PRDM9: meiosis and recombination: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 65: | Line 65: | ||
=== Online References === | === Online References === | ||
Open [http://www.ncbi.nlm.nih.gov/pubmed/21334090,21388701,21159001,21343182,20044539,20818382,21170346,20150474,20334833,20981099,20877321,20044541,20210982,20044538,20948797,20961408,19168450,19074312,19997497,20041164,19150836,18941885,17916234,17032681,16582607,10656784,18325330,20961408,20346131,20478078,19261184,19175294 | Open [http://www.ncbi.nlm.nih.gov/pubmed/21334090,21388701,21159001,21343182,20044539,20818382,21170346,20150474,20334833,20981099,20877321,20044541,20210982,20044538,20948797,20961408,19168450,19074312,19997497,20041164,19150836,18941885,17916234,17032681,16582607,10656784,18325330,20961408,20346131,20478078,19261184,19175294 | ||
[http://www.ncbi.nlm.nih.gov/pubmed/10656784 10656784 .......... 2000] Laity DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol Biol. 2000 Jan 28;295(4):719-27. | |||
[http://www.genetics.iastate.edu/Phillips_Dernburg_2006.pdf 17141157 .......... 2006] Phillips A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis. Dev Cell. 2006 Dec;11(6):817-29. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/18286240 18286240 .......... 2008] Brayer The protein-binding potential of C2H2 zinc finger domains. Cell Biochem Biophys. 2008;51(1):9-19. | |||
[http://www.sciencemag.org/content/323/5912/350.full?rss=1 19150836 .......... 2009] Willis Origin of species in overdrive. Science. 2009 Jan 16;323(5912):350-1. | |||
[http://www.sciencemag.org/content/327/5967/791.full?rss=1 20150474 .......... 2010] Cheung Genetic control of hotspots. Science. 2010 Feb 12;327(5967):791-2. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/20334833 20334833 .......... 2010] Hochwagen Meiosis: a PRDM9 guide to the hotspots of recombination. Curr Biol. 2010 Mar 23;20(6):R271-4. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/20478078 20478078 .......... 2010] Klug The discovery of zinc fingers and practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010 Feb;43(1):1-21. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/20818382 20818382 .......... 2010] Berg PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010 Oct;42(10):859-63. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/20877321 20877321 .......... 2010] McVean PRDM9 marks the spot. Nat Genet. 2010 Oct;42(10):821-2. | |||
[http://eebweb.arizona.edu/nachman/Suggested%20Papers/Lab%20papers%20fall%202010/Kong_et_al_2010.pdf20981099 20981099 .......... 2010] Kong Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010 Oct 28;467(7319):1099-103. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/21334090 21334090 .......... 2011] Neaves Unisexual reproduction among vertebrates. Trends Genet. 2011 Mar;27(3):81-8. | |||
[http://www.ncbi.nlm.nih.gov/pubmed/21388701 21388701 .......... 2011] Ponting What are the genomic drivers of the rapid evolution of PRDM9? Trends Genetics (2011) 1–7 | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1817654 17352536 PMC1817654 2007] Coop Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007 Mar 9;3(3):e35. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2082429 17916234 PMC2082429 2007] Fumasoni Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007 Oct 4;7:187. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2346554 18464896 PMC2346554 2008] Duret The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008 May 9;4(5):e1000071. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2593767 18941885 PMC2593767 2008] Miyamoto Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 2008 Nov-Dec;25(11-12):553-7. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2631073 19175294 PMC2631073 2009] Berglund Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009 Jan 27;7(1):e26. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2667407 19261184 PMC2667407 2009] Thomas Evolution of C2H2-zinc finger genes revisited. BMC Evol Biol. 2009 Mar 4;9:51. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2779102 19997497 PMC2779102 2009] Oliver Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009 Dec;5(12):e1000753. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2794550 20041164 PMC2794550 2009] Thomas Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One. 2009 Dec 30;4(12):e8505. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2821451 20044538 PMC2821451 2010] Parvanov Prdm9 controls activation of mammalian recombination hotspots. Science. 2010 Feb 12;327(5967):835. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2865497 20346131 PMC2865497 2010] Lorenz The ancient mammalian KRAB zinc finger gene cluster on human chromosome 8q24.3 BMC Genomics. 2010 Mar 26;11:206. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2872867 20210982 PMC2872867 2010] Neale PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010;11(2):104. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2950028 20948797 PMC2950028 2010] Sandovici PRDM9 sticks its zinc fingers into recombination hotspots and between species. F1000 Biol Rep. 2010 May 24;2. | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2999565 21170346 PMC2999565 2010] Billings Patterns of recombination activity on mouse chromosome 11 revealed by high resolution mapping. PLoS One. 2010 Dec 8;5(12):e15340. | |||
[http://bioinformatics.oxfordjournals.org/content/22/23/2841.long 17032681 .......... 2006] Birtle Meisetz and the birth of the KRAB motif. Bioinformatics. 2006 Dec 1;22(23):2841-5. | |||
[http://www.landesbioscience.com/journals/cc/hayashiCC5-6.pdf 16582607 .......... 2006] Hayashi Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell Cycle. 2006 Mar;5(6):615-20. | |||
[http://bioinfo.hanyang.ac.kr/ZIFIBI/frameset.php 18325330 .......... 2008] Cho Prediction of DNA binding sites for zinc finger proteins. BBRC 2008 May 9;369(3):845-8. | |||
[http://www.andrologyjournal.org/cgi/content/full/30/4/426 19168450 .......... 2009] Irie Single-nucleotide polymorphisms of the PRDM9 (MEISETZ) gene in patients with nonobstructive azoospermia. J Androl. 2009 Jul-Aug;30(4):426-3 1. | |||
[http://www.sciencemag.org/content/323/5912/373.full 19074312 .......... 2009] Mihola A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009 Jan 16;323(5912):373-5. | |||
[http://www.med.upenn.edu/timm/documents/Highlyefficientendogenous.pdf 15806097 .......... 2010] Urnov Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005 Jun 2;435(7042):646-51. | |||
[http://genomebiology.com/2010/11/10/R103 20961408 .......... 2010] Zheng Detecting sequence polymorphisms associated with meiotic recombination hotspots in the human genome. Genome Biol. 2010;11(10):R103. | |||
[http://www.sciencemag.org/content/327/5967/836.full 20044539 .......... 2010] Baudat PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010 Feb 12;327(5967):836-40. | |||
[http://www.sciencemag.org/content/327/5967/876.full 20044541 .......... 2010] Myers Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010 Feb 12;327(5967):876-9. | |||
[http://nar.oxfordjournals.org/content/early/2011/02/21/nar.gkr048.full 21343182 .......... 2011] Yanover Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2 zinc fingers. Nucleic Acids Res. 2011 Feb 22 | |||
[http://www.tiem.utk.edu/~fubeda/papers/U&W11JEB.pdf 21159001 .......... 2011] Ubeda Red Queen theory of recombination hotspots. J Evol Biol. 2011 Mar;24(3):541-53. | |||
=== Curated reference sequences === | === Curated reference sequences === | ||
Revision as of 22:41, 15 March 2011
Introduction
(to be continued)
Comparative genomics of PRDM9 and PRDM7
The human PRDM9 sequence below is annotated in color for domains relative to its exon breaks. The protein can be best understood in terms of its concatenated domains, not all of which may be present in antecedent and descendant homologs. The first two domains KRAB and SSXRD (together called the PR domain) interact with transcription factors. The SET domain (and some flanking sequence) form an enzyme, which uses S-adenosyl methionine to place the third methyl group on a specific lysine of histone H3(K4). The C2H2 domains -- so named for two cysteines and two histidines binding a structural zinc ion -- each recognize a specific trinucleotide and so together provide specific binding sites along the genome, though ambiguity and synergistic effects between adjacent units make it difficult to read out these sites precisely.
The repetitive C2H2 domains, necessarily similar at the dna level, are prone to replication slippage. This can give rise to point mutations as well as causing the number of repeats to form a distribution rather than to be a fixed number. Many other unrelated genes with internal repeats (such as the octapeptide region of the prion gene PRNP) are also affected by replication slippage. These regions are readily identified genomewide by mRNA dot plots.
The C2H2 domains always reside in a long distinctive terminal exon of splicing phase 2 that has been shuffled over mammalian evolutionary time into various contexts. Concepts such as paralogy and orthology need piecewise definitions in composite proteins. Synteny plays an important role in deconstructing events in specific lineages. Here the unrelated single-copy conserved gene GAS8 plays an important role. PRDM7 occurs immediately distal to it on the negative strand (the genes are convergently transcribed). PRDM7 is otherwise the last gene on the q arm of its chromosome telomere which may predispose it to copy number dispersal events. PRDM9 is not consistently located within placental mammals, suggesting independent relocation events.
