Personal genomics: ACTN3
Introduction to ACTN3 comparative genomics
The alpha actinin gene ACTN3 is a coding gene on human chromosome 11, quite interesting in its own right but best known as ground zero in the debate over frivolity and unexpected consequences of personal genomics. This gene first of all needs careful and exhaustive re-annotation before considering this controversy because its existing peer-reviewed scientific literature (some 22 papers) is a mixture of pre-genomic era obsolescence and gross factual errors such as expression said specific to skeletal muscle.
Some unfortunate historic terminology needs to be explained. Actinins were erroneously thought similar to actin in early studies of myofibrillar components; instead they are homologically unrelated proteins that happen to bind actin. These 'actinins' were then improperly divided into 3 classes (alpha, beta, gamma) before it became known that their respective gene families were wholly unrelated (not homologous). For example, 'beta' actinins refer to heterodimers functioning as actin barbed-end capping proteins in skeletal muscle; they are comprised of the distinct gene families CAPZA and CAPZB themselves non-homologous and thus further misnamed. 'Gamma' actinins refer to yet other unrelated genes.
In this article, actinin shall mean alpha actinin, ie a protein encoded by one of the four paralogous genes ACTN1-ACTN4. Gene names are used for both gene or gene product (as this is always clear from context). Genus and species are indicated with standard 6-letter code (eg ACTN3_homSap). Care still must be taken with published articles and genBank entries that may fail to specify the alpha actinin under consideration despite a 2003 article calling for adherence to HGNC international nomenclature standards followed here.
The comparative genomics situation is further confused by high sequence conservation within the ACTN gene family, by paralog loss in some clades, by possible independent duplication events, and by pre-duplication parental genes only in early deuterostomes. It is not easy to assign transcripts or genomic fragments to correct orthology class by methods such as best reciprocal Blast, especially when the query itself is a fragment (eg third spectrin domain of ACTN3). Many genBank entries are unlabelled, mislabelled, or ambiguously labelled as to correct ortholog family.
However a reliable actinin classifier can be built by requiring flanking gene synteny, diagnostic signature residues and indels in building the reference sequence seed collection that focus on signature regions in which ortholog classes differ significantly from each other. For example ACTN2/3 share a five-residue deletion in exon 19 relative to (ancestral-length) ACTN1/4.
The human ortholog of ACTN3 is unusual (but not unique) in having a fairly abundant null allele, R577x, meaning the arginine at position 577 of the 901 residue protein has been replaced by a stop codon. Worldwide 18% of the human population is homozygous 577x 577x. It is very unlikely that a functional truncated protein can be produced because even if the promoter region is still functional, the mRNA would be degraded by nonsense-mediated decay (with no possibility of selenocysteine substitution) and the stable quaternary dimer necessary for function cannot form (as explained below).
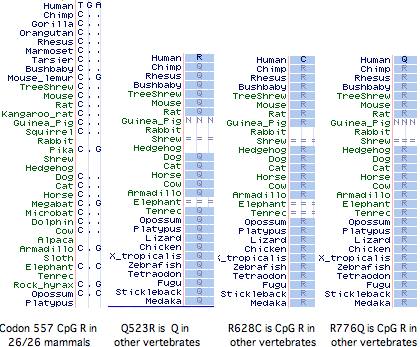
It has not been established whether the 577x was the initial inactivating mutation, as 3 additional amino acid changes (Q523R, R628C, R776Q) have also accrued at otherwise invariant sites in this allele (ie, in the dna donor to the public human genome relative to genBank reference sequence NM_001104). The latter two substitutions are also CpG mutational hotspots (the entire mRNA has 131 such sites). It is not known whether these other changes became widespread before or after R577x nor whether they affect ACTN3 function. However it is not easy to inactivate a large structural protein comprised of independent modules by single substitutions.
Curiously Q523R, R577x, and R628C all occur in the third spectrin repeat despite this region constituting only 11% of the gene, yet not a single nonsynonymous base change has occured in the 2706 bp coding region. With the advent of HapMap and similar projects, the phenotypic associations of these changes, possible co-occurence with wildtype R577, and the date(s) of 577x founder mutations could be resolved.
All mammals with assembled genomes encode a CpG hotspot at codon 577. This has transitioned to TpG in the human 577x allele but is not a polymorphic site in any other known mammal, though the search has been restricted to the individual animals used in genome projects (since transcripts rarely extend this far), plus 36 unrelated baboons and 33 chimpanzees all genotyping to ‘wild-type’ 577R. Thus there is no support for 577x as balanced polymorphism in any mammal other than human even though Z-line skeletal muscle structures may be very similar.
