Selenoprotein evolution: SECIS: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 2,245: | Line 2,245: | ||
>SELI_monDom Monodelphis domestica (opossum) score: 26.46 | >SELI_monDom Monodelphis domestica (opossum) score: 26.46 | ||
uucuacugaa ugcaauu ugugcuuga augaa gagugugucuu aa auccuuuauauggac aggcugcacuu ggag aga auaagca caaccauguu | uucuacugaa ugcaauu ugugcuuga augaa gagugugucuu aa auccuuuauauggac aggcugcacuu ggag aga auaagca caaccauguu | ||
>SELI_macEug Macropus eugenii (wallaby) trace score: 26 | |||
UUUCUACUGA AUGCAGU UUGUGCUUGA AUGAA GAGUGUGUCUU AA ACCCUUUUUAUGGAC AGGCUGCACUU GGAG AGA AUAAGCA CAACCAUGUU | |||
>SELI_ornAna Ornithorhynchus anatinus (platypus) score: 19.25 | >SELI_ornAna Ornithorhynchus anatinus (platypus) score: 19.25 | ||
Revision as of 16:58, 1 July 2008
Introduction to selenocysteine insertion into selenoproteins
The SECIS element in 3' UTR of selenoproteins allows the insertion of selenocysteine for the appropriate TGA codon at time of translation. Auxillary gene products are needed for this to happen, including a selenocysteine-specific translation elongation factor (eEFSec), Sec-tRNA[Ser]Sec, ribosomal protein L30, and a binding protein (SBP2).
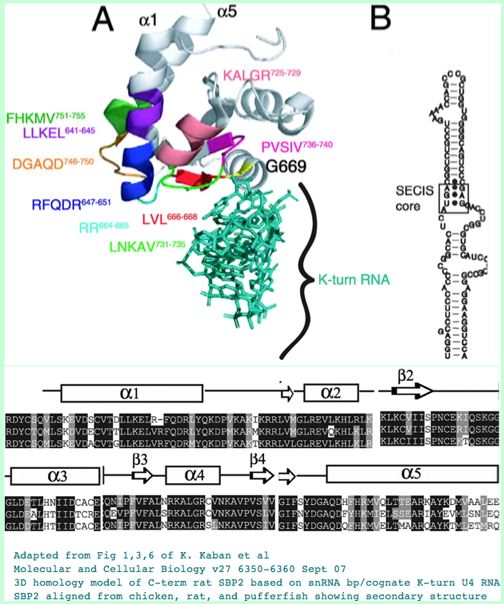
The SBP2 protein has three domains, including a conserved L7Ae RNA binding motif for SECIS RNA binding, a not completely overlapping ribosome binding site and a redox-sensitive cysteine-rich domain. SBP2 shuttles between the nucleus and the cytoplasm via nuclear localization motif SELEQNESSKKNKKKKEKSKSSYEVLPVQE and nuclear export signal motifs, the latter using the CRM1 pathway.
The kink-turn tertiary structure in the SECIS element is at the core of binding recognition. A mutation in that of SEPN or in SB2 lead to rigid spine muscular dystrophy or abnormal thyroid hormone function, the latter through inadequate production of DIO2 needed to activate thyroid hormone T4.
A strong paralog of BP2 was noticed and briely studied earlier. That protein, KIAA0256, also has 17 coding exons with near-identical intron locations and phases and full-length alignability, establishing KIAA0256 and BP2 as resulting from an ancestral gene duplication. The percent identity overall is only 42% but reaches 72% (human to human) in the 3 exons defining the L7Ae kink-turn binding motif.
Since both genes can be recovered from chondrichthyes, lamprey, and amphioxus, the duplication event preceded the emergence of chordates. The roots of the selenoproteome and the insertion mechanism lie much deeper in pre-metazoa so it follows that the parent gene must have assumed the key SECIS recognition roles (as did both descendent genes, at least initially).
We could ask if KIAA0256 still plays a role in SECIS in contemporary organisms. One possibility is that some of the 30-odd selenoproteins preferentially use KIAA0256 over BP2, correlating to the two observed structural classes of SECIS elements (which needs to be revisited in the genomics era). Another possibility is differential use during development or in various cell types. It's worth noting that KIAA0256 is considerably more conserved across its length than BP2, which has only a few exons conserved back to fish) suggesting the latter has a more limited role.
A literature review establishes that DIO, GPX, and SEPP1 were generally taken as proxies for all selenoprotein SECIS elements in terms of establishing a role for BP2. Yet the timing of expansion of these and specialization of the former to thyroid during the period of gene duplication may have tainted their representativeness. Possibly many other selenoproteins utilize KIAA0256 at least to some extent. That would account for the greater conservation of KIAA0256 and the difficulty finding SECIS elements in expected position for certain selenoproteins, ie their elements may be adapted to a different receptor and so have low Cove scores to the extent that tool is based on the BP2 fold paradigm.
Ribosomal protein L30 appears also to bind the SECIS kink-turn and play a role in selenocysteine insertion despite its meagre length of 114 amino acids. Likely many ribosomal proteins, L30 has remarkable sequence conservation, here still 94% at human-lamprey.
L30 has weak homology and secondary structure parallels to the pertinent region of BP2 and KIAA0256. These latter proteins might be viewed as domain composites that have accreted the kink-turn binding domain long ago from this undoubtedly ancient ribosomal protein. This opens a window on the origin of selenocysteine insertion in early eukaryotes.
KIAA0256: the forgotten, misannotated SECIS binding protein
>SECISBP2_homSap Homo sapiens (human) full length 0 MASEGPREPESE 0 0 GIKLSADVKPFVPRFAGLNVAWLESSEACVFPSSAATYYPFVQEPPVTE 2 1 QKIYTEDMAFGASTFPPQYLSSEITLHPYAYSPYTLDSTQNVYSVPGSQYLYNQPSCYRGFQTVKHRNENTCPLPQEMKALFK 0 0 KKTYDEKKTYDQQKFDSERADGTISSEIKSARGSHHLSIYAENSLKS 1 2 DGYHKRTDRKSRIIAKNVSTSKPEFEFTTLDFPELQGAENNMSEIQKQPKWGPVHSVSTDISLLREVVKPAAVLSK 0 0 GEIVVKNNPNESVTANAATNSPSCTR 1 2 ELSWTPMGYVVRQTLSTELSAAPKNVTSMINLKTIASSADPKNVSIPSSEALSSDPSYNKEKHIIHPTQK 0 0 SKASQGSDLEQNEASRKNKKKKEKSTSKYEVLTVQEPPRIE 0 0 DAEEFPNLAVASERRDRIETPKFQSKQQPQ 0 0 DNFKNNVKKSQLPVQLDLGGMLTALEKKQHSQHAKQSSKPVVVS 1 2 VGAVPVLSKECASGERGRRMSQMKTPHNPLDSSAPLMKKGKQREIPKAKKPTSLKK 0 0 IILKERQERKQRLQENAVSPAFTSDDTQDGESGGDDQFPEQAELS 1 2 GPEGMDELISTPSVEDKSEEPPGTELQRDTEASHLAPNHTTFPKIHSRRFRD 2 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLKKLKCVIISPNCEKIQSK 1 2 GGLDDTLHTIIDYACEQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTVAARQAYKTMLENVQQELVGEPRPQAPPSLPTQGPSCPAEDGPPALKEKEEPHY 1 2 IEIWKKHLEAYSGCTLELEESLEASTSQMMNLNL* 0 407–525 domain required for U insertion but not SECIS binding (399–516 in rat) 540 R540Q allele of SBP2 decreases GPX1 and DIO2 650–752 L7Ae motif kink-turn binding motif 676 invariant glycine (669 in rat) >KIAA0256_homSap Homo sapiens (human) length=1101 0 MDRAPTEQ 0 0 NVKLSAEVEPFIPQKKSPDTFMIPMALPNDNGSVSGVEPTPIPSYLITCYPFVQENQSNR 2 1 QFPLYNNDIRWQQPNPNPTGPYFAYPIISAQPPVSTEYTYYQLMPAPCAQVMGFYHPFPTPYSNTFQAANTVNAITTECTERPSQLGQVFPLSSHRSRNSNRGSVVPK 0 0 QQLLQQHIKSKRPLVKNVATQKETNAAGPDSRSKIVLLVDASQQT 1 2 DFPSDIANKSLSETTATMLWKSKGRRRRASHPTAESSSEQGASEADIDSDSGYCSPKHSNNQPAAGALRNPDSGTMN 0 0 HVESSMCA 1 2 GGVNWSNVTCQATQKKPWMEKNQTFSRGGRQTEQRNNSQ 0 0 VGFRCRGHSTSSERRQNLQKRPDNKHLSSSQSHRSDPNSESLYFE 0 0 DEDGFQELNENGNAKDENIQQKLSSKV 0 0 LDDLPENSPINIVQTPIPITTSVPKRAKSQKKKALAAALATAQEYSEISMEQKKLQ 0 0 EALSKAAGKKNKTPVQLDLGDMLAALEKQQQAMKARQITNTRPLSYT 1 2 VVTAASFHTKDSTNRKPLTKSQPCLTSFNSVDIASSKAKKGKEKEIAKLKRPTALKK 0 0 VILKEREEKKGRLTVDHNLLGSEEPTEMHLDFIDDLPQEIVSQE 1 2 DTGLSMPSDTSLSPASQNSPYCMTPVSQGSPASSGIGSPMASSTITKIHSKRFRE 2 1 YCNQVLCKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 0 SLFNKLVELTEEARKAYKDMVAAMEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 2 ETNWRNMVETSDGLEASENEKEVSCKHSTSEKPSKLPFDTPPIGKQPSLVATGSTTSATSAGKSTASDKEEVKPDDLEWASQQSTETGSLDGSCRDLLNSSITSTTSTLVP GMLEEEEDEDEEEEEDYTHEPISVEVQLNSRIESWVSETQRTMETLQLGKTLNGSEEDNVEQSGEEEAEAPEVLEPGMDSEAWTADQQASPGQQKSSNCSSLNKEHSDSNYTTQTT* 0 phosphoserines predicted at SwissProt; no counterparts in SECISBP2 exon 8 skipped in RefSeq KIAA0256 exon 11 lacks counterpart in SECISBP2
Note the Irish proband with compound mutation K438stop/IVS8ds+29G/A (paternal allele inactive, maternal allele a splice donor mutation leading to early truncation) has been incorrectly described as a SECISBP2 knockout; in fact 48% production of wildtype maternal allele still occurs. Additionally, KIAA0256 may be able to partially compensate for reduction in level or loss of SECISBP2. Knockout mice for tRNA(Sec), unable to make any selenoproteins, die in utero.
In two earlier papers, the same authors in twice-published Figure 1B aligned full length SECISBP2 to an intermediate region of the KIAA0256 gene that happened to begin with a methionine, namely residues 422-849 of the 1056 residue protein (which includes the motif-bearing residues 632-829 of exons 14-16). Worse, the model KIAA0256 transcript was an exon-skipping minor splice variant and contains an additional exon 11 without counterpart in SECISBP2:
KIAA0256 originally arose in a GenBank submission package from a large-scale mRNA project at the Kaluza Institute. It skips over highly conserved exon 8 (which does not alter downstream reading frame since it resides in a series of consecutive phase 00 spliced exons). Modelling on this oddity, NCBI later confused the record by mixing in its own predicted genes (from genome assemblies without significant transcript programs) and labelling them mRNAs.
While baboon also has experimental transcripts skipping this exon (FC178616, FC178616), mammalian transcripts almost always retain the exon, for example human (AK307480), macaque (CJ457866), mouse (AK145135), rat (CK602552), dog (CO708934), horse (CX604216), cow (CK846448), sheep (EE864720), and even chicken (DR417186). SwissProt provides full length protein Q93073 without providing a supporting accession.
Transcription and its processing in mammals is a noisy process and artefacts abound. While exon-skipping in some cases may have physiological significance, lacking significant comparative genomics support, the null hypothesis is that they do not. Here the full length gene must contain exon 8 since it is quite conserved in amino acid sequence throughout tetrapods and assuredly ancestral. As all SECIS binding experiments to date used protein lacking exon 8; consequently we know nothing about the SECIS binding properties of intact protein.
Exon 8 comparative genomics: VGFRCRGHSTSSERRQNLQKRPDNKHLSSSQSHRSDPNSESLYFE Homo sapiens VGFRCRGHSTSSERRQNLQKRPDNKHLSSSQSHRSDPNSESLYFE Macaca fascicularis VGFRCRGHSTSSERRQNLQKRQDNKQLNPSQSHRSDSNSESLYFE Tupaia belangeri VGFRCRGHSTSSERRQNLQKRQDNKHLNSTQSHRSDPNSESLYFE Mus musculus VGFRCRGHSTSSERRQNLQKRQDNKHLNSTQSHRSDPNSESLYFE Rattus norvegicus VGFRCRGHSTSSERRQNLPKRQDNNKQLNASQSHRGDSNSESLYFE Canis familiaris VGFRCRGHSTSSERRQNLQKRQDNKQLNPSQSHRGNPNSESLYFE Equus caballus VGFKCRGHSTSSERRQNLQKRQDNKQLNPNQSHRSDPNSESLYFE Myotis lucifugus VGFRCRGHSTSSERRQNLQKRQDNKQLNPSQSHRGDPNSESLYFE Bos taurus VGFRCRGHSTSSERRQNLQKRQDNKQLNPSQSHRGDPNSESLYFE Ovis aries VGFRCRGHSTSSERRQNLQKKQDNKQLNSSQSHRGDPNSESLYFE Dasypus novemcinctus VGFRCRGHSTSSERRQNLQKRQDNKQLNPIQSQRGDPNSESLYFE Loxodonta africana VGFRCRGHSTSSERRQSLQKRQDNKPL-GNHSHRVETSSDPLYFE Monodelphis domestica SGFRCRGHSTSSERRQNLQKRHE-KPLTTSQSSRAEQSPEPLYFE Gallus gallus PAFRCRGHSTSSERRQNLQKKPE-KPVSSSQSSKREQSPGSLYFE Anolis carolinensis LGYRLRGQSTSSERRHNLQRKQDNKTGTPASSNKSGQSPDHLYFE Xenopus tropicali
KIAA0256 and SECISBP2 actually align moderately well over their entire lengths and have 17 near perfectly comparable exons (trillions:one odds for coincidence), meaning they reflect a segmental gene duplication. It is imperative to enforce exon boundaries to achieve true homological alignment of two proteins this diverged and so gappy N-terminally; structure-based alignment has different rules (allowing convergent evolution) and different goals.
The teleost fish Pimephales promelas has sufficient transcript coverage to allow recover of an accurate nearly full length KIAA0256 gene with a respectable 62% identity to human. (No fish has sufficient transcripts to recover full length SECISBP2.) That gene is shown below as homologically gapped exon-by-exon to human. Some early exons are quite well conserved over many billions of years of branch length, strong evidence that they retain an unknown function under fairly strong selection. However the gaps in other early exons are incompatible with retention of tertiary protein structure. No early pfam domain can be found.
>KIAA0256_pimPro Pimephales promelas (minnow) based on transcript tiling; exons by homology 0 MD-AGERK 0 0 DVKLSAEVEPFIPQKKGVEASLLPMSLCGEGGA----EPTQIPSYLITCYPFVQENQSNSR 2 1 QLPMYNGGDQRWQQLNPSPGGPYLAYPILSSPQPPVTSDYATYYHAIMPTPCPPVMGFYQPFPGPFAGPVPAG-VLNPVS-DCSDRPT---------SQRGRGVPRTPVLHK 0 0 QPMAQP-MRAKRPVMRSVAVQKEVCATGPDGRTKTVLLVDAAQQT 1 2 DFPGEASGSGAVRCVSDXASPQLWSNKARRXRTSQQ--ESSSEQGVSEADIDSDSGYCSPKH--NQGANNTSTNQHTPAA 0 0 A-VDAGVMT 1 2 A-VSWGNVSSQAVQK-PWPDRNTPFFRGSRTPERSYTQDFQ 0 0 MSFGCRA---AGPRRSTPPETP-NTHLT--------P--EPLYFQ 0 0 DEDEFPDLATGGAAQRNKPDPVQPKLPKTL 0 0 LDNLPENSPISIVQTPIPITSSVPKRAKSQRKKALAAALATAQEYSEISMEQKKLQ 0 0 EALSKAAGKKSRTPVQLDLGDMLAALEKQQQAMRARQLNNTKPLSYT 1 2 VGTVSALHSKDCGSRVTGLKNTHT-PPHNILDSSAPRIKRGKEREIPKVKKTTAMKK 0 0 IILQEREVKKGKSSADQGVSGADEQRDS-LSFTDTLTQE---QD 1 2 ENGLSMPSDASLSPASQNSPYSITPVSQGSPASSGIGSPMAASAITKIHSRRFRE 2 1 YCNQVLSKDIDESVTLLLQELVRFQERVYQNEPSKAKAKRRLVMGLREVTKHMKLHKIKCVIISPNCEKIQAK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYSGAE 0 0 ALFNTLVSLTEEARRAYKEMVSALEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 2 ETNWRTMVENADAPEPPDSEP-ISRGNNRDQREVVSP---PP---QPT--ANQSLTP--SPGVARAPD--ESRTDDRLEWASL-STETGSLDGSGRDRLNSSHHSTTSTLVPGMLEEE...* 0
There is no reason to believe an odd fragment studied on a few SECIS sequences could accurately represent binding properties of full length protein in regards to the 25-odd orthology classes of SECIS elements. However these results became accepted folklore within the selenocysteine research community and the properties of full length KIAA0256 were never experimentally determined.
We have to wonder how sea urchin, which has a full length apparent ortholog of KIAA0256 on Scaffold18963:101,648-115,302 but nothing clustering to SECISBP2, can insert selenocysteine into its numerous selenoproteins (SEPHS1, SELU1, SELU2, SELM, SELO, SELW, SELN1, GPX3, GPX2, GPX4, GPX7,...). Unless a second copy has been lost, all SECIS interaction at the ribosome at sea urchin divergence appears to have been handled by KIAA056.
Using the kink-turn binding motifs of the two human proteins in turn as blastp query against the both collections of deuterostome KIAA0256 and SECISBP2 sequences, establishes KIAA0256 as the slower evolving protein by a wide margin. This fits KIAA0256 retaining ancestral function and its gene duplicate SECISBP2 specializing to a neofunctionalization. It's difficult to extend the table into invertebrates because of high divergence even in the L7ae region. The table shows Blastp score ratios KIAA0256/SECISBP2 relative to human query:
galGal 1.41 72% identity anoCar 1.35 xenTro 1.41 68% identity danRer 1.44 tetNig 1.59 takRub 1.45 64% identity gasAcu 1.60 oryLat 1.52 calMil 1.43 65% identity
It has not been previously noted that both proteins bristle with potential NxT/S x not P glycosylation sites, 13 for KIAA0256 and 6 for SECISBP2, with implications for cellular localization. These do not lie in homologous positions, unsurprisingly in view of the deep divergence of these genes and volatility of glycosylation sites as seen in other gene families, eg the 17 mammalian sulfatases. Even within orthologs of one gene here, they are conserved only to moderate depth (and that could be for reasons unrelated to glycosylation). Hence these site do not provide reliable anchors in region of poor sequence conservation.
Potential for phosphoserine conservation in exon 5 of KIAA0256: DFPSDIANKSLSETTATMLWKSKGRRRRASHPTAESSSEQGASEADIDSDSGYCSPKHSNNQPAAGALRNPDSGTMN homSap .FPSDIANKSLSESTATMLWKAKGRRRRASHPAVESSSEQGASEADIDSDSGYCSPKH-NNQSAPGALRDPASGTMN musMus DFPSDIANKSLSESSATMLWKSKGRRRRASHPTAESSSEQGASEADIDSDSGYCSPKHSNNQPAAGALRNPDSSTMN canFam DFPSDIANKSLSESSSTMLWKSKGRRRRSSHPTAESSSEQGASEADIDSDSGYCSPKHSNNQATAMTSRNTDSGSIN monDom DFPLDIANKSLSESAATVLWKSKGRRRRASHPAAESSSEQGASEADIDSDSGYCSPKHGNNQAAGPAARSADSGPAN ornAna G insertion DFPSDIANKSLSESASTMLWKSKGRRRRASHPAAESSSEQGASEADIDSDSGYCSPKHGNNQAAAVTSRNADSCAMN galGal DFPSEIASKSLSESMSTMHWKPKTRRRRSSHP-AESSSEQGASEADIDSDSGYCSPKHS-NQAAAVTSRSVESAAGN anoCar DFPNEIANKTICESVGATPWKSKVRRRRLSHPAAESSSEQGASEADIDSDSGYCSPKHC--QAAAMCTRHADCGAV. xenTro DFPGEASGGVRCVSDQVSPQQWKNKPRRRRTSQQESSSEQGASEADIDSDSGYCSPKH--NQGAA............ danRer DFPGEVSGRCAAERASPQLWKNKTKRRRASHP-AENYSEQGASEADIDSDSGYCSPKH--NQAAGVTQR........ gasAcu DFPGEAAVRCVSDQASPQLWSNKARRRRTSQ--QESSSEQGVSEADIDSDSGYCSPKHSTNQPAAAV----DAGVM pimPro SGSG NQGANNT HT insertions DFPDDIADKSLRDKPSPLLRKSKARRLASRRPQDPSSTDSEEDEGGIDSDSGYSSPKHGRNQSA..............braFlo DFPEAIANKPLSDKTSNLTSRSKAKTRKKSQGNASSSSDSEVENTPHDSDSGYYSPLHAQQ................ strPur QTGRD insertion Comparative genomics of 4 glycosylation sites in exon 7 of KIAA0256: GGVNWSNVTCQATQKKPWMEKNQTFSRGGRQTEQRNNSQ Homo sapiens (human) GGVNWSNVTCQATQKKPWMEKNQTFSRGGRQTEQRNNSQ Macaca mulatta (rhesus) GGVNWPKVTCQATQKRPWMEKNQAFSRGGRQTEQRNNLQ Mus musculus (mouse) GGVNWPKVTCQATQKRPWMEKNQAFSRGGRQTEQRNNSQ Rattus norvegicus (rat) GSVNWSNVTCQATQKKPWMEKNQTFSRGGRQTEQRNNSQ Canis familiaris (dog) GGVNWSNVTSQATQKKPWMEKNQTFSRGGRQAEQRNNSQ Sus scrofa (pig) GGVNWSNVTCQATQKKPWMEKNQTFSRGGRQTEQRNNSQ Equus caballus (horse) GGVNWSNVTCQGTQKKPWLEKNQTFSKGGRQMEQRNNSQ Dasypus novemcinctus (armadillo) GHVNWSNVTCQATQKKPWMEKHQTFSRGGRQTEQRNNAQ Loxodonta africana (elephant) GGASWSNVTSQATQKKPWMEKSQPFSRGGRQTEQRNNSQ Monodelphis domestica (opossum) .GVSWTNVNSQATQKKPWIEKTQTFIRGGRQAEQRNSSQ Gallus gallus (chicken) AGATWANVSSQATQKKPWMERTPAFSRGGRQAEQHNSSQ Anolis carolinensis (lizard)
Reference set of 27 vertebrate SECIS BP2 L7Ae motif exons
>SECISBP2_homSap Homo sapiens (human) full length 0 MASEGPREPESE 0 0 GIKLSADVKPFVPRFAGLNVAWLESSEACVFPSSAATYYPFVQEPPVTE 2 1 QKIYTEDMAFGASTFPPQYLSSEITLHPYAYSPYTLDSTQNVYSVPGSQYLYNQPSCYRGFQTVKHRNENTCPLPQEMKALFK 0 0 KKTYDEKKTYDQQKFDSERADGTISSEIKSARGSHHLSIYAENSLKS 1 2 DGYHKRTDRKSRIIAKNVSTSKPEFEFTTLDFPELQGAENNMSEIQKQPKWGPVHSVSTDISLLREVVKPAAVLSK 0 0 GEIVVKNNPNESVTANAATNSPSCTR 1 2 ELSWTPMGYVVRQTLSTELSAAPKNVTSMINLKTIASSADPKNVSIPSSEALSSDPSYNKEKHIIHPTQK 0 0 SKASQGSDLEQNEASRKNKKKKEKSTSKYEVLTVQEPPRIE 0 0 DAEEFPNLAVASERRDRIETPKFQSKQQPQ 0 0 DNFKNNVKKSQLPVQLDLGGMLTALEKKQHSQHAKQSSKPVVVS 1 2 VGAVPVLSKECASGERGRRMSQMKTPHNPLDSSAPLMKKGKQREIPKAKKPTSLKK 0 0 IILKERQERKQRLQENAVSPAFTSDDTQDGESGGDDQFPEQAELS 1 2 GPEGMDELISTPSVEDKSEEPPGTELQRDTEASHLAPNHTTFPKIHSRRFRD 2 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLKKLKCVIISPNCEKIQSK 1 2 GGLDDTLHTIIDYACEQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTVAARQAYKTMLENVQQELVGEPRPQAPPSLPTQGPSCPAEDGPPALKEKEEPHY 1 2 IEIWKKHLEAYSGCTLELEESLEASTSQMMNLNL* 0 >SECISBP2_panTro Pan troglodytes (chimp) 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLKKLKCVIISPNCEKIQSK 1 2 GGLDDTLHTIIDYACEQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTMAARQAYKTMLENVQQELVGEPRPQAPPSLPTQGPSCPAEDGPPALTEKEEPHY 1 >SECISBP2_macMul Macaca mulatta (rhesus) 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLKKLKCVIISPNCEKIQSK 1 2 GGLDDTLHTIIDYACEQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTMAARQAYKTMLENVQQELAGEPRPQAPPSPPTQGPSCPAEDGPPALTEKEEPHY 1 >SECISBP2_otoGar Otolemur garnettii (bushbaby) 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKERRLVLGLREVLKHLKLKKLICVISPNCERQSK 1 2 GGLDDTLHTIIDYACEQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTMAARQAYKTMLENVQRELAGEPGPQVPSSLPMEGPSCSVEDSPPAPTEKEEPHY 1 >SECISBP2_tupBel Tupaia belangeri (treeShrew) 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRVVLGLREVLKHLKLKKLKCVIISPIZEKIQSK 1 2 GGLDDTLHTIIAYACAQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTMEARQAYRSMLESARQELAGEPGLQAPPQPPVQGPRASSEGSAPAPTGRQEPHC 1 >SECISBP2_musMus Mus musculus (mouse) 1 YCSQMLSKEVDACVTGLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLRKLKCIIISPNCEKTQSK 1 2 GGLDDTLHTIIDCACEQNIPFVFALNRKALGRSLNKAVPVSIVGIFSYDGAQ 0 0 DQFHKMVELTMAARQAYKTMLETMRQEQAGEPGPQSPPSPPMQDPIPSTEEGTLPSTGEEPHY 1 >SECISBP2_ratNor Rattus norvegicus (rat) exons 1416 1 YCSQMLSKEVDACVTGLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLRKLKCIIISPNCEKTQSK 1 2 GGLDDTLHTIIDCACEQNIPFVFALNRKALGRSLNKAVPVSIVGIFSYDGAQDQ 0 0 FHKMVELTMAARQAYKTMLETMRQEQAGEPGPQTPPSPPMQDPIQSTDEGTLASTGEEPHY 1 >SECISBP2_cavPor Cavia porcellus (guineaPig) 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLKKLKCIIISP 1 2 GLDDTLHTIIDYACAQNIPFVFALNRKALGRSLNKTVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTMAARQAYKTMLENVRQELAGEPRPQMPPDPPSEGPSSSLEDTAPDPSAEEPHY 1 >SECISBP2_oryCun Oryctolagus cuniculus (rabbit) 1 YCSQMLSKEVDACVTDLFKELVRFHDLMYQDPVKATTKCQFELRVGKALDHLRLKKLKCIIVFPKHKKQS 1 2 TIIDYACEQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTMAARQAYKTMLENMRHELAGEPGPPTPQPVQGPSCSAEDGPPAPTEGEVPHY 1 >SECISBP2_canFam Canis familiaris (dog) 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLRKLKCIIISPNCEKIQSK 1 2 GGLDDTLHTIIDYACEQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHRMVELTMAARQAYKTMLENVRQELAGEPGTPALANPPMQGLGCSTQDSPPAPTEKEEPHY 1 >SECISBP2_felCat Felis catus (cat) 1 YCSQMLSKEVDACVTDLLRELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLRKLKCIIISPNCEKIQSK 1 2 GGLDDTLHTIIGYACEQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHRMVELTMAARQAYKTMLENARQELAGEPGPPAPGSPPPQPPAPAGRDEPRY >SECISBP2_equCab Equus caballus (horse) 1 YCSQILSKEVDACVTELLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLRKLKCIIISPNCEKIQSK 1 2 GGLDDTLHTIIDYACEQNIPCVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTKAARQAYKAMLENVHQELAGEPGPQAPASPPAQGPSCSTEGAPPAPTGKEEPHY 1 >SECISBP2_bosTau Bos taurus (cow) 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKAKRRLVLGLREVLKHLKLRKLKCIIISPNCEKIQSK 1 2 GGLDDTLHTIIDYACDQNIPFVFALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTMAARQAYRTMLENARQELPGELGPCAPVGPPSQGPGCPVEDSPLAPTEKEEPHY 1 >SECISBP2_eriEur Erinaceus europaeus (hedgehog) 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLKKLKCIIISPNCEKIQSK 1 2 GGLDETLHTIIDCACEQNIPFVFALNRKALGRSLNKGVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTMAARQAYKALLENMRQELAEESGSPAPSSPPVQSPSEDGPPAPAEKEEPHY 1 >SECISBP2_dasNov Dasypus novemcinctus (armadillo) 1 YCSQVLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLKKLKCVIISPNCEKIQSK 1 2 GGELDDTLHTIIDYAASRHSICVALNRKALGRSLNKAVPVSVVGIFSYDGAQ 0 0 DQFHKMVELTMAARQAYKAMLENVRKELAGEPGPRSPPSPPALGPHSSAGDVHPTSAGKEEPHY 1 >SECISBP2_loxAfr Loxodonta africana (elephant) 1 YCSQMLSKEVDACVTDLLKELVRFQDRMYQKDPVKAKTKRRLVLGLREVLKHLKLKKLKCVIISPNCEKIQSK 1 2 GGLDDTLHTIIDYACEQNIPFVFALHRKALGRSLNKPVPVSVVGIFSYDRAQ 0 0 DQFHKMVELTMAARQEYKTMLESVRQELAEEPRAGSPPSPPTQGPGCSAEVPRPAPTEKEEPRY 1 >SECISBP2_monDom Monodelphis domestica (opossum) 1 YCSQMLSKEVDDCVMDLLKELVRFQDRMYQKDPVKAKTKRRLVMGLREVLKHLKLKKLKCVIISPNCEKSKSK 1 2 GGLDETLHTIIDYACEQNVPFVFALNRKALGRSVNKVVPVSVVGIFSYDGAQ 0 0 DQFHKMIALTMEARQAYKIMLSTLKEEPALETENPPSPSLPRPSESCPSELGQTDPTQEEEPNY 1 >SECISBP2_triVul Trichosurus vulpecula (possum) 1 YCSQMLSKEVDDCVMDLLKELVRFQDRMYQKDPVKAKTKRRLVMGLREVLKHLKLKKLKCVIISPNCEKSKSK 1 2 GGLDETLHTIIDYACEQNVPFVFALNRKALGRSVNKVVPVSVVGIFSYDGAQ 0 0 DQFRKMIELTMEARQAYKVMLATLKEGAEALQTENPLPTSLTPQGQGCSSELSKTTDPTKEEEPNY 1 >SECISBP2_galGal Gallus gallus (chicken) 1 YCSQVLSKEVDSCVTDLLKELVRFQDRLYQKDPVKAKIKRRLVMGLREVLKHLRLKKLKCVIISPNCEKIQSK 1 2 GGLDETLHNIIDCACEQNIPFVFALNRKALGRCVNKAVPVSVVGIFSYDGAQ 0 0 DHFHRMVQLTTEARKAYKDMVAALEEELKELSKPLNZKSCLSETGKTSSTKEDIPNY 1 >SECISBP2_anoCar Anolis carolinensis (lizard) 1 YCTQVLSKEVDSCVTDLLKELVRFQDRLYQKDPVKAKTKRRLVMGLREVLKHLKLKKLKCVIISPNCEKIQSK 1 2 GGLDETLHLIIDSACEQNIPFVFALNRKALGRCLNKAVPVSVVGIFSYDGAQ 0 0 DYFHKMVELTMEARQAYKDMISALERELKKKTVRKKPLQSRPLDTVEASSTEEDVPDY 1 >SECISBP2_xenTro Xenopus tropicalis (frog) NM_001097262 1 YCSQVLSKDVDNCVMELLKELVRFQDRLFLKEPAKAKSKRRLVMGLREVLKHLKLQKLKCIIISPNCEKIQSK 1 2 GGLDDTLQTIISHACEQNVPFVFALNRKALGRCLNKAVPVSVVGVFSYDGAQ 0 0 DHFHKLCELTVQARQAYKDMIAAAQEQQSETEAGKNEEDPVAVNGQNKSDDMREESKAEEPDEPNY 1 >SECISBP2_danRer Danio rerio (zebrafish) 1 YCNQVLSKDVDECVSNLLKELVRFQDRLYQKDPMKARMKRRLVMGLREVLKHLKLKKVKCVIISPNCERIQSK 1 2 GGLDEALHNIIDTCRDQSVPFVFALSRKALGRCVNKAVPVSLVGIFNYDGAQ 0 0 DFYHKMIELSSEARTAYEVMLLNLEQTDAEEAQQTSPLAEKVETSSGDPQPEEPEY 1 >SECISBP2_tetNig Tetraodon nigroviridis (pufferfish) 1 YCNQVLSKEIDESVTLLLQELVRFQERVYQKDPTKAKSKRRLVMGLREVTKHMKLQTIKCVIISPNCEKIQAK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYSGAE 0 0 DFYHKMIELSSEARIAYEVMLSNLEQTSAEEEPQTCTLAEKINTSSEDAQPEEPEY 1 >SECISBP2_takRub Takifugu rubripes (fugu) 1 YCTQMLSKDVDECVTTLLKELVRFQDRLYQKDPIKARMKRRIVMGLREVQKHLKLRKLKCVIISPNCERIQSK 1 2 GGLDEALHTIIDTCREQAVPFVFALSRRALGRCVNKAVPVSLVGIFNYDGAQ 0 0 DFYHKMIELSSEARTAYEVMLLNLEQTDAEEAQQTSPLAEKVETSSGDPQPEEPEY 1 >SECISBP2_gasAcu Gasterosteus aculeatus (stickleback) 1 YCNQVLSKEIDESVTMLLQELVRFQERIYQKDPTKAKTKRRLVMGLREVTKHMKLNKIKCVLISPNCEKIQAK 1 2 GGLDEALYNVIAMARDQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYSGAE 0 0 DFYHKMIELSSEARRAYEVMVSSLEQTGQADPESVEEKLQISSAAEEAELGRDITPPEEPEY 1 >SECISBP2_oryLap Oryzias latipes (medaka) 1 YCSQMLRKDVDECVTVLLKELVRFQDRLYHKDPIKARMKRRLVMGLREVLKHLKLRKVKCVIISPNCEQIQSK 1 2 GGLDEALHTIIQTCREQAVPFVFALSRKALGHCVNKAVPVSLVGIFNYDGAQ 0 0 DHYHKMIELSAEARKAYEVLVSSLERDQQEESHPDRGTCFGSVTAEPEKPHY 1 >SECISBP2_calMil Callorhinchus milii (elephantfish) AAVX01044988 1 YCSQVLSKDVDSCVTDLLKELVRFQDRLYQKDPIKAKKKRRIVMGLREVLKHLKLKRLKCIIISPNCEKIQSR 1 2 GGLDDALHNIISIACEQEIPFVFALNRKALGQCVNKPVPVSVLGIFSYDGAE 0 0 NQFHQMVEITEEARKAYQEMLDALQQELEADEEKGDSEEQPLISSESSTIHFNNVTSQPFSEADEPEY 1 >SECISBP2_braFlo Branchiostoma floridae (amphioxus) extra exon 1 YCNQVLDKEIDATVTMLLQDLVRFQDRQYHK 00 DPIKAKAKRRIVMGLREVTKHLKLRKLKCIIIAPNLEKIQSK 1 2 GGLDDAIETILNLCMEQDVPFVFALGRKALGRAVNKLVPVSVVGVFNYDGAE 0 0 1
Reference set of 23 deuterostome SECIS KIAA0256 L7Ae motif exons
>KIAA0256_homSap Homo sapiens (human) length=1101 13 glycosylation sites 0 MDRAPTEQ 0 0 NVKLSAEVEPFIPQKKSPDTFMIPMALPNDNGSVSGVEPTPIPSYLITCYPFVQENQSNR 2 1 QFPLYNNDIRWQQPNPNPTGPYFAYPIISAQPPVSTEYTYYQLMPAPCAQVMGFYHPFPTPYSNTFQAANTVNAITTECTERPSQLGQVFPLSSHRSRNSNRGSVVPK 0 0 QQLLQQHIKSKRPLVKNVATQKETNAAGPDSRSKIVLLVDASQQT 1 2 DFPSDIANKSLSETTATMLWKSKGRRRRASHPTAESSSEQGASEADIDSDSGYCSPKHSNNQPAAGALRNPDSGTMN 0 0 HVESSMCA 1 2 GGVNWSNVTCQATQKKPWMEKNQTFSRGGRQTEQRNNSQ 0 0 VGFRCRGHSTSSERRQNLQKRPDNKHLSSSQSHRSDPNSESLYFE 0 0 DEDGFQELNENGNAKDENIQQKLSSKV 0 0 LDDLPENSPINIVQTPIPITTSVPKRAKSQKKKALAAALATAQEYSEISMEQKKLQ 0 0 EALSKAAGKKNKTPVQLDLGDMLAALEKQQQAMKARQITNTRPLSYT 1 2 VVTAASFHTKDSTNRKPLTKSQPCLTSFNSVDIASSKAKKGKEKEIAKLKRPTALKK 0 0 VILKEREEKKGRLTVDHNLLGSEEPTEMHLDFIDDLPQEIVSQE 1 2 DTGLSMPSDTSLSPASQNSPYCMTPVSQGSPASSGIGSPMASSTITKIHSKRFRE 2 1 YCNQVLCKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 0 SLFNKLVELTEEARKAYKDMVAAMEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 2 ETNWRNMVETSDGLEASENEKEVSCKHSTSEKPSKLPFDTPPIGKQPSLVATGSTTSATSAGKSTASDKEEVKPDDLEWASQQSTETGSLDGSCRDLLNSSITSTTSTLVP GMLEEEEDEDEEEEEDYTHEPISVEVQLNSRIESWVSETQRTMETLQLGKTLNGSEEDNVEQSGEEEAEAPEVLEPGMDSEAWTADQQASPGQQKSSNCSSLNKEHSDSNYTTQTT* 0 >KIAA0256_panTro Pan troglodytes (chimp) 2 YCNQVLCKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 0 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 1 SLFNKLVELTEEARKAYKDMVAAMEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_macMul Macaca mulatta (rhesus) 2 YCNQVLCKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 0 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 1 SLFNKLVELTEEARKAYKDMVAAMEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_tupBel Tupaia belangeri (treeShrew) 1 YCNQVLCKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 0 SLFNKLVELTEEARKAYKDMVAAMEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_musMus Mus musculus (mouse) 0 YCNQVLSKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 1 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 2 SLFNRLVELTEEARKAYKDMVAATEQEQAEEALRSVKTVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_ratNor Rattus norvegicus (rat) 2 YCNQVLSKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 0 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 1 SLFNRLVELTEEARKAYKDMVAATEQEQAEEALRSVKAVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_canFam Canis familiaris (dog) 1 YCNQVLCKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 0 SLFNKLVELTEEARKAYKDMVAAMEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_equCab Equus caballus (horse) 1 YCNQVLCKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 0 SLFNKLVALTEEARRAYKDMVAALEQEQAEEASKNVKKGPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_dasNov Dasypus novemcinctus (armadillo) 1 YCNQVLCKEIDECVTLLLQELVSFQERIYQKDPVRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSKG 1 2 GLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 0 SLFNKLVELTEEARKAYKDMVAAMEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_monDom Monodelphis domestica (opossum) 0 YCNQVLCKEIDECVTLLLQELVSFQERIYQKDPVKAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 1 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYCGAE 0 2 SLFNKLVELTEEARKAYKDMVAAMEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_galGal Gallus gallus (chicken) 1 YCNQVLSKEIDECVTLLLQELVSFQERIYQKDPMRAKARRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYSGAE 0 0 DLFNKLVSLTEEARKAYRDMVAAMEQEQAEEALKNVKKAPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_anoCar Anolis carolinensis (lizard) 1 YCNQVLSKEIDECVTLLLQELVSFQEQIYQKDPMRAKAKRRLVMGLREVTKHMKLSKIKCVIISPNCEKIQSK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYSGAE 0 0 NLFNKLVSLTEEARKAYRDMVAAMEQEQEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_xenTro Xenopus tropicalis (frog) 1 YCNQVLSKEIDECVTVLLQELVSFQERVYQKDPVKAKSKRRLVMGLREVTKHMKLNKIKCVIISPNCEKIQSK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFSYSGAE 0 0 SLFHNLVSLTEEARKAYKDMVSSMEQEQAEEALKNIKKVHMGHSRNPSAASAISFCSVISEPISEVNEKDY 1 >KIAA0256_danRer Danio rerio (zebrafish) 1 YCNQVLSKEIDESVTLLLQELVRFQERVYQKEPSKAKAKRRLVMGLREVTKHMKLHKIKCVIISPNCEKIQAK 1 2 GGLDEALHNIIDTCRDQSVPFVFALSRKALGRCVNKAVPVSLVGIFNYDGAQ 0 0 GLFNKLVSLTEEARRAYKEMVSALEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 >KIAA0256_tetNig Tetraodon nigroviridis (pufferfish) 1 YCNQVLSKEIDESVTLLLQELVRFQERVYQKDPTKAKSKRRLVMGLREVTKHMKLQTIKCVIISPNCEKIQAK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYSGAE 0 0 SLFNQLVSLTEEARKAYKDMVSALEQEQTEEALKNEKKVPHQMGHYRNHSAASAVSFCSIFSEPISEVNEKEY 1 >KIAA0256_takRub Takifugu rubripes (fugu) 1 YCNQVLSKEIDESVTLLLQELVRFQERVYQKDPTKAKSKRRLVMGLREVTKHMKLQTIKCVIISPNCEKIQAK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNFSGAE 0 0 SLFNQLVSLTEEARKAYKDMVSALEQEQTEEALKNEKKVPHQMGHYRNHSAASAVSFCSIFSEPISEVNEKEY 1 >KIAA0256_gasAcu Gasterosteus aculeatus (stickleback) 1 YCNQVLSKEIDESVTMLLQELVRFQERIYQKDPTKAKTKRRLVMGLREVTKHMKLNKIKCVLISPNCEKIQAK 1 2 GGLDEALYNVIAMARDQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYSGAE 0 0 GLFNRLVSLTEEARKAYKDMVSALEQEQAEEAQKNDKKLPHHMGHSRNHSAASAISFCSIFSEPISEVNEKEY 1 >KIAA0256_oryLap Oryzias latipes (medaka) 0 YCNQVLSKEIDESVTLLLQELVRFQERVYQKDPSKAKSKRRLVMGLREVTKHMKLHKIKCVIISPNCEKIQAK 1 1 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYSGAE 0 2 GLFNQLVSLTEEARKAYKEMVSALEQEQAEEALKHDKKVPHHMGHSRNHSAASAISFCSILSEPISEVNEKEY 1 >KIAA0256_pimPro Pimephales promelas (minnow) based on transcript tiling; exons by homology; 62% identity 0 MDAGERK 0 0 DVKLSAEVEPFIPQKKGVEASLLPMSLCGEGGAEPTQIPSYLITCYPFVQENQSNSR 2 1 QLPMYNGGDQRWQQLNPSPGGPYLAYPILSSPQPPVTSDYATYYHAIMPTPCPPVMGFYQPFPGPFAGPVPAGVLNPVSDCSDRPTSQRGRGVPRTPVLHK 0 0 QPMAQPMRAKRPVMRSVAVQKEVCATGPDGRTKTVLLVDAAQQT 1 2 DFPGEASGSGAVRCVSDXASPQLWSNKARRXRTSQQESSSEQGVSEADIDSDSGYCSPKHNQGANNTSTNQHTPAA 0 0 AVDAGVMT 1 2 AVSWGNVSSQAVQKPWPDRNTPFFRGSRTPERSYTQDFQ 0 0 MSFGCRAAGPRRSTPPETPNTHLTPEPLYFQ 0 0 DEDEFPDLATGGAAQRNKPDPVQPKLPKTL 0 0 LDNLPENSPISIVQTPIPITSSVPKRAKSQRKKALAAALATAQEYSEISMEQKKLQ 0 0 EALSKAAGKKSRTPVQLDLGDMLAALEKQQQAMRARQLNNTKPLSYT 1 2 VGTVSALHSKDCGSRVTGLKNTHTPPHNILDSSAPRIKRGKEREIPKVKKTTAMKK 0 0 IILQEREVKKGKSSADQGVSGADEQRDSLSFTDTLTQEQD 1 2 ENGLSMPSDASLSPASQNSPYSITPVSQGSPASSGIGSPMAASAITKIHSRRFRE 2 1 YCNQVLSKDIDESVTLLLQELVRFQERVYQNEPSKAKAKRRLVMGLREVTKHMKLHKIKCVIISPNCEKIQAK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYSGAE 0 0 ALFNTLVSLTEEARRAYKEMVSALEQEQAEEALKNVKKVPHHMGHSRNPSAASAISFCSVISEPISEVNEKEY 1 2 ETNWRTMVENADAPEPPDSEPISRGNNRDQREVVSPPPQPTANQSLTPSPGVARAPDESRTDDRLEWASLSTETGSLDGSGRDRLNSSHHSTTSTLVPGMLEEE * 0 >KIAA0256_calMil Callorhinchus milii (elephantfish) AAVX01105236 1 YCNQVLSKDIDECVTLLLQELVRFQERVYQKDPIKAKMKRRLVMGLREVTKHMKLRKIKCVIISPNCEKIQSK 1 2 GGLDEALYNVIAMAREQEIPFVFALGRKALGRCVNKLVPVSVVGIFNYFGAE 0 0 1 >KIAA0256_petMar Petromyzon marinus (lamprey) 1 LHKLRALIISPNCEKIQAK 1 2 GGLDEALQTVIALASEQSVPFVFALNRKALGHCLNKKVPVSVVGVFHYGGAE 0 0 THFQRLVALTEEARSAYRNMVSSLQRQEAAATSEPTGHTEDPLEASASPPSVPAHDPTALLHLLRPQQGPREDDPAEASGRSPGRNA 1 >KIAA0256_cioInt Ciona intestinalis (tunicate) 1 YCCQVLDKRVDEMSNQMLQRLVYFQDR 21 RLYKTDPAKAKRKRRVVLGFREVTKHLKMKKLRCVIISPNLEKIESK 1 2 GGLDDVLHEILDLCKEQNIPYVFALGKKALGRAVSKTVPVSIVGVFDYSGAE 0 0 1 >KIAA0256_strPur Strongylocentrotus purpuratus (sea_urchin) 1 YCNQVLDKDIDGCCTTLLQTLVKFQDRQYHKDPAK 00 AKMKRRLVMGLREVTKHLKLKKIKCVVVSPNLERIQSK 1 2 GGLDEAMDRISSLASEQNVPLIFALGRKALGRAVNKVVPVSVVGIFNYDGAE 0 0 DTYKQLLDLSTRARNAYADMVRKFQQELEAANAASAARMAKHRHHMGHNRNLFKG 1
Reference set of 10 deuterostome ribosomal L30 L7Ae motifs
>L30_homSap Homo sapiens (human) 4 exons numerous pseudogenes 0 MVAAKKT 0 0 KKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALR 2 1 KSEIEYYAMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDP 1 2 GDSDIIRSMPEQTGEK* 0 >L30_tupBel Tupaia belangeri (treeShrew) 0 MVAAKKT 0 0 KKSLESINSQLQLAMKDGKYVLGYKQTLKMIRQGKAKLVILANNCPALR 2 1 KSEIEYYAMLAKTGVHHYSGNNIELGTACGKYYRACTLAIMDP 1 2 GDSDIIRSMPEQTGEK* 0 >L30_ratNor Rattus norvegicus (rat) Sep15 Gpx4 Gpx1 Dio1 quite weak homology 35% with BP2 exons 0 MVAAKKT 0 0 KKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALR 2 1 KSEIEYYAMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDP 1 2 GDSDIIRSMPEQTGEK* 0 >L30_myoLuc Myotis lucifugus (microbat) 0 MVAAKKT 0 0 KKSLESINSRLQLVMKSGKYLLGYKQTLKMIRQGKAKLVILANNCPALR 2 1 ISEIEYYAMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDP 1 2 GDSD-IRSMPEQTGEK* 0 >L30_echTel Echinops telfairi (tenrec) 0 MVAAKKT 0 0 KNSLESINSRLQLVMKSGKYMLGYKQMLKMIRQGKAKLVVLANNCPALR 2 1 KSEIEYYAMLAKTGVHHYSGHNIELGTACGKSCRVCTLAITDP 1 2 GDADIIRSMPEQTGEK* 0 >L30_anoCar Anolis carolinensis (lizard) 0 MVAAKKT 0 0 KKSLESINSRLQLVMKSGKYVLGYKQTLKMIQQGKAKLVILANNCPALG 2 1 KSEIEYYAMLAKTGVHHYSGNNIEMGTACGKYYRVCTLAIIDP 1 2 GDSDIIRSMQEQTAEK* 0 >L30_danRer Danio rerio (zebrafish) 94% 0 MVAAKKT 0 0 KKSLESINSRLQLVMKSGKYVLGYKQSQKMIRQGKAKLVILANNCPALR 2 1 KSEIEYYAMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDP 1 2 GDSDIIRSMPDQQQGGEK* 0 >L30_squAca Squalus acanthias (spiny dogfish) 97% 0 MVAAKKT 0 0 KKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALR 2 1 KSEIEYYAMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDP 1 2 GDSDIIRSMPEQISEK* 0 >L30_petMar Petromyzon marinus (lamprey) 94% 0 MSAKKT 0 0 KKAIESINSRLQLVMKSGKYCLGYRQTLKMIRQGKAKLVLLANNCPALR 2 1 KSEIEYYAMLAKTGVHHYSGNNIEMGTACGKYYRVCTLAIIDP 1 2 GDSDIIRSMPEQQQPQPGDK* 0 >L30_braFlo Branchiostoma floridae (amphioxus) 84% to homSap 0 MKQK 0 0 RKTMESINSRLQLVMKSGKYVLGLKETLKVLRQGKAKLIIIANNTPALR 2 1 KSEIEYYAMLAKTGVHHYSGNNIELGTACGKYFRVCTLAITDP 1 2 GDSDIIRSMPAEDKGESK* 0
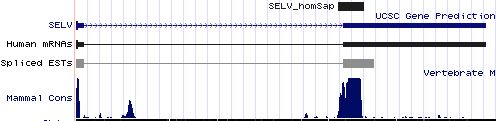
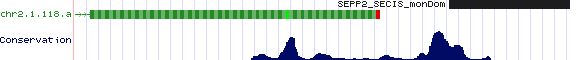
Positioning of SECIS elements in 3' UTR
Comparative genomic analysis of SECIS elements
Key residues for SECIS functioning
Below the residues considered key to SECIS element functionality have been extracted from the 358 sequences in the reference set collection. The first column gives gene name, genus, species, and COVE score rounded to the nearest integer. The second thru fourth columns give the traditional critical regions in SECIS structure as provided by SECIS tool in its output spacing. The fifth column, when all is pasted in a spreadsheet, re-sorts back into gene name in phylogenetic order.
The table is currently sorted by col4, col3, then col2 to bring together similar anomalies. Some but not by no means all of the departures from conventional wisdom are artefacts from genome assemblies and comp genomics methods. However many anomalies are consistent by gene and correlate with adjacent column anomalies, so are verly likely valid). It is clear from comparing this to the alignment conservation of SECIS elements that our picture of what is important in a SECIS element needs considerable revision -- the many billion years of comparative genomics branch length swamp out local errors even before curation.
>TXNRD1_ratNor_28 atgaa aa agaa 99 >TXNRD1_musMus_28 atgaa aa agaa 98 >DIO2_galGal_25 atgaa aa agat 349 >DIO2_anoCar_27 atgaa aa agat 348 >DIO2_ornAna_25 atgaa aa agat 347 >SELT_ornAna_36 atgaa aa agat 166 >SELT_monDom_37 atgaa aa agat 165 >SELT_loxAfr_36 atgaa aa agat 164 >SELT_eriEur_39 atgaa aa agat 162 >SELT_bosTau_39 atgaa aa agat 161 >SELT_felCat_39 atgaa aa agat 159 >SELT_canFam_39 atgaa aa agat 158 >SELT_oryCun_39 atgaa aa agat 157 >SELT_cavPor_39 atgaa aa agat 156 >SELT_ratNor_39 atgaa aa agat 155 >SELT_musMus_39 atgaa aa agat 154 >SELT_otoGar_39 atgaa aa agat 153 >SELT_macMul_39 atgaa aa agat 152 >SELT_panTro_39 atgaa aa agat 151 >SELT_homSap_39 atgaa aa agat 150 >SELM1_macMul_28 atgaa aa cagat 64 >SELM1_panTro_28 atgaa aa cagat 63 >SELM1_homSap_27 atgaa aa cagat 62 >SEPP1b_danRer_24 atgaa aa ccgag 36 >SELM1_tupBel_25 atgaa aa ccgat 66 >TXNRD1_monDom_23 atgaa aa cgaa 107 >GPX1_cavPor_15 atgaa aa cgat 281 >GPX3_galGal_31 atgaa aa cgat 261 >SELT_equCab_38 atgaa aa cgat 160 >SEPN_dasNov_27 atgaa aa cgat 48 >TXNRD1_echTel_20 atgaa aa cgat 106 >SELV_sorAra_26 atgaa aa ctgac 183 >SELM1_loxAfr_27 atgaa aa ctgat 77 >SELM1_dasNov_24 atgaa aa ctgat 76 >SELM1_bosTau_24 atgaa aa ctgat 75 >SELM1_equCab_29 atgaa aa ctgat 74 >SELM1_felCat_30 atgaa aa ctgat 73 >SELM1_canFam_28 atgaa aa ctgat 72 >SELM1_eriEur_27 atgaa aa ctgat 71 >SELM1_sorAra_26 atgaa aa ctgat 70 >SELM1_otoGar_28 atgaa aa ctgat 65 >SELI_anoCar_22 atgaa aa ggaa 224 >TXNRD1_galGal_27 atgaa aa ggaa 110 >TXNRD1_anoCar_33 atgaa aa ggaa 109 >TXNRD1_ornAna_19 atgaa aa ggaa 108 >TXNRD1_loxAfr_23 atgaa aa ggaa 105 >TXNRD1_equCab_22 atgaa aa ggaa 104 >TXNRD1_felCat_24 atgaa aa ggaa 103 >TXNRD1_eriEur_26 atgaa aa ggaa 101 >TXNRD1_tupBel_25 atgaa aa ggaa 97 >TXNRD1_otoGar_23 atgaa aa ggaa 96 >TXNRD1_panTro_25 atgaa aa ggaa 95 >TXNRD1_homSap_25 atgaa aa ggaa 94 >SELI_galGal_25 atgaa aa ggag 225 >SELI_ornAna_19 atgaa aa ggag 223 >SELI_monDom_26 atgaa aa ggag 222 >TXNRD1_canFam_25 atgaa aa ggag 102 >SELI_loxAfr_26 atgaa aa ggat 221 >SELI_dasNov_18 atgaa aa ggat 220 >SELI_equCab_20 atgaa aa ggat 219 >SELI_canFam_14 atgaa aa ggat 218 >SELI_eriEur_14 atgaa aa ggat 217 >SELI_oryCun_19 atgaa aa ggat 216 >SELI_cavPor_17 atgaa aa ggat 215 >SELI_ratNor_25 atgaa aa ggat 214 >SELI_musMus_25 atgaa aa ggat 213 >SELI_otoGar_21 atgaa aa ggat 212 >SELI_macMul_21 atgaa aa ggat 211 >SELI_panTro_22 atgaa aa ggat 210 >SELI_homSap_22 atgaa aa ggat 209 >SELT_tetNig_33 atgaa aa ggat 170 >SELT_danRer_35 atgaa aa ggat 169 >SELT_galGal_38 atgaa aa ggat 168 >SELT_anoCar_34 atgaa aa ggat 167 >SELT_sorAra_36 atgaa aa ggat 163 >MSRB1_galGal_22 atgaa aa ggat 93 >SEPP1a_monDom_31 atgaa aa ggat 17 >SEPP1a_canFam_33 atgaa aa ggat 11 >SEPP1a_sorAra_33 atgaa aa ggat 9 >SEPP1a_cavPor_35 atgaa aa ggat 7 >SEPP1a_tupBel_31 atgaa aa ggat 4 >SELK_galGal_31 atgaa aa tgaa 245 >GPX4_musMus_36 atgaa aa tgag 314 >GPX3_bosTau_31 atgaa aa tgag 255 >GPX3_ratNor_33 atgaa aa tgag 251 >GPX3_musMus_33 atgaa aa tgag 250 >TXNRD2_echTel_37 atgaa aa tgag 129 >DIO3_bosTau_23 atgaa aa tgat 358 >GPX1_bosTau_30 atgaa aa tgat 287 >SELS_echTel_38 atgaa aa tgat 145 >SEPP1a_danRer_33 atgaa aa tgat 20 >SEPP1a_anoCar_33 atgaa aa tgat 19 >SELO_monDom_24 atgaa cc cgac 174 >GPX3_felCat_30 atgaa ga tgag 253 >GPX3_canFam_30 atgaa ga tgag 252 >SEPHS2_otoGar_12 atgac aa agaa 54 >GPX4_sorAra_37 atgac aa agac 318 >SEPHS2_bosTau_22 atgac aa agac 60 >GPX6_panTro_23 atgac aa agag 263 >SEPHS2_equCab_22 atgac aa agag 59 >SEPHS2_felCat_22 atgac aa agag 58 >SEPHS2_canFam_20 atgac aa agag 57 >SEPHS2_ratNor_19 atgac aa agag 56 >SEPHS2_macMul_21 atgac aa agag 53 >SEPHS2_panTro_21 atgac aa agag 52 >SEPHS2_homSap_21 atgac aa agag 51 >DIO3_cavPor_24 atgac aa ccgaa 355 >DIO3_loxAfr_22 atgac aa cgaa 359 >GPX4_galGal_39 atgac aa cgaa 323 >GPX4_anoCar_39 atgac aa cgaa 322 >GPX4_macMul_33 atgac aa cgaa 312 >GPX3_ornAna_31 atgac aa cgaa 259 >SELS_anoCar_25 atgac aa cgaa 148 >SEPP2_tetNig_17 atgac aa cgac 208 >GPX4_canFam_31 atgac aa cgag 317 >GPX4_ratNor_34 atgac aa cgag 315 >GPX2_ornAna_28 atgac aa cgag 309 >TXNRD2_equCab_31 atgac aa cgag 128 >TXNRD2_canFam_34 atgac aa cgag 127 >TXNRD2_panTro_31 atgac aa cgag 121 >TXNRD2_homSap_31 atgac aa cgag 120 >SEPW_ratNor_27 atgac aa cgat 329 >GPX4_monDom_41 atgac aa cgat 320 >GPX4_echTel_41 atgac aa cgat 319 >GPX1_eriEur_19 atgac aa cgat 283 >SELS_sorAra_36 atgac aa cgat 143 >MSRB1_ornAna_26 atgac aa cgat 92 >SELV_echTel_26 atgac aa ctgag 185 >SELV_equCab_33 atgac aa ctgat 182 >SELV_otoGar_27 atgac aa ctgat 178 >DIO3_felCat_29 atgac aa tgaa 357 >DIO3_ratNor_26 atgac aa tgaa 354 >DIO3_musMus_26 atgac aa tgaa 353 >DIO3_macMul_30 atgac aa tgaa 352 >DIO3_panTro_31 atgac aa tgaa 351 >DIO3_homSap_31 atgac aa tgaa 350 >GPX1_dasNov_29 atgac aa tgaa 288 >SELH_loxAfr_26 atgac aa tgaa 200 >SELH_bosTau_28 atgac aa tgaa 197 >SELH_equCab_26 atgac aa tgaa 196 >SELH_felCat_25 atgac aa tgaa 195 >SELH_canFam_24 atgac aa tgaa 194 >SELH_otoGar_23 atgac aa tgaa 189 >TXNRD2_ratNor_35 atgac aa tgaa 124 >TXNRD2_tupBel_37 atgac aa tgaa 122 >SEPP2a_ornAna_34 atgac aa tgac 206 >DIO3_canFam_26 atgac aa tgag 356 >GPX2_cavPor_20 atgac aa tgag 299 >SEPP1b_ornAna_18 atgac aa tgag 34 >DIO1_sorAra_24 atgac aa tgat 337 >DIO1_bosTau_28 atgac aa tgat 336 >DIO1_cavPor_24 atgac aa tgat 335 >GPX1_ratNor_26 atgac aa tgat 280 >GPX1_tupBel_21 atgac aa tgat 278 >SEPN_loxAfr_32 atgac aa tgat 49 >SEPP2_monDom_30 atgac aa tgat 204 >MSRB1_oryCun_28 atgac aa tgat 87 >GPX4_cavPor_34 atgac ca agac 316 >GPX4_panTro_29 atgac ca cgaa 311 >GPX4_homSap_29 atgac ca cgaa 310 >GPX4_ornAna_34 atgac ca tgaa 321 >SELO_takRub_20 atgac ca tgaa 175 >GPX6_echTel_31 atgac ga cgaa 273 >GPX3_echTel_13 atgag aa ccgag 257 >SELM1_cavPor_31 atgag aa ctgat 69 >SELM1_ratNor_30 atgag aa ctgat 68 >SELM1_musMus_35 atgag aa ctgat 67 >SEPP2b_ornAna_23 atgag aa ggaa 207 >TXNRD1_cavPor_24 atgag aa ggat 100 >SEPP1a_ornAna_29 atgag aa ggat 18 >SEPP1a_echTel_33 atgag aa ggat 16 >SEPP1a_loxAfr_33 atgag aa ggat 15 >SEPP1a_dasNov_33 atgag aa ggat 14 >SEPP1a_bosTau_31 atgag aa ggat 13 >SEPP1a_equCab_29 atgag aa ggat 12 >SEPP1a_eriEur_33 atgag aa ggat 10 >SEPP1a_ratNor_33 atgag aa ggat 6 >SEPP1a_musMus_33 atgag aa ggat 5 >SEPP1a_macMul_31 atgag aa ggat 3 >SEPP1a_panTro_31 atgag aa ggat 2 >SEPP1a_homSap_31 atgag aa ggat 1 >SELK_loxAfr_35 atgag aa tgac 241 >SELK_dasNov_29 atgag aa tgac 240 >SELK_bosTau_39 atgag aa tgac 239 >SELK_felCat_37 atgag aa tgac 238 >SELK_canFam_36 atgag aa tgac 237 >SELK_otoGar_32 atgag aa tgac 229 >SELK_macMul_36 atgag aa tgac 228 >SELK_panTro_35 atgag aa tgac 227 >SELK_homSap_35 atgag aa tgac 226 >GPX3_anoCar_28 atgag aa tgag 260 >GPX3_tupBel_29 atgag aa tgag 249 >GPX3_macMul_28 atgag aa tgag 248 >GPX3_panTro_28 atgag aa tgag 247 >GPX3_homSap_28 atgag aa tgag 246 >DIO1_ornAna_24 atgag aa tgat 339 >SEPW_dasNov_29 atgag aa tgat 331 >GPX1_loxAfr_25 atgag aa tgat 289 >GPX1_oryCun_24 atgag aa tgat 282 >GPX1_macMul_26 atgag aa tgat 276 >GPX1_panTro_20 atgag aa tgat 275 >GPX1_homSap_20 atgag aa tgat 274 >SELK_eriEur_37 atgag aa tgat 236 >SELK_sorAra_41 atgag aa tgat 235 >SELK_cavPor_34 atgag aa tgat 233 >SELK_ratNor_33 atgag aa tgat 232 >SELK_musMus_32 atgag aa tgat 231 >SELK_tupBel_37 atgag aa tgat 230 >GPX6_loxAfr_19 atgat aa agaa 272 >GPX6_dasNov_20 atgat aa agaa 271 >GPX6_cavPor_20 atgat aa agaa 265 >GPX6_sorAra_23 atgat aa agag 270 >GPX6_bosTau_23 atgat aa agag 268 >GPX6_macMul_22 atgat aa agag 264 >GPX6_homSap_18 atgat aa agag 262 >GPX3_monDom_23 atgat aa agag 258 >GPX3_loxAfr_16 atgat aa agag 256 >GPX3_equCab_26 atgat aa agag 254 >GPX6_equCab_25 atgat aa cagaa 267 >SEPW_musMus_34 atgat aa ctgat 328 >SELV_loxAfr_34 atgat aa ctgat 184 >SELV_cavPor_34 atgat aa ctgat 181 >SELV_ratNor_33 atgat aa ctgat 180 >SELV_musMus_33 atgat aa ctgat 179 >SELV_panTro_34 atgat aa ctgat 177 >SELV_homSap_34 atgat aa ctgat 176 >SELM1_anoCar_32 atgat aa ctgat 79 >SELM1_ornAna_27 atgat aa ctgat 78 >SEPHS2_dasNov_18 atgat aa ggag 61 >DIO2_monDom_29 atgat aa ggat 346 >DIO2_dasNov_27 atgat aa ggat 345 >DIO2_canFam_27 atgat aa ggat 344 >DIO2_ratNor_28 atgat aa ggat 343 >DIO2_musMus_29 atgat aa ggat 342 >DIO2_macMul_30 atgat aa ggat 341 >DIO2_homSap_30 atgat aa ggat 340 >MSRB1_macMul_22 atgat aa ggat 82 >MSRB1_panTro_23 atgat aa ggat 81 >MSRB1_homSap_26 atgat aa ggat 80 >SEPP1a_oryCun_33 atgat aa ggat 8 >GPX2_equCab_28 atgat aa tgaa 304 >SELK_anoCar_34 atgat aa tgaa 244 >SELH_anoCar_27 atgat aa tgaa 203 >SELH_monDom_27 atgat aa tgaa 202 >SELH_echTel_27 atgat aa tgaa 201 >SELH_dasNov_26 atgat aa tgaa 199 >SELH_sorAra_24 atgat aa tgaa 198 >SELH_ratNor_24 atgat aa tgaa 192 >SELH_musMus_25 atgat aa tgaa 191 >SELH_tupBel_27 atgat aa tgaa 190 >SELH_macMul_27 atgat aa tgaa 188 >SELH_panTro_24 atgat aa tgaa 187 >SELH_homSap_24 atgat aa tgaa 186 >TXNRD2_cavPor_34 atgat aa tgaa 125 >TXNRD2_musMus_37 atgat aa tgaa 123 >SEPHS2_musMus_22 atgat aa tgaa 55 >SEPP1b_anoCar_16 atgat aa tgaa 35 >SEPP1b_monDom_16 atgat aa tgaa 33 >SEPP1b_echTel_18 atgat aa tgaa 32 >SEPP1b_loxAfr_14 atgat aa tgaa 31 >SEPP1b_dasNov_14 atgat aa tgaa 30 >SEPP1b_bosTau_13 atgat aa tgaa 29 >SEPP1b_equCab_16 atgat aa tgaa 28 >SEPP1b_canFam_16 atgat aa tgaa 27 >SEPP1b_sorAra_15 atgat aa tgaa 26 >SEPP1b_oryCun_16 atgat aa tgaa 25 >SEPP1b_tupBel_16 atgat aa tgaa 24 >SEPP1b_macMul_20 atgat aa tgac 23 >SEPP1b_panTro_20 atgat aa tgac 22 >SEPP1b_homSap_20 atgat aa tgac 21 >GPX2_echTel_30 atgat aa tgag 307 >GPX2_loxAfr_27 atgat aa tgag 306 >GPX2_bosTau_26 atgat aa tgag 305 >GPX2_felCat_22 atgat aa tgag 303 >GPX2_canFam_23 atgat aa tgag 302 >GPX2_eriEur_25 atgat aa tgag 301 >GPX2_sorAra_29 atgat aa tgag 300 >GPX2_ratNor_31 atgat aa tgag 298 >GPX2_musMus_34 atgat aa tgag 297 >GPX2_tupBel_29 atgat aa tgag 296 >GPX2_otoGar_28 atgat aa tgag 295 >GPX2_macMul_30 atgat aa tgag 294 >GPX2_panTro_30 atgat aa tgag 293 >GPX2_homSap_30 atgat aa tgag 292 >SELK_oryCun_39 atgat aa tgag 234 >MSRB1_equCab_24 atgat aa tgag 90 >TXNRD2_eriEur_35 atgat aa tgag 126 >DIO1_eriEur_26 atgat aa tgat 338 >DIO1_macMul_29 atgat aa tgat 334 >DIO1_ponPyg_29 atgat aa tgat 333 >DIO1_homSap_29 atgat aa tgat 332 >SEPW_macMul_34 atgat aa tgat 327 >SEPW_ponPyg_31 atgat aa tgat 326 >SEPW_panTro_36 atgat aa tgat 325 >SEPW_homSap_36 atgat aa tgat 324 >GPX2_monDom_26 atgat aa tgat 308 >GPX1_ornAna_26 atgat aa tgat 291 >GPX1_monDom_26 atgat aa tgat 290 >GPX1_equCab_27 atgat aa tgat 286 >GPX1_felCat_28 atgat aa tgat 285 >GPX1_canFam_22 atgat aa tgat 284 >GPX1_musMus_30 atgat aa tgat 279 >GPX1_otoGar_25 atgat aa tgat 277 >SELK_ornAna_34 atgat aa tgat 243 >SELK_monDom_22 atgat aa tgat 242 >SELS_galGal_36 atgat aa tgat 149 >SELS_ornAna_33 atgat aa tgat 147 >SELS_monDom_41 atgat aa tgat 146 >SELS_dasNov_36 atgat aa tgat 144 >SELS_eriEur_37 atgat aa tgat 142 >SELS_bosTau_41 atgat aa tgat 141 >SELS_equCab_41 atgat aa tgat 140 >SELS_felCat_41 atgat aa tgat 139 >SELS_canFam_39 atgat aa tgat 138 >SELS_cavPor_40 atgat aa tgat 137 >SELS_ratNor_40 atgat aa tgat 136 >SELS_musMus_42 atgat aa tgat 135 >SELS_tupBel_40 atgat aa tgat 134 >SELS_otoGar_38 atgat aa tgat 133 >SELS_macMul_37 atgat aa tgat 132 >SELS_panTro_37 atgat aa tgat 131 >SELS_homSap_37 atgat aa tgat 130 >SEPN_monDom_27 atgat aa tgat 50 >SEPN_bosTau_32 atgat aa tgat 47 >SEPN_equCab_38 atgat aa tgat 46 >SEPN_felCat_29 atgat aa tgat 45 >SEPN_canFam_35 atgat aa tgat 44 >SEPN_sorAra_34 atgat aa tgat 43 >SEPN_cavPor_35 atgat aa tgat 42 >SEPN_ratNor_35 atgat aa tgat 41 >SEPN_musMus_35 atgat aa tgat 40 >SEPN_macMul_32 atgat aa tgat 39 >SEPN_panTro_32 atgat aa tgat 38 >SEPN_homSap_32 atgat aa tgat 37 >SEPP2_macEug_26 atgat aa tgat 205 >MSRB1_bosTau_28 atgat aa tgat 91 >MSRB1_felCat_32 atgat aa tgat 89 >MSRB1_canFam_29 atgat aa tgat 88 >MSRB1_cavPor_26 atgat aa tgat 86 >MSRB1_ratNor_22 atgat aa tgat 85 >MSRB1_musMus_24 atgat aa tgat 84 >MSRB1_otoGar_25 atgat aa tgat 83 >GPX6_eriEur_24 atgat ca agag 269 >GPX6_canFam_24 atgat ca agag 266 >SELO_macMul_26 atgat cc tgag 173 >SELO_panTro_26 atgat cc tgag 172 >SELO_homSap_26 atgat cc tgag 171 >SEPW_canFam_34 atgat ga ctgat 330 >SELH_cavPor_19 gtgat ca tgaa 193 >GPX4_tupBel_31 ttgac aa cgag 313 >TXNRD3_bosTau_13 ttgac ag cgac 119 >TXNRD3_calJac_25 ttgac ga tgat 114 >TXNRD3_ratNor_25 ttgac gg cgac 116 >TXNRD3_musMus_22 ttgac gg cgac 115 >TXNRD3_macMul_18 ttgac gg cgac 113 >TXNRD3_felCat_20 ttgac gg cgat 118 >TXNRD3_canFam_27 ttgac gg cgat 117 >TXNRD3_homSap_19 ttgac gg tgac 111
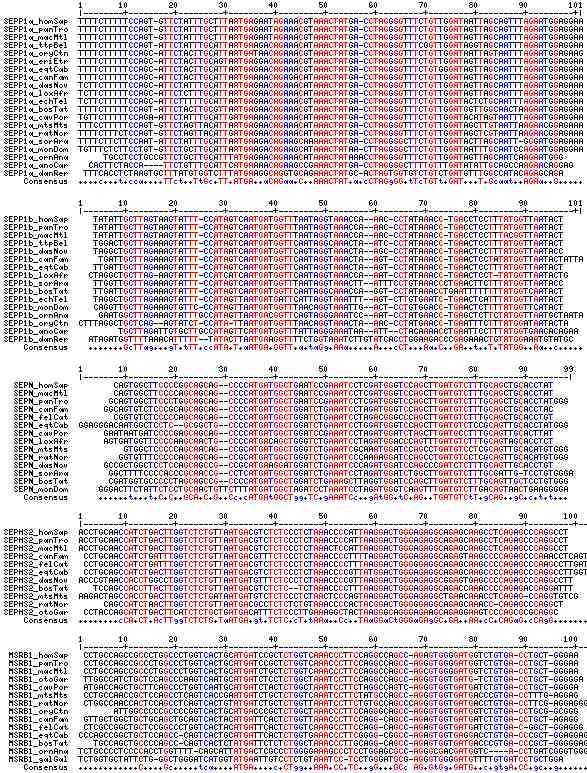
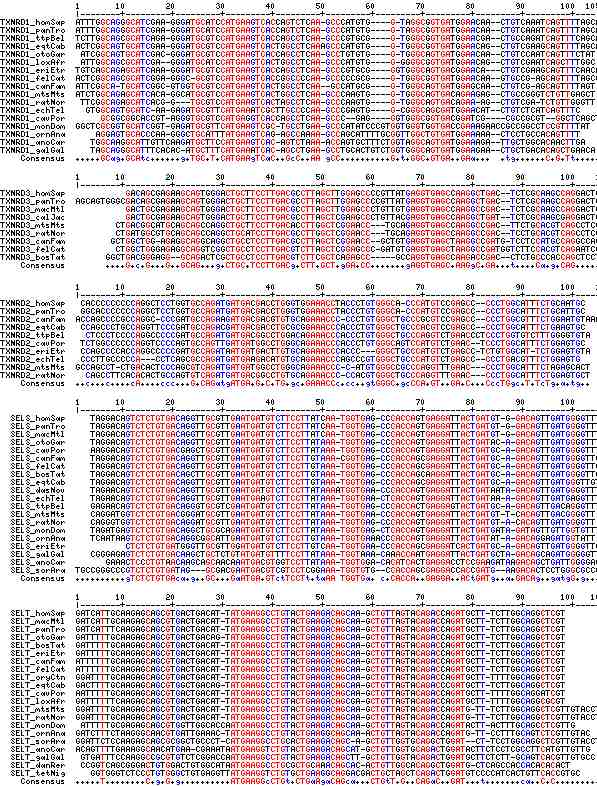
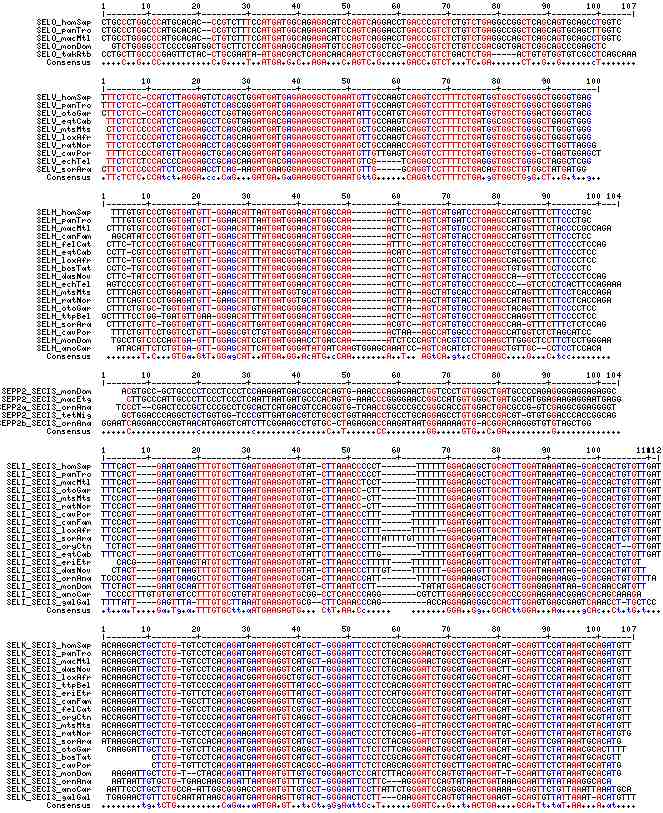
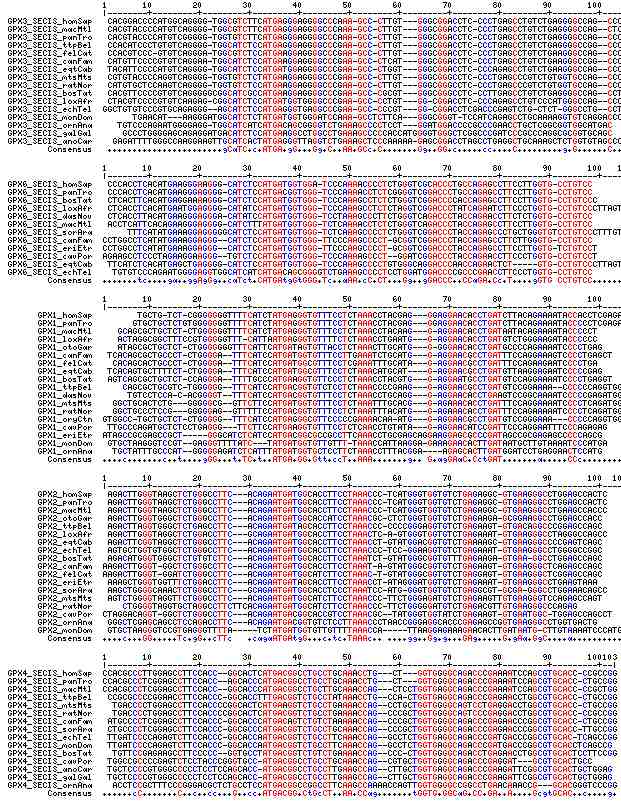
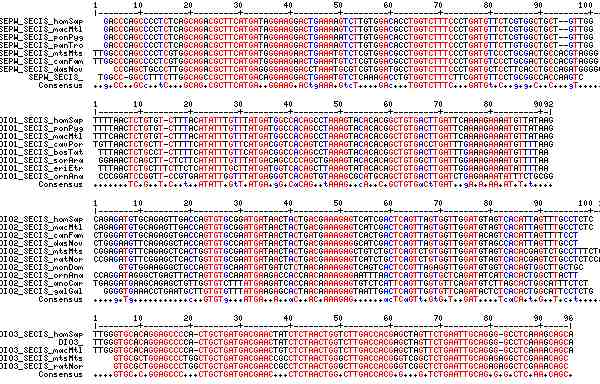
Gene-by-gene colored alignment of SECIS elements
Here the SECIS elements are colored red when identical at 90% of the input sequences and blue when 80%. These alignments would be more easily compared if the sequencs were trimmed to a common length and sporadic small insertions removed. It may be possible, especially within a given gene family, to find conserved regions in common that extend beyond the regions traditionally considered important to SECIS recognition.
Gene-by-gene text alignment of SECIS elements
Here all SECIS sequences (except SELM) are aligned using the 3 key regions identified by SECIS web tool (output spacing) as alignment anchors. To trim the sequences to a common length, 20 flanking residues were taken on the 5' and 3' sides (rather than the full-length element reported by the SECIS tool). Dots are used as spacers to bring sequence divisions to common lengths. Numbers appended to the gene name provide COVE scores.
***** ** **** * = key regions >SEPP1a_homSap_31 ccagtgttctatttgcttta atgag aatagaaacgt .. aa actatgacctaggggtttctgtt ........ ggat aattagcagtttagaatgg >SEPP1a_panTro_31 ccagtgttctatttgcttta atgag aatagaaacgt .. aa actatgacctaggggtttctgtt ........ ggat agttagcagtttagaatgg >SEPP1a_macMul_31 ccagtgttctatttgcttta atgag aatagaaacgt .. aa actatgacctaggggtttctgtt ........ ggat agttagcaatttagaatgg >SEPP1a_tupBel_31 ccagtattctatttgcatta atgaa gacagaaacat .. aa actatgacctaggggtttcggtt ........ ggat aggtagcaatttagaatgg >SEPP1a_musMus_33 ccagtgttctagttacatta atgag aacagaaacat .. aa actatgacctaggggtttctgtt ........ ggat agcttgtaattaagaacgg >SEPP1a_ratNor_33 ccagtgttctagttacattg atgag aacagaaacat .. aa actatgacctaggggtttctgtt ........ ggat agctcgtaattaagaacgg >SEPP1a_cavPor_35 ccagaattctatttgcattg atgaa aacagaaacat .. aa actatgacctaggggtttctgtt ........ ggat acatagtaatttagaatgg >SEPP1a_oryCun_33 ccagcattctacttgcatta atgat aacagaaacgt .. aa actatgacctaggggtttctgtt ........ ggat aattagtaatttagaatgg >SEPP1a_sorAra_33 ccaatattctatttgcatta atgaa gacagaagcaa .. aa actatgacctaggggcttctgtt ........ ggat acttagcaattgggatgga >SEPP1a_eriEur_33 ccagcattctacttgcagta atgag gacagaaacgt .. aa actatgacctaggggtttctgtt ........ ggat agttagcaatttagaatgg >SEPP1a_canFam_33 ccagcattctacttgcatta atgaa aacagagacat .. aa actatgacctaggggtttctgtt ........ ggat agttagcaatttagaatgg >SEPP1a_equCab_29 ccagcattctacttgcatta atgag aacagaaacgt .. aa actatgacctaggggtttctgtg ........ ggat agttagcaatttagaatgg >SEPP1a_bosTau_31 ccagtattctacttgcgtta atgag aacagaaacgt .. aa actataacctaggggtttctgtt ........ ggat ggttggcaactaagaatgg >SEPP1a_dasNov_33 ccagcattctatttgcatta atgag aacagaaacat .. aa actatgacctaggggtttctgtt ........ ggat agttagcaatttagaatgg >SEPP1a_loxAfr_33 ccagcattctctttgcatta atgag aacagaaacat .. aa actatgacctaggggtttctgtt ........ ggat agttagcaatttagaatgg >SEPP1a_echTel_33 ccagcattctttttgcatta atgag aacagaaacgt .. aa actatgacctaggggtttctgtt ........ ggat actctgcaatttagaatgg >SEPP1a_monDom_31 cctgtgttctgcttgcatta atgaa gacagaagcat .. aa actataacttaggggcttctgtt ........ ggat aatttgcaatccagaatgg >SEPP1a_ornAna_29 ctgccgttctgcttgcattt atgag aacagaaacat .. aa actataaacctaggggtttctgtt ....... ggat agttagcaatcaagaatgg >SEPP1a_anoCar_33 ctaccattctgtttgcattc atgaa aacagaagccg .. aa actatgacctaggggcttttgtt ........ tgat atattgcagccaagaacag >SEPP1a_danRer_33 aagtgctttatgtggtcttt atgaa ggcaggtgcag .. aa actatgcactagtggtgtctgtc ........ tgat gtttggccatacagagcag >SEPP1b_homSap_20 agtaagtatttccatagtca atgat ggtttaataggt . aa accaaaccctataaacc .............. tgac ctcctttatggttaatact >SEPP1b_panTro_20 agtaagtatttccatagtca atgat ggtttaataggt . aa accaaaccctataaacc .............. tgac ctcctttatggttaatact >SEPP1b_macMul_20 agtaagtatttccatagtca atgat ggtttaataggt . aa accaaaccctataaacc .............. tgac ctcctttacggttaatact >SEPP1b_tupBel_16 agaaagtatttccatagtca atgat ggttcaataggc . aa actaatccctataaacc .............. tgaa ctcctttatggttaatact >SEPP1b_oryCun_16 ctcagagtatctccatatta atgat ggtttaacaggt . aa actaaaccctatgaacc .............. tgaa tttctttatggataatact >SEPP1b_sorAra_15 agaaagtatttccatagtca atgat ggtttaataggt . aa acttatttcctgtaaaccc ............ tgaa cttctttatggttaatacc >SEPP1b_canFam_16 agaaagtatttccatagtca atgat ggttcaataggt . aa actaagtcctataaacc .............. tgaa ctcctatatggttaatact >SEPP1b_equCab_16 agaaagtatttccatagtca atgat ggttcaataggt . aa actaagtcctataaacc .............. tgaa ctcctttatggttaatact >SEPP1b_bosTau_13 agaaagtatttccatagtca atgat ggttcaataggc . aa accaggtcctataaacc .............. tgaa ttttttttatggtcaatac >SEPP1b_dasNov_14 agaaagtatttccataatca atgat ggttcagtaggt . aa actaagccctataaacc .............. tgaa ctcctttatggttaatacc >SEPP1b_loxAfr_14 tagaagtatttccatcatca atgat ggttcaataggt . aa accaaatcctataaacc .............. tgaa ctccttatggttaatactg >SEPP1b_echTel_18 agaaagtatttccatagtta atgat gattcattaggt . aa atttagtcttgtgaatc .............. tgaa ctcatttatggttaatact >SEPP1b_monDom_16 agaaagtgttcccatagtta atgat ggtttaacaggt . aa attgactcctgtggacc .............. tgaa ctcttttatggttcatact >SEPP1b_ornAna_18 gaaagtatttgccatagtta atgac ggttcagtaggg . aa atccaatcctatgaacc .............. tgag ctcttctatggttaatgct >SEPP1b_anoCar_16 gattgtgcttgccatagttc atgat ggtttattaggg . aa ataaattctacaaaacc .............. tgaa ttcctttatggtgaacaca >SEPP1b_danRer_24 ttaaacattttttatactta atgaa ggttttctggt .. aa atcttgtatcacctggaagac .......... ccgag aaactgtatggaaatgtat >SEPN_homSap_32 .. gcttccccggcagcagcccc atgat ggctgaatccg .. aa atcctcgatgggtccagct ............ tgat gtctttgcagctgcaccta >SEPN_panTro_32 .. gcttccctggcagcagcccc atgat ggctgaatccg .. aa atcctcgatgggtccagct ............ tgat gtctttgcagctgcaccta >SEPN_macMul_32 .. gcttccccagcagcagcccc atgat ggctgaatccg .. aa atcctcgatgggtccagct ............ tgat gtctttgcagctgcaccta >SEPN_musMus_35 .. gctcccccagcaacagccgc atgat ggctgggtctg .. aa atcgcaaatggatccagcc ............ tgat gtcctcgcagttgcacatg >SEPN_ratNor_35 .. tttcccccagcagcagccgc atgat ggctgggtctg .. aa atcccaaaaggatccagcc ............ tgat gtcctcgcagttgcacatg >SEPN_cavPor_35 .. gatccccgagcagcagcccc atgat ggctggatccg .. aa atcctagatggatctagct ............ tgat gcctttgcagttgcactta >SEPN_sorAra_34 .. tccccacccgcaaccgcctc atgat ggctggatctg .. aa atcctagatggatctggct ............ tgat gtctttgcgattgtcctgt >SEPN_canFam_35 .. tctcccgcagcagcagcccc atgat ggctggatctg .. aa atcctagatgggtccagct ............ tgat gtctttgcagctgcaccta >SEPN_felCat_29 .. tctcccccagcagcagcccc atgat ggctggatctg .. aa atcctagacgggcccagct ............ tgat gtctttgcagctgcacctg >SEPN_equCab_38 .. tggctcctcgcggctgcccc atgat ggctggatctg .. aa atcctggatgggtccagct ............ tgat gtctccgcagttgcaccta >SEPN_bosTau_32 .. tgccccctagcagccgcccc atgat ggctggatctg .. aa agcttaggtggatccagct ............ tgat gtctttgcagttgctcctg >SEPN_dasNov_27 .. gctcctccagcagccgccgc atgaa ggatggatctg .. aa atcccagatggatccagcc ............ cgat ctctttgcagttgcacctg >SEPN_loxAfr_32 .. gttccccaagcaactgcccc atgac agctgggtctg .. aa atcctagatggacccagtt ............ tgat gtctttgcagtagcacctc >SEPN_monDom_27 .. ttctcctgcaactgttcttt atgat ggctagatcct .. aa atcctagatgggtcaagtt ............ tgat gtcttgacagtaactgaag >SEPHS2_homSap_21 atctgacttggtctctgtta atgac gtctctccctct . aa accccattaaggactgggagaggc ....... agag caagcctcagagcccaggc >SEPHS2_panTro_21 atctgacttggtctctgtta atgac gtctctccctct . aa accccattaaggactgggagaggc ....... agag caagcctcagagcccaggc >SEPHS2_macMul_21 atctgacttggtctctgtta atgac gtctctccctct . aa actccattaaggactgggagaggc ....... agag caagcctcagagcccaggc >SEPHS2_otoGar_12 atctgacttcatctctgctg atgac atttctcccttg . aa aggctactaagggcaggg ............. agaa gcagaggaagtcccagcgc >SEPHS2_musMus_22 acctgacttggtctctgata atgat gtctctccctct . aa ctcccagtaaggactgggagaggc ....... tgaa caaacctcagagccaggtg >SEPHS2_ratNor_19 atctaacttggtctctgtta atgac gcctctctctgt . aa accccactacggactgggggaggc ....... agag caaacccagagcccaggct >SEPHS2_canFam_20 atctgatttggtctctgtta atgac gtttctccctct . aa accccttttaggactgggagaggc ....... agag taagccccagagcccaaac >SEPHS2_felCat_22 atctgatttggtctctgtta atgac gtctctccctct . aa acccctttaaggactgggggaggc ....... agag caagccccagagcccaagc >SEPHS2_equCab_22 atctgacttggtctctgtta atgac gtctctccctct . aa accccgttatggactgggagaggc ....... agag caagccccagagcccaggc >SEPHS2_bosTau_22 accttacttggtctctgtta atgac gtctctctcta .. aa cccctttaaggactgggagaggc ........ agac caaaccccagagaccagga >SEPHS2_dasNov_18 acctggcctggtctctgtta atgat gtttctccctct . aa accccgttaaggactgggagaggc ....... ggag tgagccccagagcccaggc >SELM1_homSap_27 . attcacaaagatttgcgtta atgaa gactacacaga .. aa acctttctagggatttgtgtggat ....... cagat acatacttggcaaattttt >SELM1_panTro_28 . attcacaaagatttgcgtta atgaa gactacacaga .. aa acctttctaaggatttgtgtggat ....... cagat acatacttggcaaattttt >SELM1_macMul_28 . attcacaaagatttgcgtta atgaa gactacacaga .. aa accttcctaaggatttgtgtggat ....... cagat acatacttggcaaattttt >SELM1_otoGar_28 . attcacaaaaatttgcatta atgaa gactacacaga .. aa acctttcagaggatttgtgtggat ....... ctgat acttggcaaattttgagtt >SELM1_tupBel_25 . attcacaaagatttgcgtta atgaa gactgcacaga .. aa acctttctagggatttgtgtggat ....... ccgat aattggcaaatttttgtat >SELM1_musMus_35 . acttataaaggtttgcatta atgag gattacacaga .. aa acctttgttaaggacttgtgtagat ...... ctgat aattggcaaatttttattt >SELM1_ratNor_30 . acttataaaggtttgcatta atgag gattacacaga .. aa acctttgttaagggtttgtgtcgat ...... ctgat aattggcaaatttttattt >SELM1_cavPor_31 . ggccacaaaaacttgcatta atgag gactatacaga .. aa accttcctaaggatttgtatagat ....... ctgat gtatggcaaatttttgttt >SELM1_sorAra_26 . ttgcacaaagattcgcatta atgaa ggctacacagg .. aa accttactaaggatttgtgtggat ....... ctgat aactagcaaatttttgtgc >SELM1_eriEur_27 . actcacaaaggtttgcatcc atgaa ggctacataga .. aa accttactgaggatttgtgtagat ....... ctgat gctcagcaaacttctgtgt >SELM1_canFam_28 . cttcacagagatttgcatta atgaa gactacacaga .. aa accttcaggaggatttgtgtggat ....... ctgat atttagcaaaattttgtgt >SELM1_felCat_30 . attcacaacgatttgcatta atgaa gactacacaga .. aa accttcatgaggatttgtgtggat ....... ctgat gatatttagcaaatttttg >SELM1_equCab_29 . attcacaaagattcgcgtta atgaa gactacacaga .. aa acctttctgaggatttgtgtggat ....... ctgat atccggcaaatttttgtgc >SELM1_bosTau_24 . tcacacaaagatttgcatta atgaa gactacacaga .. aa accttcctgaggatttgtgtggac ....... ctgat acttagcaaatttttgtgc >SELM1_dasNov_24 . atttacaaagatttgcacta atgaa gactacacaga .. aa acctttctaaggatttgtatggat ....... ctgat atttggcaaatttttgtgt >SELM1_loxAfr_27 . attcacaaagatttgcatta atgaa gactgcacaga .. aa acctttgtaaggatttgtgtggat ....... ctgat atttggcaagtttttgtgt >SELM1_ornAna_27 . catgaagacaatgcgcatta atgat gactacacaga .. aa acctatcatggatttgtgtatat ........ ctgat gtttcgtgtatttcctcag >SELM1_anoCar_32 . tgccagcaaaatactcttta atgat gactacatgta .. aa gcctttaacaaaaggcatgtgtaggt ..... ctgat gtcagatgtacttttgtta >MSRB1_homSap_26 . gccctggccctggtcactgc atgat ccgctctggtc .. aa acccttccaggccagccagagtgg ....... ggat ggtctgtgacctgctggga >MSRB1_panTro_23 . gccctggccctggtcactgc atgat ccgctctggtc .. aa acccttccaggccagccagggtgg ....... ggat ggtctgtgacctgctggga >MSRB1_macMul_22 . gccctggccctggtcactgc atgat ctgctctggtc .. aa acccttccaggccagccagcgtgg ....... ggat ggtctgtgacctgctggga >MSRB1_otoGar_25 . gctccagcccaagtcaatgc atgat ctcctctggct .. aa atccttccaggccagccagggtgg ....... tgat gtctgtgacctgctggggg >MSRB1_musMus_24 . gctccagcctcagtcaccga atgat ctgctctggtc .. aa atccttctatgccagccagggtgg ....... tgat gacccgtgacctttgagga >MSRB1_ratNor_22 . actccagcctcagtcactgt atgat ctgctctggtt .. aa atccttctgtgccagcccagggtgg ...... tgat gacctgtgaccttcgagga >MSRB1_cavPor_26 . gcttcagccctggtcaatgc atgat ctactctggct .. aa atccttctaggccagtcagggtgg ....... tgat gatctgtgacctgctgggg >MSRB1_oryCun_28 . gcccccgccccggtcactgc atgac ccgctctggct .. aa acccttccagggcagccagggtgg ....... tgat ggtctgtgacctgcggcgg >MSRB1_canFam_29 . gctgcagctgcagtcactac atgat tcactctggtt .. aa acccttccaggcagccagagtgg ........ tgat gatctgtgacctgctagag >MSRB1_felCat_32 . gctccggccctggtcactac atgat tcactctggtc .. aa agccttccaggcagccagagtgg ........ tgat gatctgtgacggctggagg >MSRB1_equCab_24 . tgctccagcccagtcactgc atgat tcactctggtg .. aa acccttcggggcggccagagtgg ........ tgag gacctgtgacccgctggag >MSRB1_bosTau_28 . tgccccagcccagtcactct atgat tctctctggct .. aa acccttgcaggcagccagagtgg ........ tgat gacccgtgacccgctgggg >MSRB1_ornAna_26 . cccacctggttttcagcatt atgac tcgccctgggg .. aa agccctcctggagcgccagggcga ....... cgat ggtcacctgatcgggtggg >MSRB1_galGal_22 . ttctgggctgggatcatggt atgaa ttgtcctctgt .. aa atcctcctgggatgcgagggtgg ........ ggat gttttgtgatcctgctgga >TXNRD1_homSap_25 ggcatcgaagggatgcatcc atgaa gtcaccagtctc . aa gcccatgtggtaggcggtgat .......... ggaa caactgtcaaatcagtttt >TXNRD1_panTro_25 ggcatcgaagggatgcatcc atgaa gtcaccagtctc . aa gcccatgtggtaggcggtgat .......... ggaa caactgtcaaatcagtttt >TXNRD1_otoGar_23 tgcatcgacgggatgcgtcc atgaa gtcaccagcctc . aa gcctgtgtggtgggcagtgat .......... ggaa caactgtccaatcagtttc >TXNRD1_tupBel_25 cgcatcagagggatgcgtcc atgaa gtcaccagcctc . aa gcccgtgcggtgggcggtgat .......... ggaa cgactgccagatcagtttc >TXNRD1_musMus_28 agcatcacaggcatgcgtcc atgaa gtcactggcctc . aa gcccaagtggtgggcagtgac .......... agaa gagctgccgggtctgttga >TXNRD1_ratNor_28 cagagcatcacggtgcgtcc atgaa gtcactagcctc . aa gcccaagtggtgggcagtgac .......... agaa agctgtcgatctgttgggt >TXNRD1_cavPor_24 cggcaccgtagggtgcgtcc atgag gtcaccagcctc . aa gcccgagggtgggcggtgac ........... ggat cgcgccgcgtggctcagct >TXNRD1_eriEur_26 agcatcaaagggatgcgtcc atgaa gtcaccagcctc . aa gcccgtgcgggtgggcagtgac ......... ggaa cactgtcgaagcagtttca >TXNRD1_canFam_25 tgcatcggcgtggtgcgtcc atgaa gtcactggcctc . aa gccatgcggtgggcagtgat ........... ggag caactgtcgagcagtttta >TXNRD1_felCat_24 gcgcatcggagggcgcgtcc atgaa gtcaccggcccc . aa gcccccgcggtgggcggtgat .......... ggaa caagtgccgagcagtttta >TXNRD1_equCab_22 tgcatcgaagggatgcgtcc atgaa gtcactggcctc . aa agcccatgtggtgggcggtgat ......... ggaa cagctgtcgaagcagtttt >TXNRD1_loxAfr_23 gcgcatcgagggatgcatcc atgaa gtcactggcctc . aa gcccatgtggggggcggtgat .......... ggaa cagctgtcgaatcagcttt >TXNRD1_echTel_20 gtgcatcaagagatgcgttc atgaa atcgcttgcccc . aa gcccgagtggcgggcag .............. cgat ggaacatctgtctcatcag >TXNRD1_monDom_23 tgcatcggtgagatgcgttc atgaa gtcgctgcctg .. aa gcccatatcccgtggtgggtggtgac ..... cgaa agaaccgccggcctccgtt >TXNRD1_ornAna_19 tgcacccaagggctgcattt atgaa gtcagagccaa .. aa gccagcattttgcggttggctgtgat ..... ggaa aaactcctgccacagtttt >TXNRD1_anoCar_33 gcattgttcaagatgcttcc atgaa gtcacagtcta .. aa accagtgctttctggtaggcagtgat ..... ggaa agattgctggcacaacttg >TXNRD1_galGal_27 ggcatttcacacatgctttc atgaa atcacagcctg .. aa gcctgcactgtctggtgggcagtgat ..... ggaa gaactgctgacacagctga >TXNRD3_homSap_19 gaagcagtgggactgcttcc ttgac gccttagctt ... gg agccccgttatgaggtgagccaaggc ..... tgac tctcgcaagccaggactga >TXNRD3_macMul_18 gaagcagtgggactgcttcc ttgac gccttagctt ... gg agccctgttgtgaggtgagccaaggc ..... cgac tctcgcaagccaggactca >TXNRD3_calJac_25 gaagcagtgggactgcttcc ttgac gccttagctc ... ga agccctgttacgaggtgagccaaggc ..... tgat tctcgcaagcgaggactga >TXNRD3_musMus_22 gcagcagccaggctgcttcc ttgac accttggctc ... gg aacctgcagaggtgagccaaggc ........ cgac ttctgcacgtcagcctcga >TXNRD3_ratNor_25 gcagcagccaggctgcatcc ttgac gccttggctc ... gg aacctgcagaggtgagccaaggc ........ cgac tcctgcacgtcagcctcga >TXNRD3_canFam_27 gaggcaggcaggctgcctcc ttgac gccttagctc ... gg aaccgctgtgaggtgagctaaggc ....... cgat gtcctccatgccaggccag >TXNRD3_felCat_20 gaggcaggtcggctgcctcc ttgac gtcttagctc ... gg agcccgatgtgaggtgagctaaggc ...... cgat ggtcttccacgtcagaatc >TXNRD3_bosTau_13 agggcagactcgctgcctcc ttgac gtcttcgctc ... ag agccgccaggtgagccaagac .......... cgac ctctgcccaccagctcctc >TXNRD2_homSap_31 caggctcctggtgccagatg atgac gacctgggtgg .. aa acctaccctgtgggcacccatgtc ....... cgag ccccctggcatttctgcaa >TXNRD2_panTro_31 caggctcctggtgccagatg atgac gacctgggtgg .. aa acctaccctgtgggcacccatgtc ....... cgag ccccctggcatttctgcat >TXNRD2_tupBel_37 caggcccccgatgccagatg atgac ggcctggacag .. aa acccaccctgtgggctgcccaggtc ...... tgaa ccctccctggtgtctttgg >TXNRD2_musMus_37 gacactcccagcgtcagatg atgat ggcctgggcag .. aa accccatgtgggccgcccaggtt ........ tgaa cccctggcatttctagagc >TXNRD2_ratNor_35 cacactgccagtgtcagatg atgac ggcctgtgcag .. aa acccccacgtgggctgcccaggtt ....... tgaa cccctggcatttctggagt >TXNRD2_cavPor_34 caggtccccagtgccagttg atgat ggcctgggcag .. aa acccaccctgtgggcagtccatgtc ...... tgaa ctccctggcatttctggag >TXNRD2_eriEur_35 caggcccccgatgccagata atgat gacttgtgcag .. aa acccacccgggctgcccatgtc ......... tgag cctctgtggcattctggag >TXNRD2_canFam_34 ccaggccccgatgccagaag atgac gacgtgtgcag .. aa acccccctgtgggctgcccgcgtc ....... cgag ccccctggcgtttctggaa >TXNRD2_equCab_31 caggttcccgatgccagacg atgac gacctgcgcgg .. aa acccaccctgtgggctgcccacgtc ...... cgag ccccctggcatttctgaag >TXNRD2_echTel_37 ccccacctcagcgccagatg atgaa gacatgtgcag .. aa acccagcccgtgggctgcccatgtc ...... tgag ccccctgacgtttctggag >SELS_homSap_37 .. tctgtgacaggttgcgttga atgat gtcttccttatc . aa tggtgagcccaccagtgaggattac ...... tgat gtggacagttgatggggtt >SELS_panTro_37 .. tctgtgacaggttgcgttga atgat gtcttccttatc . aa tggtgagcccaccagtgaggattac ...... tgat gtggacagttgatggggtt >SELS_macMul_37 .. tctgtgacaggttgcgttga atgat gtcttccttatc . aa tggtgagcccaccagtgaggattac ...... tgat gtggacagttgatggggtt >SELS_otoGar_38 .. tctgtgacaggttgcgttga atgat gtcttccttat .. aa atggtgagcccaccagtgaggattac ..... tgat acagacagttgatggggtt >SELS_tupBel_40 .. tctgtgacagggtgcgtcga atgat gtcttccttat .. aa atggtgagcccaccactgaggagtac ..... tgat gcagacagttgacagggtt >SELS_musMus_42 .. tctgtgacgggatgcgttga atgat gtcttccttat .. aa atggtgaacccaccagtgaggattac ..... tgat gttcacagttgacggggtt >SELS_ratNor_40 .. tctgtgacaggatgcgttga atgat gtcttccttat .. aa atggtgagcccaccagtgaggattac ..... tgat gtacacagttgatggggtt >SELS_cavPor_40 .. tctgtgacgagctgcgttga atgat gtcttccttat .. aa atggtgagcccaccagtgaggattac ..... tgat gcagacagttgatggggtt >SELS_canFam_39 .. tctgtgacaggttgcgttga atgat gtcttccttgt .. aa acggtgagcccaccagcgaggattac ..... tgat gcagacagttgatggggtt >SELS_felCat_41 .. tctgtgacaggttgcgttga atgat gtcttccttgt .. aa atggtgagcccaccagcgaggattac ..... tgat gcagacagttgatggggtt >SELS_equCab_41 .. tctgtgacaggttgcgttga atgat gtcttccttgt .. aa atggtgagcccaccagcgaggattac ..... tgat gcagacagttgatgggttg >SELS_bosTau_41 .. tctgtgacagcttgcgttga atgat gtcttccttgt .. aa atggtgagcccaccagcaaggattac ..... tgat gcagacagttgatggggtt >SELS_eriEur_37 .. tctgtgatgggttgcgttgg atgat gtcttccttgtc . aa tggtgaacccaccagcgaggatcac ...... tgat gcagacagttgatggggtt >SELS_sorAra_36 .. gtctctgtgataggcgacga atgac gtcgtcctcgg .. aa atggtgtgccaccagcgaggaccac ...... cgat gaagacactcctgggcgcc >SELS_dasNov_36 .. tctgtgacaggttgcgttga atgat gtcttccttat .. aa aatggtgaacccaccagtgaggattac .... tgat aatagacagttgatggggt >SELS_echTel_38 .. tctgtgacaggttgcgttga atgaa gtcttccttat .. aa atggtgaactcaccagtgaggattac ..... tgat aaagacagttgatgaggtt >SELS_monDom_41 .. tctgtgacaggctgcgcaga atgat gtcttccttat .. aa atggtgagctcaccagtgaggattac ..... tgat gatagatagttgttggtgt >SELS_ornAna_33 .. tctgtgacaggcggcattga atgat gtcttccttgt .. aa atggtgaaacccaccagtgaggattac .... tgat atagacaggagatggtatt >SELS_anoCar_25 .. cctgtaacaagcagcaacaa atgac gtggtccttat .. aa atggtggacacatcactgaggacctc ..... cgaa gataagacagctgattggg >SELS_galGal_36 .. tctgtgacaagctgctctgt atgat gttttccttat .. aa atggtaaacaaaccaatgaggattac ..... tgat gctagacagcagatggggg >SELT_homSap_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gctttcttggcaggctcgt >SELT_panTro_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gctttcttggcaggctcgt >SELT_macMul_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gctttcttggcaggctcgt >SELT_otoGar_39 .. agagcagcgtgactgacagt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gctttcttggcaggctcgt >SELT_musMus_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gtttcttggcaggctcgtt >SELT_ratNor_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gtttcttggcaggctcgtt >SELT_cavPor_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gcttttttggcaggatcgt >SELT_oryCun_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gcttttttggcaggctcgt >SELT_canFam_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gctttcttggcaggctcgt >SELT_felCat_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gctttcttggcaggctcgt >SELT_equCab_38 .. agagcagcgtggctgacagt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... cgat gctttcttggcaggctcgt >SELT_bosTau_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gctttcttggcaggctcgt >SELT_eriEur_39 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat gctttcttggcaggctcgt >SELT_sorAra_36 .. agagcagcgcggctgccctc atgaa ggcctgcactg .. aa gacagcagctgttggtgcaggct ........ ggat cttcctggcaggctcgttg >SELT_loxAfr_36 .. agagcagcgtgactgacatt atgaa ggcctgtactg .. aa gacagcgagctgttagtacagacc ....... agat gcttttttggcaggcgcgt >SELT_monDom_37 .. gagcagcgtggttggcacca atgaa ggcctgtactg .. aa gacagcaagctgttagtacagacc ....... agat acttactttgcagcctcgt >SELT_ornAna_36 .. agggcaacgtgattgaaact atgaa ggcctgtactg .. aa ggcagcaaactgttagtacagacc ....... agat gttcctttgcagtctcgtt >SELT_anoCar_34 .. aaggcaacatgaacgaaata atgaa ggtctgtactg .. aa gacagcatgctgttggtgcagact ....... ggat acttctcctcgccttcatg >SELT_galGal_38 .. aggccgcgtgtctcggacca atgaa ggcctgtactg .. aa gacagcttgctgttggtacagact ....... ggat gcttctcttgcagtcacgt >SELT_danRer_35 .. ggactgtggactgtggcata atgaa ggcctgcgctgc . aa acagcacactgttggcacaggct ........ ggat gctcagccaccacacacac >SELT_tetNig_33 .. tccctgtgggctgtgaggtt atgaa ggtctgtgctg .. aa ggcaggacgactgctagctcagact ...... ggat gtccccatcactgttcacc >SELO_homSap_26 .. ccatgcacacccgtctttcc atgat ggcagagacat .. cc agtcaggacctgacccgtctctgtc ...... tgag gccggctcagcagtgcagc >SELO_panTro_26 .. ccatgcacacccgtctttcc atgat ggcagagacat .. cc agtcaggacctgacccgtctctgtc ...... tgag gccggctcagcagtgcagc >SELO_macMul_26 .. ccatgcacacctgtctttcc atgat ggcagagacat .. cc agtcaggacctgacccgtctctgtc ...... tgag gccagctcagcagtgcagc >SELO_monDom_24 .. tccccgatggctgcttctcc atgaa ggcagagatgt .. cc agtcggctccgacccgtctctgtc ....... cgac gctgactcggcagcccgag >SELO_takRub_20 .. ccgagttctacctgcgaata atgac gactcagacaa .. ca agtctgccagtgacctgtctgactc ...... tgaa ctgtgtggtgtcgcctcag >SELV_homSap_34 .. atcttaggagtctcagctgg atgat gagaagggctg .. aa atgttgccaagtcaggtcctttt ........ ctgat ggtggctggggctggggtg >SELV_panTro_34 .. atcttaggagtctcagcggg atgat gagaagggctg .. aa atgttgccaagtcaggtcctttt ........ ctgat ggtggctggggctggggtg >SELV_otoGar_27 .. atctcaggagcctcggtagg atgac gagaagggctg .. aa atattgccatgtcaggtcctttt ........ ctgat ggtggctgggactggggta >SELV_musMus_33 .. atctcaggagcctcagcagg atgat gagaagggctg .. aa atgctgccaaaccaggtcctttt ........ ctgat ggtggctggggcttgggtg >SELV_ratNor_33 .. gtctcaggaacctcagcggg atgat gagaagggctg .. aa atgctgccaaaccaggtcctttt ........ ctgat ggtggctggggcttggtta >SELV_cavPor_34 .. atgttaggagctgcagcagg atgat gagaagggctg .. aa atgttgttgagtcaggtcctttt ........ ctgat ggtggctgggctgagtgga >SELV_equCab_33 .. atctcaggagcctcggtgag atgac gagaagggctg .. aa atgttgccaagtcaggtcctttt ........ ctgat ggcggctggggctgaggtg >SELV_sorAra_26 .. catctcaggaacctcagaag atgaa gggaagggctg .. aa atgttggcaggtcctttt ............. ctgac agtggctgtggctatgatg >SELV_loxAfr_34 .. atctcaggagcctcagcaag atgat gagaagggctg .. aa atgttgccaagtcaggtcctttt ........ ctgat ggtggctggggctggggtg >SELV_echTel_26 .. accccaggagccgcagcaag atgac gagaagggctg .. aa atgtcgtcaggccctttt ............. ctgag ggtggctggggctaggctc >SELH_homSap_24 .. ctggtgatgttggaacatta atgat ggaacatggcc .. aa acttcagtcatgatcc ............... tgaa gccatggtttcttccctgc >SELH_panTro_24 .. ctggtgatgttggaacatta atgat ggaacatggcc .. aa acttcagtcatgatcc ............... tgaa gccatggtttcttccctgc >SELH_macMul_27 .. ctggtgatgctggaacatta atgat ggaacatggcc .. aa acttcagtcatgtgcc ............... tgaa gccatggtttctaccccgc >SELH_otoGar_23 .. ctggtgatgttggagcatgt atgac gggacatggcc .. aa acttaagtcatgtgcc ............... tgaa gctacagtttcttcccctc >SELH_tupBel_27 .. gtgatgttgaagggacattt atgat gggacatggcc .. aa acttcagtcatgtgcc ............... tgaa gccaaggtttcttcccctc >SELH_musMus_25 .. ctggagatgttgaagcattt atgat ggtgcatggcc .. aa acttaagctatgcacc ............... tgaa gccatagtttcttcctcac >SELH_ratNor_24 .. ctggagatgttgaagcattt atgat ggtgcatggcc .. aa acttaagctatgtacc ............... tgaa gccatagtttcttcctcac >SELH_cavPor_19 .. ctggtcctgttggggcgtct gtgat gggacatggc ... ca aactaaagccatggccc .............. tgaa gccatggtctctagcatcc >SELH_canFam_24 .. ctggtgatgttggagcattt atgac ggaacatggcc .. aa acttcagtcatgtacc ............... tgaa gccatggtttcttccctcc >SELH_felCat_25 .. tggtgacgtttggagcattt atgac gggacatggcc .. aa atttcagtcatgtgcc ............... tgaa gccctggtttcttcccctc >SELH_equCab_26 .. ctggtgttgttggagcattt atgac ggtacatggcc .. aa acatcagtcatgtgcc ............... tgaa gctgtggtttcttcccctc >SELH_bosTau_28 .. ctggtgatgttggagcattt atgac gggacatggcc .. aa acttcagtcatgtccc ............... tgaa gctgtggtttcctcccctc >SELH_sorAra_24 .. ctggtgatgttggagcattg atgat ggaacatgacc .. aa acgtcagtcatgtgcc ............... tgaa gccaagtttctttctctcc >SELH_dasNov_26 .. ttggtgatgttggagcattt atgat gggacatggcc .. aa acttcagtcatgtacc ............... tgaa gccagtttctccccctcca >SELH_loxAfr_26 .. ctggtgatgttggagcattt atgac ggaacatggcc .. aa acctcagtcatgtgcc ............... tgaa gccacggtttcttcccctc >SELH_echTel_27 .. ctggtgatgttggagcattt atgat ggaacatggcc .. aa acttcagtcatgtgcc ............... tgaa gcccgtctcctcacttcca >SELH_monDom_27 .. ccagtgagtttggagcatcc atgat ggaacctgacc .. aa atctcccagtcacgtccc ............. tgaa gcttgggctccttctcctg >SELH_anoCar_27 .. ctctgtgagttggagcattc atgat gggatatgatc .. aa gtggagcaaatccagtcacatctc ....... tgaa gctgttgccctcctccaca >SEPP2_monDom_30 . gcccctccctccctccaaga atgac gcccacagtga .. aa cccagagaactggtccctgtgggc ....... tgat gccccagaggggaggagag >SEPP2_macEug_26 . gcccttccctccctcaatta atgat gcccacagtga .. aa cccggggaaccggccatggtgggc ....... tgat gccatggagaagaggaatg >SEPP2a_ornAna_34 cccgctcccgcctcgcactc atgac gtccacggtgtc . aa ccggcccgccgggcaccgtggac ........ tgac gccggtcgaggcggagggg >SEPP2b_ornAna_23 ggaatcaggaacccagtaac atgag gtcatcttcgg .. aa gcctgtgcctagaggaccaagataat ..... ggaa aaagtgacggacaagggtg >SEPP2_tetNig_17 . ggctgctggtggtcccgttg atgac gtctgcgctggt . aa acctgcctgcaggagcctgtggac ....... cgac gtgtgtggacccaccggca >SELI_homSap_22 .. tgaatgaagtttgtgcttga atgaa gagtgtatctta . aa ccccctttttttggacaggctgcactt .... ggat aaaataggcaccactgtgt >SELI_panTro_22 .. tgaatgaagtttgtgcttga atgaa gagtgtatctta . aa ccccctttttttggacaggctgcactt .... ggat aaaataggcaccactgtgt >SELI_macMul_21 .. tgaatgaagtttgtgcttga atgaa gagtgtatctta . aa cctcctttttttggacaggctgcactt .... ggat aacataggcaccactgtgt >SELI_otoGar_21 .. taagtgaagtttgtgcttga atgaa gagtgtatctt .. aa acccttttttttggacaggttgcactt .... ggat aaaataggcaccattgtgt >SELI_musMus_25 .. tgaatgaagtttgtgcttaa atgaa gagtgtgtctt .. aa acccttttttttggacaggttgcactt .... ggat aaaataggcaccactgtgt >SELI_ratNor_25 .. tgaatgaagtttgtgcttaa atgaa gagtgtgtctt .. aa acccttttttttggacaggttgcactt .... ggat aacataggcaccgctgtgt >SELI_cavPor_17 .. tgaatgaagtttgtgcttga atgaa gagtgtatctta . aa cccttttttttttggacaggttgcactt ... ggat aaaataggcaccactgtgt >SELI_oryCun_19 .. tgaatgaagtttgtgcttga atgaa gagtgtatctta . aa ccctttttttttggacaggttgcactt .... ggat aaaataggcaccactgttg >SELI_eriEur_14 .. ggaatgaagtatgtgcttga atgaa gagtgtatctt .. aa acccttttttttttttggacaggttgcactt ggat ataataggcaccactctgt >SELI_canFam_14 .. tgaatgaagtttgtgctcga atgaa gagtgtatctta . aa cccttttttttttggatggattgcactt ... ggat aaaataggcaccactgtgt >SELI_equCab_20 .. tgagtgaagtttgtgctcga atgaa gagtgtattctt . aa acccttgtttttggatggattgcactt .... ggat aaaataagcaccactgtgt >SELI_dasNov_18 .. tgaattaagtttgtgcttga atgaa gagtgtatctt .. aa accctttgtttttttggacaggttgcactt . ggat aaaataggcaccactatgt >SELI_loxAfr_26 .. tgaatgaagtttgtgcttga atgaa gagtgtatctt .. aa acccttttttttggacaggttgcactt .... ggat aaagtaggcaccactgtgt >SELI_monDom_26 .. tgaatgcaatttgtgcttga atgaa gagtgtgtctt .. aa atcctttatatggacaggctgcactt ..... ggag agaataagcacaaccatgt >SELI_ornAna_19 .. tgaatgaagcttgtgcttga atgaa gagtgcatctta . aa cccattttttttggaaaagctgcactt .... ggag agaaagggcacgactgtgt >SELI_anoCar_22 .. tgtgtgtcctttgtgcgtgt atgaa gagtgcggcctc . aa cccaggcgtcttggaagggccgcaccc .... ggaa gaaacggagcacagcaaag >SELI_galGal_25 .. ttgagtttatttgtgcttaa atgaa gagtgcgcttc .. aa acccagaccaggagagggcgcactt ...... ggag tgagcgagtcaaaccttgc >SELK_homSap_35 .. ctctgtgtcctcacagatga atgag gtcatgctggg .. aa ttccctctgcagggaactggcctgac ..... tgac atgcagttccataaatgca >SELK_panTro_35 .. ctctgtgtcctcacagatga atgag gtcatgctggg .. aa ttccctctgcagggaactggcctgac ..... tgac atgcagttccataaatgca >SELK_macMul_36 .. ctctgtgtcctcacagatga atgag gtcatgctagg .. aa ttccctctacagggaactggcctgac ..... tgac atgcagttccataaatgca >SELK_otoGar_32 .. ctctgtgtcttcacagatga atgag gtcaggctggg .. aa ttctctcttcagggaactggcctgac ..... tgac atgcagttctataaacgca >SELK_tupBel_37 .. ctctgtgtccccacagatga atgag gttatgccggg .. aa ttccctccacagggatctggcctgac ..... tgat acgcagttctataaatgca >SELK_musMus_32 .. ctctgtgtccccacagatga atgag gtcatgctggg .. aa ttccctctgcaggatctagcctgac ...... tgat acgcagttctataaatgta >SELK_ratNor_33 .. ctctgtgtcctcacagaaga atgag gtcatgctggg .. aa ctccctctgcaggatctggcctgac ...... tgat gtgcagttctataaatgta >SELK_cavPor_34 .. ctctgtgttctcacagataa atgag gtcacgccagg .. aa ttctctcagcagggatctggcttgac ..... tgat acgcagttctctaaatgca >SELK_oryCun_39 .. ctctgtgtccccacagatga atgat gtcaggctggg .. aa ttccctccacagggatctggcctgat ..... tgag atgcagttctataaatgcg >SELK_sorAra_41 .. ttctgtgtccacacagatga atgag gtcatgctggg .. aa ttccctctacggggatctggcatgac ..... tgat atgcagttcgataaatgca >SELK_eriEur_37 .. ctctgtgttctcacaggtga atgag gttatgctggg .. aa ttccctccatggggatctggcatgac ..... tgat atgcagttctataaatgca >SELK_canFam_36 .. ctctgtgccttcacagacgg atgag gttgtgctagg .. aa ttccctccccagggatctggcatgac ..... tgac atgcagttctataaatgca >SELK_felCat_37 .. ctctgtgtcctcacagacag atgag gtcgtgctggg .. aa ttccctccccagggatctggcatgac ..... tgac atgcagttctataaatgca >SELK_bosTau_39 .. ctctgtgtcctcacagacga atgag gtcatgctggg .. aa ttccctccgcagggatctggcatgac ..... tgac atgcagttctataaatgca >SELK_dasNov_29 .. ctctgtgtcctcacagatga atgag gtcatgtttggg . aa ttccctctgcagggatctggcatgac ..... tgac ttgcagttccataaatgca >SELK_loxAfr_35 .. ctctgtgtccccacagacgg atgag gctgtgctggg .. aa ttccctctgcagggatctggcatggc ..... tgac atgcagttccataaatgca >SELK_monDom_22 .. tgctctgtctacacagatta atgat gttgtgctggg .. aa ctcccatcttacaggatccagtgtaac .... tgat tgcaattgtataaatgcac >SELK_ornAna_34 .. tgctgtgaacaagcagatta atgat gttttgctggg .. aa ttccttcagggatccagtataac ........ tgat aaagcaattatataaaggc >SELK_anoCar_34 .. ctctgccaattggcgggacc atgat gttgtcctggg .. aa ttccttattctgggatccagggcaac ..... tgaa aagcagttctgttaaatta >SELK_galGal_31 .. tctgcaatataagcagatga atgaa gttgtactggg .. aa ctccttcaaggatccagtgtaac ........ tgaa gtgcagtgttattaaatac >GPX3_homSap_28 .. atggcaggggtggcgtcttc atgag ggaggggccca .. aa gcccttgtgggcggacctcccc ......... tgag cctgtctgaggggccagcc >GPX3_panTro_28 .. atgtcgggggtggtgtcttc atgag ggaggggccca .. aa gcccttgtgggcggacctcccc ......... tgag cctgtctgaggggccagcc >GPX3_macMul_28 .. atgtcaggggtggcgtcttc atgag ggaggggccca .. aa gcccttgtgggcggacctcccc ......... tgag cctgtctgaggggccagct >GPX3_tupBel_29 .. gtgtcaggggtggcatctcc atgag ggaggggcccg .. aa gcccttgtgggcggacctcccc ......... tgag cctgtctgaggggccggcc >GPX3_musMus_33 .. aggtcaggggtggtgtctct atgaa ggaggggcccg .. aa gcccttgtgggcgggcctcccc ......... tgag cccgtctgtggtgccagcc >GPX3_ratNor_33 .. aagtcaggggtggtgtctcc atgaa ggaggggcccg .. aa gcccttgtgggcgggcctcccc ......... tgag cccgtctgtggtgccagcc >GPX3_canFam_30 .. gtgtcaggaatggcatctcc atgaa ggaggggccc ... ga agccctcatgggcggacctcccc ........ tgag cctgtctgaagggccggcc >GPX3_felCat_30 .. gtgtcagggatggcgtctcc atgaa ggaggggccc ... ga agcccttgtgggcggacctcccc ........ tgag cctgtctgaagggccagcc >GPX3_equCab_26 .. atgtcaggggtggcatctcc atgat ggaggggcccg .. aa gccctggtgggcggacctcccc ......... agag cctgtctgaagggccagcc >GPX3_bosTau_31 .. tgtcagggggcggcatcgcc atgaa ggaggggcccg .. aa gcccgcgtgggcgggcctccct ......... tgag cctgtctgaggggccagcc >GPX3_loxAfr_16 .. gtgtcaagagcggcatctcc atgat ggtggggcccg .. aa gcccctgtggcggacctcccc .......... agag cctgtcccatgggccagcc >GPX3_echTel_13 .. cgtgcagagggagcatctcc atgag ggtgaggcccg .. aa gcccccgtggcggacctcgac .......... ccgag tctgctctgggcctgcctt >GPX3_monDom_23 .. acataagggatggcatctct atgat ggtgggatcca .. aa gcctcttcagggcgggttccatc ........ agag cctgcaaaaggtgtcagga >GPX3_ornAna_31 .. aatggggaggtggcatcatc atgac agcggggtctg .. aa agcccctcctggatggaccccgcc ....... cgaa cctgctcggcggtggcatg >GPX3_anoCar_28 .. gccaaggaagttgcatcact atgag ggttaggtctg .. aa agctcccaaaaagagcggacctagcc ..... tgag gctgcaaagctctggtgta >GPX3_galGal_31 .. gagcagaggatgacatctcc atgaa ggcctggcctg .. aa agcccccaccatggggtgggctcggcc .... cgat cccgcccaggcgcggtgca >GPX6_homSap_18 .. atgaagggaagggcatctcc atgat ggtggatccc ... aa aacccctctgggtcgcaccctgcc ....... agag ccttccttggtgcctgtcc >GPX6_panTro_23 .. atgaagggaggggcatctcc atgac ggtgggtccc ... aa aacctctcggggtcggaccctgcc ....... agag ccttccttggtgcctgtcc >GPX6_macMul_22 .. cagaagggaggggcatcttt atgat ggtgggtctc ... aa aacctctctgggtcagaccctacc ....... agag ccttccttggtgcctgtcc >GPX6_cavPor_20 .. ctagaaggaagggtgtctcc atgat ggtgggtccc ... aa aagccctggatcggaccctacc ......... agaa ccttccctggtgcctgtcc >GPX6_canFam_24 .. tatgaaaggagggcatctcc atgat ggtgggttcc ... ca agcccctgcggtcggaccctacc ........ agag ccttcttgggtgcctgtcc >GPX6_equCab_25 .. atgagctgaggggcatctcc atgat ggtgggtccc ... aa agcccctgtgggcaggacccaac ........ cagaa ctctgtgcctgtcccttag >GPX6_bosTau_23 .. atgaggaaaagggcatctcc atgat ggtgggtccc ... aa agcctctctgggtcggaccccacc ....... agag ccttccttggtgcctgtcc >GPX6_eriEur_24 .. tatgaaaggagggcatctcc atgat ggtgggttcc ... ca agcccctgcggtcggaccctacc ........ agag ccttccttggtgtctgtcc >GPX6_sorAra_23 .. tgaaaggagggggcatctcc atgat ggtgggtctc ... aa agcccctctgggtcggaccctacc ....... agag ccctgctgggtgtcctgtc >GPX6_dasNov_20 .. atgaagggagggacatatcc atgat ggtgggtcct ... aa agcccttctgggtcagaccctacc ....... agaa ccttctctggtgcctgtcc >GPX6_loxAfr_19 .. atgaatggaggggcatatcc atgat ggtgggtccc ... aa agcctctctaggtcggaccctatc ....... agaa tcttccctggtgcctgtcc >GPX6_echTel_31 .. aatggggaggtggcatcatc atgac agcggggtct ... ga aagcccctcctggatggaccccgcc ...... cgaa ccttccctggtgcctgtcc >GPX1_homSap_20 .. tctcgggggggttttcatct atgag ggtgtttcctct . aa acctacgagggaggaacacc ........... tgat cttacagaaaataccacct >GPX1_panTro_20 .. ctgtgggggggttttcatct atgag ggtgtttcctct . aa acctacgagggaggaacacc ........... tgat cttacagaaaataccccct >GPX1_macMul_26 .. tctctggggggttttcatct atgag ggtgtttcctct . aa acctacaaggaggaacacc ............ tgat aatacagaaaataccccct >GPX1_otoGar_25 .. tctctgggggggtttcattc atgat agtgttacctct . aa acttgcatgggggaacacc ............ tgat gccccagaaaatcccctga >GPX1_tupBel_21 .. gcgtctggggggtttcatcc atgac ggtgtctcctct . aa accccgaaggaggaacgcc ............ tgat gtccggaaaaccccccagg >GPX1_musMus_30 .. ctctggggggcggttcttcc atgat ggtgtttcctct . aa atttgcacggagaaacacc ............ tgat ttccaggaaaatcccctca >GPX1_ratNor_26 .. ctccggggggaggtttttcc atgac ggtgtttcctct . aa atttacatggagaaacacc ............ tgat ttccagaaaaatcccctca >GPX1_cavPor_15 .. gctctcctgagggttcttcc atgaa ggtgtttcctctc aa cctgtatagaggaacatc ............. cgat tcccaggaatttcccagag >GPX1_oryCun_24 .. gctctctgggggtttcatcc atgag ggcgttcccccg . aa aacaaatggaggaacgcc ............. tgat gtccgggaaacccccaggt >GPX1_eriEur_19 .. agccgctgggcatctcatcc atgac ggcgccgccttc . aa acctgcgagcaggaaggagcgcc ........ cgat agccgcgagagcccccagc >GPX1_canFam_22 .. gccctctggggatttcatcc atgat ggtgtttccttg . aa atctgcatggaggaacgcc ............ tgat ttccaggaaagtcccctga >GPX1_felCat_28 .. gccctctggggatttcatcc atgat ggcgtttcctcg . aa atttgcatagaggaacgcc ............ tgat ttccagaagaatcccctga >GPX1_equCab_27 .. tttctctggggatttcatcc atgat ggcgtttcctct . aa acatgcatgaggaacgcc ............. tgat gttaaggagaatcccccga >GPX1_bosTau_30 .. gctctccagggattttgccc atgaa ggtgttccctct . aa acctacgtggaggaatgcc ............ tgat gtccaggaaaatcccctga >GPX1_dasNov_29 .. tccacacggggttttcatcc atgac ggtgtttcctct . aa acctgcagggaggaacacc ............ tgaa gtccggcaaaatcccccga >GPX1_loxAfr_25 .. tttccgtgggggtttcatta atgag ggtgttttctct . aa acctgaatggaggaacacc ............ tgat gtctgggaagatacccccc >GPX1_monDom_26 .. ggtccgtgagggttttatct atgat ggtgttgtttt .. aa accattaaggagaaagaacact ......... tgat aatgcttgtaaaatcccat >GPX1_ornAna_26 .. ccatgggggagatctcattt atgat ggtgctccttct . aa acctttacggaagagcact ............ tgat ggatcctgaggaactccat >GPX2_homSap_30 .. aagctctgggccttcacaga atgat ggcaccttcct .. aa accctcatgggtggtgtc ............. tgag aggcgtgaagggcctggag >GPX2_panTro_30 .. aagctctgggccttcacaga atgat ggcaccttcct .. aa accctcatgggtggtgtc ............. tgag aggcgtgaagggcctggag >GPX2_macMul_30 .. aagctctgggccttcacaga atgat ggcaccttcct .. aa accctcatgggtggtgtc ............. tgag aggcgtgaagggcctgaag >GPX2_otoGar_28 .. gggctctgggccttcacaga atgat ggcaccatcct .. aa acgcctctgggtggtgtc ............. tgag aagagcggaaggcctggag >GPX2_tupBel_29 .. gggctctgagccttcacaga atgat agcaccttcct .. aa acccccccgggaggtgtc ............. tgag aagtgtgacaggcccggag >GPX2_musMus_34 .. aggttctgggccttcacaga atgat ggcatcttcct .. aa acccttctgggagatgtc ............. tgag aagttgtgaagggtccaga >GPX2_ratNor_31 .. tgctaggccttcttcacaga atgat ggcatcttcct .. aa acccttctgggggatgtc ............. tgag acgttgtgaagggcccaga >GPX2_cavPor_20 .. tggctctgggccttcacaga atgac ggcaccgtcct .. aa acgctatgggtggtatc .............. tgag aagtgtgaatggctggagc >GPX2_sorAra_29 .. aaactctgggccttcgcaga atgat ggcacctccct .. aa atccatggggtggtgtc .............. tgag gcgtgcgagggcctggaaa >GPX2_eriEur_25 .. ggtttctggaccttcacaga atgat agcaccttcct .. aa acctatagggatggtgtc ............. tgag aaatgtgaagggcctgaag >GPX2_canFam_23 .. tggctctgggccttcacaga atgat ggcaccttcct .. aa atagtatgggcggtgtc .............. tgag aagtgtgaagggctcagag >GPX2_felCat_22 .. tggatctgggccttcacaga atgat ggcatcttcct .. aa actgtatgggcggtgtc .............. tgag gagtgtgaagggctcggag >GPX2_equCab_28 .. aggctctgggccttcacaga atgat ggcaccttcct .. aa acctgtatggacggtgtc ............. tgaa aagcgtgaagggccccgag >GPX2_bosTau_26 .. gggctctgtgtcttcacaga atgat ggcaccttcct .. aa atctgtatgggcggtgtt ............. tgag aagagtgaaggcctggagc >GPX2_loxAfr_27 .. aggctctggaccttcgcaga atgat ggcaccttcct .. aa actcagtgggtggtgtc .............. tgag aaatgtgaagggcctaggg >GPX2_echTel_30 .. tggctctgggccttcacaga atgat ggcaccttcct .. aa accctccggaaggtgtc .............. tgag aaatgtgaagggcctgggg >GPX2_monDom_26 .. ggtccgtgagggttttatct atgat ggtgttgtttt .. aa accattaaggagaaagaacact ......... tgat aatgcttgtaaaatcccat >GPX2_ornAna_28 .. agcctccagaccttcacaga atgac ggtgtctcctt .. aa accctaaccgggaggcacc ............ cgag agccggtgaagggcctggt >GPX4_homSap_29 .. ggagccttccaccggcactc atgac ggcctgcctg ... ca aacctgctggtggggcagacc .......... cgaa aatccagcgtgcaccccgc >GPX4_panTro_29 .. ggagccttccaccagcaccc atgac ggcctgcctg ... ca aacctgctggtggggcagacc .......... cgaa aatccagcgtgcaccctgc >GPX4_macMul_33 .. ggagccttccaccggcaccc atgac ggcctgcttgc .. aa accagctcctggtgaggcagacc ........ cgaa aatccagcgtgcaccccgc >GPX4_tupBel_31 .. gaaccttccacccggcacct ttgac ggtctgcctat .. aa acctgccactggtgaggcagacc ........ cgag aacctggcgtgcaccctgc >GPX4_musMus_36 .. agccttccaccccggcactc atgaa ggtctgcctg ... aa aaccagcctgctggtggggcagtcc ...... tgag gacctggcgtgcaccctgc >GPX4_ratNor_34 .. agccttccaccccggcactc atgac ggtctgcctg ... aa aaccagcccgctggtggggcagtcc ...... cgag gacctggcgtgcaccccgc >GPX4_cavPor_34 .. gagtctcctacccgggtgcc atgac ggcctgcctg ... ca aaccagcgtgctggtggggc ........... agac ccgaggatgcgtgcactgc >GPX4_canFam_31 .. gagccttccacccggcaccc atgac agtctgtctaa .. aa accagcccgctggtggggcagacc ....... cgag aacccggcgtgcaccctgc >GPX4_sorAra_37 .. gagccttccacccggcgccc atgac ggtctgcctgc .. aa accagcccgctggtggggc ............ agac ccgagaacccggcgtgcac >GPX4_echTel_41 .. gagtcttccacccggcacca atgac ggtctgccttc .. aa accaggcctctggtgaggcagacc ....... cgat gacccggcgtgcactcagc >GPX4_monDom_41 .. gagtcttccacccggcacca atgac ggtctgccttc .. aa accaggcctctggtgaggcagacc ....... cgat gacccggcgtgcacctcag >GPX4_ornAna_34 .. tcccgggacgctctgcctcc atgac ggccggcctt ... ca agccaaaaccagttggtggggccggcc .... tgaa caaaccggcacgggtcccg >GPX4_anoCar_39 .. gggccccctcctccagcacc atgac ggcctgccttg .. aa gccagcttgctggtgaggcagacc ....... cgaa gattcggcgtgcactgctg >GPX4_galGal_39 .. gggccccctcctccagcacc atgac ggcctgccttg .. aa gccagcttgctggtgaggcagacc ....... cgaa gattcggcgtgcactgctg >SEPW_homSap_36 .. cccctctcagcagacgcttc atgat aggaaggactg .. aa aagtcttgtggacacctggtctttccc .... tgat gttctcgtggctgctgttg >SEPW_panTro_36 .. cccctctcagcagacgcttc atgat aggaaggactg .. aa aagtcttgtggacacctggtctttccc .... tgat gttcttgtggctgctgttg >SEPW_ponPyg_31 .. cccctctcagcagacgcttc atgat aggaaggactg .. aa aaatcttgtggacacctggtctttccc .... tgat gttctcgtggctgctgttg >SEPW_macMul_34 .. cccctctcagcagacgcttc atgat aggaaggactg .. aa aagtcttgtggacgcctggtctttccc .... tgat gttctcgtggctgctgttg >SEPW_musMus_34 .. cccctcgtggcagacgcttc atgat gggaagaactg .. aa atgtctcgtggacgcctggtctttcc ..... ctgat gtccctgcgactgccacgt >SEPW_ratNor_27 .. cctttcttggcagccgcttc atgac aggaaggactg .. aa atgtctcaaagacctgtggtctttctt .... cgat gttcctgcggccaccaagt >SEPW_canFam_34 .. cccctcgtggcagacgcttc atgat gggaagaact ... ga aatgtctcgtggacgcctggtctttcc .... ctgat gtccctgcgactgccacgt >SEPW_dasNov_29 .. ctgcccttggcagacgcttc atgag gggaaggacct .. aa atgcgtcgtggatgcctggtctttccc .... tgat gctccttcacctgccagat >DIO1_homSap_29 .. gtgtctttacatatttgttt atgat ggccacagcct .. aa agtacacacggctgtgact ............ tgat tcaaaagaaaatgttataa >DIO1_ponPyg_29 .. gtgtctttacatatttgttt atgat ggccacagcct .. aa agtacacacggctgtgact ............ tgat tcaaaagaaaatgttataa >DIO1_macMul_29 .. gtgtctttacatatttgttt atgat ggccacagcct .. aa agtacacacggctgtgact ............ tgat tcaaaagaaaatgttataa >DIO1_cavPor_24 .. gcttcttttcatatttgttc atgac ggtcacagtct .. aa agtacacacagctgtgacc ............ tgat ttgaaagaaaatgttttaa >DIO1_bosTau_28 .. gcctcttttcatatttgttc atgac ggccacagcct .. aa agtacacacggctgtgact ............ tgat ttgaaagaaaatgttttaa >DIO1_sorAra_24 .. gcttctcttcatatttgttt atgac agccccagctg .. aa agtacacacagctgtggct ............ tgat tggaaagaaaatgttttaa >DIO1_eriEur_26 .. ctttcttctcatatttgctt atgat ggtcacagctt .. aa agtatacacagctgtgact ............ tgat tggaaagaaaatattttaa >DIO1_ornAna_24 .. ggttccgtgaatattggttt atgag ggtcacagtgt .. aa agcgcatgcagctgtgact ............ tgat ctgagaaaatatttctgcg >DIO2_homSap_30 .. cagagttgaccagtgtgcgg atgat aactactgacg .. aa agagtcatcgactcagttagtggtt ...... ggat gtagtcacattagtttgcc >DIO2_macMul_30 .. cagagttgaccagtgtgcgg atgat aactactgacg .. aa agagtcatcgactcagttagtggtt ...... ggat gtagtcacattagtttgcc >DIO2_musMus_29 .. cagagctcactggtgtgcga atgat aactactgacg .. aa agagctgtctgctcagtctgtggtt ...... ggat gtagtcacacgagtctgcc >DIO2_ratNor_28 .. cggagctcactggtgtgcga atgat aactactgacg .. aa agagtcatctgctcagtctgtggtt ...... ggat gtagtcacacgagtctgcc >DIO2_canFam_27 .. cagaggtgaccagtgtgcga atgat aactactgatg .. aa agagtcactgactcagttagtggtt ...... ggat acagtcacattagttttcc >DIO2_dasNov_27 .. cagaggctaccagtgtgcca atgat aactactgacg .. aa agaggcatcgactcagttagtggtt ...... ggat gtagccacattagtttgcc >DIO2_monDom_29 .. gaagggctgccagtgtgcaa atgat gatctctaaca .. aa agagtcagtcactccgttagaggtt ...... ggat gtggtcacagtggcttgct >DIO2_ornAna_25 .. gctgagttactagtgtgcaa atgaa gaccaccaaca .. aa agagaatttaactcagttggtgctc ...... agat atcatcacactggcttact >DIO2_anoCar_27 .. gcagagctgttggtgtcttt atgaa gatcaccaaca .. aa agagtgtctcattcagttggtgttc ...... agat gtcttagcactggcatttc >DIO2_galGal_25 .. acctgaatgcttgtgtgttt atgaa gagcactaaca .. aa agagtaattgactcagttggtgttc ...... agat actctcacactggcattcc >DIO3_homSap_31 .. gcacaggagccccactgctg atgac gaactatctct .. aa ctggtcttgaccacgagctagttc ....... tgaa ttgcaggggcctcaaagca >DIO3_panTro_31 .. gcacaggagccccactgctg atgac gaactatctct .. aa ctggtcttgaccacgagctagttc ....... tgaa ttgcaggggcctcaaagca >DIO3_macMul_30 .. gcacaggagccccactgctg atgac gaactgtctct .. aa ctggtcttgaccacgagctagttc ....... tgaa ttgcaggggcctcaaaaca >DIO3_musMus_26 .. cgctggagccctggctgctg atgac gaaccgcctct .. aa ctgggcttgaccacgggtcggctc ....... tgaa ttgcagagaggctcgaaac >DIO3_ratNor_26 .. cgctggagccctggctgctg atgac gaaccgcctct .. aa ctgggcttgaccacgggtcggctc ....... tgaa ttgcagagaggctcgaaac >DIO3_cavPor_24 .. gcgcatgagcccctctgctg atgac gaactgtccct .. aa ctggtctcgaccacgggcgggtt ........ ccgaa attgcaggatggctcgaat >DIO3_canFam_26 .. gctggcgagccccactgctg atgac gagccgcctct .. aa ctggtcttgaccacgagctggttc ....... tgag ttgcaggggggcttgcagc >DIO3_felCat_29 .. gctcacgagccccactgctg atgac gagctatctct .. aa ctggtcttgaccacgagctggttc ....... tgaa ccgcagggggcttgcagca >DIO3_bosTau_23 .. gctcacgagccccactgctg atgaa gagctgtctct .. aa ctggcctcgaccacgagctggttc ....... tgat ttgcaggaggctcgcagca >DIO3_loxAfr_22 .. gcgctagagccccactgctg atgac gaactgtctct .. aa ctggtcttgaccacgagctgattc ....... cgaa ttgcagggaactcgcagca
Reference sets of 380 vertebrate SECIS elements
Introduction
The comparative genomics of SECIS elements can be investigated at much greater depth in the era of 50 completed vertebrate genomes. The situation currently is unsatisfactory since hardly any SECIS elements have rarely been submitted to GenBank, requiring manual entry from journal graphics prior to the approach described here.
The initial goal is obtaining 2-3 dozen phylogenetically dispersed orthologous vertebrate sequences for each SECIS element in each selenoprotein gene. After that, their evolution can be studied. To date, 380 SECIS elements have been obtained primarily from mammals, tetrapods, and fish. These collections can readily be leveraged to earlier diverging species by locating terminal coding exons and examining downstream sequence via uBlast against the seed set, hopefully in congruence to SECIS web tool motif recognition.
Functionality of a SECIS element depends not so much on its linear nucleotide sequence as its secondary and kink-turn tertiary fold which are experimentally established as critical to recognition by KIAA0256/SECISBP2 binding proteins. As long as hairpins retain complementary bases (perhaps straying intermittantly) and loops retain similar length, this constraint may be satisified without conservation of sequences (except at a handful of key residues).
This poses difficulties in recovering reliably orthologous sequences even in mammalian species (often wholly lacking in experimental transcript data) by blast query alone of an experimentally established fiducial query. Prospective elements might be validated by the SECIS web tool but this can yield both false positives and false negatives.
The approach here exploits the availability of a 30-species multiz whole-genome alignment which also provides a statistically sound PhastCons track quantitating peaks of cross-species conservation, notably in non-coding 3' UTR sequence where metazoan SECIS elements reside. Whole genome alignment methods provide more reliable truly syntenic alignment than ad hoc methods especially in non-coding regions.
It turns out that a rather small fraction of 3' mRNA has any notable PhastCons conservation and that SECIS elements of all human selenoprotein encoding genes reside at a PhastCons peak, whether or not (eg SELM) they are canonical. However the registration is sometimes imperfect, suggesting when the peak is broader that the SECIS element might be larger than the region favored by the SECIS web tool. Typically, 3' UTR of these genes contain other unexplained PhastCons peaks; these may or may not have any specific connection to the selenocysteine aspect of the gene.
Because human -- where the 30-species alignment track resides -- has an exhaustive set of selenoprotein transcripts, it is feasible to retrieve full length post-coding sequence and identify putative SECIS elements using the SECIS web tool. Blatting the identified SECIS elements back into the whole genome establishes that essentially all give unique matches not only within the expected 3' UTR position of the expected gene in correct strand orientation but also to a PhastCons peak.
Further, nucleotide sequences from species other than human, when recovered from the alignment details page for the PhastCons peak, are almost always identified as SECIS elements by the SECIS web tool which is based on PatScan and RNAfold rather than linear sequence homology. This provides independent cross-validation of the validity of the two methods.
Consequently the compilations below are orthologous in the classical sense within each gene. Within gene families, the SECIS elements are then paralogs (related by vertical genetic descent from last common ancestor preceding the gene duplication). However, different selenoprotein gene families may bear no detectable resemblance to each other even at the fold level even when sharing a CxxC motif. Conversely, the SECIS elements of SELM1 and diverged paralog SELM2 are radically diverged at supposedly invariant structural nucleotides.
SECIS elements of non-homologous coding regions could still themselves be homologous provided say an ancestral mobile element distributed a master element to unrelated genes. Alternatively, these elements arose from convergent evolution (perhaps kink-turn starting points were common in 3' UTR for other reasons) and were never orthologous.
This issue cannot be resolved by examining vertebrate (or even metazoan) selenoprotein genes because selenocysteine genes have far more ancient origins. Vertebrates are completely devoid of de novo originated selenoproteins. Their new selenocysteine genes appear to have arisen exclusively by segmental and retrogene duplication mechanisms which bring along the parental SECIS element. Their new selenocysteine sites within genes have arisen exclusively in existing selenocysteine genes that already have a functional SECIS element.
Cautionary note on sequence errors
Cautionary note: using the 30-species genomic alignment track on the UCSC genome browser as sequence source can result in occasional errors, notably when indels occur in other species relative to human and the details page does not provide them. Sequences for other species can be checked directly by Blat in the individual browser for that species.
A second type of error, no SECIS sequence for a given species, can arise when the genomic alignment is missing or incorrect. That data might well be available within the individual browser for that species or within its transcripts. An incorrect alignment results in an irrelevent sequence being offered to the SECIS tool, resulting in no SECIS element being reported. That situation is difficult to distinguish from false negatives associated with the recognition tool.
A third type of error, ascertainment bias, arises from the choice of human genome as reference point for the 30-species alignment. This results in the sequence collection consistenting primarily of placental mammals because other tetrapods and teleost fish are often too diverged to genomically align well.
In other words, despite 4 quality controls currently provided by appropriate location, strand, orthology, and SECIS tool validation, the sequence set would benefit from additional quality controls and supplementation to greater phylogenetic depth. The latter is best done by tblastn of two terminal coding exons of each selenoprotein against NCBI transcripts, followed by SECIS tool applied to the longest 3'UTR. The supplemented set can then be extended, for example to species such as lizard without meaningful transcript collections by Blat of the nearest related species chicken.
It is also feasible to use the trace archives provided a sufficiently close species can provide the SECIS query. The best matches can be validated by blastx against the terminal exon if the trace is sufficiently long (which it often isn't). Here to boost outgroups, a complete set of elephant SECIS elements was obtained from the genome as augmented by traces. Those elephant SECIS elements were then used against sloth, armadillo, tenrec, and hyrax trace archives to obtain fairly reliable sets in those species, totally 85 SECIS sequences with at least two for each selenoprotein. These satisfactorily determine the Atlantogenata node; the rate of evolution of each gene's SECIS element relative to human can then be averaged for the node. These rates are by no means uniform.
Making a comprehensive effort to find SECIS elements in cartilaginous fish results in confirmable SECIS elements in SELS, TXNRD2, and DIO1 for elephantshark (where genomic contigs sometimes extend far enough into 3'utr) and DIO3 in tropical shark. However none of 11 transcripts from 8 other chondrichthyes selenoproteins gave results, in 8 cases because the mRNA was too short and in 3 cases where the mRNA seemed long enough but the presumptive SECIS element could not be recognized by the SECIS tool or by blastn to reference collections because it was too diverged or did not follow the rules:
SEPP1_torCal EH115677 too short 694 714 SEPP2_leuEri EE987575 too short SELO_calMil AAVX01333182 too short 556-928 TXNRD2_squAca ES606786 too short SELU1_squAca too short CX663017 686-693 SELU1_calMil too short AAVX01548610 114-243 DIO2_calMil AAVX01619547 too short 1-187 DIO3_calMil AAVX01619547 too short 1-147 SEPW_squAca EE722256 not too short 314-669 SEPW_calMil AAVX01059582 not too short 1482-2187 SEPW_torCal EH115680 not too short 319-635
The easiest approach to finding SECIS elements in teleost fish exploits the presence of a 5-species track in zebrafish that includes the other 4 genomic teleost fish as well as human. This quickly leverages any locatable fish SECIS element to the others. Fish SECIS elements could furnish an important outgroup to tetrapods or alternatively be so diverged they constitute a 'red herring" relative to more conserved earlier diverging chondricthyhians.
