Opsin evolution: RGR phyloSNPs: Difference between revisions
Tomemerald (talk | contribs) No edit summary |
Tomemerald (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:Opsin_RGR_earlySpl.png|left]] | |||
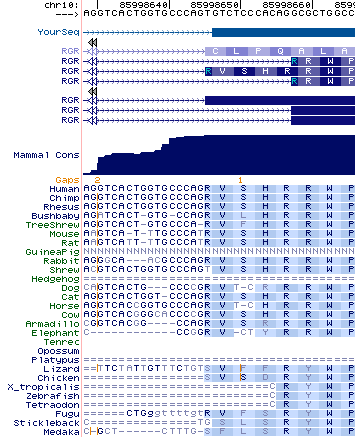
RGR in primates may contain an earlier splice acceptor in the second intron resulting in insertion of four amino acids in the extracellular loop EX1 between TM2 and TM3. This is observed in a number of human transcripts but all appear to originate from a single brain tumor sample (eg BC011349). The intronic region is conserved in the UCSC 28-way track but it cannot be a splice acceptor in tarsier, mouse lemur, or tree shrew much less other placentals. For example, in dog, horse and armadillo, translation would cause loss of reading frame -- and horse even has the AG acceptor. Thus it is difficult to say whether the insert is simply tolerated or has acquired a secondary function. The nucleotide sequence might be conserved as part of a splice enhancer. | |||
A second defective splice isoform has also been [http://www.molvis.org/molvis/v13/a131/ studied] that entirely skips the 6th exon. While exons don't correspond cleanly to transmembrane domains, TM7 is lost -- while the altered has been proven to be produced abundantly by mass spectroscopy (unlike the vast majority of supposed alternative splices), surely it cannot function as an opsin. It is observed to accumulate extracellularly basal boundary of RPE cells and primarily in the extracellular areas of Bruch's membrane, adjacent choriocapillaris, and intercapillary region. The carboxy terminus has been cleaved as well. A role in degenerative eye disease has neither been established nor ruled out. | |||
the | |||
<br clear="all" /> | |||
<pre> | <pre> | ||
| Line 37: | Line 35: | ||
Below set of RGR opsin reference sequences is greatly expanded to 50 species in fish and mammals from the more limited but phylogenetically representative set found in the opsin classifier. Notably absent are marsupial RGR which for other genes are commonly available from three species. By locating syntenic genes in placental mammals and platypus, the appropriate region of the opossum genome can be searched. However the adjacent genes are now on separate opossum chromosomes, suggesting a translocation has disrupted the RGR genes. None of the individual exons can be found by tblastn of the genome or blastn of the trace archives. This lack of even pseudogene debris supports the idea of gene loss and decay sometime after marsupial divergence. | |||
The loss of RGR in marsupials challenges the usual functional explanation given to RGR in placental mammals (replenishing cis-retinal at cone and rod opsins). Since marsupials are known from experiment to have normal color and rod vision, either other pathways for replenishment exist or the role was somewhat different in the ancestor. | |||
>RGR_homSap Homo sapiens (human) | >RGR_homSap Homo sapiens (human) | ||
Revision as of 16:56, 20 February 2008
RGR in primates may contain an earlier splice acceptor in the second intron resulting in insertion of four amino acids in the extracellular loop EX1 between TM2 and TM3. This is observed in a number of human transcripts but all appear to originate from a single brain tumor sample (eg BC011349). The intronic region is conserved in the UCSC 28-way track but it cannot be a splice acceptor in tarsier, mouse lemur, or tree shrew much less other placentals. For example, in dog, horse and armadillo, translation would cause loss of reading frame -- and horse even has the AG acceptor. Thus it is difficult to say whether the insert is simply tolerated or has acquired a secondary function. The nucleotide sequence might be conserved as part of a splice enhancer.
A second defective splice isoform has also been studied that entirely skips the 6th exon. While exons don't correspond cleanly to transmembrane domains, TM7 is lost -- while the altered has been proven to be produced abundantly by mass spectroscopy (unlike the vast majority of supposed alternative splices), surely it cannot function as an opsin. It is observed to accumulate extracellularly basal boundary of RPE cells and primarily in the extracellular areas of Bruch's membrane, adjacent choriocapillaris, and intercapillary region. The carboxy terminus has been cleaved as well. A role in degenerative eye disease has neither been established nor ruled out.
