Opsin evolution: key critters (cnidaria): Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| (93 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
== <span style="color: #990099;">Cnidaria .. | '''See also:''' [[Opsin_evolution|Curated Sequences]] | [[Opsin_evolution:_key_critters_%28deuterostomes%29|Deuterostomes]] | [[Opsin_evolution:_key_critters_%28ecdysozoa%29|Ecdysozoa]] | [[Opsin_evolution:_key_critters_%28lophotrochozoa%29|Lophotrochozoa]] | [[Opsin_evolution:_update_blog|Update Blog]] | ||
== <span style="color: #990099;">Cnidaria .. 5+ opsins established</span> == | |||
Biologists have belatedly realized that many molecular and morphological innovations attributed to chordates (or grudgingly to bilatera) actually track back much earlier to the common ancestor with cnidaria (Eumetazoa) if not earlier still to sponges | Biologists have belatedly realized that many molecular and morphological innovations attributed to chordates (or grudgingly to bilatera) actually track back much earlier to the common ancestor with cnidaria (Eumetazoa) if not earlier still to placazoa, sponges, and choanoflagellates. That's certainly true of photoreception. Two cnidarian genome projects have been more or less finished (Nematostella and Hydra) but that selection needs to be seriously expanded. Hydra especially repeats the whole mistake of sequencing a hugely derived genome with relatively little applicability to Bilatera based on a shallow 'model organism' approach (Hydra has only 66 publications in the last 30 years). | ||
A scientifically neutral definition of eye needs to embrace the full variety of photoreceptors, including those with fewer "features" than the most complex. Probably the cutoff should be based on use of bona fide opsins classifying to the root of the encephalopsin, melanopsin, and RGR families covalently binding retinal and variants as agonist. Some purposes for an eye can be fully met by just perceiving and acting upon one "pixel", that is a simple photoreceptor eye with no pigment cup (that could provide directionality, two pixels resolution). Far too much emphasis has been given to the distinctness of lensed vision whereas such systems are evidently easily evolved and only a part of a broader photoreceptor continuum. | A scientifically neutral definition of eye needs to embrace the full variety of photoreceptors, including those with fewer "features" than the most complex. Probably the cutoff should be based on use of bona fide opsins classifying to the root of the encephalopsin, melanopsin, and RGR families covalently binding retinal and variants as agonist. Some purposes for an eye can be fully met by just perceiving and acting upon one "pixel", that is a simple photoreceptor eye with no pigment cup (that could provide directionality, two pixels resolution). Far too much emphasis has been given to the distinctness of lensed vision whereas such systems are evidently easily evolved and only a part of a broader photoreceptor continuum. | ||
| Line 9: | Line 11: | ||
We don't say humans lack eyes just because a redtailed hawk has more pixels; we don't say humans lack color vision just because a turtle sees richer, sharper colors. When a simpler photostructure already suffices to distinguish day from night for gamete release, up from down for settlement, towards or away for predator evasion, cornea, lens, retina, and centralized nervous system are just baggage that can't be developed or maintained under darwinian selection. Cnidarian eyes exemplify this full range of possibilities. | We don't say humans lack eyes just because a redtailed hawk has more pixels; we don't say humans lack color vision just because a turtle sees richer, sharper colors. When a simpler photostructure already suffices to distinguish day from night for gamete release, up from down for settlement, towards or away for predator evasion, cornea, lens, retina, and centralized nervous system are just baggage that can't be developed or maintained under darwinian selection. Cnidarian eyes exemplify this full range of possibilities. | ||
Sponges and cnidarians have operated for immense timescales under selective pressure on huge population numbers on a steady body plan. Rather than frozen in time at some primitive condition as often portrayed ( | Sponges and cnidarians have operated for immense timescales under selective pressure on huge population numbers on a steady body plan. Rather than frozen in time at some primitive condition as often portrayed (bio-bigotry), quite the opposite, they are fast evolvers that have had eons to perfect their genes and expression systems even as mammals played evolutionary catchup (eg human knee or defective LWS opsin duplication). | ||
No living animal represents a long-gone ancestral node -- evolution never stops at the dna level even if outward morphology seems constant. All extant species have proven equally successful at survival. Evolution is not a story book progressing to human-- if cnidarians are so dumb and their vision so bad, how then are they able to chase, catch, kill, and eat advanced vertebrates? | No living animal represents a long-gone ancestral node -- evolution never stops at the dna level even if outward morphology seems constant. All extant species have proven equally successful at survival. Evolution is not a story book progressing to human-- if cnidarians are so dumb and their vision so bad, how then are they able to chase, catch, kill, and eat advanced vertebrates? | ||
| Line 19: | Line 21: | ||
This new gene has the [http://www.cbs.dtu.dk/services/TMHMM/ expected 7-transmembrane topology], conserved disulfide, ERY domain (as ERF), and conserved lysine for covalent chromophore attachment (with counterion predicted here at the ancestral 'E181' position EGV). Only the lysine and counterion are specific properties of opsins relative to generic rhodopsin-class GPCR. Other conserved residues specific to ciliary opsins serve to distinguish it from rhabodomeric opsins and contribute to its unequivocal blastp clustering within vertebrate ciliary opsins. These latter are slow-evolving and consequently the distance is less than to protostome ciliary opsins. | This new gene has the [http://www.cbs.dtu.dk/services/TMHMM/ expected 7-transmembrane topology], conserved disulfide, ERY domain (as ERF), and conserved lysine for covalent chromophore attachment (with counterion predicted here at the ancestral 'E181' position EGV). Only the lysine and counterion are specific properties of opsins relative to generic rhodopsin-class GPCR. Other conserved residues specific to ciliary opsins serve to distinguish it from rhabodomeric opsins and contribute to its unequivocal blastp clustering within vertebrate ciliary opsins. These latter are slow-evolving and consequently the distance is less than to protostome ciliary opsins. | ||
The authors suggest the NKQ motif (NRS in Tripedalia) at the start of the last cytoplasmic region is a reliable signature for ciliary transducin interaction (this was established experimentally [http://www.jbc.org/cgi/content/full/275/3/1930? as a contributing factor] but only for bovine RHO1 and its co-evolving transducin Gt) but comparative genomics of 300 phylogenetically dispersed opsins shows this cannot hold in general as TMT opsins lack the basic middle residue and encephalopsins do not conserve the motif at all. Note rod and cone transducins cannot be tracked back orthologously even to tunicates. Melanopsins are not known from pre-Bilatera; at some point they coalesce with ciliary opsins, perhaps with this motif | The authors suggest the NKQ motif (NRS in Tripedalia) at the start of the last cytoplasmic region is a reliable signature for ciliary transducin interaction (this was established experimentally [http://www.jbc.org/cgi/content/full/275/3/1930? as a contributing factor] but only for bovine RHO1 and its co-evolving transducin Gt) but comparative genomics of 300 phylogenetically dispersed opsins shows this cannot hold in general as TMT opsins lack the basic middle residue and encephalopsins do not conserve the motif at all. Note rod and cone transducins cannot be tracked back orthologously even to tunicates. Melanopsins are not known from pre-Bilatera; at some point they coalesce with ciliary opsins, perhaps with this motif coalescing to the latter's pattern. Opsins can also trigger multiple signaling systems. | ||
[[Image:OpsinActivation.png|center|]] | [[Image:OpsinActivation.png|center|]] | ||
| Line 26: | Line 28: | ||
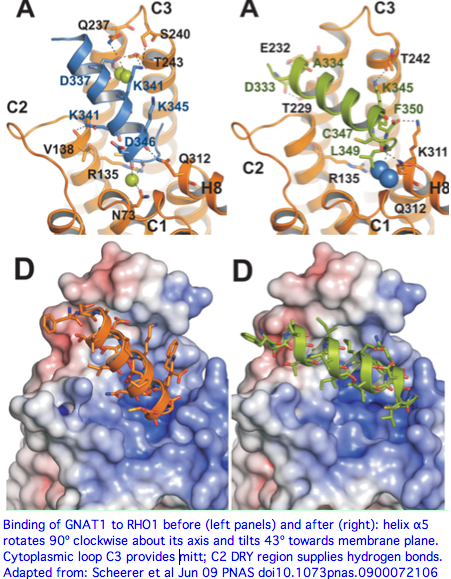
Tripedalia and its still-hypothetical Gt transducin have been co-evolving on a separate trajectory from the bovine pair for perhaps 1.2 billion years. The histories of gene duplication in the heterotrimeric G proteins may differ, along with the constraints in vertebrates of transducin having to simultaneously interact with multiple ciliary opsin genes. However as initial gene duplicates, Gt and Gq would have had the same binding site; [http://www.ijdb.ehu.es/web/paper.php?doi=14756332&a=f even as the proteins differentiated to serve distinct opsin sets,] the binding site (at the juncture of last transmembrane and last cytoplasmic sections) would likely remain conserved because of homologic inertia of the binding pocket and the implausibility of creating a new transduction mechanism. | Tripedalia and its still-hypothetical Gt transducin have been co-evolving on a separate trajectory from the bovine pair for perhaps 1.2 billion years. The histories of gene duplication in the heterotrimeric G proteins may differ, along with the constraints in vertebrates of transducin having to simultaneously interact with multiple ciliary opsin genes. However as initial gene duplicates, Gt and Gq would have had the same binding site; [http://www.ijdb.ehu.es/web/paper.php?doi=14756332&a=f even as the proteins differentiated to serve distinct opsin sets,] the binding site (at the juncture of last transmembrane and last cytoplasmic sections) would likely remain conserved because of homologic inertia of the binding pocket and the implausibility of creating a new transduction mechanism. | ||
It is easy enough to identify candidates for the photocascade alpha subunit of heterotrimeric G protein in cnidaria -- simple Blast of human GNAT2 protein (cone transducin) calls up two strong 64% identity matches in Nematostella, Hydra, and others (meaning the binding site could be accurately | It is easy enough to identify candidates for the photocascade alpha subunit of heterotrimeric G protein in cnidaria -- simple Blast of human GNAT2 protein (cone transducin) calls up two strong 64% identity matches in Nematostella, Hydra, and others (meaning the binding site could be accurately modeled). These could be close relatives of the implied Tripedalia ciliary opsin transducin (for which there is no data); extensive staining st would needed to make the case. | ||
Actual ciliary opsin conservation in this boundary region looks like yNP.IY..mNkqFr.c (YNPVIYCLLNRSFRKM in Tripedalia) with the serine of NRS not observed in other species; the [http://www.cbs.dtu.dk/~gorodkin/appl/plogo.html logos] graphic shown below makes this quantitative. Note while this region looks very different in melanopsins (typically HPK in HNPIIYAITHPKYRM), there was never any potential for confusion given the much worse alignment there. The cubomedusan opsin also appears oddly truncated at the carboxy terminus; it appears to lack the cysteines need for palmitoylation and the serines/threonines for kinase activation. | Actual ciliary opsin conservation in this boundary region looks like yNP.IY..mNkqFr.c (YNPVIYCLLNRSFRKM in Tripedalia) with the serine of NRS not observed in other species; the [http://www.cbs.dtu.dk/~gorodkin/appl/plogo.html logos] graphic shown below makes this quantitative. Note while this region looks very different in melanopsins (typically HPK in HNPIIYAITHPKYRM), there was never any potential for confusion given the much worse alignment there. The cubomedusan opsin also appears oddly truncated at the carboxy terminus; it appears to lack the cysteines need for palmitoylation and the serines/threonines for kinase activation. | ||
| Line 44: | Line 46: | ||
xenTro YTGGTNNN--SYIMALFLTCFIMPLSTIIF-SYSNL----LMALRAVAAQQKDSETT---QRAEKEVTRMVIAMVLAFLICWLPYASFAVVVAVNKDVVIEPTVASLPSYFSKTATVYNPIIYVFMNKQFRNCLMTLLCCGRS 316 | xenTro YTGGTNNN--SYIMALFLTCFIMPLSTIIF-SYSNL----LMALRAVAAQQKDSETT---QRAEKEVTRMVIAMVLAFLICWLPYASFAVVVAVNKDVVIEPTVASLPSYFSKTATVYNPIIYVFMNKQFRNCLMTLLCCGRS 316 | ||
The low overall percent identity (37%) of its best matches to any known Bilateran opsin, attributable to the great evolutionary time spans involved, disappointingly does not open any new doors. The sequence here does not have striking | The low overall percent identity (37%) of its best matches to any known Bilateran opsin, attributable to the great evolutionary time spans involved, disappointingly does not open any new doors. The sequence here does not have striking homology to cnidarian opsins recently proposed by several other groups, does not elicit dramatic new ones in sequenced cnidarian genomes or transcript programs, nor serve to locate opsins in sponge genome. | ||
Note the other phototransduction cascade proteins co-located in the rhopalia by the authors have much more striking percent identity to both Nematostella and human homologs than the opsin, most astonishingly PDE6D at 80%. In terms of human gene names, function, and associated disease: | Note the other phototransduction cascade proteins co-located in the rhopalia by the authors have much more striking percent identity to both Nematostella and human homologs than the opsin, most astonishingly PDE6D at 80%. In terms of human gene names, function, and associated disease: | ||
| Line 54: | Line 56: | ||
GUCY2F 48% EU310503 rod outer membrane guanyl cyclase resynthesis of cGMP for recovery of the dark state | GUCY2F 48% EU310503 rod outer membrane guanyl cyclase resynthesis of cGMP for recovery of the dark state | ||
Most curiously, the authors observe that the major Tripedalia lens crystallin J1 protein is also strongly expressed in the nominally lensless slit and pit eyes. This raises the question of whether our concept of lens is too anthropomorphic and whether other anatomical configurations of high refractive index proteins can accomplish the same ends, possibly requiring an 'upgrade' for slit and pit eye functional assessment. For example, cone | Most curiously, the authors observe that the major Tripedalia lens crystallin J1 protein is also strongly expressed in the nominally lensless slit and pit eyes. This raises the question of whether our concept of lens is too anthropomorphic and whether other anatomical configurations of high refractive index proteins can accomplish the same ends, possibly requiring an 'upgrade' for slit and pit eye functional assessment. For example, cone mega-mitochondria in treeshrews (refractive index 1.4) may have some [http://www.ncbi.nlm.nih.gov/pubmed/9135860,12511073 lensing or waveguide function.] | ||
[[Image:CoboTax.png|left|]] | [[Image:CoboTax.png|left|]] | ||
| Line 60: | Line 62: | ||
The opsins of slit and pit and larval eyes (implied by their photoreceptor cell structures) need to be determined with high priority. What's needed here too is a massive ortholog sequencing effort in the 19 extant species of cubozoan to break up isolated opsin long branch to show how such sequences are evolving, allow estimation of when they were recruited to imaging vision and reconstruct the earliest such ancestor. If a rhabdomeric opsin also occurs in larval or other eyes, that too needs comparative genomics. | The opsins of slit and pit and larval eyes (implied by their photoreceptor cell structures) need to be determined with high priority. What's needed here too is a massive ortholog sequencing effort in the 19 extant species of cubozoan to break up isolated opsin long branch to show how such sequences are evolving, allow estimation of when they were recruited to imaging vision and reconstruct the earliest such ancestor. If a rhabdomeric opsin also occurs in larval or other eyes, that too needs comparative genomics. | ||
It's unclear whether the last common eumetazoan ancestor of Tripedalia and Bilatera had imaging vision (note the hydozoan Cladonema radiatum has eyes and sponge larva have photoreceptor structures), yet clearly that ancestor contained one or more ciliary-type rhodopsin-class 7TM GPCR from which ciliary opsins descended in both clades, not always to be recruited for imaging. The fossil record for cubozoan cnidaria [http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0001121 predates the Cambrian,] though when eyes and statocysts (both regulated by PAXB) first appeared is unclear. Note further that planaria larva of Tripedalia have a | It's unclear whether the last common eumetazoan ancestor of Tripedalia and Bilatera had imaging vision (note the hydozoan Cladonema radiatum has eyes and sponge larva have photoreceptor structures), yet clearly that ancestor contained one or more ciliary-type rhodopsin-class 7TM GPCR from which ciliary opsins descended in both clades, not always to be recruited for imaging. The fossil record for cubozoan cnidaria [http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0001121 predates the Cambrian,] though when eyes and statocysts (both regulated by PAXB) first appeared is unclear. Note further that planaria larva of Tripedalia have a rhabdomeric photoreceptor, suggesting melanopsin photoreception is also very ancient. | ||
It's worth noting that the phylogenetic tree for early metazoans has entered a state of turmoil. Sponges may be secondarily simplified in the adult stage; [http://www.ncbi.nlm.nih.gov/pubmed/18322464? ctenophores may be basal,] and so forth. It's even been suggested that [http://www.ncbi.nlm.nih.gov/pubmed/18315817? ancestral sponge larva represent the central object] from which complex metazoan are descended. | It's worth noting that the phylogenetic tree for early metazoans has entered a state of turmoil. Sponges may be secondarily simplified in the adult stage; [http://www.ncbi.nlm.nih.gov/pubmed/18322464? ctenophores may be basal,] and so forth. It's even been suggested that [http://www.ncbi.nlm.nih.gov/pubmed/18315817? ancestral sponge larva represent the central object] from which complex metazoan are descended. | ||
These considerations suggest a very early evolutionary origin for the basic genes of photoreception and their regulation, with a great many lineage-specific subsequent upgrades and downgrades of the details, deuterostomes being the last to get on board with imaging vision. Thus the question Darwin asked, how many times did vision originate, requires a more nuanced answer than just a number. Most likely, the basic package of photoreceptor genes and their developmental regulation of expression arose just once, with all subsequent systems descended from that. However that package was subjected to numerous gene duplications and morphological variations in deployment, and | These considerations suggest a very early evolutionary origin for the basic genes of photoreception and their regulation, with a great many lineage-specific subsequent upgrades and downgrades of the details, deuterostomes being the last to get on board with imaging vision. Thus the question Darwin asked, how many times did vision originate, requires a more nuanced answer than just a number. Most likely, the basic package of photoreceptor genes and their developmental regulation of expression arose just once, with all subsequent systems descended from that. However that package was subjected to numerous gene duplications and morphological variations in deployment, and outside recruitment in the case of crystallins and pigments. | ||
Protostomes recruited melanopsin-class opsins for their imaging vision (despite available retained ciliary opsins), whereas early deuterostomes lacked imaging vision per se but retained ciliary opsins in related | Protostomes recruited melanopsin-class opsins for their imaging vision (despite available retained ciliary opsins), whereas early deuterostomes lacked imaging vision per se but retained ciliary opsins in related photoreception roles. Later post-amphioxus, post-tunicate deuterostomes independently recruited a descendent ciliary opsin (despite an available retained melanopsin-class opsin), moving from pineal to bilateral imaging eyes in the third and latest invention of imaging vision. | ||
The spectral sensitivity of neritic (near-shore) lens eyes of a box jellyfish, Tripedalia cystophora [http://jeb.biologists.org/cgi/content/full/209/19/3758#REF17 previously considered] by M Coates et al was interpreted as a single vitamin A-1 based opsin with peak sensitivity near 500 nm (blue-green). However nothing was sequenced. This species was most helpfully [http://www.ijdb.ehu.es/web/paper.php?doi=041851jp&a=f reviewed earlier] by Piatigorsky and Kozkmik who note Eakin already commented on seemingly ciliary photoreceptors in 1962. However, 45 years later we still didn't know if opsins in cnidarians would classify with vertebrate ciliary opsins. They could even share conserved intron positions though that cannot be determined from transcript data. | The spectral sensitivity of neritic (near-shore) lens eyes of a box jellyfish, Tripedalia cystophora [http://jeb.biologists.org/cgi/content/full/209/19/3758#REF17 previously considered] by M Coates et al was interpreted as a single vitamin A-1 based opsin with peak sensitivity near 500 nm (blue-green). However nothing was sequenced. This species was most helpfully [http://www.ijdb.ehu.es/web/paper.php?doi=041851jp&a=f reviewed earlier] by Piatigorsky and Kozkmik who note Eakin already commented on seemingly ciliary photoreceptors in 1962. However, 45 years later we still didn't know if opsins in cnidarians would classify with vertebrate ciliary opsins. They could even share conserved intron positions though that cannot be determined from transcript data. | ||
| Line 72: | Line 74: | ||
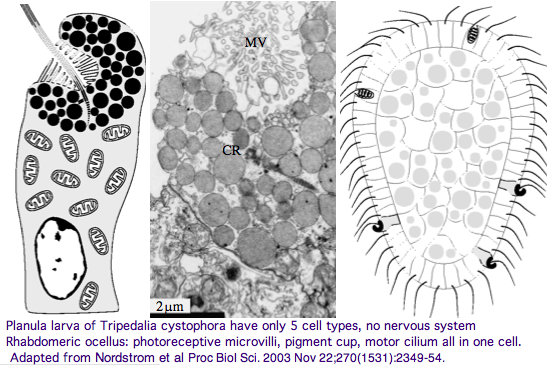
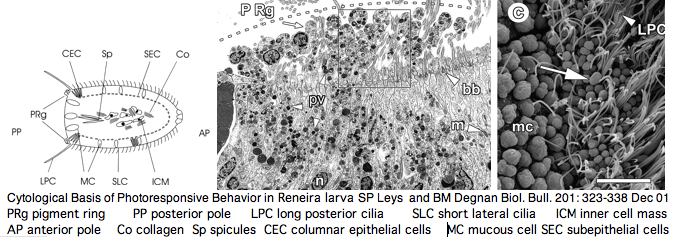
[[Image:Opsin_cnid_larva.png]] | [[Image:Opsin_cnid_larva.png]] | ||
Furthermore, as [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=14667350 noted] by Nordstrom et al, planula larvae of Tripedalia have a series of single-cell pigment cup rhabdomeric-like photoreceptors directly connected to motor cilia. These lack neural connections in line with Gehring's notion of the eye preceding the brain in evolution, rather than being a later add-on. So cnidaria might actually retain | Furthermore, as [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=14667350 noted] by Nordstrom et al, planula larvae of Tripedalia have a series of single-cell pigment cup rhabdomeric-like photoreceptors directly connected to motor cilia. These lack neural connections in line with Gehring's notion of the eye preceding the brain in evolution, rather than being a later add-on. So cnidaria might actually retain descendants of both types of ancestral opsins. No sequence is available yet for larvae, | ||
[[Image:Opsins_cubomedusae.png]] | [[Image:Opsins_cubomedusae.png]] | ||
=== <span style="color: #990099;">Cubozoa: Carybdea | === <span style="color: #990099;">Cubozoa: Carybdea rastonii (box jellyfish) .. 1 opsin</span> === | ||
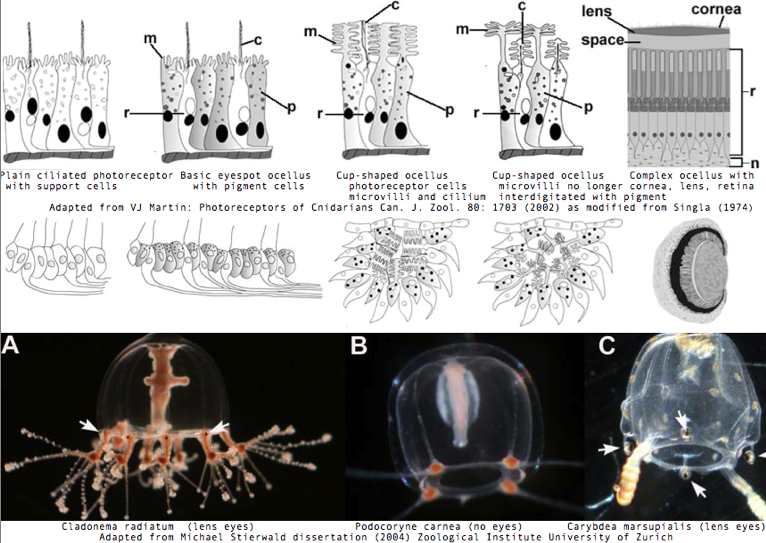
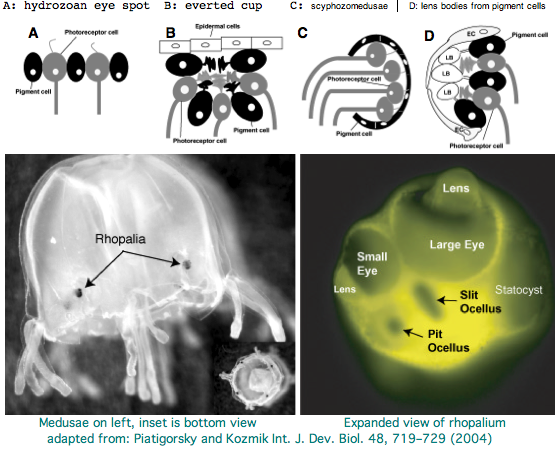
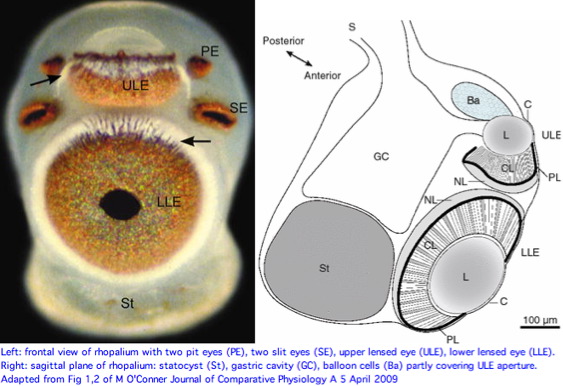
Cnidarians are the earliest diverging invertebrates with multicellular light-detecting organs. Photodetectors include simple eyespots, pigment cups, complex pigment cups with lenses, and camera-type eyes with a cornea, lens, and retina. These remarkable eyes are located on sensory clubs called rhopalia with four lining the bell. Each houses six eyes: a pair of pit ocelli, a pair of slit ocelli, and two unpaired lens eyes with counterparts to cornea, cellular lens and retina of ciliated photoreceptors. | |||
Anatomically, the ocelli have bipolar sensory photoreceptor cells interspersed among nonsensory pigment cells with the apical end making the light-receptor with the basal end forming an axon that synapses with second-order neurons to form what amounts to ocular nerves. Vision has roles in the reproduction and feeding of cubomedusae which can find each other and chase, catch, and eat teleost fish. A patch of Pelagia nocticula 10 square miles in extent and 35 feet deep recently destroyed a salmon farm off Northern Ireland. | |||
One of the most striking jellyfish from the perspective of complex eyes is Carybdea marsupialis, as [http://www.biology.appstate.edu/faculty/martinvj.htm reviewed by VJ Martin]. Antibody studies based on vertebrate cone/rod opsins are not sufficient because of possible cross reactivity to generic GPCR proteins or non-imaging photoisomerases; no opsins have been sequenced yet. Provided the retroposon and base composition are not unwieldy, Carybdea could be an instructive genome to sequence. Nematostella and Hydra, whatever their other genomic merits, sit in the Anthozoa and Hydrozoa, clades of cnidarian lacking elaborate visual systems. | |||
[http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=18832159 Carybdea rastonii] has a green-sensitive visual pigment in its ciliary-type lens eyes utilizing Gs cAMP phototransduction cascade (that is, not Gt, Go or Gq). A complete opsin-like sequence [http://www.ncbi.nlm.nih.gov/nuccore/210049957 AB435549] satisfies various opsin sequence signature requirements but does not classify clearly among known ciliary opsins. Instead, its affinities lie with opsin-like sequences from Hydra and Nematostella -- species that have 'too many' opsins for their meagre photoreceptive anatomy and photobehavioral capacities. | |||
The second problem is the lack of blastp affinity of Carybdea protein to the new validated opsin from Tripedalia cystophora, which classifies as expected within bilateran ciliary opsins. This implies the last common ancestor of box jellyfish with bilaterans possessed a conventional ciliary opsin. What then is the need for other classes of opsin-like sequences? Other interpretations of opsin-like sequences need to be considered: | |||
First, AB435549 may function more along the lines of peropsin/RGR/neuropsin (even though it does not cluster with them) as an auxiliary, possibly signaling or replenishing photoisomerase but not the primary imaging photoreceptor in Carybdea. In this scenario, AB435549 could hybridize more or less correctly in situ as would a better missing opsin -- remaining to be recovered -- more closely related to the Tripedalia opsin. | |||
This still does not explain the observed clustering of AB435549 with opsin-like proteins, especially of intronless genes of Hydra and Nematostella with dubious connection to any kind of vision. Possibly the function here instead has to do with sensing or digestion of dietary carotenoids or photo-rearrangement of double bonds for biosynthetic, energetic or regulatory purposes (eg retinoic acid metabolism). In its wildest form, this hypothesis envisions metabolic photoreception as the core ancestral property that was later co-opted during the evolution of vision. Alternately, metabolic photoreception was a later spinoff of light sensing. | |||
A third scenario just places AB435549 on another 'track' from bilateran opsins. Here it either arose from a conventional ciliary opsin after species divergence from last common ancestor or it is older and bilaterans subsequently lost all members of its gene tree class. The latter seems more plausible because AB435549 has no particular affinities to ciliary opsins relative to melanopsin or peropsin-type opsins. In this view box jellyfish have retained two systems with different retention patterns in different clades, in analogy to protostomes emphasizing melanopsins and deuterostomes ciliary in their imaging opsins. In support of this, the timing of box jellyfish divergences could be equally as old, even if they 'all look the same' from the human perspective. Here the Tripedalia group is then more relevant to the evolution of deuterostome vision whereas neither Tripedalia or Carybdea is helpful in understanding protostome vision. | |||
It would be quite practical with 2009 technology to sequence a substantial number of complete box jellyfish genomes. This has the great advantage of allowing bioinformatic recovery of complete K-rhodopsin portfolios. This would settle the question of whether Carybdea possesses a ciliary opsin clustering with that of Tripedalia. Genomes also provide homologs of all auxiliary genes such as Galpha and RPE65. With a large set of proteins and rRNA, the timing of divergences within box jellyfish could be better estimated. It remains conceivable that cnidarians are paraphyletic, ie box jellyfish share a later divergence node with bilaterans, perhaps explaining common ground in eye structures not seen in anthozoa etc. | |||
>CUBOP_carRas Carybdea rastonii sea_wasp Cnid.Cubo.Cary AB435549 cubop 18832159 | |||
MGANITEILSGFLACVVFLSISLNMIVLITFYRLRHKLAFKDALMASMAFSDVVQAIVGYPLEVFTVVDGKWTFGMELCQVAGFFITALGQVSIAHLTALAL | |||
<font color = "blue">DRY</font>FTVCRPFVATAIHGSMRNAGMVIFVCWFYASFWAVLPLVGWSNYDVEGDGMRCSINWADDSPKSYSYRVCLFVFIYLIPVLLMVATYVLVQGEMKNMRGRAAQLFGSESEAAL | |||
KNIKAEKRHTRLVFVMILSFIVAWTPYTFVAMWVSFFTKQLGPIPLYVDTLAAMLA<font color = "blue">KSSAMFNPIIYCFLHKQFR</font>RAVLRGVCGRIVGGNAIAPSSTAVEPGQTLASGTAES* | |||
(to be continued) | |||
=== <span style="color: #990099;">Cubozoa: Chiropsella bronzie (box jellyfish) .. 0 opsins</span> === | |||
=== <span style="color: #990099;">Anthozoa: Nematostella vectensis (sea anemone) .. | This box jellyfish was featured recently in a comprehensive optical and micro-anatomical [http://www.springerlink.com/content/c7244j2728444424/ study] of all four eye types. The genus Chiropsella is not currently known to GenBank taxonomy, meaning no sequence data at all is available (unless some synonym has been used). Its enveloping family Chirodropidae has barely 17 sequence entries, none relevant to vision. [http://www.ncbi.nlm.nih.gov/pubmed/18723637,17921163,17141433 Chiropsella bronzie] was first named in 2006; it occurs in knee-deep water feeding on shrimp along sandy beaches in North Queensland, Australia. | ||
A 2010 immunohistochemical and microspectrophotometry study from the same group also avoided molecular data, instead using antibodies to five zebrafish imaging opsins to locate a single ciliary type opsin candidate in the upper and lower lensed eyes. Only [http://www.ncbi.nlm.nih.gov/nuccore/AF109373 SWS1] gave a reaction -- and this [http://rspb.royalsocietypublishing.org/content/early/2010/02/08/rspb.2009.2248/F5.large.jpg predominantly in the neuronal layer] rather than the receptor cilia. The authors did not provide an accession number for the UV opsin but only a citation to a 1999 paper by TS Vihtelic et al. It is apparently AF109373 (which agrees with today's genomic). | |||
Given the great divergence of cnidarians and very rapidly evolving teleost fish, a regenerative photoisomerase (if one exists) would not be detected by this method because the best match to known ciliary cnidopsins is already very poor at 29% -- well within range of a non-opsin GPCR (for example Trichoplax XM_002114725). It's unclear which region is both exposed to soluble antibody but also sufficiently and specifically conserved to provide the observed reaction yet not stain by the other antibodies. A non-opsin GPCR would not bleach, yet spectrophotometry is uncoupled to immunohistochemistry, so it is not known if the bleached compound matches the immunoreactive entity. | |||
It would make more sense simply to sequence and assemble the entire genome (an afternoon's work), then analyze it for opsins and regenerative proteins using antibodies appropriate to the species. | |||
TMT_triCys Tripedalia cystophora (box_jelly) identities: 29% positives: 50% best match to zebrafish SWS1 | |||
Query: 30 FYLQAAFMGFVFIVGTP-----MNGIVLFVTMKYKKLRQPLNYILVNISLAGFIFDTFSV 84 | |||
FY FMG FI G+ +NG+V+ V +KY + N I++++S A + | |||
Sbjct: 29 FYNFYTFMGS-FIAGSACCSFLLNGLVIAVLIKYIRTITNTNIIVLSMSCANILIPLLGS 87 | |||
Query: 85 SQVSVCAARGYYSLGYTLCSMEAAMGSIAGLVTGWSLAVLAFERYVVICKPF--GSFKFG 142 | |||
+ + + G C+ + +++G+ + L L+FER++ I P + | |||
Sbjct: 88 PLSATSSLMRKWQFGNGGCTWYGFINTLSGISGIYHLTFLSFERFITIVLPLKRDTILST 147 | |||
Query: 143 QGQAVGAVVFTWIIGTACATPPFFGWSRYIPEGLGTACGPDWYTKSEEYNSESYTYFLLI 202 | |||
+ +G + W+ A P FGW YI EG+ T+C W +K E N SY F++ | |||
Sbjct: 148 KNIYIGLGIL-WVAAIGVAGAPVFGWCEYIKEGVRTSCSVAWSSK-ENMNVFSYNLFMIF 205 | |||
Query: 203 TCFMMPMTIIIF-SYSQL----LGALRAVAAQQAESEST---QKAEREVSRMVVVMVGSF 254 | |||
T F++PM +II+ +Y + + + RA Q +SE T KAE++++ MV+ M+ +F | |||
Sbjct: 206 TVFLLPMLVIIYCNYRFIKEVSIMSTRARGLQGGDSEMTASASKAEKQLTIMVITMIIAF 265 | |||
Query: 255 VLCYAPYAVTAMYFANSDEPNKDYRLVAIPAFFSKSSSVYNPLIYAFMNKQF 306 | |||
+ + PY V +M F ++P+ F+K+S +YNP+IY +N+ F | |||
Sbjct: 266 NIAWLPYTVVSMVFLTGYGDVVGPMGASVPSVFAKTSVIYNPVIYCLLNRSF 317 | |||
[[Image:RhopalCB.jpg|left]] | |||
[[Image:RhopalCB2.jpg|left]] | |||
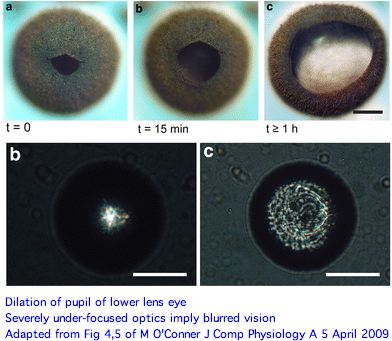
The picture of eye functions that emerges here is rather surprising. Both upper and lower lens eyes are severely under-focused (much more so than Tripedalia cystophora) with the retina so close to the lens that only blurred vision can result. And these are its best eyes. A novel long pigment cell has dark pigment moving within a white pigmented tube during light/dark adaptation for unknown advantages. | |||
Since rhopalia eyes have seemingly had several hundred million years to evolve deeper vitreal space (which seems simple enough), an eye tuned to detect large structures at short range (spatial low-pass filter) evidently suits Chiropsella. The primary function may be visual avoidance of obstacles or detection of prey within range. Higher spatial resolution and rapid refreshing entail a concomitant expansion of the nervous system and higher ongoing energetic costs to adaptively process massive extra information. | |||
The skyward pointing upper lens eye has various peculiar features from our perspective. The ellipsoid lens lacks focusing refractive power, has a cataract-like inclusion casting a shadow on the retina, a hole in connection with the pigment layer exposing the retina to direct sunlight, balloon cells partly covering the lens aperture, gastric cells contacting the posterior lens side, and less of a pupillary response. Although little is known on the biological side, the upper lens-eye shadow line capability could be suited for detecting sun or moon position. | |||
The two smaller pit and slit eye types have an epithelium/cornea covering but do not contain a lens. The photoreceptors are pigmented and organized into ciliary, pigment and neural layers. These eyes are capable at best of monitoring ambient light intensity, perhaps guiding overall phototactic behavior or orientation. Yet this proposed function could seemingly be accomplished as a byproduct of lens eye functionality and would not require paired pit and slit eyes, much less four sets of them around the full quadrant of rhopalia. | |||
While vertebrates too have anatomically separated photoreceptors for diverse functions, there are meagre prospects for homologizing here to melanocyte, ganglial, pineal or other deep brain structures. In fact even the main lower lens eye may not descend from an ancestral structure that, in another clade, became a bilateran eye because apparent common ground can originate multiple times just from convergent considerations of optical physics. Thus a 'cornea' is merely a protective epithelial layer and a lens just a thickening filled with overproduced protein providing refractive index. | |||
Even if the old mystery of the origin of the eye can been pushed pack through homologization to the mystery of the origin of the rhopalium, no 'intermediate states' are likely be found in the [http://www.plosone.org/article/info:doi/10.1371/journal.pone.0001121 Cambrian jellyfish fossil record] and seemingly not in other living species of box jellyfish (all of which have the (2+2+1+1)x4=24 eye pattern). | |||
It is not clear whether distinct opsin genes are utilized in these various eyes and if so, what their gene tree might look like, eg ((pit,slit),(upper,lower)) vs (((pit,slit),upper),lower). We are left wondering too about the origin of rhabdomeric melanopsins if ultrastructure in pre-bilaterans is always specialized to modified cilia. | |||
<br clear ="all"> | |||
=== <span style="color: #990099;">Anthozoa: Nematostella vectensis (sea anemone) .. 3 opsins</span> === | |||
The Nematostella genome has been [http://genome.jgi-psf.org/Nemve1/Nemve1.home.html released] along with [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=17615350,18054114,17982171,17893746,17921482,17716645,17503371,17335570,16867185,16837574,15128674 major papers] and an upgrade to [http://stellabase.org/SNP Stellabase.] Not all 6.1 million traces were used up by the assembly, so any gene missing from the assembly should be sought directly in the trace archives. | The Nematostella genome has been [http://genome.jgi-psf.org/Nemve1/Nemve1.home.html released] along with [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=17615350,18054114,17982171,17893746,17921482,17716645,17503371,17335570,16867185,16837574,15128674 major papers] and an upgrade to [http://stellabase.org/SNP Stellabase.] Not all 6.1 million traces were used up by the assembly, so any gene missing from the assembly should be sought directly in the trace archives. | ||
The sea anemone, an anthozoan within Cnidaria having epithelial cells, neurons, stem cells, complex extra-cellular matrix, muscle fibers, and symmetry axis, is emerging as a high-profile evo-devo model species to elucidate the emergence and deployment of genes that determine animal body plans. However those plans don't seem to include eyes or overt photoreceptor structures such as pigment cells -- for that cubomedusae would be far better. PAX6 and RX are especially | The sea anemone, an anthozoan within Cnidaria having epithelial cells, neurons, stem cells, complex extra-cellular matrix, muscle fibers, and symmetry axis, is emerging as a high-profile evo-devo model species to elucidate the emergence and deployment of genes that determine animal body plans. However those plans don't seem to include eyes or overt photoreceptor structures such as pigment cells -- for that cubomedusae would be far better. PAX6 and RX are especially relevant to photoreceptor structures; their expression has been thoroughly studied in Nematostella without uncovering any sensory system though they contribute to patterning specific components of the ectodermal nerve net. | ||
The JGI annotation pipeline produced a number of extensively annotated gene models for Nematostella opsins. These are available simply by keyword lookup, tblastn of various queries the best of which turn out to be -- unsurprisingly -- an encephalopsin subclass from Branchiostoma. It is important to <span style="color: #990099;">credit the JGI staff</span> for providing the | The JGI annotation pipeline produced a number of extensively annotated gene models for Nematostella opsins. These are available simply by keyword lookup, tblastn of various queries the best of which turn out to be -- unsurprisingly -- an encephalopsin subclass from Branchiostoma. It is important to <span style="color: #990099;">credit the JGI staff</span> for providing the relevant bioinformatic track computations because they were first to characterize and release these opsins into the public domain (eg GenBank NR and Entrez Gene). It does not constitute independent "discovery" to perform keyword lookup and copy out other peoples' work. Without proper citation, that's [http://en.wikipedia.org/wiki/Plagiarism plagiarism.] | ||
I extended improperly truncated JGI gene models (ie those lacking iMet and stop codon), validated the extensions still lacked introns (GT-Ag splice junctions missing at positions expected from closest homologs), placed the best 3 (of a half dozen) in the Opsin | I extended improperly truncated JGI gene models (ie those lacking iMet and stop codon), validated the extensions still lacked introns (GT-Ag splice junctions missing at positions expected from closest homologs), placed the best 3 (of a half dozen) in the Opsin Classifier with fasta headers, noted their best matches below, and validated lysine and counterion glutamate in the expected positions. All this is consistent with (but does not prove) a role for ciliary Gt opsins in pre-Bilateran photoreception. | ||
We expect cnidarians (maybe not this particular anthozoan) to have both melanopsins and encephalopsins. Our tendency is to think that imaging eye opsins, whether insect rhabdomeric or vertebrate ciliary, are the main attraction, with the other opsins playing out obscure roles in secondary functions like timing of gamete release . That's quite wrong-headed. Deeper gene family trees show that the melanopsin and encephalopsin | We expect cnidarians (maybe not this particular anthozoan) to have both melanopsins and encephalopsins. Our tendency is to think that imaging eye opsins, whether insect rhabdomeric or vertebrate ciliary, are the main attraction, with the other opsins playing out obscure roles in secondary functions like timing of gamete release. That's quite wrong-headed. Deeper gene family trees show that the melanopsin and encephalopsin constitute the primary photoreceptors. Over vast evolutionary time scales, they gave rise to various spin-offs in various clades at various times through gene duplication and subsequent neofunctionalization. At even greater phylogenetic depth, melanopsin and encephalopsin are themselves related by gene duplication of an ur-opsin, which itself arose as a duplication of an established non-opsin GPCR. As noted by [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=15514158.4756332,11874910 Arendt,] that exploited prior gene duplication within the alpha subunit of heteromeric G protein and profound diversification in signaling system second messaging. | ||
The odd thing about all these cnidarian encephalopsins is their lack of introns (three ancestrals are expected). That's very unlikely to be the Eumetazoan ancestral state for encephalopsin because Nematostella is no rogue organism when it comes to [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=17615350,17503371 intron conservation.] A common explanation for this within eukaryotic bioinformatics is gene duplication of a master gene via fully processed retrogenes (rather than through tandem, segmental, chromosomal, or whole genome duplications -- all of which preserve introns). Mixed mechanisms are also common (as in olfactory receptors): an initial intronless retrogene is duplicated tandemly etc. These paralogs can even displace the master gene by taking over its function, causing it subsequently to be displaced or even lost. That scenario played out within [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=12618404 zebrafish opsins.] | The odd thing about all these cnidarian encephalopsins is their lack of introns (three ancestrals are expected). That's very unlikely to be the Eumetazoan ancestral state for encephalopsin because Nematostella is no rogue organism when it comes to [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=17615350,17503371 intron conservation.] A common explanation for this within eukaryotic bioinformatics is gene duplication of a master gene via fully processed retrogenes (rather than through tandem, segmental, chromosomal, or whole genome duplications -- all of which preserve introns). Mixed mechanisms are also common (as in olfactory receptors): an initial intronless retrogene is duplicated tandemly etc. These paralogs can even displace the master gene by taking over its function, causing it subsequently to be displaced or even lost. That scenario played out within [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=12618404 zebrafish opsins.] | ||
| Line 104: | Line 176: | ||
Given an finished genome, the mode of gene amplification can be explored by looking at flanking genes. Perhaps ENCEPHa_nemVec and ENCEPHa_nemVec are adjacent (ie tandem duplication) or perhaps their flanking genes are paralogous (syntenic segmental duplication). However the Nematostella genome is currently unfinished and the (gapless) contigs containing the encephalopsins run about 10 kbp. Depending on gene density that can be too small to establish synteny. These contigs, separated by strings of N's of unknown length, are further assembled into larger scaffolds (ample for synteny), a process usually trustworthy at highly experienced JGI but sometimes confounded by issues such as repeats, compositional simplicity, very recent duplicative regions, and clonability. | Given an finished genome, the mode of gene amplification can be explored by looking at flanking genes. Perhaps ENCEPHa_nemVec and ENCEPHa_nemVec are adjacent (ie tandem duplication) or perhaps their flanking genes are paralogous (syntenic segmental duplication). However the Nematostella genome is currently unfinished and the (gapless) contigs containing the encephalopsins run about 10 kbp. Depending on gene density that can be too small to establish synteny. These contigs, separated by strings of N's of unknown length, are further assembled into larger scaffolds (ample for synteny), a process usually trustworthy at highly experienced JGI but sometimes confounded by issues such as repeats, compositional simplicity, very recent duplicative regions, and clonability. | ||
