Opsin evolution: key critters (deuterostomes)
Chondrichthyes: Callorhinchus milii (elephantshark) .. 13 opsins
Five ray-finned fish genomes and massive transcript studies in yet other Clupeomorpha are available but the utility is this data is complicated by lineage-specific expansions and rapid evolvution of sequences. Little data is available from lobed-finned fish; whereas coelocanth genome has been proposed, that has stalled at 169,000 traces as of May 2008. This makes the preliminary genome assembly of the much earlier diverging Callorhinchus (oft-misspelled) and numerous skate transcipts very special because it is the "last stop" before the very difficult lamprey genome.
This large-eyed cartilaginous fish lives to depths to 200m on the continental shelf of southern Australia and New Zealand but migrates into coastal estuaries to lay egg cases in sand and muddy substrates. The distinctively-shaped egg cases are sometimes found washed ashore after storms. They are up to 25cm long, 10cm wide, and take up to eight months to hatch. The one member of the genus studied has a vitamin A1-based photopigment with maximum absorbance at 499 nm presumably adapted to the overall photic environment at that depth.
Given that sequencing to 6x will not resume at WUGSC until January 2009, I made an exhaustive search of elephantfish data in WGS and Trace divisions of GenBank on 5 Nov 2007, recovering many complete exons but mostly fragmentary genes. The opsin classifier reliably assigns these fragments to their ortholog class.
Overall, Callorhinchus appears to have a full complement of vertebrate opsin genes. The exceptions are RHO2, SWS1, SWS2 (oddly also missing in skate and dogfish ESTs) apparently leaving elephantshark with only RHO1 and LWS rod/cone pigments. Parietopsin is also missing to date. Two encephalopsin- and two melanopsin-class opsins were found. The RGR, peropsin, and neuropsin genes will prove important in better determining their unresolved overall gene tree placement (which an October 2007 opsin phylogeny paper placed deeply within rhabdopsins).
Agnatha: Petromyzon marinus (lamprey) .. 9 opsins
Lamprey are the immediate outgroup to jawed vertebrates. The genera Geotria and Petromyzon split 280-220 myr ago (helpful in breaking up a long branch) whereas Lethenteron/Petromyzon split much later at 20 myr (which is helpful in locating orthologs from the former in the sequenced genome of the latter). Photoreceptor systems have been studied extensively in both larva and adults.
Geotria australis has a full complement of imaging opsins LWS, SWS1, SWS2, RHO2, and RHO1, though its non-pineal opsin classes are less characterized. This implies that the ancestral vertebrate possessed already possessed full-on photopic (bright light) cone-based color vision with the potential for pentachromacy and likely circadian rhythm, pupilary reflex, and pineal and parapineal functions. Photoreceptor morphology and spectral sensitivity change dramatically during various phases of the lamprey lifecycle, presumably adaptively; lineage-specific issues are not a central focus here.
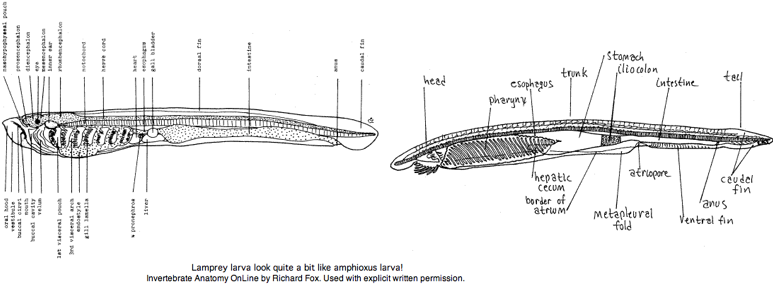
The larvae (ammocoetes or 'sand dweller') are often said blind. It is more accurate to say larva have developmentally arrested eyes but with a small region of differentiated retina. That potentially allows for limited imaging vision -- opsin expression has not been examined. The larval stage, which can last up to 17 years, was considered a separate species by early workers. The anatomical resemblance of lamprey larva to (adult) amphioxus is quite remarkable, a case perhaps of ontogeny recapitulating phylogeny.