>RGR_ext_homSap VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCCATCGCATGGGGGCGTTATCACCACTACTGCACCCGT >RGR_ext_panTro VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCCATCGCATGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_gorGor VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCCATCGCATGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_ponPyg VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGTTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCCATCGCGTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_nomLeu VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCATTAGCCAGCATCTGCAGCAGTGCAGCCATCGCATGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_macMul VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCTATCGCCTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_papHam VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCAGATGGCTGCCAGGCTCATGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCTATCGCCTGGGGGCGCTATCATCACTACTGCACCCGT >RGR_ext_calJac VSHRRWPYGSDGCQIHGFQGFVTALASICGSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGATTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCGGCAGTGCAGCCATCGCCTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_tarSyr GGTGTCTCCCACAGGCGCTGGCCGTACGGCTTGGACGGCTGCCAGGCTCACGGCTTCCAAGGCTTTGTGACAGCTTTGGCCAGCATCGGCGGCAGCGCAGCCATCGCCTGGGGGCGCTATCATCACTACTGCACTCGT >RGR_ext_otoGar VLHRRWPYGSGGCQAHGFQGFTTALASICGSAAIAWGRYHHYCTR AGTGTTCTCCACAGGCGCTGGCCCTACGGCTCAGGTGGCTGCCAGGCTCACGGCTTCCAGGGCTTCACGACGGCATTGGCCAGCATCTGCGGCAGCGCAGCCATCGCCTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_micMur CGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGATGGCTGCCAGGCTCACGGCTTCCAGGCTGCGTGACGGTGCTGGCCAGCATCTGCAGCAGCGCGGCCATCGCCTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_tupBel CATGTCTTCCACAGGCGCTGGCCCTACGGCTCAGACGGCTGCAAGGTTCATGGCTTCCAGGGCTTCGCAACAGCGTTGGCCAGCATCTCTGGCAGCGCGGCCATCGCCTGGGGGCGCTATCACCAGTACTGCACTCGT
Below set of RGR opsin reference sequences is greatly expanded to 50 species in fish and mammals from the more limited but phylogenetically representative set found in the opsin classifier. Notably absent are marsupial RGR which for other genes are commonly available from three species. By locating syntenic genes in placental mammals and platypus, the appropriate region of the opossum genome can be searched. However the adjacent genes are now on separate opossum chromosomes, suggesting a translocation has disrupted the RGR genes. None of the individual exons can be found by tblastn of the genome or blastn of the trace archives. This lack of even pseudogene debris supports the idea of gene loss and decay sometime after marsupial divergence.
The loss of RGR in marsupials challenges the usual functional explanation given to RGR in placental mammals (replenishing cis-retinal at cone and rod opsins). Since marsupials are known from experiment to have normal color and rod vision, either other pathways for replenishment exist or the role was somewhat different in the ancestor.
