Opsin evolution: Cytoplasmic face: Difference between revisions
Tomemerald (talk | contribs) |
m (fix regardless) |
||
| (9 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
'''See also:''' [[Opsin_evolution|Curated Sequences]] | [[Opsin_evolution:_ancestral_introns|Ancestral Introns]] | [[Opsin_evolution:_informative_indels|Informative Indels]] | [[Opsin_evolution:_ancestral_sequences|Ancestral Sequences]] | [[Opsin_evolution:_alignment|Alignment]] | [[Opsin_evolution:_update_blog|Update Blog]] | |||
=== Comparative genomics of the cytoplasmic face of GPCR proteins === | === Comparative genomics of the cytoplasmic face of GPCR proteins === | ||
| Line 29: | Line 31: | ||
MEL2_anoCa DRYCVITKPLQSIKRTSKKR TCIIIVFVW 20 T P Gq | MEL2_anoCa DRYCVITKPLQSIKRTSKKR TCIIIVFVW 20 T P Gq | ||
While it might seem straightforward to thread any opsin onto its best fit among the five newly available crystallographic structures, that does not work for distantly related paralogs beyond the universal 7-transmembrane feature because loop regions can be of quite different length and so lack | While it might seem straightforward to thread any opsin onto its best fit among the five newly available crystallographic structures, that does not work for distantly related paralogs beyond the universal 7-transmembrane feature because loop regions can be of quite different length and so lack discernible alignability, having diverged greatly in amino acid sequence (even though they are all ultimately homologous). | ||
While these structures entail various compromises (such as | While these structures entail various compromises (such as replacements of C3 by lysozyme and deletion of carboxy tail to enable stable crystallization), they are hugely important to annotation transfer of sequence/function relationships via comparative genomics. Yet most of the 18 vertebrate opsin orthology classes have only remote models to date and even these can be indeterminate for mid-loop C2 residues (indicative of flexible conformation). | ||
Gene PDB Protein PubMed Best human opsin Next Best Signaling | Gene PDB Protein PubMed Best human opsin Next Best Signaling | ||
| Line 51: | Line 53: | ||
The squid melanopsin structure, submitted online to SwissModel, could otherwise predict the structure of the cytoplasmic loops of all opsins of melanopsin class, of which 48 vertebrate sequences, 9 lophotrochozoan, 43 arthropod, and 1 cnidarian sequences are available [[Opsin evolution|here]]. | The squid melanopsin structure, submitted online to SwissModel, could otherwise predict the structure of the cytoplasmic loops of all opsins of melanopsin class, of which 48 vertebrate sequences, 9 lophotrochozoan, 43 arthropod, and 1 cnidarian sequences are available [[Opsin evolution|here]]. | ||
The Gq signalling partner will be used throughout these melanopsins, yet what features the Galpha protein specifically recognizes in the cytoplasmic face remain obscure. It cannot really be the terminal helical extension per se because squid Gq protein will prove | The Gq signalling partner will be used throughout these melanopsins, yet what features the Galpha protein specifically recognizes in the cytoplasmic face remain obscure. It cannot really be the terminal helical extension per se because squid Gq protein will prove structuraly homologous to its 16 paralogs (in vertebrates) of different signaling types, meaning some universally conserved feature must be utilized instead. | ||
<br clear="all"> | <br clear="all"> | ||
| Line 58: | Line 60: | ||
=== The first cytoplasmic loop === | === The first cytoplasmic loop === | ||
This can be defined from bovine RHO1 and squid melanopsin structures or by bioinformatic calculation of transmembrane helices. Note the three online tools for that seldom agree with each other or xray structures (which have | This can be defined from bovine RHO1 and squid melanopsin structures or by bioinformatic calculation of transmembrane helices. Note the three online tools for that seldom agree with each other or xray structures (which have interpretive artifacts of their own). Here best representatives for each opsin class were found by blastp against SwissProt and the cytoplasmic loop taken from SwissProt annotation. It emerges that that a highly GPCR-conserved glutamate in transmembrane helix 2 must be a fixed number of residues in (namely 10) to conserve its helical wheel position with respect to the overall membrane structure and residues with which it interacts. This aspartate is known to hydrogen bond to Asn55 on TM1 (GFPIN) and main chain Ala299 in TMH7 (AKTSA), thus organizing the relationship of TM1,2,7 in the vicinity of the Schiff base. | ||
Consequently, cytoplasmic loop 1 must end at the PLN motif of RHO1 and hence all other opsins. The beginning of the cytoplasmic loop can be defined by similar considerations. It emerges from a mega-alignment that every opsin is indel-free in this region. Thus all CL1 must be of the same length (12 amino acids). Some sequence conservation, notably the proline at position 9, is universal. This proline may break the continuation of membrane alpha helix from the cytoplasmic domain into the cytoplasm. Internal basic residues are also found consistently. | Consequently, cytoplasmic loop 1 must end at the PLN motif of RHO1 and hence all other opsins. The beginning of the cytoplasmic loop can be defined by similar considerations. It emerges from a mega-alignment that every opsin is indel-free in this region. Thus all CL1 must be of the same length (12 amino acids). Some sequence conservation, notably the proline at position 9, is universal. This proline may break the continuation of membrane alpha helix from the cytoplasmic domain into the cytoplasm. Internal basic residues are also found consistently. | ||
| Line 363: | Line 365: | ||
On the basis of length (19 to rhodopsin, 20 to melanopsin), all the opsins except encephalopsin and RGR (both 16 residues) and TMT (18 residues subsequent to a deletion in amniote stem) have a structural model. This model is further constrained by predictable helical extensions of transmembrane helices into the cytoplasm, leaving only the mid-loop region to be predicted. It's not clear whether observed residue conservation -- both within and across orthology classes -- derives from structural importance or instead to Galpha binding specificity requirements. | On the basis of length (19 to rhodopsin, 20 to melanopsin), all the opsins except encephalopsin and RGR (both 16 residues) and TMT (18 residues subsequent to a deletion in amniote stem) have a structural model. This model is further constrained by predictable helical extensions of transmembrane helices into the cytoplasm, leaving only the mid-loop region to be predicted. It's not clear whether observed residue conservation -- both within and across orthology classes -- derives from structural importance or instead to Galpha binding specificity requirements. | ||
The adenosine and adrenergic receptor structures -- however useful they might be for annotation transfer to the other 350 non-oderant human GPCR -- ultimately will not prove helpful in modeling the second cytoplasmic loop of opsins (squid melanopsin does that better already). Note C2 in these three structures is consistently | The adenosine and adrenergic receptor structures -- however useful they might be for annotation transfer to the other 350 non-oderant human GPCR -- ultimately will not prove helpful in modeling the second cytoplasmic loop of opsins (squid melanopsin does that better already). Note C2 in these three structures is consistently stabilized by a mid-loop hydrogen bond to the DRY residues. This constraint is not observed in squid melanopsins or other metazoan opsin classes; indeed it is not feasible because no hydrogen bond-capable residue consistently occurs there (in the comparative genomics sense of conserved residue). Ancestrally, this mid-loop bridge might be a derived feature fairly early in the stem of non-opsin GPCR. | ||
[[Image:OpsinCyto2Five.jpg]] | [[Image:OpsinCyto2Five.jpg]] | ||
| Line 375: | Line 377: | ||
== The third cytoplasmic loop in 83 melanopsins == | == The third cytoplasmic loop in 83 melanopsins == | ||
This loop may be an important contributer to the Gq specificity. The structure has been determined for [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2Z73 squid melanopsin], denoted MEL_todPac below. It is a typical 'HEK' extended-helix CL3 found in vast majority of protostome melanopsins. However deuterostome melanopsins never have this feature, yet also appear to signal through Gq. | This loop may be an important contributer to the Gq specificity. The structure has been determined for [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2Z73 squid melanopsin], denoted MEL_todPac below. It is a typical 'HEK' extended-helix CL3 found in vast majority of protostome melanopsins. However deuterostome melanopsins never have this feature, yet also appear to signal through Gq. Melanopsin introns within this motif are considered [[Opsin_evolution:_ancestral_introns#Ancestral_melanopsin_intronation|elsewhere]]. | ||
The orphan Drosophila opsin RH7, which has not yet been associated with an anatomical structure, also lacks the HEK feature and is considerably shorter. However, as the lower sequences in the alignment below show, length variability is by no means unprecedented in this melanopsin loop. Indeed, the one cnidarian opsin available also lacks the HEK motif and also the length of those motifs. | The orphan Drosophila opsin RH7, which has not yet been associated with an anatomical structure, also lacks the HEK feature and is considerably shorter. However, as the lower sequences in the alignment below show, length variability is by no means unprecedented in this melanopsin loop. Indeed, the one cnidarian opsin available also lacks the HEK motif and also the length of those motifs. | ||
| Line 556: | Line 558: | ||
== The carboxy-terminal tail and VxPx motif == | == The carboxy-terminal tail and VxPx motif == | ||
This distinctive region has quite baffling length variation across -- and sometimes within -- opsin classes. The extent of conservation also differs greatly, with no real universally conserved residues past the end of the seventh transmembrane helix. The observed terminal conservation pattern for a given opsin must be indicative of its functional importance, even as that stands today insufficiently explained by arrestin phosphoserine or cysteine | This distinctive region has quite baffling length variation across -- and sometimes within -- opsin classes. The extent of conservation also differs greatly, with no real universally conserved residues past the end of the seventh transmembrane helix. The observed terminal conservation pattern for a given opsin must be indicative of its functional importance, even as that stands today insufficiently explained by arrestin phosphoserine or cysteine palmityolation sites, opsin dimerization or other membrane macro organization, or interaction with Galpha proteins. Some interactions would seem to require commonality across all orthology classes (or larger assemblages such as ciliary opsins) while others do not. | ||
Several studies have implicated the carboxy terminal motif VxPx of ciliary opsins as the intra-cellular targeting motif for proteins that function within cilia (or modified apical cilia such as rod and cone outer segments). The phylogenetic origin or age of this motif function has not been established nor its lineage-specific variations, though cilia themselves are pre-metazoan and the need to direct opsins specifically to outer segments would have been present already prior to lamprey divergence. | Several studies have implicated the carboxy terminal motif VxPx of ciliary opsins as the intra-cellular targeting motif for proteins that function within cilia (or modified apical cilia such as rod and cone outer segments). The phylogenetic origin or age of this motif function has not been established nor its lineage-specific variations, though cilia themselves are pre-metazoan and the need to direct opsins specifically to outer segments would have been present already prior to lamprey divergence. | ||
The description of the recognition pattern as VxPx is unsatisfactory | The description of the recognition pattern as VxPx alone is unsatisfactory: it is too short and vapid to serve this purpose. The residues valine and proline are all but inert and valine would be hard for the recognition apparatus to distinguish from leucine and isoleucine. Valine and proline would occur by random in this pattern in 4 proteins per thousand; mis-targeting would arise frequently from de novo substitutions in situations where one of V or P was already present. Thus the motif must reflect the end-of-gene position, ie VxPx* properly describes the motif and internal VxPx cannot. | ||
In opsins, we see from cytoplasmic tail alignments below that RGR, | In opsins, we see from cytoplasmic tail alignments below that RGR, peropsin, neuropsins, melanopsins, PPINb and TMT all lack any sign of a terminal VxPx motif. Here TMT is surprising in its total lack of any distal conservation whereas its nearest relative encephalopsin does have a strongly conserved VxPA motif VxPL, x:RK). RHO1 (VAPA), RHO2 (VSPA), SWS2 (VxPy, x:SAG, y:AS), LWS (VxPA X:AS), PPIN (VxPy x:AS, Y:ASLV), PARIE (VxPy x:AST, y:AVL), PIN VxPy x:MTA, y:AS), and VAOP (VxPy x:CY, y:ILM; motif lost in Aves). | ||
Thus the motif is really quite constrained in second and fourth position to a non-bulky uncharged side chain; VxPx does not accurately describe the observed reduced alphabet at these positions. However the carboxy terminus might have other functionalities in addition to | Thus the motif is really quite constrained in second and fourth position to a non-bulky uncharged side chain; VxPx does not accurately describe the observed reduced alphabet at these positions. However the carboxy terminus might have other functionalities in addition to ciliary targeting at least in opsins. Conversely it is not so clear that PPIN, PIN, PARIE, VAOP and encephalopsin are specifically targeted to modified pineal, brain, and melanocyte cilia in the same sense that rod and cone opsins are. | ||
Photoreceptor retinol dehydrogenase RDH8, another enzyme of the cis-retinal regeneration cycle located in the outer segments, also terminates in a similar motif VRPR. This is not the case for RDH11, RDH12 or RDH16 [http://www.jneurosci.org/cgi/content/full/24/11/2623 nor] in arrestin, transducin subunits, cGMP phosphodiesterase subunits, cGMP-gated channel subunits, Na/K/Ca exchanger, RGS9, R9AP, guanylate cyclases 2D and 2F, guanylate cyclase activating protein, phosducin, and recoverin. | Photoreceptor retinol dehydrogenase RDH8, another enzyme of the cis-retinal regeneration cycle located in the outer segments, also terminates in a similar motif VRPR. This is not the case for RDH11, RDH12 or RDH16 [http://www.jneurosci.org/cgi/content/full/24/11/2623 nor] in arrestin, transducin subunits, cGMP phosphodiesterase subunits, cGMP-gated channel subunits, Na/K/Ca exchanger, RGS9, R9AP, guanylate cyclases 2D and 2F, guanylate cyclase activating protein, phosducin, and recoverin. | ||
=== RGR === | === RGR === | ||
The first hand-gapped alignment below illustrates these issues using RGR from 53 species. The alignment begins inside the last transmembrane segment with the Schiff base lysine K and continues past the NAxxY motif at a deeply invariant length ( | The first hand-gapped alignment below illustrates these issues using RGR from 53 species. The alignment begins inside the last transmembrane segment with the Schiff base lysine K and continues past the NAxxY motif at a deeply invariant length (totaling 19 residues) to the "YR" motif found in almost all GPCR. This marks the beginning of the carboxy terminal cytoplasmic tail, which in RGR is fairly fixed at 23 residues, remain alignable and may extend the transmembrane helix but bear no resemblance to any other opsin or GPCR. | ||
The degree of conservation establishes selection is at work. It appears that RGR must terminate in several charged (characteristically basic) residues | The degree of conservation establishes selection is at work. It appears that RGR must terminate in several charged (characteristically basic) residues regardless of length indels. These could possibly associate electrostatically with membrane phospholipid or be important to initial establishment of topology. Mammals have in effect lost the YR motif though most have an R one residue later. This does not quite coincide with the advent of ERY or GRY mammals in cytoplasmic loop C2. | ||
Conservation of G.WQ.L..Q has persisted for tens of billions of years and cannot be explained by helix or beta sheet per se -- possibly it is constrained by interaction with parts of the other cytoplasmic face. It appears that arrestin could recognize | Conservation of G.WQ.L..Q has persisted for tens of billions of years and cannot be explained by helix or beta sheet per se -- possibly it is constrained by interaction with parts of the other cytoplasmic face. It appears that arrestin could recognize phosphoserine or threonine in almost all species but palmityolation cannot be widespread. A few species, such as guinea pig, microbat and armadillo may be exhibiting early stages of pseudogenization or at least partial loss of function. | ||
Absent any experimental information or | Absent any experimental information or relevant 3D structure or capacity for annotation transfer from homologous regions, the specifics of individual residue and residue patch conservation will remain difficult to explain. | ||
K..PT.NA..YaLG.E.yr .G.Wq.L..q..........k.K | K..PT.NA..YaLG.E.yr .G.Wq.L..q..........k.K | ||
<font color="blue">>RGR_homSap KMVPTINAINYALGNEMVC RGIWQCLSPQKRE-----KDRTK RGR_homSap KMVPTINAINYALGNEMVC RGIWQCLSPQKREKDRTK | <font color="blue">>RGR_homSap KMVPTINAINYALGNEMVC RGIWQCLSPQKRE-----KDRTK RGR_homSap KMVPTINAINYALGNEMVC RGIWQCLSPQKREKDRTK | ||
| Line 630: | Line 632: | ||
=== Peropsin === | === Peropsin === | ||
Peropsin exhibits greater conservation both in its post-K helix and in its cytoplasmic tail than RGR. The FR motif is perfectly conserved throughout vertebrates. Length, ancestrally 32 residues, experienced an era of variability in amniotes but then settled down to a fixed 35 residues in mammals. The | Peropsin exhibits greater conservation both in its post-K helix and in its cytoplasmic tail than RGR. The FR motif is perfectly conserved throughout vertebrates. Length, ancestrally 32 residues, experienced an era of variability in amniotes but then settled down to a fixed 35 residues in mammals. The difference alignment shows that a central motif EITISN conserved in early vertebrates changed character completely (to TMPVTS) in mammals, though the earlier motif still appears faded in platypus. A cysteine conserved back to invertebrates might be palmitoylated; conserved serines and threonines offer potential phosphorylation sites. | ||
The cytoplasmic tail of peropsin is completely unalignable to RGR. Unlike RGR, tblastn of peropsin tail against whole human genome elicits matches to imaging opsins and a GPCR (neuropeptide Y receptor). While these matches are weak and largely driven by the last transmembrane section alone, 3 early tail residues (*) emerge as possible conserved residues. Whether or not homologically valid, this suggests modeling of the first 9 residues of peropsin tail by known bovine rhodopsin structure. | The cytoplasmic tail of peropsin is completely unalignable to RGR. Unlike RGR, tblastn of peropsin tail against whole human genome elicits matches to imaging opsins and a GPCR (neuropeptide Y receptor). While these matches are weak and largely driven by the last transmembrane section alone, 3 early tail residues (*) emerge as possible conserved residues. Whether or not homologically valid, this suggests modeling of the first 9 residues of peropsin tail by known bovine rhodopsin structure. | ||
| Line 781: | Line 783: | ||
The cytoplasmic tail in melanopsin can be quite variable in length and sequence. No strongly conserved residues exist in bilateran melanopsins beyond the P.L beginning at position 8; consequently very little can be learned about the cytoplasmic tail of vertebrate or even arthropod melanopsins from study of molluscan melanopsins. Its contribution to structure and function of the cytoplasmic face must be quite variable. Note the FR motif is almost always YR outside of lophotrochozoans. | The cytoplasmic tail in melanopsin can be quite variable in length and sequence. No strongly conserved residues exist in bilateran melanopsins beyond the P.L beginning at position 8; consequently very little can be learned about the cytoplasmic tail of vertebrate or even arthropod melanopsins from study of molluscan melanopsins. Its contribution to structure and function of the cytoplasmic face must be quite variable. Note the FR motif is almost always YR outside of lophotrochozoans. | ||
Within just vertebrates, the cytoplasmic tail of melanopsin exhibits much more extensive conservation of 11 residues extending out to position 66 (human numbering). The two conserved serines might be cyclically phosphorylated and the single cysteine at position 9 palmitoylated (as it cannot be in a disulfide residing in the reduced cytoplasmic | Within just vertebrates, the cytoplasmic tail of melanopsin exhibits much more extensive conservation of 11 residues extending out to position 66 (human numbering). The two conserved serines might be cyclically phosphorylated and the single cysteine at position 9 palmitoylated (as it cannot be in a disulfide residing in the reduced cytoplasmic milieu). | ||
While the remaining residues are very likely stably structured, it's not clear whether they interact primarily with the other cytoplasmic loops or with | While the remaining residues are very likely stably structured, it's not clear whether they interact primarily with the other cytoplasmic loops or with auxiliary proteins. The latter is more likely recalling that melanopsins signal via Gq and the inositol triphosphate cascade rather than the very different cyclic nucleotide pathway. | ||
[[Image:MelCytoTail.jpg|left]] | [[Image:MelCytoTail.jpg|left]] | ||
| Line 910: | Line 912: | ||
=== Encephalopsin === | === Encephalopsin === | ||
This opsin class, despite its phylogenetically erratic pattern of tetrapod gene loss, is exceedingly conserved in its carboxy terminus in both length and sequence back to | This opsin class, despite its phylogenetically erratic pattern of tetrapod gene loss, is exceedingly conserved in its carboxy terminus in both length and sequence back to lamprey. This conservation is unprecedented in this region and must reflect mission-critical binding to another protein. | ||
The cytoplasmic tail of encephalopsin has no detectable homology to other ciliary opsins for more that 6 residues beyond the FR motif (FRRSLLQL) even though it shares the same very ancient terminal exon break as other ciliary opsins (phase 0, just prior to the FR). | The cytoplasmic tail of encephalopsin has no detectable homology to other ciliary opsins for more that 6 residues beyond the FR motif (FRRSLLQL) even though it shares the same very ancient terminal exon break as other ciliary opsins (phase 0, just prior to the FR). The VxPx* motif can be recognized in the conserved pattern VRPL*; if this primarily drives cell targeting to cilia, it may or may not have arisen independently from similar motifs in other ciliary opsins. | ||
An interesting phyloSNP can be seen in the difference alignment in the primate stem (S-->N) two residues after the critical Schiff lysine. This may slightly shift the chemical environment of the chromophore. | |||
ENCEPH_hom KSNTVYNPVIYVFMIRKFR RSLLQLLCLRLLRCQRPAKDLPA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-NGSKVDVIQVRPL | ENCEPH_hom KSNTVYNPVIYVFMIRKFR RSLLQLLCLRLLRCQRPAKDLPA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-NGSKVDVIQVRPL | ||
| Line 985: | Line 989: | ||
=== TMT opsin === | === TMT opsin === | ||
TMT predominantly exhibits FY for its FR motif though perhaps the conserved FYK/R motif accomplishes the same end. Within the whole TMT family, no observable conservation past the first 9 residues though some 35 residues are alignable within the TMT | TMT predominantly exhibits FY for its FR motif though perhaps the conserved FYK/R motif accomplishes the same end. Within the whole TMT family, no observable conservation occurs past the first 9 residues, though some 35 residues are alignable within the sole TMT locus tracking into mammals (marsupials). The conserved pair of cysteines might be palmitoylated. Opossum has acquired an upstream stop codon recently -- the 22 residues following are still alignable to wallaby. GenBank lacks any tetrapod transcripts of this TMT locus as of Jan 09. The last exon of this gene is curiously intertwined with that of the opposing strand gene, the sialyltransferase ST6GAL2. | ||
TMT_monDom KSSTVCNPIIYVLMNKQFY KCFLILFHCQPAQSGPDVS LCPSNVTVIQLGQRKNKDA PGSI*DFPEVSEKQLCLLS PEVWPQP | TMT_monDom KSSTVCNPIIYVLMNKQFY KCFLILFHCQPAQSGPDVS LCPSNVTVIQLGQRKNKDA PGSI*DFPEVSEKQLCLLS PEVWPQP | ||
| Line 1,022: | Line 1,026: | ||
The cytoplasmic tails of these opsins begin and end with highly conserved motifs but the middle sections have been subject to numerous indels, suggesting that absolute length is unimportant for binding site recognition. The VAPA terminal motif can be recognized in all but the secondary parapinopsin group PPINb (found only in some teleost fish and apparently reflecting differential survival of gene duplication and in avian VAOP where chicken and finch have recent changes in stop codon. | The cytoplasmic tails of these opsins begin and end with highly conserved motifs but the middle sections have been subject to numerous indels, suggesting that absolute length is unimportant for binding site recognition. The VAPA terminal motif can be recognized in all but the secondary parapinopsin group PPINb (found only in some teleost fish and apparently reflecting differential survival of gene duplication and in avian VAOP where chicken and finch have recent changes in stop codon. | ||
LWS is shown [[Opsin_evolution:_LWS_PhyloSNPs#Indels_in_the_cytoplasmic_tail|elsewhere]] greatly expanded to 82 species to illustrate the issues. Four indels, all deletions, have | LWS is shown [[Opsin_evolution:_LWS_PhyloSNPs#Indels_in_the_cytoplasmic_tail|elsewhere]] greatly expanded to 82 species to illustrate the issues. Four indels, all deletions, have occurred during vertebrate history: a 2 residue loss in mammals, a 1 residue loss in birds but not lizards, and a 1 and 5 residue loss in teleost fish. Otherwise, LWS has been remarkably constant -- its key features and almost every residue past FR were already firmly settled prior to lamprey divergence. | ||
This region cannot be important to Galpha binding because it is too highly variable just within cone opsins which all use the same transducin. Cysteines are conserved to depth but palmitoylation could be universal exclusive of VAOP. LWS also lacks the distal cysteine (CCGK motif has been LFGK since lamprey stem) found in other ciliary opsins. Serines and threonines (for arrestin) are common but are not a deeply conserved feature. | This region cannot be important to Galpha binding because it is too highly variable just within cone opsins which all use the same transducin. Cysteines are conserved to depth but palmitoylation could be universal exclusive of VAOP. LWS also lacks the distal cysteine (CCGK motif has been LFGK since lamprey stem) found in other ciliary opsins. Serines and threonines (for arrestin) are common but are not a deeply conserved feature. | ||
| Line 1,724: | Line 1,728: | ||
petMa DRYLVLTRPLASIGAMSKRRAMYITAAVW | petMa DRYLVLTRPLASIGAMSKRRAMYITAAVW | ||
</pre> | </pre> | ||
'''See also:''' [[Opsin_evolution|Curated Sequences]] | [[Opsin_evolution:_ancestral_introns|Ancestral Introns]] | [[Opsin_evolution:_informative_indels|Informative Indels]] | [[Opsin_evolution:_ancestral_sequences|Ancestral Sequences]] | [[Opsin_evolution:_alignment|Alignment]] | [[Opsin_evolution:_update_blog|Update Blog]] | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 23:59, 3 December 2010
See also: Curated Sequences | Ancestral Introns | Informative Indels | Ancestral Sequences | Alignment | Update Blog
Comparative genomics of the cytoplasmic face of GPCR proteins
The cytoplasmic face of an opsin (or any GPCR) is comprised of three disjoint connecting loops and the carboxy terminus. It is presumably responsible for all interactions with downstream signal relaying partners because these latter are cytoplasmic proteins having no physical access to the extracellular loops or transmembrane segments. Here it must be noted that photoisomerization and retinal release from Schiff base deep within the transmembrane region must drive a significant change in conformation in the cytoplasmic face that differentiates its inactive from active states.
For bioinformatic purposes, it is convenient to 'reorganize' each linear protein sequence into its intracellular, membrane and outer regions for separate consideration. This is done below for the cytoplasmic face for 500 curated opsins from each of the 20 vertebrate opsin genetic loci using multiple representatives for each phylogenetic node and intense bracketing at eras of functional transition (eg between DRY and GRY opsins of RGR class). A range of non-opsin GPCR are included to define properties common to all members of this large gene family (not specific to opsins).
The two critical goals in GPCR research are finding the natural ligands (which largely concerns the extracellular and transmembrane regions) notably for orphan receptors and to determining their specific Galpha signaling partner among the 17 such paralogs in the vertebrate genome. For vertebrate opsins, the ligand is known (11-cis retinal or related) but the signaling partner generally is not. For example, does RGR opsin signal at all (most are predicted to signal with both Gi/o and Gq/11), to what regulatory effect, and what is the meaning of the abrupt shift in the DRY motif to GRY at boreoeuthere divergence?
DRY loop motif transmemb aa 7 9 signaling ENCEPH_hom ERYIRVVHARVINFSW AWRAITYIW 16 V A G? RGR_homSap GRYHHYCTRSQLAWNS AVSLVLFVW 16 C R G? RGR2_gasAc DRYHQYCTRQKLFWST TLTMSAIIW 16 C R G? RHO1_homSa ERYVVVCKPMSNFRFGENH AIMGVAFTW 19 C P GNAT1 RHO2_galGa ERYIVVCKPMGNFRFSATH AMMGIAFTW 19 C P GNAT2 SWS2_ornAn ERFLVICKPLGNLSFRGTH AIFGCAATW 19 C P GNAT2 PIN_galGal ERYVVVCRPLGDFQFQRRH AVSGCAFTW 19 C P G? SWS1_homSa ERYIVICKPFGNFRFSSKH ALTVVLATW 19 C P GNAT2 LWS_homSap ERWMVVCKPFGNVRFDAKL AIVGIAFSW 19 C P GNAT2 VAOP_galGa ERYIVICRPVGNMRLRGKH AAQGIAFVW 19 C P Gt PARIE_utaS ERYNVVCQPLGTLQMSTKR GYQLLGFIW 19 C P Gd+Go PPIN_xenTr DRVFVVCKPMGTLTFTPKQ ALAGIAASW 19 C P Gt PER_homSap DRYLTICLPDVGRRMTTNT YIGLILGAW 19 C P Go NEUR1_homS DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L G? NEUR2_galG VCCLKICFPAYGNRFRRKH GQILIACAW 19 C P G? NEUR3_galG IRFLVTNSSKSNSNKISKNT VHILITFIW 20 N S G? NEUR4_ornA TRYIKGCHPHRGHFINTAN ISVALILIW 19 C P G? TMT_monDom ERYRTL-TLCPGQGADYQK ALLAVAGSW 19 - L G? MEL1_homSa DRYLVITRPLATFGVASKRR AAFVLLGVW 20 T P Gq MEL2_anoCa DRYCVITKPLQSIKRTSKKR TCIIIVFVW 20 T P Gq
While it might seem straightforward to thread any opsin onto its best fit among the five newly available crystallographic structures, that does not work for distantly related paralogs beyond the universal 7-transmembrane feature because loop regions can be of quite different length and so lack discernible alignability, having diverged greatly in amino acid sequence (even though they are all ultimately homologous).
While these structures entail various compromises (such as replacements of C3 by lysozyme and deletion of carboxy tail to enable stable crystallization), they are hugely important to annotation transfer of sequence/function relationships via comparative genomics. Yet most of the 18 vertebrate opsin orthology classes have only remote models to date and even these can be indeterminate for mid-loop C2 residues (indicative of flexible conformation).
Gene PDB Protein PubMed Best human opsin Next Best Signaling RHO1_bosTau 1JFP 3C9M 2J4Y bovine rod rhodopsin 17825322 RHO1_homSap 93% SWS1_homSap 45% Gt GNAT1 raises cGMP MEL1_todPac 2Z73 2ZIY squid melanopsin 18480818 MEL1_homSap 43% PER1_homSap 30% Gq GNAQ? inositol trisphosphate ADORA2A_homSap 3EML adenosine receptor 2A 18832607 MEL1_homSap 27% ENCEPH_homSap 27% Gs GNAT3 raises cAMP ADRB1_melGal 2VT4 beta 1 adrenergic receptor 18594507 MEL1_homSap 29% ENCEPH_homSap 25% Gs GNAT3 raises cAMP ADRB2_homSap 2R4R beta 2 adrenergic receptor 17962520 MEL1_homSap 28% PER1_homSap 29% Gs GNAT3 raises cAMP
It has not proven feasible to predict loop conformations ab initio or from peptide libraries; it is folly to consider individual loop structure in isolation (rather than the cytoplasmic face in its entirety) or fail to specify the activation state being computed. Any predicted structure and special roles for individual residues must be consistent with the comparative genomics of close and even distant orthologs because binding relationships to Galpha and other proteins do not change rapidly in evolutionary time (as seen from heterologous substitution experiments). Even when a cytoplasmic loop seems to lack a definable structure, individual residues can be conserved over vast branch length times. That conservation must ultimately be explained.
