Phospholipases PLBD1 and PLBD2
Introduction
(to be continued shortly)
Conservation at critical sites
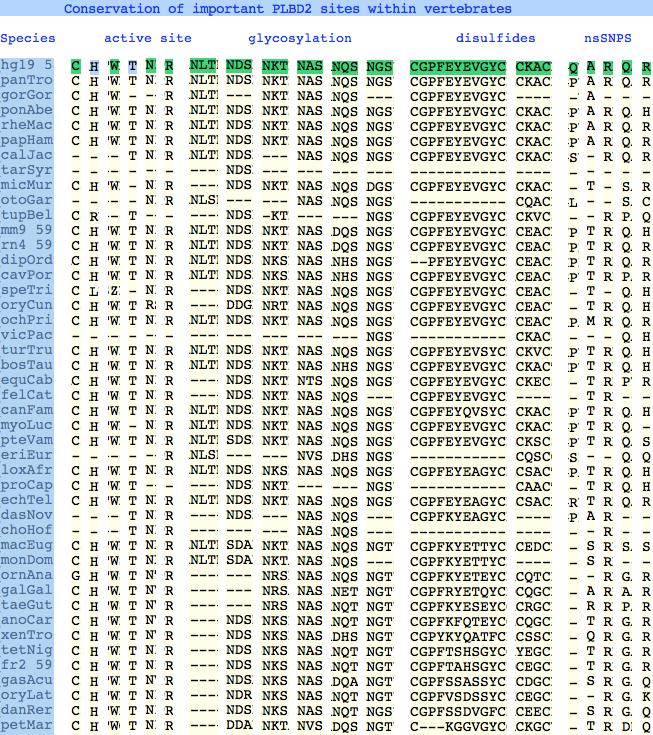
The six residues of PLBD2 associated with the active site are completely conserved within vertebrates to within genomic sequencing error. These same six residues are also completely conserved within PLBD1. Indeed 3 of the residues are conserved in the broader class of NTN hydrolases.
This is perhaps unsurprising since the active site was established a billion years earlier in the bacterial ancestor. However if PLBD2 and PLBD1 have different substrates, this establishes that these six residues are insufficient to distinguish the two active sites. Note H266 and T330 do not contribute their side chain, leaving them and W269 to separate phospholipases from the other NTN hydrolases.
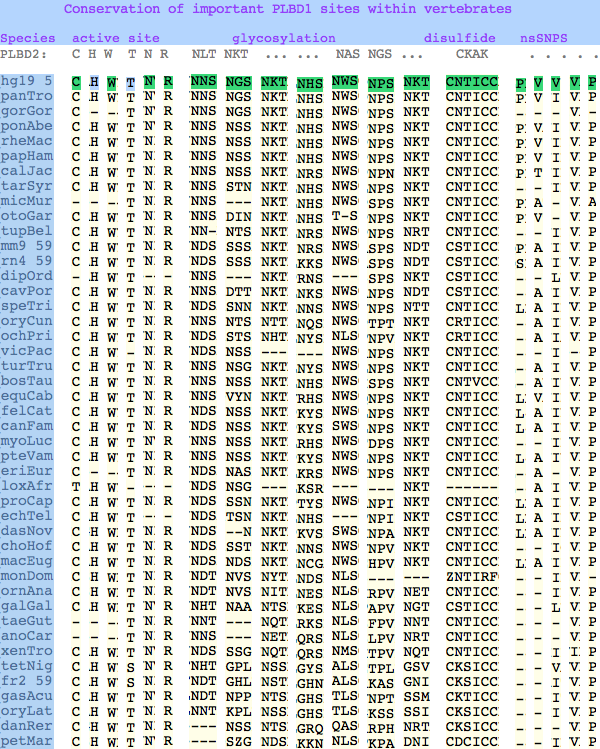
The glycosylation sites are surprisingly conserved both within and between PLBD2 and PLBD1. Some of the motifs may be either recently acquired within later vertebrates or spurious glycosylation motifs with N and D both acceptable (or similar small amino acids). Glycosylation is important in targeting of lysosomal proteins, more so than in generic endoplasmic reticulum proteins where motifs are often poorly conserved (as in sulfatases).
PLBD2 has two established disulfides. Strict sequence conservation of these throughout vertebrates (indeed, throughout metazoa) suggests both play an important role in protein structure and stability.
