Opsin evolution: Difference between revisions
Tomemerald (talk | contribs) mNo edit summary |
Tomemerald (talk | contribs) |
||
| (216 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
'''See also:''' [[Opsin_evolution:_LWS_PhyloSNPs|LWS]] | [[Opsin_evolution:_Encephalopsin_gene_loss|Encephalopsins]] | [[Opsin_evolution:_Melanopsin_gene_loss|Melanopsins]] | [[Opsin_evolution:_Neuropsin_phyloSNPs|Neuropsin]] | [[Opsin_evolution:_Peropsin_phyloSNPs|Peropsin]] | [[Opsin_evolution:_RGR_phyloSNPs|RGR phyloSNPs]] | [[Opsin_evolution:_update_blog|Update Blog]] | |||
== Introduction to Opsin Evolution == | == Introduction to Opsin Evolution == | ||
| Line 5: | Line 7: | ||
Below is the largest set of phylogenetically dispersed hand-curated opsin sequences ever assembled. The sequences are organized into true orthology classes using coding indels, intron location and phase, synteny of flanking genes, diagnostic residues, blast clustering, and experimental characterization when available. The reference set of opsin sequences includes selected GenBank entries but mostly new opsins extracted from dozens of newly completed genome projects. | Below is the largest set of phylogenetically dispersed hand-curated opsin sequences ever assembled. The sequences are organized into true orthology classes using coding indels, intron location and phase, synteny of flanking genes, diagnostic residues, blast clustering, and experimental characterization when available. The reference set of opsin sequences includes selected GenBank entries but mostly new opsins extracted from dozens of newly completed genome projects. | ||
The set serves as a gene family classifier ... just [http://www.proweb.org/proweb/Tools/WU-blast.html uBlast] an unknown candidate opsin against the full database below and look for consistent | The set serves as a gene family classifier ... just [http://www.proweb.org/proweb/Tools/WU-blast.html uBlast] an unknown candidate opsin against the full database below and look for consistent labeling of the top hits from the <span style="color: #990099;">Opsin Classifier</span>. Then validate the apparent orthology class using three independent classes of rare genomic events (indels, introns, signature residues) by including the new sequence in a full-width [http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html multalignment], being sure to include a couple dozen non-opsin GPCRs as controls. Then check for residual bilateral flanking gene adjacency when a genome assembly is available. The whole process takes only 15 seconds per query! | ||
The set of reference sequences is deliberately not exhaustive -- that seriously | The set of reference sequences is deliberately not exhaustive -- that seriously over-samples in popular experimental clades such as vertebrates and insects. When a given clade has many similar sequences available, those in species with genome assemblies are chosen to represent the group, for example anole is preferred to gecko, and (rightly or wrongly) any experimental results from gecko transfered over. This avoids the uninformative clutter of near-identical sequences. However if a clade reflects a very deep divergence especially important to opsin evolution (such as lamprey or amphioxus), all available sequences are needed to maximally break up long branches. | ||
Sequences not available from GenBank were culled from trace archives, tiled contigs, and genome assemblies, typically by uBlastx against the growing set of reference sequences, as described in the [[Opsin_evolution:_annotation_tricks|annotation section]]. The level of error in the curated sequences is very low, declines with time as anomalies are revisited and repaired, but never reaches zero because of problems inherent to experimental data, imperfect sequencing reads, less-than-complete assemblies, and sequence manipulation. | Sequences not available from GenBank were culled from trace archives, tiled contigs, and genome assemblies, typically by uBlastx against the growing set of reference sequences, as described in the [[Opsin_evolution:_annotation_tricks|annotation section]]. The level of error in the curated sequences is very low, declines with time as anomalies are revisited and repaired, but never reaches zero because of problems inherent to experimental data, imperfect sequencing reads, less-than-complete assemblies, and sequence manipulation. | ||
| Line 13: | Line 15: | ||
[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=17984227,17975064 Rare genomic changes] can supplement (and even displace) traditional maximal likelihood and bayesian inference in resolving polytomic divergence nodes in gene and species tree topologies. Rare genomic changes applicable to opsins include coding indels (deletions and insertions), intron placement (position and phase comparison), synteny (gene order along the chromosome), and gene copy number change (gene gain from retropositional, tandem, segmental, and whole genome duplications; gene loss from pseudogenization or deletion). Results from these methods must be evaluated for their susceptibility to homoplasy (misleading recurrent independent events that mimic a single event) and incomplete penetration in the population level at the time of speciation (lineage sorting). | [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=17984227,17975064 Rare genomic changes] can supplement (and even displace) traditional maximal likelihood and bayesian inference in resolving polytomic divergence nodes in gene and species tree topologies. Rare genomic changes applicable to opsins include coding indels (deletions and insertions), intron placement (position and phase comparison), synteny (gene order along the chromosome), and gene copy number change (gene gain from retropositional, tandem, segmental, and whole genome duplications; gene loss from pseudogenization or deletion). Results from these methods must be evaluated for their susceptibility to homoplasy (misleading recurrent independent events that mimic a single event) and incomplete penetration in the population level at the time of speciation (lineage sorting). | ||
Opsins are more informatively stored as proteins since nucleotide sequences are far too diverged at metazoan evolutionary distances and do not explicitly manifest residue properties that experience selection. These protein sequences are parsed into constituent exons using genomic information when available -- fortunately splicing mechanisms are exceeding well conserved across animal phyla. When not directly available (eg the opsin originated as a cDNA in a species lacking a genome project), exon breaks have been inferred from the phylogenetically closest neighbors via | Opsins are more informatively stored as proteins since nucleotide sequences are far too diverged at metazoan evolutionary distances and do not explicitly manifest residue properties that experience selection. These protein sequences are parsed into constituent exons using genomic information when available -- fortunately splicing mechanisms are exceeding well conserved across animal phyla. When not directly available (eg the opsin originated as a cDNA in a species lacking a genome project), exon breaks have been inferred from the phylogenetically closest neighbors via heterologous alignment (but not in insects where intron turnover is too high). As an example, lamprey opsins from Geotria australis and Lethenteron japonicum can work as blastn queries to locate orthologs within the Petromyzon maritimus genome project (which consists solely of 19 million traces as of mid-December 2007).Numbers flanking exons, namely 0 1 2, show the [[Opsin_evolution:_annotation_tricks|phasing]] of each intron, eg 12 means an overhang of 1 bp at the 3' end of an exon with fragmentary codon completed by a 2 bp overhang at the beginning of the next exon. | ||
Intron position and phasing are conserved over vast evolutionary distances -- human to sponge and beyond -- with the exception of certain rogue lineages (that unfortunately includes two major model organisms). Informative conservation is still available long after protein percent identity has slid into the twilight zone of uncertainty (critical in opsins because they are readily confused with generic GPCR receptors). For example, no variation whatsoever in intron pattern has | Intron position and phasing are conserved over vast evolutionary distances -- human to sponge and beyond -- with the exception of certain rogue lineages (that unfortunately includes two major model organisms). Informative conservation is still available long after protein percent identity has slid into the twilight zone of uncertainty (critical in opsins because they are readily confused with generic GPCR receptors). For example, no variation whatsoever in intron pattern has occurred in any vertebrate opsin class since lamprey divergence, no events in many billions of years of branch length. Prior to that, rare events are observed, for example LWS opsins gained an extra intron of phase 12. In a protein having 333 locations with 3 possible phases at each location, convergent evolution (homoplasy) is uncommon, that is, [[Opsin_evolution:_ancestral_introns|close examination]] establishes opsin introns are still highly informative even when sequences have greatly diverged. | ||
Insertions and deletions ([[Opsin_evolution:_informative_indels|coding indels]]) are sufficiently uncommon in opsins that they are a potentially phylogenetically informative class of rare genomic events. We'll harvest these from a [[Opsin_evolution:_alignment|massive opsin alignment]] and stratify by location and phylogenetic depth. Indels are more subject to homoplasy than intron gain or loss but the risk of that varies greatly by region. Indels are less affected by lineage-specific rates than other event classes. | Insertions and deletions ([[Opsin_evolution:_informative_indels|coding indels]]) are sufficiently uncommon in opsins that they are a potentially phylogenetically informative class of rare genomic events. We'll harvest these from a [[Opsin_evolution:_alignment|massive opsin alignment]] and stratify by location and phylogenetic depth. Indels are more subject to homoplasy than intron gain or loss but the risk of that varies greatly by region. Indels are less affected by lineage-specific rates than other event classes. | ||
Syntentic relationships can have great value in determining orthology relationships, though gene order is not often conserved over great evolutionary timescales due to chromosomal rearrangements. As with introns, certain rogue lineages lose almost all this information. Tracking synteny is nomenclaturally confused because many genes are unnamed and simply numbered by annotation pipeline procedures in each genome without homological consistency. Here, HUGO-convention names are used for two genes flanking each side of a given human opsin. Strand orientation is noted relative to a fixed convention of plus strand for the human opsin. Then each genome assembly is visited to determine the extent of conservation of these flanking genes and orientation. In the event humans lack the gene, synteny is defined by the nearest diverging species, typically platypus or chicken, that has the gene. Sometimes the original synteny is only partly retained (left or right half-synteny). For deeply diverging species such as amphioxus, flanking genes can be are pushed forward into 'nearby' species. This can also be done without an assembly in species with sufficiently large contigs containing the opsin. Blast clustering can be uncertain because of diminishing percent identity or even loss of these flanking genes; common | Syntentic relationships can have great value in determining orthology relationships, though gene order is not often conserved over great evolutionary timescales due to chromosomal rearrangements. As with introns, certain rogue lineages lose almost all this information. Tracking synteny is nomenclaturally confused because many genes are unnamed and simply numbered by annotation pipeline procedures in each genome without homological consistency. Here, HUGO-convention names are used for two genes flanking each side of a given human opsin. Strand orientation is noted relative to a fixed convention of plus strand for the human opsin. Then each genome assembly is visited to determine the extent of conservation of these flanking genes and orientation. In the event humans lack the gene, synteny is defined by the nearest diverging species, typically platypus or chicken, that has the gene. Sometimes the original synteny is only partly retained (left or right half-synteny). For deeply diverging species such as amphioxus, flanking genes can be are pushed forward into 'nearby' species. This can also be done without an assembly in species with sufficiently large contigs containing the opsin. Blast clustering can be uncertain because of diminishing percent identity or even loss of these flanking genes; common chromosomal displacements such as inversions eventually disrupt most syntentic arrangements. | ||
The fasta header of each sequence serves as a miniature database that conveniently collects this and other basic information, with fields showing the opsin type, genus, species and common name, accession number, best PubMed citation, indels, intron pattern, sequence length, lambda max adsorption, flanking synteny, and G protein type with which it interacts (all subject to availability and work-in-progress). These fasta headers by themselves provide a quick over view the opsin reference set collection -- simply paste into a blank document and pull lines containing '>'. | The fasta header of each sequence serves as a miniature database that conveniently collects this and other basic information, with fields showing the opsin type, genus, species and common name, accession number, best PubMed citation, indels, intron pattern, sequence length, lambda max adsorption, flanking synteny, and G protein type with which it interacts (all subject to availability and work-in-progress). These fasta headers by themselves provide a quick over view the opsin reference set collection -- simply paste into a blank document and pull lines containing '>'. | ||
| Line 25: | Line 27: | ||
Thus the usual querying at GenBank does not remotely compare to the <span style="color: #990099;">Opsin Classifier</span>. Those sequences -- often mis-annotated by an unattended pipeline or well-intentioned individual with no qualifications in comparative genomics -- are spread out over separate databases not accessible by any single method, with difficult-to-interpret edge creep of genomic blast matches, uncorrected frameshifts, missing stop codons and erroneous amino terminals. | Thus the usual querying at GenBank does not remotely compare to the <span style="color: #990099;">Opsin Classifier</span>. Those sequences -- often mis-annotated by an unattended pipeline or well-intentioned individual with no qualifications in comparative genomics -- are spread out over separate databases not accessible by any single method, with difficult-to-interpret edge creep of genomic blast matches, uncorrected frameshifts, missing stop codons and erroneous amino terminals. | ||
As worst-case scenario, half-baked annotation of the sea urchin genome by pipelines and casual procedures has left a awful legacy of bogus opsin gene structures at GenBank, journal alignments, and genome browsers -- often mis-classified because of an inadequate set of reference sequences and non-opsin GPCR controls, chimeric confusion in tandem duplicates, and non-consideration of intron structure, indels, and synteny. These errors may ripple downstream to errors by subsequent scientists trusting GenBank nr as classifier and lead to a whole subculture taken up with non-existent "virtual" opsins and | As worst-case scenario, half-baked annotation of the sea urchin genome by pipelines and casual procedures has left a awful legacy of bogus opsin gene structures at GenBank, journal alignments, and genome browsers -- often mis-classified because of an inadequate set of reference sequences and non-opsin GPCR controls, chimeric confusion in tandem duplicates, and non-consideration of intron structure, indels, and synteny. These errors may ripple downstream to errors by subsequent scientists trusting GenBank nr as classifier and lead to a whole subculture taken up with non-existent "virtual" opsins and attendant vacuous speculation on echinoderm photoreception. | ||
Melanopsins, the unexpected rhabdomeric-class Gq-coupled opsin recently found in upper deuterostomes, illustrate some of the difficulties of accurate annotation. They can be confused homologically due to various expansions and contractions. Mammals, human through platypus, have a single melanopsin. However a common ancestor to chicken, lizard, frog, teleost fish, and possibly | Melanopsins, the unexpected rhabdomeric-class Gq-coupled opsin recently found in upper deuterostomes, illustrate some of the difficulties of accurate annotation. They can be confused homologically due to various expansions and contractions. Mammals, human through platypus, have a single melanopsin. However a common ancestor to chicken, lizard, frog, teleost fish, and possibly cartilaginous fish had a multi-gene segmental duplication with both resulting melanopsins retained (though substantially diverged). In ray-finned fish, a processed retrogene arose that may be functional in zebrafish though lost in fugu and stickleback. After whole genome duplication, zebrafish also retained two copies of the original melanopsin. Chondrichthyes also have a second copy of the primary melanopsin but synteny -- which is essential for analysis since intron placement is uninformative in duplications and sequence alignment too dependent on unknown rates -- is not available in the current contig-level assembly. | ||
Amphioxus contains two melanopsins from an apparently independent duplication. Flanking gene order today bears no relation to any vertebrate gene order. The lamprey situation awaits assembly of its traces or targeted transcript studies. At this time, only a four-exon fragmentary melanopsin can be recovered (however with high percent identity, 80%). Possibly orthologs of this melanopsin locus could be tracked into the highly derived tunicates, acorn worm, and sea urchins. The distinctive intron pattern may even allow melanopsin antecedents to be identified in Cnidaria and Protostomia. At this point, the best blastp match to insects stands at 37% with no evident syntenic or intronic support. | Amphioxus contains two melanopsins from an apparently independent duplication. Flanking gene order today bears no relation to any vertebrate gene order. The lamprey situation awaits assembly of its traces or targeted transcript studies. At this time, only a four-exon fragmentary melanopsin can be recovered (however with high percent identity, 80%). Possibly orthologs of this melanopsin locus could be tracked into the highly derived tunicates, acorn worm, and sea urchins. The distinctive intron pattern may even allow melanopsin antecedents to be identified in Cnidaria and Protostomia. At this point, the best blastp match to insects stands at 37% with no evident syntenic or intronic support. | ||
While clade-specific proliferations of melanopsins -- and implied role subfunctionalizations -- confounds the situation for chordates, it has little impact on the opsin classifier described here. Unknown sequences readily find their place because of the extensive phylogenetic representation of reference sequence orthology classes and the inherent distance of melanopsins from the ciliary | While clade-specific proliferations of melanopsins -- and implied role subfunctionalizations -- confounds the situation for chordates, it has little impact on the opsin classifier described here. Unknown sequences readily find their place because of the extensive phylogenetic representation of reference sequence orthology classes and the inherent distance of melanopsins from the ciliary sub-collection. At that level of alignment, the melanopsins serve as outgroup to ciliary opsins and so help define motifs specific to Gt-coupled signaling and other structure/function issues. | ||
It appears however that far too much 'lumping' has taken place in nearly all non-imaging opsins, for example encephalopsins, melanopsins, and peropsins. The taxonomic counterpart is too much lumping of species outside of mammal. In all likelihood, additional opsin orthology classes need definition and distinctive naming. These opsins were belatedly discovered through whole-genome homology searches; they may differ one from another just as much as cone and rod opsins. However, it is currently difficult to disentangle phylogenetically short-lived expansions within known families from deep-rooted parallel-evolving subfamilies concentrated (superficially) within 'secondary' photoreception systems. Genome sampling density and completion efforts are overwhelmingly concentrated in a few model species and near human. | |||
We should not be too hasty to write off peropsins as mere auxiliary retinal isomerases that replenish cis-retinal for 'real' opsins. This reaction is not a simple isomerization (like glucose isomerase) but photoisomerization with an evolutionarily tuned and conserved visual light action spectrum. There is no reason that trans-retinal cannot be the signaling agonist with conversion to all-cis retinal the waste product. Pairing two opposing types of opsins in a single cell or nearby cells then completes a full visual cycle with interesting opportunities for sensory capabilities. Of all the so-called retinal isomerases, peropsins retain more of the diagnostic residues of ciliary opsins; possibly they function similarly but in balance with another opsin class co-expressed in the same cell. Melanopsin, surprisingly, is fully capable of self-replenishment, so perhaps the first-studied ciliary opsins are the true anomaly with their need for an auxiliary replenishment cycle in the retinal epithelium. | |||
It's abundantly clear from distinct ancestral introns and alignment clustering that peropsins together with neuropsins and RGR opsins comprise a distinctive subclade within the overall opsin gene tree. The comparative genomics of RGR illustrates the danger of phylogenetic under-sampling: with much deeper sampling, we can observe that the E/DRY motif -- conserved across all classes of opsins (indeed GPCR) and critical to maintaining non-signaling conformation -- has become GRY in all boreoeutheran mammals (it's ERY in afrotheres, lost without residual debris in marsupials, and DRY from platypus through shark and even tunicate, and DRY in neuropsins and peropsins). | |||
In other words, after several trillion years of branch length conservation as charged amino acid, a radical amino acid substitution has taken hold -- to glycine with its tiny non-interactive side chain. Yet this subclade of placental mammals, this glycine has been conserved without exception for over two billion years of branch length. Given the importance of this motif for maintaining the non-signaling state, this suggests a major change in functional properties of RGR opsins within boreoeutheres, a change that does not tolerate a reduced alphabet at this site. That change might be breakdown to non-signaling isomerase within that clade. It remains unclear how marsupials cope without the gene at all or how Afrothera and Xenarthra visual cycles differ from other placental mammals. | |||
Comparative Genomics of DRY motifs in exon 3 of RGR Opsins: | Comparative Genomics of DRY motifs in exon 3 of RGR Opsins: | ||
1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAW<font color="magenta">G</font>RYHHYCT 1 human | 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAW<font color="magenta">G</font>RYHHYCT 1 human | ||
| Line 78: | Line 81: | ||
Excellent recent publications have greatly added to our understanding of the evolutionary origin of light reception capabilities, yet only with the advent of genomic sequencing have we begun to get a full grip on opsins and a few other evo-devo photoreceptor components such as PAX6. Yet critical early-diverging metazoan genomes that might retain information on ancestral characters are greatly under-represented given the branch lengths involved. No purpose is served by elaborate gene tree computations that include a single token peropsin or parietopsin among hundreds of cone opsins; if opsins are central to the endeavor yet so haphazardly annotated, what is the status of dozens of lesser but still critical photoreceptor gene families? | Excellent recent publications have greatly added to our understanding of the evolutionary origin of light reception capabilities, yet only with the advent of genomic sequencing have we begun to get a full grip on opsins and a few other evo-devo photoreceptor components such as PAX6. Yet critical early-diverging metazoan genomes that might retain information on ancestral characters are greatly under-represented given the branch lengths involved. No purpose is served by elaborate gene tree computations that include a single token peropsin or parietopsin among hundreds of cone opsins; if opsins are central to the endeavor yet so haphazardly annotated, what is the status of dozens of lesser but still critical photoreceptor gene families? | ||
The question of how many times eyes independently arose is ill-posed. We can only wonder how people with no grasp whatsoever of the underlying molecular biology of photoreception could pontificate so boldly on its evolution prior to even sequencing of the first opsin in 1980. First collect comprehensive comparative genomic data sets on all the components in all the | The question of how many times eyes independently arose is ill-posed. We can only wonder how people with no grasp whatsoever of the underlying molecular biology of photoreception could pontificate so boldly on its evolution prior to even sequencing of the first opsin in 1980. First collect comprehensive comparative genomic data sets on all the components in all the relevant clades; then ask what it could mean to homologenize two contemporary systems comprised of partially overlapping sets of heterogeneous paralogs from assorted ancient gene families having long complex histories of clade-specific expansion and contraction cycles. If four arrestins serve a thousand GPCR, if even specialized photoreceptor cells express dozens of barely related 7TM signaling molecules, it's difficult to imagine what a cognate arrestin or ectopic evo-devo cassette might be. It's difficult enough to reconstruct an ancestral gene or two; reconstructing ancestral systems biology interactions lies in the future. | ||
After reviewing topics such as ciliary opsin in protostomes, rhabdomeric opsins in deuterostomes,the rich opsin repertoires in cnidarians and probable opsins in sponges, we will consider special topics such as the origin of image-forming eyes | After reviewing topics such as ciliary opsin in protostomes, rhabdomeric opsins in deuterostomes,the rich opsin repertoires in cnidarians and probable opsins in sponges, we will consider special topics such as the origin of image-forming eyes between amphioxus and lamprey divergences, noting throughout that our notion of 'eye' is much more nuanced than earlier. The reconstruction of Ur-bilateran and Urmetazoan eyes awaits additional cnidarian genomes but no new ones are currently underway. However the plethora of new arthropod and lophotrochozoan genome assemblies has opened up new avenues of research as the realization grows that fly and nematode model species are exceedingly derived, with better ancestral characters retained in other lineages. | ||
Numerous conflicting gene trees have been published for ciliary opsins. Some methodologies have bordered on the preposterous -- thin phylogenetic coverage, dimly related outgroups such as adrenergic receptors, and naive underlying mutational models assumed for maximal likelihood despite great diversity of species and many billions of years of branch length. Nonetheless the resultant trees have only moderate conflict, suggesting that a definitive opsin gene tree is not far off. We'll do this using the multiple types of evidence discussed above. | Numerous conflicting gene trees have been published for ciliary opsins. Some methodologies have bordered on the preposterous -- thin phylogenetic coverage, dimly related outgroups such as adrenergic receptors, and naive underlying mutational models assumed for maximal likelihood despite great diversity of species and many billions of years of branch length. Nonetheless the resultant trees have only moderate conflict, suggesting that a definitive opsin gene tree is not far off. We'll do this using the multiple types of evidence discussed above. | ||
The first point to be understood in deuterostome ciliary opsin evolution is jawless fish such as lamprey already exhibit a full-blown set of modern rod and cone opsins whereas earlier diverging clades as represented by hemichordate, echinoderm, amphioxus and tunicate genomes totally lack them and indeed lack imaging eyes altogether, while using their rhabdomeric opsins in a very distinct signaling system for their own | The first point to be understood in deuterostome ciliary opsin evolution is jawless fish such as lamprey already exhibit a full-blown set of modern rod and cone opsins whereas earlier diverging clades as represented by hemichordate, echinoderm, amphioxus and tunicate genomes totally lack them and indeed lack imaging eyes altogether, while using their rhabdomeric opsins in a very distinct signaling system for their own photoreception systems. Despite 7 sequenced opsin mRNAs in the amphioxus Branchiostoma belcheri and an initial assembly in Branchiostoma floridae providing counterparts there, no rod/cone opsin can be located there or in earlier diverging deuterostomes with genome projects (3 tunicates, 2 urchins, 1 acorn worm). These species may have larval eye spots, ocelli, pigment cells, and related photoreceptors but lack imaging eyes. | ||
Characters in extant (living) species should never be confused with ancestral characters present at the time of divergence nodes (last common ancestors); conceivably these early diverging deuterostomes have lost opsin genes, perhaps due to a habitat shift to deep water, burrowing habitat, or nocturnal lifestyle. However the molecular evidence is quite clear that full-blown pentachromatic color vision and most other modern ciliary opsin classes first appeared during the evolutionary stem preceding lamprey divergence. These are demonstrably not 'new' genes but derived from gene duplication and divergence of still older opsins of ciliary class.. | Characters in extant (living) species should never be confused with ancestral characters present at the time of divergence nodes (last common ancestors); conceivably these early diverging deuterostomes have lost opsin genes, perhaps due to a habitat shift to deep water, burrowing habitat, or nocturnal lifestyle. However the molecular evidence is quite clear that full-blown pentachromatic color vision and most other modern ciliary opsin classes first appeared during the evolutionary stem preceding lamprey divergence. These are demonstrably not 'new' genes but derived from gene duplication and divergence of still older opsins of ciliary class.. | ||
| Line 90: | Line 93: | ||
The fossil record is unsatisfactory: less than 1 bilateran in 10,000 in Chengjiang and Burgess Shale fossils is even a candidate for deuterostomy. Low numbers of specimens and poor preservation conspire with career pressures and impact-seeking journals in egregiously misinterpreted data in the view of Hou, discoverer of the Chinese lagerstaette. Myllokunmingia is the best situation with 500 specimens; however Haikouichthys as supposed stem deuterostome, Metaspreggina as supposed post-Ediacaran, and Yunnanozoan generally are problematic, valid only in the eye of some beholder. | The fossil record is unsatisfactory: less than 1 bilateran in 10,000 in Chengjiang and Burgess Shale fossils is even a candidate for deuterostomy. Low numbers of specimens and poor preservation conspire with career pressures and impact-seeking journals in egregiously misinterpreted data in the view of Hou, discoverer of the Chinese lagerstaette. Myllokunmingia is the best situation with 500 specimens; however Haikouichthys as supposed stem deuterostome, Metaspreggina as supposed post-Ediacaran, and Yunnanozoan generally are problematic, valid only in the eye of some beholder. | ||
While signs of | While signs of bilaterally disposed eyes are sometimes documentable, it does not follow these were image-forming eyes. Indeed contemporary Branchiostoma and tunicate larva have an eye-spot (ocellus); the genomes contain ciliary opsins but only clustering to ENCEPH and PPIN -- still a long ways from any imaging opsin. Echinoderms and hemichordates genomes also have opsins but even further diverged. Sea urchin genome encodes at least six opsins, four of these cluster classify to rhabdomeric, ciliary and Go-type. Tube feet are apparently the photosensory organ in adult urchins. | ||
The oldest known fossil lamprey, Priscomyzon, dates at 360 myr to the Devonian. Molecular clocks place lamprey appearance at approximately 430 myr, some 100 million years after Chengjiang and Burgess Shales fossil | The oldest known fossil lamprey, Priscomyzon, dates at 360 myr to the Devonian. Molecular clocks place lamprey appearance at approximately 430 myr, some 100 million years after Chengjiang and Burgess Shales fossil Lagerstaette formed. Like most soft tissues, eyes seldom leave a good fossil record, though bilateral placement might be reflected in bone orbits. Hagfish, sister group to lamprey, have imaging eyes but have not been studied; their opsins situation may be derived due to deepwater marine habitat (similarly deepwater coelacanth opsins are adapted to less scattered wavelengths, centered at420 nm). | ||
The next-diverging | The next-diverging chondrichthyes have inadequate data at GenBank -- only a few rhodopsin genes from skates and dogfish. This makes even fragmentary opsins from the partially sequenced elephantfish Callorhyncus milii quite valuable. Those 9 fragments and 3 from the lamprey genome are provided in the data section -- the opsin classifier tool can reliably type a fragment from a single mid-sized exon. Full length genes are preferable but these fragments serve to prove existence of various gene class at the time of a given divergence node. Further, they can validate certain rare genomic events provided the fragment happens to overlap the region of interest. | ||
On the other hand, thousands of high-quality Cambrian arthropod fossils unmistakably show stalked paired eyes. Fossil trilobite eyes have been much studied; these are better preserved due to calcite as lens crystalin. Imaging eyes of contemporary arthropods and lophotrochozoa are rhabdomeric, utilizing depolarizing Gq-type receptor, phospholipase C, phosphoinositol, diacylglycerol, and transient receptor potential TRP and TRPL channel signaling. However their genomes can also contain ciliary opsins, using hyperpolarizing Gt-type transducins and phosphodiesterase cGMP second-messaging (as well as Go-type gustducin ciliary opsins in other types of photoreceptors). | On the other hand, thousands of high-quality Cambrian arthropod fossils unmistakably show stalked paired eyes. Fossil trilobite eyes have been much studied; these are better preserved due to calcite as lens crystalin. Imaging eyes of contemporary arthropods and lophotrochozoa are rhabdomeric, utilizing depolarizing Gq-type receptor, phospholipase C, phosphoinositol, diacylglycerol, and transient receptor potential TRP and TRPL channel signaling. However their genomes can also contain ciliary opsins, using hyperpolarizing Gt-type transducins and phosphodiesterase cGMP second-messaging (as well as Go-type gustducin ciliary opsins in other types of photoreceptors). | ||
| Line 112: | Line 115: | ||
=== Opsin Gene and Species Trees === | === Opsin Gene and Species Trees === | ||
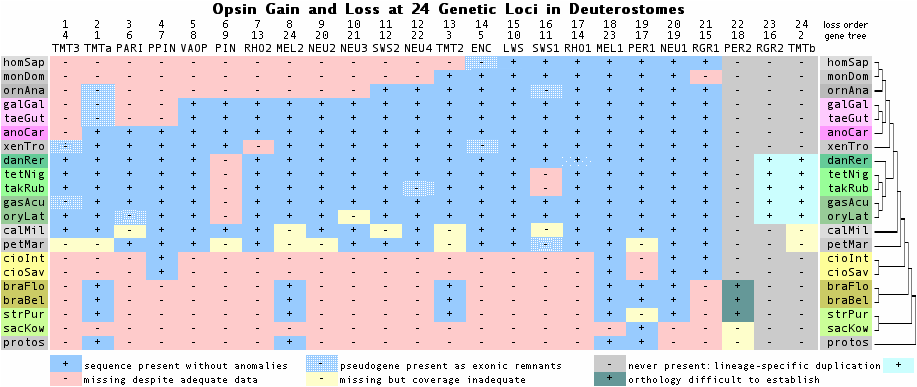
The phylogenetic tree below shows the presence or absence of various opsin genes in clade-representative species, as reflected in the collected reference sequences. The purpose is timing appearance (or disappearance) of a given class of opsin gene. For example, cone and rod opsins first appeared before lamprey divergence; otherwise they are absent from urochordates, cephalochordates, and earlier deuterostomes. Note however a given gene might appear absent because of a genome project gap, lack of experimental effort, insufficient or outdated bioinformatics, or species | The phylogenetic tree below shows the presence or absence of various opsin genes in clade-representative species, as reflected in the collected reference sequences. The purpose is timing appearance (or disappearance) of a given class of opsin gene. For example, cone and rod opsins first appeared before lamprey divergence; otherwise they are absent from urochordates, cephalochordates, and earlier deuterostomes. Note however a given gene might appear absent because of a genome project gap, lack of experimental effort, insufficient or outdated bioinformatics, or species choice (ie present in a different representative of that clade). In other cases (eg platypus SWS1) pseudogene remnants or a syntenically proven deletion establish a gene as definitely absent. | ||
[[Image: | [[Image:TMTsurvivors.png]] | ||
<br clear="all" /> | <br clear="all" /> | ||
Opsin retention at 18 genetic loci over 540 million years (Jan 2009 update) | Opsin retention at 18 genetic loci over 540 million years (Jan 2009 update) | ||
| Line 143: | Line 146: | ||
Gene loss is a mixture of deep stem loss (mammals) and recent lineage-specific loss. | Gene loss is a mixture of deep stem loss (mammals) and recent lineage-specific loss. | ||
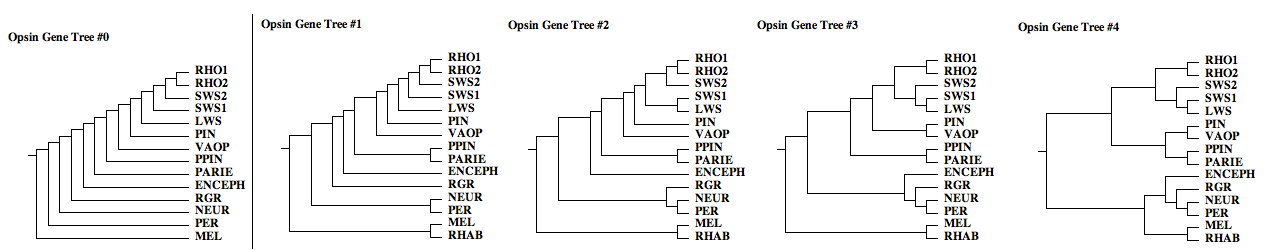
The opsin gene trees below illustrate only a few of the myriad possibilities, even beginning with commonsense ordering (blast nearest neighbors). Because these gene families originated long ago and are only known from remotely related representatives in extant species with wildly differently mutational mechanisms and histories, the true tree cannot be reliably | The opsin gene trees below illustrate only a few of the myriad possibilities, even beginning with commonsense ordering (blast nearest neighbors). Because these gene families originated long ago and are only known from remotely related representatives in extant species with wildly differently mutational mechanisms and histories, the true tree cannot be reliably inferred from maximal likelihood. Indeed no two such attempts have ever come up with the same gene tree! Instead, we'll keep this set of gene trees in view until analysis of rare genomic events is completed | ||
[[Image:opsin_gene_trees.png|{left}|]] | [[Image:opsin_gene_trees.png|{left}|]] | ||
| Line 151: | Line 154: | ||
=== Using the Opsin Classifier === | === Using the Opsin Classifier === | ||
Below is the primary collection of | Below is the primary collection of opsin protein sequences. Here "fields" in the fasta header show gene name, genus, species, common name, heterotrimeric G protein alpha subunit used in signaling, intron structure, synteny (2 flanking genes on each side of the opsin), indel status, sequence length, lambda max, and comment field. The 230-odd sequences are now organized into deuterostomes, lophotrochozoans, and ecdysozoan divisions, further broken refined into ciliary, rhabdomeric, or neither. Even with the full set copied into the [http://www.proweb.org/proweb/Tools/WU-blast.html uBlast], a novel opsin candidate can be classified in 6 seconds over just a conventional DSL internet connection. | ||
On 26 Nov 07, I added 41 new sequences, mostly arthropod rhabdomeric imaging opsins, extracting them from a 2007 [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17684563 pancrustacean opsin] paper, using the much-studied accessions in their Table 1, as ordered phylogenetically according to their Fig.3, with subsampling to avoid too-close sequences and narrow lineage-specific expansions. This involved replacing a few defective accessions and partial sequences with comparable complete ones, favoring sequences with completed or planned genome projects which can be directly intronated and their synteny determined. Lambda max values were helpfully compiled for all these opsins by the original authors. | On 26 Nov 07, I added 41 new sequences, mostly arthropod rhabdomeric imaging opsins, extracting them from a 2007 [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17684563 pancrustacean opsin] paper, using the much-studied accessions in their Table 1, as ordered phylogenetically according to their Fig.3, with subsampling to avoid too-close sequences and narrow lineage-specific expansions. This involved replacing a few defective accessions and partial sequences with comparable complete ones, favoring sequences with completed or planned genome projects which can be directly intronated and their synteny determined. Lambda max values were helpfully compiled for all these opsins by the original authors. | ||
| Line 157: | Line 160: | ||
This significantly upgrades the resolving power of the Opsin Classifier vis-a-vis these new classes of protostome opsins. It raises thorny nomenclature issues because of short-sighted historic uses such as rhodopsin or LWS for both fruitfly and human genes. These are indeed vaguely homologous in the distant pre-Bilateran GPCR past but are certainly not orthologous as implied by the same common name. The <span style="color: #990099;">definition of orthology requires genes under comparison in extant species to descend from a single parent gene in their last common ancestor.</span> If this LCA requirement is relaxed, the entire set of a thousand GPCR all become orthologous. | This significantly upgrades the resolving power of the Opsin Classifier vis-a-vis these new classes of protostome opsins. It raises thorny nomenclature issues because of short-sighted historic uses such as rhodopsin or LWS for both fruitfly and human genes. These are indeed vaguely homologous in the distant pre-Bilateran GPCR past but are certainly not orthologous as implied by the same common name. The <span style="color: #990099;">definition of orthology requires genes under comparison in extant species to descend from a single parent gene in their last common ancestor.</span> If this LCA requirement is relaxed, the entire set of a thousand GPCR all become orthologous. | ||
Additional ecdysozoan and lophotrochozoan opsins are needed, whatever new that can be extracted from invertebrate genome projects; some of these will prove ciliary and conversely some deuterostome opsins rhabdomeric. Melanopsin/ | Additional ecdysozoan and lophotrochozoan opsins are needed, whatever new that can be extracted from invertebrate genome projects; some of these will prove ciliary and conversely some deuterostome opsins rhabdomeric. Melanopsin/encephalopsin appear at the heart of the Big Switchover that took place in chordates -- their imaging opsins did not arise from gene duplication and divergence of anything we see among contemporary protostomal imaging opsins or any reconstructed ancestor. | ||
In fact, none of the opsin genes in | In fact, none of the opsin genes in Ur-bilatera destined to become rhabdomeric imaging opsins in living arthropods (even all of protostomia) seems to have descended directly to any deuterostome. It may turn out that none of the opsin genes in Ur-Bilatera destined to become ciliary imaging opsins in living vertebrates (even all of Deuterostomia) survived in any protostome. The pool of GPCR genes was already large and signaling diversified across gene copies. However lophotrochozoa and basal ecdysozoan ciliary opsins are still largely unexplored. A similarly 'bad' Venn diagram could hold in Ur-eumetazoa. Here the only two cnidarians with sequence data (Hydra and Nematostella) were not the best choices for studying opsin evolution. We may have to wait for genome sequencing of a full-featured cubomedusan. | ||
Please do not add text or edit sequences at this time, even though genomeWiki encourages participation; consider starting a fresh page to hold your contributions. After finishing the first round of articles, everything will be revisited and reorganized at a second pass. | Please do not add text or edit sequences at this time, even though genomeWiki encourages participation; consider starting a fresh page to hold your contributions. After finishing the first round of articles, everything will be revisited and reorganized at a second pass. | ||
=== | === Curated opsin reference sequences === | ||
The set | |||
The 460 manually curated opsin sequences below provides an optimal set for phylogenetic research in opsin evolution. These sequences have been collected from over 75 metazoan genome projects in addition to those extracted comprehensively from GenBank. Errors in genome assembly, flawed or missing annotation and mistakes in gene models have largely been corrected by advanced bioinformatic methods. (GenBank data cannot be used as it stands.) An opsin sequences is included only if it provides more information than clutter -- gene expansions limited to narrow clades and over-sampled loci are restricted to proxy representatives. | |||
However eight opsin genetic loci -- some new -- have greatly expanded reference sequence collections within separate articles. These explore more fine-grained opsin evolution restricted to the 48 vertebrate genomes available on 10 Jan 2010. Data from complete genomes offers four advantages over transcripts, namely it can provide regulatory regions, introns, flanking gene synteny, and exhaustive opsin paralog sets present in the organism. | |||
* [[Opsin_evolution:_LWS_PhyloSNPs|LWS reference sequences from 95 species]] | * [[Opsin_evolution:_LWS_PhyloSNPs|LWS reference sequences from 95 species]] | ||
* [[Opsin_evolution:_Encephalopsin_gene_loss|Encephalopsin and TMT: yet more opsins lost in mammals]] | * [[Opsin_evolution:_Encephalopsin_gene_loss|Encephalopsin and TMT: yet more opsins lost in mammals]] | ||
* [[Opsin_evolution:_RGR_phyloSNPs|RGR opsin: abrupt change in DRY motif in boreoeutheres]] | * [[Opsin_evolution:_RGR_phyloSNPs|RGR opsin: abrupt change in DRY motif in boreoeutheres]] | ||
* [[Opsin_evolution:_Peropsin_phyloSNPs|Peropsin: phyloSNPs in | * [[Opsin_evolution:_Peropsin_phyloSNPs|Peropsin: phyloSNPs in peropsin evolution]] | ||
* [[Opsin_evolution:_Neuropsin_phyloSNPs|Neuropsin (OPN5): comparative genomics of 52 deuterostome genes and | * [[Opsin_evolution:_Neuropsin_phyloSNPs|Neuropsin (OPN5): comparative genomics of 52 deuterostome genes and three new paralogs NEUR2, NEUR3, NEUR4]] | ||
The fasta header lines below are structured as a flatfile database. Fields are separated by spaces rather than tabs (which display poorly). The first field (all that will be retained by most web alignment tools) gives the gene name together with a 3+3 genus-species latin name code. | |||
These gene names mostly comply with international conventions and historic first-use but do not perpetuate poor terminology (use of already-assigned gene names, obtuse acronyms, greek letters, roman numerals, hyphens, subscripts, calling everything rhodopsin and so forth). Paralog subfamilies are numbered sequentially; recent gene duplicates (limited in phylogenetic distribution) are distinguished by lower case letters with different letters denoting those demonstrably arising from whole genome duplications. The nomenclature closely follows the opsin gene tree, indeed implicitly provides it (other than deep branching). | |||
Each major branch of the metazoan opsin gene tree is assigned a distinct short gene name that evokes its common name without need for explanatory table. Some names require periodic revision as more information emerges on their phylogenetic origin. While a later field of the fasta header does supply synonomy, should any confusion over sequence nomenclature ever arise, it is best resolved by text query of a sequence snippet using web browser search capability. | |||
The next three fields provide the full genus and species names (per GenBank taxonomy) and then a one-word common name. The latter can provide helpful orientation when the genus/species is unfamiliar. Next comes a field providing three units of higher order taxonomic nomenclature that allow phylogenetic subsetting of the database. For example chicken receives Deut.Saur.Arch as a deuterostome, sauropod like lizard, but archeosaur like finch but unlike lizard. This field implicitly places all the sequences on a phylogenetic tree. It assumes, unlike GenBank, that ecdysozoa and lophotrochozoa are the correct partition of protostomes. | |||
The next two fields provide a GenBank accession and PubMed identifier if any exist. The reference sequences here do not follow GenBank when it errs (as it often does, eg by retaining introns, omitting exons, extending the 'first' exon to a false initiating methionine, extending the 'final' exon to a false stop codon, leapfrogging across tandem genes to form chimeric genes). Many uncurated gene models are poor quality. However even these entries can provide historic credits and supplement the protein entries below with underlying dna. | |||
The PubMed identifier links to a recent journal article with substantive experimental data specifically relevant to the opsin sequence at hand. Such papers review earlier literature and most hubs also provide subsequent papers citing the initial paper. This allows rapid collection of all work pertaining to a given opsin gene. Papers are not cited that simply sequenced tens of thousands of transcripts without characterization or comment on fortuitously included opsins. | |||
<br clear="all"> | |||
The next field provides a measure of sequence completeness. About 1 sequence in 7 is fragmentary (but often not affecting the 7-transmembrane core). While important in proving that a given orthology class exists in a given clade (as are recent pseudogenes) and helpful in defining conserved residues in local alignments, these sequences cannot currently be completed. Reasons include too-rapid divergence of non-membrane regions, gaps in assembly coverage of individual exons, uncertainty in exon boundaries, and incomplete transcripts. | |||
[[Image:GabgComplexity.jpg|left]] | |||
The following field suggests a Galpha signaling partner for each opsin, important to understanding both structure and function. This is more wish-list than established fact because the rare cases known from direct experimental work have merely been extended below using homology to not-too-divergent orthologs in fairly closely related species -- even though the reliability of this procedure remains untested. | |||
Indeed, the opsin literature is confused over what this classification should even mean, the options being (1) the specific Galpha gene product(s) physically docking to the cytoplasmic face of the opsin, (2) merely a gene tree subclade (eg Gs) of interacting Galpha protein whatever that might mean within a [[Opsin_evolution:_transducins|complicated gene duplication tree]] itself scarcely comparable over metazoa and already inadequate to resolve rod and cone GNAT transducins, (3) the small molecule produced that does the actual signaling (eg cyclic nucleotide or phosphoinositol), or (4) the character of downstream signaling ultimately produced (eg inhibitory). | |||
2 | |||
Here the issue revolves around what is experimentally measurable -- opsins are not tested in a biochemically defined reconstituted system against individual Galpha proteins purified from the cognate species but rather signaling is characterized indirectly via the effect of some inhibitor or output of small molecule. While it appears that all opsin loci retain the universal features of GPCR signaling, it hasn't actually been established that all do in fact signal (rather than just serve as non-signaling photoisomerases in intermediary metabolism of retinoids). | |||
The next fields provide syntenic gene order (when known and relevant to a research issue), alternative gene names, tissues of expression, percent identity to nearest neighbor, and unstructured miscellaneous comments. Lambda-max peak adsorption could be added though it has been measured by a mix of techniques and is not necessarily reliably calculated from classical tuning residues in less-familiar opsin classes. Length in amino acids is no longer provided because it does not satisfactorily explain fragmentary entries nor the run-on amino and carboxy termini in some opsin classes. | |||
Adherence to rigid database format drops off after the early fields because the information is just not available or predictable for many entries. However the structure still suffices for routine queries such as 'how many opsins do tunicates have' or 'which protostomes have ciliary opsins'. Note the fasta header lines are easily isolated by pasting the entire database (including sequence lines) into a spreadsheet and sorting to collect lines beginning with '>'. The fields can be separated cleanly into columns by replacing spaces with tabs. | |||
The protein sequences themselves are broken into their constituent exons. The numbering indicates [[Opsin_evolution:_ancestral_introns|phase]] (ie basepair overhang), a deeply conserved invariant useful in distinguishing orthology classes and gene tree divisions. All opsins in all species use standard GT-AG splice junctions. Alternate splicing within the core protein may occur but never produces functional protein. The exon structures in genomic species have been determined individually by tblastn and those of non-genomic species predicted by homology. | |||
Full length genes beginning with an initial methionine and ending with a stop codon are the most useful. However the initial methionine cannot always be disambiguated, a problem not restricted to opsins. In run-on genes lacking 3' transcripts, the stop codon is taken as the first encountered in extending the last homologically reliable exon; however the true stop codon could lie in a different reading frame (ie in a cryptic terminal exon). | |||
Fragmentary genes are included if the species occupies a critical position in the phylogenetic tree (eg lamprey) and the fragment is long enough to reliably classify it. Fragmentary data often takes the form of entire exons missing from genomic coverage (rather than incomplete exons). When these exons lie in unconnected contigs, their combination into a single gene is not entirely reliable. | |||
For example the Callorhinchus genome project, currently with 0.6 exon coverage, still suffices to establish that many gene duplications important to vertebrate photoreceptive diversity had already occurred by the chondrichthyian divergence node. However without transcripts, a lineage-specific duplication in LWS might have result in a single chimeric gene assembled from scattered individual LWS exons. | |||
> | Fragmentary genes for isolated fast-evolving invertebrates are difficult to distinguish from non-opsin GPCR which share many motifs such as DRY. To balance the need for maximal phylogenetic coverage against the imperative of not contaminating the opsin reference collection with non-opsin GPCR, partial genes are required to contain the K296 Schiff base and other diagnostic residues and motifs specific to opsins. | ||
0 MNGTEGDNFYVPFSNKTGLARSPYEYPQYYLAEPWKYSALAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAMANLFMVLFGFTVTMYTSMNGYFVFGPTMCSIEGFFATLG 1 | <font color ="blue">>RHO1_bosTau Bos taurus (cow) Deut.Euth.Laur NM_001014890 2145276 full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 +H1FOO -PLXND1) | ||
2 GEVALWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVAFTWIMALACAAPPLVGWSR 2 | 0 MNGTEGPNFYVPFSNKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1 | ||
1 YIPEGMQCSCGPDYYTLNPNFNNESYVVYMFVVHFLVPFVIIFFCYGRLLCTVKE 0 | 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSR 2 | ||
0 AAAAQQESASTQKAEKEVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ 0 | 1 YIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKE 0 | ||
0 FRNCMITTLCCGKNPLGDDESGASTSKTEVSSVSTSQVSPA* 0 | 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQ 0 | ||
0 FRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA* 0 | |||
>RHO1_homSap Homo sapiens (human) Deut.Euth.Euar NM_000539 16565402 full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 +H1FOO -PLXND1) | |||
0 MNGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLEGFFATLG 1 | |||
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSR 2 | |||
1 YIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQ 0 | |||
0 FRNCMLTTICCGKNPLGDDEASATVSKTETSQVAPA* 0 | |||
>RHO1_monDom Monodelphis domesticus (opossum) Deut.Meta.Dide genomic full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 +H1FOO -PLXND1) | |||
0 MNGTEGPNFYVPFSNKTGTVRSPFEEPQYYLADPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAIADLFMVFGGFTMTLYTSLHGYFVFGPTGCNLEGFFATLG 1 | |||
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIIGVAFTWVMALACAFPPLIGWSR 2 | |||
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGQLVFTVKE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSNFGPIFMTIPAFFAKSSSVYNPVIYIMMNKQ 0 | |||
0 FRTCMITTLCCGKNPLGDDEASATASKTETSQVAPA* 0 | |||
>RHO1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono ABN43074 17339011 full Gt RH1 RHO rhodopsin rod opsin syn(+IFT122 - -PLXND1) | |||
0 MNGTEGQDFYIPMSNKTGVVRSPFEYPQYYLAEPWQYSVLAAYMFMLIMLGFPINFLTLYVTIQHKKLRTPLNYILLNLAFANHFMVLGGFTTTLYTSLHGYFVFGPTGCNIEGFFATLG 1 | |||
2 GEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACALPPLVGWSR 2 | |||
1 YIPEGMQCSCGIDYYTLRPEVNNESFVIYMFVVHFTIPMTIIFFCYGRLVFTVKE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTVPAFFAKSSAIYNPVIYIMMNKQ 0 | |||
0 FRNCMLTTICCGKNPLGDDEASATASKTEQSSVSTSQVSPA* 0 | |||
>RHO1_galGal Gallus gallus (chicken) Deut.Saur.Arch NM_205490 1385866 full Gt RH1 RHO rhodopsin rod syn(--MBD4 syn(+IFT122 +H1FOO -PLXND1) | |||
0 MNGTEGQDFYVPMSNKTGVVRSPFEYPQYYLAEPWKFSALAAYMFMLILLGFPVNFLTLYVTIQHKKLRTPLNYILLNLVVADLFMVFGGFTTTMYTSMNGYFVFGVTGCYIEGFFATLG 1 | |||
2 GEIALWSLVVLAVERYVVVCKPMSNFRFGENHAIMGVAFSWIMAMACAAPPLFGWSR 2 | |||
1 YIPEGMQCSCGIDYYTLKPEINNESFVIYMFVVHFMIPLAVIFFCYGNLVCTVKE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTNQGSDFGPIFMTIPAFFAKSSAIYNPVIYIVMNKQ 0 | |||
0 FRNCMITTLCCGKNPLGDEDTSAGKTETSSVSTSQVSPA* 0 | |||
>RHO1_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 - -PLXND1) | |||
0 MNGTEGQNFYVPMSNKTGVVRNPFEYPQYYLADPWQFSALAAYMFLLILLGFPINFLTLFVTIQHKKLRTPLNYILLNLAVANLFMVLMGFTTTMYTSMNGYFIFGTVGCNIEGFFATLG 1 | |||
2 GEMGLWSLVVLAVERYVVICKPMSNFRFGETHALIGVSCTWIMALACAGPPLLGWSR 2 | |||
1 YIPEGMQCSCGVDYYTPTPEVHNESFVIYMFLVHFVTPLTIIFFCYGRLVCTVKA 0 | |||
0 AAAQQQESATTQKAEREVTRMVVIMVISFLVCWVPYASVAFYIFTHQGSDFGPVFMTIPAFFAKSSAIYNPVIYILMNKQ 0 | |||
0 FRNCMIMTLCCGKNPLGDEDTSAGTKTETSTVSTSQVSPA* 0 | |||
>RHO1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 - -PLXND1) | |||
0 MNGTEGPNFYIPMSNKTGVVRSPFDYPQYYLAEPWKYSALAAYMFLLILLGFPINFMTLYVTIQHKKLRTPLNYILLNLVFANHFMVLCGFTVTMYTSMHGYFIFGQTGCYIEGFFATLG 1 | |||
2 GEMALWSLVVLAIERYVVVCKPMANFRFGENHAIMGVVFTWIMALSCAAPPLFGWSR 2 | |||
1 YIPEGMQCSCGVDYYTLKPEVNNESFVVYMFIVHFTIPLCVIFFCYGRLLCTVKE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVVMMVIFFLICWVPYAYVAFYIFTHQGSDFGPVFMTVPAFFAKSSAIYNPVIYIVLNKQ 0 | |||
0 FRNCLITTLCCGKNPFGDEEGSSAASSKTEASSVSSSQVSPA* 0 | |||
>RHO1_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGPNFYVPMTNKTGVVRSPFEYPQYYLADPWKYSALAAYMFFLILTGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTMYTAMNGYFVFGVVGCNLEGFFATFG 1 | |||
2 GIIALWCLVVLAIERYIVVCKPISNFRFGENHAIMGVVFTWIMALACAGPPLFGWSR 2 | |||
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFIVHFTIPLIIIFFCYGRLMCTVKE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVYIMVISYLVCWLPYASVSFYIFTHQGSDFGPVFMTVPAFFAKTASVYNPVIYILMNKQ 0 | |||
0 FRNCMITTLCCGKNPFGDEETTSAGTSKTEASSVSSSQVSPA* 0 | |||
>RHO1_latCha Latimeria chalumnae (coelacanth) Deut.Sarc.Coel AAD30519 10339578 full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGPNFYVPMSNKTGVVRNPFEYPQYYLADPWKYSALAAYMFFLILVGFPINFLTLFVTIQHKKLRTPLNYILLDLAVADLCMVFGGFFVTMYSSMNGYFVLGPTGCNIEGFFATLG 1 | |||
2 GQVALWALVVLAIERYVVVCKPMSNFRFGENHAIMGVIFTWIMALSCAVPPLFGWSR 2 | |||
1 YIPEGMQSSCGVDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGRLVCTVKD 0 | |||
0 AAAQQQESATTQKAEKEVTRMVIVMVISFLVCWVPYASVAAYIFFNQGSEFGPVFMTAPSFFAKSASFYNPVIYILLNKQ 0 | |||
0 FRNCMITTLCCGKNPFGDEDATSAAGSSKTEASSVSSSSVSPA* 0 | |||
>RHO1_angAng Anguilla anguilla (european_eel) Deut.Acti.Elop L78008 full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGPNFYIPMSNITGVVRSPFEYPQYYLAEPWAYTILAAYMFTLILLGFPVNFLTLYVTIEHKKLRTPLNYILLNLAVANLFMVFGGFTTTVYTSMHGYFVFGETGCNLEGYFATLG 1 | |||
2 GEISLWSLVVLAIERWVVVCKPMSNFRFGENHAIMGLAFTWIMANSCAMPPLFGWSR 2 | |||
1 YIPEGMQCSCGVDYYTLKPEVNNESFVIYMFIVHFSVPLTIISFCYGRLVCTVKE 0 | |||
0 AAAQQQESETTQRAEREVTRMVVIMVIAFLVCWVPYASVAWYIFTHQGSTFGPVFMTVPSFFAKSSAIYNPLIYICLNSQ 0 | |||
0 FRNCMITTLFCGKNPFQEEEGASTTASKTEASSVSSVSPA* 0 | |||
>RHO1_conMyr Conger myriaster (conger_eel) Deut.Acti.Elop AB043817 full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGPNFYIPMSNATGVVRSPFEYPQYYLAEPWAFSALSAYMFFLIIAGFPINFLTLYVTIEHKKLRTPLNYILLNLAVADLFMVFGGFTTTMYTSMHGYFVFGPTGCNIEGFFATLG 1 | |||
2 GEIALWCLVVLAIERWMVVCKPVTNFRFGESHAIMGVMVTWTMALACALPPLFGWSR 2 | |||
1 YIPEGLQCSCGIDYYTRAPGINNESFVIYMFTCHFSIPLAVISFCYGRLVCTVKE 0 | |||
0 AAAQQQESETTQRAEREVTRMVVIMVISFLVCWVPYASVAWYIFTHQGSTFGPIFMTIPSFFAKSSALYNPMIYICMNKQ 0 | |||
0 FRHCMITTLCCGKNPFEEEDGASATSSKTEASSVSSSSVSPA* 0 | |||
>RHO1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_131212 18597743 full Gt RH1 RHO rhodopsin rod opsin extra-ocular rhodopsin | |||
0 MNGTEGPNFYVPMSNRTGLVRSPFEEPQYYLAEPWQFSLLAAYMLFLILGSFPINALTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTVTLYTALHGYFLLGVTGCNIEGFFATLG 1 | |||
2 GEIALWSLVVLAIERYIVVCKPMSTFRFGEKHAIIGVGFTWVMALTCAVPPLLGWSR 2 | |||
1 YIPEGMQCSCGIDYYTPKPEVHNTSFVIYMFILHFSIPLLIIFFCYSRLLCTVRA 0 | |||
0 AAAQQQESETTQRAEREVTRMVVVMVIAFLVCWVPYASVAWYIFANQGAEFGPVFMTVPAFFAKSAALYNPVIYIMLNRQ 0 | |||
0 FRNCMLSTVCCGKNPLAEDESSSAVSSKTQSSVVSSAQVSPA* 0 | |||
>RHO1_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGPNFYVPMSNKTGVVRSPFEYPQYYLAEPWKYSLVAAYMLFLIVTAFPVNFLTLFVTVKHKKLRTPLNYVLLNLAVADLFMVIGGFTVTLYTALHAYFVLGVTGCNIEGFFATLG 1 | |||
2 GEIALWSLVVLAIERYIVVCKPVTNFRFGEKHAIAGLAFTWIMALSCAVPPLLGWSR 2 | |||
1 YIPEGMQCSCGIDYYTPKPEIHNTSFVIYMFVLHFCIPLAIIFFCYSRLLCTVRA 0 | |||
0 AAALQQESETTQRAEKEVTRMVIVMVISFLVCWVPYASVAWYIFANQGTEFGPVFMTAPAFFAKSAALYNPVIYILLNRQ 0 | |||
0 FRNCMITTVCCGKNPFGDDDATTSVSKTQSSSVSTSQVAPA* 0 | |||
>RHO1_takRub Takifugu rubripes (fugu) Deut.Acti.Perc AF201472 10692583 full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 - -PLXND1) | |||
0 MNGTEGPNFYIPMSNKTGVVRSPFEYPQYYLAEPWKYSLVAAYMLFLIITAFPVNFLTLFVTVKHKKLRTPLNYVLLNLAVADLFMVIGGFTVTLYTALHAYFVLGVTGCNIEGFFATLG 1 | |||
2 GEIALWSLVVLAVERYIVVCKPMTNFRFGEKHAIAGLVFTWIMALTCATPPLLGWSR 2 | |||
1 YIPEGMQCSCGIDYYTPKPEINNTSFVIYMFILHFSIPLAIIFFCYSRLLCTVRA 0 | |||
0 AAALQQESETTQRAEKEVTRMVIVMVISFLVCWVPYASVAWYIFANQGTEFGPVFMTAPAFFAKSAALYNPVIYILLNRQ 0 | |||
0 FRNCMITTVCCGKNPFGDDDAATTVSKTQSSSVSSSQVAPA* 0 | |||
>RHO1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc BT028623 full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGPYFYVPMMNTTGIVRSPYEYPQYYLVNPAAYAALGAYMFLLILVGFPVNFLTLYVTLEHKKLRTPLNYILLNLAVANLFMVLGGFTTTMYTSMHGYFVLGRLGCNVEGFFATLG 1 | |||
2 GEIALWSLVVLAVERWVVVCKLIANFRFGEDHAIMGLALTWVMASACAVPPLVGWSR 2 | |||
1 YIPEGMQCSCGVDYYTRAEGFNNESFVIYMFVCHFMIPMIIIFFCYGRLLCAVKEA 0 | |||
0 AAAQQESETTQRAEREVSRMVVIMVIAFLVCWVPYASVAWFIFCNQGSEFGPVFMTIPAFFAKSSSVYNPLIYICMNKQ 0 | |||
0 FRHCMITTLCCGKNPFEEEEGASSTKTEASSVSTSSVSPA* 0 | |||
>RHO1_oryLat Oryzias latipes (medaka) Deut.Acti.Perc AB180742 15964232 full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGPYFYVPMVNTTGIVRSPYEYPQYYLVNPAAYAALGAYMFFLILVGFPINFLTLYVTLEHKKLRTPLNYILLNLAVADLFMVFGGFTTTMYTSMHGYFVLGRLGCNLEGFFATLG 1 | |||
2 GEIGLWSLVVLAIERWVVVCKPISNFRFGENHAIMGLVFTWIMAASCAVPPLVGWSR 2 | |||
1 YIPEGMQCSCGVDYYTRAEGFNNESFVVYMFVCHFLIPLIVVFFCYGRLLCAVKEA 0 | |||
0 AAAQQESETTQRAEREVTRMVVIMVIGFLVCWLPYASVAWYIFTNQGSEFGPLFMTIPAFFAKSSSIYNPAIYICMNKQ 0 | |||
0 FRNCMITTLCCGKNPFEEEEGASTTASKTEASSVSSSSVSPA* 0 | |||
>RHO1_leuEri Leucoraja erinacea (skate) Deut.Chon.Elas U81514 9256070 full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGENFYVPMSNKTGVVRSPFDYPQYYLGEPWMFSALAAYMFFLILTGLPVNFLTLFVTIQHKKLRQPLNYILLNLAVSDLFMVFGGFTTTIITSMNGYFIFGPAGCNFEGFFATLG 1 | |||
2 GEVGLWCLVVLAIERYMVVCKPMANFRFGSQHAIIGVVFTWIMALSCAGPPLVGWSR 2 | |||
1 YIPEGLQCSCGVDYYTMKPEVNNESFVIYMFVVHFTIPLIVIFFCYGRLVCTVKE 0 | |||
0 AAAQQQESESTQRAEREVTRMVIIMVVAFLICWVPYASVAFYIFINQGCDFTPFFMTVPAFFAKSSAVYNPLIYILMNKQ 0 | |||
0 FRNCMITTICLGKNPFEEEESTSASASKTEASSVSSSQVAPA* 0 | |||
>RHO1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGENFYIPMSNKTGVVRSPFEYPQYYLAEPWQFSILAAYMFFLIITCFPVNFLTLYVTFEHKKLRQPLNFILLNLAVADLFMVFGGFFITVYTSLHGYFVFGVTGCNFEGFFATLG 1 | |||
2 GEIGLWSLVVLAIERYVVVCKPMSNFRFGTNHAIMGVAFTWVMALACAVPPLMGWSR 2 | |||
1 YIPEGLQCSCGVDYYTLKPEINNESFVIYMFVVHFLIPLIIIFFCYGRLVCTVKE 0 | |||
0 AAAQQQESESTQRAEREVTRMVIIMVIFFLICWVPYASVAFFIFTNQGSEFGPIFMAVPAFFAKSSALYNPLIYILLNKQ 0 | |||
0 FRNCMITTLCCGKNPFEEDESTSAAASKTEASSVSSSQVSPA* 0 | |||
>RHO1_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGENFYIPFSNKTGLARSPFEYPQYYLAEPWKYSVLAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVANLFMVLFGFTLTMYSSMNGYFVFGPTMCNFEGFFATLG 1 | |||
2 GEMSLWSLVVLAIERYIVICKPMGNFRFGSTHAYMGVAFTWFMALSCAAPPLVGWSR 2 | |||
1 YLPEGMQCSCGPDYYTLNPNFNNESFVIYMFLVHFIIPFIVIFFCYGRLLCTVKE 0 | |||
0 AAAAQQESASTQKAEKEVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTVPAFFAKTSALYNPIIYILMNKQ 0 | |||
0 FRNCMITTLCCGKNPLGDEDSGASTSKTEVSSVSTSQVSPA* 0 | |||
>RHO1_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366493 17463225 full Gt RhA | |||
0 MNGTEGQNFYIPFSNKTDVARSPFEYPQYYLAEPWKFSALAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVSNLFMILFGFTTTMYTSMNGYFVFGPTMCSIEGFFATLG 1 | |||
2 GEVSLWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVALTWVMALSCAAPPLLGWSR 2 | |||
1 YLPEGMQCSCGPDYYTMNPTYNNESFVIYMFIVHFTIPFVIIFFSYGRLLCTVKE 0 | |||
0 AAAAQQESASTQKAEKEVTRMVVLMVVGFLVCWVPYASVAFYIFTNQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ 0 | |||
0 FRNCMITTLCCGKNPLGDDDSGASTSKTEVSSVSTSQVAPA* 0 | |||
>RHO1_letJap Lethenteron japonicum (lamprey) Deut.Agna.Hype AB116382 15096614 full Gt RH1 RHO rhodopsin rod opsin | |||
0 MNGTEGDNFYVPFSNKTGLARSPYEYPQYYLAEPWKYSALAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAMANLFMVLFGFTVTMYTSMNGYFVFGPTMCSIEGFFATLG 1 | |||
2 GEVALWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVAFTWIMALACAAPPLVGWSR 2 | |||
1 YIPEGMQCSCGPDYYTLNPNFNNESYVVYMFVVHFLVPFVIIFFCYGRLLCTVKE 0 | |||
0 AAAAQQESASTQKAEKEVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ 0 | |||
0 FRNCMITTLCCGKNPLGDDESGASTSKTEVSSVSTSQVSPA* 0 | |||
>RHO2_galGal Gallus gallus (chicken) Deut.Saur.Arch NP_990771 2268324 full Gt syn(-IHPK3 -LEMD2 +RHO2 -GRM4 -NUDT3) | |||
0 MNGTEGINFYVPMSNKTGVVRSPFEYPQYYLAEPWKYRLVCCYIFFLISTGLPINLLTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFVFGPVGCAVEGFFATLG 1 | |||
2 GQVALWSLVVLAIERYIVVCKPMGNFRFSATHAMMGIAFTWVMAFSCAAPPLFGWSR 2 | |||
1 YMPEGMQCSCGPDYYTHNPDYHNESYVLYMFVIHFIIPVVVIFFSYGRLICKVRE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVILMVLGFMLAWTPYAVVAFWIFTNKGADFTATLMAVPAFFSKSSSLYNPIIYVLMNKQ 0 | |||
0 FRNCMITTICCGKNPFGDEDVSSTVSQSKTEVSSVSSSQVSPA* 0 | |||
>RHO2_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch NM_001076696 full G? | |||
0 MNGTEGINFYVPMSNKTGVVRSPFEYPQYYLAEPWKYRLVCCYIFFLISTGFPINFLTLLVTFKHKKLRQPLNYILVNLAVADLCMACFGFTVTFYTAWNGYFVFGPIGCAVEGFFATLG 1 | |||
2 GQVALWSLVVLAIERYIVICKPMGNFRFSASHALMGIAFTWVMAISCAAPPLFGWSR 2 | |||
1 YIPEGMQCSCGPDYYTHNPDFHNESYVLYMFVIHFIIPVVIIFFSYGRLVCKVRE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVILMVLGFMLAWTPYAVVAFWIFTNKGADFTATLMAVPAFFSKSSSLYNPIIYVLMNKQ 0 | |||
0 FRNCMITTICCGKNPFGDEETSSTVSQSKTEVSSVSSSQVSPA* 0 | |||
>RHO2_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt syn(-IP6K3 -LEMD2 -MLN +RHO2 -GRM4 -NUDT3) | |||
0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLAEPWKYKVVCCYIFFLIFTGLPINILTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFIFGPIGCAIEGFFATLG 1 | |||
2 GQVALWSLVVLAIERYIVVCKPMGNFRFSATHALMGISFTWFMSFSCAAPPLLGWSR 2 | |||
1 YIPEGMQCSCGPDYYTLNPDYHNESYVLYMFGVHFVIPVVVIFFSYGRLICKVRE 0 | |||
0 AAAQQQESASTQKAEREVTRMVILMVLGFLLAWTPYAMVAFWIFTNKGVDFSATLMSVPAFFSKSSSLYNPIIYVLMNKQ 0 | |||
0 FRNCMITTICCGKNPFGDEDVSSSVSQSKTEVSSVSSSQVSPA* 0 | |||
>RHO2_gekGek Gekko gekko (gecko) Deut.Saur.Lepi AY024356 11591478 full Gt in pure rod retina | |||
0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLADPWKFKVLSFYMFFLIAAGMPLNGLTLFVTFQHKKLRQPLNYILVNLAAANLVTVCCGFTVTFYASWYAYFVFGPIGCAIEGFFATIG 1 | |||
2 GQVALWSLVVLAIERYIVICKPMGNFRFSATHAIMGIAFTWFMALACAGPPLFGWSR 2 | |||
1 FIPEGMQCSCGPDYYTLNPDFHNESYVIYMFIVHFTVPMVVIFFSYGRLVCKVRE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVILMVLGFLLAWTPYAATAIWIFTNRGAAFSVTFMTIPAFFSKSSSIYNPIIYVLLNKQ 0 | |||
0 FRNCMVTTICCGKNPFGDEDVSSSVSQSKTEVSSVSSSQVAPA* 0 | |||
>RHO2_podSic Podarcis sicula (lizard) Deut.Saur.Lepi AY941829 12887418 full G? | |||
0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLAEPWKYKMVCCYIFFLISTGLPINLLTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFIFGPIGCAIEGFFATLG 1 | |||
2 GQVALWSLVVLAIERYIVVCKPMGNFRFSSSHALMGIAFTWVMSLSCACPPLFGWSR 2 | |||
1 YIPEGMQCSCGPDYYTLNPDYHNESYVVYMFVIHFVIPVVVIFFSYGRLICKVRE 0 | |||
0 AAAQQQESASTQKAEKEVTRMVILMVLGFMLAWTPYAVVAFWIFTNKGADFSATLMSVPAFFSKSSSLYNPIIYVLMNKQ 0 | |||
0 FRNCMITTICCGKNPFGDDDVSSTVSQSKTEVSSISSSQVSPA* 0 | |||
>RHO2_pheMad Phelsuma madagascariensis (lizard) Deut.Saur.Lepi AF074044 10069370 full G? | |||
0 MNGTEGFNFYVPVSNRTGLVRSPYEYPQYYLAEPWKFKALSLYMFFLILVGLPLNGLTLFVTFQHKKLRQPLNYILVNLAVANLLMVICGFTVTFYTSWYGYFVFGPMGCAFEGFFATIG 1 | |||
2 GQVALWSLVVLAIERYIVICKPMGNFRFSSSHAMMGISFTWFMALCCGGPPLFGWSR 2 | |||
1 FIPEGMQCSCGPDYYTLNPDFHNESYVIYLFTVHFLTPMIIIFFSYGRLVCKVRE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVILMVMGFLVAWTPYATVACWIFNNKGAEFSVTFMTVPAFFSKSSCIYNPIIYVLLNKQ 0 | |||
0 FRNCMVTTICCGKNPFGDEDASSSVSQSKTEVSSVSSSQVAPA* 0 | |||
>RHO2_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt | |||
0 MNGTEGINFYVPHSNKTGVVRSPFEYPQYYLADPWKYSIVCAYMFFLIITGLPINLLTLVVTFKHKKLRQPLNYILVNLAVADLFMVCFGFTVTFSTAINGYFIFGPRGCAIEGFMATLG 1 | |||
2 GEVALWSLVVLAIERYIVVCKPMGNFRFSNNHSIIGIVFTWLAALSCAAPPLFGWSR 2 | |||
1 YLPEGMQCSCGPDYYTMNPDYHNESFVIYMFVVHFFIPVIVIFVSYGRLICKVKE 0 | |||
0 AAAQQQESASTQKAEREVTRMVILMVIGFMTAWTPYATVAFWIFMNKGAEFGATFMAAPAFFSKSSALYNPIIYVLMNKQ 0 | |||
0 FRNCMVTTLCCGKNPFGDDDVSSSVSAGKTEVSSVSSSQVSPA* 0 | |||
>RHO2_latCha Latimeria chalumnae (coelacanth) Deut.Sarc.Coel AH007713 10339578 full Gt RH2 | |||
0 MNGTEGMNFYVPLSNRTGLVRSPFEYTQYYLAEPWKFSVLCAYMFLLIILGFPINFLTLLVTFKHKKLRQPLNYILVNLAVASLFMVVFGFTVTFYSSLNGYFVLGPMGCAMEGFFATLG 1 | |||
2 GQVALWSLVVLAIERYIVVCKPMGNFRFASSHAIMGIAFTWIMALACAAPPLVGWSR 2 | |||
1 YIPEGLQCSCGPDYYTLNPDFHNESYVMYLFLVHFLLPIIIIFFTYGRLICKVKE 0 | |||
0 AAAQQQESASTQKAEKEVTRMVILMVIGFLTAWVPYASAAFWIFCNRGAEFTATLMTVPAFFSKSSCLFNPIIYVLLNKQ 0 | |||
0 FRNCMITTLCCGKNPLGDDDTSSAVSQSKTDVSSVSSSQVSPA* 0 | |||
>RHO2_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic 15647516 full G? opn1mw1 ancestral representing 4x expansion | |||
0 MNGTEGNNFYIPMSNRTGLVRSPYEYPQYYLAEPWQFKLLAVYMFFLICLGFPINGLTLLVTAQHKKLRQPLNFILVNLAVAGTIMVCFGFTVTFYSAINGYFVLGPTGCAIEGFMATLG 1 | |||
2 GEVALWSLVVLAIERYIVVCKPMGSFKFSSSHAMAGIAFTWVMAMACAAPPLFGWSR 2 | |||
1 YIPEGMQCSCGPDYYTLNPEYNNESYVLYMFICHGIVPVTIIFFTYGRLVCTVKA 0 | |||
0 AAAQQQESESTQKAEREVTRMVILMVLGFLVAWTPYATVAAWIFFNKGAAFSAQFMAVPAFFSKSSALFNPIIYVLLNKQ 0 | |||
0 FRNCMLTTLFCGKNPLGDDESSTVSTSKTEVSSVSPA* 0 | |||
>RHO2a_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_131253 15647516 full G? opn1mw1 | |||
0 MNGTEGSNFYIPMSNRTGLVRSPYDYTQYYLAEPWKFKALAFYMFLLIIFGFPINVLTLVVTAQHKKLRQPLNYILVNLAFAGTIMVIFGFTVSFYCSLVGYMALGPLGCVMEGFFATLG 1 | |||
2 GQVALWSLVVLAIERYIVVCKPMGSFKFSANHAMAGIAFTWFMACSCAVPPLFGWSR 2 | |||
1 YLPEGMQTSCGPDYYTLNPEYNNESYVMYMFSCHFCIPVTTIFFTYGSLVCTVKA 0 | |||
0 AAAQQQESESTQKAEREVTRMVILMVLGFLFAWVPYASFAAWIFFNRGAAFSAQAMAVPAFFSKTSAVFNPIIYVLLNKQ 0 | |||
0 FRSCMLNTLFCGKSPLGDDESSSVSTSKTEVSSVSPA* 0 | |||
>RHO2b_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_182891 15647516 full G? opn1mw2 highest expression | |||
0 MNGTEGNNFYIPMSNRTGLVRSPYEYTQYYLADPWQFKALAFYMFFLICFGLPINVLTLLVTAQHKKLRQPLNYILVNLAFAGTIMAFFGFTVTFYCSINGYMALGPTGCAIEGFFATLG 1 | |||
2 GQVALWSLVVLAIERYIVVCKPMGSFKFSSNHAMAGIAFTWVMASSCAVPPLFGWSR 2 | |||
1 YIPEGMQTSCGPDYYTLNPEFNNESYVLYMFSCHFCVPVTTIFFTYGSLVCTVKA 0 | |||
0 AAAQQQESESTQKAEREVTRMVILMVLGFLVAWVPYASFAAWIFFNRGAAFSAQAMAIPAFFSKASALFNPIIYVLLNKQ 0 | |||
0 FRSCMLNTLFCGKSPLGDDESSSVSTSKTEVSSVSPA* 0 | |||
>RHO2c_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_182892 15647516 full G? opn1mw3 | |||
0 MNGTEGNNFYIPMSNRTGLVRSPYEYPQYYLAEPWQFKLLAVYMFFLMCFGFPINGLTLVVTAQHKKLRQPLNFILVNLAVAGTIMVCFGFTVTFYTAINGYFVLGPTGCAIEGFMATLG 1 | |||
2 GQISLWSLVVLAIERYIVVCKPMGSFKFSSNHAFAGIGFTWIMALSCAAPPLVGWSR 2 | |||
1 YIPEGMQCSCGPDYYTLNPDYNNESYVLYMFCCHFIFPVTTIFFTYGRLVCTVKA 0 | |||
0 AAAQQQESESTQKAEREVTRMVILMVLGFLVAWTPYASVAAWIFFNRGAAFSAQFMAVPAFFSKSSSIFNPIIYVLLNKQ 0 | |||
0 FRNCMLTTLFCGKNPLGDDESSTVSTSKTEVSSVSPA* 0 | |||
>RHO2d_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_131254 15647516 full G? opn1mw4 | |||
0 MNGTEGNNFYIPLSNRTGLARSPYEYPQYYLAEPWQFKLLAVYMFFLICLGFPINGLTLLVTAQHKKLRQPLNFILVNLAVAGTIMVCFGFTVTFYTAINGYFVLGPTGCAIEGFMATLG 1 | |||
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSASHAFAGCAFTWVMAMACAAPPLVGWPR 2 | |||
1 YIPEGMQCSCGPDYYTLNPEYNNESYVLYMFICHFILPVTIIFFTYGRLVCTVKA 0 | |||
0 AAAQQQESESTQKAEREVTRMVILMVLGFLIAWTPYATVAAWIFFNKGAAFSAQFMAVPAFFSKTSALYNPVIYVLLNKQ 0 | |||
0 FRNCMLTTLFCGKNPLGDDESSTVSTSKTEVSSVSPA* 0 | |||
>RHO2_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? RH2 | |||
0 MVWDGGIEPNGTEGKNFYIPMSNRTGIVRSPFEYPQYYLVDPIMFKMLALYMFFLICTGTPINGLTLLVTAQNKKLRQPLNYILVNLAVAGLIMCAFGFTITITSAINGYFILGATACAVEGFMATLG 1 | |||
2 GEVALWSLVVLAIERYIVVCKPMGSFKFTGTHAAVGVLFTWIMAFACAGPPLFGWSR 2 | |||
1 YLPEGMQCSCGPDYYTLAPGYNNESYVIYMFVVHFFVPVFLIFFTYGSVVLTVka 0 | |||
0 AAAQQQESESTQKAQREVTRMCILMVLGFLVAWTPYATFSGWIFMNKGAAFHPLTAALCAFFAKSSALYNPVIYVLM 0 | |||
0 FRNCMLSTIGMGGAVDDETSVSASKTEVSSVS* 0 | |||
>RHO2_takRub Takifugu rubripes (fugu) Deut.Acti.Perc AF226989 10692583 full G? | |||
0 MAWDGGIEPNGTEGKNFYIPMSNRTGIVRSPFEYPQYYLADPIMFKILALYMFFLICTGTPINGLTLLVTAQNKKLRQPLNYILVNLAVAGLIMCAFGFTITITSAVNGYFILGATACAVEGFMATLG 1 | |||
2 GEIALWSLVVLAVERYVVVCKPMGSFKFTGTHAAVGVAFTWIMAFACAAPPLFGWSR 2 | |||
1 YLPEGMQCSCGPDYYTLAPGYNNESYVIYMFSCHFFVPVITIFFTYGSLVLTVKA 0 | |||
0 AAAQQQESESTQKAQKEVTRMCILMVFGFLMAWTPYATFSAWIFMNKGAAFHPLTAAVCAFFAKSSALYNPVIYVLLNKQ 0 | |||
0 FRNCMLSTIGMGGAVDDETSVSASKTEVSSVS* 0 | |||
>RHO2_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? 2nd near-identical tandem copy | |||
0 MAWEGGLEPNGTEGKNFYIPMSNRTGVVRSPFEYQQYYLADPIMFKILALYMFFLICTGTPINGLTLLVTAQNKKLRQPLNYILVNLAVAGLIMCAFGFTITITSAVNGYFILGATACAVEGFMATLG 1 | |||
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSGTHAGAGVLFTWIMAMACAAPPLFGWSR 2 | |||
1 YLPEGMQCSCGPDYYTLAPGFNNESYVIYMFVVHFFTPVFIIFFTYGSLVLTVKA 0 | |||
0 AAAQQQESESTQKAEREVTRMCILMVFGFLVAWVPYASFAGWIFLNKGAPFSALTAAIPAFFAKSSALYNPVIYVLLNKQ 0 | |||
0 FRNCMLTTIGMGGMVEDETSVSASKTEVSSVS* 0 | |||
>RHO2_oryLat Oryzias latipes (medaka) Deut.Acti.Perc AB001603 9463691 full G? | |||
0 MENGTEGKNFYIPMNNRTGLVRSPYEYPQYYLADPWQFKLLGIYMFFLILTGFPINALTLVVTAQNKKLRQPLNFILVNLAVAGLIMVCFGFTVCIYSCMVGYFSLGPLGCTIEGFMATLG 1 | |||
2 GQVSLWSLVVLAIERYIVVCKPMGSFKFTATHSAAGCAFTWIMASSCAVPPLVGWSR 2 | |||
1 YIPEGIQVSCGPDYYTLAPGFNNESFVMYMFSCHFCVPVFTIFFTYGSLVMTVKA 0 | |||
0 AAAQQQDSASTQKAEKEVTRMCFLMVLGFLLAWVPYASYAAWIFFNRGAAFSAMSMAIPSFFSKSSALFNPIIYILLNKQ 0 | |||
0 FRNCMLATIGMGGMVEDETSVSTSKTEVSTAA* 0 | |||
>RHO2_oreNil Oreochromis niloticus (tilapia) Deut.Acti.Perc AF247124 11470845 full G? | |||
0 MAWEGGIEPNGTEGKNFYIPMSNRTGIVRNPFEYSQYYLADPIFFKLLAFYMFFLICTGTPINGLTLFVTAQNKKLRQPLNYILVNLAVAGLIMCCFGFTITITSAINGYFVLGTTFCAIEGFMATLG 1 | |||
2 GEVALWSLVVLAVERYIVVCKPMGSFKFTGAHAGAGVLFTWIMAMACAAPPLFGWSR 2 | |||
1 YIPEGMQCSCGPDYYTLAPGFNNESYVIYMFVVHFFVPVFIIFFTYGSLVMTVRA 0 | |||
0 AAAQQQDSASTQKAEKEVTRMCVLMVMGFLIAWTPYASFAGWIFLNKGAAFSALTAAIPAFFAKSSALYNPIIYVLMNKQ 0 | |||
0 FRNCMLSTIGMGGMVEDETSVSTSKTEVSSVS* 0 | |||
>RHO2_hipHip Hippoglossus hippoglossus (halibut) Deut.Acti.Perc AF156263 11925012 full G? | |||
0 MVWDGGIEPNGTEGKNFYIPMSNRTGIVRSPFEYPQFYMVDSMMFKFLAFYMFFLVCTGTPINGLTLFVTAQNKKLRQPLNYILVNLAVAGLIMCCFGFTITITSAFNGYFILGATFCTIEGFMATLG 1 | |||
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSGTHAGIGVLFTWVMAFACAGPPLFGWSR 2 | |||
1 YIPEGMQCSCGPDYYTLAPGFNNESYVIYMFVVHFFLPVFVIFFTYGSLVLTVKA 0 | |||
0 AAAQQQESESTQKAEKEVTRMCILMVFGFLFAWTPYATFAGWIFMNKGAAFTALTASIPAFFAKSSALYNPVIYVLLNKQ 0 | |||
0 FRNCMLSTIGMGGMVEDESSVSASKTEVSSVS* 0 | |||
>RHO2_mulSur Mullus surmuletus (mullet) Deut.Acti.Perc Y18680 full G? | |||
0 MNGTEGKNFYIPMSNRTGIVRSPFEYPQYYMVDPMIYKLLAFYMFFLICTGTPINGLTLLVTFQNKKLQQPLNYILVNLAVVGLIMCAFGFTITITSALNGYFILGPTFCAIEGFMATLG 1 | |||
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSGTHAGAGVAFTWIMAFACAGPPLFGWSR 2 | |||
1 YLPEGMQCSCGPDYYTLAPGFNNESYVIYMFVVHFFVPVFVIFFTYGSLVLTVKA 0 | |||
0 AAAQQQESESTQKAEREVTRMCILMVIGFLVAWVPYATFAGWIFLNKGAAFTALTAALPAFFAKSSALYNPVIYVMMNKQ 0 | |||
0 FRNCILSAIGMGGMVEDETSVSTSKTEVSTAS* 0 | |||
>RHO2_pomMin Pomatoschistus minutus (sand_goby) Deut.Acti.Perc Y18679 full G? | |||
0 MNGTEGKNFYIPMSNRTGIVRSPYEYPQYYMVDPWIYKLLAFYMFFLICTGTPINALTLLVTFQNKKLRQPLNFILVNLAVAGLIMCAFGFTITITSALNGYFILGATFCAIEGFMATLG 1 | |||
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSGAHAGAGVALTWIMAMACAAPPLFGWSR 2 | |||
1 YLPEGMQCSCGPDYYTLAPGFNNESYVMYMFVVHFFIPVFLIFFTYGSLVLTVKA 0 | |||
0 AAAQQQDSASTQKAEKEVTRMCFLMVMGFLVAWVPYASFAGWIFLNKGAAFTAMTAAIPAFFAKSSALYNPVIYVLMNKQ 0 | |||
0 FRNCMLSAVGMGGMVDDETSVSASKTEVSSVS* 0 | |||
>RHO2_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo EF565168 19196633 full G? | |||
0 MNGTKGSNFYIPMSNRTGVVRNPFEYPQYYLADRWLFSSISAYMFLLICAGLPINGLTLLVTVKHKKLRQPLNFILLNLAVADLFMVFGGFFITVYTSLHGYFVFGVTGCNFEGFFATLG 1 | |||
2 GEIGLWSLVVLAIERYVVVCKPMSNFRFGTSHALMGMGFTWFMALTAAVPPLVGWSR 2 | |||
1 FIPEGFQCSCTPDFYTTNPLYNNDSYLMYLFSVHFAFPVTLIFFSYGRLICKVKE 0 | |||
0 AAAQQQESATTQKAEKEVTRMVILMVIGFLTAWLPYASLSIWIFTHQGAWISPLLMTIPSFFSKSSVLYNPIIYILMNKQ 0 | |||
0 FRSSMITTVCCGKNPFGDDDSSSVTSQSKTEVSSVSTSQVSPA* 0 | |||
>RHO2_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366494 17463225 full Gt RhB | |||
0 MNGTEGANFYIPFHNRTGVVRSPYEYPQYYLADPWMYSAISAYVFTLILIGFPVNFMTLFVTFKLKKLRQPLNFILVNLCVADLLMIMFGFTTTFYTAMNGYFVFGPTGCNIEGFFATLG 1 | |||
2 GEVSLWSLVMLAIERYIVVCKPMGNFRFATTHAALGVVFTWVMASACAVPPLVGWSR 2 | |||
1 YIPEGMQCSCGPDYYTLNPKYYNESYVIYLFLVHFLLPVTIIFFTYGRLICTVKE 0 | |||
0 AAAQQQESASTQKAEREVTRMVIIMVVGFLVCWVPYASFAFYLFMNKGILFSATAMTVPAFFSKSSVLYNPIIYVLLNKQ 0 | |||
0 FRTCMVTTLFCGKNPFGEDDSSMVSTSKTEVSSVSSSQVSPS* 0 | |||
>SWS2_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono ABN43074 17339011 full Gt syn(-IRAK1 -MECP2 - +TKTL1) blue tandem with MWS1 | |||
0 MHKTHRNLQNELPEDFFIPLPLDTDNITSLSPFLVPQTHLGGSGIFMSLAAFMFLLITLGFPINLLTVICTIKYKKLRSHLNYILVNLAVSNMLVVCVGSATAFYSFAHMYFVLGPTACKIEGFAATLG 1 | |||
2 GMVSLWSLAVIAFERFLVICKPLGNLSFRGTHAIFGCAATWVFGLAASLPPLFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFSFCFGVPLSIIIFSYGRLLLTLRA 0 | |||
0 VAKQQEQSATTQKAEREVTKMVIVMVLGFLVCWLPYASFSLWVVTNRGQVFDLRMASIPSVFSKASTIYNPIIYVFMNKQ 0 | |||
0 FRSCMLKLVFCGKSPFGDEDEISGSSQATQVSSVSSSQVSPA* 0 | |||
>SWS2_galGal Gallus gallus (chicken) Deut.Saur.Arch NP_990848 7975342 full Gt | |||
0 MHPPRPTTDLPEDFYIPMALDAPNITALSPFLVPQTHLGSPGLFRAMAAFMFLLIALGVPINTLTIFCTARFRKLRSHLNYILVNLALANLLVILVGSTTACYSFSQMYFALGPTACKIEGFAATLG 1 | |||
2 GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCVATWVLGFVASAPPLFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTTDNKWHNESYVLFLFTFCFGVPLAIIVFSYGRLLITLRA 0 | |||
0 VARQQEQSATTQKADREVTKMVVVMVLGFLVCWAPYTAFALWVVTHRGRSFEVGLASIPSVFSKSSTVYNPVIYVLMNKQ 0 | |||
0 FRSCMLKLLFCGRSPFGDDEDVSGSSQATQVSSVSSSHVAPA* 0 | |||
>SWS2_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full Gt | |||
0 MPKPREMRDELPEDFYIPMSLETPNLTALSPFLVPQTHLGSPGIFKAMAAFMFLLVLLGVPINALTVLCTAKYKKLRSHLNYILVNLAVANLLVVCVGSTTAFYSFSQMYFALGPLACKIEGFTATLG 1 | |||
2 GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCAITWIFGLIASLPPLFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTTDNKWNNESYVIFLFCFCFGFPLTVIVFSYGRLLLTLRA 0 | |||
0 VAKQQEQSASTQKAEREVTKMVVVMVLGFLVCWLPYCSFALWVVTHRGHPFDLGLASIPSVFSKASTVYNPIIYVFMNKQ 0 | |||
0 FRSCMLKLVFCGRSPFGDEDDVSGSSQATQVSSVSSSQVSPA* 0 | |||
>SWS2_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G? | |||
0 MQKSRPDSRDNLPEDFFIPVPLDVANITTLSPFLVPQTHLGNPSLFMGMAAFMFILIVLGVPINVLTIFCTFKYKKLRSHLNYILVNLSVSNLLVVCVGSTTAFYSFSNMYFSLGPTACKIEGFSATLG 1 | |||
2 GMVSLWSLAVVAFERYLVICKPLGNFTFRGTHAIIGCAVTWMFGLAASLPPLFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTTENKWNNESYVIFLFCFCFGVPLSVIIFSYGRLLLTLRA 0 | |||
0 VAKQQEQSATTQKAEREVTKMVVVMVMGFLVCWLPYASFALWVVTHRGEPFDVRLASIPSVFSKASTVYNPVIYVLMNKQ 0 | |||
0 FRSCMLKLIFCGKSPFGDEDDVSGSSQATQVSSVSSSQVSPA* 0 | |||
>SWS2_utaSta Uta stansburiana (lizard) Deut.Saur.Lepi DQ100326 16543463 full Gt | |||
0 MHNSRPHSRDDLPEDFFIPMPLDVANITTLSPFLVPQTHLGSPALFMGMAAFMFLLIILGVPINVLTIFCTFKYKKLRSHLNYILVNLAVSNLLVVCIGSTTAFYSFAQMYFSLGPTACKIEGFAATLG 1 | |||
2 GMVSLWSLAVVAFERFLVICKPLGNFSFRGTHAIIGCIITWVFGLVASLPPLFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTTNNKWNNESYVLFLFSFCFGVPLSVIIFSYGRLLLTLRA 0 | |||
0 VAKQQEQSATTQKAEREVTKMVVVMVMGFLVCWLPYASFALWVVTHRGEPFDVRLATIPSVFSKASSVYNPVIYVFMNKQ 0 | |||
0 FRSCMLKLVFCGKSPFGDEDDVSGSSQTTQVSSVSSSQVSPA* 0 | |||
>SWS2_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt syn(-IRAK1 -MECP2 -) | |||
0 MSKGRPDLRMEMPDEFYVPIPLETTNISSLSPFLVPQTHLGTPGIFMSISAFMLFTIIFGFPLNLLTIICTVKYKKLRSHLNYILVNLAVANLIVICFGSTTAFYSFSQMYFSLGTLACKIEGFTATLG 1 | |||
2 GIIGLWSLAVVAFERFLVICKPMGNFTFRESHAVLGCILTWVIGLVAAIPPLLGWSR 2 | |||
1 YIPEGLQCSCGPDWYTVNNKWNNESYVLFLFCFCFGFPLAIIVFSYGRLLLALHA 0 | |||
0 VAKQQEQSATTQKAEREVTRMVIVMVVGFLVCWLPYASFALWAVTHRGELFDLRMSSVPSVFSKASTVYNPFIYIFMNRQ 0 | |||
0 FRSCMMKMIFCGKNPLGDDEETSVSGSTQVSSVSSSQIAPS* 0 | |||
>SWS2_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt | |||
0 MHRTKPDPQEDLPDDFYIPVSLNTNNITMLSPFLVPQTHLGSPSVFMVLSVFMFFLLITGIPINVLTIICTFKYKKLRSHLNYILVNLAVANLIVVGFGSTTAFYSFSQMYFAWGPLACKIEGFAATLG 1 | |||
2 GMVSLWSLAVVAFERFLVICKPLGNFTFRSTHAIIGCVATWVFGLISSAPPLFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFCFCFGFPLSVIIFSYGRLLMTLRA 0 | |||
0 VAKQQEQSASTQKAEREVTKMVVVMVLGFLVCWLPYTVFSLWVVTHRGESFELALGSIPAVFSKSSTVYNPLIYVFMNKQ 0 | |||
0 FRSCMMKLIFCGKSPFGDEDDASSASQSTQVSSVSSSQVAPA* 0 | |||
>SWS2_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? | |||
0 MRGNRQHEFQEDFFIPIPLEVDNITALSPFLVPQDHLGSPALFYGMAAFMFCLFVSGTGINALTIACTVKYKKLRSHLNYILVNLAVSNLLVTCVGSFTCFCCFTVRYMFVGPLGCKIEGFAATLG 1 | |||
2 GMVSLWSLAVVAFERWLVVCKPLGNFIFKPDHAIACCAATWLFALVVSGPPLFGWSR 2 | |||
1 YIPEGFQCSCGPDWYTTGNKYNNESYVMFIFGFGFAVPFFVIVFCYSQLLFVLKS 0 | |||
0 AQAESASTQKAEKEVTRMVVVMIFGFLVCWLPYASFALWAVNNRGTPFDLRLATIPACFSKSSAVYNPVIYVVLNKQ 0 | |||
0 FRSCMKKMLGMSGGDDEDSSASQSVTEVSKVSPS* 0 | |||
>SWS2_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gt | |||
0 MRGVRQHEFQEDFYIPIPLDVDNITALSPFLVPQDHLGSPAVFYGMSAFMFFLFVAGTGINVLTIACTIQYKKLRSHLNYILVNLAFSNLLVTTVGSFTCFCCFFVRYMIVGPLGCKIEGFAATLG 1 | |||
2 GMVSLWSLAVVAFERWLVVCKPLGNFIFKPDHAIVCCIFTWFFALIISAPPLFGWSR 2 | |||
1 YIPEGFQCSCGPDWYTTGNKYNNESYVWFIFGFGFAVPLFVIVFCYSQLLVMLKS 0 | |||