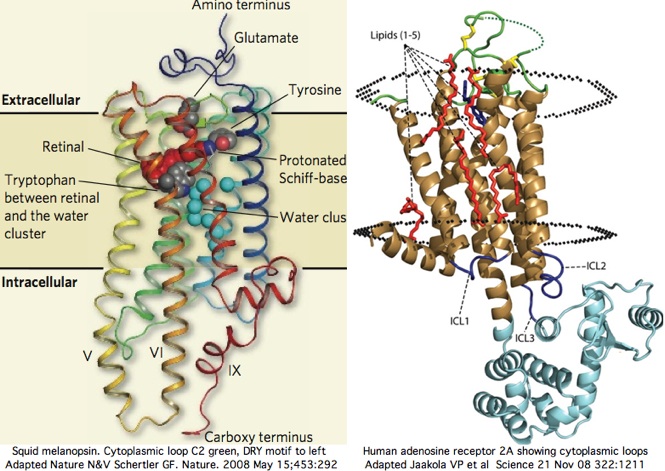
Two new high resolution structures of squid melanopsin establish that the cytoplasmic face is not structurally homologous even within melanopsins. We knew this already from comparative genomics alignment but not specifically why. The xray structure exhibits unprecedented rigid extensions of transmembrane helices 5 and 6 of order 25 angstroms out into the cytoplasm, greatly constraining the intermediate residues of cytoplasmic loop C3. The proximal carboxy terminus also contributes importantly to the overall structure here.
This structure cannot be replicated in non-cephalopod melanopsins because conservation is observed only out to a proline 8 within the 127 residue FR motif. Even central conservation rapidly drops below 45% even within other lophotrochozoans. Consequently the 25 angstrom cytoplasmic knob of squid melanopsin has no value for annotation transfer but rather represents a lineage-specific innovation. Thus it likely has very little to do with Galpha signaling specificity.
The squid melanopsin structure, submitted online to SwissModel, could otherwise predict the structure of the cytoplasmic loops of all opsins of melanopsin class, of which 48 vertebrate sequences, 9 lophotrochozoan, 43 arthropod, and 1 cnidarian sequences are available here.
The Gq signalling partner will be used throughout these melanopsins, yet what features the Galpha protein specifically recognizes in the cytoplasmic face remain obscure. It cannot really be the terminal helical extension per se because squid Gq protein will prove structuraly homologous to its 16 paralogs (in vertebrates) of different signaling types, meaning some universally conserved feature must be utilized instead.
The first cytoplasmic loop
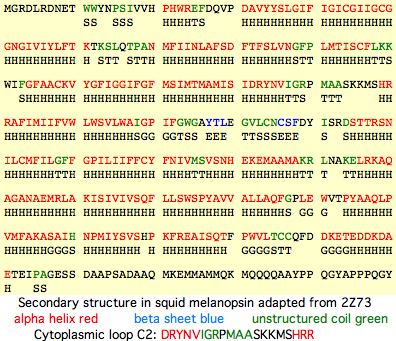
This can be defined from bovine RHO1 and squid melanopsin structures or by bioinformatic calculation of transmembrane helices. Note the three online tools for that seldom agree with each other or xray structures (which have interpretive artifacts of their own). Here best representatives for each opsin class were found by blastp against SwissProt and the cytoplasmic loop taken from SwissProt annotation. It emerges that that a highly GPCR-conserved glutamate in transmembrane helix 2 must be a fixed number of residues in (namely 10) to conserve its helical wheel position with respect to the overall membrane structure and residues with which it interacts. This aspartate is known to hydrogen bond to Asn55 on TM1 (GFPIN) and main chain Ala299 in TMH7 (AKTSA), thus organizing the relationship of TM1,2,7 in the vicinity of the Schiff base.
Consequently, cytoplasmic loop 1 must end at the PLN motif of RHO1 and hence all other opsins. The beginning of the cytoplasmic loop can be defined by similar considerations. It emerges from a mega-alignment that every opsin is indel-free in this region. Thus all CL1 must be of the same length (12 amino acids). Some sequence conservation, notably the proline at position 9, is universal. This proline may break the continuation of membrane alpha helix from the cytoplasmic domain into the cytoplasm. Internal basic residues are also found consistently.
The question of Galpha binding here must address how opsins using different signaling partners could still be so similar across orthology classes, yet have a fair amount of variation within.
SwissProt predictions RHO1_homSa TVQHKKLRTPLN SWS1_homSa TLRYKKLRQPLN ENCEPH_hom YYKFQRLRTPTH TMT_monDom FCKFKVLRNPVN MEL1_homSa FCRSRSLRTPAN PER1_homSa FIKYKELRTPTN Alignment of CL1 (with early residues of TM2 also shown up to the registration residue D) RHO1_homSa TVQHKKLRTPLN YILLNLAVAD RHO1_monDo TIQHKKLRTPLN YILLNLAIAD RHO1_bosTa TVQHKKLRTPLN YILLNLAVAD RHO1_conMy TIEHKKLRTPLN YILLNLAVAD RHO1_ornAn TIQHKKLRTPLN YILLNLAFAN RHO1_angAn TIEHKKLRTPLN YILLNLAVAN RHO1_galGa TIQHKKLRTPLN YILLNLVVAD RHO1_neoFo TVQHKKLRTPLN YILLNLAVAD RHO1_takRu TVKHKKLRTPLN YVLLNLAVAD RHO1_leuEr TIQHKKLRQPLN YILLNLAVSD RHO1_calMi TFEHKKLRQPLN FILLNLAVAD RHO2_calMi TVKHKKLRQPLN FILLNLAVAD RHO1_latCh TIQHKKLRTPLN YILLDLAVAD RHO1_anoCa TIQHKKLRTPLN YILLNLAVAN RHO1_petMa TVQHKKLRTPLN YILLNLAVAN RHO1_letJa TVQHKKLRTPLN YILLNLAMAN RHO1_geoAu TVQHKKLRTPLN YILLNLAVSN RHO1_xenTr TIQHKKLRTPLN YILLNLVFAN RHO2_galGa TFKHKKLRQPLN YILVNLAVAD RHO2_podSi TFKHKKLRQPLN YILVNLAVAD RHO2_anoCa TFKHKKLRQPLN YILVNLAVAD RHO2_taeGu TFKHKKLRQPLN YILVNLAVAD RHO2_neoFo TFKHKKLRQPLN YILVNLAVAD RHO2_latCh TFKHKKLRQPLN YILVNLAVAS RHO2_gekGe TFQHKKLRQPLN YILVNLAAAN RHO2_pheMa TFQHKKLRQPLN YILVNLAVAN RHO2_geoAu TFKLKKLRQPLN FILVNLCVAD RHO2_ancDa TAQHKKLRQPLN FILVNLAVAG RHO2d_danR TAQHKKLRQPLN FILVNLAVAG RHO2c_danR TAQHKKLRQPLN FILVNLAVAG RHO2a_danR TAQHKKLRQPLN YILVNLAFAG RHO2b_danR TAQHKKLRQPLN YILVNLAFAG RHO2_oryLa TAQNKKLRQPLN FILVNLAVAG RHO2_takRu TAQNKKLRQPLN YILVNLAVAG RHO2_gasAc TAQNKKLRQPLN YILVNLAVAG RHO2_oreNi TAQNKKLRQPLN YILVNLAVAG RHO2_hipHi TAQNKKLRQPLN YILVNLAVAG RHO2_mulSu TFQNKKLQQPLN YILVNLAVVG RHO2_pomMi TFQNKKLRQPLN FILVNLAVAG SWS2_ornAn TIKYKKLRSHLN YILVNLAVSN SWS2_anoCa TFKYKKLRSHLN YILVNLSVSN SWS2_utaSt TFKYKKLRSHLN YILVNLAVSN SWS2_taeGu TAKYKKLRSHLN YILVNLAVAN SWS2_neoFo TFKYKKLRSHLN YILVNLAVAN SWS2_xenTr TVKYKKLRSHLN YILVNLAVAN SWS2_galGa TARFRKLRSHLN YILVNLALAN SWS2_geoAu TIKYKKLRSHLN YILVNLAIAN SWS2_takRu TIQYKKLRSHLN YILVNLAFSN SWS2_gasAc TVQNKKLRSHLN YILVNLAVSN SWS1_homSa TLRYKKLRQPLN YILVNVSFGG SWS1_monDo TLRYKKLRQPLN YILVNVSLCG SWS1_smiCr TLRYKKLRQPLN YILVNISLAG SWS1_tarRo TLRYKKLRQPLN YILVNISLAG SWS1_anoCa TVKYKKLRQPLN YILVNISFAG SWS1_utaSt TVKYKKLRQPLN YILVNISFAG SWS1_neoFo TIKYKKLQQPLN YILVNISLAG SWS1_taeGu TIKYKKLRQPLN YILVNISVSG SWS1_xenLa TIKYKKLRQPLN YILVNITVGG SWS1_galGa TVRYKRLRQPLN YILVNISASG SWS1_petMa TVKCKKLRQPLT YMLVNISAAG SWS1_geoAu TIKYKKLRQPLN YILVNISAAG SWS1_danRe TMKYKKLRQPLN YILVNISLAG SWS1_oryLa TAKYKKLRVPLN YILVNITFAG LWS_homSap TMKFKKLRHPLN WILVNLAVAD LWS_monDom TMKFKKLRHPLN WILVNLAVAD LWS_ornAna TMKFKKLRHPLN WILVNLAVAD LWS_galGal TWKFKKLRHPLN WILVNLAVAD LWS_anoCar TAKFKKLRHPLN WILVNLAIAD LWS_neoFor TYKFKKLRHPLN WILVNLAIAD LWS_xenTro TLKFKKLRHPLN WILVNMAIAD LWS_takRub TAKFKKLRHPLN WILVNLAIAD LWS_gasAcu TAKFKKLQHPLN WILVNLAIAD LWS2_calMi TWKFKKLRHPLN WILVNLAIAD LWS_geoAus TLKFKKLRHPLN WILVNLAIAD LWS_petMar TVKFKKLRHPLN WIIVNLAIAD LWS_letJap TMKFKKLRHPLN WILVNLAIAD LWS1_calMi TVRFKKLRHPLN WILVNMALAD PIN_galGal SICYKKLRSPLN YILVNLAVAD PIN_colLiv SIRYKKLRSPLN YILVNLAMAD PIN_taeGut SVRHKRLRSPLN YILLNLAVAN PIN_utaSta SIQYKKLRSPLN YILVNLAIAD PIN_podSic SVQFKKLRSPLN YVLVNLAVAD PIN_pheMad SVRFKRLRSPLN YILVNLATAD PIN_xenTro TLKYKKLRSPLN YILVNLAIAN PIN_bufJap SLKYKKLRSPLN YILVNLAVAD VAOP_galGa TFKFKQLRQPVN YVIVNLSVAD VAOP_taeGu TFKFKQLRQPIN YIIVNLSVAD VAOP_anoCa TIKFKQLRQPLN YVIVNLSVAD VAOP_danRe TFRFQQLRQPLN YIIVNLSLAD VAOP_rutRu TFRFTQLRKPLN YIIVNLSLAD VAOP_takRu TFKFKQLRQPLN YIIVNLAIAD VAOP_xenTr TAKFKQLRQPLN YIIVNLSVAD VAOP_petMa TARFRQLRQPLN YVLVNLAAAD PPIN_anoCa TIKYRQLRQPIN YSLVNLAIAD PPIN_xenTr TFKYRQLRHPIN YSLVNLAIAD PPINa_petM TLRHRQLRHPLN FSLVNLAVAD PPIN_letJa TLRHRQLRHPLN FSLVNLAVAD PPIN_danRe TLKYKQLRQPLN FALVNLAVAD PPIN_ictPu TVRYKQLRQPLN YALVNLAVAD PPIN_oncMy TMRHRKLRQPLN YALVNLAVAD PPINb_takR TMKHRQLRQPLS YALVNLAICD PPINb_tetN TLKHRQLRQPLN YALVNLAICD PPINb_gasA TARHRQLRQPLS YALVNLAVCD PPINa_gasA TLMHKQLRQPLN YALVNMALAD PPINa_takR SLMHKQLRQPLN YALVNMAVAD PPINa_tetN SLMHKQLRQPLN YALVNMAAAD PPINa_cioI TLKNKVLRQPLN YIIVNLAVVD PPINa_cioS TLKNKVLRQPLN YIIVNLAVVD PPINb_cioI TMKNKKLRQPLN YIIINLSIAD PPINb_cioS TYKNKDLRRPIN YIIVNLAVAD PARIE_utaS TLKNPQLRNPIN IFILNLSFSD PARIE_anoC TLKNPQLRNPIN IFILNLSFSD PARIE_xenT TLKHPQLRNPIN IFILNLSFSD PARIE_takR MLKNPSLLQPIN IFILSLAVSD PARIE_tetN MLKNPALLQPIN IFILSLAVSD PARIE_gasA LVRNPSLLQPMN VFILSLAVSD PARIE_danR MVKNLHFLNAMT VIIFSLAVSD ENCEPH_hom YYKFQRLRTPTH LLLVNISLSD ENCEPH_oto YYKFPRLRTPTH LFLVNISLSD ENCEPH_lox YYKFQRLRTPTH LFLVNISLSD ENCEPH_mon YYKFQRLRTPTH LFLVNISFND ENCEPH_can FLEFQRLRTPTH LLLVNLSLSD ENCEPH_mus YSKFPRLRTPTH LFLVNLSLGD ENCEPH_pte YYKFQQVRTPFY LFLVNISFSD ENCEPH_ano YAKFKRLRTPTH LFLVNISLSD ENCEPH_gal YYKFKRLRTPTN LFLVNISLSD ENCEPH_dan YSRYKRLRTPTN LLIVNISVSD ENCEPH_tak YCRFKRLRTPTN LLLVNISLSD ENCEPH_ory YCKFKRLRTPTS LLLVNISLSD ENCEPH_gas YCKFKRLRTPTN LLVVNISLSD ENCEPH_squ YCKFKRLRTPTN LFLVNISISD ENCEPH_pet FVGFKRLQTPTN LLLVNISLSD ENCEPH_cal YYKFKRLRTPTN LLLVNISVSD ENCEPH_xen YCKFKRLQTPTN LLFFNTSLCH ENCEPH4_br IGCHRQLRTPFN LLLLNMSVAD TMT_braFlo FLKFRQLRTPFN MLLLNMSVAD TMT_braBel FLKFPQLRTPFN LLLLNMAVAD TMT_monDom FCKFKVLRNPVN MLLLNISISD TMT_macEug FCKFKVLRNPVN MLLLNISISD TMT_galGal FCKFKTLRNPVN MLLLNISISD TMT_anoCar FCKFKTLRNPVN MLLLNISASD TMT_taeGut FCKFKTLRNPVN MLLLNISVSD TMT_xenTro FCKFKTLRTPVN MMLLNISASD TMT_ornAna FCKFKALRNPVN MIMLNISASD TMT_danRer FCKFKTLRTPVN MLLLNISISD TMT_takRub FCKFKKLRTPVN MLLLNISVSD TMT_oryLat FCKFKKLRTPVN MLLLNISVSD TMT_gasAcu FCKFKKLRTPVN MLLLNISVSD TMT_tetNig FCKFKKLRTPVN VLLLNISVSD TMTa1_danR FGRYKVLRSPIN FLLVNICLSD TMTa_takRu FCRYKMLRSPIN LLLMNISISD TMTa_tetNi FCRFKVLRSPIN LLLVNISVSD TMTa_gasAc FCRYKMLRSPIN LLLINISISD TMTa_oryLa FCRYKILRSPIN LLLINISISD TMTa_pimPr FCRYKVLRSPMN YLLVSIAVSD TMTb_danRe FCRYKVLRSPMN CLLISISVSD TMTa1_calM FCKYKVLRSPMN MLLLNISVSD TMTb_takRu FCRYRALRTPMN LMLVSISASD TMTb_tetNi FCRFRALRTPMN LMLVSISASD TMTb_oryLa FCRYRALRTPMN LLLVSISVSD TMTb_gasAc FCRYRALRTPMN LLLVSISASD TMTa_braFl VGRYKQLRTPFN ILMVNLSVSD MEL2_strPu FLRFKKLHSPIN LLIVNLSASD MEL1_homSa FCRSRSLRTPAN MFIINLAVSD MEL1_panTr FCRSRSLRTPAN MFIINLAVSD MEL1_gorGo FCRSRSLRTPXN MFIINLAVSD MEL1_ponAb FCRSRGLRTPAN MFIINLAVSD MEL1_rheMa FCRSRGLRTPAN MFIINLAISD MEL1_calJa FCRSRGLRTPAN MFIINLAVSD MEL1_bosTa FCRSRGLRTPAN MFIINLAVSD MEL1_susSc FCRSRGLRTPAN MFIINLAVSD MEL1_equCa FCRSRGLRTPAN MFIINLAVSD MEL1_eriEu FCRSRSLRTPAN MFIINLAVSD MEL1_echTe FCRSRSLRTPAN MLIINLAVSD MEL1_otoGa FCRVRGLRTPAN MFVINLAVSD MEL1_micMu FCRSRSLRTPAN MFVINLAVSD MEL1_myoLu FCRSRGLRTPAN MFIINLAVSD MEL1_pteVa FCRSRGLRTPAN MFIINLAVSD MEL1_felCa FCRSRGLRTPAN MFIINLAVSD MEL1_canFa FCRTRGLRTPSN MFIINLAVSD MEL1_proCa FFRSRGLRTPAN MFIINLAISD MEL1_loxAf FFRSRGLRTPAN MFIINLAVSD MEL1_musMu FCRNRGLRTPAN MFIINLAVSD MEL1_ratNo FCRNRGLRTPAN MLIINLAVSD MEL1_phoSu FCRSRSLRTPAN MLIINLAVSD MEL1_nanEh FCRSRGLRTRAN MFTVNLAVSD MEL1_smiCr FCRSRSLRTPAN MFIINLAISD MEL1_monDo FCRSHSLRTPAN MFIINLAISD MEL1_xenTr FCRSRSLRSPAN MFIINLAITD MEL1_ornAn FCRSRSLRTPAN MFIINLSISD MEL1_taeGu FCRSRSLQTPAN ILIINLAISD MEL1_galGa FCRSRTLQKPAN IFIINLAVSD MEL1_danRe FSRSRTLRTPAN LFIINLAITD MEL1_gasAc FSKSRSLRTPAN MFIINLAITD MEL1_oryLa FSRSRSLRTPAN MFIINLAITD MEL1_takRu FCRSRSLRTPAN MFIINLAVTD MEL1_calMi FLRSRSLRTPAN TFIINLAATD MEL1_petMa FSKSKSLRSPAN IFIINLAFAD MEL2_galGa FYSNKKLRTPQN FFIMNLAVSD MEL2_anoCa FYSNKRLRTPPN YFIMNLAVSD MEL2_xenLa FYRNKKLRTAPN YFIINLAISD MEL2_tetNi FYSNKKLRSLPN YFIVNLAVSD MEL2_danRe FYRNKKLRSLPN YFIMNLAVSD MEL2_gasAc VYSNKKLRNLPN YFIMNLAVSD MEL1a_braF FIKSKGLRTPAN FFIINLALSD MEL1a_braB FIKSKGLRTPAN FFIINLALSD TMTPIN_sto FARFPSLRHPIN SFLFNVSLSD PER2_patYe FAKRRSVRRPIN FFVLNLAVSD PER1_homSa FIKYKELRTPTN AIIINLAVTD PER1_monDo FVKYKALRTATN TIIINLAVTD PER1_ornAn FVKFEELRTATN AIIINLAVTD PER1_xenTr FVKYKELRTATN AIIINLAFTD PER1_gasAc FWKFKELRTATN FIIINLAFTD PER1a_braF FTKFRSLRSPTT MLLVHLAIAD PER1a_braB FSKFRSLRSPTT MLLVHLAIAD NEUR_strPu SLRKREKLKPID LLTINLAIAD NEUR1_homS SSRRKKKLRPAE IMTINLAVCD NEUR1_calJ SSRRKKKLRPAE IMTINLAVCD NEUR1_canF SSRRKKKLRPAE IMTINLAICD NEUR1_bosT SSRRKKKLRPAE IMTVNLAICD NEUR1_dasN SSKRKKKLRPAE IMTINLAVCD NEUR1_musM SSRRKKKLRPAE IMTINLAVCD NEUR1_ornA SSRRKKKLRPAE IMTVNLAVCD NEUR1_loxA SCRRKKKLRPAE IMTINLAVCD NEUR1_monD SSKRKKKLRPAE IMTVNLAVCD NEUR1_ochP SSRRKKKLRPAE IMTINLAVCD NEUR1_galG SSKRKKKLRPAE IMTVNLAVCD NEUR1_xenT ACSRKKKLRPAE IMTINLAVCD NEUR1_danR TFKRKTKLKPPE IMTLNLAIFD NEUR1_calM SITQKRKLKPPE ILITNLAISD NEUR1a_bra SYRCRARLRPVE MFVVSLAAAD NEUR1b_bra SYRNWAKLRPVE LFVVSLAVTD NEUR2_galG SYKKKHLLKPAE YFIINLAISD NEUR2_anoC SYKKKNLLKPAE YFMINLAISD NEUR2_xenT AYRKRSILKPAE FFIVNLSISD NEUR2_danR AYRKRSSLKPAE FFVVNLSVSD NEUR3_galG AVKRSSLLKSPE LLTVNLAVAD NEUR3_taeG AVKRSSLLKPPE LLTVNLAVAD NEUR3_anoC AVKRSSCLRSPE LLTVNLAATD NEUR3_xenT AVKCSSHLKAPD LLSINLAVAD NEUR3b_dan AYKRSNHMKPPE LLSVNLAVTD NEUR3a_dan AAWRHSVLKAPE LLTVNLAVTD NEUR3a_tet ASRRLTPLKAPE LLTVNLAVTD NEUR3_petM AARRWAKLKAPE LLSVNLALTD PER2a_strP RYRTFRKRSINL LLINMAASDL PER2b_strP RYGTFRKRSVNI LLMNMAVSDL PER1b_braF WRQLCRKAPNLL IINLAAVDLC PER1b_braB WRQLCRKAPNLL VINLAAANLC PER2_braFl TEKEFRKKEHNS FALNLAIADL PER2_braBe TEKEFRKKQQNG FVLNLAIADL PER1a_sacK SNPDYCSKAGN- FFLSLAVTDL RGR1_homSa FCKTPELRTPCH LLVLSLALAD RGR1_ornAn FRKIKELRTPSN LLVVSLALAD RGR1_galGa FRKIKELRTPSN LLVLSIALAD RGR1_xenTr FYKIRELRTPSN LFIISLAVAD RGR1_gasAc FLKVRELRTPSN FLVFSLAVAD RGR1_calMi FYKIKELRTPSN LLITSLALSD RGR2_danRe FLRVREMQTPNN FFIFNLAVAD RGR2_pimPr FLRVREIQTPNN FFIFNLAVAD RGR2_tetNi FLTVKEMRNPSN FFVFNLALAD RGR2_gasAc FLRVKEMWNPSN FFVFNLAVAD RGR2_oryLa FLRVKEMRSPSS FLVFNLALAD MEL1b_braF FCRSRSLRRPKN YLIANLCLTD MEL1b_braB FCRTRSLRRPKN YVVANLCLTD PER1_lotGi EKGLFKYGRAWL HISLAIANVG PER1_aplCa DTKLTKGSQPWL HILLALANVG PER1_todPa ARQSPKPRRKYA ILIHVLITAM NEUR4_ornA LHRQRGILNPTD YLTFNLAVSD NEUR4_galG LYKQRHLLQPTD YLTFNLAVSD NEUR4_taeG LYKQRHVLQPTD YLTFNLAVSD NEUR4_anoc LYRQRAGLQPTD YLTFNLAVSD NEUR4_xenT LYKQRANLLPTD YLTFNLAVSD NEUR4_danR LFRQRSTLQPTD YLTLNLAVSD NEUR4_tetN LVRQRSSLQPTD LLTFNLAVSD NEUR4_gasA LYRQRASLQSTD FLTLNLAISD NEUR4_calM LYRQRLSLQPPD YLTLNLAVSD MEL1_anoCa FFRIRGLRTPAN MFVINLAVSD
(to be continued)
The second cytoplasmic loop
In squid melanopsin, first six residues of cytoplasmic loop C2 also form an extensional helix in squid melanopsin beginning with the DRY motif and surprisingly terminating three residues before the deeply conserved proline (normally a helix breaker as in adrenergic receptors). This proline alone cannot define the two states through its cis and trans configurations because glycine or leucine can also characterize whole opsin orthology classes at this position. The last 3 residues of basic character HRR of loop C2 also preface a transmembrane helix as RAR do in distantly related turkey adrenergic receptor.
Cytoplasmic loop C2 has conserved length of 16-20 in all opsins with much more rigid constraint within individual opsin classes (eg all vertebrate imaging opsins have length 19. The structure of the C2 loop of over 100 melanopsins can readily be modelled based on its closest match among the determined structures, currently squid melanopsin or bovine rhodopsin, with adenosine and adrenergic receptors serving as 'structural outgroup'.
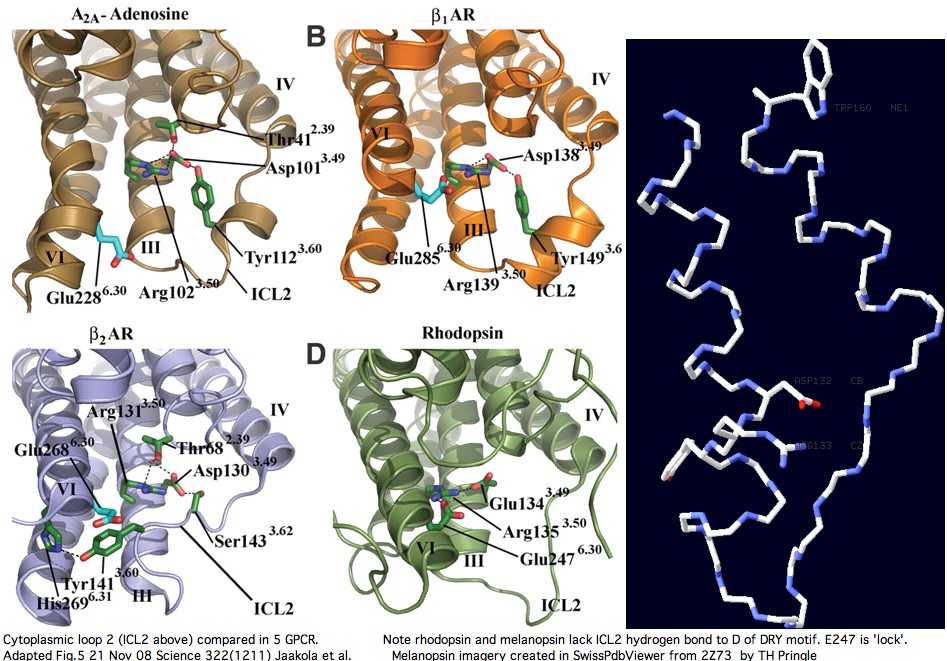
On the basis of length (19 to rhodopsin, 20 to melanopsin), all the opsins except encephalopsin and RGR (both 16 residues) and TMT (18 residues subsequent to a deletion in amniote stem) have a structural model. This model is further constrained by predictable helical extensions of transmembrane helices into the cytoplasm, leaving only the mid-loop region to be predicted. It's not clear whether observed residue conservation -- both within and across orthology classes -- derives from structural importance or instead to Galpha binding specificity requirements.
The adenosine and adrenergic receptor structures -- however useful they might be for annotation transfer to the other 350 non-oderant human GPCR -- ultimately will not prove helpful in modeling the second cytoplasmic loop of opsins (squid melanopsin does that better already). Note C2 in these three structures is consistently stabilized by a mid-loop hydrogen bond to the DRY residues. This constraint is not observed in squid melanopsins or other metazoan opsin classes; indeed it is not feasible because no hydrogen bond-capable residue consistently occurs there (in the comparative genomics sense of conserved residue). Ancestrally, this mid-loop bridge might be a derived feature fairly early in the stem of non-opsin GPCR.
(to be continued)
The third cytoplasmic loop in 83 melanopsins
This loop may be an important contributer to the Gq specificity. The structure has been determined for squid melanopsin, denoted MEL_todPac below. It is a typical 'HEK' extended-helix CL3 found in vast majority of protostome melanopsins. However deuterostome melanopsins never have this feature, yet also appear to signal through Gq. Melanopsin introns within this motif are considered elsewhere.
The orphan Drosophila opsin RH7, which has not yet been associated with an anatomical structure, also lacks the HEK feature and is considerably shorter. However, as the lower sequences in the alignment below show, length variability is by no means unprecedented in this melanopsin loop. Indeed, the one cnidarian opsin available also lacks the HEK motif and also the length of those motifs.