In regards to the supposed evolutionary advantage of various allele combinations (proxied by sprinting or endurance sports prowess), humans remain slow and weak relative to other mammals irregardless of their codon 577 status. The fastest human sprinter cannot outrun a dog with heartworms, much les a rabbit or chubby grizzly bear. A wild male chimp -- without any training or drug enhancement -- has the strength and aggression to rip apart the fittest human cage fighter.
Complete loss of ACTN3 does not give rise to a disease state or even observable phenotype in humans, despite a fallacious initial association with dystrophinopathy. Double knockouts of the orthologous gene in mouse are quite viable but exhibit various measurable effects. Over evolutionary time, the gene has been lost by natural genomic deletion in chicken and finch without known impact, yet retained in lizard, snake, and frog and even doubled in zebrafish (but not other rayfinned fish), again without known effect.
This suggested to some that human ACTN3 had an inessential or inconsequential physiological role to begin with (or become so since divergence with chimps), with its loss readiliy compensatable by other genes (presumably the 80% paralog ACTN3 with which it forms a heterodimer). In this view, ACTN3 may be on its way out the door, to disappear over time as a biallelic pseudogene. Given that over 80 other human genes have been lost, some very conserved over long evolutionary spans] since chimp divergence, it would not be surprising to catch a loss-in-progress.
The primary hurdle to clear here is the extraordinary conservation (proteome: 90th percentile) over 450 million years of ACTN3 amino acid sequence. It appears that this gene arose by segmental duplication from ancestral ACTN2 after the divergence of chondrichthyes (where all matches are ambiguous with respect to ACTN2/3). Gene duplicates are not usually retained over such a time frame unless they have a distinct functional niche that provides selective protection from constantly accruing deleterious mutations ('use it or lose it').
It is quite possible that earlier loss of a companion gene essential to the functional chain of ACTN3 has triggered its subsequent degeneration. The skeletal myosin gene MYH16 is potentially an attractive candidate, though actinins do not bind myosins directly but rather via actin. If ACTN3 functionality overlapped with ACTN2 apart from a critical role involving MYH16, then the loss of the latter gene would leave ACTN2 with no role that could not be compensated for by ACTN3. MYH16 was also lost to an internal stop codon after a half-billiion years of conservation following its separation from other myosins.
Another explanation is balanced polymorphism ('somewhat less is more') along the lines of allele proportions maintained in sickle-cell hemoglobins (heterozygosity can be selectively advantageous in malarial resistance). The idea here that human groups are benefited if some individuals have exceptional speed or endurance.
For ACTN3, frequencies of the three possible diploid states (R577/R577, R577/577x, 577x/R577x) vary by ethnic group and supposedly correlate -- imperfectly but predictively -- with athletic prowess. While single-locus genetic determinism is preposterous as determinative of a complex and vaguely specified phenotype, this has nonetheless gained traction in the popular mindset.
Correlations per se have a poor track record in establishing causality -- for example, a low IQ might also correlate quite strongly with interest and participation in sports. Perhaps R577x (ACTN3 is in fact expressed in brain) merely contributes to low IQ.