0 AKAQAESASTQKAEREVTRMVVVMILGFLVCWLPYASFALWVVNNRGTPFDLRLATIPACFSKASTVYNPIIYVVLNKQ 0 | |||
0 FRSCMKKMLGMSGGDDEESSSQSVTEVSKVSPS* 0 | |||
>SWS2_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gt | |||
0 MKHGRVPEIPEDFYIPISLDTDNITSLSPFLVPQDHLASKATFYSLAFYMFFILIVGTFINALTVACTVQNKKLRSHLNYILVNLAVSNLLVSGVGAFTAFLSFAARYFVLGTLACKVEGFLATLG 1 | |||
2 GMVSLWSLAVIAFERWLVICKPLGNFIFKPDHALVCCAFTWVFALAASAPPLVGWSR 2 | |||
1 YIPEGLQCSCGPDWYTTNNKYNNESYVLFLFGFCFAVPFCTICFCYSQLLFTMKMA 0 | |||
0 AKAQAESASTQKAEREVTRMVVLMVMGFLVCWMPYASFALWVVNNRGQTFDLRFASIPSVFSKSSAVYNPVIYVLLNKQ 0 | |||
0 FRSCMMKMLGMGGGDDEESSTSSVTEVSKVGPA* 0 | |||
>SWS2_oryLat Oryzias latipes (medaka) Deut.Acti.Perc AB223056 16460888 full Gt | |||
0 MRFISGGELPDDFWIPIPLDTNNLSLLSPFLVPQDHLGSSGTFYAMAAFMFFLFVFGTSINSLTIACTFQNKKLRSHLNYILVNLSVANLLVSGVGSSTAFCSFACRYFVFGSLACKIEGFAATLG 1 | |||
2 GMVGLWSLAVIAFERWLVICKPLGNFTFKPEHALACCLVTWVCALAAAAPPLAGWSR 2 | |||
1 YIPEGLQCSCGPDWYTTNNKYNNESYVMFLFCFCFAVPFATIVFCYSQLLVTLKMA 0 | |||
0 AKAQAESASTQKAEREVTRMVVVMVLGFLVCWMPYASFALWVVNNRGHSFDLRLATIPSCLSKASTVYNPVIYVLLNKQ 0 | |||
0 FRSCMLTMLGMGGGEEEDSTSVTEVSKVGPA* 0 | |||
>SWS2_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366492 17463225 full Gt | |||
0 MYQGKSTQVDDLPEDFYIPIALNVKNMSELSPFLVPQVHLGDSFIFYGMSAFMLFLVLAGFPLNFLTVFVTIKYKKLRSHLNYILVNLAIANLIVVCCGSTLAFYSFMHKYFILGPLFCKMEGFTATLG 1 | |||
2 GMLSLWSLAVLAFERCLVICKPFGNIAFRGTHALIRCGFAWAAAIAASTPPLFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTTNNKYNNESYVMFLFIFCFGTPFTIIIVSYSKLILTLRA 0 | |||
0 AAAQQQESASTQKAEKEVSRMVVIMVGGFLVCWLPYASLALWIVFNRGSPFDLRLATIPSVFSKASTVYNPVIYIFLNKQ 0 | |||
0 FRSCMMKTIFCGKNPLGDDEDATSTTTQVSSVSTSQVAPA* 0 | |||
>SWS1_homSap Homo sapiens (human) Deut.Euth.Euar NM_001708 1385866 full Gt OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC) | |||
0 MRKMSEEEFYLFKNISSVGPWDGPQYHIAPVWAFYLQAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVA 1 | |||
2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWSR 2 | |||
1 FIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKA 0 | |||
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQ 0 | |||
0 FQACIMKMVCGKAMTDESDTCSSQKTEVSTVSSTQVGPN* 0 | |||
>SWS1_monDom Monodelphis domesticus (opossum) Deut.Meta.Dide genomic full Gt OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC) | |||
0 MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHFQTVFMGFVFCAGTPLNAVVLVATLRYKKLRQPLNYILVNVSLCGFIFCIFAVFTVFISSSQGYFIFGRHVCAMEAFLGSVA 1 | |||
2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWVIGIGVSIPPFFGWSR 2 | |||
1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIMPLFLICFSYSQLLRALRA 0 | |||
0 VAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNQNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 | |||
0 FHACIMEMVCRKPMTDDSDVSSSQKTEVSAVSSSQVGPT* 0 | |||
>SWS1_smiCra Sminthopsis crassicaudata (dunnart) Deut.Meta.Dasy AY442173 full G? OPN1SW | |||
0 MSGDEEFYLFKNISLVGPWDGPQYHLAPAWAFHFQTAFMGFVFFAGTSLNGVVLIATLRYKKLRQPLNYILVNISLAGFIFCVFSVFTVFVSSSQGYFVFGRHVCAMEGFLGSVA 1 | |||
2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWIIGIGVSIPPFFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLICFSYSQLLGALRA 0 | |||
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAAMAMYMVNNRNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 | |||
0 FHACIMEMICKKPMTDDSETTSSQKTEVSTVSSSQVGPS* 0 | |||
>SWS1_tarRos Tarsipes rostratus (honey_possum) Deut.Meta.Dipr AY772472 18426754 full G? OPN1SW | |||
0 MSGDEEFYLFKDISSVGPWDGPQYHIAPAWAFHFQTTFMGFVFFAGTPLNAVVLIATLRYKKLRQPLNYILVNISLAGFIFCVISVFTVFISSSQGYFIFGRHVCAMEAFLGSVA 1 | |||
2 GLVTGWSLAFLAFERFIVICKPFGNFRFSSKHAMMVVLATWVIGIGVSIPPFFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTVGTKYHSEYYTGFLFIFCFIVPLSLICFSYSQLLGALRA 0 | |||
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSACVYNPIVYWFMNKQ 0 | |||
0 FHACIMEMVCRKPMTDDSEISSSQKTEVSTVSSSQVGPS* 0 | |||
>SWS1_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full Gt OPN1SW violet | |||
0 MSSDDDFYLFTNGSVPGPWDGPQYHIAPPWAFYLQTAFMGIVFAVGTPLNAVVLWVTVRYKRLRQPLNYILVNISASGFVSCVLSVFVVFVASARGYFVFGKRVCELEAFVGTHG 1 | |||
2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSRHALLVVVATWLIGVGVGLPPFFGWSR 2 | |||
1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0 | |||
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 | |||
0 FRACIMETVCGKPLTDDSDASTSAQRTEVSSVSSSQVGPT* 0 | |||
>SWS1_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full Gt OPN1SW | |||
0 MDEEEFYLFKNQSSVGPWDGPQYHIAPMWAFYLQTIFMGLVFVAGTPLNAIVLIVTIKYKKLRQPLNYILVNISVSGLMCCVFCIFTVFIASSQGYFVFGKHMCAFEGFAGATG 1 | |||
2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSRHALLVVAATWIIGVGVAIPPFFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0 | |||
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCMCYVPYAALAMYMVNNREHGIDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0 | |||
0 FRACIMETVCGRPMTDDSEVSSSAQRTEVSSVSSSQVGPS* 0 | |||
>SWS1_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt OPN1SW syn(+SWS1 -CALU) | |||
0 MSGQEDFYLFENISSVGPWDGPQYHIAPMWAFYFQTAFMGFVFFAGTPLNAIILIVTVKYKKLRQPLNYILVNISFAGFLFCTFSVFTVFMASSQGYFFFGRHVCAMEAFLGSVA 1 | |||
2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSRHALLVVAATWIIGVGVAIPPFFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLTLIIFSYSQLLGALRA 0 | |||
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYASLAMYMVNNRDHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0 | |||
0 FRACILETVCGKPMSDESDVSSSAQKTEVSSVSSSQVSPS* 0 | |||
>SWS1_utaSta Uta stansburiana (lizard) Deut.Saur.Lepi DQ100325 16543463 full Gt OPN1SW | |||
0 MSGEEDFYLFENISSVGPWDGPQYHIAPMWAFYFQTAFMGFVFFAGTPLNAIILIVTVKYKKLRQPLNYILVNISFAGFLFCVFSVFTVFLASSQGYFFFGRHICALEAFLGSVA 1 | |||
2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSKHALLVVAATWFIGIGVSIPPFFGWSR 2 | |||
1 FIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLTLIIFSYSQLLGALRA 0 | |||
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGIDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 | |||
0 FRACIMETVCGKPMTDESDVSSSAQKTEVSSVSSSQVSPS* 0 | |||
>SWS1_xenLae Xenopus laevis (frog) Deut.Amph.Anur genomic full Gt OPN1SW syn(+SWS1 -CALU) | |||
0 MLEEEDFYLFKNVSNVSPFDGPQYHIAPKWAFTLQAIFMGMVFLIGTPLNFIVLLVTIKYKKLRQPLNYILVNITVGGFLMCIFSIFPVFVSSSQGYFFFGRIACSIDAFVGTLT 1 | |||
2 GLVTGWSLAFLAFERYIVICKPMGNFNFSSSHALAVVICTWIIGIVVSVPPFLGWSR 2 | |||
1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFIFIFCFVIPLSLICFSYGRLLGALRA 0 | |||
0 VAAQQQESASTQKAEREVSRMVIFMVGSFCLCYVPYAAMAMYMVTNRNHGLDLRLVTIPAFFSKSSCVYNPIIYSFMNKQ 0 | |||
0 FRGCIMETVCGRPMSDDSSVSSTSQRTEVSTVSSSQVSPA* 0 | |||
>SWS1_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt OPN1SW | |||
0 MSGEEEFYLFKNISSVGPWDGPQYHIAPKWAFFLQAAFMGFVLFVGTPLNAIVLFVTIKYKKLQQPLNYILVNISLAGFIFCFFGVFAVFIASCQGYFIFGKTVCALEGFTGSVA 1 | |||
2 GLVTGWSLAILAFERYLVICKPIGNFRFGSKHSMIAVVAAWVIGVGVSIPPFFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIIPLFIICFSYSQLLGALRA 0 | |||
0 VAAQQQESATTQKAEREVSRMIIVMVGSFCVCYVPYAALAMYMVNNRDHGIDLRLVTIPAFFSKSSFVYNPIIYCFMNKQ 0 | |||
0 FRACIMQTVFGKPMTDDSDISSSGKTEVSSVSSSQVNPS* | |||
>SWS1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full Gt OPN1SW syn(-PNPLAS +IRG5 -TNPO3 +SWS1 -CALU -DYRK2) | |||
0 MDAWAVQFGNASKVSPFEGEQYHIAPKWAFYLQAAFMGFVFIVGTPMNGIVLFVTMKYKKLRQPLNYILVNISLAGFIFDTFSVSQVSVCAARGYYSLGYTLCSMEAAMGSIA 1 | |||
2 GLVTGWSLAVLAFERYVVICKPFGSFKFGQGQAVGAVVFTWIIGTACATPPFFGWSR 2 | |||
1 YIPEGLGTACGPDWYTKSEEYNSESYTYFLLITCFMMPMTIIIFSYSQLLGALRA 0 | |||
0 VAAQQAESESTQKAEREVSRMVVVMVGSFVLCYAPYAVTAMYFANSDEPNKDYRLVAIPAFFSKSSSVYNPLIYAFMNKQ 0 | |||
0 FNACIMETVFGKKIDESSEVSSKTETSSVSA* 0 | |||
>SWS1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc DW624108 full G? OPN1SW syn(ARF4 +IRG5 -TNPO3 +SWS1) BT028514 brain transcript | |||
0 MGKHFHLYENISKVDPFEGPQYYLAPMWAFYLQTAFMGFVFFAGTPLNFTVLLVTAKYKKLRVPLNYILVNITLAGFIFVTFSVSQVFVASARGYYFLGYTLCALEAAMGSVA 1 | |||
2 GLVTAWSLAVLSFERYLIICKPFGAFKFTSNHALGAVAFTWFMGICCSSPPFFGWSR 2 | |||
1 YIPEGLGCSCGPDWYTHNVEFNCTSYLYFLIVTCFIMPLSIIIFSYSQLLGALRA 0 | |||
0 VAAQQAESESTQKAEKEVSKMIIVMVGSFVACYAPYAITGLWFAFSPDENKDYRLVTVPAYFSKSSCVYNPLIYAFMNKQ 0 | |||
0 FNACIMETVFGKPMDESSEVSSKTEVSSVSTAS* 0 | |||
>SWS1_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full Gt OPN1SW syn(-TNPO3 +SWS1 +SOCS2 -CRADD +ARHGAP8) | |||
0 MGKYFYLYENISKVGPYDGPQYYLAPTWAFYLQAAFMGFVFFVGTPLNFVVLLATAKYKKLRVPLNYILVNITFAGFIFVTFSVSQVFLASVRGYYFFGQTLCALEAAVGAVA 1 | |||
2 GLVTSWSLAVLSFERYLVICKPFGAFKFGSNHALAAVIFTWFMGVGCACPPFFGWSR 2 | |||
1 YIPEGLGCSCGPDWYTNCEEFSCASYSKFLLVTCFICPITIIIFSYSQLLGALRA 0 | |||
0 VAAQQAESASTQKAEKEVSRMIIVMVASFVTCYGPYALTAQYYAYSQDENKDYRLVTIPAFFSKSSCVYNPLIYAFMNKQ 0 | |||
0 FNGCIMEMVFGKKMEEASEVSSKTEVSTDS*0 | |||
>SWS1_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic pseu G? OPN1SW exonic pseudogene frameshifts stops no synteny homSap | |||
0 MSGDEEFYLFKNISKVGPLDGPHFHIATKWAFDFQAAFMGFVFLCGtPLNAIVLIVTVKCKKLRQPLTYMLVNISAAGLVFCLFSISTVFLFSTQGYFVFGPTVCALESLFGSMA 1 | |||
2 GLVTGWSLAFLAAERYIVICKPFGNFRFGSIHSLFAFCLTWVLGLGVALPPFFGWSR 2 | |||
1 YIPeGLQCSCSPDWNTVGTKYESEYCTYFLFVFCFFVQLSIIIFSYGKLLNTLra 0 | |||
0 VAVQqQESSLSSTQKAEREMSRMVIVMVGSFCTCYVAALALYVVTNRDHNIDLRFVTVPAFFSKASCVYNPLIYSFMNKQ 0 | |||
0 FRARIMETVCGKFITDESETSSSRTAVSSVSTSQVSPG* 0 | |||
>SWS1_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366495 17463225 full Gt OPN1SW | |||
0 MSGDEEFYLFKNISKVGPWDGPQFHIAPKWAFYLQAAFMGFVFICGTPLNAIVLVVTIKYKKLRQPLNYILVNISAAGLVFCLFSISTVFVASMQGYFFLGPTICALEAFFGSLA 1 | |||
2 GLVTGWSLAFLAAERYIVICKPFGNFRFGSKHALVAVGLTWMLGLSVALPPFFGWSR 2 | |||
1 YIPEGLQCSCGPDWYTVGTKYKSEYYTYFLFVFCFVVPLSIIIFSYGSLLGTLRA 0 | |||
0 VAAQQQESASTQKAEREVSRMVIMMVASFCTCYVPYAALAVYMVTNRDHNIDLRFVTVPAFFSKASCVYNPLIYSFMNKQ 0 | |||
0 FRACILETVCGKPITDESETSSSRTEVSSVSTTQMIPG* 0 | |||
>LWS_homSap Homo sapiens (human) Deut.Euth.Euar NP_000504 12853434 full Gt OPN1LW long wavelength syn(-IRAK1 -MECP2 -TEX28) OPN1MW deutan | |||
0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 | |||
2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 | |||
2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRA 0 | |||
0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 | |||
0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 | |||
>LWS_monDom Monodelphis domesticus (opossum) Deut.Meta.Dide genomic full G? OPN1LW long wavelength exon 1:4 residue insert | |||
0 MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNNTR 1 | |||
2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 | |||
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTAPPLFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYVQVWLAIRA 0 | |||
0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYSFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRTCILQLFGKKVDDGSEVSSTSKTEGSSVSSVAPA* 0 | |||
>LWS_macEug Macropus eugenii (wallaby) Deut.Meta.Dipr genomic full G? OPN1LW long wavelength | |||
0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 | |||
2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | |||
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPLFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGNSDPGVQSYMIVLMSTCCILPLSVIFLCYIQVWLAIRS 2 | |||
0 VAKQQKESESTQKAEKEVSRMVVVMILAFCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 | |||
>LWS_smiCra Sminthopsis crassicaudata (dunnart) Deut.Meta.Dasy EU232013 18426754 full G? OPN1LW long wavelength | |||
0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1 | |||
2 GPFEGPNYHIAPRWVYNLTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1 | |||
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIILCYIQVWLAIRA 0 | |||
0 VAKQQKESESTQKAEKEVSRMVMVMILAFCFCWGPYALFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 | |||
>LWS_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono ABN43074 17339011 full Gt OPN1LW long wavelength syn(-IRAK1 -MECP2 - -) LWS green | |||
0 MTPAWNSGVYAARRRFEDEEDTTRTSVFVYTNSNNTR 1 | |||
2 DPFEGPNYHIAPRWAYNVTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC 1 | |||
2 GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIVLCYLQVWLAIRA 0 | |||
0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA* 0 | |||
>LWS_galGal Gallus gallus (chicken) Deut.Saur.Arch NM_205438 12716987 full Gt OPN1LW iodopsin missing in assembly | |||
0 MAAWEAAFAARRRHEEEDTTRDSVFTYTNSNNTR 1 | |||
2 GPFEGPNYHIAPRWVYNLTSLWMIFVVAASVFTNGLVLVATWKFKKLRHPLNWILVNLAVADLGETVIASTISVINQISGYFILGHPMCVVEGYTVSAC 1 | |||
2 GITALWSLAIISWERWFVVCKPFGNIKFDGKLAVAGILFSWLWSCAWTAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFFPLAIIILCYLQVSLAIRA 0 | |||
0 VAAQQKESESTQKAEKEVSRMVVVMIVAYCFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRNCILQLFGKKVDDGSEVSTSRTEVSSVSNSSVSPA* 0 | |||
>LWS_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt OPN1LW long wavelength syn(- - -TEX28 +TKTL1) | |||
0 MAGTVTEAWDVAVFAARRRNDEDDTTRDSLFTYTNSNNTR 1 | |||
2 GPFEGPNYHIAPRWVYNITSVWMIFVVIASIFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVIASTISVINQISGYFILGHPMCVLEGYTVSTC 1 | |||
2 GISALWSLAVISWERWVVVCKPFGNVKFDAKLAVAGIVFSWVWSAVWTAPPVFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSDDPGVLSYMIVLMITCCFIPLAVILLCYLQVWLAIRA 0 | |||
0 VAAQQKESESTQKAEKEVSRMVVVMIIAYCFCWGPYTVFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSNSSVSPA* 0 | |||
>LWS_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt OPN1LW long wavelength syn(-IRAK1 -MECP2 - -) | |||
0 MASHWNEAVFAARRRNDDDDTTRSSVFTYTNSNNTR 1 | |||
2 GPFEGPNYHIAPRWVYNISSLWMIFVVLASVFTNGLVLVATLKFKKLRHPLNWILVNMAIADLGETVIASTISVCNQIFGYFVLGHPMCILEGYTVSVC 1 | |||
2 GIAALWSLTVIAWERWFVVCKPFGNIKFDGKLAATGIIFSWVWAAGWCAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMLVLMITCCIIPLAIIVLCYMHVWLTIRQ 0 | |||
0 VAQQQKESESTQKAEREVSRMVVVMIIAYIFCWGPYTFFACFAAFNPGYNFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRNCIYQLFGKKVDDGSEVSSTSRTEVSSVSNSSVSPA* 0 | |||
>LWS_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt OPN1LW long wavelength | |||
0 MAEPWDAVLAARRRHQDEETTRSTIFVYTNSNNTR 1 | |||
2 GPFEGPNYHIAPRWVYNLTSLWMIFVVFASCFTNGLVLMATYKFKKLRHPLNWILVNLAIADLGETLIASTISVTNQIFGYFILGHPMCMLEGFTVATC 1 | |||
2 GITGLWSLTIIAWERWVVVCKPFGNIKFDGKWAAGGIIFSWVWSAFWCAMPLFGWSR 2 | |||
1 FWPHGLKTSCGPDVFSGEDKYGTRSFMIALMITCCIIPLGVIILCYIQVWWAIRT 0 | |||
0 VAKQQKESESTQKAEKEVSRMVVVMIFAYCFCWGPYTFMACFGAAYPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRNCIYQLLGKKVDDGSELSSTSKTEVSSVSNSSVSPA* 0 | |||
>LWS_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full Gt | |||
0 MAEHWGDAIYAARRKGDETTREAMFTYTNSNNTK 1 | |||
2 DPFEGPNYHIAPRWVYNVATVWMFFVVVASTFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETLFASTISVINQFFGYFILGHPMCIFEGYTVSVC 1 | |||
2 GIAALWSLTVISWERWVVVCKPFGNVKFDAKWASAGIIFSWVWAAAWCAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSEDPGVQSYMVVLMITCCIIPLAIIILCYIAVYLAIHA 2 | |||
0 VAQQQKDSESTQKAEKEVSRMVVVMIFAYCFCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 | |||
0 FRVCIMQLFGKKVDDGSEVSTSKTEVSSVAPA* 0 | |||
>LWS_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? | |||
0 MAEEWGKQSFAARRYHEDSTRGSAFVYTNSNHTR 1 | |||
2 DPFEGPNYHIAPRWVYNLATLWMFFVVVLSVFTNGLVLVATAKFKKLRHPLNWILVNLAVADLGETLFASTISVCNQFFGYFILGHPMCIFEGYVVSVC 1 | |||
2 GIAGLWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWIWPICWCAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCIIPLAIIVLCYLAVWMAIRA 2 | |||
0 VAMQQKESESTQKAEREVSRMVVVMILAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRVCIMKLFGKEVDDGSEVSTSKTEVSSVAPA* 0 | |||
>LWS_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gt OPN1LW long wavelength | |||
0 MAEEWGKQSFAARRYHEDTTRGSAFVYTNSNHTR 1 | |||
2 DPFEGPNYHIAPRWVYNVATVWMFIVVVLSVFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYTVSTC 1 | |||
2 GIAALWSLTIISWERWVVVCKPFGNVKFDAKWATGGIVFSWVWAAVWCAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCIIPLAIIILCYLAVWLAIRS 0 | |||
0 VAMQQKESESTQKAEKEVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 | |||
0 FRVCIMKLFGKEVDDGSEVSTSKTEVSSVAPA* 0 | |||
>LWS_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gt OPN1LW long wavelength | |||
0 MAEEWGKQAFAARRYNEDTTRGSMFVYTNSNNTK 1 | |||
2 DPFEGPNYHIAPRWVYNLSTLWMFIVVALSVFTNGLVLVATAKFKKLQHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSVC 1 | |||
2 GITALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWIWSAVWCAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCLIPLAIIILCYLAVWLAIRA 0 | |||
0 VAMQQKESESTQKAERDVSRMVVVMIVAYIVCWGPYTTFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 | |||
0 FRSCIMQLFGKEVDDGSEVSTsKTEVSSVAPA* 0 | |||
>LWS_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full Gt | |||
0 MAEQWGKQVFAARRQNEDTTRGSAFTYTNSNHTR 1 | |||
2 DPFEGPNYHIAPRWVYNLATLWMFFVVVLSVFTNGLVLVATAKFKKLRHPLNWILvNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSTC 1 | |||
2 GIAALWSLTIISWERWVVVCKPFGNVKFDAKWAIGGIVFSWVWSAVWCAPPVFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSDDPGVQSYMIVLMITCCIIPLAIIILCYLAVWLAIRA 0 | |||
0 VAMQQKESESTQKAEREVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 | |||
0 FRTCIMQLFGKQVDDGSEVSTSKTEVSSVAPA* 0 | |||
>LWS1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo EF565165 19196633 full G? OPN1LW long wavelength | |||
0 MTQSWELVAPAARRGFKYDEPTHSGIFVYTNSNQTR 1 | |||
2 GPFEGPNYHIAPRWAYNLTSVWMVGVVVASVFTNGLVLVATVRFKKLRHPLNWILVNMALADLGETVLASTVSVANQFFGYFILGHPLCVFEGFVVSLC 1 | |||
2 GITALWSLTIIAWERWVVVCKPFGNMKFDSKMAVAGIVFSWVWSAGWCLPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGNEDPGVQSYMVALTLSCAVLPLLIIILCYFQVWWAIRA 0 | |||
0 VALQQKESESTQKAEKEVSRMVVVMVAAFCLCWGPYACFAMFSALNPGYAFHPLVASIPSYFAKSSTIYNPIIYVFMNRQ 0 | |||
0 FRNCILQLFGKKVDDGSELSSTSKTDVSSVSNSSVSPA* 0 | |||
>LWS2_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo EF565166 19196633 full G? OPN1LW long wavelength | |||
0 MAEPRGSVAFAARRWNDHEGTTVGEFTYTNSNSTR 1 | |||
2 DPFEGPNYHIAPRWTYNLTSLWMVVVVILSVFTNGLVLVATWKFKKLRHPLNWILVNLAIADLGETLFASTISICNQVFGYFILGHPMCVFEGFTVSAC 1 | |||
2 GITALWSLTIIAWERWVVVCKPFGNVKFDGKWAAFGIIFSWVWSIGWCLPPVFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSDPGVKSYMVTLVITCAALPLTIIIVCLYQVWLAIRA 0 | |||
0 VAMQQKESESTQKAEKEVSRMVVVMIIAFCFCWGPYTSFAVFSALNPGYSFHPLMAALPAYFAKSSTIYNPIIYVFMNRQ 0 | |||
0 FRNCILQLFGKKVDDGSELSSTSKTDVSSVSNSSVSPA* 0 | |||
>LWS_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic full Gt OPN1LW long wavelength traces key to intron 3 position and gapping | |||
0 MTASWQGAMFAARRRQDDEDTTMESLFRYTNENNTK 1 | |||
2 DPFEGPNYHIAPRWVFNLTSVWMIIVVVLSLFSNGLVLVATVKFKKLRHPLNWIIVNLAIADILETIFASTISVCNQVYGYFILGHPMCVFEGYVVSTC 1 | |||
2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIATILIVFSWVWPASWCSLPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFLPLSIIILCYLQVWLAIHS 0 | |||
0 VAQQQKESETTQKAERDVSRMVVVMILAYVFCWGPYTFFACFAAANPGYSFHPIAAALPAYFAKGATIYNPIIYVFMNRQ 0 | |||
0 FRNCILQLFGKKVDDGSEVSSSSRTEVSSVSNSSVSPA* 0 | |||
>LWS_letJap Lethenteron japonicum (lamprey) Deut.Agna.Hype AB116381 15096614 full Gt OPN1LW long wavelength | |||
0 MTASWHGAVFAARRRNDDEDTTKDSIFRYTNENNTR 1 | |||
2 DPFEGPNYHIAPRWMFNLTSVWMIIVVVLSLFTNGLVLVATMKFKKLRHPLNWILVNLAIADILETIFASTISVCNQVFGYFILGHPMCVFEGYVVSTC 1 | |||
2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIAIILIVFSWVWPACWCSLPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFLPLSVIILCYLQVWLAIHS 0 | |||
0 VAQQQKESETTQKAERDVSRMVVVMILAYIFCWGPYTFFACYAAANPGYAFHPLTAALPAYFAKSATIYNPVIYVFMNRQ 0 | |||
0 FRNCIMQLFGKKVDDGSEVSSASRTEVSSVSNSSISPA* | |||
>LWS_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366491 17463225 full Gt OPN1LW long wavelength | |||
0 MAQSWERAMFAARRRQDEDTTKGDLFRYTNENNTR 1 | |||
2 DPFEGPNYHIAPRWMYNLTSFWMIIVVILSLFTNGLVLVATLKFKKLRHPLNWILVNLAIADIGETIFASTVSVVNQIFGYFILGHPLCVFEGFTVSVC 1 | |||
2 GITALWSLAIISFERWMVVCKPFGNLKFDGKVAIVLIIFSWAWSAGWCAPPIFGWSR 2 | |||
1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFIPLALIIICYLQVWLAIHT 0 | |||
0 VAQQQKESETTQKAERDVSRMVVVMIFAYIFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 | |||
0 FRNCIMQLFGKKVDDGSEVSSSARTEVSSVSNSSVSPA* 0 | |||
>PIN_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full Gt pinopsin pineal non-visual | |||
0 MSSNSSQAPPNGTPGPFDGPQWPYQAPQSTYVGVAVLMGTVVACASVVNGLVIVVSICYKKLRSPLNYILVNLAVADLLVTLCGSSVSLSNNINGFFVFGRRMCELEGFMVSLT 1 | |||
2 GIVGLWSLAILALERYVVVCRPLGDFQFQRRHAVSGCAFTWGWALLWSTPPLLGWSSYVPE 1 | |||
2 GLRTSCGPNWYTGGSNNNSYILSLFVTCFVLPLSLILFSYTNLLLTLRA 0 | |||
0 AAAQQKEADTTQRAEREVTRMVIVMVMAFLLCWLPYSTFALVVATHKGIIIQPVLASLPSYFSKTATVYNPIIYVFMNKQ 0 | |||
0 FQSCLLEMLCCGYQPQRTGKASPGTPGPHADVTAAGLRNKVMPAHPV* 0 | |||
>PIN_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full G? pinopsin | |||
0 MDSTQEPPNSSTPGPFDGPQWPHQAPRATYLVVAVLMGLVVASASLLNGLVIVVSVRHKRLRSPLNYILLNLAVANLLVTLCGSSVSLSNNISGFFVFGERLCQLEGFMVSLT 1 | |||
2 GIVGLWSLAILALERYLVVCRPLGDFRFQQQHAASGCAFTWGWSLLWTTPPLLGWSSYVPE 1 | |||
2 GLRTSCGPNWYTGGSNNSSYILALFVTCFVMPLSLILFSYTNLLLTLRA 0 | |||
0 AAARQQESDTTQQAEREVTRMVVAMVVAFLTCWLPYATFALVVATHKDIVIQPALASLPSYFSKTATVYNPIIYVFMNKQ 0 | |||
0 SCLLGMLCCGHHPRGMGKTSPAAPSPQVAAEGLRNKVTPSHPV* 0 | |||
>PIN_colLiv Columba livia (pigeon) Deut.Saur.Arch CLU50598 8982090 full G? pinopsin | |||
0 MDPTNSPQEPPHTSTPGPFDGPQWPHQAPRGMYLSVAVLMGIVVISASVVNGLVIVVSIRYKKLRSPLNYILVNLAMADLLVTLCGSSVSFSNNINGFFVFGKRLCELEGFMVSLT 1 | |||
2 GIVGLWSLAILALERYVVVCRPLGDFRFQHRHAVTGCAFTWVWSLLWTTPPLLGWSSYVPE 1 | |||
2 GLRTSCGPNWYTGGSNNNSYILTLFVTCFVMPLSLILFSYANLLMTLRA 0 | |||
0 AAAQQQESDTTQQAERQVTRMVVAMVMAFLICWLPYTTFALVVATNKDIAIQPALASLPSYFSKTATVYNPIIYVFMNKQ 0 | |||
0 FQSCLLKMLCCGHHPRGTGRTAPAAPASPTDGLRNKVTPSHPV* 0 | |||
>PIN_utaSta Uta stansburiana (lizard) Deut.Saur.Lepi DQ100321 16543463 full Gt pinopsin missing genome | |||
0 MVNEWSNATPGPFDGPQWPYLAPRSIYTSVAVLMGLVVVSAAFVNGLVIVVSIQYKKLRSPLNYILVNLAIADLLVTSFGSTLSFANNIYGFFVLGQTACEFEGFMVSLT 1 | |||
2 GIVGLWSLAILAFERYLVICKPVGDFRFQQRHAVFGCVFTWMWSLVWTLPPLFGWSSYVPE 1 | |||
2 GLRTSCGPNWYTGGSGNNSYIMALFVTCFALPLGMIIFSYASLLLTLRA 0 | |||
0 VATQQKEVETTQQAEKEVTRRVIAMVMAFLVCWLPYASFAMVVATNKDLVIQPALASLPSYFSKTATVYNPIIYVFMNKQ 0 | |||
0 FRSCLLSTMSCGHRPRGAQETTPAMISIPQGPTSALQGSRNKVTPSASEGSGNEAIPS* 0 | |||
>PIN_pheMad Phelsuma madagascariensis (gecko) Deut.Saur.Lepi AB022881 full Gt pinopsin no_ref AB022881 | |||
0 MHVQMANASQASLKNGTLSPFDGPQWPHRASRRVYTSLAALMGVVVLSASLANGLVIAVSVRFKRLRSPLNYILVNLATADLLVTFFGSIISFVNNAVGFFVFGKTACRFEGFMVSLT 1 | |||
2 GIVGLWSLAILAFERYLVICKPVGDFQFQRRHAVIGCLYTWGWSLIWTVPPLFGWSSYVPE 1 | |||
2 GLGTSCGPNWYMGGTNNNSYIVALFVTCFALPLSMILFSYANLLLTLRA 0 | |||
0 VAAQQKEQETTQRAEKEVTRMVITMVMAFLVCWLPYATFAMVVATTKDLSIQPGLASLPSYFSKTATVYNPIIYVFMNKQ 0 | |||
0 FRSCLLNTVSCGRIPQTMPGTPATTAVRGGFVLTSEGRGNKVASTELHS* 0 | |||
>PIN_podSic Podarcis sicula (lizard) Deut.Saur.Lepi DQ013042 16688437 full Gt pinopsin | |||
0 MQASNASWVEVRNRTPGPFEGPQWPYLAPQSTYISVAVLMGLVVISATLVNGLVIVVSVQFKKLRSPLNYVLVNLAVADLLVTFFGSTISFVNNAQGFFIFGQATCEFEGFMVSLT 1 | |||
2 GIVGLWSLAILAFERYLVICKPVGDFRFPARHAVLGCAFTWGWSFVWTVPPLLGWSSYVPE 1 | |||
2 GLRTSCGPNWYSGGSSNNSYIMTLFVTCFAMPLSTILFSYANLLMTLRT 0 | |||
0 VAAQQKEQETTQRAEREVTRMVVAMVAAFLVCWLPYASFAMVVATHKDLAIRPALASLPSYFSKTATVYNPIIYVFMNKQ 0 | |||
0 FRSCLLYKMSCGHRALSSQDTTPAGISLPGRLTTSASKGSRNQVSPS* 0 | |||
>PIN_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt pinopsin | |||
0 MRAGNMSAYEAPGPYDGPQWPHLAPRSTFLTVAAVMCMVVILAFFVNGLVIVVTLKYKKLRSPLNYILVNLAIANLLVTIFGSSVSFSNNVVGYFFMGKTMCEFEGFMVSLT 1 | |||
2 GIVGLWSLAILAFERYLVICKPMGDFRFQQKHAILGCSFTWVWSFIWTSPPLFGWCSYVPE 1 | |||
2 GLRTSCGPNWYTGGTNNNSYIMALFLTCFIMPLSTIIFSYSNLLMALRA 0 | |||
0 VAAQQKDSETTQRAEKEVTRMVIAMVLAFLICWLPYASFAVVVAVNKDVVIEPTVASLPSYFSKTATVYNPIIYVFMNKQ 0 | |||
0 FRNCLMTLLCCGRSFGDDETSSASGRTDVTSVSEAGGNKVTPA* 0 | |||
>PIN_bufJap Bufo japonicus (toad) Deut.Amph.Anur AF200433 9537517 full Gt pinopsin classifies oddly | |||
0 MHSANMSALETPGPFEGPQWPHVAPRSTYLTVAVLMGMVVFLAFFVNGMVIVVSLKYKKLRSPLNYILVNLAVADILVTMFGSTVSFHNNIFGFFTLGKLVCELEGFVVSLT 1 | |||
2 GIVGLWSLAILAFERYIVICKPMGDFRFQQRHAVMGCAFTWIWAFLWTSPPLIGWCSYVPE 1 | |||
2 GLGTSCGPNWYTGGTNNNSYILALFTTCFMMPLTTIIFSYSNLLLALRA 0 | |||
0 VAAQQKESETTQRAEREVTRMVIAMVLAFLICWLPYAVFAIVMASNKNVVIDPTLASMPSYFSKTATVYNPVIYVFMNKQ 0 | |||
0 FRDCLTKLLCCGRNPFGEDETSTTSGRTDVTSVSEGGGNKVTPA* 0 | |||
>PIN_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gt pinopsin | |||
0 FGSTVSFSNNINGYFVLGETVCQFEGFMVSLT 1 | |||
2 GIVGLWSLAILAFERYIVICKPMGDFRFQQKHAVWGCLFTWLWSLFWTLPPLFGWCSYVPE 1 | |||
2 GLRTSCGPNWYTGGANNSSYVVALFITCFTLPLSLIIFSYASLLVVLRA 0 | |||
>VAOP_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic 19664923 full Gt vertebrate ancient opsin exon 1 genbank error new stop *RKNGDEH | |||
0 MDVFRALGNESLLSNSSGPARWDPFHHPLDSIQPWHFRLVAAVMFVVTSLSLAENLAVILVTFKFKQLRQPVNYVIVNLSVADFLVSLTGGTISFLANLKGYFYMGHWACVLEGFAVTFF 1 | |||
2 GIVALWSLALLAFERYIVICRPVGNMRLRGKHAAQGIAFVWTFSFIWTIPPTMGWSSYTTSKIGTTCEPNW 2 | |||
1 YSGAYNDRSYIIAFFTTCFIVPLLVILVSYGKLLQKLRK 0 | |||
0 VSNTQGRLRTARKPERQVTRMVVVMIIAFLICWMPYAVFSILATAYPSIELDPHLAAIPAFFSKTATVYNPIIYVFMNKQ 0 | |||
0 FRMCLIQMFKCSAIETAESNMNPTSERATLTQDKRDSQLSVMAVRSTIS* 0 | |||
>VAOP_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full G? vertebrate ancient opsin last exon newly truncated | |||
0 MHLEYLKMGHFCPVTLLLPDGDPFHRPLDSIQPWQFKLLAAVMLLVTSLSLAENLAVILVTFKFKQLRQPINYIIVNLSVADFLVSLTGGTISFLTNLKGYFFMGYWACVLEGFAVTFF | |||
2 GIVALWSLALLAFERYIVICRPLRNARLRGKHAALGIVFVWSFSFIWTIPPTTGWSSYTTSKIGTTCEPNW 2 | |||
1 YSGAYADHTYIITFFTTCFIVPLLVILVSYGKLVWKLKK 0 | |||
0 VSDAQGRLGAARRPERQVTRMVVFMIVAFLICWMPYATFSILVTAYPSIELDPCVAAIPAFFSKTATVYNPVIYVFMNKQ 0 | |||
0 FRQCLIQMFSCSAIGTAESNMKLTSERAVLMQGRRGSKRTPMAVHSTVLKRKTGDEHRADDLWLF* 0 | |||
>VAOP_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt vertebrate ancient opsin syn(+INPP5A -NXK6 +GPR125 +KNDC1) | |||
0 MAGLRREAENDSWLFDPSSSSAPFDPFLQPLDIIEPWNFHLISALMFVVTLFSLSENFTVILVTIKFKQLRQPLNYVIVNLSVADFLVSLIGGTISFSTNLKGYFYMGHWACVLEGFAVTFF 1 | |||
2 GIVALWSLALLAFERYVVICRPLGNMRLNGKHAALGVAFVWIFSFIWTVPPTMGWSSYTTSKIGTTCEPNW 2 | |||
1 YSGDYNDHTFIITFFTTCFILPLLVILVSYGKLMRKLRK 0 | |||
0 VSDTQGRLGTTRKPERQVTGMVVIMILAFLICWSPYAAFSILVTACPSIELDPRLAAIPAFFSKTATVYNPVIYVFMNNQ 0 | |||
0 FRKCLVQLFQCSSQETMDANVNPISEKDTLTHTKHCGEMSTVAAHVIVFNPRSEDEQGSCQSFAQLAISENKVYPL* 0 | |||
>VAOP_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt vertebrate ancient opsin syn(- +GSTO2 -C10orf92 -) new | |||
0 MPTNVSLLATPENSTVWNPFTGPLKTIEAWNFHLLAALMFVVTSLSIAENFIVILVTAKFKQLRQPLNYIIVNLSVADFLVSVIGGTISIATNSRGYFYLGSWACVLEGFAVTFF 1 | |||
2 GIVALWSLSVLAFERYIVICRPLGNLRLQGKHSALAIIFVWVFSFVWTIPPTMGWSSYTTSKIGTTCEPNW 2 | |||
1 YSGEMRDHTYIITFLTTCFVFPLLVIFMSYGKLMRKLRK 0 | |||
0 VSDTQGRLGSTRKPEKEVTRMVVIMILAFLICWTPYAAFSILITAHPTIDLDPRLAAIPAFFAKTASMYNPIIYVYMNKQ 0 | |||
0 FRRCLYQMFNINDPEAKESNLNPTSERGVLTRNNNGGEMLAIATHITSSAVTNREEEKSSSNSFAHIPVSDNKVCPM* 0 | |||
>VAOP_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_131586 17067577 full Gt valop assembly missing exon 3 | |||
0 MEASSAAVNAVSPAEDPFSAPLSSIAPWNYSVLAALMFVVTALSLSENFTVMLVTFRFQQLRQPLNYIIVNLSLADFLVSLTGGSISFLTNYHGYFFLGKWACVLEGFAVTFF 1 | |||
2 GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLVFVWSFSFIWTVPPVLGWSSYTVSRIGTTCEPNW 2 | |||
1 YSGNFHDHTFIITLFSTCFIFPLGVIIVCYCKLIRKLRK 0 | |||
0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIIITAHPSMHVDPRLAAIPAFVAKTAAVYNPIIYVFMNKQ 0 | |||
0 FRKCLVQLLSCSKVTVVEGNNNQTTERAGMTSGSNTGEMSAIAARVSVPKTEENPGDRSTFSHIPIPENKVCPM* 0 | |||
>VAOP_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? vertebrate ancient opsin syn(INPP5A NKX6 +VAOP GABRP KNDC1) | |||
0 MESLSLSVNGVSHTVAAELAPTNDPFSGPINNIAQWNFTILAVLMFVVTALSLSENFLVMFITFKFKQLRQPLNYIIVNLAIADFLVSITGGLISFLTNARGYFFLGRWACVLEGFAVTYF 1 | |||
2 GIVAMWSLAVLSFERFFVICRPLGNMRLQAKHAVIGLLFVWTFSFVWTFPPVLGWNRYTVSKIGTTCEPDW 2 | |||
1 YSNNMTSHSYIITFFVTCFILPLGVIFFCYGKLLRKLRK 0 | |||
0 VSHGRLATARKPERQVTRMVVVMIVAFMVAWTPYATFAILVTIYPTIELDPRLASIPAFFSKTAAVYNPIIYVFMNKQ 0 | |||
0 FRKCLMQHFIGGGGTIESNMNPTSERPGITADSHTGEMSAIAARVPVGASASPLSDDSPTAQLPIPENKVCP* 0 | |||
>VAOP_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gt vertebrate ancient opsin syn(+INPP5A -NXK6 - +KNDC1) | |||
0 MESLSLSVNGVSYTVAAELAPTNDPFTGPINNIAQWNFTILAVLMFVVTSLSLCENFLVMFITFKFKQLRQPLNYIIVNLAIADFLVSLTGGLISFLTNARGYFFLGRWACVLEGFAVTYF 1 | |||
2 GIVAMWSLAVLSFERFFVICRPLGNMRLQAKHAAIGLLFVWTFSFVWTFPPVLGWNRYTVSKIGTTCEPDW 2 | |||
1 YSNNMTSHSYIITFFSTCFILPLGIIFFCYGKLLRKLRK 0 | |||
0 VSHGRLATARKPERQVTRMVVVMIVAFMVAWTPYATFAILVTIHPTIELDPRLASIPAFFSKTAAVYNPIIYVFMNKQ 0 | |||
0 FRKCLIQHFIGMGVMAESNMNPTSERPGITAESQTGEMSAIAARVPVGATAALHSDGSPTDCGSLAQLPIPENKVCPI* 0 | |||
>VAOP_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gt vertebrate ancient opsin | |||
0 MDTLSLSVNAVSHTVAAELPATSDPFNGPIKNIAPWNFTILAVLMFVVTSLSLSENFLVMFVTFKYKQLRQPLNYIIVNLAIADFLVSLTGGVISFLTNARGYFFLGKGACVLEGFAVTFF 1 | |||
2 GIVALWSLAVLSFERFFVICRPLGNIRLQPKHAILGLLFVWTFSFIWTFPPVLGWNRYTISKIGTTCEPDW 2 | |||
1 YSTNMIDHSYIITFFATCFILPLGVIFFCYGKLLRKLRK 0 | |||
0 VSHGRLATAKKPERQVTRMVVVMIVAFMVGWTPYAVFSLLVTSHPSIKLDPRLASIPAFFSKTAAVYNPIIYVFMNKQ 0 | |||
0 FRKCLIQLFSGRGTIPDGHLDPTSERPGMSQTGEMSAIATRIPMSSTVSPRSDDSPTDCVSFAQLPIPENKVCPM* 0 | |||
>VAOP_oryLat Oryzias latipes (medaka) Deut.Acti.Perc NM_001136515 full Gt vertebrate ancient opsin DK020629 brain DK098095 embryo | |||
0 MDSLSLSVNAVSFTVATEIFSTDDPFTGPIKNIAPWNFTVLAVLMFVITSLSLTENFLVMFVTFKFKQLRQPLNYIIVNLAIADFLVSLTGGLISFLTNARGYFFLGKWACILEGFAVTFF 1 | |||
2 GIVALWSLAVLSFERFFVICRPLGNIRLQSKHAMLGLLFVWTFSFIWTFPPVLGWNRYTVSKIGTTCEPDW 2 | |||
1 YSTNMSDHSYIITFFATCFILPLGVIFYCYGRLLRKLRKVS 0 | |||
0 VSHGRLATARKPERQVTRMVVVMIVAFMVGWTPYAVFSILVTAHPTIKLDPRLASIPAFFSKTAAVYNPIIYVFMNKQ 0 | |||
0 FRKCLIQHFTGMGTIPESNLNQTSERPGITAESHTGEMSAIAARIPMGGTTSPRSDDSQTDCGSFAQLPIPENKVCPL* 0 | |||
>VAOP_salSal Salmo salar (salmon) Deut.Acti.Salm NM_001123626 9136902 frag Gt vertebrate ancient opsin first VAOP seq, last exon wrong AF001499 | |||
0 MDTLRIAVNGVSYNEASEIYKPHADPFTGPITNLAPWNFAVLATLMFVITSLSLFENFTVMLATYKFKQLRQPLNYIIVNLSLADFLVSLTGGTISFLTNARGYFFLGNWACVLEGFAVTYF 1 | |||
2 GIVAMWSLAVLSFERYFVICRPLGNVRLRGKHAALGLLFVWTFSFIWTIPPVFGWCSYTVSKIGTTCEPNW 2 | |||
1 YSNNIWNHTYIITFFVTCFIMPLGMIIYCYGKLLQKLRK 0 | |||
0 VSHDRLGNAKKPERQVSRMVVVMIVAYLVGWTPYAAFSIIVTACPTIYLDPRLAAAPAFFSKTAAVYNPVIYVFMNKQ 0 | |||
>VAOP_pleAlt Plecoglossus altivelis (smelt) Deut.Acti.Salm AB074483 11779166 full Gt vertebrate.ancient.opsin brain and amacrine cells of retina | |||
0 MDTLRIPVNGVSYTEATEILKPEDPFTGPLSNIAPWNFTFLAVLMFFVSFLSLCENFTVMFVTFKFKQLRQPLNYIIVNLSIADFLVSLSGGLISFLTNAKGYFFLGKWACVLEGFAVTYF 1 | |||
2 GIVAMWSLAVLSFERFFVICRPLGNIRLRGKHAVLGLLFVWTFSFIWTIPPVLGWSSYTVSKIGTTCEPNW 2 | |||
1 YSTKLSDHIYIITFFFTCFILPLGVIIFCYGKLLRKLRK 0 | |||
0 VSHGRLGNARKPERQVTRMVVVMIVAFMVGWTPYAAFSIVVTACPHIYIDPRMAAIPAFFSKTAACYNPVIYVFMNKQ 0 | |||
0 FRKCLIKLFTGKGTIPEGNLGQTSERPGMTAESQTGEMSAIAARIPVGGTAATKSDEQPSECNSFAQLPIPENKVCPI* 0 | |||
>VAOP_rutRut Rutilus rutilus (minnow) Deut.Acti.Otoc AY116411 12906786 full Gt vertebrate ancient opsin | |||
0 MELFPVAVNGVSHAEDPFSGPLTFIAPWNYKVLATLMFVVTAASLSENFAVMLVTFRFTQLRKPLNYIIVNLSLADFLVSLTGGTISFLTNYHGYFFLGKWACVLEGFAVTYF 1 | |||
2 GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLLFVWTFSFIWTIPPVLGWSSYTVSKIGTTCEPNW 2 | |||
1 YSGNFHDHTFIIAFFITCFILPLGVIVVCYCKLIKKLRK 0 | |||
0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIVVTAHPSIHLDPRLAAAPAFFSKTAAVYNPVIYVFMNKQ 0 | |||
0 FRKCLVQLLRCRDVTIIEGNINQTSERQGMTNESHTGEMSTIASRIPKDGSIPEKTQEHPGERRSLAHIPIPENKVCPM* 0 | |||
>VAOP_cypCar Cyprinus carpio (carp) Deut.Acti.Otoc AF233520 -------- full Gt vertebrate.ancient.opsin --- | |||
0 MESLAAAVNGVSHTEDPFSGPLSSIAPWNYRVLAALMFVVTSVSLCENFTVMLVTFRFKQLRQPLNYIIVNLSLADFLVSLSGGTISFLTNFHGYFFLGRWACVLEGFAVTF 1 | |||
2 FGIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLLFVWSFSFVWTIPPVLGWSSYTVSKIGTTCEPNW 2 | |||
1 YSGNFQDHTFIITFFTTCFIFPLAVIIVCYCKLLRKLRK 0 | |||
0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIVVTALPSIHLDPRLAAAPAFFSKTAAVYNPIIYVFMNKQ 0 | |||
0 FRKCVVQLLSCRDVTVVEGNINQTTDRAGLTNESNTGEMSAIAARIPAAGTVPPKTEEPPNERSSFTQIPIPENKVCPM* 0 | |||
>VAOP_ictPun Ictalurus punctatus (catfish) Deut.Acti.Otoc FJ839436 -------- full Gt vertebrate.ancient.opsin --- | |||
0 MGSFGTAVRGVNYTEAADVIQPEDPFSGPVKSIAPWNFKVLAAWMVVITSLSLSENFLVMLVTYKFKQLRQPLNFLIVNLSLADFFMSLIGGTLSVVTNYRGYFFLGSWACVLEGFAVTF 1 | |||
2 FGIVGMWSLATLAFERYFVICRPLGNVRLQVKHAVLGLLFVWTFSFIWTIPPVLGWSSYTVSKIGTTCEPNWYSGDPHDHTFIITFFLTCFILPLAVIIVCYSKLLRKLRK 0 | |||
0 VSNSHGRLVNARKPDRQVSHMVVVMIVAFVVAWTPYALFSFIVTVHPTIYLDPIMAAVPAFFAKTAAVYNPIIYVFMNKQ 0 | |||
0 FRKCLIQLLSCSGNTTVEVNQSTDRAGMIGESNTGEMSAIAARIPAASTTSPPKANEQHSNSCSYSQLPIPENKVCPM* 0 | |||
>VAOP_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gt vertebrate ancient opsin | |||
0 VASTQGRLGVARKPEKQVTRMVIVMILAFLFCWTPYAAFSITVTACPTIKLDPRLAAIPAFFSKTATVYNPIIYVFMNKQ 0 | |||
>VAOP_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype U90667 9427550 full Gt vertebrate ancient opsin exons 123 in traces pineal gland-specific | |||
0 MDALQESPPSHHSLPSALPSATGGNGTVATMHNPFERPLEGIAPWNFTMLAALMGTITALSLGENFAVIVVTARFRQLRQPLNYVLVNLAAADLLVSAIGGSVSFFTNIKGYFFLGVHACVLEGFAVTYF 1 | |||
2 GVVALWSLALLAFERYFVICRPLGNFRLQSKHAVLGLAVVWVFSLACTLPPVLGWSSYRPSMIGTTCEPNW 2 | |||
1 YSGELHDHTFILMFFSTCFIFPLAVIFFSYGKLIQKLKK 0 | |||
0 ASETQRGLESTRRAEQQVTRMVVVMILAFLVCWMPYATFSIVVTACPTIHLDPLLAAVPAFFSKTATVYNPVIYIFMNKQ 0 | |||
0 FRDCFVQVLPCKGLKKVSATQTAGAQDTEHTASVNTQSPGNRHNIALAAGSLRFTGAVAPSPATGVVEPTMSAAGSMGAPPNKSTAPCQQQGQQQQQQGTPIPAITHVQPLLTHSESVSKICPV* 0 | |||
>PPIN_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt parapinopsin syn(-CPEB2 -CACNA2D3 +SELK) syntenic deleted chicken | |||
0 MDSLDTNTLSPNASTVRVVLMPRIGYTIIAIIMATSCTLSVILNTAVIAITIKYRQLRQPINYSLVNLAIADLGAALLGGSLNVETNAVGYYNLGRVGCVTEGFAMAFF 1 | |||
2 GIVALCTIAVIAVDRAIVIAKPMGTITFTTRKAMIGVAVSWIWSLVWNTPPLFGWGGYQMEGVMTSCAPDWANSDPINVSYIICYFLFCFTIPFITILASYGYLIWTLRQ 0 | |||
0 VAKVGLAQRGSTTKAEAQVSRMVIVMVMAFLICWLPYATFALVVVGNPQIYINPIIATIPMYMAKSSTFYNPIIYIFMNKQ 0 | |||
0 FRDCLVRCLLCGRNPCASEQTDEDDLEVSTIAPAPSSRRGKVAPV* 0 | |||
>PPIN_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt parapinopsin para bistable UV lamprey pineal broken contigs | |||
0 MADEALLPPMMNVTNEEMHPGKVLMPRIGYTILALIMAVFCAAALFLNVTVIVVTFKYRQLRHPINYSLVNLAIADLGVTVLGGALTVETNAVGYFNLGRVGCVIEGFAVAFF 1 | |||
2 GIAALCTIAVIALDRVFVVCKPMGTLTFTPKQALAGIAASWIWSLIWNTPPLFGWGSYELEGVMTSCAPNWYSADPVNMSYIVCYFSFCFAIPFLIIVGSYGYLMWTLRQ 0 | |||
0 VAKLGVAEGGTTSKAEVQVSRMVIVMILAFLVCWLPYAAFAMTVVANPGMHIDPIIATVPMYLTKTSTVYNPIIYIFMNKQ 0 | |||
0 FQECVIPFLFCGRNPWAAEKSSSMETSISVTSGTPTKRGQVAPA* 0 | |||
>PPIN_danRer Danio rerio (zebrafish) Deut.Acti.Otoc XM_681591 full Gt parapinopsin syn(- - +SELK -) | |||
0 MESETSTAASGSIAEVMPRMGYTILAVIIGVFSVCGVILNVTVITVTLKYKQLRQPLNFALVNLAVADLGCAVFGGLPTVVTNAMGYFSLGRVGCVLEGFAVAFF 1 | |||
2 GIAALCSVAVIALERCMVVCRPVGSISFQTRHAVFGVAVSWLWSFIWNTPPLFGWGRLQLEGVRTSCAPDWYSRDLANVSFIVCYFLLCFALPFSVIVYSYTRLLWTLRQ 0 | |||
0 VSRLQVCEGGSAARAEAQVSCMVVVMILAFLLTWLPYASFALCVILIPELYIDPVIATVPMYLTKSSTVFNPIIYIFMNRQ 0 | |||
0 FRDRALPFLLCGRNPWAAEAEEEEEETTVSSVSRSTSVSPA* 0 | |||
>PPINa_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? parapinopsin | |||
0 MKPSVYLNTSLYLGPPEEPPLPRSGFIVLSILMALVTGPAIVLNATVIIVSLMHKQLRQPLNYALVNMAAADLGTAVSGGLLSVVNNAQGHFSLGRTGCVLEGFAVSLC 1 | |||
2 GIASLCTVALIAVERMFVICKPLGQMQFQKQHALAGISLSWLWSLTWNLPPLFGWGRYELEGVGTSCAPDWQSREPHNVSYVLAYFTVCFAAPFAIILVSYAKLMWTLHK 0 | |||
0 VTKMTCLEGGAVAKSEMKVAYMVVLMVATFLLSWLPYAGLSMLVVFKPDVEINPLVGTVPVYLAKSSTAYNPIIYIYLNKQ 0 | |||
0 FRKYALPFLLCRRALEAEDEVSETTVESSRRVSPS* 0 | |||
>PPINa_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? parapinopsin | |||
0 MKPSAFYLNASLYLGPQGEPPLPRSGFIALSVIMALLTGPAIVLNATVIIVSLMHKQLRQPLNYALVNMAVADLGTAMTGGLLSVVNNAQGYFSLGRTGCVLEGFAVSLC 1 | |||
2 GIASLCTVALIAVERMFVICKPLGQMQFQKQHALGGIALAWLWSLTWNLPPLFGWGRYELEGVGTSCAPDWHSREPQNVSYVLAYFTVCFAAPFVIILVSYSKLMWTLHK 0 | |||
0 VTKMACMEGGAVAKSEMTVAYMVILMVVIFLISWLPYAGLSMLVVLSPDVKIHPLVGTVPVYLAKSSTVYNPIIYIYLNKQ 0 | |||
0 FRKYAVPFLLCGRELEMEDELSMTTVETSNRVSPA* 0 | |||
>PPINa_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc DN691174 full G? parapinopsin adult eyes | |||
0 MQPSHTFSNSSAYTGPHGEPPLSRTGFIILSIIMAGFTGPAIVLNATVIIVTLMHKQLRQPLNYALVNMALADLGTAMTGGVLSVVNNAQGYFSLGRSGCVMEGFAVSLF 1 | |||
2 GITSLCTVALIAVERMFVVCKPLGQIIFQKKHAVGGIAISWLWSLSWNLPPLFGWGRYELEGVGTSCAPDWHNQDPKNVSYIVAYFAVCFAVPFALILASYTKLMWTLHQ 0 | |||
0 VSKMTCLEGGAVAKGEMKVASMVVLMVLTFLISWLPYASLAMLVVYNPKVEIHALVGTVPVYLAKSSTVFNPIIYIYLNKQ 0 | |||
0 FRKYAVPFLLCCKEPLDDEEASEAATTVEISPSKVSPA* 0 | |||
>PPIN_ictPun Ictalurus punctatus (catfish) Deut.Acti.Otoc genomic full Gt parapinopsin index sequence | |||
0 MASIILINFSETDTLHLGSVNDHIMPRIGYTILSIIMALSSTFGIILNMVVIIVTVRYKQLRQPLNYALVNLAVADLGCPVFGGLLTAVTNAMGYFSLGRVGCVLEGFAVAFF 1 | |||
2 GIAGLCSVAVIAVDRYMVVCRPLGAVMFQTKHALAGVVFSWVWSFIWNTPPLFGWGSYQLEGVMTSCAPNWYRRDPVNVSYILCYFMLCFALPFATIIFSYMHLLHTLWQ 0 | |||
0 VAKLQVADSGSTAKVEVQVARMVVIMVMAFLLTWLPYAAFALTVIIDSNIYINPVIGTIPAYLAKSSTVFNPIIYIFMNRQ 0 | |||
0 FRDYALPCLLCGKNPWAAKEGRDSDTNTLTTTVSKNTSVSPL* 0 | |||
>PPIN_oncMyk Oncorhynchus mykiss (trout) Deut.Acti.Salm genomic full Gt parapinopsin | |||
0 MDHQQLLPNLHGNISSSPGSVSEALLSRTGFTILAVIIGVFSVSGVCMNVLVIMVTMRHRKLRQPLNYALVNLAVADLGCALFGGLPTMVTNAMGYFSMGRLGCVLEGFAVAFF 1 | |||
2 GIAGLCSVAVIAVDRYVVVCRPMGAVMFQTRHAVGGVVLSWVWSFLWNTPPLFGWGSFELEGVRTSCSPNWYSREPGNMSYIILYFLLCFAIPFSIIMVSYARILFTLHQ 0 | |||
0 VSKLKVLEGNSTTRVEIQVVRMVVVMVMAFLLSWLPYAAFALSVILDPSLHINPLIATVPMYLAKSSTVYNPIIYVFMNRQ 0 | |||
0 FRDCAVPFLLCGLNPWASEPVGSEADTALSSVSKNPRVSPQ* | |||
>PPINb_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? parapinopsin syn(+PCTK1 -PPIN +DDX20 -KCND2) absent medaka zfish | |||
0 MEQGIQEGNSSSLSSVSSGTLSRTGYTVLAFIMGVLSAGGIILNVLVIVVTMKHRQLRQPLSYALVNLAICDLGCALFGGIPTTITSAMGYFSLGRVGCVLEGFAVAFF 1 | |||
2 GIASLCTIGVISVERYVVVSNPMGAVLFQTR 2 | |||
1 HAVAGVVFSWVWSFVWNTPPLFGWGSFDLEGVRTSCAPNWYSRDVGIMSYIVIYLLFCFAVPFTIITVSYSRLLWTLRQ 0 | |||
0 VTGLQVAEGGSTNRVEVQVARMVVVMVLAFLLTWLPYAAMALAVVMDSTLYINPIIATIPVYLAKSSTVYNPIIYIFMNRQ 0 | |||
0 FRGCAINTVLCGRRAWITDLQTSEGETTVASTSKSQKISPKGSLN* 0 | |||
>PPINb_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? parapinopsin syn(-PPIN +DDX20) | |||
0 MEQEIQDGNSSSLNSVSPGILSQTGFTVLAFIMGMLSVGGIILNVLVIVVTLKHRQLRQPLNYALVNLAICDLGCALFGGIPTTVTSAMGYFSLGRLGCVLEGFAVAFF 1 | |||
2 GIASLCTIGIISVERYIVVSNPMGAVLFQTR 2 | |||
1 HAVAGVVFSWVWSFVWNTPPLFGWGSFELEGVGTSCAPNWYSRDMGNMSYIIIYLLFCFAVPFSIIMVSYSRLLWTLHQ 0 | |||
0 VTKLHVAEGGSTNRVEVHVARMVVLMVLAFLLTWLPYASMALAVVMDSTLYIDPVTATIPVYLAKSSTVYNPIIYIFMNRQ 0 | |||
0 FRGYAINTILCGRRAWVSEQQTSEGETTVVSVSKSQKISPKGSLQ * 0 | |||
>PPINb_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc DW621258 full G? parapinopsin syn(+DNMT2 -PPIN +DDX20 -TAZ) 21 day old larvae | |||
0 MERDGNASADAQLLSPSGYAALAAVMGAFSVAGILLNALVIVVTARHRQLRQPLSYALVNLAVCDLGCAACGGLPTTVTSAMGYFSLGRAGCVLEAFAVAFF 1 | |||
2 GIASLCTIGVISVERYVVVCYPMGAVLFQTR 1 | |||
2 HAVAGVVLSWVWSFVWNTPPLFGWGSYELEGVKISCAPNWYSRDPGNVSYVTIYFLLCFAVPFSVIMVSYSRLLWTLRQ 0 | |||
0 VTKLQVSETGSTNRVEVQVARMIVVMVLAFLVTWLPYAAMALAVITDSTLHIDPVIATIPVYLAKSSTVYNPIIYIFMNRQ 0 | |||
0 FRGYAVPSILCGWNPWAEEQTSEEETVGSVMKSQRVSPKGSLQE* 0 | |||
>PPINb_mayZeb Maylandia zebra (cichlid) Deut.Acti.Perc genomic frag G? parapinopsin | |||
0 1 | |||
2 GIASLCTVAVISVERYVVVCYPMGAVLFQTR 2 | |||
1 HAVAGVVLSWVWSFVWSTPPLFGWGTFELEGVKTSCAPKWHSRDVGDMSYMIIFFFLCFALPFSVVMVSYSRLLWTLRQ 0 | |||
0 GMIVVMVLAFLVTWLPYAALALAVMIDSSLYVDPVIATIPVYFAKSSTVYNPIIYIFMNRQ 0 | |||
0 FRGYTVAAVLCGWDPWSSEPQTSENETTVPFFIKTPKKIVPKKSLE*0 | |||
>PPIN_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gt parapinopsin | |||
0 MDPHNRSANLSEGPGLGGGGAVPGWGPSVRAPLSLVMAVISLSSIVLNSLAIAVVLRFQVLQQPLNYALLSLASADLGTAATGGVLSTVCTALGSFVLGRHSCVAEGFF 1 | |||
>PPINa_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic full Gt parapinopsin bistable pineal UV/green | |||
0 MENLTSLDLLPNGEVPLMPRYGFTILAVIMAVFTLASLVLNSTVIIVTLRHRQLRHPLNFSLVNLAVADLGVTVFGASLVVETNAVGYFNLGRVGCVIEGFAVAFF 1 | |||
2 GIAALCTIAVIAVDRFVVVCKPLGTLMFTRRHALLGITWAWLWSFVWNTPPLFGWGSYKLEGVRTSCAPDWYSRDPANVSYIVSFFSFCFAIPFLVIVVAYGRLLWTLHQ 0 | |||
0 VAKLGMGESGSTAKAEAQVSRMVVVMVVAFLVCWLPYALFAMIVVAKPGVYIDPVIATLPMYLTKTSTVYNPIIYIFMNRQ 0 | |||
0 FRDCAVPFLLCGRNPWAEPSSESATTASTSATSVTLASVPGQVSPS* 0 | |||
>PPINb_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag G? parapinopsin odd genomic | |||
2 GMTALITVCVLAVERYVVVCKPLGGVHFGTQHGLCGVAISWTWALAWSAPPLFGWGRYHYEGVGTSCAPDWADSSPSGRSYMTTYFIFCFALPMIIILFCYTKLMIAIHK 0 | |||
0 VSKLGLSANDTAERKVGIMVVVMVFAFFLCWLPYAALAIAVVIKPDLK 0 | |||
0 VSPVTASIPVYLAKSSGAYNPIIYIFMHRQ 0 | |||
>PPIN_letJap Lethenteron japonicum (lamprey) Deut.Agna.Hype AB116380 14981504 full Gt parapinopsin bistable pineal UV/green | |||
0 MENLTSLDLLPNGEVPLMPRYGFTILAVIMAVFTIASLVLNSTVVIVTLRHRQLRHPLNFSLVNLAVADLGVTVFGASLVVETNAVGYFNLGRVGCVIEGFAVAFF 1 | |||
2 GIAALCTIAVIAVDRFVVVCKPLGTLMFTRRHALLGIAWAWLWSFVWNTPPLFGWGSYELEGVRTSCAPDWYSRDPANVSYITSYFAFCFAIPFLVIVVAYGRLMWTLHQ 0 | |||
0 VAKLGMGESGSTAKAEAQVSRMVVVMVVAFLVCWLPYALFAMIVVTKPDVYIDPVIATLPMYLTKTSTVYNPIIYIFMNRQ 0 | |||
0 FRDCAVPFLLCGRNPWAEPSSESATAASTSATSVTLASAPGQVSPS* 0 | |||
>PPINa_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni NM_001032555 11591373 full Gt Ci-opsin1 syn(-HOXB1 +HHEX +CUL4A -) odd exons larval ocellus | |||
0 MDHDVTPTVDLTDGVPQCKDLNPYVLKGDGWVPQHISRANRSTYSFLCVYMTFVFLLSCSLNILVIVATLKNK 0 | |||
0 VLRQPLNYIIVNLAVVDLLSGFVGGFISIAANGAGYFFWGKTMCQIEGYFVSNF 1 | |||
2 GVTGLLSIAVMAFERYFVICKPFGPVRFEEKHSIF 1 | |||
2 GIVITWVWSMFWNTPPLIFWDGYDTEGLGTSCAPNWFVKEKRERLFIILYFVFCFVIPLAVIMICYGKLILTLRQ 0 | |||
0 IAKESSLSGGTSPEGEVTKMVVVMVTAFVFCWLPYAAFAMYNVVNPEAQ 0 | |||
0 IDYALGAAPAFFAKTATIYNPLIYIGLNRQ 0 | |||
0 FRDCVVRMIFNGRNPWVDELVGSQVSSTGSQLTAVSSNKVAPA* 0 | |||
>PPINb_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni genomic full Gt parapinopsin syn(-TMEM165 +FUT4 - -) jgi gene model wrong both ends | |||
0 MTTAETTTECYEKNPYIRNEMGWVPKHILIAERHIYTILAVYMTFIFLLAVSLNGFVIIATMKNK 0 | |||
0 KLRQPLNYIIINLSIADFLSGLVGGFIGMISNSAGYFYFGKTVCILEGYIVSVA 1 | |||
2 GVCGLMSISVMAFERYFVVCKPYGPFTLTNTHAAL 1 | |||
2 GIGFTWTWSVLWSTPGLIWLDGYVPEGLGTSCAPNWFSKNK 2 | |||
1 SERIFIFVYFVFCFFIPLLVIIICYGKIVLFLKQ 0 | |||
0 ATRQSSASSNRQADNKVTKMVLVMISAFLICWTPYGVLSLYNAINPDKQ 0 | |||
0 LDYGLGAVPVFFAKTANIYNPLIYIGLNKQ 0 | |||
0 FRDGVIKMVFRGRNPWAEEMSTQQRQRSTEAGQPIVSNEV* 0 | |||
>PPINa_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni genomic full G? parapinopsin | |||
0 MPTEASIAVDVSPTMGIPQCKDINPYVLKGDGWVPQHISRADRSVYSFLAVYMTFICLISCSLNILVITATLKNK 0 | |||
0 VLRQPLNYIIVNLAVVDLLSGLVGGVISIFANGAGYFFWGKFMCQVEGYTVSNF 1 | |||
2 GVTGLLSIAVMAFERYFVICKPFGPVRFEEKHAVI 1 | |||
2 GIAVTWIWAMFWNTPPLIFWDGYDTEGLGTSCAPNWFVKGNTERLFIILYFVFCFLIPLAIIVLCYGKLILQLRQ 0 | |||
0 IAKESSLSGGTSPEGEVTKMVVVMVTAFVICWLPYAAFAMYNVVNPEAQ 0 | |||
0 IDYALGAAPAFFAKTATIYNPLIYIGLNRQ 0 | |||
0 FRDCVVRMIFNGRNPWVDEMVGSQVSSSASQMTAVSSNKVAPA* 0 | |||
>PPINb_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni genomic full G? parietopsin | |||
0 MSSIPQNYSNGNPYATTDSGWVPEHIEIANRSTYSGLCVFMSFVFVLAVPLNLLVIVATYKNK 0 | |||
0 DLRRPINYIIVNLAVADLTCSVVGGLLGVLNNGAGYYFLGKSVCIFEGYVMSVT 1 | |||
2 GVCGILSITVMAFERYFVVCKPFGQTNLKWSHAIT 1 | |||
2 GIVFTWTWSVIWHTPGLFFWNGYEPEGFGTSCAPNWFSQQK 2 | |||
1 SERIFIFAYFAFCFLTPLTIIFACYLKLILFIRK 0 | |||
0 VSKKSMVNEADRRDFEVTRMVFVMIAAFLICWLPYGCLSMYNAIHPDNL 0 | |||
0 LSYGIGSVPAFFAKTATIYNPIIYMGLNKK 0 | |||
0 FRDGVIRMLFKGRNPWLDGRNTTSSTSTRAQGSLINREVDI* 0 | |||
>PARIE_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gd+Go parietopsin syn(+EEA1 -FLJ46688 +BTG1 -) gusducin not transducin | |||
0 MENESSLVLEGAEGYIVRPTIFPRAGYGVLAFLMFINALFSLFNNFLVIAVTLKNPQLRNPINIFILNLSFSDLMMSICGTTIVIATNYHGYFYLGRRFCIFQGFAVNYF 1 | |||
2 GIVSLWSLTILAYERYNVVCQPLGTLQMSTQRAYQLLGFIWVFCLFWAVVPLFGWSSYGPEGVQTSCSIGWEERSWNNYSYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG 0 | |||
0 LNKKVEQLGGKSNPEEEFRAVIMVLVMVVAFLICWLPYTLFALTVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKE 0 | |||
0 FRECAVEFITCGKVVLTSPEEDISTSAISDEGIAPCKINQVTPV* 0 | |||
>PARIE_utaSta Uta stansburiana (lizard) Deut.Saur.Lepi DQ100320 16543463 full Gd+Go parietopsin shift in counterion | |||
0 MENDSSLATELAEGAIVKPTIFPKAGYGVLAFLMFLNALFSIFNNSLVIAVTLKNPQLRNPINIFILNLSFSDLMMSLCGTTIVIATNYYGYFYLGRKFCIFQGFAVNYF 1 | |||
2 GIVSLWSLTILAYERYNVVCQPLGTLQMSTKRGYQLLGFIWVFCLFWAVVPLFGWSSYGPEGVQTSCSIGWEERSWSNYSYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG 0 | |||
0 LNKKVEQLGGKSSPEEEFRAVIMVLVMVVAFLICWLPYTVFALIVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKQ 0 | |||
0 FRDCAVEFITCGQVVLTSPEEDISTSAIPVEGKGPCKINQVTPV* 0 | |||
>PARIE_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur DN070761 16543463 full Gd+Go parietopsin syn(-lum -DCN) lung | |||
0 MDGNSTTPGIAVNLTVMPTIFPRSGYSILSFLMFLNAVFSICNNAIVILVTLKHPQLRNPINIFILNLSFSDLMMALCGTTIVVSTNYHGYFYLGKQFCIFQGFAVNYF 1 | |||
2 GIVSLWSLTLLAYERYNVVCEPIGALKLSTKRGYQGLVFIWLFCLFWAIAPLFGWSSYGPEGVQTSCSIGWEERSWSNYSYIISYFLTCFIIPVGIIGFSYGSILRSLHQ 0 | |||
0 LNRKIEQQGGKTNPREEKRVVIMVLFMVLAFLICWLPYTVFALIVVINPQLYISPLAATLPTYFAKTSPVYNPIIYIFLNKQ 0 | |||
0 FRTYAVQCLTCGHINLDSLEEDTESVSAQAENMLTPKTNQVAPA* 0 | |||
>PARIE_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic 16543463 full Gd+Go parietopsin syn(- +NT5DC2 +FBXL13) | |||
0 MENFAKTELTMMVQPTIFPRVGYSILSYLMFINTTLSVFNNVLVIAVMVKNLHFLNAMTVIIFSLAVSDLLIATCGSAIVTVTNYEGSFFLGDAFCVFQGFAVNYF 1 | |||
2 GLVSLCTLTLLAYERYNVVCKPMAGFKLNVGRSCQGLLLVWLYCLFWAVAPLLGWSSYGPEGVQTSCSLGWEERSWRNYSYLILYTLMCFILPTVIITYCYSNVLLTMRK 0 | |||
0 INKSIECQGGKNCAEDNEHAVRMVLAMIIAFFICWLPYTAISVLVVVNPEISIPPLIATMPMYFAKTSPVYNPIIYFLTNKR 0 | |||
0 FRESSLEVLSCGRYISRETGGPLMGSSMQRGQSRVNPV* 0 | |||
>PARIE_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? parietopsin genomic frameshift error | |||
0 MDSASTPWSPHPASGQAEAVTAAPTIFPRVGYSILSFLMFINTVLSIFNNGLAITVMLKNPALLQPINIFILSLAVSDLMIGLCGSLVVTITNYQGSFFIGHTACVFQGFAVNYF 1 | |||
2 GLVSLCTLTLLAYERYNVVCKPRAGLKLNMRRSLVGLLFVWTFCLFWAVTPLLGWSSYGPEGVQTSCSLAWEERSWNNYSYLILYTLLCFILPVGVIIYCYTKVLTSMNK 0 | |||
0 LNKSVELQGGRSCQKENDHAISMVLAMIIAFFVCWLPYTALSVVVVVDPELRIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0 | |||
0 FRDATLEVLSCGRYIPHASTRVTFNMCAFNRRSRLPSLSRSINTHSKVSPL* 0 | |||
>PARIE_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic 16543463 full Gd+Go parietopsin syn(-HSP90B1 +NT5DC2 -KCND3 -FLNC) | |||
0 MDSNSTPWSSPPAPLQAEAVTVAPTIFPRVGYSILSFLMFINTVLSVFNNSLAIAVMLKNPSLLQPINIFILSLAVSDLMIGLCGSLVVTITNYHGSFFIGHTACVFQGFAVNYF 1 | |||
2 GLVSLCTLTLLAYERYNVVCKPRAGLKLTMRRSIIGLLFVWTFCLFWAVTPLLGWSSYGPEGVQTSCSLAWEERSWNNYSYLILYTLLCFIFPVGVIIYCYCKVLTSMNK 0 | |||
0 LNKSVELQGGLSCRRENKHAINMVLAMIIAFFVCWLPYTALSVVVVVDPELHIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0 | |||
0 FRDATLEVLSCSRYIPHASSRVSINMRSLNRRSVNTHSKVSPL* 0 | |||
>PARIE_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? parietopsin syn(-HSP90B1 +NT5DC2 -KCND3 -FLNC) | |||
0 MDSNSTLWSSGSPPPSIHGKMLTITPTIFPRVGYSILSFLMFINTVLTVFNNVLVITVLVRNPSLLQPMNVFILSLAVSDLMIGLCGSLVVTITNYHGSFFIGHTACIFQGFAVNYF 1 | |||
2 GLVSLCTLTLLSYERYNVVCRPRNALKLSMRRSIHGLLIVWTFCLFWAVAPLFGWSGYGPEGVQTSCSLAWEERSWSNYSYLVLYTLLCFIVPVAVIIYCYAKVLTSMNT 0 | |||
0 LNRSVEVQGGRSSQKENDHAVSMVLAMIIAFFSCWLPYTALSVVVVVDPTLYIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0 | |||
0 FRDAALEMLSCGRYIAHMPNTVSINMRSLNRRSRLSSLSRNVNSHSKVLPL* 0 | |||
>PARIE_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag Gd+Go parietopsin | |||
0 LNKKIKRVGGHPDPREEMRATVMVLAMVGAFLACWLPYTVLALCVVLAPGTQIPPLVATLPMYFAKTSPIYNPIIYFFLNRQ 0 | |||
>ENC_homSap Homo sapiens (human) Deut.Euth.Euar NM_014322 12242008 full Gt OPN3 encephalopsin syn(-EXO1 -WDR64 -KMO +FH) with intron loss | |||
0 MYSGNRSGGHGYWDGGGAAGAEGPAPAGTLSPAPLFSPGTYERLALLLGSIGLLGVGNNLLVLVLYYKFQRLRTPTHLLLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSGSLF 1 | |||
2 GIVSIATLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPLGVIAHCYGHILYSIRM 0 | |||
0 LRCVEDLQTIQVIKILKYEKKLAKMCFLMIFTFLVCWMPYIVICFLVVNGHGHLVTPTISIVSYLFAKSNTVYNPVIYVFMIRK 0 | |||
0 FRRSLLQLLCLRLLRCQRPAKDLPAAGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDKTNGSKVDVIQVRPL* 0 | |||
>ENC_otoGar Otolemur garnettii (lemur) Deut.Euth.Euar genomic full G? OPN3 encephalopsin | |||
0 MYSGNRSGGQGFWEGGGAAGAEEPTPEGTLSPAPLFSPSAYERLALLLGSIGLLGVANNLLVLVLYYKFPRLRTPTHLFLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSGSLF 1 | |||
2 GIVSIATLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPVGVVAHCYGHILYSIRM 0 | |||
0 LRCVEDLQTTQVIKILKYEKKVAKMCFFMIFTFLVCWMPLIVICFLVVNGQGHLVTPTVSIVSYLLAKSNTVYNPVIYIFMLRK 0 | |||
0 FRRSLLQLLCFRLLRCQRPAKDLPAAESEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDNSDKTNGSKVDVIQVRPL* 0 | |||
>ENC_musMus Mus musculus (mouse) Deut.Euth.Euar NM_010098 full G? OPN3 encephalopsin panopsin 2aa del | |||
0 MYSGNRSGDQGYWEDGAGAEGAAPAGTRSPAPLFSPTAYERLALLLGCLALLGVGGNLLVLLLYSKFPRLRTPTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1 | |||
2 GFVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWRSKDANDSSFVLFLFLGCLVVPVGIIAHCYGHILYSVRM 0 | |||
0 LRCVEDLQTIQVIKMLRYEKKVAKMCFLMAFVFLTCWMPYIVTRFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMNRK 0 | |||
0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMHIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVEDSDRSSASKVDVIQVRPL* 0 | |||
>ENC_canFam Canis familiaris (dog) Deut.Euth.Laur DN422921 full early gap G? OPN3 encephalopsin XP_854433 grossly wrong | |||
0 MYSGNRSGGQGHWEGGGAAGAEGPGPAGTLSPAPLFSPGTYERLALLLGSVGLLGVGNNLLVLVLYSKFQRLRTPTHLLLVNLSLSDLLVSLFGVTFTFVSCLRNGWVWDSVGCVWDGFSSSLF 1 | |||
2 GIVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWSGAPLLGWNRYILDVHGLGCTVDWKSKDANDSFFVLFLFLGCLVVPMGVIVHCYGHILYSIRM 0 | |||
0 LRCVEDLQTIQVIKILRYEKKVAKMCFLMIFIFLIFWMPYIVICFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIIMIRK 0 | |||
0 FRRSLLQLLCFRPLRCQRPAKDLPANGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESVSIDDSDKTSVSKVDVIQVRPL* 0 | |||
>ENC_pteVam Pteropus vampyrus (macrobat) Deut.Euth.Laur genomic full G? OPN3 encephalopsin | |||
0 MHSGNRSGGLDSWEGGGAAGAEGPGLAGTLSPGSVFNPSTYERLALLLGSIGLLGVANNLLVLVFYYKFQQVRTPFYLFLVNISFSDLLVSFFGVTFTFVSCLRNGWVWDTVGCVWDGFSSSLF 1 | |||
2 GTVSMTTLTVLAYERYIRVVQARAIDFSWAWRTITYIWLYSLGWSGAPLLGWNRYILDVHGLGCAVDWKSKDANDSSFVLFLFLGCLVVPVVVIAHCYGHILYSVQM 0 | |||
0 LRCVEDLQTIQVIKILRYEKKMAKMCFLMIFTFLISWMPYIVICFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMIRK 0 | |||
0 FRRFVLQLLCFRPLRCRRPATDLPAGGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFVITSDESLSVDDSDKINGSKADGIQVRPL* 0 | |||
>ENC_loxAfr Loxodonta africana (elephant) Deut.Euth.Afro genomic full G? OPN3 encephalopsin 2 exons in browser 1 2x | |||
0 MYSGNRSGGQDLWEGGGGSGGAGPAGTLSPAPVFRSGTYERLALLVGSIGLLGVGNNLLVLVLYYKFQRLRTPTHLFLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSSSLF 1 | |||
2 GIASITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWSGAPLLGWNRYILDTHGLACTVDWKSNNSSDSSFVLFLFLGCLVVPVGVIAHCYGHILYSIRM 0 | |||
0 LRCVEDLQTIQVIKILRHEKKLAKMCLFMIFTFLICWMPYIVICFLVVNGYGHLVTPTISIVSYLFAKSSTVYNPVIYTFMIRK 0 | |||
0 FRRSLLQLLCFRLLRCQRPAKDLPVVGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVNNIDKTNGSKADVIQIRPL* 0 | |||
>ENC_monDom Monodelphis domestica (opossum) Deut.Meta.Dide genomic full G? OPN3 encephalopsin syn(-EXO1 -WDR64 +ENC -KMO +FH +RGS7) | |||
0 MYSDNSSDDGGGGYWGSGRAGGASGTGVTGEPGPEGSPRQAPLFSPGTYELLALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLFLVNISFNDLLVSLFGVTFTFVSCLRSGWVWDSVGCAWDGFSNTLF 1 | |||
2 GIVSIMTLTVLAYERYNRIVHAKVINFSWAWRAITYIWLYSLVWTGAPLLGWNRYTLEIHGLGCSVDWKSKDPNDSSFVIFLFFGCLMLPVGVMAYCYGHILYAIRM 0 | |||
0 LRCVEELQTIQVIKILRYEKKVAKMCFLMIAIFLFCWMPYAVICLLVANGYGSLVTPTVAIIASLFAKSSTAYNPIIYIFMSRK 0 | |||
0 FRRCLLQLLCFRLLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDETQMIDENDKNSGTKVNVIQVRPL* 0 | |||
>ENC_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full Gt OPN3 encephalopsin syn(-EXO1 -WDR64 +ENC -PIGM +R | |||
0 MHSGNGTGATSRPQLAAAGHEVPGERPLFSAGTYELLALLIATIGTLGVCNNLLVLVLYYKFKRLRTPTNLFLVNISLSDLLVSVCGVSLTFMSCLRSRWVWDAAGCVWDGFSNSLF 1 | |||
2 GIVSIMTLTVLAYERYIRVVHAKVIDFSWSWRAITYIWLYSLAWTGAPLLGWNRYTLEIHGLGCSMDWKSKDPNDTSFVLLFFLGCLVAPVVIMAYCYGHILYAVRM 0 | |||
0 LRCVEDFQTSQVIKLLKYEKKVAKMCFLMISTFLICWMPYAVVSLLVTYGYSNLVTPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0 | |||
0 FRQCLLQLLCFRLMRFQRIMKEPSGAGNVKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIIASDDTQQIDDNSKHNGTKVNVIQVKPL* 0 | |||
>ENC_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt OPN3 encephalopsin syn(-EXO1 -WDR64 +ENC -PIGM +RGS7) | |||
0 MFSANGTRSGAGSDLEPGPGQQQQQREASEEEERGAGLSPFSAGTYELLALLVAAIGLLGLCNNLLVLVLYAKFKRLRTPTHLFLVNISLSDLLVSLFGVSFTFGSCLRHRWVWDAAGCVWDGFSNSLF 1 | |||
2 GIVSIMTLTVLAYERYIRVVHARVIDFSWSWRAITYIWLYSLAWTGAPLLGWNHYTLEIHGLGCSVDWQSKEPSDSSFVLFFFLGCLAAPVGIMAYCYGHILHAIRM 0 | |||
0 LRCVEDLQSIQVIKILRYEKKVAKMCFLMVTTFLICWMPYAVVSLLIAYGYGHLITPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0 | |||
0 FRRCLVQLFCVQFLRFKRTLKEQPAIESNKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDDTEQIDVSTKCSDTKINVIQVKPL* 0 | |||
>ENC_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic pseu G? OPN3 encephalopsin recent pseudogene I for Schiff K C-term residues lost | |||
0 MPVTNGSHNNSISWLHSKDMFTEDTYHFLALIVATVGFLGLVNNLLVLILYCKFKRLQTPTNLLFFNTSLCHFVFSLLAITFTFMSCVRGSWAFSVEMCVFHGFSKNLL 1 | |||
2 GIVSFGTLTVVAYERYARVVYGKYVNSSWSKRSITFVWVYSLAWTGFPLIGWNLYTFETHKLDCSFEWTATDPKDTAFVLLFFLACITLPLSIMAYCYGYILYEIQK 0 | |||
0 LRSVKNIQNFQEITILDYEIKMAKMCLLMMLTFLIGWMPYTILSLLVTSGYSKFITPTITVMPSLLAIASAAYNPVIHIFTIKK 0 | |||
0 FRQCLVQLLFHNFWRLLKNLNGRLAMKKVKPVLGKGRSHNRPEKKVFSSSDFFTRTTSDTGTHGITESTKGKRTNVRLIQVHPLYP* 0 | |||
>ENC_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_001111164 full G? OPN3 encephalopsin | |||
0 MNSFNETPTEAHLENYNYIFADETYKLLTFTIGSIGVLGFCNNIIVIILYSRYKRLRTPTNLLIVNISVSDLLVSLTGVNFTFVSCVKRRWVFNSATCVWDGFSNSLF 1 | |||
2 GIVSIMTLSGLAYERYIRVVHAKVVDFPWAWRAITHIWLYSLAWTGAPLLGWNRYTLEVHQLGCSLDWASKDPNDASFILFFLLGCFFVPVGVMVYCYGNILYTVKM 0 | |||
0 LRSIQDLQTVQTIKILRYEKKVAVMFLMMISCFLVCWTPYAVVSMLEAFGKKSVVSPTVAIIPSLFAKSSTAYNPVIYAFMSRK 0 | |||
0 FRRCMLQMLCSRLTSLQHTIKDRPLSRIEHPIRPIVMSQSRTDRPKKRVTFSSSSIVFIIASHDTHPLDITSKCNDEPDINVIQVRPL* 0 | |||
>ENC_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? OPN3 encephalopsin | |||
0 MSSADDSRSARSGEPSLFAVHTYRLLAAAIGAIGVLGFCNNLAVAALYWRFRRLRTPTNLLLLNISLSDLLVSLLGVNFTFAACVQGRWTWNQATCVWDGFSNSLF 1 | |||
2 GIVSIMTLAALAYERYIRVVHAQVVDFPWAWRAIGHIWLYSLAWTGAPLLGWNRYTLEIHRLGCSLDWASKDPNDASFILLFLLACFFVPVGIMIYCYGNILYAVHM 0 | |||
0 IRSIQDLQTVQIIKILRYEKKVSVMFFLMISCFLLCWTPYAVVSMMVAFGRKSMVSPTVAIIPSFFAKSSTAYNPVIYVFMSRK 0 | |||
0 FRRCLLQLLCSRLSWLQRGLKERPLAPVQRPIRPIVVSRPCGKGTRPKKKVTFSSSSIVFIITSDDFRQLDVTSRAGDSADVNAIQVRPL* 0 | |||
>ENC_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? OPN3 encephalopsin | |||
0 MNPANGSRSERSAEQLLFSGDTYRVLAFTIGTIGAFGFCNNFVVLALYCRFKRLRTPTNLLLVNISLSDLLVSLFGINFTFAACVQGRWTWTQATCVWDGFSNSLF 1 | |||
2 GIVSIMTLAALAYERYIRVVHAQVVDFPWAWRAIGHIWLYALAWTGAPLLGWNRYTLEIHRLGCSLDWASKDPNDASFILLFLLACFFVPVGIMIYCYGNILYAVQM 0 | |||
0 IRSIQDLQTVQIIKILRYEKKVSVMFFLMISCFLLCWTPYAVVSMMVAFGRRSMVSPTMAIIPSFFAKSSTAYNPLIYVFMSRK 0 | |||
0 FRHCLLQLLCSRLSWLQRSLKERPLAPVQRPIRPIVMSRPCGKGNRPKKKVTFSSSSIVFIITSDDFGQLDVTSKSGDSADVNAIQVRPL* 0 | |||
>ENC_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? OPN3 encephalopsin | |||
0 MNPDNGTREERSTDHSIFAVGTYKLLAFAIGTIGVFGFCNNVVVIVLYCKFKRLRTPTNLLVVNISLSDLLVSVIGINFTFVSCIRGGWTWSRATCIWDGFSNSLF 1 | |||
2 GIVSIMTLASLAYERYIRVVHAQVVDFPWAWRAIGHIWLYSLVWTGAPLLGWNRYTLEIHRLGCSLDWASKDPNDASFILLFLLACFFVPVGIMIYCYGNILYAVQM 0 | |||
0 LRSIQDLQTVQIIKILRYEKKVAVMFLLMISCFLLCWTPYAVVSMMEAFGRKNMVSPTVAIIPSFFAKSSTAYNPLICVFMSRK 0 | |||
0 FRRCLMQLLCSRVTCLQCNLKERPLAPVQRPIRPIVVSAACGGGRVRPKKRVTFSSSSIVFIITRNDIRHTDVTSNTRESSEANVFQVRPL* 0 | |||
>ENC_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full G? OPN3 encephalopsin | |||
0 MNPANESRAGRHEERSVFAVGTYKLLTVIIGTIGVFGFCNNLLVILLYCKFKRLRTPTSLLLVNISLSDLLVSVVGINFTLASCVKGRWMWSQATCVWDGFSNSLF 1 | |||
2 GIVSIMTLAALAYERYIRVVHAQVVDFPWAWRAIGHIWLYSLAWTGAPLLGWNRYTLEIHQLGCSLDWASKDPNDAAFILLFLLGCFFVPVGIMIYCYGNILYAVRM 0 | |||
0 LRSIEDLQTVQIIKILRYEKKVAAMFLLMISCFLVCWTPYAVVSMMEAFGKKSMVSPTVAIVPSFFAKSSTAYNPLIYVFMNRK 0 | |||
0 FRRCFLQLLGSRLCSKISWLQCTLKEHPLTPVERPIRPIVASTSCGSRHRPKKRVTFNSSSIVFMITGDEFQQLDVTSKSRNSSEANVFHVRPL* 0 | |||
>ENC_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? OPN3 encephalopsin | |||
0 MNPTNSTEPQEEHLFSPNTYKLLAVIIGTIGIVGFCNNILVLLLYYKFKRLRTPTNLLLVNISVSDLLVSVFGLSFTFVSCTQGRWGWDSAACVWDGfSHSLF 1 | |||
2 GTVSIVTLTVLAYERYIRVVNAKATNFPWAWRAITYTWFYSLAWSGAPLV | |||
0 yRRCLSQLFCSHLMSLQWSIKDPSSKARNDMPVKPIVLSQKGDRPKKRVTFSSSSIVFIITSDDTQELGSIAGSNATQISIVQVQPL* 0 | |||
>ENC_squAca Squalus acanthias (dogfish) EB687868 frag Gt OPN3 encephalopsin | |||
0 MNAANSTDTREESLFSPGTYQVLAVIIGTIGVVGFCNNLLMLVLYCKFKRLRTPTNLFLVNISISDLLLSVFGVIFTFVSCVKGRWVWDSAACVWDGFSNCLF 1 | |||
2 GISSIMSLTVLAYERYIRVVNATAIDFSWAWRAITYIWLYSLAWTGAPLIGWNSYTLELHRLGCSVNWDSRNPSDTSFVLFLFLGCLLCPIGVIAYCYG | |||
>ENC_leuEri Leucoraja erinacea (skate) Deut.Chon.Elas FL592692 frag | |||
0 MNTANSTEIPAETLFGAGAYKVLAVVVGTVGAVGFCNNLLMLVLYCKFKRLRSPANLFLVNISISDLLLSVFGIFFTLVSCVKGGWVWDNSACIWDGFSNTLF 1 | |||
2 GISSIMSLTVLAYERYLRMVNATAINFTWAWRAVTYIWLYSLAW | |||
>ENC_torCal Torpedo californica (ray) Deut.Chon.Elas EW690209 17309107 frag electric organ | |||
2 WSGAPLLgwNRYTLEMHRLGCSVTwELEKPSDTSFILFLFLGSLLLIPVGVIAYCYGNIFYTIRM 0 | |||
0 LQSIEDFQTARFAKTLTNEMNSSKMCFFMISVAFSCWLPYAVTSFMVVYGCTDVITPTITIIFSLLAKSSAISYPIIYIFMSRK 0 | |||
0 FRWCLMQLLCFRLVRIKWAIKDRTVRIRSDKPIKPIVIFEKVVNRPKKRVTFSSFSIIFIANDETQDIGSTAELNATNQNVVQVRPLRSERAER* 0 | |||
>ENC_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic full Gt OPN3 encephalopsin rough sequence in two contigs | |||
0 MQSPKQDSLHYAGDTGAKAAPDSAQGNASALGSNFLLHGGDLGEGSTAFSAATFRLLAGVVGTIGVAGFLNNLLLVALFVGFKRLQTPTNLLLVNISLSDLLVSVFGNTLTLVSCVRRRWVWGNGGCVWDGFSNSLF 1 | |||
2 GIVSISTLTALSYERYARLIKAQVLDFSWAWRAVTYTWLYSAAWTGAPLLGWSRYVLEKHGLGCSIDWASSNPPDAAFVLFFFLGCLAAPLLVMGFCFGRIALAITQ 0 | |||
0 FRKLDRLQTPRVLKARCSERKVSAVCLLMMLLFLLCWSPYAVASLFVASGFEHLVSPPVSIVPSLLAKSNAVCNPLLFLLMSGN 0 | |||
0 FFRCLRTMFFTLRWRVNLSRVGIVCPQSQRRPRQREAARPKKKATFNSSSIVFIISSSEEDPVEPADAEGEKSDALAAPADTCVVHVKPT* 0 | |||
>ENC4_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran AB050608 12435605 full Gt Amphiop4 syn(-ZFYVE1 +RTF1 -CES1 -POMT2) new exon 12 and 34 | |||
0 MALYNNTSSPSQDLLWDAPYSQGHIWDNSSASNSSEDVMDQGKVELQDFSDAGYTAIATCLALI 1 | |||
2 GFVGFTNNFVVILLIGCHRQLRTPFNLLLLNMSVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANSLF 1 | |||
2 GIVSLVTLSALAFERYCVVVRSSDMLTYKSSLVVITFIWLYSLLWTSLPLLGWSSYQFEGHN 0 | |||
0 VGCSVNWVQHNPDNVSYIVTLMVTCFFVPMVVVCWSYAWIWRTVRM 0 | |||
0 SSEAKPECGNSQNAGRLVTTMVVVMIICFLVCWTPYAVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ 0 | |||
0 FREFLLARLQRVCCRQQAVPRVTPMDDNVHVRLGGEGPSQSQQFLPAGENVENVDMLEYVQENCKPKADSLSTISE* 0 | |||
>ENC4_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050608 12435605 full G? Amphiop4 introns from braFlo | |||
0 MPLYNTSSGPTQGLPWDTPYSQDPIWNDSSPSNSSEDAVVDQGRGELQDFSDAGYTAIATGLALI 1 | |||
2 GLVGSMNNFVVILLIGCHRQLRTPFNLLLLNVSVADLLVSVCGNTLSFASAVQHRWLWGRPGCVWYGFANSLF 1 | |||
2 GIVSLVTLSALAFERYCVVVRSSEMLTYKSSLGMIAFIWMYSLLWTSLPLLGWSSYQFEGHS 0 | |||
0 VGCSVNWVKHNVNNVSYIITLMVTCFFVPMVVVCWSYACIWRTVRM 0 | |||
0 SAEMKSEFGNPQNTGRLVTTMVVVMIVCFLVCWTPYTVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ 0 | |||
0 FREFLLARLRTFCCRQPRMLRVTPMDDNAHARLVGEGPSHAQQVIPSEENGENVEMRKVQGNQLKADSLSTISE* 0 | |||
>ENC_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu AAGJ02133080 full G? OPN3 encephalopsin terminal exon extended to stop codon | |||
0 MENFTSIVTDGTNEENTDGDAWPGYAHLLAGSFLTLVFIISIIGNSVVLFLFAWDRHLRTPTNMFLLSLTISDWLVTVVGIPFVTASIYAHRWLFAHVGCIS 2 | |||
1 YAFIMTFLGLNSLMSHAVIAVDRYLVITKPHF 1 | |||
2 GIVVTYPKAFLMISIPWVFSFAWAVFPLAGWGEFTYEGTGAWCSVRWDSDQPQIMSYVLAMMFLTFISSIVIMMYCYICIFLTTRRMPRWATSNSIKTHERNRRRR 2 | |||
1 EQKLLKTLIAIAIAFLVAWSPYAITSMIVVFGGSELLSLTATTLPSLFAKSSVMINPIIYAVTSRVFRKSLKK 0 | |||
0 MLTSFFPGCMTYIMTDKSPPSSSRPIQLGLCKYHFLY* 0 | |||
>TMT5_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002589165 12435605 full G? Amphiop5 extra 0 intron | |||
0 MLGMHNVMNATDYDNNNATFAAWNFQRNGTTEEEVEFSGFDTVAVVIAAIGIAGFLSNGAVVLLFLKFRQLRTPFNMLLLNMSVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANHLF 1 | |||
2 GLVSLISLAVISYERYRMVVKPKGPGSSYLTYNKVGLAIIFIYLYCLLWTTLPIVGWSSYQLE 0 | |||
0 GPKISCSVAWEEHSLSNTSYIVAIFIMCLLLPLLIIIYSYCRLWYKVKK 0 | |||
0 GSQNLPPAIRKSSQKEQKIARMVVVMITCFLVCWLPYGAMALVVSFGGESLISPTAAVVPSLLAKSSTCYNPLVYFAMNNQ 0 | |||
0 FRRYFQDLLCCGRRLFDASASVNTCNTSAMPRHSPVFQKPDSDQYNGIQKSREPQMRTTGQNAPYRQWIEMQTIAVVVKADEVNNKFGEVKT* 0 | |||
>TMT5_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050609 12435605 full G? Amphiop5 introns from braFlo | |||
0 MLGIYNVVNATEYGNNTTFAAWDFKRNGTGGEEEVEFFGYDAVAGVIAIIGVVGFVSNGAVVVLFLKFPQLRTPFNLLLLNMAVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANHLF 1 | |||
2 GLVSLISLAVISFLRYRMVVKPKGPGSSYLTYTKVGLAILFIYLYCLLWTTLPIAGWSSYQLE 0 | |||
0 GPKIGCSVAWEEHSWSNTSYIVVLFITCLFAPLLIIVYSYYRLWHKVKQ 0 | |||
0 GSRNLPAAMRKSSQKEQKIAMMVIVMITCFMVCWLPYGAMALVVTFGGERLISHTAAVVPSLLAKSSTCYNPVVYFAMNSQ 0 | |||
0 FRRYFQDLLCCGRRLFDVSQSVVTGNTAMPRNNSQGFRKDDSDQKQDNGLPKQSEGPMCDHSSNESQMEGSRHNTAASQQWIEMQTIAVVVKAVEVDTSAANEP* 0 | |||
>TMT_monDom Monodelphis domestica (opossum) Deut.Meta.Dide XM_001372110 full G? syn(+NCK2 -UXS1 +TMT -ST6 GAL2_overlap) -RALY shortened final exon | |||
0 MSNNLTTNLSLEALLSASEDKQRNGLSRTGHTIVAVFLGIILIFGSISNFIVLVLFCKFKVLRNPVNMLLLNISISDMLVCLSGTTLSFASSIQGRWIGGKHGCRWYGFANSCF 1 | |||
2 GIVSLISLAILSYERYRTLTLCPGQGADYQKALLAVAGSWLYSLVWTVPPLIGWSSYGTEGAGTSCSVHWTSKSVESVSYIMCLFIFCLVIPILVMVYFYGRLLYAVKQ 0 | |||
0 VGKIRKTAARKREYHVLFMVVTAVICYLICWVPYGMIALLATFGPPGVVSPVANVVPSILAKSSTVCNPIIYVLMNKQ 0 | |||
0 FYKCFLILFHCQPAQSGPDVSLCPSNVTVIQLGQRKNKDAPGSI* 0 | |||
>TMT_macEug Macropus eugenii (wallaby) Deut.Meta.Dipr genomic full G? teleost multiple tissue | |||
0 MSINLTANLSFGTLLPDSEEKQRSGLSRTGHTVTAVFLGLILILGVINNFIVLVLFCKFKVLRNPVNMLLLNISISDMLVCLTGTTLSFASSIRGRWIAGYHGCRWYGFANSCF 1 | |||
2 GIVSLISLAVLSYERYRTLTLCPRQGTDYHKALLAVAGSWLYSLIWTVPPLIGWSSYGTEGAGTSCSVHWTSKSVESVSYIMCLFIFCLVIPILFMVYFYGRLLYTVKQ 0 | |||
0 VGKIRKSAARKREYHVLFMVVTAVICYLICWVPYGMIALLATFGPPGVVSPVANVVPSILAKSSTVCNPIIYILMNKQ 0 | |||
0 FYKCFLILFHCQPASSASDASLCPSKMTVIQLGQRKDKEVPCAIQDLPEVSKKQLCLLSPESNVAPSSGHPQEKMEEKPLSE* 0 | |||
>TMT_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono genomic frag G? teleost multiple tissue | |||
0 GLSRTGHTMVAVFLGIILVFGFMNNLIVLILFCKFKALRNPVNMIMLNISASDMLVCVSGTTLSFASNISGRWIGGDPGCRWYGFVNSCL 1 | |||
2 GIVSLISLAVLSYERYRTLTLHPKQSTDYQKAVLAVGASWIYSLIWTIPPLLGWSSYGTEGAGTSCSVHWSSKSPVSVSYIVCLFIFCLVIPVLVMIYCYGRLLYAVKQ 0 | |||
0 IGKARKTAARKREYHVLFMVITTVICYLVCWMPYGVTALLATFGQPGTVSPEASVIPSILAKSSTVCNPIIYILMNKQ 0 | |||
0 FYKCFLILFHCQPPRAADAPSTYPSQVMVIQLNQRRSRETAGAPQVLLEMKHQTLHLLGPQLHETPSWERSTPVHPE* 0 | |||
>TMT_galGal Gallus gallus (chicken) Deut.Saur.Arch XM_001234388 full G? syn(+NCK2 -UXS1 +TMT -ST6 GAL2_overlap +SLC5A7 +SULT1C4) | |||
0 MNHTWTYNLSFGAPTDPVEPRAGLSRNGHTVVAVFLGFILFFGFLNNLIVLILFCKFKTLRNPVNMLLLNISISDMLVCISGTTLSFASNIHGKWIGGEHGCRWYGFVNSCF 1 | |||
2 GIVSLISLAVLSYERYSTLTLCNKRSDDYRKALLAVGGSWVYSLLWTVPPLLGWSSYGIEGAGTSCSVRWSSETAESTSYIICLFIFCLVIPVMVMMYCYGRLLYAVKQ 0 | |||
0 VGKIHKNTARKREYHVLFMVITTVICYLVCWIPYGVIALLATFGKPGVVTPVASIIPSILAKSSTVCNPIIYILMNKQ 0 | |||
0 FYKCFRQLFHCQPPSSTDGEPTCHSKVTVIQLNQKTDGGKLCNNKPRPETDNKVTSLLHPEPGLEPAAKTVPPM* 0 | |||
>TMT_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch XM_002192719 full G? teleost multiple tissue | |||
0 MNHTWMYNLSFGAPAHPVEPRAGLSRSGHTVVAVFLGLILFFGFLNNLIVLILFCKFKTLRNPVNMLLLNISVSDMLVCISGTTLSFASNIRGKWIGGDHACRWYGFVNSCF 1 | |||
2 GVVSLISLAVLSYERYNTLTLCHKRSDDFRKALLAVAGSWIYSLVWTVPPLLGWSSYGVEGAGTSCSVRWSSESAESTSYIICLFVFCLVVPVMVMMYCYGRLLYAVKQ 0 | |||
0 VGKIHKNAARKREYHVLFMVIPTVICYLVCWIPYGVIALLATFGKPGAVTPITSIIPSILAKSSTVCNPIIYILMNKQ 0 | |||
0 FYKCFRQLFHCQPPSSTDGEPTCHSKVTVIQLDQRADGGNMCNNEPHPETDSKMTSLLCPETTSKATPPTS* 0 | |||
>TMT_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G? teleost multiple tissue syn(+TMT -ST6 GAL2_overlap +SLC5A7) | |||
0 MSELSSNLTFNMSTSIEEPGSGLSRMGHNIVAVFLGLILVFGFLNNLVVLILFCKFKTLRNPVNMLLLNISASDMLVCISGTTLSFVSNIYGRWIGGEHGCRWYGFVNSCF 1 | |||
2 GIVSLISLAILSYERYSTLTQTNKRGSDYQKALLGVGGSWLYSLIWTVPPLIGWSSYGLEGAGTSCSVRWTSETLESVTYIICLFIFCLAIPVLVMIYCYARLFYAVKQ 0 | |||
0 VGKLRKTSARKREFHVLFMIITTIICYLICWMPYGVIALLATFGRPGLVSPVASVIPSILAKSSTVFNPIIYILMNKQ 0 | |||
0 FYKCFLMLLHCQPSSVADGETICQSKVMAIHQNQKAQGGVILKSQVVPQMDEKAICLLSPESSLDPVLESTPQLSKENSFL* 0 | |||
>TMT_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur AL773998 full G? syn(-UXS1 +TMT -ST6 GAL2_overlap +SLC5A7) short gastrula frag | |||
0 MSTIKNWTTNISVENSMSYIENDLSLPTEAVLSRTGHTVVAIFLGFILIFGFLNNFVVLILFCKFKTLRTPVNMMLLNISASDMLVCVSGTTLSFTSSIKGKWIGGEYGCQWYGFVNSCF 1 | |||
2 GIVSLISLAILSYERYSTLTLYNKGGPNFKKALLAVASSWLYSLVWTVPPLLGWSSYGREGAGTSCSVRWTSESVESVSYIICLFIFCLALPVFVMLYCYGRLLYAVKQ 0 | |||
0 VGKIRKIAARKREYHVLFMVITTVICYLLCWLPYGVVALLATFGRPGVISPVASVVPSILAKSSTVFNPIIYILMNKQ 0 | |||
0 FYKCFLILFHCHPTSSADGKSICQSNYTVIQLNQKLNNIVAIPGQTQIPESVDKMPCIHRQNNESPSDQMPQSTTEHLISGT* 0 | |||
>TMTa_danRer Danio rerio (zebrafish) Deut.Acti.Otoc XM_002192719 full G? syn(-UXS1 +TMT -ST6 GAL2_overlap +GPR89A -pdzk1l) | |||
0 MFFEQADLNYSFNMSEEDRLTLLDEDWSDSPMETLSRAGFIALSVFLGFIMTFGFFNNLVVLVLFCKFKTLRTPVNMLLLNISISDMLVCMFGTTLSFASSVRGRWLLGRHGCMWYGFINSCF 1 | |||
2 GIVSLISLVVLSYDRYSTLTVYHKRAPDYRKPLLAVGGSWLYSLIWTVPPLLGWSSYGLEGAGTSCSVSWTQRTAESHAYIICLFVFCLGLPVLVMVYCYGRLLYAVKQ 0 | |||
0 VGKIRKTAARKREYHVLFMVITTVVCYLLCWMPYGVVAMMATFGRPGIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0 | |||
0 FYRCFRILFCCQRSLLQNGHSSMPSKTTVIQLNRRVNSNAVACTAQISTGTHNHDCSTHVTERSNPPEVIP* 0 | |||
>TMTb_danRer Danio rerio (zebrafish) Deut.Acti.Otoc CN506730 full G? teleost multiple tissue syn(-FHL2 +NCK2 +TMT -ST6GAL2_lap) seg dup | |||
0 MFPEETNMSYISNGTDDDLLSALEDWSDTPAEKLSRTGHNVVAVILGSILIFGTLNNLVVLVLFCKFKTLRTPVNMLLLNISVSDMLVCLFGTTLSFAASIRGRWLVGRHGCMWYGFVNSCF 1 | |||
2 GIVSLISLAILSYDRYSTLTVYNKRAPDYSKPLLAVGGSWLYSLFWTVPPLLGWSSYGLEGAGTSCSVTWTANTPQSHSYIICLFIFCLGIPVLVMVYCYSRLICAVKQ 0 | |||
0 VGRIRKTAARRREYHILFMVITTVVCYLLCWMPYGVVAMMATFGRPGIISPIASVVPSLLAKSSTVINPLIYILMNKQ 0 | |||
0 FYRCFLILIHCKHSSLENGQSSMPSRTTGIQLNRRPYSNPVADNAPPSIDLQNDCSTPVSG* 0 | |||
>TMT_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? syn(-UXS1 +TMT -ST6 GAL2) multiply frameshifted assembly | |||
0 MFSGQAGLNSSFNLSDGRGLEDAPAGRGRLSPTGFVVLSVVLGFIITFGFLNNFIVLLLFCKFKKLRTPVNVLLLNISVSDMLVCLFGTTLSFASSLRGRWLLGRSGCNWYGFINSCF 1 | |||
2 GIVSLISLVILSHDRYSTLTVYNKQGINYRKPLLAVGGTWLYSLLWTVPPLLGWSSYGIEGAGTSCSVSWTVQTAQSHAYIICLFIFCLGLPVLVMVYCYSRLLWAVKQ 0 | |||
0 VGKIRKTSARKREYHILFMVVTTAACYLVCWMPYGVVAMMATFGPPNIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0 | |||
0 FYKCFLILFHCSHWSADNGTTSVPSKITVIQLNRRAYSNTVACADPLSTDALKQCCSAKNASTIEVKLS* 0 | |||
>TMT_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? teleost multiple tissue | |||
0 MFSGQAGLNYSFNLSDDRELLDAPAGRAKLSPTGFVVLSVVLGFIMTFGFLNNFVVLLLFCKFKKLRTPVNMLLLNISVSDMLVCLFGTTLSFASSIRGRWLLGRIGCSWYGFINSCF 1 | |||
2 GIVSLISLVILSYDRYSTLTVYNKQGINYRKPLLAVGGTWLYSLFWTVPPLLGWSSYGIEGAGTSCSVSWTVQTAQSHAYIICLFTFCLGIPILVMIYCYSRLLWAVKQ 0 | |||
0 VGRIRKTAARKREYHILFMVVTTAACYLVCWMPYGVVAMMATFGPPNIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0 | |||
0 FYKCFLILFHCGHWSADNGNTSMPSKTTAIQLNRRVYSNTVACADQLSTDALKQCCSANTISTKNTSTVEGKLS* 0 | |||
>TMT_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? teleost multiple tissue | |||
0 MVFGQAGLNHSFNLSDDRELLDTSAGRAKLSPTGFVVLSVMLGFIMTFGFVNNLVVLLLFCKFKKLRTPVNMLLLNISVSDMLVCLFGTTLSFASSLRGKWLLGRSGCSWYGFINSCF 1 | |||
2 GIVSLISLVILSYDRYSTLTVYNKAGPDYRKPLLAIGGSWLYSLFWTVPPLLGWSSYGIEGAGTSCSVSWTVQTAQSHAYIICLFTFCLGLPMLVMIYCYSRLLLAVKQ 0 | |||
0 VGRIRKTAARRREYHILFMVLTTAACYMLCWMPYGVVAMMATFGPPNIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0 | |||
0 FYRCFLILFHCKHWSAENHNTSMPSKTTVIHLNRRVCSNTLPCTAQASTDAANHFCSTSATKHTSPPLQGHGLSLNVLNMIRQENHSHDEAAKNQLDCLT* 0 | |||
>TMT_oryLat Oryzias latipes (medaka) Deut.Acti.Perc DK170580 full G? teleost multiple tissue whole embryo | |||
0 MFSGQTGLNFSFNQSDDRELEDTPAGSAKLSQAGFVVLSVVLGFIMTFGFLNNFVVLILFCKFKKLRTPVNMLLLNISVSDMLVCLFGTTLSFASSIRGRWLLGRGGCSWYGFINSCF 1 | |||
2 GIVSLISLVILSYDRYSTLTVYNKGGLNYRKPLLAVGGSWLYSLFWTVPPLLGWSSYGLEGAGTSCSVSWTANTAQSHAYIICLFIFCLGLPILVMIYCYSRLLLAVKQ 0 | |||
0 VGKIRKTAARKREYHILFMVLTTAACYLLCWMPYGVVAMMATFGPPNIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0 | |||
0 FYRCFLILFHCDHWSSENGNTSVPSKTTVIPLNRRIYTNTVAQISTDNAN* 0 | |||
>TMT_ictPun Ictalurus punctatus (catfish) Deut.Acti.Otoc FD287269 frag G? teleost multiple tissue 2 transcripts from whole fry | |||
0 WLLGRTGCMWYGFINSCF 1 | |||
2 GIVSLISLMILSYERYSTMTVYNNQGPNYRKHLLAVGGSWLYSLIWTVPPLLGWSSYGLEGAGTSCSVSWTDHSPKSHAYIICLFIFCLGLPVLLMVYSYGRLLYAVKQ 0 | |||
0 LGKIHKTARRRDYHLLFMITTTVVCYLLCWTPYSVVALMASFGRPGIITPVASIIPSLLAKSSTVINPVIYIFMNKQ 0 | |||
0 FYRCFRTLLGYKERSAVPDDHSLMATKNTAIQLKCIMHNNPVPSPAHTPPPFF | |||
>TMTc_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? ciliary uropsin syn(+GPR112 +BRS3 +VGLL1 +TMTx +CD40LG -ARGHGEF7) | |||
0 MLLALNLQNESFLLNLSHGETRDSLPPATGLSRTAHSVVAVCLGCILVLGSLYNSFVLLIFVKFTAIRTPINMILLNISVSDLLVCIFGTPFSFVSSVSGGWLLGQQGCKWYGFCNSLF 1 | |||
2 GLVSMISLSMLSYERYLTVLKCTKADMTDYKKSWLCIIVSWLYSLCWTLPPLIGWSSYGLESSGTTCSVVWHSKSSNNISYIVCLFLFCLVLPLFIMIFCYGHIVRVIRG 0 | |||
0 VCRINMTTAQKREHRLLFMVVCMVTCYLLCWMPYGLVSLMTAFGKPGMITPTVSIIPSILAKSSTFINPLIYIFMNKQ 0 | |||
0 FYRCFIALIKCESGPHHNENISTSRQSREMRLETRCKVQSSIFYTHKARRTKDTTSSPEMQRKLTLTVHYRDDR* 0 | |||
>TMTc_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G? ciliary uropsin syn(+FHL1 +GPR64/112 +TMTx -ARGHGEF7 -RBMX) | |||
0 MVVYIWSLNISSKDTSALNQSGNVSSGDPLEPHDSPPGLSRTGHTVTAVCLGAILLLGCLNNLFVLLVFARFRTLWTPINLILLNISVSDILVCLFGTPFSFASSLYGKWLLGHHGCKWYGFANSLF | |||
2 GIVSLMSLSILSYERYAALLRATKADVSDFRRAWLCVAGSWLYSLLWTLPPFLGWSNYGPEGPGTTCSVQWHLRSTSSISYVMCLFIFCLLLPLVLMIFCYGKILLLIKG 0 | |||
0 VTKINLLTAQRRENHILLMVVTMVSCYLLCWMPYGVVALLATFGRTGLITPVTSIVPSVLAKSSTVVNPVIYVLFNNQ 0 | |||
0 FYRCFVAFLKCQGEPSVHGQNPQHSSKEDPHVFRPCDGASIHRSAEGPQKKEQHSLSLVVHYTP* 0 | |||
>TMTc_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? ciliary uropsin syn(-TAF7L +SLC9A7 +FHL1 +TMTx -ARGHGEF7 -RBMX) | |||
0 MVVQPRDCNFSTPDSSVSIFGPSIDARGPWDPQCPGGLSRSSHTAVAVLLGVILVAGILSNSLVLLLFVKYRSLWTPINLILLNINLSDILVCVFGTPFSFAASLQGRWLIGEGGCMWYGFANSLF 1 | |||
2 GIVSLVSLSVLSYERCTVVLQPSQVDVSDFRKARFCVGGSWLYALLWTSPPLLGWSSYGPEGAGTTCSVQWQLRSPASVSYVLCLLVFCLLLPFLVMVYSYGRILVAIRR 0 | |||
0 VGRINQLTAQRREQHILLMVLSMVSCYMLCWMPYGIMALVATFGKLGLVTPMVSVVPSILAKFSTVVNPIIYMFFNNQ 0 | |||
0 FYRCFMAFIRCQKEPEPIQEE* 0 | |||
>TMTc_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? ciliary uropsin syn(-TAF7L +SLC9A7 +FHL1 +TMTx -ARGHGEF7 -RBMX) | |||
0 MVLQTRECNFSTPDTSVRGPWAPQGPGGMSRTGHTVVAVMLGTILLAGVFGNSVVFLVFVKYRSLRTPINLILLNISLSDILVCVFGTPLSFAASLKGRWLLGERGCEWYGFANSLF 1 | |||
2 GIVSLVSLSVLSYERYTVVLQPTQVDVSYFRKAWFCVGGSWLYALFWTLPPLLGWSRYGPEGPGTMCSVQWHLRSPANISYVLCLFIFCLLLPLVVMVYSYGRIWVAVRR 0 | |||
0 AGRINLLTAQRREQHILWMVLSMVSCYMLCWMPYGIIALVATLGRLGPISPAVSVVPSILAKFSTVVNPVIYMFFNNQ 0 | |||
0 FYRCFMAFVRCQKEPESIQGE* 0 | |||
>TMTc_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full G? ciliary uropsin syn(+SLC9A7 +FHL1 +GPR56 +TMTx -ARGHGEF7 -RBMX) | |||
0 MVLHSHNCNFSTKDSSPCSTNSTEPHNPDGLSRTGHTAVAVCLGFILVAGILNNFLTLLVFAKFRSLWTPINLILLNISLSDILVCVLGTPFSFAASVRGRWLIGESGCKWYAFANSLF 1 | |||
2 GIVSLVSLSVLSYERYITVLHSSQADLSNFRKAWFCVGGSWLYSLLWTLPPFLGWSSYGPEGPGTTCSIQWHLRSPTSVSYVLCLFIFCLVLPLVLMVYSYGRILVALRR 0 | |||
0 VGKINLLAAQRREQRILVMVFSMVSCYILCWMPYGIVALMATFGRKGLVTPLTSVIPSILAKFSTVVNPVIYVFFNSQ 0 | |||
0 FYRCLVAFVRCSGDPEEHPTPRTQLSDVVRVQKQNSVSSSSPRLLSDPTDRMLCNHHPDRHVLFVHYTP* 0 | |||
>TMTa_oncMyk Oncorhynchus mykiss (trout) Deut.Acti.Salm CU062745 full G? teleost multiple tissue testis new | |||
0 MVVESGNLNFSTDDSSGTNLSPKDSVRDTLTSRQSDLGRTGHTVVAVFLGVIFLLGFLSNLFVLLVFARFQVLRTPINLILLNISVSDMLVCIFGTPFSFAASLYGRWLIGAHGCKWYGFANSLF 1 | |||
2 GIVSLVSLAILSYERYSTILCYTKADPSDYKKAWLAIAGAWLYSLVWTVPPFFGWSSYGPEGPGTTCSVQWHQRSSGNISYVTCLFIFCLLLPLLLMMFCYGKILFAIRG 0 | |||
0 VAKINQSSAQRRETHVLVMVVSMVSCYLLCWMPYGVVALLATFGQVGLVSPTTSIVPSILAKSSTFLNPVIYGLLNNQ 0 | |||
0 FYRCFLAFMSCGSEAAGSHTLHTLPSSRVVGPYGNSPAPEEPGTREPLDGSGTSSKTQTRGPQREKRDLVLVVHYTP* 0 | |||
>TMTa_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G? teleost multiple tissue | |||
0 MTVPSISPSCIGVANGVWCSNGGSSSNSHHRHGQEQSLSPTGHLITAICLGVIGSLGFLNNLLVLVLFCRNKVLRSPINLLLMNISLSDLMICIVGTPFSFAASTQGKWLIGPAGCVWYGFANTFF 1 | |||
2 GTVSLISLAVLSYERYCTMMGTTEADATNYKKVWMGIFLSWIYSLFWSLPPLFGWSSYGPEGPGTTCSVNWHSRDANNISYIICLFIFCLVIPFIVIVYCYGKLLCAIKK 0 | |||
0 VSGVTQGMAQTREQRVLIMVVVMIICFLLCWLPYGIVALIATFGKPGLITPSASIIPSVLAKSSTVYNPVIYIFLNKQ 0 | |||
0 FYRCFCALLKCGKKSIASSNKCSSRSTRVCRSIRKQDNFTFVAASAGPPSSEHQDAILSIQNPEPPTDNSPKAKQRVLLVAHYSV* 0 | |||
>TMTa_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? teleost multiple tissue scaffold_55 | |||
0 MAFHSSSPSCSDSSSVTCRSSIQQYGPENHPNLSPTGHLLVAVFLGVIGSLGFFNNLVVLILFCQYKVLRSPINMLLMNISLSDLMVCILGTPFSFAASTQGHWLIGEIGCIWYGFVNTLF 1 | |||
2 GTVSLVSLAVLSYERYCTMLRSTEADLTNYKKAWLGILVSWIYSLVWTLPPLFGWSKYGPEGPGTTCSVNWHSRDANNISYIVCLFIFCLALPFAVIVYCYGRLLFAIKQ 0 | |||
0 VSGVSKSSSRAREQRVLIMVIVMVVCFLLCWLPYGVMALVATFGKPGIISPSASIIPSVLAKSSTVYNPIIYIFLNKQ 0 | |||
0 FYRCFTALIHCNKHPQVSSNKGSSKTTKIMLTARKLPDANFTVNAASNPPSSSVVKYEADSKTHNGDTKPFKTLVANYVI* 0 | |||
>TMTa1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc AF349947 12670711 full G? teleost multiple tissue syn(+PBX3 +TNK2 +TMTa1 -PAP2D +LPPR4) | |||
0 MIVSNLSVLSCRRNSALCLGAVEGHLEASSSYRTLSPTGHILVAVSLGFIGTFGFLNNLLVLVLFGRYKVLRSPINFLLVNICLSDLLVCVLGTPFSFAASTQGRWLIGDTGCVWYGFANSLL 1 | |||
2 GIVSLISLAVLSYERYCTMMGSTEADATNYKKVIGGVLMSWIYSLIWTLPPLFGWSRYGPEGPGTTCSVDWTTKTANNISYIICLFIFCLIVPFLVIIFCYGKLLHAIKQ 0 | |||
0 VSSVNTSVSRKREHRVLLMVITMVVFYLLCWLPYGIMALLATFGAPGLVTAEASIVPSILAKSSTVINPVIYIFMNKQ 0 | |||
0 FYRCFRALLNCDKPQRGSSLKSSSKTKPFRPGRRTDNFTFMVASVGPNQTNPVEDGPPSADNTKPAVLSLVAHYNG* 0 | |||
>TMTa_takRub Takifugu rubripes (fugu) Deut.Acti.Perc AF402774 12670711 full G? teleost multiple tissue syn(-CALD1 +TNK2 -RAB18 +ABI1 12670711) | |||
0 MIVSNVSLSGCAGVNGAVCAAEGHQAGGSDRSTLTPTGNLVVSVFLGFIGTFGLVNNLLVLVLFCRYKMLRSPINLLLMNISISDLLVCVLGTPFSFAASTQGRWLIGEAGCVWYGFANSLF 1 | |||
2 GVVSLISLAVLSFERYSTMMTPTEADPSNYCKVCLGITLSWVYSLVWTVPPLFGWSSYGPEGPGTTCSVNWTAKTTNSISYIICLFVFCLIVPFLVIVFCYGKLLCAIRQ 0 | |||
0 VSGINASTSRKREQRVLCMVVIMVICYLLCWLPYGVVALLATFGPPDLVTPEASIIPSVLAKSSTVINPIIYVFMNKQ 0 | |||
0 FYRCFLALLCCQDPRSGSSMKSSSKVATKAKGVTPTGQRRTDFLYMVASLGRPAATIPQLGPSFDATNDFTKPPSSDTIKPVVVSLAAHCDG* 0 | |||
>TMTa_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? teleost multiple tissue | |||
0 MIASNASVSGCAGVHGAACAADAPPAGGSHRSSSSLTPTGNLVVSVFLGLIGTSGLVSNLLVLVLFCRFKVLRSPINLLLVNISVSDLLVCVLGTPFSFAASTQGRWLIGAAGCVWYGFVNSLF 1 | |||
2 GIVSLISLAVLSFERYSTMMTPTEADSSNYCKVCLGIGLSWVYSLLWTVPPLLGWSSYGPEGPGTTCSVNWTAKTANSVSYIICLFVFCLILPFLVIVFCYGKLLCAIRQ 0 | |||
0 VSGVNASMSRRREQRVLFMVVVMVICYLLCWLPYGVVALLATFGPPGLVTPAASIIPSILAKSSTVINPVIYVFMNKQ 0 | |||
0 FSRCFLSLLCCEDPRSSTSLRSSSRVTTKAVRGGTLTGQRRTNHLLYMVAALGRPVATAMPQLGPSFDATYDITKAPSSDNHQPVVVSLEAHG* 0 | |||
>TMTa_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? teleost multiple tissue syn(+TNK2 +ENC) | |||
0 MIVSNLSLSGCAGVSSALCAAAGEGHLSGGSHRNTLTPTGHLVVAVCLGFIGTLGLMNNLLVLVLFCRYKMLRSPINLLLINISISDLLVCVLGTPFSFAASTQGRWLIGEGGCVWYGFANSLF 1 | |||
2 GIVSLISLAVLSYERYSTMVAPTEADSSNYHKISLGITLSWVYSLIWTAPPLFGWSHYGPEGPGTTCSVDWTARTANSISYIICLFVFCLIVPFLVIVFCYGKLLCAIRQ 0 | |||
0 VSGINASLSRKREQRVLFMVVIMVVCYLLCWLPYGIMALMATFGPPGLITPVASIIPSVLAKTSTVINPVIYVFMNKQ 0 | |||
0 FYRCFKALLRCEAPRPSSSLKSSSKVPTKAMRGAAVTGPRHTNNFLFVVASLGRPVATIPQLGPSVEPTIDVTGGPSSDNNKPVIVSLVAQCDG* 0 | |||
>TMTa_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic frag G? teleost multiple tissue syn(SLC12A3) two contigs frag | |||
0 MLVSNVSLGGCAEFNSALCAGAGEEHLGGGSYRTTLTPTGHLIVAVCLGFIGTFGLVNNLLVLVLFCRYKILRSPINLLLINISISDLLVCVLGTPFSFAASTQGRWLIGEGGCVWYGFANSLC 1 | |||
2 GIVSLISLAVLSYERYSTMMTPAEADSSNYRKISLGIILSWGYSLLWTLPPLFGWSHYGPEGPGTTCSVDWTAKTANNISYIICLFVFCLIVPFMVIVFCYGKLLYAIKQ 0 | |||
0 VSGINVSVSRKREQRVLFMVVIMVICYLLCWLPYGIMALLATFGPPDLVTPEASIIPSVLAKTSTAINPVIYVFMNKQ 0 | |||
>TMTa_pimPro Pimephales promelas (minnow) Deut.Acti.Otoc DT200813 frag G? teleost multiple tissue | |||
0 GHLVVAVCLGFIGTfGFLNNTLVLILFCRYKVLRSPMNYLLVSIAVSDLLVCVLGTPFSFAASTQGRWLIGRAGCVWYGFINSCL 1 | |||
2 GVVSLISLAVLSYERYCTMMGATQADSTNYKKVAMGIAFSWIYSMVWTLPPLFGWSCYGPEGPGTTCSVNWAARTANNVSYIICLFFFCLILPFIVIVYSYGRLLQAITQ 0 | |||
0 VSRINTVVSRKREQRVLFMVITMVVCYLLCWLPYGIMALLAAFGRPGLVTPAASIVPSVLAKTSTVINPIIYIFMNKQ 0 | |||
0 FCRCFHALIMCTTPQRGSSFKNSSKVTKTLRTVRRANGQNVTFAVASAGHPTICAPH | |||
>TMTc_danRer Danio rerio (zebrafish) Deut.Acti.Otoc CO927631 full G? teleost multiple tissue syn(+TNK1 +TMTb -MYEOV2) | |||
0 MIESNVSRSCEWCAGGGEGTGAHLDENHSDHSLSPTGHLVVAVCLGFIGTFGFLNNTLVLVLFCRYKVLRSPMNCLLISISVSDLLVCVLGTPFSFAASTQGRWLIGRAGCVWYGFINSFL 1 | |||
2 GVVSLISLAVLSYERYCTMMGSTQADSTNYRKVVIGIAFSWIYSMVWTLPPLFGWSCYGPEGPGTTCSVNWAARTPNNVSYIVCLFVFCLILPFIVIVYSYGRLLQAITQ 0 | |||
0 VSRINTVVSRKREQRVLFMVVTMVVCYLLCWLPYGIMALLATFGHPGLVTPAASIVPSLLAKSSTVINPIIYIFMNKQ | |||
0 FCRCFHALIMCTTPERGSSFKNSSKVTKTLRTVRRANGQNVTFAVASAVHRTPYSDRQKSSSEGEKLPPATGQGTSKPVVSLVAYYNG* 0 | |||
>TMTb_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? teleost multiple tissue syn(+TFRC +TMTb +CHES1 -MYEOV2 -ARHGAP21) | |||
0 MIVCNVSLSCAHCPGEGTAANDAYAQASGSLATPTLSQRGHLVVAVCLGFIGTVGFLSNFLVLALFCRYRALRTPMNLMLVSISASDLLVSVLGTPFSFAASTQGRWLIGRAGCVWYGFVNACL 1 | |||
2 GIVSLISLAVLSYERYCTMVSSTIASNRDYRPVLGGICFSWFYSLAWTVPPLLGWSRYGPEGPGTTCSVDWRTQTPNNISYIVCLFTFCLLLPFFVILYSYGKLLHTIRQ 0 | |||
0 VRRVSSTVTRRREHRVLVMVVAMVVCYLICWLPYGVTALLATFGPPNLLTPEATITPSLLAKFSTVINPFIYIFMNKQ 0 | |||
0 FYRCFRAFLNCSTPKRDSTVRTFTRISLRALRQDQQQKGSALAPSSARPTPNSIHESSLKGSHSTPSNGGAAAAKSPAANRSKPKLILVAHYRE* 0 | |||
>TMTb_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? teleost multiple tissue | |||
0 MIVCNLSLSCAHCPGGGAAATDAYAYAEAPGSLAPPTLSQRGHLVVAVCLGAIGTVGFLSNLLVLALFCRFRALRTPMNLMLVSISASDLLVSVLGTPFSFAASTQGRWLLGRAGCVWYGFVNACL 1 | |||
2 GIVSLISLAVLSYERYCTMMASTMASNRDYRPVLLGICFSWFYSLAWTVPPLLGWSRYGPEGPGTTCSVDWRTQTPNNISYIVCLFAFCLLLPFCVILYSYGKLLHTIRQ 0 | |||
0 VSSVSSAVTRRREHRVLVMVVAMVVCYLICWLPYGVTALLATFGPPNLLTPEATITPSLLAKFSTVINPFIYIFMNKQ 0 | |||
0 FYRCFRAFLSCSSPERGSTVRTFTRISLRAVCQRKQQRVSAPAASSACPTPNSIHHSSRKGSHSASSNSGTAAAAKTPAANSSKPKLILVVHYRE* 0 | |||
>TMTb_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? teleost multiple tissue | |||
0 MIVCNVSLSCVHCPGGGAGGTAATATGAYEEVSDSLPAPSLSPKGHLVVAVCLGFIGTFGFLSNFLVLALFCRYRALRTPMNLLLVSISASDLLVSMVGTPFSFAASTQGRWLIGRAGCVWYGFVNACL 1 | |||
2 GIVSLISLAVLSFERYSTMVKPTVADGRDFRPALGGIAFSWLYSVAWTVPPLLGWSEYGPEGPGTTCSVDWKTQTANNISYIVCLFVFCLVLPFCVILYSYSRLLQAIRQ 0 | |||
0 VSVVSSVVTRHREQRVLAMVVVMVACYLVCWLPYGVAALLATFGPRDLLSPEASITPSLLAKFSTVVNPFIYIFMNKQ 0 | |||
0 FYRCFRAFLSCSTPERGSTLKTFSRPTKTLRAGRHEKGRRVSAAAPSTAQPTRNSAPRSSQGANHASATPPPSPADGRCAAAGAAKPKRTLVAHYRE* 0 | |||
>TMTb_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full G? teleost multiple tissue | |||
0 MIVPNASLSCAHCDGDAAEQDAPGSAAAPSLSPTGHLVVAVCLGLIGTCGFLSNLLVLALFCRYRALRTPMNLLLVSISVSDLLVSVLGTPFSFAASTQGRWLIGRAGCVWYGFINACL 1 | |||
2 GIVSLISLAVLSYERYSTVMTPNMADGRDFRPALGGICFSWLYSVAWTVPPLLGWSRYGPEGPGTTCSVDWKTQTPNNISYIICLFTFCLLLPFGVIVYSYGKMLRVIRQ 0 | |||
0 VRSMSSVVTRRREQRVLVMVVTMVVCYLVCWLPYGIAALLATFGPRDLLTPAASITPSLLAKFSTVINPLIYIFMNKQ 0 | |||
0 FYRCFWAFFCCSTPEQVSTLRTFSRVTKTIRTFRQERELHVSAPAPSSGLPTPNSIQKGNNHVDPSSINQACAASDSPDSRKPKVVLVAHYQE* 0 | |||
>TMTa1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? teleost multiple tissue exons 1,2,3 | |||
0 MLNSSPNSSPSLPLSQVGWTGLSRTGLTVVAVCLGIIMVLGFLNNLLVLVLFCKYKVLRSPMNMLLLNISVSDMLVCICGTPFSFAASVQGRWLVGEQGCKWYGFANSLF 1 | |||
2 GIVSLMSLTILSYDRYITITGTTEADITNYNKTIVGIALSWIYSLMWTLPPLFGWSNYGPEGPGTTCSVNWQSKEVSSKSYIICLFIFCLLMPFLVIVYCYGKLVLAVRK 0 | |||
0 VSANNSMGRTRENKLLIMVTFMIICFLLCWLPYGIVALLATFGSPGLITPTASIIPSVLAKTSTVYNPIIYIFMNKQ 0 | |||
>TMTa2_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? teleost multiple tissue exons 1 and 3 | |||
0 MMAHSANISTTLNASDHAPNLAGLSQSGHTTVAVFLGIILVLGCVNNLLVLLLFVCFKEIRTPLNMILLNISLSDLSVCVFGTPFSFAASIYRRWLIGHKGCKWYGFANSLF 1 | |||
0 VGKINQMTAQTREHRILLMVISMVTFYLLCWLPYGTVALIGTFGNADLITPTCSVIPSILAKSSTVINPVIYVIMNKQ 0 | |||
>TMTx_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002207814 frag G? assembly duplication no N-term even in v2 | |||
0 VAAILALIGVLGIVNNSTTLYLVGRYKQLRTPFNILMVNLSVSDLLMCVLGTPFSFVSSLHGRWMFGHSGCEWYGFICNFL 1 | |||
2 GIVSLITLTVISYERYLLMKRLPNERILSYRAVALAVVFIWCYSLLWTAPPLVGWSSYGPE 0 | |||
0 GYGISCSVNWESRTANDTSYIVAYFVGCLVFPVAIIVISYTRLILYMRQ 0 | |||
0 APSAPMQMLVRREKRVTKMVVVMIMGFTICWTPYTIVALIVTCGGEGIITPAAATVPALFAKSSVVYNAAIYVAMNNQ 0 | |||
0 FRKCFLRSLNCRSQPRDPSSQQYTLKTNQVGMSTSGSQAARTADRIKTVHVATANPQDHRSSSGQAVEDNGGFRKSLTHSLPLNSISTLLEAEK* 0 | |||
>TMTy_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran FE572481 full G? teleost multiple tissue gastrula XM_002222645 flawed allele dup new | |||
0 MASAGQNVTFPAIDTMAPTPEALTSDPTTPAYFTTEQHLLMAVWLGFIGSFGFVTNLLTVLVFWCFKSLRTPFHLYLGGIALSDLLVAALGSPFAVASAVGERWLFGRAVCVWYAFVNYFL 1 | |||
2 SIVSIVTMATMSFSRYWVIIRPQSAPRLDTVYGACVVNALAWCYSFFWTIMPVLGWSRFTQ 0 | |||
0 VAAMTVCSLDWDHHTPLSKSYIPVAFLTCLFLPLGVIIFSVFKTTMHLRR 0 | |||
0 AAEVEDEVPNEVRAGRKTTRITLVMAGCWLVAWLPYACMALVIAAGGRVSPTVEVLATKFAKTSYIVNTIIYLVMEKE 0 | |||
0 FRKSLVLLLFCGRDPFDIQIEQPAYEKADVYVERLVTAEPMVEMEAVNVRPAQQEPARAPFGTPL* 0 | |||
>TMT1_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XM_001177470 16311335 full G? teleost multiple tissue PIN-type introns no cdna no sacKow | |||
0 MSNLMTGLVTNVNALSGIGNETPTTIGLSSLVVPVSRTTYNYLTVYTGFLTIFGILNNGIVMILFARFPSLRHPINSFLFNVSLSDLIISCLASPFTFASNFAGRWLFGDLGCTLYAFLVFVA 1 | |||
2 GTEQIVILAALSIQRCMLVVRPFTAQKMTHRWALFFISLTWIYSLIICVPPLFGWNRYTYEGPGT 1 | |||
2 ACSVAWNSPSPGDTSYIIFIFVLVLVIPFGIIIFCYGLLVYAVKK 0 | |||
0 ISRTQAALSSEAKADRKVSKMIFIMILFFLIAWTPYTGFSLYVTFGKNVVITPLAGTFPPFFAKLCTIHNPIIYFLLNKQ 0 | |||
0 FKDALIQLFCCGENPFDRDESEHEGRGGRHRHRTAPSATAHIGGRGRASSLPTATSMLDIPQAASTAASSSGKTQNKESLEKGPSTSETTNKRVFELSSKIQKFEISEKNNTPSSSELPGASSLSGALMPPRRAMKNQVGCLPPVDN* 0</font> | |||
<font color ="#0066CC">>TMT1_plaDum Platynereis dumerilii (ragworm) Ecdy.Anne.Poly CT030681 16311335 full G? 5 exons | |||
0 MDDLGFLGNSSVNYTVPLLQEDPLLLRILYFGPTSYVITAIYLCIVGVIGTLSNGVIMYLYFKDKSLRSPMNLLFVNLAMSDFTVAFFGAMFQFGLTCTRKYMSPGMALCDFYGFITFLG 1 | |||
2 GLASEMNLFIISVERYLAVVRPFDVGNLTNRRVIAGG 1 | |||
2 VFVWLYSLVFAGGPLVGWSSYRPEGLGTWCSISWQDRSMNTMSYVTAVFLGCYFFPVSIIIFCYFNVWRKVKE 0 | |||
0 AADAQGGAGTAGKAEKSIFRMSVIMVTCYLTAWTPYAIVCLIASYGPPNGLPIYAEVLPSLFAKSSQVYNPIIYVLMNKP 0 | |||
0 YRSALVSLVCRGRNPFDEAGGTAGGTTAKDETLGKGNKVAAA* 0 | |||
>TMT2_plaDum Platynereis dumerilii (ragworm) Ecdy.Anne.Poly AY692353 15514158 full G? new genomic | |||
0 MDGENLTIPNPVTELMDTPINSTYFQNLNAETDGGNHYIYNAFTATDYNICAAYLFFIACLGVSLNVLVLVLFIKDRKLRSPNNFLYVSLALGDLLVAVFGTAFKFIITARKTLLREEDGFCKWYGFITYLG 1 | |||
2 GLAALMTLSVIAFVRCLAVLRLGSFTGLTTRMGVAAMA 1 | |||
2 FIWIYSLAFTLAPLLGWNHYIPEGLATWCSIDWLSDETSDKSYVFAIFIFCFLVPVLIIVVSYGLIYDKVRK 0 | |||
0 VAKTGGSVAKAEREVLRMTLLMVSLFMLAWSPYAVICMLASFGPKDLLHPVATVIPAMFAKSSTMYNPLIYVFMNKQ 0 | |||
0 FRRSLKVLLGMGVEDLNSESERATGGTATNQVAAT* 0 | |||
>TMT_terTra Terebratalia transversa (lampshell) Loph.Brac.Rhyn HQ679623 full 21362157 | |||
MSTASSGTVTTVMESNITTLSPFTTRAPMFSDGANVIIVLVLFVTAILGFFMNGSVLIIFYRKKELRSPTNMFIISMAFNDLMMSILGNPFAAVSNLHHR | |||
WIFGDAACVWYGFIMFFLGLNGIYHLTAIAFDRYIVIAKPLLSSKITTRVAALAISVCWFGAFLWSVFPVFGWSSYVEEGEHSSCSINWISQSIVDSSYI | |||
ISIFIFCFFLPILLVIFSYFHVYMTVRTIAKSNTFGKDSKITKKNEAVERKMAKMVIIMITVFFGSWSPYAIVSMFVAFGDGSSLPAAAIAAPAIIAKSA | |||
MIWNPIIYVLMNVQFRSGFAEMYPCFAVCANTDGGKVKVERPPTVQDNIGDDNTEMSDVRSAVPPAPTSNILKVAPAPDTTKADVHTSETGEQSIAMSIA | |||
RMSVRHKVESRTPVEQMNAVEV*</font> | |||
<font color ="#6699FF">>TMT1_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_312503 full G? GPROP11 pteropsin GPROP11 head-to-head tandem GPROP12 | |||
0 MYDVTDAAAINSDHQELMAPWAYNGAAVTLFFIGFFGFFLNIFVIALMYKDVQ 0 | |||
0 LWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWLYGKSICVAYGFFMSLL 1 | |||
2 GIASITTLTVLSYERFCLISRPFAAQNRSKQGACLAVLFIWSYSFALTSPPLFGWGAYVNEAANIS 2 | |||
1 CSVNWESQTANATSYIIFLFIFGLILPLAVIIYSYINIVLEMRK 0 | |||
0 NSARVGRVNRAERRVTSMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFWRIRRSNGVAGQPDSNNTNNSNRDKESARHTAKEGL | |||
ECSLDFCHWTVRGTRVSISSAERNVPAPAARERSGGHSVTGSREESRDRHVTLKTMLSVGPRSPSSVAPVAADCSTTDVPTSGDGSVRIVRQDSELSVIHDGGGGGGGSSSRVLVIKSQKPRSNML* 0 | |||
>TMT2_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_312502 full G? GPROP12 pteropsin encephalopsin GPROP12 | |||
0 MNDAPNDVAASAVDYEDLMAPWAYNASAVTLFFIGFFGFFLNLFVIALMCKDMQ 0 | |||
0 LWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWIFGRTLCVAYGFFMSLL 1 | |||
2 GITSITTLTVLSYERYCLISRPFSSRNLTRRGAFLAIFFIWGYSFALTSPPLFGWGAYVQEAANIS 2 | |||
1 CSVNWESQTKNATTYIIFLFVFGLVVPLIVIVYSYTNIIVNMRE 0 | |||
0 NSARVGRINRAEQRVTSMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFSRVRNKGQQAAADQNTTTMQRELTKSSRDMVECSF | |||
DFCRKKSRFKISLVKPTAPLAVVDVSSTSHRDKGTSRSPLDQTVLNETNEDVGRERSGGGGGGGAYAGTRFVRPDFELSVINSGKSILIKSKNFRSNLL* 0 | |||
>TMT_aedAeg Aedes aegypti (mosquito) Ecdy.Inse.Dipt XM_001650752 full G? pteropsin genome frameshifted | |||
0 MESWAYVASAVTLFFIGFFGFFLNLFVIALMCKDVQ 0 | |||
0 LWTPINIILFNLVCSDFSVSIIGNPFTLTSAISRHWIFGRTVCIAYGFFMSLL 1 | |||
2 GITSITTLTVLSYERFCLISHPFSSRSLSRRGAVFAILFIWSYSFALTSPPLFGWGAYVNEAANIS 2 | |||
1 CSVNWESQTLNATSYIIFLFVFGLVVPLVVIVYSYTNIVVNMKR 0 | |||
0 NAARVGRINRAEKRVTRMVFVMVLAFMIAWTPYAVFALIEQFGPTDIISPALGVLPALIAKSSICYNPIIYVGMNTQFRAAFNRVRNNESVDNNTITNQKDITMNTSK | |||
EIVECSFDFCRKKRLKIKLQSNAKSKNNNNNSRNQSIADPSSTSNGDDLDQPSPAQTVLNSTVANSGSVASFGPKKRLRSDFELSVISSGKSILIKSNTFRSNLV* 0 | |||
>TMT_culPip Culex pipiens (mosquito) Ecdy.Inse.Dipt genomic full G? pteropsin | |||
0 MPPWAYVATAVVLFFIGFFGFFLNLFVIALMCKEVQ 0 | |||
0 LWTPMNIILLNLVCSDFSVSIVGNPFTLSSAISHRWLFGRKLCVAYGFFMSLL 1 | |||