The HEK motif is not specific to wavelength or ommatidia position as the full gamut of drosophila opsins RH1-RH6 have the feature. The motif specifically co-occurs with conserved A.K and more distal A..A whereas a more distal E....K motif are almost universal to all melanopsins -- indeed the E is universal to all opsins (except RGR and peropsin) but not other GPCR. Curiously RH7 has phenylalanine in place of K here. Alanine is inert in terms of side chain potential for interactions, so its conservation is a bit puzzling.
gene transmembrane helix 5 cytoplasmic loop CL3 transmem helix 5 RH1_droMel YYIPLFLICYSYWFIIAAVSA HEKAMREQAKKMN--VKSLRSSEDAE---KSA-EGKLAK VALVTITLWFMAWTPY RH2_droMel YYTPLFLICYSYWFIIAAVAA HEKAMREQAKKMN--VKSLRSSEDCD---KSA-EGKLAK VALTTISLWFMAWTPY LWS1_apiMe YYTPLFTIIYSYYFIVSAVAA HEKAMKEQAKKMN--VTSLRSGDNQN---TSA-EAKLAK VALTTISLWFMAWTPY LWS2_apiMe YFVPLFLIIYSYWFIIQAVAA HEKNMREQAKKMN--VASLRSSENQN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_bomTer YFFPLFLIIWSYWFIiQAVAA HEKNMREQAKKMN--VASLRSSENQN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_catBom YFLPLFLIIYSYFFIIQAVAA HEKNMREQAKKMN--VASLRSAENQS---TSA-ECKLAK VALMTISLWFMAWTPY LWS_papXut YYTPLLLIIYSYFFIVQAVAA HEKAMREQAKKMN--VASLRSSEAAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_manSex YFLPLLLIIYSYFFIVQAVAA HEKAMREQAKKMN--VASLRSSEAAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_vanCar YFSPLFLIIYSYFFIVQAVAA HEKAMREQAKKMN--VASLRSSDAAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_helSar YYAPLFLIIYSYFFIVQAVAA HEKAMREQAKKMN--VASLRSSDAAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_pieRap YFLPLFLIVYSYWFIVQAVAA HERAMREQAKKMN--VASLRSSEQAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_triCas YFVPLFTIIYSYWFIVQAVAA HEKSMREQAKKMN--VASLRSSEAAQ---TSA-ECKLAK IALMTITLWFFAWTPY LWS_rhoPro YFLPLFTIIYSYFFILQAVSA HEKQMREQAKKMN--VASLRSAEAAN---TSA-EAKLAK VALMTISLWFMAWTPY LWS_schGre YLLPLGTIIYSYFFILQAVSA HEKQMREQRKKMN--VASLRSAEASQ---TSA-ECKLAK VALMTISLWFFGWTPY LWS_meoOer YIGPLALIIYCYFHIVSAVAT HEKQMRDQAKKMG--VKSLRTEEAKK---TSA-ECRLAK VALTTVSLWFMAWTPY LWS_neoOer YIGPLALIIYCYFHIVSAVAT HEKQMRDQAKKMG--VKSLRTEEAKK---TSA-GCRLAK VALTTVSLWFMAWTPY LWS_camLud YFLPLAITIYCYVFIIKAVAA HEKGMRDQAKKMG--IKSLRNEEAQK---TSA-ECRLAK IAMTTVALWFIAWTPY LWS_proMil YFLPLTITIYCYVFIIKAVAA HEKGMRDQAKKMG--IKSLRNEEAQK---TSA-ECRLAK IAMTTVALWFIAWTPY LWS_eupSub YLFPFFIIVYCYTYIVSAVFA HEKGMRDQAKKMG--VKSLRNEEAQK---TSA-ECRLAK VALVTVSLWFIAWTPY LWS_homGam YFLPLVIIVYCYTYIVAAVSA HERQMREQAKKMG--VKSLRSEESKK---TSN-ECRLAK VALTTVSLWFIAWTPY LWS_arcGre YYTPLLYIIYAYTFIVQAVSA HEKGMREQAKKMG--VKSLRNEEAQK---TSA-ECRLAK VALMTVSLWFMAWTPY LWS_holCos YLFPLAYIIYSYTFIVKAVAA HEKGMREQAKKMG--VKSLRSEEAQK---TSA-ECRLCK VALMTVTLWFMAWTPY LWS_neoAme YIFPLFLNIYLYTFIIKAVAN HEKQMREQAKKMG--VKSLRSEESQK---TSA-ECRLAK VALMTVSLWFMAWTPY LWS_mysDil YFIPLGITIYCYSYIVHAVAN HEKSMKEQAKKMG--VKSFRNEETQR---TSA-EFRLAK IALMTVSLWFIAWTPY LWS_pedHum YFLPLFIIIYSYIFIIQAVID HENNMRMQAKKME--VASLRSQDDKK---KSV-EIKLAK IALMTIALWFFAWTPY RH6_droMel YLTPLLTIIFSYWHIMKAVAA HEKAMREQAKKMN--VASLRNSEADK---SKAIEIKLAK VALTTISLWFFAWTPY MWS_limPol YALPLMVIIYCYIFIVKAVCD HERHLREQAKKMN--VASLRSNVDTQ---KASAEMRIAK VALVNVLLWVVSWTPY BCR_limPol YALPLMVIIYCYIFIVKAVCD HERHLREQAKKMN--VASLRSNVDTQ---KASAEMRIAK VALVNVLLWVVSWTPY BCR_dapPul YCVPLIIIIFCYYHIVRAIVH HEDALRDQAKKMN--VSSLRSNADQK---SQSAEIRVAK IAMMNITLWVAAWTPY LWS_limPol YFLPLITMIYCYFFIVHAVAE HEKQLREQAKKMN--VASLRANADQQ---KQSAECRLAK VAMMTVGLWFMAWTPY LWS2_plePa YFIPLFTLIYNYTFIVRAVSI HEDNLREQAKKMN--VTSLRANADQQ---KQSAECRLAK IALMTVGLWFIAWTPY LWS2_hasAd YFTPLFTLIYNYTFIVRSVSI HENNLREQAKKMN--VSSLRANADQQ---KQSAECRLAK IALMTVGLWFIAWTPY LWS_ixoSca YWTPLFINIYCYSKIVRAVAQ HEKQLRLQARKMN--VASLRANAEQT---KTSAEARLAK IALMTVGLWFMAWTPY LWS1_plePa YFVPLFIIIYCYTYIVMQVAA HEKSLREQAKKMN--IKSLRSNEDNK---KASAEFRLAK VALMTICLWFMAWTPY LWS1_hasAd YFVPLFIIIYCYAFIVMQVAA HEKSLREQAKKMN--IKSLRSNEDNK---KASAEFRLAK VAFMTICCWFMAWTPY MWS_hemSan FFLPASVIVFSYVFIVKAIFA HEAAMRAQAKKMN--VTNLRSNEAET---QRA-EIRIAK TALVNVSLWFICWTPY RH3_droMel FVCPTTMITYYYSQIVGHVFS HEKALRDQAKKMN--VESLRSNVDKN---KETAEIRIAK AAITICFLFFCSWTPY RH4_droMel FVCPTLMILYYYSQIVGHVFS HEKALREQAKKMN--VESLRSNVDKS---KETAEIRIAK AAITICFLFFVSWTPY UVV_camAbd YCVPMLLIIYYYSQIVGHVVS HEKALREQAKKMN--VESLRSNVNTN---AQSAEIRIAK AAITICFLFVLSWTPY UVV_catBom YCIPMSLIIYYYSQIVSHVVN HEKALREQAKKMN--VESLRSNTNTN---AQSAEIRIAK AAITICFLFVLSWTPY UVV_apiMel YCIPMILIIYYYSQIVSHVVN HEKALREQAKKMN--VDSLRSNANTS---SQSAEIRIAK AAITICFLYVLSWTPY UVV_rhoPro YVIPMSLIIYFYSQIVSHVII HEHNLREQAKKMN--VESLRSNANMH---TQSAEIRIAK AAITICFLFVASWTPY UVV_manSex YVFPMSLIIYFYSGIVKQVFA HEAALREQAKKMN--VESLRANQGGS---SESAEIRIAK AALTVCFLFVASWTPY UVV_papXut YIFPMIAILYFYSGIVKQVFA HEAALREQAKKMN--VDSLRSNQNAA---AESAEIRIAK AALTVCFLYVASWTPY UVV_pedHum YVLPLSLIIYFYTKIVLHVIN HEKSLKAQAKKMN--VESLRSDGNKN----YAVEIRITK VAIAMCFLFVISWTPY UVV_dapPul YVIPLAMLIFYYSKIVRSVGD HEKTLRDQAKKMN--VTSLRSNRDQN---EKSAEVRIAK VAIALATLFVFAWTPY BLU_manSex YCIPMALICYFYSQLFGAVRL HERMLQEQAKKMN--VKSLASNKEDN---SRSVEIRIAK VAFTIFFLFICAWTPY BLU_apiMel YVIPLIFIILFYSRLLSSIRN HEKMLREQAKKMN--VKSLVSN-QDK---ERSAEVRIAK VAFTIFFLFLLAWTPY RH5_droMel YVIPMTMILVSYYKLFTHVRV HEKMLAEQAKKMN--VKSLSANANAD---NMSVELRIAK AALIIYMLFILAWTPY UVV_plePay WFIPVAAIVFFYVQIFLAVKD HEEKIKEQARKMN--VDSIRSNEAVK---NSSAEVRIAK TAMCVFLMFLSSWAPY UVV_hasAda WFIPVAAIIFFYAQIFLAVKD HEEKIKEQARKMN--VDSFRSNEALK---NSSAEVRIAK TAMCVVLLFLTSWVPY MEL_plaDum FIFPVAIIFFCYLGIVRAIFA HHAEMMATAKRMG--A-N--TGKADA---DKKSEIQIAK VAAMTIGTFMLSWTPY MEL_lotGig FVVPLGVIIFCYVFIIKSVMN HEKEMAKMADKLD--AKD--VRSTKE---KAKAEIKIAK VSMTIILLYLMSWTPY MEL_sepOff FCFPILIIFFCYFNIVMAVSN HEKEMAAMAKRLN--AKE--LRKAQA---GASAEMKLAK ISIVIVTQFLLSWSPY MEL_todPac FFGPILIIFFCYFNIVMSVSN HEKEMAAMAKRLN--AKE--LRKAQA---GANAEMRLAK ISIVIVSQFLLSWSPY MEL_entDof FMLPIIIIAFCYFNIVMSVSN HEKEMAAMAKRLN--AKE--LRKAQA---GASAEMKLAK ISMVIITQFMLSWSPY MEL_schMed FIIPVGIIIFCYYQIVKAVRV HELEMLKMAQKMN--ASHPTSMKTGA----KKADVQAAK ISVIIVFLYMLSWTPY MEL_patYes FLIPLIIIGVCYVLIIRGVRR HDQKMLTITRS----MKTEDARANNK---RARSELRISK IAMTVTCLFIISWSPY MEL_schMan FLCPVFIIIFSYYQIVKTVRL NELELMKMAQSLD--LQNPSAMKTGG---DKKADIEAAK TSIILVLLYLMSWSPY MEL_homSap FFLPLLIIIYCYIFIFRAIRE TGRALQTFGAC----KGNGESLWQRQ---RLQSECKMAK IMLLVILLFVLSWAPY MEL_rheMac FFLPLLIIIYCYIFIFRAIRE TGRALQTFGAC----KGSGESLWQRQ---RLQSECKMAK IMLLVILLFVLSWAPY MEL_bosTau FFLPLLIIIYCYIFIFKAIRE TGQALQTFGTC----EGGSECPRQRQ---RLQNEWKMAK IELLVILLFVLSWAPY MEL_proCap FFLPLLVIIYCYVFIFKAIRE TGRALQTFGAC----EGASETPRQWQ---RLQSEWKMAK IALLAILLYVLSWAPY MEL_galGal FFIPLIAIIYSYVFIFEAIKK ANKSVQTFGCK----HGNRELQKQYH---RMKNEWKLAK IALIVILLYVISWSPY MEL_monDom FFIPLIVIIYCYIFIFRAIQD TNKAVHSIGSG-----ESTASPRHCQ---RMKNEWKMAK IALVVILLYVLSWAPY MEL_xenTro FFIPLFIIIYCYIFIFKAIKN TNRAVQKIGTD-----NNKESHKQYQ---KMKNEWKMAK IALIVILLYVVSWSPY MEL_danRer FFIPLIVIIYCYFFIFRSIRT TNEAVGKINGD-----NKRDSMKRFQ---RLKNEWKMAK IALIVILMYVISWSPY MEL_gasAcu FFLPLFIIIYCYFFIFRAIRV TNRAVGKMNGSIHSHGSGRDSTKNFH---RLQNEWKMAK IALIVILLYVVSWSPY MEL_braFlo YFIPMGVIIYCYYNIFATVKS GDKQFGKAVKEMAHE-DVKNKAQQER---QRKNEIKTAK IAFIVITLFLSAWTPY MEL_strPur FVVPVTIIIVCFTRIAITVRA HRHELNKMRTKLTEDKDKKHKSSIRR-ANKAKTEFQIAK VGFQVTIFYVLSWMPY MEL_dapPul FFLPVSVLTFCYAAIFRFILR SSKEITRLIMTSDGTTSFSKSTVSFR-KRRRQTDVRTAL IILSLAILCFTAWTPY BLU_dapPul WVCPLTIITFCYAAIVRAVYR VRQNVTRV---PSQPIDNKHLHQCIN---QPNVEIAIPK IVAGLVLSWIIAWTPY MEL2_schMa FLCPLFLSLFCYARIILIVRS RGKDFIEM---AASSKGTNQKEKSAN-VSSSKSDTFVSK SSAILLGVYLICWTPY MEL3_schMa FMFPVLLCIYCYVNLLKIVRN NERVVLIS---LSNDGASKQRESVRN---RKRLDIEATK SVILSLLFYLMSWTPY MEL_aplCal FVLPFALMVFSYFRIWVAVRK VKSGNVFCAIRHNYNLALGSTLFVKQHRYRLHCEQKTVK IIMFLLIAFTVSWSPY MEL2_lotGi FVLPLCFILFAYSRILHLISS HSR--EMKSYRSAVIISKGKASIPKRFR----SERKTAI TLLITVVVFCLSWVPY MEL_helRob FGMPVSVIILSYIGIIRSIAK NRKEFSSLTAENSS---------------RARQEIKIAK VFAVCMTAFILCWVPY MEL_acrMil YFVPLAIIVYCYVFMIRSVRF MTKNAQKIW--------GVRSAAALE---TVQATWKMAK IGLIMVVGFFVAWTPY RH7_droMel YCIPLTSIVYSYFYILKVVFT ASRIQS-----NKD---------------KAKTEQKLAF IVAAIIGLWFLAWSPY RH7_droYak YCIPLTSIVYSYFYILKVVFT ASRIQS-----NKD---------------KAKTEQKLAF IVAAIIGLWFLAWSPY RH7_droPse YCVPLTTIVYSYFYILKVVFT ASRIQS-----NKD---------------KAKTEQKLAF IVAAIIGLWFLAWSPY RH7_droGri YCIPLTCIVYSYFYILKVVFT ANRIQS-----SKD---------------KAKTEQKLTF IVAAIIGLWFIAWSPY UVV_ixoSca WCVPLVFVTTCYSGILVTVIR SRKALA-----QES---------------R-RSELRVAK VSLALVLLWTVAWTPY RH1_droMel YYIPLFLICYSYWFIIAAVSA HEKAMREQAKKMN--VKSLRSSEDAE---KSA-EGKLAK VALVTITLWFMAWTPY RH2_droMel YYTPLFLICYSYWFIIAAVAA HEKAMREQAKKMN--VKSLRSSEDCD---KSA-EGKLAK VALTTISLWFMAWTPY LWS1_apiMe YYTPLFTIIYSYYFIVSAVAA HEKAMKEQAKKMN--VTSLRSGDNQN---TSA-EAKLAK VALTTISLWFMAWTPY LWS2_apiMe YFVPLFLIIYSYWFIIQAVAA HEKNMREQAKKMN--VASLRSSENQN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_bomTer YFFPLFLIIWSYWFIXQAVAA HEKNMREQAKKMN--VASLRSSENQN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_catBom YFLPLFLIIYSYFFIIQAVAA HEKNMREQAKKMN--VASLRSAENQS---TSA-ECKLAK VALMTISLWFMAWTPY LWS_papXut YYTPLLLIIYSYFFIVQAVAA HEKAMREQAKKMN--VASLRSSEAAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_manSex YFLPLLLIIYSYFFIVQAVAA HEKAMREQAKKMN--VASLRSSEAAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_vanCar YFSPLFLIIYSYFFIVQAVAA HEKAMREQAKKMN--VASLRSSDAAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_helSar YYAPLFLIIYSYFFIVQAVAA HEKAMREQAKKMN--VASLRSSDAAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_pieRap YFLPLFLIVYSYWFIVQAVAA HERAMREQAKKMN--VASLRSSEQAN---TSA-ECKLAK VALMTISLWFMAWTPY LWS_triCas YFVPLFTIIYSYWFIVQAVAA HEKSMREQAKKMN--VASLRSSEAAQ---TSA-ECKLAK IALMTITLWFFAWTPY LWS_rhoPro YFLPLFTIIYSYFFILQAVSA HEKQMREQAKKMN--VASLRSAEAAN---TSA-EAKLAK VALMTISLWFMAWTPY LWS_schGre YLLPLGTIIYSYFFILQAVSA HEKQMREQRKKMN--VASLRSAEASQ---TSA-ECKLAK VALMTISLWFFGWTPY LWS_meoOer YIGPLALIIYCYFHIVSAVAT HEKQMRDQAKKMG--VKSLRTEEAKK---TSA-ECRLAK VALTTVSLWFMAWTPY LWS_neoOer YIGPLALIIYCYFHIVSAVAT HEKQMRDQAKKMG--VKSLRTEEAKK---TSA-GCRLAK VALTTVSLWFMAWTPY LWS_camLud YFLPLAITIYCYVFIIKAVAA HEKGMRDQAKKMG--IKSLRNEEAQK---TSA-ECRLAK IAMTTVALWFIAWTPY LWS_proMil YFLPLTITIYCYVFIIKAVAA HEKGMRDQAKKMG--IKSLRNEEAQK---TSA-ECRLAK IAMTTVALWFIAWTPY LWS_eupSub YLFPFFIIVYCYTYIVSAVFA HEKGMRDQAKKMG--VKSLRNEEAQK---TSA-ECRLAK VALVTVSLWFIAWTPY LWS_homGam YFLPLVIIVYCYTYIVAAVSA HERQMREQAKKMG--VKSLRSEESKK---TSN-ECRLAK VALTTVSLWFIAWTPY LWS_arcGre YYTPLLYIIYAYTFIVQAVSA HEKGMREQAKKMG--VKSLRNEEAQK---TSA-ECRLAK VALMTVSLWFMAWTPY LWS_holCos YLFPLAYIIYSYTFIVKAVAA HEKGMREQAKKMG--VKSLRSEEAQK---TSA-ECRLCK VALMTVTLWFMAWTPY LWS_neoAme YIFPLFLNIYLYTFIIKAVAN HEKQMREQAKKMG--VKSLRSEESQK---TSA-ECRLAK VALMTVSLWFMAWTPY LWS_mysDil YFIPLGITIYCYSYIVHAVAN HEKSMKEQAKKMG--VKSFRNEETQR---TSA-EFRLAK IALMTVSLWFIAWTPY LWS_pedHum YFLPLFIIIYSYIFIIQAVID HENNMRMQAKKME--VASLRSQDDKK---KSV-EIKLAK IALMTIALWFFAWTPY RH6_droMel YLTPLLTIIFSYWHIMKAVAA HEKAMREQAKKMN--VASLRNSEADK---SKAIEIKLAK VALTTISLWFFAWTPY MWS_limPol YALPLMVIIYCYIFIVKAVCD HERHLREQAKKMN--VASLRSNVDTQ---KASAEMRIAK VALVNVLLWVVSWTPY BCR_limPol YALPLMVIIYCYIFIVKAVCD HERHLREQAKKMN--VASLRSNVDTQ---KASAEMRIAK VALVNVLLWVVSWTPY BCR_dapPul YCVPLIIIIFCYYHIVRAIVH HEDALRDQAKKMN--VSSLRSNADQK---SQSAEIRVAK IAMMNITLWVAAWTPY LWS_limPol YFLPLITMIYCYFFIVHAVAE HEKQLREQAKKMN--VASLRANADQQ---KQSAECRLAK VAMMTVGLWFMAWTPY LWS2_plePa YFIPLFTLIYNYTFIVRAVSI HEDNLREQAKKMN--VTSLRANADQQ---KQSAECRLAK IALMTVGLWFIAWTPY LWS2_hasAd YFTPLFTLIYNYTFIVRSVSI HENNLREQAKKMN--VSSLRANADQQ---KQSAECRLAK IALMTVGLWFIAWTPY LWS_ixoSca YWTPLFINIYCYSKIVRAVAQ HEKQLRLQARKMN--VASLRANAEQT---KTSAEARLAK IALMTVGLWFMAWTPY LWS1_plePa YFVPLFIIIYCYTYIVMQVAA HEKSLREQAKKMN--IKSLRSNEDNK---KASAEFRLAK VALMTICLWFMAWTPY LWS1_hasAd YFVPLFIIIYCYAFIVMQVAA HEKSLREQAKKMN--IKSLRSNEDNK---KASAEFRLAK VAFMTICCWFMAWTPY MWS_hemSan FFLPASVIVFSYVFIVKAIFA HEAAMRAQAKKMN--VTNLRSNEAET---QRA-EIRIAK TALVNVSLWFICWTPY RH3_droMel FVCPTTMITYYYSQIVGHVFS HEKALRDQAKKMN--VESLRSNVDKN---KETAEIRIAK AAITICFLFFCSWTPY RH4_droMel FVCPTLMILYYYSQIVGHVFS HEKALREQAKKMN--VESLRSNVDKS---KETAEIRIAK AAITICFLFFVSWTPY UVV_camAbd YCVPMLLIIYYYSQIVGHVVS HEKALREQAKKMN--VESLRSNVNTN---AQSAEIRIAK AAITICFLFVLSWTPY UVV_catBom YCIPMSLIIYYYSQIVSHVVN HEKALREQAKKMN--VESLRSNTNTN---AQSAEIRIAK AAITICFLFVLSWTPY UVV_apiMel YCIPMILIIYYYSQIVSHVVN HEKALREQAKKMN--VDSLRSNANTS---SQSAEIRIAK AAITICFLYVLSWTPY UVV_rhoPro YVIPMSLIIYFYSQIVSHVII HEHNLREQAKKMN--VESLRSNANMH---TQSAEIRIAK AAITICFLFVASWTPY UVV_manSex YVFPMSLIIYFYSGIVKQVFA HEAALREQAKKMN--VESLRANQGGS---SESAEIRIAK AALTVCFLFVASWTPY UVV_papXut YIFPMIAILYFYSGIVKQVFA HEAALREQAKKMN--VDSLRSNQNAA---AESAEIRIAK AALTVCFLYVASWTPY UVV_pedHum YVLPLSLIIYFYTKIVLHVIN HEKSLKAQAKKMN--VESLRSDGNKN----YAVEIRITK VAIAMCFLFVISWTPY UVV_dapPul YVIPLAMLIFYYSKIVRSVGD HEKTLRDQAKKMN--VTSLRSNRDQN---EKSAEVRIAK VAIALATLFVFAWTPY BLU_manSex YCIPMALICYFYSQLFGAVRL HERMLQEQAKKMN--VKSLASNKEDN---SRSVEIRIAK VAFTIFFLFICAWTPY BLU_apiMel YVIPLIFIILFYSRLLSSIRN HEKMLREQAKKMN--VKSLVSN-QDK---ERSAEVRIAK VAFTIFFLFLLAWTPY RH5_droMel YVIPMTMILVSYYKLFTHVRV HEKMLAEQAKKMN--VKSLSANANAD---NMSVELRIAK AALIIYMLFILAWTPY UVV_plePay WFIPVAAIVFFYVQIFLAVKD HEEKIKEQARKMN--VDSIRSNEAVK---NSSAEVRIAK TAMCVFLMFLSSWAPY UVV_hasAda WFIPVAAIIFFYAQIFLAVKD HEEKIKEQARKMN--VDSFRSNEALK---NSSAEVRIAK TAMCVVLLFLTSWVPY MEL_plaDum FIFPVAIIFFCYLGIVRAIFA HHAEMMATAKRMG--A-N--TGKADA---DKKSEIQIAK VAAMTIGTFMLSWTPY MEL_lotGig FVVPLGVIIFCYVFIIKSVMN HEKEMAKMADKLD--AKD--VRSTKE---KAKAEIKIAK VSMTIILLYLMSWTPY MEL_sepOff FCFPILIIFFCYFNIVMAVSN HEKEMAAMAKRLN--AKE--LRKAQA---GASAEMKLAK ISIVIVTQFLLSWSPY MEL_todPac FFGPILIIFFCYFNIVMSVSN HEKEMAAMAKRLN--AKE--LRKAQA---GANAEMRLAK ISIVIVSQFLLSWSPY MEL_entDof FMLPIIIIAFCYFNIVMSVSN HEKEMAAMAKRLN--AKE--LRKAQA---GASAEMKLAK ISMVIITQFMLSWSPY MEL_schMed FIIPVGIIIFCYYQIVKAVRV HELEMLKMAQKMN--ASHPTSMKTGA----KKADVQAAK ISVIIVFLYMLSWTPY MEL_schMan FLCPVFIIIFSYYQIVKTVRL NELELMKMAQSLD--LQNPSAMKTGG---DKKADIEAAK TSIILVLLYLMSWSPY MEL_patYes FLIPLIIIGVCYVLIIRGVRR HDQKMLTITRS----MKTEDARANNK---RARSELRISK IAMTVTCLFIISWSPY MEL_homSap FFLPLLIIIYCYIFIFRAIRE TGRALQTFGAC----KGNGESLWQRQ---RLQSECKMAK IMLLVILLFVLSWAPY MEL_rheMac FFLPLLIIIYCYIFIFRAIRE TGRALQTFGAC----KGSGESLWQRQ---RLQSECKMAK IMLLVILLFVLSWAPY MEL_bosTau FFLPLLIIIYCYIFIFKAIRE TGQALQTFGTC----EGGSECPRQRQ---RLQNEWKMAK IELLVILLFVLSWAPY MEL_proCap FFLPLLVIIYCYVFIFKAIRE TGRALQTFGAC----EGASETPRQWQ---RLQSEWKMAK IALLAILLYVLSWAPY MEL_galGal FFIPLIAIIYSYVFIFEAIKK ANKSVQTFGCK----HGNRELQKQYH---RMKNEWKLAK IALIVILLYVISWSPY MEL_monDom FFIPLIVIIYCYIFIFRAIQD TNKAVHSIGSG-----ESTASPRHCQ---RMKNEWKMAK IALVVILLYVLSWAPY MEL_xenTro FFIPLFIIIYCYIFIFKAIKN TNRAVQKIGTD-----NNKESHKQYQ---KMKNEWKMAK IALIVILLYVVSWSPY MEL_danRer FFIPLIVIIYCYFFIFRSIRT TNEAVGKINGD-----NKRDSMKRFQ---RLKNEWKMAK IALIVILMYVISWSPY MEL_gasAcu FFLPLFIIIYCYFFIFRAIRV TNRAVGKMNGSIHSHGSGRDSTKNFH---RLQNEWKMAK IALIVILLYVVSWSPY MEL_braFlo YFIPMGVIIYCYYNIFATVKS GDKQFGKAVKEMAHE-DVKNKAQQER---QRKNEIKTAK IAFIVITLFLSAWTPY MEL_strPur FVVPVTIIIVCFTRIAITVRA HRHELNKMRTKLTEDKDKKHKSSIRR-ANKAKTEFQIAK VGFQVTIFYVLSWMPY MEL_dapPul FFLPVSVLTFCYAAIFRFILR SSKEITRLIMTSDGTTSFSKSTVSFR-KRRRQTDVRTAL IILSLAILCFTAWTPY BLU_dapPul WVCPLTIITFCYAAIVRAVYR VRQNVTRV---PSQPIDNKHLHQCIN---QPNVEIAIPK IVAGLVLSWIIAWTPY MEL2_schMa FLCPLFLSLFCYARIILIVRS RGKDFIEM---AASSKGTNQKEKSAN-VSSSKSDTFVSK SSAILLGVYLICWTPY MEL3_schMa FMFPVLLCIYCYVNLLKIVRN NERVVLIS---LSNDGASKQRESVRN---RKRLDIEATK SVILSLLFYLMSWTPY MEL_aplCal FVLPFALMVFSYFRIWVAVRK VKSGNVFCAIRHNYNLALGSTLFVKQHRYRLHCEQKTVK IIMFLLIAFTVSWSPY MEL2_lotGi FVLPLCFILFAYSRILHLISS HSR--EMKSYRSAVIISKGKASIPKRFR----SERKTAI TLLITVVVFCLSWVPY MEL_helRob FGMPVSVIILSYIGIIRSIAK NRKEFSSLTAENSS---------------RARQEIKIAK VFAVCMTAFILCWVPY RH7_droMel YCIPLTSIVYSYFYILKVVFT ASRIQS-----NKD---------------KAKTEQKLAF IVAAIIGLWFLAWSPY RH7_droYak YCIPLTSIVYSYFYILKVVFT ASRIQS-----NKD---------------KAKTEQKLAF IVAAIIGLWFLAWSPY RH7_droPse YCVPLTTIVYSYFYILKVVFT ASRIQS-----NKD---------------KAKTEQKLAF IVAAIIGLWFLAWSPY RH7_droGri YCIPLTCIVYSYFYILKVVFT ANRIQS-----SKD---------------KAKTEQKLTF IVAAIIGLWFIAWSPY UVV_ixoSca WCVPLVFVTTCYSGILVTVIR SRKALA-----QES---------------R-RSELRVAK VSLALVLLWTVAWTPY MEL_acrMil YFVPLAIIVYCYVFMIRSVRF MTKNAQKIW--------GVRSAAALE---TVQATWKMAK IGLIMVVGFFVAWTPY
(continued shortly)
The carboxy-terminal tail and VxPx motif
This distinctive region has quite baffling length variation across -- and sometimes within -- opsin classes. The extent of conservation also differs greatly, with no real universally conserved residues past the end of the seventh transmembrane helix. The observed terminal conservation pattern for a given opsin must be indicative of its functional importance, even as that stands today insufficiently explained by arrestin phosphoserine or cysteine palmityolation sites, opsin dimerization or other membrane macro organization, or interaction with Galpha proteins. Some interactions would seem to require commonality across all orthology classes (or larger assemblages such as ciliary opsins) while others do not.
Several studies have implicated the carboxy terminal motif VxPx of ciliary opsins as the intra-cellular targeting motif for proteins that function within cilia (or modified apical cilia such as rod and cone outer segments). The phylogenetic origin or age of this motif function has not been established nor its lineage-specific variations, though cilia themselves are pre-metazoan and the need to direct opsins specifically to outer segments would have been present already prior to lamprey divergence.
The description of the recognition pattern as VxPx alone is unsatisfactory: it is too short and vapid to serve this purpose. The residues valine and proline are all but inert and valine would be hard for the recognition apparatus to distinguish from leucine and isoleucine. Valine and proline would occur by random in this pattern in 4 proteins per thousand; mis-targeting would arise frequently from de novo substitutions in situations where one of V or P was already present. Thus the motif must reflect the end-of-gene position, ie VxPx* properly describes the motif and internal VxPx cannot.
In opsins, we see from cytoplasmic tail alignments below that RGR, peropsin, neuropsins, melanopsins, PPINb and TMT all lack any sign of a terminal VxPx motif. Here TMT is surprising in its total lack of any distal conservation whereas its nearest relative encephalopsin does have a strongly conserved VxPA motif VxPL, x:RK). RHO1 (VAPA), RHO2 (VSPA), SWS2 (VxPy, x:SAG, y:AS), LWS (VxPA X:AS), PPIN (VxPy x:AS, Y:ASLV), PARIE (VxPy x:AST, y:AVL), PIN VxPy x:MTA, y:AS), and VAOP (VxPy x:CY, y:ILM; motif lost in Aves).
Thus the motif is really quite constrained in second and fourth position to a non-bulky uncharged side chain; VxPx does not accurately describe the observed reduced alphabet at these positions. However the carboxy terminus might have other functionalities in addition to ciliary targeting at least in opsins. Conversely it is not so clear that PPIN, PIN, PARIE, VAOP and encephalopsin are specifically targeted to modified pineal, brain, and melanocyte cilia in the same sense that rod and cone opsins are.
Photoreceptor retinol dehydrogenase RDH8, another enzyme of the cis-retinal regeneration cycle located in the outer segments, also terminates in a similar motif VRPR. This is not the case for RDH11, RDH12 or RDH16 nor in arrestin, transducin subunits, cGMP phosphodiesterase subunits, cGMP-gated channel subunits, Na/K/Ca exchanger, RGS9, R9AP, guanylate cyclases 2D and 2F, guanylate cyclase activating protein, phosducin, and recoverin.
RGR
The first hand-gapped alignment below illustrates these issues using RGR from 53 species. The alignment begins inside the last transmembrane segment with the Schiff base lysine K and continues past the NAxxY motif at a deeply invariant length (totaling 19 residues) to the "YR" motif found in almost all GPCR. This marks the beginning of the carboxy terminal cytoplasmic tail, which in RGR is fairly fixed at 23 residues, remain alignable and may extend the transmembrane helix but bear no resemblance to any other opsin or GPCR.
