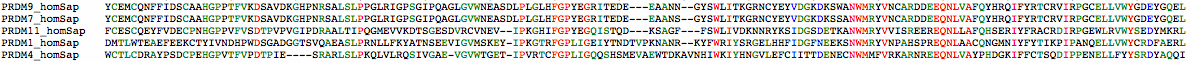PRDM9: meiosis and recombination
Introduction
PRDM9 is a gene on human chromosome 5 with a very peculiar history. Its primary function -- after many false starts -- has only recently become clear: scanning the genome with its terminal zinc finger array to locate and mark recombination hotspots with its histone methylase where its transcription factor domain can direct additional proteins to initiate the double stranded breaks needed for meiosis. Recombination between homologous chromosomes is essential to proper alignment and separation into daughter cells (as well as for juxaposing favorable alleles for adaptive evolution).
Such a mission-critical protein is typically highly conserved. However this is not the case here at all. Indeed, it proves exceedingly difficult to find a comprehensive set of PRDM9 orthologs in sequenced mammalian genomes, with immense confusion in the literature over paralogs, lost copies, pseudogenes, and other composite domain proteins overlapping in domain content but having no immediate homology.
Syntenic relationships help resolve events during mammalian evolution. Here TUBB3+ AFG3L1+ GAS8+ has stably existed since the stem amniote, with the arrangement TUBB3+ AFG3L1+ GAS8+ PRDM7- qTer found in nearly all placental mammals. PRDM9 however is found in many syntentic contexts, depending on clade.
From the perspective of comparative genomics, PRDM7 is the fundamental gene, not the disparate collection of genes mis-labelled PRDM9 genes. At different times in different placental clades, PRDM7 spun off segmental duplications of itself to other sites in other chromosomes, probably because of its susceptible location at the extreme q arm of an autosomal chromosome.
These paralogous copies -- despite all being called PRDM9 -- are not orthologous outside their narrow clade. Orthology requires by definition vertical descent from a common gene in the last common ancestor of two species. Here the PRDM9 are descended from a common gene but at different stages in its evolution. While an unresolved terminological muddle, these copies are sometimes called in-paralogs within a species and co-orthologous across them.
In euarchontoglires, a segmental duplication of PRDM7 occurred in a stem catarrhine primate and descended through speciation events to contemporary old world monkeys and great apes. This second copy (PRDM9) relocated to and stayed within a cadherin gene complex on a different chromosome. PRDM7 persisted at its original ancestral location but became an overt pseudogene in some lineages (rhesus, gibbon, gorilla, chimp and human) but not others (orangutan). Earlier diverging primates such as lemurs, tarsier and new world monkeys have a single PRMR7 gene adjacent to GAS8.
Rodents and lagomorphs also have no counterpart to PRDM9, though the situation is confused by chromosomal rearrangements (no homolog or even debris adjacent to GAS8 or CDH). The mouse gene may well be orthologous to primate PRDM7, not PRDM9. The rat gene occurs in the same syntenic context. Rabbit has two apparent PRDM7, called here PRDM7a and PRDM7b; neither copy is syntenic to mouse/rat or any other mammal.
Although an obvious pseudogene, human PRDM7 is often treated as a functional gene with 'isoforms'. However exon 9 of the reference sequence hg18 contains an internal direct tandem repeat of 88 nucleotides that throws off the reading frame and subsequent splice to exon 10, which itself has a frameshift (GGGG to GGG) in the second of its three zinc fingers. The protein is incorrectly described at NCBI, SwissProt and UCSC -- zinc fingers translated into the wrong reading frame cannot form a meaningful fold. Given the comparative genomics context of duplication followed by subsequent pseudogenization, this feature is unquestionably a pseudogene whether it is still transcribed or not.
However the confusion doesn't stop there. After separate segmental duplication in afrotheres and pecoran ruminants, still other retention and loss scenarios have played out. Both copies can seemingly functions for long periods, but the parental gene PRDM7 can also be lost completely. In others, pseudogene remnants in various degrees of decay can still be detected.
Rapid evolution of this gene subfamily occurs at the amino acid level as well, especially in zinc finger number and substitutions at the 4 dna-recognizing residues. All this may be directly related to the role in meiosis: the process tends to destroy its recombination hotspots by biased gene conversion. Since recombination is essential, new hotspots must emerge. The race is then on for PRDM7 and its spun-off PRDM9s to rapidly evolve new histone markup sites.
This rapid evolution may cause breeding incompatibility between populations in the F1 generation (meiosis arrest for lack of cross-overs, notably between chrX and chrY). However it takes very different forms in different lineages. In effect each major clade of placentals is evolving a qualitatively different mating system, with its most extreme form in ruminants with 6 PRDM9 genes. This follows upon the very different structures of sex chromosomes between monotremes, marsupials and placentals.
Comparative genomics: sequence availability
Classification of available placental mammal sequences: 56 genes from 34 species. These genes all have ten identically intronated exons but 72 exons of the 560 expected are missing because many genomes are incomplete. The number of zinc fingers is shown in the second column, phylogenetic clade in the third, and an adjacent gene (synteny) in the fifth.
The number of zinc fingers is quite variable in human and likely so in all species; the table provides that of the individuals selected for genome projects. These zinc finger arrays have been corrected in low coverage genomes for minor frameshifts and premature stop codons arising from nucleotide run length errors (eg, ggggg misread as gggg).
Pseudgenes are sometimes obvious (large deletions, multiple reading frame errors, stop codon in early exons, amino acid substitutions disrespecting the conservation profile) but otherwise can be difficult to distinguish from sequencing error or a bad allele of a usually intact gene in the population. A pseudogene can continue being transcribed for tens of millions of years after losing all functionality at the protein level. However no confusion arises here because PRDM7 nor PRDM9 are represented only by a half dozen transcripts among the tens of million mammalian transcripts at GenBank.
The PRDM7 genes are all orthologous in the classical sense but the PRDM9 genes are not unless syntentic and in the same phylogenetic cluster.
- PRDM7: genes with ancestral location GAS8 synteny
- PRDM9: lineage-specific segmental duplications of PRDM7
- Pseudogenes: multiple disabling frameshifts and stop codons
>PRDM9_homSap 13 prim gene CDH12 Homo sapiens (human) NM_020227 >PRDM9_panTro 19 prim gene CDH12 Pan troglodytes (chimp) GU166820 >PRDM9_gorGor - prim gene cdh12 Gorilla gorilla (gorilla) CABD02290264 >PRDM9_ponAbe 10 prim gene CDH12 Pongo abelii (orangutan) XR_093432 >PRDM9_nomLeu 10 prim gene cdh12 Nomascus leucogenys (gibbon) ADFV01015315 >PRDM9_macMul 9 prim gene CDH12 Macaca mulatta (rhesus) XM_001083675 >PRDM9_papHam 11 prim gene cdh12 Papio hamadryas (baboon) genome >PRDM7_homSap 3 prim gene GAS8+ Homo sapiens (human) genome >PRDM7_panTro 2 prim pseu GAS8+ Pan troglodytes (chimp) genome >PRDM7_gorGor 3 prim pseu GAS8+ Gorilla gorilla (gorilla) genome >PRDM7_ponAbe 4 prim gene GAS8+ Pongo abelii (orangutan) genome >PRDM7_nomLeu 5 prim pseu gas8+ Nomascus leucogenys (gibbon) ADFV01125891 >PRDM7_macMul 2 prim pseu GAS8+ Macaca mulatta (rhesus) genome >PRDM7_papHam 2 prim pseu gas8+ Papio hamadryas (baboon) genome >PRDM7_calJac 12 prim gene GAS8+ Callithrix jacchus (marmoset) XR_090591 >PRDM7_micMur 8 prim gene gas8+ Microcebus murinus (lemur) ABDC01433247 >PRDM7_otoGar 7 prim gene GAS8+ Otolemur garnettii (galago) genome >PRDM7_tarSyr - prim pseu gas8+ Tarsius syrichta (tarsier) ABRT011082008 >PRDM9_oryCun 8 glir gene other Oryctolagus cuniculus (rabbit) genome >PRDM7_oryCun 4 glir gene other Oryctolagus cuniculus (rabbit) genome >PRDM7_ochPri - glir gene noDet Ochotona princeps (pika) AAYZ01312269 >PRDM7_ratNor 10 glir gene PDCD2 Rattus norvegicus (rat) NM_001108903 >PRDM7_musMus 12 glir gene PDCD2 Mus musculus (mouse) NM_144809 >PRDM7_musMol 11 glir gene noDet Mus molossinus (wild_mouse) GU216230 >PRDM7_dipOrd - glir gene noDet Dipodomys ordii (kangaroo_rat) genome >PRDM7_speTri - glir gene noDet Spermophil tridecemlin (squirrel) AAQQ01308561 >PRDM9a_bosTau 7 laur gene noDet Bos taurus (cattle) NW_003053109 >PRDM9b_bosTau 5 laur gene noDet Bos taurus (cattle) DAAA02065087 >PRDM9c_bosTau - laur gene noDet Bos taurus (cattle) XM_002699750 >PRDM9d_bosTau 9 laur gene noDet Bos taurus (cattle) genome >PRDM9e_bosTau 9 laur gene noDet Bos taurus (cattle) genome >PRDM9e_oviAri - laur pseu noDet Ovis aries (sheep) genome >PRDM9d_oviAri - laur gene noDet Ovis aries (sheep) genome >PRDM9c_oviAri 4 laur pseu noDet Ovis aries (sheep) genome >PRDM9b_oviAri 2 laur pseu noDet Ovis aries (sheep) genome >PRDM9a_oviAri 9 laur gene noDet Ovis aries (sheep) genome >PRDM9d_munMun 4 laur gene noDet Muntiacus muntjak (muntjac) AC216498 >PRDM9c_munMun 15 laur gene noDet Muntiacus muntjak (muntjac) AC154919 >PRDM9b_munMun 13 laur gene noDet Muntiacus muntjak (muntjac) AC218859 >PRDM9a_munMun 7 laur gene noDet Muntiacus muntjak (muntjac) AC225653 >PRDM7_bosTau - laur pseu GAS8+ Bos taurus (cattle) genome >PRDM7_turTru 9 laur gene gas8+ Tursiops truncatus (dolphin) ABRN01441536 >PRDM7_susScr 9 laur gene GAS8+ Sus scrofa (pig) FP476134 >PRDM7_canFam 5 laur pseu GAS8+ Canis familiaris (dog) genome >PRDM7_felCat 11 laur gene GAS8+ Felis catus (cat) genome >PRDM7_ailMel 6 laur gene GAS8+ Ailuropoda melanoleuca (panda) GL193502 >PRDM9_pteVam 15 laur pseu noDet Pteropus vampyrus (bat) ABRP01232219 >PRDM7_pteVam 7 laur gene GAS8+ Pteropus vampyrus (bat) ABRP01250178 >PRDM7_myoLuc 6 laur gene gas8+ Myotis lucifugus (bat) AAPE02062260 >PRDM7_equCab 4 laur gene GAS8+ Equus caballus (horse) genome >PRDM7_sorAra 8 laur gene noDet Sorex araneus (shrew) AALT01000095 >PRDM9a_loxAfr 12 afro gene noDet Loxodonta africana (elephant) genome >PRDM9b_loxAfr 3 afro pseu noDet Loxodonta africana (elephant) genome >PRDM7_loxAfr 5 afro pseu GAS8+ Loxodonta africana (elephant) genome >PRDM7_echTel 5 afro pseu noDet Echinops telfairi (tenrec) genome >PRDM7_proCap 13 afro gene noDet Procavia capensis (hyrax) ABRQ01392668
Comparative genomics of PRDM9 and PRDM7
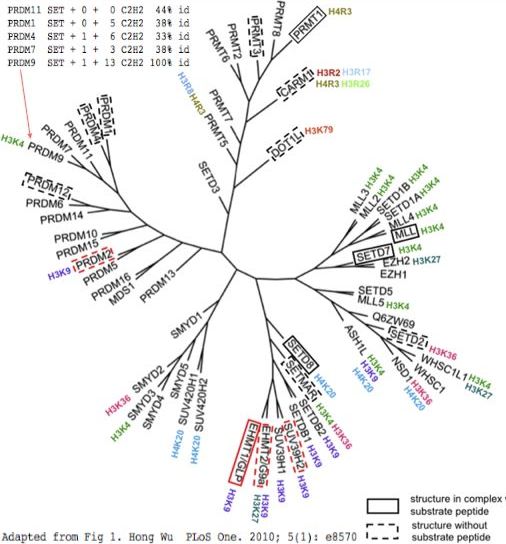
PRDM9 is one of many human proteins sharing a set of common domains, as well as various multiplicities of the zinc finger domain C2H2. The diagram at left shows an effort at organizing these into phylogenetic tree according to structural considerations of the SET domain these proteins all share.
The traditional SET domain is too small for an enzyme with distinctive substrates so flanking sequence must be added despite its lack of apparent conservation. Using S-adenosyl methionine, PRDM9 places the third methyl group only on the fourth position arginine in histone H3, one of many such epigenetic methylases in the human genome. The histone recognized by such methylases correlates poorly with evolutionary grouping by SET domain (figure).
The upper left corner shows the variability in domain structure. While PRDM9 and PRDM7 share the same domains (an upstream KRAB domain is not shown), of PR-class homologs, PRDM11 shares only the SET domain despite nesting deep within the PRDM9 subtree. PRDM4 has both the SET and C2H2 domains, possibly sharing the early C2H2 domain in an exon beginning with a phase 2 splice acceptor (as shown in reference sequence section). Overall however, PRDM9 and PRDM7 have no full length homologs with matching exon structure. Even the SET domain is intronated differently within PR-class proteins (with the sole exception of PRDM11), suggesting either ancient divergence or unusual evolution. These incongruities may have arisen from domain shuffling, gain and loss.
The human PRDM9 sequence below is annotated in color for domains relative to exon breaks. The protein can be best understood in terms of concatenated domains, not all of which may be present in antecedent and descendant homologs. The first two domains KRAB and SSXRD interact with transcription factors.
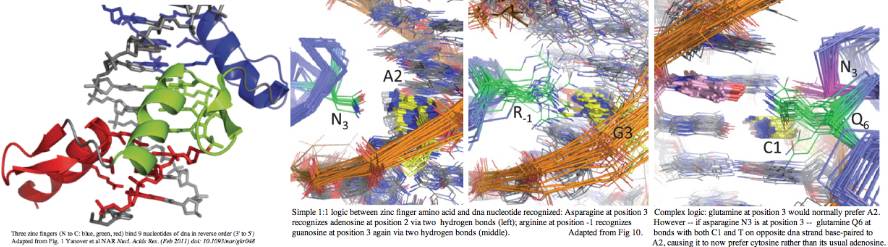
Each C2H2 domain -- so named for two cysteines and two histidines liganding to a structural zinc ion -- recognizes a specific trinucleotide (more or less) and so concatenated in a large array recognize specific binding sites along the genome, though tolerance of nucleotide variability and synergistic effects between adjacent units make it difficult to read out these sites precisely, despite immense efforts.
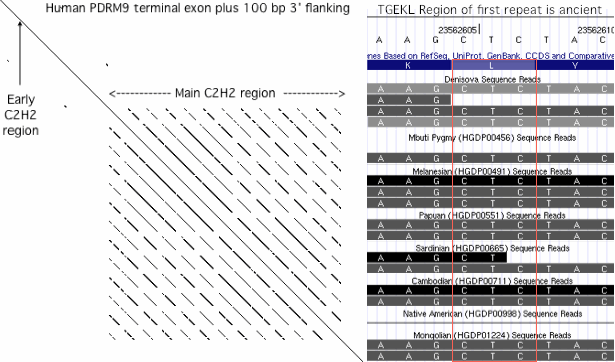
The concatenated C2H2 domains, conserved at the amino acid level so necessarily similar at the dna level, are prone to replication slippage. This process can give rise to point mutations as well as leading to a peaked distribution of repeat number rather than to a single number. Many other unrelated genes with internal repeats (such as the octapeptide region of the prion gene PRNP) are also affected by replication slippage. Such proteins regions are conveniently identified genomewide by mRNA dot plots.
The C2H2 domains generally reside in a long distinctive terminal exon of splicing phase 2 that has been shuffled over mammalian evolutionary time into various contexts. Concepts such as paralogy and orthology need piecewise definitions in these composite proteins. Synteny (gene adjacency) plays a major role in reliably deconstructing events in specific lineages.
Here the unrelated single-copy conserved gene GAS8 plays an important role. PRDM7 occurs immediately distal to it on the negative strand, making the two genes are convergently transcribed). PRDM7 is otherwise the last gene on the q arm of its chromosome in many species which may predispose it to copy number dispersal events. PRDM9 is not consistently located within placental mammals, suggesting independent relocation events.
Both PRDM9 and PRDM7 contain a seldom-mentioned C2H2 domain early in the exon annotated by SwissProt and readily found by the online domain tools regardless of species. This domain conserves the four critical residues needed for zinc binding (and so the associated fold) but lacks the terminal cap TGEKP which otherwise serves to lock down a C2H2 zinc finger after it has scanned along genomic dna to an appropriate trinucleotide. The function of this early domain and the following 112 residues are unknown -- no homologous 3D structure has ever been determined.
The first C2H2 of the main repeat region is proximaly degenerate, beginning in VKY in all species (instead of YCE). The tyrosine cannot plausibly replace the usual cysteine for zinc binding though the other three needed residues are present. This domain ends in a typical cap region TGEKP. Humans are the exception here where the conserved helix-ending proline has been replaced with leucine in the reference human genome with unknown functional consequences.