ACTN3 transcription IS NOT restricted to skeletal muscle
(section to be added shortly)
R577x as ACTN3 phyloSNP
(section to be added shortly)
R577x and co-evolution of actinin spectrin repeats
(section to be added shortly)
Actinin reference sequence collection and classifier tool
>ACTN1_homSap Homo sapiens (human) length=892 MDHYDSQQTNDYMQPEEDWDRDLLLDPAWEKQQRKTFTAWCNSHLRKAGTQIENIEEDFR DGLKLMLLLEVISGERLAKPERGKMRVHKISNVNKALDFIASKGVKLVSIGAEEIVDGNV KMTLGMIWTIILRFAIQDISVEETSAKEGLLLWCQRKTAPYKNVNIQNFHISWKDGLGFC ALIHRHRPELIDYGKLRKDDPLTNLNTAFDVAEKYLDIPKMLDAEDIVGTARPDEKAIMT YVSSFYHAFSGAQKAETAANRICKVLAVNQENEQLMEDYEKLASDLLEWIRRTIPWLENR VPENTMHAMQQKLEDFRDYRRLHKPPKVQEKCQLEINFNTLQTKLRLSNRPAFMPSEGRM VSDINNAWGCLEQVEKGYEEWLLNEIRRLERLDHLAEKFRQKASIHEAWTDGKEAMLRQK DYETATLSEIKALLKKHEAFESDLAAHQDRVEQIAAIAQELNELDYYDSPSVNARCQKIC DQWDNLGALTQKRREALERTEKLLETIDQLYLEYAKRAAPFNNWMEGAMEDLQDTFIVHT IEEIQGLTTAHEQFKATLPDADKERLAILGIHNEVSKIVQTYHVNMAGTNPYTTITPQEI NGKWDHVRQLVPRRDQALTEEHARQQHNERLRKQFGAQANVIGPWIQTKMEEIGRISIEM HGTLEDQLSHLRQYEKSIVNYKPKIDQLEGDHQLIQEALIFDNKHTNYTMEHIRVGWEQL LTTIARTINEVENQILTRDAKGISQEQMNEFRASFNHFDRDHSGTLGPEEFKACLISLGY DIGNDPQGEAEFARIMSIVDPNRLGVVTFQAFIDFMSRETADTDTADQVMASFKILAGDK NYITMDELRRELPPDQAEYCIARMAPYTGPDSVPGALDYMSFSTALYGESDL* >ACTN2_homSap Homo sapiens (human) length=894 MNQIEPGVQYNYVYDEDEYMIQEEEWDRDLLLDPAWEKQQRKTFTAWCNSHLRKAGTQIE NIEEDFRNGLKLMLLLEVISGERLPKPDRGKMRFHKIANVNKALDYIASKGVKLVSIGAE EIVDGNVKMTLGMIWTIILRFAIQDISVEETSAKEGLLLWCQRKTAPYRNVNIQNFHTSW KDGLGLCALIHRHRPDLIDYSKLNKDDPIGNINLAMEIAEKHLDIPKMLDAEDIVNTPKP DERAIMTYVSCFYHAFAGAEQAETAANRICKVLAVNQENERLMEEYERLASELLEWIRRT IPWLENRTPEKTMQAMQKKLEDFRDYRRKHKPPKVQEKCQLEINFNTLQTKLRISNRPAF MPSEGKMVSDIAGAWQRLEQAEKGYEEWLLNEIRRLERLEHLAEKFRQKASTHETWAYGK EQILLQKDYESASLTEVRALLRKHEAFESDLAAHQDRVEQIAAIAQELNELDYHDAVNVN DRCQKICDQWDRLGTLTQKRREALERMEKLLETIDQLHLEFAKRAAPFNNWMEGAMEDLQ DMFIVHSIEEIQSLITAHEQFKATLPEADGERQSIMAIQNEVEKVIQSYNIRISSSNPYS TVTMDELRTKWDKVKQLVPIRDQSLQEELARQHANERLRRQFAAQANAIGPWIQNKMEEI ARSSIQITGALEDQMNQLKQYEHNIINYKNNIDKLEGDHQLIQEALVFDNKHTNYTMEHI RVGWELLLTTIARTINEVETQILTRDAKGITQEQMNEFRASFNHFDRRKNGLMDHEDFRA CLISMGYDLGEAEFARIMTLVDPNGQGTVTFQSFIDFMTRETADTDTAEQVIASFRILAS