The metamorphosis into adult lamprey is every bit as profound as that of amphibians, involving a radical rearrangement of internal organs, continued development of eyes and transformation from sediment-dwellling filter feeder into pelagic marine vertebrate predator. Adult lampreys, like many early-diverging vertebrates, have three eyes: a pair of lateral eyes on the sides of the head and a single, unpaired median eye on top of the head. Each lateral eye is a spherical black structure lying beside the posterior prosencephalon that marks its boundary to the mesencephalon.
According to a 1936 study, larva respond to light in the green-blue by swimming avoidance, perhaps via lateral line photoreceptors. More recently Dickson and Collard, Am J Anat. 1979 Mar;154(3):321-36 write:
"Development of the retina of the ammocoete begins early in embryogenesis, with the formation of the optic vesicle, but development of the rudimentary eye is suspended and remains arrested during larval life. Prior to the onset of metamorphosis, the retina of the ammocoete is completely undifferentiated, with the exception of a small area (Zone II) surrounding the optic nerve head, where all of the adult retinal layers are found. The photoreceptors in this area have developed to include synaptic contacts as well as inner and outer segments. The pigment epithelium in this area, too, has differentiated to include well-formed melanin granules, myeloid bodies and endoplasmic reticulum and is closely associated with the receptor cell outer segments. "
"With the approach of metamorphosis, differentiation of the remainder of the retina (Zone I) begins, taking place in a radial fashion from the optic nerve head. Differentiating pigment epithelial cells adjacent to the differentiated retinal zone begin to accumulate melanin granules. In the neural retina, junctional complexes are established in the form of an external limiting membrane, and connecting cilia project into the optic ventricle. Photoreceptor differentiation begins with the formation of a mitochondria-filled ellipsoid within the inner segment."
The lamprey genome project has resulted in an anemic assembly despite accruing 19 million traces. It seems a 16 kbp retroposon has expanded enormously, which in conjunction with very high AT content and high leveles heterozygosity makes assembly of the 2.4 Gbp genome quite unsatisfactory. However the blast page at WUSTL allows a Petromyzon "3.0 supercontig" tblastn option. UCSC provided a lamprey browser In January 2008. Gene name searches work better than Blat because of very considerable divergence.
New sequencing technologies make affordable an immense collection of cDNA a potential solution in lamprey. It would permit a pseudo-assembly of exons flanked by some at least genomic dna. That is, transcripts are aligned into the trace archives to obtain non-coding context for exons. This allows restricted topics to be studied (intron retention, invariant non-coding) but not others (upstream regulatory, chromosomal gene order). Labelling techniques such as FISH could provide some linkages but not gene-level order; perhaps this could be done at the level of BACs using exons from all possible gene pairs. Alternatively, the genome of Geotria might present fewer issues.
GenBank opsin data for Geotria and Lethenteron can be mapped into Petromyzon using these traces to obtain a supplemental set of sequences parsed for intron breaks and phases. Opsin classes not available in these other lamprey can be collected using chondrichthyes genes as query, with most genes only recoverable as fragments because of gaps in coverage (rather than divergence). In all cases, intron placement remains perfectly conserved from lamprey to mammal, indeed to amphioxus with some complications.
In evolutionary accounts, imaging opsins are often claimed to have appeared 'abruptly' in lamprey as a complete color vision set orthologous to fish and amniotes (and actually exceeding a far more restricted mammalian set). It is fair to say that the lamprey opsin complement establishes that the ancestral node animal had full-blown color vision and auxillary photoreception (ie the gene family had already fully finished expanding).
It's also accurate to say complete genomes of living urochordates (eg Ciona), cephalochordates (Branchiostoma) and hemichordates (Strongylocentrotus) lack imaging opsins within their various opsin-based photoreception systems. However, parapinopsins in Ciona today establish their much older ancestral presence, meaning invention of these opsins cannot be accredited to lamprey, somewhat dissipating the supposed abruptness.
Branchiostoma photoreception today revolves about an expanded set of encephalopsins, melanopsins, and neuropsin-group opsins; we know nothing about what opsins might have been lost during its 500 myr long branch. Imagine the inferential nonsense that would emerge from using humans -- which have lost 9 of 16 genes from the vertebrate opsin complement -- to pontificate about photoreceptor capacities in ancestral species.