>PRDM9_homSap Homo sapiens (human) Q9NQV7 10 exons chr5:23,509,579 span 18,301 bp KRAB SSXRD SET C2H2 cap
0 MSPEKSQEESPEEDTERTERKPM 0
0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1
2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1
2 VKPPWMALRVEQRKHQK 0
0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1
2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1
2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0
0 ITKGRNCYEYVDGKDKSWANWMR 2
1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1
2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK
VKYGECGQGFSVKSDVITHQRTHTGEKL
YVCRECGRGFSWKSHLLIHQRIHTGEKP
YVCRECGRGFSWQSVLLTHQRTHTGEKP
YVCRECGRGFSRQSVLLTHQRRHTGEKP
YVCRECGRGFSRQSVLLTHQRRHTGEKP
YVCRECGRGFSWQSVLLTHQRTHTGEKP
YVCRECGRGFSWQSVLLTHQRTHTGEKP
YVCRECGRGFSNKSHLLRHQRTHTGEKP
YVCRECGRGFRDKSHLLRHQRTHTGEKP
YVCRECGRGFRDKSNLLSHQRTHTGEKP
YVCRECGRGFSNKSHLLRHQRTHTGEKP
YVCRECGRGFRNKSHLLRHQRTHTGEKP
YVCRECGRGFSDRSSLCYHQRTHTGEKP YVCREDE* 0
-1 23 6 traditional numbering of dna recognizing amino acids
HPCPSCCLAFSSQKFLSQHVERNH alignment of early C2H2 domain
* * * * zinc liganding positions
Only in PRDM11 (and PRDM1 to a lesser extent) is the SET domain intronated like PRDM9 and PRDM7: >PRDM9_homSap 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 >PRDM11_homSap intronation of SET domain 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGSDETKANWMR 2 1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR 1 >PRDM1_homSap intronation of SET domain 0 AAPKCNSSTVRFQGLAEGTKGTMKMDMEDADMTLWTEAEFEEKCTYIVNDHPWDSGADGGTSVQAEASLPRNLLFKYATNSEE 0 0 VIGVMSKEYIPKGTRFGPLIGEIYTNDTVPKNANRKYFWR 0 0 IYSRGELHHFIDGFNEEKSNWMRYVNPAHSPREQNLAACQNGMNIYFYTIKPIPANQELLVWYCRDFAERLHYPYPGELTMMNL 1 >PRDM4_homSap intronation of SET domain 2 WCTLCDRAYPSDCPEHGPVTFVPDTPIESRARLSLPKQLVLRQSIVGAEV 1 2 GVWTGETIPVRTCFGPLIGQQSHSMEVAEWTDKAVNHIWK 0 0 IYHNGVLEFCIITTDENECNWMMFVRKAR 2 1 NREEQNLVAYPHDGKIFFCTSQDIPPENELLFYYSRDYAQQI 1
Divergence of SET domains:
Different segmental duplications relate PRDM9 and PRDM7
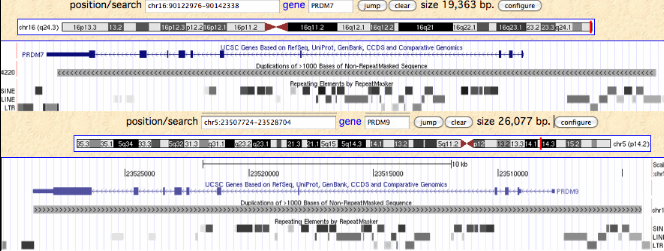
In humans, PRDM9 and PRDM7 are related by a 26 kbp segmental duplication that begins about 8 kbp upstream of the start codon and continues through most of the 3' UTR. Since the retroposon patterns are nearly identical, the duplication must be fairly recent. The overall percent identity of non-coding dna is about 93%, again inconsistent with either early (within stem placental or late divergence (post-chimpanzee). The duplication contains a potentially diagnostic 1845 bp retroposon-free region upstream of the first coding exon.
Note PRDM7 is situated at the extreme tip of chromosome 16q, perhaps predisposing it to chromosomal copy number rearrangements. The syntenic context is TUBB3+ DEFB+ AFG3L1+ DBNDD1- GAS8+ PRDM7- qTel, meaning it is transcribed convergently with GAS8, a non-homologous highly conserved single copy gene often detectable even in low coverage genomes in the small contig containing PRDM7. This association has been extremely stable over boreoeutheran placental mammal evolutionary time and so serves to reliably define PRDM7 orthologs and their spin-off copies. Elephants also have a gene pair similar to human PRDM9 and PRDM7. The former is at a syntenically novel site but the latter is an old pseudogene but still detectably adjacent to GAS8 in opposite orientation. It thus follows that 'PRDM9' in elephant is an independent earlier spin-off of its conventional PRDM7 gene. This is consistent with telomeric susceptibility to repeated rearrangements.
Recall here the actual definition of gene orthology: two genes in two species are orthologous if they are vertically descended from the same gene in their last common ancestor. Here the LCA of human and elephant is ur-placental mammal which had PRDM7 but no PRDM9. The two PRDM9 genes are thus not descended from a common ancestral PRDM9 gene but from parallel gene duplications of a common PRDM7 gene at different times in different clades during the course of mammalian speciation. Such genes are called in-paralogs within a given species and co-orthologs across them.
The syntenic context of PRDM9 is quite variable, supporting the scenario of multiple origins. This context can be used to count the number of distinct segmental duplications of PRDM7. For example, in humans, PRDM9 basically lies in a retroposon-rich gene desert but is eventually flanked by two pairs of cadherin genes at the much larger scale of 7 mbp. In rhesus, these same genes are seen (with some minor rearrangements), establishing that this PRDM9 segmental duplication preceded the divergence of old world monkeys.
Marmoset has a seemingly functional PRDM7 in the usual position facing GAS8, still at the extreme end of chromosome 20. The cadherin cluster is intact on chr2:178,954,165-180,696,523. However Blastx of the intervening dna -- which is similar in size to rhesus and human so not suggesting large deletions -- shows not even a suggestion of an old PRDM9 pseudogene. The assembly is gapless here. and Blastx is sensitive enough to detect very old pseudogenes provided they decayed by small indels and nucleotide substitutions. Thus it appears that PRDM7 never duplicated in marmoset -- placing that even in the stem to old world monkeys (or prior to tarsier divergence -- that assembly has poor coverage). Note that the marmoset PRDM7 has a respectable terminal zinc finger array of twelve units, enough to specify 36 bp.
Gene Strand Protein Start Species CDH18 - cadherin 18 19981287 homSap ponAbe macMul CDH12 - cadherin 12 22853731 homSap ponAbe macMul calJac PRDM9 + human PRDM9 23528704 homSap ponAbe macMul calJac CDH10 - cadherin 10 24644911 homSap ponAbe macMul calJac CDH9 - cadherin 9 27038689 homSap ponAbe macMul
Lemurs present a new complication. The Otolemur assembly has two distinct and seemingly functional PRDM7 copies (each with seven zinc fingers) containing GAS8 end-sequence in expected opposite orientation. One of the GAS8 copies appears to be a pseudogene. This represents a new type of lineage-specific segmental duplication. There is no sign of PRDM9. The other lemur with an assembly, Microcebus murinus, has but a single copy, again with seven zinc fingers. The only relevant contigs (ABDC01433247 and ABDC01371462) contain no coding syntenic information so this gene cannot be assigned to PRDM7 with certainty.
The tree shrew assembly, like tarsier, has low coverage and only blast matches to zinc finger arrays that cannot be assigned to the PRDM family. This cannot be totally attributed to low coverage because many ordinary genes are satisfactorily represented in these species. Other issues such as telomeric position, gene copy number (mobility), pseudogenization, deletional loss, chimerization, and individual heterozygosity must be affecting recovery of PRDM9 gene models in these species.
Moving on to laurasiatheres, Bos taurus presents a much more complicated situation. First, the GAS8 locus on chr18 contains the first two exons of a PRDM7 pseudogene in expected orientation but distal regions of the gene are completely deleted. The cadherin locus on chr20 is also intact but the 2.6 mbp region between CDH12 and CDH10 contains no indication of PRDM9, consistent with that segmental duplication being primate-specific and PRDM7 being the older parental location. This holds in the Baylor 4.0 assembly carried at UCSC, the Baylor 4.2 assembly, and the alternative assembly of the same data, UMD3.1. The latter two can be queried by the genomic blast server at NCBI.
A third locus on chr 1 hosts an unreviewed GenBank pipline entry called PRDM9, derived as NW_003053109 from the alternative bovine assembly UMD3.1 Staff corrected an unspecified frameshift to fix the reading frame -- a dangerous practise in a gene family so prone to pseudogenization. The gene, called PRDM9a here, resides on the extreme end of chromosome 1 and differs from the Baylor 4.0 assembly at two amino acids outside the zinc finger region. The syntenic context here is novel: EFHB- RAB5A+ PCAF+ ZNF596- PRDM9a- which corresponds overall to human chr 3. The juxtapositioning of two zinc finger proteins on the same strand causes PRDM9 alignments to extend spuriously into the 12 zinc fingers of ZNF596, jumping over its 5 earlier coding exons.
ZNF596 contains a KRAB domain but no SET methylase. Humans encode a best-blast protein of the same assigned name on chr 8 (77% identity). Note the early exons of ZNF596 can be added to end of PRDM9a to form an artificial probe for this association in other species, though the two genes have a 43,400 bp spacer in cow, which is large relative to contig size in low coverage assemblies. The sole fragmentary transcript from yak testis (EF432551) is nearly identical to this PRDM9a, suggesting that the gene -- and perhaps its syntenic location -- became established prior to yak-cow divergence and is still functional. However its array of seven zinc fingers could recognize at most a region of 21 bp.
ZNF596 did not arise from a PRDM9-like gene through loss of the SET domain, though it is one of the better matches within the large zinc finger family. Excluding the zinc finger domain, ZNF343, ZNF133 and ZNF169 provide much higher blastp scores, as they also do just comparing the zinc finger arrays. The juxtaposition of ZNF596 and PRDM9a is likely coincidental rather than a consequence of inhomogeneous recombination between zinc fingers bringing PRDM9 to this site.
The fourth PRDM9 locus of interest, called here PRDM9b, is still not mapped to any bovine chromosome. It resides in contig DAAA02065087 in the UMD3.1 assembly and is temporarily assigned to chr Un.004.649 at Baylor assembly. Here the reading frame in exon two can be restored if a run of 5 A's is corrected to 6 A's. That is done here in the reference sequences because this is typically just sequencing error. The protein has a full set of domains KRAB SSXRD SET C2H2 with a moderate zinc finger array of five. Synteny cannot be determined in chr Un features which can simply pool unrelated unplaceable contigs into a manageable unit. Flanking dna in DAAA02065087map to several places in the cow genome, suggesting this feature has copy number attributes, perhaps of telomeric repeat type. PRDM9b is not a recent feature because it differs at a considerable number of amino acids from other PRDM9 in the cow genome. These substitutions avoid highly conserved residues, not consistent with early pseudogenization. PRDM9b is capable of histone marking but it is not clear whether that has functional significance to meiosis.
Yet another locus in the Baylor 4.0 assembly, called PRDM9c here, could not initially be placed on a cow chromosome. While such features are often assembly artefacts, this one is supported by a transcript from 4-cell embryos (GO353654) consistent with a role in or after meiosis. In UMD3.1, this gene has been placed on chr X. Despite a very large contig, no zinc fingers occur in any reading frame, suggesting that the gene was transferred here without the last exon (or it subsequently got deleted). In any event, the penultimate exon does not have a phase 1 splice donor in expected position and so terminates at the next stop codon downstream. The protein retains the KRAB, SSXRD and SET domains but does not possess the ability to scan or bind dna. It has accrued various amino acid substitutions relative to other bovine that rule out recent establishment.
Finally, two additional genes, denoted PRDM9d and PRDM9e here, are located as a parallel tandem pair in a higher quality region of bovine chr X. These are 96% identical as proteins, consistent with one being derived fairly recently from the other. Synteny here will not be informative until other ruminant genomes become available.
Overall the situation in cow is very different from primates and rodents. Results there about the function of single-copy autosomal PRDM9 gnes in meiosis markup can scarcely be carried over to a species with five seemingly intact genes, three of which are on chr X (which intriguingly has the very limited pseudoautosomal region on chr Y where it can cross over).
The cow situation cannot be limited to the Hereford breed used for the genome project because the PRDM9 are too diverged from one another outside the zinc finger region. Indeed there is some suggestion from non-NCBI sheep genome that it too has many of these copies. However other cetartiodactyl genomes (dolphin, pig and alpaca) and other laurasiatheres (panda, dog, cat, shrew, bats) do not show these copies, suggesting that this complexity could be limited to pecoran ruminants. All-vs-all blastp percent identities are consistent with this, though rates of evolution in this gene family are hardly typical.This cannot be resolved with cow genome alone -- there is no good candidate still present for parent gene to all these copies. These results are summarized in the table below:
Gene #ZNF Status Chr Synteny cDNA Accession 9a_bosTau 9b_bosTau 9e_bosTau 9a_oviAri 9a_turTru 7_ailMel PRDM7 - pseudo 18 GAS8 no none -- -- -- -- -- -- PRDM9a 7 ok 1 ZNF596 yes NW_003053109 100% 85% 81% 82% 76% 72% PRDM9b 5 ok ? not det no DAAA02065087 81% 100% 78% 79% 72% 68% PRDM9c 0 ok X not det yes XM_002699750 80% 80% 82% 83% 74% 73% PRDM9d 9 ok X --- no none 80% 78% 96% 93% 73% 67% PRDM9e 9 ok X --- no none 81% 78% 100% 93% 73% 68%
The role of CpG mutations
Human PRDM9 has 39 CpG sites in its coding exons, potentially mutational hotspots. After attempted dna repair, these usually resolve to CpA or TpG. If not at a synonymous site, these changes alter the encoded amino acid. Some 28 of the CpG sites are at arginine CGn codons (of which the protein has 90 overall). These always result in a substitution: for G -> A, histidine for CGT and CGC and glutamine for CGG and CGA; for C -> T, cysteine for CGT and CGC and tryptophan and stop codon for CGG and CGA. These changes are in fact seen in many of the reference sequences. The display below shows wildtype human PRDM9 in the top lines and the effects of G -> A and C -> T in the next. The zinc finger motif is highlighted.
PRDM9_homSapWT MSPEKSQEESPEEDTERTERKPMVKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITIGLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQVKPPWMALRVEQRKHQKGMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKLELRKKETERKM PRDM9_homSapCA Q H Q Q H PRDM9_homSapTG W C * * C V PRDM9_homSapWT YSLRERKGHAYKEVSEPQDDDYLYCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWLITKGRNCYEYVDGKDKSWANWMRYVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDE PRDM9_homSapCA Q K H Q K Q Q PRDM9_homSapTG * L C L * W * PRDM9_homSapWT YGQELGIKWGSKWKKELMAGREPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAKVKYGECGQGFSVKSDVIP PRDM9_homSapCA S H H PRDM9_homSapTG C C PRDM9_homSapWT THQRTHTGEKLYVCRECGRGFSWKSHLLIHQRIHTGEKPYVCRECGRGFSWQSVLLTHQRTHTGEKPYVCRECGRGFSRQSVLLTHQRRHTGEKPYVCRECGRGFSRQSVLLTHQRRHTGEKPYVCRECGRGFSWQSVLLTHQRTHTGEKPYVCRECGRGFSWQSVLLTHQRTHTGEKPY PRDM9_homSapCA I Q Q Q Q Q Q Q Q PRDM9_homSapTG W W W W W W W W PRDM9_homSapWT VCRECGRGFSNKSHLLRHQRTHTGEKPYVCRECGRGFRDKSHLLRHQRTHTGEKPYVCRECGRGFRDKSNLLSHQRTHTGEKPYVCRECGRGFSNKSHLLRHQRTHTGEKPYVCRECGRGFRNKSHLLRHQRTHTGEKPYVCRECGRGFSDRSSLCYHQRTHTGEKPYVCREDE PRDM9_homSapCA Q Q HN Q Q Q H I Q N I PRDM9_homSapTG W W C W W W C W
Structural considerations in C2H2 zinc fingers
High resolution structures of C2H2 zinc finger domains have been available for decades. As the name suggests, the divalent zinc atom locks the two cysteines and two histidines into a rigid geometry providing a core conformation that a small peptide of 28 residues could not otherwise stably assume. Note in the unbound state, finger tips must retain flexibility while the domain ensemble scans its genome for specific dna sequences appropriate to its function. Each finger binds a trinucleotide -- in effect making a zinc finger the protein counterpart to tRNA anticodon. However overall binding is not a simple read-off code because adjacent fingers alter each other's specificities in subtle ways.
The linker region TGEKP plays a key role when the correct DNA sequence is encountered, snap-locking its finger down onto its target by capping the C-terminus of its alpha helix. A hydrogen bond between the first threonine and middle glutamate is key to this binding-induced conformational shift. From comparative genomics, it appears that a serine in first position can also form this hydrogen bond. The role of the glycine is to stay out of the way; the lysine counterbalances the negative charge of the glutamate; the proline terminates any helical propensity, allowing a fresh start in the adjacent finger.
While this motif is immensely conserved within C2H2 zinc finger of PDRM9 homologs, exceptions do occur. It is important to understand these because these loss of dna lock-down could loosen or even eliminate trinucleotide binding specificity. Such steps might represent initial stages of pseudogenization. However many exceptions occur within the first or last fingers. It is also common for fragmentary and imperfect motifs to end the protein, sometimes continuing on in another reading frame past the current stop codon.
Note in aligning zinc finger motifs, the breaks should always be put at the end of the linker region. It is completely illogical to break at the first cysteine as some authors do because capping by the linker region is specific to its zinc finger, not the following one.
Predicting dna binding sites of zinc finger domains
Curated reference sequences
The sequences below have been compiled from genome projects -- only rarely do validating transcripts exist at GenBank. Sequences with a single frameshift or other glitch have been edited to allow full length proteins on the theohry that the error either reflects an aberrant atypical individual chosen for sequencing or simple error in low coverage projects within a difficult repeat region. However such sequences may instead reflect early stages of pseudogenization. Many sequences are in fact clearly pseudogenes; here recognizable exons have been collected to allow rough dating of loss of function.
In the case of more intensively studied species such as human and mouse, the number of C2H2 repeats varies widely. Only the most common representative is shown here. This variation likely occurs in all species but the individual animal chosen for sequencing may or may not be typical. Many clades have distinctive patterns of gene amplification and gene loss, making both orthologous and functional comparisons problematic.
Other useful sequences such as the GAS8 synteny neighbor, other zinc finger quasi-homologs having similar exon and domain structures, and bogus orthologs outside of mammals are also included for reference purposes.
