Opsin evolution: key critters (lophotrochozoa)
Lophotrochozoa: 17 opsins
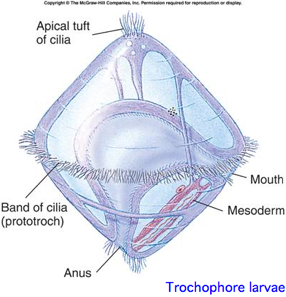
This is a proposed monophyletic group of bilaterans defined on molecular and developmental grounds (Perrier 1897; Seurat 1920) reflecting a basal split deep within protostomes. The classification is based both on molecular considerations and a shared larval form with ciliated wheel, in contrast to characters of adult animals such as segmentation. The placement of nematodes is not relevent because -- as of June 2009 -- the whole clade appears to have lost all opsins.
Lophotrochozoa is not recognized at GenBank so blast searches cannot be restricted to Lophotrochozoa (the remedy being two searches for Annelida and Mollusca as almost all sequence data resides there). However Entrez and PubMed searches can be so restricted using boolean queries. In terms of genome projects Lophotrochozoa currently consists of 7 species of flatworms, molluscs, and annelids. However, it also contains Brachiopoda, Bryozoa, Entoprocta, Nemertea, Sipuncula, etc which collectively account for less than 3,000 of the 5.7 million nucleotide sequences at GenBank and no annotated opsins.
The Lophotrochozoa have not been surveyed as a whole for those that might be 'living fossils' in terms of opsins and photoreceptor structures. Even those would not necessarily make good genome projects because of potentially large genome size and base compositional issues. However Annelida has been thoroughly considered by Purschke, Arendt et al in a recent offline, off-Pubmed review (Arthropod Structure & Development 35(2006) 211-230).
Most of the genome projects have not yet resulted in publications. However assembled contigs have generally been released to the wgs division of GenBank where they can now by queried by tblastn. This is actually preferable to using uncurated pipeline gene models which have far too high an error rate to be used in comparative genomics. Still, the sampling of lophotrochozoa today is far too sparse to allow ancestral opsins to be reconstructed or to write a history of gene expansion and contraction.
Annelida: Platynereis dumerilii (ragworm) .. 3 opsins
This small annelid may be an emerging model organism, though plans for genome sequencing in France have apparently collapsed. (Indeed all metazoan genomic sequencing in Europe has ceased.) Three recent papers have established that Platynereis qualifies as living fossil, at least with respect to ancestral anatomy and development, slowly evolving protein sequences and retention of genes and ancestral introns, and further has retained ciliary opsins.
That is to say, fruit fly and nematode have proven unfortunate choices because of so many lost genes and signalling pathways, rapid evolution and highly derived characters. Lophotrochozoa may thus give us very significant insight into the bilateran ancestor that had appeared lost from consideration of just Arthropoda. It should be noted though that not all insects should be written off, Anopheles has also retained ciliary opsins.
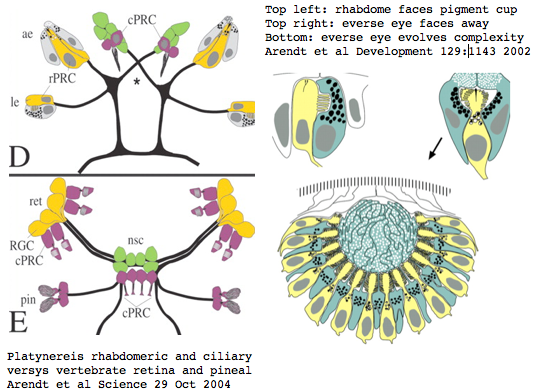
Platynereis develops various pairs of eyes going by localization of opsin expression: inverse larval eyes used in phototaxis (just one pigment cell and one photoreceptor cell) and two pairs of everse adult eyes needed for adult vision. These originate from an initially unsplit single anlage. These eyes use exclusively rhabdomeric photoreceptor cells and corresponding rhabdomeric-class opsins as expected from phylogenetic position. However two paired structures in the developing median brain dorsal to the apical organ express an opsin that unambiguously classifies as ciliary. Further, a retinal homeobox (specific to ciliary pineal eyes) and circadian rhythm regulator bmal are also expressed at this location in Platynereis. However the pigment cells necessary for directional photoreception are missing. This all fits with a role for the ciliary opsin as the primary receptor underlying circadian rhythm which does not require directionality.
The emerging picture is Urbilatera having both ciliary and rhabdomeric structures. The later specialized structure was lost but the photoreceptor component retained in vertebrates in the form of melanopsins expressed in retinal ganglion cells.
Remarkably, Platynereis contains a second ciliary opsin next to alpha tubulin: Using the initial ciliary opsin (a transcript with unknown intronation) as probe at various GenBank databases, a genomeWiki contributer found a 171,779 bp survey sequence in the high throughput genomic sequence HTGS division (meaning it would be overlooked using Blast of the nucleotide division) had a good match in the unannotated contig CT030681, submitted 05-DEC-2005 by Genoscope as 6 ordered contigs (the last of which proves reverse-complemented).
