Opsin evolution: key critters (cnidaria)
Cnidaria .. ? opsins
Biologists have belatedly realized that many molecular and morphological innovations attributed to chordates or grudgingly to bilatera actually track back much earlier to the common ancestor with cnidaria (Eumetazoa) if not earlier still to sponges or choanoflagellates. That's certainly true of photoreception. Two cnidarian genome projects have been funded but that selection needs to be seriously expanded.
A scientifically neutral definition of eye needs to embrace the full variety of photoreceptors, including those with fewer "features" than the most complex. Probably the cutoff should be based on use of bona fide opsins classifying to the root of the encephalopsin and melanopsin families and covalently binding retinal as agonist. Some purposes for an eye can be fully met by just perceiving and acting upon one "pixel", that is a simple photoreceptor eye with no pigment cup to provide directionality (two pixels). Sponges and cnidarians have operated for immense timescales under selective pressure on a steady body plan, far longer than mammals.
We don't say humans lack eyes just because a redtailed hawk has more pixels; we don't say humans lack color vision just because a turtle sees richer, sharper colors. Cornea, lens, retina, and CNS are just baggage that can't be maintained under darwinian selection when a simpler photostructure already suffices to distinguish day from night for gamete release, up from down for settlement, towards or away for predator evasion. Cnidarian eyes exemplify this full range of possibilities.
No living animal represents an ancestral node -- those are long gone, evolution never stops. All extant species have proven equally adept at survival. Evolution is not a story book progressing to human-- if cnidarians are so dumb and their vision so bad, how then are they able to chase, catch, kill, and eat advanced vertebrates such as fish?
Cubozoa: Carybdea marsupialis (jellyfish) .. 0 opsins
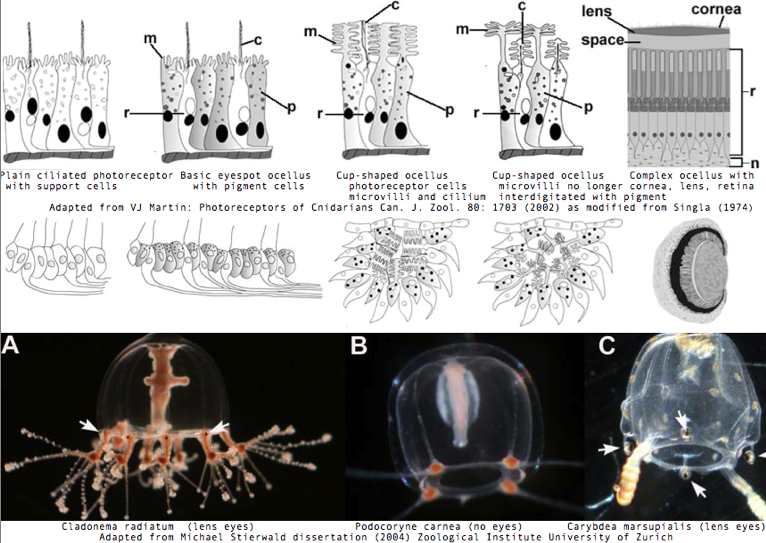
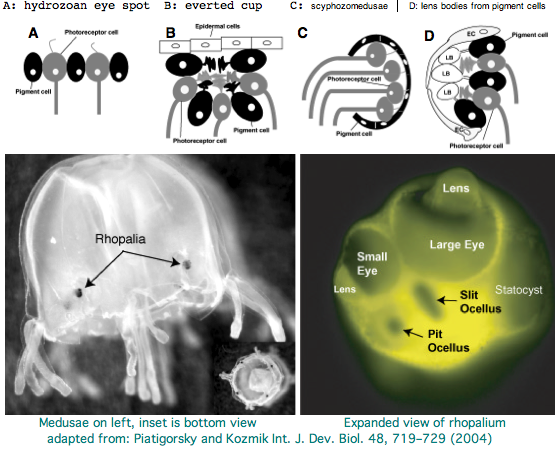
Cnidarians are the earliest diverging invertebrates with multicellular light-detecting organs, called ocelli. Photodetectors include simple eyespots, pigment cups, complex pigment cups with lenses, and camera-type eyes with a cornea, lens, and retina. These remarkable eyes are located on sensory clubs called rhopalia with four lining the bell. Each houses six eyes: a pair of pit ocelli, a pair of slit ocelli, and two unpaired lens eyes with counterparts to cornea, cellular lens and retina of ciliated photoreceptors. Anatomically, ocelli have bipolar sensory photoreceptor cells interspersed among nonsensory pigment cells with the apical end making the light-receptor and the basal end forming an axon that synapses with second-order neurons to form what amounts to ocular nerves.
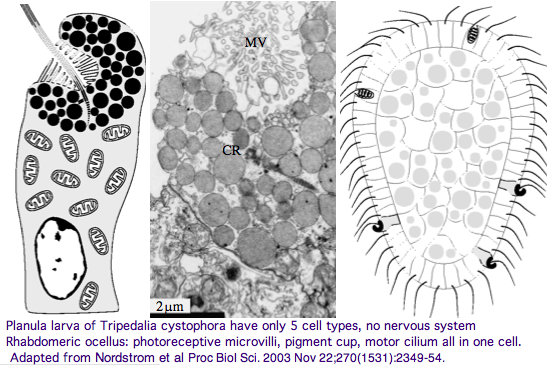
The spectral sensitivity of neritic (near-shore) lens eyes of a box jellyfish, Tripedalia cystophora recently considered by M Coates et al was interpreted as a single vitamin A-1 based opsin with peak sensitivity near 500 nm (blue-green). However nothing was sequenced. This species was most helpfully reviewed by Piatigorsky and Kozkmik who note Eakin already commented on seemingly ciliary photoreceptors in 1962. However, 45 years later we still don't know if opsins in cnidarians would classify with vertebrate ciliary opsins. They might even share conserved intron positions.