2 GITSITTLTVLSYERFYLISRPFSSRSLSRRGALGAVLLIWCYSFALTSPPLFGWGAYVNEAANIS 2 | |||
1 CSVNWETQTLNATTYIIYLFVFGLVVPLTVIVYSYTNIIVNMKK 0 | |||
0 NAARVGRINRAEKRVTTMVAVMVIAFMVAWTPYSVFALMEQFGPPDVIGPGLAVLPALIAKSSICYNPIIYVGMNTQFRAAFNRVRHDPGDMANTTTNQKELT | |||
RSSRDVASVDCSFDFCRKKNRLKMKLHNSIHRSAAIKAGRSQSPHSESERSSSGATQERSTRVQTMLNATVDSRSISGSTGKLMKSDFELSVINSGKSILIKSNTFRSNLV* 0 | |||
>TMT_triCas Tribolium castaneum (flour_beetle) Ecdy.Inse.Cole genomic full G? pteropsin | |||
0 MKNFNSTEIGDELLIPVEGYIAAAVVLFCIGFFGFSLNLTVIIFMLKERQ 0 | |||
0 LWSPLNIILFNLVVSDFLVSVLGNPWTFFSAINYGWIFGETGCTIYGFIMSLL 1 | |||
2 SITSITTLTVLAFERYLLIARPFRNNALNFHSAALSVFSIWLYSLSLTIPPLIGWGEYVHEAANLS 2 | |||
1 CSVNWEEKSPNSTSYILYLFAFGLFLPLVIITFSYVNIILTMRR 0 | |||
0 NAAFRVGQVSKAENKVAYMIFIMIIAFLTAWSPYAIMALIVQFGDAALVTPGMAVIPALLAKSSICYNPVIYIGLNAQVKGAKWVSGLIYLFQFQQAWMQKWKKNRR | |||
GSDALGTSRVMLETIHQACRDEKTDKLLEKKTKFCKDFETDVSML* 0 | |||
>TMT_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme NM_001039968 16291092 full G? AmLop2 pteropsin ciliary compound eye not ocelli clock | |||
0 MSLNRSTMEHVIYEDQVSPVMYIGAAIALGFIGFFGFTANLLVAIVIVKDAQILWTPVNVILFNLV 0 | |||
0 FGDFLVSIFGNPVAMVSAATGGWYWGYKMCLW 2 | |||
1 YAWFMSTLGFASIGNLTVMAVERWLLVARPMQALSIR 2 | |||
1 HAVILASFVWIYALSLSLPPLFGWGSYGPEAGNVSCSVSWEVHDPVTNSDTYIGFLFVLGLIVPVFTIVSSYAAIVLTLKKVRKRA 1 | |||
2 GASGRREAKITKMVALMITAFLLAWSPYAALAIAAQYFN 0 | |||
0 AKPSATVAVLPALLAKSSICYNPIIYAGLNNQFSRFLKKIFDARGSRTAVPDSQHTALTALNRQEQRK* 0 | |||
>TMT_bomTer Bombus terrestris (bumblebee) Ecdy.Inse.Hyme 454SRA full G? pteropsin presumed ciliary compound eye not ocelli clock | |||
0 MSLNRSTVDNVVYDAEDQVSPMVYVAAAIALGFIGFFGFTMNLLVAIVIVKDAQILWTPVNVILVNLV 0 | |||
0 VGDFLVAAFGNPVAMISAITGGWYWSYEMCLW 2 | |||
1 YAWFMSTLGFASIGNLTVMAVERWLLVARPMKALSIR 2 | |||
1 HAITLGIFVWFYALCLSLPPLFGWGSYGPEAGNVSCSVSWEVHDPLTKSDSYIAFLFVFGLIIPVLVISSSYVAIIVTLRKVRKRA 1 | |||
2 GASGRREAKITKMVALMITAFLLAWSPYAALAIAAQYFD 0 | |||
0 AKPSASVAVLPALLAKSSICYNPIIYAGLNSQFPRSLKKIFDVRIARSSLPDSQNTALTLLNRQEQRN* 0 | |||
>TMT_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01001608 full 63% bee | |||
0 MSLNLSVEEQLSIDIYIFAAIALGFIGFL 21 GFFLNLLVIITVLKNANVLWTPNNVVLINMV 0 | |||
0 IGDFLVAALGNPFTMTSAIAGEWFWSHEVCLW 2 | |||
1 YAWFMTTMGFASIGNLTVMAMERFLLVTCPMKTLSIR 2 | |||
1 HAYILAILVWMYALSLSLPPFFNWGIYGPEAGNISCSVSWEIHDPYMHNDTYIGFLFIVGFFLPVTIIISSYYGIIKTLRKIRKRV 1 | |||
2 GAKNRREKKVTKMVYLMILAFLIAWSPYAVLALATQYFYV 0 | |||
0 QTSHILAVLPALLAKSSICYNPIIYASMTAQFPTWKKMFSINRNNTKSKQRHGSELQIK* 0 | |||
>TMT_rhoPro Rhodnius prolixus (kissing_bug) Ecdy.Inse.Hemi ACPB01038515 full G? pteropsin ACPB01038514 | |||
0 MLMPSAGFLAASIILFLIGFLGFFGNLIVIIIMCRDKN 0 | |||
0 LWTPVNFILFNVIVSDFSVAALGNPFTLASAIAKRWFFGQSMCVAYGFFMALL 1 | |||
2 GITSINSLTVLALERYLIVSQPVSHGSLSRPTASDIVGSIWLYSFVITiPPLVGWGEYGLEAANIS 2 | |||
1 CSINWETRSHSSTSYILFLFTFGFFIPIIVISYSYMNIILTMKK 0 | |||
0 STMNAGRVNKAESRVTWMIFVMIFAFFLAWTPYAILALMIAFFDSNVSPAIATIPAIFAKTSICYNPFIYAGLNTQ 0 | |||
0 FRQSWRRVLGGKREDSTTMATATSFGLNSKRYKEVSCVIDVKGKDKIKLSALNKSTATETAI* 0 | |||
>TMT_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi genomic full G? pteropsin two contigs, 1st exon very wrong in XM_001952259 | |||
0 MNGEYAQPQHISDAIYLGAAIVLSIIGIVGFIFNTCVIFIMIRDTR 0 | |||
0 LWTPQNVIIFNLATSDLAVSVLGNPVTLAAAITKGWIFGQTICVIYGFFMALF 1 | |||
2 GIASITTLTVLAYDRYLMIRYPFSSSRLTKETALYAIAGIWIYAFAVTGPPLFGWNRYVNESANIS 2 | |||
1 CSIDWESGEHSNYVIYIFVFGLFLPVTVIIYSYVSLVVTVRK 0 | |||
0 AEKIIGQATKAECRVAIMVAVMILAFLTAWMPYSVLALMIAFGGVHISPVVSIIPALCAKSSICWNPIIYIGLNTQ 0 | |||
0 FRSAWKRFLNIQDTLSEVSLDADITTGMTKLMTGHQELPAHPMNNGDASHPPGLIMCCLAHDEHRQSATYADRYECNLEMKSCNPQTLGRRPETDIGDVSL* 0 | |||
>TMT_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BY930435 frag G? ovary cDNA | |||
0 MPRWGYVASAFVLFLIGFFGFFLNLMVILLMFKDRQ 0 | |||
0 LWTPLNIILFNLVCSDFSVSVLGNPFTLISALFHRWIFGHTMCVLYGFFMALL 1 | |||
2 GITSITTLTVISFERYLMVTRPLTSRHLSSKGAVLSIMFIWTYSLALTTPPLLGWGNYVNEAANIQ 2 | |||
1 CSVNWHEQSTNTLTYIMFLFAMGQILPLSVITFSYVNIIRTLKR 0 | |||
0 NSQRLGRVSRAEARATAMVFIMIIAFTVAWTPYSLFALMEQFATEGI 0 | |||
0 VSPGASVIPALVAKSSICYDPLIYVGMNTQ 0 | |||
0 FR | |||
>TMT_helVir Heliothis virescens (moth) Ecdy.Inse.Lepi GR968759 frag G? pteropsin pheromone gland | |||
0 MPRWGYVVSAFVLFLIGFFGFFLNLMVILLMFKDRQ 0 | |||
0 LWTPLNIILFNLVCSDFSVSVLGNPFTLISALFHRWIFGKTMCVLYGFFMALL 1 | |||
2 GITSITTLTVISFERYMMVTRPLNSRHLSSKGAIMSIVF | |||
>TMTa_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic full G? pteropsin retained intron? | |||
0 MPVWVYWSASAYLLFISIAGLFMNIVVVVIILNDSQ 0 | |||
0 KMTPLNWMLLNLACSDGAIAGFG 2 | |||
1 TPISAAAALKFTWPFSHELCVAYAMIMSTA 1 | |||
2 GIGSITTLTVLALWRCQHVVWCPTNRNSNFTDPNGRLDRRQGALLLTFIWTYTLIVTCPPLFGWGRYDREAAHIS 2 | |||
1 CSVNWESKMDNNRSYILYMFAMGLFIPLMAIFVSYISILLFIHK 0 | |||
0 SQQTSNNSDTVEKRVTFMVAVMIGAFLTAWTPYSIMALVETFTGDNVTNDSVSSEIKFYAGTISPAVATVPSLFAKTSAVLNPLIYGLLNTQ 0 | |||
0 FRTAWEKFSSRFLGRKKRHQRSQMAMGVSHKRRRDYLRTLLNRPASDEPAIVQHPSTKEMASSQAVSCVVVSNLDVPRAPNNSYVTVNDE* 0 | |||
>TMTb_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic full G? pteropsin long tail IGQDNHSTYYSSRINNATNFSSAFPDGDLSY intron? | |||
0 MPTWAYRLTAAYLLLISVLGLIMNVVVVIVILNDSQ 0 | |||
0 RMTPLNWMLLNLACSDGAIAGFG 2 | |||
1 TPISTAAALEFGWPFSQELCVAYAMIMSTA 1 | |||
2 GIGSITTLTALAIWRCQLVVCCPAKRKSAFTNHSGRLGCRQGVILLVIIWIYALAITCPPLFGWGRYDREAAHIs 2 | |||
1 CSVNWESKTNNNRSYILYMFCMGLVVPLAVIIISYVRILRVVQK 0 | |||
0 NQQQSGNVHRHRRDAAEKRVTMMVACMIAAFMAAWTPYSILALFETFIGQDNHSTYYSSRINNATNFSSAFPDGDLSYVGTISPAFATIPSLFAKTSAVLNPLIYGLLNTQ 0 | |||
0 FRLAWERFSLRFLGRFQCHRTQGVSGQHGANHHKTRRNVRKYLPNCYGDSRSLKPTPTVHLPMKEMVVSHAEQKVKTAQEQASSSVTKITTIPLISSDNQTIVSCPSSIMAN | |||
CQQHETNQANHQQAARPDKVVDHQHLLQPNRLSSLLSLSLPSVLISTPNLPCSAQRQSAAEDQAMATCQQMTSGRIRDQQQQSDSFVVVGLLSRSADCYHHHTGDVEQFVFLDSTVDELGLTARSASP* 0</font> | |||
<font color ="#990099">>MEL1_homSap Homo sapiens (human) Deut.Euth.Euar NM_033282 full Gq OPN4 melanopsin syn(-GRID1 -WAPAL +LDB3 +BMPR1A) | |||
0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0 | |||
0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1 | |||
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2 | |||
1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2 | |||
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLTSHTSNLSWISIRRRQESLGSESEV 0 | |||
0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGK 0 | |||
0 TKGLIPSQDPRM* 0 | |||
>MEL1_panTro Pan troglodytes (chimpanzee) Deut.Euth.Euar genomic full G? OPN4 melanopsin | |||
0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0 | |||
0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1 | |||
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2 | |||
1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2 | |||
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLISHTSNLSWISIRRRQESLGSESEV 0 | |||
0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGK 0 | |||
0 TKGLIPSQDPRM* 0 | |||
>MEL1_gorGor Gorilla gorilla (gorilla) Deut.Euth.Euar genomic full G? OPN4 melanopsin | |||
0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0 | |||
0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRSLRTPXNMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1 | |||
2 GCEFYAFCGALFGISSMITLXAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAyVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2 | |||
1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2 | |||
1 YAHVLTPYMSSVPaVIAKAsAIHNPIIYAITHPKYR 2 | |||
1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLISHTSNLSWISIRRRQESLGSESEV 0 | |||
0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGK 0 | |||
0 TKGLIPSQDPRM* 0 | |||
>MEL1_ponAbe Pongo abelii (orangutan) Deut.Euth.Euar genomic full G? OPN4 melanopsin | |||
0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGQLPSISPT 0 | |||
0 APGTWAAALVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLMVIYTFCR 2 | |||
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1 | |||
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATIGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2 | |||
1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2 | |||
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTMISHTSNLSWISGRRRQESLGSESEV 0 | |||
0 GWTHMEAAAVWGAAQQANGRFLYDQGLEDLEAKAPPRPQGEEAETPGK 0 | |||
0 TKGLIPSQDPRM* 0 | |||
>MEL1_rheMac Rhesus macaca (rhesus) Deut.Euth.Euar genomic full G? OPN4 melanopsin fill-in traces | |||
0 MNPPSGPRVPPSPTQEPSCMATPAPPSRWDSSQSSISSLGQLPSVSPT 0 | |||
0 AAGTWAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRGLRTPANMFIINLAISDFLMSFTQAPVFFASSLYKHWLFGET 1 | |||
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATIGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGr 2 | |||
1 ALQTFGACKGSGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2 | |||
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 VAIAQHLPCLGVLLGVSRRHSHPYPSYRSTHRSTLISHTSNLSWISGRRRQESLGSESEV 0 | |||
0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGQEAETPGK 0 | |||
0 TKGLLPCKDSRM* 0 | |||
>MEL1_calJac Callithrix jacchus (marmoset) Deut.Euth.Euar genomic full G? OPN4 melanopsin fill-in traces | |||
0 MNLPTGSRVLPSPTQEPSCMTTPAPPSRWDSSQSSISSLSQLPSISPT 0 | |||
0 AAGTWAAAWIPFPTVDVPDHAHYTLGTVILLVGVTGMLGNLTVIYTFCR 2 | |||
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKRWLFGET 1 | |||
2 GCEFYAFCGALFGISSMITLMAIALDRYLVITRPLATIGVASTKRAAFVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2 | |||
1 ALQTFGACKGSGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2 | |||
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 VAIAQHLPCLGVLLGVSRRHSHPYPSYRSTHRSTLISHTSNLSWISGRRRQESLGSESEV 0 | |||
0 GWTHMEAAAAWGAAQQANGRSLYGHGLEDLEAKAPPRPQRQEAETPGK 0 | |||
0 tkGLLPSLDARW* 0 | |||
>MEL1_micMur Microcebus murinus (mouse_lemur) Deut.Euth.Euar genomic full G? OPN4 melanopsin fill-in traces 4aa del exon 9 confirmed | |||
0 MNPPSGPRMPPSPAQEPSCVATPALASSGDSSQNSVSSLGQLLPASPT 0 | |||
0 ATGAWAAAWVPFPTADVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCr 2 | |||
1 SRSLRTPANMFVINLAVSDFLMSVTQAPVFFASSLYKRWLFGEA 1 | |||
2 gCEFYAFCGALFGISSMITLTAIALDRYLVITRPLASVGTASKRRAGLVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLCCFVFFLPLLVIIYCYIFIFRAIRETGR 2 | |||
1 ALQTFGASKGTSESPRQQQRLQNEWKMAKIMLLVILLFLLSWAPYTAVALVAFAG 2 | |||
1 YAHILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 VAIAQHLPCVGVLLGVSRQHSRPYPSYRSTHRSTLSSQASDLSWISGRRRQESLGSENDM 0 | |||
0 GWTDMEVAAAWGAAPRVRGRCPYGQGPEDMEVKV QGQEAETPGK 0 | |||
0 AEGLPPCMDPRM* 0 | |||
>MEL1_otoGar Otolemur garnettii (lemur) Deut.Euth.Euar genomic full G? OPN4 melanopsin fill-in traces | |||
0 mnLPSAPRVPPSPAQASSCVATPAPHSRWDSSQTSISSLGQLRPVSPT 0 | |||
0 ASGAWAAGWIPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 VRGLRTPANMFVINLAVSDFLMSVTQAPVFFASSLYKQWLFGET 1 | |||
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLTTVGVASKRRAALVLLGVWLYSLAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYTSFTPSVRTYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2 | |||
1 ALQTFGACKGSSESPRQRQRLQNEWKMAKITLLVILFFVLSWAPYTTVALVAFAG 2 | |||
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 VAIAQHLPCLGLLLGVSRQHSRPYPSYRFTHHSTLSSQASDLSWISGRRRQESLGSESEV 0 | |||
0 GWTDMEAAATWGAALQVSGQCPYSQGLEDMEAKGPLRPQGPETKTSGK 0 | |||
0 TKGLLPSLDPRM* 0 | |||
>MEL1_musMus Mus musculus (mouse) Deut.Euth.Euar AF147789 10632589 full G? OPN4 melanopsin GenBank wrong readthru | |||
0 MDSPSGPRVLSSLTQDPSFTTSPALQGIWNGTQNVSVRAQLLSVSPT 0 | |||
0 TSAHQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 NRGLRTPANMFIINLAVSDFLMSVTQAPVFFASSLYKKWLFGET 1 | |||
2 GCEFYAFCGAVFGITSMITLTAIAMDRYLVITRPLATIGRGSKRRTALVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMTFTPQVRAYTMLLFCFVFFLPLLIIIFCYIFIFRAIRETGR 2 | |||
1 ACEGCGESPLRQRRQWQRLQSEWKMAKVALIVILLFVLSWAPYSTVALVAFAG 2 | |||
1 YSHILTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 VAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGRKRQESLGSESEV 0 | |||
0 GWTDTETTAAWGAAQQASGQSFCSQNLEDGELKASSSPQVQRSKTPK 0 | |||
0 TKGHLPSLDLGM* 0 | |||
>MEL1_ratNor Rattus norvegicus (rat) Deut.Euth.Euar AY072689 11834834 full G? OPN4 melanopsin | |||
0 MNSPSESRVPSSLTQDPSFTASPALLQGIWNSTQNISVRVQLLSVSPT 0 | |||
0 TPGLQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 NRGLRTPANMLIINLAVSDFLMSFTQAPVFFASSLYKKWLFGET 1 | |||
2 GCKFYAFCGAVFGIVSMITLTAIAMDRYLVITRPLATIGMRSKRRTALVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYVTFTPLVRAYTMLLFCFVFFLPLLIIIFCYIFIFRAIRETGR 2 | |||
1 ACEGCGESPLRRRQWQRLQSEWKMAKVALIVILLFVLSWAPYSTVALVGFAG 2 | |||
1 YSHILTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 AAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGQKRQESLGSESEV 0 | |||
0 GWTDTETTAAWGAAQQASGQSFCSHDLEDGEVKAPSSPQEQKSKTPK 0 | |||
0 TKRHLPSLDRRM* 0 | |||
>MEL1_nanEhr Nannospalax ehrenbergi (molerat) Deut.Euth.Euar AM748539 full G? OPN4 melanopsin | |||
0 MNSPSGPRVPPGLAQKPSFMVTPVLPNQWISFQKNVSVGIQLPPASAT 0 | |||
0 ATGAQAASWVPFPTVDVPVHAHYTLGTVILLVGLTGMLGNLIVIYTFCR 2 | |||
1 SRGLRTRANMFTVNLAVSDFLMSFTQAPVFFASSLYKRWLFGEA 2 | |||
2 GCEFYAFCGAVSGITSMTTLTAIALDRYLVITRPLATIGVASKRRTALVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFLPLLIIIFCYIFIFKAIRETGR 2 | |||
1 ACEGCGESPQRRRQWQRLQNEWKMAKVALLVIFLFVLSWAPYSTVALVAFAG 2 | |||
1 YSHILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 LAISQHLPCLGVLIGVSSQRSHPSLSYRSTHRSTLSSQASDLSWISGRKRQESLGSESEV 0 | |||
0 GWTDTEVTAAWGVAQEASGWSPYRHSLEDGEVKASPSPQGQEAKTSR 0 | |||
0 TKGQLPSLNLRM* 0 | |||
>MEL1_phoSun Phodopus sungorus (hamster) Deut.Euth.Euar AY726733 15698924 frag G? OPN4 melanopsin 3D ends in error: AKGQLPSLDLGMQDAP | |||
0 MDSPPGPTAPPGLTQGPSFMASTTLHSHWNSTQKVSTRAQLLAVSPT 0 | |||
0 ASGPEAAAWVPFPTVDVPDHAHYILGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRSLRTPANMLIINLAVSDFLMSFTQAPVFFASSLYKKWLFGET 2 | |||
1 GCEFYAFCGAVLGITSMITLTAIALDRYLVITRPLATIGMGSKRRTALVLLGIWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYVTFTPQVRAYTMLLFCFVFFLPLLVIIFCYISIFRAIRETGR 2 | |||
1 ACEGWSESPQRRRQWHRLQSEWKMAKVALIVILLFVLSWAPYSTVALVAFAG 2 | |||
1 YSHILTPYMSSVPAVIAKASAIHNPIVYAITHPKYR 2 | |||
1 AAIAQHLPCLGVLLGVSSQRNRPSLSYRSTHRSTLSSQSSDLSWISAPKRQESLGSESEV 0 | |||
0 GWTDTEATAVWGAAQPASGQSSCGQNLEDGMVKAPSSPQ | |||
0 * 0 | |||
>MEL1_bosTau Bos taurus (cow) Deut.Euth.Laur genomic full G? OPN4 melanopsin | |||
0 MNPPSGPRAPLGPVQESSCLATPASSSRWDSSRSSASSLGHPPSISPT 0 | |||
0 AVRAQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKQWLFGEA 1 | |||
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATVGMVSKRRAALVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYVSFTPSVRAYTMLLFCFVFFLPLLIIIYCYIFIFKAIRETGQ 2 | |||
1 ALQTFGTCEGGSECPRQRQRLQNEWKMAKIELLVILLFVLSWAPYSTVALMGFAG 2 | |||
1 YAHILTPYMNSVPAVIAKASAIYNPIIYAITHPKYR 2 | |||
1 LAIAQHLPCLGVLLGVSGQRTGLYTSYRSTHRSTLSSQASDLSWISGRRRQASLGSESEV 0 | |||
0 GWMDTEATAAWGAGQQVSGWSPCSQRLDDVEAKALPRPQGRDSEAPGK 0 | |||
0 AKGLLPNLDARM* 0 | |||
>MEL1_susScr Sus scrofa (pig) Deut.Euth.Laur genomic full G? OPN4 melanopsin | |||
0 MNPPSRPSAPPDPALESSCMATPASPSRWDSSQSSTSSLGHPLPISPT 0 | |||
0 AARVRAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKQWLFGEA 2 | |||
1 GCEFYAFCGAVFGITSMITLTAIALDRYLVITHPLATVGMVSKRRAALVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYVSFTPKVRAYTMLLFCFVFLLPLLVIIYSYIFIFKAIRETGQ 2 | |||
1 ALQTFGACERRGKCPRQRQRLQNEWKMAKIELLVILLFVLSWAPYSTVALMGFAG 2 | |||
1 YGHVLTPYVNSVPAVIAKASAIYNPIIYAITHPKYR 2 | |||
1 MAIAQHLPCLGVLLGVSGQRTGLYTSYRSTHRSTLSSQASDLSWISGRRRQASLGSESEV 0 | |||
0 GWTDTEATAAWGAAQQVSRWSPCGQGLEDMEAKTSLGRLGWEAEAPGK 0 | |||
0 TKGLLPSLDPRM* 0 | |||
>MEL1_equCab Equus caballus (horse) Deut.Euth.Laur genomic full G? OPN4 melanopsin bad GenBank | |||
0 MNPPSEPQVPLGLAQEPGCVATPASPSRWSGSRSSTSSLGQPLPVGPT 0 | |||
0 AAGAQADAWVPFPTVDVPDHAHYTMGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKQWLFGKA 1 | |||
1 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATVGVVSKRWAALVLLGIWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLMCFVFFLPLLVIVYCYVFIFRAIRETGR 2 | |||
1 ALQTFGAWEGGGECPRQRQRLQSEWKMAKIVLLVILLFVLSWAPYSVVALVAFAG 2 | |||
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAIIHPKYR 2 | |||
1 MAIAQHLPCLGVLLGVSSQRTRPYTSYRSTHRSTLSSQGSDLSWISGRRRQASLGSESEV 0 | |||
0 GWMDTEAAAVWGAAQQMSGWSPCGQGLEDMEAKAPPRPQGWEGEALRK 0 | |||
0 IKGLLPSLDPRM* 0 | |||
>MEL1_felCat Felis catus (cat) Deut.Euth.Laur AY382594 15842746 full G? OPN4 melanopsin wrong readthru | |||
0 MNPPSGPRTQEPSCVATPASPSRWDGYRSSTSSLDQPLPISPT 0 | |||
0 AARAQAAAWIPFPTVDVPDHAHYTLGTVILLVGLTGILGNLMVIYTFCR 2 | |||
1 SRGLRTPANMFIINLAVSDFFMSFTQAPVFFASSLHKRWLFGEA 1 | |||
2 GCEFYAFCGALFGITSMITLMAIALDRYLVITHPLATIGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMSFTPSVRAYTMLLFCFVFFLPLLVIVYCYIFIFRAIRETGQ 2 | |||
1 ALQTFRACEGGGRSPRQRQRLQREWKMAKIELLVILLFVLSWAPYSIVALMAFAG 2 | |||
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 MAIAQHLPCLGVLLGVSGQHTGPYASYRSTHRSTLSSQASDLSWISGRRRQASLGSESEV 0 | |||
0 GWMDTEAAAVWGAAQQVSGRFPCSQGLEDREAKAPVRPQGREAETPGQ 0 | |||
0 TKGLLPSQDPRM* 0 | |||
>MEL1_canFam Canis familiaris (dog) Deut.Euth.Laur genomic full G? OPN4 melanopsin fixed 44-way | |||
0 MNPPSGPGAQEPGCVATAASPGRWHGSPRSTVGLDQALPTGPT 0 | |||
0 AAGARAAAWAPFPTVDVPDHAHYILGTVILLVGLTGMLGNLMVIYTFCR 2 | |||
1 TRGLRTPSNMFIINLAVSDFFMSFTQAPVFFASSLHKRWLFGEA 1 | |||
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITHPLAAVGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMSFTPSVRAYTMLLFCFVFFLPLLVIVYCYVFIFRAIRETGQ 2 | |||
1 ALQTFRACEGGARSPRQRQRLQREWKMAKMELLVILLFVLSWAPYSAVALTAFAG 2 | |||
1 YSHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 MAIAQHLPCLGVLLGVSGQRTGPYASYRSTHRSTLSSQASDLSWISGRRRQASLGSESEV 0 | |||
0 GWMDTEAAAVWGAAQPAGGRFLCTQGLEDAEAKAPLRPRGQAVETPGK 0 | |||
0 TKGRLPSLDPSRE* 0 | |||
>MEL1_myoLuc Myotis lucifugus (microbat) Deut.Euth.Laur genomic full G? OPN4 melanopsin 7 aa del in exon 1 confirmed 1 aa del in DRY cyto loop | |||
0 MNPPSGPRGPLDPAREPGCVATPVSP SSTSSVDPPLPTSPT 0 | |||
0 AAEAQAAAWVSFPTVDVPAHAHYTLGIVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRGLRTPANMFIINLAVSDFLMCFTQAPVVFASSIYKRWLFGEA 1 | |||
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPL AIGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMSFTPAVRSYTMLLFCFVFFLPLLVIIYCYVFIFRAIRETGQ 2 | |||
1 ALQTFGACEGRSELPRQRQGLQNEWKMAKIVLLVILLFVLSWAPYSTVALMAFAG 2 | |||
1 YAHVLTPYMNSVPAIIAKASAIHNPIIYAITHPKYR 2 | |||
1 MAIAQHLPCLGLLLGVSGQRTGPYASYRSTHRSTLSSQASDLSWVSRRRRQESLGSESEM 0 | |||
0 GWTDTEAAAMWGAAQQVSGPPPCSQGLEDVETKAPPKSQGHEAEDPRK 0 | |||
0 TKGLLPSPDPRM* 0 | |||
>MEL1_pteVam Pteropus vampyrus (macrobat) Deut.Euth.Laur genomic full G? OPN4 melanopsin | |||
0 MNLPVGPRVPLDPAQEPSCMATPASPS WDSSPSSASSLDQPLPISPT 0 | |||
0 AAGSQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVVFISSLYKRWLFGQA 1 | |||
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLAAIGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMSFTPSVRAYTMLLFCFVFFLPLLIIIYCYIFIFRAIRETGQ 2 | |||
1 ALQTFGACEGLSELPRQRQRLQSEWKMAKIVLLVILLFVLSWAPYSTVALMAFAG 2 | |||
1 YSHVLTPYMNSVPAIIAKASAIHNPIIYAITHPKYR 2 | |||
1 MAIAQHLPCLGVLLGMSGQHTGPYTSYRSTHRSTLSSQASDLSWISRRRRQASLGSESEM 0 | |||
0 GWTDTEAAATWGTAQQVSGPSLWGQDLEDVEAKAPPKPQGREAEAPRK 0 | |||
0 TKELLPSLDPRM* 0 | |||
>MEL1_eriEur Erinaceus europaeus (hedgehog) Deut.Euth.Laur genomic full G? OPN4 melanopsin | |||
0 MALSLGPRVPTSQALDPSCMDTPASPSRWDSSQNSTSSLAQLPLISLT 0 | |||
0 SVSPQATASAPFPTVNVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 | |||
1 SRSLRTPANMFIINLAVSDFLMSFTQTPVFFASSLYKQWLFGEA 1 | |||
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRVALVLLGVWLYSLAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMRFTPSFRAYTMLLFCFVFFLPLLVIIYCYIFIFRAIRETGQ 2 | |||
1 ALQTFRACEGSCDFPRQQQRLQSEWKMAKIILLVILLFVLSWAPYSTVALMAFAG 2 | |||
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 MAIAQHLPCLRVLLGVSGQRDRPYTSYRSTHRSTLSSQISDLSWVSRRRRQASLGSESEV 0 | |||
0 GWTDTEVAAVWGT MSGHFPCGQGLDDMEAKAAHNPRGLEAETPGK 0 | |||
0 IKGLLPSLDPQM* 0 | |||
>MEL1_loxAfr Loxodonta africana (elephant) Deut.Euth.Afro genomic frag G? OPN4 melanopsin | |||
0 MNPPWGPRVPSGRAQEPSCVATPASASRWNSSRASASSLGELPPSSPT 0 | |||
0 AARAHTAAWDPFPTVDVPDHAHYTLGAVILLVGLMGMLGNLMVIYIFFR 2 | |||
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKRWLFGEA 1 | |||
2 GCKFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRAALVLLGIWLYALAWSLPPFFGW 1 | |||
2 SAYVPDGLLTSC 2 | |||
1 ALQTFGACEGASEPPRQWQRLQSEWKMAKIALLAILLYVLSWAPYSTVALVAFAG 2 | |||
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 MAIAQHLPCLGVMLGVSGQRTRPYTSYHSTLHSTLSSQASDLSWISGRRRQASLGSESEV 0 | |||
0 GWTDTEAAAAWEGAQQVSGQASCSQALQNLEANTPPRPQGWGPETPRK 0 | |||
0 * 0 | |||
>MEL1_proCap Procavia capensis (rock_hyrax) Deut.Euth.Afro genomic full G? OPN4 melanopsin | |||
0 MNPPWGPRVPSRPAQEPSCMSTPASAGRWDSSQATASSLAELPPSSPT 0 | |||
0 EARTQTADWVPFPTVDVPDYAHYTLGTVILLVGLTGVLGNLMVIYIFFR 2 | |||
1 SRGLRTPANMFIINLAISDFLMSLTQAPVFFASSLYKRWLFGEA 1 | |||
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRTALVLLGTWLYALAWSLPPFFGW 1 | |||
2 SAYVPDGLLTSCSWDYKSFMPSARTYTMLLCCFVFFLPLLVIIYCYVFIFKAIRETGR 2 | |||
1 ALQTFGACEGASETPRQWQRLQSEWKMAKIALLAILLYVLSWAPYSTVALVGFAG 2 | |||
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 MAIAQHLPCLGVLLGVSDQHTRPYTSYRSTHHSTLSSQASDISWISGRRRQASLGSESEV 0 | |||
0 GWTDTEAAAAWEGAQQVSGRASCSQVLESMEANTPPRPQGWGPETPRK 0 | |||
0 VKGLPLLDPRA* 0 | |||
>MEL1_echTel Echinops telfairi (hedgehog) Deut.Euth.Laur genomic frag G? OPN4 melanopsin | |||
0 MNLPSGSRVPSGPAQEPSHVATAASASRWNS GSSLDAPPPSSPT 0 | |||
0 AARAPTAASAPFPIVDVPDHVHYTLGTVILLVGLTGMLGNLMVIYTFCR 2 | |||
1 SRSLRTPANMLIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGEA 1 | |||
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRAALVLLVIWLYALAWSLPPFFGW 1 | |||
2 SAYVPDGLLTSCSWDYMSFTPSVRAYTMLLFCFVFFLPLLVIIYCYIFIFRAIRETGR 2 | |||
1 ALQTFGACEGSRDSPRQRQRLQSEWKMAKIALLVISLFVLSWAPYSTVVLVAFAG 2 | |||
1 2 | |||
1 0 | |||
0 GWTDTEVTATSGYTQQVSGRCPRGQDLESMDANPTPRPRGWETETAQK 0 | |||
0 IKGLPLSLNPQA* 0 | |||
>MEL1_monDom Monodelphis domestica (opossum) Deut.Meta.Dide genomic full Gq OPN4 melanopsin syn(-GRID1 -WAPAL +LDB3 +BMPR1A) | |||
0 MNPSPMLRGLSCPAQDTNCTKIMASMSEWNNTEEDAYHLVDLPSIAPT 0 | |||
0 AVVLPPSSQNIFPTVDVPDHAHYTIGAIILAVGITGMLGNFLVIYTFCR 2 | |||
1 SHSLRTPANMFIINLAISDFFMSFTQAPVFFASSMYKRWIFGEK 1 | |||
2 ACEFYAFCGALFGITSMITLMAIALDRYFVITRPLASIGVISKKKTGFILLGVWLYSLAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYTTFTPSVRAYTMLLFCFVFFIPLIVIIYCYIFIFRAIQDTNK 2 | |||
1 AVHSIGSGESTASPRHCQRMKNEWKMAKIALVVILLYVLSWAPYSTVALVAFAG 2 | |||
1 YSHILTPYMNSVPAIIAKASAIHNPIIYAISHPKYR 2 | |||
1 MAIAQNFPCLRALLCVRHPRTRSFSSYRFTRRSTMTSQASDISWLPRGRRQLSLGSESEI 0 | |||
0 GWNNMEAGTTSLTSRNQQGSCRMDQETMETRELAAIAKAKGRSWETLEK 0 | |||
0 TLEEMDDSSLLEVSVDMEQ* 0 | |||
>MEL1_smiCra Sminthopsis crassicaudata (dunnart) Deut.Meta.Dasy DQ383281 17785267 full G? OPN4 melanopsin | |||
0 MNPSPMLRHLSCPAQDSNCTKIMASISEWNNTEVDAYHLVDLPPITPT 0 | |||
0 AVVLPPYSQKVFPTADVPDYAHYTIGATILVVGFTGVLGNLLVIYTFCR 2 | |||
1 SRSLRTPANMFIINLAISDFFMSFTQAPVFFASSLYERWIFGEK 2 | |||
1 GCEFYAFCGALFGITSMITLMVIALDRYFVITRPLASIGMISKKKTGLILLGVWLYSLAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYTTFTPSVRAYTILLFCFVFFIPLTVIIYCYIFIFRAIKDTNK 2 | |||
1 AVQNIGSSEHTPSLRHFQRMKNEWKMAKIALVVILLFVLSWAPYSTVALVAFAG 2 | |||
1 YSHVLTPYMNSVPAIIAKASAIHNPIIYAISHPKYR 2 | |||
1 MAIAQNFPCLRAVLGIRHPRTQSFSSYRFTHRSTTASQASDISWQSRGRRQLSLGSESEA 0 | |||
0 GWNNIETGLTLRSLEGSCGMDEETMDTRELSASTKAKGQSWETLAKTLEE 0 | |||
0 MDDLSLLEAGTLLSSLDLQI* 0 | |||
>MEL1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono genomic frag Gq OPN4 melanopsin | |||
0 0 | |||
0 FPTADVPDHAHYTIGATILAVGFTGVLGNLLVIYTFCR 2 | |||
1 SRSLRTPANMFIINLSISDFFMSLTQAPVFFASSLHKRWIFGEK 1 | |||
2 GCQLYAFCGALFGITSMITLTVIALDRYFVITRPLASIGVISKKRALLILTGVWFYSLAWSLPPFFGW 1 | |||
2 sAYVPEGLLTSCSWDYMTFTPPVRAYTMLLFCFVFFIPLIMIIYCYFFIFRAIRGTNK 2 | |||
1 AVETIGSDDCRGSQRQCQRMKNEWKTAKIALMVILLYVISWCPYSVVALVAFAG | |||
1 YSHLLTPYMNSVPAVIAKSSAIHNPIIYAITHPKYR 2 | |||
1 MAITKYIPCLGPLLRVSRQDSRSSSHYASSRRSTVTSQSLDGSWLPGRRRPLSSASDSES 0 | |||
0 0 | |||
0 * 0 | |||
>MEL1_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic frag G? OPN4 melanopsin diverged | |||
0 0 | |||
0 ERTMFNLPDPFPTVDVPTHAHYTIGAVILVVGITGTLGNLLVIYVFFR 2 | |||
1 IRGLRTPANMFVINLAVSDFL 1 | |||
2 GCELYAFCGALFGIASMITLTVIALDRYFVITRPLASIGAMSTKKALLILSGVWLYSLAWSLPPFFGW 1 | |||
2 sAYVPEGLLTSCSWDYITFTPSVRAYTMLLFCFVFFIPLIAIIYSYVFIFIAIKNSNR 2 | |||
1 AVQRTNSDNSKEGQKLYQKLKNEWKMAKVALIVILLVISWSPYSVVALVAFAG 2 | |||
1 YSHLLTPYMNSVPAVIAKASVIHNPIIYAIVHPKYR 2 | |||
1 MAIAKFLPCLGSLLRVPRKDSSYPSTRRPTVTSQSSDINGVPRGHRRLSSVSDSES 0 | |||
0 DWTDTEADISSQNSRVASGSISYRIYEDTTETIKVKSKMRSHDSGIFER 0 | |||
0 0 | |||
0 TGEDLNAFGWRREESYSGPSTSSQIPSIIVTFSNVQRTDLPLESSSGALCSRNSSYSWEKDSNS* 0 | |||
>MEL1_galGal Gallus gallus (chicken) Deut.Saur.Arch AY88294 16856781 full Gq OPN4 melanopsin syn(-GRID1 -WAPAL +LDB3 +BMPR1A) short exon 1 | |||
0 MDLPPRAPT 0 | |||
0 KMTVKDVRGAFPTVDVPDHAHYTIGTVILIVGITGTLGNFLVIYAFCR 2 | |||
1 SRTLQKPANIFIINLAVSDFLMSITQSPVFFTNSLHKRWIFGEK 1 | |||
2 GCELYAFCGALFGITSMITLMVIALDRYFVITKPLASVRVMSKKKALIILVGVWLYSLAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFIPLIAIIYSYVFIFEAIKKANK 2 | |||
1 SVQTFGCKHGNRELQKQYHRMKNEWKLAKIALIVILLYVISWSPYSVVALVAFAG 2 | |||
1 YSHVLTPFMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 TAIATYVPCLGFLLRVSPKESRSFSSYPSSRRTTITSQSSETSGLQKGKRRLSSISDSES 0 | |||
0 GCTDTETDITSMISRPASSQVSYEMGEDTTQTSDLGGKPKVKSHDSGIFRK 0 | |||
0 TVVDADEIPMVEINDTEHSATSTCKTSEKCNVEEIQ 0 | |||
0 RSESLSGIGLREGESRHRTSASQIPSIIITYSNVQGVELHSGYSAGFLHPKNKSHKQNKSSNS* 0 | |||
>MEL1_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full G? OPN4 melanopsin short exon 1 -GRID1 +LDB3 synteny | |||
0 MDLLLRAPT 0 | |||
0 KMTVQDVPRAFPTVDVPDHAHYTIGVVILIVGITGTLGNFLVFYAFCR 2 | |||
1 SRSLQTPANILIINLAISDFLMSITQSPVFFTSSLYKHWIFGEK 1 | |||
2 GCELYAFCGALFGITSMITLMVIALDRYFVITKPLASVGVTSKKKALIILVGVWLYSLAWSLPPFFGW 1 | |||
2 sAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFIPLIAIIYSYVSIFEAIKKANK 2 | |||
1 SIQTFGCKRGNREFQKQYQRMKNEWKMAKIALIVILFFVISWSPYSVVALVAFAG 2 | |||
1 YSHVLTPFMNSIPAVIAKASVIHNPIIYAITHPKYR 2 | |||
1 KAIATYVPCLGPLLRVSPKDSRSFSSYHSSRRATISSQSSEISGLQERKRRLSSLSDSES 0 | |||
0 GCTETETDTPSMFSRLARRQISYKTDKDTTQTSDIRAKLTSQDSGNCGK 0 | |||
0 TAVDADDILMVELNVTEYMATPTVRTILLIFGNKN 0 | |||
0 KSESLNSIGQRREFHQGSSSAQIPSITITCSSVQGIELPSRYNSGFLYPKSSSHKQNKKSSS* 0 | |||
>MEL1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur DQ384639 16856781 full Gq OPN4 melanopsin syn(-GRID1 -WAPAL +LDB3 +BMPR1A) | |||
0 MNYQSVRKGITCPPQDANCSRILESLNSWNNSEVNSYKLVELPPIVTT 0 | |||
0 ETPQYEIHHVYPTVDVPDHVHYVVGAVILAVGITGMLGNFLVIYAFCR 2 | |||
1 SRSLRSPANMFIINLAITDFLMSVTQAPVFFATSLHKRWIFGEK 1 | |||
2 GCELYAFCGALFGITSMITLMVIAVDRYFVITRPLTSIGVMSKKRAVLILSGVWLYSLAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFCFVFFIPLFIIIYCYIFIFKAIKNTNR 2 | |||
1 AVQKIGTDNNKESHKQYQKMKNEWKMAKIALIVILLYVVSWSPYSTVALLAFAG 2 | |||
1 YASILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 MAIAKYIPCLGSLLRVKRRDSRSYSSYPSSRRSTVTSHCSQSSDVGGHPKLKNHLPSVSDSES 0 | |||
0 GWTDTEADSSVNSRPASRQVSYEMGKDTTETNDLKSKAKLKSHDSGIFEK 0 | |||
0 TSMDADDISLVELGTVDRSSPIM 0 | |||
0 ANKHLNGLGQRKGDSFTRRSPSSRIPSIVVTHSNHQGSPAAVRHNSTLPGIKVSNSQDREKELKRQIEKVKQYVPIVTITSDTENSTGGFSNELLPANTS* 0 | |||
>MEL1a_danRer Danio rerio (zebrafish) Deut.Acti.Otoc AY078161 12487121 full Gq OPN4 melanopsin syn(- +USP54 +LDB3 +BMPR1A) | |||
0 MMSGAAHSVRKGISCPTQDPNCTRIVESLSAWNDSVMSAYRLVDLPPTTTTTTSVA 0 | |||
0 MVEESVYPFPTVDVPDHAHYTIGAVILTVGITGMLGNFLVIYAFSR 2 | |||
1 SRTLRTPANLFIINLAITDFLMCATQAPIFFTTSMHKRWIFGEK 1 | |||
2 GCELYAFCGALFGICSMITLMVIAVDRYFVITRPLASIGVLSQKRALLILLVAWVYSLGWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFIPLIVIIYCYFFIFRSIRTTNE 2 | |||
1 AVGKINGDNKRDSMKRFQRLKNEWKMAKIALIVILMYVISWSPYSTVALTAFAG 2 | |||
1 YSDFLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 LAIAKYIPCLRLLLCVPKRDLHSFHSSLMSTRRSTVTSQSSDMSGRFRRTSTGKSRLSSASDSES 0 | |||
0 GWTDTEADLSSMSSRPASRQVSCDISKDTAEMPDFKPCNSSSFKSKLKSHDSGIFEK 0 | |||
0 SSSDVDDVSVAGIIQPDRTLTN 0 | |||
0 AGDITDVPISRGAIGRIPSIVITSESSSLLPSVRPTYRISRSNVSTVGTNPARRDSRGGVQQGAAHLSNAAETPESGHIDNHRPQYL* 0 | |||
>MEL1b_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic frag Gq OPN4 melanopsin syn(- +USP54 +LDB3 +BMPR1A) | |||
0 QVAMVQDVRHPFPTVDVPDHAHYTIGSVILAVGITGMVGNLLVMYAFCK 2 | |||
1 SRSLRTPANMFIINLAVTDFLMCVTQTPIFFTTSLHKRWIFGEK 1 | |||
2 GCELYAFCGALFGICSMITLMIIAVDRYFVITRPLASIGVMSRKRALLILSAAWAYSMGWSLPPFFGW 1 | |||
2 SGAYVPEGLLTSCSWDYMTFSPSVRAYTMLLFTFVFFIPLFVIIYCYFFIFKAIRETNR 2 | |||
1 AVGKINGEGGPRDSIKKIHRMKNEWKMAKIALIVILLYVISWSPYSCVALTAF 2 | |||
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 SAIAKYIPCLGVLLCVPRRDRFSSSSFISTRRSTLTSQSSETSSNLHRAGKARLSSVSDSES 0 | |||
0 GWTDTEADLSTASSRPASRQVSSEIRKDLCDIKHSSSLRLKVKSRDSGIFDR 0 | |||
0 0 | |||
0 QNDVSEKADEKRPLVRIPSIIVTSETCPAVLPAGHSSRLIPGAPAVTDS* 0 | |||
>MEL1_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? OPN4 melanopsin first exon uncertain | |||
0 MGEVGVTVGPACSAAAAYQSGVSCGGELEPSGSAVVMTSLRLAELQLPAST 0 | |||
0 LQVAMARPFPTVDVPDHAHYTIGSVILAIGITGMIGNFLVIYAFCR 2 | |||
1 SRSLRTPANIFIINLAVADLLMCVTQTPIFFTTSMHKRWIFGEK 1 | |||
2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRAFIILMTVWVYSLGWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIFCYIFIFRAIRSTNK 2 | |||
1 AVGKVNGSVHSHSRRQESLKKLQRLQSEWRMAKVALIVILLYVISWSPYSCVALTAFAG 2 | |||
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 LALARYIPCLGLLLCVSPQELRSTSSSFMSMRRSTVTSQTSDISGQFRRQSKPRLSTASDSES 0 | |||
0 CLTDTEADLTSTASRPASRQVSCDIGRDTADLPEFQPAASFKSKAKSPDSGVFEK 0 | |||
0 TSFDFDASMAAPRQHSSIPN 0 | |||
0 NGDFSEGHVTRVALGRIPSIVITSESSFLPHGRKGSASTSAQTATSSDGKAGLG* 0 | |||
>MEL1_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gq OPN4 melanopsin syn(- +USP54 +LDB3 +BMPR1A) | |||
0 MNFGKSALQPPAQQSVVSCGGGGPEPNCTLRLAVTVMMSVRLAELQLHAST 0 | |||
0 LQVAMVRPFPTVDVPDHAHYTIGSVILVIGITGMIGNFLVIYAFCR 2 | |||
1 SRSLRTPANMFIINLAVTDLLMCVTQTPIFFTTSMYKRWIFGEK 1 | |||
2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRAFVILMTVWIYSLGWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCTWDYMTFSPSVRAYTMLLFIFVFFLPLFIIIYCYFFIFRAIRATNK 2 | |||
1 AVGKVNGSVHSHSRRRESVKNFQRLQNEWKMAKIALMVILLYVISWSPYSCVALTAFAG 2 | |||
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 LALAKYIPCLGFLLCISPHELQSTSSSFMSLRRSTVTSQTSDISGQFRPQSKPRRSSASDSES 0 | |||
0 CLTDTEADLSSMGSRPASRQVSCDISRDTTELPEYKPASSFNSKVKSPDSGIFEK 0 | |||
0 TSFDFDASMAASRERSSIPN 0 | |||
0 SGEFPEGHVMRRTLARIPSIIITSESSHFLPNGRKASSTTCIANGSDIKVGPR* 0 | |||
>MEL1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gq OPN4 melanopsin syn(- - +LDB3 +BMPR1A) | |||
0 MNAGESELLLPTQQSILPCGDHEPNCPVAQAETLALSAASANGSA 0 | |||
0 VQVAMVSRAPHPYPTVDVPDHAHYTIGSVILAIGITGIIGNVLVIYAFSK 2 | |||
1 SRSLRTPANMFIINLAITDLLMCVTQAPIFFTTSMHKRWIFGEK 1 | |||
2 GCELYAFCGALFGICSMITLTVIALDRYFVITRPLTSIGMMSRRRALLILMGAWTYSLGWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIYCYFFIFRAIRVTNR 2 | |||
1 AVGKMNGSIHSHGSGRDSTKNFHRLQNEWKMAKIALIVILLYVVSWSPYSAVALTAFAG 2 | |||
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 IALAKYIPFLGVLLCVPPRELRSASSSFRSTRRSTVTSQTSDVSSQQRRQGSRNSRLSSASDSES 0 | |||
0 CLTDTEADGSSVGSRPASRQVSCDIGRDTAELPEFKPSSSFKSKMKSHDSGIFEK 0 | |||
0 SYDTDISMAGVSERGSIPN 0 | |||
0 QTDFAEGRDRRSTIGRIPSIVITSETSPFLPTGRNGSCNGRPKTANSSHPGAGSG* 0 | |||
>MEL1_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full Gq OPN4 melanopsin syn(- +USP54 +LDB3 +BMPR1A) | |||
0 LQVAMVPQTFHPFPTVDVPDHAHYTIGSVILAIGITGIIGNFLVIYAFSR 2 | |||
1 SRSLRTPANMFIINLAITDLLMCVTQSPIFFTTSMHKRWIFGEK 1 | |||
2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRALLILSAAWAYSLGWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIYCYVFIFRAIRSTNR 2 | |||
1 AVGKINGNTRDAVKSFNRLQNEWKMAKIALIVILLYVISWSPYSTVALTAFAG 2 | |||
1 YADMLTPYMNSIPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 MALAKYIPGLGVLLCIHPKDLRSASSSFVSTRRSTVTSQSSDISSQLRRQSTFKSRLSSLSDSES 0 | |||
0 GLTDTEADLSSLSSRPASRQVSCEISRDTAELPDFKHTSSFKAKLKNNDSGIFEK 0 | |||
0 TSFDTVSIGGVSEHNSIPS 0 | |||
0 NRDFGDGNVTRATIGRIPSIVVTSEMSPFLPVGRNGSRTNRSKMANSSAGAGPV* 0 | |||
>MEL1a_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gq OPN4 melanopsin | |||
0 ASVTDAQHHHMFPTVDVPDHAHYIIGATILAVGVTGMVGNFLVIYAFLR 2 | |||
1 SRSLRTPANTFIINLAATDFLMSVTQSPIFFITSIHKRWIFGEK 1 | |||
2 GCELYAFCGALFGITSMITLMVIALDRYFVITRPLASIGVLSHRRAGLIILSLWLYSLAWSLPPFFGW 1 | |||
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFCFVFFIPLGVIIYCYIFIFRAIKSTNK 2 | |||
1 KVGGSTNRESQKQHQRMKNEWKMAKIALIVILLFVISWSPYSTVALTAFAG 2 | |||
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 | |||
1 MAIAKYVPLLGLLLRVSRRDSRTSGQYYSTRRSTLTSQTSDLSGYPRGKGRLSSASDSES 0 | |||
>MEL1b_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gq OPN4 melanopsin based on Squalus | |||
1 SKSLRTPANMFIINLAISDFFMSATQPPVFFVTSLHKRWIFGEK | |||
2 GCKLYAFCGALFGITSMITLMAISIDRYWVITKPLQSISSTTTKKNTLKVIILVWLYSLAWSLPPLLGW 1 | |||
>MEL1_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag G? OPN4 melanopsin first exon uncertain | |||
0 MAPSIVPLMRKLSCPLSRKTPPKHNVSTSMFDGGDGVLGVIGSIP 0 | |||
0 VSFLPSQREKFPMLDIPHHVHHTIGSVVIAIGFTGIIGNFLVIYVFCR 2 | |||
0 VQVAMVSRAPHPYPTVDVPDHAHYTIGSVILAIGITGIIGNVLVIYAFSK 2 | |||
1 SKSLRSPANIFIINLAFADFFMSITQTPIFFVTSLHKRWIFGEK 1 | |||
2 GCELYAFCGALFGIASMVTLMVIATDRYLVLTRPLASIGAMSKRRAMYITAAVWFYSLAWSLPPFFGW 1 | |||
2 SAYVPEGLMTSCTWDYVTFTPAVRSYTMLLFCFVFFIPLIVIIFCYVRIFAAIKNTNR 2 | |||
1 AVKTLGDAHDSKESQKQQQRMNAEWKLAKIALIVILLYVVSWSPYSCVALVAWAG 2 | |||
1 YADMLTPYMNSVPAIIAKASAIHNPIVYAITHPKYR 2 | |||
>MEL1_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni AABS01000008 frag G? OPN4 melanopsin transcripts BW447434 BW019524 BW048729 rough seq | |||
0 DRRYPSCYKGEQVTFIYLIPIFLFRTLAFLSSVFVTMTPSAVLPLVTTEARPVHPINDVAMYTFGGLMLTAGTVAVVGNIMVMYTFLRR 0 | |||
1 PLHWFIVQLAVADFFVGLIVLWIGTFSSLFLDTVSLMSGIATYGVLAAATSTSTLGVLFIAVDRHFYILRHRRYKQIMTRLRVGTAIVVACVVPATFFVVVPAFGWN 1 | |||
2 PEYIEEPLIPSCIFDRFTNSLSNRLYIITMCTFVFFIPLVFICYCLYRIFWAVKSSS 0 | |||
2 SLSRAGSRKSQSSGKLSKSNSSRSKRGIQTIEFQILKSAVLLVVLFVSSWMPFTVAAIISIGSNQVSPYVILVSYLFAKASCVHSSFAYITNAHFRATIGLIRCKHTHRA* 0 | |||
>MEL1_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni AABS01000008 frag G? OPN4 melanopsin 0 transcripts rough | |||
FTTLATLAVIPPTMVLNNSTHPIKVSALLAFGSLMLCAGIIAIVGNLVVMYTFLRPLNWFILQLAVADFFVGLIVLWIGVFSSLLL | |||
ETVSLMSGVATYGVLAAATSTSTLGVLFIAVDRHFYILKHRRYKSIMTRVKVGVAIFFACVTPLAFFVAAPAAGWN 1 | |||
DYIAEPLIPSCIFDRFTTSLANQTYIITMCAFVFFVPLLFICYCLVRIYKAVKTSSQTVEFQILKSAVLLMVLFVS | |||
SWMPFTVAAMISVVAEQVDPYVILVSYLFAKASCVHSSFAYITNAHFRATLGVLRCKKRRSV* 0 | |||
>MELx_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XP_002586120 full Gq | |||
0 MDRMSPNLTNTSLLPNRTDRPELTPADVTMQLVFGSMMLVFGLIGVVGNAVALYAFCS 2 | |||
1 TRKLRRPKNYVVANLCLTDLIMCIVYCPVIVISSFSGR 2 | |||
1 IPTDGACTMEGFVVGMASIASVGSL 0 | |||
0 VAIAVERFFSITRPMKSLTILTKRTFLGGVAVVWLYSLILVIPPLLGWGRYVREETKLSCSFDYLSTDDANRSYVIWLVIVAFGLPLLVIAYCYISVFITVKKCTKKRKLMSPHKKSRSEVKTAVNAFIMTTAFCLCWCPY | |||
AVVATMGISGSSVQGTVVFGAALLAKLSVLINPVAYVFSIPSFRKALFGHRKRGYGTSDGLANDSSSEKRRGKKHESDANSATEYLRLTVFYSMYSRTGQLSPWASKRLAGKTKSMLDLTTEYGRLERTAHKESWV* 0 | |||
>MEL_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran AB205400 15936279 full Gq Amphi-mop 12 exons | |||
0 MTELPSFQPPTNSTEEENAVFPTALTEWISE 0 | |||
0 VGNQVGEAALKLLSGEGDGMEVTPTPGCTGNASVCNGTDSGGGVVWDIPPLAHYIVGTAVFCVGCCGMFGNAVVVYSFIK 2 | |||
1 SKGLRTPANFFIINLALSDFLMNLTNMPIFAVNSAFQRWLLSDF 1 | |||
2 ACELYGFAGGLFGCLSINTLMAISMDRYLVITKPFLVMRIVTKQR 0 | |||
0 VMFAILLLWIWSLVWALPPLFGWSAYVPEGF 1 | |||
2 GTSCTFDYMTPKLSYHIFTYIIFFTMYFIPMGVIIYCYYNIFATVKSGDKQFGKAVKEMAHEDVKNK 0 | |||
0 AQQERQRKNEIKTAKIAFIVITLFLSAWTPYAVVSALGTLGYQDLVTPYLQSIPAVFAKSSAVYNPI 1 | |||
2 VYAITHPKFRAAVKKHIPCLSGCLPADEEETKTKTRGATTTASMSMTQTTAPTV 0 | |||
0 HDPQASVHSGSSVSVDDSSGVSRQDTMMVK 0 | |||
0 VEVDNRMEKAGGGAADTAPKDGTSVPTVSAQIEVRPSGNVNTKAEVIPSPQSAAVAHGASASPVPK 0 | |||
0 VAELSSSVSLESAAIPGKIPTPLPSQPIAAPIERHMAAMADDPPPKPRGVATTVNVRRSESGYERSQDSLRKK 0 | |||
0 AVSETRSRSFNSTKDHFASERQTSTTLNQPRDMYSGDMVKKTRQSPEKQEYDNPAFDAGIAEIDTDSENETEGSYDMLSVRFQAMAEEPPVETYRKASDMSINLGKASLMLTEAHDETVL* 0 | |||
>MEL_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB205400 15936279 full Gq Amphiop6 | |||
0 MTEIPSFQPPINATEVEEENAVFPTALTEWFSE 0 | |||
0 VGNQVGEVALKLLSGEGDGMEVTPTPGCTGNGSVCNGTDSGGVVWDIPPLAHYIVGTAVFCIGCCGMFGNAVVVYSFIK 2 | |||
1 SKGLRTPANFFIINLALSDFLMNLTNMPIFAVNSAFQRWLLSDF 1 | |||
2 ACELYGFAGGLFGCLSINTLMAISMDRYLVITKPFLVMRIVTKQR 0 | |||
0 VMFAILLLWIWSLVWALPPLFGWSAYVSEGF 1 | |||
2 GTSCTFDYMTPKLSYHIFTYIIFFTMYFIPGGVMIYCYYNIFATVKSGDKQFGKAVKEMAHEDVKNK 0 | |||
0 AQQERQRKNEIKTAKIAFIVISLFMSAWTPYAVVSALGTLGYQDLVTPYLQSIPAMFAKSSAVYSPI 1 | |||
2 VYAITYPKFREAVKKHIPCLSGCLPASEEETKTKTRGQSSASASMSMTQTTAPV 0 | |||
0 HDPQASVDSGSSVSVDDSSGVSRQDTMMVK 0 | |||
0 VEVDKRMEKAGGGAADAAPQEGASVSTVSAQIEVRPSGKVTTKADVISTPQTAHGLSASPVPK 0 | |||
0 VAELGSSATLESAAIPGKIPTPLPSQPIAAPIERHMAAMADEPPPKPRGVATTVNVRRTESGYDRSQDSQRKK 0 | |||
0 VVGDTHRSRSFNTTKDHFASEQPAALIQPKELYSDDTTKKMARQSSEKHEYDNPAFDEGITEVDTDSENETEGSYDMLSVRFQAMAEEPPVETYRKASDLAINLGKASLMLSEAHDETVL* 0 | |||
>MEL6_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran genomic 12435605 full Gq Amphiop6 | |||
0 MSPNLTNTSLLPNRTDRPELSPADVTMQLVFGSMMLVFGLIGVVGNAVALYAFCR 2 | |||
1 SRSLRRPKNYLIANLCLTDMVVCLVYSPIIVTRSLSHG 2 | |||
1 LPSKESCIVEGFVVGLGSIVSICSLAGIAVERYVTITQPIKSLSILTHRALLGAVSAVWVYAFLLAFPPLVGWGRYVSEESKISCTFDYLSTDDATRAHVIVLVIGAFGLPFSVITYCYV | |||
RSFATVRKCTKERKQMSPLAKSDSRSEVKAAVNSFVITTSFCLCWCPYAVVATMGVSGFTVHSHAVFIAALLAKLSVLFNPVAYVLSIPSFRKALFSSSNDRTKYQTAFTFESLAKTSPV | |||
ERKWCADSIERYRSSNVNIESTELTVPYSASRESCLLSRAATERLAGRSPSLTDIVREFGLQQTASHRETWV* 0 | |||
>MEL6_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050611 12435605 full Gq | |||
0 MSSNLTNVSLVANRTDQTELSPTDVTMQLIFGSMMLVFGLIGVVGNVVALYAFCR 2 | |||
1 TRSLRRPKNYVVANLCLTDMFVCLVYCPIVVSRSFSHG 2 | |||
1 FPSKESCIVEGFMVGVGSIASICSLAAIAVERYLSVTQPLKSLTILTQRKLLVAVLTVWVYSLLLAFPPLVGWGRYVREETYISCTFDYLSTDDATRAYVITLVMGAFGFPLLTIAYCY | |||
IRVFTTARKHAEERKFMSPLKRPESRTEIKTAVTACVITTSFCLCWCPYAVVATLGISGVSVQQQTVFSAALLAKLTVIINPIVYVLSIPNFRKALFAQEREKYASEDVVLTSLPGKTRRMK | |||
KVERSQSSNSNVVIEVKESSMAYSTSRESCLLSRAATKRLAGKTKSIVDLVDEFGLQETAPHKESLV* 0 | |||
>MEL2_galGal Gallus gallus (chicken) Deut.Saur.Arch AY882944 17977531 full Gq syn(+GRID2+SMARCAD1 -PGDS -SEC24B +COL25A1) | |||
0 MGTQPHSVTKSEIPDHVLYTVGTCVLVIGSIGIIGNLLVLYAFYS 2 | |||
1 NKKLRTPQNFFIMNLAVSDFLMSASQAPICFVNSLHREWILGDI 1 | |||
2 GCDLYAFCGALFGITSMMTLLAISVDRYLVITKPLRSIQWTSKKRTIQIIAAVWLYSLGWSVAPLLGW 1 | |||
2 SSYVPEGLMISCTWDYVTYSPANRSYTMILCCCVFFIPLIIILHCYLFMFLAIRSTGR 2 | |||
1 DVQKLGSCSRKSFLSQSMKNEWKLAKIAFVVIIVYVLSWSPYACVTLIAWAG 2 | |||
1 RGNTLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYR 2 | |||
1 KTIHNAVPCLRFLIRISKNDLLRGSINESSFRTSLSSHQSLAGRTKNTCVSSVSTGEA 0 | |||
0 NWSDVELDTVEPAHEKLQPRRSHSFSSSLRQKRDLLPDSYSCSEETEEK 0 | |||
0 VSLSSSYLEKVLGRSAFPSSPVALVTSSLRAASLPVGLNSSSASRGAGSDISQMKTEESHNNGGLDSIVSNTVPQIIIIPTSETNLFQEEPEEEETELFHFHDKKNNLLDLEGLSSSTEFLEAVEKFLS* 0 | |||
>MEL2_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gq syn(+GRID2+ SMARCAD1 -ATOH1 +PDLIM5 +BMPR1B) | |||
0 MGPHHRTKVDVPDHVLYTVGSCVLVIGCIGITGNLLVLYAFYS 2 | |||
1 NKRLRTPPNYFIMNLAVSDFLMSATQAPICFLNSMHKEWVLGDI 1 | |||
2 GCNLYAFCGALFGITSMITLLAISVDRYCVITKPLQSIKRTSKKRTCIIIVFVWLYSLGWSVCPLFGW 1 | |||
2 SSYIPEGLMISCTWDYVTYSPANRSYTMMLCCCVFFIPLVIIFHCYIFMFLAIRSTGR 2 | |||
1 RKSSISHSIKSEWKLAKIAFVAIVVFVLSWSPYACVTLISWAG 2 | |||
1 YARTLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYR 2 | |||
1 RTIRSAVPCLRFLIPISKSDLSTSSMSESSFRASVSSRHSFSYRNKSTYISSISAKET 0 | |||
0 TWCDVELDPVESGHKKLQAYRSNSFSAKGVAEEESGLLLRTNNCNVPARKK 0 | |||
>MEL2_xenLae Xenopus laevis (frog) Deut.Amph.Anur genomic full Gq Xmop syn(SMARCAD1 +PDLIM5 +BMPR1B) | |||
0 MDLGKTVEYGTHRQDAIAQIDVPDQVLYTIGSFILIIGSVGIIGNMLVLYAFYR 2 | |||
1 NKKLRTAPNYFIINLAISDFLMSATQAPVCFLSSLHREWILGDI 1 | |||
2 GCNVYAFCGALFGITSMMTLLAISINRYIVITKPLQSIQWSSKKRTSQIIVLVWMYSLMWSLAPLLGW 1 | |||
2 SSYVPEGLRISCTWDYVTSTMSNRSYTMMLCCCVFFIPLIVISHCYLFMFLAIRSTGR 2 | |||
1 NVQKLGSYGRQSFLSQSMKNEWKMAKIAFVIIIVFVLSWSPYACVTLIAWAG 2 | |||
1 HGKSLTPYSKTVPAVIAKASAIYNPIIYGIIHPKYR 2 | |||
1 ETIHKTVPCLRFLIREPKKDIFESSVRGSIYGRQSASRKKNSFISTVSTAET 0 | |||
0 VSSHIWDNTPNGHWDRKSLSQTMSNLCSPLLQDPNSSHTLEQTLTWPDDPSPKEILLPSSLKSVTYPIGLESIVKDEHTNNSCVR | |||
NHRVDKSGGLDWIINATLPRIVIIPTSESNISETKEEHDNNSEEKSKRTEEEEDFFNFHVDTSLLNLEGLNSSTDLYEVVERFLS* 0 | |||
>MEL2_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full Gq syn(- +FLJ39155 +PDLIM5 -) | |||
0 MEPQRQIYKRLDVPDHVHYIIAFLILIIGTLGVSGNALVMFAFYR 2 | |||
1 NKKLRSLPNYFIMNLAVSDFLMAITQSPIFFINCLYKEWMFGEL 1 | |||
2 GCKIYAFCGALFGITSMINLLAISIDRYLVITKPLQTIQWNSKRRTGLAILCIWLYSLAWSLAPLIGW 1 | |||
2 GSYIPEGLMTSCTWDYVSPSPANKSYTMMLCCFVFFIPLSIILYCYLFMFLSVRQASR 2 | |||
1 QKSSFVKQQSMRSEWKLAKIAAVVIVVYVLSWAPYACVTLVAWAG 2 | |||
1 HQDVLTPYSKTLPAVLAKSSAIYNPFIYAIIHNKYR 2 | |||
1 RTLAEKVPGLSCLSRSQKDGLSSSTNSDASAQDSSVSRQSSVSKNRLHSTMVQ* 0 | |||
>MEL2_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full Gq syn(- - - +BMPR1B) | |||
0 MEPKDTHITSSFFSKVDVPDHVHYIIAFFVFVIGILGITGNVLVIFAFYS 2 | |||
1 NKKLRSLPNYFIVNLAVSDLLMASTQSPIFFINLYKEWMFGET 1 | |||
2 ACKMYAFCGALFGITSMINLLAISVDRYVVITKPLQTIRRSSKRRTALAILMVWLYSLAWSLAPLVGW 1 | |||
2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLAIILCCYLLMFLAIRKTSR 2 | |||
1 RKSTLIQQKSIRSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2 | |||
1 YGSTLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYR 2 | |||
1 KTIRSAVPCLRFLIPISKSDLSTSSMSDSSFRSALSCRHSYRSRSTYISSISAKET 0 | |||
>MEL2_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gq | |||
0 MEPEDAHSPGSFINKVDVPDHVHYIVAFFVFVIGVLGMTGNVLVIFAFYS 2 | |||
1 NKKLRSLPNYFIVNLAVSDFLMASTQSPIFFINCLYKEWVFGEM 1 | |||
2 CKMYAFCGALFGITSMINLLAISIDRYVVITKPLQTIHWSSKRRTTLAILMVWLYSLAWSLAPLVGW 1 | |||
2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLAIILSCYLLMFLAVRKTSR 2 | |||
1 RKSTLIQQKSIRSEWKLAKIAFVVIIVYVLSWSPYACVTLISWAG 2 | |||
1 HANILTPYSKAVPAIIAKASTIYNPFIYAIIHNKYR 2 | |||
1 MTLVEKFACLWFLSPTARKDCMSSSSISESSIRDSIISRQSTTSRSHFIAGSTRMVNVKSEMSLILNGP* 0 | |||
>MEL2_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gq syn(KNTC2 +FLJ39155 +PDLIM5 +BMPR1B) | |||
0 MEPDNAHTQRSFINKVDVPDHAHYIVAVFVVVIGTLGITGNALVMLAVYS 2 | |||
1 NKKLRNLPNYFIMNLAVSDFLMAFTQSPIFFINCLYKEWAFGET 1 | |||
2 GCKIYAFCGALFGIASMINLLAISIDRYLVITKPLQAIHWGSKRRTTLAILLVWLYSLAWSLAPLVGW 1 | |||
2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLGIILYCYLFMFLAIRKTSR 2 | |||
1 RKSTLIKQKSMKSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2 | |||
1 HADILSPYSKAVPAIIAKASAIYNPFIYAIIHNKYR 2 | |||
1 MTLAAKFPCLRFLSPTPRKDTSSSISESSYRDSVISRQSTASRTHFITACPDTVNFFFSFSSRLGCH* 0 | |||
>MEL2_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full Gq | |||
0 MDPESTQGSLLSKVNVPDHAHYIVAMFVFVIGALGITGNALVMFAFYS 2 | |||
1 NKKLRNLPNYFIMNLAISDFLMAFTQSPIFFINCLYREWMFGET 1 | |||
2 GCKVYAFCGALFGISSMINLLAISVERYLVITKPLQTIHWSSKRKTTLAIFLVWFYSLAWSLAPLVGW 1 | |||
2 SSYIPEGLMTSCTWDYVTYTPANRSYTMMLCCFVFFIPLGIILYCYLRMFLSIRKTTr 2 | |||
1 RKSTVIQQKTIRSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2 | |||
1 HADILSPYSKAVPAVIAKASTIYNPIIYAIIHNKYR 2 | |||
1 MTLAEKVSCLRFLSPNPPKECTSYSFSESSFRDSIISRQSSVSRSHFINTLPSRADMVNDHKLVFI* 0 | |||
>MEL1_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XR_026330 frag G? opsin4 no cdna losing introns larval postoral arm ancestral intron | |||
0 2 | |||
1 WTKSLRTPPNMLIVNLAISDFGMVITNFPLMFASTIYNRWLFGDA 1 | |||
2 GCQFYAFCGALFGIMSIANMTAIALDR 2 | |||
1 YYVICWSLEAVRSVTHRRSMIIIIIVWCYAIFWSIPPFFGVGSYVLEGYGLGCTFDFMTKDLNHYLHV | |||
SFLFASSFVVPVTIIIVCFTRIAITVRAHRHELNKMRTKLTEDKDKKHKSSIRRANKAKTEFQIAKVGFQVTIFYVLSWM | |||
PYSIVAVIGQYFDSDLLTPLGTVVPVIFAKCSAIWNPIIYCLSHEKFNAALKEKLMGMCGIEIPSKHRSMGSQESSVTGR | |||
RGMHRQNSSTLSESSVTSTVDQDAIELKDRKQGPATVKVQQEKVEGGTYRRNPGDVTFSKDAGVEVDEKRRGDQGQRDDR | |||
VRPQGEGQMDQWSQPPPAPASASAPTPGVNDKEYLTKM* 0 | |||
>MEL2_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu DQ285097 full G? cdna: S droebachiensis retained intron: tube feet | |||
0 MPTTLMENSTPGWMADDSQMEETHPAFPLIGGYLLVVVLLGTAGNSLVIYTFLRFKKLHSPINLLIVNLSASDLLVATTG | |||
TPLSMVSSFYGRWLFGTNACAFYGFVNYYCGCISLNSLAAISVFRYIIVVRGQAQNNKLSLRSSIYAILVIHLYTLIFST | |||
PPLYGWNRFVLAGYHTSCDIDFHTKTPLFVSYICYMFFFLFFLPLGLISWSYFKIYQRVSKHSNSMRTSFTGVTKEINSDEKHA 2 | |||
1 NHRRTASTLFVTIVVFLFAWFPYCIVSLWVLIGDANSISKLSTTIPSLFAKSSVIYNPLIYVVLNSKFRKALIQTLSFLKCLSKHELSESS* 0</font> | |||
<font color ="brown">>MEL1_plaDum Platynereis dumerilii (ragworm) Ecdy.Anne.Poly AJ316544 11874910 full Gq rhabdomeric melanopsin unavailable genomically | |||
0 MSRSEVLVPGSMSLDGLLTTAHPIGNDSI 0 | |||
0 ETILHPYWQQFDIENTIPDSWHYAVAAWMTFFGILGVSGNLLVVWTFLK 2 | |||
1 TKSLRTAPNMLLVNLAIGDMAFSAINGFPLLTISSINKRWVWGKL 1 | |||
2 WRELYAFVGGIFGLMSINTLAWIAIDRFYVITNPLGAAQTMTKKRAFIILTIIWANASLWALAPFFGW 1 | |||
2 GAYIPEGFQTSCTYDYLTQDMNNYTYVLGMYLFGFIFPVAIIFFCYLGIVRAIFAHHA 2 | |||
1 EMMATAKRMGANTGKADADKKSEIQIAKVAAMTIGTFMLSWTPYAVVGVFGMIK 2 | |||
1 PHSEMFIHPLLAEIPVMMAKASARYNPIIYALSHPKFR 2 | |||
1 AEIDKHFPWLLCCCKPKPKAQLPSSTTKGSIASKTEADTSV* 0 | |||
>MEL1_capCap Capitella capitata (polychaete) Ecdy.Anne.Poly genomic full G? jgi 119596 distally uncertain shares 2 introns with melanopsins | |||
0 MMSYADGYLDNSTAPIEESPYLPHGTFFHPHWRPYREMLLNMNPLIYYGLGLYMAVVGIVGTLGNLVVITLFIK 2 | |||
1 TRSLRTPPNMFIINLALSDMGFCATNGFPLMTVASFQKLWRWGPV 1 | |||
2 ACELYALAGSITGFNSIATLALISMDRYMVIAKPFYAMKHVSHKRSLIQIILAWTWAFIWSAPPLLRMGYGRYIP | |||
EGFQVSCTFDYLSRDLKNLIFVWCLFVFGFFIPVLAIACSYVGIIRAVGAQSKEMRKTAEKMGAKTGKSDKEKKQDIAMAKVA | |||
AGTIGLFLMSWTPYAAVSMIGIAGNRSWITPYVSQIPVMFAKASAMWNPILYALSHPKFRAALEDHMPWLLVC* 0 | |||
>MEL1_helRob Helobdella robusta (leech) Ecdy.Anne.Clit genomic full G? | |||
0 MDSVTWYKDFHPHWWKYHDIIVNAPMAFYYFLGTFFAVVGFLGVFGNIIVVWVFSR 2 | |||
1 TPSLRTPSNVLVINLAICDILFSALIGFPMSALSCFQRHWIWGNF 1 | |||
2 YCQFYSFVAGITGLASINCLAVIAVDRYLVVGQPLAMLNQSHFRRSFYHVLIIWTWACVWSAMPLIGWGEYILEGFGVSCTFD | |||
YLTRTTWNISFNVCLFTFCFGMPVSVIILSYIGIIRSIAKNRKEFSSLTAENSSRARQEIKIAKVFAVCMTAFILCWVPYAT | |||
VAQLGIYGYDQMVSPYTAELPVMLAKTSALWNPIIYAFSHPKYRKCLKELPIFRQKRKFRFLGSKSHTKSSDGVGTIALTSTINRTATRH* 0 | |||
>MEL2_helRob Helobdella robusta (leech) Ecdy.Anne.Clit genomic frag G? from scaffold_391 | |||
1 TPILRTHANVLIINLALCDLIFSSLIGFPMTALSCFKRHWIWGDL 1 | |||
2 GCDFYGFVAGWTGLGSITCLAFISIDRYMAIVHPFYMLNKKSSSVLTLLQIGAVWSWALIWSVMPLFGWGRFIPEAFGVSCTFDYLTRTWSNICFNYVLITCGFFLPVVIIVTSYIGIVIEVTKS 1 | |||
2 VEEDGMKDHMDAQNARCYVFVAVLSM 0 | |||
0 LKTAKVLACCFGAFLICWTPYAIVAQLGINGFAHLVTPFTSEVPVLFAKTSSIWNPLIYALSHPRYRRAV 0 | |||
>MEL1_schMed Schmidtea mediterranea (planaria) Loph.Plat.Turb AF112361 10220416 frag G? | |||
0 eVYHYLVGVYISIVGISGVLGNLLVLYIFAR 2 | |||
1 AKSLRTPPNMFIMSLAIGDLTFSAVNGFPLLTISSFNTRWAWGKL 1 | |||
2 TCEIYGFIGGLFGFISINTMALISLDRYFVIAQPFQTMKSLTIKRAIIMLVFVWLYSLIWSTPPFFGY 1 | |||
2 GNYVPEGFQTSCTFDYLTQSKGNIIFNIGMYIGNFIIPVGIIIFCYYQIVKAVRVHELEMLKMAQKMNASHPTSMKTG 1 | |||
2 AKKADVQAAKISVIIVFLYMLSWTPYAIIALMALTGRRDHLNPYTAELPVLFAKTSAMYNPFIYAINHPKFRIQLEKKFPCLICCCPPKPK 0 | |||
0 * 0 | |||
>MEL1_schMan Schistosoma mansoni (trematode_worm) Loph.Plat.Trem AF155134 11166392 full G? 6 exons | |||
0 MKQNLTFATLWPDDNDFASIVHSHWHKFIQPDPLYYYLVGIYIGIVGILAVMGNSLVITLFLL 2 | |||
1 CKQLRTPPNMLIVSLAISDFSFALINGFPLKTIAAFNHRWGWGKL 1 | |||
2 ACELYGFAGSIFGFISLTTMAFIALDRYLVIVQPFETFSRITYGKVIVMIFITWIWSALWSIPPFFGY 1 | |||
2 GSYIPEGFHTSCTFDYLSTDLPNLIFNAGLYILGFLCPVFIIIFSYYQIVKTVRLNELELMKMAQSLDLQNPSAMKTg 1 | |||
2 GDKKADIEAAKTSIILVLLYLMSWSPYAIVCLMTLIGSRDSLTPFHSELPVLFAKTSAVYNPIVYAVKHPKFRMEIEKRFPFLICCCPPKPK 0 | |||
0 ERLQNTIVSKIQVSQIGIGTVSGGNENTLNTVKRED* 0 | |||
>MEL2_schMan Schistosoma mansoni (trematode_worm) Loph.Plat.Trem CD096414 12973350 full G? | |||
0 MSSNRTIEMLRPYMKDFDSIVLPYWYKFEQPNPYYQYAIGLFIAVVGITGMCLNLLVIVFFTM 2 | |||
1 FKSLRTPSNILVVNLAISDFGFSAVIGFPLKTMAAFNNFWPWGKL 1 | |||
2 ACDLYGLAGGLFGFVSLSTIAAVALDRYLVIATPFESVFQTTPRRTLLLMLFLWMWSLMWTIPPLFGF 1 | |||
2 GRYVTEGYQTSCTMDYISTDLNNRLFNIGLFGFGFLCPLFLSLFCYARIILIVRSRGKDFIEMAASSKGTNQKEKSANV 1 | |||
2 SSSKSDTFVSKSSAILLGVYLICWTPYSFVCLMALIGYADYITPLMVEIPCLCAKTANPCIYAFRYPKFRSLLQQRFGFLRLTKNRVSY 0 | |||
0 ERSQHAILSTIHVVTDCQYGTVSGGNENTLNTFILLD* 0 | |||
>MEL3_schMan Schistosoma mansoni (trematode_worm) Loph.Plat.Trem XM_002581128 full G? | |||
0 MSEIKNFTRSLLLYNRTFSMIKNNIHDSDIIMLNHWIKYTQPDPIYNYLVAIFVALIGIFGTITNLLVIFVFL 2 | |||
1 TPKSSISLQCALIINLAISDFGFSAVIGFPLKTIAAFNQYWPWGSV 1 | |||
2 ACQLYGFISATFGFLSLTTIAAISFDRYLVIVKDHKTTNFRVICTVIGFLWIWSIIWTIPPFFGF 1 | |||
2 GRYVLEGYQTSCTFDYISNDMPSLLFSGGMYIFGFMFPVLLCIYCYVNLLKIVRNNERVVLISLSNDGASKQRESVR 1 | |||
2 NRKRLDIEATKSVILSLLFYLMSWTPYAMVCLISILGQSYFLTPTIAEMPHIFAKMAAIYNPILYAFTNRKFKNALGIRKTSSVIMQQQRLLSKGQLKPLVSLLFLVN* 0 | |||
>MEL1_lotGig Lottia gigantea (limpet) Loph.Moll.Gast FC774055 full G? ests long tail omitted 3 exons best: molluscan melanopsins | |||
0 MTTLPPKENTTHLFDTWDDMFVHPHWKNFPPVSAAWHNFIGIFITFVGITGVIGNFVVIYTFSR 2 | |||
1 TKSLRTASNMFVVNLALSDLTFSAVNGFPLFSLSSFSHKWIFGRV 1 | |||
2 ACELYGLIGGIFGLMSINTMAMISIDRYLVITSPFTAMRNMTHKRAFLMIVGVWIWSILWAIPPIFGWGAYIPEGFQTSCTFDYLTRGDNRRSYIMCLYICGFVVPLGVIIFCYVFIIKSVMNHEKE | |||
MAKMADKLDAKDVRSTKEKAKAEIKIAKVSMTIILLYLMSWTPYAIVALIAQWGPALVVTPYVSEIPVLFAKASAMHNPVIYALSHPKFRDAVSKLMPWFLCCCGLTDAEKKARDELAKSKFTRQT* 0 | |||
>MEL1_sepOff Sepia officinalis (octopus) Loph.Moll.Ceph AF000947 900420 full G? similar X56788 Loligo forbesi | |||
0 MGRDIPDNETWWYNPTMEVHPHWKQFNQVPDAVYYSLGIFIGICGIIGCTGNGIVIYLFTK 2 | |||
1 TKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFIKKWVFGMA 1 | |||
2 ACKVYGFIGGIFGLMSIMTMSMISIDRYNVIGRPMAASKKMSHRRAFLMIIFVWMWSTLWSIGPIFGWGAYVLEGVLCNCSFDYITRDSATRSNIVCMYIFAFCFPILIIFF | |||
CYFNIVMAVSNHEKEMAAMAKRLNAKELRKAQAGASAEMKLAKISIVIVTQFLLSWSPYAVVALLAQFGPIEWVTPYAAQLPVMFAKASAIHNPLIYSVSHPKFREAIAENFPWII | |||
TCCQFDEKEVEDDKDAETEIPATEQSGGESADAAQMKEMMAMMQKMQQQQAAYPPQGAYPPQGGYPPQGYPPPPAQGGYPPQGYPPPPQGYPPAQGYPPQGYPPPQGAPPQGAPPQ | |||
AAPPQGVDNQAYQA* 0 | |||
>MEL1_todPac Todarodes pacificus (squid) Loph.Moll.Ceph X70498 11106382 full Gq squid rhodopsin 3D Cys 337 palmitoyled | |||
0 MGRDLRDNETWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTK 2 | |||
1 TKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWIFGFA 1 | |||
2 ACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFF | |||
CYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVL | |||
TCCQFDDKETEDDKDAETEIPAGESSDAAPSADAAQMKEMMAMMQKMQQQQAAYPPQGYAPPPQGYPPQGYPPQGYPPQGYPPQGYPPPPQGAPPQGAPPAAPPQGVDNQAYQA* 0 | |||
>MEL1_entDof Enteroctopus dofleini (octupus) Loph.Moll.Ceph X07797 full Gq | |||
0 MVESTTLVNQTWWYNPTVDIHPHWAKFDPIPDAVYYSVGIFIGVVGIIGILGNGVVIYLFSK 2 | |||
1 TKSLQTPANMFIINLAMSDLSFSAINGFPLKTISAFMKKWIFGKV 1 | |||
2 ACQLYGLLGGIFGFMSINTMAMISIDRYNVIGRPMAASKKMSHRRAFLMIIFVWMWSIVWSVGPVFNWGAYVPEGILTSCSFDYLSTDPSTRSFILCMYFCGFMLPIIIIA | |||
FCYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGASAEMKLAKISMVIITQFMLSWSPYAIIALLAQFGPAEWVTPYAAELPVLFAKASAIHNPIVYSVSHPKFREAIQTTFPWL | |||
LTCCQFDEKECEDANDAEEEVVASERGGESRDAAQMKEMMAMMQKMQAQQAAYQPPPPPQGYPPQGYPPQGAYPPPQGYPPQGYPPQGYPPQGYPPQGAPPQVEAPQGAPPQGVDNQAYQA* 0 | |||
>MEL1_aplCal Aplysia californica (sea_hare) Loph.Moll.Gast AASC01108363 full Gq 4 exons uncertainties | |||
0 MNVSSSLTSQPYHELLHPHWLEHEEAPEGVHLSVGVFITLVGVLAVCGNSLVIITCIR 2 | |||
1 FKDLRTRSNILIINLAVGDLLMCLIDFPLLAAASFYGEWPYGRQ 1 | |||
2 VCQMYAFLTAIAGLVTINTLAVIAADRYWAVVRRPTPGQKLPKCVTSIAVASVWAYSISWALCPILGWGAYVLDGIRTTCTFDFLTRTWENRSFVIGMMIGNFVLPFALMVFSYFRIWVAVRKVKSG 2 | |||
1 NVFCAIRHNYNLALGSTLFVKQHRYRLHCEQKTVKIIMFLLIAFTVSWSPYLAVSIIGLFGDRSQLTYQNTLTASLIAKTSMVFNPILYSISHPKVRKRIANLACCYSVRRHQQQTSRIKTGRRSTSSATPSRS* 0 | |||
>MEL2_aplCal Aplysia californica (sea_hare) Loph.Moll.Gast AASC02005512 frag G? 1st exon missing; new 3rd intron | |||
0 2 | |||
1 HSSLRTSSNLLVVNLTVADLVMSSLDFPILAISSYKGCWVMGFL 1 | |||
2 GCQVYGVSSGVAGLVTINTLAAISVDRFVVVVHRLSPMHQMGKSTT 1 | |||
2 GVIIIAIWALSVIWAVLPITGVSSYRLEGMGTSCTFDYASRTSSNRWFFIALVIFNFFIPLALIIFSYWRIYASVRAVKRELKLLQSARTSILRKRFEVQAEI | |||
KTAITALVIISIFCLAWTPYVIIAFVGLYGPTTAIDPLVSMFPNILAKISTVSNPILYSIGHPEVRKKMKKLFLPGQQDSSWRATSATAGNSSPAQSENMNNISLPTYL* 0 | |||
>MEL2_lotGig Lottia gigantea (limpet) Loph.Moll.Gast genomic full G? | |||
0 MSIASHVWTNSSTNHFNFSVLHQHWQNQTPLSTACQYTIGIFISTVAVIAVIGNSIVIWAHVR 2 | |||
1 IKSLSTTSNMLILNLCVGCLIMCIVDFPLYATSSFLQKWIFGHK 1 | |||
2 VCEIYATITGTAGLLIMNSYSAIAFDRFITVTRYNNPNYPRSKSATMCISGFVWIYSLSWSMAPVVGWSRYQLDGSGTT | |||
CTFDYLSTTWTNRSFILSIAFFNFVLPLCFILFAYSRILHLISSHSREMKSYRSAVIISKGKASIPKRFRSERKTAITLLI | |||
TVVVFCLSWVPYVIIALIGQFGNQSFITPQISVIPQLVAKLSTVTNPILYSLSHPVVRNKLFLRLRHELYRRPSDSVSSSRGIQMKNIEFI* 0 | |||
>MEL1_patYes Patinopecten yessoensis (scallop) Loph.Moll.Gast AB006454 9287291 full Gq retina | |||
0 MADNKSTLPGLPDINGTLNRSMTPNTGWEGPYDMSVHLHWTQFP PVTEEWHYIIGVYITIVGLLGIMGNTTVVYIFSN 2 | |||
1 TKSLRSPSNLFVVNLAVSDLIFSAVNGFPLLTVSSFHQKWIFGSL 1 | |||
2 FCQLYGFVGGVFGLMSINTLTAISIDRYVVITKPLQASQTMTRRKVHLMIVIVWVLSILLSIPPFFGWGAYIPEGFQTSCTFDYLTKTARTRTYIVVLYLFGFLIPLIIIGVC | |||
YVLIIRGVRRHDQKMLTITRSMKTEDARANNKRARSELRISKIAMTVTCLFIISWSPYAIIALIAQFGPAHWITPLVSELPMMLAKSSSMHNPVVYALSHPKFRKALYQRVPWLF | |||
CCCKPKEKADFRTSVCSKRSVTRTESVNSDVSSVISNLSDSTTTLGLTSEGATRANRETSFRRSVSIIKGDEDPCTHPDTFLLAYKEVEVGNLFDMTDDQNRRDSNLHSLYIPTRVQHRPTTQSLGTTPGGVYIVDNGQRVNGLTFNS* 0</font> | |||
<font color ="#996633">>MEL1_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic full G? no close homologs | |||
0 MTSSNDSAGYLWAINATIWIIDDSNETLGIDWDDWDVSLWTQEQRQLLEHGGIPRQVHVALGVLLSFIVLFGFAANSTILYVFSR 2 | |||
1 FKRLRTPANVFIINLTICDFLACCLHPLAVYSAFRGRWSFGQT 1 | |||
2 GCNWYGMGVAFFGLNSIVTLSAIACERYIVITSSSCRPVVAKWRITRRQAQK 0 | |||
0 VCAGIWLHCAALVSPPLLFGWSSYLPEGVLVTCSWDYTSRTLSNRLYYFYLLFFGFFLPVSVLTFCYAAIFRFILRSSKEITRLIMTSDGTTSFSKSTVSFRKRRRQTDVRTALI | |||
ILSLAILCFTAWTPYTIVSLIGQFGPVDEDGELKLSPMVTSIPAFLAKTAIVFDPLVYGFSSPQFRNSVRQILRQQSISSSGNAGNRAGPNNMAMARTAIQNSRASSHATVSSF | |||
SRNARMFPKDPLSKKTPNDPFVSTPLAVQQIPHFRLPTDVDINEQQFRRGIYANKSVSYWIDIIVLLQLGENLRKSCMKRKNSFKIPAGSIPQKNKLSNSRCSLLEDVSTHSLA | |||
LRQMIFRKEGELYLFHHQPSHNAELAANKMDHQGNNKRIRRRFSEADMMHRSGKCRKNLPVSTSFDQ* 0 | |||
>UV7_ixoSca Ixodes scapularis (tick) Ecdy.Chel.Arac genomic frag G? exon 1 missing exon 2 disjunct K90 at EIP XM_002408275 wrong | |||
0 2 | |||
1 RRRIRSQANLLVFNLALSDLLMVLEIPLLVYNSLKLRPALGVW 1 | |||
2 GCQLYGLMGGLSGTSAIFSIAALSLERYLALGRPRDPFARLTRSRAFALSLSSWIYALCFSAWPLLGVTSPYVPEGFLTSCSFHFLSDATSDRCFVWIFFVAAWCVPLVFVTTCYSGILVTVIRSRKALAQES | |||
RRSELRVAKVSLALVLLWTVAWTPYAIVALLGITGRRNLLTPWGSMAPAMFCKSAAVLDPFVYGLSHPSFRRELAIMLPCLRPRQRPVSLTLRAVVQLPKRPGPRSAGSSTSVPVTAPGTTKDNHCPTPPNVSR* 0 | |||
>UV7_tetUrt Tetranychus urticae (spider_mite) Ecdy.Chel.Arac GW013907 frag G? larval transcripts | |||
AGCKIYGFFGGLSGTSAIMTIAMMAMERLNSISKPLSPHSRLSMRRALVISTLIWIYSLIFSTPPVFHLFNRYVPEGYLTSCSFDYLSKDLVSRIYIFAFFTAAWIVPLTLIVTSYVGIIIAVERNETMFQKTQDRLCERAS | |||
SVYRASLRDSRGRSVKESGKMEMRLVKIVLSLIAFWILSWTPYAIIALLGLLDQSYITPTSSMIPAIFAKLSSVVNPYLYGLGNPRFKRELKRKLCFCVANKASVERRTVIQSTVTLGDPKEGTQFVHISDGNLSEIELSDYENR* 0 | |||
>UV7a_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran frag ACJG01004266 tandem1 | |||
0 MLNHTHSISRETIHGNAICDQHRKFVNLYPVDVWKSSGFFDDIDLMDINCHWLQFEPPTPSSHFMLAIIYSVVFVIGFVSNTAIVYILLR 2 | |||
1 SKKLLTPANILLVNLAISDLLIISMIPIFLYNSFLQGPAIGGL 1 | |||
2 GCKVYGFLSGFSGTSTILTLAAIAIDRYLVISRPLDLAKKPTRTWAYATIAITWLYSATFASLPFLGAGKYVPEGYLTSCSFDYLSEDTGTRVFIFIFFVAAWVCPLT IITFCYAAITYVNICSTNTARPLIIIQHKYAGWRQCNIEIAIPKIVAGLVLSWIIAWTPYSLISLLGISGHTDLLTPLSSMLPALFAKTAACVDPFIYSLNHPKIRQEII | |||
>UV7b_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic ACJG01004266 full tandem2 | |||
0 MNAINMCNQTNCATDVVTDDIIERWKLCGFFEDDDFLDINCHWLQFEPAPLVNHLLLVCIFVFILLTGCLCNIIVIYIFAR 2 | |||
1 SRHLRTPANIIIMNLAISDFFMLIKMPIFLYNSLLQGPALGIK 1 | |||
2 GCQIYGFMSGLTGTTSIMSLATVAVDRYLVISQPLNVNRKSTRTRAYLTTCFIWLYSAIFASLPLFGIGKYVPEGYLTSCSFDYLSNNVKTRSFILAFFLAAWLFPFCIITTCY | |||
TAITYVNICSTNTARPLIIIQHKYAGWRQCNIEVVIAKIVVVLVAMWMISWTPYAFVALLGISGTQTRLTPGMTMFPALFAKFSACVNPIIYTLTHPKIKKEILRRWYCFMSSGTS | |||
>UV7a_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD4798 full G? 3 exons altered HEK K90 | |||
0 MIDFKTKYPVNLWKDHGLYTDDYIKLINSHWLKFMPPNPTSHYVLGLLYTVIMVFGCTGNSLVIFMYFK 2 | |||
1 CRSLQTPANMLIINLAVSDFIMLAKASVFIYNSYYLGPALGKL 1 | |||
2 GCQVCGFLGGLTGTVSIMTLAAISLDRYYVIVCPLKAAVKTTKQRARIWIGLIWIYGFSFSIVPVLDLGYSRYVSEGYLTSCSFDYLSDNDQDKRFI | |||
LVFFTAAWCIPFTIILYCYVNILMAVWMTTEIVTSRVGQQEEKRKTDIRLGYMVIGALALWFVSWTPYAVVALLGVFDLKEYISPLSSMIPALFCKAAS | |||
CTDPWFYAITHPRFKKELMKLLTKSKSRKLVRNYGMKKGWVGSHLNKNGSVDFDNCLKTEYKEENTTIFMLESDDNNLHCQGSTSGHKTESTKEPETKFTASASQETLKYMLPS* 0 | |||
>UV7b_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD14504 full G? altered HEK CL3 K in in K90 | |||
0 MSDFKTKYPIDTWKEHGFYTDDYMKLINSHWFKFMPPNATSHYILGFLYSVIMVLGCFGNSLVIFMYIK 2 | |||
1 CKSLQTPANVLIMNLAVSDFIMLAKTPVFIYNSFYQGPTLGKL 1 | |||
2 GCQIYGFFGGLTGTVSIMTLAAISLDRYYVIVHPLNAAVKTTKQRARVWIGLIWIYGFLFSIIPVMDLGYNRYVPEGYLTSCSFDYLSDDNQEKGFILVFFTAAWCIPFTTISYCYIKI | |||
LRAVWMTSEMAASRFGQEEEKRKTEIRLGYVVVGVIMLWFVSWTPYAMVALLGVFDRKDYITPLSSMIPAVLCKAASCMDPWIYAITHPRFKNELTKLMSRKKTRKLERDYGMKKNWGGQ | |||
SYSNKSGAGLRNLSSSEDECVEEVIVVIDPDDKKMKRQGSTSSHKTEETKALETKFPPTRQESLKYMPPSWYKLPRTTSKSSIMLDPKLTGDDNNK* 0 | |||
>UV7_rhoPro Rhodnius prolixus (kissing_bug) Ecdy.Inse.Hemi genomic full G? K90 at KMP ortholog RH7 of droMel | |||
0 mKYFHLYPIEQWKMHRFFTEEYLKLVNTHWFEYPPPNKQIHYIFAAVYFLVMLVGVSGNLLVIFMILR 2 | |||
1 FRTLRTSSNILILNLAVSDFLMVAKMPVFIYNSFYFGPVLGEM 1 | |||
2 GCHFYGFIGGLSGTASILTLAAIAMDRYLGIAHPLNFNQGRAKKRTIVWITFIWVYSITFASIPLSHIGVKTYVPEGFLTSCSFDYLSTDIQNRCFIFIYFVAAWCLPLLVIITSYVGICREVLRVSLIRKGQE | |||
REQRKREAKLSAILALATFLWFLSWTPYAAVALLGIFGYKNHITQLASMIPALFCKTAACVNPFIYGLNHPRLRQQLLKLCCKKRYNLEKTHFSRSWRNTSCSFKLKEQSLCNVSQSRLRRTSTVASEPSEHSTHFM* 0 | |||
>UV7_pedHum Pediculus humanus (louse) Ecdy.Inse.Phth AAZO01007270 full G? | |||
0 mKTFKLKWPEEWKKLGLFDDEYLYKINKYWMKFPPPSPMSHYFMGIIYSVIMVVGVFGNFLIIYLFLR 2 | |||
1 KRSLRTPSNVFIFNLAVSDSLLLLKMPVFIINSFYLGPALGNL 1 | |||
2 GCSAYGFVGGLTGTVSIMTLAAIAFDRYQVIVHPLERKTKAAVYFQILLIWIYAIFFSIIPLLDVGLNKYVPEGYLTSCSFDYLTQDTASRLTIFVFFVAAWIVPLSIILGSYM | |||
ALYKVVLKARGTHFNTVMTRHCKDIEIQRPELKAAVTVICIVCLWTLSWTPYAVVALLGITGNEKYISPMSSMIPALFCKTASCIDPFVYAATNRRFRNELKRKYRKRSRYQPSLKTE | |||
RKDFFTLSEDNNDRGKGNTIRIREK* 0 | |||
>UV7_spoLit Spodoptera littorali (butterfly) Ecdy.Inse.Lepi FQ016398 FQ021555 Cfrag male antenna | |||
MFSTIGCWGNIIVLLMYLRCRTLRTPGNILVANLALSDFLMLAKTPIFIFNSFNLGPALGKTGCVIYGFVGGLTGTTSIATLSAIALDRYWAVVRPLEPL | |||
RALTAIRARLLAIGAWLYACVFAIIPALDIGYGSYVPEGYLTSCSFDYLTEDFSPRCFIFVFFCAAWLAPFCTITFCYISIFRVVVFNTSMSYGNQDQRL | |||
SSRHVKERTKRKAEIKLAFLVMAVIALFFISWTPYAIVALLGITGKKDVLTPITSMIPALFC | |||
>UV7_mayDes Mayetiola destructor (hessian_fly) Ecdy.Inse.Dipt AEGA01002257 frag genomic | |||
1 VYFLHSSYNGYPDTKVLESINIHWIGFNPPKPWEHYTLAIMISIIMTIGLIGNFLVICM 2 | |||
1 SRILKRASNILLLNLAFADFFMLAKSPIYIYNCLKFGPASGDL 1 | |||
2 ACRAYGFIGGLTGTVSIMTLSAISFDRYMGIMYPFNTEKFGNKKIRAFILVAFVWFYSFTFAIMPALNIGLSKYTPEGFLTSCGFDYLDNQTSARIFMFTFFIGAWMIPFLTITYCYIQIFRV | |||
VNLTRQIQSNRKRQRAEQKLAIVVMNLIAIWFFAWTPYAIVALLGIAGQQQYITPWASMIPALFCKASACINPYIYSIRYPEFKQEIFRILSKYFKFADEHLQQNATRTMSVHFVTRTRVQINEQY* 0 | |||
>UV7_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_308329 full G? K90=EAP | |||
0 MGRQGSGNAVRISPSSRNQPYFSSAHLSFVVPFPVHSKYVVRSGYVLPVDPLFVAKINPFWLRFDPPSAGEHYGLAVFYFLMMLFGVIGNALVVFMFYR 2 | |||
1 YRSLRTPANYLVINLAVADFIIMMEAPMFIYNSIHQGPALGSI 1 | |||
2 GCTVYALMGAVGGTVAIATLTVISIDRYNVVVYPLNPNRSTTKLKCYFLIAFTWAYGLLFASFPALEIGLSRYTAEGYLTACSFDYLDRTYKARVFMFVYFVFAW | |||
LIPFAIISYCYARILIAVINANAIQSSKSKNKTEVKLAGVVVGIIGLWFAAWTPYAVVAMMGVFGYEQYLTPLNSMIPAVFAKIAASIDPYFYAMNHPRYRQMLER | |||
MFCNRGADQGNSQYQTSHYTRGASRGGDSEGGGGEESGGGGGVGRAPGGGNAGLGRGGTVRGGGGGGRLIAGKGGGGANATGSTGGGGVKALKKQISNGDETSLEVSLEM* 0 | |||
>UV7_aedAeg Aedes aegypti (mosquito) Ecdy.Inse.Dipt XM_001650694 frag G? wrong exon 1 K90=EAP | |||
0 FPPNSRYMALSGYSGPTIEDAFRDRINPFWLQFDPPSRTAHYILGFIYFMMMMFGLCGNLLVILMFFR 2 | |||
1 FKSLRTPANYLVINLAIADFIIMLEAPLFVYNSYHQGPATGNVWCTIYALLGAVGGTVAIVTLTMISIDRYNVVVYPLNPKRSTTRLKVALMIVF | |||
AWIYGLVFSVIPALDIGLSRYTPEGFLTACSFDYLERTRDARLFMFLYFIFAWVVPIIAITFCYIQILRVVIGANSIQSSKNKSKTEVKLAGVVIGIIGLWFIAWTPYAIVAMMGV | |||
FGYESLLSPLGSMVPAILAKTAACIDPYFYAMNHPRYRQELRKMFGLNQQDLGNSQYQTSRYTRNASRMDDSEGGASERVTIGRQPGKTTTDEPEPSQQTEQGPQPTYSKNLAANS | |||
RGALQRAQSSISAADDTSLSVSIDLTETNPNSNH* 0 | |||
>UV7_culQui Culex quinquefasciatus (mosquito) Ecdy.Inse.Dipt XM_001861603 frag G? wrong exon 1 K90=EAP | |||
0 PEPVHPASKYISLSGYDGPPVEDAFRDRINPFWLQFEPPSPVAHYALGFVYFLMMVWGLFGNVLVIFMFFK 2 | |||
1 FKSLRTPANYLVINLAVADFLIMLEAPIFVYNSYHLGPAFGNTL 1 | |||
2 CTIYSLLGAIGGTVAIMTLTMISVDRYNVVVYPLNPNRSTTRLKVMLMIVFTWIYALVFSL | |||
MPALEIGLSRYTPEGFLTACSFDYLDRGWDARVFMFMYFVFAWVIPFLTISYCYVAILRVVVGAGSIQSSKNKNKQEVKLAGVVIGIIGLWFIAWTPYAVVAMLGVFGYEHLLTPL | |||
GSMIPAILAKTASCIDPYFYAMNHPRFRQELRKMFGKEQEMNHSQYQTSRYTRNASRNDSEAGPSERVQLGRAPGKDADPIPAVSSSVAQPNYSQNLASNRKGGLQRAQSSISAAD | |||
DTSLSCSIDLTETQPNNH* 0 | |||
>UV7_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BGIBMGA012539 full G? kaikoblast | |||
0 MSEILDRRSNVSGEVKETRLPIKYSKYEVNELVSSEANNEVLIKKFKELWPVNLWRRFGLFTDDYLHLVNPYWLSFAPPHPLLHYGLGAFYIMMTTVGSTGNALVMFMYFR 2 | |||
1 CRSLRTPGNILVANLALSDFMMLAKTPIFIFNSFNLGPALGKT 1 | |||
2 GCVIYGFVGGLTGTTSIAILTAISLDRYWAVVRPLEPLRALTAIRARLMAIAAWIYATLFAIIPALDIGYGRYVPEGFLTSCSFDYLTEELPPRYFIFVFFCAAWLAPFCTISYC | |||
YISIFRVVVCSRNITTKNQEQRLSTRHVKERTKRKAEIKLAFLVIVVIALFFISWTPYAIVALLGIFGKKDLIKPLTSMIPALFCKTAACINPFIYIITHPKFRKEFKRMLSRDKTE | |||
RSGTIKTIGYYTEASKFHRPSKDFSDTEVEIVEMKDIPYRNEDTTNTVEEKINTVSAKLAKREGSQKSQSSISMKSFEENVVSPPSWYCKPKFAKKKSFHRRSTHSMVSQNTDPTV* 0 | |||
>UV7_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_079311 full G? RH7 long N-terminal has M comp genomics support 2 of 3 exons novel | |||
0 MEAIIMTTLPNLTTDAGDSSFWLTGALSLSEMLANSSHSHSTGSTTSTAGSSATESSAVNVGKDHDKHVNDSVSTGLS 2 | |||
1 NYSNYPSYIHYRDKYDLSYIAKVNPFWLQFEPPKSSTFLIMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDI 1 | |||
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKEMPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQ | |||
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGEGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNSISSVMEQSKF* 0 | |||
>UV7_droYak Drosophila yakuba (fruitfly) Ecdy.Inse.Dipt genomic 12209654 full G? RH7 chr3L:12207286 12209654 | |||
0 MEAIIMTTLPALTTDAGDSSSFWLTGALSLSEMLANSSHGHSTGSTSSTAGSSATESSTVNVGKDHDVTKHVNDSVSTGLS 2 | |||
1 NYSNYPSYIHYRDKYDLSYIAKVNPFWLQFEPPKSSTFLVMAALYFLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDI 1 | |||
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQ | |||
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGDGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNSVSSVMEQSKF* 0 | |||
>UV7_droAna Drosophila ananassae (fruitfly) Ecdy.Inse.Dipt genomic full G? RH7 scaffold_13337 frameshifted | |||
0 MEAIILSTLPSLTTNASGSSSHWLTGALSLPEILANSSGSPNTSSADTGSGINLSARDADRHFNISTEAR 2 | |||
1 NYSYYPGYIHYRDKYDLSYIAKVNPFWLQFEPPHSSTFLAMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDV 1 | |||
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCFPLTAIVYSYFYILKVVFSAGRIQSNKDKAKTEQ | |||
KLAFIVAAIIGLWFLAWSPYAVVAMMGVFGLEKHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGILRRVSTTRSSYMTRSRSSFTHPAGRADGGTGRDHRMETYLMNNNLMMVPEETEENEEIVVVAEINNSVSSAIEQSKF* 0 | |||
>UV7_droPse Drosophila pseudoobscura (fruitfly) Ecdy.Inse.Dipt genomic full G? RH7 chrXR_group6:2491547 2493151 | |||
0 MEALMAALPTLTTEAAGSSLWLTSALSLSEMLANSSTSPNASLVAATTSSAAVATASTTSAAEAVGKVPDKHEVNDNVSTVLS 2 | |||
1 TSSSYPGYIHYRDKYDLSYIARVNPFWLQFEPPKSSTFYLMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDA 1 | |||
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIIFLIWSYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCVPLTTIVYSYFYILKVVFTASRIQSNKDKAKTEQ | |||
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGQERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFFGRGVLRRVSTTRSSYMTRSRSSFNRRLRPTPDAEHRVESYLMNNNLMMVPEETEENEEIVVVAEFNNSSYSGMEQSKF* 0 | |||
>UV7_droWil Drosophila willistoni (fruitfly) Ecdy.Inse.Dipt genomic full G? RH7 scaffold_180949 | |||
0 MDMDMALDMNDAATTTSLWITSAALSLSEILVNTTSHVVTTSPASTSTVETTAVAAVTATGKVVHDDEKHHHHHHHHHQDEVNDNNVTTVLR 2 | |||
1 NFSSYPGYIHYRDKYDLSYIAKVNPFWLQFEPPRSSTFYIMAALYCLISVVGCIGNAFVIFMFSNRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDI 1 | |||
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLVIVMIWCYSFLFAIMPALDVGLSVYVPEGYLTTCSFDYLNKETPARIFMALFFVAAYCVPLTCIMFSYFYILKVVFTANRIQSNKDKAKTEQ | |||
KLTFIVAAIIGLWFLAWSPYAVVAMMGVFGLEQHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLIYGRGVLRRVSTTRSSYITRSRSSFTRRLRTGSELDMRTEPYIMNNNLMMVPEETEENEEIVVVAEINNPSRCVSMHEHTSKF* 0 | |||
>UV7_droMoj Drosophila mojavensis (fruitfly) Ecdy.Inse.Dipt genomic full G? RH7 scaffold_6680 | |||
0 METIMSTLPTLTADDGSLWITSALTELLASGANSSSGSSSVVADGTQNATFVAAATTTTTTVAAAAAAAAAAAVNASTATTANATKGHHKHPHGVNDSETDLR 2 | |||
1 LCSSYPGYIHYRDKYDLTYIAKVNPFWLQFEPPDTSTFYIMAALYCLISVVGCVGNAFVIFMFGSRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIQEGPALGDA 1 | |||
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRYSRLRSYLIIFAIWCYSFLFAVMPALDVGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLASIVYSYFYILKVVFTANRIQSSKDKAKTEQ | |||
KLTFIVAAIIGLWFIAWSPYAIVAMMGVFGQEQHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLMYGRGVLRRVSTTRSSYMTRSRSSFTHRLRPSSGDCENRAEPYTLNNNLMMVPEETEENEEIIVVAEINNSISGVMEQSKF* 0 | |||
>UV5_plePay Plexippus paykulli (jumping_spider) Ecdy.Chel.Arac AB251851 18217181 full G? PpRh3 kumopsin3 | |||
MLNNTIPGPATLDDIGPPSWCYETRFNGWNAAPDIYVPDYWKQFRAPAPYLHYMLGFFYICLMSIAVV | |||
GNAIVMYIFFSAKTLRTPTNMFVIGLAMADLLMMSKTPVFIYNCFHLGPVFGQIGCDIYGIVGTYSGIGSAFCNAIIAYDRYRVIVHP | |||
FSKSGMSITKAIAFLVIIYLYITPFAILPALKIWSRYVPEGFLTSCSADFFMQDFNGRSYIV | |||
GTWFFGWFIPVAAIVFFYVQIFLAVKDHEEKIKEQARKMNVDSIRSNEAVKNSSAEVRIAKTAMCVFLMFLSSWAPYILVAFITGFSDPKLKRITPVISMVPAMTIKASACFDPFF | |||
YALSHPRYRLELQNRMPWLCINEKAEASGPADDSVSKTTEHVA* | |||
>UV5_hasAda Hasarius adansoni (jumping_spider) Ecdy.Chel.Arac AB251848 18217181 full G? HaRh3 kumopsin3 | |||
MLNNTALQPAVLDDIGPPSWCYETRFNGWNAAPDILVPDYWKQFRAPAPYLHYILGCLYICLMSVALI | |||
GNAIVIYIFSVSKSLRTPTNMFVIGLAMADLLMMSKTPVFIYNCFHLGPVFGQLGCDIYAIVGTYSGIGSAFCNAVIAYDRYRVIVHP | |||
FSKSGMTMTKAIAILVIVYLYITPFAILPALKIWSRFVPEGFLTSCSSDFYMQDFNGRSYIV | |||
GTWFFGWFIPVAAIIFFYAQIFLAVKDHEEKIKEQARKMNVDSFRSNEALKNSSAEVRIAKTAMCVVLLFLTSWVPYILVAFIAGFSDPKLKRVTPVISMIPAMTIKGSACFDPFF | |||
YALSHPRYRLELQNKLPWLCINEKAEASGPADDSVSKTTEHVA* | |||
>UV5_braKug Branchinella kugenumaensis (fairy_shrimp) Ecdy.Crus.Bran AB293436 18984904 full G? | |||
MANVTGFDYYRYERRELGWNTPAEYMEFVHPHWKQFEAPNPFLHYMLGVFYIIFMFCSLIGNGVVIWVFASAKSLRTPSNLFVINLAVLDFLMMLKTPVFIVNSFNEGPIWGKTGCDFFALLGSYAGIGGATTNAAIAFD | |||
RYRTIAHPFDGKLSRGQAITLCMLCWLYATPFSLMPFFGIWGRFVPEGFLTTCSFDYITEDSSTRAFVGTIFFTSYVLPMILIIYFYSQIVGHVRQHEETLRAQAKKMNVATLRSGKDDQEQSAEVRIAKVCIGLFSMFV | |||
ISWTPYAAVALLCAFGNRAAVTPLVSMIPALTCKAVACIDPWIYAINHPRYRLELQKRLPWFCIHEPEPNNDSASVNSEKTVATTTPS* | |||
>UV5_triLon Triops longicaudatus (tadpole_shrimp) Ecdy.Crus.Bran AB293435 18984904 full G? | |||
MAFLQNQTYDGTSPSFSFFRTSERIMLGHNTPKDYMEYVHPHWQTFEAPNPFLHYLLGVLYIGFMFCALVGNGVVIWIFSSAKSLRTPSNMFVINLAVLDFIMMMKTPVFIVNSFNEGPIWGKFGCDLFALMGSYSGIGG | |||
AMTNAAIAFDRYRTIARPFDGKLSRGKVLTICAGIWLWATPFSLMPLFGIWGRFVPEGFLTTCSFDYMTETSSIRWFVGCIFTYSYIIPLGLIIYYYSKIVGHVQEHERILREQARKMNVESLRSGKDQQEKSAEIRIAK | |||
VAIGLSLMFVVAWTPYALVALIAAFGNRAVLTPLVSMIPACCCKAVACIDPWIYAINHPRYRLELQKRMPWFCVHEPEPEMIDNSSAITEKTST* | |||
>UV5_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran AB293430 18984904 full G? | |||
MAFLQNQTHDGTSPSFSFYRTSERVMLGQYTPKDYMDYVHPHWQTFEAPNPFLHYLLGVLYIGFMFCALVGNGVVIWIFSSAKSLRTPSNMFVINLAVLDFIMMMKTPVFIVNSFNEGPIWGKFGCDMFALMGSYSGIGG | |||
AMTNAAIAFDRYRTIARPFDGKLSRGKVLTICAGIWLWATPFSLMPLFGIWGRFVPEGFLTTCSFDYMTETSSIRWFVGCVFTYSYIIPLGLIVYYYSKIVGHVQEHERILREQARKMNVESLRSGRDHQEKSAEIRIAK | |||
VAIGLSLMFVVAWTPYALVALIAAFGNRAVLTPLVSMIPACCCKAVACIDPWIYAINHPRYRLELQKRMPWFCVHEPEPEIIDNSSAITEKTST* | |||
>UV5a_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran FE384049 full G? | |||
0 MLGWNTPEDYMSYVHP 2 | |||
1 YWKTFEAPNPFLLYMIGFLYTIFMFCCVAGNGVVIWIFTN 2 | |||
1 CKSLRTPSNMLVVNLAILDMLMMLKSPVMIINSYNEGPIWGKLGCDVFGLMGSYNGIGSAVNNAAIAYDRHR 2 | |||
1 TISRPLDGKLSRKQVTLMIVAIWAWATPFSVMPFLGIWGRYVP 1 | |||
2 EGFLTTCTFDYMTEDASTRFFVGSIFVYSYVIPLAMLIFYYSKIVRSVGDHEKTLRDQAKKMNVTSLRSNRDQNEKSAEVRIAKVAIALATLFVFAWTPYAFVALTAAFGNR 2 | |||
1 SVLTPLLSTVPACCCKLVSCINPWIYAINHPRYR 2 | |||
1 MELQKKMPWFCIHEPVPTNDDSSVGSATTEMSGVSKETSS* 0 | |||
>UV5b_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic full G? penultimate intron lost last intron has slid back 2 aa | |||
0 MNGWNTPADYKSYVHPHWLSYEEPNPMLHHLLGVLYIFFMIASCLGNGIVIYIFST 2 | |||
1 TKELKTPSNILILNLAICDFIMMIKTPIFIVNSFNEGPVFGRLGCSIFGLLGAYVGPCSAVTNAAIAYDRYR 2 | |||
1 CISDPMGKRWSKSQASLIVLGCWVYASPVSLLPFTEIVNRFVP 1 | |||
2 EGYLTSCTFDYMTDNLETKMFVFILWIWCWIMPLGVIIFSYGKITTQVMTHEARLKEQAKKMNVESLRSGANKDARNEIRVAKVGISLTTLFLLSWTPYFAIAFIGCYGNR SLLTPGLSMIPACTCKMAACVDPFVYAINHPK 2 | |||
1 YRLELMKRFPWLCVHEKDDSTRSENSTNATIASEAESRT* 0 | |||
>UV5_triCas Tribolium castaneum (flour_beetle) Ecdy.Inse.Cole genomic full G? | |||
0 MYVVHPFKIIRNKVTILRTMETMANHLGWNVPKDELIHIPQHWLVYPEPEASMHFLLALIYIGFFIMATIGNGLVIWIFST 2 | |||
1 SKSLRTASNMFVVNLAICDFAMMIKTPIFIYNSFYRGFALGHLGCQIFAFIGSLSGIGAGMTNACIAYDRYT TITRPFDGKITRTKALVMIIFVWGYTIPWAVMPLLEIWGRFAP 1 | |||
2 EGFLTACSFDYLTDTFDNHMFVTSIFICSYVIPMSMIIYFYSQIVSKVFSHEKALREQ 0 | |||
0 AKKMNVESLRSNQSQQASQSAELRIAKAAIAICSLFVASWTPYAVLALIGAFGDQSLLTPGVTMVPACACKFVACLDPYVYAISHPKYR 2 | |||
1 LELQKRLPWLAIKETAASETQSTTTENTTTQSATTTT* 0 | |||
>UV5_lucCru Luciola cruciata (firefly) Ecdy.Inse.Cole AB300329 19232386 full G? | |||
MILHNATVFAAAQTQDDPDSIVHLLGWNVPKSELHHIPEHWLVYPEPEASIHYLLGIVYIFICFMGIVGNGLVLWIFSTSKSLKTASNMFVVNLAFCDFIMM | |||
MKMPIFVYNSFNRGYALGHIGCQIFGFVGSLSGIGAGMTNAFIAYDRYATISNPLEGKLTRTKALIMIFIIWGYTFPWAVLPMFEVWCRFVPEGFLTSCTFDYLTDTFDNDMFVAV | |||
IFICSYVIPMSMIIYFYSQIVKHVMHHEKALRDQAKKMNVESLRSNQSLQSQSIEIKIAKVAIMVCFLFVASWTPYAVLALIGGFGDQSLLTPGVTMVPALACKFVACLDPYVYAL | |||
SHPRYRMELQKRLPWLAIKEDAVSDAQSMVTTTTAAATPAATEQAPTA* | |||
>UV5_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_556823 full G? novel short exon | |||
0 MGLVQLDNQTAYRPEALIGADQSGLRYLGWNVPPEELVHIPEHWLQFPEPEASLHYLLGLLYIAFTIFSLVGNGLVIWIFIA 2 | |||
1 AKSLRTPSNVFVINLAICDFFMMAKTPIFIYNSFTKGFTLGNLGCQIFGFVGSLT 1 | |||
2 GIGAGATNALIAYDR 2 | |||
1 YNTITRPFEGRLTQTKAIIFICLIWAYTIPWGVLPLLEIWGRYVP 1 | |||
2 EGFLTSCTFDYLSGTFDTRLFVASIFTFSYVLPMSLIIYYYSQIVSHVVNHEKSLREQAKKMNVESLRSNQNQKDASVEIRIAKAAITVC | |||
FLFVASWTPYAVLALIGAFGDKSLLTPGVTMFPACACKFVACLDPYVYAISHPRYRIELQKRLPWLAITETLPAENASTCTEQQDGNATTQS* 0 | |||
>UVB_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_312478 full G? | |||
0 MFLGNESISEGAMLMPMARTAGEMPKLLGWNLPPEEQYLVHDHWKGFPSPPYYMHLMLAMIYFVLMNTSLIGNGIVLWIFGT 2 | |||
1 SKSLRNGSNMFIINLAIFDLLMMCEMPMFLVNSFSERLVGYGVGCSVYAALGSMSGIGGAISNAVIAFDRYRTISNPLDGRLSRVQAGLLICLTWLWTMPFTLLPLFEIWGRY | |||
IPEGYLTTCSFDYLTDDPDTRVFVGCIFTWAYVIPMIFICYFYARLFGHVRQHEMMLKNQARKMNVESLTANRSEKAQAVEMRIAKAAFTIFFLFVCAWTPYAIVTMIGAFGDR 2 | |||
1 TMLTPFVTMVPAVCCKIVSCLDPWVYAISHPKYRQELERRLPWMGIKEADDSVSTTES* 0 | |||
>UV5B_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_057748 19398933 full G? RH5 two small introns also seen in Apis and Daphnia | |||
0 MHINGPSGPQAYVNDSLGDGSVFPMGHGYPAEYQHMVHAHWRGFREAPIYYHAGFYIAFIVLMLSSIFGNGLVIWIFST 2 | |||
1 SKSLRTPSNLLILNLAIFDLFMCTNMPHYLINATVGYIVGGDLGCDIYALNGGISGMGASITNAFIAFDRYKTISNPIDGRLSYGQIVLLILFTWLWATPFSVLPLFQIWGRYQP 1 | |||
2 EGFLTTCSFDYLTNTDENRLFVRTIFVWSYVIPMTMILVSYYKLFTHVRVHEKMLAEQAKKMNVKSLSANANADNMSVELRIAKAALIIYMLFILAWTPYSVVALI | |||
GCFGEQQLITPFVSMLPCLACKSVSCLDPWVYATSHPKYRLELERRLPWLGIREKHATSGTSGGQESVASVSGDTLALSVQN* 0 | |||
>UV4_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_057353 19949290 full G? RH4 one ancestral intron with intercolated genes | |||
0 MEPLCNASEPPLRPEARSSGNGDLQFLGWNVPPDQIQYIPEHWLTQLEPPASMHYMLGVFYIFLFCASTVGNGMVIWIFST | |||
SKSLRTPSNMFVLNLAVFDLIMCLKAPIFIYNSFHRGFALGNTWCQIFASIGSYSGIGAGMTNAAIGYDRYNVITKPMNRNMTFTKAVIMNIIIWLYCTPWVVLPLTQFWDRFVP 1 | |||
2 EGYLTSCSFDYLSDNFDTRLFVGTIFFFSFVCPTLMILYYYSQIVGHVFSHEKALREQAKKMNVESLRSNVDKSKETAEIRIAKAAITICFLFFVSWTPYGVMSLI | |||
GAFGDKSLLTPGATMIPACTCKLVACIDPFVYAISHPRYRLELQKRCPWLGVNEKSGEISSAQSTTTQEQQQTTAA* 0 | |||
>UV3_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_079687 9165115 full G? RH3 single exon | |||
0 MESGNVSSSLFGNVSTALRPEARLSAETRLLGWNVPPEELRHIPEHWLTYPEPPESMNYLLGTLYIFFTLMSMLGNGLVIWVFSAAKSLRTPSNILVINLAFCDFMMMVKTPIFIYNSFH | |||
QGYALGHLGCQIFGIIGSYTGIAAGATNAFIAYDRFNVITRPMEGKMTHGKAIAMIIFIYMYATPWVVACYTETWGRFVPEGYLTSCTFDYLTDNFDTRLFVACIFFFSFVCPTTMITYY | |||
YSQIVGHVFSHEKALRDQAKKMNVESLRSNVDKNKETAEIRIAKAAITICFLFFCSWTPYGVMSLIGAFGDKTLLTPGATMIPACACKMVACIDPFVYAISHPRYRMELQKRCPWLALNEKAPESSAVASTSTTQEPQQTTAA* 0 | |||
>UV5_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD14509 full G? 8 exons K90 | |||
0 MDFNRTVSRPLAQLGs 2 | |||
1 SLMENEVGETHLLGWNLQAEDLIHIPEHWLKYQEPSSLQHYYLAFMYTIFMFVALFGNGLVIWVFCV 2 | |||
1 AKPLRTPSNIFVINLALCDFVMMAKAPIFILGSINRGYQ GHFLCQLFGTAGAFSGIGASATNAAIAYDRFS 2 | |||
1 TIAKPFDGRMTYGRAFFLIICIWTYTLPWGLLPLTEKWNRYVP 1 | |||
2 EGYLTSCTFDYLSPTDETRAFVGIMFVICYVIPVSLVIFFYSQIVSHVFNHEKALREQ 0 | |||
0 AKKMNVESLRSNQDANAQSAEVRIAKAAITICCLFIASWTPYAVVAMIGAFGDR 2 | |||
1 SLLTPGITMIPAIFCKTVACFDPYVYAISHPRYR 2 | |||
1 LELSKRVPCLGISEKPPPTASETQSTTTAA* 0 | |||
>UV5_rhoPro Rhodnius prolixus (kissing_bug) Ecdy.Inse.Hemi genomic frag G? exon 1 missing K90 at KTP | |||
0 0 | |||
1 ASTSGNIRTLGWNLSPEDLKHIPEHWLSYPEPEPILNYALGVLYIFFMLIALIGNGLVIWIFST 2 | |||
1 AKTLRTPSNIFVVNLAICDFLMMSKTPIFIYNSFKLGYALGHRACQIFALLGSFSGIGASATNAVIAYDRYR 2 | |||
1 VIATPFAPKLSRTKAVLYLALVWAYVTPWALLPLFEQWSRFVP 1 | |||
2 EGFLTSCTFDYLTPTSEIRNFVTVMFFICYVFPMSLIIYFYSQIVSHVIIHEHNLREQ 0 | |||
0 AKKMNVESLRSNANMHTQSAEIRIAKAAITICFLFVASWTPYAVLALIGAYGNQ 2 | |||
1 DLLTPAVTMIPACACKAVACVDPYVYAISHPRYR 2 | |||
1 QELSKKFPWLDIKEAPAPSSVDANSTATEMTLPTQTSPAEA* 0 | |||
>UV5_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme AF004169 9502802 full G? 5 exons | |||
0 MSNDSIHWEARYLPAGPPRLLGWNVPAEELIHIPEHWLVYPEPNPSLHYLLALLYILFTFLALLGNGLVIWIFCA 2 | |||
1 AKSLRTPSNMFVVNLAICDFFMMIKTPIFIYNSFNTGFALGNLGCQIFAVIGSLTGIGAAITNAAIAYDRYS 2 | |||
1 TIARPLDGKLSRGQVILFIVLIWTYTIPWALMPVMGVWGRFVPEGFLTSCSFDYLTDTNEIRIFVATIFTFSYCIPMILIIYYYSQIVSHVVNHEKALREQAKKMNVDSLRSNANTSSQSAEIRIAK 0 | |||
0 AAITICFLYVLSWTPYGVMSMIGAFGNKALLTPGVTMIPACTCKAVACLDPYVYAISHPKYR 2 | |||
1 LELQKRLPWLELQEKPISDSTSTTTETVNTPPASS* 0 | |||
>UV5_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01002850 full 83% bee | |||
0 MVNESIHWEARVLSAEPIRLLGWNVPAEELIHIPEHWLIYPEPNPSLHYLLAIVYIIFTFVALLGNGLVIWIFCS 2 | |||
1 AKSLRTSSNLFVVNLAFCDFFMMIKAPIFIYNSFNTGFATGHLGCQIFAFIGSLSGIGAGMTNAAIAYDRYS 2 | |||
1 VIARPLDGKLSRGQVILFIVLIWAYTIPWALMPIMHVWGRFVPEGFLTSCTFDYLTDTPEIQYFVATIFTFSYCIPMSLIIYYYSQIVNHVIDHEKALREQAKKMNVESLRSNVSANTQTMEIRIAK 0 | |||
0 AAITVCFLFVLSWTPYGVLAMIGAFGDKTLLTPGITMIPACTCKFVACLDPYVYAISHPRYR 2 | |||
1 LELQKRLPWLELQEKSTDTQSSTSEVVNVPSS* 0 | |||
>UV5_nasVit Nasonia vitripennis (jewel_wasp) Ecdy.Inse.Hyme XM_001608024 full G? gene wrong transcripts GE436449 GE390962 | |||
0 MPYYNWNGTDQTAGWPEARIQPAGAPRLLGWNVPPEELVHIPEHWLVYPEPNPALHYLLALLYILFTFVALLGNGLVIWIFCA 2 | |||
1 AKSLRTPSNMFVVNLAICDFMMMLKTPIFIYNSFHTGFALGNLGCQIFSFIGSLSGIGASITNAAIAYDRYS 2 | |||
1 TIARPLDGKLSRGQVMMLIVLIWMYTIPWALMPSMGVWGRFVP EGFLTSCTFDYITDSDEIRYFVGTIFTFSYAIPMTLIIYFYSQIVGHVVNHEKALREQAKKMNVESLRSGQNKDQASAEVRIAK 0 | |||
0 VALTICFLFVAAWTPYGVMSLIGAFGNK SLLTPGVTMIPACCCKAVACLDPYVYAISHPRYR 2 | |||
1 LELQKRMPWLELQEKPPASDATSTTTEAVPASS* 0 | |||
>UV5_papXut Papilio xuthus (butterfly) Ecdy.Inse.Lepi AB028218 full G? Rh5 | |||
0 MIPAAVMDNHTENNYNYGAYFAPYRLEGVELLGAGLTGEDLAAIPEHWLSYPAPPASAHTMLALVYVFFTAAALIGNGLVIFIFSASKSLRTPSNLLVVQLAVLDFLMMLKAPIFIYNSIK | |||
RGFASGVIGCQIFAFMGSVSGTAAGLTNACIAYDRHSTITRPLDGRLSRGKVLLMMVCVWLYTAPWAILPQLQIWGRYVPEGFLTSCTFDYLTTTFD | |||
NKLFVASMFVCVYIFPMIAILYFYSGIVKQVFAHEAALREQAKKMNVDSLRSNQNAAAESAEIRIAKAALTVCFLYVASWTPYGVMSLIGAFGDQNLLTPGVTMIPALACKGVACI | |||
DPWVYAISHPKYRQELQKRMPWLQIDEPDDNASNTTSNTANSSAPA* 0 | |||
>UV5_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BABH01012019 -------- frag G? --- | |||
0 1 | |||
2 VEMLGDGLSGDDLAAVPEHWLTFPAPPASAHTALALLYIFFTAAALLGNGLVIFVFST 2 | |||
1 TKSLRTSSNLLILQLAILDFIMMAKAPIFIYNSAMRGFATGTIGCQIFSVMGAYSGIGAGMTNACIAYDR 2 | |||
1 HSTITRPLDGRLSKGKVLLMMAFVWIYCTPWALLPLLKIWGRYVP 1 | |||
2 EGYLTSCTFDYLTNTFDTKLFVACIFVCSYVFPMSMIIYFYSGIVKQVFAHEAALR 2 | |||
1 EQAKKMNVESLRANQSGASQSAEIRIAKAALTVCFLFVASWTPYGVMALIGAFGDQQLLTPG 0 | |||
0 VTMIPAVACKAVACIDPWVYAISHPKYR 2 | |||
1 QELQRRMPWLQINEPDDTTSTATSNTVSSAPPAAPA* 0 | |||
>UV5_manSex Manduca sexta (moth) Ecdy.Inse.Lepi L78081 9343857 full G? Manop2 | |||
0 MNNQSENYYHGAQFEALKSAGAIEMLGDGLTGDDLAAIPEHWLSYPAPPASAHTALALLYIFFTFAALVGNGMVIFIFSTTKSLRTSSNFLVLNLAILDFIM | |||
MAKAPIFIYNSAMRGFAVGTVGCQIFALMGAYSGIGAGMTNACIAYDRHSTITRPLDGRLSEGKVLLMVAFVWIYSTPWALLPLLKIWGRYVPEGYLTSCSFDYLTNTFDTKLFVA | |||
CIFTCSYVFPMSLIIYFYSGIVKQVFAHEAALREQAKKMNVESLRANQGGSSESAEIRIAKAALTVCFLFVASWTPYGVMALIGAFGNQQLLTPGVTMIPAVACKAVACISPWVYA | |||
IRHPMYRQELQRRMPWLQIDEPDDTVSTATSNTTNSAPPAATA* 0 | |||
>UV5_pedHum Pediculus humanus (louse) Ecdy.Inse.Phth AAZO01000117 full G? exon 1 uncertain | |||
0 MKITTESENNISLSYYQPF 1 | |||
2 IDKEESLIWNVDPSELVHIPDHWFNFSAPHPLSNYLLGFLYFIFFVISCTGNGIVIWIFTT 2 | |||
1 SKNLRTASNVFVVNLAIFDFIMMAKTPIMIYNSMNLGFECGFVWCQIFASAGALSGIGASITNTCIAYDRCE 2 | |||
1 TITNPLQ KSGKKKAFLLAAFTWIYALPWAVLPFLEIWGKFAPEGYLTTCTVDYLTDTSQTRMFIVTIFFAAYVLPLSLIIYFYTKIVLHVINHEKSLKAQ 0 | |||
0 AKKMNVESLRSDGNKNYAVEIRITKVAIAMCFLFVISWTPYAVVALIGCFGNK 2 | |||
1 HLITPLVSMIPACACKAVACIDPYIYAISHPRFR 2 | |||
1 VEVNKRFACLAGCLQEKELQDDAVSKNTVNAENVDT* 0 | |||
>UV5_diaNig Dianemobius nigrofasciatus (cricket) Ecdy.Inse.Orth AB458852 full G? | |||
MELQGSNVSNLSVWRPEARLATRLLGWNVPAEELIHIPEHWLTYPAPDAFSYYILGMLYVAFCFIALIGNGLVIWVFSSAKTLRTPSNIFVINLALYDFIMM | |||
LKTPIFIYNSFNLGFGLGQLGCQIFAFMGSVSGIGAAATNACIAYDRYRVIARPFDSKMSIKGATLLVLLVWMWALPWAILPLLEIWGRYAPEGYLTSCSFDYLTDTPENHMFVLC | |||
IFICSYVIPMSLIIYFYSQIVSHVVNHEKALKEQAKKMNVDSLRSNQQQNQTSAEIRIAKVAIGICFLFVASWTPYAVLALIGAFGNKALLTPGVTMIPACTCKAVACLDPYVYAI | |||
SHPRYRAELQKRLPWLCIKEESASDTTSNATTTSTNAGATST* 0 | |||
>UVB_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD14509 full G? 8 exons K90 | |||
0 MDFNRSVSRPLSQLGS 2 | |||
1 SFMENEEELQLMGWNLTPEDLTHIPEHWLSYPEVRSLYHYILAFSYTILFCLGVIGNGLVLWIFCV 2 | |||
1 SKPLRTPSNLFVLNLALCDFSMVLVLPILIYDSIDHKYP GHLQCQIFALCGSISGIGAGATNAAIAYDRYS 2 | |||
1 TIAKPFEGRMTYGKALILIICIWIYVLPWCLLPLTEKWNRFVP 1 | |||
2 EGFLTSCSFDYLTPTEETKAFVGTMFVICYVIPMSFIIYFYSQIVCHVFNHEKALREQ 0 | |||
2 AKKMNVESLRSNQDANAQSAEVRIAKAAITICFLFVAAWTPYAVVAMIGAFGDQ 2 | |||
1 SLLTPIASMLPAVFAKTVACFDPYVYAISHPKYR 2 | |||
1 LELSKRVPCLGITEKPLATSDTQSITTAA* 0 | |||
>UVB_megVic Megoura viciae (vetch_aphid) Ecdy.Inse.Hemi AF189715 10762427 full G? | |||
MDFNRSVSRPLSQLGSSFMENEDELQLMGWNLTPEDLTHIPEHWLSYPEVRSLYHYILAFSYTILFCIGVI | |||
GNGLVLWIFCVSKPLRTPSNLFVLNLALCDFSMVLVLPILIYDSIDHKYPGHLQCQIFALCGSISGIGAGATNAAIAYDRYSTIAKP | |||
FEGRMTYGKALILIICIWIYVLPWCLLPLTEKWNRFVPEGFLTSCSFDYLTPTEETKAFVGTMFVICYVIPMSFIIYFYSQIVCHVFNHEKALREQAKKMNVESLRSNQDANAQ | |||
SAEVRIAKAAITICFLFVAAWTPYAVVAMIGAFGDQSLLTPIASMLPAVFAKTVACFDPYVYAISHPKYRLELSKRVPCLGITEKPPAASDTQSITTAA* 0 | |||
>UVB_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme AF004168 9502802 full G? 8 exons | |||
0 MLLHNKTLAGKALAFIAEEG 2 | |||
1 YVPSMREKFLGWNVPPEYSDLVHPHWRAFPAPGKHFHIGLAIIYSMLLIMSLVGNCCVIWIFST 2 | |||
1 SKSLRTPSNMFIVSLAIFDIIMAFEMPMLVISSFMERMIGWEIGCDVYSVFGSISGMGQAMTNAAIAFDRYR 2 | |||
1 TISCPIDGRLNSKQAAVIIAFTWFWVTPFTVLPLLKVWGRYTT 1 | |||
2 EGFLTTCSFDFLTDDEDTKVFVTCIFIWAYVIPLIFIILFYSRLLSSIRNHEKMLREQ 0 | |||
0 AKKMNVKSLVSNQDKERSAEVRIAKVAFTIFFLFLLAWTPYATVALIGVYGNR 2 | |||
1 ELLTPVSTMLPAVFAKTVSCIDPWIYAINHPR 2 | |||
1 YRQELQKRCKWMGIHEPETTSDATSAQTEKIKTDE* 0 | |||
>UVB_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01025579 ADTU01025580 frag 74% bee | |||
0 2 | |||
1 DVQPLQEEHFLGWNFPPEHADLVHPHWRVFLAPGRLWHVALALIYFLLLMLSLLGNGIVIWIFST 2 | |||
1 SKSLRTPSNIFILNLAVFDLLMATEMPMLIINSFLERTIGWETGCDVYALLGAISGMGQAITNAAIAFDRYR 2 | |||
1 TISCPIDGRLNGKQAMVLALFTWFWALPFSILPMTGLWGRYVA 1 | |||
2 EGFLTTCSFDYLTDDEDTRTFVITIFFWAYVLPMLLIIYFYSQLLKSIRSHEKMLRDQ 0 | |||
0 AKKMNVKSLVSNQNKERSVEMRIAKVAFTIFFLFVLAWTPYAVVALIGAYGNR 2 | |||
1 EALTPFSTMLPAIFAKTVSCIDPWIYAINHPR 2 | |||
1 YRQELEKRCKWMGIREKVAEDTVSAQTEKVNAEET* 0 | |||
>UVB_nasVit Nasonia vitripennis (jewel_wasp) Ecdy.Inse.Hyme XM_001604572 full G? | |||
0 MAFVGLNGAMGGMGPA 1 | |||
2 EKPLQRYSQGPQMQEHLLGWNHPPEHIDIVHPHWRGFLAPGKYWHIGLALIYFMLLVLSFVGNGCVVWIFST 2 | |||
1 SKVLRTPSNLFIINLALFDLVMALEIPMLIINSFIERMIGWGLGCDIYAALGSVSGIGSAITNAAIAYDRYR 2 | |||
1 TISCPIDGRLNGKQAAVMVAFTWFWTMPFTILPFAKIWGRYTT 1 | |||
2 EGFLTTCSFDFLSDDQDTKVFVAAIFSWSYCFPMVLIIYFYSQLIKSVRRHEKMLREQ 0 | |||
0 AKKMNVKSLSAQDKERSVEMRIAKVAFTIFFLFVCSWTPYAVVTMIAAFGNR 2 | |||
1 ELVTPFSSMLPAVFAKTVSCIDPWVYAINHPR 2 | |||
1 YRQELTKRCQWMGIHEPDSGPSQNNAEAVSVTTEKLKSDDA* 0 | |||
>UVB_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BABH01016294 -------- full G? --- | |||
0 MALNFTQEISDIGPMAYPLKM 2 | |||
1 VSREMVEHMLGWNIPEEHQDLVHEHWRNFPSVSKYWHYCLALIYAMLMVTSLVGNGIVIWIFGT 2 | |||
1 SKSLRSASNMFVINLAIFDLMMMLEMPLLIINSFYQRLVGYQLGCDIYAVLGSLSGIGGAITNAVIAFDRYK 2 | |||
1 TISCPLDGRINKVQASLLIAFTWFWALPFTILPAFRIWGRYVP 1 | |||
2 EGFLTTCSFDYFTDDQDTKVFVACIFVWSYCIPMTLICYFYSQLFGA 0 | |||
0 VRLHERMLQEQ AKKMNVKSLAANKEDASRSVEIRIAKVAFTIFFLFVCAWTPYAFVTMTGAFGDR 2 | |||
1 NLLTPIATMVPAVCCKVVSCIDPWVYAINHPR 2 | |||
1 YRAELTKRLPWLGVRESDPDAVSTTTSVGTAQSQATAEA* 0 | |||
>UVB_manSex Manduca sexta (moth) Ecdy.Inse.Lepi AD001674 9343857 full G? exons from 454 | |||
0 MATNFTQELYEIGPMAYPLKM 2 | |||
1 ISKDVAEHMLGWNIPEEHQDLVHDHWRNFPAVSKYWHYVLALIYTMLMVTSLTGNGIVIWIFST 2 | |||
1 SKSLRSASNMFVINLAVFDLMMMLEMPLLIMNSFYQRLVGYQLGCDVYAVLGSLSGIGGAITNAVIAFDRYK 2 | |||
1 TISSPLDGRINTVQAGLLIAFTWFWALPFTILPAFRIWGRFVP 1 | |||
2 EGFLTTCSFDYFTEDQDTEVFVACIFVWSYCIPMALICYFYSQLFGA 1 | |||
0 VRLHERMLQEQAKKMNVKSLASNKEDNSRSVEIRIAKVAFTIFFLFICAWTPYAFVTMTGAFGDR | |||
1 TLLTPIATMIPAVCCKVVSCIDPWVYAINHPR 2 | |||
1 RAELQKRLPWMGVREQDPDAVSTTTSVATAGFQPPAAEA* 0 | |||
>UVB_diaNig Dianemobius nigrofasciatus (cricket) Ecdy.Inse.Orth AB291232 full G? | |||
MNSSVGLQGAPIALPYESYVAQMLGWNIPAEHIELVHSHWRGYEAPSKYWHYWFAFMYFCIMIMSCLGNGIVLWIFATTKSLRTPSNMFVVNQALLDLLMMI | |||
EMPMFVLNSLFYQRPIGWEMGCDIYALLGAVSGIGSAINNAAIAYDRYRTISFPLDGRLQFGHALAFIVGVWSWAMPFSLLPLLKVWGRYVPEGLLTTCSFDYLTDDEDTKVFTAS | |||
IFTWSYAFPLCLIVFFYCKLFKQVRLHEKMLQEQARKMNVKSLQTNQDVAQKSVEIRIAKVAFTIFFLFLCSWTPYATVAMIGAFGNRALLTPMSTMIPALFSKIVSCIDPWIYAI | |||
NHPRFRGELLKRAPWFGVEELKSSDVSSIGTDRTTATAAIETPAA* 0 | |||
>LMS_ixoSca Ixodes scapularis (tick) Ecdy.Chel.Arac EW898719 full G? ocellar transcripts | |||
0 MGSEGQRTNMSLLDELASPYMKNGTLVESVPDEMLYMVHPHWYNFKPMNPLWHSLLGFAMVILGVISVVGNSMVIYIMTTSKSLRSPTNMLVVNLAFSDW 2 | |||
1 CMMAFMMPTMAANCFAETWILGPFMCEVYGMVGSLFGCGSIWSMVMITLDRYNVIVRGVAAAPLTHKRAALMIFFVWFWALTWTLLPFFGWSR 2 | |||
1 YVPEGNMTSCTIDYLTKALWSASYVVAYAGGVYWTPLFINIYCYSKIVRAVAQHEKQLRLQARKMNVASLRANAEQTKTSAEARLAK 0 | |||
0 IALMTVGLWFMAWTPYLTIAWAGIFSDGSKLTPLATIWGSVFAKANACYNPIVYGISHPKYRAALARRFPSLVCMPPGGDQLDTRSEASGITTIEDKVMTTET* 0 | |||
>LMS_tetUrt Tetranychus urticae (spider_mite) Ecdy.Chel.Arac GW028305 frag G? larval transcripts | |||
CEFYGMWGSLFGCGSVWSLVFITRDRYNVIVRGMSAGRLTIKKSLAQVLFIWFYSVAWTIIPFFGWSRYVPEGNMTSCTIDYLSKDLWNASYTVVYAVAVYFLPLFTLVYHYYYIVCAVATHERTLREQA | |||
KKMNVASLRANADANKESADIRIAKVAMMTVGLWFMAWTPYCTLAFLGIFTDGDYLTPMVSIWGAVFAKSAACYNPIVYALSHPKYRSALFTRFPSLACGVGAVEPVVDDDAKSEMSVATTFSDHTSAP* 0 | |||
>LMS1_plePay Plexippus paykulli (jumping_spider) Ecdy.Chel.Arac AB251849 18217181 full G? PpRh1 kumopsin1 | |||
MLPQAAKMAARASSGVDGKNISIVDLLPEDMLYMIHEHWYKYPPMESTMHYLLGITIILIGIISVS | |||
GNSIVIYLMLSVKSLRTPANFLVTSLAVSDGGMLAFMAPTMPINCFAQTWVLGPFMCELYGMVGSLFGSASIWNMVMITLDRYNVIVRG | |||
MSGKPLTKVGALLRIIFVWVWSLGWTIAPMYGWSSYAPEGSMTGCTVDYLHTDISTMSYLIVY | |||
AIFVYFVPLFIIIYCYTYIVMQVAAHEKSLREQAKKMNIKSLRSNEDNKKASAEFRLAKVALMTICLWFMAWTPYLILSLLGIFSDREWLTPLTSIWGAVFAKAASAYNPIVYGIS | |||
HPKYRAALHEKFPCLNCATESPKGDSASTVAESDKGGD* | |||
>LMS2_plePay Plexippus paykulli (jumping_spider) Ecdy.Chel.Arac AB251850 18217181 full G? PpRh2 kumopsin2 | |||
MSSQIINGAYMVSRDALGLHLPTNLGGPLPQDNSYYPYLRNTTVVDTVPKEILHMIHDHWYQFPPLNPLWHSLLGIAMILLGIVSVI | |||
GNGMVMYLMNTTKSLKTPTNMLIVNLAFSDFCMMAFMMPTMAANCFAETWILGPFMCEIYGMAGSLFGCVSIWSMVMIAFDRYNVIVRG | |||
MNAEPLTTKKAAAQIFLIWAWAIMWTVLPFFGWSRYVPEGNM | |||
TSCTVDYLSEDLKSSSYVLIYGCAVYFIPLFTLIYNYTFIVRAVSIHEDNLREQAKKMNVTSLRANADQQKQSAECRLAKIALMTVGLWFIAWTPYLCIAWSGIFSSRKHLTPLAT | |||
IWGAVFAKAVAVYNPIVYGISHPKYRAALFQKFPSLACTTESDVIDNKSEVTFVTDEKPPKTQEA* | |||
>LMS1_hasAda Hasarius adansoni (jumping_spider) Ecdy.Chel.Arac AB251846 18217181 full G? HaRh1 kumopsin1 | |||
MLPHAAKMAARVAGDHDGRNISIVDLLPEDMLPMIHEHWYKFPPMETSMHYILGMLIIVIGIISVS | |||
GNGVVMYLMMTVKNLRTPGNFLVLNLALSDFGMLFFMMPTMSINCFAETWVIGPFMCELYGMIGSLFGSASIWSLVMITLDRYNVIVKG | |||
MAGKPLTKVGALLRMLFVWIWSLGWTIAPMYGWSRYVPEGSMTSCTIDYIDTAINPMSYLIAY | |||
AIFVYFVPLFIIIYCYAFIVMQVAAHEKSLREQAKKMNIKSLRSNEDNKKASAEFRLAKVAFMTICCWFMAWTPYLTLSFL | |||
GIFSDRTWLTPMTSVWGAIFAKASACYNPIVYGISHPKYRAALHDKFPCLKCGSDSPKGDSASTVAESEKAGE* | |||
>LMS2_hasAda Hasarius adansoni (jumping_spider) Ecdy.Chel.Arac AB251847 18217181 full G? HaRh2 kumopsin2 | |||
MSSHTINSAFMVPRDVLGLHLPNNLGGPLPHDNSYYPYLRNATVVDTVPKEILHMIHDHWYQFAPLNPLWHSLLGIAMIILGIVSVI | |||
GNGMVIYLMSTTKSLKTPTNMLIVNLAFSDFCMMAFMMPTMAANCFAETWILGPLMCEIYGMAGSLFGCVSIWSMVMIAFDRYNVIVRG | |||
MSAEPLTTKKAAAQIFFIWTWATTWTLFPFFGWSRYVPEGNMTSCTVDYLTEDLKSSSYVLIYGCAVYFTPLFTLIYNYTFIVRSVSIHENNLREQAKKM | |||
NVSSLRANADQQKQSAECRLAKIALMTVGLWFIAWTPYLSIAWSGIFSSRKHLTPLATIWGAVFAKAVAVYNPIVYGISHPKYRAALFEKFPSLACTTESDVTDNKSEVTLVTDEKPPKTQEA* | |||