This second opsin, being genomic, after difficult recovery of full length gene from a moderate match, could be intronated (unlike the original transcript) assuming GT-AG splice junctions (like 99% of all genes and 100% of all known opsins). These introns had positions and phases identical to ciliary -- but not Go or Gq -- deuterostome opsins. Assuming the first opsin is not derived as a processed retrogene from the first, it can be intronated via homological alignment. These are stored in the Opsin Classifier as CILI1_plaDum and CILI2_plaDum, resp.
Using the second opsin as blastp query against our phylogenetically dispersed collection of 225 hand-curated Eumetazoan opsins (including new cnidarian ciliary opsins), it classifies in the encephalopsin-to-pinopsin area in accord with independent classification by intron pattern and close homology with the experimentally characterized Platynereis first opsin. The percent identity to deuterostome opsins is not only quite high (considering the immense round-trip time since common ancestor) but also overwelmingly concentrated on invariant and near-invariant amino acids characteristic of ciliary opsins. Thus this second Platynereis opsin cannot be a pseudogene (unless that happened yesterday or so).
For purposes of conserved synteny [eg establishing orthology to related opsins in other lochotrophoran genomes], other coding genes on this contig using blastx vs metazoan proteins) can be considered. The only other gene is alpha-tubulin, at positions 124517-122811, downstream from the second ciliary opsin at 46848-87956 using original contig ordering.
Recall the Arendt group used antibody to acetylated alpha-tubulin was used as marker for stabilized microtubules in cilia and axons. They needed the sequence for that. Probably the larger contig was then sequenced as part of the genome feasibility survey. There was no particular reason to look at this contigs for opsins at that time, which would be hard to distinguish from abundant non-photoreceptor rhodopsin-superfamily genes or generic GPCR.
Supposing Platynereis has 15,000 coding genes, this is quite a coincidence to have two genes adjacent that might be critical to the same photoreceptor structure. If these two genes are transcribed divergently (lie on different strands) after fixing (reverse-complementing) the last contig piece, then symmetric transcriptional regulatory element DNA (read the same whichever strand), this could mean the second opsin is tethered to alpha-tubulin production in terms of co-expression in some cell types. Transcribed in the same direction is less attractive as operons are rare in eukaryotes, though read-through is not unheard of and that too could be developmentally regulated in extent.
Re-assembly CT030681 using multi-exon bridging is possible. It turned out pieces 1 and 2 were irrelevent, piece 3 had exons 1,2,3 of the opsin on the plus strand, piece 4 had opsin exon 4 and 5 on the minus strand to piece-coordinate 41,899 for the stop codon. This piece also contains the first three exons of alpha tubulin also on the minus strand beginning at 36,767. Its initial methionine is stranded as a solitary phase 0 codon on the end of 5' UTR, 36,707-05. The remaining two exons of alpha tubulin are on the minus strand of piece 5.
Joining piece 3 with reverse-complemented pieces 4 and 5 then fixes orientations to the plus strand and establishes intron sizes subject to the two strings of Ns. This results in parallel gene order CILI2_plaDum+ TUBA_plaDum+, that is tubilin downstream of the opsin with an intergenic gap of 5,132 bp. If there is any coordination of expression by read-thru, on the upstream end it would have to involve the regulatory regions of the opsin.
The fifth exon of CILI2_plaDum has too weak match with that of CILI1_plaDum to be found by conventional searching. However the dna where it has to be located is squeezed between exon 4 and the start of tubulin, reducing query size. Blastx of that dna against the full-blown set of opsins turns up a consistent match candidate in frog and skate opsins. Looking at the intron phasing validates the match since the splice acceptor AG is 1 of 16 dinucleotides, the phase 0 required by exon 4 (and ancestral ciliary phase) is 1 of 3 possible phases, and 1 of 2 strand requirement have together a 1 in 96 chance of random occurence, more than sufficient in conjunction with the blast expectation of 1.1e-06.
This opsin if co-expressed with CILI1_plaDum would amount to 'circadian rhythm color vision'. Alternately it might be expressed at a different developmental stage or in an unsuspected auxillary photoreceptor.