The most convenient approach here is tblastn of ENCEPHa_nemVec against the wgs menu item at NCBI Blast, specifying Nematostella. The three genes here are on different scaffolds altogether, ruling out tandem position. The nearest flanking genes can be extracted by blastx of the enveloping contig (or whole scaffold) against GenBank protein. JGI has in effect already done this, as could be seen by expanding out from the | The most convenient approach here is tblastn of ENCEPHa_nemVec against the wgs menu item at NCBI Blast, specifying Nematostella. The three genes here are on different scaffolds altogether, ruling out tandem position. The nearest flanking genes can be extracted by blastx of the enveloping contig (or whole scaffold) against GenBank protein. JGI has in effect already done this, as could be seen by expanding out from the initial browser view. Comparing 3 browser views is complicated by the fact that flanking paralogs might be named differently, but that is readily overcome by collecting sequences (noting strand orientation) into a mini-database and comparing within [http://www.proweb.org/proweb/Tools/WU-blast.html uBlast.] | ||
Notice the Opsin Classifier [ | Notice the Opsin Classifier [[Opsin_evolution|collection]] already contains the outcome of this process as a fasta header field (for deuterostome opsins). It is conceivable that orthology of a Nematostella opsin to say a Branchiostoma opsin could be established in this way (synteny). However gene order in both genomes has been independently scrambled over immense time scales and orthology would have been to the Nematostella master gene (with introns) that appears lost. It's better to build out from a local synteny chain but that requires data from additional cnidaria. Note the irony here in that the farther removed the genome from human, the more densely they must be sampled. | ||
It's evident from a casual ClustalW alignment, after marking up columns for membrane-spanning sections and considering hydrophobicity, that Nematostella opsins conform to the standard central pattern. That's unsurprising since proteins retain 3D structure at far lower percent identity and the pattern here cuts much deeper, into the overall rhodopsin superfamily and beyond to generic GPCR. However encephalopsins can have very considerable extensions at their amino and especially carboxy termini that need separate consideration. | It's evident from a casual ClustalW alignment, after marking up columns for membrane-spanning sections and considering hydrophobicity, that Nematostella opsins conform to the standard central pattern. That's unsurprising since proteins retain 3D structure at far lower percent identity and the pattern here cuts much deeper, into the overall rhodopsin superfamily and beyond to generic GPCR. However encephalopsins can have very considerable extensions at their amino and especially carboxy termini that need separate consideration. | ||
For now, sequences can be trimmed to whatever is alignable across the full spectrum of ciliary opsins. Recall that by design the Opsin Classifier [ | For now, sequences can be trimmed to whatever is alignable across the full spectrum of ciliary opsins. Recall that by design the Opsin Classifier [[Opsin_evolution|collection]] seeks maximal phylogenetic dispersion to mitigate over-weighting by over-studied species that might introduce clade-specific interpretive bias. That could also be done by distilling the dataset down to ancestral sequences at lamprey divergence, the risk there being co-evolution of non-adjacent | ||
residues (eg different alpha helices) can be lost in residue-by-residue ancestral reconstructions. | residues (eg different alpha helices) can be lost in residue-by-residue ancestral reconstructions. | ||
| Line 135: | Line 207: | ||
Four putative opsins have been [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17940617 proposed] by Plachetzki et al. Accessions of the supporting gene models are given in the JGI protein ID system (non-GenBank) as Nematostella1 219988, Nematostella2 85309, Nematostella3 130042, and Nematostella4 108738 (or fragments in the alignment graphic allow recovery of the respective cdnas by tblastn of GenBank WGS). As noted in the Hydra section, multiple lines of evidence are necessary to establish the first bona fide opsins in cnidarians. | Four putative opsins have been [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17940617 proposed] by Plachetzki et al. Accessions of the supporting gene models are given in the JGI protein ID system (non-GenBank) as Nematostella1 219988, Nematostella2 85309, Nematostella3 130042, and Nematostella4 108738 (or fragments in the alignment graphic allow recovery of the respective cdnas by tblastn of GenBank WGS). As noted in the Hydra section, multiple lines of evidence are necessary to establish the first bona fide opsins in cnidarians. | ||
There appear to be 2 Nematastella opsin-like cdnas at GenBank that cannot be found in the genome assembly or trace archives, DV091537 and DV087469. While genes can be missing from first assemblies, it is bizarre for both to be missing considering coverage is 6x. Upon back-blast to GenBank nr or the Opsin Classifier, very strong matches are seen consistently within crustacea. Thus it appears that these Sars Institute products are contaminants from another species, possibly a brine shrimp widely used in aquarium food. It is not unusual to see transcript (at issue here) and genome projects contaminated with dna from other species such as commensals, parasites, and food source -- this is reminiscent of [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=12931184 Xenoturbella] being confused with a | There appear to be 2 Nematastella opsin-like cdnas at GenBank that cannot be found in the genome assembly or trace archives, DV091537 and DV087469. While genes can be missing from first assemblies, it is bizarre for both to be missing considering coverage is 6x. Upon back-blast to GenBank nr or the Opsin Classifier, very strong matches are seen consistently within crustacea. Thus it appears that these Sars Institute products are contaminants from another species, possibly a brine shrimp widely used in aquarium food. It is not unusual to see transcript (at issue here) and genome projects contaminated with dna from other species such as commensals, parasites, and food source -- this is reminiscent of [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=12931184 Xenoturbella] being confused with a mollusk in its diet. | ||
New Nematostella transcripts continue to be posted by JGI into mid-Dec 2007. Using proxies for all possible queries, I located a possible melanopsin and possible rhabdomeric LWS counterpart, The former had two coding exons but not at a melanopsin position; the latter had but one. These are fairly weak matches and further characterization is needed. They're stored in the Opsin Classifier as MEL_nemVec and LWS_nemVec2. | New Nematostella transcripts continue to be posted by JGI into mid-Dec 2007. Using proxies for all possible queries, I located a possible melanopsin and possible rhabdomeric LWS counterpart, The former had two coding exons but not at a melanopsin position; the latter had but one. These are fairly weak matches and further characterization is needed. They're stored in the Opsin Classifier as MEL_nemVec and LWS_nemVec2. | ||
| Line 141: | Line 213: | ||
A third group has taken a serious look at photoreception in Nematostella. No paper or dissertation has emerged as yet; no cnidarian opsins have been posted to GenBank. | A third group has taken a serious look at photoreception in Nematostella. No paper or dissertation has emerged as yet; no cnidarian opsins have been posted to GenBank. | ||
The claim of orthology will prove exceedingly difficult to establish in a 700 million year long branch. It is not a property of a gene tree per se. By definition, two genes in species A and species B are orthologous if and only if they have descended vertically from the same single parental gene '''in their last common ancestor'''. The last component is exceedingly important because all opsins -- indeed all GPCR -- are ultimately descended from a single gene. However that single gene was not to be found in the common | The claim of orthology will prove exceedingly difficult to establish in a 700 million year long branch. It is not a property of a gene tree per se. By definition, two genes in species A and species B are orthologous if and only if they have descended vertically from the same single parental gene '''in their last common ancestor'''. The last component is exceedingly important because all opsins -- indeed all GPCR -- are ultimately descended from a single gene. However that single gene was not to be found in the common ancestor of cnidarian and bilaterans because sponges already appear to have classical opsins and perhaps hundreds of GPCR. | ||
Most ancestral introns in human genes were established in unicellular eukaryotes well prior to fungal and green plant divergence. For example the distinct introns in close paralogs SUMF1 and SUMF2 were in place before human/diatom separation. It's very difficult to imagine how the introns in neuropsins, rgropsins, peropsins, melanopsins, encephalopsins, pteropsins, and ciliary opsins could have descended from a single gene in Eumetazoa. | Most ancestral introns in human genes were established in unicellular eukaryotes well prior to fungal and green plant divergence. For example the distinct introns in close paralogs SUMF1 and SUMF2 were in place before human/diatom separation. It's very difficult to imagine how the introns in neuropsins, rgropsins, peropsins, melanopsins, encephalopsins, pteropsins, and ciliary opsins could have descended from a single gene in Eumetazoa. | ||
| Line 153: | Line 225: | ||
"Eyes have evolved numerous times within the animals, yet there has been surprising convergence in | "Eyes have evolved numerous times within the animals, yet there has been surprising convergence in | ||
the morphology, function and molecular basis of development in these structures. Although these diverse | the morphology, function and molecular basis of development in these structures. Although these diverse | ||
eye types have arisen independently, many taxa utilize similar cassettes of genes to specify them. These | eye types have arisen independently, many taxa utilize similar cassettes of genes to specify them. These developmental genes include members of the SIX class of homeodomain proteins (sine oculis and optix), eyes | ||
absent, dachshund and famously, the Pax genes (Pax6). Additionally, all animals in which photoreception has | absent, dachshund and famously, the Pax genes (Pax6). Additionally, all animals in which photoreception has | ||
been investigated use the opsin family, a class of seven transmembrane receptors, to detect light. Cnidarians | been investigated use the opsin family, a class of seven transmembrane receptors, to detect light. Cnidarians | ||
are an early branching lineage that are likely to have diverged from the rest of the animals before the | are an early branching lineage that are likely to have diverged from the rest of the animals before the evolution of discrete eye structures. | ||
The ancestral cnidarian did not posses eyes, however like the extant anthozoan cnidarians (sea anemones, | The ancestral cnidarian did not posses eyes, however like the extant anthozoan cnidarians (sea anemones, | ||
| Line 187: | Line 257: | ||
</pre> | </pre> | ||
=== <span style="color: #990099;"> | === <span style="color: #990099;">Anthozoa: Anemonia viridis (symbiotic anemone) .. 1 opsin</span> === | ||
This ciliary opsin fragment is associated with an October 2009 [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=19627569 metagenomic article] on the symbiotic coral reef organism Anemonia viridis and its dinoflagellate algae but was not annotated in the EST collection ([http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=Nucleotide&list_uids=219278581&dopt=GenBank&RID=EYFU57YH015 FK729339]). By back-tblastn, it is clearly a rhodopsin-type GPCR with Schiff base lysine (but unknown chromophore). | |||
It is most closely allied with ENCEPHb_nemVec of Nematostella vectensis in the currently known set of cnidarian opsins -- about 49% identity but not enough is known about Anemonia opsin multiplicity to say it is an ortholog. High identity might be due either to sequence conservation or close relatedness of these two genera -- their time since divergence is an unknown. Conservation (of sequence and hence function) seems more likely since -- at least at GenBank taxonomy -- the divergence is quite deep: | |||
Cnidaria; Anthozoa; Hexacorallia; Actiniaria; Nynantheae; Actiniidae; Anemonia | |||
Cnidaria; Anthozoa; Hexacorallia; Actiniaria; Edwardsiidae; Nematostella | |||
Cnidaria; Anthozoa; Hexacorallia; Scleractinia; Astrocoeniina; Acroporidae; Acropora | |||
>ENCEPH_aneVir Anemonia viridis (symbiotic sea anemone) frag:202-338 pubmed:19627569 | |||
KDAVARWGNKSPPTQQTMQAQKKTIRMSLVMVFAYLLAWTPYALTSLYSSFIASDITPLLSVMPALFAKLSSCYNPIIYFFMYSKFR | |||
KAAKKMIRRNLVGHDSNSGQGVSNTFATSFPRPISFLRYKRSAVAPLSDIPQVSSVDLPQVGRENDVTVQQDKASEINT* | |||
aneVir KDAVARWGNKSPPTQQTMQAQKKTIRMSLVMVFAYLLAWTPYALTSLYSSFIASDITP-LLSVMPALFA<span style="color: red">K</span>LSSCYNPIIYFFMYSKFR | |||
+D++ RWG S TQQT+ A++KT MS +MV A+L AW PYA+ SLY+SF P L+S +PA+ A<span style="color: red">K</span> S+CYNPIIYFFMYSKFR | |||
nemVec RDSMRRWGASSDSTQQTLAAERKTAWMSFIMVLAFLFAWVPYAVVSLYASFGGVTTIPKLMSTLPAMLA<span style="color: red">K</span>TSACYNPIIYFFMYSKFR | |||
=== <span style="color: #990099;">Anthozoa: Acropora millepora (stony coral) .. 4 opsins</span> === | |||
A new cnidarian opsin appeared at GenBank on 13 May 2009 based on the [http://www.dtd.nlm.nih.gov/Traces/sra/sra.cgi?cmd=show&f=main&m=main&s=main 454 transcriptome survey SRA003728] of 5-day-old planulae larva in Acropora millepora. The entry [http://www.ncbi.nlm.nih.gov/nuccore/222785024 EZ013658] generates 244 alignable residues (after a few N's are manually corrected using genetic code redundancy) of a melanopsin-type protein; unfortunately it terminates 20 residues short of the expected Schiff lysine (so 38 residues short of a [[Opsin_evolution:_Cytoplasmic_face#Melanopsins|full motif]]). | |||
Extending this sequence to the end of the seventh transmembrane segment is a high priority. That would likely raise the percent identity (excluding tails) over 40%, far beyond agreement of opsins to generic GPCRs. Even as it stands, the conserves early signature residues of melanopsins within opsins and opsins within GPCR and cleanly clusters with melanopsins at the opsin blastp classifier. With additional orthologs from other cnidaria (Nematostella genome lacks one), a better ancestral sequence could be worked out at the divergence node with bilatera. This sequence in turn may be as close as we get to the origin of melanopsic photoreception. | |||
On 29 May 2009, [http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=MegaBlast&PROGRAM=blastn&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch&BLAST_SPEC=SRA blastn of the 454 Short Read Archive] became available at NCBI. While EZ013658 matched 3 reads quite well, it was not fully tiled. These reads allowed various errors to be corrected but best of all, 56 extra amino acids to be added C-terminally. These included a standard Schiff motif KTASVYNPIIYFFSYKSFR. It remains a bit mysterious as to where the central region of EZ013658 came from in its assembly. | |||
The best matches within cnidaria are to Nematostella K-rhodopsins classifying to TMT/ENCEPH, even while being more distant to Nematostella opsins classifying as melanopsins (eg [http://www.ncbi.nlm.nih.gov/nuccore/163914296 BR000662], suggesting these opsin homolog classes converge in cnidaria. This means their signaling partners cannot be safely inferred from sequence alone; indeed Galpha programs themselves have complicated lineage-specific expansions. | |||
A sub-sequence of EZ013658 was correctly identified as melanopsin-related, though not linked to circadian rhythm, in a brief April 2009 study of [http://www.biolbull.org/cgi/content/full/216/2/131 24 circadian rhythm genes] and the extraordinarily light-driven reproductive timing of broadcast-spawning corals (which may utilize cryptochrome photoreceptors rather than opsins). | |||
Special care must be taken in cnidarians to maintain a rigorous definition of opsin photoreceptor candidates and not digress to [http://www.biolbull.org/cgi/external_ref?access_num=17196770&link_type=MED deeply diverged non-opsin GPCRs] lacking ability to bind chromophore and -- as far as we know -- any relevance to photoreception. Even with K-rhodopsins, it remains quite possible that some are merely involved in light-driven catabolism or rearrangement of dietary beta-carotenoids. | |||
The four sequence fragments below can only be evaluated over their coverage, though the ones here all include a standard K296 NP FR motif and gave only recognized opsins upon backblast to GenBank nr. Two of them, upon blastp to the 485 curated opsin classifier, give best matches to deuterostome TMT and encephelopsins, suggesting a closer relationship to bilateran ciliary opsins than most cnidarian K296 opsin-like proteins do. | |||
Alignment of Acropora 454 transcript with human melanopsin | |||
<font color ="blue">Blue</font> shows the first two cytoplasmic domains; <font color ="red">red</font> invariant disulfide and trigger motif; <font color ="magenta">magenta</font> Schiff lysine end motif. | |||
acrMil 1 HHTISFLYFLLALFSFSLNSVVILT<font color ="blue">FLLDRSLLFPAN</font>LIILSIAISDWLMSVVPNIMGGVANASNDLPFTDWS<font color ="red">C</font>TVFAFVATLLGLSNMLHHAAFAL<font color ="red">DRY</font><font color ="blue">MVITRPMRANH--SMTR</font>ILA 118 | |||
H+T+ + L+ L N VI T<font color ="blue">F RSL PAN</font>+ I+++A+SD+LMS + ++ F + <font color ="red">C</font> +AF L G+S+M+ A AL<font color ="red">DRY</font><font color ="blue">+VITRP+ S R</font> | |||
homSap 70 HYTLGTVILLVGLTGMLGNLTVIYT<font color ="blue">FCRSRSLRTPAN</font>MFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGETG<font color ="red">C</font>EFYAFCGALFGISSMITLTAIAL<font color ="red">DRY</font><font color ="blue">LVITRPLATFGVASKRR</font>AAF 189 | |||
acrMil 119 VIAFLWCFALTWSLFPLVGWSAYVREAGDIA<font color ="red">C</font>SVNWQSDNPSDSSYMVCLFFFFYFVPLAIIVYCYVFMIRSVRFMTKNAQKIWGV-----RSAAALETVQATWKMAKIGLIMVLRFFVAWTPYAVVSFIIAF 244 | |||
V+ +W +AL WSL P GWSAYV E +<font color ="red">C</font>S ++ S P R+Y + L F +F+PLII+ YCY+F+ R++R T A + +G S + +Q+ KMAKI L+++L F ++W PY+ V+ ++AF | |||
homSap 190 VLLGVWLYALAWSLPPFFGWSAYVPEGLLTS<font color ="red">C</font>SWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIR-ETGRALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVA-LVAF | |||
acrMil DSVKDIPT-IAEIVPSMFA<font color ="magenta">KTASVYNPIIYFFSYKSFR</font>ESLVK 288 | |||
+ T VP++ A<font color ="magenta">K ++++NPIIY ++ +R</font> ++ + | |||
homSap AGYAHVLTPYMSSVPAVIA<font color ="magenta">KASAIHNPIIYAITHPKYR</font>VAIAQ 363 | |||
>MEL1_acrMil Acropora millepora (stony coral) 454 transcriptome shotgun assembly EZ013658 + 454 blastn, frag 40%/63% ENCEPHc_nemVec; 35%/57% MEL1_homSap | |||
HHTISFLYFLLALFSFSLNSVVILTFLLDRSLLFPANLIILSIAISDWLMSVVPNIMGGVANASNDLPFTDWSCTVFAFVATLLGLSNMLHHAAFALDRYMVITRPMRANHSMTRILAVIAFLWCFALTWSLFPLVGWSAYVREAGDVACSVNW | |||
QSDNPSDTSYMVCLFFFFYFVPLAIIVYCYVFMIRSVRFMTKNAQKIWGVRSAAALETVQATWKMAKIGLIMVVGFFVAWTPYAVVSFIIAFDSVKDIPTIAEIVPSMFA<font color ="red">KTASVYNPIIYFFSYKSFR</font>ESLVKSWRRYRNRNNVWPL | |||
>ENC1_acrMil Acropora millepora (stony coral) EZ018307 frag 454 transcriptome most like CNOPa1_monFav Montastraea faveolata | |||
IAFHLRKVSKTAKRTWGKNSQITKDGVIVKRKAVTHGAIMVTxVMITWLPYSMVAFYTLLGFEKVKLSALAYTITSMFA<font color ="red">KTSTLVNPVICFFWYRRFR</font>EGTKKLCNRVMDFFLRKHRRASNT | |||
>ENC2_acrMil Acropora millepora (stony coral) EZ007079 EZ005208 frag 454 transcriptome most like ENC_aneVir Anemonia viridis | |||
RAKSTWGRQSPFTKRTFVMERKMMALFGVMTVAFLVAWTPYAVVSLISMIGGPDVISDVTASIPAYFA<font color ="red">KSSSCYNPVIYVFLYKRLR</font>RQMRFAVRRD | |||
>CNOP_acrMil Acropora millepora (stony coral) EZ007080 frag 454 transcriptome | |||
TLVSLTSIFRGNSVLSTGEAEVPALMA<font color ="red">KASVIYNPIVYTVMNRRFR</font>RTLRHIVSCMTCRLLSFVWPTMHREKQETKKR | |||
With the | === <span style="color: #990099;">Anthozoa: Acropora digitifera (stony coral) .. 13 opsins</span> === | ||
An excellent new cnidarian genome assembly (Acropora digitifera) became [http://www.nature.com/nature/journal/vaop/ncurrent/full/nature10249.html available] on 24 July 2011. This species diverged from sea anemone Nematostella vectensis approximately 500 million years ago so naturally the opsin-like proteins are quite diverged and by no means in 1:1 correspondence. | |||
This coral has 7 complete, intronless opsins. Three of these have clear orthologs in the Acropora millepora transcriptome (all fragmentary genes). Coral also has a long open reading frame with an opsin sequence in front apparently fused to something else (not a known domain). This corresponds to a similar long open reading frame in Nematostella. | |||
Acropora digitifera has five additional opsins called CNOP1-5 below. Four of these are full length. All have a phase 21 intron break at the same position. This does not match any of the ancestral introns in position. Nematostella has this intron too though contig misassembly -- homopolymer run error leading to frameshift -- creates a necessity for manual curation. This intron was thus present at the divergence of Nematostella and Acropora. | |||
CNOP2 has an earlier phase 12 intron as well. This matches -- in position and phase -- the first intron of bovine rhodopsin (position 120 (ATLG 12 GEIA) and one found in numerous arthropod ciliary opsins. It thus appears that this intron was already present in an opsin in the last common ancestor of bilaterans and cnidarians. | |||
These are the first introns to be located in cnidarians. They are moderately short (eg 533 bp and 788 bp in CNOP2) and have conventional GT-AG donors and acceptors. Because of extreme divergence, it is unclear whether the much more common intronless cnidarian opsins arose via retrogene processing from them. | |||
CNOP1-5 have conventional K-296 motifs but lack conserved counterparts to the DRY motif. This is not wholly unprecedented in bilateran opsins or in GPCR but does imply unusual functioning (perhaps something other than vision). It should be stressed that CNOP1-5 all give best-blast matches at GenBank to known opsins rather than to other GPCR. Nothing is known experimentally about sites of expression or photobiological anatomical correlates. | |||
Within the opsin collection, CNOP1-5 give best matches either to melanopsins or ciliary opsins, so have no bearing on the origin of the peropsin/neuropsin/rgropsin group. The table below shows that CNOP2 gives consistently better matches to melanopsins than to TMT, though no dramatic scoring shelf separates the two. Branchiostoma opsins again occupy an unusual position. | |||
CNOP2_acrDig Acropora digitifera (stony_coral) BACK01019215 1867 1.4e-195 100% | |||
CNOP4_acrDig Acropora digitifera (stony_coral) BACK01002540 766 6.7e-79 47% | |||
CNOP3_acrDig Acropora digitifera (stony_coral) BACK01018... 733 2.1e-75 45% | |||
CNOP5_acrDig Acropora digitifera (stony_coral) BACK01002513 729 5.5e-75 46% | |||
CNOP2_nemVec Nematostella vectensis ABAV01018948 688 1.2e-70 41% | |||
TMTx_braFlo Branchiostoma floridae (amphioxus) Deut.Cep... 382 3.3e-38 31% | |||
MEL1_otoGar Otolemur garnettii (lemur) Deut.Euth.Euar g... 381 4.2e-38 31% | |||
MEL1_phoSun Phodopus sungorus (hamster) Deut.Euth.Euar ... 380 5.3e-38 30% | |||
TMT_triCas Tribolium castaneum (flour_beetle) Ecdy.Ins... 377 1.1e-37 | |||
MEL1_ponAbe Pongo abelii (orangutan) Deut.Euth.Euar gen... 375 1.8e-37 | |||
MEL1_micMur Microcebus murinus (mouse_lemur) Deut.Euth.... 370 6.1e-37 | |||
MEL1_myoLuc Myotis lucifugus (microbat) Deut.Euth.Laur ... 369 7.8e-37 | |||
MEL1_rheMac Rhesus macaca (rhesus) Deut.Euth.Euar genom... 368 1.0e-36 | |||
MEL1_musMus Mus musculus (mouse) Deut.Euth.Euar AF14778... 367 1.3e-36 | |||
MEL1_canFam Canis familiaris (dog) Deut.Euth.Laur genom... 367 1.3e-36 | |||
TMTa1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc AF34... 366 1.6e-36 | |||
MEL1_homSap Homo sapiens (human) Deut.Euth.Euar NM_0332... 366 1.6e-36 | |||
MEL1_panTro Pan troglodytes (chimpanzee) Deut.Euth.Euar... 366 1.6e-36 | |||
MEL1_ratNor Rattus norvegicus (rat) Deut.Euth.Euar AY07... 365 2.1e-36 | |||
MEL1_proCap Procavia capensis (rock_hyrax) Deut.Euth.Af... 364 2.6e-36 | |||
MEL1_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch ... 364 2.6e-36 | |||
MEL1_pteVam Pteropus vampyrus (macrobat) Deut.Euth.Laur... 362 4.3e-36 | |||
MEL1_gorGor Gorilla gorilla (gorilla) Deut.Euth.Euar ge... 361 5.5e-36 | |||
MEL1_eriEur Erinaceus europaeus (hedgehog) Deut.Euth.La... 360 7.0e-36 | |||
MEL1_felCat Felis catus (cat) Deut.Euth.Laur AY382594 1... 359 9.0e-36 | |||
NEUR_strPur Strongylocentrotus purpuratus (sea_urchin) ... 359 9.0e-36 | |||
MEL1_galGal Gallus gallus (chicken) Deut.Saur.Arch AY88... 359 1.1e-35 27% | |||
MEL1_nanEhr Nannospalax ehrenbergi (molerat) Deut.Euth.... 358 1.1e-35 30% | |||
MEL1_bosTau Bos taurus (cow) Deut.Euth.Laur genomic ful... 358 1.1e-35 29% | |||
ENC4_braFlo Branchiostoma floridae (amphioxus) Deut.Cep... 357 1.5e-35 27% | |||
>MEL1_acrDig Acropora digitifera (stony_coral) BACK01045931 | |||
METASGVPIVIEASVHHTISFLYFLLALFSFSLNSVVILTFLLDRSLLFPANLIILSIAISDWLMSVVPNIMGGVANASNDLPFTDWSCTVFAFVATLLG | |||
LSNMLHHAAFAL<font color ="red">DRY</font>MVITRPMRANHSMKRILAVIAFLWCFALTWSLFPLVGWSAYVREAGDIACSVNWQSDNPSDTSYMVCLFFFFYFVPLAVIVYCYA | |||
FMIKSVRFMTKNAQKIWGVRSAAALETVQATWKMAKIGLIMVVGFFVAWTPYAVVSFIIAFDLVKDIPTVAEIVPSMFA<font color ="red">KTASVYNPIIYFFSFAYCK</font>NQ | |||
SVNKCINQSTVQSKNFKLS* | |||
>MEL2_acrDig Acropora digitifera (stony_coral) BACK01017283 | |||
MPGFQNKSEEPQPTEGISSQDRFIVASFYALLAVTALGLNTPVLITFMKDRKLRVLSNRIILSITIGDWLHAFLAYPVAVFANASHDSQGLTGVVCSWYG | |||
FITVFLSFGIMLHHATFAV<font color ="red">ERA</font>IVIQFATTSLTNAKTINFMVASLWVFALLWSSFPLFGWSAYVPHVVLCSLDWQSRDLRDVVFVYCIFFVFFLVPIVVM | |||
VTSYYKIFQTVKKMTQNARDLWGEKAAPTREAFESQKKTARMACVMSFCFLFAWTPYAAVSLYVFLWKPQSMAPSISIVPALFA<font color ="red">KTSACFNPVIYFLLFR | |||
KFR</font>ESLKQTMCGFFMDPKTGTNETNKNKRHVLKNTESKCACVKINNLSPRDPCNLHQRSDCRSVGNSKSHITVD* | |||
>ENC1_acrDig Acropora digitifera (stony_coral) BACK01046894 | |||
MLTFACGSTHIYNMSAVFYGFLAAGTVFLNVIVILTFFKVRALLIPASFPILSLAMADVFLASAVMPLGIASCASGHWIFGAVGCNWYAFMHTTVGLSSI | |||
LHHAILAL<font color ="red">ERC</font>LTIFEPMKKHFDRHTMTRTLAFLWTLVSLWSLCPILGWSAYVPEGTGTICSIEWHSKNSLDVIFVVSTFVFFCFVPFVFIAVCYTAIAF | |||
HLRKVSKTAKRTWGKNSQITKDGVIVKRKAVTHGAIMVTTVMITWLPYSMVAFYTLLGFEKVKLSALAYAITAMFA<font color ="red">KTSTLVNPIICFFWYRRFR</font>EGTKK | |||
LCNRIMDFILRKHRRASNTSHSSKTVWYDGKTIVFRRSVVDSCSTRTQNDRGRSS* | |||
>ENC2_acrDig Acropora digitifera (stony_coral) BACK01016849 11738 bp INV 28-JUL-2011 DOI:10.1038/nature10249 | |||
MTALIYSVFAIIAAVICVLGVPLNGLVCWVFYNNRELLNAPNIFIASVAFSDFLYCISCLPLLVISNAYGKWIYGPIGCKATAFIATWSGLTSLMNLSVA | |||
SY<font color ="red">ERY</font>STLAFLCTKNRTFAKRTATCYSAAMWLYALFWSLMPLCGWSGFELEGIGTSCSVRWKSKNMLDMSYNLCLIIACYVLPVSVLVTSYYKCYREIAK | |||
STWRAKSTWGRQSPFTKRTFVMERKMMALFGVMTVAFLVAWTPYAVVSLISMIGGPDVISDVTASIPAYFA<font color ="red">KSSSCYNPVIYVFLYKRLR</font>RQMRFAVRRD | |||
NCSSSRKGTSSDSTMAHHSETKRLRSTIELTDNYSTRNRPENI* | |||
>ENC3_acrDig Acropora digitifera (stony_coral) BACK01015014 possibly N-incomplete | |||
MVAPHFAGFAIAASLFGSCGIVLNFTVCLTYILNRRLLDASNIFILNISAGDFLYSITALPMLVTSNALGKWSFGEGGCVAYGFLTTFFALGAMMNLAGA | |||
AY<font color ="red">ERY</font>VTMCKLYESGECQFSRRKAAFLCSVLWTYALIWSIAPIFGWSSYKQEGIGTSCSTDWKSRDVKSLSYGVVLIITCFVVPVAVILYCHIEAYKVTR | |||
KLGKQAQQNWGSHTRATRETLKAQKKMGEIAVVITVGFVVAWTPYTVASVIGMYDPNLVSDVGASIPAYFA<font color ="red">KSSSCYNPFIYLFMYKKLR</font>IGMVRLLCCM | |||
KKQVYPSSSGLTRDPTCQEIPMVVPVNNPS* | |||
>ENC4_acrDig Acropora digitifera (stony_coral) BACK01020962 possibly N-incomplete | |||
MTKTAAWSTVFLIIEFVVCLLGIFFNCLVISIIFKNANRLAAPGFLVLSMALSDVLSCSVAVPFSIVAHFQKKWPFGMAGCEAHAFMVFLFALVSITHLA | |||
AISA<font color ="red">GKY</font>LTISRSLTRESYFDRRQVLLVVIACWLYSLAFSVAPLVGWSRYGLEGTNATCSIKWDSSEPSDKAYFGVVFIACFFLPMGVITLCYYKIHKVS | |||
KRIVENIQGQSIATSAMSRTHALVKKHRRSAMYFLAIVAAFMLAWSPYAIVSLIVVLSGTMDPIATSACSVFA<font color ="red">KTSFLVNPFLYAIFSRSFR</font>RRLALVFP | |||
MPRQNRQEGRQERNCNALPPSQSQPFVL* | |||
>ENC5_acrDig Acropora digitifera (stony_coral) BACK01011015 | |||
MDQIKIWKMLFVVIKFIVSIFGGFFNGLVVLTIRKNLRRLPSSSYLILSIAFSDFIASVVAIPFSIVIHFVGSWPLRLCRAHAFMVFFLGIVTITHLTCF | |||
AV<font color ="red">EKY</font>LTITRSLSKLSFFSKKQTLVVVMACWFYSLCFSLAPLLGWASYGLEGSNDTCSIKWDSSLAKDHIYFILVFLACYLLPIVLITSSYFKILRISKR | |||
ILKATPRVGGIGETMAQALMRKHRRGALYFLSVIAVFLMSWTPYAIISVLVIFKGVNLFPLALSACGVFA<font color ="red">KMSFMLNPIMYFAFSHNFR</font>LLVKRTFCICD | |||
RLDSNSEGKKR* | |||
>CNOP1_acrDig Acropora digitifera (stony_coral) BACK01019215 tandem fragment last exon at beginning of contig | |||
1 LLRRDVVTEDSRIKLRRRHWQMKLLRLTAVAITAFMLSWSPYSLVSLTSIFRGNSVLSTGEAEVPALMA<font color ="red">KASVIYNPIVYTVMNRRFR</font>RTLRHIVSCMTCRLLSFVWPTMHGEKQETKKRVTSTMTVTSSTPAPEGNFPEILVSLHQVPF* | |||
>CNOP2_acrDig Acropora digitifera (stony_coral) BACK01019215 MEL | |||
0 MQFSREEDPPSGFNWNNAWFIAYGVVMIAVLSIGFLGNTMTVLILRKHEHASKSLTPLMINLAIASLIIIVLGYPLVISLVVRGSHVTKEDPTCRWSAFINGTV 1 | |||
2 GISSIATLTEMSL<font color ="red">VIN</font>YSLHRMNPNVRLTKRNMALLIAGAWLYGLVSMFPPLVGWNRFVPGAVRISCGPDWTDKSASGVSYNLVLVVLGFFLPLCVMIKAYYEIYRS 2 | |||
1 LLRSREMLISANGSFQLRQKLYIKKLVRMTVLAIAAFMLSWAPYCFVSIVAIFKGSHIITSGEAEIPELMA<font color ="red">KASVIYNPVVYLITNSSYR</font>ASFWKVISCQKQTMIVHVRENLMPNGPSRRTLRSRRMAVLLKPLNEVSVYNGGYVVAMGFASVRDL* 0 | |||
>CNOP3_acrDig Acropora digitifera (stony_coral) BACK01018578 TMT | |||
0 MLESSNQSAEETILISKASYYAYGVVMFFILTVGFLGNVMTLLVLFQHEHRKKAMTPYMVNIALADIFIILFGYPVAMRANLRGKILESSHCSWGGFVNGAV | |||
GISSIFTLTEMSF<font color ="red">VSY</font>HGLRQVNRSSRFSPFQVVCSVAVAWLYGVLCMLPPFLGWNRFVVSASRISCCPDWSGKSISDAAYNLLLVFFGFVAPLTAMTVCYYKIYR 2 | |||
1 SLVHHAVVPGNLPQIQLRRRQSELKVAKVTAMNVIAFLLSWAPYCCVSLAAVFTKQFVLVDWEAEIPELLA<font color ="red">KASVIYGPIIYSTMHSRFQ</font>ATLFRILHCRRRVILAPQFITDATRNNCSARMATDRTQGTSHIDQRFLQLPLQAMKRRVAVSYGGTAICKRESSAFVSG* 0 | |||
>CNOP4_acrDig Acropora digitifera (stony_coral) BACK01002540 TMT | |||
0 MTTRSDDNPNSARRPQQIAFIVYVVIMTVILIVGFLGNLFTIIVLRCPEHKKKIITPLMMNLAFADIIIIVFGYPVVMASNFTEHNVLTNPMLCVWSGFINGSV | |||
GIASIANLTMMSL<font color ="red">VMF</font>KSLNKVGTVKKIPWKKMLAMILFTWVYGVLSMFPPLVGWNKFVPGASGISCCPDWSPDTKAAVGYNILLVFVGFVLPLTIIIFCYYRMYR 2 | |||
1 FIHTQQPVTGNASIQASRRRSEIKIVRMIALAIAAFVLSWSPYCFVSIAGTIRGSSLLTAGEAEVPDLLA<font color ="red">KASVIYNPIVYTVMNDRFR</font>GTLLRVIPCRRRWFNGDINPTVSVSAPNSQDGNSHNFRERPQSVKGNSTV* 0 | |||
>CNOP5_acrDig Acropora digitifera (stony_coral) BACK01002513 and near identical BACK01007224 | |||
0 MTTSFRSNMTDEWGTHKYFSAGYIAYGVVMFCILSLGFVGNTLTVLVLTKREHRWRGVTPLMINLAVADLLIIIFGYPTQVHANFTGDGVPNDSHCNYRGFIN | |||
GIVGITCIFTLTEMGV<font color ="red">VSY</font>SGLRRVSGNVRLSSYQVACLIGAAWCYGALCMLPPLLGWSKFVISASKLSCSPNWSGQSTADKAYTLLLVTFGFFAPLTVMIISYYKTFW 2 | |||
1 VVRRHFVPGNTAVQLRQRKSMLRIVHVTLMSVISMMLSWAPYCFISLASVVRGKPVIENWEAEIPELLA<font color ="red">KASVVYNPLIYTFMNKSFR</font>MTLFRIVGLHRCFGAGHEVAPTAAIQKRPNVLQIQFHRHSSEVEVRTPRVPPIE* 0 | |||
>MEL3_acrDig Acropora digitifera (stony_coral) BACK01025991 resembles long intronless Nematostella FAA00396 | |||
VFSITSTLHGRWLFGRAGCVTYGFFHYFLICCTVSTLAAISV<font color ="red">DRF</font>FFITRPSQARTAQMITPRKAVAMLAIIYFYTLMFTFPPCIGWNNFEVEESFYSGC | |||
YINYADQRASSIAYSIIAPFFLFLVPLTIMIFCYARIFAVVRRSTQRTINRPFGAQSLKRYPLLKRTHIQTAKIIVVVILFSMIVWVPYVVVSFVKAANG | |||
KVNPLLSHITVLVA<font color ="red">KSCVVYNVLIYAVLNRKLK</font>ASILDVICCGKQNFRMVGSPSSGNSLQRNRNSRFLSQREVPSDIAESLKNRASTLSVQEGNGNVHDQ | |||
YTDNISPSQLTQLRNGVLKSIDHVDGRESGKVVGKKEVDVVRNGDKVQVGESSGFSETRMVIATQERDSKILGPSDARSSDRGTNRFLARLETDTLPEIA | |||
PQDTHLKPEAVSNASSLSRNETPRVDCKVDTPLCNSSSSNRQLNRLHHMYSSKSSRHARLQNLSSPSQLYKKGARPASAQRVTGHIASDSRTSRAGVSSG | |||
TESDVSTRASHRPATAQRPKKRRAADRTNSTHTSERRKSSSYINYGRKDSVDPLRTSPKELWEIQNYWKRMSLCLDDIDVDEVTREIQL* | |||
>MEL3_nemVec ABAV01020362 'Serpentine type 7TM GPCR chemoreceptor Srsx' | |||
MSATTTAANISTDVYQTSRQVSVAYLLIVGILGTLGNSLVVFVLWKYPKLRTATNLFILNLAFCDLVMALLDLSLSVASALSNRWLWGKSGCLGYGFLHY | |||
FFISNSVSTLAAISV<font color ="red">DRF</font>FYITKPGQIKAWRITRTRAGIILIIIYIYVLLFTFPPIAGWSSFTEEKFYFSGCFIKYSDQNPASMAYSVIASVFLYLMPLI | |||
LMICCYSKIFMAVRKSTRRTMTNSNAVRKKFPIMKRTHIQTAKMIVVVIFFSMIVWVPYVAVSLIQAFTADSIITPTASHITVLVA<font color ="red">KSCVIYNVLIYVVL | |||
NRKLK</font>AVILNLLCCGRQINFSGSNMPTSNLRISRILSETDREIAPDAVRNRLNVLQNNDNASAVDASVERSSPIPTHTTFKPDLIETPKGTKQRHGNVKT | |||
VEFKVVEQDGNVITLVNDAANGTGSGGNLPKIRRKKLPQIPRDNQETNGNLVGQFGDDLNSVGVSNKDFSSCEETSPAKTEKLLLTETVTRAEARRRALR | |||
KLNDRKVSNSVPSIEKRDSTSSGATVATGNESHDRSISPSTPNGRILPEIPNHVGDKHEKKRNGNAKTNEADRIGADSSSTAIPSSCTSTLSNHSRAWSE | |||
KDTGNPAAPARSMTSPRRNGRRKTRKHMDRNASLRARLQRCSVDVLQASPSELHEIQNYWKRMSLCLDDIDLDEMV* | |||
>CNOP4_nemVec Nematostella vectensis ABAV01018948 | |||
0 MSTANSPWGSEGLVAYGVVMSLILTTGLTGNILTIIVLMRREHRSKVITPFMINVAVLDLIIIALGYPVAIIANFTGIRMKEGDSRCQWTAFLNGAT | |||
GIAAIITLTAMSI<font color ="red">VMR</font>NTITHPIVKKRLSCKYLFGSLCAIWTYGILTMLPPLVGWSKFVPGAAKVSCGPDWETQNASTLSYNIVLLIVGFVIPVGIILPCYFHIFR 2 | |||
1 FLRSQPVPPAFISITSRRRRAHLRIARMILVSIISFVISWSPYCVVSVIAMVQRAPALSQGFAEIPELMA<font color ="red">KASVVYNPLVFTVMNRGFR</font>RSLCLIVPCLGCSVGHGISGRDGLYIWLGMKDLTDRSSVISNVAVKKR* 0 | |||
=== <span style="color: #990099;">Anthozoa: Anemonia viridis (symbiotic anemone)) .. 1 opsin</span> === | |||
This ciliary opsin fragment is associated with an October 2009 [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=19627569 metagenomic article] on the symbiotic coral reef organism Anemonia viridis and its dinoflagellate algae but was not annotated in the EST collection ([http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=Nucleotide&list_uids=219278581&dopt=GenBank&RID=EYFU57YH015 FK729339]). | |||
It is most closely allied with ENCEPHb_nemVec of Nematostella vectensis in the currently known set of cnidarian opsins -- about 49% identity but not enough is known about Anemonia opsin multiplicity to say it is an ortholog. High identity might be due either to sequence conservation or close relatedness of these two genera -- their time since divergence is an unknown. Conservation (of sequence and hence function) seems more likely since -- at least at GenBank taxonomy -- the divergence is quite deep: | |||
Cnidaria; Anthozoa; Hexacorallia; Actiniaria; Nynantheae; Actiniidae; Anemonia | |||
Cnidaria; Anthozoa; Hexacorallia; Actiniaria; Edwardsiidae; Nematostella | |||
Cnidaria; Anthozoa; Hexacorallia; Scleractinia; Astrocoeniina; Acroporidae; Acropora | |||
>ENCEPH_aneVir Anemonia viridis (symbiotic sea anemone) frag:202-338 pubmed:19627569 | |||
KDAVARWGNKSPPTQQTMQAQKKTIRMSLVMVFAYLLAWTPYALTSLYSSFIASDITPLLSVMPALFAKLSSCYNPIIYFFMYSKFR | |||
KAAKKMIRRNLVGHDSNSGQGVSNTFATSFPRPISFLRYKRSAVAPLSDIPQVSSVDLPQVGRENDVTVQQDKASEINT* | |||
=== <span style="color: #990099;">Hydrozoa: Hydra magnipapillata (hydra) .. 41 opsin-like genes</span> === | |||
Because opsin photoreception is quite ancient, clearly pre-Bilaterans have a major role to play in illuminating the origins of photoreception systems. What's not so clear is that the two cnidarians chosen so far for genome projects are optimal in this regard. The excellent [http://www.nature.com/nature/journal/v464/n7288/full/nature08830.html Hydra genome article] of 25 Mar 10 does not specifically address opsins but a 10 Mar 10 [http://www.ncbi.nlm.nih.gov/pubmed/20219739 article] considers them in a signalling context. | |||
Hydra does not have overt photoreceptive structures or cells obviously specialized for light detection yet it exhibits marked behavioral photosensitivity (noted by Trembley in 1744). [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16843587,11318381,10942072,17940617 Studies] beginning in 2000 flagged the ectoderm (using antibodies to squid rhodopsin), known to contain epidermal sensory neurons, as responsible for extraocular photoreception. [http://www.ncbi.nlm.nih.gov/pubmed/16843587,11318381,10942072 Musio and coworkers] sought to recover opsins using degenerate primers, targeting melanopsin and peropsin as the most plausible in Hydra because the latter opsin seems not to require advanced relationships with neighboring cells or auxiliary enzymes (ie, acts as photosensor and its own photoisomerase). | |||
The Hydra cdna CB073527 was proposed as a peropsin based on best-blast to mouse peropsin. However using a much larger collection of demonstrably orthologous chordate peropsins in the [[Opsin_evolution|Opsin Classifier]] conflicts with this interpretation: the putative cnidarian gene needs to consistently associate with this gene family (equivalently, have best match to it among all reconstructed ancestral opsins) but does not. Furthermore the best match is very weak at 31%. This is the signature of generic non-opsin rhodopsin superfamily members (which we expect any eumetazoan to have by the hundreds). | |||
With the availability of the diploid Hydra genome assembly (ACZU vs ABRM accessions), the 161 amino acids of the fragmentary transcript can be extended for example with trace 1121878952 to apparent full length (309 aa) and its introns determined (none). This does not improve its best-blast score nor family coherence. It does not cluster consistently with ciliary opsins -- what would the signaling partner be when the matches are scattered between Gt, Go, and Gq opsins? The blast probability of 1.6e-34 does not mean much under these circumstances. | |||
[[Image:Opsin_hydra_doubtful.png|left|]] | [[Image:Opsin_hydra_doubtful.png|left|]] | ||
Two putative hydra opsin fragments can be extracted from Fig.1 of an [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17940617#pone.0001054.s001 Oct 2007 paper], AKSSTIINPTISCIIYKE and AKLSAVLNALVNCYINKS. These too fail to extend to | Two putative hydra opsin fragments can be extracted from Fig.1 of an [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17940617#pone.0001054.s001 Oct 2007 paper], AKSSTIINPTISCIIYKE and AKLSAVLNALVNCYINKS (reclassified below as CNOPa5 and CNOPe4). These too fail to extend to convincing opsins. Expression centers around the hydropore, a better fit to GPCR chemoreceptor localization. There is meagre behavioral evidence for photoreception near the hydropore and no possibility of a pigment-backed eye. An ultrastructure study is needed to demonstrate that the putative opsin is expressed in specialized photoreceptor cells. It is critical to include non-opsin GPCR as alignment controls -- all GPCR proteins bind heteromeric G proteins and many have lysine without binding of retinal. The cdna accession numbers are Hydra1 CN554949 and Hydra2 CV151648. | ||
These papers highlight the special difficulties in working with cnidarian opsin candidates. We know from the outset that they will be quite diverged. Multiple forms of supporting data are needed, preferably in the form of diagnostic introns, alignments demonstrating conservation of critical residues and structures, in situ hybridization to anatomically plausible neuronal photoreceptors, and specific loss of photobehavior upon knockdown. | These papers highlight the special difficulties in working with cnidarian opsin candidates. We know from the outset that they will be quite diverged from bilateran opsins. Multiple forms of supporting data are needed, preferably in the form of diagnostic introns, alignments demonstrating conservation of critical residues and structures, in situ hybridization to anatomically plausible neuronal photoreceptors, and specific loss of photobehavior upon knockdown. | ||
A higher standard of proof is needed for the first cnidarian opsins because validated ones will surely be used to pull in further homologs via annotation transfer. There is a definite risk in admitting inadequately documented opsins to the Opsin Classifier because once that database is tainted, it could draw in even more non-opsins from the GPCR world. | A higher standard of proof is needed for the first cnidarian opsins because validated ones will surely be used to pull in further homologs via annotation transfer. There is a definite risk in admitting inadequately documented opsins to the Opsin Classifier because once that database is tainted, it could draw in even more non-opsins from the GPCR world. | ||
[http://cnidbase.bu.edu/ CnidBase] provides a blast service to cnidarians including hydra but this appears restricted to ESTs and so only duplicates GenBank. GenBank | [http://cnidbase.bu.edu/ CnidBase] provides a blast service to cnidarians including hydra but this appears restricted to ESTs and so only duplicates GenBank. GenBank carries contigs of the genome assembly on 25 May 2009 in the wgs division. Some 10.2 million Hydra traces have been provided by JCVI, ample for the 1290 Mbp estimated genome size. However the hydra genome project was dropped from that website. The draft genome [http://polyp.biochem.uci.edu/wiki/index.php/Resources expected in Dec 2005] was delayed because of high AT content (71%) and lack of an inbred strain. A [http://hydrazome.metazome.net/search.php?show=blast&db=hydra_072606 'hydrazome' blast server] and [http://hydrazome.metazome.net/cgi-bin/gbrowse/hydra/ funky browser for assembly v2.0.4] has surfaced along with Gnomon gene predictions at NCBI nr. | ||
The best current opsin search strategy simply uses tblastn of the usual three NCBI (nr, est_others and wgs) restricted to Hydra. After genes are recovered, their best-blast to all of nr must be considered. The outcome first lists other cnidarian 'opsins' and then mixed bona fide bilateran opsins. That's encouraging because generic GPCR come later but does not prove by any means that any of the Hydra genes are true opsins, even those with lysine in Schiff base position and having other signature residues. | |||
The basic problem is Hydra has '''far too many''' opsin-like genes (41 genes, all K296) for its complete lack of anatomical photoreceptive structures and minimalist phototactic behavior. It is not plausible that such a simple organism could have a larger opsin repertoire than an amniote with four color imaging vision and numerous pineal and deep brain photoreception. All the Hydra genes are intronless, suggesting they have as arisen, much like the olfactory gene expansion in mammals, as processed retrogenes. | |||
If not opsins, if their function is chemoreception, why then is the Schiff lysine conserved? Note first that it is not currently known whether a retinal species occupies any of the Schiff base sites nor whether any cis-trans photoisomerization with ligand release takes place. These opsin-like genes have retained many key residues for signaling so very likely follow standard GPCR mechanisms (though with unknown Galpha partnering) and are certainly not pseudogenes. | |||
Among the many possible scenarios: the opsin-like genes have other primary agonists. The genes were perhaps derived long ago from an opsin and retain the Schiff lysine and (undetermined) counterion through evolutionary inertia. That is, mutational loss of the lysine + counterion is a two-step process. Initial loss of either leaves a defective salt bridge in the extreme hydrophilic milieu -- negative selection would inactivate the gene before the second component could be favorably mutated. Here though it needs to be recognized that generic GPCR do not have these charged internal residues (ie transitioning is possible since all GPCR presumably coalesce to a single ancestral gene). This argues for non-retinal agonist binding -- after all, what is so special about the terminal aldehyde in a retinal? Indeed assimilation of beta-carotene and its variants generates a large number of them. | |||
> | |||
[[Image:HydraOpBlast.jpg]] | |||
The 41 Hydra opsin-like intronless genes with disulfide, questionable DRY motif, Schiff lysine at 296 and weak FR switch motif. Of these only CNOPa1, CNOPa5 and CNOPc1 have (unlocalized) transcripts. These genes cluster into discrete blocks by blastp as shown above; this classification agrees with the alignment-derived tree and classification by indels and signature residues. | |||
>CNOPa1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01000679 KSSTILNPIIYCLMYKKFR 37% CB073527 CB271253 CN554455 CN554795 no tissue 65% single exon | |||
MAFVFIIVFLSFLCGFSVILNVTVVLTILAKGNTKNTRDVILMSLAICDGVQCTIGYPVELFGYANYKNPSLSEKFCKPSGFIVMYLALTAIAHLVCLCIYRYLTIVYPLKLQIFLTKSNWSACGC | |||
IAFcWIYGLFWSLSPLLGWNEIVREKQDTYRCSINLYPDNEIKSSYLYALAIFCYLIPLIIIIYCSLKVRSELRNMLKMCKQISGVEANITKVTYRIEKQDFISVSFIIASFFTVWTPYA | |||
VCVFYLTIGKKLPPSFLTYCALFAKSSTILNPIIYCLMYKKFRQTLQSKFGKLFNNPTVTPAV* | |||
>CNOPa2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01004988 KSSTILNPIIYCIMYKEFR single exon XM_002163173 | |||
MAAPVIGLLSFLCGLSVSLNVTVVVTIISKKNTKDMRDIIIMSLAICDGVECTLGYPVELYGYANIDSPSQNEYLCKTNGFIVMYLALTALTHLVFLCVHRYLVIVHPMKVQKFFTDTKTCALYFIIPSWI | |||
YGLFWSITPLIGWSEIVREKEDTHRCTINMYPDDLLKRSYLYALTLFCYFLPAAIIIYCITKVHFELRNMLKLCKQISGEDAAITRATYKLERQNFLSVSLIITSFFLIWTPYTICVCYL | |||
ILGRELPVGILTYSALFAKSSTILNPIIYCIMYKEFRQTVRSKVRKLFRGPTVAPITE* | |||
>CNOPa3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01055709 KSSTILNPVIYCLMYKEYR single exon | |||
MASVIILLSFLCGFSVVLNATVVLAIFLKRKSKGMRDIILMSLAICDGVQCTLGFPVELYGFSNLNSPLQNEYLCKANGFIVMYLALTAISHL | |||
VCLCVYRFLSIVYPLKIQKFFLDSRREALLFILFCWIYGFFWSVTPLLGWNEIIREKEDTHRCSINLNPEDNNKRSYLYSLMVFCYFIPLVIIIFCSLRVHFELSKMLKLCKHISGREAS | |||
ITKETYKLERQDFISISLIVSSFFIVWTPYTICVIFSSLRSSLPTGLLTYSALFAKSSTILNPVIYCLMYKEYRETLQSEYRKIFKSSVVAPFTERSQTAALSTLSSHGDASFT* | |||
>CNOPa4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01004994 KSSTIINPIVYCIVYKEFR single exon XM_002163322 | |||
MALVSIIVLLSFFCGLSFVLNATVIFVVLSKSSIDARETILMSLAVCDGLQSIIGFPNEIYGYVNKDNPFKSESLCKTSGFVVMCLAITAISHLVSLCIYRYAAIVYPMETQVFV | |||
TSPIKVTLYLVLPNWIYGLFWSITPFLGWNEIVREKQDTYRCSINLYPNDQIKQSYLYALTIFCYFLPLSIIIYCIIKVHLELRCMLKLGKQISGADASITKATYKLEKEHFISVFLIVV | |||
SFFTVWTPYFICVCFLVYGRSLPSGFLTYCALFAKSSTIINPIVYCIVYKEFRQTAISKVRKLLRRPAVAPMLVSQLSTAAAPTLALSGL* | |||
>CNOPa5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01004993 KSSTIINPTISCIIYKEYF single exon CN554602 CN554949 XM_002163291 3 glitches | |||
MASLFIIVLLSCLCGLSVTLNVTAVIAIISTKKNKDVCDVILMSLAVSDGVECIFGFSVELYGYATKGKTLQNETLCKINGFIVMYLALTSISHLVCLCLYRYILIVHSLKAQRYLTNVKQSALYFIIPSW | |||
IYGLFWSIAAISGWNEIIREKVDTHRCTINMSPDDELKRSYLYSLTVFCFLVPVVIIIYCSLKVHLKLQNMWKLCVQISGEYAAITKATYKLERKHFIFLGLIIGSFFVVWTPYALCVFF | |||
LALQIKLPRVLLTYSALFAKSSTIINPTISCIIYKEYFQILRIKVQKLFRNNIVSPANL* | |||
