Phospholipases PLBD1 and PLBD2: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 866: | Line 866: | ||
* Fraud: It is not unusual for annotational hoaxes to be perpetrated in the scientific literature, complete with bogus submissions to GenBank. Here however simple tBlastn searches show that the most recent assembly of the drosophila genome does contain a letter-perfect match to the LAMA entry and further that this protein has a more or less a full length match to conventional vertebrate PLBD2. This is beyond anyone's ability to manipulate. | * Fraud: It is not unusual for annotational hoaxes to be perpetrated in the scientific literature, complete with bogus submissions to GenBank. Here however simple tBlastn searches show that the most recent assembly of the drosophila genome does contain a letter-perfect match to the LAMA entry and further that this protein has a more or less a full length match to conventional vertebrate PLBD2. This is beyond anyone's ability to manipulate. | ||
* Pseudogene: This is a large protein of almost 600 amino acids. Should it have lost its function, rapid divergence would result (as seen), with accrual of frameshifts and stop codons (not seen). The divergence would occur equally rapidly at ultra-conserved residues as at ones not under selection (not seen), either in D. melanogaster or one of the other 11 drosophilids with sequenced genome (not seen). | * Pseudogene: This is a large protein of almost 600 amino acids. Should it have lost its function, rapid divergence would result (as seen), with accrual of frameshifts and stop codons (not seen). The divergence would occur equally rapidly at ultra-conserved residues as at ones not under selection (not seen), either in D. melanogaster or one of the other 11 drosophilids with sequenced genome (not seen). However recent pseudogenization in certain lineages cannot be ruled out as comparative genomics becomes insufficient. | ||
* Horizontal transfer: The LAMA protein might fail to nest within the gene tree as expected from the fruitfly position in the species tree. This happens rarely in metazoan from gene transfer from other species, notably parasites, commensals, and intra-cellular symbionts. In this case, tBlastn would locate the appropriate clade, even if the exact species had no data because genomic coverage is so pervasive today. However the LAMA gene has no outside affinities. | * Horizontal transfer: The LAMA protein might fail to nest within the gene tree as expected from the fruitfly position in the species tree. This happens rarely in metazoan from gene transfer from other species, notably parasites, commensals, and intra-cellular symbionts. In this case, tBlastn would locate the appropriate clade, even if the exact species had no data because genomic coverage is so pervasive today. However the LAMA gene has no outside affinities. | ||
| Line 873: | Line 873: | ||
* Role loss: Along the lines of [http://en.wikipedia.org/wiki/Gene_sharing Piatigorsky], a single gene product may have multiple roles, sometimes quite distinct (eg aldehyde dehydrogenase ALDH3A1 also serving as a lens crystallin). Should the gene subsequently be duplicated, these pre-existing roles can be split up in the two copies (rather than one copy continue with the parental role and the other copy scramble to acquire a new adaptive function). Selection initially acts on all the functions in proportion to their importance. | * Role loss: Along the lines of [http://en.wikipedia.org/wiki/Gene_sharing Piatigorsky], a single gene product may have multiple roles, sometimes quite distinct (eg aldehyde dehydrogenase ALDH3A1 also serving as a lens crystallin). Should the gene subsequently be duplicated, these pre-existing roles can be split up in the two copies (rather than one copy continue with the parental role and the other copy scramble to acquire a new adaptive function). Selection initially acts on all the functions in proportion to their importance. | ||
[[Image:DroMelPLDB2X.png|left]] | |||
In this last scenario, phospolipase activity was lost, leaving selection to act solely on the residual regulatory function. However within the 12 drosophilid genomes, the PLBD2 are quite diverged, with percent identity quickly ranging from 75% down to 45%. This suggests the protein is not yet well adapted to what was initially a secondary function or that few of its features are needed. | In this last scenario, phospolipase activity was lost, leaving selection to act solely on the residual regulatory function. However within the 12 drosophilid genomes, the PLBD2 are quite diverged, with percent identity quickly ranging from 75% down to 45%. This suggests the protein is not yet well adapted to what was initially a secondary function or that few of its features are needed. | ||
This raises the question whether PLBD2 also serves a cryptic non-catalytic function in other species (mammals | This raises the question whether PLBD2 also serves a cryptic non-catalytic function in other species (eg mammals). It is thus important to date the onset of loss of phospholipase activity (coupled with retention of within the PLBD2 locus) within arthropods because this also provides some dating of the secondary function origin. This can be dated to the last tens of millions of years because only a few other insects (eg tsetse Glossina morsitans, [http://www.ncbi.nlm.nih.gov/nucleotide/289740208 EZ422576]) outside drosophilids carry a comparable gene. | ||
Within insects, some species such as Acyrthosiphon carry both a normal and derived PLBD2. However the normal form has been lost in other insects with derived PLBD2. Although no chelicerate has the derived PLBD2, Ixodes has multiple copies of normal PLBD2, some closely related and others fairly diverged but still clearly classifying as PLBD2. Molluscs, annelids, and trematodes have PLBD2 but not PLBD1 or PLBD2x. | Within insects, some species such as Acyrthosiphon carry both a normal and derived PLBD2. However the normal form has been lost in other insects with derived PLBD2. Although no chelicerate has the derived PLBD2, Ixodes has multiple copies of normal PLBD2, some closely related and others fairly diverged but still clearly classifying as PLBD2. Molluscs, annelids, and trematodes have PLBD2 but not PLBD1 or PLBD2x. | ||
| Line 882: | Line 884: | ||
The loss of active site residues characteristic of NTN hydratases and more specifically those shared by both PLBD1 and PLBD2 phospholipases is taken here as a proxy for loss of phospholipase (and indeed all) catalytic activity. | The loss of active site residues characteristic of NTN hydratases and more specifically those shared by both PLBD1 and PLBD2 phospholipases is taken here as a proxy for loss of phospholipase (and indeed all) catalytic activity. | ||
Active site residues are lost in derived PLBD2 though the first disulfide is retained: | |||
<---- human active site ----> <-- human disulfides --> <-- glycosylation--> | <---- human active site ----> <-- human disulfides --> <-- glycosylation--> | ||
<font color=blue>PLBD2_homSap C244 H261 W264 T325 N427 R458 C142-C152 C492-C495 N436S N515S</font> | |||
PLBD2x_droMel A216 H238 A241 G305 G438 ? C98-C108 | PLBD2x_droMel A216 <font color=blue>H238</font> A241 G305 G438 ? <font color=blue>C98-C108</font> | ||
PLBD2x_glomor A210 H231 A234 G298 G424 ? C94-C104 | PLBD2x_glomor A210 <font color=blue>H231</font> A234 G298 G424 ? <font color=blue>C94-C104</font> | ||
PLBD2x_acrPis A233 Q250 S253 V315 S413 | PLBD2x_acrPis A233 Q250 S253 V315 S413 V436 <font color=blue>C132-C139</font> | ||
PLBD2_acrPis C208 H225 W228 T288 N390 R421 C112-C119 C434-C437 N399S N477T | <font color=blue>PLBD2_acrPis C208 H225 W228 T288 N390 R421 C112-C119 C434-C437 N399S N477T</font> | ||
Unless there is some counterpart to the secondary function, the tempo and mode of evolution of the LAMA protein makes it largely irrelevant to its human ortholog and lysosomal storage diseases. Indeed, fruitflies have in effect lost both PLBD1 and PLBD2, proving they are non-essential to lysosome function. | Unless there is some counterpart to the secondary function, the tempo and mode of evolution of the LAMA protein makes it largely irrelevant to its human ortholog and lysosomal storage diseases. Indeed, fruitflies have in effect lost both PLBD1 and PLBD2, proving they are non-essential to lysosome function. | ||
Drosophila PLDB2x at SwissModel: little of the original fold remains: | |||
PLDB2x 45 CATAL WTKQV-GFQI ENWKQQNDLV NIPTGVGRIC YKDSVYENGW | |||
3fgrA 63 vsr--trsll ldaasgqlrl ed-----g-- fhpdavawan ltnairetgw | |||
ssss ss ssss s sssss sssss ss | |||
sss sssss ss ssss ss s sssss sssss ss | |||
PLDB2x 89 AQIEVETQRT YPDWVQAYAA GMLEGSLTWR NIYNQWSNTI SSSCERDEST | |||
3fgrA 104 ayldlstngr yndslqayaa gvveasvsee liymhwmntv vnycgpfeye | |||
ssssssss hhhhhhhh hhhhhh h hhhhhhhh | |||
ssssssss hhhhhhhh hhhhh hh hhhhhhhh | |||
PLDB2x 139 QKFCGWLRDL LTTNYHRLKR QTEKAENDHY WHQLHLFITQ LEGLETGYKR | |||
3fgrA 154 vgyceklknf leanlewmqr emelnpdspy whqvrltllq lkgledsyeg | |||
hhhhhhhhhh hhhhhhhhhh hhhh hh hhhhhhhhhh hhhhhhhhh | |||
hhhhhhhhhh hhhhhhhhhh hhhh hh hhhhhhhhhh hhhhhhhhh | |||
PLDB2x 189 GASRARSDLE EEIPFSDFLL MNAAADIQDL KIYYENY -- - | |||
3fgrA 204 rltfptgr-- ftikplgfll lqisgdledl epalnktgsg s | |||
hh h hhhhhh hhh | |||
hh h hhhhhh hhh | |||
=== Nematodes (C. elegans) === | === Nematodes (C. elegans) === | ||
Potentially a better model organism than fruitfly, C. elegans has 3 phospholipase B genes, with the best match to human | Potentially a better model organism than fruitfly, C. elegans has 3 phospholipase B genes, with the best match to human PLBD2 having 44% identity. These appear to be a lineage-specific expansion of PLBD2, whereas PLBD1 has been lost from this and other nematodes. (The C. elegans genome is assuredly complete and the detection methods are sufficiently sensitive.) | ||
>PLBD2a_caeEle Caenorhabditis elegans (nematode) NP_499668 Y37D8A | >PLBD2a_caeEle Caenorhabditis elegans (nematode) NP_499668 Y37D8A | ||
| Line 924: | Line 950: | ||
Below, only a few key sequences are shown -- ones with an experimental literature and some from deeply diverged pre-bilaterans. These have been intronated by blast of protein against genome assemblies. | Below, only a few key sequences are shown -- ones with an experimental literature and some from deeply diverged pre-bilaterans. These have been intronated by blast of protein against genome assemblies. | ||
A separate section holds anomalous PLBD2x insect sequences. These resulted from a duplication of PLBD2, followed bu severe specialization of one copy and often loss of the original. These nonetheless will retain most features of the fold, though use of SwissModel here would require considerable intervention after the fact. Despite an experimental literature, these are largely irrelevant to human considerations. | |||
In the last section, sequences have been trimmed to the region covered by the xray structural determination. Species with one-off insertions that confuse gap placement have been adjusted. This helps avoid artifacts in alignment. | In the last section, sequences have been trimmed to the region covered by the xray structural determination. Species with one-off insertions that confuse gap placement have been adjusted. This helps avoid artifacts in alignment. | ||
| Line 1,197: | Line 1,225: | ||
=== Derived PLBD2 sequences === | === Derived PLBD2 sequences === | ||
>PLBD2x_droMel Drosophila melanogaster (fruitfly) | >PLBD2x_droMel Drosophila melanogaster (fruitfly) Q9VRK8 retinal lamina neuron ancestor (lama) PMID: 16077094,8892229 | ||
0 MERPEYDGTYCATALWTKQVGFQIENWKQQNDLVNIPTGVGRICYKDSVYENGW 0 | 0 MERPEYDGTYCATALWTKQVGFQIENWKQQNDLVNIPTGVGRICYKDSVYENGW 0 | ||
0 AQIEVETQRTYPDWVQAYAAGMLEGSLTWRNIYNQWSN 2 | 0 AQIEVETQRTYPDWVQAYAAGMLEGSLTWRNIYNQWSN 2 | ||
| Line 1,260: | Line 1,288: | ||
=== Trimmed alignable sequences === | === Trimmed alignable sequences === | ||
It is useful for various purposes to trim protein sequences to their conserved core and matured length. Here, since compilations of signal peptides have been previously considered, they can be discarded, greatly simplifying acquisition of | It is useful for various purposes to trim protein sequences to their conserved core and matured length. Here, since compilations of signal peptides have been previously considered, they can be discarded, greatly simplifying acquisition of reliably alignable sequence. | ||
Note too that exon boundaries mostly differ between the two paralogs and so limit the matched pairs from a fixed species that be collected from genomic contigs using tblastn from a necessarily diverged query. However the final exons do match and are already provided above from a very large and phylogenetically disperse species set. In some cases, there is a benefit to removing single-species insertions that introduce a gap in the rest of the alignment because then other gapping can be done more accurately. | |||
Finally, as the xray structural determination did not extend over the whole protein, flanking sequence needs be included only to the extent it is strongly conserved. Thus the boundaries of the protein are established, allowing comparative genomics on the alignable core. | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 14:03, 15 November 2010
Introduction
A surprising number of orphan human enzymes (unknown substrate) still exist ten years after the completion of the human genome project. PLBD1 and PLBD2 are semi-orphans in the sense of being probable phospholipases of B class but with uncertain physiological substrates and thus functionalities. This is especially important in the case of PLBD2 which localizes to the lysosome, as its absence could plausibly lead to a serious yet unrecognized lysosomal storage disease.
No bioinformatic algorithm or experimental protocol leads with any certainty to determination of function. The gene pair here has eight targeted publications but cases exist where protein function remains unknown after ten thousand papers (eg PRNP).
PLBD1 and PLBD2 constitute a small gene family (sequence homology class) within vertebrates though one that occurs expanded in some early diverging eukaryotes. However, the Pfam clan NTN (N-terminal nucleophile aminohydrolases) may have, among its ten family members, additional representatives in humans diverged beyond recognizability in primary sequence. These establish the great antiquity of the fold and certain of its features but are not likely to shed additional light on phospholipases specifically.
PLBD2 presents a special difficulty in that a sequence of post-translational steps are apparently necessary for its activation. Without these, potential substrates can hardly be assayed. These steps include removal of the signal peptide, mannosylation appropriate to the lysosome targeting receptor, and self-catalytic proteolytic activation (into 28k and 42k fragments which remain associated) to expose the substrate binding site as this becomes appropriate.
Because PLBD1 and PLBD2 are full length paralogs, the bioinformatic approach below considers both on an equal footing. PLBD1 has been more amenable to activation whereas PLBD2 has a high-resolution structural determination. Thus comparative genomics allows for annotation transfer, first from PLBD2 to a structural model for PLBD1 (already provided by the SwissModel pipeline), then perhaps transfer of PLBD1 experimental protocols to PLBD2.
However the gene duplication event occurred some 650 million years ago and the two genes are quite diverged today. It is not known whether substrates have diverged or merely their cell type of expression. Increased gene dosage per se is seldom an explanation. Yet certain core features remain conserved, including the fold, active site residues, signature motifs, certain glycosylation sites and even the fragmentation pattern, implying these are essential functional features under long-range strong selective pressure for their maintenance.
Disulfides are only separately conserved within each paralog but this fortuitously provides a reliable signature for assigning deeply diverged proteins from early eukaryotes to their orthology class. As the respective functions become better known, we can hope to understand how the gene duplication event contributed advantageously to increasing evolutionary complexity, leading to persistence of both enzymes in most species over immense time spans.
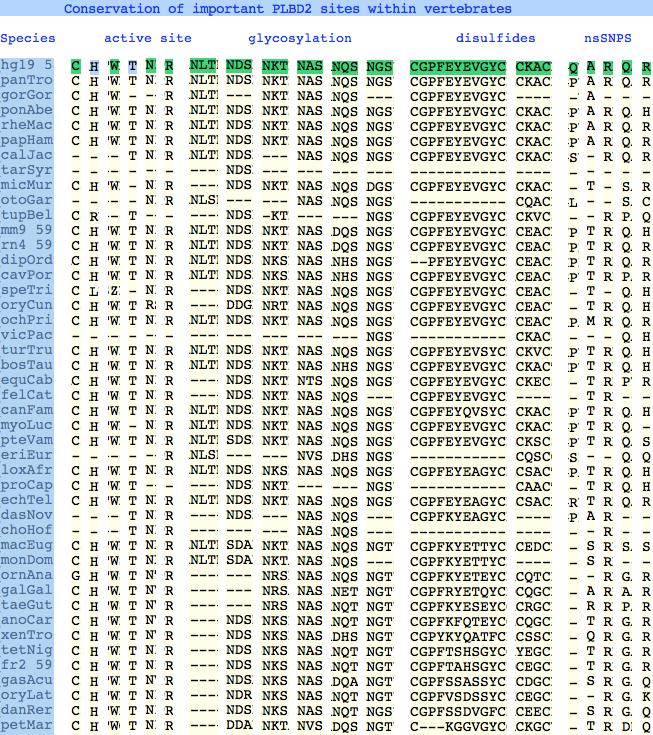
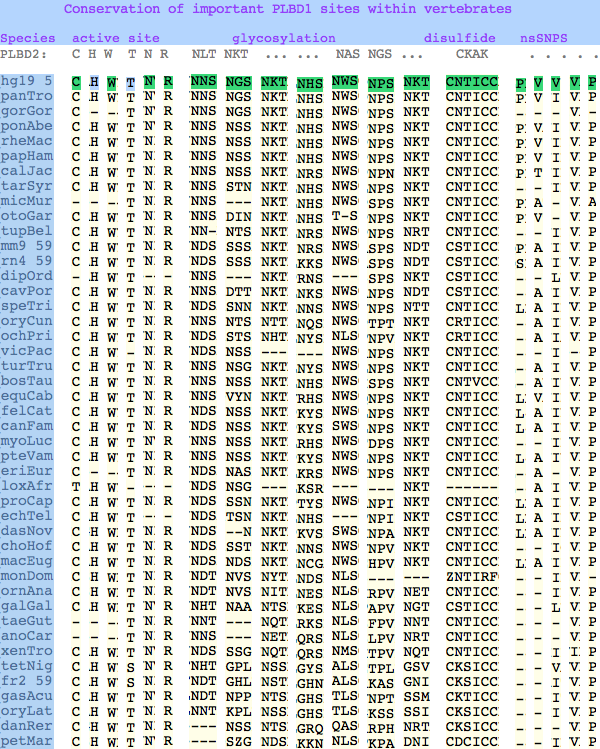
Conservation at critical sites
The six residues of PLBD2 associated with the active site are completely conserved within vertebrates to within genomic sequencing error. These same six residues are also completely conserved within PLBD1. Indeed 3 of the residues are conserved in the broader NTN hydrolase clan.
This is perhaps unsurprising since the active site was established a couple billion years earlier in the bacterial ancestor. However if PLBD2 and PLBD1 have different substrates, this establishes that these six residues are insufficient to distinguish the two active sites. Note H266 and T330 do not contribute their side chain, leaving them and W269 to separate phospholipases from the other NTN hydrolases.
The glycosylation sites are surprisingly conserved both within and between PLBD2 and PLBD1. Some of the motifs may be either recently acquired within later vertebrates or spurious glycosylation motifs with N and D both acceptable (or similar small amino acids) in the first slot of the NxS/T motif. Glycosylation is important in correct targeting of lysosomal proteins, more so than in generic endoplasmic reticulum proteins where motifs are often poorly conserved (as in sulfatases).
PLBD2 has two established disulfides. Strict sequence conservation of these throughout vertebrates (indeed, throughout metazoan) suggests both play an important role in protein structure and stability.
In PLBD1 however, the first disulfide is not a possibility and while an opportunity exists for a disulfide homologous to the the second disulfide of PLBD2, indels cloud the alignment and spacing would have to be different. There is additionally ambiguity given C...CC as to the cysteines involved. Indeed a second distal disulfide may occur utilizing C...CC.............C which has no counterpart in PLBD2. While cysteines can be conserved for many reasons other than disulfide (as in the nucleophile cysteine here), suitably proximity and side chain orientation in the SwissModel of PLBD1 would argue for disulfide. Comparative genomics suggests that C2 and C4 may form an ancient disulfide whereas C1 and C3 might represent a deuterostome innovation.
homSap CNTICCREDLNSPNPSPGGC human PLBD1 braFlo CSAICCRKDLAKVGAKPDGC Branchiostoma floridae strPur SKSICMRGDLM-TSPMPNGC Strongylocentrotus purpuratus XM_001192029 nemVec MNAICSRGDLIADGPRASGC Nematostella vectensis XM_001638165 monBre YNAICSRGDLESDSPSPGGC Monosiga brevicollis XM_001745398 SwissModel coordinates for PLBD1 show the 2nd and 4th sulfur atoms separated by 2.03 angstroms: ATOM 3552 SG CYS 471 49.680 -13.769 -12.461 ATOM 3579 SG CYS 475 49.273 -14.310 -4.881 ATOM 3585 SG CYS 476 51.067 -9.716 -9.172 ATOM 3678 SG CYS 490 50.737 -13.198 -5.750
The known human SNPs of PLBD2 are in some cases quite radical substitutions in terms of both physical qualities of the substituted amino acid and the degree of observed phylogenetic conservation at that site. These likely result in unstable and/or inactive enzyme. Both enzymes are autosomal so compensation might occur in the recessive state, or alternately, PLBD2 and PLBD1 could fill for each other to some extent. In either case, lysosomal storage disease might not be clinically observable.
Here Q54P may actually be a deleterious mutation in the reference sequence individual (with the SNP representing wildtype) as proline is highly conserved throughout mammals. The neanderthal genome also has proline at this location (UCSC track: 1C>0A(CCG>CAG)).In A204V, valine is quite a bulky substituent for a site normally restricted to small amino acids; R354C is definitely a serious mutation, no doubt attributable to a CpG hotspot; Q521K appears milder as does R524C.
The known human SNPs of PLBD1 can be analyzed similarly. P26Q and V30L may be inconsequential as they occur in the rather unconstrained primary sequence of the N-terminus; V265I occurs at an ILV reduced alphabet; V377A and P534A are much more serious despite the aliphatic nature of alanine and likely give rise to dysfunctional protein.
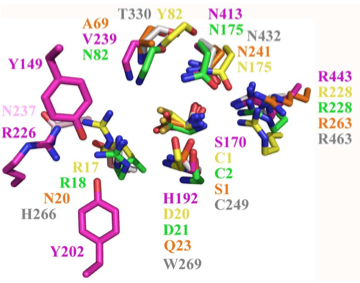
Structural superposition of active sites from five NTN hydrolases showing conserved side chains (*) and relevant main chains (....) (adapted from Fig 6 of Lakomek et al. BMC Struct Biol.2009;9:56:) * (*) * * PLBD2 phospholipase B-like gray 3FGR C244 H261 W264 T325 N427 R458 human numbering PLBD1 phospholipase B-like .... pred C228 H245 W248 T303 N402 R433 human numbering SwissModel PLBD2 phospholipase B-like .... .... C225 H242 W246 T302 N401 R432 Dictyostelium numbering Cephalosporin acylase pink 1OQZ S170 .... H192 .... N413 R443 Conjugated bile acid hydrolase green 2BJF C2 .... D21 .... N175 R228 Penicillin V acylase yellow 3PVA C1 .... D20 .... N175 R228 Penicillin G acylase orange 1K5S S1 .... Q23 .... N241 R263 Human SNPs resulting in amino acid substitutions: PLBD2: PLBD1: Q54P rs7965471 P26Q rs1141509 A204V rs12231990 V30L rs12296104 R354C rs56935204 V265I rs7957558 Q521K rs17852787 V377A rs2287541 R524C rs12425042 P534A rs1600
Intron evolution
PLBD1 and PLBD2, being full length paralogs, clearly indicate an early gene duplication and subsequent divergence to the current low percent identity. Segmental duplications preserve any introns present at the time of the event and these generally persist in both position and phase into living species.
However PLBD1 and PLBD2 -- despite having similar numbers of introns -- exhibit very little in common in terms of location as the diagram below shows. One possibility is that a second copy arose as a retroprocessed gene (a mechanism erasing existing introns) and was subsequently intronated at random positions. This is unlikely here given that 10-11 relatively rare events would be needed.
