Medical Sequencing Data: Difference between revisions
Jaspersaris (talk | contribs) |
|||
| (10 intermediate revisions by 5 users not shown) | |||
| Line 50: | Line 50: | ||
http://www.pharmgkb.org/ - Requires registration for much data. Fan & Jim registered<BR> | http://www.pharmgkb.org/ - Requires registration for much data. Fan & Jim registered<BR> | ||
http://globin.bx.psu.edu/genphen/ - Belinda, Ross and Webb's work, mostly covers hemoglobins<BR> | http://globin.bx.psu.edu/genphen/ - Belinda, Ross and Webb's work, mostly covers hemoglobins<BR> | ||
http://www.hgvs.org/dblist/dblist.html - Variation Databases and Related Sites, updated 3/31/06 on HGVS (Human Genome Variation Society) | http://www.hgvs.org/dblist/dblist.html - Variation Databases and Related Sites, updated 3/31/06 on HGVS (Human Genome Variation Society)<br> | ||
http://www.ncbi.nlm.nih.gov/geo/ - NCBI GEO Gene Expression Omnibus [Gene expression is a phenotype]<br> | |||
* http://www.ncbi.nlm.nih.gov/geo/gds/gds_browse.cgi?gds=1332 Study on Huntington's Disease<br> | |||
http://genome-www5.stanford.edu/ - SMD Stanford Microarray Database.<br> | |||
== Some Large Scale Studies Currently in Progress == | |||
http://www.framinghamheartstudy.org/heartstudy.asp - 500,000 SNP genotype data added to a 5000 person heart study that's been going on for three generations. | |||
http://www.fnih.org/GAIN/GAIN_home.shtml - A public/private partnership Francis Collins and the NHGRI folks have been hard at work organizing. The data access model the propose is very clear. | |||
http://www.wtccc.org.uk/ - The Welcome Trust Case Control Consortium. | |||
== Some Relevant Papers == | == Some Relevant Papers == | ||
http://www.pubmedcentral.gov/picrender.fcgi?artid=1271381&blobtype=pdf - Recent large Parkinson's SNP association study. | http://www.pubmedcentral.gov/picrender.fcgi?artid=1271381&blobtype=pdf - Recent large Parkinson's SNP association study. | ||
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16642434&query_hl=11&itool=pubmed_DocSum - Some statistical techniques for studies involving related and unrelated people. | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16642434&query_hl=11&itool=pubmed_DocSum - Some statistical techniques for studies involving related and unrelated people. | ||
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16607626&query_hl=11&itool=pubmed_DocSum - Some theory of two stage study design for full-genome genotyping screens. | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16607626&query_hl=11&itool=pubmed_DocSum - Some theory of two stage study design for full-genome genotyping screens. | ||
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16607624&query_hl=11&itool=pubmed_DocSum - More statistical techniques, addressing in particular non-normal distribution of quantitative traits in patient populations. | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16607624&query_hl=11&itool=pubmed_DocSum - More statistical techniques, addressing in particular non-normal distribution of quantitative traits in patient populations. | ||
http://www.pubmedcentral.gov/picrender.fcgi?artid=1435944&blobtype=pdf - A web based tools for picking candidate SNPs to study for a particular disease. | http://www.pubmedcentral.gov/picrender.fcgi?artid=1435944&blobtype=pdf - A web based tools for picking candidate SNPs to study for a particular disease. | ||
http://www.biomedcentral.com/content/pdf/1471-2156-6-S1-S89.pdf - Examines utility of genome-wide linkage vs. SNP association studies in the context of a scan for alcoholism susceptibility. | http://www.biomedcentral.com/content/pdf/1471-2156-6-S1-S89.pdf - Examines utility of genome-wide linkage vs. SNP association studies in the context of a scan for alcoholism susceptibility. | ||
http://www.pubmedcentral.gov/picrender.fcgi?artid=1182008&blobtype=pdf - Looks at SNP linkage vs. microsatelites in a genomic scan for genes involve in rheumitoid arthritis. | http://www.pubmedcentral.gov/picrender.fcgi?artid=1182008&blobtype=pdf - Looks at SNP linkage vs. microsatelites in a genomic scan for genes involve in rheumitoid arthritis. | ||
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=15069025&query_hl=25&itool=pubmed_DocSum - 2 stage study of 552 affected/unaffected sibling pairs for multiple sclerosis. Stage 1 - 498 microsatilite markers, Stage 2 - more detailed genotyping in regions suggested by stage 1. | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=15069025&query_hl=25&itool=pubmed_DocSum - 2 stage study of 552 affected/unaffected sibling pairs for multiple sclerosis. Stage 1 - 498 microsatilite markers, Stage 2 - more detailed genotyping in regions suggested by stage 1. | ||
| Line 84: | Line 101: | ||
[[image:GenPhenTrack.gif]] | [[image:GenPhenTrack.gif]] | ||