>RGR_homSap Homo sapiens (human) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSQLAWNSAVSLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKREKDRTK * 0
>RGR_panTro Pan troglodytes (chimp) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSQLAWNSAISLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAIPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSEKDRTK * 0
>RGR_gorGor Gorilla gorilla (gorilla) 0 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLAPAESGISLNALVGATSTLLG 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 GSTLACKSAVSLVLSGRMSSAFWADLPLLGWGPYDYEPLRTCCTLDYSEADR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSKKDRTK * 0
>RGR_ponPyg Pongo pygmaeus (orang_abelii) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 GSQLAWNSAISLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAVLYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSEKDRTK * 0
>RGR_nomLeu Nomascus leucogenys (gibbon) 0 MAETSVLPTGFGELKLLAGGMGLLAE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 GSQLAWNSAISLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAVNYALGNEMVCRGIWQCLSPQKSEKDRAK * 0
>RGR_macMul Macaca mulatta (rhesus) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALIAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSQLAWNSAISLVLFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSEKDRAK * 0
>RGR_papHam Papio hamadryas (baboon) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALIAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSEKDRAK * 0
>RGR_calJac Callithrix jacchus (marmoset) 0 MAESSTLPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNSLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQIHGFQGFVTALASICGSAAIAWGRYHHYCT 1 2 GSQLAWNSAISLVLFVWLSSTFWAAFPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYRLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTIDAINYALGNEMICRGIWQCLSPQKSEKDRTK * 0
>RGR_tarSyr Tarsius syrichta (tarsier) 0 MAEAGALPAGFGELEVLAVGMVLLVE 1 2 ALSGLSLNSLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALIGATSSLLR 2 1 RWPYGLDGCQAHGFQGFVTALASIGGSAAIAWGRYHHYCT 1 2 GSQLAWNTAISLVLFVWLSYAFWAALPLLGWGHYDYEPLGTCCTLEYSKGDR 2 1 0 0 VNTTLPIRTLMLGWGPYALLYLCAVIADVTSISPKLQM 0 0 VPALIAKTVPTINAYHYALGSEMVCRGIWQCLSPHSSE * 0
>RGR_otoGar Otolemur garnettii (bushbaby) 0 MAEPGTLPAGFGEIEVLAVGTVLLVE 1 2 2 1 RWPYGSGGCQAHGFQGFTTALASICGSAAIAWGRYHHYCT 1 2 GRPLAWSTAISLVLFVWLSSAFWAALPLLGWGHYDYEPLRTCCTLDYSRGDR 2 1 NFTSFLFTMAFFNFLTPLFITLTSYQLMEQKLRRSGHLQ 0 0 VNTTLPARTLLLGWGPYALLYLYATIADVTSISPKLQM 0 0 VPALIAKTVPTINAVNYALGSEMVCRGIWQCLSLQRSKQDGAK * 0
>RGR_micMur Microcebus murinus (mouse_lemur) 0 MAEPGTLPTGFRELEVLAVGTVLLVE 1 2 2 1 RWPYGSGGCQAHGFQGFTTALASICGSAAIAWGRYHHYCT 1 2 GSPLAWSTAISLVLFVWLSSAFWAALPLLGWGHYNYEPLGTCCTLDYSRGDR 2 1 NVTSFLFTMAFFNFLIPLFITHTSYQLMEQKLKKSGHLQ 0 0 VNTTLPARTLLLGWGPYALLYLYATVADVTSISPKLQM 0 0 VPALIAKTVPTINAINYALGSETVCRGIWQCLSPQRSEQDRAK * 0
