Selenoprotein evolution: introduction: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 68: | Line 68: | ||
Like many genes on the densely packed human chromosome 19, SEPW1 seems to have given rise to a segmental duplication during the boreoeuthere stem (ie, it is not present in any xenarthra, afrothere, marsupial or earlier vertebrate). The second copy, called SELV, retains the same intron placements and phases but with greatly expanded and compositionally anomalous exon 1. | Like many genes on the densely packed human chromosome 19, SEPW1 seems to have given rise to a segmental duplication during the boreoeuthere stem (ie, it is not present in any xenarthra, afrothere, marsupial or earlier vertebrate). The second copy, called SELV, retains the same intron placements and phases but with greatly expanded and compositionally anomalous exon 1. | ||
This | This long exon retains some conservation at its distal end to the last 5 residues of SEPW1 (unsurprising because the cysteine of the CxxU motif is at the end). There is observable conservation also at the amino terminus but otherwise the exon is evolving unconstrained. Possibly the initial methionine of SEPW1 was lost at the time of duplication but the terminal residues and splice donor were retained. Transcription far upstream lead to a remote methionine serving as replacement. | ||
This is a very odd gene but transcripts in a half dozen species and excellent conservation in exons 2-5 over hundreds of millions of years witin boreoeutheres proves that it is evolving under selective constraint and so is functional. Distal exons however retain only 50% conservation to those of SEPW1 | This is a very odd gene but transcripts in a half dozen species and excellent conservation in exons 2-5 over hundreds of millions of years witin boreoeutheres proves that it is evolving under selective constraint and so is functional. Distal exons however retain only 50% conservation to those of SEPW1 | ||
Revision as of 15:21, 23 April 2008
Introduction to selenoprotein evolution
(more selenoprotein classes shortly)
SEPP2: lost in placentals
SEPP2 is a newly discovered paralog of SEPP1 with a UxxC motif (plus a later conserved cysteine and distal CxxC motif) and high-scoring putative SECIS element in 3' UTR that is quite conserved in vertebrates but only through marsupials. No corresponding gene -- or even decayed debris -- can be found in syntentic position in any placental mammal, including Atlantogenata. Also, cysteine has displaced the selenocysteine in Xenopus, a species much depleted in selenoproteins. SEPP2 does not appear at GenBank non-redundant other than mis-annotated SEPP1 chicken mRNAs. SEPP2 transcripts are available only in skate, zebrafish, and echidna (a short 454 read) .
The two paralogs have 4 coding exons with identical intron phases so clearly represent a segmental gene duplication. The gene duplication can be tracked back to before chondricthyes divergence but only one copy can be located in earlier diverging deuterostomes (sea urchin and acorn worm). SEPP2 retains the selenocysteine TGA only in its first exon and lacks the cluster of them in exon 4 of SEPP1. Exon 4 in SEPP2 is diverging very rapidlly, about neutrally, and is difficult to recover from blast searches without transcripts.
It can be surmised the first seleoncysteine in both SEPP1 and SEPP2 has a different functional role from terminal selenocysteines in SEPP1, possibly a conserved redox role distinct from selenostorage function of exon 4 in SEPP1. SEPP2 is very likely functional in all the species in which it occurs because of its conservation. There is no indication it is "on its way out" in marsupials -- the SECIS would be obliterated very quickly if the gene were non-functional.
Selenoprotein SELU: 3 paralogs, variable timing losses
SELU: This family consists of three deeply diverged (distinct exon patterns) paralogs. The encoding gene has 5 average exons with anomalously short introns like many selenoproteins. In the SELU1 group, selenocysteine occurs in a UxxC motif already in the earliest deuterostome but drops out in mammals after monotremes, being replaced by CxxC in marsupials and placentals. Amphibia separately lost selenocysteine.
The second paralog SELU2 has selenocysteine in bilaterans only to the node of sea urchin, suggesting it was lost early in the deuterostome ancestor. It is the closer paralog of SelU1, 36% vs 27% percent identity. No vestigal SECIS element persists in living species that encode cysteine. (The decayed SECIS elements still identifiable in 3' UTR of cysteine-containing GPX6 genes in rodents and human GPX5 represent much more recent loss of selenocysteine.)
The third paralog SELU3 has cysteine in all species for which a sequence is available. It might be called virtual selenoprotein supposing orthologs in early diverging eukaryotes could be located that contained selenocysteine. This would suggest a scenario in which selenocysteine was present in an ancestral gene prior to gene duplications followed by conversion to cysteine in different phylogenetic patterns within each gene subfamily.
This family exhibits the "selenocysteine rachet": if selenocysteine happens to be replaced by ordinary cysteine (despite catalytic inferiority) in some stem lineage, the unselected 3' UTR SECIS element then deteriorates over a few million years from accrued mutations, for the same reason (lack of purifying selection) the crayfish in the cave loses its imaging opsins. Consequently the whole following clade will contain cysteine -- a reversion to TGA at the cystein codon might occur but it would simultaneously require a multi-step reversion or de novo evolution of a SECIS element, ie all SECIS elements are ancient and selenocysteines cannot wink back on paraphyletically. (However the overall selenoproteome can still increase over time because of gene duplications elsewhere.)
A phylogenetic overview of the occurence of selenocysteine in SELU1 in 38 vertebrates:
.........................*..... C Homo sapiens genome EPRTFKAKELWEKNGAVIMAVRRPGcFLCRE C Pan troglodytes AACZ02115591 EPRTFKAKELWEKNGAVIMAVRRPGCFLCRE C Pongo abelii ABGA01228099 EPRTLKAKELWEKNGAVIMAVRRPGCFLCRE C Macaca mulatta AANU01282766 EPRTLKAKELWEKNGAVIMAVRRPGCFLCRE C Microcebus murinus ABDC01489848 EPRTFKAKELWEKNGAVIMAVRRPGCFLCRE C Otolemur garnettii AAQR01538573 EPRTFKAKELWEKNGAVIMAVRRPGCFLCRE C Tupaia belangeri AAPY01309022 EPRTFKAKELWGERGAVIMAVRRPGCFLCRE C Mus musculus AAHY01113156 EPRTFKAKELWEKNGAVIMAVRRPGCFLCR. C Rattus norvegicus AAHX01086750 EPRTFKAKELWEKNGAVIMAVRRPGCFLCR. C Spermophilus tridec AAQQ01288000 EPRTFKAKELWEKSGAVIMAVRRPGCFLCRE C Cavia porcellus AAKN02044618 EPRTFKAKELWEKNGAVIMAVRRPGCFLCRE C Oryctolagus cuniculus AAGW01591660 EPRTFKAKELWEKNGAVIMAVRRPGCFLCRE C Canis familiaris AAEX02011808 EPRTFKAKELWEKNGAVIMAVRRPGCFLCRE C Bos taurus AAFC03065652 ...TFKAKALWEKNGAVIMAVRRPGCFLCRE C Equus caballus AAWR02000382 EPRTFKAKELWEKNGAVIMAVRRPGCFLCRE C Myotis lucifugus AAPE01631988 EPRTFKAKELWEEKGAVIMAVRRPGCFLCRE C Sorex araneus AALT01607337 zPKTFKAKELWSKSGAVIMAVRRPGCFLCRE C Boreoeuthere ancestralis ancestral EPRTFKAKELWEKNGAVIMAVRRPGcFLCRE C Echinops telfairi AAIY01623759 ...TFQSKGALGKNGAVIMAVRRPGCFLCRE C Dasypus novemcinctus AAGV01392885 EPRTFKAKELWEKNGAVIMAVRRPGCFLCRE C Monodelphis domestica AAFR03024314 SPKTFKARELWEHRGAVIMAVRRPGCFLCRE C Trichosurus vulpecula transcript SPKTFKARELWEHRGAVIMAVRRPGCFLCRE C Macropus eugenii genome ..KTFKARELWEHRGAVIMAVRRPGCFLCRE U Ornithorhynchus anatin AAPN01249400 EPRTFKARELWQRNGAVIMAVRRPGUFLCRE U Tachyglossus aculeatus genome EPRTFKARELWQRNGAVIMAVRRPGUFLCRE U Anolis carolinensis AAW.01013574 ..RTFKAEELWKKNGAVIMAVRRPGUFLCRE U Gallus gallus AADN02035315 EPRTFKASELWKKNGAVIMAVRRPGUFLCRE U Taeniopygia guttata genome EKRTFKAGELWKQNGAVIMAVRRPGUFLCRE C Xenopus tropicalis genome EPKSFKAKDLWEKNGAVVMAVRRPGCFLCRE C Xenopus laevis transcript EPRLFKAKDLWERDGAVIMAVRRPGCFLCRE U Danio rerio CAAK04015812 DDRVFKARELWESSGAVIMAVRRPGUFMCRE U Tetraodon nigroviridis CAAE01014976 ETKTFKAKTLWEKCGAVVMAVRRPGUFLCRE U Fugu rubripes CAAB01000016 ETKTFKAKSLWENSGAVVMAVRRPGUFLCRE U Gasterosteus aculeatus AANH01005113 ...VIKGRSLWDKNGAVVMAVRRPGUFLCRE U Oryzias latipes BAAE01190338 DTKIIKAKSLWDKNGAVVMAVRRPGUFLCRE U Fundulus heteroclitus transcript .....KAKSLWEKNGAVVMAVRRPGUFLCRE U Oncorhynchus mykiss CR369769 .....KAKALWEKTGAVVMAVRRPGUFLCRE U Callorhinchus milii AAVX01258517 ENRTFRASELWAGRGAVIMAVRRPGUFLCRE C Gasterosteus aculeatus AANH01005113 ......AKTLWDKTGAVVMVVRRPGCLLCRE (anomalous gene duplication with cysteine)
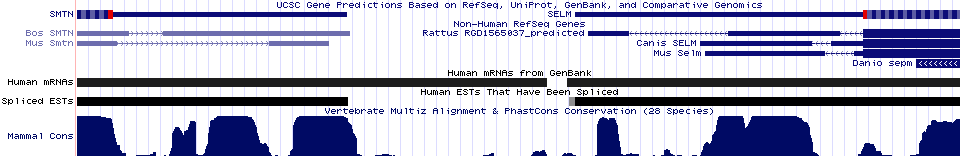
Selenoprotein SEPW1: small protein with an odd paralog
Selenoprotein SEPW1 is one of the shortest known mammalian proteins at 87 aa. With its CxxU motif, it is likely limited to simple redox reactions. Curiously, despite the small size protein still has 5 coding exons. One of these is of relatively recent origin because chondrichtyes and telost fish have the second and third exons fused (which, given the tree and extreme rarity of intron gain/loss must be the ancestral condition.
Like many genes on the densely packed human chromosome 19, SEPW1 seems to have given rise to a segmental duplication during the boreoeuthere stem (ie, it is not present in any xenarthra, afrothere, marsupial or earlier vertebrate). The second copy, called SELV, retains the same intron placements and phases but with greatly expanded and compositionally anomalous exon 1.
This long exon retains some conservation at its distal end to the last 5 residues of SEPW1 (unsurprising because the cysteine of the CxxU motif is at the end). There is observable conservation also at the amino terminus but otherwise the exon is evolving unconstrained. Possibly the initial methionine of SEPW1 was lost at the time of duplication but the terminal residues and splice donor were retained. Transcription far upstream lead to a remote methionine serving as replacement.
This is a very odd gene but transcripts in a half dozen species and excellent conservation in exons 2-5 over hundreds of millions of years witin boreoeutheres proves that it is evolving under selective constraint and so is functional. Distal exons however retain only 50% conservation to those of SEPW1
Selenoprotein SELH: rapid evolution
SELH is another small selenoprotein with a conserved CxxU redox motif split by a phase 21 intron. The introns in mammalian SELH are exceedingly short (eg, 93 and 162 bp in human with gene coding span very short at 619 bp) but the level of transcription is not remarkable so provides no explanation for the lack of retroposons. Some species like orangutan have processed pseudogenes, implying transcription in germ-line tissues. Zebrafish has a diverged duplicate gene with tryptophan at the seleocysteine site in addition to an intronless transcribed gene with TGA selenocysteine -- it appears that this has displaced the normal three-intron gene (seen in other fish). That can work in selenoproteins since retropositioning begins at the 3' end of transcripts, meaning the SECIS likely accompanies the coding region.
Protein conservation is below average -- SELH percent identities (to human) drop to the low 80's within Laurasiatheres, to 72% with marsupial, and 57% with chicken. Further, rodents exhibit significant residue loss upstream of the CxxU motif, very unusual in such a short protein but indicative of an inessential structural region. Indeed observed conservation is primarily centered in the middle of the protein. No phylogenetic conversion of selenocysteine to cysteine is observed within vertbrates
Selenoprotein SELM: retained in ER
Another selenoprotein of unknown function, SELM with CxxU motif, surfaces during Blastp searches using selenoprotein SEP15 (which oddly has a CxU motif) as query. A 3' UTR SECIS element cannot be located with SECISearch 2.19 yet it seemingly must be located in the comparative genomic peaks of conservation lying between SELM and the neighboring gene.
Since the protein is quite short with 5 introns, complete SELM sequences are best recovered from cDNAs and genomic alignments in the UCSC 28way. This is an ancient protein recoverable from vertebrates, amphioxus, sea urchin, shrimp, mite, moths, and hydra, and plants. Moths (silkworm and hawkmoth) -- but not their outgroups -- encode cysteine in a CxxC motif in place of CxxU. No loss of selenocysteine is seen in 22 species of phylogenetically dispersed vertebrates.
