Marsupial phyloSNPs
Introduction to Marsupial phyloSNPs
In this project, new genomic data from the Tasmanian devil (Sarcophilus harrisii), Tasmanian tiger (Thylacinus cynocephalus), and echidna (Tachyglossus aculeatus) are analyzed for significant changes at the protein coding level. The goal is to find single amino acid changes in one of these species at a highly invariant residue in a well-conserved exon in a gene with known or predictable tertiary structure. Such changes are thought to enrich for genetic changes with significant, adaptive biochemical or phenotypic consequences (1,2,3,4), in contrast to ordinary SNPs at positions of low conservation. Thus phyloSNPs are informative to the distinctive biology of the species carrying them and suggest a focus for subsequent experiment.
Marsupial genomic and cDNA data to date has been quite limited compared to placental mammal. Yet as outgroup, metatheran animals provide important context to placentals and represent important context in understanding human protein evolution. The monotheres are inevitably limited by the paucity of extant species (basically platypus and echidna) and dim prospects for fossil DNA. Consequently echidna provides an important adjunct to the existing but incomplete platypus assembly. While extant birds and reptiles -- the preceding divergence node -- are abundant it must be remembered that a very considerable time elapsed (from 310 mry to 175 mry) prior to divergence of mammals with living representatives. This gap of 135 myr is comparable to the whole evolutionary record of theran mammals.
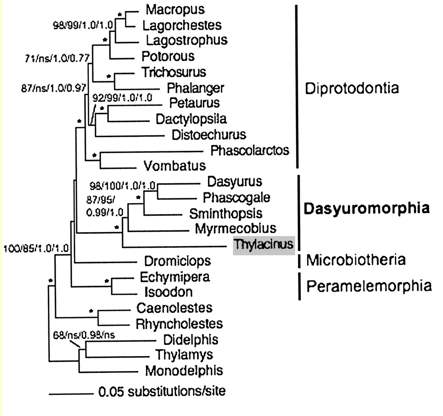
Assumed vertebrate phylogenetic tree
Marsupial relationships taken from 2009 paper establishing the mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus):
Newick tree that generates vertebrate phylogenetic tree used in the analysis here: ((((((((((((((((((homSap,panTro),gorGor),ponPyg),macMul),calJac),tarSyr),(micMur,otoGar)),tupBel), (((((musMus,ratNor),dipOrd),cavPor),speTri),(oryCun,ochPri))), (((((vicPac,susScr),turTru),bosTau),((equCab,(felCat,canFam)),(myoLuc,pteVam))),(eriEur,sorAra))), (((loxAfr,proCap),echTel),(dasNov,choHof))), (monDom,((macEug,triVul),(sarHar,thyCyn)))), (ornAna,tacAcu)), ((galGal,taeGut),anoCar)), xenTro), (((tetNig,takRub),(gasAcu,oryLap)),danRer)), calMil), petMar);
Phylo-sorting data
- - - - - - - (((((((((((((((((( - - - - 10 26 10 > 27 gene homSap , Homo sapiens (human) hg181 11 38 11 > 40 gene panTro ), Pan troglodytes (chimp) panTro 12 25 12 > 26 gene gorGor ), Gorilla gorilla (gorilla) gorGor 13 40 13 > 42 gene ponPyg ), Pongo pygmaeus (orang) ponAbe 14 28 14 > 30 gene macMul ), Macaca mulatta (rhesus) rheMac 15 12 15 > 12 gene calJac ), Callithrix jacchus (marmoset) calJac 16 48 16 > 53 gene tarSyr ),( Tarsius syrichta (tarsier) tarSyr 17 29 17 > 31 gene micMur , Microcebus murinus (mouse_lemur) micMur 18 37 18 > 39 gene otoGar )), Otolemur garnettii (bushbaby) otoGar 19 50 19 > 57 gene tupBel ),((((( Tupaia belangeri (tree_shrew) tupBel 20 31 20 > 33 gene musMus , Mus musculus (mouse) mm91 21 43 21 > 45 gene ratNor ), Rattus norvegicus (rat) rn41 22 18 22 > 19 gene dipOrd ), Dipodomys ordii (kangaroo_rat) dipOrd 23 14 23 > 15 gene cavPor ), Cavia porcellus (guinea_pig) cavPor 24 45 24 > 48 gene speTri ),( Spermophilus tridecemlineatus (squirrel) speTri 25 35 25 > 37 gene oryCun , Oryctolagus cuniculus (rabbit) oryCun 26 33 26 > 35 gene ochPri ))),((((( Ochotona princeps (pika) ochPri 27 52 27 > 59 gene vicPac , Vicugna pacos (lama) vicPac 54 57 28 > 49 gene susScr ), Sus scrofa (pig) 28 51 29 > 58 gene turTru ), Tursiops truncatus (dolphin) turTru 29 11 30 > 11 gene bosTau ),(( Bos taurus (cow) bosTau 30 20 31 > 21 gene equCab ,( Equus caballus (horse) equCab 31 22 32 > 23 gene felCat , Felis catus (cat) felCat 32 13 33 > 14 gene canFam )),( Canis familiaris (dog) canFam 33 32 34 > 34 gene myoLuc , Myotis lucifugus (microbat) myoLuc 34 42 35 > 44 gene pteVam ))),( Pteropus vampyrus (macrobat) pteVam 35 21 36 > 22 gene eriEur , Erinaceus europaeus (hedgehog) eriEur 36 44 37 > 47 gene sorAra ))),((( Sorex araneus (shrew) sorAra 37 27 38 > 28 gene loxAfr , Loxodonta africana (elephant) loxAfr 38 41 39 > 43 gene proCap ), Procavia capensis (hyrax) proCap 39 19 40 > 20 gene echTel ),( Echinops telfairi (tenrec) echTel 40 17 41 > 18 gene dasNov , Dasypus novemcinctus (armadillo) dasNov 41 15 42 > 16 gene choHof ))),( Choloepus hoffmanni (sloth) choHof 42 30 43 > 32 gene monDom ,(( Monodelphis domestica (opossum) monDom 55 55 44 > 29 gene macEug , Macropus eugenii (wallaby) 56 56 45 > 46 gene sarHar ),( Sarcophilus harrisii (tasmanian_devil) 57 60 46 > 56 gene triVul , Trichosurus vulpecula (bushytail_possum) 58 59 47 > 55 gene thyCyn )))),( Thylacinus cynocephalus (tasmanian_tiger) 43 34 48 > 36 gene ornAna , Ornithorhynchus anatinus (platypus) ornAna 59 58 49 > 50 gene tacAcu )),(( Tachyglossus aculeatus (echidna) 44 23 50 > 24 gene galGal , Gallus gallus (chicken) galGal 45 46 51 > 51 gene taeGut ), Taeniopygia guttata (finch) taeGut 46 10 52 > 10 gene anoCar )), Anolis carolinensis (lizard) anoCar 47 53 53 > 60 gene xenTro ),((( Xenopus tropicalis (frog) xenTro 48 49 54 > 54 gene tetNig , Tetraodon nigroviridis (pufferfish) tetNig 49 47 55 > 52 gene takRub ),( Takifugu rubripes (fugu) fr21 50 24 56 > 25 gene gasAcu , Gasterosteus aculeatus (stickleback) gasAcu 51 36 57 > 38 gene oryLap )), Oryzias latipes (medaka) oryLat 52 16 58 > 17 gene danRer )), Danio rerio (zebrafish) danRer 60 54 59 > 13 gene calMil ), Callorhinchus milii (elephantfish) 53 39 60 > 41 gene petMar ) Petromyzon marinus (lamprey) petMar 44 44 51 f 51 gene fasta tree_syntax genus species common ucsc phy alp phy alp
Candidate analysis
(methods explained here shortly)
Case of ERN2
chr6_5971 ERN2 4
contig00001 length=355 numreads=5
KLPFTIPELVHASPCRSSDGVLYT
.....................F..