Both PRDM9 and PRDM7 contain an early C2H2 domain noted in SwissProt annotations and readily found by the online PFAM tool irregardless of species. This domain conserves the four critical residues needed for zinc binding (and so locally that structure) but lacks the terminal cap TGEKP which otherwise serves to lock down a C2H2 zinc finger after it has scanned along genomic dna to an appropriate trinucleotide. The function of this early domain and the following 112 residues are otherwise unknown -- no relevant 3D structure has ever been determined.
The first C2H2 of the repeat region is proximally degenerate, beginning in VKY in all species. The tyrosine probably cannot serve in place of the usual cysteine in terms of zinc binding though the other three needed residues are present. This domain ends in a normal cap region except in humans where the conserved helix-ending proline has been replaced with leucine TGEKL (in the reference human genome) with unknown functional consequences.
>PRDM9_homSap Homo sapiens (human) Q9NQV7 10 exons chr5:23,509,579 span 18,301 bp KRAB SSXRD SET C2H2 cap
0 MSPEKSQEESPEEDTERTERKPM 0
0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1
2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1
2 VKPPWMALRVEQRKHQK 0
0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1
2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1
2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0
0 ITKGRNCYEYVDGKDKSWANWMR 2
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1
2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK
VKYGECGQGFSVKSDVITHQRTHredTGEKL
YVCRECGRGFSWKSHLLIHQRIHTGEKP
YVCRECGRGFSWQSVLLTHQRTHTGEKP
YVCRECGRGFSRQSVLLTHQRRHTGEKP
YVCRECGRGFSRQSVLLTHQRRHTGEKP
YVCRECGRGFSWQSVLLTHQRTHTGEKP
YVCRECGRGFSWQSVLLTHQRTHTGEKP
YVCRECGRGFSNKSHLLRHQRTHTGEKP
YVCRECGRGFRDKSHLLRHQRTHTGEKP
YVCRECGRGFRDKSNLLSHQRTHTGEKP
YVCRECGRGFSNKSHLLRHQRTHTGEKP
YVCRECGRGFRNKSHLLRHQRTHTGEKP
YVCRECGRGFSDRSSLCYHQRTHTGEKP YVCREDE* 0
-1 23 6 traditional numbering of dna recognizing amino acids
HPCPSCCLAFSSQKFLSQHVERNH alignment of early C2H2 domain
* * * * zinc ligating residues
Gene duplication, loss and syntenic dispersion
(to be continued)
Structural considerations in C2H2 zinc fingers
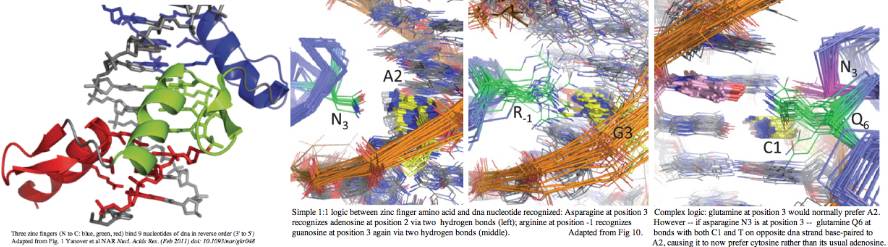
High resolution structures of C2H2 zinc finger domains have been available for decades. As the name suggests, the divalent zinc atom locks the two cysteines and two histidines into a rigid geometry providing a core conformation that a small peptide of 28 residues could not otherwise stably assume. Note in the unbound state, finger tips must retain flexibility while the domain ensemble scans its genome for specific dna sequences appropriate to its function. Each finger binds a trinucleotide -- in effect making a zinc finger the protein counterpart to tRNA anticodon. However overall binding is not a simple read-off code because adjacent fingers alter each other's specificities in subtle ways.
The linker region TGEKP plays a key role when the correct DNA sequence is encountered, snap-locking its finger down onto its target by capping the C-terminus of its alpha helix. A hydrogen bond between the first threonine and middle glutamate is key to this binding-induced conformational shift. From comparative genomics, it appears that a serine in first position can also form this hydrogen bond. The role of the glycine is to stay out of the way; the lysine counterbalances the negative charge of the glutamate; the proline terminates any helical propensity, allowing a fresh start in the adjacent finger.
While this motif is immensely conserved within C2H2 zinc finger of PDRM9 homologs, exceptions do occur. It is important to understand these because these loss of dna lock-down could loosen or even eliminate trinucleotide binding specificity. Such steps might represent initial stages of pseudogenization. However many exceptions occur within the first or last fingers. It is also common for fragmentary and imperfect motifs to end the protein, sometimes continuing on in another reading frame past the current stop codon.
Note in aligning zinc finger motifs, the breaks should always be put at the end of the linker region. It is completely illogical to break at the first cysteine as some authors do because capping by the linker region is specific to its zinc finger, not the following one.
Predicting dna binding sites of zinc finger domains
Online References
10656784 .......... 2000 Laity DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol Biol. 2000 Jan 28;295(4):719-27. 17141157 .......... 2006 Phillips A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis. Dev Cell. 2006 Dec;11(6):817-29. 18286240 .......... 2008 Brayer The protein-binding potential of C2H2 zinc finger domains. Cell Biochem Biophys. 2008;51(1):9-19. 19150836 .......... 2009 Willis Origin of species in overdrive. Science. 2009 Jan 16;323(5912):350-1. 20150474 .......... 2010 Cheung Genetic control of hotspots. Science. 2010 Feb 12;327(5967):791-2. 20334833 .......... 2010 Hochwagen Meiosis: a PRDM9 guide to the hotspots of recombination. Curr Biol. 2010 Mar 23;20(6):R271-4. 20478078 .......... 2010 Klug The discovery of zinc fingers and practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010 Feb;43(1):1-21. 20818382 .......... 2010 Berg PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010 Oct;42(10):859-63. 20877321 .......... 2010 McVean PRDM9 marks the spot. Nat Genet. 2010 Oct;42(10):821-2. 20981099 .......... 2010 Kong Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010 Oct 28;467(7319):1099-103. 21334090 .......... 2011 Neaves Unisexual reproduction among vertebrates. Trends Genet. 2011 Mar;27(3):81-8. 21388701 .......... 2011 Ponting What are the genomic drivers of the rapid evolution of PRDM9? Trends Genetics (2011) 1–7 17352536 PMC1817654 2007 Coop Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007 Mar 9;3(3):e35. 17916234 PMC2082429 2007 Fumasoni Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007 Oct 4;7:187. 18464896 PMC2346554 2008 Duret The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008 May 9;4(5):e1000071. 18941885 PMC2593767 2008 Miyamoto Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 2008 Nov-Dec;25(11-12):553-7. 19175294 PMC2631073 2009 Berglund Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009 Jan 27;7(1):e26. 19261184 PMC2667407 2009 Thomas Evolution of C2H2-zinc finger genes revisited. BMC Evol Biol. 2009 Mar 4;9:51. 19997497 PMC2779102 2009 Oliver Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009 Dec;5(12):e1000753. 20041164 PMC2794550 2009 Thomas Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One. 2009 Dec 30;4(12):e8505. 20044538 PMC2821451 2010 Parvanov Prdm9 controls activation of mammalian recombination hotspots. Science. 2010 Feb 12;327(5967):835. 20346131 PMC2865497 2010 Lorenz The ancient mammalian KRAB zinc finger gene cluster on human chromosome 8q24.3 BMC Genomics. 2010 Mar 26;11:206. 20210982 PMC2872867 2010 Neale PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010;11(2):104. 20948797 PMC2950028 2010 Sandovici PRDM9 sticks its zinc fingers into recombination hotspots and between species. F1000 Biol Rep. 2010 May 24;2. 21170346 PMC2999565 2010 Billings Patterns of recombination activity on mouse chromosome 11 revealed by high resolution mapping. PLoS One. 2010 Dec 8;5(12):e15340. 17032681 .......... 2006 Birtle Meisetz and the birth of the KRAB motif. Bioinformatics. 2006 Dec 1;22(23):2841-5. 16582607 .......... 2006 Hayashi Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell Cycle. 2006 Mar;5(6):615-20. 18325330 .......... 2008 Cho Prediction of DNA binding sites for zinc finger proteins. BBRC 2008 May 9;369(3):845-8. 19168450 .......... 2009 Irie Single-nucleotide polymorphisms of the PRDM9 (MEISETZ) gene in patients with nonobstructive azoospermia. J Androl. 2009 Jul-Aug;30(4):426-3 1. 19074312 .......... 2009 Mihola A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009 Jan 16;323(5912):373-5. 15806097 .......... 2010 Urnov Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005 Jun 2;435(7042):646-51. 20961408 .......... 2010 Zheng Detecting sequence polymorphisms associated with meiotic recombination hotspots in the human genome. Genome Biol. 2010;11(10):R103. 20044539 .......... 2010 Baudat PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010 Feb 12;327(5967):836-40. 20044541 .......... 2010 Myers Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010 Feb 12;327(5967):876-9. 21343182 .......... 2011 Yanover Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2 zinc fingers. Nucleic Acids Res. 2011 Feb 22 21159001 .......... 2011 Ubeda Red Queen theory of recombination hotspots. J Evol Biol. 2011 Mar;24(3):541-53.
