Opsin evolution: RGR phyloSNPs
Introduction to RGR Opsin Evolution
A Curated Set of 50 RGR Opsins
The collection of RGR opsin reference sequences is greatly expanded below to 50 species of vertebrates from the more limited yet phylogenetically representative set found in the opsin classifier. This expanded set brings much needed branch length to detailed studies of evolutionary change such as reduced alphabets at given residues, phyloSNPs, alternative splice forms, and gene loss in marsupials.
It has not proven possible to date to find an ortholog of RGR in any species diverging prior to chondrichthyes (other than tunicate). tBlastn searches of the new lamprey and amphioxus genomes yield nothing, as do blastn searches of the trace archives. Similarly, no closely related homologs can be found in earlier deuterostomes and protostomes, even though neuropsins and peropsins are readily traced back further. Since RGR is an ancient gene family divergence -- rather than derived in Bilateran times from these nearest relatives, the genes must have been retained along the stem but not in terminal leaves.
Comparative genomics of RGR at E/DRY
The comparative genomics of RGR illustrates the danger of generalizing from phylogenetic undersampling (just studying humans and mice): with much deeper sampling, it emerges that the E/DRY motif -- conserved across all classes of opsins (including generic GPCR) and critical to maintaining non-signalling state -- has become GRY in all boreoeutheran mammals (it's ERY in afrotheres and xenarthrans and DRY from platypus back to shark and even tunicate divergence, and DRY consistently in neuropsins and peropsins).
In other words, after several trillion years of branch length conservation as charged amino acid, a radical amino acid substitution has taken hold in laurasiatheres and euarchontoglires -- to glycine with tiny inert side chain. And in this subclade of placental mammals, glycine has been conserved without exception for over two billion years of branch length. Given the importance of this motif for maintaining the non-signalling state, this suggests a major change in functional properties of RGR opsins within boreoeutheres. That change might be breakdown to mere isomerase within that clade.
We might ask whether any correlated residue change took place in another residue, either adjacent in the linear sequence or adjacent after the 3D structure is considered. To do this, the phylogenetic depth must be increased to the maximum possible today (50 vertebrate genomes) and every residue scored for clade-congruent changes.
It emerges that no contemporaneous coevolutionary changes accompanied the D to G transition. The bulk of such changes occured on the 75 myr stem between the platypus divergence node and placental node. These cannot be further resolved because marsupials have lost RGR due to a chromosomal rearrangement. The other cluster of clade-coherent events occurs on the stem dividing ray-finned fish and tetrapods. (Here fish are not assumed primitive -- they simply share the ancestral value with earlier diverging cartilaginous fish.)
PhyloSNPs in RGR
All significant phylogenetic information for RGR -- SNPs that stuck in all descendent clades -- can be extracted with simple cladesheet methods (described shortly). This involves disentangling sporadic non-adaptve variants from neutral flop-flopping within a reduced alphabet from clade-coherent change. Clade-coherent change is what makes a boreoeuthere a boreoeuthere (or a whatever a whatever).
PhyloSNPs are residues fixed for considerable branch length that subsequently experience an enduring change on one descendant branch while maintaining the ancestral value in all other descendant species. Because billions of years of branch length are involved (given the current 50 vertebrate genomes available), this change has to have been and continue to be functionally adaptive. Collectively, the set of phyloSNPs on a given internode stem defines at the protein level what is distinctive about the clade and common to it.
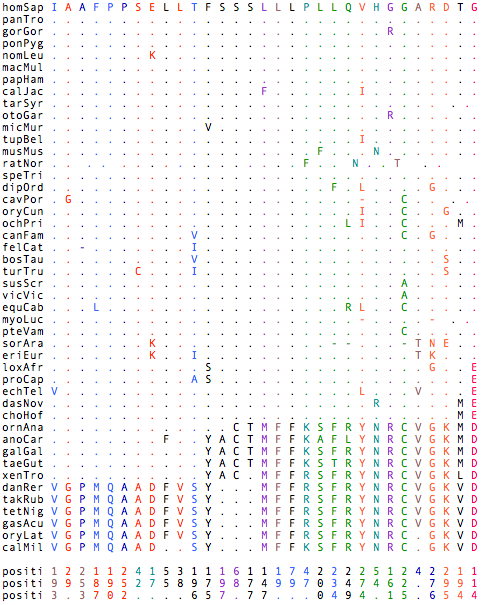
The array at left is time-sorted horizontally (with subsorting for noise) and phylo-ordered vertically as indicated by the coloring. Dots mean the amino acid residue in the indicated species agrees with homSap. Starting at the lower left corner, 11 positions in RGR where, at the phylogenetic depth of shark to fish, a signficant change took place in the tetrapod stem prior to tetrapod divergence from lobe-finned fishes and held in tetrapod clade and its outgroups to the present day. For example V>I, G>A, P>A, M>F for the first four.
These phyloSNPs do not assume or support teleost fish proteins being more 'primitive' or more ancestral than mammal -- indeed fish in general are much more evolved from ancestral state than human, meaning a random outgroup protein from chondrichthyes will have higher percent identity to mammal. Cartilaginous fish and lamprey nodes play a key role in allocating phyloSNPs: in parsimony it suffices to have the two preceding nodes the same to establish the ancestral state on their interstem. Skate and dogfish ESTs can validate elephantshark genome; some opsin dara exists for multiple species of lamprey. There will be phyloSNPs that just make fish fish but that is not the focus here.
