Opsin evolution: orgins of opsins
Introduction: the origin of opsins
The origin of the first opsins is intriguing. Opsins are operationally defined here as 7-transmembrane proteins structurally and sequentially homologous to GPCR with (Schiff base) lysine in TM7 in alignment with K296 of bovine rhodopsin (or any established opsin).
This section moves forward in time from the parental gene content of the immediate ancestral genome (greatly facilitated by the new Trichoplax and Monosiga assemblies) that gave rise to the first opsin via gene duplication and neofunctionalization of one copy to photoreception. Subsequent sections work backwards in time, first coalescing separate gene trees of ciliary, melanopsic and other opsins to their respective ur-opsins and ultimately deducing properties of the crown group opsin.
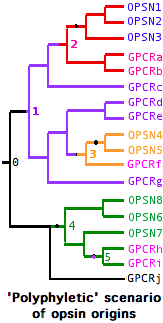
The opsin origination event was not necessarily unique -- GPCR always retain many essential properties via their own evolutionary constraints amd conceivably could have given rise to opsins at widely scattered intervals from rather different parental genes.
In the case of multiple such opsins surviving to the present day, branches will coelesce first to separate parental non-lysine GPCRs, which in turn eventually coelesce -- as all GPCR must do -- to a master parental gene.
In this type of history, the opsin gene tree will not be not 'monophyletic' but instead contain embedded non-opsin GPCR. A gene tree illustrated these concepts is at left. It is generated by Newick string ((((((OPSN1,OPSN2),OPSN3),(GPCRa,GPCRb)),GPCRc),(((GPCRv,GPCRw),((GPCRx,OPSNy),GPCRz)),GPCR6)),(GPCRp,GPCRq));
This scenario -- the molecular version of whether 'vision' arose once vs multiple times -- can be ruled out for bilateran opsins (provided the relevent GPCR outgroups have left descendent genes) but still must be considered seriously in the case of cnidarian opsins and perhaps ctenophores and sponges as well.
Events 600 million years ago may seem hopelessly inaccessible and indeed many uncertainties will remain even after every relevent genome has been sequenced. However sequencing to date has been phylogenetically lopsided with far too little effort expended on early diverging non-model organisms with strategic tree positions. Yet comparative genomics has already provided substantial insights into certain aspects of opsin evolution:
- The first opsins were not associated with gross morphological structures (such as stalked eyes) that could possibly leave a fossil record (as in trilobites) -- key events took place strictly at the molecular subcellular level. Genomes of extant species (some more than others) are not exactly living fossils because the evolutionary accrual of mutations never ceases.
Cases exist of opsins demonstrably obliterated both by gradual pseudogenization and large scale deletions, confusing the record. Yet opsin genes and even their regulatory regions, when compared across the entire metazoan tree, can furnish reliable reconstructions of opsin content and even sequence at ancestral species divergence nodes.
- Opsins are definitely not the 'original' GPCR because these were already widely deployed at much earlier divergence nodes -- yeast, protozoa, choanflagellates, trichoplax have GPCR but lack opsins. Nor are opsins the prototype for the 'rhodopsin class' R of the GRAFS classification of GPCR which again was established far earlier. Indeed, even the Ralpha subgroup with of rhodopsin class GPCR was well-established prior to the first metazoan opsin.
- Opsins are thus latecomers, not pioneers, to a rapidly expanding paralogous gene clade within already full-featured GPCR. Judging by their closest extant blastp relatives among tens of thousands of GPCR at GenBank, opsins specifically arose as a gene duplication within the peptide receptor subgroup PEP. Indeed, certain of these proteins list opsins among their top ten best back-blast matches (ie have better matches than to almost all non-opsin GPCR). Note here that blast scores can be misleading because the 'floor' of percent identity is about 25% just due to universal conserved residues plus accidental matches.
- Note an 'intermediate' GPCR does not exist: either lysine is present at K296 or it isn't. Reconstructing ancestral states from the best contemporary set of GPCR proteins lacking K296 cannot produce a lysine there by any rational methodology. The 20 encoded amino acids can be clustered into subgroups (eg by polarity or bulk) but ultimately form a unorderable discrete set not furnishing continuum transitional states.
- Most likely the parental gene had several introns and the original opsins inherited this pattern (ie the duplication was segmental rather than retroprocessional as in some cnidarian opsins). The history of introns within opsins is already complex and becomes quite problematic within the enveloping GPCR gene family. Opsins (with the exception of a fragmentary sea urchin melanopsin) lack the ubiquitious phase 21 intron breaking the DRY motif arginine.
