Selenoprotein evolution: GPX
Introduction to glutathione peroxidases
The GPX family of proteins can be traced back to great phylogenetic depth. It has experienced various expansions, some narrowly lineage-specific but others retained very broadly. Gene family contractions are more difficult to establish since absence of evidence in 2x genomic assemblies rarely constitutes evidence of absence. In vertebrates, the most notable recent expansion is of GPX3 in placental mammals into the divergently tandem pair GPX7 and GPX8. Marsupials and earlier diverging species lack these latter genes while retaining GPX3 in syntenically established orthologous position (flanking genes TNIP1 and ANXA6).
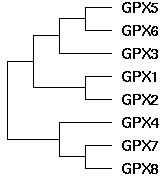
GPX gene tree topology
The topology of the GPX gene tree can be unambiguously established as ((((GPX5,GPX6),GPX3),(GPX1,GPX2)),(GPX4,(GPX7,GPX8))) using blastp clustering, chromosomal locations, rare genomic events such as indels, signature residues, and intron positions and phases. The main technical issue is the placement of GPX4, which can be resolved using the large indels it shares with GPX7 and GPX8 in addition to its sequence clustering. GPX4 is by far the most conserved member of this gene family, suggesting its function has not strayed far from its role in ancestral metazoa, even as its gene duplicates have subfunctionalized and sublocalized.
Alignment of GPX sequences
The alignment shows 103 of the 240 available deuterostome GPX sequences evenly distributed (more or less) over both the eight members of the gene tree and the chordate species tree. The reddish color shows residues conserved at the 90% level; the bluish less-conserved at 70%. The selenocysteine is represented by Z because U is not etained by the alignment tool used here.
Note the three large deletions in GPX4, GPX7, and GPX8 relative to other GPX. The latter have ancestral length judging by the GPX4-classifying sequence in the metazoan outgroup species Monosiga brevicollis. Parsimonously, the deletions occured once in the common ancestral sequence to GPX4 and GPX7/8. It is already present in the tubeworm metazona Ridgeia. Thus the pattern of indels suggests grouping GPX4 with GPX7 and GPX8 to the exclusion of GPX1 and GPX2.
Comparative genomics of GPX selenocysteine residues
It can be seen that most proteinwide cysteines occur sporadically or align just within a particular paralog group, the exceptions being the univeral selenocysteine site (occupied by cysteine in some paralog families and serine anomalously in teleost fish) and a following cysteine 30 residues distal (eg UGKTEVNYTQLVDLHARYAECGLRILAFPC in GPX4_homSap). These two residues very likely form a mixed diselenide (resp. disulfide) with an essential role in the redox reactions carried out by glutathione peroxidases. However some exceptions occur in CPX3 and GPX2.
Other cysteines might be in structural disulfides yet in some GPX an odd number occur, meaning they could not all pair off. This, in conjunction with non-homologous positions, argues against structural disulfides in most cases.
Note GPX7 and GPX8 have classical KDEL endoplasmic reticulum retention signals. This implies an interaction with the protein systems responsible for retrograde translocation and retention. This subcellular localization apparently arose in GPX7 post-amphioxus divergence since the motif is missing there, in sea urchin, and early eukaryotes. GPX8 arose as a subsequent duplication of this ancestral GPX7 and so inherited the motif.