>RGR_tupBel Tupaia belangeri (treeshrew) 0 MAESGALPSGFGELEVLAVGTVLLVE 1 2 ALSGLSLNSLTVFSFCKSPELRTPSHLLVLSVALADSGISLNALIAATSSLLR 2 1 RWPYGSDGCKVHGFQGFATALASISGSAAIAWGRYHQYCT 1 2 2 1 NFTSFLFTMAFFNFLMPLFITLTSYWLMEEKLRKGGRLQ 0 0 VNTTLPSRTLLLGWGPYALLYLYAAFADVTPLSPKLQM 0 0 VPALVAKMVPTVNAVNYALGSETICRGIWGCLSPKRERDRAR * 0
>RGR_musMus Mus musculus (mouse) 0 MAATRALPAGLGELEVLAVGTVLLME 1 2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2 1 RWPHGSEGCQVHGFQGFATALASICGSAAVAWGRYHHYCT 1 2 GRQLAWDTAIPLVLFVWMSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2 1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKFSRSGHLP 0 0 VNTTLPGRMLLLGWGPYALLYLYAAIADVSFISPKLQM 0 0 VPALIAKTMPTINAINYALHREMVCRGTWQCLSPQKSKKDRTQ * 0
>RGR_ratNor Rattus norvegicus (rat) 0 MTATRALPAGFGELEVLAIGIVLLME 1 2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2 1 RWPHGSEGCRVHGFQGFATALASICGSAAIAWGRYHHYCT 1 2 GRQLAWDTAIPLVLFVWLSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2 1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKLSRSGHLQ 0 0 VNTTLPGRMLLLGWGPYALLYLYAAVADVSFISPKLQM 0 0 VPALIAKTMPTINAINYALRSEMVCRGTWQCRSAQKSKQDRTQ * 0
>RGR_speTri Spermophilus tridecemlineatus (ground_squirrel) 0 MAETAALPAGFGELEVLAVGTVLLVE 1 2 ALSGLSLNGLTIFSFCKTPELRTPNHLLLLSLAVADSGISLNALIAAISSLLR 2 1 RWPYGSDGCQAHGFQGFVTALSSICGSAAIAWGRYHHYCT 1 2 GSQLAWNTAIPLVLFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFISFLFTMAFFNFFVPLFITLTSYRLMEQKLARSGHLQ 0 0 0 0 * 0
>RGR_dipOrd Dipodomys ordii (kangaroo_rat) 0 MATSGDLPTGFGELEVLTVGTVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPIHLLDLSLAVADSGISLNALIAAISSLEW 2 1 HWPYGLEGCQAHGFQGFVTALASISGSAAIAWGRCHHHCT 1 2 GSLLGWDTAVSLVIFVWLSSAFWAALPLLGWGHYNYEPLGTCCTLDYSRGDR 2 1 NFTSFLFTMAFFNFLVPLFITLTSYQLMKQKFARSGRLQ 0 0 VNTTLPTRTLLLGWGPYALLYFYAAIMDVNSISPKLQM 0 0 VPALIAKMVPTVNAINYALCNELLCGGFSLGLLPQKGKQDRTQ * 0
>RGR_cavPor Cavia porcellus (guinea_pig) 0 MATSEALPAGFGELEVLAVGTVLLLE 1 2 GLCGLSLNGLTVVSFWKSPALRTPNHLLVLSLALADSGLSLNALVAAGSSLLR 2 1 HWPGSGHCQALGFQGFTTALASISGTAALSWGRHQQCCT 1 2 RGRLTWSTAVPLVLFVWLSSAFWAALPLLGWGRYDYEPLGTCCTLDYSTGDR 2 1 NFTSFLFTMAFFNFLVPLFITVTSCQLMERHLARSSRLQ 0 0 VSVRQPARTLLLCWSPYALLYLYAVLADAHTLSPRLQM 0 0 VPALIAKTVPTIYSLGRGPWQSLEMQRSKQD * 0
>RGR_oryCun Oryctolagus cuniculus (rabbit) 0 MAEPGTLPPGFEELEVLAVGTVLLVE 1 2 ALSGLSLNGLTIFSFCKTPELWTPSHLLVLSLAVADSGISLNALIAAVSSLLR 2 1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1 2 GSQLAWNTAVLLVLFVWLSSVFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFISFLITMAFFNFLMPLFITLTSYSLMEQKLSKSGRLQ 0 0 VNTTLPGRTLLFCWGPYAVLYLCAAVADMSSITLKLQM 0 0 VPALIAKTVPTVNAVNYALGSEVIRRGIWQCLLPQRSVRGRAQ * 0
>RGR_ochPri Ochotona princeps (pika) 0 MAEPGTLPPGFEELEVLAVGTVLLVE 1 2 ALTGLSLNSLTIFSFCTSPELRTPSHLLVLSLALADSGVSLNALAAATASLLR 2 1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1 2 GSQLAWNTAVLLVLFVWLSSVFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFISFLVTMAFFNFLMPLFIMLTSYSLMEQKLAKSGRLQ 0 0 VNTTLPARTLLFCWGPYAILCLCATVMDMSTVSPKLLM 0 0 VPALIAKAVPTVNAINYALGSEVIRRGIWQCLLPQRSVRDRAQ * 0