>CNOPa6_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01000199 KLSTIFDPIIYCLVYKNFR 9 glitches | |||
MISVIVLSSILCGCSVVLNLIVILTVSKKRSKDLRHLILMSLALCEGMSITVWYLNYVYSYFHKHEPLTKKLCGISGFIELYLTITAVSHLVWLCIYRFISISHPLKIHYFFVDTKRRVLYF | |||
ILPCWIYGLFWSSAPLFGWIKLGEENSSYYRCTVIYYPDEFLKRSYLFVFIVFFYFMPLSAISFFTLKVHLELRSMLKQCKKVSGNNALITKDTYKLARQDFINACLIVASFAVVWSPYA | |||
LCVFALSLGYKKLKGFLFFCALFSKLSTIFDPIIYCLVYKNFRETLRRDIKTIFKVAAVAPQ* | |||
>CNOPb1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01027146 KLSTVSNVLVNCFINKSFK XM_002160412 | |||
MTAYDVAYKVFLSLILVLSVMLNTTACYTILIKVKKKELAHLFILSISFTNLMESIIGVLPHVITSGEILSGSNSLCIASGFAVFGFAITSISHLCIHSLNRTVAIIYPIYYFKYRKRFWYRSALISLC | |||
YAYGFMWASFPAIGWSKYNLDVDKKRCSLDWKLTRSNSLSYILSVLIFCNIFPGILIVIGLFRSKKVIYRRRTRSLGQKDDNQTDFLEKHYLKVFSLSSAMFFVTWTPYALVSILALLKI | |||
APPPLLVTTSALLSKLSTVSNVLVNCFINKSFKKHVFSIRFIQDITKKGSELRKTAYVIFLYKD* | |||
>CNOPb2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01051946 KLSAVSNVLVNCFINKSFI | |||
MEPYEVAYKVFLSLILTLSVVLNTTAFYTISIKVKKKELAHLFILSISFTNLLESIIGLLPHIITSGEILSGSNSLCIASGFAVFGFATTSITHLCVHSLSRTVAIK | |||
YPIYYFKNRKRFWYRTALISVCYVYGFIWATFPAISWSKYDLDLDKRRCSLDWNLTRSDSLSYILSVLIFCNIFPGILITIGLFKSKKVIYRRRTRNLGQKDDNQPDIFEKHYLKVFSLS | |||
SVLFFVAWTPYAVVSILALLKIAPPPLLVTTAALFSKLSAVSNVLVNCFINKSFIKHLFSIRVIQVVAKRGTELKIIGHAAFLHKEKSLSY* | |||
>CNOPb3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01043783 KLSTISNVLINCFINKSFQ | |||
MNKSQITSSIFLSLILIVSVVLNTTASYIMLFKVKKKELMHLFIISLSTTNLLESIVGITPQIIFSEQSLLRKTPLCIFSGFAVFGFAVTSITHMS | |||
AFSLIRTVIVKYPIYYFQRYKAFHYKAALILSCYVYGFSWATFPFIGWSKYEVDLDKKRCSLDWKLSQSESPSFFLAILIFCNILPGTVIALGLYFSTKIIHRRKACKTRQDKNKTLDIL | |||
EMEYLKVNFLSAILYFVIWTPYAIVSILTLLKITVPAYLVTVSALFSKLSTISNVLINCFINKSFQKHLLNLRLIRFLANTNFKRRLKNHSSTSCHFFKLEQK* | |||
>CNOPb4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01065240 KLSTISNVLINCFINRYFQ | |||
MTCYRITSSIFLSLILIVSVVLNTTASYIMLFKVKKVELTHLFIVSLSITNLLESIIGLIPQIIISDESVMRKTPMCIVSGFAVFGFAVTSITHMSLFSLI | |||
RTIIVKYPFFYFQRCKAFYYKVALILSCYVYGFSWATFPFIGWSKYEIDLDKKRCSLDWKLSQPESRSFFLAVLIFCNILPGIVIALGLYYSTKIIHRRKACKFRQDKKKRLVDILEIEY | |||
LKVNFLSAAIYFVIWTPYAVVSILTLLKITVPAHVVTVCALFSKLSTISNVLINCFINRYFQKHLLNLGLIKFLANSTFKRRLKNHPSTIFFKLERK* | |||
>CNOPb5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01078635 KISTISNVLTNCFINTSFR | |||
MTSYEIAANIFYSTVLVVSVILNTTACYIILFKVKKKELTHIFIISLSVTNLLESVIGLIPQIIISSEAKLEKTPLCITGSFAVFGFAVTSITHLSIFSLVRTVAIKYPIYYFKNSKRFWCRAT | |||
LILICYCYGFSWAIFPVIGWSKYEIDLDKKRCSLDWRMTRPDSLSYILSILIFCNFLPGTLIAIGLYISKKVIYRRRTCKLRKNENNRSIDLLEKDYLRVCFLSSILFFAIWTPYAVVGI | |||
LALSKIVPPPLLVTTAAMFSKISTISNVLTNCFINTSFRKHLLNLKIVRVFLNKKIESPRIEDV* | |||
>CNOPc1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01005587 CX830752 KLSTITNVIINCFIVKSFK | |||
MTNNNSVLSSFFFIILLTSVMLNATACYIILLKVKKKEITHLFIISISLINMLESIVGFIPDILTSDGSLINRTPLCISSGFTVFGCAITSITHITVLSFIRTLVIKYPFFYHKTCKKMWVKATLIAMCYLYGFSWATF | |||
PLIGWSKYELDLDKKRCSLDWKLTQSDSASYFLTILIFCNLLPAIVISTTLYTSRKIISKRNAREDCQNHQASLDILENDYLNVCLLSTGTFFFIWTPYAVIGFLTILKIVIPTQLVTAS | |||
AIFAKLSTITNVIINCFIVKSFKNQLLELRLIQYIRNINNKNSAVHPCDDSKINYS* | |||
>CNOPc2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01005585 KLSTITNVVINCFIVKSFK | |||
MALNLFLFTILFTSVILNATACYIILLKIKKKEITHLFITSISLTNMLESIVGFIPQITTSENFLLERTPLCISSGFAVFGCAITSITHITALSFMRTIVMKYPFFYYKTCKKMWVKATLIAMCYFYGFSWAT | |||
FPLIGWSKYELDLDKKRCSLDWKLTQSDSASYFLTILIFCNLLPAIVISTTLYTSRKIISKRNAREDCQNHQASLDILENDYLNVCLLSTATFFFIWTPYAVIGVLTILKIVIPTQLVTA | |||
GAMFAKLSTITNVVINCFIVKSFKNQLLELRLIQYIRNINKKISAVHPYHDSTLN* | |||
>CNOPc3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01005579 KLSTITNVIINCFIIESFK | |||
MINNYSVSNVLLSTILLISVILNATACYIILLKVKKKEITHLFIASISFTNMLESMVGFIPQIVTSDKILLERTPLCILSGFVVFGCAVTSITHITILSSIRTAVI | |||
KYPFFYYETSKKVWFKTTLITICYIYGFSWATFPLIGWSKYEVDVDKKRCSLDWKLTKSNSLSYFAAVLVFCNIMPCLVISTTLYTSRKLISERKARKASQSQSHQVSLDILENDYLKVC | |||
LLSAIAFFITWAPYAVIGILTVLKIVIPTQLVTAAALFAKfSTITNVIINCFIIESFKNQVFDLRFIKYIRSINKITSKVQPQTINYNC* | |||
>CNOPd1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01091217 KLSTITNTLINCFIIKSFR | |||
MTNKLLKVFLFTILFTSVVLNGTAIYILLFKVKKKEITHLFITSISLVNLLESLVEFIPLLLLVYSDASLIERTSLCIASSFVVFGFAMSSITQQSMLSLI | |||
KIIVIKYPIFYHKTFKKMWIKAALITICYLYGFCWATFPLIGWSKYQLDIDKMRCSLDWNLTHPGSFFYFLTITIFCYLLPVIVISSTFYTGNKIIYGRKVSIARQNKNNQYINILEKDY | |||
LKVSFLSVTTFLIIWTPYLVTGILATLKIVIPKQLVTTAAMFSKLSTITNTLINCFIIKSFRNHILDLKIVQCIRNKKNAAKIKPQPYYINCNYLRQT* | |||
>CNOPd2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01027487 KLSTISNVLINCYVIKSFR | |||
MNTNKLVQSVLSTVLVTSVVLNMTACYVILVKVKRKEITHLFVVSISMTNLLETMIGLTPQILIADQSLLEKTPLCIASGFIVLGFAITNISHLAMLSFI | |||
RTFAIKYPMRYLQLHKMFWCKVASILVCYANGFFWAILPLLGWSKYELDLDKKRCSLDWSLTKANSFSYILTIFIFCNILPGIIIVLTLYFSKKTVFRRKTCKDCQNRETNILEKEYLRV | |||
CFLSATAYFLFWTTYAIVSVLTLLKVFVPVELTTTTALVAKLSTISNVLINCYVIKSFRKQLLNLMLIRFIKKYLWVLIACKKSSVNDDIN* | |||
>CNOPd3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01075622 KLSTISNVLINCYAIKSFR | |||
MSSIKVIQSVISTVLLTSIILNMTACYIILFKVKKKEITHLFIVSISITNLLETTIGLTPQLMMANEFLLERTPLCIVSGFSVLGFAITNITHLTILSFIRTVAIKYPLQFLRYNKMFWCR | |||
ATLIFVCYANGFFWAILPIIGWSKYELDLDKQRCSLDWRLTKLNSFSYILSIFICCNILPGIVIVLMLYFSKKTIYRRKGRKICQSRETGFLEKEYLRVCLLSTVTFFFFWSFYALFSLL | |||
TLMKFAIPTNLATTSALFAKLSTISNVLINCYAIKSFRKQLLDICVIRFMKRCCWVLIACQKSSVNDQIN* | |||
>CNOPd4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01076129 KLSTISNVLVNCYAIKSFQ | |||
MNTNKVIQTVISTVLLTSVVLNMTACYIILVKVKRKEITHLFVVSISITNLLETTIGLTPQLAMADESLLESTPLCIVGGFAVLGFAITNITHLTMLSFIRTVAIKYPRRYFQYHKMFWCKVTLILVCYAYGFFW | |||
ATLPIIGWSKYELDLDKKRCSLDWKLTKANSFSYILAIFICCNILPGIVIALTLYYSAKEIRHRKACKFPQNKKTDHLEKEYFQVCLLSAFAYFFFWTSYAIVGVLTLLKFNIPMHLATT | |||
TAMFSKLSTISNVLVNCYAIKSFQKQLLNLKIVQIMKSYLRILIVCKKV* | |||
>CNOPd5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01038272 KLSTISNVVVNCFVLKSFR | |||
MNIEKIVSKVYLSAVLVTSLALNITAFYITLLKVKKKELTHFFIVSISITNLLESTVGIIPQLVISNDSLLERTPLCISSGFAVLGFAITNITHLAVLSFIRTVAIKYPML | |||
YLKYHKMIWCRVTLILVCYAYGFVWATLPLIGWSKYDLDLDKKRCSLDWKLTKSNSFSYILAIYICCNILPGIVVALTLYFSKKTIYRRQSCKLRkNFKKGRLEREYLKICYLSAiAYFF | |||
FWTTYAVIGALTLLQIQISPILATFTALITKLSTISNVVVNCFVLKSFRNHFINLKFIRIIKKYLEVLKVKKHSKLDHTV* | |||
>CNOPd6_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01049360 KLSTISNVLINCFINKPFR WRHF removed | |||
MLNNKRASAVLLSTIFIVSMILNTIACYIIVYKVKRKKLTHWVVISISIANLLETIIGLLPEIVMSDKPLLKKTPLCVTGSYAVFGLAITSITHLLVLSLIRTAAIKYPIFYLKISSTVWCK | |||
ATLIIFCYSYGFSWATFPVLGWSKYEIDLNKKHCSLDWKLTQSNSLSYILTILIFCSIFPGLVITCASYICKKAITLQKTSYIRNKNSRPMYTLKKEHLKVCFLSALLFFVTWTPYTVVG | |||
VLTLFRIIIPTELVTIASLFSKLSTISNVLINCFINKPFRKHLLNLRLVRLVINK* | |||
>CNOPd7_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01040368 KLSTISNVLINCFINKSFQ | |||
MAASETANIFLSTIILASVVLNTTACYIILFKVKKKKLTHLFIISISVANLIESIVGLTPQLFISNELILVKTPLCVINSFAIMGLAITSITHISVLSLIRIVVIKYPIFYYKTCKKMWCKVI | |||
LISICYIYGFGWATVPLIGWSKYVLDLDKMRFSLDWKLTQADSLSFILTLLVFCNILPGAVIALTTYIGTKTISKQKSNEVSQTEKEKPVDILEKYYLRVFTLSALMFFVIWTPYAIGILTLSK | |||
FVIPTHFVTVAALFAKLSTISNVLINCFINKSFQNQLLSLGIIQAFINKSKRSPVFPEK* | |||
>CNOPe1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01068872 KLSAISNAMTNCYFNKYFR | |||
MDVLVSAYFVCVLVIIILSILLNALLIYIIVYKIKVTKLPHVFILSISISDVIHTLIGYTAEVWVLFEVYSLKIRFVCIGASFLTVFTSVSNILQTVITSIMRAVAIKWPFFYIIYCKTM | |||
KMKMFLLLFCYFYGFLWPFIPLIGWSKYELDLDKMRCSFDWKLKHSDSLSYLIVLFLFCYFLPVIAMIFAYVAIKKTVFNNCTIKRAQVGKNRNIQEMVYLKLAFWSAIFYFIIWTPYAG | |||
VSLLSVFKIKSPSVIYTFCALFSKLSAISNAMTNCYFNKYFRNHLKKIKLFQYFIKRNRRSSDNVML* | |||
>CNOPe2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01085554 KLSSISNALTNCYFNKYFR | |||
MDVLVSAYFVCVLVIIILSILLNALLIYIIVYKIKVTKLPHVFILSISISDVIHTLIGYIAEVWVLFEVHSLKKSFVCIGPSFITVFTSVSNILQTVTISIMRAIAIKWPFFYIIYCKTMKMKILLLFFCYFYGF | |||
FWPFIPLIGWSKYELDLDELRCSFDWKLMHSDSLSYLIVLLIFCYILPVIALIFAYITLKKTVVDSSTVKRAQLGNNRNIQEMVYLKLAFWSAIFYFIVWTPYASASLLSIFKIKSPNVI | |||
YTLCALFSKLSSISNALTNCYFNKYFRNHLKKIKLFQYFVKWNRRSCDNLIL* | |||
>CNOPe3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01057385 KSSAISNALVNCYMNKSFQ | |||
MELLVAVYSLYVLALIILSTTLNLIICYIILSKIKAIEMPHLFILSISISDILHTSIGLVSELCVLHGATSLKKSYTCIGASFLTYSFTVSNIMQMVMISIVRVIALKFPI | |||
FYFNNCKKMKFRLGCLFLCYFYGFLWPSFPLLGWSTYEEDLDKRRCSLDWNLTNSNSFSYLMFAFIFCYIFPAIVLIWSLNVTKKTVDEQRESSFRKNRPNEQMEILEKVYLKIFLCSAI | |||
AYFVIWTPYACATLLSIFRIKTPSVVFTFCALFAKSSAISNALVNCYMNKSFQVHLNEIRVFSCFIKENRVAPRITNASVTEETQA* | |||
>CNOPe4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01037497 CV151648 | |||
MVLVALYTTYIVVLIALSTSLNLIICYVIIRKIKAIEIPHLFILSISISDIVHTVVGLVSEVCVLHKVTSLKNSYISVGASFLTYSISVSNIMQIVMISAIRVVALKRPMFYFKNCDKVKYKMASLCCCYFYGILWASFPLLGWSKYEEDLDKKRCSLD | |||
WNLTKSNTFSYLMFAFIFCYKLPTTLLIWALRVKHQTVDEHRRFRENRPNQQIEILEKAYLKVLLCSAIAYFIVWTPYSVVSLLSIFKVKIPSIIFTFCALFAKLSAVLNALVNCYINKSFQAHLHDLRIFQCFLKGKRIEHKNISWAEETQVAETGFT* | |||
>CNOPe5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01037501 KLTTVLNALVNCYFNKSFQ | |||
MEKALYTIYILLLIIISTLLNLFICYVIIYKIKTFEITHLFIISISISDIVHAIIGLVSEIYVLQNFTIFDKSYICFEASFLTYSISVSNIVHTVIISVIRLIALKWPIYYFNNCKKLRYK | |||
MGSLFFCYFYGLLWALFPLLGWSKYEKDLDGMRCSLDWNLTKYNSFTYLMFTFIFCYILPAIFLFWSLKVVHQTVDCHRSFRENYPNQLIEILEKSYLNVLICSAVAYFVVWTPYSVVSL | |||
LSIFQIKFPGFVITLCALFAKLTTVLNALVNCYFNKSFQVHLYKLKIFEYFLKGNKIAHNSNSITGETQA* | |||
>CNOPe6_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01013912 KLSAVSNALMNCYFNKYFR | |||
MDVLISAYSACVLAIIILSTLLNALLIYIIIYKIKVTKLPHVFILSISISDVIHTLIGYTAEVWVLHEVYSLKKSFICIGASFLMVFITVSNILQTVIISIMKVIAIRWPFFYIIY | |||
CKTIKMKILLLFFCYFYGFLWPFIPLFGWSKYELDLDKMRCSFDWKLIHSGSLSYLIVLLSFCYILPIVALVFAYIAIKKTVSDRCTVKRAQLGKNRGIQEMFYLKLALWSAILYFIIWT | |||
PYAGASLLSVFKIKSPNVIYTLCALFSKLSAVSNALMNCYFNKYFRNHLKKIKLFQYFIKWNRRSNDKVL* | |||
>CNOPf1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01095006 KLSALINPIVNVWYNLEFR | |||
MSKALESTYICLVLIFTLTMNILLLTFLKKRYSYINISILFNASISICEILQVTLGYIPDLLTQLDVIKVTTSGNGSWICIGSSLLTFIFAVIVLMHFLVLSVSRIVAIKYPVFYVGTFSKKKLAIFLMILCYIYGIIWPVFPLFK | |||
WSKYAKDIDEKRCSLDWRLEQKNSFSYIICVILFAFILPFVTIFILLFCKKYAALANQNSRCRKRHSSIEYQEKIKGLEKKHFNMCAASTILYTLAWSPYAIVGFLSVFNSNIPPTISTS | |||
AALFAKLSALINPIVNVWYNLEFRNFLFNLKSIRFFEKNFCRKVESQESMTSSKVQCTNV* | |||
>CNOPf2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01063269 KLSALINPFVNVWFNLEFR | |||
MSKALESTYICLLLIFTLTMNVLLLTFLKKRYSYINISILFNASISVCEILQVTLGYVPDLLAQLDFIKVTVSGNGSWVCIGSSLLTFIFAVVVLMHFLVLSVSRIVA | |||
IKYPVFYVATFSKKKLSIILLILCYIYGIIWPVFPLFKWSKYAKDIDEKRCSLDWRLEQKNSFSYIICVILFAFILPFVTIFILLFCKKYATIVNQSSPSRRRHSSIEYQKKIKGLEKKY | |||
FNMCAASTILYTLAWSPYAVVGFLSVFNIIIPPMVSTSAALFAKLSALINPFVNVWFNLEFRNFIFNLKSIRFFKINLCIKVDPQESMTSSKVQCTNV* | |||
>CNOPf3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01063264 KLSASINPIVNVWYNREFR | |||
MSKALESTFICLVLIFTLTMNTLLLTFLKKRYSYINISNLFNASISVCEILQVTLGYVPDLLAQLDFIKVTTSGNGSWICIGSSLLTFIFAVVVLMHFLVLSVSRIVAIKYPVFYVTTFSKKKLSIILLILCYIYGI | |||
IWPVFPLFKWSKYAKDIDEKRCSLDWRLEQKDSFSYIICVILFVFILPFISILTLLLCKKYATLANQNSSRRRRHSSVEYQKKIKELEIKYFNMCAASTILYTLAWSPYAVVGFLSVFNS | |||
SIPPMVSTSAALFAKLSASINPIVNVWYNREFRTFLFNLKSNWFLKKIFCIKADRQKLLTFSRIQCTKL* | |||
>CNOPf4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01049000 KLSASINPIVNIWFNWEFR | |||
MINALESTYICLVLTFTLTMNVLLLTFLKNRYSYINISILFNASISLCEILQVTLGYVPDLLAQLGVIKVTTS | |||
ESGSWVCIGSSMLTFILAVVILMHFLVLSVSRILALKYPVFYISTFSKRWLAILLLILCYIYGVSWPVLPLFKWSKYAKDIDGKRCSLDWRLEQKDSFSYIICVILFAFILPLVTIFVLL | |||
FCKKYAIIVNQNSPSRKRHSSIEYQKKIKGLEKKYFNMAAASTILYTIAWSPYAVIGFLSILNRTIPSLVSTSAALFAKLSASINPIVNIWFNWEFRNFIFSLKSIRFLKKFFCKNTERQESLTSSRNQCTNV* | |||
>CNOPg1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01024987 KLSAVVNPFIYYWKDGLFK XM_002157121 | |||
MDGVKFTLITIISVAIFTNSVSLYFLFKKQKKNNYVILCINLSLSDLLQSIAGYIPALFIDKLQKATMLCKLSAFFIAFPSFTTIAMLTSMALSRMVLLSTCFHCNQINYKILFRKIGFI | |||
SWIYGFIWAVFPLFGFSSYTLEGTHSRCSIDFSPKTIADKVYLIMIVAFGFLIPVMSILISCIYTAKVMRSKYKFFYVTYGKENVETKRYKEKEKKAFSSFVLMVTSFIICWSPYATIGC | |||
LSAFTLTRIPKWLLHSAAFLGKLSAVVNPFIYYWKDGLFKKRFTSTTWNASYFISKSQQNIEDNQKNSRGALRNYMNVFCG* | |||
>CNOPg2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01083619 KLSALINPFIYYWKDGLFK | |||
MDAVKIILLIITSIAVLTNGISFYFLYKKQKKNNYVILCINLCVSDILQSVAGYIPALFFDINMQKATTLCKLSAFFIAFPSFTTIAMLSAMALSRMVLLSLCFQSNQINYKKLFIRIATASWIYGFIWAVLPFLGFSSY | |||
TLENTRARCSINFSPKKNVEKLYLILLMAFGFVIPLISIIASCLFTARVITTKYKYFCVTYGKDNDETIRYKEKEKKTFFSFLIMVLSFAVCWTPYATIGCFSAFTSFKIPQWLLHVAAF | |||
FGKLSALINPFIYYWKDGLFKKCFLNKRLKATKTFLIKNQETNQVDK* | |||
>CNOPg3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01007423 KLSALVNPFIYYWKDGLFK | |||
MDAVKTTLLIITSIAVITNSVSLYFLYKKQKKNNYVILCINLSASDLLQSIAGYIPALFFDANMQKATTLCKLSAFFIAFPSFTTIAMLSAMAI | |||
SRMVLLGTCFHSTQINYKKLFIRIGIASWIYGFIWAVLPLLGFSSYTVENTRSRCSINFSPKTSVEKLYLIILMAFGFFIPIISILASCLFTARVINTKYKYFCVTYGKENVETKRYKKK | |||
EKKTFLSFIIMVLSFVVCWTPYATVGCFSAFTSLKIPKWLLHVAAFFGKLSALVNPFIYYWKDGLFKKCFLNIRFKTTKLLIRKSQENSQK* | |||
>CNOPg4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01103264 KLSALVNPVIYYWKDGLFK | |||
MDPVKITLVIIITIAIFTNTISLYFLYKKRKSNYVILCINLSFADLLQSIAGYIPALFLDTNIQKATTLCKLTAFFVAFPSFSAIAMLTGMALS | |||
RMVLLSSCFSSNLINYKKLFIKIGIFSWIYGLFWAFLPLIGFSSYTVEATHSRCSMNFSPKNIIEKAYLILIFAFG | |||
FCIPVTIIITSCLFTAHVIVTKYNYFYVTYGKENVETKLFKEKEKKAISSFLLMVLSFIVCWTPYATIGCFTAFTSVKTPNWLLHVAAFFAKLSALVNPVIYYWKDGLFKKRSVRKKSITKSLLIIKSEINKKVEHII* | |||
>CNOPg5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01014309 KLSALVNPVIYYWKDGIFK | |||
MDPVKINLVIIITIAIFTNTTSLYFLCKKRKSNYVILCINLTFADILQSIAGYIPDLFLDTNLQKATTFCKLTAFFVAFPSFATIAMHAGIALSRMVLLSSCFSSNLINYKKLFIKIGIFSWIYGLFWATL | |||
PLVGFSSYTLEDTQSRCSINFSPKTIIEKAYLILIFAFGFCIPVTIIITSCIFTAHVIVTKYNYFYVTYGKENVETKLFKEKEKKAISSFLLMVLSFIVCWTPYATIGCLTAFTSVKTPK | |||
WLLQVAALFAKLSALVNPVIYYWKDGIFKKRSAREKSFTKSLLIIKSEINRSVDHIA* | |||
>CNOPg6_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01086515 KCSTIVNPVIYIWKDGLFK YK removed | |||
MNIEKICFFILSTTSTILNALSLYFSYRKMHKSNYVILCINLSFSDLLRSIAGYIPSLFIEEHINTATMLCKLSAFFIALTAYTAIAMITAIALSRMVLLSTCILSNKISYKALF | |||
IKIGVLSWIYGFTLAVMPFVGFSSYTLEETRARCSINFSPKTRVEKVYLILLIAFGFFIPVIIISISCLYTAYVMRTKYKYFNATYGRENIETKRYKEKEKKAFLSFVLMVLSFIVCWTPYAI | |||
IGFFSAFTSIKIFKWLLHLAVYFGKCSTIVNPVIYIWKDGLFKKSFISKRAQLIQLLTRKTRVSNRPMI* | |||
>CNOPh1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01018050 KCSALVNPIIYCWKDSLLS lacks FR motif | |||
MNAVKTALLIVSSIATITNAVSLFFLYRKHKNNYVILCINLSLSDLVRSIAGYIPALFLETNLNRASTLCKLSAFFIAFSSFTTIAMLTA | |||
IALSRMILLSTCFLHSQINYKTLFIKIGFFAWIYGFTWAVMPFFGFSSYTLENTCTRCSIDFFPKTKVEKVYLILLIAFGFLIPIITILSSCLYTANVMRSQYNYFYMIYGKNNVETKKY | |||
KVKEKKAFSSFILMVLSFVVCWTPYATVGCFSAFTSLEIPKWLLHFAPFFGKCSALVNPIIYCWKDSLLSWYIKSKRSLIIKLLFKRSEDNLFTCQIEKI* | |||
>CNOPh2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01085207 KCSALVNPVIYYWKDSLLN lacks FR motif | |||
MNAVKTALLVISSTATITNAISLYFLYRKHKNNYVILCINLSFSDLLRSIAGYIPALFLETNLNRASTLCKLSAFFIAFSSFTTIAMLTAIALSRMVLLST | |||
CFLHSQVNYKTLFIKIGLLSWIYGFTWATMPFLGFSSYTLENTNSRCSIDFFPKTKTEKVYLILLIAFGFLIPIITIITSCLYTANVMRSQYNYFYMIYGKNNVETKKYKVKEKKAFSSF | |||
LLMVLSFIVCWTPYATVGCFSAFTSLVIPNWLLHFAAFFGKCSALVNPVIYYWKDSLLNHYFKSKRLQIMKLLFRRSEDNLTILPIKEN* | |||
>CNOPh3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01011197 KSSALVNPLIYYWKDGLIA lacks FR motif | |||
MDSIKLPLLIISLVSMITNGTSLFFLLKTKHKSNYIVLCINLSISDLLRSTVGYTTELFLDPNLQSASKMCKLSAFLVAFPSLTSIALLAEISLFRMALLNAPYLRNQVANKMLFRRNGILSWLYAFVWTVLPL | |||
IGFSSYSLETTKYRCSINFLTKTVIDKVYLLSIMCFGFIIPSMIVLISCLFTARIIAEKFKYFSRTYGIKNKKTISFKEKRNKAFTSFSLMFFFFVFCWLPYATISNLSGFTSIKLPMWL | |||
YHTAAYVAKSSALVNPLIYYWKDGLIAKKKIKAAGNNIFLYSR* | |||
> | >CNOPh4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01090363 KLSAMVNPLLYWQKEDFKK lacks FR motif | ||
MDAAKVIVTIVTILSTIFNALALFNLCKKRRKTNYIFFCIFLSVSDLLQTLLGYLPAIFLKKRMENATKWCQISAFFVAFPSFTSIALLTAISFIKAYLINEHFLIGDMIYRKVVKVTILCCWIYGLFWSTFPL | |||
IGVSSYTLESTKARCSIKWYTNTNTEKLYLILLLVFCYTIPVSIVLASTMISARFVRSSFKISSSSLGMGESFVKIFKLREAKVKFSLTLMCLSFIMFWTPYAIIGLLSGFARVKHPKWL | |||
LETAALFAKLSAMVNPLLYWQKEDFKKKVRKAFLVFRTYVFYS* | |||
=== <span style="color: #990099;">Hydrozoa: Cladonema radiatum (jellyfish) .. | === <span style="color: #990099;">Hydrozoa: Cladonema radiatum (jellyfish) .. 18 opsin-like genes</span> === | ||
[[Image:Opsin_cladonema.png]] | [[Image:Opsin_cladonema.png]] | ||
| Line 242: | Line 722: | ||
</pre> | </pre> | ||
The number of 'opsins' is excessive given numbers of validated opsins that occur in complete bilatera genomes, even allowing for eyespot developmental stage variations and | The number of 'opsins' is excessive given numbers of validated opsins that occur in complete bilatera genomes, even allowing for eyespot developmental stage variations and auxiliary functions such as gonad photoreceptive gamete release. Recent gene family expansions might make more sense for chemosensory or chemokine recepters than opsins. | ||
In this view, 1-2 bona fide opsins might lurk among the collection -- but which ones?. Opsins evidently experienced various gene duplications which appear subsequently co-opted to (unknown) non-opsin, non-photoreceptive roles. This causes them to nest confusingly within the opsins even though they are no longer photoreceptors. There may have been selection to maintain the buried lysine because it worked structurally at the time of duplication (offset perhaps by chloride ion) or is used for a new but chemically related | In this view, 1-2 bona fide opsins might lurk among the collection -- but which ones?. Opsins evidently experienced various gene duplications which appear subsequently co-opted to (unknown) non-opsin, non-photoreceptive roles. This causes them to nest confusingly within the opsins even though they are no longer photoreceptors. There may have been selection to maintain the buried lysine because it worked structurally at the time of duplication (offset perhaps by chloride ion) or is used for a new but chemically related signaling agonist. The same phenomenon (gene duplication and neofunctionalization) may have occurred within Nematostella and Daphnia, which both have 'too many' opsins for their imaging needs. The Daphnia non-opsin opsin gene family expansion is not even broadly shared within the Crustacean clade. | ||
The confusion really arises from the notion of "terminally diverged" opsins first studied within Bilatera. That is, within deuterostomes, we observe a sequence of gene duplication and divergence say encephalopsin --> pinopsin --> LWS --> other cone opsins but that stays within opsins. Even that sequence had largely terminated 500 million years ago at time of lamprey divergence (primate color vision recovery is an exception). An analogous sequence is familiar within Arthropoda for example melanopsin --> MWS --> UVS. We don't observe the sequence encephalopsin --> pinopsin --> LWS --> bradykinin receptor in human nor melanopsin --> MWS --> glycoprotein hormone receptor in drosophila. Nor do we expect any such 500 million years in the future. | The confusion really arises from the notion of "terminally diverged" opsins first studied within Bilatera. That is, within deuterostomes, we observe a sequence of gene duplication and divergence say encephalopsin --> pinopsin --> LWS --> other cone opsins but that stays within opsins. Even that sequence had largely terminated 500 million years ago at time of lamprey divergence (primate color vision recovery is an exception). An analogous sequence is familiar within Arthropoda for example melanopsin --> MWS --> UVS. We don't observe the sequence encephalopsin --> pinopsin --> LWS --> bradykinin receptor in human nor melanopsin --> MWS --> glycoprotein hormone receptor in drosophila. Nor do we expect any such 500 million years in the future. | ||
| Line 254: | Line 734: | ||
Opsins seem unique in that cis-retinal is covalently attached, whereas other GPCR agonists diffuse in transiently to their binding site. This makes it difficult to see what the 'next' agonist could be in a duplicated non-opsin opsin (other than something very similar like vitamin A variants seen in some teleosts). However it's been argued that the photoisomerization product, trans-retinal, is really the agonist. That's non-covalently bound similarly to other GPCR effectors. | Opsins seem unique in that cis-retinal is covalently attached, whereas other GPCR agonists diffuse in transiently to their binding site. This makes it difficult to see what the 'next' agonist could be in a duplicated non-opsin opsin (other than something very similar like vitamin A variants seen in some teleosts). However it's been argued that the photoisomerization product, trans-retinal, is really the agonist. That's non-covalently bound similarly to other GPCR effectors. | ||
If so, that would make agonist shift in a duplicated ancestral metazoan opsin no different from other shifts taking place in other duplicated GPCR. Indeed, opsins arose from other GPCR; the ur-GPCR was not necessarily a retinal binder. Still, we wonder what the new agonists could be in this cloud of | If so, that would make agonist shift in a duplicated ancestral metazoan opsin no different from other shifts taking place in other duplicated GPCR. Indeed, opsins arose from other GPCR; the ur-GPCR was not necessarily a retinal binder. Still, we wonder what the new agonists could be in this cloud of cnidarian opsin-like proteins and what signaling is accomplished where. Hybridization in Cladodema suggests the site of signaling has not moved appreciably. | ||
It's worth re- | It's worth re-examining the notion of "once an opsin, always an opsin" even in Bilateran history. First we wonder about the cloud of lineage-specific opsin-like duplications in the crustacean genome of Daphnia. Using bovine RHO1 as query against human genome turns up a dozen non-opsin GPCR exhibiting better matches than the bona fide opsin RGR. Very likely these would nest within opsins with respect to RGR. This suggests that certain non-opsins occur inside the broader photo-opsin family. However here it is not so certain that RGR is a degenerate photoreceptor opsin, today 'merely' a retinal photoisomerase in boreoeuthere placentals retaining the Schiff base lysine but not covalently binding cis-retinal. A similar question arises with neuropsin and peropsin. | ||
<pre> | <pre> | ||
| Line 280: | Line 760: | ||
No genome project is planned so the mRNA cannot be examined for diagnostic intronation; this could be done indirectly if candidates help locate orthologs in other cnidaria. However they are already very diverged from Nematostella and Hydra opsins. Introns would not be informative anyway in discriminating opsins from co-opted non-opsin, non-photoreceptors arising as segmental duplications. Indels would be similar. This presents a very difficult bioinformatic problem because truly diagnostic residues of functioning opsins could be quite subtle. | No genome project is planned so the mRNA cannot be examined for diagnostic intronation; this could be done indirectly if candidates help locate orthologs in other cnidaria. However they are already very diverged from Nematostella and Hydra opsins. Introns would not be informative anyway in discriminating opsins from co-opted non-opsin, non-photoreceptors arising as segmental duplications. Indels would be similar. This presents a very difficult bioinformatic problem because truly diagnostic residues of functioning opsins could be quite subtle. | ||
Similarly on the experimental side, antibodies would likely cross-react. The derived non-opsins likely still signal with a transducin-type G-protein and are quenched by the same arrestin, so no help there. Knockdowns might have | Similarly on the experimental side, antibodies would likely cross-react. The derived non-opsins likely still signal with a transducin-type G-protein and are quenched by the same arrestin, so no help there. Knockdowns might have unforeseen consequences in these non-opsins, indirectly disrupting photosensitive behaviors. Thus the best way forward in identifying the true opsins within the collection is probably in vitro expression, reconstitution with cis-retinal, and demonstration of photoisomerization. | ||
The set below contains 18 Cladonema 'opsins' plus miscellaneous other K296 cnidarian sequences | |||
>CNOPa1_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332416 Cladonema radiatum CropB1 | |||
MAVKTTYPVLIIAFLIVILNSILLLGFKKKYRRGFTTYDFFLVSFTIANLIQGVFTYPFVLYTLVSEQNKNHWQCIIAAVLMLNCSVSTIWHLIILSFERFF | |||
SLKYPLFYAKHNKSRLWRGIGVSIPWLSGCFWSFPPLFGWNRYDIEFQGRPDCNIDLRDHSKSGLSYLYSLFVFNYIVPIMAFAFTFYHLRIELKKISKMARRAIGVGSKIAKASM | |||
KTEKTQTILIIIMVVSFMVAWTPYSVLVFYRTAQENEIEVYIYELSAILAKTSCIFNPIIYGLVYKDIRDAILKSIVGKKRRNNRVSILATETDILPNYSVAMVSATPKVCSLDGCYN* | |||
>CNOPa2_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332417 CropB4 | |||
MAVETTYPVLIIAFLIVIFNSLLLLGLKKKYRRGFTTYDFLLVSFTFANLIQGVFTYPFVLYTLVSEQNESHWQCITSAVLMLNCSISTIWHLIMLSFERFF | |||
SLKYPIFYAKHNKSWLWRGVGVSIPWLSGCFWSFPPLFGWNRYDIEFQGRPNCNIDLRDHSKSGLSYLYSLFVFNYIVPIMAFAFTFYHLRIELKKIFEMARKAIGVESKIAKASK | |||
KTEKTQTILIIIMVVSFMVAWTPYSVLVFYRTAQENEIEVYIYELSAILAKTSCIFNPIIYGLVYRDIRDAILKSVVWKKRRNNRVSAIATKTEILRHNNVAVVNASSLV* | |||
>CNOPa3_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332420 CropC | |||
MTGPLVFIFLLAGIILNLLVIFVVLKKQNRTVKHILVISFACCLLFEIFFGFIWEGYGRYIDDSSLELCKMAGFGATFSSLASIMHLVDMALERYISIIHPM | |||
TAYQYFNNYMLGIYMVIPSWIIGFVWAFFPFLGWGEYMREANHSYRCSIVGTSNSVNAQSYNYTMLIVFFFLPIIIITYLCVRIQMQLVKTRKKAAETFKGATKMIDGRRKYEKQH | |||
FVLVCINIIAYFVTWAPYAISTCWYTFFNSAPDDVIGYSALFAKSSLVTNPIIYVFFYREFREILKATLFKKNTTPSVEMPTIEQSTEQ* | |||
>CNOPa4_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332422 CropD | |||
MDAVIGSLVLLYLIASVILNILVIVAVLKKGNHTFKDILVISLSCSLLSEIGFGFAWEGYGRFIDDSSLELCKFAGFGATFSAFISILQLMGLAFERYVSII | |||
HPVTAHSHFKGYTLAMLFVIPSWAFCIIWASLPFYGWGEYMREAVHSYRCSVVGTGKSYDAKSYNYSLLILFFLAPVLISTYLLVRVHMELVKTRKRAGERSTGTTSITSRIWKYE | |||
RQHFFLTCFVIILFFLTWTPYALSAGWYTFFDSNPDHVVNYSALFAKSGLVTNPIIYVLFNKELRKTLKRKILERKPLITTDIQLDSITH* | |||
>CNOPa5_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332421 CropE | |||
MDELTGTLVFLFLLVGIILNLLVIATVLKKKNRKVKDVLIISLACAMISEICFGFVWEGYGRYIDDTSLELCKMAGFGSAFSAFVSILHLIAMALERYISIV | |||
HPLKAYQHFENHMLAMYMVVPAWVIGLAWALFPFFGWGVYLREEVHTYRCSAIGFIKPSNSQSYSYGLLIFFFFLPSIIIIYLCVRVQMELIKTRKNTAEKFQGATMMVADSKKCE | |||
KQHFVLVCFIIITYFLTWTPYALSACWYTFFDSKPDHVVSYSAIFAKSGLVTNPIIYVFFYREFRETLITKVFKRKAPSPSNPSTSVEMKTRSTTNN* | |||
>CNOPa6_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332426 CropF | |||
MAEVTAPIVGLFLIAGIFLNVLVVIGMLKKQSLVVKDFLVVSFACCSLVEICFGFIWEAYGRYIDDSSLTLCKMAGFGTTFPAFTSIIHLVAMAFERYISIL | |||
HPMTAHKYFVSSKVAFYFIAPSWLFGFVWAFLPIFGWGGYIREKLHTYRCSVIATDNSAVAQSYNYSLLVLYYFLPSSLIIFCCVCVQLEIQKMTRRISNGLSSAMKERTKKTERN | |||
HFILVCIVIISYFLSWTPYAISACWFTFFNTAPDELVSYSALFAKSCMITNPLVYAFFYKEFRETLKNKVFSRNIRVQTVEAPTSSLQDTKETNH* | |||
>CNOPa7_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332427 CropG1 | |||
MANLKHVEGGIIMSFIVASVSLNSLVIVGMLKKPNRALRDLILVSLACSDLVETCVGIMWEAFARYIDSSSFILCQLAGFGITFTALVSITHLAALAIERYL | |||
SMVHALKTYEFFADKKKSLLFTIPSWIHGTFWGVVPFFGWGGYIREPYHTHRCSVMMTDNSLIAQSYNYALLVFCYLIPIFIIFYCCYKVQKELKKMTERSEEIAGSSSSITQTTK | |||
KAEKQHFIMICVVMGAYFIAWTPYAVVACWFAFFDTVPGKLIAYSAVFAKFSVIANPIVYVIFYKDFRQTVKRMFCGRGRVVPA* | |||
>CNOPa8_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332428 CropG2 | |||
MANLKHVEGGIIMSFIVASVSLNSLVIVGMLKKPNRALRDLILVSLACSDLVETCVGIMWEAFARYIDSSSFILCQLAGFGITFTALVSITHLAALAIERYL | |||
SMVHALKTYEFFADKKKSLLFTIPSWIHGTFWGVVPFFGWGGYIREPYHTHRCSVMMTDNSLIAQSYNYALLVFCYLIPIFIIFYCCYKVQRELKKMTERSEEIAGSSSSITQTTK | |||
KAEKQHFIMICVVMGAYFIAWTPYAVVACWFAFFDTVPGKLIAYSAVFAKFSVIANPIVYVIFYKDFRQTVKRMFCGRGRVVPA* | |||
>CNOPa9_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332423 CropH | |||
MDAVIGCLVSLYLLAGVILNLLVIVAVLKKGNHTFKDVLMISLACSLLSEICFGFVWEGYGRFIDDSSLELCKIAGFGATFSAFVSIIQLMGLVLERYVSII | |||
HPVTAQSHFRGHTVAMLFVIPSWVFCIVWASLPFYGWGEYMREAVYTYRCSVVGTGKSYNARSYNYSMLIFFFFAPILIITYLLVRAQIRLTMTRKMAAARSTGTTATTARIYKYE | |||
RHHFILTCFVIIMFFLTWTPYAVSGCGYTFFDSSPQHVVNYSALFAKSGLVTNPIIYVLLNKELRKTLKRTIFGRKPLTTTNMQFDSITQ* | |||
>CNOPa10_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332424 CropI | |||
MDAVMGSLFLLFLIAGTTLNILVIVAVLKKRKHTFKDIMVISLTCSLLSEIYFGFVWEGYGRFIDDSSLELCKFAAYGATFSALASIMQLMGMVLERYVSII | |||
YPVTAHSYFKGNTLAMLFVLPSWAFCIVWAFLPFIGWGEYKREAVHTYRCSVIGTGKSYNARSYNYSLLILFFFAPIIIITYLIVRVQMELAIIRKMAAARSTGTTATTTRIWKYE | |||
RQHFILTCIVIVLFFITWTPYALSACWYALFDSTPDKFVSYSALFAKSSLVTNPIIFVFFNKEFYKTLKRNIFQRNPLANTDIQLDSITQ* | |||
>CNOPa11_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332433 CropJ | |||
MFNAGLVIGVVIAIGFSCFLNLLVVIGMLKKSTPLSLRDILLVSLSVADFCQTGLGYTTELYGHLSDDYSPIFLCRASGFAVTFTALVAITHLMALAVERYL | |||
SLVHALKAHEFFSEPKRALYFIIPAWIWGLFWSGAPLLKWGGYEREPHAPHRCSVVLAEKTPDVLSYSYGLLIACFAFPVLVITWCCFRVQRELKGMTDRSKEIAGQESAITAATQ | |||
KAEKQHFIMICCVVIAFFIAWTPYAGCVVYFTLVGNLPANLMSTSAFFAKFSVLANPIIYAIFYKDFRRSVKAMLCGDSAVAPSE* | |||
>CNOPa12_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332431 CropK1 | |||
MLLEIESIMVSLFAANIVLGFSMNLVVISALCRVSGKSVRDILLISLALCGLLETGLGYAIELYGRLNVRASSLLCQMAGFSVTFFALSSIFHIVGLAIERY | |||
LSIVCAVNARGYFSMKHRALYIIFPAWINGLVWSIAPFLGWGAYKREPIDKYRCSVLMSDVSFVSWSYTIMLLTFAYAIPLLVILYCCLKVQCTLNAMSKNISDPENRTRDSGLEA | |||
TKQAEKRHYYMVCAIIVTFFVAWTPYATSALLFSFLENVPNTLITYSAFFAKFSVVANPVIYVIFYKDCRKQVKKMVCVDTAVAPIEA* | |||
>CNOPa13_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332432 CropK2 | |||
MLLEIESIMVSLFAANIVLGFSMNLVVISALCRVSEKSVRDILLISLALCGLLETGLGYAIELYGRLNVRASSLICQMAGFSVTFFALSSIFHIVGLAIERY | |||
LSIVCAVNARGYFSMKHRALYIIFPAWINGLVWSIAPFLGWGAYKREPIDKYRCSVLMSDVSFVSWSYTIMLLTFAYAIPLLVILYCCLKVQCTLNAMSKNISDPENRTRDSGLEA | |||
TKQAEKRHYYMVCAIIVSFFVAWTPYATSALLFSILENVPNTLITYSAFFAKFSVVANPVIYVIFYKDCRKQVKKMVCVDTAVAPLEA* | |||
>CNOPa14_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332425 CropL | |||
MDDSSMDAITGILVVLFLTTGIILNTLVILAVMKKSSHTFKDVLVISFTCCLLGEICFGIIWEGYGRYIDDSSLALCKMAGFGATFTTLVSIIHLIGMALER | |||
YISIIHPMTAYQYFKDHMLAMYFLIPSWVFGIAWACFPFFGWGAYMREPVHTYRCSVVASSNSNDAQSYNYSLLIFFFFLPIILIIYLCACVQIELAKTRKEAIETLTGDSAMIDS | |||
RKKHEKEHFVLVCFVIITYLVTWTPYAISACWFAFFKSQSNEMVGYSALFAKSALVTNPIIYVFFYKEFRETLKTKVFTRKASTSVDMPIRSSLSQKIDIHE* | |||
>CNOPa15_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332418 CropM | |||
MTVKGEDISIIVIVIFSTFLNICLILALVQKTSKPNFMPCLLLNMALCSLLQGLFAYPLELNNIFKASVTSDVTDISCVVSAVMVYGLSLIVIFTITLLSIE | |||
RVLAIKFPYLYRKCERNHKCFLVFSIMIIWALGAFWACAPLFGWSNYQKETDDSHRCNINFTRKDANNASYLCTLLVSCYVIPVSIMVSSYTLVKNSMRRMSVYADQRVGKNATLS | |||
KNSKKSEKQQTLLTILMIFAFLYAWTPYTTVVFFLMIDKRVSPTIYTIAALNAKTSSFYCPLLYGFVYKRFRRRLKDVVAKIFV* | |||
>CNOPa16_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332429 CropN1 | |||
MADEVGGPVILVFIVGSVFLNLLVIVGMLKKPNIALRDLILVSLAISDFIETVFGSLWEAYAKYIDSSSLFLCKMAGFGITFTALTSISHLAALAIERYLSM | |||
VHALKTYEFFADKKKAIIFIIPAWTYGTFWGGVPLLGWGGYFREPIHTFRCAVLMTSKSFLSMSYNYCLLAFCYLLPISIISICCYKVQRELKNMTERSEAIAGSSSAITESTKKA | |||
EKQHFLMICIVLVAFFIAWTPYAFTACWFAFFDNPPDELVTYSATFAKFSVVSNPIVYVIFYKDFRQTVKKMFCGRGRVAPV* | |||
>CNOPa17_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332430 CropN2 | |||
MADEVGGPVILVFIVGSVFLNLLVIVGMLKKPNIALRDLILVSLAISDFIETVFGSLWEAYAKYIDSSSLFLCKMAGFGITFTALTSISHLAALAIERYLSM | |||
VHALKTYEFFADKKKAIIFIIPAWTYGTFWGGVPLLGWGGYFREPIHTFRCAVLMTSKSFLSMSYNYCLLAFCYLLPISIISICCYKVQRELKNMTERSEAIAGSSSAITESTKKA | |||
EKQHFLMICIVLVAFFIAWTPYAFTACWFAFFDNPPDELVTYSATFAKFSVVSNPIVYVIFYKDFRQTVKKMFCGRGRVAPV* | |||
>CNOPa18_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332419 CropO | |||
MKVENITIVVIVALSVVLNICLIAVLLQKTLRYTFMHYFLFNMALCSLFQGLLAYPLELSSLLKGRKKMDFECLMGAILVFGLALVVIWTIALLSIDRVLAM | |||
KFPYLYKRCIRNKKRFVFLSITFTWLIGGFWALTPLLGWSSYQKEMKPSHRCNINFRSKDPVNMSYLCSLMVTCYVVPVTIMISSLTLVKKSLRKMSIYAEKRVGRRASLSLDAQR | |||
CEKRQTLLTLLMIFSFFYAWTPYTIVVTFLMLDKSVPPTVYTIAALNAKTSSFYCPLLYGFVYKPLRSTFKSAVSKIMV* | |||
>CNOPa1_claRad Podocoryna carnea (jellyfish) Cnid.Hydr.Anth AB332435 PcopC | |||
MDRTTFTAFLTIIVALSASLNTAIIIALIRTWNKLSSYDVTLLFLTTVELLQAVIGYPIELTTNLREGGPLCIIAGYSTTTLALTSISILVALSFMRLLSVT | |||
QPLKAVEWIKTRRYSIYLILPSCIYGFMWGIFPLIGWSEYAPELGSNKRCTIDLHSEDNATRSYLYSLLLFCYVLPLTTMTINFYKIRKKIIEQSVKLSQNRERIRTNRIKRERRL | |||
AIATFVMTVVFLVAWTPYAVAIFIIGFQKRVDEAWFFMDYSSLLAKSSTFFNPLIYVLMHRKCKTAVVDMFQYIICLIRRRDSQASVSYNGSAQRVQFLHIASMSSQKPDTCSPCV | |||
HIQRSEYLVAKDAANGPNYSFTATNI* | |||
>CNOPg1_claRad Podocoryna carnea (jellyfish) Cnid.Hydr.Anth AB332434 PcopB | |||
MDPLKIILSSIVVSSIILNTATCFILIRKKRKTKFVILCIHLAISDLLQAVCGYVPELIIQKNNFPTLLCKASACTVAVMALASISLITLISITRIISIKYP | |||
FTSVSTSKVTVNILAIISWLYAAFWAICPLLGWSQYVLEPTHRRCSLDWNLKTANSKSYIYAIVLFCYILPITIHVISFFMVRRTVENHARFCADTYGRRNSATLIVERKKLKSFF | |||
LSLSMFSIFIVAWTPYTLVGISAQYVTVPTWSMDSAAMLAKSSTLFNPLVYCYRTKFRRKNIFREDNVTEDNITMLRTSLRRNQCDNNGNVLNGESVNVCYDKKSLSISFGVIEESQT* | |||
>CNOPa1_clyHem Clytia hemisphaerica (jellyfish) Cnid.Hydr.Anth CU434354 | |||
MVLEVTYEHIFLLVMVTLGTILNLLVIITLLFMMKKGNHELRNVILMSLAVSDMLPALFGFSLELTSTDDIMFCKISAFAVTSSSLSSIMH | |||
MVLLMIERAYTIKYPYRAHIVLKRKSIMACFVSIAWIYGTVLGVCPLLGWGNYTHEGVSMKRCSIDINMNTWSVTTYNYSLLLLGYLIPIAASVACYVVIKKEHGKLSEKAVRYQGKSPL | |||
MTaHTMDIQRSFTLMFLVMLLAFLISWT | |||
>CNOPa1_monFav Montastraea faveolata Cnid.Anth.Hexa GW273473 fragment | |||
ASFIICIFVIFFLFPVVSLVVSYAIILQQISQMAKRAKSKWDDEAQQTLEILLARKKVVWTGFIMVASFVVVWTPYAILSFY | |||
AAFGKPQSIPPLAATIPALFAKTSTVLNPIIYVIRYKRFRKGVKKLFKMI* | |||
== <span style="color: #990099;"> Ctenophora .. 2 opsins</span> == | |||
The authors identify 4 genes as photoreceptive opsins, based on GPCR blast character, lysine in expected position, and unpublished accounts of phototactic behavior. However notation used in the document (scaffold8_1356180_1396180 = Opsin 6, scaffold13_1543556_1583556 = Opsin 26, scaffold8_1308974_1348974 = Opsin 30. scaffold8_1467859_1507859 = Opsin 31) does not match that provided at GenBank nor JGI genome. Gaps in assembly are indicated by N's which may or may not be counted in coordinate numbers. Opsins with these scaffold numberings do not surface by tBlastn of GenBank WGS where the Trichoplax contigs reside. | Ctenophores are an early diverging metazoan group, though it is not quite clear whether they should be sistered with cnidarians or split from the metazoan stem earlier. The data situation has greatly improved in the last year with transcript programs and a completed genome project with quite large contigs (relative to gene size). | ||
=== <span style="color: #990099;"> Ctenophora: Pleurobrachia pileus (sea gooseberry) .. 2 opsins</span> === | |||
The first bona fide ctenophore opsin has surfaced in an unannotated cDNA ([http://www.ncbi.nlm.nih.gov/nucest/167802800 CU419614]) deposited to GenBank in February 2008 in the course of a never-published phylogenetic study of sponges (Manuel, Houliston: "The sponges reunited: implications for early animal evolution"). This planned manuscript may have been subsumed in a [http://www.ncbi.nlm.nih.gov/pubmed/19345102 different article] that concluded calcisponges, homoscleromorphs, demosponges and hexactinellids form a basal monophyletic metazoan group. | |||
This paper also sistered Ctenophora + Cnidaria, a tree topology [http://www-2.cs.cmu.edu/~durand/03-711/2009/Homework/Telford09.pdf reducing] the number of mainstem metazoan nodes by one. It may take complete genome sequencing to determine this relationship reliably -- it has been debated for over a century. The [http://www.ncbi.nlm.nih.gov/pubmed/20920349,20920347 Mnemiopsis genome] is 150 Mbp so easily within an afternoon's reach and indeed became available in Oct 2010. Haeckelia rubra, the species whose ingested and recycled cnidarian nematocysts once confused its taxonomy, is 10x this size. | |||
Genomic sequencing must disentangle ctenophore dna from that of the parasitic cnidarian Edwardsiella lineata whose planula larvae enter the ctenophore mouth as fake food but then bore into the mesoglea and help themselves to ingested pharyngeal food. Up to 60% of a Mnemiopsis population can host these larva. | |||
Ctenophora full length genes are poorly represented at GenBank with only 156 nucleotide entries for the whole phylum, so vision-related entries cannot be expected there. However transcripts are in better shape with 24,292 as of Jan 2010. These consist entirely of the lobate Mnemiopsis leidyi gastrula library (15,752) and cydippid Pleurobrachia pileus (8,540 cDNA, undescribed library) despite ~150 [http://en.wikipedia.org/wiki/Ctenophora known species]. A negligible number of reads (3,360) from early cleavage Mnemiopsis can be found in the trace archives. These two at least represent the two main groups of ctenophores at according to GenBank taxonomy, Cyclocoela and Typhlocoela. | |||
The sensory system of ctenophores is located at the rear (aboral pole, ie head is not around the mouth) and can connect to the non-centralized nerve net. The statocysts, suspended by four fibers like those of Ciona or for that matter human otoliths, report to nerves coordinating cilia locomotory beat. Statocysts would deflect according to gravitational field lines and acceleration, allowing orientation relative to the vertical. Water current velocities could be detected in a distinct system of stiff cilia projecting out into the water that bend in proportion to current. | |||
The ectodermal sensory region also contains four crescent shaped groups of round bodies, interpreted as photoreceptors on page 166 of a 1880 monograph by Chun. Ctenophores lack an overt response to shadows; these eyespots could not plausibly provide imaging vision or function in predation or avoidance. They could however track length of day length, distinguish up from down and serve as depth detectors. Elsewhere, epithelial walls of food distribution canals have osmoregulatory rosettes, light-inhibited bioluminescent photocytes utilizing the original green fluorescent protein and testis and ovary on opposite canal walls. | |||
The eyespots, statocysts, photocytes and gonads are the most plausible sites for opsin based photoreception in ctenophores. No data is available on ctenopsin anatomical expression as all cDNAs are tied to a whole-organism library. Nothing has been done to date to localize opsin expression because until now, no sequence has been available. An unfortunate term 'mnemiopsin' was introduced and abandoned in the 1970's. It apparently references the bioluminescent protein which completely lacks any resemblance much less homology to opsins. | |||
[[Image:CtenOpsinPics.jpg]] | |||
The initial Pleurobrachia opsin sequence was not full length as it starts 27 residues after a presumptive DRY motif and continues past the Schiff base lysine and an altered NPxxY motif but not to a stop codon. However 5 cDNAs posted in May 2010 extend it to full length. While the sequence is quite diverged from any known opsin (low 30's for best percent blastp identity), the evidence is nonetheless overwhelming that this is an opsin: | |||
* the candidate sequence turns up as best tblastn match of any opsin query at GenBank restricted to Ctenophora | |||
* when translated and aligned to 430 curated opsins, residue matches are overwhelmingly concentrated in conserved sites (graphic below) | |||
* the sequence has lysine at homologous position K296 (bovine rhodopsin numbering) to Schiff base lysine, unlike any non-opsin GPCR | |||
* opsin-diagnostic regions (absent in the most closely related GPCR) are observed, notably the [[Opsin_evolution:_informative_indels#Indels_in_the_TM4-EC2-TM5_region|GWSR motif]] retinal plug in EC3: PLVGWCEYGPEGY | |||
* best back-blast to all of GenBank returns opsins ahead of all other GPCR (in contrast to candidate CU422164 whose best back-blasts are serotonins) | |||
* the sequence roots in the tree of all opsins as expected for a ctenophore photoreceptor (tree below) | |||
* the sequence classifies evenhandedly among ciliary, rhabdomeric and cnidopsins as expected for an early diverging opsin (list below) | |||
While establishing the presence of a K-rhodopsin in this ctenophore, this analysis hardly proves that this protein functions directly in photoreception (as opposed say to photocycle regeneration, bioluminescence, carotenoid metabolism, gamete release or statolith orientation). The anatomical characterization of potential photoreceptors appears limited to an offline 1979 Russian text "Electron microscopic study of presumptive photoreceptor cells in the aboral organ of the ctenophore, Beroë cucumis" by MZ Aronova. It is not clear whether these classify as either ciliary or rhabdomeric ("presumptive photoreceptor cells which have a short central projection and form in their basal part the synaptic contacts..."). Elsewhere it is [http://reefkeeping.com/issues/2004-03/rs/index.php said] ctenophores do not react to shadows and while the eyespots cannot form an image they may track changing day length and depth in the water column. | |||
There are also significant deviations from near-universally conserved residues believed critical to the Galpha signaling partner interaction, notably in the NP..Y......FR region which here is WI..Y......IK. While Galpha proteins (and especially their fifth helix which binds to the C2/C3 mitt of opsins) are conserved to still greater phylogenetic depth, the protein fragment here is difficult to [http://athina.biol.uoa.gr/bioinformatics/PRED-COUPLE2/ evaluate] without more of its upstream residues. It could be chimerized to full length using a known opsin (but which one?). As it stands however, the fragment gives highly significant positive scores to both Gi/o and Gq type signaling partners. The 3rd cytoplasmic loop lacks the HEK motif common to most but not all melanopsins, so provides no further clues. | |||
A second apparent ctenophore opsin appears as an unannotated cDNA fragment [http://www.ncbi.nlm.nih.gov/nucleotide/295255996?report=genbank&log$=nuclalign&blast_rank=1&RID=7D48R0AV013 FQ011385] that aligns only up to residue 203 in RHO1_bosTau numbering, thus its putative lysine at residue 296 remains speculative. Its ERY motif is more conventional than the other ctenopsin. | |||
The phylogenetic position of ctenophores has been subject to endless back-and-forth for two centuries. This discussion has intensified very recently with the advent of substantial molecular data. While can hardly be resolved by the one protein fragment here, for an opsin it has many primitive features that do not favor backsliding from cnidarian and bilateran opsins sharing immensely conserved contemporary features. Thus it is not consistent with ctenophores branching later than cnidarians but rather earlier (or sistering). | |||
This opsin and its role could be greatly clarified by determination of the full length sequence and its sites of expression. It potentially represents an opsin descended from an era when melanopsins, cilopsins and cnidopsins were coalesced into a single gene. Another way of saying this is all these opsins are orthologous relative to the last common ancestor of sea combs, anemone, fruitfly, squid, and human. Peropsins, neuropsins and RGRopsins are still excluded and represent even earlier gene duplication and divergence. | |||
Best blastp matches to reference opsin collection: | |||
ENCEPHc_nemVec Nematostella vectensis (anemone) Anthozoa 8.0e-25 | |||
ENCEPHb_nemVec Nematostella vectensis (anemone) anthozoa 4.5e-23 | |||
ENCEPHa_nemVec Nematostella vectensis (anemone) anthozoa 8.2e-23 | |||
PARIE_takRub Takifugu rubripes (fugu) Gd+Go -HSP90B1 4.2e-20 | |||
PARIE_danRer Danio rerio (zebrafish) Gd+Go - +NT5DC2 4.2e-20 | |||
MEL1_acrMil Acropora millepora (stony_coral) anthozoa 6.0e-20 | |||
MEL1_lotGig Lottia gigantea (limpet) FC774055 ests 1.1e-19 | |||
CUBOP_carRas Carybdea rastonii (sea_wasp) cubomedusae 1.5e-19 | |||
LMS1_hasAda Hasarius adansoni (jumping_spider) Chelice 1.6e-19 | |||
TMTx_braFlo Branchiostoma floridae (amphioxus) XM_0022 1.9e-19 | |||
ENCEPH_strPur Stronglyocentrotus purpuratus GLEAN3_ 3.3e-19 | |||
Sample alignment to the ciliary parietopsin of Takifugu (with <font color="blue">cytoplasmic loop C3</font> and <font color="red">Schiff lysine region</font> colored): | |||
plePil 6 FFGIAPLVGWCEYGPEGYGVSNSLMWNNLNDNNASYIICAMIIGYFFPLIIIMFCYRAIY 65 | |||
F+ + PL+GW YGPEG S SL W + NN SY+I ++ + FP+ +I++CY + | |||
takRub 61 FWAVTPLLGWSSYGPEGVQTSCSLAWEERSWNNYSYLILYTLLCFIFPVGVIIYCYCKVL 220 | |||
plePil 66 RLVQA<font color="blue">QLNTSVVKMTVSANVTSLSTNRNHIMLVQE</font>RQLASTITFVILSFFFAWTPYVFVN 125 | |||
+ <font color="blue">+LN SV + ++ N++ I +V </font> LA +I++FF W PY ++ | |||
takRub 221 TSMN-<font color="blue">KLNKSV---ELQGGLSCRRENKHAINMV--</font>--LA-----MIIAFFVCWLPYTALS 267 | |||
plePi l26 IINMFSTLILKYRILATIPALFA<font color="red">KSSTLWWIIVYCLMDVRIK</font>KACRK--TCLRF 177 | |||
++ + + ++AT+P FA<font color="red">K+S ++ I+Y L + + +</font> A + +C R+ | |||
takRub 268 VVVVVDPELHIPPLVATMPMYFA<font color="red">KTSPVYNPIIYFLSNKQFR</font>DATLEVLSCSRY 321 | |||
>CTENOP1_plePil Pleurobrachia pileus (sea_gooseberry) Cten.Typh.Pleu FP995093 CU419614 full G? ctenop EQY intronated via Mnemiopsis ortholog | |||
0 MELRVSLSLIMACVCTGSVF 1 | |||
2 GNMLVFISLVKERPLPNPAVEMIFRHLSLANLVLAGIGEPLVVVSLAA 1 | |||
2 GKWVWGESMRYFEAFWVTTFGLFSMMILGLISF<font color="red">EQY</font>SRFVRQKQLKTHQ 0 | |||
0 VAVLLSGIYFLAFFFGIAPLVGWCEYGPE 1 | |||
2 GYGVSNSLMWNNLNDNNASYIICAMIIGYFFPLIIIMFCYRAIYRLVQ 0 | |||
0 A<font color="blue">QLNTSVVKMTVSANVTSLSTNRNHIML 0 | |||
0 VQE</font>RQLASTITFVILSFFFAWTPYVFVNIINMFSTLILKYRILAT 0 | |||
0 IPALFA<font color="red">KSSTLWWIIVYCLMDVRIK</font>KACRKTCLRFKRSFRVWYFS* 0 | |||
>CTENOP2_plePil Pleurobrachia pileus (sea_gooseberry) Cten.Typh.Pleu FQ011385 ERY intronated via Mnemiopsis ortholog | |||
0 MTTAVDDEMMGDEEPKVVYTLASLLAVNALCSIA 1 | |||
2 GNALVIYTFLKEKPLGAPLRVILTHLAVTNLIIASIGEPMVVISGYS 1 | |||
2 MRWVFGEMGRGFEAFIVTSCGLMAMSLLALVSI<font color="red">ERY</font>LR 2 | |||
1 VTPRQRFPFTEPTAVKCCSFLWVFNVIYASLPFYGIAAWEGE 1 | |||
2 GIGISNSITWTTEHTQDFVYIICMMSTGYFIPLFIIAFCY 2 | |||
[[Image:CtenOps.jpg]] | |||
=== <span style="color: #990099;"> Ctenophora: Mnemiopsis leidyi (sea walnut) .. 2 opsins</span> === | |||
The first ctenophore genome assembly became available to NCBI's wgs tBlastn in Sep 2011 after being held back nearly a full year. This species has two opsins, one each of ciliary and melanopsin type, that are clearly orthologous to the two Pleurobrachia opssin available from transcripts. These opsins, unlike those of cnidarians, prove quite rich in exons with 8 each, matching each other in three but not at all with bovine rhodopsin. The two opsins are about 39% identical. Neither have close matches to cnidarian or bilateran opsins though CTENOP1 classifies clearly with ciliary opsins and CTENOP2 less clearly with melanopsins. | |||
The ctenophore opsins are not closely related to any non-opsin GPCR either in ctenophores or human. Within the human proteome, CTENOP2_mneLei matches peropsin, melanopsin and rhodopsin equally poorly. The first GPCR encountered are somatostatin, GPCR136 and chemokine receptor 6. CTENOP1_mneLei has similar affinities. Neither CTENOP elicits a K296 match in sponge, the best matches being to sponge proteins resembling human TSHR (thyroid stimulating hormone receptor). These non-opsin GPCR are among those already identified as parental to opsins. | |||