Carnivores -- but not bats or horses -- have an intervening cadherin gene before GAS8: PRDM7_ailMel 1724 1 579 579 100.0% GL193502.1 +- 628987 644235 15249 CAD1_homSap 185 133 334 882 72.9% GL193502.1 +- 620344 624223 3880 GAS8_homSap 1110 2 478 478 91.0% GL193502.1 ++ 594843 609901 15059 PRDM7_canFam 681 141 880 884 82.3% 5 ++ 66560684 66567275 6592 CAD1_homSap 368 134 521 882 74.7% 5 ++ 66571832 66581008 9177 GAS8_homSap 1188 2 478 478 93.4% 5 +- 66587321 66604940 17620 PRDM7_felCat 707 337 572 572 100.0% Un_ACBE01450414 +- 10493 13105 2613 CAD1_homSap 130 133 223 882 74.7% Un_ACBE01450414 +- 3902 4280 379 PRDM7_equCab 1294 1 435 435 100.0% 3 +- 36378853 36387224 8372 GAS8_homSap 1176 2 478 478 93.0% 3 ++ 36348528 36361906 13379
>PRDM9_homSap Homo sapiens (human) genome Prim gene 13 CDH12 chr5 10 exon size 18,301 bp KRAB SSXRD SET C2H2 0 MSPEKSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKL YVCRECGRGFSWKSHLLIHQRIHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRDKSHLLRHQRTHTGEKP YVCRECGRGFRDKSNLLSHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRNKSHLLRHQRTHTGEKP YVCRECGRGFSDRSSLCYHQRTHTGEKP YVCREDE..................... >PRDM9_panTro Pan troglodytes (chimp) genome Prim gene 19 CDH12 chr5 frag assembly glitch in mid C2H2 0 MSPERSQEESPEGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWgKTRYRiVKMNYNALITi 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRGEQSKHQK 0 0 GMPKASFNNESSLkELSGmPNLLNTSgSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRRKETvGKMYSLRERKGHAYKEISEPQDDDYL 1 2 yCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLRVWNEASDPPLGLHSGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSwANWMR 2 1 YENCARDDEEQNLVSFQYHRQSFYRTCRVIRPGCELLVWYGDE GQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPRSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTAKLFVGVGISRIAK VKYGECGQGFSDKSDVITHQRTHTGGKP YVCRECGRGFSWKSHLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHRTTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSQQSNLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSKQSHLLSHQRTHTGEKP YVCRECGRGFSVQSNLLSHQRTHTGEKL YVCRECGRGFSQQSHLLRHQRTHTGEKP YVCRecgrgfsqqshLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSKQSHLLSHQRTHTGEKP YVCRECGRGFSQQSHLLSHQRTHTGEKP YVCRECGRGFSQQSHLLRHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECERGFSQQSHLLRHQRTHTGEKP YVCRECGRGFSRQSALLIHQRTHTGEKP VCREDE...................... >PRDM9_gorGor Gorilla gorilla (gorilla) CABD02290264 Prim gene -- cdh12 chr5 several contigs needed, most of ZNF domain missing 0 MSPERSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPCMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRKKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 yCEMCQNFFIDSCAAHGPPIFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYKGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARTLLQPENPCPGDQNQEQQYPDPRSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESR TGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKP YVC......................... >PRDM9_ponAbe Pongo abelii (orangutan) genome Prim gene 10 CDH12 chr5 frameshift extra a penultimate ZNF 0 MSPERSQEESPkGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWTEMGDWEKTRYRNVKRNYKTLITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRGEQSKHQK 0 0 GMPKASFNNESSLKELSGTQNLLNTSGSEQAQKPVSPPGEASTSGQHSTLKI 1 2 ELRRKETEGKTYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCAWDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMPGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNHEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVITHQRTHTGGRS YVCRECGRGFSRQSVLLIHQRTHTGEKP YVCRECGRGFSRRSVLLIHQRTHTGEKP YVCRECGRGFSQQSVLLIHQRTHTGEKP YVCRECGRGFSRRSVLLIHQRTHTGEKP YVCRECGRGFSWKSVLLRHQRTHTGEKP YVCRECGRGFSQQSVVFIHQRTHTGEKP YVCRECGRGFSGKSVLFRHQRTHTGEKP YVCRECGRGFSDKSGVCYHQRTHTRGEA YVCRECGRGFSVKSNLLSHQRTHTEEKL YVCREDE..................... >PRDM9_nomLeu Nomascus leucogenys (gibbon) ADFV01015315 Prim gene 10 cdh12 ADFV01015317 ADFV01015319 no CDH but best blastn favors PRDM9 0 MSPERSQEESPEEDTERTEQKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 - 0 0 - 1 2 - 1 2 AAHGPPTFIKDSTVGKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 - 1 2 EPKAEIHPCPSCCLAFSSQKFLSQHVARHHSSQNFPGPSARKFLQPENPCPGDQNQEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWQREISRAFSSSPKVQMGSCRVGKRIIEESRTGQKVNPGNTGQLFVGVGISRIAE VKYGECGQGFSVKSDVITHQRTHTGEKL YLCRECGRGFSVKSSLLSHQRTHTGEKP YVCRECGRGFSKKSNLLSHQRTHTGEKP YVCRECGRGFSDKSSLLRHQRTHTGEKP YVCRECGRGFSQKSSLLSHQRTHTGEKP YVCRECGRGFSQKSSLLSHQRTHTGEKP YVCRECGRGFSDKSSLLRHQRTHTGEKP YVCRECGRGFSQKSSLLSHQRTHTGEKP YVCRECGRGFSVKSNLLSHQRTHTGEKP YVCRECGRGFSDKSSLLRHQRTHTGEKP >PRDM9_macMul Macaca mulatta (rhesus) genome Prim gene 9 CDH12 chr6 exon 4 lost to Ns 0 MSPERSQEESPEEDTERTERKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 - 0 0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITQDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSTQNFPGPSARRLFQPENLCSGDQNQEQQYSDPRSCNDKTKGQEIKERSKLLNKRTWPKEISRAFSSPPKGQMGSSRVGERMMEEEYRTGQKVNPENTGKLFVGVGISRIAK VKYGECGQGFSDKSDVIIHQRTHTGEKP YLCRECGRGFSQKSSLRRHQRTHTGEKP YLCRECGRGFRDNSSLRYHQRTHTGEKP YLCRECGRGFSNNSGLCYHQRTHTGEKP YLCRECGRGFSDNSSLHRHQRTHTGEKP YLCRECGRGFSNNSGLRYHQRTHTGEKP YLCRECGRGFSNNSGLRHHQRTHTGEKP YLCRECGRGFSQKANLLRHQRTHTGEKP YLCRECGRGFSQKADLLSHQRTHTGEKP VCRKDE...................... >PRDM9_papHam Papio hamadryas (baboon) genome Prim gene 11 cdh12 contigs scattered 0 - 0 0 - 1 2 - 1 2 VKPPWMAFRVEQSKHQK 0 0 EMPKTSFSNESSLKELSGTPNLLSTSGSEQAQKPASPPGEASTSGQHSRLKL 1 2 ELRRKEAEGKMYSLRERKGHAYKEVSELQDDDYL 1 2 ycEMCQNFFIDSCAAHGPPTFVKDSAVNKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 - 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSTQNFPGPSARRLLQPENLCSGDQNQEQQYSDPCSCNDKTKGQEIKERSKLLNKRTWQKEISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPENIGKLFVEVGISRIAK VKYGECGQGFSGKSDVITHQRTHTEGKP YLCRECGRGFSQKSNLLRHQRTHTGEKP YLCRECGRGFRDNSSLRCHQRTHTGEKP YLCRECGRGFRDNSSLRCHQRTHTGEKP YLCRECGRGFSDNSSLRYHQRTHTGEKP YLCRECGRGFRDNSSLRYHQRTHTGEKP YLCRECGRGFSVKSNLLSHQRTHTGEKP YVCRECGRGFSDNSSLRCHQRTHTGEKP YLCRECGRGFSQMSHLRCHQRTHTGEKP YLCRECGRGFSVKSNLLSHQRTHTGEKP YVCRECGRGFSRKANLLSHQRTHTGEKP >PRDM7_homSap Homo sapiens (human) genome Prim gene 3 GAS8+ chr16 TUBB3+ DEFB+ AFG3L1+ DBNDD1- GAS8+ PRDM7- 92% id 0 MSPERSQEESPEGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKMNYNALITV 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAFRGEQSKHQK 0 0 GMPKASFNNESSLRELSGTPNLLNTSDSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEISEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSSANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQERQYSDPRCCNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVITHQRTHTGGKP YVCRECGRgFSRKSDLLSHQRTHTGEKP YVCRECERGFSRKSVLLIHQRTHRGDAP VCRKDE...................... >PRDM7_panTro Pan troglodytes (chimp) genome Prim pseu 2 GAS8+ chr16 0 MSPERSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPLMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELKKKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYKGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLQPENP PGDQNQERQYSDPRCCNDKTKGQEVKERSKLLNKWTWQREISRAFSSLPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKP YVCRECGQGFSRKSVLLIHQRTHRGEKP VCRKDE...................... >PRDM7_gorGor Gorilla gorilla (gorilla) genome Prim pseu 3 GAS8+ chr15730 numerous frameshifts in terminal ZNF domain 0 MSPERSQEESPEGDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATQPVFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 - 0 0 GMPKASFNNESSLKELSGTPNLLNTSGSEQAQKPVSPPGEASTSGQHSRRKL 1 2 ELRRKETEGKMYSLRERKGHAYKEISKPQDDDYL 1 2 yCEMCQNFFIDSCAAHGPPTFVKDSAVDKRHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDKEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVALQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQERQYSDPRCCNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSWKSNLLRHQRTHTGGKP YVCRECGRGFSWKSDLLSHQRTHTGEKP YVCRECGRGFSWKSNLLSHQRTHTGEKP >PRDM7_ponAbe Pongo abelii (orangutan) genome Prim gene 4 GAS8+ chr16 0 MSPERSQEESPEDDTERTERKPT 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 0 0 GMPKASFNNESSLKELSETANLLNASGSEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRSKETEGNTYSLRERKGHAYKEISEPQDDDYL 1 2 yCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALTLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITKDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARHLLQAENPCPGDQNQEQQYSDPDCCNDKTKGQEIKERSKLLNKRTWQREISRAFSSSAKGQMGSSRVGERMMEEESGTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKP YICRESGRGFTQKSGLLSHQRTHTGEKP YVCRECGWGFSQKSNLLRHQRTHTGEKP YVCRECGRGFSRKSVLLIHQRTHTGEKP VCRKDE...................... >PRDM7_nomLeu Nomascus leucogenys (gibbon) ADFV01125891 Prim pseu 5 gas8+ synteny implied by non-coding 0 - 0 0 - 1 2 - 1 2 IKSPWMAVRVEQSKHQK 0 0 GMPKASFNNESGLKELSGTQNLLNTSG EQARKPVSPPGEASTSGQHSRQKL 1 2 ELRRKETEGKMYSL ERKGHAYKEVSEPQDDDYL 1 2 yCEMCQNFFTDSCAAHGPPTFVKDSAVDKGHPNHSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKS ANWMK 2 1 YVNCARDHEEQNLVAFQYHRQIFYRTCQVIRPGCEPLVWYGDEYGQELGIKWGSKWKKELTAER 1 2 EPKPEIHPCPSCCLVFTSQKFLSQHVECNHSSQNFPGPSARKLLQRENPCPGDQNQEQQYSDSRSCNDKTKGQEIKERSKL NKRIWQRKISRAFSSLPKGQMGSSRVGERMMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSDKSDVIAHQGTHTGGKS .ICRECGWGFSQESHLLIHQRTHTGEKL YVCRECGQGFSQKSDLLSHQRTHTGEKP YVRRECGRGFSQKSNLLSHQRTHTEEKP YVCRECGWGFSQKSHLLIHQRTHTGKKP VCRKDE...................... >PRDM7_macMul Macaca mulatta (rhesus) genome Prim pseu 2 GAS8+ chr20 frameshifts exon 5 and 10, exon 10 a to aa restores frame 0 - 0 0 - 1 2 - 1 2 VKPPWMAFRVEQSKHQK 0 0 EMPKTSFNNESSLKELSGTPNLLSTSDSE AQKPASPPGEASTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKRHAYKEASELQHDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDNAVNKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPCEGRITEDKEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWAKWMR 2 1 - 1 2 EPKPEIYPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQSENPCPGDQNQEQQYSDPSSCNDKTKGQEIKERSKLLNKRTWQREILRAFTSPPKGQMGSSRVGERMMEEEFRTGQKANPGNTGKLFVGVEISRIAK VKYGECGQGFSGKSDVITHQRTHTEGKP YVCRGCGRRFSQKSSLLRHQRTHTGEKP VCKKNE...................... >PRDM7_papHam Papio hamadryas (baboon) genome Prim pseu 2 gas8+ contigs scattered 0 MSPERSQEESPEEDTERTEWKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMAVRVEQSKHQK 0 0 GMPKASFNNESSLKEVSGMANLLNTSGSEQAQKPVSPPGEARTSGQHSRLKL 1 2 ELRRKETEGKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFIKDSAVEKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITQDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 EPKPEIYPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQSENPCPGDQNQEQQYSDPSSCNDKTKGQEIKERSKLLNKRTRQRQILRAFTSPPKGQMGSSRVGERMMKEEFRTGQKANPGNTGKLFVGVEISRIAK VKYGECGQGFSDKSDVVIHQRTHTREKP YVYRgCGQGFSIKSNLLRHQRIHTGEKP >PRDM7_calJac Callithrix jacchus (marmoset) genome Prim gene 12 GAS8+ chr20 one frameshift in repeat area chr20 terminus 0 MSPERSQEESPEGDTGRTEQKPM 0 0 VKDAFKDISMYFSKEEWAEMGDWEKTRYRNMKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPGMAFRVGQSKHQK 0 0 GMPKASFGNESSLKKLSGTANVLNTSGPEQAQKPVSPPGEASTSGQHSRLKL 1 2 ELRRKDTEEKMYSLRERKGLAYKEVSEPQDDDYL 1 2 yCEICQNFFIDSCAAHGPPTFVKDSAVDKGHPNHAALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRVTEDEEAASSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 