Opsin evolution: Difference between revisions
Tomemerald (talk | contribs) No edit summary |
Tomemerald (talk | contribs) No edit summary |
||
| Line 50: | Line 50: | ||
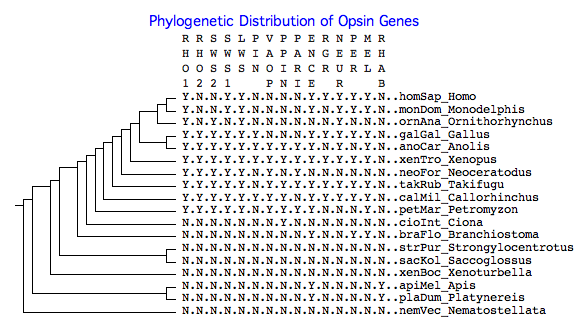
The phylogenetic tree below shows the presence or absence of various opsin genes in clade-representative species, as reflected in the collected reference sequences. The purpose is timing appearance (or disappearance) of a given class of opsin gene. For example, cone and rod opsins first appeared before lamprey divergence; otherwise they are absent from urochordates, cephalochordates, and earlier deuterostomes. Note however a given gene might appear absent because of a genome project gap, lack of experimental effort, insufficient or outdated bioinformatics, or species idiosyncracies (ie be present in a different species of that clade). In other cases (eg platypus SWS1) pseudogene remnants or a syntenically proven deletion establish the gene is definitely absent. Y means yes (present), N means no (absent). | The phylogenetic tree below shows the presence or absence of various opsin genes in clade-representative species, as reflected in the collected reference sequences. The purpose is timing appearance (or disappearance) of a given class of opsin gene. For example, cone and rod opsins first appeared before lamprey divergence; otherwise they are absent from urochordates, cephalochordates, and earlier deuterostomes. Note however a given gene might appear absent because of a genome project gap, lack of experimental effort, insufficient or outdated bioinformatics, or species idiosyncracies (ie be present in a different species of that clade). In other cases (eg platypus SWS1) pseudogene remnants or a syntenically proven deletion establish the gene is definitely absent. Y means yes (present), N means no (absent). | ||
[[Image:Opsin phylo.png]] | |||
[[Image:Opsin phylo.png|{left}|]] | |||
Revision as of 17:56, 19 November 2007
Below is a large set of phylogenetically representative hand-curated opsin sequences that serves as a gene family classifier ... just blast an unknown sequence against the database below (using http://www.proweb.org/proweb/Tools/WU-blast.html) and look for consistent labelling of the top hits.
The set of sequences is not intended to be exhaustive. Rather, if a given clade has many available similar sequences, those with genome assemblies are chosen to represent the group, for example anole is preferred to gecko, and (rightly or wrongly) any experimental results transfered over. This avoids uninformative clutter from near-identical sequences. If the clade reflects a very deep divergence such as lamprey or amphioxus, all available sequences are provided so as to break up long branches. About half the sequences are not available from GenBank but rather are culled from trace archives, genomic contigs, and genome assemblies, typically by blastx against the full (and growing) set of reference sequences. The level of error is very low, declines with time as anomalies are revisited and fixed, but never reaches zero because of problems inherent to experimental data, incomplete assemblies, and sequence manipulation.
The fasta header of each sequence is a miniature database, with fields showing the opsin type, genus, species and common name, accession number or other source, indels, introns, sequence length, lambda max adsorption, and G protein type with which it interacts. These new standardized fasta headers also serve to summarize the collection.
The protein sequences are broken into their constituent exons using genomic information when available. When not available (eg the opsin originated as a cDNA in a species lacking a genome project), the exons are inferred from the phylogenetically closest opsins. The numbers flanking exons, 012, show the phasing of each intron, eg 12 means an overhang of 1 bp at the 3' end of an exon with that fragmentary codon completed by a 2 bp overhang at the beginning of the next exon. Intron position and phasing are generally conserved over great evolutionary distances -- note here lamprey eel has identical intronation of its opsin genes orthologous to human. Cone and rod opsin paralogs are intronated identically in all species with the exception of LWS opsins which have an extra early intron of phase 12. LWS must have acquired this prior to divergence of lamprey.
Syntentic relationships are also shown. The nearest flanking HUGO-named genes are first chosen for the human opsin, two on each side. The strand orientation noted relative to a fixed convention of plus strand for the human opsin. Then each assembly is revisited to determine the extent of conservation of these flanking genes. In the event humans lack the gene, synteny is defined by the nearest diverging species, typically platypus, that has the gene. Sometimes the original synteny is only partly retained (left- or right-synteny). For deeply diverging species such as amphioxus with an assembly, flanking genes there are pushed forward into other species to help define orthologous opsins (blast clustering can be uncertain because of the diminishing percent identity).
Melanopsins, the unexpected rhabdomeric-class Gq-coupled opsin recently found in upper deuterostomes, are readily confused homologically due to various expansions and contractions. Mammals, human through platypus, have a single melanopsin. However chicken, lizard, frog, and teleost fish experienced a multi-gene segmental duplication and the resulting melanopsins were both retained (though diverged substantially). In ray-finned fish, a processed retrogene arose that may be functional in zebrafish though lost in fugu and stickleback. After its whole genome duplication, zebrafish also retained two copies of the original melanopsin. Chondrichthyes also have a second copy of the primary melanopsin but synteny -- which is essential for analysis since intron placement is uninformative in duplications and sequence alignment is too dependent on unknown rates -- is not available in the current contig-level assembly.
Amphioxus also contains two melanopsins from an apparently independent duplication. Flanking gene order today bears no relation to vertebrate gene order. The lamprey situation awaits assembly of its traces or targeted transcript studies. At this time, only a four exon fragmentary melanopsin can be recovered (however with high percent identity, 80%). Possibly orthologs of this melanopsin locus could be tracked into the highly derived tunicates, acorn worm, and sea urchins. The distinctive intron pattern may even allow melanopsin antecedents to be identified in Cnidaria and Protostomia. At this point, the best blastp match to insects stands at 37% with no evident syntenic or intronic support
While clade-specific proliferation of melanopsins -- and implied role subfunctionalization -- confounds the situation for chordates, it really has little impact on the opsin classifier described here. Unknown sequences will readily find their place because of excellent phylogenetic distribution of reference sequences and the inherent distance of melanopsins from the ciliary collection. The main utility at the level of opsin classifier is the ability to identify other rhabdomeric opsins in later deuterostomes should they occur. At the level of alignment, the melanopsis serve as outgroup to ciliary opsins and so help define motifs specific to Gt-coupled signaling and other structure/function issues.
A dozen very recent publications have shaken our understanding of the evolution of light reception capabilities. After reviewing topics such as ciliary opsin in protostomes, rhabdomeric opsins in deuterostomes, rich opsin repertoires in cnidarians, and other novel opsin classes, I will consider topics such as the origin of image-forming eyes beween amphioxus and lamprey divergences, noting however that our notion of 'eye' is much more nuanced today. The reconstruction of the ur-bilateran eye probably awaits additional cnidarian genomes -- no new ones are being undertaken unfortunately. However the plethora of new arthopod and lophotrochozoan genome assemblies has opened up new avenues of research as the realization grows that fly and nematode are exceedingly derived, with better ancestral characters retained in other species.
Numerous conflicting gene trees have been published for ciliary opsins. Some methodologies have bordered on the preposterous -- thin phylogenetic coverage, dimly related outgroups such as drosophila rhabdomeric opsin, and naive fixed underlying mutational models assumed for maximal likelihood software despite the great diversity of species and many billions of years of branch length. Nonetheless, the resultant trees have only moderate conflict, suggesting that a definitive opsin tree might not be far off.
Rare genomic changes have lately come into vogue as a supplement to traditional maximal likelihood methods, primarily to resolve polytomies (divergence nodes tightly spaced close in time) and otherwise uncertain gene or species tree topologies. The rare genomic changes applicable to opsins include coding indels (deletions and insertions), intron placement (position and phase comparison), synteny (gene order along the chromosome), and gene copy number change (gene gain from retropositional, tandem, segmental, and whole genome duplications; gene loss from pseudogenization or deletion). Results from these methods must be evaluated for their susceptibility to homoplasy (misleading recurrent independent events that mimic a single event) and incomplete penetration in the population level at the time of speciation (lineage sorting).
Among other phylogenetically informative rare genomic events, we'll be looking at a 6 bp amino acid insert, a novel 12 upstream intron in LWS, and post-GWSR introns in rod/cone opsins, all events located between transmembrane helices TM2 and TM3, ie in extracellular loop 2. Their lack of homoplasy can be seen in the massive alignments below.
Because not all cDNA sequences takes place in species having genome projects and not all species having genome projects have cDNAs, existing cDNAs had to be aligned within the heterologous genome project in order to determine their intron placement. As an example, lamprey opsins from Geotria australis and Lethenteron japonicum worked as queries to locate orthologs within the Petromyzon maritimus genome project (which consists solely of 19 million traces as of mid-November 2007).
The first point to be understood in ciliary opsin evolution is jawless fish such as lamprey exhibit a full-blown set of modern rod and cone opsins whereas early deuterostomes such as hemichordates, echinoderms, amphioxus and tunicates genomes totally lack them (Xenoturbella is not available yet) and indeed altogether lack conventional imaging eyes while using protostome-like rhabdomeric opsins with their disjunct signaling system for photorecepton. Of course, characters in extant (living) species should never be confused with ancestral characters at the time of divergence nodes (last common ancestors); conceivably these early diverging deuterostomes have lost opsin genes, perhaps due to a habitat shift to deep water or burrowing habitat.
However the molecular evidence is quite clear that full-blown pentachromatic color vision and most other modern ciliary opsin classes first appeared during the evolutionary stem preceding lamprey divergence. The oldest known fossil lamprey, Priscomyzon, dates at 360 myr to the Devonian. Molecular clocks place lamprey appearance at approximately 430 myr, some 100 million years after Chengjiang and Burgess Shales fossil Lagerstatte formed. Like most soft tissues, eyes seldom leave a good fossil record, though bilateral placement might be reflected in bone orbits.
Hagfish, sister group to lamprey, have imaging eyes but have not been studied; their opsins situation may be derived due to deepwater marine habitat (similarly deepwater coelocanth opsins are adapted to 420 nm). The next-diverging chondrichtyes have inadequate data at GenBank -- only a few rhodopsin genes from skates and dogfish.
This makes even fragments from the partially sequenced elephantfish Callorhyncus milii quite valuable. Those 9 fragments and 3 from the lamprey genome are provided in the data section. The opsin classifier tool can reliably type a fragment from a single mid-sized exon. While full length genes are always preferable, these fragments serve to prove existence of that gene class at the time of a given divergence node. Further, they can validate certain rare genomic events provided the fragment happens to overlap the region of interest.
Despite 6 sequenced opsin mRNAs in the amphioxus Branchiostoma belcheri and an initial assembly in Branchiostoma floridae, no rod/cone opsin can be located there or in earlier diverging deuterostomes with genome projects (3 unicates, 2 urchins, 1 acorn worm). These species may have larval eye spots, ocelli, pigment cells, and related photoreceptors but lack imaging eyes.
The fossil record is unsatisfactory: less than 1 bilateran in 10,000 in Chengjiang and Burgess Shale fossils is even a candidate for deuterostomy. Low numbers of specimens and poor preservation conspire with career pressure and cite-seeking journals to egregiously misinterpret data in the analysis of Hou, discoverer of the Chinese lagerstaette. Myllokunmingia is in the best situation with 500 specimens but Haikouichthys as stem deuterostome, Metaspreggina as post-ediacaran, and Yunnanozoan are all problematic (in the eye of the beholder). While signs of bilaterily disposed eyes are sometimes inferred, it does not follow these were image-forming eyes. Indeed contemporary Branchiostoma and tunicate larva have an eye-spot (ocellus); the genomes contain ciliary opsins clustering to approximately ENCEPH and PPIN -- still a long long road to imaging opsins. Echinoderms and hemichordates genomes have opsins but even more remote. Sea urchin genome encodes at least six opsins, four of these cluster classify to rhabdomeric, ciliary and Go-type. Tube feet are apparently the photosensory organ in adult urchins.
Meanwhile, thousands of high-quality Cambrian arthropod fossils unmistakably show stalked paired eyes. Fossil trilobite eyes are much studied, due to use of calcite as lens crystalin. Imaging eyes of contemporary arthropods and lophotrochozoa are rhabdomeric, utilizing depolarizing Gq-type receptor, phospholipase C, phosphoinositola, diacylglycerol, and transient receptor potential TRP and TRPL channel signaling. However their genomes can also contain ciliary opsins, using hyperpolarizing Gt-type transducins and phosphodiesterase cGMP second-messaging (as well as Go-type gustducin ciliary opsins in other types of photoreceptors).
Vertebrates are just the opposite, having crossed over to a ciliary opsin-based imaging system, while retaining rhabdomeric signaling in melanopsin retinal ganglion cells. Cnidarian opsins are available from Hydra and Nematostella genomes. Hydra expresses a ciliary-type opsin in ectodermal sensory nerve cells whereas Nematostella has opsins classifying between melanopsin and encephalopsin.
It must not be thought that bilaterans invented imaging eyes because earlier diverging cubomedusan jellyfish Carybdea marsupialis has 4 eyestalks each with 6 photoreceptors of 4 types: simple eyespots, pigment cups, complex pigment cups with lenses, and camera-type eyes with a cornea, lens, and retina. This jellyfish tracks, captures, and eats teleost fish. The species very much needs a genome project.
Thus there is no evidence whatsoever -- and every reason to doubt from genomic analysis -- that deuterostomes had imaging eyes during the Cambrian. Despite this, a BBC series, Walking With Monsters, portrayed a school of 25 mm Haikouichthys attacking and wounding an Anomalocaris twenty times their size. It is easy to guess at the scientific advisory panel. This recurrent anthropocentric theme is echoed by fantastic museum imagery of early mammals nimbly predating on dinosaur nests -- dioramas quietly dismantled after Yucatan meteriorite discovery.
Imaging eyes are not essential to survival; even today subterranean mammals such as blind mole rat flourish without them. Discounting ray-finned fish numbers, a very substantial proportion of extant animal species lack imaging eyes 525 myr after the Cambrian. Of 33 animal phyla, a one-third have no specialized organ for detecting light, one-third have light-sensitive organs, and the remaining 6 have imaging eyes (Cnidaria, Mollusca, Annelida, Onychophora, Arthropoda, and Chordata). Thus 82% of animal phyla have survived well over 500 myr without imaging eyes despite the supposedly unrelenting competition/predation from animals with them.