SEPP1a: 20 SECIS insertion sequences
>SEPP1a_homSap Homo sapiens (human) score: 30.80 uuuucuuuuu ccagugu ucuauuugcuuua augag aauagaaacgu aa acuaugaccuag ggguuucuguu ggau aauuagc aguuuag aauggaggaa >SEPP1a_panTro Pan troglodytes (chimp) score: 30.80 uuuucuuuuu ccagugu ucuauuugcuuua augag aauagaaacgu aa acuaugaccuag ggguuucuguu ggau aguuagc aguuuag aauggaggaa >SEPP1a_macMul Macaca mulatta (rhesus) score: 30.80 uuuucuuuuu ccagugu ucuauuugcuuua augag aauagaaacgu aa acuaugaccuag ggguuucuguu ggau aguuagc aauuuag aauggaggaa >SEPP1a_tupBel Tupaia belangeri (treeShrew) score: 31.04 uuuucuuuuu ccaguau ucuauuugcauua augaa gacagaaacau aa acuaugaccuag ggguuucgguu ggau agguagc aauuuag aauggaggaa >SEPP1a_musMus Mus musculus (mouse) score: 32.57 uuuccuuuuu ccagugu ucuaguuacauua augag aacagaaacau aa acuaugaccuag ggguuucuguu ggau agcuugu aauuaag aacggagaaa >SEPP1a_ratNor Rattus norvegicus (rat) score: 32.57 uuuucuuucu ccagugu ucuaguuacauug augag aacagaaacau aa acuaugaccuag ggguuucuguu ggau agcucgu aauuaag aacggagaaa >SEPP1a_cavPor Cavia porcellus (guineaPig) score: 34.57 uguucuuuuu ccagaau ucuauuugcauug augaa aacagaaacau aa acuaugaccuag ggguuucuguu ggau acauagu aauuuag aauggaggaa >SEPP1a_oryCun Oryctolagus cuniculus (rabbit) score: 33.28 uuuucuuuuu ccagcau ucuacuugcauua augau aacagaaacgu aa acuaugaccuag ggguuucuguu ggau aauuagua auuuag aauggagaag >SEPP1a_sorAra Sorex araneus (shrew) score: 33.08 uuuucuucuu ccaauau ucuauuugcauua augaa gacagaagcaa aa acuaugaccuag gggcuucuguu ggau acuua gcaauugg gauggagaaa >SEPP1a_eriEur Erinaceus europaeus (hedgehog) score: 33.29 uuuucuuuuu ccagcau ucuacuugcagua augag gacagaaacgu aa acuaugaccuag ggguuucuguu ggau aguuagca auuuag aauggaggaa >SEPP1a_canFam Canis familiaris (dog) score: 33.38 uuuucuuuuu ccagcau ucuacuugcauua augaa aacagagacau aa acuaugaccuag ggguuucuguu ggau aguuagca auuuag aauggaggaa >SEPP1a_equCab Equus caballus (horse) score: 29.34 uuuucuuuuu ccagcau ucuacuugcauua augag aacagaaacgu aa acuaugaccuag ggguuucugug ggau aguuagca auuuag aauggaggaa >SEPP1a_bosTau Bos taurus (cow) score: 30.75 uuuucuuuuu ccaguau ucuacuugcguua augag aacagaaacgu aa acuauaaccuag ggguuucuguu ggau gguuggc aacuaag aauggaggaa >SEPP1a_dasNov Dasypus novemcinctus (armadillo) score: 32.57 ucuucuuuuu ccagcau ucuauuugcauua augag aacagaaacau aa acuaugaccuag ggguuucuguu ggau aguuagca auuuag aauggagaaa >SEPP1a_loxAfr Loxodonta africana (elephant) score: 32.57 ucuucuuuuu ccagcau ucucuuugcauua augag aacagaaacau aa acuaugaccuag ggguuucuguu ggau aguuagca auuuag aauggaggaa >SEPP1a_echTel Echinops telfairi (tenrec) score: 32.80 ucuucuuuuu ccagcau ucuuuuugcauua augag aacagaaacgu aa acuaugaccuag ggguuucuguu ggau acucugca auuuag aauggaggaa >SEPP1a_monDom Monodelphis domestica (opossum) score: 30.77 uguuucucuu ccugugu ucugcuugcauua augaa gacagaagcau aa acuauaacuuag gggcuucuguu ggau aauuugca auccag aauggaggaa >SEPP1a_macEug Macropus eugenii (wallaby) trace score: 24 UGUUUCUCUU CCUGUGU UCUGUUUGCAUUA AUGAA GAGAGAAGCUG AA ACUAUAACCUAG GGGCUUCUGUU GGAA AAU UUGCAAG UCAGAAUGGA >SEPP1a_triVul Trichosurus vulpecula mRNA score: 25 ugguucucuu ccugcgu ucuguuugcauua augaa gagagaagcug aa acuauaaccuag gggcuucuguu ggau aauuugca auucag aauggaagaa >SEPP1a_ornAna Ornithorhynchus anatinus (platypus) score: 29.28 ugccuccugc cguucug cuugcauuu augag aacagaaacau aa acuauaaaccuag ggguuucuguu ggau agu uagcaau caagaauggg >SEPP1a_galGal Gallus gallus (chicken) score: 30 T2 UUCUAAUGUU CUGCUUG CAUUU GUGAG GACAGGAGCAU AA ACUAUGACCUA GGGGCUCCUGUU UGAU UUUUUG CAAUCA AGAACAGAAG >SEPP1a_anoCar Anolis carolinensis (lizard) score: 32.99 cacuucuacc auucugu uugcauuc augaa aacagaagccg aa acuaugaccuag gggcuuuuguu ugau auauu gcagcca agaacaggag >SEPP1a_danRer Danio rerio (zebrafish) score: 32.87 uuucaccucu aagugcu uuauguggucuuu augaa ggcaggugcag aa acuaugcacuag uggugucuguc ugau guuu ggccau acagagcaga
SEPP1b: 16 SECIS insertion sequences
>SEPP1b_homSap Homo sapiens (human) score 20.32 uauauugcuu aguaagu auuuccauaguca augau gguuuaauaggu aa accaaa cccuauaaacc ugac cuc cuuuau gguuaauacu >SEPP1b_panTro Pan troglodytes (chimp) score 20.32 uauauugcuu aguaagu auuuccauaguca augau gguuuaauaggu aa accaaa cccuauaaacc ugac cuc cuuuau gguuaauacu >SEPP1b_macMul Macaca mulatta (rhesus) score 20.32 uauauugcuu aguaagu auuuccauaguca augau gguuuaauaggu aa accaaa cccuauaaacc ugac cuc cuuuac gguuaauacu >SEPP1b_tupBel Tupaia belangeri (treeShrew) score 16.22 uggacugcuu agaaagu auuuccauaguca augau gguucaauaggc aa acuaau cccuauaaacc ugaa cuc cuuuau gguuaauacu >SEPP1b_oryCun Oryctolagus cuniculus (rabbit) score 15.69 cuuuaggcug cucagag uaucuccauauua augau gguuuaacaggu aa acuaaa cccuaugaacc ugaa uuu cuuuaug gauaauacua >SEPP1b_sorAra Sorex araneus (shrew) score 15.19 uugguugcuu agaaagu auuuccauaguca augau gguuuaauaggu aa acuuauuu ccuguaaaccc ugaa cuu cuuuau gguuaauacc >SEPP1b_canFam Canis familiaris (dog) score 16.12 ugauugcuua gaaagua uuuccauaguca augau gguucaauaggu aa acuaag uccuauaaacc ugaa cuccua uaugguu aauacuauua >SEPP1b_equCab Equus caballus (horse) score 16.12 uugauugcuu agaaagu auuuccauaguca augau gguucaauaggu aa acuaag uccuauaaacc ugaa cuc cuuuau gguuaauacu >SEPP1b_bosTau Bos taurus (cow) score 13.38 uugauugcuu agaaagu auuuccauaguca augau gguucaauaggc aa accagg uccuauaaacc ugaa uuu uuuuuau ggucaauacu >SEPP1b_dasNov Dasypus novemcinctus (armadillo) score 14.48 uaggcugcuu agaaagu auuuccauaauca augau gguucaguaggu aa acuaag cccuauaaacc ugaa cuc cuuuau gguuaauacc >SEPP1b_choHof score: 16 T2 GCTGCTTAGAAAGTATTTCCATAAT AATGATGGTTCAATAGGTAACCCAAGCCCTATAAACCTGAA CTCCTTTTATGGTTAATAC >SEPP1b_loxAfr Loxodonta africana (elephant) score 14.36 cuaggcugcu uagaagu auuuccaucauca augau gguucaauaggu aa accaaa uccuauaaacc ugaa cuc cuuaug guuaauacug >SEPP1b_echTel Echinops telfairi (tenrec) score 17.68 uaggcugcuu agaaagu auuuccauaguua augau gauucauuaggu aa auuua gucuugugaauc ugaa cuc auuuau gguuaauacu >SEPP1b_monDom Monodelphis domestica (opossum) score 16.17 cagguugcuu agaaagu guucccauaguua augau gguuuaacaggu aa auugac uccuguggacc ugaa cuc uuuuau gguucauacu >SEPP1b_macEug Macropus eugenii (wallaby) trace score: 18 CAGGUUACUU AGAAAGU AUUCCCAUAGUCA AUGAU GGUUCAGUAGGU AA AUUUAC UUCUAUGGACC UGAA CUC UUUUAU GGUUAAUACU >SEPP1b_ornAna Ornithorhynchus anatinus (platypus) score 17.91 gaauggcuua gaaagua uuugccauaguua augac gguucaguaggg aa auccaa uccuaugaacc ugag cucuuc uaugguu aaugcuaaua >SEPP1b_anoCar Anolis carolinensis (lizard) score 16.09 ugcuuagauu gugcuug ccauaguuc augau gguuuauuaggg aa auaaa uucuacaaaacc ugaa uuccu uuauggu gaacacagaa >SEPP1b_danRer Danio rerio (zebrafish) score 24.26 auagaugguu uuaaaca uuuuuuauacuua augaa gguuuucuggu aa aucuuguauc accuggaagac ccgag aaac uguaugg aaauguaugc
SEPN: 14 SECIS insertion sequences
>SEPN_homSap Homo sapiens (human) score: 32.06 caguggcuuc cccggca gcagcccc augau ggcugaauccg aa auccucga uggguccagcu ugau guc uuugcag cugcaccuau >SEPN_panTro Pan troglodytes (chimp) score: 32.06 gcaguggcuu cccuggc agcagcccc augau ggcugaauccg aa auccucga uggguccagcu ugau gucuuu gcagcug caccuauggg >SEPN_macMul Macaca mulatta (rhesus) score: 32.06 caguggcuuc cccagca gcagcccc augau ggcugaauccg aa auccucga uggguccagcu ugau guc uuugcag cugcaccuau >SEPN_musMus Mus musculus (mouse) score: 34.87 guggcucccc cagcaac agccgc augau ggcugggucug aa aucgcaaa uggauccagcc ugau guccuc gcaguug cacauguggg >SEPN_ratNor Rattus norvegicus (rat) score: 35.19 gguguuuccc ccagcag cagccgc augau ggcugggucug aa aucccaaa aggauccagcc ugau guccucg caguug cacauguggg >SEPN_cavPor Cavia porcellus (guineaPig) score: 34.85 gaauaaugau ccccgag cagcagcccc augau ggcuggauccg aa auccuaga uggaucuagcu ugau gcc uuugcag uugcacuuau >SEPN_sorAra Sorex araneus (shrew) score: 33.75 ggcuuuuccc cacccgc aaccgccuc augau ggcuggaucug aa auccuaga uggaucuggcu ugau gucuuu gcgauug uccuguggga >SEPN_canFam Canis familiaris (dog) score: 34.73 ggcagugucu cccgcag cagcagcccc augau ggcuggaucug aa auccuaga uggguccagcu ugau guc uuugcag cugcaccuac >SEPN_felCat Felis catus (cat) score: 28.82 cggugucucc cccagca gcagcccc augau ggcuggaucug aa auccuaga cgggcccagcu ugau guc uuugcag cugcaccugu >SEPN_equCab Equus caballus (horse) score: 37.53 ggagggacaa uggcucc ucgcggcugcccc augau ggcuggaucug aa auccugga uggguccagcu ugau gucuc cgcaguug caccuauggg >SEPN_bosTau Bos taurus (cow) score: 31.65 cgauggugcc cccuagc agccgcccc augau ggcuggaucug aa agcuuagg uggauccagcu ugau gucuuu gcaguug cuccuguggg >SEPN_dasNov Dasypus novemcinctus (armadillo) score: 26.61 gccgcuggcu ccuccag cagccgccgc augaa ggauggaucug aa aucccaga uggauccagcc cgau cuc uuugcag uugcaccugu >SEPN_loxAfr Loxodonta africana (elephant) score: 31.75 agugaugguu ccccaag caacugcccc augac agcugggucug aa auccuaga uggacccaguu ugau guc uuugcag uagcaccucu >SEPN_monDom Monodelphis domestica (opossum) score: 26.99 gggacuucua uucuccu gcaacuguucuuu augau ggcuagauccu aa auccuaga ugggucaaguu ugau gucuug acaguaa cugaagggga >SEPN_macEug Macropus eugenii (wallaby) trace score: 34 AUUUCUGUUC UCUGCCU GCAGCUGUUCUUU AUGAU GGCUUGGUCCU AA AUCCCAGA UGGGUCAAGCU UGAU GUCUUCUU GACAGUA ACUGGAAAGG
SEPHS2: 11 SECIS insertion sequences
>SEPHS2_homSap Homo sapiens (human) score: 20.81 accugcaacc aucugac uuggucucuguua augac gucucucccucu aa accccauuaagg acugggagaggc agag caag ccucaga gcccaggccu >SEPHS2_panTro Pan troglodytes (chimp) score: 20.81 accugcaacc aucugac uuggucucuguua augac gucucucccucu aa accccauuaagg acugggagaggc agag caag ccucaga gcccaggccu >SEPHS2_macMul Macaca mulatta (rhesus) score: 20.90 accugcaacc aucugac uuggucucuguua augac gucucucccucu aa acuccauuaagg acugggagaggc agag caag ccucaga gcccaggccu >SEPHS2_otoGar Otolemur garnettii (bushbaby) score: 11.55 ccuaccagca ucugacu ucaucucugcug augac auuucucccuug aa aggcuac uaagggcaggg agaa gcagagga aguccca gcgcgcaggc >SEPHS2_musMus Mus musculus (mouse) score: 22.00 aagacuagcc accugac uuggucucugaua augau gucucucccucu aa cucccaguaagg acugggagaggc ugaa caaac cucagag ccaggugucg >SEPHS2_ratNor Rattus norvegicus (rat) score: 18.62 cagccaucua acuuggu cucuguua augac gccucucucugu aa accccacuacgg acugggggaggc agag caa acccag agcccaggcu >SEPHS2_canFam Canis familiaris (dog) score: 20.27 ccugcaacca ucugauu uggucucuguua augac guuucucccucu aa accccuuuuagg acugggagaggc agag uaagccccag agcccaa accucagugg >SEPHS2_felCat Felis catus (cat) score: 22.33 ccugcgacca ucugauu uggucucuguua augac gucucucccucu aa accccuuuaagg acugggggaggc agag caagccccag agcccaa gccuugaugg >SEPHS2_equCab Equus caballus (horse) score: 21.57 ccugcagcca ucugacu uggucucuguua augac gucucucccucu aa accccguuaugg acugggagaggc agag caagccccag agcccagg ccuugguggu >SEPHS2_bosTau Bos taurus (cow) score: 22.19 uccagccacc uuacuug gucucuguua augac gucucucucua aa ccccuuuaagga cugggagaggc agac caaac cccagag accaggauuu >SEPHS2_dasNov Dasypus novemcinctus (armadillo) score: 17.60 acccguaacc accuggc cuggucucuguua augau guuucucccucu aa accccguuaagg acugggagaggc ggag ugagc cccagag cccaggccuu >SEPHS2_choHof score: 16 ACCUGUAACC AUCUGGC CUGGUCUCUGUUA AUGAU GUUUCUCCCUUUG AA CCCCAUUCA GGAUGGGGAGAGGC AGAC CGAG CCCCAGA ACCCAGGCCU >SEPHS2_loxAfr Loxodonta africana (elephant) score: 20 traces lacking coding AUCUGUAACC AUCUGAC UUGGUCUCUGUUA AUGAU GUUUCUCCCUCU AA ACCUCAUUAAG GACUGGGAGAGGC AGAG CAAG CCUCAGA GCCCAGGCCU >SEPHS2_echTel Echinops telfairi (tenrec) score: 22 T2 ATCCGGAACCATCTGACGTGGTCTCTGTT AATGATGTTTCTCCCTCAAAACCCCGTGAAGGACCGGGAGAGACGGAG CAAATGCCAGAGCCCAGGCCT >SEPHS2a_monDom Monodelphis domestica (opossum) score: 32 UGCUAACCAC AAGCCUU GUCUCUGUUU AUGAU GCUUCUCCCAU AA AACCCUAUUAAGG GCUGGGAGAGGC UGAG CAAAACCUC AGAGUCU AGGCCUUGGU >SEPHS2b_monDom Monodelphis domestica (opossum) score: 23 T2 inconsistent CATCTCGCTGATCATAGGCCTTGG CTCTGTTTATGATGCTTCTCCCtcta aacctctttgagggctggatgaagc >SEPHS2ab_macEug Macropus eugenii (wallaby) trace score: 30 id issues UCUGCUAACC UUGCCUU GUCUCUAUUU AUGAU GCCUCUCCCAU AA AACCCUAUCAAGG GCUGGAAGAGGC UGAG CAAAACUUCA GAGCCUA GGCCUUGGUG
SELM1: 18 SECIS insertion sequences
>SELM1_homSap Homo sapiens (human) score 27.34 agagugaaac auucaca aagauuugcguua augaa gacuacacaga aa accuuucuaggga uuuguguggau cagau acauac uuggcaa auuuuugagu >SELM1_panTro Pan troglodytes (chimp) score 27.93 agagugaaac auucaca aagauuugcguua augaa gacuacacaga aa accuuucuaagga uuuguguggau cagau acauac uuggcaa auuuuugagu >SELM1_macMul Macaca mulatta (rhesus) score 27.71 agggugaaac auucaca aagauuugcguua augaa gacuacacaga aa accuuccuaagga uuuguguggau cagau acauac uuggcaa auuuuugagu >SELM1_otoGar Otolemur garnettii (bushbaby) score 28.3 gaaacauuca caaaaau uugcauua augaa gacuacacaga aa accuuucagagga uuuguguggau cugau acuuggc aaauuuug aguuuuauau >SELM1_tupBel Tupaia belangeri (treeShrew) score 24.54 agagggaaac auucaca aagauuugcguua augaa gacugcacaga aa accuuucuaggga uuuguguggau ccgau aau uggcaaa uuuuuguauu >SELM1_musMus Mus musculus (mouse) score 35.23 aauuuaauac uuauaaa gguuugcauua augag gauuacacaga aa accuuuguuaagga cuuguguagau cugau aauuggcaaa uuuuua uuuuaaaagu >SELM1_ratNor Rattus norvegicus (rat) score 30.11 aauguaauac uuauaaa gguuugcauua augag gauuacacaga aa accuuuguuaaggg uuugugucgau cugau aauuggcaaa uuuuua uuuuuaaaau >SELM1_cavPor Cavia porcellus (guineaPig) score 31 gaaugaaaug gccacaa aaacuugcauua augag gacuauacaga aa accuuccuaagga uuuguauagau cugau guauggcaaa uuuuugu uuuuacauuc >SELM1_sorAra Sorex araneus (shrew) score 25.76 uaagaauaau uugcaca aagauucgcauua augaa ggcuacacagg aa accuuacuaagga uuuguguggau cugau aac uagcaa auuuuugugc >SELM1_eriEur Erinaceus europaeus (hedgehog) score 26.92 uagcaguacu cacaaag guuugcaucc augaa ggcuacauaga aa accuuacugagga uuuguguagau cugau gcucagcaaa cuucug uguuuuacag >SELM1_canFam Canis familiaris (dog) score 27.8 agaguaucuu cacagag auuugcauua augaa gacuacacaga aa accuucaggagga uuuguguggau cugau auuuagcaa aauuuug uguuucacac >SELM1_felCat Felis catus (cat) score 30.14 gagggaaaca uucacaa cgauuugcauua augaa gacuacacaga aa accuucaugagga uuuguguggau cugau gaua uuuagca aauuuuugug >SELM1_equCab Equus caballus (horse) score 28.65 agugaaacau ucacaaa gauucgcguua augaa gacuacacaga aa accuuucugagga uuuguguggau cugau auccggcaaa uuuuug ugcuuuacau >SELM1_bosTau Bos taurus (cow) score 23.97 gugaaauauu cacacaa agauuugcauua augaa gacuacacaga aa accuuccugagga uuuguguggac cugau acuuagcaaa uuuuug ugcuuuacau >SELM1_dasNov Dasypus novemcinctus (armadillo) score 23.56 agagugaaac auuuaca aagauuugcacua augaa gacuacacaga aa accuuucuaagga uuuguauggau cugau auu uggcaaau uuuuguguuu >SELM1_loxAfr Loxodonta africana (elephant) score 27.21 agagugaaac auucaca aagauuugcauua augaa gacugcacaga aa accuuuguaagga uuuguguggau cugau auu uggcaag uuuuuguguu >SELM1_echTel Echinops telfairi (tenrec) score: 20 T2 AGAATGAAACATTCACAAAGATCGCGT AATGAAGACTACCCAGAAAACTTTGTAGATTTGTGTGGATCTGAT ATTTGGCAAATTTTGTTTT >SELM1_monDom Monodelphis domestica (opossum) score: 22 T2 TGTGAAACATGAAGACAATGCGCAT TAATGATGACTACACAGAAAACCTATCATGGGCTTGTGTAAATCTGA TGTTTAGTGTAT >SELM1_ornAna Ornithorhynchus anatinus (platypus) score 26.78 gacugugaaa caugaag acaaugcgcauua augau gacuacacaga aa accuaucaugga uuuguguauau cugau guuu cguguau uuccucaguu >SELM1_anoCar Anolis carolinensis (lizard) score 31.51 gauuggagga ugccagc aaaauacucuuua augau gacuacaugua aa gccuuuaacaaaagg cauguguaggu cugau guca gaugua cuuuuguuaa
MSRB1: 14 SECIS insertion sequences
>MSRB1_homSap Homo sapiens (human) score: 26.23 ccugccagcc gcccugg cccuggucacugc augau ccgcucugguc aa acccuuccaggcc agccagagugg ggau ggu cugugac cugcugggaa >MSRB1_panTro Pan troglodytes (chimp) score: 23.19 ccugccagcc gcccugg cccuggucacugc augau ccgcucugguc aa acccuuccaggcc agccagggugg ggau ggu cugugac cugcugggaa >MSRB1_macMul Macaca mulatta (rhesus) score: 22.02 ccugccagcc gcccugg cccuggucacugc augau cugcucugguc aa acccuuccaggcc agccagcgugg ggau ggu cugugac cugcugggaa >MSRB1_otoGar Otolemur garnettii (bushbaby) score: 25.09 uuggccaucu gcuccag cccaagucaaugc augau cuccucuggcu aa auccuuccaggcc agccagggugg ugau guc ugugacc ugcuggggga >MSRB1_musMus Mus musculus (mouse) score: 23.75 ccugccaacc gcuccag ccucagucaccga augau cugcucugguc aa auccuucuaugcc agccagggugg ugau gac ccgugac cuuugaggag >MSRB1_ratNor Rattus norvegicus (rat) score: 21.64 cuggccaacc acuccag ccucagucacugu augau cugcucugguu aa auccuucugugcca gcccagggugg ugau gaccugugac cuucgag gagggaggcg >MSRB1_cavPor Cavia porcellus (guineaPig) score: 26.14 augaccagcu gcuucag cccuggucaaugc augau cuacucuggcu aa auccuucuaggcc agucagggugg ugau gau cugugac cugcugggga >MSRB1_oryCun Oryctolagus cuniculus (rabbit) score: 28.41 auuggccccc gccccgg ucacugc augac ccgcucuggcu aa acccuuccagggc agccagggugg ugau ggu cugugac cugcggcggg >MSRB1_canFam Canis familiaris (dog) score: 28.62 guugcuggcu gcugcag cugcagucacuac augau ucacucugguu aa acccuuccaggc agccagagugg ugau gau cugugac cugcuagagg >MSRB1_felCat Felis catus (cat) score: 31.99 cucgccggcu gcuccgg cccuggucacuac augau ucacucugguc aa agccuuccaggc agccagagugg ugau gau cugugac ggcuggaggg >MSRB1_equCab Equus caballus (horse) score: 24.02 cccagccggc ugcucca gcccagucacugc augau ucacucuggug aa acccuucggggc ggccagagugg ugag gaccug ugacccg cuggaggagc >MSRB1_bosTau Bos taurus (cow) score: 27.78 ugccagcugc cccagcc cagucacucu augau ucucucuggcu aa acccuugcaggc agccagagugg ugau gacccgugac ccgcuggg ggagcaggcg >MSRB1_loxAfr Loxodonta africana (elephant) score: 16 genomic 3' called SEPX1 CCGACCAGCU GCUCCAG CCCUGGUCACUGC AUGAU UCGCUCUCGUU AA ACCGUUCCAGGCC CACCAGGGUG CUGAU GGU UCGUGAU CCACUAGGAG >MSRB1_echTel Echinops telfairi (tenrec) score: 14 CCCAGUCAGC GGCUCCA GCCCGGUCACUGC AUGAA UCGCUCUGGUC AA AUCCUUCCAGGCUC ACCAGGGUAC UGAU GGUC UGUGACC UGCUGGGGCA >MSRB1_monDom Monodelphis domestica (opossum) score: 21 UUUCCUGUUA CUUUGGU UUGGGGUCACAGU AUGAA UUGCCCUGGUU AA ACCCUUAUAGGAU GGCCAUGGUGA UGAA GGUUAGU GACCAGG CCAGAAAACG >MSRB1_ornAna Ornithorhynchus anatinus (platypus) score: 26.32 ucugccccuc cccaccu gguuuucagcauu augac ucgcccugggg aa agcccuccuggag cgccagggcga cgau ggucaccu gaucggg ugggcaggga >MSRB1_galGal Gallus gallus (chicken) score: 22 ucuggugcua uucuggg cugggaucauggu augaa uuguccucugu aa auccuccugggau gcgagggugg ggau guu uuguga uccugcugga
TXNRD1: 17 SECIS insertion sequences
>TXNRD1_homSap Homo sapiens (human) score: 25.36 auuuggcagg gcaucga agggaugcaucc augaa gucaccagucuc aa gcccaugugg uaggcggugau ggaa caacug ucaaauc aguuuuagca >TXNRD1_panTro Pan troglodytes (chimp) score: 25.36 auuuggcagg gcaucga agggaugcaucc augaa gucaccagucuc aa gcccaugugg uaggcggugau ggaa caacug ucaaauc aguuuuagca >TXNRD1_otoGar Otolemur garnettii (bushbaby) score: 23.42 aucggcagug caucgac gggaugcgucc augaa gucaccagccuc aa gccugugugg ugggcagugau ggaa caacu guccaau caguuucuau >TXNRD1_tupBel Tupaia belangeri (treeShrew) score: 25.13 ucuuggcagc gcaucag agggaugcgucc augaa gucaccagccuc aa gcccgugcgg ugggcggugau ggaa cgacug ccagauc aguuucagca >TXNRD1_musMus Mus musculus (mouse) score: 27.9 aucuggcaga gcaucac aggcaugcgucc augaa gucacuggccuc aa gcccaagug gugggcagugac agaa gagcu gccgggu cuguugagcu >TXNRD1_ratNor Rattus norvegicus (rat) score: 28.03 uucggcagag caucacg gugcgucc augaa gucacuagccuc aa gcccaagugg ugggcagugac agaa agc ugucgau cuguuggguu >TXNRD1_cavPor Cavia porcellus (guineaPig) score: 23.88 gcggcggcac cguaggg ugcgucc augag gucaccagccuc aa gcccgaggg ugggcggugac ggau cgc gccgcg uggcucagcu >TXNRD1_eriEur Erinaceus europaeus (hedgehog) score: 26.25 ugucagcaga gcaucaa agggaugcgucc augaa gucaccagccuc aa gcccgugcggg ugggcagugac ggaa cacug ucgaagc aguuucaaca >TXNRD1_canFam Canis familiaris (dog) score: 24.54 auucggcaug caucggc guggugcgucc augaa gucacuggccuc aa gccaugcg gugggcagugau ggag caac ugucgag caguuuuagu >TXNRD1_felCat Felis catus (cat) score: 24.14 acucggcagc gcaucgg agggcgcgucc augaa gucaccggcccc aa gcccccgcg gugggcggugau ggaa caagu gccgagc aguuuuagcg >TXNRD1_equCab Equus caballus (horse) score: 21.76 acucggcagu gcaucga agggaugcgucc augaa gucacuggccuc aa agcccaugug gugggcggugau ggaa cagcug ucgaagc aguuuuagca >TXNRD1_choHof score: 16 GUUUGGCAGU GCAUCUU UGGGAUGCGUCC AUGAU GUCACCGGCCUC AA GCCCGUGAGG AUGGGCAGUGAU GGAA CAUCU GUCGAGU CUUUUUUUAG >TXNRD1_loxAfr Loxodonta africana (elephant) score: 22.55 auuggcagcg caucgag ggaugcaucc augaa gucacuggccuc aa gcccaugug gggggcggugau ggaa cagcu gucgaau cagcuuuggc >TXNRD1_echTel Echinops telfairi (tenrec) score: 19.5 guggcagugc aucaaga gaugcguuc augaa aucgcuugcccc aa gcccga guggcgggcag cgau ggaaca ucugucu caucaguuuc >TXNRD1_monDom Monodelphis domestica (opossum) score: 23.46 ggcucgcggu gcaucgg ugagaugcguuc augaa gucgcugccug aa gcccauaucccgugg uggguggugac cgaa agaaccg ccggcc uccguuuuau >TXNRD1_macEug Macropus eugenii (wallaby) trace score: 21 GGUCUCGUGG UGCAUCC AACAGAUGCGUUA AUGAA GUCACUACCUAG AA GCCUGUAUCUUGUG GUGGGUGGUGAC CGAA GAGCCACU GCCAUCA GUUUUAUCAA >TXNRD1_ornAna Ornithorhynchus anatinus (platypus) score: 18.89 aggagugcac ccaaggg cugcauuu augaa gucagagccaa aa gccagcauuuugcgg uuggcugugau ggaa aaa cuccug ccacaguuuu >TXNRD1_anoCar Anolis carolinensis (lizard) score: 32.53 uggcaaggca uuguuca agaugcuucc augaa gucacagucua aa accagugcuuucugg uaggcagugau ggaa aga uugcugg cacaacuuga >TXNRD1_galGal Gallus gallus (chicken) score: 26.64 uagcagggca uuucaca caugcuuuc augaa aucacagccug aa gccugcacugucugg ugggcagugau ggaa gaacu gcugaca cagcugaaca
TXNRD3: 9 SECIS insertion sequences (problematic)
>TXNRD3_homSap Homo sapiens (human) score: 19.10 gacagcgaga agcagug ggacugcuucc uugac gccuuagcuu gg agccccguuaugaggu gagccaaggc ugac ucu cgcaagc caggacugag >TXNRD3_panTro Pan troglodytes (chimp) score: 19.10 agcagugggc GACAGCGAGA AGCAGUG GGACUGCUUCC UUGAC GCCUUAGCUU GG AGCCCCGUUAUGAGGU GAGCCAAGGC UGAC UCU CGCAAGC CAGGACUGAG >TXNRD3_macMul Macaca mulatta (rhesus) score: 17.86 GACuGCGAGA AGCAGUG GGACUGCUUCC UUGAC GCCUUAGCUU GG AGCCCuGUUgUGAGGU GAGCCAAGGC cGAC UCU CGCAAGC CAGGACUcAG >TXNRD3_calJac Callithrix jacchus (marmoset) score: 25.26 GACuGCGAGA AGCAGUG GGACUGCUUCC UUGAC GCCUUAGCUc Ga AGCCCuGUUAcGAGGU GAGCCAAGGC UGAu UCU CGCAAGC gAGGACUGAG >TXNRD3_musMus Mus musculus (mouse) score: 21.86 CUGACGGCAU GCAGCAG CCAGGCUGCUUCC UUGAC ACCUUGGCUC GG AACCUGCAGAGGU GAGCCAAGGC CGAC UUC UGCACGU CAGCCUCGAC >TXNRD3_ratNor Rattus norvegicus (rat) score: 25.37 cugauggcgu gcagcag ccaggcugcaucc uugac gccuuggcuc gg aaccugcagagg ugagccaaggc cgac ucc ugcacgu cagccucgac >TXNRD3_canFam Canis familiaris (dog) score: 26.93 GCUGGCUGGA GAGGCAG GCAGGCUGCCUCC UUGAC GCCUUAGCUC GG AACCGCUGUGAGG UGAGCUAAGGC CGAU GUC CUCCAU GCCAGGCCAG >TXNRD3_felCat Felis catus (cat) score: 20.14 CUGGCUgGGA GAGGCAG GucGGCUGCCUCC UUGAC GuCUUAGCUC GG AgcccgaUGUGAGG UGAGCUAAGGC CGAU Ggu cuuccac gucagaaucg >TXNRD3_bosTau Bos taurus (cow) score: 12.55 GGCUGACGGG AGGGCAG ACUCGCUGCCUCC UUGAC GUCUUCGCUC AG AGCCGCCAGGU GAGCCAAGAC CGAC CUC UGCCCA CCAGCUCCUC >TXNRD3_choHof score: 24 GACTTGGAGAAGCAGTTAGACTGCTTG CTTGACGTCTTAGCTCGGAACCCCAGCGTGAGGTGAGCTAAGACTGAC TCTCCTGAAGTCAG >TXNRD3_dasNov Dasypus novemcinctus (armadillo) score: 21 T2 GACTTGGAGAAGCAGTTAGACTGCTTT CTTGACGTCTTAGCTCGGAGCCTGCATGAGGTGAGCTAAGACTGAC TCTCCTGAAGTCAG >TXNRD3_loxAfr Loxodonta africana (elephant) score: 21 T2 extended trace of last exon but no genomic GACTTGGAGAAGCAGTTA GACTGCTTTCTTGACGTCTTAGCTCGGAGCCTGCATGAGGTGAGCT AAGACTGACTCTCCTGAAGTCAG >TXNRD3_monDom Monodelphis domestica (opossum) score: 28 CCUUGGAaAA GCAuUaA ucCUGCUuUC UUGAc GUCAUAGCUCAG AA CCCUAUUGUGA GGUGAGCUAUGaC UGAU GAUUuCCU UGCAAGU CAGGACcccu >TXNRD3_macEug Macropus eugenii mRNA score: 28 cuuccuugga gaagcag ugacgcugcuguc uugau gucauagcucag aa cccuauuguga ggugagcuauggc ugau gauu cccuugc aagucaggac
TXNRD2: 11 SECIS insertion sequences