- Intracellular targeting of early opsins was likely to cytoplasmic or endoplasmic reticulum membranes as isolated monomers, with limited microvillar or especially ciliary specialization (to motile larva) also plausible. These opsins were the first eyes to the world but only in the sense of indicating the intensity (and later directionality) of sunlight striking the cell utilizing already refined GPCR second messenger signal transduction.
- Opsin creation does not imply saltatory evolution because the basics had been established far earlier -- the 7-transmembrane helical structure with fixed topology, the TM1-TM2 salt bridge N55-D83 that could serve as initial counterion, the DRY ionic lock, the NPxxY terminal helix, the conformational shift upon binding of ligand that could trigger signaling, the Galpha protein binding site needed for the signaling cascade, and an arrestin-type mechanism signaling termination. The earliest opsins contained and continued all of these features from the get-go, adapting them over the course of time to various photoreceptive functions.
- Opsins are unique among GPCR in several respects: they catalyze a mild in-situ enxymatic reaction -- cis-trans photoismorization -- that furnishes the signaling agonist. (This reaction also occurs thermally without enzyme but so does carbon dioxide dissolution in water yet humans have 15 carbonic anhydrases). Cis-retinal, being lipid soluble, does not diffuse through the extracellular mileau to reach its receptor binding site as in all other GPCR. Instead it is covalently bound to a lysine deeply internal to TM7, again unprecedented among GPCR (though other internal charged amino acids can occur, notably the D83 glutamate salt bridge and K90 of ultraviolet opsins).
- Opsins did not arise from flavinoid-based cryptochromes, mechanistically different photoreceptors that evolved much earlier to establish circadian rhythm and eventually magneto-sensing. Cryptochromes are homologous to DNA photolyase repair enzymes, not GPCR.
- Although literature searches turn up scattered assertion about 'opsins' in species such as Chlamydemonas ('chlamyopsin' Z48968) and 'volvoxopsin', not to mention bacterial 'rhodopsins', these amount to abusive terminological metaphors, unwelcome additions to an already complex gene family. These proteins do not have seven transmembrane helices in the same arrangement as GPCR nor possess the slightest sequence homology at deeply conserved GPCR residues, so represent independent evolution of photobiology (along the lines of bat and butterfly wings representing independent origins of flying).
- Conceivably forerunners of opsins bound a related chromophore non-covalently, perhaps an all-trans retinoid in the manner of peropsins. Retinoic acid is sometimes proposed as ancestral ligand but retinoic acid receptors (RAR and RXR) are non-GPCR nuclear hormone receptors that bind all trans-RA or 9-cis-RA but not 13-cis-RA. Furthermore, the GPCR receptors inducible by retinoic acid -- RAIG1 proteins (GPRC5C etc) belong elsewhere in the GRAFS classification, have no particular affiliation with opsins and again do not bind retinoids themselves. The fact that pseudo-opsin chromophores are similar retinoids may be coincidence arising from the ubiquity of metabolic carotenoids (availability) and the restricted number of biochemicals (isoprenoids but not amino acids) with tunable adsorption in the visual range (suitability).
- In principle, GPCRs could continue to spawn new clades of opsins from time to time. However, they did not in bilaterans. That is, no gene tree of a bilateran opsin coalesces with a GPCR gene later than the bilateran common ancestor. All bilateran opsins are descended from one of six opsins classes present in the ur-bilateran. Indeed gene tree comprised of all opsins excludes all GPCR, consistent with a unique K296 origination event. However, it remains possible that some cnidarian or ctenophoran opsins arose from a second wing of GPCR with no representative of this opsin surviving in bilaterans.
Two genes in separate species are by definition orthologous only when descended vertically from a single gene in their last common ancestor. It appears that all bilateran opsins -- after accounting for later clade-specific expansions and losses -- are orthologous to either a cilopsin, melanopsin, peropsin, rgropsin, or neuropsin at the bilateran common ancestor. ('Rhabdomeric' protostome opsins do not define a separate class but instead coelesce with vertebrate melanopsins.)
These 5 opsin classes appear not fully coelesced even at the last common ancestor of bilaterans with cnidarians -- while sequence data is woefully limited today in early taxa, it seems both melanopsins and cilopsins classes existed in this ancestor, perhaps in addition other opsin classes no longer represented in bilaterans. Conversely, peropsins have been retained in lophotrochozoan, ecdysozoan, and deuterstome lineages but not in any cnidarian sequence to date. Neuropsins survived solely in chordates, whereas rgropsins are even more restricted to vertebrates, even though they could not have originated there. These latter genes are conceptual analogs of cnidarian-only opsin classes.