>LMS_limPol Limulus polyphemus (horseshoe_crab) Ecdy.Chel.Mero AY190512S1 14977331 full G? intronated lateral_eye | |||
0 MANQLSYSSLGWPYQPNASVVDTMPKEMLYMIHEHWYAFPPMNPLWYSILGVAMIILGIICVLGNGMVIYLMMTTKSLRTPTNLLVVNLAFSDFCMMAFMMPTMTSNCFAETW | |||
ILGPFMCEVYGMAGSLFGCASIWSMVMITLDRYNVIVRGMAAAPLTHKKATLLLLFVWIWSGGWTILPFFGWSR 2 | |||
1 YVPEGNLTSCTVDYLTKDWSSASYVVIYGLA 0 | |||
0 VYFLPLITMIYCYFFIVHAVAEHEKQLREQAKKMNVASLRANADQQKQSAECRLAK 0 | |||
0 VAMMTVGLWFMAWTPYLIISWAGVFSSGTRLTPLATIWGSVFAKANSCYNPIVYGISHPRYKAALYQRFPSLACGSGESGSDVKSEASATTTMEEKPKIPEA* 0 | |||
>LMS_neoOer Neogonodactylus oerstedii (mantis_shrimp) Ecdy.Crus.Mala DQ646869 17053049 full G? Rh1 | |||
MSYWNSNKIVEEYSLPSTNPYGNFTVVDTVPENMLHMIHSHWYQFPPLNPMWYGILAFVVTVVGLCSICGNFVVIWVFMNTKALRSPANTLVVSLAVSDFIM | |||
MACMFPPLVLNCYWGTWIFGPLFCEVYAFIGNTVGCASIGNMIFITFDRYNVIVKGISGTPLSQKNTTLQVLFVWICSIMWCVFPFFGWNRYVPRGDMTACGTDYLTEDEFSRSYL | |||
YVYSVWVYIGPLALIIYCYFHIVSAVATHEKQMRDQAKKMGVKSLRTEEAKKTSAECRLAKVALTTVSLWFMAWTPYLIINWAGMFYPSVVSPLFSIWGSVFAKANAVYNPIVYAI | |||
SHPKYRAALYKKLPCLACSTESADEGSATNSATTTTAEKYESA* 0 | |||
>LMS_lucCru Luciola cruciata (firefly) Ecdy.Inse.Cole AB300328 19232386 full G? | |||
MSVLGEPSFAAWASQAGVMSSRFGGGNITVVDKVPPDMLHLIDAHWYQYPPLNPLWHAILGFMIGVLGCISVTGNGMVIYIFSTTKSLRSPSNLLVVNLAFS | |||
DFLMMFTMAPPMVINCYNETWVWGPLFCQIYGMLGSLFGCTSIWTMTMIALDRYNVIVKGLSAKPLTKQGALIRIFLVWVFSIGWTIAPVFGWNRYVPEGNMTACGTDYLSTGWFS | |||
RSYILFYSWFVYFIPLFAIIYSYWFIVQAVSAHEKAMREQAKKMNVASLRSSEAAQTSAECKLAKVALMTISLWFLAWTPYLVTNYAGIFDGSKISPLATIWSSLFAKANAVYNPI | |||
VYGISHPKYRQALQKKFPSLVCAAEPDDTVSQTTAATAASEEKAAA* 0 | |||
>LMS_triCas Tribolium castaneum (flour_beetle) Ecdy.Inse.Cole ES544655 18342244 full G? 3 exons from AAJJ01000967 5 fusion relative to bee | |||
0 MSVMGEPNFIAWAAQRSGYGGGNLTVVDKVLPDMLHLVDAHWYQFPPMNPLWHGILGFVIGVLGFVSIVGNGMVIYIFSSTKALRTPSNLL | |||
VVNLAFSDFLMMlCMSPAMVINCYNETWVLGPLVCELYGMSGSLFGCASIWTMTFIALDRYNVIVKGLSAQPLTKKGAMLRILIIWVFSTLW | |||
TIAPFFGWNRYVPEGNMTACGTDYLTKDWVSRSYILVYAVWVYFVPLFTIIYSYWFIVQ 0 | |||
0 AVAAHEKSMREQAKKMNVASLRSSEAAQTSAECKLAKIALMTITLWFFAWTPYLVTNFTGIFEGAKISPLATIWCSLFAKANAVYNPIVYGIS 2 | |||
1 HPKYRQALQKKFPSLVCAGEPDDTTSTASGVTNVTTDEKPATA* 0 | |||
>LMS1_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_079683 19398933 full G? RH1 ninaE | |||
0 ME 0 | |||
0 SFAVAAAQLGPHFAPLSNGSVVDKVTPDMAHLISPYWNQFPAMDPIWAKILTAYMIMIGMISWCGNGVVIYIFATTKSLRTPANLLVINLAISDFGIMITNTPMM | |||
GINLYFETWVLGPMMCDIYAGLGSAFGCSSIWSMCMISLDRYQVIVKGMAGRPMTIPLALGKIAYIWFMSSIWCLAPAFGWSR 2 | |||
1 YVPEGNLTSCGIDYLERDWNPRSYLIFYSIFVYYIPLFLICYSYWFIIA 0 | |||
0 AVSAHEKAMREQAKKMNVKSLRSSEDAEKSAEGKLAKVALVTITLWFMAWTPYLVINCMGLFKFEGLTPLNTIWGACFAKSAACYNPIVYGIS 2 | |||
1 HPKYRLALKEKCPCCVFGKVDDGKSSDAQSQATASEAESKA* 0 | |||
>LMS6_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_079644 19398933 full G? RH6 gross genomic misassembly exon1 | |||
0 MASLHPPSFAYMRDGRNLSLAESVPAEIMHMVDPYWYQWPPLEPMWFGIIGFVIAILGTMSLAGNFIVMYIFTSSKGLRTPSNMFVVNLAFSDFMMMFTMFPPVVLNGFYGT | |||
WIMGPFLCELYGMFGSLFGCVSIWSMTLIAYDRYCVIVKGMARKPLTATAAVLRLMVVWTICGAWALM | |||
PLFGWNRYVPEGNMTACGTDYFAKDWWNRSYIIVYSLWVYLTPLLTIIFSYWHIMK 0 | |||
0 AVAAHEKAMREQAKKMNVASLRNSEADKSKAIEIKLAKVALTTISLWFFAWTPYTIINYAGIFESMHLSPLSTICGSVFAKANAVCNPIVYGLS 2 | |||
1 HPKYKQVLREKMPCLACGKDDLTSDSRTQATAEISESQA* 0 | |||
>LMS_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_319247 full G? most introns obliterated | |||
0 MPYYGPMQQPGLWGQPVANLTVVDKVPPEIMHLVDPHWSQFPPMNPLWHSIIGFVIFVLGVVSIIGNGMVIYIFSTAKSLRTPSNLFIVNLALSDFLMMGTNAFTMVYNCWFETWSLG | |||
LLMCDLYAFFGSLFGCCSIWTMTMIALDRHNVIVHGLSGKPLTNTGAILRILLCWLIGVVWGILPMLGWNRYVPEGNMTACGTDYLTDDWFHKSYILVYSVFVYYTPLFTIIYAYFFIIK 0 | |||
0 AVSAHEKNMREQAKRMNVQSLRSSDDGKSTEMKLAKVALVTISLWFMAWTPYTVINYTGVFKTASITPLATIWGSVFAKANAVYNPIVYGISHPKY | |||
RAALLRRFPSLACSDGPPADDKSLASEASGITSAGNPTTA* 0 | |||
>LMS2_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt M12896 full G? RH2 CG16740-RA Rh2 ocellar-specific | |||
0 MERSHLPETPFDLAHSGPRFQAQSSGNGSVLDN 0 | |||
0 VLPDMAHLVNPYWSRFAPMDPMMSKILGLFTLAIMIISCCGNGVVVYIFGGTKSLRTPANLLVLNLAFSDFCMMASQSPVMIINFYYETWVLGPLWCDIYAGCGSLFGCVSIWSMC | |||
MIAFDRYNVIVKGINGTPMTIKTSIMKILFIWMMAVFWTVMPLIGWSAYVPEGNLTACSIDYMTRMWNPRSYLITYSLFVYYTPLFLICYSYWFIIAAVAAHEKAMREQAKKMNVKSL | |||
RSSEDCDKSAEGKLAKVALTTISLWFMAWTPYLVICYFGLFKIDGLTPLTTIWGATFAKTSAVYNPIVYGIS 2 | |||
1 HPKYRIVLKEKCPMCVFGNTDEPKPDAPASDTETTSEADSKA* 0 | |||
>LMS_rhoPro Rhodnius prolixus (kissing_bug) Ecdy.Inse.Hemi genomic full G? | |||
0 MAQPIGPSFAAYQWGQSANPSANRSVVDMVPPEMLSMVDAHW 2 | |||
1 YQFPPLNPLWHGILGFVIGVLGIISIVGNGMVIFIFSSTKTLRTPSNLLVVNLAFSDFLMMFTMSPPMVINCYNETWVL 1 | |||
2 GPLMCELYGMLGSLFGCASIWTMTMIALDRYNVIVK 0 | |||
0 GISAKPMTNKTAMLRILLVWAFSIMWTVFPFFGWNR 2 | |||
1 YVPEGNMTACGTDYLTKNWVSRSYILVYSVFVYFLPLFTIIYSYFFILQ 0 | |||
0 AVSAHEKQMREQAKKMNVASLRSAEAANTSAEAKLAK VALMTISLWFMAWTPYLVINYSGIFETISISPLFTIWGSLFAKANAVYNPIVYAIR 2 | |||
1 HPKYKQALEKKFPSLSCASPQDDTTSVATGVTTSTDDKAPSA* 0 | |||
>LMS_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD6053 full G? | |||
0 MLNKIGSHYERQENWVAEGGFGNETVVDRVPADMMHLIDPSW 2 | |||
1 YQFPPMESMWYKWLGVTIFFLGILSVVGNGMVIYIFTCTKNLRTPSNLLIVNLAFSDFCLMFTMCPAMVWNCFYETWMF 1 | |||
2 GPFACELYAMFGSLFGVTSIWTMVFIALDRYNVIVK 0 | |||
0 GLSAKPMTTKLALLQIFCIYLHGLFWTLTPFFGWSR 2 | |||
1 YVPEANMTACGTDYLTLAWHSRSYVLVYAIFAYYLPLLVIIYAYYFIVK 0 | |||
0 AVASHEKSMREQAKKMNVSSLRSGDQSNTSAEFKLAKVALMTISLWFMAWTPYMVINFAGIFQLMTIDPLFTIWGSVFAKANAVYNPIVYAIS 2 | |||
1 HPKYRLALDKKFPCLVCGKLEDDRSDSKSVASAQTTISEDKV* 0 | |||
>LMS_homCoa Homalodisca coagulata (sharpshooter) Ecdy.Inse.Hemi AY588065 full G? | |||
MSLISEPSFSAYSWASQGGFGNQTVVDKVPPEMLYLVDAHWYQFPPMNPLWHSLLGFAMVVLGFIAVTGNGMVVYIFSCTKALRTPSNLLVVNLAFSDFLMM | |||
FTMAPPMVLNCYYETWVLGPFMCELYAMFGSILGCTSIWTMVMIANDRYNVIVKGLSAKPMTIKSALARILFCWAHSLIWCLAPFLGWGRYVPEGNMTACGTDYLTPDWISKSYIL | |||
VYSLFCYFMPLFLIIYSYWFIVQAVSAHEKAMREQAKKMNVASLRSSDAANTSAEHKLAKVALMTISLWFCAWTPYLVINYAGIFQALTISPLFTIWGSVFAKANACYNPIVYAIS | |||
HPKYRAALNKKFPSLVCGATEAPASTSDGASVASGATTLTEDKSAAA* | |||
>LMSa_nasVit Nasonia vitripennis (jewel_wasp) Ecdy.Inse.Hyme XM_001606013 full G? 22063-23541 - strand of AAZX01007316 -- | |||
0 MGPSFLTLTAMAQRGGYGGGGGFGGGFNNQTVVDKAPPEIHHMIDPYWYQFPPMNPLWYGILGFVIGCLGCISVAGNGMVVYIFASTKSLRTPSNLLVINLAFSDFCMMFTMSPPM 0 | |||
0 VINCYYETWVFGPLMCEIYALCGSIFGCGSIWTMCMIAFDRYNVIVKGLSAKPMTINGSLLRILGIWLMASIWTIAPMFGWNR 2 | |||
1 YVPEGNLTACGTDYFSKDWVSRSYIVVYSFFVYFLPLFMIIYSYYFIIKAVSAHEKNMREQAKKMNVASLRQGDSQSAENKLAK 0 | |||
0 IALMTISLWFMAWTPYLVINWAGIFDLARLTPLFTIWGSVFAKANAVYNPIVYGIS 2 | |||
1 HPKYRAALFARFPSLACAGDAPAGAASDAVSTTSGVTTLTDHDKSNA* 0 | |||
>LMSb_nasVit Nasonia vitripennis (jewel_wasp) Ecdy.Inse.Hyme AAZX01007316 full G? tandem pair to LWSa fairly diverged | |||
0 MEHPIVAAGVNATGEFDASSGSASSTTTMVTTAAVQVASTIGPHFARQVMRGFGNLTVVDKVPPEMLHLVGPHW 2 | |||
1 YQFPPLWPIWHKLLGVVMIFIGVLGWCGNGMVVYIFLVTPSLRTPSNLLVINLAFSDFVMMIIMSPPMVVNCWYETWIL 1 | |||
2 GPLMCDIYALIGSLCGGASIWTMTAIAYDRYNVIVK 0 | |||
0 GMSGTPLTIPRALVQIVLIWTHGLIWAMLPLFGWNR 2 | |||
1 YVPEGNMTSCGTDYVSDDWLGKSYILVYSIFVYYTPLFSIILCYWHIVS 0 | |||
0 AVAAHERGMREQAKKMNVASLRSGDQSGESAEVKLAK 0 | |||
0 VAVTTISLWFLAWTPYLVTNYMGIFAKQHVSPLFTIWASLFAKTNACYNPIVYGIS 2 | |||
1 HPKYRAGLKVKCPCLVFGDTEDKPKPAAATPAADAASTHSKA* 0 | |||
>LMSa_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme NM_001077825 16291092 full Gq rhabdomeric long wavelength ocelli not compound | |||
0 MDTLNITTSFFIEVMPSNISTLTTTGPQFARQLMRFNNQTVVSKVPEEMLHLIDLYW 2 | |||
1 YQFPPLDPLWHKILGLVMIILGIMGWCGNGVVVYVFIMTPSLRTPSNLLVVNLAFSDFIMMGFMCPPMVICCFYETWVL 1 | |||
2 GSLMCDIYAMVGSLCGCASIWTMTAIALDRYNVIVK 0 | |||
0 GMSGTPLTIKRAMLQILGIWLFGLIWTILPLVGWNR 2 | |||
1 YVPEGNMTACGTDYLSQDWTFKSYILVYSFFVYYTPLFTIIYSYYFIVS 0 | |||
0 AVAAHEKAMKEQAKKMNVTSLRSGDNQNTSAEAKLAK 0 | |||
0 VALTTISLWFMAWTPYLVINYIGIFNRSLITPLFTIWGSLFAKANAIYNPIVYGIS 2 | |||
1 HPKYRAALKEKLPFLVCGSTEDQTAATAGDKASEN* 0 | |||
>LMSa_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01020830 16601190 full iMet uncertain 80% bee | |||
0 MAFADVTRTIGPRAMQQIMSFNNKTVVNKVPPDMMHLIDPHW 2 | |||
1 YQFPPMNPMWHKILGLVMIVLGFMGWCGNGVVVYIFLMTPSLRTPSNLLIVNLAFSDFIMMIIMTPHMMINCYYETWIL 1 | |||
2 GPLMCDMHAMVGSLCGCASIWTMTAIALDRYNVIVK 0 | |||
0 GMAGKPLTIKKALLEILIIWLFASIWAILPIVGWNR 2 | |||
1 YVPEGNMTACGTDYLTKDWSSKSYILVYSIFVYYMPLLIIIYSYYFIVS 0 | |||
0 TVAAHERTMREQAKKMNVQSLRSGDNQNISAEAKLAK 0 | |||
0 VALMTISLWFMAWTPYLVINYVGIFGIAKISPLFTIWGSLFAKANAIYNPIVYGIR 2 | |||
1 HPKYRAALKEKLPFFVCGSTEDPSVAAKTAETSSTTTT* 0 | |||
>LMSb_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme U26026 full G? 5 exonsArthropoda Insecta 540 complete genNow | |||
0 MIAVSGPSYEAFSYGGQARFNNQTVVDKVPPDMLHLIDANWYQYPPLNPMWHGILGFVIGMLGFVSVMGNGMVVYIFLSTKSLRTPSNLFVINLAISDFLMMFCMSPPM 0 | |||
0 VINCYYETWVLGPLFCQIYAMLGSLFGCGSIWTMTMIAFDRYNVIVKGLSGKPLSINGALIRIIAIWLFSLGWTIAPMFGWNR 2 | |||
1 YVPEGNMTACGTDYFNRGLLSASYLVCYGIWVYFVPLFLIIYSYWFIIQAVAAHEKNMREQAKKMNVASLRSSENQNTSAECKLAK 0 | |||
0 VALMTISLWFMAWTPYLVINFSGIFNLVKISPLFTIWGSLFAKANAVYNPIVYGIS 2 | |||
1 HPKYRAALFAKFPSLACAAEPSSDAVSTTSGTTTVTDNEKSNA* 0 | |||
>LMSb_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01020829 18362345 full 78% bee | |||
0 MSFVSGPSHAAYTWAAQGGGFGNQTVVDKVPPEMLHMVDVHWYQFPPMNPLWHGLLGFAIGVLGTISIIGNGMVIYIFTTTKSLRTPSNLLVVNLAISDFLMMLWMSPAM 0 | |||
0 VINCYYETWVLGPLFCELYGMMGSLFGCGSIWTMTMIAFDRYNVIVKGLSAKPMTIKGALIRIFAIWLFTILWTIAPLFGWNR 2 | |||
1 YVPEGNMTACGTDYLTKDIVSRSYILFYSIFVYFMPLFLIIYSYFFIIQAVAAHEKNMREQAKKMNVASLRSAENQSTSAECKLAK 0 | |||
0 VALMTISLWFMAWTPYLIINFAGIFETTKISPLFTIWGSLFAKANAVYNPIVYGIR 2 | |||
1 HPKYRAALFQRFPSLACATEPAPGADTMSTTTTVTEGEKSVA* 0 | |||
>LMS_manSex Manduca sexta (moth) Ecdy.Inse.Lepi L78080 full G? | |||
MDPGPGLAALQAWAAKSPAYGAANQTVVDKVPPDMMHMIDPHWYQFPPMNPLWHALLGFTIGVLGFVSISGNGMVIYIFMSTKSLKTPSNLLVVNLAFSDFL | |||
MMCAMSPAMVVNCYYETWVWGPFACELYACAGSLFGCASIWTMTMIAFDRYNVIVKGIAAKPMTSNGALLRILGIWVFSLAWTLLPFFGWNRYVPEGNMTACGTDYLSKSWVSRSY | |||
ILIYSVFVYFLPLLLIIYSYFFIVQAVAAHEKAMREQAKKMNVASLRSSEAANTSAECKLAKVALMTISLWFMAWTPYLVINYTGVFESAPISPLATIWGSLFAKANAVYNPIVYG | |||
ISHPKYQAALYAKFPSLQCQSAPEDAGSVASGTTAVSEEKPAA* | |||
>LMS_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BAAB01068714 11549248 full G? Bcop boceropsin Bomopsin1 tandem misassembly CK529859 hemocyte larva CK552244 fat body (pupa) | |||
0 MISLDPGPGMAALQAWGGQVAAYGAANQTVVDKVPPDMLHMVDPHW 2 | |||
1 YQFPPMNPLWHALLGFTIGVLGFISISGNGMVIYIFMSTK 0 | |||
0 SLKTPSNLLVVNLAFSDFLMMCAMSPAMVVNCYNETWVF 1 | |||
2 GPFACELYACAGSLFGCASIWTMTMIAFDRYNVIVKGIAAKPMTNNGALLRILGIWAFSLAWTLAPFFGWNR 2 | |||
1 YVPEGNMTACGTDYLSKDWFSRSYILIYSVFVYFAPLLLIIYSYFFIVQ 0 | |||
0 AVAAHEKAMREQAKKMNVASLRSSEAANTSAECKLAK 0 | |||
0 VALMTISLWFMAWTPYLVINYTGVFESAPISPLATIWGSLFAKANAVYNPIVYGIR 2 | |||
1 HPKYQAALYARFPSLQCQSAPPDDGGSVASGATAVSEEKPAA* 0 | |||
>LMS_papXut Papilio xuthus (butterfly) Ecdy.Inse.Lepi AB007424 9547302 full G? Rh2 | |||
MAIANLEPGMGASEAWGGQAAAFGSNQTVVDKVTPDMMHLIDPHWYQFPPMNPMWHGLLGFTIGVLGFISITGNGMVVYIFTSTKSLKTPSNLLVVNLAFSD | |||
FLMMLCMAPPMLINCYYETWVFGPLACELYACAGSLFGSISIWTMTMIAFDRYNVIVKGIAAKPMTINGALLRILGIWLFSLAWTIAPMLGWNRYVPEGNMTACGTDYLSKSWLSR | |||
SYILVYSIFVYYTPLLLIIYSYFFIVQAVAAHEKAMREQAKKMNVASLRSSEAANTSAECKLAKVALMTISLWFMAWTPYLVINYTGVFETAPISPLATIWGSVFAKANAVYNPIV | |||
YGISHPKYRAALYQKFPSLACQPSAEETGSVASGATTACEEKPSA* | |||
>LMS_schGre Schistocerca gregaria (locust) Ecdy.Inse.Orth X80071 full G? | |||
MASASLISEPSFSAYWGGSGGFANQTVVDKVPPEMLYLVDPHWYQFPPMNPLWHGLLGFVIGVLGVISVIGNGMVIYIFSTTKSLRTPSNLLVVNLAFSDFL | |||
MMFTMSAPMGINCYYETWVLGPFMCELYALFGSLFGCGSIWTMTMIALDRYNVIVKGLSAKPMTNKTAMLRILFIWAFSVAWTIMPLFGWNRYVPEGNMTACGTDYLTKDWVSRSY | |||
ILVYSFFVYLLPLGTIIYSYFFILQAVSAHEKQMREQRKKMNVASLRSAEASQTSAECKLAKVALMTISLWFFGWTPYLIINFTGIFETMKISPLLTIWGSLFAKANAVFNPIVYG | |||
ISHPKYRAALEKKFPSLACASSSDDNTSVASGATTVSDEKSEKSASA* | |||
>BCR_limPol Limulus polyphemus (horseshoe_crab) Ecdy.Chel.Mero FJ791252 full G? ventral eye | |||
MSTGSYFIGNSTAPRSSGWWSYDPGLSVRDTAPENIKHLISDHWSKFPAVNPMWHYLLGLIYIVLGIASLTGQSVVLYLFAKTKPLRTPANMLIVNLAFSDF | |||
MMMITQFPVFIINCLGGGAWQLGPLLCEITGFAGGLFGYGSIVTLAVISIDRYNVIVRGFSASPLTHARSAVFILVIWAWTLGWALPPFFGWGRYVPEGILNSCSFDYLTRDWATV | |||
SYIMGCWICEYALPLMVIIYCYIFIVKAVCDHERHLREQAKKMNVASLRSNVDTQKASAEMRIAKVALVNVLLWVVSWTPYAAIAMIGIAGDQMLITPLRSALPALAGKAASVYNP | |||
IVYAISHPKFRLAMQKEIPCCCINEPQPQSDTSSEMSTKTSVATVNGEDSTAGGTTNN* | |||
>BCR_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran BAG80976 18984904 full G? RhA | |||
MAAYTEAWNASEEILVRMARAVPSVAWGYPAGVSIADLVPSDMKTMVHSHWNKFPPVNPMWHYLLGMVYIILGTVSIAGNSLVISLFTKTKELRTPANMFVVNLAFSDLCMMITQFPMFVYNCFNGGMWLFGPFLCELYA | |||
ATGAVFGLCSICTLACIAFDRYNLIVKGMSGPKMTSKRATILIAFCWAYAIGWSLPPFFGWGRYIPEGILDSCSFDYLTRDSSTKSFGLCLFFFDYVTPLSIIVFAYFHIVRAIFEHEKILREQAKKMNVTSLRSNADQN | |||
AQSAEIRIAKVALINISLWVAMWTPYATIVLQGLLGNQENITPLVSILPALIAKSASIYNPVIYAISHPRYRVALQQKLPWFCIHEEEKKPISDTDSAKTEASSS* | |||
>BCR2_triLon Triops longicaudatus (tadpole_shrimp) Ecdy.Crus.Bran BAG80981 18984904 full G? RhA | |||
MATYTEAWNASEEILVRMVRAAPSVAWGYPTGVSIVDLVPSDMKTMVHSHWSKFPPVNPMWHYLLGLVYIVLGTVSIAGNSLVISLFTKTKELRTPANMFVVNLAFSDLCMMITQFPMFVYNCFNGGMWLFGPFLCELYA | |||
ATGAVFGLCSICTLACIAYDRYNLIVKGMSGPKMTSKRATILIAFCWSYAIGWSLPPFFGWGRYIPEGILDSCSFDYLTRDSSTKSFGLCLFFFDYITPLSIIVFAYFHIVRAIFEHEKILREQAKKMNVTSLRSNADQN | |||
AQSAEIRIAKVALINISLWVAMWTPYATIVLQGLLGNQENITPLVSILPALIAKSASIYNPVIYAISHPRYRIALQQKLPWFCIHEEEKKPISISDTDSAKTETSSS* | |||
>BCR1_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran AB293431 18984904 full G? | |||
MANASHYEALQQEFNPWALPESFTLYAYAPEDVRAFLHPHWHNFPATHPAIYYLFGLVYLVLGVTSVG | |||
GNYLVLRIFTKFQELRRPSNVLVINLALSDMLLMLTLFPECVYNFLSGGPWRFGDLGCQIHAFCGALFGYNQ | |||
ITTLVFISYDRFNVIVRGMGGTPLTYARVSAMVAFSWLWATGWSVAPLVGWGGYALDGMLGTCSFDYVTR | |||
TWNNRSHILAATAFMWVIPVLIIAGCYWFIVQAVFKHEAELKAQAKKMNVASLRSNADQQQVSAEIRIAK | |||
VAITNVVLWLSAWTPFMVISNLGIWADPQQVTPLVSSLPVLLSKTSCSYNPLVYAISHPKYRECLKTLVPWICIVLPNDRRGGDNVSSSSSRTEASGKAETVDA* | |||
>BCR2_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran AB293429 18984904 full G? | |||
MSSGVFNSTDPIALARVSAGSNAHQQVGYNILIKTDGLSVRDVAPLDMHHLLHSHWDAYPPADPRIHYLL | |||
GMLYFFLGIAACMGNVLVLHIFGKHKNLRSPTNTLLMNLAFCDLMIFIGLYPEMLGNIFMNDGTWMWGDV | |||
ACRIHAWFGLVFGFGQMQTLMYMSIDRYNVIVKGLSAQPLTYKKVTQWLAQVWIVSLFWGTAPFFGFGNF | |||
ALDGILNTCSFDYFSRDMLSMSYIVSACVWAYVIPLIVIIFCYTFIVRAVFEHEETLRQQAAKMNVTSLR | |||
SSANSEDTSAEFRIAKIAMINVCLWLWAWSPFTIVSFIGIFGNQAIITPYLSSLPVILAKTSSVYNPIVYALSHPRYQAALKEEFAWLCVKTNSGNSGSSDTKSSVTMESSQPA* | |||
>BCR3_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran AB293432 18984904 full G? | |||
MMHNFSEPRYEAQVVRYGDFAPGVSVRDMAPENVRYMVHLHWEKFPPPDPRVHTALGALYLIMGVMSAVG | |||
NVLVLYIFGKYKSLRSPTNVLVMNLAFCDLGLFVGLYPELLGNIFINNGPWMWGDVACKIHAWCGLAFGF | |||
GQMQTLMFVSMDRYYVIVKGLKAPPLTYWKVSVWLAMVWIVSIFWATSPFFGFGNLSVDGLLNTCSYDYY | |||
TRDLPTVAYIVGSCVHAYVLPLAVIIFCYSYIVQAVFHHERQLREQAAKMNVASLRSSGGKQDEMSAEFR | |||
IAKIALINCCLWLWAWTPFTVISFMGVLHDDQSIINPYVSSLPVLLAKTSAVYNPIVYGLSHPKFQQCLREEFGWNIGLPKKKDNDSKSVTSVETAMT* | |||
>BCR1_triLon Triops longicaudatus (tadpole_shrimp) Ecdy.Crus.Bran AB293434 18984904 full G? | |||
MSSSGFNSTDPIALARVSAGSNAHQQVGYNILIKTDGLSVRDVAPLDMHHLLHSHWDSYPPADPRIHYLL | |||
GMLYFFLGIAACVGNVLVLHIFGKHKNLRSPTNTLLMNLAFCDLMIFIGLYPEMLGNIFMNDGTWMWGDI | |||
ACRLHAWFGLVFGFGQMQTLMYMSIDRYNVIVKGLSAQPLTYKKVTQWLAQVWIVSLFWGTAPFFGFGNF | |||
ALDGILNTCSFDYFTRDMPAMSYIVGACVSAYVIPLIVIIVCYTFIVRAVFEHEETLRQQAAKMNVTSLR | |||
SSASAEDTSAEFRIAKIAMINVCLWLWAWSPFTIVSFIGIFGNQAIITPYLSSLPVILAKTSSVYNPIVYALSHPKYQAALKEEFAWLCVKTNAGNSGSSDTKSSVTMESNQPA* | |||
>BCR2_braKug Branchinella kugenumaensis (fairy_shrimp) Ecdy.Crus.Bran AB293438 18984904 full G? Rhd | |||
MLNNSEPSFAAYSVADGIWYPAGTKQIDGAPADVIAMTHAHWKQFPPSNPAWNYLFGVIYFFLWIVNHI | |||
GNGLVIWIFLKTKSLRTPSNMLIVNLAIADFFMMLTQSPLYIISAFTSRWWIWGHFWCRFYGYTGGITGIA | |||
AIFTMVFIGYDRYNVIVKGMNGTKITKGMAFIMILWTWIYANAFCLPAMLEVWGNFSPEGLLSTCSFDYL | |||
NDNKFHGYFYTMYIFTGAYCVPMLLLMFFYSQIVKAVWAHEASSRAQAKKMNVESLRSNADANAESAEMR | |||
IAKVALTNVLLWVCIWTPYAFVAVTGAFGNRQILTPLVAQLPSLICKMASCLNPLVYAISHPKYRQVLQK | |||
ELPWFCIHEPEDKKSDATSVGSATTTATA* | |||
>BCR3_braKug Branchinella kugenumaensis (fairy_shrimp) Ecdy.Crus.Bran AB293437 18984904 full G? | |||
MLNFSEPRFAAYSVAEGVWYPPGTTQIDGAPADIVALTHAHWKKFPPSNPAWNYLFACLYFFLWVINHI | |||
GNGLVIKIFLKTKSLRTPSNMLIVNLAIADFFMMLTQSPLFIISAFSSRWWIWGHFWCRFYGYTGGITGIA | |||
AIFTLVFIGYDRYNVIVKGMSGKRISKGMAFGMIVWTWVYANVFCLPPMLQVWGDFSPEGMLSTCSFDYL | |||
NENRLHGPIFTGYIFFGAYCVPMFLLFFFYSQIVKAVWAHEAALKAQAKKMNVESLRSNADANAESAEVR | |||
IAKVALTNVLLWICIWTPYAFVAVTGAFGNRQILTPLVAQLPSLICKCASSLNPIVYAISHPKFRQVIQK | |||
DYPWFCIHEPESSADTKSVTSGQTQVAA* | |||
>BCRa_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran AB293433 18984904 full G? RhA | |||
0 MSNNLSSGYSSVAYRSEGASVLWGYPPGLSIVDLVPDDMKEFIHPHWNKFPPVNPMWHYL 2 | |||
1 LGVIYVILGITSVT 1 | |||
2 GNSLVVHLFAKTRDLRTPANMFVINLAFSDLCMMITQFPMFVFNCFNGGVWLFGPLFCELYACTGSIFGLCSICTMAAISYDRYNVIVNGMNRRRMTY 1 | |||
2 GRAGGLILFCWIYAIGWSIPPFVGWGKYIPEGILDSCSFDYLTRDTM 0 | |||
0 TISFTCCLFAFDYCVPLIIIIFCYYHIVRAIVHHEDALRDQAKKMNVSSLRSNADQKSQSAEIRVAKIAMMNITLWVAAWTPYAAICLQGAVGNQDKITPLVTILPALIAKSASIFNPVVYAISHPKYRL 0 | |||
0 ALQKALPWFCIHEKEEKEPPQDRREDSQSIATTNTNSSDVSLP* 0 | |||
>BCRa_hemSan Hemigrapsus sanguineus (crab) Ecdy.Crus.Mala D50583 9318091 full G? BcRh1 compound eye R1-R7 blue-green 480nm | |||
MANVTGPQMAFYGSGAATFGYPEGMTVADFVPDRVKHMVLDHWYNYPPVNPMWHYLLGVVYLFLGVISIAGNGLVIYLYMKSQALKTPANMLIVNLALSDLI | |||
MLTTNFPPFCYNCFSGGRWMFSGTYCEIYAALGAITGVCSIWTLCMISFDRYNIICNGFNGPKLTQGKATFMCGLAWVISVGWSLPPFFGWGSYTLEGILDSCSYDYFTRDMNTIT | |||
YNICIFIFDFFLPASVIVFSYVFIVKAIFAHEAAMRAQAKKMNVTNLRSNEAETQRAEIRIAKTALVNVSLWFICWTPYAAITIQGLLGNAEGITPLLTTLPALLAKSCSCYNPFV | |||
YAISHPKFRLAITQHLPWFCVHEKDPNDVEENQSSNTQTQEKS* | |||
>BCRb_hemSan Hemigrapsus sanguineus (crab) Ecdy.Crus.Mala D50584 9318091 full G? BcRh2 compound eye R1-R7 blue-green 480nm | |||
MTNATGPQMAYYGAASMDFGYPEGVSIVDFVRPEIKPYVHQHWYNYPPVNPMWHYLLGVIYLFLGTVSIFGNGLVIYLFNKSAALRTPANILVVNLALSDLI | |||
MLTTNVPFFTYNCFSGGVWMFSPQYCEIYACLGAITGVCSIWLLCMISFDRYNIICNGFNGPKLTTGKAVVFALISWVIAIGCALPPFFGWGNYILEGILDSCSYDYLTQDFNTFS | |||
YNIFIFVFDYFLPAAIIVFSYVFIVKAIFAHEAAMRAQAKKMNVSTLRSNEADAQRAEIRIAKTALVNVSLWFICWTPYALISLKGVMGDTSGITPLVSTLPALLAKSCSCYNPFV | |||
YAISHPKYRLAITQHLPWFCVHETETKSNDDSQSNSTVAQDKA* | |||
>BCR_porPel Portunus pelagicus (sand_crab) Ecdy.Crus.Mala EF110527 19040762 full G? horrible distal frameshifts | |||
MANSTGPQMAFYGSQDMTYGYPEGVSIVDFVRPEIKPYVHQHWYNYPPVNPMWHYLLGVIYLCLGFISIIGNGMVIYLFAKCQALRTPANILVVNLALSDLI | |||
MLTTNVPFFTYNCFNGGVWMFSATYCEIYGCLGAITGVTSTWLLCMISFDRYNIICNGFNGPKLTNGKAIILAFISWAISVGFGIAPLFGWGKYILEGILTSCSYDYLTQDFNTRS | |||
YNIIIFVFDYFLPAAIIIFSYVFIVKAIFAHEAAMRAQAKKMNVTnLRSGEAESQRAEIRIARTALVNVSLWFICwTPYALISLQgvlgdlsginlLVTTLPALLARSCSW*</font> | |||
<font color ="red">>RGR1_homSap Homo sapiens (human) Deut.Euth.Euar NM_001012720 18474598 full G? syn(+PCDH21 -LRIT1 -GRID1 -WAPAL) RPE Mueller exon-skipping splice | |||
0 MAETSALPTGFGELEVLAVGMVLLVE 1 | |||
2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 | |||
1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 | |||
2 RSQLAWNSAVSLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 | |||
1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 | |||
0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 | |||
0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKREKDRTK* 0 | |||
>RGR1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono genomic frag G? last D in DRY motif Atlantogenata ERY other placentals GRY | |||
0 1 | |||
2 ALLGLCLNGLTIASFRKIKELRTPSNLLVVSLALADSGICLNALMAALSSFLR 2 | |||
1 HWPYGAEGCRLHGFQGFATALASISLSAAIGWDRYLRHCS 1 | |||
2 RSKPQWGTAVSTVLFAWGFSAFWSMMPILGWGQYDYEPLRTCCTLDYSKGDR 2 | |||
1 NFTTYLFAVAFFNFVIPLFIMLTSYQSIEQRFKKSGLFK 0 | |||
0 LNTRLPTRTLLFCWGPYALLCFYATVENVTFISPKLRM 0 | |||
0 IPALIAKTVPVIDAFTYALRNEDYRGGIWQFLTGQKIERVEVENKIK* 0 | |||
>RGR1_galGal Gallus gallus (chicken) Deut.Saur.Arch NM_001031216 14985289 full G? syn(+PCDH21 -LRIT1 +CHAT -PARG) retinal ganglia | |||
0 MVTSHPLPEGFTEIEVFAIGTALLVE 1 | |||
2 ALLGFCLNGLTIISFRKIKELRTPSNLLVLSIALADCGICINAFIAAFSSFLR 2 | |||
1 YWPYGSEGCQIHGFQGFLTALASISSSAAVAWDRYHHYCT 1 | |||
2 RSKLQWSTAISMMVFAWLFAAFWATMPLLGWGEYDYEPLRTCCTLDYSKGDR 2 | |||
1 NYITFLFALSIFNFMIPGFIMMTAYQSIHQKFKKSGHYK 0 | |||
0 FNTGLPLKTLVICWGPYCLLSFYAAIENVMFISPKYRM 0 | |||
0 IPAIIAKTVPTVDSFVYALGNENYRGGIWQFLTGQKIEKAEVDSKTK* 0 | |||
>RGR1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur BC135113 full G? retina G protein receptor syn(+PCDH21 -LRIT1 +CHAT -PARG) | |||
0 MVTSYPLPEGFTETEVFAIGTTLLVE 1 | |||
2 ALLGLLLNGLTLLSFYKIRELRTPSNLFIISLAVADTGLCLNAFVAAFSSFLR 2 | |||
1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1 | |||
2 RSKLHWSTAVSVVFFIWGFSAFWSAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 | |||
1 NYISYLFTMAFFEFLVPLFILMTAYQSIYQKMKKSGQIR 0 | |||
0 FNTSMPVKSLVFCWGPYCLLCFYAVIQDATILSPKLRM 0 | |||
0 IPALLAKTSPAVNAYVYGLGNENYRGGIWQYLTGQKLEKAETDNKTK* 0 | |||
>RGR1_cynPyr Cynops pyrrhogaster (newt) Deut.Amph.Batr FS290827 frag G? 20090923 lens regenerating iris | |||
0 GFTEIEVFGLGTALLIE 1 | |||
2 ALLGFILNGLTLLSFYKIRSLRTPHNFLIVSLALADTGVCINAFIAAFSSFLR 2 | |||
1 YWPYGSEGCQIHGFQGFVTALSSISSCGVIAWDRYNQYCT 1 | |||
2 RTKLQWGTAISLVSFVWAFSAFWSVMPLLGWGQYDYEPLRTCCTLDYTKGDK 2 | |||
1 NFISYLFPLAFFEFVIPLFIMLTAYQSVEQKFKKTGQHK 0 | |||
0 FNTGLPVKTLVMCWGPYSLLCFYATIENATTISPKIRM 0 | |||
0 LPAILAKTAPAINAFLYGMGNESYRGGIWQFlTGQKIEKAEVDNKTK* 0 | |||
>RGR1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? syn(+PCDH21 -LRIT1 +CHAT -PARG) retinal ganglia | |||
0 MVSSYPLPDGFTDFDVFSLGSCLLVE 1 | |||
2 GLLGILLNAVTIAAFLKVRELRTPSNFLVFSLAVADIGISMNATIAAFSSFLR 2 | |||
1 YWPYGSDGCQTHGFQGFVTALASIHFIAAIAWDRYHQYCT 1 | |||
2 RTKLQWSSAITLAVFVWLFTAFWSAMPLIGWGEYDYEPLRTCCTLDYTKGDR 2 | |||
1 NYVSYLIPMAIFNMAIQVFVVMSSYQSIAQKFKKTGNPR 0 | |||
0 FNPNTPLKAMLFCWGPYGILAFYAAVENATLVSTKLRM 0 | |||
0 MAPILAKTSPTFNVFLYALGNENYRGGIWQLLTGEKIDVPQIENKSK* 0 | |||
>RGR1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? retina G protein receptor | |||
0 EGFTDFEVFGLGTALLVE 1 | |||
2 GLVGLLLNGLTLLAFYKIKELRTPSNLLITSLALSDFGISMNAFIAAFSSFLR 2 | |||
1 YWPYGSEGCQTHGFHGFLMALASINACAAIAWDRYHQNCS 1 | |||
2 RSRLQWSSAITVTVFIWGIAAFWSAMPLLGWGVYDYEPLRTCCTLDYSKGDR 2 | |||
1 NFISFFIIMGSFEFIFPIFIMLSSYQSCKSKFKKNGQVK 0 | |||
0 FNTGLPVKTLIFCWGPYSLLCFYATIENITILSPKLRM 0 | |||
0 * 0 | |||
>RGRa_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni AB079882 12687683 full G? Ci-opsin3 syn(PPT1 NOMO2+ FOXC2- FAHD1-) larval photoisomerase | |||
0 MEVNDKRVYGVLMGLL 1 | |||
2 GLLTITGYSLLFVIFAKRPDLKKKNKFLLSLATSDLLITVHVFASTIAAFAPQWPFGDLGCQ 0 | |||
0 VDAFIGMAPTFISIAGAALIAKDKYYRFCKPKM 1 | |||
2 VGRNYSFHVYLTWTMGIIGGALPFIGFGRYGFETDDVTWRTGCLLDFKSISA 2 | |||
1 KYSFYIILISTVWFVWPVYKLVSSYMKISTKINKFYP 0 | |||
0 LLFVVPVQMAIGLLPYAIYAMVSITIGVSAVPYFCVVINN 0 | |||
0 LAAKVFVGSNPFIYIYFDPELRESCKQIFCSPPAPTNDKISEDSKDE* 0 | |||
>RGRa_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni genomic full G? Ci-opsin3 larval visual cycle photoisomerase | |||
0 MDFSSKRTYGIAMAAL 1 | |||
2 GFIAWVGYGLLFVIFAKSPDLKKKNRFLFSLAVSDLLITIHVVASVVASFQSEWPFGSIGCQ 0 | |||
0 LDAFIGMAPTFISIAGAALVAKDKYYRICKPKM 1 | |||
2 LVGRNYSFSIYANWTLGIIGGLLPFFGFGQYGFETDDLSLRTGCLLDFKTVSA 2 | |||
1 KYRFYIVFISLVWFVWPLYKLTSHYIKISAKLDRFHP 0 | |||
0 LMFVVPLQMLVSLLPYAIYAMISITVGVSSAPYYLVAVNN 0 | |||
0 IAAKVFIGTNPFIYIYFDPELRLACKNLFKYSSTPVQDQIQDKKDE* 0 | |||
>RGRb1_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni AB078611 12373590 full G? CiNut entire neural tube | |||
0 MEIDFGFARTVYGVALLLM 1 | |||
2 VFITLLGYAVYFGAIWRSKTLQTRHIWLTSLACGDIIMMVHLILESLSSLGMGHRPRQNFECQ 0 | |||
0 VGALVGLFSGYVTIASITWIAIDRYYRQCKPEK 1 | |||
2 VGVNYCFYVIIVWAMSFLAASGPALGFGAYESAEENTVKCLIDLNKKDT 2 | |||
1 NSRLYIILVSAVWFVYPFVKMILYNKKLVQEAKEPQP 0 | |||
0 MAFAVPLTFFLCYLPFAIYASLKITVGLPPLNSMVVASIY 0 | |||
0 MLPKVISVVNPYLYMRSDPELLAACRHVVGLTDGKKAV* 0 | |||
>RGRb2_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni genomic full G? retina G protein receptor | |||
0 MELEFGITRTAYGIAMLMM 1 | |||
2 AAVATFGYSVYILAIWSSKKLQTKHIWLTSLACADLLMMVHLFMDGLSSFHQGRRPKGIFECQ 0 | |||
0 VSAHMGLFSGFVSIASMTWICIDRYYRKFKPEK 1 | |||
2 VGVNYCFYVIIVWAMSFLAASGPALGFGAYESAEENTVKCLIDLENTDM 2 | |||
1 NTIKYFVVVGFLFFFYPIFKMIKYNTKFAYKSEEEKA 0 | |||
0 VVIAAPVSFVLGYLPYLVYACLKLTIGLPPLNQASIAFL 0 | |||
0 YLLPKFISVMNPYMYMRSDPELLRAAKRVVNFDFDQKIE* 0 | |||
>RGRb3_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni genomic full G? retina G protein receptor | |||
0 MELEFGIIRTAYGIAMLLM 1 | |||
2 AVVATFGYSIYLRAIWSSRKLQTRHIWLTSLACADLIMMVHLFMDGLSSFHQGRRPKGNFECQ 0 | |||
0 ASALMGLFSGFVSIGSVTWICIDRYYRRYKPEK 1 | |||
2 AGLNYCFYVIIVWGLSFLAASGPAMGFGTYESAQENTVKCLIDLDNTDG 2 | |||
1 NTIKYFMVIGVLYFFYPIFKMVKYNTLAQATEVEKT 0 | |||
0 VVIAAPATFVIGYLPYLSYAILKLTIGLPPVNEAFIAFV 0 | |||
0 ILPKFISVLNPYMYMRSDPELLRAAKDVVNFNFYTKMD* 0 | |||
>RGR2_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_001024436 full G? retina G protein receptor embryonic RPE brain gut embryo PA | |||
0 MASYPLPEGFTDFDMFAFGSALLVG 1 | |||
2 GLLGFFLNAISVLAFLRVREMQTPNNFFIFNLAVADLSLNINGLVAAYACYLR 2 | |||
1 HWPFGSEGCQLHAFQGMVSILAAISFLGAVAWDRYHQYCT 1 | |||
2 KQKMFWSTSITISCLIWILAVFWAAMPLPAIGWGVFDFEPLRTCCTLDYSQGDR 2 | |||
1 GYITYMLTITVLYLAFPVLVLQSSYSAIHAYFKKTHHYR 0 | |||
0 FNTGLPLKALLFCWGPYVVVCSLACFEDVSVLSPRLRM 0 | |||
0 VLPVLAKTSPIFHAVLYAYGNEFYRGGVWQFLTGQKSADKKK* 0 | |||
>RGR2_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? retina G protein receptor eye | |||
0 MAAYTLPEGFTEFDMFTFGTALLVG 1 | |||
2 GVLGFFLNAISIVSFLTVKEMRNPSNFFVFNLALADISLNVNGLIAAYASYLR 2 | |||
1 YWPFGQDGCSYHAFHGMISVLASISFMAAIAWDRYHQYCT 1 | |||
2 RQKLFWSTTLTMSSIIWILSIFWSAVPLMGWGVYDFEPMRTCCTLDYTRGDR 2 | |||
1 DYVTYMLTLVVLYLTFPAATMWSCYDSIYKHFKKVHQHR 0 | |||
0 FNTSMPLRVLLVCWGPYVVMCVYACFENVKVVSPKLRM 0 | |||
0 LLPVIAKTNPIFNALLYTFGNEFYRGGVWHFLTGHKIVDPVLKKSK* 0 | |||
>RGR2_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? retina G protein receptor eye | |||
0 MAAFALPEGFTEFDMFTFGSALLVG 1 | |||
2 GLIGFFLNAISIASFLRVKEMWNPSNFFVFNLAVADICLNVNGLTAAYASYLR 2 | |||
1 YWPFGQDGCTFHAFQGMIAVLASISFMGVIAWDRYHQYCT 1 | |||
2 RQKLFWSTTLTMSAIIWILSIFWAAVPLMGWGVYDFEPMRTCCTLDYTKGDR 2 | |||
1 DYVTYMLTLVFLYLMFPALTMWSCYDAIHKHFKKIHLHK 0 | |||
0 FNTSTPLRVLLMCWGPYVLMCIYACFENVKVVSPKLRM 0 | |||
0 LLPVVAkTNPIFNALLYSFGNEFYRGGVWHFLTGQKMVDPVVKKSK* 0 | |||
>RGR2_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full G? retina G protein receptor whole embryo | |||
0 MGTHTLPEGFTDFDMFTFGSALLVG 1 | |||
2 GLLGFFLNAISILAFLRVKEMRSPSSFLVFNLALADISLNINGLTAAYASYLR 2 | |||
1 YWPFGQEGCDYHGFQGMISVLASISFMAAIAWDRYHQYCT 1 | |||
2 RQKLFWSTSITISLIIWILSILWSAFPLMGWGVYDFEPMRIGCTLDYTKGDR 2 | |||
1 DYITYMLSLVFFYLMFPAFIMLSCYDAIYKHFKKIHYYR 0 | |||
0 FNTSLPLRVMLMCWGPYVLMCIYACFENVKLVSPKLRM 0 | |||
0 LPVIAKTNPFFNALLYSFGNEFYRGGVWNFLTGQKIVEPDVKKSKQK* 0 | |||
>RGR2_pimPro Pimephales promelas (minnow) Deut.Acti.Otoc genomic full G? retina G protein receptor liver brain PA | |||
0 MASYALPEGFSDFDMFAFGSALLVG 1 | |||
2 GLLGFFLNLISVLAFLRVREIQTPNNFFIFNLAVADLSLNINGLVAAYASYLR 2 | |||
1 YWPFGSEGCQIHGFQGMVSILASISFLGAIAWDRYHLYCT 1 | |||
2 KQKMFWSTSGTISALIWILAVFWAALPLPAIGWGVFDFEPMRTCCTLDYTIGDR 2 | |||
1 NYISYMLTITVLYLAFPVLIMQSSYNGIYAHFKKTHHFK 0 | |||
0 FNTGLPLKMLLFCWGPYVLMCTYACFENASLVSPKLRM 0 | |||
0 VLPVLAKTSPIFHAAMYAYGNEFYRGGIWQFLTGQKPADKKK* 0</font> | |||
<font color ="#9900FF">>PER1_homSap Homo sapiens (human) Deut.Euth.Euar NM_006583 17167409 full G? RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6) | |||
0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 | |||
2 GMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 | |||
0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDV 1 | |||
2 GRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 | |||
1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTK 0 | |||
0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 | |||
1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 | |||
>PER1_monDom Monodelphis domestica (opossum) Deut.Meta.Dide genomic full G? RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6) | |||
0 MFKNNSVKTLAPEKEGPSVFSPIEHKIVAAYLITA 1 | |||
2 GVISIVSNVIVLGIFVKYKALRTATNTIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYDGCQ 0 | |||
0 IYAGLNIFFGMASIGLLTAVAIDRYLTICQPDL 1 | |||
2 GRMTSYNYTLMILTAWVNGFFWALMPIVGWAGYAPDPTGATCTINWRKNDV 2 | |||
1 SFVSYTMTVITINFAMPLGVMFYCYYNVSQKMKQYSPSNCPDHINRDWSNQVAVTK 0 | |||
0 MSVVMILMFLLAWSPYSIVCLWASFGDPKEIPPAMAIVAPLFAKSSTFYNPCIYVAANKK 2 | |||
1 FRRAISAMIRCQTHQSMPISNALPMN* 0 | |||
>PER1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono XM_001506366 full G? RRH peropsin | |||
0 MRRNDSANLLESEHHDRSAFSQTDHNIVAAYLITA 1 | |||
2 GIMSIVSNVIVLGIFVKFEELRTATNAIIINLAVTDIGVSGIGYPMSAASDLHGSWKFGHAGCQ 0 | |||
0 IYAGLNIFFGMSSIGLLTVVAVDRYLTICRPAI 1 | |||
2 GRKMTRSNYTAMILAAWMNGFFWASMPLLGWASYASDPTGATCTINWRKNDA 2 | |||
1 SFISYTMTVIAVNFAVPLIVMFYCYYNVSKAMRQYPASRVLENLNIDWSEQVDVTK 0 | |||
0 MSVVMILMFLMAWSPYSIVCLWSSFGDPKKISPAVAIMAPLFAKSSTFYNPCIYVVANKK 2 | |||
1 FRRAMLSMVQCQTHREITITDVLPMNRSRSPH* 0 | |||
>PER1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6) | |||
0 METLAEVSTLLPAGTGTVNISDASSEVHSVFSQSEHNIVAAYLITA 1 | |||
2 GVISILSNIIVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYVGCQ 0 | |||
0 IYAGLNIFFGMASIGLLTVVAIDRYLTICRPDI 1 | |||
2 GRRISGRHYTAMILAAWINAVFWSVMPVVGWSSYAPDPTGATCTINWRKNDV 2 | |||
1 SFVSYTMSVVAVNFVVPLMVMFYCYYNVSRTMKGYGSRSSLGGINADWSDQTDVTK 0 | |||
0 MSMVMIVMFLVAWSPYSIVCLWSSFGDPRKIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 | |||
1 FRRAILSMVQCKSRQEVTLDNHFPMNVSQSTLTT* 0 | |||
>PER1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? RRH peropsin syn(+GPR68 -GNPDA1 -ENPEP -C14orf100) | |||
0 MGIDPEVNVTDDVTLYGGKSAFTQLEHNIVAGYLITA 1 | |||
2 GVISLFSNIVVLLMFWKFKELRTATNFIIINLAFTDIGVAGIGYPMSAASDIHGSWKFGYAGCQ 0 | |||
0 IYAALNIFFGMASIGLLTVVAIDRYLTICRPDI 1 | |||
2 GQKMTMQSYNLLILAAWLNAVFWSSMPVVGWASYAPDPTGATCTINWRQNDV 2 | |||
1 SFISYTMAVIAVNFVLPLSAMFYCYYNVSATVKRYKASNCLDSANIDWSDQMDVTK 0 | |||
0 MSIVMIIMFLVAWSPYSIVCLWASFGDPKTIPAPMAIIAPLFAKSSTFYNPCIYVIANKK 2 | |||
1 FRRAIIGMVRCQTRQRITINSQVPMTTSQQPLTQ* 0 | |||
>PER1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? RRH peropsin | |||
1 LFVSYTMTVIAVNFVVPLSVMFFCYYNVSKTMSRFISSPSPENINLDWSDQLDVTK 0 | |||
0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 | |||
1 FRKAIMAMICCQNRQEITINHTLPMTISRVPLTE* 0 | |||
>PER1_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002228504 12435605 full Go RRH peropsin Amphiop1 | |||
0 MNASPSSWLPSGELFTDSPENSSEWPWTDGPTDTAWHHHQTVDPVTYGGYLASAVYLTIT 1 | |||
2 GLIAFVGNIFAIIVFLTEKEFRKKEHNSFALNLAIADLSVCVFAYPSSTIS 1 | |||
2 GYAGEWMLGDVGCTIYGFLCFTFSLTSMVTLCAISVYRYIVICKPQY 1 | |||
2 AHLLTHRRTNYVILGIWLYALVFSVPPLFGVNRYTYEPIS 2 | |||
1 ITCSLDWNVQHVGETIYTAAVIIIVYVLNVSIMCFCYFNIIFKSANLKFAALASEKTRTAAKKDIWKTSM 0 | |||
0 MCLAMVVSFLIAWTPYAVSSTWDILTEEDLPIIATILPTMFAKSSCMMNPIIYSCCNGKFRQAALKTFSK 0 | |||
0 VGSSNKQNGQAQVEPRDPGFAVEPAGHQAFQMRVLPSSSAMTL* 0 | |||
>PER1_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050606 12435605 full G? RRH peropsin Amphiop1 introns from braFlo | |||
0 MNASPSSWLSSGEFFTDSPENSSEWPWTDGPTDTTWRHHQSVDSVSYEGYLASAIYITLT 1 | |||
2 GLIAFFGNVITITVFLTEKEFRKKQQNGFVLNLAIADLSVCVFAYPSSAIA 1 | |||
2 GYAGRWVLGDVGCTIYGFLCFTFALVSMVTLCVISIYRYILICKPQY 1 | |||
2 AHLLTHRRTVYVIIGTWLYALVFTVPPLVGVKRYTYEPMQ 2 | |||
1 ITCSLDWNVQHPGEKAYIAAVLVIVYVLQVLIMCFCYFNIIFKSANLKFAALASEKTKMAAKKDTWKTSV 0 | |||
0 MCLTMVVSFLIAWTPYAVSSTWDILSAEDLPIIATILPSLFAKSSCMMNPIIYACCNTKFRQAAVKSFRK 0 | |||
0 LCGMCKQKVPLSTPQVVLAMQRNTEFTSTVEPTGQAFPMRVLPSISATHTAL* 0 | |||
>PER2_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002209058 12435605 frag G? RRH peropsin Amphiop3 | |||
0 MIPTNNNTENNDLEWGLEKEHGVSATIMGVYLTIV 1 | |||
2 GLVSTVGNATVVLMFMLKWRQLCRKAPNLLIINLAAVDLCISVFGYPFSASSGFANQWLFSDAICT 0 | |||
0 LYGFSCFLLSMVSMHTLCLISAHRYITICRPEH 1 | |||
2 ASKLTMTRTILAVVGAWVYGISVAVPPLFGIA 1 | |||
2 GYTYESFGLSCTIDFHGTTVADMVYLSILIILCYVINVAVMGTCYFKIIRK 2 | |||
1 FSKHRFREVRDVRTSHQHSFERGVTL 0 | |||
0 RCILMTLFYLISWTPYTAVAVWTMVGPPPPVQLGMVAALTAKTHCAFNPILYMLMSE 0 | |||
0 VYRKLVLRTMCPCCFNKISNKLVRLPADDSKHSGNLDIFTVGYNTRDQAVQINKNAARRFCFVMET 0 | |||
0 ASDDLGIDDEVFAGQLGLCSRVKATEPGVEGFGGSEVPQSPSGTESEWSLSLLDFLPKRSSSKTAKASSLSETCSDNTVLSSAARK | |||
MAFLESSHQQSDREVCCIENRQAPEDTKPCKFAIESLGVRLPHKCCTASQVAGAPSRYAGMIETFTDSKGKTKKKAAVSLSEIDVKKP | |||
PPASKTWERRKTSKNTSRGQRVKRTFGKSRKHAYIVDC* 0 | |||
>PER2_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050607 12435605 full G? RRH peropsin Amphiop2 introns from braFlo | |||
0 MIPTNNNTENNDLEWGLEKEHGVSATIMGVYLTIV 1 | |||
2 GLVATVGNATVVLMFIMKWRQLCRKAPNLLVINLAAANLCITIFGYPFSASSGYAHQWLFPDAICT 0 | |||
0 LYGFSCFLLSMVSMHTLCLISAHRYITICRPEH 1 | |||
2 ASKLTMNRTVLAVIGTWLYAIAVAVPPLFNIA 1 | |||
2 RYTYEPSGLSCTIDFRVTTVADLVYLGSLIVLCYVIHVAVMATCYFKIIRK 2 | |||
1 FSRHRFRQVRDIRTSHQRSFEMGVTM 0 | |||
0 RCILMTLFYLLSWTPYTAVCIWTMVGPPPPVVVSMAAALIAKTHCAFNPILYAFMSE 0 | |||
0 VYRKLVFRTMCPCCFNRISCKFVGTPTGGSKVSANPDIFTVDYNSRDQAVQINKAPSRRFCFVMET 0 | |||
0 SEDLGSDDTGLTGHSGLWRSGAEVEGLGGLQVTQSPSVSGSELSLSLLDFLPPKPSGRAVSAKLPSPPALNSERATCP | |||
ESSQQPSDRPATGLRQYQKGDTTRSSVGDLILTEDDVTNLPPASETWGRKKSENPLSYRQTTRRTFGRSRKHSYIVD* 0 | |||
>PER3_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran AB050610 12435605 full G? RRH peropsin Amphiop3 | |||
0 MDIPTETPYGAGDDPAGTGWRWAETDQNGFHKYDHLIVGLYLFVI 1 | |||
2 GIIGTVENGITLATFTKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLR 0 | |||
0 SHWLFGGVGCQWYGFNGMFFGMANIGLLTCVAVDRYLVICRQDL 1 | |||
2 VDKVNYNTYGVMAALGWLFAAFWAALPLVGWAEYSLEPS 1 | |||
2 GTACTINWQKNDSLYISYVTSCFILGFALPLAVMMFCYWQ 0 | |||
0 ASCFVNKVLKGDISGDLTFPVAVNVDWEYQNHFSK 0 | |||
0 MCLAMVAAFVVAWTPYSVLFLFAAFGNPADIPAWITLLPPLIAKSSALYNPIIYIIANRRFRSAIFSMVKGQNPDVE 0 | |||
0 TLFARDFRISPIEDTGKEMSSMGNANA* 0 | |||
>PER3_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050610 12435605 full G? RRH peropsin Amphiop3 introns from braFlo | |||
0 MDIPTETPYGAEEDIGESAGWRWTETDKNGFHKYDHLIVGLYLFVI 1 | |||
2 GIIGTIENGITLATFSKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLR 0 | |||
0 SHWLFGGVGCQWYGFNGMFFGMANIGLLTCVAVDRYLVICRHDL 1 | |||
2 VDKVNYNTYGVMAALGWLFAAFWAALPLVGWAEYALEPS 1 | |||
2 GTACTINFQKNDSLYISYVTSCFVLGFVVPLAVMAFCYWQ | |||
0 ASCFVSKVLKGDIAGDLTFPVAANVDWEYQNHFSK 0 | |||
0 MCLAMVAAFVVAWTPYSVLFLFAAFWNPADIPAWLTLLPPLIAKSSALYNPIIYIIANRRFRNAICSMMKGQDPDVE 0 | |||
0 DDEHADEHRVRSIEDNDKEIISMVNLNMTV* 0 | |||
>PER2a_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XM_778236 full Go RRH peropsin overshoots iMet spread across tandem | |||
0 MAASVTESSATEAISRLEPEYMVPLTRTGYLLTAIYLTIV 1 | |||
2 GSIATVGNITVICVLCRYRTFRKRSINLLLINMAASDLGVSVAGYPLTTVSGYWGRWLFGDVGCQFYAFCVYTLSCSTISTHAAIAVYRYIYIVKTDL 1 | |||
2 RPKLTANFTSGVIVVIWVYAFFWTVTPFVGWSSYIYEPFGTSCSVNWVGRTISDISYMVACTIGVYLLQIFIMLYCYIRVAKK 2 | |||
1 IRGVDPGRTEEKDAGVVVFGRLRKREAKIDTHVTK 0 | |||
0 MCFMMMLTFIVVWAPYAVECLRAAHVHRISALSSVLPTMFAKSSCMVNPIIFLTSSSKFRQDLGKLWSRPSSQDSLQLEER | |||
NKTQRSLYVRHSELGSAHGNDTASVYYEKERIYIGEMRATSIQKEAELLQRDPELLSIASSTNSDVKFVVRDRPKRYTKR | |||
PVKPQGPRGPEMFTASGVTNKGSSTSDSGGQSTSSGTTGSKPKRSGRKASRQYSMKSQSEDTGEIFTLDGSALEMMSLRKL* 0 | |||
>PER2b_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XM_778236 17067569 full Go RRH peropsin no cdna inline tandem PER2_strPur | |||
0 MNSFSEESYVTDPTTTQPTLFLTPLSQTGYLLTALYLTLV 1 | |||
2 GIVSTIGNITVLCVLCRYGTFRKRSVNILLMNMAVSDLGVSVAGYPLTAISGYRGRWVFADIGCQFSGFCVYALSCSTISTHAVVAIYRYIYIVKPYH 1 | |||
2 RPRLSSSTSCLAILCIWTFTLFWTITPFFGWSSYTYEPFGTSCSINWYGKSLGDLTYIICCVVFVFILPIIIMLYCYIGVAKK 2 | |||
1 IKGIDPLRTEERDIAVVFGRLRKHETKIDTRVTK 0 | |||
0 ICFMMMASFIVVWTPYAVGSIWASKIGKISASASVLPTMFAKSSCMINPIIFLTSSSKFRADLGKLWNRPSSLEHTIRVEERSREQRSFF | |||
VRQSALPDAMVSRSASVYYDKERIYIGEMRAASIQKEADLLHRDPEAISIASSTSSSLQFVLKDRQNRYKKKAGEASKKGSNILHFPYDDTE | |||
GSMINNLMRPRSHSVTSDNISRVFAPSLKRPTKKRSMSHPDIPSTSADIFTVSPTTIKNLQKQ* 0 | |||
>PER1a_sacKol Saccoglossus kowalevskii (acornworm) Deut.Hemi.Ente ACQM01133041 full G? RRH peropsin 7 ests + ACQM01133041 | |||
0 MVTTDSLANSTDEPVPSILTLQQHYAASVTLLAL 1 | |||
2 AVIGTVLSSVNFRMLLSNPDYCSKAGNFFLSLAVTDL 1 | |||
2 CVCIFETPFSAFSHHAGFWIFGDTACQLYAFFGIFFGLVNIFMVTFISLDRYWATCSPVE 1 | |||
2 VELKSKYYTRMTALGWMVALFWAAAPVFGWSRYaMEPSMASCSIDYMTNDFSYVTYITCLTLTCYVVPIVVMVYCYVKASKNIKYTGKVTEWAHENNATK 0 | |||
0 ISRLCVLQLVFCWSLYGFNCMWTVVADDVETLPKMLTVLAPILAKTTPILNSGLYFLHNKKFRGAAVDMFKAKEE* 0 | |||
>PER1b_sacKol Saccoglossus kowalevskii (acornworm) Deut.Hemi.Ente ACQM01067921 frag G? RRH peropsin 0 ests | |||
0 1 | |||
2 gVLSVIGNSVVLEMFRRYKELLSPSAILLISLALADL 1 | |||
2 GLTIFGMSLSCVSSFAGRWLFGKFGCYFHGFAGMLFGLGSIGNLTVISIDRYIITCKRsL 1 | |||
2 QWSYRHYYALLAVAWSNALFWSMMPLFGWSSYALEPEGTSCTIDWMNNDNQYISYVSCVTVTCFILPCAVMTYDYLAAYMKMVKAGYTLSEETEKPNND 0 | |||
0 MCIALVAAFLLSWFPSATVFLWAAFGNPGNIPLSFTGVADAFTKIPAVFNPVIYVALNPEFRKYFGKTIGCRRKRKKPIAVRLNGSEQNVENTI* 0</font> | |||
<font color="magenta">>PER1_lotGig Lottia gigantea (limpet) Loph.Moll.Gast genomic full G? retinochrome | |||
0 MTAAEFSSFEHSIVGITYMVI 1 | |||
2 GISGTLLSLLVALTFIREKGLFKYGRAWLHISLAIANVGVVGAFPFSGSSSFSGR 2 | |||
1 WLYGSGMCTFYGFIGMFFGIAAIGNVFALCVERYLVSKKKDS 1 | |||
2 VDKVSNQFYWMITALVWINAFFWGIMPALGWTS 2 | |||
1 YDIEPSGTSCTIKWQNYDSGYPSFMAMLSLTCFLIPLPVALICLILSGTDKITEDKEEKTYFREDQLRS 0 | |||
0 TCTFLLILALIGWGPYCFICIWALFADTTQVSMLAAVIPPLAAKTMVLLYPVAYCQGNKRFKNAFLGMFIFNESPKQQ* 0 | |||
>PER1_aplCal Aplysia californica (sea_slug) Loph.Moll.Gast EB338056 17190607 full G? retinochrome from 8 8k contig to traces | |||
0 MNDDGNALDGAGVDPAATAAHVSTALGFTKFEHASVGALYMLF 1 | |||
2 CIVGVTLNLLTALTFYKDTKLTKGSQPWLHILLALANVGVVAPSPFPASSSFSGR 2 | |||
1 WLYGSTMCQIYAFEGMFIGIAAIGAVIALCIERYIACQRSGA 1 | |||
2 NDTQGWFYGWSITLVLGNALFWAIMPLLGWSR 2 | |||
1 YSVEHTGTSCSIDWKNPDESFVSYIMTLEVFSFGIPMMSAFFCLISASPRPQPQGGATAAAGSGQEQQGEDTACKGCFSEDQLRL 0 | |||
0 LCYVFIGFVLVGWGPFAYLCTLAVFSDARGISMLAAAIPPLACKAMVSAYPLAYAVVSPRFRQSFLALIGGGEKKKE* 0 | |||
>PER1_todPac Todarodes pacificus (squid) Loph.Moll.Ceph genomic 2226795 full G? retinochrome retinochrome 1991 seq dubious before GDPAH | |||
0 MFGNPAMTGLHQFTMWEHYFTGSIYLVL 1 | |||
2 GCVVFSLCGMCIIFLARQSPKPRRKYAILIHVLITAMAVNGGDPAHASSSIVGR 2 | |||
1 WLYGSVGCQLMGFWGFFGGMSHIWMLFAFAMERYMAVCHREF 1 | |||
2 YQQMPSVYYSIIVGLMYTFGTFWATMPLLGWAS 2 | |||
1 YGLEVHGTSCTINYSVSDESYQSYVFFLAIFSFIFPMVSGWYAISKAWSGLSAIPDAEKEKDKDILSEEQLTA 0 | |||
0 LAGAFILISLISWSGFGYVAIYSALTHGGAQLSHLRGHVPPIMSKTGCALFPLLIFLLTARSLPKSDTKKP* 0 | |||
>PER2_patYes Patinopecten yessoensis (scallop) Loph.Moll.Biva AB006455 9287291 full Go retinochrome depolarizing scop2 inner retina layer | |||
0 MPFPLNRTDTALVISPSEFRIIGIFISIC 1 | |||
2 CIIGVLGNLLIIIVFAKRRSVRRPINFFVLNLAVSDLIVALLGYPMTAASAFSNR 2 | |||
1 WIFDNIGCKIYAFLCFNSGVISIMTHAALSFCRYIIICQYGY 1 | |||
2 RKKITQTTVLRTLFSIWSFAMFWTLSPLFGWSSYVIEVVPVSCSVNWYGHGLGDVSYTISVIVAVYVFPLSIIVFSYGMILQEKVCKDSRKNGIRAQQRYTPRFIQDIEQRVTF 0 | |||
0 ISFLMMAAFMVAWTPYAIMSALAIGSFNVENSFAALPTLFAKASCAYNPFIYAFTNANFRDTVVEIMAPWTTRRVGVSTLPWPQVTYYPRRRTS | |||
AVNTTDIEFPDDNIFIVNSSVNGPTVKREKIVQRNPINVRLGIKIEPRDSRAATENTFTADFSVI* 0</font> | |||
<font color="#CC00FF">>PER_hasAda Hasarius adansoni (spider) Ecdy.Chel.Arac AB525082 19960196 full Go peropsin first ecdysozoan peropsin | |||
0 MDDNMSEIALADDMSTLSTQEPSENVYPYVFPLSTHTIVGTYLIII 1 | |||
2 GILGTLGNGLVLVTFLRFRVLVTPTTLLLVNLAVSDLGLILFGFPFSASSSLSAK 2 | |||
1 WIFGEGGCQWYAFMGFLFGSAHIGTLALLALDRYLIACRISL 1 | |||
2 RGKLTFKRYTQMITVVWTYAFFWALMPLLGWGR 2 | |||
1 YGLEPSVTTCTIDWQHNDSSYKSFLIVYFVLGFMVPFAIIAVSYIAIARRVGKKSKERPVVRDLWTNERSVTL 0 | |||
0 MAFILIVTFFVAWSPYAVLCLWTIFAEPNTVPPFLTLIPPLFAKSSTVVNPLIYFLSNPKLRTAILSTLSCCNEAPIQNIELPDSPERAANNDAI* 0 | |||
>PER_ixoSca Ixodes scapularis (tick) Ecdy.Chel.Arac ABJB010816023 frag Go peropsin mRNAs XM_002409761 XM_002435011 embryonated eggs | |||
1 WLFGATGCQAYAFMGFLFGSAHIGTLTLLALDRYLATCRIGF 1 | |||
2 RSKPTFKRYFQLLLLVWLYGLFWAVMPLLGWAR 2 | |||
1 YGLEPSFQSCTIDWRHNDSSYKSFTLVYFVLGFLVPACIVLVCYRTSAIHIRAPKPKTVRRDVTDDYWASEEMVTV 0 | |||
0 MVALIVVTFFFAWTPYAVLCLWAVFADTKSVPHLLAMVPPLFAKTASTINPFIYFLSNPCIRADVLQLLGCRARSSPHMAISSDAVEEERCCQDQA* 0</font> | |||
<font color ="#CC3366">>NEUR1_homSap Homo sapiens (human) Deut.Euth.Euar NM_181744 14623103 full G? OPN5 neuropsin index sequence | |||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | |||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | |||
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | |||
2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2 | |||
1 LHTVTTVRKSSAVLEIHEEV* 0 | |||
>NEUR1_calJac Callithrix jacchus (marmoset) Deut.Euth.Euar genomic full G? OPN5 neuropsin | |||
0 MALNHTSLPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | |||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | |||
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | |||
2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 | |||
1 LHTVTTVRKSSAVLEIHEEV* 0 | |||
>NEUR1_musMus Mus musculus (mouse) Deut.Euth.Euar genomic full G? OPN5 neuropsin | |||
0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 | |||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | |||
2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | |||
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQAGGLRGTKKKSLEDFR 2 | |||
1 LHTVTTVRKSSAVLEIHQEV* 0 | |||
>NEUR1_ochPri Ochotona princeps (pika) Deut.Euth.Euar genomic full G? OPN5 neuropsin | |||
0 MALNDTALPQDEHLPHYFRDGDPFASKLSWEADLVAGFYLTII 1 | |||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | |||
2 VVGKPFTIISCFCHRWVFGWIGCRLYGWADFFFGCGSLITMTAVSLDRYLK 1 | |||
2 GVWLKRRHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHGSHVLEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFSCCRTGGLKQTKKKSLEDFR 2 | |||
1 LHTVTTVRKSSAVLEIHQEv* 0 | |||
>NEUR1_canFam Canis familiaris (dog) Deut.Euth.Laur genomic full G? OPN5 neuropsin | |||
0 MALNHTARPQDERLPHYLREGDPFASKLSWEADLVAGFYLTII 1 | |||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | |||
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | |||
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASLGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGRLKATKKKSLEDFR 2 | |||
1 LNTVTTVRKSSAVLEIhQEV* 0 | |||
>NEUR1_bosTau Bos taurus (cow) Deut.Euth.Laur genomic full G? OPN5 neuropsin | |||
0 MALNHTAPPPDERRPPYLRDGDPFASKLSWEADLVAGFYLTII 1 | |||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAICDLGIS 1 | |||
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | |||
2 GIWLKRKHAYICLAVIWAYAAFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 | |||
1 LHTVTTVRKSSAVLEVHQEv* 0 | |||
>NEUR1_loxAfr Loxodonta africana (elephant) Deut.Euth.Afro genomic full G? OPN5 neuropsin | |||
0 MTLNHTAPPQDDRLPQYLQDGDPFTSKLSWEADLVAGFYLTII 1 | |||
2 GILSTFGNGYVLYMSCRRKKKLRPAEIMTINLAVCDLGIS 1 | |||
2 VVGKPFVIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | |||
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHMLEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLRATKKKSLEGFR 2 | |||
1 LHTVTTVKKSSAVLEVHQEv* 0 | |||
>NEUR1_dasNov Dasypus novemcinctus (armadillo) Deut.Euth.Xena genomic full G? OPN5 neuropsin | |||
0 MALNHTALPQDDRLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | |||
2 gILSTFGNGYVLYMSSKRKKKLRPAEIMTINLAVCDLGIS 1 | |||
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | |||
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLRATKKKSLEDFR 2 | |||
1 LHTVTTVRESSAVLEVHQEV* 0 | |||
>NEUR1_monDom Monodelphis domesticus (opossum) Deut.Meta.Dide genomic full G? OPN5 neuropsin | |||
0 MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | |||
2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | |||
2 VVGKPFTIISCFSHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 | |||
2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPVQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCQSGGQKAAKKESLRTYR 2 | |||
1 LHTVTTVRRSSAVLEIHQEv* 0 | |||
>NEUR1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono genomic full G? OPN5 neuropsin | |||
0 MTNYSAPQLGDYLPHYLREGDPFVSKLSWEADLVAGVYLVII 1 | |||
2 GVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 | |||
2 VVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 | |||
2 GTWLKRHHAYICLAIIWAYASFWATMPLVGLGNYAPEPFGTSCTLDWWLAQASVAGQAFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQNSHVLEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPIQFSVVPTLLAKSAAMYNPIIYQVIDCRISCCRLGGPKTGKKESLKNSR 2 | |||
1 SHSMSTIRKPSAVSGPHQEV* 0 | |||
>NEUR1_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full G? OPN5 neuropsin | |||
0 MASDCNSSSQEEYLPHYMQQEDPFASKLSREADIIAGFYLTVI 1 | |||
2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | |||
2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 | |||
2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGPKTLQKKSSLKESR 2 | |||
1 MYTISSHRDSAALSGTQLEV* 0 | |||
>NEUR1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? OPN5 neuropsin | |||
0 MAGNSSYREESGYIPHYERDSDPFASKLSREADIFAGVYLMAI 1 | |||
2 GILSTLGNGYVIYMACSRKKKLRPAEIMTINLAVCDLGIS 1 | |||
2 VTGKPFAIVSCFSHRWVFGWNACRWYGWAGFFFGCGSLITLTVVSLDRYLKICHLRY 1 | |||
2 GTWLKRRHAFIALAVIWAYATLWATLPLVGVGNYAPEPFGTTCTLDWWLAQASVKGQIFVLSMLFFCLLFPTMVIVFSYAKIIAKVKSSAKEVAHFDTRNQNNHTLEIKLTK 0 | |||
0 VAMLICAGFLIAWFPYAVVSVWSAFGQPDSIPIELSVVPTMMAKSASMYNPIIYQVIDCKPACCKKDKSLQNTTSR 2 | |||
1 VYTISTFRKSTTSAR* 0 | |||
>NEUR1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic frag G? OPN5 neuropsin | |||
0 MENETSISSGYIPHYLLRGDPFASKLSKEADIVAAFYILVI 1 | |||
2 GILSATGNGYVMYMTFKRKTKLKPPEIMTLNLAIFDFGIS 1 | |||
2 VSGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSFDRYLKICHLRY 1 | |||
2 gTWLKRHHAFLSVVFIWAYAAFWATMPVVGWGNYAPEPFGTSCTLDWWLTQASVSGQSFVMCMLFFCLIFPTVIIVFSYVMIIFKVKSSAKEVSHFDTRNKNNHSLEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVM | |||
>NEUR1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? OPN5 neuropsin | |||
0 MTAFDNSTALYSGYWLHDSLHGDPFVSKLSWEADIISACYLIVT 1 | |||
2 GLLSTLGNGYVIYLSITQKRKLKPPEILITNLAISDFGMS 1 | |||
2 VGGQPFLIISCFSHRWIFGWVGCRWHGWAGFFFGCGSLITMTVVSLDRYLKICHLQY 1 | |||
2 GSWLQRRHVFMSLAFIWFYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLARVSVSGLIFVLTILFFCLLLPIIIIVFSYIKIIAKVKSSAKEVAHFDSRIQNHHSLEMNLTK 0 | |||
>NEUR1_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag G? OPN5 neuropsin | |||
0 VAMMICAGFLLAWIPYAVVSVWSAFGAPDSVPVAVSMVPTMFAKSAAMYNPLIYQLLSRRGTGAHCCRCRKARGTLRRPR 2 | |||
>NEUR1a_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran FE548698 frag G? OPN5 neuropsinframeshifted cDNA FE548698 last exons missing | |||
0 MATTPADRLDGLTPAGRGATTAETHADDFASKLSREADIVIGVYLILI 1 | |||
2 GTGAILGNGRVLWLSYRCRARLRPVEMFVVSLAVADVGLSLVGHPFAAASSLMGRWSFGSAGCTW 1 | |||
2 YGFVVFFLGIASIASMTLMSIMRFMIVYKRYP 1 | |||
2 GQYPTRRASCVLVTAAWLYGLFWACAPLA 1 | |||
2 GWSQYQPEPYGLSCSVDWGGFSSDAGGSSFIICMLLFCTAVPVVIMVTSYAAIFVIYRQAQKGVVLNLQVNATFGGKRQRTERKLTL 0 | |||
0 IALAVCGGFLLAWLPYAVVGLWASVAGVDAVPLALASAAPLFAKSNSLWNPIIYLGMNERFR 2 | |||
>NEUR1b_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002202511 full G? OPN5 neuropsin from tracesseparate allele | |||
0 MATTPGLPLDGLAPTGRGVTAADTLDDDFASKLSREADIVIGVYLLLI 1 | |||
2 GTGSILGNGRVLWLSYRNWAKLRPVELFVVSLAVTDVGISVFGYPFAASSSLLGRWSFGSAGCTW 1 | |||
2 YGFTGFFFGLTSIANMALMSIMRFMIVYKGYP 1 | |||
2 GPYPSRRATSGLIAAAWLYGLFWACAPLA 1 | |||
2 GWSQYHVEPFGLSCTVDWGSFSRDAGGMSFIICLLVFCVAIPVTAIMASYVAISAIYRQAKKSIAGHLQDNSAMCKKRNKLERSITL 0 | |||
0 MALAVCGGFLLAWLPYAVVGLWSAVAGVDAVPLALASAAPLFAKSSSLWNPIIYLGMNDRFS 2 | |||
1 AELSREHSYLPATGVRTTGPPAEDNQAEGDDVSSPSLLFFRRLFWTSKT 1 | |||
2 SNAMRTLVVLTWISTIFPLLLAAPQADPRTTYKVSRWDEAWRPQRFGRSGRGDTKDGWRPQRFGRGRYEQGWRPQRFGRNEGLAGLREVLDGEAFPLLQMTR | |||
TDLHHDLPGIGYTPGTAHGSLRALPLLRLYDRGALQLINGAAKQSSTNPPSLRMIRAAAESLQGFARGGDQQDEEEMFAPPRSTDDWLAEIQRLGLRGRKRRDVS* 0 | |||
>NEUR_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XM_001197837 full G? CX694910 CX690664 | |||
0 MDVNAKWWTNETLRTRDQFSDDHYTSVLSYEGDIWAGVYLMFI 1 | |||
2 SLIAFIGNISVIVISLRKREKLKPIDLLTINLAIADFLICVVSYPLPMISAFRHR 0 | |||
0 WSFGKFGCVWYGFTSFLFAVGSMATLMVIALLRYAKLCRENV 1 | |||
2 DQYQSRPFVIKVIVAIWGFAFFTTAPPLFGWS 2 | |||
1 SYVPEPYHLSCTIDFADTSPSGLSYTYFTTIVVFFMPLMIIVLCYVAIARKMIHHNRRINVGHNAGRMLLEIRLLK 0 | |||
0 TACMITMAYTISWTPYAVIAMWVTYIPVNQIPDAFRILPAFCAKTSSVYNPIIYCIFNKSFRQDLSSLICCCACQCYTITINLDINSHAQQQFRRIEERR | |||
DEVGTYKRRPLMICSNPFAWSRDFHETWRQRRIRGIHRNCRNNVRVENINVNFRRDTDMVELNAPTPAEIHRPELNTASTRSGARTKSMATHLPALEEVPSG | |||
APQCSALLHNTPIPRSLQGTPLPYQPQPSTSDLHDEFLNPSVVSRNMCVIVVKPNIEEELSTD* 0 | |||
>NEUR2_galGal Gallus gallus (chicken) Deut.Saur.Arch AB368181 18570255 full G? cOpn5L1 syn(-B4GALT6 -NEUR2_galGal -KIAA1012) GenBank 5'UTR error | |||
0 MDPSFANSTFQSKITEAADIVVGTCYMVF 1 | |||
2 GICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHR 2 | |||
1 WLYGKHICLFYAFCGLFFGICSLSTLTLLSVVCCLKICFPAY 1 | |||
2 GNRFRRKHGQILIACAWTYAAIFACSPLAHWGEYGEEPYGTACCIDWQSTNVDVMSMSYTVVLFVLCFILPCGVIVTSYSLILVTVKESRKAVEQHVSGPTRINNVQTITAK 0 | |||
0 LSIAVCIGFFAAWSPYAIIAMWAAFGSIDKIPPLAFAIPAVFAKSSTLYNPIIHLLLKPNFRSNIAKDFTVIQQLCVRCCFCVKELQTYRSTFNTGLRTFKGKNESSCNALPIMEG | |||
CSYFPSEKGSHTFECFKSYPNCFQERLSTMGCHLQDCESLENDLQVEVTQGSRNSMKVVEQEEKSTELDNLEITLEAVPVSCTFTDL* 0 | |||
>NEUR2_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G? | |||
0 MESYFANTTFHSKITEAADVIVGVFYIVF 1 | |||
2 GICSFCGNSILLYVSYKKKNLLKPAEYFMINLAISDLGMTLTLYPLAVTSSLAHR 2 | |||
1 WLFGQQVCLFYAFCGVFFGVCSLTTLTLLSIVCCLKICFPVY 1 | |||
2 GNRFRPGHGWILIACAWVYAAIFAFSPLAHWGEYGAEPYGTACCIDWRISNMKKTAMSYTTALFVFCYIIPCGIIITSYTLILITVKDSRKAVEQHALGPTRMSSVHTITAK 0 | |||
0 LSIAVCIGFFVAWSPYAIIAMWAAFGSIDMIPPLAFAVPAVFAKSSTLYNPAMYLFLKPNFRSTIAKDLTVLHRLCLKSCFCPRGMQNCSYRSALEAPLKSFKGRNESSSNSVQIVGGCS | |||
YFPCEKCHDPFECFKNYPKCCQGRLNVMDHTPRESISVENNMQSKTKHASEKYIKVVIRGEKNTDIDNLEITLEHIPTDIKFANL* 0 | |||
>NEUR2_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? abundant transcripts | |||
0 MGNKSDASAFYSSISETDDIVLGVLYSVF 1 | |||
2 GLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHR 2 | |||
1 WLFDKVTCNYYAFCGMLFGLCSLTNLTVLSSVCCLKVCYPAY 1 | |||
2 GNKFSTAHSRILLLGIWAYAGLFATAPLADWGKYGPEPYGTACCLDWEASYRERKALSYTISLFVFCYLIPSSLIFISYTLIFVTVKGARRAVQQHLSPQAKGSSIHSLIIK 0 | |||
0 LSIAVCIGFLIAWTPYAIVAMMAAFGDPTKIPSLVFALAAAFAKSSTIYNPVVYLLLKPNFLNVVTKDLTLFQTMCAVVCGWCRTPAVKTPCPHKD | |||
LKTTSKPPSSFKKSQGVCRNCVDTFECFRNYPRCCSVGNVDAAQPMAASLVRIPPANGAPQQTVQLVVSSSRTRSGVETVEVSTEAPMSDFIKDFI* 0 | |||
>NEUR2_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G? acquired new intron | |||
0 MGNVSKTALFMSTISRQHDILMGSLYSVF 1 | |||
2 FVLSLLGNGMLLFVAYRKRSSLKPAEFFVVNLSVSDLGMTLSLFPLAIPSALAHR 2 | |||
1 WLFGEITCLCYAVCGVLFGLCSLTNLTALSSVCCLKVCFPNY 1 | |||
2 GNKFSSSHACVMVIGVWCYASVFAVGPLVHWGSFGPEPYGTACCINW 2 | |||
1 YTPSHDALAMSYIISLFIFCYVVPCTIIILSYTFILVTVRGSQQAVQQHVSPQTKVTNAHALIVK 0 | |||
0 LSVAVCIGFLTAWSPYAIVAMWAAFSANEQVPPTAFALAAIMAKSSTIYNPMVYLLFKPNFRKSLSQDTQMFRHRICLSHSKASPSPGMKDQERQS | |||
SQQCNNKDGSISTPFSSGQAESYGACHVYAEAGPHYQQISRQITARVLEGSVQSEIPVKQLTEKMQNDLL* 0 | |||
>NEUR2_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? novel paralog of neuropsin | |||
0 1 | |||
2 GILSLVGNSVLLFVAYRKRQILKPAEYFVANLAVSDISMTVTLLPLAISSNFSHR 2 | |||
1 WLFVSKpCMYYGFCSMLFGICSLTNLTVLSTVCCMKVCFPAY 1 | |||
2 0 | |||
0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVISYTMTVIAVNFVVPLSVMFFCYYNV | |||
>NEUR3_galGal Gallus gallus (chicken) Deut.Saur.Arch AB368183 18570255 full G? cOpn5L2 testis exon 2 fused exon 3 rel NEUR1/2 | |||
0 MEEQYISKLHPVVDYGAGVFLLII 1 | |||
2 AILTILGNSAVLATAVKRSSLLKSPELLTVNLAVADIGMAISMYPLAIASAWNHAWLGGDASCIYYALMGFLFGVCSMMTLCAMAVIRFLVTNSSKSN 1 | |||
2 SNKISKNTVHILITFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHSSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNINRIPHAAKLEKKLTL 0 | |||
0 MAVLISVGFLSAWTPYAAASFWSIFNSSDSLQPIVTLLPCLFAKSSTAYNPFIYYIFSKTFRHEIKQLQCCWGWRVHFFSADNSAENSVSMMWSGRDNIRLSPTAKVESQGAARH* 0 | |||
>NEUR3_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch ABQF01025032 full G? | |||
0 MEEQYISKLHPVVDYGAGVFLLII 1 | |||
2 AILTILGNSAVLATAVKRSSLLKPPELLTVNLAVADIGMALSMYPLAIASAWSHAWLGGDASCVYYALMGFLLGVCSMMTLCAMAVIRFLVTNSPKSN 1 | |||
2 sNKITKNTVCILIAFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHNSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNIDRIPNAAKLEKKLTL 0 | |||
0 MAVLISVGFLSSWTPYAATSFWSIFNSSHSLQPVVTLLPCLFAKSSTAYNPFIYYVFSKTFRCEVKRLQCCCAWRVHYFSSDNSVENPLSTMWSGRDNIRLSAAPQVQNPGAAAP* 0 | |||
>NEUR3_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi AAWZ01001057 full G? | |||
0 MEEHYISKVHPVWDYGMGVFLLII 1 | |||
2 AILTILGNSMVLAVAVKRSSCLRSPELLTVNLAATDLGMGLSMYPLAIASAWNHAWLGGEATCIYYALMGFLFGVSSIMTLSAMAVIRFLVTFSSKPA 1 | |||
2 GHKINRKVMHICIMLIWAYAVLWAILPLLGWGHYGPEPFGTSCTIAWGQFHNSQKGFAFILSMFILCTFLPAITIIMCYLGIAWKFHKTHQEMQNLNRISSAAKLEKKLIL 0 | |||
0 VAVLISVGFLGAWTPYAIVSFWSVFHSSESIPYIVTLLPCLFAKSSTAYNPFIYYTFSKTFRHEVKHLRCYSGQRAQENMKNSINSNVSFMWHGGGNICLSTRQIEMREIPNQ* 0 | |||
>NEUR3_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? cdna ovary embryo | |||
0 MEERYLSKLHPLVDFGSGVFLLLV 1 | |||
2 AILTVLGNCAVLATAVKCSSHLKAPDLLSINLAVADLGMAISMYPLAIASAWNHAWLGGDASCLYYALMGFFFGVSSMMTLTVMAIIRYRVTSSFKYS 1 | |||
2 GCTIEKKAVCILIMCIWLYALLWAVLPLLGWGRYGPEPFGTSCTIAWGDFHHSSNGFSFIISMFILCTISPAVTIVVCYSGIAWKLHKAYQEIKNQDKIPNSTKVEKKLTL 0 | |||
0 LAILVSFGFLISWTPYAAVSFWSLFHSSKYIPPVVSLLPCLFAKSSTAFNPMIYYAFSKTFRRKVKHLKCCCGWRVHFLQSENSVENPRVSVIWTGKENVMVSSVPKLMKGVPGTPTGTQ* 0 | |||
>NEUR3a_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G? | |||
0 MDRYTSKLSPAVDYSAGTFLLVI 1 | |||
2 AILSILGNAAVLLTAAWRHSVLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWIGGDPSCLYYGLMGMIFSVASIMTLAVMGLVRYLVTGNPPK 1 | |||
2 SGSKFRRKTISILIGVIWMYSLLWAVFPILGWGGYGPEPFGLACSVDWMGYQHSLNRSSFIMALAILCTLMPCVVILFSYSGIAWKLHKAYQSIQSNDNLPNSGAVERKVTL 0 | |||
0 MGILISTGFIVSWAPYVFVSLWTMFRSEGEDSVVPIVSLLPCLFAKCSTVYNPLVYYVFRKSFRREIHQIRICCFQGCWDAVSKMTRGDGPEETSGTHETDNI* 0 | |||
>NEUR3b_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G? | |||
0 MDIYSSKLSSAVDYGIGAFLLLI 1 | |||
2 TILSILGNLMVLVMAYKRSNHMKPPELLSVNLAVTDLGAAVTMYPLAVASAWNHHWIGGDVSCVYYGLMGFLFGAASMMTLTIMAIVRFIVSLTLQSP 1 | |||
2 KEKISKRNAKILVATTWLYALLWAIFPLIGWGKYGPEPFGLSCTLDWRDMKEHSQSFVITIFLMNLILPAIIIVSCYCGIALRLYVTYKSMDDSNHVPNMIKMQRRLMV 0 | |||
0 IAVLISIGFVGCWAPYGIVSLWSIYRPGDSIPAEVSMLPCLFAKTSTVYNPFIYYIFSKTFKREVNQLSRFCGRSNICRPTDAKNRPENTIYLVCDVNKSKPGVEDLSLARSKENETQMLPNQDLHE* 0 | |||
>NEUR3a_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? | |||
0 MDDKYMSKLSPPVDLWAGIYLVVI 1 | |||
2 ALLSVLGNASVLFSASRRLTPLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWMGGDTACLYYGLMGMIFSITSIMTLAVMGMIRYLVTGSPPR 1 | |||
2 SGVQFQKKTICVVICAIWLYACLWAAFPLLGWGSYGPEPFGTACSIDWTGYGDSLNNATFIVAMSVLCTFLPCLVIFFTYFGIAWKLHKAYKSIKSSDFQYASVERRITL 0 | |||
0 IAVLISVGFLGSWAPYGLVSLWSILKDSSSIPPQVSLLPCLFAKSSTVYNPVIYYIFSQSFKLEVQQLFLCC* 0 | |||
>NEUR3_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag G? new exon | |||
0 MAEQGEDDQFRSKLSPTADIAAGTFLLAV 1 | |||
2 AVLSLAGNGAVLGVAARRWAKLKAPELLSVNLALTDLGIAASIYPLAVASAWNHRWLGGQPVCTYYAFAGFFFGTASMGTLTAMAGVRYKGTSTQVH 1 | |||
2 VKQITKRAMLAVIVAVWAYALLWSCLPLLGWGR 2 1 YGVEPFGVSCTLAWAELQLTPGGVAFLYAMFVLCLLLPAIAIGLCYAGIVCKLRRAYREGRSKRRTPTARHVESRLTK 0 | |||
>NEUR4_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono XM_001508128 full G? | |||
0 MSLSHSLQVPWRNNLTFLNKEAQVSEQGETIIGIYLLAL 1 | |||
2 GWMSWFGNSMVIFILHRQRGILNPTDYLTFNLAVSDASVSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLAMISVTRYIKGCHPHR 1 | |||
2 GHFINTANISVALILIWVSALFWSAGPVLGWGSYT 1 | |||
2 DRMYGTCEIDWAEANFSSICKSYIISIFFCCFFLPVSIMFFSYVSIIKMVKSSHTLAGADDPTDRQRRLDRDVTR 0 | |||
0 VSVVICTAFIVAWSPYAVISMWSAFGHSVPNLTSVLASLFAKSASFYNPIIYFGMNSKFRKDILVLLPCAKESKEPVKLKKFKNLRQKQGFTLQKPEKAHVLQVPDSGPMSLINTPPLGNRNSFDLACDNSDFECVRL* 0 | |||
>NEUR4_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full G? genome gappy | |||
0 MSLQLSPQAPWRNNNISFLSREAAVTEQGETIIGFYLLAL 1 | |||
2 GWMSWFGNSVVIFVLYKQRHLLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPER 1 | |||
2 AHCISNSSMTVAMVLIWIAAFFWSAAPLLGWGSYT 1 | |||
2 DRMYGTCEIDWAKANFSTIYKSYIISIFICCFFLPVTVMVFSYVSIINTVKLSHALTGLSDPTERQRRMERDVTR 0 | |||
0 IVICTAFIIAWSPYAVLLLWSAYGHPVPNLPLYLSSLFAKSASFYNPIIYFGMSSKFRRDIFILFHCAKEVKDPVKLKRFKNLKQKQEPSQKEEKYAAEMHPAPSPDSGVGSPTNTPPPANREEYFGILDTPSNSPDIECDRL* 0 | |||
>NEUR4_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full G? | |||
0 MSVQFSAQAPWRNNNISFLTREAAVTEQGETIIGFYLLAL 1 | |||
2 GWLSWFGNSIVIFVLYKQRHVLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPER 1 | |||
2 GHCISNSSMSVALVLIWVAAFFWSAAPLLGWGSYT 1 | |||
2 DRMYGTCEIDWAKASFSTIYKSYIVSIFICCFFLPVTVMVFSYVSIINTVKLSHT LTGLGDPTDRQRRIERDVTR 0 | |||
0 VSLCTAFIIAWSPYAVISIWSAYGHPVPNLTSILASLFAKSASFYNPIIYFGMSSKFRRDIFIFHCAKELKDPVKLKRFKNLKPKQPQPSQKEEKYAPEMHPAPSPDSGVGSPTNSPPPANREVYFGILDTPSNNPNIECDRL* 0 | |||
>NEUR4_anocar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G? | |||
0 MSLQVSPQAPWRNNNVTFSNKEVPVSEQGETIIGFYLLAL 1 | |||
2 GWMSWFGNSIVIFVLYRQRAGLQPTDYLTFNLAVSDASVSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPDR 1 | |||
2 GKCISNSSISVALFLIWIAAFFWSVAPVLGWGSYr 1 | |||
2 DRMYGTCEIDWAKANFSTIYKSYIVSIFICCFFLPVSVMVFSYVSIINTVKSSHALSGVGDPTERQRRMERSVTR 0 | |||
0 VSLVVICTAFITAWSPYAVISMWSAYGYTVPNLTSILASLFAKSASFYNPIIYFGMSSKFRKDIFVLLHCAKEIKDPVKLKRFKNLKQKQEVSPSQREEKYAADVQPALSPDSGVGRSNTPPPVNREVYFGAFDTFSNNPDVECDRL* 0 | |||
>NEUR4_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? | |||
0 MSLQFPRPAPWRNNNLTLLQKENPLTEQGETIIGIYLLAL 1 | |||
2 GWLSWFGNSIVIFVLYKQRANLLPTDYLTFNLAVSDASTSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLTLISVTRYIKGCHPQR 1 | |||
2 ANCISNGSITISLALIWIAALFWSVAPLLGWGSYR 1 | |||
2 DRMYGTCEIDWTKASFSTIYKSYIISIFICCFFLPVMVMVFCYVSIINTVKSSRALTSEGDLSERQRKMERDVTR 0 | |||
0 VSVVICTAFIVAWSPYAVISMWSACGYYVPSLTSILAALFAKSASFYNPLIYFGMSSKFRKDLCVVLPCAKAQKDPVKLKRYKDKKQGSAPRAREQTEIEQPVQLQPAPSQDSGVGSPSNTPPLRTKDVHIVDIDLVSDNPSYECDRL* 0 | |||
>NEUR4_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G? | |||
0 MSAQNPLQVVNIPWRNNNFSLMSRDPPLSDQGETIIGVYLLIL 1 | |||
2 GWLSWFGNSIVIFVLFRQRSTLQPTDYLTLNLAVSDASISVFGYSRGILEIFNIFKDSGYIISSVWTCQ 0 | |||
0 VDGFFTLVFGLSSINTLTVISITRFIKGCHPHK 1 | |||
2 AHCITNSTVAVCVVFIWIGAFFWSAAPVLGWGSYT 1 | |||
2 DRGYGTCEIDWVKANYSTIHKSYIISIFIFCFLVPVLLMLFCYISIINTVKRGNAMNADGDLSDRQRKIERDVTI 0 | |||
0 VSIVICTAFILAWSPYAVVSMWSAWGFHVPNLTSIFTRLFAKSASFYNPLIYFGLSSKFRKDVSVLLPCGREGRDPVRLKRFKRLRGRAEPPGAPAHTPHPQIALKNYNNHSKPHAGPAHCTGHAPSPDSGVGSHHETPPPQPRPQLFFIDVPEPEAESECVRL* 0 | |||
>NEUR4_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? | |||
0 MEPSRPWRNSSVLGGGAEPPLSEQGETIIGVYLLLL 1 | |||
2 GWLSWFGNTVVLFVLVRQRSSLQPTDLLTFNLAVSDASISVFGYSRGIIQIFNVFQDSGFIISSIWTCE 0 | |||
0 VDGFLTLIFGLSSINTLTVISITRYIKGCQPSR 1 | |||
2 AALISRSSVSVCLLLIWTTAGFWSGAPLLGWGSYT 1 | |||
2 DRGYGTCEIDWSKAASSGVYRSYIISIFIFCFFIPVFIMLFCYISIINTVKRGNALAADGHLSHRQRTMERDVTV 0 | |||
0 ISVVICTAFIMAWSPYAVVSMWSAWGFHVPSTTSIVTRLFAKSASFYNPLIYFGMSSKFRKDVSLILPCAKERREVVLLQRFKNIKPKAAAAPPPPPLPVYRPKEKNEDEPKLSVHDNDSGVNSPPETPPSDAQEVFPVDPPSQIETSEYWSDRL* 0 | |||
>NEUR4_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic frag G? | |||
0 PVKVVNIPWRNNNLSNLNTDPPLSEQGETFIGVYLLVL 1 | |||
2 GWLSWFGNSLVMFVLYRQRASLQSTDFLTLNLAISDASISIFGYSRGILEIFNIFNDDGYLINWIWTCQ 0 | |||
0 VDGFFTLLFGLASINTLTVISVTRYIKGCHPNK 1 | |||
2 AYCISTNTIAVSLICIWTGAVFWSVAPLLGWGSFT 1 | |||
2 DRGYGTCEVDWSKANYSTIHKSYIISILIFCFFIPVMIMLFSYVSIINTVKSTNAMSADGFLSTRQRKVERDVTRV 0 | |||
0 ISIVICTAFITAWSPYAVVSMWSAWGFHVPSTTSIITRLFAKSASFYNPLIYFGMSSKFRKDVSVLVPCTRERREVVHLQHFKNIKPKAEAPPTPASLPVQKLGAKYAVPNPDADSGVNNPPQRPATDPQGDLNIDLPSHIETSEYWCDRL* 0 | |||
>NEUR4_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? | |||
0 MGCSLGWKVLLWFLHGILICPRPWRNHNSTFQPKEHPISEQGETIIGVYLLIL 1 | |||
2 GWLSWFGNSIVIFILYRQRLSLQPPDYLTLNLAVSDASISIFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 | |||
0 1 | |||
2 AVSISAGSIAASLVLIWIAAIFWSGAPLFNWGSYT 1 | |||
2 DRMYGTCEIDWSRASFSTIYKSYIISIFICCFFLPVFVMLFSYISIINTVKSSHAFAGNADLSDRQRRMEKDVTR 0 | |||
0 VSMVICTAFIIAWSPYAVISMWSASGYTVPQLTGIFASLFAKSASFYNPMIYFGLNSKFRKDIYILLPCVKEPKESVKLKRFKHLRHRPEQQQANKDRYAEELQQVASPDSGMGSPSKSPPLHNKDVFFVLWLRGLKK</font> | |||
<font color ="green">>TMT_triCys Tripedalia cystophora (box_jelly) Cnid.Cubo.Cary EU310498 18577593 full G? c-opsin-like Schiff lysine mRNA with predicted introns | |||
0 MADHGRNTTSNDTNPISKIPLDDFDPQRFYNFYTFMGSFIAGSACCSFLLNGLVIAVLIKYIRTITNTNIIVLSMSCANILIPLLGSPLSATSSLMRKWQFGNGGCTWYGFINTLS 1 | |||
2 GISGIYHLTFLSFERFITIVLPLKRDTILSTKNIYIGLGILWVAAIGVAGAPVFGWCEYIKE 1 | |||
2 GVRTSCSVAWSSKENMNVFSYNLFMIFTVFLLPMLVIIYCNYRFIKEVSIMSTRA 0 | |||
0 RGLQGGDSEMTASASKAEKQLTIMVITMIIAFNIAWLPYTVVSMVFLTGYGDVVGPMGASVPSVFAKTSVIYNPVIYCLLNRS 0 | |||
0 FRKMLCGNSVEPE* 0 | |||
>CUBOP_carRas Carybdea rastonii (sea_wasp) Cnid.Cubo.Cary AB435549 18832159 full G? cubop | |||
MGANITEILSGFLACVVFLSISLNMIVLITFYRLRHKLAFKDALMASMAFSDVVQAIVGYPLEVFTVVDGKWTFGMELCQVAGFFITALGQVSIAHLTALAL | |||
DRYFTVCRPFVATAIHGSMRNAGMVIFVCWFYASFWAVLPLVGWSNYDVEGDGMRCSINWADDSPKSYSYRVCLFVFIYLIPVLLMVATYVLVQGEMKNMRGRAAQLFGSESEAAL | |||
KNIKAEKRHTRLVFVMILSFIVAWTPYTFVAMWVSFFTKQLGPIPLYVDTLAAMLAKSSAMFNPIIYCFLHKQFRRAVLRGVCGRIVGGNAIAPSSTAVEPGQTLASGTAES* | |||
>ENCa_nemVec Nematostella vectensis (anemone) Cnid.Anth.Hexa genomic full G? Schiff lysine no cdna complete 1 exon | |||
0 MIDNALGRHEANIVLGYYIAIFVIGFVTNTIVVIIFISSQRLHTTPNLILFSMSVCDWLMATMAKSVGIYGNARYWPTVGKVTCDYYAFATSAIGYASILHLAALAVEKRMAVVSPMTNSFNGRRMLVIIATLWGFAILW | |||
AVFPLIGWSSYGPEPGYVSCSITWYTTDHNNVSYIICVSVLFFLIPIVTMTFCFASIYHTIRNLSHEATARWGSDARATQETIRAKAKTAKMAFLMVMCFLFAWTPYAVVSLWTTFGDTHRIPALLGVLPSLFAKLSSCY | |||
NPIIYFFMYTKFRCAGKALLYQEHH* 0 | |||
>ENCb_nemVec Nematostella vectensis (anemone) Cnid.Anth.Hexa genomic full G? Schiff lysine NC-extended 1 exon | |||
0 MSNEALRRHEANIVLGYYIAIFVIGFVTNTIVVITFIFSKRLHTTPNLILFSMSVCDWLMAAMAKSVGIYGNARYWPTVGKVTCDYYAFATSAIGYASILHLAALAVEKRMAVASP | |||
MTNSLNERRMLVIIATLWGFAILWAVFPLIGWSSYGPEPGYVSCSITWYTTDHNNVSYIICISVLFFFVPIVTMTFSFASIYKAIRNISHEAIARWGSHARATQETIKAKAKTAKMAF | |||
LMVMCFLFAWTPYAVVSLWTTFGGTHRNPALLGVLPSLFAKLSSCYNPIIYFFMYTKFRRAAKLLFIKKVIRPTEAERSRVLSGIRTRAASTFAVKLTVHAEKGQQIAPNITPQGAVKGPVESIEDNLQKIETITPCTSV* 0 | |||
>ENCc_nemVec Nematostella vectensis (anemone) Cnid.Anth.Hexa genomic full G? Schiff lysine C-extended 1 exo | |||
0 MELFTSYHAITVMYSLLAAGAFVLNGIVLIIFLATRSLRTIPNMILLSMAWADWLMACLADAVGAYANANNWPSMVGGLCVYYGFITTALGLTSMIHLTALSVERFVTVTIPMTRPITETQMLLVVTFLWAFSFLWAIFP | |||
LVGWSSYGPEPGYAACSIAWYRQDLNNMSYILCLFMFFFFLPIVIMIACFSSIYFTVRKLTRDSMRRWGASSDSTQQTLAAERKTAWMSFIMVLAFLFAWVPYAVVSLYASFGGVTTIPKLMSTLPAMLAKTSACYNPII | |||
YFFMYSKFRKAFQRFFFKNVITPSQTGGSSTINSASVIPTSFPRASYAVRSSSVLPSSTASHSFRSRE* 0 | |||
>ENC_aneVir Anemonia viridis (anemone) Cnid.Anth.Hexa FK729339 19627569 frag G? symbiotic | |||
KDAVARWGNKSPPTQQTMQAQKKTIRMSLVMVFAYLLAWTPYALTSLYSSFIASDITPLLSVMPALFAKLSSCYNPIIYFFMYSKFR | |||
KAAKKMIRRNLVGHDSNSGQGVSNTFATSFPRPISFLRYKRSAVAPLSDIPQVSSVDLPQVGRENDVTVQQDKASEINT* 0 | |||
>MEL1_acrDig Acropora digitifera (stony_coral) BACK01045931 | |||
METASGVPIVIEASVHHTISFLYFLLALFSFSLNSVVILTFLLDRSLLFPANLIILSIAISDWLMSVVPNIMGGVANASNDLPFTDWSCTVFAFVATLLG | |||
LSNMLHHAAFALDRYMVITRPMRANHSMKRILAVIAFLWCFALTWSLFPLVGWSAYVREAGDIACSVNWQSDNPSDTSYMVCLFFFFYFVPLAVIVYCYA | |||
FMIKSVRFMTKNAQKIWGVRSAAALETVQATWKMAKIGLIMVVGFFVAWTPYAVVSFIIAFDLVKDIPTVAEIVPSMFAKTASVYNPIIYFFSFAYCKNQ | |||
SVNKCINQSTVQSKNFKLS* | |||
>MEL2_acrDig Acropora digitifera (stony_coral) BACK01017283 | |||
MPGFQNKSEEPQPTEGISSQDRFIVASFYALLAVTALGLNTPVLITFMKDRKLRVLSNRIILSITIGDWLHAFLAYPVAVFANASHDSQGLTGVVCSWYG | |||
FITVFLSFGIMLHHATFAVERAIVIQFATTSLTNAKTINFMVASLWVFALLWSSFPLFGWSAYVPHVVLCSLDWQSRDLRDVVFVYCIFFVFFLVPIVVM | |||
VTSYYKIFQTVKKMTQNARDLWGEKAAPTREAFESQKKTARMACVMSFCFLFAWTPYAAVSLYVFLWKPQSMAPSISIVPALFAKTSACFNPVIYFLLFR | |||
KFRESLKQTMCGFFMDPKTGTNETNKNKRHVLKNTESKCACVKINNLSPRDPCNLHQRSDCRSVGNSKSHITVD* | |||
>ENC1_acrDig Acropora digitifera (stony_coral) BACK01046894 | |||
MLTFACGSTHIYNMSAVFYGFLAAGTVFLNVIVILTFFKVRALLIPASFPILSLAMADVFLASAVMPLGIASCASGHWIFGAVGCNWYAFMHTTVGLSSI | |||
LHHAILALERCLTIFEPMKKHFDRHTMTRTLAFLWTLVSLWSLCPILGWSAYVPEGTGTICSIEWHSKNSLDVIFVVSTFVFFCFVPFVFIAVCYTAIAF | |||
HLRKVSKTAKRTWGKNSQITKDGVIVKRKAVTHGAIMVTTVMITWLPYSMVAFYTLLGFEKVKLSALAYAITAMFAKTSTLVNPIICFFWYRRFREGTKK | |||
LCNRIMDFILRKHRRASNTSHSSKTVWYDGKTIVFRRSVVDSCSTRTQNDRGRSS* | |||
>ENC2_acrDig Acropora digitifera (stony_coral) BACK01016849 11738 bp INV 28-JUL-2011 DOI:10.1038/nature10249 | |||
MTALIYSVFAIIAAVICVLGVPLNGLVCWVFYNNRELLNAPNIFIASVAFSDFLYCISCLPLLVISNAYGKWIYGPIGCKATAFIATWSGLTSLMNLSVA | |||
SYERYSTLAFLCTKNRTFAKRTATCYSAAMWLYALFWSLMPLCGWSGFELEGIGTSCSVRWKSKNMLDMSYNLCLIIACYVLPVSVLVTSYYKCYREIAK | |||
STWRAKSTWGRQSPFTKRTFVMERKMMALFGVMTVAFLVAWTPYAVVSLISMIGGPDVISDVTASIPAYFAKSSSCYNPVIYVFLYKRLRRQMRFAVRRD | |||
NCSSSRKGTSSDSTMAHHSETKRLRSTIELTDNYSTRNRPENI* | |||
>ENC3_acrDig Acropora digitifera (stony_coral) BACK01015014 possibly N-incomplete | |||
MVAPHFAGFAIAASLFGSCGIVLNFTVCLTYILNRRLLDASNIFILNISAGDFLYSITALPMLVTSNALGKWSFGEGGCVAYGFLTTFFALGAMMNLAGA | |||
AYERYVTMCKLYESGECQFSRRKAAFLCSVLWTYALIWSIAPIFGWSSYKQEGIGTSCSTDWKSRDVKSLSYGVVLIITCFVVPVAVILYCHIEAYKVTR | |||
KLGKQAQQNWGSHTRATRETLKAQKKMGEIAVVITVGFVVAWTPYTVASVIGMYDPNLVSDVGASIPAYFAKSSSCYNPFIYLFMYKKLRIGMVRLLCCM | |||
KKQVYPSSSGLTRDPTCQEIPMVVPVNNPS* | |||
>ENC4_acrDig Acropora digitifera (stony_coral) BACK01020962 possibly N-incomplete | |||
MTKTAAWSTVFLIIEFVVCLLGIFFNCLVISIIFKNANRLAAPGFLVLSMALSDVLSCSVAVPFSIVAHFQKKWPFGMAGCEAHAFMVFLFALVSITHLA | |||
AISAGKYLTISRSLTRESYFDRRQVLLVVIACWLYSLAFSVAPLVGWSRYGLEGTNATCSIKWDSSEPSDKAYFGVVFIACFFLPMGVITLCYYKIHKVS | |||
KRIVENIQGQSIATSAMSRTHALVKKHRRSAMYFLAIVAAFMLAWSPYAIVSLIVVLSGTMDPIATSACSVFAKTSFLVNPFLYAIFSRSFRRRLALVFP | |||
MPRQNRQEGRQERNCNALPPSQSQPFVL* | |||
>ENC5_acrDig Acropora digitifera (stony_coral) BACK01011015 | |||
MDQIKIWKMLFVVIKFIVSIFGGFFNGLVVLTIRKNLRRLPSSSYLILSIAFSDFIASVVAIPFSIVIHFVGSWPLRLCRAHAFMVFFLGIVTITHLTCF | |||
AVEKYLTITRSLSKLSFFSKKQTLVVVMACWFYSLCFSLAPLLGWASYGLEGSNDTCSIKWDSSLAKDHIYFILVFLACYLLPIVLITSSYFKILRISKR | |||
ILKATPRVGGIGETMAQALMRKHRRGALYFLSVIAVFLMSWTPYAIISVLVIFKGVNLFPLALSACGVFAKMSFMLNPIMYFAFSHNFRLLVKRTFCICD | |||
RLDSNSEGKKR* | |||
>CNOP1_acrDig Acropora digitifera (stony_coral) BACK01019215 tandem fragment last exon at beginning of contig | |||
1 LLRRDVVTEDSRIKLRRRHWQMKLLRLTAVAITAFMLSWSPYSLVSLTSIFRGNSVLSTGEAEVPALMAKASVIYNPIVYTVMNRRFRRTLRHIVSCMTCRLLSFVWPTMHGEKQETKKRVTSTMTVTSSTPAPEGNFPEILVSLHQVPF* | |||
>CNOP2_acrDig Acropora digitifera (stony_coral) BACK01019215 MEL | |||
0 MQFSREEDPPSGFNWNNAWFIAYGVVMIAVLSIGFLGNTMTVLILRKHEHASKSLTPLMINLAIASLIIIVLGYPLVISLVVRGSHVTKEDPTCRWSAFINGTV 1 | |||
2 GISSIATLTEMSLVINYSLHRMNPNVRLTKRNMALLIAGAWLYGLVSMFPPLVGWNRFVPGAVRISCGPDWTDKSASGVSYNLVLVVLGFFLPLCVMIKAYYEIYRS 2 | |||
1 LLRSREMLISANGSFQLRQKLYIKKLVRMTVLAIAAFMLSWAPYCFVSIVAIFKGSHIITSGEAEIPELMAKASVIYNPVVYLITNSSYRASFWKVISCQKQTMIVHVRENLMPNGPSRRTLRSRRMAVLLKPLNEVSVYNGGYVVAMGFASVRDL* 0 | |||
>CNOP3_acrDig Acropora digitifera (stony_coral) BACK01018578 TMT | |||
0 MLESSNQSAEETILISKASYYAYGVVMFFILTVGFLGNVMTLLVLFQHEHRKKAMTPYMVNIALADIFIILFGYPVAMRANLRGKILESSHCSWGGFVNGAV | |||
GISSIFTLTEMSFVSYHGLRQVNRSSRFSPFQVVCSVAVAWLYGVLCMLPPFLGWNRFVVSASRISCCPDWSGKSISDAAYNLLLVFFGFVAPLTAMTVCYYKIYR 2 | |||
1 SLVHHAVVPGNLPQIQLRRRQSELKVAKVTAMNVIAFLLSWAPYCCVSLAAVFTKQFVLVDWEAEIPELLAKASVIYGPIIYSTMHSRFQATLFRILHCRRRVILAPQFITDATRNNCSARMATDRTQGTSHIDQRFLQLPLQAMKRRVAVSYGGTAICKRESSAFVSG* 0 | |||
>CNOP4_acrDig Acropora digitifera (stony_coral) BACK01002540 TMT | |||
0 MTTRSDDNPNSARRPQQIAFIVYVVIMTVILIVGFLGNLFTIIVLRCPEHKKKIITPLMMNLAFADIIIIVFGYPVVMASNFTEHNVLTNPMLCVWSGFINGSV | |||
GIASIANLTMMSLVMFKSLNKVGTVKKIPWKKMLAMILFTWVYGVLSMFPPLVGWNKFVPGASGISCCPDWSPDTKAAVGYNILLVFVGFVLPLTIIIFCYYRMYR 2 | |||
1 FIHTQQPVTGNASIQASRRRSEIKIVRMIALAIAAFVLSWSPYCFVSIAGTIRGSSLLTAGEAEVPDLLAKASVIYNPIVYTVMNDRFRGTLLRVIPCRRRWFNGDINPTVSVSAPNSQDGNSHNFRERPQSVKGNSTV* 0 | |||
>CNOP5_acrDig Acropora digitifera (stony_coral) BACK01002513 and near identical BACK01007224 | |||
0 MTTSFRSNMTDEWGTHKYFSAGYIAYGVVMFCILSLGFVGNTLTVLVLTKREHRWRGVTPLMINLAVADLLIIIFGYPTQVHANFTGDGVPNDSHCNYRGFIN | |||
GIVGITCIFTLTEMGVVSYSGLRRVSGNVRLSSYQVACLIGAAWCYGALCMLPPLLGWSKFVISASKLSCSPNWSGQSTADKAYTLLLVTFGFFAPLTVMIISYYKTFW 2 | |||
1 VVRRHFVPGNTAVQLRQRKSMLRIVHVTLMSVISMMLSWAPYCFISLASVVRGKPVIENWEAEIPELLAKASVVYNPLIYTFMNKSFRMTLFRIVGLHRCFGAGHEVAPTAAIQKRPNVLQIQFHRHSSEVEVRTPRVPPIE* 0</font> | |||
<font color ="purple">>CTENOP1_plePil Pleurobrachia pileus (sea_gooseberry) Cten.Typh.Pleu FP995093 CU419614 full G? ctenop EQY | |||
0 MELRVSLSLIMACVCTGSVF 1 | |||
2 GNMLVFISLVKERPLPNPAVEMIFRHLSLANLVLAGIGEPLVVVSLAA 1 | |||
2 GKWVWGESMRYFEAFWVTTFGLFSMMILGLISFEQYSRFVRQKQLKTHQ 0 | |||
0 VAVLLSGIYFLAFFFGIAPLVGWCEYGPE 1 | |||
2 GYGVSNSLMWNNLNDNNASYIICAMIIGYFFPLIIIMFCYRAIYRLVQ 0 | |||
0 AQLNTSVVKMTVSANVTSLSTNRNHIML 0 | |||
0 VQERQLASTITFVILSFFFAWTPYVFVNIINMFSTLILKYRILAT 0 | |||
0 IPALFAKSSTLWWIIVYCLMDVRIKKACRKTCLRFKRSFRVWYFS* 0 | |||
>CTENOP1_mneLei Mnemiopsis leidyi AGCP01013851 AGCP01013853 77% identical CTENOP1_plePil | |||
0 MGLRISLSIAMGCVCVGSVF 1 | |||
2 GNLLVLISLVRERPLPNPAVQLIFRHLSLANFLLACIGEPLVVISLAS 1 | |||
2 GKWVWGESMRLFEAFWVTTFGLLSMMILALISLEQYSRFVRQKQLRTVQ 0 | |||
0 VAVLLSGIYFLAFFFGIAPLVGWCEYGPE 1 | |||
2 GYGVSNSLLWNELTDNSASYIVTVMIVGYFFPLVIITYCYRSIYQLVQ 0 | |||
0 AQSNVAIVRTMVSVNVTSLAVNRNHSLI 0 | |||
0 VQERKLATTITFVILSFFFAWTPYVLINLLNMFTTWVVKYKILAT 0 | |||
0 IPALFAKSSTLWWIIIYSMMDKRIKKACRKTCEHFQRALRIWYFSS* 0 | |||
>CTENOP2_plePil Pleurobrachia pileus (sea_gooseberry) Cten.Typh.Pleu FQ011385 ERY 33-203 range in bosTau rhodopsin | |||
0 MTTAVDDEMMGDEEPKVVYTLASLLAVNALCSIA 1 | |||
2 GNALVIYTFLKEKPLGAPLRVILTHLAVTNLIIASIGEPMVVISGYS 1 | |||
2 MRWVFGEMGRGFEAFIVTSCGLMAMSLLALVSIERYLR 2 | |||
1 VTPRQRFPFTEPTAVKCCSFLWVFNVIYASLPFYGIAAWEGE 1 | |||
2 GIGISNSITWTTEHTQDFVYIICMMSTGYFIPLFIIAFCY 2 | |||
>CTENOP2_mneLei Mnemiopsis leidyi AGCP01013109 AGCP01013108 AGCP01013106 | |||
0 MSSPNDEPTVDSDPKAVYILTSLLCMNCIFSLV 1 | |||
2 GNMLVIYTFLRERPLGAPLRTILCHLAITNLMIAGIGEPMVVISGFN 1 | |||
2 VRWVFGEMGRKIEAYTVTASGLMAMSLLAFVSIERYLR 2 | |||
1 VTPGQKFLIRQTTSVRVCTLMWIYNVIYASLPFYGIAAWRGE 1 | |||
2 GIGISNSLSWDTNNKIDVAYVMVMMATGYFIPLAVITFCYNRYVEIQQRLSTFL 2 | |||
SKLKMTMETNITSAANSQN | |||
0 AAEKKISNVVAIIIVSFFISWTPYTIVNLLVTFDKAGR 2 | |||
1 GINATIPAFLAKTSTVWSPIIYCFMNGEVSIFVLEGGHVMKYLYL* 0</font> | |||
=== GPCR outgroup sequences === | |||
[[Image:OpsinOrigins.jpg|left]] | |||
It is sometimes convenient to have a close-in outgroup to opsins selected from the roughly 100,000 GPCR receptors available at GenBank. The set below of 29 non-opsin GPCRs serves satisfactorily as proxy for an exhaustive GPCR compilation. It was constructed by taking best-blast in turn of each human opsin against all other human GPCR then collating the lists, winnowing out repeated entries and too-recent gene family expansions. TACR2 (tachykinin receptor) and SSTR1 (somatostatin receptor) are the best single representatives. | |||
These are usefully supplemented with non-opsin GPCR having determined 3D structures, some nearest neighbors in recent GRAFS classification trees, and two astonishingly close pre-opsins (called UROPS1 and UROPS2 here) from Trichoplax, an early diverging eukaryote lacking opsins. Note TSHR binding of heterotrimeric G protein has recently been [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0009745 modeled] in great detail. | |||
The aligned sequences below have been trimmed from full length at both ends to the earliest indications of conservation. This alignable region begins at the [[Opsin_evolution:_informative_indels#Alignment_in_TM2_region:_420_curated_opsins|GN region]] of TM1 and extends to the [[Opsin_evolution:_Cytoplasmic_face#The_carboxy-terminal_tail_and_VxPx_motif|FR motif]] just beyond TM7. Highly conserved residues are shown in red and less conserved residues in blue. The Schiff base lysine (position -16 relative to the FR end of TM7) does not occur outside of opsins. Note many conserved patches in these GPCR are very similar to those of opsins, implying those residues have no utility in distinguishing opsins from non-opsins. | |||
Conserved outgroup residues shared with opsins evidently describe commonalities needed for generic GPCR structure and signaling but not specifically for photobiology. Departures from the norm in certain opsin classes might indicate they are no longer signaling or are signaling constituitively. Opsins also have conserved diagnostic residues and even [[Opsin_evolution:_informative_indels#Indels_in_the_TM4-EC2-TM5_region|regions]] not found in any GPCR. | |||
These close-in outgroup sequences illuminate, as discussed elsewhere, the [[Opsin_evolution:_orgins_of_opsins#Introduction:_the_origin_of_opsins|origin of opsins]]. | |||
[[Image:OpsinOutgroup.jpg]] | |||
> | Exon count refers to trimmed alignable sequences. Some idiosyncratic loops have been deleted. Intron positions and phases are indicated relative to bovine | ||
rhodopsin numbering (sometimes uncertain due to gapping issues). These GPCR [[Opsin_evolution:_ancestral_introns#Deep_ancestral_introns|do not]] significantly conserve major opsin introns, so parental relations remain obscure. | |||
1 | >GPR21_homSap Homo sapiens (human) orphan receptor good match mollusk opsin 1 exon | ||
IVFLTVLIISGNIIVIFVFHCAPLLNHHTTSYFIQTMAYADLFVGVSCVVPSLSLLHHPLPVEESLTCQIFGFVVSVLKSVSMASLACISIDRYIAITKPLTYNTLVTPWRLRLCIFLIWLYSTLVFLPSFFHWGKPGYH | |||
GDVFQWCAESWHTDSYFTLFIVMMLYAPAALIVCFTYFNIFRICQQHTKDISERQARFSSQSGETGEVQACPDKRYAMVLFRITSVFYILWLPYIIYFLLESSTGHSNRFASFLTTWLAISNSFCNCVIYSLSNSVFQR | |||
> | >GPR52_homSap Homo sapiens (human) orphan receptor immediate outgroup to opsins 1 exon | ||
IVLLTFLIIAGNLTVIFVFHCAPLLHHYTTSYFIQTMAYADLFVGVSCLVPTLSLLHYSTGVHESLTCQVFGYIISVLKSVSMACLACISVDRYLAITKPLSYNQLVTPCRLRICIILIWIYSCLIFLPSFFGWGKPGYH | |||
2 | GDIFEWCATSWLTSAYFTGFIVCLLYAPAAFVVCFTYFHIFKICRQHTKEINDRRARFPSHEVDSSRETGHSPDRRYAMVLFRITSVFYMLWLPYIIYFLLESSRVLDNPTLSFLTTWLAISNSFCNCVIYSLSNSVFRL | ||
>MTNR1A_homSap Homo sapiens (human) melatonin receptor circadian rhythm 2 exons 71-12 | |||
LIFTIVVDILGNLLVILSVYRNKKLRNA 1 | |||
2 GNIFVVSLAVADLVVAIYPYPLVLMSIFNNGWNLGYLHCQVSGFLMGLSVIGSIFNITGIAINRYCYICHSLKYDKLYSSKNSLCYVLLIWLLTLAAVLPNLRAGTLQYDPRIYSCTFAQ | |||
> | SVSSAYTIAVVVFHFLVPMIIVIFCYLRIWILVLQVRQRVKPDRKPKLKPQDFRNFVTMFVVFVLFAICWAPLNFIGLAVASDPASMVPRIPEWLFVASYYMAYFNSCLNAIIYGLLNQNFRK | ||
>HRH2_homSap Homo sapiens (human) histamine receptor 1 exon | |||
LAVLILITVAGNVVVCLAVGLNRRLRNLTNCFIVSLAITDLLLGLLVLPFSAIYQLSCKWSFGKVFCNIYTSLDVMLCTASILNLFMISLDRYCAVMDPLRYPVLVTPVRVAISLVLIWVISITLSFLSIHLGWNS | |||
RNETSKGNHTTSKCKVQVNEVYGLVDGLVTFYLPLLIMCITYYRIFKVARDQAKRINHISSWKAATIREHKATVTLAAVMGAFIICWFPYFTAFVYRGLRGDDAINEVLEAIVLWLGYANSALNPILYAALNRDFRT | |||
>SSTR1_homSap Homo sapiens (human) somatostatin receptor SOG Rgamma class 1 exon | |||
> | YSVVCLVGLCGNSMVIYVILRYAKMKTATNIYILNLAIADELLMLSVPFLVTSTLLRHWPFGALLCRLVLSVDAVNMFTSIYCLTVLSVDRYVAVVHPIKAARYRRPTVAKVVNLGVWVLSLLVILPIVVFSRT | ||
AANSDGTVACNMLMPEPAQRWLVGFVLYTFLMGFLLPVGAICLCYVLIIAKMRMVALKAGWQQRKRSERKITLMVMMVVMVFVICWMPFYVVQLVNVFAEQDDATVSQLSVILGYANSCANPILYGFLSDNFKR | |||
>OPRM1_homSap Homo sapiens (human) opioid receptor mu 3 exons 64-21 182-12 ? | |||
YSIVCVVGLFGNFLVMYVIVR 2 | |||
1 YTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIINVCNWILSSAIGLPVMFMATTKYRQ 1 | |||
2 GSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALVTIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKR | |||
> | |||
>GALR1_homSap Homo sapiens (human) galanin receptor SOG Rgamma 3 exons 225-00 251-00 ? | |||
2 | FGLIFALGVLGNSLVITVLARSKPGKPRSTTNLFILNLSIADLAYLLFCIPFQATVYALPTWVLGAFICKFIHYFFTVSMLVSIFTLAAMSVDRY | ||
VAIVHSRRSSSLRVSRNALLGVGCIWALSIAMASPVAYHQGLFHPRASNQTFCWEQWPDPRHKKAYVVCTFVFGYLLPLLLICFCYAK 0 | |||
0 VLNHLHKKLKNMSKKSEASKKK 0 | |||
0 TAQTVLVVVVVFGISWLPHHIIHLWAEFGVFPLTPASFLFRITAHCLAYSNSSVNPIIYAFLSENFRK | |||
> | >CCR4_homSap Homo sapiens (human) chemokine (C-C motif) receptor 1 exon | ||
YSLVFVFGLLGNSVVVLVLFKYKRLRSMTDVYLLNLAISDLLFVFSLPFWGYYAADQWVFGLGLCKMISWMYLVGFYSGIFFVMLMSIDRYLAIVHAVFSLRARTLTYGVITSLATWSVAVFASLPGFLFSTCY | |||
TERNHTYCKTKYSLNSTTWKVLSSLEINILGLVIPLGIMLFCYSMIIRTLQHCKNEKKNKAVKMIFAVVVLFLGFWTPYNIVLFLETLVELEVLQDCTFERYLDYAIQATETLAFVHCCLNPIIYFFLGEKFRK | |||
0 | >BDKRB2_homSap Homo sapiens (human) bradykinin receptor 1 exon | ||
0 | LWVLFVLATLENIFVLSVFCLHKSSCTVAEIYLGNLAAADLILACGLPFWAITISNNFDWLFGETLCRVVNAIISMNLYSSICFLMLVSIDRYLALVKTMSMGRMRGVRWAKLYSLVIWGCTLLLSSPMLVFRTMKEYS | ||
DEGHNVTACVISYPSLIWEVFTNMLLNVVGFLLPLSVITFCTMQIMQVLRNNEMQKFKEIQTERRATVLVLVVLLLFIICWLPFQISTFLDTLHRLGILSSCQDERIIDVITQIASFMAYSNSCLNPLVYVIVGKRFRK | |||
> | |||
>GPR17_homSap Homo sapiens (human) uracil/cys-leukotriene dual receptor 1 exon | |||
YLLDFILALVGNTLALWLFIRDHKSGTPANVFLMHLAVADLSCVLVLPTRLVYHFSGNHWPFGEIACRLTGFLFYLNMYASIYFLTCISADRFLAIVHPVKSLKLRRPLYAHLACAFLWVVVAVAMAPLLVSPQT | |||
VQTNHTVVCLQLYREKASHHALVSLAVAFTFPFITTVTCYLLIIRSLRQGLRVEKRLKTKAVRMIAIVLAIFLVCFVPYHVNRSVYVLHYRSHGASCATQRILALANRITSCLTSLNGALDPIMYFFVAEKFRH | |||
>CYSLTR1_homSap Homo sapiens (human) cys-leukotriene receptor 1 exon | |||
YSMISVVGFFGNGFVLYVLIKTYHKKSAFQVYMINLAVADLLCVCTLPLRVVYYVHKGIWLFGDFLCRLSTYALYVNLYCSIFFMTAMSFFRCIAIVFPVQNINLVTQKKARFVCVGIWIFVILTSSPFLMAKPQKDE | |||
> | KNNTKCFEPPQDNQTKNHVLVLHYVSLFVGFIIPFVIIIVCYTMIILTLLKKSMKKNLSSHKKAIGMIMVVTAAFLVSFMPYHIQRTIHLHFLHNETKPCDSVLRMQKSVVITLSLAASNCCFDPLLYFFSGGNFRK | ||
>P2RY8_homSap Homo sapiens (human) purinergic receptor AMIN Ralpha class 1 exon | |||
YSLVAAVSIPGNLFSLWVLCRRMGPRSPSVIFMINLSVTDLMLASVLPFQIYYHCNRHHWVFGVLLCNVVTVAFYANMYSSILTMTCISVERFLGVLYPLSSKRWRRRRYAVAACAGTWLLLLTALSPLARTDLTY | |||
PVHALGIITCFDVLKWTMLPSVAMWAVFLFTIFILLFLIPFVITVACYTATILKLLRTEEAHGREQRRRAVGLAAVVLLAFVTCFAPNNFVLLAHIVSRLFYGKSYYHVYKLTLCLSCLNNCLDPFVYYFASREFQL | |||
>HCRTR1_homSap Homo sapiens (human) orexin receptor Rbeta class 7 exons 57-12 117-00 187-12 226-00 276-21 310-12 | |||
YVAVFVVALVGNTL 1 | |||
2 VCLAVWRNHHMRTVTNYFIVNLSLADVLVTAICLPASLLVDITESWLFGHALCKVIPYLQ 0 | |||
0 AVSVSVAVLTLSFIALDRWYAICHPLLFKSTARRARGSILGIWAVSLAIMVPQAAVMECSSVLPELANRTRLFSVCDERWA 1 | |||
2 DDLYPKIYHSCFFIVTYLAPLGLMAMAYFQIFRKLWGRQ 0 | |||
0 IPGTTSALVRNWKRPSDQLGDLEQGLSGEPQPRGRAFLAEVKQMRARRKTAKMLMVVLLVFALCYLPISVLNVLKR 2 | |||
1 VFGMFRQASDREAVYACFTFSHWLVYANSAANPIIYNFLS 1 | |||
2 GKFRE | |||
> | |||
>TACR2_homSap Homo sapiens (human) tachykinin receptor Gq-coupled SOG Rgamma class first gc-ag 5 exons 135-21 202-21 250-00 ? 312-21 | |||
YLALVLVAVTGNAIVIWIILAHRRMRTVTNYFIVNLALADLCMAAFNAAFNFVYASHNIWYFGRAFCYFQNLFPITAMFVSIYSMTAIAADR 2 | |||
1 YMAIVHPFQPRLSAPSTKAVIAGIWLVALALASPQCFYSTVTMDQGATKCVVAWPEDSGGKTLLL 2 | |||
1 YHLVVIALIYFLPLAVMFVAYSVIGLTLWRRAVPGHQAHGANLRHLQAMKK 0 | |||
0 FVKTMVLVVLTFAICWLPYHLYFILGSFQEDIYCHKFIQQVYLALFWLAMSSTMYNPIIYCCLNHR 2 | |||
1 FRS | |||
> | |||
>NMUR2_homSap Homo sapiens (human) neuromedin U receptor 4 exons 229-00 ? 256-12 291-12 | |||
YVPIFVVGVIGNVLVCLVILQHQAMKTPTNYYLFSLAVSDLLVLLLGMPLEVYEMWRNYPFLFGPVGCYFKTALFETVCFASILSITTVSVERYVA | |||
ILHPFRAKLQSTRRRALRILGIVWGFSVLFSLPNTSIHGIKFHYFPNGSLVPGSATCTVIKPMWIYNFIIQVTSFLFYLLPMTVISVLYYLMALR 0 | |||
0 LKKDKSLEADEGNANIQRPCRKSVNKML 1 | |||
2 FVLVLVFAICWAPFHIDRLFFSFVEEWSESLAAVFNLVHVVS 1 | |||
2 GVFFYLSSAVNPIIYNLLSRRFQA | |||
> | |||
>QRFPR_homSap Homo sapiens (human) pyroglutamylated RFamide peptide receptor MEC Ralpha class 6 exons 105-12 157-12 180-00 247-21 277-12 | |||
2 | GVLIFALALFGNALVFYVVTRSKAMRTVTNIFICSLALSDLLITFFCIPVTMLQNISDNWLG 1 | ||
1 | 2 GAFICKMVPFVQSTAVVTEILTMTCIAVERHQGLVHPFKMKWQYTNRRAFTML 1 | ||
2 GVVWLVAVIVGSPMWHVQQLE 0 | |||
0 | 0 IKYDFLYEKEHICCLEEWTSPVHQKIYTTFILVILFLLPLMVMLILYSKIGYELWIKKRVGDGSVLRTIHGKEMSKIAR 2 | ||
1 KKKRAVIMMVTVVALFAVCWAPFHVVHMMIEY 1 | |||
2 SNFEKEYDDVTIKMIFAIVQIIGFSNSICNPIVYAFMNENFKK | |||
>GPR19_homSap Homo sapiens (human) orphan receptor 19 1 exon | |||
FGILWLFSIFGNSLVCLVIHRSRRTQSTTNYFVVSMACADLLISVASTPFVLLQFTTGRWTLGSATCKVVRYFQYLTPGVQIYVLLSICIDRFYTIVYPLSFKVSREKAKKMIAASWIFDAGFVTPVLFFYGS | |||
> | NWDSHCNYFLPSSWEGTAYTVIHFLVGFVIPSVLIILFYQKVIKYIWRIGTDGRTVRRTMNIVPRTKVKTIKMFLILNLLFLLSWLPFHVAQLWHPHEQDYKKSSLVFTAITWISFSSSASKPTLYSIYNANFRR | ||
>PPYR1_homSap Homo sapiens (human) pancreatic polypeptide receptor MEC Ralpha class 1 exon | |||
1 | YSIETVVGVLGNLCLMCVTVRQKEKANVTNLLIANLAFSDFLMCLLCQPLTAVYTIMDYWIFGETLCKMSAFIQCMSVTVSILSLVLVALERHQLIINPTGWKPSISQAYLGIVLIWVIACVLSLPFLANSILENVFHKNHS | ||
0 | KALEFLADKVVCTESWPLAHHRTIYTTFLLLFQYCLPLGFILVCYARIYRRLQRQGRVFHKGTYSLRAGHMKQVNVVLVVMVVAFAVLWLPLHVFNSLEDWHHEAIPICHGNLIFLVCHLLAMASTCVNPFIYGFLNTNFKK | ||
>NPY1R_homSap Homo sapiens (human) neuropeptide Y receptor Rbeta class 2 exons 225-00 | |||
> | YGAVIILGVSGNLALIIIILKQKEMRNVTNILIVNLSFSDLLVAIMCLPFTFVYTLMDHWVFGEAMCKLNPFVQCVSITVSIFSLVLIAVERHQ | ||
0 | LIINPRGWRPNNRHAYVGIAVIWVLAVASSLPFLIYQVMTDEPFQNVTLDAYKDKYVCFDQFPSDSHRLSYTTLLLVLQYFGPLCFIFICYFK 0 | ||
2 | 0 IYIRLKRRNNMMDKMRDNKYRSSETKRINIMLLSIVVAFAVCWLPLTIFNTVFDWNHQIIATCNHNLLFLLCHLTAMISTCVNPIFYGFLNKNFQR | ||
1 | |||
>PRLHR_homSap Homo sapiens (human) prolactin releasing hormone receptor 1 exon | |||
YSVVVVVGLVGNCLLVLVIARVRRLHNVTNFLIGNLALSDVLMCTACVPLTLAYAFEPRGWVFGGGLCHLVFFLQPVTVYVSVFTLTTIAVDRYVVLVHPLRRRISLRLSAYAVLAIWALSAVLALPAAVHTYHVELKP | |||
HDVRLCEEFWGSQERQRQLYAWGLLLVTYLLPLLVILLSYVRVSVKLRNRVVPGCVTQSQADWDRARRRRTFCLLVVVVVVFAVCWLPLHVFNLLRDLDPHAIDPYAFGLVQLLCHWLAMSSACYNPFIYAWLHDSFRE | |||
> | |||
>GPR161_homSap Homo sapiens (human) orphan receptor 161 2 exons 135-21 | |||
2 | IIVITIFVCLGNLVIVVTLYKKSYLLTLSNKFVFSLTLSNFLLSVLVLPFVVTSSIRREWIFGVVWCNFSALLYLLISSASMLTLGVIAIDR 2 | ||
1 YYAVLYPMVYPMKITGNRAVMALVYIWLHSLIGCLPPLFGWSSVEFDEFKWMCVAAWHREPGYTAFWQIWCALFPFLVMLVCYGFIFRVARVKARKVHCGTVV | |||
IVEEDAQRTGRKNSSTSTSSSGSRRNAFQGVVYSANQCKALITILVVLGAFMVTWGPYMVVIASEALWGKSSVSPSLETWATWLSFASAVCHPLIYGLWNKTVRK | |||
>ADRA1D_homSap Homo sapiens (human) alpha-1D-adrenergic receptor PUR Ralpha class 2 exons 274-12 | |||
> | LAAFILMAVAGNLLVILSVACNRHLQTVTNYFIVNLAVADLLLSATVLPFSATMEVLGFWAFGRAFCDVWAAVDVLCCTASILSLCTISVDRYVGVRHSLKYPAIMTERKAAAILALLWVVALVVSVGPLLGWKEPV | ||
PPDERFCGITEEAGYAVFSSVCSFYLPMAVIVVMYCRVYVVARSTTRSLEAG VKRERGKASE VVLRIHCRGAATGADGAHGMRSAKGHTFRSSLSVRLLKFSREKKAAKTLAIVVGVFVLCWFPFFFVLPL 1 | |||
2 GSLFPQLKPSEGVFKVIFWLGYFNSCVNPLIYPCSSREFKR | |||
>TRHR_homSap Homo sapiens (human) thyrotropin-releasing hormone receptor 2 exons 251-00 | |||
VLIICGLGIVGNIMVVLVVMRTKHMRTPTNCYLVSLAVADLMVLVAAGLPNITDSIYGSWVYGYVGCLCITYLQYLGINASSCSITAFTIERYIAICHPIKAQFLCTFSRAKKIII | |||
FVWAFTSLYCMLWFFLLDLNISTYKDAIVISCGYKISRNYYSPIYLMDFGVFYVVPMILATVLYGFIARILFLNPIPSDPKENSKTWKNDSTHQNTNLNVNTSNRCFNSTVSSRKQ 0 | |||
> | 0 VTKMLAVVVILFALLWMPYRTLVVVNSFLSSPFQENWFLLFCRICIYLNSAINPVIYNLMSQKFRA | ||
2 | >TSHR_homSap Homo sapiens (human) thyrotropin receptor 1 exon PM:20305779 | ||
VWFVSLLALLGNVFVLLILLTSHYKLNVPRFLMCNLAFADFCMGMYLLLIASVDLYTHSEYYNHAIDWQTGPGCNTAGFFTVFASELSVYTLTVITLERWYAITFAMRLDRKIRLRHACAIMVGGWVCCFLL | |||
ALLPLVGISSYAKVSICLPMDTETPLALAYIVFVLTLNIVAFVIVCCCYVKIYITVRNPQYNPGDKDTKIAKRMAVLIFTDFICMAPISFYALSAILNKPLITVSNSKILLVLFYPLNSCANPFLYAIFTKAFQR | |||
0 | |||
>ADRB2_homSap Homo sapiens (human) beta2 adrenoceptor pdb:2R4R PUR Ralpha class 1 exon | |||
> | MSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYAN | ||
ETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRFHVQNLSQVEQDGRTGHGLRRSSKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRI | |||
2 | |||
1 | >ADORA2A_homSap Homo sapiens (human) beta2 adenosine receptor pdb:3EML PUR Ralpha class 2 exons 144-21 | ||
ELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLR 2 | |||
1 YNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKNHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLA | |||
ARRQLKQMESQPLPGERARSTLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQ | |||
> | |||
>ADRB1_melGal Meleagris gallopavo (turkey) beta1 adrenergic receptor pdb:2VT4 1 exon | |||
2 | MALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRD | ||
EDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRKIDRASKRKRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCRSPDFRK | |||
> | |||
0 | |||
> | |||
1 | |||
> | |||
1 | |||
> | |||
> | >UROPS1_triAdh Trichoplax adhaerens (trichoplax) XM_002114542 ends trimmed 1 exon | ||
YSVLSLITILGNMLVFLTFYKHASLRTTSNLFIINLAITDLLTGGIKDTLFIYGLTSYNWPKSAILCSFVGFINCVCYVATVYTLTVIAVFRYLAIVCNLGRKIKRKHSILTIACVWIYSSACCLMPIIGWSRY | |||
IYEPTECTCVRSLDKKYYSYTIYILVADFLLPLSVVSFCYGNIFAKMKRNHHHIKRDLSDGQKLDVAEKLSRREEIVARRMFIILAEHVVCFLPYTIIVTMLAANGVDIEPVWYFIVGYLLNLNSALNPVTYIVVNPRLKK | |||
1 | |||
> | >UROPS2_triAdh Trichoplax adhaerens (trichoplax) XM_002112401 ends trimmed 2 exons 241-21 ? | ||
0 MTLFMLMSCIGNGAVLLVLRYHHDDIKSASNYFITNLALTDFLLGVLCMPCILISCLNGQWVFGQTLCSLTGFANSFFCINSMITLAAVSVEKYCAIASPLTY | |||
HHYMSKSKVTCVISIIWIHSAINASLPFLGWGEYVYLPFETICTVAWWSFPNYVGFIVGINFGLPTVIMSCTYFLILKIARKHSRRIGVST 2 | |||
1 RRIHYKTHIKATLMLLIVIGSFIVCWLPHLISMIYLTIYEISPLPCSFHQITTWLAMANSAFNPIIYGAMDTSIRK | |||
2 | |||
'''See also:''' [[Opsin_evolution:_LWS_PhyloSNPs|LWS]] | [[Opsin_evolution:_Encephalopsin_gene_loss|Encephalopsins]] | [[Opsin_evolution:_Melanopsin_gene_loss|Melanopsins]] | [[Opsin_evolution:_Neuropsin_phyloSNPs|Neuropsin]] | [[Opsin_evolution:_Peropsin_phyloSNPs|Peropsin]] | [[Opsin_evolution:_RGR_phyloSNPs|RGR phyloSNPs]] | [[Opsin_evolution:_update_blog|Update Blog]] | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 15:13, 30 September 2011
See also: LWS | Encephalopsins | Melanopsins | Neuropsin | Peropsin | RGR phyloSNPs | Update Blog
Introduction to Opsin Evolution
The Curated Set of Metazoan Opsins
Below is the largest set of phylogenetically dispersed hand-curated opsin sequences ever assembled. The sequences are organized into true orthology classes using coding indels, intron location and phase, synteny of flanking genes, diagnostic residues, blast clustering, and experimental characterization when available. The reference set of opsin sequences includes selected GenBank entries but mostly new opsins extracted from dozens of newly completed genome projects.
