Opsin evolution: Neuropsin phyloSNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| (79 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
'''See also:''' [[Opsin_evolution|Curated Sequences]] | [[Opsin_evolution:_Peropsin_phyloSNPs|Peropsins]] | [[Opsin_evolution:_RGR_phyloSNPs|RGR phyloSNPs]] | [[Opsin_evolution:_LWS_PhyloSNPs|LWS]] | [[Opsin_evolution:_Encephalopsin_gene_loss|Encephalopsins]] | [[Opsin_evolution:_Melanopsin_gene_loss|Melanopsins]] | [[Opsin_evolution:_update_blog|Update Blog]] | |||
== Neuropsin backgrounder == | == Neuropsin backgrounder == | ||
Neuropsin (OPN5) is a deeply diverged member of the opsin family with a [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=14623103 | Neuropsin (OPN5, GPR136, NEUR1) is a deeply diverged member of the opsin family with a mere [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=14623103,20679218,18570255 three experimental publications] and considerable confusion over the name (mysteriously used for an unrelated kalikrein serine protease not remotely related to opsins). OPN5 itself is a poor nomenclatural choice, having no mneumonic value and not extendable to the four vertebrate paralogs, so NEUR1-4 are used here for these loci. | ||
There are no known disease associations or described knockout phenotypes; in mouse, neuropsin is expressed primarily in brain, spinal chord, and testes. In chicken, neuropsin is expressed in [http://onlinelibrary.wiley.com/doi/10.1002/dvdy.21611/full embryonic and early posthatching] neural retina (but not pineal gland) in subsets of differentiating ganglion cells and amacrine cells, ie prior to functioning of ciliary opsins thus ruling out a retinal replenishment role ("photoisomerase"). | |||
A 2010 [http://www.pnas.org/content/107/34/15264.long study] in quail brain mapped NEUR1 to the paraventricular organ, a region of the diencephalon of nonmammalian vertebrates containing aminergic neuronal cell bodies beneath the epithelial membrane lining the [http://en.wikipedia.org/wiki/Ventricular_system third ventricle]. The vertebrate forebrain's diencephalon contains the thalamus, hypothalamus and posterior pituitary. | |||
The specific model here envisions light detected by NEUR1 in paraventricular neurons signaling via cerebrospinal fluid to the pars tuberalis of the pituitary gland, inducing there thyroid-stimulating hormone, in turn inducing [[Selenoprotein_evolution:_introduction|deiodinase DIO2]] in tanycytes lining the third ventricle leading to long-day–induced T3 in the mediobasal hypothalamus ultimately inducing gonadotropin-releasing hormone and so testis growth associated with seasonal reproduction. Peak adsorption observed at 420 nm is not readily transfered to orthologs in other species via alignment as tuning residue formulas have not been described for this class of opsin. | |||
A [http://www.pnas.org/content/107/36/15662.full commentary piece] assigns the paraventricular organ to the hypothalamus so even though no exact mammalian [http://www.ncbi.nlm.nih.gov/pubmed/6635521,9550137 anatomical counterpart] exists, the Allen Brain Atlas might show mouse expression of NEUR1 there, assuming the photoperiodic reproduction hormomal control mechanism is retained from the ancestral situation. (The paraventricular organ is known from teleost fish and frog as well as birds.) Photoentrainment of daily (circadian) activity rhythms is then quite distinct in terms of opsin use from photoreceptor control of seasonal reproduction. This fits with human NEUR1 transcripts most commonly recovered from testis, for example DB097202, and also with the phylogenetic range (back to amphioxus and sea urchin where unfortunately nothing is known). | |||
The other 3 deeply conserved neuropsins found in nonmammalian vertebrates including birds were not considered in the quail study, leaving it unclear what functional roles might be left for them. NEUR4 even persists into some mammals. Much more work needs to be done on non-imaging opsins, given that they comprise the vast majority of the 24 vertebrate opsin loci. The neuropsins collapsed in mammals along with many other opsins (including imaging) in good agreement with the [[Opsin_evolution:_trichromatic_ancestral_mammal|GT Wall hypothesis]]. | |||
Neuropsin has all the classical attributes of a rhodopsin-class GPCR and indeed opsin photoreceptor: Schiff base lysine at expected position, standard tyrosine counterion and DRY motif, seven transmembrane configuration, disulfide at expected position, proximal glycosylation and distal palmitoylation and kinase sites. It is most closely related to peropsin and rgropsin in terms of blast clustering and intron positioning. | |||
Its G-protein signaling partner is not really known though GNAQ is a good candidate among the 16 paralogs and [http://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006393107/-/DCSupplemental/pnas.201006393SI.pdf?targetid=nameddest=SF5 is expressed] in quail paraventricular organ. However the [http://genomewiki.ucsc.edu/images/c/c0/GabgComplexity.jpg closest GNAQ paralogs], namely GNA11, GNA14 and GNA15, were not considered in the quail study. | |||
Its evolution is illuminated by the massive comparative genomics study described here, which extracts and compares over 50 full length NEUR1 vsequences from various genomics projects. Neuropsin can be located outside chordates but not outside deuterostomes. However, like peropsin and rgropsin, it must have originated much earlier in pre-Bilaterans. Thus its absence in earlier diverging species must be due to gene loss because unrecognizability can be ruled out given test sensitivity. Some role in deuterostomes has persisted over many billions of years of branch length, perhaps correlating with their ciliary imaging vision and anti-correlating to protostomal rhabdomeric imaging. | |||
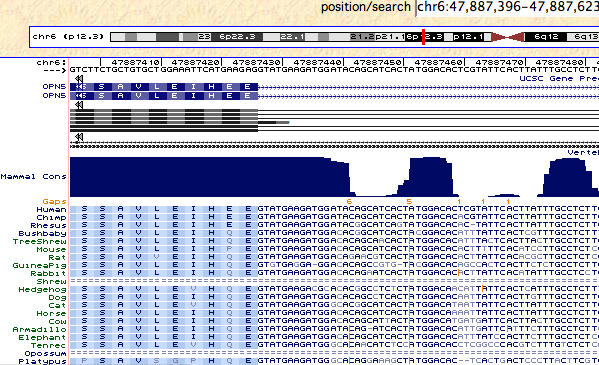
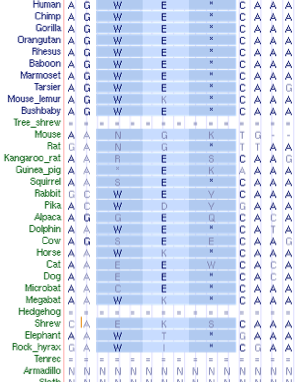
Within placental mammals, neuropsin is extraordinarily conserved, with percent identity relative to human protein 96% averaged over 31 species (exceeding the 95% percentile of all coding genes proteomewide). That conservation drops considerably at marsupials and monotremes (86%), is less striking at tetrapods (78%), and not especially remarkable at teleost fish (68%). This pattern suggests neuropsin acquired significant new adaptive functionality on the placental mammal stem, leading to marked resilience to fixation of any further variation. | |||
[[Image:Opsin_NeurStop.png]][[Image:NEUR1we*.png]] | |||
The structure of the neuropsin gene is rather odd at the 3' end. In human, a weak splice may have developed that results, after an intron of 12,244 bp, in a seventh very short coding exon continued by a long 3'UTR. However a stop codon is soon encountered if this splice is not taken (which it is in 6 of 7 transcripts). This results in two slightly different alternative carboxy termini sequences EEV* vs EEWE*. Very few transcripts exist in this region for any species but it appears that the ancestral form of the protein only utilized the initial stop codon in exon 6. | |||
This feature GTatga is conserved in all species back to platypus; indeed an unexplained conservation in nucleotide sequence extends well beyond this. However the splice acceptor and WE* appear an option only back to lemurs, whereas the QEV* option (EEV* from tarsier to human) is available in all 38 available mammal genomic sequences. Oddly the carboxy terminus first becomes conserved in mammals. | |||
A single mouse transcript also terminates early; four others continue on. No species other than rat has an available transcript in this region. No other rodent genomes could have an orthologous split exon: kangaroo rat and ground squirrel have frame shifts, pika lacks the splice acceptor, and rabbit and guinea pig have no homologous sequence. | |||
This can be explained by independent origins of splice acceptors in various clades. However far more comparative transcripts are needed to understand the 3' end of this gene. It may be that the conserved intronic sequence following exon 6 is an accident waiting to happen as it conserves -- for whatever reason -- half of a splice site. | |||
It must be said here that GenBank has a very muddled policy in the nr nucleotide division, not distinguishing real experimental transcript data from predictions, gene mRNA models, synthetic clones, the seemingly meaningless term cDNA, staff interpretations of genomic data, and poorly documented third-party submissions. | |||
In summary, UniProt, NCBI, UCSC, and Ensembl take the atypical primate-specific splice variant as the canonical form of the gene. This gene model cannot however serve as ancestral. | |||
The 5' end has its oddities as well. It seems that in gallinaceous birds, the initial methionine could be further upstream by 22 residues. However the good agreement of amino acids could be an artefact of good conservation of good coding promoter or initiation sequence. There is no support for this extension in finch, lizard, frog or platypus or in the chicken transcript NM_001130743. The experimental transcript here, which maps accurately into the chicken genome, shows an upstream exon separated from the first coding exon by a 9181 bp intron with 14 bp of bp preceding the iMet. Thus the gene model assumed for quail is likely in error. A smaller extension of 3 residues is possible in all birds plus platypus. This extension has been adapted in the reference sequences below. | |||
Note that the new turkey genome is now available for [http://birdbase.net/cgi-bin/turkeyBlat Blat searching]. This provides an important addition to neuropsin comparative genomics. Turkey contains all four neuropsin paralogs as expected. | |||
== Novel neuropsins in amphioxus and sea urchin == | == Novel neuropsins in amphioxus and sea urchin == | ||
The genome of Branchiostoma (amphioxus, lancelet) contains two distinct neuropsins about 75% identical to each other and 42% to human. These cluster unambiguously with vertebrate | The genome of Branchiostoma (amphioxus, lancelet) contains two distinct neuropsins about 75% identical to each other and 42% to human. These cluster unambiguously with vertebrate neuropsins and share critical conserved residues. An extra intron distinguishes them from the vertebrate neuropsin pattern. Recall Branchiostoma species has three rather diverged (and well-studied) peropsins but no evident Rgr opsin. These raises the question whether neuropsin and peropsin developed substantial photoreceptor roles in this species as an alternative to the ciliary imaging opsin pathway seen already at lamprey divergence. Sea urchins, but not acornworm Saccoglossus, contain a single neuropsin that is quite diverged. | ||
These neuropsins are newly reported here, meaning they were not localized in recent in situ hybridization studies. That's especially unfortunate in view of the antecedent role the Branchiostoma ancestral node plays in the evolution of chordate eye and the complexities of photoreceptor tissues in the extant species. | These neuropsins are newly reported here, meaning they were not localized in recent in situ hybridization studies. That's especially unfortunate in view of the antecedent role the Branchiostoma ancestral node plays in the evolution of chordate eye and the complexities of photoreceptor tissues in the extant species. | ||
| Line 148: | Line 168: | ||
</pre> | </pre> | ||
== | == Neuropsin (NEUR1 compared to newropsin (NEUR2) == | ||
Newropsins are a new opsin gene family -- first reported here -- most closely related to neuropsins (42% percent identity) and next to melanopsins and peropsins. Like so many opsin families, they persist from chondrichthyes to archeosaurs but vanish without a trace in platypus, marsupials, and placentals. (The syntenic order B4GALT6 NEUR2 KIAA1012 remains conserved in mammals but no NEUR2 debris remains.) Newropsins retain many key attributes of GPCR signaling proteins and indeed opsins such as the seven transmembrane arrangement, Schiff base lysine, counterion tyrosine, amino terminal glycosylation site, and disulfide but have a very odd replacement of the G-protein binding site DRY with (invariantly conserved) VCC. | |||
This motif must be an ancient derived feature that followed the gene duplication event with neuropsin since the much older DRY could not plausibly have re-evolved in neuropsin from VCC. Newropsins very likely link covalently via their orthologous Schiff base lysine with a retinal and interact with light according to some action spectrum. The VCC motif has been conserved over billions of years of branch length so cannot reflect simple loss of functionality; however its signaling capabilities if any are unclear. | |||
It seems feasible that non-signaling opsins have become photoisomerases. Their substrate, while evidently involving an aldehyde capable of forming a Schiff base with the conserved lysine and likely interconverting cis/trans double bonds, need not be part of a ciliary opsin replenishment cycle. Other metabolic derivatives of beta-carotenes and lincopenes such as retinoic acid intermediates might be the substrates. | |||
It must be recalled too that quite a diversity of photoreceptive retinoids are used in non-mammalian species, for example 9-cis isorhodopsin, porphyropsin (or 3-dehydroretinal vitamin A2) in freshwater fishes and some frogs, 3-dehydroretinal in freshwater crayfish, all-trans-5,6-dihydroretinal in cottoid fish in Lake Baikal and so forth. These are [http://www.cell.com/biophysj/fulltext/S0006-3495(99)76953-5 spectrally influenced] by the surrounding opsin. Here too the negatively charged counterion, Glu113 in bovine rhodopsin, an alternative glutamate in melanopsins, or a special bound chloride ion. The counterparts of these are not known in neuropsins. | |||
Below, conserved residues are shown for NEUR2 relative to human neuropsin (which represents that family accurately). Newropsin orthologs are rather rapidly diverging, especially in teleost fish. Transmembrane domains on newropsins were assigned by homology to neuropsin (taken from SwissProt ab initio annotation consistent with experimentally determined bovine rod rhodopsin). Newropsin is relatively truncated amino terminally but very extended in a highly variable manner carboxy terminally. That extension would lie in the cytoplasm and possibly be removed endoproteolytically. The early glycosylation is present in all species but appears shifted distally by four residues in tetrapods relative to fish. | |||
According to transcript annotations, newropsin is expressed in zebrafish anterior segment (minus lens), fish brain and testes (Pimephales and Oncorhynchus), embryo, oviduct and fat body (Xenopus). These, while familiar sites from other opsins, provide only meagre constraints on possible newropsin functionality and association with photoreceptive tissues. | |||
Newropsin is further evidence that the neuropsin/peropsin/rgropsin group played a much | The intronation pattern is not a perfect match to neuropsin as might be expected. Some of the difference is lineage-specific, such as a gain in zebrafish, but other differences may be much older. Unless homologs have been retained recognizably in earlier diverging species, it won't be feasible to date the original gene duplication. No NEUR2 could be located in [http://genome.wustl.edu/tools/blast/index.cgi?gsc_link_id=112 lamprey], tunicate, or [http://genome.jgi-psf.org/Amphioxus lancelet.] | ||
Newropsin is further evidence that the neuropsin/peropsin/rgropsin group played a much greater role in ancestral vertebrate photoreception (which persisted into contemporary species), roles which were lost in stem mammals. That is quite similar to the ciliary opsin story. Overall mammals have retained less than half (7 of 17) of the vertebrate opsin repertoire. Such widespread gene loss is fully consistent with an old inference of a nocturnal era during which no selective pressure existed to maintain these photoreceptors. | |||
[[Image:Opsin_NEWR.png]] | [[Image:Opsin_NEWR.png]] | ||
| Line 167: | Line 190: | ||
<br clear="all" /> | <br clear="all" /> | ||
<pre> | <pre> | ||
position ...................................................................................................1.........1.........1.........1.........1.........1.........1.........1........1 | position ...................................................................................................1.........1.........1.........1.........1.........1.........1.........1........1. | ||
position .........1.........2.........3.........4.........5.........6.........7.........8.........9.........0.........1.........2.........3.........4.........5.........6.........7........ | position .........1.........2.........3.........4.........5.........6.........7.........8.........9.........0.........1.........2.........3.........4.........5.........6.........7........8. | ||
position | position 123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890 | ||
excMemCy | excMemCy eeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMcccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeeeee | ||
keyResid ...GLC.........glc.glc.................................................................................diS..cIon.................DRY?.............................................. | keyResid ....GLC.........glc.glc.................................................................................diS..cIon.................DRY?.............................................. | ||
NEUR2_galG MDPSFANST-FQSKITEAADIVVGTCYMVFGICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHRWLYGKHICLFYAFCGLFFGICSLSTLTLLSVVCCLKICFPAYGNRFRRKHGQILIACAWTYAAIFACSPLAHWGEYGEEPY | |||
NEUR2_anoC MESYFANTT-FHSKITEAADVIVGVFYIVFGICSFCGNSILLYVSYKKKNLLKPAEYFMINLAISDLGMTLTLYPLAVTSSLAHRWLFGQQVCLFYAFCGVFFGVCSLTTLTLLSIVCCLKICFPVYGNRFRPGHGWILIACAWVYAAIFAFSPLAHWGEYGAEPY | |||
NEUR2_xenT MGNKSDASA-FYSSISETDDIVLGVLYSVFGLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHRWLFDKVTCNYYAFCGMLFGLCSLTNLTVLSSVCCLKVCYPAYGNKFSTAHSRILLLGIWAYAGLFATAPLADWGKYGPEPY | |||
NEUR2_danR MGNVSKTAL-FMSTISRQHDILMGSLYSVFFVLSLLGNGMLLFVAYRKRSSLKPAEFFVVNLSVSDLGMTLSLFPLAIPSALAHRWLFGEITCLCYAVCGVLFGLCSLTNLTALSSVCCLKVCFPNYGNKFSSSHACVMVIGVWCYASVFAVGPLVHWGSFGPEPY | |||
NEUR2_pimP MGNVSETAL-FVSTISRQHDILMGSLYSVFCVLSLLGNGMLLFVAYRKRSSLKPAEFFVINLSVSDLGMTLSLFPLAIPSALAHRWLFGEVVCLCYAVCGVLFGLCSLTNLTALSSVCCLKVCCPNYGNKFSSNHACVMVIGVWCYASVFAVGPLIRWGSFAPEPY | |||
NEUR2_tetN MGNASDTSDAFNSKISKEHDFLIGSIYSVFCVLSLMGNCILLLVAHHKRSTLKPAEFFIVNLSISDLGMTLTLFPLAIPSSFSHRWLFGEIACQLYATCGVLFGLCSLTNLTVLSSVCCLKVCLPNLGSKFSSSHARLLVAGVWGYASVFAVGPLVQWGHYGPEPY | |||
NEUR2_takR MGNASEASDIFLSKISKEHDILIGSIYSVFGLLSLAGNCILLLVAYHKRSMLKPAEFFIINLSISDLGMTLTLFPLAIPSSFSHRWLFGEITCQLYAMCGVLFGLCSLTNLTALSLVCCLKVCFPNHGSRFSSSHARLLVVGVWCYASVFAVGPLVQWGHYGPEPY | |||
NEUR2_gasA MGNASDTSAVFASTISKERDILMGSLYSVFGVLSLVGNCILLLVAYHKRSTLKPAEFFIINLSISDLGMTLSLFPLAIPSAFKHRWLFGELTCQLYAMCGVLFGLCSLTNLTALSFVCCLKVCFPNHGNRFSSSHARLLVVAVWGYASVFAVGPLARWGRYSPEPY | |||
NEUR2_oryL MGNVSDTSSLFASSISREHDILMGSLYSVFGLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHRWLFGEITCQLYAMCGVLFGLSSLTNLTALSLVCCLKVCFPNHGNKFSFSHARLLVAGVWCYASVFAVGPLARWGRYSAEPY | |||
NEUR2_calM GILSLVGNSVLLFVAYRKRQILKPAEYFVANLAVSDISMTVTLLPLAISSNFSHRWLFVSKPCMYYGFCSMLFGICSLTNLTVLSTVCCMKVCFPAYMSVVMIV-MFLLAWSPYSIVCLWASFGNPKLIPPAMAII | |||
NEUR1_homSa MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGISVVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPF | |||
NEUR1_canFa MALNHTARPQDERLPHYLREGDPFASKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGISVVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPF | |||
NEUR1_musMu MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGISVVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPF | |||
NEUR1_loxAf MTLNHTAPPQDDRLPQYLQDGDPFTSKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSCRRKKKLRPAEIMTINLAVCDLGISVVGKPFVIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPF | |||
NEUR1_monDo MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTIIGVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGISVVGKPFTIISCFSHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSYGTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPF | |||
NEUR1_ornAn MTNYSAPQLGDYLPHYLREGDPFVSKLSWEADLVAGVYLVIIGVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGISVVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSYGTWLKRHHAYICLAIIWAYASFWATMPLVGLGNYAPEPF | |||
NEUR1_calMi MTAFDNSTALYSGYWLHDSLHGDPFVSKLSWEADIISACYLIVTGLLSTLGNGYVIYLSITQKRKLKPPEILITNLAISDFGMSVGGQPFLIISCFSHRWIFGWVGCRWHGWAGFFFGCGSLITMTVVSLDRYLKICHLQYGSWLQRRHVFMSLAFIWFYAAFWATMPLVGWGNYAPEPF | |||
NEUR1_galGa MASDCNSSSQEEYLPHYMQQEDPFASKLSREADIIAGFYLTVIGILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGISVVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAYGTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPF | |||
NEUR1_xenTr MAGNSSYREESGYIPHYERDSDPFASKLSREADIFAGVYLMAIGILSTLGNGYVIYMACSRKKKLRPAEIMTINLAVCDLGISVTGKPFAIVSCFSHRWVFGWNACRWYGWAGFFFGCGSLITLTVVSLDRYLKICHLRYGTWLKRRHAFIALAVIWAYATLWATLPLVGVGNYAPEPF | |||
NEUR1_danRe MENETSISSGYIPHYLLRGDPFASKLSKEADIVAAFYILVIGILSATGNGYVMYMTFKRKTKLKPPEIMTLNLAIFDFGISVSGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSFDRYLKICHLRYGTWLKRHHAFLSVVFIWAYAAFWATMPVVGWGNYAPEPF | |||
NEUR1_takRu MENDTSIPSGYVPHYLLRGDPFASKLSKEADIVAAFYILVIGVLSATGNGYVIYQTIKRKTKLKPPEFMTLNLAVFDFGISVTGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSLDRYLKICHLRYGTWFKRHHAFLCLVFTWLYAAFWATMPVVGWGNYAPEPF | |||
NEUR1_tetNi MENETWTHSSYVPHYLLRGDPFASRLSKEADIVAALYICIIGLMSATGNGYVLYMTFKRKTKLKPPELMTLNLAIFDFGISVTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRYGAWLKRHHAFLCLASVWAYAAFWATMPLVGWGSYAPEPF | |||
NEUR1_gasAc MDNETRSHPSYVPHYLLRGDPFASRLSKEADIVAAFYIFIIGVMSATGNGYVLYMTFKRKTKLKPPELMTVNLAIFDFGISVTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRYGTWLKRHHAFVCLALVWAYAAFWATMPLVGWGSYAPEPF | |||