The first table below shows the reference opsin sequences at a glance, grouped by class. Below that is the primary collection of opsing protein sequences. Here the "fields" in the fasta header show gene name, genus, species, common name, heterotrimeric G protein alpha subunit used in signaling, intron structure, synteny (2 flanking genes on each side of the opsin), indel status, sequence length, lambda max, and comment field.
The phylogenetic tree below shows the presence or absence of various opsin genes in clade-representative species, as reflected in the collected reference sequences. The purpose is timing appearance (or disappearance) of a given class of opsin gene. For example, cone and rod opsins first appeared before lamprey divergence; otherwise they are absent from urochordates, cephalochordates, and earlier deuterostomes. Note however a given gene might appear absent because of a genome project gap, lack of experimental effort, insufficient or outdated bioinformatics, or species idiosyncracies (ie be present in a different species of that clade). In other cases (eg platypus SWS1) pseudogene remnants or a syntenically proven deletion establish the gene is definitely absent. Y means yes (present), N means no (absent).
Please do not add or edit sequences at this time -- email me instead with suggestions. tom @ cyber-dyne. com (no spaces). After upgrading the Cnidarian and Protostome opsin content, I will update the alignments, fasta headers, rare genomic event sectors (indels and introns), provide some ancestral sequences at the common ancestor to lamprey, and provide a definitive gene tree. Some sections, such as vision in the ancestral mammal may be split off into separate articles.
RHO1_homSap PIN_galGal RGR_homSap RHO1_monDom PIN_utaSta RGR_galGal RHO1_ornAna PIN_pheMad RGR_xenTro RHO1_galGal PIN_podSic RGR_gasAcu RHO1_anoCar PIN_xenTro RGR_calMil RHO1_xenTro PIN_bufJap .......... RHO1_neoFor PIN_calMil PER_homSap RHO1_latCha VAOP_galGal PER_monDom RHO1_takRub VAOP_galGal PER_galGal RHO1_leuEri VAOP_anoCar PER_xenTro RHO1_calMil VAOP_xenTro PER_gasAcu RHO1_petMar VAOP_danRer PER_calMil RHO1_geoAus VAOP_takRub PERa_braBel RHO1_letJap VAOP_rutRut PERb_braBel ........... VAOP_calMil PERc_braBel RHO2_galGal VAOP_petMar ........... RHO2_anoCar PPIN_anoCar NEUR_homSap RHO2_gekGek PPIN_xenTro NEUR_monDom RHO2_neoFor PPIN_ictPun NEUR_ornAna RHO2_latCha PPIN_danRer NEUR_galGal RHO2_geoAus PPIN_oncMyk NEUR_anoCar ........... PPIN_calMil NEUR_xenTro SWS2_ornAna PPIN_petMar NEUR_gasAcu SWS2_galGal PPIN_letJap NEUR_calMil SWS2_taeGut PPINa_cioInt ........... SWS2_utaSta PPINb_cioInt MEL1_homSap SWS2_xenTro PARIE_utaSta MEL1_monDom SWS2_neoFor PARIE_anoCar MEL1_galGal SWS2_takRub PARIE_xenTro MEL1_xenTro SWS2_gasAcu PARIE_takRub MEL1_danRer SWS2_geoAus PARIE_gasAcu MEL1D_danRer ........... PARIE_danRer MEL1_takRub SWS1_homSap PARIE_petMar MEL1_gasAcu SWS1_monDom ........... MEL1_oryLat SWS1_galGal ENCEPH_homSap MEL1_calMil SWS1_taeGut ENCEPH_monDom MEL1b_calMil SWS1_anoCar ENCEPH_galGal MEL1_petMar SWS1_utaSta ENCEPH_anoCar MEL1a_braFlo SWS1_xenLae ENCEPH_xenTro MEL1a_braBel SWS1_neoFor ENCEPH_takRub MEL1b_braFlo SWS1_danRer ENCEPH4a_takRub MEL1b_braBel SWS1_oryLat ENCEPH4b_takRub MEL2_galGal SWS1_geoAus ENCEPH_gasAcu MEL2_anoCar ........... ENCEPH_calMil MEL2_xenLae LWS_homSap ENCEPH4_calMil MEL2_danRer LWS_monDom ENCEPH5_calMil MEL2_tetNig LWS_ornAna ENCEPH_squAca MEL2_gasAcu LWS_galGal ENCEPH_petMar RHAB_plaDum LWS_anoCar ENCEPH4_braFlo RHAB1_apiMel LWS_xenTro ENCEPH4_braBel RHAB2_apiMel LWS_neoFor ENCEPH5_braFlo RHAB3_apiMel LWS_takRub ENCEPH5_braBel RHAB4_apiMel LWS_gasAcu ENCEPH_apiMel ........... LWS_calMil ENCEPH1_anoGam ........... LWS_petMar ENCEPH2_anoGam ........... LWS_letJap CILL2_plaDum ........... LWS_geoAus CILL1_plaDum ...........
>RHO1_homSap Homo sapiens (human) Gt 0...2.1.0.0 indel -MBD4 +IFT122 +H1FOO -PLXND1 349 aa 497 nm 16565402 NM_000539 rod rhodopsin RHO ciliary all GT-AG 0 MNGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSR 2 1 YIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQ 0 0 FRNCMLTTICCGKNPLGDDEASATVSKTETSQVAPA* 0 >RHO1_monDom Monodelphis domesticus (opossum) Gt 0...2.1.0.0 indel -MBD4 +IFT122 +H1FOO -PLXND1 349 aa 000 nm no_ref genome rod rhodopsin 0 MNGTEGPNFYVPFSNKTGTVRSPFEEPQYYLADPWQFSCLAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAIADLFMVFGGFTMTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIIGVAFTWVMALACAFPPLIGWSR 2 1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSNFGPIFMTIPAFFAKSSSVYNPVIYIMMNKQ 0 0 FRTCMITTLCCGKNPLGDDEASATASKTETSQVAPA* 0 >RHO1_ornAna Ornithorhynchus anatinus (platypus) Gt 0...2.1.0.0 indel - +IFT122 - -PLXND1 354 aa 000 nm ABN43074 17339011 rod rhodopsin 0 MNGTEGQDFYIPMSNKTGVVRSPFEYPQYYLAEPWQYSVLAAYMFMLIMLGFPINFLTLYVTIQHKKLRTPLNYILLNLAFANHFMVLGGFTTTLYTSLHGYFVFGPTGCNIEGFFATLG 1 2 GEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACALPPLVGWSR 2 1 YIPEGMQCSCGIDYYTLRPEVNNESFVIYMFVVHFTIPMTIIFFCYGRLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTVPAFFAKSSAIYNPVIYIMMNKQ 0 0 FRNCMLTTICCGKNPLGDDEASATASKTEQSSVSTSQVSPA* 0 >RHO1_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel -MBD4 +IFT122 +H1FOO -PLXND1 352 aa 000 nm 1385866 NM_205490 rod rhodopsin RH1 0 MNGTEGQDFYVPMSNKTGVVRSPFEYPQYYLAEPWKFSALAAYMFMLILLGFPVNFLTLYVTIQHKKLRTPLNYILLNLVVADLFMVFGGFTTTMYTSMNGYFVFGVTGCYIEGFFATLG 1 2 GEIALWSLVVLAVERYVVVCKPMSNFRFGENHAIMGVAFSWIMAMACAAPPLFGWSR 2 1 YIPEGMQCSCGIDYYTLKPEINNESFVIYMFVVHFMIPLAVIFFCYGNLVCTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTNQGSDFGPIFMTIPAFFAKSSAIYNPVIYIVMNKQ 0 0 FRNCMITTLCCGKNPLGDEDTSAGKTETSSVSTSQVSPA* 0 >RHO1_anoCar Anolis carolinensis (lizard) Gt 0...2.1.0.0 indel -MBD4 +IFT122 - -PLXND1 343 aa 000 nm no_ref genome rod rhodopsin 0 MNGTEGQNFYVPMSNKTGVVRNPFEYPQYYLADPWQFSALAAYMFLLILLGFPINFLTLFVTIQHKKLRTPLNYILLNLAVANLFMVLMGFTTTMYTSMNGYFIFGTVGR 2 2 GEMGLWSLVVLAVERYVVICKPMSNFRFGETHALIGVSCTWIMALACAGPPLLGWSR 2 1 YIPEGMQCSCGVDYYTPTPEVHNESFVIYMFLVHFVTPLTIIFFCYGRLVCTVKA 0 0 AAAQQQESATTQKAEREVTRMVVIMVISFLVCWVPYASVAFYIFTHQGSDFGPVFMTIPAFFAKSSAIYNPVIYILMNKQ 0 0 FRNCMIMTLCCGKNPLGDEDTSAGTKTETSTVSTSQVSPA* 0 >RHO1_xenTro Xenopus tropicalis (frog) Gt 0...2.1.0.0 indel -MBD4 +IFT122 - -PLXND1 355 aa 000 nm no_ref genome rod rhodopsin 0 MNGTEGPNFYIPMSNKTGVVRSPFDYPQYYLAEPWKYSALAAYMFLLILLGFPINFMTLYVTIQHKKLRTPLNYILLNLVFANHFMVLCGFTVTMYTSMHGYFIFGQTGCYIEGFFATLG 1 2 GEMALWSLVVLAIERYVVVCKPMANFRFGENHAIMGVVFTWIMALSCAAPPLFGWSR 2 1 YIPEGMQCSCGVDYYTLKPEVNNESFVVYMFIVHFTIPLCVIFFCYGRLLCTVKE 0 0 AAAQQQESATTQKAEKEVTRMVVMMVIFFLICWVPYAYVAFYIFTHQGSDFGPVFMTVPAFFAKSSAIYNPVIYIVLNKQ 0 0 FRNCLITTLCCGKNPFGDEEGSSAASSKTEASSVSSSQVSPA* 0 >RHO1_neoFor Neoceratodus forsteri (lungfish) Gt 0...2.1.0.0 indel x x x x 355 aa 000 nm 17961206 EF526299 rod rhodopsin 0 MNGTEGPNFYVPMTNKTGVVRSPFEYPQYYLADPWKYSALAAYMFFLILTGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTMYTAMNGYFVFGVVGCNLEGFFATFG 1 2 GIIALWCLVVLAIERYIVVCKPISNFRFGENHAIMGVVFTWIMALACAGPPLFGWSR 2 1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFIVHFTIPLIIIFFCYGRLMCTVKE 0 0 AAAQQQESATTQKAEKEVTRMVYIMVISYLVCWLPYASVSFYIFTHQGSDFGPVFMTVPAFFAKTASVYNPVIYILMNKQ 0 0 FRNCMITTLCCGKNPFGDEETTSAGTSKTEASSVSSSQVSPA* 0 >RHO1_latCha Latimeria chalumnae (coelacanth) Gt 0...2.1.0.0 indel x x x x 354 aa 478 nm 10339578 AAD30519 rod rhodopsin 0 MNGTEGPNFYVPMSNKTGVVRNPFEYPQYYLADPWKYSALAAYMFFLILVGFPINFLTLFVTIQHKKLRTPLNYILLDLAVADLCMVFGGFFVTMYSSMNGYFVLGPTGCNIEGFFATLG 1 2 GQVALWALVVLAIERYVVVCKPMSNFRFGENHAIMGVIFTWIMALSCAVPPLFGWSR 2 1 YIPEGMQSSCGVDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGRLVCTVKD 0 0 AAAQQQESATTQKAEKEVTRMVIVMVISFLVCWVPYASVAAYIFFNQGSEFGPVFMTAPSFFAKSASFYNPVIYILLNKQ 0 0 FRNCMITTLCCGKNPFGDEDATSAAGSSKTEASSVSSSSVSPA* 0 >RHO1_takRub Takifugu rubripes (pufferfish) Gt 0...2.1.0.0 indel -MBD4 +IFT122 - -PLXND1 355 aa 000 nm 12783465 AF201472 rod rhodopsin 0 MNGTEGPNFYIPMSNKTGVVRSPFEYPQYYLAEPWKYSLVAAYMLFLIITAFPVNFLTLFVTVKHKKLRTPLNYVLLNLAVADLFMVIGGFTVTLYTALHAYFVLGVTGCNIEGFFATLG 1 2 GEIALWSLVVLAVERYIVVCKPMTNFRFGEKHAIAGLVFTWIMALTCATPPLLGWSR 2 1 YIPEGMQCSCGIDYYTPKPEINNTSFVIYMFILHFSIPLAIIFFCYSRLLCTVRA 0 0 AAALQQESETTQRAEKEVTRMVIVMVISFLVCWVPYASVAWYIFANQGTEFGPVFMTAPAFFAKSAALYNPVIYILLNRQ 0 0 FRNCMITTVCCGKNPFGDDDAATTVSKTQSSSVSSSQVAPA* 0 >RHO1_leuEri Leucoraja erinacea (skate) Gt 0...2.1.0.0 indel x x x x 355 aa 000 nm 9256070 U81514 rod rhodopsin 0 MNGTEGENFYVPMSNKTGVVRSPFDYPQYYLGEPWMFSALAAYMFFLILTGLPVNFLTLFVTIQHKKLRQPLNYILLNLAVSDLFMVFGGFTTTIITSMNGYFIFGPAGCNFEGFFATLG 1 2 GEVGLWCLVVLAIERYMVVCKPMANFRFGSQHAIIGVVFTWIMALSCAGPPLVGWSR 2 1 YIPEGLQCSCGVDYYTMKPEVNNESFVIYMFVVHFTIPLIVIFFCYGRLVCTVKE 0 0 AAAQQQESESTQRAEREVTRMVIIMVVAFLICWVPYASVAFYIFINQGCDFTPFFMTVPAFFAKSSAVYNPLIYILMNKQ 0 0 FRNCMITTICLGKNPFEEEESTSASASKTEASSVSSSQVAPA* 0 >RHO1_calMil Callorhinchus milii (elephantfish) Gt 0...2.1.0.0 indel x x x x 355 aa 000 nm no_ref genome rod rhodopsin complete wgs 0 MNGTEGENFYIPMSNKTGVVRSPFEYPQYYLAEPWQFSILAAYMFFLIITCFPVNFLTLYVTFEHKKLRQPLNFILLNLAVADLFMVFGGFFITVYTSLHGYFVFGVTGCNFEGFFATLG 1 2 GEIGLWSLVVLAIERYVVVCKPMSNFRFGTNHAIMGVAFTWVMALACAVPPLMGWSR 2 1 YIPEGLQCSCGVDYYTLKPEINNESFVIYMFVVHFLIPLIIIFFCYGRLVCTVKE 0 0 AAAQQQESESTQRAEREVTRMVIIMVIFFLICWVPYASVAFFIFTNQGSEFGPIFMAVPAFFAKSSALYNPLIYILLNKQ 0 0 FRNCMITTLCCGKNPFEEDESTSAAASKTEASSVSSSQVSPA* 0 >RHO1_petMar Petromyzon marinus (lamprey) Gt 0...2.1.0.0 indel x x x x 354 aa 000 nm no_ref genome rod rhodopsin 0 MNGTEGENFYIPFSNKTGLARSPFEYPQYYLAEPWKYSVLAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVANLFMVLFGFTLTMYSSMNGYFVFGPTMCNFEGFFATLG 1 2 GEMSLWSLVVLAIERYIVICKPMGNFRFGSTHAYMGVAFTWFMALSCAAPPLVGWSR 2 1 YLPEGMQCSCGPDYYTLNPNFNNESFVIYMFLVHFIIPFIVIFFCYGRLLCTVKE 0 0 AAAAQQESASTQKAEKEVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTVPAFFAKTSALYNPIIYILMNKQ 0 0 FRNCMITTLCCGKNPLGDEDSGASTSKTEVSSVSTSQVSPA* 0 >RHO1_geoAus Geotria australis (lamprey) Gt 0...2.1.0.0 indel x x x x 354 aa 497 nm 17463225 AY366493 rod rhodopsin rodRhA 0 MNGTEGQNFYIPFSNKTDVARSPFEYPQYYLAEPWKFSALAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVSNLFMILFGFTTTMYTSMNGYFVFGPTMCSIEGFFATLG 1 2 GEVSLWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVALTWVMALSCAAPPLLGWSR 2 1 YLPEGMQCSCGPDYYTMNPTYNNESFVIYMFIVHFTIPFVIIFFSYGRLLCTVKE 0 0 AAAAQQESASTQKAEKEVTRMVVLMVVGFLVCWVPYASVAFYIFTNQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ 0 0 FRNCMITTLCCGKNPLGDDDSGASTSKTEVSSVSTSQVAPA* 0 >RHO1_letJap Lethenteron japonicum (lamprey) Gt 0...2.1.0.0 indel x x x x 354 aa 000 nm 15096614 AB116382 cone rhodopsin 0 MNGTEGDNFYVPFSNKTGLARSPYEYPQYYLAEPWKYSALAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAMANLFMVLFGFTVTMYTSMNGYFVFGPTMCSIEGFFATLG 1 2 GEVALWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVAFTWIMALACAAPPLVGWSR 2 1 YIPEGMQCSCGPDYYTLNPNFNNESYVVYMFVVHFLVPFVIIFFCYGRLLCTVKE 0 0 AAAAQQESASTQKAEKEVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ 0 0 FRNCMITTLCCGKNPLGDDESGASTSKTEVSSVSTSQVSPA* 0 >RHO2_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel -IHPK3 -LEMD2 -GRM4 +HMGA1 356 aa 000 nm 2268324 NP_990771 cone rhodopsin 0 MNGTEGINFYVPMSNKTGVVRSPFEYPQYYLAEPWKYRLVCCYIFFLISTGLPINLLTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFVFGPVGCAVEGFFATLG 1 2 GQVALWSLVVLAIERYIVVCKPMGNFRFSATHAMMGIAFTWVMAFSCAAPPLFGWSR 2 1 YMPEGMQCSCGPDYYTHNPDYHNESYVLYMFVIHFIIPVVVIFFSYGRLICKVRE 0 0 AAAQQQESATTQKAEKEVTRMVILMVLGFMLAWTPYAVVAFWIFTNKGADFTATLMAVPAFFSKSSSLYNPIIYVLMNKQ 0 0 FRNCMITTICCGKNPFGDEDVSSTVSQSKTEVSSVSSSQVSPA* 0 >RHO2_anoCar Anolis carolinensis (lizard) Gt 0...2.1.0.0 indel -IHPK3 -LEMD2 -GRM4 +HMGA1 356 aa 000 nm no_ref genome cone rhodopsin 0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLAEPWKYKVVCCYIFFLIFTGLPINILTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFIFGPIGCAIEGFFATLG 1 2 GQVALWSLVVLAIERYIVVCKPMGNFRFSATHALMGISFTWFMSFSCAAPPLLGWSR 2 1 YIPEGMQCSCGPDYYTLNPDYHNESYVLYMFGVHFVIPVVVIFFSYGRLICKVRE 0 0 AAAQQQESASTQKAEREVTRMVILMVLGFLLAWTPYAMVAFWIFTNKGVDFSATLMSVPAFFSKSSSLYNPIIYVLMNKQ 0 0 FRNCMITTICCGKNPFGDEDVSSSVSQSKTEVSSVSSSQVSPA* 0 >RHO2_gekGek Gekko gekko (gecko) Gt 0...2.1.0.0 indel x x x x 356 aa 000 nm 11591478 AY024356 cone rhodopsin in pure rod-retina 0 MNGTEGINFYVPLSNKTGLVRSPFEYPQYYLADPWKFKVLSFYMFFLIAAGMPLNGLTLFVTFQHKKLRQPLNYILVNLAAANLVTVCCGFTVTFYASWYAYFVFGPIGCAIEGFFATIG 1 2 GQVALWSLVVLAIERYIVICKPMGNFRFSATHAIMGIAFTWFMALACAGPPLFGWSR 2 1 FIPEGMQCSCGPDYYTLNPDFHNESYVIYMFIVHFTVPMVVIFFSYGRLVCKVRE 0 0 AAAQQQESATTQKAEKEVTRMVILMVLGFLLAWTPYAATAIWIFTNRGAAFSVTFMTIPAFFSKSSSIYNPIIYVLLNKQ 0 0 FRNCMVTTICCGKNPFGDEDVSSSVSQSKTEVSSVSSSQVAPA* 0 >RHO2_neoFor Neoceratodus forsteri (lungfish) Gt 0...2.1.0.0 indel x x x x 356 aa 000 nm 17961206 EF526299 cone rhodopsin 0 MNGTEGINFYVPHSNKTGVVRSPFEYPQYYLADPWKYSIVCAYMFFLIITGLPINLLTLVVTFKHKKLRQPLNYILVNLAVADLFMVCFGFTVTFSTAINGYFIFGPRGCAIEGFMATLG 1 2 GEVALWSLVVLAIERYIVVCKPMGNFRFSNNHSIIGIVFTWLAALSCAAPPLFGWSR 2 1 YLPEGMQCSCGPDYYTMNPDYHNESFVIYMFVVHFFIPVIVIFVSYGRLICKVKE 0 0 AAAQQQESASTQKAEREVTRMVILMVIGFMTAWTPYATVAFWIFMNKGAEFGATFMAAPAFFSKSSALYNPIIYVLMNKQ 0 0 FRNCMVTTLCCGKNPFGDDDVSSSVSAGKTEVSSVSSSQVSPA* 0 >RHO2_latCha Latimeria chalumnae (coelacanth) Gt 0...2.1.0.0 indel x x x x 355 aa 485 nm 10339578 AH007713 cone rhodopsin RH2 0 MNGTEGMNFYVPLSNRTGLVRSPFEYTQYYLAEPWKFSVLCAYMFLLIILGFPINFLTLLVTFKHKKLRQPLNYILVNLAVASLFMVVFGFTVTFYSSLNGYFVLGPMGCAMEGFFATLG 1 2 GQVALWSLVVLAIERYIVVCKPMGNFRFASSHAIMGIAFTWIMALACAAPPLVGWSR 2 1 YIPEGLQCSCGPDYYTLNPDFHNESYVMYLFLVHFLLPIIIIFFTYGRLICKVKE 0 0 AAAQQQESASTQKAEKEVTRMVILMVIGFLTAWVPYASAAFWIFCNRGAEFTATLMTVPAFFSKSSCLFNPIIYVLLNKQ 0 0 FRNCMITTLCCGKNPLGDDDTSSAVSQSKTDVSSVSSSQVSPA* 0 >RHO2_geoAus Geotria australis (lamprey) Gt 0...2.1.0.0 indel x x x x 355 aa 492 nm 17463225 AY366494 cone rhodopsin RhB 0 MNGTEGANFYIPFHNRTGVVRSPYEYPQYYLADPWMYSAISAYVFTLILIGFPVNFMTLFVTFKLKKLRQPLNFILVNLCVADLLMIMFGFTTTFYTAMNGYFVFGPTGCNIEGFFATLG 1 2 GEVSLWSLVMLAIERYIVVCKPMGNFRFATTHAALGVVFTWVMASACAVPPLVGWSR 2 1 YIPEGMQCSCGPDYYTLNPKYYNESYVIYLFLVHFLLPVTIIFFTYGRLICTVKE 0 0 AAAQQQESASTQKAEREVTRMVIIMVVGFLVCWVPYASFAFYLFMNKGILFSATAMTVPAFFSKSSVLYNPIIYVLLNKQ 0 0 FRTCMVTTLFCGKNPFGEDDSSMVSTSKTEVSSVSSSQVSPS* 0 >SWS2_ornAna Ornithorhynchus anatinus (platypus) Gt 0...2.1.0.0 indel -IRAK1 -MECP2 - +TKTL1 364 aa 000 nm 17339011 ABN43074 cone short blue tandem -FLNB--+MECP2 with MWS1 0 MHKTHRNLQNELPEDFFIPLPLDTDNITSLSPFLVPQTHLGGSGIFMSLAAFMFLLITLGFPINLLTVICTIKYKKLRSHLNYILVNLAVSNMLVVCVGSATAFYSFAHMYFVLGPTACKIEGFAATLG 1 2 GMVSLWSLAVIAFERFLVICKPLGNLSFRGTHAIFGCAATWVFGLAASLPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFSFCFGVPLSIIIFSYGRLLLTLRA 0 0 VAKQQEQSATTQKAEREVTKMVIVMVLGFLVCWLPYASFSLWVVTNRGQVFDLRMASIPSVFSKASTIYNPIIYVFMNKQ 0 0 FRSCMLKLVFCGKSPFGDEDEISGSSQATQVSSVSSSQVSPA* 0 >SWS2_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel x x x x 362 aa 000 nm 7975342 NP_990848 cone short2 blue 0 MHPPRPTTDLPEDFYIPMALDAPNITALSPFLVPQTHLGSPGLFRAMAAFMFLLIALGVPINTLTIFCTARFRKLRSHLNYILVNLALANLLVILVGSTTACYSFSQMYFALGPTACKIEGFAATLG 1 2 GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCVATWVLGFVASAPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTDNKWHNESYVLFLFTFCFGVPLAIIVFSYGRLLITLRA 0 0 VARQQEQSATTQKADREVTKMVVVMVLGFLVCWAPYTAFALWVVTHRGRSFEVGLASIPSVFSKSSTVYNPVIYVLMNKQ 0 0 FRSCMLKLLFCGRSPFGDDEDVSGSSQATQVSSVSSSHVAPA* 0 >SWS2_taeGut Taeniopygia guttata (finch) Gt 0...2.1.0.0 indel x x x x 363 aa 000 nm no_ref genome cone short2 0 MPKPREMRDELPEDFYIPMSLETPNLTALSPFLVPQTHLGSPGIFKAMAAFMFLLVLLGVPINALTVLCTAKYKKLRSHLNYILVNLAVANLLVVCVGSTTAFYSFSQMYFALGPLACKIEGFTATLG 1 2 GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCAITWIFGLIASLPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTDNKWNNESYVIFLFCFCFGFPLTVIVFSYGRLLLTLRA 0 0 VAKQQEQSASTQKAEREVTKMVVVMVLGFLVCWLPYCSFALWVVTHRGHPFDLGLASIPSVFSKASTVYNPIIYVFMNKQ 0 0 FRSCMLKLVFCGRSPFGDEDDVSGSSQATQVSSVSSSQVSPA* 0 >SWS2_utaSta Uta stansburiana (lizard) Gt 0...2.1.0.0 indel x x x x 364 aa 000 nm 16543463 DQ100326 cone short 0 MHNSRPHSRDDLPEDFFIPMPLDVANITTLSPFLVPQTHLGSPALFMGMAAFMFLLIILGVPINVLTIFCTFKYKKLRSHLNYILVNLAVSNLLVVCIGSTTAFYSFAQMYFSLGPTACKIEGFAATLG 1 2 GMVSLWSLAVVAFERFLVICKPLGNFSFRGTHAIIGCIITWVFGLVASLPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKWNNESYVLFLFSFCFGVPLSVIIFSYGRLLLTLRA 0 0 VAKQQEQSATTQKAEREVTKMVVVMVMGFLVCWLPYASFALWVVTHRGEPFDVRLATIPSVFSKASSVYNPVIYVFMNKQ 0 0 FRSCMLKLVFCGKSPFGDEDDVSGSSQTTQVSSVSSSQVSPA* 0 >SWS2_xenTro Xenopus tropicalis (frog) Gt 0...2.1.0.0 indel -IRAK1 -MECP2 - - 363 aa 000 nm no_ref genome cone short 0 MSKGRPDLRMEMPDEFYVPIPLETTNISSLSPFLVPQTHLGTPGIFMSISAFMLFTIIFGFPLNLLTIICTVKYKKLRSHLNYILVNLAVANLIVICFGSTTAFYSFSQMYFSLGTLACKIEGFTATLG 1 2 GIIGLWSLAVVAFERFLVICKPMGNFTFRESHAVLGCILTWVIGLVAAIPPLLGWSR 2 1 YIPEGLQCSCGPDWYTVNNKWNNESYVLFLFCFCFGFPLAIIVFSYGRLLLALHA 0 0 VAKQQEQSATTQKAEREVTRMVIVMVVGFLVCWLPYASFALWAVTHRGELFDLRMSSVPSVFSKASTVYNPFIYIFMNRQ 0 0 FRSCMMKMIFCGKNPLGDDEETSVSGSTQVSSVSSSQIAPS* 