The existence of ctenophore opsins shows that this gene family was already well-established by the time of ctenophore divergence from other metazoans. If no opsin can ever be located in sponges, then the timing of opsin origin is quite well defined. | |||
>CTENOP1_mneLei Mnemiopsis leidyi AGCP01013851 AGCP01013853 77% identical CTENOP1_plePil | |||
0 MGLRISLSIAMGCVCVGSVF 1 | |||
2 GNLLVLISLVRERPLPNPAVQLIFRHLSLANFLLACIGEPLVVISLAS 1 | |||
2 GKWVWGESMRLFEAFWVTTFGLLSMMILALISL<font color="red">EQY</font>SRFVRQKQLRTVQ 0 | |||
0 VAVLLSGIYFLAFFFGIAPLVGWCEYGPE 1 | |||
2 GYGVSNSLLWNELTDNSASYIVTVMIVGYFFPLVIITYCYRSIYQLVQ 0 | |||
0 AQSNVAIVRTMVSVNVTSLAVNRNHSLI 0 | |||
0 VQERKLATTITFVILSFFFAWTPYVLINLLNMFTTWVVKYKILAT 0 | |||
0 IPALFA<font color="red">KSSTLWWIIIYSMMDKRIK</font>KACRKTCEHFQRALRIWYFSS* 0 | |||
>CTENOP2_mneLei Mnemiopsis leidyi AGCP01013109 AGCP01013108 AGCP01013106 6th exon uncertain | |||
0 MSSPNDEPTVDSDPKAVYILTSLLCMNCIFSLV 1 | |||
2 GNMLVIYTFLRERPLGAPLRTILCHLAITNLMIAGIGEPMVVISGFN 1 | |||
2 VRWVFGEMGRKIEAYTVTASGLMAMSLLAFVSI<font color="red">ERY</font>LR 2 | |||
1 VTPGQKFLIRQTTSVRVCTLMWIYNVIYASLPFYGIAAWRGE 1 | |||
2 GIGISNSLSWDTNNKIDVAYVMVMMATGYFIPLAVITFCYNRYVEIQQRLSTFL 2 | |||
1 0 | |||
0 AAEKKISNVVAIIIVSFFISWTPYTIVNLLVTFDKAGR 2 | |||
1 GINATIPAFLA<font color="red">KTSTVWSPIIYCFMNGEVS</font>IFVLEGGHVMKYLYL* 0 | |||
Comparison of CTENOP1 with CTENOP2: numbers denote exon boundaries and phases | |||
1 2 0 1 | |||
CV-CVGSVFGNLLVLISLVRERPLPNPAVQLIFRHLSLANFLLACIGEPLVVISLASGKWVWGESMRLFEAFWVTTFGLLSMMILALISLEQYSRFVR-QKQL--RTVQVAVLLSGIYFLAFFFGIAPLVGWCEYGPEGYGV | |||
C+ C+ S+ GN+LV+ + +RERPL P ++ I HL++ N ++A IGEP+VVIS + +WV+GE R EA+ VT GL++M +LA +S+E+Y R QK L +T V V L IY+ + P G + EG G+ | |||
CMNCIFSLVGNMLVIYTFLRERPLGAP-LRTILCHLAITNLMIAGIGEPMVVISGFNVRWVFGEMGRKIEAYTVTASGLMAMSLLAFVSIERYLRVTPGQKFLIRQTTSVRVCTLMWIYNV--IYASLPFYGIAAWRGEGIGI | |||
1 1 2 1 | |||
0 0 0 | |||
SNSLLWNELTDNSASYIVTVMIVGYFFPLVIITYCYRSIYQLVQAQSN-VAIVRTMVSVNVTSLAVNRNHSLIVQERKLATTITFVILSFFFAWTPYVLINLLNMFTTWVVKYKILATIPALFAKSSTLWWIIIYSMMDKRI | |||
SNSL W+ +Y++ +M GYF PL +IT+CY ++ Q S ++ ++ + N+TS A ++N + E+K++ + +I+SFF +WTPY ++NLL F I ATIPA AK+ST+W IIY M+ + | |||
SNSLSWDTNNKIDVAYVMVMMATGYFIPLAVITFCYNRYVEIQQRLSTFL AAEKKISNVVAIIIVSFFISWTPYTIVNLLVTFDK--AGRGINATIPAFLAKTSTVWSPIIYCFMNGEV | |||
2 0 2 | |||
== <span style="color: #990099;">Placozoa: Trichoplax adhaerens .. 0 opsins</span> == | |||
The sequenced genome of Trichoplax adhaerens appears in the [http://www.nature.com/nature/journal/v454/n7207/full/nature07191.html 21 August 08 Nature] and associated [98 pages of supplemental], an excellent treatment as first assembly articles go but still insufficient detail to replicate various bioinformatic assertions. The genome and browser are hosted at JGI but it is equally convenient for most purposes to simply use NCBI tblastn targeted to the WGS contig division restricted to Trichoplax or [http://www.ncbi.nlm.nih.gov//genomes/geblast.cgi?bact=off&gi=6008 assembly blast]. | |||
The authors identify 4 genes as photoreceptive opsins, based on GPCR blast character, Schiff base lysine in expected position (though that position was not described), and unpublished accounts of phototactic behavior. However notation used in the document (scaffold8_1356180_1396180 = Opsin 6, scaffold13_1543556_1583556 = Opsin 26, scaffold8_1308974_1348974 = Opsin 30. scaffold8_1467859_1507859 = Opsin 31) does not match that provided at GenBank nor JGI genome. Gaps in assembly are indicated by N's which may or may not be counted in coordinate numbers. Opsins with these scaffold numberings do not surface by tBlastn of GenBank WGS where the Trichoplax contigs reside. The authors have not responded to an email request to furnish the fasta sequences. | |||
In light of similar claims made for Hydra, Cladonema and Nematostella opsins that didn't bear up to scrutiny, given the lack of photoreceptor structures and cell-specific expression staining in Trichoplax, the burden of proof for opsins has not yet been met. Nothing in the Trichoplax genome meets the best-blastp threshold (exceeding the 25-35 percent identity range of generic GPCR matches) of the opsin classifier. Bona fide opsins also require a counterion E at ancestral position, a DRY motif, a binding motif for alpha subunit of heteromeric G protein, and about 150 semi-invariant residues that distinguish photoreceptors by ortholog class (eg ciliary vs rhabdomeric) and separate them from non-photoreceptive 'EK-rhodopsin-class' generic GPCR. | In light of similar claims made for Hydra, Cladonema and Nematostella opsins that didn't bear up to scrutiny, given the lack of photoreceptor structures and cell-specific expression staining in Trichoplax, the burden of proof for opsins has not yet been met. Nothing in the Trichoplax genome meets the best-blastp threshold (exceeding the 25-35 percent identity range of generic GPCR matches) of the opsin classifier. Bona fide opsins also require a counterion E at ancestral position, a DRY motif, a binding motif for alpha subunit of heteromeric G protein, and about 150 semi-invariant residues that distinguish photoreceptors by ortholog class (eg ciliary vs rhabdomeric) and separate them from non-photoreceptive 'EK-rhodopsin-class' generic GPCR. | ||
The only validated pre-Bilateran opsin to date occurs in the imaging eye of the cubozoa Tripedalia cystophora where a single ciliary opsin sequence meets all expression and classification tests. This opsin has no noteworthy matches in Trichoplax. Cnidarian larva also have rhabdomeric | The only validated pre-Bilateran opsin to date occurs in the imaging eye of the cubozoa Tripedalia cystophora where a single ciliary opsin sequence meets all expression and classification tests. This opsin has no noteworthy matches in Trichoplax. Cnidarian larva also have a rhabdomeric structure implying that class of opsin as well. | ||
Examples exist of non-opsin non-photoreceptive GPCR that contain lysine at the retinal position, ie this lysine is necessary for an opsin but far from sufficient. They are easy to collect using blast of consensus opsin sequence ending somewhat past that lysine. Cnidaria are especially rich in these lysine-containing non-photoreceptors. Indeed the 'opsins' reported for Hydra, Cladonema and Nematostella greatly exceed the (non-observed) photoreceptive structures and organismal needs -- which surely aren't twice those of the most advanced bilatera. | |||
Nematostella: no photoreceptor structures but 16+ GPCR genes with lysine in 'retinal position': | |||
>tpd|BR000664 ID: 44/121 36 FLFAWTPYAVVSLWTTFGDTHRIPALLGVLPSLFA K LSSCYNPIIYFFMYTKFR 39% ENCEPH4_braFlo * | |||
>tpd|BR000663 ID: 48/121 39 FLFAWVPYAVVSLYASFGGVTTIPKLMSTLPAMLA K TSACYNPIIYFFMYSKFR 35% TMT5_braFlo * | |||
>ref|XM_001637473 ID: 35/115 30 FLACWLPYVIVSTCMSLGRRPQISLLTLEITLLVA K SGVIYNPFIYAALNLRFR 33% MEL_schMed * | |||
>tpd|BR000666 ID: 32/81 39 FLIAWLPYAIVSLYSAITGE.RVSPEAATIPGMLA K SASCYNPLIYVFLYSRFR 33% TMT_ornAna * | |||
>tpd|BR000667 ID: 38/121 31 FLVAWSPYAITSLYISWTGIQTIPDIARILPPMFA K AYSCYNPVIYYGMSRKFR 29% MEL2_xenLae * | |||
>tpd|BR000671 ID: 28/66 42 FVISWSPYCVVSVMAMVHRAPSLPRGLAEIPELMA K ASVIYNPLVFTVMNIEFR 32% VAOP_takRub | |||
>tpd|BR000685 ID: 30/75 40 FTLSWSPYAIVCLKSMVVGEQKLAPFTSEVTALMA K ASAVYNPIVYVVLSKRFR 35% MEL1_schMan | |||
>ref|XM_001634921 ID: 37/107 34 FLVAWTPYAVISFYSITGHARDLSFISVVLPAIFA K TSAFYNPIITLVFWFCLR 37% RHO1_letJap frag | |||
>tpd|BR000662 ID: 36/107 33 FVFAWTPYAVVSIYS.ALLKPKLPLIAGILPPLFA K TSTLYNPLLCFIGSPPIR 31% ENCEPH4_braFlo | |||
>tpd|BR000672 ID: 25/75 33 FVISWSPYCVVSVIAMVQRAPALSQGFAEIPELMA K ASVVYNPLVFTVMNRGFR 30% MEL_TMT | |||
>ref|XM_001627232 ID: 37/115 32 FFISWSPYCIVSLIESAKGEVVLSPGVSMIPELMA K ASVMYNPVVYTLMNARFR 31% MEL_TMT | |||
>ref|XM_001639117 ID: 37/118 31 FTLSWSPDAILSVI....SMVTGRPIIHVVPSLMA K SSVIYTPIVYIAFSRSFR 34% VAOP_danRer | |||
>tpd|BR000661 ID: 38/121 31 FLIAWTPYAVVSFYYSLRGPTGVPLMAAMLPSLFA K ASSLFNPIIYFAMSREFR 31% MEL2_galGal | |||
>tpd|BR000686 ID: 35/114 30 FAITWLPYAVYVLIAAFGGSHLFDAVMSVVPAMVA K MSILYNPFVYALVNPRFR 31% TMT_triCas | |||
>tpd|BR000674 ID: 36/121 29 FLACWLPYVIVSTCMSLGRRPQISLLTLEITLLVA K SGVIYNPFIYAALNLRFR 35% MEL2_strPur | |||
>tpd|BR000669 ID: 35/121 28 FSM.WVPYVAVSLIQAFTADSIITPTASHITVLVA K SCVIYNVLIYVVLNRKLK 34% TMT5_braFlo | |||
<br clear="all"> | |||
[[Image:KopsinTree.jpg|left|]] | |||
The tree at left (which would benefit from a few dozen outgroup genes such as beta adrenergic receptor) shows how some related sequences clustered in a recent [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17940617opsin evolution study by Plachetzki et al] who sought but did not identify opsins in sponge, Trichoplax or Monosiga. It's not known if any of their 'cnidops' bind retinal or have a photoreceptive role. It is possible that retinal is bound in Schiff base position but requires a secondary agonist to signal. Alternatively, signaling compounds related to but distinct from retinal assume the binding site. | |||
A third scenario conserves the lysine and its counterion through evolutionary inertia (the salt bridge avoids unfavorable burying of charged residues in hydrophobic membrane and maintains the trigger) without Schiff base, with the diversity of genes perhaps serving chemoreception. Such a class of GPCR in early cnidaria could have been the foundational source for later recruitment to opsins in cubomedusa. | |||
An analogous situation in sea urchin (rapid expansion of an intronless opsin-like class of genes) was noted by Raibl and coworkers (who denoted them specific rapidly expanded lineages of GPCRs or surreal-GPCRs) and noted a parallel to olfactory gene expansion in amniotes. In other words, prior to dead-end specialization as imaging opsins, earlier gene duplicates could find novel but useful roles. That's not so different from even earlier diverging 'rhodopsin-class' GPCR taking on roles such as beta adrenergic receptors. | |||
In this scenario, photoreceptive opsins have a basal outgroup originating in cnidaria (or Trichoplax) of EK-rhodopsin class non-opsins and a distinct class of EK-rhodopsin class non-opsins paraphyletically nested within them in sea urchin and possibly amphioxus GPCR (where genes are still conservatively intronated). These latter two species also have 'too many' opsins versus a paucity of photoreceptor cell morphologies especially in urchin. | |||
Gene duplication (and loss) of opsins continues as an active process to the present day in many species. We're not surprised to see LWS duplicate to MWS in primate cones. However we don't expect LWS to give rise to processed retrogenes serving olfaction or non-retinal signaling. Those days could be behind it. LWS may be terminally differentiated and too many mutational steps away from more versatile deployment. If so, evolutionary processes do not remain constant; some genes mature over time into locked-down roles with less potential for novelty in their duplicates than their ancestral predecessors. We see from the context of Ciona that GPCR have a very long history of acquiring new agonists for novel expanded gene family branches: | |||
[[Image:RhodCiona.jpg|center|]] | |||
To recapitulate, photoreceptive opsins did not originate out of thin air by miraculous multiple mutations providing overnight EK, signature motifs and retinal binding pocket. More plausibly, opsins specialized from a pre-existing gene expansion pool of EK-class GPCR that itself evolved from generic group 1 GPCR. That class persists today in pre-bilaterans with unknown agonists and function though it apparently winked out in bilatera. Genuine opsins may nest paraphyletically (recruited by independent events, eg melanopsin, encephalopsin, neuropsin) within EK-rhodopsin class GPCR; a diversity of cnidarian opsin sequences are needed. | |||
Even if this phylum lacks opsins, the genome will still prove important in determining the state of signaling systems. The 98 mbp genome is 3% the size of a mammalian genome but the estimated 11,514 coding genes are not that different from Drosophila and 55% of the 20,000 genes of human | The unpublished observation of phototaxis in Trichoplax is only weak supportive evidence for opsins because demosponge larva also exhibit phototaxis (shadow seeking under coral rubble) whereas the action spectrum fits a flavin or carotenoid chromophore better than retinal. Larval photoreceptors don't reside in a rhabdomeric or ciliary morphological setting but rather in ring of columnar monociliated epithelial cells. No expression staining has been conducted for the putative opsins -- Trichoplax has only 4 cell types, none specialized in appearance to photoreceptor. | ||
Cryptochromes are another well-known alternate photoreceptive system in bilatera with no homology at the protein level to opsins. In all kingdoms of life, [http://www.ncbi.nlm.nih.gov/pubmed/14730990? only seven known families of proteins] transduce light into signal (opsins, cryptochromes, phytochromes, xanthopsins, phototropins, blue flavin and lite1); Trichoplax would have to be evaluated for each of these too. For example, nematode C. elegans lacks opsins and cryptochromes but has a shorter wavelength photoresponse bypassing cAMP and diacylglycerol (DAG) downstream signaling that neurons commonly use to control behavior. The [http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0060198 primary photoreceptor LITE1 (NP_509043)] is a 8-transmembrane member of the invertebrate gustatory receptor family (non-GPCR) without known chromophore. | |||
Trichoplax could have an unobserved larva and metamorphosis in view of the proposed post-sponge position. Alternatively, it may have be a direct developer or have lost the ability to reproduce other than by fission (which is contradicted by meiosis genes, oocytes and population genetics despite non-observation of male gametes). Trichoplax, being too small, has never been observed directly in the wild -- instead it is collected over weeks on submerged alga-coated microscope slides (or aquarium walls). Whole aspects of its life cycle may go undetected in laboratory culture. The current situation (assuming post-sponge divergence) -- highly conserved genome yet highly derived morphology -- is an odd but not impossible mix. | |||
Even if this phylum lacks opsins, the genome will still prove important in determining the state of signaling systems at its ancestral node. The 98 mbp genome is 3% the size of a mammalian genome but the estimated 11,514 coding genes are not that different from Drosophila and 55% of the 20,000 genes of human. Pseudogenes, which outnumber genes in mammals, are difficult to detect in isolated genomes such as Trichoplax. | |||
This large gene set if assumed approximately ancestral is not consistent with subsequent subsequent 2R whole genome duplications (giving 45,056 genes in human) without massive gain in Trichoplax and/or loss in human). In other words, even if 2R occurred, it was an inconsequential mechanism in the overall scheme of gene duplication and retention; the complement of genes was in large measure already established in early metazoa. That has special relevance to understanding expansion and specialization of opsins and associated G protein alpha subunits which primarily exhibit tandem duplication followed by translocation. | |||
[[Image:TrichoplaxSEM.jpg|left|]] | [[Image:TrichoplaxSEM.jpg|left|]] | ||
Trichoplax is an odd amoeboid-shaped animal without overt symmetry, body plan, recognizable organs, or internal digestive gut but clear dorsal/ventral sides. Only a single species has received a Linnean name; it is assigned a whole phylum, Placozoa. However the taxonomy section of GenBank lists 87 other placozoan isolates; these have ~200 deposited sequences and [http://www.biolbull.org/cgi/external_ref?access_num=15556848 previous publications] have established quite diverged mitochondrial genes, compatible with a whole order (even as external morphologies remain indistinguishable). Thus the 600 myr long branch of the Red Sea isolate could readily subdivided by 454 sequencing to provide supplementing glimpses of ancestral characters and better reconstruction of ancestral proteins. | Trichoplax is an odd amoeboid-shaped animal without overt symmetry axes, body plan, recognizable organs, or internal digestive gut but clear dorsal/ventral sides. Only a single species has received a Linnean name to date; it is assigned a whole phylum, Placozoa. However the taxonomy section of GenBank lists 87 other placozoan isolates; these have ~200 deposited sequences and [http://www.biolbull.org/cgi/external_ref?access_num=15556848 previous publications] have established quite diverged mitochondrial genes, compatible with a whole order (even as external morphologies remain indistinguishable). Thus the 600 myr long branch of the Red Sea isolate could readily subdivided by 454 sequencing to provide supplementing glimpses of ancestral characters and better reconstruction of ancestral proteins. | ||
The authors place the divergence node between sponge and cnidaria, surprising given the advanced sponge larva and its similarity to later-diverging metazoan. Post-sponge divergence (but sistered with Cnidaria) was proposed earlier in a 1993 Science article based on [http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=8469985 small subunit ribosomal RNA sequences.] The place of Ctenophores still cannot be resolved for lack of an available genome, though just about every topology has received support. Thus the number of early metazoan nodes discussed in "The Ancestor's Tale" is still uncertain. | The authors place the divergence node between sponge and cnidaria, surprising given the advanced sponge larva and its similarity to later-diverging metazoan. Post-sponge divergence (but sistered with Cnidaria) was proposed earlier in a 1993 Science article based on [http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=8469985 small subunit ribosomal RNA sequences.] The place of Ctenophores still cannot be resolved for lack of an available genome, though just about every topology has received support. Thus the number of early metazoan nodes discussed in "The Ancestor's Tale" is still uncertain. | ||
| Line 312: | Line 1,072: | ||
<blockquote>"Our analysis shows that the Trichoplax mitochondrion possesses the largest known metazoan mtDNA genome, at 43,079 bp, more than twice the size of the typical metazoan mtDNA. Its large size is due not to secondary expansion but to features shared with metazoan outgroups, such as intragenic spacers, several introns [cox1 introns share identical positions with choanoflagellate Monosiga and fungus], ORFs of unknown function, and protein-coding regions that are generally larger than that found in animals. The large Trichoplax mtDNA is the least derived mitochondrial genome of any animal. Moreover, the Trichoplax mitochondrion shares unique derived features with other lower metazoans, notably the loss of all ribosomal protein genes. These structural features of the Trichoplax mitochondrial genome, along with Bayesian and maximum-likelihood (ML) analyses of mitochondrial proteins from metazoans and outgroups, provide robust support for the phylogenetic placement of the phylum Placozoa at the root of the Metazoa.... the basal phylogenetic position of Placozoa within the lower metazoans is robust, with P values between 0.924 and 1.000 for the various statistical tests."</blockquote> | <blockquote>"Our analysis shows that the Trichoplax mitochondrion possesses the largest known metazoan mtDNA genome, at 43,079 bp, more than twice the size of the typical metazoan mtDNA. Its large size is due not to secondary expansion but to features shared with metazoan outgroups, such as intragenic spacers, several introns [cox1 introns share identical positions with choanoflagellate Monosiga and fungus], ORFs of unknown function, and protein-coding regions that are generally larger than that found in animals. The large Trichoplax mtDNA is the least derived mitochondrial genome of any animal. Moreover, the Trichoplax mitochondrion shares unique derived features with other lower metazoans, notably the loss of all ribosomal protein genes. These structural features of the Trichoplax mitochondrial genome, along with Bayesian and maximum-likelihood (ML) analyses of mitochondrial proteins from metazoans and outgroups, provide robust support for the phylogenetic placement of the phylum Placozoa at the root of the Metazoa.... the basal phylogenetic position of Placozoa within the lower metazoans is robust, with P values between 0.924 and 1.000 for the various statistical tests."</blockquote> | ||
It appears that an unpublished, unreleased assembly of proteins of the sponge Amphimedon was used yet again | It appears that an unpublished, unreleased assembly of proteins of the sponge Amphimedon was used (yet again). This genome project is 100% publicly funded yet its assembly seem to have been privatized to a shortlisted clique of insiders. Lottia gene models are also used without bibliographic citation. Both were evidently analyzed by unknown parties as unpublished [http://www.biomedcentral.com/1471-2105/5/59 SNAP] ab initio predictions (Table S7.1). This raises some questions because neither reviewers nor other scientists can independently evaluate the evidence supporting the unexpected phylogenetic relationships. It is reminiscent of the recent uproar over non-release of dinosaur collagen mass spectroscopy. | ||
The JGI site shows sponge [http://www.jgi.doe.gov/sequencing/why/3161.html still in draft assembly on 25 Aug 08] whereas this paper was submitted nine months earlier, on 4 Dec 07. Trace reads were completed and made available in June 2005 but these cannot be used for genes or proteins without assembly as individual read typically cover but single exons. Collagen researchers have [[http://www.ncbi.nlm.nih.gov/pubmed/18697744? assembled genes] using tiling from mate pairs and ESTs. By custom, assembly and first publication is normally reserved for the sequencing laboratory but that was never intended to apply to holding back data for 3-4 years. | |||
Trichoplax appears to retain a goodly share of ancestral characters, including some chromosomal gene associations and a large number of apparent 1:1 orthologs to human and anemone. However it must be noted that the authors have significantly altered the traditional definition of synteny in making their dot plot (Oxford grid) by discarding strand orientation altogether, allowing weak partial length blastp hits (perhaps just to common domains or pseudogenes) to count as orthologous matches, and accepting departures of 10 genes from adjacency (which can amount to several million bp in mammalian genomes). | Trichoplax appears to retain a goodly share of ancestral characters, including some chromosomal gene associations and a large number of apparent 1:1 orthologs to human and anemone. However it must be noted that the authors have significantly altered the traditional definition of synteny in making their dot plot (Oxford grid) by discarding strand orientation altogether, allowing weak partial length blastp hits (perhaps just to common domains or pseudogenes) to count as orthologous matches, and accepting departures of 10 genes from adjacency (which can amount to several million bp in mammalian genomes). | ||
The assumption here is small and very localized intra-chromosomal inversions have been more frequent than inter-chromosomal | The assumption here is small and very localized intra-chromosomal inversions have been more frequent than inter-chromosomal arrangements over the vast 1.2 billion year roundtrip span of evolutionary time (but see gibbon, fly, tunicate etc). As described a decade ago, dot plots rendered in Photoshop better retain orientation and quality of fit using grayscale and tint, rather than the all-or-none as here. | ||
The 11,514 coding gene models arise largely from an informatics pipeline tool fGenesh. Like all gene predictors, even when parameterized with transcripts, this tool makes numerous errors in human (where it can be thoroughly evaluated by comparative genomics) and is no longer included in the 35 predictive gene tracks at UCSC (though still valued for [http://www.wormbase.org/wiki/index.php/Results nematode] and other species). The tool acted after RepeatMasker masking of 665 transposons; if that library were incomplete on low copy elements, the gene count could be too high. | |||
Manual curation is mentioned briefly but could not have been extensive given the number of genes. Gene models missing start or stop codons were simply extended out from available sequence (though in some cases transcript extension was feasible). While a downstream stop codon can always be found, an initial methionine candidate does not always occur before an upstream stop codon is encountered. This is not a good annotation practice as it assumes that partial first and last exons are at hand; better to do this only when accurate gene models from other species warrant these two assumptions. These extensions will seldom have homological support even when correct unless the termini are important (eg signal peptides; G protein C-terminal opsin interaction). | |||
Only 58 ESTs are provided by GenBank (from an unpublished immunology study by different authors); in the nucleotide division, it appears that GenBank does not distinguish experimental mRNAs from those simply inferred from genome models. Transcripts have proven very helpful in quantitating gene prediction accuracy. In assessing completeness of the genome, the authors state 85% of 14,571 T. adhaerens ESTs (later described as 2,506 when clustered of 21% coverage) have quality matches (a more nuanced account appears in Supplemental). If 15% did not make the cut -- assuming them to be representative of missing or partial genes or absent from transcripts -- that would bring the total coding gene count to 13,241. | |||
These 2,506 assembled transcripts could be downloaded at the [http://www.jgi.doe.gov/trichoplax indicated JGI url] and tested to see how and if they are stored at GenBank as this affects queries of the average user. The article does not discuss how discrepancies between transcripts and pipeline gene models were resolved. These can involve spliced-out exons with or without comparative genomics support and non-support of called exon boundaries (here the transcripts will have it right). | |||
On a species so distant and little studied biochemically as Trichoplax (27 articles since 1974, mainly field work or rDNA sequencing), when protein alignment dips below the 40% protein-level identity on partial matches (notably GPCR putative opsins), homological transfer of annotation from functionally characterized proteins won't be reliable beyond rough domain-level concepts. Best-blastp percent identity to known genes is not noted at GenBank model entries. | |||
Here signalP, InterPro, and TMHMM were used to predict signal peptides, domains, and transmembrane peptides; while not validated with pre-bilateran metazoan protein chemistry to any extent, these tools provide a good start to localized functional elements. KEGG and KOG are not sufficiently developed to be worthwhile; indeed, their development appears abandoned (2004-05 versions were used). EC numbers and GO nonsense add little value. Biosynthetic pathways were not analyzed and so essential amino acids, cofactors, and intermediary metabolism were not determined. | |||
What is needed here is an ability to filter or grade annotation transfer quality vis-a-vis match quality, match extent, domain structure, and gene family extent along the lines of GeneSorter at UCSC. For example, Trichoplax/human matches can be very high as in the 77% identity agreement seen in G protein alpha subunits (below) or very low 29% in best matches of some opsins to Trichoplax GPCR. | |||
Conversely, Trichoplax and the secret sponge annotation will now propagate all over. This can become a self-reinforcing paradigm for error propagation, eg as later authors see corroborating annotation in their top homology matches. | |||
None of this has to affect opsin analysis (other than missing assembly) because the genome sequence can be directly searched by tblastn without reference to JGI gene models or annotation. Any assertion of opsins must establish the location and phasing of introns in a gene model as these are exceedingly conserved in opsins and diagnostic of orthology classes (and so especially important when protein sequences are so diverged as to blur into generic GPCR). | |||
At this late date, it might not be expected that a metazoan would contain many authentic coding genes with no blast matches in all of GenBank. These may prove greatly enriched for artifacts, just as the Ensembl human gene set used contains nearly 2,000 pseudogenes, related debris and mispredictions not supported by the 28way comparative genomics track to any phylogenetic depth. It's not clear what fraction had supporting transcripts. Dropping these will roughly offset valid genes missing from the assembly or missed by gene prediction. | |||
Similarly, introns will not be located correctly if this is homologically forced in regions of poor identity (self-fulfilling prophecy); phase agreement can help but not so much if based simply on GT-AG which occur every 16 bp approximately, nor even with nuanced splice rules if merely assumed from remotely related species. The practical impact is that exceptions and imperfect matches are missed -- the authors report excellent overall conservation of introns with respect to human and Nematostella, which fits the modern picture of intronation being deeply ancestral and profoundly conserved outside a few rogue species (such as fly, nematode, and tunicate) with high turnover. Some 150 known splice sites that use rare alternatives to GT-AG may also be as deeply conserved though this has not yet been investigated. | Similarly, introns will not be located correctly if this is homologically forced in regions of poor identity (self-fulfilling prophecy); phase agreement can help but not so much if based simply on GT-AG which occur every 16 bp approximately, nor even with nuanced splice rules if merely assumed from remotely related species. The practical impact is that exceptions and imperfect matches are missed -- the authors report excellent overall conservation of (high quality flanking) introns with respect to human and Nematostella , which fits the modern picture of intronation being deeply ancestral and profoundly conserved outside a few rogue species (such as fly, nematode, and tunicate) with high turnover. Some 150 known splice sites that use rare alternatives to GT-AG may also be as deeply conserved though this has not yet been investigated. | ||
Manual curation of selected genes of signal transduction (alpha subunits of Trichoplax heteromeric G proteins) shows that cGMP Gt transducin-type alpha subunits (presumably still hyperpolarizing) share the identical 8-exon structure and phasing of human and other bilatera, including the anomalously short 15 residue exon 2 that distinguishes this class of genes from the otherwise identically intronated 7-exon Gq alpha subunits that utilize phospholipase C hydrolysis of PIP2 and IP3 signaling. This strongly suggests that, though the requisite gene duplication and divergence took place much earlier, the distinctive exon 2 emergence (or fusion) predates metazoa. The presence of Gt and Gq subunits does not imply the existence of melanopsin or encephalopsin homologs because these alpha subunits must service hundreds of other unrelated GPCR. | Manual ab initio curation here at genomeWiki of selected genes of signal transduction (alpha subunits of Trichoplax heteromeric G proteins) shows that cGMP Gt transducin-type alpha subunits (presumably still hyperpolarizing) share the identical 8-exon structure and phasing of human and other bilatera, including the anomalously short 15 residue exon 2 that distinguishes this class of genes from the otherwise identically intronated 7-exon Gq alpha subunits that utilize phospholipase C hydrolysis of PIP2 and IP3 signaling. This strongly suggests that, though the requisite gene duplication and divergence took place much earlier, the distinctive exon 2 emergence (or fusion) predates metazoa. The presence of Gt and Gq subunits does not imply the existence of melanopsin or encephalopsin homologs because these alpha subunits must service hundreds of other unrelated GPCR. | ||
>GNAi1_triAdh Trichoplax adhaerens (placazoa) Gt XM_002115978 77% GNAi1_homSap 8 exons | >GNAi1_triAdh Trichoplax adhaerens (placazoa) Gt XM_002115978 77% GNAi1_homSap 8 exons | ||
| Line 356: | Line 1,124: | ||
== <span style="color: #990099;">Porifera: Amphimedon queenslandica (sponge) .. 0 opsins</span> == | == <span style="color: #990099;">Porifera: Amphimedon queenslandica (sponge) .. 0 opsins</span> == | ||
One marine demosponge genome is available | One marine demosponge genome, called Reniera sp. at JGI and trace archives blastn but subsequently reclassified to Amphimedon queenslandica at GenBank and PubMed, is available as 2,917,892 traces dating to July 2005 or as finally assembled on 28 May 2010 in the wgs division of GenBank where they can be queried with tBlastn. Importantly, not all traces made it into the assembly, so tBlastn offered at [http://compagen.zoologie.uni-kiel.de/datasets.html Compagen] is still critically important. A publication on the genome will no doubt appear shortly. | ||
The | The genomic DNA used came from embryos derived from a single parent sponge collected from the Great Barrier Reef. However GenBank contig entries say the 8-fold assembly comes from a sponge population with more than two haplotypes. Contig sizes appear respectable, with many dozen over 100kbp in size but internally many have gaps. | ||
Sponges lie at the base of multicellular animals and are not notable for a nervous system. However demosponge larva do exhibit phototaxis (shadow seeking under coral rubble) but the action spectrum is [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=11976887 supposedly] a better fit to a flavin or carotenoid chromophore. Sponges also can respond to gravity, current, and chemical cues. | |||
The basis for sponge responsiveness to light has been carefully studied from an [http://www.biolbull.org/cgi/content/full/201/3/323 ultrastructural perspective] -- for an animal lacking nerves and cell junctions, the parenchymella larva are quite capable of responding effectively to light and other stimuli. Larval photoreceptors may lie in a posterior ring of columnar monociliated epithelial cells. A pigment cell occurs but the pigment itself has not been chemically characterized -- the issue is whether it is a homologously derived (melanin via tyrosine hydroxylase) or novel. | |||
Negative larval phototaxis there has been [http://www.biolbull.org/cgi/content/full/201/3/323 attributed] to pigment-filled protrusions in a posterior ring of columnar monociliated epithelial cells. This species may prove insufficient to explain the full range of photoresponsive responses in sponge larva such as [http://circadiana.blogspot.com/2006/03/do-sponges-have-circadian-clocks.html circadian rhythm] and hexactinellid photoreception (notably the role of [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=15935634 stalk spicules]). | |||
Jacobs et al have proposed a far more [http://icb.oxfordjournals.org/cgi/content/full/icm094v1 sweeping view] of early evolution of sensory (and other!) organs in sponges. A related view, that of Gehring, proposes that the eye (and other sensory systems) came before the brain, indeed that the nervous system arose later to coordinate a response to all these inputs. There is support for that in simple photoreceptor cells controlling their own cilia. Consequently we should not be too quick to dismiss sponge photoreception for lack of neurons. | |||
In | In an article entitled [http://www.ncbi.nlm.nih.gov/pubmed/18315817?2008 "Six major steps in animal evolution: are we derived sponge larvae?"] C. Nielsen writes: | ||
<blockquote>A scenario for the early evolution of the metazoans. The metazoan ancestor "choanoblastaea" was a pelagic sphere consisting of choanocytes. The evolution of multicellularity enabled division of labor between cells and an "advanced choanoblastaea" consisting of choanocytes and nonfeeding cells. Polarity became established, and an adult, sessile stage developed. Choanocytes of the upper side became arranged in a groove with the cilia pumping water along the groove. Cells overarched the groove so that a choanocyte chamber was formed, establishing the body plan of an adult sponge; the pelagic larval stage was retained but became lecithotrophic [yolk-supplied]. The sponges radiated into monophyletic Silicea, Calcarea, and Homoscleromorpha. Homoscleromorph larvae show cell layers resembling true, sealed epithelia. </blockquote> | <blockquote>A scenario for the early evolution of the metazoans. The metazoan ancestor "choanoblastaea" was a pelagic sphere consisting of choanocytes. The evolution of multicellularity enabled division of labor between cells and an "advanced choanoblastaea" consisting of choanocytes and nonfeeding cells. Polarity became established, and an adult, sessile stage developed. Choanocytes of the upper side became arranged in a groove with the cilia pumping water along the groove. Cells overarched the groove so that a choanocyte chamber was formed, establishing the body plan of an adult sponge; the pelagic larval stage was retained but became lecithotrophic [yolk-supplied]. The sponges radiated into monophyletic Silicea, Calcarea, and Homoscleromorpha. Homoscleromorph larvae show cell layers resembling true, sealed epithelia. </blockquote> | ||
| Line 378: | Line 1,145: | ||
[[Image:Opsin_sponge.png|left|]] | [[Image:Opsin_sponge.png|left|]] | ||
The resulting picture of | The resulting picture of Amphimedon larva -- numerous differentiated and pluripotential cell types arranged in stereotypic patterns along central-lateral and anterior-posterior axes -- is not one typically conjured up of parazoan ("almost metazoan") in the view of Leys and Degnan. Indeed the common ancestor humans shared with sponge may have been rather advanced. | ||
The concept here is that a [http://www.springerlink.com/content/u1wtjec1um512rw4/ photoreceptor cell can control its associated cilium] without the baggage of a CNS, either as a passive rudder or more actively directing phototactic motion. In effect the single photocell is a self-sufficient brain that processes external environmental inputs, asseses them and acts appropriately. Chemoreception, a very similar GPCR signaling system, might work the same way. In this view, the nervous system evolved as a secondary system to coordinate these stand-alone sensory effectors. | |||
A [http://www.plosone.org/article/info:doi/10.1371/journal.pone.0001054 futile search] for sponge opsins turned up only non-opsin, rhodopsin-class GPCR genes from Amphimedon. Similarly, no bone fida opsins have been located in even earlier diverging placozoan Trichoplax, choanoflagellate Monosiga, and fungal genomes. This fits a picture of photoreceptor opsins first appearing subsequent to sponge in eumetazoa cnidarians. However these were hardly de novo genetic innovations but rather evolved out of the already-rich cauldron of GPCR gene expansions in the sponge ancester. | |||
Some later diverging species such as the model organism C. elegans lost all of their opsin genes, making them useless in Urbilateran ancestor reconstruction. This argues for much more intensive genomic sampling of sponges and cnidarians so as to sidestep inference mislead by gene loss in model organisms chosen for historic reasons. | |||
However on 31 May 2010, a fragmentary Schiff K296 opsin was found here among residual traces not used in the assembly, namely in ti|922429579 (also called BAYB198960.b1). This trace, despite a length of 1018 bp and the overall 8x shotgun coverage, has no overlap with any other trace as it is likely Lottia gigantea contamination (see below). No additional K296 sequences occur in the assembly using a wide variety of probes. | |||
The candidate (possibly Lottia contaminant) opsin sequence within trace ti|922429579 is shown below aligned to its best match among the thousands of established opsins, a peropsin from amphioxus(with next-best an undocumented opsin from the cnidarian Nematostella). Observe not only is the Schiff base lysine conserved in precise homogous position upstream of the NPIIY ... FR but the entire conservation profile is an excellent match to that of known opsins. While some of this profile is also conserved within generic rhodopsin-class GPCR, the fragment here elicits only much weaker matches to non-opsin GPCR (the best being bradykinin receptor). All top matches from tBlastn to GenBank are opsins. Thus homological evidence for this fragment being an opsin is much stronger than just a lysine at Schiff position 296. | |||
<---------TM6--------><--EC3---><---------TM7--------> | |||
sponge: 1 VCVVVIFSFMICWSPYATFSLWYSYSDTSGIPLWVTALPVLAA<font color="red">K</font>LFCCINPVIYLVVSKRFR | |||
+C+ ++ +F++ W+PY+ L+ ++ + + IP W+T LP L A<font color="red">K NP+IY++ ++RFR</font> | |||
perop: 257 MCLAMVAAFVVAWTPYSVLFLFAAFGNPADIPAWITLLPPLIAKSSALYNPIIYIIANRRFR PER3_braFlo Branchiostoma floridae | |||
cons profile m...mv..F...W.PYa..............p......P..fA<font color="red">K.s...NP!IY......FR</font> | |||
>PER3_braFlo Branchiostoma floridae (amphioxus) AB050610 12435605 | |||
0 MDIPTETPYGAGDDPAGTGWRWAETDQNGFHKYDHLIVGLYLFVI 1 | |||
2 GIIGTVENGITLATFTKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLR 0 | |||
0 SHWLFGGVGCQWYGFNGMFFGMANIGLLTCVAVDRYLVICRQDL 1 | |||
2 VDKVNYNTYGVMAALGWLFAAFWAALPLVGWAEYSLEPS 1 | |||
2 GTACTINWQKNDSLYISYVTSCFILGFALPLAVMMFCYWQ 0 | |||
0 ASCFVNKVLKGDISGDLTFPVAVNVDWEYQNHFSK 0 | |||
<font color="blue">0 MCLAMVAAFVVAWTPYSVLFLFAAFGNPADIPAWITLLPPLIA<font color="red">K</font>SSALYNPIIYIIANRRFR</font>SAIFSMVKGQNPDVE 0 | |||
0 TLFARDFRISPIEDTGKEMSSMGNANA* 0 | |||
>OPS_nemVec Nematostella vectensis (anemone) no introns | |||
MADASGNTTSVVVTRVYSAYSAITLVLNTLLILVFYKKSLLRRSYRFYIFSLALVDILMAVLVDSIGAYANSSPGRALSSRGCQYYGAVASFVGFTSVHHLTAIYA | |||
DRWYSIRFVMRRKPSKRVTLRVILALWISGLIWAIPPLVGWSSYAHQGSFSHCSVKWFSVDPKDYSYTVVIFTKFYFFPIAFMTFSYVHVFVITRRMAKNATQLWGE | |||
KAAATAKSIKSESKAAE<font color="blue">IALVMVLSFLIAWTPYAVVSFYYSLRGPTGVPLMAAMLPSLFA<font color="red">K</font>ASSLFNPIIYFAMSREFR</font>NSCRDFARR* | |||
ttttttc<font color="red">ag</font>gtttgtgtggttgtcatattttcctttatgatatgctggagtccttatgcc phase 00 splice acceptor preceding sponge exon | |||
F F Q V C V V V I F S F M I C W S P Y A | |||
Blast of the Lottia gigantea genome yields an opsin sequence identical to the one recovered from the Amphimedon traces: >jgi|Lotgi1|152675|fgenesh2_pg.C_sca_2000095. This implies one or both genome project had dna contamination. | |||
The trace read can be extended somewhat beyond the last region of homology but truncates prior to reaching a stop codon. Similarly, it can be extended N-terminally for a half dozen residues preceding the start of homology but then reaches a stop codon within the reading frame. Since a standard GT-AG splice acceptor immediately precedes the start of homology, this region very likely represents the start of the final coding exon. This is supported by both the 00 phase and position of the exon (252 EVTR in bovine rhodopsin homology numbering) exactly matching those of its best match. | |||
This exon was [[Opsin_evolution:_ancestral_introns#Ancestral_peropsin.2C_neuropsin_and_RGRopsin_intronation|previously recognized]] as ultra-conserved within the peropsin-neuropsin-rgropsin group of opsins. This provides strong independent support for the identification of this fragment as an opsin. It also implies a very ancient origin of intronation and subsequent immense conservation for this gene family. This is borne out by the nearly perfect match of sponge transducins GNAQ, GNAS, GNA13 to those of human. | |||
However, despite an extra 729 bp preceding the opsin-homologous region, no earlier exons can be detected by blastx even with the most tolerant settings. This could be explained either by a moderately long intron or by divergence so extreme that the preceding exon -- which is predicted from homology to be short -- does not exhibit homology. More likely, however, is the possibility that it represents contamination from the Lottia gigantea genome sequencing project, which occured in the same facility. There is no indication of compositional simplicity or presence of a retroposon here. The data also exclude processed pseudogene and indeed all forms of pseudogene (which decay without respect to conservation profile). Introns in sponge tranducin GNAS range from 42, 54, 69, 69, 84, 108 to 401 bp, short and not indicative of massive retroposon invasion. GNAS is [http://www.pnas.org/cgi/pmidlookup?view=long&pmid=18832159 surprisingly associated] with photoreceptor transduction in jellyfish. | |||
The exon begins just inside transmembrane helix VI and continues through the third extracellular loop to transmembrane helix VII and the final cytoplasmic helix VIII. Thus the preceding exon missing in the trace would primarily consist of the third cytoplasmic loop whose length is highly variable within peropsin-neuropsin-rgropsin group. There are no reliable anchor residue patches for many dozens of residues upstream. Thus should introns in the genome assembly generally prove to be short, divergence would be the likeliest interpretation. Introns in sponge transducins also well-conserved and of moderate size. | |||