The remaining possibility is that the gene duplication took place prior to the main era in early eukaryotes during which the bulk of introns were established. This fits the current state of high divergence despite fairly slow rates of evolution during metazoan times.
The last five amino acids of each PLBD1 exon are colored below. Then using an alignment of PLBD1 to PLBD2, the colors are mapped to the homologous five residues within PLBD2. There they fall on the ends of exons only when these correspond to those of PLBD1. The outcome here -- despite uncertainties in alignment gapping -- shows intron positions do not correspond with the exception of the terminal intron (which also is phase 0).
While this merely compares human PLBD1 and PLBD2, the collected reference sequences (intronated against their respective genome assemblies) confirm that introns in both genes are deeply conserved.
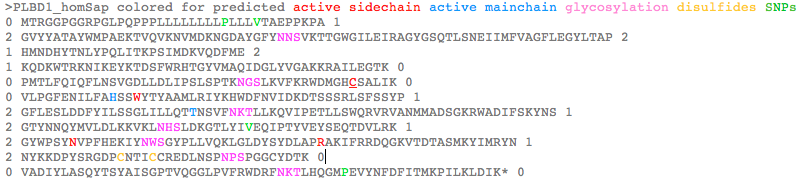
PLBD1 introns do not correspond well to those of PLBD2: >PLBD1_homSap Homo sapiens (human) first and last introns are not mappable 0 MTRGGPGGRPGLPQPPPLLLLLLLLPLLLVTAEPPKPA 1 2 GVYYATAYWMPAEKTVQVKNVMDKNGDAYGFYNNSVKTTGWGILEIRAGYGSQTLSNEIIMFVAGFLEGYLTAP 2 1 HMNDHYTNLYPQLITKPSIMDKVQDFME 2 1 KQDKWTRKNIKEYKTDSFWRHTGYVMAQIDGLYVGAKKRAILEGTK 0 0 PMTLFQIQFLNSVGDLLDLIPSLSPTKNGSLKVFKRWDMGHCSALIK 0 0 VLPGFENILFAHSSWYTYAAMLRIYKHWDFNVIDKDTSSSRLSFSSYP 1 2 GFLESLDDFYILSSGLILLQTTNSVFNKTLLKQVIPETLLSWQRVRVANMMADSGKRWADIFSKYNS 1 2 GTYNNQYMVLDLKKVKLNHSLDKGTLYIVEQIPTYVEYSEQTDVLRK 1 2 GYWPSYNVPFHEKIYNWSGYPLLVQKLGLDYSYDLAPRAKIFRRDQGKVTDTASMKYIMRYN 1 2 NYKKDPYSRGDPCNTICCREDLNSPNPSPGGCYDTK 0 0 VADIYLASQYTSYAISGPTVQGGLPVFRWDRFNKTLHQGMPEVYNFDFITMKPILKLDIK* 0 >PLBD2_homSap Homo sapiens (human) 0 MVGQMYCYPGSHLARALTRALALALVLALLVGPFLSGLAGAIPAPGGRWARDGQVPPASRSRSVLLDVSAGQLLMVDGRHPDAVAWANLTNAIRETG 2 1 WAFLELGTSGQYNDSLQAYAAGVVEAAVSEE 0 0 LIYMHWMNTVVNYCGPFEYEVGYCERLKSFLEANLEWMQEEMESNPDSPYWHQ 0 0 VRLTLLQLKGLEDSYEGRVSFPAGKFTIKPLGFL 2 1 LLQLSGDLEDLELALNKTKIKPSLGSGSCSALIKLLPGQSDLLVAHNTWNNYQHMLRVIKKYWLQFREGPW 1 2 GDYPLVPGNKLVFSSYPGTIFSCDDFYILGSGL 0 0 VTLETTIGNKNPALWKYVRPRGCVLEWVRNIVANRLASDGATWADIFKRFNSGT 2 1 YNNQWMIVDYKAFIPGGPSPGSRVLTILEQIP 2 1 GMVVVADKTSELYQKTYWASYNIP 2 1 SFETVFNASGLQALVAQYGDWFSYDGSPRAQIFRRNQSLVQDMDSMVRLMR 2 1 YNDFLHDPLSLCKACNPQPNGENAISARSDLNPANGSYPFQALRQRSHGGIDVK 0 0 VTSMSLARILSLLAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFAPVKVSWD* 0
Signal peptide compositional anomaly
The first exon of both PLBD1 and PLBD2 are ill-behaved in alignments. The explanation can be see in their compositional distortion (very high GC content) that specialized masking tools such as seg and gnu recognize. Such dna manifests itself at the protein level by high levels of the amino acids, such as GPL that use those codons in the three reading frames.
Such regions are prone to repeated expansions and contractions via replication slippage. Not only do we expect such alleles in human but also that inter-species comparisons will be difficult and alignments problematic (as homology by definition is lost even if the sequences still align).
This matters very little to the mature protein since this region is trimmed off during maturation but the question still arises as to how signal peptide variations continue to be recognized efficiently by the signal receptor complex. Indeed a class of mutations could exist in which the signal peptide cannot be processed correctly and the protein never reaches the lysosomal compartment, in effect a knockout mutation.
This compositional anomaly may have caused vertebrate-wide sequencing problems. Many assemblies had difficulty sequencing back to the initial methionine and alignment programs also fell short. A set of reliable sequences could only be obtained after careful hand-curation and only then from fewer species than usual in comparative genomics.
Even then the set of first exons raises more questions than it answers as it seems to be evolving quite chaotically in fish. Mammals also exhibit a peculiar conserved insertion as placentals diverged from marsupials. And using SignalP 3.0 separately on each sequence, it emerges that the marsupial signal peptide and those of earlier diverging species are much shorter. That isn't a problem per se because signal peptide lengths are quite variable.
PLBD1 also exhibits a shift in the location of the signal peptide cleavage site over evolutionary time, crossing the boundary into exon 2 in some clades. (Since the exon break is extremely conserved, this conclusion is independent of alignment gapping.) Here again this would be functionally irrelevant since the co-processing of nascent chain takes place well after mRNA splicing. However this does provide an interesting case of homologous residues not being functionally homologous and so evolving under the different functional constraints.
PLBD1: ATGAcccgcggcggtccgggcgggcgcccggggctgccacagccgccaccgcttctgctgctgctgctgctgctgccgctgttgTTAGTCACCGCGGAGCCGCCGAAACCTGCAG
MTRxxxxxxxxxxxxxxxxxxxxxxxxxxVTAEPPKPA
MTRGGPGGRPGLPQPPPLLLLLLLLPLLLVTAEPPKPA
PLBD2: ATGGTGGGCCAGATGTACTGCTACCCCGGCAGCCACCTGGCCCGGGCGCTGACGCGGGCGCTGGCGCTGGCCCTGGTGCTGGCCCTGCTGGTCGGGCCGTTCCTGAGCGGCCTGGCGGGGGCGATCCCAGCGCCGGGGGGCCGCT...
MVGQMYCYPGSHxxxxxxxxxxxxxxxxxxxGPFLSGLAGAIPAPGGR...
MVGQMYCYPGSHLARALTRALALALVLALLVGPFLSGLAGAIPAPGGR...
Phylogenetic variation in first exon signal peptide of PLBD2:
<------ signal peptide -----------------> <---- start of 3FGW 3FGR 3FGT------------->
>PLBD2_homSap MVGQMYCYPGSHLARALTRALALALVLALLVGPFLSGLAGAIPAPGGRWARDGQVPPASRSRSVLLDVSAGQLLMVDGRHPDAVAWANLTNAIRETG
>PLBD2_panTro MVGQMYCSPGSHLARALTRALALALVLALLVGPFLSGLAGAIPAPGGRWARDGPVPPASRSRSVLLDVSAGQLLMVDGRHPDAVAWANLTNAIRETG
>PLBD2_ponAbe MVGQMYGSSGSHLA----RALALALVLALLVGPFLSGLAGAIPAPGGRWARDGPVTPASRSRSVLLDASAGQLLLVDGRHPDAVAWANLTNAIRETG
>PLBD2_rheMac MVGQMYCSSGSPLARALTRALALALVLALLVGLFLSGLAGAIPAPGGRWAHDGPVTPASRSRSVLLHAATGQLLLVDGRQPDAVAWANLTNSIHETG
>PLBD2_papHam MVGQMYCSSGSPLARALTRALALALVLALLVGLFLSGLAGAIPAPGGRWAHDGPVTPASRSRSVLLDAATGQLLLVDGRHPDAVAWANLTNAIRETG
>PLBD2_calJac MVGKMYSSPSSRLAQALTRALALALVLALLAGLFLSGLSGAIPAPGGRWARDGSVPSGSGSRSVVLDAAAGQLLLVDGRHPDAVAWANLTNAIHETG
>PLBD2_otoGar MvGPMYGSPGGRLARALTRALALALVLaLLIGLFLSCLAGAiPPPGSGRARDGLITPASRSSSVLLDATTDQLRLVDGRHPDAVAWANLSNAIHETG
>PLBD2_musMus MAAPVDGSSGGWAARALRRALALTSLLASLTGLLLSGPAGALPTLGPGWQRQNPDPPVSRTRSLLLDAASGQLRLEDGFHPDAVAWANLTNAIRETG
>PLBD2_ratNor MAAPMDRTHGGRAARALRRALA----LASLAGLLLSGLAGALPTLGPGWRRQNPEPPASRTRSLLLDAASGQLRLEYGFHPDAVAWANLTNAIRETG
>PLBD2_dipOrd MAAPPYGSRGGRPAGSLSRALV----LAVLVGLSPSGPAGAVPSPGDRWGRHKPEPPVSRSRSVLVDAASGQLRLVDGLHPGAVAWANLTNAIRETG
>PLBD2_cavPor MAAPTYVSLDGRPVRARALALA--PALCLLVGLSLGRLAGAVPAPGPRGARDGPVPAA--CRSVLLDAASGQLRLVDGLQPGAVAWANLTNAIPETG
>PLBD2_oryCun MVAPRDGCAGGRLARALALALL--------TGLLLGGLAGAAPAPGGGEQRDPPSPPASCCRSALLDAATGQLRLVDGRHPDAVAWANLTNAIHETG
>PLBD2_ochPri MAATRDSSAGCRLARVLTRALAL---LALPTGLFLSGPAGAIPVRGDGEERGRPAPSGSRCRSVLVDAESGQLRLVDGRHPAAVAWANLTNAIHETG
>PLBD2_turTru MVDPMYGCPGGRLARALTRALALALVLALLVGLFLSGLTGAIPTPRGHRGPGRPVPPASRCRSVLLDPEtGQLRLVDGRHPDAVAWANLTNAIRETG
>PLBD2_bosTau MVAPMYGSPGGRLARAVTRALALALVLALLVGLFLSGLTGAIPTPRGQRGRGMPVPPASRCRSLLLDPETGQLRLVDGRHPDAVAWANLTNAIRETG
>PLBD2_oviAri MVAPMYGSPGGRLARAVTRALALALVLALLVGLFLSGLTGAIPTPRGQRGRGMPVPPASRCRSLLLDPETGQLSLVDGRHPDAVAWANLTNAIRETG
>PLBD2_susScr MVAPMYGSPGGRLARALTRALALALVLALLVGLFLSGLTSAIPTPKGYRGSGRSVPPASRSRSVLLDTETGQLRLVDGRHPDAVAWANLTNAIHENG
>PLBD2_ursAme MAAPMYGSPGGRLARALTRALALALVLALLVGLFLSGLTGAIPISGRQWGPNGPVPPDSRSRSVLLDAETGQLRLVDGRHPEAVAWANLTNAIRETG
>PLBD2_musPut GS-GGRLARALTRALALALVLALLVGLFLSGLTGAIPISGRQWGPKGPVPPDSRSRSVLLDAETGQLRLVDGRHPDAVAWANLTNAIRETG
>PLBD2_canFam ...................................SGLTGATPVSGRRWGPSGPVPPASRSRSVRLDPQTGQFQLVDGRNPDAVAWANLTNAIRDTG
>PLBD2_myoLuc MVAPPSRSPGGRLTPALSRAPALAPGLALLAGLFLSGWTGAIPTPRDPWGPNGPVPPASRSRSVVLDARTGQLQLVDGRQPDAVAWANLTNAIHETG
>PLBD2_pteVam MVAPMDRSPGGRLAGALTRTLELTLVLAPLAGLFLSGRTSAIQTPGSRWGSEGPVSPASRSRSVLLDPQTGQLRLVDGRHPDAVAWANLTNAIHETG
>PLBD2_eriEur MVAPMCGSPGGRPARALTRALALAPALALLVGLFLSSLAGAIPPPEDNWGRNGSFPPVSRCRSVLLDSETGQLRLVDGRHPDAVAWANLSNAIHETG
>PLBD2_loxAfr MVAPVYGSPGGRLARALTQALAVALVLALLVGLFLSGLTGAISLTGHRWGPDGPAPPASRSRSVLLDTATGQLRLVDGRHPDAVAWANLTNAIRETG
>PLBD2_echTel MVATEYGSPGGRLARALTRAPALALMLALLVGLFLSGLTGAISPAGGRREPNGRVPPASSSRSALLDPATGQLRLADGRHPEAVAWANLTNAIHETG
>PLBD2_macEug mVATMYQ--GGCLALGLALGLGLVLVLSLP--------------------QPSLPPPPSRTRSVVMDSATGQLNVVEGWEAGAIAWANLTNAIAETG
>PLBD2_monDom MVATMCQ--GSSLALGLALALGLALGLR-------------------PPQPSLPPPAPSRSCSVVLDEASGQLKVVEGAQAGAVAWANLTNAIGETG
>PLBD2_anoCar MAPAWLLRFFGLALLLARSPARR------------------------PPPFPDPAAVPTRSCSVVLEPGSAALKLVNGWAPGAVAWANLTEGIRQNG
>PLBD2_galGal MAVVRALLVAAAVAAWVPGVASGP-------------------------------TPPPRSASVLLEPGSGRLRVLPGRQPAAVAWAELTDHIQAVG
>PLBD2_melGaL MAVVRALLVAAAVAAWVPGVASGP-------------------------------TPPPRSASVLLEPGSGRLRVLPGRQPAAIAWAELTDHIQAVG
>PLBD2_xenTro MGAQLLLIFMLFSLGAAQQAV---------------------------------------VSVLFDPATGNITTVEEKRVVGAVAWAELKDSILENG
>PLBD2_xenLae MAPWQLFIFSLFCVGAAQQQA--------------------------------------VVSVLFDPATGNITTVAEKKVAGAAAWAELTDSIQENG
>PLBD2_oryLat MAFRQNKTVCAKMSTFMKSLLVLGLFWGCGRAEI---------------------------RSAVIDKGSGKLTVVEGYHEGFVAWANFTNDIETSG
>PLBD2_dicLab MASRLNKTSAVGGFSKVLNVLAVLSGLCLLFASVGAE-----------------------IRTAVIDKQTGQLSVVDGYREGFVAWANFTDDIKTSG
>PLBD2_hipHip masrlnktDGVQDKQDVFCGEFSSASVAFYVLCLTCVRAEI--------------------KSAVIDGQSGELSVVDGFQKDFVAWANFTDDIQTSG
>PLBD2_parOli MASRINKMGVEDKQDVSCVEFCVRAEI----------------------------------KSAVIDAQSGDLCVRDGFHQDLVAWANFTDDIQTSG
>PLBD2_gasAcu MASRQNTTVTLRHFKAVLSALFVMCACVQAEI-----------------------------RSAVIDKQTGKLSVVEGYREGFVAWSNFTDDINTSG
>PLBD2_oreNil MACRRNGADRVRSFTEVLGLLKMFLLLFCLFAVRAEI-----------------------SRTAVIDKQTGQLSVIEGYQEDFVAWANFTNDIETSG
>PLBD2_sebCau MASRHNKMFAVGRFKVALSVLSTLCFMCASVGAEV--------------------------RTAVVNKQTGQLSVVEGYREDFVAWSNFTDDIKTSG
>PLBD2_osmMor MAFRLLRLSTTLHLAVFLHVLFLSCSSIKAEI-----------------------------STIVLDEKTGQLTILEGYRDDYVAWANFTDDIEHSG
>PLBD2_onyTsh MADRRTQMSLTTEKMFMFSCVFYLSWTSVRAEI----------------------------PSKILDKQTGQLSLEEGFRDDYVAWANFTDDIKNSg
>PLBD2_salSal madrrtqMSVTTEKMFMFLCVFYLSWTSVGAEI----------------------------HSAVLDKQTGQLSLEEGFRDDFVAWANFTDDIKNSG
>PLBD2_danRer MAHLQLLVSAVCVLLSVCQAQI---------------------------------------YSAIYEEETAQLLLIEGARTHSVAEANFTDHINTTG
>PLBD2_calMil MCVGVRGQGLGLGLPLLLVLAAVGVSPSARGHL---------------------------LRSVVLDEHSGRLRVVGGLNPHSIAWANLTDRIRATG
>PLBD2_braFlo MAACRNIFCGRMLSCLLLFSFVFSAV-----------------------------SDGSKLASVRYDEAAKTYQITDKLDPSAAAWANFTDRISSTG
>PLBD2_acyPis MLSIRCILLSLLFVWALQCSATQK------------------------------NQTLLAVKTDNNRITIQPKHYSVKDKEIIIGKGKFIDRINSTG
>PLBD2_triAdh MAQCGKFLIYFSIFIITLATLCSCQS-------------------------------------GSVIYKDGLYTFSKGINKRAASYGTFTDKIASSG
The two paralogs do not align at all in the signal region and only poorly thereafter:
PLBD2_homSap: 67 DVSAGQLLMVDGRHPDAVAWANLTNAIRETGWAFLEL--GTSGQ-YNDSLQAYAAGVVEAAVSEE 128
Q+ V ++ DA + N N+++ TGW LE+ G Q ++ + + AG +E ++
PLBD1_homSap: 50 AEKTVQVKNVMDKNGDAYGFYN--NSVKTTGWGILEIRAGYGSQTLSNEIIMFVAGFLEGYLTAP 112
Phylogenetic variation signal peptide location in first two exons of PLBD1:
<------ signal peptide ---------> <-------------------------- second exon ---------------------------------->
>PLBD1_homSap MTRGGPGGRPGLPQPPPLLLLLLLLPLLLVTAEPPKPA:GVYYATAYWMPAEKTVQVKN-VMDKNGDAYGFYNNSVKTTGWGILEIRAGYGSQTLSNEIIMFVAGFLEGYLTAP
>PLBD1_panTro MTRGGPGGRPGLPQPPPLLLLLLLLPLLLVTAEPPKPA:GVYYATAYWMPAEKTVQVKN-VMDKNGDAYGFYNNSVKTTGWGILEIRAGYGSQTLSNEIIMFVAGFLEGYLTAP
>PLBD1_ponAbe MTRGGPGGRPGLPPPPPLLLLLLLPPLLLVAAEPANSA:GVYYATAYWMPTEKTVQVKN-VMDKNGDAYGFYNNSVKTTGWGILEIRAGYGSQALSNEIIMFVAGFLEGYLTAP
>PLBD1_rheMac MTRGGPGGCPGLPPPLPLLLRLLLPPLLLVTAESPNPA:GVYYATAYWMPAEMTVEVKN-IMDKNGDAYGFYNNSVETTGWGILEIRAGYGSQALSNEIIMFVAGFLEGYLTAP
>PLBD1_papHam MTRGGPGGCPGLPPQLPLLLRLLLPPLLLVTAESPNPA:GVYYATAYWMPAEMTVEVKN-IMDKNGDAYGFYNNSVETTGWGILEIRAGYGSQALSNEIIMFVAGFLEGYLTAP
>PLBD1_calJac MTRGGPGGRLGLPPPPLLLLLLLLLPPLPTTAEPPTPA:GISYATAYWMPAEKTVQVKN-VMDKNGDAYGFYNNSVKTTGWGILEIRAGYGSQALSNEIIMFVAGFLEGYLTAL
>PLBD1_otoGar MANRTLDRRLGLPPPPLLLLLLLPPPPLLVTAARKNPP:GVYYATAYWKPAEKTVEVKK-VIDKNGDAYGFYNNSMNATGWGILEIRAGYGSQALSNEMTMFVAGVLEGYLTAP
>PLBD1_musMus MCHRSPGRSLRPPSPLLLLLPLLLQPP-WAAALPASPT:GVHCATAYWSPESKKVEIKT-VLDKNGDAYGYYNDSIKTTGWGILEIRAGYGSQVLSNEIIMFLAGYLEGYLTAL
>PLBD1_ratNor MCHRSHGRSLRPPSPLLLLLPLLLQSP-WAAAPLRSSA:GVHYATAYWLPDTKAVEIKM-VLDKKGDAYGFYNDSIQTTGWGVLEIKAGYGSQILSNEIIMFLAGYLEGYLTAL
>PLBD1_cavPor MALCGPGCSPGLPPSPLLLLPLLL----LAAAWSPSPP:GIHYATAYWIPDTKTVEVKD-ILDKDGDAYGYYNNSMEATGWGILEIKAGYGSQELTNEIIMFVAGFLEGYLTAL
>PLBD1_speTri MSRRSLGCGRW-PPPPLQLLPLLLLLLPLAAAQP----:EVYYATAYWIPSEKSIKVKH-VMDKSGDAYGYYNDSMETTGWSILEIRAGYGSQALSNEIIMFVAGFLEGYLTAP
>PLBD1_oryCun MALWLPPLLFPLL---------------LAAAEPPSPE:GVSYATAYWMDAEKKVQVRN-VLDKNGDAYGFYNNSVKTTGWGILEIRAGYGSQALSNEIIMFVAGFLEGYLTAP
>PLBD1_turTru MSRRSPDGSLGLLSPPALLLLLL------AAVVPSGLA:GVYYATAYWMPTEKRIQVQN-VLDRNGDAYGFYNNSVKTTGWGILEIRAGYGSRSLSNEIVMFAAGFLEGYLTAP
>PLBD1_bosTau MSRHSQDERLGLPQPPALLPLLLLL----AVAVPLSQA:GVYYATAYWMPTEKTIQVKN-VLDRKGDAYGFYNNSVKTTGWGILEIKAGYGSQSLSNEIIMFAAGFLEGYLTAP
>PLBD1_oviAri MPRHRRDERLGLPPPPARLPLLLLLL---AAAVPLSQA:GVYYATAYWMPTEKRIQVKN-VLDRKGDAYGFYNNSVKTTGWGILEIKAGYGSQSLSNEIIMFAAGFLEGYLTAP
>PLBD1_susScr MSRRSRDGRLGLPAPPAPL-LLLLLL---AAAVPPSLA:GVYYATAYWMPTEKRMLVKN-VLDRNGDAYGFYNDSMKTTGWGILEIRAGYGSQSLSNNIIMFAAGYLEGYLTAP
>PLBD1_equCab MARHRPDGRLGLPAPPAPPLPPLLLLLLV-AAVSPSQA:VVYSATAYWMPAEKTVQVKN-VMDRNGDAYGFYNNSVKTTGWGILEIRAGYGSQTLSNDITMFVAGFLEGYLTAL
>PLBD1_felCat MARRSRDGRPGLSAPPTPPLLPLLLL---AAAVSPSLA:EVHYATVYWMPAEKTIQVKN-VLDRNGDAYGFYNDSVKTTGWGVLEIRAGYGSQALSNEIIMFVAGFLEGYLTAP
>PLBD1_canFam MPRRARDARLEPCPPLLPLLLLLL-----AAAVPQGRA:EVYYATAYWIPDEKTIQVKN-VLDRNGDAYGFYNDSVKTTGWGILEIRAGYGSQILSNEITMFVAGFLEGYLTAP
>PLBD1_pteVam MSRRSLDGRLGLPATSAPPLLLLLLL---AAAVPPSLA:evyYATAYWMPAEKTVNVKN-LLDKNGDAYGFYNNSMNTTGWGILEIKAGYGSQTLSNDIIMFVAGYLEGYLTAP
>PLBD1_eriEur MSRRSRDGRLGLLLSPPLLLLLLLL-----AAAPPSLQ:EIYYATAYWMPEEEEIQVKN-VLDKNGDAYGFYNDSMLTTGWGILEIKAGYGSHQLSNDVVMFVAGFLEGYLTAP
>PLBD1_sorAra MARGGGDGPPALLPLPLLSLLLALL----AAAVPPSLA:EVHYATAYWMPDEQRVEIKT-TLDKKGDAYGYYNDSVLTTGWGILEIRAGYGSQDLTDEITMFVAGALEGYLTAP
>PLBD1_loxAfr MSSRSRGRHHGPAPQLPQLLLLLLLLLLVAAAAPPSLA:EVHYATVYWMSSEKTMQVKD-VLDKKGDAYGYYNDSVLTTGWGVLEIKAGYGSQALSNDIIMFAAGYLEGYLTAL
>PLBD1_proCap MCSRSV--PCRLSPPLSPPLSLPLLLLLLAAAAPPSLA:EVHYATVYWMSSEKTMQVKD-TLDKNGDAYGFYNDSMQTTGWGVLEIKAGYGSQGLSNDVIMYAAGYLEGYLTAp
>PLBD1_echTel MSTHSRGGR--PAPPLSPSLSLTPLLLL-AALVAPSLA:EIHYATAYWMSSEKTIQIKD-VLDKSGDAYGFYNDSVNATGWGILEIRAGYGSQNLSNDIIMFAAGFLEGYLTAP
>PLBD1_choHof MSRSCQAERLGPVPRRRLLLLLL-----VASAAPPSVA:EVFYATAYWIPSEKKIVVKD-ILDQNGDAYGFYNDSMKTTGWGILEIKAGYGSHIPSNEIIMFTAGFLEGYLTAE
>PLBD1_triVul MSRRSRDGRLGLPAPPAPLLLLLLL----AAAVPPSLA:GVYYATAYWMPTEKRMLVKN-VLDRNGDAYGFYNDSMKTTGWGILEIRAGYGSQSLSNNIIMFAAGYLEGYLTAP
>PLBD1_monDom MTRFSCFGRLQLW--PLQVLLLLLL----TFGAPVTQA:GIHYATVYWNSSTSSAEVKD-SLDPDGDAYGFYNDTIQTTGWGILEIRAGYGANSLTDEIIMFVAGFLEGYLTAQ
>PLBD1_ornAna MSRTCRGGRSGPPQPAPTPAGLLLLLL--TVASPLLQS:HVRYATAYWESATQTVRVKD-VLDWDGDAYGFYNHTVQTTGWGTLEIRAGYGAQALSDEVVMFVAGFLEGYLTAP
>PLBD1_taeGut MARAGGGVCRCCCWALVLLWAAAGGRA-----------:ELRYATVYWNRAEKILQVKN-TLDRSGDAYGFYNNSLQTTGWGVLEIRAGYGSQTLSNEDIMYVAGFLEGYLTAP
>PLBD1_galGal MARLGGGALCCCWGLVLLWAVAGGRA------------:EMRYATLYWNKAQKILQVKN-ILDRSGDAYGFYNNTVQTTGWGVLEIKAGYGHQTLSNEDIMYAAGFLEGYLTAP
>PLBD1_melGal MARLGGGPLCCCWGLVLLWAVAGGRA------------:EMRYATLYWNKAQKILQVKN-ILDRSGDAYGFYNNTVQKTGWGVLEIKAGYGHQTLSNEDIMYAAGFLEGYLTAP
>PLBD1_sisCat MIRFGNPSSSDTRRQRCRSWYWGGLLLLWAVAETRA--:DIHYATVYWLEAEKSFQIKD-VLDKNGDAYGYYNDTIQSTGWGILEIKAGYGNQPISNEILMYAAGFLEGYLTAS
>PLBD1_ambMex MGGLRQLLPLCALLLLQPLGAR----------------:AIRYATVYWTD-RKTVLVKE-VLDKGGDAYGFYNDTIQSTGWGVLEIRAGYAPTSRTNEEIMFAAGYLEGYLTAL
>PLBD1_takRub MFLLTSTCAFVLLTLPATSSTADG--------------:GTAAATVYWDPQHKTVLLKEGVLEQEGDAYGYFNDTLSSTGWSVLEIRAGYGTTPETDEVIFFLAGYLEGFLTAQ
>PLBD1_danRer MPDFSFCVLFLIGFLFSSRSD-----------------:KLK-ATVYWDATHKSAVLKQGVLDPAGASYGYYDNVLLSTGWGVLEVRAGYGDTTQTDDITMFTAGYLEGFLTAP
>PLBD1_ictPun MTEFMVCVCMFLCAVIAVRTDS----------------:VHK-ATAYWDPDSKTVLLKDGVLEDTGDAYGFYNDSFSETGWGVMEVRAGYGQTPRADERTFFLAGYLEGFLTAR
>PLBD1_perFla MEKQSIKLCVLLSTLAASVQTY----------------:QLQEATVYWDGAQKSVILKEGVMETEGGAYGYFNDTLLLSGWGVLEICAGHGGITQEDETTFFLAGYLEGYLTAG
>PLBD1_gasAcu MFLEKTLYVLLLCSVSTTSSAD----------------:KMTAATVYWDPQHKVVLLKEGVLEKEGDAYGYLNDTLSSTGWSVLEIRAGYGETPETDEVTFFLAGYLEGFLTAQ
>PLBD1_oryLat MKLEVFLLLHVIATFASSQ-------------------:KLTAATVYWDAQHKLVLLKEGVLETEGDAYGYLNNTLSTSGWSILEIRAGYGKTPEDDEITFFLAGYLEGFLTAQ
>PLBD1_pimPro MDTNSICVLLLLCSVSTTSSAD----------------:KMTAATVYWDPQHKVVLLKEGVLEKEGDAYGYLNDTLSSTGWSVLEIRAGYGETPETDEVTFFLAGYLEGFLTAQ
>PLBD1_dicLab MPLVTRLYVFLLFTVVTSFASAD---------------:KMTAATVYWDPLHKLVKLKEGVLETEGDAYGYLNDTLSSSGWSILEIRAGYGKTPETDELTFFLAGYLEGYLTAQ
>PLBD1_salSal MKRVCLLFFFYVAASFASAD------------------:EMKAATVYWDATHKTVQLKEGVIEKEGDAYGYLNDTLSQTGWSVLEIRAGYGETLEHDEVTYFLAGYLEGFLTAP
Difference alignment of exon 1 from placental mammals:
PLBD2_homSap MVGQMYCYPGSHLARALTRALALALVLALLVGPFLSGLAGAIPAPGGRWARDGQVPPASRSRSVLLDVSAGQLLMVDGRHPDAVAWANLTNAIRETG
PLBD2_panTro .......S.............................................P...........................................