>RGR_canFam Canis familiaris (dog) 0 MADSGALPAGFGELEVLAVGTVLLVE 1 2 ALTGLCLNGLTILSFCKTPELRTPTHLLVLSLAVADTGISLNALVAAISSLLR 2 1 RWPYGPDGCQAHGFQGFATALASICSSAALAWGRYHHYCT 1 2 RGQLAWNTAISLVLCVWLSSVFWAALPLLGWGRYDYEPLGTCCTLDYSRVDR 2 1 NFTSYLFTMAFFNFFLPLLITLVSYRLMEQKLKKPGHLQ 0 0 VSTTVPARTLLLCWGPYALLYLYATVADVRSVPPKLQM 0 0 VPALIAKAAPTINAIHYALGGDMVHGGLWQCLSPQRSQPDRAR * 0
>RGR_felCat Felis catus (cat) 0 MAESGSLPTGFGELEVLAVGMVLLVE 1 2 2 1 RWPYGSNGCQAHGFQGFVTALASICSSAAIAWGRYHHYCS 1 2 GSQLAWNTAISLVICVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFTSFLFTMAFFNFFMPLFITFISYRLMEQKLRKTGHLQ 0 0 VNTTLPARTLLFGWGPYALLYLYATIADVSSVSPKLQM 0 0 VPTINAINYALGSEMVHRGIWQCLSPQGSGLDRAR * 0
>RGR_bosTau Bos taurus (cow) 0 MAESGTLPTGFGELEVLAVGTVLLVE 1 2 ALSGLSLNILTILSFCKTPELRTPSHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSEGCQAHGFQGFVTALASICSSAAVAWGRYHHFCT 1 2 GSRLDWNTAVSLVFFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFTSFLFTMAFFNFLLPLFITVVSYRLMEQKLGKTSRPP 0 0 VNTVLPARTLLLGWGPYALLYLYATIADATSISPKLQM 0 0 VPALIAKAVPTVNAMNYALGSEMVHRGIWQCLSPQRREHSREQ * 0
>RGR_turTru Tursiops truncatus (dolphin) 0 MAESGALPSGFGELEVLAVGTVLLVE 1 2 ALSGLSLNSLTILCFCKNPELRTPSHLLVLSLALSDSGISLNALMAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 GSRLDWNTAVSLVFFVWLSSAFWATLPLLGWGHYDREPLGTCCTLDYSRRDR 2 1 NFTSFLFTMAFFNFLLPLFITVISYRLMEQKLGKTGRPP 0 0 VNTVLPARTLLFGWGPYALLYLYAAVADVTSISPKLQM 0 0 * 0
>RGR_susScr Sus scrofa (pig) 0 MAEPGALPTGFGELEVLAVGTLLLVE 1 2 ALSGLSLNSLTILSFCKTPELRTPSHLLVLSLALADSGISLNAFVAATSSLLR 2 1 RWPYGSEGCQAHGFQGFATALASICSSAAIAWGRYHHYCT 1 2 RSRLDWNTAVSLVFFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSRVDR 2 1 NFTSFLFTMAFFNFLLPLFITVTSYRLMEQKLGKTGRPP 0 0 VNTILPARTLMLAWGPYALLYLYATFADVTSISPKLQM 0 0 VPALIAKMVPTVNAINYALGGEMVHRGIWQCLSPQRRERDREQ * 0
>RGR_vicVic Vicugna vicugna (vicugna) 0 MAESRALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNSLTILSFCKTPELRTPNHLLVLSLALADSGISLNALVAATSSLLR 2 1 1 2 GSRLDWNTAVSLVFFVWLSSTCWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 0 0 0 0 * 0
>RGR_equCab Equus caballus (horse) 0 MAESGSLPTGFRELEVLAVGTVLLVE 1 2 ALAGLSLNSLTILSFCKTPELRTPSHLLVLSLAVADSGLSLNALVAATSSLLR 2 1 RWPYGSEGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSRLAWNTAVFLVFFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLLTMAFFNLLLPLLITLTSYRLMEQKLGKTGQLQ 0 0 VNTTLPARTLLLCWGPYALLYLYATVADATSISPKLRM 0 0 VPALVAKTVPTINAVNYALGSEMLHRGIWQCLSPQKSERDRAQ * 0
>RGR_myoLuc Myotis lucifugus (microbat) 0 MAEAGSLPTGFGELEVLAVGVVLLVE 1 2 ALTGLSLNSLTIFSFCTSPELRTPSHLLVLSLALADSGVSLNALAAATASLLR 2 1 RWPYGSGGCQAHGFQGFAAALASICGSAAVAWGRYHHYCT 1 2 GSRLAWRTAASLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCSLGYARGSR 2 1 NFTSFLFLMAFFNFLLPLFITFTSYRLMEQKLGRTRPPQ 0 0 VNTTLPARTLLLGWGPYALLHLCAALAGTALIPPRLQV 0 0 VPALIAKMVPTVNAVNYALGSGIWQRLSLQ * 0