Earlier exons still might be detectable in this trace via de novo gene prediction tools such as GeneScan. If these had expected lengths and intron phasing, that would lend support even if homology remains weak. For blastp extensional matching, it would be necessary to get far enough N-terminally to encounter a patch of conserved residues. However blastx already excludes this. None of the ab initio tools tested worked satisfactorily on this trace, unsurprisingly since all are optimized to vertebrate (eg human) gene-finding parameters. | |||
In summary, the sponge genome does not appear to contain any opsins. | |||
From homology and intronation, it appears most closely related to the peropsin-neuropsin-rgropsin group, with markedly less affinity to melanopsins or cilopsins. The best affinity might change if full-length sequence were available. Note too that all opsins may coalesce at this phylogenetic depth. Since sponge is basal among metazoans with opsins, this might suggest that the peropsin group is ancestral and the others more derived. However this cannot be safelyl concluded from just one species of sponge because its opsin content might reflect gene loss from the ancestral state. | |||
>PER_lotGig Amphimedon queenslandica (sponge) contaminating fragment apparently from Lottia instead | |||
VCVVVIFSFMICWSPYATFSLWYSYSDTSGIPLWVTALPVLAA<font color="red">K</font>LFCCINPVIYLVVSKRFR QRYIEVVCRSYKYCGKRNSITPERTQV | |||
>GNAQ_ampQue Amphimedon queenslandica (sponge) 70% | |||
0 MACCLSEEAKEARRINDEIERQLRRDKRDARRELKLLLL 1 | |||
2 GAGESGKSTFIKQMRIIHGSGYSVEDRKQFVTLIYRNMYTSLQALTTAMESLKIPYT | |||
DPQSQSYAESISTVNPETVSEIPDDHKEAFKCLWSDDGIQQCYQRRREFQLSDSTKY 2 | |||
1 YLDDAERLLAQDFVPTTQDVLRVRIPTTGIIEYHFDIRNVIFR 2 | |||
1 MVDVGGQRSERRKWIHCFENVTSVMFLVAISEYDQILVETDE 0 | |||
0 NRMEESLALFETISRYPWFRTSSVILFLNKKDLLEEKIMISDLSDYFPEYK 1 | |||
2 GPPKDAEKAKEFIKEMFLSVNPDEKKHIFPHFTTATDTENIQRVFEAVKTTVLEENLRDYNLI* 0 | |||
>GNA13_ampQue Amphimedon queenslandica (sponge) 54% | |||
0 MAFNCLKTEEEKEGERRSRQIDRQLKSLDKEKDWKREMKVLLLGAGESGKSTFLKQMRIIHGEDYSDSDRLEFRPLIYHNILK | |||
GMKVLVEACRRLQIPFSEPDNDSKGDIVSSGYQHSQELSPEEFAPYVEPLKSLWKDTGIQTTLARSNEFQL 0 | |||
0 GDSVPYFLSKLEVIGAV 0 | |||
0 DYLPSKDDILNSRKATRGVVEYTTAIRNQQFRFVDVGGQRSQRQKWFQCFDEVTAILFIVASSAFNQTLLEDRVTNRLIESLNIFETIVNNRCFR | |||
NVAIMVFFNKSDLLETKVQNPKVDISKYFPEFNGDPHNLDHVKGFMVGMFDSVRRNQRQDFYHYFTTAVNTRNIHMVFDIVKDIILSGHIGQILQI* 0 | |||
>GNAS_ampQue Amphimedon queenslandica (sponge) 62% | |||
0 MSALGCFGEPLTEEERQRKEKSKEIDQQLRKEKVEYKSTHRLLLL 1 | |||
2 GAGESGKSTVVKQMKILHVNGFSKE 2 | |||
1 ERLQNVDDIRSNVREVIQ 0 | |||
0 TVTSQMDKLDPPVTLKDERNQLRLSYIQNVITPLPEYETNFSP 0 | |||
0 EFYEHAEALWNDEGVQEAFRRSNEYQLIDCAPY 2 | |||
1 FMNSKLREVANPDYLPSDD 00 DILRARRMTKGIYETHFIIEKVHFQ 2 | |||
1 MFDVGGQRVERSKWIQCFN 1 | |||
2 DVTAIIYVVASSSFNMVIREDNRT 00 NRLRESLNLFEQIWNNR 2 | |||
1 WLRNVSVILFLNKMDILKEKIEGKHFHLEDYFPEFSSYRPPAES 1 | |||
2 EKREQGDTETVLRARFFIRDQFL 0 | |||
0 RISSRGANDGRHFCYPHFTTAVDTENIRRVFESCRDIIQRIHLHRYELL* 0 | |||
== <span style="color: #990099;">Choanoflagellates: Monosiga brevicollis .. 0 opsins</span> == | |||
[[Image:MonoBrev.jpg|left|]] | |||
The genome sequence of Monosiga brevicollis appears in the [http://www.nature.com/nature/journal/v451/n7180/full/nature06617.html 14 Feb 08 issue of Nature.] It contains a reported 9,200 genes in 42 Mbp. These are densely intronated but reflect net loss since common ancestor with metazoans. Domain orders are [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=18599463 markedly shuffled] relative to eumetazoan counterparts and some key proteins such as Notch have only partial-length matches. | |||
The 14 extant genera of choanoflagellates have been minimally studied overall with only 59 non-Monosigna sequences at GenBank so additional representative genomes are needed to fully understand ancestral characters (ie, retained by at least one species of choanoflagellate). | |||
Separate studies have considered the emergence of [http://www.jbc.org/cgi/pmidlookup?view=long&pmid=18697744 the three collagen clades], [http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=18276888 cell adhesion via cadherins], the greatly elaborated [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=18621719 tyrosine kinase signaling network] with 128 tyrosine kinases, 38 tyrosine phosphatases, [http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=12869759 signalling origins], and 123 phosphotyrosine-binding SH2 proteins, and even [http://www.ncbi.nlm.nih.gov/pubmed/18621583? transcriptionally active LTR retrotransposons.] | |||
Opsins, though not really expected from known behavior or morphology, have been sought directly [http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0001054 without success] in Monosiga. Indeed, no GPCR with lysine in retinal position can be detected either. Monosiga has either lost this class gene or more likely never had them. This puts the spotlight on sponge, yet blastn of traces is not sufficiently sensitive. | |||
Alpha heterotrimeric G protein subunits that could bind opsin-like receptors have not been directly studied. Here we see the first alpha protein below has two introns identically located and phased to human to Gi class proteins but lacks the short exon and downstream introns. The second Gq class subunit has altogether different intronation despite 55% identity to the apparent human ortholog. These gene structures imply an era of active intron gain/loss in the era of the ancestor. Encephalopsin and melanopsin could have been supported had they been present but G alpha signalling there is a specialization of a much earlier evolved process. | |||
<br clear="all"> | |||
>GNAi_monBre Monosiga brevicollis 3 exons XM_001747738 55% GNAi_homSapheteromeric G protein alpha subunit Gi | |||
<span style="color: #990099;">0 MGICMSAEQKAQQARTAAVEAQLERDAQLASRTIKLLLL 1 | |||
2 GAGESGKSTLVKQMKIIHGDGFSNEELKSYKPTICDNLVHSMRAVLEAMGPLVIDIGDQVRPP 0 | |||
0</span> HAKVVLSYIELGTSGGLTPELTEALKALWADSGVQECFRRSNEYQLNDSAEYFFNNIDRIAQSNYLPTQEDVLRARVRTTGV | |||
IETTFRYKDLIYRMFDVGGQRSERRKWIHCFNDVTAVLFVAALSGYDMKLFEDQETNRIHESLTLFDAICNNSFFINTAIILFL | |||
NKTDLFSQKIARTPLKDYFPEYDGPPNNASEAKKFIAGMFKRLNKNPNKPVYEHFVCATETQNIRYVFDAVK* 0+ | |||
>GNAQ_monBre Monosiga brevicollis 7 exon form XM_001745795 55% GNAQ_homSap heteromeric G protein alpha subunit Gq | |||
0 MPCGPPDETRRRSLAIDRQLRKERMSKQREYKILLL GTGESGKSTIIKQMRIIYGQGFNESDRLAYKPLVYRNIITSMKRMLDALDQLSLQLADSSLEEDAYDK | |||
LDVDVNTVDAIEPYYPLLKKLWNDNGIQQVFQRRNEYQLSDSTAYYYNRLDAVAAADYIPTVDDVLRSRQATTGIHEFEFDLDSVVFRMMDVGGQRSERRKWIHSFE 0 | |||
0 GVTSIIFIAACNEYDQVLAEDTNVNRMQESLALFGQIIQYHW 2 | |||
1 FANSSFILFLNKQDLLEEKVKTHPIKPFFPDYTGQE 0 | |||
0 GDYENIKKFIETMYRSRKPAGKDLYTHFTMATDTSNIQFVFNAVRSTLLRIHLKDYNLF* 0 | |||
'''See also:''' [[Opsin_evolution|Curated Sequences]] | [[Opsin_evolution:_key_critters_%28deuterostomes%29|Deuterostomes]] | [[Opsin_evolution:_key_critters_%28ecdysozoa%29|Ecdysozoa]] | [[Opsin_evolution:_key_critters_%28lophotrochozoa%29|Lophotrochozoa]] | [[Opsin_evolution:_update_blog|Update Blog]] | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 15:40, 31 December 2011
See also: Curated Sequences | Deuterostomes | Ecdysozoa | Lophotrochozoa | Update Blog
Cnidaria .. 5+ opsins established
Biologists have belatedly realized that many molecular and morphological innovations attributed to chordates (or grudgingly to bilatera) actually track back much earlier to the common ancestor with cnidaria (Eumetazoa) if not earlier still to placazoa, sponges, and choanoflagellates. That's certainly true of photoreception. Two cnidarian genome projects have been more or less finished (Nematostella and Hydra) but that selection needs to be seriously expanded. Hydra especially repeats the whole mistake of sequencing a hugely derived genome with relatively little applicability to Bilatera based on a shallow 'model organism' approach (Hydra has only 66 publications in the last 30 years).
A scientifically neutral definition of eye needs to embrace the full variety of photoreceptors, including those with fewer "features" than the most complex. Probably the cutoff should be based on use of bona fide opsins classifying to the root of the encephalopsin, melanopsin, and RGR families covalently binding retinal and variants as agonist. Some purposes for an eye can be fully met by just perceiving and acting upon one "pixel", that is a simple photoreceptor eye with no pigment cup (that could provide directionality, two pixels resolution). Far too much emphasis has been given to the distinctness of lensed vision whereas such systems are evidently easily evolved and only a part of a broader photoreceptor continuum.
We don't say humans lack eyes just because a redtailed hawk has more pixels; we don't say humans lack color vision just because a turtle sees richer, sharper colors. When a simpler photostructure already suffices to distinguish day from night for gamete release, up from down for settlement, towards or away for predator evasion, cornea, lens, retina, and centralized nervous system are just baggage that can't be developed or maintained under darwinian selection. Cnidarian eyes exemplify this full range of possibilities.
Sponges and cnidarians have operated for immense timescales under selective pressure on huge population numbers on a steady body plan. Rather than frozen in time at some primitive condition as often portrayed (bio-bigotry), quite the opposite, they are fast evolvers that have had eons to perfect their genes and expression systems even as mammals played evolutionary catchup (eg human knee or defective LWS opsin duplication).
No living animal represents a long-gone ancestral node -- evolution never stops at the dna level even if outward morphology seems constant. All extant species have proven equally successful at survival. Evolution is not a story book progressing to human-- if cnidarians are so dumb and their vision so bad, how then are they able to chase, catch, kill, and eat advanced vertebrates?
Cubozoa: Tripedalia cystophora (jellyfish) .. 1 opsin
A landmark paper by Kozmik et al in the 24 June 089 PNAS has found the first convincing camera-eye imaging opsin sequence in pre-Bilatera, accompanied by homologous genes needed for the signaling cascade and melanin formation. That opsin classifies deeply within deuterostome TMT- and encephalopsin-class cilary opsins, just as expected (should such an opsin even exist in cnidarian photoreception) because pineal and retinal opsins descended via gene duplications from this ancient opsin class (which also has non-imaging representatives in protostomes) prior to lamprey divergence.
This new gene has the expected 7-transmembrane topology, conserved disulfide, ERY domain (as ERF), and conserved lysine for covalent chromophore attachment (with counterion predicted here at the ancestral 'E181' position EGV). Only the lysine and counterion are specific properties of opsins relative to generic rhodopsin-class GPCR. Other conserved residues specific to ciliary opsins serve to distinguish it from rhabodomeric opsins and contribute to its unequivocal blastp clustering within vertebrate ciliary opsins. These latter are slow-evolving and consequently the distance is less than to protostome ciliary opsins.
The authors suggest the NKQ motif (NRS in Tripedalia) at the start of the last cytoplasmic region is a reliable signature for ciliary transducin interaction (this was established experimentally as a contributing factor but only for bovine RHO1 and its co-evolving transducin Gt) but comparative genomics of 300 phylogenetically dispersed opsins shows this cannot hold in general as TMT opsins lack the basic middle residue and encephalopsins do not conserve the motif at all. Note rod and cone transducins cannot be tracked back orthologously even to tunicates. Melanopsins are not known from pre-Bilatera; at some point they coalesce with ciliary opsins, perhaps with this motif coalescing to the latter's pattern. Opsins can also trigger multiple signaling systems.
Tripedalia and its still-hypothetical Gt transducin have been co-evolving on a separate trajectory from the bovine pair for perhaps 1.2 billion years. The histories of gene duplication in the heterotrimeric G proteins may differ, along with the constraints in vertebrates of transducin having to simultaneously interact with multiple ciliary opsin genes. However as initial gene duplicates, Gt and Gq would have had the same binding site; even as the proteins differentiated to serve distinct opsin sets, the binding site (at the juncture of last transmembrane and last cytoplasmic sections) would likely remain conserved because of homologic inertia of the binding pocket and the implausibility of creating a new transduction mechanism.
It is easy enough to identify candidates for the photocascade alpha subunit of heterotrimeric G protein in cnidaria -- simple Blast of human GNAT2 protein (cone transducin) calls up two strong 64% identity matches in Nematostella, Hydra, and others (meaning the binding site could be accurately modeled). These could be close relatives of the implied Tripedalia ciliary opsin transducin (for which there is no data); extensive staining st would needed to make the case.
Actual ciliary opsin conservation in this boundary region looks like yNP.IY..mNkqFr.c (YNPVIYCLLNRSFRKM in Tripedalia) with the serine of NRS not observed in other species; the logos graphic shown below makes this quantitative. Note while this region looks very different in melanopsins (typically HPK in HNPIIYAITHPKYRM), there was never any potential for confusion given the much worse alignment there. The cubomedusan opsin also appears oddly truncated at the carboxy terminus; it appears to lack the cysteines need for palmitoylation and the serines/threonines for kinase activation.
Alignment of new cnidarian opsin with best Blastp in reference collection, compared to consensus sequence of all known ciliary opsins. triCys 47 SFLLNGLVIAVLIKYIRTITNTNIIVLSMSCANILIPLLGSPLSATSSLMRKWQFGNGGCTWYGFINTLSGISGIYHLTFLSFERFITIVLPLKRDTILSTKNIYIGLGILWVAAIGVAGAPVFGWCEYIKEGVRTSCSVAW bestBl +F +NGLVI V +KY + + N I+++++ AN+L+ + GS +S +++++ + G C + GF+ +L+GI G++ L +FER++ I P+ D K+ +G WV + P+FGWC Y+ EG+RTSC consen n............Lr.p.n..l.n...........g..........g....g...C...gf.....G......l.....eR......p...........a.......W........pPl.GW..y..eg....C.... xenTro 43 AFFVNGLVIVVTLKYKKLRSPLNYILVNLAIANLLVTIFGSSVSFSNNVVGYFFMGKTMCEFEGFMVSLTGIVGLWSLAILAFERYLVICKPMG-DFRFQQKHAILGCSFTWVWSFIWTSPPLFGWCSYVPEGLRTSCGPNW triCys -SSKENMNVFSYNLFMIFTVFLLPMLVIIYCNYRFIKEVSIMSTRARGLQGGDSEMTASASKAEKQLTIMVITMIIAFNIAWLPYTVVSMVFLTGYGDVVGPMGASVPSVFAKTSVIYNPVIYCLLNRSFRKML-----CGNS 325 bestBl + N N SY + + T F++P+ II+ +Y + +M+ RA Q DSE T +AEK++T MVI M++AF I WLPY ++V+ P AS+PS F+KT+ +YNP+IY +N+ FR L CG S consen ..........sy.....f.....P...i....Y..............................E..v..Mv..Mv..f...w.PY......................P....K....yNP.IY..mn.qfr........CG.. xenTro YTGGTNNN--SYIMALFLTCFIMPLSTIIF-SYSNL----LMALRAVAAQQKDSETT---QRAEKEVTRMVIAMVLAFLICWLPYASFAVVVAVNKDVVIEPTVASLPSYFSKTATVYNPIIYVFMNKQFRNCLMTLLCCGRS 316
The low overall percent identity (37%) of its best matches to any known Bilateran opsin, attributable to the great evolutionary time spans involved, disappointingly does not open any new doors. The sequence here does not have striking homology to cnidarian opsins recently proposed by several other groups, does not elicit dramatic new ones in sequenced cnidarian genomes or transcript programs, nor serve to locate opsins in sponge genome.
Note the other phototransduction cascade proteins co-located in the rhopalia by the authors have much more striking percent identity to both Nematostella and human homologs than the opsin, most astonishingly PDE6D at 80%. In terms of human gene names, function, and associated disease:
OCA2 49% EU310502 melanocyte membrane transporter of melanin precursor tyrosine (ocular and cutaneous albinism) MITF 48% EU310499 basic helix-loop-helix and leucine zipper transcription factor (auditory+pigmentary syndromes) PDE5A 48% EU310500 cGMP-specific phosphodiesterase 5A PDE6D 80% EU310501 cGMP-specific phosphodiesterase subunit delta recognizing prenylation GUCY2F 48% EU310503 rod outer membrane guanyl cyclase resynthesis of cGMP for recovery of the dark state
Most curiously, the authors observe that the major Tripedalia lens crystallin J1 protein is also strongly expressed in the nominally lensless slit and pit eyes. This raises the question of whether our concept of lens is too anthropomorphic and whether other anatomical configurations of high refractive index proteins can accomplish the same ends, possibly requiring an 'upgrade' for slit and pit eye functional assessment. For example, cone mega-mitochondria in treeshrews (refractive index 1.4) may have some lensing or waveguide function.
The opsins of slit and pit and larval eyes (implied by their photoreceptor cell structures) need to be determined with high priority. What's needed here too is a massive ortholog sequencing effort in the 19 extant species of cubozoan to break up isolated opsin long branch to show how such sequences are evolving, allow estimation of when they were recruited to imaging vision and reconstruct the earliest such ancestor. If a rhabdomeric opsin also occurs in larval or other eyes, that too needs comparative genomics.
It's unclear whether the last common eumetazoan ancestor of Tripedalia and Bilatera had imaging vision (note the hydozoan Cladonema radiatum has eyes and sponge larva have photoreceptor structures), yet clearly that ancestor contained one or more ciliary-type rhodopsin-class 7TM GPCR from which ciliary opsins descended in both clades, not always to be recruited for imaging. The fossil record for cubozoan cnidaria predates the Cambrian, though when eyes and statocysts (both regulated by PAXB) first appeared is unclear. Note further that planaria larva of Tripedalia have a rhabdomeric photoreceptor, suggesting melanopsin photoreception is also very ancient.
It's worth noting that the phylogenetic tree for early metazoans has entered a state of turmoil. Sponges may be secondarily simplified in the adult stage; ctenophores may be basal, and so forth. It's even been suggested that ancestral sponge larva represent the central object from which complex metazoan are descended.
These considerations suggest a very early evolutionary origin for the basic genes of photoreception and their regulation, with a great many lineage-specific subsequent upgrades and downgrades of the details, deuterostomes being the last to get on board with imaging vision. Thus the question Darwin asked, how many times did vision originate, requires a more nuanced answer than just a number. Most likely, the basic package of photoreceptor genes and their developmental regulation of expression arose just once, with all subsequent systems descended from that. However that package was subjected to numerous gene duplications and morphological variations in deployment, and outside recruitment in the case of crystallins and pigments.
Protostomes recruited melanopsin-class opsins for their imaging vision (despite available retained ciliary opsins), whereas early deuterostomes lacked imaging vision per se but retained ciliary opsins in related photoreception roles. Later post-amphioxus, post-tunicate deuterostomes independently recruited a descendent ciliary opsin (despite an available retained melanopsin-class opsin), moving from pineal to bilateral imaging eyes in the third and latest invention of imaging vision.
The spectral sensitivity of neritic (near-shore) lens eyes of a box jellyfish, Tripedalia cystophora previously considered by M Coates et al was interpreted as a single vitamin A-1 based opsin with peak sensitivity near 500 nm (blue-green). However nothing was sequenced. This species was most helpfully reviewed earlier by Piatigorsky and Kozkmik who note Eakin already commented on seemingly ciliary photoreceptors in 1962. However, 45 years later we still didn't know if opsins in cnidarians would classify with vertebrate ciliary opsins. They could even share conserved intron positions though that cannot be determined from transcript data.
Furthermore, as noted by Nordstrom et al, planula larvae of Tripedalia have a series of single-cell pigment cup rhabdomeric-like photoreceptors directly connected to motor cilia. These lack neural connections in line with Gehring's notion of the eye preceding the brain in evolution, rather than being a later add-on. So cnidaria might actually retain descendants of both types of ancestral opsins. No sequence is available yet for larvae,
Cubozoa: Carybdea rastonii (box jellyfish) .. 1 opsin
Cnidarians are the earliest diverging invertebrates with multicellular light-detecting organs. Photodetectors include simple eyespots, pigment cups, complex pigment cups with lenses, and camera-type eyes with a cornea, lens, and retina. These remarkable eyes are located on sensory clubs called rhopalia with four lining the bell. Each houses six eyes: a pair of pit ocelli, a pair of slit ocelli, and two unpaired lens eyes with counterparts to cornea, cellular lens and retina of ciliated photoreceptors.
Anatomically, the ocelli have bipolar sensory photoreceptor cells interspersed among nonsensory pigment cells with the apical end making the light-receptor with the basal end forming an axon that synapses with second-order neurons to form what amounts to ocular nerves. Vision has roles in the reproduction and feeding of cubomedusae which can find each other and chase, catch, and eat teleost fish. A patch of Pelagia nocticula 10 square miles in extent and 35 feet deep recently destroyed a salmon farm off Northern Ireland.
One of the most striking jellyfish from the perspective of complex eyes is Carybdea marsupialis, as reviewed by VJ Martin. Antibody studies based on vertebrate cone/rod opsins are not sufficient because of possible cross reactivity to generic GPCR proteins or non-imaging photoisomerases; no opsins have been sequenced yet. Provided the retroposon and base composition are not unwieldy, Carybdea could be an instructive genome to sequence. Nematostella and Hydra, whatever their other genomic merits, sit in the Anthozoa and Hydrozoa, clades of cnidarian lacking elaborate visual systems.
Carybdea rastonii has a green-sensitive visual pigment in its ciliary-type lens eyes utilizing Gs cAMP phototransduction cascade (that is, not Gt, Go or Gq). A complete opsin-like sequence AB435549 satisfies various opsin sequence signature requirements but does not classify clearly among known ciliary opsins. Instead, its affinities lie with opsin-like sequences from Hydra and Nematostella -- species that have 'too many' opsins for their meagre photoreceptive anatomy and photobehavioral capacities.
The second problem is the lack of blastp affinity of Carybdea protein to the new validated opsin from Tripedalia cystophora, which classifies as expected within bilateran ciliary opsins. This implies the last common ancestor of box jellyfish with bilaterans possessed a conventional ciliary opsin. What then is the need for other classes of opsin-like sequences? Other interpretations of opsin-like sequences need to be considered:
First, AB435549 may function more along the lines of peropsin/RGR/neuropsin (even though it does not cluster with them) as an auxiliary, possibly signaling or replenishing photoisomerase but not the primary imaging photoreceptor in Carybdea. In this scenario, AB435549 could hybridize more or less correctly in situ as would a better missing opsin -- remaining to be recovered -- more closely related to the Tripedalia opsin.
This still does not explain the observed clustering of AB435549 with opsin-like proteins, especially of intronless genes of Hydra and Nematostella with dubious connection to any kind of vision. Possibly the function here instead has to do with sensing or digestion of dietary carotenoids or photo-rearrangement of double bonds for biosynthetic, energetic or regulatory purposes (eg retinoic acid metabolism). In its wildest form, this hypothesis envisions metabolic photoreception as the core ancestral property that was later co-opted during the evolution of vision. Alternately, metabolic photoreception was a later spinoff of light sensing.
A third scenario just places AB435549 on another 'track' from bilateran opsins. Here it either arose from a conventional ciliary opsin after species divergence from last common ancestor or it is older and bilaterans subsequently lost all members of its gene tree class. The latter seems more plausible because AB435549 has no particular affinities to ciliary opsins relative to melanopsin or peropsin-type opsins. In this view box jellyfish have retained two systems with different retention patterns in different clades, in analogy to protostomes emphasizing melanopsins and deuterostomes ciliary in their imaging opsins. In support of this, the timing of box jellyfish divergences could be equally as old, even if they 'all look the same' from the human perspective. Here the Tripedalia group is then more relevant to the evolution of deuterostome vision whereas neither Tripedalia or Carybdea is helpful in understanding protostome vision.
It would be quite practical with 2009 technology to sequence a substantial number of complete box jellyfish genomes. This has the great advantage of allowing bioinformatic recovery of complete K-rhodopsin portfolios. This would settle the question of whether Carybdea possesses a ciliary opsin clustering with that of Tripedalia. Genomes also provide homologs of all auxiliary genes such as Galpha and RPE65. With a large set of proteins and rRNA, the timing of divergences within box jellyfish could be better estimated. It remains conceivable that cnidarians are paraphyletic, ie box jellyfish share a later divergence node with bilaterans, perhaps explaining common ground in eye structures not seen in anthozoa etc.
>CUBOP_carRas Carybdea rastonii sea_wasp Cnid.Cubo.Cary AB435549 cubop 18832159 MGANITEILSGFLACVVFLSISLNMIVLITFYRLRHKLAFKDALMASMAFSDVVQAIVGYPLEVFTVVDGKWTFGMELCQVAGFFITALGQVSIAHLTALAL DRYFTVCRPFVATAIHGSMRNAGMVIFVCWFYASFWAVLPLVGWSNYDVEGDGMRCSINWADDSPKSYSYRVCLFVFIYLIPVLLMVATYVLVQGEMKNMRGRAAQLFGSESEAAL KNIKAEKRHTRLVFVMILSFIVAWTPYTFVAMWVSFFTKQLGPIPLYVDTLAAMLAKSSAMFNPIIYCFLHKQFRRAVLRGVCGRIVGGNAIAPSSTAVEPGQTLASGTAES*
(to be continued)
Cubozoa: Chiropsella bronzie (box jellyfish) .. 0 opsins
This box jellyfish was featured recently in a comprehensive optical and micro-anatomical study of all four eye types. The genus Chiropsella is not currently known to GenBank taxonomy, meaning no sequence data at all is available (unless some synonym has been used). Its enveloping family Chirodropidae has barely 17 sequence entries, none relevant to vision. Chiropsella bronzie was first named in 2006; it occurs in knee-deep water feeding on shrimp along sandy beaches in North Queensland, Australia.
A 2010 immunohistochemical and microspectrophotometry study from the same group also avoided molecular data, instead using antibodies to five zebrafish imaging opsins to locate a single ciliary type opsin candidate in the upper and lower lensed eyes. Only SWS1 gave a reaction -- and this predominantly in the neuronal layer rather than the receptor cilia. The authors did not provide an accession number for the UV opsin but only a citation to a 1999 paper by TS Vihtelic et al. It is apparently AF109373 (which agrees with today's genomic).
Given the great divergence of cnidarians and very rapidly evolving teleost fish, a regenerative photoisomerase (if one exists) would not be detected by this method because the best match to known ciliary cnidopsins is already very poor at 29% -- well within range of a non-opsin GPCR (for example Trichoplax XM_002114725). It's unclear which region is both exposed to soluble antibody but also sufficiently and specifically conserved to provide the observed reaction yet not stain by the other antibodies. A non-opsin GPCR would not bleach, yet spectrophotometry is uncoupled to immunohistochemistry, so it is not known if the bleached compound matches the immunoreactive entity.
It would make more sense simply to sequence and assemble the entire genome (an afternoon's work), then analyze it for opsins and regenerative proteins using antibodies appropriate to the species.
TMT_triCys Tripedalia cystophora (box_jelly) identities: 29% positives: 50% best match to zebrafish SWS1
Query: 30 FYLQAAFMGFVFIVGTP-----MNGIVLFVTMKYKKLRQPLNYILVNISLAGFIFDTFSV 84
FY FMG FI G+ +NG+V+ V +KY + N I++++S A +
Sbjct: 29 FYNFYTFMGS-FIAGSACCSFLLNGLVIAVLIKYIRTITNTNIIVLSMSCANILIPLLGS 87
Query: 85 SQVSVCAARGYYSLGYTLCSMEAAMGSIAGLVTGWSLAVLAFERYVVICKPF--GSFKFG 142
+ + + G C+ + +++G+ + L L+FER++ I P +
Sbjct: 88 PLSATSSLMRKWQFGNGGCTWYGFINTLSGISGIYHLTFLSFERFITIVLPLKRDTILST 147
Query: 143 QGQAVGAVVFTWIIGTACATPPFFGWSRYIPEGLGTACGPDWYTKSEEYNSESYTYFLLI 202
+ +G + W+ A P FGW YI EG+ T+C W +K E N SY F++
Sbjct: 148 KNIYIGLGIL-WVAAIGVAGAPVFGWCEYIKEGVRTSCSVAWSSK-ENMNVFSYNLFMIF 205
Query: 203 TCFMMPMTIIIF-SYSQL----LGALRAVAAQQAESEST---QKAEREVSRMVVVMVGSF 254
T F++PM +II+ +Y + + + RA Q +SE T KAE++++ MV+ M+ +F
Sbjct: 206 TVFLLPMLVIIYCNYRFIKEVSIMSTRARGLQGGDSEMTASASKAEKQLTIMVITMIIAF 265
Query: 255 VLCYAPYAVTAMYFANSDEPNKDYRLVAIPAFFSKSSSVYNPLIYAFMNKQF 306
+ + PY V +M F ++P+ F+K+S +YNP+IY +N+ F
Sbjct: 266 NIAWLPYTVVSMVFLTGYGDVVGPMGASVPSVFAKTSVIYNPVIYCLLNRSF 317
The picture of eye functions that emerges here is rather surprising. Both upper and lower lens eyes are severely under-focused (much more so than Tripedalia cystophora) with the retina so close to the lens that only blurred vision can result. And these are its best eyes. A novel long pigment cell has dark pigment moving within a white pigmented tube during light/dark adaptation for unknown advantages.
Since rhopalia eyes have seemingly had several hundred million years to evolve deeper vitreal space (which seems simple enough), an eye tuned to detect large structures at short range (spatial low-pass filter) evidently suits Chiropsella. The primary function may be visual avoidance of obstacles or detection of prey within range. Higher spatial resolution and rapid refreshing entail a concomitant expansion of the nervous system and higher ongoing energetic costs to adaptively process massive extra information.
The skyward pointing upper lens eye has various peculiar features from our perspective. The ellipsoid lens lacks focusing refractive power, has a cataract-like inclusion casting a shadow on the retina, a hole in connection with the pigment layer exposing the retina to direct sunlight, balloon cells partly covering the lens aperture, gastric cells contacting the posterior lens side, and less of a pupillary response. Although little is known on the biological side, the upper lens-eye shadow line capability could be suited for detecting sun or moon position.
The two smaller pit and slit eye types have an epithelium/cornea covering but do not contain a lens. The photoreceptors are pigmented and organized into ciliary, pigment and neural layers. These eyes are capable at best of monitoring ambient light intensity, perhaps guiding overall phototactic behavior or orientation. Yet this proposed function could seemingly be accomplished as a byproduct of lens eye functionality and would not require paired pit and slit eyes, much less four sets of them around the full quadrant of rhopalia.
While vertebrates too have anatomically separated photoreceptors for diverse functions, there are meagre prospects for homologizing here to melanocyte, ganglial, pineal or other deep brain structures. In fact even the main lower lens eye may not descend from an ancestral structure that, in another clade, became a bilateran eye because apparent common ground can originate multiple times just from convergent considerations of optical physics. Thus a 'cornea' is merely a protective epithelial layer and a lens just a thickening filled with overproduced protein providing refractive index.
Even if the old mystery of the origin of the eye can been pushed pack through homologization to the mystery of the origin of the rhopalium, no 'intermediate states' are likely be found in the Cambrian jellyfish fossil record and seemingly not in other living species of box jellyfish (all of which have the (2+2+1+1)x4=24 eye pattern).
It is not clear whether distinct opsin genes are utilized in these various eyes and if so, what their gene tree might look like, eg ((pit,slit),(upper,lower)) vs (((pit,slit),upper),lower). We are left wondering too about the origin of rhabdomeric melanopsins if ultrastructure in pre-bilaterans is always specialized to modified cilia.
Anthozoa: Nematostella vectensis (sea anemone) .. 3 opsins
The Nematostella genome has been released along with major papers and an upgrade to Stellabase. Not all 6.1 million traces were used up by the assembly, so any gene missing from the assembly should be sought directly in the trace archives.
The sea anemone, an anthozoan within Cnidaria having epithelial cells, neurons, stem cells, complex extra-cellular matrix, muscle fibers, and symmetry axis, is emerging as a high-profile evo-devo model species to elucidate the emergence and deployment of genes that determine animal body plans. However those plans don't seem to include eyes or overt photoreceptor structures such as pigment cells -- for that cubomedusae would be far better. PAX6 and RX are especially relevant to photoreceptor structures; their expression has been thoroughly studied in Nematostella without uncovering any sensory system though they contribute to patterning specific components of the ectodermal nerve net.
The JGI annotation pipeline produced a number of extensively annotated gene models for Nematostella opsins. These are available simply by keyword lookup, tblastn of various queries the best of which turn out to be -- unsurprisingly -- an encephalopsin subclass from Branchiostoma. It is important to credit the JGI staff for providing the relevant bioinformatic track computations because they were first to characterize and release these opsins into the public domain (eg GenBank NR and Entrez Gene). It does not constitute independent "discovery" to perform keyword lookup and copy out other peoples' work. Without proper citation, that's plagiarism.
I extended improperly truncated JGI gene models (ie those lacking iMet and stop codon), validated the extensions still lacked introns (GT-Ag splice junctions missing at positions expected from closest homologs), placed the best 3 (of a half dozen) in the Opsin Classifier with fasta headers, noted their best matches below, and validated lysine and counterion glutamate in the expected positions. All this is consistent with (but does not prove) a role for ciliary Gt opsins in pre-Bilateran photoreception.
We expect cnidarians (maybe not this particular anthozoan) to have both melanopsins and encephalopsins. Our tendency is to think that imaging eye opsins, whether insect rhabdomeric or vertebrate ciliary, are the main attraction, with the other opsins playing out obscure roles in secondary functions like timing of gamete release. That's quite wrong-headed. Deeper gene family trees show that the melanopsin and encephalopsin constitute the primary photoreceptors. Over vast evolutionary time scales, they gave rise to various spin-offs in various clades at various times through gene duplication and subsequent neofunctionalization. At even greater phylogenetic depth, melanopsin and encephalopsin are themselves related by gene duplication of an ur-opsin, which itself arose as a duplication of an established non-opsin GPCR. As noted by Arendt, that exploited prior gene duplication within the alpha subunit of heteromeric G protein and profound diversification in signaling system second messaging.
The odd thing about all these cnidarian encephalopsins is their lack of introns (three ancestrals are expected). That's very unlikely to be the Eumetazoan ancestral state for encephalopsin because Nematostella is no rogue organism when it comes to intron conservation. A common explanation for this within eukaryotic bioinformatics is gene duplication of a master gene via fully processed retrogenes (rather than through tandem, segmental, chromosomal, or whole genome duplications -- all of which preserve introns). Mixed mechanisms are also common (as in olfactory receptors): an initial intronless retrogene is duplicated tandemly etc. These paralogs can even displace the master gene by taking over its function, causing it subsequently to be displaced or even lost. That scenario played out within zebrafish opsins.
If so, we might expect Nematostella encephalopsins to be more closely related to each other than any known opsin from any species. Indeed ENCEPHa_nemVec is 90% identical to ENCEPHb_nemVec and 52% with ENCEPHc_nemVec, whereas only 39% identical to the best bilateran opsin, ENCEPH4_braFlo of amphioxus. Those are profound differences -- mammalian proteins typically take 100 myr to lose 10% of their percent identity. Here though we know next to nothing about clade-specific rates and have very long branches indeed. Of course, a seven-transmembrane protein has very different evolutionary constraints from the generic globular cytoplasmic protein to which off-the-shelf phylogenetic software is tuned, so no purpose is served applying that.
It appears the three Nematostella proteins may share a distinctive rare genetic event, an indel in a loop region. That would favor a common history. It will prove difficult to resolve indels as to insertion or deletion for lack of suitable outgroup.
Given an finished genome, the mode of gene amplification can be explored by looking at flanking genes. Perhaps ENCEPHa_nemVec and ENCEPHa_nemVec are adjacent (ie tandem duplication) or perhaps their flanking genes are paralogous (syntenic segmental duplication). However the Nematostella genome is currently unfinished and the (gapless) contigs containing the encephalopsins run about 10 kbp. Depending on gene density that can be too small to establish synteny. These contigs, separated by strings of N's of unknown length, are further assembled into larger scaffolds (ample for synteny), a process usually trustworthy at highly experienced JGI but sometimes confounded by issues such as repeats, compositional simplicity, very recent duplicative regions, and clonability.
The most convenient approach here is tblastn of ENCEPHa_nemVec against the wgs menu item at NCBI Blast, specifying Nematostella. The three genes here are on different scaffolds altogether, ruling out tandem position. The nearest flanking genes can be extracted by blastx of the enveloping contig (or whole scaffold) against GenBank protein. JGI has in effect already done this, as could be seen by expanding out from the initial browser view. Comparing 3 browser views is complicated by the fact that flanking paralogs might be named differently, but that is readily overcome by collecting sequences (noting strand orientation) into a mini-database and comparing within uBlast.
Notice the Opsin Classifier collection already contains the outcome of this process as a fasta header field (for deuterostome opsins). It is conceivable that orthology of a Nematostella opsin to say a Branchiostoma opsin could be established in this way (synteny). However gene order in both genomes has been independently scrambled over immense time scales and orthology would have been to the Nematostella master gene (with introns) that appears lost. It's better to build out from a local synteny chain but that requires data from additional cnidaria. Note the irony here in that the farther removed the genome from human, the more densely they must be sampled.
It's evident from a casual ClustalW alignment, after marking up columns for membrane-spanning sections and considering hydrophobicity, that Nematostella opsins conform to the standard central pattern. That's unsurprising since proteins retain 3D structure at far lower percent identity and the pattern here cuts much deeper, into the overall rhodopsin superfamily and beyond to generic GPCR. However encephalopsins can have very considerable extensions at their amino and especially carboxy termini that need separate consideration.
For now, sequences can be trimmed to whatever is alignable across the full spectrum of ciliary opsins. Recall that by design the Opsin Classifier collection seeks maximal phylogenetic dispersion to mitigate over-weighting by over-studied species that might introduce clade-specific interpretive bias. That could also be done by distilling the dataset down to ancestral sequences at lamprey divergence, the risk there being co-evolution of non-adjacent residues (eg different alpha helices) can be lost in residue-by-residue ancestral reconstructions.
As noted, Nematostella opsins are at best 39% identical. These had better be strongly concentrated at invariant and near-invariant ciliary opsin positions rather than randomly distributed. Blastp of course doesn't know the difference. We know at the outset this strong association will occur for any GPCR to the extent that it is reliably alignable, so the question really is whether conservation is concentrated at the conserved positions specific to ciliary opsins (ie conservation not shared with Go and Gq opsins). This has all been studied before but not nearly at the phylogenetic depth made possible by comparative genomics. There is always a need in remote opsins for independent support (here stratified signature residues) of candidates suggested by blast searches.
For that, it is most convenient to cut conservation tranches with Corpet's Multalign because user-specifiable line width can set breaks after structurally meaningful locations. Here the cutoff for invariant is set variously at 100%, 95%, 90%,... (with Nematostella omitted) and the stack of consensus lines retrieved. That results in a nuanced version of invariance that can be set off against the Nematostella sequence at those positions. For "controls" rhabodomeric opsins, rhodopsin superfamily, and generic GPCR generate their own stacks. (Alternatives such as logos or the misnamed evolutionary trace would give similar outcomes. None of the methods make use of the known phylogenetic tree relating the sequences.) The bottom line here will be that these new cnidarian opsins will have conserved residue signatures specific to a conventionally functioning ciliary opsin, though ultimately that can only be tested by experiment.
>ENCEPHa_nemVec Nematostella vectensis (anemone) no cdna complete 1 exon 306 aa best:ENCEPH4_braFlo scaffold_465_Cont27987 alt: Nemve1:219988 Nem1 >ENCEPHb_nemVec Nematostella vectensis (anemone) NC-extended 1 exon 275 aa best:ENCEPH4_braFlo scaffold_273_Cont21871 alt:Nemve1:130042 Nem3 >ENCEPHc_nemVec Nematostella vectensis (anemone) C-extended 1 exon 289 aa best: ENCEPH5_braFlo scaffold_11_Cont2404alt: Nemve1:85309 Nem2 ENCEPH4_braFlo Branchiostoma floridae (amphioxus) Gt 0.... 470 7.0e-48 39% identity to ENCEPH4_braFlo ENCEPH4_braBel Branchiostoma belcheri (amphioxus) Gt 0.... 449 1.2e-45 PER_xenTro Xenopus tropicalis (frog) ?? 0.2.0.2.1.0... 438 1.7e-44 ENCEPH4a_takRub Takifugu rubripes (teleost) Gt 0...2...0... 435 3.6e-44 PER_homSap Homo sapiens (human) ?? 0.2.0.2.1.0.1 in... 426 3.2e-43 ENCEPH4b_takRub Takifugu rubripes (teleost) Gt 0...2...0... 418 2.3e-42 ENCEPH5_braFlo Branchiostoma floridae (amphioxus) Gt 0.... 418 2.3e-42 ENCEPH_gasAcu Gasterosteus aculeatus (stickleback) Gt ... 415 4.7e-42 PER_monDom Monodelphis domestica (opossum) ?? 0.2.0... 411 1.2e-41
Four putative opsins have been proposed by Plachetzki et al. Accessions of the supporting gene models are given in the JGI protein ID system (non-GenBank) as Nematostella1 219988, Nematostella2 85309, Nematostella3 130042, and Nematostella4 108738 (or fragments in the alignment graphic allow recovery of the respective cdnas by tblastn of GenBank WGS). As noted in the Hydra section, multiple lines of evidence are necessary to establish the first bona fide opsins in cnidarians.
There appear to be 2 Nematastella opsin-like cdnas at GenBank that cannot be found in the genome assembly or trace archives, DV091537 and DV087469. While genes can be missing from first assemblies, it is bizarre for both to be missing considering coverage is 6x. Upon back-blast to GenBank nr or the Opsin Classifier, very strong matches are seen consistently within crustacea. Thus it appears that these Sars Institute products are contaminants from another species, possibly a brine shrimp widely used in aquarium food. It is not unusual to see transcript (at issue here) and genome projects contaminated with dna from other species such as commensals, parasites, and food source -- this is reminiscent of Xenoturbella being confused with a mollusk in its diet.
New Nematostella transcripts continue to be posted by JGI into mid-Dec 2007. Using proxies for all possible queries, I located a possible melanopsin and possible rhabdomeric LWS counterpart, The former had two coding exons but not at a melanopsin position; the latter had but one. These are fairly weak matches and further characterization is needed. They're stored in the Opsin Classifier as MEL_nemVec and LWS_nemVec2.
A third group has taken a serious look at photoreception in Nematostella. No paper or dissertation has emerged as yet; no cnidarian opsins have been posted to GenBank.
The claim of orthology will prove exceedingly difficult to establish in a 700 million year long branch. It is not a property of a gene tree per se. By definition, two genes in species A and species B are orthologous if and only if they have descended vertically from the same single parental gene in their last common ancestor. The last component is exceedingly important because all opsins -- indeed all GPCR -- are ultimately descended from a single gene. However that single gene was not to be found in the common ancestor of cnidarian and bilaterans because sponges already appear to have classical opsins and perhaps hundreds of GPCR.
Most ancestral introns in human genes were established in unicellular eukaryotes well prior to fungal and green plant divergence. For example the distinct introns in close paralogs SUMF1 and SUMF2 were in place before human/diatom separation. It's very difficult to imagine how the introns in neuropsins, rgropsins, peropsins, melanopsins, encephalopsins, pteropsins, and ciliary opsins could have descended from a single gene in Eumetazoa.