In PLBD1 however, the first disulfide is not a possibility and while an opportunity exists for a disulfide homologous to the the second disulfide of PLBD2, indels cloud the alignment and spacing would have to be different. There is additionally ambiguity given C...CC as to the cysteines involved. Indeed a second distal disulfide may occur utilizing C...CC.............C which has no counterpart in PLBD2. While cysteines can be conserved for many reasons other than disulfide (as in the nucleophile cysteine here), suitably proximity and side chain orientation in the SwissModel of PLBD1 would argue for disulfide. Comparative genomics suggests that C2 and C4 may form an ancient disulfide whereas C1 and C3 might represent a deuterostome innovation.
homSap CNTICCREDLNSPNPSPGGC human PLBD1 braFlo CSAICCRKDLAKVGAKPDGC Branchiostoma floridae strPur SKSICMRGDLM-TSPMPNGC Strongylocentrotus purpuratus XM_001192029 nemVec MNAICSRGDLIADGPRASGC Nematostella vectensis XM_001638165 monBre YNAICSRGDLESDSPSPGGC Monosiga brevicollis XM_001745398
The known human SNPs of PLBD2 are in some cases quite radical substitutions in terms of both physical qualities of the substituted amino acid and the degree of observed phylogenetic conservation at that site. These likely result in unstable and/or inactive enzyme. Both enzymes are autosomal so compensation might occur in the recessive state, or alternately, PLBD2 and PLBD1 could fill for each other to some extent. In either case, lysosomal storage disease might not be clinically observable.
Here Q54P may actually be a mutation in the reference sequence individual (with the SNP representing wildtype) as proline is quite well conserved throughout mammals. In A204V, valine is quite a bulky substituent for a site normally restricted to small amino acids; R354C is definitely a serious mutation, no doubt attributable to a CpG hotspot; Q521K appears milder as does R524C.
The known human SNPs of PLBD1 can be analyzed similarlly. P26Q and V30L may be inconsequential as they occur in the rather unconstrained primary sequence of the N-terminus; V265I occurs at an ILV reduced alphabet; V377A and P534A are much more serious despite the aliphatic nature of alanine and likely give rise to dysfunctional protein.
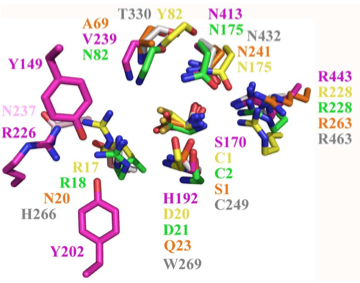
Structural superposition of active sites from five NTN hydrolases showing conserved side chains (*) and relevant main chains (....) (adapted from Fig 6 of Lakomek et al. BMC Struct Biol.2009;9:56:) * (*) * * PLBD2 phospholipase B-like gray 3FGR C249 H266 W269 T330 N432 R463 mouse numbering PLBD1 phospholipase B-like .... pred C228 H245 W248 T303 N402 R433 SwissModel Cephalosporin acylase pink 1OQZ S170 .... H192 .... N413 R443 Conjugated bile acid hydrolase green 2BJF C2 .... D21 .... N175 R228 Penicillin V acylase yellow 3PVA C1 .... D20 .... N175 R228 Penicillin G acylase orange 1K5S S1 .... Q23 .... N241 R263 Human SNPs resulting in amino acid substitions: PLBD2: PLBD1: Q54P rs7965471 P26Q rs1141509 A204V rs12231990 V30L rs12296104 R354C rs56935204 V265I rs7957558 Q521K rs17852787 V377A rs2287541 R524C rs12425042 P534A rs1600
Intron evolution
PLBD1 and PLBD2, being full length paralogs, clearly indicate an early gene duplication and subsequent divergence to the current low percent identity. Segmental duplications preserve any introns present at the time of the event and these generally persist in both position and phase into living species.
However PLBD1 and PLBD2 -- despite having similar numbers of introns -- exhibit very little in common in terms of location as the diagram below shows. One possibility is that a second copy arose as a retroprocessed gene (a mechanism erasing existing introns) and was subsequently intronated at random positions. This is unlikely here given that 10-11 relatively rare events would be needed.
The remaining possiblity is that the gene duplication took place prior to the main era in early eukaryotes during which the bulk of introns were established. This fits the current state of high divergence despite fairly slow rates of evolution during metazoan times.