The degree of conservation establishes selection is at work. It appears that RGR must terminate in several charged (characteristically basic) residues regardless of length indels. These could possibly associate electrostatically with membrane phospholipid or be important to initial establishment of topology. Mammals have in effect lost the YR motif though most have an R one residue later. This does not quite coincide with the advent of ERY or GRY mammals in cytoplasmic loop C2.
Conservation of G.WQ.L..Q has persisted for tens of billions of years and cannot be explained by helix or beta sheet per se -- possibly it is constrained by interaction with parts of the other cytoplasmic face. It appears that arrestin could recognize phosphoserine or threonine in almost all species but palmityolation cannot be widespread. A few species, such as guinea pig, microbat and armadillo may be exhibiting early stages of pseudogenization or at least partial loss of function.
Absent any experimental information or relevant 3D structure or capacity for annotation transfer from homologous regions, the specifics of individual residue and residue patch conservation will remain difficult to explain.
K..PT.NA..YaLG.E.yr .G.Wq.L..q..........k.K >RGR_homSap KMVPTINAINYALGNEMVC RGIWQCLSPQKRE-----KDRTK RGR_homSap KMVPTINAINYALGNEMVC RGIWQCLSPQKREKDRTK >RGR_panTro KMVPTINAINYALGNEMVC RGIWQCLSPQKSE-----KDRTK RGR_panTro ................... ...........S...... >RGR_gorGor KMVPTINAINYALGNEMVC RGIWQCLSPQKSK-----KDRTK RGR_ponPyg ................... ...........S...... >RGR_ponPyg KMVPTINAINYALGNEMVC RGIWQCLSPQKSE-----KDRTK RGR_gorGor ................... ...........SK..... >RGR_nomLeu KMVPTINAVNYALGNEMVC RGIWQCLSPQKSE-----KDRAK RGR_nomLeu ........V.......... ...........S....A. >RGR_macMul KMVPTINAINYALGNEMVC RGIWQCLSPQKSE-----KDRAK RGR_macMul ................... ...........S....A. >RGR_papHam KMVPTINAINYALGNEMVC RGIWQCLSPQKSE-----KDRAK RGR_papHam ................... ...........S....A. >RGR_calJac KMVPTIDAINYALGNEMIC RGIWQCLSPQKSE-----KDRTK RGR_calJac ......D..........I. ...........S...... >RGR_tarSyr KTVPTINAYHYALGSEMVC RGIWQCLSPHSSE-----..... RGR_tarSyr .T......YH....S.... .........HSS. >RGR_otoGar KTVPTINAVNYALGSEMVC RGIWQCLSLQRSK-----QDGAK RGR_otoGar .T......V.....S.... ........L.RSKQ.GA. >RGR_micMur KTVPTINAINYALGSETVC RGIWQCLSPQRSE-----QDRAK RGR_micMur .T............S.T.. ..........RS.Q..A. >RGR_tupBel KMVPTVNAVNYALGSETIC RGIWGCLSP-KRE-----RDRAR RGR_tupBel .....V..V.....S.TI. ....G....KR-.R..AR >RGR_musMus KTMPTINAINYALHREMVC RGTWQCLSPQKSK-----KDRTQ RGR_musMus .TM..........HR.... ..T........SK....Q >RGR_ratNor KTMPTINAINYALRSEMVC RGTWQCRSAQKSK-----QDRTQ RGR_ratNor .TM..........RS.... ..T...R.A..SKQ...Q >RGR_cavPor KTVPTINAINYSLG----- RGPWQSLEMQRSK-----QD RGR_cavPor .T.........S..R---- -.P..S.EM.RSKQ. >RGR_dipOrd KMVPTVNAINYALCNELLC GGFSLGLLPQKGK-----QDRTQ RGR_dipOrd .....V.......C..LL. G.FSLG.L...GKQ...Q >RGR_oryCun KTVPTVNAVNYALGSEVIR RGIWQCLLPQRSV-----RGRAQ RGR_oryCun .T...V..V.....S.VIR .......L..RSVRG.AQ >RGR_ochPri KAVPTVNAINYALGSEVIR RGIWQCLLPQRSV-----RDRAQ RGR_ochPri .A...V........S.VIR .......L..RSVR..AQ >RGR_bosTau KAVPTVNAMNYALGSEMVH RGIWQCLSPQRRE-----HSREQ RGR_bosTau .A...V..M.....S...H ..........R..HS.EQ >RGR_susScr KMVPTVNAINYALGGEMVH RGIWQCLSPQRRE-----RDREQ RGR_susScr .....V........G...H ..........R..R..EQ >RGR_canFam KAAPTINAIHYALGGDMVH GGLWQCLSPQRSQ-----PDRAR RGR_canFam .AA......H....GD..H G.L.......RSQP..AR >RGR_felCat kaVPTINAINYALGSEMVH RGIWQCLSPQGSG-----LDRAR RGR_felCat .A............S...H ..........GSGL..AR >RGR_equCab KTVPTINAVNYALGSEMLH RGIWQCLSPQKSE-----RDRAQ RGR_equCab .T......V.....S..LH ...........S.R..AQ >RGR_myoLuc KMVPTVNAVNYALGS---- -GIWQRLSLQ............. RGR_myoLuc .....V..V.....S---- -....R..L. >RGR_pteVam KMAPTINAVNYALGSEMVQ RGIWQCLSPQRSE-----RDHAQ RGR_pteVam ..A.....V.....S...Q ..........RS.R.HAQ >RGR_sorAra KTVPTVNALHYGLGSGMVQ NGFRKGLWLQRRE-----RERAL RGR_sorAra .T...V..LH.G..SG..Q N.FRKG.WL.R..RE.AL >RGR_eriEur ktVPTVNAVHYVLGSEKVH KGFWQCFSPQRSE-----QDRAR RGR_eriEur .T...V..VH.V..S.K.H K.F...F...RS.Q..AR >RGR_loxAfr KAVPVINACHYALGSEVVR GGIWQYLSRQRGESPLRARDRTH RGR_loxAfr .A..V...CH....S.V.R G....Y..R.RG.SPLRAR DRTH >RGR_proCap KAVPIVNACHYALGSETVH RGIWQCLSRQRGESPPRTRDRTQ RGR_proCap .A..IV..CH....S.T.H ........R.RG.SPPRTR DRTQ >RGR_echTel KAVPIVNACHYALGSETVH RGIWQCLSRQRGESPPRTRDRTQ RGR_echTel .A..IV..CH....S.T.H ........R.RG.SPPRTR DRTQ >RGR_choHof KTMPTINAFQYALGSETVC RDIWQCLPRLRSMGRSSGHD RGR_choHof .TM.....FQ....S.T.. .D.....PRLRSMGRSSGH D >RGR_dasNov KTMPTVNALYYALGRESVH RNA RGR_dasNov .TM..V..LY....R.S.H .NA >RGR_ornAna KTVPVIDAFTYALRNEDYR GGIWQFLTGQKIERV-EVENKIK RGR_ornAna .T..V.D.FT...R..DYR G....F.TG..I.RVEVEN KIK >RGR_xenTro KTSPAVNAYVYGLGNENYR GGIWQYLTGQKLEKA-ETDNKTK RGR_xenTro .TS.AV..YV.G....NYR G....Y.TG..L..AE.DN KTK >RGR_xenLae KISPAVNAYVYGLGNENYR GGIWLYLTGQKLEKA-ETDSRTK RGR_xenLae .IS.AV..YV.G....NYR G...LY.TG..L..AE.DS RTK >RGR1_danRer KTSPTFNVFVYALGNENYR GGIWQLLTGQKIESP-AIENKSK RGR1_danRe .TS..F.VFV......NYR G....L.TG..I.SPAIEN KSK >RGR1_takRub KTCPTINVFLYALGNENYR GGIWQFLTGEKIEAP-QIENKSK RGR1_gasAc .TS..F.VFL......NYR G....L.TGE.IDVPQIEN KSK >RGR1_tetNig KTCPTVNVFLYALGNENYR GGIWQFLTGEKIETP-QLENKTK RGR1_gadMo .TA..F.VFL......NYR G....L.TGE.I.VPQIEN KSK >RGR1_gasAcu KTSPTFNVFLYALGNENYR GGIWQLLTGEKIDVP-QIENKSK RGR1_takRu .TC....VFL......NYR G....F.TGE.I.APQIEN KSK >RGR1_oryLat KTSPTFNPLLYALGNENYR GGIWQFLTGEKIHVP-QDDNKSK RGR1_tetNi .TC..V.VFL......NYR G....F.TGE.I.TPQLEN KTK >RGR1_gadMor KTAPTFNVFLYALGNENYR GGIWQLLTGEKIEVP-QIENKSK RGR1_oryLa .TS..F.PLL......NYR G....F.TGE.IHVPQDDN KSK >RGR2_danRer KTSPIFHAVLYAYGNEFYR GGVWQFLTGQK-----SAD-KKK RGR2_danRe .TS.IFH.VL..Y...FYR G.V..F.TG..SADKKK >RGR2_pimPro KTSPIFHAAMYAYGNEFYR GGIWQFLTGQK-----PAD-KKK RGR2_pimPr .TS.IFH.AM..Y...FYR G....F.TG..PADKKK >RGR2_tetNig KTNPIFNALLYTFGNEFYR GGVWHFLTGHKIVDP-VLK-KSK RGR2_tetNi .TN.IF..LL.TF...FYR G.V.HF.TGH.IVDPVL.K SK >RGR2_gasAcu kTNPIFNALLYSFGNEFYR GGVWHFLTGQKMVDP-VVK-KSK RGR2_gasAc .TN.IF..LL.SF...FYR G.V.HF.TG..MVDPVV.K SK >RGR2_oryLat KTNPFFNALLYSFGNEFYR GGVWNFLTGQKIVEP-DVK-KSKQK RGR2_hipHi .TN.IF..LL.SF...FYR G.V.HF.TG..IVDPVV.K SK >RGR2_oncMyk KTNPISNAWLYSFGNEFYR GGVWQFLTGQKFTEP-VVV-KLKGR RGR2_oryLa .TN.FF..LL.SF...FYR G.V.NF.TG..IVEPDV.K SKQK >RGR2_espLuc KMNPIFNALLYSFGNEFYR GGVWQFLTGQKFTEL-VVV-KLKGR RGR2_poeRe .TN.IF..FL.SF...FYR G.V.NF.TG..IVEPDV.K SK >RGR2_gadMor KTNPISNALLYSFGNESYR SGVWHFLTGQKFVEP-SFK-KIK RGR2_oncMy .TN.IS..WL.SF...FYR G.V..F.TG..FTEPVVVK LKGR >RGR2_poeRet KTNPIFNAFLYSFGNEFYR GGVWNFLTGQKIVEP-DVK-KSK RGR2_espLu ..N.IF..LL.SF...FYR G.V..F.TG..FTELVVVK LKGR >RGR2_hipHip KTNPIFNALLYSFGNEFYR GGVWHFLTGQKIVDP-VVK-KSK RGR2_gadMo .TN.IS..LL.SF...SYR S.V.HF.TG..FVEPSF.K IK
Peropsin
Peropsin exhibits greater conservation both in its post-K helix and in its cytoplasmic tail than RGR. The FR motif is perfectly conserved throughout vertebrates. Length, ancestrally 32 residues, experienced an era of variability in amniotes but then settled down to a fixed 35 residues in mammals. The difference alignment shows that a central motif EITISN conserved in early vertebrates changed character completely (to TMPVTS) in mammals, though the earlier motif still appears faded in platypus. A cysteine conserved back to invertebrates might be palmitoylated; conserved serines and threonines offer potential phosphorylation sites.
The cytoplasmic tail of peropsin is completely unalignable to RGR. Unlike RGR, tblastn of peropsin tail against whole human genome elicits matches to imaging opsins and a GPCR (neuropeptide Y receptor). While these matches are weak and largely driven by the last transmembrane section alone, 3 early tail residues (*) emerge as possible conserved residues. Whether or not homologically valid, this suggests modeling of the first 9 residues of peropsin tail by known bovine rhodopsin structure.
* * *
peropsin KSSTFYNPCIYVVANKKFR RAMLAMFKC
KS+T YNP IYV N++FR +L +F C
LWS opsin KSATIYNPVIYVFMNRQFR NCILQLF
RHO opsin KSAAIYNPVIYIMMNKQFR NCMLTTICC
NPY2R GPCR ..STFANPLLYGWMNSNYR KAFLSAFRC
Conserved ksstfynpciyv.ankkFR rAm.aMfkCqthq.mpvts.lpm.vsq.pl.sgr.
PER_homSap KSSTFYNPCIYVVANKKFR RAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI PER_homSap KSSTFYNPCIYVVANKKFR RAMLAMFKCQTHQTMPVTS ILPMDVSQNPLASGRI
PER_panTro KSSTFYNPCIYVVANKKFR RAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI PER_panTro ................... ................... ................
PER_gorGor ksstfynpciyvvankKFR RAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI PER_gorGor ................... ................... ................
PER_ponPyg KSSTFYNPCIYVVANKKFR RAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI PER_ponPyg ................... ................... ................
PER_nomLeu KSSTFYNPCIYVVANKKFR KAMLAMFKWPNHQTMPGTSILPMDVSQNPLTSGKI PER_nomLeu ................... K.......WPN.....G.. ...........T..K.
PER_macMul KSSTFYNPCIYVVANKKFR RAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI PER_macMul ................... ................... ................
PER_papHam KSSTFYNPCIYMVANKKFR RAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI PER_papHam ...........M....... ................... ................
PER_calJac KSSTFYNPCIYVVANKKFR RAMLAMLKCQTHQTMPVTSVLPMDISQNPLASGRI PER_calJac ................... ......L............ V....I..........
PER_tarSyr ksstfynpciyvvankKFR RAMFAMLKCQTYQAMPATSSLPMNVSQNPLTSGKN PER_tarSyr ................... ...F..L....Y.A..A.. S...N......T..KN
PER_otoGar KSSTFYNPCIYVVANKKFR RAMFAMFKCQTHQAMAVTSILPMDISQNPLASRRI PER_otoGar ................... ...F.........A.A... .....I.......R..
PER_micMur KSSTFYNPCIYVIANKKFR RAMFAMFKCQTHQAMPVTSIFPMGVSQNPLPSGRT PER_micMur ............I...... ...F.........A..... .F..G......P...T
PER_tupBel KSSTFYNPCIYVLANKKFR KAMCAMFKCQTHQAMSVTSVLPMASSPRPLAPARV PER_tupBel ............L...... K..C.........A.S... V...AS.PR...PA.V
PER_musMus KSSTFYNPCIYVAAHKKFR KAMLAMFKCQPHLAVPEPSTLPMDMPQSSLAPVRI PER_musMus ............A.H.... K.........P.LAV.EP. T....MP.SS..PV..
PER_ratNor KSSTFYNPCIYVAANKKFR KAMFAMLKCQPHQAMPEPSTLAMGVPHSPLAPARI PER_ratNor ............A...... K..F..L...P..A..EP. T.A.G.PHS...PA..
PER_ochPri KSSTFYNPCIYVAANKRSR RAMFAMFKCQIPQAKPVTSLSPRDVSQSPLSSGRT PER_cavPor ............I...... ...F...Q.....AV..A. .....A..S.......
PER_cavPor KSSTFYNPCIYVIANKKFR RAMFAMFQCQTHQAVPVASILPMDASQSPLASGRI PER_dipOrd ................... ......L......A..... ................
PER_speTri KSSTFYNPCIYVAANKRFR RAMFAMFKCQTHQAMPVTSVLPMDVSQSPRASGRI PER_speTri ............A...R.. ...F.........A..... V.......S.R.....
PER_oryCun KSSTFYNPCIYVAANKRFR RAMFAMFKCQTHQAMPVTSVLPMDVSQNPLPSGII PER_ochPri ............A...RS. ...F......IP.AK.... LS.R....S..S...T
PER_dipOrd KSSTFYNPCIYVVANKKFR RAMLAMLKCQTHQAMPVTSILPMDVSQNPLASGRI PER_oryCun ............A...R.. ...F.........A..... V..........P..I.
PER_bosTau KSSTFYNPCIYVIANKKFR RAMLAMFKCQTTQAMPVTSVLPMDVPQNPLTSGKV PER_bosTau ............I...... ...........T.A..... V.....P....T..KV
PER_turTru KSSTFYNPCIYVIANKKFR RAMLAMFKCQTHQAMPMESILPMDVPQNPLTSGKV PER_turTru ............I...... .............A..ME. ......P....T..KV
PER_susScr KSSTFYNPCIYVIANKKFR RAMLAMFKCQTHQAMPLESTLPMDVPQNPLASGRV PER_vicVic ............I...... .............A..M.. ......P....T...L
PER_vicVic KSSTFYNPCIYVIANKKFR RAMLAMFKCQTHQAMPMTSILPMDVPQNPLTSGRL PER_susScr ............I...... .............A..LE. T.....P........V
PER_canFam KSSTFYNPCIYVVANKKFR KAIFAMFKCQTHQAMPGTSILPMDVSQNPLASGRN PER_canFam ................... K.IF.........A..G.. ...............N
PER_felCat ksstfynpciyvvankKFR KAMFAMFKCENRQPMPVTSILPMDVSQNPLTSGRK PER_felCat ................... K..F.....ENR.P..... ...........T...K
PER_equCab KSSTFYNPCIYVVANKKFR RAMFAMFKCQTHRAMPVTSILPMDVPQNQLASGRI PER_equCab ................... ...F........RA..... ......P..Q......
PER_myoLuc KSSTFYNPCIYVVANKKFR RAMFAMFKCQTHQTMTTMSFLPMDVPQNPLTSGRI PER_myoLuc ................... ...F...........TTM. F.....P....T....
PER_pteVam KSSTFYNPCIYVVANKKFR RAMFAMFKCQDHQSMPVTSVLPMDVPQNPLTSGRI PER_pteVam ................... ...F......D..S..... V.....P....T....
PER_eriEur KSSTFYNPCIYVLANKKFR RAMFAMFKCQTHQAMPVTNTLPMDIPQK-LDSRRN PER_eriEur ............L...... ...F.........A....N T....IP.K-.D.R.N
PER_sorAra KSSTFYNPCIYVVANKKFR RAMSAMLTCRAQGAMPAASTLPMDAAHSPQASGRN PER_sorAra ................... ...S..LT.RAQGA..AA. T....AAHS.Q....N
PER_loxAfr KSSTFYNPCIYVVANKKFR RAMFAMFKCQTHQAEPVTCILPMNVSQNPLAAGRI PER_loxAfr ................... ...F.........AE...C ....N.......A...
PER_echTel ksstfynpciyvvankKFR RAMFALLQCQPQEARRVTSILPMNVSQNPMASGRL PER_echTel ................... ...F.LLQ..PQEARR... ....N.....M....L
PER_proCap KSSTFYNPCIYVVANKKFR RAMLAMFKCQTHQAVPVTNILPMTVSQNSSASGRI PER_proCap ................... .............AV...N ....T....SS.....
PER_choHof KSSTFYNPCIYVVANKKFR TIMFAMLKCQTHQAVPVTSILPMNVSENPLASGRI PER_choHof ................... TI.F..L......AV.... ....N..E........
PER_dasNov KSSTFYNPCIYVVANKKFR RAIFAMLKCQTHQAMPVMSILPMNVSENPLASGRI PER_dasNov ................... ..IF..L......A...M. ....N..E........
PER_monDom KSSTFYNPCIYVAANKKFR RAISAMIRCQTHQSMPISNALPMN PER_monDom ............A...... ..IS..IR.....S..ISN A...N
PER_macEug KSSTFYNPCIYVAANKKFR RAISAMMRCETHQSMPVSNALPLNLT PER_macEug ............A...... ..IS..MR.E...S...SN A..LNLT
PER_ornAna KSSTFYNPCIYVVANKKFR RAMLSMVQCQTHREITITDVLPMNRSRSPLTL PER_ornAna ................... ....S.VQ....REITI.D V...NR.RS..TL
PER_galGal KSSTFYNPCIYVIANKKFR RAILAMVRCQTRQEITISNALPMTVSLSALTS PER_galGal ............I...... ..I...VR...R.EITISN A...T..LSA.T.
PER_taeGut KSSTFYNPCIYVIANKKFR RAILAMVRCQTRQEITINNALPMSVSQSALTSQNSSHLPA PER_taeGut ............I...... ..I...VR...R.EITINN A...S...SA.T.QNSSHL PA
PER_anoCar KSSTFYNPCIYVIANKRFR RAILAMIRCQTRQEITINNVLPMSVSQSTIA PER_anoCar ............I...R.. ..I...IR...R.EITINN V...S...STI.
PER_xenTro KSSTFYNPCIYVIANKKFR RAILSMVQCKSRQEVTLDNHFPMNVSQSTLTT PER_xenTro ............I...... ..I.S.VQ.KSR.EVTLDN HF..N...ST.TT
PER_danRer KSSTFYNPCIYVIANKKFR RAIIGMIRCQTRQRVTINNQLPMMASSVPLNP PER_danRer ............I...... ..IIG.IR...R.RVTINN Q...MA.SV..NP
PER_gasAcu KSSTFYNPCIYVIANKKFR RAIIGMVRCQTRQRITINSQVPMTTSQQPLTQ PER_gasAcu ............I...... ..IIG.VR...R.RITIN. QV..TT..Q..TQ
PER_oryLat KSSTFYNPCIYVIANKKFR RAIIGMIRCQTRQRITISTQVPMTISQQPLTQ PER_oryLat ............I...... ..IIG.IR...R.RITIST QV..TI..Q..TQ
PER_takRub KSSTFYNPCIYVIANKKFR RAIIGMIRCQTRQQMTINTEIPMTTSQQTATQ PER_takRub ............I...... ..IIG.IR...R.Q.TINT EI..TT..QTATQ
PER_tetNig KSSTFYNPCIYVITNKKFR QAIIGMIRCQTRQQITINTDIPMTASQQTLTQ PER_tetNig ............IT..... Q.IIG.IR...R.QITINT DI..TA..QT.TQ
PER_calMil KSSTFYNPCIYVIANKKFR KAIMAMICCQNRQEITINHTLPMTISRVPLTE PER_calMil ............I...... K.IM..IC..NR.EITINH T...TI.RV..TE
PER1b_sacK KIPAVFNPVIYVALNPEFR KYFGKTIGCRRKRKKPIAVRLNGSEQNVENTI PER1b_sacK .IPAVF..V...AL.PE.. KYFGKTIG.RRKRKK.IAV R.NGSEQNVENTI
Neuropsins
Here NEUR1 is a bit unusual in the last transmembrane helix terminating in FA instead of the FR found in the other neuropsin classes. It is not clear how a neutral alanine affects signaling at this key residue. Conceivably the helix is longer and a more distal conserved FR plays this role 19 aminio acids later. The length and sequence of the carboyx terminus is strongly conserved out to the stop codon in available species implying functional significance.
However this region is completely unalignable to other neuropsins past the FR motif. In other neuropsins the carboxy terminus is poorly conserved and alignable past the FR motif only by 6, 10, and 24 residues in NEUR2-4. Some termini are quite extended in seemingly random sequence.
NEUR1_homS KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEGFR LHTVTTVRKSSAVLEIHEEV NEUR1_nomL KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEGFR LHTVTSVRKSSAVLEIHEEV NEUR1_panT KSAAMYNPIIYQVIDYKFA CCQTGGLKET-KKKSLEGFR LHTVTTVRKSSAVLEIHEEV NEUR1_ponP KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEGFR LHTVTTVRKSSAVLEIHEEV NEUR1_macM KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEGFR LHTVTTVRKSSAVLEIHEEV NEUR1_papH KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEGFR LHTVTTVRKSSAVLEIHEEV NEUR1_calJ KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEDFR LHTVTTVRKSSAVLEIHEEV NEUR1_tarS KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEDFR LHTVTTVRKSSAVLEIHEEV NEUR1_equC KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEDFR LHTVTTVRKSSAVLEIHQEV NEUR1_oryC KSAAMYNPIIYQVIDYKFS CCRTSGLKAT-KKKSLEDFR LHTVTTVRKSSAVLEIHQEV NEUR1_canF KSAAMYNPIIYQVIDYKFA CCQTGRLKAT-KKKSLEDFR LNTVTTVRKSSAVLEIHQEV NEUR1_sorA KSAAMYNPIIYQVIDYRFA CCQSGGLRAT-KKKSLDDFR LHTVTTVRESSAVLEIHQEV NEUR1_bosT KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEDFR LHTVTTVRKSSAVLEVHQEV NEUR1_susS KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEDFR LHTVTTVRKSSAVLEIRQEV NEUR1_pteV KSAAMYNPIIYQVIDYKFA CCQTSGLRAT-KKKSLEDFR LHTITTVREASAVLEIHQEV NEUR1_musM KSAAMYNPIIYQVIDYRFA CCQAGGLRGT-KKKSLEDFR LHTVTTVRKSSAVLEIHQEV NEUR1_ratN KSAAMYNPIIYQVIDYRFA CCQTGGLRAT-KKKSLEDFR LHTVTAVRKSSAVLEIHPEV NEUR1_cavP KSAAMYNPIIYQVIDSRFA CCQNAGLKAT-KKKSLEDFR LHTVTTDRKS-AVLEIHQEV NEUR1_ochP KSAAMYNPIIYQVIDYKFS CCRTGGLKQT-KKKSLEDFR LHTVTTVRKSSAVLEIHQEV NEUR1_speT KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEDFR LHTVTAVRKSSAVVEIHQEV NEUR1_myoL KSSAMYNPIIYQVIDYKLA CCQTGGLRAT-KKKSLENFR LHTVTTVRKSSAVLEIHQEV NEUR1_felC KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEDFR LHTVTTVRKSSAVLEIHQEV NEUR1_eriE KSAAMYNPIIYQVIDYKFA CCQTGGLKAN-KKKSLKDYR LHTVTTVRRSSAVLEIHQEV NEUR1_otoG KSAAMYNPIIYQVIDYKFA CCQTGGLKTT-KKKSLEDFR LHTVTTVRKSSAVLEIHQEV NEUR1_turT KSAAVYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEDFR LHTVTTVRKSSAVLEIHQEV NEUR1_vicV KSAAMYNPIIYQVIDYKFA CCQTGGLKAT-KKKSLEDFR LHAVTTVRKSSAVLEIHQEV NEUR1_loxA KSAAMYNPIIYQVIDYKFA CCQTGGLRAT-KKKSLEGFR LHTVTTVKKSSAVLEVHQEV NEUR1_dasN KSAAMYNPIIYQVIDYKFA CCQTGGLRAT-KKKSLEDFR LHTVTTVRESSAVLEVHQEV NEUR1_proC KSAAMYNPIIYQVIDYKFA CCRTRGLRAT-KEKSLEGVR LHTVTTVRKSSAVLEIHQEV NEUR1_choH KSAAMYNPIIYQVIDYKFA CCRTGGLRAT-KKKSFEGFR LHTVTTVRKSSAVLEIHQEV NEUR1_monD KSAAMYNPIIYQVIDCKFA CCQSGGQKAA-KKESLRTYR SHSMSTIRKPSAVSGPHQEV NEUR1_ornA KSAAMYNPIIYQVIDCRIS CCRLGGPKTG-KKESLKNSR LHTVTTVRKSSAVLEIHEEV NEUR1_galG KSAAMYNPIIYQVIDCKFA CCRSGGPKTLQKKSSLKESR MYTISSHRDSAALSGTQLEV NEUR1_taeG KSAAMYNPIIYQVIECRLA CCRPGG---LKAKSSLKKSR TYTISAHRDSTAMNETQLEA NEUR1_anoC KSAAMYNPVIYQVIDCKSA CCRPGNLQPLQKK----NSR ... NEUR1_xenT KSASMYNPIIYQVIDCKPA CCKKDKS--LQNT----TSR VYTISTFRKSTTSAR NEUR1_danR KSSAMYNPIIYQVIDCKKK CVKSCCFQAWRKKKPSKTSR FYTISGSIKQRPGDEASIEI NEUR1_takR KSSAMYNPIIYQVVDVKTS CTNFSCCKALKERIHFRKSR FYSISASMKKRPANEVPTEI NEUR1_tetN KSSAMYNPIIYQVADLKTS CTSSSCCKALKERVLFRKSR YTISGSLRDTLPPKEAHIEM NEUR1_gasA KSSAMYNPIIYQVLDLKNS CMKSSCFKGLKKPRHFRKSR YTISGSLRDTLPPKEAHIEM NEUR1_oryL KSSAMYNPIIYQVLDLKNS CMKSSCFKGLKKPRH---FR YTISGSLKDTAPAKEAHIEI NEUR1_pimP KSSAMYNPIIYQVIDCKKN CAKLSCFQAWSKRKHYKTSR FYSISASMKKRPANEVPTEI NEUR1_petM KSAAMYNPLIYQLLSRRGT GAHCCRCRKARGTLRR--PR ... NEUR2_galG KSSTLYNPIIHLLLKPNFR SNIAKD FTVIQQLCVRCCFCVKELQTYRSTFNTGLRTFKGK--NESSCNALPIMEGCSYFP... NEUR2_anoC KSSTLYNPAMYLFLKPNFR STIAKD LTVLHRLCLKSCFCPRGMQNCSYRSALEAPLKSFKGRNESSSNSVQIVGGCSYFP... NEUR2_xenT KSSTIYNPVVYLLLKPNFL NVVTKD LTLFQTMCAVVCGWCRTPAVKTPCPHKDLKTTSKPPSSFKKSQGVHRICLSHSKASP... NEUR2_oncM KSSTIYNPIIYLLLRPNFR RVMYRD LVSLCRAFLKGCLCSCSQGAVGKCHSHLVVRVSLQSFCRLPGHGQSCSPTSSARQALGESRG NEUR2_danR KSSTIYNPMVYLLFKPNFR KSLSQD TQMFRHRICLSHSKASPSPGMKDQERQSSQQCNNKDGSISTPFSSGQAESYGA NEUR2_pimP KSSTIYNPMVYLLFKPNFR KILSQD TQNIRHRMCVSHSKASPTPEIKAQSSQQCKDATISTPFSSGQAESYGT NEUR2_tetN KSSTIYNPLVYLLCKPNFR ECLYKD TSTLRQRIYRGSPLSGPRDRSGGV-TQRHKDLSVSTRLSNGQQDSYGT NEUR2_takR KSSTIYNPVVYLLCKPNFR ECLYKD TSTLRQRIYRGSPQSEPRERFGGT-SQRHKDLSISTRLSNGQQDSYGT NEUR2_gasA KSSTIYNPVVYLLCKPNFR ACLYRD TTLLRQRIYRGSPRSEPKAHFGST-SQRNKDMSVSVRSSNGQQDSYGA NEUR2_oryL KSSTIYNPMVYLLCKPNFR ECLCRD TSLLRHMIYRGSPQPQER--FGSD-SRRNKDITASTRFSNGQQESYGA NEUR3_galG KSSTAYNPFIYYIFSKTFR HEIKQLQCCW GWRVHFFSADNSAENSVSMMWSGRDNIRLSPTAKVESQGAARH NEUR3_taeG KSSTAYNPFIYYVFSKTFR CEVKRLQCCC AWRVHYFSSDNSVENPLSTMWSGRDNIRLSAAPQVQNPGAAAP NEUR3_xenT KSSTAFNPMIYYAFSKTFR RKVKHLKCCC GWRVHFLQSENSVENPRVSVIWTGKENVMVSSVPKLMKGVPGTPTGTQ NEUR3_anoC KSSTAYNPFIYYTFSKTFR HEVKHLRCYS GQRAQENMKNSINSNVSFMWHGGGNICLSTRQIEMREIPNQ NEUR3a_dan KCSTVYNPLVYYVFRKSFR REIHQIRICC FQGCWDAVSKMTRGDGPEETSGTHETDNI NEUR3a_tet KSSTVYNPVIYYIFSQSFK LEVQQLFLCC LSFRSSRTNNCKSNESSIFMVSNGKNLTPALTQQNTSHAVIMN NEUR3a_tak KSSTAYNPFIYFFFQRNTG HKLLPFHRHAFSCSDRADSSREGEKEESKVSKNLGFTCFGAGTYETCPGLAGDQSQREMAELG NEUR3a_gas KSSTVYNPFIYFIFQRSSW RELLRLHRHLLCCWHRASPPAEGRRSQRGSEGGSWGGACESDDAFGLVHVMKSNATCQTISWA NEUR3b_dan KTSTVYNPFIYYIFSKTFK REVNQLSRFC GRSNICRPTDAKNRPENTIYLVCDVNKSKPGVEDLSLARSKENETQMLPNQDLHE NEUR3b_ory KSSTVYNPMIYYFFSKSFQ REVKQLSWLC VGSNPCHVSNSVNDNNIYMVSVNVKSKETRRETLQEITESRQEDITNERVER NEUR3b_tet KSSTVYNPVIYYIFSQSFK LEVQQLFLCC LSFRSSRTNNCKSNESSIFMVSNGKNLTPALTQQNTSHAVIMN NEUR3b_tak KSSTVYNPIIYYMFSQSFK MEVQQLFLWC PSFEFCRTSSNNGNETTIYMVSTGKT NEUR3b_gas KSSTVYNPLIYYIFSQSFR REVKQLWRHL GSTLCSVSNSVNDAAVSNTGKSN NEUR4_ornA KSASFYNPIIYFGMNSKFR KDILVLLPCAKESKEPVKLKKFKNLR QKQGFTLQKPEKAHVLQV NEUR4_galG KSASFYNPIIYFGMSSKFR RDIFILFHCAKEVKDPVKLKRFKNLK QKQEPSQKEEKYAAEMHPA NEUR4_taeG KSASFYNPIIYFGMSSKFR RDIFIFHCAKELKDPVKLKRFKNLKP KQPQPSQKEEKYAPEMHPA NEUR4_anoc KSASFYNPIIYFGMSSKFR KDIFVLLHCAKEIKDPVKLKRFKNLK QKQEVSPSQREEKYAADVQPA NEUR4_xenT KSASFYNPLIYFGMSSKFR KDLCVVLPCAKAQKDPVKLKRYKDKK QGSAPRAREQTEIEQPVQLQPA NEUR4_danR KSASFYNPLIYFGLSSKFR KDVSVLLPCGREGRDPVRLKRFKRLR GRAEPPGAPAHTPHPQIALKNYNNHSKPHAGPAHCTGH NEUR4_tetN KSASFYNPLIYFGMSSKFR KDVSLILPCAKERREVVLLQRFKNIK PKAAAAPPPPPLPVYRPKEKNEDEPKLSV NEUR4_gasA KSASFYNPLIYFGMSSKFR KDVSVLVPCTRERREVVHLQHFKNIK PKAEAPPTPASLPVQKLGAKYAVPN NEUR4_calM KSASFYNPMIYFGLNSKFR KDIYILLPCVKEPKESVKLKRFKHLR HRPEQQQANKDRYAEELQQV NEUR4_petM KSASFYNPFIYFGMSGKFR ADVRAMLPCRATSVKAPRDAVRLKRY RTHVDPERASHRAAVAAREQPAPRAAAPRPASPAP
Melanopsins
The cytoplasmic tail in melanopsin can be quite variable in length and sequence. No strongly conserved residues exist in bilateran melanopsins beyond the P.L beginning at position 8; consequently very little can be learned about the cytoplasmic tail of vertebrate or even arthropod melanopsins from study of molluscan melanopsins. Its contribution to structure and function of the cytoplasmic face must be quite variable. Note the FR motif is almost always YR outside of lophotrochozoans.
