIILAVGITGMLGNFLVIYTFCR 2 1 SHSLRTPANMFIINLAISDFFMSFTQAPVFFASSMYKRWIFGEK 1 2 ACEFYAFCGALFGITSMITLMAIALDRYFVITRPLASIGVISKKKTGFILLGVWLYSLAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYTTFTPSVRAYTMLLFCFVFFIPLIVIIYCYIFIFRAIQDTNK 2 1 AVHSIGSGESTASPRHCQRMKNEWKMAKIALVVILLYVLSWAPYSTVALVAFAG 2 1 YSHILTPYMNSVPAIIAKASAIHNPIIYAISHPKYR 2 1 MAIAQNFPCLRALLCVRHPRTRSFSSYRFTRRSTMTSQASDISWLPRGRRQLSLGSESEI 0 0 GWNNMEAGTTSLTSRNQQGSCRMDQETMETRELAAIAKAKGRSWETLEK 0 0 TLEEMDDSSLLEVSVDMEQ* 0 >MEL1_ornAna Ornithorhynchus anatinus (platypus) fragment 0 0 0 FPTADVPDHAHYTIGATILAVGFTGVLGNLLVIYTFCR 2 1 SRSLRTPANMFIINLSISDFFMSLTQAPVFFASSLHKRWIFGEK 1 2 GCQLYAFCGALFGITSMITLTVIALDRYFVITRPLASIGVISKKRALLILTGVWFYSLAWSLPPFFGW 1 2 sAYVPEGLLTSCSWDYMTFTPPVRAYTMLLFCFVFFIPLIMIIYCYFFIFRAIRGTNK 2 1 AVETIGSDDCRGSQRQCQRMKNEWKTAKIALMVILLYVISWCPYSVVALVAFAG 1 YSHLLTPYMNSVPAVIAKSSAIHNPIIYAITHPKYR 2 1 MAITKYIPCLGPLLRVSRQDSRSSSHYASSRRSTVTSQSLDGSWLPGRRRPLSSASDSES 0 0 0 0 * 0 >MEL1_anoCar Anolis carolinensis diverged frag 0 0 0 ERTMFNLPDPFPTVDVPTHAHYTIGAVILVVGITGTLGNLLVIYVFFR 2 1 IRGLRTPANMFVINLAVSDFL 1 2 GCELYAFCGALFGIASMITLTVIALDRYFVITRPLASIGAMSTKKALLILSGVWLYSLAWSLPPFFGW 1 2 sAYVPEGLLTSCSWDYITFTPSVRAYTMLLFCFVFFIPLIAIIYSYVFIFIAIKNSNR 2 1 AVQRTNSDNSKEGQKLYQKLKNEWKMAKVALIVILLVISWSPYSVVALVAFAG 2 1 YSHLLTPYMNSVPAVIAKASVIHNPIIYAIVHPKYR 2 1 MAIAKFLPCLGSLLRVPRKDSSYPSTRRPTVTSQSSDINGVPRGHRRLSSVSDSES 0 0 DWTDTEADISSQNSRVASGSISYRIYEDTTETIKVKSKMRSHDSGIFER 0 0 0 0 TGEDLNAFGWRREESYSGPSTSSQIPSIIVTFSNVQRTDLPLESSSGALCSRNSSYSWEKDSNS* 0 >MEL1_galGal Gallus gallus (chicken) Gq short exon 1 -GRID1 -WAPAL +LDB3 +BMPR1A 529 aa 16856781 AY88294 melanopsin OPN4m 0 MDLPPRAPT 0 0 KMTVKDVRGAFPTVDVPDHAHYTIGTVILIVGITGTLGNFLVIYAFCR 2 1 SRTLQKPANIFIINLAVSDFLMSITQSPVFFTNSLHKRWIFGEK 1 2 GCELYAFCGALFGITSMITLMVIALDRYFVITKPLASVRVMSKKKALIILVGVWLYSLAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFIPLIAIIYSYVFIFEAIKKANK 2 1 SVQTFGCKHGNRELQKQYHRMKNEWKLAKIALIVILLYVISWSPYSVVALVAFAG 2 1 YSHVLTPFMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 TAIATYVPCLGFLLRVSPKESRSFSSYPSSRRTTITSQSSETSGLQKGKRRLSSISDSES 0 0 GCTDTETDITSMISRPASSQVSYEMGEDTTQTSDLGGKPKVKSHDSGIFRK 0 0 