In summary, differentiation of ciliary opsins (encephalopsin --> parapinopsins --> cone opsins) was dispersed over a very considerable evolutionary time frame. Note the vast majority of major deuterostome clades have gotten along just fine without them, undercutting the victim-to-predation argument with 500 myr of successful evolution. However, further intermediate details in the sequential gene expansion of cone opsins may elude us if informative additional species prove unavailable. The situation is not nearly as bad as the internode gap between bird-lizard and platypus divergences, yet lies much further in the past.
Care must be taken in discussing "vision" not to confuse low resolution with no resolution. No lensing does not equate to no image of the outside world. Up/down, forward/back are already resolution seen in sponge larva. Today we view images on a color computer monitor with 3 million pixels; ten years ago that was a tenth the pixels and in rod opsin monotone, yet that primitive monitor image was already a big step forward from linux command lines, not to mention teletypewriter return output (no monitor at all). Newton/Gauss/Einstein lacked even that -- were they primitive evolutionary dead-ends, are we more successful at math today with our big color monitors?
It's sometimes argued that oxygen levels in the sea rose markedly in the early Cambrian.This terminal electron acceptor allows 38 ATP to be produced from a single glucose molecule, a big energy gain over 2 ATP possible with fermentative glycolysis under anaerobic conditions. Since eukaryotes developed aerobic pathway enzymes much earlier, prior to the emergence of sponges, the discussion is really about intermediate oxygenation levels in the pre-Cambrian and the rate of oxygen consumption that enables.
Energy-intensive processes such as swimming muscles and imaging vision (with its intensive consumption of ATP via cyclic GMP, nerve conduction, and CNS analysis) may have first become feasible at some threshold of oxygen partial pressure. While accompanying caloric intake must sustain the energy drawdown, that is readily provided without predation (eg cows). We don't have an energy budget for photoreception in non-cone species such as Branchiostoma to assess whether its 7 opsins use more or less ATP than 3 retinal opsins in mammals. We don't have an energy budget for jellyfish bell-flexing swimming to compare cnidarian oxygen consumption with myotome swimming -- recall the predation direction here is cnidarian-on-vertebrate.
The available lamprey opsin sequence data as of April 2008 consists of 15 lamprey sequences from 3 species. Petromyzon notably has a SWS1 intronic transcribed pseudogene, apparently its only copy (Geotria has a functional copy). RHO2 and SWS2 are also missing at least at this level of coverage but again were located in Geotria by PCR. A number of opsin family members are newly reported here, including neuropsin. No trace of RGR opsin or peropsin can be found in this species, despite an earlier allusion to them in L. japonicum that provided no data.
Summary of lamprey sequence data and lambda max available in the opsin reference collection: RHO1_geoAus Geotria australis full P497 AY366493 PMed:17463225 RHO2_geoAus Geotria australis full P492 AY366494 PMed:17463225 SWS2_geoAus Geotria australis full P439 AY366492 PMed:17463225 SWS1_geoAus Geotria australis full P359 AY366495 PMed:17463225 LWS_geoAus Geotria australis full P560 AY366491 PMed:17463225 RHO1_letJap Lethenteron japonicum full P--- AB116382 PMed:15096614 LWS_letJap Lethenteron japonicum full P--- AB116381 PMed:15096614 PPIN_letJap Lethenteron japonicum full P370 AB116380 PMed:14981504 RHO1_petMar Petromyzon marinus full P--- AB116382 PMed: SWS1_petMar Petromyzon maritimus full ---- EB722598 transcribed unprocessed pseudogene LWS_petMar Petromyzon maritimus full P--- genome PMed:no_ref PPIN_petMar Petromyzon maritimus full P370 genome PMed:15096614 VAOP_petMar Petromyzon marinus full P--- U90671 PMed:9427550 PARI_petMar Petromyzon marinus frag1:4 P--- genome PMed:no_ref ENCE_petMar Petromyzon marinus frag2:4 P--- genome PMed:no_ref MEL1_petMar Petromyzon marinus frag5:10 P--- genome PMed:no_ref NEUR_petMar Petromyzon marinus frag1:6 P--- genome PMed:no_ref
Agnatha: Eptatretus burgeri (hagfish) .. 0 opsins
Hagfish, after decades of back-and-forth, are now sistered with lamprey, news not accepted yet by the Taxonomy division of GenBank. However including rogue taxa such as Oikapleura can severely skew results in molecular phylogeny studies. Monophyly of Cyclostoma is unfortunate if true -- an earlier node would very much help in understanding the origin of the eye. It would be better to put aside tree topology until rare genomic events can be developed and simply procede with opsin sequencing.