ESKPEIHPCPSCCLAFSSQKFLSHHVERNHSSQNFPGTSTRKLLQPENPCPGKQKEEQQYFDPCNSNDKTKGQETKERSKLLNIRTWQREMARAFSNPPKGQMGSSRVEERMMEEESRTGQKVNPVDTGKLFVGVGISRIAK AKYGECGQGFSDMSDVTGHQRTHTGEKP YVCRECGRGFSQKSALLSHQRTHTGEKP YVCRECGRGFSQKSHLLSHQRTHTGEKP YVCTECGRGFSQKSVLLSHQRTHTGEKP YVCTECGRGFSRKSNLLSHQRTHTGEKP YVCRECGRGFSRKSALLSHQRTHTGEKP YVCRKCGRGFSQKSNLLSHQGTHTGEKP YVCTECGRGFSQKSHLLSHQRTHTGEKP YVCRKCGRGFSQKSNLLSHQRTHTGEKP YVCRECGRGFSFKSALLRHQRTHTGEKP YVCRECGRGFSRKSHLLSHQGTHIGEKP YVCRECGRGFSRKSNLLSHQRIHTGEKP YVRREDE..................... >PRDM7_micMur Microcebus murinus (lemur) ABDC01433247 Prim gene 8 gas8+ weak coverage 0 MSPEKSQEESPEEDTERTERKPM 0 0 vKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 0 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEKCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLKIRPSGIPQAGLGVWNEASELPLGLHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDDSWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCQVIRPGCELLVWYGDEYGQELGIKWGSKWKEELTIRQ 1 2 EPKPEIHPCPSCSLAFSSQKFLSQHVKHTHSSQISPRTSGRKHLQPENPCPGDQNQEQQHSDPHSCNDKAKDQEVKERPKPFHKKTQQRGISRAFSSPPKGKMGSCREGKRIMEEEPRTGQKVGPGDTDKLCAAGGISRISR VKYGDSGQSFSDKSNVIIHQRTHTGEKP YVCRECGRGFSQKSDLLKHQRTHTGEKP YVCRECGRGFSQKSHLLRHQRTHTGEKP YVCRECGRGFSQKSDLLIHQRTHTGEKP YVCRECGRGFSCKSHLLIHQRTHTGEKP YVCRECGRGFSCKSSLLIHQRTHTGEKP YVCRGVWGEALAESQTSSYTRGHTQGRS PVFAGRVSKSLALNYISTATGGHLLTSH LPTPALGGASKGSLLTLYISQECKETRN >PRDM7_otoGar Otolemur garnettii (galago) genome Prim gene 7 GAS8+ good coverage 0 MSPEKSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKHPWMAFRMEQSKRQK 0 0 ILKKCMLSFNMHLKELSGPASLPNISGSEQHQKHMSSPREASTSGQHSGRKS 1 2 DLRIKEIEVRMYSLRERKGHAYKEVSEPQDDDYL 1 2 yCEKCQNFFIDNCAVHGPPTFVKDTAVEKGHPNRSVLSLPSGLGIRTSGIPQAGFGVWNEASDLQLGLHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDESQGNWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGQ 1 2 EPKPEIHPCPSCSLAFSTQKFLSQHVERTHPSQISQGTSGRKNLRPQTPCPRDENQEQQHSDPNSRNDKTKGQEVKEMSKTSHKKTQQSRISRIFSCPPKGQMGSSREGERMIEEEPRPDQKVGPGDTEKFCVAIGISGIVK VKNRECVQSFSNKS NLRHQRTHTGEKP YMCRDCGRGFSHKSSLFRHQRTHTGEKP YVCRDCGRGFSLKANLLTHQRTHTGEKP YVCRDCGQGFSQKAHLLRHQRTHTGEKP YMCRDCGQGFSRKAYLLTHQRTHTGEKP YVCRDCGQGFSQKAHLLTHQRTHTGEKP YVCRDCGRGFSHKSSLFRHQRTHTGEKP YICRDCG >PRDM7_tarSyr Tarsius syrichta (tarsier) ABRT011082008 Prim pseu -- gas8+ double frameshift in exon 5, ABRT010499286 0 - 0 0 - 1 2 GLRAPRPAFMCHRKRAIKPLVDDTEDSDEEWTPRQQ 1 2 - 0 0 GMPRAPLSIVSSLKELSEMANLLNTSDSEQAWKPVSPSREASTSEQHSRKKL 1 2 EFRKKEIEVNMYSLRERKDCAYKEVNEPQDDDYL 1 2 YCEQCQNFFIDSCATHGIPTFINDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASELPLGLHFGPYEGQITDDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRIIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 1 2 - - >PRDM9_oryCun Oryctolagus cuniculus (rabbit) genome Glir gene 8 other Un0161 exon 2 ttt to tt restores frame; ZNF717+ DCAF4+ YAP1+ PRDM9- qTer 0 MSAAAPAEPSPGADAGQARGKPE 0 0 VQDAFRDISIYFSKEEWAEMGEWEKIRYRNVKRNYCALVAI 1 2 GLRAPRPAFMCHRRLAVRARADDTEDSDEEWTPRQQ 1 2 VKPPWMAFRTEHSKHQK 0 0 GMPRLPVNNESSLKELSGTANLLKTTGSEEDQKPSFPPKETRTSGQHSTRKL 1 2 GLRRKNIEVKMYSFRKRKSQAYKECSEPQDDDYL 1 2 YCEKCQNFFLDSCAVHGPPIFVKDSAVDKGHPNRSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEEEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDRSWANWMR 2 1 YVNCARNDEEQNLVAFQYHKQIFYRTCQVIKPGCELLVWYGDEYGQELGIKWGSKWKEELTAGR 1 2 EPKPEIHPCPSCSLAFSSHKFLSQHMERSHSSQIFPGAPARNHLQPANPCPGKEHQKLSDPQSWNDKNEGQDVKEKSRFSSKRTRQKAISRSFSSLPKGQVETSREGERMIEEEPRIGQELNPEDTGKSSVGAGLSRIAG VKYRDCRQGLSDKSHLINGQRAHTGEKP YACRECERGFTVKSNLISHQRTHTGEKP YACRECGRGFTVKSALTTHQRTHTGEKP YACRECGRGFTVKSHLISHQRTHTGEKP YACRECGRGFTVKSALITHQRTHTGEKP YACRECGQGFTVKSNLISHQRTHTGEKP YACRECGRGFTQKSHLINHLRAHTGEKP YACRECGRGFTVKSDLISHQRTHTGEKP YACRVDE..................... >PRDM7_oryCun Oryctolagus cuniculus (rabbit) genome Glir gene 4 other synteny novel 0 - 0 0 - 1 2 GLRAPRPAFMCHRRLAVRARADDTEDSDEEWTPRQQ 1 2 VKPPWMAFRTEHSKHQK 0 0 GMPRLPVNNESSLKELSGIANLLNTTGSEEDQKPSFPPKETRTSGQHSTRKL 1 2 GLRRKNIEVKMYSFRKRKSQAYKECSEPQDDDYL 1 2 YCEKCQNFFLDSCAVHGPPIFVKDSAVDKGHPNRSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEEEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDRSWANWMR 2 1 YVNCARNDEEQNLVAFQYHKQIFYRTCQVIKPGCELLVWYGDEYGQELGIKWGSKWKEELTAGR 1 2 EPKPEIHPCPSCSLAFSSHKFLSQHMECSHSSQIFPGAPARNHLQPANPCPGKEHQKLSDPQSWNDKNEGQDVKEKSRFSSKRTRQKAISRSFSSLPKGQVETSREGERMIEEEPRIGQELNPEDTGKSSVGAGLSRIAG VKYRDCRQGLSDKSHLINGQRAHTGEKP YACRECGQSFTVKSNLISHQRTHTGEKP YACRECGRGFTQKSHLIRHQRTHTGEKP YACRECGQSFTWKSNLISHQRTHTGEKP YACRVDE..................... >PRDM7_ochPri Ochotona princeps (pika) AAYZ01312269 Glir gene -- noDet dubious fragment, no orthologous terminal exon 0 - 0 0 - 1 2 - 1 2 - 0 0 - 1 2 - 1 2 yCEMCQNFFIESCAVHGSPTFVKD GHPHRSVLSLPSGLRIGPSGIPEAGLGVWNETTDLPLGLHFGPYEGQVTEEEEATNSGYSWL 0 0 ITKGRNRYEYVDGKDPSQANWMR 2 1 YVNCARNDEEQNLVAFQYHRQIFYRTCRAVRQGCELLVWYGDEYGQELGIKWGSKWKEELTAGR 1 2 - - >PRDM7_ratNor Rattus norvegicus (rat) P0C6Y7 Glir gene 10 PDCD2 chr1 FM103467 single transcript from body fat 0 MNTNKPEENSTEGDAGKLEWKPK 0 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 2 GLRAPRPAFMCYQRQAIKPQINDNEDSDEEWTPKQQ 1 2 VSSPWVPFRVKHSKQQK 0 0 ETPRMPLSDKSSVKEVFGIENLLNTSGSEHAQKPVCSPEEGNTSGQHFGKKL 1 2 KLRRKNVEVNRYRLRERKDLAYEEVSEPQDDDYL 1 2 YCEKCQNFFIDSCPNHGPPVFVKDSVVDRGHPNHSVLSLPPGLRIGPSGIPEAGLGVWNEASDLPVGLHFGPYKGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGQDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGRELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 2 ELRTEIHPCFLCSLAFSSQKFLTQHVEWNHRTEIFPGASARINPKPGDPCPDQLQEHFDSQNKNDKASNEVKRKSKPRHKWTRQRISTAFSSTLKEQMRSEESKRTVEEELRTGQTTNIEDTAKSFIASETS RIERQCGQCFSDKSNVSEHQRTHTGEKP YICRECGRGFSQKSDLIKHQRTHTEEKP YICRECGRGFTQKSDLIKHQRTHTEEKP YICRECGRGFTQKSDLIKHQRTHTGEKP YICRECGRGFTQKSDLIKHQRTHTEEKP YICRECGRGFTQKSSLIRHQRTHTGEKP YICRECGLGFTQKSNLIRHLRTHTGEKP YICRECGLGFTRKSNLIQHQRTHTGEKP YICRECGQGLTWKSSLIQHQRTHTGEKP YICRECGRGFTWKSSLIQHQRTHTVEK. >PRDM7_musMus Mus musculus (mouse) Q96EQ9 Glir gene 12 PDCD2 chr17 CN723438 eight transcripts, four from retina 0 MNTNKLEENSPEEDTGKFEWKPK 0 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 2 GLRAPRPAFMCYQRQAMKPQINDSEDSDEEWTPKQQ 1 2 VSPPWVPFRVKHSKQQK 0 0 ESSRMPFSGESNVKEGSGIENLLNTSGSEHVQKPVSSLEEGNTSGQHSGKKL 1 2 KLRKKNVEVKMYRLRERKGLAYEEVSEPQDDDYL 1 2 YCEKCQNFFIDSCPNHGPPLFVKDSMVDRGHPNHSVLSLPPGLRISPSGIPEAGLGVWNEASDLPVGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGQDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 2 ELRTEIHPCLLCSLAFSSQKFLTQHMEWNHRTEIFPGTSARINPKPGDPCSDQLQEQHVDSQNKNDKASNEVKRKSKPRQRISTTFPSTLKEQMRSEESKRTVEELRTGQTTNTEDTVKSFIASEIS SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQNSHLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTQKSVLIKHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTAKSVLIQHQRTHTGEKP YVCRECGRGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREK. >PRDM7_musMol Mus molossinus (wild_mouse) GU216230 Glir gene 11 noDet full length deposit 0 MNTNKLEENSPEEDTGKFEWKPK 0 0 VKDEFKDISIYFSKEEWAEMGEWEKIRYRNVKRNYKMLISI 1 2 GLRAPRPAFMCYQRQAMKPQINDSEDSDEEWTPKQQ 1 2 VSPPWVPFRVKHSKQQK 0 0 ESSRMPFSGESNVKEGSGIENLLNTSGSEHVQKPVSSLEEGNTSGQHSGKKL 1 2 KLRKKNVEVKMYRLRERKGLAYKEVSEPQDDDYL 1 2 YCEKCQNFFIDSCPNHGPPLFVKDSMVDRGHPNHSVLSLPPGLRISPSGIPEAGLGVWNEASDLPVGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGQDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRKIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKMKKGFTAGR 1 2 ELRTEIHPCLLCSLAFSSQKFLTQHMEWNHRTEIFPGTSARINPKPGDPCSDQLQEQHVDSQNKNDKASNEVKRKSKPRQRISTTFPSTLKEQMRSEESKRTVEELRTGQTTNTEDTVKSFIASEIS SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSVLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTEKSSLIKHQRTHTGEKP YVCRECGWGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSSLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGWGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTR.EK >PRDM7_dipOrd Dipodomys ordii (kangaroo_rat) genome Glir gene -- noDet dubious fragment, no orthologous terminal exon 0 - 0 0 - 1 2 GLKAPRPVFMCHRRQAIKPQVDDTDDSDEEWTPGRQ 1 2 - 0 0 - 1 2 elRTKEVKMRMYSLRERKSYAYEEISEPQDDDYL 1 2 yCEQCQNFFINSCTVHGPPIFVRDNVVDKGHYDRSVLSLPPGLRIRQSSIPEAGLGVWNEESDLPLGLHFGPYEGQITEDEDAANSGYSWM 0 0 ITKGRNCYVYVDGKDKSQANWMR 2 1 YVNCARYDEEQNLVAFQYHRQIFYRTCRVIKAGCELLVWYGDEYGQELGIKWGSKWKRELTAgr 1 2 - - >PRDM7_speTri Spermophil tridecemlin (squirrel) AAQQ01308561 Glir gene -- noDet plus exon by exon traces 0 - 0 0 - 1 2 GFRAPRPAFMCHQRQTIKLQMDDTEDSDEEWTPRQQ 1 2 - 0 0 LKPEVLLSNESSLKELSGTANLLNTSGSEQVQKPVSPLREASASRQHSRRKL 1 2 ELRTKEVEVKMYSLRERKGHAYKEVSEPQDDDYL 1 2 yCDKCQNFFMDSCPVHGPPTFIKDSVVNKDHSNHSTLSLPLGLRIGPSSIPEAGLGVWNEATDLPLGLHFGPYRGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDESQANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELSAGR 1 2 EPKPEIHPCPSCSLAFSSQKFLSQHVDRSHPSQIFPGTSMRKKLIPGDSSPRDQLQEQQHPDPHGWNDKARGQEVQGSLKPTHKGTRQRGISSPPKGQMGRSEESERMMEDDLKADQEINPEDTDKILVGVEMSRI - >PRDM9a_bosTau Bos taurus (cattle) NW_003053109 Laur gene 7 noDet chr1 0 MSQNRSPEERTKGDAGRTEWKLT 0 0 AKDAFKDISIYFSKEEWAEMGEWEKTGYRNVKRNYEVLIAI 1 2 GLRATQPAFMHHRRQVIKPQGDDTEDSDEEWTPQHQ 1 2 GKPSRKAFRMEHRKHQK 0 0 GKSRGPLSKVSSLKKLQGAAKLLNTSGSKWAQKPANPPRETRTLEQHSRQKV 1 2 ELRRKETDMKRYSLRERKGHVYQEVSEPQDDDYL 1 2 YCQECQNFFIDSCDAHGPPTFVKDSAVEKGHANRSVLTLPPGLSIKLSGIPEAGLGVWNEASHLPLGLHFGPYEGQITDDKEAINSGYSWL 0 0 ITKGRNSYEYVDGKDTSLaNWMR 2 1 YVNCARHYEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGEKLGIKCESRGKSMFAAGr 1 2 ESKPKIHPCASCSLAFSSQKFLSQHVQHNHPSQTLLRPSARDYLQPEDPCPGSQNQQQRYSDPHSPSDKPEGREVKDRPQPLLKSIRLKRISRASSYSPRGQMGASGVHERITEEPSTSQKPNPEDTGKLFMGAGVSGIIK VKYGECGQGSKDRSSLITNQRTHTGEKP YVCGECGQSFNQKSTLITHQRTHTGEKP YVCGECGRSFNQKSTLITHQRTHTGEKP YVCGECGRSFSQKSTLIKHQRTHTGEKP YVCGECGQSFNQKSTLITHQRTHTGEKP YVCGECGQSFNQKSTLITHQRTHTGEKP YVCGECGRSFSRKSTLITHQRTHRGEKL CLQGV...................... >PRDM9b_bosTau Bos taurus (cattle) DAAA02065087 Laur gene 5 noDet chrU aaaaa fixed to aaaaaa in exon 2 KRAB SSXRD SET C2H2 0 MSPNRSPENSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIQYRNVKRNYEALIAI 1 2 GFRATQPGFMHHGRQVLKSQVDDTEDSDEEWTPRQQ 1 2 GKPSGMAFRGEPSKHPK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 2 ELRRKETEVKRYSVRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSNSGYCWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 2 AKMHPCASCSLAFSSQKFLSQHVQRNHPSQTLLRPSARDHLQPEDPCPGNQNQQQRYSDPHSPSDKPEGRKAKDRPQPLLKSIKLKRISRASSYSPRGQVGRSGVHERITEEPSTSQKLNPEDTGKLFMGAGVSGIIK VKYRECGQGSKDRSSLITHERTHRAEAL CLRRVWAKLQSEVPLLVMHQRTHTGEKL YVCGECGKSFSQKSPLIRHQRTHTGEKP YVCGECGKSFSQKSPLIRHQRTHTGKKP YVCRECGRSFSDKSH.HTPEYTHRGEAL HLRGVWA..................... >PRDM9c_bosTau Bos taurus (cattle) XM_002699750 Laur gene -- noDet chrX GO353654 4-cell embryo transcript no zinc downstream despite 43k bp 0 MSPNRSPENSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATQPGFMHHRRQVLKPQVDDTEDSDEEWTPRQQ 1 2 GKPSGMAFRGERSKHQK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKSGSKQAQKPVPPPREARTPGKHPRHKV 1 2 ELRRKETKVKRYSVRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQIIYNEEDSHSGYCWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCRVVRPGCELLVWYGDEYGEELGIKQDKRGKSKLSAQR 1 2 - - >PRDM9d_bosTau Bos taurus (cattle) genome Laur gene 9 noDet chrX proximal tandem 0 MRPNTSPEESTERDAGRTEWKPT 0 0 AKDAFKDISVYFSKEEWEEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATRPAFMHHRRQVIKLQADDTEDSDEEWTPRQQ 1 2 GKLSSMAFRVEHNKHQN 0 0 TMSRAPLSKEFSLKELPGAAKLLKTSGSKQAQKLVPPPGKARTPGQHPRQKV 1 2 ELRRKETEVKRYSLRERKGHVYQEVSEPQDDDYL 1 2 