NEUR1_oryLa MENETWTHPSYIPHYLLRGDPFASRLSKEADIIAAFYICIIGIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGISVTGKPFFVVSSFAHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRYGTWLKRQHAFLCLVFVWMYAAFWATMPLVGWGNYAPEPF | |||
NEUR1_pimPr MENTSWPHSSYVPHYLLRGDPFASRLSKEADIVAAFYILIIGIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGISVTGKPFFVVSSFSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRYGTWLKRQHIFLCLVFVWIYAAFWATMPLVGWGSYAPEPF | |||
NEUR1_anoCa MEQGQNISSQDDNQQEEDPFASKLSVEADIVAGVYLLVIGILSTLGNGYVIYMSTQRKKKLKPAEIMTVNLAVCDLGISVVGKPFSIIAFFSHRWIFGWSGCRWYGWAGFFFGIGSLITMTAVSLDRYFKICHLSYGTWLKRHHVFICLGIIWSYAAFWATIPFAGFGNYAPEPF | |||
position 1.........1.........2.........2.........2.........2.........2.........2.........2.........2.........2.........2.........3........3..........3.........3.........3.........3.....3 | position 1.........1.........2.........2.........2.........2.........2.........2.........2.........2.........2.........2.........3........3..........3.........3.........3.........3.....3 | ||
| Line 204: | Line 227: | ||
excMemCy eeeeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMcccccccccccccccccccccccccccccccccccccccccccccc* | excMemCy eeeeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMcccccccccccccccccccccccccccccccccccccccccccccc* | ||
keyResid .diS................................................................................................................K............................................................ | keyResid .diS................................................................................................................K............................................................ | ||
NEUR2_galG GTACCIDWQSTNVDVMSMSYTVVLFVLCFILPCGVIVTSYSLILVTVKESRKAVEQHVSGPTRINNVQTITAKLSIAVCIGFFAAWSPYAIIAMWAAFGSIDKIPPLAFAIPAVFAKSSTLYNPIIHLLLKPNFRSNIAKDFTVIQQLCVR---CCFCVKELQ--TYRSTFNTGLRTFKG | |||
NEUR2_anoC GTACCIDWRISNMKKTAMSYTTALFVFCYIIPCGIIITSYTLILITVKDSRKAVEQHALGPTRMSSVHTITAKLSIAVCIGFFVAWSPYAIIAMWAAFGSIDMIPPLAFAVPAVFAKSSTLYNPAMYLFLKPNFRSTIAKDLTVLHRLCLK---SCFCPRGMQNCSYRSALEAPLKSFKG | |||
NEUR2_xenT GTACCLDWEASYRERKALSYTISLFVFCYLIPSSLIFISYTLIFVTVKGARRAVQQHLSPQAKGSSIHSLIIKLSIAVCIGFLIAWTPYAIVAMMAAFGDPTKIPSLVFALAAAFAKSSTIYNPVVYLLLKPNFLNVVTKDLTLFQTMCAV---VCGWCR-----TPAVKTPCPHKDLKT | |||
NEUR2_danR GTACCINWYTPSHDALAMSYIISLFIFCYVVPCTIIILSYTFILVTVRGSQQAVQQHVSPQTKVTNAHALIVKLSVAVCIGFLTAWSPYAIVAMWAAFSANEQVPPTAFALAAIMAKSSTIYNPMVYLLFKPNFRKSLSQDTQMFRHRICLSHSKASPSPGMKDQERQSSQQCNNKDGSI | |||
NEUR2_pimP GTACCINWYIPSHDALAMSYIISLFIFCYVVPCTIIILSYTFILLRVRGSRQAVQKHVSPKTKETNAHTLIVKLSVAVCIGFVTAWSPYAVVAMWAAFSANEPVPPTAFALAAILAKSSTIYNPMVYLLFKPNFRKILSQDTQNIRHRMCVSHSKASPTPEIK---AQSSQQC--KDATI | |||
NEUR2_tetN GTACCINWQAPNHELSSLSYIVCLFLFCYVLPCAIIILSYTCILMTVRGSRQAIQQHVSPQTKTANAHALIVKLSVAVCIGFLGAWSPYAVVAMWASFGDATWVPPDAFAIAAILAKSSTIYNPLVYLLCKPNFRECLYKDTSTLRQRIY----RGSPLSGPRDRSGGVTQR--HKDLSV | |||
NEUR2_takR GTACCIDWRAPNHELSSLSYIVCLFFFCYVLPCATIILSYTCILMTVRGSRQAIQQHVSPQTKTANAHSLIVKLSVAVCIGFLGAWSPYAIVAMWAAFGDATWVPPDAFAIAAILAKSSTIYNPVVYLLCKPNFRECLYKDTSTLRQRIY----RGSPQSEPRERFGGTSQR--HKDLSI | |||
NEUR2_gasA GTACCIDWHAPNHELAALSYIVCLFVFCYALPCATIFLSYTFILLTVRGSRQAVQQHVSPQTKTTNTHALIVKLSVAVCIGFLGAWTPYAVVAIWAAFGDATLVPPDAFALAAMFAKSSTIYNPVVYLLCKPNFRACLYRDTTLLRQRIY----RGSPRSEPKAHFGSTSQR--NKDMSV | |||
NEUR2_calM APLFAKSSTFYNPCIYVISYTMTVIAVNFVVPLSVMFFCYYNV | |||
NEUR2_oryL GTACCIDWHAPNHELWALSYILCLFIFCYALPCTIIFLSYAFILLTVRGSRQAVQQHVSPQTKTTNAHTLIVKLSVAVCIGFLGAWTPYAVIAMWAAFGDATQVPPTAFALAAVFAKSSTIYNPMVYLLCKPNFRECLCRDTSLLRHMIY----RGSP--QPQERFGSDSRR--NKDITA | |||
NEUR1_homSa GTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKA-TKKKSLEGFRLHTVT-TVRKSSAVLEIHEEV | |||
NEUR1_calJa GTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKA-TKKKSLEDFRLHTVT-TVRKSSAVLEIHEEV | |||
NEUR1_canFa GTSCTLDWWLAQASLGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGRLKA-TKKKSLEDFRLNTVT-TVRKSSAVLEIHQEV | |||
NEUR1_musMu GTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQAGGLRG-TKKKSLEDFRLHTVT-TVRKSSAVLEIHQEV | |||
NEUR1_loxAf GTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHMLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLRA-TKKKSLEGFRLHTVT-TVKKSSAVLEVHQEV | |||
NEUR1_monDo GTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPVQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCQSGGQKA-AKKESLRTYRLHTVT-TVRRSSAVLEIHQEV | |||
NEUR1_ornAn GTSCTLDWWLAQASVAGQAFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQNSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPIQFSVVPTLLAKSAAMYNPIIYQVIDCRISCCRLGGPKT-GKKESLKNSRSHSMS-TIRKPSAVSGPHQEV | |||
NEUR1_calMi GTSCTLDWWLARVSVSGLIFVLTILFFCLLLPIIIIVFSYIKIIAKVKSSAKEVAHFDSRIQNHHSLEMNLTK | |||
NEUR1_galGa GTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGPKTLQKKSSLKESRMYTIS-SHRDSAALSGTQLEV | |||
NEUR1_xenTr GTTCTLDWWLAQASVKGQIFVLSMLFFCLLFPTMVIVFSYAKIIAKVKSSAKEVAHFDTRNQNNHTLEIKLTKVAMLICAGFLIAWFPYAVVSVWSAFGQPDSIPIELSVVPTMMAKSASMYNPIIYQVIDCKPACCKK------DKSLQNTTSRVYTIS-TFRKSTTSAR | |||
NEUR1_danRe GTSCTLDWWLTQASVSGQSFVMCMLFFCLIFPTVIIVFSYVMIIFKVKSSAKEVSHFDTRNKNNHSLEMKLTKVAMLICAGFLIAWIPYAVVSVMSAFGEPDSVPIPVSVVPTLLAKSSAMYNPIIYQVIDCKKKCVKSCCFQAWRKKKPSKTSRFYTISGSIKQR-PGDEASIEI | |||
NEUR1_takRu GTSCTLDWWLAQASVSGQSFVMCMLIFCLVLPTGVIVFSYVMIILQVKSSAQEVSHFDTQNKNKHHLEMKLTKVAMLICAGFLIAWIPYAVVSVVSAFGDPDSVPISISVVPTLLAKSSAMYNPIIYQVIDCKKNCAKLSCFQAWSKRKHYKTSRFYSISASMKKR-PANEVPTEI | |||
NEUR1_tetNi GTSCTLDWWLAQASVSGQSFVMAILFFCLILPTGIIVFSYVMIIFKVKSSAKEISHFDARIRNSHDLEIKLTKVAMLICAGFLIAWIPYAVVSVISAFGEPDSVPIPVSVIPTLLAKSSAMYNPIIYQVVDVKTSCTNFSCCKALKERIHFRKSRLYTISGSLRDPLPPKEAHIEM | |||
NEUR1_gasAc GTACTLDWWLAQASVSGQSFVMAILFFCLVLPTGIIVFSYIMIIFKVKSSAKEISHFDARIKNSHSLEIKLTKVAMLICAGFLIAWIPYAVVSVVSAFGEPDSVPIPVSVIPTLLAKSSAMYNPIIYQVADLKTSCTSSSCCKALKERVLFRKARLYTISGSLRDTLPPKEAHIEM | |||
NEUR1_oryLa GTSCTLDWWLAQASVSGQSFVVAILFFCLVLPAGIIVFSYVMIIFKVKSSAKEISNFDARIKNSHNLEIKLTKVAMLICAGFLIAWIPYAVVSVVSAFGEPDSVPISVSVIPTLLAKSSAMYNPIIYQVLDLKNSCMKSSCFKGLKKPRHFRKSRFYTISGSVKDNTTAKEAQIEM | |||
NEUR1_pimPr GTSCTLDWWLAQASVSGQSFVMSILFFCLVLPAGIIVFSYVMIICKVKSSSKEVSSFDARIKNSHTLEIKLTKVAMLICAGFLIAWIPYAVVSVVSAFGEPDSIPIPVSVIPTLLAKSSAMYNPIIYQLVDLKNSC-STCCAKVIRKRTHFRNSRFYTISGSLKDTAPAKEAHIEI | |||
NEUR1_anoCa GTSCTLDWWLAQGSVAGQAFILNILFFCLVLPTAVIMFCYVKIIAKVQSSTKEVAHYDTRIQNQHVLEMKLTKVAMLICAGFMFAWIPYAVVSVWSAFGRPDSVPIKVSVIPTLLAKSAAMYNPVIYQVIDCKSACCRPGNLQPLQKKNSR | |||
</pre> | </pre> | ||
== Curated | == Neuropsin (NEUR1 compared to newtopsin (NEUR3) == | ||
A third paralogous family NEUR3 was [http://www.ncbi.nlm.nih.gov/pubmed/18570255 reported in July 2008] and characterized by syntenic relations and expression in chicken. However in chicken, NEUR1 actually is most abundant of the three paralogs in developing and early post-hatch neural retina, notably in differentiating ganglion cells and amacrine cells. | |||
Recoverable sequences range from lamprey to shark to fish (where a further lineage-specific tandem duplication has occurred) to frog to sauropsids, with the gene again lost in all mammals including platypus. This locus exhibits an ancestral fusion of exon 2-3 relative to neuropsin and newropsin. The tandem duplication in ray-finned fish could easily be mistaken for a whole genome duplication effect -- however there is no sign of such an event for any of the 3 paralogs in any of the 5 fish with assembled genomes. | |||
The NEUR3 group could possibly signal through heteromeric G protein. The DRY motif is a bit unusual, consisting of V/I R F/Y whereas the YNPxIY x aliphatic is fairly conventional. The Schiff base K is preserved in both NEUR3a and NEUR3b sequences. The counterion glutamate or chloride ion has not been determined. | |||
The other oddity is NEUR1 and NEUR3 are on the same chromosome for all species examinable. They are separated by a million or so bp as well as other coding genes. This possibly represents an old tandem duplication that experienced subsequent rearrangements. On the whole, synteny has been quite well preserved for all NEUR genes: | |||
NEUR3_galGal +CRISP +RHAG ..... +MUT -NEUR3 ..... +CDC5L -SUPT3H +RUNX2 | |||
NEUR3_anoCar -CRISP2 +RHAG ..... +Mut -NEUR3 ..... +CDC5L -SUPT3H +RUNX2 | |||
NEUR3_xenTro -CRISP2 -RHAG -PPHLN +MUT -NEUR3 ..... +CDC5L ..... ..... | |||
NEUR3a_danRer +XRN2 -TSTA3 +MGST3 +MUT -NEUR3a -NEUR3b -NAPB +DNMT3A ..... | |||
NEUR3a_tetNig ..... ..... ..... +MUT -NEUR3a -NEUR3b ..... ..... +RUNX2 | |||
NEUR1_galGal ..... +TNFRSF2 +CD2AP +GPR111 +NEUR1 ..... +MRPL19 ..... ..... | |||
NEUR1_anoCar -GPR111 -TNFRSF2 +CD2AP ..... +NEUR1 -SPATS1 +MRPL19 ..... +ITSN2 | |||
NEUR1_xenTro ..... +TNFRSF21 +CD2AP +GPR111 +NEUR1 -PTCHD1 +MRPL19 ..... +ITSN2 | |||
NEUR1_danRer +GPR111 -TNFRSF21 +CD2AP ..... +NEUR1 +CNIH2 -LBR -ENAH +ITSN2 | |||
NEUR2_galGal -DSC1 +DSG2 +TTR -B4GALT6 -NEUR2 -K1012 +RNF138 ..... ..... | |||
NEUR2_anoCar -DSC1 +DSG2 +TTR -B4GALT6 -NEUR2 -K1012 +RNF138 ..... ..... | |||
[[Image:NeurAll.jpg]] | |||
A proposed revised terminology for this family follows. Note NEUR2 and NEUR3 will never receive official HGCN nomenclature because (like thousands of amniote ancestral genes) they are absent in human and mouse. Here lower case a and b are used in the case of lineage-specific duplications, with a reserved for the copy with higher blastp score to human (or, if absent, nearest species). | |||
Gene Protein HGCN Synonyms Lineage-specific duplicate DRY YNPxxY K Accessions | |||
NEUR1 neuropsin OPN5 cOpn5m NEUR1a/b cephalochordate DRY YNPIIY K NM_181744 NM_001130743 | |||
NEUR2 newropsin ---- cOpn5L1 VCC YNPxIY K XM_419178 | |||
NEUR3 newtopsin ---- cOpn5L2 NEUR3a/b actinopterygii vRf YNPxIY K XM_420056 | |||
== NEUR4: a fourth neuropsin from lamprey to platypus == | |||
Yet another new opsin in this group! These genes were first described here on 29 Jan 2009 (note GenBank had frog gene correctly predicted but chicken gene chimerized in a misassembly). Like many opsins, NEUR4 orthologs range throughout the vertebrates with the exception of theran mammals. Platypus is thus again distinguished by its retention of this ancient gene, whereas it is long gone from marsupials and placentals. This pattern of retention is consistent with platypus being more bird-like than mammal and supports the Wall 'dark era' apparently experienced by mammals during which Monotremata were somehow less dramatically affected. | |||
This opsin is unusual for its TRY at the DRY motif, though its Schiff base lysine region is standard (KSASFYNPIIYFGMNSKFR). Its best match within opsins is to NEUR1; outside of neuropsins to peropsins, melanopsins and various ancestral ciliary opsins (it has no special affinities to RGR). Transcripts are abundant in frog and fish and the former demonstrate expression in adult eye (ES678087). This opsin is clearly capable of signaling through some heterotrimeric Galpha protein but its function is unknown. | |||
Zebrafish and frog have an additional neuropsin, NEUR4b, discovered here in Oct 2010. It shares some synteny with NEUR4a indicating a block duplication. No counterpart can be located in other fish genomes. These seemed to have retained the extra copy from the ancestral whole genome duplication but evidently lost NEUR4b or never had it. It is possible that zebrafish and frog NEUR4b represent separate duplication events as they do not cluster dramatically together to the exclusion of the more numerous NEUR4a. The rate of evolution cannot be currently determined without further NEUR4b sequences and its function is equally mysterious as that of NEUR4a. | |||
NEUR4 shares some early intron positions and phases with NEUR1 but otherwise the pattern differs significantly, suggesting rather ancient divergence after origination by segmental duplication (not as processed pseudogene subsequently re-intronated). This is consistent with a very low percent identity (38%) to NEUR1, considering that a 'floor' of about 25% identity relates any pair of GPCR (eg NEUR4 is 23% identical to its best non-opsin match in human, tachykinin receptor). | |||
The phylogenetic distribution of the neuropsin gene swarm is puzzling. Only NEUR1 is found in non-vertebrate deuterostomes today yet family origins must be far more ancient as it is basal to ciliary opsins already found in cnidaria. Clearly neuropsins persisted for a very long period in pre-bilateran and bilateran ancestors prior to being lost (perhaps in a few stem events). Neuropsins may be important only in lineages where ciliary imaging opsins are important, yet all but NEUR1 have been lost in placentals despite persistence of the other three classes for several hundred million years. | |||
Alternatively, the neuropsin gene family expanded in the lamprey stem, diverged rapidly in primary sequence, and experienced multiple intron reorganization events. Because even one is rare, two independent events is rare-squared, and three is effectively impossible. Because the minimal number of events needed to synchronize intronation is five, this hypothesis appears unsustainable. Each orthology class has the same intronation pattern in all its members back to the earliest divergence for which sequence is available. | |||
Introns compared in four neuropsins 1^2 etc indicate relative intron gain. | |||
Exons 1 is omitted (same in all 4 genes). Irrelevant amino acids not shown (...) | |||
NEUR1 GIL...GIS 1^2 VVG...SHR WIF...YGW AGF...LAY GTW...SYA PEP...LTK ESR 2^1 MYT...LEV* | |||
NEUR3 AIL...GMA ISM...NHA WLG...YAL MGF...KSN SNK...YYG PEP...LTL SAD NSA...ARH* | |||
NEUR2 GIC...AMT LTL...SHR 2^1 WLY...YAF CGL...PAY GNR...EYG EEP...TAK RCC FCV...TDL* | |||
NEUR4 GWM...SIS VFG...NVF RDD...DGF 0^0 LTL...PER AHC...SYT 1^2 DRM...VTR KLK RFK...DRL* | |||
The most parsimonious explanation here -- assuming the gene tree (((NEUR1, NEUR3), NEUR2), NEUR4) -- is that NEUR3 represents the ancestral intronation pattern. This correlates well with its relative lack of retroposon events which may facilitate intron gain -- a single unshared CR1 LINE -- and the gene span is 1/3 to 1/8 of the others. NEUR1 has 7 LINE elements for comparison. NEUR1, NEUR2 and NEUR4 separately acquired new introns (different locations and phases) within the second exon. NEUR1 also acquired a new intron in the terminal exon (which curiously is alignable to the end only in NEUR3). Intron gain is both rare in vertebrate coding genes and considerably less common than intron loss, yet the events here are more economically placed on terminal gene tree leaves as intron gains. | |||
NEUR1 and NEUR3 both lie on the minus strand of chicken chromosome 3, separated by 1,310,977 bp and a half dozen coding genes. While not adjacent, this still suggests recent tandem duplication followed by local inversions, which is supported by the deeper match of the run-on terminal exon. This arrangement is readily tracked back to teleost fish, indicating the last expansion of neuropsin occurred prior to this, yet not by whole genome duplication. Perhaps the genes will be directly tandem in the upcoming revised Callorhinchus and Petromyzon genomes. | |||
In chicken but not lizard, NEUR4 also lies on this same chromosome, though greatly distant. This possibly indicates that it too arose from tandem duplication of NEUR1 though at a much earlier date. More likely its current position is coincidental because, while chickens have 39 chromosomes, only 6 are macro-sized. | |||
Indels are another type of potentially informative rare genomic event. Upon alignment of the reference sequence set below, five phylogenetically coherent indels emerge. Amphioxus and sea urchin genes can be used as outgroup to determine ancestral length (eg, to resolve each indel as deletion or insertion); this gives the same outcome as alignment to all ciliary opsins and indeed many GPCR. Each indel affects every member of its orthology class and none affects more than one class (other than in fish where the event affected the parent NEUR3 gene prior to its tandem duplication). Thus, as with introns, indels are predominantly ancient and do not provide internal clustering of neuropsins genes to guide the gene tree. Possibly the pre-lamprey indels were fixed shortly after gene duplication as the new paralogs re-functionalized. | |||
Indel #res type affected_genes timing location | |||
A 5 insert NEUR4 pre-lamprey 2 residues before first disulfide cysteine | |||
B 1 insert NEUR3 NEUR3a NEUR3b pre-teleost 2 residues after DRY motif, cytoplasmic loop C2 | |||
C 2 deletion NEUR4 pre-lamprey 1 residue before second disulfide cysteine | |||
D 2 deletion NEUR3b post-fish tandem 9 residues after second disulfide | |||
E 3 insert NEUR4 pre-lamprey 16 residues before Schiff lysine | |||
It appears very likely NEUR1 is the parent gene providing the core (unknown) function: | |||
* NEUR1 is best-blastp to amphioxus and sea urchin homologs whereas the others are undetectable outside vertebrates; | |||
* NEUR1 survived the longest in mammals; | |||
* NEUR1 is the best-Blastp for each of the others; | |||
* NEUR1 alone retains the ancestral DRY signaling motif. | |||
* NEUR1 has ancestral length (no indels relative to broader opsins or GPCR | |||
== Curated collection of neuropsins == | |||
These genes, almost all full length, have been extracted from various genome projects and cDNA data sets. In a few instances, accurate gene models had been previously computed by bioinformatic pipelines but these are so mixed with erroneous and mislabeled predictions as to be worthless for comparative genomics. The UCSC 44-species alignment is very helpful for rapid collection of individual exons taking care to note small insertions relative to human sequence are suppressed. The proteins below are parsed into exons whose coding phase is also shown. Because intronation is exceedingly conservative, genomically deduced introns can be reliable transfered to cDNA-only species. | |||
Despite the incomplete nature of many genome assemblies and lack of transcripts in specialized cell types, the absence of a given neuropsin in a given clade is rarely attributable to lack of data. For example, while sloth assembly alone might have only 2x mean coverage (and thus lack many coding genes), overall coverage of its clade (Atlantogenata: armadillo, sloth, elephant, mammoth, hyrax, and tenrec) approaches 30x (90 million 1 kbp traces). Here coding genes can be missing only for the compositionally oddest or most extreme chromosomal locations. | |||
For early diverging deuterostomes, the situation is somewhat different. Here only one species per divergence node is generally available and assemblies encountered extreme diploid heterozygosity and retroposon issues with little outside support from transcript programs (except for Ciona and sea urchin). The species chosen may be quite specialized within its clade and have experienced very extensive gene loss, much as Drosophila is wholly unrepresentative of the protostome genome. Lamprey, despite 19 million traces, has poor coverage with contigs rarely encompassing more than an exon or two. | |||
However gene divergence (to the point of unrecognizability) is not an issue because the divergence floor to GPCR is well within tblastn reporting capabilities. Even in short contig assemblies, individual exons can be identified and reliably assigned to opsin orthology class using the classifier, provided the exon is reasonably conserved. Even poorly conserved N- and C-terminal exons can be extended outward to an initial methionine or stop codon with some reliability. | |||
Consequently the phylogenetic 'end points' of a given gene are fairly certain even on the early-diverging side, though that remains somewhat muddled because lineage-specific loss is not at all uncommon in opsins or specialized species. | |||
=== NEUR1: 56 deuterostome neuropsins === | |||
<pre> | <pre> | ||
> | >NEUR1_homSap Homo sapiens (human) | ||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 244: | Line 359: | ||
1 LHTVTTVRKSSAVLEIHEEV* 0 | 1 LHTVTTVRKSSAVLEIHEEV* 0 | ||
> | >NEUR1_panTro Pan troglodytes (chimp) | ||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 252: | Line 367: | ||
1 LHTVTTVRKSSAVLEIHEEV* 0 | 1 LHTVTTVRKSSAVLEIHEEV* 0 | ||
> | >NEUR1_gorGor Gorilla gorilla (gorilla) | ||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 12 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 12 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | ||
| Line 259: | Line 374: | ||
1 LHTVTTVRKSSAVLEIHEEv* 0 | 1 LHTVTTVRKSSAVLEIHEEv* 0 | ||
> | >NEUR1_ponPyg Pongo pygmaeus (orang_sumatran) | ||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 267: | Line 382: | ||
1 LHTVTTVRKSSAVLEIHEEV* 0 | 1 LHTVTTVRKSSAVLEIHEEV* 0 | ||
> | >NEUR1_nomLeu Nomascus leucogenys (gibbon) | ||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 275: | Line 390: | ||
1 LHTVTSVRKSSAVLEIHEEv* 0 | 1 LHTVTSVRKSSAVLEIHEEv* 0 | ||
> | >NEUR1_macMul Macaca mulatta (rhesus) | ||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADIVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADIVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 283: | Line 398: | ||
1 LHTVTTVRKSSAVLEIHEEV* 0 | 1 LHTVTTVRKSSAVLEIHEEV* 0 | ||
> | >NEUR1_papHam Papio hamadryas (baboon) | ||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADIVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADIVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 291: | Line 406: | ||
1 LHTVTTVRKSSAVLEIHEEv* 0 | 1 LHTVTTVRKSSAVLEIHEEv* 0 | ||
> | >NEUR1_calJac Callithrix jacchus (marmoset) | ||
0 MALNHTSLPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTSLPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 299: | Line 414: | ||
1 LHTVTTVRKSSAVLEIHEEV* 0 | 1 LHTVTTVRKSSAVLEIHEEV* 0 | ||
> | >NEUR1_tarSyr Tarsius syrichta (tarsier) | ||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 307: | Line 422: | ||
1 LHTVTTVRKSSAVLEIHEEv* 0 | 1 LHTVTTVRKSSAVLEIHEEv* 0 | ||
> | >NEUR1_otoGar Otolemur garnettii (bushbaby) | ||
0 MALNHTALPQDELRPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDELRPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 | ||
| Line 315: | Line 430: | ||