The Genotype/Phenotype track would show information from a variety of studies. It would not (at least in the default modes) show subject-by-subject information. Instead it would show the 'marker association probability' at each of the positions assayed in the study. As depicted here it is showing the probability in a little bar graph. The horizontal baseline of the graph serves to link together all the positions assayed. I've grouped studies together using background, but I'm not sure if this will actually work in the genome browser context, and perhaps we could dispense with this. I'm sure it will be a struggle, as it has been with ENCODE, to come up with good 16 letter labels, but if we are able to do this, it should be clear enough. | The Genotype/Phenotype track would show information from a variety of studies. It would not (at least in the default modes) show subject-by-subject information. Instead it would show the 'marker association probability' at each of the positions assayed in the study. As depicted here it is showing the probability in a little bar graph. The horizontal baseline of the graph serves to link together all the positions assayed. I've grouped studies together using background, but I'm not sure if this will actually work in the genome browser context, and perhaps we could dispense with this. I'm sure it will be a struggle, as it has been with ENCODE, to come up with good 16 letter labels, but if we are able to do this, it should be clear enough. | ||
== Some Thoughts from Fan == | |||
=== Goal === | |||
Develop the UCSC website to become a major public resource that integrate the following data: | |||
* reference genomes | |||
** with associated genes, proteins, regulatory elements, etc. | |||
* genomic variations | |||
* genotype data | |||
* phenotype data | |||
** especially medically related data | |||
* associations of the above | |||
and provide and enable needed applications associated with the data above. | |||
=== Scope === | |||
UCSC’s role is mainly to integrate data from 3rd party sources. We do not have the resource to produce or manually curate primary data. The data we will host and create are top-level summary data and the associations between various data. For more detailed and domain specific data, we connect our users to the locus/domain/disease specific websites and databases via URL links. | |||
The focus is genotype and phenotype data and the associations among the data. We do not intend to build an all-encompassing general biological knowledge base. | |||
=== In a few years ... === | |||
In a few years, I envision UCSC website will have a comprehensive set of public known genomic variations of human and model organisms and their associations to known diseases, risk factors, and other quantitative and qualitative phenotype measures. | |||
Medical researchers (and possibly clinical doctors as well) can compare their own genotyping data of their patients or experimental subjects against our integrated genotype and phenotype databases to identify potential diseases/risk factors and clues for further research, experiments, or tests. Our VBLAT (Variation BLAT, coined by Heather) program will do just that, similar to BLAT today can instantaneously locate the genomic position of any given sequence. | |||
=== Data Model === | |||
This chapter will/should be substantially improved. | |||
==== General Approach ==== | |||
Because of our historical root, most of our existing data are designed to support the tracks of UCSC Genome Browser. Entities are typically defined by its genomic positions. | |||
As we move to represent and integrate genotype/phenotype/disease data, we need a data schema that enables broader aspects and other views of the biomedical data. The biological entities aligned to a base genome would be just one of the many views we will be enabling. Give some examples here: | |||
… | |||
==== E/R (Object Orientated) based data model ==== | |||
Based on my previous experience of data modeling to support CASE (Computer Aided Software Engineering), representing and automation of two major medical clinical guidelines, discussion with Heather on her idea of an object oriented data model, Jim’s simple and elegant data model that covers the entire Swiss-Prot database, reviewing the data model of pharmGKB, MGI, genPhen, etc., and our early prototype effort of DV/hgMut/GenPhen, I feel that at this early stage, we need to lay down a good data modeling foundation with simplicity and extensibility. | |||
I propose to develop our schema with an EAR (Entity, Attribute, and Relationship) approach, which seems quite similar to an object oriented approach. I firmly believe this foundation will bring clarity and efficiency to our database and application development efforts. | |||
The data will be stored in a relational database. MySQL is our choice. | |||
AutoSQL (improved) will be used to define the tables and generate data accessing templates in C (and possibly additional languages in the future). | |||
==== Key Entities ==== | |||
There will be a few key entities: | |||
· Individual | |||