DKPYILAEELRRELPPDQAQYCIKRMPAYSGPGSVPGALDYAAFSSALYGESDL* >ACTN3_homSap Homo sapiens (human) 21 exons 901aa NM_001104 Q523R R577x R628C R776Q 0 MMMVMQPEGLGAGEGRFAGGGGGGEYMEQEEDWDRDLLLDPAWEKQQRK 0 0 TFTAWCNSHLRKAGTQIENIEEDFRNGLKLMLLLEVIS 1 2 GERLPRPDKGKMRFHKIANVNKALDFIASKGVKLVSIGAE 1 2 EIVDGNLKMTLGMIWTIILRFAIQDISVE 1 2 ETSAKEGLLLWCQRKTAPYRNVNVQNFHTS 2 1 WKDGLALCALIHRHRPDLIDYAKLR 0 0 KDDPIGNLNTAFEVAEKYLDIPKMLDAE 1 2 DIVNTPKPDEKAIMTYVSCFYHAFAGAEQ 0 0 AETAANRICKVLAVNQENEKLMEEYEKLASE 0 0 LLEWIRRTVPWLENRVGEPSMSAMQRKLEDFRDYRRLHKPPRIQEKCQLEINFNTLQTKLRLSHRPAFMPSEGKLVS 0 0 DIANAWRGLEQVEKGYEDWLLSEIRRLQRLQHLAEKFRQKASLHEAWTR 1 2 GKEEMLSQRDYDSALLQEVRALLRRHEAFESDLAAHQDRVEHIAALAQELN 2 1 ELDYHEAASVNSRCQAICDQWDNLGTLTQKRRDALE 0 0 RMEKLLETIDQLQLEFARRAAPFNNWLDGAVEDLQDVWLVHSVEETQ 0 0 SLLTAHDQFKATLPEADRERGAIMGIQGEIQKICQTYGLRPCSTNPYITLSPQDINTKWDM 0 0 VRKLVPSRDQTLQEELARQQVNERLRRQFAAQANAIGPWIQAKVE 0 0 EVGRLAAGLAGSLEEQMAGLRQQEQNIINYKTNIDRLEGDHQLLQESLVFDNKHTVYSME 0 0 HIRVGWEQLLTSIARTINEVENQVLTRDAKGLSQEQLNEFRASFNHFDR 0 0 KRNGMMEPDDFRACLISMGYDL 0 0 GEVEFARIMTMVDPNAAGVVTFQAFIDFMTRETAETDTTEQVVASFKILAGDK 0 0 NYITPEELRRELPAKQAEYCIRRMVPYKGSGAPAGALDYVAFSSALYGESDL* 0 >ACTN4_homSap Homo sapiens (human) length=911 MVDYHAANQSYQYGPSSAGNGAGGGGSMGDYMAQEDDWDRDLLLDPAWEKQQRKTFTAWC NSHLRKAGTQIENIDEDFRDGLKLMLLLEVISGERLPKPERGKMRVHKINNVNKALDFIA SKGVKLVSIGAEEIVDGNAKMTLGMIWTIILRFAIQDISVEETSAKEGLLLWCQRKTAPY KNVNVQNFHISWKDGLAFNALIHRHRPELIEYDKLRKDDPVTNLNNAFEVAEKYLDIPKM LDAEDIVNTARPDEKAIMTYVSSFYHAFSGAQKAETAANRICKVLAVNQENEHLMEDYEK LASDLLEWIRRTIPWLEDRVPQKTIQEMQQKLEDFRDYRRVHKPPKVQEKCQLEINFNTL QTKLRLSNRPAFMPSEGKMVSDINNGWQHLEQAEKGYEEWLLNEIRRLERLDHLAEKFRQ KASIHEAWTDGKEAMLKHRDYETATLSDIKALIRKHEAFESDLAAHQDRVEQIAAIAQEL NELDYYDSHNVNTRCQKICDQWDALGSLTHSRREALEKTEKQLEAIDQLHLEYAKRAAPF NNWMESAMEDLQDMFIVHTIEEIEGLISAHDQFKSTLPDADREREAILAIHKEAQRIAES NHIKLSGSNPYTTVTPQIINSKWEKVQQLVPKRDHALLEEQSKQQSNEHLRRQFASQANV VGPWIQTKMEEIGRISIEMNGTLEDQLSHLKQYERSIVDYKPNLDLLEQQHQLIQEALIF DNKHTNYTMEHIRVGWEQLLTTIARTINEVENQILTRDAKGISQEQMQEFRASFNHFDKD HGGALGPEEFKACLISLGYDVENDRQGEAEFNRIMSLVDPNHSGLVTFQAFIDFMSRETT DTDTADQVIASFKVLAGDKNFITAEELRRELPPDQAEYCIARMAPYQGPDAVPGALDYKS FSTALYGESDL* >ACTN2_galGal Gallus gallus