The last five amino acids of each PLBD1 exon are colored below. Then using an alignment of PLBD1 to PLBD2, the colors are mapped to the homologous five residues within PLBD2. There they fall on the ends of exons only when these correspond to those of PLBD1. The outcome here -- despite uncertainties in alignment gapping -- shows intron positions do not correspond with the exception of the terminal intron (which also is phase 0).
While this merely compares human PLBD1 and PLBD2, the collected reference sequences (intronated against their respective genome assemblies) confirm that introns in both genes are deeply conserved.
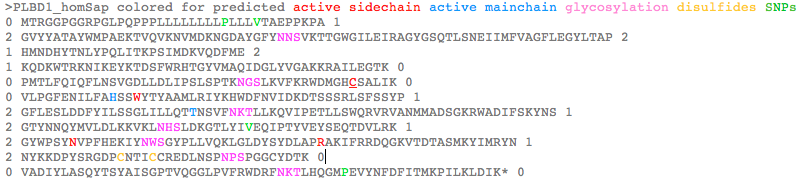
>PLBD1_homSap Homo sapiens (human) first and last introns are not mappable 0 MTRGGPGGRPGLPQPPPLLLLLLLLPLLLVTAEPPKPA 1 2 GVYYATAYWMPAEKTVQVKNVMDKNGDAYGFYNNSVKTTGWGILEIRAGYGSQTLSNEIIMFVAGFLEGYLTAP 2 1 HMNDHYTNLYPQLITKPSIMDKVQDFME 2 1 KQDKWTRKNIKEYKTDSFWRHTGYVMAQIDGLYVGAKKRAILEGTK 0 0 PMTLFQIQFLNSVGDLLDLIPSLSPTKNGSLKVFKRWDMGHCSALIK 0 0 VLPGFENILFAHSSWYTYAAMLRIYKHWDFNVIDKDTSSSRLSFSSYP 1 2 GFLESLDDFYILSSGLILLQTTNSVFNKTLLKQVIPETLLSWQRVRVANMMADSGKRWADIFSKYNS 1 2 GTYNNQYMVLDLKKVKLNHSLDKGTLYIVEQIPTYVEYSEQTDVLRK 1 2 GYWPSYNVPFHEKIYNWSGYPLLVQKLGLDYSYDLAPRAKIFRRDQGKVTDTASMKYIMRYN 1 2 NYKKDPYSRGDPCNTICCREDLNSPNPSPGGCYDTK 0 0 VADIYLASQYTSYAISGPTVQGGLPVFRWDRFNKTLHQGMPEVYNFDFITMKPILKLDIK* 0 >PLBD2_homSap Homo sapiens (human) 0 MVGQMYCYPGSHLARALTRALALALVLALLVGPFLSGLAGAIPAPGGRWARDGQVPPASRSRSVLLDVSAGQLLMVDGRHPDAVAWANLTNAIRETG 2 1 WAFLELGTSGQYNDSLQAYAAGVVEAAVSEE 0 0 LIYMHWMNTVVNYCGPFEYEVGYCERLKSFLEANLEWMQEEMESNPDSPYWHQ 0 0 VRLTLLQLKGLEDSYEGRVSFPAGKFTIKPLGFL 2 1 LLQLSGDLEDLELALNKTKIKPSLGSGSCSALIKLLPGQSDLLVAHNTWNNYQHMLRVIKKYWLQFREGPW 1 2 GDYPLVPGNKLVFSSYPGTIFSCDDFYILGSGL 0 0 VTLETTIGNKNPALWKYVRPRGCVLEWVRNIVANRLASDGATWADIFKRFNSGT 2 1 YNNQWMIVDYKAFIPGGPSPGSRVLTILEQIP 2 1 GMVVVADKTSELYQKTYWASYNIP 2 1 SFETVFNASGLQALVAQYGDWFSYDGSPRAQIFRRNQSLVQDMDSMVRLMR 2 1 YNDFLHDPLSLCKACNPQPNGENAISARSDLNPANGSYPFQALRQRSHGGIDVK 0 0 VTSMSLARILSLLAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFAPVKVSWD* 0