YCEECQSFFIDSCAAHGPPIFVKDCAVEKGHANRSALTLPPGLSIRESSIPEAGLGVWNEVSDLPLGLHFGPYEGQITDDEEAANSGYSWL 0 0 ITKRRNCYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCQVVRPGCELLVWYGDEYGQDLGIKRESSRKSELAGPR 1 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNQQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRPKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR VNYGDHEQGSKDRSSLITHEKIHTGEKP YVCKECGKSFNGRSDLTKHKRTHTGEKP YACGECGRSFSFKKNLITHKRTHTREKP YVCRECGRSFNEKSRLTIHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCGECGRSFNEKSRLTIHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCGECGQSFNEKSRLTIHKRTHTGEKP YACGDCGQSFSLKSVLITHQRTHTGEKP YVCMECE..................... >PRDM9e_bosTau Bos taurus (cattle) genome Laur gene 9 noDet chrX distal tandem 0 MRPNRSPEESTEGDAGRTEWKPM 0 0 AKDAFKDISIYFSKEEWEEMGEWEKIRYRNVKRNYEVLITI 1 2 GFRAARPAFMHHRRQVIKPQVNDIKDSDEEWTPRQQ 1 2 GKPFSMAFRVEHSKHQK 0 0 GMSRAPLSKESSLKELPGAAKLLKTSGCKQAQKLVPPPRKARTPEQHPRQKV 1 2 ERRRKETGVKRYSLREREGLVYQEVSEPLDDDYL 1 2 YCEECQSFFIDICAAHRPPTFVKDCAVEKGHANCSALTLPPGLSIRLSGIPEAGLGVWNEASDLPLGLHFGPYEGQITDDKEAAHSRYSWL 0 0 ITKGRNCYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYQGQIFYRTCQVVRPGCELLVWYGDEYGWDLSIKQDSRGKNKLAAGR 1 2 EPKPKIYPCASCCLSFSSQKFLSQHVQRNHPSQILLRPSIGDHLQPEDPCPGSQNEQQRYSDPHSLSDKPEGREPKERPHPLLKGPKLCIRLKRISTASSYPPKGQMGGSEVHERMTEEPSTSQKLNPEDTGKLFMEAGVSGIVR VKYGEHEQDSKDKSSLITHEKIHTGEKP YVCTECGKSFNWKSDLTKHKRTHSEEKP YACGECGRSFSFKKNLIIHQRTHTGEKP YVCGECGRSFSEKSNLTKHKRTHTGEKP YACGECGQSFSFKKNLITHQRTHTGEKP YVCGECGRSFSEKSRLTTHKRTHTGEKP YVCGDCGQSFSLKSVLITHQRTHTGEKP YVCRECGRSFSVISNLIRHQRTHTGEKP YVCRECEQSFREKSNLVRHQRTHTGEKP YVCMECE..................... >PRDM9e_oviAri Ovis aries (sheep) genome Laur pseu -- noDet chr 18 cow has PDRM7 pseudogene; sheep GAS8 is on sheep chr14 0 - 0 0 - 1 2 GLRAP PPFMYHRRQVIKPQVDDIEDSDEEWTPRQQ 1 2 - 0 0 - 1 2 ELRRKETEMKIYSLQKRKGHMYQEVSDPQDDNYL 1 2 ycEKCQNF INSCAAHGPPTFVKDCVVEKGHASCSALtLSPGLSIRPSGIPEAGLRVWNEASDLPLGLHFGPYKGQITDDEEVANSRYFWL 0 0 - 2 1 YVNCAQDDEEQNLVAFQYHRQIFS TCWVVRPGCELLVWYRDEYGQELSIK GSRHKSELTVRR 1 2 - - >PRDM9d_oviAri Ovis aries (sheep) genome Laur gene -- noDet chr1 near end chr1 0 - 0 0 - 1 2 GLRATRLAFMHHCRQVIKPQVDDIEDSDEEWTPRQQ 1 2 - 0 0 - 1 2 - 1 2 - 0 0 ITKGRNCYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYQGQIFYRTCQVVRPGCELLVWYGDEYGQDLGIKRDSSGKSELAAGR 1 2 - - >PRDM9c_oviAri Ovis aries (sheep) genome Laur pseu 4 noDet chr5 middle of 108,514,869 bp 0 - 0 0 - 1 2 GLRATRLAFMHHCRQVIKPQVDDIEDSDEEWTPRQQ 1 2 - 0 0 GMSKALVSNKSSLKEMPGASKLLKTRGPKQAQIPVPAPREPSTSEQHPRQKV 1 2 - 1 2 HGLPTLVKDCAVEKGHANHSALSLSPGSSIRPSGIPEAGLGVWNKVSDLLLGLHFGSYVGQITDDEEAAKSGYSWL 0 0 - 2 1 YVNGAQD KEQNLVAFLTHRQIFY TCRVVRPGCELLVWYRDTYSQELSIKCGSRWKSELTASR 1 2 PMCSCSLAFSSQKFLSQHVKCNHPSQILLKTSARDRLQPEDPCPGNPNQQQQYSDLHSWSDKPESRESKEKPQPLLKSIRLRRISRASSYSSRGQMGGFRVHKRMREEPSTGKEVSPEDAGKLFMGEGVSRIMR VKYGDCG GSKDRSSLMTHQRTHTGENP YVCREYE.SFSEKSSLIKHQRTHTGEKP YVCRECWQSFGRKSTLITHQRMHTREKP CVCRECGRSFSKKSTLITHQRTHTGQKP >PRDM9b_oviAri Ovis aries (sheep) genome Laur pseu 2 noDet chrX not tandem: 62 mbp separation 0 MSPNRSPENSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATQPAFMHHHRQVIKPQVDDTEDSEEEWTPRQQ 1 2 GKPSGMAFRGERSKHQK 0 0 RLSRGPLNKVSSLKKLPGAAKLLKKTGSKQAQKPVPPPREARTPGQHPRHKV 1 2 ELRRKETEVKRYSLRERKGHVYQEVSELQDDDYL 1 2 yCEECQNFFIDSCAAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQVIYNEEASHSGYSWL 0 0 VTKGRNSYEYVDGKDTSLANWMR 2 1 YVNCARDDEEQNLVALQYHGQIFYRTCQVVRPGCELLVWYGDEYGEELGIKQDSRGKSKLSAQR 1 2 ELKPKIHPCASCSPAFSSQKFLSQYVQPNHPSQILLRPSARDHLQPEDPCPGNQNEQQ YSDPHSPSDKPEGCKAKERPPWLLKSMSVRISMASSYSPKGQMRGSETHYRMTEEPSTSQKLNPEDIGKLFMGTGVSGIIK IKYEECGQVSKDRSSLITHEGTHTREQS YVCRECGQSFSVKSSLIRLQRTHTGEKP Y........................... >PRDM9a_oviAri Ovis aries (sheep) genome Laur gene 9 noDet chrX not tandem 0 MSPNRSPENSTEGDAGRTEWKPM 0 0 AKDAFKDISIYFTKEEWAEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATQPAFMHHHRQVIKPQVDDTEDSEEEWTPRQQ 1 2 GKPSGMAFRGERSKHQK 0 0 GMSRGPLSKVSSLKKLPGTTKLLKTSGSKQAQKPVPSSREARTSG HTRQKV 1 2 ELGRKETDMKRYSLRERKGHVYQEVSEPQDDDYL 1 2 yCQECQNFFINSCDAHGPPTFVKDSAVEKGHANRSALTLPPGLSIRLSGIPEAGLGVWNEASHLPLGLHFGPYEGQITDDKEAVNSGYSWL 0 0 - 2 1 YVNCARHYEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGEKLGIRCESRGKSMLAAGR 1 2 EPKPKIHPCASCSLSFSSQKFLSQHVQRSHPSQILLRPSPRDHLQPEDPCPGKQNQQQRYSDPHSPSDKPEGQEPKERPHPLLKGPKLCIRLKRISTASSYTPKGQMGGSEVHEKMTEEPSTSQKLNPENTGKLFMEAGVSGIVR VKYGEHEQGSKDKSSLITHERIHTGEKP YVCKECGKSFNGRSNLTRHKRTHTGEKP YVCRECGQSFSLKSILITHQRTHTGEKP YVCGECGQSFSEKSNLTRHKRTHTGEKP YVCRECGQSFSLKSILITHQRTHTGEKP YVCRECGRSFSVKSNLTRHKMTHTGEKP YVCGECGQSFSQKPHLIKHQRTHTGEKP YVCRECGRSFSAMSNLIRHQRTHTGEKP YVCRECGRSFSAMSNLIRHQRTHTGEKP YVCREC...................... >PRDM9d_munMun Muntiacus muntjak (muntjac) AC216498 Laur gene 4 noDet frameshift exon 9 no syntenic loci; identities: 92%b 89%a 90%c 0 MRPNRSQEESTEGNAGRTERKPT 0 0 GKDAFKDISVYFSKEEWEEMGEWEKIRYRNMKRNYEALIAI 1 2 GFRATQPTFMHHRRQVIKSQVDDTEDSDEEWTPRQQ 1 2 GKPSSMAFRVEHSKNQK 0 0 RMSRAPLSNESGLKELPGAAKSLKTSDSKQARNPVPHHRKARTPGQLPRQKV 1 2 ELRRKETGVKRYSLRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFINSCAAHGPpTFVKDCAVEKGHANRSALTLPHGLSIRLSGIPDAGLGVWNKVSDLALGLHFGPYKGQITDNEEAANSGYAWL 0 0 ITKGRNCYEYVDGKDTSWANWMR 2 1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVVRPGCELLVWYGDEYGQDFGIKRNSRGKSELAAGR 1 2 EPKPKIHPCASCSLTFSSQKFLSQHIQCSHPPQTLLRPSERDLLQPEDPCPGNQNQQQRYSDPHSPSDKPEGHEAKDRPQPLLKSIRLKRISRASSCSPRGQMGGSGVHERMTEEPSTSQKLNPGDTGTLLTGAGVSGIMK VKYGECGQGSKDRSSLSTHERTHTGEKP YVCRECGQSFSGKPVLIRHQRTHTGEKP YVCMECGRSFSAKSVLMTHHRTHTGEKP YICRECGQSFSQKIHLIRHQRIHTGE.P SVFRECE..................... >PRDM9c_munMun Muntiacus muntjak (muntjac) AC154919 Laur gene 15 noDet no syntenic loci AC204173 99% identical 0 MRPNRSPEESTEGDAGRTEQKPT 0 0 AKDAFKDISVYFSKEEWEEMGDWEKIRYRNMKRNYEVLIAI 1 2 GFRATRPDFMHHRRQVIKPQVDDTEDSDEEWAPRQQ 1 2 GKPSSVAFRVEHSKHQK 0 0 RMSRAPLSNESGLKELPGAAKPLKTSGSKQAQNPVPHHRKARTPGQLPRQKV 1 2 ELRRKETGVKRYSLRERKGHVYQEVSKPQDDDYL 1 2 YCEKCQNFFIDSCAAHGPPTFVKDCAVEKGHANRSLLTLPPGLSIRLSGIPDAGLGVWNEASDLPLGLHFGPYEGQITDDEEAANSGYAWL 0 0 ITKGRDCYQYVDGKDTSWANWMR 2 1 YVNCARDDEEQNLVAFQYHGQIFYQTCQVVRPGCELLVWCGDEYGQDLGIKRNSRGKSELVAGR 1 2 EPKPKIHPCASCSLAFSSQKFLSQHIQRSHPSQTLLRPSERDLLQPEDPCPGNQNQRFSDPHRPSDRPQPLLKSIRLKRISRASSYSPRGQMGGSGVHELMTEEPSTSHKLNPEDTGTLLMGAGVSGIMR VTYGECGQGSKDRSSLTTHERTYTGEKP YVCGECGRSFCQKAHLITHQRTHTGEKP YVCRECGQSFSRNSLLIRHQRIHTGEKP YVCGECGRSFRDKSNLISHRRTHTGEKP YVCGECGQSFSDKSNLIRHQRTHAGEKP YVCGECGRSFNRKSHLITHQRTHTGEKP YACRECGQSFSQKSILITHQRTHTGEKP YACRECG.SFSQKSILITHQRTHTGEKP YVCGECGRSFSQKSLLITHQRTHTGEKP YVCMECGRSFSQKTHLITHQRTHTGEKP YVCGECGRSFSQKSLLITHQRTHTGEKP YVCGECGRSFSQKSLLITHQRTHTGEKP YICMECGRSFSQKTHLITHQRTHTGEKP YVCGKCGQSFSDKSNLISHKRTHTGEKP YVCRECGRSFNRKSLLITHQRTHT.E.P YVCRECE..................... >PRDM9b_munMun Muntiacus muntjak (muntjac) AC218859 Laur gene 13 noDet no syntenic loci 0 MRPNTSPEESTEGDAGRTERKPT 0 0 AKDAFKDISVYFSKEEWEEMGDWEKSRYRNMKRNYEVLIAI 1 2 GFRATRPDFMHHRRQVIKPQVDDTEDSDEEWAPRQQ 1 2 GKPSSMAFRVEHSKHQK 0 0 RMSRAPLSNESGLKELPGAAKPLKTSGSKQAQNPVPHHRKARTPGQLPRQKV 1 2 ELRRKETGVKRYSLRERKGHVYQEVSKPQDDDYL 1 2 YCEECQNFFIDSCAAHGPPTFVKDCAVEKGHANRSALTLPPGLSIRLSGIPDAGLGVWNETSDLPLGLHFGPYEGQITDDEEAANSGYAWL 0 0 ITKGRNCYQYVDGKDTSWANWMR 2 1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVIRPGCELLVWYGDEYGQDLGIKRNSRGKSELATGR 1 2 EPKPKIHPCASCSLAFSSQKFLSQHIQRSHPSQTLLRPSERDLLQPEDPCPGSQNQRYSDPHSPSDKPEGQEAKDRPQQLLKSIRLKRISRASSYSPGGQMGGSGVHERMTEEPSTSQKLNPEDTGTLLTGAGVSGIMR VTYGECWKGSKDRSSLTTHERTHTGEKP YVCGECGQSFHHGSVLIRHQRTHTGEKP YVCGECGRSFSQKSVLIRHQRTHTGEKP YVCGECGRSFSQKSVLIRHQRTHTGEKP YVCGECGRSFSQKAHLITHQRTHTGEKP YVCGECGRSFSQKTHLISHKRTHTGEKP YVCGECGRSFCQKSALIRHQRAHTGEKP YVCGECGRSFIQKSDFIRHQRTHTGEKP YVCRECGQSYSDKTVLITHERTHTGEKP YVCGECGRSYSDKTVLITHERTHTGEKP YVCGECGRSFLWKSALIRHQRTHTGEKP YACGDCGRSFNQKSNFIRHQRTHTGEKP YVCGECWRSFSQKSSSSDTRGHTQGRRP VCRECG..SFSQKSHLISHQRTHTEEKP YVCRECE..................... >PRDM9a_munMun Muntiacus muntjak (muntjac) AC225653 Laur gene 7 noDet unordered contigs htgs; no synteny tag stop instead of aag K 0 MRPNRSPEESTEGDAGRTEQKPT 0 0 AKDAFKDISVYFSKEEWEEMGEWEKIRYRNVKRNYEALIAI 1 2 GFRATRPDFMHHCRQVIKPQVDDTEDSDEEWTPRQQ 1 2 GKPSSMAFRVKHSKHQK 0 0 GMSRAPLIKESSLKELLGAAKLMKTSGSKQAQNPVPHPRKARTPGQHPRQKV 1 2 ELTRKETGVKRYSLRERKGHVYQEVSEPQDDDYL 1 2 YCEECQNFFIDSCAAHGLPTFVKDCAVEKGHANRSALTLPPGLSIRLSGIPDAGLGVWNEESDLPLGLHFGPYEGQITDDEEAANSGYAWL 0 0 ITKGRNCYQYVDGKDTSWANWMR 2 1 YVNCARDDEEQNLVAFQYHGQIFYRTCQVIRPGCELLVWYGDEYGQDLGIKRNSRGKSELAAGR 1 2 EPKPKIHPCASCSLAFTSQKFLSQHIQRSHPAQTLLRPSERNLLQPEHPCPGSQNQRYSDPHSLSDKPEGQEAKDRPQPLLKSIRLKRISRASSYSPGGQMGGSGVHERMKDEPSTSQKLNPEDTGTLLTGAGVSGIMR VTYGECGKGSKDRSSLTTHERTHTGEKP YACRECGRSFRQKSDFITHQRTHTGEKP YVCGQCGRSFGRKFALIRHQRIHTGEKP YVCRECGQSFSQKTHLSSHQRTHTGEKP YVCGECGRSFSQKSVLIRHQRTHTGEKP YVCQECGRSFSDKSNLISHKRTHMGEKP YVCRECGRSFIRKSVLIRHQRTHTGE.P YVCRECE..................... >PRDM7_bosTau Bos taurus (cattle) genome Laur pseu -- GAS8+ missing C2H2 0 MSPNRSPEESIEGDTGRTEWKPT 0 0 AKDAFKDISIYFCKEEWAQMG WEKIRYRNVKRNYEALITL 1 2 - 1 2 - 0 0 - 1 2 - 1 2 - 0 0 - 2 1 - 1 2 - - >PRDM7_turTru Tursiops truncatus (dolphin) ABRN01441536 Laur gene 9 gas8+ no useful synteny 0 MSTDRWPEDSTEGDAGRTAWKPT 0 0 VKDAFKDISIYFSKEEWTEMGEWEKIRYRNVKKNYEALVTL 1 2 GLRAPRPAFMCHRRQAIKAQVGDPEDSDEEWTPRQQ 1 2 VKPSWVAFRVEHSKHQK 0 0 AVPPVPLSNESSLKKLPGAAQLQKASGPAQAQSPAPPPGAASTSAWHTRQKL 1 2 ERRAKQIEVKMYSLRERKGHVYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAAHGAPTFVKDSAVEKGHPNRSALTLPPGLSIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDTSWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYSQELGIPWGSGWKSQLVaGR 1 2 DPKPKIQPCGSCSLAFSSQKILSQHVECSHPSQVLPRTSARDRVQPEDPCPGYQNRQQQYSDPHSWSNKPECQEVKERSKPLLKRIRLGRISRAFSSSPKGQMGSSRAHERMMEAGPSTGQKVNPEATGKLLIGAGVSRVVK VKYRSSGQGSKDRSSLTKHQRTHTGEKP YVCGECGRDFSLKSDLIRHQRTHTGEKP YVCGECGRDFSLKSGLISHQRTHTGEKP YVCGECGRDFSQKSGLIRHQRTHTGEKP YVCGECGRDFSLKSGLISHQRTHTGEKP YVCGECGRDFSQKSGLIRHQRTHTGEKP YVCGECGRDFSLKSGLITHQRTHTGEKP YVCGECGRDFSQKSNLITHQRTHTGEKP YVCGECGRDFSRKSSYI........... >PRDM7_susScr Sus scrofa (pig) FP476134 Laur gene 9 GAS8+ unordered HTGS not wgs misassembly or inversion; not in genome browser 0 MRPDRRPEESPDPAAGSTERKAA 0 0 ATDAFKDISIYFSKEEWTEMGEWEKIRYRNVKRNYEALTTI 1 2 GLRAPRPAFMCHRRQAIKPQVDDTEDSDEEWTPRQQ 1 2 VKPCRVAFRVEHNKHQK 0 0 SDSRVPLSNKSSLKELLTTAEVPETSGSEQAQEPVSPPGEASTSRRRSGQEL 1 2 ARRRKDTEARMYSLRERKGHAYQEVGEPQDDDYL 1 2 yCEKCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALTLPPGLRIRPSGIPEAGLGVWNEAHDLPLGLHFGPYEGQVTEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYGQELGIKWGSKWKKELTAGI 1 2 EPKPKIHPCPSCSLAFSSQRFLSQHVERSHPSQSLPRASARRGLQPEGPCPDNQQQQQPYPDPHSWDGTSESQDVKEGSKPFLERRRLRKTSRASSYAPEGQMRSSRVRERMTEEEPSAGQKVNPEDTGTLFTVAGES GILRVENRGYGPDSGLTRHPRTHTGEKP HVCSECGRGFSVKSHLIRHQRTHTGEKP YVCRECGRGFSVKSHLIRHQRTHTGEKP YVCRECGRGFSVKSSLITHQRTHTGEKP YVCRECGRGFSVKSHLIRHQRTHTGEKP YVCRECGRGFSEKSSLVTHQRTHTGEKP FVCRECGRGFSVKSSLVTHQRTHTGEKP YVCRECGRGFSVKSNFITHQRTHTGEKP YVCRECGRGFSEKSSLVTHQRTHTGEKP YVCREGE..................... >PRDM7_canFam Canis familiaris (dog) genome Laur pseu 5 GAS8+ frameshift fixed to 6 ZNF; synteny MNS1 K1F1B intervening CDH3 oddity 0 - 0 0 - 1 2 - 1 2 VKPSWVAFRMEQSKHQK 0 0 GIPRVPLSNKSSLKELSETAKLLNTSSPEQGQKSVSLPGKASTSGHHTRQKL 1 2 ELRRKDVEVKMYSLQERKGLAYQEVSEPQDDDYL 1 2 yCEK QTFFIDSCTVHGPPTFVKDSEVDKGQPNHSALTLPPGLRIRTSSIPQAGLGVWN ASDLPLGLHFGPYKGQITEDEEAANSGYSCL 0 0 ITKGRNCYEYVDGKDkSWANWMR 2 1 YMNCARDDEEQS LVAFQYHRQIFYRTPGHQASCELLVWYGDEYSQELGIKWGSKWKSELTAGK 1 2 EPNPEIHPCPSCSL