All opsins are homologous so any given pair is ultimately orthologous at some earlier common ancestor -- but which one? The species tree itself is confused here on sistering vs independent nodes at cnidarian/ctenophore. The single ctenophore opsin available -- regretably just a distal fragment -- is difficult to classify. The fact that its best blast matches cluster about equally well with melanopsins and cilopsins (to the exclusion of other bilateran classes) suggests that their merger is not far off.
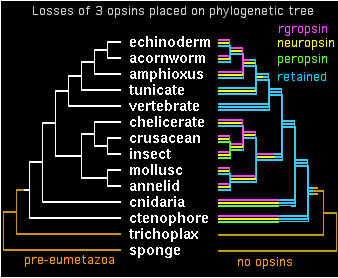
The opsin gene tree can largely be worked out and coordinated with species tree divergences. Despite many efforts at this, some deeper topology remains problematic. It appears from sequence clustering, indel analysis, and especially intron conservation that ((peropsin, rgropsin),neuropsin) is a valid subgroup. Further, this assemblage associates more closely with cilopsins, leaving a final topology to be superimposed on the phylogenetic tree:
gene tree ((((cilopsin,((peropsin,rgropsin),neuropsin)),melanopsin),cnidopsin),GPCRpep); species tree (((((((((echinoderm,acornworm),amphioxus),tunicate),vertebrate),((chelicerate,(crusacean,insect)),(mollusc,annelid))),cnidaria),ctenophore),trichoplax),sponge);
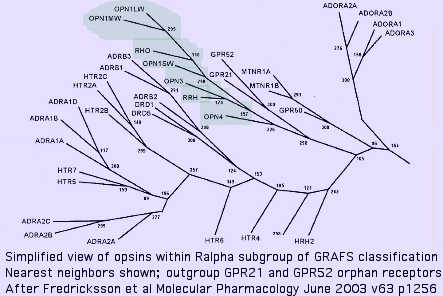
Nearest neighbors of opsins among GPCR
The immediate outgroup of opsins lie among a vast number GPCR receptors. The set of 29 non-opsin GPCRs in the reference gene collection was constructed by taking best-blasts separately for each opsin class to all human GPCR (because these have the best prospects for determined ligand), then collating the lists, winnowing out of repeated entries and too-recent gene family expansions. TACR2 (tachykinin receptor) and SSTR1 (somatostatin receptor) are the best single representatives.
To these are added the 3 non-opsin GPCR with determined 3D structures, as well as the closest two non-opsins in an early diverging eukaryote (Trichoplax), and a few nearest neighbors claimed in a May 2009 update of the GRAFS classification tree. Finally, a few human GPCR arising as best-blast of cnidarian and ctenophore opsins are included to complete the mix.
None of these GPCR represent the actually parental gene to opsin (even if they are directly descended from it) because they have themselves evolved forward some 600 million years from the putative opsin creation event. The consensus line perhaps represents a better approximation to the desired ancestral sequence. It is difficult to reconstruct an ancesteral sequence accurately because opsins residues widely separated in primary sequence co-evolve, creating algorithmic errors in methods that neglect this.
The aligned sequences below have been trimmed at both ends to the earliest indications of conservation. Highly conserved residues are shown in red and less conserved residues in blue. Observe the Schiff base lysine (position -16 relative to the FR end of TM7) does not occur in these GPCR which lie outside of opsins.
Many conserved patches in these GPCR are highly similar to those of opsins, implying those residues have no utility in distinguishing opsins from non-opsins. Not being diagnostic for opsins, they cannot be used to determine whether gene fragments (not covering the Schiff lysine) are indeed opsins. These shared conserved residues describe commonalities needed for generic GPCR structure and signaling so cannot define photobiological specializations. Departures in certain opsin or GPCR classes may indicate they are constituitively signaling or no longer signaling.
The expansion of the Ralpha class within GRAFS had already taken place by early-diverging metazoan such as Monosiga and Trichoplax, species which do not contain opsins, implying the ancestral metazoan lacked them as well. The orphan receptors GPR21 and GPR52 form the immediate outgroup (within the 800 human GPCR) in anhttp://genomewiki.ucsc.edu/index.php?title=Opsin_evolution:_orgins_of_opsins&action=submit oft-cited [http://www.ncbi.nlm.nih.gov/pubmed/12761335,15862553,17428229 2003 study and its sequels. These GPCR have isoleucine at K296; their ligands still remain unknown as of Dec 2009. Conservation is high throughout deuterostomes; blast matches within GenBank nr are restricted (within opsins) to molluscan melanopsins, suggesting Gq signaling.