Annelida: Capitella sp (marine worm) .. 2 opsins
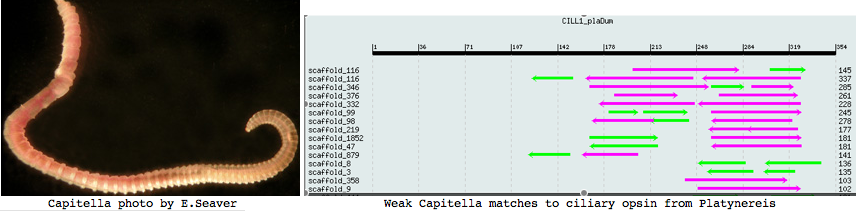
Capitella is a small segmented benthic marine worm most closely related, in the genome project sense, to its fellow annelid Platynereis. The taxonomy of the genus Capitella was thoroughly muddled by a quaint 1976 starch gel electrophoresis allozyme study; Linnean nomenclature has never been developed for the 6 alleged species defined there. The isolate used in the JGI genome project is called Capitella sp. I ES-2005 instead of Capitella capitata.
The last of 3,709,316 trace reads were taken in Nov 2005. As with Lottia, a multi-year lag ensued in release of the assembly, deposition in GenBank, and publication of central paper. As of Dec 2008, the only access to the genome is through JGI Blast. The genome is small at 240 Mb and distributed across 10 chromosomes.
This is a subsurface deposit feeder associated with organic-rich mud, seemingly not conducive to an extensive visual system. However an extensive 1993 study of both larval and adult eyes was published in the now-defunct Journal of Morphology (online acces $25). Developing larva hava a pair of eyespots consisting of one sensory cell, one pigment cell, and one support cell. The photoreceptor cell has an array of parallel microvilli with cisternae. It is surrounded by a diaphragm formed by a pigment cell ring of microvilli-like structures. These last but a few days because at metamorphosis the larval eyespots are greatly reduced. Adults have one pair of eyes built of 2-3 pigment cells and one sensory cell in juveniles increased by 2-3 more in adults.
Unusual morphological aspects of Capitella eyes can be placed within the overall context of photoreceptor cells and eyes in Annelida, whose ultrastructural issues were carefully reviewed by Purschke in an off-PubMed journal "Arthropod Structure & Development" v35:4, 2006 (viewing issue full text costs $175). In addition to rhabdomeric and ciliary types, less-known phaosomous photosensory cells are discussed. Phaosomes (Greek: phaos = light, soma = body) were first described in the earthworm dermal photoreceptors as a central intracellular cavity (phaosome) filled with microvilli but may represent a derived form. They occur at various extraocular sites such as dermus and genitalia (in butterflies). Multiple types of photoreceptors thus provide a potential role for the diversity of opsins observed in the genome.
It's clear from Purschke's review that photoreceptors require a combination of ultrastructure, transcript expression mapping, and genomics. In other words, it's necessary to account for all the opsins found in the genome. Many photoreceptors have been overlooked entirely, notably the undirected type (no pigment cell backing); many others have stalled out in controversy for lack of gene availability.
I found a number of related opsin fragments in Capitella using various queries but surprisingly no counterpart to Platynereis ciliary opsins. One, stored as MEL1_capCap, clusters consistently with melanopsins and shares two exon breaks. It may be an ortholog of the rhabdomeric Platynereis opsin. The second MEL2_capCap is more distantly related. Reliable full length genes will require a cdna program which so far is totally lacking.
Annelida: Helobdella robusta (leech) .. 2 opsins
The JGI genome project for the leech Helobdella robusta is well along with 3,168,749 traces, a very recent assembly to blast, but no cdna. The genome is fairly small at 300 Mb but does not appear reduced in terms of gene count. Fifteen unannotated 100 kbp contigs are available at the HTG division of GenBank; these do not contain opsins but might otherwise suggest gene and retroposon densities and extent of synteny retention. The genome had not been submitted to GenBank by Dec 07.
Helobdella could be considered a promising emerging experimental system because techniques such as large-scale whole-mount in situ hybridization screening, RNA interference, and morpholino knock-down are established. It's not clear however that leech retains the degree of ancestral characters as nereid polychaetes. Until a cdna program is established, it will prove very difficult to annotate complete coding genes. The nearest species with a transcript program is the earthworm Lumbricus rubellus with 19,934 ests (but no opsins).
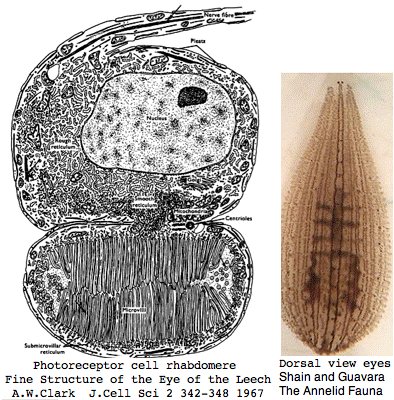
Helobdella is a rhynchobdellid, which is to say (ελεο marsh, ῥύγχος snout, βδελλα leech) a California marsh leech with a muscular straw-like proboscis in a retractable sheah for puncturing prey. Thus it is not closely related to the medicinal leech, Hirudo medicinalis. The anatomy of the closely-spaced single pair of eyes was intensively studied 40 years ago. An eye in this group consists of 30-100 photoreceptive cells in a deep pigment cup providing directional vision. Larvae are not free swimming but stay in the albuminous fluid of a cocoon. The 88 Pubmed articles include many on body plan gene expression but only two on eyes and these tangentially. We can only hope the genome project will stimulate additional studies of leech photoreceptors. It seems that every lab uses a different strain if not a different species.