PLBD1 reference sequences
>PLBD1_homSap Homo sapiens (human) FLJ22662 PMID: 19019078,20093120 0 MTRGGPGGRPGLPQPPPLLLLLLLLPLLLVTAEPPKPA 1 2 GVYYATAYWMPAEKTVQVKNVMDKNGDAYGFYNNSVKTTGWGILEIRAGYGSQTLSNEIIMFVAGFLEGYLTAP 2 1 HMNDHYTNLYPQLITKPSIMDKVQDFME 2 1 KQDKWTRKNIKEYKTDSFWRHTGYVMAQIDGLYVGAKKRAILEGTK 0 0 PMTLFQIQFLNSVGDLLDLIPSLSPTKNGSLKVFKRWDMGHCSALIK 0 0 VLPGFENILFAHSSWYTYAAMLRIYKHWDFNVIDKDTSSSRLSFSSYP 1 2 GFLESLDDFYILSSGLILLQTTNSVFNKTLLKQVIPETLLSWQRVRVANMMADSGKRWADIFSKYNS 1 2 GTYNNQYMVLDLKKVKLNHSLDKGTLYIVEQIPTYVEYSEQTDVLRK 1 2 GYWPSYNVPFHEKIYNWSGYPLLVQKLGLDYSYDLAPRAKIFRRDQGKVTDTASMKYIMRYN 1 2 NYKKDPYSRGDPCNTICCREDLNSPNPSPGGCYDTK 0 0 VADIYLASQYTSYAISGPTVQGGLPVFRWDRFNKTLHQGMPEVYNFDFITMKPILKLDIK* 0 >PLBD1_braFlo Branchiostoma floridae (lancelet) XM_002595538 0 MEGRACRSCRLHHLSAVFLLFLVTIAA 1 2 GAEIQATAYLQAQGKVQVKLGVLDKQNGDAVATYDDR 2 1 LTENGWGVLNVVSGFGPKKLSDNDIMYLAGYLEGVLTQE 2 1 RIYQHYLNLYGIFFMGKSEDLVGK 0 0 VKKFYTAQDTWVRAQVKQSTDPVMKHLSYILSQYDGLVKGYNDN 0 0 LFPHVSFFQKLDIFAFQLLNGNGDTFDIIPAVNPSSRPDFSNMSRVEIDDWVSAHSHCSALVK 0 0 VLGAYENVYMSHSSWFNYAATMRIYKHYNFNIANPATATRKMSFSSYP 1 2 GYLESLDDFYLMDSGLVMLQTTNNVFNGTLYDLVKPESILAWQRVRTANMLARNGDQWGAIMNVHNS 1 2 GTYNNQYMIIDLNLIELGKTIHDGALYVVEQIPGLVMSADQTDILRA 1 2 GYWPSYNIPFYEKVYNLSGYPEFAKSQGLDYTYQLAPRAKIFRRDAGKVKDMESMKAIMRYN 1 2 DYLHDPYSKGNPCSAICCRKDLAKVGAKPDGCYDTK 0 0 VSDYYLARNLTSFAINGPTLGTGLEPFSWSDKFKISHIGLPKVYNFSFVTMTPAEL* 0 >PLBD1_strPur Strongylocentrotus purpuratus (urchin) XM_001192029 MANKFRMFKILTAFLVLVLVNLSTGELLQGTVYKQEDGTFTVSS GIIDKQGVAYGSYNNTLFQTGWGELHLFAGYSTADNVALSDADRMYAAGILEGALTAK QISQTLRNINISQTLQNINVTFFSAESDPEIWRRVADFFETQDAWMKGMIIERADEDP FWEGVGLVLAQFEGLIKGYEMSQFSNASTSNGFLAMQVLNSCGDLLDLKSAVMPSLIP DWDKLTKKEFLKFIRTSGHCSALVKICAALVKVGRFAPPFQSLLYSISSYFKSQAILK LNSPSCQLFGIEGFLESLDDFYIMSSGLSMLQTTNNIFNKTLYKYVKPQSLLAWQRVR VANMMARSGKDWARIVARYNSGTYNNQYMVIDRTKIKPNVAILDDALWVVEQVPTLVA SGDQTNILRAGYWPSYNVPFYEEIYNISGYPEYAYKGGADISYQLAPRAKIFRRDQGN VVDMESFKKIMRFNDYKNDPYSEGDPSKSICMRGDLMTSPMPNGCYDTKVTNLAMAAK