Looking at the lower middle, at 14 positions after the phylogenetic run of shark to platypus, a signficant change took place in the theran stem and held in both sister clades to the present day. For example M>L, F>L, F>L, K>P for the first four. In the lower right, at 1 position in RGR where, at the phylogenetic depth of shark to Afrothere + Xenarthra, a signficant change took place in the boreotheran stem and held in the descendent clade and outgroups to the present day, E/D>G. As RGR is a GPCR signalling protein and the E/DRY motif (which helps prevent false signalling) has been conserved for many trillions of years of branch length. This is an example of an interpretable clade-coherent functional change making a boreoeuthere distinct from Afrotheres, Xenarthra, and everything else.
Colors group residues nearby in the linear sequence that changed at about the same geological time. These are candidates for co-evolving residues where after the first changes, a second makes a compensatory shift. The interactions of residues in different transmembrane helices that touch are another form of adjacency. These are not yet displayed but likely will suggest further co-evolving candidate pairs.
Some phyloSNP positions exhibit more background sporadic variation than others. These can be filtered to only the most pristine cases for purposes of additional bioinformatics or experimental investment. Doubling the number of species with sequenced genomes would have the effect of further refining these distinctions.
RGR has an unusuall high proportion of phyloSNPS at 32 out of 292 residues. Proteins such as PRNP, of the same length but twice the sequence density and more rapidly evolving, appear not to have a single phyloSnp of depth beyond euarchontoglires.
Local splice migration and exon-skipping in RGR Opsins
RGR in primates may contain an earlier splice acceptor in the second intron resulting in insertion of four amino acids in the extracellular loop EX1 between TM2 and TM3. This is observed in a number of human transcripts but all appear to originate from a single brain tumor sample (eg BC011349). The intronic region is conserved in the UCSC 28-way track but it cannot be a splice acceptor in tarsier, mouse lemur, or tree shrew much less other placentals. For example, in dog, horse and armadillo, translation would cause loss of reading frame -- and horse even has the AG acceptor. Thus it is difficult to say whether the insert is simply tolerated or has acquired a secondary function. The nucleotide sequence might be conserved as part of a splice enhancer.
A second defective splice isoform has also been studied that entirely skips the 6th exon. While exons don't correspond cleanly to transmembrane domains, TM7 is lost -- while the altered has been proven to be produced abundantly by mass spectroscopy (unlike the vast majority of supposed alternative splices), surely it cannot function as an opsin. It is observed to accumulate extracellularly basal boundary of RPE cells and primarily in the extracellular areas of Bruch's membrane, adjacent choriocapillaris, and intercapillary region. The carboxy terminus has been cleaved as well. A role in degenerative eye disease has neither been established nor ruled out.
Possibilities for upstream splice migration in Euarchonta: >RGR_ext_homSap VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCCATCGCATGGGGGCGTTATCACCACTACTGCACCCGT >RGR_ext_panTro VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCCATCGCATGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_gorGor VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCCATCGCATGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_ponPyg VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGTTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCCATCGCGTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_nomLeu VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCATTAGCCAGCATCTGCAGCAGTGCAGCCATCGCATGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_macMul VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGGCTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCTATCGCCTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_papHam VSHRRWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCAGATGGCTGCCAGGCTCATGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCAGCAGTGCAGCTATCGCCTGGGGGCGCTATCATCACTACTGCACCCGT >RGR_ext_calJac VSHRRWPYGSDGCQIHGFQGFVTALASICGSAAIAWGRYHHYCTR AGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGACGGCTGCCAGATTCACGGCTTCCAGGGCTTTGTGACAGCGTTGGCCAGCATCTGCGGCAGTGCAGCCATCGCCTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_tarSyr GGTGTCTCCCACAGGCGCTGGCCGTACGGCTTGGACGGCTGCCAGGCTCACGGCTTCCAAGGCTTTGTGACAGCTTTGGCCAGCATCGGCGGCAGCGCAGCCATCGCCTGGGGGCGCTATCATCACTACTGCACTCGT >RGR_ext_otoGar VLHRRWPYGSGGCQAHGFQGFTTALASICGSAAIAWGRYHHYCTR AGTGTTCTCCACAGGCGCTGGCCCTACGGCTCAGGTGGCTGCCAGGCTCACGGCTTCCAGGGCTTCACGACGGCATTGGCCAGCATCTGCGGCAGCGCAGCCATCGCCTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_micMur CGTGTCTCCCACAGGCGCTGGCCCTACGGCTCGGATGGCTGCCAGGCTCACGGCTTCCAGGCTGCGTGACGGTGCTGGCCAGCATCTGCAGCAGCGCGGCCATCGCCTGGGGGCGCTATCACCACTACTGCACCCGT >RGR_ext_tupBel CATGTCTTCCACAGGCGCTGGCCCTACGGCTCAGACGGCTGCAAGGTTCATGGCTTCCAGGGCTTCGCAACAGCGTTGGCCAGCATCTCTGGCAGCGCGGCCATCGCCTGGGGGCGCTATCACCAGTACTGCACTCGT
The Loss of RGR in Marsupials
Notably absent are RGR genes in marsupials; for other genes, marsupial representatives are typically available from 2-3 species. By locating syntenic genes in placental mammals and platypus, the appropriate region of the opossum genome can be identified. However the adjacent orthologs are now on separate autosomal opossum chromosomes, suggesting a translocation has disrupted the RGR genes. None of the individual exons can be found by tblastn of the genome or blastn of the very extensive trace archive coverage. This lack of even pseudogene debris supports the idea of gene loss and decay sometime after marsupial divergence.