The set serves as a gene family classifier ... just uBlast an unknown candidate opsin against the full database below and look for consistent labeling of the top hits from the Opsin Classifier. Then validate the apparent orthology class using three independent classes of rare genomic events (indels, introns, signature residues) by including the new sequence in a full-width multalignment, being sure to include a couple dozen non-opsin GPCRs as controls. Then check for residual bilateral flanking gene adjacency when a genome assembly is available. The whole process takes only 15 seconds per query!
The set of reference sequences is deliberately not exhaustive -- that seriously over-samples in popular experimental clades such as vertebrates and insects. When a given clade has many similar sequences available, those in species with genome assemblies are chosen to represent the group, for example anole is preferred to gecko, and (rightly or wrongly) any experimental results from gecko transfered over. This avoids the uninformative clutter of near-identical sequences. However if a clade reflects a very deep divergence especially important to opsin evolution (such as lamprey or amphioxus), all available sequences are needed to maximally break up long branches.
Sequences not available from GenBank were culled from trace archives, tiled contigs, and genome assemblies, typically by uBlastx against the growing set of reference sequences, as described in the annotation section. The level of error in the curated sequences is very low, declines with time as anomalies are revisited and repaired, but never reaches zero because of problems inherent to experimental data, imperfect sequencing reads, less-than-complete assemblies, and sequence manipulation.
Rare genomic changes can supplement (and even displace) traditional maximal likelihood and bayesian inference in resolving polytomic divergence nodes in gene and species tree topologies. Rare genomic changes applicable to opsins include coding indels (deletions and insertions), intron placement (position and phase comparison), synteny (gene order along the chromosome), and gene copy number change (gene gain from retropositional, tandem, segmental, and whole genome duplications; gene loss from pseudogenization or deletion). Results from these methods must be evaluated for their susceptibility to homoplasy (misleading recurrent independent events that mimic a single event) and incomplete penetration in the population level at the time of speciation (lineage sorting).
Opsins are more informatively stored as proteins since nucleotide sequences are far too diverged at metazoan evolutionary distances and do not explicitly manifest residue properties that experience selection. These protein sequences are parsed into constituent exons using genomic information when available -- fortunately splicing mechanisms are exceeding well conserved across animal phyla. When not directly available (eg the opsin originated as a cDNA in a species lacking a genome project), exon breaks have been inferred from the phylogenetically closest neighbors via heterologous alignment (but not in insects where intron turnover is too high). As an example, lamprey opsins from Geotria australis and Lethenteron japonicum can work as blastn queries to locate orthologs within the Petromyzon maritimus genome project (which consists solely of 19 million traces as of mid-December 2007).Numbers flanking exons, namely 0 1 2, show the phasing of each intron, eg 12 means an overhang of 1 bp at the 3' end of an exon with fragmentary codon completed by a 2 bp overhang at the beginning of the next exon.
Intron position and phasing are conserved over vast evolutionary distances -- human to sponge and beyond -- with the exception of certain rogue lineages (that unfortunately includes two major model organisms). Informative conservation is still available long after protein percent identity has slid into the twilight zone of uncertainty (critical in opsins because they are readily confused with generic GPCR receptors). For example, no variation whatsoever in intron pattern has occurred in any vertebrate opsin class since lamprey divergence, no events in many billions of years of branch length. Prior to that, rare events are observed, for example LWS opsins gained an extra intron of phase 12. In a protein having 333 locations with 3 possible phases at each location, convergent evolution (homoplasy) is uncommon, that is, close examination establishes opsin introns are still highly informative even when sequences have greatly diverged.
Insertions and deletions (coding indels) are sufficiently uncommon in opsins that they are a potentially phylogenetically informative class of rare genomic events. We'll harvest these from a massive opsin alignment and stratify by location and phylogenetic depth. Indels are more subject to homoplasy than intron gain or loss but the risk of that varies greatly by region. Indels are less affected by lineage-specific rates than other event classes.
Syntentic relationships can have great value in determining orthology relationships, though gene order is not often conserved over great evolutionary timescales due to chromosomal rearrangements. As with introns, certain rogue lineages lose almost all this information. Tracking synteny is nomenclaturally confused because many genes are unnamed and simply numbered by annotation pipeline procedures in each genome without homological consistency. Here, HUGO-convention names are used for two genes flanking each side of a given human opsin. Strand orientation is noted relative to a fixed convention of plus strand for the human opsin. Then each genome assembly is visited to determine the extent of conservation of these flanking genes and orientation. In the event humans lack the gene, synteny is defined by the nearest diverging species, typically platypus or chicken, that has the gene. Sometimes the original synteny is only partly retained (left or right half-synteny). For deeply diverging species such as amphioxus, flanking genes can be are pushed forward into 'nearby' species. This can also be done without an assembly in species with sufficiently large contigs containing the opsin. Blast clustering can be uncertain because of diminishing percent identity or even loss of these flanking genes; common chromosomal displacements such as inversions eventually disrupt most syntentic arrangements.
The fasta header of each sequence serves as a miniature database that conveniently collects this and other basic information, with fields showing the opsin type, genus, species and common name, accession number, best PubMed citation, indels, intron pattern, sequence length, lambda max adsorption, flanking synteny, and G protein type with which it interacts (all subject to availability and work-in-progress). These fasta headers by themselves provide a quick over view the opsin reference set collection -- simply paste into a blank document and pull lines containing '>'.
Thus the usual querying at GenBank does not remotely compare to the Opsin Classifier. Those sequences -- often mis-annotated by an unattended pipeline or well-intentioned individual with no qualifications in comparative genomics -- are spread out over separate databases not accessible by any single method, with difficult-to-interpret edge creep of genomic blast matches, uncorrected frameshifts, missing stop codons and erroneous amino terminals.
As worst-case scenario, half-baked annotation of the sea urchin genome by pipelines and casual procedures has left a awful legacy of bogus opsin gene structures at GenBank, journal alignments, and genome browsers -- often mis-classified because of an inadequate set of reference sequences and non-opsin GPCR controls, chimeric confusion in tandem duplicates, and non-consideration of intron structure, indels, and synteny. These errors may ripple downstream to errors by subsequent scientists trusting GenBank nr as classifier and lead to a whole subculture taken up with non-existent "virtual" opsins and attendant vacuous speculation on echinoderm photoreception.
Melanopsins, the unexpected rhabdomeric-class Gq-coupled opsin recently found in upper deuterostomes, illustrate some of the difficulties of accurate annotation. They can be confused homologically due to various expansions and contractions. Mammals, human through platypus, have a single melanopsin. However a common ancestor to chicken, lizard, frog, teleost fish, and possibly cartilaginous fish had a multi-gene segmental duplication with both resulting melanopsins retained (though substantially diverged). In ray-finned fish, a processed retrogene arose that may be functional in zebrafish though lost in fugu and stickleback. After whole genome duplication, zebrafish also retained two copies of the original melanopsin. Chondrichthyes also have a second copy of the primary melanopsin but synteny -- which is essential for analysis since intron placement is uninformative in duplications and sequence alignment too dependent on unknown rates -- is not available in the current contig-level assembly.
Amphioxus contains two melanopsins from an apparently independent duplication. Flanking gene order today bears no relation to any vertebrate gene order. The lamprey situation awaits assembly of its traces or targeted transcript studies. At this time, only a four-exon fragmentary melanopsin can be recovered (however with high percent identity, 80%). Possibly orthologs of this melanopsin locus could be tracked into the highly derived tunicates, acorn worm, and sea urchins. The distinctive intron pattern may even allow melanopsin antecedents to be identified in Cnidaria and Protostomia. At this point, the best blastp match to insects stands at 37% with no evident syntenic or intronic support.
While clade-specific proliferations of melanopsins -- and implied role subfunctionalizations -- confounds the situation for chordates, it has little impact on the opsin classifier described here. Unknown sequences readily find their place because of the extensive phylogenetic representation of reference sequence orthology classes and the inherent distance of melanopsins from the ciliary sub-collection. At that level of alignment, the melanopsins serve as outgroup to ciliary opsins and so help define motifs specific to Gt-coupled signaling and other structure/function issues.
It appears however that far too much 'lumping' has taken place in nearly all non-imaging opsins, for example encephalopsins, melanopsins, and peropsins. The taxonomic counterpart is too much lumping of species outside of mammal. In all likelihood, additional opsin orthology classes need definition and distinctive naming. These opsins were belatedly discovered through whole-genome homology searches; they may differ one from another just as much as cone and rod opsins. However, it is currently difficult to disentangle phylogenetically short-lived expansions within known families from deep-rooted parallel-evolving subfamilies concentrated (superficially) within 'secondary' photoreception systems. Genome sampling density and completion efforts are overwhelmingly concentrated in a few model species and near human.
We should not be too hasty to write off peropsins as mere auxiliary retinal isomerases that replenish cis-retinal for 'real' opsins. This reaction is not a simple isomerization (like glucose isomerase) but photoisomerization with an evolutionarily tuned and conserved visual light action spectrum. There is no reason that trans-retinal cannot be the signaling agonist with conversion to all-cis retinal the waste product. Pairing two opposing types of opsins in a single cell or nearby cells then completes a full visual cycle with interesting opportunities for sensory capabilities. Of all the so-called retinal isomerases, peropsins retain more of the diagnostic residues of ciliary opsins; possibly they function similarly but in balance with another opsin class co-expressed in the same cell. Melanopsin, surprisingly, is fully capable of self-replenishment, so perhaps the first-studied ciliary opsins are the true anomaly with their need for an auxiliary replenishment cycle in the retinal epithelium.
It's abundantly clear from distinct ancestral introns and alignment clustering that peropsins together with neuropsins and RGR opsins comprise a distinctive subclade within the overall opsin gene tree. The comparative genomics of RGR illustrates the danger of phylogenetic under-sampling: with much deeper sampling, we can observe that the E/DRY motif -- conserved across all classes of opsins (indeed GPCR) and critical to maintaining non-signaling conformation -- has become GRY in all boreoeutheran mammals (it's ERY in afrotheres, lost without residual debris in marsupials, and DRY from platypus through shark and even tunicate, and DRY in neuropsins and peropsins).
In other words, after several trillion years of branch length conservation as charged amino acid, a radical amino acid substitution has taken hold -- to glycine with its tiny non-interactive side chain. Yet this subclade of placental mammals, this glycine has been conserved without exception for over two billion years of branch length. Given the importance of this motif for maintaining the non-signaling state, this suggests a major change in functional properties of RGR opsins within boreoeutheres, a change that does not tolerate a reduced alphabet at this site. That change might be breakdown to non-signaling isomerase within that clade. It remains unclear how marsupials cope without the gene at all or how Afrothera and Xenarthra visual cycles differ from other placental mammals.
Comparative Genomics of DRY motifs in exon 3 of RGR Opsins: 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 human 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 macaque 1 RWPYGSGGCQAHGFQGFTTALASICGSAAIAWGRYHHYCT 1 lemur 1 RWPYGSDGCKVHGFQGFATALASISGSAAIAWGRYHQYCT 1 treeshrew 1 RWPHGSEGCQVHGFQGFATALASICGSAAVAWGRYHHYCT 1 mouse 1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1 rabbit 1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1 pika 1 RWPYGSEGCQAHGFQGFVTALASICSSAAVAWGRYHHFCT 1 cow 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 horse 1 RWPYGSNGCQAHGFQGFVTALASICSSAAIAWGRYHHYCS 1 cat 1 RWPYGPDGCQAHGFQGFATALASICSSAALAWGRYHHYCT 1 dog 1 RWPYGSGGCQAHGFQGFAAALASICGSAAVAWGRYHHYCT 1 bat 1 RWPFGPDGCQAHGFQGFATALASICSSAAIAWGRYHHYCT 1 shrew 1 RWPYGSDGCQAHGFQGFVTALASICSCAAIAWERYHHYCT 1 elephant 1 RWPYGSDGCQAHGFQGFVMALTSICSCAAIAWERYHHYCT 1 hyrax 1 HWPYGSGGCQAHGFQGFTVALASICSCAAIAWERYHHYCT 1 tenrec 1 RWPHGSDSCQAHSFQGFATALASISSSAAIAWERYRHHCT 1 sloth 1 RWPYGSGGCQAHGFQGFVTALASISSSAAIAWERCHRHCI 1 armadillo 1 HWPYGAEGCRLHGFQGFATALASISLSAAIGWDRYLRHCS 1 platypus 1 YWPYGSDGCQIHGFHGFLTALTSISSAAAVAWDRHHQYCT 1 lizard 1 YWPYGSEGCQIHGFQGFLTALASISSSAAVAWDRYHHYCT 1 chicken 1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1 frog 1 YWPYGSDGCQTHGFQGFVTALASIHFIAAIAWDRYHQYCT 1 stickleback 1 YWPYGSDGCQTHGFQGFMTALASIHFIAAIAWDRYHQYCT 1 zfish1 1 HWPFGSEGCQLHAFQGMVSILAAISFLGAVAWDRYHQYCT 1 zfish2 1 YWPYGSEGCQTHGFQGFVTALASIHFVAAIAWDRYHQYCT 1 tetraodon 1 YWPYGSDGCQTHGFQGFVTALASIHFVAAIAWDRYHQYCT 1 fugu 1 YWPYGSEGCQTHGFHGFLTALASIHFIAAIAWDRYHQYCT 1 medaka 1 YWPYGSDGCQTHGFQGFMTALASIHFIAAIAWDRYHQYCT 1 pimephales 1 YWPYGSDGCQTHGFQGFMTALASIHFVAAIAWDRYHQYCT 1 osmerus 1 YWPYGSEGCQTHGFHGFLMALASINACAAIAWDRYHQNCS 1 elephantshark 1 EWPFGSIGCQLDAFIGMAPTFISIAGAALVAKDKYYRICK 1 tunicate 1 1 RWLFGKFGCYFHGFAGMLFGLGSIGNLTVISIDRYIITCK 1 tunicate 2 1 QWPFGDLGCQVDAFIGMAPTFISIAGAALIAKDKYYRFCK 1 tunicate 2
Excellent recent publications have greatly added to our understanding of the evolutionary origin of light reception capabilities, yet only with the advent of genomic sequencing have we begun to get a full grip on opsins and a few other evo-devo photoreceptor components such as PAX6. Yet critical early-diverging metazoan genomes that might retain information on ancestral characters are greatly under-represented given the branch lengths involved. No purpose is served by elaborate gene tree computations that include a single token peropsin or parietopsin among hundreds of cone opsins; if opsins are central to the endeavor yet so haphazardly annotated, what is the status of dozens of lesser but still critical photoreceptor gene families?
The question of how many times eyes independently arose is ill-posed. We can only wonder how people with no grasp whatsoever of the underlying molecular biology of photoreception could pontificate so boldly on its evolution prior to even sequencing of the first opsin in 1980. First collect comprehensive comparative genomic data sets on all the components in all the relevant clades; then ask what it could mean to homologenize two contemporary systems comprised of partially overlapping sets of heterogeneous paralogs from assorted ancient gene families having long complex histories of clade-specific expansion and contraction cycles. If four arrestins serve a thousand GPCR, if even specialized photoreceptor cells express dozens of barely related 7TM signaling molecules, it's difficult to imagine what a cognate arrestin or ectopic evo-devo cassette might be. It's difficult enough to reconstruct an ancestral gene or two; reconstructing ancestral systems biology interactions lies in the future.
After reviewing topics such as ciliary opsin in protostomes, rhabdomeric opsins in deuterostomes,the rich opsin repertoires in cnidarians and probable opsins in sponges, we will consider special topics such as the origin of image-forming eyes between amphioxus and lamprey divergences, noting throughout that our notion of 'eye' is much more nuanced than earlier. The reconstruction of Ur-bilateran and Urmetazoan eyes awaits additional cnidarian genomes but no new ones are currently underway. However the plethora of new arthropod and lophotrochozoan genome assemblies has opened up new avenues of research as the realization grows that fly and nematode model species are exceedingly derived, with better ancestral characters retained in other lineages.
Numerous conflicting gene trees have been published for ciliary opsins. Some methodologies have bordered on the preposterous -- thin phylogenetic coverage, dimly related outgroups such as adrenergic receptors, and naive underlying mutational models assumed for maximal likelihood despite great diversity of species and many billions of years of branch length. Nonetheless the resultant trees have only moderate conflict, suggesting that a definitive opsin gene tree is not far off. We'll do this using the multiple types of evidence discussed above.
The first point to be understood in deuterostome ciliary opsin evolution is jawless fish such as lamprey already exhibit a full-blown set of modern rod and cone opsins whereas earlier diverging clades as represented by hemichordate, echinoderm, amphioxus and tunicate genomes totally lack them and indeed lack imaging eyes altogether, while using their rhabdomeric opsins in a very distinct signaling system for their own photoreception systems. Despite 7 sequenced opsin mRNAs in the amphioxus Branchiostoma belcheri and an initial assembly in Branchiostoma floridae providing counterparts there, no rod/cone opsin can be located there or in earlier diverging deuterostomes with genome projects (3 tunicates, 2 urchins, 1 acorn worm). These species may have larval eye spots, ocelli, pigment cells, and related photoreceptors but lack imaging eyes.
Characters in extant (living) species should never be confused with ancestral characters present at the time of divergence nodes (last common ancestors); conceivably these early diverging deuterostomes have lost opsin genes, perhaps due to a habitat shift to deep water, burrowing habitat, or nocturnal lifestyle. However the molecular evidence is quite clear that full-blown pentachromatic color vision and most other modern ciliary opsin classes first appeared during the evolutionary stem preceding lamprey divergence. These are demonstrably not 'new' genes but derived from gene duplication and divergence of still older opsins of ciliary class..
The fossil record is unsatisfactory: less than 1 bilateran in 10,000 in Chengjiang and Burgess Shale fossils is even a candidate for deuterostomy. Low numbers of specimens and poor preservation conspire with career pressures and impact-seeking journals in egregiously misinterpreted data in the view of Hou, discoverer of the Chinese lagerstaette. Myllokunmingia is the best situation with 500 specimens; however Haikouichthys as supposed stem deuterostome, Metaspreggina as supposed post-Ediacaran, and Yunnanozoan generally are problematic, valid only in the eye of some beholder.
While signs of bilaterally disposed eyes are sometimes documentable, it does not follow these were image-forming eyes. Indeed contemporary Branchiostoma and tunicate larva have an eye-spot (ocellus); the genomes contain ciliary opsins but only clustering to ENCEPH and PPIN -- still a long ways from any imaging opsin. Echinoderms and hemichordates genomes also have opsins but even further diverged. Sea urchin genome encodes at least six opsins, four of these cluster classify to rhabdomeric, ciliary and Go-type. Tube feet are apparently the photosensory organ in adult urchins.
The oldest known fossil lamprey, Priscomyzon, dates at 360 myr to the Devonian. Molecular clocks place lamprey appearance at approximately 430 myr, some 100 million years after Chengjiang and Burgess Shales fossil Lagerstaette formed. Like most soft tissues, eyes seldom leave a good fossil record, though bilateral placement might be reflected in bone orbits. Hagfish, sister group to lamprey, have imaging eyes but have not been studied; their opsins situation may be derived due to deepwater marine habitat (similarly deepwater coelacanth opsins are adapted to less scattered wavelengths, centered at420 nm).
The next-diverging chondrichthyes have inadequate data at GenBank -- only a few rhodopsin genes from skates and dogfish. This makes even fragmentary opsins from the partially sequenced elephantfish Callorhyncus milii quite valuable. Those 9 fragments and 3 from the lamprey genome are provided in the data section -- the opsin classifier tool can reliably type a fragment from a single mid-sized exon. Full length genes are preferable but these fragments serve to prove existence of various gene class at the time of a given divergence node. Further, they can validate certain rare genomic events provided the fragment happens to overlap the region of interest.
On the other hand, thousands of high-quality Cambrian arthropod fossils unmistakably show stalked paired eyes. Fossil trilobite eyes have been much studied; these are better preserved due to calcite as lens crystalin. Imaging eyes of contemporary arthropods and lophotrochozoa are rhabdomeric, utilizing depolarizing Gq-type receptor, phospholipase C, phosphoinositol, diacylglycerol, and transient receptor potential TRP and TRPL channel signaling. However their genomes can also contain ciliary opsins, using hyperpolarizing Gt-type transducins and phosphodiesterase cGMP second-messaging (as well as Go-type gustducin ciliary opsins in other types of photoreceptors).
Vertebrates are just the opposite, having crossed over to a near-exclusively ciliary opsin-based imaging system, while retaining rhabdomeric signaling in retinal ganglion cells and elsewhere. (A very recent report demonstrates very rare human and mouse cone cells expressing exclusively melanopsin; however melanopsin cannot have given rise to ciliary cone opsins.)
It must not be thought that bilaterans invented imaging eyes because much earlier diverging cubomedusan jellyfish such as Carybdea marsupialis has 4 eyestalks each with 6 photoreceptors of 4 types: simple eyespots, pigment cups, complex pigment cups with lenses, and camera-type eyes with a cornea, lens, and retina. This jellyfish chases, captures, and eats 'more advanced' teleost fish. This wing of cnidarians very much needs a genome project.
Cnidarian opsins are becoming available from Hydra and Nematostella genomes and targeted hybridization experiments. Hydra may express a ciliary-type opsin in ectodermal sensory nerve cells whereas Nematostella has opsins classifying between melanopsin and encephalopsin. These distant opsin are very difficult to distinguish bioinformatically from non-opsins in the rhodopsin superfamily within GPCR, so it is exceedingly important to include controls because a somewhat unspecialized photoreceptor cell may also express other sensory system signaling non-opsins that are nonetheless genetically homologous.
In summary, there is no evidence whatsoever -- and every reason to doubt from genomic analysis -- that deuterostomes had imaging eyes during the Cambrian. Despite this, a BBC series "Walking With Monsters" portrayed a school of 25 mm Haikouichthys attacking and wounding an Anomalocaris twenty times their size. It is easy to guess at a scientific advisory panel that envisions deuterostomes triumphing over protostomes. This recurrent anthropocentric fantasy is echoed in museum imagery of early mammals nimbly predating on dinosaur nests -- dioramas quietly dismantled after Yucatan meteorite discovery.
Imaging eyes are not essential to survival; even today subterranean mammals such as blind mole rat flourish without them. Discounting ray-finned fish numbers, a very substantial proportion of all extant animal species lack imaging eyes 525 myr after the Cambrian. Of 33 animal phyla, one-third have no specialized organ for detecting light, one-third have light-sensitive organs, and the remaining 6 have imaging eyes (Cnidaria, Mollusca, Annelida, Onychophora, Arthropoda, and Chordata). Thus 82% of animal phyla have survived well over 500 myr without imaging eyes despite the supposedly unrelenting competition/predation from animals having them.
Opsin Gene and Species Trees
The phylogenetic tree below shows the presence or absence of various opsin genes in clade-representative species, as reflected in the collected reference sequences. The purpose is timing appearance (or disappearance) of a given class of opsin gene. For example, cone and rod opsins first appeared before lamprey divergence; otherwise they are absent from urochordates, cephalochordates, and earlier deuterostomes. Note however a given gene might appear absent because of a genome project gap, lack of experimental effort, insufficient or outdated bioinformatics, or species choice (ie present in a different representative of that clade). In other cases (eg platypus SWS1) pseudogene remnants or a syntenically proven deletion establish a gene as definitely absent.
Opsin retention at 18 genetic loci over 540 million years (Jan 2009 update)
1 RHO1 petMar calMil takRub xenTro galGal anoCar ornAna monDom bosTau homSap RHO1
2 RHO2 geoAus calMil danRer ...... galGal anoCar ...... ...... ...... ...... RHO2
3 SWS1 geoAus ...... danRer xenLae galGal anoCar :::::: monDom bosTau homSap SWS1
4 SWS2 geoAus ...... gasAcu xenTro galGal anoCar ornAna ...... ...... ...... SWS2
5 LWS petMar calMil danRer xenTro galGal anoCar ornAna monDom bosTau homSap LWS
6 VAOP petMar calMil danRer xenTro galGal anoCar ...... ...... ...... ...... VAOP
7 PIN ------ calMil ...... xenTro galGal utaSta...... ...... ...... ...... PIN
8 PPIN petMar calMil danRer xenTro ...... anoCar ...... ...... ...... ...... PPIN
9 PARIE petMar ------ danRer xenTro ...... anoCar ...... ...... ...... ...... PARIE
10 ENCEPH petMar calMil danRer xenTro galGal anoCar ...... monDom :::::: homSap ENCEPH
11 TMT ------ calMil danRer xenTro galGal anoCar ornAna monDom ...... ...... TMT
12 MEL1 petMar calMil danRer xenTro galGal anoCar ornAna monDom bosTau homSap MEL1
13 MEL2 ------ calMil danRer xenLae galGal anoCar ...... ...... ...... ...... MEL2
14 NEUR1 petMar calMil danRer xenTro galGal anoCar ornAna monDom bosTau homSap NEUR1
15 NEUR2 ------ calMil danRer xenTro galGal anoCar ...... ...... ...... ...... NEUR2
16 NEUR3 petMar calMil danRer xenTro galGal anoCar ...... ...... ...... ...... NEUR3
17 PER ------ calMil danRer xenTro galGal anoCar ornAna monDom bosTau homSap PER
18 RGR petMar calMil danRer xenTro galGal anoCar ornAna ...... bosTau homSap RGR
1 3 15 17 17 16 18 7 8 7 8
Notes: in low coverage genomes, missing data was sought in closely related species.
Absence of data (---) is distinguished from strong evidence of absence (...).
In rare cases, pseudogene debris is still detectable at syntenic location (:::)
Gene loss is a mixture of deep stem loss (mammals) and recent lineage-specific loss.
The opsin gene trees below illustrate only a few of the myriad possibilities, even beginning with commonsense ordering (blast nearest neighbors). Because these gene families originated long ago and are only known from remotely related representatives in extant species with wildly differently mutational mechanisms and histories, the true tree cannot be reliably inferred from maximal likelihood. Indeed no two such attempts have ever come up with the same gene tree! Instead, we'll keep this set of gene trees in view until analysis of rare genomic events is completed
Using the Opsin Classifier
Below is the primary collection of opsin protein sequences. Here "fields" in the fasta header show gene name, genus, species, common name, heterotrimeric G protein alpha subunit used in signaling, intron structure, synteny (2 flanking genes on each side of the opsin), indel status, sequence length, lambda max, and comment field. The 230-odd sequences are now organized into deuterostomes, lophotrochozoans, and ecdysozoan divisions, further broken refined into ciliary, rhabdomeric, or neither. Even with the full set copied into the uBlast, a novel opsin candidate can be classified in 6 seconds over just a conventional DSL internet connection.
On 26 Nov 07, I added 41 new sequences, mostly arthropod rhabdomeric imaging opsins, extracting them from a 2007 pancrustacean opsin paper, using the much-studied accessions in their Table 1, as ordered phylogenetically according to their Fig.3, with subsampling to avoid too-close sequences and narrow lineage-specific expansions. This involved replacing a few defective accessions and partial sequences with comparable complete ones, favoring sequences with completed or planned genome projects which can be directly intronated and their synteny determined. Lambda max values were helpfully compiled for all these opsins by the original authors.
This significantly upgrades the resolving power of the Opsin Classifier vis-a-vis these new classes of protostome opsins. It raises thorny nomenclature issues because of short-sighted historic uses such as rhodopsin or LWS for both fruitfly and human genes. These are indeed vaguely homologous in the distant pre-Bilateran GPCR past but are certainly not orthologous as implied by the same common name. The definition of orthology requires genes under comparison in extant species to descend from a single parent gene in their last common ancestor. If this LCA requirement is relaxed, the entire set of a thousand GPCR all become orthologous.
Additional ecdysozoan and lophotrochozoan opsins are needed, whatever new that can be extracted from invertebrate genome projects; some of these will prove ciliary and conversely some deuterostome opsins rhabdomeric. Melanopsin/encephalopsin appear at the heart of the Big Switchover that took place in chordates -- their imaging opsins did not arise from gene duplication and divergence of anything we see among contemporary protostomal imaging opsins or any reconstructed ancestor.
In fact, none of the opsin genes in Ur-bilatera destined to become rhabdomeric imaging opsins in living arthropods (even all of protostomia) seems to have descended directly to any deuterostome. It may turn out that none of the opsin genes in Ur-Bilatera destined to become ciliary imaging opsins in living vertebrates (even all of Deuterostomia) survived in any protostome. The pool of GPCR genes was already large and signaling diversified across gene copies. However lophotrochozoa and basal ecdysozoan ciliary opsins are still largely unexplored. A similarly 'bad' Venn diagram could hold in Ur-eumetazoa. Here the only two cnidarians with sequence data (Hydra and Nematostella) were not the best choices for studying opsin evolution. We may have to wait for genome sequencing of a full-featured cubomedusan.
Please do not add text or edit sequences at this time, even though genomeWiki encourages participation; consider starting a fresh page to hold your contributions. After finishing the first round of articles, everything will be revisited and reorganized at a second pass.
Curated opsin reference sequences
The 460 manually curated opsin sequences below provides an optimal set for phylogenetic research in opsin evolution. These sequences have been collected from over 75 metazoan genome projects in addition to those extracted comprehensively from GenBank. Errors in genome assembly, flawed or missing annotation and mistakes in gene models have largely been corrected by advanced bioinformatic methods. (GenBank data cannot be used as it stands.) An opsin sequences is included only if it provides more information than clutter -- gene expansions limited to narrow clades and over-sampled loci are restricted to proxy representatives.
However eight opsin genetic loci -- some new -- have greatly expanded reference sequence collections within separate articles. These explore more fine-grained opsin evolution restricted to the 48 vertebrate genomes available on 10 Jan 2010. Data from complete genomes offers four advantages over transcripts, namely it can provide regulatory regions, introns, flanking gene synteny, and exhaustive opsin paralog sets present in the organism.
- LWS reference sequences from 95 species
- Encephalopsin and TMT: yet more opsins lost in mammals
- RGR opsin: abrupt change in DRY motif in boreoeutheres
- Peropsin: phyloSNPs in peropsin evolution
- Neuropsin (OPN5): comparative genomics of 52 deuterostome genes and three new paralogs NEUR2, NEUR3, NEUR4
The fasta header lines below are structured as a flatfile database. Fields are separated by spaces rather than tabs (which display poorly). The first field (all that will be retained by most web alignment tools) gives the gene name together with a 3+3 genus-species latin name code.
These gene names mostly comply with international conventions and historic first-use but do not perpetuate poor terminology (use of already-assigned gene names, obtuse acronyms, greek letters, roman numerals, hyphens, subscripts, calling everything rhodopsin and so forth). Paralog subfamilies are numbered sequentially; recent gene duplicates (limited in phylogenetic distribution) are distinguished by lower case letters with different letters denoting those demonstrably arising from whole genome duplications. The nomenclature closely follows the opsin gene tree, indeed implicitly provides it (other than deep branching).
Each major branch of the metazoan opsin gene tree is assigned a distinct short gene name that evokes its common name without need for explanatory table. Some names require periodic revision as more information emerges on their phylogenetic origin. While a later field of the fasta header does supply synonomy, should any confusion over sequence nomenclature ever arise, it is best resolved by text query of a sequence snippet using web browser search capability.
The next three fields provide the full genus and species names (per GenBank taxonomy) and then a one-word common name. The latter can provide helpful orientation when the genus/species is unfamiliar. Next comes a field providing three units of higher order taxonomic nomenclature that allow phylogenetic subsetting of the database. For example chicken receives Deut.Saur.Arch as a deuterostome, sauropod like lizard, but archeosaur like finch but unlike lizard. This field implicitly places all the sequences on a phylogenetic tree. It assumes, unlike GenBank, that ecdysozoa and lophotrochozoa are the correct partition of protostomes.
The next two fields provide a GenBank accession and PubMed identifier if any exist. The reference sequences here do not follow GenBank when it errs (as it often does, eg by retaining introns, omitting exons, extending the 'first' exon to a false initiating methionine, extending the 'final' exon to a false stop codon, leapfrogging across tandem genes to form chimeric genes). Many uncurated gene models are poor quality. However even these entries can provide historic credits and supplement the protein entries below with underlying dna.
The PubMed identifier links to a recent journal article with substantive experimental data specifically relevant to the opsin sequence at hand. Such papers review earlier literature and most hubs also provide subsequent papers citing the initial paper. This allows rapid collection of all work pertaining to a given opsin gene. Papers are not cited that simply sequenced tens of thousands of transcripts without characterization or comment on fortuitously included opsins.
The next field provides a measure of sequence completeness. About 1 sequence in 7 is fragmentary (but often not affecting the 7-transmembrane core). While important in proving that a given orthology class exists in a given clade (as are recent pseudogenes) and helpful in defining conserved residues in local alignments, these sequences cannot currently be completed. Reasons include too-rapid divergence of non-membrane regions, gaps in assembly coverage of individual exons, uncertainty in exon boundaries, and incomplete transcripts.
The following field suggests a Galpha signaling partner for each opsin, important to understanding both structure and function. This is more wish-list than established fact because the rare cases known from direct experimental work have merely been extended below using homology to not-too-divergent orthologs in fairly closely related species -- even though the reliability of this procedure remains untested.
Indeed, the opsin literature is confused over what this classification should even mean, the options being (1) the specific Galpha gene product(s) physically docking to the cytoplasmic face of the opsin, (2) merely a gene tree subclade (eg Gs) of interacting Galpha protein whatever that might mean within a complicated gene duplication tree itself scarcely comparable over metazoa and already inadequate to resolve rod and cone GNAT transducins, (3) the small molecule produced that does the actual signaling (eg cyclic nucleotide or phosphoinositol), or (4) the character of downstream signaling ultimately produced (eg inhibitory).
Here the issue revolves around what is experimentally measurable -- opsins are not tested in a biochemically defined reconstituted system against individual Galpha proteins purified from the cognate species but rather signaling is characterized indirectly via the effect of some inhibitor or output of small molecule. While it appears that all opsin loci retain the universal features of GPCR signaling, it hasn't actually been established that all do in fact signal (rather than just serve as non-signaling photoisomerases in intermediary metabolism of retinoids).
The next fields provide syntenic gene order (when known and relevant to a research issue), alternative gene names, tissues of expression, percent identity to nearest neighbor, and unstructured miscellaneous comments. Lambda-max peak adsorption could be added though it has been measured by a mix of techniques and is not necessarily reliably calculated from classical tuning residues in less-familiar opsin classes. Length in amino acids is no longer provided because it does not satisfactorily explain fragmentary entries nor the run-on amino and carboxy termini in some opsin classes.
Adherence to rigid database format drops off after the early fields because the information is just not available or predictable for many entries. However the structure still suffices for routine queries such as 'how many opsins do tunicates have' or 'which protostomes have ciliary opsins'. Note the fasta header lines are easily isolated by pasting the entire database (including sequence lines) into a spreadsheet and sorting to collect lines beginning with '>'. The fields can be separated cleanly into columns by replacing spaces with tabs.
The protein sequences themselves are broken into their constituent exons. The numbering indicates phase (ie basepair overhang), a deeply conserved invariant useful in distinguishing orthology classes and gene tree divisions. All opsins in all species use standard GT-AG splice junctions. Alternate splicing within the core protein may occur but never produces functional protein. The exon structures in genomic species have been determined individually by tblastn and those of non-genomic species predicted by homology.
Full length genes beginning with an initial methionine and ending with a stop codon are the most useful. However the initial methionine cannot always be disambiguated, a problem not restricted to opsins. In run-on genes lacking 3' transcripts, the stop codon is taken as the first encountered in extending the last homologically reliable exon; however the true stop codon could lie in a different reading frame (ie in a cryptic terminal exon).
Fragmentary genes are included if the species occupies a critical position in the phylogenetic tree (eg lamprey) and the fragment is long enough to reliably classify it. Fragmentary data often takes the form of entire exons missing from genomic coverage (rather than incomplete exons). When these exons lie in unconnected contigs, their combination into a single gene is not entirely reliable.
For example the Callorhinchus genome project, currently with 0.6 exon coverage, still suffices to establish that many gene duplications important to vertebrate photoreceptive diversity had already occurred by the chondrichthyian divergence node. However without transcripts, a lineage-specific duplication in LWS might have result in a single chimeric gene assembled from scattered individual LWS exons.
Fragmentary genes for isolated fast-evolving invertebrates are difficult to distinguish from non-opsin GPCR which share many motifs such as DRY. To balance the need for maximal phylogenetic coverage against the imperative of not contaminating the opsin reference collection with non-opsin GPCR, partial genes are required to contain the K296 Schiff base and other diagnostic residues and motifs specific to opsins.
>RHO1_bosTau Bos taurus (cow) Deut.Euth.Laur NM_001014890 2145276 full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 +H1FOO -PLXND1)
0 MNGTEGPNFYVPFSNKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSR 2
1 YIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQ 0
0 FRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA* 0
>RHO1_homSap Homo sapiens (human) Deut.Euth.Euar NM_000539 16565402 full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 +H1FOO -PLXND1)
0 MNGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSR 2
1 YIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEASATVSKTETSQVAPA* 0
>RHO1_monDom Monodelphis domesticus (opossum) Deut.Meta.Dide genomic full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 +H1FOO -PLXND1)
0 MNGTEGPNFYVPFSNKTGTVRSPFEEPQYYLADPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAIADLFMVFGGFTMTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIIGVAFTWVMALACAFPPLIGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSNFGPIFMTIPAFFAKSSSVYNPVIYIMMNKQ 0
0 FRTCMITTLCCGKNPLGDDEASATASKTETSQVAPA* 0
>RHO1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono ABN43074 17339011 full Gt RH1 RHO rhodopsin rod opsin syn(+IFT122 - -PLXND1)
0 MNGTEGQDFYIPMSNKTGVVRSPFEYPQYYLAEPWQYSVLAAYMFMLIMLGFPINFLTLYVTIQHKKLRTPLNYILLNLAFANHFMVLGGFTTTLYTSLHGYFVFGPTGCNIEGFFATLG 1
2 GEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACALPPLVGWSR 2
1 YIPEGMQCSCGIDYYTLRPEVNNESFVIYMFVVHFTIPMTIIFFCYGRLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTVPAFFAKSSAIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEASATASKTEQSSVSTSQVSPA* 0
>RHO1_galGal Gallus gallus (chicken) Deut.Saur.Arch NM_205490 1385866 full Gt RH1 RHO rhodopsin rod syn(--MBD4 syn(+IFT122 +H1FOO -PLXND1)
0 MNGTEGQDFYVPMSNKTGVVRSPFEYPQYYLAEPWKFSALAAYMFMLILLGFPVNFLTLYVTIQHKKLRTPLNYILLNLVVADLFMVFGGFTTTMYTSMNGYFVFGVTGCYIEGFFATLG 1
2 GEIALWSLVVLAVERYVVVCKPMSNFRFGENHAIMGVAFSWIMAMACAAPPLFGWSR 2
1 YIPEGMQCSCGIDYYTLKPEINNESFVIYMFVVHFMIPLAVIFFCYGNLVCTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTNQGSDFGPIFMTIPAFFAKSSAIYNPVIYIVMNKQ 0
0 FRNCMITTLCCGKNPLGDEDTSAGKTETSSVSTSQVSPA* 0
>RHO1_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 - -PLXND1)
0 MNGTEGQNFYVPMSNKTGVVRNPFEYPQYYLADPWQFSALAAYMFLLILLGFPINFLTLFVTIQHKKLRTPLNYILLNLAVANLFMVLMGFTTTMYTSMNGYFIFGTVGCNIEGFFATLG 1
2 GEMGLWSLVVLAVERYVVICKPMSNFRFGETHALIGVSCTWIMALACAGPPLLGWSR 2
1 YIPEGMQCSCGVDYYTPTPEVHNESFVIYMFLVHFVTPLTIIFFCYGRLVCTVKA 0
0 AAAQQQESATTQKAEREVTRMVVIMVISFLVCWVPYASVAFYIFTHQGSDFGPVFMTIPAFFAKSSAIYNPVIYILMNKQ 0
0 FRNCMIMTLCCGKNPLGDEDTSAGTKTETSTVSTSQVSPA* 0
>RHO1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 - -PLXND1)
0 MNGTEGPNFYIPMSNKTGVVRSPFDYPQYYLAEPWKYSALAAYMFLLILLGFPINFMTLYVTIQHKKLRTPLNYILLNLVFANHFMVLCGFTVTMYTSMHGYFIFGQTGCYIEGFFATLG 1
2 GEMALWSLVVLAIERYVVVCKPMANFRFGENHAIMGVVFTWIMALSCAAPPLFGWSR 2
1 YIPEGMQCSCGVDYYTLKPEVNNESFVVYMFIVHFTIPLCVIFFCYGRLLCTVKE 0
0 AAAQQQESATTQKAEKEVTRMVVMMVIFFLICWVPYAYVAFYIFTHQGSDFGPVFMTVPAFFAKSSAIYNPVIYIVLNKQ 0
0 FRNCLITTLCCGKNPFGDEEGSSAASSKTEASSVSSSQVSPA* 0
>RHO1_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGPNFYVPMTNKTGVVRSPFEYPQYYLADPWKYSALAAYMFFLILTGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTMYTAMNGYFVFGVVGCNLEGFFATFG 1
2 GIIALWCLVVLAIERYIVVCKPISNFRFGENHAIMGVVFTWIMALACAGPPLFGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFIVHFTIPLIIIFFCYGRLMCTVKE 0
0 AAAQQQESATTQKAEKEVTRMVYIMVISYLVCWLPYASVSFYIFTHQGSDFGPVFMTVPAFFAKTASVYNPVIYILMNKQ 0
0 FRNCMITTLCCGKNPFGDEETTSAGTSKTEASSVSSSQVSPA* 0
>RHO1_latCha Latimeria chalumnae (coelacanth) Deut.Sarc.Coel AAD30519 10339578 full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGPNFYVPMSNKTGVVRNPFEYPQYYLADPWKYSALAAYMFFLILVGFPINFLTLFVTIQHKKLRTPLNYILLDLAVADLCMVFGGFFVTMYSSMNGYFVLGPTGCNIEGFFATLG 1
2 GQVALWALVVLAIERYVVVCKPMSNFRFGENHAIMGVIFTWIMALSCAVPPLFGWSR 2
1 YIPEGMQSSCGVDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGRLVCTVKD 0
0 AAAQQQESATTQKAEKEVTRMVIVMVISFLVCWVPYASVAAYIFFNQGSEFGPVFMTAPSFFAKSASFYNPVIYILLNKQ 0
0 FRNCMITTLCCGKNPFGDEDATSAAGSSKTEASSVSSSSVSPA* 0
>RHO1_angAng Anguilla anguilla (european_eel) Deut.Acti.Elop L78008 full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGPNFYIPMSNITGVVRSPFEYPQYYLAEPWAYTILAAYMFTLILLGFPVNFLTLYVTIEHKKLRTPLNYILLNLAVANLFMVFGGFTTTVYTSMHGYFVFGETGCNLEGYFATLG 1
2 GEISLWSLVVLAIERWVVVCKPMSNFRFGENHAIMGLAFTWIMANSCAMPPLFGWSR 2
1 YIPEGMQCSCGVDYYTLKPEVNNESFVIYMFIVHFSVPLTIISFCYGRLVCTVKE 0
0 AAAQQQESETTQRAEREVTRMVVIMVIAFLVCWVPYASVAWYIFTHQGSTFGPVFMTVPSFFAKSSAIYNPLIYICLNSQ 0
0 FRNCMITTLFCGKNPFQEEEGASTTASKTEASSVSSVSPA* 0
>RHO1_conMyr Conger myriaster (conger_eel) Deut.Acti.Elop AB043817 full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGPNFYIPMSNATGVVRSPFEYPQYYLAEPWAFSALSAYMFFLIIAGFPINFLTLYVTIEHKKLRTPLNYILLNLAVADLFMVFGGFTTTMYTSMHGYFVFGPTGCNIEGFFATLG 1
2 GEIALWCLVVLAIERWMVVCKPVTNFRFGESHAIMGVMVTWTMALACALPPLFGWSR 2
1 YIPEGLQCSCGIDYYTRAPGINNESFVIYMFTCHFSIPLAVISFCYGRLVCTVKE 0
0 AAAQQQESETTQRAEREVTRMVVIMVISFLVCWVPYASVAWYIFTHQGSTFGPIFMTIPSFFAKSSALYNPMIYICMNKQ 0
0 FRHCMITTLCCGKNPFEEEDGASATSSKTEASSVSSSSVSPA* 0
>RHO1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_131212 18597743 full Gt RH1 RHO rhodopsin rod opsin extra-ocular rhodopsin
0 MNGTEGPNFYVPMSNRTGLVRSPFEEPQYYLAEPWQFSLLAAYMLFLILGSFPINALTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTVTLYTALHGYFLLGVTGCNIEGFFATLG 1
2 GEIALWSLVVLAIERYIVVCKPMSTFRFGEKHAIIGVGFTWVMALTCAVPPLLGWSR 2
1 YIPEGMQCSCGIDYYTPKPEVHNTSFVIYMFILHFSIPLLIIFFCYSRLLCTVRA 0
0 AAAQQQESETTQRAEREVTRMVVVMVIAFLVCWVPYASVAWYIFANQGAEFGPVFMTVPAFFAKSAALYNPVIYIMLNRQ 0
0 FRNCMLSTVCCGKNPLAEDESSSAVSSKTQSSVVSSAQVSPA* 0
>RHO1_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? RH1 RHO rhodopsin rod opsin
0 MNGTEGPNFYVPMSNKTGVVRSPFEYPQYYLAEPWKYSLVAAYMLFLIVTAFPVNFLTLFVTVKHKKLRTPLNYVLLNLAVADLFMVIGGFTVTLYTALHAYFVLGVTGCNIEGFFATLG 1
2 GEIALWSLVVLAIERYIVVCKPVTNFRFGEKHAIAGLAFTWIMALSCAVPPLLGWSR 2
1 YIPEGMQCSCGIDYYTPKPEIHNTSFVIYMFVLHFCIPLAIIFFCYSRLLCTVRA 0
0 AAALQQESETTQRAEKEVTRMVIVMVISFLVCWVPYASVAWYIFANQGTEFGPVFMTAPAFFAKSAALYNPVIYILLNRQ 0
0 FRNCMITTVCCGKNPFGDDDATTSVSKTQSSSVSTSQVAPA* 0
>RHO1_takRub Takifugu rubripes (fugu) Deut.Acti.Perc AF201472 10692583 full Gt RH1 RHO rhodopsin rod opsin syn(-MBD4 +IFT122 - -PLXND1)
0 MNGTEGPNFYIPMSNKTGVVRSPFEYPQYYLAEPWKYSLVAAYMLFLIITAFPVNFLTLFVTVKHKKLRTPLNYVLLNLAVADLFMVIGGFTVTLYTALHAYFVLGVTGCNIEGFFATLG 1
2 GEIALWSLVVLAVERYIVVCKPMTNFRFGEKHAIAGLVFTWIMALTCATPPLLGWSR 2
1 YIPEGMQCSCGIDYYTPKPEINNTSFVIYMFILHFSIPLAIIFFCYSRLLCTVRA 0
0 AAALQQESETTQRAEKEVTRMVIVMVISFLVCWVPYASVAWYIFANQGTEFGPVFMTAPAFFAKSAALYNPVIYILLNRQ 0
0 FRNCMITTVCCGKNPFGDDDAATTVSKTQSSSVSSSQVAPA* 0
>RHO1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc BT028623 full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGPYFYVPMMNTTGIVRSPYEYPQYYLVNPAAYAALGAYMFLLILVGFPVNFLTLYVTLEHKKLRTPLNYILLNLAVANLFMVLGGFTTTMYTSMHGYFVLGRLGCNVEGFFATLG 1
2 GEIALWSLVVLAVERWVVVCKLIANFRFGEDHAIMGLALTWVMASACAVPPLVGWSR 2
1 YIPEGMQCSCGVDYYTRAEGFNNESFVIYMFVCHFMIPMIIIFFCYGRLLCAVKEA 0
0 AAAQQESETTQRAEREVSRMVVIMVIAFLVCWVPYASVAWFIFCNQGSEFGPVFMTIPAFFAKSSSVYNPLIYICMNKQ 0
0 FRHCMITTLCCGKNPFEEEEGASSTKTEASSVSTSSVSPA* 0
>RHO1_oryLat Oryzias latipes (medaka) Deut.Acti.Perc AB180742 15964232 full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGPYFYVPMVNTTGIVRSPYEYPQYYLVNPAAYAALGAYMFFLILVGFPINFLTLYVTLEHKKLRTPLNYILLNLAVADLFMVFGGFTTTMYTSMHGYFVLGRLGCNLEGFFATLG 1
2 GEIGLWSLVVLAIERWVVVCKPISNFRFGENHAIMGLVFTWIMAASCAVPPLVGWSR 2
1 YIPEGMQCSCGVDYYTRAEGFNNESFVVYMFVCHFLIPLIVVFFCYGRLLCAVKEA 0
0 AAAQQESETTQRAEREVTRMVVIMVIGFLVCWLPYASVAWYIFTNQGSEFGPLFMTIPAFFAKSSSIYNPAIYICMNKQ 0
0 FRNCMITTLCCGKNPFEEEEGASTTASKTEASSVSSSSVSPA* 0
>RHO1_leuEri Leucoraja erinacea (skate) Deut.Chon.Elas U81514 9256070 full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGENFYVPMSNKTGVVRSPFDYPQYYLGEPWMFSALAAYMFFLILTGLPVNFLTLFVTIQHKKLRQPLNYILLNLAVSDLFMVFGGFTTTIITSMNGYFIFGPAGCNFEGFFATLG 1
2 GEVGLWCLVVLAIERYMVVCKPMANFRFGSQHAIIGVVFTWIMALSCAGPPLVGWSR 2
1 YIPEGLQCSCGVDYYTMKPEVNNESFVIYMFVVHFTIPLIVIFFCYGRLVCTVKE 0
0 AAAQQQESESTQRAEREVTRMVIIMVVAFLICWVPYASVAFYIFINQGCDFTPFFMTVPAFFAKSSAVYNPLIYILMNKQ 0
0 FRNCMITTICLGKNPFEEEESTSASASKTEASSVSSSQVAPA* 0
>RHO1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGENFYIPMSNKTGVVRSPFEYPQYYLAEPWQFSILAAYMFFLIITCFPVNFLTLYVTFEHKKLRQPLNFILLNLAVADLFMVFGGFFITVYTSLHGYFVFGVTGCNFEGFFATLG 1
2 GEIGLWSLVVLAIERYVVVCKPMSNFRFGTNHAIMGVAFTWVMALACAVPPLMGWSR 2
1 YIPEGLQCSCGVDYYTLKPEINNESFVIYMFVVHFLIPLIIIFFCYGRLVCTVKE 0
0 AAAQQQESESTQRAEREVTRMVIIMVIFFLICWVPYASVAFFIFTNQGSEFGPIFMAVPAFFAKSSALYNPLIYILLNKQ 0
0 FRNCMITTLCCGKNPFEEDESTSAAASKTEASSVSSSQVSPA* 0
>RHO1_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGENFYIPFSNKTGLARSPFEYPQYYLAEPWKYSVLAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVANLFMVLFGFTLTMYSSMNGYFVFGPTMCNFEGFFATLG 1
2 GEMSLWSLVVLAIERYIVICKPMGNFRFGSTHAYMGVAFTWFMALSCAAPPLVGWSR 2
1 YLPEGMQCSCGPDYYTLNPNFNNESFVIYMFLVHFIIPFIVIFFCYGRLLCTVKE 0
0 AAAAQQESASTQKAEKEVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTVPAFFAKTSALYNPIIYILMNKQ 0
0 FRNCMITTLCCGKNPLGDEDSGASTSKTEVSSVSTSQVSPA* 0
>RHO1_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366493 17463225 full Gt RhA
0 MNGTEGQNFYIPFSNKTDVARSPFEYPQYYLAEPWKFSALAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVSNLFMILFGFTTTMYTSMNGYFVFGPTMCSIEGFFATLG 1
2 GEVSLWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVALTWVMALSCAAPPLLGWSR 2
1 YLPEGMQCSCGPDYYTMNPTYNNESFVIYMFIVHFTIPFVIIFFSYGRLLCTVKE 0
0 AAAAQQESASTQKAEKEVTRMVVLMVVGFLVCWVPYASVAFYIFTNQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ 0
0 FRNCMITTLCCGKNPLGDDDSGASTSKTEVSSVSTSQVAPA* 0
>RHO1_letJap Lethenteron japonicum (lamprey) Deut.Agna.Hype AB116382 15096614 full Gt RH1 RHO rhodopsin rod opsin
0 MNGTEGDNFYVPFSNKTGLARSPYEYPQYYLAEPWKYSALAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAMANLFMVLFGFTVTMYTSMNGYFVFGPTMCSIEGFFATLG 1
2 GEVALWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVAFTWIMALACAAPPLVGWSR 2
1 YIPEGMQCSCGPDYYTLNPNFNNESYVVYMFVVHFLVPFVIIFFCYGRLLCTVKE 0
0 AAAAQQESASTQKAEKEVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ 0
0 FRNCMITTLCCGKNPLGDDESGASTSKTEVSSVSTSQVSPA* 0
>RHO2_galGal Gallus gallus (chicken) Deut.Saur.Arch NP_990771 2268324 full Gt syn(-IHPK3 -LEMD2 +RHO2 -GRM4 -NUDT3)
0 MNGTEGINFYVPMSNKTGVVRSPFEYPQYYLAEPWKYRLVCCYIFFLISTGLPINLLTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFVFGPVGCAVEGFFATLG 1
2 GQVALWSLVVLAIERYIVVCKPMGNFRFSATHAMMGIAFTWVMAFSCAAPPLFGWSR 2
1 YMPEGMQCSCGPDYYTHNPDYHNESYVLYMFVIHFIIPVVVIFFSYGRLICKVRE 0
0 AAAQQQESATTQKAEKEVTRMVILMVLGFMLAWTPYAVVAFWIFTNKGADFTATLMAVPAFFSKSSSLYNPIIYVLMNKQ 0
0 FRNCMITTICCGKNPFGDEDVSSTVSQSKTEVSSVSSSQVSPA* 0
>RHO2_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch NM_001076696 full G?
0 MNGTEGINFYVPMSNKTGVVRSPFEYPQYYLAEPWKYRLVCCYIFFLISTGFPINFLTLLVTFKHKKLRQPLNYILVNLAVADLCMACFGFTVTFYTAWNGYFVFGPIGCAVEGFFATLG 1
2 GQVALWSLVVLAIERYIVICKPMGNFRFSASHALMGIAFTWVMAISCAAPPLFGWSR 2
1 YIPEGMQCSCGPDYYTHNPDFHNESYVLYMFVIHFIIPVVIIFFSYGRLVCKVRE 0
0 AAAQQQESATTQKAEKEVTRMVILMVLGFMLAWTPYAVVAFWIFTNKGADFTATLMAVPAFFSKSSSLYNPIIYVLMNKQ 0
0 FRNCMITTICCGKNPFGDEETSSTVSQSKTEVSSVSSSQVSPA* 0
>RHO2_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt syn(-IP6K3 -LEMD2 -MLN +RHO2 -GRM4 -NUDT3)
0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLAEPWKYKVVCCYIFFLIFTGLPINILTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFIFGPIGCAIEGFFATLG 1
2 GQVALWSLVVLAIERYIVVCKPMGNFRFSATHALMGISFTWFMSFSCAAPPLLGWSR 2
1 YIPEGMQCSCGPDYYTLNPDYHNESYVLYMFGVHFVIPVVVIFFSYGRLICKVRE 0
0 AAAQQQESASTQKAEREVTRMVILMVLGFLLAWTPYAMVAFWIFTNKGVDFSATLMSVPAFFSKSSSLYNPIIYVLMNKQ 0
0 FRNCMITTICCGKNPFGDEDVSSSVSQSKTEVSSVSSSQVSPA* 0
>RHO2_gekGek Gekko gekko (gecko) Deut.Saur.Lepi AY024356 11591478 full Gt in pure rod retina
0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLADPWKFKVLSFYMFFLIAAGMPLNGLTLFVTFQHKKLRQPLNYILVNLAAANLVTVCCGFTVTFYASWYAYFVFGPIGCAIEGFFATIG 1
2 GQVALWSLVVLAIERYIVICKPMGNFRFSATHAIMGIAFTWFMALACAGPPLFGWSR 2
1 FIPEGMQCSCGPDYYTLNPDFHNESYVIYMFIVHFTVPMVVIFFSYGRLVCKVRE 0
0 AAAQQQESATTQKAEKEVTRMVILMVLGFLLAWTPYAATAIWIFTNRGAAFSVTFMTIPAFFSKSSSIYNPIIYVLLNKQ 0
0 FRNCMVTTICCGKNPFGDEDVSSSVSQSKTEVSSVSSSQVAPA* 0
>RHO2_podSic Podarcis sicula (lizard) Deut.Saur.Lepi AY941829 12887418 full G?
0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLAEPWKYKMVCCYIFFLISTGLPINLLTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFIFGPIGCAIEGFFATLG 1
2 GQVALWSLVVLAIERYIVVCKPMGNFRFSSSHALMGIAFTWVMSLSCACPPLFGWSR 2
1 YIPEGMQCSCGPDYYTLNPDYHNESYVVYMFVIHFVIPVVVIFFSYGRLICKVRE 0
0 AAAQQQESASTQKAEKEVTRMVILMVLGFMLAWTPYAVVAFWIFTNKGADFSATLMSVPAFFSKSSSLYNPIIYVLMNKQ 0
0 FRNCMITTICCGKNPFGDDDVSSTVSQSKTEVSSISSSQVSPA* 0
>RHO2_pheMad Phelsuma madagascariensis (lizard) Deut.Saur.Lepi AF074044 10069370 full G?
0 MNGTEGFNFYVPVSNRTGLVRSPYEYPQYYLAEPWKFKALSLYMFFLILVGLPLNGLTLFVTFQHKKLRQPLNYILVNLAVANLLMVICGFTVTFYTSWYGYFVFGPMGCAFEGFFATIG 1
2 GQVALWSLVVLAIERYIVICKPMGNFRFSSSHAMMGISFTWFMALCCGGPPLFGWSR 2
1 FIPEGMQCSCGPDYYTLNPDFHNESYVIYLFTVHFLTPMIIIFFSYGRLVCKVRE 0
0 AAAQQQESATTQKAEKEVTRMVILMVMGFLVAWTPYATVACWIFNNKGAEFSVTFMTVPAFFSKSSCIYNPIIYVLLNKQ 0
0 FRNCMVTTICCGKNPFGDEDASSSVSQSKTEVSSVSSSQVAPA* 0
>RHO2_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt
0 MNGTEGINFYVPHSNKTGVVRSPFEYPQYYLADPWKYSIVCAYMFFLIITGLPINLLTLVVTFKHKKLRQPLNYILVNLAVADLFMVCFGFTVTFSTAINGYFIFGPRGCAIEGFMATLG 1
2 GEVALWSLVVLAIERYIVVCKPMGNFRFSNNHSIIGIVFTWLAALSCAAPPLFGWSR 2
1 YLPEGMQCSCGPDYYTMNPDYHNESFVIYMFVVHFFIPVIVIFVSYGRLICKVKE 0
0 AAAQQQESASTQKAEREVTRMVILMVIGFMTAWTPYATVAFWIFMNKGAEFGATFMAAPAFFSKSSALYNPIIYVLMNKQ 0
0 FRNCMVTTLCCGKNPFGDDDVSSSVSAGKTEVSSVSSSQVSPA* 0
>RHO2_latCha Latimeria chalumnae (coelacanth) Deut.Sarc.Coel AH007713 10339578 full Gt RH2
0 MNGTEGMNFYVPLSNRTGLVRSPFEYTQYYLAEPWKFSVLCAYMFLLIILGFPINFLTLLVTFKHKKLRQPLNYILVNLAVASLFMVVFGFTVTFYSSLNGYFVLGPMGCAMEGFFATLG 1
2 GQVALWSLVVLAIERYIVVCKPMGNFRFASSHAIMGIAFTWIMALACAAPPLVGWSR 2
1 YIPEGLQCSCGPDYYTLNPDFHNESYVMYLFLVHFLLPIIIIFFTYGRLICKVKE 0
0 AAAQQQESASTQKAEKEVTRMVILMVIGFLTAWVPYASAAFWIFCNRGAEFTATLMTVPAFFSKSSCLFNPIIYVLLNKQ 0
0 FRNCMITTLCCGKNPLGDDDTSSAVSQSKTDVSSVSSSQVSPA* 0
>RHO2_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic 15647516 full G? opn1mw1 ancestral representing 4x expansion
0 MNGTEGNNFYIPMSNRTGLVRSPYEYPQYYLAEPWQFKLLAVYMFFLICLGFPINGLTLLVTAQHKKLRQPLNFILVNLAVAGTIMVCFGFTVTFYSAINGYFVLGPTGCAIEGFMATLG 1
2 GEVALWSLVVLAIERYIVVCKPMGSFKFSSSHAMAGIAFTWVMAMACAAPPLFGWSR 2
1 YIPEGMQCSCGPDYYTLNPEYNNESYVLYMFICHGIVPVTIIFFTYGRLVCTVKA 0
0 AAAQQQESESTQKAEREVTRMVILMVLGFLVAWTPYATVAAWIFFNKGAAFSAQFMAVPAFFSKSSALFNPIIYVLLNKQ 0
0 FRNCMLTTLFCGKNPLGDDESSTVSTSKTEVSSVSPA* 0
>RHO2a_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_131253 15647516 full G? opn1mw1
0 MNGTEGSNFYIPMSNRTGLVRSPYDYTQYYLAEPWKFKALAFYMFLLIIFGFPINVLTLVVTAQHKKLRQPLNYILVNLAFAGTIMVIFGFTVSFYCSLVGYMALGPLGCVMEGFFATLG 1
2 GQVALWSLVVLAIERYIVVCKPMGSFKFSANHAMAGIAFTWFMACSCAVPPLFGWSR 2
1 YLPEGMQTSCGPDYYTLNPEYNNESYVMYMFSCHFCIPVTTIFFTYGSLVCTVKA 0
0 AAAQQQESESTQKAEREVTRMVILMVLGFLFAWVPYASFAAWIFFNRGAAFSAQAMAVPAFFSKTSAVFNPIIYVLLNKQ 0
0 FRSCMLNTLFCGKSPLGDDESSSVSTSKTEVSSVSPA* 0
>RHO2b_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_182891 15647516 full G? opn1mw2 highest expression
0 MNGTEGNNFYIPMSNRTGLVRSPYEYTQYYLADPWQFKALAFYMFFLICFGLPINVLTLLVTAQHKKLRQPLNYILVNLAFAGTIMAFFGFTVTFYCSINGYMALGPTGCAIEGFFATLG 1
2 GQVALWSLVVLAIERYIVVCKPMGSFKFSSNHAMAGIAFTWVMASSCAVPPLFGWSR 2
1 YIPEGMQTSCGPDYYTLNPEFNNESYVLYMFSCHFCVPVTTIFFTYGSLVCTVKA 0
0 AAAQQQESESTQKAEREVTRMVILMVLGFLVAWVPYASFAAWIFFNRGAAFSAQAMAIPAFFSKASALFNPIIYVLLNKQ 0
0 FRSCMLNTLFCGKSPLGDDESSSVSTSKTEVSSVSPA* 0
>RHO2c_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_182892 15647516 full G? opn1mw3
0 MNGTEGNNFYIPMSNRTGLVRSPYEYPQYYLAEPWQFKLLAVYMFFLMCFGFPINGLTLVVTAQHKKLRQPLNFILVNLAVAGTIMVCFGFTVTFYTAINGYFVLGPTGCAIEGFMATLG 1
2 GQISLWSLVVLAIERYIVVCKPMGSFKFSSNHAFAGIGFTWIMALSCAAPPLVGWSR 2
1 YIPEGMQCSCGPDYYTLNPDYNNESYVLYMFCCHFIFPVTTIFFTYGRLVCTVKA 0
0 AAAQQQESESTQKAEREVTRMVILMVLGFLVAWTPYASVAAWIFFNRGAAFSAQFMAVPAFFSKSSSIFNPIIYVLLNKQ 0
0 FRNCMLTTLFCGKNPLGDDESSTVSTSKTEVSSVSPA* 0
>RHO2d_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_131254 15647516 full G? opn1mw4
0 MNGTEGNNFYIPLSNRTGLARSPYEYPQYYLAEPWQFKLLAVYMFFLICLGFPINGLTLLVTAQHKKLRQPLNFILVNLAVAGTIMVCFGFTVTFYTAINGYFVLGPTGCAIEGFMATLG 1
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSASHAFAGCAFTWVMAMACAAPPLVGWPR 2
1 YIPEGMQCSCGPDYYTLNPEYNNESYVLYMFICHFILPVTIIFFTYGRLVCTVKA 0
0 AAAQQQESESTQKAEREVTRMVILMVLGFLIAWTPYATVAAWIFFNKGAAFSAQFMAVPAFFSKTSALYNPVIYVLLNKQ 0
0 FRNCMLTTLFCGKNPLGDDESSTVSTSKTEVSSVSPA* 0
>RHO2_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? RH2
0 MVWDGGIEPNGTEGKNFYIPMSNRTGIVRSPFEYPQYYLVDPIMFKMLALYMFFLICTGTPINGLTLLVTAQNKKLRQPLNYILVNLAVAGLIMCAFGFTITITSAINGYFILGATACAVEGFMATLG 1
2 GEVALWSLVVLAIERYIVVCKPMGSFKFTGTHAAVGVLFTWIMAFACAGPPLFGWSR 2
1 YLPEGMQCSCGPDYYTLAPGYNNESYVIYMFVVHFFVPVFLIFFTYGSVVLTVka 0
0 AAAQQQESESTQKAQREVTRMCILMVLGFLVAWTPYATFSGWIFMNKGAAFHPLTAALCAFFAKSSALYNPVIYVLM 0
0 FRNCMLSTIGMGGAVDDETSVSASKTEVSSVS* 0
>RHO2_takRub Takifugu rubripes (fugu) Deut.Acti.Perc AF226989 10692583 full G?
0 MAWDGGIEPNGTEGKNFYIPMSNRTGIVRSPFEYPQYYLADPIMFKILALYMFFLICTGTPINGLTLLVTAQNKKLRQPLNYILVNLAVAGLIMCAFGFTITITSAVNGYFILGATACAVEGFMATLG 1
2 GEIALWSLVVLAVERYVVVCKPMGSFKFTGTHAAVGVAFTWIMAFACAAPPLFGWSR 2
1 YLPEGMQCSCGPDYYTLAPGYNNESYVIYMFSCHFFVPVITIFFTYGSLVLTVKA 0
0 AAAQQQESESTQKAQKEVTRMCILMVFGFLMAWTPYATFSAWIFMNKGAAFHPLTAAVCAFFAKSSALYNPVIYVLLNKQ 0
0 FRNCMLSTIGMGGAVDDETSVSASKTEVSSVS* 0
>RHO2_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? 2nd near-identical tandem copy
0 MAWEGGLEPNGTEGKNFYIPMSNRTGVVRSPFEYQQYYLADPIMFKILALYMFFLICTGTPINGLTLLVTAQNKKLRQPLNYILVNLAVAGLIMCAFGFTITITSAVNGYFILGATACAVEGFMATLG 1
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSGTHAGAGVLFTWIMAMACAAPPLFGWSR 2
1 YLPEGMQCSCGPDYYTLAPGFNNESYVIYMFVVHFFTPVFIIFFTYGSLVLTVKA 0
0 AAAQQQESESTQKAEREVTRMCILMVFGFLVAWVPYASFAGWIFLNKGAPFSALTAAIPAFFAKSSALYNPVIYVLLNKQ 0
0 FRNCMLTTIGMGGMVEDETSVSASKTEVSSVS* 0
>RHO2_oryLat Oryzias latipes (medaka) Deut.Acti.Perc AB001603 9463691 full G?
0 MENGTEGKNFYIPMNNRTGLVRSPYEYPQYYLADPWQFKLLGIYMFFLILTGFPINALTLVVTAQNKKLRQPLNFILVNLAVAGLIMVCFGFTVCIYSCMVGYFSLGPLGCTIEGFMATLG 1
2 GQVSLWSLVVLAIERYIVVCKPMGSFKFTATHSAAGCAFTWIMASSCAVPPLVGWSR 2
1 YIPEGIQVSCGPDYYTLAPGFNNESFVMYMFSCHFCVPVFTIFFTYGSLVMTVKA 0
0 AAAQQQDSASTQKAEKEVTRMCFLMVLGFLLAWVPYASYAAWIFFNRGAAFSAMSMAIPSFFSKSSALFNPIIYILLNKQ 0
0 FRNCMLATIGMGGMVEDETSVSTSKTEVSTAA* 0
>RHO2_oreNil Oreochromis niloticus (tilapia) Deut.Acti.Perc AF247124 11470845 full G?
0 MAWEGGIEPNGTEGKNFYIPMSNRTGIVRNPFEYSQYYLADPIFFKLLAFYMFFLICTGTPINGLTLFVTAQNKKLRQPLNYILVNLAVAGLIMCCFGFTITITSAINGYFVLGTTFCAIEGFMATLG 1
2 GEVALWSLVVLAVERYIVVCKPMGSFKFTGAHAGAGVLFTWIMAMACAAPPLFGWSR 2
1 YIPEGMQCSCGPDYYTLAPGFNNESYVIYMFVVHFFVPVFIIFFTYGSLVMTVRA 0
0 AAAQQQDSASTQKAEKEVTRMCVLMVMGFLIAWTPYASFAGWIFLNKGAAFSALTAAIPAFFAKSSALYNPIIYVLMNKQ 0
0 FRNCMLSTIGMGGMVEDETSVSTSKTEVSSVS* 0
>RHO2_hipHip Hippoglossus hippoglossus (halibut) Deut.Acti.Perc AF156263 11925012 full G?
0 MVWDGGIEPNGTEGKNFYIPMSNRTGIVRSPFEYPQFYMVDSMMFKFLAFYMFFLVCTGTPINGLTLFVTAQNKKLRQPLNYILVNLAVAGLIMCCFGFTITITSAFNGYFILGATFCTIEGFMATLG 1
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSGTHAGIGVLFTWVMAFACAGPPLFGWSR 2
1 YIPEGMQCSCGPDYYTLAPGFNNESYVIYMFVVHFFLPVFVIFFTYGSLVLTVKA 0
0 AAAQQQESESTQKAEKEVTRMCILMVFGFLFAWTPYATFAGWIFMNKGAAFTALTASIPAFFAKSSALYNPVIYVLLNKQ 0
0 FRNCMLSTIGMGGMVEDESSVSASKTEVSSVS* 0
>RHO2_mulSur Mullus surmuletus (mullet) Deut.Acti.Perc Y18680 full G?
0 MNGTEGKNFYIPMSNRTGIVRSPFEYPQYYMVDPMIYKLLAFYMFFLICTGTPINGLTLLVTFQNKKLQQPLNYILVNLAVVGLIMCAFGFTITITSALNGYFILGPTFCAIEGFMATLG 1
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSGTHAGAGVAFTWIMAFACAGPPLFGWSR 2
1 YLPEGMQCSCGPDYYTLAPGFNNESYVIYMFVVHFFVPVFVIFFTYGSLVLTVKA 0
0 AAAQQQESESTQKAEREVTRMCILMVIGFLVAWVPYATFAGWIFLNKGAAFTALTAALPAFFAKSSALYNPVIYVMMNKQ 0
0 FRNCILSAIGMGGMVEDETSVSTSKTEVSTAS* 0
>RHO2_pomMin Pomatoschistus minutus (sand_goby) Deut.Acti.Perc Y18679 full G?
0 MNGTEGKNFYIPMSNRTGIVRSPYEYPQYYMVDPWIYKLLAFYMFFLICTGTPINALTLLVTFQNKKLRQPLNFILVNLAVAGLIMCAFGFTITITSALNGYFILGATFCAIEGFMATLG 1
2 GEVALWSLVVLAVERYIVVCKPMGSFKFSGAHAGAGVALTWIMAMACAAPPLFGWSR 2
1 YLPEGMQCSCGPDYYTLAPGFNNESYVMYMFVVHFFIPVFLIFFTYGSLVLTVKA 0
0 AAAQQQDSASTQKAEKEVTRMCFLMVMGFLVAWVPYASFAGWIFLNKGAAFTAMTAAIPAFFAKSSALYNPVIYVLMNKQ 0
0 FRNCMLSAVGMGGMVDDETSVSASKTEVSSVS* 0
>RHO2_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo EF565168 19196633 full G?
0 MNGTKGSNFYIPMSNRTGVVRNPFEYPQYYLADRWLFSSISAYMFLLICAGLPINGLTLLVTVKHKKLRQPLNFILLNLAVADLFMVFGGFFITVYTSLHGYFVFGVTGCNFEGFFATLG 1
2 GEIGLWSLVVLAIERYVVVCKPMSNFRFGTSHALMGMGFTWFMALTAAVPPLVGWSR 2
1 FIPEGFQCSCTPDFYTTNPLYNNDSYLMYLFSVHFAFPVTLIFFSYGRLICKVKE 0
0 AAAQQQESATTQKAEKEVTRMVILMVIGFLTAWLPYASLSIWIFTHQGAWISPLLMTIPSFFSKSSVLYNPIIYILMNKQ 0
0 FRSSMITTVCCGKNPFGDDDSSSVTSQSKTEVSSVSTSQVSPA* 0
>RHO2_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366494 17463225 full Gt RhB
0 MNGTEGANFYIPFHNRTGVVRSPYEYPQYYLADPWMYSAISAYVFTLILIGFPVNFMTLFVTFKLKKLRQPLNFILVNLCVADLLMIMFGFTTTFYTAMNGYFVFGPTGCNIEGFFATLG 1
2 GEVSLWSLVMLAIERYIVVCKPMGNFRFATTHAALGVVFTWVMASACAVPPLVGWSR 2
1 YIPEGMQCSCGPDYYTLNPKYYNESYVIYLFLVHFLLPVTIIFFTYGRLICTVKE 0
0 AAAQQQESASTQKAEREVTRMVIIMVVGFLVCWVPYASFAFYLFMNKGILFSATAMTVPAFFSKSSVLYNPIIYVLLNKQ 0
0 FRTCMVTTLFCGKNPFGEDDSSMVSTSKTEVSSVSSSQVSPS* 0
>SWS2_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono ABN43074 17339011 full Gt syn(-IRAK1 -MECP2 - +TKTL1) blue tandem with MWS1
0 MHKTHRNLQNELPEDFFIPLPLDTDNITSLSPFLVPQTHLGGSGIFMSLAAFMFLLITLGFPINLLTVICTIKYKKLRSHLNYILVNLAVSNMLVVCVGSATAFYSFAHMYFVLGPTACKIEGFAATLG 1
2 GMVSLWSLAVIAFERFLVICKPLGNLSFRGTHAIFGCAATWVFGLAASLPPLFGWSR 2
1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFSFCFGVPLSIIIFSYGRLLLTLRA 0
0 VAKQQEQSATTQKAEREVTKMVIVMVLGFLVCWLPYASFSLWVVTNRGQVFDLRMASIPSVFSKASTIYNPIIYVFMNKQ 0
0 FRSCMLKLVFCGKSPFGDEDEISGSSQATQVSSVSSSQVSPA* 0
>SWS2_galGal Gallus gallus (chicken) Deut.Saur.Arch NP_990848 7975342 full Gt
0 MHPPRPTTDLPEDFYIPMALDAPNITALSPFLVPQTHLGSPGLFRAMAAFMFLLIALGVPINTLTIFCTARFRKLRSHLNYILVNLALANLLVILVGSTTACYSFSQMYFALGPTACKIEGFAATLG 1
2 GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCVATWVLGFVASAPPLFGWSR 2
1 YIPEGLQCSCGPDWYTTDNKWHNESYVLFLFTFCFGVPLAIIVFSYGRLLITLRA 0
0 VARQQEQSATTQKADREVTKMVVVMVLGFLVCWAPYTAFALWVVTHRGRSFEVGLASIPSVFSKSSTVYNPVIYVLMNKQ 0
0 FRSCMLKLLFCGRSPFGDDEDVSGSSQATQVSSVSSSHVAPA* 0
>SWS2_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full Gt
0 MPKPREMRDELPEDFYIPMSLETPNLTALSPFLVPQTHLGSPGIFKAMAAFMFLLVLLGVPINALTVLCTAKYKKLRSHLNYILVNLAVANLLVVCVGSTTAFYSFSQMYFALGPLACKIEGFTATLG 1
2 GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCAITWIFGLIASLPPLFGWSR 2
1 YIPEGLQCSCGPDWYTTDNKWNNESYVIFLFCFCFGFPLTVIVFSYGRLLLTLRA 0
0 VAKQQEQSASTQKAEREVTKMVVVMVLGFLVCWLPYCSFALWVVTHRGHPFDLGLASIPSVFSKASTVYNPIIYVFMNKQ 0
0 FRSCMLKLVFCGRSPFGDEDDVSGSSQATQVSSVSSSQVSPA* 0
>SWS2_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G?
0 MQKSRPDSRDNLPEDFFIPVPLDVANITTLSPFLVPQTHLGNPSLFMGMAAFMFILIVLGVPINVLTIFCTFKYKKLRSHLNYILVNLSVSNLLVVCVGSTTAFYSFSNMYFSLGPTACKIEGFSATLG 1
2 GMVSLWSLAVVAFERYLVICKPLGNFTFRGTHAIIGCAVTWMFGLAASLPPLFGWSR 2
1 YIPEGLQCSCGPDWYTTENKWNNESYVIFLFCFCFGVPLSVIIFSYGRLLLTLRA 0
0 VAKQQEQSATTQKAEREVTKMVVVMVMGFLVCWLPYASFALWVVTHRGEPFDVRLASIPSVFSKASTVYNPVIYVLMNKQ 0
0 FRSCMLKLIFCGKSPFGDEDDVSGSSQATQVSSVSSSQVSPA* 0
>SWS2_utaSta Uta stansburiana (lizard) Deut.Saur.Lepi DQ100326 16543463 full Gt
0 MHNSRPHSRDDLPEDFFIPMPLDVANITTLSPFLVPQTHLGSPALFMGMAAFMFLLIILGVPINVLTIFCTFKYKKLRSHLNYILVNLAVSNLLVVCIGSTTAFYSFAQMYFSLGPTACKIEGFAATLG 1
2 GMVSLWSLAVVAFERFLVICKPLGNFSFRGTHAIIGCIITWVFGLVASLPPLFGWSR 2
1 YIPEGLQCSCGPDWYTTNNKWNNESYVLFLFSFCFGVPLSVIIFSYGRLLLTLRA 0
0 VAKQQEQSATTQKAEREVTKMVVVMVMGFLVCWLPYASFALWVVTHRGEPFDVRLATIPSVFSKASSVYNPVIYVFMNKQ 0
0 FRSCMLKLVFCGKSPFGDEDDVSGSSQTTQVSSVSSSQVSPA* 0
>SWS2_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt syn(-IRAK1 -MECP2 -)
0 MSKGRPDLRMEMPDEFYVPIPLETTNISSLSPFLVPQTHLGTPGIFMSISAFMLFTIIFGFPLNLLTIICTVKYKKLRSHLNYILVNLAVANLIVICFGSTTAFYSFSQMYFSLGTLACKIEGFTATLG 1
2 GIIGLWSLAVVAFERFLVICKPMGNFTFRESHAVLGCILTWVIGLVAAIPPLLGWSR 2
1 YIPEGLQCSCGPDWYTVNNKWNNESYVLFLFCFCFGFPLAIIVFSYGRLLLALHA 0
0 VAKQQEQSATTQKAEREVTRMVIVMVVGFLVCWLPYASFALWAVTHRGELFDLRMSSVPSVFSKASTVYNPFIYIFMNRQ 0
0 FRSCMMKMIFCGKNPLGDDEETSVSGSTQVSSVSSSQIAPS* 0
>SWS2_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt
0 MHRTKPDPQEDLPDDFYIPVSLNTNNITMLSPFLVPQTHLGSPSVFMVLSVFMFFLLITGIPINVLTIICTFKYKKLRSHLNYILVNLAVANLIVVGFGSTTAFYSFSQMYFAWGPLACKIEGFAATLG 1
2 GMVSLWSLAVVAFERFLVICKPLGNFTFRSTHAIIGCVATWVFGLISSAPPLFGWSR 2
1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFCFCFGFPLSVIIFSYGRLLMTLRA 0
0 VAKQQEQSASTQKAEREVTKMVVVMVLGFLVCWLPYTVFSLWVVTHRGESFELALGSIPAVFSKSSTVYNPLIYVFMNKQ 0
0 FRSCMMKLIFCGKSPFGDEDDASSASQSTQVSSVSSSQVAPA* 0
>SWS2_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G?
0 MRGNRQHEFQEDFFIPIPLEVDNITALSPFLVPQDHLGSPALFYGMAAFMFCLFVSGTGINALTIACTVKYKKLRSHLNYILVNLAVSNLLVTCVGSFTCFCCFTVRYMFVGPLGCKIEGFAATLG 1
2 GMVSLWSLAVVAFERWLVVCKPLGNFIFKPDHAIACCAATWLFALVVSGPPLFGWSR 2
1 YIPEGFQCSCGPDWYTTGNKYNNESYVMFIFGFGFAVPFFVIVFCYSQLLFVLKS 0
0 AQAESASTQKAEKEVTRMVVVMIFGFLVCWLPYASFALWAVNNRGTPFDLRLATIPACFSKSSAVYNPVIYVVLNKQ 0
0 FRSCMKKMLGMSGGDDEDSSASQSVTEVSKVSPS* 0
>SWS2_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gt
0 MRGVRQHEFQEDFYIPIPLDVDNITALSPFLVPQDHLGSPAVFYGMSAFMFFLFVAGTGINVLTIACTIQYKKLRSHLNYILVNLAFSNLLVTTVGSFTCFCCFFVRYMIVGPLGCKIEGFAATLG 1
2 GMVSLWSLAVVAFERWLVVCKPLGNFIFKPDHAIVCCIFTWFFALIISAPPLFGWSR 2
1 YIPEGFQCSCGPDWYTTGNKYNNESYVWFIFGFGFAVPLFVIVFCYSQLLVMLKS 0
0 AKAQAESASTQKAEREVTRMVVVMILGFLVCWLPYASFALWVVNNRGTPFDLRLATIPACFSKASTVYNPIIYVVLNKQ 0
0 FRSCMKKMLGMSGGDDEESSSQSVTEVSKVSPS* 0
>SWS2_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gt
0 MKHGRVPEIPEDFYIPISLDTDNITSLSPFLVPQDHLASKATFYSLAFYMFFILIVGTFINALTVACTVQNKKLRSHLNYILVNLAVSNLLVSGVGAFTAFLSFAARYFVLGTLACKVEGFLATLG 1
2 GMVSLWSLAVIAFERWLVICKPLGNFIFKPDHALVCCAFTWVFALAASAPPLVGWSR 2
1 YIPEGLQCSCGPDWYTTNNKYNNESYVLFLFGFCFAVPFCTICFCYSQLLFTMKMA 0
0 AKAQAESASTQKAEREVTRMVVLMVMGFLVCWMPYASFALWVVNNRGQTFDLRFASIPSVFSKSSAVYNPVIYVLLNKQ 0
0 FRSCMMKMLGMGGGDDEESSTSSVTEVSKVGPA* 0
>SWS2_oryLat Oryzias latipes (medaka) Deut.Acti.Perc AB223056 16460888 full Gt
0 MRFISGGELPDDFWIPIPLDTNNLSLLSPFLVPQDHLGSSGTFYAMAAFMFFLFVFGTSINSLTIACTFQNKKLRSHLNYILVNLSVANLLVSGVGSSTAFCSFACRYFVFGSLACKIEGFAATLG 1
2 GMVGLWSLAVIAFERWLVICKPLGNFTFKPEHALACCLVTWVCALAAAAPPLAGWSR 2
1 YIPEGLQCSCGPDWYTTNNKYNNESYVMFLFCFCFAVPFATIVFCYSQLLVTLKMA 0
0 AKAQAESASTQKAEREVTRMVVVMVLGFLVCWMPYASFALWVVNNRGHSFDLRLATIPSCLSKASTVYNPVIYVLLNKQ 0
0 FRSCMLTMLGMGGGEEEDSTSVTEVSKVGPA* 0
>SWS2_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366492 17463225 full Gt
0 MYQGKSTQVDDLPEDFYIPIALNVKNMSELSPFLVPQVHLGDSFIFYGMSAFMLFLVLAGFPLNFLTVFVTIKYKKLRSHLNYILVNLAIANLIVVCCGSTLAFYSFMHKYFILGPLFCKMEGFTATLG 1
2 GMLSLWSLAVLAFERCLVICKPFGNIAFRGTHALIRCGFAWAAAIAASTPPLFGWSR 2
1 YIPEGLQCSCGPDWYTTNNKYNNESYVMFLFIFCFGTPFTIIIVSYSKLILTLRA 0
0 AAAQQQESASTQKAEKEVSRMVVIMVGGFLVCWLPYASLALWIVFNRGSPFDLRLATIPSVFSKASTVYNPVIYIFLNKQ 0
0 FRSCMMKTIFCGKNPLGDDEDATSTTTQVSSVSTSQVAPA* 0
>SWS1_homSap Homo sapiens (human) Deut.Euth.Euar NM_001708 1385866 full Gt OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC)
0 MRKMSEEEFYLFKNISSVGPWDGPQYHIAPVWAFYLQAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVA 1
2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWSR 2
1 FIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKA 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQ 0
0 FQACIMKMVCGKAMTDESDTCSSQKTEVSTVSSTQVGPN* 0
>SWS1_monDom Monodelphis domesticus (opossum) Deut.Meta.Dide genomic full Gt OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC)
0 MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHFQTVFMGFVFCAGTPLNAVVLVATLRYKKLRQPLNYILVNVSLCGFIFCIFAVFTVFISSSQGYFIFGRHVCAMEAFLGSVA 1
2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWVIGIGVSIPPFFGWSR 2
1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIMPLFLICFSYSQLLRALRA 0
0 VAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNQNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0
0 FHACIMEMVCRKPMTDDSDVSSSQKTEVSAVSSSQVGPT* 0
>SWS1_smiCra Sminthopsis crassicaudata (dunnart) Deut.Meta.Dasy AY442173 full G? OPN1SW
0 MSGDEEFYLFKNISLVGPWDGPQYHLAPAWAFHFQTAFMGFVFFAGTSLNGVVLIATLRYKKLRQPLNYILVNISLAGFIFCVFSVFTVFVSSSQGYFVFGRHVCAMEGFLGSVA 1
2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWIIGIGVSIPPFFGWSR 2
1 YIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLICFSYSQLLGALRA 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAAMAMYMVNNRNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0
0 FHACIMEMICKKPMTDDSETTSSQKTEVSTVSSSQVGPS* 0
>SWS1_tarRos Tarsipes rostratus (honey_possum) Deut.Meta.Dipr AY772472 18426754 full G? OPN1SW
0 MSGDEEFYLFKDISSVGPWDGPQYHIAPAWAFHFQTTFMGFVFFAGTPLNAVVLIATLRYKKLRQPLNYILVNISLAGFIFCVISVFTVFISSSQGYFIFGRHVCAMEAFLGSVA 1
2 GLVTGWSLAFLAFERFIVICKPFGNFRFSSKHAMMVVLATWVIGIGVSIPPFFGWSR 2
1 YIPEGLQCSCGPDWYTVGTKYHSEYYTGFLFIFCFIVPLSLICFSYSQLLGALRA 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSACVYNPIVYWFMNKQ 0
0 FHACIMEMVCRKPMTDDSEISSSQKTEVSTVSSSQVGPS* 0
>SWS1_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full Gt OPN1SW violet
0 MSSDDDFYLFTNGSVPGPWDGPQYHIAPPWAFYLQTAFMGIVFAVGTPLNAVVLWVTVRYKRLRQPLNYILVNISASGFVSCVLSVFVVFVASARGYFVFGKRVCELEAFVGTHG 1
2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSRHALLVVVATWLIGVGVGLPPFFGWSR 2
1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0
0 FRACIMETVCGKPLTDDSDASTSAQRTEVSSVSSSQVGPT* 0
>SWS1_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full Gt OPN1SW
0 MDEEEFYLFKNQSSVGPWDGPQYHIAPMWAFYLQTIFMGLVFVAGTPLNAIVLIVTIKYKKLRQPLNYILVNISVSGLMCCVFCIFTVFIASSQGYFVFGKHMCAFEGFAGATG 1
2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSRHALLVVAATWIIGVGVAIPPFFGWSR 2
1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCMCYVPYAALAMYMVNNREHGIDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0
0 FRACIMETVCGRPMTDDSEVSSSAQRTEVSSVSSSQVGPS* 0
>SWS1_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt OPN1SW syn(+SWS1 -CALU)
0 MSGQEDFYLFENISSVGPWDGPQYHIAPMWAFYFQTAFMGFVFFAGTPLNAIILIVTVKYKKLRQPLNYILVNISFAGFLFCTFSVFTVFMASSQGYFFFGRHVCAMEAFLGSVA 1
2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSRHALLVVAATWIIGVGVAIPPFFGWSR 2
1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLTLIIFSYSQLLGALRA 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYASLAMYMVNNRDHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0
0 FRACILETVCGKPMSDESDVSSSAQKTEVSSVSSSQVSPS* 0
>SWS1_utaSta Uta stansburiana (lizard) Deut.Saur.Lepi DQ100325 16543463 full Gt OPN1SW
0 MSGEEDFYLFENISSVGPWDGPQYHIAPMWAFYFQTAFMGFVFFAGTPLNAIILIVTVKYKKLRQPLNYILVNISFAGFLFCVFSVFTVFLASSQGYFFFGRHICALEAFLGSVA 1
2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSKHALLVVAATWFIGIGVSIPPFFGWSR 2
1 FIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLTLIIFSYSQLLGALRA 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGIDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0
0 FRACIMETVCGKPMTDESDVSSSAQKTEVSSVSSSQVSPS* 0
>SWS1_xenLae Xenopus laevis (frog) Deut.Amph.Anur genomic full Gt OPN1SW syn(+SWS1 -CALU)
0 MLEEEDFYLFKNVSNVSPFDGPQYHIAPKWAFTLQAIFMGMVFLIGTPLNFIVLLVTIKYKKLRQPLNYILVNITVGGFLMCIFSIFPVFVSSSQGYFFFGRIACSIDAFVGTLT 1
2 GLVTGWSLAFLAFERYIVICKPMGNFNFSSSHALAVVICTWIIGIVVSVPPFLGWSR 2
1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFIFIFCFVIPLSLICFSYGRLLGALRA 0
0 VAAQQQESASTQKAEREVSRMVIFMVGSFCLCYVPYAAMAMYMVTNRNHGLDLRLVTIPAFFSKSSCVYNPIIYSFMNKQ 0
0 FRGCIMETVCGRPMSDDSSVSSTSQRTEVSTVSSSQVSPA* 0
>SWS1_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt OPN1SW
0 MSGEEEFYLFKNISSVGPWDGPQYHIAPKWAFFLQAAFMGFVLFVGTPLNAIVLFVTIKYKKLQQPLNYILVNISLAGFIFCFFGVFAVFIASCQGYFIFGKTVCALEGFTGSVA 1
2 GLVTGWSLAILAFERYLVICKPIGNFRFGSKHSMIAVVAAWVIGVGVSIPPFFGWSR 2
1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIIPLFIICFSYSQLLGALRA 0
0 VAAQQQESATTQKAEREVSRMIIVMVGSFCVCYVPYAALAMYMVNNRDHGIDLRLVTIPAFFSKSSFVYNPIIYCFMNKQ 0
0 FRACIMQTVFGKPMTDDSDISSSGKTEVSSVSSSQVNPS*
>SWS1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full Gt OPN1SW syn(-PNPLAS +IRG5 -TNPO3 +SWS1 -CALU -DYRK2)
0 MDAWAVQFGNASKVSPFEGEQYHIAPKWAFYLQAAFMGFVFIVGTPMNGIVLFVTMKYKKLRQPLNYILVNISLAGFIFDTFSVSQVSVCAARGYYSLGYTLCSMEAAMGSIA 1
2 GLVTGWSLAVLAFERYVVICKPFGSFKFGQGQAVGAVVFTWIIGTACATPPFFGWSR 2
1 YIPEGLGTACGPDWYTKSEEYNSESYTYFLLITCFMMPMTIIIFSYSQLLGALRA 0
0 VAAQQAESESTQKAEREVSRMVVVMVGSFVLCYAPYAVTAMYFANSDEPNKDYRLVAIPAFFSKSSSVYNPLIYAFMNKQ 0
0 FNACIMETVFGKKIDESSEVSSKTETSSVSA* 0
>SWS1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc DW624108 full G? OPN1SW syn(ARF4 +IRG5 -TNPO3 +SWS1) BT028514 brain transcript
0 MGKHFHLYENISKVDPFEGPQYYLAPMWAFYLQTAFMGFVFFAGTPLNFTVLLVTAKYKKLRVPLNYILVNITLAGFIFVTFSVSQVFVASARGYYFLGYTLCALEAAMGSVA 1
2 GLVTAWSLAVLSFERYLIICKPFGAFKFTSNHALGAVAFTWFMGICCSSPPFFGWSR 2
1 YIPEGLGCSCGPDWYTHNVEFNCTSYLYFLIVTCFIMPLSIIIFSYSQLLGALRA 0
0 VAAQQAESESTQKAEKEVSKMIIVMVGSFVACYAPYAITGLWFAFSPDENKDYRLVTVPAYFSKSSCVYNPLIYAFMNKQ 0
0 FNACIMETVFGKPMDESSEVSSKTEVSSVSTAS* 0
>SWS1_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full Gt OPN1SW syn(-TNPO3 +SWS1 +SOCS2 -CRADD +ARHGAP8)
0 MGKYFYLYENISKVGPYDGPQYYLAPTWAFYLQAAFMGFVFFVGTPLNFVVLLATAKYKKLRVPLNYILVNITFAGFIFVTFSVSQVFLASVRGYYFFGQTLCALEAAVGAVA 1
2 GLVTSWSLAVLSFERYLVICKPFGAFKFGSNHALAAVIFTWFMGVGCACPPFFGWSR 2
1 YIPEGLGCSCGPDWYTNCEEFSCASYSKFLLVTCFICPITIIIFSYSQLLGALRA 0
0 VAAQQAESASTQKAEKEVSRMIIVMVASFVTCYGPYALTAQYYAYSQDENKDYRLVTIPAFFSKSSCVYNPLIYAFMNKQ 0
0 FNGCIMEMVFGKKMEEASEVSSKTEVSTDS*0
>SWS1_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic pseu G? OPN1SW exonic pseudogene frameshifts stops no synteny homSap
0 MSGDEEFYLFKNISKVGPLDGPHFHIATKWAFDFQAAFMGFVFLCGtPLNAIVLIVTVKCKKLRQPLTYMLVNISAAGLVFCLFSISTVFLFSTQGYFVFGPTVCALESLFGSMA 1
2 GLVTGWSLAFLAAERYIVICKPFGNFRFGSIHSLFAFCLTWVLGLGVALPPFFGWSR 2
1 YIPeGLQCSCSPDWNTVGTKYESEYCTYFLFVFCFFVQLSIIIFSYGKLLNTLra 0
0 VAVQqQESSLSSTQKAEREMSRMVIVMVGSFCTCYVAALALYVVTNRDHNIDLRFVTVPAFFSKASCVYNPLIYSFMNKQ 0
0 FRARIMETVCGKFITDESETSSSRTAVSSVSTSQVSPG* 0
>SWS1_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366495 17463225 full Gt OPN1SW
0 MSGDEEFYLFKNISKVGPWDGPQFHIAPKWAFYLQAAFMGFVFICGTPLNAIVLVVTIKYKKLRQPLNYILVNISAAGLVFCLFSISTVFVASMQGYFFLGPTICALEAFFGSLA 1
2 GLVTGWSLAFLAAERYIVICKPFGNFRFGSKHALVAVGLTWMLGLSVALPPFFGWSR 2
1 YIPEGLQCSCGPDWYTVGTKYKSEYYTYFLFVFCFVVPLSIIIFSYGSLLGTLRA 0
0 VAAQQQESASTQKAEREVSRMVIMMVASFCTCYVPYAALAVYMVTNRDHNIDLRFVTVPAFFSKASCVYNPLIYSFMNKQ 0
0 FRACILETVCGKPITDESETSSSRTEVSSVSTTQMIPG* 0
>LWS_homSap Homo sapiens (human) Deut.Euth.Euar NP_000504 12853434 full Gt OPN1LW long wavelength syn(-IRAK1 -MECP2 -TEX28) OPN1MW deutan
0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1
2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1
2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0
0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0
>LWS_monDom Monodelphis domesticus (opossum) Deut.Meta.Dide genomic full G? OPN1LW long wavelength exon 1:4 residue insert
0 MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNNTR 1
2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTAPPLFGWSR 2
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYVQVWLAIRA 0
0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYSFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0
0 FRTCILQLFGKKVDDGSEVSSTSKTEGSSVSSVAPA* 0
>LWS_macEug Macropus eugenii (wallaby) Deut.Meta.Dipr genomic full G? OPN1LW long wavelength
0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1
2 GPFEGPNYHIAPRWVFNLTSLWMIFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPLFGWSR 2
1 YWPHGLKTSCGPDVFSGNSDPGVQSYMIVLMSTCCILPLSVIFLCYIQVWLAIRS 2
0 VAKQQKESESTQKAEKEVSRMVVVMILAFCFCWGPYAIFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0
>LWS_smiCra Sminthopsis crassicaudata (dunnart) Deut.Meta.Dasy EU232013 18426754 full G? OPN1LW long wavelength
0 MTQAWDPAGFLAWRRDENEETTRASLFVYTNSNNTK 1
2 GPFEGPNYHIAPRWVYNLTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETIIASTISVINQIYGYFILGHPMCVLEGYTVSLC 1
2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIILCYIQVWLAIRA 0
0 VAKQQKESESTQKAEKEVSRMVMVMILAFCFCWGPYALFACFAAANPGYAFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0
0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0
>LWS_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono ABN43074 17339011 full Gt OPN1LW long wavelength syn(-IRAK1 -MECP2 - -) LWS green
0 MTPAWNSGVYAARRRFEDEEDTTRTSVFVYTNSNNTR 1
2 DPFEGPNYHIAPRWAYNVTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC 1
2 GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIVLCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0
0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA* 0
>LWS_galGal Gallus gallus (chicken) Deut.Saur.Arch NM_205438 12716987 full Gt OPN1LW iodopsin missing in assembly
0 MAAWEAAFAARRRHEEEDTTRDSVFTYTNSNNTR 1
2 GPFEGPNYHIAPRWVYNLTSLWMIFVVAASVFTNGLVLVATWKFKKLRHPLNWILVNLAVADLGETVIASTISVINQISGYFILGHPMCVVEGYTVSAC 1
2 GITALWSLAIISWERWFVVCKPFGNIKFDGKLAVAGILFSWLWSCAWTAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFFPLAIIILCYLQVSLAIRA 0
0 VAAQQKESESTQKAEKEVSRMVVVMIVAYCFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVDDGSEVSTSRTEVSSVSNSSVSPA* 0
>LWS_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt OPN1LW long wavelength syn(- - -TEX28 +TKTL1)
0 MAGTVTEAWDVAVFAARRRNDEDDTTRDSLFTYTNSNNTR 1
2 GPFEGPNYHIAPRWVYNITSVWMIFVVIASIFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVIASTISVINQISGYFILGHPMCVLEGYTVSTC 1
2 GISALWSLAVISWERWVVVCKPFGNVKFDAKLAVAGIVFSWVWSAVWTAPPVFGWSR 2
1 YWPHGLKTSCGPDVFSGSDDPGVLSYMIVLMITCCFIPLAVILLCYLQVWLAIRA 0
0 VAAQQKESESTQKAEKEVSRMVVVMIIAYCFCWGPYTVFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0
0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSNSSVSPA* 0
>LWS_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt OPN1LW long wavelength syn(-IRAK1 -MECP2 - -)
0 MASHWNEAVFAARRRNDDDDTTRSSVFTYTNSNNTR 1
2 GPFEGPNYHIAPRWVYNISSLWMIFVVLASVFTNGLVLVATLKFKKLRHPLNWILVNMAIADLGETVIASTISVCNQIFGYFVLGHPMCILEGYTVSVC 1
2 GIAALWSLTVIAWERWFVVCKPFGNIKFDGKLAATGIIFSWVWAAGWCAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMLVLMITCCIIPLAIIVLCYMHVWLTIRQ 0
0 VAQQQKESESTQKAEREVSRMVVVMIIAYIFCWGPYTFFACFAAFNPGYNFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0
0 FRNCIYQLFGKKVDDGSEVSSTSRTEVSSVSNSSVSPA* 0
>LWS_neoFor Neoceratodus forsteri (lungfish) Deut.Sarc.Dipn EF526299 17961206 full Gt OPN1LW long wavelength
0 MAEPWDAVLAARRRHQDEETTRSTIFVYTNSNNTR 1
2 GPFEGPNYHIAPRWVYNLTSLWMIFVVFASCFTNGLVLMATYKFKKLRHPLNWILVNLAIADLGETLIASTISVTNQIFGYFILGHPMCMLEGFTVATC 1
2 GITGLWSLTIIAWERWVVVCKPFGNIKFDGKWAAGGIIFSWVWSAFWCAMPLFGWSR 2
1 FWPHGLKTSCGPDVFSGEDKYGTRSFMIALMITCCIIPLGVIILCYIQVWWAIRT 0
0 VAKQQKESESTQKAEKEVSRMVVVMIFAYCFCWGPYTFMACFGAAYPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0
0 FRNCIYQLLGKKVDDGSELSSTSKTEVSSVSNSSVSPA* 0
>LWS_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full Gt
0 MAEHWGDAIYAARRKGDETTREAMFTYTNSNNTK 1
2 DPFEGPNYHIAPRWVYNVATVWMFFVVVASTFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETLFASTISVINQFFGYFILGHPMCIFEGYTVSVC 1
2 GIAALWSLTVISWERWVVVCKPFGNVKFDAKWASAGIIFSWVWAAAWCAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSEDPGVQSYMVVLMITCCIIPLAIIILCYIAVYLAIHA 2
0 VAQQQKDSESTQKAEKEVSRMVVVMIFAYCFCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0
0 FRVCIMQLFGKKVDDGSEVSTSKTEVSSVAPA* 0
>LWS_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G?