TVVDADEIPMVEINDTEHSATSTCKTSEKCNVEEIQ 0 0 RSESLSGIGLREGESRHRTSASQIPSIIITYSNVQGVELHSGYSAGFLHPKNKSHKQNKSSNS* 0 >MEL2_galGal Gallus gallus (chicken) Gq 0.0.1.2.2.1.1.1.0.0 indel +GRID2 +SMARCAD1 -PGDS -SEC24B +COL25A1 544 aa 000 nm 17977531 NM_204625 full 0 MGTQPHSVTKSEIPDHVLYTVGTCVLVIGSIGIIGNLLVLYAFYS 2 1 NKKLRTPQNFFIMNLAVSDFLMSASQAPICFVNSLHREWILGDI 1 2 GCDLYAFCGALFGITSMMTLLAISVDRYLVITKPLRSIQWTSKKRTIQIIAAVWLYSLGW 1 2 SVAPLLGWSSYVPEGLMISCTWDYVTYSPANRSYTMILCCCVFFIPLIIILHCYLFMFLAIRSTGR 2 1 DVQKLGSCSRKSFLSQSMKNEWKLAKIAFVVIIVYVLSWSPYACVTLIAWAG 2 1 RGNTLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYR 2 1 KTIHNAVPCLRFLIRISKNDLLRGSINESSFRTSLSSHQSLAGRTKNTCVSSVSTGEA 0 0 NWSDVELDTVEPAHEKLQPRRSHSFSSSLRQKRDLLPDSYSCSEETEEK 0 0 VSLSSSYLEKVLGRSAFPSSPVALVTSSLRAASLPVGLNSSSASRGAGSDISQMKTEESHNNGGLDSIVSNTVPQIIIIPTSETNLFQEEPEEEETELFHFHDKKNNLLDLEGLSSSTEFLEAVEKFLS* 0
PRNP (3+ marsupials)
The Sarcophilus repeat region is of considerable interest -- the high GC content of this region makes it difficult to sequence and so provides a test of the 454 technology and Newbler assembler. This region consists in placentals a five octapeptide repeat, in marsupials and platypus a five nona- or decapeptide residue repeat that may resolve fine details of the marsupial phylogenetic tree, which in birds, lizards, turtles, frogs and fish is a hexapeptide repeat with trimeric internal substructure. Even though the single exon gene is clearly orthologous in all these species, the repeat regions within it are not directly comparable because they have expanded and contracted through replication slippage, plus experienced the odd repeat length change in marsupials and another in placentals.
The Sarcophilus prion gene has very high coverage that overcomes the occasional problem with frameshifts and allows the gene to be accurately tiled. However familiarity with the gene and reliable fiducial sequences are key to rapid assembly of the full length gene. No sequencing difficulties were observed in the high GC repeat region. The gene is very normal and has no indications whatsoever of abnormal numbers of repeats (4) or prion disease disposition.