Evolution of photoreception: the eyeless anthozoan Nematostella vectensis as a model Poster talk March 22-23, 2007 Heather Q. Marlow, Daniel I. Speiser, David Q. Matus and Mark Q. Martindale (Email: marlow@hawaii.edu) "Eyes have evolved numerous times within the animals, yet there has been surprising convergence in the morphology, function and molecular basis of development in these structures. Although these diverse eye types have arisen independently, many taxa utilize similar cassettes of genes to specify them. These developmental genes include members of the SIX class of homeodomain proteins (sine oculis and optix), eyes absent, dachshund and famously, the Pax genes (Pax6). Additionally, all animals in which photoreception has been investigated use the opsin family, a class of seven transmembrane receptors, to detect light. Cnidarians are an early branching lineage that are likely to have diverged from the rest of the animals before the evolution of discrete eye structures. The ancestral cnidarian did not posses eyes, however like the extant anthozoan cnidarians (sea anemones, corals, and sea pens), it was likely to have had photoreceptive cells. In order to determine the level at which cnidarian photoreceptive cells may share homology with bilaterian eyes, we have examined the expression of these “eye” genes during development in the anthozoan cnidarian model Nematostella vectensis through in situ hybridization. We have also identified, cloned, and studied the expression of many members of the visual opsin class of receptors in N. vectensis. Our data indicate that N. vectensis possesses putative photoreceptive cells which express several orthologs to the visual opsins, that the organization of photoreceptor cells differs between different life history stages of the animal, and that presumptive photoreceptor cells express many of the same developmental molecules that specify eye development in bilaterian animals. These findings support the hypothesis that eyes may share homology only at the level of the photoreceptor, and that additional “eye” genes may have been co-opted into the eye specification pathway from more general neural roles in bilaterians."
A fourth group published a 19 Dec 07 paper on putative anthozoan and hydrozoan opsins, releasing 54 full length sequences to GenBank. These include 31 full length intronated predicted genes for Nematostella, 21 mRNA for lens-eyed Cladonema radiatum, and 2 for eyeless Podocoryne carnea. The latter two species are hydrozoa without genome projects meaning the transcripts cannot be intronated. Many of the 54 proteins have best-blastp below 30% identity within the 230 validated opsins of the phylogenetically comprehensive reference collection. This is worse than some generic non-opsin GPCR, so almost all of the residue matching will be non-specifically exhausted. All have lysine in homologous position, so potentially covalently bound retinal (though that was not established chemically). The counterion situation does not work out at either E113 or E181. Five are missing the universally conserved early asparagine and two others are truncated.
Conserved residues in putative cnidarian opsins relative to bovine rhodopsin and consensus sequences for ciliary, melanopsins, pteropsins, peropsins, and all validated opsins.
..........................................................*...................................................................*..................................................................................................................*..................
rho1 NFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFR
cili N.lv...t.k.k..LrPlN.ilvNla.a#l.....g..........gyfG.....C..eG%...l.G.v.lwsl.vla.dRy.v!ckp.g..f.a.g.........f.....W..pPl.GWs.Y.peg...sC...w.....s%..f.c...........Pl.i....Y..l.........aE..v.rM!..M!..%l...........cW.PYaa........p..P...........faKss.%NPi.IY.f$Nk#fr
cnid N..vi..................s.a..d..........................C...gf........si.hl.....ery........................W.....w...Pl.GW..y..e.....C...w.....sY............l..%P.................m....i..%......................aWtPYa..............l.........fAK.s..nP...%......fr
vali N..V.......k..LRP.N...vNLA..Dl...................g.....C..yg%.....G..s...$..ia.dRY.v!..P......a...........W.....w...Pl.GW..Y.pEg..tsC..#w.....s%.............f.%Pl!I.%..Y..i..........E.....m...m!..F.............W.PYa.........p..P...........fAK.s.%NP!.IY......%R
mela N.lv...f...ksLrtp.N.fIiNLA.sDf.ms....P....s.....W.fG...C.lYaF.g.lfG..S..t$..Ia.DRY.v!t.Pl..s.r.i.v........W.ysl.Ws.pP.fGwg.YvpEG..tsCt.D%.t..r.%.$.f.FPl.i..cY..if.a!r....#.k.ak.........%.......................sW.PYa.!.lG..ltpy.P............AKSai.NPi.iYa..hpkfR
pter Ng.V!.!F..tKsLRTPsN$lV!NLA.sDf.MM..m.Ppm.nc%.t..w.lG...C#.Ya..Gsl.Gc.siwtm..Ia.DRYnvIvkg.p$t..Ali.........W.....W...P.fgwnRYVPEGn$TaCgtDYLt.srs%.ys.vYP$.I!%.Y.fIv.aV.aHEkE.rlAK.vAl.t.sLwf......................aWTPY..!n.G...tPl.ti............k.a...p..vy.ishp.yr
pero N..v...f.k........#....nLA..D.g!s..g.p....S.....W.%G.G.Cq.ygf.gf.fg..Si...t.!a.DRY..iC......$.............W...afWa..Pl.Gwg.YEP.g..t.Ctl#w......%...............%P.!m....Y..!..K.k.....tk............%l...........aW.PYa!..w..f..p.ip.$..........AKs...NP..!Y...#..fr
rho1.NFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFR
Anthozoa: Anemonia viridis (symbiotic anemone) .. 1 opsin
This ciliary opsin fragment is associated with an October 2009 metagenomic article on the symbiotic coral reef organism Anemonia viridis and its dinoflagellate algae but was not annotated in the EST collection (FK729339). By back-tblastn, it is clearly a rhodopsin-type GPCR with Schiff base lysine (but unknown chromophore).
It is most closely allied with ENCEPHb_nemVec of Nematostella vectensis in the currently known set of cnidarian opsins -- about 49% identity but not enough is known about Anemonia opsin multiplicity to say it is an ortholog. High identity might be due either to sequence conservation or close relatedness of these two genera -- their time since divergence is an unknown. Conservation (of sequence and hence function) seems more likely since -- at least at GenBank taxonomy -- the divergence is quite deep:
Cnidaria; Anthozoa; Hexacorallia; Actiniaria; Nynantheae; Actiniidae; Anemonia Cnidaria; Anthozoa; Hexacorallia; Actiniaria; Edwardsiidae; Nematostella Cnidaria; Anthozoa; Hexacorallia; Scleractinia; Astrocoeniina; Acroporidae; Acropora >ENCEPH_aneVir Anemonia viridis (symbiotic sea anemone) frag:202-338 pubmed:19627569 KDAVARWGNKSPPTQQTMQAQKKTIRMSLVMVFAYLLAWTPYALTSLYSSFIASDITPLLSVMPALFAKLSSCYNPIIYFFMYSKFR KAAKKMIRRNLVGHDSNSGQGVSNTFATSFPRPISFLRYKRSAVAPLSDIPQVSSVDLPQVGRENDVTVQQDKASEINT* aneVir KDAVARWGNKSPPTQQTMQAQKKTIRMSLVMVFAYLLAWTPYALTSLYSSFIASDITP-LLSVMPALFAKLSSCYNPIIYFFMYSKFR +D++ RWG S TQQT+ A++KT MS +MV A+L AW PYA+ SLY+SF P L+S +PA+ AK S+CYNPIIYFFMYSKFR nemVec RDSMRRWGASSDSTQQTLAAERKTAWMSFIMVLAFLFAWVPYAVVSLYASFGGVTTIPKLMSTLPAMLAKTSACYNPIIYFFMYSKFR
Anthozoa: Acropora millepora (stony coral) .. 4 opsins
A new cnidarian opsin appeared at GenBank on 13 May 2009 based on the 454 transcriptome survey SRA003728 of 5-day-old planulae larva in Acropora millepora. The entry EZ013658 generates 244 alignable residues (after a few N's are manually corrected using genetic code redundancy) of a melanopsin-type protein; unfortunately it terminates 20 residues short of the expected Schiff lysine (so 38 residues short of a full motif).
Extending this sequence to the end of the seventh transmembrane segment is a high priority. That would likely raise the percent identity (excluding tails) over 40%, far beyond agreement of opsins to generic GPCRs. Even as it stands, the conserves early signature residues of melanopsins within opsins and opsins within GPCR and cleanly clusters with melanopsins at the opsin blastp classifier. With additional orthologs from other cnidaria (Nematostella genome lacks one), a better ancestral sequence could be worked out at the divergence node with bilatera. This sequence in turn may be as close as we get to the origin of melanopsic photoreception.
On 29 May 2009, blastn of the 454 Short Read Archive became available at NCBI. While EZ013658 matched 3 reads quite well, it was not fully tiled. These reads allowed various errors to be corrected but best of all, 56 extra amino acids to be added C-terminally. These included a standard Schiff motif KTASVYNPIIYFFSYKSFR. It remains a bit mysterious as to where the central region of EZ013658 came from in its assembly.
The best matches within cnidaria are to Nematostella K-rhodopsins classifying to TMT/ENCEPH, even while being more distant to Nematostella opsins classifying as melanopsins (eg BR000662, suggesting these opsin homolog classes converge in cnidaria. This means their signaling partners cannot be safely inferred from sequence alone; indeed Galpha programs themselves have complicated lineage-specific expansions.
A sub-sequence of EZ013658 was correctly identified as melanopsin-related, though not linked to circadian rhythm, in a brief April 2009 study of 24 circadian rhythm genes and the extraordinarily light-driven reproductive timing of broadcast-spawning corals (which may utilize cryptochrome photoreceptors rather than opsins).
Special care must be taken in cnidarians to maintain a rigorous definition of opsin photoreceptor candidates and not digress to deeply diverged non-opsin GPCRs lacking ability to bind chromophore and -- as far as we know -- any relevance to photoreception. Even with K-rhodopsins, it remains quite possible that some are merely involved in light-driven catabolism or rearrangement of dietary beta-carotenoids.
The four sequence fragments below can only be evaluated over their coverage, though the ones here all include a standard K296 NP FR motif and gave only recognized opsins upon backblast to GenBank nr. Two of them, upon blastp to the 485 curated opsin classifier, give best matches to deuterostome TMT and encephelopsins, suggesting a closer relationship to bilateran ciliary opsins than most cnidarian K296 opsin-like proteins do.
Alignment of Acropora 454 transcript with human melanopsin
Blue shows the first two cytoplasmic domains; red invariant disulfide and trigger motif; magenta Schiff lysine end motif.
acrMil 1 HHTISFLYFLLALFSFSLNSVVILTFLLDRSLLFPANLIILSIAISDWLMSVVPNIMGGVANASNDLPFTDWSCTVFAFVATLLGLSNMLHHAAFALDRYMVITRPMRANH--SMTRILA 118
H+T+ + L+ L N VI TF RSL PAN+ I+++A+SD+LMS + ++ F + C +AF L G+S+M+ A ALDRY+VITRP+ S R
homSap 70 HYTLGTVILLVGLTGMLGNLTVIYTFCRSRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGETGCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAF 189
acrMil 119 VIAFLWCFALTWSLFPLVGWSAYVREAGDIACSVNWQSDNPSDSSYMVCLFFFFYFVPLAIIVYCYVFMIRSVRFMTKNAQKIWGV-----RSAAALETVQATWKMAKIGLIMVLRFFVAWTPYAVVSFIIAF 244
V+ +W +AL WSL P GWSAYV E +CS ++ S P R+Y + L F +F+PLII+ YCY+F+ R++R T A + +G S + +Q+ KMAKI L+++L F ++W PY+ V+ ++AF
homSap 190 VLLGVWLYALAWSLPPFFGWSAYVPEGLLTSCSWDYMSFTPAVRAYTMLLCCFVFFLPLLIIIYCYIFIFRAIR-ETGRALQTFGACKGNGESLWQRQRLQSECKMAKIMLLVILLFVLSWAPYSAVA-LVAF
acrMil DSVKDIPT-IAEIVPSMFAKTASVYNPIIYFFSYKSFRESLVK 288
+ T VP++ AK ++++NPIIY ++ +R ++ +
homSap AGYAHVLTPYMSSVPAVIAKASAIHNPIIYAITHPKYRVAIAQ 363
>MEL1_acrMil Acropora millepora (stony coral) 454 transcriptome shotgun assembly EZ013658 + 454 blastn, frag 40%/63% ENCEPHc_nemVec; 35%/57% MEL1_homSap
HHTISFLYFLLALFSFSLNSVVILTFLLDRSLLFPANLIILSIAISDWLMSVVPNIMGGVANASNDLPFTDWSCTVFAFVATLLGLSNMLHHAAFALDRYMVITRPMRANHSMTRILAVIAFLWCFALTWSLFPLVGWSAYVREAGDVACSVNW
QSDNPSDTSYMVCLFFFFYFVPLAIIVYCYVFMIRSVRFMTKNAQKIWGVRSAAALETVQATWKMAKIGLIMVVGFFVAWTPYAVVSFIIAFDSVKDIPTIAEIVPSMFAKTASVYNPIIYFFSYKSFRESLVKSWRRYRNRNNVWPL
>ENC1_acrMil Acropora millepora (stony coral) EZ018307 frag 454 transcriptome most like CNOPa1_monFav Montastraea faveolata
IAFHLRKVSKTAKRTWGKNSQITKDGVIVKRKAVTHGAIMVTxVMITWLPYSMVAFYTLLGFEKVKLSALAYTITSMFAKTSTLVNPVICFFWYRRFREGTKKLCNRVMDFFLRKHRRASNT
>ENC2_acrMil Acropora millepora (stony coral) EZ007079 EZ005208 frag 454 transcriptome most like ENC_aneVir Anemonia viridis
RAKSTWGRQSPFTKRTFVMERKMMALFGVMTVAFLVAWTPYAVVSLISMIGGPDVISDVTASIPAYFAKSSSCYNPVIYVFLYKRLRRQMRFAVRRD
>CNOP_acrMil Acropora millepora (stony coral) EZ007080 frag 454 transcriptome
TLVSLTSIFRGNSVLSTGEAEVPALMAKASVIYNPIVYTVMNRRFRRTLRHIVSCMTCRLLSFVWPTMHREKQETKKR
Anthozoa: Acropora digitifera (stony coral) .. 13 opsins
An excellent new cnidarian genome assembly (Acropora digitifera) became available on 24 July 2011. This species diverged from sea anemone Nematostella vectensis approximately 500 million years ago so naturally the opsin-like proteins are quite diverged and by no means in 1:1 correspondence.
This coral has 7 complete, intronless opsins. Three of these have clear orthologs in the Acropora millepora transcriptome (all fragmentary genes). Coral also has a long open reading frame with an opsin sequence in front apparently fused to something else (not a known domain). This corresponds to a similar long open reading frame in Nematostella.
Acropora digitifera has five additional opsins called CNOP1-5 below. Four of these are full length. All have a phase 21 intron break at the same position. This does not match any of the ancestral introns in position. Nematostella has this intron too though contig misassembly -- homopolymer run error leading to frameshift -- creates a necessity for manual curation. This intron was thus present at the divergence of Nematostella and Acropora.
CNOP2 has an earlier phase 12 intron as well. This matches -- in position and phase -- the first intron of bovine rhodopsin (position 120 (ATLG 12 GEIA) and one found in numerous arthropod ciliary opsins. It thus appears that this intron was already present in an opsin in the last common ancestor of bilaterans and cnidarians.
These are the first introns to be located in cnidarians. They are moderately short (eg 533 bp and 788 bp in CNOP2) and have conventional GT-AG donors and acceptors. Because of extreme divergence, it is unclear whether the much more common intronless cnidarian opsins arose via retrogene processing from them.
CNOP1-5 have conventional K-296 motifs but lack conserved counterparts to the DRY motif. This is not wholly unprecedented in bilateran opsins or in GPCR but does imply unusual functioning (perhaps something other than vision). It should be stressed that CNOP1-5 all give best-blast matches at GenBank to known opsins rather than to other GPCR. Nothing is known experimentally about sites of expression or photobiological anatomical correlates.
Within the opsin collection, CNOP1-5 give best matches either to melanopsins or ciliary opsins, so have no bearing on the origin of the peropsin/neuropsin/rgropsin group. The table below shows that CNOP2 gives consistently better matches to melanopsins than to TMT, though no dramatic scoring shelf separates the two. Branchiostoma opsins again occupy an unusual position.
CNOP2_acrDig Acropora digitifera (stony_coral) BACK01019215 1867 1.4e-195 100% CNOP4_acrDig Acropora digitifera (stony_coral) BACK01002540 766 6.7e-79 47% CNOP3_acrDig Acropora digitifera (stony_coral) BACK01018... 733 2.1e-75 45% CNOP5_acrDig Acropora digitifera (stony_coral) BACK01002513 729 5.5e-75 46% CNOP2_nemVec Nematostella vectensis ABAV01018948 688 1.2e-70 41% TMTx_braFlo Branchiostoma floridae (amphioxus) Deut.Cep... 382 3.3e-38 31% MEL1_otoGar Otolemur garnettii (lemur) Deut.Euth.Euar g... 381 4.2e-38 31% MEL1_phoSun Phodopus sungorus (hamster) Deut.Euth.Euar ... 380 5.3e-38 30% TMT_triCas Tribolium castaneum (flour_beetle) Ecdy.Ins... 377 1.1e-37 MEL1_ponAbe Pongo abelii (orangutan) Deut.Euth.Euar gen... 375 1.8e-37 MEL1_micMur Microcebus murinus (mouse_lemur) Deut.Euth.... 370 6.1e-37 MEL1_myoLuc Myotis lucifugus (microbat) Deut.Euth.Laur ... 369 7.8e-37 MEL1_rheMac Rhesus macaca (rhesus) Deut.Euth.Euar genom... 368 1.0e-36 MEL1_musMus Mus musculus (mouse) Deut.Euth.Euar AF14778... 367 1.3e-36 MEL1_canFam Canis familiaris (dog) Deut.Euth.Laur genom... 367 1.3e-36 TMTa1_danRer Danio rerio (zebrafish) Deut.Acti.Otoc AF34... 366 1.6e-36 MEL1_homSap Homo sapiens (human) Deut.Euth.Euar NM_0332... 366 1.6e-36 MEL1_panTro Pan troglodytes (chimpanzee) Deut.Euth.Euar... 366 1.6e-36 MEL1_ratNor Rattus norvegicus (rat) Deut.Euth.Euar AY07... 365 2.1e-36 MEL1_proCap Procavia capensis (rock_hyrax) Deut.Euth.Af... 364 2.6e-36 MEL1_taeGut Taeniopygia guttata (finch) Deut.Saur.Arch ... 364 2.6e-36 MEL1_pteVam Pteropus vampyrus (macrobat) Deut.Euth.Laur... 362 4.3e-36 MEL1_gorGor Gorilla gorilla (gorilla) Deut.Euth.Euar ge... 361 5.5e-36 MEL1_eriEur Erinaceus europaeus (hedgehog) Deut.Euth.La... 360 7.0e-36 MEL1_felCat Felis catus (cat) Deut.Euth.Laur AY382594 1... 359 9.0e-36 NEUR_strPur Strongylocentrotus purpuratus (sea_urchin) ... 359 9.0e-36 MEL1_galGal Gallus gallus (chicken) Deut.Saur.Arch AY88... 359 1.1e-35 27% MEL1_nanEhr Nannospalax ehrenbergi (molerat) Deut.Euth.... 358 1.1e-35 30% MEL1_bosTau Bos taurus (cow) Deut.Euth.Laur genomic ful... 358 1.1e-35 29% ENC4_braFlo Branchiostoma floridae (amphioxus) Deut.Cep... 357 1.5e-35 27%
>MEL1_acrDig Acropora digitifera (stony_coral) BACK01045931
METASGVPIVIEASVHHTISFLYFLLALFSFSLNSVVILTFLLDRSLLFPANLIILSIAISDWLMSVVPNIMGGVANASNDLPFTDWSCTVFAFVATLLG
LSNMLHHAAFALDRYMVITRPMRANHSMKRILAVIAFLWCFALTWSLFPLVGWSAYVREAGDIACSVNWQSDNPSDTSYMVCLFFFFYFVPLAVIVYCYA
FMIKSVRFMTKNAQKIWGVRSAAALETVQATWKMAKIGLIMVVGFFVAWTPYAVVSFIIAFDLVKDIPTVAEIVPSMFAKTASVYNPIIYFFSFAYCKNQ
SVNKCINQSTVQSKNFKLS*
>MEL2_acrDig Acropora digitifera (stony_coral) BACK01017283
MPGFQNKSEEPQPTEGISSQDRFIVASFYALLAVTALGLNTPVLITFMKDRKLRVLSNRIILSITIGDWLHAFLAYPVAVFANASHDSQGLTGVVCSWYG
FITVFLSFGIMLHHATFAVERAIVIQFATTSLTNAKTINFMVASLWVFALLWSSFPLFGWSAYVPHVVLCSLDWQSRDLRDVVFVYCIFFVFFLVPIVVM
VTSYYKIFQTVKKMTQNARDLWGEKAAPTREAFESQKKTARMACVMSFCFLFAWTPYAAVSLYVFLWKPQSMAPSISIVPALFAKTSACFNPVIYFLLFR
KFRESLKQTMCGFFMDPKTGTNETNKNKRHVLKNTESKCACVKINNLSPRDPCNLHQRSDCRSVGNSKSHITVD*
>ENC1_acrDig Acropora digitifera (stony_coral) BACK01046894
MLTFACGSTHIYNMSAVFYGFLAAGTVFLNVIVILTFFKVRALLIPASFPILSLAMADVFLASAVMPLGIASCASGHWIFGAVGCNWYAFMHTTVGLSSI
LHHAILALERCLTIFEPMKKHFDRHTMTRTLAFLWTLVSLWSLCPILGWSAYVPEGTGTICSIEWHSKNSLDVIFVVSTFVFFCFVPFVFIAVCYTAIAF
HLRKVSKTAKRTWGKNSQITKDGVIVKRKAVTHGAIMVTTVMITWLPYSMVAFYTLLGFEKVKLSALAYAITAMFAKTSTLVNPIICFFWYRRFREGTKK
LCNRIMDFILRKHRRASNTSHSSKTVWYDGKTIVFRRSVVDSCSTRTQNDRGRSS*
>ENC2_acrDig Acropora digitifera (stony_coral) BACK01016849 11738 bp INV 28-JUL-2011 DOI:10.1038/nature10249
MTALIYSVFAIIAAVICVLGVPLNGLVCWVFYNNRELLNAPNIFIASVAFSDFLYCISCLPLLVISNAYGKWIYGPIGCKATAFIATWSGLTSLMNLSVA
SYERYSTLAFLCTKNRTFAKRTATCYSAAMWLYALFWSLMPLCGWSGFELEGIGTSCSVRWKSKNMLDMSYNLCLIIACYVLPVSVLVTSYYKCYREIAK
STWRAKSTWGRQSPFTKRTFVMERKMMALFGVMTVAFLVAWTPYAVVSLISMIGGPDVISDVTASIPAYFAKSSSCYNPVIYVFLYKRLRRQMRFAVRRD
NCSSSRKGTSSDSTMAHHSETKRLRSTIELTDNYSTRNRPENI*
>ENC3_acrDig Acropora digitifera (stony_coral) BACK01015014 possibly N-incomplete
MVAPHFAGFAIAASLFGSCGIVLNFTVCLTYILNRRLLDASNIFILNISAGDFLYSITALPMLVTSNALGKWSFGEGGCVAYGFLTTFFALGAMMNLAGA
AYERYVTMCKLYESGECQFSRRKAAFLCSVLWTYALIWSIAPIFGWSSYKQEGIGTSCSTDWKSRDVKSLSYGVVLIITCFVVPVAVILYCHIEAYKVTR
KLGKQAQQNWGSHTRATRETLKAQKKMGEIAVVITVGFVVAWTPYTVASVIGMYDPNLVSDVGASIPAYFAKSSSCYNPFIYLFMYKKLRIGMVRLLCCM
KKQVYPSSSGLTRDPTCQEIPMVVPVNNPS*
>ENC4_acrDig Acropora digitifera (stony_coral) BACK01020962 possibly N-incomplete
MTKTAAWSTVFLIIEFVVCLLGIFFNCLVISIIFKNANRLAAPGFLVLSMALSDVLSCSVAVPFSIVAHFQKKWPFGMAGCEAHAFMVFLFALVSITHLA
AISAGKYLTISRSLTRESYFDRRQVLLVVIACWLYSLAFSVAPLVGWSRYGLEGTNATCSIKWDSSEPSDKAYFGVVFIACFFLPMGVITLCYYKIHKVS
KRIVENIQGQSIATSAMSRTHALVKKHRRSAMYFLAIVAAFMLAWSPYAIVSLIVVLSGTMDPIATSACSVFAKTSFLVNPFLYAIFSRSFRRRLALVFP
MPRQNRQEGRQERNCNALPPSQSQPFVL*
>ENC5_acrDig Acropora digitifera (stony_coral) BACK01011015
MDQIKIWKMLFVVIKFIVSIFGGFFNGLVVLTIRKNLRRLPSSSYLILSIAFSDFIASVVAIPFSIVIHFVGSWPLRLCRAHAFMVFFLGIVTITHLTCF
AVEKYLTITRSLSKLSFFSKKQTLVVVMACWFYSLCFSLAPLLGWASYGLEGSNDTCSIKWDSSLAKDHIYFILVFLACYLLPIVLITSSYFKILRISKR
ILKATPRVGGIGETMAQALMRKHRRGALYFLSVIAVFLMSWTPYAIISVLVIFKGVNLFPLALSACGVFAKMSFMLNPIMYFAFSHNFRLLVKRTFCICD
RLDSNSEGKKR*
>CNOP1_acrDig Acropora digitifera (stony_coral) BACK01019215 tandem fragment last exon at beginning of contig
1 LLRRDVVTEDSRIKLRRRHWQMKLLRLTAVAITAFMLSWSPYSLVSLTSIFRGNSVLSTGEAEVPALMAKASVIYNPIVYTVMNRRFRRTLRHIVSCMTCRLLSFVWPTMHGEKQETKKRVTSTMTVTSSTPAPEGNFPEILVSLHQVPF*
>CNOP2_acrDig Acropora digitifera (stony_coral) BACK01019215 MEL
0 MQFSREEDPPSGFNWNNAWFIAYGVVMIAVLSIGFLGNTMTVLILRKHEHASKSLTPLMINLAIASLIIIVLGYPLVISLVVRGSHVTKEDPTCRWSAFINGTV 1
2 GISSIATLTEMSLVINYSLHRMNPNVRLTKRNMALLIAGAWLYGLVSMFPPLVGWNRFVPGAVRISCGPDWTDKSASGVSYNLVLVVLGFFLPLCVMIKAYYEIYRS 2
1 LLRSREMLISANGSFQLRQKLYIKKLVRMTVLAIAAFMLSWAPYCFVSIVAIFKGSHIITSGEAEIPELMAKASVIYNPVVYLITNSSYRASFWKVISCQKQTMIVHVRENLMPNGPSRRTLRSRRMAVLLKPLNEVSVYNGGYVVAMGFASVRDL* 0
>CNOP3_acrDig Acropora digitifera (stony_coral) BACK01018578 TMT
0 MLESSNQSAEETILISKASYYAYGVVMFFILTVGFLGNVMTLLVLFQHEHRKKAMTPYMVNIALADIFIILFGYPVAMRANLRGKILESSHCSWGGFVNGAV
GISSIFTLTEMSFVSYHGLRQVNRSSRFSPFQVVCSVAVAWLYGVLCMLPPFLGWNRFVVSASRISCCPDWSGKSISDAAYNLLLVFFGFVAPLTAMTVCYYKIYR 2
1 SLVHHAVVPGNLPQIQLRRRQSELKVAKVTAMNVIAFLLSWAPYCCVSLAAVFTKQFVLVDWEAEIPELLAKASVIYGPIIYSTMHSRFQATLFRILHCRRRVILAPQFITDATRNNCSARMATDRTQGTSHIDQRFLQLPLQAMKRRVAVSYGGTAICKRESSAFVSG* 0
>CNOP4_acrDig Acropora digitifera (stony_coral) BACK01002540 TMT
0 MTTRSDDNPNSARRPQQIAFIVYVVIMTVILIVGFLGNLFTIIVLRCPEHKKKIITPLMMNLAFADIIIIVFGYPVVMASNFTEHNVLTNPMLCVWSGFINGSV
GIASIANLTMMSLVMFKSLNKVGTVKKIPWKKMLAMILFTWVYGVLSMFPPLVGWNKFVPGASGISCCPDWSPDTKAAVGYNILLVFVGFVLPLTIIIFCYYRMYR 2
1 FIHTQQPVTGNASIQASRRRSEIKIVRMIALAIAAFVLSWSPYCFVSIAGTIRGSSLLTAGEAEVPDLLAKASVIYNPIVYTVMNDRFRGTLLRVIPCRRRWFNGDINPTVSVSAPNSQDGNSHNFRERPQSVKGNSTV* 0
>CNOP5_acrDig Acropora digitifera (stony_coral) BACK01002513 and near identical BACK01007224
0 MTTSFRSNMTDEWGTHKYFSAGYIAYGVVMFCILSLGFVGNTLTVLVLTKREHRWRGVTPLMINLAVADLLIIIFGYPTQVHANFTGDGVPNDSHCNYRGFIN
GIVGITCIFTLTEMGVVSYSGLRRVSGNVRLSSYQVACLIGAAWCYGALCMLPPLLGWSKFVISASKLSCSPNWSGQSTADKAYTLLLVTFGFFAPLTVMIISYYKTFW 2
1 VVRRHFVPGNTAVQLRQRKSMLRIVHVTLMSVISMMLSWAPYCFISLASVVRGKPVIENWEAEIPELLAKASVVYNPLIYTFMNKSFRMTLFRIVGLHRCFGAGHEVAPTAAIQKRPNVLQIQFHRHSSEVEVRTPRVPPIE* 0
>MEL3_acrDig Acropora digitifera (stony_coral) BACK01025991 resembles long intronless Nematostella FAA00396
VFSITSTLHGRWLFGRAGCVTYGFFHYFLICCTVSTLAAISVDRFFFITRPSQARTAQMITPRKAVAMLAIIYFYTLMFTFPPCIGWNNFEVEESFYSGC
YINYADQRASSIAYSIIAPFFLFLVPLTIMIFCYARIFAVVRRSTQRTINRPFGAQSLKRYPLLKRTHIQTAKIIVVVILFSMIVWVPYVVVSFVKAANG
KVNPLLSHITVLVAKSCVVYNVLIYAVLNRKLKASILDVICCGKQNFRMVGSPSSGNSLQRNRNSRFLSQREVPSDIAESLKNRASTLSVQEGNGNVHDQ
YTDNISPSQLTQLRNGVLKSIDHVDGRESGKVVGKKEVDVVRNGDKVQVGESSGFSETRMVIATQERDSKILGPSDARSSDRGTNRFLARLETDTLPEIA
PQDTHLKPEAVSNASSLSRNETPRVDCKVDTPLCNSSSSNRQLNRLHHMYSSKSSRHARLQNLSSPSQLYKKGARPASAQRVTGHIASDSRTSRAGVSSG
TESDVSTRASHRPATAQRPKKRRAADRTNSTHTSERRKSSSYINYGRKDSVDPLRTSPKELWEIQNYWKRMSLCLDDIDVDEVTREIQL*
>MEL3_nemVec ABAV01020362 'Serpentine type 7TM GPCR chemoreceptor Srsx'
MSATTTAANISTDVYQTSRQVSVAYLLIVGILGTLGNSLVVFVLWKYPKLRTATNLFILNLAFCDLVMALLDLSLSVASALSNRWLWGKSGCLGYGFLHY
FFISNSVSTLAAISVDRFFYITKPGQIKAWRITRTRAGIILIIIYIYVLLFTFPPIAGWSSFTEEKFYFSGCFIKYSDQNPASMAYSVIASVFLYLMPLI
LMICCYSKIFMAVRKSTRRTMTNSNAVRKKFPIMKRTHIQTAKMIVVVIFFSMIVWVPYVAVSLIQAFTADSIITPTASHITVLVAKSCVIYNVLIYVVL
NRKLKAVILNLLCCGRQINFSGSNMPTSNLRISRILSETDREIAPDAVRNRLNVLQNNDNASAVDASVERSSPIPTHTTFKPDLIETPKGTKQRHGNVKT
VEFKVVEQDGNVITLVNDAANGTGSGGNLPKIRRKKLPQIPRDNQETNGNLVGQFGDDLNSVGVSNKDFSSCEETSPAKTEKLLLTETVTRAEARRRALR
KLNDRKVSNSVPSIEKRDSTSSGATVATGNESHDRSISPSTPNGRILPEIPNHVGDKHEKKRNGNAKTNEADRIGADSSSTAIPSSCTSTLSNHSRAWSE
KDTGNPAAPARSMTSPRRNGRRKTRKHMDRNASLRARLQRCSVDVLQASPSELHEIQNYWKRMSLCLDDIDLDEMV*
>CNOP4_nemVec Nematostella vectensis ABAV01018948
0 MSTANSPWGSEGLVAYGVVMSLILTTGLTGNILTIIVLMRREHRSKVITPFMINVAVLDLIIIALGYPVAIIANFTGIRMKEGDSRCQWTAFLNGAT
GIAAIITLTAMSIVMRNTITHPIVKKRLSCKYLFGSLCAIWTYGILTMLPPLVGWSKFVPGAAKVSCGPDWETQNASTLSYNIVLLIVGFVIPVGIILPCYFHIFR 2
1 FLRSQPVPPAFISITSRRRRAHLRIARMILVSIISFVISWSPYCVVSVIAMVQRAPALSQGFAEIPELMAKASVVYNPLVFTVMNRGFRRSLCLIVPCLGCSVGHGISGRDGLYIWLGMKDLTDRSSVISNVAVKKR* 0
Anthozoa: Anemonia viridis (symbiotic anemone)) .. 1 opsin
This ciliary opsin fragment is associated with an October 2009 metagenomic article on the symbiotic coral reef organism Anemonia viridis and its dinoflagellate algae but was not annotated in the EST collection (FK729339).
It is most closely allied with ENCEPHb_nemVec of Nematostella vectensis in the currently known set of cnidarian opsins -- about 49% identity but not enough is known about Anemonia opsin multiplicity to say it is an ortholog. High identity might be due either to sequence conservation or close relatedness of these two genera -- their time since divergence is an unknown. Conservation (of sequence and hence function) seems more likely since -- at least at GenBank taxonomy -- the divergence is quite deep:
Cnidaria; Anthozoa; Hexacorallia; Actiniaria; Nynantheae; Actiniidae; Anemonia Cnidaria; Anthozoa; Hexacorallia; Actiniaria; Edwardsiidae; Nematostella Cnidaria; Anthozoa; Hexacorallia; Scleractinia; Astrocoeniina; Acroporidae; Acropora >ENCEPH_aneVir Anemonia viridis (symbiotic sea anemone) frag:202-338 pubmed:19627569 KDAVARWGNKSPPTQQTMQAQKKTIRMSLVMVFAYLLAWTPYALTSLYSSFIASDITPLLSVMPALFAKLSSCYNPIIYFFMYSKFR KAAKKMIRRNLVGHDSNSGQGVSNTFATSFPRPISFLRYKRSAVAPLSDIPQVSSVDLPQVGRENDVTVQQDKASEINT*
Hydrozoa: Hydra magnipapillata (hydra) .. 41 opsin-like genes
Because opsin photoreception is quite ancient, clearly pre-Bilaterans have a major role to play in illuminating the origins of photoreception systems. What's not so clear is that the two cnidarians chosen so far for genome projects are optimal in this regard. The excellent Hydra genome article of 25 Mar 10 does not specifically address opsins but a 10 Mar 10 article considers them in a signalling context.
Hydra does not have overt photoreceptive structures or cells obviously specialized for light detection yet it exhibits marked behavioral photosensitivity (noted by Trembley in 1744). Studies beginning in 2000 flagged the ectoderm (using antibodies to squid rhodopsin), known to contain epidermal sensory neurons, as responsible for extraocular photoreception. Musio and coworkers sought to recover opsins using degenerate primers, targeting melanopsin and peropsin as the most plausible in Hydra because the latter opsin seems not to require advanced relationships with neighboring cells or auxiliary enzymes (ie, acts as photosensor and its own photoisomerase).
The Hydra cdna CB073527 was proposed as a peropsin based on best-blast to mouse peropsin. However using a much larger collection of demonstrably orthologous chordate peropsins in the Opsin Classifier conflicts with this interpretation: the putative cnidarian gene needs to consistently associate with this gene family (equivalently, have best match to it among all reconstructed ancestral opsins) but does not. Furthermore the best match is very weak at 31%. This is the signature of generic non-opsin rhodopsin superfamily members (which we expect any eumetazoan to have by the hundreds).
With the availability of the diploid Hydra genome assembly (ACZU vs ABRM accessions), the 161 amino acids of the fragmentary transcript can be extended for example with trace 1121878952 to apparent full length (309 aa) and its introns determined (none). This does not improve its best-blast score nor family coherence. It does not cluster consistently with ciliary opsins -- what would the signaling partner be when the matches are scattered between Gt, Go, and Gq opsins? The blast probability of 1.6e-34 does not mean much under these circumstances.
Two putative hydra opsin fragments can be extracted from Fig.1 of an Oct 2007 paper, AKSSTIINPTISCIIYKE and AKLSAVLNALVNCYINKS (reclassified below as CNOPa5 and CNOPe4). These too fail to extend to convincing opsins. Expression centers around the hydropore, a better fit to GPCR chemoreceptor localization. There is meagre behavioral evidence for photoreception near the hydropore and no possibility of a pigment-backed eye. An ultrastructure study is needed to demonstrate that the putative opsin is expressed in specialized photoreceptor cells. It is critical to include non-opsin GPCR as alignment controls -- all GPCR proteins bind heteromeric G proteins and many have lysine without binding of retinal. The cdna accession numbers are Hydra1 CN554949 and Hydra2 CV151648.
These papers highlight the special difficulties in working with cnidarian opsin candidates. We know from the outset that they will be quite diverged from bilateran opsins. Multiple forms of supporting data are needed, preferably in the form of diagnostic introns, alignments demonstrating conservation of critical residues and structures, in situ hybridization to anatomically plausible neuronal photoreceptors, and specific loss of photobehavior upon knockdown.
A higher standard of proof is needed for the first cnidarian opsins because validated ones will surely be used to pull in further homologs via annotation transfer. There is a definite risk in admitting inadequately documented opsins to the Opsin Classifier because once that database is tainted, it could draw in even more non-opsins from the GPCR world.
CnidBase provides a blast service to cnidarians including hydra but this appears restricted to ESTs and so only duplicates GenBank. GenBank carries contigs of the genome assembly on 25 May 2009 in the wgs division. Some 10.2 million Hydra traces have been provided by JCVI, ample for the 1290 Mbp estimated genome size. However the hydra genome project was dropped from that website. The draft genome expected in Dec 2005 was delayed because of high AT content (71%) and lack of an inbred strain. A 'hydrazome' blast server and funky browser for assembly v2.0.4 has surfaced along with Gnomon gene predictions at NCBI nr.
The best current opsin search strategy simply uses tblastn of the usual three NCBI (nr, est_others and wgs) restricted to Hydra. After genes are recovered, their best-blast to all of nr must be considered. The outcome first lists other cnidarian 'opsins' and then mixed bona fide bilateran opsins. That's encouraging because generic GPCR come later but does not prove by any means that any of the Hydra genes are true opsins, even those with lysine in Schiff base position and having other signature residues.
The basic problem is Hydra has far too many opsin-like genes (41 genes, all K296) for its complete lack of anatomical photoreceptive structures and minimalist phototactic behavior. It is not plausible that such a simple organism could have a larger opsin repertoire than an amniote with four color imaging vision and numerous pineal and deep brain photoreception. All the Hydra genes are intronless, suggesting they have as arisen, much like the olfactory gene expansion in mammals, as processed retrogenes.
If not opsins, if their function is chemoreception, why then is the Schiff lysine conserved? Note first that it is not currently known whether a retinal species occupies any of the Schiff base sites nor whether any cis-trans photoisomerization with ligand release takes place. These opsin-like genes have retained many key residues for signaling so very likely follow standard GPCR mechanisms (though with unknown Galpha partnering) and are certainly not pseudogenes.
Among the many possible scenarios: the opsin-like genes have other primary agonists. The genes were perhaps derived long ago from an opsin and retain the Schiff lysine and (undetermined) counterion through evolutionary inertia. That is, mutational loss of the lysine + counterion is a two-step process. Initial loss of either leaves a defective salt bridge in the extreme hydrophilic milieu -- negative selection would inactivate the gene before the second component could be favorably mutated. Here though it needs to be recognized that generic GPCR do not have these charged internal residues (ie transitioning is possible since all GPCR presumably coalesce to a single ancestral gene). This argues for non-retinal agonist binding -- after all, what is so special about the terminal aldehyde in a retinal? Indeed assimilation of beta-carotene and its variants generates a large number of them.
The 41 Hydra opsin-like intronless genes with disulfide, questionable DRY motif, Schiff lysine at 296 and weak FR switch motif. Of these only CNOPa1, CNOPa5 and CNOPc1 have (unlocalized) transcripts. These genes cluster into discrete blocks by blastp as shown above; this classification agrees with the alignment-derived tree and classification by indels and signature residues.