Within just vertebrates, the cytoplasmic tail of melanopsin exhibits much more extensive conservation of 11 residues extending out to position 66 (human numbering). The two conserved serines might be cyclically phosphorylated and the single cysteine at position 9 palmitoylated (as it cannot be in a disulfide residing in the reduced cytoplasmic milieu). While the remaining residues are very likely stably structured, it's not clear whether they interact primarily with the other cytoplasmic loops or with auxiliary proteins. The latter is more likely recalling that melanopsins signal via Gq and the inositol triphosphate cascade rather than the very different cyclic nucleotide pathway.
just vertebrates: full length cytoplasmic tail, mammals MEL1_homSa KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLTSHTSNLSWISIRRRQESLGSESEVGWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGKTKGLIPSQDPRM MEL1_panTr KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTLISHTSNLSWISIRRRQESLGSESEVGWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGHEAETPGKTKGLIPSQDPRM MEL1_ponpy KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSRRHSRPYPSYRSTHRSTMISHTSNLSWISGRRRQESLGSESEVGWTHMEAAAVWGAAQQANGRFLYDQGLEDLEAKAPPRPQGEEAETPGKTKGLIPSQDPRM MEL1_rheMa KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSRRHSHPYPSYRSTHRSTLISHTSNLSWISGRRRQESLGSESEVGWTHMEAAAVWGAAQQANGRSLYGQGLEDLEAKAPPRPQGQEAETPGKTKGLLPCKDSRM MEL1_calJa KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSRRHSHPYPSYRSTHRSTLISHTSNLSWISGRRRQESLGSESEVGWTHMEAAAAWGAAQQANGRSLYGHGLEDLEAKAPPRPQRQEAETPGKTKGLIPSQDPRM MEL1_otoGa KASAIHNPIIYAITHPKYR VAIAQHLPCLGLLLGVSRQHSRPYPSYRFTHHSTLSSQASDLSWISGRRRQESLGSESEVGWTDMEAAATWGAALQVSGQCPYSQGLEDMEAKGPLRPQGPETKTSGKTKGLLPSLDPRM MEL1_musMu KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGRKRQESLGSESEVGWTDTETTAAWGAAQQASGQSFCSQNLEDGELKASSSPQVQRSKTPKTKGHLPSLDLGM MEL1_ratNo KASAIHNPIIYAITHPKYR AAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGQKRQESLGSESEVGWTDTETTAAWGAAQQASGQSFCSHDLEDGEVKAPSSPQEQKSKTPKTKRHLPSLDRRM MEL1_nanSp KASAIHNPIIYAITHPKYR LAISQHLPCLGVLIGVSSQRSHPSLSYRSTHRSTLSSQASDLSWISGRKRQESLGSESEVGWTDTEVTAAWGVAQEASGWSPYRHSLEDGEVKASPSPQGQEAKTSRKTKGQLPSLNLRM MEL1_phoSu KASAIHNPIVYAITHPKYR AAIAQHLPCLGVLLGVSSQRNRPSLSYRSTHRSTLSSQSSDLSWISAPKRQESLGSESEVGWTDTEATAVWGAAQPASGQSSCGQNLEDGMVKAPSSPQAKGQLPSLDLGM MEL1_bosTa KASAIYNPIIYAITHPKYR LAIAQHLPCLGVLLGVSGQRTGLYTSYRSTHRSTLSSQASDLSWISGRRRQASLGSESEVGWMDTEATAAWGAGQQVSGWSPCSQRLDDVEAKALPRPQGRDSEAPGKAKGLLPNLDARM MEL1_canfa KASAIHNPIIYAITHPKYR MAIAQHLPCLGVLLGVSGQRTGPYASYRSTHRSTLSSQASDLSWISGRRRQASLGSESEVGWMDTEAAAVWGAAQPAGGRFLCTQGLEDAEAKAPLRPRGQAVETPGKTKGRLPSLDPSR MEL1_felCa KASAIHNPIIYAITHPKYR MAIAQHLPCLGVLLGVSGQHTGPYASYRSTHRSTLSSQASDLSWISGRRRQASLGSESEVGWMDTEAAAVWGAAQQVSGRFPCSQGLEDREAKAPVRPQGREAETPGQTKGLLPSQDPRM MEL1_equCa KASAIHNPIIYAIIHPKYR MAIAQHLPCLGVLLGVSSQRTRPYTSYRSTHRSTLSSQGSDLSWISGRRRQASLGSESEVGWMDTEAAAVWGAAQQMSGWSPCGQGLEDMEAKAPPRPQGWEGEALRKIKGLLPSLDPRM MEL1_micMa KASAIHNPIIYAITHPKYR VAIAQHLPCVGVLLGVSRQHSRPYPSYRSTHRSTLSSQASDLSWISGRRRQESLGSE MEL1_eriEu KASAIHNPIIYAITHPKYR MAIAQHLPCLRVLLGVSGQRDRPYTSYRSTHRSTLSSQISDLSWVSRRRRQASLGSESEVGWTDTEVAAVWGTMSGHFPCGQGLDDMEAKAAHNPRGLEAETPGKIKGLLPSLDPQM MEL1_loxAf KASAIHNPIIYAITHPKYR MAIAQHLPCLGVMLGVSGQRTRPYTSYHSTLHSTLSSQASDLSWISGRRRQASLGSESEVGWTDTEAAAAWEGAQQVSGQASCSQALQNLEANTPPRPQGWGPETPRK MEL1_proCa KASAIHNPIIYAITHPKYR MAIAQHLPCLGVLLGVSDQHTRPYTSYRSTHHSTLSSQASDISWISGRRRQASLGSESEVGWTDTEAAAAWEGAQQVSGRASCSQVLESMEANTPPRPQGWGPETPRK MEL1_dasNo KASAIHNPIVYAITHPEYR MAIAQHLPCLGLLLGVLGHRPRPGSSPGSTRCSAHSGQASGLSWISRQRRRASLGSKDEVGWEDVEAAAASGAAGQESGRSPRAQDLEHMEAEAARWPSWEAEPEK MEL1_monDo KASAIHNPIIYAISHPKYR MAIAQNFPCLRALLCVRHPRTRSFSSYRFTRRSTMTSQASDISWLPRGRRQLSLGSESEIGWNNMEAGTTSLTSRNQQGSCRMDQETMETRELAAIAKAKGRSWETLEKTLEEMDDSSLLE MEL1_smiCr KASAIHNPIIYAISHPKYR MAIAQNFPCLRAVLGIRHPRTQSFSSYRFTHRSTTASQASDISWQSRGRRQLSLGSESEAGWNNIETGLTLRSLEGSCGMDEETMDTRELSASTKAKGQSWETLAKTLEEMDDLSLLE MEL1_ornAn KSSAIHNPIIYAITHPKYR MAITKYIPCLGPLLRVSRQDSRSSSHYASSRRSTVTSQSLDGSWLPGRRRPLSSASDSESGWTDTAADAGSASSRAASRQVSYRMSQGPTEHCDLRAKVKPKSWEVGSFQK MEL1_taeGu KASVIHNPIIYAITHPKYR KAIATYVPCLGPLLRVSPKDSRSFSSYHSSRRATISSQSSEISGLQERKRRLSSLSDSESGCTETETDTPSMFSRLARRQISYKTDKDTTQTSDIRAKLTSQDSGWGVA MEL1_galGa KASAIHNPIIYAITHPKYR TAIATYVPCLGFLLRVSPKESRSFSSYPSSRRTTITSQSSETSGLQKGKRRLSSISDSESGCTDTETDITSMISRPASSQVSYEMGEDTTQTSDLGGKPKVKSHDSGIFGKAVVDADEIPM MEL1_xenTr KASAIHNPIIYAITHPKYR MAIAKYIPCLGSLLRVKRRDSRSYSSYPSSRRSTVTSHCSQSSDVGGHPKLKNHLPSVSDSESGWTDTEADSSVNSRPASRQVSYEMGKDTTETNDLKSKAKLKSHDSGIFEKTSMDADDISL MEL1_anoCa KASVIHNPIIYAIVHPKYR MAIAKFLPCLGSLLRVPRKDSSYPSTRRPTVTSQSSDINGVPRGHRRLSSVSDSESDWTDTEADISSQNSRVASGSISYRIYEDTTETIKVKSKMRSHDSGIFERTSVDADDISM MEL1a_danR KASAIHNPIIYAITHPKYR LAIAKYIPCLRLLLCVPKRDLHSFHSSLMSTRRSTVTSQSSDMSGRFRRTSTGKSRLSSASDSESGWTDTEADLSSMSSRPASRQVSCDISKDTAEMPDFKPCNSSSFKSKLKSHDSGIFEKSSSDVDDVSV MEL1_takRu KASAIHNPIIYAITHPKYR LALAKYIPCLGFLLCISPHELQSTSSSFMSLRRSTVTSQTSDISGQFRPQSKPRRSSASDSESCLTDTEADLSSMGSRPASRQVSCDISRDTTELPEYKPASSFNSKVKSPDSGIFEKTSFDFDASM MEL1_gasAc KASAIHNPIIYAITHPKYR IALAKYIPFLGVLLCVPPRELRSASSSFRSTRRSTVTSQTSDVSSQQRRQGSRNSRLSSASDSESCLTDTEADGSSVGSRPASRQVSCDIGRDTAELPEFKPSSSFKSKMKSHDSGIFEKSYDTDISM MEL1_oryLa KASAIHNPIIYAITHPKYR MALAKYIPGLGVLLCIHPKDLRSASSSFVSTRRSTVTSQSSDISSQLRRQSTFKSRLSSLSDSESGLTDTEADLSSLSSRPASRQVSCEISRDTAELPDFKHTSSFKAKLKNNDSGIFEKTSFDTVSI MEL1b_danR KASAIHNPIIYAITHPKYR SAIAKYIPCLGVLLCVPRRDRFSSSSFISTRRSTLTSQSSETSSNLHRAGKARLSSVSDSESGWTDTEADLSTASSRPASRQVSSEIRKDLCDIKHSSSLRLKVKSRDSGIFDRQNDVS MEL1_rutRu KASAIHNPIIYAITHPKYR AAIARYIPVLRTILRVKEKELRSSFSSGSVSSRRPTLSSQCSLGVSIGNNGRWGKKRLSSASDSDSCWTESEADGSSVSSLTFGRRVSTEISTDTVILSPGSSNSTASGQKSEKAHKVVSVPVPSITFETDSA MEL1_astBu KASAIHNPIIYAITHPKYR AAIGHYVPFLRSVLRLQEKDLRSSFSSSATSSRCTTFTSSPKGRLNANGHQAQNRLSSVSDSKSCWMESDADGSSRRSERQAFSEATANPLDSTTPRQHVGHTDASSSDGAVLEAKLPL MEL2_galGa KASAIYNPIIYAIIHPRYR KTIHNAVPCLRFLIRISKNDLLRGSINESSFRTSLSSHQSLAGRTKNTCVSSVSTGEANWSDVELDTVEPAHEKLQPRRSHSFSSSLRQKRDLLPDSYSCSEETEEKVSLSSSY MEL2_taeGu KASAIYNPIIYAIIHPRYR KTIHQAVPCLRFLIRISKNDLLRGSINESSFRTSLCSHHSLAGKTKSICVSSISTGEATWSNVELDPVEPAQEKLKPRRSNSFSTSLRQEKRDLLPKTCSYDAATAQKVSLSSSC MEL2_anoCa KASAIYNPIIYAIIHPRYR RTIRSAVPCLRFLIPISKSDLSTSSMSESSFRASVSSRHSFSYRNKSTYISSISAKETTWCDVELDPVESGHKKLQAYRSNSFSAKGVAEEESGLLLRTNNCNVPARKKVALSSIS MEL2_podSi KASAIYNPIIYAIIHPRYR RTIRSAVPCLRFLIRISPSDLSTSSVNESSFRASMSSRHSFAARNKSSCVSSISAAETTWSDMELEPVEAARKKQQPHRSRSFSKQAEEETGLLLKTQSCNVLTGEKVAVSSIS MEL2_tetNi KASAIYNPIIYAIIHPRYR KTIRSAVPCLRFLIPISKSDLSTSSMSDSSFRSALSCRHSYRSRSTYISSISAKETTWCDVELDPVESGHKKLQAYRSNSFSAKGVAEEESGLLLRTNNCNVPARKK MEL2_gadMo KASAIYNPFIYAIIHSKYR DTLAEHVPCLYFLRQPPRKVSMSRAQSECSFRDSMVSRQSSASKTKFHRVSSTSTADTQVWSDVELDPMNHEGQSLRTSHSLGVLGRSKEHRGPPAQQNRQTRSSDTLEQATVADWRPPL MEL2_xenLa KASAIYNPIIYGIIHPKYR ETIHKTVPCLRFLIREPKKDIFESSVRGSIYGRQSASRKKNSFISTVSTAETVSSHIWDNTPNGHWDRKSLSQTMSNLCSPLLQDPNSSHTLEQTLTWPDDPSPKEILL MEL1a_calM KASAIHNPIIYAITHPKYR MAIAKYVPLLGLLLRVSRRDSRTSGQYYSTRRSTLTSQTSDLSGYPRGKGRLSSASDSES MEL2_danRe KSSAIYNPFIYAIIHNKYR RTLAEKVPGLSCLSRSQKDGLSSSTNSDASAQDSSVSRQSSVSKNRLHSTMVQ MEL2_gasAc KASAIYNPFIYAIIHNKYR MTLAAKFPCLRFLSPTPRKDTSSSISESSYRDSVISRQSTASRTHFITACPDTVN all eumetazoan taxon consensus KASA..NPI.YAI.HPKYR .......P.L......... loph MEL1_todPa KASAIHNPMIYSVSHPKFR EAISQTFPWVLTCCQFDDK loph MEL1_plaDu KASARYNPIIYALSHPKFR AEIDKHFPWLLCCCKPKPK loph MEL1_lotGi KASAMHNPVIYALSHPKFR DAVSKLMPWFLCCCGLTDA loph MEL1_sepOf KASAIHNPLIYSVSHPKFR EAIAENFPWIITCCQFDEK loph MEL1_entDo KASAIHNPIVYSVSHPKFR EAIQTTFPWLLTCCQFDEK loph MEL1_patYe KSSSMHNPVVYALSHPKFR KALYQRVPWLFCCCKPKEK loph MEL1_capCa KASAMWNPILYALSHPKFR AALEDHMPWLLVC loph MEL_schMed KTSAMYNPFIYAINHPKFR IQLEKKFPCLICCCPPKPK loph MEL1_schMa KTSAVYNPIVYAVKHPKFR MEIEKRFPFLICCCPPKPK loph MEL3_schMa KMAAIYNPILYAFTNRKFK NALGIRKTSSVIMQQQRLL loph MEL_aplCal KTSMVFNPILYSISHPKVR KRIANLACCYSVRRHQQQT loph MEL2_lotGi KLSTVTNPILYSLSHPVVR NKLFLRLRHELYRRPSDSV arth CHEL_LWS_l KANSCYNPIVYGISHPRYK AALYQRFPSLAC-GSGESG arth CHEL_LWS_i KANACYNPIVYGISHPKYR AALARRFPSLVCMPPGGDQ arth INSE_LWS1_ KANAIYNPIVYGISHPKYR AALKEKLPFLVCGSTEDQT arth INSE_LWS2_ KANAVYNPIVYGISHPKYR AALFAKFPSLACAAEPSSD arth CRUS_LWS_m KANAVYNPIVYAISHPKYR AALYKKLPCLACSTESADE arth CRUS_LWS_n KSNAVYNPIVYAISHPKYR AALYKKLPCLACSTESADE arth INSE_LWS_d KANAVCNPIVYGLSHPKYK QVLREKMPCLACGKDDLTS arth INSE_MWS1_ KSAACYNPIVYGISHPKYR LALKEKCPCCVFGKVDDGK arth INSE_MWS_c KSAACYNPIVYGISHPKYG IALKEKCPCCVFGKVDDGK arth INSE_MWS2_ KTSAVYNPIVYGISHPKYR IVLKEKCPMCVFGNTDEPK arth INSE_UVV_c KFVACLDPYVYAISHPRYR LELQKRLPWLE--LQEKPV arth INSE_BLU_m KVVSCIDPWVYAINHPRYR AELQKRLPWMGVREQDPDA arth INSE_BLU_a KTVSCIDPWIYAINHPRYR QELQKRCKWMGIHE--PET arth INSE_BLU_d KSVSCLDPWVYATSHPKYR LELERRLPWLGIREKHATS arth INSE_UVV_r KAVACVDPYVYAISHPRYR KAFQRFFFKNVITPSQTGG arth INSE_UVV2_ KTASCIDPFVYAATNRRFR NELKRKYRKRSRYQPSLKT cnid ENC_nemVec KTSACYNPIIYFFMYSKFR QELSKKFPWLDIKEAPAPS mamm MEL1_homSa KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSRR mamm MEL1_rheMa KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSRR mamm MEL1_calJa KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSRR mamm MEL1_otoGa KASAIHNPIIYAITHPKYR VAIAQHLPCLGLLLGVSRQ mamm MEL1_micMu KASAIHNPIIYAITHPKYR VAIAQHLPCVGVLLGVSRQ mamm MEL1_bosTa KASAIYNPIIYAITHPKYR LAIAQHLPCLGVLLGVSGQ mamm MEL1_susSc KASAIYNPIIYAITHPKYR MAIAQHLPCLGVLLGVSGQ mamm MEL1_equCa KASAIHNPIIYAIIHPKYR MAIAQHLPCLGVLLGVSSQ mamm MEL1_myoLu KASAIHNPIIYAITHPKYR MAIAQHLPCLGLLLGVSGQ mamm MEL1_pteVa KASAIHNPIIYAITHPKYR MAIAQHLPCLGVLLGMSGQ mamm MEL1_felCa KASAIHNPIIYAITHPKYR MAIAQHLPCLGVLLGVSGQ mamm MEL1_canFa KASAIHNPIIYAITHPKYR MAIAQHLPCLGVLLGVSGQ mamm MEL1_proCa KASAIHNPIIYAITHPKYR MAIAQHLPCLGVLLGVSDQ mamm MEL1_eriEu KASAIHNPIIYAITHPKYR MAIAQHLPCLRVLLGVSGQ mamm MEL1_musMu KASAIHNPIIYAITHPKYR VAIAQHLPCLGVLLGVSGQ mamm MEL1_ratNo KASAIHNPIIYAITHPKYR AAIAQHLPCLGVLLGVSGQ mamm MEL1_nanEh KASAIHNPIIYAITHPKYR LAISQHLPCLGVLIGVSSQ mamm MEL1_phoSu KASAIHNPIVYAITHPKYR AAIAQHLPCLGVLLGVSSQ mamm MEL1_smiCr KASAIHNPIIYAISHPKYR MAIAQNFPCLRAVLGIRHP mamm MEL1_monDo KASAIHNPIIYAISHPKYR MAIAQNFPCLRALLCVRHP mamm MEL1_loxAf KASAIHNPIIYAITHPKYR MAIAQHLPCLGVMLGVSGQ mamm MEL1_ornAn KSSAIHNPIIYAITHPKYR MAITKYIPCLGPLLRVSRQ tetr MEL1_anoCa KASVIHNPIIYAIVHPKYR MAIAKFLPCLGSLLRVPRK tetr MEL1_taeGu KASVIHNPIIYAITHPKYR KAIATYVPCLGPLLRVSPK tetr MEL1_galGa KASAIHNPIIYAITHPKYR TAIATYVPCLGFLLRVSPK tetr MEL1_xenTr KASAIHNPIIYAITHPKYR MAIAKYIPCLGSLLRVKRR tetr MEL1_danRe KASAIHNPIIYAITHPKYR LAIAKYIPCLRLLLCVPKR tetr MEL1_takRu KASAIHNPIIYAITHPKYR LALAKYIPCLGFLLCISPH tetr MEL1_gasAc KASAIHNPIIYAITHPKYR IALAKYIPFLGVLLCVPPR tetr MEL1_oryLa KASAIHNPIIYAITHPKYR MALAKYIPGLGVLLCIHPK vert MEL1_calMi KASAIHNPIIYAITHPKYR MAIAKYVPLLGLLLRVSRR vert MEL1_petMa KASAIHNPIVYAITHPKYR deut MEL1a_braF KSSAVYNPIVYAITHPKFR AAVKKHIPCLSGCLPADEE deut MEL1a_braB KSSAVYSPIVYAITYPKFR EAVKKHIPCLSGCLPASEE deut MEL1_strPu KCSAIWNPIIYCLSHEKFN AALKEK---LMGMCGIEIP deut MEL1b_braB KLTVIINPIVYVLSIPNFR KALFAQEREKYASEDVVLT tetr MEL2_galGa KASAIYNPIIYAIIHPRYR KTIHNAVPCLRFLIRISKN tetr MEL2_taeGu KASAIYNPIIYAIIHPRYR KTIHQAVPCLRFLIRISKN tetr MEL2_anoCa KASAIYNPIIYAIIHPRYR RTIRSAVPCLRFLIPISKS tetr MEL2_podSi KASAIYNPIIYAIIHPRYR RTIRSAVPCLRFLIRISPS tetr MEL2_xenLa KASAIYNPIIYGIIHPKYR ETIHKTVPCLRFLIREPKK fish MEL2_gadMo KASAIYNPFIYAIIHSKYR DTLAEHVPCLYFLRQPPRK fish MEL2_tetNi KASAIYNPIIYAIIHPRYR KTIRSAVPCLRFLIPISKS fish MEL2_danRe KSSAIYNPFIYAIIHNKYR RTLAEKVPGLSCLSRSQKD fish MEL2_gasAc KASAIYNPFIYAIIHNKYR MTLAAKFPCLRFLSPTPRK
Encephalopsin
This opsin class, despite its phylogenetically erratic pattern of tetrapod gene loss, is exceedingly conserved in its carboxy terminus in both length and sequence back to lamprey. This conservation is unprecedented in this region and must reflect mission-critical binding to another protein.
The cytoplasmic tail of encephalopsin has no detectable homology to other ciliary opsins for more that 6 residues beyond the FR motif (FRRSLLQL) even though it shares the same very ancient terminal exon break as other ciliary opsins (phase 0, just prior to the FR). The VxPx* motif can be recognized in the conserved pattern VRPL*; if this primarily drives cell targeting to cilia, it may or may not have arisen independently from similar motifs in other ciliary opsins.
An interesting phyloSNP can be seen in the difference alignment in the primate stem (S-->N) two residues after the critical Schiff lysine. This may slightly shift the chemical environment of the chromophore.
ENCEPH_hom KSNTVYNPVIYVFMIRKFR RSLLQLLCLRLLRCQRPAKDLPA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-NGSKVDVIQVRPL
ENCEPH_pan KSNTVYNPVIYVFMIRKFR RSLLQLLCLRLLRCQRPAKDLPA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-NGSKVDVIQVRPL
ENCEPH_mac KSNTVYNPVIYVFMIRKFR RSLLQLLCLRLLRCQRPAKDLPA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-NGSKVDVIQVRPL
ENCEPH_pap KSNTVYNPVIYVFMIRKFR RSLLQLLCLRLLRCQRPAKDLPA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-NGSKVDVIQVRPL
ENCEPH_pon KSNTVYNPVIYVFMIRKFR RSLLQLLCLRLLRCQRPAKDLPA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-NGSKVDVIQVRPL
ENCEPH_nom KSNTVYNPVIYVLMIRKFR RSLLQLLCLRLLRCQRPAKDLPA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-NGSKVDVIQVRPL
ENCEPH_cal KSNTVYNPVIYVFMIRKFR RSLLQLLCLRMLRCQQPAKDLSA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-NGSKVDVIQVRPL
ENCEPH_tar KSNTVYNPVIYIFMIRKFR RSLLQFLCLRLLRCQQPAKDLPA-AENEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKT-SGSKVDVIQVRPL
ENCEPH_mic KSNTVYNPIIYIFMIRKFR RSLLQLLCFRLLRCQRPAKDLPA-SESEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDNSDKT-SGSKVDVIQVRPL
ENCEPH_oto KSNTVYNPVIYIFMLRKFR RSLLQLLCFRLLRCQRPAKDLPA-AESEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDNSDKT-NGSKVDVIQVRPL
ENCEPH_tup KSSTVYNPVIYIFMIRKFR RSLLQLLCFRLLRYQRPAKDLPA-AGSEMQIRPIVMSQKDGD---KPKKKVTFNSSSIIFIITSDESLSVDDSDKT-SGSKVDVIQVRPL
ENCEPH_dip KSSTIYNPVIYIFMIRKFR RSLLQLLCFRLLRCQRPAKDLPA-AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSVRS-SGSKADVIQVRPL
ENCEPH_ory KSSTAYNPIIYIFMIRKFR RSLLQLLCFQPLRCQQPPKDLPT-VGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIIASDESLAVDDNEKA-SGPKVDVIQVRPL
ENCEPH_mus KSSTVYNPVIYIFMNRKFR RSLLQLLCFRLLRCQRPAKNLPA-AESEMHIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVEDSDRS-SASKVDVIQVRPL
ENCEPH_rat KSSTVYNPVIYIFMIRKFR RSLLQLLCFRLLRCQRPAKNLPA-AESEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVEDSDRS-SASKVDVIQVRPL
ENCEPH_cav KSSTVYNPVIYVLMIRKFR RSLLQLHCLRLLRCQQPAKDLPA-VEREMHIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDRT-SGSKVDTIQVRPL
ENCEPH_spe KSSTVYNPVIYIFMIRKFR RSLLQLLCSRLLRCQQPAKDLPA-VGNEMQIRPIVISQKDGE---RPKKKVTFNSSSIVFIITSDESLSVDDSNRT-SGSKADVIQVRPL
ENCEPH_fel KSSTVYNPVIYIFMIRKFR RSLLQLLCFRLLRCQRPAKDLPT-NGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVEDSDKT-SVSKVDVIQVRPL
ENCEPH_can KSSTVYNPVIYIIMIRKFR RSLLQLLCFRPLRCQRPAKDLPA-NGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESVSIDDSDKT-SVSKVDVIQVRPL
ENCEPH_pte KSSTVYNPVIYIFMIRKFR RFVLQLLCFRPLRCRRPATDLPA-GGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFVITSDESLSVDDSDKI-NGSKADGIQVRPL
ENCEPH_equ KSSTIYNPIIYIFTIRKFR RSLSQLLCFRLLRCQRPAKDQPP-VGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVHDSDKI-NGSKVEVIQVRPL
ENCEPH_lox KSSTVYNPVIYTFMIRKFR RSLLQLLCFRLLRCQRPAKDLPV-VGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVNNIDKT-NGSKADVIQIRPL
ENCEPH_pro KSSTVYNPVIYTFMIRKFR RSLFQLLCFRLLRCQRPAKNKPE-VGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVNDTDKI-NGSKADVIQVRPL
ENCEPH_cho KSSTVYNLVIYIFMLRKFR RSLLQLLCFRLLRCQRPAKDLPV-VGCEMQIRPIVMSQKEGH---RPKKKVTFNSSSIIFIITSDESISVDGSDKT-NGPKVDVIQVRPL
ENCEPH_mon KSSTAYNPIIYIFMSRKFR RCLLQLLCFRLLKFQQPKKDRPV-IRTEKQIRPIVMSQKVGD---RPKKKVTFSSSSIIFIITSDETQMIDENDKN-SGTKVNVIQVRPL
ENCEPH_mac KSSTAYNPIIYIFMSRKFR RCLLQLLCFRQLKFQQPKKDRPV-IRTEKQIRPIVMSQKVGD---RPKKKVTFSSSSIIFIITSDETQMIDDNDKN-NGTKVNVIQVRPL
ENCEPH_gal KSSTAYNPVIYIFMSRKFR QCLLQLLCFRLMRFQRIMKEPSG-AGNVKPIRPIVMSQKVGD---RPKKKVTFSSSSIIFIIASDDTQQIDDNSKH-NGTKVNVIQVKPL
ENCEPH_tae KSSTAYNPVIYIFMSRKFR RCLLQLLCFRLMRFQRTMRETPA-TGSDKPIRPIVLSQKAGD---RPKKKVTFSSSSVIFIITSDDAEQIEDSSKH-NETKVNAIQVKPL
ENCEPH_ano KSSTAYNPVIYIFMSRKFR RCLVQLFCVQFLRFKRTLKEQPA-IESNKPIRPIVMSQKVGD---RPKKKVTFSSSSIIFIITSDDTEQIDVSTKC-SDTKINVIQVKPL
ENCEPH_dan KSSTAYNPVIYAFMSRKFR RCMLQMLCSRLTSLQHTIKDRPL-SRIEHPIRPIVMSQS--RTD-RPKKRVTFSSSSIVFIIASHDTHPLDITSKCNDEPDINVIQVRPL
ENCEPH_tak KSSTAYNPLIYVFMSRKFR HCLLQLLCSRLSWLQRSLKERPL-APVQRPIRPIVMSRPCGKGN-RPKKKVTFSSSSIVFIITSDDFGQLDVTSKSGDSADVNAIQVRPL
ENCEPH_gas KSSTAYNPLICVFMSRKFR RCLMQLLCSRVTCLQCNLKERPL-APVQRPIRPIVVSAACGGGRVRPKKRVTFSSSSIVFIITRNDIRHTDVTSNTRESSEANVFQVRPL
ENCEPH_ory KSSTAYNPLIYVFMNRKFR RCFLQLLCSKISWLQCTLKEHPL-TPVERPIRPIVASTSCGSRH-RPKKRVTFNSSSIVFMITGDEFQQLDVTSKSRNSSEANVFHVRPL
ENCEPH_cal KSSTAYNPLIYVFMNRKYR RCLSQLFCSHLMSLQWSIKDPSSKARNDMPVKPIVLSQKGD----RPKKRVTFSSSSIVFIITSDDTQELGSIAGS-NATQISIVQVQPL
Consensus KSsT.YNPvIyifMiRKFR r.$lQLlC.rllr.qrpaK#.p....emq!rPIVmSqk.gd....RPKKkVTFnSSS!!FiITSD#s.s.dd.dk...gskvdv!QVrPL
*
ENCEPH_hom KSNTVYNPVIYVFMIRKFR RSLLQLLCLRLLRCQRPAKDLPA AGSEMQIRPIVMSQKDGD---RPKKKVTFNSSSIIFIITSDESLSVDDSDKTNGS-KVDVIQVRPL
ENCEPH_pan ................... ....................... ..................---..................................-..........