PLBD2_ponAbe ......GSS.....----...................................P.T...........A......L......................
PLBD2_rheMac .......SS..P....................L.................H..P.T..........HAAT....L....Q...........S.H...
PLBD2_papHam .......SS..P....................L.................H..P.T...........AAT....L......................
PLBD2_calJac ...K..SS.S.R..Q...............A.L.....S..............S..SG.G....V..AA.....L..................H...
PLBD2_otoGar ...P..GS..GR..................I.L...C......P..SGR....LIT.....S.....ATTD..RL..............S...H...
PLBD2_musMus .AAPVDGSS.GWA....R.....TSL..S.T.LL...P...L.TL.PG.Q.QNPD..V..T..L...AAS...RLE..F..................
PLBD2_ratNor .AAP.DRTH.GRA....R.....----.S.A.LL.......L.TL.PG.R.QNPE.....T..L...AAS...RLEY.F..................
PLBD2_dipOrd .AAPP.GSR.GRP.GS.S...V.----.V...LSP..P...V.S..D..G.HKPE..V.......V.AAS...RL...L..G...............
PLBD2_cavPor .AAPT.VSLDGRPV..--......PA.C....LS.GR....V....P.G....P..A.--C......AAS...RL...LQ.G...........P...
PLBD2_oryCun ..APRDGCA.GR.....A--------....T.LL.G.....A.....GEQ..PPS....CC..A...AAT...RL..................H...
PLBD2_ochPri .AATRDSSA.CR...V.......---...PT.L....P.....VR.DGEE.GRPA.SG..C....V.AES...RL......A...........H...
PLBD2_turTru ..DP..GC..GR....................L.....T....T.R.HRGPGRP......C......PET...RL......................
PLBD2_bosTau ..AP..GS..GR....V...............L.....T....T.R.QRG.GMP......C..L...PET...RL......................
PLBD2_oviAri ..AP..GS..GR....V...............L.....T....T.R.QRG.GMP......C..L...PET...SL......................
PLBD2_susScr ..AP..GS..GR....................L.....TS...T.K.YRGSGRS.............TET...RL..................H.N.
PLBD2_ursAme .AAP..GS..GR....................L.....T....IS.RQ.GPN.P...D.........AET...RL......E...............
PLBD2_myoLuc ..APPSRS..GR.TP..S..P...PG....A.L....WT....T.RDP.GPN.P..........V..ART...QL....Q.............H...
PLBD2_pteVam ..AP.DRS..GR..G....T.E.T....P.A.L....RTS..QT..S..GSE.P.S...........PQT...RL..................H...
PLBD2_eriEur ..AP.CGS..GRP...........PA......L...S......P.EDN.G.N.SF..V..C......SET...RL..............S...H...
PLBD2_loxAfr ..APV.GS..GR......Q...V.........L.....T...SLT.H..GP..PA............TAT...RL......................
PLBD2_echTel ..ATE.GS..GR........P....M......L.....T...SPA...REPN.R.....S...A...PAT...RLA.....E...........H...
Consensus MVAPMYGSPGGRLARALTRALALALVLALLVGLFLSGLAGAIPaPGGRWGRDGPVPPASRSRSVLLDAATGQLRLVDGRHPDAVAWANLTNAIRETG
Alignment of first two exons of PLBD1 from vertebrates showing onset of conservation:
Understanding conserved residues in PLBD1 and PLBD2
Although the gene duplication creating these paralogs took place in early unicellular eukaryotes with PLBD1 quite diverged in primary sequence from PLBD2 today, it is nonetheless instructive to compare individual residues and residue patches that are still conserved, given the folds have diverged rather little. Here we wish to exploit the situation that more is known about the maturation and substrates of PLBD1 whereas excellent crystallographic structures exist for PLBD2 and certain of its ancient homologs.
Localization of conserved residues within compared secondary structures: s = beta sheet, h = alpha helix
38 PTGVHCATAYWSPESKKVEIKTVLDKNGDAYGYYNDSIKTTGWGILEIRAGYGSQVLSNEIIMFLAGYLEGYLTALHMYDHFTNLYPQLIKN----PSIV PLBD1
61 PPVSRTRSLLLDAASGQLRLEDGFHPDAVAWANLTNAIRETGWAYLDLST---NGRYNDSLQAYAAGVVEASVSEELIYMHWMNTVVNYCGPFEYEVGYC PLBD2
ssssssssss sssssss ssssssssss ssssssssss hhhhhhhhhhhhhh hhhhhhhhh hh
ssssssssss sssssss ssssssssss sssssssss s hhhhhhhhhhhhhh hhhhhhhhhh hhhh
134 KKVQDFMEKQEMWTRQNIKAQKDDPFWRHTGYVVTQLDGLYLGAQKRASEE-KIKPMTMFQIQFLNAVGDLLDLIPSLSPTKSSSMMKFKIWEMGHCSAL PLBD1
158 EKLKNFLEANLEWMQREMELNPDSPYWHQVRLTLLQLKGLEDSYEGRLTFPTGRFTIKPLGFLLLQISGDLEDLEPALNKTN----------GSGSCSAL PLBD2
hhhhhhhhhhhhhhhhhhhh hhhhhhhhhhhhhhhhhhhhh hhhhhhhhh sss
hhhhhhhhhhhhhhhhhhhh hhhhhhhhhhhhhhhhhhhhh hhhhhhhhh sss
233 IKVLPGFENIYFAHSSWYTYAAMLRIYKHWDFNIKD------KYTLSKRLSFSSYPGFLESLDDFYILSSGLILLQTTNSVYNKTLLKQVVPK-TLLAWQ PLBD1
253 IKLLPGGHDLLVAHNTWNSYQNMLRIIKKYRLQFREGPQEEYPLVAGNNLVFSSYPGTIFSGDDFYILGSGLVTLETTIGNKNPALWKYVQPQGCVLEWI PLBD2
sssss sssssssssssss ssssssss sssssss ssss sssssssss hhhh hh
sssss sssssssssssss ssssssss sss sss sssssss ssss sssssssss hhhh hh
326 RVRVANMMAEGGKEWAQIFSKHNSGTYNNQYMVLDLKKVTINRSL-DKGTLYIVEQIPTYVEYSDQTNV-LRKGYWASYNIPFHKTIYNWSGYPLLVHKL PLBD1
353 RNVVANRLALDGATWADVFKRFNSGTYNNQWMIVDYKAFLPNGPSPGSRVLTILEQIPGMVVVADKTAELYKTTYWASYNIPYFETVFNASGLQALVAQY PLBD2
hhhhhhhh hhhhhhhh sssssssss ssssssss sssssss hh sssss hhhhhh hhhhhhh
hhhhhhhh hhhhhhhh sssssssss ssssssss ssssssshhhhhhhsssss hhhhhh hhhhhh
424 GLDYSYDLAPRAKIFRRDQGNVTDMASMKYIMRYNNYKEDPYSKGDPC-------STICCREDLNGAS---------PSPGGCYDTKVADIFLASQYKAYAISGPTVQDGLPPFNWNRF--NETLHRGMPEVFDFNFVTMK -
453 GDWFSYTKNPRAKIFQRDQSLVEDMDAMVRLMRYNDFLHDPLSLCEACNPKPNAENAISARSDLNPANGSYPFQALHQRAHGGIDVKVTSFTLAKYMSMLAASGPTW-DQCPPFQWSKSPFHSMLHMGQPDLWMFSPIRVPWD
hhhhhhh hhhhhhhhh sss ssssssssss hhhhh ssssss sss sss sss
hhhhhhhh hhhhhhhhh sss sss sss ssssssssss hhhhh ssssss sss sss sss
Suppose a phylogenetically broad set of curated PLBD1 and PLBD2 are aligned together. After careful consideration of gap placement, a restricted number of residues will prove very deeply conserved in both proteins throughout eukaryotes. Of these, some are universal localizational, modificational, structural, or catalytic features basic to the entire NTN clan of 12 protein families and so not particular to phospholipases.
This class of residues must be found by structural alignment of crystallographic structures, as primary sequences are too diverged for these to be accurately located by ClustalW or similar tools. Since the fold of PLBD2 was originally recognized by the fold comparison (via Dali) to all of PDB, these are known already: the autocatalytic cysteine residue at the N-terminus of the 40 kDa fragment and the three active site residues noted above.
An additional 6%-14% of structurally equivalent amino acids (themselves only half of the chain) are identical as amino acids, with IMPC (inosine monophosphate cyclohydrolase) being the highest but PVA (penicillin acylase V) and CBAH (conjugated bile acid hydrolase) also significant. Dali provides a primary sequence multiple alignment and so the super-invariant amino acids (plus those with narrow reduced alphabets, while eliminating accidental matches) not specific to phospholipases PLBD1 and PLBD2. These largely lie within the beta strands of the core αββα sandwich as they are better conserved than alpha helices within NTN hydralases but it is fair to say that this fold class is not understood until an explanation can be given for each of the universally conserved residues.
1oqz CA cephalosporin acylase 3pva PVA penicillin V acylase 1k5s PGA penicillin G acylase 2bjf CBAH choloylglycine hydrolase 2ntm IMP cyclohydrolase 1ryp --- a chain among 28 of yeast proteasome Dali report on 3fgr vs 1oqzB: AyaA is a candidate super-invariant region but only second A occurs in both PLBD1 and PLBD2 DSSP leeeeeeeeelllleeeeeelllllLLEEEEEEEEhhhhleEEEEEEELllllhhhHHHHH Query vsrtrsllldaasgqlrledgfhpdAVAWANLTNAiretgwAYLDLSTNgryndslQAYAA 61 ident A A Sbjct ---------------pqapiaaykpRSNEILWDGY------GVPHIYGV-------DAPSA 33 DSSP ---------------llllllllllLLLEEEEELL------LLEEEELL-------LHHHH DSSP HHHHHHHHHHHHHHHHHhhLLLL--LLLL------LLLH-HHHH--HHHHHHHHHHHHH Query GVVEASVSEELIYMHWMntVVNY--CGPF------EYEV-GYCE--KLKNFLEANLEWM 109 ident Sbjct FYGYGWAQARSHGDNIL-rLYGEarGKGAeywgpdYEQTtVWLLtnGVPERAQQWYAQQ 91 DSSP HHHHHHHHHHHHHHHHH-hHHHHhlLLHHhhhlhhHHHHhHHHHhlLHHHHHHHHHHLL DSSP hhhhhhllllhhhhhHHHHHHHHHHHHHHHHL---------llllllllllLLLLlLHHHHL-HHHH-HHHHhhlLLL Query qremelnpdspywhqVRLTLLQLKGLEDSYEG---------rltfptgrftIKPLgFLLLQI-SGDL-EDLEpalNKT ident L Sbjct ---------------SPDFRANLDAFAAGINAyaqqnpddispdvrqvlpvSGAD-VVAHAHrLMNFlYVAS---PGR DSSP ---------------LHHHHHHHHHHHHHHHHhhhhlhhhllhhhhlllllLHHH-HHHHAHrLMNFlYVAS---PGR
In effect, these residues must be 'subtracted off' the larger set of phylogenetically invariant residues between PLBD1 and PLBD2 to reach conserved residues defining what is specifically important to phospholipases. These sites developed subsequent to the divergence of phospholipases from generic NTN hydralases but prior to the gene duplication giving rise to PLBD1 and PLBD2 because convergent evolution later is implausible. They are evidently critical to structure/function, being retained in both paralogs in all surviving species up until the present day. Several active site residues fall into this category as described earlier.
Conserved residues determining mannosylation
PLBD1 and PLBD2 are among the 2500 human proteins carrying signal peptides targeting them initially to the endoplasmic reticulum and subsequently to the golgi, where they are sorted and packaged according to final destination (lysosome, plasma membrane, extracellular secretion). Many of these proteins are initially glycosylated via a limited repertoire but few of the soluble members are ultimately targeted to the lysozome (about 50 of 2500).
These receive an additional post-translational modification in the golgi of terminal mannose residues to mannose 6-phosphate making them recognizable to the two lysosomal mannose receptors (IGF2R and M6PR). Bizarrely, this simple phosphorylation is not accomplished by a kinase but rather requires 3 separate gene products, GNPTAB (UDP GlcNAc-1-phosphotransferase, internally cleaved into α,β catalytic and recognition subunits), GNPTG (γ regulatory subunit of the α2β2γ2 hexamer) and NAGPA (N-acetylglucosaminidase, or uncovering enzyme).
Overall, trafficking is very imperfect, with some protein molecules [eluding the uncovering enzyme yet http://www.ncbi.nlm.nih.gov/pubmed/20615935 still ending up in the lysosome] and while others even with M6P markers are improperly secreted.
An alternate pathway exists: β-glucocerebrosidase (of Gaucher disease) localizes to the lysozyme via the coiled coil of theLIMP2 protein encoded by SCARB2.) This presumably continues despite mutations (inclusion-cell disease) in GNPTAB, GNPTG, NAGPA, IGF2R and M6PR that disable the main pathway. Conversely, mutations in SCARB2 (causing action myoclonus renal failure syndrome) do not appear to affect proteins using the M6P lysosomal trafficking route. The SORT1 gene product sortilin may also let proteins sidestep the M6P pathway to the lysosome.
PLBD1 and PLBD2 likely use the conventional M6P pathway, nonetheless raising difficult bioinformatic issues. The 50 proteins using it have unrelated folds. They share the generic glycosylation NxT/S sequence motif but this is completely insufficient to differentiate them from proteins not targeted to the lysozome. There is no support for a larger motif enveloping NxT/S or a supplementary contiguous motif elsewhere.
Recognition must reside within individual GNPTAB/GNPTG/NAGPA subunits but their roles are complex, providing no clear bioinformatic handle, only two separated lysines forming a critical triad with M6P at 34 angstrom distance.
However in homologous lysosomal sulfatases ARSA and ARSB, the two identified lysines did not occupy homologous positions, perhaps because sulfatases duplicated and diverged prior to the evolutionary appearance of the lysosome.
This then illustrates convergent evolution rather than conservation of homology. This applies too to the many signal peptides and NxT/S motifs that must have arisen independently a great many times (as no such mobile elements could have provided them).
PLBD2 targeting is then fairly complex from a predictive standpoint as its 40 lysines and 6 M6P would yield a great many candidate pairs, depending on the softness of the 34 angstrom requirement. As PLBD1 and PLBD2 may have diverged later than sulfatases (ie, after recruitment to the lysosome), the lysine pairs might still be homologous. Looking at human gene alignment, this still gives an unwieldy set of 7 homologous pairs (some poor quality because of gap uncertainty).
However these lysines need to be invariant within phylogenetically dispersed PLBD2 and PLBD1 orthologs. Under that assumption, only 3 of the lysines are strong candidates (red), 1 is so-so (magenta) and the rest are implausible (blue). The lysine in CSALIK may be part of the autocleavage motif, leaving two candidates to be compared to the six M6P sites for the required 34 angstrom geometry.
PLBD2: 189 LKGLEDSYEGRVSFPAGK-FTIKPLGFLLLQLSGDLEDLELALNKTK---IK--PSLGSG 242
+ GL + R K T+ + FL GDL DL +L+ TK +K G
PLBD1: 169 IDGLYVGAKKRAILEGTKPMTLFQIQFL--NSVGDLLDLIPSLSPTKNGSLKVFKRWDMG 226
PLBD2: 243 SCSALIKLLPGQSDLLVAHNTWNNYQHMLRVIKKYWLQFREGPWGDYPLVPGNKLVFSSY 302
CSALIK+LPG ++L AH++W Y MLR+ K +W F D ++L FSSY
PLBD1: 227 HCSALIKVLPGFENILFAHSSWYTYAAMLRIYK-HW-DFNVID-KD---TSSSRLSFSSY 280
PLBD2: 303 PGTIFSCDDFYILGSGLVTLETTIGNKNPALWKYVRPRGCVLEWVRNIVANRLASDGATW 362
PG + S DDFYIL SGL+ L+TT N L K V P +L W R VAN +A G W
PLBD1: 281 PGFLESLDDFYILSSGLILLQTTNSVFNKTLLKQVIPE-TLLSWQRVRVANMMADSGKRW 339
PLBD2: 363 ADIFKRFNSGTYNNQWMIVDYKAFIPGGPSPGSRVLTILEQIPGMVVVADKTSELYQKTY 422
ADIF ++NSGTYNNQ+M++D K + S L I+EQIP V +++T L +K Y
PLBD1: 340 ADIFSKYNSGTYNNQYMVLDLKK-VKLNHSLDKGTLYIVEQIPTYVEYSEQTDVL-RKGY 397
PLBD2: 483 DFLHDPLSLCKACNPQPNGENAISARSDLNPANGSYPFQALRQRSHGGIDVKVTSMSLAR 542
++ DP S CN I R DLN N S P G D KV + LA
PLBD1: 458 NYKKDPYSRGDPCN-------TICCREDLNSPNPS-P--------GGCYDTKVADIYLAS 501
The phylogenetic emergence of these features could time the onset of targeting to the lysozyme during the evolutionary history of phospholipases PLBD1 and PLBD2. Mutations localizing at M6P receptor targeting sites would lead to non-recognition, non-arrival, and perhaps lysosomal storage disease (as the proteins would then be secreted).