SELM may begin with a signal peptide. There are no glycosylation sites nor additional conserved cysteines beyond the CxxU motif. SuperFamily finds no similarity to proteins of known 3D structure. The terminal residues appear to be a phylogenetically conserved KDAL-class endoplasmic retention signal
In whole-mount zebrafish embryos, SELM is expressed within the notochord and anterior somites, axial fin fold, dorsal spinal chord neurons, then in lateral line neuromasts. SelM is located in the ER/Golgi as is its distant homolog Sep15 (found associated with UDP-glucose glycoprotein glucosyltransferase, an ER-resident protein involved in quality control of protein folding).
Selenoprotein MSRB123: methionine sulfoxidases
MSRB1 is a short odd protein, rich in cysteines, with two CxxC motifs, a near-amino terminal cysteine and a more distal selenocysteine in a motif with serine. MSRB is now known to be a stereospecific methionine-R-sulfoxide reductase repairing oxidative damage to methionine in native proteins. The two pairs of cysteines bind a zinc atom. The all-beta structure has been determined in a bacterial homolog with a internal structural duplication so weak it needs an xray determination to be revealed. Humans have 9 such domains in 9 proteins according to SuperFamily. These could potentially encode other selenoproteins at least in some species.
The fold exists as a small family of paralogs, three in mammals, with specialization to cell compartment -- cytosol, mitochondria and endoplasmic reticulum (via KAEL* signal), respectively.). All contain catalytic zinc. This multiplicity of MSRB contrasts with a single non-homologous methionine-S-sulfoxide reductase (not a selenoprotein in any species).
MSRB1 has selenocysteine in its active site in conserved motif UxxS, whereas MSRB2 and MSRB3 contain Cys in the well-conserved motif CINS. The cysteine in the Ux53xC motif of prokaryotes is replaced by serine or threonine in all eukaryotic MSRBs as U/CxxS/T. While serine and threonine have polar hydroxyl groups reminiscent of a cysteine, whether they form a covalent bond with the selenocysteine or otherwise contribute to catalysis remains unresolved. Thus this protein has a defunct CxxU motif that was unusual among selenoproteins in having 53 intervening residues.
The exon structures, relative to reliably locatable CxxC anchors, show MSRB2 and MSRB3 more closely related (having a distinctive break within the second CxxC motif), whereas MSRB1 introns are placed altogether differently. This suggests two rounds of gene duplication with intronation of MSRB2/3 prior to the second round. A tree based on the alignable conserved core of these proteins indicates the same result.
The phylogenetic distribution of orthologs of MSRB1 is orderly, with selenocysteine back through fish, with no counterpart currently locatable in amphioxus or tunicates (which otherwise have 3 MRSB genes). Further, there is no sporadic appearance of selenocysein in MRSB2/3 -- cysteine is always found, even prior to bilatera, suggesting though selenocysteine is ancestral (viz prokaryotes) but became cysteine prior to the second duplication giving rise to MRSB2/3.
Care must be taken at greater depth because of possible confusion among paralog members (and lineage-specific expansions and contractions). Synteny is lost so intron phasing and siting must be used along with blast clustering. The selenoprotein rachet predicts as more species are sequenced, some clades may exhibit lost selenocysteine in MRSB1 but none will acquire it in MRSB2 or MRSB3 lineages.
This raises the question that if selenocysteine is not necessary, why is it retained in MSRB1? Possibly that has some connection to localization in the reduced cytosol whereas more oxidizing mitochondrial and ER compartments utilize cysteines. In this view, selenocysteine will not be lost in any vertebrate clade no matter how many species are sequenced for MSRB1; whereas in MSRB23 cysteine was an adaptive change rather than mere rift, allowing wider intracellular distribution. Evidently methionine sulfoxide forms in all three compartments.
32 vertebrate MSRB1 aligned in exon 3: VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK homSap Homo sapiens (human) VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK panTro Pan troglodytes (chimp) VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK ponPyg Pongo pygmaeus (orang_sumatran) VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK macMul Macaca mulatta (rhesus) VSCGRCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFIPK otoGar Otolemur garnettii (bushbaby) VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK micMur Microcebus murinus (mouse_lemur) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFVPK musMus Mus musculus (mouse) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK ratNor Rattus norvegicus (rat) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK cavPor Cavia porcellus (guinea_pig) VSCGKCGHGLGHEFLNDGPKPGQSRFuIFSSSLKFIPK oryCun Oryctolagus cuniculus (rabbit) VFCGKCGHRFGHEFLNDGLKPGQSRFuIFSNTLKFVPK ochPri Ochotona princeps (pika) VSCGKCGNGLGHEFLNDGPKPGKSRFuIFSSSLKFIPK canFam Canis familiaris (dog) VSCGRCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFIPK felCat Felis catus (cat) VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK bosTau Bos taurus (cow) VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK susScr Sus scrofa (pig) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFVPK equCab Equus caballus (horse) VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPR eriEur Erinaceus europaeus (hedgehog) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK loxAfr Loxodonta africana (elephant) VSCGKYGHGLGHEFLNDGPNWGQSRFuIFSSSLKFIPK echTel Echinops telfairi (tenrec) VSCGKCGNGLGHEFLNDGPKKGQSRFuIFSNTLKFVPK triVul Trichosurus vulpecula VSCGKCGNGLGHEFLNDGPRRGQSRFuIFSSSLKFIPK ornAna Ornithorhynchus anatinus (platypus) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK galGal Gallus gallus (chicken) VLCGKCGNGLGHEFINDGPKKGQSRFuIFSSSLKFVPK anoCar Anolis carolinensis (lizard) VSCGKCGNGLGHEFINDGPKKGQSRFuIFSSSLKFIPK xenTro Xenopus tropicalis (frog) VRCGKCGNGLGHEFVNDGPKHGLSRFuIFSSSLKFIPK danRer Danio rerio (zebrafish) VRCGKCGNGLGHEFLNDGPSRGLSRFuIFSSSLKFIPK tetNig Tetraodon nigroviridis (pufferfish) VRCGKCGNGLGHEFVNDGPSRGLSRFuIFSSSLRFIPK takRub Fugu rubripes (fugu) VRCGKCGNGLGHEFVNDGPAKGVSRFuIFSSSLKFIPK gasAcu Gasterosteus aculeatus (stickleback) VRCGKCGNGLGHEFVNDGPSKGLSRFuIFSSSLKFIPK oryLap Oryzias latipes (medaka) IRCGKCNNGLGHEFLNDGPKHGLSRFuIFSSSLKFV.. ictFur Ictalurus furcatus (fish) VRCGKCGNGLGHEFVGDGPKKGLSRFuIFSSSLKFV.. oncMyk Oncorhynchus mykiss (trout) VRCGKCGNGLGHEFVGDGPKKGLSRFuIFSSSLKFV.. salSal Salmo salar (salmon) .SCGKCGNGLGHEFLNDGLKAGQSRYuIFSNSLKFVPK calMil Callorhinchus milii (elephantfish)
Reference sets of vertebrate selenoproteins
SEPP2: 12 vertebrate sequences