>TXNRD2_homSap Homo sapiens (human) score: 31.00 cacccccccc caggcuc cuggugccagaug augac gaccugggugg aa accuacccugugg gcacccauguc cgag ccccc uggcauu ucugcaaugc >TXNRD2_panTro Pan troglodytes (chimp) score: 31.00 ggcacccccc caggcuc cuggugccagaug augac gaccugggugg aa accuacccugugg gcacccauguc cgag ccccc uggcauu ucugcauugc >TXNRD2_tupBel Tupaia belangeri (treeShrew) score: 36.55 cucccucccc aggcccc cgaugccagaug augac ggccuggacag aa acccacccuguggg cugcccagguc ugaa cccuccc uggugucu uuggggugua >TXNRD2_musMus Mus musculus (mouse) score: 36.94 gccagccucu gacacuc ccagcgucagaug augau ggccugggcag aa accccauguggg ccgcccagguu ugaa ccccu ggcauuu cuagagcacu >TXNRD2_ratNor Rattus norvegicus (rat) score: 34.71 cagccuucac acacugc cagugucagaug augac ggccugugcag aa acccccacguggg cugcccagguu ugaa ccccug gcauuu cuggagugcu >TXNRD2_cavPor Cavia porcellus (guineaPig) score: 34.41 ucuggccccc caggucc ccagugccaguug augau ggccugggcag aa acccacccuguggg caguccauguc ugaa cuccc uggcauu ucuggagugc >TXNRD2_eriEur Erinaceus europaeus (hedgehog) score: 34.68 ccagccccac caggccc ccgaugccagaua augau gacuugugcag aa acccacccggg cugcccauguc ugag ccucug uggcauu cuggagugua >TXNRD2_canFam Canis familiaris (dog) score: 34.36 accagccccg ccaggcc ccgaugccagaag augac gacgugugcag aa accccccuguggg cugcccgcguc cgag cccccuggc guuucugg aauguaaaua >TXNRD2_equCab Equus caballus (horse) score: 31.37 ccagcccugc cagguuc ccgaugccagacg augac gaccugcgcgg aa acccacccuguggg cugcccacguc cgag ccccc uggcauu ucugaagugc >TXNRD2_loxAfr Loxodonta africana (elephant) score: 31 genomic 3' AGCCAGCCCA GCGGCCC CCAGUGCCAGAUG AUGAC GACCUGCGCAG AA ACCCACCCUGUGGG CUGCCCACGUU UGAG CCCCC UGGCAUU UCUGGAGUGC >TXNRD2_echTel Echinops telfairi (tenrec) score: 37.41 cccuugcccc caccuca gcgccagaug augaa gacaugugcag aa acccagcccguggg cugcccauguc ugag ccccc ugacgu uucuggagug >TXNRD2_monDom Monodelphis domestica (opossum) score: 34 CAGCCCUCCU CCGUGUC CACAUGCCAGAUC AUGAC GACUUGCGCAG AA ACCCACCCGUGGG CCGCGCAAGGC CGAG CUCUCCU GGCAUU UCCAGUGCAU >TXNRD2_macEug Macropus eugenii (wallaby) trace score: 31 CAACUCCCAU UUGUGUC CAAAUGCCAGAAC AUGAC GACUUGAACAG AA ACCCACCCUAUGGG CUGUGCAAGGC CGAG CUCCCCU GGCAUU UCUUAUGUGU >TXNRD2_calMil Callorhinchus milii (elephantfish) AAVX01015557 position 193 after stop score: 30.15 caugaccaac acuuugc cguuua augac gagcuguuugu aa acccagcgacguggg agagacagccc ugau gacaa ucggcau cgagaucuaa
SELS: 21 SECIS insertion sequences
>SELS_homSap Homo sapiens (human) score 36.61 uaggacaguc ucuguga cagguugcguuga augau gucuuccuuauc aa uggugagcccacca gugaggauuac ugau gugga caguuga ugggguuugu >SELS_panTro Pan troglodytes (chimp) score 36.61 uaggacaguc ucuguga cagguugcguuga augau gucuuccuuauc aa uggugagcccacca gugaggauuac ugau gugga caguuga ugggguuugu >SELS_macMul Macaca mulatta (rhesus) score 36.61 uaggacaguc ucuguga cagguugcguuga augau gucuuccuuauc aa uggugagcccacca gugaggauuac ugau gugga caguuga ugggguuugu >SELS_otoGar Otolemur garnettii (bushbaby) score 38.23 uaggacaguc ucuguga cagguugcguuga augau gucuuccuuau aa auggugagcccacca gugaggauuac ugau acaga caguuga ugggguuugu >SELS_tupBel Tupaia belangeri (treeShrew) score 40.14 gagaacaguc ucuguga cagggugcgucga augau gucuuccuuau aa auggugagcccacca cugaggaguac ugau gcaga caguuga caggguuugu >SELS_musMus Mus musculus (mouse) score 41.68 caggaugguc ucuguga cgggaugcguuga augau gucuuccuuau aa auggugaacccacca gugaggauuac ugau guu cacagu ugacgggguu >SELS_ratNor Rattus norvegicus (rat) score 40.42 cagggugguc ucuguga caggaugcguuga augau gucuuccuuau aa auggugagcccacca gugaggauuac ugau gua cacagu ugaugggguu >SELS_cavPor Cavia porcellus (guineaPig) score 40.42 aaggacaguc ucuguga cgagcugcguuga augau gucuuccuuau aa auggugagcccacca gugaggauuac ugau gcaga caguuga ugggguuuau >SELS_canFam Canis familiaris (dog) score 39.26 uaggacaguc ucuguga cagguugcguuga augau gucuuccuugu aa acggugagcccacca gcgaggauuac ugau gcaga caguuga ugggguuguu >SELS_felCat Felis catus (cat) score 41.01 uaggacaguc ucuguga cagguugcguuga augau gucuuccuugu aa auggugagcccacca gcgaggauuac ugau gcaga caguuga ugggguuguu >SELS_equCab Equus caballus (horse) score 41.01 uaggacaguc ucuguga cagguugcguuga augau gucuuccuugu aa auggugagcccacca gcgaggauuac ugau gcaga caguuga uggguuguuu >SELS_bosTau Bos taurus (cow) score 41.15 uaggacaguc ucuguga cagcuugcguuga augau gucuuccuugu aa auggugagcccacca gcaaggauuac ugau gcaga caguuga ugggguuguu >SELS_eriEur Erinaceus europaeus (hedgehog) score 36.87 cucucuguga uggguug cguugg augau gucuuccuuguc aa uggugaacccacca gcgaggaucac ugau gcag acaguug augggguugu >SELS_sorAra Sorex araneus (shrew) score 35.70 ugccgggccc gucucug ugauaggcgacga augac gucguccucgg aa auggugugccacca gcgaggaccac cgau gaagacacu ccugggc gccccccccc >SELS_dasNov Dasypus novemcinctus (armadillo) score: 35.89 uagaacaguc ucuguga cagguugcguuga augau gucuuccuuau aa aauggugaacccacca gugaggauuac ugau aauaga caguuga ugggguuugu >SELS_loxAfr Loxodonta africana (elephant) score: 39 traces lacking coding UGGAAUAGUC UCUGUGA CAGGGUGCGUUGA AUGAU GUCUUCCUUAU AA AUGGUGAACCCACCA GUGAGGAUUAC UGAU AAUAGA CAGUUGA UGGGGUUUGU >SELS_echTel Echinops telfairi (tenrec) score 38.43 uagaacaguc ucuguga cagguugcguuga augaa gucuuccuuau aa auggugaacucacca gugaggauuac ugau aaaga caguuga ugagguuugu >SELS_monDom Monodelphis domestica (opossum) score 40.64 uagaugaguc ucuguga caggcugcgcaga augau gucuuccuuau aa auggugagcucacca gugaggauuac ugau gauagauag uuguugg uguuuuuuuc >SELS_macEug Macropus eugenii (wallaby) trace score: 42 UAGAUGAGUC UCUGUGA CAGGCUGCGCAGA AUGAU GUCUUCCUUAU AA AUGGUGAACUCACCA GUGAGGAUUAC UGAU GAUAGA UAGUUGG UGGUGUUUUU >SELS_triVul Trichosurus vulpecula mRNA score: 42 uagaugaguc ucugugg caggcugcgcaga augau gucuuccuuau aa auggugaacucacca gugaggauuac ugau gauaga uaguugg ugguguuuuu >SELS_ornAna Ornithorhynchus anatinus (platypus) score 33.47 ucaauaaguc ucuguga caggcggcauuga augau gucuuccuugu aa auggugaaacccacca gugaggauuac ugau aua gacagg agaugguauu >SELS_anoCar Anolis carolinensis (lizard) score 25.44 gaaacucccu guaacaa gcagcaacaa augac gugguccuuau aa augguggacacauca cugaggaccuc cgaa gauaagacag cugauug gggacaguuc >SELS_galGal Gallus gallus (chicken) score 35.60 cgggagaguc ucuguga caagcugcucugu augau guuuuccuuau aa augguaaacaaacca augaggauuac ugau gcua gacagca gauggggguu >SELS_calMil Callorhinchus milii (elephantfish) AAVX01138705 position 244 after stop COVE score: 33.09 ugaagcaaga ugaaauu gacagcuauga augac gccauccuugu aa augguuccccaacca augaggauguc ugaa auuggacagg gauagug cgagauguau
SELT: 21 SECIS insertion sequences
>SELT_homSap Homo sapiens (human) score: 39.15 gaucauugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uucuug gcaggcucgu >SELT_panTro Pan troglodytes (chimp) score: 39.15 gaucauuuca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uucuug gcaggcucgu >SELT_macMul Macaca mulatta (rhesus) score: 39.15 gaucauugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uucuug gcaggcucgu >SELT_otoGar Otolemur garnettii (bushbaby) score: 39.15 gauuuuugca agagcag cgugacugacagu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uucuug gcaggcucgu >SELT_musMus Mus musculus (mouse) score: 39.15 ggauuuugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau guuucuugg caggcuc guuguaccuc >SELT_ratNor Rattus norvegicus (rat) score: 39.15 ggauuuugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau guuucuugg caggcuc guuguaccuc >SELT_cavPor Cavia porcellus (guineaPig) score: 39.15 aauuuuugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uuuuug gcaggaucgu >SELT_oryCun Oryctolagus cuniculus (rabbit) score: 39.15 ggauuuugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uuuuug gcaggcucgu >SELT_canFam Canis familiaris (dog) score: 39.15 auuuuuugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uucuug gcaggcucgu >SELT_felCat Felis catus (cat) score: 39.15 auuuuuugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uucuug gcaggcucgu >SELT_equCab Equus caballus (horse) score: 38.49 gacuuuugca agagcag cguggcugacagu augaa ggccuguacug aa gacagcaagcugu uaguacagacc cgau gcu uucuug gcaggcucgu >SELT_bosTau Bos taurus (cow) score: 39.15 gauuuuugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uucuug gcaggcucgu >SELT_eriEur Erinaceus europaeus (hedgehog) score: 39.15 gauuuuugca agagcag cgugacugacauu augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau gcu uucuug gcaggcucgu >SELT_sorAra Sorex araneus (shrew) score: 35.80 ggauucucca agagcag cgcggcugcccuc augaa ggccugcacug aa gacagcagcugu uggugcaggcu ggau cuuccugg caggcuc guuguaccuc >SELT_loxAfr Loxodonta africana (elephant) score: 36.15 gauuuuuuca agagcag cgugacugacauu augaa ggccuguacug aa gacagcgagcugu uaguacagacc agau gcu uuuuug gcaggcgcgu >SELT_echTel Echinops telfairi (tenrec) score: 21 UCUUUUGCAA GAGCAGC GAGACUGACCUC AUGAA GGCCUGUACUG AA GACAGCCCUGCUG UUAGUACAGGCC AGAU GCUUCUCCUG GCAGGCUC GUUUGUACCU >SELT_monDom Monodelphis domestica (opossum) score: 36.95 auuuuugcga gagcagc gugguuggcacca augaa ggccuguacug aa gacagcaagcugu uaguacagacc agau acuu acuuugc agccucguug >SELT_macEug Macropus eugenii mRNA score: 38 auuuuugcga gagcagc gugguuggcacca augaa ggccuguacug aa gacagcaugcugu uaguacagacc agau gcu uuauuu gcagccucgu >SELT_ornAna Ornithorhynchus anatinus (platypus) score: 36.17 gaucuuucua agggcaa cgugauugaaacu augaa ggccuguacug aa ggcagcaaacugu uaguacagacc agau guuccu uugcagu cucguuguac >SELT_anoCar Anolis carolinensis (lizard) score: 34.03 acaguuuuga aaggcaa caugaacgaaaua augaa ggucuguacug aa gacagcaugcugu uggugcagacu ggau acuucuc cucgccuu cauguuguug >SELT_galGal Gallus gallus (chicken) score: 38.05 gugauuucca aggccgc gugucucggacca augaa ggccuguacug aa gacagcuugcugu ugguacagacu ggau gcuucucuu gcaguc acguugugcc >SELT_danRer Danio rerio (zebrafish) score: 34.86 ccggucagcg ggacugu ggacuguggcaua augaa ggccugcgcugc aa acagcacacug uuggcacaggcu ggau gcu cagccac cacacacacu >SELT_tetNig Tetraodon nigroviridis (pufferfish) score: 33.32 ggugggucuc ccugugg gcugugagguu augaa ggucugugcug aa ggcaggacgacugc uagcucagacu ggau gucccca ucacug uucaccgugc
SELO: 9 SECIS insertion sequences
>SELO_homSap Homo sapiens (human) score: 25.77 cugcccuggc ccaugca cacccgucuuucc augau ggcagagacau cc agucaggaccuga cccgucucuguc ugag gccggcuc agcagug cagccugguc >SELO_panTro Pan troglodytes (chimp) score: 25.77 cugcccuggc ccaugca cacccgucuuucc augau ggcagagacau cc agucaggaccuga cccgucucuguc ugag gccggcuc agcagug cagcuugguc >SELO_macMul Macaca mulatta (rhesus) score: 25.77 cugccugggc ccaugca caccugucuuucc augau ggcagagacau cc agucaggaccuga cccgucucuguc ugag gccagcuc agcagug cagccugguc >SELO_loxAfr Loxodonta africana (elephant) score: 25 T2 genomic 3' CTGCCGTGTCCTGAGACACTGTCTCTC CATGGCGGCAGAGACGTCCAGTCAGGCCTGACCCGTCTCTGTCTGAT GTCGACTCTGCAGTGTGCC >SELO_proCap score: 25 CTGCTGTGTCTCGAGACCCCGTCTCTC CATGGCGGCAGAGACGCCCAGTCAGGCCTGACCTGTCTCTGCCCGAT GCTGACCTGGCAGTGTGCC >SELO_monDom Monodelphis domestica (opossum) score: 24.43 gucuggggcc uccccga uggcugcuucucc augaa ggcagagaugu cc agucggcuccga cccgucucuguc cgac gcugac ucggca gcccgagcuc >SELO_macEug Macropus eugenii (wallaby) trace score: 27 AGCCCCCCAA AUCGCUG UUUUUCC AUGAA GGCAAAGACAU CC AGUCAGUCCUGAC CUGUCUCUGUC UGAU GCUGACU CAGCAG CCUAACCCUA >SELO_danRer Danio rerio (zebra fish) score: 17.80 UGUAAGUGAA AUGUGAC UGGUUUUUA AUGAU GACUUUGACC AG CAGUCUUUGCAUGACA GGUCUCAGUC GGAA AGACAGAA GUUAAG CCUCUCUAUG >SELO_takRub Takifugu rubripes (fugu) score: 20.04 ccugcuugcc ccgaguu cuaccugcgaaua augac gacucagacaa ca agucugccagugac cugucugacuc ugaa cugugu ggugucg ccucagcaaa
SELV: 10 SECIS insertion sequences
>SELV_homSap Homo sapiens (human) score: 34.00 uuucucuccc aucuuag gagucucagcugg augau gagaagggcug aa auguugccaagu cagguccuuuu cugau ggug gcugggg cuggggugag >SELV_panTro Pan troglodytes (chimp) score: 34.00 uuucucuccc aucuuag gagucucagcggg augau gagaagggcug aa auguugccaagu cagguccuuuu cugau ggug gcugggg cuggggugag >SELV_otoGar Otolemur garnettii (bushbaby) score: 27.28 cuucucuccc aucucag gagccucgguagg augac gagaagggcug aa auauugccaugu cagguccuuuu cugau ggug gcuggga cugggguacg >SELV_musMus Mus musculus (mouse) score: 33.37 cucucucccc aucucag gagccucagcagg augau gagaagggcug aa augcugccaaac cagguccuuuu cugau ggug gcugggg cuuggguggg >SELV_ratNor Rattus norvegicus (rat) score: 33.37 uucucucccu gucucag gaaccucagcggg augau gagaagggcug aa augcugccaaac cagguccuuuu cugau ggug gcuggggc uugguuaggg >SELV_cavPor Cavia porcellus (guineaPig) score: 34.34 uuuucucccc auguuag gagcugcagcagg augau gagaagggcug aa auguuguugagu cagguccuuuu cugau ggugg cugggcu gaguggagcu >SELV_equCab Equus caballus (horse) score: 32.67 uucucucucc aucucag gagccucggugag augac gagaagggcug aa auguugccaagu cagguccuuuu cugau ggcg gcugggg cugagguggg >SELV_sorAra Sorex araneus (shrew) score: 26.16 cuucucuccc caucuca ggaaccucagaag augaa gggaagggcug aa auguugg cagguccuuuu cugac agu ggcugug gcuaugaugg >SELV_dasNov Dasypus novemcinctus (armadillo) score: 22 T2 CTTTCCCCATCTCAGGAGCCTCAGCAG GATGGCGAGAAGGGCTGAAATGTTGCAGTCAGGTCCTTTTCTGAT GGTGGCTGGGGCTAGGGTGGG >SELV_loxAfr Loxodonta africana (elephant) score: 34.00 uucucucccc aucucag gagccucagcaag augau gagaagggcug aa auguugccaagu cagguccuuuu cugau ggug gcugggg cugggguggg >SELV_echTel Echinops telfairi (tenrec) score: 26.31 uucucucucc accccag gagccgcagcaag augac gagaagggcug aa augucgu caggcccuuuu cugag ggug gcugggg cuaggcucgg
SELH: 18 SECIS insertion sequences
>SELH_homSap Homo sapiens (human) score: 23.94 UUUGUGUCCC UGGUGAU GUUGGAACAUUA AUGAU GGAACAUGGCC AA ACUUC AGUCAUGAUCC UGAA GCC AUGGUU UCUUCCCUGC >SELH_panTro Pan troglodytes (chimp) score: 23.94 uuuguguccc uggugau guuggaacauua augau ggaacauggcc aa acuuc agucaugaucc ugaa gcc augguu ucuucccugc >SELH_macMul Macaca mulatta (rhesus) score: 26.97 cuuugugucc cugguga ugcuggaacauua augau ggaacauggcc aa acuuc agucaugugcc ugaa gccaugg uuucua ccccgccaga >SELH_otoGar Otolemur garnettii (bushbaby) score: 22.59 cuuucugugc uggugau guuggagcaugu augac gggacauggcc aa acuua agucaugugcc ugaa gcu acaguuu cuuccccucc >SELH_tupBel Tupaia belangeri (treeShrew) score: 26.55 gcuuuuccug gugaugu ugaagggacauuu augau gggacauggcc aa acuuc agucaugugcc ugaa gcc aagguuu cuuccccucc >SELH_musMus Mus musculus (mouse) score: 24.55 cuuucagucc cuggaga uguugaagcauuu augau ggugcauggcc aa acuua agcuaugcacc ugaa gccauag uuucuu ccucaccaga >SELH_ratNor Rattus norvegicus (rat) score: 23.97 cuuucagucc cuggaga uguugaagcauuu augau ggugcauggcc aa acuua agcuauguacc ugaa gccauag uuucuu ccucaccaga >SELH_cavPor Cavia porcellus (guineaPig) score: 19.25 uuucuguucc ugguccu guuggggcgucu gugau gggacauggc ca aacuaaag ccauggccc ugaa gcc augguc ucuagcaucc >SELH_canFam Canis familiaris (dog) score: 23.64 agcauauccc uggugau guuggagcauuu augac ggaacauggcc aa acuuc agucauguacc ugaa gcc augguu ucuucccucc >SELH_felCat Felis catus (cat) score: 24.80 cuucucuccc uggugac guuuggagcauuu augac gggacauggcc aa auuuc agucaugugcc ugaa gcccug guuucu uccccuccag >SELH_equCab Equus caballus (horse) score: 25.73 ccuucguccc ugguguu guuggagcauuu augac gguacauggcc aa acauc agucaugugcc ugaa gcu gugguuu cuuccccucc >SELH_bosTau Bos taurus (cow) score: 28.39 ccucuguccc uggugau guuggagcauuu augac gggacauggcc aa acuuc agucauguccc ugaa gcu gugguu uccuccccuc >SELH_sorAra Sorex araneus (shrew) score: 24.10 cuuucuguuc uggugau guuggagcauug augau ggaacaugacc aa acguc agucaugugcc ugaa gccaa guuucu uucucuccag >SELH_dasNov Dasypus novemcinctus (armadillo) score: 25.97 cuucuauccu uggugau guuggagcauuu augau gggacauggcc aa acuuc agucauguacc ugaa gcca guuucu cccccuccag >SELH_choHof score: 22 UAUCCUUGGG GAUGUUG GAGCAUUU AUGAU GGGACACGGCC AA AUUUCA GUCAUGUACC UGAA GCC ACUGUU UCUUCCCCUC >SELH_loxAfr Loxodonta africana (elephant) score: 25.83 cuucuguccc uggugau guuggagcauuu augac ggaacauggcc aa accuc agucaugugcc ugaa gcc acgguuu cuuccccucc >SELH_echTel Echinops telfairi (tenrec) score: 26.97 agucccgucc cugguga uguuggagcauuu augau ggaacauggcc aa acuuc agucaugugcc ugaa gcccguc uccuca cuuccagaaa >SELH_monDom Monodelphis domestica (opossum) score: 26.74 ugccuguccc cagugag uuuggagcaucc augau ggaaccugacc aa aucuccca gucacguccc ugaa gcuuggg cuccuu cuccugggaa >SELH_anoCar Anolis carolinensis (lizard) score: 27.10 auacauucuc ugugagu uggagcauuc augau gggauaugauc aa guggagcaaaucc agucacaucuc ugaa gcu guugcc cuccuccaca
SEPP2: 5 SECIS sequences
>SEPP2_monDom Monodelphis domestica (opossum) score: 30 ACGUGCCGCU GCCCCUC CCUCCCUCCAAGA AUGAC GCCCACAGUGA AA CCCAGAGAACUGG UCCCUGUGGGC UGAU GCCC CAGAGGG GAGGAGAGGC >SEPP2_macEug Macropus eugenii (wallaby) score: 26.38 CUUGCCCAUU GCCCUUC CCUCCCUCAAUUA AUGAU GCCCACAGUGA AA CCCGGGGAACCGG CCAUGGUGGGC UGAU GCCAU GGAGAAG AGGAAUGAGG >SEPP2a_ornAna Ornithorhynchus anatinus (platypus) score: 34.28 UCCCUCGACU CCCGCUC CCGCCUCGCACUC AUGAC GUCCACGGUGUC AA CCGGCCCGCCG GGCACCGUGGAC UGAC GCC GGUCGAG GCGGAGGGGU >SEPP2b_ornAna Ornithorhynchus anatinus (platypus) score: 22.82 GGAAUCAGGA ACCCAGU AAC AUGAG GUCAUCUUCGG AA GCCUGUGCCUAGAGGA CCAAGAUAAU GGAA AAAGUGACGG ACAAGGGU GUGUAGCUGG >SEPP2_tetNig Tetraodon nigroviridis (pufferfish) score: 17 GCUGGACCCA GGCUGCU GGUGGUCCCGUUG AUGAC GUCUGCGCUGGU AA ACCUGCCUGCAGG AGCCUGUGGAC CGAC GUG UGUGGAC CCACCGGCAG
SELL: 17 SECIS sequences
>SELL_danRer Danio rerio (zebrafish) Score2: 23 no score1 gctccccctgAGGTTGCATGG ATGAA GATTTGACTCG AA TTCTTATATTTACGAAGAGTCTTTTC TGAT GGGAATTTACACCCTCAG
SELI: 17 SECIS sequences
>SELI_homSap Homo sapiens (human) score: 21.61 UUUCACUGAA UGAAGUU UGUGCUUGA AUGAA GAGUGUAUCUUA AA CCCCCUUUUUUUGGA CAGGCUGCACUU GGAU AAAAUA GGCACCA CUGUGUUGAU >SELI_panTro Pan troglodytes (chimp) score: 21.61 uuucacugaa ugaaguu ugugcuuga augaa gaguguaucuua aa cccccuuuuuuugga caggcugcacuu ggau aaaaua ggcacca cuguguugau >SELI_macMul Macaca mulatta (rhesus) score: 21.25 uuucacugaa ugaaguu ugugcuuga augaa gaguguaucuua aa ccuccuuuuuuugga caggcugcacuu ggau aacaua ggcacca cuguguugau >SELI_otoGar Otolemur garnettii (bushbaby) score: 21.25 uuucacuaag ugaaguu ugugcuuga augaa gaguguaucuu aa acccuuuuuuuuggac agguugcacuu ggau aaaaua ggcacca uuguguugau >SELI_musMus Mus musculus (mouse) score: 24.62 uuccacugaa ugaaguu ugugcuuaa augaa gagugugucuu aa acccuuuuuuuuggac agguugcacuu ggau aaaaua ggcacca cuguguugau >SELI_ratNor Rattus norvegicus (rat) score: 24.62 uuccacugaa ugaaguu ugugcuuaa augaa gagugugucuu aa acccuuuuuuuuggac agguugcacuu ggau aacaua ggcaccg cuguguugau >SELI_cavPor Cavia porcellus (guineaPig) score: 17.42 uuccagugaa ugaaguu ugugcuuga augaa gaguguaucuua aa cccuuuuuuuuuugga cagguugcacuu ggau aaaaua ggcacca cuguguugau >SELI_oryCun Oryctolagus cuniculus (rabbit) score: 19.47 ucuacugaau gaaguuu gugcuuga augaa gaguguaucuua aa cccuuuuuuuuugga cagguugcacuu ggau aaaau aggcac cacuguugau >SELI_eriEur Erinaceus europaeus (hedgehog) score: 14.43 cacggaauga aguaugu gcuuga augaa gaguguaucuu aa acccuuuuuuuuuuuugga cagguugcacuu ggau aua auaggc accacucugu >SELI_canFam Canis familiaris (dog) score: 14.04 uuccacugaa ugaaguu ugugcucga augaa gaguguaucuua aa cccuuuuuuuuuugga uggauugcacuu ggau aaaaua ggcacca cuguguugau >SELI_equCab Equus caballus (horse) score: 19.57 uuucacugag ugaaguu ugugcucga augaa gaguguauucuu aa acccuuguuuuugga uggauugcacuu ggau aaaaua agcacca cuguguugau >SELI_dasNov Dasypus novemcinctus (armadillo) score: 17.76 cuacugaauu aaguuug ugcuuga augaa gaguguaucuu aa acccuuuguuuuuuugga cagguugcacuu ggau aaaa uaggca ccacuauguu >SELI_loxAfr Loxodonta africana (elephant) score: 25.81 uuccacugaa ugaaguu ugugcuuga augaa gaguguaucuu aa acccuuuuuuuuggac agguugcacuu ggau aaagua ggcacca cuguguugau >SELI_monDom Monodelphis domestica (opossum) score: 26.46 uucuacugaa ugcaauu ugugcuuga augaa gagugugucuu aa auccuuuauauggac aggcugcacuu ggag aga auaagca caaccauguu >SELI_macEug Macropus eugenii (wallaby) trace score: 26 UUUCUACUGA AUGCAGU UUGUGCUUGA AUGAA GAGUGUGUCUU AA ACCCUUUUUAUGGAC AGGCUGCACUU GGAG AGA AUAAGCA CAACCAUGUU >SELI_ornAna Ornithorhynchus anatinus (platypus) score: 19.25 ucccagugaa ugaagcu ugugcuuga augaa gagugcaucuua aa cccauuuuuuuugga aaagcugcacuu ggag agaaag ggcacg acuguguuua >SELI_anoCar Anolis carolinensis (lizard) score: 22.02 uccccuuugu guguguc cuuugugcgugu augaa gagugcggccuc aa cccaggcgucuugga agggccgcaccc ggaa gaa acggagc acagcaaaga >SELI_galGal Gallus gallus (chicken) score: 24.62 uuuuauugag uuuauuu gugcuuaa augaa gagugcgcuuc aa acccagaccaggag agggcgcacuu ggag ugagc gagucaa accuugcucc
SELK: 19 SECIS sequences
>SELK_homSap Homo sapiens (human) score: 34.53 acaaggacug cucugug uccucacagauga augag gucaugcuggg aa uucccucugcaggga acuggccugac ugac augcaguuc cauaaa ugcagauguu >SELK_panTro Pan troglodytes (chimp) score: 34.53 acaaggacug cucugug uccucacagauga augag gucaugcuggg aa uucccucugcaggga acuggccugac ugac augcaguuc cauaaa ugcagauguu >SELK_macMul Macaca mulatta (rhesus) score: 36.14 acaaggacug cucugug uccucacagauga augag gucaugcuagg aa uucccucuacaggga acuggccugac ugac augcaguuc cauaaa ugcagauguu >SELK_otoGar Otolemur garnettii (bushbaby) score: 31.99 caaggauugc ucugugu cuucacagauga augag gucaggcuggg aa uucucucuucaggga acuggccugac ugac augcagu ucuauaa acgcacuuuu >SELK_tupBel Tupaia belangeri (treeShrew) score: 36.65 acaaggauug cucugug uccccacagauga augag guuaugccggg aa uucccuccacaggga ucuggccugac ugau acgcaguuc uauaaa ugcacauguu >SELK_musMus Mus musculus (mouse) score: 31.73 acaaggauug cucugug uccccacagauga augag gucaugcuggg aa uucccucugcagga ucuagccugac ugau acgcaguuc uauaaa uguacauguu >SELK_ratNor Rattus norvegicus (rat) score: 33.02 acaaggacug cucugug uccucacagaaga augag gucaugcuggg aa cucccucugcagga ucuggccugac ugau gugcaguuc uauaaa uguacaugug >SELK_cavPor Cavia porcellus (guineaPig) score: 33.72 cucuguguuc ucacaga uaa augag gucacgccagg aa uucucucagcaggga ucuggcuugac ugau acgcagu ucucua aaugcauaug >SELK_oryCun Oryctolagus cuniculus (rabbit) score: 38.80 accaggauug cucugug uccccacagauga augau gucaggcuggg aa uucccuccacaggga ucuggccugau ugag augcaguuc uauaaa ugcguauguu >SELK_sorAra Sorex araneus (shrew) score: 40.91 auaaggacug uucugug uccacacagauga augag gucaugcuggg aa uucccucuacgggga ucuggcaugac ugau augcag uucgaua aaugcacaug >SELK_eriEur Erinaceus europaeus (hedgehog) score: 37.30 acaaggauug cucugug uucucacagguga augag guuaugcuggg aa uucccuccaugggga ucuggcaugac ugau augcaguuc uauaaa ugcacauguu >SELK_canFam Canis familiaris (dog) score: 36.30 acaaggauug cucugug ccuucacagacgg augag guugugcuagg aa uucccuccccaggga ucuggcaugac ugac augcaguuc uauaaa ugcacauguu >SELK_felCat Felis catus (cat) score: 37.35 acgagaauug cucugug uccucacagacag augag gucgugcuggg aa uucccuccccaggga ucuggcaugac ugac augcaguuc uauaaa ugcacauguu >SELK_bosTau Bos taurus (cow) score: 39.16 cucugugucc ucacaga cga augag gucaugcuggg aa uucccuccgcaggga ucuggcaugac ugac augcagu ucuaua aaugcacguu >SELK_dasNov Dasypus novemcinctus (armadillo) score: 29.00 acaaggauug cucugug uccucacagauga augag gucauguuuggg aa uucccucugcaggg aucuggcaugac ugac uugcaguuc cauaaa ugcacauguu >SELK_loxAfr Loxodonta africana (elephant) score: 34.85 acaaagacug cucugug uccccacagacgg augag gcugugcuggg aa uucccucugcaggga ucuggcauggc ugac augcaguuc cauaaa ugcacauguu >SELK_echTel Echinops telfairi (tenrec) score: 33 ACAAGAAUUG CUCUGUA UCCCCACAGACGA AUGAG GUCGUGCUGGG AA UUCCCUCUGCAGGGA UCUGGCGUGGC UGAC AUGCAGUUC UAUAAA UGCACAUUUU >SELK_monDom Monodelphis domestica (opossum) score: 21.75 aagaauugcu cugucua cacagauua augau guugugcuggg aa cucccaucuuacagga uccaguguaac ugau ugcaa uuguaua aaugcacaug >SELK_macEug Macropus eugenii (wallaby) trace score: 23 AAAAGGAUUG CUCUGUA UCUCCACAGAUUA AUGAU GUUGUGCUGGG AA CUCCCUUCUGCCGGGA UCCGGUGUAAC UGAU UGCAGU UGUAUA AAUGCACAUG >SELK_ornAna Ornithorhynchus anatinus (platypus) score: 33.68 aauaauugug cugugaa caagcagauua augau guuuugcuggg aa uuccuucaggga uccaguauaac ugau aaagc aauuaua uaaaggcaca >SELK_anoCar Anolis carolinensis (lizard) score: 33.87 aauucccugc ucugcca auuggcgggacc augau guuguccuggg aa uuccuuauucuggga uccagggcaac ugaa aagcaguuc uguuaa auuaaaugca >SELK_galGal Gallus gallus (chicken) score: 30.90 ugagaacugu ucugcaa uauaagcagauga augaa guuguacuggg aa cuccuucaagga uccaguguaac ugaa gugcagug uuauuaa auacauguuu
GPX3: 15 SECIS sequences