>RGR_pteVam Pteropus vampyrus (macrobat) 0 MAESRSLPTVFWELEVLAVGTVLMVE 1 2 ALSGLSLNSLTILSFCKNPELRTPIHLLVLSLALADSGISLNALIAATSSLLR 2 1 RWPFGPDGCQAHGFQGFATALASICSSAAIAWGRYHHYCT 1 2 GSRLAWNTAVSLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSRRDR 2 1 NFTSFLFTMAFFNFVLPLFITLTSYQLMEQKLGKTGHPQ 0 0 VNTTLPARTLMLCWGPYALLYLYAAVMDVASISPKLQM 0 0 VPALIAKMAPTINAVNYALGSEMVQRGIWQCLSPQRSERDHAQ * 0
>RGR_sorAra Sorex araneus (shrew) 0 MTESGALPAGFKELKMLAVGTLLLWG 1 2 2 1 RWPFGPDGCQAHGFQGFATALASICSSAAIAWGRYHHYCT 1 2 GRQLAWDVAIALVIFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSRGGR 2 1 NFVSFLFTMAFFNFLLPLFITVTSYRLMEQKLGKMGQPQ 0 0 0 0 VPALIAKTVPTVNALHYGLGSGMVQNGFRKGLWLQRRERERAL * 0
>RGR_eriEur Erinaceus europaeus (hedgehog) 0 1 2 2 1 RWPYGSDGCQAHGFQGFVMALASICSSAAIAWGRYHHHCT 1 2 RSRLAWNTAVFLVFFVWVSSVFWAALPLLGWGHYDYEPLGTCCTLDYSSGDR 2 1 NFISFLFTMAFFNFLLPLFITLISYQLMEQKLRKTGHPQ 0 0 VNTTLPARTLLLGWGPYALLYLYAVIADVALLSPKLQM 0 0 VPALIAMVPTVNAVHYVLGSEKVHKGFWQCFSPQRSEQDRAR * 0
>RGR_loxAfr Loxodonta africana (elephant) 0 MAEPGHLPAGFQELEVLTVGTVLLLE 1 2 ALSGLSLNGLTILSFCKIPELRTPGHLLVLSLALADSGISLNALVAAMSSLRR 2 1 RWPYGSDGCQAHGFQGFVTALASICSCAAIAWERYHHYCT 1 2 RSRLAWSSASALVLFVWLSSAFWAALPLLGWGRYNYEPLGTCCTLDYSRGDR 2 1 NSTSFLLTMAFFNFLLPLFITLTSYRLMEQKLKKKGPLQ 0 0 VNTTLPARTLLLGWGPYALLYLCAAATDMTSISPRLQM 0 0 VPALVAKAVPVINACHYALGSEVVRGGIWQYLSRQRGESPLRARDRTH * 0
>RGR_proCap Procavia capensis (hyrax) 0 MADPRPLPTGFGELEVLTVGTVLLVE 1 2 ALSGLSLNGLTILSFYKIPELRTPGHLLVLNLALADSGMSLNALVAAVSSLRG 2 1 RWPYGSDGCQAHGFQGFVMALTSICSCAAIAWERYHHYCT 1 2 GSKLAWSSAGALVLFMWLSSAFWAALPLLGWGRYNYEPLGTCCTLDYSRGDR 2 1 NSTSFLFTMAFFNFLLPLFITLASYRLMEQKLKKEGPLQ 0 0 VNTTLPARTLLLGWGPYALLYLYTAITDVNSISPKLQM 0 0 VPALIAKAVPIVNACHYALGSETVHRGIWQCLSRQRGESPPRTRDRTQ * 0
>RGR_echTel Echinops telfairi (tenrec) 0 MVEPRTLPPGFGELEVLAVGTVLLVE 1 2 2 1 HWPYGSGGCQAHGFQGFTVALASICSCAAIAWERYHHYCT 1 2 GSQFTWSSASTLVLFMWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMTFFNFSMPILVTLTSYQLMQQKLKKSGPLQ 0 0 VNTTLPTRTLLLGWGPYALLYLCAACTDVTGISPKLQM 0 0 VPAIVAKAVPIVNACHYALGNKVLRRGIWQFLSQQSGRQDRTQ * 0
>RGR_dasNov Dasypus novemcinctus (armadillo) 0 MAGSGVLPPGFGELEVLAVGTVLLVE 1 2 ALSGLVLNGLAIISFCKTPELRSPSRLLVLSLALADSGVSLNALVAATSSLLR 2 1 RWPYGSGGCQAHGFQGFVTALASISSSAAIAWERCHRHCI 1 2 GRRLAWSTAGCLVLCLWMAAAFWAALPLLGWGLYDYEPLGTCCTLDYSRGDR 2 1 NFISFLVTLALFNFFLPLLIMLTSYRLMAQKLKRSGHVQ 0 0 VSTALPGRLLLLGWGPYALLYLYAAVADATSLSPRLQM 0 0 VPALIAKTMPTVNALYYALGRESVHRNA * 0
>RGR_choHof Choloepus hoffmanni (sloth) 0 MAESRVLPTGFGELEVLAVGIVLLVE 1 2 ALSGLTLNGLTLFSFCKTPELQRPSHLLVLSLALADSGVSLNALLAATASLIG 2 1 RWPHGSDSCQAHSFQGFATALASISSSAAIAWERYRHHCT 1 2 GSQLSWSTAGSLVLCVWLSSVFWATLPILGWGHYDYEPLGTCCTLGYSRGDR 2 1 0 0 0 0 VPALIAKTMPTINAFQYALGSETVCRDIWQCLPRLRSMGRSSGHD * 0