>SEPP2_monDom Monodelphis domestica (opossum) chr2:9,451,774-9,455,977 1 selenocys tga stop taa 0 MPPPGLSLAVLLGLLGASLAFENRTRFCQPAPPWQVGGGRAPMEEALGNVTVVALLKASuHFCLKQAAS 2 1 MGSLQERLARMGAPDVRFIIVNEKSPQSRALHGELELHAPPGVPVYGQPELGPDIWSILGGAKDDFLIYDR 2 1 CGRLTFHIRLPFSFLHFPYVESAIRFTHRQDSCGNCSFYPAQ 0 0 VNSTDERKGESKHSPGLEGEGQEPLGEKPDSRGLTLGSSAPTAHAHDPVHGGMEKPSPSLPSPALEPSLHEGEAPG* 0 >SEPP2_macEug Macropus eugenii (wallaby) trace archive tga stop taa 0 MGLPGLAAALLLGLVGATLASENGTRICQPAPKWQVDRGASPMEEALGQVTVVALLKASuHFCLKQAAS 2 1 IGNLQERLARVGATGVRFIIINEKSPLSQALYGELELHAPSGVLVYNQQGPGPDIWSILGGAKDDFLIYDR 2 1 CGRLTFHLPLPLSFLYFPYVESAIRFTHQQDHCGNCSFYPAQ 0 0 VNNTDKGKAESLTQSPRLEGEGQESLAEGPSTHGPTLWPSTPHPAHARGPVHSPGSKRLSPSLPPPSLEWPPHEEANK* 0 >SEPP2_ornAna platypus Ultra131:348583-348720 Zswim5 synteny last exon uncertain 0 MAGSGLLGPALTLATLLAAAGALPDLENGTRICQPAPRWTVNGVAPMEGTEGQVIVVALLKASuHFCLKQAAR 2 1 LAGLRERLAGHGAGNVSFLIVNQRDPTAQLLHTELERHAPPGVPVYAQDGPDPDVWSVLGGDKDDFFVYDR 2 1 CGRLTFHIQLPFSFLHFPYVEAAVRFTHRRDFCGNCSYYFPQVGT 0 0 VNDTTTQESELEKSPGAPGEEPEGSPVREPDRPQSQDPTGPFSGVLLQGKENKIIPWKTPLQAAPRKPSHPPGAHD* 0 >SEPP1_tacAcu Tachyglossus aculeatus (echidna) EUEMSW408ERBZB length=227 run=R_2007_08_22_12_11_10_ 59% 1 VPVYAQDGPDPDVWSILGGGKDDFLVYDR 2 1 CGRLTFHIRLPFSFLHFPYVEAAVLFSHRHDFCGNCS >SEPP2_galGal Gallus gallus (chicken) taa stop synteny Zswim5 HPDL MUTYH 0 MGSLLLALASCLGLAVASEGATNGSRLCHEAPAWRINGSSPMEGAAGQVTVVALLKASuHFCLLQARS 2 1 LGALRERLGQQGVSDVRYVIVNEQAPLSRAMFGELQRHAPPGVPVLQQQPHEPDVWQLLGGDKDDFLVYDR 2 1 CGRLAFHIQLPYSFLHLPYVESAIRFTHRKDFCGNCSLYPNSTQE 0 0 ANSTMEVPATLTPLPKQEEKESETPAHHQPNHLHPHHRAVGNGTAPEPSGDHRPAHAHHHHGAHGKLHPKGQTPEGRDP* 0 >SEPP2_taeGut Taeniopygia guttata (finch) 0 MGLLVLALATWLGLGLASASEETANSSRICQEAPAWTINGSSPMEGAAGQVTVVALLKASuQFCLRQAHS 2 1 LGGLRERLARQGMADVRYMIVNEKAPLSRAMLPELQRHAPPGVPVFQPEREDPDVWQVLGGDKDDFLVYDR 2 1 CGRLAFHIQLPFSFLHFPYVEAAIRSSHIKDFCGNCSLYPNTTQE 0 0 ANSTMEGPATPSSLPEHEGMESETPVHQHKPLHPHRHHEVNSERDTNPSEDHKPATHAHHHHGDHGQLHHKGRKQKEGDEH* 0 >SEPP2_anoCar Anolis carolinensis (lizard) scaffold_4:5,993,109-5,994,846 0 MDYSLATRILLLGLVVISATQAEVTENKTRICQPAPLWKINGTAPMAGMEGQVTVVALLKASuPFCLKQAAK 2 1 IGGLQKKLSNEGVANVSFLIVNEKTPLSRAMYWELKRNAPQGIPVYQQQILEPDVWQILDGDKDDFLIYDR 2 1 CSRLTFHIQLPYSFLHLPYVEAAVHYTHRKDYCGNCSRYYSE 0 0 * 0 >SEPP2_xenTro Xenopus tropicalis (frog) NM_001006907 misannotated, no selenocysteine 0 MHNLALTVSILMGLLGQVSSSEQTNSSICKPPPKWSIEGEVPMAEALGKVTVVALLQASCGFCLVQAAR 2 1 MGPLRYKLSLQGMTDIKYMIVNDQSLHSANMFPELKRWAPEGIPVYQQTPGQDDVWELLDGNKDDFLIYDR 2 1 CGRLTFHVRLPLSFLHFPYVEAAILSTYNESFCGNCSFTSNSTLIPM 0 0 NGTTVSPSGDDSSSPLQNKDEPVNKEPSPTLEKHNDQRKLDSELRLHDHSQHHPINSHKRQENQNNHPRNLIKNGKQN* 0 >SEPP2_tetNig Tetraodon nigroviridis (pufferfish) from cDNA, genome misassembled 0 MSSPWLLWLQVALTGLLWASQGQSATSRICKAAPRWEVGGQAPMEALVGRVVVVALLKASuQFCHIQASK 2 1 IGPLREKLSRRNVTEVSFVIVNEQEPVSRALYWQLRRRAPPGVPVYQQAPLQDDVWEALDGDKDDFLIYDR 2 1 CGQLTFHIGLPYSSLRYTYVEAAVIATHQGNICNCS 0 0 ANFTSLSISNSSGSGGMPSQTNQTVTAETDGPHTTHHHPHPHRHHHHHQHLSPEQPTPTAMPGQATPTPA* >SEPP2_gasAcu Gasterosteus aculeatus (stickleback0 chrIII:3,915,059-3,917,020 0 MTPPGSASRPPHWTIKERAPMQELLGNVVVVALLKASuQFCLTQASK 2 1 IGGLRDKLNRSNLTDVSFMIVNEREPHSRAMYWELKRRAPPGVPVYQQAALQSDVWEALDGDKDDFLVYDR 2 1 CGLLTFHIVLPYSFLHNVYVEAAIRATYLKNICNCT 0 0 VDSVVSSLNNSVMNNETDFNVSQTNATPRIQPDTNDPEGAGTPPPPT >SEPP2_leuEri Leucoraja erinacea (skate) cDNA 0 MGLQRSLLIILVGAAITLAAANNDTRICQVAPHWEIGNQSPMEQLSGQLVVVALLKASuQFCLTQAAKLGILRDKLSLQGLKNIHYIIVNEKT LESRAMFWKLKLKTPKNITVYQQSAFQPDVWRILRGNKDDFLIYDRCGKLTFHITSPYSYLNFRYVEAAIMATYNTDYCGNCMGSSTTLEATS >SEPP2_calMil elephant fish Callorhinchus milii (elephantfish) 1 LGSLQDKLARQGLKDVHYMIVNEKAPESRAMLWELKRHVPNNVSVYQQSPIQPDVWHSLQGGKDDFLIYDR 2 1 CGRLTFHVVLPYSSLQYPYIEAAIRATHKRDICGECTIT 0
SELU1: 13 vertebrate sequences
>SELU1_homSap Homo sapiens (human) processed pseudogenes chr8 and chr12 0 MSFLQDPSFFTMGMWSIGAGALGAAALALLLANTDVFLSKPQKAALEYLEDIDLKTLEK 1 2 EPRTFKAKELWEKNGAVIMAVRRPGcFLCREE 0 0 AADLSSLKSMLDQLGVPLYAVVKEHIRTEVKDFQPYFKGEIFLDEK 0 0 KKFYGPQRRKMMFMGFIRLGVWYNFFRAWNGGFSGNLEGEGFILGGVFVVGSGKQ 0 0 GILLEHRENEFGDKVNLLSVLEAAKMIKPQTLASEKK* 0 >SELU2_homSap Homo sapiens (human) 7 exons chr1p36.32 36% id NM_152371 0 MSTVDLARVGACILKHAVTGE 0 0 AVELRSLWREHACVVAGLRRFGCVVCRWIAQDLSSLAGLLDQHGVRLVGVGPEALGLQEFLDGDYFAG 1 2 ELYLDESKQLYKELGFKR 2 1 YNSLSILPAALGKPVRDVAAK 0 0 AKAVGIQGNLSGDLLQSGGLLVVSK 1 2 GGDKVLLHFVQKSPGDYVPKEHILQVLGISAEVCASDPPQ 0 0 CDREV* 0 >SELU3_homSap Homo sapiens (human) 6 exons chr9q22.32 25% id processed pseudogene chrX 0 MAAPAPVTRQVSGAAALVPAPSGPDSGQPLAAAVAELPVLDARGQRVPFGALFRERRAVVVFVR 0 0 HFLCYICKEYVEDLAKIPRSFLQ 0 0 EANVTLIVIGQSSYHHIE 0 0 PFCKLTGYSHEIYVDPEREIYKRLGMKRGEEIASS 1 2 GQSPHIKSNLLSGSLQSLWRAVTGPLFDFQGDPAQQGGTLILGP 1 2 GNNIHFIHRDRNRLDHKPINSVLQLVGVQHVNFTNRPSVIHV* 0 >SELU1_borAnc Boreoeuthere ancestralis (northern beast) 5 exons no selenocyseine 0 MSFLQDPSFFTMGMWSIGAGALGAAALALLLANTDVFLSKPQKAALEYLEDIDLKTLEK 1 2 EPRTFKAKELWEKNGAVIMAVRRPGcFLCREV 0 0 AADLSSLKPKLDELGVPLYAVVKEHIRTEVKDFQPYFKGEIFLDEK 0 0 KKFYGPQRRKMMFMGFVRLGVWYNFFRAWNGGFSGNLEGEGFILGGVFVVGPGKQ 0 0 GILLEHREKEFGDKVNPVSVLEAARKIKPQTSASEKK* 0 >SELU1_triVul Trichosurus vulpecula (brushtail opossum) EC360881 0 MSFLDLSFFSMGMWSLGAGALGAAVLSLILANTNLFLTKSVTATLEFLEEIELKTLDN 1 2 ESPKTFKARELWEHRGAVIMAVRRPGCFLCREE 0 0 AAELSALKPQLDQLGIPLYAVVKEKIGSEVENFQPYFKGKIFLDER 0 0 KKFYGPQKRKMMFMGFVRLGVWQNFFRARSKGFSGNLEGEGFILGGVYVIGPGKQ 0 0 GILLEHREKEFGDKVDPASVLEAA * 0 >SELU1_macEug Macropus eugenii (tammar wallaby) EX196548 full 0 MSFLDLSFLSMGMWSLGAGALGAAVLSLILANTDVFLTKSVTATLEFLEDIELKTLDN 1 2 KTFKARELWEHRGAVIMAVRRPGCFLCREE 0 0 AADLSALKPQLDQLGIPLYAVVKEKIGSEVEDFQPYFKGKIFLDER 0 0 KKFYGPQKRKMMFMGFVRLGVWQNFFRARSKGFSGNLEGEGFILGGVYVIGPRKQ 0 0 GILLDHREKELGDKVNPASVLEACKKIKLHA* 0 >SELU1_monDom Monodelphis domestica (opossum) tgt-cys 0 MSFLDLNFFSMSMWSLGAGALGAAALSLILANTDLFLTKSVDATLEFLEEIQLKTLDN 1 2 ESPKTFKARELWEHRGAVIMAVRRPGCFLCREV 0 0 AADLSALKPQLDLLGVPLYAVVKEKIGSEVENFQPYFKGKIFLDER 0 0 KKFYGPQKRKMMFMGFVRLGVWQNFFRARSKGFSGNLEGEGFVLGGVYVIGPGKQ 0 0 GILLEHREKEFGDKVNPASVLEAAKKIKPHTSTSEGK* 0 >SELU1_oan data Ornithorhynchus anatinus (platypus) taa early stop full 0 MPLPPDLGLFNLGMWSVGVGALGAAAVGLLLANTDLLLTKPEKATLEYLEDTELKTLGK 1 2 EPRTFKARELWQRNGAVIMAVRRPGuFLCREE 0 0 AAELSSLKPQLDRLGVPLYAVVKEKIGTEVEDFQPYFKGEIFLDER 0 0 KKFYGPHKRKMLFLGFIRLGVWQNFLRARNRGFSGNLEGEGLILGGVYVLGAGKQ 0 0 GILLEHREREFGDKVSPASVLEAAQRIKPQPL* 0 >SELU1_tacAcu Tachyglossus aculeatus (echidna) 454:EUEMSW405C31QQ (74%) tSASEKK terminus? frag 0 1 2 EPRTFKARELWQRNGAVIMAVRRPGuFLCREE 0 0 AAELSSLKPQLDQLGVPLYAVVKENIGTEVEDFQPYFKGEIFLDER 0 0 KRFYGPHKRKMLFLGLIRLGVWQNFIRARNKGFPPVTWEGEG 0 0 GVLLEHREREFGDKVSPASVLEAAQKIKPQ* 0 >SELU1_gga Gallus gallus (chicken) 0 MSFLPDFGIFTMGMWSVGLGAVGAAITGIVLANTDLFLSKPEKATLEFLEAIELKTLGS 1 2 EPRTFKASELWKKNGAVIMAVRRPGuFLCREE 0 0 ASELSSLKPQLSKLGVPLYAVVKEKIGTEVEDFQHYFQGEIFLDEK 0 0 RSFYGPRKRKMMLSGFFRXGVWQNFFRAWKNGYSGNLEGEGFTLGGVYVIGAGRQ 0 0 GVLLEHREKEFGDKVSLPSVLEAAEKIKPQAS* 0 >SELU1_tgu Taeniopygia guttata (finch) 0 msflpdfgiFTMGMWSVGLGAIGAAVTGIVLANTDLFLSKPEKATLEFLEEIELKTLGS 1 2 EKRTFKAGELWKQNGAVIMAVRRPGuFLCREE 0 0 ASELSSLKPQLSKLGVPLYAVVKENIGTEVEDFQHYFKGEIFLDEK 0 0 KGFYGPRRRKMMLSGFFRLGVWQNFVRAWRSGYSGNLEGEGFTLGGVYVIGAGRQ 0 0 GVLLEHREKEFGDKVSLPSVLEAAEKIKPQAS* 0 >SELU1_anoCar Anolis carolinensis (lizard) 0 MWTIGLGAIGAAVTGIILANTDLFLSKAEQASLDFLEAIDLKTLGE 1 2 NQRTFKAEELWKKNGAVIMAVRRPGuFLCREV 0 0 AAELSSLKPQLDKLGVPLYAVVKENLGTEVMDFQPYFKGEIFLDEK 0. 0 KQFYGPQKRKMLFMGFIRCSVWRNFFRAWKSGYTGNIDGEGFVLGGVFVVGPGKQ 0 0 GVLLEHREKEFGDKVSLDAVLEAVKNIQPQPSEKDK* 0 >SelU1_fugRer Fugu rubripes (fugu) 0 MGLLAKLLAAVGGFVTAVMNSVTDAFLTPPLRATLEHLEETDLKTLSG 1 2 ALVIRLIPTRTETKTFKAKSLWENSGAVVMAVRRPGuFLCRE 0 0 EAAELSSLKPRLDQLGVPLYAVVKEDVGTEIQNFRPYFQGEIFLDEK 0 0 RRFYGPRERKMGLLGFLRVGVWMNGLRAFRSGFMGNVLGEGFVLGGVFVIGREQQ 0 0 GILLEHREREFGDKVNIEDVIQAVDRIAQELMPVTQN* 0 >SELU1_gasAcu Gasterosteus aculeatus (stickleback) chrVI.790.1 length=214 MGMWSLGLGAVGAALAGIFLANTDLCLPKAASASLENLEDADLRS KGRSLWDKNGAVVMAVRRPGuFLCREV ASGLSSLKPQLEELGVPLVAVVKEDVGTEIRDFRPHFAGDIFIDEK SFYGPLQRKMGGLGFIRLGVWQNFMRAWRSGYQGNMNGEGFILGGVFVFGAGNQ GILLEHREKEFGDKVQIADVLEAVKKIVPAK* >SELU1_calMil Callorhinchus milii (elephantfish) frag 2 ENRTFRASELWAGRGAVIMAVRRPGuFLCRE 0 0 AAALSSLRPSLAQLGVPL 0 GHLLEHREKEFGDAVNLTAVMEAAGKISPRQSAE* 0 >SELU1_squAca Squalus acanthias (spiny dogfish) also selenocysteine 0 MVVVVEDFHMGLWTLGLGALGAAITGVILANTDLLLPKAETASLAYLSGAELRTLDR 1 2 EERTLKAGDLWSRSGAVIMVVRRPGuFLCREE 0 0 AAEISSLRPQLDELGVPLYGVIKENINNELKNFQPFFKGEIFLDVE 0 0 MRFYGPKPRTMGLMGFMRLGVWKNFVRAWQKGFSGNTDGEGFILgGVFVIGAGQQ 0 0 GVLLEHREKEFGDVVNISSVLEARRKIETQRTEP* 0
SELV: 13 Boreoeuthere sequences