0 >SWS2_neoFor Neoceratodus forsteri (lungfish) Gt 0...2.1.0.0 indel x x x x 364 aa 000 nm 17961206 EF526299 cone short 0 MHRTKPDPQEDLPDDFYIPVSLNTNNITMLSPFLVPQTHLGSPSVFMVLSVFMFFLLITGIPINVLTIICTFKYKKLRSHLNYILVNLAVANLIVVGFGSTTAFYSFSQMYFAWGPLACKIEGFAATLG 1 2 GMVSLWSLAVVAFERFLVICKPLGNFTFRSTHAIIGCVATWVFGLISSAPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKWNNESYVIFLFCFCFGFPLSVIIFSYGRLLMTLRA 0 0 VAKQQEQSASTQKAEREVTKMVVVMVLGFLVCWLPYTVFSLWVVTHRGESFELALGSIPAVFSKSSTVYNPLIYVFMNKQ 0 0 FRSCMMKLIFCGKSPFGDEDDASSASQSTQVSSVSSSQVAPA* 0 >SWS2_takRub Takifugu rubripes (pufferfish) Gt 0...2.1.0.0 indel x x x x 351 aa 000 nm no_ref genome cone short2 0 MRGVRQHEFQEDFYIPIPLDVDNITALSPFLVPQDHLGSPAVFYGMSAFMFFLFVAGTGINVLTIACTIQYKKLRSHLNYILVNLAFSNLLVTTVGSFTCFCCFFVRYMIVGPLGCKIEGFAATLG 1 2 GMVSLWSLAVVAFERWLVVCKPLGNFIFKPDHAIVCCIFTWFFALIISAPPLFGWSR 2 1 YIPEGFQCSCGPDWYTTGNKYNNESYVWFIFGFGFAVPLFVIVFCYSQLLVMLKS 0 0 AKAQAESASTQKAEREVTRMVVVMILGFLVCWLPYASFALWVVNNRGTPFDLRLATIPACFSKASTVYNPIIYVVLNKQ 0 0 FRSCMKKMLGMSGGDDEESSSQSVTEVSKVSPS* 0 >SWS2_gasAcu Gasterosteus aculeatus (stickleback) Gt 0.2.2.1.0.0 indel x x x x 359 aa 000 nm no_ref genome cone short 0 MKHGRVPEIPEDFYIPISLDTDNITSLSPFLVPQDHLASKATFYSLAFYMFFILIVGTFINALTVACTVQNKKLRSHLNYILVNLAVSNLLVSGVGAFTAFLSFAARYFVLGTLACKVEGFLATLG 1 2 GMVSLWSLAVIAFERWLVICKPLGNFIFKPDHALVCCAFTWVFALAASAPPLVGWSR 2 1 YIPEGLQCSCGPDWYTTNNKYNNESYVLFLFGFCFAVPFCTICFCYSQLLFTMKMA 0 0 AKAQAESASTQKAEREVTRMVVLMVMGFLVCWMPYASFALWVVNNRGQTFDLRFASIPSVFSKSSAVYNPVIYVLLNKQ 0 0 FRSCMMKMLGMGGGDDEESSTSSVTEVSKVGPA* 0 >SWS2_geoAus Geotria australis (lamprey) Gt 0...2.1.0.0 indel x x x x 362 aa 439 nm 17463225 AY366492 cone short2 blue retinal 0 MYQGKSTQVDDLPEDFYIPIALNVKNMSELSPFLVPQVHLGDSFIFYGMSAFMLFLVLAGFPLNFLTVFVTIKYKKLRSHLNYILVNLAIANLIVVCCGSTLAFYSFMHKYFILGPLFCKMEGFTATLG 1 2 GMLSLWSLAVLAFERCLVICKPFGNIAFRGTHALIRCGFAWAAAIAASTPPLFGWSR 2 1 YIPEGLQCSCGPDWYTTNNKYNNESYVMFLFIFCFGTPFTIIIVSYSKLILTLRA 0 0 AAAQQQESASTQKAEKEVSRMVVIMVGGFLVCWLPYASLALWIVFNRGSPFDLRLATIPSVFSKASTVYNPVIYIFLNKQ 0 0 FRSCMMKTIFCGKNPLGDDEDATSTTTQVSSVSTSQVAPA* 0 >SWS1_homSap Homo sapiens (human) Gt 0.2.2.1.0.0 indel -FAM137A -CALU -NAG6 -FLNC 348 aa 000 nm 1385866 NP_990769 cone short 0 MRKMSEEEFYLFKNISSVGPWDGPQYHIAPVWAFYLQAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVA 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQ 0 0 FQACIMKMVCGKAMTDESDTCSSQKTEVSTVSSTQVGPN* 0 >SWS1_monDom Monodelphis domesticus (opossum) Gt 0...2.1.0.0 indel -FAM137A -CALU -NAG6 -FLNC 347 aa 000 nm no_ref genome cone short 0 MSGDEEFYLFKNISSVGPWDGPQYHIAPAWAFHFQTVFMGFVFCAGTPLNAVVLVATLRYKKLRQPLNYILVNVSLCGFIFCIFAVFTVFISSSQGYFIFGRHVCAMEAFLGSVA 1 2 GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWVIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIMPLFLICFSYSQLLRALRA 0 0 VAAQQQESATTQKAEREVSRMVVMMVGSFCLCYVPYAALAMYMVNNQNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FHACIMEMVCRKPMTDDSDVSSSQKTEVSAVSSSQVGPT* 0 >SWS1_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel x x x x 348 aa 000 nm no_ref genome cone short1 violet 0 MSSDDDFYLFTNGSVPGPWDGPQYHIAPPWAFYLQTAFMGIVFAVGTPLNAVVLWVTVRYKRLRQPLNYILVNISASGFVSCVLSVFVVFVASARGYFVFGKRVCELEAFVGTHG 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFSSRHALLVVVATWLIGVGVGLPPFFGWSR 2 1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FRACIMETVCGKPLTDDSDASTSAQRTEVSSVSSSQVGPT* 0 >SWS1_taeGut Taeniopygia guttata (finch) Gt 0...2.1.0.0 indel x x x x 347 aa 000 nm no_ref genome cone short1 0 MDEEEFYLFKNQSSVGPWDGPQYHIAPMWAFYLQTIFMGLVFVAGTPLNAIVLIVTIKYKKLRQPLNYILVNISVSGLMCCVFCIFTVFIASSQGYFVFGKHMCAFEGFAGATG 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSRHALLVVAATWIIGVGVAIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCMCYVPYAALAMYMVNNREHGIDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0 0 FRACIMETVCGRPMTDDSEVSSSAQRTEVSSVSSSQVGPS* 0 >SWS1_anoCar Anolis carolinensis (lizard) Gt 0.2.2.1.0.0 indel - -CALU - - 347 aa 000 nm no_ref genome cone short 0 MSGQEDFYLFENISSVGPWDGPQYHIAPMWAFYFQTAFMGFVFFAGTPLNAIILIVTVKYKKLRQPLNYILVNISFAGFLFCTFSVFTVFMASSQGYFFFGRHVCAMEAFLGSVA 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSRHALLVVAATWIIGVGVAIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLTLIIFSYSQLLGALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYASLAMYMVNNRDHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0 0 FRACILETVCGKPMSDESDVSSSAQKTEVSSVSSSQVSPS* 0 >SWS1_utaSta Uta stansburiana (lizard) Gt 0...2.1.0.0 indel x x x x 348 aa 000 nm 16543463 DQ100325 cone short 0 MSGEEDFYLFENISSVGPWDGPQYHIAPMWAFYFQTAFMGFVFFAGTPLNAIILIVTVKYKKLRQPLNYILVNISFAGFLFCVFSVFTVFLASSQGYFFFGRHICALEAFLGSVA 1 2 GLVTGWSLAFLAFERYIVICKPFGNFRFNSKHALLVVAATWFIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLTLIIFSYSQLLGALRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGIDLRLVTIPAFFSKSACVYNPIIYCFMNKQ 0 0 FRACIMETVCGKPMTDESDVSSSAQKTEVSSVSSSQVSPS* 0 >SWS1_xenLae Xenopus laevis (frog) Gt 0...2.1.0.0 indel - -CALU - - 348 aa 000 nm no_ref genome cone short 0 MLEEEDFYLFKNVSNVSPFDGPQYHIAPKWAFTLQAIFMGMVFLIGTPLNFIVLLVTIKYKKLRQPLNYILVNITVGGFLMCIFSIFPVFVSSSQGYFFFGRIACSIDAFVGTLT 1 2 GLVTGWSLAFLAFERYIVICKPMGNFNFSSSHALAVVICTWIIGIVVSVPPFLGWSR 2 1 YMPEGLQCSCGPDWYTVGTKYRSEYYTWFIFIFCFVIPLSLICFSYGRLLGALRA 0 0 VAAQQQESASTQKAEREVSRMVIFMVGSFCLCYVPYAAMAMYMVTNRNHGLDLRLVTIPAFFSKSSCVYNPIIYSFMNKQ 0 0 FRGCIMETVCGRPMSDDSSVSSTSQRTEVSTVSSSQVSPA* 0 >SWS1_neoFor Neoceratodus forsteri (lungfish) Gt 0...2.1.0.0 indel x x x x 347 aa 000 nm 17961206 EF526299 cone short 0 MSGEEEFYLFKNISSVGPWDGPQYHIAPKWAFFLQAAFMGFVLFVGTPLNAIVLFVTIKYKKLQQPLNYILVNISLAGFIFCFFGVFAVFIASCQGYFIFGKTVCALEGFTGSVA 1 2 GLVTGWSLAILAFERYLVICKPIGNFRFGSKHSMIAVVAAWVIGVGVSIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIIPLFIICFSYSQLLGALRA 0 0 VAAQQQESATTQKAEREVSRMIIVMVGSFCVCYVPYAALAMYMVNNRDHGIDLRLVTIPAFFSKSSFVYNPIIYCFMNKQ 0 0 FRACIMQTVFGKPMTDDSDISSSGKTEVSSVSSSQVNPS* >SWS1_danRer Danio rerio (zebrafish) Gt 0...2.1.0.0 indel - -CALU - - 337 aa 000 nm no_ref genome cone short1 0 MDAWAVQFGNASKVSPFEGEQYHIAPKWAFYLQAAFMGFVFIVGTPMNGIVLFVTMKYKKLRQPLNYILVNISLAGFIFDTFSVSQVSVCAARGYYSLGYTLCSMEAAMGSIA 1 2 GLVTGWSLAVLAFERYVVICKPFGSFKFGQGQAVGAVVFTWIIGTACATPPFFGWSR 2 1 YIPEGLGTACGPDWYTKSEEYNSESYTYFLLITCFMMPMTIIIFSYSQLLGALRA 0 0 VAAQQAESESTQKAEREVSRMVVVMVGSFVLCYAPYAVTAMYFANSDEPNKDYRLVAIPAFFSKSSSVYNPLIYAFMNKQ 0 0 FNACIMETVFGKKIDESSEVSSKTETSSVSA* 0 >SWS1_oryLat Oryzias latipes (medaka) Gt 0...2.1.0.0 indel - - - - 336 aa 000 nm no_ref genome cone short1 0 MGKYFYLYENISKVGPYDGPQYYLAPTWAFYLQAAFMGFVFFVGTPLNFVVLLATAKYKKLRVPLNYILVNITFAGFIFVTFSVSQVFLASVRGYYFFGQTLCALEAAVGAVA 1 2 GLVTSWSLAVLSFERYLVICKPFGAFKFGSNHALAAVIFTWFMGVGCACPPFFGWSR 2 1 YIPEGLGCSCGPDWYTNCEEFSCASYSKFLLVTCFICPITIIIFSYSQLLGALRA 0 0 VAAQQAESASTQKAEKEVSRMIIVMVASFVTCYGPYALTAQYYAYSQDENKDYRLVTIPAFFSKSSCVYNPLIYAFMNKQ 0 0 FNGCIMEMVFGKKMEEASEVSSKTEVSTDS*0 >SWS1_geoAus Geotria australis (lamprey) Gt 0...2.1.0.0 indel x x x x 346 aa 359 nm 17463225 AY366495 cone short1 UV retinal 0 MSGDEEFYLFKNISKVGPWDGPQFHIAPKWAFYLQAAFMGFVFICGTPLNAIVLVVTIKYKKLRQPLNYILVNISAAGLVFCLFSISTVFVASMQGYFFLGPTICALEAFFGSLA 1 2 GLVTGWSLAFLAAERYIVICKPFGNFRFGSKHALVAVGLTWMLGLSVALPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYKSEYYTYFLFVFCFVVPLSIIIFSYGSLLGTLRA 0 0 VAAQQQESASTQKAEREVSRMVIMMVASFCTCYVPYAALAVYMVTNRDHNIDLRFVTVPAFFSKASCVYNPLIYSFMNKQ 0 0 FRACILETVCGKPITDESETSSSRTEVSSVSTTQMIPG* 0 >LWS_homSap Homo sapiens (human) Gt 0.2.2.1.0.0 indel -IRAK1 -MECP2 -TEX28 +TKTL1 364 aa 530 nm 12853434 NP_000504 cone long OPN1MW deutan 0 MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTSVWMIFVVIASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPMCVLEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLAIVGIAFSWIWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSYPGVQSYMIVLMVTCCITPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVTRMVVVMVLAFCFCWGPYAFFACFAAANPGYPFHPLMAALPAFFAKSATIYNPVIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA* 0 >LWS_monDom Monodelphis domesticus (opossum) Gt 0.2.2.1.0.0 indel -IRAK1 -MECP2 - +TKTL1 368 aa 000 nm no_ref genome cone long 0 MTQAWDPAGFLARRRDVNEDDNDETTRSSLFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMVFVVIASIFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETVIASTISVINQIYGYFILGHPLCVLEGYTVSLC 1 2 GITGLWSLAIISWERWVVVCKPFGNVKFDAKLAMVGIIFSWVWAAVWTAPPLFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMATCCIFPLSIILLCYVQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTLFACFAAANPGYSFHPLTASLPAYFAKSATIYNPIIYVFMNRQ 0 0 FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA* 0 >LWS_ornAna Ornithorhynchus anatinus (platypus) Gt 0.2.2.1.0.0 indel -IRAK1 -MECP2 - - 365 aa 000 nm 17339011 ABN43074 cone long LWS green 0 MTPAWNSGVYAARRRFEDEEDTTRTSVFVYTNSNNTR 1 2 DPFEGPNYHIAPRWAYNVTSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC 1 2 GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIVLCYLQVWLAIRA 0 0 VAKQQKESESTQKAEKEVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA* 0 >LWS_galGal Gallus gallus (chicken) Gt 0.2.2.1.0.0 indel x x x x 363 aa 000 nm 12716987 NM_205438 cone long green iodopsin missing in assembly 0 MAAWEAAFAARRRHEEEDTTRDSVFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVAASVFTNGLVLVATWKFKKLRHPLNWILVNLAVADLGETVIASTISVINQISGYFILGHPMCVVEGYTVSAC 1 2 GITALWSLAIISWERWFVVCKPFGNIKFDGKLAVAGILFSWLWSCAWTAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFFPLAIIILCYLQVSLAIRA 0 0 VAAQQKESESTQKAEKEVSRMVVVMIVAYCFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSEVSTSRTEVSSVSNSSVSPA* 0 >LWS_anoCar Anolis carolinensis (lizard) Gt 0.2.2.1.0.0 indel - - -TEX28 +TKTL1 366 aa 000 nm no_ref genome cone long 0 MAGTVTEAWDVAVFAARRRNDEDDTTRDSLFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNITSVWMIFVVIASIFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVIASTISVINQISGYFILGHPMCVLEGYTVSTC 1 2 GISALWSLAVISWERWVVVCKPFGNVKFDAKLAVAGIVFSWVWSAVWTAPPVFGWSR 2 1 YWPHGLKTSCGPDVFSGSDDPGVLSYMIVLMITCCFIPLAVILLCYLQVWLAIRA 0 0 VAAQQKESESTQKAEKEVSRMVVVMIIAYCFCWGPYTVFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSNSSVSPA* 0 >LWS_xenTro Xenopus tropicalis (frog) Gt 0.2.2.1.0.0 indel -IRAK1 -MECP2 - - 370 aa 000 nm no_ref genome cone long 0 MASHWNEAVFAARRRNDDDDTTRSSVFTYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNISSLWMIFVVLASVFTNGLVLVATLKFKKLRHPLNWILVNMAIADLGETVIASTISVCNQIFGYFVLGHPMCILEGYTVSVC 1 2 GIAALWSLTVIAWERWFVVCKPFGNIKFDGKLAATGIIFSWVWAAGWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMLVLMITCCIIPLAIIVLCYMHVWLTIRQ 0 0 VAQQQKESESTQKAEREVSRMVVVMIIAYIFCWGPYTFFACFAAFNPGYNFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQLFGKKVDDGSEVSSTSRTEVSSVSNSSVSPA* 0 >LWS_neoFor Neoceratodus forsteri (lungfish) Gt 0.2.2.1.0.0 indel x x x x 365 aa 000 nm 17961206 EF526299 cone long 0 MAEPWDAVLAARRRHQDEETTRSTIFVYTNSNNTR 1 2 GPFEGPNYHIAPRWVYNLTSLWMIFVVFASCFTNGLVLMATYKFKKLRHPLNWILVNLAIADLGETLIASTISVTNQIFGYFILGHPMCMLEGFTVATC 1 2 GITGLWSLTIIAWERWVVVCKPFGNIKFDGKWAAGGIIFSWVWSAFWCAMPLFGWSR 2 1 FWPHGLKTSCGPDVFSGEDKYGTRSFMIALMITCCIIPLGVIILCYIQVWWAIRT 0 0 VAKQQKESESTQKAEKEVSRMVVVMIFAYCFCWGPYTFMACFGAAYPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIYQLLGKKVDDGSELSSTSKTEVSSVSNSSVSPA* 0 >LWS_takRub Takifugu rubripes (pufferfish) Gt 0...2.1.0.0 indel x x x x 358 aa 000 nm no_ref genome cone long 0 MAEEWGKQSFAARRYHEDTTRGSAFVYTNSNHTR 1 2 DPFEGPNYHIAPRWVYNVATVWMFIVVVLSVFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYTVSTC 1 2 GIAALWSLTIISWERWVVVCKPFGNVKFDAKWATGGIVFSWVWAAVWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCIIPLAIIILCYLAVWLAIRS 0 0 VAMQQKESESTQKAEKEVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 FRVCIMKLFGKEVDDGSEVSTSKTEVSSVAPA* 0 >LWS_gasAcu Gasterosteus aculeatus (stickleback) Gt 0.2.2.1.0.0 indel - - - - 358 aa 000 nm no_ref genome cone long 0 MAEEWGKQAFAARRYNEDTTRGSMFVYTNSNNTK 1 2 DPFEGPNYHIAPRWVYNLSTLWMFIVVALSVFTNGLVLVATAKFKKLQHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSVC 1 2 GITALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWIWSAVWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCLIPLAIIILCYLAVWLAIRA 0 0 VAMQQKESESTQKAERDVSRMVVVMIVAYIVCWGPYTTFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ 0 0 FRSCIMQLFGKEVDDGSEVSTsKTEVSSVAPA* 0 >LWS_calMil Callorhinchus milii (elephantfish) Gt 0.2.2.1.0.0 indel x x x x 262 aa 000 nm no_ref genome fragment exon break 2 dPFEGPNYHIAPRWAYNLTSVWMVGVVVASVFTNGLVLVATVRFKKLRHPLNWILVNMALADLGETVLASTVSVANQFFGYFILGHPLCVFEGFVVSLC 1 2 GITALWSLTIIAWERWVVVCKPFGNVKFDGKWAAFGIIFSWVWSIGWCLPPVFGWSR 2 0 AEKEVSRMVVVMVAAFCLCWGPYACFAMFSALNPGYAFHPLVASIPSYFAKSSTIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSELSSTSKTDVSSVSNSSVSPA* 0 >LWS_petMar Petromyzon maritimus (lamprey) Gt 0.2.2.1.0.0 indel x x x x 366 aa 000 nm no_ref genome cone traces key to intron 3 position and gapping 0 MTASWQGAMFAARRRQDDEDTTMESLFRYTNENNTK 1 2 DPFEGPNYHIAPRWVFNLTSVWMIIVVVLSLFSNGLVLVATVKFKKLRHPLNWIIVNLAIADILETIFASTISVCNQVYGYFILGHPMCVFEGYVVSTC 1 2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIATILIVFSWVWPASWCSLPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFLPLSIIILCYLQVWLAIHS 0 0 VAQQQKESETTQKAERDVSRMVVVMILAYVFCWGPYTFFACFAAANPGYSFHPIAAALPAYFAKGATIYNPIIYVFMNRQ 0 0 FRNCILQLFGKKVDDGSEVSSSSRTEVSSVSNSSVSPA* 0 >LWS_letJap Lethenteron japonicum (lamprey) Gt 0.2.2.1.0.0 indel x x x x 365 aa 000 nm 15096614 AB116381 cone long 0 MTASWHGAVFAARRRNDDEDTTKDSIFRYTNENNTR 1 2 DPFEGPNYHIAPRWMFNLTSVWMIIVVVLSLFTNGLVLVATMKFKKLRHPLNWILVNLAIADILETIFASTISVCNQVFGYFILGHPMCVFEGYVVSTC 1 2 GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIAIILIVFSWVWPACWCSLPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFLPLSVIILCYLQVWLAIHS 0 0 VAQQQKESETTQKAERDVSRMVVVMILAYIFCWGPYTFFACYAAANPGYAFHPLTAALPAYFAKSATIYNPVIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSEVSSASRTEVSSVSNSSISPA* >LWS_geoAus Geotria australis (lamprey) Gt 0.2.2.1.0.0 indel x x x x 365 aa 560 nm 17463225 AY366491 cone long red retinal 0 MAQSWERAMFAARRRQDEDTTKGDLFRYTNENNTR 1 2 DPFEGPNYHIAPRWMYNLTSFWMIIVVILSLFTNGLVLVATLKFKKLRHPLNWILVNLAIADIGETIFASTVSVVNQIFGYFILGHPLCVFEGFTVSVC 1 2 GITALWSLAIISFERWMVVCKPFGNLKFDGKVAIVLIIFSWAWSAGWCAPPIFGWSR 2 1 YWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFIPLALIIICYLQVWLAIHT 0 0 VAQQQKESETTQKAERDVSRMVVVMIFAYIFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FRNCIMQLFGKKVDDGSEVSSSARTEVSSVSNSSVSPA* 0 >PIN_galGal Gallus gallus (chicken) Gt 0...2.2.0.0 indel x x x x 352 aa 000 nm no_ref genome pinopsin pineal non-visual 0 MSSNSSQAPPNGTPGPFDGPQWPYQAPQSTYVGVAVLMGTVVACASVVNGLVIVVSICYKKLRSPLNYILVNLAVADLLVTLCGSSVSLSNNINGFFVFGRRMCELEGFMVSLT 1 2 GIVGLWSLAILALERYVVVCRPLGDFQFQRRHAVSGCAFTWGWALLWSTPPLLGWSSYVPE 1 2 GLRTSCGPNWYTGGSNNNSYILSLFVTCFVLPLSLILFSYTNLLLTLRA 0 0 AAAQQKEADTTQRAEREVTRMVIVMVMAFLLCWLPYSTFALVVATHKGIIIQPVLASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FQSCLLEMLCCGYQPQRTGKASPGTPGPHADVTAAGLRNKVMPAHPV* 0 >PIN_utaSta Uta stansburiana (lizard) Gt 0...2.2.0.0 indel x x x x 359 aa 000 nm 16543463 DQ100321 pinopsin pinopsin missing Anole genome 0 MVNEWSNATPGPFDGPQWPYLAPRSIYTSVAVLMGLVVVSAAFVNGLVIVVSIQYKKLRSPLNYILVNLAIADLLVTSFGSTLSFANNIYGFFVLGQTACEFEGFMVSLT 1 2 GIVGLWSLAILAFERYLVICKPVGDFRFQQRHAVFGCVFTWMWSLVWTLPPLFGWSSYVPE 1 2 GLRTSCGPNWYTGGSGNNSYIMALFVTCFALPLGMIIFSYASLLLTLRA 0 0 VATQQKEVETTQQAEKEVTRRVIAMVMAFLVCWLPYASFAMVVATNKDLVIQPALASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FRSCLLSTMSCGHRPRGAQETTPAMISIPQGPTSALQGSRNKVTPSASEGSGNEAIPS* 0 >PIN_pheMad Phelsuma madagascariensis (gecko) Gt 0...2.2.0.0 indel x x x x 358 aa 000 nm no_ref AB022881 pinopsin 0 MHVQMANASQASLKNGTLSPFDGPQWPHRASRRVYTSLAALMGVVVLSASLANGLVIAVSVRFKRLRSPLNYILVNLATADLLVTFFGSIISFVNNAVGFFVFGKTACRFEGFMVSLT 1 2 GIVGLWSLAILAFERYLVICKPVGDFQFQRRHAVIGCLYTWGWSLIWTVPPLFGWSSYVPE 1 2 GLGTSCGPNWYMGGTNNNSYIVALFVTCFALPLSMILFSYANLLLTLRA 0 0 VAAQQKEQETTQRAEKEVTRMVITMVMAFLVCWLPYATFAMVVATTKDLSIQPGLASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FRSCLLNTVSCGRIPQTMPGTPATTAVRGGFVLTSEGRGNKVASTELHS* 0 >PIN_podSic Podarcis sicula (lizard) Gt 0...2.2.0.0 indel x x x x 354 aa 000 nm 16688437 DQ013042 pinopsin pinopsin mRNA 0 MQASNASWVEVRNRTPGPFEGPQWPYLAPQSTYISVAVLMGLVVISATLVNGLVIVVSVQFKKLRSPLNYVLVNLAVADLLVTFFGSTISFVNNAQGFFIFGQATCEFEGFMVSLT 1 2 GIVGLWSLAILAFERYLVICKPVGDFRFPARHAVLGCAFTWGWSFVWTVPPLLGWSSYVPE 1 2 GLRTSCGPNWYSGGSSNNSYIMTLFVTCFAMPLSTILFSYANLLMTLRT 0 0 VAAQQKEQETTQRAEREVTRMVVAMVAAFLVCWLPYASFAMVVATHKDLAIRPALASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FRSCLLYKMSCGHRALSSQDTTPAGISLPGRLTTSASKGSRNQVSPS* 0 >PIN_xenTro Xenopus tropicalis (frog) Gt 0...2.2.0.0 indel x x x x 346 aa 000 nm no_ref genome pinopsin 0 MRAGNMSAYEAPGPYDGPQWPHLAPRSTFLTVAAVMCMVVILAFFVNGLVIVVTLKYKKLRSPLNYILVNLAIANLLVTIFGSSVSFSNNVVGYFFMGKTMCEFEGFMVSLT 1 2 GIVGLWSLAILAFERYLVICKPMGDFRFQQKHAILGCSFTWVWSFIWTSPPLFGWCSYVPE 1 2 GLRTSCGPNWYTGGTNNNSYIMALFLTCFIMPLSTIIFSYSNLLMALRA 0 0 VAAQQKDSETTQRAEKEVTRMVIAMVLAFLICWLPYASFAVVVAVNKDVVIEPTVASLPSYFSKTATVYNPIIYVFMNKQ 0 0 FRNCLMTLLCCGRSFGDDETSSASGRTDVTSVSEAGGNKVTPA* 0 >PIN_bufJap