^
15 R=3(75) H=2(50
Read data format: the top row gives project gene name, HGNC gene name and exon number from ENSEMBL monDom5
and human orthology predictions, then Monodelphis amino-acid segment, then sequence differences in
tasmanian devil (in this case, both individuals differ from Monodelphis by L->F), then differences between the two thylacines
(here one individual has R at position 15, the other has H), and finally the number of experimental reads that confirm the nucleotide
difference and the sum of the quality scores. The sequences were assembled by Newbler (the official 454 assembler) which uses
lower-case letters for less confident calls.
Pseudogene issues: ERN2 has not generated potentially confusing recent processed pseudogenes in mammals (lack of human, opossum or platypus genome Blat matches to ERN2 query). The variation observed here between individual tasmanian devils is implausibly an early stage in the loss of parent gene because of ERN2 functional essentiality; the exon cannot come from a decaying segmental duplication because coverage is high enough to also detect the main gene.
Paralog issues: The GeneSorter tool at UCSC shows a single significant full-length paralog in human, ERN1, also with 22 coding exons. The genes reside on different chromosomes but in regions with local homology of synteny. However this particular exon is a good match (3 differences out of 23), so there is potential for experimental difficulties in distinguishing them in short reads (including the following exon readily resolves them bioinformatically). In any event, at positions 15 and 20, ERN1 is identical at the amino acid level to ERN2.
Homoplasy (recurrent mutation) issues: This exon is very conserved and does not exhibit repetitive sequence, compositional simplicity, or indels in any species in either paralog that could foster experimental error or alignment ambiguity. At position 15, the ancestral value is arginine in both paralogs. The G--> A transition to histidine in one individual is conservative under most circumstances (still basic) and arises from an arginine codon CpG hotspot conserved back to lamprey in 30 of 32 species with available data, yet histidine is not observed part of a reduced alphabet (ie R/H) at this position over many billions of years of branch length. Consequently R-->H is a significant change in this individual tasmanian devil.
Known variations: No human disease variants have been reported for either ERN2 or ERN1, probably attributable to essentiality. Site-specific mutation close to the exon here have been generated for K121P, D123P, W125A, and Q105E but only for ERN1. Naturally occuring coding SNPs in the human population relevent to the ERN2 exon are not known but low frequency alleles could emerge from the 1000 Genomes Project.
Side issues: a very ancient conserved leucine at position 21 appears to be transitioning to phenylalanine at marsupial node but has not been fixed, so settles out as L or F depending on lineage-sorting on each terminal marsupial leaf whereas placentals are all changed to phenylalanine (a phyloSNP caught in mid-air). While L and F might seem about the 'same' as amino acids, the branch length conservation totals say both are important but for different reasons: this is not a waffle codon nor reduced alphabet situation. This raises the question -- given the extreme conservation of this exon otherwise -- of whether the L-->F change at position 21 in both individuals has 'enabled' (made neutral or adaptive) an otherwise unfavorable R-->H change at position 15 in one individual.
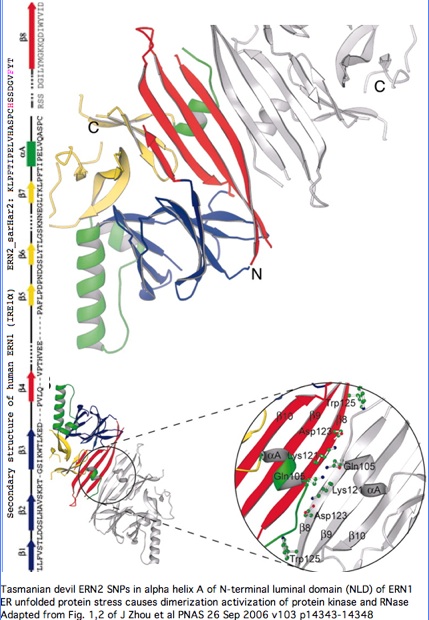
Structural significance: By good fortune, the crystal structure of ERN1 (alternately called IRE1) has been published. The PDB 2HZ6 structure has good coverage of this particular exon. Consequently the marsupial ERN2 could be very accurately modelled and the structural effects of L-->F with or without R-->H computed by submission to online SwissProt modelling service.