QTSFVINGPTRGDGSLPPFKWVAPFTGWSHVGLPTVYDFNFVEMCPKEL* >PLBD1_nemVec Nematostella vectensis (anemone) XM_001638165 MTLIRNSVMITVTFVLILFVFGCHGSQKSATVYYNRGQGYSLKF GVVDKLMGVAYGTFEDSLNTTGWYELNIVSGTGIEPYNDDVIMHAAGYLEGALTASQI NDNYANLYGVFFKSEDDPMVAKVEKFFIEQDIWMRKMIALKSSNSSFWRQMGNIIAQF DGLVEGYQKYPATDKALGVFAFQMLNGVGDLLDLTKALMPERMADWDHMTEKEILEKV AMDGHCSALIKVLPAYENVFASHVSWFTYSAMLRVYKHYHLNLKDETTAAQRMSFSSY PGFLESLDDFYIMDSKLVMLQTTNNVFNKSLYEQVVPESLFSWQRVRLANLVASSGRQ WADIVGQYNSGTYNNQYMVLDLKLIQLNNTIQDNALWVVEQIPTLVASGDQTAILRAG YWPSYNVPFYELVYNLSGYPDFVARHGVQFSHELAPRAKIFRRDQSMVHDLDSMKHIM RYNDFQHDPYSQGNPMNAICSRGDLIADGPRASGCYDGKVTDFTMAQSLISHAINGPT HEQQVPFHWSQYQFKNKHEGQPDLFNFDFVEMKPKF* >PLBD1_monBre Monosiga brevicollis (choanoflagellate) XM_001745398 MSSLNNGIPEPLLKFLAAQFNWTRSQVAANQDDVFWQQVGLIMA QYDGLRAGYGANVYDKHVLPEFAFQLLNGNGDFFDIIPKAVDVTKMSSREFHDWRMRN GRCSALIKLTGDFSDLFMSHSAWYIYQAMNRIYKHCASYNFQATITHAKKISFSSYPG YLESLDDFYLMSSGLVMLQTTNNVFNTDLQQYIQPESLQSWIRIRTATALAQTSEDWA ELAGRHNSGTYNNQYMVMDLNKFTPGQPLLDGTLYVAEQIPGTWEYADVTKMLSLGYW PSYNVPFFEKIYNLSGYPAVVKQHGTDDSYELAPRAKIFRRDQTTVVDLDSFKAIMRY NDYKNDPYAKGDPYNAICSRGDLESDSPSPGGCYDTKVTTYSMALKLQSQVINGPTTS HGLPPFSWSQFPNASHLGMPEVFNFTFETMDAGW*
PLBD2 reference sequences
>PLBD2_homSap Homo sapiens (human) PMID: 19706171,19237744,17007843 0 MVGQMYCYPGSHLARALTRALALALVLALLVGPFLSGLAGAIPAPGGRWARDGQVPPASRSRSVLLDVSAGQLLMVDGRHPDAVAWANLTNAIRETG 2 1 WAFLELGTSGQYNDSLQAYAAGVVEAAVSEE 0 0 LIYMHWMNTVVNYCGPFEYEVGYCERLKSFLEANLEWMQEEMESNPDSPYWHQ 0 0 VRLTLLQLKGLEDSYEGRVSFPAGKFTIKPLGFL 2 1 LLQLSGDLEDLELALNKTKIKPSLGSGSCSALIKLLPGQSDLLVAHNTWNNYQHMLRVIKKYWLQFREGPW 1 2 GDYPLVPGNKLVFSSYPGTIFSCDDFYILGSGL 0 0 VTLETTIGNKNPALWKYVRPRGCVLEWVRNIVANRLASDGATWADIFKRFNSGT 2 1 YNNQWMIVDYKAFIPGGPSPGSRVLTILEQIP 2 1 GMVVVADKTSELYQKTYWASYNIP 2 1 SFETVFNASGLQALVAQYGDWFSYDGSPRAQIFRRNQSLVQDMDSMVRLMR 2 1 YNDFLHDPLSLCKACNPQPNGENAISARSDLNPANGSYPFQALRQRSHGGIDVK 0 0 VTSMSLARILSLLAASGPTWDQVPPFQWSTSPFSGLLHMGQPDLWKFAPVKVSWD* 0 >PLBD2_braFlo Branchiostoma floridae (lancelet) XM_002612057 0 MAACRNIFCGRMLSCLLLFSFVFSAVSDGSKLASVRYDEAAKTYQITDKLDPSAAAWANFTDRISSTG 