MNSMNQIETNMQYTYNYEEDEYMTQEEEWDRDLLLDPAWEKQQR KTFTAWCNSHLRKAGTQIENIEEDFRNGLKLMLLLEVISGERLPKPDRGKMRFHKIAN VNKALDYIASKGVKLVSIGAEEIVDGNVKMTLGMIWTIILRFAIQDISVEETSAKEGL LLWCQRKTAPYRNVNIQNFHLSWKDGLGLCALIHRHRPDLIDYSKLNKDDPIGNINLA MEIAEKHLDIPKMLDAEDIVNTPKPDERAIMTYVSCFYHAFAGAEQAETAANRICKVL AVNQENERLMEEYERLASELLEWIRRTIPWLENRTPEKTMQAMQKKLEDFRDYRRKHK PPKVQEKCQLEINFNTLQTKLRISNRPAFMPSEGKMVSDIAGAWQRLEQAEKGYEEWL LNEIRRLERLEHLAEKFRQKASTHEQWAYGKEQILLQKDYESASLTEVRAMLRKHEAF ESDLAAHQDRVEQIAAIAQELNELDYHDAASVNDRCQKICDQWDSLGTLTQKRREALE RTEKLLETIDQLHLEFAKRAAPFNNWMEGAMEDLQDMFIVHSIEEIQSLISAHDQFKA TLPEADGERQAILSIQNEVEKVIQSYSMRISASNPYSTVTVEEIRTKWEKVKQLVPQR DQSLQEELARQHANERLRRQFAAQANVIGPWIQTKMEEIARSSIEMTGPLEDQMNQLK QYEQNIINYKHNIDKLEGDHQLIQEALVFDNKHTNYTMEHIRVGWELLLTTIARTINE VETQILTRDAKGITQEQMNDFRASFNHFDRRKNGLMDHDDFRACLISMGYDLGEAEFA RIMSLVDPNGQGTVTFQSFIDFMTRETADTDTAEQVIASFRILASDKPYILADELRRE LPPEQAQYCIKRMPQYTGPGSVPGALDYTSFSSALYGESDL* >ACTN1_galGal Gallus gallus MDHHYDPQQTNDYMQPEEDWDRDLLLDPAWEKQQRKTFTAWCNS HLRKAGTQIENIEEDFRDGLKLMLLLEVISGERLAKPERGKMRVHKISNVNKALDFIA SKGVKLVSIGAEEIVDGNVKMTLGMIWTIILRFAIQDISVEETSAKEGLLLWYQRKTA PYKNVNIQNFHISWKDGLGFCALIHRHRPELIDYGKLRKDDPLTNLNTAFDVAEKYLD IPKMLDAEDIVGTARPDEKAIMTYVSSFYHAFSGAQKAETAANRICKVLAVNQENEQL MEDYEKLASDLLEWIRRTIPWLENRAPENTMQAMQQKLEDFRDYRRLHKPPKVQEKCQ LEINFNTLQTKLRLSNRPAFMPSEGKMVSDINNAWGGLEQAEKGYEEWLLNEIRRLER LDHLAEKFRQKASIHESWTDGKEAMLQQKDYETATLSEIKALLKKHEAFESDLAAHQD RVEQIAAIAQELNELDYYDSPSVNARCQKICDQWDNLGALTQKRREALERTEKLLETI DQLYLEYAKRAAPFNNWMEGAMEDLQDTFIVHTIEEIQGLTTAHEQFKATLPDADKER QAILGIHNEVSKIVQTYHVNMAGTNPYTTITPQEINGKWEHVRQLVPRRDQALMEEHA RQQQNERLRKQFGAQANVIGPWIQTKMEEIGRISIEMHGTLEDQLNHLRQYEKSIVNY KPKIDQLEGDHQQIQEALIFDNKHTNYTMEHIRVGWEQLLTTIARTINEVENQILTRD AKGISQEQMNEFRASFNHFDRKKTGMMDCEDFRACLISMGYNMGEAEFARIMSIVDPN RMGVVTFQAFIDFMSRETADTDTADQVMASFKILAGDKNYITVDELRRELPPDQAEYC IARMAPYNGRDAVPGALDYMSFSTALYGESDL* >ACTN4_galGal Gallus gallus MVDYHSAGQPYPYGGNGPGPNGDYMAQEDDWDRDLLLDPAWEKQ QRKTFTAWCNSHLRKAGTQIENIDEDFRDGLKLMLLLEVISGERLPKPERGKMRVHKI NNVNKALDFIASKGVNVVSIGAEEIVDGNAKMTLGMIWTIILRFAIQDISVEETSAKE GLLLWCQRKTAPYKNVNVQNFHISWKDGLAFNALIHRHRPELIEYDKLRKDDPVTNLN NAFEVAEKYLDIPKMLDAEDIVNTARPDEKAIMTYVSSFYHAFSGAQKAETAANRICK