Alignment of Monodelphis ERN2 (key exon replaced by that of sarHar2) with crytallograph human ERN1 luminal domain
Expect = 5.8e-65 Identities = 109/180 (60%), Positives = 141/180 (78%)
ERN2_monDom 1 PESLLFISTLDGSLHAVSKKTGDIQWTLKDDPIIQGPVYATEPAFLPDPSDGSLYILGEE 60
PE+LLF+STLDGSLHAVSK+TG I+WTLK+DP++Q P + EPAFLPDP+DGSLY LG +
ERN1_homSap 8 PETLLFVSTLDGSLHAVSKRTGSIKWTLKEDPVLQVPTHVEEPAFLPDPNDGSLYTLGSK 67
ERN2_monDom 61 SKQGLMKLPFTIPELVHASPCHSSDGVFYTGRKQDTWFMVDPKSGKKQTMLSTETWDGLY 120
+ +GL KLPFTIPELV ASPCRSSDG+LY G+KQD W+++D +G+KQ LS+ D L
ERN1_homSap 68 NNEGLTKLPFTIPELVQASPCRSSDGILYMGKKQDIWYVIDLLTGEKQQTLSSAFADSLC 127
ERN2_monDom 121 PSAPLLYIGRTQYTVTMYDPRSQALRWNTTYRGYSAPLLDHLPGYQVGHFTCSGEGLVVT 180
PS LLY+GRT+YT+TMYD +++ LRWN TY Y+A L + Y++ HF +G+GLVVT
ERN1_homSap 128 PSTSLLYLGRTEYTITMYDTKTRELRWNATYFDYAASLPEDDVDYKMSHFVSNGDGLVVT 187
Functional significance: A considerable amount is known about the paralog ERN1. Annotation transfer is likely applicable to ERN2. The two gene products differ primarily in expression -- ERN1 ubiquitious but ERN2 restricted to intestinal epithelial cells:
"The unfolded protein response (UPR) is an evolutionarily conserved mechanism by which all eukaryotic cells adapt to the accumulation of unfolded proteins in the endoplasmic reticulum (ER). Inositol-requiring kinase 1 (IRE1 or ERN1) and PKR-related ER kinase (PERK) are two type I transmembrane ER-localized protein kinase receptors that signal the UPR through a process that involves homodimerization and autophosphorylation... The monomer of the luminal domain comprises a unique fold of a triangular assembly of beta-sheet clusters. Structural analysis identified an extensive dimerization interface stabilized by hydrogen bonds and hydrophobic interactions... Mutations that disrupt the dimerization interface produced ERN1 protein that failed to either dimerize or activate the UPR upon ER stress."
"ERN1 is a type I transmembrane protein kinase receptor that also has a site-specific RNase activity that, upon activation, initiates a site-specific unconventional splicing reaction. The substrate for IRE1 RNase in metazoans is Xbp1 mRNA, which encodes a basic leucine zipper transcription factor of the ATF/CREB family. XBP1 controls expression of genes containing an X-box element or a UPR element in their promoter regions. The IRE1-mediated splicing reaction introduces into XBP1 an alternative C terminus, thereby generating an XBP1 molecule that is a more potent transcriptional activator. Therefore, activation of IRE1 and its RNase increases the transcription of genes encoding ER chaperones and folding catalysts... the ERN1 N-terminal luminal domain (NLD) functions as an ER stress sensor... under normal conditions IRE1 is maintained in a monomeric state through interaction of the NLD with the ER resident chaperone BiP. Upon ER stress, Grp78 binds to unfolded proteins as they accumulate, permitting the released NLD to form homodimers. Dimerization of the NLD in turn leads to the activation of the protein kinase and RNase activities in the cytosolic domain of ERN1."