Curated reference sequences
The sequences below have been compiled from genome projects -- only rarely do validating transcripts exist at GenBank. Sequences with a single frameshift or other glitch have been edited to allow full length proteins on the theory that the error either reflects an aberrant atypical individual chosen for sequencing or simple error in low coverage projects within a difficult repeat region. However such sequences may instead reflect early stages of pseudogenization. Many sequences are in fact clearly pseudogenes; here recognizable exons have been collected to allow rough dating of loss of function.
In the case of more intensively studied species such as human and mouse, the number of C2H2 repeats varies widely. Only the most common representative is shown here. This variation likely occurs in all species but the individual animal chosen for sequencing may or may not be typical. Many clades have distinctive patterns of gene amplification and gene loss, making both orthologous and functional comparisons problematic.
Other useful sequences such as the GAS8 synteny neighbor, other zinc finger quasi-homologs having similar exon and domain structures, and bogus orthologs outside of mammals are also included for reference purposes.
>PRDM9_homSap Homo sapiens (human) Q9NQV7 10 exons chr5:23,509,579 size 18,301 bp KRAB SSXRD SET C2H2 0 MSPEKSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKL YVCRECGRGFSWKSHLLIHQRIHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRDKSHLLRHQRTHTGEKP YVCRECGRGFRDKSNLLSHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRNKSHLLRHQRTHTGEKP YVCRECGRGFSDRSSLCYHQRTHTGEKP YVCREDE* 0 >PRDM9_homNea Homo sapiens (neanderthal) variants R HDL S R 0 MSPEKSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 1 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 1 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 2 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKL YVCRECGRGFSWKSHLLIHQRIHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRDKSHLLRHQRTHTGEKP YVCRECGRGFRDKSNLLSHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRNKSHLLRHQRTHTGEKP YVCRECGRGFSDRSSLCYHQRTHTGEKP >PRDM9_panTro Pan troglodytes (chimp) frag assembly glitch *VCREDE* 0 MSPERSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPLMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELKKKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYKGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPRSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTAKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKP YVCRECGRGFSWKSHLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHRTTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSQQSNLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSKQSHLLSHQRTHTGEKP YVCRECGRGFSVQSNLLSHQRTHTGEKL YVCRECGRGFSQQSHLLRHQRTHTGEKP YVCR LLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSKQSHLLSHQRTHTGEKP YVCRECGRGFSQQSHLLSHQRTHTGEKP YVCRECGRGFSQQSHLLRHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECERGFSQQSHLLRHQRTHTGEKP YVCRECGRGFSRQSALLIHQRTHTGEKP* 0 >PDRM9_ponAbe Pongo abelii (orangutan) GEKPYVCRECGRGFSVKSNLLSHQRTHTEEKLYVCREDE* 0 MSPERSQEESPEDDTERTERKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 0 0 GMPKASFNNESSLKELSETANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRSKETEGNTYSLRERKGHAYKEISEPQDDDYL 1 2 CEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALTLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITKDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNHEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKP YVCRECGRGFSRQSVLLIHQRTHTGEKP YVCRECGRGFSRRSVLLIHQRTHTGEKP YVCRECGRGFSQQSVLLIHQRTHTGEKP YVCRECGRGFSRRSVLLIHQRTHTGEKP YVCRECGRGFSWKSVLLRHQRTHTGEKP YVCRECGRGFSQQSVVFIHQRTHTGEKP YVCRECGRGFSGKSVLFRHQRTHTGEKP YVCRECGRGFSDKSGVCYHQRTHTRGEA LCLQGVWAGL* 0 >PRDM9_macMul Macaca mulatta (rhesus) 0 MSPERSQEESPEEDTERTERKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAVRVEQSKHQK 0 0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITQDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSTQNFPGPSARRLFQPENLCSGDQNQEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWPKEISRAFSSPPKGQMGSSRVGERMMEEEYRTGQKVNPENTGKLFVGVGISRIAK VKYGECGQGFSDKSDVIIHQRTHTGEKP YLCRECGRGFSQKSSLRRHQRTHTGEKP YLCRECGRGFRDNSSLRYHQRTHTGEKP YLCRECGRGFSNNSGLCYHQRTHTGEKP YLCRECGRGFSDNSSLHRHQRTHTGEKP YLCRECGRGFSNNSGLRYHQRTHTGEKP YLCRECGRGFSNNSGLRHHQRTHTGEKP YLCRECGRGFSQKANLLRHQRTHTGEKP YLCRECGRGFSQKADLLSHQRTHTGEKP* >PRDM9_calJac Callithrix jacchus (marmoset) one frameshift in repeat area chr20 terminus 0 MSPERSQEESPEGDTGRTEQKPM 0 0 VKDAFKDISMYFSKEEWAEMGDWEKTRYRNMKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPGMAFRVGQSKHQK 0 0 GMPKASFGNESSLKKLSGTANVLNTSGPEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRRKDTEEKMYSLRERKGLAYKEVSEPQDDDYL 1 2 yCEICQNFFIDSCAAHGPPTFVKDSAVDKGHPNHAALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRVTEDEEAASSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 ESKPEIHPCPSCCLAFSSQKFLSHHVERNHSSQNFPGTSTRKLLQPENPCPGKQKEEQQYFDPCNSNDKTKGQETKERSKLLNIRTWQREMARAFSNPPKGQMGSSRVEERMMEEESRTGQKVNPVDTGKLFVGVGISRIAK AKYGECGQGFSDMSDVTGHQRTHTGEKP