Furthermore, as noted by Nordstrom et al, planula larvae of Tripedalia have a series of single-cell pigment cup rhabdomeric-like photoreceptors directly connected to motor cilia. These lack neural connections in line with Gehring's notion of the eye preceding the brain in evolution, rather than being a later add-on. So cnidaria might actually retain descendents of both types of ancestral opsins.
The most striking jellyfish from the perspective of a complex set of eyes is Carybdea marsupialis, as reviewed by VJ Martin. Antibody studies based on vertebrate cone/rod opsins are doubtful because of cross reactivity to generic GPCR proteins; again no opsins have been sequenced yet. This would make a great genome to study provided the retroposon and base composition are not unwieldy. Nematostella and Hydra, whatever their other genomic merits, sit in the Anthozoa and Hydrozoa respectively, types of cnidarian lacking elaborate visual systems.
Vision has roles in the reproduction and feeding of cubomedusae which can find each other and chase, catch, and eat teleost fish. A patch of Pelagia nocticula 10 square miles in extent and 35 feet deep recently destroyed a salmon farm off Northern Ireland.
Anthozoa: Nematostella vectensis (starlet sea anemone) .. ? opsins
The Nematostella genome has been released along with major papers and an upgrade to Stellabase. Not all 6.1 million traces were used up by the assembly, so any gene missing from the assembly should be sought directly in the trace archives.
The sea anemone, an anthozoan within Cnidaria having epithelial cells, neurons, stem cells, complex extra-cellular matrix, muscle fibers, and symmetry axis, is emerging as a high-profile evo-devo model species to elucidate the emergence and deployment of genes that determine animal body plans. However those plans don't seem to include eyes or overt photoreceptor structures such as pigment cells -- for that cubomedusae would be far better. PAX6 and RX are especially relevent to photoreceptor structures; their expression has been thoroughly studied in Nematostella without uncovering any sensory system though they contribute to patterning specific components of the ectodermal nerve net.
The JGI annotation pipeline produced a number of extensively annotated gene models for Nematostella opsins. These are available simply by keyword lookup, tblastn of various queries the best of which turn out to be -- unsurprisingly -- an encephalopsin subclass from Branchiostoma. It is important to credit the JGI staff for providing the relevent bioinformatic track computations because they were first to characterize and release these opsins into the public domain (eg GenBank NR and Entrez Gene). It does not constitute independent "discovery" to perform keyword lookup and copy out other peoples' work. Without proper citation, that's plagiarism.
I extended improperly truncated JGI gene models (ie those lacking iMet and stop codon), validated the extensions still lacked introns (GT-Ag splice junctions missing at positions expected from closest homologs), placed the best 3 (of a half dozen) in the Opsin Classifer with fasta headers, noted their best matches below, and validated lysine and counterion glutamate in the expected positions. All this is consistent with (but does not prove) a role for ciliary Gt opsins in pre-Bilateran photoreception. Nematostella transcripts contain
We expect cnidarians (maybe not this particular anthozoan) to have both melanopsins and encephalopsins. Our tendency is to think that imaging eye opsins, whether insect rhabdomeric or vertebrate ciliary, are the main attraction, with the other opsins playing out obscure roles in secondary functions like timing of gamete release . That's quite wrong-headed. Deeper gene family trees show that the melanopsin and encephalopsin constititue the primary photoreceptors. Over vast evolutionary time scales, they gave rise to various spin-offs in various clades at various times through gene duplication and subsequent neofunctionalization. At even greater phylogenetic depth, melanopsin and encephalopsin are themselves related by gene duplication. As noted by Arendt, that exploited gene duplication within the alpha subunit of heteromeric G protein and profound diversification in signalling system second messaging.
The odd thing about all these cnidarian encephalopsins is their lack of introns (three ancestrals are expected). That's very unlikely to be the Eumetazoan ancestral state for encephalopsin because Nematostella is no rogue organism when it comes to intron conservation. A common explanation for this within eukaryotic bioinformatics is gene duplication of a master gene via fully processed retrogenes (rather than through tandem, segmental, chromosomal, or whole genome duplications -- all of which preserve introns). Mixed mechanisms are also common (as in olfactory receptors): an initial intronless retrogene is duplicated tandemly etc. These paralogs can even displace the master gene by taking over its function, causing it subsequently to be displaced or even lost. That scenario played out within zebrafish opsins.
If so, we might expect Nematostella encephalopsins to be more closely related to each other than any known opsin from any species. Indeed ENCEPHa_nemVec is 90% identical to ENCEPHb_nemVec and 52% with ENCEPHc_nemVec, whereas only 39% identical to the best bilateran opsin, ENCEPH4_braFlo of amphioxus. Those are profound differences -- mammalian proteins typically take 100 myr to lose 10% of their percent identity. Here though we know next to nothing about clade-specific rates and have very long branches indeed. Of course, a seven-transmembrane protein has very different evolutionary constraints from the generic globular cytoplasmic protein to which off-the-shelf phylogenetic software is tuned, so no purpose is served applying that.