· Variation | |||
· Phenotype | |||
· … | |||
Each entity will be implemented by a simple table, e.g. | |||
create table disease | |||
( | |||
id varchar(40), | |||
label varchar(40), | |||
description varchar(255), | |||
) | |||
The id field is mainly for system/programming usage purpose; it could either be automatically generated by the system, or using an industrial standard ID, e.g. MESH term ID. | |||
The label field is like the short label of our track, it is meant for human programmers (and users) to read. | |||
The description field has longer text that could be used for presentation for various applications, e.g. web pages or data mining reports. | |||
We may want to include an additional field, lastUpdated, to keep track of the time dimension. This will be further discussed in another section. | |||
==== Attributes ==== | |||
A simple design that is open-ended and can support unlimited possibilities. PhenCode already has a working example. Details of this section to be written. | |||
==== Views ==== | |||
The underlying schema of the new system will be quite normalized. To support application development, various VIEWs will be defined. MySQL 5.0 support VIEWS. | |||
Since VIEWs are defined by a selection statement, before we migrate to MySQL 5.0 from 4.0, we could proceed with our initial prototype development effort with tables generated by the same select statements that define the VIEWs . | |||
=== The Time Dimension === | |||
One main challenge of any modeling system is the time dimension. Knowledge changes as time goes by. | |||
The PharmGKB has a fairly elaborate design to track the time dimension. It seems to enable a user to get a view of the database at any specific time the user specifies. This may not be feasible for us because of the complexity to implement such a system and our current resource constraint. | |||
I propose that we follow our current approaches of the browser databases, i.e. our system represent a best effort snapshot of what are known. As time goes, we build another snapshot as a new release and keep an archive of previous releases to support our users community and minimize impact of version (time) changes. | |||
=== Data Security and Privacy === | |||
==== A showstopper or an opportunity? ==== | |||
When medical or genomic data of individual human beings are involved, the security and privacy issues become a major concern. Some of us felt that this challenge alone is such a huge hurdle that we should not pursue medical related projects. | |||
I recognize this issue as one of the major challenges in brining about the personalized medicine. But I also believe that this challenge will be resolved in time, just like the public fear and anxiety of Internet commerce a few years ago, which are mostly subsided. Like millions of others, I routinely buy books from Amazon; manage my bank accounts and medical insurance claims over the Internet. | |||
UCSC could actually lead an effort in this arena, if the group is willing to pursue it. Getting Paul excited, getting the group committed, and then get necessary funding seems to be three pieces we need to have to give it a shot. | |||
==== Adopt Existing Approaches ==== | |||
Should we choose not to lead, we could still follow. Stanford’s PharmGKB is a system that restricts user access to data related to individual patients. Its design is relatively simple that we may want to consider adopting, for our initial implementation. | |||
I heard NCBI has some more restrictive implementation/procedure. We need to understand how they are/will be doing it. | |||
The Framingham Heart Study project must have tons of relevant data. They must have been distributing their data to research groups for decades. We should try to get their data (at least as a test bed) to make sure that our system would be able to support data like theirs. We certainly can also learn a lot by understanding how they implement their controls. | |||
==== Be Careful ==== | |||
Before we have our own data security system up and running, we need to be very careful on how we handle sensitive data. A mistake in this arena could seriously damage our reputation and credibility. | |||
==== A secure server and a few authorized users ==== | |||
One possible short-term solution might be a separate secure server, with a small number of authorized users; each educated and signed necessary legal documents to gain appropriate rights to access sensitive data. | |||
On non-restrictive system(s), we could adopt Jim’s approach by generating fake data to enable the development effort. | |||
==== If we have more hands ==== | |||
If we get enough funding, it would be great to design and implement an integrated architecture that not only supports the following two: | |||
1. For non-restrictive public data, a completely open public server. | |||
2. A semi-public server that hosts somewhat restrictive data, accessible by legitimate and authorized users. | |||