ENCEPH_pon ................... ....................... ..................---..................................-..........
ENCEPH_nom ............L...... .........................................---..................................-..........
ENCEPH_mac ................... ....................... ..................---..................................-..........
ENCEPH_pap ................... ....................... ..................---..................................-..........
ENCEPH_cal ................... ..........M....Q.....S. ..................---..................................-..........
ENCEPH_tar ...........I....... .....F.........Q....... .EN...............---...............................S..-..........
ENCEPH_mic ........I..I....... ........F.............. SE................---..........................N....S..-..........
ENCEPH_oto ...........I..L.... ........F.............. .E................---..........................N.......-..........
ENCEPH_tup ..S........I....... ........F....Y......... ..................---K..............................S..-..........
ENCEPH_mus ..S........I..N.... ........F..........N... .E...H............---.........................E...RSSA.-..........
ENCEPH_rat ..S........I....... ........F..........N... .E................---.........................E...RSSA.-..........
ENCEPH_spe ..S........I....... ........S......Q....... V.N........I.....E---.............V..............NR.S..-.A........
ENCEPH_cav ..S.........L...... ......H........Q....... VER..H............---.............................R.S..-...T......
ENCEPH_dip ..S.I......I....... ........F.............. ..................---............................VRSS..-.A........
ENCEPH_ory ..S.A...I..I....... ........FQP....Q.P....T V.................---.................A.....A...NE.AS.P-..........
ENCEPH_fel ..S........I....... ........F.............T N.................---.........................E.....SV.-..........
ENCEPH_can ..S........II...... ........F.P............ N.................---......................V.I......SV.-..........
ENCEPH_pte ..S........I....... .FV.....F.P...R...T.... G.................---...............V..............I...-.A.G......
ENCEPH_equ ..S.I...I..I.T..... ...S....F...........Q.P V.................---.........................H....I...-..E.......
ENCEPH_lox ..S........T....... ........F.............V V.................---.........................NNI......-.A....I...
ENCEPH_pro ..S........T....... ...F....F..........NK.E V.................---.........................N.T..I...-.A........
ENCEPH_cho ..S....L...I..L.... ........F.............V V.C............E.H---......................I...G......P-..........
ENCEPH_mon ..S.A...I..I..S.... .C......F...KF.Q.K..R.V IRT.K..........V..---........S............TQMI.EN..NS.T-..N.......
ENCEPH_gal ..S.A......I..S.... QC......F..M.F..IM.EPSG ..NVKP.........V..---........S........A..DTQQI..NS.H..T-..N....K..
ENCEPH_tae ..S.A......I..S.... .C......F..M.F..TMRET.. T..DKP.....L...A..---........S...V.......DAEQIE..S.H.ET-..NA...K..
ENCEPH_ano ..S.A......I..S.... .C.V..F.VQF..FK.TL.EQ.. IE.NKP.........V..---........S...........DTEQI.V.T.CSDT-.IN....K..
ENCEPH_dan ..S.A......A..S.... .CM..M..S..TSL.HTI..R.L SRI.HP........SRT.---....R...S....V...A.HDTHPL.ITS.C.DEPDIN.......
ENCEPH_tak ..S.A...L.....S.... HC......S..SWL..SL.ER.L .PVQRP.......RPC.KGN-........S....V......DFGQL.VTS.SGD.AD.NA......
ENCEPH_gas ..S.A...L.C...S.... .C.M....S.VTCL.CNL.ER.L .PVQRP.....V.AAC.GGRV....R...S....V....RNDIRHT.VTSN.RE.SEAN.F.....
ENCEPH_cal ..S.A...L.....N..Y..C.S..F.SH.MSL.WSI..PSSK.RND.PVK...L.. .-..---....R...S....V......DTQELGSIAGS.AT-QISIV..Q..
TMT opsin
TMT predominantly exhibits FY for its FR motif though perhaps the conserved FYK/R motif accomplishes the same end. Within the whole TMT family, no observable conservation occurs past the first 9 residues, though some 35 residues are alignable within the sole TMT locus tracking into mammals (marsupials). The conserved pair of cysteines might be palmitoylated. Opossum has acquired an upstream stop codon recently -- the 22 residues following are still alignable to wallaby. GenBank lacks any tetrapod transcripts of this TMT locus as of Jan 09. The last exon of this gene is curiously intertwined with that of the opposing strand gene, the sialyltransferase ST6GAL2.
TMT_monDom KSSTVCNPIIYVLMNKQFY KCFLILFHCQPAQSGPDVS LCPSNVTVIQLGQRKNKDA PGSI*DFPEVSEKQLCLLS PEVWPQP
TMT_macEug KSSTVCNPIIYILMNKQFY KCFLILFHCQPASSASDAS LCPSKMTVIQLGQRKDKEV PCAIQDLPEVSKKQLCLLS PESNVAPSSGHPQEKMEEKPLSE
TMT_ornAna KSSTVCNPIIYILMNKQFY KCFLILFHCQPPRAADAPS TYPSQVMVIQLNQRRSRET AGAPQVLLEMKHQTLHLLG PQLHETPSWERSTPVHPE
TMT_taeGut KSSTVCNPIIYILMNKQFY KCFRQLFHCQPPSSTDGEP TCHSKVTVIQLDQRADGGN MCNNEPHPETDSKMTSLLC PETTSKATPPTS
TMT_galGal KSSTVCNPIIYILMNKQFY KCFRQLFHCQPPSSTDGEP TCHSKVTVIQLNQKTDGGK LCNNKPRPETDNKVTSLLH PEPGLEPAAKTVPPM
TMT_anoCar KSSTVFNPIIYILMNKQFY KCFLMLLHCQPSSVADGET ICQSKVMAIHQNQKAQGGV ILKSQVVPQMDEKAICLLS PESSLDPVLESTPQLSKENSFL
TMT_xenTro KSSTVFNPIIYILMNKQFY KCFLILFHCHPTSSADGKS ICQSNYTVIQLNQKLNNIV AIPGQTQIPESVDKMPCIH RQNNESPSDQMPQSTTEHLISGT
TMT_danRer KSSTVINPLIYILMNKQFY RCFRILFCCQRSLLQNGHS SMPSKTTVIQLNRRVNSNA VACTAQISTGTHNHDCSTH VTERSNPPEVIP*
TMT_tetNig KSSTVINPLIYILMNKQFY KCFLILFHCSHWSADNGTT SVPSKITVIQLNRRAYSNT VACADPLSTDALKQCCSAK NASTIEVKLS*
TMT_takRub KSSTVINPLIYILMNKQFY KCFLILFHCGHWSADNGNT SMPSKTTAIQLNRRVYSNT VACADQLSTDALKQCCSAN TISTKNTSTVEGKLS*
TMT_gasAcu KSSTVINPLIYILMNKQFY RCFLILFHCKHWSAENHNT SMPSKTTVIHLNRRVCSNT LPCTAQASTDAANHFCSTS ATKHTSPPLQGHGLSLNVLNMIRQENHSHDEAAKNQLDCLT*
TMT_oryLat KSSTVINPLIYILMNKQFY RCFLILFHCDHWSSENGNT SVPSKTTVIPLNRRIYTNT VAQISTDNAN*
TMT_ictPun KSSTVINPVIYIFMNKQFY RCFRTLLGYKERSAVPDDH SLMATKNTAIQLKCIMHNN PVPSPAHTPPPFF...
TMT_oncMyk KSSTVINPLIYILMNKQFY RCFLILFHCKRPSSENGVS SMPSKTTVIQLNRRGHSNN VALTPQLSTGANHHNHNHT VECSTNNREVTTPIGLPHSGWL* 0
TMTa_danRe KSSTVINPVIYIFMNKQFY RCFRALLNCDKPQRGSSLK SSSKTKPFRPGRRTDNFTF MVASVGPNQTNPVEDGPPSADNTKPAVLSLVAHYNG
TMTa_takRu KSSTVINPIIYVFMNKQFY RCFLALLCCQDPRSGSSMK SSSKVATKAKGVTPTGQRR TDFLYMVASLGRPAATIPQLGPSFDATNDFTKPPSSDTIKPVVVSLAAHCDG
TMTa_tetNi KSSTVINPVIYVFMNKQFS RCFLSLLCCEDPRSSTSLR SSSRVTTKAVRGGTLTGQR RTNHLLYMVAALGRPVATAMPQLGPSFDATYDITKAPSSDNHQPVVVSLEAHG
TMTa_gasAc KTSTVINPVIYVFMNKQFY RCFKALLRCEAPRPSSSLK SSSKVPTKAMRGAAVTGPR HTNNFLFVVASLGRPVATIPQLGPSVEPTIDVTGGPSSDNNKPVIVSLVAQCDG
TMAa_ictPu KSSTVINPVIYIFMNKQFY RCFRTLLGYKERSAVPDDS LMATKNTAIQLKCIMHNNP
TMTb_takRu KFSTVINPFIYIFMNKQFY RCFRAFLNCSTPKRDSTVR TFTRISLRALRQDQQQKGS ALAPSSARPTPNSIHESSLKGSHSTPSNGGAAAAKSPAANRSKPKLILVAHYRE
TMTb_tetNi KFSTVINPFIYIFMNKQFY RCFRAFLSCSSPERGSTVR TFTRISLRAVCQRKQQRVS APAASSACPTPNSIHHSSRKGSHSASSNSGTAAAAKTPAANSSKPKLILVVHYRE
TMTb_gasAc KFSTVVNPFIYIFMNKQFY RCFRAFLSCSTPERGSTLK TFSRPTKTLRAGRHEKGRR VSAAAPSTAQPTRNSAPRSSQGANHASATPPPSPADGRCAAAGAAKPKRTLVAHYRE
TMTb_oryLa KFSTVINPLIYIFMNKQFY RCFWAFFCCSTPEQVSTLR TFSRVTKTIRTFRQERELH VSAPAPSSGLPTPNSIQKNNHVDPSSINQACAASDSPDSRKPKVVLVAHYQE
TMTb_pimPr KTSTVINPIIYIFMNKQFC RCFHALIMCTTPQRGSSFK NSSKVTKTLRTVRRANGQN VTFAVASAGHPTICAPH
TMTb_danRe KSSTVINPIIYIFMNKQFC RCFHALIMCTTPERGSSFK NSSKVTKTLRTVRRANGQN VTFAVASAVHRTPYSDRQKSSSEGEKLPPATGQGTSKPVVSLVAYYNG*
Imaging ciliary opsins
The cytoplasmic tails of these opsins begin and end with highly conserved motifs but the middle sections have been subject to numerous indels, suggesting that absolute length is unimportant for binding site recognition. The VAPA terminal motif can be recognized in all but the secondary parapinopsin group PPINb (found only in some teleost fish and apparently reflecting differential survival of gene duplication and in avian VAOP where chicken and finch have recent changes in stop codon.
LWS is shown elsewhere greatly expanded to 82 species to illustrate the issues. Four indels, all deletions, have occurred during vertebrate history: a 2 residue loss in mammals, a 1 residue loss in birds but not lizards, and a 1 and 5 residue loss in teleost fish. Otherwise, LWS has been remarkably constant -- its key features and almost every residue past FR were already firmly settled prior to lamprey divergence.
This region cannot be important to Galpha binding because it is too highly variable just within cone opsins which all use the same transducin. Cysteines are conserved to depth but palmitoylation could be universal exclusive of VAOP. LWS also lacks the distal cysteine (CCGK motif has been LFGK since lamprey stem) found in other ciliary opsins. Serines and threonines (for arrestin) are common but are not a deeply conserved feature.
RHO1_homSa KSAAIYNPVIYIMMNKQFR NCMLTTICCGKNPLGDDE--ASATVSKTETS -----QVAPA RHO1_bosTa KTSAVYNPVIYIMMNKQFR NCMVTTLCCGKNPLGDDE--ASTTVSKTETS -----QVAPA RHO1_monDo KSSSVYNPVIYIMMNKQFR TCMITTLCCGKNPLGDDE--ASATASKTETS -----QVAPA RHO1_ornAn KSSAIYNPVIYIMMNKQFR NCMLTTICCGKNPLGDDE--ASATASKTEQS SVSTSQVSPA RHO1_galGa KSSAIYNPVIYIVMNKQFR NCMITTLCCGKNPLGDEDTSAG----KTETS SVSTSQVSPA RHO1_anoCa KSSAIYNPVIYILMNKQFR NCMIMTLCCGKNPLGDEDTSAGT---KTETS TVSTSQVSPA RHO1_xenTr KSSAIYNPVIYIVLNKQFR NCLITTLCCGKNPFGDEEGSSAA-SSKTEAS SVSSSQVSPA RHO1_neoFo KTASVYNPVIYILMNKQFR NCMITTLCCGKNPFGDEETTSA-GTSKTEAS SVSSSQVSPA RHO1_latCh KSASFYNPVIYILLNKQFR NCMITTLCCGKNPFGDEDATSAAGSSKTEAS SVSSSSVSPA RHO1_takRu KSAALYNPVIYILLNRQFR NCMITTVCCGKNPFGDDDAATTV--SKTQSS SVSSSQVAPA RHO1_angAn KSSAIYNPLIYICLNSQFR NCMITTLFCGKNPFQEEE-GASTTASKTEAS SVSS--VSPA RHO1_conMy KSSALYNPMIYICMNKQFR HCMITTLCCGKNPFEEED-GASATSSKTEAS SVSSSSVSPA RHO1_calMi KSSALYNPLIYILLNKQFR NCMITTLCCGKNPFEEDE-STSAAASKTEAS SVSSSQVSPA RHO1_leuEr KSSAVYNPLIYILMNKQFR NCMITTICLGKNPFEEEE-STSASASKTEAS SVSSSQVAPA RHO1_petMa KTSALYNPIIYILMNKQFR NCMITTLCCGKNPLGDEDSGASTS--KTEVS SVSTSQVSPA RHO1_letJa KSSALYNPVIYILMNKQFR NCMITTLCCGKNPLGDDESGASTS--KTEVS SVSTSQVSPA RHO1_geoAu KSSALYNPVIYILMNKQFR NCMITTLCCGKNPLGDDDSGASTS--KTEVS SVSTSQVAPA RHO2_galGa KSSSLYNPIIYVLMNKQFR NCMITTICCGKNPFGDEDVSSTVSQSKTEVS SVSSSQVSPA RHO2_taeGu KSSSLYNPIIYVLMNKQFR NCMITTICCGKNPFGDEETSSTVSQSKTEVS SVSSSQVSPA RHO2_podSi KSSSLYNPIIYVLMNKQFR NCMITTICCGKNPFGDDDVSSTVSQSKTEVS SISSSQVSPA RHO2_anoCa KSSSLYNPIIYVLMNKQFR NCMITTICCGKNPFGDEDVSSSVSQSKTEVS SVSSSQVSPA RHO2_gekGe KSSSIYNPIIYVLLNKQFR NCMVTTICCGKNPFGDEDVSSSVSQSKTEVS SVSSSQVAPA RHO2_pheMa KSSCIYNPIIYVLLNKQFR NCMVTTICCGKNPFGDEDASSSVSQSKTEVS SVSSSQVAPA RHO2_neoFo KSSALYNPIIYVLMNKQFR NCMVTTLCCGKNPFGDDDVSSSVSAGKTEVS SVSSSQVSPA RHO2_latCh KSSCLFNPIIYVLLNKQFR NCMITTLCCGKNPLGDDDTSSAVSQSKTDVS SVSSSQVSPA RHO2_takRu KSSALYNPVIYVLLNKQFR NCMLSTIGMGGAV--DDE--TSVSASKTEVS -------SVS RHO2_gasAc KSSALYNPVIYVLLNKQFR NCMLTTIGMGGMV--EDE--TSVSASKTEVS -------SVS RHO2_oreNi KSSALYNPIIYVLMNKQFR NCMLSTIGMGGMV--EDE--TSVSTSKTEVS -------SVS RHO2_hipHi KSSALYNPVIYVLLNKQFR NCMLSTIGMGGMV--EDE--SSVSASKTEVS -------SVS RHO2_mulSu KSSALYNPVIYVMMNKQFR NCILSAIGMGGMV--EDE--TSVSTSKTEVS -------TAS RHO2_pomMi KSSALYNPVIYVLMNKQFR NCMLSAVGMGGMV--DDE--TSVSASKTEVS -------SVS RHO2_oryLa KSSALFNPIIYILLNKQFR NCMLATIGMGGMV--EDE--TSVSTSKTEVS -------TAA RHO2a_danR KTSAVFNPIIYVLLNKQFR SCMLNTLFCGKSPLGDDE-SSSVSTSKTEVS -----SVSPA RHO2b_danR KASALFNPIIYVLLNKQFR SCMLNTLFCGKSPLGDDE-SSSVSTSKTEVS -----SVSPA RHO2c_danR KSSSIFNPIIYVLLNKQFR NCMLTTLFCGKNPLGDDE-SSTVSTSKTEVS -----SVSPA RHO2d_danR KTSALYNPVIYVLLNKQFR NCMLTTLFCGKNPLGDDE-SSTVSTSKTEVS -----SVSPA RHO2_calMi KSSVLYNPIIYILMNKQFR SSMITTVCCGKNPFGDDD-SSSVTSQSKTEVSSVSTSQVSPA RHO2_geoAu KSSVLYNPIIYVLLNKQFR TCMVTTLFCGKNPFGEDD-SSMVSTSKTEVS SVSSSQVSPS SWS2_ornAn KASTIYNPIIYVFMNKQFR SCMLKLVFCGKSPFGDEDE-ISGSSQATQVS SVSSSQVSPA SWS2_anoCa KASTVYNPVIYVLMNKQFR SCMLKLIFCGKSPFGDEDD-VSGSSQATQVS SVSSSQVSPA SWS2_utaSt KASSVYNPVIYVFMNKQFR SCMLKLVFCGKSPFGDEDD-VSGSSQTTQVS SVSSSQVSPA SWS2_taeGu KASTVYNPIIYVFMNKQFR SCMLKLVFCGRSPFGDEDD-VSGSSQATQVS SVSSSQVSPA SWS2_neoFo KSSTVYNPLIYVFMNKQFR SCMMKLIFCGKSPFGDEDD-ASSASQSTQVS SVSSSQVAPA SWS2_galGa KSSTVYNPVIYVLMNKQFR SCMLKLLFCGRSPFGDDED-VSGSSQATQVS SVSSSHVAPA SWS2_xenTr KASTVYNPFIYIFMNRQFR SCMMKMIFCGKNPLGDDEE--TSVSGSTQVS SVSSSQIAPS SWS2_takRu KASTVYNPIIYVVLNKQFR SCMKKML---GMSGGDDEE-------SSSQS VTEVSKVSPS SWS2_gasAc KSSAVYNPVIYVLLNKQFR SCMMKML---GMGGGDDEE-------SSTSS VTEVSKVGPA SWS2_geoAu KASTVYNPVIYIFLNKQFR SCMMKTIFCGKNPLGDDED---ATSTTTQVS SVSTSQVAPA SWS1_homSa KSACIYNPIIYCFMNKQFQ ACIMKM-VCGKAMT--DESDTCSS-QKTEVS TVSSTQVGPN SWS1_monDo KSACVYNPIIYCFMNKQFH ACIMEM-VCRKPMT--DDSDVSSS-QKTEVS AVSSSQVGPT SWS1_smiCr KSACVYNPIIYCFMNKQFH ACIMEM-ICKKPMT--DDSETTSS-QKTEVS TVSSSQVGPS SWS1_tarRo KSACVYNPIVYWFMNKQFH ACIMEM-VCRKPMT--DDSEISSS-QKTEVS TVSSSQVGPS SWS1_taeGu KSSCVYNPIIYCFMNKQFR ACIMET-VCGRPMT--DDSEVSSSAQRTEVS SVSSSQVGPS SWS1_anoCa KSSCVYNPIIYCFMNKQFR ACILET-VCGKPMS--DESDVSSSAQKTEVS SVSSSQVSPS SWS1_utaSt KSACVYNPIIYCFMNKQFR ACIMET-VCGKPMT--DESDVSSSAQKTEVS SVSSSQVSPS SWS1_neoFo KSSFVYNPIIYCFMNKQFR ACIMQT-VFGKPMT--DDSDISSSG-KTEVS SVSSSQVNPS SWS1_galGa KSACVYNPIIYCFMNKQFR ACIMET-VCGKPLT--DDSDASTSAQRTEVS SVSSSQVGPT SWS1_xenLa KSSCVYNPIIYSFMNKQFR GCIMET-VCGRPMS--DDSSVSSTSQRTEVS TVSSSQVSPA SWS1_petMa KASCVYNPLIYSFMNKQFR ARIMET-VCGKFIT--DESETSSS--RTAVS SVSTSQVSPG SWS1_geoAu KASCVYNPLIYSFMNKQFR ACILET-VCGKPIT--DESETSSS--RTEVS SVSTTQMIPG SWS1_danRe KSSSVYNPLIYAFMNKQFN ACIMET-VFGKKI---DES--------SEVS SKTETSSVSA SWS1_oryLa KSSCVYNPLIYAFMNKQFN GCIMEM-VFGKKM---EEA--------SEVS SKTE-VSTDS LWS_homSap KSATIYNPVIYVFMNRQFR NCILQL--FGKKV---DDGSELSSASKTEVS --SVSSVSPA LWS_monDom KSATIYNPIIYVFMNRQFR TCILQL--FGKKV---DDGSEVSSTSRTEVS --SVSSVAPA LWS_ornAna KSATIYNPIIYVFMNRQFR NCIMQL--FGKKV---DDGSELSSTSRTEVS --SVSSVSPA LWS_galGal KSATIYNPIIYVFMNRQFR NCILQL--FGKKV---DDGSEV-STSRTEVS SVSNSSVSPA LWS_anoCar KSATIYNPIIYVFMNRQFR NCIMQL--FGKKV---DDGSELSSTSRTEVS SVSNSSVSPA LWS_xenTro KSATIYNPIIYVFMNRQFR NCIYQL--FGKKV---DDGSEVSSTSRTEVS SVSNSSVSPA LWS_neoFor KSATIYNPIIYVFMNRQFR NCIYQL--LGKKV---DDGSELSSTSKTEVS SVSNSSVSPA LWS_takRub KSATIYNPVIYVFMNRQFR VCIMKL--FGKEV---DDGSEV-STSKTEVS -----SVAPA LWS_gasAcu KSATIYNPVIYVFMNRQFR SCIMQL--FGKEV---DDGSEV-STSKTEVS -----SVAPA LWS1_calMi KSSTIYNPIIYVFMNRQFR NCILQL--FGKKV---DDGSELSSTSKTDVS SVSNSSVSPA LWS2_calMi KSSTIYNPIIYVFMNRQFR NCILQL--FGKKV---DDGSELSSTSKTDVS SVSNSSVSPA LWS_petMar KGATIYNPIIYVFMNRQFR NCILQL--FGKKV---DDGSEVSSSSRTEVS SVSNSSVSPA LWS_letJap KSATIYNPVIYVFMNRQFR NCIMQL--FGKKV---DDGSEVSSASRTEVS SVSNSSISPA LWS_geoAus KSATIYNPIIYVFMNRQFR NCIMQL--FGKKV---DDGSEVSSSARTEVS SVSNSSVSPA PPIN_anoCa KSSTFYNPIIYIFMNKQFR DCLVRCLLCGRNPCA-SEQTDEDDLEVSTIAPAP SSRRGKVAPV* PPIN_xenTr KTSTVYNPIIYIFMNKQFQ ECVIPFLFCGRNPWA--AEKSSSMETSISVTSGT PTKRGQVAPA* PPIN_ictPu KSSTVFNPIIYIFMNRQFR DYALPCLLCGKNPWA----AKEGRDSDTNTLTTT VSKNTSVSPL* PPIN_oncMy KSSTVYNPIIYVFMNRQFR DCAVPFLLCGLNPWA-----SEPVGSEADTALSS VSKNPRVSPQ* PPIN_oryLa KSSTVYNPVIYIYLNNQFR RYAVPFLLCGREP---------RDEDEASETTTT IEITNKVSPS* PPIN_danRe KSSTVFNPIIYIFMNRQFR DRALPFLLCGRNPWA-----AEAEEEEEETTVSS VSRSTSVSPA* PPINa_takR KSSTVYNPIIYIYLNKQFR KYAVPFLLCGRELEM----------EDELSMTTV -ETSNRVSPA* PPINa_tetN KSSTAYNPIIYIYLNKQFR KYALPFLLCRRALEA----------EDEVSETTV -ESSRRVSPS* PPINa_gasA KSSTVFNPIIYIYLNKQFR KYAVPFLLCCKEPLD--------DEEASEAATTV EISPSKVSPA* PPIN_petMa KTSTVYNPIIYIFMNRQFR DCAVPFLLCGRNPWAEPSSESATTASTSATSVTL ASVPGQVSPS* PPIN_letJa KTSTVYNPIIYIFMNRQFR DCAVPFLLCGRNPWAEPSSESATAASTSATSVTL ASAPGQVSPS* PPINa_cioI KTATIYNPLIYIGLNRQFR DCVVRMIFNGRNPWV---DELVGSQVSSTGSQLT AVSSNKVAPA* PPINa_cioS KTATIYNPLIYIGLNRQFR DCVVRMIFNGRNPWV---DEMVGSQVSSSASQMT AVSSNKVAPA* PPINb_gasA KSSTVYNPIIYIFMNRQFR GYAVPSILCGWNPWA--EEQTSEEETVGSVMKSQ RVSPKGSLQE* PPINb_tetN KSSTVYNPIIYVFMNRQFR GYAINTILCGRRAWVSEQQTSEGETTVVSVSKSQ KISPKGSLQ* PPINb_takR KSSTVYNPIIYIFMNRQFR GCAINTVLCGRRAWITDLQTSEGETTVASTSKSQ KISPKGSLN* PPINb_mayZ KSSTVYNPIIYIFMNRQFR GYTVAAVLCGWDPWSSEPQTSENETTVPFFIKTPKKIVPKKSLE* PARIE_anoC KTSPVYNPIIYIFLNKEFR ECAVEFITCGKVVLTSPEEDISTSAISDEGIA-- PCKINQVTPV* PARIE_utaS KTSPVYNPIIYIFLNKQFR DCAVEFITCGQVVLTSPEEDISTSAIPVEGKG-- PCKINQVTPV* PARIE_xenT KTSPVYNPIIYIFLNKQFR TYAVQCLTCGHINLDSLEEDTESVSAQAENML-- TPKTNQVAPA* PARIE_takR KTSPVYNPIIYFLSNKQFR DATLEVLSCSRYIPHASSRVSINMRSLNRRS--- VNTHSKVSPL* PARIE_tetN KTSPVYNPIIYFLSNKQFR DATLEVLSCGRYIPHASTRVTFNMCAFNRRSRLPSLSRSINTHSKVSPL* PARIE_gasA KTSPVYNPIIYFLSNKQFR DAALEMLSCGRYIAHMPNTVSINMRSLNRRSRLSSLSRNVNSHSKVLPL* PARIE_danR KTSPVYNPIIYFLTNKRFR ESSLEVLSCGRYISRETGGPLMGSSM-------- QRGQSRVNPV* PIN_galGal KTATVYNPIIYVFMNKQFQ SCLLEMLCCGYQPQRTGKASPGTPGPHA-DVT--AA GLRNKVMPA HPV* PIN_colLiv KTATVYNPIIYVFMNKQFQ SCLLKMLCCGHHPRGTGRTAPAAPASPT-D------ GLRNKVTPS HPV* PIN_taeGut FQ SCLLGMLCCGHHPRGMGKTSPAAPSP-----QVAAE GLRNKVTPS HPV* PIN_utaSta KTATVYNPIIYVFMNKQFR SCLLSTMSCGHRPRGAQETTPAMISIPQGP-TSALQ GSRNKVTPS ASEGSGNEAIPS* PIN_podSic KTATVYNPIIYVFMNKQFR SCLLYKMSCGHRALSSQDTTPAGISLPGRLTTSASK GSRNQVSPS* PIN_pheMad KTATVYNPIIYVFMNKQFR SCLLNTVSCGRIPQTMPGTPATTAVRGGFVLTSE-- GRGNKVAST ELHS* PIN_xenTro KTATVYNPIIYVFMNKQFR NCLMTLLCCGRS-FGDDETSSA---SGRTDVTSVSE AGGNKVTPA* PIN_xenlae KTATVYNPIIYVFMNKQFR NCLMTLLCCGRSPFGDDETSTS---SARTDVTSVSK AGGNKVTPA* PIN_bufJap KTATVYNPVIYVFMNKQFR DCLTKLLCCGRNPFGEDETSTT---SGRTDVTSVSE GGGNKVTPA* VAOP_galGa KTATVYNPIIYVFMNKQFR MCLIQMFKCSAIETAESNMNPTSERATLTQDKRDSQLSVMAVRSTIS* VAOP_taeGu KTATVYNPVIYVFMNKQFR QCLIQMFSCSAIGTAESNMKLTSERAVLMQGRRGSKRTPMAVHSTVLKRKTGDEHRADDLWLF* VAOP_anoCa KTATVYNPVIYVFMNNQFR KCLVQLFQCSSQETMDANVNPISEKDTLTHTKHCGEMSTVAAHVI---VFNPRSEDEQGSCQSFAQLAISENKVYPL* VAOP_xenTr KTASMYNPIIYVYMNKQFR RCLYQMFNINDPEAKESNLNPTSERGVLTRNNNGGEMLAIATHIT--SSAVTNREEEKSSSNSFAHIPVSDNKVCPM* VAOP_danRe KTAAVYNPIIYVFMNKQFR KCLVQLLSCSKVTVVEGNNNQTTERAGMTSGSNTGEMSAIAARVS-----VPKTEENPGDRSTFSHIPIPENKVCPM* VAOP_takRu KTAAVYNPIIYVFMNKQFR KCLIQHFIGMGVMA-ESNMNPTSERPGITAESQTGEMSAIAARVPVGATAALHSDGSPTDCGSLAQLPIPENKVCPI* VAOP_rutRu KTAAVYNPVIYVFMNKQFR KCLVQLLRCRDVTIIEGNINQTSERQGMTNESHTGEMSTIASRIPKDGSIPEKTQEHPGERRSLAHIPIPENKVCPM* VAOP_petMa KTATVYNPVIYIFMNKQFR DCFVQVLPCKGLKKVSATQTAGAQDTEHTASVNTQSPGNRHNIALAAGSLRFTGAVAPSPATGVVEPTMSAAGSMGAPPNKSTAPCQQQGQQQQQQGTPIPAITHVQPLLTHSESVSKICPV*