It appears the three Nematostella proteins may share a distinctive rare genetic event, an indel in a loop region. That would favor a common history. It will prove difficult to resolve indels as to insertion or deletion for lack of suitable outgroup.
Given an finished genome, the mode of gene amplification can be explored by looking at flanking genes. Perhaps ENCEPHa_nemVec and ENCEPHa_nemVec are adjacent (ie tandem duplication) or perhaps their flanking genes are paralogous (syntenic segmental duplication). However the Nematostella genome is currently unfinished and the (gapless) contigs containing the encephalopsins run about 10 kbp. Depending on gene density that can be too small to establish synteny. These contigs, separated by strings of N's of unknown length, are further assembled into larger scaffolds (ample for synteny), a process usually trustworthy at highly experienced JGI but sometimes confounded by issues such as repeats, compositional simplicity, very recent duplicative regions, and clonability.
The most convenient approach here is tblastn of ENCEPHa_nemVec against the wgs menu item at NCBI Blast, specifying Nematostella. The three genes here are on different scaffolds altogether, ruling out tandem position. The nearest flanking genes can be extracted by blastx of the enveloping contig (or whole scaffold) against GenBank protein. JGI has in effect already done this, as could be seen by expanding out from the inital browser view. Comparing 3 browser views is complicated by the fact that flanking paralogs might be named differently, but that is readily overcome by collecting sequences (noting strand orientation) into a mini-database and comparing within uBlast.
Notice the Opsin Classifier collection already contains the outcome of this process as a fasta header field (for deuterostome opsins). It is conceivable that orthology of a Nematostella opsin to say a Branchiostoma opsin could be established in this way (synteny). However gene order in both genomes has been independently scrambled over immense time scales and orthology would have been to the Nematostella master gene (with introns) that appears lost. It's better to build out from a local synteny chain but that requires data from additional cnidaria. Note the irony here in that the farther removed the genome from human, the more densely they must be sampled.
It's evident from a casual ClustalW alignment, after marking up columns for membrane-spanning sections and considering hydrophobicity, that Nematostella opsins conform to the standard central pattern. That's unsurprising since proteins retain 3D structure at far lower percent identity and the pattern here cuts much deeper, into the overall rhodopsin superfamily and beyond to generic GPCR. However encephalopsins can have very considerable extensions at their amino and especially carboxy termini that need separate consideration.
For now, sequences can be trimmed to whatever is alignable across the full spectrum of ciliary opsins. Recall that by design the Opsin Classifier collection seeks maximal phylogenetic dispersion to mitigate over-weighting by over-studied species that might introduce clade-specific interpretive bias. That could also be done by distilling the dataset down to ancestral sequences at lamprey divergence, the risk there being co-evolution of non-adjacent residues (eg different alpha helices) can be lost in residue-by-residue ancestral reconstructions.
As noted, Nematostella opsins are at best 39% identical. These had better be strongly concentrated at invariant and near-invariant ciliary opsin positions rather than randomly distributed. Blastp of course doesn't know the difference. We know at the outset this strong association will occur for any GPCR to the extent that it is reliably alignable, so the question really is whether conservation is concentrated at the conserved positions specific to ciliary opsins (ie conservation not shared with Go and Gq opsins). This has all been studied before but not nearly at the phylogenetic depth made possible by comparative genomics. There is always a need in remote opsins for independent support (here stratified signature residues) of candidates suggested by blast searches.
For that, it is most convenient to cut conservation tranches with Corpet's Multalign because user-specifiable line width can set breaks after structurally meaningful locations. Here the cutoff for invariant is set variously at 100%, 95%, 90%,... (with Nematostella omitted) and the stack of consensus lines retrieved. That results in a nuanced version of invariance that can be set off against the Nematostella sequence at those positions. For "controls" rhabodomeric opsins, rhodopsin superfamily, and generic GPCR generate their own stacks. (Alternatives such as logos or the misnamed evolutionary trace would give similar outcomes. None of the methods make use of the known phylogenetic tree relating the sequences.) The bottom line here will be that these new cnidarian opsins will have conserved residue signatures specific to a conventionally functioning ciliary opsin, though ultimately that can only be tested by experiment.
>ENCEPHa_nemVec Nematostella vectensis (anemone) no cdna complete 1 exon 306 aa best:ENCEPH4_braFlo scaffold_465_Cont27987 alt: Nemve1:219988 Nem1 >ENCEPHb_nemVec Nematostella vectensis (anemone) NC-extended 1 exon 275 aa best:ENCEPH4_braFlo scaffold_273_Cont21871 alt:Nemve1:130042 Nem3 >ENCEPHc_nemVec Nematostella vectensis (anemone) C-extended 1 exon 289 aa best: ENCEPH5_braFlo scaffold_11_Cont2404alt: Nemve1:85309 Nem2 ENCEPH4_braFlo Branchiostoma floridae (amphioxus) Gt 0.... 470 7.0e-48 39% identity to ENCEPH4_braFlo ENCEPH4_braBel Branchiostoma belcheri (amphioxus) Gt 0.... 449 1.2e-45 PER_xenTro Xenopus tropicalis (frog) ?? 0.2.0.2.1.0... 438 1.7e-44 ENCEPH4a_takRub Takifugu rubripes (teleost) Gt 0...2...0... 435 3.6e-44 PER_homSap Homo sapiens (human) ?? 0.2.0.2.1.0.1 in... 426 3.2e-43 ENCEPH4b_takRub Takifugu rubripes (teleost) Gt 0...2...0... 418 2.3e-42 ENCEPH5_braFlo Branchiostoma floridae (amphioxus) Gt 0.... 418 2.3e-42 ENCEPH_gasAcu Gasterosteus aculeatus (stickleback) Gt ... 415 4.7e-42 PER_monDom Monodelphis domestica (opossum) ?? 0.2.0... 411 1.2e-41
Four putative opsins have been proposed by Plachetzki et al. Accessions of the supporting gene models are given in the JGI protein ID system (non-GenBank) as Nematostella1 219988, Nematostella2 85309, Nematostella3 130042, and Nematostella4 108738 (or fragments in the alignment graphic allow recovery of the respective cdnas by tblastn of GenBank WGS). As noted in the Hydra section, multiple lines of evidence are necessary to establish the first bona fide opsins in cnidarians.