>SELV_homSap Homo sapiens (human) chr 19 uc002oly.1 66 pro + 32 thr +21 ser first exon 0 MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTLTPVLTPSPAGTSPLVLTPAPAQIPTLVPTPALARIPRLVPPPAPAWIPTPVPT PVPVRNPTPVPTPARTLTPPVRVPAPAPAQLLAGIRAALPVLDSYLAPALPLDPPPEPAPELPLLPEEDPEPAPSLKLIPSVSSEAGPAPGPLPTRTPL AANSPGPTLDFTFRADPSAIGLADPPIPSPVPSPILGTIPSAISLQNCTETFPSSSENFALDKRVLIRVTYC 2 1 GLuSYSLR 0 0 YILLKKSLEQQFPNHLLF 0 0 EEDRAAQATGEFEVFVNGRLVHSKK 0 0 RGDGFVNESRLQKIVSVIDEEIKKR* 0 >SELV_panTro Pan troglodytes (chimp) essentially identical to humna MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTLTPVLTPSPAGTPPLVLTPAPAQIPTLVPTPALARIPRLVPPPAPAWIPTPVPTPV PVRNPTPVPTPARTLTPPVRVPAPAPAQLLAGIRAALPVLDSYLAPALPLDPPPEPAPELPLLPEEDPEPAPSLKLIPSVSSEAGPAPGPLPTRTPLAANS PGPTLDFTFRADPSATGLADPPIPSPVPSPILGTIPSAISLQNSTETFPSSSENFALDKRVLIRVTYC 2 1 GLuSYSLR 0 0 YILLKKSLEQQFPNHLLF 0 0 EEDRAAQATGEFEVFVNGRLVHSKK 0 0 RGDGFVNESRLQKIVSVIDEEIKKR* 0 >SELV_ponPyg Pongo abelii (orang) 0 MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTLTPVLTPSPAGTPPLVLTPAPAQIPTLVPTPALARIPRLVPPPAPAWIPTPVPTPVPVRNPTPVPTPARTLTTPV RVPAPAPAPPPAQVLAGIRAALPILDSYLAPALRLHPPPEPAPELPLSPEEDPEPAPSLKLIPSVSSEAGPAPGPLPARTPQAANSPGPTLDFTFRADLSATGLADPPIPSPVPSPILGT TSSAISLQNSTENFASSSENFALDKRVLIRVTYc 2 1 GLuSYSLR 0 0 YILLKKSLEQQFPNHLLF 0 0 EEDRAAQATGEFEVFVNGRLVHSKK 0 0 RGDGFVNESTLQKIVSVIDEEIKKR* 0 >SELV_macMul Macaca mulatta (rhesus) distal seq from M. nemestrina transcript 0 MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTQTPVLTPSPARLPALVQPPAPAHIPTQVPTPALARIPGLVPPPARAWIPTPVPTPARVRNPTPVPTPARTLTPPV RVPAPAPDPAQVLAGIRTALPALDSYPAPALSLDSPPEPAQEPPLSPEEDPEPAPSLKLIPSVSSEAGPALGPLPAHTPLAAKSSGPTLDFTFRADPSATGLADPHIPSPVPAPILGTIP SALSLQNFTETLVSTSENFALDKRVLIRVTYC 2 1 GLuSYSLR 0 0 YILLKNSLEQQFPSHLLF 0 0 EEDRAAQATGEFEVFVNGRLVHSKK 0 >SELV_calJac Callithrix jacchus (marmoset) 0 MNNQARTPAPSSARTSTSVRASTPTRTPTPVRTPTAVRIRTPIRTLTPSLAGTPALVPTPTPARISRLVPTPAPARTPTPIPTLVRTLTPVPLPAPARIPAPAPAPAPAPAPAPSPALVP AGIRATLPVLDSYPALALPWDPPPEPVPEPLVSVSSEEDPEPAPSLKLVPSVSGETGPAPGPLPACTPLATNPPEPTLDFTADSSATELAVPTIPGSVPAPILGTIPLAASLLNSTESFL SASENFALDKRVPIRVTYc 2 1 GLuSYSLR 0 0 YILLKKSLEQRFPDCLLF 0 0 EEDRAAQATGDFEVFVDGRLVHSKK 0 0 RGDGFVDEASLQKIVSVIDEEIKKR* 0 >SELV_otoGar Otolemur garnettii (bushbaby) 0 MNSQARASVHPSRTSTAVRASIPARVHPRARTTPVQPRTLITQDRIPAPVRVPP 1 GLuSYGLQ 0 0 YILLRQNLEHHFPNRLLF 0 0 EEGRAAQATGEFEVFVDGKLVHSKK 0 0 NGDGFVDEIRLQKIVNIIDEEIKKRQ 0 >SELV_musMus Mus musculus (mouse) AV279316 58 Pro, 25 Thr, 25 Ser syntenic chr 7 46% hsa! MNNKARVPAPSSVRANTPARTPAPIRTATPVRAPNPAHNSTPVRTSIRVRAPAQVPNPVPIRFPTPAPVPAPTLTPAPTPAPVRHAAPVRTPAPVRAPNLGRV FPKISPGRFFPSLASPTAQPLSSRAASALLKDPTLAQNQKPSIHSLAEAIQGPLPVLTPSSSKTQGSIPDTASPIDSLASTAMASSTLGPIPGPNPTLEFLAS PLKETPGLGKLSTISPAPSFGSTKEIPSTSEDVPTPNRILIRVMYC 2 1 GLuSYGLR 0 0 YIILKRTLEHQFPNLLEF 0 0 EEERATQVTGEFEVFVDGKLIHSKK 0 0 KGDGFVDESGLKKLVGAIDEEIKKR* 0 >SELV_ratNor Rattus norvegicus (rat) chr 1 synteny 83% mmu MNNKARNPAPSSVRANTPSRTPTPVRTATPVRASTPAHNRSPVRTSIRVRTPANPVPIRFPTPAPAPAPTPTPAPTPAPVPAAAPVRTPAPVRASIQGRSFPTIS PVRFLRNLALPAAQPLSSGGAGSLSKDLTLAQKQKPSIHSLAEAIQGPFPVLTPSASSETHGSIPDPAPPTDSLASTAMASSTLDPIPGPKPTLEFLASPLKETP DLGKLSTISPAQNFVSTKEVPSTSEDVPTANRILIRVMYC 2 1 GLuSYGLR 0 0 YILLKKTLEHQFPNLLEF 0 0 EEERATQVTGEFEVFVDGKLIHSKK 0 0 KGDGFVDETSLKKLVGAIDEEIKKR* 0 >SELV_cavPor Cavia porcellus (guinea_pig) 1 GLuSYGFQ 0 0 YTLLKMSLKQQFPNLLRF 0 0 EDERATQVTGEFEVFVEGKLVHSKK 0 0 QGDGFVDDNRMQTIVNAINEEIKRR* 0 >SELV_oryCun Oryctolagus cuniculus (rabbit) 0 MNNQARTPAPAQARTSSVVRASAPTRVSTQIRTPATGWTPTPVQASTPVRTQTPVRTPTLVQASTPVRTPTLGQASTPVRTPTPVQALTPVRTPTPVSGPDSGRTPTPVGTPVGTL 1 GLuSYGLR 0 0 YILLKKNLEELFPDCLLF 0 0 EEERATQASGEFEVFVNGKLVHSKK 0 0 KGDGFVDEVKLRKIVTAINEEIK... 0 >SELV_canFam Canis familiaris (dog) CO599097 0 MNNQARAPPRTSARVLAWVRASTPVRTSIPVRTPTPARIPTSSRAPTSVQTPAPARTPNPVQIPTPVQTSTSARIPNPARTLTPVQTPASAWTPNPVQMLTPARTPTPVPTP VPTPIPARTPTPARTPTPVEAAAAAPASESFGSSALPLEPPPEPASEPTTSPHQDLSPTPSVKPLPSVTNGFGSTQEPLPDLTPPATDFLGPTLGSTSRADSSATKLTDSSESVR VPIPGTPSATALATSTNTFAPVGESCSVKIAVRVIYC 1 GLuSYGLR 0 0 YILLKKSLEQQFPNCLLF 0 0 EEERAAQATGEFEVFVDGKLVHSKK 0 0 KGDGFVDEARLQKIMNVIEEEVRKR* 0 >SELV_felCat Felis catus (cat) 0 MNNQARAPNPSPARVLALVRASTPVRSSIPVRIPTPARIPARTRNPQTPVRAPNPVQIPNPVQISARASNPARAPRPVQIPARTPNPPQTLTPGRAPNPVPTPVQTPTLVQTP TPVQTPTPVWTETPVQTPTAVGTPTRVQTLTPVIPRVRTPTPIRPLWPDPSPNLIPSGPDHPAPESSLRNSAPSFWINSPDSLPAPILETPSTAFASTFENLPEDSKILIRVIYC 1 GLuSYSLR 0 0 YILLRKSLELQFPNCLRF 0 0 EEERSAQATGEFEVFVDGKLVHSKK 0 0 KGDGFVDEARLQKIVSTIDEEIRKR* 0 >SELV_bosTau Bos taurus (cow) DT829759 0 MNNQPRTPAPTPARASTPVRGSTLHRTSIQVRTPTPTLTPALIRTLTPIRTPTPVRTPTPVPPSTPVRSPIPVRTPTPVRTPTPVPLSTPVRPPTPVRPPTPVRPPTPVPTPIGTPTP VQIPIPEPIPTPIPSRVLIPPLESFPDSALPSGPPLELEPTLTVSPAKNLEPSPRVKQVSSAANGFPPIQEPLPALTPLATDLSSLGSPLRTDTSTTNLIASSGHVRTPILGAIQAILPV PATALASISGNLKEENKIMIRVVYC 1 GLuSYSLR 0 0 YILLRKSLEQQFPNSLIF 0 0 EEEISAQATGEFEVFVDGKLIHSKK 0 0 NGDGFVDEVKLQTIVNLLNEEFKKR* 0 >SELV_equCab Equus caballus (horse) 1 GLuSYGLR 0 0 YILLRKSLDQQFPNRLVF 0 0 EEDVGAQASGEFEVFVEGKLVHSKK 0 0 KGDGFVDEARLRKIVSAINEEIKRR* 0
SEPW1: 26 vertebrate sequences
>SEPW1_homSap Homo sapiens (human) Selenoprotein W chr19 87 aa uc002phn.1 has retroprocessed pseudogene 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_panTro Pan troglodytes (chimp) 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 RGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_ponPyg Pongo pygmaeus (orang_sumatran) CR926472 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_macMul Macaca mulatta (rhesus) 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_macFas Macaca fascicularis (cynomolgus_monkey) AB169486 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_papAnu Papio anubis (baboon) EY285690 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_calJac Callithrix jacchus (marmoset) 0 MALTVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 SGEGTPQATGFFEVTVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_micMur Microcebus murinus (mouse_lemur) 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCLDI 0 0 CGEGTPQATGFFEVMVAGKLVHSKK 0 0 GDGYVDTESKFLKLV >SEPW1_musMus Mus musculus (mouse) 0 MALAVRVVYC 2 1 GAuGYKPK 0 0 YLQLKEKLEHEFPGCLDI 0 0 CGEGTPQVTGFFEVTVAGKLVHSKK 0 0 RGDGYVDTESKFRKLVTAIKAALAQCQ* 0 >SEPW1_ratNor Rattus norvegicus (rat) BC087625 0 MALAVRVVYC 2 1 GAuGYKPK 0 0 YLQLKEKLEHEFPGCLDI 0 0 CGEGTPQVTGFFEVTVAGKLVHSKK 0 0 RGDGYVDTESKFRKLVTAIKAALAQCQ* 0 >SEPW1_cavPor Cavia porcellus (guinea_pig) 0 MALAVRVVYC 2 1 GAuGYKPK 0 0 YLQLKEKLEDEFPGCLDI 0 0 CGEGTPQTTGFFEVTVAGKLVHSKK 0 0 GGDGFVDTEGKFRKLVAAIKAALAQG* 0 >SEPW1_oryCun Oryctolagus cuniculus (rabbit) 0 MALAVRVVYC 2 1 GAuGYKPK 0 0 YLQLKKKLEDEFPGCLDI 0 0 CGEGTPQVTGFFEVTVAGKLVHSKK 0 0 RGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_ochPri Ochotona princeps (pika) 0 MALSVRVVYW 2 1 GAuGYKPK 0 0 YLQLKKRLEDEFPGCLDI 0 0 GEGTPQVTGFFEVMVAGKLVHSKK 0 0 SGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_canFam Canis familiaris (dog) 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCLDI 0 0 RGEGTPQATGFFEVTVAGKLVHSKK 0 0 RGDGYVDTESKFLRLVAAIKTALAQG* 0 >SEPW1_felCat Felis catus (cat) 0 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCLDI 0 0 RGEGTPQATGFFEVMVGGKLVHSKK 0 0 RGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_ bosTau Bos taurus (cow) 0 MAVVVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPSRLDI 0 0 RGEGTPQVTGFFEVFVAGKLVHSKK 0 0 GGDGYVDTESKFLKLVAAIKAALAQA* 0 >SEPW1_oviAri Ovis aries (sheep) 0 MAVVVRVVYC 2 1 GAuGYKPK 0 0 YLQLKKKLEDEFPSRLDI 0 0 CGEGTPQVTGFFEVFVAGKLVHSKK 0 0 GGDGYVDTESKFLKLVAAIKAALAQA* 0 >SEPW1_susScr Sus scrofa (pig) AF380118 0 MGVAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQVTGFFEVLVAGKLVHSKK 0 0 GGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_eriEur Erinaceus europaeus (hedgehog) 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCLDI 0 0 RGEGTPQGTGFFEVLVAGKLVHSKK 0 0 KGDGYVDTETKFLKLVTAIKAALAQG* 0 >SEPW1_sorAra Sorex araneus (shrew) 0 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCVDV 0 0 CGEGTPQVTGFFEVMVAGKLVHSKK 0 0 RGDGYVDSESKYVRLVTAIKTALAQA* 0 >SEPW1_Choloepus