Mm9 multiple alignment: Difference between revisions
mNo edit summary |
|||
| Line 686: | Line 686: | ||
Z(1) increment between successive words in sequence 1. | Z(1) increment between successive words in sequence 1. | ||
</PRE> | </PRE> | ||
<TABLE BORDER=0> | |||
<TR><TD>The default scoring matrix is:</TD><TD WIDTH=42> </TD> | |||
<TD>The HoxD55 scoring matrix is:</TD></TR> | |||
<TR><TD> | |||
<TABLE BORDER=1> | |||
<TR><TH> </TH><TH>A</TH><TH>C</TH><TH>G</TH><TH>T</TH></TR> | |||
<TR><TH>A</TH><TD>91</TD><TD>-114</TD><TD>-31</TD><TD>-123</TD></TR> | |||
<TR><TH>C</TH><TD>-114</TD><TD>100</TD><TD>-125</TD><TD>-31</TD></TR> | |||
<TR><TH>G</TH><TD>-31</TD><TD>-125</TD><TD>100</TD><TD>-114</TD></TR> | |||
<TR><TH>T</TH><TD>-123</TD><TD>-31</TD><TD>-114</TD><TD>91</TD></TR> | |||
</TABLE> | |||
</TD><TD WIDTH=42> </TD><TD><TABLE BORDER=1> | |||
<TR><TH> </TH><TH>A</TH><TH>C</TH><TH>G</TH><TH>T</TH></TR> | |||
<TR><TH>A</TH><TD>91</TD><TD>-90</TD><TD>-25</TD><TD>-100</TD></TR> | |||
<TR><TH>C</TH><TD>-90</TD><TD>100</TD><TD>-100</TD><TD>-25</TD></TR> | |||
<TR><TH>G</TH><TD>-25</TD><TD>-100</TD><TD>100</TD><TD>-90</TD></TR> | |||
<TR><TH>T</TH><TD>-100</TD><TD>-25</TD><TD>-90</TD><TD>91</TD></TR> | |||
</TABLE> | |||
</TD></TR></TABLE> | |||
==matrix parameters== | ==matrix parameters== | ||
Revision as of 21:26, 20 August 2007
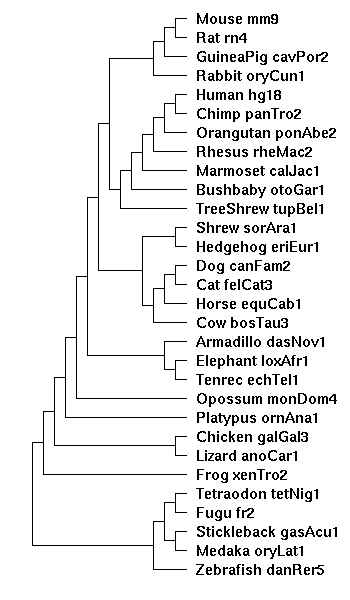
Mouse Mm9 multiple alignment/conservation track
|
To avoid artifacts in downstream processing of the UCSC multiple alignments, it is important to be careful on the use of the parameters used in the blastz processing pipeline. There are a number of steps in the pipeline and a variety of tunable parameters involved. This page will track the various parameters used in the alignments as they proceed toward the completion of a multiple alignment conservation track on the mm9 mouse (NCBI build 37) assembly |
axtChain parameters and end results
| name db | chrom count (*) |
genome size |
tree distance |
axtChain minScore |
axtChain linearGap |
% of mm9 matched |
% of other matched by mm9 |
done |
|---|---|---|---|---|---|---|---|---|
| rat rn4 | 21 | 2702 Mb | 0.160657 | 3000 | medium | 68.357 | 69.541 | 16 August |
| human hg18 | 24 | 2963 Mb | 0.452619 | 3000 | medium | 38.499 | 35.201 | 16 August |
| Rhesus rheMac2 | 22 | 2731 Mb | 0.452745 | 3000 | medium | xx.123 | xx.456 | tbd |
| Orangutan ponAbe1 | 79553 | 3090 Mb | 0.453809 | 3000 | medium | xx.123 | xx.456 | tbd |