ENR2 is readily distinguished from its ERN1 paralog at tBlastn by including the two following exons which bring percent identity to 62%:
ERN2_monDom KLPFTIPELVHASPCRSSDGVLYTGRKQDTWFMVDPKSGKKQTMLSTETWDGLYPSAPLLYIGRTQYTVTMYDPRSQALRWNTTYRGYSA
KLPFTIPELV ASPCRSSDG+LY G+KQD W++VD +G+KQ LS+ + L PS LLY+GRT+YT+TM+D +S+ LRWN TY Y+A
ERN1_monDom KLPFTIPELVQASPCRSSDGILYMGKKQDIWYVVDLMTGEKQQTLSSAFAESLCPSTSLLYLGRTEYTITMFDTKSRELRWNATYFDYAA
^ *
ERN2_homSap KLPFTIPELVHASPCRSSDGVFYT
ERN2_panTro KLPFTIPELVHASPCRSSDGVFYT
ERN2_ponAbe KLPFTIPELVHASPCRSSDGVFYT
ERN2_rheMac KLPFTIPELVHASPCRSSDGVFYT
ERN2_calJac KLPFTIPELVHASPCRSSDGVFYT
ERN2_tarSyr KLPFTIPELVHASPCRSSDGVFYT
ERN2_micMur KLPFTIPELVHASPCRSSDGVFYT
ERN2_tupBel KLPFTIPELVHASPCRSSDGVFYT
ERN2_musMus KLPFTIPELVHASPCRSSDGVFYT
ERN2_ratNor KLPFTIPELVHASPCRSSDGVFYT
ERN2_cavPor KLPFTIPELVHTSPCRSSDGVFYT
ERN2_speTri KLPFTIPELVHASPCRSSDGVFYT
ERN2_oryCun KLPFTIPELVHASPCRSSDGVFYT
ERN2_ochPri KLPFSIPELVHASPCRSSDGVFYT
ERN2_turTru RLPFTIPELVHASPCRSSDGVFYT
ERN2_bosTau RLPFTIPELVHASPCRSSDGVFYT
ERN2_equCab KLPFTIPELVHASPCRSSDGVFYT
ERN2_felCat RLPFTIPELVHASPCRSSDGVFYT
ERN2_canFam KLPFTIPELVHASPCRSSDGVFYT
ERN2_myoLuc KLPFTIPELVHASPCRSSDGVFYT
ERN2_eriEur KLPFTVPELVHTSPCRSSDGVFYT
ERN2_sorAra KLPFTIPELVHASPCRSSDGVFYT
ERN2_loxAfr KLPFTIPELVHASPCRSSDGVFYT
ERN2_echTel KLPFTIPELVLASPCRSSDGVFYT
ERN2_dasNov KLPFTIPELVHTSPCRSSDGIFYT
ERN2_monDom KLPFTIPELVHASPCRSSDGVLYT
ERN2_macEug KLPFTIPELVHASPCRSSDGVFYT
ERN2_sarHar1 KLPFTIPELVQASPCRSSDGIFYM
ERN2_sarHar2 KLPFTIPELVQASPCHSSDGIFYM
ERN2_ornAna KLPFTIPELVQSSPCRSSDGILYT
ERN2_anoCar KLPFTIPELVQSSPCRSSDGIIYT
ERN2_taeGut KLPFTIPELVQSSPCRSSDGVLYT
ERN2_galGal KLPFTIPELVQASPCRSSDGILYM
ERN2_xenTro KLPFTIPELVQSSPCRSSDGILYT
ERN2_xenLae KLPFTIPELVQSSPCRSSDGILYT
ERN2_tetNig KLPFTIPELVQASPCRSSDGVLYM
ERN2_takRub KLPFTIPELVQASPCRSSDGVLYM
ERN2_gasAcu KLPFTIPDLVQSAPCRSSDGILYT
ERN2_oryLat KLPFTIPELVQSAPCRSSDGILYT
ERN2_calMil KLPFTIPELVQSSPCRSSDGILYT
ERN2_petMar KLPFTIPELVHASPCRTSDGVLYT
ERN1_homSap KLPFTIPELVQASPCRSSDGILYM