1 LHTVTTVRKSSAVLEIHQEV* 0 | 1 LHTVTTVRKSSAVLEIHQEV* 0 | ||
> | >NEUR1_micMur Microcebus murinus (mouse_lemur) | ||
0 MALNHTVLPQDERLPHYLRDGDPFASKLSWEADLVAGFYLIII 1 | 0 MALNHTVLPQDERLPHYLRDGDPFASKLSWEADLVAGFYLIII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 323: | Line 438: | ||
1 LHTVTAVRKSSAVLEIHQEv* 0 | 1 LHTVTAVRKSSAVLEIHQEv* 0 | ||
> | >NEUR1_tupBel Tupaia belangeri (tree_shrew) | ||
0 MALNHTALPQDESLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDESLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 331: | Line 446: | ||
1 LHTVTTVRKSSAVLEIHQEV* 0 | 1 LHTVTTVRKSSAVLEIHQEV* 0 | ||
> | >NEUR1_musMus Mus musculus (mouse) | ||
0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 339: | Line 454: | ||
1 LHTVTTVRKSSAVLEIHQEV* 0 | 1 LHTVTTVRKSSAVLEIHQEV* 0 | ||
> | >NEUR1_ratNor Rattus norvegicus (rat) | ||
0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 347: | Line 462: | ||
1 LHTVTAVRKSSAVLEIHPEv* 0 | 1 LHTVTAVRKSSAVLEIHPEv* 0 | ||
> | >NEUR1_speTri Spermophilus tridecemlineatus (squirrel) | ||
0 MALNHTALPQDEHLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDEHLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 355: | Line 470: | ||
1 LHTVTAVRKSSAVVEIHQEv* 0 | 1 LHTVTAVRKSSAVVEIHQEv* 0 | ||
> | >NEUR1_dipOrd Dipodomys ordii (kangaroo_rat) | ||
0 MAFNHTAGTQGQGLPHYLPEEDPFTSKLSWEADIVAGFYLTII 1 | 0 MAFNHTAGTQGQGLPHYLPEEDPFTSKLSWEADIVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 363: | Line 478: | ||
1 * 0 | 1 * 0 | ||
> | >NEUR1_cavPor Cavia porcellus (guinea_pig) | ||
0 MALNHTAPPQNEHLPRYLQDEDPFVSKLSWEADLVAGFYLTII 1 | 0 MALNHTAPPQNEHLPRYLQDEDPFVSKLSWEADLVAGFYLTII 1 | ||
2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | 2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | ||
| Line 371: | Line 486: | ||
1 LHTVTTDRKSAVLEIHQEV* 0 | 1 LHTVTTDRKSAVLEIHQEV* 0 | ||
> | >NEUR1_oryCun Oryctolagus cuniculus (rabbit) | ||
0 MALNHTALPQDEHLPHYLREGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDEHLPHYLREGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 379: | Line 494: | ||
1 LHTVTTVRKSSAVLEIHQEv* 0 | 1 LHTVTTVRKSSAVLEIHQEv* 0 | ||
> | >NEUR1_ochPri Ochotona princeps (pika) | ||
0 MALNDTALPQDEHLPHYFRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNDTALPQDEHLPHYFRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 387: | Line 502: | ||
1 LHTVTTVRKSSAVLEIHQEv* 0 | 1 LHTVTTVRKSSAVLEIHQEv* 0 | ||
> | >NEUR1_canFam Canis familiaris (dog) | ||
0 MALNHTARPQDERLPHYLREGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTARPQDERLPHYLREGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | ||
| Line 395: | Line 510: | ||
1 LNTVTTVRKSSAVLEIhQEV* 0 | 1 LNTVTTVRKSSAVLEIhQEV* 0 | ||
> | >NEUR1_ailMel Ailuropoda melanoleuca (panda) XM_002919006 | ||
0 MALNHTAPPQEEHLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | |||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | |||
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 | |||
2 GVWLKRKHAYICLAFIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRVHSSHVLEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 | |||
1 LHTVTTVRKSSAVLEIHQEV* 0 | |||
>NEUR1_felCat Felis catus (cat) | |||
0 MALNHTAPPQDERLPHYLREGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTAPPQDERLPHYLREGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAE 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAE 1 | ||
| Line 403: | Line 526: | ||
1 LHTVTTVRKSSAVLEIHQEv* 0 | 1 LHTVTTVRKSSAVLEIHQEv* 0 | ||
> | >NEUR1_bosTau Bos taurus (cow) | ||
0 MALNHTAPPPDERRPPYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTAPPPDERRPPYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAICDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAICDLGIS 1 | ||
| Line 411: | Line 534: | ||
1 LHTVTTVRKSSAVLEVHQEv* 0 | 1 LHTVTTVRKSSAVLEVHQEv* 0 | ||
> | >NEUR1_turTru Tursiops truncatus (dolphin) | ||
0 1 | 0 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLKPAEIMTINLAICDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLKPAEIMTINLAICDLGIS 1 | ||
| Line 419: | Line 542: | ||
1 LHTVTTVRKSSAVLEIHQEv* 0 | 1 LHTVTTVRKSSAVLEIHQEv* 0 | ||
> | >NEUR1_susScr Sus scrofa (pig) | ||
0 MALNHTAPPPDERRPHYLREGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTAPPPDERRPHYLREGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAICDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAICDLGIS 1 | ||
| Line 427: | Line 550: | ||
1 LHTVTTVRKSSAVLEIRQEV* 0 | 1 LHTVTTVRKSSAVLEIRQEV* 0 | ||
> | >NEUR1_vicVic Vicugna vicugna (vicugna) | ||
0 MALNHTAPPPDERRPRHLRDGdPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTAPPPDERRPRHLRDGdPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTLGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTLGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 435: | Line 558: | ||
1 LHAVTTVRKSSAVLEIHQEV* 0 | 1 LHAVTTVRKSSAVLEIHQEV* 0 | ||
> | >NEUR1_equCab Equus caballus (horse) | ||
0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | ||
| Line 443: | Line 566: | ||
1 LHTVTTVRKSSAVLEIHQEV* 0 | 1 LHTVTTVRKSSAVLEIHQEV* 0 | ||
> | >NEUR1_myoLuc Myotis lucifugus (microbat) | ||
0 MALNHTALPQDEGLPHYLQDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDEGLPHYLQDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 | 2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 | ||
| Line 451: | Line 574: | ||
1 LHTVTTVRKSSAVLEIHQEv* 0 | 1 LHTVTTVRKSSAVLEIHQEv* 0 | ||
> | >NEUR1_pteVam Pteropus vampyrus (macrobat) | ||
0 MALNHTVLPQDEHLPHYVRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTVLPQDEHLPHYVRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | 2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 | ||
| Line 459: | Line 582: | ||
1 LHTITTVREASAVLEIHQEV* 0 | 1 LHTITTVREASAVLEIHQEV* 0 | ||
> | >NEUR1_sorAra Sorex araneus (shrew) | ||
0 MALNHTALPQDENLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDENLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 467: | Line 590: | ||
1 LHTVTTVRESSAVLEIHQEV* 0 | 1 LHTVTTVRESSAVLEIHQEV* 0 | ||
> | >NEUR1_eriEur Erinaceus europaeus (hedgehog) | ||
0 MSLNQTALPQDEGLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MSLNQTALPQDEGLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 475: | Line 598: | ||
1 * 0 | 1 * 0 | ||
> | >NEUR1_loxAfr Loxodonta africana (elephant) | ||
0 MTLNHTAPPQDDRLPQYLQDGDPFTSKLSWEADLVAGFYLTII 1 | 0 MTLNHTAPPQDDRLPQYLQDGDPFTSKLSWEADLVAGFYLTII 1 | ||
2 GILSTFGNGYVLYMSCRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSCRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 483: | Line 606: | ||
1 LHTVTTVKKSSAVLEVHQEv* 0 | 1 LHTVTTVKKSSAVLEVHQEv* 0 | ||
> | >NEUR1_proCap Procavia capensis (hyrax) | ||
0 MTLNHTVLPEDDRLSHYLRDGDPFTSKLSWEADLVAGFYLTVI 1 | 0 MTLNHTVLPEDDRLSHYLRDGDPFTSKLSWEADLVAGFYLTVI 1 | ||
2 GILSTCGNGYVLYMSYRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTCGNGYVLYMSYRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 491: | Line 614: | ||
1 LHTVTTVRKSSAVLEIHQEv* 0 | 1 LHTVTTVRKSSAVLEIHQEv* 0 | ||
> | >NEUR1_echTel Echinops telfairi (tenrec) | ||
0 MALNHTAPPQDNSLPHYLRDGDPFVSKLSWEADLGAGFYLIII 1 | 0 MALNHTAPPQDNSLPHYLRDGDPFVSKLSWEADLGAGFYLIII 1 | ||
2 GILSTFGNGYVLYMSYRRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSYRRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 499: | Line 622: | ||
1 LHTITTVRKSSAVLEIHQEV* 0 | 1 LHTITTVRKSSAVLEIHQEV* 0 | ||
> | >NEUR1_dasNov Dasypus novemcinctus (armadillo) | ||
0 MALNHTALPQDDRLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHTALPQDDRLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 gILSTFGNGYVLYMSSKRKKKLRPAEIMTINLAVCDLGIS 1 | 2 gILSTFGNGYVLYMSSKRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 507: | Line 630: | ||
1 LHTVTTVRESSAVLEVHQEV* 0 | 1 LHTVTTVRESSAVLEVHQEV* 0 | ||
> | >NEUR1_choHof Choloepus hoffmanni (sloth) | ||
0 MALNHTGLPQDDSLPHYFRDGDPFASKLSWEADLVAGFYLIII 1 | 0 MALNHTGLPQDDSLPHYFRDGDPFASKLSWEADLVAGFYLIII 1 | ||
2 GILSTFGNGYVLYMSSRRRKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTFGNGYVLYMSSRRRKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 515: | Line 638: | ||
1 LHTVTTVRKSSAVLEIHQEv* 0 | 1 LHTVTTVRKSSAVLEIHQEv* 0 | ||
> | >NEUR1_monDom Monodelphis domestica (opossum) not extendable N-terminally | ||
0 MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | 0 MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTII 1 | ||
2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | 2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | ||
| Line 523: | Line 646: | ||
1 LHTVTTVRRSSAVLEIHQEv* 0 | 1 LHTVTTVRRSSAVLEIHQEv* 0 | ||
> | >NEUR1_macEug Macropus eugenii (wallaby) | ||
0 1 | 0 1 | ||
2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | 2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | ||
| Line 531: | Line 654: | ||
1 RHTVSTIRKSSSVSETYQEV* 0 | 1 RHTVSTIRKSSSVSETYQEV* 0 | ||
> | >NEUR1_ornAna Ornithorhynchus anatinus (platypus) no further possible upstream extension of exon 1 | ||
0 | 0 MNSMTNYSAPQLGDYLPHYLREGDPFVSKLSWEADLVAGVYLVII 1 | ||
2 GVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 | 2 GVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 | ||
2 VVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 | 2 VVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 | ||
| Line 539: | Line 662: | ||
1 SHSMSTIRKPSAVSGPHQEV* 0 | 1 SHSMSTIRKPSAVSGPHQEV* 0 | ||
> | >NEUR1_galGal Gallus gallus (chicken) MGAIVCSVGFVCLFVFSDTELD possible upstream extension of exon 1 | ||
0 | 0 MSGMASDCNSSSQEEYLPHYMQQEDPFASKLSREADIIAGFYLTVI 1 | ||
2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | 2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | ||
2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 | 2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 | ||
| Line 547: | Line 670: | ||
1 MYTISSHRDSAALSGTQLEV* 0 | 1 MYTISSHRDSAALSGTQLEV* 0 | ||
> | >NEUR1_melGal Meleagris gallopavo (turkey) MGAVVYSLGFVCLFVFSDTELD possible upstream extension of exon 1 | ||
0 | 0 MSGMASDRNSSSQEEYLPHYVQQEDPFASKLSREADIIAGFYLTVI 1 | ||
2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | |||
2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 | |||
2 gTWLKRHHAFICLALIWTYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFILSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGLKALQKKSSLKESR 2 | |||
1 MYTISSHRDSAAPSETQLEV* 0 | |||
>NEUR1_cotJap Coturnix japonica (quail) AB547151 20679218 MGAVVCSVRSVCLFVFSDTELD possible upstream extension of exon 1 | |||
0 MSGMASDCNSSQEEYLPHHVQQEDPFASKLSREADIIAGFYLTVI 1 | |||
2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | |||
2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 | |||
2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 | |||
0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFSCCRSGGLKTLQKKSSLKDSR 2 | |||
1 MYTISSHRDSAALSETQLEV* 0 | |||
>NEUR1_taeGut Taeniopygia guttata (finch) not extendable N-terminally | |||
0 MSGMASEYNNSSQEEYIPHYLQEEDPFASKLSREADIIAGFYLTII 1 | |||
2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | 2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 | ||
2 VVGKPFSIISFFSHRWMFGWIGCCWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 | 2 VVGKPFSIISFFSHRWMFGWIGCCWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 | ||
2 GTWLKRHHAFICLAIIWAYAMFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQVFVLSILFFCLLLPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 | 2 GTWLKRHHAFICLAIIWAYAMFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQVFVLSILFFCLLLPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 | ||
0 | 0 VAMLICAGFLLAWIPYAVVSVWSAFGRPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIECRLACCRPGGCCRPGGLKAKSSLKKSR 2 | ||
1 | 1 TYTISAHRDSTAMNETQLEA* 0 | ||
> | >NEUR1_anoCar Anolis carolinensis (lizard) no N-terminal extension possible | ||
0 MEQGQNISSQDDNQQEEDPFASKLSVEADIVAGVYLLVI 1 | 0 MEQGQNISSQDDNQQEEDPFASKLSVEADIVAGVYLLVI 1 | ||
2 GILSTLGNGYVIYMSTQRKKKLKPAEIMTVNLAVCDLGIS 1 | 2 GILSTLGNGYVIYMSTQRKKKLKPAEIMTVNLAVCDLGIS 1 | ||
| Line 561: | Line 700: | ||
2 GTWLKRHHVFICLGIIWSYAAFWATIPFAGFGNYAPEPFGTSCTLDWWLAQGSVAGQAFILNILFFCLVLPTAVIMFCYVKIIAKVQSSTKEVAHYDTRIQNQHVLEMKLTK 0 | 2 GTWLKRHHVFICLGIIWSYAAFWATIPFAGFGNYAPEPFGTSCTLDWWLAQGSVAGQAFILNILFFCLVLPTAVIMFCYVKIIAKVQSSTKEVAHYDTRIQNQHVLEMKLTK 0 | ||
0 VAMLICAGFMFAWIPYAVVSVWSAFGRPDSVPIKVSVIPTLLAKSAAMYNPVIYQVIDCKSACCRPGNLQPLQKKNSR 2 | 0 VAMLICAGFMFAWIPYAVVSVWSAFGRPDSVPIKVSVIPTLLAKSAAMYNPVIYQVIDCKSACCRPGNLQPLQKKNSR 2 | ||
1 | 1 LYIIPTGKKSEVVQETQLDSV* 0 | ||
> | >NEUR1_xenTro Xenopus tropicalis (frog) no N-terminal extension possible | ||
0 MAGNSSYREESGYIPHYERDSDPFASKLSREADIFAGVYLMAI 1 | 0 MAGNSSYREESGYIPHYERDSDPFASKLSREADIFAGVYLMAI 1 | ||
2 GILSTLGNGYVIYMACSRKKKLRPAEIMTINLAVCDLGIS 1 | 2 GILSTLGNGYVIYMACSRKKKLRPAEIMTINLAVCDLGIS 1 | ||
| Line 571: | Line 710: | ||
1 VYTISTFRKSTTSAR* 0 | 1 VYTISTFRKSTTSAR* 0 | ||
> | >NEUR1_danRer Danio rerio (zebrafish) | ||
0 MENETSISSGYIPHYLLRGDPFASKLSKEADIVAAFYILVI 1 | 0 MENETSISSGYIPHYLLRGDPFASKLSKEADIVAAFYILVI 1 | ||
2 GILSATGNGYVMYMTFKRKTKLKPPEIMTLNLAIFDFGIS 1 | 2 GILSATGNGYVMYMTFKRKTKLKPPEIMTLNLAIFDFGIS 1 | ||
| Line 579: | Line 718: | ||
1 FYTISGSIKQRPGDEASIEI* 0 | 1 FYTISGSIKQRPGDEASIEI* 0 | ||
> | >NEUR1_takRub Takifugu rubripes (fugu) | ||
0 MENDTSIPSGYVPHYLLRGDPFASKLSKEADIVAAFYILVI 1 | 0 MENDTSIPSGYVPHYLLRGDPFASKLSKEADIVAAFYILVI 1 | ||
2 GVLSATGNGYVIYQTIKRKTKLKPPEFMTLNLAVFDFGIS 1 | 2 GVLSATGNGYVIYQTIKRKTKLKPPEFMTLNLAVFDFGIS 1 | ||
2 VTGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSLDRYLKICHLRY 1 | 2 VTGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSLDRYLKICHLRY 1 | ||
2 GTWFKRHHAFLCLVFTWLYAAFWATMPVVGWGNYAPEPFGTSCTLDWWLAQASVSGQSFVMCMLIFCLVLPTGVIVFSYVMIiLQVKSSAQEVSHFDTQNKNKHHLEMKLTK 0 | 2 GTWFKRHHAFLCLVFTWLYAAFWATMPVVGWGNYAPEPFGTSCTLDWWLAQASVSGQSFVMCMLIFCLVLPTGVIVFSYVMIiLQVKSSAQEVSHFDTQNKNKHHLEMKLTK 0 | ||
0 | 0 VAMLICAGFLIAWIPYAVVSVVSAFGDPDSVPISISVVPTLLAAKSSAMYNPIIYQVVDVKTSCTNFSCCKALKERIHFRKSR 2 | ||
1 FYSISASMKKRPANEVPTEI* 0 | 1 FYSISASMKKRPANEVPTEI* 0 | ||
> | >NEUR1_tetNig Tetraodon nigroviridis (pufferfish) | ||
0 MENETWTHSSYVPHYLLRGDPFASRLSKEADIVAALYICII 1 | 0 MENETWTHSSYVPHYLLRGDPFASRLSKEADIVAALYICII 1 | ||
2 gLMSATGNGYVLYMTFKRKTKLKPPELMTLNLAIFDFGIS 1 | 2 gLMSATGNGYVLYMTFKRKTKLKPPELMTLNLAIFDFGIS 1 | ||
2 VTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 | 2 VTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 | ||
2 GAWLKRHHAFLCLASVWAYAAFWATMPLVGWGSYAPEPFGTSCTLDWWLAQASVSGQSFVMAILFFCLILPTGIIVFSYVMIIFKVKSSAKEISHFDARIRNSHDLEIKLTK 0 | 2 GAWLKRHHAFLCLASVWAYAAFWATMPLVGWGSYAPEPFGTSCTLDWWLAQASVSGQSFVMAILFFCLILPTGIIVFSYVMIIFKVKSSAKEISHFDARIRNSHDLEIKLTK 0 | ||
0 | 0 VAMLICAGFLIAWIPYAVVSVISAFGEPDSVPIPVSVIPTLLAKSSAMYNPIIYQVADLKTSCTSSSCCKALKERVLFRKSR 2 | ||
1 | 1 YTISGSLRDTLPPKEAHIEM* 0 | ||
> | >NEUR1_gasAcu Gasterosteus aculeatus (stickleback) | ||
0 MDNETRSHPSYVPHYLLRGDPFASRLSKEADIVAAFYIFII 1 | 0 MDNETRSHPSYVPHYLLRGDPFASRLSKEADIVAAFYIFII 1 | ||
2 GVMSATGNGYVLYMTFKRKTKLKPPELMTVNLAIFDFGIS 1 | 2 GVMSATGNGYVLYMTFKRKTKLKPPELMTVNLAIFDFGIS 1 | ||
2 VTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 | 2 VTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 | ||
2 GTWLKRHHAFVCLALVWAYAAFWATMPLVGWGSYAPEPFGTACTLDWWLAQASVSGQSFVMAILFFCLVLPTGIIVFSYIMIIFKVKSSAKEISHFDARIKNSHSLEIKLTK 0 | 2 GTWLKRHHAFVCLALVWAYAAFWATMPLVGWGSYAPEPFGTACTLDWWLAQASVSGQSFVMAILFFCLVLPTGIIVFSYIMIIFKVKSSAKEISHFDARIKNSHSLEIKLTK 0 | ||
0 | 0 VAMLICAGFLIAWIPYAVVSVVSAFGEPDSVPIPVSVIPTLLAKSSAMYNPIIYQVLDLKNSCMKSSCFKGLKKPRHFRKSR 2 | ||
1 | 1 YTISGSLRDTLPPKEAHIEM* 0 | ||
> | >NEUR1_oryLat Oryzias latipes (medaka) | ||
0 MENETWTHPSYIPHYLLRGDPFASRLSKEADIIAAFYICII 1 | 0 MENETWTHPSYIPHYLLRGDPFASRLSKEADIIAAFYICII 1 | ||
2 gIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGIS 1 | 2 gIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGIS 1 | ||
| Line 609: | Line 748: | ||
2 GTWLKRQHAFLCLVFVWMYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLAQASVSGQSFVVAILFFCLVLPAGIIVFSYVMIIFKVKSSAKEISNFDARIKNSHNLEIKLTK 0 | 2 GTWLKRQHAFLCLVFVWMYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLAQASVSGQSFVVAILFFCLVLPAGIIVFSYVMIIFKVKSSAKEISNFDARIKNSHNLEIKLTK 0 | ||
0 VAMLICAGFLIAWIPYAVVSVVSAFGEPDSVPISVSVIPTLLAKSSAMYNPIIYQVLDLKNSCMKSSCFKGLKKPRHFRKSR 2 | 0 VAMLICAGFLIAWIPYAVVSVVSAFGEPDSVPISVSVIPTLLAKSSAMYNPIIYQVLDLKNSCMKSSCFKGLKKPRHFRKSR 2 | ||
1 | 1 YTISGSLKDTAPAKEAHIEI* 0 | ||
> | >NEUR1_pimPro Pimephales promelas (minnow) | ||
0 MENTSWPHSSYVPHYLLRGDPFASRLSKEADIVAAFYILII 1 | 0 MENTSWPHSSYVPHYLLRGDPFASRLSKEADIVAAFYILII 1 | ||
2 GIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGIS 1 | 2 GIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGIS 1 | ||
2 VTGKPFFVVSSFSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 | 2 VTGKPFFVVSSFSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 | ||
2 GTWLKRQHIFLCLVFVWIYAAFWATMPLVGWGSYAPEPFGTSCTLDWWLAQASVSGQSFVMSILFFCLVLPAGIIVFSYVMIICKVKSSSKEVSSFDARIKNSHTLEIKLTK 0 | 2 GTWLKRQHIFLCLVFVWIYAAFWATMPLVGWGSYAPEPFGTSCTLDWWLAQASVSGQSFVMSILFFCLVLPAGIIVFSYVMIICKVKSSSKEVSSFDARIKNSHTLEIKLTK 0 | ||
0 | 0 VAMLICAGFLIAWIPYAVVSVVSAFGEPDSIPIPVSVIPTLLAKSSAMYNPIIYQVIDCKKNCAKLSCFQAWSKRKHYKTSR 2 | ||
1 | 1 FYSISASMKKRPANEVPTEI* 0 | ||
> | >NEUR1_calMil Callorhinchus milii (elephantfish) | ||
0 MTAFDNSTALYSGYWLHDSLHGDPFVSKLSWEADIISACYLIVT 1 | 0 MTAFDNSTALYSGYWLHDSLHGDPFVSKLSWEADIISACYLIVT 1 | ||
2 GLLSTLGNGYVIYLSITQKRKLKPPEILITNLAISDFGMS 1 | 2 GLLSTLGNGYVIYLSITQKRKLKPPEILITNLAISDFGMS 1 | ||
| Line 627: | Line 766: | ||
1 * 0 | 1 * 0 | ||
> | >NEUR1_petMar Petromyzon marinus (lamprey) frag | ||
2 GTWVRRRHAFLCVLAVWLYAAFWATMPLLGWGSYAPEPFGTSCTLDWWLAQSSAAGRSFVLCMLLFCLLLPAAAILFAYARIVGAVRRSARDLAHFERRARGGGGGGGGGGVALELRITK 0 | |||
0 VAMMICAGFLLAWIPYAVVSVWSAFGAPDSVPVAVSMVPTMFAKSAAMYNPLIYQLLSRRGTGAHCCRCRKARGTLRRPR 2 | 0 VAMMICAGFLLAWIPYAVVSVWSAFGAPDSVPVAVSMVPTMFAKSAAMYNPLIYQLLSRRGTGAHCCRCRKARGTLRRPR 2 | ||
> | >NEUR1a_braFlo Branchiostoma floridae (amphioxus) FE548698 | ||
0 MATTPADRLDGLTPAGRGATTAETHADDFASKLSREADIVIGVYLILI 1 | 0 MATTPADRLDGLTPAGRGATTAETHADDFASKLSREADIVIGVYLILI 1 | ||
2 GTGAILGNGRVLWLSYRCRARLRPVEMFVVSLAAADVGLSLVGHPFSAASSLMGRWSFGSAGCTW 1 | 2 GTGAILGNGRVLWLSYRCRARLRPVEMFVVSLAAADVGLSLVGHPFSAASSLMGRWSFGSAGCTW 1 | ||
| Line 637: | Line 777: | ||
2 GWSQYHPEPYGLSCSVDWGGFSRGAGGSSFIICMLLFCTAVPVVVMVTSYAAIFALYRQAQKGVVLNLQVNATFGGKRQRTER | 2 GWSQYHPEPYGLSCSVDWGGFSRGAGGSSFIICMLLFCTAVPVVVMVTSYAAIFALYRQAQKGVVLNLQVNATFGGKRQRTER | ||
0 IALAVCGGFLLAWLPYAVVGLWASVAGVDAVPLALASAAPLFAKSNSLWNPIIYLGMNERFR 2 | 0 IALAVCGGFLLAWLPYAVVGLWASVAGVDAVPLALASAAPLFAKSNSLWNPIIYLGMNERFR 2 | ||
1 * 0 | |||
> | >NEUR2b_braFlo from traces and genome chrUn ++ 187375671 187384042 8372 nearly identical chrUn ++ 32271780 32281075 9296 | ||
0 MATTPGLPLDGLAPTGRGVTAADTLDDDFASKLSREADIVIGVYLLLI 1 | 0 MATTPGLPLDGLAPTGRGVTAADTLDDDFASKLSREADIVIGVYLLLI 1 | ||
2 GTGSILGNGRVLWLSYRNWAKLRPVELFVVSLAVTDVGISVFGYPFAASSSLLGRWSFGSAGCTW 1 | 2 GTGSILGNGRVLWLSYRNWAKLRPVELFVVSLAVTDVGISVFGYPFAASSSLLGRWSFGSAGCTW 1 | ||
| Line 645: | Line 786: | ||
2 GWSQYHVEPFGLSCTVDWGSFSRDAGGMSFIICLLVFCVAIPVTAIMASYVAISAIYRQAKKSIAGHLQDNSAMCKKRNKLE 0 | 2 GWSQYHVEPFGLSCTVDWGSFSRDAGGMSFIICLLVFCVAIPVTAIMASYVAISAIYRQAKKSIAGHLQDNSAMCKKRNKLE 0 | ||
0 MALAVCGGFLLAWLPYAVVGLWSAVAGVDAVPLALASAAPLFAKSSSLWNPIIYLGMNDRFR 2 | 0 MALAVCGGFLLAWLPYAVVGLWSAVAGVDAVPLALASAAPLFAKSSSLWNPIIYLGMNDRFR 2 | ||
1 * 0 | |||
> | >NEUR1_strPur Strongylocentrotus purpuratus (sea_urchin) XM_001197837 CX694910 CX690664 | ||
0 MDVNAKWWTNETLRTRDQFSDDHYTSVLSYEGDIWAGVYLMFI 1 | 0 MDVNAKWWTNETLRTRDQFSDDHYTSVLSYEGDIWAGVYLMFI 1 | ||
2 SLIAFIGNISVIVISLRKREKLKPIDLLTINLAIADFLICVVSYPLPMISAFRHR 0 | 2 SLIAFIGNISVIVISLRKREKLKPIDLLTINLAIADFLICVVSYPLPMISAFRHR 0 | ||
| Line 655: | Line 797: | ||
DEVGTYKRRPLMICSNPFAWSRDFHETWRQRRIRGIHRNCRNNVRVENINVNFRRDTDMVELNAPTPAEIHRPELNTASTRSGARTKSMATHLPALEEVPSG | DEVGTYKRRPLMICSNPFAWSRDFHETWRQRRIRGIHRNCRNNVRVENINVNFRRDTDMVELNAPTPAEIHRPELNTASTRSGARTKSMATHLPALEEVPSG | ||
APQCSALLHNTPIPRSLQGTPLPYQPQPSTSDLHDEFLNPSVVSRNMCVIVVKPNIEEELSTD* 0 | APQCSALLHNTPIPRSLQGTPLPYQPQPSTSDLHDEFLNPSVVSRNMCVIVVKPNIEEELSTD* 0 | ||
</pre> | </pre> | ||
== | === NEUR2: 12 vertebrate newropsins === | ||
<pre> | <pre> | ||
> | >NEUR2_galGal Gallus gallus AB368181 18570255 synteny: -B4GALT6 -NEUR2-KIAA1012 cOpn5L1 | ||
0 MDPSFANSTFQSKITEAADIVVGTCYMVF 1 | 0 MDPSFANSTFQSKITEAADIVVGTCYMVF 1 | ||
2 GICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHR 2 | 2 GICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHR 2 | ||
| Line 668: | Line 809: | ||
CSYFPSEKGSHTFECFKSYPNCFQERLSTMGCHLQDCESLENDLQVEVTQGSRNSMKVVEQEEKSTELDNLEITLEAVPVSCTFTDL* 0 | CSYFPSEKGSHTFECFKSYPNCFQERLSTMGCHLQDCESLENDLQVEVTQGSRNSMKVVEQEEKSTELDNLEITLEAVPVSCTFTDL* 0 | ||
> | >NEUR2_melGal Meleagris gallopavo (turkey) genomic | ||
0 MDPSFANSTFQSKITEAADIVVGTCYMVF 1 | |||
2 GICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHR 2 | |||
1 WLYGKHICLFYAFCGVFFGICSLSTLTLLSVVCCLKICFPAY 1 | |||
2 GNRFRRKHGQILIACAWTYAAIFACSPLAHWGEYGEEPYGTACCIDWQSTNVEITSMSYTVVLFIFCFILPCGVILTSYSLILVTVKESRRAVEQHVSGPTRINNVQTITVK 0 | |||
0 LSIAVCIGFFAAWSPYAIIAMWAAFGSIDKIPPLAFAIPAVFAKSSTLYNPIIHLLLKPNFRSIVAKDFSVLQQLCVRCCFCVKELQIYRSTFNAGLRTFKGRNEFSCNALPDMEG | |||
CSYFPSEKGNHTFECFKSYPKCCQERLSTMGCHPQERESLENDLQVEMTEGSRNSMKVVDQEEKSTELDNLEITLEAVPVHCTFTDL* 0 | |||
>NEUR2_anoCar Anolis carolinensis (lizard) | |||
0 MESYFANTTFHSKITEAADVIVGVFYIVF 1 | 0 MESYFANTTFHSKITEAADVIVGVFYIVF 1 | ||
2 GICSFCGNSILLYVSYKKKNLLKPAEYFMINLAISDLGMTLTLYPLAVTSSLAHR 2 | 2 GICSFCGNSILLYVSYKKKNLLKPAEYFMINLAISDLGMTLTLYPLAVTSSLAHR 2 | ||
| Line 676: | Line 825: | ||
YFPCEKCHDPFECFKNYPKCCQGRLNVMDHTPRESISVENNMQSKTKHASEKYIKVVIRGEKNTDIDNLEITLEHIPTDIKFANL* 0 | YFPCEKCHDPFECFKNYPKCCQGRLNVMDHTPRESISVENNMQSKTKHASEKYIKVVIRGEKNTDIDNLEITLEHIPTDIKFANL* 0 | ||
> | >NEUR2_xenTro Xenopus tropicalis (frog) abundant transcripts | ||
0 MGNKSDASAFYSSISETDDIVLGVLYSVF 1 | 0 MGNKSDASAFYSSISETDDIVLGVLYSVF 1 | ||
2 GLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHR 2 | 2 GLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHR 2 | ||
| Line 684: | Line 833: | ||
LKTTSKPPSSFKKSQGVCRNCVDTFECFRNYPRCCSVGNVDAAQPMAASLVRIPPANGAPQQTVQLVVSSSRTRSGVETVEVSTEAPMSDFIKDFI* 0 | LKTTSKPPSSFKKSQGVCRNCVDTFECFRNYPRCCSVGNVDAAQPMAASLVRIPPANGAPQQTVQLVVSSSRTRSGVETVEVSTEAPMSDFIKDFI* 0 | ||
> | >NEUR2_danRer Danio rerio (zebrafish) acquired new intron | ||
0 MGNVSKTALFMSTISRQHDILMGSLYSVF 1 | 0 MGNVSKTALFMSTISRQHDILMGSLYSVF 1 | ||