The melatonin receptor MLTNR1A emerges as a close relative to opsins. Curiously it plays a key role in circadian rhythms and so could coordinate with an opsin photosensors (ie one arising from a gene duplication and divergence). N-acetyl-5-methoxytryptamine, the ligand, bears no obvious relationship to cis-retinal however and K296 is lacking, not decisively supporting a parent gene relationship. Here the (2,6,6-trimethylcyclohexen-1-yl) ring moiety of retinal only superficially resembles the fused ring of melatonin.
Closeness in the GRAFS tree does not fully accord with closeness of opsin blastp match, suggesting (unsurprisingly) that its topology is slightly wrong at some internal nodes. On average rank in blastp top scores (or by average 5 best blast expectation values), as representatives of all opsin classes are aligned with the GPCR below, the highest scoring ones by far are are the Trichoplax opsins followed by various peptide receptors:
Rank Gene Exp Exons Receptor Ligand 4.2 UROPS2_triAd e-29 2 orphan histamine? (HRH2: best non-opsin blast human) 5.4 UROPS1_triAd e-28 1 orphan peptide? (SSTR1: best non-opsin blast human) 5.6 SSTR1_homSap e-26 1 somatostatin peptide 7.2 TACR2_homSap e-25 5 tachykinin peptide 8.1 GALR1_homSap e-24 3 galanin peptide 8.9 MTNR1A_homSa e-23 2 melatonin N-acetyl-5-methoxytryptamine
Trichoplax has two very curious 7-transmembrane protein that emerges as its best genomic matches to opsin queries. While lacking K296 for a Schiff base, their best back-blast to GenBank nr returns almost entirely opsins (rather than other GPCR receptors). While Trichoplax is 600+ million years removed from a common ancestor, this gene could still offer clues about the immediate GPCR ancestor to opsins. It is not plausibly descended from an opsin gene expansion followed by loss of K296 because Trichoplax ancestors all lack opsins.
In summary, the parent GPCR gene for opsins can be localized to the PEP subgroup of R class GPCR within GRAFS but no particular gene there stands out as a pre-opsin. The time span is immense and the gene class has experienced much churning through expansion and contraction cycles.
Another clue to the origin of opsins might be provided by comparing slowly evolving GPCR intron positions and phases to reconstructed ancient introns in opsins. Many non-olfactory GPCR with sequence similarity to opsins have either no introns or just one, suggesting the genes duplication by retroprocessing followed by later intron acquisition at non-historic position. UROPS2 of trichoplax has one intron but it does not correspond to any in opsins. Cnidarian opsins to date have been either intronless (Nematostellata) or not determined (known only from processed transcripts).
Origin of contemporary opsin classes
Traceback of opsins can begin by selecting certain 'index sequences'. It ultimately does not matter which or how many, but for historical reasons bovine rhodopsin, frog melanopsin, human peropsin, mouse neuropsin and so forth might be used.
Each index sequence is then built out to a larger class of orthologs in nearby species using flanking gene synteny to confirm best-blast. Lineage-specific gene duplications with close affinities (eg from recent clade-specific paralogous expansions such as teleost fish whole genome duplications) are added. Eventually the set collides with an expanding set of another index sequence and all bilateran opsin sequences are uniquely placed in one of five clusters.
Ciliary opsins (generated from RHO1) forms a cohesive gene clade that does not coalesce with melanopsins, peropsins, neuropsins, or rgropsins within vertebrates, deuterostomes, or even bilatera. The index gene picks up rod and cone imaging opsins, pinopsin, parapinopsin, parietopsin, very ancient opsin, encephalopsin, teleost multiple tissue, and certain ciliary opsins from protostomes.
Hardly a vertebrate innovation, include early deuterostomes lacking imaging eyes, both branches of protostomes (initially bee and ragworm), pre-bilateran cnidarians and possibly ctenophores. Sponges are still uncertain (because of a 5 year wait on the assembly) but the very earliest metazoan genomes (Monosiga and Trichoplax) definitely lack ciliary and (all other) opsins. If those genomes are representative, then ciliary opsins emerged on the post-Trichoplax stem. Certain cnidarian opsins -- but not all -- already exhibit some sequence specializations of ciliary opsins.