YVCRECGRGFSQKSALLSHQRTHTGEKP YVCRECGRGFSQKSHLLSHQRTHTGEKP YVCTECGRGFSQKSVLLSHQRTHTGEKP YVCTECGRGFSRKSNLLSHQRTHTGEKP YVCRECGRGFSRKSALLSHQRTHTGEKP YVCRKCGRGFSQKSNLLSHQGTHTGEKP YVCTECGRGFSQKSHLLSHQRTHTGEKP YVCRKCGRGFSQKSNLLSHQRTHTGEKP YVCRECGRGFSFKSALLRHQRTHTGEKP YVCRECGRGFSRKSHLLSHQGTHIGEKP YVCRECGRGFSRKSNLLSHQRIHTGEKP YVRREDE* >PRDM9_musMus Mus musculus (mouse) Q96EQ9 0 MNTNKLEENSPEEDTGKFEWKPK 0 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 2 GLRAPRPAFMCYQRQAMKPQINDSEDSDEEWTPKQQ 1 2 VSPPWVPFRVKHSKQQK 0 0 ESSRMPFSGESNVKEGSGIENLLNTSGSEHVQKPVSSLEEGNTSGQHSGKKLKL 1 2 RKKNVEVKMYRLRERKGLAYEEVSEPQDDDYL 1 2 YCEKCQNFFIDSCPNHGPPLFVKDSMVDRGHPNHSVLSLPPGLRISPSGIPEAGLGVWNEASDLPVGLHFGPYEGQITEDEEAANSGYSWLITKGRNC 1 2 YEYVDGQDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 2 ELRTEIHPCLLCSLAFSSQKFLTQHMEWNHRTEIFPGTSARINPKPGDPCSDQLQEQHVDSQNKNDKASNEVKRKSKPRQRISTTFPSTLKEQMRSEESKRTVEELRTGQTTNTEDTVKSFIASEIS SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQNSHLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREK* 0 >PRDM9_ratNor Rattus norvegicus (rat) P0C6Y7 0 MNTNKPEENSTEGDAGKLEWKPK 0 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 2 GLRAPRPAFMCYQRQAIKPQINDNEDSDEEWTPKQQ 1 2 VSSPWVPFRVKHSKQQK 0 0 ETPRMPLSDKSSVKEVFGIENLLNTSGSEHAQKPVCSPEEGNTSGQHFGKKLKL 1 2 RRKNVEVNRYRLRERKDLAYEEVSEPQDDDYL 1 2 YCEKCQNFFIDSCPNHGPPVFVKDSVVDRGHPNHSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPVGLHFGPYKGQITEDEEAANSGYSWLITKGRNC 1 2 YEYVDGQDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGRELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 2 ELRTEIHPCFLCSLAFSSQKFLTQHVEWNHRTEIFPGASARINPKPGDPCPDQLQEHFDSQNKNDKASNEVKRKSKPRHKWTRQRISTAFSSTLKEQMRSEESKRTVEEELRTGQTTNIEDTAKSFIASETS RIERQCGQCFSDKSNVSEHQRTHTGEKP YICRECGRGFSQKSDLIKHQRTHTEEKP YICRECGRGFTQKSDLIKHQRTHTEEKP YICRECGRGFTQKSDLIKHQRTHTGEKP YICRECGRGFTQKSDLIKHQRTHTEEKP YICRECGRGFTQKSSLIRHQRTHTGEKP YICRECGLGFTQKSNLIRHLRTHTGEKP YICRECGLGFTRKSNLIQHQRTHTGEKP YICRECGQGLTWKSSLIQHQRTHTGEKP YICRECGRGFTWKSSLIQHQRTHTVEK* 0 >PRDM9_turTru Tursiops truncatus (dolphin) 0 MSTDRWPEDSTEGDAGRTAWKPT 0 0 VKDAFKDISIYFSKEEWTEMGEWEKIRYRNVKKNYEALVTL 1 2 GLRAPRPAFMCHRRQAIKAQVGDPEDSDEEWTPRQQ 1 2 VKPSWVAFRVEHSKHQK 0 0 AVPPVPLSNESSLKKLPGAAQLQKASGPAQAQSPAPPPGAASTSAWHTRQKL 1 2 ERRAKQIEVKMYSLRERKGHVYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAAHGAPTFVKDSAVEKGHPNRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDTSWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYSQELGIPWGSGWKSQLVAAGR 1 2 DPKPKIQPCGSCSLAFSSQKILSQHVECSHPSQVLPRTSARDRVQPEDPCPGYQNRQQQYSDPHSWSNKPECQEVKERSKPLLKRIRLGRISRAFSSSPKGQMGSSRAHERMMEAGPSTGQKVNPEATGKLLIGAGVSRVVK VKYRSSGQGSKDRSSLTKHQRTHTGEKP YVCGECGRDFSLKSDLIRHQRTHTGEKP YVCGECGRDFSLKSGLISHQRTHTGEKP YVCGECGRDFSQKSGLIRHQRTHTGEKP YVCGECGRDFSLKSGLISHQRTHTGEKP YVCGECGRDFSQKSGLIRHQRTHTGEKP YVCGECGRDFSLKSGLITHQRTHTGEKP YVCGECGRDFSQKSNLITHQRTHTGEKP YVCGECGRDFSRKSSYI* 0 >PRDM9_pteVam Pteropus vampyrus (bat) pseudogene 2 LRRKGVEVKMDSLRERMGRVYQEVSEPQDDDYL 1 2 CEKCQNFFIDSCAAHGSPIFVKDSEVDIGHPNHSALTLPPGLRIGPSGIPEAGLGVWNEASNLPLGLLFGPYEGQVTEDEEAANSKYS*M 0 0 spKGETAEYVDGKDESRANWMR 2 1 YVNCARDDEDQNLVAFQFRRQIFYRTCRVIMPGCELLVWYGDEYGQGLGIKWGSKWKREFTAGR 1 2 EPKPEIHPCPSCSLAFSSRKFLSQHMKRSHPSQSLPGISARKHLQSKEPHPEDQSQQQQQQQHTDPCSWNDKAEGQEVKERSKPMLERNGQRKISRAFSKPPKGQMGSPRECERMMEAEPSTSQKVNPENTGKSSVGVGASRIVR VKYGGCGHGFDDGSHFIRHQRTHSGEKP FVCRECERGFNEKSSLTMHQRTHSGEKP FVCRECE*GFSVKSSLIRHQRTYSGEKP FVCRECEQGFNEKSSLTMHQRTHSGEKP FFCRECE*GFSVKSSLIRHQRTHSGQKP FVCRECKRGFTQKSHLITHQRTHSGEKP FAGSVSEALHKSHISSSTRGHTQGRSPLFAGSVRESLALNCISTATGQM* >PRDM9_bosTau1 Bos taurus (cattle) NW_003053109 SRSAHILVAQLLSPKVKACDGSHAHRHQQHRPPGSWPNCEVQQRPVDTL YVCRECE* 0 MRPNTSPEESTERDAGRTEWKPT 0 0 AKDAFKDISVYFSKEEWEEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATRPAFMHHRRQVIKLQADDTEDSDEEWTPRQQ 1 2 GKLSSMAFRVEHNKHQN 0 0 TMSRAPLSKEFSLKELPGAAKLLKTSGSKQAQKLVPPPGKARTPGQHPRQKV 1 2 ELRRKETEVKRYSLRERKGHVYQEVSEPQDDDYL 1 2 YCEECQSFFIDSCAAHGPPIFVKDCAVEKGHANRSALTLPPGLSIRESSIPEAGLGVWNEVSDLPLGLHFGPYEGQITDDEEAANSGYSWL 0 0 ITKRRNCYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCQVVRPGCELLVWYGDEYGQDLGIKRESSRKSELAGPR 1 2 ESKPKIHPCASCSLAFSSQKFLSQHVQHNHPSQTLLRPSARDYLQPEDPCPGSQNQQQRYSDPHSPSDKPEGREVKDRPQPLLKSIRLKRISRASSYSPRGQMGASGVHERITEEPSTSQKPNPEDTGKLFMGAGVSGIIK VKYGECGQGSKDRSSLITNQRTHTGEKP YVCGECGQSFNQKSTLITHQRTHTGEKP YVCGECGRSFNQKSTLITHQRTHTGEKP YVCGECGRSFSQKSTLIKHQRTHTGEKP YVCGECGQSFNQKSTLITHQRTHTGEKP YVCGECGQSFNQKSTLITHQRTHTGEKP YVCGECGRSFSRKSTLITHQRTHRGEKL CLQGV* 0 >PRDM9_bosTauX1 Bos taurus (cattle) chrX 0 MSPNRSPENSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIQYRNVKRNYEALIAI 1 2 GFRATQPGFMHHGRQVLKSQVDDTEDSDEEWTPRQQ 1 2 GKPSGMAFRGEPSKHPK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 2 ELRRKETEVKRYSVRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSNSGYCWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNQQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRPKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR VNYGDHEQGSKDRSSLITHEKIHTGEKP YVCKECGKSFNGRSDLTKHKRTHTGEKP YACGECGRSFSFKKNLITHKRTHTREKP YVCRECGRSFNEKSRLTIHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCGECGRSFNEKSRLTIHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCGECGQSFNEKSRLTIHKRTHTGEKP