Dasypus MVRSRVGCWLLLLFVATWSELGLC KK.RPKPGGGWNTGG SRYPGQ GSPGG NRYP PQGGG WGQ PHGGG WGQ PHGGG WGQ PHGGG WGQ PHGGG WGQ GGAHGQ Trichosurus MGKIQLGYWILVLFIVTWSDLGLC KKPKPRPGGGWNSGGS NRYPGQPGSPGG NRYPGWGH PQGGGTNWGQ PHPGGSNWGQ PHPGGSSWGQ PH GGSNWGQ GG YN Sarcophilus MGKIRLGYWILALFIVTWSDLGLC KKPKPRPGGGWNSGGS NRYPGQPGSAGG NRYPGWGH PQGGGTNWGQ PHPGGSSWGQ PHAGGSNWGQ PH.GGSNWGQ SGSSYNQ Monodelphis MGKIHLGYWFLALFIMTWSDLTLC KKPKPRPGGGWNSGG NRYPGQ SG GWGH PQGGGTNWGQ PHAGGSNWGQ PRPGGSNWGQ PHPGGSNWGQ PHPGGSNWGQ AGSSYNQ Macropus MAKIQLGYWILALFIVTWSELGLC KKPKTRPGGGWNSGGS NRYPGQPGSPGG NRYPGWGH PQGGGTNWGQ PHPGGSSWGQ PHAGGSNWGQ PH.GGSNWGQ GGGSYG Ornithorhynchus ------------------------ -------GGGWNSG NRYPGQPANPG GWGH PQGGGASWGH PQGGGASWGH PQGGGSNWGH PQGGGASWGH PQ GGGYS Dasypus WNKPSKPKTNM KHVAGAAAAGAVVG LGGYLVGSAMSRPLIHFGNDYEDRYYRENMYRYPNQVYYRSVEQYSSEKNFVHD CV MERVVEQMCITQYQ Trichosurus KWKPDKPKTNL KHVAGAAAAGAVVGGLGGYMLGSAMSRPVIHFGNEYEDRYYRENQYRYPNQVMYRPIDQYSSQNNFVHD CVNITVKQHTTTTTTKGENFTETDIKIMERVVEQMCITQYQN Sarcophilus KWKPDKPKTNM KHMAGAAAAGAVLGSLGGYVLGSAMSRPIMHFGNDYEDRYYRENQYRYPNQVMYRPIDQYSSQNNFVHD CVNITVKQHTTTTTTKGENFTETDIKIMERVVEQMCITQYQN Monodelphis KWKPDKPKTNM KHVAGAAAAGAVVGGLGGYMLGSAMSRPIMHFGNDYEDRYYRENQYRYPNQVMYRPIDQYNNQNNFVHD CVNITVKQHTTTTTTKGENFTETDIKIMERVVEQMCITQYQN Macropus KWKPDKPKTNL KHVAGAAAAGAVVGGLGGYMLGSAMSRPVMHFGNEYEDRYYRENQYRYPNQVMYRPIDQYGSQNSFVHD CVNITVKQHTTTTTTKGENFTETDIKIMERVVEQMCITQYQN Ornithorhynchus KYKPDKPKTGM KHVAGAAAAGAVVGGLGGYMIGSAMSRPPMHFGNEFEDRYYRENQNRYPNQVYYRPVDHFCSQDGFVRD CVNITVTQHTVTTT.EGKNLNETDVKIMTRVLEQMC
The signal region of Sarcophilus PRNP is expected to show the same length as the other 3 known marsupial sequences, which is confirmed by the sequence. Placentals exhibit a one residue deletion relative to this ancestral length.
MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Homo sapiens MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Pan troglodytes MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Gorilla gorilla MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Pongo pygmaeus MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Nomascus leucogenys MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Hylobates lar MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Symphalangus syndactylus MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca arctoides MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca fascicularis MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca fuscata MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca mulatta MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca nemestrina MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Papio hamadryas MA--NLGCWMLFLFVATWSDLGLCKK--RPKPG Callithrix jacchus MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Cebus apella MA--NLGCWMLVVFVATWSDLGLCKK--RPKPG Cercopithecus aethiops MA--NLGCWMLVVFVATWSDLGLCKK--RPKPG Cercopithecus dianae MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Colobus guereza MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Presbytis francoisi MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Saimiri sciureus MA--KLGYWLLVLFVATWSDVGLCKK--RPKPG Tarsius syrichta MA--NLGCWMLVVFVATWSDVGLCKK--RPKPG Microcebus murinus MA--RLGCWMLVLFVATWSDIGLCKK--RPKPG Otolemur garnettii ME--NLGCWMLILFVATWSDIGLCKK--RPKPG Cynocephalus variegatus MA--QLGCWLMVLFVATWSDVGLCKK--RPKPG Tupaia belangeri MA--NLGYWLLALFVTMWTDVGLCKK--RPKPG Mus musculus MA--NLGYWLLALFVTTCTDVGLCKK--RPKPG Rattus norvegicus MA--NAGCWLLVLFVATWSDTGLCKK--RPKPG Cavia porcellus MA--NLGCWLLVLFVATWSDLGLCKK--RTKPG Dipodomys ordii MV--NPGCWLLVLFVATLSDVGLCKK--RPKPG Spermophilus tridecemlineatus MA--HLGYWMLLLFVATWSDVGLCKK--RPKPG Oryctolagus cuniculus MA--HLSYWLLVLFVAAWSDVGLCKK--RPKPG Ochotona princeps MVKSHIGSWILVLFVAMWSDVGLCKK--RPKPG Bos taurus MVKSHIGGWILVLFVAAWSDIGLCKK--RPKPG Sus scrofa MVKSHMGSWILVLFVVTWSDMGLCKK--RPKPG Vicugna vicugna MVKSHVGGWILVLFVATWSDVGLCKK--RPKPG Equus caballus MVRSHVGGWILVLFVATWSDVGLCKK--RPKPG Diceros bicornis MVKSLVGGWILLLFVATWSDVGLCKK--RPKPG Myotis lucifugus MVKNYIGGWILVLFVATWSDVGLCKK--RPKPG Pteropus vampyrus MVKSHIANWILVLFVATWSDMGFCKK--RPKPG Tursiops truncatus MVKSHIGGWILLLFVATWSDVGLCKK--RPKPG Canis lupus familiaris MVKSHIGSWILVLFVAMWSDVGLCKK--RPKPG Felis catus MVKSHIGSWLLVLFVATWSDIGFCKK--RPKPG Mustela putorius MVKSHIGSWLLVLFVATWSDIGFCKK--RPKPG Mustela vison MVKSHIGSWILVLFVAMWSDVGLCKK--RPKPG Ailuropoda melanoleuca MVKNHVGCWLLVLFVATWSEVGLCKK--RPKPG Erinaceus europaeus MVTGHLGCWLLVLFMATWSDVGLCKK--RPKPG Sorex araneus MVKSHLGCWIMVLFVATWSEVGLCKK--RPKPG Cyclopes didactylus MVRSRVGCWLLLLFVATWSELGLCKK--RPKPG Dasypus novemcinctus MVKGTVSCWLLVLVVAACSDMGLCKK--RPKPG Echinops telfairi MVKSSLGCWILVLFVATWSDMGLCKK--RPKPG Loxodonta