Bufo japonicus (toad) Gt 0...2.2.0.0 indel x x x x 347 aa 000 nm 9537517 AF200433 pinopsin classifies oddly 0 MHSANMSALETPGPFEGPQWPHVAPRSTYLTVAVLMGMVVFLAFFVNGMVIVVSLKYKKLRSPLNYILVNLAVADILVTMFGSTVSFHNNIFGFFTLGKLVCELEGFVVSLT 1 2 GIVGLWSLAILAFERYIVICKPMGDFRFQQRHAVMGCAFTWIWAFLWTSPPLIGWCSYVPE 1 2 GLGTSCGPNWYTGGTNNNSYILALFTTCFMMPLTTIIFSYSNLLLALRA 0 0 VAAQQKESETTQRAEREVTRMVIAMVLAFLICWLPYAVFAIVMASNKNVVIDPTLASMPSYFSKTATVYNPVIYVFMNKQ 0 0 FRDCLTKLLCCGRNPFGEDETSTTSGRTDVTSVSEGGGNKVTPA* 0 >PIN_calMil Callorhinchus milii (elephantfish) Gt 0...2.2.0.0 indel x x x x 093 aa 000 nm no_ref genome fragment 0 FGSTVSFSNNINGYFVLGETVCQFEGFMVSLT 1 2 GIVGLWSLAILAFERYIVICKPMGDFRFQQKHAVWGCLFTWLWSLFWTLPPLFGWCSYVPE 1 >VAOP_galGal Gallus gallus (chicken) Gt 0...2.1.0.0 indel +INPP5A -NXK6 +C10orf61 +ALDH18A1 393 aa 000 nm no_ref genome TCTN3 exon 1 genbank error 0 MDVFRALGNESLLSNSSGPARWDPFHHPLDSIQPWHFRLVAAVMFVVTSLSLAENLAVILVTFKFKQLRQPVNYVIVNLSVADFLVSLTGGTISFLANLKGYFYMGHWACVLEGFAVTFF 1 2 GIVALWSLALLAFERYIVICRPVGNMRLRGKHAAQGIAFVWTFSFIWTIPPTMGWSSYTTSKIGTTCEPNW 2 1 YSGAYNDRSYIIAFFTTCFIVPLLVILVSYGKLLQKLRK 0 0 VSNTQGRLRTARKPERQVTRMVVVMIIAFLICWMPYAVFSILATAYPSIELDPHLAAIPAFFSKTATVYNPIIYVFMNKQ 0 0 FRMCLIQMFKCSAIETAESNMNPTSERATLTQDKRDSQLSVMAVRSTILKRKTGDEHRADDLWLFRQLQKPKCVPCRAGDGS* 0 >VAOP_anoCar Anolis carolinensis (lizard) Gt 0...2.1.0.0 indel +INPP5A -NXK6 +GPR125 +KNDC1 389 aa 000 nm no_ref genome vertebrate ancient 0 MAGLRREAENDSWLFDPSSSSAPFDPFLQPLDIIEPWNFHLISALMFVVTLFSLSENFTVILVTIKFKQLRQPLNYVIVNLSVADFLVSLIGGTISFSTNLKGYFYMGHWACVLEGFAVTFF 1 2 GIVALWSLALLAFERYVVICRPLGNMRLNGKHAALGVAFVWIFSFIWTVPPTMGWSSYTTSKIGTTCEPNW 2 1 YSGDYNDHTFIITFFTTCFILPLLVILVSYGKLMRKLRK 0 0 VSDTQGRLGTTRKPERQVTGMVVIMILAFLICWSPYAAFSILVTACPSIELDPRLAAIPAFFSKTATVYNPVIYVFMNNQ 0 0 FRKCLVQLFQCSSQETMDANVNPISEKDTLTHTKHCGEMSTVAAHVIVFNPRSEDEQGSCQSFAQLAISENKVYPL* 0 >VAOP_xenTro Xenopus tropicalis (frog) Gt 0...2.1.0.0 indel - +GSTO2 -C10orf92 - 383 aa 000 nm no_ref genome vertebrate ancient new 0 MPTNVSLLATPENSTVWNPFTGPLKTIEAWNFHLLAALMFVVTSLSIAENFIVILVTAKFKQLRQPLNYIIVNLSVADFLVSVIGGTISIATNSRGYFYLGSWACVLEGFAVTFF 1 2 GIVALWSLSVLAFERYIVICRPLGNLRLQGKHSALAIIFVWVFSFVWTIPPTMGWSSYTTSKIGTTCEPNW 2 1 YSGEMRDHTYIITFLTTCFVFPLLVIFMSYGKLMRKLRK 0 0 VSDTQGRLGSTRKPEKEVTRMVVIMILAFLICWTPYAAFSILITAHPTIDLDPRLAAIPAFFAKTASMYNPIIYVYMNKQ 0 0 FRRCLYQMFNINDPEAKESNLNPTSERGVLTRNNNGGEMLAIATHITSSAVTNREEEKSSSNSFAHIPVSDNKVCPM* >VAOP_danRer Danio rerio (zebrafish) Gt 0...2.1.0.0 indel - - - - 378 aa 000 nm 17067577 NM_131586 vertebrate ancient valop vertebrate assembly missing exon 3 0 MEASSAAVNAVSPAEDPFSAPLSSIAPWNYSVLAALMFVVTALSLSENFTVMLVTFRFQQLRQPLNYIIVNLSLADFLVSLTGGSISFLTNYHGYFFLGKWACVLEGFAVTFF 1 2 GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLVFVWSFSFIWTVPPVLGWSSYTVSRIGTTCEPNW 2 1 YSGNFHDHTFIITLFSTCFIFPLGVIIVCYCKLIRKLRK 0 0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIIITAHPSMHVDPRLAAIPAFVAKTAAVYNPIIYVFMNKQ 0 0 FRKCLVQLLSCSKVTVVEGNNNQTTERAGMTSGSNTGEMSAIAARVSVPKTEENPGDRSTFSHIPIPENKVCPM* >VAOP_takRub Takifugu rubripes (teleost) Gt 0...2.1.0.0 indel +INPP5A -NXK6 - +KNDC1 362 aa 000 nm no_ref genome vertebrate ancient 0 MESLSLSVNGVSYTVAAELAPTNDPFTGPINNIAQWNFTILAVLMFVVTSLSLCENFLVMFITFKFKQLRQPLNYIIVNLAIADFLVSLTGGLISFLTNARGYFFLGRWACVLEGFAVTYF 1 2 GIVAMWSLAVLSFERFFVICRPLGNMRLQAKHAAIGLLFVWTFSFVWTFPPVLGWNRYTVSKIGTTCEPDW 2 1 YSNNMTSHSYIITFFSTCFILPLGIIFFCYGKLLRKLRK 0 0 VSHGRLATARKPERQVTRMVVVMIVAFMVAWTPYATFAILVTIHPTIELDPR 0 FRKCLIQHFIGMGVMAESNMNPTSERPGITAESQTGEMSAIAARVPVGATAALHSDGSPTDCGSLAQLPIPENKVCPI* 0 >VAOP_rutRut Rutilus rutilus (minnow) Gt 0...2.1.0.0 indel x x x x 383 aa 000 nm 12906786 AY116411 vertebrate ancient vertebrate 0 MELFPVAVNGVSHAEDPFSGPLTFIAPWNYKVLATLMFVVTAASLSENFAVMLVTFRFTQLRKPLNYIIVNLSLADFLVSLTGGTISFLTNYHGYFFLGKWACVLEGFAVTYF 1 2 GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLLFVWTFSFIWTIPPVLGWSSYTVSKIGTTCEPNW 2 1 YSGNFHDHTFIIAFFITCFILPLGVIVVCYCKLIKKLRK 0 0 VSNTHGRLGNARKPERQVTRMVVVMIVAFMVAWTPYAAFSIVVTAHPSIHLDPRLAAAPAFFSKTAAVYNPVIYVFMNKQ 0 0 FRKCLVQLLRCRDVTIIEGNINQTSERQGMTNESHTGEMSTIASRIPKDGSIPEKTQEHPGERRSLAHIPIPENKVCPM* 0 >VAOP_calMil Callorhinchus milii (elephantfish) Gt 0...2.1.0.0 indel x x x x 080 aa 000 nm no_ref genome fragment 0 VASTQGRLGVARKPEKQVTRMVIVMILAFLFCWTPYAAFSITVTACPTIKLDPRLAAIPAFFSKTATVYNPIIYVFMNKQ 0 >VAOP_petMar Petromyzon marinus (lamprey) Gt 0...2.1.0.0 indel x x x x 445 aa 000 nm 9427550 U90667 vertebrate ancient exons 123 in traces pineal gland-specific 0 MDALQESPPSHHSLPSALPSATGGNGTVATMHNPFERPLEGIAPWNFTMLAALMGTITALSLGENFAVIVVTARFRQLRQPLNYVLVNLAAADLLVSAIGGSVSFFTNIKGYFFLGVHACVLEGFAVTYF 1 2 GVVALWSLALLAFERYFVICRPLGNFRLQSKHAVLGLAVVWVFSLACTLPPVLGWSSYRPSMIGTTCEPNW 2 1 YSGELHDHTFILMFFSTCFIFPLAVIFFSYGKLIQKLKK 0 0 ASETQRGLESTRRAEQQVTRMVVVMILAFLVCWMPYATFSIVVTACPTIHLDPLLAAVPAFFSKTATVYNPVIYIFMNKQ 0 0 FRDCFVQVLPCKGLKKVSATQTAGAQDTEHTASVNTQSPGNRHNIALAAGSLRFTGAVAPSPATGVVEPTMSAAGSMGAPPNKSTAPCQQQGQQQQQQGTPIPAITHVQPLLTHSESVSKICPV* 0 >PPIN_anoCar Anolis carolinensis (lizard) Gt 0...2...0.0 indel -CPEB2 -CACNA2D3 +SELK +ACTR8 346 aa 000 nm no_ref genome parapinopsin syntenic deleted in chicken 0 MDSLDTNTLSPNASTVRVVLMPRIGYTIIAIIMATSCTLSVILNTAVIAITIKYRQLRQPINYSLVNLAIADLGAALLGGSLNVETNAVGYYNLGRVGCVTEGFAMAFF 1 2 GIVALCTIAVIAVDRAIVIAKPMGTITFTTRKAMIGVAVSWIWSLVWNTPPLFGWGGYQMEGVMTSCAPDWANSDPINVSYIICYFLFCFTIPFITILASYGYLIWTLRQ 0 0 VAKVGLAQRGSTTKAEAQVSRMVIVMVMAFLICWLPYATFALVVVGNPQIYINPIIATIPMYMAKSSTFYNPIIYIFMNKQ 0 0 FRDCLVRCLLCGRNPCASEQTDEDDLEVSTIAPAPSSRRGKVAPV* 0 >PPIN_xenTro Xenopus tropicalis (frog) Gt 0...2...0.0 indel - - +SELK - 349 aa 000 nm no_ref genome parapinopsin bistable UV lamprey pineal broken contigs 0 MADEALLPPMMNVTNEEMHPGKVLMPRIGYTILALIMAVFCAAALFLNVTVIVVTFKYRQLRHPINYSLVNLAIADLGVTVLGGALTVETNAVGYFNLGRVGCVIEGFAVAFF 1 2 GIAALCTIAVIALDRVFVVCKPMGTLTFTPKQALAGIAASWIWSLIWNTPPLFGWGSYELEGVMTSCAPNWYSADPVNMSYIVCYFSFCFAIPFLIIVGSYGYLMWTLRQ 0 0 VAKLGVAEGGTTSKAEVQVSRMVIVMILAFLVCWLPYAAFAMTVVANPGMHIDPIIATVPMYLTKTSTVYNPIIYIFMNKQ 0 0 FQECVIPFLFCGRNPWAAEKSSSMETSISVTSGTPTKRGQVAPA* 0 >PPIN_ictPun Ictalurus punctatus (catfish) Gt 0...2...0.0 indel x x x x 347 aa 000 nm no_ref genome parapinopsin parapinopsin index sequence 0 MASIILINFSETDTLHLGSVNDHIMPRIGYTILSIIMALSSTFGIILNMVVIIVTVRYKQLRQPLNYALVNLAVADLGCPVFGGLLTAVTNAMGYFSLGRVGCVLEGFAVAFF 1 2 GIAGLCSVAVIAVDRYMVVCRPLGAVMFQTKHALAGVVFSWVWSFIWNTPPLFGWGSYQLEGVMTSCAPNWYRRDPVNVSYILCYFMLCFALPFATIIFSYMHLLHTLWQ 0 0 VAKLQVADSGSTAKVEVQVARMVVIMVMAFLLTWLPYAAFALTVIIDSNIYINPVIGTIPAYLAKSSTVFNPIIYIFMNRQ 0 0 FRDYALPCLLCGKNPWAAKEGRDSDTNTLTTTVSKNTSVSPL* 0 >PPIN_danRer Danio rerio (zebrafish) Gt 0...2...0.0 indel - - +SELK - 338 aa 000 nm no_ref XM_681591 parapinopsin parapinopsin 0 MESETSTAASGSIAEVMPRMGYTILAVIIGVFSVCGVILNVTVITVTLKYKQLRQPLNFALVNLAVADLGCAVFGGLPTVVTNAMGYFSLGRVGCVLEGFAVAFF 1 2 GIAALCSVAVIALERCMVVCRPVGSISFQTRHAVFGVAVSWLWSFIWNTPPLFGWGRLQLEGVRTSCAPDWYSRDLANVSFIVCYFLLCFALPFSVIVYSYTRLLWTLRQ 0 0 VSRLQVCEGGSAARAEAQVSCMVVVMILAFLLTWLPYASFALCVILIPELYIDPVIATVPMYLTKSSTVFNPIIYIFMNRQ 0 0 FRDRALPFLLCGRNPWAAEAEEEEEETTVSSVSRSTSVSPA* 0 >PPIN_oncMyk Oncorhynchus mykiss (trout) Gt 0...2...0.0 indel x x x x 347 aa 000 nm no_ref genome parapinopsin 0 MDHQQLLPNLHGNISSSPGSVSEALLSRTGFTILAVIIGVFSVSGVCMNVLVIMVTMRHRKLRQPLNYALVNLAVADLGCALFGGLPTMVTNAMGYFSMGRLGCVLEGFAVAFF 1 2 GIAGLCSVAVIAVDRYVVVCRPMGAVMFQTRHAVGGVVLSWVWSFLWNTPPLFGWGSFELEGVRTSCSPNWYSREPGNMSYIILYFLLCFAIPFSIIMVSYARILFTLHQ 0 0 VSKLKVLEGNSTTRVEIQVVRMVVVMVMAFLLSWLPYAAFALSVILDPSLHINPLIATVPMYLAKSSTVYNPIIYVFMNRQ 0 0 FRDCAVPFLLCGLNPWASEPVGSEADTALSSVSKNPRVSPQ* >PPIN_calMil Callorhinchus milii (elephantfish) Gt 0...2...0.0 indel x x x x 109 aa 000 nm no_ref genome fragment 0 MDPHNRSANLSEGPGLGGGGAVPGWGPSVRAPLSLVMAVISLSSIVLNSLAIAVVLRFQVLQQPLNYALLSLASADLGTAATGGVLSTVCTALGSFVLGRHSCVAEGFF 1 >PPIN_petMar Petromyzon maritimus (lamprey) Gt 0...2...0.0 indel x x x x 344 aa 000 nm no_ref genome parapinopsin bistable pineal UV/green 0 MENLTSLDLLPNGEVPLMPRYGFTILAVIMAVFTLASLVLNSTVIIVTLRHRQLRHPLNFSLVNLAVADLGVTVFGASLVVETNAVGYFNLGRVGCVIEGFAVAFF 1 2 GIAALCTIAVIAVDRFVVVCKPLGTLMFTRRHALLGITWAWLWSFVWNTPPLFGWGSYKLEGVRTSCAPDWYSRDPANVSYIVSFFSFCFAIPFLVIVVAYGRLLWTLHQ 0 0 VAKLGMGESGSTAKAEAQVSRMVVVMVVAFLVCWLPYALFAMIVVAKPGVYIDPVIATLPMYLTKTSTVYNPIIYIFMNRQ 0 0 FRDCAVPFLLCGRNPWAEPSSESATTASTSATSVTLASVPGQVSPS* 0 >PPIN_letJap Lethenteron japonicum lamprey Gt 0...2...0.0 indel x x x x 344 aa 000 nm 14981504 AB116380 parapinopsin bistable pineal UV/green 0 MENLTSLDLLPNGEVPLMPRYGFTILAVIMAVFTIASLVLNSTVVIVTLRHRQLRHPLNFSLVNLAVADLGVTVFGASLVVETNAVGYFNLGRVGCVIEGFAVAFF 1 2 GIAALCTIAVIAVDRFVVVCKPLGTLMFTRRHALLGIAWAWLWSFVWNTPPLFGWGSYELEGVRTSCAPDWYSRDPANVSYITSYFAFCFAIPFLVIVVAYGRLMWTLHQ 0 0 VAKLGMGESGSTAKAEAQVSRMVVVMVVAFLVCWLPYALFAMIVVTKPDVYIDPVIATLPMYLTKTSTVYNPIIYIFMNRQ 0 0 FRDCAVPFLLCGRNPWAEPSSESATAASTSATSVTLASAPGQVSPS* 0 >PPINa_cioInt Ciona intestinalis (tunicate) Gt 0...2...0.0 indel -HOXB1 +HHEX +CUL4A - 391 aa 000 nm 11591373 NM_001032555 parapinopsin Ci-opsin odd exons larval ocellus 0 MNQWLETIMNERKFEIVNSSLQG 2 1 LLKPDSLGMDHDVTPTVDLTDGVPQCKDLNPYVLKGDGWVPQHISRANRSTYSFLCVYMTFVFLLSCSLNILVIVATLKNK 0 0 VLRQPLNYIIVNLAVVDLLSGFVGGFISIAANGAGYFFWGKTMCQIEGYFVSNFGVTGLL 0 0 SIAVMAFERYFVICKPFGPVRFEEKHSIFGIV 0 0 ITWVWSMFWNTPPLIFWDGYDTEGLGTSCAPNWFVKEKRERLFIILYFVFCFVIPLAVIMICYGKLILTLRQ 0 0 IAKESSLSGGTSPEGEVTKMVVVMVTAFVFCWLPYAAFAMYNVVNPEAQ 0 0 IDYALGAAPAFFAKTATIYNPLIYIGLNRQ 0 0 FRDCVVRMIFNGRNPWVDELVGSQVSSTGSQLTAVSSNKVAPA* 0 >PPINb_cioInt Ciona intestinalis (tunicate) Gt 0...2...0.0 indel -TMEM165 +FUT4 - - 353 aa 000 nm no_ref genome parapinopsin jgi gene model wrong both ends 0 MTTAETTTECYEKNPYIRNEMGWVPKHILIAERHIYTILAVYMTFIFLLAVSLNGFVIIATMKNK 0 0 KLRQPLNYIIINLSIADFLSGLVGGFIGMISNSAGYFYFGKTVCILEGYIVSVA 1 2 GVCGLMSISVMAFERYFVVCKPYGPFTLTNTHAAL 1 2 GIGFTWTWSVLWSTPGLIWLDGYVPEGLGTSCAPNWFSKNK 2 1 SERIFIFVYFVFCFFIPLLVIIICYGKIVLFLKQVSLY 0 0 ATRQSSASSNRQADNKVTKMVLVMISAFLICWTPYGVLSLYNAINPDKQ 0 0 LDYGLGAVPVFFAKTANIYNPLIYIGLNKQ 0 0 FRDGVIKMVFRGRNPWAEEMSTQQRQRSTEAGQPIVSNEV* 0 >PARIE_utaSta Uta stansburiana (lizard) Gd+Go 0...2...0.0 indel x x x x 347 aa 522 nm 16543463 DQ100320 parietopsin shift in counterion Gt + Go 0 MENDSSLATELAEGAIVKPTIFPKAGYGVLAFLMFLNALFSIFNNSLVIAVTLKNPQLRNPINIFILNLSFSDLMMSLCGTTIVIATNYYGYFYLGRKFCIFQGFAVNYF 1 2 GIVSLWSLTILAYERYNVVCQPLGTLQMSTKRGYQLLGFIWVFCLFWAVVPLFGWSSYGPEGVQTSCSIGWEERSWSNYSYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG 0 0 LNKKVEQLGGKSSPEEEFRAVIMVLVMVVAFLICWLPYTVFALIVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKQ 0 0 FRDCAVEFITCGQVVLTSPEEDISTSAIPVEGKGPCKINQVTPV* 0 >PARIE_anoCar Anolis carolinensis (lizard) Gd+Go 0...2...0.0 indel +EEA1 -FLJ46688 +BTG1 - 347 aa 000 nm no_ref genome parietopsin Go like scallop, gusducin not transducin 0 MENESSLVLEGAEGYIVRPTIFPRAGYGVLAFLMFINALFSLFNNFLVIAVTLKNPQLRNPINIFILNLSFSDLMMSICGTTIVIATNYHGYFYLGRRFCIFQGFAVNYF 1 2 GIVSLWSLTILAYERYNVVCQPLGTLQMSTQRAYQLLGFIWVFCLFWAVVPLFGWSSYGPEGVQTSCSIGWEERSWNNYSYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG 0 0 LNKKVEQLGGKSNPEEEFRAVIMVLVMVVAFLICWLPYTLFALTVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKE 0 0 FRECAVEFITCGKVVLTSPEEDISTSAISDEGIAPCKINQVTPV* 0 >PARIE_xenTro Xenopus tropicalis (frog) Gd+Go 0...2...0.0 indel -lum -DCN - - 346 aa 000 nm 16543463 NM_001045791 parietopsin 0 MDGNSTTPGIAVNLTVMPTIFPRSGYSILSFLMFLNAVFSICNNAIVILVTLKHPQLRNPINIFILNLSFSDLMMALCGTTIVVSTNYHGYFYLGKQFCIFQGFAVNYF 1 2 GIVSLWSLTLLAYERYNVVCEPIGALKLSTKRGYQGLVFIWLFCLFWAIAPLFGWSSYGPEGVQTSCSIGWEERSWSNYSYIISYFLTCFIIPVGIIGFSYGSILRSLHQ 0 0 LNRKIEQQGGKTNPREEKRVVIMVLFMVLAFLICWLPYTVFALIVVINPQLYISPLAATLPTYFAKTSPVYNPIIYIFLNKQ 0 0 FRTYAVQCLTCGHINLDSLEEDTESVSAQAENMLTPKTNQVAPA* 0 >PARIE_takRub Takifugu rubripes (teleost) Gd+Go 0...2...0.0 indel -HSP90B1 +NT5DC2 -KCND3 -FLNC 351 aa 000 nm 16543463 genome parietopsin 0 MDSNSTPWSSPPAPLQAEAVTVAPTIFPRVGYSILSFLMFINTVLSVFNNSLAIAVMLKNPSLLQPINIFILSLAVSDLMIGLCGSLVVTITNYHGSFFIGHTACVFQGFAVNYF 1 2 GLVSLCTLTLLAYERYNVVCKPRAGLKLTMRRSIIGLLFVWTFCLFWAVTPLLGWSSYGPEGVQTSCSLAWEERSWNNYSYLILYTLLCFIFPVGVIIYCYCKVLTSMNK 0 0 LNKSVELQGGLSCRRENKHAINMVLAMIIAFFVCWLPYTALSVVVVVDPELHIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0 0 FRDATLEVLSCSRYIPHASSRVSINMRSLNRRSVNTHSKVSPL* 0 >PARIE_gasAcu Gasterosteus aculeatus (stickleback) Gd+Go 0...2...0.0 indel -HSP90B1 +NT5DC2 -KCND3 -FLNC 361 aa 000 nm no_ref genome parietopsin 0 MDSNSTLWSSGSPPPSIHGKMLTITPTIFPRVGYSILSFLMFINTVLTVFNNVLVITVLVRNPSLLQPMNVFILSLAVSDLMIGLCGSLVVTITNYHGSFFIGHTACIFQGFAVNYF 1 2 GLVSLCTLTLLSYERYNVVCRPRNALKLSMRRSIHGLLIVWTFCLFWAVAPLFGWSGYGPEGVQTSCSLAWEERSWSNYSYLVLYTLLCFIVPVAVIIYCYAKVLTSMNT 0 0 LNRSVEVQGGRSSQKENDHAVSMVLAMIIAFFSCWLPYTALSVVVVVDPTLYIPPLVATMPMYFAKTSPVYNPIIYFLSNKQ 0 0 FRDAALEMLSCGRYIAHMPNTVSINMRSLNRRSRLSSLSRNVNSHSKVLPL* 0 >PARIE_danRer Danio rerio (zebrafish) Gd+Go 0...2...0.0 indel - +NT5DC2 +FBXL13 - 337 aa 000 nm 16543463 genome parietopsin 0 MENFAKTELTMMVQPTIFPRVGYSILSYLMFINTTLSVFNNVLVIAVMVKNLHFLNAMTVIIFSLAVSDLLIATCGSAIVTVTNYEGSFFLGDAFCVFQGFAVNYF 1 2 GLVSLCTLTLLAYERYNVVCKPMAGFKLNVGRSCQGLLLVWLYCLFWAVAPLLGWSSYGPEGVQTSCSLGWEERSWRNYSYLILYTLMCFILPTVIITYCYSNVLLTMRK 0 0 INKSIECQGGKNCAEDNEHAVRMVLAMIIAFFICWLPYTAISVLVVVNPEISIPPLIATMPMYFAKTSPVYNPIIYFLTNKR 0 0 FRESSLEVLSCGRYISRETGGPLMGSSMQRGQSRVNPV* 0 >PARIE_petMar Petromyzon marinus (lamprey) Gd+Go 0...2...0.0 indel x x x x 082 aa 000 nm no_ref genome fragment 0 LNKKIKRVGGHPDPREEMRATVMVLAMVGAFLACWLPYTVLALCVVLAPGTQIPPLVATLPMYFAKTSPMYNPIIYFFLNPQ 0 >ENCEPH_homSap Homo sapiens (human) Gt 0...2...0.0 indel -EXO1 -WDR64 -KMO +FH 403 aa 000 nm 12242008 NM_014322 parietopsin OPN3 with intron loss 0 MYSGNRSGGHGYWDGGGAAGAEGPAPAGTLSPAPLFSPGTYERLALLLGSIGLLGVGNNLLVLVLYYKFQRLRTPTHLLLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSGSLF 1 2 GIVSIATLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPLGVIAHCYGHILYSIRM 0 0 LRCVEDLQTIQVIKILKYEKKLAKMCFLMIFTFLVCWMPYIVICFLVVNGHGHLVTPTISIVSYLFAKSNTVYNPVIYVFMIRK 0 0 FRRSLLQLLCLRLLRCQRPAKDLPAAGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDKTNGSKVDVIQVRPL* 0 >ENCEPH_monDom Monodelphis domestica (opossum) Gt 0...2...0.0 indel -EXO1 -WDR64 -KMO +FH 411 aa 000 nm no_ref genome encephalopsin OPN3 extra intron alt splicing 0 MYSDNSSDDGGGGYWGSGRAGGASGTGVTGEPGPEGSPRQAPLFSPGTYELLALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLFLVNISFNDLLVSLFGVTFTFVSCLRSGWVWDSVGCAWDGFSNTLF 1 2 GIVSIMTLTVLAYERYNRIVHAKVINFSWAWRAITYIWLYSLVWTGAPLLGWNRYTLEIHGLGCSVDWKSKDPNDSSFVIFLFFGCLMLPVGVMAYCYGHILYAIRM 0 LRCVEELQTIQVIKILRYEKKVAKMCFLMIAIFLFCWMPYAVICLLVANGYGSLVTPTVAIIASLFAKSSTAYNPIIYIFMSRK 0 0 FRRCLLQLLCFRLLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDETQMIDENDKNSGTKVNVIQVRPL* 0 >ENCEPH_galGal Gallus gallus (chicken) Gt 0...2...0.0 indel -EXO1 -WDR64 -PIGM +RGS7 396 aa 000 nm no_ref genome encephalopsin OPN3 0 MHSGNGTGATSRPQLAAAGHEVPGERPLFSAGTYELLALLIATIGTLGVCNNLLVLVLYYKFKRLRTPTNLFLVNISLSDLLVSVCGVSLTFMSCLRSRWVWDAAGCVWDGFSNSLF 1 2 GIVSIMTLTVLAYERYIRVVHAKVIDFSWSWRAITYIWLYSLAWTGAPLLGWNRYTLEIHGLGCSMDWKSKDPNDTSFVLLFFLGCLVAPVVIMAYCYGHILYAVRM 0 0 LRCVEDFQTSQVIKLLKYEKKVAKMCFLMISTFLICWMPYAVVSLLVTYGYSNLVTPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0 0 FRQCLLQLLCFRLMRFQRIMKEPSGAGNVKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIIASDDTQQIDDNSKHNGTKVNVIQVKPL* 0 >ENCEPH_anoCar Anolis carolinensis (lizard) Gt 0...2...0.0 indel -EXO1 -WDR64 -PIGM +RGS7 408 aa 000 nm no_ref genome encephalopsin OPN3 0 MFSANGTRSGAGSDLEPGPGQQQQQREASEEEERGAGLSPFSAGTYELLALLVAAIGLLGLCNNLLVLVLYAKFKRLRTPTHLFLVNISLSDLLVSLFGVSFTFGSCLRHRWVWDAAGCVWDGFSNSLF 1 2 GIVSIMTLTVLAYERYIRVVHARVIDFSWSWRAITYIWLYSLAWTGAPLLGWNHYTLEIHGLGCSVDWQSKEPSDSSFVLFFFLGCLAAPVGIMAYCYGHILHAIRM 0 0 LRCVEDLQSIQVIKILRYEKKVAKMCFLMVTTFLICWMPYAVVSLLIAYGYGHLITPTVAIIPSFFAKSSTAYNPVIYIFMSRK 0 0 FRRCLVQLFCVQFLRFKRTLKEQPAIESNKPIRPIVMSQKVGDRPKKKVTFSSSSIIFIITSDDTEQIDVSTKCSDTKINVIQVKPL* 0 >ENCEPH_takRub Takifugu rubripes (teleost) Gt 0...2...0.0 indel -ABLIM1 +PTK7 -KMO +IDE 388 aa 000 nm no_ref genome encephalopsin TMT multiple tissue circadian clock 0 MPVTNGSHNNSISWLHSKDMFTEDTYHFLALIVATVGFLGLVNNLLVLILYCKFKRLQTPTNLLFFNTSLCHFVFSLLAITFTFMSCVRGSWAFSVEMCVFHGFSKNLL 1 2 GIVSFGTLTVVAYERYARVVYGKYVNSSWSKRSITFVWVYSLAWTGFPLIGWNLYTFETHKLDCSFEWTATDPKDTAFVLLFFLACITLPLSIMAYCYGYILYEIQK 0 0 LRSVKNIQNFQEITILDYEIKMAKMCLLMMLTFLIGWMPYTILSLLVTSGYSKFITPTITVMPSLLAIASAAYNPVIHIFTIKK 0 0 FRQCLVQLLPPINFHPPINPPINNFWRLLKNLNGRLAMKKVKPVLGKGRSHNRPEKKVPPINFSSSDFFTRTTSDTGTHGITESTKGKRTNVRLIQVHPL* 0 >ENCEPH4a_takRub Takifugu rubripes (teleost) Gt 0...2...0.0 indel -CALD1 +TNK2 -RAB18 +ABI1 403 aa 000 nm 12670711 AF402774 encephalopsin TMT multiple tissue circadian clock 0 MIVSNVSLSGCAGVNGAVCAAEGHQAGGSDRSTLTPTGNLVVSVFLGFIGTFGLVNNLLVLVLFCRYKMLRSPINLLLMNISISDLLVCVLGTPFSFAASTQGRWLIGEAGCVWYGFANSLF 1 2 GVVSLISLAVLSFERYSTMMTPTEADPSNYCKVCLGITLSWVYSLVWTVPPLFGWSSYGPEGPGTTCSVNWTAKTTNSISYIICLFVFCLIVPFLVIVFCYGKLLCAIRQ 0 0 VSGINASTSRKREQRVLCMVVIMVICYLLCWLPYGVVALLATFGPPDLVTPEASIIPSVLAKSSTVINPIIYVFMNKQ 0 0 FYRCFLALLCCQDPRSGSSMKSSSKVATKAKGVTPTGQRRTDFLYMVASLGRPAATIPQLGPSFDATNDFTKPPSSDTIKPVVVSLAAHCDG* >ENCEPH4b_takRub Takifugu rubripes (teleost) Gt 