YACGDCGQSFSLKSVLITHQRTHTGEKP YVCMECE* 0 >PRDM9_bosTauX2 Bos taurus (cattle) chrX 93% 81% 0 MSPNRSPENSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATQPGFMHHRRQVLKPQVDDTEDSDEEWTPRQQ 1 2 GKPSGMAFRGERSKHQK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 2 ELRRKETKVKRYSVRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSHSGYCWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNEQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRLKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR VKYGEHEQDSKDKSSLITHEKIHTGEKP YVCTECGKSFNWKSDLTKHKRTHSEEKP YACGECGRSFSFKKNLIIHQRTHTGEKP YVCGECGRSFSEKSNLTKHKRTHTGEKP YACGECGQSFSFKKNLITHQRTHTGEKP YVCGECGRSFSEKSRLTTHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCRECGRSFSVISNLIRHQRTHTGEKP YVCRECEQSFREKSNLVRHQRTHTGEKP YVCMECE* 0 >PRDM9_bosGru1 Bos grunniens (yak) transcript EF432551 99% 2 NRSALTLPPGLSIRESSIPEAGLGVWNEVSDLPLGLHFGPYEGQITDDEEAANSGYSWL 0 0 ITKRRNCYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCQVVRPGCELLVWYGDEYGQDLGIKRESSRKSELAAPR 2 ESKPKIHPCASCSLAFSSQKFLSQHVQHNHPSQTLLRPSARDYLQPEDPCPGSQNQQPRYSDPHSPSDKPEGREVK >PRDM9_oviAriX1 Ovis aries (sheep) 0 MSPNRSPENSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATQPAFMHHHRQVIKPQVDDTEDSEEEWTPRQQ 1 2 GKPSGMAFRGERSKHQK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKTGSKQAQKPVPPPREARTPGQHPRHKV 1 2 ELRRKETEVKRYSLRERKGHVYQEVSELQDDDYL 1 2 CEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQVIYNEEASHSGYSWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCQVVRPGCELLVWYGDEYGEELGIKQDSRGKSKLSAQR 1 2 EPKPKIHPCASCSLSFSSQKFLSQHVQRSHPSQILLRPSPRDHLQPEDPCPGKQNQQQRYSDPHSPSDKPEGQEPKERPHPLLKGPKLCIRLKRISTASSYTPKGQMGGSEVHEKMTEEPSTSQKLNPENTGKLFMEAGVSGIVR VKYGEHEQGSKDKSSLITHERIHTGEKP YVCKECGKSFNGRSNLTRHKRTHTGEKP YVCRECGQSFSLKSILITHQRTHTGEKP YVCGECGQSFSEKSNLTRHKRTHTGEKP YVCRECGQSFSLKSILITHQRTHTGEKP YVCRECGRSFSVKSNLTRHKMTHTGEKP YVCGECGQSFSQKPHLIKHQRTHTGEKP YVCRECGRSFSAMSNLIRHQRTHTGEKP YVCRECGRSFSAMSNLIRHQRTHTGEKP YVCREC* 0 >PRDM9_oviAriX2 pseudogene 0 MSPNRSPENSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATQPAFMHHHRQVIKPQVDDTEDSEEEWTPRQQ 1 2 GKPSGMAFRGERSKHQK 0 0 GMSRGPLSKVSSLKKLPGTTKLLKTSGSKQAQKPVPSSREARTSG*HTRQKV 1 2 ELGRKETDMKRYSLRERKGHVYQEVSEPQDDDYL 1 2 CQECQNFFINSCDAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRLSGIPEAGLGVWNEASHLPLGLHFGPYEGQITDDKEAVNSGYSWL 2 1 YVNCARHYEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGEKLGIRCESRGKSMLAAGR 1 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNEQQ*YSDPHSPSDKPEGCKAKERPPWLLKSMSV-----RISMASSYSPKGQMRGSETHYRMTEEPSTSQKLNPEDIGKLFMGTGVSGIIK IKYEECGQVSKDRSSLITHEGTHTREQ >PRDM9_sorAra Sorex araneus (shrew) AALT01000095 0 MSLNRPAEMNTQGKARKLMLKPM 0 0 SKDAFKDISMYFSKEEWAEMGDWEKIRHRNVKRNYEELISI 1 2 GLRAARPAFMSHRRQAIKTQLDDTEESDEEWTPNQQ 1 2 VKSLRVAFRAEQSKHQK 0 0 GRSRTPISNESSSKELSGTRTLLNTKCTKQAQKPLFPPGEASTSGHYSKPKL 1 2 ELRRKEPEVKMYSLRERKGRAYQEVSEPQDDDYL 1 2 YCENCQNFFINKCSAHGSPIFVKDNAVAKGHSNRSALTLPHGLRIGPSGIPEAGLGIWNEASDLPLGLHFGPYEGQITNDEEAANSGYSWL 0 0 ITKGRNCYEYVDGVDESLANWMR 2 1 YVNCARDYEEQNLVAFQYHRQIFYRTCRIIKPGCELLVWYGDEYGQELGIKWGSKWKSELTADK 1 2 EPKPEIYPCPCCSLAFSNQKFLSRHVEHSHPSLILPGTSARTHPKSVNFCPGDQNQWQQHSDACNDKPDEPWNDKLENHKSKGRSKPLPKRMGQKRISTAFPNLRSSKMGSSNKHETIMDKINTGQKENPKDTYRVFAGIGMPRIIR DKHVTLRRSFTNRSSPLTHQRTHTGEKP YVCRECGRGFSQKSHLLTHQRTHTGEKP YVCRECGRGFTDRSSLLTHQRTHTGEKP YVCRECGRGFSLKSSLLRHQRTHTGEKP YVCRECGRGFSLKSSLLTHQRTHTGEKP YVCRECGRGFTDRSSLLTHQRTHTGEKP YVCRECGRGFSLKSSLLTHQRTHTGEKP YVCRECGRGFSRKSSLLRHQRTHTGEKPYVCES* 0 >PRDM9_loxAfr Loxodonta africana (elephant) 0 MSPARAAKKNPRGDVGSAGRTPT 0 0 aKDTFRDISIYFSKEEWAEMGEWEKFRYRNVKRNYEALVTI 1 2 GLRAPRPAFMCHRRQAIKAQVDNTEDSDEEWTPRQQ 1 2 VKPPSVASRAEQSRHQK 0 0 GTPKALLGNESSLKEVSGTAILLNTTGSEQAQKPVSSPGEASTSDQPSRWKL 1 2 EPRRNEVEVKMYNLRERKGLEYQEVSEPQDDDYL 1 2 yCEKCQNFFIDTCAVHGAPMFVKDSPVDRGHPNHSALTLPPGLRIGPSSIPKAGLGVWNEASELPLGLHFGPYEGQVTEDKEAANSGYSWL 0 0 ITKGKNCYEYVDGKDESWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRTIQPDCELLVWYGDEYGQELGIKWGSRWKKELTSGR 1 2 EPKPEIHPCPSCRLAFSSQKFLSQHMKHSHPSPPFPGTPERKYLQPEDPRPGGRRQQRSEQHMWSDKAEDPEAGDGSRLVFERTRRGCISKACSSLPKGQIGSSREGNRMMETKPSPGQKANPEDAEKLFLGVGTSRIAK VRCGECGQGFSQKSVLIRHQKTHSGEKP YVCGECGRGFSVKSVLIKHQRTHSGEKP YVCGECGRGFSVKSVLITHQRTHSGEKP YVCGECGRGFSVKSVLITHQRTHSGEKP YVCGECGRGFSQKSDLIKHQRTHSGEKP YSCRECGRGFSRKSVLITHQRTHSGEKP YVCGECGRGFSQKSNLITHQRTHSGEKP YVCGECGRGFSRKSVLITHQRTHSGEKP YVCGECGRGFSQKSNLITHQRTHSGEKP YVCGECGRGFSQKSDLITHQRTHSGEKP YVCRECGRGFSRKSNLITHQRTHSGEKP YVCRECRRGFSVKSALIGHGRRKCSKSAEPLHFPRVSRDQK* 0 >PRDM9_macEug Macropus eugenii (wallaby) fragment 2 gFSAPRPTFMCHGKQNKEAKVEESGDFDEEWIRKQP 1 2 CEECQTFFLETCAVHGPPKFVQDSVMVKGHPYCSAITLPPGLRIGLSGIPGAGLGIWNEASNLPLGLHFGPYEGQMTEDDEAANSGYSWM 0 1 YVNCARDEEEQNLVAFQYHRKIFYRTCQIIRPGCELLVWYGDEYGQELGIKWGSKWKRPPITLT 1 >PRDM9_monDom Monodelphis domestica (opossum) not at GAS8 weak C2H2 domain fragment 0 GEDAFKDISTYFSKKQWVKLKEWEKVRLKNVKRNYEAMIKI 1 2 GLSVPRPAFMCRGRQNKKVKVEESGDSDEEWIPKQL 1 2 DCRRKDVEVHIYSLRERKYQVYQEMWDPQDDDYL 1 2 yCEECQIFFLDSCPLHGPPTFVQDSAMVKGHPYCSAITLPPGLRIGLSGIPGAGLGVWNEASTLPLGLHFGPYKGKMTEDDEAANSGYSWM 0 0 ITKGRNCYEYVDGKEESCSNWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKRPLPELTGE 1 2 EARNLSLPFHPLGFSTHTFLNQHTLLKTAPLSVLGFRDRKIMSWEIYSRTSYLQPVPTVK