>CNOPa1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01000679 KSSTILNPIIYCLMYKKFR 37% CB073527 CB271253 CN554455 CN554795 no tissue 65% single exon MAFVFIIVFLSFLCGFSVILNVTVVLTILAKGNTKNTRDVILMSLAICDGVQCTIGYPVELFGYANYKNPSLSEKFCKPSGFIVMYLALTAIAHLVCLCIYRYLTIVYPLKLQIFLTKSNWSACGC IAFcWIYGLFWSLSPLLGWNEIVREKQDTYRCSINLYPDNEIKSSYLYALAIFCYLIPLIIIIYCSLKVRSELRNMLKMCKQISGVEANITKVTYRIEKQDFISVSFIIASFFTVWTPYA VCVFYLTIGKKLPPSFLTYCALFAKSSTILNPIIYCLMYKKFRQTLQSKFGKLFNNPTVTPAV* >CNOPa2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01004988 KSSTILNPIIYCIMYKEFR single exon XM_002163173 MAAPVIGLLSFLCGLSVSLNVTVVVTIISKKNTKDMRDIIIMSLAICDGVECTLGYPVELYGYANIDSPSQNEYLCKTNGFIVMYLALTALTHLVFLCVHRYLVIVHPMKVQKFFTDTKTCALYFIIPSWI YGLFWSITPLIGWSEIVREKEDTHRCTINMYPDDLLKRSYLYALTLFCYFLPAAIIIYCITKVHFELRNMLKLCKQISGEDAAITRATYKLERQNFLSVSLIITSFFLIWTPYTICVCYL ILGRELPVGILTYSALFAKSSTILNPIIYCIMYKEFRQTVRSKVRKLFRGPTVAPITE* >CNOPa3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01055709 KSSTILNPVIYCLMYKEYR single exon MASVIILLSFLCGFSVVLNATVVLAIFLKRKSKGMRDIILMSLAICDGVQCTLGFPVELYGFSNLNSPLQNEYLCKANGFIVMYLALTAISHL VCLCVYRFLSIVYPLKIQKFFLDSRREALLFILFCWIYGFFWSVTPLLGWNEIIREKEDTHRCSINLNPEDNNKRSYLYSLMVFCYFIPLVIIIFCSLRVHFELSKMLKLCKHISGREAS ITKETYKLERQDFISISLIVSSFFIVWTPYTICVIFSSLRSSLPTGLLTYSALFAKSSTILNPVIYCLMYKEYRETLQSEYRKIFKSSVVAPFTERSQTAALSTLSSHGDASFT* >CNOPa4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01004994 KSSTIINPIVYCIVYKEFR single exon XM_002163322 MALVSIIVLLSFFCGLSFVLNATVIFVVLSKSSIDARETILMSLAVCDGLQSIIGFPNEIYGYVNKDNPFKSESLCKTSGFVVMCLAITAISHLVSLCIYRYAAIVYPMETQVFV TSPIKVTLYLVLPNWIYGLFWSITPFLGWNEIVREKQDTYRCSINLYPNDQIKQSYLYALTIFCYFLPLSIIIYCIIKVHLELRCMLKLGKQISGADASITKATYKLEKEHFISVFLIVV SFFTVWTPYFICVCFLVYGRSLPSGFLTYCALFAKSSTIINPIVYCIVYKEFRQTAISKVRKLLRRPAVAPMLVSQLSTAAAPTLALSGL* >CNOPa5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01004993 KSSTIINPTISCIIYKEYF single exon CN554602 CN554949 XM_002163291 3 glitches MASLFIIVLLSCLCGLSVTLNVTAVIAIISTKKNKDVCDVILMSLAVSDGVECIFGFSVELYGYATKGKTLQNETLCKINGFIVMYLALTSISHLVCLCLYRYILIVHSLKAQRYLTNVKQSALYFIIPSW IYGLFWSIAAISGWNEIIREKVDTHRCTINMSPDDELKRSYLYSLTVFCFLVPVVIIIYCSLKVHLKLQNMWKLCVQISGEYAAITKATYKLERKHFIFLGLIIGSFFVVWTPYALCVFF LALQIKLPRVLLTYSALFAKSSTIINPTISCIIYKEYFQILRIKVQKLFRNNIVSPANL* >CNOPa6_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01000199 KLSTIFDPIIYCLVYKNFR 9 glitches MISVIVLSSILCGCSVVLNLIVILTVSKKRSKDLRHLILMSLALCEGMSITVWYLNYVYSYFHKHEPLTKKLCGISGFIELYLTITAVSHLVWLCIYRFISISHPLKIHYFFVDTKRRVLYF ILPCWIYGLFWSSAPLFGWIKLGEENSSYYRCTVIYYPDEFLKRSYLFVFIVFFYFMPLSAISFFTLKVHLELRSMLKQCKKVSGNNALITKDTYKLARQDFINACLIVASFAVVWSPYA LCVFALSLGYKKLKGFLFFCALFSKLSTIFDPIIYCLVYKNFRETLRRDIKTIFKVAAVAPQ* >CNOPb1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01027146 KLSTVSNVLVNCFINKSFK XM_002160412 MTAYDVAYKVFLSLILVLSVMLNTTACYTILIKVKKKELAHLFILSISFTNLMESIIGVLPHVITSGEILSGSNSLCIASGFAVFGFAITSISHLCIHSLNRTVAIIYPIYYFKYRKRFWYRSALISLC YAYGFMWASFPAIGWSKYNLDVDKKRCSLDWKLTRSNSLSYILSVLIFCNIFPGILIVIGLFRSKKVIYRRRTRSLGQKDDNQTDFLEKHYLKVFSLSSAMFFVTWTPYALVSILALLKI APPPLLVTTSALLSKLSTVSNVLVNCFINKSFKKHVFSIRFIQDITKKGSELRKTAYVIFLYKD* >CNOPb2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01051946 KLSAVSNVLVNCFINKSFI MEPYEVAYKVFLSLILTLSVVLNTTAFYTISIKVKKKELAHLFILSISFTNLLESIIGLLPHIITSGEILSGSNSLCIASGFAVFGFATTSITHLCVHSLSRTVAIK YPIYYFKNRKRFWYRTALISVCYVYGFIWATFPAISWSKYDLDLDKRRCSLDWNLTRSDSLSYILSVLIFCNIFPGILITIGLFKSKKVIYRRRTRNLGQKDDNQPDIFEKHYLKVFSLS SVLFFVAWTPYAVVSILALLKIAPPPLLVTTAALFSKLSAVSNVLVNCFINKSFIKHLFSIRVIQVVAKRGTELKIIGHAAFLHKEKSLSY* >CNOPb3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01043783 KLSTISNVLINCFINKSFQ MNKSQITSSIFLSLILIVSVVLNTTASYIMLFKVKKKELMHLFIISLSTTNLLESIVGITPQIIFSEQSLLRKTPLCIFSGFAVFGFAVTSITHMS AFSLIRTVIVKYPIYYFQRYKAFHYKAALILSCYVYGFSWATFPFIGWSKYEVDLDKKRCSLDWKLSQSESPSFFLAILIFCNILPGTVIALGLYFSTKIIHRRKACKTRQDKNKTLDIL EMEYLKVNFLSAILYFVIWTPYAIVSILTLLKITVPAYLVTVSALFSKLSTISNVLINCFINKSFQKHLLNLRLIRFLANTNFKRRLKNHSSTSCHFFKLEQK* >CNOPb4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01065240 KLSTISNVLINCFINRYFQ MTCYRITSSIFLSLILIVSVVLNTTASYIMLFKVKKVELTHLFIVSLSITNLLESIIGLIPQIIISDESVMRKTPMCIVSGFAVFGFAVTSITHMSLFSLI RTIIVKYPFFYFQRCKAFYYKVALILSCYVYGFSWATFPFIGWSKYEIDLDKKRCSLDWKLSQPESRSFFLAVLIFCNILPGIVIALGLYYSTKIIHRRKACKFRQDKKKRLVDILEIEY LKVNFLSAAIYFVIWTPYAVVSILTLLKITVPAHVVTVCALFSKLSTISNVLINCFINRYFQKHLLNLGLIKFLANSTFKRRLKNHPSTIFFKLERK* >CNOPb5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01078635 KISTISNVLTNCFINTSFR MTSYEIAANIFYSTVLVVSVILNTTACYIILFKVKKKELTHIFIISLSVTNLLESVIGLIPQIIISSEAKLEKTPLCITGSFAVFGFAVTSITHLSIFSLVRTVAIKYPIYYFKNSKRFWCRAT LILICYCYGFSWAIFPVIGWSKYEIDLDKKRCSLDWRMTRPDSLSYILSILIFCNFLPGTLIAIGLYISKKVIYRRRTCKLRKNENNRSIDLLEKDYLRVCFLSSILFFAIWTPYAVVGI LALSKIVPPPLLVTTAAMFSKISTISNVLTNCFINTSFRKHLLNLKIVRVFLNKKIESPRIEDV* >CNOPc1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01005587 CX830752 KLSTITNVIINCFIVKSFK MTNNNSVLSSFFFIILLTSVMLNATACYIILLKVKKKEITHLFIISISLINMLESIVGFIPDILTSDGSLINRTPLCISSGFTVFGCAITSITHITVLSFIRTLVIKYPFFYHKTCKKMWVKATLIAMCYLYGFSWATF PLIGWSKYELDLDKKRCSLDWKLTQSDSASYFLTILIFCNLLPAIVISTTLYTSRKIISKRNAREDCQNHQASLDILENDYLNVCLLSTGTFFFIWTPYAVIGFLTILKIVIPTQLVTAS AIFAKLSTITNVIINCFIVKSFKNQLLELRLIQYIRNINNKNSAVHPCDDSKINYS* >CNOPc2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01005585 KLSTITNVVINCFIVKSFK MALNLFLFTILFTSVILNATACYIILLKIKKKEITHLFITSISLTNMLESIVGFIPQITTSENFLLERTPLCISSGFAVFGCAITSITHITALSFMRTIVMKYPFFYYKTCKKMWVKATLIAMCYFYGFSWAT FPLIGWSKYELDLDKKRCSLDWKLTQSDSASYFLTILIFCNLLPAIVISTTLYTSRKIISKRNAREDCQNHQASLDILENDYLNVCLLSTATFFFIWTPYAVIGVLTILKIVIPTQLVTA GAMFAKLSTITNVVINCFIVKSFKNQLLELRLIQYIRNINKKISAVHPYHDSTLN* >CNOPc3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01005579 KLSTITNVIINCFIIESFK MINNYSVSNVLLSTILLISVILNATACYIILLKVKKKEITHLFIASISFTNMLESMVGFIPQIVTSDKILLERTPLCILSGFVVFGCAVTSITHITILSSIRTAVI KYPFFYYETSKKVWFKTTLITICYIYGFSWATFPLIGWSKYEVDVDKKRCSLDWKLTKSNSLSYFAAVLVFCNIMPCLVISTTLYTSRKLISERKARKASQSQSHQVSLDILENDYLKVC LLSAIAFFITWAPYAVIGILTVLKIVIPTQLVTAAALFAKfSTITNVIINCFIIESFKNQVFDLRFIKYIRSINKITSKVQPQTINYNC* >CNOPd1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01091217 KLSTITNTLINCFIIKSFR MTNKLLKVFLFTILFTSVVLNGTAIYILLFKVKKKEITHLFITSISLVNLLESLVEFIPLLLLVYSDASLIERTSLCIASSFVVFGFAMSSITQQSMLSLI KIIVIKYPIFYHKTFKKMWIKAALITICYLYGFCWATFPLIGWSKYQLDIDKMRCSLDWNLTHPGSFFYFLTITIFCYLLPVIVISSTFYTGNKIIYGRKVSIARQNKNNQYINILEKDY LKVSFLSVTTFLIIWTPYLVTGILATLKIVIPKQLVTTAAMFSKLSTITNTLINCFIIKSFRNHILDLKIVQCIRNKKNAAKIKPQPYYINCNYLRQT* >CNOPd2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01027487 KLSTISNVLINCYVIKSFR MNTNKLVQSVLSTVLVTSVVLNMTACYVILVKVKRKEITHLFVVSISMTNLLETMIGLTPQILIADQSLLEKTPLCIASGFIVLGFAITNISHLAMLSFI RTFAIKYPMRYLQLHKMFWCKVASILVCYANGFFWAILPLLGWSKYELDLDKKRCSLDWSLTKANSFSYILTIFIFCNILPGIIIVLTLYFSKKTVFRRKTCKDCQNRETNILEKEYLRV CFLSATAYFLFWTTYAIVSVLTLLKVFVPVELTTTTALVAKLSTISNVLINCYVIKSFRKQLLNLMLIRFIKKYLWVLIACKKSSVNDDIN* >CNOPd3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01075622 KLSTISNVLINCYAIKSFR MSSIKVIQSVISTVLLTSIILNMTACYIILFKVKKKEITHLFIVSISITNLLETTIGLTPQLMMANEFLLERTPLCIVSGFSVLGFAITNITHLTILSFIRTVAIKYPLQFLRYNKMFWCR ATLIFVCYANGFFWAILPIIGWSKYELDLDKQRCSLDWRLTKLNSFSYILSIFICCNILPGIVIVLMLYFSKKTIYRRKGRKICQSRETGFLEKEYLRVCLLSTVTFFFFWSFYALFSLL TLMKFAIPTNLATTSALFAKLSTISNVLINCYAIKSFRKQLLDICVIRFMKRCCWVLIACQKSSVNDQIN* >CNOPd4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01076129 KLSTISNVLVNCYAIKSFQ MNTNKVIQTVISTVLLTSVVLNMTACYIILVKVKRKEITHLFVVSISITNLLETTIGLTPQLAMADESLLESTPLCIVGGFAVLGFAITNITHLTMLSFIRTVAIKYPRRYFQYHKMFWCKVTLILVCYAYGFFW ATLPIIGWSKYELDLDKKRCSLDWKLTKANSFSYILAIFICCNILPGIVIALTLYYSAKEIRHRKACKFPQNKKTDHLEKEYFQVCLLSAFAYFFFWTSYAIVGVLTLLKFNIPMHLATT TAMFSKLSTISNVLVNCYAIKSFQKQLLNLKIVQIMKSYLRILIVCKKV* >CNOPd5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01038272 KLSTISNVVVNCFVLKSFR MNIEKIVSKVYLSAVLVTSLALNITAFYITLLKVKKKELTHFFIVSISITNLLESTVGIIPQLVISNDSLLERTPLCISSGFAVLGFAITNITHLAVLSFIRTVAIKYPML YLKYHKMIWCRVTLILVCYAYGFVWATLPLIGWSKYDLDLDKKRCSLDWKLTKSNSFSYILAIYICCNILPGIVVALTLYFSKKTIYRRQSCKLRkNFKKGRLEREYLKICYLSAiAYFF FWTTYAVIGALTLLQIQISPILATFTALITKLSTISNVVVNCFVLKSFRNHFINLKFIRIIKKYLEVLKVKKHSKLDHTV* >CNOPd6_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01049360 KLSTISNVLINCFINKPFR WRHF removed MLNNKRASAVLLSTIFIVSMILNTIACYIIVYKVKRKKLTHWVVISISIANLLETIIGLLPEIVMSDKPLLKKTPLCVTGSYAVFGLAITSITHLLVLSLIRTAAIKYPIFYLKISSTVWCK ATLIIFCYSYGFSWATFPVLGWSKYEIDLNKKHCSLDWKLTQSNSLSYILTILIFCSIFPGLVITCASYICKKAITLQKTSYIRNKNSRPMYTLKKEHLKVCFLSALLFFVTWTPYTVVG VLTLFRIIIPTELVTIASLFSKLSTISNVLINCFINKPFRKHLLNLRLVRLVINK* >CNOPd7_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01040368 KLSTISNVLINCFINKSFQ MAASETANIFLSTIILASVVLNTTACYIILFKVKKKKLTHLFIISISVANLIESIVGLTPQLFISNELILVKTPLCVINSFAIMGLAITSITHISVLSLIRIVVIKYPIFYYKTCKKMWCKVI LISICYIYGFGWATVPLIGWSKYVLDLDKMRFSLDWKLTQADSLSFILTLLVFCNILPGAVIALTTYIGTKTISKQKSNEVSQTEKEKPVDILEKYYLRVFTLSALMFFVIWTPYAIGILTLSK FVIPTHFVTVAALFAKLSTISNVLINCFINKSFQNQLLSLGIIQAFINKSKRSPVFPEK* >CNOPe1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01068872 KLSAISNAMTNCYFNKYFR MDVLVSAYFVCVLVIIILSILLNALLIYIIVYKIKVTKLPHVFILSISISDVIHTLIGYTAEVWVLFEVYSLKIRFVCIGASFLTVFTSVSNILQTVITSIMRAVAIKWPFFYIIYCKTM KMKMFLLLFCYFYGFLWPFIPLIGWSKYELDLDKMRCSFDWKLKHSDSLSYLIVLFLFCYFLPVIAMIFAYVAIKKTVFNNCTIKRAQVGKNRNIQEMVYLKLAFWSAIFYFIIWTPYAG VSLLSVFKIKSPSVIYTFCALFSKLSAISNAMTNCYFNKYFRNHLKKIKLFQYFIKRNRRSSDNVML* >CNOPe2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01085554 KLSSISNALTNCYFNKYFR MDVLVSAYFVCVLVIIILSILLNALLIYIIVYKIKVTKLPHVFILSISISDVIHTLIGYIAEVWVLFEVHSLKKSFVCIGPSFITVFTSVSNILQTVTISIMRAIAIKWPFFYIIYCKTMKMKILLLFFCYFYGF FWPFIPLIGWSKYELDLDELRCSFDWKLMHSDSLSYLIVLLIFCYILPVIALIFAYITLKKTVVDSSTVKRAQLGNNRNIQEMVYLKLAFWSAIFYFIVWTPYASASLLSIFKIKSPNVI YTLCALFSKLSSISNALTNCYFNKYFRNHLKKIKLFQYFVKWNRRSCDNLIL* >CNOPe3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01057385 KSSAISNALVNCYMNKSFQ MELLVAVYSLYVLALIILSTTLNLIICYIILSKIKAIEMPHLFILSISISDILHTSIGLVSELCVLHGATSLKKSYTCIGASFLTYSFTVSNIMQMVMISIVRVIALKFPI FYFNNCKKMKFRLGCLFLCYFYGFLWPSFPLLGWSTYEEDLDKRRCSLDWNLTNSNSFSYLMFAFIFCYIFPAIVLIWSLNVTKKTVDEQRESSFRKNRPNEQMEILEKVYLKIFLCSAI AYFVIWTPYACATLLSIFRIKTPSVVFTFCALFAKSSAISNALVNCYMNKSFQVHLNEIRVFSCFIKENRVAPRITNASVTEETQA* >CNOPe4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01037497 CV151648 MVLVALYTTYIVVLIALSTSLNLIICYVIIRKIKAIEIPHLFILSISISDIVHTVVGLVSEVCVLHKVTSLKNSYISVGASFLTYSISVSNIMQIVMISAIRVVALKRPMFYFKNCDKVKYKMASLCCCYFYGILWASFPLLGWSKYEEDLDKKRCSLD WNLTKSNTFSYLMFAFIFCYKLPTTLLIWALRVKHQTVDEHRRFRENRPNQQIEILEKAYLKVLLCSAIAYFIVWTPYSVVSLLSIFKVKIPSIIFTFCALFAKLSAVLNALVNCYINKSFQAHLHDLRIFQCFLKGKRIEHKNISWAEETQVAETGFT* >CNOPe5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01037501 KLTTVLNALVNCYFNKSFQ MEKALYTIYILLLIIISTLLNLFICYVIIYKIKTFEITHLFIISISISDIVHAIIGLVSEIYVLQNFTIFDKSYICFEASFLTYSISVSNIVHTVIISVIRLIALKWPIYYFNNCKKLRYK MGSLFFCYFYGLLWALFPLLGWSKYEKDLDGMRCSLDWNLTKYNSFTYLMFTFIFCYILPAIFLFWSLKVVHQTVDCHRSFRENYPNQLIEILEKSYLNVLICSAVAYFVVWTPYSVVSL LSIFQIKFPGFVITLCALFAKLTTVLNALVNCYFNKSFQVHLYKLKIFEYFLKGNKIAHNSNSITGETQA* >CNOPe6_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01013912 KLSAVSNALMNCYFNKYFR MDVLISAYSACVLAIIILSTLLNALLIYIIIYKIKVTKLPHVFILSISISDVIHTLIGYTAEVWVLHEVYSLKKSFICIGASFLMVFITVSNILQTVIISIMKVIAIRWPFFYIIY CKTIKMKILLLFFCYFYGFLWPFIPLFGWSKYELDLDKMRCSFDWKLIHSGSLSYLIVLLSFCYILPIVALVFAYIAIKKTVSDRCTVKRAQLGKNRGIQEMFYLKLALWSAILYFIIWT PYAGASLLSVFKIKSPNVIYTLCALFSKLSAVSNALMNCYFNKYFRNHLKKIKLFQYFIKWNRRSNDKVL* >CNOPf1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01095006 KLSALINPIVNVWYNLEFR MSKALESTYICLVLIFTLTMNILLLTFLKKRYSYINISILFNASISICEILQVTLGYIPDLLTQLDVIKVTTSGNGSWICIGSSLLTFIFAVIVLMHFLVLSVSRIVAIKYPVFYVGTFSKKKLAIFLMILCYIYGIIWPVFPLFK WSKYAKDIDEKRCSLDWRLEQKNSFSYIICVILFAFILPFVTIFILLFCKKYAALANQNSRCRKRHSSIEYQEKIKGLEKKHFNMCAASTILYTLAWSPYAIVGFLSVFNSNIPPTISTS AALFAKLSALINPIVNVWYNLEFRNFLFNLKSIRFFEKNFCRKVESQESMTSSKVQCTNV* >CNOPf2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01063269 KLSALINPFVNVWFNLEFR MSKALESTYICLLLIFTLTMNVLLLTFLKKRYSYINISILFNASISVCEILQVTLGYVPDLLAQLDFIKVTVSGNGSWVCIGSSLLTFIFAVVVLMHFLVLSVSRIVA IKYPVFYVATFSKKKLSIILLILCYIYGIIWPVFPLFKWSKYAKDIDEKRCSLDWRLEQKNSFSYIICVILFAFILPFVTIFILLFCKKYATIVNQSSPSRRRHSSIEYQKKIKGLEKKY FNMCAASTILYTLAWSPYAVVGFLSVFNIIIPPMVSTSAALFAKLSALINPFVNVWFNLEFRNFIFNLKSIRFFKINLCIKVDPQESMTSSKVQCTNV* >CNOPf3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01063264 KLSASINPIVNVWYNREFR MSKALESTFICLVLIFTLTMNTLLLTFLKKRYSYINISNLFNASISVCEILQVTLGYVPDLLAQLDFIKVTTSGNGSWICIGSSLLTFIFAVVVLMHFLVLSVSRIVAIKYPVFYVTTFSKKKLSIILLILCYIYGI IWPVFPLFKWSKYAKDIDEKRCSLDWRLEQKDSFSYIICVILFVFILPFISILTLLLCKKYATLANQNSSRRRRHSSVEYQKKIKELEIKYFNMCAASTILYTLAWSPYAVVGFLSVFNS SIPPMVSTSAALFAKLSASINPIVNVWYNREFRTFLFNLKSNWFLKKIFCIKADRQKLLTFSRIQCTKL* >CNOPf4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01049000 KLSASINPIVNIWFNWEFR MINALESTYICLVLTFTLTMNVLLLTFLKNRYSYINISILFNASISLCEILQVTLGYVPDLLAQLGVIKVTTS ESGSWVCIGSSMLTFILAVVILMHFLVLSVSRILALKYPVFYISTFSKRWLAILLLILCYIYGVSWPVLPLFKWSKYAKDIDGKRCSLDWRLEQKDSFSYIICVILFAFILPLVTIFVLL FCKKYAIIVNQNSPSRKRHSSIEYQKKIKGLEKKYFNMAAASTILYTIAWSPYAVIGFLSILNRTIPSLVSTSAALFAKLSASINPIVNIWFNWEFRNFIFSLKSIRFLKKFFCKNTERQESLTSSRNQCTNV* >CNOPg1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01024987 KLSAVVNPFIYYWKDGLFK XM_002157121 MDGVKFTLITIISVAIFTNSVSLYFLFKKQKKNNYVILCINLSLSDLLQSIAGYIPALFIDKLQKATMLCKLSAFFIAFPSFTTIAMLTSMALSRMVLLSTCFHCNQINYKILFRKIGFI SWIYGFIWAVFPLFGFSSYTLEGTHSRCSIDFSPKTIADKVYLIMIVAFGFLIPVMSILISCIYTAKVMRSKYKFFYVTYGKENVETKRYKEKEKKAFSSFVLMVTSFIICWSPYATIGC LSAFTLTRIPKWLLHSAAFLGKLSAVVNPFIYYWKDGLFKKRFTSTTWNASYFISKSQQNIEDNQKNSRGALRNYMNVFCG* >CNOPg2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01083619 KLSALINPFIYYWKDGLFK MDAVKIILLIITSIAVLTNGISFYFLYKKQKKNNYVILCINLCVSDILQSVAGYIPALFFDINMQKATTLCKLSAFFIAFPSFTTIAMLSAMALSRMVLLSLCFQSNQINYKKLFIRIATASWIYGFIWAVLPFLGFSSY TLENTRARCSINFSPKKNVEKLYLILLMAFGFVIPLISIIASCLFTARVITTKYKYFCVTYGKDNDETIRYKEKEKKTFFSFLIMVLSFAVCWTPYATIGCFSAFTSFKIPQWLLHVAAF FGKLSALINPFIYYWKDGLFKKCFLNKRLKATKTFLIKNQETNQVDK* >CNOPg3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01007423 KLSALVNPFIYYWKDGLFK MDAVKTTLLIITSIAVITNSVSLYFLYKKQKKNNYVILCINLSASDLLQSIAGYIPALFFDANMQKATTLCKLSAFFIAFPSFTTIAMLSAMAI SRMVLLGTCFHSTQINYKKLFIRIGIASWIYGFIWAVLPLLGFSSYTVENTRSRCSINFSPKTSVEKLYLIILMAFGFFIPIISILASCLFTARVINTKYKYFCVTYGKENVETKRYKKK EKKTFLSFIIMVLSFVVCWTPYATVGCFSAFTSLKIPKWLLHVAAFFGKLSALVNPFIYYWKDGLFKKCFLNIRFKTTKLLIRKSQENSQK* >CNOPg4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01103264 KLSALVNPVIYYWKDGLFK MDPVKITLVIIITIAIFTNTISLYFLYKKRKSNYVILCINLSFADLLQSIAGYIPALFLDTNIQKATTLCKLTAFFVAFPSFSAIAMLTGMALS RMVLLSSCFSSNLINYKKLFIKIGIFSWIYGLFWAFLPLIGFSSYTVEATHSRCSMNFSPKNIIEKAYLILIFAFG FCIPVTIIITSCLFTAHVIVTKYNYFYVTYGKENVETKLFKEKEKKAISSFLLMVLSFIVCWTPYATIGCFTAFTSVKTPNWLLHVAAFFAKLSALVNPVIYYWKDGLFKKRSVRKKSITKSLLIIKSEINKKVEHII* >CNOPg5_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01014309 KLSALVNPVIYYWKDGIFK MDPVKINLVIIITIAIFTNTTSLYFLCKKRKSNYVILCINLTFADILQSIAGYIPDLFLDTNLQKATTFCKLTAFFVAFPSFATIAMHAGIALSRMVLLSSCFSSNLINYKKLFIKIGIFSWIYGLFWATL PLVGFSSYTLEDTQSRCSINFSPKTIIEKAYLILIFAFGFCIPVTIIITSCIFTAHVIVTKYNYFYVTYGKENVETKLFKEKEKKAISSFLLMVLSFIVCWTPYATIGCLTAFTSVKTPK WLLQVAALFAKLSALVNPVIYYWKDGIFKKRSAREKSFTKSLLIIKSEINRSVDHIA* >CNOPg6_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01086515 KCSTIVNPVIYIWKDGLFK YK removed MNIEKICFFILSTTSTILNALSLYFSYRKMHKSNYVILCINLSFSDLLRSIAGYIPSLFIEEHINTATMLCKLSAFFIALTAYTAIAMITAIALSRMVLLSTCILSNKISYKALF IKIGVLSWIYGFTLAVMPFVGFSSYTLEETRARCSINFSPKTRVEKVYLILLIAFGFFIPVIIISISCLYTAYVMRTKYKYFNATYGRENIETKRYKEKEKKAFLSFVLMVLSFIVCWTPYAI IGFFSAFTSIKIFKWLLHLAVYFGKCSTIVNPVIYIWKDGLFKKSFISKRAQLIQLLTRKTRVSNRPMI* >CNOPh1_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01018050 KCSALVNPIIYCWKDSLLS lacks FR motif MNAVKTALLIVSSIATITNAVSLFFLYRKHKNNYVILCINLSLSDLVRSIAGYIPALFLETNLNRASTLCKLSAFFIAFSSFTTIAMLTA IALSRMILLSTCFLHSQINYKTLFIKIGFFAWIYGFTWAVMPFFGFSSYTLENTCTRCSIDFFPKTKVEKVYLILLIAFGFLIPIITILSSCLYTANVMRSQYNYFYMIYGKNNVETKKY KVKEKKAFSSFILMVLSFVVCWTPYATVGCFSAFTSLEIPKWLLHFAPFFGKCSALVNPIIYCWKDSLLSWYIKSKRSLIIKLLFKRSEDNLFTCQIEKI* >CNOPh2_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01085207 KCSALVNPVIYYWKDSLLN lacks FR motif MNAVKTALLVISSTATITNAISLYFLYRKHKNNYVILCINLSFSDLLRSIAGYIPALFLETNLNRASTLCKLSAFFIAFSSFTTIAMLTAIALSRMVLLST CFLHSQVNYKTLFIKIGLLSWIYGFTWATMPFLGFSSYTLENTNSRCSIDFFPKTKTEKVYLILLIAFGFLIPIITIITSCLYTANVMRSQYNYFYMIYGKNNVETKKYKVKEKKAFSSF LLMVLSFIVCWTPYATVGCFSAFTSLVIPNWLLHFAAFFGKCSALVNPVIYYWKDSLLNHYFKSKRLQIMKLLFRRSEDNLTILPIKEN* >CNOPh3_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01011197 KSSALVNPLIYYWKDGLIA lacks FR motif MDSIKLPLLIISLVSMITNGTSLFFLLKTKHKSNYIVLCINLSISDLLRSTVGYTTELFLDPNLQSASKMCKLSAFLVAFPSLTSIALLAEISLFRMALLNAPYLRNQVANKMLFRRNGILSWLYAFVWTVLPL IGFSSYSLETTKYRCSINFLTKTVIDKVYLLSIMCFGFIIPSMIVLISCLFTARIIAEKFKYFSRTYGIKNKKTISFKEKRNKAFTSFSLMFFFFVFCWLPYATISNLSGFTSIKLPMWL YHTAAYVAKSSALVNPLIYYWKDGLIAKKKIKAAGNNIFLYSR* >CNOPh4_hydMag Hydra magnipapillata (hydra) Cnid.Hydr.Anth ACZU01090363 KLSAMVNPLLYWQKEDFKK lacks FR motif MDAAKVIVTIVTILSTIFNALALFNLCKKRRKTNYIFFCIFLSVSDLLQTLLGYLPAIFLKKRMENATKWCQISAFFVAFPSFTSIALLTAISFIKAYLINEHFLIGDMIYRKVVKVTILCCWIYGLFWSTFPL IGVSSYTLESTKARCSIKWYTNTNTEKLYLILLLVFCYTIPVSIVLASTMISARFVRSSFKISSSSLGMGESFVKIFKLREAKVKFSLTLMCLSFIMFWTPYAIIGLLSGFARVKHPKWL LETAALFAKLSAMVNPLLYWQKEDFKKKVRKAFLVFRTYVFYS*
Hydrozoa: Cladonema radiatum (jellyfish) .. 18 opsin-like genes
A Dec 2007 paper reports 20 mRNA opsin candidates for the lens-eyed hydrozoan jellyfish, Cladonema radiatum. These generally classify as ciliary opsins and the ones tested are expressed somewhat appropriately. However even the best alignment to validated opsins is very weak, no better in percent identity than many non-opsin GPCR (here 13 were used as outgroup without rationalization). However back-blastp to GenBank nr shows no non-opsin GPCR among the best matches.
The authors did not establish the existence of retinal in this species nor show 11-cis retinal covalently bound to any candidate. They note Schiff base lysine occurs in correct homologous position but do not comment on the absence of counterion at traditional positions 113 or 181 (bovine RHO1 numbering). That lysine is necessary for an opsin but not sufficient (it might arise here from primer bias). Non-opsins such as GPCR176 can have lysine at this position as well, making it only semi-diagnostic:
CropN1 KFSVVSNPIVYVIFYKDFR
K S+++NP++++ K R
GPCR176 KVSLLANPVLFLTVNKSVR NP_009154
CropN1 KFSVVSNPIVYVIFYKDFR
K + + NP++YV + FR
LWS_homSap KSATIYNPVIYVFMNRQFR
The number of 'opsins' is excessive given numbers of validated opsins that occur in complete bilatera genomes, even allowing for eyespot developmental stage variations and auxiliary functions such as gonad photoreceptive gamete release. Recent gene family expansions might make more sense for chemosensory or chemokine recepters than opsins.
In this view, 1-2 bona fide opsins might lurk among the collection -- but which ones?. Opsins evidently experienced various gene duplications which appear subsequently co-opted to (unknown) non-opsin, non-photoreceptive roles. This causes them to nest confusingly within the opsins even though they are no longer photoreceptors. There may have been selection to maintain the buried lysine because it worked structurally at the time of duplication (offset perhaps by chloride ion) or is used for a new but chemically related signaling agonist. The same phenomenon (gene duplication and neofunctionalization) may have occurred within Nematostella and Daphnia, which both have 'too many' opsins for their imaging needs. The Daphnia non-opsin opsin gene family expansion is not even broadly shared within the Crustacean clade.
The confusion really arises from the notion of "terminally diverged" opsins first studied within Bilatera. That is, within deuterostomes, we observe a sequence of gene duplication and divergence say encephalopsin --> pinopsin --> LWS --> other cone opsins but that stays within opsins. Even that sequence had largely terminated 500 million years ago at time of lamprey divergence (primate color vision recovery is an exception). An analogous sequence is familiar within Arthropoda for example melanopsin --> MWS --> UVS. We don't observe the sequence encephalopsin --> pinopsin --> LWS --> bradykinin receptor in human nor melanopsin --> MWS --> glycoprotein hormone receptor in drosophila. Nor do we expect any such 500 million years in the future.
In Bilatera, it seems once an opsin, always an opsin. Gene duplication still occurs but opsins are apparently so deeply dug into their hole of specialization of function and tissue expression that a gene duplicate cannot be retained unless it can carve out a niche for itself as limited variant of photoreceptor opsin, say new color sensitivity or polarization detector. Lens crystalins prove that genes often have pre-existing multiple disjoint functions (glycolytic enzyme, refractive index supplier) and so at duplication the niche is already there, merely awaiting partitioning of expression after which sequences can optimize to their respective niches or just drift. Opsins in contrast are single-purpose.
Opsins may not have been so committed in early diverging ancestral cnidaria with less elaborated photoreceptive systems and metazoan cell type specializations. Not so terminally diverged, opsin gene duplicates may have retained overall GPCR signaling capacity but for some other agonist than cis-retinal. After all, shifts in outside molecular trigger happened frequently in generic GPCR evolution, accounting for their vast diversity of functionality despite minimal departures from the universal hepta-transmembrane structure. Ready shifts in agonist are not a design flaw but rather a design feature. Variation in agonist may be tolerated, especially in ancestral GPCR, and GPCR gene duplication and divergence coupled to that of a peptide agonist.
Opsins seem unique in that cis-retinal is covalently attached, whereas other GPCR agonists diffuse in transiently to their binding site. This makes it difficult to see what the 'next' agonist could be in a duplicated non-opsin opsin (other than something very similar like vitamin A variants seen in some teleosts). However it's been argued that the photoisomerization product, trans-retinal, is really the agonist. That's non-covalently bound similarly to other GPCR effectors.
If so, that would make agonist shift in a duplicated ancestral metazoan opsin no different from other shifts taking place in other duplicated GPCR. Indeed, opsins arose from other GPCR; the ur-GPCR was not necessarily a retinal binder. Still, we wonder what the new agonists could be in this cloud of cnidarian opsin-like proteins and what signaling is accomplished where. Hybridization in Cladodema suggests the site of signaling has not moved appreciably.
It's worth re-examining the notion of "once an opsin, always an opsin" even in Bilateran history. First we wonder about the cloud of lineage-specific opsin-like duplications in the crustacean genome of Daphnia. Using bovine RHO1 as query against human genome turns up a dozen non-opsin GPCR exhibiting better matches than the bona fide opsin RGR. Very likely these would nest within opsins with respect to RGR. This suggests that certain non-opsins occur inside the broader photo-opsin family. However here it is not so certain that RGR is a degenerate photoreceptor opsin, today 'merely' a retinal photoisomerase in boreoeuthere placentals retaining the Schiff base lysine but not covalently binding cis-retinal. A similar question arises with neuropsin and peropsin.
RHO1_bosTau KTSAVYNPVIYIMMNQKFR query NP_001044 somatostatin receptor 5 [Homo sapiens] 1e-26 NSCA--NPVLYGFLSDNFR non-opsin GPCR NP_001048 tachykinin receptor 2 [Homo sapiens] 2e-25 MSSTMYNPIIYCCLNDRFR non-opsin GPCR NP_000900 neuropeptide Y receptor Y1 [Homo sapiens] 1e-23 MISTCVNPIFYGFLNKNFQ non-opsin GPCR NP_000721 cholecystokinin A receptor [Homo sapiens] 1e-23 YTSSCVNPIIYCFMNKRFR non-opsin GPCR NP_000903 opioid receptor, mu 1 isoform MOR-1 [Homo sapie 3e-22 YTNSCLNPVLYAFLDENFK non-opsin GPCR NP_004212 G protein-coupled receptor 50 [Homo sapiens] 1e-21 YFNSCLNAVIYGLLNENFR non-opsin GPCR NP_006047 neuromedin U receptor 1 [Homo sapiens] 1e-20 LGSAA-NPVLYSLMSSRFR non-opsin GPCR NP_001471 galanin receptor 1 [Homo sapiens] 1e-20 YSNSSVNPIIYAFLSENFR non-opsin GPCR NP_005949 melatonin receptor 1A [Homo sapiens] 1e-20 YFNSCLNAIIYGLLNQNFR non-opsin GPCR NP_000614 bradykinin receptor B2 [Homo sapiens] 4e-19 YSNSCLNPLVYVIVGKRFR non-opsin GPCR NP_002912 retinal G-protein coupled receptor RGR_homsap 4e-18 KMVPTINAINYALGNEMVC opsin RGR_homsap NP_003292 thyrotropin-releasing hormone receptor [Homo 3e-18 YLNSAINPVIYNLMSQKFR non-opsin GPCR NP_005152 angiotensin II receptor-like 1 [Homo sapiens] 7e-18 YVNSCLNPFLYAFFDPRFR non-opsin GPCR NP_000901 neuropeptide Y receptor Y2 [Homo sapiens] 9e-18 MCSTFANPLLYGWMNSNYR non-opsin GPCR NP_000570 chemokine (C-C motif) receptor 5 [Homo sapie 2e-17 MTHCCINPIIYAFVGEKFR non-opsin GPCR NP_000670 alpha-1B-adrenergic receptor [Homo sapiens] 3e-16 YFNSCLNPIIYPCSSKEFK non-opsin GPCR
No genome project is planned so the mRNA cannot be examined for diagnostic intronation; this could be done indirectly if candidates help locate orthologs in other cnidaria. However they are already very diverged from Nematostella and Hydra opsins. Introns would not be informative anyway in discriminating opsins from co-opted non-opsin, non-photoreceptors arising as segmental duplications. Indels would be similar. This presents a very difficult bioinformatic problem because truly diagnostic residues of functioning opsins could be quite subtle.
Similarly on the experimental side, antibodies would likely cross-react. The derived non-opsins likely still signal with a transducin-type G-protein and are quenched by the same arrestin, so no help there. Knockdowns might have unforeseen consequences in these non-opsins, indirectly disrupting photosensitive behaviors. Thus the best way forward in identifying the true opsins within the collection is probably in vitro expression, reconstitution with cis-retinal, and demonstration of photoisomerization.
The set below contains 18 Cladonema 'opsins' plus miscellaneous other K296 cnidarian sequences >CNOPa1_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332416 Cladonema radiatum CropB1 MAVKTTYPVLIIAFLIVILNSILLLGFKKKYRRGFTTYDFFLVSFTIANLIQGVFTYPFVLYTLVSEQNKNHWQCIIAAVLMLNCSVSTIWHLIILSFERFF SLKYPLFYAKHNKSRLWRGIGVSIPWLSGCFWSFPPLFGWNRYDIEFQGRPDCNIDLRDHSKSGLSYLYSLFVFNYIVPIMAFAFTFYHLRIELKKISKMARRAIGVGSKIAKASM KTEKTQTILIIIMVVSFMVAWTPYSVLVFYRTAQENEIEVYIYELSAILAKTSCIFNPIIYGLVYKDIRDAILKSIVGKKRRNNRVSILATETDILPNYSVAMVSATPKVCSLDGCYN* >CNOPa2_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332417 CropB4 MAVETTYPVLIIAFLIVIFNSLLLLGLKKKYRRGFTTYDFLLVSFTFANLIQGVFTYPFVLYTLVSEQNESHWQCITSAVLMLNCSISTIWHLIMLSFERFF SLKYPIFYAKHNKSWLWRGVGVSIPWLSGCFWSFPPLFGWNRYDIEFQGRPNCNIDLRDHSKSGLSYLYSLFVFNYIVPIMAFAFTFYHLRIELKKIFEMARKAIGVESKIAKASK KTEKTQTILIIIMVVSFMVAWTPYSVLVFYRTAQENEIEVYIYELSAILAKTSCIFNPIIYGLVYRDIRDAILKSVVWKKRRNNRVSAIATKTEILRHNNVAVVNASSLV* >CNOPa3_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332420 CropC MTGPLVFIFLLAGIILNLLVIFVVLKKQNRTVKHILVISFACCLLFEIFFGFIWEGYGRYIDDSSLELCKMAGFGATFSSLASIMHLVDMALERYISIIHPM TAYQYFNNYMLGIYMVIPSWIIGFVWAFFPFLGWGEYMREANHSYRCSIVGTSNSVNAQSYNYTMLIVFFFLPIIIITYLCVRIQMQLVKTRKKAAETFKGATKMIDGRRKYEKQH FVLVCINIIAYFVTWAPYAISTCWYTFFNSAPDDVIGYSALFAKSSLVTNPIIYVFFYREFREILKATLFKKNTTPSVEMPTIEQSTEQ* >CNOPa4_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332422 CropD MDAVIGSLVLLYLIASVILNILVIVAVLKKGNHTFKDILVISLSCSLLSEIGFGFAWEGYGRFIDDSSLELCKFAGFGATFSAFISILQLMGLAFERYVSII HPVTAHSHFKGYTLAMLFVIPSWAFCIIWASLPFYGWGEYMREAVHSYRCSVVGTGKSYDAKSYNYSLLILFFLAPVLISTYLLVRVHMELVKTRKRAGERSTGTTSITSRIWKYE RQHFFLTCFVIILFFLTWTPYALSAGWYTFFDSNPDHVVNYSALFAKSGLVTNPIIYVLFNKELRKTLKRKILERKPLITTDIQLDSITH* >CNOPa5_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332421 CropE MDELTGTLVFLFLLVGIILNLLVIATVLKKKNRKVKDVLIISLACAMISEICFGFVWEGYGRYIDDTSLELCKMAGFGSAFSAFVSILHLIAMALERYISIV HPLKAYQHFENHMLAMYMVVPAWVIGLAWALFPFFGWGVYLREEVHTYRCSAIGFIKPSNSQSYSYGLLIFFFFLPSIIIIYLCVRVQMELIKTRKNTAEKFQGATMMVADSKKCE KQHFVLVCFIIITYFLTWTPYALSACWYTFFDSKPDHVVSYSAIFAKSGLVTNPIIYVFFYREFRETLITKVFKRKAPSPSNPSTSVEMKTRSTTNN* >CNOPa6_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332426 CropF MAEVTAPIVGLFLIAGIFLNVLVVIGMLKKQSLVVKDFLVVSFACCSLVEICFGFIWEAYGRYIDDSSLTLCKMAGFGTTFPAFTSIIHLVAMAFERYISIL HPMTAHKYFVSSKVAFYFIAPSWLFGFVWAFLPIFGWGGYIREKLHTYRCSVIATDNSAVAQSYNYSLLVLYYFLPSSLIIFCCVCVQLEIQKMTRRISNGLSSAMKERTKKTERN HFILVCIVIISYFLSWTPYAISACWFTFFNTAPDELVSYSALFAKSCMITNPLVYAFFYKEFRETLKNKVFSRNIRVQTVEAPTSSLQDTKETNH* >CNOPa7_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332427 CropG1 MANLKHVEGGIIMSFIVASVSLNSLVIVGMLKKPNRALRDLILVSLACSDLVETCVGIMWEAFARYIDSSSFILCQLAGFGITFTALVSITHLAALAIERYL SMVHALKTYEFFADKKKSLLFTIPSWIHGTFWGVVPFFGWGGYIREPYHTHRCSVMMTDNSLIAQSYNYALLVFCYLIPIFIIFYCCYKVQKELKKMTERSEEIAGSSSSITQTTK KAEKQHFIMICVVMGAYFIAWTPYAVVACWFAFFDTVPGKLIAYSAVFAKFSVIANPIVYVIFYKDFRQTVKRMFCGRGRVVPA* >CNOPa8_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332428 CropG2 MANLKHVEGGIIMSFIVASVSLNSLVIVGMLKKPNRALRDLILVSLACSDLVETCVGIMWEAFARYIDSSSFILCQLAGFGITFTALVSITHLAALAIERYL SMVHALKTYEFFADKKKSLLFTIPSWIHGTFWGVVPFFGWGGYIREPYHTHRCSVMMTDNSLIAQSYNYALLVFCYLIPIFIIFYCCYKVQRELKKMTERSEEIAGSSSSITQTTK KAEKQHFIMICVVMGAYFIAWTPYAVVACWFAFFDTVPGKLIAYSAVFAKFSVIANPIVYVIFYKDFRQTVKRMFCGRGRVVPA* >CNOPa9_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332423 CropH MDAVIGCLVSLYLLAGVILNLLVIVAVLKKGNHTFKDVLMISLACSLLSEICFGFVWEGYGRFIDDSSLELCKIAGFGATFSAFVSIIQLMGLVLERYVSII HPVTAQSHFRGHTVAMLFVIPSWVFCIVWASLPFYGWGEYMREAVYTYRCSVVGTGKSYNARSYNYSMLIFFFFAPILIITYLLVRAQIRLTMTRKMAAARSTGTTATTARIYKYE RHHFILTCFVIIMFFLTWTPYAVSGCGYTFFDSSPQHVVNYSALFAKSGLVTNPIIYVLLNKELRKTLKRTIFGRKPLTTTNMQFDSITQ* >CNOPa10_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332424 CropI MDAVMGSLFLLFLIAGTTLNILVIVAVLKKRKHTFKDIMVISLTCSLLSEIYFGFVWEGYGRFIDDSSLELCKFAAYGATFSALASIMQLMGMVLERYVSII YPVTAHSYFKGNTLAMLFVLPSWAFCIVWAFLPFIGWGEYKREAVHTYRCSVIGTGKSYNARSYNYSLLILFFFAPIIIITYLIVRVQMELAIIRKMAAARSTGTTATTTRIWKYE RQHFILTCIVIVLFFITWTPYALSACWYALFDSTPDKFVSYSALFAKSSLVTNPIIFVFFNKEFYKTLKRNIFQRNPLANTDIQLDSITQ* >CNOPa11_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332433 CropJ MFNAGLVIGVVIAIGFSCFLNLLVVIGMLKKSTPLSLRDILLVSLSVADFCQTGLGYTTELYGHLSDDYSPIFLCRASGFAVTFTALVAITHLMALAVERYL SLVHALKAHEFFSEPKRALYFIIPAWIWGLFWSGAPLLKWGGYEREPHAPHRCSVVLAEKTPDVLSYSYGLLIACFAFPVLVITWCCFRVQRELKGMTDRSKEIAGQESAITAATQ KAEKQHFIMICCVVIAFFIAWTPYAGCVVYFTLVGNLPANLMSTSAFFAKFSVLANPIIYAIFYKDFRRSVKAMLCGDSAVAPSE* >CNOPa12_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332431 CropK1 MLLEIESIMVSLFAANIVLGFSMNLVVISALCRVSGKSVRDILLISLALCGLLETGLGYAIELYGRLNVRASSLLCQMAGFSVTFFALSSIFHIVGLAIERY LSIVCAVNARGYFSMKHRALYIIFPAWINGLVWSIAPFLGWGAYKREPIDKYRCSVLMSDVSFVSWSYTIMLLTFAYAIPLLVILYCCLKVQCTLNAMSKNISDPENRTRDSGLEA TKQAEKRHYYMVCAIIVTFFVAWTPYATSALLFSFLENVPNTLITYSAFFAKFSVVANPVIYVIFYKDCRKQVKKMVCVDTAVAPIEA* >CNOPa13_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332432 CropK2 MLLEIESIMVSLFAANIVLGFSMNLVVISALCRVSEKSVRDILLISLALCGLLETGLGYAIELYGRLNVRASSLICQMAGFSVTFFALSSIFHIVGLAIERY LSIVCAVNARGYFSMKHRALYIIFPAWINGLVWSIAPFLGWGAYKREPIDKYRCSVLMSDVSFVSWSYTIMLLTFAYAIPLLVILYCCLKVQCTLNAMSKNISDPENRTRDSGLEA TKQAEKRHYYMVCAIIVSFFVAWTPYATSALLFSILENVPNTLITYSAFFAKFSVVANPVIYVIFYKDCRKQVKKMVCVDTAVAPLEA* >CNOPa14_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332425 CropL MDDSSMDAITGILVVLFLTTGIILNTLVILAVMKKSSHTFKDVLVISFTCCLLGEICFGIIWEGYGRYIDDSSLALCKMAGFGATFTTLVSIIHLIGMALER YISIIHPMTAYQYFKDHMLAMYFLIPSWVFGIAWACFPFFGWGAYMREPVHTYRCSVVASSNSNDAQSYNYSLLIFFFFLPIILIIYLCACVQIELAKTRKEAIETLTGDSAMIDS RKKHEKEHFVLVCFVIITYLVTWTPYAISACWFAFFKSQSNEMVGYSALFAKSALVTNPIIYVFFYKEFRETLKTKVFTRKASTSVDMPIRSSLSQKIDIHE* >CNOPa15_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332418 CropM MTVKGEDISIIVIVIFSTFLNICLILALVQKTSKPNFMPCLLLNMALCSLLQGLFAYPLELNNIFKASVTSDVTDISCVVSAVMVYGLSLIVIFTITLLSIE RVLAIKFPYLYRKCERNHKCFLVFSIMIIWALGAFWACAPLFGWSNYQKETDDSHRCNINFTRKDANNASYLCTLLVSCYVIPVSIMVSSYTLVKNSMRRMSVYADQRVGKNATLS KNSKKSEKQQTLLTILMIFAFLYAWTPYTTVVFFLMIDKRVSPTIYTIAALNAKTSSFYCPLLYGFVYKRFRRRLKDVVAKIFV* >CNOPa16_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332429 CropN1 MADEVGGPVILVFIVGSVFLNLLVIVGMLKKPNIALRDLILVSLAISDFIETVFGSLWEAYAKYIDSSSLFLCKMAGFGITFTALTSISHLAALAIERYLSM VHALKTYEFFADKKKAIIFIIPAWTYGTFWGGVPLLGWGGYFREPIHTFRCAVLMTSKSFLSMSYNYCLLAFCYLLPISIISICCYKVQRELKNMTERSEAIAGSSSAITESTKKA EKQHFLMICIVLVAFFIAWTPYAFTACWFAFFDNPPDELVTYSATFAKFSVVSNPIVYVIFYKDFRQTVKKMFCGRGRVAPV* >CNOPa17_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332430 CropN2 MADEVGGPVILVFIVGSVFLNLLVIVGMLKKPNIALRDLILVSLAISDFIETVFGSLWEAYAKYIDSSSLFLCKMAGFGITFTALTSISHLAALAIERYLSM VHALKTYEFFADKKKAIIFIIPAWTYGTFWGGVPLLGWGGYFREPIHTFRCAVLMTSKSFLSMSYNYCLLAFCYLLPISIISICCYKVQRELKNMTERSEAIAGSSSAITESTKKA EKQHFLMICIVLVAFFIAWTPYAFTACWFAFFDNPPDELVTYSATFAKFSVVSNPIVYVIFYKDFRQTVKKMFCGRGRVAPV* >CNOPa18_claRad Cladonema radiatum (jellyfish) Cnid.Hydr.Anth AB332419 CropO MKVENITIVVIVALSVVLNICLIAVLLQKTLRYTFMHYFLFNMALCSLFQGLLAYPLELSSLLKGRKKMDFECLMGAILVFGLALVVIWTIALLSIDRVLAM KFPYLYKRCIRNKKRFVFLSITFTWLIGGFWALTPLLGWSSYQKEMKPSHRCNINFRSKDPVNMSYLCSLMVTCYVVPVTIMISSLTLVKKSLRKMSIYAEKRVGRRASLSLDAQR CEKRQTLLTLLMIFSFFYAWTPYTIVVTFLMLDKSVPPTVYTIAALNAKTSSFYCPLLYGFVYKPLRSTFKSAVSKIMV* >CNOPa1_claRad Podocoryna carnea (jellyfish) Cnid.Hydr.Anth AB332435 PcopC MDRTTFTAFLTIIVALSASLNTAIIIALIRTWNKLSSYDVTLLFLTTVELLQAVIGYPIELTTNLREGGPLCIIAGYSTTTLALTSISILVALSFMRLLSVT QPLKAVEWIKTRRYSIYLILPSCIYGFMWGIFPLIGWSEYAPELGSNKRCTIDLHSEDNATRSYLYSLLLFCYVLPLTTMTINFYKIRKKIIEQSVKLSQNRERIRTNRIKRERRL AIATFVMTVVFLVAWTPYAVAIFIIGFQKRVDEAWFFMDYSSLLAKSSTFFNPLIYVLMHRKCKTAVVDMFQYIICLIRRRDSQASVSYNGSAQRVQFLHIASMSSQKPDTCSPCV HIQRSEYLVAKDAANGPNYSFTATNI* >CNOPg1_claRad Podocoryna carnea (jellyfish) Cnid.Hydr.Anth AB332434 PcopB MDPLKIILSSIVVSSIILNTATCFILIRKKRKTKFVILCIHLAISDLLQAVCGYVPELIIQKNNFPTLLCKASACTVAVMALASISLITLISITRIISIKYP FTSVSTSKVTVNILAIISWLYAAFWAICPLLGWSQYVLEPTHRRCSLDWNLKTANSKSYIYAIVLFCYILPITIHVISFFMVRRTVENHARFCADTYGRRNSATLIVERKKLKSFF LSLSMFSIFIVAWTPYTLVGISAQYVTVPTWSMDSAAMLAKSSTLFNPLVYCYRTKFRRKNIFREDNVTEDNITMLRTSLRRNQCDNNGNVLNGESVNVCYDKKSLSISFGVIEESQT* >CNOPa1_clyHem Clytia hemisphaerica (jellyfish) Cnid.Hydr.Anth CU434354 MVLEVTYEHIFLLVMVTLGTILNLLVIITLLFMMKKGNHELRNVILMSLAVSDMLPALFGFSLELTSTDDIMFCKISAFAVTSSSLSSIMH MVLLMIERAYTIKYPYRAHIVLKRKSIMACFVSIAWIYGTVLGVCPLLGWGNYTHEGVSMKRCSIDINMNTWSVTTYNYSLLLLGYLIPIAASVACYVVIKKEHGKLSEKAVRYQGKSPL MTaHTMDIQRSFTLMFLVMLLAFLISWT >CNOPa1_monFav Montastraea faveolata Cnid.Anth.Hexa GW273473 fragment ASFIICIFVIFFLFPVVSLVVSYAIILQQISQMAKRAKSKWDDEAQQTLEILLARKKVVWTGFIMVASFVVVWTPYAILSFY AAFGKPQSIPPLAATIPALFAKTSTVLNPIIYVIRYKRFRKGVKKLFKMI*
Ctenophora .. 2 opsins
Ctenophores are an early diverging metazoan group, though it is not quite clear whether they should be sistered with cnidarians or split from the metazoan stem earlier. The data situation has greatly improved in the last year with transcript programs and a completed genome project with quite large contigs (relative to gene size).
Ctenophora: Pleurobrachia pileus (sea gooseberry) .. 2 opsins
The first bona fide ctenophore opsin has surfaced in an unannotated cDNA (CU419614) deposited to GenBank in February 2008 in the course of a never-published phylogenetic study of sponges (Manuel, Houliston: "The sponges reunited: implications for early animal evolution"). This planned manuscript may have been subsumed in a different article that concluded calcisponges, homoscleromorphs, demosponges and hexactinellids form a basal monophyletic metazoan group.
This paper also sistered Ctenophora + Cnidaria, a tree topology reducing the number of mainstem metazoan nodes by one. It may take complete genome sequencing to determine this relationship reliably -- it has been debated for over a century. The Mnemiopsis genome is 150 Mbp so easily within an afternoon's reach and indeed became available in Oct 2010. Haeckelia rubra, the species whose ingested and recycled cnidarian nematocysts once confused its taxonomy, is 10x this size.
Genomic sequencing must disentangle ctenophore dna from that of the parasitic cnidarian Edwardsiella lineata whose planula larvae enter the ctenophore mouth as fake food but then bore into the mesoglea and help themselves to ingested pharyngeal food. Up to 60% of a Mnemiopsis population can host these larva.
Ctenophora full length genes are poorly represented at GenBank with only 156 nucleotide entries for the whole phylum, so vision-related entries cannot be expected there. However transcripts are in better shape with 24,292 as of Jan 2010. These consist entirely of the lobate Mnemiopsis leidyi gastrula library (15,752) and cydippid Pleurobrachia pileus (8,540 cDNA, undescribed library) despite ~150 known species. A negligible number of reads (3,360) from early cleavage Mnemiopsis can be found in the trace archives. These two at least represent the two main groups of ctenophores at according to GenBank taxonomy, Cyclocoela and Typhlocoela.
The sensory system of ctenophores is located at the rear (aboral pole, ie head is not around the mouth) and can connect to the non-centralized nerve net. The statocysts, suspended by four fibers like those of Ciona or for that matter human otoliths, report to nerves coordinating cilia locomotory beat. Statocysts would deflect according to gravitational field lines and acceleration, allowing orientation relative to the vertical. Water current velocities could be detected in a distinct system of stiff cilia projecting out into the water that bend in proportion to current.
The ectodermal sensory region also contains four crescent shaped groups of round bodies, interpreted as photoreceptors on page 166 of a 1880 monograph by Chun. Ctenophores lack an overt response to shadows; these eyespots could not plausibly provide imaging vision or function in predation or avoidance. They could however track length of day length, distinguish up from down and serve as depth detectors. Elsewhere, epithelial walls of food distribution canals have osmoregulatory rosettes, light-inhibited bioluminescent photocytes utilizing the original green fluorescent protein and testis and ovary on opposite canal walls.
The eyespots, statocysts, photocytes and gonads are the most plausible sites for opsin based photoreception in ctenophores. No data is available on ctenopsin anatomical expression as all cDNAs are tied to a whole-organism library. Nothing has been done to date to localize opsin expression because until now, no sequence has been available. An unfortunate term 'mnemiopsin' was introduced and abandoned in the 1970's. It apparently references the bioluminescent protein which completely lacks any resemblance much less homology to opsins.
The initial Pleurobrachia opsin sequence was not full length as it starts 27 residues after a presumptive DRY motif and continues past the Schiff base lysine and an altered NPxxY motif but not to a stop codon. However 5 cDNAs posted in May 2010 extend it to full length. While the sequence is quite diverged from any known opsin (low 30's for best percent blastp identity), the evidence is nonetheless overwhelming that this is an opsin:
- the candidate sequence turns up as best tblastn match of any opsin query at GenBank restricted to Ctenophora
- when translated and aligned to 430 curated opsins, residue matches are overwhelmingly concentrated in conserved sites (graphic below)
- the sequence has lysine at homologous position K296 (bovine rhodopsin numbering) to Schiff base lysine, unlike any non-opsin GPCR
- opsin-diagnostic regions (absent in the most closely related GPCR) are observed, notably the GWSR motif retinal plug in EC3: PLVGWCEYGPEGY
- best back-blast to all of GenBank returns opsins ahead of all other GPCR (in contrast to candidate CU422164 whose best back-blasts are serotonins)
- the sequence roots in the tree of all opsins as expected for a ctenophore photoreceptor (tree below)
- the sequence classifies evenhandedly among ciliary, rhabdomeric and cnidopsins as expected for an early diverging opsin (list below)
While establishing the presence of a K-rhodopsin in this ctenophore, this analysis hardly proves that this protein functions directly in photoreception (as opposed say to photocycle regeneration, bioluminescence, carotenoid metabolism, gamete release or statolith orientation). The anatomical characterization of potential photoreceptors appears limited to an offline 1979 Russian text "Electron microscopic study of presumptive photoreceptor cells in the aboral organ of the ctenophore, Beroë cucumis" by MZ Aronova. It is not clear whether these classify as either ciliary or rhabdomeric ("presumptive photoreceptor cells which have a short central projection and form in their basal part the synaptic contacts..."). Elsewhere it is said ctenophores do not react to shadows and while the eyespots cannot form an image they may track changing day length and depth in the water column.
There are also significant deviations from near-universally conserved residues believed critical to the Galpha signaling partner interaction, notably in the NP..Y......FR region which here is WI..Y......IK. While Galpha proteins (and especially their fifth helix which binds to the C2/C3 mitt of opsins) are conserved to still greater phylogenetic depth, the protein fragment here is difficult to evaluate without more of its upstream residues. It could be chimerized to full length using a known opsin (but which one?). As it stands however, the fragment gives highly significant positive scores to both Gi/o and Gq type signaling partners. The 3rd cytoplasmic loop lacks the HEK motif common to most but not all melanopsins, so provides no further clues.