0...2...0.0 indel +TFRC +CHES1 -MYEOV2 -ARHGAP21 407 aa 000 nm no_ref genome encephalopsin 0 MIVCNVSLSCAHCPGEGTAANDAYAQASGSLATPTLSQRGHLVVAVCLGFIGTVGFLSNFLVLALFCRYRALRTPMNLMLVSISASDLLVSVLGTPFSFAASTQGRWLIGRAGCVWYGFVNACL 1 2 GIVSLISLAVLSYERYCTMVSSTIASNRDYRPVLGGICFSWFYSLAWTVPPLLGWSRYGPEGPGTTCSVDWRTQTPNNISYIVCLFTFCLLLPFFVILYSYGKLLHTIRQ 0 0 VRRVSSTVTRRREHRVLVMVVAMVVCYLICWLPYGVTALLATFGPPNLLTPEATITPSLLAKFSTVINPFIYIFMNKQ 0 0 FYRCFRAFLNCSTPKRDSTVRTFTRISLRALRQDQQQKGSALAPSSARPTPNSIHESSLKGSHSTPSNGGAAAAKSPAANRSKPKLILVAHYRE* 0 >ENCEPH_gasAcu Gasterosteus aculeatus (stickleback) Gt 0...2...0.0 indel -LDOC1L +CDC42EP3 -KMO +IDE 389 aa 000 nm no_ref genome encephalopsin OPN3 0 MNPDNGTREERSTDHSIFAVGTYKLLAFAIGTIGVFGFCNNVVVIVLYCKFKRLRTPTNLLVVNISLSDLLVSVIGINFTFVSCIRGGWTWSRATCIWDGFSNSLF 1 2 GIVSIMTLASLAYERYIRVVHAQVVDFPWAWRAIGHIWLYSLVWTGAPLLGWNRYTLEIHRLGCSLDWASKDPNDASFILLFLLACFFVPVGIMIYCYGNILYAVQM 0 0 LRSIQDLQTVQIIKILRYEKKVAVMFLLMISCFLLCWTPYAVVSMMEAFGRKNMVSPTVAIIPSFFAKSSTAYNPLICVFMSRK 0 0 FRRCLMQLLCSRVTCLQCNLKERPLAPVQRPIRPIVVSAACGGGRVRPKKRVTFSSSSIVFIITRNDIRHTDVTSNTRESSEANVFQVRPL* 0 >ENCEPH_calMil Callorhinchus milii (elephantfish) Gt 0...2...0.0 indel x x x x 097 aa 000 nm no_ref genome fragment 0 MNPTNSTEPQEEHLFSPNTYKLLAVIIGTIGIVGFCNNILVLLLYYKFKRLRTPTNLLLVNISVSDLLVSVFGLSFTFVSCTQGRWGWDSAACVWDG >ENCEPH4_calMil Callorhinchus milii (elephantfish) Gt 0...2...0.0 indel x x x x 177 aa 000 nm no_ref genome fragment 0 MLNSSPNSSPSLPLSQVGWTGLSRTGLTVVAVCLGIIMVLGFLNNLLVLVLFCKYKVLRSPMNMLLLNISVSDMLVCICGTPFSFAASVQGRWLVGEQGCKWYGFANSLF 1 0 REHRILLMVISMVTFYLLCWLPYGTVALIGTFGNADLITPTCSVIPSILAKSSTVINPVIYVIMNKQ 0 >ENCEPH5_calMil Callorhinchus milii (elephantfish) Gt 0...2...0.0 indel x x x x 070 aa 000 nm no_ref genome fragment AQTREHRILLMVISMVTFYLLCWLPYGTVALIGTFGNADLITPTCSVIPSILAKSSTVINPVIYVIMNKQ 0 >ENCEPH_squAca Squalus acanthias (dogfish) Gt 0...2...0.0 indel x x x x 202 aa 000 nm no_ref genome fragment 0 MNAANSTDTREESLFSPGTYQVLAVIIGTIGVVGFCNNLLMLVLYCKFKRLRTPTNLFLVNISISDLLLSVFGVIFTFVSCVKGRWVWDSAACVWDGFSNCLF 1 2 GISSIMSLTVLAYERYIRVVNATAIDFSWAWRAITYIWLYSLAWTGAPLIGWNSYTLELHRLGCSVNWDSRNPSDTSFVLFLFLGCLLCPIGVIAYCYG >ENCEPH_petMar Petromyzon marinus (lamprey) Gt 0...2...0.0 indel x x x x 293 aa 000 nm no_ref genome fragment 0 MQSPKQDSLHYAGDTGAKAAPDSAQGNASALGSNFLLHGGDLGEGSTAFSAATFRLLAGVVGTIGVAGFLNNLLLVALFVGFKRLQTPTNLLLVNISLSDLLVSVFGNTLTLVSCVRRRWVWGNGGCVWDGFSNSLF 1 2 GIVSISTLTALSYERYARLIKAQVLDFSWAWRAVTYTWLYSAAWTGAPLLGWSRYVLEKHGLGCSIDWASSNPPDAAFVLFFFLGCLAAPLLVMGFCFGRIALAITQ 0 0 CWSPYAVASLFVASGFEHLVSPPVSIVPSLLAKSNAVCNPLLFLLMSGN 0 >ENCEPH4_braFlo Branchiostoma floridae (amphioxus) Gt 0...2...0.0 indel -ZFYVE1 +RTF1 -CES1 -POMT2 402 aa 000 nm 12435605 AB050608 encephalopsin Amphiop4 new exon 12 and 34 + perfect fit 0 MALYNNTSSPSQDLLWDAPYSQGHIWDNSSASNSSEDVMDQGKVELQDFSDAGYTAIATCLALI 1 2 GFVGFTNNFVVILLIGCHRQLRTPFNLLLLNMSVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANSLF 1 2 GIVSLVTLSALAFERYCVVVRSSDMLTYKSSLVVITFIWLYSLLWTSLPLLGWSSYQFEGHN 0 0 VGCSVNWVQHNPDNVSYIVTLMVTCFFVPMVVVCWSYAWIWRTVRM 0 0 SSEAKPECGNSQNAGRLVTTMVVVMIICFLVCWTPYAVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ 0 0 FREFLLARLQRVCCRQQAVPRVTPMDDNVHVRLGGEGPSQSQQFLPAGENVENVDMLEYVQENCKPKADSLSTISE* 0 >ENCEPH4_braBel Branchiostoma belcheri (amphioxus) Gt 0...2...0.0 indel x x x x 401 aa 000 nm no_ref genome encephalopsin Amphiop4 introns from braFlo 0 MPLYNTSSGPTQGLPWDTPYSQDPIWNDSSPSNSSEDAVVDQGRGELQDFSDAGYTAIATGLALI 1 2 GLVGSMNNFVVILLIGCHRQLRTPFNLLLLNVSVADLLVSVCGNTLSFASAVQHRWLWGRPGCVWYGFANSLF 1 2 GIVSLVTLSALAFERYCVVVRSSEMLTYKSSLGMIAFIWMYSLLWTSLPLLGWSSYQFEGHS 0 0 VGCSVNWVKHNVNNVSYIITLMVTCFFVPMVVVCWSYACIWRTVRM 0 0 SAEMKSEFGNPQNTGRLVTTMVVVMIVCFLVCWTPYTVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ 0 0 FREFLLARLRTFCCRQPRMLRVTPMDDNAHARLVGEGPSHAQQVIPSEENGENVEMRKVQGNQLKADSLSTISE* 0 >ENCEPH5_braFlo Branchiostoma floridae (amphioxus) Gt 0...2...0.0 indel -ZFYVE1 +RTF1 +ATP6V0E1 -Etf1 409 aa 000 nm no_ref genome encephalopsin extra 0 intron 0 MLGMHNVMNATDYDNNNATFAAWNFQRNGTTEEEVEFSGFDTVAVVIAAIGIAGFLSNGAVVLLFLKFRQLRTPFNMLLLNMSVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANHLF 1 2 GLVSLISLAVISYERYRMVVKPKGPGSSYLTYNKVGLAIIFIYLYCLLWTTLPIVGWSSYQLE 0 0 GPKISCSVAWEEHSLSNTSYIVAIFIMCLLLPLLIIIYSYCRLWYKVKK 0 0 GSQNLPPAIRKSSQKEQKIARMVVVMITCFLVCWLPYGAMALVVSFGGESLISPTAAVVPSLLAKSSTCYNPLVYFAMNNQ 0 0 FRRYFQDLLCCGRRLFDASASVNTCNTSAMPRHSPVFQKPDSDQYNGIQKSREPQMRTTGQNAPYRQWIEMQTIAVVVKADEVNNKFGEVKT* 0 >ENCEPH5_braBel Branchiostoma belcheri (amphioxus) Gt 0...2...0.0 indel x x x x 421 aa 000 nm 12435605 AB050609 encephalopsin Amphiop5 extra Nfrag in mrna 0 MLGIYNVVNATEYGNNTTFAAWDFKRNGTGGEEEVEFFGYDAVAGVIAIIGVVGFVSNGAVVVLFLKFPQLRTPFNLLLLNMAVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANHLF 1 2 GLVSLISLAVISFLRYRMVVKPKGPGSSYLTYTKVGLAILFIYLYCLLWTTLPIAGWSSYQLE 0 0 GPKIGCSVAWEEHSWSNTSYIVVLFITCLFAPLLIIVYSYYRLWHKVKQ 0 0 GSRNLPAAMRKSSQKEQKIAMMVIVMITCFMVCWLPYGAMALVVTFGGERLISHTAAVVPSLLAKSSTCYNPVVYFAMNSQ 0 0 FRRYFQDLLCCGRRLFDVSQSVVTGNTAMPRNNSQGFRKDDSDQKQDNGLPKQSEGPMCDHSSNESQMEGSRHNTAASQQWIEMQTIAVVVKAVEVDTSAANEP* 0 >ENCEPH_apiMel Apis mellifera (bee) Gt 0...2...0.0 indel x x x x 329 aa 000 nm 16291092 NM_001039968 encephalopsin ciliary Gt pteropsin clock 0 MSLNRSTMEHVIYEDQVSPVMYIGAAIALGFIGFFGFTANLLVAIVIVKDAQILWTPVNVILFNLV 0 0 FGDFLVSIFGNPVAMVSAATGGWYWGYKMCLW 2 1 YAWFMSTLGFASIGNLTVMAVERWLLVARPMQALSIR 2 1 HAVILASFVWIYALSLSLPPLFGWGSYGPEAGNVSCSVSWEVHDPVTNSDTYIGFLFVLGLIVPVFTIVSSYAAIVLTLKKVRKRA 1 2 GASGRREAKITKMVALMITAFLLAWSPYAALAIAAQYFN 0 0 AKPSATVAVLPALLAKSSICYNPIIYAGLNNQFSRFLKKIFDARGSRTAVPDSQHTALTALNRQEQRK* 0 >ENCEPH1_anoGam Anopheles gambiae (mosquito) Gt 0...2...0.0 indel x x x x 461 aa 000 nm no_ref XM_312503 encephalopsin GPROP11 adjacent head-to-head tandem GPROP12 0 MYDVTDAAAINSDHQELMAPWAYNGAAVTLFFIGFFGFFLNIFVIALMYKDVQ 0 0 LWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWLYGKSICVAYGFFMSLL 1 2 GIASITTLTVLSYERFCLISRPFAAQNRSKQGACLAVLFIWSYSFALTSPPLFGWGAYVNEAANI 1 2 SCSVNWESQTANATSYIIFLFIFGLILPLAVIIYSYINIVLEMRK 0 0 NSARVGRVNRAERRVTSMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFWRIRRSNGVAGQPDSNNTNNSNRDKESARHTAKEGL ECSLDFCHWTVRGTRVSISSAERNVPAPAARERSGGHSVTGSREESRDRHVTLKTMLSVGPRSPSSVAPVAADCSTTDVPTSGDGSVRIVRQDSELSVIHDGGGGGGGSSSRVLVIKSQKPRSNML* 0 >ENCEPH2_anoGam Anopheles gambiae (mosquito) Gt 0...2...0.0 indel x x x x 434 aa 000 nm no_ref XM_312502 encephalopsin GPROP12 0 MNDAPNDVAASAVDYEDLMAPWAYNASAVTLFFIGFFGFFLNLFVIALMCKDMQ 0 0 LWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWIFGRTLCVAYGFFMSLL 1 2 GITSITTLTVLSYERYCLISRPFSSRNLTRRGAFLAIFFIWGYSFALTSPPLFGWGAYVQEAANI 1 2 SCSVNWESQTKNATTYIIFLFVFGLVVPLIVIVYSYTNIIVNMRE 0 0 NSARVGRINRAEQRVTSMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFSRVRNKGQQAAADQNTTTMQRELTKSSRDMVECSF DFCRKKSRFKISLVKPTAPLAVVDVSSTSHRDKGTSRSPLDQTVLNETNEDVGRERSGGGGGGGAYAGTRFVRPDFELSVINSGKSILIKSKNFRSNLL* 0 >CILL2_plaDum Platynereis dumerilii (ragworm) Gt 0...2...0.0 indel x x x x 310 aa 000 nm 16311335 CT030681 proto cilliary htgs new 5 exons 1 missing 0 MDDLGFLGNSSVNYTVPLLQEDPLLLRILYFGPTSYVITAIYLCIVGVIGTLSNGVIMYLYFKDKSLRSPMNLLFVNLAMSDFTVAFFGAMFQFGLTCTRKYMSPGMALCDFYGFITFLG 1 2 GLASEMNLFIISVERYLAVVRPFDVGNLTNRRVIAGG 1 2 VFVWLYSLVFAGGPLVGWSSYRPEGLGTWCSISWQDRSMNTMSYVTAVFLGCYFFPVSIIIFCYFNVWRKVKE 0 0 AADAQGAGTAGKAEKSIFRMSVIMVTCYLTAWTPYAIVCLIASYGPPNGLPIYAEVLPSLFAKSSQVYNPIIYVLMNKP 0 0 0* 0 >CILL1_plaDum Platynereis dumerilii (ragworm) Gt 0...2...0.0 indel x x x x 355 aa 000 nm 15514158 AAV63834 lophotrochozoa ciliary polychaeta new genomic 0 MDGENLTIPNPVTELMDTPINSTYFQNLNAETDGGNHYIYNAFTATDYNICAAYLFFIACLGVSLNVLVLVLFIKDRKLRSPNNFLYVSLALGDLLVAVFGTAFKFIITARKTLLREEDGFCKWYGFITYLG 1 2 GLAALMTLSVIAFVRCLAVLRLGSFTGLTTRMGVAAMA 1 2 FIWIYSLAFTLAPLLGWNHYIPEGLATWCSIDWLSDETSDKSYVFAIFIFCFLVPVLIIVVSYGLIYDKVRK 0 0 VAKTGGSVAKAEREVLRMTLLMVSLFMLAWSPYAVICMLASFGPKDLLHPVATVIPAMFAKSSTMYNPLIYVFMNKQ 0 0 FRRSLKVLLGMGVEDLNSESERATGGTATNQVAAT* >RGR_homSap Homo sapiens (human) ?? 0.2.1.2.1.0.0 indel +PCDH21 -LRIT1 -GRID1 -WAPAL 296 aa 000 nm 17679941 NM_001012720 RGR retinal epithelium Mueller exon-skipping splice isoform 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSQLAWNSAVSLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKREKDRTK* 0 >RGR_galGal Gallus gallus (chicken) ?? 0.2.1.2.1.0.0 indel +PCDH21 -LRIT1 +CHAT -PARG 296 aa 000 nm 14985289 NM_001031216 retinal ganglia RGR 0 MVTSHPLPEGFTEIEVFAIGTALLVE 1 2 ALLGFCLNGLTIISFRKIKELRTPSNLLVLSIALADCGICINAFIAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFLTALASISSSAAVAWDRYHHYCT 1 2 RSKLQWSTAISMMVFAWLFAAFWATMPLLGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYITFLFALSIFNFMIPGFIMMTAYQSIHQKFKKSGHYK 0 0 FNTGLPLKTLVICWGPYCLLSFYAAIENVMFISPKYRM 0 0 IPAIIAKTVPTVDSFVYALGNENYRGGIWQFLTGQKIEKAEVDSKTK* 0 >RGR_xenTro Xenopus tropicalis (frog) ?? 0.2.1.2.1.0.0 indel +PCDH21 -LRIT1 +CHAT -PARG 296 aa 000 nm no_ref BC135113 retinal ganglia RGR 0 MVTSYPLPEGFTETEVFAIGTTLLVE 0 0 ALLGLLLNGLTLLSFYKIRELRTPSNLFIISLAVADTGLCLNAFVAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1 2 RSKLHWSTAVSVVFFIWGFSAFWSAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYISYLFTMAFFEFLVPLFILMTAYQSIYQKMKKSGQIR 0 0 FNTSMPVKSLVFCWGPYCLLCFYAVIQDATILSPKLRM 0 0 IPALLAKTSPAVNAYVYGLGNENYRGGIWQYLTGQKLEKAETDNKTK* 0 >RGR_gasAcu Gasterosteus aculeatus (stickleback) ?? 0.2.1.2.1.0.0 indel +PCDH21 -LRIT1 +CHAT -PARG 296 aa 000 nm no_ref genome retinal ganglia RGR 0 MVSSYPLPDGFTDFDVFSLGSCLLVE 0 0 GLLGILLNAVTIAAFLKVRELRTPSNFLVFSLAVADIGISMNATIAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFVTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSSAITLAVFVWLFTAFWSAMPLIGWGEYDYEPLRTCCTLDYTKGDR 2 1 NYVSYLIPMAIFNMAIQVFVVMSSYQSIAQKFKKTGNPR 0 0 FNPNTPLKAMLFCWGPYGILAFYAAVENATLVSTKLRM 0 0 MAPILAKTSPTFNVFLYALGNENYRGGIWQLLTGEKIDVPQIENKSK* 0 >RGR_calMil Callorhinchus milii (elephantfish) ?? 0.2.1.2.1.0.0 indel x x x x 227 aa 000 nm no_ref genome fragment 0 EGFTDFEVFGLGTALLVE 0 0 GLVGLLLNGLTLLAFYKIKELRTPSNLLITSLALSDFGISMNAFIAAFSSFLR 2 1 YWPYGSEGCQTHGFHGFLMALASINACAAIAWDRYHQNCS 1 2 SRLQWSSAITVTVFIWGIAAFWSAMPLLGWGVYDYEPLRTCCTLDYSKGDR 2 1 EFIFPIFIMLSSYQSCKSKFKKTGQVK 0 0 FNTGLPVKTLIFCWGPYSLLCFYATIENITILSPKLRM 0 >PER_homSap Homo sapiens (human) ?? 0.2.0.2.1.0.1 indel -CFI +NOLA1 +EGF -ELOVL6 338 aa 000 nm 17167409 NM_006583 peropsin RRH RRH retinal photoisomerase Retinal epithelium 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDV 1 2 GRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 >PER_monDom Monodelphis domestica (opossum) ?? 0.2.0.2.1.0.1 indel -CFI +NOLA1 +EGF -ELOVL6 326 aa 000 nm no_ref genome peropsin RRH 0 MFKNNSVKTLAPEKEGPSVFSPIEHKIVAAYLITA 1 2 GVISIVSNVIVLGIFVKYKALRTATNTIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYDGCQ 0 0 IYAGLNIFFGMASIGLLTAVAIDRYLTICQPDL 1 2 GRMTSYNYTLMILTAWVNGFFWALMPIVGWAGYAPDPTGATCTINWRKNDV 2 1 SFVSYTMTVITINFAMPLGVMFYCYYNVSQKMKQYSPSNCPDHINRDWSNQVAVTK 0 0 MSVVMILMFLLAWSPYSIVCLWASFGDPKEIPPAMAIVAPLFAKSSTFYNPCIYVAANKK 2 1 FRRAISAMIRCQTHQSMPISNALPMN* 0 >PER_galGal Gallus gallus (chicken) ?? 0.2.0.2.1.0.1 indel -CFI +NOLA1 +EGF -ELOVL6 335 aa 000 nm 14985289 NM_001079759 peropsin RRH 0 MHWNDSANSSESDAEAHSVFTQTEHNIVAAYLITA 1 2 GVISIFSNIVVLGIFVKYKEFRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYTGCQ 0 0 IYAALNIFFGMASIGLLTVVAVDRYLTICRPDI 1 2 GRRMTTRNYAALILAAWINAVFWASMPTVGWAGYASDPTGATCTANWRKNDV 2 1 PFVSYTMSVIAVNFVVPLTVMFYCYYNVSRTMKQYTSSNCLESINMDWSDQVDVTK 0 0 MSVVMIVMFLVAWSPYSIVCLWSSFGDPKKISPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILAMVRCQTRQEITISNALPMTVSLSALTS* 0 >PER_xenTro Xenopus tropicalis (frog) ?? 0.2.0.2.1.0.1 indel -CFI +NOLA1 +EGF -ELOVL6 347 aa 000 nm no_ref genome peropsin RRH 0 METLAEVSTLLPAGTGTVNISDASSEVHSVFSQSEHNIVAAYLITA 1 2 GVISILSNIIVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYVGCQ 0 0 IYAGLNIFFGMASIGLLTVVAIDRYLTICRPDIG 1 2 GRRISGRHYTAMILAAWINAVFWSVMPVVGWSSYAPDPTGATCTINWRKNDV 2 1 SFVSYTMSVVAVNFVVPLMVMFYCYYNVSRTMKGYGSRSSLGGINADWSDQTDVTK 0 0 MSMVMIVMFLVAWSPYSIVCLWSSFGDPRKIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILSMVQCKSRQEVTLDNHFPMNVSQSTLTT* 0 >PER_gasAcu Gasterosteus aculeatus (stickleback) ?? 0.2.0.2.1.0.1 indel +GPR68 -GNPDA1 -ENPEP -C14orf100 338 aa 000 nm no_ref genome peropsin RRH 0 MGIDPEVNVTDDVTLYGGKSAFTQLEHNIVAGYLITA 1 2 GVISLFSNIVVLLMFWKFKELRTATNFIIINLAFTDIGVAGIGYPMSAASDIHGSWKFGYAGCQ 0 0 IYAALNIFFGMASIGLLTVVAIDRYLTICRPDIG 1 2 GQKMTMQSYNLLILAAWLNAVFWSSMPVVGWASYAPDPTGATCTINWRQNDV 2 1 SFISYTMAVIAVNFVLPLSAMFYCYYNVSATVKRYKASNCLDSANIDWSDQMDVTK 0 0 MSIVMIIMFLVAWSPYSIVCLWASFGDPKTIPAPMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAIIGMVRCQTRQRITINSQVPMTTSQQPLTQ* 0 >PER_calMil Callorhinchus milii (elephantfish) ?? 0.2.0.2.1.0.1 indel x x x x 151 aa 000 nm no_ref genome fragment 1 LFVSYTMTVIAVNFVVPLSVMFFCYYNVSKTMSRFISSPSPENINLDWSDQLDVTK 0 0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRKAIMAMICCQNRQEITINHTLPMTISRVPLTE* 0 >PERa_braBel Branchiostoma belcheri (amphioxus) ?? 0.2.0.2.1.0.1 indel x x x x 365 aa 000 nm 12435605 AB050610 peropsin Amphiop3 0 MDIPTETPYGAEEDIGESAGWRWTETDKNGFHKYDHLIVGLYLFVI 0 GIIGTIENGITLATFSKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLRSHWLFGGVGCQ 0 0 WYGFNGMFFGMANIGLLTCVAVDRYLVICRHDLV 2 DKVNYNTYGVMAALGWLFAAFWAALPLVGWAEYALEPSGTACTINFQKNDS 2 1 LYISYVTSCFVLGFVVPLAVMAFCYWQASCFVSKVLKGDIAGDLTFPVAANVDWEYQ 0 NHFSKMCLAMVAAFVVAWTPYSVLFLFAAFWNPADIPAWLTLLPPLIAKSSALYNPIIYIIANRR 0 0 FRNAICSMMKGQDPDVEDDEHADEHRVRSIEDNDKEIISMVNLNMTV* 0 >PERb_braBel Branchiostoma belcheri (amphioxus) ?? 0.2.0.2.1.0.1 indel x x x x 522 aa 000 nm 12435605 AB050607 peropsin Amphiop2 RRH MIPTNNNTENNDLEWGLEKEHGVSATIMGVYLTIVGLVATVGNATVVLMFIMKWRQLCRKAPNLLVINLAAANLCITIFG YPFSASSGYAHQWLFPDAICTLYGFSCFLLSMVSMHTLCLISAHRYITICRPEHASKLTMNRTVLAVIGTWLYAIAVAVP PLFNIARYTYEPSGLSCTIDFRVTTVADLVYLGSLIVLCYVIHVAVMATCYFKIIRKFSRHRFRQVRDIRTSHQRSFEMG VTMRCILMTLFYLLSWTPYTAVCIWTMVGPPPPVVVSMAAALIAKTHCAFNPILYAFMSEVYRKLVFRTMCPCCFNRISC KFVGTPTGGSKVSANPDIFTVDYNSRDQAVQINKAPSRRFCFVMETSEDLGSDDTGLTGHSGLWRSGAEVEGLGGLQVTQ SPSVSGSELSLSLLDFLPPKPSGRAVSAKLPSPPALNSERATCPESSQQPSDRPATGLRQYQKGDTTRSSVGDLILTEDD VTNLPPASETWGRKKSENPLSYRQTTRRTFGRSRKHSYIVD* >PERc_braBel Branchiostoma belcheri (amphioxus) Go 0.2.0.2.1.0.1 indel x x x x 391 aa 000 nm 12435605 AB050606 peropsin Amphiop1 RRH MNASPSSWLSSGEFFTDSPENSSEWPWTDGPTDTTWRHHQSVDSVSYEGYLASAIYITLTGLIAFFGNVITITVFLTEKE FRKKQQNGFVLNLAIADLSVCVFAYPSSAIAGYAGRWVLGDVGCTIYGFLCFTFALVSMVTLCVISIYRYILICKPQYAH LLTHRRTVYVIIGTWLYALVFTVPPLVGVKRYTYEPMQITCSLDWNVQHPGEKAYIAAVLVIVYVLQVLIMCFCYFNIIF KSANLKFAALASEKTKMAAKKDTWKTSVMCLTMVVSFLIAWTPYAVSSTWDILSAEDLPIIATILPSLFAKSSCMMNPII YACCNTK FRQAAVKSFRKLCGMCKQKVPLSTPQVVLAMQRNTEFTSTVEPTGQAFPMRVLPSISATHTAL* >NEUR_homSap Homo sapiens (human) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 -MUT 355 aa 000 nm 15774036 NM_181744 neuropsin OPN5 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2 1 LHTVTTVRKSSAVLEIHEEV* 0 >NEUR_monDom Monodelphis domestica (opossum) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 -MUT 352 aa 000 nm no_ref genome neuropsin OPN5 0 MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIISCFSHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAYICLVIIWAYATFWATMPLAGLGNYAPEPFGTSCTLDWWLAQASVTGQTFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQSSHVLEMKLTK 0 0 AMLICAGFLIAWIPYAVVSVWSAFGQPDSIPVQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCQSGGQKAAKKESLRTYR 2 1 HTVATIRKSSAVSETHQEV* 0 >NEUR_ornAna Ornithorhynchus anatinus (platypus) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 - 351 aa 000 nm no_ref genome neuropsin OPN5 0 MTNYSAPQLGDYLPHYLREGDPFVSKLSWEADLVAGVYLVII 1 2 GVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAYICLAIIWAYASFWATMPLVGLGNYAPEPFGTSCTLDWWLAQASVAGQAFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQNSHVLEMKLTK 0 0 AMLICAGFLIAWIPYAVVSVWSAFGQPDSIPIQFSVVPTLLAKSAAMYNPIIYQVIDCRISCCRLGGPKTGKKESLKNSR 2 1 HSMSTIRKPSAVSGPHQEV* 0 >NEUR_galGal Gallus gallus (chicken) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 - 352 aa 000 nm no_ref genome neuropsin OPN5 0 MASDCNSSSQEEYLPHYMQQEDPFASKLSREADIIAGFYLTVI 1 2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGPKTLQKKSSLKES 2 1 YTISSHRDSAALSGTQLEV* 0 >NEUR_anoCar Anolis carolinensis (lizard) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 +ITSN2 340 aa 000 nm no_ref genome neuropsin OPN5 0 MEQGQNISSQDDNQQEEDPFASKLSVEADIVAGVYLLVI 1 2 GILSTLGNGYVIYMSTQRKKKLKPAEIMTVNLAVCDLGIS 1 2 VGKPFSIIAFFSHRWIFGWSGCRWYGWAGFFFGIGSLITMTAVSLDRYFKICHLSY 1 2 GTWLKRHHVFICLGIIWSYAAFWATIPFAGFGNYAPEPFGTSCTLDWWLAQGSVAGQAFILNILFFCLVLPTAVIMFCYVKIIAKVQSSTKEVAHYDTRIQNQHVLEMKLTK 0 0 VAMLICAGFMFAWIPYAVVSVWSAFGRPDSVPIKVSVIPTLLAKSAAMYNPVIYQVIDCKSACCRPGNLQPLQKKNSRYVF 2 1 MLQWDKGHDEV* 0 >NEUR_xenTro Xenopus tropicalis (frog) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 - 340 aa 000 nm no_ref genome neuropsin OPN5 0 MAGNSSYREESGYIPHYERDSDPFASKLSREADIFAGVYLMAI 1 2 GILSTLGNGYVIYMACSRKKKLRPAEIMTINLAVCDLGIS 1 2 VTGKPFAIVSCFSHRWVFGWNACRWYGWAGFFFGCGSLITLTVVSLDRYLKICHLRY 1 2 GTWLKRRHAFIALAVIWAYATLWATLPLVGVGNYAPEPFGTTCTLDWWLAQASVKGQIFVLSMLFFCLLFPTMVIVFSYAKIIAKVKSSAKEVAHFDTRNQNNHTLEIKLTK 0 0 AMLICAGFLIAWFPYAVVSVWSAFGQPDSIPIELSVVPTMMAKSASMYNPIIYQVIDCKPACCKKDKSLQNTTSRYVFVVYIPFHHYR 2 >NEUR_gasAcu Gasterosteus aculeatus (stickleback) ?? 