Ciliary opsins have been totally lost on numerous occasions in numerous lineages, notably 'model' organisms like drosophila and worse, nematodes, which have lost all types of opsins. Hemichordates and non-annelid lophotrochozoans lost ciliary opsins independently. Other explanations (such as multiple re-emergences of ciliary-like opsins) are manifestly impossible.
The earliest deuterostome ciliary ur-opsin is best represented by the TMT class of opsins, in particular by the TMT1 subgroup that has retained important ancestral characteristics in the diagnostic TM2 region. Sequential expansion of TMT1 gave rise to all the other ciliary opsins found in vertebrates, including all rod and cone opsins. This fundamental gene, though retained through ampibian and amniote, curiously was eventually lost in birds and mammals. Transcripts are often annotated as testis libraries suggesting a function in gamete release timing. Its immediate descendent gene TMT2, whose subfunctionalization is unknown, is retained in monotremes and marsupials but lost in all placentals. The best experimental organism for studying TMT1 is probably Xenopus.
Melanopsins, discovered in 1998 in frog lateral line dermal melanophores (as well as hypothalamus, iris, and retinal horizontal cells) form another ancient opsin class. Melanopsins include rhabdomeric arthropod opsins (which have an unnecessary dual nomenclature -- they're melapsins by multiple independent criteria) and lophotrochozoan melanopsins (which other than scallop, squid and octopus sit undocumented within genome projects). One cnidarian opsin from coral classifies as a melanopsin yet closely shares other properties with cnidarian opsins that don't.
Peropsins are a third major class of opsins in the sense of broad but not universal retention.. They are expanded in deuterostomes but rarely occur in arthropods yet are of major importance in lophotrochozoa. They are the only opsin class retained in hemichordates. There is nothing resembling them in cnidaria though that could very well reflect gene loss in the two genomes available as no coalescence with other opsin classes is evident.
Neuropsins are a much expanded but little studied group of opsins restricted to living deuterostomes though they did not originate there (unless divergence from another opsin class was exceeding abrupt and then immensely slowed). The neuropsin expansion to 4 genes in the lamprey stem continued unchanged to the amniote ancestor but subsequently contracted to 2 in monotremes and only 1 in marsupials and placentals.
Rgropsins are another little-studied group that is represented today only on from the tunicate-vertebrate last common ancestor. Again these opsins must have originated far earlier in pre-bilaterans because their ancestral reconstructed sequence is still far from coelescence with other ancestral opsin classes.
Rgropsins and neuropsins are conceivably retained in other bilatera but diverged to the point of unrecognizability. This scenario has to be discarded because analysis of complete genomes is adequately sensitive to locate all K296-containing GPCR. This reasoning is applicable to peropsins as well -- they have been definitely lost in all insect and molluscan genomes as well as the chelicerate Ixodes though fortunately retained in arachnids. This illustrates the incredible interpretive importance of a taxonomically broad sequencing program.
Peropsin, neuropsins and rgropsins are unified by their intronation, sharing 3 ancestral introns despite numerous differences. This indicate -- given the slow rate of intron gain and loss in most deuterostomes -- that they share deep roots in pre-bilateran, implying near total loss of neuropsins and rgropsins in invertebrates.. None of these introns are shared with cilopsins or melanopsins or for that matter known GPCR.
In hindsight, large scale loss of opsin classes should not come as a surprise -- humans lost 12 of 20 opsin loci present in the amniote ancestor. This is characteristic of GPCR evolution as well: collapse or near-collapse of a large gene clade, followed by later massive expansion but retention only in scattered lineages. This can result in two species have similar number of GPCR genes but very poor correspondences among them. This is a very different history from genes such as ribosomal proteins or catabolic enzymes (eg homogenistate oxygenase) that are retained in all species as single copies. Other genes like globins exhibit moderate expansion to several copies accompanying a trend to organismal specializational complexity with little evidence of contraction cycles.
Once over this conceptual hurdle, these cycles of expansion and contraction can be repeatedly invoked on various branches of the phylogenetic tree to explain many aspects of opsin classification. Vertebrates have a remarkable history of largely terminal expansion in the lamprey stem, followed by retention in most lineages, clade-specific expansions in teleost fish, and major attrition in mammals but with some recovery in cone genes in primates.
Cnidopsins are a taxonomically based collection of opsins that largely do not classify unambiguously within bilateran opsins. Much more intensive sampling is needed here because neither of the two genome projects to date was favorable for opsins. Ctenophores currently have a single unexpected opsin gene obtained as accidental byproduct of another project -- obviously much greater sequencing effort is needed given their currently basal position of opsin-containing species.