Jawless fish first appeared in the Ordovician. Hagfish and lamprey split well after the Cambrian, roughly 430 myr ago according to molecular clocks. That's a time span comparable to divergence of human from shark. The oldest fossil hagfishes are Late Carboniferous (330 myr). The two extant hagfish groups split some 75 myr ago (human from mouse). Only recently has it been possible to obtain hagfish eggs and embryos and revisit the neural crest issue. Hagfish experienced an extra round of HOX gene expansion, undercutting both HOX cluster copy number as a hallmark of supposed vertebrate body plan innovation and premature speculation on 1R and 2R whole genome duplication.
Hagfish are nocturnal in aquaria and deep-sea in their natural habitat -- a new Eptatretus species was even captured at a hydrothermal vent. This lifestyle is not conducive to a well-preserved imaging opsin portfolio, though hagfish still have circadian rhythm (based in the preoptic nucleus) and dermal photoreceptors though no pineal gland. The non-imaging paired eyes lack cornea, lens, vitreous body, and extrinsic eye muscle but nonetheless the retina and optic nerve react with opsin antibody. The eyes are larger in Eptatretus than in Myxine, where they are partly covered by the trunk musculature. However 1.3 mm is still quite small for an eye. After comparison of all extant genera, Fernholm and Holmberg concluded in 1975 that the hagfish eyes are secondarily degenerated from more conventional eyes adapted for shallow water (for example an early lens placode disappears). The comparative anatomy of hagfish eyes has an excellent discussion in The Biology of Hagfishes (JM Jorgensen), pages 542-543 available by google book search.
No hagfish opsins have been sequenced; no genome project is scheduled. Even if hagfish imaging opsins are mostly gone, other ciliary and rhabdomeric opsins could be quite informative. Hagfish may have information about a critical era in deuterostome imaging opsin evolution.
Urochordata: Ciona intestinalis (tunicate) .. 4 opsins
Tunicates occupy the strategic urochordate position in the phylogenetic tree. Three tunicate genomes have been sequenced. These proved disappointing for comparative genomics due to their derived nature, which adversely impacts coding sequence divergence, gain and loss of genes, overwriting of ancestral introns, almost total loss of gene order, and high positional heterozygosity. It may not be possible to find more conservatively evolving tunicates if rapid generation time and free spawning are characteristics of all extant urochordates. Yet in other aspects, such as reconstructing the evolutionary trajectory of the vertebrate eye, contemporary tunicates may have retained critical information.
The most useful of these rogue genomes is Ciona intestinalis; Oikopleura dioica and Ciona savignyi have been all but abandoned as model organisms (Max: Yet, the savignyi genome is better assembled than the intestinalis one. The best bioinformatics guys in ascidians (sidow lab) and some biologists stick to savignyi as in some areas of the world they are easier to culture. Savignyi is not really abandoned.) Halocynthia roretzi has many cdna but has not been evaluated for genomic characters whereas Ciona intestinalis has been developed extensively as an experimental system; its massive cdna coverage allow recovery of complete coding gene models which would be nearly impossible from mere homology alignments.
Fortunately, both larval and adult photoreception have been thoroughly studied. Ciona lacks imaging eyes and thus any counterparts to rod and cone opsins like the cephalochordate Branchiostoma. The relative topology of these two with respect to the vertebrates has tilted in recent years towards amphioxus as outgroup -- we'll check later if rare genomic events in opsins support that picture.