The loss of RGR in marsupials further challenges the usual functional explanation given to RGR in placental mammals (a complex with 11-cis-retinol dehydrogenase RDH5 that replenishes cis-retinal at cone and rod opsins). Since marsupials are known from experiment to have normal color and rod vision, either other pathways for replenishment exist or the role of RGR was somewhat different in the ancestor.
If RGR has 1/5th the quantum efficiency of rod rhodopsin and 1/100th its abundance, the question arises how it keeps up with bright light replenishment needs. Furthermore, knockout mice develop a normal retina and RP epithelium without any morphological phenotype (unlike those lacking trans-retinol isomerase RPE65), whereas humans even heterozygous in RGR for Ser66Arg exhibit ERG abnormalities and eventually retinitis pigmentosa and RPE degeneration.
Most experimental work has unfortunately been done on GRY species whereas platypus and earlier diverging species have the more conventional DRY or ERY. Thus the role of RGR could have drastically shifted at the time of marsupial divergence. They lost the gene altogether while placentals retained it but likely with a rather altered signaling properties and functionalities.
RGR reference sequences from 50 vertebrates
>RGR_homSap Homo sapiens (human) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSQLAWNSAVSLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKREKDRTK * 0 >RGR_panTro Pan troglodytes (chimp) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSQLAWNSAISLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAIPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSEKDRTK * 0 >RGR_gorGor Gorilla gorilla (gorilla) 0 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLAPAESGISLNALVGATSTLLG 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 GSTLACKSAVSLVLSGRMSSAFWADLPLLGWGPYDYEPLRTCCTLDYSEADR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSKKDRTK * 0 >RGR_ponPyg Pongo pygmaeus (orang_abelii) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 GSQLAWNSAISLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAVLYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSEKDRTK * 0 >RGR_nomLeu Nomascus leucogenys (gibbon) 0 MAETSVLPTGFGELKLLAGGMGLLAE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 GSQLAWNSAISLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAVNYALGNEMVCRGIWQCLSPQKSEKDRAK * 0 >RGR_macMul Macaca mulatta (rhesus) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALIAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSQLAWNSAISLVLFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSEKDRAK * 0 >RGR_papHam Papio hamadryas (baboon) 0 MAETSALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALIAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 2 1 NFTSFLFTMSFFNFAMPLFITITSYSLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTINAINYALGNEMVCRGIWQCLSPQKSEKDRAK * 0 >RGR_calJac Callithrix jacchus (marmoset) 0 MAESSTLPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNSLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSDGCQIHGFQGFVTALASICGSAAIAWGRYHHYCT 