hoffmanni (sloth) 0 MALAVRVVYW 2 1 GAuGYKPK 0 0 YVQLKKKLEDEFPGCLDI 0 0 SGEGTPQTTGFFEVMVAGKLVHSKK 0 0 QKGDGFVDTESKFLRLVAAIKAALAQG* 0 >SEPW1_monDom Monodelphis domestica (opossum) diverged 0 MAIQVRVVYW 2 1 GAuGYKPK 0 0 YLLLKKKLEDEYPGLLRH 0 0 NGEGTPEVTGFFEVTVAGKLVHSKK 0 0 AGHGFVDTADKYLQIVAEIKAALA* 0 >SEPW1_ornAna Ornithorhynchus anatinus (platypus) 0 MASLEAFPRGVVPVHVVYC 2 1 GAuGYKPK 0 0 FLQLKKKLENEFPGQVEI 0 0 SGEGTPQVTGWFEVTVAGKLVHSKK 0 0 EGDGFVDSESKFAKIRMAIKAALVPGY* 0 >SELW_galGal Gallus gallus (chicken) tga confirmed 0 MPLRVTVLYC 2 1 GAuGYKPK 0 0 YERLRAELEKRFPGALEM 0 0 RGQGTQEVTGWFEVTVGSRLVHSKK 0 0 NGDGFVDTNAKLQRIVAAIQAALP* 0 >SELW_anoCar Anolis carolinensis (lizard) 1 GAuGYSPK 0 0 YQQLKRGLEKEFPGKLEI 0 0 TGEGTPQVTGWFEVTVAGKLVHSKK 0 0 NGDGFVDNDTKLHKILMAIKAALA* 0 >SELW_xenTro Xenopus tropicalis (frog) tga confirmed 0 MPDTMVKVNVVYC 2 1 GAuGYLSK 0 0 FRRLKKELEQRFPGKLSI 0 0 DGEGTERMTGWFEVSINGKLVHSKK 0 0 NGDGYVDNDAKLQKIILAIEAALKQ* 0 >SELW_danRer Danio rerio (zebrafish) tga confirmed 0 MTVKVHVVYC 2 1 GGuGYRPK 0 0 FIKLKTLLEDEFPNELEI 0 0 TGEGTPSTTGWLEVEVNGKLVHSKK 0 0 NGDGFVDSDSKMQKIVTAIEQAMGK* 0 >SEPW1_takRub 0 MGVTIRVEYC 2 1 GGuGYGPR 0 0 YEELARVVRAEFPDADVSGFVGRM 1 2 GSFEIQINEQLIFSKLETGGFPYEDD 0 >SEPW2_calMil Callorhinchus milii (elephantfish) 1 GAuGYEPRYQKLAIVIKDEFPDADVSGKVGRT 1 2 GSFEIEINGQLIFSKLETGGFPYEND 0 0 ISEAVQKANNGEELQKIENSRPPCVIL* 0 0 VMHAIQCVSDGKPVEKITKSRPPCVIM* 0
DIO123: 24 vertebrate deiodinases
>DIO1_homSap Homo sapiens (human) iodothyronine deiodinase type I; 4 exons on chr1p32.3 0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWEFMQ 1 2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLVIYIEEAHAS 1 2 DGWAFKNNMDIRNHQNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYIIQEGRILYK 0 0 GKSGPWNYNPEEVRAVLEKLHS 0* >DIOI_ratNor Rattus norvegicus (rat) 7-258 thioredoxin-like MGLSQLWLWLKRLVIFLQVALEVATGKVLMTLFPERVKQNILAMGQKTGMTRNPRFAPDNWVPTFFSIQY FWFVLKVRWQRLEDRAEYGGLAPNCTVVRLSGQKCNVWDFIQGSRPLVLNFGSCTuPSFLLKFDQFKRLV DDFASTADFLIIYIEEAHATDGWAFKNNVDIRQHRSLQDRLRAAHLLLARSPQCPVVVDTMQNQSSQLYA ALPERLYVIQEGRICYKGKPGPWNYNPEEVRAVLEKLCIPPGHMPQF >DIO1_susScr Sus scrofa (pig) MELPLPGLWLKRLWVLFQVALHVAMGKVLMTLFPGRVKQDILAMSQKTGMAKNPHFSHENWIPTFFSAQY FWFVLKVRWQRLEDKTEEGGLAPNCPVVSLSGQRCHIWDFMQGNRPLVLNFGSCTUPSFIFKFDQFKRLI EDFSSIADFLIIYIEEAHASDGWAFKNNVDIKNHQNLQDRLRAAHLLLDRSPQCPVVVDTMKNQSSRLYA ALPERLYVLQAGRILYKGKPGPWNYHPEEVRAVLEKLHS >DIO1_sunMur Suncus murinus (shrew) MGLPGLGLLLKRFGVLVRVALKVAVGKVLLTLWPSAIRPHLLAMSEKTGMAKNPRFTYEDWAPTFFSTQY FWFVLKVNWQQLEDRTKQGDIAPDSPVVHLSGQRARLWDFMQGNRPLVLNFGSCSuPSFLFKFDQFKRLV EDFSSVADFLTVYIEEAHASDGWAFKNNVDIRRHRDLQERLQAARLLLDRNPGCPVVVDTMENRSSQLYA ALPERLYVLQEGRILYKGGPGPWNYHPEEVHAVLEQLCRSSAQSPRL >DIO1_galGal Gallus gallus (chicken) Type I iodothyronine deiodinase 4 exons chr8 MLSIRVLLHKLLILLQVTLSVVVGKTMMILFPDTTKRYILKLGEKSRMNQNPKFSYENWGPTFFSFQYLLF VLKVKWRRLEDEAHEGRPAPNTPVVALNGEMQHLFSFMRDNRPLILNFGSCTuPSFMLKFDEFNKLVKDF SSIADFLIIYIEEAHAVDGWAFRNNVVIKNHRSLEDRKTAAQFLQQKNPLCPVVLDTMENLSSSKYAALP ERLYILQAGNVIYKGGVGPWNYHPQEIRAVLEKLK >DIO1_xenTro Xenopus laevis (frog) tga confirmed MLRYIQKALILFFLFLYVVVGKVLMFLFPQTMASVLKSRFEISGVH-DPKFQYEDWGPTFFTYKFLRSVLEIMWMRLE DEAFVGHSAPNTPVVDLSGELHHIWDYLQGTRPLVLSFGSCT*PPFLFRLG EFNKLVNEFNSIADFLIIYIDEAHAADEWALKNNLHIKKHRSLQDRLAAAKRLME ESPSCPVVLDTMSNLCSAKYAA LPERLYILQEGKIIYKGKMGPWGYKPEEVCSVLEKKK* >DIO1_danRer Danio rerio (zebrafish) MGSAVGFALRKLFVYISAVLMVCAAILQMSMLKLLSFISPGRMRKIHMKMGERSTMTQNPKFRYEDWGPA FFSLAFIKTLFFVNWCSLGLEAFEGHAAPDSALITLDRQKTSVHRFLKGNRPLVLSFGSCTuPPFLYKLD EFKQLVKDFSNVADFLIVYLAEAHATDAWAFKNNVDISVHKNLEERLAAARTLLKEDPPCPVVVDEMNNI TASKYGALPERLYVIQSGKVIYQASDLGGQA >DIO1_tru fugu 4 exons genome glitches 0 MLLQKLAMYLSTAGLFCFMITLNVVLWILNIVAPALAKKIALKMGEKATMTQDPLFKYEDWGLTFASTALVKTASRHMWLSLGQEAFAGLEAPDSPVVTMERKRSSIGEFMK 1 2 TNRPLVLNFGSCTuPPFMFKLEEFKQLVRDFSDVADFLVVYIAEAHST 1 2 DGWAFKNNFDINQHRNLEDRLSAAQILVQKDPLCPVVVDDMNNSCAIKYGALPERLYVLQAGKVLYK 0 0 GAVGPWGYDPREVRSYLEKMK *0 >DIO2_homSap Homo sapiens (human) iodothyronine deiodinase type II; 2 exons size 8,781 bp on chr14q31.1 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGLRCVWKSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSSAEGGDNSGNGTQEKIAEGATCHLLDFASPERPLVVNFGSATuPPFTSQLPAFRKLVEEFSSVADFLLVYIDEAHPSDGWAI PGDSSLSFEVKKHQNQEDRCAAAQQLLERFSLPPQCRVVADRMDNNANIAYGVAFERVCIVQRQKIAYLGGKGPFSYNLQEVRHWLEKNFSKRuKKTRLAG 0* >DIO2_ratNor Rattus norvegicus (rat) 85-253 thioredoxin-like MGLLSVDLLITLQILPVFFSNCLFLALYDSVILLKHVALLLSRSKSTRGEWRRMLTSEGLRCVWNSFLLD AYKQVKLGEDAPNSSVVHVSNPEAGNNCASEKTADGAECHLLDFACAERPLVVNFGSATuPPFTRQLPAF RQLVEEFSSVADFLLVYIDEAHPSDGWAVPGDSSMSFEVKKHRNQEDRCAAAHQLLERFSLPPQCQVVAD RMDNNANVAYGVAFERVCIVQRRKIAYLGGKGPFSYNLQEVRSWLEKNFSKRXILD >DIO2_susScr Sus scrofa (pig) MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGMRCIWKSFLLD AYKQVKLGEDAPNSSVVHVSNPEGSNNHGHGTQEKTVDGAECHLLDFANPERPLVVNFGSATUPPFTSQL PAFSKLVEEFSSVADFLLVYIDEAHPSDGWAVPGDSSLSFEVKKHQNQEDRCAAAHQLLERFSLPPQCRV VADRMDNNANVAYGVAFERVCIVQRQKIAYLGGKGPFYYNLQEVRRWLEKNFSKR >DIO2_galGal Gallus gallus (chicken) deiodinase, iodothyronine, type II 2 exons chr5 MGLLSADLLITLQILPVFFSNCLFLALYDSVILLKHMVLFLSRSKSARGEWRRMLTSEGLRCVWNSFLLD AYKQVKLGGEAPNSSVIHIAKGNDGSNSSWKSVGGKCGTKCHLLDFANSERPLVVNFGSATuPPFTSQLS AFSKLVEEFSGVADFLLVYIDEAHPSDGWAAPGISPSSFEVKKHRNQEDRCAAAHQLLERFSLPPQCQVV ADCMDNNANVAYGVSFERVCIVQRQKIAYLGGKGPFFYNLQEVRLWLEQNFSKRUNPLSTEDLSTDVSL >DIO2_xenTro Xenopus laevis (frog) GTRRERERLSVDLLITLQILPGFFSNCLFLALYDSVVLVKHVLLQLNRSKSSQSQWRRMLTPEGLRCVWN SFLLDAYKQVKLGQDAPNSNVIQVSNNRTSKSVQRKFAGKCHLLDFASSERPLVVNFGSATuPPFISQLP AFSKLVEEFSSVADFVLVYIDEAHPSDGWAAPGTASYEVKKHRSQEERCAAASKLLQHFSIPPQCQVVAD CMDNNANVAYGVSFERVCIVQRQKIVYLGGKGPFFYNIQEIRRWLELSFGKR >DIO2_ranCat Rana catesbiana (bullfrog) MGLLSVDLLITLQILPGFFSNCLFLALYDSVVLVKHVLLQLNRSKSSHGQWRRMLTPEGLRCVWNSFLLD AYKQVKLGGDAPNSNVIHVTDKNSSSGKPGTPCHLLDFASSERPLVVNFGSATuPPFISQLPAFSKMVEE FSAVADFLLVYIDEAHPSDGWAAPGISSYEVKKHRNQEDRCAAANKLLEQYSLPPQCQVVADCMDNNTNA AYGVSFERVCIVQRQKIVYLGGKGPFFYNLQEVRQWLELTFGKKAESGQTGTEK >DIO2_nfo lungfish Neoceratodus forsteri exons unknown MGLLSVDLLITLQILPWFFSNCLFLALYDSVVLLKHVILLLSCSKSSRGEWRRMLTSEGLRTVWNSFLLD AYKQVKLGGDAPNSKVVRVTSGCCRRRSFSGKGESECHLLDFASSNRPLVVNFGSATUPPFISQLPTFRK LVEEFSDVADFLLVYIDEAHPADGWAAPGVATKSFEVKKHRSQEERCVAAHKLLEHFSLPPQCQVVADCM DNNTNVAYGVSFERVCIVQRQKIAYLGGKGPFFYNLKEVRHWLEQTYRKRUVPTCELIM >DIO2_danRer Danio rerio (zebrafish) MGLLSVDLLVTLQILPGFFSNCLFFVLYDSIVLVKRVVSLLSCSGSTGEWQRMLTTAGVRSIWNSFLLDA YKQVKLGEAAPNSKVVKVTGINRCWSISGKTHNQCHLLDFESPDRPLVVNFGSATuPPFISQLPVFRRMV EEFSDVADFLLVYIDEAHPSNGWVGPP MENFSFEVRKHRNLEERM FAARTLLEHFSLPPQCQLVADCM DNNANIAYGVSYERVCIVQKNKIAYLGGKGPFFYNLKDVRRWLEKC >DIO3_homSap Homo sapiens (human) size iodothyronine deiodinase type III; 1 exon 2,502 bp chr14q32.31 0 MLRSLLLHSLRLCAQTASCLVLFPRFLGTAFMLWLLDFLCIRKHFLGRRRRGQPEPEVEL NSEGEEVPPDDPPICVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVLPDGF QSQHILDYAQGNRPLVLNFGSCTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDG WVTTDSPYIIPQHRSLEDRVSAARVLQQGAPGCALVLDTMANSSSSAYGAYFERLYVIQS GTIMYQGGRGPDGYQVSELRTWLERYDEQLHGARPRRV 0* >DIO3_ratNor Rattus norvegicus (rat) 103-266 thioredoxin-like MLRSLLLHSLRLCAQTASCLVLFPRFLGTAFMLWLLDFLCIRKHFLRRRHPDHPEPEVELNSEGEEMPPD DPPICVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVRPDGFQSQRILDYAQGTRPLVLNFG SCTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYVIPQHRSLEDRVSAARVLQQGA PGCALVLDTMANSSSSAYGAYFERLYVIQSGTIMYQGGRGPDGYQVSELRTWLERYDEQLHGTRPRRL >DIO3_susScr Sus scrofa (pig) MLHSLLLHSLRLCAQTASCLVLFPRFLGTACMLWLLDFLCIRKHLLGRRRRGEPETEVELNSDGDEVPPD DPPICVSDDNRLCTLASLRAVWHGQKLDFFKQAHEGGPAPNSEVVLPDGFQNQHILDYARGNRPLVLNFG SCTUPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYSIPQHRSLEDRVSAARVLQQGA PECSLVLDTMANSSSSAYGAYFERLYVIQSGTIMYQGGRGPDGYQVSELRTWLERYDQQLHGPQPRRV >DIO3_galGal Gallus gallus (chicken)type III iodothyronine deiodinase 1 exon chr5 11mbp separation AACILLFPRFLLTAVMLWLLDFLCIRKKMLTMPTAEEAAGAGEGPPPDDPPVCVSDSNRMFTLESLKAVW