KECE*GFTHQTNLVTHRWTHSGERPYVCV* >PRDM9a_ornAna Ornithorhynchus anatinus (platypus) fragment RIGKKPQVRDFNLRKQKRKIYNENYRPEDDDYL CEICQTFFLEKCVLHGPPVFVQDLPVEKWRPNRSTITLPPGMQIKVSGIPNAGLGVWNQATSLPRGLHFGPYMGIRTKNEKESHSGYSWMI IVRGKNYEYLDGKDKAFSNWMR YVNCARSEREQNLVAIQYQGEIYYRTCRVIPPGQELLVWYGLEYGRHLG EARPSRYRDVVFSSGVYGSSVTLLTSGVP >PRDM9b_ornAna Ornithorhynchus anatinus (platypus) tandem fragment RSGKKPQVRDFNLRKQKRKMYTEESEPEDDDYL CEDCQTFFLEKCSVHGPPVFVQDCEAKRCQQNRSEVTLPPGLLIKMSGIPNAGLGVWNQATSLPRGLYFGPFVGIRKNNVKDSLSGYSWAV ILRGRNYEYLDGKNTSFSNWMR ASDSWTGRERLILLPSWGVGGAASGGKRVREDSGKGGWRESR KPVSALTTSWDSDGWERAADEVTVSLLPGERPHSSGGSFAPSARSGGVKQRIWSKRRSAA LQRTRERRNSTHDFPPKHEDTAARQDERQCPDRGRAKQRGVRKSEQIERAKAMGRKKALG GLSPPRRERLSDEAGQRKKSGHEQFWQKPGPSEAWAGPAEGSTIPRR HCCDVCGKAFNRLSRLKQHKRVHTGEKP LVCKICKRAFSDPSNLNRHAKRHTGEKP FVCRVCGRSFNRSDNMNEHRWKHTSNNIIP NTGHMSATVVENASLCINRNYQIYKERATYL- HCCDVCRKAFKRLSHLRQHKRIHTGEKP LVCKVCRRTFSDPSNLNRHSRIHTGLRP YVCKLCRKAFADPSNLKRHVFSHTGHKP FVCEKCGKGFNRCDNLKDHSAKHSEDNSTPKP* >PRDM7_homSap Homo sapiens (human) chr16:90,122,976 TUBB3+ DEFB+ AFG3L1+ DBNDD1- GAS8+ PRDM7- 92% identical 0 MSPERSQEESPEGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKMNYNALITV 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRGEQSKHQK 0 0 GMPKASFNNESSLRELSGTPNLLNTSDSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEISEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSSANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQERQYSDPRCCNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVITHQRTHTGGKP YVCRECGR FSRKSDLLSHQRTHTGEKP YVCRECERGFSRKSVLLIHQRTHRGDAP VCRKDE* >PRDM7_panTro Pan troglodytes (chimp) pseudogene GAS8 0 MSPERSQEESPEGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWgKTRYRiVKMNYNALITi 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRGEQSKHQK 0 0 GMPKASFNNESSLkELSGmPNLLNTSgSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRRKETvGKMYSLRERKGHAYKEISEPQDDDYL 1 2 yCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLRVWNEASDPPLGLHSGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSwANWMR 2 1 YENCARDDEEQNLVSFQYHRQS*FYRTCRVIRPGCELLVWYGDE*GQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLQPENP*PGDQNQERQYSDPRCCNDKTKGQEVKERSKLLNKWTWQREISRAFSSLPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVITHQRTHTGGKP YVCRECGQGFSRKSVLLIHQRTHRGEKP* 0 VCRKDE >PRDM7_ponAbe Pongo abelii (orangutan) GAS* 0 MSPERSQEESPkGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWTEMGDWEKTRYRNVKRNYKTLITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRGEQSKHQK 0 0 GMPKASFNNESSLKELSGTQNLLNTSGSEQAQKPVSPPGEASTSGQHSTLKI 1 2 ELRRKETEGKTYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCAWDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMPGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARHLLQAENPCPGDQNQEQQYSDPDCCNDKTKGQEIKERSKLLNKRTWQREISRAFSSSAKGQMGSSRVGERMMEEESGTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVITHQRTHTGGRS YICRESGRGFTQKSGLLSHQRTHTGEKP YVCRECGWGFSQKSNLLRHQRTHTGEKP YVCRECGRGFSRKSVLLIHQRTHTGEKP* VCRKDE* >PRDM7_nomLeu Nomascus leucogenys (gibbon) ADFV01125891 no info GAS8 pseudogene 0 MSPERSQEESPkGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWTEMGDWEKTRYRNVKRNYKTLITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 IKSPWMAVRVEQSKHQK 0 0 GMPKASFNNESGLKELSGTQNLLNTSG*EQARKPVSPPGEASTSGQHSRQKL 1 2 ELRRKETEGKMYSL*ERKGHAYKEVSEPQDDDYL 1 2 yCEMCQNFFTDSCAAHGPPTFVKDSAVDKGHPNHSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKS*ANWMK 2 1 YVNCARDHEEQNLVAFQYHRQIFYRTCQVIRPGCEPLVWYGDEYGQELGIKWGSKWKKELTAER 1 2 EPKPEIHPCPSCCLVFTSQKFLSQHVECNHSSQNFPGPSARKLLQRENPCPGDQNQEQQYSDSRSCNDKTKGQEIKERSKL*NKRIWQRKISRAFSSLPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVIAHQGTHTGGKS *ICRECGWGFSQESHLLIHQRTHTGEKL YVCRECGQGFSQKSDLLSHQRTHTGEKP YVRRECGRGFSQKSNLLSHQRTHTEEKP YVCRECGWGFSQKSHLLIHQRTHTGKKP* VCRKDE >PRDM7_macMul Macaca mulatta (rhesus) pseudogene 0 MSPERSQEESPEEDTERTERKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRVEQSKHQK 0 0 EMPKTSFNNESSLKELSGTPNLLSTSDSE*AQKPASPPGEASTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKRHAYKEASELQHDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDNAVNKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPCEGRITEDKEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWAKWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIYPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQSENPCPGDQNQEQQYSDPSSCNDKTKGQEIKERSKLLNKRTWQREILRAFTSPPKGQMGSSRVGERMMEEEFRTGQKANPGNTGKLFVGVEISRIAK VKYGECGQGFSGKSDVITHQRTHTEGKP YVCRGCGRRFSQKSSLLRHQRTHTGEKP* >PRDM7_papHam Papio hamadryas (baboon) GAS* end of scaffold 0 MSPERSQEESPEEDTERTEWKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAVRVEQSKHQK 0 0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITQDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSTQNFPGPSARRLLQPENLCSGDQNQEQQYSDPCSCNDKTKGQEIKERSKLLNKRTWQKEISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPENIGKLFVEVGISRIAK VKYGECGQGFSDKSDVVIHQRTHTREKP YLCRECGRGFSQKSNLLRHQRTHTGEKP YLCRECGRGFRDNSSLRCHQRTHTGEKP YLCRECGRGFRDNSSLRCHQRTHTGEKP YLCRECGRGFSDNSSLRYHQRTHTGEKP YLCRECGRGFRDNSSLRYHQRTHTGEKP YLCRECGRGFSVKSNLLSHQRTHTGEKP YVCRECGRGFSDNSSLRCHQRTHTGEKP