1 2 GSQLAWNSAISLVLFVWLSSTFWAAFPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMSFFNFAMPLFITITSYRLMEQKLGKSGHLQ 0 0 VNTTLPARTLLLGWGPYAILYLYAVIADVTSISPKLQM 0 0 VPALIAKMVPTIDAINYALGNEMICRGIWQCLSPQKSEKDRTK * 0 >RGR_tarSyr Tarsius syrichta (tarsier) 0 MAEAGALPAGFGELEVLAVGMVLLVE 1 2 ALSGLSLNSLTIFSFCKTPELRTPCHLLVLSLALADSGISLNALIGATSSLLR 2 1 RWPYGLDGCQAHGFQGFVTALASIGGSAAIAWGRYHHYCT 1 2 GSQLAWNTAISLVLFVWLSYAFWAALPLLGWGHYDYEPLGTCCTLEYSKGDR 2 1 0 0 VNTTLPIRTLMLGWGPYALLYLCAVIADVTSISPKLQM 0 0 VPALIAKTVPTINAYHYALGSEMVCRGIWQCLSPHSSE * 0 >RGR_otoGar Otolemur garnettii (bushbaby) 0 MAEPGTLPAGFGEIEVLAVGTVLLVE 1 2 2 1 RWPYGSGGCQAHGFQGFTTALASICGSAAIAWGRYHHYCT 1 2 GRPLAWSTAISLVLFVWLSSAFWAALPLLGWGHYDYEPLRTCCTLDYSRGDR 2 1 NFTSFLFTMAFFNFLTPLFITLTSYQLMEQKLRRSGHLQ 0 0 VNTTLPARTLLLGWGPYALLYLYATIADVTSISPKLQM 0 0 VPALIAKTVPTINAVNYALGSEMVCRGIWQCLSLQRSKQDGAK * 0 >RGR_micMur Microcebus murinus (mouse_lemur) 0 MAEPGTLPTGFRELEVLAVGTVLLVE 1 2 2 1 RWPYGSGGCQAHGFQGFTTALASICGSAAIAWGRYHHYCT 1 2 GSPLAWSTAISLVLFVWLSSAFWAALPLLGWGHYNYEPLGTCCTLDYSRGDR 2 1 NVTSFLFTMAFFNFLIPLFITHTSYQLMEQKLKKSGHLQ 0 0 VNTTLPARTLLLGWGPYALLYLYATVADVTSISPKLQM 0 0 VPALIAKTVPTINAINYALGSETVCRGIWQCLSPQRSEQDRAK * 0 >RGR_tupBel Tupaia belangeri (treeshrew) 0 MAESGALPSGFGELEVLAVGTVLLVE 1 2 ALSGLSLNSLTVFSFCKSPELRTPSHLLVLSVALADSGISLNALIAATSSLLR 2 1 RWPYGSDGCKVHGFQGFATALASISGSAAIAWGRYHQYCT 1 2 2 1 NFTSFLFTMAFFNFLMPLFITLTSYWLMEEKLRKGGRLQ 0 0 VNTTLPSRTLLLGWGPYALLYLYAAFADVTPLSPKLQM 0 0 VPALVAKMVPTVNAVNYALGSETICRGIWGCLSPKRERDRAR * 0 >RGR_musMus Mus musculus (mouse) 0 MAATRALPAGLGELEVLAVGTVLLME 1 2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2 1 RWPHGSEGCQVHGFQGFATALASICGSAAVAWGRYHHYCT 1 2 GRQLAWDTAIPLVLFVWMSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2 1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKFSRSGHLP 0 0 VNTTLPGRMLLLGWGPYALLYLYAAIADVSFISPKLQM 0 0 VPALIAKTMPTINAINYALHREMVCRGTWQCLSPQKSKKDRTQ * 0 >RGR_ratNor Rattus norvegicus (rat) 0 MTATRALPAGFGELEVLAIGIVLLME 1 2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2 1 RWPHGSEGCRVHGFQGFATALASICGSAAIAWGRYHHYCT 1 2 GRQLAWDTAIPLVLFVWLSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2 1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKLSRSGHLQ 0 0 VNTTLPGRMLLLGWGPYALLYLYAAVADVSFISPKLQM 0 0 VPALIAKTMPTINAINYALRSEMVCRGTWQCRSAQKSKQDRTQ * 0 >RGR_speTri Spermophilus tridecemlineatus (ground_squirrel) 0 MAETAALPAGFGELEVLAVGTVLLVE 1 2 ALSGLSLNGLTIFSFCKTPELRTPNHLLLLSLAVADSGISLNALIAAISSLLR 2 1 RWPYGSDGCQAHGFQGFVTALSSICGSAAIAWGRYHHYCT 1 2 GSQLAWNTAIPLVLFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFISFLFTMAFFNFFVPLFITLTSYRLMEQKLARSGHLQ 0 0 0 0 * 0 >RGR_dipOrd Dipodomys ordii (kangaroo_rat) 0 MATSGDLPTGFGELEVLTVGTVLLVE 1 2 ALSGLSLNTLTIFSFCKTPELRTPIHLLDLSLAVADSGISLNALIAAISSLEW 2 1 HWPYGLEGCQAHGFQGFVTALASISGSAAIAWGRCHHHCT 1 2 GSLLGWDTAVSLVIFVWLSSAFWAALPLLGWGHYNYEPLGTCCTLDYSRGDR 2 1 NFTSFLFTMAFFNFLVPLFITLTSYQLMKQKFARSGRLQ 0 0 VNTTLPTRTLLLGWGPYALLYFYAAIMDVNSISPKLQM 0 0 VPALIAKMVPTVNAINYALCNELLCGGFSLGLLPQKGKQDRTQ * 0 >RGR_cavPor Cavia porcellus (guinea_pig) 0 MATSEALPAGFGELEVLAVGTVLLLE 1 2 