2 FVLSLLGNGMLLFVAYRKRSSLKPAEFFVVNLSVSDLGMTLSLFPLAIPSALAHR 2 | 2 FVLSLLGNGMLLFVAYRKRSSLKPAEFFVVNLSVSDLGMTLSLFPLAIPSALAHR 2 | ||
| Line 693: | Line 842: | ||
SQQCNNKDGSISTPFSSGQAESYGACHVYAEAGPHYQQISRQITARVLEGSVQSEIPVKQLTEKMQNDLL* 0 | SQQCNNKDGSISTPFSSGQAESYGACHVYAEAGPHYQQISRQITARVLEGSVQSEIPVKQLTEKMQNDLL* 0 | ||
> | >NEUR2_tetNig Tetraodon nigroviridis (pufferfish) gene mix | ||
0 MGNASDTSDAFNSKISKEHDFLIGSIYSVF 1 | 0 MGNASDTSDAFNSKISKEHDFLIGSIYSVF 1 | ||
2 CVLSLMGNCILLLVAHHKRSTLKPAEFFIVNLSISDLGMTLTLFPLAIPSSFSHR 2 | 2 CVLSLMGNCILLLVAHHKRSTLKPAEFFIVNLSISDLGMTLTLFPLAIPSSFSHR 2 | ||
| Line 701: | Line 850: | ||
LSNGQQDSYGTCLHCAEDAELGHVTGSRRTACILTGSTFTEVTLSQLSATPADLL* 0 | LSNGQQDSYGTCLHCAEDAELGHVTGSRRTACILTGSTFTEVTLSQLSATPADLL* 0 | ||
> | >NEUR2_takRub Takifugu rubripes (fugu) | ||
0 MGNASEASDIFLSKISKEHDILIGSIYSVF 1 | 0 MGNASEASDIFLSKISKEHDILIGSIYSVF 1 | ||
2 GLLSLAGNCILLLVAYHKRSMLKPAEFFIINLSISDLGMTLTLFPLAIPSSFSHR 2 | 2 GLLSLAGNCILLLVAYHKRSMLKPAEFFIINLSISDLGMTLTLFPLAIPSSFSHR 2 | ||
| Line 709: | Line 858: | ||
LSNGQQDSYGTCLHCADDAERGHVTTSQRTACILTGSTFTEVTVGQLSAAPADLL* | LSNGQQDSYGTCLHCADDAERGHVTTSQRTACILTGSTFTEVTVGQLSAAPADLL* | ||
> | >NEUR2_gasAcu Gasterosteus aculeatus (stickleback) | ||
0 MGNASDTSAVFASTISKERDILMGSLYSVF 1 | 0 MGNASDTSAVFASTISKERDILMGSLYSVF 1 | ||
2 GVLSLVGNCILLLVAYHKRSTLKPAEFFIINLSISDLGMTLSLFPLAIPSAFKHR 2 | 2 GVLSLVGNCILLLVAYHKRSTLKPAEFFIINLSISDLGMTLSLFPLAIPSAFKHR 2 | ||
| Line 717: | Line 866: | ||
APCHVMTPQRTACILTESTNREVTVSRLADKPQADFL* | APCHVMTPQRTACILTESTNREVTVSRLADKPQADFL* | ||
> | >NEUR2_oryLat Oryzias latipes (medaka) | ||
0 MGNVSDTSSLFASSISREHDILMGSLYSVF 1 | 0 MGNVSDTSSLFASSISREHDILMGSLYSVF 1 | ||
2 GLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHR 2 | 2 GLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHR 2 | ||
| Line 725: | Line 874: | ||
TGLCQLASPQNTACILTGSTYAEVTVQQLVDKQQPDFL* 0 | TGLCQLASPQNTACILTGSTYAEVTVQQLVDKQQPDFL* 0 | ||
> | >NEUR2_pimPro Pimephales promelas (minnow) | ||
0 MGNVSETALFVSTISRQHDILMGSLYSVF 1 | 0 MGNVSETALFVSTISRQHDILMGSLYSVF 1 | ||
2 CVLSLLGNGMLLFVAYRKRSSLKPAEFFVINLSVSDLGMTLSLFPLAIPSALAHR 2 | 2 CVLSLLGNGMLLFVAYRKRSSLKPAEFFVINLSVSDLGMTLSLFPLAIPSALAHR 2 | ||
| Line 732: | Line 881: | ||
0 LSVAVCIGFVTAWSPYAVVAMWAAFSANEPVPPTAFALAAILAKSSTIYNPMVYLLFKPNFRKILSQDTQNIRHRMCVSHSKASPTPEIK-AQSSQQCKDATISTPFSSGQAESYGTCHIYAEAEPHFQQISPQRTVRILEGIIQSEISVRHMTDRMQNDLL* 0 | 0 LSVAVCIGFVTAWSPYAVVAMWAAFSANEPVPPTAFALAAILAKSSTIYNPMVYLLFKPNFRKILSQDTQNIRHRMCVSHSKASPTPEIK-AQSSQQCKDATISTPFSSGQAESYGTCHIYAEAEPHFQQISPQRTVRILEGIIQSEISVRHMTDRMQNDLL* 0 | ||
> | >NEUR2_oncMyk Oncorhynchus mykiss (trout) no glycosylation site, anomalous agreement with chicken | ||
0 MGVLASIDDIAFLSNIPVAADITVAIVYAVF 1 | 0 MGVLASIDDIAFLSNIPVAADITVAIVYAVF 1 | ||
2 GMCSLFSNSTLLYISYKKKHLLKPAEFFIINLAISDMSLTLSLYPMAITSSIYHR 2 | 2 GMCSLFSNSTLLYISYKKKHLLKPAEFFIINLAISDMSLTLSLYPMAITSSIYHR 2 | ||
| Line 740: | Line 889: | ||
SCSPTSSARQALGESRGCTSPGEKCSDAFECFRHYPRGCHGGTNIPSSSARVYAPQDQLSTEPQLQSMTQKQMRKQEACHKKSLRATKHSKRTSEIDNLRINFEMVPGHAKVAWP* 0 | SCSPTSSARQALGESRGCTSPGEKCSDAFECFRHYPRGCHGGTNIPSSSARVYAPQDQLSTEPQLQSMTQKQMRKQEACHKKSLRATKHSKRTSEIDNLRINFEMVPGHAKVAWP* 0 | ||
> | >NEUR2_calMil Callorhinchus milii (elephantfish) frag | ||
0 1 | 0 1 | ||
2 GILSLVGNSVLLFVAYRKRQILKPAEYFVANLAVSDISMTVTLLPLAISSNFSHR 2 | 2 GILSLVGNSVLLFVAYRKRQILKPAEYFVANLAVSDISMTVTLLPLAISSNFSHR 2 | ||
| Line 747: | Line 896: | ||
0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVISYTMTVIAVNFVVPLSVMFFCYYNV | 0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVISYTMTVIAVNFVVPLSVMFFCYYNV | ||
</pre> | </pre> | ||
== | |||
=== NEUR3: 16 vertebrate newtopsins === | |||
<pre> | <pre> | ||
> | >NEUR3_galGal Gallus gallus cOpn5L2 AB368183 chr3 XM_420056 CN231992 testis exon 2^3 rel NEUR1/2 | ||
0 MEEQYISKLHPVVDYGAGVFLLII 1 | 0 MEEQYISKLHPVVDYGAGVFLLII 1 | ||
2 AILTILGNSAVLATAVKRSSLLKSPELLTVNLAVADIGMAISMYPLAIASAWNHAWLGGDASCIYYALMGFLFGVCSMMTLCAMAVIRFLVTNSSKSN 1 | 2 AILTILGNSAVLATAVKRSSLLKSPELLTVNLAVADIGMAISMYPLAIASAWNHAWLGGDASCIYYALMGFLFGVCSMMTLCAMAVIRFLVTNSSKSN 1 | ||
2 SNKISKNTVHILITFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHSSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNINRIPHAAKLEKKLTL 0 | 2 SNKISKNTVHILITFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHSSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNINRIPHAAKLEKKLTL 0 | ||
0 MAVLISVGFLSAWTPYAAASFWSIFNSSDSLQPIVTLLPCLFAKSSTAYNPFIYYIFSKTFRHEIKQLQCCWGWRVHFFSADNSAENSVSMMWSGRDNIRLSPTAKVESQGAARH* | 0 MAVLISVGFLSAWTPYAAASFWSIFNSSDSLQPIVTLLPCLFAKSSTAYNPFIYYIFSKTFRHEIKQLQCCWGWRVHFFSADNSAENSVSMMWSGRDNIRLSPTAKVESQGAARH* 0 | ||
> | >NEUR3_taeGut Taeniopygia guttata ABQF01025032 | ||
0 MEEQYISKLHPVVDYGAGVFLLII 1 | 0 MEEQYISKLHPVVDYGAGVFLLII 1 | ||
2 AILTILGNSAVLATAVKRSSLLKPPELLTVNLAVADIGMALSMYPLAIASAWSHAWLGGDASCVYYALMGFLLGVCSMMTLCAMAVIRFLVTNSPKSN 1 | 2 AILTILGNSAVLATAVKRSSLLKPPELLTVNLAVADIGMALSMYPLAIASAWSHAWLGGDASCVYYALMGFLLGVCSMMTLCAMAVIRFLVTNSPKSN 1 | ||
| Line 761: | Line 911: | ||
0 MAVLISVGFLSSWTPYAATSFWSIFNSSHSLQPVVTLLPCLFAKSSTAYNPFIYYVFSKTFRCEVKRLQCCCAWRVHYFSSDNSVENPLSTMWSGRDNIRLSAAPQVQNPGAAAP* 0 | 0 MAVLISVGFLSSWTPYAATSFWSIFNSSHSLQPVVTLLPCLFAKSSTAYNPFIYYVFSKTFRCEVKRLQCCCAWRVHYFSSDNSVENPLSTMWSGRDNIRLSAAPQVQNPGAAAP* 0 | ||
> | >NEUR3_melGal Meleagris gallopavo (turkey) | ||
0 MEEQYISKLHPVVDYGAGVFLLI 1 | |||
2 PILTILGNSAVLATAVKRSSLLKSPELLTVNLAVADIGMAISMYPLAIASAWNHAWLGGDASCVYYALMGFLFGVCSMMTLCAMAVIRFLVTNSSKSN 1 | |||
2 SNKISKNTVHILITFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHSSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNINRIPHAAKLEKKLTL 0 | |||
0 MAVLISVGFLSAWTPYAAASFWSIFNSSDSLQPIVTLLPCLFAKSSTAYNPFIYYIFSKTFRHEIKQLQCCWAWRVRFFSTDNSADNSVSMMWSGRDNARLSSNAKVESQGAAMH* 0 | |||
>NEUR3_anoCar Anolis carolinensis (lizard) AAWZ01001057 | |||
0 MEEHYISKVHPVWDYGMGVFLLII 1 | 0 MEEHYISKVHPVWDYGMGVFLLII 1 | ||
2 | 2 AILTILGNSMVLAVAVKRSSCLRSPELLTVNLAATDLGMGLSMYPLAIASAWNHAWLGGEATCIYYALMGFLFGVSSIMTLSAMAVIRFLVTFSSKPA 1 | ||
2 | 2 GHKINRKVMHICIMLIWAYAVLWAILPLLGWGHYGPEPFGTSCTIAWGQFHNSQKGFAFILSMFILCTFLPAITIIMCYLGIAWKFHKTHQEMQNLNRISSAAKLEKKLIL 0 | ||
0 VAVLISVGFLGAWTPYAIVSFWSVFHSSESIPYIVTLLPCLFAKSSTAYNPFIYYTFSKTFRHEVKHLRCYSGQRAQENMKNSINSNVSFMWHGGGNICLSTRQIEMREIPNQ* 0 | 0 VAVLISVGFLGAWTPYAIVSFWSVFHSSESIPYIVTLLPCLFAKSSTAYNPFIYYTFSKTFRHEVKHLRCYSGQRAQENMKNSINSNVSFMWHGGGNICLSTRQIEMREIPNQ* 0 | ||
> | >NEUR3_xenTro Xenopus tropicalis (frog) cdna ovary embryo | ||
0 MEERYLSKLHPLVDFGSGVFLLLV 1 | 0 MEERYLSKLHPLVDFGSGVFLLLV 1 | ||
2 AILTVLGNCAVLATAVKCSSHLKAPDLLSINLAVADLGMAISMYPLAIASAWNHAWLGGDASCLYYALMGFFFGVSSMMTLTVMAIIRYRVTSSFKYS 1 | 2 AILTVLGNCAVLATAVKCSSHLKAPDLLSINLAVADLGMAISMYPLAIASAWNHAWLGGDASCLYYALMGFFFGVSSMMTLTVMAIIRYRVTSSFKYS 1 | ||
| Line 773: | Line 929: | ||
0 LAILVSFGFLISWTPYAAVSFWSLFHSSKYIPPVVSLLPCLFAKSSTAFNPMIYYAFSKTFRRKVKHLKCCCGWRVHFLQSENSVENPRVSVIWTGKENVMVSSVPKLMKGVPGTPTGTQ* 0 | 0 LAILVSFGFLISWTPYAAVSFWSLFHSSKYIPPVVSLLPCLFAKSSTAFNPMIYYAFSKTFRRKVKHLKCCCGWRVHFLQSENSVENPRVSVIWTGKENVMVSSVPKLMKGVPGTPTGTQ* 0 | ||
> | >NEUR3a_danRer Danio rerio (zebrafish) | ||
0 MDRYTSKLSPAVDYSAGTFLLVI 1 | 0 MDRYTSKLSPAVDYSAGTFLLVI 1 | ||
2 AILSILGNAAVLLTAAWRHSVLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWIGGDPSCLYYGLMGMIFSVASIMTLAVMGLVRYLVTGNPPK 1 | 2 AILSILGNAAVLLTAAWRHSVLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWIGGDPSCLYYGLMGMIFSVASIMTLAVMGLVRYLVTGNPPK 1 | ||
| Line 779: | Line 935: | ||
0 MGILISTGFIVSWAPYVFVSLWTMFRSEGEDSVVPIVSLLPCLFAKCSTVYNPLVYYVFRKSFRREIHQIRICCFQGCWDAVSKMTRGDGPEETSGTHETDNI* 0 | 0 MGILISTGFIVSWAPYVFVSLWTMFRSEGEDSVVPIVSLLPCLFAKCSTVYNPLVYYVFRKSFRREIHQIRICCFQGCWDAVSKMTRGDGPEETSGTHETDNI* 0 | ||
> | >NEUR3a_tetNig Tetraodon nigroviridis (pufferfish) | ||
0 MDDKYMSKLSPPVDLWAGIYLVVI 1 | 0 MDDKYMSKLSPPVDLWAGIYLVVI 1 | ||
2 ALLSVLGNASVLFSASRRLTPLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWMGGDTACLYYGLMGMIFSITSIMTLAVMGMIRYLVTGSPPR 1 | 2 ALLSVLGNASVLFSASRRLTPLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWMGGDTACLYYGLMGMIFSITSIMTLAVMGMIRYLVTGSPPR 1 | ||
| Line 785: | Line 941: | ||
0 IAVLISVGFLGSWAPYGLVSLWSILKDSSSIPPQVSLLPCLFAKSSTVYNPVIYYIFSQSFKLEVQQLFLCC* 0 | 0 IAVLISVGFLGSWAPYGLVSLWSILKDSSSIPPQVSLLPCLFAKSSTVYNPVIYYIFSQSFKLEVQQLFLCC* 0 | ||
> | >NEUR3a_takRub Takifugu rubripes (fugu) | ||
0 MDDKFTSKLSPAVDLWAGTYLVFI 1 | 0 MDDKFTSKLSPAVDLWAGTYLVFI 1 | ||
2 ALLSVLGNASVLFSAGRRLSMLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWMGGDASCLYYGLMGMIFSITSIMTLAVMGMIRFLVTGTPPR 1 | 2 ALLSVLGNASVLFSAGRRLSMLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWMGGDASCLYYGLMGMIFSITSIMTLAVMGMIRFLVTGTPPR 1 | ||
2 SGIKFQKKTISVVISAIWLYACLWAVFPILGWGSYGPEPFGIACSVDWMGYGESLNNATFISTLSVLCTFLPYLVIVFTYFGIAWKLHRAYRSIKSSDIQYTNVERRITL 0 | 2 SGIKFQKKTISVVISAIWLYACLWAVFPILGWGSYGPEPFGIACSVDWMGYGESLNNATFISTLSVLCTFLPYLVIVFTYFGIAWKLHRAYRSIKSSDIQYTNVERRITL 0 | ||
0 | 0 MAVMISSGFLIAWTPYVAVSFWSMRNSQRQGHMAPSVTLLPCLFAKSSTAYNPFIYFFFQRNTGHKLLPFHRHAFSCSDRADSSREGEKEESKVSKNLGFTCFGAGTYETCPGLAGDQSQREMAELG* 0 | ||
> | >NEUR3a_gasAcu Gasterosteus aculeatus (stickleback) | ||
0 MEDKYVSKLSPAVDFWAGTYLIII 1 | 0 MEDKYVSKLSPAVDFWAGTYLIII 1 | ||
2 AVLSIFGNTAILVSAARRSGPLKAPELLTVNLAVTDLGMALSMYPLSIASAFNHAWIGGDASCLYYSLMGMIFSITSIVTLAVMGMVRYLVTGNPPR 1 | 2 AVLSIFGNTAILVSAARRSGPLKAPELLTVNLAVTDLGMALSMYPLSIASAFNHAWIGGDASCLYYSLMGMIFSITSIVTLAVMGMVRYLVTGNPPR 1 | ||
| Line 797: | Line 953: | ||
0 MAAMISSGFLFSWTPYVAVSLWSMFRSREHIPPLVALLPCLFAKSSTVYNPFIYFIFQRSSWRELLRLHRHLLCCWHRASPPAEGRRSQRGSEGGSWGGACESDDAFGLVHVMKSNATCQTISWA* 0 | 0 MAAMISSGFLFSWTPYVAVSLWSMFRSREHIPPLVALLPCLFAKSSTVYNPFIYFIFQRSSWRELLRLHRHLLCCWHRASPPAEGRRSQRGSEGGSWGGACESDDAFGLVHVMKSNATCQTISWA* 0 | ||
> | >NEUR3_calMil Callorhinchus milii (elephantfish) frag | ||
2 AILSIFGNSVVLLVAAKKSSQLKPPELLTVNLAITDFCSAVTMYPLAVGSAWKHTWLGGDASCKYYGFMDFFFGIASIGTLTVMAIVRFLVTSTTQN 1 | 2 AILSIFGNSVVLLVAAKKSSQLKPPELLTVNLAITDFCSAVTMYPLAVGSAWKHTWLGGDASCKYYGFMDFFFGIASIGTLTVMAIVRFLVTSTTQN 1 | ||
> | >NEUR3_petMar Petromyzon marinus (lamprey) exon frag | ||
0 MAEQGEDDQFRSKLSPTADIAAGTFLLAV 1 | 0 MAEQGEDDQFRSKLSPTADIAAGTFLLAV 1 | ||
2 AVLSLAGNGAVLGVAARRWAKLKAPELLSVNLALTDLGIAASIYPLAVASAWNHRWLGGQPVCTYYAFAGFFFGTASMGTLTAMAGVRYKGTSTQVH 1 | 2 AVLSLAGNGAVLGVAARRWAKLKAPELLSVNLALTDLGIAASIYPLAVASAWNHRWLGGQPVCTYYAFAGFFFGTASMGTLTAMAGVRYKGTSTQVH 1 | ||
2 | 2 sVKQITKRAMLAVIVAVWAYALLWSCLPLLGWGR 2 1 YGVEPFGVSCTLAWAELQLTPGGVAFLYAMFVLCLLLPAIAIGLCYAGIVCKLRRAYREGRSKRRTPTARHVESRLTK 0 | ||
> | >NEUR3b_danRer Danio rerio (zebrafish) | ||
0 MDIYSSKLSSAVDYGIGAFLLLI 1 | 0 MDIYSSKLSSAVDYGIGAFLLLI 1 | ||
2 TILSILGNLMVLVMAYKRSNHMKPPELLSVNLAVTDLGAAVTMYPLAVASAWNHHWIGGDVSCVYYGLMGFLFGAASMMTLTIMAIVRFIVSLTLQSP 1 | 2 TILSILGNLMVLVMAYKRSNHMKPPELLSVNLAVTDLGAAVTMYPLAVASAWNHHWIGGDVSCVYYGLMGFLFGAASMMTLTIMAIVRFIVSLTLQSP 1 | ||
2 KEKISKRNAKILVATTWLYALLWAIFPLIGWGKYGPEPFGLSCTLDWRDMKEHSQSFVITIFLMNLILPAIIIVSCYCGIALRLYVTYKSMDDSNHVPNMIKMQRRLMV 0 | 2 KEKISKRNAKILVATTWLYALLWAIFPLIGWGKYGPEPFGLSCTLDWRDMKEHSQSFVITIFLMNLILPAIIIVSCYCGIALRLYVTYKSMDDSNHVPNMIKMQRRLMV 0 | ||
0 IAVLISIGFVGCWAPYGIVSLWSIYRPGDSIPAEVSMLPCLFAKTSTVYNPFIYYIFSKTFKREVNQLSRFCGRSNICRPTDAKNRPENTIYLVCDVNKSKPGVEDLSLARSKENETQMLPNQDLHE* | 0 IAVLISIGFVGCWAPYGIVSLWSIYRPGDSIPAEVSMLPCLFAKTSTVYNPFIYYIFSKTFKREVNQLSRFCGRSNICRPTDAKNRPENTIYLVCDVNKSKPGVEDLSLARSKENETQMLPNQDLHE* 0 | ||
>NEUR3b_etNig Tetraodon nigroviridis (pufferfish) assembly errs in exon 2 frameshift, used traces | |||
0 MDMYTSALSPALDIGTGCYLLVI 1 | |||
2 AVLSFIGNLLVIITAVKKSSKMKPPELLCVNLAVTDLGAAVTMYPLSVASAWSHRWIGGDVTCVYYGLVGFLFEVASIMNLTVLAIVRFTVSLNLQSP 1 | |||
2 EEKISWKSVKIMCLLIWLYGVIWAMFPVLGWGRYGPEPFGISCSLAWGQMKNEGFSFVVAMFSFNLAVPALIIVSCYFGMAINLYFTHKKMVNTGNRIPAVIKLHRRLLR 0 | |||
0 IAVLISVGFLGSWAPYGLVSLWSILKDSSSIPPQVSLLPCLFAKSSTVYNPVIYYIFSQSFKLEVQQLFLCCLSFRSSRTNNCKSNESSIFMVSNGKNLTPALTQQNTSHAVIMN* 0 | |||
>NEUR3b_takRub | |||
0 MDIYSSTLSPALDIGTGCFILVV 1 | |||
2 GVLSIIGNLLVIITAVKRSSKMKPPELLCVNLAVTDLGAAVTMYPLSVASAWSHRWIGGDATCIYYGLVGFLFGVASIMNLTILAIVRFTVSLNLQSP 1 | |||
2 eEKITWKSVKIMCMWVWLYSIMWAMFPILGWGRYGPEPFGISCSLAWGQMKDEGFSFVVTIFSLNFAVPAVIIICCYFGIAIKLYFTYKKTVNTNQIPVIIKLHRRLLM 0 | |||
0 IAVLISVGFLGCWAPYGLVSLWSILKDSSSIPPEVSLLPCMFAKSSTVYNPIIYYMFSQSFKMEVQQLFLWCPSFEFCRTSSNNGNETTIYMVSTGKT* 0 | |||
>NEUR3b_gasAcu | |||
0 MDIYASTLSPAVDVGAGCYLLFV 1 | |||
2 AVFSIVGNLLVLVMAVKRSSRMKPPELLSVNLAVTDLGAAVTMYPLAVASAWRHRWLGGDATCVYYAVAGFFFGLASIMSLTGLAIVRFIVSLNLQSP 1 | |||
2 NEKISWRKVKLLCACTWLYALAWAAFPFLGWGRYGPEPYGLSCSLAWGQMKHEGFSFVVSMFSLNLVLPCVIIAGCYFGIAFKLYFTYRKSNNNSNRLPNVVRRHRRLLA 0 | |||
0 IAVLISLGFVVCWSPYAVVSLWSIFHDSGSIPPEVSLLPCMFAKSSTVYNPLIYYIFSQSFRREVKQLWRHLGSTLCSVSNSVNDAAVSNTGKSN* 0 | |||
>NEUR3b_oryLat Oryzias latipes (medaka) | |||
0 MDIYASALSPALDIGTGCYLLVL 1 | |||
2 TVLSIIGNLLVVIMAFKRSSRMKPPELLSVNLALTDLGAAVFMYPLAVASAWSHHWLGGDVSCIYYGLAGFFFGSASVMNLTALAVVRFIVSLNLHSP 1 | |||
2 KEKVSWRKVKILCLWSWLYALIWALFPILGWGRYGPEPFGLSCSLAWGEMKQEGPSFVISLFSFNLVLPSVVIICCYFGIAMKLYFTYKKSANSNHVPNIIKLHRRLLIIA 0 | |||
0 ILISIGFIGCWTPYGLVSLWSIFNDSSKIPPEVSLLPCMFAKSSTVYNPMIYYFFSKSFQREVKQLSWLCVGSNPCHVSNSVNDNNIYMVSVNVKSKETRRETLQEITESRQ* 0 | |||
</pre> | </pre> | ||
=== NEUR4: 11 vertebrate newwopsins === | |||
<pre> | |||
>NEUR4_ornAna Ornithorhynchus anatinus hypothetical protein XM_001508128 | |||
0 MSLSHSLQVPWRNNLTFLNKEAQVSEQGETIIGIYLLAL 1 | |||
2 GWMSWFGNSMVIFILHRQRGILNPTDYLTFNLAVSDASVSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLAMISVTRYIKGCHPHR 1 | |||
2 GHFINTANISVALILIWVSALFWSAGPVLGWGSYT 1 | |||
2 DRMYGTCEIDWAEANFSSICKSYIISIFFCCFFLPVSIMFFSYVSIIKMVKSSHTLAGADDPTDRQRRLDRDVTR 0 | |||
0 VSVVICTAFIVAWSPYAVISMWSAFGHSVPNLTSVLASLFAKSASFYNPIIYFGMNSKFRKDILVLLPCAKESKEPVKLKKFKNLRQKQ | |||
GFTLQKPEKAHVLQVPDSGPMSLINTPPLGNRNSFDLACDNSDFECVRL* 0 | |||
>NEUR4_galGal Gallus gallus (chicken) genome gappy | |||
0 MSLQLSPQAPWRNNNISFLSREAAVTEQGETIIGFYLLAL 1 | |||
2 GWMSWFGNSVVIFVLYKQRHLLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPER 1 | |||
2 AHCISNSSMTVAMVLIWIAAFFWSAAPLLGWGSYT 1 | |||
2 DRMYGTCEIDWAKANFSTIYKSYIISIFICCFFLPVTVMVFSYVSIINTVKLSHALTGLSDPTERQRRMERDVTR 0 | |||
0 VSIVICTAFIIAWSPYAVLLLWSAYGHPVPNLPLYLSSLFAKSASFYNPIIYFGMSSKFRRDIFILFHCAKEVKDPVKLKRFKNLKQKQ | |||
EPSQKEEKYAAEMHPAPSPDSGVGSPTNTPPPANREEYFGILDTPSNSPDIECDRL* 0 | |||
>NEUR4_melGal Meleagris gallopavo (turkey) | |||
0 MSLQLSPQAPWRNNNISFLSREAAVTEQGETIIGFYLLAL 1 | |||
2 GWMSWFGNSIVIFVLYKQRHLLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPDR 1 | |||
2 AHCISNSSMTVAMVLIWIAAFFWSAAPLLGWGSYT 1 | |||
2 DRMYGTCEIDWAKANFSTIYKSYIISIFICCFFLPVTVMVFSYVSIINTVKLSHALTGFSDPTDRQRRMERDVTR 0 | |||
0 VSIVICTAFIIAWSPYAVISIWSAYGHPVPNLTSILASLFAKSASFYNPIIYFGMSSKFRRDIFILFHCAKEVKDPVKLKRFKNLKQKQ | |||
EPSQKEEKYAPEMHPAPSPDSGVGSPTNTPPPAKREEYFGILDTPSNNPDIECDRL* 0 | |||
>NEUR4_taeGut Taeniopygia guttata (finch) | |||
0 MSVQFSAQAPWRNNNISFLTREAAVTEQGETIIGFYLLAL 1 | |||
2 GWLSWFGNSIVIFVLYKQRHVLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPER 1 | |||
2 GHCISNSSMSVALVLIWVAAFFWSAAPLLGWGSYT 1 | |||
2 DRMYGTCEIDWAKASFSTIYKSYIVSIFICCFFLPVTVMVFSYVSIINTVKLSHTLTGLGDPTDRQRRIERDVTR 0 | |||
0 VSIVICTAFIIAWSPYAVISIWSAYGHPVPNLTSILASLFAKSASFYNPIIYFGMSSKFRRDIFIFHCAKELKDPVKLKRFKNLKPKQ | |||
PQPSQKEEKYAPEMHPAPSPDSGVGSPTNSPPPANREVYFGILDTPSNNPNIECDRL* 0 | |||
>NEUR4_anocar Anolis carolinensis (lizard) | |||
0 MSLQVSPQAPWRNNNVTFSNKEVPVSEQGETIIGFYLLAL 1 | |||
2 GWMSWFGNSIVIFVLYRQRAGLQPTDYLTFNLAVSDASVSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPDR 1 | |||
2 GKCISNSSISVALFLIWIAAFFWSVAPVLGWGSYr 1 | |||
2 DRMYGTCEIDWAKANFSTIYKSYIVSIFICCFFLPVSVMVFSYVSIINTVKSSHALSGVGDPTERQRRMERSVTR 0 | |||
0 VSLVVICTAFITAWSPYAVISMWSAYGYTVPNLTSILASLFAKSASFYNPIIYFGMSSKFRKDIFVLLHCAKEIKDPVKLKRFKNLKQKQ | |||
EVSPSQREEKYAADVQPALSPDSGVGRSNTPPPVNREVYFGAFDTFSNNPDVECDRL* 0 | |||
>NEUR4a_xenTro Xenopus tropicalis (frog) numerous transcripts TTC13 FAM89A COCH/VIT NEUR4a AKAP MTHFD1 | |||
0 MSLQFPRPAPWRNNNLTLLQKENPLTEQGETIIGIYLLAL 1 | |||
2 GWLSWFGNSIVIFVLYKQRANLLPTDYLTFNLAVSDASTSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 | |||
0 VDGFLTLLFGLASINTLTLISVTRYIKGCHPQR 1 | |||
2 ANCISNGSITISLALIWIAALFWSVAPLLGWGSYR 1 | |||
2 DRMYGTCEIDWTKASFSTIYKSYIISIFICCFFLPVMVMVFCYVSIINTVKSSRALTSEGDLSERQRKMERDVTR 0 | |||
0 VSVVICTAFIVAWSPYAVISMWSACGYYVPSLTSILAALFAKSASFYNPLIYFGMSSKFRKDLCVVLPCAKAQKDPVKLKRYKDKKQ | |||
GSAPRAREQTEIEQPVQLQPAPSQDSGVGSPSNTPPLRTKDVHIVDIDLVSDNPSYECDRL* 0 | |||
>NEUR4b_xenTro Xenopus tropicalis (frog) XM_002932842 no transcripts, not tandem 54% identity COCH/VIT NEUR4b SCFD1 | |||
0 MDGLLMDSSSLLPNSSSGARVLEEGETAIGAYLLLL 1 | |||
2 GWLSWLGNGAVICLMCKRRRLLDSHDLLTLNLAVSDAGISIFGYSRGIVELFHGLGKDGFLANNLWTCQ 0 | |||
0 VGGFLILLFGLMSISTLTAISLLRYIKGCQPHK 1 | |||
2 AHMVDQRHVTMAIVFIWISSIFWSGSPVLGWGSFT 1 | |||
2 ERKYGTCEIDWVQAASSTVYKSYVIGVFIWGFVLPVSIMVFCYVSIIRTVHKSHRNSRGGEISQRQLTMERDITR 0 | |||
0 VSFVICTAFLLAWSPYAVISMWSACGYQVPGLTGVAATLLAKSASFYNPIIYLGMSPKFRQELRALLCCLRQSGDSPQSFEKPVIT | |||
HEPKMKQCNSPSNSLAAKMEQPVLEAQGIQESTLIKGAADSLTVNSQTSDPVKNIDISLDFPMESHQI* 0 | |||
>NEUR4a_danRer Danio rerio (zebrafish) | |||
0 MSAQNPLQVVNIPWRNNNFSLMSRDPPLSDQGETIIGVYLLIL 1 | |||
2 GWLSWFGNSIVIFVLFRQRSTLQPTDYLTLNLAVSDASISVFGYSRGILEIFNIFKDSGYIISSVWTCQ 0 | |||
0 VDGFFTLVFGLSSINTLTVISITRFIKGCHPHK 1 | |||
2 AHCITNSTVAVCVVFIWIGAFFWSAAPVLGWGSYT 1 | |||
2 DRGYGTCEIDWVKANYSTIHKSYIISIFIFCFLVPVLLMLFCYISIINTVKRGNAMNADGDLSDRQRKIERDVTI 0 | |||
0 VSIVICTAFILAWSPYAVVSMWSAWGFHVPNLTSIFTRLFAKSASFYNPLIYFGLSSKFRKDVSVLLPCGREGRDPVRLKRFKRLRGRA | |||
EPPGAPAHTPHPQIALKNYNNHSKPHAGPAHCTGHAPSPDSGVGSHHETPPPQPRPQLFFIDVPEPEAESECVRL* 0 | |||
>NEUR4b_danRer Danio rerio (zebrafish) COCH/VIT NEUR4b SCFD1 retinal transcript: DN901362 | |||
0 MDIHSIPPTNITVYRVSDGGETAIGVYLVIL 1 | |||
2 GWLSWIGNGTVILLLTKQRKALEPQDFLTLNLAISDASISIFGYSRGILEVFDVFRDEGYLIKTFWTCK 0 | |||
0 VDGFLILLFGLISINTLTAISVIRYIKGCHPHH 1 | |||
2 AHHINKRNICLVITAVWLFCLFWAGAPLLGWGSYR 1 | |||
2 ARGYGTCEIDWTRALYSIPFKLYVIGIFFFNFFVPLFIIVFAYVSIIRTVNSSHKSSQGGDVSERQKKIERSITR 0 | |||
0 VSLILCAAFLLAWSPYAVISMWSALGYQIPTLNGILASLFAKSASFYNPFIYIGMSKFRKDLQALFYCLRKDQVMRCFRCNSVPFLMQTSLKVGNSTGTLF* 0 | |||
>NEUR4_tetNig Tetraodon nigroviridis (pufferfish) | |||
0 MEPSRPWRNSSVLGGGAEPPLSEQGETIIGVYLLLL 1 | |||
2 GWLSWFGNTVVLFVLVRQRSSLQPTDLLTFNLAVSDASISVFGYSRGIIQIFNVFQDSGFIISSIWTCE 0 | |||
0 VDGFLTLIFGLSSINTLTVISITRYIKGCQPSR 1 | |||
2 AALISRSSVSVCLLLIWTTAGFWSGAPLLGWGSYT 1 | |||
2 DRGYGTCEIDWSKAASSGVYRSYIISIFIFCFFIPVFIMLFCYISIINTVKRGNALAADGHLSHRQRTMERDVTV 0 | |||
0 ISVVICTAFIMAWSPYAVVSMWSAWGFHVPSTTSIVTRLFAKSASFYNPLIYFGMSSKFRKDVSLILPCAKERREVVLLQRFKNIKPKAA | |||
AAPPPPPLPVYRPKEKNEDEPKLSVHDNDSGVNSPPETPPSDAQEVFPVDPPSQIETSEYWSDRL* 0 | |||
>NEUR4_takRub recent pseudogene 8/8 traces support stop codon indel too 94% identity | |||
0 MADSIPPWRNSSVLGGGAEPPLSEQGETIIGVYLLLLG 1 | |||
2 GWLSWFGNTVVLFVLYRQRSTLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVF*DSGFIISSIWTCE 0 | |||
0 VDGFFTLVFGLSSINTLTVISITRYIKGCQPSR 1 | |||
2 AGHINRTFVSVCLLLIWIMAGFWSGSPLLGWGSYT 1 | |||
2 DRGYGTCEIDWSKAAYSTAYRSYIISIFIFCYFIPVFIMLFCYISIINRVKRGNALAA-GDLTDRQRKMERDVTI 0 | |||
0 VSIVICTAFILAWSPYAVVSMWSAWGFHVPNLTSIFTRLFAKSASFYNPLIYFGLSSKFRKDVAVLLPCTKDAKDTVKVKRFK NIKPKAAAAPPPPPLPVYRPKEKNEDEPKLSVHDNDSGVNSPPETPPSDAQEVFPVDPPSQIETSEYWSDRL* 0 | |||
>NEUR4_gasAcu Gasterosteus aculeatus (stickleback) | |||
0 PVKVVNIPWRNNNLSNLNTDPPLSEQGETFIGVYLLVL 1 | |||
2 GWLSWFGNSLVMFVLYRQRASLQSTDFLTLNLAISDASISIFGYSRGILEIFNIFNDDGYLINWIWTCQ 0 | |||
0 VDGFFTLLFGLASINTLTVISVTRYIKGCHPNK 1 | |||
2 AYCISTNTIAVSLICIWTGAVFWSVAPLLGWGSFT 1 | |||
2 DRGYGTCEVDWSKANYSTIHKSYIISILIFCFFIPVMIMLFSYVSIINTVKSTNAMSADGFLSTRQRKVERDVTRV 0 | |||
0 ISIVICTAFITAWSPYAVVSMWSAWGFHVPSTTSIITRLFAKSASFYNPLIYFGMSSKFRKDVSVLVPCTRERREVVHLQHFKNIKPKAEAPPTPASLPVQKLGAKYAVPNPDADSGVNNPPQRPATDPQGDLNIDLPSHIETSEYWCDRL* 0 | |||
>NEUR4_oryLat Oryzias latipes (medaka) frag | |||
0 MEITLKAFPLKVVNIPWRNNNLSTLHSEPPLSEQGETVIGVYLLVL 1 | |||
2 GWLSWFGNSLVIFVLCKQRASLQPTDFFTLNLAVSDASISVFGYSRGILEIFNILKDDGYLITWIWTCQ 0 | |||
0 VDGFLTLLFGLVSINTLTVISVTRYIKGCHPHK 1 | |||
2 AHCISSSTIAVSLIIVWAAALFWSVAPLLGWGSYT 1 | |||
2 DRGYGTCEVDWSKANYSTFYKSYIISILIFCFFIPVVIMLFSYVSIINTVKSTNAMSAVGFLSARQRKMERDVTRV 0 | |||
0 ISIVICTAFITAWSPYAVVSMWSAWGFHVPSTTSIITRLFAKSASFYNPLIYFGMSSKFRKDVSVLVPCTRERREVVHLQHFKNIKPKAEAPPTPASLPVQKLGAKYAVPNPDADSGVNNPPQRPATDPQGDLNIDLPSHIETSEYWCDRL* 0 | |||
0 SASFYNPLIYFGMSSKFRKDISVLLPCAAEGREVVHLQRFQNIKPKADTPLTAAPHPPPAKPLAAEMNQTNADGDPGVNNPPHTPPQIFHIDVPSHIETSEFWCDRL* 0 | |||
>NEUR4_calMil Callorhinchus milii (elephantfish) frag | |||
0 MGCSLGWKVLLWFLHGILICPRPWRNHNSTFQPKEHPISEQGETIIGVYLLIL 1 | |||
2 GWLSWFGNSIVIFILYRQRLSLQPPDYLTLNLAVSDASISIFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 | |||
0 1 | |||
2 AVSISAGSIAASLVLIWIAAIFWSGAPLFNWGSYT 1 | |||
2 DRMYGTCEIDWSRASFSTIYKSYIISIFICCFFLPVFVMLFSYISIINTVKSSHAFAGNADLSDRQRRMEKDVTR 0 | |||
0 VSMVICTAFIIAWSPYAVISMWSASGYTVPQLTGIFASLFAKSASFYNPMIYFGLNSKFRKDIYILLPCVKEPKESVKLKRFKHLRHRPEQQQANKDRYAEELQQVASPDSGMGSPSKSPPLHNKDVFFVLWLRGLKK | |||
>NEUR4_petMar Petromyzon marinus (lamprey) frag | |||
0 1 | |||
2 GWLSWLGNGLVIFVLTRQWSSLQPPDLLTLNLALSDASIAVFGYSRGIIEIFNVFQDDGYIIKSTWTCQ 0 | |||
0 1 | |||
2 PTKVTSTSMVVSLALVWAASLFWSAAPLLGWGSYT 1 | |||
2 DRRYGTCEIDWMKATFSTIYKSYIISIFICCFFMPISTMLFAYISIINTVKSSHVTARMGDVSERQRNMERDITRI 0 | |||
0 VSIVICCAFILAWSPYAVISMYSACGHRVPALTSLLAALFAKSASFYNPFIYFGMSGKFRADVRAMLPCRATSVKAPRDAVRLKRYRTHVDPERASHRAAVAAREQPAPRAAAPRPASPAPSAARDRDPELDEREFDPEGRASALAEVAAVESRDSGIACTRGKRRASRGDDVEVRNDV* 0 | |||
</pre> | |||
'''See also:''' [[Opsin_evolution|Curated Sequences]] | [[Opsin_evolution:_Peropsin_phyloSNPs|Peropsins]] | [[Opsin_evolution:_RGR_phyloSNPs|RGR phyloSNPs]] | [[Opsin_evolution:_LWS_PhyloSNPs|LWS]] | [[Opsin_evolution:_Encephalopsin_gene_loss|Encephalopsins]] | [[Opsin_evolution:_Melanopsin_gene_loss|Melanopsins]] | [[Opsin_evolution:_update_blog|Update Blog]] | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
VDGFLTLLFGLASINTLTVISVTRYIKGCHPDRAHCISNSSMTVAMVLIWIAAFFWSAAP | |||
LLGWGSYT | |||
Latest revision as of 11:20, 7 October 2010
See also: Curated Sequences | Peropsins | RGR phyloSNPs | LWS | Encephalopsins | Melanopsins | Update Blog
Neuropsin backgrounder
Neuropsin (OPN5, GPR136, NEUR1) is a deeply diverged member of the opsin family with a mere three experimental publications and considerable confusion over the name (mysteriously used for an unrelated kalikrein serine protease not remotely related to opsins). OPN5 itself is a poor nomenclatural choice, having no mneumonic value and not extendable to the four vertebrate paralogs, so NEUR1-4 are used here for these loci.