YLCRECGRGFSQMSHLRCHQRTHTGEKP YLCRECGRGFSVKSNLLSHQRTHTGEKP YVCRECGRGFSRKANLLSHQRTHTGEKP* 0 >PRDM7_micMur Microcebus murinus (lemur) ABDC01433247 0 MSPEKSQEESPEEDTERTERKPM 0 0 vKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEKCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLKIRPSGIPQAGLGVWNEASELPLGLHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDDSWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCQVIRPGCELLVWYGDEYGQELGIKWGSKWKEELTIRQ 1 2 EPKPEIHPCPSCSLAFSSQKFLSQHVKHTHSSQISPRTSGRKHLQPENPCPGDQNQEQQHSDPHSCNDKAKDQEVKERPKPFHKKTQQRGISRAFSSPPKGKMGSCREGKRIMEEEPRTGQKVGPGDTDKLCAAGGISRISR VKYGDSGQSFSDKSNVIIHQRTHTGEKP YVCRECGRGFSQKSDLLKHQRTHTGEKP YVCRECGRGFSQKSHLLRHQRTHTGEKP YVCRECGRGFSQKSDLLIHQRTHTGEKP YVCRECGRGFSCKSHLLIHQRTHTGEKP YVCRECGRGFSCKSSLLIHQRTHTGEKP YVCRGVWGEALAESQTSSYTRGHTQGRSP VFAGRVSKSLALNYISTATGGHLLTSHLP TPALGGASKGSLLTLYISQECKETRNN* >PRDM7_otoGar Otolemur garnettii (galago) GAS8 0 MSPEKSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKHPWMAFRMEQSKRQK 0 0 ILKKCMLSFNMHLKELSGPASLPNISGSEQHQKHMSSPREASTSGQHSGRKS 1 2 DLRIKEIEVRMYSLRERKGHAYKEVSEPQDDDYL 1 2 yCEKCQNFFIDNCAVHGPPTFVKDTAVEKGHPNRSVLSLPSGLGIRTSGIPQAGFGVWNEASDLQLGLHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDESQGNWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGQ 1 2 EPKPEIHPCPSCSLAFSTQKFLSQHVERTHPSQISQGTSGRKNLRPQTPCPRDENQEQQHSDPNSRNDKTKGQEVKEMSKTSHKKTQQSRISRIFSCPPKGQMGSSREGERMIEEEPRPDQKVGPGDTEKFCVAIGISGIVK VKNRECVQSFSNKS NLRHQRTHTGEKP YMCRDCGRGFSHKSSLFRHQRTHTGEKP YVCRDCGRGFSLKANLLTHQRTHTGEKP YVCRDCGQGFSQKAHLLRHQRTHTGEKP YMCRDCGQGFSRKAYLLTHQRTHTGEKP YVCRDCGQGFSQKAHLLTHQRTHTGEKP YVCRDCGRGFSHKSSLFRHQRTHTGEKP YICRDCG* >PRDM7_bosTau chr Un.004.251 bad XM_598831 + transcript GO353654 no zinc downstream 0 MSQNRSPEERTKGDAGRTEWKLT 0 0 AKDAFKDISIYFSKEEWAEMGEWEKTGYRNVKRNYEVLIAI 1 2 GLRATQPAFMHHRRQVIKPQGDDTEDSDEEWTPQHQ 1 2 GKPSRKAFRMEHRKHQK 0 0 GKSRGPLSKVSSLKKLQGAAKLLNTSGSKWAQKPANPPRETRTLEQHSRQKV 1 2 ELRRKETDMKRYSLRERKGHVYQEVSEPQDDDYL 1 2 YCQECQNFFIDSCDAHGPPTFVKDSAVEKGHANRSVLTLPPGLSIKLSGIPEAGLGVWNEASHLPLGLHFGPYEGQITDDKEAINSGYSWL 0 0 ITKGRNSYEYVDGKDTSLNWMR 2 1 YVNCARHYEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGEKLGIKCESRGKSMFAAGGV 1 2 EGHPSSSTPPHSGELPR* 0 >PRDM7_bosTau Bos taurus (cow) adjacent to GAS8 and in correct orientation pseudogene but missing C2H2. 0 MSPNRSPEESIEGDTGRTEWKPT 0 0 AKDAFKDISIYFCKEEWAQMG*WEKIRYRNVKRNYEALITL 1 >PRDM7_ailMel Ailuropoda melanoleuca (panda) GAS8 synteny no 9 0 MSLNTSPEETPERDSGRTGWKPT 0 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYEALITI 1 2 GLRAPRPAFMCHRRQAIKPQVDDTEDSDEEWTPRRQ 1 2 VRPSWVAFRMEQSKHQR 0 0 GIPRAPLRNESSLKELSETAKLLNTSGSELGQKPVSLPGEASTSGHDSLQKL 1 2 GFRRKDVEVKMYSLRERKSLAYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAVHGPPTFVKDSAVDKGQPNRSALTLPPGLRIRPSGIPQAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDNSWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKSELAAGK 1 2 EPKPEIHPCPSCSLAFSSQKFLSQHLEHNHPSQILSRKSASEHFQQEDPCPGHQNQQQQQHSDPHRWNDKAKGQEVKERFKPLLKSIRQRRISRAFSSPCKGQTRSSTVCEGMVEEEPSAGQKLNPEETGKLFMGVGMSGIIR VKYRGCGRDFSDRSHQSGHQRRHQKKP SVCKKVKREFSHKSVLITHQRTHTGEKP YVCRECGRGFTQRSNLIRHQRTHTGEKP YVCRECGRGFTQRSNLIRHQRTHTGEKP YVCRECGRGFTQRSSLIRHQRTHTGEKP YVCRECGRGFTLRPNLIGHQRTHTEALP INYISTTKEQM* 0 >PRDM7_felCat fragment no GAS8 chrUn_ACBE01450406:1-11,793 0 MEPSPASESARGQPGGPGTTSPLRFPEQSAERGSRKARWKPT 0 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYEALMTI 1 2 gLRAPRPAFMCHRRQAIKPQVDVTEDSDEEWTPRQQ 1 2 VKPSWVASRVDQNKQHKV 0 0 GTHRVPLSKESSLKDFSETAKLLNTSGSEQGQKPVSLPGEASTSGHHSRRKL 1 2 frRRKEIGVKMYSLRERKGFAYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAVHGPPTFVKDNAVGKGHPNRSALTLPPGLRIRPSSIPEAGLGVWNEASDLPLGTHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDNSWANWMR 2 >PRDM7_canFam pseudgene 2 VKPSWVAFRMEQSKHQK 0 2 yCEK*QTFFIDSCTVHGPPTFVKDSEVDKGQPNHSALTLPPGLRIRTSSIPQAGLGVWN*ASDLPLGLHFGPYKGQITEDEEAANSGYSCL 0 0 ITKGRNCYEYVDGKDKDNSWANWMR 2 1 YMNCARDDEEQS*WPFNITGR--YSTEPVEQPGHQASCELLVWYGDEYSQELGIKWGSKWKSELTAGK 1 2 EPNPEIHPCPSCSL AFSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECGQALYRGQISAYIRGHTQGRSPM >PRDM7_equCab missing front exons, some frameshifts 2 VKPSWVAFRVEQSKQQK 0 2 SGTAKLLKTSSSEQVQKPVSPLGEASSSEQHSRRKLELRRKEVGVKMYSLRERKGHAYQEVSEPQDDDYL 1 2 yCENCQNFFIDSCAAHGPPIFVKDSAVDKGHPNRSALTLPLGLRIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDISWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYGQELGIKWGSKWKRELTAGR 1 2 EPKLEIHPCPSCSLAFSSQKFLSQHVERNHPSQILPGTSARNHLQPEDPSPGDQNQQQQHSDPHSWKDKAHSQEVKERSKPLLKKIRQRRIPRAFSYPPKGQMENFRMRERIMEEKPSIGRKVNPEDTGKLFLEMRMSRNVR VQYGGCGRGFNDRASLIKHQRTHTGEKP YVCRECEQGFTQKSSLIAHQRTHTGEKP YVCRECEQGFSEKSHLIRHQRTHTGEKP YVCRECE* >PRDM7_loxAfr (elephant) pseudogene adjacent to GAS8 in standard orientation many frameshifts 0 MSLNTSPEETPERDSGRTGWKPT 0 0 AKDAFRDIFIYFSKEGYVEMGEWEKLCYRNLKMNYKALVTT 1 2 GLRASHPAFTCHCMQAIKAQMDDTEDSNEEQTPRQ 1 2 VRPSWVAFRMEQSKHQR 0 0 GMLRVPRSNESSLKNLSGTSIMLSRAGSEQAQKLVLPPGKASTSDEHSRQKP 1 2 EHRRKGVEVKMYSF*ERKGLVYQEIS*PQDDDYL 1 2 YCEKCQNFFIDTCESHGVPTFVKNSTTDSGHPNHLALTPSSGLRTRPSSIPKAWLRLWNKAFELLLGLPFSPCEGQVIEDEAVDNSGYSWL 0 0 ITKGRNCYEYVDGKDNSWANWMR 2 1 YVNGTQDEKEQNLVFFQYHRQIFYQTCYAVWPGCQLLVWYRDECGQELGIKWDNRGKKEFe 1 2 