but also support a 3rd scenario: | |||
3. The user has his restricted data either on his own PC or his own secure data server. The search/analysis results will process and present both his own data and public data from our server. | |||
It would be similar to our current custom track function, but hopefully the user’s private data stays at the local computer or the private secure server and would never make their trips to our central public server. | |||
=== How we go about it? === | |||
* Look at other existing systems to get better ideas on this domain and the schemas used. | |||
* Lay down a solid practical data model and | |||
** define additional necessary attributes and relationship between major entities. | |||
* Identify and integrate publicly available data. | |||
** solicitate collaborations with major DBs and LSDBs | |||
** consider providing a facility for public submission | |||
* Build necessary presentation and application (html, graphics and analysis) layer. | |||
In parallel of the above and for the short term, continue to develop the PhenCode prototype to generate public interest/awareness and enhance our chance of getting additional funding. PhenCode could also be a good testbed for the new architecture we are contemplating/implementing. | |||
=== Other Considerations === | |||
==== Medical Domain Expert ==== | |||
Currently our group does not have any medical domain expert. We need to identify/recruit a competent (an hopefully well-known) medical domain expert in residence. | |||
If this is not possible in near future, we should at least find MD(s) from UCSF and/or Stanford to collaborate closely. QB3 and our scientific advisory board members might help us establish some connections and/or recommend candidates. | |||
==== OMIM Enhancement ==== | |||
OMIM is currently the most comprehensive and authoritative public source (not in a traditional structured database format) that connects genomic variations to diseases. | |||
One of its major weaknesses is that a substantial part of the genomic positions of variations are imprecise or outdated. If this aspect of OMIM can be improved, together with its other content be extracted into a structured database, OMIM’s great value would be amplified even much more. | |||
It would require a huge manual effort to enhance OMIM to turn it into a structured format that is machine-readable and place all its variations accurately on the genomic map. I believe given enough funding, this could be done. UCSC might want to consider initiating a pilot project to prove its feasibility and then seek sufficient funding to complete the job. | |||
My understanding is that Belinda had done something on the OMIM data along this line for one or two LSDB. We might leverage her experience and insights. | |||
(Jasper Saris:) From a clinical molecular geneticists point the locus-specific-databases (LSdB) are more fruitfull to incorporate. Often they are moderated. The variants (normal variation, unclassified or proven pathogenic) are often pinpointed to the original article or submitter. In practice, one needs the original article(s) to be certain hoe seriously the pathogenecity has been proven. Quite often we need to set up correspondence on a certain mutation. The variatns in OMIM are mere to make the point why that gene is believed to be linked to a certain disease. The LUMC group of Johan den Dunnen has developed a free database to stimulate LSdB's in relation to his interest in HUGO and its HGVD site (Human Genome Variation in Disease). | |||
?? (Jasper Saris:) Where might I a propose a OMIM track witch lists genetic diseases and proven genes and/or regions. As like the QTL and GAP tracks. However, OMIM contains quite some genes without disease like from the period it was trying to incorporate all genes. These should be left out/filtered and the absence of disease causing variants could be used for that. | |||
==== A genomic standard as part of the U.S. personal electronic medical record standard ==== | |||
U.S. government is pushing to implement a nation wide electronic medical record standard. This effort is long overdue and seems quite promising. As a major player of the genomics world and as we tackle the challenge of integrating genotype and phenotype data, we should keep in mind and initiate some collaboration with the group(s) that shapes the emerging standard. | |||
A previous bioinformatics student of mine is working for a major medical record software company in the Bay area. He claims that his company is the dominating player with the largest market share (25%). We might want to establish further contact to explore the possibility that UCSC to develop the standard for the genotype data sub-section to enable the coming era of personalized medicine. | |||
[[Category:Technical FAQ]] | |||
Latest revision as of 10:22, 30 May 2008
Medical Sequencing Data
Some combination of phenotype and genotype ... Details unknown. Worthwhile to design ?