There are no known disease associations or described knockout phenotypes; in mouse, neuropsin is expressed primarily in brain, spinal chord, and testes. In chicken, neuropsin is expressed in embryonic and early posthatching neural retina (but not pineal gland) in subsets of differentiating ganglion cells and amacrine cells, ie prior to functioning of ciliary opsins thus ruling out a retinal replenishment role ("photoisomerase").
A 2010 study in quail brain mapped NEUR1 to the paraventricular organ, a region of the diencephalon of nonmammalian vertebrates containing aminergic neuronal cell bodies beneath the epithelial membrane lining the third ventricle. The vertebrate forebrain's diencephalon contains the thalamus, hypothalamus and posterior pituitary.
The specific model here envisions light detected by NEUR1 in paraventricular neurons signaling via cerebrospinal fluid to the pars tuberalis of the pituitary gland, inducing there thyroid-stimulating hormone, in turn inducing deiodinase DIO2 in tanycytes lining the third ventricle leading to long-day–induced T3 in the mediobasal hypothalamus ultimately inducing gonadotropin-releasing hormone and so testis growth associated with seasonal reproduction. Peak adsorption observed at 420 nm is not readily transfered to orthologs in other species via alignment as tuning residue formulas have not been described for this class of opsin.
A commentary piece assigns the paraventricular organ to the hypothalamus so even though no exact mammalian anatomical counterpart exists, the Allen Brain Atlas might show mouse expression of NEUR1 there, assuming the photoperiodic reproduction hormomal control mechanism is retained from the ancestral situation. (The paraventricular organ is known from teleost fish and frog as well as birds.) Photoentrainment of daily (circadian) activity rhythms is then quite distinct in terms of opsin use from photoreceptor control of seasonal reproduction. This fits with human NEUR1 transcripts most commonly recovered from testis, for example DB097202, and also with the phylogenetic range (back to amphioxus and sea urchin where unfortunately nothing is known).
The other 3 deeply conserved neuropsins found in nonmammalian vertebrates including birds were not considered in the quail study, leaving it unclear what functional roles might be left for them. NEUR4 even persists into some mammals. Much more work needs to be done on non-imaging opsins, given that they comprise the vast majority of the 24 vertebrate opsin loci. The neuropsins collapsed in mammals along with many other opsins (including imaging) in good agreement with the GT Wall hypothesis.
Neuropsin has all the classical attributes of a rhodopsin-class GPCR and indeed opsin photoreceptor: Schiff base lysine at expected position, standard tyrosine counterion and DRY motif, seven transmembrane configuration, disulfide at expected position, proximal glycosylation and distal palmitoylation and kinase sites. It is most closely related to peropsin and rgropsin in terms of blast clustering and intron positioning.
Its G-protein signaling partner is not really known though GNAQ is a good candidate among the 16 paralogs and is expressed in quail paraventricular organ. However the closest GNAQ paralogs, namely GNA11, GNA14 and GNA15, were not considered in the quail study.
Its evolution is illuminated by the massive comparative genomics study described here, which extracts and compares over 50 full length NEUR1 vsequences from various genomics projects. Neuropsin can be located outside chordates but not outside deuterostomes. However, like peropsin and rgropsin, it must have originated much earlier in pre-Bilaterans. Thus its absence in earlier diverging species must be due to gene loss because unrecognizability can be ruled out given test sensitivity. Some role in deuterostomes has persisted over many billions of years of branch length, perhaps correlating with their ciliary imaging vision and anti-correlating to protostomal rhabdomeric imaging.
Within placental mammals, neuropsin is extraordinarily conserved, with percent identity relative to human protein 96% averaged over 31 species (exceeding the 95% percentile of all coding genes proteomewide). That conservation drops considerably at marsupials and monotremes (86%), is less striking at tetrapods (78%), and not especially remarkable at teleost fish (68%). This pattern suggests neuropsin acquired significant new adaptive functionality on the placental mammal stem, leading to marked resilience to fixation of any further variation.
The structure of the neuropsin gene is rather odd at the 3' end. In human, a weak splice may have developed that results, after an intron of 12,244 bp, in a seventh very short coding exon continued by a long 3'UTR. However a stop codon is soon encountered if this splice is not taken (which it is in 6 of 7 transcripts). This results in two slightly different alternative carboxy termini sequences EEV* vs EEWE*. Very few transcripts exist in this region for any species but it appears that the ancestral form of the protein only utilized the initial stop codon in exon 6.
This feature GTatga is conserved in all species back to platypus; indeed an unexplained conservation in nucleotide sequence extends well beyond this. However the splice acceptor and WE* appear an option only back to lemurs, whereas the QEV* option (EEV* from tarsier to human) is available in all 38 available mammal genomic sequences. Oddly the carboxy terminus first becomes conserved in mammals.
A single mouse transcript also terminates early; four others continue on. No species other than rat has an available transcript in this region. No other rodent genomes could have an orthologous split exon: kangaroo rat and ground squirrel have frame shifts, pika lacks the splice acceptor, and rabbit and guinea pig have no homologous sequence.
This can be explained by independent origins of splice acceptors in various clades. However far more comparative transcripts are needed to understand the 3' end of this gene. It may be that the conserved intronic sequence following exon 6 is an accident waiting to happen as it conserves -- for whatever reason -- half of a splice site.
It must be said here that GenBank has a very muddled policy in the nr nucleotide division, not distinguishing real experimental transcript data from predictions, gene mRNA models, synthetic clones, the seemingly meaningless term cDNA, staff interpretations of genomic data, and poorly documented third-party submissions.
In summary, UniProt, NCBI, UCSC, and Ensembl take the atypical primate-specific splice variant as the canonical form of the gene. This gene model cannot however serve as ancestral.
The 5' end has its oddities as well. It seems that in gallinaceous birds, the initial methionine could be further upstream by 22 residues. However the good agreement of amino acids could be an artefact of good conservation of good coding promoter or initiation sequence. There is no support for this extension in finch, lizard, frog or platypus or in the chicken transcript NM_001130743. The experimental transcript here, which maps accurately into the chicken genome, shows an upstream exon separated from the first coding exon by a 9181 bp intron with 14 bp of bp preceding the iMet. Thus the gene model assumed for quail is likely in error. A smaller extension of 3 residues is possible in all birds plus platypus. This extension has been adapted in the reference sequences below.
Note that the new turkey genome is now available for Blat searching. This provides an important addition to neuropsin comparative genomics. Turkey contains all four neuropsin paralogs as expected.
Novel neuropsins in amphioxus and sea urchin
The genome of Branchiostoma (amphioxus, lancelet) contains two distinct neuropsins about 75% identical to each other and 42% to human. These cluster unambiguously with vertebrate neuropsins and share critical conserved residues. An extra intron distinguishes them from the vertebrate neuropsin pattern. Recall Branchiostoma species has three rather diverged (and well-studied) peropsins but no evident Rgr opsin. These raises the question whether neuropsin and peropsin developed substantial photoreceptor roles in this species as an alternative to the ciliary imaging opsin pathway seen already at lamprey divergence. Sea urchins, but not acornworm Saccoglossus, contain a single neuropsin that is quite diverged.
These neuropsins are newly reported here, meaning they were not localized in recent in situ hybridization studies. That's especially unfortunate in view of the antecedent role the Branchiostoma ancestral node plays in the evolution of chordate eye and the complexities of photoreceptor tissues in the extant species.
PhyloSNPs in vertebrate neuropsins
Alignment analysis coming shortly. Neuropsin has rather few of them.
position ...................................................................................................1.........1.........1.........1.........1.........1.........1.........1......1.. position .........1.........2.........3.........4.........5.........6.........7.........8.........9.........0.........1.........2.........3.........4.........5.........6.........7......7.. position 123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567.. excMemCy eeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMcccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeee.. keyResid ...GLC.................................................................................................diS..cIon.................DRY............................................... exonNumb 111111111111111111111111111111111111111111122222222222222222222222222222222222222223333333333333333333333333333333333333333333333333333333334444444444444444444444444444444444444.. 10homSap MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGISVVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPE.. 11panTro ................................................................................................................................................................................... 12gorGor ............................................................................................................................................------------------..................... 13ponPyg ................................................................................................................................................................................... 14nomLeu ................................................................................................................................................................................... 15rheMac .................................I............................................................................................................................................A.... 16papHam .................................I............................................................................................................................................A.... 17calJac ......S.......................................................................................................................................................................A.... 18tarSyr ....................................................................................I.........................................................................................A.... 19otoGar ............LR..........................................................V...................................................................----------------------..M......E....... 20micMur ......V.................................I..........................................---------------------------------------------------------..............L....V..............A.... 21tupBel ............S.............................................................................................................................................V...................A.... 22musMus ....................E..................................................................................F..................................................V...................A.... 23ratNor ....................E..................................................................................F..................................................V...................A.... 24speTri ............H.......E................................................................................................................................F....V...................A.... 25dipOrd ..F....GT.GQG.....PEE...T........I........................................................................................................................V........................ 26cavPor .......P..N.H..R..Q.E...V.......................V...........................I..................R.............................V.................................V..............A.... 27oryCun ............H......E..............................................................................................................................R.......L...................A.... 28ochPri ....D.......H....F.........................................................................................L....D.....................------......R.......V...................A.... 29canFam .......R...........E........................................................I.............................................................................V...................A.... 30felCat .......P...........E.................................................--------------.......................................................................V...................A.... 31bosTau .......P.P...R.P........................................................V...I................................................................I............V.....A.............A.... 32turTru ......................K..........I.............................................................................V...................A.... 33susScr .......P.P...R.....E....................................................V...I.............................................................................V.....A.............A.... 34vicVic .......P.P...R.RH...............................L..................................................M........................................-------------------------------------.. 35equCab ............................................................................I.............................................................................V........................ 36myoLuc ............G.....Q.............................V.......................V...........................................................................T.....F........................ 37pteVam ......V.....H....V..............................V...........................I.............................................................................V........................ 38sorAra ............N..........................................................................................M.................................................VV...................A.... 39eriEur .S..Q.......G.........................................................................................................................................L...V...................A.... 40loxAfr .T.....P...D...Q..Q.....T.................................C..............................V................................................................V...................A.... 41proCap .T....V..E.D..S.........T................V......C.........Y..............................I...S..........................................H...-------------------------------------.. 42echTel .......P...NS...........V.........G.....I.................Y....................................S.......T..................................................V........................ 43dasNov ...........D...............................................K..............................................................................................V........................ 44choHof ......G....DS....F......................I....................R............................................................................................V.............L.......... 45monDom .....SVS...DYI..............................V...L.....I....K............V......................S.......V................................H....T....H..F....L.....T..A.V.FA.V.S.A.... 46macEug .V...L.....I....K............V..............................V................................H....T....H......VI.....T..A....A...N.A.... 47ornAna MT.YS.PQLGDY......E....V............V..V...V...L.....I.................V..................V...........M................................H....T....H.......I........A........N.A.... 48galGal ..SDCNSSS.E.Y....MQQE........R...II......V......L.....IF...K............V................S...F.S...I...M................................H.A..T....H..F....L.....T..A.V.FA.V.S.A.... 49taeGut ..SEYNNSS.E.YI....QEE........R...II.............L.....IF...K............V................S...F.S...M......C.............................H....T....H..F....I.....M..A.V.FA.V.S.A.... 50anoCar .EQGQNISS..DN----QQEE........V...I...V..LV......L.....I...TQ.....K......V................S..AF.S...I...S.............I..............F...H....T....H.VF...GI..S..A..A.I.FA.F.N.A.... 51xenTro ..G.SSYREESGYI...E..S........R...IF..V..MA......L.....I..ACS........................T....A.V...S.......NA..................L.V..........H.R..T....R..F.A..V.....TL.A.L....V.N.A.... 52danRer .E-NET-SISSGYI....LR.........K...I..A..ILV.....AT.....M..TFK..T..K.P....L...IF.F....S....F.V.S.S...L...Q...Y.................I..F.......H.R..T....H..FLSVVF.....A..A...V..W.N.A.... 53pimPro .E-NDT-SIPSGYV....LR.........K...I..A..ILV..V..AT.....I.QTIK..T..K.P.F..L....F.F....T....F.V.S.S...L...Q...Y.................I..........H.R..T.F..H..FL..VFT.L..A..A...V..W.N.A.... 54takRub .E-NET-WTHSSYV....LR......R..K...I..AL.IC...LM.AT........TFK..T..K.P.L..L...IF.F....T....F.V.SLS...L...E...F.................V..........H.R..A....H..FL...SV....A..A......W.S.A.... 54tetNig .D-NET-RSHPSYV....LR......R..K...I..A..IF...VM.AT........TFK..T..K.P.L..V...IF.F....T....F.V.SLS...L...E...F.................V..........H.R..T....H..FV...LV....A..A......W.S.A.... 56gasAcu .E-NET-WTHPSYI....LR......R..K...II.A..IC....M.AT.....I..TIK..S..K.P.L..V....F.F....T....FVV.S.A...L...E...F.................V..........H.R..T....Q..FL..VFV.M..A..A......W.N.A.... 57oryLat .E-N.S-W.HSSYV....LR......R..K...I..A..IL....M.AT.....I..TIK..S..K.P.L..V....F.F....T....FVV.S.S...L...E...F.................V..........H.R..T....Q.IFL..VFV.I..A..A......W.S.A.... 59calMil .TFDNSTALYSGYWL.DSLH....V........IISAC..IVT.L...L.....I.L.ITQ.R..K.P..LIT...IS.F.M..G.Q..L.....S...I...V....H................V..........H.Q..S..Q.R.VFMS..F..F..A..A......W.N.A.... 10homSap MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGISVVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPE.. phyloSNP ..................AA......B......A..A..B.....B.BB.........B......C.A.........B.A....A..........B......................................A...B..A.......A.............A............... .. .. position 1.1.........1.........2.........2.........2.........2.........2.........2.........2.........2.........2.........2.........3........3..........3.........3.........3.........3.....3 position 7.8.........9.........0.........1.........2.........3.........4.........5.........6.........7.........8.........9.........0........1..........2.........3.........4.........5.....6 position 89012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456 excMemCy eeeeeeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMcccccccccccccccccccccccccccccccccccccccccccccc* keyResid ...diS................................................................................................................K........................................... exonNumb 44444444444444444444444444444444444444444444444444444444444444444444444444455555555555555555555555555555555555555555555555555555555555555555555555555555555556666666666666666666666 10homSap PFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKS-LEGFRLHTVT-TVRKSSAVLEIHEEV* 11panTro .................................................................................................................................................E.....-..........-...............* 12gorGor ...............................................................P.....................................K.................................................-....Q.....-...............* 13ponPyg .....................................................................................................S.................................................-..........-...............* 14nomLeu .......................................................................................................................................................-..........-S..............* 15rheMac .......................................................................................................................................................-..........-...............* 16papHam .......................................................................................................................................................-..........-...............* 17calJac .......................................................................................................................................................-..D.......-...............* 18tarSyr ......................................................G.........T......................................................................................-..D.......-...............* 19otoGar ..-...........-.L...............................................G................................................................................T.....-..D.......-............Q..* 20micMur .......................................................I.............................................................................----------------------H......-A...........Q..* 21tupBel ....................I..................................I..........................................................................................................-............Q..* 22musMus ................G.......S.................A...........................V...............................................................R.....A...RG.....-..D.......-............Q..* 23ratNor ................G.......S.................A...........................V................................N..............................R.........R......-..D.......-A...........Q..* 24speTri .......................-..............E..............E..............-------.......................................S....................................-..D.......-A.......V...Q..* 25dipOrd ................LA.................S...........................P.......................................................................................-.......................Q..* 26cavPor ................A...I...H...........M..................I.............................................................................SR.....NA.........-..D.......-.D...-......Q..* 27oryCun ........................................................................................................................................S..R.S.........-..D.......-............Q..* 28ochPri ................................................................G.......................................................................S..R.....Q.....-..D.......-............Q..* 29canFam ................L...I.........................................................................................................................R........-..D...N...-............Q..* 30felCat ....................I..................................................................................................................................-..D.......-............Q..* 31bosTau ....................I.................................................V................................................................................-..D.......-..........V.Q..* 32turTru ....................I..........................................P..........................................................V............................-..D.......-............Q..* 33susScr .....................................................................................V.................................................................-..D.......-...........RQ..* 34vicVic ---------------------------------------------------------------------------............................................................................-..D....A..-............Q..* 35equCab ....................I...........................................G.....V................................................................................-..D.......-............Q..* 36myoLuc ...............T....I.................................K.........-----------.............................................S..............L........R......-..N.......-............Q..* 37pteVam ....................I...........................................G..M.........................................................................S..R......-..D.....I.-...EA.......Q..* 38sorAra ....................I..................................................................................N..............................R.....S...R......-.DD.......-...E........Q..* 39eriEur ................L...I.................................K............M..............................................................................N....-.KDY...................q..* 40loxAfr ....................I..................................I...........M............................................................................R......-..........-..K.......V.Q..* 41proCap ---------------------------------------------------------------------------..............................V.................................R.R..R...E..-...V......-............Q..* 42echTel .......................................................I...........M............................................................................................I.-...........HQ..* 43dasNov ................................................................................................................................................R......-..D.......-...E......V.Q..* 44choHof ...................................................................M.......................................................................R....R......-F.........-............Q..* 45monDom .................A..A.V.S.......F.............L.....T.....Y.T..QN..I.................................Q.....V.F.......................C......S..Q..A..E.-.RTY......-...R........Q..* 46macEug .................T..T...............................T..........Q..............................................................................................RHTVSTIRKSSSVSETYQ..* 47ornAna .................A..A...............................T..........QN....................................Q.......F.......................CRIS..RL..P.TG..E.-.KNS.S.SMS-.I..P...SGP.Q..* 48galGal .................A..A.V.S.......F.............L.....T.....Y.T..QN..I.................................Q...V...F.......................C.....RS..P.TLQ...S.KES.MY.IS-SH.D.A.LSGTQL..* 49taeGut .................A....V.S.....................L.....T.....Y.T..QN..I.................L...................V...F......................ECRL...RP..* 50anoCar ..............G..A..A..........V......M.C........Q..T.....Y.T..QNQ..................MF...................V..KV..I.............V......C.S...RP.N.QPLQ..NSR* 51xenTro ....T............K..I.V.SM......F..M......A.........A.......T.NQNN.T..I.................F............Q......E......MM....S...........C.P...KKD--.SLQNTT----S.VY.IS-.F...TTSAR* 52danRer ............T....S..S.VMCM.....IF..VI......M..F.....A...S...T.NKNN.S............................M....E...V..PV..........S............C.KK.VKSCCFQ.WR..KPSKTS.FY.ISGSIKQRPGD-.ASI.I* 53pimPro .................S..S.VMCM.I...V...G.......M..LQ....AQ..S...TQNKNK.H............................V....D...V..SI..........S............C.KN.AKLSCFQ.WS.RKHYKTS.FYSISASMK.RP.N-.VPT.I* 54takRub .................S..S.VMA......I...GI......M..F.....A..IS...A..RN..D..I.........................I....E...V..PV..I.......S..........V.V.TS.TNFSCC..L.ERIHFRKS..Y.ISGSL.DPLPPK.A.I.M* 54tetNig ....A............S..S.VMA......V...GI.....IM..F.....A..IS...A..KN..S..I.........................V....E...V..PV..I.......S..........A.L.TS.TSSSCC..L.ERVLFRKA..Y.ISGSL.DTLPPK.A.I.M* 56gasAcu .................S..S.VVA......V..AGI......M..F.....A..ISN..A..KN..N..I.........................V....E...V..SV..I.......S..........L.L.NS.MKSSCF.GL..PRHFRKS.FY.ISGS.KDNTTAK.AQI.M* 57oryLat .................S..S.VMS......V..AGI......M..C.........SS..A..KN..T..I.........................V....E......PV..I.......S.........LV.L.NS.-S.CCA.VIR.RTHFRNS.FY.ISGSLKDTAPAK.A.I.I* 59calMil .............RV..S.LI.V.T.........III.....I.........A..........QNH.S...N..........................................................................................................* 10homSap PFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKS-LEGFRLHTVT-TVRKSSAVLEIHEEV* phyloSNP ...........................................B............B......CA.......................................................B.................................B...B.BBA............A..*
Neuropsin (NEUR1 compared to newropsin (NEUR2)
Newropsins are a new opsin gene family -- first reported here -- most closely related to neuropsins (42% percent identity) and next to melanopsins and peropsins. Like so many opsin families, they persist from chondrichthyes to archeosaurs but vanish without a trace in platypus, marsupials, and placentals. (The syntenic order B4GALT6 NEUR2 KIAA1012 remains conserved in mammals but no NEUR2 debris remains.) Newropsins retain many key attributes of GPCR signaling proteins and indeed opsins such as the seven transmembrane arrangement, Schiff base lysine, counterion tyrosine, amino terminal glycosylation site, and disulfide but have a very odd replacement of the G-protein binding site DRY with (invariantly conserved) VCC.
This motif must be an ancient derived feature that followed the gene duplication event with neuropsin since the much older DRY could not plausibly have re-evolved in neuropsin from VCC. Newropsins very likely link covalently via their orthologous Schiff base lysine with a retinal and interact with light according to some action spectrum. The VCC motif has been conserved over billions of years of branch length so cannot reflect simple loss of functionality; however its signaling capabilities if any are unclear.
It seems feasible that non-signaling opsins have become photoisomerases. Their substrate, while evidently involving an aldehyde capable of forming a Schiff base with the conserved lysine and likely interconverting cis/trans double bonds, need not be part of a ciliary opsin replenishment cycle. Other metabolic derivatives of beta-carotenes and lincopenes such as retinoic acid intermediates might be the substrates.
It must be recalled too that quite a diversity of photoreceptive retinoids are used in non-mammalian species, for example 9-cis isorhodopsin, porphyropsin (or 3-dehydroretinal vitamin A2) in freshwater fishes and some frogs, 3-dehydroretinal in freshwater crayfish, all-trans-5,6-dihydroretinal in cottoid fish in Lake Baikal and so forth. These are spectrally influenced by the surrounding opsin. Here too the negatively charged counterion, Glu113 in bovine rhodopsin, an alternative glutamate in melanopsins, or a special bound chloride ion. The counterparts of these are not known in neuropsins.
Below, conserved residues are shown for NEUR2 relative to human neuropsin (which represents that family accurately). Newropsin orthologs are rather rapidly diverging, especially in teleost fish. Transmembrane domains on newropsins were assigned by homology to neuropsin (taken from SwissProt ab initio annotation consistent with experimentally determined bovine rod rhodopsin). Newropsin is relatively truncated amino terminally but very extended in a highly variable manner carboxy terminally. That extension would lie in the cytoplasm and possibly be removed endoproteolytically. The early glycosylation is present in all species but appears shifted distally by four residues in tetrapods relative to fish.
According to transcript annotations, newropsin is expressed in zebrafish anterior segment (minus lens), fish brain and testes (Pimephales and Oncorhynchus), embryo, oviduct and fat body (Xenopus). These, while familiar sites from other opsins, provide only meagre constraints on possible newropsin functionality and association with photoreceptive tissues.
The intronation pattern is not a perfect match to neuropsin as might be expected. Some of the difference is lineage-specific, such as a gain in zebrafish, but other differences may be much older. Unless homologs have been retained recognizably in earlier diverging species, it won't be feasible to date the original gene duplication. No NEUR2 could be located in lamprey, tunicate, or lancelet.