AFSSQKFLSQHLEHNHPSQILPRISVREHFRPKDPCPGCQNQQQQQHSDPQRWNDRAKGQEGKERFKPLPKSIRQRRISRAFSTPCKGQTTCEGIVKEEPSAGSQKLNPEDTGKLFKGVGMTRIIR VKYRGCGRGFNDRSHLSRHQRTHTGENP YVCRECGRGFIHRTNLIIHQRTHTGEKP YVCRECGtGFIQRSNLSIHQRTHTGEKP YVCRECGRGFTQRSTLNEHQRTHTEEKP YVCRECGRSFTRRSTLITHQRTHTGEKP YVCRECGRSFT................. KRSTWDPWVAQRFGACLWP......... >PRDM7_felCat Felis catus (cat) genome Laur gene 11 GAS8+ two contigs GAS8 implied by downstream CAD1 0 mSPLRFPEQSAERGSRKARWKPT 0 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYEALMTI 1 2 gLRAPRPAFMCHRRQAIKPQVDVTEDSDEEWTPRQQ 1 2 VKPSWVASRVDQNKQHK 0 0 GTHRVPLSKESSLKDFSETAKLLNTSGSEQGQKPVSLPGEASTSGHHSRRKL 1 2 frRRKEIGVKMYSLRERKGFAYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAVHGPPTFVKDNAVGKGHPNRSALTLPPGLRIRPSSIPEAGLGVWNEASDLPLGTHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDNSWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKSELSTGK 1 2 EPQPDIHRCPSCSLAFSSQKFLSQHVECKHSSQSLPQISARKHFQPENPCPGDQNQQQQQHSDPHSWNDKAKCQEVKERSRPLLKSIKQRRISRAFSTPCKGQMGSSRVCEGMVEEGPSMGQNLNSEDTGKLFMGVGMSRIVR IKNRGCEQGFNDRSHFSRHQRTHKEEKP SVCNEFRRDFSHKSALITHQRTHTGEKP YVCRECGRGFTQRSNLFRHQRTHTGEKP YVCRECGRGFTQRSDLFTHQRTHTGEKP YVCRECGRGFTRRSNLFTHQRTHTGEKP YVCRECGRGFTRRSHLFTHQRTHTGEKP YVCRECGRGFTQRSNLFTHQRTHTGEKP YVCRECGRGFTQRSDLFRHQRTHTGEKP YVCRECGRGFTQRSHLFTHQRTHTGEKP YVCRECGRGFTQRSNLFRHQRTHTGEKP YVCRECGRGFTWRSNLFTHQRTHTGEKP YVCRKDGQGFTNKLHL.SYQRT...... NVATTHSIPQL................. >PRDM7_ailMel Ailuropoda melanoleuca (panda) GL193502 Laur gene 6 GAS8+ first three exons from different contig 0 MSLNTSPEETPERDSGRTGWKPT 0 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYEALITI 1 2 GLRAPRPAFMCHRRQAIKPQVDDTEDSDEEWTPRRQ 1 2 VRPSWVAFRMEQSKHQR 0 0 GIPRAPLRNESSLKELSETAKLLNTSGSELGQKPVSLPGEASTSGHDSLQKL 1 2 GFRRKDVEVKMYSLRERKSLAYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAVHGPPTFVKDSAVDKGQPNRSALTLPPGLRIRPSGIPQAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDNSWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKSELAAGK 1 2 EPKPEIHPCPSCSLAFSSQKFLSQHLEHNHPSQILSRKSASEHFQQEDPCPGHQNQQQQQHSDPHRWNDKAKGQEVKERFKPLLKSIRQRRISRAFSSPCKGQTRSSTVCEGMVEEEPSAGQKLNPEETGKLFMGVGMSGIIR VKYRGCGRDFSDRSHQSGHQRRHQ KKP SVCKKVKREFSHKSVLITHQRTHTGEKP YVCRECGRGFTQRSNLIRHQRTHTGEKP YVCRECGRGFTQRSNLIRHQRTHTGEKP YVCRECGRGFTQRSSLIRHQRTHTGEKP YVCRECGRGFTLRPNLIGHQRTHTEALP INYISTTKEQM................. >PRDM9_pteVam Pteropus vampyrus (bat) ABRP01232219 Laur pseu 15 noDet frameshift ttt to tttt fixed in last zinc finger; no blastx synteny 0 - 0 0 - 1 2 - 1 2 vQPSWVAFGVEQSKHQK 0 0 AMPRVPLSNESSLKELSVIANPLKASGSEQNQQPVFPPGKASASRQHSRRKL 1 2 eLRRKGVEVKMDSLRERMGRVYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAAHGSPIFVKDSEVDIGHPNHSALTLPPGLRIGPSGIPEAGLGVWNEASNLPLGLLFGPYEGQVTEDEEAANSKYSwM 0 0 spKGETAEYV DGKDESRANWMR 2 1 YVNCARDDEDQNLVAFQFRRQIFYRTCRVIMPGCELLVWYGDEYGQGLGIKWGSKWKREFTAGR 1 2 EPKPEIHPCPSCSLAFSSRKFLSQHMKRSHPSQSLPGISARKHLQSKEPHPEDQSQQQQQQQHTDPCSWNDKAEGQEVKERSKPMLERNGQRKISRAFSKPPKGQMGSPRECERMMEAEPSTSQKVNPENTGKSSVGVGASRIVR VKYGGCGHGFDDGSHFIRHQRTHSGEKP FVCRECERGFNEKSSLTMHQRTHSGEKP FVCREC.EGFSVKSSLIRHQRTYSGEKP FVCRECEQGFNEKSSLTMHQRTHSGEKP FFCRECEGFSVK.SSLIRHQRTHSGQKP FVCRECKRGFTQKSHLITHQRTHSGEKP FCRECER.GFTQKSHLIKHQRTHSGEKP FVCRECA..................... >PRDM7_pteVam Pteropus vampyrus (bat) ABRP01250178 Laur gene 7 GAS8+ 4 distal exons of GAS8+-; unique F sweep in zinc finger; 15 ZNF dotplot no CAD1 0 MRPDRSPEEAPEGDTRRTGCKPK 0 0 AKDAFKDISIYFSKEEWTEMGDWEKIRYRNVKRNYDALQAI 1 2 GLRAPRPAFMCRRRQAIKPQVDDSEDSDEEWTPRQQ 1 2 - 0 0 AMPRVPLSNEPSLKELSVIANLLKASGSEQDQKPVFPPGKASASRQHSRQKL 1 2 GLRRKGVEVKMYSLRERTGRVYQEVSEPQDDDYL 1 2 yCEKCQNFFIDSCAAHGSPIFVKDSEVDIRHPNRSALTLPPGLRIGPSGIPEAGLGVWNEASDLPLGLLFGPYEGQVTEDEEAANSGYSWL 0 0 QGKGRNCYEYVDGKDESRANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKRELTAGR 1 2 EPKPAIHPCPSCSLAFSGQKFLSQHMKRSHPSQSLPGISARKHLQSKEPHPEDQSQQQHNDPRSWNDKAEGQEVKERSKPLLERNRQRKIFRAFSKPPKGQMGSPREYERMMEAEPSTSQKVNPENTGKSSVGVGASRIVI VKYGGCEHGFDDGSHLIMHQRTHSGEKP FVCRECERGFSKKSNLITHQRTHSGEKP FVCRECERGFTRKSSLITHQRTHSGEKP FVCRECERGFTQKSHLITHQRTHSGEKP FVCRECERGFSEKSSLIKHQRTHSGEKP FVCRECERGFTRKSSLITHQRTHSGEKP FVCRECERGFTQKSSLIKHQRTHSGEKP FVCRECERGFTQKSSLIKHQRTHSGEKP FVCRECERGFTQKSSLIKHQRTHSGEKP FVCRECERGFTQKSSLITHQRTHSGEKP FVCRECERGFTQKSHLITHQRTHSGEKP FVCRECERGFSKKSNLITHQRTHSGEKP FVCRECERGFTRKSLLITHQRTHSGEKP FVFRECERGFTQKSSLITHQRTHSGEKP FVCRECERGFTRKSYLITHQRTHSGEKP FVGRECE..................... >PRDM7_myoLuc Myotis lucifugus (bat) AAPE02062260 Laur gene 6 gas8+ TGA stop codon; CpG hotspot for R CGA; SXXRD implies missing KRAB no CAD1 0 - 0 0 - 1 2 - 1 2 - 0 0 AKSRAPLSNESSLKELSGTANLLTTSGSEQTQKTVPPPGEASTSGQHPRSKL 1 2 dLRRKEIEVKMYSLRERKCRVYQEISEPQDDDYL 1 2 YCEKCQNFFIDSCAVHGPPTFVKDSAVDKGHANRSALTLPPGLRIGPSGIPEAGLGVWNEECDLPVGLHYGPYEGQITEDEAIANSGYSWL 0 0 ITKGRNCYEYVDGKDTSQANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRKGCELLVWYGEEYGQELGIKWGSKWKTEPVAGR 1 2 EPKPEIHPCPSCSVAFSSQTFLSQHGKRNHPSEILPGAPAGNHLQSEEPGPERQNQQQQQQTGPHGWNDKAEGQEVKGRSKPLLKRIRQRGTSRASFKPPNRHMGSSSERERIREEEPSTGQNVNHKNTGKLFVGVKRSKSVT IKHGGCGQGFNDGSHIDTHQRTHSGEKP YICRECGGFTHKSDL.IRHQRTHSQENP YVCRECGRGFRDRSTLITHQRTHSGEKP YVCRECGRGLTEKSTLITHQRTHSGEKP YVCRECGRGFTRKSTLITHQRTHSGEKP YVCRECGRGSRVKSNLIRHQRTHSGEK SGVCIEGE.................... >PRDM7_equCab Equus caballus (horse) genome Laur gene 4 GAS8+ missing front exons, pre-terminal stop GAS8+- flanked right by EMR2- 0 - 0 0 - 1 2 - 1 2 VKPSWVAFRVEQSKQQK 0 0 RMRTAPLSNESRLKELSGTAKLLKTSSSEQVQKPVSPLGEASSSEQHSRRKL 1 2 ELRRKEVGVKMYSLRERKGHAYQEVSEPQDDDYL 1 2 yCENCQNFFIDSCAAHGPPIFVKDSAVDKGHPNRSALTLPLGLRIRPSGIPEAGLGVWNEASDLPLGLHFGPYEGQITEDEEAANSGYSWL 0 0 ITKGRNCYEYVDGKDISWANWMR 2 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVVRPGCELLVWYGDEYGQELGIKWGSKWKRELTAGR 1 2 EPKLEIHPCPSCSLAFSSQKFLSQHVERNHPSQILPGTSARNHLQPEDPSPGDQNQQQQHSDPHSWKDKAHSQEVKERSKPLLKKIRQRRIPRAFSYPPKGQMENFRMRERIMEEKPSIGRKVNPEDTGKLFLEMRMSRNVR VQYGGCGRGFNDRASLIKHQRTHTGEKP YVCRECEQGFTQKSSLIAHQRTHTGEKP YVCRECEQGFSEKSHLIRHQRTHTGEKP YVCRECEQGFSVKSNLIRHQRTHTGEKL .FCREGK..................... >PRDM7_sorAra Sorex araneus (shrew) AALT01000095 Laur gene 8 noDet no useful synteny; upstream spectrin, IgG; GAS8 contig has no sign of pseudogene 0 MSLNRPAEMNTQGKARKLMLKPM 0 0 SKDAFKDISMYFSKEEWAEMGDWEKIRHRNVKRNYEELISI 1 2 GLRAARPAFMSHRRQAIKTQLDDTEESDEEWTPNQQ 1 2 VKSLRVAFRAEQSKHQK 0 0 GRSRTPISNESSSKELSGTRTLLNTKCTKQAQKPLFPPGEASTSGHYSKPKL 1 2 ELRRKEPEVKMYSLRERKGRAYQEVSEPQDDDYL 1 2 YCENCQNFFINKCSAHGSPIFVKDNAVAKGHSNRSALTLPHGLRIGPSGIPEAGLGIWNEASDLPLGLHFGPYEGQITNDEEAANSGYSWL 0 0 ITKGRNCYEYVDGVDESLANWMR 2 1 YVNCARDYEEQNLVAFQYHRQIFYRTCRIIKPGCELLVWYGDEYGQELGIKWGSKWKSELTADK 1 2 EPKPEIYPCPCCSLAFSNQKFLSRHVEHSHPSLILPGTSARTHPKSVNFCPGDQNQWQQHSDACNDKPDEPWNDKLENHKSKGRSKPLPKRMGQKRISTAFPNLRSSKMGSSNKHETIMDKINTGQKENPKDTYRVFAGIGMPRIIR DKHVTLRRSFTNRSSPLTHQRTHTGEKP YVCRECGRGFSQKSHLLTHQRTHTGEKP YVCRECGRGFTDRSSLLTHQRTHTGEKP YVCRECGRGFSLKSSLLRHQRTHTGEKP YVCRECGRGFSLKSSLLTHQRTHTGEKP YVCRECGRGFTDRSSLLTHQRTHTGEKP YVCRECGRGFSLKSSLLTHQRTHTGEKP YVCRECGRGFSRKSSLLRHQRTHTGEKP YVCES....................... >PRDM9a_loxAfr Loxodonta africana (elephant) genome Afro gene 12 noDet chr 153 novel synteny THEG+ MIER2+ PPAP2C PRDM9- ZNF699- 0 MSPARAAKKNPRGDVGSAGRTPT 0 0 aKDTFRDISIYFSKEEWAEMGEWEKFRYRNVKRNYEALVTI 1 2 GLRAPRPAFMCHRRQAIKAQVDNTEDSDEEWTPRQQ 1 2 VKPPSVASRAEQSRHQK 0 0 GTPKALLGNESSLKEVSGTAILLNTTGSEQAQKPVSSPGEASTSDQPSRWKL 1 2 EPRRNEVEVKMYNLRERKGLEYQEVSEPQDDDYL 1 2 yCEKCQNFFIDTCAVHGAPMFVKDSPVDRGHPNHSALTLPPGLRIGPSSIPKAGLGVWNEASELPLGLHFGPYEGQVTEDKEAANSGYSWL 0 0 ITKGKNCYEYVDGKDESWANWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRTIQPDCELLVWYGDEYGQELGIKWGSRWKKELTSGR 1 2 EPKPEIHPCPSCRLAFSSQKFLSQHMKHSHPSPPFPGTPERKYLQPEDPRPGGRRQQRSEQHMWSDKAEDPEAGDGSRLVFERTRRGCISKACSSLPKGQIGSSREGNRMMETKPSPGQKANPEDAEKLFLGVGTSRIAK VRCGECGQGFSQKSVLIRHQKTHSGEKP YVCGECGRGFSVKSVLIKHQRTHSGEKP YVCGECGRGFSVKSVLITHQRTHSGEKP YVCGECGRGFSVKSVLITHQRTHSGEKP YVCGECGRGFSQKSDLIKHQRTHSGEKP YSCRECGRGFSRKSVLITHQRTHSGEKP YVCGECGRGFSQKSNLITHQRTHSGEKP YVCGECGRGFSRKSVLITHQRTHSGEKP YVCGECGRGFSQKSNLITHQRTHSGEKP YVCGECGRGFSQKSDLITHQRTHSGEKP YVCRECGRGFSRKSNLITHQRTHSGEKP YVCRECRRGFSVKSALI........... GHGRRKCSKSAEPLHFPRVSRDQK.... >PRDM9b_loxAfr Loxodonta africana (elephant) genome Afro pseu 3 noDet approx seq after frameshift correction 0 - 0 0 - 1 2 - 1 2 - 0 0 GTPKVLLSNESSLKEVSGTAILLSTMGSEQAQKPVSSPGEASTSDQPSRRKQ 1 2 EPRRKEVEVNMYSLRERKGLVYQEVGEPQDDDYL 1 2 yCEKCQNFFIHTCAVHGAPMFVKDSHVDRGHLNHSALTLPPGLRIGPSSIPEAGLRVR EVSEQLLGLHIGPYEGQVTEDkEAAHSGYSWL 0 0 ITKGRNCYKYVDGKDDPWANRMR 2 1 YVNCIQD KEQNLVAFQYHRQIFHWTCCTIRPGCELLVWYGDNYSQELGIKWGSR KKEL 1 2 EPKPEIHPCPSCPLAISSQKFLDQHTKHSHPSPPFPGTPERKHLQPEDPHPGGRRQQHSEQHLNDKAEDPETGDGSKPVFERARLVGGGAGGVSKVCSSLPKGQMGSSREGNRMMETEGQKVNPEDTEKLFLGVGISRLAK VRCGEYGQGFSQKSVLIRHQRTYSGEEH YVCGECGRGFSWKSQLTRHQRSHSWEKP YVCRECGGFSVKSTLI............ GTGEGNAATIHLHLPS............ >PRDM7_loxAfr Loxodonta africana (elephant) genome Afro pseu 5 GAS8+ scaffold_57 several frameshifts; ZNF540 opposite strand upstream of N-terminus 0 - 0 0 - 1 2 GLRASHPAFTCHCMQAIKAQMDDTEDSNEEQTPRQq 1 2 VRPSWVAFRMEQSKHQR 0 0 GMLRVPRSNESSLKNLSGTSIMLSRAGSEQAQKLVLPPGKASTSDEHSRQKP 1 2 EHRRKGVEVKMYSF ERKGLVYQEIS PQDDDYL 1 2 YCEKCQNFFIDTCESHGVPTFVKNSTTDSGHPNHLALTPSSGLRTRPSSIPKAWLRLWNKAFELLLGLPFSPCEGQVIEDEAVDNSGYSWL 0 0 - 2 1 YVNGTQDEKEQNLVFFQYHRQIFYQTCYAVWPGCQLLVWYRDECGQELGIKWDNRGKKEFe 1 2 EPKPEAHPCPSCPLAFSSEKFLSQHMKHNHPSQSSPETPERKHLQPEDPHPGHQNQQQQQHSDPHRWNDKAEGQQTGDRSKPMFENIRQEVTSRAFSSLPKGQMVCSREGNRMMETEPSPGLKVNPEVTGKLFLGVESSRIAK VKYRGCGRDFSDRSHQSGHQRRHQ KKP SVCKKVKREFSHKSVLITHQRTHSGEKS YVCKESGRGFSAKSNLIRPRRTHTGEKP YVCGERGG.FSVSGLII.HQRAHSPEKP YVCREGRRGFGDKSSFIKHQRATLGEKS YVCKESGRGFS................. AKSNLIRPRRKKCRHDTTPHPQL..... >PRDM7_echTel Echinops telfairi (tenrec) genome Afro pseu 5 noDet 2 frameshifts plus stop codon 0 - 0 0 - 1 2 GLRAPRPAFMCHHRPAAKGQVEDSEDSDEEWTPRQR 1 2 - 0 0 GMPGVSLRNESNLKVLSGTAILLTAAEPEQPH PGSPPGEATTSHEHLRQKV 1 2 epELRRRAVMMNSLRERKNLMYQEVSTPCDDNCL 1 2 YGERCHNFFIDTHIAHGATTFVKDS PMDRSNCSILPPGLRIGPSGIPEAGLGVWNEASELPLGLHFVPYEGQVTKDEAATNSGYSWM 0 0 ITKGRNCYEYVDGKDKSWANwMr 2 1 - 1 2 EPKPEVNPCPSCPLALSSQQLKHSHPFQSLPGTPAEKHLQAEDFHPRGQKLHHFEHHIRNERAEGLETGDGSKPMLERTRLGKMSKTTYNSPKGQTRSSGETNRIREADLNPGQGVNAEDTRNLFLGIGISRIAK VRCRECGHGFSVKSSLITHQRIHTGEKP YVCSECGQGFSQKSVLIRHQRIHTGEKP YICRECDRGFSRKSHLIKHQRTHSGEKP YVCRECGQGFSQKSVLITHHRTHSGEKP YVCRECGRGFSQKSDLIKHERTHS.... >PRDM7_proCap Procavia capensis (hyrax) ABRQ01392668 Afro gene 13 noDet CpG stop in ZNF1, frameshift exon 5 c to cc, blastn favors PDRM9 0 MSPTNAEEWSPGGDTASMGTKPK 0 0 AKDAFRDISIYFSKEEWAEMGEWEKSRYRNVKRNYEALVAI 1 2 GIRVFHPAFMIHPRKTIKAQMDDSEDSDEDWTARQQ 1 2 AKPPSVASREELRKPQK 0 0 GPSRAPLRIKSSLKRVSEPAIVWSTADSEQAQERVQKPVEASASDQPLRRKV 1 2 EPRRHEAEDKRYSLRGGTGPACQEVGEPQDDDYL 1 2 yCEECRNFFIDTCVAHGTPVFIKDISVERGHPNRLALTLPTGLRIGPSSIPDAGLGVWNEASELPPGLHFGPCEGQVTEDEEAANSGYSWL 0 0 VTKGRSCFEYVDGKNEALANWMR 2 1 YVRRARDTEERNLVAFQYHRQIFYRTCCTVRPGCELLVWRGAEDSQALG SRRTMEELTSQK 1 2 EARPEIHPCPSCPLAFSTQKFLSYHVNHSHSSEPFPGTHARRHLPREDPRPGYERDQRSEQHNWNDSTGGPERDVSRPVIERTWEGEISEACSSLPRGHMGRSREGERMAETQSSPGLKVTLAK VRWDEYGQGFGPKSHHITQQTKHSGKKP .CVCKECGGFRVKSLLKSHQMTHSGEKP YVCRECGRGFSVKSTLITHQRTHSGEKP YVCRECGRGFSVKSFLISHQRTHSGEKP YVCRECGRGFSWKSGLITHQRTHTGEKR YVCRECGHGFNRPSRLIRHQRTHSGEQP YVCRECGHGFNRRSQLIRHQRTHTGEQP YVCRECGQGFSGKSGLNRHQRTHSGEKP YVYKECGRGFSVKSTLIKHQRGHSGEKP YVCKECGRGFSRNSGLITHQRTHSGEQP YVCRECGRGFNQKSGVISHQRIHSGEKP FVCGECGRRFSWQSNLITHQRTHSGEKP FVCRECGRGFSAKTSLINHQRIH.....