VLAVNQENEHLMEDYEKLASDLLEWIRRTIPWLEDRSPQKTIQEMQQKLEDFRDYRRV HKPPKVQEKCQLEINFNTLQTKLRLSNRPAFMPSEGRMVSDINTGWQHLEQAEKGYEE WLLNEIRRLEPLDHLAEKFRQKASIHEAWTEGKEAMLKQKDYETATLSDIKALIRKHE AFESDLAAHQDRVEQIAAIAQELNELDYYDSPSVNARCQKICDQWDVLGSLTHSRREA LEKTEKQLETIDELHLEYAKRAAPFNNWMESAMEDLQDMFIVHTIEEIEGLIAAHDQF KATLPDADREREAILGIQREAQRIADLHSIKLSGNNPYTSVTPQVINSKWERVQQLVP TRDRALQDEQSRQQCNERLRRQFAGQANIVGPWMQTKMEEIGRISIEMHGTLEDQLQH LKHYEQSIVDYKPNLELLEHEHQLVEEALIFDNKHTNYTMEHIRVGWEQLLTTIARTI NEVENQILTRDAKGISQEQMQEFRASFNHFDKDHCGALGPEEFKACLISLGYDVENDR QGDAEFNRIMSLVDPNGSGSVTFQAFIDFMSRETTDTDTADQVIASFKVLAGDKNYIT AEELRRELPPEQAEYCIARMAPYRGPDAAPGALDYKSFSTALYGESDL* >ACTN3_echOce Echis ocellatus (snake) fragment not covering codon 577 EYMAQEEDWDRDLLLDPAWEKQQRKTFTAWCNSHLRKAGTQIENIEEDFRNGLKLMLLLE VISGERLPKPDKGKMRFHKIANVNKALDFIASKGVKLVSIGAEEIVDGNLKMTLGMIWTI ILRFAIQDISVEETSAKEGLLLWCQRKTAPYRNVNVQNFHISWKDGLALCALIHRHRPDL DYAKLRKDDPIGNLNTAFEVAEKYLDIPKMLDAEDIVNTPKP >ACTN3_anoCar Anolis carolinensis (lixard) 19 of 22 exons scaffold_1178:54057-80055 weak synteny ZDHHC24 MTTQIETHVQYNHSYMTTEDYMAQEEDWDRDLLLDPAWEKQQRK ETSAKEGLLLWCQRKTAPYRNVNVQNFHIR WKDGLALCALIHRHRPDLIDYAKLRK DDPIGNLNTAFEVAEKYLDIPKMLDAE DIVNTPKPDEKAIMTYVSCFYHAFAGAEQ AETAANRICKVLAVNQENEKLMQEYEKLASE LLEWIRRTIPWLENRVPEKTMSAMQRKLEDFRDYRRVHKPPRVQEKCQLEINFNTLQTKLRLSNRPAFMPSEGKMVS DIANAWKGLEQVEKGYEEWLLTEIRRLERLDHLAEKFKQKATLHENWTR GKEDMLTQKDYESASLYEIRALMRKHEAFESDLAAHQDRVEQIAAIAQELn ELDYHDAASVNSRCQAICDQWDTLGTLTQKRRDSLE RVEKLLETIDQLYLEFAKRAAPFNNWMDGAIEDLQDMFIVHSIEEIQ SLITAHEQFKATLPEADKERMAILGIQNEIQKIAQTYGIKLSGINPYTNLSHLDIANKWDT VKQLVPHRDQTLQEELARQQANERLRRQFAAQANIIGPWVQTKME EIGHISVDISGSLEDQMNHLKQHEQNIINYKSNIDKLEGDHQLIQEALVFDNKHTNYTME HIRVGWEQLLTTIARTINEIENQILTRDAKGISQEQMNEFRASFNHFDR KRNGMMDPDDFRACLISMGYD GEVEFARIMTLVDPNNTGVVTFQAFIDFMTRETAETDTAEQVMASFKILASDK SYITIEELRRELPPEQAEYCISRMTKYTGADAVSGALDYVSFSSALYGESDL* >ACTN3_xenTro NP_001135513 length=896 solid right synteny homSap MTTIETHIQYSNSYNMTEEDYMVQEEDWDRDLLLDPAWEKQQRKTFTAWC NSHLRKAGTQIENIEEDFRNGLKLMLLLEVISGERLPKPDKGKMRFHKIA NVNKALDFIASKGVKLVSIGAEEIVDGNLKMTLGMIWTIILRFAIQDISV EETSAKEGLLLWCQRKTAPYRNVNVQNFHISWKDGLALCALIHRHRPDLI DYSKLRKDDPIGNLNTAFDVAEKYLDIPKMLDAEDIVNTPKPDEKAIMTY