The evolution of the lysozyme itself, preposterously claimed a vertebrate innovation as late as 2006, instead uses components that readily trace back to slime mold. These surface as the eight loci responsible for flawed lysosome related organelle biogenesis (Hermansky-Pudlak syndrome).
Ragged carboxy terminus in PLBD1
The deuterostome orthologs of PLBD1 display an unusual pattern of extensions and contractions of the carboxy terminus. These do not correspond to subclades, implying numerous separate events affecting the stop codon in fairly recent times (which is supported by read-through past the stop codon, lower case). The precise location of the stop codon may not be at all important but conservation does continue on to the proline near the end (which is strongly invariant. Proline is a common helix terminating residue but here the structural model suggests it is terminating a beta strand.
PLBD2 is much more orderly at its carboxy terminus, though some extensions are seen in early deuterostomes. Again an almost-terminal proline is conserved. However past it, a tryptophan is also universally conserved. The xray structure fortunately includes both residues -- the goal for every protein is to provide an explanation for each conserved residue (for rational interpretation of possible disease SNPs) though here the focus initially is on activation and catalysis.
PLBD1_homSap VADIYLASQYTSYAISGPTVQGGLPVFRWDRFNKTLHQGMPEVYNFDFITMKPILKLDIK* PLBD1_panTro VADIYLASQYTSYAISGPTVQGGLPVFRWDRFNKTLHQGMPEVYNFDFITMKPILKLDIK* PLBD1_ponAbe VADIYLASQYTSYAVSGPTVQGGLPVFRWDRFNKTLHQGMPEVYNFDFITMKPILKLDIK* PLBD1_rheMac VADIYLASQYTSYAISGPTVQGGLPVFHWNRFNKTLHQGMPEVYNFDFITMKPILKRDMK* PLBD1_papHam VADIYLASQYTSYAISGPTVQGGLPVFHWNRFNKTLHQGMPEVYNFDFITMKPILKRDMK* PLBD1_calJac VSDIYLASQYTSYAISGPTVQGGLPVFRWNRFNKTLHQGMPEVYNFDFITTKPILK*hkmk PLBD1_tarSyr VADIYLASQYTAYAISGPTVQDGLPVFHWNRFNKTLHQGMPEVYNFDFVTMKPILKLDIK* PLBD1_micMur VsDIFPASQFTGHAINGPTVPSGlPVFYRPPFNKTPHQGIAEAYHFDFISKKPILKPDIK* PLBD1_otoGar VADIYLASQYTAYAISGPTVQGGLPVFHWHRFNKTLHHGMPEAYNFDFITMKPVLKLDIK* PLBD1_tupBel VADIYLASQYTAYAISGPTVQDGLPVFHWNRFNRTVHQGMPEAYNFDFITMKPVLKLDIK* PLBD1_musMus VADIFLASQYKAYAISGPTVQDGLPPFNWNRFNDTLHRGMPEVFDFNFVTMKPILS*dkk* PLBD1_ratNor VADIFLASQYKAYAISGPTVQNGLPPFNWNRFNDTLHQGMPDVFDFDFVTMKPILT*dkn* PLBD1_perMan VADIFLAFQYTAYAISGPTVQDGLPAFDWKHFNKTLHEGMPDVFNFDFVTMKPILTEDIK* PLBD1_dipOrd VSDIFLASKYIAYAISGPTVQDGLPAFSWRLFNKTLHQGMPEIYNFDFVLMKPFFND*qk PLBD1_cavPor VADIHLASEYTAYAISGPTVQGGLPVFRWNRFNDTLHQGMPEVYNFDFITMKPILKPNVKRRRKMRE* PLBD1_speTri VSDIYLASQYTAYAISGPTVQGGLPVFRWNRFNTTLHQGMPEAYNFDFITMKPVLKIDIK* PLBD1_oryCun VSDIYLASRYTAYAISGPTVQGGLPVFHWNRFNKTLHQGMPEVYNFDFITTKPILKLDKR* PLBD1_ochPri VSDVHLASQYTAYAISGPTVQGKLPVFHWSQFNKTLHQGMPDAYNFDFITMKPILKKMREDEAEGNRMK* PLBD1_vicPac VADIYLASQSTAHAISGPTAEDGLPVFHWNRFNKTLHSGMPEVYNFDFITMKPIL*ldik* PLBD1_turTru VADIHLASAYTAYAISGPTVQGGLPVFHWSRFNKTLHEGMPEAYNFDFITMKPIL*ldmk PLBD1_bosTau VADIYLASKYKAYAISGPTVQGGLPVFHWSRFNKTLHEGMPEAYNFDFITMKPIL*ldik PLBD1_oviAri VADIYLASKYKAYAISGPTVQGGLPVFHWSRFNKTLHEGLPEAYNFDFITMKPIL*ldik PLBD1_lamPac VADIYLASQSTAHAISGPTAEDGLPVFHWNRFNKTLHSGMPEVYNFDFITMKPIL*LDIK PLBD1_susScr VADIHLASTYTAYAISGPTVQDGLPVFHWNHFNKTLHEGMPEAYNFDFITMKPTL*LD PLBD1_equCab VADIYLASKYTAYAISGPTVQGGLPVFHWNRFNKTLHEGMPEAYNFDFITMKPILKPYVKGRR* PLBD1_ailMel VADIYLASEYTAYAISGPTVQGGLPIFHWNRFNKTLHKGMPETYDFDFITMKPILKRDKK PLBD1_felCat VADIYLASAYTAHAISGPTVQDGLPVFHWNRFNKTLHQGMPETYNFDFIIMKPILKQDIK* PLBD1_canFam VADIYLASEYTAYAISGPTTQGGLPVFHWNRFNKTLHKGMPEIYNFDFMTMKPILKHDRK* PLBD1_myoLuc VADMYLALEYTAHAISGPTVQGGLPVFHWKRFNKTLHEGMPEAYNFDFITMKPILKPDIK* PLBD1_pteVam VADIYLASQYTAHAISGPTVQGALPVFHWNQFNKTLHEGMPEAYNFDFVTMQPILKPDKK* PLBD1_eriEur VADFYLTFKYTAYAISGPTVQDGLPAFHWNRFNKTLHKGMPEVYNFDFVTMKPVL*ldrk PLBD1_sorAra VADIYLAAKFTAYAISGPTVQGGLPVFRWDPFNKTLHRGMPESFDFDFITVKPTL*qdkk PLBD1_loxAfr VADMYLASEYTAYAISGPTVQNGLPVFHWNRFNKTLHHGMPEAYNFDFVTMRPILKPDRN* PLBD1_proCap VSDMFLASEFIAYAISGPTVQNGLPVFHWNNFNKTLHQGMPEAYNFDFVTMQPILKLDRKL PLBD1_echTel VADMWLASKYRAYAISGPTVQDGLPVFRWGSFNKTVHQGMPEAYNFDFTHMKPILT*gr* PLBD1_dasNov VADIYLASQYTAYAISGPTVQGGLPVFHWNRFNKTLHEGMPEAYNFDFITMKPSLNSDIK* PLBD1_choHof VADIYLASQYTAYAISGPTVQGGLPVFHWNRFNKTLHRGMPETYNFDFITMKPILT*ne* PLBD1_monDom VADMFLASQFTAYAINGPTVDDGLPVFEWKKFNETIHKGLPEAYNFDFVTMKPLLEFCELHKEKKKRCGKQVRRWKRRN* PLBD1_ornAna VSDMALAARLTAHAISGPTVQGGLPVFRWSRFNGTVHRGLPEAYDFDFVTMRPVLRPPWPREAGGR* PLBD1_galGal VSDFRLASAFTATAINGPPVQGGLPVFTWRRFNNTRHQGLPESYNFKFVTMRPIL* PLBD1_taeGut VSDFRLAAAFTASAINGPPVQGGLPAFSWRRFNRTRHQGLPESYNFDFVTMRPIL* PLBD1_anoCar VADINMAMKFTSYAINGPPVEEGLPIFTWSRFNQTKHQGLPDSYNFDFITMKPVL* PLBD1_botJar VADISMAAKFTAYAISGPTVEKGLPVFSWVHFNKTKHQGLPESYNFDFVTMKPVL PLBD1_ambMex VSDFYLAATYTAHAINGPPVADGLPPFSWSPFHETIHEGLPEHYNFSFILTKPVL PLBD1_tetNig VTDFLMAGKFRAEAINGPTTQSGLPPFVWDRFGSVSHQGLPQSYNFTFVPMQPLLF* PLBD1_takRub VTDFFMAGKFRAEAINGPTTQNGLPPFAWDGFGNISHEGLPKYYNFTFVQMQPILFQP* PLBD1_gasAcu VTDFHMAGDFRAEAVNGPTTQDGLPPFFWDKFSSMSHQGLPQFYNFTFIRMQPVLFEP* PLBD1_oryLat VTDFFMAGDFTAEAVNGPTTQDGLPPFYWDKFSSISHQGLPRFYNFTFVTMKPLMFKP* PLBD1_salSal VTDFHMAQEFRAEAVNGPTTQGDLPPFSWEDFNSTAHQGLPDHYDFPFISMQPALFMP* PLBD1_danRer VADYRMAQMFTAEAVNGPTSQNGLPLFSWSRFNRTAHQGLPQTYNFTFITMQPLLFAFRDQAKTER... PLBD1_ictPun VTDLNMAQQFVSEALNGPSTDGDLPPFTWDAFNRTTHQGLPRLYNYTFVTMHPVLFSP PLBD1_petMar VADMRMAKKFMTSAVNGPTVEGKLPAFSWSPFDNIKHEGLPNTYKFPFVTMQPTLFTIP* PLBD1_braFlo VSDYYLARNLTSFAINGPTLGTGLEPFSWSDKFKISHIGLPKVYNFSFVTMTPAEL* PLBD1_strPur VTNLAMAAKQTSFVINGPTRGDGSLPPFKWVAPFTGWSHVGLPTVYDFNFVEMCPKEL* PLBD2_homSap VTSMSLARILSLLAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFAPVKVSWD* PLBD2_gorGor VTSMSLARILSLLAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFAPVKVSWD* PLBD2_ponAbe VTSMSLARILSLLAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFAPVKVSWD* PLBD2_nomLeu VTSMSLARILSLLAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFAPVKVSWD* PLBD2_papHam VTSMSLARILGLLAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFAPVKVSWD* PLBD2_calJac VTSMSLAKILSLLAASGPTWDQVPPFQWSTSPFSTLLHMGQPDLwKFCAPKVSWD* PLBD2_micMur VTSMSLAKALSLLAVSGPTWDQVPPFQWSASPFSSLLHMGQPDLWKFSPIRVWWQ* PLBD2_otoGar VTSMSLAKALSLLAVSGPTWDQVPPFQWSTSPFSSLLHMGHPDLWKFLPIEVWWD* PLBD2_tupBel VTSMSLAKTLSLVAASGPTWDQVPPFQWSTSAFSHLLHMGHPDLWRFSPIQVSWD* PLBD2_musMus VTSFTLAKYMSMLAASGPTWDQCPPFQWSKSPFHSMLHMGQPDLWMFSPIRVPWD* PLBD2_ratNor VTSVALAKYMSMLAASGPTWDQLPPFQWSKSPFHNMLHMGQPDLWMFSPVKVPWD* PLBD2_dipOrd VTSMSLAKALGLLAVSGPTWDQVPPFQWSSSPFPDVLHMGQPDLWKFLPVEVLWGL PLBD2_cavPor VTSMSMAKTLSLQAVSGPTWDQVPAFQWSTSPFRDMLHMGHPDLWKFAPVEVSWG* PLBD2_oryCun VTSKSLAKAMSLLAASGPTWDQVPPFQWSTSPFRDQLHMGHPDLWKFLPIRVLWD* PLBD2_ochPri VTSMSLAKALSLLADSGSTWDQVPPFQWSASPFRDKLHMGHPDLWKFLPFKVLWD* PLBD2_vicPac VTNMALAKALRLLAASGPTWDQLPPFQWSTSPFSRLLHMGQPDLWKFSPIDVWWD* PLBD2_turTru VTSTALAKALRLLAVSGPTWDQLPPFQWSSSPFSSLLHMGQPDLWKFSPIEVWWD* PLBD2_bosTau VTSTALAKALRLLAVSGPTWDQLPPFQWSTSPFSGMLHMGQPDLWKFSPIEVSWD* PLBD2_oviAri VTSTALAKALRLLAVSGPTWDQLPPFQWSTSPFSGMLHMGQPDLWKFSPIEVSWD* PLBD2_lamPac VTNMALAKALRLLAASGPTWDQLPPFQWSTSPFSRLLHMGQPDLWKFSPIDVWWD* PLBD2_susScr VTSMALARVFGLLAASGPTWDQLPPFQWSTSPFSHLLHMGQPDLWKFSPIEVSWD* PLBD2_felCat VTSMALAKAFQLVAASGPTWDQLPPFQWSASPFSGLLHMGQPDLWKFSPIEVRWD* PLBD2_canFam MTSMALAKAFHIIAVSGPTWDQVPPFQWSSSPFSGLLHMGQPDVWKFLPIETWWD* PLBD2_ailMel VTSMALARAFHIIAVSGPTWDQLPPFQWSSSPFSSLLHMGQPDLWKFSPIEVWWD* PLBD2_myoLuc VTSMALAKALRLVAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFSPVKVSWD* PLBD2_pteVam VTSAALAKALRFLAASGPTWDQLPPFQWSTSPFSGLLHMGQPDLWKFSPIEVWWD* PLBD2_eriEur VTNVSLVRALGLLAVSGPTWDQLPPFQWSTSPFSGLLHMGQPDLWKFspievwwd* PLBD2_loxAfr VTSLAMAKALRLLAVSGPTWDQLPPFQWSTSPFRSLLHMGQPDLWKFLPIEVWWD* PLBD2_proCap VTNLAMAKALRLRAVSGPTWDQLPPFQWSTSPFQSLLHMGQPDLWKFLPIEVWWN* PLBD2_echTel VTSSGLAKSLRLWAVSGPTWDQLPPFQWSSSPFHNLLHMGQPDLWKFSPVEFGWD* PLBD2_macEug VTSYELSKDLRLIAVSGPTWDQLPPFQWSSSPFDKLLHMGHPDLWKFFPIKVSWE* PLBD2_ornAna VTSSQLAKDFRFVAASGPTWDQVPAFRWSSSPFKGLVHMGHPDLWRFSPVHVRWD* PLBD2_galGal VTSFGMARTFGLVAASGPTWDDVPPFRWSTSPCSHLLRMGHPDLWRFPPVKVRWD* PLBD2_taeGut VTSSAMVPTFGLVAVSGPAWDDVPPFRWSASPCSSLLHMGHPDLWTFPPVKVHWD* PLBD2_anoCar VTSFEMAKLYSFVATSGPTWDDLPAFEWSSSPYRNLLHMGHPDLWRFSPIQVHWG* PLBD2_xenTro LTSYEMAKKYEMVVVNGPTWDQVPPFQWSTSPFSSLMHMGHPDLWKFSPITIRWH* PLBD2_ranCat VTSYEFAKEYMMFAVNGPTWDQVPPFQWSTSPFSNLMHMGHPDLWKFDPILIRWK* PLBD2_cynPyr VTSFNMARVYGMVAVSGPTWDDLPPFQWSTSPFSVQLHMGHPDLWQFDPVEVLWWQ* PLBD2_tetNig LTSYQMFRDYAMIAVSGPTWDQVPPFQWSTSPYKDLLHMGHPGTWTFKPVKVTWKP* PLBD2_takRub LTSYKMFRDYGLIAVSGPTWDQVPPFQWSTSPYKDLLHMGHPDTWTFKPITVIWTP* PLBD2_gasAcu LTSFEMFRDYAMLAVSGPTWDQVPPFQWSTSPYSDLMHMGHPDSWAFKPVKVSWNP* PLBD2_oryLat MTSFGMFKEYGMIAVSGPTWDQLPPFQWSTSPYKDLVHMGHPDVWNFKPIKVTWTP* PLBD2_salSal VTSYGLWREFGFLAASGPTWDQVPAFQWSSSPYSDLMHMGHPDTWAFTPIHVTWST* PLBD2_danRer MTSSSMFRQWELLAASGPSCEQTPVFQWSRSPYSSLMHMGQPDRWDFPTVHVRWAT* PLBD2_ictPun MTSYGLFKQYELLAVSGPTWDQVPAFEWSTSPYSSLTHIGHPDRWDFPTVHIRWSE PLBD2_squAca VTSFELHSTYQMIAVNGPTWDEVPAFQWSKSPFSSLMHMGHPDLWRFLPVMVQWK* PLBD2_calMil VTSSEMYKTFEMIAISGPAWDQVPPFQWSKSSYSGLIHMGHPDLWKFPPVMVRWS* PLBD2_petMar ITSKAMVPRLEMVAQSGPTWDQQPPFQWSKSPFSSLSHVGQPDLWSFLPEHISWCKHSGQ* PLBD2_braFlo MTSYSMHESHQMMAVSGPTHDQQQPFQWSTSDYDKQFYHLGHPDLFNFDPIHVIWFDQSDN* PLBD2_cioInt VVGYSMMKNFEILAECGPTHDQQPPFVWSKSPFSHVSHKGMPDKYDFKPTLIIWDKFSPLKMLDKIHKSVNL* PLBD2_halRor VTSYTLHKTLQMVAEAGPTHDQQPVFQWSTSPYASKSHEGHPDRFDFLPVLIKWDGEILPK* PLBD2_strPur VTTSSMVKSLSMVAVCGPTTDQQPPFQWSKSDFNQTLHLGHPDLFNFKPINVQWYD* PLBD2_sacKow LTNYVMHKDLSFVAISGPTQDQQPVFQWSTSPYLDVLHLGHPDKFDFGPVQVNWKND* PLBD2_aplCal LISYDLFKSLSFLAISSPTYDDLPPFQWSKSDYNYMSHLGHPDVWKFPRILFKGTDPLA* PLBD2_creFor LTSAEMVKDLTFIAVGGPSWDQQPPFQWSKSDFKSTSHIGQPDLWKFPPVLFNISLIF* PLBD2_eupSco LTNSSLARDLQFVAIAGPTYDQVTPFQWSKSDFKNTVSHIGHPDIFKFEPYIFGDDVMKFE* PLBD2_craGig LTNSAMFKAMQFVAISGPTYDQFAPFQWSKSDFKDNTPHMGHPDTFKFDPVVFDGTTDFKPFQR* PLBD2_helRob ITSYSMFKNFQFLAVSGPTRDQVAPFQWSKSDLKDTIRHAGHPDLWVFDPVQF* PLBD2_capCap ITSYSMFKNFQFLAVSGPTRDQVAPFQWSKSDLKDTIRHAGHPDLWVFDPVQF* PLBD2_limPol LTTFGLSQKFEFVAIGGPTHDPLPPFQWSKSDFSKDLPHYGHPDLWVFKPVTHHWK* PLBD2_ixoSca VTNFALFERQVFYAVSGPTSDDQRPFRWSTSGFDNVSHAGHPDLWDFDPILAQWRY* PLBD2_derVar LTTYQQFKEQQFFAISGPTWSQQPVFQWSTSGFNDSHVGHPDRWEFGPVLNYWGSCR* PLBD2_dapPul VTSHQLMTSLDFIAVGGPTFDSLPAFRWSESDFVNMSHIGHPDLWKFEPVQTEWTL* PLBD2_calCle VTSFDLLLRGSFIAGGGPTYDSVEPFQWSKADFEKDTPHFGHPDKWDFKPMRVEWDNVL* PLBD2_lepSal ITSFDLFMKGSFIAGGGPTYDSVEPFQWSKTDFGKTTPHFGHPDKWAFKPIKVDWENAFNRDEDVV* PLBD2_hydMag ATSFQLSKLMSQFIVGGPTYDQQPPFQWSKTEWNRPLGHPDIFKFNPELLDWRKEEWIYSKLNI* PLBD2_nemVec ITSFELFQKFQCIAVSGPTHDQQPAFQWSTSEWEKPLGHPDKFDFEPVKVSWDNKD* PLBD2_merSen ITSSELFKKFQCQAVSGPTHDQQPVFQWSKSDWKRPLGLPDKFDFSPVMVSWENEE* PLBD2_acrPal ITNSEMVKSLECVAVSGPTHDQQPVFKWSASGWDTPLGHPDAWDFEPIVVKWQEN* PLBD2_porAst ATSHQLVQQLSTIATCGPTHAQQPVFKWSESGETKPLGHPDAFDFPTVQIKWNKQ* PLBD2_monFav VTNSELIKELQCMAVSGPTHDQQPVFKWSTSGWKRPTGHPDAWDFEPIKVTWE* PLBD2_ampQue LTNSEMVKSLSCLATSGPTHSQQPVFKWSTSGFQDTPPLGHPDEFDFAPIVIKWGEIN* PLBDa_dicDis VVSADMVAALLVNAQSGPSHDNETPFTWNSQWNQKYTYAGQPTTWNFDWMTMSLQSMKPASPSSDSSSDSTTFN*
Signature conservation in the final exon
Using the above collection of sequences for the final exon of PLBD1 and PLBD2, it is possible to compare the same 45 vertebrate species for both genes. When these are aligned retaining phylogenetic order and allowing 0,1,2,...,10 departures from absolute invariance at each site, the two tables below result.
From them, residues conserved in PLBD1 but not PLBD2, conserved in PLBD2 but not PLBD1 or conserved in both can be extracted. The former serve quite well to classify phospholipases in the very earliest diverging eukaryotes which otherwise might have poor overall Blast scores to their mammalian ortholog. That classification is usually supported by an indel location: PLBD2 has lost one residue relative to PLBD1 just before the conserved SGPT motif and PLBD1 has lost two residues relative to PLBD2 further down.
The two proteins have been conserved equally well over the 500 million years of vertebrate evolution, quantitated simply by a dot count (.) in the two tables relative to the total number of residues (45 spp x 55 sites = 2475). Oddly, this conservation often takes place at non-homologous (different) sites in the two proteins despite the overwhelming similarity in their folds. At two sites near the carboxy terminus, a residue is conserved but it differs for the two protein (F/P and P/W). Otherwise 11 residues are conserved in both proteins -- this conservation may date back to the original gene duplicated (and perhaps earlier in NTN hydratases).
Vertebrate conservation of last exon of PLBD1 with increasing tolerance for exceptions: 0 V.D..........A..GP.....LP.F....--F....H.G........F....P.. 1 V.D...A......A..GP.....LP.F.W..--F....H.G.P....F.F....P.. 2 V.D...A......A..GP.....LP.F.W..--F....H.G.P..Y.F.F....P.L 3 V.D...A......A..GP.....LP.F.W..--F....H.G.P..Y.F.F....P.L 4 V.D...A......A..GP....GLP.F.W..--F..T.H.G.P..Y.F.F..M.P.L 5 V.D...A......A..GPT...GLP.F.W..--FN.T.H.G.P..Y.F.F..M.P.L 6 V.D...A......AI.GPT...GLP.F.W..--FN.T.H.G.P..YNF.F..M.P.L 7 V.D...A......AI.GPT.Q.GLP.F.W..--FN.T.H.G.P..YNF.F..M.P.L 8 V.D...A....a.AI.GPT.Q.GLP.F.W..--FN.T.H.G.P..YNF.F.TM.P.L 9 V.D..LA....a.AI.GPTVQ.GLP.F.W..--FN.T.H.G.PE.YNFDF.TM.P.L 10 V.D..LA....a.AI.GPTVQ.GLP.F.W..--FN.T.H.G.PE.YNFDF.TM.P.L V.D...A...............gL.......--Fn.t............F..m.P.L conserved PLBD1 not PLBD2 .............A..GPt.....P.F.W.........H.G.P..w.F........ conserved PLBD1 and PLBD2 .Ts............S...-WDq......S.SP......m...D.....P..v.W.. conserved PLBD2 not PLBD1 Vertebrate conservation of last exon of PLBD2 with increasing tolerance for exceptions: 0 .T..............G..-....P.F.WS.S........G.P..W.F......W.. 1 .T..............G..-....P.F.WS.S........G.P..W.F......W.. 2 .T...........A.SGP.-WD..P.F.WS.SP.....H.G.PD.W.F......W.. 3 .T...........A.SGPT-WD..P.F.WS.SP.....HMG.PD.W.F.P....W.. 4 .TS..........A.SGPT-WDQ.P.F.WS.SP.....HMG.PD.W.F.P....W.. 5 .TS..........A.SGPT-WDQ.P.FQWS.SP.....HMG.PD.W.F.P..V.W.. 6 .TS..........A.SGPT-WDQ.P.FQWS.SP.....HMG.PD.W.F.P..V.WD. 7 .TS..........A.SGPT-WDQ.PPFQWS.SP.....HMG.PD.W.F.P..V.WD. 8 .TS..........A.SGPT-WDQ.PPFQWS.SP....LHMG.PDLW.F.P..V.WD. 9 VTS..........A.SGPT-WDQ.PPFQWS.SP...LLHMG.PDLW.F.P..V.WD. 10 VTS..........A.SGPT-WDQ.PPFQWS.SP...LLHMG.PDLW.F.P..V.WD.