0.2.2.2.0.1 indel +CD2AP +GPR115 -PTCHD1 - 331 aa 000 nm no_ref genome neuropsin OPN5 0 MENETWTHPSYIPHYLLRGDPFASRLSKEADIIAAFYICII 1 2 GIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGIS 1 2 VTGKPFFVVSSFAHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 2 GTWLKRQHAFLCLVFVWMYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLAQASVSGQSFVVAILFFCLVLPAGIIVFSYVMIIFKVKSSAKEISNFDARIKNSHNLEIKLTK 0 0 AMLICAGFLIAWIPYAVVSVVSAFGEPDSVPISVSVIPTLLAKSSAMYNPIIYQVLDLKNSCMKSSCFKGLKKPRHFRKSR 2 >NEUR_calMil Callorhinchus milii (elephantfish) ?? 0.2.2.2.0.1 indel x x x x 209 aa 000 nm no_ref genome fragment 2 GLLSTLGNGYVIYLSITQKRKLKPPEILITNLAISDFGMS 1 2 VGGQPFLIISCFSHRWIFGWVGCRWHGWAGFFFGCGSLITMTVVSLDRYLKICHLQY 1 2 GSWLQRRHVFMSLAFIWFYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLARVSVSGLIFVLTILFFCLLLPIIIIVFSYIKIIAKVKSSAKEVAHFDSRIQNHHSLEMNLTK 0 >MEL1_homSap Homo sapiens (human) Gq 0.0.1.2.2.1.1.1.0.0 indel -GRID1 -WAPAL +LDB3 +BMPR1A 483 aa 000 nm 16961436 NM_033282 melanopsin OPN4 0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0 0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIRETGR 2 1 ALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVALVAFAG 2 1 YAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2 1 VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLTSHTSNLSWISIRRRQESLGSESEV 0 0 GWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGK 0 0 TKGLIPSQDPRM* 0 >MEL1_monDom Monodelphis domestica (opossum) Gq 0.0.1.2.2.1.1.1.0.0 indel -GRID1 -WAPAL +LDB3 +BMPR1A 483 aa 000 nm no_ref genome melanopsin OPN4 0 MNPSPMLRGLSCPAQDTNCTKIMASMSEWNNTEEDAYHLVDLPSIAPT 0 0 AVVLPPSSQNIFPTADVPDHAHYTIGATILAVGFTGVLGNLLVIYTFCR 2 1 LRTPANMFIINLAISDFFMSFTQAPVFFASSMYKRWIFGEK 1 2 ACEFYAFCGALFGITSMITLMAIALDRYFVITRPLASIGVISKKKTGFILLGVWLYSLAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYTTFTPSVRAYTMLLFCFVFFIPLIVIIYCYIFIFRAIQDTNK 2 1 AVHSIGSGESTASPRHCQRMKNEWKMAKIALVVILLYVLSWAPYSTVALVAFAG 2 1 YSHILTPYMNSVPAIIAKASAIHNPIIYAISHPKYR 2 1 MAIAQNFPCLRALLCVRHPRTRSFSSYRFTRRSTMTSQASDISWLPRGRRQLSLGSESEI 0 0 GWNNMEAGTTSLTSRNQQGSCRMDQETMETRELAAIAKAKGRSWETLEK 0 0 TLEEMDDSSLLEVSVDMEQ* 0 >MEL1_galGal Gallus gallus (chicken) Gq 0.0.1.2.2.1.1.1.0.0 indel -GRID1 -WAPAL +LDB3 +BMPR1A 529 aa 000 nm 16856781 AY88294 melanopsin OPN4m 0 MDLPPRAPT 0 0 KMTVKDVRGAFPTVDVPDHAHYTIGTVILIVGITGTLGNFLVIYAFCR 2 1 SRTLQKPANIFIINLAVSDFLMSITQSPVFFTNSLHKRWIFGEK 1 2 GCELYAFCGALFGITSMITLMVIALDRYFVITKPLASVRVMSKKKALIILVGVWLYSLAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFIPLIAIIYSYVFIFEAIKKANK 2 1 SVQTFGCKHGNRELQKQYHRMKNEWKLAKIALIVILLYVISWSPYSVVALVAFAG 2 1 YSHVLTPFMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 TAIATYVPCLGFLLRVSPKESRSFSSYPSSRRTTITSQSSETSGLQKGKRRLSSISDSES 0 0 GCTDTETDITSMISRPASSQVSYEMGEDTTQTSDLGGKPKVKSHDSGIFRK 0 0 TVVDADEIPMVEINDTEHSATSTCKTSEKCNVEEIQ 0 0 RSESLSGIGLREGESRHRTSASQIPSIIITYSNVQGVELHSGYSAGFLHPKNKSHKQNKSSNS* 0 >MEL1_xenTro Xenopus tropicalis (frog) Gq 0.0.1.2.2.1.1.1.0.0 indel -GRID1 -WAPAL +LDB3 +BMPR1A 596 aa 000 nm 16856781 DQ384639 melanopsin OPN4m 0 MNYQSVRKGITCPPQDANCSRILESLNSWNNSEVNSYKLVELPPIVTT 0 0 ETPQYEIHHVYPTVDVPDHVHYVVGAVILAVGITGMLGNFLVIYAFCR 2 1 SRSLRSPANMFIINLAITDFLMSVTQAPVFFATSLHKRWIFGEK 1 2 GCELYAFCGALFGITSMITLMVIAVDRYFVITRPLTSIGVMSKKRAVLILSGVWLYSLAWSLPPFFGW 1 2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFCFVFFIPLFIIIYCYIFIFKAIKNTNR 2 1 AVQKIGTDNNKESHKQYQKMKNEWKMAKIALIVILLYVVSWSPYSTVALLAFAG 2 1 YASILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 MAIAKYIPCLGSLLRVKRRDSRSYSSYPSSRRSTVTSHCSQSSDVGGHPKLKNHLPSVSDSES 0 0 GWTDTEADSSVNSRPASRQVSYEMGKDTTETNDLKSKAKLKSHDSGIFEK 0 0 TSMDADDISLVELGTVDRSSPIM 0 0 ANKHLNGLGQRKGDSFTRRSPSSRIPSIVVTHSNHQGSPAAVRHNSTLPGIKVSNSQDREKELKRQIEKVKQYVPIVTITSDTENSTGGFSNELLPANTS* 0 >MEL1_danRer Danio rerio (zebrafish) Gq 0.0.1.2.2.1.1.1.0.0 indel - +USP54 +LDB3 +BMPR1A 594 aa 000 nm no_ref AY078161 melanopsin OPN4m 0 MMSGAAHSVRKGISCPTQDPNCTRIVESLSAWNDSVMSAYRLVDLPPTTTTTTSVA 0 0 MVEESVYPFPTVDVPDHAHYTIGAVILTVGITGMLGNFLVIYAFSR 2 1 SRTLRTPANLFIINLAITDFLMCATQAPIFFTTSMHKRWIFGEK 1 2 GCELYAFCGALFGICSMITLMVIAVDRYFVITRPLASIGVLSQKRALLILLVAWVYSLGWSLPPFFGW 1 2 SAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFIPLIVIIYCYFFIFRSIRTTNE 2 1 AVGKINGDNKRDSMKRFQRLKNEWKMAKIALIVILMYVISWSPYSTVALTAFAG 2 1 YSDFLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 LAIAKYIPCLRLLLCVPKRDLHSFHSSLMSTRRSTVTSQSSDMSGRFRRTSTGKSRLSSASDSES 0 0 GWTDTEADLSSMSSRPASRQVSCDISKDTAEMPDFKPCNSSSFKSKLKSHDSGIFEK 0 0 SSSDVDDVSVAGIIQPDRTLTN 0 0 AGDITDVPISRGAIGRIPSIVITSESSSLLPSVRPTYRISRSNVSTVGTNPARRDSRGGVQQGAAHLSNAAETPESGHIDNHRPQYL* 0 >MEL1D_danRer Danio rerio (zebrafish) Gq 0.0.1.2.2.1.1.1.0.0 indel - +USP54 +LDB3 +BMPR1A 473 aa 000 nm no_ref genome melanopsin OPN4m 0 QVAMVQDVRHPFPTVDVPDHAHYTIGSVILAVGITGMVGNLLVMYAFCK 2 1 SRSLRTPANMFIINLAVTDFLMCVTQTPIFFTTSLHKRWIFGEK 1 2 GCELYAFCGALFGICSMITLMIIAVDRYFVITRPLASIGVMSRKRALLILSAAWAYSMGWSLPPFFGW 1 2 SGAYVPEGLLTSCSWDYMTFSPSVRAYTMLLFTFVFFIPLFVIIYCYFFIFKAIRETNR 2 1 AVGKINGEGGPRDSIKKIHRMKNEWKMAKIALIVILLYVISWSPYSCVALTAF 2 1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 SAIAKYIPCLGVLLCVPRRDRFSSSSFISTRRSTLTSQSSETSSNLHRAGKARLSSVSDSES 0 0 GWTDTEADLSTASSRPASRQVSSEIRKDLCDIKHSSSLRLKVKSRDSGIFDR 0 0 0 0 QNDVSEKADEKRPLVRIPSIIVTSETCPAVLPAGHSSRLIPGAPAVTDS* 0 >MEL1_takRub Takifugu rubripes (teleost) Gq 0.0.1.2.2.1.1.1.0.0 indel - +USP54 +LDB3 +BMPR1A 555 aa 000 nm no_ref genome melanopsin OPN4m 0 MNFGKSALQPPAQQSVVSCGGGGPEPNCTLRLAVTVMMSVRLAELQLHAST 0 0 LQVAMVRPFPTVDVPDHAHYTIGSVILVIGITGMIGNFLVIYAFCR 2 1 SRSLRTPANMFIINLAVTDLLMCVTQTPIFFTTSMYKRWIFGEK 1 2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRAFVILMTVWIYSLGWSLPPFFGW 1 2 SGAYVPEGLLTSCTWDYMTFSPSVRAYTMLLFIFVFFLPLFIIIYCYFFIFRAIRATNK 2 1 AVGKVNGSVHSHSRRRESVKNFQRLQNEWKMAKIALMVILLYVISWSPYSCVALTAFAG 2 1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 LALAKYIPCLGFLLCISPHELQSTSSSFMSLRRSTVTSQTSDISGQFRPQSKPRRSSASDSES 0 0 CLTDTEADLSSMGSRPASRQVSCDISRDTTELPEYKPASSFNSKVKSPDSGIFEK 0 0 TSFDFDASMAASRERSSIPN 0 0 SGEFPEGHVMRRTLARIPSIIITSESSHFLPNGRKASSTTCIANGSDIKVGPR* 0 >MEL1_gasAcu Gasterosteus aculeatus (stickleback) Gq 0.0.1.2.2.1.1.1.0.0 indel - - +LDB3 +BMPR1A 556 aa 000 nm no_ref genome melanopsin OPN4m 0 MNAGESELLLPTQQSILPCGDHEPNCPVAQAETLALSAASANGSA 0 0 VQVAMVSRAPHPYPTVDVPDHAHYTIGSVILAIGITGIIGNVLVIYAFSK 2 1 SRSLRTPANMFIINLAITDLLMCVTQAPIFFTTSMHKRWIFGEK 1 2 GCELYAFCGALFGICSMITLTVIALDRYFVITRPLTSIGMMSRRRALLILMGAWTYSLGWSLPPFFGW 1 2 SGAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIYCYFFIFRAIRVTNR 2 1 AVGKMNGSIHSHGSGRDSTKNFHRLQNEWKMAKIALIVILLYVVSWSPYSAVALTAFAG 2 1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 IALAKYIPFLGVLLCVPPRELRSASSSFRSTRRSTVTSQTSDVSSQQRRQGSRNSRLSSASDSES 0 0 CLTDTEADGSSVGSRPASRQVSCDIGRDTAELPEFKPSSSFKSKMKSHDSGIFEK 0 0 SYDTDISMAGVSERGSIPN 0 0 QTDFAEGRDRRSTIGRIPSIVITSETSPFLPTGRNGSCNGRPKTANSSHPGAGSG* 0 >MEL1_oryLat Oryzias latipes (medaka) Gq 0.0.1.2.2.1.1.1.0.0 indel - +USP54 +LDB3 +BMPR1A 504 aa 000 nm no_ref genome melanopsin OPN4m 0 LQVAMVPQTFHPFPTVDVPDHAHYTIGSVILAIGITGIIGNFLVIYAFSR 2 1 SRSLRTPANMFIINLAITDLLMCVTQSPIFFTTSMHKRWIFGEK 1 2 GCELYAFCGALFGICSMITLTVIAIDRYFVITRPLTSIGVLSRKRALLILSAAWAYSLGWSLPPFFGW 1 2 SGAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFIFVFFLPLFIIIYCYVFIFRAIRSTNR 2 1 AVGKINGNTRDAVKSFNRLQNEWKMAKIALIVILLYVISWSPYSTVALTAFAG 2 1 YADMLTPYMNSIPAVIAKASAIHNPIIYAITHPKYR 2 1 MALAKYIPGLGVLLCIHPKDLRSASSSFVSTRRSTVTSQSSDISSQLRRQSTFKSRLSSLSDSES 0 0 GLTDTEADLSSLSSRPASRQVSCEISRDTAELPDFKHTSSFKAKLKNNDSGIFEK 0 0 TSFDTVSIGGVSEHNSIPS 0 0 NRDFGDGNVTRATIGRIPSIVVTSEMSPFLPVGRNGSRTNRSKMANSSAGAGPV* 0 >MEL1_calMil Callorhinchus milii (elephantfish) Gq 0.0.1.2.2.1.1.1.0.0 indel - - - - 369 aa 000 nm no_ref genome melanopsin OPN4m 0 ASVTDAQHHHMFPTVDVPDHAHYIIGATILAVGVTGMVGNFLVIYAFLRYH 2 1 SRSLRTPANTFIINLAATDFLMSVTQSPIFFITSIHKRWIFGEK 1 2 GCELYAFCGALFGITSMITLMVIALDRYFVITRPLASIGVLSHRRAGLIILSLWLYSLAWSLPPFFGW 1 2 SGAYVPEGLLTSCTWDYMTFTPSVRAYTMLLFCFVFFIPLGVIIYCYIFIFRAIKSTNK 2 1 KVGGSTNRESQKQHQRMKNEWKMAKIALIVILLFVISWSPYSTVALTAFAG 2 1 YADMLTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 MAIAKYVPLLGLLLRVSRRDSRTSGQYYSTRRSTLTSQTSDLSGYPRGKGRLSSASDSES 0 >MEL1b_calMil Callorhinchus milii (elephantfish) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 113 aa 000 nm no_ref EB687868 melanopsin OPN4m 1 SKSLRTPANMFIINLAISDFFMSATQPPVFFVTSLHKRWIFGEK 2 GCKLYAFCGALFGITSMITLMAISIDRYWVITKPLQSISSTTTKKNTLKVIILVWLYSLAWSLPPLLGW 1 >MEL1_petMar Petromyzon marinus (lamprey) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 205 aa 000 nm no_ref genome fragment 1 SKSLRSPANIFIINLAFADFFMSITQTPIFFVTSLHKRWIFGEK 1 2 GCELYAFCGALFGIASMVTLMVIATDRYLVLTRPLASIGAMSKRRAMYITAAVWFYSLAWSLPPFFGW 1 2 AYVPEGLMTSCTWDYVTFTPAVRSYTMLLFCFVFFIPLIVIIFCYVRIFAAIKNTNR 2 1 YADMLTPYMNSVPAIIAKASAIHNPIVYAITHPKYR 2 >MEL1a_braFlo Branchiostoma floridae (amphioxus) Gq 0.0.1.2.2.1.1.1.0.0 indel - - - - 709 aa 000 nm no_ref genome melanopsin Amphi-mop 12 exons +tandem dup assembly error 0 MTELPSFQPPTNSTEEENAVFPTALTEWISE 0 0 VGNQVGEAALKLLSGEGDGMEVTPTPGCTGNASVCNGTDSGGGVVWDIPPLAHYIVGTAVFCVGCCGMFGNAVVVYSFIK 2 1 SKGLRTPANFFIINLALSDFLMNLTNMPIFAVNSAFQRWLLSDF 1 2 ACELYGFAGGLFGCLSINTLMAISMDRYLVITKPFLVMRIVTKQR 0 0 VMFAILLLWIWSLVWALPPLFGWSAYVPEGF 1 2 GTSCTFDYMTPKLSYHIFTYIIFFTMYFIPMGVIIYCYYNIFATVKSGDKQFGKAVKEMAHEDVKNK 0 0 AQQERQRKNEIKTAKIAFIVITLFLSAWTPYAVVSALGTLGYQDLVTPYLQSIPAVFAKSSAVYNPI 1 2 VYAITHPKFRAAVKKHIPCLSGCLPADEEETKTKTRGATTTASMSMTQTTAPTV 0 0 HDPQASVHSGSSVSVDDSSGVSRQDTMMVK 0 0 VEVDNRMEKAGGGAADTAPKDGTSVPTVSAQIEVRPSGNVNTKAEVIPSPQSAAVAHGASASPVPK 0 0 VAELSSSVSLESAAIPGKIPTPLPSQPIAAPIERHMAAMADDPPPKPRGVATTVNVRRSESGYERSQDSLRKK 0 0 AVSETRSRSFNSTKDHFASERQTSTTLNQPRDMYSGDMVKKTRQSPEKQEYDNPAFDAGIAEIDTDSENETEGSYDMLSVRFQAMAEEPPVETYRKASDMSINLGKASLMLTEAHDETVL* 0 >MEL1a_braBel Branchiostoma belcheri (amphioxus) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 707 aa 000 nm 15936279 AB205400 melanopsin Amphi-mop 0 MTEIPSFQPPINATEVEEENAVFPTALTEWFSE 0 0 VGNQVGEVALKLLSGEGDGMEVTPTPGCTGNGSVCNGTDSGGVVWDIPPLAHYIVGTAVFCIGCCGMFGNAVVVYSFIK 2 1 SKGLRTPANFFIINLALSDFLMNLTNMPIFAVNSAFQRWLLSDF 1 2 ACELYGFAGGLFGCLSINTLMAISMDRYLVITKPFLVMRIVTKQR 0 0 VMFAILLLWIWSLVWALPPLFGWSAYVSEGF 1 2 GTSCTFDYMTPKLSYHIFTYIIFFTMYFIPGGVMIYCYYNIFATVKSGDKQFGKAVKEMAHEDVKNK 0 0 AQQERQRKNEIKTAKIAFIVISLFMSAWTPYAVVSALGTLGYQDLVTPYLQSIPAMFAKSSAVYSPI 1 2 VYAITYPKFREAVKKHIPCLSGCLPASEEETKTKTRGQSSASASMSMTQTTAPV 0 0 HDPQASVDSGSSVSVDDSSGVSRQDTMMVK 0 0 VEVDKRMEKAGGGAADAAPQEGASVSTVSAQIEVRPSGKVTTKADVISTPQTAHGLSASPVPK 0 0 VAELGSSATLESAAIPGKIPTPLPSQPIAAPIERHMAAMADEPPPKPRGVATTVNVRRTESGYDRSQDSQRKK 0 0 VVGDTHRSRSFNTTKDHFASEQPAALIQPKELYSDDTTKKMARQSSEKHEYDNPAFDEGITEVDTDSENETEGSYDMLSVRFQAMAEEPPVETYRKASDLAINLGKASLMLSEAHDETVL* 0 >MEL1b_braFlo Branchiostoma floridae (amphioxus) Gq 0.0.1.2.2.1.1.1.0.0 indel - - - - 402 aa 000 nm no_ref genome melanopsin Amphiop6 0 MSPNLTNTSLLPNRTDRPELSPADVTMQLVFGSMMLVFGLIGVVGNAVALYAFCR 2 1 SRSLRRPKNYLIANLCLTDMVVCLVYSPIIVTRSLSHG 2 1 LPSKESCIVEGFVVGLGSIVSICSLAGIAVERYVTITQPIKSLSILTHRALLGAVSAVWVYAFLLAFPPLVGWGRYVSEESKISCTFDYLSTDDATRAHVIVLVIGAFGLPFS VITYCYVRSFATVRKCTKERKQMSPLAKSDSRSEVKAAVNSFVITTSFCLCWCPYAVVATMGVSGFTVHSHAVFIAALLAKLSVLFNPVAYVLSIP 1 2 NSNVNIESTELTVPYSASRESCLLSRAATERLAGRSPSLTDIVREFGLQQTASHRE >MEL1b_braBel Branchiostoma belcheri (amphioxus) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 402 aa 000 nm 12435605 AB050611 melanopsin Amphiop6 0 MSSNLTNVSLVANRTDQTELSPTDVTMQLIFGSMMLVFGLIGVVGNVVALYAFCR 2 1 TRSLRRPKNYVVANLCLTDMFVCLVYCPIVVSRSFSHG 2 1 FPSKESCIVEGFMVGVGSIASICSLAAIAVERYLSVTQPLKSLTILTQRKLLVAVLTVWVYSLLLAFPPLVGWGRYVREETYISCTFDYLSTDDATRAYVITLVMGAFGFPLL TIAYCYIRVFTTARKHAEERKFMSPLKRPESRTEIKTAVTACVITTSFCLCWCPYAVVATLGISGVSVQQQTVFSAALLAKLTVIINPIVYVLSIPNFRKALFAQEREKYASED VVLTSLPGKTRRMKKVERSQSSNSNVVIEVKESSMAYSTSRESCLLSRAATKRLAGKTKSIVDLVDEFGLQETAPHKESLV* >MEL2_galGal Gallus gallus (chicken) Gq 0.0.1.2.2.1.1.1.0.0 indel +GRID2+SMARCAD1 -PGDS -SEC24B +COL25A1 544 aa 000 nm 17977531 AY882944 melanopsin 0 MGTQPHSVTKSEIPDHVLYTVGTCVLVIGSIGIIGNLLVLYAFYS 2 1 NKKLRTPQNFFIMNLAVSDFLMSASQAPICFVNSLHREWILGDI 1 2 GCDLYAFCGALFGITSMMTLLAISVDRYLVITKPLRSIQWTSKKRTIQIIAAVWLYSLGW 1 2 SVAPLLGWSSYVPEGLMISCTWDYVTYSPANRSYTMILCCCVFFIPLIIILHCYLFMFLAIRSTGR 2 1 DVQKLGSCSRKSFLSQSMKNEWKLAKIAFVVIIVYVLSWSPYACVTLIAWAG 2 1 RGNTLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYRK 2 1 TIHNAVPCLRFLIRISKNDLLRGSINESSFRTSLSSHQSLAGRTKNTCVSSVSTGEA 0 0 NWSDVELDTVEPAHEKLQPRRSHSFSSSLRQKRDLLPDSYSCSEETEEK 0 0 VSLSSSYLEKVLGRSAFPSSPVALVTSSLRAASLPVGLNSSSASRGAGSDISQMKTEESHNNGGLDSIVSNTVPQIIIIPTSETNLFQEEPEEEETELFHFHDKKNNLLDLEGLSSSTEFLEAVEKFLS* 0 >MEL2_anoCar Anolis carolinensis (lizard) Gq 0.0.1.2.2.1.1.1.0.0 indel +GRID2+SMARCAD1 -ATOH1 +PDLIM5 +BMPR1B 290 aa 000 nm no_ref genome melanopsin 0 MGPHHRTKVDVPDHVLYTVGSCVLVIGCIGITGNLLVLYAFYS 2 1 NKRLRTPPNYFIMNLAVSDFLMSATQAPICFLNSMHKEWVLGDI 1 2 GCNLYAFCGALFGITSMITLLAISVDRYCVITKPLQSIKRTSKKRTCIIIVFVWLYSLGWSVCPLFGW 1 2 SSYIPEGLMISCTWDYVTYSPANRSYTMMLCCCVFFIPLVIIFHCYIFMFLAIRSTGR 2 1 RKSSISHSIKSEWKLAKIAFVAIVVFVLSWSPYACVTLISWAG 2 1 TLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYRK 2 1 TIRSAVPCLRFLIPISKSDLSTSSMSESSFRASVSSRHSFSYRNKSTYISSISAKET 0 0 TWCDVELDPVESGHKKLQAYRSNSFSAKGVAEEESGLLLRTNNCNVPARKK 0 >MEL2_xenLae Xenopus laevis (frog) Gq 0.0.1.2.2.1.1.1.0.0 indel +SMARCAD1 +PDLIM5 +BMPR1B 535 aa 000 nm no_ref genome melanopsin Xmop 21 0 0 0 MDLGKTVEYGTHRQDAIAQIDVPDQVLYTIGSFILIIGSVGIIGNMLVLYAFYR 2 1 NKKLRTAPNYFIINLAISDFLMSATQAPVCFLSSLHREWILGDI 1 2 GCNVYAFCGALFGITSMMTLLAISINRYIVITKPLQSIQWSSKKRTSQIIVLVWMYSLMWSLAPLLGW 1 2 SSYVPEGLRISCTWDYVTSTMSNRSYTMMLCCCVFFIPLIVISHCYLFMFLAIRSTGR 2 1 NVQKLGSYGRQSFLSQSMKNEWKMAKIAFVIIIVFVLSWSPYACVTLIAWAG 2 1 HGKSLTPYSKTVPAVIAKASAIYNPIIYGIIHPKYRE 2 1 TIHKTVPCLRFLIREPKKDIFESSVRGSIYGRQSASRKKNSFISTVSTAET 0 0 VSSHIWDNTPNGHWDRKSLSQTMSNLCSPLLQDPNSSHTLEQTLTWPDDPSPKEILLPSSLKSVTYPIGLESIVKDEHTNNSCVR NHRVDKSGGLDWIINATLPRIVIIPTSESNISETKEEHDNNSEEKSKRTEEEEDFFNFHVDTSLLNLEGLNSSTDLYEVVERFLS* 0 >MEL2_danRer Danio rerio (zebrafish) Gq 0.0.1.2.2.1.1.1.0.0 indel - +FLJ39155 +PDLIM5 - 346 aa 000 nm no_ref genome melanopsin 0 MEPQRQIYKRLDVPDHVHYIIAFLILIIGTLGVSGNALVMFAFYR 2 1 NKKLRSLPNYFIMNLAVSDFLMAITQSPIFFINCLYKEWMFGEL 1 2 GCKIYAFCGALFGITSMINLLAISIDRYLVITKPLQTIQWNSKRRTGLAILCIWLYSLAWSLAPLIGW 1 2 GSYIPEGLMTSCTWDYVSPSPANKSYTMMLCCFVFFIPLSIILYCYLFMFLSVRQASR 2 1 QKSSFVKQQSMRSEWKLAKIAAVVIVVYVLSWAPYACVTLVAWAG 2 1 LTPYSKTLPAVLAKSSAIYNPFIYAIIHNKYRA 2 1 TLAEKVPGLSCLSRSQKDGLSSSTNSDASAQDSSVSRQSSVSKNRLHSTMVQ* 0 >MEL2_tetNig Tetraodon nigroviridis (pufferfish) Gq 0.0.1.2.2.1.1.1.0.0 indel - - - +BMPR1B 404 aa 000 nm no_ref genome melanopsin 0 MEPKDTHITSSFFSKVDVPDHVHYIIAFFVFVIGILGITGNVLVIFAFYS 2 1 NKKLRSLPNYFIVNLAVSDLLMASTQSPIFFINLYKEWMFGET 1 2 ACKMYAFCGALFGITSMINLLAISVDRYVVITKPLQTIRRSSKRRTALAILMVWLYSLAWSLAPLVGW 1 2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLAIILCCYLLMFLAIRKTSR 2 1 RKSTLIQQKSIRSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2 1 TLTPYSKSVPAVIAKASAIYNPIIYAIIHPRYRK 2 1 TIRSAVPCLRFLIPISKSDLSTSSMSDSSFRSALSCRHSYRSRSTYISSISAKET 0 0 TWCDVELDPVESGHKKLQAYRSNSFSAKGVAEEESGLLLRTNNCNVPARKK 0 >MEL2_gasAcu Gasterosteus aculeatus (stickleback) Gq 0.0.1.2.2.1.1.1.0.0 indel KNTC2 +FLJ39155 +PDLIM5 +BMPR1B 353 aa 000 nm no_ref genome melanopsin 0 MEPDNAHTQRSFINKVDVPDHAHYIVAVFVVVIGTLGITGNALVMLAVYS 2 1 NKKLRNLPNYFIMNLAVSDFLMAFTQSPIFFINCLYKEWAFGET 1 2 GCKIYAFCGALFGIASMINLLAISIDRYLVITKPLQAIHWGSKRRTTLAILLVWLYSLAWSLAPLVGW 1 2 GSYIPEGLMTSCTWDYVTYTLANRSYTMMLCCFVFFIPLGIILYCYLFMFLAIRKTSR 2 1 RKSTLIKQKSMKSEWKLAKIAFVVIVVYVLSWSPYACVTLISWAG 2 1 ILSPYSKAVPAIIAKASAIYNPFIYAIIHNKYRM 2 1 TLAAKFPCLRFLSPTPRKDTSSSISESSYRDSVISRQSTASRTHFITACPDTVN 0 >RHAB_plaDum Platynereis dumerilii (polychaete) Gq 0.0.1.2.2.1.1.1.0.0 indel x x x x 383 aa 000 nm 11874910 AJ316544 rhabdomeric melanopsin unavailable genomically 0 MSRSEVLVPGSMSLDGLLTTAHPIGNDSI 0 0 ETILHPYWQQFDIENTIPDSWHYAVAAWMTFFGILGVSGNLLVVWTFLK 2 1 TKSLRTAPNMLLVNLAIGDMAFSAINGFPLLTISSINKRWVWGKL 1 2 WRELYAFVGGIFGLMSINTLAWIAIDRFYVITNPLGAAQTMTKKRAFIILTIIWANASLWALAPFFGW 1 2 GAYIPEGFQTSCTYDYLTQDMNNYTYVLGMYLFGFIFPVAIIFFCYLGIVRAIFAHHA 2 1 EMMATAKRMGANTGKADADKKSEIQIAKVAAMTIGTFMLSWTPYAVVGVFGMIK 2 1 PHSEMFIHPLLAEIPVMMAKASARYNPIIYALSHPKFR 2 1 AEIDKHFPWLLCCCKPKPKAQLPSSTTKGSIASKTEADTSV* 0 >RHAB1_apiMel Apis mellifera (bee) Gq 0.0.1.0.1 indel x x x x 378 aa 000 nm 16291092 NM_001011639 rhabdomeric Lop1 long wavelength compound eyes 0 MIAVSGPSYEAFSYGGQARFNNQTVVDKVPPDMLHLIDANWYQYPPLNPMWHGILGFVIGMLGFVSVMGNGMVVYIFLSTKSLRTPSNLFVINLAISDFLMMFCMSPPM 0 0 VINCYYETWVLGPLFCQIYAMLGSLFGCGSIWTMTMIAFDRYNVIVKGLSGKPLSINGALIRIIAIWLFSLGWTIAPMFGWNR 2 1 YVPEGNMTACGTDYFNRGLLSASYLVCYGIWVYFVPLFLIIYSYWFIIQAVAAHEKNMREQAKKMNVASLRSSENQNTSAECKLAK 0 0 VALMTISLWFMAWTPYLVINFSGIFNLVKISPLFTIWGSLFAKANAVYNPIVYGIs2 1 HPKYRAALFAKFPSLACAAEPSSDAVSTTSGTTTVTDNEKSNA* 0 >RHAB2_apiMel Apis mellifera (bee) Gq 0.1.0.0.1.0.0.1 indel x x x x 386 aa 000 nm 16291092 NM_001077825 rhabdomeric Lop2 long wavelength ocelli 0 MDTLNITTSFFIEVMPSNISTLTTTGPQFARQLMRFNNQTVVSKVPEEMLHLIDLYW 2 1 YQFPPLDPLWHKILGLVMIILGIMGWCGNGVVVYVFIMTPSLRTPSNLLVVNLAFSDFIMMGFMCPPMVICCFYETW 0 0 VLGSLMCDIYAMVGSLCGCASIWTMTAIALDRYNVIVK 0 0 GMSGTPLTIKRAMLQILGIWLFGLIWTILPLVGWNR 2 1 YVPEGNMTACGTDYLSQDWTFKSYILVYSFFVYYTPLFTIIYSYYFIVS 0 0 AVAAHEKAMKEQAKKMNVTSLRSGDNQNTSAEAKLAK 0 0 VALTTISLWFMAWTPYLVINYIGIFNRSLITPLFTIWGSLFAKANAIYNPIVYGIS 2 1 HPKYRAALKEKLPFLVCGSTEDQTAATAGDKASEN* 0 >RHAB3_apiMel Apis mellifera (bee) Gq 0.1.1.0.1 indel x x x x 372 aa 000 nm 16291092 BK005513 rhabdomeric UV 0 MSNDSIHWEARYLPAGPPRLLGWNVPAEELIHIPEHWLVYPEPNPSLHYLLALLYILFTFLALLGNGLVIWIFCA 2 1 AKSLRTPSNMFVVNLAICDFFMMIKTPIFIYNSFNTGFALGNLGCQIFAVIGSLTGIGAAITNAAIAYDRYS 2 1 TIARPLDGKLSRGQVILFIVLIWTYTIPWALMPVMGVWGRFVPEGFLTSCSFDYLTDTNEIRIFVATIFTFSYCIPMILIIYYYSQIVSHVVNHEKALREQAKKMNVDSLRSNANTSSQSAEIRIAK 0 0 AAITICFLYVLSWTPYGVMSMIGAFGNKALLTPGVTMIPACTCKAVACLDPYVYAISHPKYR 2 1 LELQKRLPWLELQEKPISDSTSTTTETVNTPPASS* 0 >RHAB4_apiMel Apis mellifera (bee) Gq 0.1.1.1.2.0.1.1 indel x x x x 378 aa 000 nm 16291092 NM_001011606 rhabdomeric Blop blue 9502802 0 MLLHNKTLAGKALAFIAEEG 2 1 YVPSMREKFLGWNVPPEYSDLVHPHWRAFPAPGKHFHIGLAIIYSMLLIMSLVGNCCVIWIFST 2 1 SKSLRTPSNMFIVSLAIFDIIMAFEMPMLVISSFMERMIGWEIGCDVYSVFGSISGMGQAMTNAAIAFDRYR 2 1 TISCPIDGRLNSKQAAVIIAFTWFWVTPFTVLPLLKVWGRYTT 1 2 EGFLTTCSFDFLTDDEDTKVFVTCIFIWAYVIPLIFIILFYSRLLSSIRNHEKMLREQ 0 0 AKKMNVKSLVSNQDKERSAEVRIAKVAFTIFFLFLLAWTPYATVALIGVYGNR 2 1 ELLTPVSTMLPAVFAKTVSCIDPWIYAINHPR 2 1 YRQELQKRCKWMGIHEPETTSDATSAQTEKIKTDE* 0