VSCFYHAFAGAEQAETAANRICKVLAVNQENEKLMEEYEKLASELLEWIR RTIPWLENRTPEKNMGAMQKKLEDFRDYRRVHKPPRVQEKCQLEINFNTL QTKLRLSNRPAFMPSEGKMVSDIANAWKGLEQVEKGYEEWLLLEIRRLER LEHLAEKFKQKASLHESWTRGKEELLTQRDYESASLMEIRALVRKHEAFE SDLAAHQDRVEQIAAIAQELNELDYHDAASVNSRCQAICDQWDNLGTLTQ KRRDALERVEKLLETIDQLYLEFAKRAAPFNNWMDGAVEDLQDMFIVHSI EEIQSLITAHEQFKATLPEADKEKMSILGIQAEIQKIAQTYGIKLSGINP YTNLTHLDISNKWETVKQLVPHRDQTLQEELARQQANERLRRQFAAQANV IGPWIQSKMEEIGRISVDISASLEDQMNHLKQYEQNIISYKSNIDKLEGD HQLIQESLIFDNKHTNYSMEHIRVGWEQLLTTIARTINEVENQILTRDAK GISQEQMNEFRASFNHFDRKRNGMMDPDDFRACLISMGYDLGEVEFARIM TLVDPNNTGVVTFQAFIDFMTRETAETDTAEQVMASFKILASDKSYITIE ELRRELPPEQAEYCITRMTKYTGADAISGALDYMSFSSALYGESDL* >ACTN2_xenTro NP_001005053 length=894 solid bilateral synteny homSap MTQAVTNETYNYCYDEEEYMNQEEEWDRDMLLDPAWEKQQRKTFTAWCNS HLRKAGTMIENIEEDFRNGLKLMLLLEVISGERLPKPDRGKMRFHKIANV NKALDFIASKGVKLVSIGAEEIVDGNVKMTLGMIWTIILRFAIQDISVEE TSAKEGLLLWCQRKTAPYRNVNIQNFHTSWKDGLGLCALIHRHRPDLIDY SKLNKDDAVGNINLAMDVAEKYLDIPKMLDAEDIVSTAKPDERAIMTYVS CFYHAFAGAEQAETAANRICKVLAVNQENERMMEEYERLASELLEWIRRT IPWLENRTSEKTMQAMLKKLEDFRDYRRKHKPPRVQEKCQLEINFNTLQT KLRISNRPAFMPSEGKMVSDIANAWQRLEQGEKGYEEWLLTELRRLERVE HLAEKFRQKASLHESWTAGKEQLLLQKDYETASLTEVRALLRKHEAFESE LAARQDRVEQIAAIAQELNELDYYDAARINERCQKICDQWDRLGTLTQKR REALERTEKLLETVDQLHLEFIKRAGPFNIWMEGAMEDLQDMFIVHNVEE IQKLITAHEQFKATLPEADSERQAILGIQNEVEKVIQSYNIKISSHNPYS SITLDEIRTKWEKVKQLVPIRDQTLQEELARQQNNERLRRQFAGQANVIG PWIQKQMEEIGRSCIDFTGTLEDQMNSLKMLEHNIVNYKHNVDKLEGDHQ MIQESLIFDNKHTNYSMEHIRVGWELMLTTVARTINEVETQILTRDAKGI TQEQINVFRSSFNHFDKKKNGLMEHDDFRACLISMGYDLGEAEFARIMAL VDPSGIGTISFQSFIDFMTRETAETDTSEQVIAAFRILAADKPYILPEEL RRELPPEQAQYCLSKMPTYTGPGAVPGALDFTCFSSALYGESDL* >ACTN3a_danRer Danio rerio (zebrafish) NP_571597 partial synteny MTAVESQVQYGSYMMTATEEYMIQEDDWDRDLFLDPAWEKQQRKTFTAWC NSHLRKAGTQIENIEEDFRNGLKLMLLLEAISGERLPKPDKGKMRFHKIA NVNKALDFISSKGVKLVSIGAEEIVDGNLKMTLGMIWTIILRFAIQDISV EETSAKEGLLLWCQRKTAPYRNVNVQNFHISWKDGLALCALIHRHRPDLI DYSKLRKDDPIGNLNTAFDIAEKFLDIPKMLDADDIVNTPKPDEKAIMTY VSCFYHAFAGAEQAETAANRICKVLAVNQENEKLMEEYEKLASELLEWIQ RTIPWLENRVAEQTMHAMQQKLEDFRDYRRVHKPPKVQEKCQLEINFNTL QTKLRLSNRPAFMPSEGKMVSDIADAWKGLEQVEKGYEEWLLTEIRRLER LDHLAEKFRQKSTLHQSWTTGKEELLSQKDYETASLMEIRALMRKHEAFE