Reference sequence collection
Cytoplasmic loop C2 from 101 melanopsins
species helix bridge area hel transmemb Le 7 9 MEL1_homSa DRYLV ITRPLATFGVAS KRR AAFVLLGVW 20 T P MEL1_panTr DRYLV ITRPLATFGVAS KRR AAFVLLGVW 20 T P MEL1_gorGo DRYLV ITRPLATFGVAS KRR AAFVLLGVW 20 T P MEL1_ponAb DRYLV ITRPLATIGVAS KRR AAFVLLGVW 20 T P MEL1_rheMa DRYLV ITRPLATIGVAS KRR AAFVLLGVW 20 T P MEL1_calJa DRYLV ITRPLATIGVAS TKR AAFVLLGVW 20 T P MEL1_micMu DRYLV ITRPLASVGTAS KRR AGLVLLGVW 20 T P MEL1_otoGa DRYLV ITRPLTTVGVAS KRR AALVLLGVW 20 T P MEL1_musMu DRYLV ITRPLATIGRGS KRR TALVLLGVW 20 T P MEL1_ratNo DRYLV ITRPLATIGMRS KRR TALVLLGVW 20 T P MEL1_nanEh DRYLV ITRPLATIGVAS KRR TALVLLGVW 20 T P MEL1_phoSu DRYLV ITRPLATIGMGS KRR TALVLLGIW 20 T P MEL1_dipOr DRYLV ITRPLATIGVTS KRR TAFVLLGVW 20 T P MEL1_cavPo DRYLV ITRPLATIGVAS KRQ AALVLLGVW 20 T P MEL1_speTr DRYLV ITRPLATIGMAS KKR AAFFLLGVW 20 T P MEL1_oryCu DRYLV ITRPLAAVGMVS KKR AGLVLLGVW 20 T P MEL1_ochPr DRYLV ITRPLAAVGMVS KRR TGLVLLGVW 20 T P MEL1_bosTa DRYLV ITRPLATVGMVS KRR AALVLLGVW 20 T P MEL1_turTr DRYLV ITRPLATVGMVS KRR AALVLLGVW 20 T P MEL1_susSc DRYLV ITHPLATVGMVS KRR AALVLLGVW 20 T P MEL1_equCa DRYLV ITRPLATVGVVS KRW AALVLLGIW 20 T P MEL1_felCa DRYLV ITHPLATIGVVS KRR AALVLLGVW 20 T P MEL1_canFa DRYLV ITHPLAAVGVVS KRR AALVLLGVW 20 T P MEL1_myoLu DRYLV ITRPLA-IGVVS KRR AALVLLGVW 19 T P MEL1_pteVa DRYLV ITRPLAAIGVVS KRR AALVLLGVW 20 T P MEL1_eriEu DRYLV ITRPLATIGVVS KRR VALVLLGVW 20 T P MEL1_loxAf DRYLV ITRPLATIGVVS KRR AALVLLGIW 20 T P MEL1_proCa DRYLV ITRPLATIGVVS KRR TALVLLGTW 20 T P MEL1_echTe DRYLV ITRPLATIGVVS KRR AALVLLVIW 20 T P MEL1_smiCr DRYFV ITRPLASIGMIS KKK TGLILLGVW 20 T P MEL1_monDo DRYFV ITRPLASIGVIS KKK TGFILLGVW 20 T P MEL1_ornAn DRYFV ITRPLASIGVIS KKR ALLILTGVW 20 T P MEL1_anoCa DRYFV ITRPLASIGAMS TKK ALLILSGVW 20 T P MEL1_taeGu DRYFV ITKPLASVGVTS KKK ALIILVGVW 20 T P MEL1_galGa DRYFV ITKPLASVRVMS KKK ALIILVGVW 20 T P MEL1_xenTr DRYFV ITRPLTSIGVMS KKR AVLILSGVW 20 T P MEL1_danRe DRYFV ITRPLASIGVLS QKR ALLILLVAW 20 T P MEL1_danRe DRYFV ITRPLASIGVMS RKR ALLILSAAW 20 T P MEL1_takRu DRYFV ITRPLTSIGVLS RKR AFVILMTVW 20 T P MEL1_gasAc DRYFV ITRPLTSIGMMS RRR ALLILMGAW 20 T P MEL1_oryLa DRYFV ITRPLTSIGVLS RKR ALLILSAAW 20 T P MEL1_calMi DRYFV ITRPLASIGVLS HRR AGLIILSLW 20 T P MEL1_petMa DRYLV LTRPLASIGAMS KRR AMYITAAVW 20 T P MEL2_galGa DRYLV ITKPLRSIQWTS KKR TIQIIAAVW 20 T P MEL2_anoCa DRYCV ITKPLQSIKRTS KKR TCIIIVFVW 20 T P MEL2_xenLa NRYIV ITKPLQSIQWSS KKR TSQIIVLVW 20 T P MEL2_danRe DRYLV ITKPLQTIQWNS KRR TGLAILCIW 20 T P MEL2_tetNi DRYVV ITKPLQTIRRSS KRR TALAILMVW 20 T P MEL2_gasAc DRYLV ITKPLQAIHWGS KRR TTLAILLVW 20 T P MEL1_plaDu DRFYV ITNPLGAAQTMT KKR AFIILTIIW 20 T P MEL1_capCa DRYMV IAKPFYAMKHVS HKR SLIQIILAW 20 A P MEL1_helRo DRYLV VGQPLAMLNQSH FRR SFYHVLIIW 20 G P MEL1_todPa DRYNV IGRPMAASKKMS HRR AFIMIIFVW 20 G P MEL1_schMe DRYFV IAQPFQTMKSLT IKR AIIMLVFVW 20 A P MEL2_schMa DRYLV IATPFESVFQTT PRR TLLLMLFLW 20 A P MEL1_lotGi DRYLV ITSPFTAMRNMT HKR AFLMIVGVW 20 T P MEL1_sepOf DRYNV IGRPMAASKKMS HRR AFLMIIFVW 20 G P MEL1_entDo DRYNV IGRPMAASKKMS HRR AFLMIIFVW 20 G P UVV_camAb DRYST IARPLDGKLS RGQ VLLLIMLIW 18 A P UVV_catBo DRYST IARPLDGKLS RGQ VILLIALIW 18 A P UVV_apiMe DRYST IARPLDGKLS RGQ VILFIVLIW 18 A P BLU_apiMe DRYRT ISCPIDGRLN SKQ AAVIIAFTW 18 S P BLU_ DRoMe DRYKT ISNPIDGRLS YGQ IVLLILFTW 18 S P BLU_manSe DRYKT ISSPLDGRIN TVQ AGLLIAFTW 18 S P UVV1_droMe DRYNV ITKPMNRNMT FTK AVIMNIIIW 18 T P UVV1_pedHu DRCET ITNPL-QKSG KKK AFLLAAFTW 18 T P UVV_manSe DRHST ITRPLDGRLS EGK VLLMVAFVW 18 T P UVV_papXu DRHST ITRPLDGRLS RGK VLLMMVCVW 18 T P UVV2_droMe DRFNV ITRPMEGKMT HGK AIAMIIFIY 18 T P UVV2_pedHu DRYQV IVHPLER-KT KAA VYFQILLIW 18 V P LWS_nemVe DRYIV IVHPMKKIMT RKK AALMIVGVW 18 V P LWS_pedHu DRYNV IVKGLSAKPMT IKM ALLNILFVW 19 V G LWS_vanCa DRYNV IVKGIAAKPLT ING AMLRVLGIW 19 V G LWS_papXu DRYNV IVKGIAAKPMT ING ALLRILGIW 19 V G LWS_helSa DRYNV IVKGIAAKPMT ING ALLRVFGIW 19 V G LWS_pieRa DRYNV IVKGIAAKPMT INS ALLRILGVW 19 V G LWS_manSe DRYNV IVKGIAAKPMT SNG ALLRILGIW 19 V G MWS2_droMe DRYNV IVKGINGTPMT IKT SIMKILFIW 19 V G LWS_rhoPr DRYNV IVKGISAKPMT NKT AMLRILLVW 19 V G LWS_meoOe DRYNV IVKGISGTPLS QKN TTLQVLFVW 19 V G LWS_catBo DRYNV IVKGLSAKPMT ING ALLRILGIW 19 V G LWS_schGr DRYNV IVKGLSAKPMT NKT AMLRILFIW 19 V G LWS_triCa DRYNV IVKGLSAQPLT KKG AMLRILIIW 19 V G LWS2_apiMe DRYNV IVKGLSGKPLS ING ALIRIIAIW 19 V G LWS_bomTe DRYNV IVKGLSGKPLT ING ALLRILGIW 19 V G MWS_calEr DRYNV IVKGMAGQPMT IKL AIMKIALIW 19 V G MWS1_droMe DRYQV IVKGMAGRPMT IPL ALGKIAYIW 19 V G LWS_droMe DRYCV IVKGMARKPLT ATA AVLRLMVVW 19 V G LWS_arcGr DRYNV IVKGVAAEPLT SKG ASIRILFVW 19 V G LWS_eupSu DRYNV IVKGVAATPLT NKG AFARNIFSW 19 V G LWS_camLu DRYNV IVKGVAGEPLS TKK ASLWILTVW 19 V G LWS_proMi DRYNV IVKGVAGEPLS TKK ASLWILIVW 19 V G LWS_holCo DRYNV IVKGVSAEPLT SGG AMMRIAGTW 19 V G LWS_homGa DRYNV IVKGVSATPLT TNG AMLRNLFSW 19 V G LWS_neoAm DRYNV IVKGVSGEPLT NSG AMTRIAGTW 19 V G LWS_neoOe DRYNV IVKGVSGKPLS QKN ATLQVLFVW 19 V G LWS_mysDi ERYNV IVKGVSSKPLS VKG AITRIVLTW 19 V G LWS1_apiMe DRYNV IVKGMSGTPLT IKR AMLQILGIW 19 V G LWS_limPo DRYNV IVRGMAAAPLT HKK ATLLLLFVW 19 V G LWS_limPo DRYNV IVRGMAAAPLT HKK ATLLLLFVW 19 V G LWS_ixoSc DRYNV IVRGVAAAPLT HKR AALMIFFVW 19 V G ADRB2_homS DRYFA ITSPFKYQSLLT KNK ARVIILMVW 20 T P ADRA2A_hom DRYWS ITQAIEYNLKRT PRR IKAIIITVW 20 T A ADRA2C_hom DRYWS VTQAVEYNLKRT PRR VKATIVAVW 20 T A HTR1A_homS DRYWA ITDPIDYVNKRT PRR AAALISLTW 20 T P CHRM1_homS DRYFS VTRPLSYRAKRT PRR AALMIGLAW 20 T P DRD2_homSa DRYTA VAMPMLYNTRYS KRR VTVMISIVW 21 A P TAAR9_homS DRYIA VTDPLTYPTKFT VSV SGICIVLSW 20 T P ADRA2B_hom DRYWA VSRALEYNSKRT PRR IKCIILTVW 20 S A
Reference collection of 377 cytoplasmic loop C2 sequences from all 20 opsin loci
The second column contains the C2 loop sequences. The third column shows the continuation into transmembrane helix 4. The end of the loop region is determined by countback from the invariant tryptophan at position 160 in squid melanopsin as well as from crystallography and transmembrane prediction tools. Other columns show loop length and values at potentially informative positions 7 and 9 (which are generally characteristic of orthology class).
RHO1_homSa ERYVVVCKPMSNFRFGENH AIMGVAFTW 19 C P RHO1_bosTa ERYVVVCKPMSNFRFGENH AIMGVAFTW 19 C P RHO1_ornAn ERYIVVCKPMSNFRFGENH AIMGVAFTW 19 C P RHO1_monDo ERYVVVCKPMSNFRFGENH AIIGVAFTW 19 C P RHO1_galGa ERYVVVCKPMSNFRFGENH AIMGVAFSW 19 C P RHO1_calMi ERYVVVCKPMSNFRFGTNH AIMGVAFTW 19 C P RHO1_xenTr ERYVVVCKPMANFRFGENH AIMGVVFTW 19 C P RHO1_latCh ERYVVVCKPMSNFRFGENH AIMGVIFTW 19 C P RHO1_neoFo ERYIVVCKPISNFRFGENH AIMGVVFTW 19 C P RHO1_angAn ERWVVVCKPMSNFRFGENH AIMGLAFTW 19 C P RHO1_takRu ERYIVVCKPMTNFRFGEKH AIAGLVFTW 19 C P RHO1_leuEr ERYMVVCKPMANFRFGSQH AIIGVVFTW 19 C P RHO1_petMa ERYIVICKPMGNFRFGSTH AYMGVAFTW 19 C P RHO1_letJa ERYIVICKPMGNFRFGNTH AIMGVAFTW 19 C P RHO1_geoAu ERYIVICKPMGNFRFGNTH AIMGVALTW 19 C P RHO2_galGa ERYIVVCKPMGNFRFSATH AMMGIAFTW 19 C P RHO2_gekGe ERYIVICKPMGNFRFSATH AIMGIAFTW 19 C P RHO2_anoCa ERYIVVCKPMGNFRFSATH ALMGISFTW 19 C P RHO2_taeGu ERYIVICKPMGNFRFSASH ALMGIAFTW 19 C P RHO2_podSi ERYIVVCKPMGNFRFSSSH ALMGIAFTW 19 C P RHO2_pheMa ERYIVICKPMGNFRFSSSH AMMGISFTW 19 C P RHO2_latCh ERYIVVCKPMGNFRFASSH AIMGIAFTW 19 C P RHO2_neoFo ERYIVVCKPMGNFRFSNNH SIIGIVFTW 19 C P RHO1_anoCa ERYVVICKPMSNFRFGETH ALIGVSCTW 19 C P RHO1_conMy ERWMVVCKPVTNFRFGESH AIMGVMVTW 19 C P RHO2_ancDa ERYIVVCKPMGSFKFSSSH AMAGIAFTW 19 C P RHO2a_danR ERYIVVCKPMGSFKFSANH AMAGIAFTW 19 C P RHO2b_danR ERYIVVCKPMGSFKFSSNH AMAGIAFTW 19 C P RHO2c_danR ERYIVVCKPMGSFKFSSNH AFAGIGFTW 19 C P RHO2d_danR ERYIVVCKPMGSFKFSASH AFAGCAFTW 19 C P RHO2_oryLa ERYIVVCKPMGSFKFTATH SAAGCAFTW 19 C P RHO2_takRu ERYVVVCKPMGSFKFTGTH AAVGVAFTW 19 C P RHO2_gasAc ERYIVVCKPMGSFKFSGTH AGAGVLFTW 19 C P RHO2_hipHi ERYIVVCKPMGSFKFSGTH AGIGVLFTW 19 C P RHO2_mulSu ERYIVVCKPMGSFKFSGTH AGAGVAFTW 19 C P RHO2_oreNi ERYIVVCKPMGSFKFTGAH AGAGVLFTW 19 C P RHO2_pomMi ERYIVVCKPMGSFKFSGAH AGAGVALTW 19 C P RHO2_calMi ERYVVVCKPMSNFRFGTSH ALMGMGFTW 19 C P RHO2_geoAu ERYIVVCKPMGNFRFATTH AALGVVFTW 19 C P SWS2_ornAn ERFLVICKPLGNLSFRGTH AIFGCAATW 19 C P SWS2_anoCa ERYLVICKPLGNFTFRGTH AIIGCAVTW 19 C P SWS2_utaSt ERFLVICKPLGNFSFRGTH AIIGCIITW 19 C P SWS2_taeGu ERFLVICKPLGNFTFRGSH AVLGCAITW 19 C P SWS2_galGa ERFLVICKPLGNFTFRGSH AVLGCVATW 19 C P SWS2_neoFo ERFLVICKPLGNFTFRSTH AIIGCVATW 19 C P SWS2_xenTr ERFLVICKPMGNFTFRESH AVLGCILTW 19 C P SWS1_homSa ERYIVICKPFGNFRFSSKH ALTVVLATW 19 C P SWS1_monDo ERFIVICKPFGNFRFNSKH AMMVVLATW 19 C P SWS1_smiCr ERFIVICKPFGNFRFNSKH AMMVVLATW 19 C P SWS1_tarRo ERFIVICKPFGNFRFSSKH AMMVVLATW 19 C P SWS1_taeGu ERYIVICKPFGNFRFNSRH ALLVVAATW 19 C P SWS1_anoCa ERYIVICKPFGNFRFNSRH ALLVVAATW 19 C P SWS1_utaSt ERYIVICKPFGNFRFNSKH ALLVVAATW 19 C P SWS1_galGa ERYIVICKPFGNFRFSSRH ALLVVVATW 19 C P SWS1_geoAu ERYIVICKPFGNFRFGSKH ALVAVGLTW 19 C P SWS1_neoFo ERYLVICKPIGNFRFGSKH SMIAVVAAW 19 C P SWS1_xenLa ERYIVICKPMGNFNFSSSH ALAVVICTW 19 C P SWS1_petMa ERYIVICKPFGNFRFGSIH SLFAFCLTW 19 C P SWS1_danRe ERYVVICKPFGSFKFGQGQ AVGAVVFTW 19 C P SWS1_oryLa ERYLVICKPFGAFKFGSNH ALAAVIFTW 19 C P SWS2_geoAu ERCLVICKPFGNIAFRGTH ALIRCGFAW 19 C P SWS2_takRu ERWLVVCKPLGNFIFKPDH AIVCCIFTW 19 C P SWS2_gasAc ERWLVICKPLGNFIFKPDH ALVCCAFTW 19 C P LWS_homSap ERWMVVCKPFGNVRFDAKL AIVGIAFSW 19 C P LWS_monDom ERWVVVCKPFGNVKFDAKL AMVGIIFSW 19 C P LWS_ornAna ERWIVVCKPFGNVKFDAKL AMVGIVFSW 19 C P LWS_anoCar ERWVVVCKPFGNVKFDAKL AVAGIVFSW 19 C P LWS_galGal ERWFVVCKPFGNIKFDGKL AVAGILFSW 19 C P LWS_xenTro ERWFVVCKPFGNIKFDGKL AATGIIFSW 19 C P LWS_neoFor ERWVVVCKPFGNIKFDGKW AAGGIIFSW 19 C P LWS_calMil ERWVVVCKPFGNVKFDGKW AAFGIIFSW 19 C P LWS_takRub ERWVVVCKPFGNVKFDAKW ATGGIVFSW 19 C P LWS_gasAcu ERWIVVCKPFGNVKFDAKW ATAGIVFSW 19 C P LWS1_calMi ERWVVVCKPFGNMKFDSKM AVAGIVFSW 19 C P LWS2_calMi ERWVVVCKPFGNVKFDGKW AAFGIIFSW 19 C P LWS_petMar ERWMVVCKPFGNIKFDGKI ATILIVFSW 19 C P LWS_letJap ERWMVVCKPFGNIKFDGKI AIILIVFSW 19 C P LWS_geoAus ERWMVVCKPFGNLKFDGKV AIVLIIFSW 19 C P PIN_galGal ERYVVVCRPLGDFQFQRRH AVSGCAFTW 19 C P PIN_pheMad ERYLVICKPVGDFQFQRRH AVIGCLYTW 19 C P PIN_utaSta ERYLVICKPVGDFRFQQRH AVFGCVFTW 19 C P PIN_xenTro ERYLVICKPMGDFRFQQKH AILGCSFTW 19 C P PIN_bufJap ERYIVICKPMGDFRFQQRH AVMGCAFTW 19 C P PIN_podSic ERYLVICKPVGDFRFPARH AVLGCAFTW 19 C P PIN_calMil ERYIVICKPMGDFRFQQKH AVWGCLFTW 19 C P VAOP_galGa ERYIVICRPVGNMRLRGKH AAQGIAFVW 19 C P VAOP_anoCa ERYVVICRPLGNMRLNGKH AALGVAFVW 19 C P VAOP_xenTr ERYIVICRPLGNLRLQGKH SALAIIFVW 19 C P VAOP_danRe ERFFVICRPLGNIRLRGKH AALGLVFVW 19 C P VAOP_rutRu ERFFVICRPLGNIRLRGKH AALGLLFVW 19 C P VAOP_takRu ERFFVICRPLGNMRLQAKH AAIGLLFVW 19 C P VAOP_petMa ERYFVICRPLGNFRLQSKH AVLGLAVVW 19 C P PPIN_anoCa DRAIVIAKPMGTITFTTRK AMIGVAVSW 19 A P PPIN_xenTr DRVFVVCKPMGTLTFTPKQ ALAGIAASW 19 C P PPIN_ictPu DRYMVVCRPLGAVMFQTKH ALAGVVFSW 19 C P PPIN_oncMy DRYVVVCRPMGAVMFQTRH AVGGVVLSW 19 C P PPIN_danRe ERCMVVCRPVGSISFQTRH AVFGVAVSW 19 C P PPIN_petMa DRFVVVCKPLGTLMFTRRH ALLGITWAW 19 C P PPIN_letJa DRFVVVCKPLGTLMFTRRH ALLGIAWAW 19 C P PPIN2_petM ERYVVVCKPLGGVHFGTQH GLCGVAISW 19 C P PARIE_utaS ERYNVVCQPLGTLQMSTKR GYQLLGFIW 19 C P PARIE_anoC ERYNVVCQPLGTLQMSTQR AYQLLGFIW 19 C P PARIE_xenT ERYNVVCEPIGALKLSTKR GYQGLVFIW 19 C P PARIE_takR ERYNVVCKPRAGLKLTMRR SIIGLLFVW 19 C P PARIE_gasA ERYNVVCRPRNALKLSMRR SIHGLLIVW 19 C P PARIE_danR ERYNVVCKPMAGFKLNVGR SCQGLLLVW 19 C P PER_homSap DRYLTICLPDVGRRMTTNT YIGLILGAW 19 C P PER_panTro DRYLTICLPDVGRRMTTNT YIGLILGAW 19 C P PER_nomLeu DRYLTICLPDVGRRMTTNT YIGLILGAW 19 C P PER_gorGor DRYLTICLPDVGRRMTTNT YIGLILGAW 19 C P PER_ponPyg DRYLTICLPDIGRRMTTNT YIGLILGAW 19 C P PER_macMul DRYLTICLPDIGRRMTTNT YIGMILGAW 19 C P PER_papHam DRYLTICLPDIGRRMTTNT YIGMILGAW 19 C P PER_otoGar DRYLTICRPDIGRRMTTNS YIGMILGAW 19 C P PER_tarSyr DRYLTICRPDIGRRMTTNT YVGMILGAW 19 C P PER_micMur DRYLTICRPDIGRRMTTHT YVGMILGAW 19 C P PER_cavPor DRYLTICRPDIGRRMTSHS YVGMILGAW 19 C P PER_ochPri DRYLTICQPDIGRRMTTHT YFGMILGAW 19 C P PER_oryCun DRYLTICHPDVGRRMTTRT YLGLILGAW 19 C P PER_calJac DRYLTICLPDIGRRMTTST YIIMILGAW 19 C P PER_canFam DRYLTICSPDTGRRMTTNT YISMILGAW 19 C P PER_felCat DRYLTICSPNSGRRMTTNT YISMILGAW 19 C P PER_susScr DRYLTICRPEAGRRMTTNT YISMILGAW 19 C P PER_vicVic DRYLTICRPDAGRRMTTNT YISMILGAW 19 C P PER_turTru DRYLTICCPGAGRRMTTNT YISMILGAW 19 C P PER_bosTau DRYLTICHPDAGRRMTANT YISMILGAW 19 C P PER_choHof DRYLTICHPDVGRRMTINT YISMILGAW 19 C P PER_dasNov DRYLTICRPDTGRRMTINT YISMILGAW 19 C P PER_echTel DRYLTICHPDRGRRMTSNT YVGMILGAW 19 C P PER_loxAfr DRYLTICHPHIGRRMTSNT YVSMILGAW 19 C P PER_sorAra DRYLTLCRPDAGRSMTTNS YVGLILGAW 19 C P PER_equCab DRYLTTCRPDAGRRMTTST YTSMILGAW 19 C P PER_dipOrd DRYLTICHPDIGRGMTTRT YVTMILGAW 19 C P PER_musMus DRYLTISCPDVGRRMTTNT YLSMILGAW 19 S P PER_ratNor DRYLTISCPDVGRRMTGNT YLSMVLGAW 19 S P PER_eriEur DRYLTICRPHTGRSMSANS YIAMILGAW 19 C P PER_tupBel DRYLTLCRPAVGRRMGSST YAAMILGAW 19 C P PER_monDom DRYLTICQPDLGGRMTSYN YTLMILTAW 19 C P PER_ornAna DRYLTICRPAIGRKMTRSN YTAMILAAW 19 C P PER_xenTro DRYLTICRPDIGRRISGRH YTAMILAAW 19 C P PER_galGal DRYLTICRPDIGRRMTTRN YAALILAAW 19 C P PER_anoCar DRYLTICKPHIGSRLTATN YTTLILAAW 19 C P PER_taeGut DRYLTICRPDIGRRMTTRS YATLILAAW 19 C P PER1_gasAc DRYLTICRPDIGQKMTMQS YNLLILAAW 19 C P PER_gasAcu DRYLTICRPDIGQKMTMQS YNLLILAAW 19 C P PER_oryLat DRYLTICRPDLGQKMTMQS YNLLILAAW 19 C P PER_takRub DRYITICRPDIGRKMTVQS YNLLILAAW 19 C P PER_tetNig DRYLTICRPDIGRKMTVQS YNLLIAAAW 19 C P PER_danRer DRYLTICRPDIGQKLTTRS YTLLIVAAW 19 C P PER1a_sacK DRYWATCSPVEVMELKSKY YTRMTALGW 19 C P NEUR1_homS DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L NEUR1_nomL DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L NEUR1_panT DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L NEUR1_ponP DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L NEUR1_macM DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L NEUR1_papH DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L NEUR1_calJ DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L NEUR1_tarS DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L NEUR1_cavP DRYLKICYLSYGVWLKRKH AYICLAAIW 19 C L NEUR1_dasN DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_equC DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_canF DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_susS DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_pteV DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_choH DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_musM DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_ratN DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_loxA DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_felC DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_turT DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_tupB DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_echT DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_dipO DRYLKICYLSYGVWLKRKH AYICLAVIW 19 C L NEUR1_bosT DRYLKICYLSYGIWLKRKH AYICLAVIW 19 C L NEUR1_eriE DRYLKICYLSYGVWLKRKH AYLCLAVIW 19 C L NEUR1_sorA DRYLKICYLSYGVWLKRKH AYICLVVIW 19 C L NEUR1_speT DRYLKICYLSYGVWLKRKH AFICLAVIW 19 C L NEUR1_oryC DRYLKICYLSYGVWLKRRH AYICLALIW 19 C L NEUR1_myoL DRYLKICYLSYGVWLKRKH TYICLAFIW 19 C L NEUR1_monD DRYLKICHLSYGTWLKRHH AFICLALIW 19 C L NEUR1_taeG DRYLKICHLSYGTWLKRHH AFICLAIIW 19 C L NEUR1_galG DRYLKICHLAYGTWLKRHH AFICLALIW 19 C L NEUR1_ornA DRYLKICHLSYGTWLKRHH AYICLAIIW 19 C L NEUR1_macE DRYLKICHLSYGTWLKRHH AYICLVIIW 19 C L NEUR1_gasA DRYLKICHLRYGTWLKRHH AFVCLALVW 19 C L NEUR1_anoC DRYFKICHLSYGTWLKRHH VFICLGIIW 19 C L NEUR1_tetN DRYLKICHLRYGAWLKRHH AFLCLASVW 19 C L NEUR1_xenT DRYLKICHLRYGTWLKRRH AFIALAVIW 19 C L NEUR1_takR DRYLKICHLRYGTWFKRHH AFLCLVFTW 19 C L NEUR1_oryL DRYLKICHLRYGTWLKRQH AFLCLVFVW 19 C L NEUR1_pimP DRYLKICHLRYGTWLKRQH IFLCLVFVW 19 C L NEUR1_danR DRYLKICHLRYGTWLKRHH AFLSVVFIW 19 C L NEUR1_calM DRYLKICHLQYGSWLQRRH VFMSLAFIW 19 C L NEUR2_galG VCCLKICFPAYGNRFRRKH GQILIACAW 19 C P NEUR2_anoC VCCLKICFPVYGNRFRPGH GWILIACAW 19 C P NEUR2_oncM VCFVKVCYPLYGNRFNAVH GRLLIACAW 19 C P NEUR2_xenT VCCLKVCYPAYGNKFSTAH SRILLLGIW 19 C P NEUR2_danR VCCLKVCFPNYGNKFSSSH ACVMVIGVW 19 C P NEUR2_pimP VCCLKVCCPNYGNKFSSNH ACVMVIGVW 19 C P NEUR2_tetN VCCLKVCLPNLGSKFSSSH ARLLVAGVW 19 C P NEUR2_takR VCCLKVCFPNHGSRFSSSH ARLLVVGVW 19 C P NEUR2_gasA VCCLKVCFPNHGNRFSSSH ARLLVVAVW 19 C P NEUR2_oryL VCCLKVCFPNHGNKFSFSH ARLLVAGVW 19 C P NEUR3_galGal IRFLVTNSSKSNSNKISKNT VHILITFIW 20 N S NEUR3_taeGut IRFLVTNSPKSNsNKITKNT VCILIAFIW 20 F P NEUR3_anoCar IRFLVTFSSKPAGHKINRKV MHICIMLIW 20 S S NEUR3_xenTro IRYRVTSSFKYSGCTIEKKA VCILIMCIW 20 G F NEUR3a_danRe VRYLVTGNPPKSGSKFRRKT ISILIGVIW 20 G P NEUR3a_tetNi IRYLVTGSPPRSGVQFQKKT ICVVICAIW 20 G P NEUR3a_takRu IRFLVTGTPPRSGIKFQKKT ISVVISAIW 20 G P NEUR3a_gasAc VRYLVTGNPPRSGLRLQRKT VSMVIGAVW 20 G P NEUR3_calMil VRFLVTSTTQN......... ......... 