2 1 WSFLTVTTNEKYDDSVQAYAAGLVEGYLTRD LMYNHWLNTVGAAFCSSRSAFCKNLESFLKTNLAWMQEQIQASGDTDDYWHQ 0 0 VKLTLQQLSGLDDGYNDDPRQPSLDINPFGFL 2 1 IFQIGGDMEDLQEALKDKDSHRVLGSGSCSALVKLLPGNADLLVAHDTWDTFQSMLRIIKKYQFPFKLGGKK 1 2 GEDKIPGHTVSFSSYPGVIYSGDDFYITSASL 0 0 VAQETTIGNSNPALWKYVQPQGQVLEWLRNIVANRLANKAMDWATIFKKYNSGT 2 1 YNNQWMIVDYKTFTPNKDLPEKGLLVVLEQLP 2 1 GMVMMDDVTSVLAKQAYWPSYNSP 2 1 YFEKIFNTSGLPAMVEKYGDWFSYEHTPRANIFRRDHGKVTDISSMIKLMR 2 1 YNDFQNDPLSKCDCTPPYSAENAISARSDLNPANGTYPFSALQHRCHGGTDMK 0 0 MTSYSMHESHQMMAVSGPTHDQQQPFQWSTSDYDKQFYHLGHPDLFNFDPIHVIWFDQSDN* 0 >PLBD2_droMel Drosophila melanogaster (fruitfly) U57314 retinal lamina neuron ancestor (lama) PMID: 16077094,8892229 MERPEYDGTYCATALWTKQVGFQIENWKQQNDLVNIPTGVGRICYKDSVYENGWAQIEVETQRTYPDWVQAYAAGMLEGSLTWRNIYNQWSNTISSSCERDESTQKFCGWLRDLLTTNYHRLKRQT EKAENDHYWHQLHLFITQLEGLETGYKRGASRARSDLEEEIPFSDFLLMNAAADIQDLKIYYENYELQNSTEHTEEPRTDQPKNFFLPSATMLTKIVQEEESPQVLQLLFGHSTAGSYSSMLRIQK RYKFHYHFSSKLRSNTVPGVDITFTGYPGILGSTDDFYTIKGRHLHAIVGGVGIKNENLQLWKTVDPKKMVPLVARVMAANRISQNRQTWASAMSRHPFTGAKQWITVDLNKMKVQDNLYNVLEGD DKHDDAPVVLNEKDRTAIQQRHDQLRDMVWIAEQLPGMMTKKDVTQGFLVPGNTSWLANGVPYFKNVLELSGVNYSEDQQLTVADEEELTSLASVDKYLRTHGFRGDLLGSQESIAYGNIDLKLFS YNARLGISDFHAFAGPVFLRFQHTQPRTLEDEGQDGGVPPAASMGDERLSVSIEDADSLAEMELITERRSVRNDMRAIAMRKIGSGPFKWSEMSPVEEGGGHEGHPDEWNFDKVSPKWAW* >PLBD2_acyPis Acyrthosiphon pisum (aphid) XM_001948827 0 MLSIRCILLSLLFVWALQCSATQKNQTLLAVKTDNNRITIQPKHYSVKDKEIIIGKGKFIDRINSTG 2 1 WAYLEIRTSQKAKDEDQAYGAGYLEGTLTADLIYSYWFNTAKGYCTDRPNVCQQLKDYMTTNKNWIKSKLNESDPYWYQ 0 0 VGLYYKQLDGLYDGYMRGKSPSTPDLTWDDLY 2 1 WLNALDDLGDLSIALYPSDISNRVLGSGSCSALIKLMPDNKDILVSHATWSG 2 1 YETMLRIQKRYSLRFRKSKKSNKLIRGFDMSFSSFPGGIQSGDDFYLISSGLTTMETTIENYNDSLWSNVKPVGQ 0 0 VLEFVRAMVANRLADNPTDWANLFKLHNSGTYNNQWMILNYAAFQPGSPLPPRDVLHVLEQIPGHVMHDDFTGHLINRTYWASYNVPYFPFIFNVSGNYEMEQIYGSW 2 1 FSYSETPRARIFARDHVKIHCDKCMLHLMRSNNYTRDPESRCDCSPPYSAENAISSR 2 1 NDLNPANGTYPIRALGHRSHGATDVKVTSSQLFQQLQFKAIAGPTQGSNNSLGPFCWSKSDFNDKVSHLGHPDCFNFKPVLHQWSL* 0 >PLBD2_triAdh Trichoplax adhaerens (trichoplax) XM_002107718 