africana MVKSSLGCWMLVLFVATWSDVGLCKK--RPKPG Procavia capensis MAKIQLGYWILALFIVTWSELGLCKKP-KTRPG Macropus eugenii MGKIHLGYWFLALFIMTWSDLTLCKKP-KPRPG Monodelphis domestica MGKIRLGYWILALFIVTWSDLGLCKKP-KPRPG Sacophilus harrisii MGKIQLGYWILVLFIVTWSDLGLCKKP-KPRPG Trichosurus vulpecular MARLLTTCCLLALLLAACTDVALSKKG-KGKPS Gallus gallus MAKLPGTSCLLLLLLLLGADLASCKKG-KGKPG Taeniopygia guttata MARLLTTCCLLALLLAACTDVALSKKG-KGKPG Meleagris gallopavo MGKHQMTCWLAIFLLLIQANVSLAKK--KPKPS Anolis carolinensis MRRFLVTCWIAVFLILLQTDVSLSKKG-KNKPG Gekko gekkko MGRYRLTCWIVVLLVVMWSDVSFSKKG-KGKGG Trachemys scripta (turtle) MGRHLISCWIIVLFVAMWSDVSLAKKG-KGKTG Pelodiscus sinensis (turtle) MPQSLWTCLVLISLICTLTVSSKKSGGGKSKTG Xenopus laevis MLRSLWTSLVLISLVCALTVSSKKSGSGKSKTG Xenopus topicalis
>PRNP_sacHar Sarcophilus harrisii (tasmanian_devil) single exon gene YVLG like Dasypus MGKIRLGYWILALFIVTWSDLGLCKKPKPRPGGGWNSGGSNRYPGQPGSAGGNRYPGWGHPQGGGTNWGQPHPGGSSWGQPHAGGSNWGQPHGGSNWGQ SGSSYNQKWKPDKPKTNMKHMAGAAAAGAVLGGVGGYVLGSAMSRPIMHFGNDYEDRYYRENQYRYPNQVMYRPIDQYSSQNNFVHDCVNITVKQHTTTTTT KGENFTETDIKIMERVVEQMCITQYQNEYRAAQYSYNMAFFSAPPVTLLLLGFLIFLIVS* >PRNP_mdo Monodelphis domestica opossum, from frameshifted genomic MGKIHLGYWFLALFIMTWSDLTLCKKPKPRPGGGWNSGGNRYPGQSGGWGHPQGGGTNWGQPHAGGSNWGQPRPGGSNWGQPHPGGSNWGQPHPGGSNWG QAGSSYNQKWKPDKPKTNMKHVAGAAAAGAVVGGLGGYMLGSAMSRPIMHFGNDYEDRYYRENQYRYPNQVMYRPIDQYNNQNNFVHDCVNITVKQHTTT TTTKGENFTETDIKIMERVVEQMCITQYQNEYRSAYSVAFFSAPPVTLLLLSFLIFLIVS* >PRNP_tvu Trichosurus vulpecular brushtail opossum MGKIQLGYWILVLFIVTWSDLGLCKKPKPRPGGGWNSGGSNRYPGQPGSPGGNRYPGWGHPQGGGTNWGQPHPGGSNWGQPHPGGSSWGQPHGGSNWGQGGY NKWKPDKPKTNLKHVAGAAAAGAVVGGLGGYMLGSAMSRPVIHFGNEYEDRYYRENQYRYPNQVMYRPIDQYSSQNNFVHDCVNITVKQHTTTTTTKGENFTETDIKIMERVVEQM CITQYQAEYEAAAQRAYNMAFFSAPPVTLLFLSFLIFLIVS* >PRNP_meu Macropus eugenii (tammar wallaby) MAKIQLGYWILALFIVTWSELGLCKKPKTRPGGGWNSGGSNRYPGQPGSPGGNRYPGWGHPQGGGTNWGQPHPGGSSWGQPHAGGSNWGQPHGGSNWGQ GGGSYGKWKPDKPKTNLKHVAGAAAAGAVVGGLGGYMLGSAMSRPVMHFGNEYEDRYYRENQYRYPNQVMYRPIDQYGSQNSFVHDCVNITVKQHTTTTTT KGENFTETDIKIMERVVEQMCITQYQNEYQAAQRYYNMAFFSAPPVTLLLLSFLIFLIVS* >PRNP_oan Ornithorhynchus anatinus platypus fragment PHWGKSPVHHWIIDICVVHLERRCRGHLHPNPCPGGRCVQQQPNRYPGQPATPGGWGHPQGGGASWGHPQGGGSNWGHPQGGGASWGHPQGGGYSKYKPDKPKTG MKHVAGAAAAGAVVGGLGGYMIGSAMSRPPMHFGNEFEDRYYRENQNRYSNQVYYRPVDQYGSQDGFVRDCVNITVTQHTVTTTEGKNLNETDVKIMTRVLEQMCVNLY
PRND (2+ marsupials)
Sarcophilus sequence for this intronless gene is a welcome addition to a limited existing set of early-diverging mammalian orthologs. With more data, the relative rates of divergence of PRND from its parental paralog PRNP could be compared in marsupial and placentals. It appears from the mere 75% identity between tasmanian devil and wallaby that doppels are diverging quite rapidly both from PRNP and from each other in the marsupial lineage, indicating some selectional pressure but not a hugely important function (that is, many residue positions have an increased reduced alphabet).