0 MAEEWGKQSFAARRYHEDSTRGSAFVYTNSNHTR 1
2 DPFEGPNYHIAPRWVYNLATLWMFFVVVLSVFTNGLVLVATAKFKKLRHPLNWILVNLAVADLGETLFASTISVCNQFFGYFILGHPMCIFEGYVVSVC 1
2 GIAGLWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWIWPICWCAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCIIPLAIIVLCYLAVWMAIRA 2
0 VAMQQKESESTQKAEREVSRMVVVMILAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0
0 FRVCIMKLFGKEVDDGSEVSTSKTEVSSVAPA* 0
>LWS_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gt OPN1LW long wavelength
0 MAEEWGKQSFAARRYHEDTTRGSAFVYTNSNHTR 1
2 DPFEGPNYHIAPRWVYNVATVWMFIVVVLSVFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYTVSTC 1
2 GIAALWSLTIISWERWVVVCKPFGNVKFDAKWATGGIVFSWVWAAVWCAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCIIPLAIIILCYLAVWLAIRS 0
0 VAMQQKESESTQKAEKEVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0
0 FRVCIMKLFGKEVDDGSEVSTSKTEVSSVAPA* 0
>LWS_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gt OPN1LW long wavelength
0 MAEEWGKQAFAARRYNEDTTRGSMFVYTNSNNTK 1
2 DPFEGPNYHIAPRWVYNLSTLWMFIVVALSVFTNGLVLVATAKFKKLQHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSVC 1
2 GITALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWIWSAVWCAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCLIPLAIIILCYLAVWLAIRA 0
0 VAMQQKESESTQKAERDVSRMVVVMIVAYIVCWGPYTTFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0
0 FRSCIMQLFGKEVDDGSEVSTsKTEVSSVAPA* 0
>LWS_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full Gt
0 MAEQWGKQVFAARRQNEDTTRGSAFTYTNSNHTR 1
2 DPFEGPNYHIAPRWVYNLATLWMFFVVVLSVFTNGLVLVATAKFKKLRHPLNWILvNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSTC 1
2 GIAALWSLTIISWERWVVVCKPFGNVKFDAKWAIGGIVFSWVWSAVWCAPPVFGWSR 2
1 YWPHGLKTSCGPDVFSGSDDPGVQSYMIVLMITCCIIPLAIIILCYLAVWLAIRA 0
0 VAMQQKESESTQKAEREVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0
0 FRTCIMQLFGKQVDDGSEVSTSKTEVSSVAPA* 0
>LWS1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo EF565165 19196633 full G? OPN1LW long wavelength
0 MTQSWELVAPAARRGFKYDEPTHSGIFVYTNSNQTR 1
2 GPFEGPNYHIAPRWAYNLTSVWMVGVVVASVFTNGLVLVATVRFKKLRHPLNWILVNMALADLGETVLASTVSVANQFFGYFILGHPLCVFEGFVVSLC 1
2 GITALWSLTIIAWERWVVVCKPFGNMKFDSKMAVAGIVFSWVWSAGWCLPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGNEDPGVQSYMVALTLSCAVLPLLIIILCYFQVWWAIRA 0
0 VALQQKESESTQKAEKEVSRMVVVMVAAFCLCWGPYACFAMFSALNPGYAFHPLVASIPSYFAKSSTIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVDDGSELSSTSKTDVSSVSNSSVSPA* 0
>LWS2_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo EF565166 19196633 full G? OPN1LW long wavelength
0 MAEPRGSVAFAARRWNDHEGTTVGEFTYTNSNSTR 1
2 DPFEGPNYHIAPRWTYNLTSLWMVVVVILSVFTNGLVLVATWKFKKLRHPLNWILVNLAIADLGETLFASTISICNQVFGYFILGHPMCVFEGFTVSAC 1
2 GITALWSLTIIAWERWVVVCKPFGNVKFDGKWAAFGIIFSWVWSIGWCLPPVFGWSR 2
1 YWPHGLKTSCGPDVFSGSSDPGVKSYMVTLVITCAALPLTIIIVCLYQVWLAIRA 0
0 VAMQQKESESTQKAEKEVSRMVVVMIIAFCFCWGPYTSFAVFSALNPGYSFHPLMAALPAYFAKSSTIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVDDGSELSSTSKTDVSSVSNSSVSPA* 0
>LWS_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic full Gt OPN1LW long wavelength traces key to intron 3 position and gapping
0 MTASWQGAMFAARRRQDDEDTTMESLFRYTNENNTK 1
2 DPFEGPNYHIAPRWVFNLTSVWMIIVVVLSLFSNGLVLVATVKFKKLRHPLNWIIVNLAIADILETIFASTISVCNQVYGYFILGHPMCVFEGYVVSTC 1
2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIATILIVFSWVWPASWCSLPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFLPLSIIILCYLQVWLAIHS 0
0 VAQQQKESETTQKAERDVSRMVVVMILAYVFCWGPYTFFACFAAANPGYSFHPIAAALPAYFAKGATIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVDDGSEVSSSSRTEVSSVSNSSVSPA* 0
>LWS_letJap Lethenteron japonicum (lamprey) Deut.Agna.Hype AB116381 15096614 full Gt OPN1LW long wavelength
0 MTASWHGAVFAARRRNDDEDTTKDSIFRYTNENNTR 1
2 DPFEGPNYHIAPRWMFNLTSVWMIIVVVLSLFTNGLVLVATMKFKKLRHPLNWILVNLAIADILETIFASTISVCNQVFGYFILGHPMCVFEGYVVSTC 1
2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIAIILIVFSWVWPACWCSLPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFLPLSVIILCYLQVWLAIHS 0
0 VAQQQKESETTQKAERDVSRMVVVMILAYIFCWGPYTFFACYAAANPGYAFHPLTAALPAYFAKSATIYNPVIYVFMNRQ 0
0 FRNCIMQLFGKKVDDGSEVSSASRTEVSSVSNSSISPA*
>LWS_geoAus Geotria australis (lamprey) Deut.Agna.Hype AY366491 17463225 full Gt OPN1LW long wavelength
0 MAQSWERAMFAARRRQDEDTTKGDLFRYTNENNTR 1
2 DPFEGPNYHIAPRWMYNLTSFWMIIVVILSLFTNGLVLVATLKFKKLRHPLNWILVNLAIADIGETIFASTVSVVNQIFGYFILGHPLCVFEGFTVSVC 1
2 GITALWSLAIISFERWMVVCKPFGNLKFDGKVAIVLIIFSWAWSAGWCAPPIFGWSR 2
1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFIPLALIIICYLQVWLAIHT 0
0 VAQQQKESETTQKAERDVSRMVVVMIFAYIFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0
0 FRNCIMQLFGKKVDDGSEVSSSARTEVSSVSNSSVSPA* 0
>PIN_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full Gt pinopsin pineal non-visual
0 MSSNSSQAPPNGTPGPFDGPQWPYQAPQSTYVGVAVLMGTVVACASVVNGLVIVVSICYKKLRSPLNYILVNLAVADLLVTLCGSSVSLSNNINGFFVFGRRMCELEGFMVSLT 1
2 GIVGLWSLAILALERYVVVCRPLGDFQFQRRHAVSGCAFTWGWALLWSTPPLLGWSSYVPE 1
2 GLRTSCGPNWYTGGSNNNSYILSLFVTCFVLPLSLILFSYTNLLLTLRA 0
0 AAAQQKEADTTQRAEREVTRMVIVMVMAFLLCWLPYSTFALVVATHKGIIIQPVLASLPSYFSKTATVYNPIIYVFMNKQ 0
0 FQSCLLEMLCCGYQPQRTGKASPGTPGPHADVTAAGLRNKVMPAHPV* 0
>PIN_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full G? pinopsin
0 MDSTQEPPNSSTPGPFDGPQWPHQAPRATYLVVAVLMGLVVASASLLNGLVIVVSVRHKRLRSPLNYILLNLAVANLLVTLCGSSVSLSNNISGFFVFGERLCQLEGFMVSLT 1
2 GIVGLWSLAILALERYLVVCRPLGDFRFQQQHAASGCAFTWGWSLLWTTPPLLGWSSYVPE 1
2 GLRTSCGPNWYTGGSNNSSYILALFVTCFVMPLSLILFSYTNLLLTLRA 0
0 AAARQQESDTTQQAEREVTRMVVAMVVAFLTCWLPYATFALVVATHKDIVIQPALASLPSYFSKTATVYNPIIYVFMNKQ 0
0 SCLLGMLCCGHHPRGMGKTSPAAPSPQVAAEGLRNKVTPSHPV* 0
>PIN_colLiv Columba livia (pigeon) Deut.Saur.Arch CLU50598 8982090 full G? pinopsin
0 MDPTNSPQEPPHTSTPGPFDGPQWPHQAPRGMYLSVAVLMGIVVISASVVNGLVIVVSIRYKKLRSPLNYILVNLAMADLLVTLCGSSVSFSNNINGFFVFGKRLCELEGFMVSLT 1
2 GIVGLWSLAILALERYVVVCRPLGDFRFQHRHAVTGCAFTWVWSLLWTTPPLLGWSSYVPE 1
2 GLRTSCGPNWYTGGSNNNSYILTLFVTCFVMPLSLILFSYANLLMTLRA 0
0 AAAQQQESDTTQQAERQVTRMVVAMVMAFLICWLPYTTFALVVATNKDIAIQPALASLPSYFSKTATVYNPIIYVFMNKQ 0
0 FQSCLLKMLCCGHHPRGTGRTAPAAPASPTDGLRNKVTPSHPV* 0
>PIN_utaSta Uta stansburiana (lizard) Deut.Saur.Lepi DQ100321 16543463 full Gt pinopsin missing genome
0 MVNEWSNATPGPFDGPQWPYLAPRSIYTSVAVLMGLVVVSAAFVNGLVIVVSIQYKKLRSPLNYILVNLAIADLLVTSFGSTLSFANNIYGFFVLGQTACEFEGFMVSLT 1
2 GIVGLWSLAILAFERYLVICKPVGDFRFQQRHAVFGCVFTWMWSLVWTLPPLFGWSSYVPE 1
2 GLRTSCGPNWYTGGSGNNSYIMALFVTCFALPLGMIIFSYASLLLTLRA 0
0 VATQQKEVETTQQAEKEVTRRVIAMVMAFLVCWLPYASFAMVVATNKDLVIQPALASLPSYFSKTATVYNPIIYVFMNKQ 0
0 FRSCLLSTMSCGHRPRGAQETTPAMISIPQGPTSALQGSRNKVTPSASEGSGNEAIPS* 0
>PIN_pheMad Phelsuma madagascariensis (gecko) Deut.Saur.Lepi AB022881 full Gt pinopsin no_ref AB022881
0 MHVQMANASQASLKNGTLSPFDGPQWPHRASRRVYTSLAALMGVVVLSASLANGLVIAVSVRFKRLRSPLNYILVNLATADLLVTFFGSIISFVNNAVGFFVFGKTACRFEGFMVSLT 1
2 GIVGLWSLAILAFERYLVICKPVGDFQFQRRHAVIGCLYTWGWSLIWTVPPLFGWSSYVPE 1
2 GLGTSCGPNWYMGGTNNNSYIVALFVTCFALPLSMILFSYANLLLTLRA 0
0 VAAQQKEQETTQRAEKEVTRMVITMVMAFLVCWLPYATFAMVVATTKDLSIQPGLASLPSYFSKTATVYNPIIYVFMNKQ 0
0 FRSCLLNTVSCGRIPQTMPGTPATTAVRGGFVLTSEGRGNKVASTELHS* 0
>PIN_podSic Podarcis sicula (lizard) Deut.Saur.Lepi DQ013042 16688437 full Gt pinopsin
0 MQASNASWVEVRNRTPGPFEGPQWPYLAPQSTYISVAVLMGLVVISATLVNGLVIVVSVQFKKLRSPLNYVLVNLAVADLLVTFFGSTISFVNNAQGFFIFGQATCEFEGFMVSLT 1
2 GIVGLWSLAILAFERYLVICKPVGDFRFPARHAVLGCAFTWGWSFVWTVPPLLGWSSYVPE 1
2 GLRTSCGPNWYSGGSSNNSYIMTLFVTCFAMPLSTILFSYANLLMTLRT 0
0 VAAQQKEQETTQRAEREVTRMVVAMVAAFLVCWLPYASFAMVVATHKDLAIRPALASLPSYFSKTATVYNPIIYVFMNKQ 0
0 FRSCLLYKMSCGHRALSSQDTTPAGISLPGRLTTSASKGSRNQVSPS* 0
>PIN_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt pinopsin
0 MRAGNMSAYEAPGPYDGPQWPHLAPRSTFLTVAAVMCMVVILAFFVNGLVIVVTLKYKKLRSPLNYILVNLAIANLLVTIFGSSVSFSNNVVGYFFMGKTMCEFEGFMVSLT 1
2 GIVGLWSLAILAFERYLVICKPMGDFRFQQKHAILGCSFTWVWSFIWTSPPLFGWCSYVPE 1
2 GLRTSCGPNWYTGGTNNNSYIMALFLTCFIMPLSTIIFSYSNLLMALRA 0
0 VAAQQKDSETTQRAEKEVTRMVIAMVLAFLICWLPYASFAVVVAVNKDVVIEPTVASLPSYFSKTATVYNPIIYVFMNKQ 0
0 FRNCLMTLLCCGRSFGDDETSSASGRTDVTSVSEAGGNKVTPA* 0
>PIN_bufJap Bufo japonicus (toad) Deut.Amph.Anur AF200433 9537517 full Gt pinopsin classifies oddly
0 MHSANMSALETPGPFEGPQWPHVAPRSTYLTVAVLMGMVVFLAFFVNGMVIVVSLKYKKLRSPLNYILVNLAVADILVTMFGSTVSFHNNIFGFFTLGKLVCELEGFVVSLT 1
2 GIVGLWSLAILAFERYIVICKPMGDFRFQQRHAVMGCAFTWIWAFLWTSPPLIGWCSYVPE 1
2 GLGTSCGPNWYTGGTNNNSYILALFTTCFMMPLTTIIFSYSNLLLALRA 0
0 VAAQQKESETTQRAEREVTRMVIAMVLAFLICWLPYAVFAIVMASNKNVVIDPTLASMPSYFSKTATVYNPVIYVFMNKQ 0
0 FRDCLTKLLCCGRNPFGEDETSTTSGRTDVTSVSEGGGNKVTPA* 0
>PIN_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gt pinopsin
0 FGSTVSFSNNINGYFVLGETVCQFEGFMVSLT 1
2 GIVGLWSLAILAFERYIVICKPMGDFRFQQKHAVWGCLFTWLWSLFWTLPPLFGWCSYVPE 1
2 GLRTSCGPNWYTGGANNSSYVVALFITCFTLPLSLIIFSYASLLVVLRA 0
>VAOP_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic 19664923 full Gt vertebrate ancient opsin exon 1 genbank error new stop *RKNGDEH
0 MDVFRALGNESLLSNSSGPARWDPFHHPLDSIQPWHFRLVAAVMFVVTSLSLAENLAVILVTFKFKQLRQPVNYVIVNLSVADFLVSLTGGTISFLANLKGYFYMGHWACVLEGFAVTFF 1
2 GIVALWSLALLAFERYIVICRPVGNMRLRGKHAAQGIAFVWTFSFIWTIPPTMGWSSYTTSKIGTTCEPNW 2
1 YSGAYNDRSYIIAFFTTCFIVPLLVILVSYGKLLQKLRK 0
0 VSNTQGRLRTARKPERQVTRMVVVMIIAFLICWMPYAVFSILATAYPSIELDPHLAAIPAFFSKTATVYNPIIYVFMNKQ 0
0 FRMCLIQMFKCSAIETAESNMNPTSERATLTQDKRDSQLSVMAVRSTIS* 0
>VAOP_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full G? vertebrate ancient opsin last exon newly truncated
0 MHLEYLKMGHFCPVTLLLPDGDPFHRPLDSIQPWQFKLLAAVMLLVTSLSLAENLAVILVTFKFKQLRQPINYIIVNLSVADFLVSLTGGTISFLTNLKGYFFMGYWACVLEGFAVTFF
2 GIVALWSLALLAFERYIVICRPLRNARLRGKHAALGIVFVWSFSFIWTIPPTTGWSSYTTSKIGTTCEPNW 2
1 YSGAYADHTYIITFFTTCFIVPLLVILVSYGKLVWKLKK 0
0 VSDAQGRLGAARRPERQVTRMVVFMIVAFLICWMPYATFSILVTAYPSIELDPCVAAIPAFFSKTATVYNPVIYVFMNKQ 0
0 FRQCLIQMFSCSAIGTAESNMKLTSERAVLMQGRRGSKRTPMAVHSTVLKRKTGDEHRADDLWLF* 0
>VAOP_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt vertebrate ancient opsin syn(+INPP5A -NXK6 +GPR125 +KNDC1)
0 MAGLRREAENDSWLFDPSSSSAPFDPFLQPLDIIEPWNFHLISALMFVVTLFSLSENFTVILVTIKFKQLRQPLNYVIVNLSVADFLVSLIGGTISFSTNLKGYFYMGHWACVLEGFAVTFF 1
2 GIVALWSLALLAFERYVVICRPLGNMRLNGKHAALGVAFVWIFSFIWTVPPTMGWSSYTTSKIGTTCEPNW 2
1 YSGDYNDHTFIITFFTTCFILPLLVILVSYGKLMRKLRK 0
0 VSDTQGRLGTTRKPERQVTGMVVIMILAFLICWSPYAAFSILVTACPSIELDPRLAAIPAFFSKTATVYNPVIYVFMNNQ 0
0 FRKCLVQLFQCSSQETMDANVNPISEKDTLTHTKHCGEMSTVAAHVIVFNPRSEDEQGSCQSFAQLAISENKVYPL* 0
>VAOP_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt vertebrate ancient opsin syn(- +GSTO2 -C10orf92 -) new
0 MPTNVSLLATPENSTVWNPFTGPLKTIEAWNFHLLAALMFVVTSLSIAENFIVILVTAKFKQLRQPLNYIIVNLSVADFLVSVIGGTISIATNSRGYFYLGSWACVLEGFAVTFF 1
2 GIVALWSLSVLAFERYIVICRPLGNLRLQGKHSALAIIFVWVFSFVWTIPPTMGWSSYTTSKIGTTCEPNW 2
1 YSGEMRDHTYIITFLTTCFVFPLLVIFMSYGKLMRKLRK 0
0 VSDTQGRLGSTRKPEKEVTRMVVIMILAFLICWTPYAAFSILITAHPTIDLDPRLAAIPAFFAKTASMYNPIIYVYMNKQ 0
0 FRRCLYQMFNINDPEAKESNLNPTSERGVLTRNNNGGEMLAIATHITSSAVTNREEEKSSSNSFAHIPVSDNKVCPM* 0
>VAOP_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_131586 17067577 full Gt valop assembly missing exon 3
0 MEASSAAVNAVSPAEDPFSAPLSSIAPWNYSVLAALMFVVTALSLSENFTVMLVTFRFQQLRQPLNYIIVNLSLADFLVSLTGGSISFLTNYHGYFFLGKWACVLEGFAVTFF 1
2 GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLVFVWSFSFIWTVPPVLGWSSYTVSRIGTTCEPNW 2
1 YSGNFHDHTFIITLFSTCFIFPLGVIIVCYCKLIRKLRK 0
0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIIITAHPSMHVDPRLAAIPAFVAKTAAVYNPIIYVFMNKQ 0
0 FRKCLVQLLSCSKVTVVEGNNNQTTERAGMTSGSNTGEMSAIAARVSVPKTEENPGDRSTFSHIPIPENKVCPM* 0
>VAOP_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? vertebrate ancient opsin syn(INPP5A NKX6 +VAOP GABRP KNDC1)
0 MESLSLSVNGVSHTVAAELAPTNDPFSGPINNIAQWNFTILAVLMFVVTALSLSENFLVMFITFKFKQLRQPLNYIIVNLAIADFLVSITGGLISFLTNARGYFFLGRWACVLEGFAVTYF 1
2 GIVAMWSLAVLSFERFFVICRPLGNMRLQAKHAVIGLLFVWTFSFVWTFPPVLGWNRYTVSKIGTTCEPDW 2
1 YSNNMTSHSYIITFFVTCFILPLGVIFFCYGKLLRKLRK 0
0 VSHGRLATARKPERQVTRMVVVMIVAFMVAWTPYATFAILVTIYPTIELDPRLASIPAFFSKTAAVYNPIIYVFMNKQ 0
0 FRKCLMQHFIGGGGTIESNMNPTSERPGITADSHTGEMSAIAARVPVGASASPLSDDSPTAQLPIPENKVCP* 0
>VAOP_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gt vertebrate ancient opsin syn(+INPP5A -NXK6 - +KNDC1)
0 MESLSLSVNGVSYTVAAELAPTNDPFTGPINNIAQWNFTILAVLMFVVTSLSLCENFLVMFITFKFKQLRQPLNYIIVNLAIADFLVSLTGGLISFLTNARGYFFLGRWACVLEGFAVTYF 1
2 GIVAMWSLAVLSFERFFVICRPLGNMRLQAKHAAIGLLFVWTFSFVWTFPPVLGWNRYTVSKIGTTCEPDW 2
1 YSNNMTSHSYIITFFSTCFILPLGIIFFCYGKLLRKLRK 0
0 VSHGRLATARKPERQVTRMVVVMIVAFMVAWTPYATFAILVTIHPTIELDPRLASIPAFFSKTAAVYNPIIYVFMNKQ 0
0 FRKCLIQHFIGMGVMAESNMNPTSERPGITAESQTGEMSAIAARVPVGATAALHSDGSPTDCGSLAQLPIPENKVCPI* 0
>VAOP_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gt vertebrate ancient opsin
0 MDTLSLSVNAVSHTVAAELPATSDPFNGPIKNIAPWNFTILAVLMFVVTSLSLSENFLVMFVTFKYKQLRQPLNYIIVNLAIADFLVSLTGGVISFLTNARGYFFLGKGACVLEGFAVTFF 1
2 GIVALWSLAVLSFERFFVICRPLGNIRLQPKHAILGLLFVWTFSFIWTFPPVLGWNRYTISKIGTTCEPDW 2
1 YSTNMIDHSYIITFFATCFILPLGVIFFCYGKLLRKLRK 0
0 VSHGRLATAKKPERQVTRMVVVMIVAFMVGWTPYAVFSLLVTSHPSIKLDPRLASIPAFFSKTAAVYNPIIYVFMNKQ 0
0 FRKCLIQLFSGRGTIPDGHLDPTSERPGMSQTGEMSAIATRIPMSSTVSPRSDDSPTDCVSFAQLPIPENKVCPM* 0
>VAOP_oryLat Oryzias latipes (medaka) Deut.Acti.Perc NM_001136515 full Gt vertebrate ancient opsin DK020629 brain DK098095 embryo
0 MDSLSLSVNAVSFTVATEIFSTDDPFTGPIKNIAPWNFTVLAVLMFVITSLSLTENFLVMFVTFKFKQLRQPLNYIIVNLAIADFLVSLTGGLISFLTNARGYFFLGKWACILEGFAVTFF 1
2 GIVALWSLAVLSFERFFVICRPLGNIRLQSKHAMLGLLFVWTFSFIWTFPPVLGWNRYTVSKIGTTCEPDW 2
1 YSTNMSDHSYIITFFATCFILPLGVIFYCYGRLLRKLRKVS 0
0 VSHGRLATARKPERQVTRMVVVMIVAFMVGWTPYAVFSILVTAHPTIKLDPRLASIPAFFSKTAAVYNPIIYVFMNKQ 0
0 FRKCLIQHFTGMGTIPESNLNQTSERPGITAESHTGEMSAIAARIPMGGTTSPRSDDSQTDCGSFAQLPIPENKVCPL* 0
>VAOP_salSal Salmo salar (salmon) Deut.Acti.Salm NM_001123626 9136902 frag Gt vertebrate ancient opsin first VAOP seq, last exon wrong AF001499
0 MDTLRIAVNGVSYNEASEIYKPHADPFTGPITNLAPWNFAVLATLMFVITSLSLFENFTVMLATYKFKQLRQPLNYIIVNLSLADFLVSLTGGTISFLTNARGYFFLGNWACVLEGFAVTYF 1
2 GIVAMWSLAVLSFERYFVICRPLGNVRLRGKHAALGLLFVWTFSFIWTIPPVFGWCSYTVSKIGTTCEPNW 2
1 YSNNIWNHTYIITFFVTCFIMPLGMIIYCYGKLLQKLRK 0
0 VSHDRLGNAKKPERQVSRMVVVMIVAYLVGWTPYAAFSIIVTACPTIYLDPRLAAAPAFFSKTAAVYNPVIYVFMNKQ 0
>VAOP_pleAlt Plecoglossus altivelis (smelt) Deut.Acti.Salm AB074483 11779166 full Gt vertebrate.ancient.opsin brain and amacrine cells of retina
0 MDTLRIPVNGVSYTEATEILKPEDPFTGPLSNIAPWNFTFLAVLMFFVSFLSLCENFTVMFVTFKFKQLRQPLNYIIVNLSIADFLVSLSGGLISFLTNAKGYFFLGKWACVLEGFAVTYF 1
2 GIVAMWSLAVLSFERFFVICRPLGNIRLRGKHAVLGLLFVWTFSFIWTIPPVLGWSSYTVSKIGTTCEPNW 2
1 YSTKLSDHIYIITFFFTCFILPLGVIIFCYGKLLRKLRK 0
0 VSHGRLGNARKPERQVTRMVVVMIVAFMVGWTPYAAFSIVVTACPHIYIDPRMAAIPAFFSKTAACYNPVIYVFMNKQ 0
0 FRKCLIKLFTGKGTIPEGNLGQTSERPGMTAESQTGEMSAIAARIPVGGTAATKSDEQPSECNSFAQLPIPENKVCPI* 0
>VAOP_rutRut Rutilus rutilus (minnow) Deut.Acti.Otoc AY116411 12906786 full Gt vertebrate ancient opsin
0 MELFPVAVNGVSHAEDPFSGPLTFIAPWNYKVLATLMFVVTAASLSENFAVMLVTFRFTQLRKPLNYIIVNLSLADFLVSLTGGTISFLTNYHGYFFLGKWACVLEGFAVTYF 1
2 GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLLFVWTFSFIWTIPPVLGWSSYTVSKIGTTCEPNW 2
1 YSGNFHDHTFIIAFFITCFILPLGVIVVCYCKLIKKLRK 0
0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIVVTAHPSIHLDPRLAAAPAFFSKTAAVYNPVIYVFMNKQ 0
0 FRKCLVQLLRCRDVTIIEGNINQTSERQGMTNESHTGEMSTIASRIPKDGSIPEKTQEHPGERRSLAHIPIPENKVCPM* 0
>VAOP_cypCar Cyprinus carpio (carp) Deut.Acti.Otoc AF233520 -------- full Gt vertebrate.ancient.opsin ---
0 MESLAAAVNGVSHTEDPFSGPLSSIAPWNYRVLAALMFVVTSVSLCENFTVMLVTFRFKQLRQPLNYIIVNLSLADFLVSLSGGTISFLTNFHGYFFLGRWACVLEGFAVTF 1
2 FGIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLLFVWSFSFVWTIPPVLGWSSYTVSKIGTTCEPNW 2
1 YSGNFQDHTFIITFFTTCFIFPLAVIIVCYCKLLRKLRK 0
0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIVVTALPSIHLDPRLAAAPAFFSKTAAVYNPIIYVFMNKQ 0
0 FRKCVVQLLSCRDVTVVEGNINQTTDRAGLTNESNTGEMSAIAARIPAAGTVPPKTEEPPNERSSFTQIPIPENKVCPM* 0
>VAOP_ictPun Ictalurus punctatus (catfish) Deut.Acti.Otoc FJ839436 -------- full Gt vertebrate.ancient.opsin ---
0 MGSFGTAVRGVNYTEAADVIQPEDPFSGPVKSIAPWNFKVLAAWMVVITSLSLSENFLVMLVTYKFKQLRQPLNFLIVNLSLADFFMSLIGGTLSVVTNYRGYFFLGSWACVLEGFAVTF 1
2 FGIVGMWSLATLAFERYFVICRPLGNVRLQVKHAVLGLLFVWTFSFIWTIPPVLGWSSYTVSKIGTTCEPNWYSGDPHDHTFIITFFLTCFILPLAVIIVCYSKLLRKLRK 0
0 VSNSHGRLVNARKPDRQVSHMVVVMIVAFVVAWTPYALFSFIVTVHPTIYLDPIMAAVPAFFAKTAAVYNPIIYVFMNKQ 0
0 FRKCLIQLLSCSGNTTVEVNQSTDRAGMIGESNTGEMSAIAARIPAASTTSPPKANEQHSNSCSYSQLPIPENKVCPM* 0
>VAOP_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gt vertebrate ancient opsin
0 VASTQGRLGVARKPEKQVTRMVIVMILAFLFCWTPYAAFSITVTACPTIKLDPRLAAIPAFFSKTATVYNPIIYVFMNKQ 0
>VAOP_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype U90667 9427550 full Gt vertebrate ancient opsin exons 123 in traces pineal gland-specific
0 MDALQESPPSHHSLPSALPSATGGNGTVATMHNPFERPLEGIAPWNFTMLAALMGTITALSLGENFAVIVVTARFRQLRQPLNYVLVNLAAADLLVSAIGGSVSFFTNIKGYFFLGVHACVLEGFAVTYF 1
2 GVVALWSLALLAFERYFVICRPLGNFRLQSKHAVLGLAVVWVFSLACTLPPVLGWSSYRPSMIGTTCEPNW 2
1 YSGELHDHTFILMFFSTCFIFPLAVIFFSYGKLIQKLKK 0
0 ASETQRGLESTRRAEQQVTRMVVVMILAFLVCWMPYATFSIVVTACPTIHLDPLLAAVPAFFSKTATVYNPVIYIFMNKQ 0
0 FRDCFVQVLPCKGLKKVSATQTAGAQDTEHTASVNTQSPGNRHNIALAAGSLRFTGAVAPSPATGVVEPTMSAAGSMGAPPNKSTAPCQQQGQQQQQQGTPIPAITHVQPLLTHSESVSKICPV* 0
>PPIN_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt parapinopsin syn(-CPEB2 -CACNA2D3 +SELK) syntenic deleted chicken
0 MDSLDTNTLSPNASTVRVVLMPRIGYTIIAIIMATSCTLSVILNTAVIAITIKYRQLRQPINYSLVNLAIADLGAALLGGSLNVETNAVGYYNLGRVGCVTEGFAMAFF 1
2 GIVALCTIAVIAVDRAIVIAKPMGTITFTTRKAMIGVAVSWIWSLVWNTPPLFGWGGYQMEGVMTSCAPDWANSDPINVSYIICYFLFCFTIPFITILASYGYLIWTLRQ 0
0 VAKVGLAQRGSTTKAEAQVSRMVIVMVMAFLICWLPYATFALVVVGNPQIYINPIIATIPMYMAKSSTFYNPIIYIFMNKQ 0
0 FRDCLVRCLLCGRNPCASEQTDEDDLEVSTIAPAPSSRRGKVAPV* 0
>PPIN_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full Gt parapinopsin para bistable UV lamprey pineal broken contigs
0 MADEALLPPMMNVTNEEMHPGKVLMPRIGYTILALIMAVFCAAALFLNVTVIVVTFKYRQLRHPINYSLVNLAIADLGVTVLGGALTVETNAVGYFNLGRVGCVIEGFAVAFF 1
2 GIAALCTIAVIALDRVFVVCKPMGTLTFTPKQALAGIAASWIWSLIWNTPPLFGWGSYELEGVMTSCAPNWYSADPVNMSYIVCYFSFCFAIPFLIIVGSYGYLMWTLRQ 0
0 VAKLGVAEGGTTSKAEVQVSRMVIVMILAFLVCWLPYAAFAMTVVANPGMHIDPIIATVPMYLTKTSTVYNPIIYIFMNKQ 0
0 FQECVIPFLFCGRNPWAAEKSSSMETSISVTSGTPTKRGQVAPA* 0
>PPIN_danRer Danio rerio (zebrafish) Deut.Acti.Otoc XM_681591 full Gt parapinopsin syn(- - +SELK -)
0 MESETSTAASGSIAEVMPRMGYTILAVIIGVFSVCGVILNVTVITVTLKYKQLRQPLNFALVNLAVADLGCAVFGGLPTVVTNAMGYFSLGRVGCVLEGFAVAFF 1
2 GIAALCSVAVIALERCMVVCRPVGSISFQTRHAVFGVAVSWLWSFIWNTPPLFGWGRLQLEGVRTSCAPDWYSRDLANVSFIVCYFLLCFALPFSVIVYSYTRLLWTLRQ 0
0 VSRLQVCEGGSAARAEAQVSCMVVVMILAFLLTWLPYASFALCVILIPELYIDPVIATVPMYLTKSSTVFNPIIYIFMNRQ 0
0 FRDRALPFLLCGRNPWAAEAEEEEEETTVSSVSRSTSVSPA* 0
>PPINa_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? parapinopsin
0 MKPSVYLNTSLYLGPPEEPPLPRSGFIVLSILMALVTGPAIVLNATVIIVSLMHKQLRQPLNYALVNMAAADLGTAVSGGLLSVVNNAQGHFSLGRTGCVLEGFAVSLC 1
2 GIASLCTVALIAVERMFVICKPLGQMQFQKQHALAGISLSWLWSLTWNLPPLFGWGRYELEGVGTSCAPDWQSREPHNVSYVLAYFTVCFAAPFAIILVSYAKLMWTLHK 0
0 VTKMTCLEGGAVAKSEMKVAYMVVLMVATFLLSWLPYAGLSMLVVFKPDVEINPLVGTVPVYLAKSSTAYNPIIYIYLNKQ 0
0 FRKYALPFLLCRRALEAEDEVSETTVESSRRVSPS* 0
>PPINa_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? parapinopsin
0 MKPSAFYLNASLYLGPQGEPPLPRSGFIALSVIMALLTGPAIVLNATVIIVSLMHKQLRQPLNYALVNMAVADLGTAMTGGLLSVVNNAQGYFSLGRTGCVLEGFAVSLC 1
2 GIASLCTVALIAVERMFVICKPLGQMQFQKQHALGGIALAWLWSLTWNLPPLFGWGRYELEGVGTSCAPDWHSREPQNVSYVLAYFTVCFAAPFVIILVSYSKLMWTLHK 0
0 VTKMACMEGGAVAKSEMTVAYMVILMVVIFLISWLPYAGLSMLVVLSPDVKIHPLVGTVPVYLAKSSTVYNPIIYIYLNKQ 0
0 FRKYAVPFLLCGRELEMEDELSMTTVETSNRVSPA* 0
>PPINa_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc DN691174 full G? parapinopsin adult eyes
0 MQPSHTFSNSSAYTGPHGEPPLSRTGFIILSIIMAGFTGPAIVLNATVIIVTLMHKQLRQPLNYALVNMALADLGTAMTGGVLSVVNNAQGYFSLGRSGCVMEGFAVSLF 1
2 GITSLCTVALIAVERMFVVCKPLGQIIFQKKHAVGGIAISWLWSLSWNLPPLFGWGRYELEGVGTSCAPDWHNQDPKNVSYIVAYFAVCFAVPFALILASYTKLMWTLHQ 0
0 VSKMTCLEGGAVAKGEMKVASMVVLMVLTFLISWLPYASLAMLVVYNPKVEIHALVGTVPVYLAKSSTVFNPIIYIYLNKQ 0
0 FRKYAVPFLLCCKEPLDDEEASEAATTVEISPSKVSPA* 0
>PPIN_ictPun Ictalurus punctatus (catfish) Deut.Acti.Otoc genomic full Gt parapinopsin index sequence
0 MASIILINFSETDTLHLGSVNDHIMPRIGYTILSIIMALSSTFGIILNMVVIIVTVRYKQLRQPLNYALVNLAVADLGCPVFGGLLTAVTNAMGYFSLGRVGCVLEGFAVAFF 1
2 GIAGLCSVAVIAVDRYMVVCRPLGAVMFQTKHALAGVVFSWVWSFIWNTPPLFGWGSYQLEGVMTSCAPNWYRRDPVNVSYILCYFMLCFALPFATIIFSYMHLLHTLWQ 0
0 VAKLQVADSGSTAKVEVQVARMVVIMVMAFLLTWLPYAAFALTVIIDSNIYINPVIGTIPAYLAKSSTVFNPIIYIFMNRQ 0
0 FRDYALPCLLCGKNPWAAKEGRDSDTNTLTTTVSKNTSVSPL* 0
>PPIN_oncMyk Oncorhynchus mykiss (trout) Deut.Acti.Salm genomic full Gt parapinopsin
0 MDHQQLLPNLHGNISSSPGSVSEALLSRTGFTILAVIIGVFSVSGVCMNVLVIMVTMRHRKLRQPLNYALVNLAVADLGCALFGGLPTMVTNAMGYFSMGRLGCVLEGFAVAFF 1
2 GIAGLCSVAVIAVDRYVVVCRPMGAVMFQTRHAVGGVVLSWVWSFLWNTPPLFGWGSFELEGVRTSCSPNWYSREPGNMSYIILYFLLCFAIPFSIIMVSYARILFTLHQ 0
0 VSKLKVLEGNSTTRVEIQVVRMVVVMVMAFLLSWLPYAAFALSVILDPSLHINPLIATVPMYLAKSSTVYNPIIYVFMNRQ 0
0 FRDCAVPFLLCGLNPWASEPVGSEADTALSSVSKNPRVSPQ*
>PPINb_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? parapinopsin syn(+PCTK1 -PPIN +DDX20 -KCND2) absent medaka zfish
0 MEQGIQEGNSSSLSSVSSGTLSRTGYTVLAFIMGVLSAGGIILNVLVIVVTMKHRQLRQPLSYALVNLAICDLGCALFGGIPTTITSAMGYFSLGRVGCVLEGFAVAFF 1
2 GIASLCTIGVISVERYVVVSNPMGAVLFQTR 2
1 HAVAGVVFSWVWSFVWNTPPLFGWGSFDLEGVRTSCAPNWYSRDVGIMSYIVIYLLFCFAVPFTIITVSYSRLLWTLRQ 0
0 VTGLQVAEGGSTNRVEVQVARMVVVMVLAFLLTWLPYAAMALAVVMDSTLYINPIIATIPVYLAKSSTVYNPIIYIFMNRQ 0
0 FRGCAINTVLCGRRAWITDLQTSEGETTVASTSKSQKISPKGSLN* 0
>PPINb_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? parapinopsin syn(-PPIN +DDX20)
0 MEQEIQDGNSSSLNSVSPGILSQTGFTVLAFIMGMLSVGGIILNVLVIVVTLKHRQLRQPLNYALVNLAICDLGCALFGGIPTTVTSAMGYFSLGRLGCVLEGFAVAFF 1
2 GIASLCTIGIISVERYIVVSNPMGAVLFQTR 2
1 HAVAGVVFSWVWSFVWNTPPLFGWGSFELEGVGTSCAPNWYSRDMGNMSYIIIYLLFCFAVPFSIIMVSYSRLLWTLHQ 0
0 VTKLHVAEGGSTNRVEVHVARMVVLMVLAFLLTWLPYASMALAVVMDSTLYIDPVTATIPVYLAKSSTVYNPIIYIFMNRQ 0
0 FRGYAINTILCGRRAWVSEQQTSEGETTVVSVSKSQKISPKGSLQ * 0
>PPINb_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc DW621258 full G? parapinopsin syn(+DNMT2 -PPIN +DDX20 -TAZ) 21 day old larvae
0 MERDGNASADAQLLSPSGYAALAAVMGAFSVAGILLNALVIVVTARHRQLRQPLSYALVNLAVCDLGCAACGGLPTTVTSAMGYFSLGRAGCVLEAFAVAFF 1
2 GIASLCTIGVISVERYVVVCYPMGAVLFQTR 1
2 HAVAGVVLSWVWSFVWNTPPLFGWGSYELEGVKISCAPNWYSRDPGNVSYVTIYFLLCFAVPFSVIMVSYSRLLWTLRQ 0
0 VTKLQVSETGSTNRVEVQVARMIVVMVLAFLVTWLPYAAMALAVITDSTLHIDPVIATIPVYLAKSSTVYNPIIYIFMNRQ 0
0 FRGYAVPSILCGWNPWAEEQTSEEETVGSVMKSQRVSPKGSLQE* 0
>PPINb_mayZeb Maylandia zebra (cichlid) Deut.Acti.Perc genomic frag G? parapinopsin
0 1
2 GIASLCTVAVISVERYVVVCYPMGAVLFQTR 2
1 HAVAGVVLSWVWSFVWSTPPLFGWGTFELEGVKTSCAPKWHSRDVGDMSYMIIFFFLCFALPFSVVMVSYSRLLWTLRQ 0
0 GMIVVMVLAFLVTWLPYAALALAVMIDSSLYVDPVIATIPVYFAKSSTVYNPIIYIFMNRQ 0
0 FRGYTVAAVLCGWDPWSSEPQTSENETTVPFFIKTPKKIVPKKSLE*0
>PPIN_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gt parapinopsin
0 MDPHNRSANLSEGPGLGGGGAVPGWGPSVRAPLSLVMAVISLSSIVLNSLAIAVVLRFQVLQQPLNYALLSLASADLGTAATGGVLSTVCTALGSFVLGRHSCVAEGFF 1
>PPINa_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic full Gt parapinopsin bistable pineal UV/green
0 MENLTSLDLLPNGEVPLMPRYGFTILAVIMAVFTLASLVLNSTVIIVTLRHRQLRHPLNFSLVNLAVADLGVTVFGASLVVETNAVGYFNLGRVGCVIEGFAVAFF 1
2 GIAALCTIAVIAVDRFVVVCKPLGTLMFTRRHALLGITWAWLWSFVWNTPPLFGWGSYKLEGVRTSCAPDWYSRDPANVSYIVSFFSFCFAIPFLVIVVAYGRLLWTLHQ 0
0 VAKLGMGESGSTAKAEAQVSRMVVVMVVAFLVCWLPYALFAMIVVAKPGVYIDPVIATLPMYLTKTSTVYNPIIYIFMNRQ 0
0 FRDCAVPFLLCGRNPWAEPSSESATTASTSATSVTLASVPGQVSPS* 0
>PPINb_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag G? parapinopsin odd genomic
2 GMTALITVCVLAVERYVVVCKPLGGVHFGTQHGLCGVAISWTWALAWSAPPLFGWGRYHYEGVGTSCAPDWADSSPSGRSYMTTYFIFCFALPMIIILFCYTKLMIAIHK 0
0 VSKLGLSANDTAERKVGIMVVVMVFAFFLCWLPYAALAIAVVIKPDLK 0
0 VSPVTASIPVYLAKSSGAYNPIIYIFMHRQ 0
>PPIN_letJap Lethenteron japonicum (lamprey) Deut.Agna.Hype AB116380 14981504 full Gt parapinopsin bistable pineal UV/green
0 MENLTSLDLLPNGEVPLMPRYGFTILAVIMAVFTIASLVLNSTVVIVTLRHRQLRHPLNFSLVNLAVADLGVTVFGASLVVETNAVGYFNLGRVGCVIEGFAVAFF 1
2 GIAALCTIAVIAVDRFVVVCKPLGTLMFTRRHALLGIAWAWLWSFVWNTPPLFGWGSYELEGVRTSCAPDWYSRDPANVSYITSYFAFCFAIPFLVIVVAYGRLMWTLHQ 0
0 VAKLGMGESGSTAKAEAQVSRMVVVMVVAFLVCWLPYALFAMIVVTKPDVYIDPVIATLPMYLTKTSTVYNPIIYIFMNRQ 0
0 FRDCAVPFLLCGRNPWAEPSSESATAASTSATSVTLASAPGQVSPS* 0
>PPINa_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni NM_001032555 11591373 full Gt Ci-opsin1 syn(-HOXB1 +HHEX +CUL4A -) odd exons larval ocellus
0 MDHDVTPTVDLTDGVPQCKDLNPYVLKGDGWVPQHISRANRSTYSFLCVYMTFVFLLSCSLNILVIVATLKNK 0
0 VLRQPLNYIIVNLAVVDLLSGFVGGFISIAANGAGYFFWGKTMCQIEGYFVSNF 1
2 GVTGLLSIAVMAFERYFVICKPFGPVRFEEKHSIF 1
2 GIVITWVWSMFWNTPPLIFWDGYDTEGLGTSCAPNWFVKEKRERLFIILYFVFCFVIPLAVIMICYGKLILTLRQ 0
0 IAKESSLSGGTSPEGEVTKMVVVMVTAFVFCWLPYAAFAMYNVVNPEAQ 0
0 IDYALGAAPAFFAKTATIYNPLIYIGLNRQ 0
0 FRDCVVRMIFNGRNPWVDELVGSQVSSTGSQLTAVSSNKVAPA* 0
>PPINb_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni genomic full Gt parapinopsin syn(-TMEM165 +FUT4 - -) jgi gene model wrong both ends
0 MTTAETTTECYEKNPYIRNEMGWVPKHILIAERHIYTILAVYMTFIFLLAVSLNGFVIIATMKNK 0
0 KLRQPLNYIIINLSIADFLSGLVGGFIGMISNSAGYFYFGKTVCILEGYIVSVA 1
2 GVCGLMSISVMAFERYFVVCKPYGPFTLTNTHAAL 1
2 GIGFTWTWSVLWSTPGLIWLDGYVPEGLGTSCAPNWFSKNK 2
1 SERIFIFVYFVFCFFIPLLVIIICYGKIVLFLKQ 0
0 ATRQSSASSNRQADNKVTKMVLVMISAFLICWTPYGVLSLYNAINPDKQ 0
0 LDYGLGAVPVFFAKTANIYNPLIYIGLNKQ 0
0 FRDGVIKMVFRGRNPWAEEMSTQQRQRSTEAGQPIVSNEV* 0
>PPINa_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni genomic full G? parapinopsin
0 MPTEASIAVDVSPTMGIPQCKDINPYVLKGDGWVPQHISRADRSVYSFLAVYMTFICLISCSLNILVITATLKNK 0
0 VLRQPLNYIIVNLAVVDLLSGLVGGVISIFANGAGYFFWGKFMCQVEGYTVSNF 1
2 GVTGLLSIAVMAFERYFVICKPFGPVRFEEKHAVI 1
2 GIAVTWIWAMFWNTPPLIFWDGYDTEGLGTSCAPNWFVKGNTERLFIILYFVFCFLIPLAIIVLCYGKLILQLRQ 0
0 IAKESSLSGGTSPEGEVTKMVVVMVTAFVICWLPYAAFAMYNVVNPEAQ 0
0 IDYALGAAPAFFAKTATIYNPLIYIGLNRQ 0
0 FRDCVVRMIFNGRNPWVDEMVGSQVSSSASQMTAVSSNKVAPA* 0
>PPINb_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni genomic full G? parietopsin
0 MSSIPQNYSNGNPYATTDSGWVPEHIEIANRSTYSGLCVFMSFVFVLAVPLNLLVIVATYKNK 0
0 DLRRPINYIIVNLAVADLTCSVVGGLLGVLNNGAGYYFLGKSVCIFEGYVMSVT 1
2 GVCGILSITVMAFERYFVVCKPFGQTNLKWSHAIT 1
2 GIVFTWTWSVIWHTPGLFFWNGYEPEGFGTSCAPNWFSQQK 2
1 SERIFIFAYFAFCFLTPLTIIFACYLKLILFIRK 0
0 VSKKSMVNEADRRDFEVTRMVFVMIAAFLICWLPYGCLSMYNAIHPDNL 0
0 LSYGIGSVPAFFAKTATIYNPIIYMGLNKK 0
0 FRDGVIRMLFKGRNPWLDGRNTTSSTSTRAQGSLINREVDI* 0
>PARIE_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gd+Go parietopsin syn(+EEA1 -FLJ46688 +BTG1 -) gusducin not transducin
0 MENESSLVLEGAEGYIVRPTIFPRAGYGVLAFLMFINALFSLFNNFLVIAVTLKNPQLRNPINIFILNLSFSDLMMSICGTTIVIATNYHGYFYLGRRFCIFQGFAVNYF 1
2 GIVSLWSLTILAYERYNVVCQPLGTLQMSTQRAYQLLGFIWVFCLFWAVVPLFGWSSYGPEGVQTSCSIGWEERSWNNYSYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG 0
0 LNKKVEQLGGKSNPEEEFRAVIMVLVMVVAFLICWLPYTLFALTVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKE 0
0 FRECAVEFITCGKVVLTSPEEDISTSAISDEGIAPCKINQVTPV* 0
>PARIE_utaSta Uta stansburiana (lizard) Deut.Saur.Lepi DQ100320 16543463 full Gd+Go parietopsin shift in counterion
0 MENDSSLATELAEGAIVKPTIFPKAGYGVLAFLMFLNALFSIFNNSLVIAVTLKNPQLRNPINIFILNLSFSDLMMSLCGTTIVIATNYYGYFYLGRKFCIFQGFAVNYF 1
2 GIVSLWSLTILAYERYNVVCQPLGTLQMSTKRGYQLLGFIWVFCLFWAVVPLFGWSSYGPEGVQTSCSIGWEERSWSNYSYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG 0
0 LNKKVEQLGGKSSPEEEFRAVIMVLVMVVAFLICWLPYTVFALIVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKQ 0
0 FRDCAVEFITCGQVVLTSPEEDISTSAIPVEGKGPCKINQVTPV* 0
>PARIE_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur DN070761 16543463 full Gd+Go parietopsin syn(-lum -DCN) lung
0 MDGNSTTPGIAVNLTVMPTIFPRSGYSILSFLMFLNAVFSICNNAIVILVTLKHPQLRNPINIFILNLSFSDLMMALCGTTIVVSTNYHGYFYLGKQFCIFQGFAVNYF 1
2 GIVSLWSLTLLAYERYNVVCEPIGALKLSTKRGYQGLVFIWLFCLFWAIAPLFGWSSYGPEGVQTSCSIGWEERSWSNYSYIISYFLTCFIIPVGIIGFSYGSILRSLHQ 0
0 LNRKIEQQGGKTNPREEKRVVIMVLFMVLAFLICWLPYTVFALIVVINPQLYISPLAATLPTYFAKTSPVYNPIIYIFLNKQ 0
0 FRTYAVQCLTCGHINLDSLEEDTESVSAQAENMLTPKTNQVAPA* 0
>PARIE_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic 16543463 full Gd+Go parietopsin syn(- +NT5DC2 +FBXL13)
0 MENFAKTELTMMVQPTIFPRVGYSILSYLMFINTTLSVFNNVLVIAVMVKNLHFLNAMTVIIFSLAVSDLLIATCGSAIVTVTNYEGSFFLGDAFCVFQGFAVNYF 1
2 GLVSLCTLTLLAYERYNVVCKPMAGFKLNVGRSCQGLLLVWLYCLFWAVAPLLGWSSYGPEGVQTSCSLGWEERSWRNYSYLILYTLMCFILPTVIITYCYSNVLLTMRK 0
0 INKSIECQGGKNCAEDNEHAVRMVLAMIIAFFICWLPYTAISVLVVVNPEISIPPLIATMPMYFAKTSPVYNPIIYFLTNKR 0
0 FRESSLEVLSCGRYISRETGGPLMGSSMQRGQSRVNPV* 0
>PARIE_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? parietopsin genomic frameshift error
0 MDSASTPWSPHPASGQAEAVTAAPTIFPRVGYSILSFLMFINTVLSIFNNGLAITVMLKNPALLQPINIFILSLAVSDLMIGLCGSLVVTITNYQGSFFIGHTACVFQGFAVNYF 1
2 GLVSLCTLTLLAYERYNVVCKPRAGLKLNMRRSLVGLLFVWTFCLFWAVTPLLGWSSYGPEGVQTSCSLAWEERSWNNYSYLILYTLLCFILPVGVIIYCYTKVLTSMNK 0
0 LNKSVELQGGRSCQKENDHAISMVLAMIIAFFVCWLPYTALSVVVVVDPELRIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0
0 FRDATLEVLSCGRYIPHASTRVTFNMCAFNRRSRLPSLSRSINTHSKVSPL* 0
>PARIE_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic 16543463 full Gd+Go parietopsin syn(-HSP90B1 +NT5DC2 -KCND3 -FLNC)
0 MDSNSTPWSSPPAPLQAEAVTVAPTIFPRVGYSILSFLMFINTVLSVFNNSLAIAVMLKNPSLLQPINIFILSLAVSDLMIGLCGSLVVTITNYHGSFFIGHTACVFQGFAVNYF 1
2 GLVSLCTLTLLAYERYNVVCKPRAGLKLTMRRSIIGLLFVWTFCLFWAVTPLLGWSSYGPEGVQTSCSLAWEERSWNNYSYLILYTLLCFIFPVGVIIYCYCKVLTSMNK 0
0 LNKSVELQGGLSCRRENKHAINMVLAMIIAFFVCWLPYTALSVVVVVDPELHIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0
0 FRDATLEVLSCSRYIPHASSRVSINMRSLNRRSVNTHSKVSPL* 0
>PARIE_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? parietopsin syn(-HSP90B1 +NT5DC2 -KCND3 -FLNC)
0 MDSNSTLWSSGSPPPSIHGKMLTITPTIFPRVGYSILSFLMFINTVLTVFNNVLVITVLVRNPSLLQPMNVFILSLAVSDLMIGLCGSLVVTITNYHGSFFIGHTACIFQGFAVNYF 1
2 GLVSLCTLTLLSYERYNVVCRPRNALKLSMRRSIHGLLIVWTFCLFWAVAPLFGWSGYGPEGVQTSCSLAWEERSWSNYSYLVLYTLLCFIVPVAVIIYCYAKVLTSMNT 0
0 LNRSVEVQGGRSSQKENDHAVSMVLAMIIAFFSCWLPYTALSVVVVVDPTLYIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0
0 FRDAALEMLSCGRYIAHMPNTVSINMRSLNRRSRLSSLSRNVNSHSKVLPL* 0
>PARIE_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag Gd+Go parietopsin
0 LNKKIKRVGGHPDPREEMRATVMVLAMVGAFLACWLPYTVLALCVVLAPGTQIPPLVATLPMYFAKTSPIYNPIIYFFLNRQ 0
>ENC_homSap Homo sapiens (human) Deut.Euth.Euar NM_014322 12242008 full Gt OPN3 encephalopsin syn(-EXO1 -WDR64 -KMO +FH) with intron loss
0 MYSGNRSGGHGYWDGGGAAGAEGPAPAGTLSPAPLFSPGTYERLALLLGSIGLLGVGNNLLVLVLYYKFQRLRTPTHLLLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSGSLF 1
2 GIVSIATLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPLGVIAHCYGHILYSIRM 0
0 LRCVEDLQTIQVIKILKYEKKLAKMCFLMIFTFLVCWMPYIVICFLVVNGHGHLVTPTISIVSYLFAKSNTVYNPVIYVFMIRK 0
0 FRRSLLQLLCLRLLRCQRPAKDLPAAGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDKTNGSKVDVIQVRPL* 0
>ENC_otoGar Otolemur garnettii (lemur) Deut.Euth.Euar genomic full G? OPN3 encephalopsin
0 MYSGNRSGGQGFWEGGGAAGAEEPTPEGTLSPAPLFSPSAYERLALLLGSIGLLGVANNLLVLVLYYKFPRLRTPTHLFLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSGSLF 1
2 GIVSIATLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPVGVVAHCYGHILYSIRM 0
0 LRCVEDLQTTQVIKILKYEKKVAKMCFFMIFTFLVCWMPLIVICFLVVNGQGHLVTPTVSIVSYLLAKSNTVYNPVIYIFMLRK 0
0 FRRSLLQLLCFRLLRCQRPAKDLPAAESEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDNSDKTNGSKVDVIQVRPL* 0
>ENC_musMus Mus musculus (mouse) Deut.Euth.Euar NM_010098 full G? OPN3 encephalopsin panopsin 2aa del
0 MYSGNRSGDQGYWEDGAGAEGAAPAGTRSPAPLFSPTAYERLALLLGCLALLGVGGNLLVLLLYSKFPRLRTPTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1
2 GFVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWRSKDANDSSFVLFLFLGCLVVPVGIIAHCYGHILYSVRM 0
0 LRCVEDLQTIQVIKMLRYEKKVAKMCFLMAFVFLTCWMPYIVTRFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMNRK 0
0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMHIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVEDSDRSSASKVDVIQVRPL* 0
>ENC_canFam Canis familiaris (dog) Deut.Euth.Laur DN422921 full early gap G? OPN3 encephalopsin XP_854433 grossly wrong
0 MYSGNRSGGQGHWEGGGAAGAEGPGPAGTLSPAPLFSPGTYERLALLLGSVGLLGVGNNLLVLVLYSKFQRLRTPTHLLLVNLSLSDLLVSLFGVTFTFVSCLRNGWVWDSVGCVWDGFSSSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWSGAPLLGWNRYILDVHGLGCTVDWKSKDANDSFFVLFLFLGCLVVPMGVIVHCYGHILYSIRM 0
0 LRCVEDLQTIQVIKILRYEKKVAKMCFLMIFIFLIFWMPYIVICFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIIMIRK 0
0 FRRSLLQLLCFRPLRCQRPAKDLPANGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESVSIDDSDKTSVSKVDVIQVRPL* 0
>ENC_pteVam Pteropus vampyrus (macrobat) Deut.Euth.Laur genomic full G? OPN3 encephalopsin
0 MHSGNRSGGLDSWEGGGAAGAEGPGLAGTLSPGSVFNPSTYERLALLLGSIGLLGVANNLLVLVFYYKFQQVRTPFYLFLVNISFSDLLVSFFGVTFTFVSCLRNGWVWDTVGCVWDGFSSSLF 1
2 GTVSMTTLTVLAYERYIRVVQARAIDFSWAWRTITYIWLYSLGWSGAPLLGWNRYILDVHGLGCAVDWKSKDANDSSFVLFLFLGCLVVPVVVIAHCYGHILYSVQM 0
0 LRCVEDLQTIQVIKILRYEKKMAKMCFLMIFTFLISWMPYIVICFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMIRK 0
0 FRRFVLQLLCFRPLRCRRPATDLPAGGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFVITSDESLSVDDSDKINGSKADGIQVRPL* 0
>ENC_loxAfr Loxodonta africana (elephant) Deut.Euth.Afro genomic full G? OPN3 encephalopsin 2 exons in browser 1 2x
0 MYSGNRSGGQDLWEGGGGSGGAGPAGTLSPAPVFRSGTYERLALLVGSIGLLGVGNNLLVLVLYYKFQRLRTPTHLFLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSSSLF 1
2 GIASITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWSGAPLLGWNRYILDTHGLACTVDWKSNNSSDSSFVLFLFLGCLVVPVGVIAHCYGHILYSIRM 0
0 LRCVEDLQTIQVIKILRHEKKLAKMCLFMIFTFLICWMPYIVICFLVVNGYGHLVTPTISIVSYLFAKSSTVYNPVIYTFMIRK 0
0 FRRSLLQLLCFRLLRCQRPAKDLPVVGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVNNIDKTNGSKADVIQIRPL* 0
>ENC_monDom Monodelphis domestica (opossum) Deut.Meta.Dide genomic full G? OPN3 encephalopsin syn(-EXO1 -WDR64 +ENC -KMO +FH +RGS7)
0 MYSDNSSDDGGGGYWGSGRAGGASGTGVTGEPGPEGSPRQAPLFSPGTYELLALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLFLVNISFNDLLVSLFGVTFTFVSCLRSGWVWDSVGCAWDGFSNTLF 1
2 GIVSIMTLTVLAYERYNRIVHAKVINFSWAWRAITYIWLYSLVWTGAPLLGWNRYTLEIHGLGCSVDWKSKDPNDSSFVIFLFFGCLMLPVGVMAYCYGHILYAIRM 0
0 LRCVEELQTIQVIKILRYEKKVAKMCFLMIAIFLFCWMPYAVICLLVANGYGSLVTPTVAIIASLFAKSSTAYNPIIYIFMSRK 0
0 FRRCLLQLLCFRLLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDETQMIDENDKNSGTKVNVIQVRPL* 0
>ENC_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full Gt OPN3 encephalopsin syn(-EXO1 -WDR64 +ENC -PIGM +R
0 MHSGNGTGATSRPQLAAAGHEVPGERPLFSAGTYELLALLIATIGTLGVCNNLLVLVLYYKFKRLRTPTNLFLVNISLSDLLVSVCGVSLTFMSCLRSRWVWDAAGCVWDGFSNSLF 1
2 GIVSIMTLTVLAYERYIRVVHAKVIDFSWSWRAITYIWLYSLAWTGAPLLGWNRYTLEIHGLGCSMDWKSKDPNDTSFVLLFFLGCLVAPVVIMAYCYGHILYAVRM 0
0 LRCVEDFQTSQVIKLLKYEKKVAKMCFLMISTFLICWMPYAVVSLLVTYGYSNLVTPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0
0 FRQCLLQLLCFRLMRFQRIMKEPSGAGNVKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIIASDDTQQIDDNSKHNGTKVNVIQVKPL* 0
>ENC_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gt OPN3 encephalopsin syn(-EXO1 -WDR64 +ENC -PIGM +RGS7)
0 MFSANGTRSGAGSDLEPGPGQQQQQREASEEEERGAGLSPFSAGTYELLALLVAAIGLLGLCNNLLVLVLYAKFKRLRTPTHLFLVNISLSDLLVSLFGVSFTFGSCLRHRWVWDAAGCVWDGFSNSLF 1
2 GIVSIMTLTVLAYERYIRVVHARVIDFSWSWRAITYIWLYSLAWTGAPLLGWNHYTLEIHGLGCSVDWQSKEPSDSSFVLFFFLGCLAAPVGIMAYCYGHILHAIRM 0
0 LRCVEDLQSIQVIKILRYEKKVAKMCFLMVTTFLICWMPYAVVSLLIAYGYGHLITPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0
0 FRRCLVQLFCVQFLRFKRTLKEQPAIESNKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDDTEQIDVSTKCSDTKINVIQVKPL* 0
>ENC_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic pseu G? OPN3 encephalopsin recent pseudogene I for Schiff K C-term residues lost
0 MPVTNGSHNNSISWLHSKDMFTEDTYHFLALIVATVGFLGLVNNLLVLILYCKFKRLQTPTNLLFFNTSLCHFVFSLLAITFTFMSCVRGSWAFSVEMCVFHGFSKNLL 1
2 GIVSFGTLTVVAYERYARVVYGKYVNSSWSKRSITFVWVYSLAWTGFPLIGWNLYTFETHKLDCSFEWTATDPKDTAFVLLFFLACITLPLSIMAYCYGYILYEIQK 0
0 LRSVKNIQNFQEITILDYEIKMAKMCLLMMLTFLIGWMPYTILSLLVTSGYSKFITPTITVMPSLLAIASAAYNPVIHIFTIKK 0
0 FRQCLVQLLFHNFWRLLKNLNGRLAMKKVKPVLGKGRSHNRPEKKVFSSSDFFTRTTSDTGTHGITESTKGKRTNVRLIQVHPLYP* 0
>ENC_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_001111164 full G? OPN3 encephalopsin
0 MNSFNETPTEAHLENYNYIFADETYKLLTFTIGSIGVLGFCNNIIVIILYSRYKRLRTPTNLLIVNISVSDLLVSLTGVNFTFVSCVKRRWVFNSATCVWDGFSNSLF 1
2 GIVSIMTLSGLAYERYIRVVHAKVVDFPWAWRAITHIWLYSLAWTGAPLLGWNRYTLEVHQLGCSLDWASKDPNDASFILFFLLGCFFVPVGVMVYCYGNILYTVKM 0
0 LRSIQDLQTVQTIKILRYEKKVAVMFLMMISCFLVCWTPYAVVSMLEAFGKKSVVSPTVAIIPSLFAKSSTAYNPVIYAFMSRK 0
0 FRRCMLQMLCSRLTSLQHTIKDRPLSRIEHPIRPIVMSQSRTDRPKKRVTFSSSSIVFIIASHDTHPLDITSKCNDEPDINVIQVRPL* 0
>ENC_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? OPN3 encephalopsin
0 MSSADDSRSARSGEPSLFAVHTYRLLAAAIGAIGVLGFCNNLAVAALYWRFRRLRTPTNLLLLNISLSDLLVSLLGVNFTFAACVQGRWTWNQATCVWDGFSNSLF 1
2 GIVSIMTLAALAYERYIRVVHAQVVDFPWAWRAIGHIWLYSLAWTGAPLLGWNRYTLEIHRLGCSLDWASKDPNDASFILLFLLACFFVPVGIMIYCYGNILYAVHM 0
0 IRSIQDLQTVQIIKILRYEKKVSVMFFLMISCFLLCWTPYAVVSMMVAFGRKSMVSPTVAIIPSFFAKSSTAYNPVIYVFMSRK 0
0 FRRCLLQLLCSRLSWLQRGLKERPLAPVQRPIRPIVVSRPCGKGTRPKKKVTFSSSSIVFIITSDDFRQLDVTSRAGDSADVNAIQVRPL* 0
>ENC_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? OPN3 encephalopsin
0 MNPANGSRSERSAEQLLFSGDTYRVLAFTIGTIGAFGFCNNFVVLALYCRFKRLRTPTNLLLVNISLSDLLVSLFGINFTFAACVQGRWTWTQATCVWDGFSNSLF 1
2 GIVSIMTLAALAYERYIRVVHAQVVDFPWAWRAIGHIWLYALAWTGAPLLGWNRYTLEIHRLGCSLDWASKDPNDASFILLFLLACFFVPVGIMIYCYGNILYAVQM 0
0 IRSIQDLQTVQIIKILRYEKKVSVMFFLMISCFLLCWTPYAVVSMMVAFGRRSMVSPTMAIIPSFFAKSSTAYNPLIYVFMSRK 0
0 FRHCLLQLLCSRLSWLQRSLKERPLAPVQRPIRPIVMSRPCGKGNRPKKKVTFSSSSIVFIITSDDFGQLDVTSKSGDSADVNAIQVRPL* 0
>ENC_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? OPN3 encephalopsin
0 MNPDNGTREERSTDHSIFAVGTYKLLAFAIGTIGVFGFCNNVVVIVLYCKFKRLRTPTNLLVVNISLSDLLVSVIGINFTFVSCIRGGWTWSRATCIWDGFSNSLF 1
2 GIVSIMTLASLAYERYIRVVHAQVVDFPWAWRAIGHIWLYSLVWTGAPLLGWNRYTLEIHRLGCSLDWASKDPNDASFILLFLLACFFVPVGIMIYCYGNILYAVQM 0
0 LRSIQDLQTVQIIKILRYEKKVAVMFLLMISCFLLCWTPYAVVSMMEAFGRKNMVSPTVAIIPSFFAKSSTAYNPLICVFMSRK 0
0 FRRCLMQLLCSRVTCLQCNLKERPLAPVQRPIRPIVVSAACGGGRVRPKKRVTFSSSSIVFIITRNDIRHTDVTSNTRESSEANVFQVRPL* 0
>ENC_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full G? OPN3 encephalopsin
0 MNPANESRAGRHEERSVFAVGTYKLLTVIIGTIGVFGFCNNLLVILLYCKFKRLRTPTSLLLVNISLSDLLVSVVGINFTLASCVKGRWMWSQATCVWDGFSNSLF 1
2 GIVSIMTLAALAYERYIRVVHAQVVDFPWAWRAIGHIWLYSLAWTGAPLLGWNRYTLEIHQLGCSLDWASKDPNDAAFILLFLLGCFFVPVGIMIYCYGNILYAVRM 0
0 LRSIEDLQTVQIIKILRYEKKVAAMFLLMISCFLVCWTPYAVVSMMEAFGKKSMVSPTVAIVPSFFAKSSTAYNPLIYVFMNRK 0
0 FRRCFLQLLGSRLCSKISWLQCTLKEHPLTPVERPIRPIVASTSCGSRHRPKKRVTFNSSSIVFMITGDEFQQLDVTSKSRNSSEANVFHVRPL* 0
>ENC_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? OPN3 encephalopsin
0 MNPTNSTEPQEEHLFSPNTYKLLAVIIGTIGIVGFCNNILVLLLYYKFKRLRTPTNLLLVNISVSDLLVSVFGLSFTFVSCTQGRWGWDSAACVWDGfSHSLF 1
2 GTVSIVTLTVLAYERYIRVVNAKATNFPWAWRAITYTWFYSLAWSGAPLV
0 yRRCLSQLFCSHLMSLQWSIKDPSSKARNDMPVKPIVLSQKGDRPKKRVTFSSSSIVFIITSDDTQELGSIAGSNATQISIVQVQPL* 0
>ENC_squAca Squalus acanthias (dogfish) EB687868 frag Gt OPN3 encephalopsin
0 MNAANSTDTREESLFSPGTYQVLAVIIGTIGVVGFCNNLLMLVLYCKFKRLRTPTNLFLVNISISDLLLSVFGVIFTFVSCVKGRWVWDSAACVWDGFSNCLF 1
2 GISSIMSLTVLAYERYIRVVNATAIDFSWAWRAITYIWLYSLAWTGAPLIGWNSYTLELHRLGCSVNWDSRNPSDTSFVLFLFLGCLLCPIGVIAYCYG
>ENC_leuEri Leucoraja erinacea (skate) Deut.Chon.Elas FL592692 frag
0 MNTANSTEIPAETLFGAGAYKVLAVVVGTVGAVGFCNNLLMLVLYCKFKRLRSPANLFLVNISISDLLLSVFGIFFTLVSCVKGGWVWDNSACIWDGFSNTLF 1
2 GISSIMSLTVLAYERYLRMVNATAINFTWAWRAVTYIWLYSLAW
>ENC_torCal Torpedo californica (ray) Deut.Chon.Elas EW690209 17309107 frag electric organ
2 WSGAPLLgwNRYTLEMHRLGCSVTwELEKPSDTSFILFLFLGSLLLIPVGVIAYCYGNIFYTIRM 0
0 LQSIEDFQTARFAKTLTNEMNSSKMCFFMISVAFSCWLPYAVTSFMVVYGCTDVITPTITIIFSLLAKSSAISYPIIYIFMSRK 0
0 FRWCLMQLLCFRLVRIKWAIKDRTVRIRSDKPIKPIVIFEKVVNRPKKRVTFSSFSIIFIANDETQDIGSTAELNATNQNVVQVRPLRSERAER* 0
>ENC_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic full Gt OPN3 encephalopsin rough sequence in two contigs
0 MQSPKQDSLHYAGDTGAKAAPDSAQGNASALGSNFLLHGGDLGEGSTAFSAATFRLLAGVVGTIGVAGFLNNLLLVALFVGFKRLQTPTNLLLVNISLSDLLVSVFGNTLTLVSCVRRRWVWGNGGCVWDGFSNSLF 1
2 GIVSISTLTALSYERYARLIKAQVLDFSWAWRAVTYTWLYSAAWTGAPLLGWSRYVLEKHGLGCSIDWASSNPPDAAFVLFFFLGCLAAPLLVMGFCFGRIALAITQ 0
0 FRKLDRLQTPRVLKARCSERKVSAVCLLMMLLFLLCWSPYAVASLFVASGFEHLVSPPVSIVPSLLAKSNAVCNPLLFLLMSGN 0
0 FFRCLRTMFFTLRWRVNLSRVGIVCPQSQRRPRQREAARPKKKATFNSSSIVFIISSSEEDPVEPADAEGEKSDALAAPADTCVVHVKPT* 0
>ENC4_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran AB050608 12435605 full Gt Amphiop4 syn(-ZFYVE1 +RTF1 -CES1 -POMT2) new exon 12 and 34
0 MALYNNTSSPSQDLLWDAPYSQGHIWDNSSASNSSEDVMDQGKVELQDFSDAGYTAIATCLALI 1
2 GFVGFTNNFVVILLIGCHRQLRTPFNLLLLNMSVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANSLF 1
2 GIVSLVTLSALAFERYCVVVRSSDMLTYKSSLVVITFIWLYSLLWTSLPLLGWSSYQFEGHN 0
0 VGCSVNWVQHNPDNVSYIVTLMVTCFFVPMVVVCWSYAWIWRTVRM 0
0 SSEAKPECGNSQNAGRLVTTMVVVMIICFLVCWTPYAVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ 0
0 FREFLLARLQRVCCRQQAVPRVTPMDDNVHVRLGGEGPSQSQQFLPAGENVENVDMLEYVQENCKPKADSLSTISE* 0
>ENC4_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050608 12435605 full G? Amphiop4 introns from braFlo
0 MPLYNTSSGPTQGLPWDTPYSQDPIWNDSSPSNSSEDAVVDQGRGELQDFSDAGYTAIATGLALI 1
2 GLVGSMNNFVVILLIGCHRQLRTPFNLLLLNVSVADLLVSVCGNTLSFASAVQHRWLWGRPGCVWYGFANSLF 1
2 GIVSLVTLSALAFERYCVVVRSSEMLTYKSSLGMIAFIWMYSLLWTSLPLLGWSSYQFEGHS 0
0 VGCSVNWVKHNVNNVSYIITLMVTCFFVPMVVVCWSYACIWRTVRM 0
0 SAEMKSEFGNPQNTGRLVTTMVVVMIVCFLVCWTPYTVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ 0
0 FREFLLARLRTFCCRQPRMLRVTPMDDNAHARLVGEGPSHAQQVIPSEENGENVEMRKVQGNQLKADSLSTISE* 0
>ENC_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu AAGJ02133080 full G? OPN3 encephalopsin terminal exon extended to stop codon
0 MENFTSIVTDGTNEENTDGDAWPGYAHLLAGSFLTLVFIISIIGNSVVLFLFAWDRHLRTPTNMFLLSLTISDWLVTVVGIPFVTASIYAHRWLFAHVGCIS 2
1 YAFIMTFLGLNSLMSHAVIAVDRYLVITKPHF 1
2 GIVVTYPKAFLMISIPWVFSFAWAVFPLAGWGEFTYEGTGAWCSVRWDSDQPQIMSYVLAMMFLTFISSIVIMMYCYICIFLTTRRMPRWATSNSIKTHERNRRRR 2
1 EQKLLKTLIAIAIAFLVAWSPYAITSMIVVFGGSELLSLTATTLPSLFAKSSVMINPIIYAVTSRVFRKSLKK 0
0 MLTSFFPGCMTYIMTDKSPPSSSRPIQLGLCKYHFLY* 0
>TMT5_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002589165 12435605 full G? Amphiop5 extra 0 intron
0 MLGMHNVMNATDYDNNNATFAAWNFQRNGTTEEEVEFSGFDTVAVVIAAIGIAGFLSNGAVVLLFLKFRQLRTPFNMLLLNMSVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANHLF 1
2 GLVSLISLAVISYERYRMVVKPKGPGSSYLTYNKVGLAIIFIYLYCLLWTTLPIVGWSSYQLE 0
0 GPKISCSVAWEEHSLSNTSYIVAIFIMCLLLPLLIIIYSYCRLWYKVKK 0
0 GSQNLPPAIRKSSQKEQKIARMVVVMITCFLVCWLPYGAMALVVSFGGESLISPTAAVVPSLLAKSSTCYNPLVYFAMNNQ 0
0 FRRYFQDLLCCGRRLFDASASVNTCNTSAMPRHSPVFQKPDSDQYNGIQKSREPQMRTTGQNAPYRQWIEMQTIAVVVKADEVNNKFGEVKT* 0
>TMT5_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050609 12435605 full G? Amphiop5 introns from braFlo
0 MLGIYNVVNATEYGNNTTFAAWDFKRNGTGGEEEVEFFGYDAVAGVIAIIGVVGFVSNGAVVVLFLKFPQLRTPFNLLLLNMAVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANHLF 1
2 GLVSLISLAVISFLRYRMVVKPKGPGSSYLTYTKVGLAILFIYLYCLLWTTLPIAGWSSYQLE 0
0 GPKIGCSVAWEEHSWSNTSYIVVLFITCLFAPLLIIVYSYYRLWHKVKQ 0
0 GSRNLPAAMRKSSQKEQKIAMMVIVMITCFMVCWLPYGAMALVVTFGGERLISHTAAVVPSLLAKSSTCYNPVVYFAMNSQ 0
0 FRRYFQDLLCCGRRLFDVSQSVVTGNTAMPRNNSQGFRKDDSDQKQDNGLPKQSEGPMCDHSSNESQMEGSRHNTAASQQWIEMQTIAVVVKAVEVDTSAANEP* 0
>TMT_monDom Monodelphis domestica (opossum) Deut.Meta.Dide XM_001372110 full G? syn(+NCK2 -UXS1 +TMT -ST6 GAL2_overlap) -RALY shortened final exon
0 MSNNLTTNLSLEALLSASEDKQRNGLSRTGHTIVAVFLGIILIFGSISNFIVLVLFCKFKVLRNPVNMLLLNISISDMLVCLSGTTLSFASSIQGRWIGGKHGCRWYGFANSCF 1
2 GIVSLISLAILSYERYRTLTLCPGQGADYQKALLAVAGSWLYSLVWTVPPLIGWSSYGTEGAGTSCSVHWTSKSVESVSYIMCLFIFCLVIPILVMVYFYGRLLYAVKQ 0
0 VGKIRKTAARKREYHVLFMVVTAVICYLICWVPYGMIALLATFGPPGVVSPVANVVPSILAKSSTVCNPIIYVLMNKQ 0
0 FYKCFLILFHCQPAQSGPDVSLCPSNVTVIQLGQRKNKDAPGSI* 0
>TMT_macEug Macropus eugenii (wallaby) Deut.Meta.Dipr genomic full G? teleost multiple tissue
0 MSINLTANLSFGTLLPDSEEKQRSGLSRTGHTVTAVFLGLILILGVINNFIVLVLFCKFKVLRNPVNMLLLNISISDMLVCLTGTTLSFASSIRGRWIAGYHGCRWYGFANSCF 1
2 GIVSLISLAVLSYERYRTLTLCPRQGTDYHKALLAVAGSWLYSLIWTVPPLIGWSSYGTEGAGTSCSVHWTSKSVESVSYIMCLFIFCLVIPILFMVYFYGRLLYTVKQ 0
0 VGKIRKSAARKREYHVLFMVVTAVICYLICWVPYGMIALLATFGPPGVVSPVANVVPSILAKSSTVCNPIIYILMNKQ 0
0 FYKCFLILFHCQPASSASDASLCPSKMTVIQLGQRKDKEVPCAIQDLPEVSKKQLCLLSPESNVAPSSGHPQEKMEEKPLSE* 0
>TMT_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono genomic frag G? teleost multiple tissue
0 GLSRTGHTMVAVFLGIILVFGFMNNLIVLILFCKFKALRNPVNMIMLNISASDMLVCVSGTTLSFASNISGRWIGGDPGCRWYGFVNSCL 1
2 GIVSLISLAVLSYERYRTLTLHPKQSTDYQKAVLAVGASWIYSLIWTIPPLLGWSSYGTEGAGTSCSVHWSSKSPVSVSYIVCLFIFCLVIPVLVMIYCYGRLLYAVKQ 0
0 IGKARKTAARKREYHVLFMVITTVICYLVCWMPYGVTALLATFGQPGTVSPEASVIPSILAKSSTVCNPIIYILMNKQ 0
0 FYKCFLILFHCQPPRAADAPSTYPSQVMVIQLNQRRSRETAGAPQVLLEMKHQTLHLLGPQLHETPSWERSTPVHPE* 0
>TMT_galGal Gallus gallus (chicken) Deut.Saur.Arch XM_001234388 full G? syn(+NCK2 -UXS1 +TMT -ST6 GAL2_overlap +SLC5A7 +SULT1C4)
0 MNHTWTYNLSFGAPTDPVEPRAGLSRNGHTVVAVFLGFILFFGFLNNLIVLILFCKFKTLRNPVNMLLLNISISDMLVCISGTTLSFASNIHGKWIGGEHGCRWYGFVNSCF 1
2 GIVSLISLAVLSYERYSTLTLCNKRSDDYRKALLAVGGSWVYSLLWTVPPLLGWSSYGIEGAGTSCSVRWSSETAESTSYIICLFIFCLVIPVMVMMYCYGRLLYAVKQ 0
0 VGKIHKNTARKREYHVLFMVITTVICYLVCWIPYGVIALLATFGKPGVVTPVASIIPSILAKSSTVCNPIIYILMNKQ 0
0 FYKCFRQLFHCQPPSSTDGEPTCHSKVTVIQLNQKTDGGKLCNNKPRPETDNKVTSLLHPEPGLEPAAKTVPPM* 0
>TMT_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch XM_002192719 full G? teleost multiple tissue
0 MNHTWMYNLSFGAPAHPVEPRAGLSRSGHTVVAVFLGLILFFGFLNNLIVLILFCKFKTLRNPVNMLLLNISVSDMLVCISGTTLSFASNIRGKWIGGDHACRWYGFVNSCF 1
2 GVVSLISLAVLSYERYNTLTLCHKRSDDFRKALLAVAGSWIYSLVWTVPPLLGWSSYGVEGAGTSCSVRWSSESAESTSYIICLFVFCLVVPVMVMMYCYGRLLYAVKQ 0
0 VGKIHKNAARKREYHVLFMVIPTVICYLVCWIPYGVIALLATFGKPGAVTPITSIIPSILAKSSTVCNPIIYILMNKQ 0
0 FYKCFRQLFHCQPPSSTDGEPTCHSKVTVIQLDQRADGGNMCNNEPHPETDSKMTSLLCPETTSKATPPTS* 0
>TMT_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G? teleost multiple tissue syn(+TMT -ST6 GAL2_overlap +SLC5A7)
0 MSELSSNLTFNMSTSIEEPGSGLSRMGHNIVAVFLGLILVFGFLNNLVVLILFCKFKTLRNPVNMLLLNISASDMLVCISGTTLSFVSNIYGRWIGGEHGCRWYGFVNSCF 1
2 GIVSLISLAILSYERYSTLTQTNKRGSDYQKALLGVGGSWLYSLIWTVPPLIGWSSYGLEGAGTSCSVRWTSETLESVTYIICLFIFCLAIPVLVMIYCYARLFYAVKQ 0
0 VGKLRKTSARKREFHVLFMIITTIICYLICWMPYGVIALLATFGRPGLVSPVASVIPSILAKSSTVFNPIIYILMNKQ 0
0 FYKCFLMLLHCQPSSVADGETICQSKVMAIHQNQKAQGGVILKSQVVPQMDEKAICLLSPESSLDPVLESTPQLSKENSFL* 0
>TMT_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur AL773998 full G? syn(-UXS1 +TMT -ST6 GAL2_overlap +SLC5A7) short gastrula frag
0 MSTIKNWTTNISVENSMSYIENDLSLPTEAVLSRTGHTVVAIFLGFILIFGFLNNFVVLILFCKFKTLRTPVNMMLLNISASDMLVCVSGTTLSFTSSIKGKWIGGEYGCQWYGFVNSCF 1
2 GIVSLISLAILSYERYSTLTLYNKGGPNFKKALLAVASSWLYSLVWTVPPLLGWSSYGREGAGTSCSVRWTSESVESVSYIICLFIFCLALPVFVMLYCYGRLLYAVKQ 0
0 VGKIRKIAARKREYHVLFMVITTVICYLLCWLPYGVVALLATFGRPGVISPVASVVPSILAKSSTVFNPIIYILMNKQ 0
0 FYKCFLILFHCHPTSSADGKSICQSNYTVIQLNQKLNNIVAIPGQTQIPESVDKMPCIHRQNNESPSDQMPQSTTEHLISGT* 0
>TMTa_danRer Danio rerio (zebrafish) Deut.Acti.Otoc XM_002192719 full G? syn(-UXS1 +TMT -ST6 GAL2_overlap +GPR89A -pdzk1l)
0 MFFEQADLNYSFNMSEEDRLTLLDEDWSDSPMETLSRAGFIALSVFLGFIMTFGFFNNLVVLVLFCKFKTLRTPVNMLLLNISISDMLVCMFGTTLSFASSVRGRWLLGRHGCMWYGFINSCF 1
2 GIVSLISLVVLSYDRYSTLTVYHKRAPDYRKPLLAVGGSWLYSLIWTVPPLLGWSSYGLEGAGTSCSVSWTQRTAESHAYIICLFVFCLGLPVLVMVYCYGRLLYAVKQ 0
0 VGKIRKTAARKREYHVLFMVITTVVCYLLCWMPYGVVAMMATFGRPGIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0
0 FYRCFRILFCCQRSLLQNGHSSMPSKTTVIQLNRRVNSNAVACTAQISTGTHNHDCSTHVTERSNPPEVIP* 0
>TMTb_danRer Danio rerio (zebrafish) Deut.Acti.Otoc CN506730 full G? teleost multiple tissue syn(-FHL2 +NCK2 +TMT -ST6GAL2_lap) seg dup
0 MFPEETNMSYISNGTDDDLLSALEDWSDTPAEKLSRTGHNVVAVILGSILIFGTLNNLVVLVLFCKFKTLRTPVNMLLLNISVSDMLVCLFGTTLSFAASIRGRWLVGRHGCMWYGFVNSCF 1
2 GIVSLISLAILSYDRYSTLTVYNKRAPDYSKPLLAVGGSWLYSLFWTVPPLLGWSSYGLEGAGTSCSVTWTANTPQSHSYIICLFIFCLGIPVLVMVYCYSRLICAVKQ 0
0 VGRIRKTAARRREYHILFMVITTVVCYLLCWMPYGVVAMMATFGRPGIISPIASVVPSLLAKSSTVINPLIYILMNKQ 0
0 FYRCFLILIHCKHSSLENGQSSMPSRTTGIQLNRRPYSNPVADNAPPSIDLQNDCSTPVSG* 0
>TMT_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? syn(-UXS1 +TMT -ST6 GAL2) multiply frameshifted assembly
0 MFSGQAGLNSSFNLSDGRGLEDAPAGRGRLSPTGFVVLSVVLGFIITFGFLNNFIVLLLFCKFKKLRTPVNVLLLNISVSDMLVCLFGTTLSFASSLRGRWLLGRSGCNWYGFINSCF 1
2 GIVSLISLVILSHDRYSTLTVYNKQGINYRKPLLAVGGTWLYSLLWTVPPLLGWSSYGIEGAGTSCSVSWTVQTAQSHAYIICLFIFCLGLPVLVMVYCYSRLLWAVKQ 0
0 VGKIRKTSARKREYHILFMVVTTAACYLVCWMPYGVVAMMATFGPPNIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0
0 FYKCFLILFHCSHWSADNGTTSVPSKITVIQLNRRAYSNTVACADPLSTDALKQCCSAKNASTIEVKLS* 0
>TMT_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? teleost multiple tissue
0 MFSGQAGLNYSFNLSDDRELLDAPAGRAKLSPTGFVVLSVVLGFIMTFGFLNNFVVLLLFCKFKKLRTPVNMLLLNISVSDMLVCLFGTTLSFASSIRGRWLLGRIGCSWYGFINSCF 1
2 GIVSLISLVILSYDRYSTLTVYNKQGINYRKPLLAVGGTWLYSLFWTVPPLLGWSSYGIEGAGTSCSVSWTVQTAQSHAYIICLFTFCLGIPILVMIYCYSRLLWAVKQ 0
0 VGRIRKTAARKREYHILFMVVTTAACYLVCWMPYGVVAMMATFGPPNIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0
0 FYKCFLILFHCGHWSADNGNTSMPSKTTAIQLNRRVYSNTVACADQLSTDALKQCCSANTISTKNTSTVEGKLS* 0
>TMT_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? teleost multiple tissue
0 MVFGQAGLNHSFNLSDDRELLDTSAGRAKLSPTGFVVLSVMLGFIMTFGFVNNLVVLLLFCKFKKLRTPVNMLLLNISVSDMLVCLFGTTLSFASSLRGKWLLGRSGCSWYGFINSCF 1
2 GIVSLISLVILSYDRYSTLTVYNKAGPDYRKPLLAIGGSWLYSLFWTVPPLLGWSSYGIEGAGTSCSVSWTVQTAQSHAYIICLFTFCLGLPMLVMIYCYSRLLLAVKQ 0
0 VGRIRKTAARRREYHILFMVLTTAACYMLCWMPYGVVAMMATFGPPNIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0
0 FYRCFLILFHCKHWSAENHNTSMPSKTTVIHLNRRVCSNTLPCTAQASTDAANHFCSTSATKHTSPPLQGHGLSLNVLNMIRQENHSHDEAAKNQLDCLT* 0
>TMT_oryLat Oryzias latipes (medaka) Deut.Acti.Perc DK170580 full G? teleost multiple tissue whole embryo
0 MFSGQTGLNFSFNQSDDRELEDTPAGSAKLSQAGFVVLSVVLGFIMTFGFLNNFVVLILFCKFKKLRTPVNMLLLNISVSDMLVCLFGTTLSFASSIRGRWLLGRGGCSWYGFINSCF 1
2 GIVSLISLVILSYDRYSTLTVYNKGGLNYRKPLLAVGGSWLYSLFWTVPPLLGWSSYGLEGAGTSCSVSWTANTAQSHAYIICLFIFCLGLPILVMIYCYSRLLLAVKQ 0
0 VGKIRKTAARKREYHILFMVLTTAACYLLCWMPYGVVAMMATFGPPNIISPVASVVPSLLAKSSTVINPLIYILMNKQ 0
0 FYRCFLILFHCDHWSSENGNTSVPSKTTVIPLNRRIYTNTVAQISTDNAN* 0
>TMT_ictPun Ictalurus punctatus (catfish) Deut.Acti.Otoc FD287269 frag G? teleost multiple tissue 2 transcripts from whole fry
0 WLLGRTGCMWYGFINSCF 1
2 GIVSLISLMILSYERYSTMTVYNNQGPNYRKHLLAVGGSWLYSLIWTVPPLLGWSSYGLEGAGTSCSVSWTDHSPKSHAYIICLFIFCLGLPVLLMVYSYGRLLYAVKQ 0
0 LGKIHKTARRRDYHLLFMITTTVVCYLLCWTPYSVVALMASFGRPGIITPVASIIPSLLAKSSTVINPVIYIFMNKQ 0
0 FYRCFRTLLGYKERSAVPDDHSLMATKNTAIQLKCIMHNNPVPSPAHTPPPFF
>TMTc_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? ciliary uropsin syn(+GPR112 +BRS3 +VGLL1 +TMTx +CD40LG -ARGHGEF7)
0 MLLALNLQNESFLLNLSHGETRDSLPPATGLSRTAHSVVAVCLGCILVLGSLYNSFVLLIFVKFTAIRTPINMILLNISVSDLLVCIFGTPFSFVSSVSGGWLLGQQGCKWYGFCNSLF 1
2 GLVSMISLSMLSYERYLTVLKCTKADMTDYKKSWLCIIVSWLYSLCWTLPPLIGWSSYGLESSGTTCSVVWHSKSSNNISYIVCLFLFCLVLPLFIMIFCYGHIVRVIRG 0
0 VCRINMTTAQKREHRLLFMVVCMVTCYLLCWMPYGLVSLMTAFGKPGMITPTVSIIPSILAKSSTFINPLIYIFMNKQ 0
0 FYRCFIALIKCESGPHHNENISTSRQSREMRLETRCKVQSSIFYTHKARRTKDTTSSPEMQRKLTLTVHYRDDR* 0
>TMTc_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G? ciliary uropsin syn(+FHL1 +GPR64/112 +TMTx -ARGHGEF7 -RBMX)
0 MVVYIWSLNISSKDTSALNQSGNVSSGDPLEPHDSPPGLSRTGHTVTAVCLGAILLLGCLNNLFVLLVFARFRTLWTPINLILLNISVSDILVCLFGTPFSFASSLYGKWLLGHHGCKWYGFANSLF
2 GIVSLMSLSILSYERYAALLRATKADVSDFRRAWLCVAGSWLYSLLWTLPPFLGWSNYGPEGPGTTCSVQWHLRSTSSISYVMCLFIFCLLLPLVLMIFCYGKILLLIKG 0
0 VTKINLLTAQRRENHILLMVVTMVSCYLLCWMPYGVVALLATFGRTGLITPVTSIVPSVLAKSSTVVNPVIYVLFNNQ 0
0 FYRCFVAFLKCQGEPSVHGQNPQHSSKEDPHVFRPCDGASIHRSAEGPQKKEQHSLSLVVHYTP* 0
>TMTc_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? ciliary uropsin syn(-TAF7L +SLC9A7 +FHL1 +TMTx -ARGHGEF7 -RBMX)
0 MVVQPRDCNFSTPDSSVSIFGPSIDARGPWDPQCPGGLSRSSHTAVAVLLGVILVAGILSNSLVLLLFVKYRSLWTPINLILLNINLSDILVCVFGTPFSFAASLQGRWLIGEGGCMWYGFANSLF 1
2 GIVSLVSLSVLSYERCTVVLQPSQVDVSDFRKARFCVGGSWLYALLWTSPPLLGWSSYGPEGAGTTCSVQWQLRSPASVSYVLCLLVFCLLLPFLVMVYSYGRILVAIRR 0
0 VGRINQLTAQRREQHILLMVLSMVSCYMLCWMPYGIMALVATFGKLGLVTPMVSVVPSILAKFSTVVNPIIYMFFNNQ 0
0 FYRCFMAFIRCQKEPEPIQEE* 0
>TMTc_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? ciliary uropsin syn(-TAF7L +SLC9A7 +FHL1 +TMTx -ARGHGEF7 -RBMX)
0 MVLQTRECNFSTPDTSVRGPWAPQGPGGMSRTGHTVVAVMLGTILLAGVFGNSVVFLVFVKYRSLRTPINLILLNISLSDILVCVFGTPLSFAASLKGRWLLGERGCEWYGFANSLF 1
2 GIVSLVSLSVLSYERYTVVLQPTQVDVSYFRKAWFCVGGSWLYALFWTLPPLLGWSRYGPEGPGTMCSVQWHLRSPANISYVLCLFIFCLLLPLVVMVYSYGRIWVAVRR 0
0 AGRINLLTAQRREQHILWMVLSMVSCYMLCWMPYGIIALVATLGRLGPISPAVSVVPSILAKFSTVVNPVIYMFFNNQ 0
0 FYRCFMAFVRCQKEPESIQGE* 0
>TMTc_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full G? ciliary uropsin syn(+SLC9A7 +FHL1 +GPR56 +TMTx -ARGHGEF7 -RBMX)
0 MVLHSHNCNFSTKDSSPCSTNSTEPHNPDGLSRTGHTAVAVCLGFILVAGILNNFLTLLVFAKFRSLWTPINLILLNISLSDILVCVLGTPFSFAASVRGRWLIGESGCKWYAFANSLF 1
2 GIVSLVSLSVLSYERYITVLHSSQADLSNFRKAWFCVGGSWLYSLLWTLPPFLGWSSYGPEGPGTTCSIQWHLRSPTSVSYVLCLFIFCLVLPLVLMVYSYGRILVALRR 0
0 VGKINLLAAQRREQRILVMVFSMVSCYILCWMPYGIVALMATFGRKGLVTPLTSVIPSILAKFSTVVNPVIYVFFNSQ 0
0 FYRCLVAFVRCSGDPEEHPTPRTQLSDVVRVQKQNSVSSSSPRLLSDPTDRMLCNHHPDRHVLFVHYTP* 0
>TMTa_oncMyk Oncorhynchus mykiss (trout) Deut.Acti.Salm CU062745 full G? teleost multiple tissue testis new
0 MVVESGNLNFSTDDSSGTNLSPKDSVRDTLTSRQSDLGRTGHTVVAVFLGVIFLLGFLSNLFVLLVFARFQVLRTPINLILLNISVSDMLVCIFGTPFSFAASLYGRWLIGAHGCKWYGFANSLF 1
2 GIVSLVSLAILSYERYSTILCYTKADPSDYKKAWLAIAGAWLYSLVWTVPPFFGWSSYGPEGPGTTCSVQWHQRSSGNISYVTCLFIFCLLLPLLLMMFCYGKILFAIRG 0
0 VAKINQSSAQRRETHVLVMVVSMVSCYLLCWMPYGVVALLATFGQVGLVSPTTSIVPSILAKSSTFLNPVIYGLLNNQ 0
0 FYRCFLAFMSCGSEAAGSHTLHTLPSSRVVGPYGNSPAPEEPGTREPLDGSGTSSKTQTRGPQREKRDLVLVVHYTP* 0
>TMTa_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G? teleost multiple tissue
0 MTVPSISPSCIGVANGVWCSNGGSSSNSHHRHGQEQSLSPTGHLITAICLGVIGSLGFLNNLLVLVLFCRNKVLRSPINLLLMNISLSDLMICIVGTPFSFAASTQGKWLIGPAGCVWYGFANTFF 1
2 GTVSLISLAVLSYERYCTMMGTTEADATNYKKVWMGIFLSWIYSLFWSLPPLFGWSSYGPEGPGTTCSVNWHSRDANNISYIICLFIFCLVIPFIVIVYCYGKLLCAIKK 0
0 VSGVTQGMAQTREQRVLIMVVVMIICFLLCWLPYGIVALIATFGKPGLITPSASIIPSVLAKSSTVYNPVIYIFLNKQ 0
0 FYRCFCALLKCGKKSIASSNKCSSRSTRVCRSIRKQDNFTFVAASAGPPSSEHQDAILSIQNPEPPTDNSPKAKQRVLLVAHYSV* 0
>TMTa_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? teleost multiple tissue scaffold_55
0 MAFHSSSPSCSDSSSVTCRSSIQQYGPENHPNLSPTGHLLVAVFLGVIGSLGFFNNLVVLILFCQYKVLRSPINMLLMNISLSDLMVCILGTPFSFAASTQGHWLIGEIGCIWYGFVNTLF 1
2 GTVSLVSLAVLSYERYCTMLRSTEADLTNYKKAWLGILVSWIYSLVWTLPPLFGWSKYGPEGPGTTCSVNWHSRDANNISYIVCLFIFCLALPFAVIVYCYGRLLFAIKQ 0
0 VSGVSKSSSRAREQRVLIMVIVMVVCFLLCWLPYGVMALVATFGKPGIISPSASIIPSVLAKSSTVYNPIIYIFLNKQ 0
0 FYRCFTALIHCNKHPQVSSNKGSSKTTKIMLTARKLPDANFTVNAASNPPSSSVVKYEADSKTHNGDTKPFKTLVANYVI* 0
>TMTa1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc AF349947 12670711 full G? teleost multiple tissue syn(+PBX3 +TNK2 +TMTa1 -PAP2D +LPPR4)
0 MIVSNLSVLSCRRNSALCLGAVEGHLEASSSYRTLSPTGHILVAVSLGFIGTFGFLNNLLVLVLFGRYKVLRSPINFLLVNICLSDLLVCVLGTPFSFAASTQGRWLIGDTGCVWYGFANSLL 1
2 GIVSLISLAVLSYERYCTMMGSTEADATNYKKVIGGVLMSWIYSLIWTLPPLFGWSRYGPEGPGTTCSVDWTTKTANNISYIICLFIFCLIVPFLVIIFCYGKLLHAIKQ 0
0 VSSVNTSVSRKREHRVLLMVITMVVFYLLCWLPYGIMALLATFGAPGLVTAEASIVPSILAKSSTVINPVIYIFMNKQ 0
0 FYRCFRALLNCDKPQRGSSLKSSSKTKPFRPGRRTDNFTFMVASVGPNQTNPVEDGPPSADNTKPAVLSLVAHYNG* 0
>TMTa_takRub Takifugu rubripes (fugu) Deut.Acti.Perc AF402774 12670711 full G? teleost multiple tissue syn(-CALD1 +TNK2 -RAB18 +ABI1 12670711)
0 MIVSNVSLSGCAGVNGAVCAAEGHQAGGSDRSTLTPTGNLVVSVFLGFIGTFGLVNNLLVLVLFCRYKMLRSPINLLLMNISISDLLVCVLGTPFSFAASTQGRWLIGEAGCVWYGFANSLF 1
2 GVVSLISLAVLSFERYSTMMTPTEADPSNYCKVCLGITLSWVYSLVWTVPPLFGWSSYGPEGPGTTCSVNWTAKTTNSISYIICLFVFCLIVPFLVIVFCYGKLLCAIRQ 0
0 VSGINASTSRKREQRVLCMVVIMVICYLLCWLPYGVVALLATFGPPDLVTPEASIIPSVLAKSSTVINPIIYVFMNKQ 0
0 FYRCFLALLCCQDPRSGSSMKSSSKVATKAKGVTPTGQRRTDFLYMVASLGRPAATIPQLGPSFDATNDFTKPPSSDTIKPVVVSLAAHCDG* 0
>TMTa_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? teleost multiple tissue
0 MIASNASVSGCAGVHGAACAADAPPAGGSHRSSSSLTPTGNLVVSVFLGLIGTSGLVSNLLVLVLFCRFKVLRSPINLLLVNISVSDLLVCVLGTPFSFAASTQGRWLIGAAGCVWYGFVNSLF 1
2 GIVSLISLAVLSFERYSTMMTPTEADSSNYCKVCLGIGLSWVYSLLWTVPPLLGWSSYGPEGPGTTCSVNWTAKTANSVSYIICLFVFCLILPFLVIVFCYGKLLCAIRQ 0
0 VSGVNASMSRRREQRVLFMVVVMVICYLLCWLPYGVVALLATFGPPGLVTPAASIIPSILAKSSTVINPVIYVFMNKQ 0
0 FSRCFLSLLCCEDPRSSTSLRSSSRVTTKAVRGGTLTGQRRTNHLLYMVAALGRPVATAMPQLGPSFDATYDITKAPSSDNHQPVVVSLEAHG* 0
>TMTa_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? teleost multiple tissue syn(+TNK2 +ENC)
0 MIVSNLSLSGCAGVSSALCAAAGEGHLSGGSHRNTLTPTGHLVVAVCLGFIGTLGLMNNLLVLVLFCRYKMLRSPINLLLINISISDLLVCVLGTPFSFAASTQGRWLIGEGGCVWYGFANSLF 1
2 GIVSLISLAVLSYERYSTMVAPTEADSSNYHKISLGITLSWVYSLIWTAPPLFGWSHYGPEGPGTTCSVDWTARTANSISYIICLFVFCLIVPFLVIVFCYGKLLCAIRQ 0
0 VSGINASLSRKREQRVLFMVVIMVVCYLLCWLPYGIMALMATFGPPGLITPVASIIPSVLAKTSTVINPVIYVFMNKQ 0
0 FYRCFKALLRCEAPRPSSSLKSSSKVPTKAMRGAAVTGPRHTNNFLFVVASLGRPVATIPQLGPSVEPTIDVTGGPSSDNNKPVIVSLVAQCDG* 0
>TMTa_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic frag G? teleost multiple tissue syn(SLC12A3) two contigs frag
0 MLVSNVSLGGCAEFNSALCAGAGEEHLGGGSYRTTLTPTGHLIVAVCLGFIGTFGLVNNLLVLVLFCRYKILRSPINLLLINISISDLLVCVLGTPFSFAASTQGRWLIGEGGCVWYGFANSLC 1
2 GIVSLISLAVLSYERYSTMMTPAEADSSNYRKISLGIILSWGYSLLWTLPPLFGWSHYGPEGPGTTCSVDWTAKTANNISYIICLFVFCLIVPFMVIVFCYGKLLYAIKQ 0
0 VSGINVSVSRKREQRVLFMVVIMVICYLLCWLPYGIMALLATFGPPDLVTPEASIIPSVLAKTSTAINPVIYVFMNKQ 0
>TMTa_pimPro Pimephales promelas (minnow) Deut.Acti.Otoc DT200813 frag G? teleost multiple tissue
0 GHLVVAVCLGFIGTfGFLNNTLVLILFCRYKVLRSPMNYLLVSIAVSDLLVCVLGTPFSFAASTQGRWLIGRAGCVWYGFINSCL 1
2 GVVSLISLAVLSYERYCTMMGATQADSTNYKKVAMGIAFSWIYSMVWTLPPLFGWSCYGPEGPGTTCSVNWAARTANNVSYIICLFFFCLILPFIVIVYSYGRLLQAITQ 0
0 VSRINTVVSRKREQRVLFMVITMVVCYLLCWLPYGIMALLAAFGRPGLVTPAASIVPSVLAKTSTVINPIIYIFMNKQ 0
0 FCRCFHALIMCTTPQRGSSFKNSSKVTKTLRTVRRANGQNVTFAVASAGHPTICAPH
>TMTc_danRer Danio rerio (zebrafish) Deut.Acti.Otoc CO927631 full G? teleost multiple tissue syn(+TNK1 +TMTb -MYEOV2)
0 MIESNVSRSCEWCAGGGEGTGAHLDENHSDHSLSPTGHLVVAVCLGFIGTFGFLNNTLVLVLFCRYKVLRSPMNCLLISISVSDLLVCVLGTPFSFAASTQGRWLIGRAGCVWYGFINSFL 1
2 GVVSLISLAVLSYERYCTMMGSTQADSTNYRKVVIGIAFSWIYSMVWTLPPLFGWSCYGPEGPGTTCSVNWAARTPNNVSYIVCLFVFCLILPFIVIVYSYGRLLQAITQ 0
0 VSRINTVVSRKREQRVLFMVVTMVVCYLLCWLPYGIMALLATFGHPGLVTPAASIVPSLLAKSSTVINPIIYIFMNKQ
0 FCRCFHALIMCTTPERGSSFKNSSKVTKTLRTVRRANGQNVTFAVASAVHRTPYSDRQKSSSEGEKLPPATGQGTSKPVVSLVAYYNG* 0
>TMTb_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full G? teleost multiple tissue syn(+TFRC +TMTb +CHES1 -MYEOV2 -ARHGAP21)
0 MIVCNVSLSCAHCPGEGTAANDAYAQASGSLATPTLSQRGHLVVAVCLGFIGTVGFLSNFLVLALFCRYRALRTPMNLMLVSISASDLLVSVLGTPFSFAASTQGRWLIGRAGCVWYGFVNACL 1
2 GIVSLISLAVLSYERYCTMVSSTIASNRDYRPVLGGICFSWFYSLAWTVPPLLGWSRYGPEGPGTTCSVDWRTQTPNNISYIVCLFTFCLLLPFFVILYSYGKLLHTIRQ 0
0 VRRVSSTVTRRREHRVLVMVVAMVVCYLICWLPYGVTALLATFGPPNLLTPEATITPSLLAKFSTVINPFIYIFMNKQ 0
0 FYRCFRAFLNCSTPKRDSTVRTFTRISLRALRQDQQQKGSALAPSSARPTPNSIHESSLKGSHSTPSNGGAAAAKSPAANRSKPKLILVAHYRE* 0
>TMTb_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? teleost multiple tissue
0 MIVCNLSLSCAHCPGGGAAATDAYAYAEAPGSLAPPTLSQRGHLVVAVCLGAIGTVGFLSNLLVLALFCRFRALRTPMNLMLVSISASDLLVSVLGTPFSFAASTQGRWLLGRAGCVWYGFVNACL 1
2 GIVSLISLAVLSYERYCTMMASTMASNRDYRPVLLGICFSWFYSLAWTVPPLLGWSRYGPEGPGTTCSVDWRTQTPNNISYIVCLFAFCLLLPFCVILYSYGKLLHTIRQ 0
0 VSSVSSAVTRRREHRVLVMVVAMVVCYLICWLPYGVTALLATFGPPNLLTPEATITPSLLAKFSTVINPFIYIFMNKQ 0
0 FYRCFRAFLSCSSPERGSTVRTFTRISLRAVCQRKQQRVSAPAASSACPTPNSIHHSSRKGSHSASSNSGTAAAAKTPAANSSKPKLILVVHYRE* 0
>TMTb_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? teleost multiple tissue
0 MIVCNVSLSCVHCPGGGAGGTAATATGAYEEVSDSLPAPSLSPKGHLVVAVCLGFIGTFGFLSNFLVLALFCRYRALRTPMNLLLVSISASDLLVSMVGTPFSFAASTQGRWLIGRAGCVWYGFVNACL 1
2 GIVSLISLAVLSFERYSTMVKPTVADGRDFRPALGGIAFSWLYSVAWTVPPLLGWSEYGPEGPGTTCSVDWKTQTANNISYIVCLFVFCLVLPFCVILYSYSRLLQAIRQ 0
0 VSVVSSVVTRHREQRVLAMVVVMVACYLVCWLPYGVAALLATFGPRDLLSPEASITPSLLAKFSTVVNPFIYIFMNKQ 0
0 FYRCFRAFLSCSTPERGSTLKTFSRPTKTLRAGRHEKGRRVSAAAPSTAQPTRNSAPRSSQGANHASATPPPSPADGRCAAAGAAKPKRTLVAHYRE* 0
>TMTb_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full G? teleost multiple tissue
0 MIVPNASLSCAHCDGDAAEQDAPGSAAAPSLSPTGHLVVAVCLGLIGTCGFLSNLLVLALFCRYRALRTPMNLLLVSISVSDLLVSVLGTPFSFAASTQGRWLIGRAGCVWYGFINACL 1
2 GIVSLISLAVLSYERYSTVMTPNMADGRDFRPALGGICFSWLYSVAWTVPPLLGWSRYGPEGPGTTCSVDWKTQTPNNISYIICLFTFCLLLPFGVIVYSYGKMLRVIRQ 0
0 VRSMSSVVTRRREQRVLVMVVTMVVCYLVCWLPYGIAALLATFGPRDLLTPAASITPSLLAKFSTVINPLIYIFMNKQ 0
0 FYRCFWAFFCCSTPEQVSTLRTFSRVTKTIRTFRQERELHVSAPAPSSGLPTPNSIQKGNNHVDPSSINQACAASDSPDSRKPKVVLVAHYQE* 0
>TMTa1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? teleost multiple tissue exons 1,2,3
0 MLNSSPNSSPSLPLSQVGWTGLSRTGLTVVAVCLGIIMVLGFLNNLLVLVLFCKYKVLRSPMNMLLLNISVSDMLVCICGTPFSFAASVQGRWLVGEQGCKWYGFANSLF 1
2 GIVSLMSLTILSYDRYITITGTTEADITNYNKTIVGIALSWIYSLMWTLPPLFGWSNYGPEGPGTTCSVNWQSKEVSSKSYIICLFIFCLLMPFLVIVYCYGKLVLAVRK 0
0 VSANNSMGRTRENKLLIMVTFMIICFLLCWLPYGIVALLATFGSPGLITPTASIIPSVLAKTSTVYNPIIYIFMNKQ 0
>TMTa2_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? teleost multiple tissue exons 1 and 3
0 MMAHSANISTTLNASDHAPNLAGLSQSGHTTVAVFLGIILVLGCVNNLLVLLLFVCFKEIRTPLNMILLNISLSDLSVCVFGTPFSFAASIYRRWLIGHKGCKWYGFANSLF 1
0 VGKINQMTAQTREHRILLMVISMVTFYLLCWLPYGTVALIGTFGNADLITPTCSVIPSILAKSSTVINPVIYVIMNKQ 0
>TMTx_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002207814 frag G? assembly duplication no N-term even in v2
0 VAAILALIGVLGIVNNSTTLYLVGRYKQLRTPFNILMVNLSVSDLLMCVLGTPFSFVSSLHGRWMFGHSGCEWYGFICNFL 1
2 GIVSLITLTVISYERYLLMKRLPNERILSYRAVALAVVFIWCYSLLWTAPPLVGWSSYGPE 0
0 GYGISCSVNWESRTANDTSYIVAYFVGCLVFPVAIIVISYTRLILYMRQ 0
0 APSAPMQMLVRREKRVTKMVVVMIMGFTICWTPYTIVALIVTCGGEGIITPAAATVPALFAKSSVVYNAAIYVAMNNQ 0
0 FRKCFLRSLNCRSQPRDPSSQQYTLKTNQVGMSTSGSQAARTADRIKTVHVATANPQDHRSSSGQAVEDNGGFRKSLTHSLPLNSISTLLEAEK* 0
>TMTy_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran FE572481 full G? teleost multiple tissue gastrula XM_002222645 flawed allele dup new
0 MASAGQNVTFPAIDTMAPTPEALTSDPTTPAYFTTEQHLLMAVWLGFIGSFGFVTNLLTVLVFWCFKSLRTPFHLYLGGIALSDLLVAALGSPFAVASAVGERWLFGRAVCVWYAFVNYFL 1
2 SIVSIVTMATMSFSRYWVIIRPQSAPRLDTVYGACVVNALAWCYSFFWTIMPVLGWSRFTQ 0
0 VAAMTVCSLDWDHHTPLSKSYIPVAFLTCLFLPLGVIIFSVFKTTMHLRR 0
0 AAEVEDEVPNEVRAGRKTTRITLVMAGCWLVAWLPYACMALVIAAGGRVSPTVEVLATKFAKTSYIVNTIIYLVMEKE 0
0 FRKSLVLLLFCGRDPFDIQIEQPAYEKADVYVERLVTAEPMVEMEAVNVRPAQQEPARAPFGTPL* 0
>TMT1_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XM_001177470 16311335 full G? teleost multiple tissue PIN-type introns no cdna no sacKow
0 MSNLMTGLVTNVNALSGIGNETPTTIGLSSLVVPVSRTTYNYLTVYTGFLTIFGILNNGIVMILFARFPSLRHPINSFLFNVSLSDLIISCLASPFTFASNFAGRWLFGDLGCTLYAFLVFVA 1
2 GTEQIVILAALSIQRCMLVVRPFTAQKMTHRWALFFISLTWIYSLIICVPPLFGWNRYTYEGPGT 1
2 ACSVAWNSPSPGDTSYIIFIFVLVLVIPFGIIIFCYGLLVYAVKK 0
0 ISRTQAALSSEAKADRKVSKMIFIMILFFLIAWTPYTGFSLYVTFGKNVVITPLAGTFPPFFAKLCTIHNPIIYFLLNKQ 0
0 FKDALIQLFCCGENPFDRDESEHEGRGGRHRHRTAPSATAHIGGRGRASSLPTATSMLDIPQAASTAASSSGKTQNKESLEKGPSTSETTNKRVFELSSKIQKFEISEKNNTPSSSELPGASSLSGALMPPRRAMKNQVGCLPPVDN* 0
>TMT1_plaDum Platynereis dumerilii (ragworm) Ecdy.Anne.Poly CT030681 16311335 full G? 5 exons
0 MDDLGFLGNSSVNYTVPLLQEDPLLLRILYFGPTSYVITAIYLCIVGVIGTLSNGVIMYLYFKDKSLRSPMNLLFVNLAMSDFTVAFFGAMFQFGLTCTRKYMSPGMALCDFYGFITFLG 1
2 GLASEMNLFIISVERYLAVVRPFDVGNLTNRRVIAGG 1
2 VFVWLYSLVFAGGPLVGWSSYRPEGLGTWCSISWQDRSMNTMSYVTAVFLGCYFFPVSIIIFCYFNVWRKVKE 0
0 AADAQGGAGTAGKAEKSIFRMSVIMVTCYLTAWTPYAIVCLIASYGPPNGLPIYAEVLPSLFAKSSQVYNPIIYVLMNKP 0
0 YRSALVSLVCRGRNPFDEAGGTAGGTTAKDETLGKGNKVAAA* 0
>TMT2_plaDum Platynereis dumerilii (ragworm) Ecdy.Anne.Poly AY692353 15514158 full G? new genomic
0 MDGENLTIPNPVTELMDTPINSTYFQNLNAETDGGNHYIYNAFTATDYNICAAYLFFIACLGVSLNVLVLVLFIKDRKLRSPNNFLYVSLALGDLLVAVFGTAFKFIITARKTLLREEDGFCKWYGFITYLG 1
2 GLAALMTLSVIAFVRCLAVLRLGSFTGLTTRMGVAAMA 1
2 FIWIYSLAFTLAPLLGWNHYIPEGLATWCSIDWLSDETSDKSYVFAIFIFCFLVPVLIIVVSYGLIYDKVRK 0
0 VAKTGGSVAKAEREVLRMTLLMVSLFMLAWSPYAVICMLASFGPKDLLHPVATVIPAMFAKSSTMYNPLIYVFMNKQ 0
0 FRRSLKVLLGMGVEDLNSESERATGGTATNQVAAT* 0
>TMT_terTra Terebratalia transversa (lampshell) Loph.Brac.Rhyn HQ679623 full 21362157
MSTASSGTVTTVMESNITTLSPFTTRAPMFSDGANVIIVLVLFVTAILGFFMNGSVLIIFYRKKELRSPTNMFIISMAFNDLMMSILGNPFAAVSNLHHR
WIFGDAACVWYGFIMFFLGLNGIYHLTAIAFDRYIVIAKPLLSSKITTRVAALAISVCWFGAFLWSVFPVFGWSSYVEEGEHSSCSINWISQSIVDSSYI
ISIFIFCFFLPILLVIFSYFHVYMTVRTIAKSNTFGKDSKITKKNEAVERKMAKMVIIMITVFFGSWSPYAIVSMFVAFGDGSSLPAAAIAAPAIIAKSA
MIWNPIIYVLMNVQFRSGFAEMYPCFAVCANTDGGKVKVERPPTVQDNIGDDNTEMSDVRSAVPPAPTSNILKVAPAPDTTKADVHTSETGEQSIAMSIA
RMSVRHKVESRTPVEQMNAVEV*
>TMT1_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_312503 full G? GPROP11 pteropsin GPROP11 head-to-head tandem GPROP12
0 MYDVTDAAAINSDHQELMAPWAYNGAAVTLFFIGFFGFFLNIFVIALMYKDVQ 0
0 LWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWLYGKSICVAYGFFMSLL 1
2 GIASITTLTVLSYERFCLISRPFAAQNRSKQGACLAVLFIWSYSFALTSPPLFGWGAYVNEAANIS 2
1 CSVNWESQTANATSYIIFLFIFGLILPLAVIIYSYINIVLEMRK 0
0 NSARVGRVNRAERRVTSMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFWRIRRSNGVAGQPDSNNTNNSNRDKESARHTAKEGL
ECSLDFCHWTVRGTRVSISSAERNVPAPAARERSGGHSVTGSREESRDRHVTLKTMLSVGPRSPSSVAPVAADCSTTDVPTSGDGSVRIVRQDSELSVIHDGGGGGGGSSSRVLVIKSQKPRSNML* 0
>TMT2_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_312502 full G? GPROP12 pteropsin encephalopsin GPROP12
0 MNDAPNDVAASAVDYEDLMAPWAYNASAVTLFFIGFFGFFLNLFVIALMCKDMQ 0
0 LWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWIFGRTLCVAYGFFMSLL 1
2 GITSITTLTVLSYERYCLISRPFSSRNLTRRGAFLAIFFIWGYSFALTSPPLFGWGAYVQEAANIS 2
1 CSVNWESQTKNATTYIIFLFVFGLVVPLIVIVYSYTNIIVNMRE 0
0 NSARVGRINRAEQRVTSMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFSRVRNKGQQAAADQNTTTMQRELTKSSRDMVECSF
DFCRKKSRFKISLVKPTAPLAVVDVSSTSHRDKGTSRSPLDQTVLNETNEDVGRERSGGGGGGGAYAGTRFVRPDFELSVINSGKSILIKSKNFRSNLL* 0
>TMT_aedAeg Aedes aegypti (mosquito) Ecdy.Inse.Dipt XM_001650752 full G? pteropsin genome frameshifted
0 MESWAYVASAVTLFFIGFFGFFLNLFVIALMCKDVQ 0
0 LWTPINIILFNLVCSDFSVSIIGNPFTLTSAISRHWIFGRTVCIAYGFFMSLL 1
2 GITSITTLTVLSYERFCLISHPFSSRSLSRRGAVFAILFIWSYSFALTSPPLFGWGAYVNEAANIS 2
1 CSVNWESQTLNATSYIIFLFVFGLVVPLVVIVYSYTNIVVNMKR 0
0 NAARVGRINRAEKRVTRMVFVMVLAFMIAWTPYAVFALIEQFGPTDIISPALGVLPALIAKSSICYNPIIYVGMNTQFRAAFNRVRNNESVDNNTITNQKDITMNTSK
EIVECSFDFCRKKRLKIKLQSNAKSKNNNNNSRNQSIADPSSTSNGDDLDQPSPAQTVLNSTVANSGSVASFGPKKRLRSDFELSVISSGKSILIKSNTFRSNLV* 0
>TMT_culPip Culex pipiens (mosquito) Ecdy.Inse.Dipt genomic full G? pteropsin
0 MPPWAYVATAVVLFFIGFFGFFLNLFVIALMCKEVQ 0
0 LWTPMNIILLNLVCSDFSVSIVGNPFTLSSAISHRWLFGRKLCVAYGFFMSLL 1
2 GITSITTLTVLSYERFYLISRPFSSRSLSRRGALGAVLLIWCYSFALTSPPLFGWGAYVNEAANIS 2
1 CSVNWETQTLNATTYIIYLFVFGLVVPLTVIVYSYTNIIVNMKK 0
0 NAARVGRINRAEKRVTTMVAVMVIAFMVAWTPYSVFALMEQFGPPDVIGPGLAVLPALIAKSSICYNPIIYVGMNTQFRAAFNRVRHDPGDMANTTTNQKELT
RSSRDVASVDCSFDFCRKKNRLKMKLHNSIHRSAAIKAGRSQSPHSESERSSSGATQERSTRVQTMLNATVDSRSISGSTGKLMKSDFELSVINSGKSILIKSNTFRSNLV* 0
>TMT_triCas Tribolium castaneum (flour_beetle) Ecdy.Inse.Cole genomic full G? pteropsin
0 MKNFNSTEIGDELLIPVEGYIAAAVVLFCIGFFGFSLNLTVIIFMLKERQ 0
0 LWSPLNIILFNLVVSDFLVSVLGNPWTFFSAINYGWIFGETGCTIYGFIMSLL 1
2 SITSITTLTVLAFERYLLIARPFRNNALNFHSAALSVFSIWLYSLSLTIPPLIGWGEYVHEAANLS 2
1 CSVNWEEKSPNSTSYILYLFAFGLFLPLVIITFSYVNIILTMRR 0
0 NAAFRVGQVSKAENKVAYMIFIMIIAFLTAWSPYAIMALIVQFGDAALVTPGMAVIPALLAKSSICYNPVIYIGLNAQVKGAKWVSGLIYLFQFQQAWMQKWKKNRR
GSDALGTSRVMLETIHQACRDEKTDKLLEKKTKFCKDFETDVSML* 0
>TMT_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme NM_001039968 16291092 full G? AmLop2 pteropsin ciliary compound eye not ocelli clock
0 MSLNRSTMEHVIYEDQVSPVMYIGAAIALGFIGFFGFTANLLVAIVIVKDAQILWTPVNVILFNLV 0
0 FGDFLVSIFGNPVAMVSAATGGWYWGYKMCLW 2
1 YAWFMSTLGFASIGNLTVMAVERWLLVARPMQALSIR 2
1 HAVILASFVWIYALSLSLPPLFGWGSYGPEAGNVSCSVSWEVHDPVTNSDTYIGFLFVLGLIVPVFTIVSSYAAIVLTLKKVRKRA 1
2 GASGRREAKITKMVALMITAFLLAWSPYAALAIAAQYFN 0
0 AKPSATVAVLPALLAKSSICYNPIIYAGLNNQFSRFLKKIFDARGSRTAVPDSQHTALTALNRQEQRK* 0
>TMT_bomTer Bombus terrestris (bumblebee) Ecdy.Inse.Hyme 454SRA full G? pteropsin presumed ciliary compound eye not ocelli clock
0 MSLNRSTVDNVVYDAEDQVSPMVYVAAAIALGFIGFFGFTMNLLVAIVIVKDAQILWTPVNVILVNLV 0
0 VGDFLVAAFGNPVAMISAITGGWYWSYEMCLW 2
1 YAWFMSTLGFASIGNLTVMAVERWLLVARPMKALSIR 2
1 HAITLGIFVWFYALCLSLPPLFGWGSYGPEAGNVSCSVSWEVHDPLTKSDSYIAFLFVFGLIIPVLVISSSYVAIIVTLRKVRKRA 1
2 GASGRREAKITKMVALMITAFLLAWSPYAALAIAAQYFD 0
0 AKPSASVAVLPALLAKSSICYNPIIYAGLNSQFPRSLKKIFDVRIARSSLPDSQNTALTLLNRQEQRN* 0
>TMT_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01001608 full 63% bee
0 MSLNLSVEEQLSIDIYIFAAIALGFIGFL 21 GFFLNLLVIITVLKNANVLWTPNNVVLINMV 0
0 IGDFLVAALGNPFTMTSAIAGEWFWSHEVCLW 2
1 YAWFMTTMGFASIGNLTVMAMERFLLVTCPMKTLSIR 2
1 HAYILAILVWMYALSLSLPPFFNWGIYGPEAGNISCSVSWEIHDPYMHNDTYIGFLFIVGFFLPVTIIISSYYGIIKTLRKIRKRV 1
2 GAKNRREKKVTKMVYLMILAFLIAWSPYAVLALATQYFYV 0
0 QTSHILAVLPALLAKSSICYNPIIYASMTAQFPTWKKMFSINRNNTKSKQRHGSELQIK* 0
>TMT_rhoPro Rhodnius prolixus (kissing_bug) Ecdy.Inse.Hemi ACPB01038515 full G? pteropsin ACPB01038514
0 MLMPSAGFLAASIILFLIGFLGFFGNLIVIIIMCRDKN 0
0 LWTPVNFILFNVIVSDFSVAALGNPFTLASAIAKRWFFGQSMCVAYGFFMALL 1
2 GITSINSLTVLALERYLIVSQPVSHGSLSRPTASDIVGSIWLYSFVITiPPLVGWGEYGLEAANIS 2
1 CSINWETRSHSSTSYILFLFTFGFFIPIIVISYSYMNIILTMKK 0
0 STMNAGRVNKAESRVTWMIFVMIFAFFLAWTPYAILALMIAFFDSNVSPAIATIPAIFAKTSICYNPFIYAGLNTQ 0
0 FRQSWRRVLGGKREDSTTMATATSFGLNSKRYKEVSCVIDVKGKDKIKLSALNKSTATETAI* 0
>TMT_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi genomic full G? pteropsin two contigs, 1st exon very wrong in XM_001952259
0 MNGEYAQPQHISDAIYLGAAIVLSIIGIVGFIFNTCVIFIMIRDTR 0
0 LWTPQNVIIFNLATSDLAVSVLGNPVTLAAAITKGWIFGQTICVIYGFFMALF 1
2 GIASITTLTVLAYDRYLMIRYPFSSSRLTKETALYAIAGIWIYAFAVTGPPLFGWNRYVNESANIS 2
1 CSIDWESGEHSNYVIYIFVFGLFLPVTVIIYSYVSLVVTVRK 0
0 AEKIIGQATKAECRVAIMVAVMILAFLTAWMPYSVLALMIAFGGVHISPVVSIIPALCAKSSICWNPIIYIGLNTQ 0
0 FRSAWKRFLNIQDTLSEVSLDADITTGMTKLMTGHQELPAHPMNNGDASHPPGLIMCCLAHDEHRQSATYADRYECNLEMKSCNPQTLGRRPETDIGDVSL* 0
>TMT_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BY930435 frag G? ovary cDNA
0 MPRWGYVASAFVLFLIGFFGFFLNLMVILLMFKDRQ 0
0 LWTPLNIILFNLVCSDFSVSVLGNPFTLISALFHRWIFGHTMCVLYGFFMALL 1
2 GITSITTLTVISFERYLMVTRPLTSRHLSSKGAVLSIMFIWTYSLALTTPPLLGWGNYVNEAANIQ 2
1 CSVNWHEQSTNTLTYIMFLFAMGQILPLSVITFSYVNIIRTLKR 0
0 NSQRLGRVSRAEARATAMVFIMIIAFTVAWTPYSLFALMEQFATEGI 0
0 VSPGASVIPALVAKSSICYDPLIYVGMNTQ 0
0 FR
>TMT_helVir Heliothis virescens (moth) Ecdy.Inse.Lepi GR968759 frag G? pteropsin pheromone gland
0 MPRWGYVVSAFVLFLIGFFGFFLNLMVILLMFKDRQ 0
0 LWTPLNIILFNLVCSDFSVSVLGNPFTLISALFHRWIFGKTMCVLYGFFMALL 1
2 GITSITTLTVISFERYMMVTRPLNSRHLSSKGAIMSIVF
>TMTa_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic full G? pteropsin retained intron?
0 MPVWVYWSASAYLLFISIAGLFMNIVVVVIILNDSQ 0
0 KMTPLNWMLLNLACSDGAIAGFG 2
1 TPISAAAALKFTWPFSHELCVAYAMIMSTA 1
2 GIGSITTLTVLALWRCQHVVWCPTNRNSNFTDPNGRLDRRQGALLLTFIWTYTLIVTCPPLFGWGRYDREAAHIS 2
1 CSVNWESKMDNNRSYILYMFAMGLFIPLMAIFVSYISILLFIHK 0
0 SQQTSNNSDTVEKRVTFMVAVMIGAFLTAWTPYSIMALVETFTGDNVTNDSVSSEIKFYAGTISPAVATVPSLFAKTSAVLNPLIYGLLNTQ 0
0 FRTAWEKFSSRFLGRKKRHQRSQMAMGVSHKRRRDYLRTLLNRPASDEPAIVQHPSTKEMASSQAVSCVVVSNLDVPRAPNNSYVTVNDE* 0
>TMTb_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic full G? pteropsin long tail IGQDNHSTYYSSRINNATNFSSAFPDGDLSY intron?