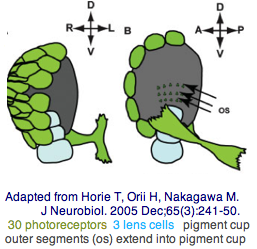
The tadpole larva CNS contains 335 cells of 13 types. These include 30 retinal photoreceptor cells in an unpaired ocellus and 5 accessory cells -- 3 for a ocellus lens-like structure, 1 for the pigment cup, and 1 pigment cell in the otolith (inconsistenly with a hydrostatic sensing role for its 19 receptors). The pigment cells of the ocellus and otolith form an equipotent developmental equivalence group -- a bilateral pair of cells in the blastula gives rise to the otolith and ocellus melanocytes whereas the retina arises from both left and right cell lineages. The observed genomic complement of opsins may largely come into play in the larva because the adult is sessile with little resemblance to vertebrates. The larva are non-feeding which scarcely fits a super-predator role for early deuterostomes opsins.
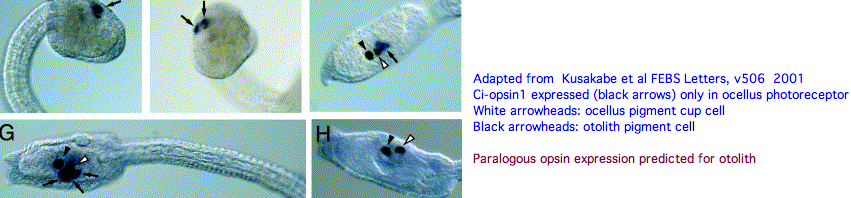
An evidently ciliary opsin called Ci-opsin1 is expressed in the larval ocellus (stored here as PPINa_cioInt). The opsin classifer places this in the PPIN/PIN/VAOP group with best match 44% identity, quite respectable given a billion years of roundtrip evolutionary time. As noted initially by Kusakabe et al in 2001, this opsin shares 3 identical introns with the vertebrate group.
Today there are 25x as many opsin sequences available with much greater phylogenetic dispersion. It appears Ci-opsin has 2 new intron insertions relative to the ancestral Gq ciliary opsin 4-intron pattern 0.2.0.0. This pattern is specific (not shared by Gt melanopsins nor Go retinal isomerases) and diagnostic (disregarding a few lineage-specific gains and losses) -- see documentation.
Three ancient ciliary introns were already established at the time of amphioxus and tunicate encephalopsin divergences. Indeed they already occur in sea urchin, ragworm, mosquito, moth, and beetle ciliary opsins. Consequently they were present in the parent ciliary opsin of Urbilatera and no doubt Cnidaria. There's nothing surprising about this because the vast majority of (human) coding introns originated far earlier in unicellular eukaryotes and have been conserved ever since. Outside of rogue lineages such as drosophila, nematode, and tunicates, event rates for intron gain and loss are perhaps 1-2 per five billion years of branch length. Convergence is not favored because 333 aa sites x 3 intron phases = 1000 distinct possibilities in an opsin-sized protein -- for an already very rare event to happen twice in the available branch length requires predisposing factors.
We will use these deep intron characters later to supplement -- and even trump -- maximal likelihood inference from primary sequence divergence which captures the broad picture but fails to resolve the issues of most interest. With opsins, alignment (at these time depths and rates of change) hits the wall of generic rhodopsin superfamily and indeed generic GPCR proteins, which numbered many hundreds at the time of Urbilatera. There are already many constraints on proteins which must have seven transmembrane helical segments, covalently bind retinal with a lysine and counterion, and interact with heterotrimeric signalling protein.
With the genome in hand, we can see Ci-opsin1 has an unstudied paralog (here called PPINb_cioInt) of 58% identity and identical introns (other than a new phase 21 intron breaking exon 4). There is no expression data for the paralog in the UCSC browser track but it cannot plausibly be a pseudogene due to the conserved nature of amino acid replacements, so we wonder about subfunctionalization. The hybridization experiment will have to be repeated at various life stages. Paralog lambda max might be computable or measurable in a construct. The 1999 experiment (which measured speed-up in swimming after light decreased, reminiscent of the pineal-mediated frog tadpole response) deduced a lambda max of 505 nm -- perhaps that was a composite action spectrum. The new paralog in fact conserves the key lysine and counterion.