>PRND_hsa Homo sapiens (human) full MRKHLSWWWLATVCMLLFSHLSAVQTRGIKHRIKWNRKALPSTAQITEAQVAENRPGAFIKQGRKLDIDFGAEGNRYYEANYWQFPDGIHYNGCSEANVTKEAFVTGCINATQAANQGEFQKPDNKLHQQVLWRLVQELCSLKHCEFWLERGAGLRVTMHQPVLLCLLALIWLTVK* >PRND_dno Dasypus novemcinctus MRKHLGGWRLAIVCVLLSGHLSMVKARGIKHRIKWNRKAAPGAAQVTEARVAEQRPGAFVRQGRRLDIDFGAEGNRYYEANYWQLPDGILYDGCAEANVTKEALVAGCVNATQLANQAELAHEGQDTLHRRVLGRLIRELCALKRCKFWPDRAAGPRLVRGAPVFGGLLLLIWLLVR* >PRND_laf Loxodonta africana African elephant Afrotheria 176 aa revised/corrected MRKHLGAWWLAIAFVLLLSHLSMVTARGIKHRIKWNRKALPNTGHVTAAQVTETRPGAFIRHGRKLDIDFGAEGNRYYEANYWQFPDGIHYDGCSEANVTKEMFVTSCINTTQAANQEEFSRKQDNKVYQRILWRLIRELCSVKHCDFWLDRGGGLRVSLDQPVMLCLLVFIWFMVK* >PRND_sacHar Sarcophilus harrisii (tasmanian_devil) single exon gene 77% macEug MRTPLETWWIAIFFTLLFSDLSLVKAKGIRQRNKSNRKSLQTNRANPTREQPSKILQGTFIRKGRKLSINFGEEGNSYYEAHYKLFPDEIHYVGCAESSVTKDVFISNCVNVTHTANKLEPPEERNSSAIYSRVLEQLIKELCALKYCEFGMQIGAGFRLSLDQSMMVYLMILAFFIVK* >PRND_mdo Monodelphis domestica doppel genomic revised +rassf2 -prnd -prnp MRRHLGICWIAIFFALLFSDLSLVKAKTTRQRNKSNRKGLQTNRTNPTTVQPSEKLQGTFIRNGRKLVIDFGEEGNSYYATHYSLFPDEIHYAGCAESNVTKEVFISNCVNATRVINKLEPLEEQNISDIYSRILEQLIKELCALNYCEFRTGKGTGLRLSLDQYVMVYLVILTCLIVK* >PRND_meu Macropus eugenii wallaby MRRHLGTWWTAIFFALLFSDLSLVKAKGTRQRNKSNRKSLQTNRVNPTTAQPSEILQGAFIRQGRKLSIDFGEEGNSYYETHYQLFPDEIHYVGCTESNVTKDIFISNCMNATHAVNNLETLEEKNASDIHSRVLEQLIKELCALKYCELETETGAGLKLSLDQSVMVYLVILTCLIVK* >PRND_oan Ornithorhynchus anatinus platypus 42% to opposum 187 aa 4 cys in register MMTVRRRRRSGGARWLLVFLVLLSGDLSSLQARGPRPRNKAGRKPPPSNAGPDSPAPRPPAGARGTFIRRGGRLSVDFGPEGNGYYQANYPLLPDAIVYPDCPTANGTREAFFGDCVNATHEANRGELTAGGNASDVHVRVLLRLVEELCALRDCGPALPTGPAPRPGPPGPPAALALLTLVLLGAQ* >PRND_aca Anolis carolinensis weak but real! scaffold_1221:78,884-117,121 syntenic, oriented like PRNP but no larger MMQRPLVVAILLTALWSEVCLCRRVSGSANRRNKKTSTTTSAPKLQSSTTATTFQGNLCRGGQMIDNMDLEPNDKVYYKANLKIFPDGLYYPNCSLLLQPNTTKEELVGECVNFTIASNKLNLSKGKDLSNTKERVMWVLIHHLCANESCGQPCPLLQNSGNLHYIGQVLTVFVGLIGCSFLSAK*