There appear to be 2 Nematastella opsin-like cdnas at GenBank that cannot be found in the genome assembly or trace archives, DV091537 and DV087469. While genes can be missing from first assemblies, it is bizarre for both to be missing considering coverage is 6x. Upon back-blast to GenBank nr or the Opsin Classifier, very strong matches are seen consistently within crustacea. Thus it appears that these Sars Institute products are contaminants from another species, possibly a brine shrimp widely used in aquarium food. It is not unusual to see transcript (at issue here) and genome projects contaminated with dna from other species such as commensals, parasites, and food source -- this is reminiscent of Xenoturbella being confused with a mollusc in its diet.
New Nematostella transcripts continue to be posted by JGI into mid-Dec 2007. Using proxies for all possible queries, I located a possible melanopsin and possible rhabdomeric LWS counterpart, The former had two coding exons but not at a melanopsin position; the latter had but one. These are fairly weak matches and further characterization is needed. They're stored in the Opsin Classifier as MEL_nemVec and LWS_nemVec2.
A third group has taken a serious look at photoreception in Nematostella. No paper or dissertation has emerged as yet; no cnidarian opsins have been posted to GenBank.
The claim of orthology will prove exceedingly difficult to establish in a 700 million year long branch. It is not a property of a gene tree per se. By definition, two genes in species A and species B are orthologous if and only if they have descended vertically from the same single parental gene in their last common ancestor. The last component is exceedingly important because all opsins -- indeed all GPCR -- are ultimately descended from a single gene. However that single gene was not to be found in the common ancester of cnidarian and bilaterans because sponges already appear to have classical opsins and perhaps hundreds of GPCR.
Most ancestral introns in human genes were established in unicellular eukaryotes well prior to fungal and green plant divergence. For example the distinct introns in close paralogs SUMF1 and SUMF2 were in place before human/diatom separation. It's very difficult to imagine how the introns in neuropsins, rgropsins, peropsins, melanopsins, encephalopsins, pteropsins, and ciliary opsins could have descended from a single gene in Eumetazoa.
Evolution of photoreception: the eyeless anthozoan Nematostella vectensis as a model Poster talk March 22-23, 2007 Heather Q. Marlow, Daniel I. Speiser, David Q. Matus and Mark Q. Martindale (Email: marlow@hawaii.edu) "Eyes have evolved numerous times within the animals, yet there has been surprising convergence in the morphology, function and molecular basis of development in these structures. Although these diverse eye types have arisen independently, many taxa utilize similar cassettes of genes to specify them. These de- velopmental genes include members of the SIX class of homeodomain proteins (sine oculis and optix), eyes absent, dachshund and famously, the Pax genes (Pax6). Additionally, all animals in which photoreception has been investigated use the opsin family, a class of seven transmembrane receptors, to detect light. Cnidarians are an early branching lineage that are likely to have diverged from the rest of the animals before the evolu- tion of discrete eye structures. The ancestral cnidarian did not posses eyes, however like the extant anthozoan cnidarians (sea anemones, corals, and sea pens), it was likely to have had photoreceptive cells. In order to determine the level at which cnidarian photoreceptive cells may share homology with bilaterian eyes, we have examined the expression of these “eye” genes during development in the anthozoan cnidarian model Nematostella vectensis through in situ hybridization. We have also identified, cloned, and studied the expression of many members of the visual opsin class of receptors in N. vectensis. Our data indicate that N. vectensis possesses putative photoreceptive cells which express several orthologs to the visual opsins, that the organization of photoreceptor cells differs between different life history stages of the animal, and that presumptive photoreceptor cells express many of the same developmental molecules that specify eye development in bilaterian animals. These findings support the hypothesis that eyes may share homology only at the level of the photoreceptor, and that additional “eye” genes may have been co-opted into the eye specification pathway from more general neural roles in bilaterians."
A fourth group published a 19 Dec 07 paper on putative anthozoan and hydrozoan opsins, releasing 54 full length sequences to GenBank. These include 31 full length intronated predicted genes for Nematostella, 21 mRNA for lens-eyed Cladonema radiatum, and 2 for eyeless Podocoryne carnea. The latter two species are hydrozoa without genome projects meaning the transcripts cannot be intronated. Many of the 54 proteins have best-blastp below 30% identity within the 230 validated opsins of the phylogenetically comprehensive reference collection. This is worse than some generic non-opsin GPCR, so almost all of the residue matching will be non-specifically exhausted. All have lysine in homologous position, so potentially covalently bound retinal (though that was not established chemically). The counterion situation does not work out at either E113 or E181. Five are missing the universally conserved early asparagine and two others are truncated.