Newropsin is further evidence that the neuropsin/peropsin/rgropsin group played a much greater role in ancestral vertebrate photoreception (which persisted into contemporary species), roles which were lost in stem mammals. That is quite similar to the ciliary opsin story. Overall mammals have retained less than half (7 of 17) of the vertebrate opsin repertoire. Such widespread gene loss is fully consistent with an old inference of a nocturnal era during which no selective pressure existed to maintain these photoreceptors.
position ...................................................................................................1.........1.........1.........1.........1.........1.........1.........1........1. position .........1.........2.........3.........4.........5.........6.........7.........8.........9.........0.........1.........2.........3.........4.........5.........6.........7........8. position 123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890 excMemCy eeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMcccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeeeee keyResid ....GLC.........glc.glc.................................................................................diS..cIon.................DRY?.............................................. NEUR2_galG MDPSFANST-FQSKITEAADIVVGTCYMVFGICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHRWLYGKHICLFYAFCGLFFGICSLSTLTLLSVVCCLKICFPAYGNRFRRKHGQILIACAWTYAAIFACSPLAHWGEYGEEPY NEUR2_anoC MESYFANTT-FHSKITEAADVIVGVFYIVFGICSFCGNSILLYVSYKKKNLLKPAEYFMINLAISDLGMTLTLYPLAVTSSLAHRWLFGQQVCLFYAFCGVFFGVCSLTTLTLLSIVCCLKICFPVYGNRFRPGHGWILIACAWVYAAIFAFSPLAHWGEYGAEPY NEUR2_xenT MGNKSDASA-FYSSISETDDIVLGVLYSVFGLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHRWLFDKVTCNYYAFCGMLFGLCSLTNLTVLSSVCCLKVCYPAYGNKFSTAHSRILLLGIWAYAGLFATAPLADWGKYGPEPY NEUR2_danR MGNVSKTAL-FMSTISRQHDILMGSLYSVFFVLSLLGNGMLLFVAYRKRSSLKPAEFFVVNLSVSDLGMTLSLFPLAIPSALAHRWLFGEITCLCYAVCGVLFGLCSLTNLTALSSVCCLKVCFPNYGNKFSSSHACVMVIGVWCYASVFAVGPLVHWGSFGPEPY NEUR2_pimP MGNVSETAL-FVSTISRQHDILMGSLYSVFCVLSLLGNGMLLFVAYRKRSSLKPAEFFVINLSVSDLGMTLSLFPLAIPSALAHRWLFGEVVCLCYAVCGVLFGLCSLTNLTALSSVCCLKVCCPNYGNKFSSNHACVMVIGVWCYASVFAVGPLIRWGSFAPEPY NEUR2_tetN MGNASDTSDAFNSKISKEHDFLIGSIYSVFCVLSLMGNCILLLVAHHKRSTLKPAEFFIVNLSISDLGMTLTLFPLAIPSSFSHRWLFGEIACQLYATCGVLFGLCSLTNLTVLSSVCCLKVCLPNLGSKFSSSHARLLVAGVWGYASVFAVGPLVQWGHYGPEPY NEUR2_takR MGNASEASDIFLSKISKEHDILIGSIYSVFGLLSLAGNCILLLVAYHKRSMLKPAEFFIINLSISDLGMTLTLFPLAIPSSFSHRWLFGEITCQLYAMCGVLFGLCSLTNLTALSLVCCLKVCFPNHGSRFSSSHARLLVVGVWCYASVFAVGPLVQWGHYGPEPY NEUR2_gasA MGNASDTSAVFASTISKERDILMGSLYSVFGVLSLVGNCILLLVAYHKRSTLKPAEFFIINLSISDLGMTLSLFPLAIPSAFKHRWLFGELTCQLYAMCGVLFGLCSLTNLTALSFVCCLKVCFPNHGNRFSSSHARLLVVAVWGYASVFAVGPLARWGRYSPEPY NEUR2_oryL MGNVSDTSSLFASSISREHDILMGSLYSVFGLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHRWLFGEITCQLYAMCGVLFGLSSLTNLTALSLVCCLKVCFPNHGNKFSFSHARLLVAGVWCYASVFAVGPLARWGRYSAEPY NEUR2_calM GILSLVGNSVLLFVAYRKRQILKPAEYFVANLAVSDISMTVTLLPLAISSNFSHRWLFVSKPCMYYGFCSMLFGICSLTNLTVLSTVCCMKVCFPAYMSVVMIV-MFLLAWSPYSIVCLWASFGNPKLIPPAMAII NEUR1_homSa MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGISVVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPF NEUR1_canFa MALNHTARPQDERLPHYLREGDPFASKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGISVVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPF NEUR1_musMu MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGISVVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPF NEUR1_loxAf MTLNHTAPPQDDRLPQYLQDGDPFTSKLSWEADLVAGFYLTIIGILSTFGNGYVLYMSCRRKKKLRPAEIMTINLAVCDLGISVVGKPFVIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSYGVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPF NEUR1_monDo MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTIIGVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGISVVGKPFTIISCFSHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSYGTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPF NEUR1_ornAn MTNYSAPQLGDYLPHYLREGDPFVSKLSWEADLVAGVYLVIIGVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGISVVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSYGTWLKRHHAYICLAIIWAYASFWATMPLVGLGNYAPEPF NEUR1_calMi MTAFDNSTALYSGYWLHDSLHGDPFVSKLSWEADIISACYLIVTGLLSTLGNGYVIYLSITQKRKLKPPEILITNLAISDFGMSVGGQPFLIISCFSHRWIFGWVGCRWHGWAGFFFGCGSLITMTVVSLDRYLKICHLQYGSWLQRRHVFMSLAFIWFYAAFWATMPLVGWGNYAPEPF NEUR1_galGa MASDCNSSSQEEYLPHYMQQEDPFASKLSREADIIAGFYLTVIGILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGISVVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAYGTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPF NEUR1_xenTr MAGNSSYREESGYIPHYERDSDPFASKLSREADIFAGVYLMAIGILSTLGNGYVIYMACSRKKKLRPAEIMTINLAVCDLGISVTGKPFAIVSCFSHRWVFGWNACRWYGWAGFFFGCGSLITLTVVSLDRYLKICHLRYGTWLKRRHAFIALAVIWAYATLWATLPLVGVGNYAPEPF NEUR1_danRe MENETSISSGYIPHYLLRGDPFASKLSKEADIVAAFYILVIGILSATGNGYVMYMTFKRKTKLKPPEIMTLNLAIFDFGISVSGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSFDRYLKICHLRYGTWLKRHHAFLSVVFIWAYAAFWATMPVVGWGNYAPEPF NEUR1_takRu MENDTSIPSGYVPHYLLRGDPFASKLSKEADIVAAFYILVIGVLSATGNGYVIYQTIKRKTKLKPPEFMTLNLAVFDFGISVTGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSLDRYLKICHLRYGTWFKRHHAFLCLVFTWLYAAFWATMPVVGWGNYAPEPF NEUR1_tetNi MENETWTHSSYVPHYLLRGDPFASRLSKEADIVAALYICIIGLMSATGNGYVLYMTFKRKTKLKPPELMTLNLAIFDFGISVTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRYGAWLKRHHAFLCLASVWAYAAFWATMPLVGWGSYAPEPF NEUR1_gasAc MDNETRSHPSYVPHYLLRGDPFASRLSKEADIVAAFYIFIIGVMSATGNGYVLYMTFKRKTKLKPPELMTVNLAIFDFGISVTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRYGTWLKRHHAFVCLALVWAYAAFWATMPLVGWGSYAPEPF NEUR1_oryLa MENETWTHPSYIPHYLLRGDPFASRLSKEADIIAAFYICIIGIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGISVTGKPFFVVSSFAHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRYGTWLKRQHAFLCLVFVWMYAAFWATMPLVGWGNYAPEPF NEUR1_pimPr MENTSWPHSSYVPHYLLRGDPFASRLSKEADIVAAFYILIIGIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGISVTGKPFFVVSSFSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRYGTWLKRQHIFLCLVFVWIYAAFWATMPLVGWGSYAPEPF NEUR1_anoCa MEQGQNISSQDDNQQEEDPFASKLSVEADIVAGVYLLVIGILSTLGNGYVIYMSTQRKKKLKPAEIMTVNLAVCDLGISVVGKPFSIIAFFSHRWIFGWSGCRWYGWAGFFFGIGSLITMTAVSLDRYFKICHLSYGTWLKRHHVFICLGIIWSYAAFWATIPFAGFGNYAPEPF position 1.........1.........2.........2.........2.........2.........2.........2.........2.........2.........2.........2.........3........3..........3.........3.........3.........3.....3 position 8.........9.........0.........1.........2.........3.........4.........5.........6.........7.........8.........9.........0........1..........2.........3.........4.........5.....6 position 012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456 excMemCy eeeeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMcccccccccccccccccccccccccccccccccccccccccccccc* keyResid .diS................................................................................................................K............................................................ NEUR2_galG GTACCIDWQSTNVDVMSMSYTVVLFVLCFILPCGVIVTSYSLILVTVKESRKAVEQHVSGPTRINNVQTITAKLSIAVCIGFFAAWSPYAIIAMWAAFGSIDKIPPLAFAIPAVFAKSSTLYNPIIHLLLKPNFRSNIAKDFTVIQQLCVR---CCFCVKELQ--TYRSTFNTGLRTFKG NEUR2_anoC GTACCIDWRISNMKKTAMSYTTALFVFCYIIPCGIIITSYTLILITVKDSRKAVEQHALGPTRMSSVHTITAKLSIAVCIGFFVAWSPYAIIAMWAAFGSIDMIPPLAFAVPAVFAKSSTLYNPAMYLFLKPNFRSTIAKDLTVLHRLCLK---SCFCPRGMQNCSYRSALEAPLKSFKG NEUR2_xenT GTACCLDWEASYRERKALSYTISLFVFCYLIPSSLIFISYTLIFVTVKGARRAVQQHLSPQAKGSSIHSLIIKLSIAVCIGFLIAWTPYAIVAMMAAFGDPTKIPSLVFALAAAFAKSSTIYNPVVYLLLKPNFLNVVTKDLTLFQTMCAV---VCGWCR-----TPAVKTPCPHKDLKT NEUR2_danR GTACCINWYTPSHDALAMSYIISLFIFCYVVPCTIIILSYTFILVTVRGSQQAVQQHVSPQTKVTNAHALIVKLSVAVCIGFLTAWSPYAIVAMWAAFSANEQVPPTAFALAAIMAKSSTIYNPMVYLLFKPNFRKSLSQDTQMFRHRICLSHSKASPSPGMKDQERQSSQQCNNKDGSI NEUR2_pimP GTACCINWYIPSHDALAMSYIISLFIFCYVVPCTIIILSYTFILLRVRGSRQAVQKHVSPKTKETNAHTLIVKLSVAVCIGFVTAWSPYAVVAMWAAFSANEPVPPTAFALAAILAKSSTIYNPMVYLLFKPNFRKILSQDTQNIRHRMCVSHSKASPTPEIK---AQSSQQC--KDATI NEUR2_tetN GTACCINWQAPNHELSSLSYIVCLFLFCYVLPCAIIILSYTCILMTVRGSRQAIQQHVSPQTKTANAHALIVKLSVAVCIGFLGAWSPYAVVAMWASFGDATWVPPDAFAIAAILAKSSTIYNPLVYLLCKPNFRECLYKDTSTLRQRIY----RGSPLSGPRDRSGGVTQR--HKDLSV NEUR2_takR GTACCIDWRAPNHELSSLSYIVCLFFFCYVLPCATIILSYTCILMTVRGSRQAIQQHVSPQTKTANAHSLIVKLSVAVCIGFLGAWSPYAIVAMWAAFGDATWVPPDAFAIAAILAKSSTIYNPVVYLLCKPNFRECLYKDTSTLRQRIY----RGSPQSEPRERFGGTSQR--HKDLSI NEUR2_gasA GTACCIDWHAPNHELAALSYIVCLFVFCYALPCATIFLSYTFILLTVRGSRQAVQQHVSPQTKTTNTHALIVKLSVAVCIGFLGAWTPYAVVAIWAAFGDATLVPPDAFALAAMFAKSSTIYNPVVYLLCKPNFRACLYRDTTLLRQRIY----RGSPRSEPKAHFGSTSQR--NKDMSV NEUR2_calM APLFAKSSTFYNPCIYVISYTMTVIAVNFVVPLSVMFFCYYNV NEUR2_oryL GTACCIDWHAPNHELWALSYILCLFIFCYALPCTIIFLSYAFILLTVRGSRQAVQQHVSPQTKTTNAHTLIVKLSVAVCIGFLGAWTPYAVIAMWAAFGDATQVPPTAFALAAVFAKSSTIYNPMVYLLCKPNFRECLCRDTSLLRHMIY----RGSP--QPQERFGSDSRR--NKDITA NEUR1_homSa GTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKA-TKKKSLEGFRLHTVT-TVRKSSAVLEIHEEV NEUR1_calJa GTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKA-TKKKSLEDFRLHTVT-TVRKSSAVLEIHEEV NEUR1_canFa GTSCTLDWWLAQASLGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGRLKA-TKKKSLEDFRLNTVT-TVRKSSAVLEIHQEV NEUR1_musMu GTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQAGGLRG-TKKKSLEDFRLHTVT-TVRKSSAVLEIHQEV NEUR1_loxAf GTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHMLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLRA-TKKKSLEGFRLHTVT-TVKKSSAVLEVHQEV NEUR1_monDo GTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPVQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCQSGGQKA-AKKESLRTYRLHTVT-TVRRSSAVLEIHQEV NEUR1_ornAn GTSCTLDWWLAQASVAGQAFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQNSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPIQFSVVPTLLAKSAAMYNPIIYQVIDCRISCCRLGGPKT-GKKESLKNSRSHSMS-TIRKPSAVSGPHQEV NEUR1_calMi GTSCTLDWWLARVSVSGLIFVLTILFFCLLLPIIIIVFSYIKIIAKVKSSAKEVAHFDSRIQNHHSLEMNLTK NEUR1_galGa GTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGPKTLQKKSSLKESRMYTIS-SHRDSAALSGTQLEV NEUR1_xenTr GTTCTLDWWLAQASVKGQIFVLSMLFFCLLFPTMVIVFSYAKIIAKVKSSAKEVAHFDTRNQNNHTLEIKLTKVAMLICAGFLIAWFPYAVVSVWSAFGQPDSIPIELSVVPTMMAKSASMYNPIIYQVIDCKPACCKK------DKSLQNTTSRVYTIS-TFRKSTTSAR NEUR1_danRe GTSCTLDWWLTQASVSGQSFVMCMLFFCLIFPTVIIVFSYVMIIFKVKSSAKEVSHFDTRNKNNHSLEMKLTKVAMLICAGFLIAWIPYAVVSVMSAFGEPDSVPIPVSVVPTLLAKSSAMYNPIIYQVIDCKKKCVKSCCFQAWRKKKPSKTSRFYTISGSIKQR-PGDEASIEI NEUR1_takRu GTSCTLDWWLAQASVSGQSFVMCMLIFCLVLPTGVIVFSYVMIILQVKSSAQEVSHFDTQNKNKHHLEMKLTKVAMLICAGFLIAWIPYAVVSVVSAFGDPDSVPISISVVPTLLAKSSAMYNPIIYQVIDCKKNCAKLSCFQAWSKRKHYKTSRFYSISASMKKR-PANEVPTEI NEUR1_tetNi GTSCTLDWWLAQASVSGQSFVMAILFFCLILPTGIIVFSYVMIIFKVKSSAKEISHFDARIRNSHDLEIKLTKVAMLICAGFLIAWIPYAVVSVISAFGEPDSVPIPVSVIPTLLAKSSAMYNPIIYQVVDVKTSCTNFSCCKALKERIHFRKSRLYTISGSLRDPLPPKEAHIEM NEUR1_gasAc GTACTLDWWLAQASVSGQSFVMAILFFCLVLPTGIIVFSYIMIIFKVKSSAKEISHFDARIKNSHSLEIKLTKVAMLICAGFLIAWIPYAVVSVVSAFGEPDSVPIPVSVIPTLLAKSSAMYNPIIYQVADLKTSCTSSSCCKALKERVLFRKARLYTISGSLRDTLPPKEAHIEM NEUR1_oryLa GTSCTLDWWLAQASVSGQSFVVAILFFCLVLPAGIIVFSYVMIIFKVKSSAKEISNFDARIKNSHNLEIKLTKVAMLICAGFLIAWIPYAVVSVVSAFGEPDSVPISVSVIPTLLAKSSAMYNPIIYQVLDLKNSCMKSSCFKGLKKPRHFRKSRFYTISGSVKDNTTAKEAQIEM NEUR1_pimPr GTSCTLDWWLAQASVSGQSFVMSILFFCLVLPAGIIVFSYVMIICKVKSSSKEVSSFDARIKNSHTLEIKLTKVAMLICAGFLIAWIPYAVVSVVSAFGEPDSIPIPVSVIPTLLAKSSAMYNPIIYQLVDLKNSC-STCCAKVIRKRTHFRNSRFYTISGSLKDTAPAKEAHIEI NEUR1_anoCa GTSCTLDWWLAQGSVAGQAFILNILFFCLVLPTAVIMFCYVKIIAKVQSSTKEVAHYDTRIQNQHVLEMKLTKVAMLICAGFMFAWIPYAVVSVWSAFGRPDSVPIKVSVIPTLLAKSAAMYNPVIYQVIDCKSACCRPGNLQPLQKKNSR
Neuropsin (NEUR1 compared to newtopsin (NEUR3)
A third paralogous family NEUR3 was reported in July 2008 and characterized by syntenic relations and expression in chicken. However in chicken, NEUR1 actually is most abundant of the three paralogs in developing and early post-hatch neural retina, notably in differentiating ganglion cells and amacrine cells.
Recoverable sequences range from lamprey to shark to fish (where a further lineage-specific tandem duplication has occurred) to frog to sauropsids, with the gene again lost in all mammals including platypus. This locus exhibits an ancestral fusion of exon 2-3 relative to neuropsin and newropsin. The tandem duplication in ray-finned fish could easily be mistaken for a whole genome duplication effect -- however there is no sign of such an event for any of the 3 paralogs in any of the 5 fish with assembled genomes.
The NEUR3 group could possibly signal through heteromeric G protein. The DRY motif is a bit unusual, consisting of V/I R F/Y whereas the YNPxIY x aliphatic is fairly conventional. The Schiff base K is preserved in both NEUR3a and NEUR3b sequences. The counterion glutamate or chloride ion has not been determined.
The other oddity is NEUR1 and NEUR3 are on the same chromosome for all species examinable. They are separated by a million or so bp as well as other coding genes. This possibly represents an old tandem duplication that experienced subsequent rearrangements. On the whole, synteny has been quite well preserved for all NEUR genes:
NEUR3_galGal +CRISP +RHAG ..... +MUT -NEUR3 ..... +CDC5L -SUPT3H +RUNX2
NEUR3_anoCar -CRISP2 +RHAG ..... +Mut -NEUR3 ..... +CDC5L -SUPT3H +RUNX2
NEUR3_xenTro -CRISP2 -RHAG -PPHLN +MUT -NEUR3 ..... +CDC5L ..... .....
NEUR3a_danRer +XRN2 -TSTA3 +MGST3 +MUT -NEUR3a -NEUR3b -NAPB +DNMT3A .....
NEUR3a_tetNig ..... ..... ..... +MUT -NEUR3a -NEUR3b ..... ..... +RUNX2
NEUR1_galGal ..... +TNFRSF2 +CD2AP +GPR111 +NEUR1 ..... +MRPL19 ..... .....
NEUR1_anoCar -GPR111 -TNFRSF2 +CD2AP ..... +NEUR1 -SPATS1 +MRPL19 ..... +ITSN2
NEUR1_xenTro ..... +TNFRSF21 +CD2AP +GPR111 +NEUR1 -PTCHD1 +MRPL19 ..... +ITSN2
NEUR1_danRer +GPR111 -TNFRSF21 +CD2AP ..... +NEUR1 +CNIH2 -LBR -ENAH +ITSN2
NEUR2_galGal -DSC1 +DSG2 +TTR -B4GALT6 -NEUR2 -K1012 +RNF138 ..... .....
NEUR2_anoCar -DSC1 +DSG2 +TTR -B4GALT6 -NEUR2 -K1012 +RNF138 ..... .....
A proposed revised terminology for this family follows. Note NEUR2 and NEUR3 will never receive official HGCN nomenclature because (like thousands of amniote ancestral genes) they are absent in human and mouse. Here lower case a and b are used in the case of lineage-specific duplications, with a reserved for the copy with higher blastp score to human (or, if absent, nearest species).
Gene Protein HGCN Synonyms Lineage-specific duplicate DRY YNPxxY K Accessions
NEUR1 neuropsin OPN5 cOpn5m NEUR1a/b cephalochordate DRY YNPIIY K NM_181744 NM_001130743
NEUR2 newropsin ---- cOpn5L1 VCC YNPxIY K XM_419178
NEUR3 newtopsin ---- cOpn5L2 NEUR3a/b actinopterygii vRf YNPxIY K XM_420056
NEUR4: a fourth neuropsin from lamprey to platypus
Yet another new opsin in this group! These genes were first described here on 29 Jan 2009 (note GenBank had frog gene correctly predicted but chicken gene chimerized in a misassembly). Like many opsins, NEUR4 orthologs range throughout the vertebrates with the exception of theran mammals. Platypus is thus again distinguished by its retention of this ancient gene, whereas it is long gone from marsupials and placentals. This pattern of retention is consistent with platypus being more bird-like than mammal and supports the Wall 'dark era' apparently experienced by mammals during which Monotremata were somehow less dramatically affected.
This opsin is unusual for its TRY at the DRY motif, though its Schiff base lysine region is standard (KSASFYNPIIYFGMNSKFR). Its best match within opsins is to NEUR1; outside of neuropsins to peropsins, melanopsins and various ancestral ciliary opsins (it has no special affinities to RGR). Transcripts are abundant in frog and fish and the former demonstrate expression in adult eye (ES678087). This opsin is clearly capable of signaling through some heterotrimeric Galpha protein but its function is unknown.
Zebrafish and frog have an additional neuropsin, NEUR4b, discovered here in Oct 2010. It shares some synteny with NEUR4a indicating a block duplication. No counterpart can be located in other fish genomes. These seemed to have retained the extra copy from the ancestral whole genome duplication but evidently lost NEUR4b or never had it. It is possible that zebrafish and frog NEUR4b represent separate duplication events as they do not cluster dramatically together to the exclusion of the more numerous NEUR4a. The rate of evolution cannot be currently determined without further NEUR4b sequences and its function is equally mysterious as that of NEUR4a.
NEUR4 shares some early intron positions and phases with NEUR1 but otherwise the pattern differs significantly, suggesting rather ancient divergence after origination by segmental duplication (not as processed pseudogene subsequently re-intronated). This is consistent with a very low percent identity (38%) to NEUR1, considering that a 'floor' of about 25% identity relates any pair of GPCR (eg NEUR4 is 23% identical to its best non-opsin match in human, tachykinin receptor).
The phylogenetic distribution of the neuropsin gene swarm is puzzling. Only NEUR1 is found in non-vertebrate deuterostomes today yet family origins must be far more ancient as it is basal to ciliary opsins already found in cnidaria. Clearly neuropsins persisted for a very long period in pre-bilateran and bilateran ancestors prior to being lost (perhaps in a few stem events). Neuropsins may be important only in lineages where ciliary imaging opsins are important, yet all but NEUR1 have been lost in placentals despite persistence of the other three classes for several hundred million years.
Alternatively, the neuropsin gene family expanded in the lamprey stem, diverged rapidly in primary sequence, and experienced multiple intron reorganization events. Because even one is rare, two independent events is rare-squared, and three is effectively impossible. Because the minimal number of events needed to synchronize intronation is five, this hypothesis appears unsustainable. Each orthology class has the same intronation pattern in all its members back to the earliest divergence for which sequence is available.
Introns compared in four neuropsins 1^2 etc indicate relative intron gain. Exons 1 is omitted (same in all 4 genes). Irrelevant amino acids not shown (...) NEUR1 GIL...GIS 1^2 VVG...SHR WIF...YGW AGF...LAY GTW...SYA PEP...LTK ESR 2^1 MYT...LEV* NEUR3 AIL...GMA ISM...NHA WLG...YAL MGF...KSN SNK...YYG PEP...LTL SAD NSA...ARH* NEUR2 GIC...AMT LTL...SHR 2^1 WLY...YAF CGL...PAY GNR...EYG EEP...TAK RCC FCV...TDL* NEUR4 GWM...SIS VFG...NVF RDD...DGF 0^0 LTL...PER AHC...SYT 1^2 DRM...VTR KLK RFK...DRL*
The most parsimonious explanation here -- assuming the gene tree (((NEUR1, NEUR3), NEUR2), NEUR4) -- is that NEUR3 represents the ancestral intronation pattern. This correlates well with its relative lack of retroposon events which may facilitate intron gain -- a single unshared CR1 LINE -- and the gene span is 1/3 to 1/8 of the others. NEUR1 has 7 LINE elements for comparison. NEUR1, NEUR2 and NEUR4 separately acquired new introns (different locations and phases) within the second exon. NEUR1 also acquired a new intron in the terminal exon (which curiously is alignable to the end only in NEUR3). Intron gain is both rare in vertebrate coding genes and considerably less common than intron loss, yet the events here are more economically placed on terminal gene tree leaves as intron gains.
NEUR1 and NEUR3 both lie on the minus strand of chicken chromosome 3, separated by 1,310,977 bp and a half dozen coding genes. While not adjacent, this still suggests recent tandem duplication followed by local inversions, which is supported by the deeper match of the run-on terminal exon. This arrangement is readily tracked back to teleost fish, indicating the last expansion of neuropsin occurred prior to this, yet not by whole genome duplication. Perhaps the genes will be directly tandem in the upcoming revised Callorhinchus and Petromyzon genomes.
In chicken but not lizard, NEUR4 also lies on this same chromosome, though greatly distant. This possibly indicates that it too arose from tandem duplication of NEUR1 though at a much earlier date. More likely its current position is coincidental because, while chickens have 39 chromosomes, only 6 are macro-sized.
Indels are another type of potentially informative rare genomic event. Upon alignment of the reference sequence set below, five phylogenetically coherent indels emerge. Amphioxus and sea urchin genes can be used as outgroup to determine ancestral length (eg, to resolve each indel as deletion or insertion); this gives the same outcome as alignment to all ciliary opsins and indeed many GPCR. Each indel affects every member of its orthology class and none affects more than one class (other than in fish where the event affected the parent NEUR3 gene prior to its tandem duplication). Thus, as with introns, indels are predominantly ancient and do not provide internal clustering of neuropsins genes to guide the gene tree. Possibly the pre-lamprey indels were fixed shortly after gene duplication as the new paralogs re-functionalized.
Indel #res type affected_genes timing location A 5 insert NEUR4 pre-lamprey 2 residues before first disulfide cysteine B 1 insert NEUR3 NEUR3a NEUR3b pre-teleost 2 residues after DRY motif, cytoplasmic loop C2 C 2 deletion NEUR4 pre-lamprey 1 residue before second disulfide cysteine D 2 deletion NEUR3b post-fish tandem 9 residues after second disulfide E 3 insert NEUR4 pre-lamprey 16 residues before Schiff lysine
It appears very likely NEUR1 is the parent gene providing the core (unknown) function:
- NEUR1 is best-blastp to amphioxus and sea urchin homologs whereas the others are undetectable outside vertebrates;
- NEUR1 survived the longest in mammals;
- NEUR1 is the best-Blastp for each of the others;
- NEUR1 alone retains the ancestral DRY signaling motif.
- NEUR1 has ancestral length (no indels relative to broader opsins or GPCR
Curated collection of neuropsins
These genes, almost all full length, have been extracted from various genome projects and cDNA data sets. In a few instances, accurate gene models had been previously computed by bioinformatic pipelines but these are so mixed with erroneous and mislabeled predictions as to be worthless for comparative genomics. The UCSC 44-species alignment is very helpful for rapid collection of individual exons taking care to note small insertions relative to human sequence are suppressed. The proteins below are parsed into exons whose coding phase is also shown. Because intronation is exceedingly conservative, genomically deduced introns can be reliable transfered to cDNA-only species.
Despite the incomplete nature of many genome assemblies and lack of transcripts in specialized cell types, the absence of a given neuropsin in a given clade is rarely attributable to lack of data. For example, while sloth assembly alone might have only 2x mean coverage (and thus lack many coding genes), overall coverage of its clade (Atlantogenata: armadillo, sloth, elephant, mammoth, hyrax, and tenrec) approaches 30x (90 million 1 kbp traces). Here coding genes can be missing only for the compositionally oddest or most extreme chromosomal locations.
For early diverging deuterostomes, the situation is somewhat different. Here only one species per divergence node is generally available and assemblies encountered extreme diploid heterozygosity and retroposon issues with little outside support from transcript programs (except for Ciona and sea urchin). The species chosen may be quite specialized within its clade and have experienced very extensive gene loss, much as Drosophila is wholly unrepresentative of the protostome genome. Lamprey, despite 19 million traces, has poor coverage with contigs rarely encompassing more than an exon or two.
However gene divergence (to the point of unrecognizability) is not an issue because the divergence floor to GPCR is well within tblastn reporting capabilities. Even in short contig assemblies, individual exons can be identified and reliably assigned to opsin orthology class using the classifier, provided the exon is reasonably conserved. Even poorly conserved N- and C-terminal exons can be extended outward to an initial methionine or stop codon with some reliability.
Consequently the phylogenetic 'end points' of a given gene are fairly certain even on the early-diverging side, though that remains somewhat muddled because lineage-specific loss is not at all uncommon in opsins or specialized species.