Major Types of information
- Subjects
- Anonymous individuals
- Family history
- Age
- Sex
- Ethnicity
- Pools of people
- Anonymous individuals
- Environmental data
- Location (zip code would be great)
- Medications they are taking
- Exercise, nutrition ...
- Genotype info
- Microarray based
- SNPs
- Haplotype blocks
- Copy number polymorphism
- Sequence based
- Random reads
- PCR products
- larger clones
- single haplotype vs. diploid
- Microarray based
- Phenotype
- Disease presence/absence or severity
- ADR - Adverse Drug Reaction
- Single physiological measure
- Enzyme activity, measure of amount of substance
- Parallel Measures
- Microarray measurements, etc...
Some Other Database Entities
- GenotypeTest - records what regions of genome probed.
- Study
- External URL
- Publications
- Contacts
- A group of subjects
- A set of phenotype and genotype tests
Existing Genotype/Phenotype Web Databases
http://www.pharmgkb.org/ - Requires registration for much data. Fan & Jim registered
http://globin.bx.psu.edu/genphen/ - Belinda, Ross and Webb's work, mostly covers hemoglobins
http://www.hgvs.org/dblist/dblist.html - Variation Databases and Related Sites, updated 3/31/06 on HGVS (Human Genome Variation Society)
http://www.ncbi.nlm.nih.gov/geo/ - NCBI GEO Gene Expression Omnibus [Gene expression is a phenotype]
- http://www.ncbi.nlm.nih.gov/geo/gds/gds_browse.cgi?gds=1332 Study on Huntington's Disease
http://genome-www5.stanford.edu/ - SMD Stanford Microarray Database.
Some Large Scale Studies Currently in Progress
http://www.framinghamheartstudy.org/heartstudy.asp - 500,000 SNP genotype data added to a 5000 person heart study that's been going on for three generations.
http://www.fnih.org/GAIN/GAIN_home.shtml - A public/private partnership Francis Collins and the NHGRI folks have been hard at work organizing. The data access model the propose is very clear.
http://www.wtccc.org.uk/ - The Welcome Trust Case Control Consortium.
Some Relevant Papers
http://www.pubmedcentral.gov/picrender.fcgi?artid=1271381&blobtype=pdf - Recent large Parkinson's SNP association study.
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16642434&query_hl=11&itool=pubmed_DocSum - Some statistical techniques for studies involving related and unrelated people.
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16607626&query_hl=11&itool=pubmed_DocSum - Some theory of two stage study design for full-genome genotyping screens.
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16607624&query_hl=11&itool=pubmed_DocSum - More statistical techniques, addressing in particular non-normal distribution of quantitative traits in patient populations.
http://www.pubmedcentral.gov/picrender.fcgi?artid=1435944&blobtype=pdf - A web based tools for picking candidate SNPs to study for a particular disease.
http://www.biomedcentral.com/content/pdf/1471-2156-6-S1-S89.pdf - Examines utility of genome-wide linkage vs. SNP association studies in the context of a scan for alcoholism susceptibility.
http://www.pubmedcentral.gov/picrender.fcgi?artid=1182008&blobtype=pdf - Looks at SNP linkage vs. microsatelites in a genomic scan for genes involve in rheumitoid arthritis.
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=15069025&query_hl=25&itool=pubmed_DocSum - 2 stage study of 552 affected/unaffected sibling pairs for multiple sclerosis. Stage 1 - 498 microsatilite markers, Stage 2 - more detailed genotyping in regions suggested by stage 1.