>RGR_ornAna Ornithorhynchus anatinus (platypus) 0 1 2 ALLGLCLNGLTIASFRKIKELRTPSNLLVVSLALADSGICLNALMAALSSFLR 2 1 HWPYGAEGCRLHGFQGFATALASISLSAAIGWDRYLRHCS 1 2 RSKPQWGTAVSTVLFAWGFSAFWSMMPILGWGQYDYEPLRTCCTLDYSKGDR 2 1 NFTTYLFAVAFFNFVIPLFIMLTSYQSIEQRFKKSGLFK 0 0 LNTRLPTRTLLFCWGPYALLCFYATVENVTFISPKLRM 0 0 IPALIAKTVPVIDAFTYALRNEDYRGGIWQFLTGQKIERVEVENKIK * 0
>RGR_anoCar Anolis carolinensis (lizard) 0 MVTTAYPVPEGFTDLEVFVIGTALLVE 1 2 ALLGFSLNMLTIVSFWKIKELRTPGNFLVFNLALSDCGICFNAFIAAFSSFLR 2 1 YWPYGSDGCQIHGFHGFLTALTSISSAAAVAWDRHHQYCT 1 2 GNKLQWGSVIPMTIFLWLFSGFWAAMPLLGWGEYDYEPLRTCCTLDYTKGDR 2 1 NYITYLIPLALFHFMIPGFIMLTAYQAIDHKFKKTGQFK 0 0 FNTGLPVKSLVICWGPYSFLCFYAAVESVTFISPKILM 0 0 IPAVIAKSSPAANALIYALGNENYQGGIWQFLTGQKIEKAEVDNKTK * 0
>RGR_galGal Gallus gallus (chicken) 0 MVTSHPLPEGFTEIEVFAIGTALLVE 1 2 ALLGFCLNGLTIISFRKIKELRTPSNLLVLSIALADCGICINAFIAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFLTALASISSSAAVAWDRYHHYCT 1 2 RSKLQWSTAISMMVFAWLFAAFWATMPLLGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYITFLFALSIFNFMIPGFIMMTAYQSIHQKFKKSGHYK 0 0 FNTGLPLKTLVICWGPYCLLSFYAAIENVMFISPKYRM 0 0 IPAIIAKTVPTVDSFVYALGNENYRGGIWQFLTGQKIEKAEVDSKTK * 0
>RGR_taeGut Taeniopygia guttata (finch) 0 MVTAHPLPEGFTEIEVFAIGTALLVE 1 2 ALLGFCLNGLTIISFRKIKELRTPSNLLVLSIALADCGICINAFIAAFSSFLR 2 1 YWPYGSDGCQIHGFQGFLTALASIGSSAAIAWDRYHHYCT 1 2 RSRLQWSTAVSMMVFAWLFAAFWSVMPLLGWGKYDYEPLRTCCTLDYSKGDR 2 1 NYVTFLFALSTFNFMIPGFIMMTAYQSIHQKFRKTGHFK 0 0 FNTGLPLKTLVICWGPYCLLCTYAAVENVMFIPPKYRM 0 0 IPALIAKTVPTVDAFIYALGNENYRGGIWQFLTGQKIEKAEVDNKTK * 0
>RGR_xenTro Xenopus tropicalis (frog) 0 MVTSYPLPEGFTETEVFAIGTTLLVE 1 2 ALLGLLLNGLTLLSFYKIRELRTPSNLFIISLAVADTGLCLNAFVAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1 2 RSKLHWSTAVSVVFFIWGFSAFWSAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYISYLFTMAFFEFLVPLFILMTAYQSIYQKMKKSGQIR 0 0 FNTSMPVKSLVFCWGPYCLLCFYAVIQDATILSPKLRM 0 0 IPALLAKTSPAVNAYVYGLGNENYRGGIWQYLTGQKLEKAETDNKTK * 0
>RGR_xenLae Xenopus laevis (frog) NM_001092855 mRNA 0 MVTSYPLPEGFTETEVFAIGTTLLIE 1 2 ALLGLLLNGLTLLSFYKIRELRTPSNLFIISLAVADTGLCLNAFVAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1 2 RSKLHWGTAVSMVLFVWGFSAFWSAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 1 NFTSFLFTMAFFEFLVPVFILLTAYQSIYQKMKKSGQIR 0 0 LNTSMPVKSLVFCWGPYCLLCFYAVIQDATILSPKLRM 0 0 MPALLAKISPAVNAYVYGLGNENYRGGIWLYLTGQKLEKAETDSRTK* 0
>RGR_danRer Danio rerio (zebrafish) 0 MVTSYPLPEGFSEFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVIAFLKIRELRTPSNFLVFSLAMADMGISTNATVAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFMTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSSAITLVLFTWLFTAFWAAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLIPMSIFNMGIQVFVVLSSYQSIDKKFKKTGQAK 0 0 FNCGTPLKTMLFCWGPYGILAFYAAVENATLVSPKLRM 0 0 IAPILAKTSPTFNVFVYALGNENYRGGIWQLLTGQKIESPAIENKSK * 0
>RGR_takRub Takifugu rubripes (fugu) 0 MVSSYPLPEGFSDFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVVAFLKVRELRTPSNFLVFSLALADMGISMNATIAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFVTALASIHFVAAIAWDRYHQYCT 1 2 RTKLQWSSAITLAVFIWLFCAFWSAMPLIGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLIPMAIFNMVIQVFVVMSSYQSIAEKFKKTGNPR 0 0 FNPSTPLKAMLLCWGPYGILAFYAAVENANLVSPKLRM 0 0 MAPILAKTCPTINVFLYALGNENYRGGIWQFLTGEKIEAPQIENKSK * 0