NEUR1: 56 deuterostome neuropsins
>NEUR1_homSap Homo sapiens (human) 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2 1 LHTVTTVRKSSAVLEIHEEV* 0 >NEUR1_panTro Pan troglodytes (chimp) 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKETKKKSLEGFR 2 1 LHTVTTVRKSSAVLEIHEEV* 0 >NEUR1_gorGor Gorilla gorilla (gorilla) 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 12 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 YASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIPSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGKPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFQ 2 1 LHTVTTVRKSSAVLEIHEEv* 0 >NEUR1_ponPyg Pongo pygmaeus (orang_sumatran) 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGSPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2 1 LHTVTTVRKSSAVLEIHEEV* 0 >NEUR1_nomLeu Nomascus leucogenys (gibbon) 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2 1 LHTVTSVRKSSAVLEIHEEv* 0 >NEUR1_macMul Macaca mulatta (rhesus) 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADIVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2 1 LHTVTTVRKSSAVLEIHEEV* 0 >NEUR1_papHam Papio hamadryas (baboon) 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADIVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEGFR 2 1 LHTVTTVRKSSAVLEIHEEv* 0 >NEUR1_calJac Callithrix jacchus (marmoset) 0 MALNHTSLPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHEEV* 0 >NEUR1_tarSyr Tarsius syrichta (tarsier) 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VIGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKGVAHFDSRIHTSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHEEv* 0 >NEUR1_otoGar Otolemur garnettii (bushbaby) 0 MALNHTALPQDELRPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 WTMMPLVGLEDYVPEPFTSCTLDWWLAQSLGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHGSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKTTKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEV* 0 >NEUR1_micMur Microcebus murinus (mouse_lemur) 0 MALNHTVLPQDERLPHYLRDGDPFASKLSWEADLVAGFYLIII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 1 2 GVWLKRKHAYICLALIWAYVSFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDHR 2 1 LHTVTAVRKSSAVLEIHQEv* 0 >NEUR1_tupBel Tupaia belangeri (tree_shrew) 0 MALNHTALPQDESLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHVLEMKLTK 0 0 2 1 LHTVTTVRKSSAVLEIHQEV* 0 >NEUR1_musMus Mus musculus (mouse) 0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQAGGLRGTKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEV* 0 >NEUR1_ratNor Rattus norvegicus (rat) 0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 gVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPNSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQTGGLRATKKKSLEDFR 2 1 LHTVTAVRKSSAVLEIHPEv* 0 >NEUR1_speTri Spermophilus tridecemlineatus (squirrel) 0 MALNHTALPQDEHLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAFICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFINILFFCLLLPTAVIEFSYVKIIAKVKSSSEEVAHFDSRIHSSHV 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPSLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHTVTAVRKSSAVVEIHQEv* 0 >NEUR1_dipOrd Dipodomys ordii (kangaroo_rat) 0 MAFNHTAGTQGQGLPHYLPEEDPFTSKLSWEADIVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASLAGQVFILNILFFCLLLPTSVIVFSYVKIIAKVKSSSKEVAHFDSRIPSSHVLEMKLTK 0 0 2 1 * 0 >NEUR1_cavPor Cavia porcellus (guinea_pig) 0 MALNHTAPPQNEHLPRYLQDEDPFVSKLSWEADLVAGFYLTII 1 2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 2 vVGKPFTIISCFRHRWVFGWIGCRWYGWAGFFFGCGSLITMTVVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYVSFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASAGGQIFILHILFFCLLLPTAMIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDSRFACCQNAGLKATKKKSLEDFR 2 1 LHTVTTDRKSAVLEIHQEV* 0 >NEUR1_oryCun Oryctolagus cuniculus (rabbit) 0 MALNHTALPQDEHLPHYLREGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRRHAYICLALIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFSCCRTSGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEv* 0 >NEUR1_ochPri Ochotona princeps (pika) 0 MALNDTALPQDEHLPHYFRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRLYGWADFFFGCGSLITMTAVSLDRYLK 1 2 GVWLKRRHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHGSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFSCCRTGGLKQTKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEv* 0 >NEUR1_canFam Canis familiaris (dog) 0 MALNHTARPQDERLPHYLREGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASLGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGRLKATKKKSLEDFR 2 1 LNTVTTVRKSSAVLEIhQEV* 0 >NEUR1_ailMel Ailuropoda melanoleuca (panda) XM_002919006 0 MALNHTAPPQEEHLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAFIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRVHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEV* 0 >NEUR1_felCat Felis catus (cat) 0 MALNHTAPPQDERLPHYLREGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAE 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEv* 0 >NEUR1_bosTau Bos taurus (cow) 0 MALNHTAPPPDERRPPYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAICDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GIWLKRKHAYICLAVIWAYAAFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEVHQEv* 0 >NEUR1_turTru Tursiops truncatus (dolphin) 0 1 2 GILSTFGNGYVLYMSSRRKKKLKPAEIMTINLAICDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIPSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAVYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEv* 0 >NEUR1_susScr Sus scrofa (pig) 0 MALNHTAPPPDERRPHYLREGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTVNLAICDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYAAFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLVAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIRQEV* 0 >NEUR1_vicVic Vicugna vicugna (vicugna) 0 MALNHTAPPPDERRPRHLRDGdPFASKLSWEADLVAGFYLTII 1 2 GILSTLGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWMFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHAVTTVRKSSAVLEIHQEV* 0 >NEUR1_equCab Equus caballus (horse) 0 MALNHTALPQDERLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHGSHVLEVKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEV* 0 >NEUR1_myoLuc Myotis lucifugus (microbat) 0 MALNHTALPQDEGLPHYLQDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHTYICLAFIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQATVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKKVAHFDSRIH 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSSAMYNPIIYQVIDYKLACCQTGGLRATKKKSLENFR 2 1 LHTVTTVRKSSAVLEIHQEv* 0 >NEUR1_pteVam Pteropus vampyrus (macrobat) 0 MALNHTVLPQDEHLPHYVRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHGSHMLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTSGLRATKKKSLEDFR 2 1 LHTITTVREASAVLEIHQEV* 0 >NEUR1_sorAra Sorex araneus (shrew) 0 MALNHTALPQDENLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLVVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPNSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQSGGLRATKKKSLDDFr 2 1 LHTVTTVRESSAVLEIHQEV* 0 >NEUR1_eriEur Erinaceus europaeus (hedgehog) 0 MSLNQTALPQDEGLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 gVWLKRKHAYLCLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASLGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKKVAHFDSRIHSSHMLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLKANKKKSLKDYR 2 1 * 0 >NEUR1_loxAfr Loxodonta africana (elephant) 0 MTLNHTAPPQDDRLPQYLQDGDPFTSKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSCRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFVIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQIFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHMLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLRATKKKSLEGFR 2 1 LHTVTTVKKSSAVLEVHQEv* 0 >NEUR1_proCap Procavia capensis (hyrax) 0 MTLNHTVLPEDDRLSHYLRDGDPFTSKLSWEADLVAGFYLTVI 1 2 GILSTCGNGYVLYMSYRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFIIISSFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSVPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCRTRGLRATKEKSLEGVR 2 1 LHTVTTVRKSSAVLEIHQEv* 0 >NEUR1_echTel Echinops telfairi (tenrec) 0 MALNHTAPPQDNSLPHYLRDGDPFVSKLSWEADLGAGFYLIII 1 2 GILSTFGNGYVLYMSYRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFSHRWVFGWTGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHMLEMKLTK 0 0 2 1 LHTITTVRKSSAVLEIHQEV* 0 >NEUR1_dasNov Dasypus novemcinctus (armadillo) 0 MALNHTALPQDDRLPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 gILSTFGNGYVLYMSSKRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCQTGGLRATKKKSLEDFR 2 1 LHTVTTVRESSAVLEVHQEV* 0 >NEUR1_choHof Choloepus hoffmanni (sloth) 0 MALNHTGLPQDDSLPHYFRDGDPFASKLSWEADLVAGFYLIII 1 2 GILSTFGNGYVLYMSSRRRKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLLGLGDYVPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHMLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFACCRTGGLRATKKKSFEGFR 2 1 LHTVTTVRKSSAVLEIHQEv* 0 >NEUR1_monDom Monodelphis domestica (opossum) not extendable N-terminally 0 MALNHSVSPQDDYIPHYLRDGDPFASKLSWEADLVAGFYLTII 1 2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIISCFSHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPVQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCQSGGQKAAKKESLRTYR 2 1 LHTVTTVRRSSAVLEIHQEv* 0 >NEUR1_macEug Macropus eugenii (wallaby) 0 1 2 GVLSTLGNGYVIYMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWVGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSy 1 2 GTWLKRHHAYICLVIIWAYATFWATMPLAGLGNYAPEPFGTSCTLDWWLAQASVTGQTFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQSSHVLEMKLTK 0 0 2 1 RHTVSTIRKSSSVSETYQEV* 0 >NEUR1_ornAna Ornithorhynchus anatinus (platypus) no further possible upstream extension of exon 1 0 MNSMTNYSAPQLGDYLPHYLREGDPFVSKLSWEADLVAGVYLVII 1 2 GVLSTLGNGYVIYMSSRRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFTIVSCFCHRWVFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAYICLAIIWAYASFWATMPLVGLGNYAPEPFGTSCTLDWWLAQASVAGQAFILNILFFCLLLPTAVIVFSYVKIIAKVKSSTKEVAHFDSRIQNSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSIPIQFSVVPTLLAKSAAMYNPIIYQVIDCRISCCRLGGPKTGKKESLKNSR 2 1 SHSMSTIRKPSAVSGPHQEV* 0 >NEUR1_galGal Gallus gallus (chicken) MGAIVCSVGFVCLFVFSDTELD possible upstream extension of exon 1 0 MSGMASDCNSSSQEEYLPHYMQQEDPFASKLSREADIIAGFYLTVI 1 2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGPKTLQKKSSLKESR 2 1 MYTISSHRDSAALSGTQLEV* 0 >NEUR1_melGal Meleagris gallopavo (turkey) MGAVVYSLGFVCLFVFSDTELD possible upstream extension of exon 1 0 MSGMASDRNSSSQEEYLPHYVQQEDPFASKLSREADIIAGFYLTVI 1 2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 2 gTWLKRHHAFICLALIWTYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFILSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFACCRSGGLKALQKKSSLKESR 2 1 MYTISSHRDSAAPSETQLEV* 0 >NEUR1_cotJap Coturnix japonica (quail) AB547151 20679218 MGAVVCSVRSVCLFVFSDTELD possible upstream extension of exon 1 0 MSGMASDCNSSQEEYLPHHVQQEDPFASKLSREADIIAGFYLTVI 1 2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFSIISFFSHRWIFGWMGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICHLAY 1 2 GTWLKRHHAFICLALIWAYATFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQAFVLSILFFCLLFPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGQPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIDCKFSCCRSGGLKTLQKKSSLKDSR 2 1 MYTISSHRDSAALSETQLEV* 0 >NEUR1_taeGut Taeniopygia guttata (finch) not extendable N-terminally 0 MSGMASEYNNSSQEEYIPHYLQEEDPFASKLSREADIIAGFYLTII 1 2 GILSTLGNGYVIFMSSKRKKKLRPAEIMTVNLAVCDLGIS 1 2 VVGKPFSIISFFSHRWMFGWIGCCWYGWAGFFFGCGSLITMTAVSLDRYLKICHLSY 1 2 GTWLKRHHAFICLAIIWAYAMFWATVPFAGVGSYAPEPFGTSCTLDWWLAQASVAGQVFVLSILFFCLLLPTAVIVFSYVKIILKVKSSTKEVAHYDTRIQNSHILEMKLTK 0 0 VAMLICAGFLLAWIPYAVVSVWSAFGRPDSVPIQFSVVPTLLAKSAAMYNPIIYQVIECRLACCRPGGCCRPGGLKAKSSLKKSR 2 1 TYTISAHRDSTAMNETQLEA* 0 >NEUR1_anoCar Anolis carolinensis (lizard) no N-terminal extension possible 0 MEQGQNISSQDDNQQEEDPFASKLSVEADIVAGVYLLVI 1 2 GILSTLGNGYVIYMSTQRKKKLKPAEIMTVNLAVCDLGIS 1 2 VVGKPFSIIAFFSHRWIFGWSGCRWYGWAGFFFGIGSLITMTAVSLDRYFKICHLSY 1 2 GTWLKRHHVFICLGIIWSYAAFWATIPFAGFGNYAPEPFGTSCTLDWWLAQGSVAGQAFILNILFFCLVLPTAVIMFCYVKIIAKVQSSTKEVAHYDTRIQNQHVLEMKLTK 0 0 VAMLICAGFMFAWIPYAVVSVWSAFGRPDSVPIKVSVIPTLLAKSAAMYNPVIYQVIDCKSACCRPGNLQPLQKKNSR 2 1 LYIIPTGKKSEVVQETQLDSV* 0 >NEUR1_xenTro Xenopus tropicalis (frog) no N-terminal extension possible 0 MAGNSSYREESGYIPHYERDSDPFASKLSREADIFAGVYLMAI 1 2 GILSTLGNGYVIYMACSRKKKLRPAEIMTINLAVCDLGIS 1 2 VTGKPFAIVSCFSHRWVFGWNACRWYGWAGFFFGCGSLITLTVVSLDRYLKICHLRY 1 2 GTWLKRRHAFIALAVIWAYATLWATLPLVGVGNYAPEPFGTTCTLDWWLAQASVKGQIFVLSMLFFCLLFPTMVIVFSYAKIIAKVKSSAKEVAHFDTRNQNNHTLEIKLTK 0 0 VAMLICAGFLIAWFPYAVVSVWSAFGQPDSIPIELSVVPTMMAKSASMYNPIIYQVIDCKPACCKKDKSLQNTTSR 2 1 VYTISTFRKSTTSAR* 0 >NEUR1_danRer Danio rerio (zebrafish) 0 MENETSISSGYIPHYLLRGDPFASKLSKEADIVAAFYILVI 1 2 GILSATGNGYVMYMTFKRKTKLKPPEIMTLNLAIFDFGIS 1 2 VSGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSFDRYLKICHLRY 1 2 gTWLKRHHAFLSVVFIWAYAAFWATMPVVGWGNYAPEPFGTSCTLDWWLTQASVSGQSFVMCMLFFCLIFPTVIIVFSYVMIIFKVKSSAKEVSHFDTRNKNNHSLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVMSAFGEPDSVPIPVSVVPTLLAKSSAMYNPIIYQVIDCKKKCVKSCCFQAWRKKKPSKTSR 2 1 FYTISGSIKQRPGDEASIEI* 0 >NEUR1_takRub Takifugu rubripes (fugu) 0 MENDTSIPSGYVPHYLLRGDPFASKLSKEADIVAAFYILVI 1 2 GVLSATGNGYVIYQTIKRKTKLKPPEFMTLNLAVFDFGIS 1 2 VTGKPFFIVSSFSHRWLFGWQGCRYYGWAGFFFGCGSLITMTIVSLDRYLKICHLRY 1 2 GTWFKRHHAFLCLVFTWLYAAFWATMPVVGWGNYAPEPFGTSCTLDWWLAQASVSGQSFVMCMLIFCLVLPTGVIVFSYVMIiLQVKSSAQEVSHFDTQNKNKHHLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVVSAFGDPDSVPISISVVPTLLAAKSSAMYNPIIYQVVDVKTSCTNFSCCKALKERIHFRKSR 2 1 FYSISASMKKRPANEVPTEI* 0 >NEUR1_tetNig Tetraodon nigroviridis (pufferfish) 0 MENETWTHSSYVPHYLLRGDPFASRLSKEADIVAALYICII 1 2 gLMSATGNGYVLYMTFKRKTKLKPPELMTLNLAIFDFGIS 1 2 VTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 2 GAWLKRHHAFLCLASVWAYAAFWATMPLVGWGSYAPEPFGTSCTLDWWLAQASVSGQSFVMAILFFCLILPTGIIVFSYVMIIFKVKSSAKEISHFDARIRNSHDLEIKLTK 0 0 VAMLICAGFLIAWIPYAVVSVISAFGEPDSVPIPVSVIPTLLAKSSAMYNPIIYQVADLKTSCTSSSCCKALKERVLFRKSR 2 1 YTISGSLRDTLPPKEAHIEM* 0 >NEUR1_gasAcu Gasterosteus aculeatus (stickleback) 0 MDNETRSHPSYVPHYLLRGDPFASRLSKEADIVAAFYIFII 1 2 GVMSATGNGYVLYMTFKRKTKLKPPELMTVNLAIFDFGIS 1 2 VTGKPFFIVSSLSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 2 GTWLKRHHAFVCLALVWAYAAFWATMPLVGWGSYAPEPFGTACTLDWWLAQASVSGQSFVMAILFFCLVLPTGIIVFSYIMIIFKVKSSAKEISHFDARIKNSHSLEIKLTK 0 0 VAMLICAGFLIAWIPYAVVSVVSAFGEPDSVPIPVSVIPTLLAKSSAMYNPIIYQVLDLKNSCMKSSCFKGLKKPRHFRKSR 2 1 YTISGSLRDTLPPKEAHIEM* 0 >NEUR1_oryLat Oryzias latipes (medaka) 0 MENETWTHPSYIPHYLLRGDPFASRLSKEADIIAAFYICII 1 2 gIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGIS 1 2 VTGKPFFVVSSFAHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 2 GTWLKRQHAFLCLVFVWMYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLAQASVSGQSFVVAILFFCLVLPAGIIVFSYVMIIFKVKSSAKEISNFDARIKNSHNLEIKLTK 0 0 VAMLICAGFLIAWIPYAVVSVVSAFGEPDSVPISVSVIPTLLAKSSAMYNPIIYQVLDLKNSCMKSSCFKGLKKPRHFRKSR 2 1 YTISGSLKDTAPAKEAHIEI* 0 >NEUR1_pimPro Pimephales promelas (minnow) 0 MENTSWPHSSYVPHYLLRGDPFASRLSKEADIVAAFYILII 1 2 GIMSATGNGYVIYMTIKRKSKLKPPELMTVNLAVFDFGIS 1 2 VTGKPFFVVSSFSHRWLFGWEGCRFYGWAGFFFGCGSLITMTVVSLDRYLKICHLRY 1 2 GTWLKRQHIFLCLVFVWIYAAFWATMPLVGWGSYAPEPFGTSCTLDWWLAQASVSGQSFVMSILFFCLVLPAGIIVFSYVMIICKVKSSSKEVSSFDARIKNSHTLEIKLTK 0 0 VAMLICAGFLIAWIPYAVVSVVSAFGEPDSIPIPVSVIPTLLAKSSAMYNPIIYQVIDCKKNCAKLSCFQAWSKRKHYKTSR 2 1 FYSISASMKKRPANEVPTEI* 0 >NEUR1_calMil Callorhinchus milii (elephantfish) 0 MTAFDNSTALYSGYWLHDSLHGDPFVSKLSWEADIISACYLIVT 1 2 GLLSTLGNGYVIYLSITQKRKLKPPEILITNLAISDFGMS 1 2 VGGQPFLIISCFSHRWIFGWVGCRWHGWAGFFFGCGSLITMTVVSLDRYLKICHLQY 1 2 GSWLQRRHVFMSLAFIWFYAAFWATMPLVGWGNYAPEPFGTSCTLDWWLARVSVSGLIFVLTILFFCLLLPIIIIVFSYIKIIAKVKSSAKEVAHFDSRIQNHHSLEMNLTK 0 0 2 1 * 0 >NEUR1_petMar Petromyzon marinus (lamprey) frag 2 GTWVRRRHAFLCVLAVWLYAAFWATMPLLGWGSYAPEPFGTSCTLDWWLAQSSAAGRSFVLCMLLFCLLLPAAAILFAYARIVGAVRRSARDLAHFERRARGGGGGGGGGGVALELRITK 0 0 VAMMICAGFLLAWIPYAVVSVWSAFGAPDSVPVAVSMVPTMFAKSAAMYNPLIYQLLSRRGTGAHCCRCRKARGTLRRPR 2 >NEUR1a_braFlo Branchiostoma floridae (amphioxus) FE548698 0 MATTPADRLDGLTPAGRGATTAETHADDFASKLSREADIVIGVYLILI 1 2 GTGAILGNGRVLWLSYRCRARLRPVEMFVVSLAAADVGLSLVGHPFSAASSLMGRWSFGSAGCTW 1 2 YGFVVFFLGIASIATMALMSIMRFMIVHKRY 1 2 GQYPSRRASCVLVAAAWLYGLFWACAPLA 1 2 GWSQYHPEPYGLSCSVDWGGFSRGAGGSSFIICMLLFCTAVPVVVMVTSYAAIFALYRQAQKGVVLNLQVNATFGGKRQRTER 0 IALAVCGGFLLAWLPYAVVGLWASVAGVDAVPLALASAAPLFAKSNSLWNPIIYLGMNERFR 2 1 * 0 >NEUR2b_braFlo from traces and genome chrUn ++ 187375671 187384042 8372 nearly identical chrUn ++ 32271780 32281075 9296 0 MATTPGLPLDGLAPTGRGVTAADTLDDDFASKLSREADIVIGVYLLLI 1 2 GTGSILGNGRVLWLSYRNWAKLRPVELFVVSLAVTDVGISVFGYPFAASSSLLGRWSFGSAGCTW 1 2 YGFTGFFFGLTSIANMALMSIMRFMIVYKGY 1 2 GPYPSRRATSGLIAAAWLYGLFWACAPLA 1 2 GWSQYHVEPFGLSCTVDWGSFSRDAGGMSFIICLLVFCVAIPVTAIMASYVAISAIYRQAKKSIAGHLQDNSAMCKKRNKLE 0 0 MALAVCGGFLLAWLPYAVVGLWSAVAGVDAVPLALASAAPLFAKSSSLWNPIIYLGMNDRFR 2 1 * 0 >NEUR1_strPur Strongylocentrotus purpuratus (sea_urchin) XM_001197837 CX694910 CX690664 0 MDVNAKWWTNETLRTRDQFSDDHYTSVLSYEGDIWAGVYLMFI 1 2 SLIAFIGNISVIVISLRKREKLKPIDLLTINLAIADFLICVVSYPLPMISAFRHR 0 0 WSFGKFGCVWYGFTSFLFAVGSMATLMVIALLRYAKLCRENV 1 2 DQYQSRPFVIKVIVAIWGFAFFTTAPPLFGWS 2 1 SYVPEPYHLSCTIDFADTSPSGLSYTYFTTIVVFFMPLMIIVLCYVAIARKMIHHNRRINVGHNAGRMLLEIRLLK 0 0 TACMITMAYTISWTPYAVIAMWVTYIPVNQIPDAFRILPAFCAKTSSVYNPIIYCIFNKSFRQDLSSLICCCACQCYTITINLDINSHAQQQFRRIEERR DEVGTYKRRPLMICSNPFAWSRDFHETWRQRRIRGIHRNCRNNVRVENINVNFRRDTDMVELNAPTPAEIHRPELNTASTRSGARTKSMATHLPALEEVPSG APQCSALLHNTPIPRSLQGTPLPYQPQPSTSDLHDEFLNPSVVSRNMCVIVVKPNIEEELSTD* 0
NEUR2: 12 vertebrate newropsins