0 MPTWAYRLTAAYLLLISVLGLIMNVVVVIVILNDSQ 0
0 RMTPLNWMLLNLACSDGAIAGFG 2
1 TPISTAAALEFGWPFSQELCVAYAMIMSTA 1
2 GIGSITTLTALAIWRCQLVVCCPAKRKSAFTNHSGRLGCRQGVILLVIIWIYALAITCPPLFGWGRYDREAAHIs 2
1 CSVNWESKTNNNRSYILYMFCMGLVVPLAVIIISYVRILRVVQK 0
0 NQQQSGNVHRHRRDAAEKRVTMMVACMIAAFMAAWTPYSILALFETFIGQDNHSTYYSSRINNATNFSSAFPDGDLSYVGTISPAFATIPSLFAKTSAVLNPLIYGLLNTQ 0
0 FRLAWERFSLRFLGRFQCHRTQGVSGQHGANHHKTRRNVRKYLPNCYGDSRSLKPTPTVHLPMKEMVVSHAEQKVKTAQEQASSSVTKITTIPLISSDNQTIVSCPSSIMAN
CQQHETNQANHQQAARPDKVVDHQHLLQPNRLSSLLSLSLPSVLISTPNLPCSAQRQSAAEDQAMATCQQMTSGRIRDQQQQSDSFVVVGLLSRSADCYHHHTGDVEQFVFLDSTVDELGLTARSASP* 0
>MEL1_homSap Homo sapiens (human) Deut.Euth.Euar NM_033282 full Gq OPN4 melanopsin syn(-GRID1 -WAPAL +LDB3 +BMPR1A)
0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0
0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2
1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLTSHTSNLSWISIRRRQESLGSESEV 0
0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGK 0
0 TKGLIPSQDPRM* 0
>MEL1_panTro Pan troglodytes (chimpanzee) Deut.Euth.Euar genomic full G? OPN4 melanopsin
0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0
0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2
1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLISHTSNLSWISIRRRQESLGSESEV 0
0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGK 0
0 TKGLIPSQDPRM* 0
>MEL1_gorGor Gorilla gorilla (gorilla) Deut.Euth.Euar genomic full G? OPN4 melanopsin
0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0
0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRSLRTPXNMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1
2 GCEFYAFCGALFGISSMITLXAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1
2 SAyVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2
1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2
1 YAHVLTPYMSSVPaVIAKAsAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLISHTSNLSWISIRRRQESLGSESEV 0
0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGK 0
0 TKGLIPSQDPRM* 0
>MEL1_ponAbe Pongo abelii (orangutan) Deut.Euth.Euar genomic full G? OPN4 melanopsin
0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGQLPSISPT 0
0 APGTWAAALVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLMVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATIGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2
1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTMISHTSNLSWISGRRRQESLGSESEV 0
0 GWTHMEAAAVWGAAQQANGRFLYDQGLEDLEAKAPPRPQGEEAETPGK 0
0 TKGLIPSQDPRM* 0
>MEL1_rheMac Rhesus macaca (rhesus) Deut.Euth.Euar genomic full G? OPN4 melanopsin fill-in traces
0 MNPPSGPRVPPSPTQEPSCMATPAPPSRWDSSQSSISSLGQLPSVSPT 0
0 AAGTWAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRGLRTPANMFIINLAISDFLMSFTQAPVFFASSLYKHWLFGET 1
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATIGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGr 2
1 ALQTFGACKGSGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSRRHSHPYPSYRSTHRSTLISHTSNLSWISGRRRQESLGSESEV 0
0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGQEAETPGK 0
0 TKGLLPCKDSRM* 0
>MEL1_calJac Callithrix jacchus (marmoset) Deut.Euth.Euar genomic full G? OPN4 melanopsin fill-in traces
0 MNLPTGSRVLPSPTQEPSCMTTPAPPSRWDSSQSSISSLSQLPSISPT 0
0 AAGTWAAAWIPFPTVDVPDHAHYTLGTVILLVGVTGMLGNLTVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKRWLFGET 1
2 GCEFYAFCGALFGISSMITLMAIALDRYLVITRPLATIGVASTKRAAFVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2
1 ALQTFGACKGSGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2
1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSRRHSHPYPSYRSTHRSTLISHTSNLSWISGRRRQESLGSESEV 0
0 GWTHMEAAAAWGAAQQANGRSLYGHGLEDLEAKAPPRPQRQEAETPGK 0
0 tkGLLPSLDARW* 0
>MEL1_micMur Microcebus murinus (mouse_lemur) Deut.Euth.Euar genomic full G? OPN4 melanopsin fill-in traces 4aa del exon 9 confirmed
0 MNPPSGPRMPPSPAQEPSCVATPALASSGDSSQNSVSSLGQLLPASPT 0
0 ATGAWAAAWVPFPTADVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCr 2
1 SRSLRTPANMFVINLAVSDFLMSVTQAPVFFASSLYKRWLFGEA 1
2 gCEFYAFCGALFGISSMITLTAIALDRYLVITRPLASVGTASKRRAGLVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLCCFVFFLPLLVIIYCYIFIFRAIRETGR 2
1 ALQTFGASKGTSESPRQQQRLQNEWKMAKIMLLVILLFLLSWAPYTAVALVAFAG 2
1 YAHILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCVGVLLGVSRQHSRPYPSYRSTHRSTLSSQASDLSWISGRRRQESLGSENDM 0
0 GWTDMEVAAAWGAAPRVRGRCPYGQGPEDMEVKV QGQEAETPGK 0
0 AEGLPPCMDPRM* 0
>MEL1_otoGar Otolemur garnettii (lemur) Deut.Euth.Euar genomic full G? OPN4 melanopsin fill-in traces
0 mnLPSAPRVPPSPAQASSCVATPAPHSRWDSSQTSISSLGQLRPVSPT 0
0 ASGAWAAGWIPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 VRGLRTPANMFVINLAVSDFLMSVTQAPVFFASSLYKQWLFGET 1
2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLTTVGVASKRRAALVLLGVWLYSLAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYTSFTPSVRTYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2
1 ALQTFGACKGSSESPRQRQRLQNEWKMAKITLLVILFFVLSWAPYTTVALVAFAG 2
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGLLLGVSRQHSRPYPSYRFTHHSTLSSQASDLSWISGRRRQESLGSESEV 0
0 GWTDMEAAATWGAALQVSGQCPYSQGLEDMEAKGPLRPQGPETKTSGK 0
0 TKGLLPSLDPRM* 0
>MEL1_musMus Mus musculus (mouse) Deut.Euth.Euar AF147789 10632589 full G? OPN4 melanopsin GenBank wrong readthru
0 MDSPSGPRVLSSLTQDPSFTTSPALQGIWNGTQNVSVRAQLLSVSPT 0
0 TSAHQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 NRGLRTPANMFIINLAVSDFLMSVTQAPVFFASSLYKKWLFGET 1
2 GCEFYAFCGAVFGITSMITLTAIAMDRYLVITRPLATIGRGSKRRTALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPQVRAYTMLLFCFVFFLPLLIIIFCYIFIFRAIRETGR 2
1 ACEGCGESPLRQRRQWQRLQSEWKMAKVALIVILLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGRKRQESLGSESEV 0
0 GWTDTETTAAWGAAQQASGQSFCSQNLEDGELKASSSPQVQRSKTPK 0
0 TKGHLPSLDLGM* 0
>MEL1_ratNor Rattus norvegicus (rat) Deut.Euth.Euar AY072689 11834834 full G? OPN4 melanopsin
0 MNSPSESRVPSSLTQDPSFTASPALLQGIWNSTQNISVRVQLLSVSPT 0
0 TPGLQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 NRGLRTPANMLIINLAVSDFLMSFTQAPVFFASSLYKKWLFGET 1
2 GCKFYAFCGAVFGIVSMITLTAIAMDRYLVITRPLATIGMRSKRRTALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYVTFTPLVRAYTMLLFCFVFFLPLLIIIFCYIFIFRAIRETGR 2
1 ACEGCGESPLRRRQWQRLQSEWKMAKVALIVILLFVLSWAPYSTVALVGFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 AAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGQKRQESLGSESEV 0
0 GWTDTETTAAWGAAQQASGQSFCSHDLEDGEVKAPSSPQEQKSKTPK 0
0 TKRHLPSLDRRM* 0
>MEL1_nanEhr Nannospalax ehrenbergi (molerat) Deut.Euth.Euar AM748539 full G? OPN4 melanopsin
0 MNSPSGPRVPPGLAQKPSFMVTPVLPNQWISFQKNVSVGIQLPPASAT 0
0 ATGAQAASWVPFPTVDVPVHAHYTLGTVILLVGLTGMLGNLIVIYTFCR 2
1 SRGLRTRANMFTVNLAVSDFLMSFTQAPVFFASSLYKRWLFGEA 2
2 GCEFYAFCGAVSGITSMTTLTAIALDRYLVITRPLATIGVASKRRTALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFLPLLIIIFCYIFIFKAIRETGR 2
1 ACEGCGESPQRRRQWQRLQNEWKMAKVALLVIFLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 LAISQHLPCLGVLIGVSSQRSHPSLSYRSTHRSTLSSQASDLSWISGRKRQESLGSESEV 0
0 GWTDTEVTAAWGVAQEASGWSPYRHSLEDGEVKASPSPQGQEAKTSR 0
0 TKGQLPSLNLRM* 0
>MEL1_phoSun Phodopus sungorus (hamster) Deut.Euth.Euar AY726733 15698924 frag G? OPN4 melanopsin 3D ends in error: AKGQLPSLDLGMQDAP
0 MDSPPGPTAPPGLTQGPSFMASTTLHSHWNSTQKVSTRAQLLAVSPT 0
0 ASGPEAAAWVPFPTVDVPDHAHYILGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRSLRTPANMLIINLAVSDFLMSFTQAPVFFASSLYKKWLFGET 2
1 GCEFYAFCGAVLGITSMITLTAIALDRYLVITRPLATIGMGSKRRTALVLLGIWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYVTFTPQVRAYTMLLFCFVFFLPLLVIIFCYISIFRAIRETGR 2
1 ACEGWSESPQRRRQWHRLQSEWKMAKVALIVILLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIVYAITHPKYR 2
1 AAIAQHLPCLGVLLGVSSQRNRPSLSYRSTHRSTLSSQSSDLSWISAPKRQESLGSESEV 0
0 GWTDTEATAVWGAAQPASGQSSCGQNLEDGMVKAPSSPQ
0 * 0
>MEL1_bosTau Bos taurus (cow) Deut.Euth.Laur genomic full G? OPN4 melanopsin
0 MNPPSGPRAPLGPVQESSCLATPASSSRWDSSRSSASSLGHPPSISPT 0
0 AVRAQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKQWLFGEA 1
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATVGMVSKRRAALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYVSFTPSVRAYTMLLFCFVFFLPLLIIIYCYIFIFKAIRETGQ 2
1 ALQTFGTCEGGSECPRQRQRLQNEWKMAKIELLVILLFVLSWAPYSTVALMGFAG 2
1 YAHILTPYMNSVPAVIAKASAIYNPIIYAITHPKYR 2
1 LAIAQHLPCLGVLLGVSGQRTGLYTSYRSTHRSTLSSQASDLSWISGRRRQASLGSESEV 0
0 GWMDTEATAAWGAGQQVSGWSPCSQRLDDVEAKALPRPQGRDSEAPGK 0
0 AKGLLPNLDARM* 0
>MEL1_susScr Sus scrofa (pig) Deut.Euth.Laur genomic full G? OPN4 melanopsin
0 MNPPSRPSAPPDPALESSCMATPASPSRWDSSQSSTSSLGHPLPISPT 0
0 AARVRAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKQWLFGEA 2
1 GCEFYAFCGAVFGITSMITLTAIALDRYLVITHPLATVGMVSKRRAALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYVSFTPKVRAYTMLLFCFVFLLPLLVIIYSYIFIFKAIRETGQ 2
1 ALQTFGACERRGKCPRQRQRLQNEWKMAKIELLVILLFVLSWAPYSTVALMGFAG 2
1 YGHVLTPYVNSVPAVIAKASAIYNPIIYAITHPKYR 2
1 MAIAQHLPCLGVLLGVSGQRTGLYTSYRSTHRSTLSSQASDLSWISGRRRQASLGSESEV 0
0 GWTDTEATAAWGAAQQVSRWSPCGQGLEDMEAKTSLGRLGWEAEAPGK 0
0 TKGLLPSLDPRM* 0
>MEL1_equCab Equus caballus (horse) Deut.Euth.Laur genomic full G? OPN4 melanopsin bad GenBank
0 MNPPSEPQVPLGLAQEPGCVATPASPSRWSGSRSSTSSLGQPLPVGPT 0
0 AAGAQADAWVPFPTVDVPDHAHYTMGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKQWLFGKA 1
1 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATVGVVSKRWAALVLLGIWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLMCFVFFLPLLVIVYCYVFIFRAIRETGR 2
1 ALQTFGAWEGGGECPRQRQRLQSEWKMAKIVLLVILLFVLSWAPYSVVALVAFAG 2
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAIIHPKYR 2
1 MAIAQHLPCLGVLLGVSSQRTRPYTSYRSTHRSTLSSQGSDLSWISGRRRQASLGSESEV 0
0 GWMDTEAAAVWGAAQQMSGWSPCGQGLEDMEAKAPPRPQGWEGEALRK 0
0 IKGLLPSLDPRM* 0
>MEL1_felCat Felis catus (cat) Deut.Euth.Laur AY382594 15842746 full G? OPN4 melanopsin wrong readthru
0 MNPPSGPRTQEPSCVATPASPSRWDGYRSSTSSLDQPLPISPT 0
0 AARAQAAAWIPFPTVDVPDHAHYTLGTVILLVGLTGILGNLMVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFFMSFTQAPVFFASSLHKRWLFGEA 1
2 GCEFYAFCGALFGITSMITLMAIALDRYLVITHPLATIGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMSFTPSVRAYTMLLFCFVFFLPLLVIVYCYIFIFRAIRETGQ 2
1 ALQTFRACEGGGRSPRQRQRLQREWKMAKIELLVILLFVLSWAPYSIVALMAFAG 2
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 MAIAQHLPCLGVLLGVSGQHTGPYASYRSTHRSTLSSQASDLSWISGRRRQASLGSESEV 0
0 GWMDTEAAAVWGAAQQVSGRFPCSQGLEDREAKAPVRPQGREAETPGQ 0
0 TKGLLPSQDPRM* 0
>MEL1_canFam Canis familiaris (dog) Deut.Euth.Laur genomic full G? OPN4 melanopsin fixed 44-way
0 MNPPSGPGAQEPGCVATAASPGRWHGSPRSTVGLDQALPTGPT 0
0 AAGARAAAWAPFPTVDVPDHAHYILGTVILLVGLTGMLGNLMVIYTFCR 2
1 TRGLRTPSNMFIINLAVSDFFMSFTQAPVFFASSLHKRWLFGEA 1
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITHPLAAVGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMSFTPSVRAYTMLLFCFVFFLPLLVIVYCYVFIFRAIRETGQ 2
1 ALQTFRACEGGARSPRQRQRLQREWKMAKMELLVILLFVLSWAPYSAVALTAFAG 2
1 YSHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 MAIAQHLPCLGVLLGVSGQRTGPYASYRSTHRSTLSSQASDLSWISGRRRQASLGSESEV 0
0 GWMDTEAAAVWGAAQPAGGRFLCTQGLEDAEAKAPLRPRGQAVETPGK 0
0 TKGRLPSLDPSRE* 0
>MEL1_myoLuc Myotis lucifugus (microbat) Deut.Euth.Laur genomic full G? OPN4 melanopsin 7 aa del in exon 1 confirmed 1 aa del in DRY cyto loop
0 MNPPSGPRGPLDPAREPGCVATPVSP SSTSSVDPPLPTSPT 0
0 AAEAQAAAWVSFPTVDVPAHAHYTLGIVILLVGLTGMLGNLTVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFLMCFTQAPVVFASSIYKRWLFGEA 1
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPL AIGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMSFTPAVRSYTMLLFCFVFFLPLLVIIYCYVFIFRAIRETGQ 2
1 ALQTFGACEGRSELPRQRQGLQNEWKMAKIVLLVILLFVLSWAPYSTVALMAFAG 2
1 YAHVLTPYMNSVPAIIAKASAIHNPIIYAITHPKYR 2
1 MAIAQHLPCLGLLLGVSGQRTGPYASYRSTHRSTLSSQASDLSWVSRRRRQESLGSESEM 0
0 GWTDTEAAAMWGAAQQVSGPPPCSQGLEDVETKAPPKSQGHEAEDPRK 0
0 TKGLLPSPDPRM* 0
>MEL1_pteVam Pteropus vampyrus (macrobat) Deut.Euth.Laur genomic full G? OPN4 melanopsin
0 MNLPVGPRVPLDPAQEPSCMATPASPS WDSSPSSASSLDQPLPISPT 0
0 AAGSQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVVFISSLYKRWLFGQA 1
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLAAIGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMSFTPSVRAYTMLLFCFVFFLPLLIIIYCYIFIFRAIRETGQ 2
1 ALQTFGACEGLSELPRQRQRLQSEWKMAKIVLLVILLFVLSWAPYSTVALMAFAG 2
1 YSHVLTPYMNSVPAIIAKASAIHNPIIYAITHPKYR 2
1 MAIAQHLPCLGVLLGMSGQHTGPYTSYRSTHRSTLSSQASDLSWISRRRRQASLGSESEM 0
0 GWTDTEAAATWGTAQQVSGPSLWGQDLEDVEAKAPPKPQGREAEAPRK 0
0 TKELLPSLDPRM* 0
>MEL1_eriEur Erinaceus europaeus (hedgehog) Deut.Euth.Laur genomic full G? OPN4 melanopsin
0 MALSLGPRVPTSQALDPSCMDTPASPSRWDSSQNSTSSLAQLPLISLT 0
0 SVSPQATASAPFPTVNVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRSLRTPANMFIINLAVSDFLMSFTQTPVFFASSLYKQWLFGEA 1
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRVALVLLGVWLYSLAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMRFTPSFRAYTMLLFCFVFFLPLLVIIYCYIFIFRAIRETGQ 2
1 ALQTFRACEGSCDFPRQQQRLQSEWKMAKIILLVILLFVLSWAPYSTVALMAFAG 2
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 MAIAQHLPCLRVLLGVSGQRDRPYTSYRSTHRSTLSSQISDLSWVSRRRRQASLGSESEV 0
0 GWTDTEVAAVWGT MSGHFPCGQGLDDMEAKAAHNPRGLEAETPGK 0
0 IKGLLPSLDPQM* 0
>MEL1_loxAfr Loxodonta africana (elephant) Deut.Euth.Afro genomic frag G? OPN4 melanopsin
0 MNPPWGPRVPSGRAQEPSCVATPASASRWNSSRASASSLGELPPSSPT 0
0 AARAHTAAWDPFPTVDVPDHAHYTLGAVILLVGLMGMLGNLMVIYIFFR 2
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKRWLFGEA 1
2 GCKFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRAALVLLGIWLYALAWSLPPFFGW 1
2 SAYVPDGLLTSC 2
1 ALQTFGACEGASEPPRQWQRLQSEWKMAKIALLAILLYVLSWAPYSTVALVAFAG 2
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 MAIAQHLPCLGVMLGVSGQRTRPYTSYHSTLHSTLSSQASDLSWISGRRRQASLGSESEV 0
0 GWTDTEAAAAWEGAQQVSGQASCSQALQNLEANTPPRPQGWGPETPRK 0
0 * 0
>MEL1_proCap Procavia capensis (rock_hyrax) Deut.Euth.Afro genomic full G? OPN4 melanopsin
0 MNPPWGPRVPSRPAQEPSCMSTPASAGRWDSSQATASSLAELPPSSPT 0
0 EARTQTADWVPFPTVDVPDYAHYTLGTVILLVGLTGVLGNLMVIYIFFR 2
1 SRGLRTPANMFIINLAISDFLMSLTQAPVFFASSLYKRWLFGEA 1
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRTALVLLGTWLYALAWSLPPFFGW 1
2 SAYVPDGLLTSCSWDYKSFMPSARTYTMLLCCFVFFLPLLVIIYCYVFIFKAIRETGR 2
1 ALQTFGACEGASETPRQWQRLQSEWKMAKIALLAILLYVLSWAPYSTVALVGFAG 2
1 YAHVLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 MAIAQHLPCLGVLLGVSDQHTRPYTSYRSTHHSTLSSQASDISWISGRRRQASLGSESEV 0
0 GWTDTEAAAAWEGAQQVSGRASCSQVLESMEANTPPRPQGWGPETPRK 0
0 VKGLPLLDPRA* 0
>MEL1_echTel Echinops telfairi (hedgehog) Deut.Euth.Laur genomic frag G? OPN4 melanopsin
0 MNLPSGSRVPSGPAQEPSHVATAASASRWNS GSSLDAPPPSSPT 0
0 AARAPTAASAPFPIVDVPDHVHYTLGTVILLVGLTGMLGNLMVIYTFCR 2
1 SRSLRTPANMLIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGEA 1
2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRAALVLLVIWLYALAWSLPPFFGW 1
2 SAYVPDGLLTSCSWDYMSFTPSVRAYTMLLFCFVFFLPLLVIIYCYIFIFRAIRETGR 2
1 ALQTFGACEGSRDSPRQRQRLQSEWKMAKIALLVISLFVLSWAPYSTVVLVAFAG 2
1 2
1 0
0 GWTDTEVTATSGYTQQVSGRCPRGQDLESMDANPTPRPRGWETETAQK 0
0 IKGLPLSLNPQA* 0
>MEL1_monDom Monodelphis domestica (opossum) Deut.Meta.Dide genomic full Gq OPN4 melanopsin syn(-GRID1 -WAPAL +LDB3 +BMPR1A)
0 MNPSPMLRGLSCPAQDTNCTKIMASMSEWNNTEEDAYHLVDLPSIAPT 0
0 AVVLPPSSQNIFPTVDVPDHAHYTIGAIILAVGITGMLGNFLVIYTFCR 2
1 SHSLRTPANMFIINLAISDFFMSFTQAPVFFASSMYKRWIFGEK 1
2 ACEFYAFCGALFGITSMITLMAIALDRYFVITRPLASIGVISKKKTGFILLGVWLYSLAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYTTFTPSVRAYTMLLFCFVFFIPLIVIIYCYIFIFRAIQDTNK 2
1 AVHSIGSGESTASPRHCQRMKNEWKMAKIALVVILLYVLSWAPYSTVALVAFAG 2
1 YSHILTPYMNSVPAIIAKASAIHNPIIYAISHPKYR 2
1 MAIAQNFPCLRALLCVRHPRTRSFSSYRFTRRSTMTSQASDISWLPRGRRQLSLGSESEI 0
0 GWNNMEAGTTSLTSRNQQGSCRMDQETMETRELAAIAKAKGRSWETLEK 0
0 TLEEMDDSSLLEVSVDMEQ* 0
>MEL1_smiCra Sminthopsis crassicaudata (dunnart) Deut.Meta.Dasy DQ383281 17785267 full G? OPN4 melanopsin
0 MNPSPMLRHLSCPAQDSNCTKIMASISEWNNTEVDAYHLVDLPPITPT 0
0 AVVLPPYSQKVFPTADVPDYAHYTIGATILVVGFTGVLGNLLVIYTFCR 2
1 SRSLRTPANMFIINLAISDFFMSFTQAPVFFASSLYERWIFGEK 2
1 GCEFYAFCGALFGITSMITLMVIALDRYFVITRPLASIGMISKKKTGLILLGVWLYSLAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYTTFTPSVRAYTILLFCFVFFIPLTVIIYCYIFIFRAIKDTNK 2
1 AVQNIGSSEHTPSLRHFQRMKNEWKMAKIALVVILLFVLSWAPYSTVALVAFAG 2
1 YSHVLTPYMNSVPAIIAKASAIHNPIIYAISHPKYR 2
1 MAIAQNFPCLRAVLGIRHPRTQSFSSYRFTHRSTTASQASDISWQSRGRRQLSLGSESEA 0
0 GWNNIETGLTLRSLEGSCGMDEETMDTRELSASTKAKGQSWETLAKTLEE 0
0 MDDLSLLEAGTLLSSLDLQI* 0
>MEL1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono genomic frag Gq OPN4 melanopsin
0 0
0 FPTADVPDHAHYTIGATILAVGFTGVLGNLLVIYTFCR 2
1 SRSLRTPANMFIINLSISDFFMSLTQAPVFFASSLHKRWIFGEK 1
2 GCQLYAFCGALFGITSMITLTVIALDRYFVITRPLASIGVISKKRALLILTGVWFYSLAWSLPPFFGW 1
2 sAYVPEGLLTSCSWDYMTFTPPVRAYTMLLFCFVFFIPLIMIIYCYFFIFRAIRGTNK 2
1 AVETIGSDDCRGSQRQCQRMKNEWKTAKIALMVILLYVISWCPYSVVALVAFAG
1 YSHLLTPYMNSVPAVIAKSSAIHNPIIYAITHPKYR 2
1 MAITKYIPCLGPLLRVSRQDSRSSSHYASSRRSTVTSQSLDGSWLPGRRRPLSSASDSES 0
0 0
0 * 0
>MEL1_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic frag G? OPN4 melanopsin diverged
0 0
0 ERTMFNLPDPFPTVDVPTHAHYTIGAVILVVGITGTLGNLLVIYVFFR 2
1 IRGLRTPANMFVINLAVSDFL 1
2 GCELYAFCGALFGIASMITLTVIALDRYFVITRPLASIGAMSTKKALLILSGVWLYSLAWSLPPFFGW 1
2 sAYVPEGLLTSCSWDYITFTPSVRAYTMLLFCFVFFIPLIAIIYSYVFIFIAIKNSNR 2
1 AVQRTNSDNSKEGQKLYQKLKNEWKMAKVALIVILLVISWSPYSVVALVAFAG 2
1 YSHLLTPYMNSVPAVIAKASVIHNPIIYAIVHPKYR 2
1 MAIAKFLPCLGSLLRVPRKDSSYPSTRRPTVTSQSSDINGVPRGHRRLSSVSDSES 0
0 DWTDTEADISSQNSRVASGSISYRIYEDTTETIKVKSKMRSHDSGIFER 0
0 0
0 TGEDLNAFGWRREESYSGPSTSSQIPSIIVTFSNVQRTDLPLESSSGALCSRNSSYSWEKDSNS* 0
>MEL1_galGal Gallus gallus (chicken) Deut.Saur.Arch AY88294 16856781 full Gq OPN4 melanopsin syn(-GRID1 -WAPAL +LDB3 +BMPR1A) short exon 1
0 MDLPPRAPT 0
0 KMTVKDVRGAFPTVDVPDHAHYTIGTVILIVGITGTLGNFLVIYAFCR 2
1 SRTLQKPANIFIINLAVSDFLMSITQSPVFFTNSLHKRWIFGEK 1
2 GCELYAFCGALFGITSMITLMVIALDRYFVITKPLASVRVMSKKKALIILVGVWLYSLAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFIPLIAIIYSYVFIFEAIKKANK 2
1 SVQTFGCKHGNRELQKQYHRMKNEWKLAKIALIVILLYVISWSPYSVVALVAFAG 2
1 YSHVLTPFMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 TAIATYVPCLGFLLRVSPKESRSFSSYPSSRRTTITSQSSETSGLQKGKRRLSSISDSES 0
0 GCTDTETDITSMISRPASSQVSYEMGEDTTQTSDLGGKPKVKSHDSGIFRK 0
0 TVVDADEIPMVEINDTEHSATSTCKTSEKCNVEEIQ 0
0 RSESLSGIGLREGESRHRTSASQIPSIIITYSNVQGVELHSGYSAGFLHPKNKSHKQNKSSNS* 0
>MEL1_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full G? OPN4 melanopsin short exon 1 -GRID1 +LDB3 synteny
0 MDLLLRAPT 0
0 KMTVQDVPRAFPTVDVPDHAHYTIGVVILIVGITGTLGNFLVFYAFCR 2
1 SRSLQTPANILIINLAISDFLMSITQSPVFFTSSLYKHWIFGEK 1
2 GCELYAFCGALFGITSMITLMVIALDRYFVITKPLASVGVTSKKKALIILVGVWLYSLAWSLPPFFGW 1
2 sAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFIPLIAIIYSYVSIFEAIKKANK 2
1 SIQTFGCKRGNREFQKQYQRMKNEWKMAKIALIVILFFVISWSPYSVVALVAFAG 2
1 YSHVLTPFMNSIPAVIAKASVIHNPIIYAITHPKYR 2
1 KAIATYVPCLGPLLRVSPKDSRSFSSYHSSRRATISSQSSEISGLQERKRRLSSLSDSES 0
0 GCTETETDTPSMFSRLARRQISYKTDKDTTQTSDIRAKLTSQDSGNCGK 0
0 TAVDADDILMVELNVTEYMATPTVRTILLIFGNKN 0
0 KSESLNSIGQRREFHQGSSSAQIPSITITCSSVQGIELPSRYNSGFLYPKSSSHKQNKKSSS* 0
>MEL1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur DQ384639 16856781 full Gq OPN4 melanopsin syn(-GRID1 -WAPAL +LDB3 +BMPR1A)
0 MNYQSVRKGITCPPQDANCSRILESLNSWNNSEVNSYKLVELPPIVTT 0
0 ETPQYEIHHVYPTVDVPDHVHYVVGAVILAVGITGMLGNFLVIYAFCR 2
1 SRSLRSPANMFIINLAITDFLMSVTQAPVFFATSLHKRWIFGEK 1
2 GCELYAFCGALFGITSMITLMVIAVDRYFVITRPLTSIGVMSKKRAVLILSGVWLYSLAWSLPPFFGW 1
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFCFVFFIPLFIIIYCYIFIFKAIKNTNR 2
1 AVQKIGTDNNKESHKQYQKMKNEWKMAKIALIVILLYVVSWSPYSTVALLAFAG 2
1 YASILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 MAIAKYIPCLGSLLRVKRRDSRSYSSYPSSRRSTVTSHCSQSSDVGGHPKLKNHLPSVSDSES 0
0 GWTDTEADSSVNSRPASRQVSYEMGKDTTETNDLKSKAKLKSHDSGIFEK 0
0 TSMDADDISLVELGTVDRSSPIM 0
0 ANKHLNGLGQRKGDSFTRRSPSSRIPSIVVTHSNHQGSPAAVRHNSTLPGIKVSNSQDREKELKRQIEKVKQYVPIVTITSDTENSTGGFSNELLPANTS* 0
>MEL1a_danRer Danio rerio (zebrafish) Deut.Acti.Otoc AY078161 12487121 full Gq OPN4 melanopsin syn(- +USP54 +LDB3 +BMPR1A)
0 MMSGAAHSVRKGISCPTQDPNCTRIVESLSAWNDSVMSAYRLVDLPPTTTTTTSVA 0
0 MVEESVYPFPTVDVPDHAHYTIGAVILTVGITGMLGNFLVIYAFSR 2
1 SRTLRTPANLFIINLAITDFLMCATQAPIFFTTSMHKRWIFGEK 1
2 GCELYAFCGALFGICSMITLMVIAVDRYFVITRPLASIGVLSQKRALLILLVAWVYSLGWSLPPFFGW 1
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFIPLIVIIYCYFFIFRSIRTTNE 2
1 AVGKINGDNKRDSMKRFQRLKNEWKMAKIALIVILMYVISWSPYSTVALTAFAG 2
1 YSDFLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 LAIAKYIPCLRLLLCVPKRDLHSFHSSLMSTRRSTVTSQSSDMSGRFRRTSTGKSRLSSASDSES 0
0 GWTDTEADLSSMSSRPASRQVSCDISKDTAEMPDFKPCNSSSFKSKLKSHDSGIFEK 0
0 SSSDVDDVSVAGIIQPDRTLTN 0
0 AGDITDVPISRGAIGRIPSIVITSESSSLLPSVRPTYRISRSNVSTVGTNPARRDSRGGVQQGAAHLSNAAETPESGHIDNHRPQYL* 0
>MEL1b_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic frag Gq OPN4 melanopsin syn(- +USP54 +LDB3 +BMPR1A)
0 QVAMVQDVRHPFPTVDVPDHAHYTIGSVILAVGITGMVGNLLVMYAFCK 2
1 SRSLRTPANMFIINLAVTDFLMCVTQTPIFFTTSLHKRWIFGEK 1
2 GCELYAFCGALFGICSMITLMIIAVDRYFVITRPLASIGVMSRKRALLILSAAWAYSMGWSLPPFFGW 1
2 SGAYVPEGLLTSCSWDYMTFSPSVRAYTMLLFTFVFFIPLFVIIYCYFFIFKAIRETNR 2
1 AVGKINGEGGPRDSIKKIHRMKNEWKMAKIALIVILLYVISWSPYSCVALTAF 2
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 SAIAKYIPCLGVLLCVPRRDRFSSSSFISTRRSTLTSQSSETSSNLHRAGKARLSSVSDSES 0
0 GWTDTEADLSTASSRPASRQVSSEIRKDLCDIKHSSSLRLKVKSRDSGIFDR 0
0 0
0 QNDVSEKADEKRPLVRIPSIIVTSETCPAVLPAGHSSRLIPGAPAVTDS* 0
>MEL1_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? OPN4 melanopsin first exon uncertain
0 MGEVGVTVGPACSAAAAYQSGVSCGGELEPSGSAVVMTSLRLAELQLPAST 0
0 LQVAMARPFPTVDVPDHAHYTIGSVILAIGITGMIGNFLVIYAFCR 2
1 SRSLRTPANIFIINLAVADLLMCVTQTPIFFTTSMHKRWIFGEK 1
2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRAFIILMTVWVYSLGWSLPPFFGW 1
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIFCYIFIFRAIRSTNK 2
1 AVGKVNGSVHSHSRRQESLKKLQRLQSEWRMAKVALIVILLYVISWSPYSCVALTAFAG 2
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 LALARYIPCLGLLLCVSPQELRSTSSSFMSMRRSTVTSQTSDISGQFRRQSKPRLSTASDSES 0
0 CLTDTEADLTSTASRPASRQVSCDIGRDTADLPEFQPAASFKSKAKSPDSGVFEK 0
0 TSFDFDASMAAPRQHSSIPN 0
0 NGDFSEGHVTRVALGRIPSIVITSESSFLPHGRKGSASTSAQTATSSDGKAGLG* 0
>MEL1_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gq OPN4 melanopsin syn(- +USP54 +LDB3 +BMPR1A)
0 MNFGKSALQPPAQQSVVSCGGGGPEPNCTLRLAVTVMMSVRLAELQLHAST 0
0 LQVAMVRPFPTVDVPDHAHYTIGSVILVIGITGMIGNFLVIYAFCR 2
1 SRSLRTPANMFIINLAVTDLLMCVTQTPIFFTTSMYKRWIFGEK 1
2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRAFVILMTVWIYSLGWSLPPFFGW 1
2 SAYVPEGLLTSCTWDYMTFSPSVRAYTMLLFIFVFFLPLFIIIYCYFFIFRAIRATNK 2
1 AVGKVNGSVHSHSRRRESVKNFQRLQNEWKMAKIALMVILLYVISWSPYSCVALTAFAG 2
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 LALAKYIPCLGFLLCISPHELQSTSSSFMSLRRSTVTSQTSDISGQFRPQSKPRRSSASDSES 0
0 CLTDTEADLSSMGSRPASRQVSCDISRDTTELPEYKPASSFNSKVKSPDSGIFEK 0
0 TSFDFDASMAASRERSSIPN 0
0 SGEFPEGHVMRRTLARIPSIIITSESSHFLPNGRKASSTTCIANGSDIKVGPR* 0
>MEL1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gq OPN4 melanopsin syn(- - +LDB3 +BMPR1A)
0 MNAGESELLLPTQQSILPCGDHEPNCPVAQAETLALSAASANGSA 0
0 VQVAMVSRAPHPYPTVDVPDHAHYTIGSVILAIGITGIIGNVLVIYAFSK 2
1 SRSLRTPANMFIINLAITDLLMCVTQAPIFFTTSMHKRWIFGEK 1
2 GCELYAFCGALFGICSMITLTVIALDRYFVITRPLTSIGMMSRRRALLILMGAWTYSLGWSLPPFFGW 1
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIYCYFFIFRAIRVTNR 2
1 AVGKMNGSIHSHGSGRDSTKNFHRLQNEWKMAKIALIVILLYVVSWSPYSAVALTAFAG 2
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 IALAKYIPFLGVLLCVPPRELRSASSSFRSTRRSTVTSQTSDVSSQQRRQGSRNSRLSSASDSES 0
0 CLTDTEADGSSVGSRPASRQVSCDIGRDTAELPEFKPSSSFKSKMKSHDSGIFEK 0
0 SYDTDISMAGVSERGSIPN 0
0 QTDFAEGRDRRSTIGRIPSIVITSETSPFLPTGRNGSCNGRPKTANSSHPGAGSG* 0
>MEL1_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full Gq OPN4 melanopsin syn(- +USP54 +LDB3 +BMPR1A)
0 LQVAMVPQTFHPFPTVDVPDHAHYTIGSVILAIGITGIIGNFLVIYAFSR 2
1 SRSLRTPANMFIINLAITDLLMCVTQSPIFFTTSMHKRWIFGEK 1
2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRALLILSAAWAYSLGWSLPPFFGW 1
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIYCYVFIFRAIRSTNR 2
1 AVGKINGNTRDAVKSFNRLQNEWKMAKIALIVILLYVISWSPYSTVALTAFAG 2
1 YADMLTPYMNSIPAVIAKASAIHNPIIYAITHPKYR 2
1 MALAKYIPGLGVLLCIHPKDLRSASSSFVSTRRSTVTSQSSDISSQLRRQSTFKSRLSSLSDSES 0
0 GLTDTEADLSSLSSRPASRQVSCEISRDTAELPDFKHTSSFKAKLKNNDSGIFEK 0
0 TSFDTVSIGGVSEHNSIPS 0
0 NRDFGDGNVTRATIGRIPSIVVTSEMSPFLPVGRNGSRTNRSKMANSSAGAGPV* 0
>MEL1a_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gq OPN4 melanopsin
0 ASVTDAQHHHMFPTVDVPDHAHYIIGATILAVGVTGMVGNFLVIYAFLR 2
1 SRSLRTPANTFIINLAATDFLMSVTQSPIFFITSIHKRWIFGEK 1
2 GCELYAFCGALFGITSMITLMVIALDRYFVITRPLASIGVLSHRRAGLIILSLWLYSLAWSLPPFFGW 1
2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFCFVFFIPLGVIIYCYIFIFRAIKSTNK 2
1 KVGGSTNRESQKQHQRMKNEWKMAKIALIVILLFVISWSPYSTVALTAFAG 2
1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 MAIAKYVPLLGLLLRVSRRDSRTSGQYYSTRRSTLTSQTSDLSGYPRGKGRLSSASDSES 0
>MEL1b_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag Gq OPN4 melanopsin based on Squalus
1 SKSLRTPANMFIINLAISDFFMSATQPPVFFVTSLHKRWIFGEK
2 GCKLYAFCGALFGITSMITLMAISIDRYWVITKPLQSISSTTTKKNTLKVIILVWLYSLAWSLPPLLGW 1
>MEL1_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag G? OPN4 melanopsin first exon uncertain
0 MAPSIVPLMRKLSCPLSRKTPPKHNVSTSMFDGGDGVLGVIGSIP 0
0 VSFLPSQREKFPMLDIPHHVHHTIGSVVIAIGFTGIIGNFLVIYVFCR 2
0 VQVAMVSRAPHPYPTVDVPDHAHYTIGSVILAIGITGIIGNVLVIYAFSK 2
1 SKSLRSPANIFIINLAFADFFMSITQTPIFFVTSLHKRWIFGEK 1
2 GCELYAFCGALFGIASMVTLMVIATDRYLVLTRPLASIGAMSKRRAMYITAAVWFYSLAWSLPPFFGW 1
2 SAYVPEGLMTSCTWDYVTFTPAVRSYTMLLFCFVFFIPLIVIIFCYVRIFAAIKNTNR 2
1 AVKTLGDAHDSKESQKQQQRMNAEWKLAKIALIVILLYVVSWSPYSCVALVAWAG 2
1 YADMLTPYMNSVPAIIAKASAIHNPIVYAITHPKYR 2
>MEL1_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni AABS01000008 frag G? OPN4 melanopsin transcripts BW447434 BW019524 BW048729 rough seq
0 DRRYPSCYKGEQVTFIYLIPIFLFRTLAFLSSVFVTMTPSAVLPLVTTEARPVHPINDVAMYTFGGLMLTAGTVAVVGNIMVMYTFLRR 0
1 PLHWFIVQLAVADFFVGLIVLWIGTFSSLFLDTVSLMSGIATYGVLAAATSTSTLGVLFIAVDRHFYILRHRRYKQIMTRLRVGTAIVVACVVPATFFVVVPAFGWN 1
2 PEYIEEPLIPSCIFDRFTNSLSNRLYIITMCTFVFFIPLVFICYCLYRIFWAVKSSS 0
2 SLSRAGSRKSQSSGKLSKSNSSRSKRGIQTIEFQILKSAVLLVVLFVSSWMPFTVAAIISIGSNQVSPYVILVSYLFAKASCVHSSFAYITNAHFRATIGLIRCKHTHRA* 0
>MEL1_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni AABS01000008 frag G? OPN4 melanopsin 0 transcripts rough
FTTLATLAVIPPTMVLNNSTHPIKVSALLAFGSLMLCAGIIAIVGNLVVMYTFLRPLNWFILQLAVADFFVGLIVLWIGVFSSLLL
ETVSLMSGVATYGVLAAATSTSTLGVLFIAVDRHFYILKHRRYKSIMTRVKVGVAIFFACVTPLAFFVAAPAAGWN 1
DYIAEPLIPSCIFDRFTTSLANQTYIITMCAFVFFVPLLFICYCLVRIYKAVKTSSQTVEFQILKSAVLLMVLFVS
SWMPFTVAAMISVVAEQVDPYVILVSYLFAKASCVHSSFAYITNAHFRATLGVLRCKKRRSV* 0
>MELx_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XP_002586120 full Gq
0 MDRMSPNLTNTSLLPNRTDRPELTPADVTMQLVFGSMMLVFGLIGVVGNAVALYAFCS 2
1 TRKLRRPKNYVVANLCLTDLIMCIVYCPVIVISSFSGR 2
1 IPTDGACTMEGFVVGMASIASVGSL 0
0 VAIAVERFFSITRPMKSLTILTKRTFLGGVAVVWLYSLILVIPPLLGWGRYVREETKLSCSFDYLSTDDANRSYVIWLVIVAFGLPLLVIAYCYISVFITVKKCTKKRKLMSPHKKSRSEVKTAVNAFIMTTAFCLCWCPY
AVVATMGISGSSVQGTVVFGAALLAKLSVLINPVAYVFSIPSFRKALFGHRKRGYGTSDGLANDSSSEKRRGKKHESDANSATEYLRLTVFYSMYSRTGQLSPWASKRLAGKTKSMLDLTTEYGRLERTAHKESWV* 0
>MEL_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran AB205400 15936279 full Gq Amphi-mop 12 exons
0 MTELPSFQPPTNSTEEENAVFPTALTEWISE 0
0 VGNQVGEAALKLLSGEGDGMEVTPTPGCTGNASVCNGTDSGGGVVWDIPPLAHYIVGTAVFCVGCCGMFGNAVVVYSFIK 2
1 SKGLRTPANFFIINLALSDFLMNLTNMPIFAVNSAFQRWLLSDF 1
2 ACELYGFAGGLFGCLSINTLMAISMDRYLVITKPFLVMRIVTKQR 0
0 VMFAILLLWIWSLVWALPPLFGWSAYVPEGF 1
2 GTSCTFDYMTPKLSYHIFTYIIFFTMYFIPMGVIIYCYYNIFATVKSGDKQFGKAVKEMAHEDVKNK 0
0 AQQERQRKNEIKTAKIAFIVITLFLSAWTPYAVVSALGTLGYQDLVTPYLQSIPAVFAKSSAVYNPI 1
2 VYAITHPKFRAAVKKHIPCLSGCLPADEEETKTKTRGATTTASMSMTQTTAPTV 0
0 HDPQASVHSGSSVSVDDSSGVSRQDTMMVK 0
0 VEVDNRMEKAGGGAADTAPKDGTSVPTVSAQIEVRPSGNVNTKAEVIPSPQSAAVAHGASASPVPK 0
0 VAELSSSVSLESAAIPGKIPTPLPSQPIAAPIERHMAAMADDPPPKPRGVATTVNVRRSESGYERSQDSLRKK 0
0 AVSETRSRSFNSTKDHFASERQTSTTLNQPRDMYSGDMVKKTRQSPEKQEYDNPAFDAGIAEIDTDSENETEGSYDMLSVRFQAMAEEPPVETYRKASDMSINLGKASLMLTEAHDETVL* 0
>MEL_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB205400 15936279 full Gq Amphiop6
0 MTEIPSFQPPINATEVEEENAVFPTALTEWFSE 0
0 VGNQVGEVALKLLSGEGDGMEVTPTPGCTGNGSVCNGTDSGGVVWDIPPLAHYIVGTAVFCIGCCGMFGNAVVVYSFIK 2
1 SKGLRTPANFFIINLALSDFLMNLTNMPIFAVNSAFQRWLLSDF 1
2 ACELYGFAGGLFGCLSINTLMAISMDRYLVITKPFLVMRIVTKQR 0
0 VMFAILLLWIWSLVWALPPLFGWSAYVSEGF 1
2 GTSCTFDYMTPKLSYHIFTYIIFFTMYFIPGGVMIYCYYNIFATVKSGDKQFGKAVKEMAHEDVKNK 0
0 AQQERQRKNEIKTAKIAFIVISLFMSAWTPYAVVSALGTLGYQDLVTPYLQSIPAMFAKSSAVYSPI 1
2 VYAITYPKFREAVKKHIPCLSGCLPASEEETKTKTRGQSSASASMSMTQTTAPV 0
0 HDPQASVDSGSSVSVDDSSGVSRQDTMMVK 0
0 VEVDKRMEKAGGGAADAAPQEGASVSTVSAQIEVRPSGKVTTKADVISTPQTAHGLSASPVPK 0
0 VAELGSSATLESAAIPGKIPTPLPSQPIAAPIERHMAAMADEPPPKPRGVATTVNVRRTESGYDRSQDSQRKK 0
0 VVGDTHRSRSFNTTKDHFASEQPAALIQPKELYSDDTTKKMARQSSEKHEYDNPAFDEGITEVDTDSENETEGSYDMLSVRFQAMAEEPPVETYRKASDLAINLGKASLMLSEAHDETVL* 0
>MEL6_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran genomic 12435605 full Gq Amphiop6
0 MSPNLTNTSLLPNRTDRPELSPADVTMQLVFGSMMLVFGLIGVVGNAVALYAFCR 2
1 SRSLRRPKNYLIANLCLTDMVVCLVYSPIIVTRSLSHG 2
1 LPSKESCIVEGFVVGLGSIVSICSLAGIAVERYVTITQPIKSLSILTHRALLGAVSAVWVYAFLLAFPPLVGWGRYVSEESKISCTFDYLSTDDATRAHVIVLVIGAFGLPFSVITYCYV
RSFATVRKCTKERKQMSPLAKSDSRSEVKAAVNSFVITTSFCLCWCPYAVVATMGVSGFTVHSHAVFIAALLAKLSVLFNPVAYVLSIPSFRKALFSSSNDRTKYQTAFTFESLAKTSPV
ERKWCADSIERYRSSNVNIESTELTVPYSASRESCLLSRAATERLAGRSPSLTDIVREFGLQQTASHRETWV* 0
>MEL6_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050611 12435605 full Gq
0 MSSNLTNVSLVANRTDQTELSPTDVTMQLIFGSMMLVFGLIGVVGNVVALYAFCR 2
1 TRSLRRPKNYVVANLCLTDMFVCLVYCPIVVSRSFSHG 2
1 FPSKESCIVEGFMVGVGSIASICSLAAIAVERYLSVTQPLKSLTILTQRKLLVAVLTVWVYSLLLAFPPLVGWGRYVREETYISCTFDYLSTDDATRAYVITLVMGAFGFPLLTIAYCY
IRVFTTARKHAEERKFMSPLKRPESRTEIKTAVTACVITTSFCLCWCPYAVVATLGISGVSVQQQTVFSAALLAKLTVIINPIVYVLSIPNFRKALFAQEREKYASEDVVLTSLPGKTRRMK
KVERSQSSNSNVVIEVKESSMAYSTSRESCLLSRAATKRLAGKTKSIVDLVDEFGLQETAPHKESLV* 0
>MEL2_galGal Gallus gallus (chicken) Deut.Saur.Arch AY882944 17977531 full Gq syn(+GRID2+SMARCAD1 -PGDS -SEC24B +COL25A1)
0 MGTQPHSVTKSEIPDHVLYTVGTCVLVIGSIGIIGNLLVLYAFYS 2
1 NKKLRTPQNFFIMNLAVSDFLMSASQAPICFVNSLHREWILGDI 1
2 GCDLYAFCGALFGITSMMTLLAISVDRYLVITKPLRSIQWTSKKRTIQIIAAVWLYSLGWSVAPLLGW 1
2 SSYVPEGLMISCTWDYVTYSPANRSYTMILCCCVFFIPLIIILHCYLFMFLAIRSTGR 2
1 DVQKLGSCSRKSFLSQSMKNEWKLAKIAFVVIIVYVLSWSPYACVTLIAWAG 2
1 RGNTLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYR 2
1 KTIHNAVPCLRFLIRISKNDLLRGSINESSFRTSLSSHQSLAGRTKNTCVSSVSTGEA 0
0 NWSDVELDTVEPAHEKLQPRRSHSFSSSLRQKRDLLPDSYSCSEETEEK 0
0 VSLSSSYLEKVLGRSAFPSSPVALVTSSLRAASLPVGLNSSSASRGAGSDISQMKTEESHNNGGLDSIVSNTVPQIIIIPTSETNLFQEEPEEEETELFHFHDKKNNLLDLEGLSSSTEFLEAVEKFLS* 0
>MEL2_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full Gq syn(+GRID2+ SMARCAD1 -ATOH1 +PDLIM5 +BMPR1B)
0 MGPHHRTKVDVPDHVLYTVGSCVLVIGCIGITGNLLVLYAFYS 2
1 NKRLRTPPNYFIMNLAVSDFLMSATQAPICFLNSMHKEWVLGDI 1
2 GCNLYAFCGALFGITSMITLLAISVDRYCVITKPLQSIKRTSKKRTCIIIVFVWLYSLGWSVCPLFGW 1
2 SSYIPEGLMISCTWDYVTYSPANRSYTMMLCCCVFFIPLVIIFHCYIFMFLAIRSTGR 2
1 RKSSISHSIKSEWKLAKIAFVAIVVFVLSWSPYACVTLISWAG 2
1 YARTLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYR 2
1 RTIRSAVPCLRFLIPISKSDLSTSSMSESSFRASVSSRHSFSYRNKSTYISSISAKET 0
0 TWCDVELDPVESGHKKLQAYRSNSFSAKGVAEEESGLLLRTNNCNVPARKK 0
>MEL2_xenLae Xenopus laevis (frog) Deut.Amph.Anur genomic full Gq Xmop syn(SMARCAD1 +PDLIM5 +BMPR1B)
0 MDLGKTVEYGTHRQDAIAQIDVPDQVLYTIGSFILIIGSVGIIGNMLVLYAFYR 2
1 NKKLRTAPNYFIINLAISDFLMSATQAPVCFLSSLHREWILGDI 1
2 GCNVYAFCGALFGITSMMTLLAISINRYIVITKPLQSIQWSSKKRTSQIIVLVWMYSLMWSLAPLLGW 1
2 SSYVPEGLRISCTWDYVTSTMSNRSYTMMLCCCVFFIPLIVISHCYLFMFLAIRSTGR 2
1 NVQKLGSYGRQSFLSQSMKNEWKMAKIAFVIIIVFVLSWSPYACVTLIAWAG 2
1 HGKSLTPYSKTVPAVIAKASAIYNPIIYGIIHPKYR 2
1 ETIHKTVPCLRFLIREPKKDIFESSVRGSIYGRQSASRKKNSFISTVSTAET 0
0 VSSHIWDNTPNGHWDRKSLSQTMSNLCSPLLQDPNSSHTLEQTLTWPDDPSPKEILLPSSLKSVTYPIGLESIVKDEHTNNSCVR
NHRVDKSGGLDWIINATLPRIVIIPTSESNISETKEEHDNNSEEKSKRTEEEEDFFNFHVDTSLLNLEGLNSSTDLYEVVERFLS* 0
>MEL2_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full Gq syn(- +FLJ39155 +PDLIM5 -)
0 MEPQRQIYKRLDVPDHVHYIIAFLILIIGTLGVSGNALVMFAFYR 2
1 NKKLRSLPNYFIMNLAVSDFLMAITQSPIFFINCLYKEWMFGEL 1
2 GCKIYAFCGALFGITSMINLLAISIDRYLVITKPLQTIQWNSKRRTGLAILCIWLYSLAWSLAPLIGW 1
2 GSYIPEGLMTSCTWDYVSPSPANKSYTMMLCCFVFFIPLSIILYCYLFMFLSVRQASR 2
1 QKSSFVKQQSMRSEWKLAKIAAVVIVVYVLSWAPYACVTLVAWAG 2
1 HQDVLTPYSKTLPAVLAKSSAIYNPFIYAIIHNKYR 2
1 RTLAEKVPGLSCLSRSQKDGLSSSTNSDASAQDSSVSRQSSVSKNRLHSTMVQ* 0
>MEL2_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full Gq syn(- - - +BMPR1B)
0 MEPKDTHITSSFFSKVDVPDHVHYIIAFFVFVIGILGITGNVLVIFAFYS 2
1 NKKLRSLPNYFIVNLAVSDLLMASTQSPIFFINLYKEWMFGET 1
2 ACKMYAFCGALFGITSMINLLAISVDRYVVITKPLQTIRRSSKRRTALAILMVWLYSLAWSLAPLVGW 1
2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLAIILCCYLLMFLAIRKTSR 2
1 RKSTLIQQKSIRSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2
1 YGSTLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYR 2
1 KTIRSAVPCLRFLIPISKSDLSTSSMSDSSFRSALSCRHSYRSRSTYISSISAKET 0
>MEL2_takRub Takifugu rubripes (fugu) Deut.Acti.Perc genomic full Gq
0 MEPEDAHSPGSFINKVDVPDHVHYIVAFFVFVIGVLGMTGNVLVIFAFYS 2
1 NKKLRSLPNYFIVNLAVSDFLMASTQSPIFFINCLYKEWVFGEM 1
2 CKMYAFCGALFGITSMINLLAISIDRYVVITKPLQTIHWSSKRRTTLAILMVWLYSLAWSLAPLVGW 1
2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLAIILSCYLLMFLAVRKTSR 2
1 RKSTLIQQKSIRSEWKLAKIAFVVIIVYVLSWSPYACVTLISWAG 2
1 HANILTPYSKAVPAIIAKASTIYNPFIYAIIHNKYR 2
1 MTLVEKFACLWFLSPTARKDCMSSSSISESSIRDSIISRQSTTSRSHFIAGSTRMVNVKSEMSLILNGP* 0
>MEL2_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full Gq syn(KNTC2 +FLJ39155 +PDLIM5 +BMPR1B)
0 MEPDNAHTQRSFINKVDVPDHAHYIVAVFVVVIGTLGITGNALVMLAVYS 2
1 NKKLRNLPNYFIMNLAVSDFLMAFTQSPIFFINCLYKEWAFGET 1
2 GCKIYAFCGALFGIASMINLLAISIDRYLVITKPLQAIHWGSKRRTTLAILLVWLYSLAWSLAPLVGW 1
2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLGIILYCYLFMFLAIRKTSR 2
1 RKSTLIKQKSMKSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2
1 HADILSPYSKAVPAIIAKASAIYNPFIYAIIHNKYR 2
1 MTLAAKFPCLRFLSPTPRKDTSSSISESSYRDSVISRQSTASRTHFITACPDTVNFFFSFSSRLGCH* 0
>MEL2_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full Gq
0 MDPESTQGSLLSKVNVPDHAHYIVAMFVFVIGALGITGNALVMFAFYS 2
1 NKKLRNLPNYFIMNLAISDFLMAFTQSPIFFINCLYREWMFGET 1
2 GCKVYAFCGALFGISSMINLLAISVERYLVITKPLQTIHWSSKRKTTLAIFLVWFYSLAWSLAPLVGW 1
2 SSYIPEGLMTSCTWDYVTYTPANRSYTMMLCCFVFFIPLGIILYCYLRMFLSIRKTTr 2
1 RKSTVIQQKTIRSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2
1 HADILSPYSKAVPAVIAKASTIYNPIIYAIIHNKYR 2
1 MTLAEKVSCLRFLSPNPPKECTSYSFSESSFRDSIISRQSSVSRSHFINTLPSRADMVNDHKLVFI* 0
>MEL1_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XR_026330 frag G? opsin4 no cdna losing introns larval postoral arm ancestral intron
0 2
1 WTKSLRTPPNMLIVNLAISDFGMVITNFPLMFASTIYNRWLFGDA 1
2 GCQFYAFCGALFGIMSIANMTAIALDR 2
1 YYVICWSLEAVRSVTHRRSMIIIIIVWCYAIFWSIPPFFGVGSYVLEGYGLGCTFDFMTKDLNHYLHV
SFLFASSFVVPVTIIIVCFTRIAITVRAHRHELNKMRTKLTEDKDKKHKSSIRRANKAKTEFQIAKVGFQVTIFYVLSWM
PYSIVAVIGQYFDSDLLTPLGTVVPVIFAKCSAIWNPIIYCLSHEKFNAALKEKLMGMCGIEIPSKHRSMGSQESSVTGR
RGMHRQNSSTLSESSVTSTVDQDAIELKDRKQGPATVKVQQEKVEGGTYRRNPGDVTFSKDAGVEVDEKRRGDQGQRDDR
VRPQGEGQMDQWSQPPPAPASASAPTPGVNDKEYLTKM* 0
>MEL2_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu DQ285097 full G? cdna: S droebachiensis retained intron: tube feet
0 MPTTLMENSTPGWMADDSQMEETHPAFPLIGGYLLVVVLLGTAGNSLVIYTFLRFKKLHSPINLLIVNLSASDLLVATTG
TPLSMVSSFYGRWLFGTNACAFYGFVNYYCGCISLNSLAAISVFRYIIVVRGQAQNNKLSLRSSIYAILVIHLYTLIFST
PPLYGWNRFVLAGYHTSCDIDFHTKTPLFVSYICYMFFFLFFLPLGLISWSYFKIYQRVSKHSNSMRTSFTGVTKEINSDEKHA 2
1 NHRRTASTLFVTIVVFLFAWFPYCIVSLWVLIGDANSISKLSTTIPSLFAKSSVIYNPLIYVVLNSKFRKALIQTLSFLKCLSKHELSESS* 0
>MEL1_plaDum Platynereis dumerilii (ragworm) Ecdy.Anne.Poly AJ316544 11874910 full Gq rhabdomeric melanopsin unavailable genomically
0 MSRSEVLVPGSMSLDGLLTTAHPIGNDSI 0
0 ETILHPYWQQFDIENTIPDSWHYAVAAWMTFFGILGVSGNLLVVWTFLK 2
1 TKSLRTAPNMLLVNLAIGDMAFSAINGFPLLTISSINKRWVWGKL 1
2 WRELYAFVGGIFGLMSINTLAWIAIDRFYVITNPLGAAQTMTKKRAFIILTIIWANASLWALAPFFGW 1
2 GAYIPEGFQTSCTYDYLTQDMNNYTYVLGMYLFGFIFPVAIIFFCYLGIVRAIFAHHA 2
1 EMMATAKRMGANTGKADADKKSEIQIAKVAAMTIGTFMLSWTPYAVVGVFGMIK 2
1 PHSEMFIHPLLAEIPVMMAKASARYNPIIYALSHPKFR 2
1 AEIDKHFPWLLCCCKPKPKAQLPSSTTKGSIASKTEADTSV* 0
>MEL1_capCap Capitella capitata (polychaete) Ecdy.Anne.Poly genomic full G? jgi 119596 distally uncertain shares 2 introns with melanopsins
0 MMSYADGYLDNSTAPIEESPYLPHGTFFHPHWRPYREMLLNMNPLIYYGLGLYMAVVGIVGTLGNLVVITLFIK 2
1 TRSLRTPPNMFIINLALSDMGFCATNGFPLMTVASFQKLWRWGPV 1
2 ACELYALAGSITGFNSIATLALISMDRYMVIAKPFYAMKHVSHKRSLIQIILAWTWAFIWSAPPLLRMGYGRYIP
EGFQVSCTFDYLSRDLKNLIFVWCLFVFGFFIPVLAIACSYVGIIRAVGAQSKEMRKTAEKMGAKTGKSDKEKKQDIAMAKVA
AGTIGLFLMSWTPYAAVSMIGIAGNRSWITPYVSQIPVMFAKASAMWNPILYALSHPKFRAALEDHMPWLLVC* 0
>MEL1_helRob Helobdella robusta (leech) Ecdy.Anne.Clit genomic full G?
0 MDSVTWYKDFHPHWWKYHDIIVNAPMAFYYFLGTFFAVVGFLGVFGNIIVVWVFSR 2
1 TPSLRTPSNVLVINLAICDILFSALIGFPMSALSCFQRHWIWGNF 1
2 YCQFYSFVAGITGLASINCLAVIAVDRYLVVGQPLAMLNQSHFRRSFYHVLIIWTWACVWSAMPLIGWGEYILEGFGVSCTFD
YLTRTTWNISFNVCLFTFCFGMPVSVIILSYIGIIRSIAKNRKEFSSLTAENSSRARQEIKIAKVFAVCMTAFILCWVPYAT
VAQLGIYGYDQMVSPYTAELPVMLAKTSALWNPIIYAFSHPKYRKCLKELPIFRQKRKFRFLGSKSHTKSSDGVGTIALTSTINRTATRH* 0
>MEL2_helRob Helobdella robusta (leech) Ecdy.Anne.Clit genomic frag G? from scaffold_391
1 TPILRTHANVLIINLALCDLIFSSLIGFPMTALSCFKRHWIWGDL 1
2 GCDFYGFVAGWTGLGSITCLAFISIDRYMAIVHPFYMLNKKSSSVLTLLQIGAVWSWALIWSVMPLFGWGRFIPEAFGVSCTFDYLTRTWSNICFNYVLITCGFFLPVVIIVTSYIGIVIEVTKS 1
2 VEEDGMKDHMDAQNARCYVFVAVLSM 0
0 LKTAKVLACCFGAFLICWTPYAIVAQLGINGFAHLVTPFTSEVPVLFAKTSSIWNPLIYALSHPRYRRAV 0
>MEL1_schMed Schmidtea mediterranea (planaria) Loph.Plat.Turb AF112361 10220416 frag G?
0 eVYHYLVGVYISIVGISGVLGNLLVLYIFAR 2
1 AKSLRTPPNMFIMSLAIGDLTFSAVNGFPLLTISSFNTRWAWGKL 1
2 TCEIYGFIGGLFGFISINTMALISLDRYFVIAQPFQTMKSLTIKRAIIMLVFVWLYSLIWSTPPFFGY 1
2 GNYVPEGFQTSCTFDYLTQSKGNIIFNIGMYIGNFIIPVGIIIFCYYQIVKAVRVHELEMLKMAQKMNASHPTSMKTG 1
2 AKKADVQAAKISVIIVFLYMLSWTPYAIIALMALTGRRDHLNPYTAELPVLFAKTSAMYNPFIYAINHPKFRIQLEKKFPCLICCCPPKPK 0
0 * 0
>MEL1_schMan Schistosoma mansoni (trematode_worm) Loph.Plat.Trem AF155134 11166392 full G? 6 exons
0 MKQNLTFATLWPDDNDFASIVHSHWHKFIQPDPLYYYLVGIYIGIVGILAVMGNSLVITLFLL 2
1 CKQLRTPPNMLIVSLAISDFSFALINGFPLKTIAAFNHRWGWGKL 1
2 ACELYGFAGSIFGFISLTTMAFIALDRYLVIVQPFETFSRITYGKVIVMIFITWIWSALWSIPPFFGY 1
2 GSYIPEGFHTSCTFDYLSTDLPNLIFNAGLYILGFLCPVFIIIFSYYQIVKTVRLNELELMKMAQSLDLQNPSAMKTg 1
2 GDKKADIEAAKTSIILVLLYLMSWSPYAIVCLMTLIGSRDSLTPFHSELPVLFAKTSAVYNPIVYAVKHPKFRMEIEKRFPFLICCCPPKPK 0
0 ERLQNTIVSKIQVSQIGIGTVSGGNENTLNTVKRED* 0
>MEL2_schMan Schistosoma mansoni (trematode_worm) Loph.Plat.Trem CD096414 12973350 full G?
0 MSSNRTIEMLRPYMKDFDSIVLPYWYKFEQPNPYYQYAIGLFIAVVGITGMCLNLLVIVFFTM 2
1 FKSLRTPSNILVVNLAISDFGFSAVIGFPLKTMAAFNNFWPWGKL 1
2 ACDLYGLAGGLFGFVSLSTIAAVALDRYLVIATPFESVFQTTPRRTLLLMLFLWMWSLMWTIPPLFGF 1
2 GRYVTEGYQTSCTMDYISTDLNNRLFNIGLFGFGFLCPLFLSLFCYARIILIVRSRGKDFIEMAASSKGTNQKEKSANV 1
2 SSSKSDTFVSKSSAILLGVYLICWTPYSFVCLMALIGYADYITPLMVEIPCLCAKTANPCIYAFRYPKFRSLLQQRFGFLRLTKNRVSY 0
0 ERSQHAILSTIHVVTDCQYGTVSGGNENTLNTFILLD* 0
>MEL3_schMan Schistosoma mansoni (trematode_worm) Loph.Plat.Trem XM_002581128 full G?
0 MSEIKNFTRSLLLYNRTFSMIKNNIHDSDIIMLNHWIKYTQPDPIYNYLVAIFVALIGIFGTITNLLVIFVFL 2
1 TPKSSISLQCALIINLAISDFGFSAVIGFPLKTIAAFNQYWPWGSV 1
2 ACQLYGFISATFGFLSLTTIAAISFDRYLVIVKDHKTTNFRVICTVIGFLWIWSIIWTIPPFFGF 1
2 GRYVLEGYQTSCTFDYISNDMPSLLFSGGMYIFGFMFPVLLCIYCYVNLLKIVRNNERVVLISLSNDGASKQRESVR 1
2 NRKRLDIEATKSVILSLLFYLMSWTPYAMVCLISILGQSYFLTPTIAEMPHIFAKMAAIYNPILYAFTNRKFKNALGIRKTSSVIMQQQRLLSKGQLKPLVSLLFLVN* 0
>MEL1_lotGig Lottia gigantea (limpet) Loph.Moll.Gast FC774055 full G? ests long tail omitted 3 exons best: molluscan melanopsins
0 MTTLPPKENTTHLFDTWDDMFVHPHWKNFPPVSAAWHNFIGIFITFVGITGVIGNFVVIYTFSR 2
1 TKSLRTASNMFVVNLALSDLTFSAVNGFPLFSLSSFSHKWIFGRV 1
2 ACELYGLIGGIFGLMSINTMAMISIDRYLVITSPFTAMRNMTHKRAFLMIVGVWIWSILWAIPPIFGWGAYIPEGFQTSCTFDYLTRGDNRRSYIMCLYICGFVVPLGVIIFCYVFIIKSVMNHEKE
MAKMADKLDAKDVRSTKEKAKAEIKIAKVSMTIILLYLMSWTPYAIVALIAQWGPALVVTPYVSEIPVLFAKASAMHNPVIYALSHPKFRDAVSKLMPWFLCCCGLTDAEKKARDELAKSKFTRQT* 0
>MEL1_sepOff Sepia officinalis (octopus) Loph.Moll.Ceph AF000947 900420 full G? similar X56788 Loligo forbesi
0 MGRDIPDNETWWYNPTMEVHPHWKQFNQVPDAVYYSLGIFIGICGIIGCTGNGIVIYLFTK 2
1 TKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFIKKWVFGMA 1
2 ACKVYGFIGGIFGLMSIMTMSMISIDRYNVIGRPMAASKKMSHRRAFLMIIFVWMWSTLWSIGPIFGWGAYVLEGVLCNCSFDYITRDSATRSNIVCMYIFAFCFPILIIFF
CYFNIVMAVSNHEKEMAAMAKRLNAKELRKAQAGASAEMKLAKISIVIVTQFLLSWSPYAVVALLAQFGPIEWVTPYAAQLPVMFAKASAIHNPLIYSVSHPKFREAIAENFPWII
TCCQFDEKEVEDDKDAETEIPATEQSGGESADAAQMKEMMAMMQKMQQQQAAYPPQGAYPPQGGYPPQGYPPPPAQGGYPPQGYPPPPQGYPPAQGYPPQGYPPPQGAPPQGAPPQ
AAPPQGVDNQAYQA* 0
>MEL1_todPac Todarodes pacificus (squid) Loph.Moll.Ceph X70498 11106382 full Gq squid rhodopsin 3D Cys 337 palmitoyled
0 MGRDLRDNETWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTK 2
1 TKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWIFGFA 1
2 ACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFF
CYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVL
TCCQFDDKETEDDKDAETEIPAGESSDAAPSADAAQMKEMMAMMQKMQQQQAAYPPQGYAPPPQGYPPQGYPPQGYPPQGYPPQGYPPPPQGAPPQGAPPAAPPQGVDNQAYQA* 0
>MEL1_entDof Enteroctopus dofleini (octupus) Loph.Moll.Ceph X07797 full Gq
0 MVESTTLVNQTWWYNPTVDIHPHWAKFDPIPDAVYYSVGIFIGVVGIIGILGNGVVIYLFSK 2
1 TKSLQTPANMFIINLAMSDLSFSAINGFPLKTISAFMKKWIFGKV 1
2 ACQLYGLLGGIFGFMSINTMAMISIDRYNVIGRPMAASKKMSHRRAFLMIIFVWMWSIVWSVGPVFNWGAYVPEGILTSCSFDYLSTDPSTRSFILCMYFCGFMLPIIIIA
FCYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGASAEMKLAKISMVIITQFMLSWSPYAIIALLAQFGPAEWVTPYAAELPVLFAKASAIHNPIVYSVSHPKFREAIQTTFPWL
LTCCQFDEKECEDANDAEEEVVASERGGESRDAAQMKEMMAMMQKMQAQQAAYQPPPPPQGYPPQGYPPQGAYPPPQGYPPQGYPPQGYPPQGYPPQGAPPQVEAPQGAPPQGVDNQAYQA* 0
>MEL1_aplCal Aplysia californica (sea_hare) Loph.Moll.Gast AASC01108363 full Gq 4 exons uncertainties
0 MNVSSSLTSQPYHELLHPHWLEHEEAPEGVHLSVGVFITLVGVLAVCGNSLVIITCIR 2
1 FKDLRTRSNILIINLAVGDLLMCLIDFPLLAAASFYGEWPYGRQ 1
2 VCQMYAFLTAIAGLVTINTLAVIAADRYWAVVRRPTPGQKLPKCVTSIAVASVWAYSISWALCPILGWGAYVLDGIRTTCTFDFLTRTWENRSFVIGMMIGNFVLPFALMVFSYFRIWVAVRKVKSG 2
1 NVFCAIRHNYNLALGSTLFVKQHRYRLHCEQKTVKIIMFLLIAFTVSWSPYLAVSIIGLFGDRSQLTYQNTLTASLIAKTSMVFNPILYSISHPKVRKRIANLACCYSVRRHQQQTSRIKTGRRSTSSATPSRS* 0
>MEL2_aplCal Aplysia californica (sea_hare) Loph.Moll.Gast AASC02005512 frag G? 1st exon missing; new 3rd intron
0 2
1 HSSLRTSSNLLVVNLTVADLVMSSLDFPILAISSYKGCWVMGFL 1
2 GCQVYGVSSGVAGLVTINTLAAISVDRFVVVVHRLSPMHQMGKSTT 1
2 GVIIIAIWALSVIWAVLPITGVSSYRLEGMGTSCTFDYASRTSSNRWFFIALVIFNFFIPLALIIFSYWRIYASVRAVKRELKLLQSARTSILRKRFEVQAEI
KTAITALVIISIFCLAWTPYVIIAFVGLYGPTTAIDPLVSMFPNILAKISTVSNPILYSIGHPEVRKKMKKLFLPGQQDSSWRATSATAGNSSPAQSENMNNISLPTYL* 0
>MEL2_lotGig Lottia gigantea (limpet) Loph.Moll.Gast genomic full G?
0 MSIASHVWTNSSTNHFNFSVLHQHWQNQTPLSTACQYTIGIFISTVAVIAVIGNSIVIWAHVR 2
1 IKSLSTTSNMLILNLCVGCLIMCIVDFPLYATSSFLQKWIFGHK 1
2 VCEIYATITGTAGLLIMNSYSAIAFDRFITVTRYNNPNYPRSKSATMCISGFVWIYSLSWSMAPVVGWSRYQLDGSGTT
CTFDYLSTTWTNRSFILSIAFFNFVLPLCFILFAYSRILHLISSHSREMKSYRSAVIISKGKASIPKRFRSERKTAITLLI
TVVVFCLSWVPYVIIALIGQFGNQSFITPQISVIPQLVAKLSTVTNPILYSLSHPVVRNKLFLRLRHELYRRPSDSVSSSRGIQMKNIEFI* 0
>MEL1_patYes Patinopecten yessoensis (scallop) Loph.Moll.Gast AB006454 9287291 full Gq retina
0 MADNKSTLPGLPDINGTLNRSMTPNTGWEGPYDMSVHLHWTQFP PVTEEWHYIIGVYITIVGLLGIMGNTTVVYIFSN 2
1 TKSLRSPSNLFVVNLAVSDLIFSAVNGFPLLTVSSFHQKWIFGSL 1
2 FCQLYGFVGGVFGLMSINTLTAISIDRYVVITKPLQASQTMTRRKVHLMIVIVWVLSILLSIPPFFGWGAYIPEGFQTSCTFDYLTKTARTRTYIVVLYLFGFLIPLIIIGVC
YVLIIRGVRRHDQKMLTITRSMKTEDARANNKRARSELRISKIAMTVTCLFIISWSPYAIIALIAQFGPAHWITPLVSELPMMLAKSSSMHNPVVYALSHPKFRKALYQRVPWLF
CCCKPKEKADFRTSVCSKRSVTRTESVNSDVSSVISNLSDSTTTLGLTSEGATRANRETSFRRSVSIIKGDEDPCTHPDTFLLAYKEVEVGNLFDMTDDQNRRDSNLHSLYIPTRVQHRPTTQSLGTTPGGVYIVDNGQRVNGLTFNS* 0
>MEL1_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic full G? no close homologs
0 MTSSNDSAGYLWAINATIWIIDDSNETLGIDWDDWDVSLWTQEQRQLLEHGGIPRQVHVALGVLLSFIVLFGFAANSTILYVFSR 2
1 FKRLRTPANVFIINLTICDFLACCLHPLAVYSAFRGRWSFGQT 1
2 GCNWYGMGVAFFGLNSIVTLSAIACERYIVITSSSCRPVVAKWRITRRQAQK 0
0 VCAGIWLHCAALVSPPLLFGWSSYLPEGVLVTCSWDYTSRTLSNRLYYFYLLFFGFFLPVSVLTFCYAAIFRFILRSSKEITRLIMTSDGTTSFSKSTVSFRKRRRQTDVRTALI
ILSLAILCFTAWTPYTIVSLIGQFGPVDEDGELKLSPMVTSIPAFLAKTAIVFDPLVYGFSSPQFRNSVRQILRQQSISSSGNAGNRAGPNNMAMARTAIQNSRASSHATVSSF
SRNARMFPKDPLSKKTPNDPFVSTPLAVQQIPHFRLPTDVDINEQQFRRGIYANKSVSYWIDIIVLLQLGENLRKSCMKRKNSFKIPAGSIPQKNKLSNSRCSLLEDVSTHSLA
LRQMIFRKEGELYLFHHQPSHNAELAANKMDHQGNNKRIRRRFSEADMMHRSGKCRKNLPVSTSFDQ* 0
>UV7_ixoSca Ixodes scapularis (tick) Ecdy.Chel.Arac genomic frag G? exon 1 missing exon 2 disjunct K90 at EIP XM_002408275 wrong
0 2
1 RRRIRSQANLLVFNLALSDLLMVLEIPLLVYNSLKLRPALGVW 1
2 GCQLYGLMGGLSGTSAIFSIAALSLERYLALGRPRDPFARLTRSRAFALSLSSWIYALCFSAWPLLGVTSPYVPEGFLTSCSFHFLSDATSDRCFVWIFFVAAWCVPLVFVTTCYSGILVTVIRSRKALAQES
RRSELRVAKVSLALVLLWTVAWTPYAIVALLGITGRRNLLTPWGSMAPAMFCKSAAVLDPFVYGLSHPSFRRELAIMLPCLRPRQRPVSLTLRAVVQLPKRPGPRSAGSSTSVPVTAPGTTKDNHCPTPPNVSR* 0
>UV7_tetUrt Tetranychus urticae (spider_mite) Ecdy.Chel.Arac GW013907 frag G? larval transcripts
AGCKIYGFFGGLSGTSAIMTIAMMAMERLNSISKPLSPHSRLSMRRALVISTLIWIYSLIFSTPPVFHLFNRYVPEGYLTSCSFDYLSKDLVSRIYIFAFFTAAWIVPLTLIVTSYVGIIIAVERNETMFQKTQDRLCERAS
SVYRASLRDSRGRSVKESGKMEMRLVKIVLSLIAFWILSWTPYAIIALLGLLDQSYITPTSSMIPAIFAKLSSVVNPYLYGLGNPRFKRELKRKLCFCVANKASVERRTVIQSTVTLGDPKEGTQFVHISDGNLSEIELSDYENR* 0
>UV7a_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran frag ACJG01004266 tandem1
0 MLNHTHSISRETIHGNAICDQHRKFVNLYPVDVWKSSGFFDDIDLMDINCHWLQFEPPTPSSHFMLAIIYSVVFVIGFVSNTAIVYILLR 2
1 SKKLLTPANILLVNLAISDLLIISMIPIFLYNSFLQGPAIGGL 1
2 GCKVYGFLSGFSGTSTILTLAAIAIDRYLVISRPLDLAKKPTRTWAYATIAITWLYSATFASLPFLGAGKYVPEGYLTSCSFDYLSEDTGTRVFIFIFFVAAWVCPLT IITFCYAAITYVNICSTNTARPLIIIQHKYAGWRQCNIEIAIPKIVAGLVLSWIIAWTPYSLISLLGISGHTDLLTPLSSMLPALFAKTAACVDPFIYSLNHPKIRQEII
>UV7b_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic ACJG01004266 full tandem2
0 MNAINMCNQTNCATDVVTDDIIERWKLCGFFEDDDFLDINCHWLQFEPAPLVNHLLLVCIFVFILLTGCLCNIIVIYIFAR 2
1 SRHLRTPANIIIMNLAISDFFMLIKMPIFLYNSLLQGPALGIK 1
2 GCQIYGFMSGLTGTTSIMSLATVAVDRYLVISQPLNVNRKSTRTRAYLTTCFIWLYSAIFASLPLFGIGKYVPEGYLTSCSFDYLSNNVKTRSFILAFFLAAWLFPFCIITTCY
TAITYVNICSTNTARPLIIIQHKYAGWRQCNIEVVIAKIVVVLVAMWMISWTPYAFVALLGISGTQTRLTPGMTMFPALFAKFSACVNPIIYTLTHPKIKKEILRRWYCFMSSGTS
>UV7a_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD4798 full G? 3 exons altered HEK K90
0 MIDFKTKYPVNLWKDHGLYTDDYIKLINSHWLKFMPPNPTSHYVLGLLYTVIMVFGCTGNSLVIFMYFK 2
1 CRSLQTPANMLIINLAVSDFIMLAKASVFIYNSYYLGPALGKL 1
2 GCQVCGFLGGLTGTVSIMTLAAISLDRYYVIVCPLKAAVKTTKQRARIWIGLIWIYGFSFSIVPVLDLGYSRYVSEGYLTSCSFDYLSDNDQDKRFI
LVFFTAAWCIPFTIILYCYVNILMAVWMTTEIVTSRVGQQEEKRKTDIRLGYMVIGALALWFVSWTPYAVVALLGVFDLKEYISPLSSMIPALFCKAAS
CTDPWFYAITHPRFKKELMKLLTKSKSRKLVRNYGMKKGWVGSHLNKNGSVDFDNCLKTEYKEENTTIFMLESDDNNLHCQGSTSGHKTESTKEPETKFTASASQETLKYMLPS* 0
>UV7b_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD14504 full G? altered HEK CL3 K in in K90
0 MSDFKTKYPIDTWKEHGFYTDDYMKLINSHWFKFMPPNATSHYILGFLYSVIMVLGCFGNSLVIFMYIK 2
1 CKSLQTPANVLIMNLAVSDFIMLAKTPVFIYNSFYQGPTLGKL 1
2 GCQIYGFFGGLTGTVSIMTLAAISLDRYYVIVHPLNAAVKTTKQRARVWIGLIWIYGFLFSIIPVMDLGYNRYVPEGYLTSCSFDYLSDDNQEKGFILVFFTAAWCIPFTTISYCYIKI
LRAVWMTSEMAASRFGQEEEKRKTEIRLGYVVVGVIMLWFVSWTPYAMVALLGVFDRKDYITPLSSMIPAVLCKAASCMDPWIYAITHPRFKNELTKLMSRKKTRKLERDYGMKKNWGGQ
SYSNKSGAGLRNLSSSEDECVEEVIVVIDPDDKKMKRQGSTSSHKTEETKALETKFPPTRQESLKYMPPSWYKLPRTTSKSSIMLDPKLTGDDNNK* 0
>UV7_rhoPro Rhodnius prolixus (kissing_bug) Ecdy.Inse.Hemi genomic full G? K90 at KMP ortholog RH7 of droMel
0 mKYFHLYPIEQWKMHRFFTEEYLKLVNTHWFEYPPPNKQIHYIFAAVYFLVMLVGVSGNLLVIFMILR 2
1 FRTLRTSSNILILNLAVSDFLMVAKMPVFIYNSFYFGPVLGEM 1
2 GCHFYGFIGGLSGTASILTLAAIAMDRYLGIAHPLNFNQGRAKKRTIVWITFIWVYSITFASIPLSHIGVKTYVPEGFLTSCSFDYLSTDIQNRCFIFIYFVAAWCLPLLVIITSYVGICREVLRVSLIRKGQE
REQRKREAKLSAILALATFLWFLSWTPYAAVALLGIFGYKNHITQLASMIPALFCKTAACVNPFIYGLNHPRLRQQLLKLCCKKRYNLEKTHFSRSWRNTSCSFKLKEQSLCNVSQSRLRRTSTVASEPSEHSTHFM* 0
>UV7_pedHum Pediculus humanus (louse) Ecdy.Inse.Phth AAZO01007270 full G?
0 mKTFKLKWPEEWKKLGLFDDEYLYKINKYWMKFPPPSPMSHYFMGIIYSVIMVVGVFGNFLIIYLFLR 2
1 KRSLRTPSNVFIFNLAVSDSLLLLKMPVFIINSFYLGPALGNL 1
2 GCSAYGFVGGLTGTVSIMTLAAIAFDRYQVIVHPLERKTKAAVYFQILLIWIYAIFFSIIPLLDVGLNKYVPEGYLTSCSFDYLTQDTASRLTIFVFFVAAWIVPLSIILGSYM
ALYKVVLKARGTHFNTVMTRHCKDIEIQRPELKAAVTVICIVCLWTLSWTPYAVVALLGITGNEKYISPMSSMIPALFCKTASCIDPFVYAATNRRFRNELKRKYRKRSRYQPSLKTE
RKDFFTLSEDNNDRGKGNTIRIREK* 0
>UV7_spoLit Spodoptera littorali (butterfly) Ecdy.Inse.Lepi FQ016398 FQ021555 Cfrag male antenna
MFSTIGCWGNIIVLLMYLRCRTLRTPGNILVANLALSDFLMLAKTPIFIFNSFNLGPALGKTGCVIYGFVGGLTGTTSIATLSAIALDRYWAVVRPLEPL
RALTAIRARLLAIGAWLYACVFAIIPALDIGYGSYVPEGYLTSCSFDYLTEDFSPRCFIFVFFCAAWLAPFCTITFCYISIFRVVVFNTSMSYGNQDQRL
SSRHVKERTKRKAEIKLAFLVMAVIALFFISWTPYAIVALLGITGKKDVLTPITSMIPALFC
>UV7_mayDes Mayetiola destructor (hessian_fly) Ecdy.Inse.Dipt AEGA01002257 frag genomic
1 VYFLHSSYNGYPDTKVLESINIHWIGFNPPKPWEHYTLAIMISIIMTIGLIGNFLVICM 2
1 SRILKRASNILLLNLAFADFFMLAKSPIYIYNCLKFGPASGDL 1
2 ACRAYGFIGGLTGTVSIMTLSAISFDRYMGIMYPFNTEKFGNKKIRAFILVAFVWFYSFTFAIMPALNIGLSKYTPEGFLTSCGFDYLDNQTSARIFMFTFFIGAWMIPFLTITYCYIQIFRV
VNLTRQIQSNRKRQRAEQKLAIVVMNLIAIWFFAWTPYAIVALLGIAGQQQYITPWASMIPALFCKASACINPYIYSIRYPEFKQEIFRILSKYFKFADEHLQQNATRTMSVHFVTRTRVQINEQY* 0
>UV7_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_308329 full G? K90=EAP
0 MGRQGSGNAVRISPSSRNQPYFSSAHLSFVVPFPVHSKYVVRSGYVLPVDPLFVAKINPFWLRFDPPSAGEHYGLAVFYFLMMLFGVIGNALVVFMFYR 2
1 YRSLRTPANYLVINLAVADFIIMMEAPMFIYNSIHQGPALGSI 1
2 GCTVYALMGAVGGTVAIATLTVISIDRYNVVVYPLNPNRSTTKLKCYFLIAFTWAYGLLFASFPALEIGLSRYTAEGYLTACSFDYLDRTYKARVFMFVYFVFAW
LIPFAIISYCYARILIAVINANAIQSSKSKNKTEVKLAGVVVGIIGLWFAAWTPYAVVAMMGVFGYEQYLTPLNSMIPAVFAKIAASIDPYFYAMNHPRYRQMLER
MFCNRGADQGNSQYQTSHYTRGASRGGDSEGGGGEESGGGGGVGRAPGGGNAGLGRGGTVRGGGGGGRLIAGKGGGGANATGSTGGGGVKALKKQISNGDETSLEVSLEM* 0
>UV7_aedAeg Aedes aegypti (mosquito) Ecdy.Inse.Dipt XM_001650694 frag G? wrong exon 1 K90=EAP
0 FPPNSRYMALSGYSGPTIEDAFRDRINPFWLQFDPPSRTAHYILGFIYFMMMMFGLCGNLLVILMFFR 2
1 FKSLRTPANYLVINLAIADFIIMLEAPLFVYNSYHQGPATGNVWCTIYALLGAVGGTVAIVTLTMISIDRYNVVVYPLNPKRSTTRLKVALMIVF
AWIYGLVFSVIPALDIGLSRYTPEGFLTACSFDYLERTRDARLFMFLYFIFAWVVPIIAITFCYIQILRVVIGANSIQSSKNKSKTEVKLAGVVIGIIGLWFIAWTPYAIVAMMGV
FGYESLLSPLGSMVPAILAKTAACIDPYFYAMNHPRYRQELRKMFGLNQQDLGNSQYQTSRYTRNASRMDDSEGGASERVTIGRQPGKTTTDEPEPSQQTEQGPQPTYSKNLAANS
RGALQRAQSSISAADDTSLSVSIDLTETNPNSNH* 0
>UV7_culQui Culex quinquefasciatus (mosquito) Ecdy.Inse.Dipt XM_001861603 frag G? wrong exon 1 K90=EAP
0 PEPVHPASKYISLSGYDGPPVEDAFRDRINPFWLQFEPPSPVAHYALGFVYFLMMVWGLFGNVLVIFMFFK 2
1 FKSLRTPANYLVINLAVADFLIMLEAPIFVYNSYHLGPAFGNTL 1
2 CTIYSLLGAIGGTVAIMTLTMISVDRYNVVVYPLNPNRSTTRLKVMLMIVFTWIYALVFSL
MPALEIGLSRYTPEGFLTACSFDYLDRGWDARVFMFMYFVFAWVIPFLTISYCYVAILRVVVGAGSIQSSKNKNKQEVKLAGVVIGIIGLWFIAWTPYAVVAMLGVFGYEHLLTPL
GSMIPAILAKTASCIDPYFYAMNHPRFRQELRKMFGKEQEMNHSQYQTSRYTRNASRNDSEAGPSERVQLGRAPGKDADPIPAVSSSVAQPNYSQNLASNRKGGLQRAQSSISAAD
DTSLSCSIDLTETQPNNH* 0
>UV7_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BGIBMGA012539 full G? kaikoblast
0 MSEILDRRSNVSGEVKETRLPIKYSKYEVNELVSSEANNEVLIKKFKELWPVNLWRRFGLFTDDYLHLVNPYWLSFAPPHPLLHYGLGAFYIMMTTVGSTGNALVMFMYFR 2
1 CRSLRTPGNILVANLALSDFMMLAKTPIFIFNSFNLGPALGKT 1
2 GCVIYGFVGGLTGTTSIAILTAISLDRYWAVVRPLEPLRALTAIRARLMAIAAWIYATLFAIIPALDIGYGRYVPEGFLTSCSFDYLTEELPPRYFIFVFFCAAWLAPFCTISYC
YISIFRVVVCSRNITTKNQEQRLSTRHVKERTKRKAEIKLAFLVIVVIALFFISWTPYAIVALLGIFGKKDLIKPLTSMIPALFCKTAACINPFIYIITHPKFRKEFKRMLSRDKTE
RSGTIKTIGYYTEASKFHRPSKDFSDTEVEIVEMKDIPYRNEDTTNTVEEKINTVSAKLAKREGSQKSQSSISMKSFEENVVSPPSWYCKPKFAKKKSFHRRSTHSMVSQNTDPTV* 0
>UV7_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_079311 full G? RH7 long N-terminal has M comp genomics support 2 of 3 exons novel
0 MEAIIMTTLPNLTTDAGDSSFWLTGALSLSEMLANSSHSHSTGSTTSTAGSSATESSAVNVGKDHDKHVNDSVSTGLS 2
1 NYSNYPSYIHYRDKYDLSYIAKVNPFWLQFEPPKSSTFLIMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDI 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKEMPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGEGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNSISSVMEQSKF* 0
>UV7_droYak Drosophila yakuba (fruitfly) Ecdy.Inse.Dipt genomic 12209654 full G? RH7 chr3L:12207286 12209654
0 MEAIIMTTLPALTTDAGDSSSFWLTGALSLSEMLANSSHGHSTGSTSSTAGSSATESSTVNVGKDHDVTKHVNDSVSTGLS 2
1 NYSNYPSYIHYRDKYDLSYIAKVNPFWLQFEPPKSSTFLVMAALYFLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLIKCPIAIYNNIKEGPALGDI 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLTSIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGLERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGVLRRVSTTRSSYMTRSRSSFTHRLRTSTTGDGGMGDHRMENYLMNNNLMMVPEETEENEEIVVVAEINNSVSSVMEQSKF* 0
>UV7_droAna Drosophila ananassae (fruitfly) Ecdy.Inse.Dipt genomic full G? RH7 scaffold_13337 frameshifted
0 MEAIILSTLPSLTTNASGSSSHWLTGALSLPEILANSSGSPNTSSADTGSGINLSARDADRHFNISTEAR 2
1 NYSYYPGYIHYRDKYDLSYIAKVNPFWLQFEPPHSSTFLAMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDV 1
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIILLIWCYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCFPLTAIVYSYFYILKVVFSAGRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAVVAMMGVFGLEKHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFYGRGILRRVSTTRSSYMTRSRSSFTHPAGRADGGTGRDHRMETYLMNNNLMMVPEETEENEEIVVVAEINNSVSSAIEQSKF* 0
>UV7_droPse Drosophila pseudoobscura (fruitfly) Ecdy.Inse.Dipt genomic full G? RH7 chrXR_group6:2491547 2493151
0 MEALMAALPTLTTEAAGSSLWLTSALSLSEMLANSSTSPNASLVAATTSSAAVATASTTSAAEAVGKVPDKHEVNDNVSTVLS 2
1 TSSSYPGYIHYRDKYDLSYIARVNPFWLQFEPPKSSTFYLMAALYCLISVVGCVGNAFVIFMFANRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDA 1
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLIIFLIWSYSFLFAVMPALDIGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCVPLTTIVYSYFYILKVVFTASRIQSNKDKAKTEQ
KLAFIVAAIIGLWFLAWSPYAIVAMMGVFGQERHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLFFGRGVLRRVSTTRSSYMTRSRSSFNRRLRPTPDAEHRVESYLMNNNLMMVPEETEENEEIVVVAEFNNSSYSGMEQSKF* 0
>UV7_droWil Drosophila willistoni (fruitfly) Ecdy.Inse.Dipt genomic full G? RH7 scaffold_180949
0 MDMDMALDMNDAATTTSLWITSAALSLSEILVNTTSHVVTTSPASTSTVETTAVAAVTATGKVVHDDEKHHHHHHHHHQDEVNDNNVTTVLR 2
1 NFSSYPGYIHYRDKYDLSYIAKVNPFWLQFEPPRSSTFYIMAALYCLISVVGCIGNAFVIFMFSNRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIKEGPALGDI 1
2 ACRIYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRCSRLRSYLVIVMIWCYSFLFAIMPALDVGLSVYVPEGYLTTCSFDYLNKETPARIFMALFFVAAYCVPLTCIMFSYFYILKVVFTANRIQSNKDKAKTEQ
KLTFIVAAIIGLWFLAWSPYAVVAMMGVFGLEQHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLIYGRGVLRRVSTTRSSYITRSRSSFTRRLRTGSELDMRTEPYIMNNNLMMVPEETEENEEIVVVAEINNPSRCVSMHEHTSKF* 0
>UV7_droMoj Drosophila mojavensis (fruitfly) Ecdy.Inse.Dipt genomic full G? RH7 scaffold_6680
0 METIMSTLPTLTADDGSLWITSALTELLASGANSSSGSSSVVADGTQNATFVAAATTTTTTVAAAAAAAAAAAVNASTATTANATKGHHKHPHGVNDSETDLR 2
1 LCSSYPGYIHYRDKYDLTYIAKVNPFWLQFEPPDTSTFYIMAALYCLISVVGCVGNAFVIFMFGSRKSLRTPANILVMNLAICDFLMLVKCPIAIYNNIQEGPALGDA 1
2 ACRLYGFVGGLSGTCAIGTLTAIALDRYNVVVHPLQPLRRYSRLRSYLIIFAIWCYSFLFAVMPALDVGLSVYVPEGFLTTCSFDYLNKETPARIFMALFFVAAYCIPLASIVYSYFYILKVVFTANRIQSSKDKAKTEQ
KLTFIVAAIIGLWFIAWSPYAIVAMMGVFGQEQHITPLGSMIPALFCKTAACVDPYLYAATHPRFRVEVRMLMYGRGVLRRVSTTRSSYMTRSRSSFTHRLRPSSGDCENRAEPYTLNNNLMMVPEETEENEEIIVVAEINNSISGVMEQSKF* 0
>UV5_plePay Plexippus paykulli (jumping_spider) Ecdy.Chel.Arac AB251851 18217181 full G? PpRh3 kumopsin3
MLNNTIPGPATLDDIGPPSWCYETRFNGWNAAPDIYVPDYWKQFRAPAPYLHYMLGFFYICLMSIAVV
GNAIVMYIFFSAKTLRTPTNMFVIGLAMADLLMMSKTPVFIYNCFHLGPVFGQIGCDIYGIVGTYSGIGSAFCNAIIAYDRYRVIVHP
FSKSGMSITKAIAFLVIIYLYITPFAILPALKIWSRYVPEGFLTSCSADFFMQDFNGRSYIV
GTWFFGWFIPVAAIVFFYVQIFLAVKDHEEKIKEQARKMNVDSIRSNEAVKNSSAEVRIAKTAMCVFLMFLSSWAPYILVAFITGFSDPKLKRITPVISMVPAMTIKASACFDPFF
YALSHPRYRLELQNRMPWLCINEKAEASGPADDSVSKTTEHVA*
>UV5_hasAda Hasarius adansoni (jumping_spider) Ecdy.Chel.Arac AB251848 18217181 full G? HaRh3 kumopsin3
MLNNTALQPAVLDDIGPPSWCYETRFNGWNAAPDILVPDYWKQFRAPAPYLHYILGCLYICLMSVALI
GNAIVIYIFSVSKSLRTPTNMFVIGLAMADLLMMSKTPVFIYNCFHLGPVFGQLGCDIYAIVGTYSGIGSAFCNAVIAYDRYRVIVHP
FSKSGMTMTKAIAILVIVYLYITPFAILPALKIWSRFVPEGFLTSCSSDFYMQDFNGRSYIV
GTWFFGWFIPVAAIIFFYAQIFLAVKDHEEKIKEQARKMNVDSFRSNEALKNSSAEVRIAKTAMCVVLLFLTSWVPYILVAFIAGFSDPKLKRVTPVISMIPAMTIKGSACFDPFF
YALSHPRYRLELQNKLPWLCINEKAEASGPADDSVSKTTEHVA*
>UV5_braKug Branchinella kugenumaensis (fairy_shrimp) Ecdy.Crus.Bran AB293436 18984904 full G?
MANVTGFDYYRYERRELGWNTPAEYMEFVHPHWKQFEAPNPFLHYMLGVFYIIFMFCSLIGNGVVIWVFASAKSLRTPSNLFVINLAVLDFLMMLKTPVFIVNSFNEGPIWGKTGCDFFALLGSYAGIGGATTNAAIAFD
RYRTIAHPFDGKLSRGQAITLCMLCWLYATPFSLMPFFGIWGRFVPEGFLTTCSFDYITEDSSTRAFVGTIFFTSYVLPMILIIYFYSQIVGHVRQHEETLRAQAKKMNVATLRSGKDDQEQSAEVRIAKVCIGLFSMFV
ISWTPYAAVALLCAFGNRAAVTPLVSMIPALTCKAVACIDPWIYAINHPRYRLELQKRLPWFCIHEPEPNNDSASVNSEKTVATTTPS*
>UV5_triLon Triops longicaudatus (tadpole_shrimp) Ecdy.Crus.Bran AB293435 18984904 full G?
MAFLQNQTYDGTSPSFSFFRTSERIMLGHNTPKDYMEYVHPHWQTFEAPNPFLHYLLGVLYIGFMFCALVGNGVVIWIFSSAKSLRTPSNMFVINLAVLDFIMMMKTPVFIVNSFNEGPIWGKFGCDLFALMGSYSGIGG
AMTNAAIAFDRYRTIARPFDGKLSRGKVLTICAGIWLWATPFSLMPLFGIWGRFVPEGFLTTCSFDYMTETSSIRWFVGCIFTYSYIIPLGLIIYYYSKIVGHVQEHERILREQARKMNVESLRSGKDQQEKSAEIRIAK
VAIGLSLMFVVAWTPYALVALIAAFGNRAVLTPLVSMIPACCCKAVACIDPWIYAINHPRYRLELQKRMPWFCVHEPEPEMIDNSSAITEKTST*
>UV5_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran AB293430 18984904 full G?
MAFLQNQTHDGTSPSFSFYRTSERVMLGQYTPKDYMDYVHPHWQTFEAPNPFLHYLLGVLYIGFMFCALVGNGVVIWIFSSAKSLRTPSNMFVINLAVLDFIMMMKTPVFIVNSFNEGPIWGKFGCDMFALMGSYSGIGG
AMTNAAIAFDRYRTIARPFDGKLSRGKVLTICAGIWLWATPFSLMPLFGIWGRFVPEGFLTTCSFDYMTETSSIRWFVGCVFTYSYIIPLGLIVYYYSKIVGHVQEHERILREQARKMNVESLRSGRDHQEKSAEIRIAK
VAIGLSLMFVVAWTPYALVALIAAFGNRAVLTPLVSMIPACCCKAVACIDPWIYAINHPRYRLELQKRMPWFCVHEPEPEIIDNSSAITEKTST*
>UV5a_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran FE384049 full G?
0 MLGWNTPEDYMSYVHP 2
1 YWKTFEAPNPFLLYMIGFLYTIFMFCCVAGNGVVIWIFTN 2
1 CKSLRTPSNMLVVNLAILDMLMMLKSPVMIINSYNEGPIWGKLGCDVFGLMGSYNGIGSAVNNAAIAYDRHR 2
1 TISRPLDGKLSRKQVTLMIVAIWAWATPFSVMPFLGIWGRYVP 1
2 EGFLTTCTFDYMTEDASTRFFVGSIFVYSYVIPLAMLIFYYSKIVRSVGDHEKTLRDQAKKMNVTSLRSNRDQNEKSAEVRIAKVAIALATLFVFAWTPYAFVALTAAFGNR 2
1 SVLTPLLSTVPACCCKLVSCINPWIYAINHPRYR 2
1 MELQKKMPWFCIHEPVPTNDDSSVGSATTEMSGVSKETSS* 0
>UV5b_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran genomic full G? penultimate intron lost last intron has slid back 2 aa
0 MNGWNTPADYKSYVHPHWLSYEEPNPMLHHLLGVLYIFFMIASCLGNGIVIYIFST 2
1 TKELKTPSNILILNLAICDFIMMIKTPIFIVNSFNEGPVFGRLGCSIFGLLGAYVGPCSAVTNAAIAYDRYR 2
1 CISDPMGKRWSKSQASLIVLGCWVYASPVSLLPFTEIVNRFVP 1
2 EGYLTSCTFDYMTDNLETKMFVFILWIWCWIMPLGVIIFSYGKITTQVMTHEARLKEQAKKMNVESLRSGANKDARNEIRVAKVGISLTTLFLLSWTPYFAIAFIGCYGNR SLLTPGLSMIPACTCKMAACVDPFVYAINHPK 2
1 YRLELMKRFPWLCVHEKDDSTRSENSTNATIASEAESRT* 0
>UV5_triCas Tribolium castaneum (flour_beetle) Ecdy.Inse.Cole genomic full G?
0 MYVVHPFKIIRNKVTILRTMETMANHLGWNVPKDELIHIPQHWLVYPEPEASMHFLLALIYIGFFIMATIGNGLVIWIFST 2
1 SKSLRTASNMFVVNLAICDFAMMIKTPIFIYNSFYRGFALGHLGCQIFAFIGSLSGIGAGMTNACIAYDRYT TITRPFDGKITRTKALVMIIFVWGYTIPWAVMPLLEIWGRFAP 1
2 EGFLTACSFDYLTDTFDNHMFVTSIFICSYVIPMSMIIYFYSQIVSKVFSHEKALREQ 0
0 AKKMNVESLRSNQSQQASQSAELRIAKAAIAICSLFVASWTPYAVLALIGAFGDQSLLTPGVTMVPACACKFVACLDPYVYAISHPKYR 2
1 LELQKRLPWLAIKETAASETQSTTTENTTTQSATTTT* 0
>UV5_lucCru Luciola cruciata (firefly) Ecdy.Inse.Cole AB300329 19232386 full G?
MILHNATVFAAAQTQDDPDSIVHLLGWNVPKSELHHIPEHWLVYPEPEASIHYLLGIVYIFICFMGIVGNGLVLWIFSTSKSLKTASNMFVVNLAFCDFIMM
MKMPIFVYNSFNRGYALGHIGCQIFGFVGSLSGIGAGMTNAFIAYDRYATISNPLEGKLTRTKALIMIFIIWGYTFPWAVLPMFEVWCRFVPEGFLTSCTFDYLTDTFDNDMFVAV
IFICSYVIPMSMIIYFYSQIVKHVMHHEKALRDQAKKMNVESLRSNQSLQSQSIEIKIAKVAIMVCFLFVASWTPYAVLALIGGFGDQSLLTPGVTMVPALACKFVACLDPYVYAL
SHPRYRMELQKRLPWLAIKEDAVSDAQSMVTTTTAAATPAATEQAPTA*
>UV5_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_556823 full G? novel short exon
0 MGLVQLDNQTAYRPEALIGADQSGLRYLGWNVPPEELVHIPEHWLQFPEPEASLHYLLGLLYIAFTIFSLVGNGLVIWIFIA 2
1 AKSLRTPSNVFVINLAICDFFMMAKTPIFIYNSFTKGFTLGNLGCQIFGFVGSLT 1
2 GIGAGATNALIAYDR 2
1 YNTITRPFEGRLTQTKAIIFICLIWAYTIPWGVLPLLEIWGRYVP 1
2 EGFLTSCTFDYLSGTFDTRLFVASIFTFSYVLPMSLIIYYYSQIVSHVVNHEKSLREQAKKMNVESLRSNQNQKDASVEIRIAKAAITVC
FLFVASWTPYAVLALIGAFGDKSLLTPGVTMFPACACKFVACLDPYVYAISHPRYRIELQKRLPWLAITETLPAENASTCTEQQDGNATTQS* 0
>UVB_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_312478 full G?
0 MFLGNESISEGAMLMPMARTAGEMPKLLGWNLPPEEQYLVHDHWKGFPSPPYYMHLMLAMIYFVLMNTSLIGNGIVLWIFGT 2
1 SKSLRNGSNMFIINLAIFDLLMMCEMPMFLVNSFSERLVGYGVGCSVYAALGSMSGIGGAISNAVIAFDRYRTISNPLDGRLSRVQAGLLICLTWLWTMPFTLLPLFEIWGRY
IPEGYLTTCSFDYLTDDPDTRVFVGCIFTWAYVIPMIFICYFYARLFGHVRQHEMMLKNQARKMNVESLTANRSEKAQAVEMRIAKAAFTIFFLFVCAWTPYAIVTMIGAFGDR 2
1 TMLTPFVTMVPAVCCKIVSCLDPWVYAISHPKYRQELERRLPWMGIKEADDSVSTTES* 0
>UV5B_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_057748 19398933 full G? RH5 two small introns also seen in Apis and Daphnia
0 MHINGPSGPQAYVNDSLGDGSVFPMGHGYPAEYQHMVHAHWRGFREAPIYYHAGFYIAFIVLMLSSIFGNGLVIWIFST 2
1 SKSLRTPSNLLILNLAIFDLFMCTNMPHYLINATVGYIVGGDLGCDIYALNGGISGMGASITNAFIAFDRYKTISNPIDGRLSYGQIVLLILFTWLWATPFSVLPLFQIWGRYQP 1
2 EGFLTTCSFDYLTNTDENRLFVRTIFVWSYVIPMTMILVSYYKLFTHVRVHEKMLAEQAKKMNVKSLSANANADNMSVELRIAKAALIIYMLFILAWTPYSVVALI
GCFGEQQLITPFVSMLPCLACKSVSCLDPWVYATSHPKYRLELERRLPWLGIREKHATSGTSGGQESVASVSGDTLALSVQN* 0
>UV4_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_057353 19949290 full G? RH4 one ancestral intron with intercolated genes
0 MEPLCNASEPPLRPEARSSGNGDLQFLGWNVPPDQIQYIPEHWLTQLEPPASMHYMLGVFYIFLFCASTVGNGMVIWIFST
SKSLRTPSNMFVLNLAVFDLIMCLKAPIFIYNSFHRGFALGNTWCQIFASIGSYSGIGAGMTNAAIGYDRYNVITKPMNRNMTFTKAVIMNIIIWLYCTPWVVLPLTQFWDRFVP 1
2 EGYLTSCSFDYLSDNFDTRLFVGTIFFFSFVCPTLMILYYYSQIVGHVFSHEKALREQAKKMNVESLRSNVDKSKETAEIRIAKAAITICFLFFVSWTPYGVMSLI
GAFGDKSLLTPGATMIPACTCKLVACIDPFVYAISHPRYRLELQKRCPWLGVNEKSGEISSAQSTTTQEQQQTTAA* 0
>UV3_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_079687 9165115 full G? RH3 single exon
0 MESGNVSSSLFGNVSTALRPEARLSAETRLLGWNVPPEELRHIPEHWLTYPEPPESMNYLLGTLYIFFTLMSMLGNGLVIWVFSAAKSLRTPSNILVINLAFCDFMMMVKTPIFIYNSFH
QGYALGHLGCQIFGIIGSYTGIAAGATNAFIAYDRFNVITRPMEGKMTHGKAIAMIIFIYMYATPWVVACYTETWGRFVPEGYLTSCTFDYLTDNFDTRLFVACIFFFSFVCPTTMITYY
YSQIVGHVFSHEKALRDQAKKMNVESLRSNVDKNKETAEIRIAKAAITICFLFFCSWTPYGVMSLIGAFGDKTLLTPGATMIPACACKMVACIDPFVYAISHPRYRMELQKRCPWLALNEKAPESSAVASTSTTQEPQQTTAA* 0
>UV5_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD14509 full G? 8 exons K90
0 MDFNRTVSRPLAQLGs 2
1 SLMENEVGETHLLGWNLQAEDLIHIPEHWLKYQEPSSLQHYYLAFMYTIFMFVALFGNGLVIWVFCV 2
1 AKPLRTPSNIFVINLALCDFVMMAKAPIFILGSINRGYQ GHFLCQLFGTAGAFSGIGASATNAAIAYDRFS 2
1 TIAKPFDGRMTYGRAFFLIICIWTYTLPWGLLPLTEKWNRYVP 1
2 EGYLTSCTFDYLSPTDETRAFVGIMFVICYVIPVSLVIFFYSQIVSHVFNHEKALREQ 0
0 AKKMNVESLRSNQDANAQSAEVRIAKAAITICCLFIASWTPYAVVAMIGAFGDR 2
1 SLLTPGITMIPAIFCKTVACFDPYVYAISHPRYR 2
1 LELSKRVPCLGISEKPPPTASETQSTTTAA* 0
>UV5_rhoPro Rhodnius prolixus (kissing_bug) Ecdy.Inse.Hemi genomic frag G? exon 1 missing K90 at KTP
0 0
1 ASTSGNIRTLGWNLSPEDLKHIPEHWLSYPEPEPILNYALGVLYIFFMLIALIGNGLVIWIFST 2
1 AKTLRTPSNIFVVNLAICDFLMMSKTPIFIYNSFKLGYALGHRACQIFALLGSFSGIGASATNAVIAYDRYR 2
1 VIATPFAPKLSRTKAVLYLALVWAYVTPWALLPLFEQWSRFVP 1
2 EGFLTSCTFDYLTPTSEIRNFVTVMFFICYVFPMSLIIYFYSQIVSHVIIHEHNLREQ 0
0 AKKMNVESLRSNANMHTQSAEIRIAKAAITICFLFVASWTPYAVLALIGAYGNQ 2
1 DLLTPAVTMIPACACKAVACVDPYVYAISHPRYR 2
1 QELSKKFPWLDIKEAPAPSSVDANSTATEMTLPTQTSPAEA* 0
>UV5_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme AF004169 9502802 full G? 5 exons
0 MSNDSIHWEARYLPAGPPRLLGWNVPAEELIHIPEHWLVYPEPNPSLHYLLALLYILFTFLALLGNGLVIWIFCA 2
1 AKSLRTPSNMFVVNLAICDFFMMIKTPIFIYNSFNTGFALGNLGCQIFAVIGSLTGIGAAITNAAIAYDRYS 2
1 TIARPLDGKLSRGQVILFIVLIWTYTIPWALMPVMGVWGRFVPEGFLTSCSFDYLTDTNEIRIFVATIFTFSYCIPMILIIYYYSQIVSHVVNHEKALREQAKKMNVDSLRSNANTSSQSAEIRIAK 0
0 AAITICFLYVLSWTPYGVMSMIGAFGNKALLTPGVTMIPACTCKAVACLDPYVYAISHPKYR 2
1 LELQKRLPWLELQEKPISDSTSTTTETVNTPPASS* 0
>UV5_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01002850 full 83% bee
0 MVNESIHWEARVLSAEPIRLLGWNVPAEELIHIPEHWLIYPEPNPSLHYLLAIVYIIFTFVALLGNGLVIWIFCS 2
1 AKSLRTSSNLFVVNLAFCDFFMMIKAPIFIYNSFNTGFATGHLGCQIFAFIGSLSGIGAGMTNAAIAYDRYS 2
1 VIARPLDGKLSRGQVILFIVLIWAYTIPWALMPIMHVWGRFVPEGFLTSCTFDYLTDTPEIQYFVATIFTFSYCIPMSLIIYYYSQIVNHVIDHEKALREQAKKMNVESLRSNVSANTQTMEIRIAK 0
0 AAITVCFLFVLSWTPYGVLAMIGAFGDKTLLTPGITMIPACTCKFVACLDPYVYAISHPRYR 2
1 LELQKRLPWLELQEKSTDTQSSTSEVVNVPSS* 0
>UV5_nasVit Nasonia vitripennis (jewel_wasp) Ecdy.Inse.Hyme XM_001608024 full G? gene wrong transcripts GE436449 GE390962
0 MPYYNWNGTDQTAGWPEARIQPAGAPRLLGWNVPPEELVHIPEHWLVYPEPNPALHYLLALLYILFTFVALLGNGLVIWIFCA 2
1 AKSLRTPSNMFVVNLAICDFMMMLKTPIFIYNSFHTGFALGNLGCQIFSFIGSLSGIGASITNAAIAYDRYS 2
1 TIARPLDGKLSRGQVMMLIVLIWMYTIPWALMPSMGVWGRFVP EGFLTSCTFDYITDSDEIRYFVGTIFTFSYAIPMTLIIYFYSQIVGHVVNHEKALREQAKKMNVESLRSGQNKDQASAEVRIAK 0
0 VALTICFLFVAAWTPYGVMSLIGAFGNK SLLTPGVTMIPACCCKAVACLDPYVYAISHPRYR 2
1 LELQKRMPWLELQEKPPASDATSTTTEAVPASS* 0
>UV5_papXut Papilio xuthus (butterfly) Ecdy.Inse.Lepi AB028218 full G? Rh5
0 MIPAAVMDNHTENNYNYGAYFAPYRLEGVELLGAGLTGEDLAAIPEHWLSYPAPPASAHTMLALVYVFFTAAALIGNGLVIFIFSASKSLRTPSNLLVVQLAVLDFLMMLKAPIFIYNSIK
RGFASGVIGCQIFAFMGSVSGTAAGLTNACIAYDRHSTITRPLDGRLSRGKVLLMMVCVWLYTAPWAILPQLQIWGRYVPEGFLTSCTFDYLTTTFD
NKLFVASMFVCVYIFPMIAILYFYSGIVKQVFAHEAALREQAKKMNVDSLRSNQNAAAESAEIRIAKAALTVCFLYVASWTPYGVMSLIGAFGDQNLLTPGVTMIPALACKGVACI
DPWVYAISHPKYRQELQKRMPWLQIDEPDDNASNTTSNTANSSAPA* 0
>UV5_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BABH01012019 -------- frag G? ---
0 1
2 VEMLGDGLSGDDLAAVPEHWLTFPAPPASAHTALALLYIFFTAAALLGNGLVIFVFST 2
1 TKSLRTSSNLLILQLAILDFIMMAKAPIFIYNSAMRGFATGTIGCQIFSVMGAYSGIGAGMTNACIAYDR 2
1 HSTITRPLDGRLSKGKVLLMMAFVWIYCTPWALLPLLKIWGRYVP 1
2 EGYLTSCTFDYLTNTFDTKLFVACIFVCSYVFPMSMIIYFYSGIVKQVFAHEAALR 2
1 EQAKKMNVESLRANQSGASQSAEIRIAKAALTVCFLFVASWTPYGVMALIGAFGDQQLLTPG 0
0 VTMIPAVACKAVACIDPWVYAISHPKYR 2
1 QELQRRMPWLQINEPDDTTSTATSNTVSSAPPAAPA* 0
>UV5_manSex Manduca sexta (moth) Ecdy.Inse.Lepi L78081 9343857 full G? Manop2
0 MNNQSENYYHGAQFEALKSAGAIEMLGDGLTGDDLAAIPEHWLSYPAPPASAHTALALLYIFFTFAALVGNGMVIFIFSTTKSLRTSSNFLVLNLAILDFIM
MAKAPIFIYNSAMRGFAVGTVGCQIFALMGAYSGIGAGMTNACIAYDRHSTITRPLDGRLSEGKVLLMVAFVWIYSTPWALLPLLKIWGRYVPEGYLTSCSFDYLTNTFDTKLFVA
CIFTCSYVFPMSLIIYFYSGIVKQVFAHEAALREQAKKMNVESLRANQGGSSESAEIRIAKAALTVCFLFVASWTPYGVMALIGAFGNQQLLTPGVTMIPAVACKAVACISPWVYA
IRHPMYRQELQRRMPWLQIDEPDDTVSTATSNTTNSAPPAATA* 0
>UV5_pedHum Pediculus humanus (louse) Ecdy.Inse.Phth AAZO01000117 full G? exon 1 uncertain
0 MKITTESENNISLSYYQPF 1
2 IDKEESLIWNVDPSELVHIPDHWFNFSAPHPLSNYLLGFLYFIFFVISCTGNGIVIWIFTT 2
1 SKNLRTASNVFVVNLAIFDFIMMAKTPIMIYNSMNLGFECGFVWCQIFASAGALSGIGASITNTCIAYDRCE 2
1 TITNPLQ KSGKKKAFLLAAFTWIYALPWAVLPFLEIWGKFAPEGYLTTCTVDYLTDTSQTRMFIVTIFFAAYVLPLSLIIYFYTKIVLHVINHEKSLKAQ 0
0 AKKMNVESLRSDGNKNYAVEIRITKVAIAMCFLFVISWTPYAVVALIGCFGNK 2
1 HLITPLVSMIPACACKAVACIDPYIYAISHPRFR 2
1 VEVNKRFACLAGCLQEKELQDDAVSKNTVNAENVDT* 0
>UV5_diaNig Dianemobius nigrofasciatus (cricket) Ecdy.Inse.Orth AB458852 full G?
MELQGSNVSNLSVWRPEARLATRLLGWNVPAEELIHIPEHWLTYPAPDAFSYYILGMLYVAFCFIALIGNGLVIWVFSSAKTLRTPSNIFVINLALYDFIMM
LKTPIFIYNSFNLGFGLGQLGCQIFAFMGSVSGIGAAATNACIAYDRYRVIARPFDSKMSIKGATLLVLLVWMWALPWAILPLLEIWGRYAPEGYLTSCSFDYLTDTPENHMFVLC
IFICSYVIPMSLIIYFYSQIVSHVVNHEKALKEQAKKMNVDSLRSNQQQNQTSAEIRIAKVAIGICFLFVASWTPYAVLALIGAFGNKALLTPGVTMIPACTCKAVACLDPYVYAI
SHPRYRAELQKRLPWLCIKEESASDTTSNATTTSTNAGATST* 0
>UVB_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD14509 full G? 8 exons K90
0 MDFNRSVSRPLSQLGS 2
1 SFMENEEELQLMGWNLTPEDLTHIPEHWLSYPEVRSLYHYILAFSYTILFCLGVIGNGLVLWIFCV 2
1 SKPLRTPSNLFVLNLALCDFSMVLVLPILIYDSIDHKYP GHLQCQIFALCGSISGIGAGATNAAIAYDRYS 2
1 TIAKPFEGRMTYGKALILIICIWIYVLPWCLLPLTEKWNRFVP 1
2 EGFLTSCSFDYLTPTEETKAFVGTMFVICYVIPMSFIIYFYSQIVCHVFNHEKALREQ 0
2 AKKMNVESLRSNQDANAQSAEVRIAKAAITICFLFVAAWTPYAVVAMIGAFGDQ 2
1 SLLTPIASMLPAVFAKTVACFDPYVYAISHPKYR 2
1 LELSKRVPCLGITEKPLATSDTQSITTAA* 0
>UVB_megVic Megoura viciae (vetch_aphid) Ecdy.Inse.Hemi AF189715 10762427 full G?
MDFNRSVSRPLSQLGSSFMENEDELQLMGWNLTPEDLTHIPEHWLSYPEVRSLYHYILAFSYTILFCIGVI
GNGLVLWIFCVSKPLRTPSNLFVLNLALCDFSMVLVLPILIYDSIDHKYPGHLQCQIFALCGSISGIGAGATNAAIAYDRYSTIAKP
FEGRMTYGKALILIICIWIYVLPWCLLPLTEKWNRFVPEGFLTSCSFDYLTPTEETKAFVGTMFVICYVIPMSFIIYFYSQIVCHVFNHEKALREQAKKMNVESLRSNQDANAQ
SAEVRIAKAAITICFLFVAAWTPYAVVAMIGAFGDQSLLTPIASMLPAVFAKTVACFDPYVYAISHPKYRLELSKRVPCLGITEKPPAASDTQSITTAA* 0
>UVB_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme AF004168 9502802 full G? 8 exons
0 MLLHNKTLAGKALAFIAEEG 2
1 YVPSMREKFLGWNVPPEYSDLVHPHWRAFPAPGKHFHIGLAIIYSMLLIMSLVGNCCVIWIFST 2
1 SKSLRTPSNMFIVSLAIFDIIMAFEMPMLVISSFMERMIGWEIGCDVYSVFGSISGMGQAMTNAAIAFDRYR 2
1 TISCPIDGRLNSKQAAVIIAFTWFWVTPFTVLPLLKVWGRYTT 1
2 EGFLTTCSFDFLTDDEDTKVFVTCIFIWAYVIPLIFIILFYSRLLSSIRNHEKMLREQ 0
0 AKKMNVKSLVSNQDKERSAEVRIAKVAFTIFFLFLLAWTPYATVALIGVYGNR 2
1 ELLTPVSTMLPAVFAKTVSCIDPWIYAINHPR 2
1 YRQELQKRCKWMGIHEPETTSDATSAQTEKIKTDE* 0
>UVB_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01025579 ADTU01025580 frag 74% bee
0 2
1 DVQPLQEEHFLGWNFPPEHADLVHPHWRVFLAPGRLWHVALALIYFLLLMLSLLGNGIVIWIFST 2
1 SKSLRTPSNIFILNLAVFDLLMATEMPMLIINSFLERTIGWETGCDVYALLGAISGMGQAITNAAIAFDRYR 2
1 TISCPIDGRLNGKQAMVLALFTWFWALPFSILPMTGLWGRYVA 1
2 EGFLTTCSFDYLTDDEDTRTFVITIFFWAYVLPMLLIIYFYSQLLKSIRSHEKMLRDQ 0
0 AKKMNVKSLVSNQNKERSVEMRIAKVAFTIFFLFVLAWTPYAVVALIGAYGNR 2
1 EALTPFSTMLPAIFAKTVSCIDPWIYAINHPR 2
1 YRQELEKRCKWMGIREKVAEDTVSAQTEKVNAEET* 0
>UVB_nasVit Nasonia vitripennis (jewel_wasp) Ecdy.Inse.Hyme XM_001604572 full G?
0 MAFVGLNGAMGGMGPA 1
2 EKPLQRYSQGPQMQEHLLGWNHPPEHIDIVHPHWRGFLAPGKYWHIGLALIYFMLLVLSFVGNGCVVWIFST 2
1 SKVLRTPSNLFIINLALFDLVMALEIPMLIINSFIERMIGWGLGCDIYAALGSVSGIGSAITNAAIAYDRYR 2
1 TISCPIDGRLNGKQAAVMVAFTWFWTMPFTILPFAKIWGRYTT 1
2 EGFLTTCSFDFLSDDQDTKVFVAAIFSWSYCFPMVLIIYFYSQLIKSVRRHEKMLREQ 0
0 AKKMNVKSLSAQDKERSVEMRIAKVAFTIFFLFVCSWTPYAVVTMIAAFGNR 2
1 ELVTPFSSMLPAVFAKTVSCIDPWVYAINHPR 2
1 YRQELTKRCQWMGIHEPDSGPSQNNAEAVSVTTEKLKSDDA* 0
>UVB_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BABH01016294 -------- full G? ---
0 MALNFTQEISDIGPMAYPLKM 2
1 VSREMVEHMLGWNIPEEHQDLVHEHWRNFPSVSKYWHYCLALIYAMLMVTSLVGNGIVIWIFGT 2
1 SKSLRSASNMFVINLAIFDLMMMLEMPLLIINSFYQRLVGYQLGCDIYAVLGSLSGIGGAITNAVIAFDRYK 2
1 TISCPLDGRINKVQASLLIAFTWFWALPFTILPAFRIWGRYVP 1
2 EGFLTTCSFDYFTDDQDTKVFVACIFVWSYCIPMTLICYFYSQLFGA 0
0 VRLHERMLQEQ AKKMNVKSLAANKEDASRSVEIRIAKVAFTIFFLFVCAWTPYAFVTMTGAFGDR 2
1 NLLTPIATMVPAVCCKVVSCIDPWVYAINHPR 2
1 YRAELTKRLPWLGVRESDPDAVSTTTSVGTAQSQATAEA* 0
>UVB_manSex Manduca sexta (moth) Ecdy.Inse.Lepi AD001674 9343857 full G? exons from 454
0 MATNFTQELYEIGPMAYPLKM 2
1 ISKDVAEHMLGWNIPEEHQDLVHDHWRNFPAVSKYWHYVLALIYTMLMVTSLTGNGIVIWIFST 2
1 SKSLRSASNMFVINLAVFDLMMMLEMPLLIMNSFYQRLVGYQLGCDVYAVLGSLSGIGGAITNAVIAFDRYK 2
1 TISSPLDGRINTVQAGLLIAFTWFWALPFTILPAFRIWGRFVP 1
2 EGFLTTCSFDYFTEDQDTEVFVACIFVWSYCIPMALICYFYSQLFGA 1
0 VRLHERMLQEQAKKMNVKSLASNKEDNSRSVEIRIAKVAFTIFFLFICAWTPYAFVTMTGAFGDR
1 TLLTPIATMIPAVCCKVVSCIDPWVYAINHPR 2
1 RAELQKRLPWMGVREQDPDAVSTTTSVATAGFQPPAAEA* 0
>UVB_diaNig Dianemobius nigrofasciatus (cricket) Ecdy.Inse.Orth AB291232 full G?
MNSSVGLQGAPIALPYESYVAQMLGWNIPAEHIELVHSHWRGYEAPSKYWHYWFAFMYFCIMIMSCLGNGIVLWIFATTKSLRTPSNMFVVNQALLDLLMMI
EMPMFVLNSLFYQRPIGWEMGCDIYALLGAVSGIGSAINNAAIAYDRYRTISFPLDGRLQFGHALAFIVGVWSWAMPFSLLPLLKVWGRYVPEGLLTTCSFDYLTDDEDTKVFTAS
IFTWSYAFPLCLIVFFYCKLFKQVRLHEKMLQEQARKMNVKSLQTNQDVAQKSVEIRIAKVAFTIFFLFLCSWTPYATVAMIGAFGNRALLTPMSTMIPALFSKIVSCIDPWIYAI
NHPRFRGELLKRAPWFGVEELKSSDVSSIGTDRTTATAAIETPAA* 0
>LMS_ixoSca Ixodes scapularis (tick) Ecdy.Chel.Arac EW898719 full G? ocellar transcripts
0 MGSEGQRTNMSLLDELASPYMKNGTLVESVPDEMLYMVHPHWYNFKPMNPLWHSLLGFAMVILGVISVVGNSMVIYIMTTSKSLRSPTNMLVVNLAFSDW 2
1 CMMAFMMPTMAANCFAETWILGPFMCEVYGMVGSLFGCGSIWSMVMITLDRYNVIVRGVAAAPLTHKRAALMIFFVWFWALTWTLLPFFGWSR 2
1 YVPEGNMTSCTIDYLTKALWSASYVVAYAGGVYWTPLFINIYCYSKIVRAVAQHEKQLRLQARKMNVASLRANAEQTKTSAEARLAK 0
0 IALMTVGLWFMAWTPYLTIAWAGIFSDGSKLTPLATIWGSVFAKANACYNPIVYGISHPKYRAALARRFPSLVCMPPGGDQLDTRSEASGITTIEDKVMTTET* 0
>LMS_tetUrt Tetranychus urticae (spider_mite) Ecdy.Chel.Arac GW028305 frag G? larval transcripts
CEFYGMWGSLFGCGSVWSLVFITRDRYNVIVRGMSAGRLTIKKSLAQVLFIWFYSVAWTIIPFFGWSRYVPEGNMTSCTIDYLSKDLWNASYTVVYAVAVYFLPLFTLVYHYYYIVCAVATHERTLREQA
KKMNVASLRANADANKESADIRIAKVAMMTVGLWFMAWTPYCTLAFLGIFTDGDYLTPMVSIWGAVFAKSAACYNPIVYALSHPKYRSALFTRFPSLACGVGAVEPVVDDDAKSEMSVATTFSDHTSAP* 0
>LMS1_plePay Plexippus paykulli (jumping_spider) Ecdy.Chel.Arac AB251849 18217181 full G? PpRh1 kumopsin1
MLPQAAKMAARASSGVDGKNISIVDLLPEDMLYMIHEHWYKYPPMESTMHYLLGITIILIGIISVS
GNSIVIYLMLSVKSLRTPANFLVTSLAVSDGGMLAFMAPTMPINCFAQTWVLGPFMCELYGMVGSLFGSASIWNMVMITLDRYNVIVRG
MSGKPLTKVGALLRIIFVWVWSLGWTIAPMYGWSSYAPEGSMTGCTVDYLHTDISTMSYLIVY
AIFVYFVPLFIIIYCYTYIVMQVAAHEKSLREQAKKMNIKSLRSNEDNKKASAEFRLAKVALMTICLWFMAWTPYLILSLLGIFSDREWLTPLTSIWGAVFAKAASAYNPIVYGIS
HPKYRAALHEKFPCLNCATESPKGDSASTVAESDKGGD*
>LMS2_plePay Plexippus paykulli (jumping_spider) Ecdy.Chel.Arac AB251850 18217181 full G? PpRh2 kumopsin2
MSSQIINGAYMVSRDALGLHLPTNLGGPLPQDNSYYPYLRNTTVVDTVPKEILHMIHDHWYQFPPLNPLWHSLLGIAMILLGIVSVI
GNGMVMYLMNTTKSLKTPTNMLIVNLAFSDFCMMAFMMPTMAANCFAETWILGPFMCEIYGMAGSLFGCVSIWSMVMIAFDRYNVIVRG
MNAEPLTTKKAAAQIFLIWAWAIMWTVLPFFGWSRYVPEGNM
TSCTVDYLSEDLKSSSYVLIYGCAVYFIPLFTLIYNYTFIVRAVSIHEDNLREQAKKMNVTSLRANADQQKQSAECRLAKIALMTVGLWFIAWTPYLCIAWSGIFSSRKHLTPLAT
IWGAVFAKAVAVYNPIVYGISHPKYRAALFQKFPSLACTTESDVIDNKSEVTFVTDEKPPKTQEA*
>LMS1_hasAda Hasarius adansoni (jumping_spider) Ecdy.Chel.Arac AB251846 18217181 full G? HaRh1 kumopsin1
MLPHAAKMAARVAGDHDGRNISIVDLLPEDMLPMIHEHWYKFPPMETSMHYILGMLIIVIGIISVS
GNGVVMYLMMTVKNLRTPGNFLVLNLALSDFGMLFFMMPTMSINCFAETWVIGPFMCELYGMIGSLFGSASIWSLVMITLDRYNVIVKG
MAGKPLTKVGALLRMLFVWIWSLGWTIAPMYGWSRYVPEGSMTSCTIDYIDTAINPMSYLIAY
AIFVYFVPLFIIIYCYAFIVMQVAAHEKSLREQAKKMNIKSLRSNEDNKKASAEFRLAKVAFMTICCWFMAWTPYLTLSFL
GIFSDRTWLTPMTSVWGAIFAKASACYNPIVYGISHPKYRAALHDKFPCLKCGSDSPKGDSASTVAESEKAGE*
>LMS2_hasAda Hasarius adansoni (jumping_spider) Ecdy.Chel.Arac AB251847 18217181 full G? HaRh2 kumopsin2
MSSHTINSAFMVPRDVLGLHLPNNLGGPLPHDNSYYPYLRNATVVDTVPKEILHMIHDHWYQFAPLNPLWHSLLGIAMIILGIVSVI
GNGMVIYLMSTTKSLKTPTNMLIVNLAFSDFCMMAFMMPTMAANCFAETWILGPLMCEIYGMAGSLFGCVSIWSMVMIAFDRYNVIVRG
MSAEPLTTKKAAAQIFFIWTWATTWTLFPFFGWSRYVPEGNMTSCTVDYLTEDLKSSSYVLIYGCAVYFTPLFTLIYNYTFIVRSVSIHENNLREQAKKM
NVSSLRANADQQKQSAECRLAKIALMTVGLWFIAWTPYLSIAWSGIFSSRKHLTPLATIWGAVFAKAVAVYNPIVYGISHPKYRAALFEKFPSLACTTESDVTDNKSEVTLVTDEKPPKTQEA*
>LMS_limPol Limulus polyphemus (horseshoe_crab) Ecdy.Chel.Mero AY190512S1 14977331 full G? intronated lateral_eye
0 MANQLSYSSLGWPYQPNASVVDTMPKEMLYMIHEHWYAFPPMNPLWYSILGVAMIILGIICVLGNGMVIYLMMTTKSLRTPTNLLVVNLAFSDFCMMAFMMPTMTSNCFAETW
ILGPFMCEVYGMAGSLFGCASIWSMVMITLDRYNVIVRGMAAAPLTHKKATLLLLFVWIWSGGWTILPFFGWSR 2
1 YVPEGNLTSCTVDYLTKDWSSASYVVIYGLA 0
0 VYFLPLITMIYCYFFIVHAVAEHEKQLREQAKKMNVASLRANADQQKQSAECRLAK 0
0 VAMMTVGLWFMAWTPYLIISWAGVFSSGTRLTPLATIWGSVFAKANSCYNPIVYGISHPRYKAALYQRFPSLACGSGESGSDVKSEASATTTMEEKPKIPEA* 0
>LMS_neoOer Neogonodactylus oerstedii (mantis_shrimp) Ecdy.Crus.Mala DQ646869 17053049 full G? Rh1
MSYWNSNKIVEEYSLPSTNPYGNFTVVDTVPENMLHMIHSHWYQFPPLNPMWYGILAFVVTVVGLCSICGNFVVIWVFMNTKALRSPANTLVVSLAVSDFIM
MACMFPPLVLNCYWGTWIFGPLFCEVYAFIGNTVGCASIGNMIFITFDRYNVIVKGISGTPLSQKNTTLQVLFVWICSIMWCVFPFFGWNRYVPRGDMTACGTDYLTEDEFSRSYL
YVYSVWVYIGPLALIIYCYFHIVSAVATHEKQMRDQAKKMGVKSLRTEEAKKTSAECRLAKVALTTVSLWFMAWTPYLIINWAGMFYPSVVSPLFSIWGSVFAKANAVYNPIVYAI
SHPKYRAALYKKLPCLACSTESADEGSATNSATTTTAEKYESA* 0
>LMS_lucCru Luciola cruciata (firefly) Ecdy.Inse.Cole AB300328 19232386 full G?
MSVLGEPSFAAWASQAGVMSSRFGGGNITVVDKVPPDMLHLIDAHWYQYPPLNPLWHAILGFMIGVLGCISVTGNGMVIYIFSTTKSLRSPSNLLVVNLAFS
DFLMMFTMAPPMVINCYNETWVWGPLFCQIYGMLGSLFGCTSIWTMTMIALDRYNVIVKGLSAKPLTKQGALIRIFLVWVFSIGWTIAPVFGWNRYVPEGNMTACGTDYLSTGWFS
RSYILFYSWFVYFIPLFAIIYSYWFIVQAVSAHEKAMREQAKKMNVASLRSSEAAQTSAECKLAKVALMTISLWFLAWTPYLVTNYAGIFDGSKISPLATIWSSLFAKANAVYNPI
VYGISHPKYRQALQKKFPSLVCAAEPDDTVSQTTAATAASEEKAAA* 0
>LMS_triCas Tribolium castaneum (flour_beetle) Ecdy.Inse.Cole ES544655 18342244 full G? 3 exons from AAJJ01000967 5 fusion relative to bee
0 MSVMGEPNFIAWAAQRSGYGGGNLTVVDKVLPDMLHLVDAHWYQFPPMNPLWHGILGFVIGVLGFVSIVGNGMVIYIFSSTKALRTPSNLL
VVNLAFSDFLMMlCMSPAMVINCYNETWVLGPLVCELYGMSGSLFGCASIWTMTFIALDRYNVIVKGLSAQPLTKKGAMLRILIIWVFSTLW
TIAPFFGWNRYVPEGNMTACGTDYLTKDWVSRSYILVYAVWVYFVPLFTIIYSYWFIVQ 0
0 AVAAHEKSMREQAKKMNVASLRSSEAAQTSAECKLAKIALMTITLWFFAWTPYLVTNFTGIFEGAKISPLATIWCSLFAKANAVYNPIVYGIS 2
1 HPKYRQALQKKFPSLVCAGEPDDTTSTASGVTNVTTDEKPATA* 0
>LMS1_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_079683 19398933 full G? RH1 ninaE
0 ME 0
0 SFAVAAAQLGPHFAPLSNGSVVDKVTPDMAHLISPYWNQFPAMDPIWAKILTAYMIMIGMISWCGNGVVIYIFATTKSLRTPANLLVINLAISDFGIMITNTPMM
GINLYFETWVLGPMMCDIYAGLGSAFGCSSIWSMCMISLDRYQVIVKGMAGRPMTIPLALGKIAYIWFMSSIWCLAPAFGWSR 2
1 YVPEGNLTSCGIDYLERDWNPRSYLIFYSIFVYYIPLFLICYSYWFIIA 0
0 AVSAHEKAMREQAKKMNVKSLRSSEDAEKSAEGKLAKVALVTITLWFMAWTPYLVINCMGLFKFEGLTPLNTIWGACFAKSAACYNPIVYGIS 2
1 HPKYRLALKEKCPCCVFGKVDDGKSSDAQSQATASEAESKA* 0
>LMS6_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt NM_079644 19398933 full G? RH6 gross genomic misassembly exon1
0 MASLHPPSFAYMRDGRNLSLAESVPAEIMHMVDPYWYQWPPLEPMWFGIIGFVIAILGTMSLAGNFIVMYIFTSSKGLRTPSNMFVVNLAFSDFMMMFTMFPPVVLNGFYGT
WIMGPFLCELYGMFGSLFGCVSIWSMTLIAYDRYCVIVKGMARKPLTATAAVLRLMVVWTICGAWALM
PLFGWNRYVPEGNMTACGTDYFAKDWWNRSYIIVYSLWVYLTPLLTIIFSYWHIMK 0
0 AVAAHEKAMREQAKKMNVASLRNSEADKSKAIEIKLAKVALTTISLWFFAWTPYTIINYAGIFESMHLSPLSTICGSVFAKANAVCNPIVYGLS 2
1 HPKYKQVLREKMPCLACGKDDLTSDSRTQATAEISESQA* 0
>LMS_anoGam Anopheles gambiae (mosquito) Ecdy.Inse.Dipt XM_319247 full G? most introns obliterated
0 MPYYGPMQQPGLWGQPVANLTVVDKVPPEIMHLVDPHWSQFPPMNPLWHSIIGFVIFVLGVVSIIGNGMVIYIFSTAKSLRTPSNLFIVNLALSDFLMMGTNAFTMVYNCWFETWSLG
LLMCDLYAFFGSLFGCCSIWTMTMIALDRHNVIVHGLSGKPLTNTGAILRILLCWLIGVVWGILPMLGWNRYVPEGNMTACGTDYLTDDWFHKSYILVYSVFVYYTPLFTIIYAYFFIIK 0
0 AVSAHEKNMREQAKRMNVQSLRSSDDGKSTEMKLAKVALVTISLWFMAWTPYTVINYTGVFKTASITPLATIWGSVFAKANAVYNPIVYGISHPKY
RAALLRRFPSLACSDGPPADDKSLASEASGITSAGNPTTA* 0
>LMS2_droMel Drosophila melanogaster (fruitfly) Ecdy.Inse.Dipt M12896 full G? RH2 CG16740-RA Rh2 ocellar-specific
0 MERSHLPETPFDLAHSGPRFQAQSSGNGSVLDN 0
0 VLPDMAHLVNPYWSRFAPMDPMMSKILGLFTLAIMIISCCGNGVVVYIFGGTKSLRTPANLLVLNLAFSDFCMMASQSPVMIINFYYETWVLGPLWCDIYAGCGSLFGCVSIWSMC
MIAFDRYNVIVKGINGTPMTIKTSIMKILFIWMMAVFWTVMPLIGWSAYVPEGNLTACSIDYMTRMWNPRSYLITYSLFVYYTPLFLICYSYWFIIAAVAAHEKAMREQAKKMNVKSL
RSSEDCDKSAEGKLAKVALTTISLWFMAWTPYLVICYFGLFKIDGLTPLTTIWGATFAKTSAVYNPIVYGIS 2
1 HPKYRIVLKEKCPMCVFGNTDEPKPDAPASDTETTSEADSKA* 0
>LMS_rhoPro Rhodnius prolixus (kissing_bug) Ecdy.Inse.Hemi genomic full G?