Lysosomal storage disease: allele prediction
Experience has shown that it is fairly simple to predict about half of the common mutations that will eventually surface as disease alleles. These are comprised of (1) rare polymorphisms in the population that occur at phylogenetically conserved sites and and (2) CpG hotspot non-synonymous substitutions at similarly conserved residues. Here there will also be replication slippage errors in the regions of anomalous GC compositions such as the signal peptide; their outcome is difficult to predict but would lead to frameshifts 2/3 of the time (if not 3n).
The reason for this is both PLBD1 and PLBD2 are autosomal, meaning that both alleles must be affected. This occurs in the consanguineous case mainly for adverse polymorphisms that have managed to attain a reasonable frequency in the population. These can otherwise combine with the dominant de novo mutations, namely CpG mutations resolving to TpG or CpA. Indeed the Q54P discussed earlier for the reference genome arose as a transversional CpG mutation CCG (pro) -> CAG (gln), as can be seen from chimpanzee sequence.
The significance of a given substitution can then be read off the 46-way vertebrate alignment -- a departure from the evolutionarily tested reduced alphabet is rarely an adaptive innovation but rather almost always an unacceptable change reducing fitness (what else could account for observed conservation?).
As a practical matter, the 1000 Human Genome Project provides rare alleles to the 1% level for every gene (though currently these are difficult to extract for a particular gene). These are supplemented with previously observed SNPs. Consequences of CpG mutations are obtained by in silico substitution, followed by Blastx against wildtype to flag differences (55 for CpA, 50 more for less frequent TpG), followed by evaluation of the change, using mammals from the 46-way with MultAlin set to maintain input order.
Neanderthal has 4 substitutions (three arising from CpG) relative to human, none of which are predicted to affect protein function:
human genome location #reads change N H comment chr12:112280948-112280949 1C>0A(CCG>CAG) P 54Q reference genome has a mutation at an ultra-conserved site chr12:112294980-112294981 0G>3C(GCT>CCT) A182P human has a phylo-acceptable change chr12:112297191-112297192 0C>2T(CGG>TGG) R286W human has a change at an unselected residue chr12:112309977-112309978 0G>3A(GAC>AAC) D496N human has a change at an unselected residue
To these should be added fatal CpG disruptions of splice donors, which PLBD2 has in 3 consecutive exons, all of phase 2 which give rise in the CpG hotspot effect either to a phylogenetically non-conservative change in 2nd letter of codon -- shown at left: T373M, P405L, P429L for CpG mutating to TpG -- and/or loss of GT-AG splice donor soon leading to a premature stop codon for for CpG transitioning to CpA:
agGTGACACTGGAGACCACCATTGGCAACAAGAACCCAGCCCTGTGGAAGTATGTGCGGCCCAGGGGCTGTGTGCTGGAGT GGGTACGCAACATCGTGGCCAACCGCCTGGCCTCGGATGGGGCCACCTGGGCAGACATCTTCAAGAGGTTCAACAGCGGCACgt 2 T373M agGTATAACAACCAGTGGATGATCGTGGACTACAAGGCGTTCATCCCGGGTGGGCCCAGCCCCGGGAGCCGGGTGCTTACCATCCTGGAGCAGATCCCgt 2 P405L agCGGCATGGTGGTGGTGGCTGACAAGACCTCGGAGCTCTACCAGAAGACCTACTGGGCCAGCTACAACATACCgt 2 P429L
No stop codons result from CpG mutations in PLBD2 but numerous non-conservative changes occur at invariant sites. A sampler of these is shown flanking the GSGSCSALIK cleavage motif below: E153K, R183Q, E197K, R199H, R272H, R282Q, G294S are all predicted to be disruptive. Dashes indicate lack of data rather than deletions -- input sequences could be greatly improved by curation as is done below -- and at much greater phylogenetic depth -- for the terminal exon.
160 170 180 190 200 210 220 230 240 250 260 270 280 290 300
| | | | | | | | | | | | | | |
CpG2CpA YCKRLKSFLEANLEWMQEEMESNPDSPYWHQVQLTLLQLKGLEDSYKGHVSFPAGKFTIKPLGFLLLQLSGDLEDLELALNKTKIKPSLGSGSCSALIKLLPGQSDLLVAHNTWNNYQHMLHVIKKYWLQFQEGPWGDYPLVPSNKLVFS
homSap ..E.............................R.............E.R........................................................................R.........R...........G......
panTro ..E.....................Y.A.....R.............E.R........................................................................R.........R...R.......G......
gorGor ..E.......................A.....R.............E.R.....................................................................Y..R.........R...RSFETVFNASG.QAL
ponAbe ..E.......................A.....R.............E.R........................................................................R.........R...R.......GS.....
rheMac ..E.......................A.....R.............E.R........................................................................R.........R...Q..S....G......
papHam ..E.......................A.....R.............E.R........................................................................R.........R...Q..S....G......
calJac ..E.................Q.....A.....----------------------------------------------------------------------------------------------------------------------
micMur ..E.......V.......QVAL.K..V.....R.............E.R.N..TE.............I........P......V.AA..A.......................HS.....R......F..R...RD.D....G..V...
tupBel ..E..................R.Q..T....KTKV.-------------------------------------------------------..GCS....---.RH.VATR----W.....R....CL.-IHQ..QE.S...........
musMus ..EK..N..........R...L..........R.............E.RLT..T.R............I........P.....NT..................GH..........S..N..RI....R...R...QEE....AG.N....
ratNor ..EK.............R...LS.........R.............E.RLT..T.R.N..........I........P.....NT...V..............SH..........S..N..RI....R...R...QEE...IAG.N.I..
dipOrd ..EK.........A...A...QQVN.T.....R.............E.R.T..TE.......X--............P....SNS..V....A....V.....HR..........A..S..R.........R...---------------
cavPor ..E.......T......QQ..LH..C......R.............E..L...T.R.S..........I.............S.TRAT.........L......R..........A..Y..R.L...Q...RV..RE.F..A.G..V...
speTri ..EK..H..........R..QWDQ..G...K.R.............E.R....T..............I........P.....RTRAV............M...R.....L.--S.S.N..-I....TS.I--..QK.AL...G.QM...
oryCun ..E...R..............R.K..T.....R.............E.R.T..T.R............I........P...R..TQAA...............HR..........S.....R......F..R...QE......G.RVI..
ochPri ..E...RY.........A.....RG.M.C...R.............E.R....M.R............I........P......T.AV................Q..........S.....RI.....F..RKD.Q..S....G.RVI..
turTru ..E...K..........K...L.NG.A.....R.............E.S.T..T..............I...............T.HAT.........................HS..Y..RIM....F..R...QEQSTRA.G.RVI..
bosTau ..E...N..........K...L.NG.A.....R.............E.S.A..T....V.........I........V......TNHAM...............R.........HS..Y..RIM....F..R...QAESTRA.G..VI..
equCab ..E......DT............K..A.....R.............E.S.A..T.R............I....G...P.....EARRT..A.A....V.....HR..........S..S..RI.....--------S.A.PA.G.R....
felCat ..EK..N...T.......Q.QR.KG.A----.R.............ESS.T..T.R.............A...G...P......S.HVM...................I.....SS..N..RI.....F..R.D.QVNS..A.G......
canFam ..E..MR...T..D..L.LIKL.K.FA..--.R.............E.SMT..T.R.................G...P......T.HIM...................I.....SS..N..RI.....F..R.D.QENS..A.G......
myoLuc ..E...G..........KQV.L.K..A.....R.............E.S.A..T..............I...............T.HT...............H...........S..N..RI.....F......QE.S..A.G......
pteVam ..EK.................L.K..A.....R.......-.....E.S.T..T..............I...............TRHT..A....V.......HG..........S..N..R.....SF..RQ..QE.S..A.G......
loxAfr ..EK.................L.R..A.....R.............G.RMT..T.......................P....S.T.RA................R..........S.....R.V...SFH.H...RENSL.A.G.SM...
proCap --------------...K...L.R..A.....R......-......G.RMT..T.N.....................P......T.RA................R..........S.....R.L...SFH.R...QE.SV.A.G.T....
echTel ..EK......T..........R.K..T.....R.A...........G...T..T.R.....................P......T.R.M...............R..........S.....RIV...SF..H...QD.SL.I.G...I..
dasNov ..E...R..........R..ALS.HDA.....R.............E.R.T.......F.........V---------------------------------------------------------------------------------
macEug ..EK..NYI.L..A...KQIA.SK.DE.....E.A...........Q.RIA..SKN...T.F....F..........P.....EQRRVM.............NN.E...S.D...S.....RI....DFR.RTL.K---..I.G..Q...
monDom ..EK...YI.M..A...KQ.A.GK.AE.....E.A...........Q.RIA..SKN...T.F....F..........P.....AHRRVM.............NRKE...S.D..SS.....RI....KFH.RTL.Q.EA..I.G.EQ...
A serious CpG mutation -- G551S -- could occur via G -> A in the terminal exon whereas other substitutions are nearly neutral:
+ - -
CpG2CpA VTSMSLARILSLLAASSPTWDQVPPFQWSTSPFS-SLLHMGQPDLWKFTPVKVSWD
PLBD2_homSap ................G..................G............A.......
PLBD2_gorGor ................G..................G............A.......
PLBD2_ponAbe ................G..................G............A.......
PLBD2_nomLeu ................G..................G............A.......
PLBD2_papHam ..........G.....G..................G............A.......
PLBD2_calJac .......K........G..................T............CAP.....
PLBD2_micMur .......KA.....V.G............A..................S.IR.W.Q
PLBD2_otoGar .......KA.....V.G........................H......L.IE.W..
PLBD2_tupBel .......KT...V...G..............A...H.....H....R.S.IQ....
PLBD2_musMus ...FT..KYM.M....G.....C......K...H..M.........M.S.IR.P..
PLBD2_ratNor ...VA..KYM.M....G.....L......K...H.NM.........M.S....P..
PLBD2_dipOrd .......KA.G...V.G............S...P.DV...........L..E.L.G
PLBD2_cavPor .....M.KT...Q.V.G.......A........R.DM....H......A..E...G
PLBD2_oryCun ...K...KAM......G................R.DQ....H......L.IR.L..
PLBD2_ochPri .......KA.....D.GS...........A...R.DK....H......L.F..L..
PLBD2_vicPac ..N.A..KA.R.....G.....L............R............S.ID.W..
PLBD2_turTru ...TA..KA.R...V.G.....L......S..................S.IE.W..
PLBD2_bosTau ...TA..KA.R...V.G.....L............GM...........S.IE....
PLBD2_oviAri ...TA..KA.R...V.G.....L............GM...........S.IE....
PLBD2_lamPac ..N.A..KA.R.....G.....L............R............S.ID.W..
PLBD2_susScr ....A...VFG.....G.....L............H............S.IE....
PLBD2_felCat ....A..KAFQ.V...G.....L......A.....G............S.IE.R..
PLBD2_canFam M...A..KAFHII.V.G............S.....G........V...L.IETW..
PLBD2_ailMel ....A...AFHII.V.G.....L......S..................S.IE.W..
PLBD2_myoLuc ....A..KA.R.V...G..................G............S.......
PLBD2_pteVam ...AA..KA.RF....G.....L............G............S.IE.W..
PLBD2_eriEur ..NV..V.A.G...V.G.....L............G............S.IE.W..
PLBD2_loxAfr ...LAM.KA.R...V.G.....L..........R..............L.IE.W..
PLBD2_proCap ..NLAM.KA.R.R.V.G.....L..........Q..............L.IE.W.N
PLBD2_echTel ...SG..KS.R.W.V.G.....L......S...H.N............S..EFG..
PLBD2_macEug ...YE.SKD.R.I.V.G.....L......S...D.K.....H......F.I....E
PLBD2_ornAna ...SQ..KDFRFV...G.......A.R..S...K.G.V...H....R.S..H.R..
PLBD2_galGal ...FGM..TFG.V...G....D....R.....C..H..R..H....R.P....R..
PLBD2_taeGut ...SAMVPTFG.V.V.G.A..D....R..A..C........H....T.P....H..
PLBD2_anoCar ...FEM.KLY.FV.T.G....DL.A.E..S..YR.N.....H....R.S.IQ.H.G
PLBD2_xenTro L..YEM.KKYEMVVVNG....................M...H......S.ITIR.H
PLBD2_ranCat ...YEF.KEYMMF.VNG..................N.M...H......D.ILIR.K
PLBD2_cynPyr ...FNM..VYGMV.V.G....DL............VQ....H....Q.D..E.L.W
PLBD2_tetNig L..YQMF.DYAMI.V.G...............YK.D.....H.GT.T.K....T.K
PLBD2_takRub L..YKMF.DYG.I.V.G...............YK.D.....H..T.T.K.IT.I.T
PLBD2_gasAcu L..FEMF.DYAM..V.G...............Y..D.M...H..S.A.K......N
PLBD2_oryLat M..FGMFKEYGMI.V.G.....L.........YK.D.V...H..V.N.K.I..T.T
PLBD2_salSal ...YG.W.EFGF....G.......A....S..Y..D.M...H..T.A...IH.T.S
PLBD2_danRer M..S.MF.QWE.....G.SCE.T.V....R..Y....M......R.D.PT.H.R.A
PLBD2_ictPun M..YG.FKQYE...V.G.......A.E.....Y....T.I.H..R.D.PT.HIR.S
PLBD2_squAca ...FE.HSTYQMI.VNG....E..A....K.......M...H....R.L..M.Q.K
PLBD2_calMil ...SEMYKTFEMI.I.G.A..........K.SY..G.I...H......P..M.R.S
PLBD2_petMar I..KAMVPR.EMV.Q.G.....Q......K.......S.V......S.L.EHI..C
PLBD2_braFlo M..Y.MHESHQMM.V.G..H..QQ.......DYDKQFY.L.H...FN.D.IH.I.F
PLBD2_cioInt .VGY.MMKNFEI..ECG..H..Q...V..K.....HVS.K.M..KYD.K.TLII..
PLBD2_halRor ...YT.HKT.QMV.EAG..H..Q.V.......YA..KS.E.H..RFD.L..LIK..
PLBD2_strPur ..TS.MVKS..MV.VCG..T..Q......K.D.N.QT..L.H...FN.K.IN.Q.Y
PLBD2_sacKow L.NYVMHKD..FV.I.G..Q..Q.V.......YL.DV..L.H..KFD.G..Q.N.K
PLBD2_aplCal LI.YD.FKS..F..I....Y.DL......K.DYN.YMS.L.H..V...PRILFKGT
PLBD2_creFor L..AEMVKD.TFI.VGG.S...Q......K.D.K..TS.I........P..LFNIS
PLBD2_eupSco L.NS....D.QFV.IAG..Y...T.....K.D.KNTVS.I.H..IF..E.YIFGD-
PLBD2_craGig L.NSAMFKAMQFV.I.G..Y..FA.....K.D.KDNTP...H..TF..D..VFDGT
PLBD2_helRob I..Y.MFKNFQF..V.G..R...A.....K.DLKDTIR.A.H....V.D..QF---
PLBD2_capCap I..Y.MFKNFQF..V.G..R...A.....K.DLKDTIR.A.H....V.D..QF---
PLBD2_limPol L.TFG.SQKFEFV.IGG..H.PL......K.D..KD.P.Y.H....V.K..THH.K
PLBD2_ixoSca ..NFA.FERQVFY.V.G..S.DQR..R....G.D.NVS.A.H....D.D.ILAQ.R
PLBD2_derVar L.TYQQFKEQQFF.I.G...S.Q.V......G.-.NDS.V.H..R.E.G..LNY.G
PLBD2_dapPul ...HQ.MTS.DFI.VGG..F.SL.A.R..E.D.V.NMS.I.H......E..QTE.T
PLBD2_calCle ...FD.LLRG.FI.GGG..Y.S.E.....KAD.EKDTP.F.H..K.D.K.MR.E..
PLBD2_lepSal I..FD.FMKG.FI.GGG..Y.S.E.....KTD.GKTTP.F.H..K.A.K.I..D.E
PLBD2_hydMag A..FQ.SKLM.QFIVGG..Y..Q......KTEWN.--RPL.H..IF..N.ELLD.R
PLBD2_nemVec I..FE.FQKFQCI.V.G..H..Q.A......EWE.--KPL.H..KFD.E.......
Four serious mutations could arise from C -> T mutations at CpG sites: A548V, T553M, P559L, and S565L
+ + + + -
CpG2TpG VTSMSLARILSLLVASGPMWDQVPLFQWSTLPFS-GLLHMGQPDLWKFVPVKVSWD
PLBD2_homSap .............A....T.....P.....S.................A.......
PLBD2_gorGor .............A....T.....P.....S.................A.......
PLBD2_ponAbe .............A....T.....P.....S.................A.......
PLBD2_nomLeu .............A....T.....P.....S.................A.......
PLBD2_papHam ..........G..A....T.....P.....S.................A.......
PLBD2_calJac .......K.....A....T.....P.....S....T............CAP.....
PLBD2_micMur .......KA....AV...T.....P....AS....S............S.IR.W.Q
PLBD2_otoGar .......KA....AV...T.....P.....S....S.....H......L.IE.W..
PLBD2_tupBel .......KT...VA....T.....P.....SA...H.....H....R.S.IQ....
PLBD2_musMus ...FT..KYM.M.A....T...C.P....KS..H.SM.........M.S.IR.P..
PLBD2_ratNor ...VA..KYM.M.A....T...L.P....KS..H.NM.........M.S....P..
PLBD2_dipOrd .......KA.G..AV...T.....P....SS..P.DV...........L..E.L.G
PLBD2_cavPor .....M.KT...QAV...T.....A.....S..R.DM....H......A..E...G
PLBD2_oryCun ...K...KAM...A....T.....P.....S..R.DQ....H......L.IR.L..
PLBD2_ochPri .......KA....AD..ST.....P....AS..R.DK....H......L.F..L..
PLBD2_vicPac ..N.A..KA.R..A....T...L.P.....S....R............S.ID.W..
PLBD2_turTru ...TA..KA.R..AV...T...L.P....SS....S............S.IE.W..
PLBD2_bosTau ...TA..KA.R..AV...T...L.P.....S.....M...........S.IE....
PLBD2_oviAri ...TA..KA.R..AV...T...L.P.....S.....M...........S.IE....
PLBD2_lamPac ..N.A..KA.R..A....T...L.P.....S....R............S.ID.W..
PLBD2_susScr ....A...VFG..A....T...L.P.....S....H............S.IE....
PLBD2_felCat ....A..KAFQ.VA....T...L.P....AS.................S.IE.R..
PLBD2_canFam M...A..KAFHIIAV...T.....P....SS.............V...L.IETW..
PLBD2_ailMel ....A...AFHIIAV...T...L.P....SS....S............S.IE.W..
PLBD2_myoLuc ....A..KA.R.VA....T.....P.....S.................S.......
PLBD2_pteVam ...AA..KA.RF.A....T...L.P.....S.................S.IE.W..
PLBD2_eriEur ..NV..V.A.G..AV...T...L.P.....S.................S.IE.W..
PLBD2_loxAfr ...LAM.KA.R..AV...T...L.P.....S..R.S............L.IE.W..
PLBD2_proCap ..NLAM.KA.R.RAV...T...L.P.....S..Q.S............L.IE.W.N
PLBD2_echTel ...SG..KS.R.WAV...T...L.P....SS..H.N............S..EFG..
PLBD2_macEug ...YE.SKD.R.IAV...T...L.P....SS..D.K.....H......F.I....E
PLBD2_ornAna ...SQ..KDFRFVA....T.....A.R..SS..K...V...H....R.S..H.R..
PLBD2_galGal ...FGM..TFG.VA....T..D..P.R...S.C..H..R..H....R.P....R..
PLBD2_taeGut ...SAMVPTFG.VAV...A..D..P.R..AS.C..S.....H....T.P....H..
PLBD2_anoCar ...FEM.KLY.FVAT...T..DL.A.E..SS.YR.N.....H....R.S.IQ.H.G
PLBD2_xenTro L..YEM.KKYEMV.VN..T.....P.....S....S.M...H......S.ITIR.H
PLBD2_ranCat ...YEF.KEYMMFAVN..T.....P.....S....N.M...H......D.ILIR.K
PLBD2_cynPyr ...FNM..VYGMVAV...T..DL.P.....S....VQ....H....Q.D..E.L.W
PLBD2_tetNig L..YQMF.DYAMIAV...T.....P.....S.YK.D.....H.GT.T.K....T.K
PLBD2_takRub L..YKMF.DYG.IAV...T.....P.....S.YK.D.....H..T.T.K.IT.I.T
PLBD2_gasAcu L..FEMF.DYAM.AV...T.....P.....S.Y..D.M...H..S.A.K......N
PLBD2_oryLat M..FGMFKEYGMIAV...T...L.P.....S.YK.D.V...H..V.N.K.I..T.T
PLBD2_salSal ...YG.W.EFGF.A....T.....A....SS.Y..D.M...H..T.A.T.IH.T.S
PLBD2_danRer M..S.MF.QWE..A....SCE.T.V....RS.Y..S.M......R.D.PT.H.R.A
PLBD2_ictPun M..YG.FKQYE..AV...T.....A.E...S.Y..S.T.I.H..R.D.PT.HIR.S
PLBD2_squAca ...FE.HSTYQMIAVN..T..E..A....KS....S.M...H....R.L..M.Q.K
PLBD2_calMil ...SEMYKTFEMIAI...A.....P....KSSY....I...H......P..M.R.S
PLBD2_petMar I..KAMVPR.EMVAQ...T...Q.P....KS....S.S.V......S.L.EHI..C
PLBD2_braFlo M..Y.MHESHQMMAV...TH..QQP.....SDYDKQFY.L.H...FN.D.IH.I.F
PLBD2_cioInt .VGY.MMKNFEI.AEC..TH..Q.P.V..KS....HVS.K.M..KYD.K.TLII..
PLBD2_halRor ...YT.HKT.QMVAEA..TH..Q.V.....S.YA.SKS.E.H..RFD.L..LIK..