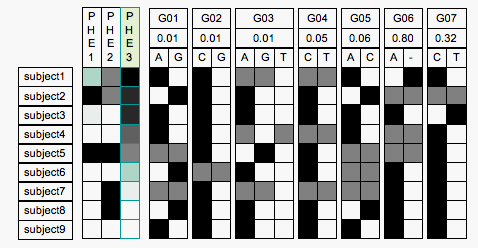
Mock Up of Phenotype Sorter
The Phenotype Sorter would be a web-based application aimed at presenting the full details of phenotype and genotype. The sorter has a line for each individual, and a column for each phenotype assayed in a study, and also a column for each genomic locus assayed in a study. The rows are sorted according the value of a selected phenotype, phe3 in the image above, which is indicated by a green hilight. (Ignore the green box in the first row of phe1, it seems to be an artifact of the conversion from TIF to GIF).
The genotype columns are divided into a subcolumn
for each allele, and at least for the simple nucleotide polymorphisms the alleles are labeled with the associated nucleotide. The number in the second row of the genotype label represents the strength of the locus as a marker for the phenotype. Possibly when sorting the rows by phenotype we should also sort the columns based on this number, though I was thinking of sorting the genotype columns just by position in genome initially.
Mock Up of Genotype/Phenotype Track
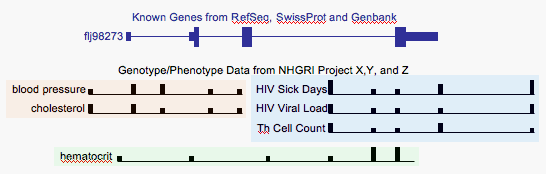 The Genotype/Phenotype track would show information from a variety of studies. It would not (at least in the default modes) show subject-by-subject information. Instead it would show the 'marker association probability' at each of the positions assayed in the study. As depicted here it is showing the probability in a little bar graph. The horizontal baseline of the graph serves to link together all the positions assayed. I've grouped studies together using background, but I'm not sure if this will actually work in the genome browser context, and perhaps we could dispense with this. I'm sure it will be a struggle, as it has been with ENCODE, to come up with good 16 letter labels, but if we are able to do this, it should be clear enough.
The Genotype/Phenotype track would show information from a variety of studies. It would not (at least in the default modes) show subject-by-subject information. Instead it would show the 'marker association probability' at each of the positions assayed in the study. As depicted here it is showing the probability in a little bar graph. The horizontal baseline of the graph serves to link together all the positions assayed. I've grouped studies together using background, but I'm not sure if this will actually work in the genome browser context, and perhaps we could dispense with this. I'm sure it will be a struggle, as it has been with ENCODE, to come up with good 16 letter labels, but if we are able to do this, it should be clear enough.
Some Thoughts from Fan
Goal
Develop the UCSC website to become a major public resource that integrate the following data:
- reference genomes
- with associated genes, proteins, regulatory elements, etc.
- genomic variations
- genotype data
- phenotype data
- especially medically related data
- associations of the above
and provide and enable needed applications associated with the data above.
Scope
UCSC’s role is mainly to integrate data from 3rd party sources. We do not have the resource to produce or manually curate primary data. The data we will host and create are top-level summary data and the associations between various data. For more detailed and domain specific data, we connect our users to the locus/domain/disease specific websites and databases via URL links.
The focus is genotype and phenotype data and the associations among the data. We do not intend to build an all-encompassing general biological knowledge base.
In a few years ...
In a few years, I envision UCSC website will have a comprehensive set of public known genomic variations of human and model organisms and their associations to known diseases, risk factors, and other quantitative and qualitative phenotype measures.
Medical researchers (and possibly clinical doctors as well) can compare their own genotyping data of their patients or experimental subjects against our integrated genotype and phenotype databases to identify potential diseases/risk factors and clues for further research, experiments, or tests. Our VBLAT (Variation BLAT, coined by Heather) program will do just that, similar to BLAT today can instantaneously locate the genomic position of any given sequence.
Data Model
This chapter will/should be substantially improved.
General Approach
Because of our historical root, most of our existing data are designed to support the tracks of UCSC Genome Browser. Entities are typically defined by its genomic positions.