We can hope that photoreception in Ciona retains ancestral characteristics that descended intact, at the same time knowing evolution of protein sequences and development have not stood still for 600 million years. Ciona photoreception may have both degenerative and innovative aspects. It is premature to homologize ocellus with pineal (or amacrine or horizontal retinal cells etc) until the role of all the opsins in the Ciona genome have assigned roles (not to mention dozens of other genes). I suggest from evo-devo equipotency that the paralog opsin functions as a photoreceptor in the otolith.
Here neural integration of hydrostatic pressure signaling with brightness directionality could advantageously inform the larva of its position and orientation even in a murky water column and help with dispersal and settlement. A pigment cell is hardly needed for hydrostatic pressure sensing -- what functionality would maintain it over evolutionary timescales? The function of pigment cells is blocking light from the back, here so the larva knows up from down. Curiously, a crystallin of definite homology to refracting vertebrate lens crystallins is expressed in the otolith but not ocellus lens-like accessory cells. The statocyte itself is sprung by its footpiece and two fibrous structures, all synaptotagmin-positive. Movement of the statocyte would be detected by these three structures and thus sense gravitational orientation.
We're left wondering if the speculative otolith/photoreception connection in urochordates has any connection to the balance sensory system (vestibular apparatus) in the vertebrate brain. The otolithic organs (utricle and saccule) detect inertial movement using tiny calcium stones (otoconia) coupled to hair cells. The Allen Brain Atlas could be explored on vestibular sections for extremely detailed expression of most opsin genes. The vestibular system coordinates extensively with the visual system via the vestibulo-ocular reflex. If true, this could radically affect homologization.
Possibly this ciliary paralog pair descended from a gene duplication already present in the last common ancester, leading after still more gene duplications to the current portfolio of vertebrate ciliary opsins. This would account for its ambivalent behavior in the Opsin Classifier with respect to the PPIN/PIN/VAOP group. Alternately the pair might represent a tunicate-specific duplication of secondary interest. Ciona savignyi has a clear ortholog (88% identity) to PPINa_cioInt but a lesser match at 59% to PPINb_cioInt, in both cases with identical introns (not an unusual pattern in gene duplications assuming PPINa_cioInt continues the original function). C. savignyi -- which is only in the same genus from a severe anthropocentric perspective -- helps gauge the rate of evolution of C. intestinalis opsins.
Photoreception in the adult ascidian, which might seem gratuitous in a sessile filter-feeder, has not been studied in quite such detail. However several non-opsin expression studies suggest that adult photoreceptors may develop about pigmented spots around oral and atrial siphons, epithelial cells of sperm duct and cerebral gangla, involving behaviors such as siphon contraction, phototroism, and gamete release. The anterior photoreceptor of the oral siphon has even been homologized to vertebrate lateral eyes.
We'll see below that exactly the same problem as above (undocumented paralog) may affect interpretation of a comprehensive experimental study of Ciona Ci-opsin3 (RGR1_cioInt at the Opsin Classifier). Here too I was able to recover a related second gene in both C. intestinalis and C. savignyi. This illustrates the power of genomics -- provided coverage is complete, a full complement of bioinformatically extracted opsins can guide experimental design from the beginning. A full set of opsin classes should be sought in the genome, even if their degree of sequence divergence and lack of transcripts makes this difficult.
Kusakabe,Tsuda and coworkers have studied the overall visual cycle -- a much better approach than considering opsins in isolation for purposes of homologization. Recall incident photon absorption by rhodopsin isomerizes 11-cis-retinal to all-trans. Without recycling or fresh cis-retinal, this would soon exhaust vision. In mammals replenishment of the visual cycle (retinal isomerase, RGR) takes place in retinal pigment epithelial cells which are distinct from the photoreceptor cells, unlike lophotrochozoa where the cycle is completed within the photoreceptor cell. What about Ciona? We might expect a mixed system since the deuterostome divide preceded the deuterostome photoreception divide with Ciona occupying a strategic phylogenetic position.
If life were simple, Ciona would have strict 1:1 orthologs to the 4 components of the mammalian visual cycle protein, RGR (Ci-opsin3), cellular retinaldehyde-binding protein CRALBP, β-carotene monooxygenase BCO, and retinal pigment epithelium RPE65. At this phylogenetic depth, we can expect a certain degree of non-parallelism between photoreceptor systems and complications from lineage-specific duplication and subfunctionalization, not to mention lack of exact mammalian counterparts to Ciona larval and adult stages.