HGQKLDFFKSAHVGSPAPNPEVIQLDGQKRLRILDFARGKRPLILNFGSCToPPFMARLRSFRRLAADFV DIADFLLVYIEEAHPSDGWVSSDAAYSIPKHQCLQDRLRAAQLMREGAPDCPLAVDTMDNASSAAYGAYF ERLYVIQEEKVMYQGGRGPEGYKISELRTWLDQYKTRLQSPGAVVIQV >DIO3_xenTro Xenopus laevis (frog) MLHCAGPHTGKLVKQVAACCLLLPRFLLTGLMLWLLDFQCIRRRVLLTAREESTAEHEDPPLCVSDSNRM CTVESLRAVWHGQKLDYFKSAHLGCSAPNTEVVMLEGRRLCKILDFSQGKRPLVVNFGSCTuPPFMARLQ AYRRLAAQHVGIADFLLVYIEEAHPSDGWLSTDASYQIPQHQCLQDRLAAAQLMLQGAPGCRVVVDTMDN SSNAAYGAYFERLYIVLEGKVVYQGGRGPEGYKISELRMWLEQYQQGLMGTKGSGQVVIQV >DIO3_ranCat Rana catesbiana (bullfrog) MLPAPHTCCRLLQQLLACCLLLPRFLLTVLLLWLLDFPCVRRRVIRGAKEEDPGAPEREDPPLCVSDTNR MCTLESLKAVWYGQKLDFFKSAHLGGGAPNTEVVTLEGQRLCRILDFSKGHRPLVLNFGSCTuPPFMARL QAYQRLAAQRLDFADFLLVYIEEAHPCDGWLSTDAAYQIPTHQCLQDRLRAAQLMLQGAPGCRVVADTMT NASNAAYGAYFERLYVILDGKVVYQGGRGPEGYKIGELRNWLDQYQTRATGNGALVIQV >DIO3_nfo lungfish Neoceratodus forsteri 0 MYQSSGVHTMNEVLKQAFACFILLPRFLVTALMLWLLDFLCVRRRVLLHMSRRQEASDLPDEPELCVSDS NRMFTLKSLRAVWHDQKLDFFKAAHIGLVAPNTEVIKLEGQRKAKILEFGGGKRPLILNFGSCTuPPFMARLKAFRGVATQYKDVADFLLIYIEEAHPSDGWVSTDAPYQIPKHQCLEDRLKAAQLMNLEIPGCLVVVDTMDNASNAAYGAFFERLYIVQQERVVYQGGRGPEGYKISELKNWLDQYKSQLQNSSAVVIQV 0* >DIO3a_danRer Danio rerio (zebrafish) SALKNAAVCVLLLPRFLLAALMLCLLDFLCIRRKLLLKMQEGAFSSPDDPPLRVSDSN KMFTLESLRAVWYGQKLDfFkSARLGGAAPNTEVFPLDGDARAAERILDYARGRRPLILN FGSCSuPPFMTRLSAFQRVARQYADIADSLLVYIEEAHPSDGWVSSDAPVQIPRHRCLED RLRAAQMLHRDAAGNAGVVDSMQNS >DIO3b_dre Danio rerio GALKNALVCLLILTRFLVAAFMLWCLDFLCVRKRVLVHLQERAYAEQEEEPL CISDSSRMFSWESLKAVFHGHKLDYMKSARLGHAAPDSEVFPLAEPRRGRVLEFARGHRP LVLSFGSCSuPPFMRRLKAFRRLVLRYADVADALLIYIEEAHPSDGWRSSDAPHQIRRHR SLEERLSAARLMEREAPGCAVVADGMENAANSAYGAYFDRLYIVQDGRVVYQ
SELH: 25 vertebrate sequences
>SELH_homSap Homo sapiens (human) NP_734467 Selenoprotein H exons chr11 80% identity musMus 0 MAPRGRKRKAEAAVVAVAEKREKLANGGEGMEEATVVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPTKPRRGSFEVTLLRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_ponPyg Pongo pygmaeus (orang_sumatran) 0 MAPRGRKCKAEATVVAVAEKrEKLTNGGEGMEEATIVIEHC 2 1 TSuRVYGRNAAALSQVLCLEAPELPVKVNPTKPRRGSFEVTLLRPDGS 1 2 SVELWTGIKKGPPCKLKFPEPQEVVEKLKKYLS* 0 >SELH_macMul Macaca mulatta (rhesus) 0 MAPRGRKRKAEAAMVAAAEKQEKLANSGEGMEETTVVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPSKPRRGSFEVTLLRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_micMur Microcebus murinus (mouse_lemur) 0 MAPRGRKRKAEASVVATAEKREKLENGGEAVEEATVVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPAKPRRGSFEVTLQRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQVVVKELKKYL.* 0 >SELH_tupBel Tupaia belangeri (tree_shrew) 0 MAPRGRKRKAEAAVVATAEKQEKLQNGGEGVKEASIVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNSAKPRRGSFEVTLLRPDGS 1 2 SVELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_musMus Mus musculus (mouse) 0 MAPHGRKRKAGAAPMETVDKREKLAEGATVVIEHC 2 1 TSuRVYGRHAAALSQALQLEAPELPVQVNPSKPRRGSFEVTLLRSDNS 1 2 RVELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_ratNor Rattus norvegicus (rat) TGA verified 77% 0 MAPLGRKRKAGAAPIESADKREKLAEGAAVVIEHC 2 1 TSuRVYRRHAAALSQALQLEAPEISVQVNRSKPRRGSFEVTLLRPDNS 1 2 RVELWTGIKKGPPRKLKFSEPQEMVEELKKYLS* 0 >SELH_speTri Spermophilus tridecemlineatus (squirrel) 0 MAPRVRKRKAEAAAVSTSEKREKLENGKEQVEEAVVIEHc 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPSKPRRGSFEVTLLRRDGTS 1 >SELH_oryCun Oryctolagus cuniculus (rabbit) 0 MAPGKRKRKAEAAPVASAEKREKLANGGQGVEEIVIEHc 2 1 tSuRVYGRNAAALSQALRLQAPELPVTVNPSKPRRGSFEVTLLRPDGS 1 2 gAELWTGIKKGPPRKLKFPEPQQVVEELKKYLS* 0 >SELH_ochPri Ochotona princeps (pika) 0 MAPNRRKRKAEAVADAAAEKREKQAKQANGVGGGEEIVIEHc 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPAKPRRGSFEVTLQRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQQVVEELKKYLS* 0 >SELH_canFam Canis familiaris (dog) 0 MASRGRKRKAEAAGVAAAEKRDKPASGRKAVEEATVVIEHC 2 1 TSuRVYGRNAAALSQALRLETPELPVEVNPAKPRRGSFEVTLLRPDGS 1 2 SVELWTGIKKGPPRKLKFPEPQEVVKALKQHLS* 0 >SELH_bosTau Bos taurus (cow) TGA verified 0 MASRGRKRKAEAALAAAAEKREKPAGGQEGGVEGPSVVIEHC 2 1 TSuRVYGRNAAALSQALRLQAPELTVKVNPARPRRGSFEVTLLRADGS 1 2 SAELWTGLKKGPPRKLKFPEPHVVLEELKKYLS* 0 >SELH_oviAri Ovis aries (sheep) 0 MASRGRKRKAEAALAAAAEKREKPAGSREGEVAGPSVVIEHC 2 1 TSuRVYGRNAAALSQALRLQAPELAVKVNPSRPRRGSFEVTLLRADGS 1 2 AELWTGLKKGPPRKLKFPEPHVVLEELKKYLS* 0 >SELH_susScr Sus scrofa (pig) 0 MASRGRKRKAETALGAAAEKQETPASGRKGMEEPSVVIEHC 2 1 TSuRVYGRNAAALSQALRVEAPELPVRVNPTKPRRGSFEVTLMRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_equCab Equus caballus (horse) 0 MASRGRKRKAEAVVAVAEKREKLTSGGKGVEEVTVVIEHc 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPAKPRRGSFEVTLLRPDGS 1 2 sAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_choHof Choloepus hoffmanni (sloth) 0 MASRGRKRKAEAVVLAAAEKQEKVASGKEGEKEAVVVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPEIPVKVNFAKPRRGSFEVTLLRPDGS 1 2 TELWTGIKKGPPRKLKFLQPLKVVEELKKYLS* 0 >SELH_loxAfr Loxodonta africana (elephant) 0 MASRGRKRKAEAAVAAAAAEKREKPVGGQAAAEEVVVIIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELSVKVNPSKPRRGSFEVTLQRPDGS 1 2 GAELWTGIKKGPPRKLKFPEPQEVVEELKKYL* 0 >SELH_monDom Monodelphis domestica (opossum) 0 MAPRGRKRKADVAAAALTEKPEKLAQGGEEGAGEARVVIEHC 2 1 QSuRVYARHAEAVGQALRLARPGLPVLLNPAKPRRSSFEVTLLRPDGS 1 2 RVELWSGIKKGPPRKLKFPEPAQVVEELKARLV* 0 >SELH_ornAna Ornithorhynchus anatinus (platypus) E5DI3CH10F50JR run=R_2008_02_11_18_03_02_ 0 DAGGAEVGEGLHVVIEHC 2 1 RSuGVYGRRAEALSRALSLAAPDLPVLLNPTKPRRNSFEVTLLRPDGT 1 2 RTELWSGIKKGRP >SELH_galGal Gallus gallus (chicken) tga confirmed 0 MAPRGRKRAARRPAEPEARADPPEKRPRDEAEGSPGDAGGPRVVIEHC 2 1 RSuRVYGRNAAALSEALRGAVASLAVEINPRQPRRNSFEVSLVKEDGS 1 2 TVQLWSGIGKGPPRKLKFPEPAAVVEALRSSLA* 0 >SELH_danRer Danio rerio (zebrafish) AI877878 BM037625 etc CKSU tga verified exons fused 0 MATRGKSARKRKADSDEKEKLDDAKKEKLEDKDEETGLRVVIEHC 21 KSuRVYGRNADVVREALADSHPELKVMINPHKPRRNSFEITLMDGERADVLWSGIKKGPPRKLKFPEPAEVVTALKQALEKE* 0 >SELH_takRub Takifugu rubripes (fugu) tga confirmed, no Ests 3 exons 0 MTPRVLMTGRRGTKRKAEEDEKPKEEKKEKQREDDQGGPRVAIEHC 2 1 KSuRVYGRNAEAVKSALLAAHPGLTVVLNPEKPRRNSFEITLLDEGK 1 2 ETSLWTGIKKGPPRKMKFPQPDVVVTALQEALKTE* 0 >SELH_ictPun Ictalurus punctatus (fish) CB940790 tga verified 0 MATRAKAGRGAKRKADVIAAAEPVAKQDKGNKGEREDDEGQRVIIEHC 2 1 KSuRVYGRNADAVREALLSAHPELHVVLNPEKPRRNSFEVTLIEGK 1 2 KELVLWTGLKKGPPRKLKFPEPAEVVTALEEALKSK* 0 >SELH_salSal Salmo salar CA043802 tga verified 0 MASRNKAGRVLKRKASVKEESVEEKRGKGEDDQPEIVTEGRRVVIEHC 2 1 KSuRVYGRNAEGVRVALLAACPDLTVVLNPQKPRSKSFEVILVEGE 1 2 KEVCLWSGIKKGPPRKLKFPEPEVVVSALEKALKTE* 0 >SELH_oryLat Oryzias latipes BJ026077 tga verified 0 MASKAGRRGTKRKVEAKKEEDKTSTEEKKARGENAHEEAGLKVLIEHC 2 1 KSuRVYGRNAEEVKSALLAARPELTVVCNPEKPRRNSFEITLLDGAK 1 2 ETSLWTGIKKGPPRKLKFPQPDDVVAAFKDALKTE* 0 >SELH_oncMyk Oncorhynchus mykiss Rainbow trout BX312781 0 MASLTKAGRVLKRKVETEESSVEGKRGKGEDDHPEIVTEGQRVVIEHC 2 1 KSuRVYGRNAEGVRVALLAACPDLTVVLNPQKPRSKSFEVILFEGE 1 2 KEVCLWSGIKKGPPRKLKFPEPEVVVSALEKALKTE* 0
MSRB1: 22 vertebrate sequences