As we move to represent and integrate genotype/phenotype/disease data, we need a data schema that enables broader aspects and other views of the biomedical data. The biological entities aligned to a base genome would be just one of the many views we will be enabling. Give some examples here:
…
E/R (Object Orientated) based data model
Based on my previous experience of data modeling to support CASE (Computer Aided Software Engineering), representing and automation of two major medical clinical guidelines, discussion with Heather on her idea of an object oriented data model, Jim’s simple and elegant data model that covers the entire Swiss-Prot database, reviewing the data model of pharmGKB, MGI, genPhen, etc., and our early prototype effort of DV/hgMut/GenPhen, I feel that at this early stage, we need to lay down a good data modeling foundation with simplicity and extensibility.
I propose to develop our schema with an EAR (Entity, Attribute, and Relationship) approach, which seems quite similar to an object oriented approach. I firmly believe this foundation will bring clarity and efficiency to our database and application development efforts.
The data will be stored in a relational database. MySQL is our choice.
AutoSQL (improved) will be used to define the tables and generate data accessing templates in C (and possibly additional languages in the future).
Key Entities
There will be a few key entities:
· Individual · Variation · Phenotype · …
Each entity will be implemented by a simple table, e.g.
create table disease (
id varchar(40), label varchar(40), description varchar(255),
)
The id field is mainly for system/programming usage purpose; it could either be automatically generated by the system, or using an industrial standard ID, e.g. MESH term ID.
The label field is like the short label of our track, it is meant for human programmers (and users) to read.
The description field has longer text that could be used for presentation for various applications, e.g. web pages or data mining reports.
We may want to include an additional field, lastUpdated, to keep track of the time dimension. This will be further discussed in another section.
Attributes
A simple design that is open-ended and can support unlimited possibilities. PhenCode already has a working example. Details of this section to be written.
Views
The underlying schema of the new system will be quite normalized. To support application development, various VIEWs will be defined. MySQL 5.0 support VIEWS.
Since VIEWs are defined by a selection statement, before we migrate to MySQL 5.0 from 4.0, we could proceed with our initial prototype development effort with tables generated by the same select statements that define the VIEWs .
The Time Dimension
One main challenge of any modeling system is the time dimension. Knowledge changes as time goes by.
The PharmGKB has a fairly elaborate design to track the time dimension. It seems to enable a user to get a view of the database at any specific time the user specifies. This may not be feasible for us because of the complexity to implement such a system and our current resource constraint.
I propose that we follow our current approaches of the browser databases, i.e. our system represent a best effort snapshot of what are known. As time goes, we build another snapshot as a new release and keep an archive of previous releases to support our users community and minimize impact of version (time) changes.
Data Security and Privacy
A showstopper or an opportunity?
When medical or genomic data of individual human beings are involved, the security and privacy issues become a major concern. Some of us felt that this challenge alone is such a huge hurdle that we should not pursue medical related projects.
I recognize this issue as one of the major challenges in brining about the personalized medicine. But I also believe that this challenge will be resolved in time, just like the public fear and anxiety of Internet commerce a few years ago, which are mostly subsided. Like millions of others, I routinely buy books from Amazon; manage my bank accounts and medical insurance claims over the Internet.
UCSC could actually lead an effort in this arena, if the group is willing to pursue it. Getting Paul excited, getting the group committed, and then get necessary funding seems to be three pieces we need to have to give it a shot.
Adopt Existing Approaches
Should we choose not to lead, we could still follow. Stanford’s PharmGKB is a system that restricts user access to data related to individual patients. Its design is relatively simple that we may want to consider adopting, for our initial implementation.
I heard NCBI has some more restrictive implementation/procedure. We need to understand how they are/will be doing it.
The Framingham Heart Study project must have tons of relevant data. They must have been distributing their data to research groups for decades. We should try to get their data (at least as a test bed) to make sure that our system would be able to support data like theirs. We certainly can also learn a lot by understanding how they implement their controls.
Be Careful
Before we have our own data security system up and running, we need to be very careful on how we handle sensitive data. A mistake in this arena could seriously damage our reputation and credibility.