0 MAQPIGPSFAAYQWGQSANPSANRSVVDMVPPEMLSMVDAHW 2
1 YQFPPLNPLWHGILGFVIGVLGIISIVGNGMVIFIFSSTKTLRTPSNLLVVNLAFSDFLMMFTMSPPMVINCYNETWVL 1
2 GPLMCELYGMLGSLFGCASIWTMTMIALDRYNVIVK 0
0 GISAKPMTNKTAMLRILLVWAFSIMWTVFPFFGWNR 2
1 YVPEGNMTACGTDYLTKNWVSRSYILVYSVFVYFLPLFTIIYSYFFILQ 0
0 AVSAHEKQMREQAKKMNVASLRSAEAANTSAEAKLAK VALMTISLWFMAWTPYLVINYSGIFETISISPLFTIWGSLFAKANAVYNPIVYAIR 2
1 HPKYKQALEKKFPSLSCASPQDDTTSVATGVTTSTDDKAPSA* 0
>LMS_acyPis Acyrthosiphon pisum (aphid) Ecdy.Inse.Hemi SCAFFOLD6053 full G?
0 MLNKIGSHYERQENWVAEGGFGNETVVDRVPADMMHLIDPSW 2
1 YQFPPMESMWYKWLGVTIFFLGILSVVGNGMVIYIFTCTKNLRTPSNLLIVNLAFSDFCLMFTMCPAMVWNCFYETWMF 1
2 GPFACELYAMFGSLFGVTSIWTMVFIALDRYNVIVK 0
0 GLSAKPMTTKLALLQIFCIYLHGLFWTLTPFFGWSR 2
1 YVPEANMTACGTDYLTLAWHSRSYVLVYAIFAYYLPLLVIIYAYYFIVK 0
0 AVASHEKSMREQAKKMNVSSLRSGDQSNTSAEFKLAKVALMTISLWFMAWTPYMVINFAGIFQLMTIDPLFTIWGSVFAKANAVYNPIVYAIS 2
1 HPKYRLALDKKFPCLVCGKLEDDRSDSKSVASAQTTISEDKV* 0
>LMS_homCoa Homalodisca coagulata (sharpshooter) Ecdy.Inse.Hemi AY588065 full G?
MSLISEPSFSAYSWASQGGFGNQTVVDKVPPEMLYLVDAHWYQFPPMNPLWHSLLGFAMVVLGFIAVTGNGMVVYIFSCTKALRTPSNLLVVNLAFSDFLMM
FTMAPPMVLNCYYETWVLGPFMCELYAMFGSILGCTSIWTMVMIANDRYNVIVKGLSAKPMTIKSALARILFCWAHSLIWCLAPFLGWGRYVPEGNMTACGTDYLTPDWISKSYIL
VYSLFCYFMPLFLIIYSYWFIVQAVSAHEKAMREQAKKMNVASLRSSDAANTSAEHKLAKVALMTISLWFCAWTPYLVINYAGIFQALTISPLFTIWGSVFAKANACYNPIVYAIS
HPKYRAALNKKFPSLVCGATEAPASTSDGASVASGATTLTEDKSAAA*
>LMSa_nasVit Nasonia vitripennis (jewel_wasp) Ecdy.Inse.Hyme XM_001606013 full G? 22063-23541 - strand of AAZX01007316 --
0 MGPSFLTLTAMAQRGGYGGGGGFGGGFNNQTVVDKAPPEIHHMIDPYWYQFPPMNPLWYGILGFVIGCLGCISVAGNGMVVYIFASTKSLRTPSNLLVINLAFSDFCMMFTMSPPM 0
0 VINCYYETWVFGPLMCEIYALCGSIFGCGSIWTMCMIAFDRYNVIVKGLSAKPMTINGSLLRILGIWLMASIWTIAPMFGWNR 2
1 YVPEGNLTACGTDYFSKDWVSRSYIVVYSFFVYFLPLFMIIYSYYFIIKAVSAHEKNMREQAKKMNVASLRQGDSQSAENKLAK 0
0 IALMTISLWFMAWTPYLVINWAGIFDLARLTPLFTIWGSVFAKANAVYNPIVYGIS 2
1 HPKYRAALFARFPSLACAGDAPAGAASDAVSTTSGVTTLTDHDKSNA* 0
>LMSb_nasVit Nasonia vitripennis (jewel_wasp) Ecdy.Inse.Hyme AAZX01007316 full G? tandem pair to LWSa fairly diverged
0 MEHPIVAAGVNATGEFDASSGSASSTTTMVTTAAVQVASTIGPHFARQVMRGFGNLTVVDKVPPEMLHLVGPHW 2
1 YQFPPLWPIWHKLLGVVMIFIGVLGWCGNGMVVYIFLVTPSLRTPSNLLVINLAFSDFVMMIIMSPPMVVNCWYETWIL 1
2 GPLMCDIYALIGSLCGGASIWTMTAIAYDRYNVIVK 0
0 GMSGTPLTIPRALVQIVLIWTHGLIWAMLPLFGWNR 2
1 YVPEGNMTSCGTDYVSDDWLGKSYILVYSIFVYYTPLFSIILCYWHIVS 0
0 AVAAHERGMREQAKKMNVASLRSGDQSGESAEVKLAK 0
0 VAVTTISLWFLAWTPYLVTNYMGIFAKQHVSPLFTIWASLFAKTNACYNPIVYGIS 2
1 HPKYRAGLKVKCPCLVFGDTEDKPKPAAATPAADAASTHSKA* 0
>LMSa_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme NM_001077825 16291092 full Gq rhabdomeric long wavelength ocelli not compound
0 MDTLNITTSFFIEVMPSNISTLTTTGPQFARQLMRFNNQTVVSKVPEEMLHLIDLYW 2
1 YQFPPLDPLWHKILGLVMIILGIMGWCGNGVVVYVFIMTPSLRTPSNLLVVNLAFSDFIMMGFMCPPMVICCFYETWVL 1
2 GSLMCDIYAMVGSLCGCASIWTMTAIALDRYNVIVK 0
0 GMSGTPLTIKRAMLQILGIWLFGLIWTILPLVGWNR 2
1 YVPEGNMTACGTDYLSQDWTFKSYILVYSFFVYYTPLFTIIYSYYFIVS 0
0 AVAAHEKAMKEQAKKMNVTSLRSGDNQNTSAEAKLAK 0
0 VALTTISLWFMAWTPYLVINYIGIFNRSLITPLFTIWGSLFAKANAIYNPIVYGIS 2
1 HPKYRAALKEKLPFLVCGSTEDQTAATAGDKASEN* 0
>LMSa_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01020830 16601190 full iMet uncertain 80% bee
0 MAFADVTRTIGPRAMQQIMSFNNKTVVNKVPPDMMHLIDPHW 2
1 YQFPPMNPMWHKILGLVMIVLGFMGWCGNGVVVYIFLMTPSLRTPSNLLIVNLAFSDFIMMIIMTPHMMINCYYETWIL 1
2 GPLMCDMHAMVGSLCGCASIWTMTAIALDRYNVIVK 0
0 GMAGKPLTIKKALLEILIIWLFASIWAILPIVGWNR 2
1 YVPEGNMTACGTDYLTKDWSSKSYILVYSIFVYYMPLLIIIYSYYFIVS 0
0 TVAAHERTMREQAKKMNVQSLRSGDNQNISAEAKLAK 0
0 VALMTISLWFMAWTPYLVINYVGIFGIAKISPLFTIWGSLFAKANAIYNPIVYGIR 2
1 HPKYRAALKEKLPFFVCGSTEDPSVAAKTAETSSTTTT* 0
>LMSb_apiMel Apis mellifera (bee) Ecdy.Inse.Hyme U26026 full G? 5 exonsArthropoda Insecta 540 complete genNow
0 MIAVSGPSYEAFSYGGQARFNNQTVVDKVPPDMLHLIDANWYQYPPLNPMWHGILGFVIGMLGFVSVMGNGMVVYIFLSTKSLRTPSNLFVINLAISDFLMMFCMSPPM 0
0 VINCYYETWVLGPLFCQIYAMLGSLFGCGSIWTMTMIAFDRYNVIVKGLSGKPLSINGALIRIIAIWLFSLGWTIAPMFGWNR 2
1 YVPEGNMTACGTDYFNRGLLSASYLVCYGIWVYFVPLFLIIYSYWFIIQAVAAHEKNMREQAKKMNVASLRSSENQNTSAECKLAK 0
0 VALMTISLWFMAWTPYLVINFSGIFNLVKISPLFTIWGSLFAKANAVYNPIVYGIS 2
1 HPKYRAALFAKFPSLACAAEPSSDAVSTTSGTTTVTDNEKSNA* 0
>LMSb_attCep Atta cephalotes (ant) Ecdy.Inse.Hyme wgs ADTU01020829 18362345 full 78% bee
0 MSFVSGPSHAAYTWAAQGGGFGNQTVVDKVPPEMLHMVDVHWYQFPPMNPLWHGLLGFAIGVLGTISIIGNGMVIYIFTTTKSLRTPSNLLVVNLAISDFLMMLWMSPAM 0
0 VINCYYETWVLGPLFCELYGMMGSLFGCGSIWTMTMIAFDRYNVIVKGLSAKPMTIKGALIRIFAIWLFTILWTIAPLFGWNR 2
1 YVPEGNMTACGTDYLTKDIVSRSYILFYSIFVYFMPLFLIIYSYFFIIQAVAAHEKNMREQAKKMNVASLRSAENQSTSAECKLAK 0
0 VALMTISLWFMAWTPYLIINFAGIFETTKISPLFTIWGSLFAKANAVYNPIVYGIR 2
1 HPKYRAALFQRFPSLACATEPAPGADTMSTTTTVTEGEKSVA* 0
>LMS_manSex Manduca sexta (moth) Ecdy.Inse.Lepi L78080 full G?
MDPGPGLAALQAWAAKSPAYGAANQTVVDKVPPDMMHMIDPHWYQFPPMNPLWHALLGFTIGVLGFVSISGNGMVIYIFMSTKSLKTPSNLLVVNLAFSDFL
MMCAMSPAMVVNCYYETWVWGPFACELYACAGSLFGCASIWTMTMIAFDRYNVIVKGIAAKPMTSNGALLRILGIWVFSLAWTLLPFFGWNRYVPEGNMTACGTDYLSKSWVSRSY
ILIYSVFVYFLPLLLIIYSYFFIVQAVAAHEKAMREQAKKMNVASLRSSEAANTSAECKLAKVALMTISLWFMAWTPYLVINYTGVFESAPISPLATIWGSLFAKANAVYNPIVYG
ISHPKYQAALYAKFPSLQCQSAPEDAGSVASGTTAVSEEKPAA*
>LMS_bomMor Bombyx mori (moth) Ecdy.Inse.Lepi BAAB01068714 11549248 full G? Bcop boceropsin Bomopsin1 tandem misassembly CK529859 hemocyte larva CK552244 fat body (pupa)
0 MISLDPGPGMAALQAWGGQVAAYGAANQTVVDKVPPDMLHMVDPHW 2
1 YQFPPMNPLWHALLGFTIGVLGFISISGNGMVIYIFMSTK 0
0 SLKTPSNLLVVNLAFSDFLMMCAMSPAMVVNCYNETWVF 1
2 GPFACELYACAGSLFGCASIWTMTMIAFDRYNVIVKGIAAKPMTNNGALLRILGIWAFSLAWTLAPFFGWNR 2
1 YVPEGNMTACGTDYLSKDWFSRSYILIYSVFVYFAPLLLIIYSYFFIVQ 0
0 AVAAHEKAMREQAKKMNVASLRSSEAANTSAECKLAK 0
0 VALMTISLWFMAWTPYLVINYTGVFESAPISPLATIWGSLFAKANAVYNPIVYGIR 2
1 HPKYQAALYARFPSLQCQSAPPDDGGSVASGATAVSEEKPAA* 0
>LMS_papXut Papilio xuthus (butterfly) Ecdy.Inse.Lepi AB007424 9547302 full G? Rh2
MAIANLEPGMGASEAWGGQAAAFGSNQTVVDKVTPDMMHLIDPHWYQFPPMNPMWHGLLGFTIGVLGFISITGNGMVVYIFTSTKSLKTPSNLLVVNLAFSD
FLMMLCMAPPMLINCYYETWVFGPLACELYACAGSLFGSISIWTMTMIAFDRYNVIVKGIAAKPMTINGALLRILGIWLFSLAWTIAPMLGWNRYVPEGNMTACGTDYLSKSWLSR
SYILVYSIFVYYTPLLLIIYSYFFIVQAVAAHEKAMREQAKKMNVASLRSSEAANTSAECKLAKVALMTISLWFMAWTPYLVINYTGVFETAPISPLATIWGSVFAKANAVYNPIV
YGISHPKYRAALYQKFPSLACQPSAEETGSVASGATTACEEKPSA*
>LMS_schGre Schistocerca gregaria (locust) Ecdy.Inse.Orth X80071 full G?
MASASLISEPSFSAYWGGSGGFANQTVVDKVPPEMLYLVDPHWYQFPPMNPLWHGLLGFVIGVLGVISVIGNGMVIYIFSTTKSLRTPSNLLVVNLAFSDFL
MMFTMSAPMGINCYYETWVLGPFMCELYALFGSLFGCGSIWTMTMIALDRYNVIVKGLSAKPMTNKTAMLRILFIWAFSVAWTIMPLFGWNRYVPEGNMTACGTDYLTKDWVSRSY
ILVYSFFVYLLPLGTIIYSYFFILQAVSAHEKQMREQRKKMNVASLRSAEASQTSAECKLAKVALMTISLWFFGWTPYLIINFTGIFETMKISPLLTIWGSLFAKANAVFNPIVYG
ISHPKYRAALEKKFPSLACASSSDDNTSVASGATTVSDEKSEKSASA*
>BCR_limPol Limulus polyphemus (horseshoe_crab) Ecdy.Chel.Mero FJ791252 full G? ventral eye
MSTGSYFIGNSTAPRSSGWWSYDPGLSVRDTAPENIKHLISDHWSKFPAVNPMWHYLLGLIYIVLGIASLTGQSVVLYLFAKTKPLRTPANMLIVNLAFSDF
MMMITQFPVFIINCLGGGAWQLGPLLCEITGFAGGLFGYGSIVTLAVISIDRYNVIVRGFSASPLTHARSAVFILVIWAWTLGWALPPFFGWGRYVPEGILNSCSFDYLTRDWATV
SYIMGCWICEYALPLMVIIYCYIFIVKAVCDHERHLREQAKKMNVASLRSNVDTQKASAEMRIAKVALVNVLLWVVSWTPYAAIAMIGIAGDQMLITPLRSALPALAGKAASVYNP
IVYAISHPKFRLAMQKEIPCCCINEPQPQSDTSSEMSTKTSVATVNGEDSTAGGTTNN*
>BCR_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran BAG80976 18984904 full G? RhA
MAAYTEAWNASEEILVRMARAVPSVAWGYPAGVSIADLVPSDMKTMVHSHWNKFPPVNPMWHYLLGMVYIILGTVSIAGNSLVISLFTKTKELRTPANMFVVNLAFSDLCMMITQFPMFVYNCFNGGMWLFGPFLCELYA
ATGAVFGLCSICTLACIAFDRYNLIVKGMSGPKMTSKRATILIAFCWAYAIGWSLPPFFGWGRYIPEGILDSCSFDYLTRDSSTKSFGLCLFFFDYVTPLSIIVFAYFHIVRAIFEHEKILREQAKKMNVTSLRSNADQN
AQSAEIRIAKVALINISLWVAMWTPYATIVLQGLLGNQENITPLVSILPALIAKSASIYNPVIYAISHPRYRVALQQKLPWFCIHEEEKKPISDTDSAKTEASSS*
>BCR2_triLon Triops longicaudatus (tadpole_shrimp) Ecdy.Crus.Bran BAG80981 18984904 full G? RhA
MATYTEAWNASEEILVRMVRAAPSVAWGYPTGVSIVDLVPSDMKTMVHSHWSKFPPVNPMWHYLLGLVYIVLGTVSIAGNSLVISLFTKTKELRTPANMFVVNLAFSDLCMMITQFPMFVYNCFNGGMWLFGPFLCELYA
ATGAVFGLCSICTLACIAYDRYNLIVKGMSGPKMTSKRATILIAFCWSYAIGWSLPPFFGWGRYIPEGILDSCSFDYLTRDSSTKSFGLCLFFFDYITPLSIIVFAYFHIVRAIFEHEKILREQAKKMNVTSLRSNADQN
AQSAEIRIAKVALINISLWVAMWTPYATIVLQGLLGNQENITPLVSILPALIAKSASIYNPVIYAISHPRYRIALQQKLPWFCIHEEEKKPISISDTDSAKTETSSS*
>BCR1_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran AB293431 18984904 full G?
MANASHYEALQQEFNPWALPESFTLYAYAPEDVRAFLHPHWHNFPATHPAIYYLFGLVYLVLGVTSVG
GNYLVLRIFTKFQELRRPSNVLVINLALSDMLLMLTLFPECVYNFLSGGPWRFGDLGCQIHAFCGALFGYNQ
ITTLVFISYDRFNVIVRGMGGTPLTYARVSAMVAFSWLWATGWSVAPLVGWGGYALDGMLGTCSFDYVTR
TWNNRSHILAATAFMWVIPVLIIAGCYWFIVQAVFKHEAELKAQAKKMNVASLRSNADQQQVSAEIRIAK
VAITNVVLWLSAWTPFMVISNLGIWADPQQVTPLVSSLPVLLSKTSCSYNPLVYAISHPKYRECLKTLVPWICIVLPNDRRGGDNVSSSSSRTEASGKAETVDA*
>BCR2_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran AB293429 18984904 full G?
MSSGVFNSTDPIALARVSAGSNAHQQVGYNILIKTDGLSVRDVAPLDMHHLLHSHWDAYPPADPRIHYLL
GMLYFFLGIAACMGNVLVLHIFGKHKNLRSPTNTLLMNLAFCDLMIFIGLYPEMLGNIFMNDGTWMWGDV
ACRIHAWFGLVFGFGQMQTLMYMSIDRYNVIVKGLSAQPLTYKKVTQWLAQVWIVSLFWGTAPFFGFGNF
ALDGILNTCSFDYFSRDMLSMSYIVSACVWAYVIPLIVIIFCYTFIVRAVFEHEETLRQQAAKMNVTSLR
SSANSEDTSAEFRIAKIAMINVCLWLWAWSPFTIVSFIGIFGNQAIITPYLSSLPVILAKTSSVYNPIVYALSHPRYQAALKEEFAWLCVKTNSGNSGSSDTKSSVTMESSQPA*
>BCR3_triGra Triops granarius (tadpole_shrimp) Ecdy.Crus.Bran AB293432 18984904 full G?
MMHNFSEPRYEAQVVRYGDFAPGVSVRDMAPENVRYMVHLHWEKFPPPDPRVHTALGALYLIMGVMSAVG
NVLVLYIFGKYKSLRSPTNVLVMNLAFCDLGLFVGLYPELLGNIFINNGPWMWGDVACKIHAWCGLAFGF
GQMQTLMFVSMDRYYVIVKGLKAPPLTYWKVSVWLAMVWIVSIFWATSPFFGFGNLSVDGLLNTCSYDYY
TRDLPTVAYIVGSCVHAYVLPLAVIIFCYSYIVQAVFHHERQLREQAAKMNVASLRSSGGKQDEMSAEFR
IAKIALINCCLWLWAWTPFTVISFMGVLHDDQSIINPYVSSLPVLLAKTSAVYNPIVYGLSHPKFQQCLREEFGWNIGLPKKKDNDSKSVTSVETAMT*
>BCR1_triLon Triops longicaudatus (tadpole_shrimp) Ecdy.Crus.Bran AB293434 18984904 full G?
MSSSGFNSTDPIALARVSAGSNAHQQVGYNILIKTDGLSVRDVAPLDMHHLLHSHWDSYPPADPRIHYLL
GMLYFFLGIAACVGNVLVLHIFGKHKNLRSPTNTLLMNLAFCDLMIFIGLYPEMLGNIFMNDGTWMWGDI
ACRLHAWFGLVFGFGQMQTLMYMSIDRYNVIVKGLSAQPLTYKKVTQWLAQVWIVSLFWGTAPFFGFGNF
ALDGILNTCSFDYFTRDMPAMSYIVGACVSAYVIPLIVIIVCYTFIVRAVFEHEETLRQQAAKMNVTSLR
SSASAEDTSAEFRIAKIAMINVCLWLWAWSPFTIVSFIGIFGNQAIITPYLSSLPVILAKTSSVYNPIVYALSHPKYQAALKEEFAWLCVKTNAGNSGSSDTKSSVTMESNQPA*
>BCR2_braKug Branchinella kugenumaensis (fairy_shrimp) Ecdy.Crus.Bran AB293438 18984904 full G? Rhd
MLNNSEPSFAAYSVADGIWYPAGTKQIDGAPADVIAMTHAHWKQFPPSNPAWNYLFGVIYFFLWIVNHI
GNGLVIWIFLKTKSLRTPSNMLIVNLAIADFFMMLTQSPLYIISAFTSRWWIWGHFWCRFYGYTGGITGIA
AIFTMVFIGYDRYNVIVKGMNGTKITKGMAFIMILWTWIYANAFCLPAMLEVWGNFSPEGLLSTCSFDYL
NDNKFHGYFYTMYIFTGAYCVPMLLLMFFYSQIVKAVWAHEASSRAQAKKMNVESLRSNADANAESAEMR
IAKVALTNVLLWVCIWTPYAFVAVTGAFGNRQILTPLVAQLPSLICKMASCLNPLVYAISHPKYRQVLQK
ELPWFCIHEPEDKKSDATSVGSATTTATA*
>BCR3_braKug Branchinella kugenumaensis (fairy_shrimp) Ecdy.Crus.Bran AB293437 18984904 full G?
MLNFSEPRFAAYSVAEGVWYPPGTTQIDGAPADIVALTHAHWKKFPPSNPAWNYLFACLYFFLWVINHI
GNGLVIKIFLKTKSLRTPSNMLIVNLAIADFFMMLTQSPLFIISAFSSRWWIWGHFWCRFYGYTGGITGIA
AIFTLVFIGYDRYNVIVKGMSGKRISKGMAFGMIVWTWVYANVFCLPPMLQVWGDFSPEGMLSTCSFDYL
NENRLHGPIFTGYIFFGAYCVPMFLLFFFYSQIVKAVWAHEAALKAQAKKMNVESLRSNADANAESAEVR
IAKVALTNVLLWICIWTPYAFVAVTGAFGNRQILTPLVAQLPSLICKCASSLNPIVYAISHPKFRQVIQK
DYPWFCIHEPESSADTKSVTSGQTQVAA*
>BCRa_dapPul Daphnia pulex (water_flea) Ecdy.Crus.Bran AB293433 18984904 full G? RhA
0 MSNNLSSGYSSVAYRSEGASVLWGYPPGLSIVDLVPDDMKEFIHPHWNKFPPVNPMWHYL 2
1 LGVIYVILGITSVT 1
2 GNSLVVHLFAKTRDLRTPANMFVINLAFSDLCMMITQFPMFVFNCFNGGVWLFGPLFCELYACTGSIFGLCSICTMAAISYDRYNVIVNGMNRRRMTY 1
2 GRAGGLILFCWIYAIGWSIPPFVGWGKYIPEGILDSCSFDYLTRDTM 0
0 TISFTCCLFAFDYCVPLIIIIFCYYHIVRAIVHHEDALRDQAKKMNVSSLRSNADQKSQSAEIRVAKIAMMNITLWVAAWTPYAAICLQGAVGNQDKITPLVTILPALIAKSASIFNPVVYAISHPKYRL 0
0 ALQKALPWFCIHEKEEKEPPQDRREDSQSIATTNTNSSDVSLP* 0
>BCRa_hemSan Hemigrapsus sanguineus (crab) Ecdy.Crus.Mala D50583 9318091 full G? BcRh1 compound eye R1-R7 blue-green 480nm
MANVTGPQMAFYGSGAATFGYPEGMTVADFVPDRVKHMVLDHWYNYPPVNPMWHYLLGVVYLFLGVISIAGNGLVIYLYMKSQALKTPANMLIVNLALSDLI
MLTTNFPPFCYNCFSGGRWMFSGTYCEIYAALGAITGVCSIWTLCMISFDRYNIICNGFNGPKLTQGKATFMCGLAWVISVGWSLPPFFGWGSYTLEGILDSCSYDYFTRDMNTIT
YNICIFIFDFFLPASVIVFSYVFIVKAIFAHEAAMRAQAKKMNVTNLRSNEAETQRAEIRIAKTALVNVSLWFICWTPYAAITIQGLLGNAEGITPLLTTLPALLAKSCSCYNPFV
YAISHPKFRLAITQHLPWFCVHEKDPNDVEENQSSNTQTQEKS*
>BCRb_hemSan Hemigrapsus sanguineus (crab) Ecdy.Crus.Mala D50584 9318091 full G? BcRh2 compound eye R1-R7 blue-green 480nm
MTNATGPQMAYYGAASMDFGYPEGVSIVDFVRPEIKPYVHQHWYNYPPVNPMWHYLLGVIYLFLGTVSIFGNGLVIYLFNKSAALRTPANILVVNLALSDLI
MLTTNVPFFTYNCFSGGVWMFSPQYCEIYACLGAITGVCSIWLLCMISFDRYNIICNGFNGPKLTTGKAVVFALISWVIAIGCALPPFFGWGNYILEGILDSCSYDYLTQDFNTFS
YNIFIFVFDYFLPAAIIVFSYVFIVKAIFAHEAAMRAQAKKMNVSTLRSNEADAQRAEIRIAKTALVNVSLWFICWTPYALISLKGVMGDTSGITPLVSTLPALLAKSCSCYNPFV
YAISHPKYRLAITQHLPWFCVHETETKSNDDSQSNSTVAQDKA*
>BCR_porPel Portunus pelagicus (sand_crab) Ecdy.Crus.Mala EF110527 19040762 full G? horrible distal frameshifts
MANSTGPQMAFYGSQDMTYGYPEGVSIVDFVRPEIKPYVHQHWYNYPPVNPMWHYLLGVIYLCLGFISIIGNGMVIYLFAKCQALRTPANILVVNLALSDLI
MLTTNVPFFTYNCFNGGVWMFSATYCEIYGCLGAITGVTSTWLLCMISFDRYNIICNGFNGPKLTNGKAIILAFISWAISVGFGIAPLFGWGKYILEGILTSCSYDYLTQDFNTRS
YNIIIFVFDYFLPAAIIIFSYVFIVKAIFAHEAAMRAQAKKMNVTnLRSGEAESQRAEIRIARTALVNVSLWFICwTPYALISLQgvlgdlsginlLVTTLPALLARSCSW*
>RGR1_homSap Homo sapiens (human) Deut.Euth.Euar NM_001012720 18474598 full G? syn(+PCDH21 -LRIT1 -GRID1 -WAPAL) RPE Mueller exon-skipping splice
0 MAETSALPTGFGELEVLAVGMVLLVE 1
2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2
1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1
2 RSQLAWNSAVSLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2
1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0
0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0
0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKREKDRTK* 0
>RGR1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono genomic frag G? last D in DRY motif Atlantogenata ERY other placentals GRY
0 1
2 ALLGLCLNGLTIASFRKIKELRTPSNLLVVSLALADSGICLNALMAALSSFLR 2
1 HWPYGAEGCRLHGFQGFATALASISLSAAIGWDRYLRHCS 1
2 RSKPQWGTAVSTVLFAWGFSAFWSMMPILGWGQYDYEPLRTCCTLDYSKGDR 2
1 NFTTYLFAVAFFNFVIPLFIMLTSYQSIEQRFKKSGLFK 0
0 LNTRLPTRTLLFCWGPYALLCFYATVENVTFISPKLRM 0
0 IPALIAKTVPVIDAFTYALRNEDYRGGIWQFLTGQKIERVEVENKIK* 0
>RGR1_galGal Gallus gallus (chicken) Deut.Saur.Arch NM_001031216 14985289 full G? syn(+PCDH21 -LRIT1 +CHAT -PARG) retinal ganglia
0 MVTSHPLPEGFTEIEVFAIGTALLVE 1
2 ALLGFCLNGLTIISFRKIKELRTPSNLLVLSIALADCGICINAFIAAFSSFLR 2
1 YWPYGSEGCQIHGFQGFLTALASISSSAAVAWDRYHHYCT 1
2 RSKLQWSTAISMMVFAWLFAAFWATMPLLGWGEYDYEPLRTCCTLDYSKGDR 2
1 NYITFLFALSIFNFMIPGFIMMTAYQSIHQKFKKSGHYK 0
0 FNTGLPLKTLVICWGPYCLLSFYAAIENVMFISPKYRM 0
0 IPAIIAKTVPTVDSFVYALGNENYRGGIWQFLTGQKIEKAEVDSKTK* 0
>RGR1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur BC135113 full G? retina G protein receptor syn(+PCDH21 -LRIT1 +CHAT -PARG)
0 MVTSYPLPEGFTETEVFAIGTTLLVE 1
2 ALLGLLLNGLTLLSFYKIRELRTPSNLFIISLAVADTGLCLNAFVAAFSSFLR 2
1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1
2 RSKLHWSTAVSVVFFIWGFSAFWSAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2
1 NYISYLFTMAFFEFLVPLFILMTAYQSIYQKMKKSGQIR 0
0 FNTSMPVKSLVFCWGPYCLLCFYAVIQDATILSPKLRM 0
0 IPALLAKTSPAVNAYVYGLGNENYRGGIWQYLTGQKLEKAETDNKTK* 0
>RGR1_cynPyr Cynops pyrrhogaster (newt) Deut.Amph.Batr FS290827 frag G? 20090923 lens regenerating iris
0 GFTEIEVFGLGTALLIE 1
2 ALLGFILNGLTLLSFYKIRSLRTPHNFLIVSLALADTGVCINAFIAAFSSFLR 2
1 YWPYGSEGCQIHGFQGFVTALSSISSCGVIAWDRYNQYCT 1
2 RTKLQWGTAISLVSFVWAFSAFWSVMPLLGWGQYDYEPLRTCCTLDYTKGDK 2
1 NFISYLFPLAFFEFVIPLFIMLTAYQSVEQKFKKTGQHK 0
0 FNTGLPVKTLVMCWGPYSLLCFYATIENATTISPKIRM 0
0 LPAILAKTAPAINAFLYGMGNESYRGGIWQFlTGQKIEKAEVDNKTK* 0
>RGR1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? syn(+PCDH21 -LRIT1 +CHAT -PARG) retinal ganglia
0 MVSSYPLPDGFTDFDVFSLGSCLLVE 1
2 GLLGILLNAVTIAAFLKVRELRTPSNFLVFSLAVADIGISMNATIAAFSSFLR 2
1 YWPYGSDGCQTHGFQGFVTALASIHFIAAIAWDRYHQYCT 1
2 RTKLQWSSAITLAVFVWLFTAFWSAMPLIGWGEYDYEPLRTCCTLDYTKGDR 2
1 NYVSYLIPMAIFNMAIQVFVVMSSYQSIAQKFKKTGNPR 0
0 FNPNTPLKAMLFCWGPYGILAFYAAVENATLVSTKLRM 0
0 MAPILAKTSPTFNVFLYALGNENYRGGIWQLLTGEKIDVPQIENKSK* 0
>RGR1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? retina G protein receptor
0 EGFTDFEVFGLGTALLVE 1
2 GLVGLLLNGLTLLAFYKIKELRTPSNLLITSLALSDFGISMNAFIAAFSSFLR 2
1 YWPYGSEGCQTHGFHGFLMALASINACAAIAWDRYHQNCS 1
2 RSRLQWSSAITVTVFIWGIAAFWSAMPLLGWGVYDYEPLRTCCTLDYSKGDR 2
1 NFISFFIIMGSFEFIFPIFIMLSSYQSCKSKFKKNGQVK 0
0 FNTGLPVKTLIFCWGPYSLLCFYATIENITILSPKLRM 0
0 * 0
>RGRa_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni AB079882 12687683 full G? Ci-opsin3 syn(PPT1 NOMO2+ FOXC2- FAHD1-) larval photoisomerase
0 MEVNDKRVYGVLMGLL 1
2 GLLTITGYSLLFVIFAKRPDLKKKNKFLLSLATSDLLITVHVFASTIAAFAPQWPFGDLGCQ 0
0 VDAFIGMAPTFISIAGAALIAKDKYYRFCKPKM 1
2 VGRNYSFHVYLTWTMGIIGGALPFIGFGRYGFETDDVTWRTGCLLDFKSISA 2
1 KYSFYIILISTVWFVWPVYKLVSSYMKISTKINKFYP 0
0 LLFVVPVQMAIGLLPYAIYAMVSITIGVSAVPYFCVVINN 0
0 LAAKVFVGSNPFIYIYFDPELRESCKQIFCSPPAPTNDKISEDSKDE* 0
>RGRa_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni genomic full G? Ci-opsin3 larval visual cycle photoisomerase
0 MDFSSKRTYGIAMAAL 1
2 GFIAWVGYGLLFVIFAKSPDLKKKNRFLFSLAVSDLLITIHVVASVVASFQSEWPFGSIGCQ 0
0 LDAFIGMAPTFISIAGAALVAKDKYYRICKPKM 1
2 LVGRNYSFSIYANWTLGIIGGLLPFFGFGQYGFETDDLSLRTGCLLDFKTVSA 2
1 KYRFYIVFISLVWFVWPLYKLTSHYIKISAKLDRFHP 0
0 LMFVVPLQMLVSLLPYAIYAMISITVGVSSAPYYLVAVNN 0
0 IAAKVFIGTNPFIYIYFDPELRLACKNLFKYSSTPVQDQIQDKKDE* 0
>RGRb1_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni AB078611 12373590 full G? CiNut entire neural tube
0 MEIDFGFARTVYGVALLLM 1
2 VFITLLGYAVYFGAIWRSKTLQTRHIWLTSLACGDIIMMVHLILESLSSLGMGHRPRQNFECQ 0
0 VGALVGLFSGYVTIASITWIAIDRYYRQCKPEK 1
2 VGVNYCFYVIIVWAMSFLAASGPALGFGAYESAEENTVKCLIDLNKKDT 2
1 NSRLYIILVSAVWFVYPFVKMILYNKKLVQEAKEPQP 0
0 MAFAVPLTFFLCYLPFAIYASLKITVGLPPLNSMVVASIY 0
0 MLPKVISVVNPYLYMRSDPELLAACRHVVGLTDGKKAV* 0
>RGRb2_cioInt Ciona intestinalis (tunicate) Deut.Uroc.Tuni genomic full G? retina G protein receptor
0 MELEFGITRTAYGIAMLMM 1
2 AAVATFGYSVYILAIWSSKKLQTKHIWLTSLACADLLMMVHLFMDGLSSFHQGRRPKGIFECQ 0
0 VSAHMGLFSGFVSIASMTWICIDRYYRKFKPEK 1
2 VGVNYCFYVIIVWAMSFLAASGPALGFGAYESAEENTVKCLIDLENTDM 2
1 NTIKYFVVVGFLFFFYPIFKMIKYNTKFAYKSEEEKA 0
0 VVIAAPVSFVLGYLPYLVYACLKLTIGLPPLNQASIAFL 0
0 YLLPKFISVMNPYMYMRSDPELLRAAKRVVNFDFDQKIE* 0
>RGRb3_cioSav Ciona savignyi (tunicate) Deut.Uroc.Tuni genomic full G? retina G protein receptor
0 MELEFGIIRTAYGIAMLLM 1
2 AVVATFGYSIYLRAIWSSRKLQTRHIWLTSLACADLIMMVHLFMDGLSSFHQGRRPKGNFECQ 0
0 ASALMGLFSGFVSIGSVTWICIDRYYRRYKPEK 1
2 AGLNYCFYVIIVWGLSFLAASGPAMGFGTYESAQENTVKCLIDLDNTDG 2
1 NTIKYFMVIGVLYFFYPIFKMVKYNTLAQATEVEKT 0
0 VVIAAPATFVIGYLPYLSYAILKLTIGLPPVNEAFIAFV 0
0 ILPKFISVLNPYMYMRSDPELLRAAKDVVNFNFYTKMD* 0
>RGR2_danRer Danio rerio (zebrafish) Deut.Acti.Otoc NM_001024436 full G? retina G protein receptor embryonic RPE brain gut embryo PA
0 MASYPLPEGFTDFDMFAFGSALLVG 1
2 GLLGFFLNAISVLAFLRVREMQTPNNFFIFNLAVADLSLNINGLVAAYACYLR 2
1 HWPFGSEGCQLHAFQGMVSILAAISFLGAVAWDRYHQYCT 1
2 KQKMFWSTSITISCLIWILAVFWAAMPLPAIGWGVFDFEPLRTCCTLDYSQGDR 2
1 GYITYMLTITVLYLAFPVLVLQSSYSAIHAYFKKTHHYR 0
0 FNTGLPLKALLFCWGPYVVVCSLACFEDVSVLSPRLRM 0
0 VLPVLAKTSPIFHAVLYAYGNEFYRGGVWQFLTGQKSADKKK* 0
>RGR2_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G? retina G protein receptor eye
0 MAAYTLPEGFTEFDMFTFGTALLVG 1
2 GVLGFFLNAISIVSFLTVKEMRNPSNFFVFNLALADISLNVNGLIAAYASYLR 2
1 YWPFGQDGCSYHAFHGMISVLASISFMAAIAWDRYHQYCT 1
2 RQKLFWSTTLTMSSIIWILSIFWSAVPLMGWGVYDFEPMRTCCTLDYTRGDR 2
1 DYVTYMLTLVVLYLTFPAATMWSCYDSIYKHFKKVHQHR 0
0 FNTSMPLRVLLVCWGPYVVMCVYACFENVKVVSPKLRM 0
0 LLPVIAKTNPIFNALLYTFGNEFYRGGVWHFLTGHKIVDPVLKKSK* 0
>RGR2_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? retina G protein receptor eye
0 MAAFALPEGFTEFDMFTFGSALLVG 1
2 GLIGFFLNAISIASFLRVKEMWNPSNFFVFNLAVADICLNVNGLTAAYASYLR 2
1 YWPFGQDGCTFHAFQGMIAVLASISFMGVIAWDRYHQYCT 1
2 RQKLFWSTTLTMSAIIWILSIFWAAVPLMGWGVYDFEPMRTCCTLDYTKGDR 2
1 DYVTYMLTLVFLYLMFPALTMWSCYDAIHKHFKKIHLHK 0
0 FNTSTPLRVLLMCWGPYVLMCIYACFENVKVVSPKLRM 0
0 LLPVVAkTNPIFNALLYSFGNEFYRGGVWHFLTGQKMVDPVVKKSK* 0
>RGR2_oryLat Oryzias latipes (medaka) Deut.Acti.Perc genomic full G? retina G protein receptor whole embryo
0 MGTHTLPEGFTDFDMFTFGSALLVG 1
2 GLLGFFLNAISILAFLRVKEMRSPSSFLVFNLALADISLNINGLTAAYASYLR 2
1 YWPFGQEGCDYHGFQGMISVLASISFMAAIAWDRYHQYCT 1
2 RQKLFWSTSITISLIIWILSILWSAFPLMGWGVYDFEPMRIGCTLDYTKGDR 2
1 DYITYMLSLVFFYLMFPAFIMLSCYDAIYKHFKKIHYYR 0
0 FNTSLPLRVMLMCWGPYVLMCIYACFENVKLVSPKLRM 0
0 LPVIAKTNPFFNALLYSFGNEFYRGGVWNFLTGQKIVEPDVKKSKQK* 0
>RGR2_pimPro Pimephales promelas (minnow) Deut.Acti.Otoc genomic full G? retina G protein receptor liver brain PA
0 MASYALPEGFSDFDMFAFGSALLVG 1
2 GLLGFFLNLISVLAFLRVREIQTPNNFFIFNLAVADLSLNINGLVAAYASYLR 2
1 YWPFGSEGCQIHGFQGMVSILASISFLGAIAWDRYHLYCT 1
2 KQKMFWSTSGTISALIWILAVFWAALPLPAIGWGVFDFEPMRTCCTLDYTIGDR 2
1 NYISYMLTITVLYLAFPVLIMQSSYNGIYAHFKKTHHFK 0
0 FNTGLPLKMLLFCWGPYVLMCTYACFENASLVSPKLRM 0
0 VLPVLAKTSPIFHAAMYAYGNEFYRGGIWQFLTGQKPADKKK* 0
>PER1_homSap Homo sapiens (human) Deut.Euth.Euar NM_006583 17167409 full G? RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6)
0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1
2 GMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0
0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDV 1
2 GRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2
1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTK 0
0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2
1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0
>PER1_monDom Monodelphis domestica (opossum) Deut.Meta.Dide genomic full G? RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6)
0 MFKNNSVKTLAPEKEGPSVFSPIEHKIVAAYLITA 1
2 GVISIVSNVIVLGIFVKYKALRTATNTIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYDGCQ 0
0 IYAGLNIFFGMASIGLLTAVAIDRYLTICQPDL 1
2 GRMTSYNYTLMILTAWVNGFFWALMPIVGWAGYAPDPTGATCTINWRKNDV 2
1 SFVSYTMTVITINFAMPLGVMFYCYYNVSQKMKQYSPSNCPDHINRDWSNQVAVTK 0
0 MSVVMILMFLLAWSPYSIVCLWASFGDPKEIPPAMAIVAPLFAKSSTFYNPCIYVAANKK 2
1 FRRAISAMIRCQTHQSMPISNALPMN* 0
>PER1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono XM_001506366 full G? RRH peropsin
0 MRRNDSANLLESEHHDRSAFSQTDHNIVAAYLITA 1
2 GIMSIVSNVIVLGIFVKFEELRTATNAIIINLAVTDIGVSGIGYPMSAASDLHGSWKFGHAGCQ 0
0 IYAGLNIFFGMSSIGLLTVVAVDRYLTICRPAI 1
2 GRKMTRSNYTAMILAAWMNGFFWASMPLLGWASYASDPTGATCTINWRKNDA 2
1 SFISYTMTVIAVNFAVPLIVMFYCYYNVSKAMRQYPASRVLENLNIDWSEQVDVTK 0
0 MSVVMILMFLMAWSPYSIVCLWSSFGDPKKISPAVAIMAPLFAKSSTFYNPCIYVVANKK 2
1 FRRAMLSMVQCQTHREITITDVLPMNRSRSPH* 0
>PER1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6)
0 METLAEVSTLLPAGTGTVNISDASSEVHSVFSQSEHNIVAAYLITA 1
2 GVISILSNIIVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYVGCQ 0
0 IYAGLNIFFGMASIGLLTVVAIDRYLTICRPDI 1
2 GRRISGRHYTAMILAAWINAVFWSVMPVVGWSSYAPDPTGATCTINWRKNDV 2
1 SFVSYTMSVVAVNFVVPLMVMFYCYYNVSRTMKGYGSRSSLGGINADWSDQTDVTK 0
0 MSMVMIVMFLVAWSPYSIVCLWSSFGDPRKIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2
1 FRRAILSMVQCKSRQEVTLDNHFPMNVSQSTLTT* 0
>PER1_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic full G? RRH peropsin syn(+GPR68 -GNPDA1 -ENPEP -C14orf100)
0 MGIDPEVNVTDDVTLYGGKSAFTQLEHNIVAGYLITA 1
2 GVISLFSNIVVLLMFWKFKELRTATNFIIINLAFTDIGVAGIGYPMSAASDIHGSWKFGYAGCQ 0
0 IYAALNIFFGMASIGLLTVVAIDRYLTICRPDI 1
2 GQKMTMQSYNLLILAAWLNAVFWSSMPVVGWASYAPDPTGATCTINWRQNDV 2
1 SFISYTMAVIAVNFVLPLSAMFYCYYNVSATVKRYKASNCLDSANIDWSDQMDVTK 0
0 MSIVMIIMFLVAWSPYSIVCLWASFGDPKTIPAPMAIIAPLFAKSSTFYNPCIYVIANKK 2
1 FRRAIIGMVRCQTRQRITINSQVPMTTSQQPLTQ* 0
>PER1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? RRH peropsin
1 LFVSYTMTVIAVNFVVPLSVMFFCYYNVSKTMSRFISSPSPENINLDWSDQLDVTK 0
0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2
1 FRKAIMAMICCQNRQEITINHTLPMTISRVPLTE* 0
>PER1_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002228504 12435605 full Go RRH peropsin Amphiop1
0 MNASPSSWLPSGELFTDSPENSSEWPWTDGPTDTAWHHHQTVDPVTYGGYLASAVYLTIT 1
2 GLIAFVGNIFAIIVFLTEKEFRKKEHNSFALNLAIADLSVCVFAYPSSTIS 1
2 GYAGEWMLGDVGCTIYGFLCFTFSLTSMVTLCAISVYRYIVICKPQY 1
2 AHLLTHRRTNYVILGIWLYALVFSVPPLFGVNRYTYEPIS 2
1 ITCSLDWNVQHVGETIYTAAVIIIVYVLNVSIMCFCYFNIIFKSANLKFAALASEKTRTAAKKDIWKTSM 0
0 MCLAMVVSFLIAWTPYAVSSTWDILTEEDLPIIATILPTMFAKSSCMMNPIIYSCCNGKFRQAALKTFSK 0
0 VGSSNKQNGQAQVEPRDPGFAVEPAGHQAFQMRVLPSSSAMTL* 0
>PER1_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050606 12435605 full G? RRH peropsin Amphiop1 introns from braFlo
0 MNASPSSWLSSGEFFTDSPENSSEWPWTDGPTDTTWRHHQSVDSVSYEGYLASAIYITLT 1
2 GLIAFFGNVITITVFLTEKEFRKKQQNGFVLNLAIADLSVCVFAYPSSAIA 1
2 GYAGRWVLGDVGCTIYGFLCFTFALVSMVTLCVISIYRYILICKPQY 1
2 AHLLTHRRTVYVIIGTWLYALVFTVPPLVGVKRYTYEPMQ 2
1 ITCSLDWNVQHPGEKAYIAAVLVIVYVLQVLIMCFCYFNIIFKSANLKFAALASEKTKMAAKKDTWKTSV 0
0 MCLTMVVSFLIAWTPYAVSSTWDILSAEDLPIIATILPSLFAKSSCMMNPIIYACCNTKFRQAAVKSFRK 0
0 LCGMCKQKVPLSTPQVVLAMQRNTEFTSTVEPTGQAFPMRVLPSISATHTAL* 0
>PER2_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002209058 12435605 frag G? RRH peropsin Amphiop3
0 MIPTNNNTENNDLEWGLEKEHGVSATIMGVYLTIV 1
2 GLVSTVGNATVVLMFMLKWRQLCRKAPNLLIINLAAVDLCISVFGYPFSASSGFANQWLFSDAICT 0
0 LYGFSCFLLSMVSMHTLCLISAHRYITICRPEH 1
2 ASKLTMTRTILAVVGAWVYGISVAVPPLFGIA 1
2 GYTYESFGLSCTIDFHGTTVADMVYLSILIILCYVINVAVMGTCYFKIIRK 2
1 FSKHRFREVRDVRTSHQHSFERGVTL 0
0 RCILMTLFYLISWTPYTAVAVWTMVGPPPPVQLGMVAALTAKTHCAFNPILYMLMSE 0
0 VYRKLVLRTMCPCCFNKISNKLVRLPADDSKHSGNLDIFTVGYNTRDQAVQINKNAARRFCFVMET 0
0 ASDDLGIDDEVFAGQLGLCSRVKATEPGVEGFGGSEVPQSPSGTESEWSLSLLDFLPKRSSSKTAKASSLSETCSDNTVLSSAARK
MAFLESSHQQSDREVCCIENRQAPEDTKPCKFAIESLGVRLPHKCCTASQVAGAPSRYAGMIETFTDSKGKTKKKAAVSLSEIDVKKP
PPASKTWERRKTSKNTSRGQRVKRTFGKSRKHAYIVDC* 0
>PER2_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050607 12435605 full G? RRH peropsin Amphiop2 introns from braFlo
0 MIPTNNNTENNDLEWGLEKEHGVSATIMGVYLTIV 1
2 GLVATVGNATVVLMFIMKWRQLCRKAPNLLVINLAAANLCITIFGYPFSASSGYAHQWLFPDAICT 0
0 LYGFSCFLLSMVSMHTLCLISAHRYITICRPEH 1
2 ASKLTMNRTVLAVIGTWLYAIAVAVPPLFNIA 1
2 RYTYEPSGLSCTIDFRVTTVADLVYLGSLIVLCYVIHVAVMATCYFKIIRK 2
1 FSRHRFRQVRDIRTSHQRSFEMGVTM 0
0 RCILMTLFYLLSWTPYTAVCIWTMVGPPPPVVVSMAAALIAKTHCAFNPILYAFMSE 0
0 VYRKLVFRTMCPCCFNRISCKFVGTPTGGSKVSANPDIFTVDYNSRDQAVQINKAPSRRFCFVMET 0
0 SEDLGSDDTGLTGHSGLWRSGAEVEGLGGLQVTQSPSVSGSELSLSLLDFLPPKPSGRAVSAKLPSPPALNSERATCP
ESSQQPSDRPATGLRQYQKGDTTRSSVGDLILTEDDVTNLPPASETWGRKKSENPLSYRQTTRRTFGRSRKHSYIVD* 0
>PER3_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran AB050610 12435605 full G? RRH peropsin Amphiop3
0 MDIPTETPYGAGDDPAGTGWRWAETDQNGFHKYDHLIVGLYLFVI 1
2 GIIGTVENGITLATFTKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLR 0
0 SHWLFGGVGCQWYGFNGMFFGMANIGLLTCVAVDRYLVICRQDL 1
2 VDKVNYNTYGVMAALGWLFAAFWAALPLVGWAEYSLEPS 1
2 GTACTINWQKNDSLYISYVTSCFILGFALPLAVMMFCYWQ 0
0 ASCFVNKVLKGDISGDLTFPVAVNVDWEYQNHFSK 0
0 MCLAMVAAFVVAWTPYSVLFLFAAFGNPADIPAWITLLPPLIAKSSALYNPIIYIIANRRFRSAIFSMVKGQNPDVE 0
0 TLFARDFRISPIEDTGKEMSSMGNANA* 0
>PER3_braBel Branchiostoma belcheri (amphioxus) Deut.Ceph.Bran AB050610 12435605 full G? RRH peropsin Amphiop3 introns from braFlo
0 MDIPTETPYGAEEDIGESAGWRWTETDKNGFHKYDHLIVGLYLFVI 1
2 GIIGTIENGITLATFSKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLR 0
0 SHWLFGGVGCQWYGFNGMFFGMANIGLLTCVAVDRYLVICRHDL 1
2 VDKVNYNTYGVMAALGWLFAAFWAALPLVGWAEYALEPS 1
2 GTACTINFQKNDSLYISYVTSCFVLGFVVPLAVMAFCYWQ
0 ASCFVSKVLKGDIAGDLTFPVAANVDWEYQNHFSK 0
0 MCLAMVAAFVVAWTPYSVLFLFAAFWNPADIPAWLTLLPPLIAKSSALYNPIIYIIANRRFRNAICSMMKGQDPDVE 0
0 DDEHADEHRVRSIEDNDKEIISMVNLNMTV* 0
>PER2a_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XM_778236 full Go RRH peropsin overshoots iMet spread across tandem
0 MAASVTESSATEAISRLEPEYMVPLTRTGYLLTAIYLTIV 1
2 GSIATVGNITVICVLCRYRTFRKRSINLLLINMAASDLGVSVAGYPLTTVSGYWGRWLFGDVGCQFYAFCVYTLSCSTISTHAAIAVYRYIYIVKTDL 1
2 RPKLTANFTSGVIVVIWVYAFFWTVTPFVGWSSYIYEPFGTSCSVNWVGRTISDISYMVACTIGVYLLQIFIMLYCYIRVAKK 2
1 IRGVDPGRTEEKDAGVVVFGRLRKREAKIDTHVTK 0
0 MCFMMMLTFIVVWAPYAVECLRAAHVHRISALSSVLPTMFAKSSCMVNPIIFLTSSSKFRQDLGKLWSRPSSQDSLQLEER
NKTQRSLYVRHSELGSAHGNDTASVYYEKERIYIGEMRATSIQKEAELLQRDPELLSIASSTNSDVKFVVRDRPKRYTKR
PVKPQGPRGPEMFTASGVTNKGSSTSDSGGQSTSSGTTGSKPKRSGRKASRQYSMKSQSEDTGEIFTLDGSALEMMSLRKL* 0
>PER2b_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XM_778236 17067569 full Go RRH peropsin no cdna inline tandem PER2_strPur
0 MNSFSEESYVTDPTTTQPTLFLTPLSQTGYLLTALYLTLV 1
2 GIVSTIGNITVLCVLCRYGTFRKRSVNILLMNMAVSDLGVSVAGYPLTAISGYRGRWVFADIGCQFSGFCVYALSCSTISTHAVVAIYRYIYIVKPYH 1
2 RPRLSSSTSCLAILCIWTFTLFWTITPFFGWSSYTYEPFGTSCSINWYGKSLGDLTYIICCVVFVFILPIIIMLYCYIGVAKK 2
1 IKGIDPLRTEERDIAVVFGRLRKHETKIDTRVTK 0
0 ICFMMMASFIVVWTPYAVGSIWASKIGKISASASVLPTMFAKSSCMINPIIFLTSSSKFRADLGKLWNRPSSLEHTIRVEERSREQRSFF
VRQSALPDAMVSRSASVYYDKERIYIGEMRAASIQKEADLLHRDPEAISIASSTSSSLQFVLKDRQNRYKKKAGEASKKGSNILHFPYDDTE
GSMINNLMRPRSHSVTSDNISRVFAPSLKRPTKKRSMSHPDIPSTSADIFTVSPTTIKNLQKQ* 0
>PER1a_sacKol Saccoglossus kowalevskii (acornworm) Deut.Hemi.Ente ACQM01133041 full G? RRH peropsin 7 ests + ACQM01133041
0 MVTTDSLANSTDEPVPSILTLQQHYAASVTLLAL 1
2 AVIGTVLSSVNFRMLLSNPDYCSKAGNFFLSLAVTDL 1
2 CVCIFETPFSAFSHHAGFWIFGDTACQLYAFFGIFFGLVNIFMVTFISLDRYWATCSPVE 1
2 VELKSKYYTRMTALGWMVALFWAAAPVFGWSRYaMEPSMASCSIDYMTNDFSYVTYITCLTLTCYVVPIVVMVYCYVKASKNIKYTGKVTEWAHENNATK 0
0 ISRLCVLQLVFCWSLYGFNCMWTVVADDVETLPKMLTVLAPILAKTTPILNSGLYFLHNKKFRGAAVDMFKAKEE* 0
>PER1b_sacKol Saccoglossus kowalevskii (acornworm) Deut.Hemi.Ente ACQM01067921 frag G? RRH peropsin 0 ests
0 1
2 gVLSVIGNSVVLEMFRRYKELLSPSAILLISLALADL 1
2 GLTIFGMSLSCVSSFAGRWLFGKFGCYFHGFAGMLFGLGSIGNLTVISIDRYIITCKRsL 1
2 QWSYRHYYALLAVAWSNALFWSMMPLFGWSSYALEPEGTSCTIDWMNNDNQYISYVSCVTVTCFILPCAVMTYDYLAAYMKMVKAGYTLSEETEKPNND 0
0 MCIALVAAFLLSWFPSATVFLWAAFGNPGNIPLSFTGVADAFTKIPAVFNPVIYVALNPEFRKYFGKTIGCRRKRKKPIAVRLNGSEQNVENTI* 0
>PER1_lotGig Lottia gigantea (limpet) Loph.Moll.Gast genomic full G? retinochrome
0 MTAAEFSSFEHSIVGITYMVI 1
2 GISGTLLSLLVALTFIREKGLFKYGRAWLHISLAIANVGVVGAFPFSGSSSFSGR 2
1 WLYGSGMCTFYGFIGMFFGIAAIGNVFALCVERYLVSKKKDS 1
2 VDKVSNQFYWMITALVWINAFFWGIMPALGWTS 2
1 YDIEPSGTSCTIKWQNYDSGYPSFMAMLSLTCFLIPLPVALICLILSGTDKITEDKEEKTYFREDQLRS 0
0 TCTFLLILALIGWGPYCFICIWALFADTTQVSMLAAVIPPLAAKTMVLLYPVAYCQGNKRFKNAFLGMFIFNESPKQQ* 0
>PER1_aplCal Aplysia californica (sea_slug) Loph.Moll.Gast EB338056 17190607 full G? retinochrome from 8 8k contig to traces
0 MNDDGNALDGAGVDPAATAAHVSTALGFTKFEHASVGALYMLF 1
2 CIVGVTLNLLTALTFYKDTKLTKGSQPWLHILLALANVGVVAPSPFPASSSFSGR 2
1 WLYGSTMCQIYAFEGMFIGIAAIGAVIALCIERYIACQRSGA 1
2 NDTQGWFYGWSITLVLGNALFWAIMPLLGWSR 2
1 YSVEHTGTSCSIDWKNPDESFVSYIMTLEVFSFGIPMMSAFFCLISASPRPQPQGGATAAAGSGQEQQGEDTACKGCFSEDQLRL 0
0 LCYVFIGFVLVGWGPFAYLCTLAVFSDARGISMLAAAIPPLACKAMVSAYPLAYAVVSPRFRQSFLALIGGGEKKKE* 0
>PER1_todPac Todarodes pacificus (squid) Loph.Moll.Ceph genomic 2226795 full G? retinochrome retinochrome 1991 seq dubious before GDPAH
0 MFGNPAMTGLHQFTMWEHYFTGSIYLVL 1
2 GCVVFSLCGMCIIFLARQSPKPRRKYAILIHVLITAMAVNGGDPAHASSSIVGR 2
1 WLYGSVGCQLMGFWGFFGGMSHIWMLFAFAMERYMAVCHREF 1
2 YQQMPSVYYSIIVGLMYTFGTFWATMPLLGWAS 2
1 YGLEVHGTSCTINYSVSDESYQSYVFFLAIFSFIFPMVSGWYAISKAWSGLSAIPDAEKEKDKDILSEEQLTA 0
0 LAGAFILISLISWSGFGYVAIYSALTHGGAQLSHLRGHVPPIMSKTGCALFPLLIFLLTARSLPKSDTKKP* 0
>PER2_patYes Patinopecten yessoensis (scallop) Loph.Moll.Biva AB006455 9287291 full Go retinochrome depolarizing scop2 inner retina layer
0 MPFPLNRTDTALVISPSEFRIIGIFISIC 1
2 CIIGVLGNLLIIIVFAKRRSVRRPINFFVLNLAVSDLIVALLGYPMTAASAFSNR 2
1 WIFDNIGCKIYAFLCFNSGVISIMTHAALSFCRYIIICQYGY 1
2 RKKITQTTVLRTLFSIWSFAMFWTLSPLFGWSSYVIEVVPVSCSVNWYGHGLGDVSYTISVIVAVYVFPLSIIVFSYGMILQEKVCKDSRKNGIRAQQRYTPRFIQDIEQRVTF 0
0 ISFLMMAAFMVAWTPYAIMSALAIGSFNVENSFAALPTLFAKASCAYNPFIYAFTNANFRDTVVEIMAPWTTRRVGVSTLPWPQVTYYPRRRTS
AVNTTDIEFPDDNIFIVNSSVNGPTVKREKIVQRNPINVRLGIKIEPRDSRAATENTFTADFSVI* 0
>PER_hasAda Hasarius adansoni (spider) Ecdy.Chel.Arac AB525082 19960196 full Go peropsin first ecdysozoan peropsin
0 MDDNMSEIALADDMSTLSTQEPSENVYPYVFPLSTHTIVGTYLIII 1
2 GILGTLGNGLVLVTFLRFRVLVTPTTLLLVNLAVSDLGLILFGFPFSASSSLSAK 2
1 WIFGEGGCQWYAFMGFLFGSAHIGTLALLALDRYLIACRISL 1
2 RGKLTFKRYTQMITVVWTYAFFWALMPLLGWGR 2
1 YGLEPSVTTCTIDWQHNDSSYKSFLIVYFVLGFMVPFAIIAVSYIAIARRVGKKSKERPVVRDLWTNERSVTL 0
0 MAFILIVTFFVAWSPYAVLCLWTIFAEPNTVPPFLTLIPPLFAKSSTVVNPLIYFLSNPKLRTAILSTLSCCNEAPIQNIELPDSPERAANNDAI* 0
>PER_ixoSca Ixodes scapularis (tick) Ecdy.Chel.Arac ABJB010816023 frag Go peropsin mRNAs XM_002409761 XM_002435011 embryonated eggs
1 WLFGATGCQAYAFMGFLFGSAHIGTLTLLALDRYLATCRIGF 1
2 RSKPTFKRYFQLLLLVWLYGLFWAVMPLLGWAR 2
1 YGLEPSFQSCTIDWRHNDSSYKSFTLVYFVLGFLVPACIVLVCYRTSAIHIRAPKPKTVRRDVTDDYWASEEMVTV 0
0 MVALIVVTFFFAWTPYAVLCLWAVFADTKSVPHLLAMVPPLFAKTASTINPFIYFLSNPCIRADVLQLLGCRARSSPHMAISSDAVEEERCCQDQA* 0
>NEUR1_homSap Homo sapiens (human) Deut.Euth.Euar NM_181744 14623103 full G? OPN5 neuropsin index sequence
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2
1 LHTVTTVRKSSAVLEIHEEV* 0
>NEUR1_calJac Callithrix jacchus (marmoset) Deut.Euth.Euar genomic full G? OPN5 neuropsin
0 MALNHTSLPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHEEV* 0
>NEUR1_musMus Mus musculus (mouse) Deut.Euth.Euar genomic full G? OPN5 neuropsin
0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQAGGLRGTKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHQEV* 0
>NEUR1_ochPri Ochotona princeps (pika) Deut.Euth.Euar genomic full G? OPN5 neuropsin
0 MALNDTALPQDEHLPHYFRDGDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRLYGWADFFFGCGSLITMTAVSLDRYLK 1
2 GVWLKRRHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHGSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFSCCRTGGLKQTKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHQEv* 0
>NEUR1_canFam Canis familiaris (dog) Deut.Euth.Laur genomic full G? OPN5 neuropsin
0 MALNHTARPQDERLPHYLREGDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASLGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGRLKATKKKSLEDFR 2
1 LNTVTTVRKSSAVLEIhQEV* 0
>NEUR1_bosTau Bos taurus (cow) Deut.Euth.Laur genomic full G? OPN5 neuropsin
0 MALNHTAPPPDERRPPYLRDGDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAICDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GIWLKRKHAYICLAVIWAYAAFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2
1 LHTVTTVRKSSAVLEVHQEv* 0
>NEUR1_loxAfr Loxodonta africana (elephant) Deut.Euth.Afro genomic full G? OPN5 neuropsin
0 MTLNHTAPPQDDRLPQYLQDGDPFTSKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSCRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFVIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHMLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLRATKKKSLEGFR 2
1 LHTVTTVKKSSAVLEVHQEv* 0
>NEUR1_dasNov Dasypus novemcinctus (armadillo) Deut.Euth.Xena genomic full G? OPN5 neuropsin
0 MALNHTALPQDDRLPHYLRDGDPFASKLSWEADLVAGFYLTII 1
2 gILSTFGNGYVLYMSSKRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLRATKKKSLEDFR 2
1 LHTVTTVRESSAVLEVHQEV* 0
>NEUR1_monDom Monodelphis domesticus (opossum) Deut.Meta.Dide genomic full G? OPN5 neuropsin
0 MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTII 1
2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1
2 VVGKPFTIISCFSHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1
2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPVQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCQSGGQKAAKKESLRTYR 2
1 LHTVTTVRRSSAVLEIHQEv* 0
>NEUR1_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono genomic full G? OPN5 neuropsin
0 MTNYSAPQLGDYLPHYLREGDPFVSKLSWEADLVAGVYLVII 1
2 GVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1
2 VVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1
2 GTWLKRHHAYICLAIIWAYASFWATMPLVGLGNYAPEPFGTSCTLDWWLAQASVAGQAFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQNSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPIQFSVVPTLLAKSAAMYNPIIYQVIDCRISCCRLGGPKTGKKESLKNSR 2
1 SHSMSTIRKPSAVSGPHQEV* 0
>NEUR1_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full G? OPN5 neuropsin
0 MASDCNSSSQEEYLPHYMQQEDPFASKLSREADIIAGFYLTVI 1
2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1
2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1
2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGPKTLQKKSSLKESR 2
1 MYTISSHRDSAALSGTQLEV* 0
>NEUR1_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? OPN5 neuropsin
0 MAGNSSYREESGYIPHYERDSDPFASKLSREADIFAGVYLMAI 1
2 GILSTLGNGYVIYMACSRKKKLRPAEIMTINLAVCDLGIS 1
2 VTGKPFAIVSCFSHRWVFGWNACRWYGWAGFFFGCGSLITLTVVSLDRYLKICHLRY 1
2 GTWLKRRHAFIALAVIWAYATLWATLPLVGVGNYAPEPFGTTCTLDWWLAQASVKGQIFVLSMLFFCLLFPTMVIVFSYAKIIAKVKSSAKEVAHFDTRNQNNHTLEIKLTK 0
0 VAMLICAGFLIAWFPYAVVSVWSAFGQPDSIPIELSVVPTMMAKSASMYNPIIYQVIDCKPACCKKDKSLQNTTSR 2
1 VYTISTFRKSTTSAR* 0
>NEUR1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic frag G? OPN5 neuropsin
0 MENETSISSGYIPHYLLRGDPFASKLSKEADIVAAFYILVI 1
2 GILSATGNGYVMYMTFKRKTKLKPPEIMTLNLAIFDFGIS 1
2 VSGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSFDRYLKICHLRY 1
2 gTWLKRHHAFLSVVFIWAYAAFWATMPVVGWGNYAPEPFGTSCTLDWWLTQASVSGQSFVMCMLFFCLIFPTVIIVFSYVMIIFKVKSSAKEVSHFDTRNKNNHSLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVM
>NEUR1_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? OPN5 neuropsin
0 MTAFDNSTALYSGYWLHDSLHGDPFVSKLSWEADIISACYLIVT 1
2 GLLSTLGNGYVIYLSITQKRKLKPPEILITNLAISDFGMS 1
2 VGGQPFLIISCFSHRWIFGWVGCRWHGWAGFFFGCGSLITMTVVSLDRYLKICHLQY 1
2 GSWLQRRHVFMSLAFIWFYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLARVSVSGLIFVLTILFFCLLLPIIIIVFSYIKIIAKVKSSAKEVAHFDSRIQNHHSLEMNLTK 0
>NEUR1_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag G? OPN5 neuropsin
0 VAMMICAGFLLAWIPYAVVSVWSAFGAPDSVPVAVSMVPTMFAKSAAMYNPLIYQLLSRRGTGAHCCRCRKARGTLRRPR 2
>NEUR1a_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran FE548698 frag G? OPN5 neuropsinframeshifted cDNA FE548698 last exons missing
0 MATTPADRLDGLTPAGRGATTAETHADDFASKLSREADIVIGVYLILI 1
2 GTGAILGNGRVLWLSYRCRARLRPVEMFVVSLAVADVGLSLVGHPFAAASSLMGRWSFGSAGCTW 1
2 YGFVVFFLGIASIASMTLMSIMRFMIVYKRYP 1
2 GQYPTRRASCVLVTAAWLYGLFWACAPLA 1
2 GWSQYQPEPYGLSCSVDWGGFSSDAGGSSFIICMLLFCTAVPVVIMVTSYAAIFVIYRQAQKGVVLNLQVNATFGGKRQRTERKLTL 0
0 IALAVCGGFLLAWLPYAVVGLWASVAGVDAVPLALASAAPLFAKSNSLWNPIIYLGMNERFR 2
>NEUR1b_braFlo Branchiostoma floridae (amphioxus) Deut.Ceph.Bran XM_002202511 full G? OPN5 neuropsin from tracesseparate allele
0 MATTPGLPLDGLAPTGRGVTAADTLDDDFASKLSREADIVIGVYLLLI 1
2 GTGSILGNGRVLWLSYRNWAKLRPVELFVVSLAVTDVGISVFGYPFAASSSLLGRWSFGSAGCTW 1
2 YGFTGFFFGLTSIANMALMSIMRFMIVYKGYP 1
2 GPYPSRRATSGLIAAAWLYGLFWACAPLA 1
2 GWSQYHVEPFGLSCTVDWGSFSRDAGGMSFIICLLVFCVAIPVTAIMASYVAISAIYRQAKKSIAGHLQDNSAMCKKRNKLERSITL 0
0 MALAVCGGFLLAWLPYAVVGLWSAVAGVDAVPLALASAAPLFAKSSSLWNPIIYLGMNDRFS 2
1 AELSREHSYLPATGVRTTGPPAEDNQAEGDDVSSPSLLFFRRLFWTSKT 1
2 SNAMRTLVVLTWISTIFPLLLAAPQADPRTTYKVSRWDEAWRPQRFGRSGRGDTKDGWRPQRFGRGRYEQGWRPQRFGRNEGLAGLREVLDGEAFPLLQMTR
TDLHHDLPGIGYTPGTAHGSLRALPLLRLYDRGALQLINGAAKQSSTNPPSLRMIRAAAESLQGFARGGDQQDEEEMFAPPRSTDDWLAEIQRLGLRGRKRRDVS* 0
>NEUR_strPur Strongylocentrotus purpuratus (sea_urchin) Deut.Echi.Eleu XM_001197837 full G? CX694910 CX690664
0 MDVNAKWWTNETLRTRDQFSDDHYTSVLSYEGDIWAGVYLMFI 1
2 SLIAFIGNISVIVISLRKREKLKPIDLLTINLAIADFLICVVSYPLPMISAFRHR 0
0 WSFGKFGCVWYGFTSFLFAVGSMATLMVIALLRYAKLCRENV 1
2 DQYQSRPFVIKVIVAIWGFAFFTTAPPLFGWS 2
1 SYVPEPYHLSCTIDFADTSPSGLSYTYFTTIVVFFMPLMIIVLCYVAIARKMIHHNRRINVGHNAGRMLLEIRLLK 0
0 TACMITMAYTISWTPYAVIAMWVTYIPVNQIPDAFRILPAFCAKTSSVYNPIIYCIFNKSFRQDLSSLICCCACQCYTITINLDINSHAQQQFRRIEERR
DEVGTYKRRPLMICSNPFAWSRDFHETWRQRRIRGIHRNCRNNVRVENINVNFRRDTDMVELNAPTPAEIHRPELNTASTRSGARTKSMATHLPALEEVPSG
APQCSALLHNTPIPRSLQGTPLPYQPQPSTSDLHDEFLNPSVVSRNMCVIVVKPNIEEELSTD* 0
>NEUR2_galGal Gallus gallus (chicken) Deut.Saur.Arch AB368181 18570255 full G? cOpn5L1 syn(-B4GALT6 -NEUR2_galGal -KIAA1012) GenBank 5'UTR error
0 MDPSFANSTFQSKITEAADIVVGTCYMVF 1
2 GICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHR 2
1 WLYGKHICLFYAFCGLFFGICSLSTLTLLSVVCCLKICFPAY 1
2 GNRFRRKHGQILIACAWTYAAIFACSPLAHWGEYGEEPYGTACCIDWQSTNVDVMSMSYTVVLFVLCFILPCGVIVTSYSLILVTVKESRKAVEQHVSGPTRINNVQTITAK 0
0 LSIAVCIGFFAAWSPYAIIAMWAAFGSIDKIPPLAFAIPAVFAKSSTLYNPIIHLLLKPNFRSNIAKDFTVIQQLCVRCCFCVKELQTYRSTFNTGLRTFKGKNESSCNALPIMEG
CSYFPSEKGSHTFECFKSYPNCFQERLSTMGCHLQDCESLENDLQVEVTQGSRNSMKVVEQEEKSTELDNLEITLEAVPVSCTFTDL* 0
>NEUR2_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G?
0 MESYFANTTFHSKITEAADVIVGVFYIVF 1
2 GICSFCGNSILLYVSYKKKNLLKPAEYFMINLAISDLGMTLTLYPLAVTSSLAHR 2
1 WLFGQQVCLFYAFCGVFFGVCSLTTLTLLSIVCCLKICFPVY 1
2 GNRFRPGHGWILIACAWVYAAIFAFSPLAHWGEYGAEPYGTACCIDWRISNMKKTAMSYTTALFVFCYIIPCGIIITSYTLILITVKDSRKAVEQHALGPTRMSSVHTITAK 0
0 LSIAVCIGFFVAWSPYAIIAMWAAFGSIDMIPPLAFAVPAVFAKSSTLYNPAMYLFLKPNFRSTIAKDLTVLHRLCLKSCFCPRGMQNCSYRSALEAPLKSFKGRNESSSNSVQIVGGCS
YFPCEKCHDPFECFKNYPKCCQGRLNVMDHTPRESISVENNMQSKTKHASEKYIKVVIRGEKNTDIDNLEITLEHIPTDIKFANL* 0
>NEUR2_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? abundant transcripts
0 MGNKSDASAFYSSISETDDIVLGVLYSVF 1
2 GLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHR 2
1 WLFDKVTCNYYAFCGMLFGLCSLTNLTVLSSVCCLKVCYPAY 1
2 GNKFSTAHSRILLLGIWAYAGLFATAPLADWGKYGPEPYGTACCLDWEASYRERKALSYTISLFVFCYLIPSSLIFISYTLIFVTVKGARRAVQQHLSPQAKGSSIHSLIIK 0
0 LSIAVCIGFLIAWTPYAIVAMMAAFGDPTKIPSLVFALAAAFAKSSTIYNPVVYLLLKPNFLNVVTKDLTLFQTMCAVVCGWCRTPAVKTPCPHKD
LKTTSKPPSSFKKSQGVCRNCVDTFECFRNYPRCCSVGNVDAAQPMAASLVRIPPANGAPQQTVQLVVSSSRTRSGVETVEVSTEAPMSDFIKDFI* 0
>NEUR2_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G? acquired new intron
0 MGNVSKTALFMSTISRQHDILMGSLYSVF 1
2 FVLSLLGNGMLLFVAYRKRSSLKPAEFFVVNLSVSDLGMTLSLFPLAIPSALAHR 2
1 WLFGEITCLCYAVCGVLFGLCSLTNLTALSSVCCLKVCFPNY 1
2 GNKFSSSHACVMVIGVWCYASVFAVGPLVHWGSFGPEPYGTACCINW 2
1 YTPSHDALAMSYIISLFIFCYVVPCTIIILSYTFILVTVRGSQQAVQQHVSPQTKVTNAHALIVK 0
0 LSVAVCIGFLTAWSPYAIVAMWAAFSANEQVPPTAFALAAIMAKSSTIYNPMVYLLFKPNFRKSLSQDTQMFRHRICLSHSKASPSPGMKDQERQS
SQQCNNKDGSISTPFSSGQAESYGACHVYAEAGPHYQQISRQITARVLEGSVQSEIPVKQLTEKMQNDLL* 0
>NEUR2_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G? novel paralog of neuropsin
0 1
2 GILSLVGNSVLLFVAYRKRQILKPAEYFVANLAVSDISMTVTLLPLAISSNFSHR 2
1 WLFVSKpCMYYGFCSMLFGICSLTNLTVLSTVCCMKVCFPAY 1
2 0
0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVISYTMTVIAVNFVVPLSVMFFCYYNV
>NEUR3_galGal Gallus gallus (chicken) Deut.Saur.Arch AB368183 18570255 full G? cOpn5L2 testis exon 2 fused exon 3 rel NEUR1/2
0 MEEQYISKLHPVVDYGAGVFLLII 1
2 AILTILGNSAVLATAVKRSSLLKSPELLTVNLAVADIGMAISMYPLAIASAWNHAWLGGDASCIYYALMGFLFGVCSMMTLCAMAVIRFLVTNSSKSN 1
2 SNKISKNTVHILITFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHSSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNINRIPHAAKLEKKLTL 0
0 MAVLISVGFLSAWTPYAAASFWSIFNSSDSLQPIVTLLPCLFAKSSTAYNPFIYYIFSKTFRHEIKQLQCCWGWRVHFFSADNSAENSVSMMWSGRDNIRLSPTAKVESQGAARH* 0
>NEUR3_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch ABQF01025032 full G?
0 MEEQYISKLHPVVDYGAGVFLLII 1
2 AILTILGNSAVLATAVKRSSLLKPPELLTVNLAVADIGMALSMYPLAIASAWSHAWLGGDASCVYYALMGFLLGVCSMMTLCAMAVIRFLVTNSPKSN 1
2 sNKITKNTVCILIAFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHNSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNIDRIPNAAKLEKKLTL 0
0 MAVLISVGFLSSWTPYAATSFWSIFNSSHSLQPVVTLLPCLFAKSSTAYNPFIYYVFSKTFRCEVKRLQCCCAWRVHYFSSDNSVENPLSTMWSGRDNIRLSAAPQVQNPGAAAP* 0
>NEUR3_anoCar Anolis carolinensis (lizard) Deut.Saur.Lepi AAWZ01001057 full G?
0 MEEHYISKVHPVWDYGMGVFLLII 1
2 AILTILGNSMVLAVAVKRSSCLRSPELLTVNLAATDLGMGLSMYPLAIASAWNHAWLGGEATCIYYALMGFLFGVSSIMTLSAMAVIRFLVTFSSKPA 1
2 GHKINRKVMHICIMLIWAYAVLWAILPLLGWGHYGPEPFGTSCTIAWGQFHNSQKGFAFILSMFILCTFLPAITIIMCYLGIAWKFHKTHQEMQNLNRISSAAKLEKKLIL 0
0 VAVLISVGFLGAWTPYAIVSFWSVFHSSESIPYIVTLLPCLFAKSSTAYNPFIYYTFSKTFRHEVKHLRCYSGQRAQENMKNSINSNVSFMWHGGGNICLSTRQIEMREIPNQ* 0
>NEUR3_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G? cdna ovary embryo
0 MEERYLSKLHPLVDFGSGVFLLLV 1
2 AILTVLGNCAVLATAVKCSSHLKAPDLLSINLAVADLGMAISMYPLAIASAWNHAWLGGDASCLYYALMGFFFGVSSMMTLTVMAIIRYRVTSSFKYS 1
2 GCTIEKKAVCILIMCIWLYALLWAVLPLLGWGRYGPEPFGTSCTIAWGDFHHSSNGFSFIISMFILCTISPAVTIVVCYSGIAWKLHKAYQEIKNQDKIPNSTKVEKKLTL 0
0 LAILVSFGFLISWTPYAAVSFWSLFHSSKYIPPVVSLLPCLFAKSSTAFNPMIYYAFSKTFRRKVKHLKCCCGWRVHFLQSENSVENPRVSVIWTGKENVMVSSVPKLMKGVPGTPTGTQ* 0
>NEUR3a_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G?
0 MDRYTSKLSPAVDYSAGTFLLVI 1
2 AILSILGNAAVLLTAAWRHSVLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWIGGDPSCLYYGLMGMIFSVASIMTLAVMGLVRYLVTGNPPK 1
2 SGSKFRRKTISILIGVIWMYSLLWAVFPILGWGGYGPEPFGLACSVDWMGYQHSLNRSSFIMALAILCTLMPCVVILFSYSGIAWKLHKAYQSIQSNDNLPNSGAVERKVTL 0
0 MGILISTGFIVSWAPYVFVSLWTMFRSEGEDSVVPIVSLLPCLFAKCSTVYNPLVYYVFRKSFRREIHQIRICCFQGCWDAVSKMTRGDGPEETSGTHETDNI* 0
>NEUR3b_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G?
0 MDIYSSKLSSAVDYGIGAFLLLI 1
2 TILSILGNLMVLVMAYKRSNHMKPPELLSVNLAVTDLGAAVTMYPLAVASAWNHHWIGGDVSCVYYGLMGFLFGAASMMTLTIMAIVRFIVSLTLQSP 1
2 KEKISKRNAKILVATTWLYALLWAIFPLIGWGKYGPEPFGLSCTLDWRDMKEHSQSFVITIFLMNLILPAIIIVSCYCGIALRLYVTYKSMDDSNHVPNMIKMQRRLMV 0
0 IAVLISIGFVGCWAPYGIVSLWSIYRPGDSIPAEVSMLPCLFAKTSTVYNPFIYYIFSKTFKREVNQLSRFCGRSNICRPTDAKNRPENTIYLVCDVNKSKPGVEDLSLARSKENETQMLPNQDLHE* 0
>NEUR3a_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G?
0 MDDKYMSKLSPPVDLWAGIYLVVI 1
2 ALLSVLGNASVLFSASRRLTPLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWMGGDTACLYYGLMGMIFSITSIMTLAVMGMIRYLVTGSPPR 1
2 SGVQFQKKTICVVICAIWLYACLWAAFPLLGWGSYGPEPFGTACSIDWTGYGDSLNNATFIVAMSVLCTFLPCLVIFFTYFGIAWKLHKAYKSIKSSDFQYASVERRITL 0
0 IAVLISVGFLGSWAPYGLVSLWSILKDSSSIPPQVSLLPCLFAKSSTVYNPVIYYIFSQSFKLEVQQLFLCC* 0
>NEUR3_petMar Petromyzon marinus (lamprey) Deut.Agna.Hype genomic frag G? new exon
0 MAEQGEDDQFRSKLSPTADIAAGTFLLAV 1
2 AVLSLAGNGAVLGVAARRWAKLKAPELLSVNLALTDLGIAASIYPLAVASAWNHRWLGGQPVCTYYAFAGFFFGTASMGTLTAMAGVRYKGTSTQVH 1
2 VKQITKRAMLAVIVAVWAYALLWSCLPLLGWGR 2 1 YGVEPFGVSCTLAWAELQLTPGGVAFLYAMFVLCLLLPAIAIGLCYAGIVCKLRRAYREGRSKRRTPTARHVESRLTK 0
>NEUR4_ornAna Ornithorhynchus anatinus (platypus) Deut.Prot.Mono XM_001508128 full G?
0 MSLSHSLQVPWRNNLTFLNKEAQVSEQGETIIGIYLLAL 1
2 GWMSWFGNSMVIFILHRQRGILNPTDYLTFNLAVSDASVSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0
0 VDGFLTLLFGLASINTLAMISVTRYIKGCHPHR 1
2 GHFINTANISVALILIWVSALFWSAGPVLGWGSYT 1
2 DRMYGTCEIDWAEANFSSICKSYIISIFFCCFFLPVSIMFFSYVSIIKMVKSSHTLAGADDPTDRQRRLDRDVTR 0
0 VSVVICTAFIVAWSPYAVISMWSAFGHSVPNLTSVLASLFAKSASFYNPIIYFGMNSKFRKDILVLLPCAKESKEPVKLKKFKNLRQKQGFTLQKPEKAHVLQVPDSGPMSLINTPPLGNRNSFDLACDNSDFECVRL* 0
>NEUR4_galGal Gallus gallus (chicken) Deut.Saur.Arch genomic full G? genome gappy
0 MSLQLSPQAPWRNNNISFLSREAAVTEQGETIIGFYLLAL 1
2 GWMSWFGNSVVIFVLYKQRHLLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPER 1
2 AHCISNSSMTVAMVLIWIAAFFWSAAPLLGWGSYT 1
2 DRMYGTCEIDWAKANFSTIYKSYIISIFICCFFLPVTVMVFSYVSIINTVKLSHALTGLSDPTERQRRMERDVTR 0
0 IVICTAFIIAWSPYAVLLLWSAYGHPVPNLPLYLSSLFAKSASFYNPIIYFGMSSKFRRDIFILFHCAKEVKDPVKLKRFKNLKQKQEPSQKEEKYAAEMHPAPSPDSGVGSPTNTPPPANREEYFGILDTPSNSPDIECDRL* 0
>NEUR4_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch genomic full G?
0 MSVQFSAQAPWRNNNISFLTREAAVTEQGETIIGFYLLAL 1
2 GWLSWFGNSIVIFVLYKQRHVLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPER 1
2 GHCISNSSMSVALVLIWVAAFFWSAAPLLGWGSYT 1
2 DRMYGTCEIDWAKASFSTIYKSYIVSIFICCFFLPVTVMVFSYVSIINTVKLSHT LTGLGDPTDRQRRIERDVTR 0
0 VSLCTAFIIAWSPYAVISIWSAYGHPVPNLTSILASLFAKSASFYNPIIYFGMSSKFRRDIFIFHCAKELKDPVKLKRFKNLKPKQPQPSQKEEKYAPEMHPAPSPDSGVGSPTNSPPPANREVYFGILDTPSNNPNIECDRL* 0
>NEUR4_anocar Anolis carolinensis (lizard) Deut.Saur.Lepi genomic full G?
0 MSLQVSPQAPWRNNNVTFSNKEVPVSEQGETIIGFYLLAL 1
2 GWMSWFGNSIVIFVLYRQRAGLQPTDYLTFNLAVSDASVSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPDR 1
2 GKCISNSSISVALFLIWIAAFFWSVAPVLGWGSYr 1
2 DRMYGTCEIDWAKANFSTIYKSYIVSIFICCFFLPVSVMVFSYVSIINTVKSSHALSGVGDPTERQRRMERSVTR 0
0 VSLVVICTAFITAWSPYAVISMWSAYGYTVPNLTSILASLFAKSASFYNPIIYFGMSSKFRKDIFVLLHCAKEIKDPVKLKRFKNLKQKQEVSPSQREEKYAADVQPALSPDSGVGRSNTPPPVNREVYFGAFDTFSNNPDVECDRL* 0
>NEUR4_xenTro Xenopus tropicalis (frog) Deut.Amph.Anur genomic full G?
0 MSLQFPRPAPWRNNNLTLLQKENPLTEQGETIIGIYLLAL 1
2 GWLSWFGNSIVIFVLYKQRANLLPTDYLTFNLAVSDASTSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0
0 VDGFLTLLFGLASINTLTLISVTRYIKGCHPQR 1
2 ANCISNGSITISLALIWIAALFWSVAPLLGWGSYR 1
2 DRMYGTCEIDWTKASFSTIYKSYIISIFICCFFLPVMVMVFCYVSIINTVKSSRALTSEGDLSERQRKMERDVTR 0
0 VSVVICTAFIVAWSPYAVISMWSACGYYVPSLTSILAALFAKSASFYNPLIYFGMSSKFRKDLCVVLPCAKAQKDPVKLKRYKDKKQGSAPRAREQTEIEQPVQLQPAPSQDSGVGSPSNTPPLRTKDVHIVDIDLVSDNPSYECDRL* 0
>NEUR4_danRer Danio rerio (zebrafish) Deut.Acti.Otoc genomic full G?
0 MSAQNPLQVVNIPWRNNNFSLMSRDPPLSDQGETIIGVYLLIL 1
2 GWLSWFGNSIVIFVLFRQRSTLQPTDYLTLNLAVSDASISVFGYSRGILEIFNIFKDSGYIISSVWTCQ 0
0 VDGFFTLVFGLSSINTLTVISITRFIKGCHPHK 1
2 AHCITNSTVAVCVVFIWIGAFFWSAAPVLGWGSYT 1
2 DRGYGTCEIDWVKANYSTIHKSYIISIFIFCFLVPVLLMLFCYISIINTVKRGNAMNADGDLSDRQRKIERDVTI 0
0 VSIVICTAFILAWSPYAVVSMWSAWGFHVPNLTSIFTRLFAKSASFYNPLIYFGLSSKFRKDVSVLLPCGREGRDPVRLKRFKRLRGRAEPPGAPAHTPHPQIALKNYNNHSKPHAGPAHCTGHAPSPDSGVGSHHETPPPQPRPQLFFIDVPEPEAESECVRL* 0
>NEUR4_tetNig Tetraodon nigroviridis (pufferfish) Deut.Acti.Perc genomic full G?
0 MEPSRPWRNSSVLGGGAEPPLSEQGETIIGVYLLLL 1
2 GWLSWFGNTVVLFVLVRQRSSLQPTDLLTFNLAVSDASISVFGYSRGIIQIFNVFQDSGFIISSIWTCE 0
0 VDGFLTLIFGLSSINTLTVISITRYIKGCQPSR 1
2 AALISRSSVSVCLLLIWTTAGFWSGAPLLGWGSYT 1
2 DRGYGTCEIDWSKAASSGVYRSYIISIFIFCFFIPVFIMLFCYISIINTVKRGNALAADGHLSHRQRTMERDVTV 0
0 ISVVICTAFIMAWSPYAVVSMWSAWGFHVPSTTSIVTRLFAKSASFYNPLIYFGMSSKFRKDVSLILPCAKERREVVLLQRFKNIKPKAAAAPPPPPLPVYRPKEKNEDEPKLSVHDNDSGVNSPPETPPSDAQEVFPVDPPSQIETSEYWSDRL* 0
>NEUR4_gasAcu Gasterosteus aculeatus (stickleback) Deut.Acti.Perc genomic frag G?
0 PVKVVNIPWRNNNLSNLNTDPPLSEQGETFIGVYLLVL 1
2 GWLSWFGNSLVMFVLYRQRASLQSTDFLTLNLAISDASISIFGYSRGILEIFNIFNDDGYLINWIWTCQ 0
0 VDGFFTLLFGLASINTLTVISVTRYIKGCHPNK 1
2 AYCISTNTIAVSLICIWTGAVFWSVAPLLGWGSFT 1
2 DRGYGTCEVDWSKANYSTIHKSYIISILIFCFFIPVMIMLFSYVSIINTVKSTNAMSADGFLSTRQRKVERDVTRV 0
0 ISIVICTAFITAWSPYAVVSMWSAWGFHVPSTTSIITRLFAKSASFYNPLIYFGMSSKFRKDVSVLVPCTRERREVVHLQHFKNIKPKAEAPPTPASLPVQKLGAKYAVPNPDADSGVNNPPQRPATDPQGDLNIDLPSHIETSEYWCDRL* 0
>NEUR4_calMil Callorhinchus milii (elephantfish) Deut.Chon.Holo genomic frag G?
0 MGCSLGWKVLLWFLHGILICPRPWRNHNSTFQPKEHPISEQGETIIGVYLLIL 1
2 GWLSWFGNSIVIFILYRQRLSLQPPDYLTLNLAVSDASISIFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0
0 1
2 AVSISAGSIAASLVLIWIAAIFWSGAPLFNWGSYT 1
2 DRMYGTCEIDWSRASFSTIYKSYIISIFICCFFLPVFVMLFSYISIINTVKSSHAFAGNADLSDRQRRMEKDVTR 0
0 VSMVICTAFIIAWSPYAVISMWSASGYTVPQLTGIFASLFAKSASFYNPMIYFGLNSKFRKDIYILLPCVKEPKESVKLKRFKHLRHRPEQQQANKDRYAEELQQVASPDSGMGSPSKSPPLHNKDVFFVLWLRGLKK
>TMT_triCys Tripedalia cystophora (box_jelly) Cnid.Cubo.Cary EU310498 18577593 full G? c-opsin-like Schiff lysine mRNA with predicted introns
0 MADHGRNTTSNDTNPISKIPLDDFDPQRFYNFYTFMGSFIAGSACCSFLLNGLVIAVLIKYIRTITNTNIIVLSMSCANILIPLLGSPLSATSSLMRKWQFGNGGCTWYGFINTLS 1
2 GISGIYHLTFLSFERFITIVLPLKRDTILSTKNIYIGLGILWVAAIGVAGAPVFGWCEYIKE 1
2 GVRTSCSVAWSSKENMNVFSYNLFMIFTVFLLPMLVIIYCNYRFIKEVSIMSTRA 0
0 RGLQGGDSEMTASASKAEKQLTIMVITMIIAFNIAWLPYTVVSMVFLTGYGDVVGPMGASVPSVFAKTSVIYNPVIYCLLNRS 0
0 FRKMLCGNSVEPE* 0
>CUBOP_carRas Carybdea rastonii (sea_wasp) Cnid.Cubo.Cary AB435549 18832159 full G? cubop
MGANITEILSGFLACVVFLSISLNMIVLITFYRLRHKLAFKDALMASMAFSDVVQAIVGYPLEVFTVVDGKWTFGMELCQVAGFFITALGQVSIAHLTALAL
DRYFTVCRPFVATAIHGSMRNAGMVIFVCWFYASFWAVLPLVGWSNYDVEGDGMRCSINWADDSPKSYSYRVCLFVFIYLIPVLLMVATYVLVQGEMKNMRGRAAQLFGSESEAAL
KNIKAEKRHTRLVFVMILSFIVAWTPYTFVAMWVSFFTKQLGPIPLYVDTLAAMLAKSSAMFNPIIYCFLHKQFRRAVLRGVCGRIVGGNAIAPSSTAVEPGQTLASGTAES*
>ENCa_nemVec Nematostella vectensis (anemone) Cnid.Anth.Hexa genomic full G? Schiff lysine no cdna complete 1 exon
0 MIDNALGRHEANIVLGYYIAIFVIGFVTNTIVVIIFISSQRLHTTPNLILFSMSVCDWLMATMAKSVGIYGNARYWPTVGKVTCDYYAFATSAIGYASILHLAALAVEKRMAVVSPMTNSFNGRRMLVIIATLWGFAILW
AVFPLIGWSSYGPEPGYVSCSITWYTTDHNNVSYIICVSVLFFLIPIVTMTFCFASIYHTIRNLSHEATARWGSDARATQETIRAKAKTAKMAFLMVMCFLFAWTPYAVVSLWTTFGDTHRIPALLGVLPSLFAKLSSCY
NPIIYFFMYTKFRCAGKALLYQEHH* 0
>ENCb_nemVec Nematostella vectensis (anemone) Cnid.Anth.Hexa genomic full G? Schiff lysine NC-extended 1 exon
0 MSNEALRRHEANIVLGYYIAIFVIGFVTNTIVVITFIFSKRLHTTPNLILFSMSVCDWLMAAMAKSVGIYGNARYWPTVGKVTCDYYAFATSAIGYASILHLAALAVEKRMAVASP
MTNSLNERRMLVIIATLWGFAILWAVFPLIGWSSYGPEPGYVSCSITWYTTDHNNVSYIICISVLFFFVPIVTMTFSFASIYKAIRNISHEAIARWGSHARATQETIKAKAKTAKMAF
LMVMCFLFAWTPYAVVSLWTTFGGTHRNPALLGVLPSLFAKLSSCYNPIIYFFMYTKFRRAAKLLFIKKVIRPTEAERSRVLSGIRTRAASTFAVKLTVHAEKGQQIAPNITPQGAVKGPVESIEDNLQKIETITPCTSV* 0
>ENCc_nemVec Nematostella vectensis (anemone) Cnid.Anth.Hexa genomic full G? Schiff lysine C-extended 1 exo
0 MELFTSYHAITVMYSLLAAGAFVLNGIVLIIFLATRSLRTIPNMILLSMAWADWLMACLADAVGAYANANNWPSMVGGLCVYYGFITTALGLTSMIHLTALSVERFVTVTIPMTRPITETQMLLVVTFLWAFSFLWAIFP
LVGWSSYGPEPGYAACSIAWYRQDLNNMSYILCLFMFFFFLPIVIMIACFSSIYFTVRKLTRDSMRRWGASSDSTQQTLAAERKTAWMSFIMVLAFLFAWVPYAVVSLYASFGGVTTIPKLMSTLPAMLAKTSACYNPII
YFFMYSKFRKAFQRFFFKNVITPSQTGGSSTINSASVIPTSFPRASYAVRSSSVLPSSTASHSFRSRE* 0
>ENC_aneVir Anemonia viridis (anemone) Cnid.Anth.Hexa FK729339 19627569 frag G? symbiotic
KDAVARWGNKSPPTQQTMQAQKKTIRMSLVMVFAYLLAWTPYALTSLYSSFIASDITPLLSVMPALFAKLSSCYNPIIYFFMYSKFR
KAAKKMIRRNLVGHDSNSGQGVSNTFATSFPRPISFLRYKRSAVAPLSDIPQVSSVDLPQVGRENDVTVQQDKASEINT* 0
>MEL1_acrDig Acropora digitifera (stony_coral) BACK01045931
METASGVPIVIEASVHHTISFLYFLLALFSFSLNSVVILTFLLDRSLLFPANLIILSIAISDWLMSVVPNIMGGVANASNDLPFTDWSCTVFAFVATLLG
LSNMLHHAAFALDRYMVITRPMRANHSMKRILAVIAFLWCFALTWSLFPLVGWSAYVREAGDIACSVNWQSDNPSDTSYMVCLFFFFYFVPLAVIVYCYA
FMIKSVRFMTKNAQKIWGVRSAAALETVQATWKMAKIGLIMVVGFFVAWTPYAVVSFIIAFDLVKDIPTVAEIVPSMFAKTASVYNPIIYFFSFAYCKNQ
SVNKCINQSTVQSKNFKLS*
>MEL2_acrDig Acropora digitifera (stony_coral) BACK01017283
MPGFQNKSEEPQPTEGISSQDRFIVASFYALLAVTALGLNTPVLITFMKDRKLRVLSNRIILSITIGDWLHAFLAYPVAVFANASHDSQGLTGVVCSWYG
FITVFLSFGIMLHHATFAVERAIVIQFATTSLTNAKTINFMVASLWVFALLWSSFPLFGWSAYVPHVVLCSLDWQSRDLRDVVFVYCIFFVFFLVPIVVM
VTSYYKIFQTVKKMTQNARDLWGEKAAPTREAFESQKKTARMACVMSFCFLFAWTPYAAVSLYVFLWKPQSMAPSISIVPALFAKTSACFNPVIYFLLFR
KFRESLKQTMCGFFMDPKTGTNETNKNKRHVLKNTESKCACVKINNLSPRDPCNLHQRSDCRSVGNSKSHITVD*
>ENC1_acrDig Acropora digitifera (stony_coral) BACK01046894
MLTFACGSTHIYNMSAVFYGFLAAGTVFLNVIVILTFFKVRALLIPASFPILSLAMADVFLASAVMPLGIASCASGHWIFGAVGCNWYAFMHTTVGLSSI
LHHAILALERCLTIFEPMKKHFDRHTMTRTLAFLWTLVSLWSLCPILGWSAYVPEGTGTICSIEWHSKNSLDVIFVVSTFVFFCFVPFVFIAVCYTAIAF
HLRKVSKTAKRTWGKNSQITKDGVIVKRKAVTHGAIMVTTVMITWLPYSMVAFYTLLGFEKVKLSALAYAITAMFAKTSTLVNPIICFFWYRRFREGTKK
LCNRIMDFILRKHRRASNTSHSSKTVWYDGKTIVFRRSVVDSCSTRTQNDRGRSS*
>ENC2_acrDig Acropora digitifera (stony_coral) BACK01016849 11738 bp INV 28-JUL-2011 DOI:10.1038/nature10249
MTALIYSVFAIIAAVICVLGVPLNGLVCWVFYNNRELLNAPNIFIASVAFSDFLYCISCLPLLVISNAYGKWIYGPIGCKATAFIATWSGLTSLMNLSVA
SYERYSTLAFLCTKNRTFAKRTATCYSAAMWLYALFWSLMPLCGWSGFELEGIGTSCSVRWKSKNMLDMSYNLCLIIACYVLPVSVLVTSYYKCYREIAK
STWRAKSTWGRQSPFTKRTFVMERKMMALFGVMTVAFLVAWTPYAVVSLISMIGGPDVISDVTASIPAYFAKSSSCYNPVIYVFLYKRLRRQMRFAVRRD
NCSSSRKGTSSDSTMAHHSETKRLRSTIELTDNYSTRNRPENI*
>ENC3_acrDig Acropora digitifera (stony_coral) BACK01015014 possibly N-incomplete
MVAPHFAGFAIAASLFGSCGIVLNFTVCLTYILNRRLLDASNIFILNISAGDFLYSITALPMLVTSNALGKWSFGEGGCVAYGFLTTFFALGAMMNLAGA
AYERYVTMCKLYESGECQFSRRKAAFLCSVLWTYALIWSIAPIFGWSSYKQEGIGTSCSTDWKSRDVKSLSYGVVLIITCFVVPVAVILYCHIEAYKVTR
KLGKQAQQNWGSHTRATRETLKAQKKMGEIAVVITVGFVVAWTPYTVASVIGMYDPNLVSDVGASIPAYFAKSSSCYNPFIYLFMYKKLRIGMVRLLCCM
KKQVYPSSSGLTRDPTCQEIPMVVPVNNPS*
>ENC4_acrDig Acropora digitifera (stony_coral) BACK01020962 possibly N-incomplete
MTKTAAWSTVFLIIEFVVCLLGIFFNCLVISIIFKNANRLAAPGFLVLSMALSDVLSCSVAVPFSIVAHFQKKWPFGMAGCEAHAFMVFLFALVSITHLA
AISAGKYLTISRSLTRESYFDRRQVLLVVIACWLYSLAFSVAPLVGWSRYGLEGTNATCSIKWDSSEPSDKAYFGVVFIACFFLPMGVITLCYYKIHKVS
KRIVENIQGQSIATSAMSRTHALVKKHRRSAMYFLAIVAAFMLAWSPYAIVSLIVVLSGTMDPIATSACSVFAKTSFLVNPFLYAIFSRSFRRRLALVFP
MPRQNRQEGRQERNCNALPPSQSQPFVL*
>ENC5_acrDig Acropora digitifera (stony_coral) BACK01011015
MDQIKIWKMLFVVIKFIVSIFGGFFNGLVVLTIRKNLRRLPSSSYLILSIAFSDFIASVVAIPFSIVIHFVGSWPLRLCRAHAFMVFFLGIVTITHLTCF
AVEKYLTITRSLSKLSFFSKKQTLVVVMACWFYSLCFSLAPLLGWASYGLEGSNDTCSIKWDSSLAKDHIYFILVFLACYLLPIVLITSSYFKILRISKR
ILKATPRVGGIGETMAQALMRKHRRGALYFLSVIAVFLMSWTPYAIISVLVIFKGVNLFPLALSACGVFAKMSFMLNPIMYFAFSHNFRLLVKRTFCICD
RLDSNSEGKKR*
>CNOP1_acrDig Acropora digitifera (stony_coral) BACK01019215 tandem fragment last exon at beginning of contig
1 LLRRDVVTEDSRIKLRRRHWQMKLLRLTAVAITAFMLSWSPYSLVSLTSIFRGNSVLSTGEAEVPALMAKASVIYNPIVYTVMNRRFRRTLRHIVSCMTCRLLSFVWPTMHGEKQETKKRVTSTMTVTSSTPAPEGNFPEILVSLHQVPF*
>CNOP2_acrDig Acropora digitifera (stony_coral) BACK01019215 MEL
0 MQFSREEDPPSGFNWNNAWFIAYGVVMIAVLSIGFLGNTMTVLILRKHEHASKSLTPLMINLAIASLIIIVLGYPLVISLVVRGSHVTKEDPTCRWSAFINGTV 1
2 GISSIATLTEMSLVINYSLHRMNPNVRLTKRNMALLIAGAWLYGLVSMFPPLVGWNRFVPGAVRISCGPDWTDKSASGVSYNLVLVVLGFFLPLCVMIKAYYEIYRS 2
1 LLRSREMLISANGSFQLRQKLYIKKLVRMTVLAIAAFMLSWAPYCFVSIVAIFKGSHIITSGEAEIPELMAKASVIYNPVVYLITNSSYRASFWKVISCQKQTMIVHVRENLMPNGPSRRTLRSRRMAVLLKPLNEVSVYNGGYVVAMGFASVRDL* 0
>CNOP3_acrDig Acropora digitifera (stony_coral) BACK01018578 TMT
0 MLESSNQSAEETILISKASYYAYGVVMFFILTVGFLGNVMTLLVLFQHEHRKKAMTPYMVNIALADIFIILFGYPVAMRANLRGKILESSHCSWGGFVNGAV
GISSIFTLTEMSFVSYHGLRQVNRSSRFSPFQVVCSVAVAWLYGVLCMLPPFLGWNRFVVSASRISCCPDWSGKSISDAAYNLLLVFFGFVAPLTAMTVCYYKIYR 2
1 SLVHHAVVPGNLPQIQLRRRQSELKVAKVTAMNVIAFLLSWAPYCCVSLAAVFTKQFVLVDWEAEIPELLAKASVIYGPIIYSTMHSRFQATLFRILHCRRRVILAPQFITDATRNNCSARMATDRTQGTSHIDQRFLQLPLQAMKRRVAVSYGGTAICKRESSAFVSG* 0
>CNOP4_acrDig Acropora digitifera (stony_coral) BACK01002540 TMT
0 MTTRSDDNPNSARRPQQIAFIVYVVIMTVILIVGFLGNLFTIIVLRCPEHKKKIITPLMMNLAFADIIIIVFGYPVVMASNFTEHNVLTNPMLCVWSGFINGSV
GIASIANLTMMSLVMFKSLNKVGTVKKIPWKKMLAMILFTWVYGVLSMFPPLVGWNKFVPGASGISCCPDWSPDTKAAVGYNILLVFVGFVLPLTIIIFCYYRMYR 2
1 FIHTQQPVTGNASIQASRRRSEIKIVRMIALAIAAFVLSWSPYCFVSIAGTIRGSSLLTAGEAEVPDLLAKASVIYNPIVYTVMNDRFRGTLLRVIPCRRRWFNGDINPTVSVSAPNSQDGNSHNFRERPQSVKGNSTV* 0
>CNOP5_acrDig Acropora digitifera (stony_coral) BACK01002513 and near identical BACK01007224
0 MTTSFRSNMTDEWGTHKYFSAGYIAYGVVMFCILSLGFVGNTLTVLVLTKREHRWRGVTPLMINLAVADLLIIIFGYPTQVHANFTGDGVPNDSHCNYRGFIN
GIVGITCIFTLTEMGVVSYSGLRRVSGNVRLSSYQVACLIGAAWCYGALCMLPPLLGWSKFVISASKLSCSPNWSGQSTADKAYTLLLVTFGFFAPLTVMIISYYKTFW 2
1 VVRRHFVPGNTAVQLRQRKSMLRIVHVTLMSVISMMLSWAPYCFISLASVVRGKPVIENWEAEIPELLAKASVVYNPLIYTFMNKSFRMTLFRIVGLHRCFGAGHEVAPTAAIQKRPNVLQIQFHRHSSEVEVRTPRVPPIE* 0
>CTENOP1_plePil Pleurobrachia pileus (sea_gooseberry) Cten.Typh.Pleu FP995093 CU419614 full G? ctenop EQY
0 MELRVSLSLIMACVCTGSVF 1
2 GNMLVFISLVKERPLPNPAVEMIFRHLSLANLVLAGIGEPLVVVSLAA 1
2 GKWVWGESMRYFEAFWVTTFGLFSMMILGLISFEQYSRFVRQKQLKTHQ 0
0 VAVLLSGIYFLAFFFGIAPLVGWCEYGPE 1
2 GYGVSNSLMWNNLNDNNASYIICAMIIGYFFPLIIIMFCYRAIYRLVQ 0
0 AQLNTSVVKMTVSANVTSLSTNRNHIML 0
0 VQERQLASTITFVILSFFFAWTPYVFVNIINMFSTLILKYRILAT 0
0 IPALFAKSSTLWWIIVYCLMDVRIKKACRKTCLRFKRSFRVWYFS* 0
>CTENOP1_mneLei Mnemiopsis leidyi AGCP01013851 AGCP01013853 77% identical CTENOP1_plePil
0 MGLRISLSIAMGCVCVGSVF 1
2 GNLLVLISLVRERPLPNPAVQLIFRHLSLANFLLACIGEPLVVISLAS 1
2 GKWVWGESMRLFEAFWVTTFGLLSMMILALISLEQYSRFVRQKQLRTVQ 0
0 VAVLLSGIYFLAFFFGIAPLVGWCEYGPE 1
2 GYGVSNSLLWNELTDNSASYIVTVMIVGYFFPLVIITYCYRSIYQLVQ 0
0 AQSNVAIVRTMVSVNVTSLAVNRNHSLI 0
0 VQERKLATTITFVILSFFFAWTPYVLINLLNMFTTWVVKYKILAT 0
0 IPALFAKSSTLWWIIIYSMMDKRIKKACRKTCEHFQRALRIWYFSS* 0
>CTENOP2_plePil Pleurobrachia pileus (sea_gooseberry) Cten.Typh.Pleu FQ011385 ERY 33-203 range in bosTau rhodopsin
0 MTTAVDDEMMGDEEPKVVYTLASLLAVNALCSIA 1
2 GNALVIYTFLKEKPLGAPLRVILTHLAVTNLIIASIGEPMVVISGYS 1
2 MRWVFGEMGRGFEAFIVTSCGLMAMSLLALVSIERYLR 2
1 VTPRQRFPFTEPTAVKCCSFLWVFNVIYASLPFYGIAAWEGE 1
2 GIGISNSITWTTEHTQDFVYIICMMSTGYFIPLFIIAFCY 2
>CTENOP2_mneLei Mnemiopsis leidyi AGCP01013109 AGCP01013108 AGCP01013106
0 MSSPNDEPTVDSDPKAVYILTSLLCMNCIFSLV 1
2 GNMLVIYTFLRERPLGAPLRTILCHLAITNLMIAGIGEPMVVISGFN 1
2 VRWVFGEMGRKIEAYTVTASGLMAMSLLAFVSIERYLR 2
1 VTPGQKFLIRQTTSVRVCTLMWIYNVIYASLPFYGIAAWRGE 1
2 GIGISNSLSWDTNNKIDVAYVMVMMATGYFIPLAVITFCYNRYVEIQQRLSTFL 2
SKLKMTMETNITSAANSQN
0 AAEKKISNVVAIIIVSFFISWTPYTIVNLLVTFDKAGR 2
1 GINATIPAFLAKTSTVWSPIIYCFMNGEVSIFVLEGGHVMKYLYL* 0
GPCR outgroup sequences
It is sometimes convenient to have a close-in outgroup to opsins selected from the roughly 100,000 GPCR receptors available at GenBank. The set below of 29 non-opsin GPCRs serves satisfactorily as proxy for an exhaustive GPCR compilation. It was constructed by taking best-blast in turn of each human opsin against all other human GPCR then collating the lists, winnowing out repeated entries and too-recent gene family expansions. TACR2 (tachykinin receptor) and SSTR1 (somatostatin receptor) are the best single representatives.
These are usefully supplemented with non-opsin GPCR having determined 3D structures, some nearest neighbors in recent GRAFS classification trees, and two astonishingly close pre-opsins (called UROPS1 and UROPS2 here) from Trichoplax, an early diverging eukaryote lacking opsins. Note TSHR binding of heterotrimeric G protein has recently been modeled in great detail.
The aligned sequences below have been trimmed from full length at both ends to the earliest indications of conservation. This alignable region begins at the GN region of TM1 and extends to the FR motif just beyond TM7. Highly conserved residues are shown in red and less conserved residues in blue. The Schiff base lysine (position -16 relative to the FR end of TM7) does not occur outside of opsins. Note many conserved patches in these GPCR are very similar to those of opsins, implying those residues have no utility in distinguishing opsins from non-opsins.
Conserved outgroup residues shared with opsins evidently describe commonalities needed for generic GPCR structure and signaling but not specifically for photobiology. Departures from the norm in certain opsin classes might indicate they are no longer signaling or are signaling constituitively. Opsins also have conserved diagnostic residues and even regions not found in any GPCR.
These close-in outgroup sequences illuminate, as discussed elsewhere, the origin of opsins.
Exon count refers to trimmed alignable sequences. Some idiosyncratic loops have been deleted. Intron positions and phases are indicated relative to bovine rhodopsin numbering (sometimes uncertain due to gapping issues). These GPCR do not significantly conserve major opsin introns, so parental relations remain obscure. >GPR21_homSap Homo sapiens (human) orphan receptor good match mollusk opsin 1 exon IVFLTVLIISGNIIVIFVFHCAPLLNHHTTSYFIQTMAYADLFVGVSCVVPSLSLLHHPLPVEESLTCQIFGFVVSVLKSVSMASLACISIDRYIAITKPLTYNTLVTPWRLRLCIFLIWLYSTLVFLPSFFHWGKPGYH GDVFQWCAESWHTDSYFTLFIVMMLYAPAALIVCFTYFNIFRICQQHTKDISERQARFSSQSGETGEVQACPDKRYAMVLFRITSVFYILWLPYIIYFLLESSTGHSNRFASFLTTWLAISNSFCNCVIYSLSNSVFQR >GPR52_homSap Homo sapiens (human) orphan receptor immediate outgroup to opsins 1 exon IVLLTFLIIAGNLTVIFVFHCAPLLHHYTTSYFIQTMAYADLFVGVSCLVPTLSLLHYSTGVHESLTCQVFGYIISVLKSVSMACLACISVDRYLAITKPLSYNQLVTPCRLRICIILIWIYSCLIFLPSFFGWGKPGYH GDIFEWCATSWLTSAYFTGFIVCLLYAPAAFVVCFTYFHIFKICRQHTKEINDRRARFPSHEVDSSRETGHSPDRRYAMVLFRITSVFYMLWLPYIIYFLLESSRVLDNPTLSFLTTWLAISNSFCNCVIYSLSNSVFRL >MTNR1A_homSap Homo sapiens (human) melatonin receptor circadian rhythm 2 exons 71-12 LIFTIVVDILGNLLVILSVYRNKKLRNA 1 2 GNIFVVSLAVADLVVAIYPYPLVLMSIFNNGWNLGYLHCQVSGFLMGLSVIGSIFNITGIAINRYCYICHSLKYDKLYSSKNSLCYVLLIWLLTLAAVLPNLRAGTLQYDPRIYSCTFAQ SVSSAYTIAVVVFHFLVPMIIVIFCYLRIWILVLQVRQRVKPDRKPKLKPQDFRNFVTMFVVFVLFAICWAPLNFIGLAVASDPASMVPRIPEWLFVASYYMAYFNSCLNAIIYGLLNQNFRK >HRH2_homSap Homo sapiens (human) histamine receptor 1 exon LAVLILITVAGNVVVCLAVGLNRRLRNLTNCFIVSLAITDLLLGLLVLPFSAIYQLSCKWSFGKVFCNIYTSLDVMLCTASILNLFMISLDRYCAVMDPLRYPVLVTPVRVAISLVLIWVISITLSFLSIHLGWNS RNETSKGNHTTSKCKVQVNEVYGLVDGLVTFYLPLLIMCITYYRIFKVARDQAKRINHISSWKAATIREHKATVTLAAVMGAFIICWFPYFTAFVYRGLRGDDAINEVLEAIVLWLGYANSALNPILYAALNRDFRT >SSTR1_homSap Homo sapiens (human) somatostatin receptor SOG Rgamma class 1 exon YSVVCLVGLCGNSMVIYVILRYAKMKTATNIYILNLAIADELLMLSVPFLVTSTLLRHWPFGALLCRLVLSVDAVNMFTSIYCLTVLSVDRYVAVVHPIKAARYRRPTVAKVVNLGVWVLSLLVILPIVVFSRT AANSDGTVACNMLMPEPAQRWLVGFVLYTFLMGFLLPVGAICLCYVLIIAKMRMVALKAGWQQRKRSERKITLMVMMVVMVFVICWMPFYVVQLVNVFAEQDDATVSQLSVILGYANSCANPILYGFLSDNFKR >OPRM1_homSap Homo sapiens (human) opioid receptor mu 3 exons 64-21 182-12 ? YSIVCVVGLFGNFLVMYVIVR 2 1 YTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIINVCNWILSSAIGLPVMFMATTKYRQ 1 2 GSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALVTIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKR >GALR1_homSap Homo sapiens (human) galanin receptor SOG Rgamma 3 exons 225-00 251-00 ? FGLIFALGVLGNSLVITVLARSKPGKPRSTTNLFILNLSIADLAYLLFCIPFQATVYALPTWVLGAFICKFIHYFFTVSMLVSIFTLAAMSVDRY VAIVHSRRSSSLRVSRNALLGVGCIWALSIAMASPVAYHQGLFHPRASNQTFCWEQWPDPRHKKAYVVCTFVFGYLLPLLLICFCYAK 0 0 VLNHLHKKLKNMSKKSEASKKK 0 0 TAQTVLVVVVVFGISWLPHHIIHLWAEFGVFPLTPASFLFRITAHCLAYSNSSVNPIIYAFLSENFRK >CCR4_homSap Homo sapiens (human) chemokine (C-C motif) receptor 1 exon YSLVFVFGLLGNSVVVLVLFKYKRLRSMTDVYLLNLAISDLLFVFSLPFWGYYAADQWVFGLGLCKMISWMYLVGFYSGIFFVMLMSIDRYLAIVHAVFSLRARTLTYGVITSLATWSVAVFASLPGFLFSTCY TERNHTYCKTKYSLNSTTWKVLSSLEINILGLVIPLGIMLFCYSMIIRTLQHCKNEKKNKAVKMIFAVVVLFLGFWTPYNIVLFLETLVELEVLQDCTFERYLDYAIQATETLAFVHCCLNPIIYFFLGEKFRK >BDKRB2_homSap Homo sapiens (human) bradykinin receptor 1 exon LWVLFVLATLENIFVLSVFCLHKSSCTVAEIYLGNLAAADLILACGLPFWAITISNNFDWLFGETLCRVVNAIISMNLYSSICFLMLVSIDRYLALVKTMSMGRMRGVRWAKLYSLVIWGCTLLLSSPMLVFRTMKEYS DEGHNVTACVISYPSLIWEVFTNMLLNVVGFLLPLSVITFCTMQIMQVLRNNEMQKFKEIQTERRATVLVLVVLLLFIICWLPFQISTFLDTLHRLGILSSCQDERIIDVITQIASFMAYSNSCLNPLVYVIVGKRFRK >GPR17_homSap Homo sapiens (human) uracil/cys-leukotriene dual receptor 1 exon YLLDFILALVGNTLALWLFIRDHKSGTPANVFLMHLAVADLSCVLVLPTRLVYHFSGNHWPFGEIACRLTGFLFYLNMYASIYFLTCISADRFLAIVHPVKSLKLRRPLYAHLACAFLWVVVAVAMAPLLVSPQT VQTNHTVVCLQLYREKASHHALVSLAVAFTFPFITTVTCYLLIIRSLRQGLRVEKRLKTKAVRMIAIVLAIFLVCFVPYHVNRSVYVLHYRSHGASCATQRILALANRITSCLTSLNGALDPIMYFFVAEKFRH >CYSLTR1_homSap Homo sapiens (human) cys-leukotriene receptor 1 exon YSMISVVGFFGNGFVLYVLIKTYHKKSAFQVYMINLAVADLLCVCTLPLRVVYYVHKGIWLFGDFLCRLSTYALYVNLYCSIFFMTAMSFFRCIAIVFPVQNINLVTQKKARFVCVGIWIFVILTSSPFLMAKPQKDE KNNTKCFEPPQDNQTKNHVLVLHYVSLFVGFIIPFVIIIVCYTMIILTLLKKSMKKNLSSHKKAIGMIMVVTAAFLVSFMPYHIQRTIHLHFLHNETKPCDSVLRMQKSVVITLSLAASNCCFDPLLYFFSGGNFRK >P2RY8_homSap Homo sapiens (human) purinergic receptor AMIN Ralpha class 1 exon YSLVAAVSIPGNLFSLWVLCRRMGPRSPSVIFMINLSVTDLMLASVLPFQIYYHCNRHHWVFGVLLCNVVTVAFYANMYSSILTMTCISVERFLGVLYPLSSKRWRRRRYAVAACAGTWLLLLTALSPLARTDLTY PVHALGIITCFDVLKWTMLPSVAMWAVFLFTIFILLFLIPFVITVACYTATILKLLRTEEAHGREQRRRAVGLAAVVLLAFVTCFAPNNFVLLAHIVSRLFYGKSYYHVYKLTLCLSCLNNCLDPFVYYFASREFQL >HCRTR1_homSap Homo sapiens (human) orexin receptor Rbeta class 7 exons 57-12 117-00 187-12 226-00 276-21 310-12 YVAVFVVALVGNTL 1 2 VCLAVWRNHHMRTVTNYFIVNLSLADVLVTAICLPASLLVDITESWLFGHALCKVIPYLQ 0 0 AVSVSVAVLTLSFIALDRWYAICHPLLFKSTARRARGSILGIWAVSLAIMVPQAAVMECSSVLPELANRTRLFSVCDERWA 1 2 DDLYPKIYHSCFFIVTYLAPLGLMAMAYFQIFRKLWGRQ 0 0 IPGTTSALVRNWKRPSDQLGDLEQGLSGEPQPRGRAFLAEVKQMRARRKTAKMLMVVLLVFALCYLPISVLNVLKR 2 1 VFGMFRQASDREAVYACFTFSHWLVYANSAANPIIYNFLS 1 2 GKFRE >TACR2_homSap Homo sapiens (human) tachykinin receptor Gq-coupled SOG Rgamma class first gc-ag 5 exons 135-21 202-21 250-00 ? 312-21 YLALVLVAVTGNAIVIWIILAHRRMRTVTNYFIVNLALADLCMAAFNAAFNFVYASHNIWYFGRAFCYFQNLFPITAMFVSIYSMTAIAADR 2 1 YMAIVHPFQPRLSAPSTKAVIAGIWLVALALASPQCFYSTVTMDQGATKCVVAWPEDSGGKTLLL 2 1 YHLVVIALIYFLPLAVMFVAYSVIGLTLWRRAVPGHQAHGANLRHLQAMKK 0 0 FVKTMVLVVLTFAICWLPYHLYFILGSFQEDIYCHKFIQQVYLALFWLAMSSTMYNPIIYCCLNHR 2 1 FRS >NMUR2_homSap Homo sapiens (human) neuromedin U receptor 4 exons 229-00 ? 256-12 291-12 YVPIFVVGVIGNVLVCLVILQHQAMKTPTNYYLFSLAVSDLLVLLLGMPLEVYEMWRNYPFLFGPVGCYFKTALFETVCFASILSITTVSVERYVA ILHPFRAKLQSTRRRALRILGIVWGFSVLFSLPNTSIHGIKFHYFPNGSLVPGSATCTVIKPMWIYNFIIQVTSFLFYLLPMTVISVLYYLMALR 0 0 LKKDKSLEADEGNANIQRPCRKSVNKML 1 2 FVLVLVFAICWAPFHIDRLFFSFVEEWSESLAAVFNLVHVVS 1 2 GVFFYLSSAVNPIIYNLLSRRFQA >QRFPR_homSap Homo sapiens (human) pyroglutamylated RFamide peptide receptor MEC Ralpha class 6 exons 105-12 157-12 180-00 247-21 277-12 GVLIFALALFGNALVFYVVTRSKAMRTVTNIFICSLALSDLLITFFCIPVTMLQNISDNWLG 1 2 GAFICKMVPFVQSTAVVTEILTMTCIAVERHQGLVHPFKMKWQYTNRRAFTML 1 2 GVVWLVAVIVGSPMWHVQQLE 0 0 IKYDFLYEKEHICCLEEWTSPVHQKIYTTFILVILFLLPLMVMLILYSKIGYELWIKKRVGDGSVLRTIHGKEMSKIAR 2 1 KKKRAVIMMVTVVALFAVCWAPFHVVHMMIEY 1 2 SNFEKEYDDVTIKMIFAIVQIIGFSNSICNPIVYAFMNENFKK >GPR19_homSap Homo sapiens (human) orphan receptor 19 1 exon FGILWLFSIFGNSLVCLVIHRSRRTQSTTNYFVVSMACADLLISVASTPFVLLQFTTGRWTLGSATCKVVRYFQYLTPGVQIYVLLSICIDRFYTIVYPLSFKVSREKAKKMIAASWIFDAGFVTPVLFFYGS NWDSHCNYFLPSSWEGTAYTVIHFLVGFVIPSVLIILFYQKVIKYIWRIGTDGRTVRRTMNIVPRTKVKTIKMFLILNLLFLLSWLPFHVAQLWHPHEQDYKKSSLVFTAITWISFSSSASKPTLYSIYNANFRR >PPYR1_homSap Homo sapiens (human) pancreatic polypeptide receptor MEC Ralpha class 1 exon YSIETVVGVLGNLCLMCVTVRQKEKANVTNLLIANLAFSDFLMCLLCQPLTAVYTIMDYWIFGETLCKMSAFIQCMSVTVSILSLVLVALERHQLIINPTGWKPSISQAYLGIVLIWVIACVLSLPFLANSILENVFHKNHS KALEFLADKVVCTESWPLAHHRTIYTTFLLLFQYCLPLGFILVCYARIYRRLQRQGRVFHKGTYSLRAGHMKQVNVVLVVMVVAFAVLWLPLHVFNSLEDWHHEAIPICHGNLIFLVCHLLAMASTCVNPFIYGFLNTNFKK >NPY1R_homSap Homo sapiens (human) neuropeptide Y receptor Rbeta class 2 exons 225-00 YGAVIILGVSGNLALIIIILKQKEMRNVTNILIVNLSFSDLLVAIMCLPFTFVYTLMDHWVFGEAMCKLNPFVQCVSITVSIFSLVLIAVERHQ LIINPRGWRPNNRHAYVGIAVIWVLAVASSLPFLIYQVMTDEPFQNVTLDAYKDKYVCFDQFPSDSHRLSYTTLLLVLQYFGPLCFIFICYFK 0 0 IYIRLKRRNNMMDKMRDNKYRSSETKRINIMLLSIVVAFAVCWLPLTIFNTVFDWNHQIIATCNHNLLFLLCHLTAMISTCVNPIFYGFLNKNFQR >PRLHR_homSap Homo sapiens (human) prolactin releasing hormone receptor 1 exon YSVVVVVGLVGNCLLVLVIARVRRLHNVTNFLIGNLALSDVLMCTACVPLTLAYAFEPRGWVFGGGLCHLVFFLQPVTVYVSVFTLTTIAVDRYVVLVHPLRRRISLRLSAYAVLAIWALSAVLALPAAVHTYHVELKP HDVRLCEEFWGSQERQRQLYAWGLLLVTYLLPLLVILLSYVRVSVKLRNRVVPGCVTQSQADWDRARRRRTFCLLVVVVVVFAVCWLPLHVFNLLRDLDPHAIDPYAFGLVQLLCHWLAMSSACYNPFIYAWLHDSFRE >GPR161_homSap Homo sapiens (human) orphan receptor 161 2 exons 135-21 IIVITIFVCLGNLVIVVTLYKKSYLLTLSNKFVFSLTLSNFLLSVLVLPFVVTSSIRREWIFGVVWCNFSALLYLLISSASMLTLGVIAIDR 2 1 YYAVLYPMVYPMKITGNRAVMALVYIWLHSLIGCLPPLFGWSSVEFDEFKWMCVAAWHREPGYTAFWQIWCALFPFLVMLVCYGFIFRVARVKARKVHCGTVV IVEEDAQRTGRKNSSTSTSSSGSRRNAFQGVVYSANQCKALITILVVLGAFMVTWGPYMVVIASEALWGKSSVSPSLETWATWLSFASAVCHPLIYGLWNKTVRK >ADRA1D_homSap Homo sapiens (human) alpha-1D-adrenergic receptor PUR Ralpha class 2 exons 274-12 LAAFILMAVAGNLLVILSVACNRHLQTVTNYFIVNLAVADLLLSATVLPFSATMEVLGFWAFGRAFCDVWAAVDVLCCTASILSLCTISVDRYVGVRHSLKYPAIMTERKAAAILALLWVVALVVSVGPLLGWKEPV PPDERFCGITEEAGYAVFSSVCSFYLPMAVIVVMYCRVYVVARSTTRSLEAG VKRERGKASE VVLRIHCRGAATGADGAHGMRSAKGHTFRSSLSVRLLKFSREKKAAKTLAIVVGVFVLCWFPFFFVLPL 1 2 GSLFPQLKPSEGVFKVIFWLGYFNSCVNPLIYPCSSREFKR >TRHR_homSap Homo sapiens (human) thyrotropin-releasing hormone receptor 2 exons 251-00 VLIICGLGIVGNIMVVLVVMRTKHMRTPTNCYLVSLAVADLMVLVAAGLPNITDSIYGSWVYGYVGCLCITYLQYLGINASSCSITAFTIERYIAICHPIKAQFLCTFSRAKKIII FVWAFTSLYCMLWFFLLDLNISTYKDAIVISCGYKISRNYYSPIYLMDFGVFYVVPMILATVLYGFIARILFLNPIPSDPKENSKTWKNDSTHQNTNLNVNTSNRCFNSTVSSRKQ 0 0 VTKMLAVVVILFALLWMPYRTLVVVNSFLSSPFQENWFLLFCRICIYLNSAINPVIYNLMSQKFRA >TSHR_homSap Homo sapiens (human) thyrotropin receptor 1 exon PM:20305779 VWFVSLLALLGNVFVLLILLTSHYKLNVPRFLMCNLAFADFCMGMYLLLIASVDLYTHSEYYNHAIDWQTGPGCNTAGFFTVFASELSVYTLTVITLERWYAITFAMRLDRKIRLRHACAIMVGGWVCCFLL ALLPLVGISSYAKVSICLPMDTETPLALAYIVFVLTLNIVAFVIVCCCYVKIYITVRNPQYNPGDKDTKIAKRMAVLIFTDFICMAPISFYALSAILNKPLITVSNSKILLVLFYPLNSCANPFLYAIFTKAFQR >ADRB2_homSap Homo sapiens (human) beta2 adrenoceptor pdb:2R4R PUR Ralpha class 1 exon MSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYAN ETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRFHVQNLSQVEQDGRTGHGLRRSSKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRI >ADORA2A_homSap Homo sapiens (human) beta2 adenosine receptor pdb:3EML PUR Ralpha class 2 exons 144-21 ELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLR 2 1 YNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKNHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLA ARRQLKQMESQPLPGERARSTLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQ >ADRB1_melGal Meleagris gallopavo (turkey) beta1 adrenergic receptor pdb:2VT4 1 exon MALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRD EDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRKIDRASKRKRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCRSPDFRK >UROPS1_triAdh Trichoplax adhaerens (trichoplax) XM_002114542 ends trimmed 1 exon YSVLSLITILGNMLVFLTFYKHASLRTTSNLFIINLAITDLLTGGIKDTLFIYGLTSYNWPKSAILCSFVGFINCVCYVATVYTLTVIAVFRYLAIVCNLGRKIKRKHSILTIACVWIYSSACCLMPIIGWSRY IYEPTECTCVRSLDKKYYSYTIYILVADFLLPLSVVSFCYGNIFAKMKRNHHHIKRDLSDGQKLDVAEKLSRREEIVARRMFIILAEHVVCFLPYTIIVTMLAANGVDIEPVWYFIVGYLLNLNSALNPVTYIVVNPRLKK >UROPS2_triAdh Trichoplax adhaerens (trichoplax) XM_002112401 ends trimmed 2 exons 241-21 ? 0 MTLFMLMSCIGNGAVLLVLRYHHDDIKSASNYFITNLALTDFLLGVLCMPCILISCLNGQWVFGQTLCSLTGFANSFFCINSMITLAAVSVEKYCAIASPLTY HHYMSKSKVTCVISIIWIHSAINASLPFLGWGEYVYLPFETICTVAWWSFPNYVGFIVGINFGLPTVIMSCTYFLILKIARKHSRRIGVST 2 1 RRIHYKTHIKATLMLLIVIGSFIVCWLPHLISMIYLTIYEISPLPCSFHQITTWLAMANSAFNPIIYGAMDTSIRK
See also: LWS | Encephalopsins | Melanopsins | Neuropsin | Peropsin | RGR phyloSNPs | Update Blog