| Marmoset calJac1 | 49724 | 2889 Mb | 0.454272 | 3000 | medium | xx.123 | xx.456 | tbd |
| Chimp panTro2 | 25 | 3028 Mb | 0.454514 | 3000 | medium | xx.123 | xx.456 | tbd |
| GuineaPig cavPor2 | 295514 | 3246 Mb | 0.479871 | 3000 | medium | xx.123 | xx.456 | tbd |
| Horse equCab1 | 32 | 1961 Mb | 0.479871 | 3000 | medium | xx.123 | xx.456 | tbd |
| TreeShrew tupBel1 | 150851 | 3491 Mb | 0.494934 | 3000 | medium | xx.123 | xx.456 | tbd |
| Bushbaby otoGar1 | 120882 | 3261 Mb | 0.498957 | 3000 | medium | xx.123 | xx.456 | tbd |
| Armadillo dasNov1 | 304391 | 3678 Mb | 0.517360 | 3000 | medium | xx.123 | xx.456 | tbd |
| Rabbit oryCun1 | 215471 | 3303 Mb | 0.519779 | 3000 | medium | xx.123 | xx.456 | tbd |
| Cat felCat3 | 217790 | 3858 Mb | 0.530610 | 3000 | medium | xx.123 | xx.456 | tbd |
| Dog canFam2 | 39 | 2331 Mb | 0.533544 | 3000 | medium | xx.123 | xx.456 | tbd |
| Elephant loxAfr1 | 233134 | 3535 Mb | 0.536627 | 3000 | medium | xx.123 | xx.456 | tbd |
| Cow bosTau3 | 30 | 2321 Mb | 0.540852 | 3000 | medium | xx.123 | xx.456 | tbd |
| Hedgehog eriEur1 | 379801 | 3211 Mb | 0.632457 | 3000 | medium | xx.123 | xx.456 | tbd |
| Shrew sorAra1 | 262057 | 2800 Mb | 0.658734 | 3000 | medium | xx.123 | xx.456 | tbd |
| Tenrec echTel1 | 325491 | 3646 Mb | 0.666303 | 3000 | medium | xx.123 | xx.456 | tbd |
| Opossum monDom4 | 9 | 3272 Mb | 0.909852 | 5000 | loose | xx.123 | xx.456 | tbd |
| Platypus ornAna1 | 201522 | 1904 Mb | 1.165888 | 5000 | loose | xx.123 | xx.456 | tbd |
| Chicken galGal3 | 33 | 984 Mb | 1.285399 | 5000 | loose | xx.123 | xx.456 | tbd |
| Lizard anoCar1 | 7233 | 1699 Mb | 1.404225 | 5000 | loose | xx.123 | xx.456 | tbd |
| X. tropicalis xenTro2 | 19759 | 1443 Mb | 1.726205 | 5000 | loose | xx.123 | xx.456 | tbd |
| Stickleback gasAcu1 | 21 | 382 Mb | 2.012649 | 5000 | loose | xx.123 | xx.456 | tbd |
| Zebrafish danRer4 | 25 | 1475 Mb | 2.027153 | 5000 | loose | xx.123 | xx.456 | tbd |
| Tetraodon tetNig1 | 21 | 207 Mb | 2.051015 | 5000 | loose | xx.123 | xx.456 | tbd |
| Fugu fr2 | 1 | 381 Mb | 2.086669 | 5000 | loose | xx.123 | xx.456 | tbd |
| Medaka oryLat1 | 24 | 690 Mb | 2.200402 | 5000 | loose | xx.123 | xx.456 | tbd |
(*) chrom count does not include haplotypes, chr*_random, chrUn or chrM unless chrUn or scaffolds are the only sequences for that assembly.
chrom size has the same limitation as the chrom count, no randoms.
Tree distances are from the hg18 28-way measurements, with ponAbe1 and calJac1 manually inserted into the tree.