ERN1_panTro KLPFTIPELVQASPCRSSDGILYM
ERN1_ponAbe KLPFTIPELVQASPCRSSDGILYM
ERN1_rheMac KLPFTIPELVQASPCRSSDGILYM
ERN1_calJac KLPFTIPELVQASPCRSSDGILYM
ERN1_tarSyr KLPFTIPELVQASPCRSSDGILYM
ERN1_micMur KLPFTIPELVQASPCRSTDGILYM
ERN1_otoGar KLPFTIPELVQASPCRSSDGILYM
ERN1_tupBel KLPFTIPELVQASPCRSSDGILYM
ERN1_musMus KLPFTIPELVQASPCRSSDGILYM
ERN1_ratNor KLPFTIPELVQASPCRSSDGILYM
ERN1_dipOrd KLPFTIPELVQASPCRSSDGILYM
ERN1_cavPor KLPFTIPELVQASPCRSSDGILYM
ERN1_speTri KLPFTIPELVQASPCRSSDGILYM
ERN1_oryCun KLPFTIPELVQASPCRSSDGILYM
ERN1_vicPac KLPFTIPELVQASPCRSSDGILYM
ERN1_turTru KLPFTIPELVQASPCRSSDGILYM
ERN1_bosTau KLPFTIPELVQASPCRSSDGILYM
ERN1_equCab KLPFTIPELVQASPCRSSDGILYM
ERN1_canFam KLPFTIPELVQASPCRSSDGILYM
ERN1_myoLuc KLPFTIPELVQASPCRSSDGILYM
ERN1_pteVam KLPFTIPELVQASPCRSSDGILYM
ERN1_eriEur KLPFTIPELVQASPCRSSDGILYM
ERN1_sorAra KLPFTIPELVQASPCRSSDGILYM
ERN1_loxAfr KLPFTIPELVQASPCRSSDGILYM
ERN1_proCap KLPFTIPELVQASPCRSSDGILYM
ERN1_echTel KLPFTIPELVQASPCRSSDGILYM
ERN1_dasNov KLPFTIPELVQASPCRSSDGILYM
ERN1_choHof KLPFTIPELVQASPCRSSDGILYM
ERN1_monDom KLPFTIPELVQASPCRSSDGILYM
ERN1_ornAna KLPFTIPELVHASPCRSSDGILYM
ERN1_galGal KLPFTIPELVQASPCRSSDGILYM
ERN1_taeGut KLPFTIPELVQASPCRSSDGILYM
ERN1_anoCar KLPFTIPELVQASPCRSSDGILYM
ERN1_xenTro KLPFTIPELVQSSPCRSSDGILYT
ERN1_tetNig KLPFTIPELVQASPCRSSDGVLYM
ERN1_takRub KLPFTIPELVQASPCRSSDGVLYM
ERN1_gasAcu KLPFTIPELVQASPCRSSDGVLYM
ERN1_oryLat KLPFTIPELVQASPCRSSDGVLYM
ERN1_danRer KLPFTIPELVQASPCRSSDGILYM
Ancient CpG in ERN2 homSap chr16:23625855-23625856
Human CG
Chimp CG
Gorilla --
Orangutan CG
Rhesus CG
Marmoset CG
Tarsier CG
Mouse lemur CG
Bushbaby --
TreeShrew CG
Mouse CG
Rat CG
Kangaroo rat --
Guinea Pig CG
Squirrel CG
Rabbit CG
Pika CG
Alpaca --
Dolphin CG
Cow CG
Horse CG
Cat CG
Dog CG
Microbat CG
Megabat --
Hedgehog CG
Shrew CG
Elephant --
Rock hyrax --
Tenrec CG
Armadillo CG
Opossum CG
Platypus CG
Lizard CG
Tetraodon CG
Fugu CG
Stickleback CT
Medaka CT
Lamprey CG
Case of XXXX
(more shortly)
Case of YYYY
(more shortly)
Case of ZZZZ
(more shortly)