>SELM_homSap Homo sapiens (human) NM_080430 Selenoprotein M (uc003ajq.1) chr22 0 MSLLLPPLALLLLLAALVAPATAATAYRPDWNRLSGLTRARVE 0 0 TCGGUQLNRLKE 0 0 VKAFVTQDIPFY 2 1 HNLVMKHLPGADPELVLLGRRYEELE 0 0 RIPLSEMTREEINALVQELGFYRKAAPDAQVPPEYVWAPAKPPEETSDHADL* 0 >SELM_musMus Mus musculus (mouse) 0 MSILLSPPSLLLLLAALVAPATSTTNYRPDWNRLRGLARGRVE 0 0 TCGGUQLNRLKE 0 0 VKAFVTEDIQLY 2 1 HNLVMKHLPGADPELVLLSRNYQELE 0 0 RIPLSQMTRDEINALVQELGFYRKSAPEAQVPPEYLWAPAKPPEEASEHDDL* 0 >SELM_ratNor Rattus norvegicus (rat) 0 MNILLSPPPLLLLLAALVAPATSITTYRPDWNRLRGLARGRVE 0 0 TCGGUQLNRLKE 0 0 VKAFVTQDIQLY 2 1 HNLVMKHLPGADPELVLLSRNYQELE 0 0 RIPLSQMTRDEINALVQELGFYRKSAPEAKVPPEYLWAPAKPPEDASDRADL* 0 >SELM_canFam Canis familiaris (dog) 0 MRLPLPPPPLLLLLAALAAAVTTFRPDWNRLHGLARARVE 0 0 TCGGUQLNRLKE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHLPGADPELVLLGHHYEELE 0 0 RIPLSEMTREEINELVQELGFYRKAAPDEAVPPEYLRAPARPAEGAPDRADL* 0 >SELM_bosTau Bos taurus (cow) 0 MHLPLPPPPLLLLLAAVAAATTTFRPDWNRLQGLARARVE 0 0 TCGGUQLNRLKE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHLPGADPELVLLGHRFEELE 0 0 RIPLSDMTREEINALVQELGFYRKASPDEPVPPEYLRAPARPAGDAPDHADL* 0 >SELM_susScr Sus scrofa (pig) 0 MHLPPLSLPLLLLLAALAAATTTFRPDWNRLQGLARARVE 0 0 TCGGUQLNRLKE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHLPGADPELVLLGHRFEELE 0 0 RIPLSDMTREEINALVQELGFYRKAAPDDPVPPEYMRAPARPAEGAPDRADL* 0 >SELM_ornAna Ornithorhynchus anatinus (platypus) 0 MLVCPERRSWVLIPPLSLLLLLPGLLAAFQPDWSRLQGLARGKVE 0 0 TCGGuQLNRLKE 0 0 VKAFVTEDIPLY 2 1 HNLVMKHLPGADPELVLLNFRYEELE 0 0 RIPLSHMTRAEINQLVQDLGFYRKAERDAPVPPEFQQAPAKTSDLREKVQPQETPKSEEQNHPDL* 0 >SELM_galGal Gallus gallus (chicken) 0 MRRAALAALLLLLAAAAGIERRPPRGLARGKVE 0 0 TCGGURLSRLPE 0 0 VKAFVSQDIPLY 2 1 HNLEMKHLPGADPELVLLSFRYEELE 0 0 RIPLSDMTREEINQLVQELGFYRKETPEAPVPEEFQFAPAKPLPTLTPRRAPAADGKTLSEQDKKDHPDL* 0 >SELM_botIns Bothrops insularis (snake) PVPDAFQMA ..PLLWLPLLLLGLLSAVAPLRAVQLDRSRLQWLARGKVE 0 0 SCGGURLNRLPE 0 0 VKAFLNEDLPLY 2 1 HNMDLKYLAGADPELILLNIKFEELQ 0 0 RIPLSDMSREEINQLMQELGFYRKDTPDSLFRCFPNGAC* 0 >SELM_xenTro Xenopus laevis (frog) 0 MWLPLPLLLGLLQLQPILSYQIDWNKLERINRGKVE 0 0 SCGGUQLNRLKE 0 0 VKGFVTEDLPLY 2 1 HNLEMKHIPGADPELVLITSRYEELE 0 0 RIPLSDMKRDEINQLLKDLGFYRKSSPDAPVPAEFKMAPARASGDTKEDL* 0 >SELM_danRer Danio rerio (zebrafish) 0 MWPLIFTALLPSVILTYEVNIEKLSGLARARVE 0 0 TCGGUQLNRMRE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHIPGADPELVLLNHYYEELD 0 0 RIPLSEMTRAEINKLLAELGFYKKDHPEDQVPEEFRFSPAKDSPFEGRQSSTAAPETTEPSDSQHTDL* 0 >SELM_ictPun Ictalurus punctatus (fish) 0 MFSTWPLLWAAFLPCISLAYEVDWKKLDGLARAKVE 0 0 SCGGUQLNRLRE 0 0 VKAFVTQDIPFY 2 1 HNLVMKHIPGADPELVLLNHYYEELD 0 0 RIPLSHMTRTDINGLLEELGFYKKARAEDDVPEEFRFSPAKDSPFKEHHTRRAPANSDLAQEPQPENESPHKDL* 0 >SELM_oncMyk Oncorhynchus mykiss (trout) 0 MWFFFFVSLLNCVSAYDVDLKKLDGLAKAKVE 0 0 SQSCGGUQLNRLRE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHIPGADPELVLLNHYYEELD 0 0 RIALSDMTRSEINELLEKLGFYKKAQAEDQVPEEFRFSPAKDSPFKATPADNASSDSDAEAKHSDL* 0
MSRB1: 17 vertebrate sequences
>MSRB1_homSap Homo sapiens (human) SEPX1 (uc002cng.1) SELX SELR human chr16 0 MSFCSFFGGEVFQNHFEP 1 2 GVYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPEHNRSEALK 0 0 VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK 1 2 GKETSASQGH* 0 >MSRB1_macMul Macaca mulatta (rhesus) 0 MSFCSFFGGEVFQNHFEP 1 2 GVYACAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPEHNRPGALK 0 0 VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK 1 2 GKETSTSQGH* 0 >MSRB1_musMus Mus musculus (mouse) exon break just at stop codon NP_038787 0 MSFCSFFGGEVFQNHFEP 1 2 GVYVCAKCSYELFSSHSKYAHSSPWPAFTETIHPDSVTKCPEKNRPEALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFVPK 1 2 GKEAAASQGH* 0 >MSRB1_ratNor Rattus norvegicus (rat) 0 MSFCSFFGGEVFQNHFEP 1 2 GVYVCAKCGYELFSSRSKYAHSSPWPAFTETIHEDSVAKCPEKNRPEALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 2 GKEAPASQGD* 0 >MSRB1_cavPor Cavia porcellus (guinea_pig) 0 MSFCSFFGGEVFQNHFES 1 2 GIYVCAKCGYELFSSRSKYAHSSPWPAFTDTIHADSVAKCPEHNRPGALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 2 DKETSASQGH* 0 >MSRB1_canFam Canis familiaris (dog) frag 0 MSFCSFFGGEVFQNHFEP 1 2 GVYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPERNSPEALK 0 0 VSCGKCGNGLGHEFLNDGPKPGKSRFuIFSSSLKFVPK 1 2 GKGTSGSQEA* 0 >MSRB1_bosTau Bos taurus (cow) 0 MSFCSFFGGEIFQNHFEP 1 2 GIYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPEHNRPGAIK 0 0 VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 2 AEETSASQGQ* 0 >MSRB1_equCab Equus caballus (horse) 0 MSFCSFFGGEIFQNHFEP 1 2 GIYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPEHNRPEALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFVPK 1 2 GKESSASQGQ* 0 >MSRB1_ loxAfr Loxodonta africana (elephant) 0 MSFCSFFRSEVFQNHFEP 1 2 GVYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVGKHPEHNRPEALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 2 GKETSASQGK* 0 >MSRB1_monDom Monodelphis domestica (opossum) frag 2 GVYVCAKCGYELFSSRSKYHHSSPWPAFTETIHADSVSKRPESGRSEALK 0 0 VSCGKCGNGLGHEFINDGPKKGQSRFuIFSSSLKFVPKG 1 >MSRB1_ornAna Ornithorhynchus anatinus (platypus) 0 1 2 GTYVCARCGYELFSSRSKYEHSSPWPAFTETIHPDSVAKREEPGRPNAFK 0 0 VSCGKCGNGLGHEFLNDGPRRGQSRFuIFSSLKFIPK 1 2 GKDSQAAQDK* 0 >MSRB1_galGal Gallus gallus (chicken) 0 MSFCSFFGGEVFKDHFEP 1 2 GVYVCARCGYELFSSRAKYEHSSPWPAFTETIHEDSVAKRKERPGALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 0 GKSPQEN* 0 >MSRB1_anoCar Anolis carolinensis (lizard) 0 MSFCAFSGGEIYQGHFEA 1 2 GMYVCSKCGFELFSSKSKYAHSSPWPAFTETIHDDSITKYLERPNAFK 0 0 VLCGKCGNGLGHEFINDGPKKGQSRFZIFSSSLKFVPK 1 >MSRB1_xenTro Xenopus tropicalis (frog) 0 MSFCSFFGGEVYKDHFKS 1 2 GIYVCSECNYELFSSRSKYQHSSPWPAFTETVHKDSISKYLERPNAYK 0 0 VSCGKCGNGLGHEFINDGPKKGQSRFuIFSSSLKFIPK 0 0 DKVDGEVQRE* 0 >MSRB1_danRer Danio rerio (zebrafish) tga confirmed 0 MSFCSFSGGEIYKDHFES 1 2 GMYVCAQCGYELFSSRSKYEHSSPWPAFTETIHEDSVSKQEERWGAYK 0 0 VRCGKCGNGLGHEFVNDGPKHGLSRFuIFSSSLKFI 0 PKVKNEQQ* 0 >MSRB1_ictFur Ictalurus furcatus (fish) 0 MAFCSFKGGEIFKDHYEP 1 2 GIYVCVKCGYELFSSTSKYKLSSPWPAFTTTIHEDSVSKQEERPGALK 0 0 IRCGKCNNGLGHEFLNDGPKHGLSRFuIFSSSLKFV 0 0 PKDKGGQ* 0 >MSRB1_oncMyk Oncorhynchus mykiss (trout) 0 MSFCSFFGGEVFKDHFKT 1 2 GLYMCAQCGHQLFSSRSKYEHSSPWPAFTETILQDSVSKHEERPGAFK 0 0 VRCGKCGNGLGHEFVGDGPKKGLSRFuIFSSSLKFV 0 0 PKDKVDGQ* 0 >MSRB1_salSal Salmo salar (salmon) 0 MSFCSFFGGEVFKDHFKT 1 2 GLYVCAQCGHQLFSSRSKYEHSSPWPAFTETVLQDSVSKHEERPGAFK 0 0 VRCGKCGNGLGHEFVGDGPKKGLSRFuIFSSSLKFV 0 0 PKDKVDGQ* 0 >MSRB2_homSap Homo sapiens (human) CBS-1 PILB 5 exons chr10 0 MARLLWLLRGLTLGTAPRRAVRGQAGGGGPGTGPGLGEA 1 2 GSLATCELPLAKSEWQKKLTPEQFYVTREKGTEP 0 0 PFSGIYLNNKEAGMYHCVCCDSPLFS 2 1 SEKKYCSGTGWPSFSEAHGTSGSDESHTGILRRLDTSLGSARTEVVCKQ 0 0 CEAHLGHVFPDGPGPNGQRFCINSVALKFKPRKH* 0 >MSRB2_musMus Mus musculus (mouse) 0 MARLLRALRGLPLLQAPGRLARGCAGS 1 2 GSKDTGSLTKSKRSLSEADWQKKLTPEQFYVTREKGTEA 0 0 PFSGMYLNNKETGMYHCVCCDSPLFSSEKKYCSGTGWPSF 2 1 SEAYGSKGSDESHTGILRRLDTSLGCPRMEVVCKQ 0 0 CEAHLGHVFPDGPKPTGQRFCINSVALKFKPSKP* 0 >MSRB2_bosTau Bos taurus (cow) 0 MARLLRALRGLTLREAPGWAVRGRADCGGFRAGA 1 2 GSPQAAHPDTFPFHRGSKSEWQKKLTPEQFHVTREKGTEP 0 0 PFSGIYLNNKEPGMYHCVCCDSPLFS 2 1 SEKKFCSGTGWPSFSEAHGTSGSDESNTGILRRADTSLGPARTEVVCKQ 0 0 CEAHLGHVFPDGPGPAGQRFCINSVALRFKPRKH* 0 >MSRB2_canFam Canis familiaris (dog) 0 MGDSSTTLRFLIRKRHSPYDFVVKIDKSKPKEERSLTKHEL 0 0 PLTKSEWQKKLTPEQFYVTREKGTEP 0 0 PFSGVYLNNKESGMYHCVCCDSPLFS 2 1 SEKKYCSGTGWPAFSEAHGTSGSDERDTGILRRVDTSLGLTRTEVVCKQ 0 0 CEAHLGHVFPDGPGPSGQRFCINSVALKFKPRKH* 0 >MSRB2_xenTro Xenopus laevis (frog) 0 MSRLLTGFRLLLRVREISFPSVTAQRLQAWSRVRMAQTGADL 1 2 GSLTRYDDSAVSTDWQKKLTPEQYYVTREKGTEL 0 0 PFSGIYLNNTEKGMYHCVCCSAPLFS 2 1 SEKKYNSGTGWPSFSEAYGAQGADESNTNVLRRLDNSLGSTGTEVICKE 0 0 CDAHLGHVFEDGPPPYGQRFCINSVALTFAPSSM* 0 >MSRB3_homSap Homo sapiens (human) ER retention signal KAEL* AY358229 0 MSPRRTLPRPLSLCLSLCLCLCLAAALGSAQS 1 2 GSCRDKKNCKVVFSQQELRKRLTPLQYHVTQEKGTES 2 1 AFEGEYTHHKDPGIYKCVVCGTPLFK 2 1 SETKFDSGS 1 2 GWPSFHDVINSEAITFTDDFSYGMHRVETSCSQ 0 0 CGAHLGHIFDDGPRPTGKRYCINSAALSFTPADSSGTAEGGSGVASPAQADKAEL* 0 >MSRB3_musMus Mus musculus (mouse) 0 MSAFNLLHLVTKSQPVAPRACGLPSGSCRDKK 1 2 NCKVVFSQQELRKRLTPLQYHVTQEKGTES 2 1 AFEGEYTHHKDPGIYKCVVCGTPLFK 1 1 SETKFDSGS 1 2 GWPAFHDVISSEAIEFTDDFSYGMHRVETSCSQ 0 0 CGAHLGHIFDDGPRPTGKRYCINSASLSFTPADSSEAEGSGIKESGSPAAADRAEL* >MSRB3_canFam Canis familiaris (dog) 0 MSAFNLLHLVTKSQPVALRACGLPSGSCRDKKNCK 1 2 VVFSQQELRKRLTPLQYHVTQEKGTESAFEGEYTHHKDPGIYKCVVCGTPLFRSESKFDSGS 1 2 GWPSFHDVISSDAITFTDDFSYGMHRVETSCSQ 0 0 CGAHLGHIFDDGPRPTGKRYCINSASLAFTPAGGGTQGSSGPGGPAAGGRAEL*
Reference sets of vertebrate SECIS elements
SEPP2: 5 SECIS sequences