>NEUR2_galGal Gallus gallus AB368181 18570255 synteny: -B4GALT6 -NEUR2-KIAA1012 cOpn5L1 0 MDPSFANSTFQSKITEAADIVVGTCYMVF 1 2 GICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHR 2 1 WLYGKHICLFYAFCGLFFGICSLSTLTLLSVVCCLKICFPAY 1 2 GNRFRRKHGQILIACAWTYAAIFACSPLAHWGEYGEEPYGTACCIDWQSTNVDVMSMSYTVVLFVLCFILPCGVIVTSYSLILVTVKESRKAVEQHVSGPTRINNVQTITAK 0 0 LSIAVCIGFFAAWSPYAIIAMWAAFGSIDKIPPLAFAIPAVFAKSSTLYNPIIHLLLKPNFRSNIAKDFTVIQQLCVRCCFCVKELQTYRSTFNTGLRTFKGKNESSCNALPIMEG CSYFPSEKGSHTFECFKSYPNCFQERLSTMGCHLQDCESLENDLQVEVTQGSRNSMKVVEQEEKSTELDNLEITLEAVPVSCTFTDL* 0 >NEUR2_melGal Meleagris gallopavo (turkey) genomic 0 MDPSFANSTFQSKITEAADIVVGTCYMVF 1 2 GICSLCGNSILLYISYKKKHLLKPAEYFIINLAISDLAMTLTLYPLAVTSSLSHR 2 1 WLYGKHICLFYAFCGVFFGICSLSTLTLLSVVCCLKICFPAY 1 2 GNRFRRKHGQILIACAWTYAAIFACSPLAHWGEYGEEPYGTACCIDWQSTNVEITSMSYTVVLFIFCFILPCGVILTSYSLILVTVKESRRAVEQHVSGPTRINNVQTITVK 0 0 LSIAVCIGFFAAWSPYAIIAMWAAFGSIDKIPPLAFAIPAVFAKSSTLYNPIIHLLLKPNFRSIVAKDFSVLQQLCVRCCFCVKELQIYRSTFNAGLRTFKGRNEFSCNALPDMEG CSYFPSEKGNHTFECFKSYPKCCQERLSTMGCHPQERESLENDLQVEMTEGSRNSMKVVDQEEKSTELDNLEITLEAVPVHCTFTDL* 0 >NEUR2_anoCar Anolis carolinensis (lizard) 0 MESYFANTTFHSKITEAADVIVGVFYIVF 1 2 GICSFCGNSILLYVSYKKKNLLKPAEYFMINLAISDLGMTLTLYPLAVTSSLAHR 2 1 WLFGQQVCLFYAFCGVFFGVCSLTTLTLLSIVCCLKICFPVY 1 1 GNRFRPGHGWILIACAWVYAAIFAFSPLAHWGEYGAEPYGTACCIDWRISNMKKTAMSYTTALFVFCYIIPCGIIITSYTLILITVKDSRKAVEQHALGPTRMSSVHTITAK 0 0 LSIAVCIGFFVAWSPYAIIAMWAAFGSIDMIPPLAFAVPAVFAKSSTLYNPAMYLFLKPNFRSTIAKDLTVLHRLCLKSCFCPRGMQNCSYRSALEAPLKSFKGRNESSSNSVQIVGGCS YFPCEKCHDPFECFKNYPKCCQGRLNVMDHTPRESISVENNMQSKTKHASEKYIKVVIRGEKNTDIDNLEITLEHIPTDIKFANL* 0 >NEUR2_xenTro Xenopus tropicalis (frog) abundant transcripts 0 MGNKSDASAFYSSISETDDIVLGVLYSVF 1 2 GLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHR 2 1 WLFDKVTCNYYAFCGMLFGLCSLTNLTVLSSVCCLKVCYPAY 1 2 GNKFSTAHSRILLLGIWAYAGLFATAPLADWGKYGPEPYGTACCLDWEASYRERKALSYTISLFVFCYLIPSSLIFISYTLIFVTVKGARRAVQQHLSPQAKGSSIHSLIIK 0 0 LSIAVCIGFLIAWTPYAIVAMMAAFGDPTKIPSLVFALAAAFAKSSTIYNPVVYLLLKPNFLNVVTKDLTLFQTMCAVVCGWCRTPAVKTPCPHKD LKTTSKPPSSFKKSQGVCRNCVDTFECFRNYPRCCSVGNVDAAQPMAASLVRIPPANGAPQQTVQLVVSSSRTRSGVETVEVSTEAPMSDFIKDFI* 0 >NEUR2_danRer Danio rerio (zebrafish) acquired new intron 0 MGNVSKTALFMSTISRQHDILMGSLYSVF 1 2 FVLSLLGNGMLLFVAYRKRSSLKPAEFFVVNLSVSDLGMTLSLFPLAIPSALAHR 2 1 WLFGEITCLCYAVCGVLFGLCSLTNLTALSSVCCLKVCFPNY 1 2 GNKFSSSHACVMVIGVWCYASVFAVGPLVHWGSFGPEPYGTACCINW 2 1 YTPSHDALAMSYIISLFIFCYVVPCTIIILSYTFILVTVRGSQQAVQQHVSPQTKVTNAHALIVK 0 0 LSVAVCIGFLTAWSPYAIVAMWAAFSANEQVPPTAFALAAIMAKSSTIYNPMVYLLFKPNFRKSLSQDTQMFRHRICLSHSKASPSPGMKDQERQS SQQCNNKDGSISTPFSSGQAESYGACHVYAEAGPHYQQISRQITARVLEGSVQSEIPVKQLTEKMQNDLL* 0 >NEUR2_tetNig Tetraodon nigroviridis (pufferfish) gene mix 0 MGNASDTSDAFNSKISKEHDFLIGSIYSVF 1 2 CVLSLMGNCILLLVAHHKRSTLKPAEFFIVNLSISDLGMTLTLFPLAIPSSFSHR 2 1 WLFGEIACQLYATCGVLFGLCSLTNLTVLSSVCCLKVCLPNL 1 2 GSKFSSSHARLLVAGVWGYASVFAVGPLVQWGHYGPEPYGTACCINWQAPNHELSSLSYIVCLFLFCYVLPCAIIILSYTCILMTVRGSRQAIQQHVSPQTKTANAHALIVK 0 0 LSVAVCIGFLGAWSPYAVVAMWASFGDATWVPPDAFAIAAILAKSSTIYNPLVYLLCKPNFRECLYKDTSTLRQRIYRGSPLSGPRDRSGGVTQRHKDLSVSTR LSNGQQDSYGTCLHCAEDAELGHVTGSRRTACILTGSTFTEVTLSQLSATPADLL* 0 >NEUR2_takRub Takifugu rubripes (fugu) 0 MGNASEASDIFLSKISKEHDILIGSIYSVF 1 2 GLLSLAGNCILLLVAYHKRSMLKPAEFFIINLSISDLGMTLTLFPLAIPSSFSHR 2 1 WLFGEITCQLYAMCGVLFGLCSLTNLTALSLVCCLKVCFPNH 1 2 GSRFSSSHARLLVVGVWCYASVFAVGPLVQWGHYGPEPYGTACCIDWRAPNHELSSLSYIVCLFFFCYVLPCATIILSYTCILMTVRGSRQAIQQHVSPQTKTANAHSLIVK 0 0 LSVAVCIGFLGAWSPYAIVAMWAAFGDATWVPPDAFAIAAILAKSSTIYNPVVYLLCKPNFRECLYKDTSTLRQRIYRGSPQSEPRERFGGTSQRHKDLSISTR LSNGQQDSYGTCLHCADDAERGHVTTSQRTACILTGSTFTEVTVGQLSAAPADLL* >NEUR2_gasAcu Gasterosteus aculeatus (stickleback) 0 MGNASDTSAVFASTISKERDILMGSLYSVF 1 2 GVLSLVGNCILLLVAYHKRSTLKPAEFFIINLSISDLGMTLSLFPLAIPSAFKHR 2 1 WLFGELTCQLYAMCGVLFGLCSLTNLTALSFVCCLKVCFPNH 1 2 GNRFSSSHARLLVVAVWGYASVFAVGPLARWGRYSPEPYGTACCIDWHAPNHELAALSYIVCLFVFCYALPCATIFLSYTFILLTVRGSRQAVQQHVSPQTKTTNTHALIVK 0 0 LSVAVCIGFLGAWTPYAVVAIWAAFGDATLVPPDAFALAAMFAKSSTIYNPVVYLLCKPNFRACLYRDTTLLRQRIYRGSPRSEPKAHFGSTSQRNKDMSVSVRSSNGQQDSYGACTENA APCHVMTPQRTACILTESTNREVTVSRLADKPQADFL* >NEUR2_oryLat Oryzias latipes (medaka) 0 MGNVSDTSSLFASSISREHDILMGSLYSVF 1 2 GLLSLSGNSMLLLVAYRKRSILKPAEFFIVNLSISDLGMTGTLFPLAIPSLFAHR 2 1 WLFGEITCQLYAMCGVLFGLSSLTNLTALSLVCCLKVCFPNH 1 2 GNKFSFSHARLLVAGVWCYASVFAVGPLARWGRYSAEPYGTACCIDWHAPNHELWALSYILCLFIFCYALPCTIIFLSYAFILLTVRGSRQAVQQHVSPQTKTTNAHTLIVK 0 0 LSVAVCIGFLGAWTPYAVIAMWAAFGDATQVPPTAFALAAVFAKSSTIYNPMVYLLCKPNFRECLCRDTSLLRHMIYRGSPQPQERFGSDSRRNKDITASTRFSNGQQESYGACLNCTEN TGLCQLASPQNTACILTGSTYAEVTVQQLVDKQQPDFL* 0 >NEUR2_pimPro Pimephales promelas (minnow) 0 MGNVSETALFVSTISRQHDILMGSLYSVF 1 2 CVLSLLGNGMLLFVAYRKRSSLKPAEFFVINLSVSDLGMTLSLFPLAIPSALAHR 2 1 WLFGEVVCLCYAVCGVLFGLCSLTNLTALSSVCCLKVCCPNY 1 2 GNKFSSNHACVMVIGVWCYASVFAVGPLIRWGSFAPEPYGTACCINWYIPSHDALAMSYIISLFIFCYVVPCTIIILSYTFILLRVRGSRQAVQKHVSPKTKETNAHTLIVK 0 0 LSVAVCIGFVTAWSPYAVVAMWAAFSANEPVPPTAFALAAILAKSSTIYNPMVYLLFKPNFRKILSQDTQNIRHRMCVSHSKASPTPEIK-AQSSQQCKDATISTPFSSGQAESYGTCHIYAEAEPHFQQISPQRTVRILEGIIQSEISVRHMTDRMQNDLL* 0 >NEUR2_oncMyk Oncorhynchus mykiss (trout) no glycosylation site, anomalous agreement with chicken 0 MGVLASIDDIAFLSNIPVAADITVAIVYAVF 1 2 GMCSLFSNSTLLYISYKKKHLLKPAEFFIINLAISDMSLTLSLYPMAITSSIYHR 2 1 WLFGKTVCLIYAFCGMLFGVCSLTTLTLLSMVCFVKVCYPLY 1 2 GNRFNAVHGRLLIACAWAWALVFACSPLAHWGEYGPEPYGTACCIDWRLSNLHPVARSYTAALFVLCYIVPCCVIVASYTGILMTVRASHKAMEHHEARQTKMSNIQDVIVK 0 0 LSVAVCIGFFAAWSPYAVVSMWAAFGHMDNIPPLAFAVPAMFAKSSTIYNPIIYLLLRPNFRRVMYRDLVSLCRAFLKGCLCSCSQGAVGKCHSHLVVRVSLQSFCRLPGHGQ SCSPTSSARQALGESRGCTSPGEKCSDAFECFRHYPRGCHGGTNIPSSSARVYAPQDQLSTEPQLQSMTQKQMRKQEACHKKSLRATKHSKRTSEIDNLRINFEMVPGHAKVAWP* 0 >NEUR2_calMil Callorhinchus milii (elephantfish) frag 0 1 2 GILSLVGNSVLLFVAYRKRQILKPAEYFVANLAVSDISMTVTLLPLAISSNFSHR 2 1 WLFVSKpCMYYGFCSMLFGICSLTNLTVLSTVCCMKVCFPAY 1 2 0 0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVISYTMTVIAVNFVVPLSVMFFCYYNV
NEUR3: 16 vertebrate newtopsins
>NEUR3_galGal Gallus gallus cOpn5L2 AB368183 chr3 XM_420056 CN231992 testis exon 2^3 rel NEUR1/2 0 MEEQYISKLHPVVDYGAGVFLLII 1 2 AILTILGNSAVLATAVKRSSLLKSPELLTVNLAVADIGMAISMYPLAIASAWNHAWLGGDASCIYYALMGFLFGVCSMMTLCAMAVIRFLVTNSSKSN 1 2 SNKISKNTVHILITFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHSSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNINRIPHAAKLEKKLTL 0 0 MAVLISVGFLSAWTPYAAASFWSIFNSSDSLQPIVTLLPCLFAKSSTAYNPFIYYIFSKTFRHEIKQLQCCWGWRVHFFSADNSAENSVSMMWSGRDNIRLSPTAKVESQGAARH* 0 >NEUR3_taeGut Taeniopygia guttata ABQF01025032 0 MEEQYISKLHPVVDYGAGVFLLII 1 2 AILTILGNSAVLATAVKRSSLLKPPELLTVNLAVADIGMALSMYPLAIASAWSHAWLGGDASCVYYALMGFLLGVCSMMTLCAMAVIRFLVTNSPKSN 1 2 sNKITKNTVCILIAFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHNSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNIDRIPNAAKLEKKLTL 0 0 MAVLISVGFLSSWTPYAATSFWSIFNSSHSLQPVVTLLPCLFAKSSTAYNPFIYYVFSKTFRCEVKRLQCCCAWRVHYFSSDNSVENPLSTMWSGRDNIRLSAAPQVQNPGAAAP* 0 >NEUR3_melGal Meleagris gallopavo (turkey) 0 MEEQYISKLHPVVDYGAGVFLLI 1 2 PILTILGNSAVLATAVKRSSLLKSPELLTVNLAVADIGMAISMYPLAIASAWNHAWLGGDASCVYYALMGFLFGVCSMMTLCAMAVIRFLVTNSSKSN 1 2 SNKISKNTVHILITFIWLYSLLWAILPLVGWGYYGPEPFGISCTIAWSKFHSSSNGFSFILSMFLLCTVLPALTIVACYLGIAWKVHKAYQEIQNINRIPHAAKLEKKLTL 0 0 MAVLISVGFLSAWTPYAAASFWSIFNSSDSLQPIVTLLPCLFAKSSTAYNPFIYYIFSKTFRHEIKQLQCCWAWRVRFFSTDNSADNSVSMMWSGRDNARLSSNAKVESQGAAMH* 0 >NEUR3_anoCar Anolis carolinensis (lizard) AAWZ01001057 0 MEEHYISKVHPVWDYGMGVFLLII 1 2 AILTILGNSMVLAVAVKRSSCLRSPELLTVNLAATDLGMGLSMYPLAIASAWNHAWLGGEATCIYYALMGFLFGVSSIMTLSAMAVIRFLVTFSSKPA 1 2 GHKINRKVMHICIMLIWAYAVLWAILPLLGWGHYGPEPFGTSCTIAWGQFHNSQKGFAFILSMFILCTFLPAITIIMCYLGIAWKFHKTHQEMQNLNRISSAAKLEKKLIL 0 0 VAVLISVGFLGAWTPYAIVSFWSVFHSSESIPYIVTLLPCLFAKSSTAYNPFIYYTFSKTFRHEVKHLRCYSGQRAQENMKNSINSNVSFMWHGGGNICLSTRQIEMREIPNQ* 0 >NEUR3_xenTro Xenopus tropicalis (frog) cdna ovary embryo 0 MEERYLSKLHPLVDFGSGVFLLLV 1 2 AILTVLGNCAVLATAVKCSSHLKAPDLLSINLAVADLGMAISMYPLAIASAWNHAWLGGDASCLYYALMGFFFGVSSMMTLTVMAIIRYRVTSSFKYS 1 2 GCTIEKKAVCILIMCIWLYALLWAVLPLLGWGRYGPEPFGTSCTIAWGDFHHSSNGFSFIISMFILCTISPAVTIVVCYSGIAWKLHKAYQEIKNQDKIPNSTKVEKKLTL 0 0 LAILVSFGFLISWTPYAAVSFWSLFHSSKYIPPVVSLLPCLFAKSSTAFNPMIYYAFSKTFRRKVKHLKCCCGWRVHFLQSENSVENPRVSVIWTGKENVMVSSVPKLMKGVPGTPTGTQ* 0 >NEUR3a_danRer Danio rerio (zebrafish) 0 MDRYTSKLSPAVDYSAGTFLLVI 1 2 AILSILGNAAVLLTAAWRHSVLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWIGGDPSCLYYGLMGMIFSVASIMTLAVMGLVRYLVTGNPPK 1 2 SGSKFRRKTISILIGVIWMYSLLWAVFPILGWGGYGPEPFGLACSVDWMGYQHSLNRSSFIMALAILCTLMPCVVILFSYSGIAWKLHKAYQSIQSNDNLPNSGAVERKVTL 0 0 MGILISTGFIVSWAPYVFVSLWTMFRSEGEDSVVPIVSLLPCLFAKCSTVYNPLVYYVFRKSFRREIHQIRICCFQGCWDAVSKMTRGDGPEETSGTHETDNI* 0 >NEUR3a_tetNig Tetraodon nigroviridis (pufferfish) 0 MDDKYMSKLSPPVDLWAGIYLVVI 1 2 ALLSVLGNASVLFSASRRLTPLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWMGGDTACLYYGLMGMIFSITSIMTLAVMGMIRYLVTGSPPR 1 2 SGVQFQKKTICVVICAIWLYACLWAAFPLLGWGSYGPEPFGTACSIDWTGYGDSLNNATFIVAMSVLCTFLPCLVIFFTYFGIAWKLHKAYKSIKSSDFQYASVERRITL 0 0 IAVLISVGFLGSWAPYGLVSLWSILKDSSSIPPQVSLLPCLFAKSSTVYNPVIYYIFSQSFKLEVQQLFLCC* 0 >NEUR3a_takRub Takifugu rubripes (fugu) 0 MDDKFTSKLSPAVDLWAGTYLVFI 1 2 ALLSVLGNASVLFSAGRRLSMLKAPELLTVNLAVTDIGMALSMYPLSIASAFNHAWMGGDASCLYYGLMGMIFSITSIMTLAVMGMIRFLVTGTPPR 1 2 SGIKFQKKTISVVISAIWLYACLWAVFPILGWGSYGPEPFGIACSVDWMGYGESLNNATFISTLSVLCTFLPYLVIVFTYFGIAWKLHRAYRSIKSSDIQYTNVERRITL 0 0 MAVMISSGFLIAWTPYVAVSFWSMRNSQRQGHMAPSVTLLPCLFAKSSTAYNPFIYFFFQRNTGHKLLPFHRHAFSCSDRADSSREGEKEESKVSKNLGFTCFGAGTYETCPGLAGDQSQREMAELG* 0 >NEUR3a_gasAcu Gasterosteus aculeatus (stickleback) 0 MEDKYVSKLSPAVDFWAGTYLIII 1 2 AVLSIFGNTAILVSAARRSGPLKAPELLTVNLAVTDLGMALSMYPLSIASAFNHAWIGGDASCLYYSLMGMIFSITSIVTLAVMGMVRYLVTGNPPR 1 2 SGLRLQRKTVSMVIGAVWLYSGLWALFPLLGWGSYGPEPFGLACSIDWSSYGESLNRSTFIMTLSVLCTFLPCLVIFFTYFGIAWKLRRAYQSIRSSDFQHGKVEQKITL 0 0 MAAMISSGFLFSWTPYVAVSLWSMFRSREHIPPLVALLPCLFAKSSTVYNPFIYFIFQRSSWRELLRLHRHLLCCWHRASPPAEGRRSQRGSEGGSWGGACESDDAFGLVHVMKSNATCQTISWA* 0 >NEUR3_calMil Callorhinchus milii (elephantfish) frag 2 AILSIFGNSVVLLVAAKKSSQLKPPELLTVNLAITDFCSAVTMYPLAVGSAWKHTWLGGDASCKYYGFMDFFFGIASIGTLTVMAIVRFLVTSTTQN 1 >NEUR3_petMar Petromyzon marinus (lamprey) exon frag 0 MAEQGEDDQFRSKLSPTADIAAGTFLLAV 1 2 AVLSLAGNGAVLGVAARRWAKLKAPELLSVNLALTDLGIAASIYPLAVASAWNHRWLGGQPVCTYYAFAGFFFGTASMGTLTAMAGVRYKGTSTQVH 1 2 sVKQITKRAMLAVIVAVWAYALLWSCLPLLGWGR 2 1 YGVEPFGVSCTLAWAELQLTPGGVAFLYAMFVLCLLLPAIAIGLCYAGIVCKLRRAYREGRSKRRTPTARHVESRLTK 0 >NEUR3b_danRer Danio rerio (zebrafish) 0 MDIYSSKLSSAVDYGIGAFLLLI 1 2 TILSILGNLMVLVMAYKRSNHMKPPELLSVNLAVTDLGAAVTMYPLAVASAWNHHWIGGDVSCVYYGLMGFLFGAASMMTLTIMAIVRFIVSLTLQSP 1 2 KEKISKRNAKILVATTWLYALLWAIFPLIGWGKYGPEPFGLSCTLDWRDMKEHSQSFVITIFLMNLILPAIIIVSCYCGIALRLYVTYKSMDDSNHVPNMIKMQRRLMV 0 0 IAVLISIGFVGCWAPYGIVSLWSIYRPGDSIPAEVSMLPCLFAKTSTVYNPFIYYIFSKTFKREVNQLSRFCGRSNICRPTDAKNRPENTIYLVCDVNKSKPGVEDLSLARSKENETQMLPNQDLHE* 0 >NEUR3b_etNig Tetraodon nigroviridis (pufferfish) assembly errs in exon 2 frameshift, used traces 0 MDMYTSALSPALDIGTGCYLLVI 1 2 AVLSFIGNLLVIITAVKKSSKMKPPELLCVNLAVTDLGAAVTMYPLSVASAWSHRWIGGDVTCVYYGLVGFLFEVASIMNLTVLAIVRFTVSLNLQSP 1 2 EEKISWKSVKIMCLLIWLYGVIWAMFPVLGWGRYGPEPFGISCSLAWGQMKNEGFSFVVAMFSFNLAVPALIIVSCYFGMAINLYFTHKKMVNTGNRIPAVIKLHRRLLR 0 0 IAVLISVGFLGSWAPYGLVSLWSILKDSSSIPPQVSLLPCLFAKSSTVYNPVIYYIFSQSFKLEVQQLFLCCLSFRSSRTNNCKSNESSIFMVSNGKNLTPALTQQNTSHAVIMN* 0 >NEUR3b_takRub 0 MDIYSSTLSPALDIGTGCFILVV 1 2 GVLSIIGNLLVIITAVKRSSKMKPPELLCVNLAVTDLGAAVTMYPLSVASAWSHRWIGGDATCIYYGLVGFLFGVASIMNLTILAIVRFTVSLNLQSP 1 2 eEKITWKSVKIMCMWVWLYSIMWAMFPILGWGRYGPEPFGISCSLAWGQMKDEGFSFVVTIFSLNFAVPAVIIICCYFGIAIKLYFTYKKTVNTNQIPVIIKLHRRLLM 0 0 IAVLISVGFLGCWAPYGLVSLWSILKDSSSIPPEVSLLPCMFAKSSTVYNPIIYYMFSQSFKMEVQQLFLWCPSFEFCRTSSNNGNETTIYMVSTGKT* 0 >NEUR3b_gasAcu 0 MDIYASTLSPAVDVGAGCYLLFV 1 2 AVFSIVGNLLVLVMAVKRSSRMKPPELLSVNLAVTDLGAAVTMYPLAVASAWRHRWLGGDATCVYYAVAGFFFGLASIMSLTGLAIVRFIVSLNLQSP 1 2 NEKISWRKVKLLCACTWLYALAWAAFPFLGWGRYGPEPYGLSCSLAWGQMKHEGFSFVVSMFSLNLVLPCVIIAGCYFGIAFKLYFTYRKSNNNSNRLPNVVRRHRRLLA 0 0 IAVLISLGFVVCWSPYAVVSLWSIFHDSGSIPPEVSLLPCMFAKSSTVYNPLIYYIFSQSFRREVKQLWRHLGSTLCSVSNSVNDAAVSNTGKSN* 0 >NEUR3b_oryLat Oryzias latipes (medaka) 0 MDIYASALSPALDIGTGCYLLVL 1 2 TVLSIIGNLLVVIMAFKRSSRMKPPELLSVNLALTDLGAAVFMYPLAVASAWSHHWLGGDVSCIYYGLAGFFFGSASVMNLTALAVVRFIVSLNLHSP 1 2 KEKVSWRKVKILCLWSWLYALIWALFPILGWGRYGPEPFGLSCSLAWGEMKQEGPSFVISLFSFNLVLPSVVIICCYFGIAMKLYFTYKKSANSNHVPNIIKLHRRLLIIA 0 0 ILISIGFIGCWTPYGLVSLWSIFNDSSKIPPEVSLLPCMFAKSSTVYNPMIYYFFSKSFQREVKQLSWLCVGSNPCHVSNSVNDNNIYMVSVNVKSKETRRETLQEITESRQ* 0
NEUR4: 11 vertebrate newwopsins
>NEUR4_ornAna Ornithorhynchus anatinus hypothetical protein XM_001508128 0 MSLSHSLQVPWRNNLTFLNKEAQVSEQGETIIGIYLLAL 1 2 GWMSWFGNSMVIFILHRQRGILNPTDYLTFNLAVSDASVSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 0 VDGFLTLLFGLASINTLAMISVTRYIKGCHPHR 1 2 GHFINTANISVALILIWVSALFWSAGPVLGWGSYT 1 2 DRMYGTCEIDWAEANFSSICKSYIISIFFCCFFLPVSIMFFSYVSIIKMVKSSHTLAGADDPTDRQRRLDRDVTR 0 0 VSVVICTAFIVAWSPYAVISMWSAFGHSVPNLTSVLASLFAKSASFYNPIIYFGMNSKFRKDILVLLPCAKESKEPVKLKKFKNLRQKQ GFTLQKPEKAHVLQVPDSGPMSLINTPPLGNRNSFDLACDNSDFECVRL* 0 >NEUR4_galGal Gallus gallus (chicken) genome gappy 0 MSLQLSPQAPWRNNNISFLSREAAVTEQGETIIGFYLLAL 1 2 GWMSWFGNSVVIFVLYKQRHLLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0 0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPER 1 2 AHCISNSSMTVAMVLIWIAAFFWSAAPLLGWGSYT 1 2 DRMYGTCEIDWAKANFSTIYKSYIISIFICCFFLPVTVMVFSYVSIINTVKLSHALTGLSDPTERQRRMERDVTR 0 0 VSIVICTAFIIAWSPYAVLLLWSAYGHPVPNLPLYLSSLFAKSASFYNPIIYFGMSSKFRRDIFILFHCAKEVKDPVKLKRFKNLKQKQ EPSQKEEKYAAEMHPAPSPDSGVGSPTNTPPPANREEYFGILDTPSNSPDIECDRL* 0 >NEUR4_melGal Meleagris gallopavo (turkey) 0 MSLQLSPQAPWRNNNISFLSREAAVTEQGETIIGFYLLAL 1 2 GWMSWFGNSIVIFVLYKQRHLLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0 0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPDR 1 2 AHCISNSSMTVAMVLIWIAAFFWSAAPLLGWGSYT 1 2 DRMYGTCEIDWAKANFSTIYKSYIISIFICCFFLPVTVMVFSYVSIINTVKLSHALTGFSDPTDRQRRMERDVTR 0 0 VSIVICTAFIIAWSPYAVISIWSAYGHPVPNLTSILASLFAKSASFYNPIIYFGMSSKFRRDIFILFHCAKEVKDPVKLKRFKNLKQKQ EPSQKEEKYAPEMHPAPSPDSGVGSPTNTPPPAKREEYFGILDTPSNNPDIECDRL* 0 >NEUR4_taeGut Taeniopygia guttata (finch) 0 MSVQFSAQAPWRNNNISFLTREAAVTEQGETIIGFYLLAL 1 2 GWLSWFGNSIVIFVLYKQRHVLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVFRDDGFIITSIWTCQ 0 0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPER 1 2 GHCISNSSMSVALVLIWVAAFFWSAAPLLGWGSYT 1 2 DRMYGTCEIDWAKASFSTIYKSYIVSIFICCFFLPVTVMVFSYVSIINTVKLSHTLTGLGDPTDRQRRIERDVTR 0 0 VSIVICTAFIIAWSPYAVISIWSAYGHPVPNLTSILASLFAKSASFYNPIIYFGMSSKFRRDIFIFHCAKELKDPVKLKRFKNLKPKQ PQPSQKEEKYAPEMHPAPSPDSGVGSPTNSPPPANREVYFGILDTPSNNPNIECDRL* 0 >NEUR4_anocar Anolis carolinensis (lizard) 0 MSLQVSPQAPWRNNNVTFSNKEVPVSEQGETIIGFYLLAL 1 2 GWMSWFGNSIVIFVLYRQRAGLQPTDYLTFNLAVSDASVSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 0 VDGFLTLLFGLASINTLTVISVTRYIKGCHPDR 1 2 GKCISNSSISVALFLIWIAAFFWSVAPVLGWGSYr 1 2 DRMYGTCEIDWAKANFSTIYKSYIVSIFICCFFLPVSVMVFSYVSIINTVKSSHALSGVGDPTERQRRMERSVTR 0 0 VSLVVICTAFITAWSPYAVISMWSAYGYTVPNLTSILASLFAKSASFYNPIIYFGMSSKFRKDIFVLLHCAKEIKDPVKLKRFKNLKQKQ EVSPSQREEKYAADVQPALSPDSGVGRSNTPPPVNREVYFGAFDTFSNNPDVECDRL* 0 >NEUR4a_xenTro Xenopus tropicalis (frog) numerous transcripts TTC13 FAM89A COCH/VIT NEUR4a AKAP MTHFD1 0 MSLQFPRPAPWRNNNLTLLQKENPLTEQGETIIGIYLLAL 1 2 GWLSWFGNSIVIFVLYKQRANLLPTDYLTFNLAVSDASTSVFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 0 VDGFLTLLFGLASINTLTLISVTRYIKGCHPQR 1 2 ANCISNGSITISLALIWIAALFWSVAPLLGWGSYR 1 2 DRMYGTCEIDWTKASFSTIYKSYIISIFICCFFLPVMVMVFCYVSIINTVKSSRALTSEGDLSERQRKMERDVTR 0 0 VSVVICTAFIVAWSPYAVISMWSACGYYVPSLTSILAALFAKSASFYNPLIYFGMSSKFRKDLCVVLPCAKAQKDPVKLKRYKDKKQ GSAPRAREQTEIEQPVQLQPAPSQDSGVGSPSNTPPLRTKDVHIVDIDLVSDNPSYECDRL* 0 >NEUR4b_xenTro Xenopus tropicalis (frog) XM_002932842 no transcripts, not tandem 54% identity COCH/VIT NEUR4b SCFD1 0 MDGLLMDSSSLLPNSSSGARVLEEGETAIGAYLLLL 1 2 GWLSWLGNGAVICLMCKRRRLLDSHDLLTLNLAVSDAGISIFGYSRGIVELFHGLGKDGFLANNLWTCQ 0 0 VGGFLILLFGLMSISTLTAISLLRYIKGCQPHK 1 2 AHMVDQRHVTMAIVFIWISSIFWSGSPVLGWGSFT 1 2 ERKYGTCEIDWVQAASSTVYKSYVIGVFIWGFVLPVSIMVFCYVSIIRTVHKSHRNSRGGEISQRQLTMERDITR 0 0 VSFVICTAFLLAWSPYAVISMWSACGYQVPGLTGVAATLLAKSASFYNPIIYLGMSPKFRQELRALLCCLRQSGDSPQSFEKPVIT HEPKMKQCNSPSNSLAAKMEQPVLEAQGIQESTLIKGAADSLTVNSQTSDPVKNIDISLDFPMESHQI* 0 >NEUR4a_danRer Danio rerio (zebrafish) 0 MSAQNPLQVVNIPWRNNNFSLMSRDPPLSDQGETIIGVYLLIL 1 2 GWLSWFGNSIVIFVLFRQRSTLQPTDYLTLNLAVSDASISVFGYSRGILEIFNIFKDSGYIISSVWTCQ 0 0 VDGFFTLVFGLSSINTLTVISITRFIKGCHPHK 1 2 AHCITNSTVAVCVVFIWIGAFFWSAAPVLGWGSYT 1 2 DRGYGTCEIDWVKANYSTIHKSYIISIFIFCFLVPVLLMLFCYISIINTVKRGNAMNADGDLSDRQRKIERDVTI 0 0 VSIVICTAFILAWSPYAVVSMWSAWGFHVPNLTSIFTRLFAKSASFYNPLIYFGLSSKFRKDVSVLLPCGREGRDPVRLKRFKRLRGRA EPPGAPAHTPHPQIALKNYNNHSKPHAGPAHCTGHAPSPDSGVGSHHETPPPQPRPQLFFIDVPEPEAESECVRL* 0 >NEUR4b_danRer Danio rerio (zebrafish) COCH/VIT NEUR4b SCFD1 retinal transcript: DN901362 0 MDIHSIPPTNITVYRVSDGGETAIGVYLVIL 1 2 GWLSWIGNGTVILLLTKQRKALEPQDFLTLNLAISDASISIFGYSRGILEVFDVFRDEGYLIKTFWTCK 0 0 VDGFLILLFGLISINTLTAISVIRYIKGCHPHH 1 2 AHHINKRNICLVITAVWLFCLFWAGAPLLGWGSYR 1 2 ARGYGTCEIDWTRALYSIPFKLYVIGIFFFNFFVPLFIIVFAYVSIIRTVNSSHKSSQGGDVSERQKKIERSITR 0 0 VSLILCAAFLLAWSPYAVISMWSALGYQIPTLNGILASLFAKSASFYNPFIYIGMSKFRKDLQALFYCLRKDQVMRCFRCNSVPFLMQTSLKVGNSTGTLF* 0 >NEUR4_tetNig Tetraodon nigroviridis (pufferfish) 0 MEPSRPWRNSSVLGGGAEPPLSEQGETIIGVYLLLL 1 2 GWLSWFGNTVVLFVLVRQRSSLQPTDLLTFNLAVSDASISVFGYSRGIIQIFNVFQDSGFIISSIWTCE 0 0 VDGFLTLIFGLSSINTLTVISITRYIKGCQPSR 1 2 AALISRSSVSVCLLLIWTTAGFWSGAPLLGWGSYT 1 2 DRGYGTCEIDWSKAASSGVYRSYIISIFIFCFFIPVFIMLFCYISIINTVKRGNALAADGHLSHRQRTMERDVTV 0 0 ISVVICTAFIMAWSPYAVVSMWSAWGFHVPSTTSIVTRLFAKSASFYNPLIYFGMSSKFRKDVSLILPCAKERREVVLLQRFKNIKPKAA AAPPPPPLPVYRPKEKNEDEPKLSVHDNDSGVNSPPETPPSDAQEVFPVDPPSQIETSEYWSDRL* 0 >NEUR4_takRub recent pseudogene 8/8 traces support stop codon indel too 94% identity 0 MADSIPPWRNSSVLGGGAEPPLSEQGETIIGVYLLLLG 1 2 GWLSWFGNTVVLFVLYRQRSTLQPTDYLTFNLAVSDASISVFGYSRGIIEIFNVF*DSGFIISSIWTCE 0 0 VDGFFTLVFGLSSINTLTVISITRYIKGCQPSR 1 2 AGHINRTFVSVCLLLIWIMAGFWSGSPLLGWGSYT 1 2 DRGYGTCEIDWSKAAYSTAYRSYIISIFIFCYFIPVFIMLFCYISIINRVKRGNALAA-GDLTDRQRKMERDVTI 0 0 VSIVICTAFILAWSPYAVVSMWSAWGFHVPNLTSIFTRLFAKSASFYNPLIYFGLSSKFRKDVAVLLPCTKDAKDTVKVKRFK NIKPKAAAAPPPPPLPVYRPKEKNEDEPKLSVHDNDSGVNSPPETPPSDAQEVFPVDPPSQIETSEYWSDRL* 0 >NEUR4_gasAcu Gasterosteus aculeatus (stickleback) 0 PVKVVNIPWRNNNLSNLNTDPPLSEQGETFIGVYLLVL 1 2 GWLSWFGNSLVMFVLYRQRASLQSTDFLTLNLAISDASISIFGYSRGILEIFNIFNDDGYLINWIWTCQ 0 0 VDGFFTLLFGLASINTLTVISVTRYIKGCHPNK 1 2 AYCISTNTIAVSLICIWTGAVFWSVAPLLGWGSFT 1 2 DRGYGTCEVDWSKANYSTIHKSYIISILIFCFFIPVMIMLFSYVSIINTVKSTNAMSADGFLSTRQRKVERDVTRV 0 0 ISIVICTAFITAWSPYAVVSMWSAWGFHVPSTTSIITRLFAKSASFYNPLIYFGMSSKFRKDVSVLVPCTRERREVVHLQHFKNIKPKAEAPPTPASLPVQKLGAKYAVPNPDADSGVNNPPQRPATDPQGDLNIDLPSHIETSEYWCDRL* 0 >NEUR4_oryLat Oryzias latipes (medaka) frag 0 MEITLKAFPLKVVNIPWRNNNLSTLHSEPPLSEQGETVIGVYLLVL 1 2 GWLSWFGNSLVIFVLCKQRASLQPTDFFTLNLAVSDASISVFGYSRGILEIFNILKDDGYLITWIWTCQ 0 0 VDGFLTLLFGLVSINTLTVISVTRYIKGCHPHK 1 2 AHCISSSTIAVSLIIVWAAALFWSVAPLLGWGSYT 1 2 DRGYGTCEVDWSKANYSTFYKSYIISILIFCFFIPVVIMLFSYVSIINTVKSTNAMSAVGFLSARQRKMERDVTRV 0 0 ISIVICTAFITAWSPYAVVSMWSAWGFHVPSTTSIITRLFAKSASFYNPLIYFGMSSKFRKDVSVLVPCTRERREVVHLQHFKNIKPKAEAPPTPASLPVQKLGAKYAVPNPDADSGVNNPPQRPATDPQGDLNIDLPSHIETSEYWCDRL* 0 0 SASFYNPLIYFGMSSKFRKDISVLLPCAAEGREVVHLQRFQNIKPKADTPLTAAPHPPPAKPLAAEMNQTNADGDPGVNNPPHTPPQIFHIDVPSHIETSEFWCDRL* 0 >NEUR4_calMil Callorhinchus milii (elephantfish) frag 0 MGCSLGWKVLLWFLHGILICPRPWRNHNSTFQPKEHPISEQGETIIGVYLLIL 1 2 GWLSWFGNSIVIFILYRQRLSLQPPDYLTLNLAVSDASISIFGYSRGIIEIFNVFRDDGFLITSIWTCQ 0 0 1 2 AVSISAGSIAASLVLIWIAAIFWSGAPLFNWGSYT 1 2 DRMYGTCEIDWSRASFSTIYKSYIISIFICCFFLPVFVMLFSYISIINTVKSSHAFAGNADLSDRQRRMEKDVTR 0 0 VSMVICTAFIIAWSPYAVISMWSASGYTVPQLTGIFASLFAKSASFYNPMIYFGLNSKFRKDIYILLPCVKEPKESVKLKRFKHLRHRPEQQQANKDRYAEELQQVASPDSGMGSPSKSPPLHNKDVFFVLWLRGLKK >NEUR4_petMar Petromyzon marinus (lamprey) frag 0 1 2 GWLSWLGNGLVIFVLTRQWSSLQPPDLLTLNLALSDASIAVFGYSRGIIEIFNVFQDDGYIIKSTWTCQ 0 0 1 2 PTKVTSTSMVVSLALVWAASLFWSAAPLLGWGSYT 1 2 DRRYGTCEIDWMKATFSTIYKSYIISIFICCFFMPISTMLFAYISIINTVKSSHVTARMGDVSERQRNMERDITRI 0 0 VSIVICCAFILAWSPYAVISMYSACGHRVPALTSLLAALFAKSASFYNPFIYFGMSGKFRADVRAMLPCRATSVKAPRDAVRLKRYRTHVDPERASHRAAVAAREQPAPRAAAPRPASPAPSAARDRDPELDEREFDPEGRASALAEVAAVESRDSGIACTRGKRRASRGDDVEVRNDV* 0
See also: Curated Sequences | Peropsins | RGR phyloSNPs | LWS | Encephalopsins | Melanopsins | Update Blog VDGFLTLLFGLASINTLTVISVTRYIKGCHPDRAHCISNSSMTVAMVLIWIAAFFWSAAP LLGWGSYT