GLCGLSLNGLTVVSFWKSPALRTPNHLLVLSLALADSGLSLNALVAAGSSLLR 2 1 HWPGSGHCQALGFQGFTTALASISGTAALSWGRHQQCCT 1 2 RGRLTWSTAVPLVLFVWLSSAFWAALPLLGWGRYDYEPLGTCCTLDYSTGDR 2 1 NFTSFLFTMAFFNFLVPLFITVTSCQLMERHLARSSRLQ 0 0 VSVRQPARTLLLCWSPYALLYLYAVLADAHTLSPRLQM 0 0 VPALIAKTVPTIYSLGRGPWQSLEMQRSKQD * 0 >RGR_oryCun Oryctolagus cuniculus (rabbit) 0 MAEPGTLPPGFEELEVLAVGTVLLVE 1 2 ALSGLSLNGLTIFSFCKTPELWTPSHLLVLSLAVADSGISLNALIAAVSSLLR 2 1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1 2 GSQLAWNTAVLLVLFVWLSSVFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFISFLITMAFFNFLMPLFITLTSYSLMEQKLSKSGRLQ 0 0 VNTTLPGRTLLFCWGPYAVLYLCAAVADMSSITLKLQM 0 0 VPALIAKTVPTVNAVNYALGSEVIRRGIWQCLLPQRSVRGRAQ * 0 >RGR_ochPri Ochotona princeps (pika) 0 MAEPGTLPPGFEELEVLAVGTVLLVE 1 2 ALTGLSLNSLTIFSFCTSPELRTPSHLLVLSLALADSGVSLNALAAATASLLR 2 1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1 2 GSQLAWNTAVLLVLFVWLSSVFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFISFLVTMAFFNFLMPLFIMLTSYSLMEQKLAKSGRLQ 0 0 VNTTLPARTLLFCWGPYAILCLCATVMDMSTVSPKLLM 0 0 VPALIAKAVPTVNAINYALGSEVIRRGIWQCLLPQRSVRDRAQ * 0 >RGR_canFam Canis familiaris (dog) 0 MADSGALPAGFGELEVLAVGTVLLVE 1 2 ALTGLCLNGLTILSFCKTPELRTPTHLLVLSLAVADTGISLNALVAAISSLLR 2 1 RWPYGPDGCQAHGFQGFATALASICSSAALAWGRYHHYCT 1 2 RGQLAWNTAISLVLCVWLSSVFWAALPLLGWGRYDYEPLGTCCTLDYSRVDR 2 1 NFTSYLFTMAFFNFFLPLLITLVSYRLMEQKLKKPGHLQ 0 0 VSTTVPARTLLLCWGPYALLYLYATVADVRSVPPKLQM 0 0 VPALIAKAAPTINAIHYALGGDMVHGGLWQCLSPQRSQPDRAR * 0 >RGR_felCat Felis catus (cat) 0 MAESGSLPTGFGELEVLAVGMVLLVE 1 2 2 1 RWPYGSNGCQAHGFQGFVTALASICSSAAIAWGRYHHYCS 1 2 GSQLAWNTAISLVICVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFTSFLFTMAFFNFFMPLFITFISYRLMEQKLRKTGHLQ 0 0 VNTTLPARTLLFGWGPYALLYLYATIADVSSVSPKLQM 0 0 VPTINAINYALGSEMVHRGIWQCLSPQGSGLDRAR * 0 >RGR_bosTau Bos taurus (cow) 0 MAESGTLPTGFGELEVLAVGTVLLVE 1 2 ALSGLSLNILTILSFCKTPELRTPSHLLVLSLALADSGISLNALVAATSSLLR 2 1 RWPYGSEGCQAHGFQGFVTALASICSSAAVAWGRYHHFCT 1 2 GSRLDWNTAVSLVFFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2 1 NFTSFLFTMAFFNFLLPLFITVVSYRLMEQKLGKTSRPP 0 0 VNTVLPARTLLLGWGPYALLYLYATIADATSISPKLQM 0 0 VPALIAKAVPTVNAMNYALGSEMVHRGIWQCLSPQRREHSREQ * 0 >RGR_turTru Tursiops truncatus (dolphin) 0 MAESGALPSGFGELEVLAVGTVLLVE 1 2 ALSGLSLNSLTILCFCKNPELRTPSHLLVLSLALSDSGISLNALMAATSSLLR 2 1 RWPYGSDGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 GSRLDWNTAVSLVFFVWLSSAFWATLPLLGWGHYDREPLGTCCTLDYSRRDR 2 1 NFTSFLFTMAFFNFLLPLFITVISYRLMEQKLGKTGRPP 0 0 VNTVLPARTLLFGWGPYALLYLYAAVADVTSISPKLQM 0 0 * 0 >RGR_susScr Sus scrofa (pig) 0 MAEPGALPTGFGELEVLAVGTLLLVE 1 2 ALSGLSLNSLTILSFCKTPELRTPSHLLVLSLALADSGISLNAFVAATSSLLR 2 1 RWPYGSEGCQAHGFQGFATALASICSSAAIAWGRYHHYCT 1 2 RSRLDWNTAVSLVFFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSRVDR 2 1 NFTSFLFTMAFFNFLLPLFITVTSYRLMEQKLGKTGRPP 0 0 VNTILPARTLMLAWGPYALLYLYATFADVTSISPKLQM 0 0 VPALIAKMVPTVNAINYALGGEMVHRGIWQCLSPQRRERDREQ * 0 >RGR_vicVic Vicugna vicugna (vicugna) 0 MAESRALPTGFGELEVLAVGMVLLVE 1 2 ALSGLSLNSLTILSFCKTPELRTPNHLLVLSLALADSGISLNALVAATSSLLR 2 1 1 2 GSRLDWNTAVSLVFFVWLSSTCWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 0 0 0 0 * 0 >RGR_equCab Equus caballus (horse) 0 