Conserved residues in putative cnidarian opsins relative to bovine rhodopsin and consensus sequences for ciliary, melanopsins, pteropsins, peropsins, and all validated opsins.
..........................................................*...................................................................*..................................................................................................................*..................
rho1 NFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFR
cili N.lv...t.k.k..LrPlN.ilvNla.a#l.....g..........gyfG.....C..eG%...l.G.v.lwsl.vla.dRy.v!ckp.g..f.a.g.........f.....W..pPl.GWs.Y.peg...sC...w.....s%..f.c...........Pl.i....Y..l.........aE..v.rM!..M!..%l...........cW.PYaa........p..P...........faKss.%NPi.IY.f$Nk#fr
cnid N..vi..................s.a..d..........................C...gf........si.hl.....ery........................W.....w...Pl.GW..y..e.....C...w.....sY............l..%P.................m....i..%......................aWtPYa..............l.........fAK.s..nP...%......fr
vali N..V.......k..LRP.N...vNLA..Dl...................g.....C..yg%.....G..s...$..ia.dRY.v!..P......a...........W.....w...Pl.GW..Y.pEg..tsC..#w.....s%.............f.%Pl!I.%..Y..i..........E.....m...m!..F.............W.PYa.........p..P...........fAK.s.%NP!.IY......%R
mela N.lv...f...ksLrtp.N.fIiNLA.sDf.ms....P....s.....W.fG...C.lYaF.g.lfG..S..t$..Ia.DRY.v!t.Pl..s.r.i.v........W.ysl.Ws.pP.fGwg.YvpEG..tsCt.D%.t..r.%.$.f.FPl.i..cY..if.a!r....#.k.ak.........%.......................sW.PYa.!.lG..ltpy.P............AKSai.NPi.iYa..hpkfR
pter Ng.V!.!F..tKsLRTPsN$lV!NLA.sDf.MM..m.Ppm.nc%.t..w.lG...C#.Ya..Gsl.Gc.siwtm..Ia.DRYnvIvkg.p$t..Ali.........W.....W...P.fgwnRYVPEGn$TaCgtDYLt.srs%.ys.vYP$.I!%.Y.fIv.aV.aHEkE.rlAK.vAl.t.sLwf......................aWTPY..!n.G...tPl.ti............k.a...p..vy.ishp.yr
pero N..v...f.k........#....nLA..D.g!s..g.p....S.....W.%G.G.Cq.ygf.gf.fg..Si...t.!a.DRY..iC......$.............W...afWa..Pl.Gwg.YEP.g..t.Ctl#w......%...............%P.!m....Y..!..K.k.....tk............%l...........aW.PYa!..w..f..p.ip.$..........AKs...NP..!Y...#..fr
rho1.NFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFR
Hydrozoa: Hydra magnipapillata (hydra) .. ? opsins
Because opsin photoreception is quite ancient, clearly pre-Bilaterans have a major role to play in illuminating the origins of photoreception systems. What's not so clear is that the two cnidarians chosen so far for genome projects are optimal in this regard.
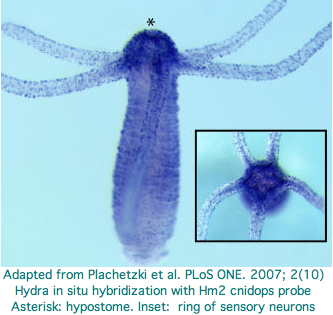
Hydra does not have overt photoreceptive structures or cells obviously specialized for light detection yet it exhibits marked behavioral photosensitivity (noted by Trembley in 1744). Studies beginning in 2000 flagged the ectoderm (using antibodies to squid rhodopsin), known to contain epidermal sensory neurons, as responsible for extraocular photoreception. Musio and coworkers sought to recover opsins using degenerate primers, targeting melanopsin as the most plausible in Hydra because that opsin seems not to require advanced relationships with neighboring cells (ie, acts as photosensor and its own photoisomerase) but no opsin sequences have been submitted to date to GenBank.
The Hydra cdna CB073527 was proposed as a peropsin based on best-blast to mouse peropsin. However using a much larger collection of demonstrably orthologous chordate peropsins in the Opsin Classifier conflicts with this interpretation: the putative cnidarian gene needs to consistently associate with this gene family (equivalently, have best match to it among all reconstructed ancestral opsins) but does not. Furthermore the best match is very weak at 31%. These are exactly the signatures of generic non-opsin rhodopsin superfamily members (which we expect any eumetazoan to have by the hundreds).
With the imminent availability of the Hydra genome assembly (or just using the trace archive), the 161 amino acids of the fragmentary transcript can be extended for example with trace 1121878952 to apparent full length (309 aa) and its introns determined (none). This does not improve its best-blast score nor family coherence. It does not cluster consistently with ciliary opsins -- what would the signaling partner be when the matches are scattered between Gt, Go, and Gq opsins? The blast probability of 1.6e-34 does not mean much under these circumstances.
Two putative hydra opsin fragments can be extracted from Fig.1 of an Oct 2007 paper, AKSSTIINPTISCIIYKE and AKLSAVLNALVNCYINKS. These too fail to extend to fully convincing opsins. Expression centers around the hydropore. Note though that fits GPCR chemoreceptor localization even better -- there is meagre behaviorial evidence for photoreception near the hydropore and no possibility of an imaging eye. No ultrastructure study was conducted -- that is needed to demonstrate that the putative opsin is expressed in specialized photoreceptor cells. No non-opsin controls were included in the alignment -- all GPCR proteins bind heteromeric G proteins and many have lysine without binding of retinal. The cdna accession numbers are Hydra1 CN554949 and Hydra2 CV15164.
