Spreadsheet links to Genome Browser views: Difference between revisions
m (fixup absolute reference) |
mNo edit summary |
||
| Line 1: | Line 1: | ||
Many users of the Genome Browser gather data of their own in Excel spreadsheets and would like to create links to the Browser using data in the spreadsheet. For example, a clinical geneticist may have lists of regions for a patient that are duplicated or deleted, as determined by comparative genomic hybridization (CGH). These regions can be the source information for a browser view allowing access to each region with a single click. | Many users of the Genome Browser gather data of their own in Excel spreadsheets and would like to create links to the Browser using data in the spreadsheet. For example, a clinical geneticist may have lists of regions or genes for a patient that are duplicated or deleted, as determined by comparative genomic hybridization (CGH). These regions can be the source information for a browser view allowing access to each region with a single click. | ||
Click to download the spreadsheet: | Click to download the spreadsheet: | ||
| Line 28: | Line 28: | ||
omimGene=pack | omimGene=pack | ||
Any track that has been open in | Any track that has been open in on your Browser will remain in the view when the new browser window opens. | ||
A new track can be added using the tableName and a visibility of choice: | A new track can be added using the tableName and a visibility of choice. e.g.: | ||
&snp131=dense | &snp131=dense | ||
Simply add to the end of the url any other desired tableName=visibility, | Simply add to the end of the url any other desired tableName=visibility, appended to the url by an ampersand (&). The simplest way to learn the name of the table underlying a track is to do a mouseover in a Genome Browser image and read the url at the bottom of the browser page. The table is shown in the url as | ||
g=tableName | g=tableName | ||
You can also see the tableName by clicking into an item in the track and then clicking "show table schema" on the next page. | |||
Visibility options include: | Visibility options include: | ||
Revision as of 03:13, 10 February 2011
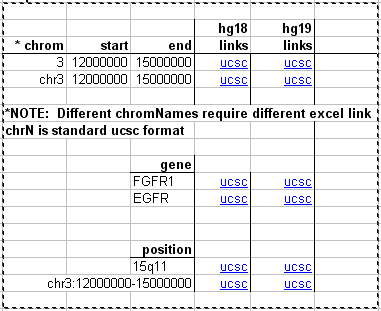
Many users of the Genome Browser gather data of their own in Excel spreadsheets and would like to create links to the Browser using data in the spreadsheet. For example, a clinical geneticist may have lists of regions or genes for a patient that are duplicated or deleted, as determined by comparative genomic hybridization (CGH). These regions can be the source information for a browser view allowing access to each region with a single click.
Click to download the spreadsheet: File:UcscLinks.xls
Careful use of Excel's "copy" and "move" functions should allow the links on this sheet to be used without modification.
Customizing the links
The contents of the last cell in the image above (cell G22 in the actual spreadsheet) are as follows:
=HYPERLINK("http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position="&E22&"&dgv=pack&knownGene=pack&omimGene=pack","ucsc")
This example shows how to create a link that turns on specific tracks of interest. In this case, three tracks are explicitly turned on:
Database of Genomic Variants (table: dgv) UCSC Genes (table: knownGene) OMIM Genes (table: omimGene)
Each track is set to "pack" in the link as follows:
dgv=pack knownGene=pack omimGene=pack
Any track that has been open in on your Browser will remain in the view when the new browser window opens.
A new track can be added using the tableName and a visibility of choice. e.g.:
&snp131=dense
Simply add to the end of the url any other desired tableName=visibility, appended to the url by an ampersand (&). The simplest way to learn the name of the table underlying a track is to do a mouseover in a Genome Browser image and read the url at the bottom of the browser page. The table is shown in the url as
g=tableName
You can also see the tableName by clicking into an item in the track and then clicking "show table schema" on the next page.
Visibility options include:
hide dense squish pack full