>RGR_tetNig Tetraodon nigroviridis (pufferfish) 0 MVSSYPLPEGFSDFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVVAFFKVRELRTPSNFLVFSLAMADMGISMNATVAAFSSFLR 2 1 YWPYGSEGCQTHGFQGFVTALASIHFVAAIAWDRYHQYCT 1 2 RTKLQWSSAITLAVFIWLFCAFWSAMPLIGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLVPMAIFNMVIQVFVVMSSYQSIAEKFKKTGNPR 0 0 FNPNTPLKAMLLCWGPYGILAFYAAVENANLVSPKLRM 0 0 MAPILAKTCPTVNVFLYALGNENYRGGIWQFLTGEKIETPQLENKTK * 0
>RGR_gasAcu Gasterosteus aculeatus (stickleback) 0 MVSSYPLPDGFTDFDVFSLGSCLLVE 1 2 GLLGILLNAVTIAAFLKVRELRTPSNFLVFSLAVADIGISMNATIAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFVTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSSAITLAVFVWLFTAFWSAMPLIGWGEYDYEPLRTCCTLDYTKGDR 2 1 NYVSYLIPMAIFNMAIQVFVVMSSYQSIAQKFKKTGNPR 0 0 FNPNTPLKAMLFCWGPYGILAFYAAVENATLVSTKLRM 0 0 MAPILAKTSPTFNVFLYALGNENYRGGIWQLLTGEKIDVPQIENKSK * 0
>RGR_oryLat Oryzias latipes (medaka) 0 MATSYPLPEGFSEFDVFSLGSCLLVE 1 2 GLLGIFLNSVTIVAFLKVRELRTPSNFLVFSLAMADIGISMNATIAAFSSFLR 2 1 YWPYGSEGCQTHGFHGFLTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSTAITLAVLVWIFTAFWAAMPLIGWGEYDYEPLRTCCTLDISKGDR 2 1 NYVSYVIPMSIFNMGIQVFVVMSSYQSIAQKFQKTGNPR 0 0 FNASTPLKTLLFCWGPYGILAFYAAVADANLVSPKIRM 0 0 IAPILAKTSPTFNPLLYALGNENYRGGIWQFLTGEKIHVPQDDNKSK * 0
>RGR_gadMor Gadus morhua (cod) ES469757 0 MVSAYPLPEGFSDFDVFSFGSFLLVE 1 2 GLLGIILNAVTIVAFCKVKELRTPSNFLVFSLAMADIGISMNASVAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFTVALASIHFVAAIAWDRYHQYCT 1 2 RTELQWSSAVTLSVFIWLFSAFWSAMPLIGWGTYDYEPLRSCCTLDYTKGDR 2 1 NYVSYLIPMTVFNMVVQIFVVMSSYQSIDQKFKKTAKPK 0 0 FNARTPLKTLLFCWGPYGILAFYAAIENASLVSPKLRM 0 0 MAPILAKTAPTFNVFLYALGNENYRGGIWQLLTGEKIVPQEVPQIENKSK* 0
>RGR_pimPro Pimephales promela DT198813 frag 0 MVTYPLPEGFSDFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVVAFLKIRELRTPSNFLVFSLAMADMGISMNATVAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFMTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSSAITLVIFIWLFTAFWSAMPLIGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLIPMSIFNMGIQVFVVLSSYQSIERKFQKSGQAK 0 0 FNCSTPLKTMLFCWGPYGILAFYAAVENATLVSPKLRM 0 0 LAPILA
>RGR_osmMor Osmerus mordax (smelt) EL524757 frag 0 MVSSYPLPDGFSDFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVVAFLKVRELRTPSNFLVFSLALADMGISSNATIAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFMTALASIHFVAAIAWDRYHQYCT 1 2 RTKLQWSSAITLVMFIWLFTAFWSAIPLIGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLIPMAIFNMAIQIFVVLSSYQSIGEK
>RGR_calMil Callorhinchus milii (elephantfish) frag 0 EGFTDFEVFGLGTALLVE 1 2 GLVGLLLNGLTLLAFYKIKELRTPSNLLITSLALSDFGISMNAFIAAFSSFLR 2 1 YWPYGSEGCQTHGFHGFLMALASINACAAIAWDRYHQNCS 1 2 RSRLQWSSAITVTVFIWGIAAFWSAMPLLGWGVYDYEPLRTCCTLDYSKGDR 2 1 NFISFFIIMGSFEFIFPIFIMLSSYQSCKSKFKKNGQVK 0 0 FNTGLPVKTLIFCWGPYSLLCFYATIENITILSPKLRM 0 0 * 0