>PRDM9_homNea Homo neanderthalus (neanderthal) gene genome CDH12- CDH10- chr5 C2H2 variants R HDL S R 0 MSPEKSQEESPEEDTERTERKPM 0 0 VKDAFKDISIYFTKEEWAEMGDWEKTRYRNVKRNYNALITI 1 2 GLRATRPAFMCHRRQAIKLQVDDTEDSDEEWTPRQQ 1 2 VKPPWMALRVEQRKHQK 1 0 GMPKASFSNESSLKELSRTANLLNASGSEQAQKPVSPSGEASTSGQHSRLKL 1 2 ELRKKETERKMYSLRERKGHAYKEVSEPQDDDYL 1 2 YCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWL 0 0 ITKGRNCYEYVDGKDKSWANWMR 1 1 YVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKKELMAGR 2 2 EPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSARKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKRTWQREISRAFSSPPKGQMGSCRVGKRIMEEESRTGQKVNPGNTGKLFVGVGISRIAK VKYGECGQGFSVKSDVITHQRTHTGEKL YVCRECGRGFSWKSHLLIHQRIHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSRQSVLLTHQRRHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSWQSVLLTHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRDKSHLLRHQRTHTGEKP YVCRECGRGFRDKSNLLSHQRTHTGEKP YVCRECGRGFSNKSHLLRHQRTHTGEKP YVCRECGRGFRNKSHLLRHQRTHTGEKP YVCRECGRGFSDRSSLCYHQRTHTGEKP >PRDM9_musCas Mus musculus castaneus ADA68112 terminal fragment ESKRTVEELRTGQTTNTEDTVKSFIASEIS SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSVLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTARSNLIQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTEKSSLIKHQRTHTGEKP YVCRECGWGFTAKSNLIQHQRTHTGEKP YVCRECGRGFTQKSSLIKHQRTHTGEKP YVCRECGRGFTAKSNLIQHQRTHTGEKP YVCRECGWGFTQKSNLIKHQRTHTGEKP YVCRECGWGFTQKSDLIQHQRTHTREK* >PRDM9_musSpi Mus spicilegus 281398541 terminal fragment ESKRTVEELRTGQTTNTEDTVKSFIASEIS SIERQCGQYFSDKSNVNEHQKTHTGEKP YVCRECGRGFTQKSNLIQHQRTHTGEKP YVCRECGRGFTQKSNLIQHQRTHTGEKP YVCRECGRGFTAKSDLIKHQRTHTGEKP YVCRECGRGFTVKSHLTQHQRTHTGEKP YVCRECGRGFTQKSDLIKHQRTHTGEKP YVCRECGRGFTAKSHLTQHQRTHTGEKP YVCRECGRGFTQKSNLIQHQRTHTGEKP YVCRECGRGFTAKSNLIKHQRTHTGEKP YVCRECGRGFTQNSHLTQHQRTHTGEKS YVCRECGWGFKQKSDLIQHQRTHTREK* >PRDM9_micAgr Microtus agrestis ADA68122 terminal fragment ESKKTMEEELRTEQKTNTEDAVRSFIGSEIS RVGGERGQCFSDKSNVNEHQRTHTGEKP YVCRECGRGFTRKSNLNVHQRTHTGEKP YVCRECGRGFTRKALLISHQRTHTGEKP YVCRECGRGFTQKALLISHQRTHTGEKP YVCRECGRGFTQKSYLILHQRTHTGEKP YVCRECGRGFTGKSNLNVHQRTHTGEKP YVCRECGRGFTQKSYLILHQRTHTGEKP YVCRECGRGFTGKSLLIRHQRTHTGEKP YVCRECGRGFTQKSYPILHQRTHTGEK* >PRDM9_arvTer Arvicola terrestris ADA68121 terminal fragment ESKKTMEEELRTDQKTNTEDAIKSFIGSEVS RVEGECGQCFNDKSNVNERQRTHTGEKP YVCRECGRGFTRKSVLILHQRTHTGEKP YVCRECGRGFTQKSVLINHQRTHTGEKP YVCRECGRGFTQKSHLIFHQRTHTGEKP YVCRECGRGFTQKSHLILHQRTHTGEKP YVCRECGRGFTWKSVLILHQRTHTGERP YVCRECGRGFTRKSHLILHQRTHTGEKP YVCRECGRGFTQKSHLILHQRTHTGEKP YVCRECGRGFTRKSVLILHQRTHTGEKP YVCRECGRGFTRKSVLINHQRTHTGEK* >PRDM9_perPol Peromyscus polionotus ADA68120 terminal fragment ESKKTMEEALRTGQKTNTKDTVKSLIGSEFS RIETECGQRFSDKSNVNESQRTHSEEKP YVCRECGQGFIQKSVLICHQRTHTGEKP YVCRECGQGFTWKSHLIRHQRTHTGEKP YVCRECGKGFIRKSHLICHQRTHTGEKP YVCRECGQGFIQKSHLICHQRTHTGEKP YVCRECGQGFTQKSVLICHQRTHTGEKP YVCRECGQGFIRKSYLICHQRTHTGEKP YVCRECGKGFTWKSVLIRHQRTHTVEK* >PRDM9_perMan Peromyscus maniculatus ADA68119 terminal fragment ESKKTMEEELRTGQKTNTKDTVKSLIGSEIS RTETECGQHFSDKSNANESQRTHSEEKP YVCRECGQGFTWKSVLIRHQRTHTGEKP YVCRECGQGFTWKSVLICHQRTHTGEKP YVCRECGQGFTWKSVLICHQRTHTGEKP YVCRECGQGFIQKSHLIRHQRTHTGEKP YVCRECGQGFIRKSHLICHQRTHTGEKP YVCRECGQGFAQKSVLIYHQRTHTGEKP YVCRECGQGFTRKSHLICHQRTHTGEKP YVCRECGQGFAQKSVLICHQRTHTGEKP YVCRECGQGFTWKSVLICHQRTHTGEKP YVCRECGQGFIQKSHLIRHQRTHTGEKP YVCRECGQGFIQKSHLIRHQRTHTGEK* >PRDM9_perLeu Peromyscus leucopus ADA68118 terminal fragment ESKKTMEEALRTGQKTNTKDTVKSLIGSEIS RIETECGQRFSDKSNANESQRTHSEEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFIQKSVLIRHQRTHTGEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFIQKSVLIRHQRTHTGEKP YVCRECGQGFTWKSVLICHQRTHTGEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFTWKSHLIRHQRTHTGEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFIQKSHLICHQRTHTGEKP YVCRECGQGFTRKSYLICHQRTHTGEKP YVCRECGQGFTWKSVLIRHQRTHTAEK* >PRDM9_merUng Meriones unguiculatus ADA68117 terminal fragment ESKRTMEELTTGQKTNTEDTVKSFIGSEIS GTGRECGQCFSDKSNVSEHQRTHTGEKP YVCRECGRGFMQRSNLISHQRTHTGEKP YVCRECGRGFMQRSNLISHQRTHTGEKP YVCRECGRGFTVKSVLISHQRTHTGEKP YVCRECGRGFTVKPHLISHQRTHTGEKP HVCRECGRGFTQRSNLIRHQRTHTGEKP YVCRECGRGFTVKPHLISHQRTHTGEKP YVCRECGRGFTVKPHLISHQRTHTGEKP YVCRECGRGFTVKSVLISHQRTHTGEKP YVCRECGRGFTVKSVLIRHQRTHTGEKP YVCRECRRGFTQRSTLIRHQRTHTGEKP HVCRECGRGFTRGSHLLRHQRTHTGEVLPFQ* >PRDM9_apoSyl Apodemus sylvaticus ADA68116 terminal fragment GGKRTVEEEIRTVQSTNTDDKVKSVIASEIS RVERQRGQCFSDKSNVSERQGTHTGEKP CVCRECGRGFTQKSHLNRHQRTHTGEKP HVCRECGRGFTQKSHLNRHQRTHTGEKP HVCRECGRGFTLKSNLNRHQRTHTGEKP CVCRECGRAFTQKSDLIQHQRTHTGEKP YVCRECGRGFTQKSNLNQHQRTHTGEKP YVCRECGRGFTRKSLLIQHQRTHTGEKP YVCRECGRGFTQKSDLNRHQRTHTGEKP YVCRECGRGLTQKSNLIQHQRTHTGEKP YVCRECGRGFTLKSDLIQHQRTHTGEKP YVCRECGRGFTRKSDLNRHQRTHTGEKP YVCRECGRGFTQKSNLIQHQRTHTGEKP YVCRECGRGFTLKSDLIQHQRTHTGEKP YVCRECGRGFTRKSDLNRHQRTHTGEK* >PRDMx_monDom Monodelphis domestica (opossum) gene genome no GAS8 fragment KRAB SSXRD SET weak C2H2 domain 0 0 0 GEDAFKDISTYFSKKQWVKLKEWEKVRLKNVKRNYEAMIKI 1 2 GLSVPRPAFMCRGRQNKKVKVEESGDSDEEWIPKQL 1 2 0 0 1 2 DCRRKDVEVHIYSLRERKYQVYQEMWDPQDDDYL 1 2 yCEECQIFFLDSCPLHGPPTFVQDSAMVKGHPYCSAITLPPGLRIGLSGIPGAGLGVWNEASTLPLGLHFGPYKGKMTEDDEAANSGYSWM 0 0 ITKGRNCYEYVDGKEESCSNWMR 2 1 YVNCARDEEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIKWGSKWKRPLPELTGE 1 2 GKPGISLCPSTLWASPLIPSSINTRCSKQPP*VFLDSGTGKL*AGRSTAGPATSNRFQLLSDKETSPKEHPSSLWGKTKQVDRREKFSLPQSQQVRGKESSSGEDLSRIQGKSTRQTTMAFQERNR KECE*GFTHQTNLVTHRWTHSGERP YVCV*GFTQKLGFSPYTWTL* 0 >PRDMx_macEug Macropus eugenii (wallaby) notDet genome ---- poor quality fragment 0 0 0 1 2 gFSAPRPTFMCHGKQNKEAKVEESGDFDEEWIRKQP 1 2 0 0 1 2 1 2 yCEECQTFFLETCAVHGPPKFVQDSVMVKGHPYCSAITLPPGLRIGLSGIPGAGLGIWNEASNLPLGLHFGPYEGQMTEDDEAANSGYSWM 0 0 2 1 YVNCARDEEEQNLVAFQYHRKIFYRTCQIIRPGCELLVWYGDEYGQELGIKWGSKWKRPPITLT 1 2 * 0 >PRDMxa_ornAna Ornithorhynchus anatinus (platypus) gene genome chrX5 fragment X5 +- 20577549 no iMet possible in first exon phase 2 0 0 0 1 2 1 2 0 0 1 2 RIGKKPQVRDFNLRKQKRKIYNENYRPEDDDYL 1 2 yCEICQTFFLEKCVLHGPPVFVQDLPVEKWRPNRSTITLPPGMQIKVSGIPNAGLGVWNQATSLPRGLHFGPYMGIRTKNEKESHSGYSWM 0 2 IVRGKNYEYLDGKDKAFSNWMR 2 1 YVNCARSEREQNLVAIQYQGEIYYRTCRVIPPGQELLVWYGLEYGRHLGILPNNNNPEP 1 2 ERAKARVRKSERIEKAMARVRKSEQIERAKARVRTSERIERAMATV RKSERIERAKVTVKKSEQIERAMGRVRKSERIERAKDMGRKKALGGLPRPCRGGLSDETQQRKGGGHEQLGQKPGPSEA RAGPAEGSATPRR HCCDVCRKAFKRLSHLRQHKRIHTGEKP LVCKVCRRTFSDPSNLNRHSRIHTGLRP YVCKLCRKAFADPSNLKRHVFSHTGHKP FVCEKCGKGFNRCDNLKDHSAKHSEDNSTPKP* 0 >PRDMxb_ornAna Ornithorhynchus anatinus (platypus) gene genome chrX5 tandem fragment slight frameshift taa to ta YVN exon X5 +- 20605294 20611704 no iMet possible in first exon phase 2 gg as expected 0 0 0 1 2 1 2 0 0 1 2 RSGKKPQVRDFNLRKQKRKMYTEESEPEDDDYL 1 2 yCEDCQTFFLEKCSVHGPPVFVQDCEAKRCQQNRSEVTLPPGLLIKMSGIPNAGLGVWNQATSLPRGLYFGPFVGIRKNNVKDSLSGYSWAV 0 0 ILRGRNYEYLDGKNTSFSNWMR 2 1 YVNCPRTKYEQNLVAIQYHREIYYRTTPCDSTRSRVAGVVWRRVRSYLGIFWKSETPKS 1 2 ERPHSSGGSFAPSARSGGVKQRIWSKRRSAALQRTRERRNSTHDFPPKHEDTAARQDERQCPDRGRAKQRGVRKSEQIERAKAMGRKKALGGLSPPRRERLSDEAGQRKKSGHEQFWQKPGPSEAWAGPAEGSTIPRR HCCDVCGKAFNRLSRLKQHKRVHTGEKP LVCKICKRAFSDPSNLNRHAKRHTGEKP FVCRVCGRSFNRSDNMNEHRWKHTSNNIIP NTGHMSATVVENASLCINRNYQIYKERATYL* 0 >PRDMx_danRer Danio rerio (zebrafish) Q6P2A1 transcript BC064665 pseudo-homologous 0 MSLSPDLPPSEEQNLEIQGSATNCYSVVIIEEQDDTFNDQPF 1 2 YCEMCQQHFIDQCETHGPPSFTCDSPAALGTPQRALLTLPQGLVIGRSSISHAGLGVFNQGQTVPLGMHFGPFDGEEISEEKALDSANSWV 0 0 ICRGNNQYSYIDAEKDTHSNWMK 2 1 FVVCSRSETEQNLVAFQQNGRILFRCCRPISPGQEFRVWYAEEYAQGLGAIWDKIWDNKCISQ 1 2 GSTEEQATQNCPCPFCHYSFPTLVYLHAHVKRTHPNEYAQFTQTHPLESEAHTPITEVEQCLVASDEALSTQTQPVTESPQEQISTQNGQPIHQTENSDEPDASDIYTAAGEISDEI HACVDCGRSFLRSCHLKRHQRTIHSKEKP YCCSQCKKCFSQATGLKRHQHTH QEQEKNIESPDRPSDI YPCTKCTLSFVAKINLHQHLKRH HHGEYLRLVESGSLTAETEEDHT EVCFDKQDPNYEPPSRGRKSTKNSLKG RGCPKKVAVGRPRGRPPKNKNLEVEVQKIS PICTNCEQSFSDLETLKTHQCPRRDDEGDNVEHPQEASQ YICGECIRAFSNLDLLKAHECIQQGEGS YCCPHCDLYFNRMCNLRRHERTIHSKEKP YCCTVCLKSFTQSSGLKRHQQSHLRRKSHRQSSALFTAAIFPCAYCPFSFTDERYLYKHIRRHHPEMSLKYLSFQEGGVLSVEKP HSCSQCCKSFSTIKGFKNHSCFKQGEKV YLCPDCGKAFSWFNSLKQHQRIHTGEKP YTCSQCGKSFVHSGQLNVHLRTHTGEKP FLCSQCGESFRQSGDLRRHEQKHSGV RPCQCPDCGKSFSRPQSLKAHQQLHVGTKL FPCTQCGKSFTRRYHLTRHHQKMHS* 0
>ZNF133_homSap Homo sapiens (human) NP_001076799 KRAB Krueppel-associated box and zinc fingers 0 MAFRDVAVDFTQDEWRLLSPAQRTLYREVMLENYSNLVSL 1 2 GISFSKPELITQLEQGKETWREEKKCSPATCP 1 2 DPEPELYLDPFCPPGFSSQKFPMQHVLCNHPPWIFTCLCAEGNIQPGDPGPGDQ EKQQQASEGRPWSDQAEGPE GEGAMPLFGRTKKRTLG AFSRPPQRQPVSSRNGLRGVELEASPAQTGNPEETDKLLKRIEVLGFGT VNCGECGLSFSKMTNLLSHQRIHSGEKP YVCGVCEKGFSLKKSLARHQKAHSGEKP IVCRECGRGFNRKSTLIIHERTHSGEKP YMCSECGRGFSQKSNLIIHQRTHSGEKP YVCRECGKGFSQKSAVVRHQRTHLEEKT IVCSDCGLGFSDRSNLISHQRTHSGEKP YACKECGRCFRQRTTLVNHQRTHSKEKP YVCGVCGHSFSQNSTLISHRRTHTGEKP YVCGVCGRGFSLKSHLNRHQNIHSGEKP IVCKDCGRGFSQQSNLIRHQRTHSGEKP MVCGECGRGFSQKSNLVAHQRTHSGERP YVCRECGRGFSHQAGLIRHKRKHSREKP YMCRQCGLGFGNKSALITHKRAHSEEKP CVCRECGQGFLQKSHLTLHQMTHTGEKP YVCKTCGRGFSLKSHLSRHRKTTSVHHR LPVQPDPEPCAGQPSDSLYSL* 0 >ZNF343_homSap Homo sapiens (human) KRAB Krueppel-associated box and zinc fingers 0 MMLPYPSALGDQYWEEILLPKNGENVETMKKLTQNHKAK 1 2 GLPSNDTDCPQKKEGKAQIV 0 0 VPVTFRDVTVIFTEAEWKRLSPEQRNLYKEVMLENYRNLLSL 1 2 AEPKPEIYTCSSCLLAFSCQQFLSQHVLQIFLGLCAENHFHPGNSSPGHWKQQGQQYSHVSCWFENAEGQERGGGSKPWSARTEERETSRAFPSPLQRQSASPRKGNMVVETEPSSAQRPNPVQLDKGLKELETLRFGA INCREYEPDHNLESNFITNPRTLLGKKP YICSDCGRSFKDRSTLIRHHRIHSMEKP YVCSECGRGFSQKSNLSRHQRTHSEEKP YLCRECGQSFRSKSILNRHQWTHSEEKP YVCSECGRGFSEKSSFIRHQRTHSGEKP YVCLECGRSFCDKSTLRKHQRIHSGEKP YVCRECGRGFSQNSDLIKHQRTHLDEKP YVCRECGRGFCDKSTLIIHERTHSGEKP YVCGECGRGFSRKSLLLVHQRTHSGEKH YVCRECRRGFSQKSNLIRHQRTHSNEKP YICRECGRGFCDKSTLIVHERTHSGEKP YVCSECGRGFSRKSLLLVHQRTHSGEKH YVCRECGRGFSHKSNLIRHQRTH* 0 >ZNF169_homSap Homo sapiens (human) KRAB Krueppel-associated box and zinc fingers 0 MSPGLLTTRKEALMAFRDVAVAFTQKEWKLLSSAQRTLYREVMLENYSHLVSL 1 2 GIAFSKPKLIEQLEQGDEPWREENEHLLDLCP 1 2 EPRTEFQPSFPHLVAFSSSQLLRQYALSGHPTQIFPSSSAGGDFQLEAPRCSSEKGESGETEGPDSSLRKRPSRISRTFFSPHQGDPVEWVEGNREGGTDLRLAQRMSLGGSDTMLKGADTSESGAVIRGNYRLGLSKKSSLFSHQKH