It turns out (using closest homologs) that Ci-BCO is predominantly expressed in larval ocellus photoreceptor cells, whereas Ci-RPE65 is not significantly expressed there nor in larval brain vesicle but rather in photoreceptor cells of the neural complex (a photoreceptor organ of the adult) right along with Ci-opsin3 and Ci-CRALBP (ie, like cephalopod). It appears the larval visual cycle uses Ci-opsin3 as restorative photoisomerase whereas the adult visual cycle Ci-RPE65. The remote paralog RGR2_cioInt was not studied and its role remains speculative. Given its degree of divergence yet persistance in a second ascidian, it is an old gene duplication maintained somewhat by selective pressure.
What about rhabdomeric opsins in Ciona? We know that melanopsin persisted into vertebrates so it must have been present at the common ancestor with ascidians. Rhabdomeres themselves as a subcellular opsin housing specialization did not persist so their apparent absence in Ciona does not imply the absence of melanopsin.
A Ciona melanopsin could be very diverged. The best possible search involves tblastn of the Ciona assembly and GenBank est_others with a variety of queries (since the best query is not known in advance; after the fact it is provided by the Opsin Classifer). Reconstructed ancestral melanopsins can improve on specific species queries by eliminated half of the roundtrip divergence.
However overly sensitive queries have the risk of merely returning generic rhodopsin-superfamily members (notably ADRA1A adrenergic receptor). While these won't receive clean approval from the Opsin Classifier, any putative melanopsin must be secondarily validated by retention of intron pattern, synteny with vertebrate melanopsins (unlikely in Ciona), and internal amino acid signatures of authentic melanopsin-type photoreceptors.
I evaluated various candidates using a wide variety of probes (such as echinoderm melanopsin) on 11 Dec 07 but none led to a convincing urochordate melanopsin. Absence of evidence is not evidence of absence but it appears that Ciona did not retain a melanopsin.
Echinodermata: Stronglyocentrotus purpuratus (sea urchin) .. 6 opsins
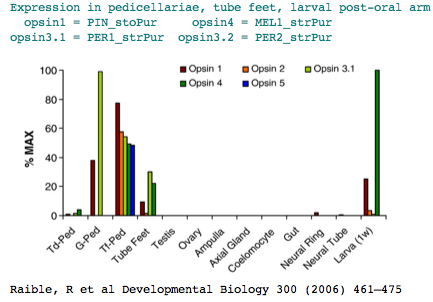
The sea urchin genome carried a big surprise: the previously dismissed echinoderm has a large set of genes for sensory and signalling capability (comparable in number to human). These include at least [1] six opsins] relevent to our purposes. Adult sea urchins exhibit a variety of responses to light intensity: shelter seeking, covering reactions, diurnal migrations, and spine defense reaction to shadowing. Various pedicellariae (jaw-like appendages around the base of spines) keep the body surface clear of encrusting organisms and aid in food capture. Larva don't have evident eyes but do express an opsin in the post-oral arm suggesting some capabilities..
Because sea urchins are seriously diverged, it is difficult to recover accurate full-length sequences by homology, especially in poorly conserved termini, without transcript evidence. At this point, only one of six urchin opsins has any cdna support -- and that from a different species of urchin! That melanopsin interestingly consists of a single exon -- evidently retroposed but still functional -- for which no parent gene can be located. It is not unusual for a descendent gene to supplant the multi-exonic parent, perhaps by accident, perhaps because of transcriptional efficiency considerations.
The two peropsin-class Go urchin sequences are adjacent in parallel tandem configuration with identical intron pattern but have only 64% amino acid identity, consistent with a moderately old tandem duplication. Despite additional weak members of this group, care must be taken not to drift off-topic into the greater rhodopsin superfamily of GPCRs (979 genes in 70 families annotated in urchin).
The sea urchin genome contains one very clear ciliary opsin (called PIN_stoPur in the sequence storage area). Here the GLEAN3_05569 prediction from Baylor appears entirely accurate whereas GenScan and GNOMON XM_778209 and XM_001177470 are impossibly flawed. The Opsin Classifier classifies this somewhat ambiguously within pinopsin-encephalopsin, suggesting it might seed a new ciliary opsin class. The intron pattern 0.2.2.0.0 is a perfect match in position and phase to pinopsins. Indels will be considered when the global alignment is revisited. It has no detectable counterpart in the Saccoglossus genome.