blastz alignment parameters details
| query | abridged repeats |
M | K | L | Q | Y |
|---|---|---|---|---|---|---|
| Rat rn4 | yes | 40M | 3K | 3K | dflt | dflt |
| Human hg18 | yes | 40M | 3K | 3K | dflt | dflt |
| Rhesus rheMac2 | no | 40M | 3K | 3K | dflt | dflt |
| Orangutan ponAbe1 | no | 50 | 3K | 3K | dflt | dflt |
| Marmoset calJac1 | no | 50 | 3K | 3K | dflt | dflt |
| Chimp panTro2 | yes | 40M | 3K | 3K | dflt | dflt |
| GuineaPig cavPor2 | no | 50 | 3K | 3K | dflt | dflt |
| Horse equCab1 | no | 40M | 3K | 3K | dflt | dflt |
| TreeShrew tupBel1 | no | 50 | 3K | 3K | dflt | dflt |
| Bushbaby otoGar1 | no | 50 | 3K | 3K | dflt | dflt |
| Armadillo dasNov1 | no | 50 | 3K | 3K | dflt | dflt |
| Rabbit oryCun1 | no | 50 | 3K | 3K | dflt | dflt |
| Cat felCat3 | no | 50 | 3K | 3K | dflt | dflt |
| Dog canFam2 | yes | 40M | 3K | 3K | dflt | dflt |
| Elephant loxAfr1 | no | 50 | 3K | 3K | dflt | dflt |
| Cow bosTau3 | no | 40M | 3K | 3K | dflt | dflt |
| Hedgehog eriEur1 | no | 50 | 3K | 3K | dflt | dflt |
| Shrew sorAra1 | no | 50 | 3K | 3K | dflt | dflt |
| Tenrec echTel1 | no | 50 | 3K | 3K | dflt | dflt |
| Opossum monDom4 | no | 50 | 2200 | 6000 | HoxD55 | 3400 |
| Platypus ornAna1 | no | 50 | 2200 | 6000 | HoxD55 | 3400 |
| Chicken galGal3 | yes | 40M | 2200 | 6000 | HoxD55 | 3400 |
| Lizard anoCar1 | no | 50 | 2200 | 6000 | HoxD55 | 3400 |
| X_tropicalis xenTro2 | no | 50 | 2200 | 6000 | HoxD55 | 3400 |
| Stickleback gasAcu1 | no | 40M | 2200 | 6000 | HoxD55 | 3400 |
| Zebrafish danRer4 | yes | 40M | 2200 | 6000 | HoxD55 | 3400 |
| Tetraodon tetNig1 | no | 40M | 2200 | 6000 | HoxD55 | 3400 |
| Fugu fr2 | no | 40M | 2200 | 6000 | HoxD55 | 3400 |
| Medaka oryLat1 | no | 40M | 2200 | 6000 | HoxD55 | 3400 |
default blastz parameters
m=80 v=0 B=2 C=0 E=30 G=0 H=0 K=3000 L=K
M=0 O=400 P=1 R=0 T=1 W=8 X=10*(A-to-A match score)
Y=O+300*E Z=1
From the blastz usage message:
Default values are given in parentheses.
m(80M) bytes of space for trace-back information
v(0) 0: quiet; 1: verbose progress reports to stderr
B(2) 0: single strand; >0: both strands
C(0) 0: no chaining; 1: just output chain; 2: chain and extend;
3: just output HSPs
E(30) gap-extension penalty.
G(0) diagonal chaining penalty.
H(0) interpolate between alignments at threshold K = argument.
K(3000) threshold for MSPs
L(K) threshold for gapped alignments
M(0) mask any base in seq1 hit this many times; 0 = no dynamic masking
O(400) gap-open penalty.
P(1) 0: entropy not used; 1: entropy used; >1 entropy with feedback.
Q load the scoring matrix from a file.
R(0) antidiagonal chaining penalty.
T(1) 0: W-bp words; 1: 12of19; 2: 12of19 without transitions.
3: 14of22; 4: 14of22 without transitions.
W(8) word size (unused unless T=0)
X(10*(A-to-A match score)) X-drop parameter for ungapped extension.
Y(O+300E) X-drop parameter for gapped extension.
Z(1) increment between successive words in sequence 1.
| The default scoring matrix is: | The HoxD55 scoring matrix is: | |||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
matrix parameters
The "medium" gap score matrix, tuned for the mouse-human distance is:
tableSize 11 smallSize 111 position 1 2 3 11 111 2111 12111 32111 72111 152111 252111 qGap 350 425 450 600 900 2900 22900 57900 117900 217900 317900 tGap 350 425 450 600 900 2900 22900 57900 117900 217900 317900 bothGap 750 825 850 1000 1300 3300 23300 58300 118300 218300 318300
The "loose" gap score matrix, tuned for the chicken-human distance is:
tablesize 11 smallSize 111 position 1 2 3 11 111 2111 12111 32111 72111 152111 252111 qGap 325 360 400 450 600 1100 3600 7600 15600 31600 56600 tGap 325 360 400 450 600 1100 3600 7600 15600 31600 56600 bothGap 625 660 700 750 900 1400 4000 8000 16000 32000 57000