These papers highlight the special difficulties in working with cnidarian opsin candidates. We know from the outset that they will be quite diverged. Multiple forms of supporting data are needed, preferably in the form of diagnostic introns, alignments demonstrating conservation of critical residues and structures, in situ hybridization to anatomically plausible neuronal photoreceptors, and specific loss of photobehavior upon knockdown.
A higher standard of proof is needed for the first cnidarian opsins because validated ones will surely be used to pull in further homologs via annotation transfer. There is a definite risk in admitting inadequately documented opsins to the Opsin Classifier because once that database is tainted, it could draw in even more non-opsins from the GPCR world.
CnidBase provides a blast service to cnidarians including hydra but this appears restricted to ESTs and so only duplicates GenBank. GenBank does not carry any contigs or genome assembly on 12 Dec 07. Some 10.2 million Hydra traces have been provided by JCVI, ample for the 1290 Mbp estimated genome size. However the hydra genome project is no longer mentioned on that website. The draft genome expected in Dec 2005 has not surfaced, possibly because high AT content complicated assembly.
Because trace archive searches require a good query, the best current opsin search strategy uses tblastn of the excellent EST collection and extends those results with good Opsin Classifier outcomes with trace reads. Using that method, I recovered an intronless melanopsin candidate could be further characterized bioinformatically but in the absence of experimental support cannot be proven definitively involved in hydra photoreception.
>MEL?_hydMag Hydra magnipapillata CV465424 MAGNDTLEKFSKEIIIIKSLYLVICIILGLIGNLVVLITILKYRKLQTITNYFVLNLSITDLLFIICCMPTIIITTIGEKWLLGNAVCNIIGFLNVLLCTNSIWNLVMISINRYLNVAKPKKIKEIYTRKKTILMI ISVWIVSGLVSVPPLLNWSSYKPGPNFCTVDRKGAKSFYLLILLIMYILPLLILVSLYSCIFFILKKKGKKILLKCNINYIEHSDCKGNASIYKNGLLSYQAG NKQISNKKIMINYFITKNNDTYKVSVKKAQNHNKKLQKCVKLYKQYQITKRLMVLVLSFF LFWTPFFIGSFLITYGVKNKKNFHFTTFGVMCGCLSSISNPFIYSMNSSFRNHLRKLSRNFFNEKNY* >Per?_hydMag magnipapillata Hydra cdna CB073527 MAFVFIIVFLSFLCGFSVILNVTVVLTILAKGNTKNTRDVILMSLAICDGVQCTIGY PVELFGYANYKNPSLSEKFCKPSGFIVMYLALTAIAHLVCLCIYRYLTIVYPLKLQIFLT KSNWSACGCIAFCWIYGLFWSLSPLLGWNEIVRENKDTYKCSINLYPDNEIKSSYLYALA IFCYLIPLIIIIYCSLKVHSELRNMLKMCKQISGVEANITKVTYRIEKQDFISVSFIIAS FFTVWTPYAVCVFYLTIGKKLPPSFLTYCALFAKSSTILNPIIYCLMYKKFRQTLQSKFG KLFNNPTVTPAV* 0
Hydrozoa: Cladonema radiatum (jellyfish) .. ? opsins
A Dec 2007 paper reports 20 mRNA opsin candidates for the lens-eyed hydrozoan jellyfish, Cladonema radiatum. These generally classify as ciliary opsins and the ones tested are expressed somewhat appropriately. However even the best alignment to validated opsins is very weak, no better in percent identity than many non-opsin GPCR (here 13 were used as outgroup without rationalization). However back-blastp to GenBank nr shows no non-opsin GPCR among the best matches.
The authors did not establish the existence of retinal in this species nor show 11-cis retinal covalently bound to any candidate. They note Schiff base lysine occurs in correct homologous position but do not comment on the absence of counterion at traditional positions 113 or 181 (bovine RHO1 numbering). That lysine is necessary for an opsin but not sufficient (it might arise here from primer bias). Non-opsins such as GPCR176 can have lysine at this position as well, making it only semi-diagnostic:
CropN1 KFSVVSNPIVYVIFYKDFR
K S+++NP++++ K R
GPCR176 KVSLLANPVLFLTVNKSVR NP_009154
CropN1 KFSVVSNPIVYVIFYKDFR
K + + NP++YV + FR
LWS_homSap KSATIYNPVIYVFMNRQFR
The number of 'opsins' is excessive given numbers of validated opsins that occur in complete bilatera genomes, even allowing for eyespot developmental stage variations and auxillary functions such as gonad photoreceptive gamete release. Recent gene family expansions might make more sense for chemosensory or chemokine recepters than opsins.
In this view, 1-2 bona fide opsins might lurk among the collection -- but which ones?. Opsins evidently experienced various gene duplications which appear subsequently co-opted to (unknown) non-opsin, non-photoreceptive roles. This causes them to nest confusingly within the opsins even though they are no longer photoreceptors. There may have been selection to maintain the buried lysine because it worked structurally at the time of duplication (offset perhaps by chloride ion) or is used for a new but chemically related signalling agonist. The same phenomonon (gene duplication and neofunctionalization) may have occured within Nematostella and Daphnia, which both have 'too many' opsins for their imaging needs. The Daphnia non-opsin opsin gene familiy expansion is not even broadly shared within the Crustacean clade.