Here are those sequences aligned (after some trimming of unalignable regions) to show rare genomic events such as indels and intron gains and losses:
>RHO1_homSap LAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLEGFFATLG GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWS >RHO1_monDom LAAYMFMLIVLGFPINFLTLYVTIQHKKLRTPLNYILLNLAIADLFMVFGGFTMTLYTSLHGYFVFGPTGCNLEGFFATLG GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIIGVAFTWVMALACAFPPLIGWS >RHO1_ornAna LAAYMFMLIMLGFPINFLTLYVTIQHKKLRTPLNYILLNLAFANHFMVLGGFTTTLYTSLHGYFVFGPTGCNIEGFFATLG GEIALWSLVVLAIERYIVVCKPMSNFRFGENHAIMGVAFTWIMALACALPPLVGWS >RHO1_galGal LAAYMFMLILLGFPVNFLTLYVTIQHKKLRTPLNYILLNLVVADLFMVFGGFTTTMYTSMNGYFVFGVTGCYIEGFFATLG GEIALWSLVVLAVERYVVVCKPMSNFRFGENHAIMGVAFSWIMAMACAAPPLFGWS >RHO1_anoCar LAAYMFLLILLGFPINFLTLFVTIQHKKLRTPLNYILLNLAVANLFMVLMGFTTTMYTSMNGYFIFGTVGCNIEGFFATLG GEMGLWSLVVLAVERYVVICKPMSNFRFGETHALIGVSCTWIMALACAGPPLLGWS >RHO1_xenTro LAAYMFLLILLGFPINFMTLYVTIQHKKLRTPLNYILLNLVFANHFMVLCGFTVTMYTSMHGYFIFGQTGCYIEGFFATLG GEMALWSLVVLAIERYVVVCKPMANFRFGENHAIMGVVFTWIMALSCAAPPLFGWS >RHO1_danRer VAAYMFFLIITGFPVNFLTLYVTIEHKKLRTPLNYILLNLAIADLFMVFGGFTTTMYTSLHGYFVFGRLGCNLEGFFATLG GEMGLKSLVVLAIERWMVVCKPVSNFRFGENHAIMGVAFTWVMACSCAVPPLVGWS >RHO1_Raja LAAYMFFLILTGLPVNFLTLFVTIQHKKLRQPLNYILLNLAVSDLFMVFGGFTTTIITSMNGYFIFGPAGCNFEGFFATLG GEVGLWCLVVLAIERYMVVCKPMANFRFGSQHAIIGVVFTWIMALSCAGPPLVGWS >RHO1_lamp1 LAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVANLFMVLFGFTLTMYSSMNGYFVFGPTMCNFEGFFATLG GEMSLWSLVVLAIERYIVICKPMGNFRFGSTHAYMGVAFTWFMALSCAAPPLVGWS >RHO1_lamp2 LAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAVSNLFMILFGFTTTMYTSMNGYFVFGPTMCSIEGFFATLG GEVSLWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVALTWVMALSCAAPPLLGWS >RHO1_lamp3 LAAYMFFLILVGFPVNFLTLFVTVQHKKLRTPLNYILLNLAMANLFMVLFGFTVTMYTSMNGYFVFGPTMCSIEGFFATLG GEVALWSLVVLAIERYIVICKPMGNFRFGNTHAIMGVAFTWIMALACAAPPLVGWS >RHO2_galGal VCCYIFFLISTGLPINLLTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFVFGPVGCAVEGFFATLG GQVALWSLVVLAIERYIVVCKPMGNFRFSATHAMMGIAFTWVMAFSCAAPPLFGWS >RHO2_anoCar VCCYIFFLIFTGLPINILTLLVTFKHKKLRQPLNYILVNLAVADLFMACFGFTVTFYTAWNGYFIFGPIGCAIEGFFATLG GQVALWSLVVLAIERYIVVCKPMGNFRFSATHALMGISFTWFMSFSCAAPPLLGWS >RHO2_Gekko LSFYMFFLIAAGMPLNGLTLFVTFQHKKLRQPLNYILVNLAAANLVTVCCGFTVTFYASWYAYFVFGPIGCAIEGFFATIG GQVALWSLVVLAIERYIVICKPMGNFRFSATHAIMGIAFTWFMALACAGPPLFGWS >RHO2_Lat LCAYMFLLIILGFPINFLTLLVTFKHKKLRQPLNYILVNLAVASLFMVVFGFTVTFYSSLNGYFVLGPMGCAMEGFFATLG GQVALWSLVVLAIERYIVVCKPMGNFRFASSHAIMGIAFTWIMALACAAPPLVGWS >RHO2_Geo ISAYVFTLILIGFPVNFMTLFVTFKLKKLRQPLNFILVNLCVADLLMIMFGFTTTFYTAMNGYFVFGPTGCNIEGFFATLG GEVSLWSLVMLAIERYIVVCKPMGNFRFATTHAALGVVFTWVMASACAVPPLVGWS >SWS1_homSap QAAFMGTVFLIGFPLNAMVLVATLRYKKLRQPLNYILVNVSFGGFLLCIFSVFPVFVASCNGYFVFGRHVCALEGFLGTVA GLVTGWSLAFLAFERYIVICKPFGNFRFSSKHALTVVLATWTIGIGVSIPPFFGWS >SWS1_macDom QTVFMGFVFCAGTPLNAVVLVATLRYKKLRQPLNYILVNVSLCGFIFCIFAVFTVFISSSQGYFIFGRHVCAMEAFLGSVA GLVTGWSLAFLAFERFIVICKPFGNFRFNSKHAMMVVLATWVIGIGVSIPPFFGWS >SWS1_galGal QTAFMGIVFAVGTPLNAVVLWVTVRYKRLRQPLNYILVNISASGFVSCVLSVFVVFVASARGYFVFGKRVCELEAFVGTHG GLVTGWSLAFLAFERYIVICKPFGNFRFSSRHALLVVVATWLIGVGVGLPPFFGWS >SWS1_Taenio QTIFMGLVFVAGTPLNAIVLIVTIKYKKLRQPLNYILVNISVSGLMCCVFCIFTVFIASSQGYFVFGKHMCAFEGFAGATG GLVTGWSLAFLAFERYIVICKPFGNFRFNSRHALLVVAATWIIGVGVAIPPFFGWS >SWS1_Gekko QTAFMGFVFFVGTPLNAIILFAIVKYKKLRQPLNYILVNISAAGFLFCVVAVFTVFISSSQGYFIFGKHICALEAFLGSLA GLVTGWSLAFLALERYIVICKPFGNFRFSAKHASLVVAATWFIGIGVSIPPYFGWS >SWS1_Utasta QTAFMGFVFFAGTPLNAIILIVTVKYKKLRQPLNYILVNISFAGFLFCVFSVFTVFLASSQGYFFFGRHICALEAFLGSVA GLVTGWSLAFLAFERYIVICKPFGNFRFNSKHALLVVAATWFIGIGVSIPPFFGWS >SWS1_Xenlae QAIFMGMVFLIGTPLNFIVLLVTIKYKKLRQPLNYILVNITVGGFLMCIFSIFPVFVSSSQGYFFFGRIACSIDAFVGTLT GLVTGWSLAFLAFERYIVICKPMGNFNFSSSHALAVVICTWIIGIVVSVPPFLGWS >SWS1_Danio QAAFMGFVFIVGTPMNGIVLFVTMKYKKLRQPLNYILVNISLAGFIFDTFSVSQVSVCAARGYYSLGYTLCSMEAAMGSIA GLVTGWSLAVLAFERYVVICKPFGSFKFGQGQAVGAVVFTWIIGTACATPPFFGWS >SWS1_Oryzia QAAFMGFVFFVGTPLNFVVLLATAKYKKLRVPLNYILVNITFAGFIFVTFSVSQVFLASVRGYYFFGQTLCALEAAVGAVA GLVTSWSLAVLSFERYLVICKPFGAFKFGSNHALAAVIFTWFMGVGCACPPFFGWS >SWS1_Geotri QAAFMGFVFICGTPLNAIVLVVTIKYKKLRQPLNYILVNISAAGLVFCLFSISTVFVASMQGYFFLGPTICALEAFFGSLA GLVTGWSLAFLAAERYIVICKPFGNFRFGSKHALVAVGLTWMLGLSVALPPFFGWS >SWS2_ornAna LAAFMFLLITLGFPINLLTVICTIKYKKLRSHLNYILVNLAVSNMLVVCVGSATAFYSFAHMYFVLGPTACKIEGFAATLG GMVSLWSLAVIAFERFLVICKPLGNLSFRGTHAIFGCAATWVFGLAASLPPLFGWS >SWS2_galGal MAAFMFLLIALGVPINTLTIFCTARFRKLRSHLNYILVNLALANLLVILVGSTTACYSFSQMYFALGPTACKIEGFAATLG GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCVATWVLGFVASAPPLFGWS >SWS2_Taenio MAAFMFLLVLLGVPINALTVLCTAKYKKLRSHLNYILVNLAVANLLVVCVGSTTAFYSFSQMYFALGPLACKIEGFTATLG GMVSLWSLAVVAFERFLVICKPLGNFTFRGSHAVLGCAITWIFGLIASLPPLFGWS >SWS2_Utasta MAAFMFLLIILGVPINVLTIFCTFKYKKLRSHLNYILVNLAVSNLLVVCIGSTTAFYSFAQMYFSLGPTACKIEGFAATLG GMVSLWSLAVVAFERFLVICKPLGNFSFRGTHAIIGCIITWVFGLVASLPPLFGWS >SWS2_Xenopu ISAFMLFTIIFGFPLNLLTIICTVKYKKLRSHLNYILVNLAVANLIVICFGSTTAFYSFSQMYFSLGTLACKIEGFTATLG GIIGLWSLAVVAFERFLVICKPMGNFTFRESHAVLGCILTWVIGLVAAIPPLLGWS >SWS2_Danio MSAFMLFLFIAGTAINVLTIVCTIQYKKLRSHLNYILVNLAISNLWVSVFGSSVAFYAFYKKYFVFGPIGCKIEGFTSTIG GMVSLWSLAVVALERWLVICKPLGNFTFKTPHAIAGCILPWCMALAAGLPPLLGWS >SWS2_Takifu MSAFMFFLFVAGTGINVLTIACTIQYKKLRSHLNYILVNLAFSNLLVTTVGSFTCFCCFFVRYMIVGPLGCKIEGFAATLG GMVSLWSLAVVAFERWLVVCKPLGNFIFKPDHAIVCCIFTWFFALIISAPPLFGWS >SWS2_Geotri MSAFMLFLVLAGFPLNFLTVFVTIKYKKLRSHLNYILVNLAIANLIVVCCGSTLAFYSFMHKYFILGPLFCKMEGFTATLG GMLSLWSLAVLAFERCLVICKPFGNIAFRGTHALIRCGFAWAAAIAASTPPLFGWS >LWS_ornAna TSLWMIFVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAVADLGETLIASTISVINQIFGYFILGHPMCVLEGYTVSLC GITGLWSLSIISWERWIVVCKPFGNVKFDAKLAMVGIVFSWVWAAVWTAPPIFGWS >LWS_galGal TSLWMIFVVAASVFTNGLVLVATWKFKKLRHPLNWILVNLAVADLGETVIASTISVINQISGYFILGHPMCVVEGYTVSAC GITALWSLAIISWERWFVVCKPFGNIKFDGKLAVAGILFSWLWSCAWTAPPIFGWS >LWS_anoCar TSVWMIFVVIASIFTNGLVLVATAKFKKLRHPLNWILVNLAIADLGETVIASTISVINQISGYFILGHPMCVLEGYTVSTC GISALWSLAVISWERWVVVCKPFGNVKFDAKLAVAGIVFSWVWSAVWTAPPVFGWS >LWS_Lithoch ATLWMFVVVVLSVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCIFEGYVVSVC GIAALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWVWAAVWCAPPIFGWS >LWS_Gastero STLWMFIVVALSVFTNGLVLVATAKFKKLQHPLNWILVNLAIADLGETVFASTISVCNQFFGYFILGHPMCVFEGYVVSVC GITALWSLTIISWERWIVVCKPFGNVKFDAKWATAGIVFSWIWSAVWCAPPIFGWS >LWS_Petrom TSVWMIIVVVLSLFSNGLVLVATVKFKKLRHPLNWIIVNLAIADILETIFASTISVCNQVYGYFILGHPMCVFEGYVVSTC GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIATILIVFSWVWPASWCSLPIFGWS >LWS_lamprey TSVWMIIVVVLSLFTNGLVLVATMKFKKLRHPLNWILVNLAIADILETIFASTISVCNQVFGYFILGHPMCVFEGYVVSTC GIAGLWSLAIISWERWMVVCKPFGNIKFDGKIAIILIVFSWVWPACWCSLPIFGWS >LWS_Geotria TSFWMIIVVILSLFTNGLVLVATLKFKKLRHPLNWILVNLAIADIGETIFASTVSVVNQIFGYFILGHPLCVFEGFTVSVC GITALWSLAIISFERWMVVCKPFGNLKFDGKVAIVLIIFSWAWSAGWCAPPIFGWS >PIN_galGal VAVLMGTVVACASVVNGLVIVVSICYKKLRSPLNYILVNLAVADLLVTLCGSSVSLSNNINGFFVFGRRMCELEGFMVSLT GIVGLWSLAILALERYVVVCRPLGDFQFQRRHAVSGCAFTWGWALLWSTPPLLGWS >PIN_UtaSta VAVLMGLVVVSAAFVNGLVIVVSIQYKKLRSPLNYILVNLAIADLLVTSFGSTLSFANNIYGFFVLGQTACEFEGFMVSLT GIVGLWSLAILAFERYLVICKPVGDFRFQQRHAVFGCVFTWMWSLVWTLPPLFGWS >PIN_pheMad LAALMGVVVLSASLANGLVIAVSVRFKRLRSPLNYILVNLATADLLVTFFGSIISFVNNAVGFFVFGKTACRFEGFMVSLT GIVGLWSLAILAFERYLVICKPVGDFQFQRRHAVIGCLYTWGWSLIWTVPPLFGWS >PIN_podSic VAVLMGLVVISATLVNGLVIVVSVQFKKLRSPLNYVLVNLAVADLLVTFFGSTISFVNNAQGFFIFGQATCEFEGFMVSLT GIVGLWSLAILAFERYLVICKPVGDFRFPARHAVLGCAFTWGWSFVWTVPPLLGWS >PIN_xenTro VAAVMCMVVILAFFVNGLVIVVTLKYKKLRSPLNYILVNLAIANLLVTIFGSSVSFSNNVVGYFFMGKTMCEFEGFMVSLT GIVGLWSLAILAFERYLVICKPMGDFRFQQKHAILGCSFTWVWSFIWTSPPLFGWC >PIN_bufJap VAVLMGMVVFLAFFVNGMVIVVSLKYKKLRSPLNYILVNLAVADILVTMFGSTVSFHNNIFGFFTLGKLVCELEGFVVSLT GIVGLWSLAILAFERYIVICKPMGDFRFQQRHAVMGCAFTWIWAFLWTSPPLIGWC >VAOP_galGal VAAVMFVVTSLSLAENLAVILVTFKFKQLRQPVNYVIVNLSVADFLVSLTGGTISFLANLKGYFYMGHWACVLEGFAVTFF GIVALWSLALLAFERYIVICRPVGNMRLRGKHAAQGIAFVWTFSFIWTIPPTMGWS >VAOP_anoCar ISALMFVVTLFSLSENFTVILVTIKFKQLRQPLNYVIVNLSVADFLVSLIGGTISFSTNLKGYFYMGHWACVLEGFAVTFF GIVALWSLALLAFERYVVICRPLGNMRLNGKHAALGVAFVWIFSFIWTVPPTMGWS >VAOP_xenTro LAALMFVVTSLSIAENFIVILVTAKFKQLRQPLNYIIVNLSVADFLVSVIGGTISIATNSRGYFYLGSWACVLEGFAVTFF GIVALWSLSVLAFERYIVICRPLGNLRLQGKHSALAIIFVWVFSFVWTIPPTMGWS >VAOP_danRer LAALMFVVTALSLSENFTVMLVTFRFQQLRQPLNYIIVNLSLADFLVSLTGGSISFLTNYHGYFFLGKWACVLEGFAVTFF GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLVFVWSFSFIWTVPPVLGWS >VAOP_rutRut LATLMFVVTAASLSENFAVMLVTFRFTQLRKPLNYIIVNLSLADFLVSLTGGTISFLTNYHGYFFLGKWACVLEGFAVTYF GIVALWSLAVLAFERFFVICRPLGNIRLRGKHAALGLLFVWTFSFIWTIPPVLGWS >VAOP_Petro LAALMGTITALSLGENFAVIVVTARFRQLRQPLNYVLVNLAAADLLVSAIGGSVSFFTNIKGYFFLGVHACVLEGFAVTYF GVVALWSLALLAFERYFVICRPLGNFRLQSKHAVLGLAVVWVFSLACTLPPVLGWS >PPIN_anaCar IAIIMATSCTLSVILNTAVIAITIKYRQLRQPINYSLVNLAIADLGAALLGGSLNVETNAVGYYNLGRVGCVTEGFAMAFF GIVALCTIAVIAVDRAIVIAKPMGTITFTTRKAMIGVAVSWIWSLVWNTPPLFGWG >PPIN_Xenop LALIMAVFCAAALFLNVTVIVVTFKYRQLRHPINYSLVNLAIADLGVTVLGGALTVETNAVGYFNLGRVGCVIEGFAVAFF GIAALCTIAVIALDRVFVVCKPMGTLTFTPKQALAGIAASWIWSLIWNTPPLFGWG >PPIN_Ictal LSIIMALSSTFGIILNMVVIIVTVRYKQLRQPLNYALVNLAVADLGCPVFGGLLTAVTNAMGYFSLGRVGCVLEGFAVAFF GIAGLCSVAVIAVDRYMVVCRPLGAVMFQTKHALAGVVFSWVWSFIWNTPPLFGWG >PPIN_Danio LAVIIGVFSVCGVILNVTVITVTLKYKQLRQPLNFALVNLAVADLGCAVFGGLPTVVTNAMGYFSLGRVGCVLEGFAVAFF GIAALCSVAVIALERCMVVCRPVGSISFQTRHAVFGVAVSWLWSFIWNTPPLFGWG >PPIN_Oncor LAVIIGVFSVSGVCMNVLVIMVTMRHRKLRQPLNYALVNLAVADLGCALFGGLPTMVTNAMGYFSMGRLGCVLEGFAVAFF GIAGLCSVAVIAVDRYVVVCRPMGAVMFQTRHAVGGVVLSWVWSFLWNTPPLFGWG >PPINa_Ciona LCVYMTFVFLLSCSLNILVIVATLKNKVLRQPLNYIIVNLAVVDLLSGFVGGFISIAANGAGYFFWGKTMCQIEGYFVSNF GVTGLLSIAVMAFERYFVICKPFGPVRFEEKHSIFGIVITWVWSMFWNTPPLIFWD >PPINb_Ciona LAVYMTFIFLLAVSLNGFVIIATMKNKKLRQPLNYIIINLSIADFLSGLVGGFIGMISNSAGYFYFGKTVCILEGYIVSVA GVCGLMSISVMAFERYFVVCKPYGPFTLTNTHAALGIGFTWTWSVLWSTPGLIWLD >PPIN_lamp LAVIMAVFTIASLVLNSTVVIVTLRHRQLRHPLNFSLVNLAVADLGVTVFGASLVVETNAVGYFNLGRVGCVIEGFAVAFF GIAALCTIAVIAVDRFVVVCKPLGTLMFTRRHALLGIAWAWLWSFVWNTPPLFGWG >PARIE_Utast LAFLMFLNALFSIFNNSLVIAVTLKNPQLRNPINIFILNLSFSDLMMSLCGTTIVIATNYYGYFYLGRKFCIFQGFAVNYF GIVSLWSLTILA YERYNVV--CQPLGTLQMSTKR GYQLLGFIWVFCLFWAVVPLFGWS >PARIE_Anole LAFLMFINALFSLFNNFLVIAVTLKNPQLRNPINIFILNLSFSDLMMSICGTTIVIATNYHGYFYLGRRFCIFQGFAVNYF GIVSLWSLTILA YERYNVV--CQPLGTLQMSTQR AYQLLGFIWVFCLFWAVVPLFGWS >PARIE_Xenop LSFLMFLNAVFSICNNAIVILVTLKHPQLRNPINIFILNLSFSDLMMALCGTTIVVSTNYHGYFYLGKQFCIFQGFAVNYF GIVSLWSLTLLA YERYNVV--CEPIGALKLSTKR GYQGLVFIWLFCLFWAIAPLFGWS >ENCEPH_braB VAGVIAIIGVVGFVSNGAVVVLFLKFPQLRTPFNLLLLNMAVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANHLF GLVSLISLAVIS FLRYRMVVKPKGPGSSYLTYTK VGLAILFIYLYCLLWTTLPIAGWS >ENCEPH_homS LALLLGSIGLLGVGNNLLVLVLYYKFQRLRTPTHLLLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSGSLF GIVSIATLTVLA YERYIRVV-----HARVINFSW AWRAITYIWLYSLAWAGAPLLGWN >ENCEPH_monD LALLIATIGLLGLCNNLLVLVLYYKFQRLRTPTHLFLVNISFNDLLVSLFGVTFTFVSCLRSGWVWDSVGCAWDGFSNTLF GIVSIMTLTVLA YERYNRIV-----HAKVINFSW AWRAITYIWLYSLVWTGAPLLGWN >ENCEPH_galG LALLIATIGTLGVCNNLLVLVLYYKFKRLRTPTNLFLVNISLSDLLVSVCGVSLTFMSCLRSRWVWDAAGCVWDGFSNSLF GIVSIMTLTVLA YERYIRVV-----HAKVIDFSW SWRAITYIWLYSLAWTGAPLLGWN >ENCEPH_anoC LALLVAAIGLLGLCNNLLVLVLYAKFKRLRTPTHLFLVNISLSDLLVSLFGVSFTFGSCLRHRWVWDAAGCVWDGFSNSLF GIVSIMTLTVLA YERYIRVV-----HARVIDFSW SWRAITYIWLYSLAWTGAPLLGWN >ENCEPH_xenT LALIVATVGFLGLVNNLLVLILYCKFKRLQTPTNLLFFNTSLCHFVFSLLAITFTFMSCVRGSWAFSVEMCVFHGFSKNLL GIVSFGTLTVVA YERYARVV-----YGKYVNSSW SKRSITFVWVYSLAWTGFPLIGWN >ENCEPH_braB IATGLALIGLVGSMNNFVVILLIGCHRQLRTPFNLLLLNVSVADLLVSVCGNTLSFASAVQHRWLWGRPGCVWYGFANSLF GIVSLVTLSALA FERYCVVV----RSSEMLTYKS SLGMIAFIWMYSLLWTSLPLLGWS >ENCEPH_braF IATCLALIGFVGFTNNFVVILLIGCHRQLRTPFNLLLLNMSVADLLVSVCGNTLSFASAVRHRWLWGRPGCVWYGFANSLF GIVSLVTLSALA FERYCVVV----RSSDMLTYKS SLVVITFIWLYSLLWTSLPLLGWS >ENCEPH2_Api AIALGFIGFFGFTANLLVAIVIVKDAQILWTPVNVILFNLVFGDFLVSIFGNPVAMVSAATGGWYWGYKMCLWYAWFMSTL GFASIGNLTVMA VERWLLVA----RPMQALSIRH AVILASFVWIYALSLSLPPLFGWG >ENCEPH1_Ano AAVTLFFIGFFGFFLNIFVIALMYKDVQLWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWLYGKSICVAYGFFMSLL GIASITTLTVLS YERFCLIS--RPFAAQNRSKQG ACLAVLFIWSYSFALTSPPLFGWG >ENCEPH2_Ano SAVTLFFIGFFGFFLNLFVIALMCKDMQLWTPMNIILFNLVCSDFSVSIIGNPLTLTSAISHRWIFGRTLCVAYGFFMSLL GITSITTLTVLS YERYCLIS--RPFSSRNLTRRG AFLAIFFIWGYSFALTSPPLFGWG >CILL2_Platy AIYLCIVGVIGTLSNGVIMYLYFKDKSLRSPMNLLFVNLAMSDFTVAFFGAMFQFGLTCTRKYMSPGMALCDFYGFITFLG GLASEMNLFIIS VERYLAVV--RPFDVGNLTNRR VIAGGVFVWLYSLVFAGGPLVGWS >CILL1_Platy AAYLFFIACLGVSLNVLVLVLFIKDRKLRSPNNFLYVSLALGDLLVAVFGTAFKFIITARKTLLREEDGFCKWYGFITYLG GLAALMTLSVIA FVR CLAV-LRLGSFTGLTTRM GVAAMAFIWIYSLAFTLAPLLGWN >MEL1_homSap LGTVILLVGLTGMLGNLTVIYTFCRSRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGETGCEFYAFCGALF GISSMITLTAIA LDRYLVIT-RPLATFGVASKRR AAFVLLGVWLYALAWSLPPFFGWS >MEL1_smiCra IGATILVVGFTGVLGNLLVIYTFCRSRSLRTPANMFIINLAISDFFMSFTQAPVFFASSLYERWIFGEKGCEFYAFCGALF GITSMITLMVIA LDRYFVIT-RPLASIGMISKKK TGLILLGVWLYSLAWSLPPFFGWS >MEL1_galGal IGTVILIVGITGTLGNFLVIYAFCRSRTLQKPANIFIINLAVSDFLMSITQSPVFFTNSLHKRWIFGEKGCELYAFCGALF GITSMITLMVIA LDRYFVIT-KPLASVRVMSKKK ALIILVGVWLYSLAWSLPPFFGWS >MEL1_xenTro VGAVILAVGITGMLGNFLVIYAFCRSRSLRSPANMFIINLAITDFLMSVTQAPVFFATSLHKRWIFGEKGCELYAFCGALF GITSMITLMVIA VDRYFVIT-RPLTSIGVMSKKR AVLILSGVWLYSLAWSLPPFFGWS >MEL1a_Bran VGTAVFCIGCCGMFGNAVVVYSFIKSKGLRTPANFFIINLALSDFLMNLTNMPIFAVNSAFQRWLLSDFACELYGFAGGLF GCLSINTLMAIS MDRYLVIT-KPFLVMRIVTKQR VMFAILLLWIWSLVWALPPLFGWS >MEL2_anoCar VGSCVLVIGCIGITGNLLVLYAFYSNKRLRTPPNYFIMNLAVSDFLMSATQAPICFLNSMHKEWVLGDIGCNLYAFCGALF GITSMITLLAIS VDRYCVIT-KPLQSIKRTSKKR TCIIIVFVWLYSLGWSVCPLFGWS >MEL2_xenLae IGSFILIIGSVGIIGNMLVLYAFYRNKKLRTAPNYFIINLAISDFLMSATQAPVCFLSSLHREWILGDIGCNVYAFCGALF GITSMMTLLAIS INRYIVIT-KPLQSIQWSSKKR TSQIIVLVWMYSLMWSLAPLLGWS >NEUR_homSap AGFYLTIIGILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGISVVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFF GCGSLITMTAVS LDRYLKIC--YLSYGVWLKRKH AYICLAAIWAYASFWTTMPLVGLG >RGR_homSap VLLVEALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLRVSHRRWPYGSDGCQAHGFQGFVT ALASICSSAAIA WGRYHHYC-----TRSQLAWNS AVSLVLFVWLSSAFWAALPLLGWG >PER_homSap VATYLIMAGMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQVYAGLNIFF GMASIGLLTVVA VDRYLTIC--LPDVGRRMTTNT YIGLILGAWINGLFWALMPIIGWA >PERa_Branc VGLYLFVIGIIGTIENGITLATFSKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLRSHWLFGGVGCQWYGFNGMFF
Here's the distal alignment of the opsins:
>RHO1_homSap RYIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE AAAQQQESATTQKAEK----EVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQ FRNCMLTTIC CGKNPLG DDEASATVS KTE TSQVAPA
>RHO1_monDom RYIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGQLVFTVKE AAAQQQESATTQKAEK----EVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSNFGPIFMTIPAFFAKSSSVYNPVIYIMMNKQ FRTCMITTLC CGKNPLG DDEASATAS KTE TSQVAPA
>RHO1_ornAna RYIPEGMQCSCGIDYYTLRPEVNNESFVIYMFVVHFTIPMTIIFFCYGRLVFTVKE AAAQQQESATTQKAEK----EVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTVPAFFAKSSAIYNPVIYIMMNKQ FRNCMLTTIC CGKNPLG DDEASATAS KTEQSSVSTSQVSPA
>RHO1_galGal RYIPEGMQCSCGIDYYTLKPEINNESFVIYMFVVHFMIPLAVIFFCYGNLVCTVKE AAAQQQESATTQKAEK----EVTRMVIIMVIAFLICWVPYASVAFYIFTNQGSDFGPIFMTIPAFFAKSSAIYNPVIYIVMNKQ FRNCMITTLC CGKNPLG DEDTSAG KTETSSVSTSQVSPA
>RHO1_anoCar RYIPEGMQCSCGVDYYTPTPEVHNESFVIYMFLVHFVTPLTIIFFCYGRLVCTVKA AAAQQQESATTQKAER----EVTRMVVIMVISFLVCWVPYASVAFYIFTHQGSDFGPVFMTIPAFFAKSSAIYNPVIYILMNKQ FRNCMIMTLC CGKNPLG DEDTSAGT KTETSTVSTSQVSPA