SDLAAHQDRVEQIAAIAQELNELDFYDAATINAQCQGICDQWDNLGTLTQ KRRESLERVEKLWETIDQLYLEFAKRAAPFNNWMDGAMEDLQDMFIVHSI EEIQSLITAHDQFKATLPEADKERMAIMGIHSEVLKIAQTYGIKIVSENP YTVLSHQNIVNKWEAVKQLVPMRDQMLQEEVARQQSNERLRRQFAAQANI IGPWIQTKMEEISHVSIDIAGSLEEQMSNLKTYEQNIINYKSNIDKLEGD HQLSQEGLIFDNKHTNYTMEHVRVGWEQLLTTIARTINEVENQILTRDAK GISQEQLNEFRASFNHFDRKRNGMMDPDDFRACLISMGYDLGEVEFARIM TLVDPNNTGVVTFQAFIDFMTRETAETDTAEQVMASFKILASDKAYITVE ELRRELPPEQAEYCISRMTKYMGGDAPPGALDYISFSSALYGESDL* >ACTN3b_danRer Danio rerio (zebrafish) NP_991107 length=890 partial synteny MQYNNTYMMSTTQDYMIQEDDWDRDLLLDPAWEKQQRKTFTAWCNSHLRK AGTQIENIEEDFRNGLKLMLLLEVISGERLPKPDKGKMRFHKIANVNKAL DFICSKGVKLVSIGAEEIVDGNVKMTLGMIWTIILRFAIQDISVEETSAK EGLLLWCQRKTAPYRNVNVQNFHISWKDGLALCALIHRHRPDLIDYSKLR KDDPIGNLNTAFEVAEKYLDIPKMLDAEDIVNTPKPDEKAIMTYVSCFYH AFAGAEQAETAANRICKVLAVNQENERLMEEYEKLASELLEWIRRTIPWL ENRAAEQTMRAMQQKLEDFRDYRRVHKPPRVQEKCQLEINFNTLQTKLRL SNRPAFMPSEGKMVSDIANAWKGLEQVEKGYEEWLLTEIRRLERLDHLAE KFKQKCSLHESWTTGKEHLLSQKDYETASLMEIRALMRKHEAFESDLAAH QDRVEQIAAIAQELNELDYHDAATVNARCQGICDQWDNLGTLTQKRRDSL ERVEKLWETIDQLYLEFAKRAAPFNNWMDGAIEDLQDMFIVHSIEEIQSL ITAHDQFKATLPEADKERMAVMGIQNEIVKIAQTYGIKLVGVNPYSVLSP QDITNKWEAVKQLVPLRDQMLQEEVARQQANERLRRSFAAQANIIGPWIQ TKMEEIGHVSVDIAGSLEEQMNNLKQYEQNIINYKSNIDKLEGDHQLIQE ALIFDNKHTNYTMEHIRVGWEQLLTTIARTINEVENQILTRDAKGISQEQ LNEFRASFNHFDRKRNGMMDPDDFRACLISMGYDLGEVEFARIMTLVDPN NTGVVTFQAFIDFMTRETAETDTAEQVMASFKILASDKPYITVEELRREL PPEQAEYCISRMTKYVGPEGALGALDYISFSSALYGESDL* >ACTN1_leuEri Leucoraja erinacea (ray) est ambiguous fragment TFTAWCNSHLRKADTQIESIEEDFRDGLKLMLLLEVISGERLAKPERGKMRVHKISNVNK ALDFIAGKGVKLVSIGAEEIVDGNAKMTLGMIWTIILRFAIQDISVEQTSAKEGLLLWCQ RKTAPYKNVNIQNFHISWKDGLGFCALIHRHRPELIDYAKLRKDDPYTNLNTAFDVAEKY LDIPKMLDAEDIVGTARPDEKAIMTYVSSFYHAFSGAQ >ACTN23_squAca Squalus acanthias (shark) est ambiguous fragment EYMLQEEDWDRDLLLDPAWEKQQRKTFTAWCNSHLRKAGTQIENIEDDFRNGLKLMLLLEVISGERLPKPDQGKMRFHKIANVNKALDYISRKGVKLVSI GAEEIVDGNVKMTLGMIWTIILRFAIQDISVEETSAKEGLLLWCQRKTAPYKNVNVQNFHTSWKDGL >ACTN23_calMil Callorhynchus milii (elephant_shark) assembly ambiguous fragment GERLPKPDKGKMRFHKIANVNKALDYIASKGVKLVSIGAEEIVDGNVKMTLGMIWTIILRFAIQDISVEETSAKEGLLLWCQRKTAPYKNVTVQNFHT