The confusion really arises from the notion of "terminally diverged" opsins first studied within Bilatera. That is, within deuterostomes, we observe a sequence of gene duplication and divergence say encephalopsin --> pinopsin --> LWS --> other cone opsins but that stays within opsins. Even that sequence had largely terminated 500 million years ago at time of lamprey divergence (primate color vision recovery is an exception). An analogous sequence is familiar within Arthropoda for example melanopsin --> MWS --> UVS. We don't observe the sequence encephalopsin --> pinopsin --> LWS --> bradykinin receptor in human nor melanopsin --> MWS --> glycoprotein hormone receptor in drosophila. Nor do we expect any such 500 million years in the future.
In Bilatera, it seems once an opsin, always an opsin. Gene duplication still occurs but opsins are apparently so deeply dug into their hole of specialization of function and tissue expression that a gene duplicate cannot be retained unless it can carve out a niche for itself as limited variant of photoreceptor opsin, say new color sensitivity or polarization detector. Lens crystalins prove that genes often have pre-existing multiple disjoint functions (glycolytic enzyme, refractive index supplier) and so at duplication the niche is already there, merely awaiting partitioning of expression after which sequences can optimize to their respective niches or just drift. Opsins in contrast are single-purpose.
Opsins may not have been so committed in early diverging ancestral cnidaria with less elaborated photoreceptive systems and metazoan cell type specializations. Not so terminally diverged, opsin gene duplicates may have retained overall GPCR signaling capacity but for some other agonist than cis-retinal. After all, shifts in outside molecular trigger happened frequently in generic GPCR evolution, accounting for their vast diversity of functionality despite minimal departures from the universal hepta-transmembrane structure. Ready shifts in agonist are not a design flaw but rather a design feature. Variation in agonist may be tolerated, especially in ancestral GPCR, and GPCR gene duplication and divergence coupled to that of a peptide agonist.
Opsins seem unique in that cis-retinal is covalently attached, whereas other GPCR agonists diffuse in transiently to their binding site. This makes it difficult to see what the 'next' agonist could be in a duplicated non-opsin opsin (other than something very similar like vitamin A variants seen in some teleosts). However it's been argued that the photoisomerization product, trans-retinal, is really the agonist. That's non-covalently bound similarly to other GPCR effectors.
If so, that would make agonist shift in a duplicated ancestral metazoan opsin no different from other shifts taking place in other duplicated GPCR. Indeed, opsins arose from other GPCR; the ur-GPCR was not necessarily a retinal binder. Still, we wonder what the new agonists could be in this cloud of cnidarion opsin-like proteins and what signalling is accomplished where. Hybridization in Cladodema suggests the site of signalling has not moved appreciably.
It's worth re-examing the notion of "once an opsin, always an opsin" even in Bilateran history. First we wonder about the cloud of lineage-specific opsin-like duplications in the crustacean genome of Daphnia. Using bovine RHO1 as query against human genome turns up a dozen non-opsin GPCR exhibiting better matches than the bona fide opsin RGR. Very likely these would nest within opsins with respect to RGR. This suggests that certain non-opsins occur inside the broader photo-opsin family. However here it is not so certain that RGR is a degenerate photoreceptor opsin, today 'merely' a retinal photoisomerase in boreoeuthere placentals retaining the Schiff base lysine but not covalently binding cis-retinal. A similar question arises with neuropsin and peropsin.
RHO1_bosTau KTSAVYNPVIYIMMNQKFR query NP_001044 somatostatin receptor 5 [Homo sapiens] 1e-26 NSCA--NPVLYGFLSDNFR non-opsin GPCR NP_001048 tachykinin receptor 2 [Homo sapiens] 2e-25 MSSTMYNPIIYCCLNDRFR non-opsin GPCR NP_000900 neuropeptide Y receptor Y1 [Homo sapiens] 1e-23 MISTCVNPIFYGFLNKNFQ non-opsin GPCR NP_000721 cholecystokinin A receptor [Homo sapiens] 1e-23 YTSSCVNPIIYCFMNKRFR non-opsin GPCR NP_000903 opioid receptor, mu 1 isoform MOR-1 [Homo sapie 3e-22 YTNSCLNPVLYAFLDENFK non-opsin GPCR NP_004212 G protein-coupled receptor 50 [Homo sapiens] 1e-21 YFNSCLNAVIYGLLNENFR non-opsin GPCR NP_006047 neuromedin U receptor 1 [Homo sapiens] 1e-20 LGSAA-NPVLYSLMSSRFR non-opsin GPCR NP_001471 galanin receptor 1 [Homo sapiens] 1e-20 YSNSSVNPIIYAFLSENFR non-opsin GPCR NP_005949 melatonin receptor 1A [Homo sapiens] 1e-20 YFNSCLNAIIYGLLNQNFR non-opsin GPCR NP_000614 bradykinin receptor B2 [Homo sapiens] 4e-19 YSNSCLNPLVYVIVGKRFR non-opsin GPCR NP_002912 retinal G-protein coupled receptor RGR_homsap 4e-18 KMVPTINAINYALGNEMVC opsin RGR_homsap NP_003292 thyrotropin-releasing hormone receptor [Homo 3e-18 YLNSAINPVIYNLMSQKFR non-opsin GPCR NP_005152 angiotensin II receptor-like 1 [Homo sapiens] 7e-18 YVNSCLNPFLYAFFDPRFR non-opsin GPCR NP_000901 neuropeptide Y receptor Y2 [Homo sapiens] 9e-18 MCSTFANPLLYGWMNSNYR non-opsin GPCR NP_000570 chemokine (C-C motif) receptor 5 [Homo sapie 2e-17 MTHCCINPIIYAFVGEKFR non-opsin GPCR NP_000670 alpha-1B-adrenergic receptor [Homo sapiens] 3e-16 YFNSCLNPIIYPCSSKEFK non-opsin GPCR
No genome project is planned so the mRNA cannot be examined for diagnostic intronation; this could be done indirectly if candidates help locate orthologs in other cnidaria. However they are already very diverged from Nematostella and Hydra opsins. Introns would not be informative anyway in discriminating opsins from co-opted non-opsin, non-photoreceptors arising as segmental duplications. Indels would be similar. This presents a very difficult bioinformatic problem because truly diagnostic residues of functioning opsins could be quite subtle.