MAESGSLPTGFRELEVLAVGTVLLVE 1 2 ALAGLSLNSLTILSFCKTPELRTPSHLLVLSLAVADSGLSLNALVAATSSLLR 2 1 RWPYGSEGCQAHGFQGFVTALASICSSAAIAWGRYHHYCT 1 2 RSRLAWNTAVFLVFFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLLTMAFFNLLLPLLITLTSYRLMEQKLGKTGQLQ 0 0 VNTTLPARTLLLCWGPYALLYLYATVADATSISPKLRM 0 0 VPALVAKTVPTINAVNYALGSEMLHRGIWQCLSPQKSERDRAQ * 0 >RGR_myoLuc Myotis lucifugus (microbat) 0 MAEAGSLPTGFGELEVLAVGVVLLVE 1 2 ALTGLSLNSLTIFSFCTSPELRTPSHLLVLSLALADSGVSLNALAAATASLLR 2 1 RWPYGSGGCQAHGFQGFAAALASICGSAAVAWGRYHHYCT 1 2 GSRLAWRTAASLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCSLGYARGSR 2 1 NFTSFLFLMAFFNFLLPLFITFTSYRLMEQKLGRTRPPQ 0 0 VNTTLPARTLLLGWGPYALLHLCAALAGTALIPPRLQV 0 0 VPALIAKMVPTVNAVNYALGSGIWQRLSLQ * 0 >RGR_pteVam Pteropus vampyrus (macrobat) 0 MAESRSLPTVFWELEVLAVGTVLMVE 1 2 ALSGLSLNSLTILSFCKNPELRTPIHLLVLSLALADSGISLNALIAATSSLLR 2 1 RWPFGPDGCQAHGFQGFATALASICSSAAIAWGRYHHYCT 1 2 GSRLAWNTAVSLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSRRDR 2 1 NFTSFLFTMAFFNFVLPLFITLTSYQLMEQKLGKTGHPQ 0 0 VNTTLPARTLMLCWGPYALLYLYAAVMDVASISPKLQM 0 0 VPALIAKMAPTINAVNYALGSEMVQRGIWQCLSPQRSERDHAQ * 0 >RGR_sorAra Sorex araneus (shrew) 0 MTESGALPAGFKELKMLAVGTLLLWG 1 2 2 1 RWPFGPDGCQAHGFQGFATALASICSSAAIAWGRYHHYCT 1 2 GRQLAWDVAIALVIFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSRGGR 2 1 NFVSFLFTMAFFNFLLPLFITVTSYRLMEQKLGKMGQPQ 0 0 0 0 VPALIAKTVPTVNALHYGLGSGMVQNGFRKGLWLQRRERERAL * 0 >RGR_eriEur Erinaceus europaeus (hedgehog) 0 1 2 2 1 RWPYGSDGCQAHGFQGFVMALASICSSAAIAWGRYHHHCT 1 2 RSRLAWNTAVFLVFFVWVSSVFWAALPLLGWGHYDYEPLGTCCTLDYSSGDR 2 1 NFISFLFTMAFFNFLLPLFITLISYQLMEQKLRKTGHPQ 0 0 VNTTLPARTLLLGWGPYALLYLYAVIADVALLSPKLQM 0 0 VPALIAMVPTVNAVHYVLGSEKVHKGFWQCFSPQRSEQDRAR * 0 >RGR_loxAfr Loxodonta africana (elephant) 0 MAEPGHLPAGFQELEVLTVGTVLLLE 1 2 ALSGLSLNGLTILSFCKIPELRTPGHLLVLSLALADSGISLNALVAAMSSLRR 2 1 RWPYGSDGCQAHGFQGFVTALASICSCAAIAWERYHHYCT 1 2 RSRLAWSSASALVLFVWLSSAFWAALPLLGWGRYNYEPLGTCCTLDYSRGDR 2 1 NSTSFLLTMAFFNFLLPLFITLTSYRLMEQKLKKKGPLQ 0 0 VNTTLPARTLLLGWGPYALLYLCAAATDMTSISPRLQM 0 0 VPALVAKAVPVINACHYALGSEVVRGGIWQYLSRQRGESPLRARDRTH * 0 >RGR_proCap Procavia capensis (hyrax) 0 MADPRPLPTGFGELEVLTVGTVLLVE 1 2 ALSGLSLNGLTILSFYKIPELRTPGHLLVLNLALADSGMSLNALVAAVSSLRG 2 1 RWPYGSDGCQAHGFQGFVMALTSICSCAAIAWERYHHYCT 1 2 GSKLAWSSAGALVLFMWLSSAFWAALPLLGWGRYNYEPLGTCCTLDYSRGDR 2 1 NSTSFLFTMAFFNFLLPLFITLASYRLMEQKLKKEGPLQ 0 0 VNTTLPARTLLLGWGPYALLYLYTAITDVNSISPKLQM 0 0 VPALIAKAVPIVNACHYALGSETVHRGIWQCLSRQRGESPPRTRDRTQ * 0 >RGR_echTel Echinops telfairi (tenrec) 0 MVEPRTLPPGFGELEVLAVGTVLLVE 1 2 2 1 HWPYGSGGCQAHGFQGFTVALASICSCAAIAWERYHHYCT 1 2 GSQFTWSSASTLVLFMWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKGDR 2 1 NFTSFLFTMTFFNFSMPILVTLTSYQLMQQKLKKSGPLQ 0 0 VNTTLPTRTLLLGWGPYALLYLCAACTDVTGISPKLQM 0 0 VPAIVAKAVPIVNACHYALGNKVLRRGIWQFLSQQSGRQDRTQ * 0 >RGR_dasNov Dasypus novemcinctus (armadillo) 0 MAGSGVLPPGFGELEVLAVGTVLLVE 1 2 ALSGLVLNGLAIISFCKTPELRSPSRLLVLSLALADSGVSLNALVAATSSLLR 2 1 RWPYGSGGCQAHGFQGFVTALASISSSAAIAWERCHRHCI 1 2 GRRLAWSTAGCLVLCLWMAAAFWAALPLLGWGLYDYEPLGTCCTLDYSRGDR 2 1 NFISFLVTLALFNFFLPLLIMLTSYRLMAQKLKRSGHVQ 0 0 VSTALPGRLLLLGWGPYALLYLYAAVADATSLSPRLQM 0 0 