introns largely conserved 0 MAQCGKFLIYFSIFIITLATLCSCQSGSVIYKDGLYTFSKGINKRAASYGTFTDKIASSG 2 1 WTYLDVHTNPQDDDFITAYAAGYVEGILTAKY IYMHWKNTVGDYCKQKSIYCQKLKSFIMKNNQWMATQIKHRPHSIYWYH 0 0 INLTLIQQKGLRDGYHKAMPHKPIDEFSFL 2 1 LIELSGDLESLETALKDEDTHHVLGSGSCSAFIKVLPDNRDLYFAHDTWTGYQTMLRIYKYYELNFSMLPKTN 1 2 VTVPGTRISFSSYPGTILSGDDYYLIGSGL 0 0 ATMETTNGNSNEKLWKYVTPSSVLEWIRTIIANRLTSSGNDWVKIFSKYNSGT 2 1 YNNQ 00 WMILDYKLFAPKRPLNPNTLWVLEQIP 2 1 GKIESADVTNVLKKQGYWASYNVP 2 1 YFSSIFNMSGNQEQAKKYGNWFTHDKCPRALIFKRDQHKVNSMESLMKLMR 2 1 YNDFKHDPLSRCNCTPPYSAENAISARSDLNPADGKYNIGALGHRCHGGTDSK STNYTMFHSGLKSYAIAGPTHEQQPPFRWSTAKFNMTKPLGHPDLFNFTRQLVSWD* 0 >PLBD2_monBre Monosiga brevicollis (choanoflagellate) introns all novel 0 MWSCGAAAAAVVAVVVLASPATATVARFVEQTDVQTTYASVFYVESDDSYVVKTENHPWDGDFEKDE 0 0 AVRIKYTPGYLVAGWDQLHVKSNSAMDDATVAYAAGYGEAQLTAEMIYNYAYNNGYDTFTPNDKLADYLAKNQAFMAASIASNRSDANGYWYHVDLILRQLQGVCDGYNSSD FAKSFPLPCESMLAINLMGDMEDLSDALASSDEWYTEDRFFRATHCSALVKLVGGASSPSDIYISQDTWSSLNSMTRIMKRYDLNFLQ 2 1 AKGADDRIAGSSIVFSSYPGSLYSGDDFYLTSAGMAVIETTIGNSNPELYQYIVPDTVLEWIRNIMANRLASNSQTWYEVYRQFNSGT 1 2 YNNMNMILDYKQFKPQEALQDELLTIVEQIP GTVTKTDVTGYLRNMTYWGS 1 2 YNVAFDQNIRELSGANQAEQLYGPW 2 1 FSYWNTSRALIFAREQKNVSSLEDLKRLMRLNQFKTDPL 2 1 YRGWTNCTPAYTAENVIATRGDLNDP 0 0 NGIYSLSSFGLRNHVATDSKISTFSTYDSNNLNVWAIS 2 1 GPTNGPPPNQPVFNWSTSYYKDTRHRGMPEAFDFDWVNFNWPF* 0 >PLBD2_dicDis Dictyostelium discoideum (slime_mold) AAFI02000019 AF411829 introns both novel 0 MRVIRSLLLLTIAIIGSVLSQSSIDDGYTVFYSQPDNYYVKPGTFSNGVAQAIFSNEMMTTGWSFMSISSSEGLYPNDIIAAGAGYLEGYISQEMIYQNWMNMYNNEYHNVIGSD VENWIQENLQYLQTMIDSAPSNDLYWQNVETVLTQITYMQRGYNQSVIDNGVDASQSLGITEFFLMNMDGDMIDLGPALNLTNGKQVTSPATATSPKQAFKEFMRRTGHCSALIKMTDDLSDLFSGHTTW 2 1 SSYYEMVRMFKVYNLKYLFNGQPPASKVTMFSGYPGTLSSIDDFYLLDTKIVVIETTNGLMNNNLYHLITSESVLSWIRVIVANRLATGGESWCQTFSLYNSGTYNNQ 0 0 WIIVDYNKFIKGYGALDGTLYILEQVPDYVEYGDQTAILRTGYWPSFNIPFYENIYGLTGFNETYAQFGNWFSYQASPRSMIFKRDANNIHSLTQFQAMLRYNNWQNDPFSQGNAGN QISSRFDLVTADDPNNQYLDPDAFGGIDSKVVSADMVAALLVNAQSGPSHDNETPFTWNSQWNQKYTYAGQPTTWNFDWMTMSLQSMKPASPSSDSSSDSTTFN* 0