I recovered two Helobedella opsin genes on 4 Dec 07 from the erratic JGI server (if no matches, close and restart with a fresh window). The full length gene, stored as MEL2_helRob has 2 conserved introns characteristic of melanopsins and its best matches there. It is likely an ortholog of a similar gene in Schistosoma, Schmidtea, Capitata, and Platynereis. The 231 aa fragment stored as MEL2_helRob has best match to octopus and chordate melanopsins and shares the first (and possibly second) intron position and phase with them. The parent scaffold 39 may contain tandem opsins or alternatively represent a misassembly. No counterpart to the ciliary opsin of Platynereis emerged. That gene -- which must have been present in the common ancestor with annelid -- could have been lost or is simply missing from the current assembly.
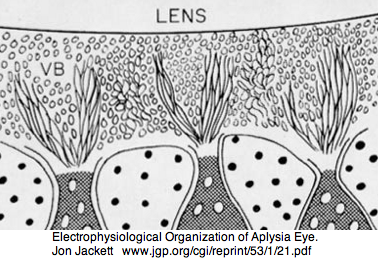
Mollusca: Aplysia californica (sea hare).. 3 opsins
Aplysia has a pair of cephalic dorsal pit eyes just anterior to the rhinophores. The eyes are quite small at 600 microns diameter, with a spherical lens and a tiny one square millimeter retina with approximately 7000 rhabdomeric photoreceptors. Despite a fair number of studies of eyes and rhinophores involved in vision, circadian rhythm and phototactic head-waving, the opsins have not been characterized beyond immunoblot (positive for etinal photoreceptors, rhinophores, cerebral ganglia and ventral abdominal ganglia giant cell R2). There is evidence for G protein alpha subunits Gq, Gi, and Go families, phospholipase C, and an inositol 1,4,5-trisphosphate receptor in the rhinophore but this may be for chemoreception.
The sea hare genome has recently be sequenced by Broad Institute. Sizeable assembled contigs are now open to tblastn at the "wgs" division of GenBank (which allows the exon pattern to be extracted). Despite the assembly, sequencing continues: 212,159 new traces were added in the last week of Nov 07. This illustrates the need to always check the primary data repository when a gene seems missing -- millions of traces might not be used in the assembly. However a close-in query is needed to get a match.
The first known Aplysia opsin was found in the 20874 bp contig AASC01108363 on 2 Dec 2007. It had a significant expectation value (e-60) but the best match percent identity within the opsin reference collection (to fellow mollusks) was only 118/319 (36%). Otherwise the best matches are consistently vertebrate melanopsins. This gene is a strong candidate for an invertebrate melanopsin ortholog. It is stored as MEL1_aplCal. In June 2009, a second melanopsin was located, again quite diverged: MEL2_aplCal.
Indeed, there are four exons but precise boundaries are difficult to locate at this low percent identity without cdna or reliably intronated guide sequence from a closely related species. However 2 introns clearly have identical position and phase to vertebrate melanosins and a 3rd quite likely; otherwise there has been intron loss in Aplysia. The contig unfortunately does not contain any information (according to blastx) on adjacent genes (synteny) despite 10 kbp still available 3'. No counterpart to the Platyerneis ciliary opsin could be found.
On 28 Dec 07, a full length peropsin was announced here, PER_aplCal, a likely ortholog (from exon breaks and best-blast) to squid retinochome which has an excellent structural model and counterion study. The Aplysia peropsin is well-represented with 11 transcripts from pedal-pleural ganglia, CNS (adult and juvenile 1), metacerebral cells, and MCC metacerebral neurons but only terminal exons are found in the assembly. However the cdna provide a window to the trace archives which allows accurate intronation of the full gene.
It is not at all clear what relationship these lophotrochozoan peropsins have to deuterostome peropsins, nor why they seem missing altogether in ecdysozoa, nor what their ancestral status is. The 3 molluscan peropsins cluster cleanly enough with vertebrate peropsins but overlap only partially in intron placement. That could result from relatively recent intron gain and loss or reflect a much deeper ancestral splitting of peropsin classes. Representatives of these may survive more completely in echinoderms, hemichordates, and cephalochordates. Peropsin may very well be capable of ciliary opsin type signaling with trans-retinal as agonist.
At this point, Aplysia is not a Rosetta stone for opsin evolution. It is however the first mollusk with a genome assembly. This may eventually allow confident transfer of orthology validated by synteny, intron pattern, and indels. The eyes appear homologous in many aspects to those of Arthropoda supporting the common ancester of Protostomia having rhabdomeric lensing eyes, though true across-the-board homology of all eye components is a very complex subject.