A second apparent ctenophore opsin appears as an unannotated cDNA fragment FQ011385 that aligns only up to residue 203 in RHO1_bosTau numbering, thus its putative lysine at residue 296 remains speculative. Its ERY motif is more conventional than the other ctenopsin.
The phylogenetic position of ctenophores has been subject to endless back-and-forth for two centuries. This discussion has intensified very recently with the advent of substantial molecular data. While can hardly be resolved by the one protein fragment here, for an opsin it has many primitive features that do not favor backsliding from cnidarian and bilateran opsins sharing immensely conserved contemporary features. Thus it is not consistent with ctenophores branching later than cnidarians but rather earlier (or sistering).
This opsin and its role could be greatly clarified by determination of the full length sequence and its sites of expression. It potentially represents an opsin descended from an era when melanopsins, cilopsins and cnidopsins were coalesced into a single gene. Another way of saying this is all these opsins are orthologous relative to the last common ancestor of sea combs, anemone, fruitfly, squid, and human. Peropsins, neuropsins and RGRopsins are still excluded and represent even earlier gene duplication and divergence.
Best blastp matches to reference opsin collection:
ENCEPHc_nemVec Nematostella vectensis (anemone) Anthozoa 8.0e-25
ENCEPHb_nemVec Nematostella vectensis (anemone) anthozoa 4.5e-23
ENCEPHa_nemVec Nematostella vectensis (anemone) anthozoa 8.2e-23
PARIE_takRub Takifugu rubripes (fugu) Gd+Go -HSP90B1 4.2e-20
PARIE_danRer Danio rerio (zebrafish) Gd+Go - +NT5DC2 4.2e-20
MEL1_acrMil Acropora millepora (stony_coral) anthozoa 6.0e-20
MEL1_lotGig Lottia gigantea (limpet) FC774055 ests 1.1e-19
CUBOP_carRas Carybdea rastonii (sea_wasp) cubomedusae 1.5e-19
LMS1_hasAda Hasarius adansoni (jumping_spider) Chelice 1.6e-19
TMTx_braFlo Branchiostoma floridae (amphioxus) XM_0022 1.9e-19
ENCEPH_strPur Stronglyocentrotus purpuratus GLEAN3_ 3.3e-19
Sample alignment to the ciliary parietopsin of Takifugu (with cytoplasmic loop C3 and Schiff lysine region colored):
plePil 6 FFGIAPLVGWCEYGPEGYGVSNSLMWNNLNDNNASYIICAMIIGYFFPLIIIMFCYRAIY 65
F+ + PL+GW YGPEG S SL W + NN SY+I ++ + FP+ +I++CY +
takRub 61 FWAVTPLLGWSSYGPEGVQTSCSLAWEERSWNNYSYLILYTLLCFIFPVGVIIYCYCKVL 220
plePil 66 RLVQAQLNTSVVKMTVSANVTSLSTNRNHIMLVQERQLASTITFVILSFFFAWTPYVFVN 125
+ +LN SV + ++ N++ I +V LA +I++FF W PY ++
takRub 221 TSMN-KLNKSV---ELQGGLSCRRENKHAINMV----LA-----MIIAFFVCWLPYTALS 267
plePi l26 IINMFSTLILKYRILATIPALFAKSSTLWWIIVYCLMDVRIKKACRK--TCLRF 177
++ + + ++AT+P FAK+S ++ I+Y L + + + A + +C R+
takRub 268 VVVVVDPELHIPPLVATMPMYFAKTSPVYNPIIYFLSNKQFRDATLEVLSCSRY 321
>CTENOP1_plePil Pleurobrachia pileus (sea_gooseberry) Cten.Typh.Pleu FP995093 CU419614 full G? ctenop EQY intronated via Mnemiopsis ortholog
0 MELRVSLSLIMACVCTGSVF 1
2 GNMLVFISLVKERPLPNPAVEMIFRHLSLANLVLAGIGEPLVVVSLAA 1
2 GKWVWGESMRYFEAFWVTTFGLFSMMILGLISFEQYSRFVRQKQLKTHQ 0
0 VAVLLSGIYFLAFFFGIAPLVGWCEYGPE 1
2 GYGVSNSLMWNNLNDNNASYIICAMIIGYFFPLIIIMFCYRAIYRLVQ 0
0 AQLNTSVVKMTVSANVTSLSTNRNHIML 0
0 VQERQLASTITFVILSFFFAWTPYVFVNIINMFSTLILKYRILAT 0
0 IPALFAKSSTLWWIIVYCLMDVRIKKACRKTCLRFKRSFRVWYFS* 0
>CTENOP2_plePil Pleurobrachia pileus (sea_gooseberry) Cten.Typh.Pleu FQ011385 ERY intronated via Mnemiopsis ortholog
0 MTTAVDDEMMGDEEPKVVYTLASLLAVNALCSIA 1
2 GNALVIYTFLKEKPLGAPLRVILTHLAVTNLIIASIGEPMVVISGYS 1
2 MRWVFGEMGRGFEAFIVTSCGLMAMSLLALVSIERYLR 2
1 VTPRQRFPFTEPTAVKCCSFLWVFNVIYASLPFYGIAAWEGE 1
2 GIGISNSITWTTEHTQDFVYIICMMSTGYFIPLFIIAFCY 2
Ctenophora: Mnemiopsis leidyi (sea walnut) .. 2 opsins
The first ctenophore genome assembly became available to NCBI's wgs tBlastn in Sep 2011 after being held back nearly a full year. This species has two opsins, one each of ciliary and melanopsin type, that are clearly orthologous to the two Pleurobrachia opssin available from transcripts. These opsins, unlike those of cnidarians, prove quite rich in exons with 8 each, matching each other in three but not at all with bovine rhodopsin. The two opsins are about 39% identical. Neither have close matches to cnidarian or bilateran opsins though CTENOP1 classifies clearly with ciliary opsins and CTENOP2 less clearly with melanopsins.
The ctenophore opsins are not closely related to any non-opsin GPCR either in ctenophores or human. Within the human proteome, CTENOP2_mneLei matches peropsin, melanopsin and rhodopsin equally poorly. The first GPCR encountered are somatostatin, GPCR136 and chemokine receptor 6. CTENOP1_mneLei has similar affinities. Neither CTENOP elicits a K296 match in sponge, the best matches being to sponge proteins resembling human TSHR (thyroid stimulating hormone receptor). These non-opsin GPCR are among those already identified as parental to opsins.
The existence of ctenophore opsins shows that this gene family was already well-established by the time of ctenophore divergence from other metazoans. If no opsin can ever be located in sponges, then the timing of opsin origin is quite well defined.
>CTENOP1_mneLei Mnemiopsis leidyi AGCP01013851 AGCP01013853 77% identical CTENOP1_plePil
0 MGLRISLSIAMGCVCVGSVF 1
2 GNLLVLISLVRERPLPNPAVQLIFRHLSLANFLLACIGEPLVVISLAS 1
2 GKWVWGESMRLFEAFWVTTFGLLSMMILALISLEQYSRFVRQKQLRTVQ 0
0 VAVLLSGIYFLAFFFGIAPLVGWCEYGPE 1
2 GYGVSNSLLWNELTDNSASYIVTVMIVGYFFPLVIITYCYRSIYQLVQ 0
0 AQSNVAIVRTMVSVNVTSLAVNRNHSLI 0
0 VQERKLATTITFVILSFFFAWTPYVLINLLNMFTTWVVKYKILAT 0
0 IPALFAKSSTLWWIIIYSMMDKRIKKACRKTCEHFQRALRIWYFSS* 0
>CTENOP2_mneLei Mnemiopsis leidyi AGCP01013109 AGCP01013108 AGCP01013106 6th exon uncertain
0 MSSPNDEPTVDSDPKAVYILTSLLCMNCIFSLV 1
2 GNMLVIYTFLRERPLGAPLRTILCHLAITNLMIAGIGEPMVVISGFN 1
2 VRWVFGEMGRKIEAYTVTASGLMAMSLLAFVSIERYLR 2
1 VTPGQKFLIRQTTSVRVCTLMWIYNVIYASLPFYGIAAWRGE 1
2 GIGISNSLSWDTNNKIDVAYVMVMMATGYFIPLAVITFCYNRYVEIQQRLSTFL 2
1 0
0 AAEKKISNVVAIIIVSFFISWTPYTIVNLLVTFDKAGR 2
1 GINATIPAFLAKTSTVWSPIIYCFMNGEVSIFVLEGGHVMKYLYL* 0
Comparison of CTENOP1 with CTENOP2: numbers denote exon boundaries and phases
1 2 0 1
CV-CVGSVFGNLLVLISLVRERPLPNPAVQLIFRHLSLANFLLACIGEPLVVISLASGKWVWGESMRLFEAFWVTTFGLLSMMILALISLEQYSRFVR-QKQL--RTVQVAVLLSGIYFLAFFFGIAPLVGWCEYGPEGYGV
C+ C+ S+ GN+LV+ + +RERPL P ++ I HL++ N ++A IGEP+VVIS + +WV+GE R EA+ VT GL++M +LA +S+E+Y R QK L +T V V L IY+ + P G + EG G+
CMNCIFSLVGNMLVIYTFLRERPLGAP-LRTILCHLAITNLMIAGIGEPMVVISGFNVRWVFGEMGRKIEAYTVTASGLMAMSLLAFVSIERYLRVTPGQKFLIRQTTSVRVCTLMWIYNV--IYASLPFYGIAAWRGEGIGI
1 1 2 1
0 0 0
SNSLLWNELTDNSASYIVTVMIVGYFFPLVIITYCYRSIYQLVQAQSN-VAIVRTMVSVNVTSLAVNRNHSLIVQERKLATTITFVILSFFFAWTPYVLINLLNMFTTWVVKYKILATIPALFAKSSTLWWIIIYSMMDKRI
SNSL W+ +Y++ +M GYF PL +IT+CY ++ Q S ++ ++ + N+TS A ++N + E+K++ + +I+SFF +WTPY ++NLL F I ATIPA AK+ST+W IIY M+ +
SNSLSWDTNNKIDVAYVMVMMATGYFIPLAVITFCYNRYVEIQQRLSTFL AAEKKISNVVAIIIVSFFISWTPYTIVNLLVTFDK--AGRGINATIPAFLAKTSTVWSPIIYCFMNGEV
2 0 2
Placozoa: Trichoplax adhaerens .. 0 opsins
The sequenced genome of Trichoplax adhaerens appears in the 21 August 08 Nature and associated [98 pages of supplemental], an excellent treatment as first assembly articles go but still insufficient detail to replicate various bioinformatic assertions. The genome and browser are hosted at JGI but it is equally convenient for most purposes to simply use NCBI tblastn targeted to the WGS contig division restricted to Trichoplax or assembly blast.
The authors identify 4 genes as photoreceptive opsins, based on GPCR blast character, Schiff base lysine in expected position (though that position was not described), and unpublished accounts of phototactic behavior. However notation used in the document (scaffold8_1356180_1396180 = Opsin 6, scaffold13_1543556_1583556 = Opsin 26, scaffold8_1308974_1348974 = Opsin 30. scaffold8_1467859_1507859 = Opsin 31) does not match that provided at GenBank nor JGI genome. Gaps in assembly are indicated by N's which may or may not be counted in coordinate numbers. Opsins with these scaffold numberings do not surface by tBlastn of GenBank WGS where the Trichoplax contigs reside. The authors have not responded to an email request to furnish the fasta sequences.
In light of similar claims made for Hydra, Cladonema and Nematostella opsins that didn't bear up to scrutiny, given the lack of photoreceptor structures and cell-specific expression staining in Trichoplax, the burden of proof for opsins has not yet been met. Nothing in the Trichoplax genome meets the best-blastp threshold (exceeding the 25-35 percent identity range of generic GPCR matches) of the opsin classifier. Bona fide opsins also require a counterion E at ancestral position, a DRY motif, a binding motif for alpha subunit of heteromeric G protein, and about 150 semi-invariant residues that distinguish photoreceptors by ortholog class (eg ciliary vs rhabdomeric) and separate them from non-photoreceptive 'EK-rhodopsin-class' generic GPCR.
The only validated pre-Bilateran opsin to date occurs in the imaging eye of the cubozoa Tripedalia cystophora where a single ciliary opsin sequence meets all expression and classification tests. This opsin has no noteworthy matches in Trichoplax. Cnidarian larva also have a rhabdomeric structure implying that class of opsin as well.
Examples exist of non-opsin non-photoreceptive GPCR that contain lysine at the retinal position, ie this lysine is necessary for an opsin but far from sufficient. They are easy to collect using blast of consensus opsin sequence ending somewhat past that lysine. Cnidaria are especially rich in these lysine-containing non-photoreceptors. Indeed the 'opsins' reported for Hydra, Cladonema and Nematostella greatly exceed the (non-observed) photoreceptive structures and organismal needs -- which surely aren't twice those of the most advanced bilatera.
Nematostella: no photoreceptor structures but 16+ GPCR genes with lysine in 'retinal position': >tpd|BR000664 ID: 44/121 36 FLFAWTPYAVVSLWTTFGDTHRIPALLGVLPSLFA K LSSCYNPIIYFFMYTKFR 39% ENCEPH4_braFlo * >tpd|BR000663 ID: 48/121 39 FLFAWVPYAVVSLYASFGGVTTIPKLMSTLPAMLA K TSACYNPIIYFFMYSKFR 35% TMT5_braFlo * >ref|XM_001637473 ID: 35/115 30 FLACWLPYVIVSTCMSLGRRPQISLLTLEITLLVA K SGVIYNPFIYAALNLRFR 33% MEL_schMed * >tpd|BR000666 ID: 32/81 39 FLIAWLPYAIVSLYSAITGE.RVSPEAATIPGMLA K SASCYNPLIYVFLYSRFR 33% TMT_ornAna * >tpd|BR000667 ID: 38/121 31 FLVAWSPYAITSLYISWTGIQTIPDIARILPPMFA K AYSCYNPVIYYGMSRKFR 29% MEL2_xenLae * >tpd|BR000671 ID: 28/66 42 FVISWSPYCVVSVMAMVHRAPSLPRGLAEIPELMA K ASVIYNPLVFTVMNIEFR 32% VAOP_takRub >tpd|BR000685 ID: 30/75 40 FTLSWSPYAIVCLKSMVVGEQKLAPFTSEVTALMA K ASAVYNPIVYVVLSKRFR 35% MEL1_schMan >ref|XM_001634921 ID: 37/107 34 FLVAWTPYAVISFYSITGHARDLSFISVVLPAIFA K TSAFYNPIITLVFWFCLR 37% RHO1_letJap frag >tpd|BR000662 ID: 36/107 33 FVFAWTPYAVVSIYS.ALLKPKLPLIAGILPPLFA K TSTLYNPLLCFIGSPPIR 31% ENCEPH4_braFlo >tpd|BR000672 ID: 25/75 33 FVISWSPYCVVSVIAMVQRAPALSQGFAEIPELMA K ASVVYNPLVFTVMNRGFR 30% MEL_TMT >ref|XM_001627232 ID: 37/115 32 FFISWSPYCIVSLIESAKGEVVLSPGVSMIPELMA K ASVMYNPVVYTLMNARFR 31% MEL_TMT >ref|XM_001639117 ID: 37/118 31 FTLSWSPDAILSVI....SMVTGRPIIHVVPSLMA K SSVIYTPIVYIAFSRSFR 34% VAOP_danRer >tpd|BR000661 ID: 38/121 31 FLIAWTPYAVVSFYYSLRGPTGVPLMAAMLPSLFA K ASSLFNPIIYFAMSREFR 31% MEL2_galGal >tpd|BR000686 ID: 35/114 30 FAITWLPYAVYVLIAAFGGSHLFDAVMSVVPAMVA K MSILYNPFVYALVNPRFR 31% TMT_triCas >tpd|BR000674 ID: 36/121 29 FLACWLPYVIVSTCMSLGRRPQISLLTLEITLLVA K SGVIYNPFIYAALNLRFR 35% MEL2_strPur >tpd|BR000669 ID: 35/121 28 FSM.WVPYVAVSLIQAFTADSIITPTASHITVLVA K SCVIYNVLIYVVLNRKLK 34% TMT5_braFlo
The tree at left (which would benefit from a few dozen outgroup genes such as beta adrenergic receptor) shows how some related sequences clustered in a recent evolution study by Plachetzki et al who sought but did not identify opsins in sponge, Trichoplax or Monosiga. It's not known if any of their 'cnidops' bind retinal or have a photoreceptive role. It is possible that retinal is bound in Schiff base position but requires a secondary agonist to signal. Alternatively, signaling compounds related to but distinct from retinal assume the binding site.
A third scenario conserves the lysine and its counterion through evolutionary inertia (the salt bridge avoids unfavorable burying of charged residues in hydrophobic membrane and maintains the trigger) without Schiff base, with the diversity of genes perhaps serving chemoreception. Such a class of GPCR in early cnidaria could have been the foundational source for later recruitment to opsins in cubomedusa.
An analogous situation in sea urchin (rapid expansion of an intronless opsin-like class of genes) was noted by Raibl and coworkers (who denoted them specific rapidly expanded lineages of GPCRs or surreal-GPCRs) and noted a parallel to olfactory gene expansion in amniotes. In other words, prior to dead-end specialization as imaging opsins, earlier gene duplicates could find novel but useful roles. That's not so different from even earlier diverging 'rhodopsin-class' GPCR taking on roles such as beta adrenergic receptors.
In this scenario, photoreceptive opsins have a basal outgroup originating in cnidaria (or Trichoplax) of EK-rhodopsin class non-opsins and a distinct class of EK-rhodopsin class non-opsins paraphyletically nested within them in sea urchin and possibly amphioxus GPCR (where genes are still conservatively intronated). These latter two species also have 'too many' opsins versus a paucity of photoreceptor cell morphologies especially in urchin.
Gene duplication (and loss) of opsins continues as an active process to the present day in many species. We're not surprised to see LWS duplicate to MWS in primate cones. However we don't expect LWS to give rise to processed retrogenes serving olfaction or non-retinal signaling. Those days could be behind it. LWS may be terminally differentiated and too many mutational steps away from more versatile deployment. If so, evolutionary processes do not remain constant; some genes mature over time into locked-down roles with less potential for novelty in their duplicates than their ancestral predecessors. We see from the context of Ciona that GPCR have a very long history of acquiring new agonists for novel expanded gene family branches:
To recapitulate, photoreceptive opsins did not originate out of thin air by miraculous multiple mutations providing overnight EK, signature motifs and retinal binding pocket. More plausibly, opsins specialized from a pre-existing gene expansion pool of EK-class GPCR that itself evolved from generic group 1 GPCR. That class persists today in pre-bilaterans with unknown agonists and function though it apparently winked out in bilatera. Genuine opsins may nest paraphyletically (recruited by independent events, eg melanopsin, encephalopsin, neuropsin) within EK-rhodopsin class GPCR; a diversity of cnidarian opsin sequences are needed.
The unpublished observation of phototaxis in Trichoplax is only weak supportive evidence for opsins because demosponge larva also exhibit phototaxis (shadow seeking under coral rubble) whereas the action spectrum fits a flavin or carotenoid chromophore better than retinal. Larval photoreceptors don't reside in a rhabdomeric or ciliary morphological setting but rather in ring of columnar monociliated epithelial cells. No expression staining has been conducted for the putative opsins -- Trichoplax has only 4 cell types, none specialized in appearance to photoreceptor.
Cryptochromes are another well-known alternate photoreceptive system in bilatera with no homology at the protein level to opsins. In all kingdoms of life, only seven known families of proteins transduce light into signal (opsins, cryptochromes, phytochromes, xanthopsins, phototropins, blue flavin and lite1); Trichoplax would have to be evaluated for each of these too. For example, nematode C. elegans lacks opsins and cryptochromes but has a shorter wavelength photoresponse bypassing cAMP and diacylglycerol (DAG) downstream signaling that neurons commonly use to control behavior. The primary photoreceptor LITE1 (NP_509043) is a 8-transmembrane member of the invertebrate gustatory receptor family (non-GPCR) without known chromophore.
Trichoplax could have an unobserved larva and metamorphosis in view of the proposed post-sponge position. Alternatively, it may have be a direct developer or have lost the ability to reproduce other than by fission (which is contradicted by meiosis genes, oocytes and population genetics despite non-observation of male gametes). Trichoplax, being too small, has never been observed directly in the wild -- instead it is collected over weeks on submerged alga-coated microscope slides (or aquarium walls). Whole aspects of its life cycle may go undetected in laboratory culture. The current situation (assuming post-sponge divergence) -- highly conserved genome yet highly derived morphology -- is an odd but not impossible mix.
Even if this phylum lacks opsins, the genome will still prove important in determining the state of signaling systems at its ancestral node. The 98 mbp genome is 3% the size of a mammalian genome but the estimated 11,514 coding genes are not that different from Drosophila and 55% of the 20,000 genes of human. Pseudogenes, which outnumber genes in mammals, are difficult to detect in isolated genomes such as Trichoplax.
This large gene set if assumed approximately ancestral is not consistent with subsequent subsequent 2R whole genome duplications (giving 45,056 genes in human) without massive gain in Trichoplax and/or loss in human). In other words, even if 2R occurred, it was an inconsequential mechanism in the overall scheme of gene duplication and retention; the complement of genes was in large measure already established in early metazoa. That has special relevance to understanding expansion and specialization of opsins and associated G protein alpha subunits which primarily exhibit tandem duplication followed by translocation.
Trichoplax is an odd amoeboid-shaped animal without overt symmetry axes, body plan, recognizable organs, or internal digestive gut but clear dorsal/ventral sides. Only a single species has received a Linnean name to date; it is assigned a whole phylum, Placozoa. However the taxonomy section of GenBank lists 87 other placozoan isolates; these have ~200 deposited sequences and previous publications have established quite diverged mitochondrial genes, compatible with a whole order (even as external morphologies remain indistinguishable). Thus the 600 myr long branch of the Red Sea isolate could readily subdivided by 454 sequencing to provide supplementing glimpses of ancestral characters and better reconstruction of ancestral proteins.
The authors place the divergence node between sponge and cnidaria, surprising given the advanced sponge larva and its similarity to later-diverging metazoan. Post-sponge divergence (but sistered with Cnidaria) was proposed earlier in a 1993 Science article based on small subunit ribosomal RNA sequences. The place of Ctenophores still cannot be resolved for lack of an available genome, though just about every topology has received support. Thus the number of early metazoan nodes discussed in "The Ancestor's Tale" is still uncertain.
In a 1996 PNAS article, four of the same authors made an equally convincing argument for Trichoplax basal to sponge using both mitochondrial protein analysis and odd ancestral features such as retained introns and 5 retained ORFs exceeding 100aa:
"Our analysis shows that the Trichoplax mitochondrion possesses the largest known metazoan mtDNA genome, at 43,079 bp, more than twice the size of the typical metazoan mtDNA. Its large size is due not to secondary expansion but to features shared with metazoan outgroups, such as intragenic spacers, several introns [cox1 introns share identical positions with choanoflagellate Monosiga and fungus], ORFs of unknown function, and protein-coding regions that are generally larger than that found in animals. The large Trichoplax mtDNA is the least derived mitochondrial genome of any animal. Moreover, the Trichoplax mitochondrion shares unique derived features with other lower metazoans, notably the loss of all ribosomal protein genes. These structural features of the Trichoplax mitochondrial genome, along with Bayesian and maximum-likelihood (ML) analyses of mitochondrial proteins from metazoans and outgroups, provide robust support for the phylogenetic placement of the phylum Placozoa at the root of the Metazoa.... the basal phylogenetic position of Placozoa within the lower metazoans is robust, with P values between 0.924 and 1.000 for the various statistical tests."
It appears that an unpublished, unreleased assembly of proteins of the sponge Amphimedon was used (yet again). This genome project is 100% publicly funded yet its assembly seem to have been privatized to a shortlisted clique of insiders. Lottia gene models are also used without bibliographic citation. Both were evidently analyzed by unknown parties as unpublished SNAP ab initio predictions (Table S7.1). This raises some questions because neither reviewers nor other scientists can independently evaluate the evidence supporting the unexpected phylogenetic relationships. It is reminiscent of the recent uproar over non-release of dinosaur collagen mass spectroscopy.
The JGI site shows sponge still in draft assembly on 25 Aug 08 whereas this paper was submitted nine months earlier, on 4 Dec 07. Trace reads were completed and made available in June 2005 but these cannot be used for genes or proteins without assembly as individual read typically cover but single exons. Collagen researchers have [assembled genes using tiling from mate pairs and ESTs. By custom, assembly and first publication is normally reserved for the sequencing laboratory but that was never intended to apply to holding back data for 3-4 years.
Trichoplax appears to retain a goodly share of ancestral characters, including some chromosomal gene associations and a large number of apparent 1:1 orthologs to human and anemone. However it must be noted that the authors have significantly altered the traditional definition of synteny in making their dot plot (Oxford grid) by discarding strand orientation altogether, allowing weak partial length blastp hits (perhaps just to common domains or pseudogenes) to count as orthologous matches, and accepting departures of 10 genes from adjacency (which can amount to several million bp in mammalian genomes).
The assumption here is small and very localized intra-chromosomal inversions have been more frequent than inter-chromosomal arrangements over the vast 1.2 billion year roundtrip span of evolutionary time (but see gibbon, fly, tunicate etc). As described a decade ago, dot plots rendered in Photoshop better retain orientation and quality of fit using grayscale and tint, rather than the all-or-none as here.
The 11,514 coding gene models arise largely from an informatics pipeline tool fGenesh. Like all gene predictors, even when parameterized with transcripts, this tool makes numerous errors in human (where it can be thoroughly evaluated by comparative genomics) and is no longer included in the 35 predictive gene tracks at UCSC (though still valued for nematode and other species). The tool acted after RepeatMasker masking of 665 transposons; if that library were incomplete on low copy elements, the gene count could be too high.
Manual curation is mentioned briefly but could not have been extensive given the number of genes. Gene models missing start or stop codons were simply extended out from available sequence (though in some cases transcript extension was feasible). While a downstream stop codon can always be found, an initial methionine candidate does not always occur before an upstream stop codon is encountered. This is not a good annotation practice as it assumes that partial first and last exons are at hand; better to do this only when accurate gene models from other species warrant these two assumptions. These extensions will seldom have homological support even when correct unless the termini are important (eg signal peptides; G protein C-terminal opsin interaction).
Only 58 ESTs are provided by GenBank (from an unpublished immunology study by different authors); in the nucleotide division, it appears that GenBank does not distinguish experimental mRNAs from those simply inferred from genome models. Transcripts have proven very helpful in quantitating gene prediction accuracy. In assessing completeness of the genome, the authors state 85% of 14,571 T. adhaerens ESTs (later described as 2,506 when clustered of 21% coverage) have quality matches (a more nuanced account appears in Supplemental). If 15% did not make the cut -- assuming them to be representative of missing or partial genes or absent from transcripts -- that would bring the total coding gene count to 13,241.
These 2,506 assembled transcripts could be downloaded at the indicated JGI url and tested to see how and if they are stored at GenBank as this affects queries of the average user. The article does not discuss how discrepancies between transcripts and pipeline gene models were resolved. These can involve spliced-out exons with or without comparative genomics support and non-support of called exon boundaries (here the transcripts will have it right).
On a species so distant and little studied biochemically as Trichoplax (27 articles since 1974, mainly field work or rDNA sequencing), when protein alignment dips below the 40% protein-level identity on partial matches (notably GPCR putative opsins), homological transfer of annotation from functionally characterized proteins won't be reliable beyond rough domain-level concepts. Best-blastp percent identity to known genes is not noted at GenBank model entries.
Here signalP, InterPro, and TMHMM were used to predict signal peptides, domains, and transmembrane peptides; while not validated with pre-bilateran metazoan protein chemistry to any extent, these tools provide a good start to localized functional elements. KEGG and KOG are not sufficiently developed to be worthwhile; indeed, their development appears abandoned (2004-05 versions were used). EC numbers and GO nonsense add little value. Biosynthetic pathways were not analyzed and so essential amino acids, cofactors, and intermediary metabolism were not determined.
What is needed here is an ability to filter or grade annotation transfer quality vis-a-vis match quality, match extent, domain structure, and gene family extent along the lines of GeneSorter at UCSC. For example, Trichoplax/human matches can be very high as in the 77% identity agreement seen in G protein alpha subunits (below) or very low 29% in best matches of some opsins to Trichoplax GPCR.
Conversely, Trichoplax and the secret sponge annotation will now propagate all over. This can become a self-reinforcing paradigm for error propagation, eg as later authors see corroborating annotation in their top homology matches.
None of this has to affect opsin analysis (other than missing assembly) because the genome sequence can be directly searched by tblastn without reference to JGI gene models or annotation. Any assertion of opsins must establish the location and phasing of introns in a gene model as these are exceedingly conserved in opsins and diagnostic of orthology classes (and so especially important when protein sequences are so diverged as to blur into generic GPCR).
At this late date, it might not be expected that a metazoan would contain many authentic coding genes with no blast matches in all of GenBank. These may prove greatly enriched for artifacts, just as the Ensembl human gene set used contains nearly 2,000 pseudogenes, related debris and mispredictions not supported by the 28way comparative genomics track to any phylogenetic depth. It's not clear what fraction had supporting transcripts. Dropping these will roughly offset valid genes missing from the assembly or missed by gene prediction.
Similarly, introns will not be located correctly if this is homologically forced in regions of poor identity (self-fulfilling prophecy); phase agreement can help but not so much if based simply on GT-AG which occur every 16 bp approximately, nor even with nuanced splice rules if merely assumed from remotely related species. The practical impact is that exceptions and imperfect matches are missed -- the authors report excellent overall conservation of (high quality flanking) introns with respect to human and Nematostella , which fits the modern picture of intronation being deeply ancestral and profoundly conserved outside a few rogue species (such as fly, nematode, and tunicate) with high turnover. Some 150 known splice sites that use rare alternatives to GT-AG may also be as deeply conserved though this has not yet been investigated.
Manual ab initio curation here at genomeWiki of selected genes of signal transduction (alpha subunits of Trichoplax heteromeric G proteins) shows that cGMP Gt transducin-type alpha subunits (presumably still hyperpolarizing) share the identical 8-exon structure and phasing of human and other bilatera, including the anomalously short 15 residue exon 2 that distinguishes this class of genes from the otherwise identically intronated 7-exon Gq alpha subunits that utilize phospholipase C hydrolysis of PIP2 and IP3 signaling. This strongly suggests that, though the requisite gene duplication and divergence took place much earlier, the distinctive exon 2 emergence (or fusion) predates metazoa. The presence of Gt and Gq subunits does not imply the existence of melanopsin or encephalopsin homologs because these alpha subunits must service hundreds of other unrelated GPCR.
>GNAi1_triAdh Trichoplax adhaerens (placazoa) Gt XM_002115978 77% GNAi1_homSap 8 exons 0 MGCAASAGDKVAAAKSKEIDKKIKSDAEKAAREVKLLLL 1 2 GAGESGKSTIVKQMR 21 IIHESGFSEEDRAQYKPVVFSNTMQSMAAIIRAMGVLRIEFGDKTSLV 0 0 GDARRLFEIMDAPGVQEFTPEIVSLLKRLWSDHGVQQCFSRSREYQLNDSAPY 2 1 YLNSIDRLGKPEYIPSEQDVLRTRVKTTGIVETHFTFKDLHFK 2 1 MFDVGGQRSERKKWIHCFEGVTAIIFCVSLSAYDLVLAEDEEM 0 0 NRMMESMKLFDSICNNKWFTETSIILFLNKKDLFQEKILKSPLTICFPEYT 1 2 GANTYEEASAYIQMKFEDLNKMKDQKEIYTHFTCATDTNNIQFVFDAVTDVIIKNNLKDCGLF* 0 >GNAQ_triAdh Trichoplax adhaerens (placazoa) Gq XM_002116172 76% GNAQ_homSap 7 exons 0 MACCLSDEAREQRRINREIEKELKKHKRDAKRELKLLLL 1 2 GTGESGKSTFIKQMRIIHGKGYTDNDRAEFTQLVFQNIFTAIQALIKAMETLNITYEHQSN 0 0 RQRVDVVRTVDPETVGSLSKEHVEAIDSIWNDSGVQECYDRRREYQLSDSAKY 2 1 YLTDLHRLAEPNYLPTQQDILRVRAPTTGIIEYDFNLDTVMFR 2 1 MVDVGGQRSERRKWIHCFENVTSIMFLVALSEYDQILAEADSQ 0 0 NRMEESKALFKTIITYPWFQNSSIILFLNKKDILEEKVQKSNIADYFPEYD 1 2 GPPRDAQAGREFILKMFVDLNPDSEKIIYSHFTCATDTENIRFVFAAVKDTILQFNLREYNLV* 0
Porifera: Amphimedon queenslandica (sponge) .. 0 opsins
One marine demosponge genome, called Reniera sp. at JGI and trace archives blastn but subsequently reclassified to Amphimedon queenslandica at GenBank and PubMed, is available as 2,917,892 traces dating to July 2005 or as finally assembled on 28 May 2010 in the wgs division of GenBank where they can be queried with tBlastn. Importantly, not all traces made it into the assembly, so tBlastn offered at Compagen is still critically important. A publication on the genome will no doubt appear shortly.
The genomic DNA used came from embryos derived from a single parent sponge collected from the Great Barrier Reef. However GenBank contig entries say the 8-fold assembly comes from a sponge population with more than two haplotypes. Contig sizes appear respectable, with many dozen over 100kbp in size but internally many have gaps.
Sponges lie at the base of multicellular animals and are not notable for a nervous system. However demosponge larva do exhibit phototaxis (shadow seeking under coral rubble) but the action spectrum is supposedly a better fit to a flavin or carotenoid chromophore. Sponges also can respond to gravity, current, and chemical cues.
The basis for sponge responsiveness to light has been carefully studied from an ultrastructural perspective -- for an animal lacking nerves and cell junctions, the parenchymella larva are quite capable of responding effectively to light and other stimuli. Larval photoreceptors may lie in a posterior ring of columnar monociliated epithelial cells. A pigment cell occurs but the pigment itself has not been chemically characterized -- the issue is whether it is a homologously derived (melanin via tyrosine hydroxylase) or novel.
Negative larval phototaxis there has been attributed to pigment-filled protrusions in a posterior ring of columnar monociliated epithelial cells. This species may prove insufficient to explain the full range of photoresponsive responses in sponge larva such as circadian rhythm and hexactinellid photoreception (notably the role of stalk spicules).
Jacobs et al have proposed a far more sweeping view of early evolution of sensory (and other!) organs in sponges. A related view, that of Gehring, proposes that the eye (and other sensory systems) came before the brain, indeed that the nervous system arose later to coordinate a response to all these inputs. There is support for that in simple photoreceptor cells controlling their own cilia. Consequently we should not be too quick to dismiss sponge photoreception for lack of neurons.
In an article entitled "Six major steps in animal evolution: are we derived sponge larvae?" C. Nielsen writes:
A scenario for the early evolution of the metazoans. The metazoan ancestor "choanoblastaea" was a pelagic sphere consisting of choanocytes. The evolution of multicellularity enabled division of labor between cells and an "advanced choanoblastaea" consisting of choanocytes and nonfeeding cells. Polarity became established, and an adult, sessile stage developed. Choanocytes of the upper side became arranged in a groove with the cilia pumping water along the groove. Cells overarched the groove so that a choanocyte chamber was formed, establishing the body plan of an adult sponge; the pelagic larval stage was retained but became lecithotrophic [yolk-supplied]. The sponges radiated into monophyletic Silicea, Calcarea, and Homoscleromorpha. Homoscleromorph larvae show cell layers resembling true, sealed epithelia.
A homoscleromorph-like larva developed an archenteron, and the sealed epithelium made extracellular digestion possible in this isolated space. This larva became sexually mature, and the adult sponge-stage was abandoned in an extreme progenesis. This eumetazoan ancestor, "gastraea," corresponds to Haeckel's gastraea.
Trichoplax represents this stage, but with the blastopore spread out so that the endoderm has become the underside of the creeping animal. Another lineage developed a nervous system; this "neurogastraea" is the ancestor of the Neuralia. Cnidarians have retained this organization, whereas the Triploblastica (Ctenophora+Bilateria), have developed the mesoderm. The bilaterians developed bilaterality in a primitive form in the Acoelomorpha and in an advanced form with tubular gut and long Hox cluster in the Eubilateria (Protostomia+Deuterostomia).... The evolution of the eumetazoan ancestor from a progenetic homoscleromorph larva implies that we, as well as all the other eumetazoans, are derived sponge larvae.
The resulting picture of Amphimedon larva -- numerous differentiated and pluripotential cell types arranged in stereotypic patterns along central-lateral and anterior-posterior axes -- is not one typically conjured up of parazoan ("almost metazoan") in the view of Leys and Degnan. Indeed the common ancestor humans shared with sponge may have been rather advanced.
The concept here is that a photoreceptor cell can control its associated cilium without the baggage of a CNS, either as a passive rudder or more actively directing phototactic motion. In effect the single photocell is a self-sufficient brain that processes external environmental inputs, asseses them and acts appropriately. Chemoreception, a very similar GPCR signaling system, might work the same way. In this view, the nervous system evolved as a secondary system to coordinate these stand-alone sensory effectors.
A futile search for sponge opsins turned up only non-opsin, rhodopsin-class GPCR genes from Amphimedon. Similarly, no bone fida opsins have been located in even earlier diverging placozoan Trichoplax, choanoflagellate Monosiga, and fungal genomes. This fits a picture of photoreceptor opsins first appearing subsequent to sponge in eumetazoa cnidarians. However these were hardly de novo genetic innovations but rather evolved out of the already-rich cauldron of GPCR gene expansions in the sponge ancester.
Some later diverging species such as the model organism C. elegans lost all of their opsin genes, making them useless in Urbilateran ancestor reconstruction. This argues for much more intensive genomic sampling of sponges and cnidarians so as to sidestep inference mislead by gene loss in model organisms chosen for historic reasons.
However on 31 May 2010, a fragmentary Schiff K296 opsin was found here among residual traces not used in the assembly, namely in ti|922429579 (also called BAYB198960.b1). This trace, despite a length of 1018 bp and the overall 8x shotgun coverage, has no overlap with any other trace as it is likely Lottia gigantea contamination (see below). No additional K296 sequences occur in the assembly using a wide variety of probes.
The candidate (possibly Lottia contaminant) opsin sequence within trace ti|922429579 is shown below aligned to its best match among the thousands of established opsins, a peropsin from amphioxus(with next-best an undocumented opsin from the cnidarian Nematostella). Observe not only is the Schiff base lysine conserved in precise homogous position upstream of the NPIIY ... FR but the entire conservation profile is an excellent match to that of known opsins. While some of this profile is also conserved within generic rhodopsin-class GPCR, the fragment here elicits only much weaker matches to non-opsin GPCR (the best being bradykinin receptor). All top matches from tBlastn to GenBank are opsins. Thus homological evidence for this fragment being an opsin is much stronger than just a lysine at Schiff position 296.
<---------TM6--------><--EC3---><---------TM7-------->
sponge: 1 VCVVVIFSFMICWSPYATFSLWYSYSDTSGIPLWVTALPVLAAKLFCCINPVIYLVVSKRFR
+C+ ++ +F++ W+PY+ L+ ++ + + IP W+T LP L AK NP+IY++ ++RFR
perop: 257 MCLAMVAAFVVAWTPYSVLFLFAAFGNPADIPAWITLLPPLIAKSSALYNPIIYIIANRRFR PER3_braFlo Branchiostoma floridae
cons profile m...mv..F...W.PYa..............p......P..fAK.s...NP!IY......FR
>PER3_braFlo Branchiostoma floridae (amphioxus) AB050610 12435605
0 MDIPTETPYGAGDDPAGTGWRWAETDQNGFHKYDHLIVGLYLFVI 1
2 GIIGTVENGITLATFTKFRSLRSPTTMLLVHLAIADLGICIFGYPFSGASSLR 0
0 SHWLFGGVGCQWYGFNGMFFGMANIGLLTCVAVDRYLVICRQDL 1
2 VDKVNYNTYGVMAALGWLFAAFWAALPLVGWAEYSLEPS 1
2 GTACTINWQKNDSLYISYVTSCFILGFALPLAVMMFCYWQ 0
0 ASCFVNKVLKGDISGDLTFPVAVNVDWEYQNHFSK 0
0 MCLAMVAAFVVAWTPYSVLFLFAAFGNPADIPAWITLLPPLIAKSSALYNPIIYIIANRRFRSAIFSMVKGQNPDVE 0
0 TLFARDFRISPIEDTGKEMSSMGNANA* 0
>OPS_nemVec Nematostella vectensis (anemone) no introns
MADASGNTTSVVVTRVYSAYSAITLVLNTLLILVFYKKSLLRRSYRFYIFSLALVDILMAVLVDSIGAYANSSPGRALSSRGCQYYGAVASFVGFTSVHHLTAIYA
DRWYSIRFVMRRKPSKRVTLRVILALWISGLIWAIPPLVGWSSYAHQGSFSHCSVKWFSVDPKDYSYTVVIFTKFYFFPIAFMTFSYVHVFVITRRMAKNATQLWGE
KAAATAKSIKSESKAAEIALVMVLSFLIAWTPYAVVSFYYSLRGPTGVPLMAAMLPSLFAKASSLFNPIIYFAMSREFRNSCRDFARR*
ttttttcaggtttgtgtggttgtcatattttcctttatgatatgctggagtccttatgcc phase 00 splice acceptor preceding sponge exon
F F Q V C V V V I F S F M I C W S P Y A
Blast of the Lottia gigantea genome yields an opsin sequence identical to the one recovered from the Amphimedon traces: >jgi|Lotgi1|152675|fgenesh2_pg.C_sca_2000095. This implies one or both genome project had dna contamination.
The trace read can be extended somewhat beyond the last region of homology but truncates prior to reaching a stop codon. Similarly, it can be extended N-terminally for a half dozen residues preceding the start of homology but then reaches a stop codon within the reading frame. Since a standard GT-AG splice acceptor immediately precedes the start of homology, this region very likely represents the start of the final coding exon. This is supported by both the 00 phase and position of the exon (252 EVTR in bovine rhodopsin homology numbering) exactly matching those of its best match.
This exon was previously recognized as ultra-conserved within the peropsin-neuropsin-rgropsin group of opsins. This provides strong independent support for the identification of this fragment as an opsin. It also implies a very ancient origin of intronation and subsequent immense conservation for this gene family. This is borne out by the nearly perfect match of sponge transducins GNAQ, GNAS, GNA13 to those of human.
However, despite an extra 729 bp preceding the opsin-homologous region, no earlier exons can be detected by blastx even with the most tolerant settings. This could be explained either by a moderately long intron or by divergence so extreme that the preceding exon -- which is predicted from homology to be short -- does not exhibit homology. More likely, however, is the possibility that it represents contamination from the Lottia gigantea genome sequencing project, which occured in the same facility. There is no indication of compositional simplicity or presence of a retroposon here. The data also exclude processed pseudogene and indeed all forms of pseudogene (which decay without respect to conservation profile). Introns in sponge tranducin GNAS range from 42, 54, 69, 69, 84, 108 to 401 bp, short and not indicative of massive retroposon invasion. GNAS is surprisingly associated with photoreceptor transduction in jellyfish.
The exon begins just inside transmembrane helix VI and continues through the third extracellular loop to transmembrane helix VII and the final cytoplasmic helix VIII. Thus the preceding exon missing in the trace would primarily consist of the third cytoplasmic loop whose length is highly variable within peropsin-neuropsin-rgropsin group. There are no reliable anchor residue patches for many dozens of residues upstream. Thus should introns in the genome assembly generally prove to be short, divergence would be the likeliest interpretation. Introns in sponge transducins also well-conserved and of moderate size.
Earlier exons still might be detectable in this trace via de novo gene prediction tools such as GeneScan. If these had expected lengths and intron phasing, that would lend support even if homology remains weak. For blastp extensional matching, it would be necessary to get far enough N-terminally to encounter a patch of conserved residues. However blastx already excludes this. None of the ab initio tools tested worked satisfactorily on this trace, unsurprisingly since all are optimized to vertebrate (eg human) gene-finding parameters.
In summary, the sponge genome does not appear to contain any opsins.
From homology and intronation, it appears most closely related to the peropsin-neuropsin-rgropsin group, with markedly less affinity to melanopsins or cilopsins. The best affinity might change if full-length sequence were available. Note too that all opsins may coalesce at this phylogenetic depth. Since sponge is basal among metazoans with opsins, this might suggest that the peropsin group is ancestral and the others more derived. However this cannot be safelyl concluded from just one species of sponge because its opsin content might reflect gene loss from the ancestral state.
>PER_lotGig Amphimedon queenslandica (sponge) contaminating fragment apparently from Lottia instead VCVVVIFSFMICWSPYATFSLWYSYSDTSGIPLWVTALPVLAAKLFCCINPVIYLVVSKRFR QRYIEVVCRSYKYCGKRNSITPERTQV >GNAQ_ampQue Amphimedon queenslandica (sponge) 70% 0 MACCLSEEAKEARRINDEIERQLRRDKRDARRELKLLLL 1 2 GAGESGKSTFIKQMRIIHGSGYSVEDRKQFVTLIYRNMYTSLQALTTAMESLKIPYT DPQSQSYAESISTVNPETVSEIPDDHKEAFKCLWSDDGIQQCYQRRREFQLSDSTKY 2 1 YLDDAERLLAQDFVPTTQDVLRVRIPTTGIIEYHFDIRNVIFR 2 1 MVDVGGQRSERRKWIHCFENVTSVMFLVAISEYDQILVETDE 0 0 NRMEESLALFETISRYPWFRTSSVILFLNKKDLLEEKIMISDLSDYFPEYK 1 2 GPPKDAEKAKEFIKEMFLSVNPDEKKHIFPHFTTATDTENIQRVFEAVKTTVLEENLRDYNLI* 0 >GNA13_ampQue Amphimedon queenslandica (sponge) 54% 0 MAFNCLKTEEEKEGERRSRQIDRQLKSLDKEKDWKREMKVLLLGAGESGKSTFLKQMRIIHGEDYSDSDRLEFRPLIYHNILK GMKVLVEACRRLQIPFSEPDNDSKGDIVSSGYQHSQELSPEEFAPYVEPLKSLWKDTGIQTTLARSNEFQL 0 0 GDSVPYFLSKLEVIGAV 0 0 DYLPSKDDILNSRKATRGVVEYTTAIRNQQFRFVDVGGQRSQRQKWFQCFDEVTAILFIVASSAFNQTLLEDRVTNRLIESLNIFETIVNNRCFR NVAIMVFFNKSDLLETKVQNPKVDISKYFPEFNGDPHNLDHVKGFMVGMFDSVRRNQRQDFYHYFTTAVNTRNIHMVFDIVKDIILSGHIGQILQI* 0 >GNAS_ampQue Amphimedon queenslandica (sponge) 62% 0 MSALGCFGEPLTEEERQRKEKSKEIDQQLRKEKVEYKSTHRLLLL 1 2 GAGESGKSTVVKQMKILHVNGFSKE 2 1 ERLQNVDDIRSNVREVIQ 0 0 TVTSQMDKLDPPVTLKDERNQLRLSYIQNVITPLPEYETNFSP 0 0 EFYEHAEALWNDEGVQEAFRRSNEYQLIDCAPY 2 1 FMNSKLREVANPDYLPSDD 00 DILRARRMTKGIYETHFIIEKVHFQ 2 1 MFDVGGQRVERSKWIQCFN 1 2 DVTAIIYVVASSSFNMVIREDNRT 00 NRLRESLNLFEQIWNNR 2 1 WLRNVSVILFLNKMDILKEKIEGKHFHLEDYFPEFSSYRPPAES 1 2 EKREQGDTETVLRARFFIRDQFL 0 0 RISSRGANDGRHFCYPHFTTAVDTENIRRVFESCRDIIQRIHLHRYELL* 0
Choanoflagellates: Monosiga brevicollis .. 0 opsins
The genome sequence of Monosiga brevicollis appears in the 14 Feb 08 issue of Nature. It contains a reported 9,200 genes in 42 Mbp. These are densely intronated but reflect net loss since common ancestor with metazoans. Domain orders are markedly shuffled relative to eumetazoan counterparts and some key proteins such as Notch have only partial-length matches.
The 14 extant genera of choanoflagellates have been minimally studied overall with only 59 non-Monosigna sequences at GenBank so additional representative genomes are needed to fully understand ancestral characters (ie, retained by at least one species of choanoflagellate).
Separate studies have considered the emergence of the three collagen clades, cell adhesion via cadherins, the greatly elaborated tyrosine kinase signaling network with 128 tyrosine kinases, 38 tyrosine phosphatases, signalling origins, and 123 phosphotyrosine-binding SH2 proteins, and even transcriptionally active LTR retrotransposons.
Opsins, though not really expected from known behavior or morphology, have been sought directly without success in Monosiga. Indeed, no GPCR with lysine in retinal position can be detected either. Monosiga has either lost this class gene or more likely never had them. This puts the spotlight on sponge, yet blastn of traces is not sufficiently sensitive.
Alpha heterotrimeric G protein subunits that could bind opsin-like receptors have not been directly studied. Here we see the first alpha protein below has two introns identically located and phased to human to Gi class proteins but lacks the short exon and downstream introns. The second Gq class subunit has altogether different intronation despite 55% identity to the apparent human ortholog. These gene structures imply an era of active intron gain/loss in the era of the ancestor. Encephalopsin and melanopsin could have been supported had they been present but G alpha signalling there is a specialization of a much earlier evolved process.
>GNAi_monBre Monosiga brevicollis 3 exons XM_001747738 55% GNAi_homSapheteromeric G protein alpha subunit Gi
0 MGICMSAEQKAQQARTAAVEAQLERDAQLASRTIKLLLL 1
2 GAGESGKSTLVKQMKIIHGDGFSNEELKSYKPTICDNLVHSMRAVLEAMGPLVIDIGDQVRPP 0
0 HAKVVLSYIELGTSGGLTPELTEALKALWADSGVQECFRRSNEYQLNDSAEYFFNNIDRIAQSNYLPTQEDVLRARVRTTGV
IETTFRYKDLIYRMFDVGGQRSERRKWIHCFNDVTAVLFVAALSGYDMKLFEDQETNRIHESLTLFDAICNNSFFINTAIILFL
NKTDLFSQKIARTPLKDYFPEYDGPPNNASEAKKFIAGMFKRLNKNPNKPVYEHFVCATETQNIRYVFDAVK* 0+
>GNAQ_monBre Monosiga brevicollis 7 exon form XM_001745795 55% GNAQ_homSap heteromeric G protein alpha subunit Gq
0 MPCGPPDETRRRSLAIDRQLRKERMSKQREYKILLL GTGESGKSTIIKQMRIIYGQGFNESDRLAYKPLVYRNIITSMKRMLDALDQLSLQLADSSLEEDAYDK
LDVDVNTVDAIEPYYPLLKKLWNDNGIQQVFQRRNEYQLSDSTAYYYNRLDAVAAADYIPTVDDVLRSRQATTGIHEFEFDLDSVVFRMMDVGGQRSERRKWIHSFE 0
0 GVTSIIFIAACNEYDQVLAEDTNVNRMQESLALFGQIIQYHW 2
1 FANSSFILFLNKQDLLEEKVKTHPIKPFFPDYTGQE 0
0 GDYENIKKFIETMYRSRKPAGKDLYTHFTMATDTSNIQFVFNAVRSTLLRIHLKDYNLF* 0
See also: Curated Sequences | Deuterostomes | Ecdysozoa | Lophotrochozoa | Update Blog