Invertebrate GPX2 and GPX4 sequences
>GPX2_litVan Litopenaeus vannamei (shrimp) Metazoa; Arthropoda; Crustacea ASSAIKSFYDLSAKALSGEMVSFKKFQGKVVLVQNTASLuGTTTRDFHQMNQLKEEFGDK LEVLAFPCNQFGHQENTTEGELLSSLRHVRPGNNFEPKMVMFGKVDVNGSTADPVFKYLK ERLPLPADDSVSFMSDPKCIIWTPVCRSDIAWNFEKFLIGKDGQPFKRFSKKYETILLKDEIANLLKA* >GPX2_eupScp Euprymna scolopes (scallop) cDNA Metazoa; Mollusca 62% KSFFDFSAKTXAGENIDFSRFKGKVVLVENVASLuGTTTRDFTQMNELVAMFADKLVVLG FPCNQFGHQENADGTEIIQSLCYVRPGNGFRPNFSIMEKVSVNGEKTHPIFDFLKDHLPA PSDDPISLMGNPQFITWKPVKRSDVSWNFEKFLVAPDGKPYMRYSRNFLTINLKADIQKLV >GPX2_capSpp Capitella spp (polychaete) cDNA Metazoa; Annelida 64% MQAAKMAKNFYQLSAELLNGKKVQMSAYKGKVVLVENVASLuGTTVRDYHQMNQLMEQFG DRLQILAFPCNQFGHQENTTNDEILKSLKYVRPGNNYTPKFDMFKKVDVNGETAHPVFQF LREQLPTPSDDTVSLMSNPKFLIWSPVCRNDVSWNFEKFLIGPDGEPVKRYSRHFETINIASDIKKLM >GPX2_helRob Helobdella robusta (leech) cDNA Metazoa; Annelida 66% KNFYQLSAELLNGKKVQMSAYKGKVVLVENVASLuGTTVRDYHQMNQLMEQFGDRLQI LAFPCNQFGHQENTTNDEILKSLKYVRPGNNYTPKFDMFKKVDVNGETAHPVFQFLREQL PTPSDDTVSLMSNPKFLIWSPVCRNDVSWNFEKFLIGPDGEPVKRYSRHFETINIASDIKKLM >GPX2_mesGib Mesobuthus gibbosus (scorpion) cDNA Metazoa; Arthropoda MAKSFYDLSAKLLLTGEKINFSQFKGKVVLIENVASLuGTTVRDYTQMNELLNKFGEEL EILGFPCNQFGHQENGNEEEIINSLKYVRPGNGFETKITLFEKIDVNGAGAHQVFQFLRN ELPYPIDDPNSLMTNPQCIIWSPVSRNDVGWNFEKFLITRDGTPFRRYSRNYLTSDIARDIQLLI >GPX4_booMic Boophilus microplus (tick) Metazoa; Arthropoda; Chelicerata TMATADDSWKDASSIYDFSAVDIDGNEVSLDKYKGHVALIVNVASKuGKTNKNYTQLVEL HEKYAESKGLRILAFPCNQFGGQEPGTETDIKKFVEKYNVKFDMFSKVNVNGDKAHPLW KYLKQKQSGFLTDAIKWNFTKFVVDKEGQPVHRYAPTTDPLDIEPD >GPX4_nasVit Nasonia vitripennis (wasp) Metazoa; Arthropoda; Hexapoda; Insecta AEVKFNQDTDWSKAKSIYEFHAKDIRGNDVSLDKYRGHVAIIVNVASQCGLTDTNYKQLQ SLFEKYGKSKGLRILAFPSNEFAGQEPGTSEEILNFVKKYNVSFDMFEKIQVNGDEAHP LYKWLKSQEEGAGTITDGIKWNFTKFLIDKNGKVVSRFAPTTEPFSMEDTITKYL* >GPX4_triCas Tribolium castaneum (red flour beetle) Metazoa; Arthropoda; Hexapoda; Insecta EKPQEAASIYEFTANDIKGEPVSLEKYKGHVCIIVNVASQCGYTKNNYAELVDLFNEYGE SKGLRILAFPCNQFAGQEPGTNEEICQFVSSKNVKFDVFEKINVNGNDAHPLWKYLKHK QGGTLGDFIKWNFTKFIIDKNGQPVERHGPSTNPKDLVKSLEKYW* >GPX4_apiMel Apis mellifera (bee) Metazoa; Arthropoda; Hexapoda; Insecta NWKSASTIYDFHAKDIHGNDVSLNKYRGHVCIIVNVASNCGLTDTNYRELVQLYEKYNEK EGLRILAFPSNEFGGQEPGTSVEILEFVKKYNVTFDLFEKINVNGDNAHPLWKWLKTQA NGFITDDIKWNFSKFIINKEGKVVSRFAPTVDPLQMESELK >GPX4_plaDum Platynereis dumerilii (flatworm) Metazoa; Annelida; Polychaeta CNMATSTDKNAYKKAGSIYEFSAKDIDGNDEVSLEKYKGEVCLIVNVASKuGLTDKNYRQ LQALHEELAGKGLRILAFPCNQFGSQEPGSDEEIKKFATEKYNVQFDMFSKIDVNGSDA HPLWKYLKHKKGGTLGDFIKWNFAKFLVDRQGQPFKRYGNSTAPFDFKKDIE