>SEPP2_SECIS_monDom Monodelphis domestica (opossum) chr2:9455773-9456962 COVE: 30 55 bp after stop codon ACGTGCCGCTGCCCCTCCCTCCCTCCAAGAATGACGCCCACAGTGAAACCCAGAGAACTGGTCCCTGTGGGCTGATGCCCCAGAGGGGAGGAGAGGC >SEPP2_SECIS_macEug Macropus eugenii (wallaby) SECIS COVE score: 26.38 trace ti|975557399 contains terminal exon CCTCATTCCTCTTCTCCATGGCATCAGCCCACCATGGCCGGTTCCCCGGGTTTCACTGTGGGCATCATTAATTGAGGGAGGGAAGGGCAATGGGCAAG >SEPP2_ornAna Ornithorhynchus anatinus (platypus)Ultra131:321101-348720 COVE: 34.28 no match to chicken TCCCTCGACTCCCGCTCCCGCCTCGCACTCATGACGTCCACGGTGTCAACCGGCCCGCCGGGCACCGTGGACTGACGCCGGTCGAGGCGGAGGGGT >SEPP2_ornAna Ornithorhynchus anatinus (platypus) Ultra131:321101-348720 COVE: 22.82 no match to chicken GGAATCAGGAACCCAGTAACATGAGGTCATCTTCGGAAGCCTGTGCCTAGAGGACCAAGATAATGGAAAAAGTGACGGACAAGGGTGTGTAGCTGG >SEPP2_tetNig Tetraodon nigroviridis (pufferfish) COVE: 17 GCTGGACCCAGGCTGCTGGTGGTCCCGTTGATGACGTCTGCGCTGGTAAACCTGCCTGCAGGAGCCTGTGGACCGACGTGTGTGGACCCACCGGCAG
SEPW1: 13 SECIS sequences
>selW_SECIS_homSap Homo sapiens (human) agggaccttgacccagcccctctcagcagacgcttcatgataggaaggactgaaaagtcttgtggacacctggtctttccctgatgttctcgtggctgctgttgggggcagagattgacgcccccggtctttgcct >selW_SECIS_panTro Pan troglodytes (chimp) AGGGACCTTGACCCAGCCCCTCTCAGCAGACGCTTCATGATAGGAAGGACTGAAAAGTCTTGTGGACACCTGGTCTTTCCCTGATGTTCTtGTGGCTGCTGTTGGGGGCAGAGATTGACGCCCCCGGTCTTTGCCT >selW_SECIS_ponPyg Pongo pygmaeus (orang_sumatran) AGTCCAGGGACCTTGACCCAGCCCCTCTCAGCAGACGCTTCATGATAGGAAGGACTGAAAAATCTTGTGGACACCTGGTCTTTCCCTGATGTTCTCGTGGCTGCTGTTGGGGGCAGAGATTGACGCCCCTGGTCTTTGCCT >selW_SECIS_macMul Macaca mulatta (rhesus) AGcGACCTTGACCCAGCCCCTCTCAGCAGACGCTTCATGATAGGAAGGACTGAAAAGTCTTGTGGACgCCTGGTCTTTCCCTGATGTTCTCGTGGCTGCTGTTGGGGGCAGAGATTGACGCCgCtGGTCTTTGCCT >selW_SECIS_musMus Mus musculus (mouse) ACTGAAATGTCTTAGACTTGGCCCAGCCCCTCGTGGCAGACGCTTCATGATGGGAAGAACTGAAATGTCTCGTGGACGCCTGGTCTTTCCCTGATGTCCCTGCGACTGCCACGTAGGGGCAGAGACTGATGCCCCTGTGGGTGCCT >selW_SECIS_ ratNor Rattus norvegicus (rat) CCTGGCCGGCCTTTCTTGGCAGCCGCTTCATGACAGGAAGGACTGAAATGTCTCAAAGACCTGTGGTCTTTCTTCGATGTTCCTGCGGCCACCAAGTCAGGCCAGAGATGGATTCTGTGTGTGGGTGCCT >selW_SECIS_oryCun Oryctolagus cuniculus (rabbit) AGTAACCTTGACCCAGCCCCTTTCATGCCTCAGCCTCGTCTCCATAGGCTAAGACTGGAGAAATGAGTCCCCTGAAGAACTGAAACTGGGGGTAGAGGGTTGGTGTTTTAAGATGTGGATGAGCTGGTCTTTAC >selW_SECIS_canFam Canis familiaris (dog) CCAGtGACCTTGgCCCAGCCCCTCgtgGCAGACGCTTCATGATgGGAAGaACTGAAAtGTCTcGTGGACgCCTGGTCTTTCCCTGATGTccctgcgactgccacgtaGGGGCAGAGAcTGAtGCCCCtGGTCTTTGCCT >selW_SECIS_susScr Sus scrofa (pig) AGTAACCTTGACCCAGCCCCTTTCATGCCTCAGCCTCGTCTCCATAGGCTAAGACTGGAGAAGTCTTGTGGACGCCTGGTCTTTCCCTGATGTTCTCGTGGCTGCTGTTGGGGGCAGAGATGGATGAGCTGGTCTTTAC >selW_SECIS_borAnc Boreoeuthere ancestralis (ancestral) AGTCCAGCAACCTTGGCCCAGCCCCTCTCAGCAGATGCTTCATGACAGGAAGGACTGAAATGTCTTGTGGACGCCTGGTCTTTCCCTGATGTTCTTGTGGCTGCTGGTTGGGGCAGAGATTGACACCCCTGGTCTTTGCCT >selW_SECIS_dasNov Dasypus novemcinctus (armadillo) CCAGCAACCTCAGCCCAGCTGCCCTTGGCAGACGCTTCATGAGGGGAAGGACCTAAATGCGTCGTGGATGCCTGGTCTTTCCCTGATGCTCCTTCACCTGCCAGATGGGGCAGAGGTCATTGCCCCTGGTCTTGGCCT >selW_SECIS_loxAfr Loxodonta africana (elephant) GGGACCTTGGCCCAGCCCCTTTCAGCAGACACTTCATGACAGGAGGACTGAAATGTCTCCCAGACGCCTGGCTCTTTCCCTGAATCTGTCGGCTGCAGGACAGGGCAGCGGTTGACTCTCTCGTTTTTTGCAT >selW_SECIS_echTel Echinops telfairi (tenrec) AGGCCAGAGACCTTGGCCCAGTCCCTCCATGACAGGCAGAACTGAAATGTCCTCTGGACAAGTGGTCTTTTCCAGAAACCCCAGGGCTGCTGGGCCGGAGCCGAGGCTGACAACCCTGGTCTTTGCCT
DIO1: 29 SECIS sequences
>DIO1_SECIS_homSap Homo sapiens (human) COVE score: 29 ttttaactctgtgtctttacatatttgtttatgatggccacagcctaaagtacacacggctgtgacttgattcaaaagaaaatgttataag >DIO1_SECIS_ponPyg Pongo pygmaeus (orang_sumatran) COVE score: 29 ttttaactctgtgtctttacatatttgtttatgatggccacagcctaaagtacacacggctgtgacttgattcaaaagaaaatgttataag >DIO1_SECIS_macMul Macaca mulatta (rhesus) COVE score: 29 tttcaactctgtgtctttacatatttgtttatgatggccacagcctaaagtacacacggctgtgacttgattcaaaagaaaatgttataag >DIO1_SECIS_cavPor Cavia porcellus (guinea_pig) COVE score: 24 tgttaactctgcttcttttcatatttgttcatgacggtcacagtctaaagtacacacagctgtgacctgatttgaaagaaaatgttttaag >DIO1_SECIS_canFam Canis familiaris (dog) COVE score: - ttttaactctgcttcttttcatgtttgtctatgacggccacagcctaaagcacacacagctgtgacttgatttgaaagaaaatgttttaag >DIO1_SECIS_felCat Felis catus (cat) COVE score: - ttttaactctgcttcttttcacgtttgtctatgacggccacagtctaaagtgcacacagctgtgacttgacttgaacgaaaatgttttaag >DIO1_SECIS_bosTau Bos taurus (cow) COVE score: 28 ttttaactctgcctcttttcatatttgttcatgacggccacagcctaaagtacacacggctgtgacttgatttgaaagaaaatgttttaag >DIO1_SECIS_sorAra Sorex araneus (shrew) COVE score: 24 cggaaactcagcttctcttcatatttgtttatgacagccccagctgaaagtacacacagctgtggcttgattggaaagaaaatgttttaag >DIO1_SECIS_eriEur Erinaceus europaeus (hedgehog) COVE score: 26 tttaactctgctttcttctcatatttgcttatgatggtcacagcttaaagtatacacagctgtgacttgattggaaagaaaatattttaag >DIO1_SECIS_borAnc Boreoeuthere ancestralis (ancestral) COVE score: 29 ttttaactctgcttcttttcatatttgttcatgatggccacagcctaaagtacacacggctgtgacttgatttgaaagaaaatgttttaag >DIO1_SECIS_dasNov Dasypus novemcinctus (armadillo) COVE score: - ttttaactctgcttcttttcatatttgtttatgatggccacagtttaaagtacatacagctgtgacttgatatgaaaaagaaatattttaag >DIO1_SECIS_loxAfr Loxodonta africana (elephant) COVE score: - ttttaactctgcttcttttttcatgtatttatgatgggccacagcctaaagtgcacaacagctgtgacttgatttgaaaaacatctttaag >DIO1_SECIS_monDom Monodelphis domestica (opossum) COVE score: - tttccatcctgcttctacaaatatttatttatgacaatcacagcctaaagctcagggcagctgggattcgacgggagaaaaagtttgtaag >DIO1_SECIS_ornAna Ornithorhynchus anatinus (platypus) COVE score: 24 ccccggatccggttccgtgaatattggtttatgagggtcacagtgtaaagcgcatgcagctgtgacttgatctgagaaaatatttctgcggc >DIO2_SECIS_homSap Homo sapiens (human) 5,630 bp utr COVE score: 30 cagagatgtgcagagttgaccagtgtgcggatgataactactgacgaaagagtcatcgactcagttagtggttggatgtagtcacattagtttgcctctc >DIO2_SECIS_macMul Macaca mulatta (rhesus) COVE score: 30 cagagatgtgcagagttgaccagtgtgcggatgataactactgacgaaagagtcatcgactcagttagtggttggatgtagtcacattagtttgcctctc >DIO2_SECIS_musMus Mus musculus (mouse) COVE score: 29 cggagatgttcagagctcactggtgtgcgaatgataactactgacgaaagagctgtctgctcagtctgtggttggatgtagtcacacgagtctgcctttctgca >DIO2_SECIS_ratNor Rattus norvegicus (rat) COVE score: 28 ccgagatgttcggagctcactggtgtgcgaatgataactactgacgaaagagtcatctgctcagtctgtggttggatgtagtcacacgagtctgcctctccatc >DIO2_SECIS_canFam Canis familiaris (dog) COVE score: 27 ctgggatgtgcagaggtgaccagtgtgcgaatgataactactgatgaaagagtcactgactcagttagtggttggatacagtcacattagttttcctct >DIO2_SECIS_borAnc Boreoeuthere ancestralis (ancestral) COVE score: 30 ctgggatgtgcagaggtgaccagtgtgcaaatgataactactgatgaaagagtcattgactcagttagtggttggatgtagtcacattagtttgcctctc >DIO2_SECIS_dasNov Dasypus novemcinctus (armadillo) iodothyronine deiodinase type II ctgggaagttcagaggctaccagtgtgccaatgataactactgacgaaagaggcatcgactcagttagtggttggatgtagccacattagtttgcctctc >DIO3_SECIS_homSap Homo sapiens (human) COVE score: 31 ttgggtgcacaggagccccactgctgatgacgaactatctctaactggtcttgaccacgagctagttctgaattgcaggggcctcaaagcagca >DIO3_SECIS_macMul Macaca mulatta (rhesus) COVE score: 30 ttgggtgcacaggagccccactgctgatgacgaactgtctctaactggtcttgaccacgagctagttctgaattgcaggggcctcaaaacagca >DIO3_SECIS_musMus Mus musculus (mouse) COVE score: 26 ttgggtgcgctggagccctggctgctgatgacgaaccgcctctaactgggcttgaccacgggtcggctctgaattgcagagaggctcgaaacagc >DIO3_SECIS_ratNor Rattus norvegicus (rat) COVE score: 26 ttgggtgcgctggagccctggctgctgatgacgaaccgcctctaactgggcttgaccacgggtcggctctgaattgcagagaggctcgaaacagc >DIO3_SECIS_canFam Canis familiaris (dog) COVE score: 26 ttgggtgctggcgagccccactgctgatgacgagccgcctctaactggtcttgaccacgagctggttctgagttgcaggggggcttgcagcggc >DIO3_SECIS_bosTau Bos taurus (cow) COVE score: 30 ttgggtgctcacgagccccactgctgatgaagagctgtctctaactggcctcgaccacgagctggttctgatttgcaggaggctcgcagcagc >DIO3_SECIS_borAnc Boreoeuthere ancestralis (ancestral) COVE score: 27 ttgggtgctcaggagccccactgctgatgacgaactgtctctaactggtcttgaccacgagctggttctgaattgcagggggctcgcagcagca >DIO3_SECIS_loxAfr Loxodonta africana (elephant) COVE score: 22 ttcggtgcgctagagccccactgctgatgacgaactgtctctaactggtcttgaccacgagctgattccgaattgcagggaactcgcagcagc >DIO3_SECIS_echTel Echinops telfairi (tenrec) COVE score: - ttcggtgctctgcagccccactgctgatgacgaactgcctctcactggtcttgaccacgagctgcttctgaaatgcaggggactcgcagccgca