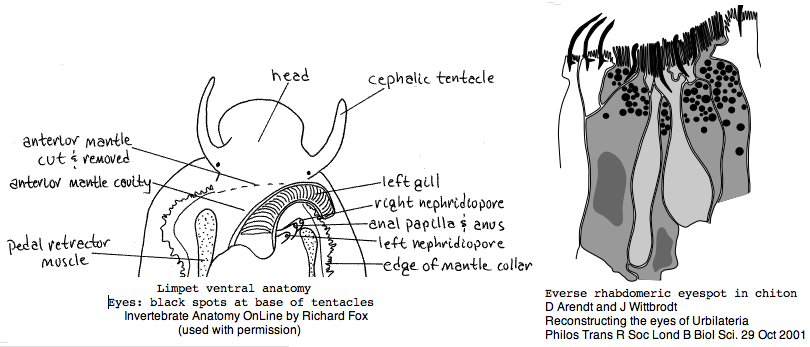
Mollusca: Lottia gigantea (limpet) .. 2 opsins
The limpet Lottia gigantea was intended to be the first lophotrochozoan for whole genome sequencing but that goal slipped. It has ancestral-like spiral cleavage and trochophore larva. The genome is small relative to other molluscs at 500 mbp. Some 5.3 million traces were sequenced by May 2005. In Jan 2007 the sequencing center presented the genome at a meeting talk. However by Dec 2007 no paper had appeared. Recently JGI enabled blast of the assembly and display on their funky browser. However nothing was submitted to Genbank. JGI predicts 4 rhodopsins for its KOG gene collection; however none are recognized by the Opsin Classifier. No transcripts are available, though other molluscs have numerous ests. A German group suggests that the genome sequenced was in fact Lottia scutum.
Under these circumstances, I annotated two Lottia melanopsin in Dec 07, MEL1_lotGig and MEL2_lotGig. Their best match is to other Gq-coupled molluscan opsins, with the first probably an ortholog. Both genes have 3 exons with the two splice positions and phases identical to those of melanopsin (which in vertebrates has numerous other introns). A long run-on carboxy terminus is also seen here. It needs to be established whether these introns are ancestral generic GPCR introns or diagnostic and informative of melanopsins as a gene class. No counterpart to the ciliary opsin of ragworm was immediately apparent.
On 28 Dec 07, I recovered a peropsin, PER_lotGig, very likely orthologous to a peropsin in squid (called retinochrome there) and Aplysia (PER_todPac, PER_aplCal). Extensive structural and experimental evidence is available for squid which likely transfers over, notably the Glu181 counterion proposed ancestral. The Lottia and Aplysia peropsins are intronated identically and by inference the squid. However these differ in some respects from chordate peropsins, suggesting either intron gain or loss or alternately a small 'cloud' of ancient peropsins that were intronated slightly differently in early metazoa.
Lottia is not emerging as a model organism. There are only a handful of studies at PubMed and none on vision. The adult limpet has a pair of eyespots at the base of its cephalic tentacles that likely house a rhabdomeric opsin, perhaps the one annotated here. There may be a second role for paired eyespots in the free-swimming larva for those five days (thoroughly reviewed for chiton trochophores by Arendt and Wittbrodt but not Lottia specifically). Circadian rhythm might involve an additional opsin. The adult is an algal gardener that clears and defends intertidal areas -- raiding limpets are sensed (visually?) and driven off. The opsin sequence found here, stored as MOLL_MEL_lotGig, suggests rapid divergence rather than living fossil character. However patellogastropods such as Lottia with symmetrical non-coiled, conical shells are sometimes taken as ancestral form.
Platyhelminthes: Schmidtea mediterranea (planaria) .. 1 opsin
The common planaria Schmidtea mediterranea has a 865 Mb genome very recently assembled from 17 million traces to 10x and placed in the wgs division of GenBank, after an initial impasse attributable to high AT (69%), repeat content (46%) and high clonal heterozygosity. The genome project is described in a white paper and has a dedicated site SmedDb. It has a strong EST collection as well.
The planarian central nervous system consists of a bilobed brain and two longitudinal ventral nerve tracts connected by commissural neurons. When planarians are decapitated they can completely regenerate a new brain, including new eyes, a boon to opsin research. During head regeneration, new eye spots are formed from precursor cells that differentiate into both cell types in a restricted area of the newly regenerated tissue or blastema. Regeneration of the nervous system is an active research area.
The structure of the planarian eye had already been described by 1915. Eye spots consist of a bipolar nerve cell with photoreceptive rhabdomere accompanied by a cup-shaped structure of pigment cells, as described by Kishida in 1967. In Bdellocephala, each eye consists of 40–50 photoreceptor cells and 6–12 pigmented eyecup cells; in Polycelis, there can be over a hundred similarly structured eyes.