>RHO1_xenTro RYIPEGMQCSCGVDYYTLKPEVNNESFVVYMFIVHFTIPLCVIFFCYGRLLCTVKE AAAQQQESATTQKAEK----EVTRMVVMMVIFFLICWVPYAYVAFYIFTHQGSDFGPVFMTVPAFFAKSSAIYNPVIYIVLNKQ FRNCLITTLC CGKNPFG DEEGSSAASS KTEASSVSSSQVSPA
>RHO1_danRer RYIPEGMQCSCGVDYYTRTPGVNNESFVIYMFIVHFFIPLIVIFFCYGRLVCTVKE AARQQQESETTQRAER----EVTRMVIIMVIAFLICWLPYAGVAWYIFTHQGSEFGPVFMTLPAFFAKTSAVYNPCIYICMNKQ FRHCMITTLC CGKNPFE EEEGASTTAS KTEASSVSSSSVSPA
>RHO1_Raja RYIPEGLQCSCGVDYYTMKPEVNNESFVIYMFVVHFTIPLIVIFFCYGRLVCTVKE AAAQQQESESTQRAER----EVTRMVIIMVVAFLICWVPYASVAFYIFINQGCDFTPFFMTVPAFFAKSSAVYNPLIYILMNKQ FRNCMITTIC LGKNPFE EEESTSASAS KTEASSVSSSQVAPA
>RHO1_lamprey1 RYLPEGMQCSCGPDYYTLNPNFNNESFVIYMFLVHFIIPFIVIFFCYGRLLCTVKE AAAAQQESASTQKAEK----EVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTVPAFFAKTSALYNPIIYILMNKQ FRNCMITTLC CGKNPLG DEDSGASTS KTEVSSVSTSQVSPA
>RHO1_lamprey2 RYLPEGMQCSCGPDYYTMNPTYNNESFVIYMFIVHFTIPFVIIFFSYGRLLCTVKE AAAAQQESASTQKAEK----EVTRMVVLMVVGFLVCWVPYASVAFYIFTNQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ FRNCMITTLC CGKNPLG DDDSGASTS KTEVSSVSTSQVAPA
>RHO1_lamprey3 RYIPEGMQCSCGPDYYTLNPNFNNESYVVYMFVVHFLVPFVIIFFCYGRLLCTVKE AAAAQQESASTQKAEK----EVTRMVVLMVIGFLVCWVPYASVAFYIFTHQGSDFGATFMTLPAFFAKSSALYNPVIYILMNKQ FRNCMITTLC CGKNPLG DDESGASTS KTEVSSVSTSQVSPA
>RHO2_galGal RYMPEGMQCSCGPDYYTHNPDYHNESYVLYMFVIHFIIPVVVIFFSYGRLICKVRE AAAQQQESATTQKAEK----EVTRMVILMVLGFMLAWTPYAVVAFWIFTNKGADFTATLMAVPAFFSKSSSLYNPIIYVLMNKQ FRNCMITTIC CGKNPFG DEDVSSTVSQSKTEVSSVSSSQVSPA
>RHO2_anoCar RYIPEGMQCSCGPDYYTLNPDYHNESYVLYMFGVHFVIPVVVIFFSYGRLICKVRE AAAQQQESASTQKAER----EVTRMVILMVLGFLLAWTPYAMVAFWIFTNKGVDFSATLMSVPAFFSKSSSLYNPIIYVLMNKQ FRNCMITTIC CGKNPFG DEDVSSSVSQSKTEVSSVSSSQVSPA
>RHO2_Gekko RFIPEGMQCSCGPDYYTLNPDFHNESYVIYMFIVHFTVPMVVIFFSYGRLVCKVRE AAAQQQESATTQKAEK----EVTRMVILMVLGFLLAWTPYAATAIWIFTNRGAAFSVTFMTIPAFFSKSSSIYNPIIYVLLNKQ FRNCMVTTIC CGKNPFG DEDVSSSVSQSKTEVSSVSSSQVAPA
>RHO2_Latime RYIPEGLQCSCGPDYYTLNPDFHNESYVMYLFLVHFLLPIIIIFFTYGRLICKVKE AAAQQQESASTQKAEK----EVTRMVILMVIGFLTAWVPYASAAFWIFCNRGAEFTATLMTVPAFFSKSSCLFNPIIYVLLNKQ FRNCMITTLC CGKNPLG DDDTSSAVSQSKTDVSSVSSSQVSPA
>RHO2_Geot RYIPEGMQCSCGPDYYTLNPKYYNESYVIYLFLVHFLLPVTIIFFTYGRLICTVKE AAAQQQESASTQKAER----EVTRMVIIMVVGFLVCWVPYASFAFYLFMNKGILFSATAMTVPAFFSKSSVLYNPIIYVLLNKQ FRTCMVTTLF CGKNPFG EDDSSMVSTS KTEVSSVSSSQVSPS
>SWS1_homSap RFIPEGLQCSCGPDWYTVGTKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKA VAAQQQESATTQKAER----EVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHGLDLRLVTIPSFFSKSACIYNPIIYCFMNKQ FQACIMKMV CGKAMTDESDTCSSQ KTEVSTVSSTQVGPN
>SWS1_macDom RFIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIMPLFLICFSYSQLLRALRA VAAQQQESATTQKAER----EVSRMVVMMVGSFCLCYVPYAALAMYMVNNQNHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ FHACIMEMV CRKPMTDDSDVSSSQ KTEVSAVSSSQVGPT
>SWS1_galGal RYMPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA VAAQQQESATTQKAER----EVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGLDLRLVTIPAFFSKSACVYNPIIYCFMNKQ FRACIMETV CGKPLTDDSDASTSAQ RTEVSSVSSSQVGPT
>SWS1_Taeni RYIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLSLIIFSYSQLLSALRA VAAQQQESATTQKAER----EVSRMVVVMVGSFCMCYVPYAALAMYMVNNREHGIDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ FRACIMETV CGRPMTDDSEVSSSAQ RTEVSSVSSSQVGPS
>SWS1_Gekko RFIPEGLQCSCGPDWYTVGTKYYSEYYTWFLFVLCFIVPLSIIVFSYSQLLSALRA VAAQQQESATTQKAER----EVSRMVVVMVGSFCLCYVPYAALAMYMVNNRNHGIDLRMVTIPAFFSKSSCVYNPIIYCFMNKQ FRGCILEMV CGKTMAEESEVSSASQ KTEVSSVSSSQVGPS
>SWS1_Uta RFIPEGLQCSCGPDWYTVGTKYKSEYYTWFLFIFCFIVPLTLIIFSYSQLLGALRA VAAQQQESATTQKAER----EVSRMVVVMVGSFCLCYVPYAALAMYMVNNRDHGIDLRLVTIPAFFSKSACVYNPIIYCFMNKQ FRACIMETV CGKPMTDESDVSSSAQ KTEVSSVSSSQVSPS
>SWS1_Xenop RYMPEGLQCSCGPDWYTVGTKYRSEYYTWFIFIFCFVIPLSLICFSYGRLLGALRA VAAQQQESASTQKAER----EVSRMVIFMVGSFCLCYVPYAAMAMYMVTNRNHGLDLRLVTIPAFFSKSSCVYNPIIYSFMNKQ FRGCIMETV CGRPMSDDSSVSSTSQ RTEVSTVSSSQVSPA
>SWS1_Danio RYIPEGLGCSCGPDWYTNCEEFSCASYSKFLLVTCFICPITIIIFSYSQLLGALRA VAAQQAESASTQKAEK----EVSRMIIVMVASFVTCYGPYALTAQYYAYSQDENKDYRLVTIPAFFSKSSCVYNPLIYAFMNKQ FNACIMETV FGKKIDESSEVSS KTETSSVSA
>SWS1_Oryzia RYIPEGLQCSCGPDWYTVGTKYKSEYYTYFLFVFCFVVPLSIIIFSYGSLLGTLRA VAAQQQESASTQKAER----EVSRMVIMMVASFCTCYVPYAALAVYMVTNRDHNIDLRFVTVPAFFSKASCVYNPLIYSFMNKQ FNGCIMEMV FGKKMEEASEVSS KTEVSTDS
>SWS1_Geotri RYIPEGLQCSCGPDWYTVGTKYKSEYYTYFLFVFCFVVPLSIIIFSYGSLLGTLRA VAAQQQESASTQKAER----EVSRMVIMMVASFCTCYVPYAALAVYMVTNRDHNIDLRFVTVPAFFSKASCVYNPLIYSFMNKQ FRACILETV CGKPITDESETSSS RTEVSSVSTTQMIPG
>SWS2_ornAna RYIPEGLQCSCGPDWYTTNNKWNNESYVIFLFSFCFGVPLSIIIFSYGRLLLTLRA VAKQQEQSATTQKAER----EVTKMVIVMVLGFLVCWLPYASFSLWVVTNRGQVFDLRMASIPSVFSKASTIYNPIIYVFMNKQ FRSCMLKLVF CGKSPFGDEDEISGSS QATQVSSVSSSQVSPA
>SWS2_galGal RYIPEGLQCSCGPDWYTTDNKWHNESYVLFLFTFCFGVPLAIIVFSYGRLLITLRA VARQQEQSATTQKADR----EVTKMVVVMVLGFLVCWAPYTAFALWVVTHRGRSFEVGLASIPSVFSKSSTVYNPVIYVLMNKQ FRSCMLKLLF CGRSPFGDDEDVSGSS QATQVSSVSSSHVAPA
>SWS2_Taeni RYIPEGLQCSCGPDWYTTDNKWNNESYVIFLFCFCFGFPLTVIVFSYGRLLLTLRA VAKQQEQSASTQKAER----EVTKMVVVMVLGFLVCWLPYCSFALWVVTHRGHPFDLGLASIPSVFSKASTVYNPIIYVFMNKQ FRSCMLKLVF CGRSPFGDEDDVSGSS QATQVSSVSSSQVSPA
>SWS2_Uta RYIPEGLQCSCGPDWYTTNNKWNNESYVLFLFSFCFGVPLSVIIFSYGRLLLTLRA VAKQQEQSATTQKAER----EVTKMVVVMVMGFLVCWLPYASFALWVVTHRGEPFDVRLATIPSVFSKASSVYNPVIYVFMNKQ FRSCMLKLVF CGKSPFGDEDDVSGSS QTTQVSSVSSSQVSPA
>SWS2_Xeno RYIPEGLQCSCGPDWYTVNNKWNNESYVLFLFCFCFGFPLAIIVFSYGRLLLALHA VAKQQEQSATTQKAER----EVTRMVIVMVVGFLVCWLPYASFALWAVTHRGELFDLRMSSVPSVFSKASTVYNPFIYIFMNRQ FRSCMMKMIF CGKNPLGDDEETSVSG STQVSSVSSSQIAPS
>SWS2_Danio RYIPEGLQCSCGPDWYTTNNKFNNESYVMFLFCFCFAVPFSTIVFCYGQLLITLKL AAKAQADSASTQKAER----EVTKMVVVMVFGFLICWGPYAIFAIWVVSNRGAPFDLRLATIPSCLCKASTVYNPVIYVLMNKQ FRSCMMKMVF NKNIEEDEASSSS QVTQVSSVAPEK
>SWS2_Takif RYIPEGFQCSCGPDWYTTGNKYNNESYVWFIFGFGFAVPLFVIVFCYSQLLVMLKS AAKAQAESASTQKAER----EVTRMVVVMILGFLVCWLPYASFALWVVNNRGTPFDLRLATIPACFSKASTVYNPIIYVVLNKQ FRSCMKKMLG MSGGDDEESSS QSVTEVSKVSPS
>SWS2_Geotria RYIPEGLQCSCGPDWYTTNNKYNNESYVMFLFIFCFGTPFTIIIVSYSKLILTLRA AAAQQQESASTQKAEK----EVSRMVVIMVGGFLVCWLPYASLALWIVFNRGSPFDLRLATIPSVFSKASTVYNPVIYIFLNKQ FRSCMMKTIF CGKNPLGDDEDATSTTT QVSSVSTSQVAPA
>LWS_ornAna RYWPHGLKTSCGPDVFSGSSDPGVQSYMIVLMSTCCILPLSIIVLCYLQVWLAIRA VAKQQKESESTQKAEK----EVSRMVVVMILAYCFCWGPYTIFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ FRNCIMQL FGKKVDDGSELSSTS RTEVSSVSS VSPA
>LWS_galGal RYWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFFPLAIIILCYLQVSLAIRA VAAQQKESESTQKAEK----EVSRMVVVMIVAYCFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ FRNCILQL FGKKVDDGSEVSTS RTEVSSVSNSSVSPA
>LWS_anoCar RYWPHGLKTSCGPDVFSGSDDPGVLSYMIVLMITCCFIPLAVILLCYLQVWLAIRA VAAQQKESESTQKAEK----EVSRMVVVMIIAYCFCWGPYTVFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ FRNCIMQL FGKKVDDGSELSSTS RTEVSSVSNSSVSPA
>LWS_Lithoch RYWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMLTCCIFPLAIIILCYLAVWMAIRA VAMQQKESESTQKAER----EVSRMVVVMIVAYCVCWGPYTFFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPIIYVFMNRQ FRTCIMQL FGKQVDDGSEVSTS KTEV SSVAPA
>LWS_Gastero RYWPHGLKTSCGPDVFSGSEDPGVQSYMIVLMITCCLIPLAIIILCYLAVWLAIRA VAMQQKESESTQKAER----DVSRMVVVMIVAYIVCWGPYTTFACFAAANPGYAFHPLAAAMPAYFAKSATIYNPVIYVFMNRQ FRSCIMQL FGKEVDDGSEVSTs KTEV SSVAPA
>LWS_Petromy RYWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFLPLSIIILCYLQVWLAIHS VAQQQKESETTQKAER----DVSRMVVVMILAYVFCWGPYTFFACFAAANPGYSFHPIAAALPAYFAKGATIYNPIIYVFMNRQ FRNCILQL FGKKVDDGSEVSSSS RTEVSSVSNSSVSPA
>LWS_lamprey RYWPHGLKTSCGPDVFSGSSDPGVQSYMVVLMVTCCFLPLSVIILCYLQVWLAIHS VAQQQKESETTQKAER----DVSRMVVVMILAYIFCWGPYTFFACYAAANPGYAFHPLTAALPAYFAKSATIYNPVIYVFMNRQ FRNCIMQL FGKKVDDGSEVSSAS RTEVSSVSNSSISPA
>LWS_Geotria RYWPHGLKTSCGPDVFSGSTDPGVQSYMVVLMITCCFIPLALIIICYLQVWLAIHT VAQQQKESETTQKAER----DVSRMVVVMIFAYIFCWGPYTFFACFAAANPGYAFHPLAAALPAYFAKSATIYNPIIYVFMNRQ FRNCIMQL FGKKVDDGSEVSSSA RTEVSSVSNSSVSPA
>PIN_galGal SYVPEGLRTSCGPNWYTGGSNNN--SYILSLFVTCFVLPLSLILFSYTNLLLTLRA AAAQQKEADTTQRAER----EVTRMVIVMVMAFLLCWLPYSTFALVVATHKGIIIQPVLASLPSYFSKTATVYNPIIYVFMNKQ FQSCLLEMLC CGYQPQRTGKASPGTPGPHADVTAAGLRNKVMPAHPV
>PIN_Uta sta SYVPEGLRTSCGPNWYTGGSGNN--SYIMALFVTCFALPLGMIIFSYASLLLTLRA VATQQKEVETTQQAEK----EVTRRVIAMVMAFLVCWLPYASFAMVVATNKDLVIQPALASLPSYFSKTATVYNPIIYVFMNKQ FRSCLLSTMS CGHRPRGAQETTPAMISIPQGPTSALQGSRNKVTPSA
>PIN_Phelsuma SYVPEGLGTSCGPNWYMGGTNNN--SYIVALFVTCFALPLSMILFSYANLLLTLRA VAAQQKEQETTQRAEK----EVTRMVITMVMAFLVCWLPYATFAMVVATTKDLSIQPGLASLPSYFSKTATVYNPIIYVFMNKQ FRSCLLNTVS CGRIPQTMPGTPATTAVRGGFVLTSEGRGNKVASTEL
>PIN_Podarcis SYVPEGLRTSCGPNWYSGGSSNN--SYIMTLFVTCFAMPLSTILFSYANLLMTLRT VAAQQKEQETTQRAER----EVTRMVVAMVAAFLVCWLPYASFAMVVATHKDLAIRPALASLPSYFSKTATVYNPIIYVFMNKQ FRSCLLYKMS CGHRALSSQDTTPAGISLPGRLTTSASKGSRNQVSPS
>PIN_xenTro SYVPEGLRTSCGPNWYTGGTNNN--SYIMALFLTCFIMPLSTIIFSYSNLLMALRA VAAQQKDSETTQRAEK----EVTRMVIAMVLAFLICWLPYASFAVVVAVNKDVVIEPTVASLPSYFSKTATVYNPIIYVFMNKQ FRNCLMTLLC CGRSFGDDETSSASGRTDVTSVSEAGGNKVTPA
>PIN_Bufo SYVPEGLGTSCGPNWYTGGTNNN--SYILALFTTCFMMPLTTIIFSYSNLLLALRA VAAQQKESETTQRAER----EVTRMVIAMVLAFLICWLPYAVFAIVMASNKNVVIDPTLASMPSYFSKTATVYNPVIYVFMNKQ FRDCLTKLLC CGRNPFGEDETSTTSGRTDVTSVSEGGGNKVTPA
>CILL2_Platyn SYRPEGLGTWCSISWQDRSMNTM--SYVTAVFLGCYFFPVSIIIFCYFNVWRKVKE AADAQGAGTAGKAEKS-----IFRMSVIMVTCYLTAWTPYAIVCLIASYGPPNGLPIYAEVLPSLFAKSSQVYNPIIYVLMNKP
>ENCEPH_braBel SYQFEGHSVGCSVNWVKHNVNNV--SYIITLMVTCFFVPMVVVCWSYACIWRTVRM SAEMKSEFGNPQNTGR----LVTTMVVVMIVCFLVCWTPYTVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ FREFLLARLRTFCCRQPRMLRVTPMDDNAHARLVGEGPSHAQQVIPSEEN
>ENCEPH_braFlo SYQFEGHNVGCSVNWVQHNPDNV--SYIVTLMVTCFFVPMVVVCWSYAWIWRTVRM SSEAKPECGNSQNAGR----LVTTMVVVMIICFLVCWTPYAVMALIVTFGADHLVTPTASVIPSLVAKSSTAYNPIIYVLMNNQ FREFLLARLQRVCCRQQAVPRVTPMDDNVHVRLGGEGPSQSQQFLPAGEN
>VAOP_galGal SYTTSKIGTTCEPNWYSGAYNDR--SYIIAFFTTCFIVPLLVILVSYGKLLQKLRK VSNTQGRLRTARKPER----QVTRMVVVMIIAFLICWMPYAVFSILATAYPSIELDPHLAAIPAFFSKTATVYNPIIYVFMNKQ FRMCLIQMFK CSAIETAESNMNPTSERATLTQDKRDSQLSVMAVRST
>VAOP_anoCar SYTTSKIGTTCEPNWYSGDYNDH--TFIITFFTTCFILPLLVILVSYGKLMRKLRK VSDTQGRLGTTRKPER----QVTGMVVIMILAFLICWSPYAAFSILVTACPSIELDPRLAAIPAFFSKTATVYNPVIYVFMNNQ FRKCLVQLFQ CSSQETMDANVNPISEKDTLTHTKHCGEMSTVAAHVI
>VAOP_xenTro SYTTSKIGTTCEPNWYSGEMRDH--TYIITFLTTCFVFPLLVIFMSYGKLMRKLRK VSDTQGRLGSTRKPEK----EVTRMVVIMILAFLICWTPYAAFSILITAHPTIDLDPRLAAIPAFFAKTASMYNPIIYVYMNKQ FRRCLYQMFN INDPEAKESNLNPTSERGVLTRNNNGGEMLAIATHIT
>VAOP_Danio SYTVSRIGTTCEPNWYSGNFHDH--TFIITLFSTCFIFPLGVIIVCYCKLIRKLRK VSNTHGRLGNARKPER----QVTRMVVVMIVAFMVAWTPYAAFSIIITAHPSMHVDPRLAAIPAFVAKTAAVYNPIIYVFMNKQ FRKCLVQLLS CSKVTVVEGNNNQTTERAGMTSGSNTGEMSAIAARVS
>VAOP_Ruti SYTVSKIGTTCEPNWYSGNFHDH--TFIIAFFITCFILPLGVIVVCYCKLIKKLRK VSNTHGRLGNARKPER----QVTRMVVVMIVAFMVAWTPYAAFSIVVTAHPSIHLDPRLAAAPAFFSKTAAVYNPVIYVFMNKQ FRKCLVQLLR CRDVTIIEGNINQTSERQGMTNESHTGEMSTIASRIP
>VAOP_Petrom SYRPSMIGTTCEPNWYSGELHDH--TFILMFFSTCFIFPLAVIFFSYGKLIQKLKK ASETQRGLESTRRAEQ----QVTRMVVVMILAFLVCWMPYATFSIVVTACPTIHLDPLLAAVPAFFSKTATVYNPVIYIFMNKQ FRDCFVQVLP CKGLKKVSATQTAGAQDTEHTASVNTQSPGNRHNIAL
>PPIN_anaCar GYQMEGVMTSCAPDWANSDPINV--SYIICYFLFCFTIPFITILASYGYLIWTLRQ VAKVGLAQRGSTTKAEA---QVSRMVIVMVMAFLICWLPYATFALVVVGNPQIYINPIIATIPMYMAKSSTFYNPIIYIFMNKQ FRDCLVRCLL CGRNPCASEQTDEDDLEVSTIAPAPSSRRGKVAPV
>PPIN_Xeno SYELEGVMTSCAPNWYSADPVNM--SYIVCYFSFCFAIPFLIIVGSYGYLMWTLRQ VAKLGVAEGGTTSKAEV---QVSRMVIVMILAFLVCWLPYAAFAMTVVANPGMHIDPIIATVPMYLTKTSTVYNPIIYIFMNKQ FQECVIPFLF CGRNPWAAEKSSSMETSISVTSGTPTKRGQVAPA
>PPIN_Icta SYQLEGVMTSCAPNWYRRDPVNV--SYILCYFMLCFALPFATIIFSYMHLLHTLWQ VAKLQVADSGSTAKVEV---QVARMVVIMVMAFLLTWLPYAAFALTVIIDSNIYINPVIGTIPAYLAKSSTVFNPIIYIFMNRQ FRDYALPCLL CGKNPWAAKEGRDSDTNTLTTTVSKNTSVSPL
>PPIN_Danio RLQLEGVRTSCAPDWYSRDLANV--SFIVCYFLLCFALPFSVIVYSYTRLLWTLRQ VSRLQVCEGGSAARAEA---QVSCMVVVMILAFLLTWLPYASFALCVILIPELYIDPVIATVPMYLTKSSTVFNPIIYIFMNRQ FRDRALPFLL CGRNPWAAEAEEEEEETTVSSVSRSTSVSPA
>PPIN_Oncorhy SFELEGVRTSCSPNWYSREPGNM--SYIILYFLLCFAIPFSIIMVSYARILFTLHQ VSKLKVLEGNSTTRVEI---QVVRMVVVMVMAFLLSWLPYAAFALSVILDPSLHINPLIATVPMYLAKSSTVYNPIIYVFMNRQ FRDCAVPFLL CGLNPWASEPVGSEADTALSSVSKNPRVSPQs
>PPIN_lamp SYELEGVRTSCAPDWYSRDPANV--SYITSYFAFCFAIPFLVIVVAYGRLMWTLHQ VAKLGMGESGSTAKAEA---QVSRMVVVMVVAFLVCWLPYALFAMIVVTKPDVYIDPVIATLPMYLTKTSTVYNPIIYIFMNRQ FRDCAVPFLL CGRNPWAEPSSESATAASTSATSVTLASAPGQVSPS
>PARIE_Uta SYGPEGVQTSCSIGWEERSWSNY--SYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG LNKKVEQLGGKSSPEEEF--RAVIMVLVMVVAFLICWLPYTVFALIVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKQ FRDCAVEFIT CGQVVLTSPEEDISTSAIPVEGKGPCKINQVTPV
>PARIE_Anole SYGPEGVQTSCSIGWEERSWNNY--SYLIVYFLSCFFIPVLIIGFSYGNVIRSLHG LNKKVEQLGGKSNPEEEF--RAVIMVLVMVVAFLICWLPYTLFALTVVFNPALNISPLAATIPTYLSKTSPVYNPIIYIFLNKE FRECAVEFIT CGKVVLTSPEEDISTSAISDEGIAPCKINQVTPV
>PARIE_Xenop SYGPEGVQTSCSIGWEERSWSNY--SYIISYFLTCFIIPVGIIGFSYGSILRSLHQ LNRKIEQQGGKTNPREEK--RVVIMVLFMVLAFLICWLPYTVFALIVVINPQLYISPLAATLPTYFAKTSPVYNPIIYIFLNKQ FRTYAVQCLT CGHINLDSLEEDTESVSAQAENMLTPKTNQVAPA
>ENCEPH_braBel SYQLEGPKIGCSVAWEEHSWSNT--SYIVVLFITCLFAPLLIIVYSYYRLWHKVKQ GSRNLPAAMRKSSQKEQ---KIAMMVIVMITCFMVCWLPYGAMALVVTFGGERLISHTAAVVPSLLAKSSTCYNPVVYFAMNSQ FRRYFQDLLC CGRRLFDVSQSVVTGNTAMPRNNSQGFRKDDSDQKQD
>ENCEPH_homSap RYILDVHGLGCTVDWKSKDANDS--SFVLFLFLGCLVVPLGVIAHCYGHILYSIRM LRCVEDLQTIQVIKILKYEKKLAKMCFLMIFTFLVCWMPYIVICFLVVNGHGHLVTPTISIVSYLFAKSNTVYNPVIYVFMIRK FRRCLLQLLC FRLLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVT
>ENCEPH_monDom RYTLEIHGLGCSVDWKSKDPNDS--SFVIFLFFGCLMLPVGVMAYCYGHILYAIRM LRCVEELQTIQVIKILRYEKKVAKMCFLMIAIFLFCWMPYAVICLLVANGYGSLVTPTVAIIASLFAKSSTAYNPIIYIFMSRK FRRCLLQLLC FRLLKFQQPKKDRPVIRTEKQIRPIVMSQKVGDRPKKKVT
>ENCEPH_galGal RYTLEIHGLGCSMDWKSKDPNDT--SFVLLFFLGCLVAPVVIMAYCYGHILYAVRM LRCVEDFQTSQVIKLLKYEKKVAKMCFLMISTFLICWMPYAVVSLLVTYGYSNLVTPTVAIIPSFFAKSSTAYNPVIYIFMSRK FRQCLLQLLC FRLMRFQRIMKEPSGAGNVKPIRPIVMSQKVGDRPKKKVT
>ENCEPH_anoCar HYTLEIHGLGCSVDWQSKEPSDS--SFVLFFFLGCLAAPVGIMAYCYGHILHAIRM LRCVEDLQSIQVIKILRYEKKVAKMCFLMVTTFLICWMPYAVVSLLIAYGYGHLITPTVAIIPSFFAKSSTAYNPVIYIFMSRK FRRCLVQLFC VQFLRFKRTLKEQPAIESNKPIRPIVMSQKVGDRPKKKVT
>ENCEPH_xenTro LYTFETHKLDCSFEWTATDPKDT--AFVLLFFLACITLPLSIMAYCYGYILYEIQK LRSVKNIQNFQEITILDYEIKMAKMCLLMMLTFLIGWMPYTILSLLVTSGYSKFITPTITVMPSLLAIASAAYNPVIHIFTIKK FRQCLVQLLP PINFHPPINPPINNFWRLLKNLNGRLAMKKVKPVLGKGRS
>ENCEPH1_Anoph AYVNEAANISCSVNWESQTANAT--SYIIFLFIFGLILPLAVIIYSYINIVLEMRK NSARVGRVNRAERRVT-------SMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFWRIRR SNGVAGQPDSNNTNNSNRDKESARHTAKEGL
>ENCEPH2_Anoph AYVQEAANISCSVNWESQTKNAT--TYIIFLFVFGLVVPLIVIVYSYTNIIVNMRE NSARVGRINRAEQRVT-------SMVAVMIVAFMVAWTPYAIFALIEQFGPPELIGPGLAVLPALVAKSSICYNPIIYVGMNTQ FRAAFSRVRN KGQQAAADQNTTTMQRELTKSSRDMVECSF
>ENCEPH2_Apis SYGPEAGNVSCSVSWEVHDPVTNSDTYIGFLFVLGLIVPVFTIVSSYAAIVLTLKK VRKRAGASGRREAKIT-------KMVALMITAFLLAWSPYAALAIAAQYFNAKPSATV-AVLPALLAKSSICYNPIIYAGLNNQ FSRFLKKIFD ARGSRTAVPDSQHTALTALNRQEQRK
>CILL1_Platyn HYIPEGLATWCSIDWLSDETSDK--SYVFAIFIFCFLVPVLIIVVSYGLIYDKVRK VAKTGGSVAKAEREVL-------RMTLLMVSLFMLAWSPYAVICMLASFGPKDLLHPVATVIPAMFAKSSTMYNPLIYVFMNKQ FRRSLKVLLG MGVEDLNSESERATGGTATNQVAAT
>PER_homSap SYAPDPTGATCTINWRKNDRSFV--SYTMTVIAINFIVPLTVMFYCYYHVTLSIKH HTTSDCTESLNRDWSD--QIDVTKMSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK FRRAMLAMFK CQTHQTMPVTSILPMDVSQNPLASGRI
>PERa_Bran EYALEPSGTACTINFQKNDSLYI--SYVTSCFVLGFVVPLAVMAFCYWQASCFVSK VLKGDIAGDLTFPVAAN.QNHFSKMCLAMVAAFVVAWTPYSVLFLFAAFWNPADIPAWLTLLPPLIAKSSALYNPIIYIIANRR FRNAICSMMK GQDPDVEDDEHADEHRVRSIEDNDKEIISMVNLNMTV
>PERc_Bran YTYETPMQITCSLDWNVQHPGEK--AYIAAVLVIVYVLQVLIMCFCYFNIIFKSAN LKFAALASEKTKMAAKKDTWKTSVMCLTMVVSFLIAWTPYAVSSTWDILSAE-DLPIIATILPSLFAKSSCMMNPIIYACCNTK FRQAAVKSFR KLCGMCKQKVPLSTPQVVLAMQRNTEFTSTVEPT
>NEUR_homSap DYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKS SSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYK FACCQTGGLK ATKKKSLEGFRLHTVTTVRKSSAVLEIHEEWE
>MEL_Platy AYIPEGFQTSCTYDYLTQDMNNY--TYVLGMYLFGFIFPVAIIFFCYLGIVRAIFA HHAEMMATAKRMGAN...EIQIAKVAAMTIGTFMLSWTPYAVVGVFGMIKPHSEMFIH.AEIPVMMAKASARYNPIIYALSHPK FRAEIDKHFPWLLCCCKPKPKAQLPSSTTKGSIASKTEADTSV
>MEL1_homSap AYVPEGLLTSCSWDYMSFTPAVR--AYTMLLCCFVFFLPLLIIIYCYIFIFRAIRE TGRALQTFGACKGNG.QSECKMAKIMLLVILLFVLSWAPYSAVALVAFAGYAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPK YRVAIAQHLP CLGVLLGVSRRHSRPYPSYRSTHRSTLTSHTSNL
>PPINa_Ciona YDTEGLGTSCAPNWFVKEKRERL--FIILYFVFCFVIPLAVIMICYGKLILTLRQ IAKESSLSGGTSPEGEVTKMVVVMVTAFVFCWLPYAAFAMYNVVNPEAQ IDYALGAAPAFFAKTATIYNPLIYIGLNRQ FRDCVVRMIF NGRNPWVDELVGSQVSSTGSQLTAVSSNKVAPA
>PPINb_Ciona GYVPEGLGTSCAPNWFSKNKSER--IFIFVYFVFCFFIPLLVIIICYGKIVLFLKQVSLY ATRQSSASSNRQADNKVTKMVLVMISAFLICWTPYGVLSLYNAINPDKQ LDYGLGAVPVFFAKTANIYNPLIYIGLNKQ FRDGVIKMVF RGRNPWAEEMSTQQRQRSTEAGQPIVSNEV
>PPIN2_Ciona SVIWHTPGLFFWNGYEPEGFGTS--CAPNWFSQQKRERIFIFAYFAFCFLTPLTIIFACYLKLILFIRKVSVSKKSMVNEADRRDFEVTRMVFVMIAAFLICWLPYGCLSMYNAIHPD FRDGVIRMLF KGRNPWLDGRNTTSSTSTRAQ
Structural and functional markers along the opsin molecule:
>RHO1_homSap rhodopsin <----------TM1---------> c1 <----------TM2-------> x1 <c--i------TM3---------> c2 <----------TM4--------> x2 <----------TM5------c-> c3
MNGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSRYIPEGLQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE
AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGKNPLGDDEASATVSKTETSQVAPA* 0
c3 <----------TM6-------> x3 <--------b-TM7---gprot> helix8 palm cyto tail