PLBD2_strPur ..TS.MVKS..MVAVC..TT..Q.P....KSD.N.QT..L.H...FN.K.IN.Q.Y
PLBD2_sacKow L.NYVMHKD..FVAI...TQ..Q.V.....S.YL.DV..L.H..KFD.G..Q.N.K
PLBD2_aplCal LI.YD.FKS..F.AI.S.TY.DL.P....KSDYN.YMS.L.H..V...PRILFKGT
PLBD2_creFor L..AEMVKD.TFIAVG..S...Q.P....KSD.K.STS.I........P..LFNIS
PLBD2_eupSco L.NS....D.QFVAIA..TY...TP....KSD.KNTVS.I.H..IF..E.YIFGD-
PLBD2_craGig L.NSAMFKAMQFVAI...TY..FAP....KSD.KDNTP...H..TF..D..VFDGT
PLBD2_helRob I..Y.MFKNFQF.AV...TR...AP....KSDLKDTIR.A.H....V.D..QF---
PLBD2_capCap I..Y.MFKNFQF.AV...TR...AP....KSDLKDTIR.A.H....V.D..QF---
PLBD2_limPol L.TFG.SQKFEFVAIG..TH.PL.P....KSD..KD.P.Y.H....V.K..THH.K
PLBD2_ixoSca ..NFA.FERQVFYAV...TS.DQRP.R...SG.D.NVS.A.H....D.D.ILAQ.R
PLBD2_derVar L.TYQQFKEQQFFAI...T.S.Q.V.....SG.-.NDS.V.H..R.E.G..LNY.G
PLBD2_dapPul ...HQ.MTS.DFIAVG..TF.SL.A.R..ESD.V.NMS.I.H......E..QTE.T
PLBD2_calCle ...FD.LLRG.FIAGG..TY.S.EP....KAD.EKDTP.F.H..K.D.K.MR.E..
PLBD2_lepSal I..FD.FMKG.FIAGG..TY.S.EP....KTD.GKTTP.F.H..K.A.K.I..D.E
PLBD2_hydMag A..FQ.SKLM.QFIVG..TY..Q.P....KTEWN.--RPL.H..IF..N.ELLD.R
PLBD2_nemVec I..FE.FQKFQCIAV...TH..Q.A.....SEWE.--KPL.H..KFD.E.......
>CpG2CpA 55 changes for PLBD2
MVGQMYCYPSSHLAQALTQALALALVLALLVRPFLSSLAGAIPAPGGHWAHNGQVPPASHSHSVLLDISAGQLLMVDRHHPDTMAWANLTNTIHKTGWAFLELGTSGQYNDSLQAYAASV
VEAAVSEELIYMHWMNTVVNYCSPFKYEVSYCKRLKSFLEANLEWMQEEMESNPDSPYWHQVQLTLLQLKGLEDSYKGHVSFPAGKFTIKPLGFLLLQLSGDLEDLELALNKTKIKPSLG
SGSCSALIKLLPGQSDLLVAHNTWNNYQHMLHVIKKYWLQFQEGPWGDYPLVPSNKLVFSSYPSTIFSCNNFYILGSGLVTLETTIGNKNPALWKYVQPRGCVLEWVHNIMANHLASDGA
TWADIFKRFNSSTYNNQWMIMDYKAFIPGGPSPRSQVLTILEQIPSMVVVADKTSELYQKTYWASYNIPSFKTVFNASGLQALVAQYGDWFSYDRSPQAQIFQQNQSLVQDMDSMVRLMR
YNDFLHDPLSLCKACNPQPNGENAISTHSNLNPANGSYPFQALHQHSHGGINVKVTSMSLARILSLLAASSPTWDQVPPFQWSTSPFSSLLHMGQPDLWKFTPVKVSWD*
>CpG2TpG 50 changes for PLBD2
MVGQMYCYPGSHLAWVLMWVLVLALVLALLVGLFLSGLVGVIPVLGGCWVCDGQVPPASCSCLVLLDVLVGQLLMVDGCHPDAVAWANLTNAICETGWAFLELGTSGQYNDSLQAYAAGV
VEAAVLEELIYMHWMNMVVNYCGPFEYEVGYCERLKSFLEANLEWMQEEMESNPDSPYWHQVWLTLLQLKGLEDSYEGCVSFPAGKFTIKPLGFLLLQLSGDLEDLELALNKTKIKPSLG
SGSCSALIKLLPGQSDLLVAHNTWNNYQHMLCVIKKYWLQFWEGPWGDYLLVPGNKLVFSSYPGTIFSCDDFYILGSGLVTLETTIGNKNPALWKYVWPRGCVLEWVCNIVANCLALDGA
TWADIFKRFNSGMYNNQWMIVDYKVFILGGPSPGSWVLTILEQIPGMVVVADKTLELYQKTYWASYNILSFETVFNASGLQALVAQYGDWFSYDGSPWAQIFWWNQSLVQDMDSMVRLMR
YNDFLHDPLSLCKACNPQPNGENAISACSDLNLANGSYPFQALCQCSHGGIDVKVTSMSLARILSLLVASGPMWDQVPLFQWSTLPFSGLLHMGQPDLWKFVPVKVSWD*
Homologs in model organisms
In this section, species are discussed that have an existing (or potential) phospholipase experimental literature for either PLBD1 or PLBD2. Their relevance to the human situation varies greatly depending on divergence and other complications. Drosophila LAMA -- despite two early papers will prove completely irrelevant. Nematodes have a reasonably comparable protein and could be quickly studied by genetic techniques. Oddly slime mold may be the most useful of the three because it has been used for decades as a model system for lysosomal targeting.
Slime molds (Dictyostelium)
Although slime molds seem an improbable model system for mammals given the immense time since divergence from the last common ancestor, Dictyostelium works fairly well for the lysosomal sorting process via the M6P pathway, establishing its great antiquity, with 220 on-topic papers dating back decades. A phospholipase B, presumably from lysosomes, was characterized in 2004. Called PLBDa here, it is only one of seven found in the newly sequenced Dictyostelium genome.
It is not immediately clear how this gene family expansion relates to mammalian PLBD1 and PLBD2. In one scenario, a single gene in the ancestral stem had expanded to the two PLBD1 and PLBD2 clades at the time of the last common ancestor. No further expansion occurred in the lineage leading to human but each clade expanded further in slime molds. This would cause the seven Dictyostelium genes to classify clearly with one branch or another of the gene tree. The data below do not support this.
A second scenario envisions a single gene at the time of species divergence. This duplicated subsequently in the lineage leading to human and expanded independently in Dictyostelium. This would give rise to the gene tree ((PLBD1, PLBD2),(dicDis subtree)). The data fit this better; it predicts a slightly later diverging species can be found in which the PLBD1/PLBD2 duplication had already occurred and the Dictyostelium-type genes are missing. The choanoflagellate Monosiga fits this description. It also predicts earlier diverging species than Dictyostelium will only have ambiguously classifying PLBDx (assuming ancestral features do not predominate in PLBD1 or PLBD2, respectively).
Note all seven genes are perfectly conserved for the six active site residues, meaning this had been established earlier. Thus any or all of them could be used as a generic structural model for mammalian PLBD1 or PLBD2, an important consideration in view of the similar processing, activation, and known substrate.
It can be seen from Blastp against SwissProt that all of the Dictyostelium proteins have been assigned accession numbers. This means they might have already been modeled by SwissModel using the mouse PLBD2 structure as template but in fact have not, though modeling requests can be submitted using the 3FGW template. Note PLBDd and PLBDg are the most favorable for PLBD1; whereas PLBDb, PLBDc, and PLBDa have better matches to PLBD2. Only PLBDa has its substrates determined.
Q550U9 PLBLA_DICDI PLBDa_dicDis SwissModel+ interactive Q55BJ6 PLBLB_DICDI PLBDb_dicDis SwissModel- Q54M94 PLBLC_DICDI PLBDc_dicDis SwissModel- Q54PS7 PLBLD_DICDI PLBDd_dicDis SwissModel- Q54ZI6 PLBLE_DICDI PLBDe_dicDis SwissModel- Q554H5 PLBLF_DICDI PLBDf_dicDis SwissModel- Q55FN1 PLBLG_DICDI PLBDg_dicDis SwissModel-
PLBD1_homSap Homo sapiens (human) FLJ22662 2968 1.2e-313 100% blastp query PLBD1 PLBD1_musMus Mus musculus (mouse) 2321 4.5e-245 79% PLBD1_braFlo Branchiostoma floridae (lancelet) 1369 3.4e-144 50% PLBD1_nemVec Nematostella vectensis (anemone) 1349 4.5e-142 51% PLBD1_strPur Strongylocentrotus purpuratus (urchin) 1122 5.1e-118 47% PLBD1_monBre Monosiga brevicollis (choanoflagellate) 1089 1.6e-114 51% PLBDd_dicDis Dictyostelium discoideum (slime_mold) 822 3.1e-86 37% d PLBDg_dicDis Dictyostelium discoideum (slime_mold) 746 1.2e-84 36% g PLBD2_triAdh Trichoplax adhaerens (trichoplax) 721 1.2e-82 38% PLBD2_braFlo Branchiostoma floridae (lancelet) 722 2.0e-80 36% PLBDb_dicDis Dictyostelium discoideum (slime_mold) 762 7.2e-80 34% b PLBDc_dicDis Dictyostelium discoideum (slime_mold) 733 8.5e-77 35% c PLBDa_dicDis Dictyostelium discoideum (slime_mold) 717 4.2e-75 33% a PLBDe_dicDis Dictyostelium discoideum (slime_mold) 717 4.2e-75 35% e PLBD2_acyPis Acyrthosiphon pisum (aphid) 667 4.7e-75 37% PLBD2_homSap Homo sapiens (human) LOC196463 639 2.0e-72 36% PLBD2_musMus Mus musculus (mouse) 631 6.0e-71 37% PLBDf_dicDis Dictyostelium discoideum (slime_mold) 640 6.1e-67 32% f PLBD2_monBre Monosiga brevicollis (choanoflagellate) 547 3.3e-66 32% PLBDx_droMel Drosophila melanogaster (fruitfly) 142 5.5e-19 32% PLBD2_homSap Homo sapiens (human) LOC196463 3142 0.0 100% blastp query PLBD2 PLBD2_musMus Mus musculus (mouse) 2571 1.4e-271 81% PLBD2_braFlo Branchiostoma floridae (lancelet) 1528 4.8e-161 54% PLBD2_triAdh Trichoplax adhaerens (trichoplax) 1293 3.9e-136 50% PLBD2_acyPis Acyrthosiphon pisum (aphid) 1129 9.2e-119 47% PLBDb_dicDis Dictyostelium discoideum (slime_mold) 873 1.2e-91 39% b PLBD2_monBre Monosiga brevicollis (choanoflagellate) 835 1.3e-87 38% PLBDc_dicDis Dictyostelium discoideum (slime_mold) 819 6.5e-86 39% c PLBDa_dicDis Dictyostelium discoideum (slime_mold) 805 2.0e-84 38% a PLBD1_nemVec Nematostella vectensis (anemone) 702 1.6e-78 38% PLBD1_musMus Mus musculus (mouse) 688 5.9e-77 38% PLBDd_dicDis Dictyostelium discoideum (slime_mold) 731 1.4e-76 37% d PLBD1_homSap Homo sapiens (human) FLJ22662 639 2.0e-72 36% PLBDg_dicDis Dictyostelium discoideum (slime_mold) 685 1.0e-71 36% g PLBDe_dicDis Dictyostelium discoideum (slime_mold) 661 3.6e-69 32% e PLBD1_braFlo Branchiostoma floridae (lancelet) 652 3.2e-68 36% PLBD1_monBre Monosiga brevicollis (choanoflagellate) 646 1.4e-67 38% PLBDf_dicDis Dictyostelium discoideum (slime_mold) 640 6.1e-67 35% f PLBD1_strPur Strongylocentrotus purpuratus (urchin) 534 7.7e-61 35% PLBDx_droMel Drosophila melanogaster (fruitfly) 218 2.9e-38 30% Alignment of PLBDa_dicDis with conserved residues of PLBD1 or PLBD2 shows no clear classification: VTDLNMAQQFVSEALNGPSTDGDLPPFTWDA--FNRTTHQGLPRLYNYTFVTM best PLBD1 match V +M + A +GPS D + P FTW++ +N+ T+ G P +N+ ++TM VVSADMVAALLVNAQSGPSHDNETP-FTWNSQWNQKYTYAGQPTTWNFDWMTM PLBDa_dicDis + S MV L + AQSGP+ D + P F W+ ++ GQP W+F ITSKAMVPRLEMVAQSGPTWDQQPP FQWSKSPFSSLSHVGQPDLWSFLPEHI best PLBD2 match V.D...A...............gL.......--Fn.t............F..m.P.L conserved PLBD1 not PLBD2 .............A..GPt.....P.F.W.........H.G.P..w.F........ conserved PLBD1 and PLBD2 .Ts............S...-WDq......S.SP......m...D.....P..v.W.. conserved PLBD2 not PLBD1
Arthropods (Drosophila)
The standard fruitfly has a very odd PLBD2-type gene product LAMA (lamina ancestor) that has been the subject of two publications. Note first that a poor choice of name has lead to widespread confusion: this protein has nothing whatsoever to do with the important vertebrate protein laminin (whose homolog in fly is called LamA). Instead it has to do with an obscure neuroanatomical structure in insects called the lamina.
The gene, call PLBD2x here, is so diverged from conventional phospholipases that a number of hypotheses have to be considered:
- Fraud: It is not unusual for annotational hoaxes to be perpetrated in the scientific literature, complete with bogus submissions to GenBank. Here however simple tBlastn searches show that the most recent assembly of the drosophila genome does contain a letter-perfect match to the LAMA entry and further that this protein has a more or less a full length match to conventional vertebrate PLBD2. This is beyond anyone's ability to manipulate.
- Pseudogene: This is a large protein of almost 600 amino acids. Should it have lost its function, rapid divergence would result (as seen), with accrual of frameshifts and stop codons (not seen). The divergence would occur equally rapidly at ultra-conserved residues as at ones not under selection (not seen), either in D. melanogaster or one of the other 11 drosophilids with sequenced genome (not seen). However recent pseudogenization in certain lineages cannot be ruled out as comparative genomics becomes insufficient.
- Horizontal transfer: The LAMA protein might fail to nest within the gene tree as expected from the fruitfly position in the species tree. This happens rarely in metazoan from gene transfer from other species, notably parasites, commensals, and intra-cellular symbionts. In this case, tBlastn would locate the appropriate clade, even if the exact species had no data because genomic coverage is so pervasive today. However the LAMA gene has no outside affinities.
- Role change: If this gene lost its original function and acquired another unrelated one, then the selection profile could change radically across the gene. For example, if the catalytic function were redirected, then the six active site residues might experience relaxed constraint. The fold itself might be conserved to assure structural stability but otherwise rapid evolution to perfect the new role might alter many otherwise conserved residues. Here the change is so rapid and so drastic that a simple change to a new substrate can be ruled out -- indeed, the two experimental papers provide evidence for a regulatory role.
- Role loss: Along the lines of Piatigorsky, a single gene product may have multiple roles, sometimes quite distinct (eg aldehyde dehydrogenase ALDH3A1 also serving as a lens crystallin). Should the gene subsequently be duplicated, these pre-existing roles can be split up in the two copies (rather than one copy continue with the parental role and the other copy scramble to acquire a new adaptive function). Selection initially acts on all the functions in proportion to their importance.
In this last scenario, phospolipase activity was lost, leaving selection to act solely on the residual regulatory function. However within the 12 drosophilid genomes, the PLBD2 are quite diverged, with percent identity quickly ranging from 75% down to 45%. This suggests the protein is not yet well adapted to what was initially a secondary function or that few of its features are needed.
This raises the question whether PLBD2 also serves a cryptic non-catalytic function in other species (eg mammals). It is thus important to date the onset of loss of phospholipase activity (coupled with retention of within the PLBD2 locus) within arthropods because this also provides some dating of the secondary function origin. This can be dated to the last tens of millions of years because only a few other insects (eg tsetse Glossina morsitans, EZ422576) outside drosophilids carry a comparable gene.
Within insects, some species such as Acyrthosiphon carry both a normal and derived PLBD2. However the normal form has been lost in other insects with derived PLBD2. Although no chelicerate has the derived PLBD2, Ixodes has multiple copies of normal PLBD2, some closely related and others fairly diverged but still clearly classifying as PLBD2. Molluscs, annelids, and trematodes have PLBD2 but not PLBD1 or PLBD2x.
The loss of active site residues characteristic of NTN hydratases and more specifically those shared by both PLBD1 and PLBD2 phospholipases is taken here as a proxy for loss of phospholipase (and indeed all) catalytic activity.
Active site residues are lost in derived PLBD2 though the first disulfide is retained:
<---- human active site ----> <-- human disulfides --> <-- glycosylation-->
PLBD2_homSap C244 H261 W264 T325 N427 R458 C142-C152 C492-C495 N436S N515S
PLBD2x_droMel A216 H238 A241 G305 G438 ? C98-C108
PLBD2x_glomor A210 H231 A234 G298 G424 ? C94-C104
PLBD2x_acrPis A233 Q250 S253 V315 S413 V436 C132-C139
PLBD2_acrPis C208 H225 W228 T288 N390 R421 C112-C119 C434-C437 N399S N477T
Unless there is some counterpart to the secondary function, the tempo and mode of evolution of the LAMA protein makes it largely irrelevant to its human ortholog and lysosomal storage diseases. Indeed, fruitflies have in effect lost both PLBD1 and PLBD2, proving they are non-essential to lysosome function.
Drosophila PLDB2x at SwissModel: little of the original fold remains:
PLDB2x 45 CATAL WTKQV-GFQI ENWKQQNDLV NIPTGVGRIC YKDSVYENGW
3fgrA 63 vsr--trsll ldaasgqlrl ed-----g-- fhpdavawan ltnairetgw
ssss ss ssss s sssss sssss ss
sss sssss ss ssss ss s sssss sssss ss
PLDB2x 89 AQIEVETQRT YPDWVQAYAA GMLEGSLTWR NIYNQWSNTI SSSCERDEST
3fgrA 104 ayldlstngr yndslqayaa gvveasvsee liymhwmntv vnycgpfeye
ssssssss hhhhhhhh hhhhhh h hhhhhhhh
ssssssss hhhhhhhh hhhhh hh hhhhhhhh
PLDB2x 139 QKFCGWLRDL LTTNYHRLKR QTEKAENDHY WHQLHLFITQ LEGLETGYKR
3fgrA 154 vgyceklknf leanlewmqr emelnpdspy whqvrltllq lkgledsyeg
hhhhhhhhhh hhhhhhhhhh hhhh hh hhhhhhhhhh hhhhhhhhh
hhhhhhhhhh hhhhhhhhhh hhhh hh hhhhhhhhhh hhhhhhhhh
PLDB2x 189 GASRARSDLE EEIPFSDFLL MNAAADIQDL KIYYENY -- -
3fgrA 204 rltfptgr-- ftikplgfll lqisgdledl epalnktgsg s
hh h hhhhhh hhh
hh h hhhhhh hhh
Nematodes (C. elegans)
Potentially a better model organism than fruitfly, C. elegans has 3 phospholipase B genes, with the best match to human PLBD2 having 44% identity. These appear to be a lineage-specific expansion of PLBD2, whereas PLBD1 has been lost from this and other nematodes. (The C. elegans genome is assuredly complete and the detection methods are sufficiently sensitive.)
>PLBD2a_caeEle Caenorhabditis elegans (nematode) NP_499668 Y37D8A MNWIFIFLAAAVAIGCEARQERTYTVCQKPEGDLHYFKEGRKTDEELCAKRLATAYFHDEVNQTGWAFLEVDVISPKIPHYLQGYAAGFAEGRATRHLID LHIINTVNGYCDGAKHFCDELGEFMVDNLKWMEQEIRENPEDEYWQQVNLTVNQLFGLIHGYENQLGAEIDFKQIAVHPIFMIQIAGDLEDLAMKFKKPE NPKKVFSGPGHCSALVKLLPKNEDILFSHVTWSSYGTMLRINKKYSFKTGDPGQIYSFSSYPASITSTDDFVLTSAKLAILETTIGNYNEKSLDLITPNT VLTWIRAEIAHRTASSGLQWAEAFGRHNSGTYNNEWVVVDYKQFHRGKEVQPETGIIHVVEQMPGHIVHSDKTAHLFRETYWPGYNQPYYKQIIRFSDTD KMVEKFGDWYSYDKTPRALIFKRDHNTVTDMSSMIALMRSNNYTKDPLSKCDCNPPYSAENAIACRSDLNPLNGTYPFKSLGFRDHGAIDVKVTNSKLIN SLQFTAVSGPPGGVTKDVPIFDWKTSPLREKVPHFGQPDRWNFAPVTYKWRKDAHRHYHLYQKLHKELSSL* >PLBD2b_caeEle Caenorhabditis elegans (nematode) NP_510509 F09B12 MTRLIRSKKQFLIRSLHSVFYYLGSLLHSTFEMNVFIGLLLATVVASQSSEGRDESYTYKQLCIVDDKPQVLDGFDCRNQVAVARWQNAVNTTGWTFLEV ETKENYCPQLQAYSAGYLEGLLSKTVLTYHLKNAQEDYCKNFTGYCSRLSDFLTENQKWIQSSLETVAPDDLYWGAVNRTYHQVSGLIDAYEGREFKPRI TYELHPILYLNLNGDFYDLEKKLNKTRDPAFEQTGGKCSGLIKVAPGNADLFISQVTMSGFQNMLRVIKLYKFGYDRQFYPGYASSFSSYPGLLYSSDDF ALQTSGLAVIETTISVFNTSLFENTKPVGQLPTWIRAIVSNQLARDAREWCKLYSLYNSGTYNNQWAVLDYKKFKPNQPLPKNGLFYVLEQMPGKIVYSD LTWFVEKYSYFPSYNIPFFKEITEISGFIGQAAKMGDWFKWGASPRAKIFERDHGNVHDLDSLTALMRYNDYKNDEFSKCKCNPPYSAEAGISARGDLNP ANGTYEFPGQGHVNHGALDYKGTNVELMKKLQFVAQGGPTWGKVPSFKWSEFDFKDKVNHVGHPDEWKFNTLVHKWETEINA* >PLBD2c_caeEle Caenorhabditis elegans (nematode) NP_497570 Y54F10AM MKLLFFLFGLIFAVEQEKPYLDNNRVPVEQILNDHSSAKFDYTYVSVCVNSTDETLLDIVYAKECKNAASRVALGKYSNQVNTTGWGILEIETFASHSYD VQAYGAGVAEGELTRLQIYYHYRNTIETMCNNHTLFCKRLYIYLQQNLDWMRSQVQANPPTDPFWRQVNLTFAQLTGIYDAYSKRNLTPEIGFDLHPIYM MQLAGDMFDLNKLLNKTADPMEYPEGGRCSGFVKLAPGNKDMFMAHVSMSSLSWMQRVLKIYKFGYDVNEVPGHIVTFSGYPGVLISTDDYTITSAGLTS IETTIAIFNQTLYTDKFMKPEGQVHCWIRSMISNLLSRTGKQWVDMFGRYNSGTYNNQWTVLDWKQFTPEKELPDKDVLWISEQTPGYYETRDMTWYLKK YTYFASYNIPFLPKVSEISGFDNKARQFAWFDWGGSPRARIFDRDHSKVTDIDSLTKLMRYNDYTHEEFARCKCTPNPYTGEGGISARGDLNTPGGTYEV ESMGFRDHAGLDFKGTNYEMFKKMRFRAWGGPPYDPLPVFDWNHTNLTNVRHFGQPDVWNFTYVDLEWQLAAQVQLTPYDN*
Reference sequences
These two genes are well represented at GenBank though some predicted gene models are erroneous. A genomic alignment of 46 species of vertebrates at UCSC (under protein fasta section of the gene details page) allows this large set of orthologs to be collected as needed. These sequences -- which are no better than the underlying assemblies -- also contain extensional errors at the termini. Insertions relative to human also are not shown. However these sequences provide a strong starting point for correction at the wgs contig division of NCBI blast.
Below, only a few key sequences are shown -- ones with an experimental literature and some from deeply diverged pre-bilaterans. These have been intronated by blast of protein against genome assemblies.
A separate section holds anomalous PLBD2x insect sequences. These resulted from a duplication of PLBD2, followed bu severe specialization of one copy and often loss of the original. These nonetheless will retain most features of the fold, though use of SwissModel here would require considerable intervention after the fact. Despite an experimental literature, these are largely irrelevant to human considerations.
In the last section, sequences have been trimmed to the region covered by the xray structural determination. Species with one-off insertions that confuse gap placement have been adjusted. This helps avoid artifacts in alignment.