A nearly complete melanopsin (including all introns) can be recovered from raw GenBank data using MEL1_schMan as query sequence. It is nearly identical to fragments from Dugesia (AJ421264) and Girardia (AJ251661). It is stored at the Opsin Classifer as MEL1_schMed and discussed in the Schistostoma section as a likely ortholog. Since the site of expression of a fragmentary transcript is known from hybridization in Girardia and no other Schmidtea opsins are apparent, this is likely the principal photoreceptor both here and in Schistostoma (which however lacks any described eyespots in any life stage).
No counterpart to lophotrochozoan ciliary opsins can be found in the current assembly, indicating (since they could hardly have been invented in Platynereis) their loss in Platyhelminthes is a derived condition. Note though the assembly must be incomplete because the melanopsin is also missing even though represented in traces and transcripts.
>MEL1_schMed Schmidtea mediterranea (planaria) AF112361 AY067648 0 eVYHYLVGVYISIVGISGVLGNLLVLYIFAR 2 1 AKSLRTPPNMFIMSLAIGDLTFSAVNGFPLLTISSFNTRWAWGKL 1 2 TCEIYGFIGGLFGFISINTMALISLDRYFVIAQPFQTMKSLTIKRAIIMLVFVWLYSLIWSTPPFFGY 1 2 GNYVPEGFQTSCTFDYLTQSKGNIIFNIGMYIGNFIIPVGIIIFCYYQIVKAVRVHELEMLKMAQKMNASHPTSMKTG 1 2 AKKADVQAAKISVIIVFLYMLSWTPYAIIALMALTGRRDHLNPYTAELPVLFAKTSAMYNPFIYAINHPKFRIQLEKKFPCLICCCPPKPK 0 0 * 0
Platyhelminthes: Schistosoma mansoni/japonicum (trematode) .. 3 opsins
Blood flukes of the genus Schistosoma are the agents of schistosomiasis, infecting more than 200 million people worldwide, exploiting as intermediate host the fresh water snail Biomphalaria glabrata (which itself has a large EST project). As an endoparasite residing deep inside lungs, hepatoportal circulation, and mesenteric veins, it would not seem a promising species for eyespots or even circadian rhythm opsins. However at least two life stages are affected by light: the hatching of the miracidium from the egg and emergence of cercaria from the snail. These swim upwards to the surface of the water and are also affected by shadows and turbulence.
GPCR proteins are the target of approximately half of all pharmaceuticals. For that reason, a Schistosoma opsin came to be studied in 2001. That gene is expressed in the miracidia and cercaria stages but down-regulated in the adult. Expression is localized to sub-tegumental structures at the front end of cercariae. Full text remains locked behind a commercial firewall 8 years later, as does a 1975 electron microscopy study of putative photoreceptor lamellae in anterior cercariae said to be extensions of modified cilia (no pigment cells mentioned).
However no ciliary opsin occurs in either S. mansoni or S. japonicum genome assemblies or EST collections which must reflect gene loss in view of Platynereis and reconstructed Urbilatera. The anatomical structure described above could not provide directional light sensing and needs to be revisited with immunostaining with all three opsin mRNA. Schistosoma cercaria hang upside-down in the water, occasionally swimming towards surface which requires directionality of photoreception. Emergence from the host snail may also involve timing relative to daylight. Cryptochromes have not been studied.
Version 4.0 of genome is readily available for blast though until 2009 not at GenBank (as with two million of the 3.8 million total traces despite NAID funding). It's unclear whether the extensive collection of 31000 assembled ESTs has been deposited. Proteins and gene synteny are most conveniently studied today by tblastn of the nr, est_other, and wgs (Schistosoma japonicum only) divisions of GenBank. No gene models have been posted as of August 2009.
The July 2009 Schistosoma mansoni genome article reports estimated 13,469 protein-coding genes (only 4% of the genome) of which 92 are GPCR. The genome is the fifth lophotrochoan genome to be sequenced (beyond opsin-less nematodes) but is treated as the first. The article wrongly reports Schistosoma as having 2 opsins (it has 3), contrasting this to a supposed 13 opsins in Drosophila (which has 7) and the 7 opsins of zebrafish (which have 8 cone opsins alone!) and an author-imagined expansion of "peropsins" in Nematostellata (whose genome contains nothing resembling a peropsin).
Four diverged alpha subunits of heterotrimeric G proteins (called transducins by the authors) are reported but not characterized further vis-a-vis the 3 opsins nor signaling chemistry. A GNAO ortholog for Schistosoma mansoni (AF540394) has previously been studied, as have GNAS (M81085) and GNAI counterparts (DQ327708); a GNAQ query using Aplysia (DQ397515) elicits Schistosoma matches of very high percent identity. These are presumably the most relevent to Schistosoma opsin signaling.