HVCPECGRGFCQRSDLIKHQRTHTGEKP YLCPECGRRFSQKASLSIHQRKHSGEKP YVCRECGRHFRYTSSLTNHKRIHSGERP FVCQECGRGFRQKIALLLHQRTHLEEKP FVCPECGRGFCQKASLLQHQSSHTGERP FLCLECGRSFRQQSLLLSHQVTHSGEKP YVCAECGHSFRQKVTLIRHQRTHTGEKP YLCPQCGRGFSQKVTLIGHQRTHTGEKP YLCPDCGRGFGQKVTLIRHQRTHTGEKP YLCPKCGRAFGFKSLLTRHQRTHSEEEL YVDRVCGQGLGQKSHLISDQRTHSGEKP CICDECGRGFGFKSALIRHQRTHSGEKP YVCRECGRGFSQKSHLHRHRRTKSGHQL LPQEVF* 0 >ZNF596_homSap Homo sapiens (human) KRAB Krueppel-associated box and zinc fingers 0 MESQESVTFQDVAVDFTQEEWALLDTSQRTLFREVMLENISHLVSV 1 2 GNQLYKSDVISHLEQGEQLSREGLGFLQGQSPVISDREDDPKKQEMLSMQHICKKDAPLISAMQWSHTQEDPLECNNFREKFTEILPLTQYVIPQVGKKPFISQDVGKAISYLPSFNIQKQIHSRSKS YECHQRRNTFIQSSAHRQHNNTQTGEKT FECHVCRKAFSKSSNLRRHEMIHTGVKP HGCHLCGKSFTHCSDLRKHERIHTGEKL YGCHLCGKAFSKSYNLRRHEVIHTKEKP NECHLCGKAFAHCSDLRKHERTHFGEKP YGCHLCGKTFSKTSYLRQHERTHNGEKP YGCHLCGKAFTHCSHLRKHERTHTGEKP YECHLCGKAFTESSVLRRHERTHTGEKP YECHLCWKAFTDSSVLKRHERTHTGEKP YECHLCGKTFNHSSVLRRHERTHTGEKP YECNICGKAFNRSYNFRLHKRIHTGEKP YKCYLCGKAFSKYFNLRQHENSCYKGNK* 0 >HKR1_homSap Homo sapiens (human) KRAB Krueppel-associated box and zinc fingers 0 MRVNHTVSTMLPTCMVHRQTMSCSGAGGITAFVAFRDVAVYFTQEEWRLLSPAQRTLHREVMLETYNHLVSL 1 2 EIPSSKPKLIAQLERGEAPWREERKCPLDLCP 1 2 ESKPEIQLSPSCPLIFSSQQALSQHVWLSHLSQLFSSLWAGNPLHLGKHYPEDQ KQQQDPFCFSGKAEWIQE GEDSRLLFGRVSKNGTSKALSSPPEEQQPAQSKEDNTVVDIGSSPERRADLEETDKVLHGLEVSGFGE IKYEEFGPGFIKESNLLSLQKTQTGETP YMYTEWGDSFGSMSVLIKNPRTHSGGKP YVCRECGRGFTWKSNLITHQRTHSGEKP YVCKDCGRGFTWKSNLFTHQRTHSGLKP YVCKECGQSFSLKSNLITHQRAHTGEKP YVCRECGRGFRQHSHLVRHKRTHSGEKP YICRECEQGFSQKSHLIRHLRTHTGEKP YVCTECGRHFSWKSNLKTHQRTHSGVKP YVCLECGQCFSLKSNLNKHQRSHTGEKP FVCTECGRGFTRKSTLSTHQRTHSGEKP FVCAECGRGFNDKSTLISHQRTHSGEKP FMCRECGRRFRQKPNLFRHKRAHSGA FVCRECGQGFCAKLTLIKHQRAHAGGKP HVCRECGQGFSRQSHLIRHQRTHSGEKP YICRKCGRGFSRKSNLIRHQRTHSG* 0 >PRDM11_homSap Homo sapiens (human) 511 aa 7 exons chr11:45115564 44% id PRDM9 SET 0 MLKMAEPIASLMIVECRACLRCSPLFLYQREK 0 0 DRMTENMKECLAQTNAAVGDMVTVVKTEVCSPLRDQEYGQPC 2 1 SRRPDSSAMEVEPKKLKGKRDLIVPKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGSDETKANWMR 2 1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR 1 2 GEKRLQREKSEQVLDNPEDLRGPIHLSVLRQGKSPYKRGFDEGDVHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLVIRKVPKYQDDAYSQCATTMTHGVQNIGQTQG EGDWKVPQGVSKEPGQLEDEEEEPSSFKADSPAEASLASDPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPPVLPPQVLELPEFSDPAGKLVWMRLLSEGRVRSGLCGG* 0 >PRDM1_homSap Homo sapiens (human) 825 aa 7 exons chr6 106,546,004 3DAL:KMDM..NLTQ SET + 5 C2H2 0 MLDICLEKRVGTTL 0 0 AAPKCNSSTVRFQGLAEGTKGTMKMDMEDADMTLWTEAEFEEKCTYIVNDHPWDSGADGGTSVQAEASLPRNLLFKYATNSEE 0 0 VIGVMSKEYIPKGTRFGPLIGEIYTNDTVPKNANRKYFWR 0 0 IYSRGELHHFIDGFNEEKSNWMRYVNPAHSPREQNLAACQNGMNIYFYTIKPIPANQELLVWYCRDFAERLHYPYPGELTMMNL 1 2 TQTQSSLKQPSTEKNELCPKNVPKREYSVKEILKLDSNPSKGKDLYRSNISPLTSEKDLDDFRRRGSPEMPFYPRVVYPIRAPLPEDFLKASLAYGIERPTYITRSPIPSSTTPSPSARSS PDQSLKSSSPHSSPGNTVSPVGPGSQEHRDSYAYLNASYGTEGLGSYPGYAPLPHLPPAFIPSYNAHYPKFLLPPYGMNCNGLSAVSSMNGINNFGLFPRLCPVYSNLLGGGSLPHPMLNPTS LPSSLPSDGARRLLQPEHPREVLVPAPHSAFSFTGAAASMKDKACSPTSGSPTAGTAATAEHVVQPKATSAAMAAPSSDEAMNLIKNKRNMTGYKTLPYPLKKQNGKIKYECNVCAKTFGQLSNLK 0 0 VHLRVHSGERPFKCQTCNKGFTQLAHLQKHYLVHTGEKPHECQ 0 0 VCHKRFSSTSNLKTHLRLHSGEKPYQCKVCPAKFTQFVHLKLHKRLHTRERPHKCSQCHKNYIHLCSLKVHLKGNCAAAPAPGLPLEDLTRINEEIEKFDISD NADRLEDVEDDISVISVVEKEILAVVRKEKEETGLKVSLQRNMGNGLLSSGCSLYESSDLPLMKLPPSNPLPLVPVKVKQETVEPMDP* >PRDM4_homSap Homo sapiens (human) 801 aa 11 exons chr12:108126644 3DB5:EHGPV..IGVPE SET + 1 + 6 C2H2 domaians 0 MHHR 2 1 MNEMNLSPVGMEQLTSSSVSNALPVSGSHLGLAASPTHSAIPAP 1 2 GLPVAIPNLGPSLSSLPSALSLMLPMGIGDRGVMCGLPERNYTLPPPPYPHLESSYFRTILP 1 2 GILSYLADRPPPQYIHPNSINVDGNTALSITNNPSALDPYQSNGNVGLEPGIVSIDSRSVNTHGAQSLHPSDGHEVALDTAITMENVSRVTSPISTDGMAEELTMDGVAGEHSQIPNGSRSHEPLSVDSVSN NLAADAVGHGGVIPMHGNGLELPVVMETDHIASRVNGMSDSALSDSIHTVAMSTNSVSVALSTSHNLASLESVSLHEVGLSLEPVAVSSITQEVAMGTGHVDVSSDSLSFVSPSLQMEDSNSNKENMATLFTI 1 2 WCTLCDRAYPSDCPEHGPVTFVPDTPIESRARLSLPKQLVLRQSIVGAEV 1 2 GVWTGETIPVRTCFGPLIGQQSHSMEVAEWTDKAVNHIWK 0 0 IYHNGVLEFCIITTDENECNWMMFVRKAR 2 1 NREEQNLVAYPHDGKIFFCTSQDIPPENELLFYYSRDYAQQI 1 2 GVPEHPDVHLCNCGKECNSYTEFKAHLTSHIHNHLPTQGHSGSHGPSHSKERKWKCSMCPQAFISPSKLHVHFMGHMGMKPHKCDFCSKAFSDPSNLRTHLKIHT 1 2 GQKNYRCTLCDKSFTQKAHLESHMVIHTGEKNLKCDYCDKLFMRRQDLKQHVLIHTQ 2 1 ERQIKCPKCDKLFLRTNHLKKHLNSHEGKRDYVCEKCTKAYLTKYHLTRHLKTCKGPTSSSSAPEEEEEDDSEEEDLADSVGTEDCRINSAVYSADESLSAHK* 0 >GAS8_homSap Homo sapiens (human) synteny marker right centromeric positive strand C16orf3- in second intron growth arrest-specific del cancer MAPKKKGKKGKAKGTPIVDGLAPEDMSKEQVEEHVSRIREELDREREERNYFQLERDKIHTFWEITRRQLEEKKAELRNKDREMEEAEERHQVEIKVYKQKVKHLLYEHQNNLTEMKAEG TVVMKLAQKEHRIQESVLRKDMRALKVELKEQELASEVVVKNLRLKHTEEITRMRNDFERQVREIEAKYDKKMKMLRDELDLRRKTELHEVEERKNGQIHTLMQRHEEAFTDIKNYYNDI TLNNLALINSLKEQMEDMRKKEDHLEREMAEVSGQNKRLADPLQKAREEMSEMQKQLANYERDKQILLCTKARLKVREKELKDLQWEHEVLEQRFTKVQQERDELYRKFTAAIQEVQQKT GFKNLVLERKLQALSAAVEKKEVQFNEVLAASNLDPAALTLVSRKLEDVLESKNSTIKDLQYELAQVCKAHNDLLRTYEAKLLAFGIPLDNVGFKPLETAVIGQTLGQGPAGLVGTPT* >CDH12_homSap Homo sapiens (human) synteny marker chr 5 794 aa MLTRNCLSLLLWVLFDGGLLTPLQPQPQQTLATEPRENVIHLPGQRSHFQRVKRGWVWNQFFVLEEYVGSEPQYVGKLHSDLDKGEGTVKYTLSGDGAGTVFTIDETTGDIHAIRSLDRE EKPFYTLRAQAVDIETRKPLEPESEFIIKVQDINDNEPKFLDGPYVATVPEMSPVGAYVLQVKATDADDPTYGNSARVVYSILQGQPYFSIDPKTGVIRTALPNMDREVKEQYQVLIQAK DMGGQLGGLAGTTIVNITLTDVNDNPPRFPKSIFHLKVPESSPIGSAIGRIRAVDPDFGQNAEIEYNIVPGDGGNLFDIVTDEDTQEGVIKLKKPLDFETKKAYTFKVEASNLHLDHRFH SAGPFKDTATVKISVLDVDEPPVFSKPLYTMEVYEDTPVGTIIGAVTAQDLDVGSSAVRYFIDWKSDGDSYFTIDGNEGTIATNELLDRESTAQYNFSIIASKVSNPLLTSKVNILINVL DVNEFPPEISVPYETAVCENAKPGQIIQIVSAADRDLSPAGQQFSFRLSPEAAIKPNFTVRDFRNNTAGIETRRNGYSRRQQELYFLPVVIEDSSYPVQSSTNTMTIRVCRCDSDGTILS CNVEAIFLPVGLSTGALIAILLCIVILLAIVVLYVALRRQKKKDTLMTSKEDIRDNVIHYDDEGGGEEDTQAFDIGALRNPKVIEENKIRRDIKPDSLCLPRQRPPMEDNTDIRDFIHQR LQENDVDPTAPPYDSLATYAYEGSGSVAESLSSIDSLTTEADQDYDYLTDWGPRFKVLADMFGEEESYNPDKVT*
Online References
Open 38 abstracts on PRDM9 and related issues. Or the reverse chronological list below provides free full text for individual articles when that is available:
abs 2011 Neaves Unisexual reproduction among vertebrates. Trends Genet. 2011 Mar;27(3):81-8. abs 2011 Ponting What are the genomic drivers of the rapid evolution of PRDM9? Trends Genetics (2011) 1–7 htm 2011 Yanover Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2 zinc fingers. Nucleic Acids Res. 2011 Feb 22 pdf 2011 Ubeda Red Queen theory of recombination hotspots. J Evol Biol. 2011 Mar;24(3):541-53. abs 2010 Hochwagen Meiosis: a PRDM9 guide to the hotspots of recombination. Curr Biol. 2010 Mar 23;20(6):R271-4. abs 2010 Klug The discovery of zinc fingers and practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010 Feb;43(1):1-21. abs 2010 Berg PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010 Oct;42(10):859-63. abs 2010 McVean PRDM9 marks the spot. Nat Genet. 2010 Oct;42(10):821-2. pdf 2010 Kong Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010 Oct 28;467(7319):1099-103. pmc 2010 Parvanov Prdm9 controls activation of mammalian recombination hotspots. Science. 2010 Feb 12;327(5967):835. pmc 2010 Lorenz The ancient mammalian KRAB zinc finger gene cluster on human chromosome 8q24.3 BMC Genomics. 2010 Mar 26;11:206. pmc 2010 Neale PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010;11(2):104. pmc 2010 Sandovici PRDM9 sticks its zinc fingers into recombination hotspots and between species. F1000 Biol Rep. 2010 May 24;2. pmc 2010 Billings Patterns of recombination activity on mouse chromosome 11 revealed by high resolution mapping. PLoS One. 2010 Dec 8;5(12):e15340. htm 2010 Cheung Genetic control of hotspots. Science. 2010 Feb 12;327(5967):791-2. pdf 2010 Urnov Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005 Jun 2;435(7042):646-51. htm 2010 Zheng Detecting sequence polymorphisms associated with meiotic recombination hotspots in the human genome. Genome Biol. 2010;11(10):R103. htm 2010 Baudat PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010 Feb 12;327(5967):836-40. htm 2010 Myers Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010 Feb 12;327(5967):876-9. pmc 2009 Berglund Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009 Jan 27;7(1):e26. pmc 2009 Thomas Evolution of C2H2-zinc finger genes revisited. BMC Evol Biol. 2009 Mar 4;9:51. pmc 2009 Oliver Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009 Dec;5(12):e1000753. pmc 2009 Thomas Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One. 2009 Dec 30;4(12):e8505. htm 2009 Willis Origin of species in overdrive. Science. 2009 Jan 16;323(5912):350-1. htm 2009 Irie Single-nucleotide polymorphisms of the PRDM9 (MEISETZ) gene in patients with nonobstructive azoospermia. J Androl. 2009 Jul-Aug;30(4):426-31. htm 2009 Mihola A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009 Jan 16;323(5912):373-5. abs 2008 Brayer The protein-binding potential of C2H2 zinc finger domains. Cell Biochem Biophys. 2008;51(1):9-19. pmc 2008 Duret The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008 May 9;4(5):e1000071. pmc 2008 Miyamoto Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 2008 Nov-Dec;25(11-12):553-7. htm 2008 Cho Prediction of DNA binding sites for zinc finger proteins. BBRC 2008 May 9;369(3):845-8. pmc 2007 Coop Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007 Mar 9;3(3):e35. pmc 2007 Fumasoni Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007 Oct 4;7:187. pdf 2006 Phillips A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis. Dev Cell. 2006 Dec;11(6):817-29. htm 2006 Birtle Meisetz and the birth of the KRAB motif. Bioinformatics. 2006 Dec 1;22(23):2841-5. pdf 2006 Hayashi Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell Cycle. 2006 Mar;5(6):615-20. pdf 2005 Hayashi A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005 Nov 17;438(7066):374-8. abs 2000 Laity DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol Biol. 2000 Jan 28;295(4):719-27.