20 S T NEUR3_petMar VRYKGTSTQVHsVKQITKRA MLAVIVAVW 20 S Q NEUR3b_danRe VRFIVSLTLQSPKEKISKRN AKILVATTW 20 L L NEUR3b_tetNi VRFTVSLNLQSPEEKISWKS VKIMCLLIW 20 L L NEUR3b_takRu VRFTVSLNLQSPeEKITWKS VKIMCMWVW 20 L L NEUR3b_gasAc VRFIVSLNLQSPNEKISWRK VKLLCACTW 20 L L NEUR3b_oryLa VRFIVSLNLHSPKEKVSWRK VKILCLWSW 20 L L NEUR4_ornAna TRYIKGCHPHRGHFINTAN ISVALILIW 19 C P NEUR4_galGal TRYIKGCHPERAHCISNSS MTVAMVLIW 19 C P NEUR4_taeGut TRYIKGCHPERGHCISNSS MSVALVLIW 19 C P NEUR4_anocar TRYIKGCHPDRGKCISNSS ISVALFLIW 19 C P NEUR4_xenTro TRYIKGCHPQRANCISNGS ITISLALIW 19 C P NEUR4_danRer TRFIKGCHPHKAHCITNST VAVCVVFIW 19 C P NEUR4_tetNig TRYIKGCQPSRAALISRSS VSVCLLLIW 19 C P NEUR4_gasAcu TRYIKGCHPNKAYCISTNT IAVSLICIW 19 C P NEUR4_calMil ...........AVSISAGS IAASLVLIW 19 . . NEUR4_petMar ...........PTKVTSTS MVVSLALVW 19 . . TMT_monDom ERYRTL-TLCPGQGADYQK ALLAVAGSW 19 - L TMT_macEug ERYRTL-TLCPRQGTDYHK ALLAVAGSW 19 - L TMT_ornAna ERYRTL-TLHPKQSTDYQK AVLAVGASW 19 - L TMT_galGal ERYSTL-TLCNKRSDDYRK ALLAVGGSW 19 - L TMT_taeGut ERYNTL-TLCHKRSDDFRK ALLAVAGSW 19 - L TMT_anoCar ERYSTL-TQTNKRGSDYQK ALLGVGGSW 19 - Q TMT_xenTro ERYSTL-TLYNKGGPNFKK ALLAVASSW 19 - L TMT_danRer ERYCTMMGSTEADATNYKK VIGGVLMSW 19 M S TMT_pimPro ERYCTMMGATQADSTNYKK VAMGIAFSW 19 M A TMTa_takRu ERYSTMMTPTEADPSNYCK VCLGITLSW 19 M P TMT_tetNig ERYSTMMTPTEADSSNYCK VCLGIGLSW 19 M P TMT_gasAcu ERYSTMVAPTEADSSNYHK ISLGITLSW 19 V P TMT_oryLat ERYSTMMTPAEADSSNYRK ISLGIILSW 19 M P TMTb_takRu ERYCTMVSSTIASNRDYRP VLGGICFSW 19 V S TMTa_calMi DRYITITGTTEADITNYNK TIVGIALSW 19 T T TMT1_plaDu ERYLAVVRPFDVGNLTNRR VIAGGVFVW 19 V P TMT2_anoGa ERYCLISRPFSSRNLTRRG AFLAIFFIW 19 S P TMT_triCas ERYLLIARPFRNNALNFHS AALSVFSIW 19 A P TMT_bomMor ERYLMVTRPLTSRHLSSKG AVLSIMFIW 19 T P ENCEPH_hom ERYIRVVHARVINFSW AWRAITYIW 16 V A TMT_aedAe ERFCLISHPFSSRSLSRRG AVFAILFIW 19 S P TMT_culPi ERFYLISRPFSSRSLSRRG ALGAVLLIW 19 S P ENCEPH_lox ERYIRVVHARVINFSW AWRAITYIW 16 V A TMT1_anoGa ERFCLISRPFAAQNRSKQG ACLAVLFIW 19 S P ENCEPH_can ERYIRVVHARVINFSW AWRAITYIW 16 V A TMT_triCa ERYLLIARPFRNNALNFHS AALSVFSIW 19 A P ENCEPH_oto ERYIRVVHARVINFSW AWRAITYIW 16 V A ENCEPH_mus ERYIRVVHARVINFSW AWRAITYIW 16 V A ENCEPH_ano ERYIRVVHARVIDFSW SWRAITYIW 16 V A ENCEPH_gal ERYIRVVHAKVIDFSW SWRAITYIW 16 V A ENCEPH_mon ERYNRIVHAKVINFSW AWRAITYIW 16 V A ENCEPH_pte ERYIRVVQARAIDFSW AWRTITYIW 16 V A ENCEPH_squ ERYIRVVNATAIDFSW AWRAITYIW 16 V A ENCEPH_xen ERYARVVYGKYVNSSW SKRSITFVW 16 V G ENCEPH_dan ERYIRVVHAKVVDFPW AWRAITHIW 16 V A ENCEPH_tak ERYIRVVHAQVVDFPW AWRAIGHIW 16 V A ENCEPH_gas ERYIRVVHAQVVDFPW AWRAIGHIW 16 V A ENCEPH_ory ERYIRVVHAQVVDFPW AWRAIGHIW 16 V A ENCEPH_cal ERYIRVVNAKATNFPW AWRAITYTW 16 V A ENCEPH_squ ERYIRVVNATAIDFSW AWRAITYIW 16 V A ENCEPH_pet ERYARLIKAQVLDFSW AWRAVTYTW 16 I A RGR_homSap GRYHHYCTRSQLAWNS AVSLVLFVW 16 C R RGR_panTro GRYHHYCTRSQLAWNS AISLVLFVW 16 C R RGR_gorGor GRYHHYCTGSTLACKS AVSLVLSGR 16 C G RGR_macMul GRYHHYCTRSQLAWNS AISLVLFVW 16 C R RGR_ponPyg GRYHHYCTGSQLAWNS AISLVLFVW 16 C G RGR_calJac GRYHHYCTGSQLAWNS AISLVLFVW 16 C G RGR_nomLeu GRYHHYCTGSQLAWNS AISLVLFVW 16 C G RGR_tarSyr GRYHHYCTGSQLAWNT AISLVLFVW 16 C G RGR_pteVam GRYHHYCTGSRLAWNT AVSLVLFVW 16 C G RGR_oryCun GRYHHYCTGSQLAWNT AVLLVLFVW 16 C G RGR_ochPri GRYHHYCTGSQLAWNT AVLLVLFVW 16 C G RGR_otoGar GRYHHYCTGRPLAWST AISLVLFVW 16 C G RGR_micMur GRYHHYCTGSPLAWST AISLVLFVW 16 C G RGR_musMus GRYHHYCTGRQLAWDT AIPLVLFVW 16 C G RGR_ratNor GRYHHYCTGRQLAWDT AIPLVLFVW 16 C G RGR_cavPor GRHQQCCTRGRLTWST AVPLVLFVW 16 C R RGR_speTri GRYHHYCTGSQLAWNT AIPLVLFVW 16 C G RGR_sorAra GRYHHYCTGRQLAWDV AIALVIFVW 16 C G RGR_myoLuc GRYHHYCTGSRLAWRT AASLVLFVW 16 C G RGR_canFam GRYHHYCTRGQLAWNT AISLVLCVW 16 C R RGR_felCat GRYHHYCSGSQLAWNT AISLVICVW 16 C G RGR_bosTau GRYHHFCTGSRLDWNT AVSLVFFVW 16 C G RGR_turTru GRYHHYCTGSRLDWNT AVSLVFFVW 16 C G RGR_susScr GRYHHYCTRSRLDWNT AVSLVFFVW 16 C R RGR_equCab GRYHHYCTRSRLAWNT AVFLVFFVW 16 C R RGR_eriEur GRYHHHCTRSRLAWNT AVFLVFFVW 16 C R RGR_dipOrd GRCHHHCTGSLLGWDT AVSLVIFVW 16 C G RGR_loxAfr ERYHHYCTRSRLAWSS ASALVLFVW 16 C R RGR_proCap ERYHHYCTGSKLAWSS AGALVLFMW 16 C G RGR_echTel ERYHHYCTGSQFTWSS ASTLVLFMW 16 C G RGR_dasNov ERCHRHCIGRRLAWST AGCLVLCLW 16 C G RGR_choHof ERYRHHCTGSQLSWST AGSLVLCVW 16 C G RGR_ornAna DRYLRHCSRSKPQWGT AVSTVLFAW 16 C R RGR_anoCar DRHHQYCTGNKLQWGS VIPMTIFLW 16 C G RGR_galGal DRYHHYCTRSKLQWST AISMMVFAW 16 C R RGR_taeGut DRYHHYCTRSRLQWST AVSMMVFAW 16 C R RGR_xenTro DRYHQYCTRSKLHWST AVSVVFFIW 16 C R RGR_xenLae DRYHQYCTRSKLHWGT AVSMVLFVW 16 C R RGR1_gasAc DRYHQYCTRTKLQWSS AITLAVFVW 16 C R RGR1_takRu DRYHQYCTRTKLQWSS AITLAVFIW 16 C R RGR1_tetNi DRYHQYCTRTKLQWSS AITLAVFIW 16 C R RGR1_pimPr DRYHQYCTRTKLQWSS AITLVIFIW 16 C R RGR1_osmMo DRYHQYCTRTKLQWSS AITLVMFIW 16 C R RGR1_gadMo DRYHQYCTRTELQWSS AVTLSVFIW 16 C R RGR1_danRe DRYHQYCTRTKLQWSS AITLVLFTW 16 C R RGR1_oryLa DRYHQYCTRTKLQWST AITLAVLVW 16 C R RGR_calMil DRYHQNCSRSRLQWSS AITVTVFIW 16 C R RGR2_gasAc DRYHQYCTRQKLFWST TLTMSAIIW 16 C R RGR2_tetNi DRYHQYCTRQKLFWST TLTMSSIIW 16 C R RGR2_oryLa DRYHQYCTRQKLFWST SITISLIIW 16 C R RGR2_danRe DRYHQYCTKQKMFWST SITISCLIW 16 C K RGR2_pimPr DRYHLYCTKQKMFWST SGTISALIW 16 C K RGR2_gadMo DRYHQYCTRQKLFWST TVTMCCIVW 16 C R RGR2_hipHi DRYHQYCTRQKLFWST TLTMSGIIW 16 C R RGR2_oncMy DRYHQYVTNQKLFWST AWTISIIIW 16 V N RGR2_esoLu DRYHQYVTNQKLFWST AWTFSIIIW 16 V N RGR2_poeRe DRYHQYCTRQKLFWST TLTMSGIIW 16 C R MEL1_homSa DRYLVITRPLATFGVASKRR AAFVLLGVW 20 T P MEL1_panTr DRYLVITRPLATFGVASKRR AAFVLLGVW 20 T P MEL1_gorGo DRYLVITRPLATFGVASKRR AAFVLLGVW 20 T P MEL1_ponAb DRYLVITRPLATIGVASKRR AAFVLLGVW 20 T P MEL1_rheMa DRYLVITRPLATIGVASKRR AAFVLLGVW 20 T P MEL1_calJa DRYLVITRPLATIGVASTKR AAFVLLGVW 20 T P MEL1_micMu DRYLVITRPLASVGTASKRR AGLVLLGVW 20 T P MEL1_otoGa DRYLVITRPLTTVGVASKRR AALVLLGVW 20 T P MEL1_musMu DRYLVITRPLATIGRGSKRR TALVLLGVW 20 T P MEL1_ratNo DRYLVITRPLATIGMRSKRR TALVLLGVW 20 T P MEL1_nanEh DRYLVITRPLATIGVASKRR TALVLLGVW 20 T P MEL1_phoSu DRYLVITRPLATIGMGSKRR TALVLLGIW 20 T P MEL1_dipOr DRYLVITRPLATIGVTSKRR TAFVLLGVW 20 T P MEL1_cavPo DRYLVITRPLATIGVASKRQ AALVLLGVW 20 T P MEL1_speTr DRYLVITRPLATIGMASKKR AAFFLLGVW 20 T P MEL1_oryCu DRYLVITRPLAAVGMVSKKR AGLVLLGVW 20 T P MEL1_ochPr DRYLVITRPLAAVGMVSKRR TGLVLLGVW 20 T P MEL1_bosTa DRYLVITRPLATVGMVSKRR AALVLLGVW 20 T P MEL1_turTr DRYLVITRPLATVGMVSKRR AALVLLGVW 20 T P MEL1_susSc DRYLVITHPLATVGMVSKRR AALVLLGVW 20 T P MEL1_equCa DRYLVITRPLATVGVVSKRW AALVLLGIW 20 T P MEL1_felCa DRYLVITHPLATIGVVSKRR AALVLLGVW 20 T P MEL1_canFa DRYLVITHPLAAVGVVSKRR AALVLLGVW 20 T P MEL1_myoLu DRYLVITRPLA-IGVVSKRR AALVLLGVW 20 T P MEL1_pteVa DRYLVITRPLAAIGVVSKRR AALVLLGVW 20 T P MEL1_eriEu DRYLVITRPLATIGVVSKRR VALVLLGVW 20 T P MEL1_loxAf DRYLVITRPLATIGVVSKRR AALVLLGIW 20 T P MEL1_proCa DRYLVITRPLATIGVVSKRR TALVLLGTW 20 T P MEL1_echTe DRYLVITRPLATIGVVSKRR AALVLLVIW 20 T P MEL1_smiCr DRYFVITRPLASIGMISKKK TGLILLGVW 20 T P MEL1_monDo DRYFVITRPLASIGVISKKK TGFILLGVW 20 T P MEL1_ornAn DRYFVITRPLASIGVISKKR ALLILTGVW 20 T P MEL1_anoCa DRYFVITRPLASIGAMSTKK ALLILSGVW 20 T P MEL1_taeGu DRYFVITKPLASVGVTSKKK ALIILVGVW 20 T P MEL1_galGa DRYFVITKPLASVRVMSKKK ALIILVGVW 20 T P MEL1_xenTr DRYFVITRPLTSIGVMSKKR AVLILSGVW 20 T P MEL1_danRe DRYFVITRPLASIGVLSQKR ALLILLVAW 20 T P MEL1_danRe DRYFVITRPLASIGVMSRKR ALLILSAAW 20 T P MEL1_takRu DRYFVITRPLTSIGVLSRKR AFVILMTVW 20 T P MEL1_gasAc DRYFVITRPLTSIGMMSRRR ALLILMGAW 20 T P MEL1_oryLa DRYFVITRPLTSIGVLSRKR ALLILSAAW 20 T P MEL1_calMi DRYFVITRPLASIGVLSHRR AGLIILSLW 20 T P MEL1_petMa DRYLVLTRPLASIGAMSKRR AMYITAAVW 20 T P MEL2_galGa DRYLVITKPLRSIQWTSKKR TIQIIAAVW 20 T P MEL2_anoCa DRYCVITKPLQSIKRTSKKR TCIIIVFVW 20 T P MEL2_xenLa NRYIVITKPLQSIQWSSKKR TSQIIVLVW 20 T P MEL2_danRe DRYLVITKPLQTIQWNSKRR TGLAILCIW 20 T P MEL2_tetNi DRYVVITKPLQTIRRSSKRR TALAILMVW 20 T P MEL2_gasAc DRYLVITKPLQAIHWGSKRR TTLAILLVW 20 T P MEL1_plaDu DRFYVITNPLGAAQTMTKKR AFIILTIIW 20 T P MEL1_capCa DRYMVIAKPFYAMKHVSHKR SLIQIILAW 20 A P MEL1_helRo DRYLVVGQPLAMLNQSHFRR SFYHVLIIW 20 G P MEL1_todPa DRYNVIGRPMAASKKMSHRR AFIMIIFVW 20 G P TMT_triCys ERFITIVLPLKRDTILSTKN IYIGLGILW 20 V P
Reference collection of structurally determined GPCR
>RHO1_bosTau cow rod rhodopsin
MNGTEGPNFYVPFSNKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAI
ERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWL
PYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA*
>MEL1_todPac Todarodes pacificus (squid) Gq X70498 480 11106382 Mollusca 'squid rhodopsin' 3D: May 2008 Cys 337 palmitoyled
MGRDLRDNETWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISI
DRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGANAEMRLAKI
SIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAETEIPAGESSDAAPSADAAQMKEMMAMMQKMQQQQAAYPPQGY
APPPQGYPPQGYPPQGYPPQGYPPQGYPPPPQGAPPQGAPPAAPPQGVDNQAYQA*
>ADRB1_melGal turkey Beta 1 adrenergic receptor with stabilising mutations And bound cyanopindolol
MGAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAI
DRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREA
KEQIRKIDRASKRKRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCRSPDFRKAFKRLLAFPRKADRRLHHHHHH*
>ADRB2_homSap beta 2 adrenergic receptor 365 aa
MGQPGNGSAFLLAPNRSHAPDHDVTQQRDEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAV
DRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYANETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRFHVQNLSQVEQDGRTGHGL
RRSSKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLCLRRSSLKAYGNGYSSNGNTGEQSG*
>ADORA2A_homSap adenosine adrenergic receptor 2A
MPIMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAI
DRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKNHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRI
FLAARRQLKQMESQPLPGERARSTLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFR
KIIRSHVLRQQEPFKAAGTSARVLAAHGSDGEQVSLRLNGHPPGVWANGSAPHPERRPNGYALGLVSGGSAQESQGNTGLPDVELLSHELKGVCPEPPGLDDPLAQDGAGVS*
The C2 loop is highly conserved within each orthology class for GPCR with determined structure:
RHO1 in vertebrates MEL1 in vertebrates ADRB1 in vertebrates ADRB2 orthologs in tetrapods ADORA2A in teleosts
homSap ERYVVVCKPMSNFRFGENHAIMGVAFTW homSa DRYLVITRPLATFGVASKRRAAFVLLGVW homSap DRYLAITSPFRYQSLLTRARARGLVCTVW homSap DRYFAITSPFKYQSLLTKNKARVIILMVW homSap DRYIAIRIPLRYNGLVTG TRAKGIIAICW
panTro ERYVVVCKPMSNFRFGENHAIMGVAFTW panTr DRYLVITRPLATFGVASKRRAAFVLLGVW panTro DRYLAITSPFRYQSLLTRARARGLVCTVW panTro DRYFAITSPFKYQSLLTKNKARVIILMVW panTro DRYIAIRIPLRYNGLVTGTRAKGIIAICW
gorGor ERYVVVCKPMSNFRFGENHAIMGVAFTW gorGo DRYLVITRPLATFGVASKRRAAFVLLGVW ponAbe DRYLAITSPFRYQSLLTRARARGLVCTVW gorGor DRYFAITSPFKYQSLLTKNKARVIILMVW gorGor DRYIAIRIPLRYNGLVTGTRAKGIIAICW
ponAbe ERYVVVCKPMSNFRFGENHAIMGVAFTW ponAb DRYLVITRPLATIGVASKRRAAFVLLGVW rheMac DRYLAITSPFRYQSLLTRARARGLVCTVW ponAbe DRYFAITSPFKYQSLLTKNKARVIILMVW ponAbe DRYIAIRIPLRYNGLVTGTRAKGIIAICW
rheMac ERYVVVCKPMSNFRFGENHAIMGVAFTW rheMa DRYLVITRPLATIGVASKRRAAFVLLGVW calJac DRYLAITSPFRYQSLLTRARARGLVCTVW rheMac DRYFAITSPFKYQSLLTKNKARVIILMVW rheMac DRYIAIRIPLRYNGLVTGTRAKGIIAICW
calJac ERYVVVCKPMSNFRFGENHAIMGVAFTW calJa DRYLVITRPLATIGVASTKRAAFVLLGVW micMur DRYLAITSPFRYQSLLTRARARALVCTVW calJac DRYFAITSPFKYQSLLTKNKARVIILMVW calJac DRYIAIRIPLRYNGLVTGTRAKGIIAICW
micMur ERYVVVCKPMSNFRFGENHAIMGVVFTW micMu DRYLVITRPLASVGTASKRRAGLVLLGVW otoGar DRYLAITSPFRYQSLLTRARARPLVCTVW micMur DRYFAITSPFKYQSLLTKNKARVVILMVW micMur DRYIAIRIPLRYNGLVTGTRAKGIIAICW
musMus ERYVVVCKPMSNFRFGENHAIMGVVFTW otoGa DRYLVITRPLTTVGVASKRRAALVLLGVW musMus DRYLAITSPFRYQSLLTRARARALVCTVW otoGar DRYFAITSPFKYQSLLTKNKARVVILMVW musMus DRYIAIRIPLRYNGLVTGMRAKGIIAICW
ratNor ERYVVVCKPMSNFRFGENHAIMGVAFTW musMu DRYLVITRPLATIGRGSKRRTALVLLGVW ratNor DRYLAITSPFRYQSLLTRARARALVCTVW tupBel DRYFAITSPFKYQSLLTKNKARVVILMVW ratNor DRYIAIRIPLRYNGLVTGVRAKGIIAICW
cavPor ERYVVVCKPMSNFRFGENHAIMGVVFTW ratNo DRYLVITRPLATIGMRSKRRTALVLLGVW cavPor DRYLAITSPFRYQSLLTRARARVLVCTVW dipOrd DRYFAITSPFKYQSLLTKNKARVVILMVW dipOrd DRYIAIRIPLRYNSLVTCTRAKGIIAICW
speTri ERYMVVCKPMSNFRFGENHAIMGVIFTW dipOr DRYLVITRPLATIGVTSKRRTAFVLLGVW oryCun DRYLAITSPFRYQSLLTRARARALVCTVW cavPor DRYFAITSPFKYQSLLTKNKARVVILMVW cavPor DRYIAIRIPLRYNGLVTCTRAKGIIAICW
oryCun ERYVVVCKPMSNFRFGENHAIMGVAFTW cavPo DRYLVITRPLATIGVASKRQAALVLLGVW ochPri DRYLAITSPFRYQSLLTRARARALVCTVW oryCun DRYFAITSPFKYQSLLTKNKARVVILMVW speTri DRYIAIRIPLRYNGLVTGMRAKGIIAICW
ochPri ERYVVVCKPMSNFRFGENHAIMGVAFTW speTr DRYLVITRPLATIGMASKKRAAFFLLGVW bosTau DRYLAITSPFRYQSLLTRARARALVCTVW ochPri DRYFAITSPFKYQSLLTKNKARVVVLMVW oryCun DRYIAIRIPLRYNGLVTGTRAKGIIAICW
bosTau ERYVVVCKPMSNFRFGENHAIMGVAFTW oryCu DRYLVITRPLAAVGMVSKKRAGLVLLGVW equCab DRYLAITSPFRYQSLLTRARARALVCTVW equCab DRYFAITSPFKYQSLLTKNKARVVILMVW ochPri DRYIAIRIPLRYNGLVTGSRAKGIIAICW
equCab ERYVVVCKPMSNFRFGENHAIMGVAFTW ochPr DRYLVITRPLAAVGMVSKRRTGLVLLGVW felCat DRYLAITSPFRYQSLLTRARARALVCTVW felCat DRYFAITSPFKYQSLLTKNKARVVILMVW turTru DRYIAIRIPLRYNGLVTGTRAKGIIAVCW
felCat ERYVVVCKPMSNFRFGENHAIMGVAFTW bosTa DRYLVITRPLATVGMVSKRRAALVLLGVW canFam DRYLAITAPFRYQSLLTRARARALVCTVW canFam DRYFAITSPFKYQSLLTKNKARVVILMVW bosTau DRYIAIRIPLRYNGLVTGTRAKGIIAVCW
canFam ERYVVVCKPMSNFRFGENHAIMGVAFTW turTr DRYLVITRPLATVGMVSKRRAALVLLGVW myoLuc DRYLAITSPFRYQSLLTRARARALVCTVW myoLuc DRYFAITSPFKYQSLLTKNKARVVILLVW canFam DRYIAIRIPLRYNGLVTGTRAKGIIAVCW
myoLuc ERYVVVCKPMSNFRFGENHAIMGLAFTW equCa DRYLVITRPLATVGVVSKRWAALVLLGIW pteVam DRYLAITSPFRYQSLLTRARARALVCTVW pteVam DRYFAITSPFKYQSLLTKNKARVVILMVW myoLuc DRYIAIRIPLRYNGLVTGARAKGIIAICW
pteVam ERYVVVCKPMSNFRFGENHAIMGLALTW felCa DRYLVITHPLATIGVVSKRRAALVLLGVW echTel DRYLAITSPFRYQSLLTRARARVLVCTVW eriEur DRYFAITSPFKYQSLLTKNKARVVILMVW eriEur DRYIAIRIPLRYNGLVTGQRAKGIIAVCW
eriEur ERYVVVCKPMSNFRFGENHAIMGVAFTW canFa DRYLVITHPLAAVGVVSKRRAALVLLGVW choHof DRYLAITSPFRYQSLLTRARARALVCTVW sorAra DRYFAITSPFKYQSLLTKNKARGVILMVW loxAfr DRYIAIRIPLRYNGLVTGTRAKGIIAVCW
dasNov ERYVVVCKPMSNFRFGENHAVMGVAFTW myoLu DRYLVITRPLA-IGVVSKRRAALVLLGVW monDom DRYIAITSPFRYQSLLTRARARALVCTVW proCap DRYFAITSPFKYQSLLTKNKARVVILMVW proCap DRYIAIRIPLRYNGLVTGTRAKGIIAVCW
monDom ERYVVVCKPMSNFRFGENHAIIGVAFTW pteVa DRYLVITRPLAAIGVVSKRRAALVLLGVW ornAna DRYIAITSPFRYRSLLTRARARGLVCGVW echTel DRYFAITSPFKYQSLLTKNKARVVILMVW galGal DRIIAIRIPLRYNGLVTGSRAKGIIAICW
ornAna ERYIVVCKPMSNFRFGENHAIMGVAFTW eriEu DRYLVITRPLATIGVVSKRRVALVLLGVW galGal DRYLAITSPFRYQSLMTRARAKGIICTVW dasNov DRYFAITSPFKYQSLLTKNKARVVILMVW taeGut DRIIAIRIPLRYNGLVTGSRAKGIIAICW
galGal ERYVVVCKPMSNFRFGENHAIMGVAFSW loxAf DRYLVITRPLATIGVVSKRRAALVLLGIW taeGut DRYLAITSPFRYQSLMTKGRAKGIICTVW monDom DRYFAITAPFRYQSMLTKGKARVVILVVW xenTro DRYIAIRIPLRYNSLVTSRRANAIIAVCW
taeGut ERYVVVCKPMSNFRFGENHAIMGVAFSW proCa DRYLVITRPLATIGVVSKRRTALVLLGTW anoCar DRYLAITSPFRYQSLMTKKRAKIIVCVVW galGal DRYFAITSPFKYQSLLTKSKARVVILVVW tetNig DRYIAIKLPLRYNGLVTGQRAQAIIAICW
anoCar ERYVVICKPMSNFRFGETHALIGVSCTW echTe DRYLVITRPLATIGVVSKRRAALVLLVIW xenTro DRYIAITSPLKYEMLVTKVRARLTVCLVW taeGut DRYFAITSPFKYQSLLTKGKARVVILVVW fugRub DRYIAIKLPLRYNSLVTGKRAQGIIAICW
xenTro ERYVVVCKPMANFRFGENHAIMGVVFTW monDo DRYFVITRPLASIGVISKKKTGFILLGVW tetNig DRYVAITSPFRYQSLLTKARARAMVCAVW anoCar DRYFAITSPFKYQSHLTKNKARVIILLVW gasAcu DRYIAIKIPLRYNGLVTGQRAQGIIAICW
tetNig ERYIVVCKPVTNFRFGEKHAIAGLAFTW ornAn DRYFVITRPLASIGVISKKRALLILTGVW fugRub DRYVAITSPFRYQSLLTKARAKAMVCAVW xenTro DRYFAITSPFRYQSLLTKCKARIVILLVW oryLat DRYIAIKIPLRYNSLVTSQRARGIIAICW
fugRub ERYIVVCKPMTNFRFGEKHAIAGLVFTW anoCa DRYFVITRPLASIGAMSTKKALLILSGVW gasAcu DRYVAITSPFRYQSLLTKARARTVVCVVW danRer DRYIAIKIPLRYNSLVTGQRARGIIAICW
gasAcu ERYVVVCKPMSNFRFGEKHAIAGLLFTW galGa DRYFVITKPLASVRVMSKKKALIILVGVW oryLat DRYVAITSPFRYQSLLTKSRAKAVVCVVW
oryLat ERYVVVCKPMTNFRFEEKHAIAGLAFSW xenTr DRYFVITRPLTSIGVMSKKRAVLILSGVW danRer DRYIAIISPFRYQSLLTKARAKVVVCAVW
danRer ERWMVVCKPVSNFRFGENHAIMGVAFTW danRe DRYFVITRPLASIGVLSQKRALLILLVAW petMar DRYIAVARPLRYETLMNKRRARFIIVAVW
petMar ERYIVICKPMGNFRFGSTHAYMGVAFTW takRu DRYFVITRPLTSIGVLSRKRAFVILMTVW
gasAc DRYFVITRPLTSIGMMSRRRALLILMGAW
oryLa DRYFVITRPLTSIGVLSRKRALLILSAAW
calMi DRYFVITRPLASIGVLSHRRAGLIILSLW
petMa DRYLVLTRPLASIGAMSKRRAMYITAAVW
See also: Curated Sequences | Ancestral Introns | Informative Indels | Ancestral Sequences | Alignment | Update Blog