Intron structure alone of the published opsin gene (called MEL1_schMan in the opsin classifier) classifies it as a melanopsin. Using this as probe, a second full length paralogous opsin MEL2_schMan is annotatable. While percent identity is moderate at 46%, the intron structure and classifier assignment are identical. MEL3_schMan is similarly intronated but more deeply diverged from other known opsins; it has no counterpart in the less complete Schistosoma japonicum assembly. Otherwise opsin orthologs across these two species are about 86% identical as proteins.
This expansion appears lineage-specific to trematodes and is independent of the melanopsin expansion in arthopods (according to clustering). Under the principle that the most conserved gene of an expansion retains ancestral function, MEL1_schMan is the parental gene:
MEL1_schMan (top group) has higher blastp scores than MEL2 (middle) and MEL3 (bottom) to other known lophotrochozoans: MEL1_schMed Schmidtea mediterranea (planaria) 2.8e-116 MEL1_entDof Enteroctopus dofleini (octupus) Gq X07797 1.6e-88 MEL1_lotGig Lottia gigantea (limpet) FC774055 ests lon 2.7e-88 MEL1_capCap Capitella capitata (polychaete_worm) jgi 1 6.6e-85 MEL1_plaDum Platynereis dumerilii (ragworm) Gq 383 aa 8.4e-85 MEL1_patYes Patinopecten yessoensis (scallop) Gq 92872 3.6e-84 MEL1_todPac Todarodes pacificus (squid) Gq X70498 480 4.2e-83 MEL1_schMed Schmidtea mediterranea (planaria) most lik 4.0e-78 MEL1_entDof Enteroctopus dofleini (octupus) Gq X07797 3.3e-74 MEL1_lotGig Lottia gigantea (limpet) FC774055 ests lon 2.4e-71 MEL1_capCap Capitella capitata (polychaete_worm) jgi 1 1.5e-69 MEL1_patYes Patinopecten yessoensis (scallop) Gq 92872 5.9e-68 MEL1_todPac Todarodes pacificus (squid) Gq X70498 480 1.6e-67 MEL1_sepOff Sepia officinalis (octopus) AF000947 PM 90 2.3e-66 MEL1_plaDum Platynereis dumerilii (ragworm) Gq 383 aa 1.0e-65 MEL1_schMed Schmidtea mediterranea (planaria) most lik 8.2e-71 MEL1_lotGig Lottia gigantea (limpet) FC774055 ests lon 2.9e-66 MEL1_entDof Enteroctopus dofleini (octupus) Gq X07797 1.1e-64 MEL1_patYes Patinopecten yessoensis (scallop) Gq 92872 3.0e-64 MEL1_capCap Capitella capitata (polychaete_worm) jgi 1 7.2e-63 MEL1_sepOff Sepia officinalis (octopus) AF000947 PM 90 2.8e-61 MEL1_todPac Todarodes pacificus (squid) Gq X70498 480 1.2e-60 MEL1_plaDum Platynereis dumerilii (ragworm) Gq 383 aa 5.4e-58
While these opsins could be specialized to developmental stages, the first gene is expressed in both, so the possibility of two or three color non-imaging photoreception needs to be considered. The first opsin MEL1_schMan is most closely related in sequence (at a striking 61% identity) to the sole known opsin (MEL1_schMed) in the planarium Schmidtea so these are likely orthologs whcih might be functionally homologized to a core function, but to no current benefit. It would be feasible to explore synteny as contigs are improved in length; MEL2_schMan may be a fairly recent tandem duplication and MEL3_schMan an earlier. As queries, these proteins turn up closest matches at GenBank EST in other platyhelminthes. Such observations do not support horizonal gene transfer of opsins from the host snail, another lophotrochozoan.
Conservation of intron position and phase can be demonstrated as shown below using blastp alignment to either MEL1_gasAcu of stickleback minnow and MEL1a_braFlo of amphioxus. Here the percent identity is fairly low (39%) but enough patches of good matching exist to reliably anchor the alignment. There is perfect agreement of the first three intron positions and phases.
This is strong evidence for a very deep connection vertical descent of these genes from a common ancestor because these introns are highly specific to melanopsin within the opsin superfamily (ie are not generic GPCR introns as seen from the total mismatch to ciliary and other opsin classes). It is not currently known if any melanopsin has a consistent functional role across all lophotrochozoa (much less all protostomes or bilaterans).