PLBD1 reference sequences
>PLBD1_homSap Homo sapiens (human) FLJ22662 PMID: 19019078,20093120 0 MTRGGPGGRPGLPQPPPLLLLLLLLPLLLVTAEPPKPA 1 2 GVYYATAYWMPAEKTVQVKNVMDKNGDAYGFYNNSVKTTGWGILEIRAGYGSQTLSNEIIMFVAGFLEGYLTAP 2 1 HMNDHYTNLYPQLITKPSIMDKVQDFME 2 1 KQDKWTRKNIKEYKTDSFWRHTGYVMAQIDGLYVGAKKRAILEGTK 0 0 PMTLFQIQFLNSVGDLLDLIPSLSPTKNGSLKVFKRWDMGHCSALIK 0 0 VLPGFENILFAHSSWYTYAAMLRIYKHWDFNVIDKDTSSSRLSFSSYP 1 2 GFLESLDDFYILSSGLILLQTTNSVFNKTLLKQVIPETLLSWQRVRVANMMADSGKRWADIFSKYNS 1 2 GTYNNQYMVLDLKKVKLNHSLDKGTLYIVEQIPTYVEYSEQTDVLRK 1 2 GYWPSYNVPFHEKIYNWSGYPLLVQKLGLDYSYDLAPRAKIFRRDQGKVTDTASMKYIMRYN 1 2 NYKKDPYSRGDPCNTICCREDLNSPNPSPGGCYDTK 0 0 VADIYLASQYTSYAISGPTVQGGLPVFRWDRFNKTLHQGMPEVYNFDFITMKPILKLDIK* 0 >PLBD1_musMus Mus musculus (mouse) NM_025806 note earlier stop codon 0 MCHRSPGRSLRPPSPLLLLLPLLLQPPWAAGAASQSDPT 1 2 GVHCATAYWSPESKKVEIKTVLDKNGDAYGYYNDSIKTTGWGILEIRAGYGSQVLSNEIIMFLAGYLEGYLTAL 2 1 HMYDHFTNLYPQLFKNPSIVKKVQDFME 2 1 KQEMWTRQNIKAQKDDPFWRHTGYVVTQLDGLYLGAQKRASEEKIK 0 0 PMTMFQIQFLNAVGDLLDLIPSLSPTKSSSMMKFKIWEMGHCSALIK 0 0 VLPGFENIYFAHSSWYTYAAMLRIYKHWDFNIKDKYTLSKRLSFSSYP 1 2 GFLESLDDFYILSSGLILLQTTNSVYNKTLLKQVVPKTLLAWQRVRVANMMAEGGKEWAQIFSKHNS 1 2 GTYNNQYMVLDLKKVTINRSLDKGTLYIVEQIPTYVEYSDQTNVLRK 1 2 GYWASYNIPFHKTIYNWSGYPLLVHKLGLDYSYDLAPRAKIFRRDQGNVTDMASMKYIMRYN 1 2 NYKEDPYSKGDPCSTICCREDLNGASPSPGGCYDTK 0 0 VADIFLASQYKAYAISGPTVQDGLPPFNWNRFNDTLHRGMPEVFDFNFVTMKPILS* 0 >PLBD1_braFlo Branchiostoma floridae (lancelet) XM_002595538 0 MEGRACRSCRLHHLSAVFLLFLVTIAA 1 2 GAEIQATAYLQAQGKVQVKLGVLDKQNGDAVATYDDR 2 1 LTENGWGVLNVVSGFGPKKLSDNDIMYLAGYLEGVLTQE 2 1 RIYQHYLNLYGIFFMGKSEDLVGK 0 0 VKKFYTAQDTWVRAQVKQSTDPVMKHLSYILSQYDGLVKGYNDN 0 0 LFPHVSFFQKLDIFAFQLLNGNGDTFDIIPAVNPSSRPDFSNMSRVEIDDWVSAHSHCSALVK 0 0 VLGAYENVYMSHSSWFNYAATMRIYKHYNFNIANPATATRKMSFSSYP 1 2 GYLESLDDFYLMDSGLVMLQTTNNVFNGTLYDLVKPESILAWQRVRTANMLARNGDQWGAIMNVHNS 1 2 GTYNNQYMIIDLNLIELGKTIHDGALYVVEQIPGLVMSADQTDILRA 1 2 GYWPSYNIPFYEKVYNLSGYPEFAKSQGLDYTYQLAPRAKIFRRDAGKVKDMESMKAIMRYN 1 2 DYLHDPYSKGNPCSAICCRKDLAKVGAKPDGCYDTK 0 0 VSDYYLARNLTSFAINGPTLGTGLEPFSWSDKFKISHIGLPKVYNFSFVTMTPAEL* 0 >PLBD1_strPur Strongylocentrotus purpuratus (urchin) XM_001192029 0 MANKFRMFKILTAFLVLVLVNLST 1 2 GELLQGTVYKQEDGTFTVSSGIIDKQGVAYGSYNNTLFQTGWGELHLFAGYSTADNVALSDADRMYAAGILEGALTAK 2 1 QISQTLRNINVTFFSAESDPEIWRRVADFFETQDAWMKGMIIERADEDPFWEGVGLVLAQFEGLIKGYEMSQFSNAST 0 0 SNGFLAMQVLNSCGDLLDLKSAVMPSLIPDWDKLTKKEFLKFIRTSGHCSALVK 0 0 ICAALVKVGRFAPPFQSLLYSIS 0 0 SYFKSQAILKLNSPSCQLFGIE 1 2 GFLESLDDFYIMSSGLSMLQTTNNIFNKTLYKYVKPQSLLAWQRVRVANMMARSGKDWARIVARYNS 1 2 GTYNNQYMVIDRTKIKPNVAILDDALWVVEQVPTLVASGDQTNILRA 1 2 GYWPSYNVPFYEEIYNISGYPEYAYKGGADISYQLAPRAKIFRRDQGNVVDMESFKKIMRFN 1 2 DYKNDPYSEGDPSKSICMRGDLMTSPMPNGCYDTK0 0 VTNLAMAAKQTSFVINGPTRGDGSLPPFKWVAPFTGWSHVGLPTVYDFNFVEMCPKEL* 0 >PLBD1_nemVec Nematostella vectensis (anemone) XM_001638165 0 MTLIRNSVMITVTFVLILFVFGCHGSQKSATVYYNRGQG 2 1 YSLKFGVVDKLMGVAYGTFEDSLNTTG 2 1 WYELNIVSGTGIEPYNDDVIMHAAGYLEGALTAS 2 1 QINDNYANLYGVFFKSEDDPMVAKVEKFFIEQ 0 0 DIWMRKMIALKSSNSSFWRQMGNIIAQFD 1 2 GLVEGYQKYPATDK 0 0 ALGVFAFQMLNGVGDLLDLTKALMPERMADWDHMTEKEILEK 0 0 VAMDGHCSALIKVLPAYENVFASHVS 2 1 WFTYSAMLRVYKHYHLNLKDETT 1 2 AAQRMSFSSYPGFLESLDDFYIMDS 2 1 KLVMLQTTNNVFNKSLYEQVVPESLFSWQRVRLANLVASSGRQWADIVGQYNS 1 2 GTYNNQYMVLDLKLIQLNNTIQDNALWVVEQIPT 2 1 LVASGDQTAILRAGYWPSYNVPFYEL 0 0 VYNLSGYPDFVARHGVQFSHELAPRAKIFRRDQSM 0 0 VHDLDSMKHIMRYNDFQHDPYSQGNPMNAICSRGDLIADGPRASGCYDGK 0 0 VTDFTMAQSLISHAINGPTHE 0 0 QQVPFHWSQYQFKNKHEGQ 0 0 PDLFNFDFVEMKPKF* 0 >PLBD1_monBre Monosiga brevicollis (choanoflagellate) XM_001745398 MSSLNNGIPEPLLKFLAAQFNWTRSQVAANQDDVFWQQVGLIMA QYDGLRAGYGANVYDKHVLPEFAFQLLNGNGDFFDIIPKAVDVTKMSSREFHDWRMRN GRCSALIKLTGDFSDLFMSHSAWYIYQAMNRIYKHCASYNFQATITHAKKISFSSYPG YLESLDDFYLMSSGLVMLQTTNNVFNTDLQQYIQPESLQSWIRIRTATALAQTSEDWA ELAGRHNSGTYNNQYMVMDLNKFTPGQPLLDGTLYVAEQIPGTWEYADVTKMLSLGYW PSYNVPFFEKIYNLSGYPAVVKQHGTDDSYELAPRAKIFRRDQTTVVDLDSFKAIMRY NDYKNDPYAKGDPYNAICSRGDLESDSPSPGGCYDTKVTTYSMALKLQSQVINGPTTS HGLPPFSWSQFPNASHLGMPEVFNFTFETMDAGW*
PLBD2 reference sequences
>PLBD2_homSap Homo sapiens (human) LOC196463 PMID: 19706171,19237744,17007843,17105447 0 MVGQMYCYPGSHLARALTRALALALVLALLVGPFLSGLAGAIPAPGGRWARDGQVPPASRSRSVLLDVSAGQLLMVDGRHPDAVAWANLTNAIRETG 2 1 WAFLELGTSGQYNDSLQAYAAGVVEAAVSEE 0 0 LIYMHWMNTVVNYCGPFEYEVGYCERLKSFLEANLEWMQEEMESNPDSPYWHQ 0 0 VRLTLLQLKGLEDSYEGRVSFPAGKFTIKPLGFL 2 1 LLQLSGDLEDLELALNKTKIKPSLGSGSCSALIKLLPGQSDLLVAHNTWNNYQHMLRVIKKYWLQFREGPW 1 2 GDYPLVPGNKLVFSSYPGTIFSCDDFYILGSGL 0 0 VTLETTIGNKNPALWKYVRPRGCVLEWVRNIVANRLASDGATWADIFKRFNSGT 2 1 YNNQWMIVDYKAFIPGGPSPGSRVLTILEQIP 2 1 GMVVVADKTSELYQKTYWASYNIP 2 1 SFETVFNASGLQALVAQYGDWFSYDGSPRAQIFRRNQSLVQDMDSMVRLMR 2 1 YNDFLHDPLSLCKACNPQPNGENAISARSDLNPANGSYPFQALRQRSHGGIDVK 0 0 VTSMSLARILSLLAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFAPVKVSWD* 0 >PLBD2_musMus Mus musculus (mouse) NM_023625 0 MAAPVDGSSGGWAARALRRALALTSLTTLALLASLTGLLLSGPAGALPTLGPGWQRQNPDPPVSRTRSLLLDAASGQLRLEDGFHPDAVAWANLTNAIRETG 2 1 WAYLDLSTNGRYNDSLQAYAAGVVEASVSEE 0 0 LIYMHWMNTVVNYCGPFEYEVGYCEKLKNFLEANLEWMQREMELNPDSPYWHQ 0 0 VRLTLLQLKGLEDSYEGRLTFPTGRFTIKPLGFL 2 1 LLQISGDLEDLEPALNKTNTKPSLGSGSCSALIKLLPGGHDLLVAHNTWNSYQNMLRIIKKYRLQFREGPQ 1 2 EEYPLVAGNNLVFSSYPGTIFSGDDFYILGSGL 0 0 VTLETTIGNKNPALWKYVQPQGCVLEWIRNVVANRLALDGATWADVFKRFNSGT 2 1 YNNQWMIVDYKAFLPNGPSPGSRVLTILEQIP 2 1 GMVVVADKTAELYKTTYWASYNIP 2 1 YFETVFNASGLQALVAQYGDWFSYTKNPRAKIFQRDQSLVEDMDAMVR 0 0 LMRYNDFLHDPLSLCEACNPKPNAENAISARSDLNPANGSYPFQALHQRAHGGIDVK 0 0 VTSFTLAKYMSMLAASGPTWDQCPPFQWSKSPFHSMLHMGQPDLWMFSPIRVPWD* 0 >PLBD2_braFlo Branchiostoma floridae (lancelet) XM_002612057 0 MAACRNIFCGRMLSCLLLFSFVFSAVSDGSKLASVRYDEAAKTYQITDKLDPSAAAWANFTDRISSTG 2 1 WSFLTVTTNEKYDDSVQAYAAGLVEGYLTRD LMYNHWLNTVGAAFCSSRSAFCKNLESFLKTNLAWMQEQIQASGDTDDYWHQ 0 0 VKLTLQQLSGLDDGYNDDPRQPSLDINPFGFL 2 1 IFQIGGDMEDLQEALKDKDSHRVLGSGSCSALVKLLPGNADLLVAHDTWDTFQSMLRIIKKYQFPFKLGGKK 1 2 GEDKIPGHTVSFSSYPGVIYSGDDFYITSASL 0 0 VAQETTIGNSNPALWKYVQPQGQVLEWLRNIVANRLANKAMDWATIFKKYNSGT 2 1 YNNQWMIVDYKTFTPNKDLPEKGLLVVLEQLP 2 1 GMVMMDDVTSVLAKQAYWPSYNSP 2 1 YFEKIFNTSGLPAMVEKYGDWFSYEHTPRANIFRRDHGKVTDISSMIKLMR 2 1 YNDFQNDPLSKCDCTPPYSAENAISARSDLNPANGTYPFSALQHRCHGGTDMK 0 0 MTSYSMHESHQMMAVSGPTHDQQQPFQWSTSDYDKQFYHLGHPDLFNFDPIHVIWFDQSDN* 0 >PLBD2_acyPis Acyrthosiphon pisum (aphid) XM_001948827 0 MLSIRCILLSLLFVWALQCSATQKNQTLLAVKTDNNRITIQPKHYSVKDKEIIIGKGKFIDRINSTG 2 1 WAYLEIRTSQKAKDEDQAYGAGYLEGTLTADLIYSYWFNTAKGYCTDRPNVCQQLKDYMTTNKNWIKSKLNESDPYWYQ 0 0 VGLYYKQLDGLYDGYMRGKSPSTPDLTWDDLY 2 1 WLNALDDLGDLSIALYPSDISNRVLGSGSCSALIKLMPDNKDILVSHATWSG 2 1 YETMLRIQKRYSLRFRKSKKSNKLIRGFDMSFSSFPGGIQSGDDFYLISSGLTTMETTIENYNDSLWSNVKPVGQ 0 0 VLEFVRAMVANRLADNPTDWANLFKLHNSGTYNNQWMILNYAAFQPGSPLPPRDVLHVLEQIPGHVMHDDFTGHLINRTYWASYNVPYFPFIFNVSGNYEMEQIYGSW 2 1 FSYSETPRARIFARDHVKIHCDKCMLHLMRSNNYTRDPESRCDCSPPYSAENAISSR 2 1 NDLNPANGTYPIRALGHRSHGATDVKVTSSQLFQQLQFKAIAGPTQGSNNSLGPFCWSKSDFNDKVSHLGHPDCFNFKPVLHQWSL* 0 >PLBD2_lepSal Lepeophtheirus salmonis (crustacean) MEILKGSIFPIFLLISTRALGTSSLKHSLSYDGKDFTLLPGHVSGSNILLHGSFVNLINQTGWSILKINTMSHFPDNIQAYGAGYIEAYMTKDLIYNHWR NTIVGYCDGKEKICEGIQGFLSKNLNYIRSMTTNSKREKSSYWHHVALFYDQIRGITEGYNKYAPPGRSISAEDIFFMNVMGDVEDLEQVFSHRLNISLP DKVLGSGSCSALIRLLPGNRDLYAAHDTWNSYQSMLRIIKKYKFGLHWVLQKFPEKDDKHTIPGRSMSFSGYPGTVFSMDDFNVIPSSGLVSIETTIGNG NASLWQYVQPIGTVLEGIRATVANRLARSGGHWVSVFSKRNSGTYNNQWMVVDYKRFKPGEKIIRPGLLWVLEQIPGRIHYTDVTGVLKKIGYWPSYNSP YFKDIFNMSGGPEAVAKFGNWFSYEKTPRALIFKRDAPKVHNMGSMMKIMRYNDFRHDPISRCNCTPPYSAENAISARCDLNPKNGTYPFGALGHRSHGG TDMKITSFDLFMKGSFIAGGGPTYDSVEPFQWSKADFGKTTPHFGHPDKWAFKPIKVDWENAFNRGEDVV* >PLBD2a_ixoSca Ixodes scapularis (chelicerate) XM_002413831 MKNSYAPRFPSHVLGGSGSEFFCSKLNVYCSSTHRNMFVTLAVSALLALQAAAETPGDASCQYAWASLGPSGGDLHIHDGKPGQGAVAWGTFQNDIRFSG WAFLDLQSNASYADDVQAYASGAVEAHLTRDLIEKHYNNMYSRYCDGQSEYCERLSKFLQENIKYSNDQEHLYYSTDPYWHMVHLQMKQLAGLSDHLENK ALNTSNEYLNVTRALYLNLDGDLMDLEAYLRRVGDENSIFQTNACSVLIKVLPDNEDILFAHNTWFLYRSMLRIEKKYTFPWHVTSESSEIIPGHTISMP SYPGKLLSLDDFYIASTGLAITETSLENNNDSLRNLIKPENAPLTWVRCMVATRLAVTGSQWVEYFGKLNSGTLNNQWMVLDYKLFTPGAAIGKDTFWIL EQMPNTTKTKDVSEYLQNQKYWASYNIPFLPEIFNLSGQPQLVKKYGNYYSHDMCPRAQLFRREQGKVEDVDSMTALMRYNDYTHDPVSGCNCTPPYNAI YAISSRYDLLDPEGSYDMPNMARRAVGATDMKLTNYGMSRRLEFIAINGPTYTNDSSVPPFQWSTSGFQDLHEGHPDKWTFGPTHHRWDSCPNF* >PLBD2b_ixoSca Ixodes scapularis (chelicerate) XM_002405205 frag 98 TGWSSIVVHTNEFYLDDQQAFAAGLVEGRLTRTLIQEQFTNEFNKYCEEDPQFCAELFKYLHANIESMVANVAEYSSTDPYWHQVRKTAFYDKPTEQRNK ACFGHSNQPVNEFFYTKRLLNLHGDLSDLETKFNRTNSRKINPTGSGSCSAIVKLAPGSRDVYFAHATWTHYNSMNRILKKYRLNYHTAPMSKDLIPGSS IVFSGYPARIFSGDDFYLINTGLAVMETTIGNENSDLWKKVTPIDTVPTSIRNMVANRLSLTGQHWTNTFARFNSGTYNNEWMVLDYKRFESTSQKLNRG FLWILEQIPGLVMSKDVTPILEEQGYWASYNSPYFTEIFNESGLPALVEKYGDWYTYDKSPRALMFKRDQEKAVDLRSVMKLMRYNDFTNDPLSRCTGCD PPYSAENAISARSDLNPANGTYPFKALGRRAHGGTDAKVTNFALFERQEFYAVSGPTSDDQRPFRWSTSGFDNVSHAGHPDLWDFDPILAQWRY* >PLBD2_plaOce Placobranchus ocellatus (mollusc) HP171048 MAIIMNSRLCVQLFVAISIFSLCNAAVTKLWVTYNTESSIFEVSDQQATDYVAVASFDNTVNQTGWAKLDVITQAGPKRKYNDSVQAYAAGFVEGHITKS LMTMHWANTGAWVCPEPLTSQCIQIKKFLESNLKWVLENIKTFSTTSPFWHHVRLFLEQTAGLQDGFAGMKGQLNLDIDVMSVYMWQVGGDMETLLNVFT GSEDNQYKTPDPLAMGMGHCSALLRLLPDYSDLYVAQDTWSDFSSMLRVLKRYSFEMLKSPQYPDIPAPGNTMTFSSYPGVLFSGDDFYIMSSGLVSQET TIGYANSSLNQYIKPTSVLEGVRTMVANRLAASGREWAQIFSLINSGTYNNEWMIVDYKLFRPGQVIPVPGLFTVLDQIPGTIVYDDLTLHLYKYKYFGS YNTPFFPEVFNKSGQPALVAKFGDWFTHDKTPRALIFKRDAPKVNDLNSMIKLMRYNDFKHDPLSRCNCTPPYSAENAIAARCDLNPANGTYPFGALGHR PHTATDMKVTTFEMAKSLSFRAVSSPPwDDLPVFQWSTSSF* >PLBD2_aplCal Aplysia californica (mollusc) est frag YPGPLMSVDDFYAISSGLVSQETTIGFSNAELVKYITSEAVMEGFRSMVANRLASSGGEGSKLFAQYNSGTYNNQWMIVDYKRFVPSRETGVPGTLYLIE QIPGTIEYADLTDYLYEKSYFGSYNVPYFPDIFNKSGLPPLVKKLGDWFTYDKNPRALIYKRECSKVTDLDSMTKLMRYNYFKNDPLSRCNCTPPYSAEN AISCRDDLNPANATYPFGALGH >PLBD2_helRob Helobdella robusta (annelid) EY564899 frag FAYLEIQTNEKVSDIEQAYTAGLAEGLLTSDLIKLHWYNTFQDYCKAPLNEFCAKLQNAMEANFNWMANQIFLEANLNPFWHQVELILVQLHGLIDGYKF EQRPRDTLTKFVFKPDVTGLLMLFLSGDISDLESVLGGNKFNVYKPFDHLSGPNTCSALIKLLPNNKDLLISQTTRSNYGTMLRILKKYDFGYHTVTGGQ LVPAKVISFSSYPGVQFSVDDFYLLSSGLVAQETTIGNSNADRWKLVKPESLLEVIRNMVANRLATTGEEWADLFVRYNSGTYNNQWMVLNYKLFTPGAK EIKDGLLYVVEQIPGLTVGQDVTFVLRNQTYWPSYNSPYFKEIFNASGCLENVKKYGDWFTYEKTPRALIFKRDHSKVVDMNSMMRLMRYNDFTHDPLSA CNCTPPYSAENAIAARCDLNPKDGKYPFDALGHRGHGATDVKITSYSMFKNFQFLAVSGPTRDQVAPFQWSKSDLKDTIRHAGHPDLWVFDPVQF >PLBD2_alvPom Alvinella pompejana (annelid) frag YCRIKVWILGLFSFFISISTAVDVYVKWDTVSNNFQISEKRLSDPVAWATFGDEIAKTGWSYLEVYTNGAYDDSKQAYAAGLAEGYITQDLITKHWYNTY KGFCTKPLNANCKKLQAFIDKNLKWMASKIATEAHKDSYWHQVQLFLEQQAGLLDGYSGQPQFPSNYSMDVTGIALLQLSGDIKDLQTALGIASANKLWFGTKFGIRGHK* >PLBD2_schJap Schistosoma japonicum (trematode) FN318739 MYFTVVFTIVISFIEIHTQHPSKALILANSHKPYLYFDYVSDDNILQNPHIIAMAVFKEEINTTGWSSLTVSTSSDFPDYLQAYWAGFLETNLTFSLTAS QWANTVRDMCPLPLSKDCQALRKYLSENMAYMLNEAYKNDKHSSFWYHVALQLWQLKGMSDAFDKRFIKRADLLNRNYLDELVDNVMGIYLLQLNGDLGD LVSALSLPTLKEGCNQNGHPFIASSSCSALIKVVDSNVYLSHVTWSPYSIMLRVLKHYNFPWKIVNNTGSQKIPGFAITFSSYPTYTSSVDDFYITSANL TITETTNNVYNKTLWEIVRNGSKNAVLTFMRGMVASRLAKTGEEWITYFKYNNSGTYNNQWMIFDAKQWPKNKGSLLIAEQLPGIVSSLDVTKILKINGY WASYNLPFIGDIYTLSGTEEMAKMFGDWYVHNKTARAKIFRRDHHKVVDFPSMLSLMRYNDFMNDPLSTCPCKPPYTSNSAISARDELNDPKGQYPIPSW SYRLHGGTDAKIVDLSMINQLNMIAISGPTYDDLPPFRWSLIKDVKKPLMHPDKWQFPPVITDFIDYPKANSNISARFLSFESLF* >PLBD2_triAdh Trichoplax adhaerens (trichoplax) XM_002107718 introns largely conserved 0 MAQCGKFLIYFSIFIITLATLCSCQSGSVIYKDGLYTFSKGINKRAASYGTFTDKIASSG 2 1 WTYLDVHTNPQDDDFITAYAAGYVEGILTAKY IYMHWKNTVGDYCKQKSIYCQKLKSFIMKNNQWMATQIKHRPHSIYWYH 0 0 INLTLIQQKGLRDGYHKAMPHKPIDEFSFL 2 1 LIELSGDLESLETALKDEDTHHVLGSGSCSAFIKVLPDNRDLYFAHDTWTGYQTMLRIYKYYELNFSMLPKTN 1 2 VTVPGTRISFSSYPGTILSGDDYYLIGSGL 0 0 ATMETTNGNSNEKLWKYVTPSSVLEWIRTIIANRLTSSGNDWVKIFSKYNSGT 2 