EPKPEAHPCPSCPLAFSSEKFLSQHMKHNHPSQSSPETPERKHLQPEDPHPGHQNQQQQQHSDPHRWNDKAEGQQTGDRSKPMFENIRQEVTSRAFSSLPKGQMVCSREGNRMMETEPSPGLKVNPEVTGKLFLGVESSRIAK VKYRGCGRDFSDRSHQSGHQRRHQKKP SVCKKVKREFSHKSVLITHQRTHSGEKS YVCKESGRGFSAKSNLIRPRRTHTGEKP YVCGERG*GFSV*SGLIIHQRAHSPEKP YVCREGRRGFGDKSSFIKHQRATLGEKS YVCKESGRGFSAKSNLIRPRRKKCRHDTTPHPQL* 0 >PRDMx_danRer Danio rerio (zebrafish) Q6P2A1 transcript BC064665 pseudo-homologous 0 MSLSPDLPPSEEQNLEIQGSATNCYSVVIIEEQDDTFNDQPF 1 2 YCEMCQQHFIDQCETHGPPSFTCDSPAALGTPQRALLTLPQGLVIGRSSISHAGLGVFNQGQTVPLGMHFGPFDGEEISEEKALDSANSWV 0 0 ICRGNNQYSYIDAEKDTHSNWMK 2 1 FVVCSRSETEQNLVAFQQNGRILFRCCRPISPGQEFRVWYAEEYAQGLGAIWDKIWDNKCISQ 1 2 GSTEEQATQNCPCPFCHYSFPTLVYLHAHVKRTHPNEYAQFTQTHPLESEAHTPITEVEQCLVASDEALSTQTQPVTESPQEQISTQNGQPIHQTENSDEPDASDIYTAAGEISDEI HACVDCGRSFLRSCHLKRHQRTIHSKEKP YCCSQCKKCFSQATGLKRHQHTH QEQEKNIESPDRPSDI YPCTKCTLSFVAKINLHQHLKRH HHGEYLRLVESGSLTAETEEDHT EVCFDKQDPNYEPPSRGRKSTKNSLKG RGCPKKVAVGRPRGRPPKNKNLEVEVQKIS PICTNCEQSFSDLETLKTHQCPRRDDEGDNVEHPQEASQ YICGECIRAFSNLDLLKAHECIQQGEGS YCCPHCDLYFNRMCNLRRHERTIHSKEKP YCCTVCLKSFTQSSGLKRHQQSHLRRKSHRQSSALFTAAIFPCAYCPFSFTDERYLYKHIRRHHPEMSLKYLSFQEGGVLSVEKP HSCSQCCKSFSTIKGFKNHSCFKQGEKV YLCPDCGKAFSWFNSLKQHQRIHTGEKP YTCSQCGKSFVHSGQLNVHLRTHTGEKP FLCSQCGESFRQSGDLRRHEQKHSGV RPCQCPDCGKSFSRPQSLKAHQQLHVGTKL FPCTQCGKSFTRRYHLTRHHQKMHS* 0 >ZNF133_homSap Homo sapiens (human) NP_001076799 0 MAFRDVAVDFTQDEWRLLSPAQRTLYREVMLENYSNLVSL 1 2 GISFSKPELITQLEQGKETWREEKKCSPATCP 1 2 DPEPELYLDPFCPPGFSSQKFPMQHVLCNHPPWIFTCLCAEGNIQPGDPGPGDQ EKQQQASEGRPWSDQAEGPE GEGAMPLFGRTKKRTLG AFSRPPQRQPVSSRNGLRGVELEASPAQTGNPEETDKLLKRIEVLGFGT VNCGECGLSFSKMTNLLSHQRIHSGEKP YVCGVCEKGFSLKKSLARHQKAHSGEKP IVCRECGRGFNRKSTLIIHERTHSGEKP YMCSECGRGFSQKSNLIIHQRTHSGEKP YVCRECGKGFSQKSAVVRHQRTHLEEKT IVCSDCGLGFSDRSNLISHQRTHSGEKP YACKECGRCFRQRTTLVNHQRTHSKEKP YVCGVCGHSFSQNSTLISHRRTHTGEKP YVCGVCGRGFSLKSHLNRHQNIHSGEKP IVCKDCGRGFSQQSNLIRHQRTHSGEKP MVCGECGRGFSQKSNLVAHQRTHSGERP YVCRECGRGFSHQAGLIRHKRKHSREKP YMCRQCGLGFGNKSALITHKRAHSEEKP CVCRECGQGFLQKSHLTLHQMTHTGEKP YVCKTCGRGFSLKSHLSRHRKTTSVHHR LPVQPDPEPCAGQPSDSLYSL* 0 >ZNF169_homSap Homo sapiens (human) 0 MSPGLLTTRKEALMAFRDVAVAFTQKEWKLLSSAQRTLYREVMLENYSHLVSL 1 2 GIAFSKPKLIEQLEQGDEPWREENEHLLDLCP 1 2 EPRTEFQPSFPHLVAFSSSQLLRQYALSGHPTQIFPSSSAGGDFQLEAPRCSSEKGESGETEGPDSSLRKRPSRISRTFFSPHQGDPVEWVEGNREGGTDLRLAQRMSLGGSDTMLKGADTSESGAVIRGNYRLGLSKKSSLFSHQKH HVCPECGRGFCQRSDLIKHQRTHTGEKP YLCPECGRRFSQKASLSIHQRKHSGEKP YVCRECGRHFRYTSSLTNHKRIHSGERP FVCQECGRGFRQKIALLLHQRTHLEEKP FVCPECGRGFCQKASLLQHQSSHTGERP FLCLECGRSFRQQSLLLSHQVTHSGEKP YVCAECGHSFRQKVTLIRHQRTHTGEKP YLCPQCGRGFSQKVTLIGHQRTHTGEKP YLCPDCGRGFGQKVTLIRHQRTHTGEKP YLCPKCGRAFGFKSLLTRHQRTHSEEEL YVDRVCGQGLGQKSHLISDQRTHSGEKP CICDECGRGFGFKSALIRHQRTHSGEKP YVCRECGRGFSQKSHLHRHRRTKSGHQL LPQEVF* 0 >ZNF596_homSap Homo sapiens (human) 0 MESQESVTFQDVAVDFTQEEWALLDTSQRTLFREVMLENISHLVSV 1 2 GNQLYKSDVISHLEQGEQLSREGLGFLQGQSPVISDREDDPKKQEMLSMQHICKKDAPLISAMQWSHTQEDPLECNNFREKFTEILPLTQYVIPQVGKKPFISQDVGKAISYLPSFNIQKQIHSRSKS YECHQRRNTFIQSSAHRQHNNTQTGEKT FECHVCRKAFSKSSNLRRHEMIHTGVKP HGCHLCGKSFTHCSDLRKHERIHTGEKL YGCHLCGKAFSKSYNLRRHEVIHTKEKP NECHLCGKAFAHCSDLRKHERTHFGEKP YGCHLCGKTFSKTSYLRQHERTHNGEKP YGCHLCGKAFTHCSHLRKHERTHTGEKP YECHLCGKAFTESSVLRRHERTHTGEKP YECHLCWKAFTDSSVLKRHERTHTGEKP YECHLCGKTFNHSSVLRRHERTHTGEKP YECNICGKAFNRSYNFRLHKRIHTGEKP YKCYLCGKAFSKYFNLRQHENSCYKGNK* 0 >HKR1_homSap Homo sapiens (human) MRVNHTVSTMLPTCMVHRQTMSCSGAGGITAFVAFRDVAVYFTQEEWRLLSPAQRTLHREVMLETYNHLVSL 1 2 EIPSSKPKLIAQLERGEAPWREERKCPLDLCP 1 2 ESKPEIQLSPSCPLIFSSQQALSQHVWLSHLSQLFSSLWAGNPLHLGKHYPEDQ KQQQDPFCFSGKAEWIQE GEDSRLLFGRVSKNGTSKALSSPPEEQQPAQSKEDNTVVDIGSSPERRADLEETDKVLHGLEVSGFGE IKYEEFGPGFIKESNLLSLQKTQTGETP YMYTEWGDSFGSMSVLIKNPRTHSGGKP YVCRECGRGFTWKSNLITHQRTHSGEKP YVCKDCGRGFTWKSNLFTHQRTHSGLKP YVCKECGQSFSLKSNLITHQRAHTGEKP YVCRECGRGFRQHSHLVRHKRTHSGEKP YICRECEQGFSQKSHLIRHLRTHTGEKP YVCTECGRHFSWKSNLKTHQRTHSGVKP YVCLECGQCFSLKSNLNKHQRSHTGEKP FVCTECGRGFTRKSTLSTHQRTHSGEKP FVCAECGRGFNDKSTLISHQRTHSGEKP FMCRECGRRFRQKPNLFRHKRAHSGA FVCRECGQGFCAKLTLIKHQRAHAGGKP HVCRECGQGFSRQSHLIRHQRTHSGEKP YICRKCGRGFSRKSNLIRHQRTHSG* 0 >PRDM11_homSap Homo sapiens (human) MLKMAEPIASLMIVECRACLRCSPLFLYQREKDRMTENMKECLAQTNAAVGDMVTVVKTEVCSPLRDQEYGQPCSRRPDSSAMEVEPKKLKGKRDLIVPKSF QQVDFWFCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWLIVDKNNRYKSIDGSDETKANWMRYVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIH RNLARGEKRLQREKSEQVLDNPEDLRGPIHLSVLRQGKSPYKRGFDEGDVHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLVIRKVPKYQDDAYSQCATTMTHGVQNIGQTQGEGDWKVPQGVSKEPGQLEDEEEEPSSFKADSPAEASLASDPHELPTTSFCPNCIR LKKKVRELQAELDMLKSGKLPEPPVLPPQVLELPEFSDPAGKLVWMRLLSEGRVRSGLCGG
>GAS8_homSap Homo sapiens (human) synteny marker right centromeric positive strand C16orf3- in second intron growth arrest-specific del cancer MAPKKKGKKGKAKGTPIVDGLAPEDMSKEQVEEHVSRIREELDREREERNYFQLERDKIHTFWEITRRQLEEKKAELRNKDREMEEAEERHQVEIKVYKQKVKHLLYEHQNNLTEMKAEG TVVMKLAQKEHRIQESVLRKDMRALKVELKEQELASEVVVKNLRLKHTEEITRMRNDFERQVREIEAKYDKKMKMLRDELDLRRKTELHEVEERKNGQIHTLMQRHEEAFTDIKNYYNDI TLNNLALINSLKEQMEDMRKKEDHLEREMAEVSGQNKRLADPLQKAREEMSEMQKQLANYERDKQILLCTKARLKVREKELKDLQWEHEVLEQRFTKVQQERDELYRKFTAAIQEVQQKT GFKNLVLERKLQALSAAVEKKEVQFNEVLAASNLDPAALTLVSRKLEDVLESKNSTIKDLQYELAQVCKAHNDLLRTYEAKLLAFGIPLDNVGFKPLETAVIGQTLGQGPAGLVGTPT*