>GPX3_homSap Homo sapiens (human) score: 27.80 CACGGACCCCA UGGCAGG GGUGGCGUCUUC AUGAG GGAGGGGCCCA AA GCCCUUGUGGGC GGACCUCCCC UGAG CCUG UCUGAG GGGCCAGCCCT >GPX3_panTro Pan troglodytes (chimp) score: 27.80 cacguacccc augucgg ggguggugucuuc augag ggaggggccca aa gcccuugugggc ggaccucccc ugag ccugu cugagg ggccagcccu >GPX3_macMul Macaca mulatta (rhesus) score: 27.80 cacguacccc augucag ggguggcgucuuc augag ggaggggccca aa gcccuugugggc ggaccucccc ugag ccugu cugagg ggccagcucu >GPX3_tupBel Tupaia belangeri (treeShrew) score: 29.38 ccacaucccu gugucag ggguggcaucucc augag ggaggggcccg aa gcccuuguggg cggaccucccc ugag ccugu cugagg ggccggcccu >GPX3_musMus Mus musculus (mouse) score: 33.37 cguguacccc aggucag ggguggugucucu augaa ggaggggcccg aa gcccuuguggg cgggccucccc ugag cccgu cuguggu gccagcccuu >GPX3_ratNor Rattus norvegicus (rat) score: 33.37 caugugcucc aagucag ggguggugucucc augaa ggaggggcccg aa gcccuuguggg cgggccucccc ugag cccgu cuguggu gccagcccuu >GPX3_canFam Canis familiaris (dog) score: 29.96 cauguucccc gugucag gaauggcaucucc augaa ggaggggccc ga agcccucaugggc ggaccucccc ugag ccugu cugaag ggccggcccu >GPX3_felCat Felis catus (cat) score: 30.08 ccacguccc gugucag ggauggcgucucc augaa ggaggggccc ga agcccuugugggc ggaccucccc ugag ccugu cugaag ggccagcccu >GPX3_equCab Equus caballus (horse) score: 26.20 uacauucccc augucag ggguggcaucucc augau ggaggggcccg aa gcccugguggg cggaccucccc agag ccugu cugaag ggccagcccu >GPX3_bosTau Bos taurus (cow) score: 30.87 cacguuccccg ugucagg gggcggcaucgcc augaa ggaggggcccg aa gcccgcguggg cgggccucccu ugag ccu gucugag gggccagccuu >GPX3_loxAfr Loxodonta africana (elephant) score: 16.32 cuacgucccc gugucaa gagcggcaucucc augau gguggggcccg aa gccccugugg cggaccucccc agag ccugucc caugggc cagcccuu >GPX3_echTel Echinops telfairi (tenrec) score: 12.78 ggcugugucc cgugcag agggagcaucucc augag ggugaggcccg aa gcccccgugg cggaccucgac ccgag ucug cucugg gccugccuuc >GPX3_monDom Monodelphis domestica (opossum) score: 22.94 ugaacauaag ggauggc aucucu augau ggugggaucca aa gccucuucaggg cggguuccauc agag ccugcaaaa gguguc aggacccuua >GPX3_ornAna Ornithorhynchus anatinus (platypus) score: 31.33 ugucccagaa uggggag guggcaucauc augac agcggggucug aa agccccuccugga uggaccccgcc cgaa ccug cucggcg guggcaugac >GPX3_anoCar Anolis carolinensis (lizard) score: 28.20 GAGAUUUUGG GCCAAGG AAGUUGCAUCACU AUGAG GGUUAGGUCUG AA AGCUCCCAAAAAGAG CGGACCUAGCC UGAG GCUGCAAA GCUCUGGU GUAGCCCUUU >GPX3_galGal Gallus gallus (chicken) score: 30.65 GCCCUGGGGA GCAGAGG AUGACAUCUCC AUGAA GGCCUGGCCUG AA AGCCCCCACCAUGGGG UGGGCUCGGCC CGAU CCCG CCCAGGC GCGGUGCAGC
GPX6: 10 SECIS sequences (most species have selenocysteine)
>GPX6_homSap Homo sapiens (human) score: 18.10 cccaccucac augaagg gaagggcaucucc augau gguggauccc aa aaccccucuggguc gcacccugcc agag ccu uccuug gugccugucc >GPX6_panTro Pan troglodytes (chimp) score: 23.45 cccaccucac augaagg gaggggcaucucc augac gguggguccc aa aaccucucgggguc ggacccugcc agag ccu uccuug gugccugucc >GPX6_macMul Macaca mulatta (rhesus) score: 21.81 accucauuca cagaagg gaggggcaucuuu augau ggugggucuc aa aaccucucuggguc agacccuacc agag ccu uccuug gugccugucc >GPX6_cavPor Cavia porcellus (guineaPig) score: 19.91 AGAAGCCUCC CUAGAAG GAAGGGUGUCUCC AUGAU GGUGGGUCCC AA AAGCCCUGGAUC GGACCCUACC AGAA CCU UCCCUGG UGCCUGUCCU >GPX6_canFam Canis familiaris (dog) score: 24.33 CCUGGCCUCA UAUGAAA GGAGGGCAUCUCC AUGAU GGUGGGUUCC CA AGCCCCUGCGGUC GGACCCUACC AGAG CCU UCUUGG GUGCCUGUCC >GPX6_equCab Equus caballus (horse) score: 25.00 uucaucucac augagcu gaggggcaucucc augau gguggguccc aa agccccugugggc aggacccaac cagaa cucug ugccugu cccuuagugc >GPX6_bosTau Bos taurus (cow) score: 23.33 cucacuucac augagga aaagggcaucucc augau gguggguccc aa agccucucuggguc ggaccccacc agag ccu uccuugg ugccuguccc >GPX6_eriEur Erinaceus europaeus (hedgehog) score: 24.33 ccuggccuca uaugaaa ggagggcaucucc augau gguggguucc ca agccccugcgguc ggacccuacc agag ccu uccuugg ugucuguccu >GPX6_sorAra Sorex araneus (shrew) score: 22.95 uuucauauga aaggagg gggcaucucc augau ggugggucuc aa agccccucuggguc ggacccuacc agag cccugcugg guguccu gucccuuugu >GPX6_dasNov Dasypus novemcinctus (armadillo) score: 19.82 cucaccuuac augaagg gagggacauaucc augau gguggguccu aa agcccuucuggguc agacccuacc agaa ccu ucucug gugccugucc >GPX6_loxAfr Loxodonta africana (elephant) score: 19.28 cucaccucac augaaug gaggggcauaucc augau gguggguccc aa agccucucuagguc ggacccuauc agaa ucuucccugg ugccugu cccuuagugc >GPX6_echTel Echinops telfairi (tenrec) score: 31.33 ugugucccag aaugggg agguggcaucauc augac agcggggucu ga aagccccuccuggau ggaccccgcc cgaa ccu ucccug gugccugucc
GPX1: 17 SECIS insertion sequences
>GPX1_homSap Homo sapiens (human) score: 20.45 UGCUGUCUCG GGGGGGU UUUCAUCU AUGAG GGUGUUUCCUCU AA ACCUACGA GGGAGGAACACC UGAU CUUACAGAAA AUACCAC CUCGAGAUGG >GPX1_panTro Pan troglodytes (chimp) score: 20.45 gugcugcucu guggggg gguuuucaucu augag gguguuuccucu aa accuacga gggaggaacacc ugau cuuacagaaa auacccc cucgagaugg >GPX1_macMul Macaca mulatta (rhesus) score: 26.45 gcagcgcugc ucucugg gggguuuucaucu augag gguguuuccucu aa accuaca aggaggaacacc ugau aau acagaa aauacccccu >GPX1_otoGar Otolemur garnettii (bushbaby) score: 25.30 auagcgcugc ucucugg ggggguuucauuc augau aguguuaccucu aa acuugcau gggggaacacc ugau gcc ccagaaa auccccugag >GPX1_tupBel Tupaia belangeri (treeShrew) score: 21.14 cagcgcugcg ucugggg gguuucaucc augac ggugucuccucu aa accccga aggaggaacgcc ugau guccggaaaa cccccca ggugggcgcc >GPX1_musMus Mus musculus (mouse) score: 29.78 ggcugcacuc ugggggg cgguucuucc augau gguguuuccucu aa auuugca cggagaaacacc ugau uuccaggaa aaucccc ucagaugggc >GPX1_ratNor Rattus norvegicus (rat) score: 25.72 ggcugcccuc cgggggg agguuuuucc augac gguguuuccucu aa auuuaca uggagaaacacc ugau uuccagaaa aaucccc ucagaugggc >GPX1_cavPor Cavia porcellus (guineaPig) score: 15.34 uugcccagau gcucucc ugaggguucuucc augaa gguguuuccucuc aa ccugua uagaggaacauc cgau uccca ggaauu ucccagagag >GPX1_oryCun Oryctolagus cuniculus (rabbit) score: 24.38 guggccugcu gcucucu ggggguuucaucc augag ggcguucccccg aa aacaaa uggaggaacgcc ugau gucc gggaaac ccccaggugg >GPX1_eriEur Erinaceus europaeus (hedgehog) score: 19.31 auagccgcga gccgcug ggcaucucaucc augac ggcgccgccuuc aa accugcgagcag gaaggagcgcc cgau agc cgcgaga gcccccagcg >GPX1_canFam Canis familiaris (dog) score: 22.40 ucacagcgcu gcccucu ggggauuucaucc augau gguguuuccuug aa aucugca uggaggaacgcc ugau uucca ggaaagu ccccugagcu >GPX1_felCat Felis catus (cat) score: 27.75 cacagcacug cccucug gggauuucaucc augau ggcguuuccucg aa auuugca uagaggaacgcc ugau uuc cagaag aauccccuga >GPX1_equCab Equus caballus (horse) score: 27.08 ucacagugcu uuucucu ggggauuucaucc augau ggcguuuccucu aa acaugc augaggaacgcc ugau guu aaggaga aucccccgag >GPX1_bosTau Bos taurus (cow) score: 29.83 agucagcgcu gcucucc agggauuuugccc augaa gguguucccucu aa accuacg uggaggaaugcc ugau gucca ggaaaau ccccugaggu >GPX1_dasNov Dasypus novemcinctus (armadillo) score: 28.92 uguccuccac acggggu uuucaucc augac gguguuuccucu aa accugca gggaggaacacc ugaa guccggcaaa aucccc cgagaugggu >GPX1_choHof score: 17 T2 frag GGGGTTTCATC CATGACGGTGTTTCCTCTAAACTATATGGAGGAACCCTGAA GCCCGGAAAAATCCCCCAAAAT >GPX1_loxAfr Loxodonta africana (elephant) score: 25.48 acuaggcggc uuuccgu ggggguuucauua augag gguguuuucucu aa accugaa uggaggaacacc ugau guc ugggaa gauacccccc >GPX1_echTel Echinops telfairi (tenrec) score: 22 CGCGGCCUGC CUUGCGU GGGGACUUCAUCA AUGAG GGUGUUUCUUCU AA ACCCGAAU GGAGGAACACC CGAU GUCC GGGAAAG CCCCCCGCAG >GPX1_monDom Monodelphis domestica (opossum) score: 26.38 gugcuaaggg uccguga ggguuuuaucu augau gguguuguuuu aa accauuaagga gaaagaacacu ugau aaugc uuguaa aaucccauga >GPX1_ornAna Ornithorhynchus anatinus (platypus) score: 25.55 ugcuauuugc ccauggg ggagaucucauuu augau ggugcuccuucu aa accuuua cggaagagcacu ugau gga uccugag gaacuccaug
GPX2: 17 SECIS insertion sequences
>GPX2_homSap Homo sapiens (human) score: 29.54 AGACUUGGGU AAGCUCU GGGCCUUCACAGA AUGAU GGCACCUUCCU AA ACCCUCA UGGGUGGUGUC UGAG AGGCGUGA AGGGCCU GGAGCCACUC >GPX2_panTro Pan troglodytes (chimp) score: 29.54 agacuugggu aagcucu gggccuucacaga augau ggcaccuuccu aa acccuca ugggugguguc ugag aggcguga agggccu ggagccacuc >GPX2_macMul Macaca mulatta (rhesus) score: 29.54 agacuugggu aagcucu gggccuucacaga augau ggcaccuuccu aa acccuca ugggugguguc ugag aggcguga agggccu gaagccaccc >GPX2_otoGar Otolemur garnettii (bushbaby) score: 27.86 agacuugggu gggcucu gggccuucacaga augau ggcaccauccu aa acgccuc ugggugguguc ugag aagagcg gaaggcc uggagccacc >GPX2_tupBel Tupaia belangeri (treeShrew) score: 28.69 agacuuaggu gggcucu gagccuucacaga augau agcaccuuccu aa acccccc cgggagguguc ugag aagugug acaggcc cggagccagc >GPX2_musMus Mus musculus (mouse) score: 33.65 agucuggggu agguucu gggccuucacaga augau ggcaucuuccu aa acccuuc ugggagauguc ugag aaguugug aaggguc cagagccagu >GPX2_ratNor Rattus norvegicus (rat) score: 30.80 cugggguagg ugcuagg ccuucuucacaga augau ggcaucuuccu aa acccuuc ugggggauguc ugag acg uugugaa gggcccagag >GPX2_cavPor Cavia porcellus (guineaPig) score: 20.08 cuaggacagg uggcucu gggccuucacaga augac ggcaccguccu aa acgcua ugggugguauc ugag aagugug aauggcug gagccagccu >GPX2_sorAra Sorex araneus (shrew) score: 29.00 aagcuggggc aaacucu gggccuucgcaga augau ggcaccucccu aa auccau ggggugguguc ugag gcgugcg agggccu ggaaacagcc >GPX2_eriEur Erinaceus europaeus (hedgehog) score: 25.06 aaagcugggu gguuucu ggaccuucacaga augau agcaccuuccu aa accuaua gggaugguguc ugag aaaugu gaagggc cugaaguaaa >GPX2_canFam Canis familiaris (dog) score: 22.57 aagacuuggg uggcucu gggccuucacaga augau ggcaccuuccu aa auagua ugggcgguguc ugag aagugug aagggcu cagagccagc >GPX2_felCat Felis catus (cat) score: 22.45 aagacuuggg uggaucu gggccuucacaga augau ggcaucuuccu aa acugua ugggcgguguc ugag gaguguga agggcu cggagccagc >GPX2_equCab Equus caballus (horse) score: 28.45 agacuucggu aggcucu gggccuucacaga augau ggcaccuuccu aa accugua uggacgguguc ugaa aagcgug aagggcc ccgagucagc >GPX2_bosTau Bos taurus (cow) score: 26.14 agacaugggu gggcucu gugucuucacaga augau ggcaccuuccu aa aucugua ugggcgguguu ugag aagagu gaaggcc uggagccagc >GPX2_loxAfr Loxodonta africana (elephant) score: 27.08 agacuugggu aggcucu ggaccuucgcaga augau ggcaccuuccu aa acucag ugggugguguc ugag aaaugug aagggccu agggccagcc >GPX2_echTel Echinops telfairi (tenrec) score: 29.95 agugcuggug uggcucu gggccuucacaga augau ggcaccuuccu aa acccuc cggaagguguc ugag aaaugug aagggcc uggggccggc >GPX2_monDom Monodelphis domestica (opossum) score: 26.38 gugcuaaggg uccguga ggguuuuaucu augau gguguuguuuu aa accauuaagga gaaagaacacu ugau aaugc uuguaa aaucccauga >GPX2_ornAna Ornithorhynchus anatinus (platypus) score: 27.55 gggcucgagc agccucc agaccuucacaga augac ggugucuccuu aa acccuaac cgggaggcacc cgag agccggu gaagggc cuggugacug
GPX4: 14 SECIS sequences
>GPX4_homSap Homo sapiens (human) score: 29.19 CCACGCCCUU GGAGCCU UCCACCGGCACUC AUGAC GGCCUGCCUG CA AACCUGCUGGU GGGGCAGACC CGAA AAUCC AGCGUGC ACCCCGCCGG >GPX4_panTro Pan troglodytes (chimp) score: 29.19 CCACGCCCUC GGAGCCU UCCACCAGCACCC AUGAc ggccugccug ca aaccugcuggu ggggcagacc cgaa aaucc agcgugc acccugccgg >GPX4_macMul Macaca mulatta (rhesus) score: 33.09 CCACGCCCUC GGAGCCU UCCACCGGCACCC AUGAc ggccugcuugc aa accagcuccuggu gaggcagacc cgaa aaucc agcgugc accccgcugg >GPX4_tupBel Tupaia belangeri (treeShrew) score: 31.05 CCGCGCCCCG GAACCUU CCACCCGGCACCU UUGAc ggucugccuau aa accugccacug gugaggcagacc cgag aaccu ggcgugc acccugccgg >GPX4_musMus Mus musculus (mouse) score: 35.62 UGACCCCUGG AGCCUUC CACCCCGGCACUC AUGAa ggucugccug aa aaccagccugcuggu ggggcagucc ugag gaccu ggcgugc acccugccgg >GPX4_ratNor Rattus norvegicus (rat) score: 34.33 UGACCCCUGG AGCCUUC CACCCCGGCACUC AUGAc ggucugccug aa aaccagcccgcuggu ggggcagucc cgag gaccu ggcgugc accccgccgg >GPX4_cavPor Cavia porcellus (guineaPig) score: 34.46 UGGCCGCCCC GAGUCUC CUACCCGGGUGCC AUGAc ggccugccug ca aaccagcgug cugguggggc agac ccg aggaugc gugcacugcc >GPX4_canFam Canis familiaris (dog) score: 31.12 AUGCCCCUCG GAGCCUU CCACCCGGCACCC AUGAc agucugucuaa aa accagcccgcugg uggggcagacc cgag aaccc ggcgugc acccugccgg >GPX4_sorAra Sorex araneus (shrew) score: 36.85 CUGCCCCUCG GAGCCUU CCACCCGGCGCCC AUGAc ggucugccugc aa accagccc gcugguggggc agac ccgagaaccc ggcgugc accuugccgg >GPX4_loxAfr Loxodonta africana (elephant) score: 34 traces with coding no genomic GCGCUUCUCA AGCCUUC CACCCGGCACCC AUGAC GGCCUGCUUGC AA ACCAGCCCGCUGGU GAGGCAGACC CGAG AACCU GGCGUGC ACCUCUGCCG >GPX4_echTel Echinops telfairi (tenrec) score: 40.88 UUGAUCCCCA GAGUCUU CCACCCGGCACCA AUGAc ggucugccuuc aa accaggccucugg ugaggcagacc cgau gaccc ggcgugc acucagccgg >GPX4_monDom Monodelphis domestica (opossum) score: 40.88 UUGAUCCCCA GAGUCUU CCACCCGGCACCA AUGAC GGUCUGCCUUC AA ACCAGGCCUCUGG UGAGGCAGACC CGAU GACCC GGCGUGC ACCUCAGCCG >GPX4_macEug Macropus eugenii mRNA score: 22.86 T2 AGTCTTCCACCCGGCACC AATGACGGTCTGCCTTCAACCAGGTCACTGGTGGGGTAGACTCGAC AACCCGGCGTGCATCTCAGCCG >GPX4_ornAna Ornithorhynchus anatinus (platypus) score: 33.89 ACCUCCGCUU UCCCGGG ACGCUCUGCCUCC AUGAc ggccggccuu ca agccaaaaccaguugg uggggccggcc ugaa caa accggca cgggucccgg >GPX4_anoCar Anolis carolinensis (lizard) score: 38.77 UGCUCCCCGU GGGCCCC CUCCUCCAGCACC AUGAc ggccugccuug aa gccagcuugcugg ugaggcagacc cgaa gauuc ggcgugc acugcuggag >GPX4_galGal Gallus gallus (chicken) score: 38.77 UGCUCCCCGU GGGCCCC CUCCUCCAGCACC AUGAc ggccugccuug aa gccagcuugcugg ugaggcagacc cgaa gauuc ggcgugc acugcuggag
SEPW: 13 SECIS sequences
>SEPW_homSap Homo sapiens (human) score: 36.01 GACCCAGCCC CUCUCAG CAGACGCUUC AUGAU AGGAAGGACUG AA AAGUCUUGUGGACACC UGGUCUUUCCC UGAU GUU CUCGUGG CUGCUGUUGG >SEPW_panTro Pan troglodytes (chimp) score: 36.01 GACCCAGCCC CUCUCAG CAGACGCUUC AUGAU AGGAAGGACUG AA AAGUCUUGUGGACACC UGGUCUUUCCC UGAU GUU CUuGUGG CUGCUGUUGG >SEPW_ponPyg Pongo pygmaeus (orang) score: 30.55 GACCCAGCCC CUCUCAG CAGACGCUUC AUGAU AGGAAGGACUG AA AAAUCUUGUGGACACC UGGUCUUUCCC UGAU GUU CUCGUGG CUGCUGUUGG >SEPW_macMul Macaca mulatta (rhesus) score: 33.55 GACCCAGCCC CUCUCAG CAGACGCUUC AUGAU AGGAAGGACUG AA AAGUCUUGUGGACgCC UGGUCUUUCCC UGAU GUU CUCGUGG CUGCUGUUGG >SEPW_musMus Mus musculus (mouse) score: 34.04 UUGGCCCAGC CCCUCGU GGCAGACGCUUC AUGAU GGGAAGAACUG AA AUGUCUCGUGGACGC CUGGUCUUUCC CUGAU GUCCCU GCGACUG CCACGUAGGG >SEPW_ratNor Rattus norvegicus (rat) score: 26.55 UGGCCGGCCU UUCUUGG CAGCCGCUUC AUGAC AGGAAGGACUG AA AUGUCUCAAAGACCUG UGGUCUUUCUU CGAU GUU CCUGCGG CCACCAAGUC >SEPW_canFam Canis familiaris (dog) score: 34.04 UUGGCCCAGC CCCUCGU GGCAGACGCUUC AUGAU GGGAAGAACU GA AAUGUCUCGUGGACGCC UGGUCUUUCC CUGAU GUCCCU GCGACUG CCACGUAGGG >SEPW_dasNov Dasypus novemcinctus (armadillo) score: 28.71 CCCAGCUGCC CUUGGCA GACGCUUC AUGAG GGGAAGGACCU AA AUGCGUCGUGGAUGCC UGGUCUUUCCC UGAU GCUCCUUCAC CUGCCAG AUGGGGCAGA >SEPW_choHof score: 38 CUUGGCCCAG CCGCCCU CGGCAGACGCUUC AUGAG GGGAAGGACUG AA AUGUCUCACGGAUCC UGGUCCUUCCC UGAU GUCCCU GCGGCUG CUGGGUGGGG >SEPW_loxAfr Loxodonta africana (elephant) score: 19 genomic 3' UGGCCCAGCC CCUUUCA GCAGACACUUC AUGAC AGGAGGACUGA AA UGUCUCCCAGACGC CUGGUCUUUCC CUGAA UCUGUCGGC UGCAGG ACAGGGCAGC >SEPW_echTel Echinops telfairi (tenrec) score: 25 T2 TGGCCCAGCCCCTCCTGGCAGACGCTT CATGACAGGAAGGACCGAAATGTCTTCTGGACGGGTGGTCTTTCCCAGAA TCCCCCGCGGCTGCTGGGC >SEPW_monDom Monodelphis domestica (opossum) score: 27 CCUCGUCUCG GCCCCUG CCCCCGAUGCUCC AUGAC GGGAAGGAUCG AA AUGUCCUCAGGAUUCC UGGUUCUUCCC UGAA AUCCUCG CAGCUGU UGGGCUGGGG >SEPW_macEug Macropus eugenii (wallaby) trace score: 8 T2 CCTTGTCTCCATCCCTGCCCATAAATGCTTTATGACCA GAATGATGGAAATGTCCTCTGGATTCCTGGTTCTTCCCTGA AATTCTTGCACTCATTGGGCTGGGG
DIO1: 9 SECIS sequences
>DIO1_homSap Homo sapiens (human) score: 29.16 uuuuaacucu gugucuu uacauauuuguuu augau ggccacagccu aa aguacaca cggcugugacu ugau ucaa aagaaa auguuauaag >DIO1_ponPyg Pongo pygmaeus (orang) score: 29.16 uuuuaacucu gugucuu uacauauuuguuu augau ggccacagccu aa aguacaca cggcugugacu ugau ucaa aagaaa auguuauaag >DIO1_macMul Macaca mulatta (rhesus) score: 29.16 uuucaacucu gugucuu uacauauuuguuu augau ggccacagccu aa aguacaca cggcugugacu ugau ucaa aagaaa auguuauaag >DIO1_cavPor Cavia porcellus (guineaPig) score: 23.71 uguuaacucu gcuucuu uucauauuuguuc augac ggucacagucu aa aguacaca cagcugugacc ugau uuga aagaaa auguuuuaag >DIO1_bosTau Bos taurus (cow) score: 27.83 uuuaacucug ccucuuu ucauauuuguuc augac ggccacagccu aa aguacaca cggcugugacu ugau uug aaagaa aauguuuuaa >DIO1_sorAra Sorex araneus (shrew) score: 23.57 ggaaacucag cuucucu ucauauuuguuu augac agccccagcug aa aguacaca cagcuguggcu ugau ugg aaagaa aauguuuuaa >DIO1_eriEur Erinaceus europaeus (hedgehog) score: 25.87 uuuaacucug cuuucuu cucauauuugcuu augau ggucacagcuu aa aguauaca cagcugugacu ugau ugg aaagaa aauauuuuaa >DIO1_Dasypus novemcinctus (armadillo) score: 28 T2 TTTTAACTCTGctTCTTTtCATATTTGTT TATGATGGCCACAGttTAAAGTACAtACaGCTGTGACTTGAT atgaAAAAGAAAtattttaa >DIO1_loxAfr Loxodonta africana (elephant) score: 22 T2 genomic 3' note pseudogene TTTTAACTCTGCTTCTTTTCATGTATTTATGATG g GCCACAGCCTAAAGTGCACAACAGCTGTGACTTGATTTGAA AAACATCTTTAAG >DIO1_echTel Echinops telfairi (tenrec) score: 25 UGCUUUAACU CUGCUUC UUUCCAUAUGUUU AUGAU GGCCACAGCUU AA AGUACACAA CAGCUGUGACU UGAG CGAA AAAUGUA UUUUAAGAUG >DIO1_monDom Monodelphis domestica (opossum) score: 12 UUUCCAUCCU GCUUCUA CAAAUAUUUAUUU AUGAC AAUCACAGCCU AA AGCUCAGGGC AGCUGGGAUU CGAC GGGAG AAAAAGU UUGUAAGAGG >DIO1_ornAna Ornithorhynchus anatinus (platypus) score: 23.83 cccggauccg guuccgu gaauauugguuu augag ggucacagugu aa agcgcaug cagcugugacu ugau cug agaaaau auuucugcgg >DIO1_calMil Callorhinchus milii (elephantfish) score: 18.25 513 stop 591-684 SECIS uaccaaagga acucucu cccuuauucaga augau uuucucagccuu aa acgcu guggcugggagc ugau guguugaau acggag aaacagcucu
DIO2: 12 SECIS insertion sequences
>DIO2_homSap Homo sapiens (human) score: 29.74 cagagaugug cagaguu gaccagugugcgg augau aacuacugacg aa agagucaucgacuc aguuagugguu ggau guaguc acauuag uuugccucuc >DIO2_macMul Macaca mulatta (rhesus) score: 29.74 cagagaugug cagaguu gaccagugugcgg augau aacuacugacg aa agagucaucgacuc aguuagugguu ggau guaguc acauuag uuugccucuc >DIO2_musMus Mus musculus (mouse) score: 29.14 cggagauguu cagagcu cacuggugugcga augau aacuacugacg aa agagcugucugcuc agucugugguu ggau guagucacac gagucug ccuuucugca >DIO2_ratNor Rattus norvegicus (rat) score: 27.88 ccgagauguu cggagcu cacuggugugcga augau aacuacugacg aa agagucaucugcuc agucugugguu ggau guagucacac gagucug ccucuccauc >DIO2_canFam Canis familiaris (dog) score: 27.29 cugggaugug cagaggu gaccagugugcga augau aacuacugaug aa agagucacugacuc aguuagugguu ggau aca gucacau uaguuuuccu >DIO2_dasNov Dasypus novemcinctus (armadillo) score: 26.93 cugggaaguu cagaggc uaccagugugcca augau aacuacugacg aa agaggcaucgacuc aguuagugguu ggau gua gccacau uaguuugccu >DIO2_choHof score: 25 CUGAGAAGUU CAGAGGC UGCCAAUGUGCGA AUGAU AACUACUGAC AA AAGAGGCAUUGACUC AGUUAGUGGUU GGAU GUA GUCACAU UAGGUUGCCU >DIO2_loxAfr Loxodonta africana (elephant) score: 25 traces lacking coding CUGGGAUGUG CAGAGGC UACUAGUGUGCAA AUGAU AACUGCUGACG AA AGAGGCACUGACU CAGUUAGUGGUU GGAU GUA GUCACAU UGGUUUGCCU >DIO2_echTel Echinops telfairi (tenrec) score: 24 CCAGGAUGUG CAGAGGC AACUAGUGUGCAA AUGAU AACUGCUGAUG AA AGAGGCACCGACU CAGUUAGUGGUU GGAU GUAGU CACACUG GUAUGCCUCU >DIO2_monDom Monodelphis domestica (opossum) score: 28.86 guguggaagg gcugcca gugugcaa augau gaucucuaaca aa agagucagucacuc cguuagagguu ggau gugg ucacagu ggcuugcugc >DIO2_macEug Macropus eugenii mRNA score: 28 ccaaguuaug gaagggc ugccagugugcaa augau gaccccuaacg aa agagucugucacuc aguuagagguu ggau guggucaca guggcuu gcuacuacau >DIO2_ornAna Ornithorhynchus anatinus (platypus) score: 24.54 ccaggauagg gcugagu uacuagugugcaa augaa gaccaccaaca aa agagaauuuaacuc aguuggugcuc agau auc aucacac uggcuuacuu >DIO2_anoCar Anolis carolinensis (lizard) score: 27.11 ugaggaugaa gcagagc uguuggugucuuu augaa gaucaccaaca aa agagugucucauuc aguugguguuc agau gucuua gcacug gcauuucucu >DIO2_galGal Gallus gallus (chicken) score: 24.57 ggggugaaac cugaaug cuuguguguuu augaa gagcacuaaca aa agaguaauugacuc aguugguguuc agau acucuca cacugg cauuccucug
DIO3: 10 SECIS insertion sequences
>DIO3_homSap Homo sapiens (human) score: 31 uugggugcac aggagcc ccacugcug augac gaacuaucucu aa cuggucuugacca cgagcuaguuc ugaa uugca ggggccu caaagcagca >DIO3_panTro Pan troglodytes (chimp) score 30.95 uugggugcac aggagcc ccacugcug augac gaacuaucucu aa cuggucuugacca cgagcuaguuc ugaa uugca ggggccu caaagcagca >DIO3_macMul Macaca mulatta (rhesus) score: 30 uugggugcac aggagcc ccacugcug augac gaacugucucu aa cuggucuugacca cgagcuaguuc ugaa uugca ggggccu caaaacagca >DIO3_musMus Mus musculus (mouse) score: 26 gugcgcugga gcccugg cugcug augac gaaccgccucu aa cugggcuugaccac gggucggcuc ugaa uugca gagaggc ucgaaacagc >DIO3_ratNor Rattus norvegicus (rat) score: 26 gugcgcugga gcccugg cugcug augac gaaccgccucu aa cugggcuugaccac gggucggcuc ugaa uugca gagaggc ucgaaacagc >DIO3_cavPor Cavia porcellus (guineaPig) score 24.06 uguuuugggc gcgcaug agccccucugcug augac gaacugucccu aa cuggucucgacc acgggcggguu ccgaa auug caggaug gcucgaaucg >DIO3_canFam Canis familiaris (dog) score 25.84 uauuuugggu gcuggcg agccccacugcug augac gagccgccucu aa cuggucuugacca cgagcugguuc ugag uugcagggg ggcuugc agcggcgccu >DIO3_felCat Felis catus (cat) score 29.2 cauuuggggu gcucacg agccccacugcug augac gagcuaucucu aa cuggucuugacca cgagcugguuc ugaa ccgc agggggc uugcagcagc >DIO3_bosTau Bos taurus (cow) score 23.46 uauuuugggu gcucacg agccccacugcug augaa gagcugucucu aa cuggccucgacca cgagcugguuc ugau uugc aggaggc ucgcagcagc >DIO3_choHof score: 23 UGUUUUUGGU GUGCACG AGCCCCGCUGCUG AUGAC GAACUGUCUCU AA CUGGUCUCGACCA CGAGGCGAUUC CGAA UUGCGGGG AGCUCGC AGCAGCGCCU >DIO3_dasNov Dasypus novemcinctus (armadillo) score: 18 UGAUUUGGGU GCGUGCG AGCCCCGCUGCUG AUGAC GAGCUGUCUCU AA CUGGUCUCGACCA AGAGGCGAGUC CGAA UUGC CGGGGGC UCGCCGCAGC >DIO3_loxAfr Loxodonta africana (elephant) score 21.95 uguuuucggu gcgcuag agccccacugcug augac gaacugucucu aa cuggucuugaccac gagcugauuc cgaa uugcagggaa cucgcagc agcgccuaaa >DIO3_echTel Echinops telfairi (tenrec) score: 15 T2 TGTTTTCGGTGCTCTGCAGCCCCACTGCT GATGACGAACTGCCTCTCACTGGTCTTGACCACGAGCTGCTTCTGAA ATGCAGGGGACTCGCAGCCGCACCTAAA >DIO3_monDom Monodelphis domestica (opossum) score: 22 GUUCUGGCAC UUCUUUG GUCCCUCUGGUA GUGAC GACCUGUUUCG AA UUGGUCCCAGACCAG GAAACCGUGUC UGAU AUUC UUAGCGA CCUAGAAGUG >DIO3_galGal Gallus gallus (chicken) score: 15 AGUGCUAACU GGGAUUU CUAGUAUUUCUUU GUGAU GACCGAUUUUG AA AUGGGUUUCUCUAAUGCCAG GAAAUCGUGUC UGAU GUUGUC AAGUACU AGAACUGCC >DIO3_chiPun Chiloscyllium punctatum (shark) score: 26 EU275162 form 2 gaauucgcag guauucu ugacacuuuuuuu augaa agccgguuuuu aa auggccucagugccag gaaaccacgcc ugau gug acaaagu guuugggaua
SELM2: 16 non-canonical SECIS insertion sequences
>SELM2_homSap Homo sapiens (human) score: non-canonical ggggacgggcatcggctctcagcaagagaagtattcccgggatgctgagcgcttcattctgtctcc >SELM2_panTro Pan troglodytes (chimp) score: non-canonical ggggacgggcatcggctctcagcaagagaagtattcccgggatgctgagcgcttcattctgtctcc >SELM2_macMul Macaca mulatta (rhesus) score: non-canonical ggggacggggcatcggccctcagcaagagaggtattcccgggatgctgagcgcttcattctgtctcc >SELM2_tupBel Tupaia belangeri (treeShrew) score: non-canonical ggggacagcacatcggcccccagcaagggaggtgctcccgggatgctgagcgcttcattcggtctcc >SELM2_musMus Mus musculus (mouse) score: non-canonical ggagactggcgtcggccctcagcaagggagatgctcccgggatactgagcgcttcattctgtctcc >SELM2_ratNor Rattus norvegicus (rat) score: non-canonical ggagaccggcgtcggccctcagcaagagagtttatgctcccgggatgctgagcgcttcattctgtctcc >SELM2_cavPor Cavia porcellus (guineaPig) score: non-canonical agacgggcatcggtcctcagcaagggagatgctcccgggatgctgagcacttcattcggtcttc >SELM2_oryCun Oryctolagus cuniculus (rabbit) score: non-canonical ggggacggggcgtcggccctcagcaagggagggggctcccgggatgctgagcgcttcatcgtgtctccc >SELM2_sorAra Sorex araneus (shrew) score: non-canonical ggggacggggcgtcgaccctcagcgagggagtgctcccgggacactgagggcttcattctgtcccc >SELM2_canFam Canis familiaris (dog) score: non-canonical tgggacgaggcatcggccctcagcgagggagacgctcccgggacgctgagcgcttcattctgtcccc >SELM2_felCat Felis catus (cat) score: non-canonical tgggacggggcgtcggccctcagcgagggaggtgttcccgggacgctgagcgcttcattctgtcccc >SELM2_equCab Equus caballus (horse) score: non-canonical cgggaccgagcgtcggccctcagcgagggaggtgctcccgggacgctgaacgcttcattctgtcccc >SELM2_bosTau Bos taurus (cow) score: non-canonical ggggacggggcgtcggccctcaacgagggaggtgctcccgggccgctgagcgcttcattctgtcccc >SELM2_dasNov Dasypus novemcinctus (armadillo) score: non-canonical taggacagggcgtcggccctcagcgagggagatgctcccgggccgctgagcgcttcattcggtcccc >SELM2_anoCar Anolis carolinensis (lizard) score: non-canonical agggacgctgtctatccaccctcggcggcaagagctttcgtccgagggcttcatcctgtcccg