Similarly on the experimental side, antibodies would likely cross-react. The derived non-opsins likely still signal with a transducin-type G-protein and are quenched by the same arrestin, so no help there. Knockdowns might have unforseen consequences in these non-opsins, indirectly disrupting photosensitive behaviors. Thus the best way forward in identifying the true opsins within the collection is probably in vitro expression, reconstitution with cis-retinal, and demonstration of photoisomerization.
Porifera, Placozoa, Choanoflagellates .. 0 opsins
One marine demosponge genome is available. Negative larval phototaxis there has been attributed to pigment-filled protrusions in a posterior ring of columnar monociliated epithelial cells. This species may prove insufficient to explain the full range of photoresponsive responses in sponge larva such as circadian rhythm and hexactinellid photoreception (notably the role of [stalk spicules). However Jacobs et al have proposed a far more sweeping view of early evolution of sensory (and other!) organs in sponges (doi:10.1093/icb/icm094 not yet on PubMed). Another view, that of Gehring, proposes that the eye (and other sensory systems) came before the brain, indeed that the nervous system arose later to coordinate a response to all these inputs. There is support for that in simple photoreceptor cells controlling their own cilia. Consequently we should not be too quick to dismiss sponges for lack of neurons.
Porifera: Amphimedon queenslandica (sponge) and earlier metazoa .. 0 opsins
Sponges lie at the base of multicellular animals. They are not noted for eyes. However demosponge larva do exhibit phototaxis (shadow seeking under coral rubble) but the action spectrum is supposedly a better fit to a flavin or carotenoid chromophore. Sponges also can respond to gravity, current, and chemical cues.
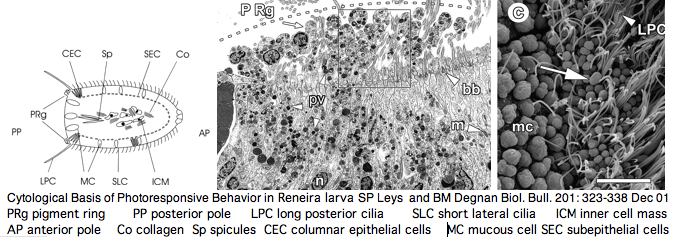
The ultrastructural basis for sponge responses to light has been carefully studied from an ultrastructural perspective -- for an animal lacking nerves and cell junctions, the parenchymella larva are quite capable of responding effectively to light and other stimuli. Larval photoreceptors may lie in a posterior ring of columnar monociliated epithelial cells. A pigment cell occurs but the pigment itself has not been chemically characterized -- the issue is whether it is a tyrosine hydroxylase homologously derived melanin.
The resulting picture of Reneira larva -- numerous differentiated and pluripotential cell types arranged in stereotypic patterns along central-lateral and anterior-posterior axes -- is not one typically conjured up of parazoan ("almost metazoan") in the view of Leys and Degnan. Indeed the common ancestor humans shared with sponge may have been rather advanced.
The concept here is that a photoreceptor cell can control its associated cilium without the baggage of a CNS, either as a passive rudder or more actively directing phototactic motion. In effect the single photocell is a self-sufficient brain that processes external environmental inputs, asseses them and acts appropriately. Chemoreception, a very similar GPCR signaling system, might work the same way. In this view, the nervous system evolvee as a secondary system to coordinate these stand-alone sensory effectors.
The genome of Amphimedon queenslandica has been available since Jun 2005 at the Trace Archives but never assembled. Consequently tblastn of contigs is not available without do-it-yourself assembly of the 2.9 million traces, a inefficient but increasingly utilized option. The species was formerly called Reniera spp. and it is still carried under that name at JGI Genome. It's also been placed in Haliclona and Adocia. Voucher specimens, here QM G315611, are needed to have everyone on the same genomics page.
A futile search for sponge opsins turned up only non-opsin, rhodopsin-class GPCR genes from Amphimedon. That needs to be revisited with tblastn after assembly with melanopsins and encephalopsins recontructed back to the eumetazoan common ancestor. Similarly, no opsins were located in even earlier diverging placozoan Trichoplax, choanoflagellate Monosiga, and fungal genomes. This fits a picture of photoreceptor opsins first appearing subsequent to sponge in eumetazoa cnidarians. However these were hardly de novo genetic innovations but rather evolved out of the already-rich cauldron of GPCR gene copies in the sponge ancester.
Some later diverging species such as the model organism C. elegans lost all of their opsin genes, making them useless in Urbilateran ancestor reconstruction. This argues for much more intensive genomic sampling of sponges and cnidarians so as to sidestep inference mislead by gene loss in model organisms chosen for historic reasons.