VPALIAKTMPTVNALYYALGRESVHRNA * 0 >RGR_choHof Choloepus hoffmanni (sloth) 0 MAESRVLPTGFGELEVLAVGIVLLVE 1 2 ALSGLTLNGLTLFSFCKTPELQRPSHLLVLSLALADSGVSLNALLAATASLIG 2 1 RWPHGSDSCQAHSFQGFATALASISSSAAIAWERYRHHCT 1 2 GSQLSWSTAGSLVLCVWLSSVFWATLPILGWGHYDYEPLGTCCTLGYSRGDR 2 1 0 0 0 0 VPALIAKTMPTINAFQYALGSETVCRDIWQCLPRLRSMGRSSGHD * 0 >RGR_ornAna Ornithorhynchus anatinus (platypus) 0 1 2 ALLGLCLNGLTIASFRKIKELRTPSNLLVVSLALADSGICLNALMAALSSFLR 2 1 HWPYGAEGCRLHGFQGFATALASISLSAAIGWDRYLRHCS 1 2 RSKPQWGTAVSTVLFAWGFSAFWSMMPILGWGQYDYEPLRTCCTLDYSKGDR 2 1 NFTTYLFAVAFFNFVIPLFIMLTSYQSIEQRFKKSGLFK 0 0 LNTRLPTRTLLFCWGPYALLCFYATVENVTFISPKLRM 0 0 IPALIAKTVPVIDAFTYALRNEDYRGGIWQFLTGQKIERVEVENKIK * 0 >RGR_anoCar Anolis carolinensis (lizard) 0 MVTTAYPVPEGFTDLEVFVIGTALLVE 1 2 ALLGFSLNMLTIVSFWKIKELRTPGNFLVFNLALSDCGICFNAFIAAFSSFLR 2 1 YWPYGSDGCQIHGFHGFLTALTSISSAAAVAWDRHHQYCT 1 2 GNKLQWGSVIPMTIFLWLFSGFWAAMPLLGWGEYDYEPLRTCCTLDYTKGDR 2 1 NYITYLIPLALFHFMIPGFIMLTAYQAIDHKFKKTGQFK 0 0 FNTGLPVKSLVICWGPYSFLCFYAAVESVTFISPKILM 0 0 IPAVIAKSSPAANALIYALGNENYQGGIWQFLTGQKIEKAEVDNKTK * 0 >RGR_galGal Gallus gallus (chicken) 0 MVTSHPLPEGFTEIEVFAIGTALLVE 1 2 ALLGFCLNGLTIISFRKIKELRTPSNLLVLSIALADCGICINAFIAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFLTALASISSSAAVAWDRYHHYCT 1 2 RSKLQWSTAISMMVFAWLFAAFWATMPLLGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYITFLFALSIFNFMIPGFIMMTAYQSIHQKFKKSGHYK 0 0 FNTGLPLKTLVICWGPYCLLSFYAAIENVMFISPKYRM 0 0 IPAIIAKTVPTVDSFVYALGNENYRGGIWQFLTGQKIEKAEVDSKTK * 0 >RGR_taeGut Taeniopygia guttata (finch) 0 MVTAHPLPEGFTEIEVFAIGTALLVE 1 2 ALLGFCLNGLTIISFRKIKELRTPSNLLVLSIALADCGICINAFIAAFSSFLR 2 1 YWPYGSDGCQIHGFQGFLTALASIGSSAAIAWDRYHHYCT 1 2 RSRLQWSTAVSMMVFAWLFAAFWSVMPLLGWGKYDYEPLRTCCTLDYSKGDR 2 1 NYVTFLFALSTFNFMIPGFIMMTAYQSIHQKFRKTGHFK 0 0 FNTGLPLKTLVICWGPYCLLCTYAAVENVMFIPPKYRM 0 0 IPALIAKTVPTVDAFIYALGNENYRGGIWQFLTGQKIEKAEVDNKTK * 0 >RGR_xenTro Xenopus tropicalis (frog) 0 MVTSYPLPEGFTETEVFAIGTTLLVE 1 2 ALLGLLLNGLTLLSFYKIRELRTPSNLFIISLAVADTGLCLNAFVAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1 2 RSKLHWSTAVSVVFFIWGFSAFWSAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYISYLFTMAFFEFLVPLFILMTAYQSIYQKMKKSGQIR 0 0 FNTSMPVKSLVFCWGPYCLLCFYAVIQDATILSPKLRM 0 0 IPALLAKTSPAVNAYVYGLGNENYRGGIWQYLTGQKLEKAETDNKTK * 0 >RGR_xenLae Xenopus laevis (frog) NM_001092855 mRNA 0 MVTSYPLPEGFTETEVFAIGTTLLIE 1 2 ALLGLLLNGLTLLSFYKIRELRTPSNLFIISLAVADTGLCLNAFVAAFSSFLR 2 1 YWPYGSEGCQIHGFQGFVAALSSIGSCAAIAWDRYHQYCT 1 2 RSKLHWGTAVSMVLFVWGFSAFWSAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 1 NFTSFLFTMAFFEFLVPVFILLTAYQSIYQKMKKSGQIR 0 0 LNTSMPVKSLVFCWGPYCLLCFYAVIQDATILSPKLRM 0 0 MPALLAKISPAVNAYVYGLGNENYRGGIWLYLTGQKLEKAETDSRTK* 0 >RGR_danRer Danio rerio (zebrafish) 0 MVTSYPLPEGFSEFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVIAFLKIRELRTPSNFLVFSLAMADMGISTNATVAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFMTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSSAITLVLFTWLFTAFWAAMPLFGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLIPMSIFNMGIQVFVVLSSYQSIDKKFKKTGQAK 0 0 FNCGTPLKTMLFCWGPYGILAFYAAVENATLVSPKLRM 0 0 IAPILAKTSPTFNVFVYALGNENYRGGIWQLLTGQKIESPAIENKSK * 0 >RGR_takRub Takifugu rubripes (fugu) 0 MVSSYPLPEGFSDFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVVAFLKVRELRTPSNFLVFSLALADMGISMNATIAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFVTALASIHFVAAIAWDRYHQYCT 1 2 RTKLQWSSAITLAVFIWLFCAFWSAMPLIGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLIPMAIFNMVIQVFVVMSSYQSIAEKFKKTGNPR 0 0 FNPSTPLKAMLLCWGPYGILAFYAAVENANLVSPKLRM 0 0 MAPILAKTCPTINVFLYALGNENYRGGIWQFLTGEKIEAPQIENKSK * 0 >RGR_tetNig Tetraodon nigroviridis (pufferfish) 0 MVSSYPLPEGFSDFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVVAFFKVRELRTPSNFLVFSLAMADMGISMNATVAAFSSFLR 2 1 YWPYGSEGCQTHGFQGFVTALASIHFVAAIAWDRYHQYCT 1 2 RTKLQWSSAITLAVFIWLFCAFWSAMPLIGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLVPMAIFNMVIQVFVVMSSYQSIAEKFKKTGNPR 0 0 FNPNTPLKAMLLCWGPYGILAFYAAVENANLVSPKLRM 0 0 MAPILAKTCPTVNVFLYALGNENYRGGIWQFLTGEKIETPQLENKTK * 0 >RGR_gasAcu Gasterosteus aculeatus (stickleback) 0 MVSSYPLPDGFTDFDVFSLGSCLLVE 1 2 GLLGILLNAVTIAAFLKVRELRTPSNFLVFSLAVADIGISMNATIAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFVTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSSAITLAVFVWLFTAFWSAMPLIGWGEYDYEPLRTCCTLDYTKGDR 2 1 NYVSYLIPMAIFNMAIQVFVVMSSYQSIAQKFKKTGNPR 0 0 FNPNTPLKAMLFCWGPYGILAFYAAVENATLVSTKLRM 0 0 MAPILAKTSPTFNVFLYALGNENYRGGIWQLLTGEKIDVPQIENKSK * 0 >RGR_oryLat Oryzias latipes (medaka) 0 MATSYPLPEGFSEFDVFSLGSCLLVE 1 2 GLLGIFLNSVTIVAFLKVRELRTPSNFLVFSLAMADIGISMNATIAAFSSFLR 2 1 YWPYGSEGCQTHGFHGFLTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSTAITLAVLVWIFTAFWAAMPLIGWGEYDYEPLRTCCTLDISKGDR 2 1 NYVSYVIPMSIFNMGIQVFVVMSSYQSIAQKFQKTGNPR 0 0 FNASTPLKTLLFCWGPYGILAFYAAVADANLVSPKIRM 0 0 IAPILAKTSPTFNPLLYALGNENYRGGIWQFLTGEKIHVPQDDNKSK * 0 >RGR_gadMor Gadus morhua (cod) ES469757 0 MVSAYPLPEGFSDFDVFSFGSFLLVE 1 2 GLLGIILNAVTIVAFCKVKELRTPSNFLVFSLAMADIGISMNASVAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFTVALASIHFVAAIAWDRYHQYCT 1 2 RTELQWSSAVTLSVFIWLFSAFWSAMPLIGWGTYDYEPLRSCCTLDYTKGDR 2 1 NYVSYLIPMTVFNMVVQIFVVMSSYQSIDQKFKKTAKPK 0 0 FNARTPLKTLLFCWGPYGILAFYAAIENASLVSPKLRM 0 0 MAPILAKTAPTFNVFLYALGNENYRGGIWQLLTGEKIVPQEVPQIENKSK* 0 >RGR_pimPro Pimephales promela DT198813 frag 0 MVTYPLPEGFSDFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVVAFLKIRELRTPSNFLVFSLAMADMGISMNATVAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFMTALASIHFIAAIAWDRYHQYCT 1 2 RTKLQWSSAITLVIFIWLFTAFWSAMPLIGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLIPMSIFNMGIQVFVVLSSYQSIERKFQKSGQAK 0 0 FNCSTPLKTMLFCWGPYGILAFYAAVENATLVSPKLRM 0 0 LAPILA >RGR_osmMor Osmerus mordax (smelt) EL524757 frag 0 MVSSYPLPDGFSDFDVFSLGSCLLVE 1 2 GLLGFFLNAVTVVAFLKVRELRTPSNFLVFSLALADMGISSNATIAAFSSFLR 2 1 YWPYGSDGCQTHGFQGFMTALASIHFVAAIAWDRYHQYCT 1 2 RTKLQWSSAITLVMFIWLFTAFWSAIPLIGWGEYDYEPLRTCCTLDYSKGDR 2 1 NYVSYLIPMAIFNMAIQIFVVLSSYQSIGEK >RGR_calMil Callorhinchus milii (elephantfish) frag 0 EGFTDFEVFGLGTALLVE 1 2 GLVGLLLNGLTLLAFYKIKELRTPSNLLITSLALSDFGISMNAFIAAFSSFLR 2 1 YWPYGSEGCQTHGFHGFLMALASINACAAIAWDRYHQNCS 1 2 RSRLQWSSAITVTVFIWGIAAFWSAMPLLGWGVYDYEPLRTCCTLDYSKGDR 2 1 NFISFFIIMGSFEFIFPIFIMLSSYQSCKSKFKKNGQVK 0 0 FNTGLPVKTLIFCWGPYSLLCFYATIENITILSPKLRM 0 0 * 0