There appears to be a second ciliary opsin (stored as ENCEPH_strPur). It is best fit to Branchiostoma and Platynereis ciliary opsins but only at 33% identity and not that distant to certain melanopsins. This opsin too is likely to be involved in some aspect of photoreception, though that won't be as closely related to vertebrate imaging as PIN_stoPur.
The two remaining opsins classify as rhadomeric melanopsins. One of them, MEL2_strPur, has a GenBank transcript DQ285097 alluding to an unpublished expression study concerning tube feet photoreceptors. The other melanopsin is expressed post-oral arm of two-week-old larvae.
Hemichordata: Saccoglossus kowalevskii (acornworm) .. 1 opsin
This surviving member of early branching deuterostomes has excellent genomic and trancript coverage, with diverse full length multi-exon genes often recoverable. Be aware that transcript data has been misplaced by GenBank to reside under Saccloglossus 'other' at the trace archives. However acorn worm may not illuminate photoreception at its ancestral node with echinoderms (Ambulacraria). Acornworm have isolated photoreceptive cells are scattered through the epidermis but no eyes or eye spots even as planktonic larva, as befits an animal that settles in its burrow on day two. Light striking epidermal photoreceptors elicits burrowing behavior.
In searching for opsins that might underlie epidermal (or other) photoreception, the best queries (for detecting diverged sequences) are likely sea urchin opsins against Saccloglossus 'other' because, being transcripts rather than short exons, these give longer matches. However opsins expressed in scattered cells may not be represented there if rarely transcibed. Promising traces must be back-blastxed against the Opsin Classifier because the initial query choice may have beeen sub-optimal. Good matches can then be intronated using the exact-matching Saccoglossus probe against genomic reads. Intron patterns are critical adjuncts to low sequence alignments in establishing opsin orthology classes because surviving synteny at this time depth is rare.
I report here the very first hemichordate opsin. The process for obtaining accurate gene models in such a remote species is a difficult exercise in bioinformatics and use of the Opsin Classifier -- a detailed procedure is given in the annotation tricks section. That Saccoglossus opsin classifies unambiguously to the peropsin/neuropsin group by similarity and intron structure and thus is not a strong candidate for the epidermal photoreceptor -- but it suggests the story is not that simple. A complete set of opsins for these species best awaits release of assembled contigs by the Baylor sequencing center.
>PER_sacKol Saccoglossus kowalevskii Expect = 2.0e-49 PERa_braFlo Identities = 97/246 (39%) IIYYFFLLSTGLTIFGMSLSCVSSF GRWLFGKFGCYFHGFAGMLFGLGSIGNLTVISIDRYIITCKRSL 1 2 WSYRHYYALLAVAWSNALFWSMMPLFGWSSYALEPEGTSCTIDWMNNDNQYISYVSCVTVTCFILPCAVMTYDYLAAYMKMVKAGYTLSEETEKPNND 0 0 MCIALVAAFLLSWFPSATVFLWAAFGNPGNIPLSFTGVADAFTKIPAVFNPVIYVALNPEFRKYFGKTIGCRRKRKKPIAVRLNGSEQNVENTI* 0
Deuterostomia: Xenoturbella bocki + Convoluta pulchra .. 0 opsins
These two taxa have recently been put forward as new phyla of basal deuterostomes, the former as outgroup to echinoderms plus hemichordates, the latter acoel flatworms as more basal still. However sequence data is extremely sparse with 3,127 sequences for all of Acoelomorpha, and Convoluta pulchra evolving far too fast for practical use, with its tree position controversial.
No genome or major transcript studies are under consideration. A quick check via tblastn of sea urchin opsins against available transcripts does not turn up good opsin candidates as of 28 Nov 2007 (other than a weak melanopsin match in Convoluta, EV602614, that might instead be generic GPCR). No information about photoreception in these species is readily available. While the above two taxa might not be ideal for opsin purposes, extant species are very limited.