>MEL1_schMan Schistosoma mansoni (trematode_worm) AF155134 11166392 381 aa 6 exons 0 MKQNLTFATLWPDDNDFASIVHSHWHKFIQPDPLYYYLVGIYIGIVGILAVMGNSLVITLFLL 2 1 CKQLRTPPNMLIVSLAISDFSFALINGFPLKTIAAFNHRWGWGKL 1 2 ACELYGFAGSIFGFISLTTMAFIALDRYLVIVQPFETFSRITYGKVIVMIFITWIWSALWSIPPFFGY 1 2 GSYIPEGFHTSCTFDYLSTDLPNLIFNAGLYILGFLCPVFIIIFSYYQIVKTVRLNELELMKMAQSLDLQNPSAMKTg 1 2 GDKKADIEAAKTSIILVLLYLMSWSPYAIVCLMTLIGSRDSLTPFHSELPVLFAKTSAVYNPIVYAVKHPKFRMEIEKRFPFLICCCPPKPK 0 0 ERLQNTIVSKIQVSQIGIGTVSGGNENTLNTVKRED* 0 >MEL2_schMan Schistosoma mansoni (trematode_worm) CD096414 egg 12973350 (46%) 0 MSSNRTIEMLRPYMKDFDSIVLPYWYKFEQPNPYYQYAIGLFIAVVGITGMCLNLLVIVFFTM 2 1 FKSLRTPSNILVVNLAISDFGFSAVIGFPLKTMAAFNNFWPWGKL 1 2 ACDLYGLAGGLFGFVSLSTIAAVALDRYLVIATPFESVFQTTPRRTLLLMLFLWMWSLMWTIPPLFGF 1 2 GRYVTEGYQTSCTMDYISTDLNNRLFNIGLFGFGFLCPLFLSLFCYARIILIVRSRGKDFIEMAASSKGTNQKEKSANV 1 2 SSSKSDTFVSKSSAILLGVYLICWTPYSFVCLMALIGYADYITPLMVEIPCLCAKTANPCIYAFRYPKFRSLLQQRFGFLRLTKNRVSY 0 0 ERSQHAILSTIHVVTDCQYGTVSGGNENTLNTFILLD* 0 >MEL3_schMan Schistosoma mansoni (trematode_worm) Smp_scaff001969 last exon uncertain 0 MSEIKNFTRSLLLYNRTFSMIKNNIHDSDIIMLNHWIKYTQPDPIYNYLVAIFVALIGIFGTITNLLVIFVFL 2 1 TPKSSISLQCALIINLAISDFGFSAVIGFPLKTIAAFNQYWPWGSV 1 2 ACQLYGFISATFGFLSLTTIAAISFDRYLVIVKDHKTTNFRVICTVIGFLWIWSIIWTIPPFFGF 1 2 GRYVLEGYQTSCTFDYISNDMPSLLFSGGMYIFGFMFPVLLCIYCYVNLLKIVRNNERVVLISLSNDGASKQRESVR 1 2 NRKRLDIEATKSVILSLLFYLMSWTPYAMVCLISILGQSYFLTPTIAEMPHIFAKMAAIYNPILYAFTNRKFKNALGIRKTSSVIMQQQRLLSKGQLKPLVSLLFLVN* 0 >MEL1_schJap Schistosoma japonicum (trematode_worm) CABF01049792 87% identical MEL1_schMan 0 MKQNLTFANLWPDDDGFTALIHPHWHKFTQPDPLYYYLVGIYIGIVGILAVMGNSLVITLFVM 2 1 CKQLRTPPNMLIVNLAISDFSFALINGFPLKTIASFNQRWGWgkl 1 2 ACELYGFAGSIFGFISLTTMAFIALDRYLVITQTFETFSHITYEKVIVMIVITWIWSALWSIPPFFGYG 1 2 GSYIPEGFHTSCTFDYLSTDLPNLIFNAGLYILGFLCPVIIIIFSYYQIVRTVRLNELELIKMAQSLNAQNLSVMKTG 1 2 GDKKADIEAAKTSVILVLLYLMSWSPYAIVCLMTLIGSRDSLTPFYSELPVLLAKTSAVYNPIVYAVKHPKFRLEIEKRFPFLICCCPPKPK 0 0 ERLQNTVISKIQASQAGVSAVGIGSENTIGAPKQEN* 0 >MEL2_schJap Schistosoma japonicum (trematode_worm) frag 86% identical MEL2_schMan 0 MHSNRTIESLRPIMKDFDSIVLPYWHKFELPSPYHQYAIGLFIAVVGITGMCLNLLVIVFFTM 2 1 FKALRTPSNILVINLAISDFGFSAVIGLPLKTMAAFNNFWPWGKT 1 2 ACTIYGLGGGLFGFVSLSTIAAIAFDRYLVIATPFESVFQTTPKRTILIMLFLWLWSLIWTIPPIFGF 1 2 GRYVTEGFQTSCTFDYISTDLKNRLFNIGMFGFGFLCPLFLSVFCYARIILIVRSRGKDFIEMAASSKAGNQKDKSANV 1 2 ASTKSDTFVLKSSAILLGVFLLCWTPYAAICLMALIGYADYITTTMVELPCLCAKTAavwdPCIYAFRYPRFRAIFQSRFGMFGKSKVSH 0 0 * 0