1 YNNQ 00 WMILDYKLFAPKRPLNPNTLWVLEQIP 2 1 GKIESADVTNVLKKQGYWASYNVP 2 1 YFSSIFNMSGNQEQAKKYGNWFTHDKCPRALIFKRDQHKVNSMESLMKLMR 2 1 YNDFKHDPLSRCNCTPPYSAENAISARSDLNPADGKYNIGALGHRCHGGTDSK STNYTMFHSGLKSYAIAGPTHEQQPPFRWSTAKFNMTKPLGHPDLFNFTRQLVSWD* 0 >PLBD2_monBre Monosiga brevicollis (choanoflagellate) introns all novel 0 MWSCGAAAAAVVAVVVLASPATATVARFVEQTDVQTTYASVFYVESDDSYVVKTENHPWDGDFEKDE 0 0 AVRIKYTPGYLVAGWDQLHVKSNSAMDDATVAYAAGYGEAQLTAEMIYNYAYNNGYDTFTPNDKLADYLAKNQAFMAASIASNRSDANGYWYHVDLILRQLQGVCDGYNSSD FAKSFPLPCESMLAINLMGDMEDLSDALASSDEWYTEDRFFRATHCSALVKLVGGASSPSDIYISQDTWSSLNSMTRIMKRYDLNFLQ 2 1 AKGADDRIAGSSIVFSSYPGSLYSGDDFYLTSAGMAVIETTIGNSNPELYQYIVPDTVLEWIRNIMANRLASNSQTWYEVYRQFNSGT 1 2 YNNMNMILDYKQFKPQEALQDELLTIVEQIP GTVTKTDVTGYLRNMTYWGS 1 2 YNVAFDQNIRELSGANQAEQLYGPW 2 1 FSYWNTSRALIFAREQKNVSSLEDLKRLMRLNQFKTDPL 2 1 YRGWTNCTPAYTAENVIATRGDLNDP 0 0 NGIYSLSSFGLRNHVATDSKISTFSTYDSNNLNVWAIS 2 1 GPTNGPPPNQPVFNWSTSYYKDTRHRGMPEAFDFDWVNFNWPF* 0 >PLBDa_dicDis Dictyostelium discoideum (slime_mold) AAFI02000019 AF411829 introns both novel 0 MRVIRSLLLLTIAIIGSVLSQSSIDDGYTVFYSQPDNYYVKPGTFSNGVAQAIFSNEMMTTGWSFMSISSSEGLYPNDIIAAGAGYLEGYISQEMIYQNWMNMYNNEYHNVIGSD VENWIQENLQYLQTMIDSAPSNDLYWQNVETVLTQITYMQRGYNQSVIDNGVDASQSLGITEFFLMNMDGDMIDLGPALNLTNGKQVTSPATATSPKQAFKEFMRRTGHCSALIKMTDDLSDLFSGHTTW 2 1 SSYYEMVRMFKVYNLKYLFNGQPPASKVTMFSGYPGTLSSIDDFYLLDTKIVVIETTNGLMNNNLYHLITSESVLSWIRVIVANRLATGGESWCQTFSLYNSGTYNNQ 0 0 WIIVDYNKFIKGYGALDGTLYILEQVPDYVEYGDQTAILRTGYWPSFNIPFYENIYGLTGFNETYAQFGNWFSYQASPRSMIFKRDANNIHSLTQFQAMLRYNNWQNDPFSQGNAGN QISSRFDLVTADDPNNQYLDPDAFGGIDSKVVSADMVAALLVNAQSGPSHDNETPFTWNSQWNQKYTYAGQPTTWNFDWMTMSLQSMKPASPSSDSSSDSTTFN* 0 >PLBDb_dicDis Dictyostelium discoideum (slime_mold) XM_640726 2 exons 0 MNKLKSNFILNIVILFTILIFNINFINCENQKQQQHQQQQQQQQQQSSSTTTTLPIYSIKFSSETGFTIYSGNDSTSIAQSGFSNEMMTMGW AYLTITTNSQFEDSLQAEAAGYIEGYLTFEMIWQCWYNILVNEYQNQTIPNQVLNWANENIAYMKQQVATNENDPYWINIGLVLTQLSGMVD GYNAANQDPSRQLSFLDFILINMDADLGDISSTFNQSTSFSEISNFKKSMDHIKKTDHCSGLIKLTDDLTELYSA 2 1 HTSWSSYINMLRIFKSYNFKFSSITNIKSKLTLFSGYPATIASLDDFYLLDTKLVVLETTNGLNNNDLYYLIKPESVLTWMRVIIANRLANGGQSWCETFERENSGTYNNQWMI VDYNKFVPGVKVRDGTLFVLEQVPGYIEFADVTNVLRTGYWPSYNIPYFETIFNMSGFNDELTDSSDYEAYEEDARSQIFRRDANKVYSLTDFQAIMRYNNFQNDPLSHGDAANQI SSRFDLNSPDSQDYDAFGGVDSKVTSFSLVNQLLVIAQSGPTHDQEPPFQWSSANWSNIYPSIGMPNLYDFGWVNFTDISYNY* >PLBDc_dicDis Dictyostelium discoideum (slime_mold) XM_632848 MNKIIILISLFLNFLFGYVVCLNENNQKSQNLIIPTYSIKLKSDGEYLVYSGNDTLSIAQASYTNEMMSIGWGYISITTNPKYNDSLQIEAAGYLEGYLSYE MIWQNWNNMMVNQNANNSFGNDIISWAKENILYMNQQIQLNQNDPYWINVNLVLQQLNGLTNGYSDANQNPDRQLSLMDFILLNMNVEIYDIMNSLKNNSSSFYQQPNNNSFDNNQ HCSALIKLTDDLTELYTGHTTWSDYYQMVRMIKSYNFRFSKLVAAKSNTTMFSGYPGVLMSVDDFYMLDSKLVVLETTNGIKDNDSELFKLIKPQSVLTWIRIIVTNRIAHSGKSW CEIFEKENSGTYNNQWMIVDYNKFIKGVRVQDGTLYVFEQLPGYVEYADVTNILRTGYWPSFNVPYFETISNMSGFNYQSSSSDSSSSSGSIAYEQYPRSQIFRRDSNKVYSISDF QAFMRYNDFQNDPLAYGDPGNQISSRFDLITPQNNASAAGGIDSKVTSLELINQFLMIAQSGPTHDQEPPFSWSSENWKNKYPTIGQPDTFDFEWVTFSTTSFGSFPSASNEKNY* >PLBDd_dicDis Dictyostelium discoideum (slime_mold) XM_633485 MIIFKNLLKLLIILLTIKLYFCIEIKREEHLTILNELNENSDVIQYSILPGNNEEYEIVKGIQEDAIVYGYYMSNVEVNGWAYLSLVSNDKYNDSTQSRAFG YLEGYLTKDLIWNSKVNYYKNAFNSSEIPNKLDDWLTENIESIHTFIVNNRKSRYWNQITLVMDQINGMLDGYNEANTNSSETLSLHDFFVLNMFGDLFDLMPALNLDKEYKYFQK DLNDIQDWFKRSQHCSALIKVSSDYSELYSGHTTWSGYYTMLRIFKSYNQQFSSDVSGTLSKRNIFSSYPGALISVDDFYLLGDTRMVVIETTNSLVTNDLYHLIRPTTVLSWMRV IVSNRMSTNGKEWCENFQRYNSGTYNNQWMIVSYNLFVPYNELKDGALYVLEQIPGYIEFSDQTQALRQGWWNSYNIPFYETIYDASGYNNYTANNYSDSTIYYMSYQTCPRAEIF RNFAGYVESLEDFQSLLRYNDFEYDPLSHKLPFYAIASRYDLSKKNPSPFGATDTKVTCNSMIDQNTIVAISGPTTSNGQPIFEWNSKIDFMESTSHLGCPEKYNFPWVSFSDTTFRNL* >PLBDe_dicDis Dictyostelium discoideum (slime_mold) MKLFILLIVIVFLISNSYSLSSSDSSSDSGSDVQYYSLTSQFQVVQGKQIPGSIAWGYFKDEMNKDGWGKLSIETVSTVSDNIAFKAAGYLEGYLTWEYIYK FSGNYFNSFFNTSNIKEIPTETLTFVSDNWEYMMERVNSSSTTDPYWIQIRNAMSQQIGLYEGYNAAAGEDYQKTFIEIYMINLYGDMGDIVTLTTTPNNEFIPMDRKEVEQLMAT TGHCTSIIKLTNNCSDLMSAHTSWADFSVMIRIYKRINIPVASTPYGSETLFSSYPGLLVSIDDFYQIRPSKLHLTETLNTILNQTLYQQINAQSFMYWVRNLVANRLANNGFQWV SIFVENNSGTNNIQFVVLDYKLFTPYSTELQSDLLWIVEQYPGGYQAADVTLTLWEQGYWPSYNRPYFEEVFDILGYPYYVEKFGDLFTYEYNPRANIFRRDHSKLETLQDMMNII DYNQYKTDPFSMGYPGNSINARFDIKGGSLPSGNPIYSWFYHGTHGGIDGKAINYDMVNSFTAVARNGPTVTSDCPPFNWNDWSLISHQYMPQIYNFTWISINI >PLBDf_dicDis Dictyostelium discoideum (slime_mold) XM_633485 MKIINSFVFIFVLLFVFNTNAIKLSEDEKYSIEEKPEWYSIDSNFAVSPGKISDALGWGYYKNEILKDGWGKLYVEMNKFMTPENSSIYYEATGYLEGYLTW STTWNYSQAYFQNYMNGTNESDIPTPLVEFLKINYDWMTSTFSNRDESVYDTQVSNVIYQFEGFARGYQQAADSDKQLTTLQLLLLQYAGDLEDVAGYLEYEMAQNKTEYINRVKS LKEIETLFAVKGRCSGLVRITPDYGELFISHTTWGSYFTAGYRIFKRIIIPDPTVPGNEILFASYAGVLTSDDDFFMIPSTEMVIIETTNDILNTSLYQYVTPNSLLYFVRSIIAN RLSNTAQEWTNNFIQYNSGTYSNQWMIVDYKLFTPYQPLQPNTFWIIEQLPGGFMSADMTEVLALGNWPSFNRPFFPEIYDAMGYSYYEKLYGDIISYDLNPRSKMFRRDVPNVMS LDGMQQIMTQNNYKSDPFSGGFPGNAIAARYDLGGGPAEPLSWSFIGLHGAIDSKITSYSLLQQNQAIAINGMTVTPDCPPFTWNSNWSTVSAHYGSPETFDFDWISITITDK >PLBDg_dicDis Dictyostelium discoideum (slime_mold) MIKSYYLFFIILIFLIFINNFILCENNNNNKYNNNNNYPFINVDKIIYTNSSNSIYLVWSNEQFNLILQSSYNEEFDDIVMIGEYFDKIETTGWGELNITFN SNSATIQISDNEQAYFTGFIESVLTGERINQMYNNFAASEFTNSDHTPSPKLIDFLNTQMEFVRDQVFENNGSSQYWYSTGLIMSQFDGLVNGYQQSPFPQLSEIQLYILTSAGDL ETLVTLFPSSSSSSSSTTNENKNKNIKISSIKPNFKDELTDCSGFIRILPDYSDVYFGHTTWRYYYALLRIYKFINLQFNFQDTPMEYKVSFSSSPGFISSKDDFYITGNKLAIME TTNNIYNESLYQYTIPQSVLVWQRAMIANMIATNSSDWVKIFSEFNSGTYQNQWMVFDYKLFIPNKQQSSSSLPPNTFWIAEQIPGQVKTADLTNILNEQGYWKSYNIPYFESIYN ISGYSEKTDLPPSYQYSYEKCPRSLIFSRNASDVLNFEDMKSLMQFNNYKTDPLSYSSPLNSISSRGDLLTIENGNRSAVAFGGVDSKITSFNQVLTLSCTAISGPSTNGGTLPPF TWSQSPLFQNITHIGVPETFNFDWQEMGPYPN*
Derived PLBD2 sequences
>PLBD2x_droMel Drosophila melanogaster (fruitfly) Q9VRK8 retinal lamina neuron ancestor (lama) PMID: 16077094,8892229 0 MERPEYDGTYCATALWTKQVGFQIENWKQQNDLVNIPTGVGRICYKDSVYENGW 0 0 AQIEVETQRTYPDWVQAYAAGMLEGSLTWRNIYNQWSN 2 1 TISSSCERDESTQKFCGWLRDLLTTNYHRLKRQTEKAENDHYWHQLHLFITQLEGLETGYKRGASRARSDLEEEIPFSD FLLMNAAADIQDLKIYYENYELQNSTEHTEEPRTDQPKNFFLPSATMLTKIVQEEESPQVLQLLFGHSTAGSYSSMLRIQK RYKFHYHFSSKLRSNTVPGVDITFTGYPGILGSTDDFYTIKGRHLHAIVGGVGIKNENLQLWKTVDPKKMVPLVARVMAANRI SQNRQTWASAMSRHPFTGAKQWITVDLNKMKVQDNLYNVLEGDDKHDDAPVVLNEKDRTAIQQRHDQLRDMVWIAEQLPGMMTKK DVTQGFLVPGNTSWLANGVPYFKNVLELSGVNYSEDQQLTVADEEELTSLASVDKYLRTHGFRGDLLGSQESIAYGNIDLKLFS YNARLGISDFHAFAGPVFLRFQHTQPRTLEDEGQDGGVPPAASMGDERLSVSIEDADSLAEMELITERRSVRNDMRAIAMRKIGSGP FKWSEMSPVEEGGGHEGHPDEWNFDKVSPKWAW* 0 >PLBD2x_gloMor Glossina morsitans (tsetse) EZ422576 59% PLBD2_droMel V for iMet, C confirmed by ests VYCATAFWTKQFGFLVRLSIEYWKQQNDLLNIPKGAARICYKDSIYENGWAQIEIETQRTSPDWVQAYGAGVIEGSLTWNQIYNQWLNTI MSSCDRDENAQHFCTWLRELLENNFNEIKETAASKSEHDHYWHQINLYFIQLKGLETGYRQGAKRARSDLEGAIPWSDFLLMNSAADIHDLKIYYENYIPNG INRTTDETEPKNFFLPSATMITKIYPTGNASTWQLLFGHSSAGSYTSMLRIQKRYKFHYHIASNQNSNTVPGMDIAFTGYPGILASTDDFYIIKGRQVHAIVAGVGIKNENLELWQ TVNVQKAMPLAARVMAANRLAQSRRSWTKALSRHPFTGSKQWITIDLNKLSPLESLYELDENNLELDEEAASIERQNHLKGLVWIAEQLPGRMHMRDVSALFLETVDNSTTWLASG IPYYKEILEASCVNNEAYITNLTAAESRELNNLEAIDKFLRTRGFRGDLLDDEQSVAYGNIDIKLFAYNARLGMSDYHAFAGPVFIRLQHVQTRSLPAAMGYRGDLAPASLIRDDR LSVVVDDAAELAELQVITERRIVRDDMRAVAMSQISSSPFKWSSVAEKMATLQHAGHPDEWNFGKVSPKWAW* >PLBD2x_aedAeg Aedes aegypti (mosquito) XM_001658072 MDRPNYSGTYCATAYWARNSGFRVEFWGQREELGEVPPGAVRACFKHSLMESGWSQLELESQPEYPDSIQAFAAGMLEGTLTWNNIYLHWSNTIESECNR DDQSEEFCHWLRRIISTNVETVKKMADMKGKNDHYWYQIGLFFDQLDGLEFGFRKGVRRSRMDYEIPMEDFLLMNSAVDIRDLKAYYMNFLDSESGMHVE PNKGMMLLKLVESVTGKVNILLGHTSDGSYASMLRMMKKYTLNYHFAEESIVERVVPNTNIVFTGYPAVLASLDDFYMLSGKHHRMVVGGIKIKNDNLNL WTKIDLVRSVSLAPRVMACNRLAHSGRVWSKYFARSPSTGAKQWLLVDLTRLNRENELLDDSAEVEEMLLSHAEYDDPKAMRKAQSTDYVEVDFEGIVRK MTSIGKISDGGLFWVVDQLPGRLHAEDMTEKIVRDGYWLGTGIPTFEELADIGQVKTNGTDSQRHSIQEKILNNITDLEGLAKFIRQSAYRGDLDQQHPT AFGNIDMKLFVETNASNGTGIFQAYSGPLYDPLTDGQAVKRSVENDGEVEAEAIPVAAAKKKRVKPFDWDSAKKLDVPHQGQPTLWNFRRQSPKWAWI* >PLBD2x_anoGam Anopheles gambiaei (mosquito) XM_316013 MLKVVGASWHKTRIGSYILIGALMLAVAALFLADMERPSYNGTYCATTYWARNSGYRLEFWGQRNDLDQVPKGAVRACFRDSILTNGWSQLELESQSTYT DTVQAYAAGIMEGALTWHNIYMHWSNTIDAVCSKDEESEEFCDWLRGLITTNVDTVKKMADMKGKHDYYWYQIALFYDQLDGLEIGFRKGVKRSRMDYEI PKQDFLLMNAAVDIRDLKIYYTNFLMENSGLKPEPAKGIMLLKLLQQPNGQSKVLLGHTSDGSYSSMLRMVKKYTLRYHLSAETARTVVSAQAAEDEDRA VVPGTNIVFTGYPGVLASLDDFYVISGRRQHRLVAAGVKMENEHVDLWRKIDLVRSVSLAPRVMAANRLGHSGRSWARYFARNPSTGVKQWLVIDMSRFV GSNETEVELLATSPASVAVDQSVEDNDIAAEGGQDVVADGEHQRIVTKRMEDYVDDDFKEHLSAIRQVRMVTIPKAPEPVAGEELVVEPSVGHTFGSGMF WVVDQLPGRLHAEDITDKIVSDGYWLTNGVPYFKEALDIGQATANATNTTAPSSTMEKILQNITDLETLARFFRQSAYRGDLDQDEPAAFGNIDLKMYTE TADGTVHFRAYSGPLYDPARSTPGQAAKRSIVKSEQLHQSTATDQKSAPASHGDSATVASHPFDWSRILPFEVRHQGHPDVWDFEHITPEWAWI* >PLBD2x_apiMel Apis mellifera (bee) HP560493 MLKVVGASWLQTRISTYILVAVALLGIGAIILGEFGHVEQDGIYSATVLWNRKGGYRIDFWGQGNDLTAVPLHAARAYYKTGIFEYGWSYIEIETSSKYP DTVQAYAAGLLEGSLTWQLIHHHWYNTIRLECEAKPIECRKLMRYLRDNTAVNRKRAELKESTDSFWHMVRLFYAQLDGLEAGWKFAVRRSRVSASLESE DFLWLALASDLCGFQQIFNISDSLVSSMIYFKSLPRENSGPLIAISHHTSASYTQMLRLLKKYTFGYHILPVTKTAALIPSRSIVMSSYPGALSSRDEFY LINGENRELVVTGTSLLMTNRTEWSFLYPDDYVMLTVRLMAANRLATNSQSWFNTLSYQNDGASALQWIIFEPRSMTLLLVEQLPSITVPINYTEEFKKI GFISYIGISNFKSTNNIVQPAKKNLNLWKTRLTRLQKNITTFEQFRNMMRGCSQEGCSAVNQKTDYLQELMFRGDLEDVPIPYGIIDTKILTVDIDGFKG FEAISGPSQSRTPFKWSQSFPNISHVGHPDTFNFESVTPKWVWI* >PLBD2x_pedHum Pediculus humanus (louse) XM_002424573 MLKVVGASWVKIRMSTWIIGIVLVFGIIVVILGQMGESEEDGNYSATAYWNEETGFEIKFWGQGLEKDKIPKGVARVYFQPHIDTTGWAIIDVETQSEYP DWVQAYAAGMLEGSLSWQLIYWHWKNTVQSVCQNQTLFCKYIKKYLEENFKSVKELYENTDDPYWHQAYLFYIQLEGMQQGFEHGVKRKGLIIEEIEFID FLWMNAASDIIDLEYKFYDSKPYSTTTTNSSSFSANNNNNSNLLLKKLIQGRSDNVESMEKLSFVGCGVTLGSALLNSFQNMRNYFSPIIPLDQTKLLVA GTPITVFNKNLWNLVSPLKQLLSGVRVVVANRLSNNASDWGSYLSKYNSGTGNKQWLIIDVDKFKRKINDSSVGERENFFWVAEQLPGHFKIDDLTENLI NSSYWASYGLPFYKDMYEMSGTNYMKENFGDAFSYENSPLAKIYRRDHNKSTSIKSILNLMRSNSFKRDPLSLGNPRCAIGARGDIPNKVSNSFPIGVID SKIISLEKKLENVTSNDTTTNNNNNIIPFNNNVSNDELIIFKAVAGPAHSLHKNSENLKGNDEIRKEKNETVEVEGIGVGGGDDDTLKPFNWQNSVFNEF PHNGHPNNWDFDAISLSP* >PLBD2x_acyPis Acyrthosiphon pisum (aphid) XM_001948052 MLKVVGASWAQTQLSWCLIIAITLLGLLAFYFGGTIRNEYDGKYAATAFWSKEFGMRIDFCGQNNDPLTVRKGVARAYYRPDLSENGWAVLEVETQAEYP DYVQAKAAGYLEGSLTWRMIYWHWKNTVENTCIGRRNFCERIRKYLEENAEEIKRMAKRRGESDPFWHQVNMFYTQLKALEDGWRFGVKRSRQDIDIPSV DFLWMNIMPDLKNFEQKWNASKDFNPDKPPLSATLVKIISTNPIDFVLAQSASGYYGSMLRIQKRYNFGFHETQSDDSALVNGKIVEFTSYPGSIYSQDD FYKVIKKGSKTETTVVGTELQNNNRQLWEKIMKTDQVLLGARIMAANRLASNSKKWYEVFSRNNSGTGNKQWLVISTNSTSIAFGVIEQMPGIVSYEELS KTLLSTGYWVSNSNPCLKETVYMSGADGRDAADHPVANILRSGQLNVTDIGTLVDLMRGTEMSLIGRTDLLGVKTNAMFRRFANQSFVEQLTAANRLQSA DATAPLKDDRTTIVTNRLPSSAASDDKLDVDKCFTGTVDLKVTSANMNGYYAASGPPFTMDRSEVEPFRWSESPIRHLPHYGHPDVWDFEVEGVVWVWK*
Trimmed alignable sequences
It is useful for various purposes to trim protein sequences to their conserved core and matured length. Here, since compilations of signal peptides have been previously considered, they can be discarded, greatly simplifying acquisition of reliably alignable sequence.
Note too that exon boundaries mostly differ between the two paralogs and so limit the matched pairs from a fixed species that be collected from genomic contigs using tblastn from a necessarily diverged query. However the final exons do match and are already provided above from a very large and phylogenetically disperse species set. In some cases, there is a benefit to removing single-species insertions that introduce a gap in the rest of the alignment because then other gapping can be done more accurately.
Finally, as the xray structural determination did not extend over the whole protein, flanking sequence needs be included only to the extent it is strongly conserved. Thus the boundaries of the protein are established, allowing comparative genomics on the alignable core.