A secure server and a few authorized users
One possible short-term solution might be a separate secure server, with a small number of authorized users; each educated and signed necessary legal documents to gain appropriate rights to access sensitive data.
On non-restrictive system(s), we could adopt Jim’s approach by generating fake data to enable the development effort.
If we have more hands
If we get enough funding, it would be great to design and implement an integrated architecture that not only supports the following two:
1. For non-restrictive public data, a completely open public server.
2. A semi-public server that hosts somewhat restrictive data, accessible by legitimate and authorized users.
but also support a 3rd scenario:
3. The user has his restricted data either on his own PC or his own secure data server. The search/analysis results will process and present both his own data and public data from our server.
It would be similar to our current custom track function, but hopefully the user’s private data stays at the local computer or the private secure server and would never make their trips to our central public server.
How we go about it?
- Look at other existing systems to get better ideas on this domain and the schemas used.
- Lay down a solid practical data model and
- define additional necessary attributes and relationship between major entities.
- Identify and integrate publicly available data.
- solicitate collaborations with major DBs and LSDBs
- consider providing a facility for public submission
- Build necessary presentation and application (html, graphics and analysis) layer.
In parallel of the above and for the short term, continue to develop the PhenCode prototype to generate public interest/awareness and enhance our chance of getting additional funding. PhenCode could also be a good testbed for the new architecture we are contemplating/implementing.
Other Considerations
Medical Domain Expert
Currently our group does not have any medical domain expert. We need to identify/recruit a competent (an hopefully well-known) medical domain expert in residence.
If this is not possible in near future, we should at least find MD(s) from UCSF and/or Stanford to collaborate closely. QB3 and our scientific advisory board members might help us establish some connections and/or recommend candidates.
OMIM Enhancement
OMIM is currently the most comprehensive and authoritative public source (not in a traditional structured database format) that connects genomic variations to diseases. One of its major weaknesses is that a substantial part of the genomic positions of variations are imprecise or outdated. If this aspect of OMIM can be improved, together with its other content be extracted into a structured database, OMIM’s great value would be amplified even much more.
It would require a huge manual effort to enhance OMIM to turn it into a structured format that is machine-readable and place all its variations accurately on the genomic map. I believe given enough funding, this could be done. UCSC might want to consider initiating a pilot project to prove its feasibility and then seek sufficient funding to complete the job.
My understanding is that Belinda had done something on the OMIM data along this line for one or two LSDB. We might leverage her experience and insights.
(Jasper Saris:) From a clinical molecular geneticists point the locus-specific-databases (LSdB) are more fruitfull to incorporate. Often they are moderated. The variants (normal variation, unclassified or proven pathogenic) are often pinpointed to the original article or submitter. In practice, one needs the original article(s) to be certain hoe seriously the pathogenecity has been proven. Quite often we need to set up correspondence on a certain mutation. The variatns in OMIM are mere to make the point why that gene is believed to be linked to a certain disease. The LUMC group of Johan den Dunnen has developed a free database to stimulate LSdB's in relation to his interest in HUGO and its HGVD site (Human Genome Variation in Disease).
?? (Jasper Saris:) Where might I a propose a OMIM track witch lists genetic diseases and proven genes and/or regions. As like the QTL and GAP tracks. However, OMIM contains quite some genes without disease like from the period it was trying to incorporate all genes. These should be left out/filtered and the absence of disease causing variants could be used for that.
A genomic standard as part of the U.S. personal electronic medical record standard
U.S. government is pushing to implement a nation wide electronic medical record standard. This effort is long overdue and seems quite promising. As a major player of the genomics world and as we tackle the challenge of integrating genotype and phenotype data, we should keep in mind and initiate some collaboration with the group(s) that shapes the emerging standard.
A previous bioinformatics student of mine is working for a major medical record software company in the Bay area. He claims that his company is the dominating player with the largest market share (25%). We might want to establish further contact to explore the possibility that UCSC to develop the standard for the genotype data sub-section to enable the coming era of personalized medicine.