Sulfatase evolution: ARSK: Difference between revisions
Tomemerald (talk | contribs) |
m (fixup absolute references) |
||
| (32 intermediate revisions by one other user not shown) | |||
| Line 40: | Line 40: | ||
=== Comparison of ARSK and IDS evolutionary rates === | === Comparison of ARSK and IDS evolutionary rates === | ||
IDS has a much more conventional phylogenetic distribution, with clear orthologs in many clades where ARSK is missing (eg tunicate, echinoderm, arthropods and cnidaria). However | IDS has a much more conventional phylogenetic distribution, with clear orthologs in many clades where ARSK is missing (eg tunicate, echinoderm, arthropods and cnidaria). Since choanoflagellates encode a distinct IDS with mediocre alignment with their ARSK, these genes diverged far earlier. However the rate of divergence of IDS (measured as divergence from human) is indistinguishable from that of ARSK over the [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC524432/ 800 million years] spanning the divergence of choanoflagellates from human. | ||
The rates at which amino acid substitutions accrue in these two proteins are somewhat above average in the context of whole proteome comparisons of these species. Note divergence of sulfatases bottoms out at about 20% since a minimal core of residues is needed to define the fold and provide activity. Although subclades evolve at various rates, the divergence curve here has remained monotonic as plotted relative to the species divergence tree, ie the earlier a lineage diverged, the more diverged its protein comparison will be. Operationally, this simply means that species appear in correct phylogenetic order on Blastp output. | |||
Blastp: <font color=blue>score exp id</font> <font color=red>score exp id</font> | Blastp: <font color=blue>score exp id</font> <font color=red>score exp id</font> | ||
| Line 51: | Line 53: | ||
<font color=blue>ARSK_braFlo Branchiostoma floridae (amphioxus) 1477 6.1e-156 54%</font> <font color=red>1704 7.5e-151 59% IDS_braFlo</font> | <font color=blue>ARSK_braFlo Branchiostoma floridae (amphioxus) 1477 6.1e-156 54%</font> <font color=red>1704 7.5e-151 59% IDS_braFlo</font> | ||
<font color=blue>ARSK_sacKow Saccoglossus kowalevskii (acornworm) 1433 2.8e-151 52%</font> <font color=red>1488 7.9e-157 53% IDS_sacKow</font> | <font color=blue>ARSK_sacKow Saccoglossus kowalevskii (acornworm) 1433 2.8e-151 52%</font> <font color=red>1488 7.9e-157 53% IDS_sacKow</font> | ||
<font color=blue>ARSK_monBre Monosiga brevicollis (choanoflagellate) 788 6.3e-83 33%</font> | <font color=blue>ARSK_monBre Monosiga brevicollis (choanoflagellate) 788 6.3e-83 33%</font> <font color=red> 448 2.8e-061 36% IDS_monBre</font> | ||
The rates at which sulfatase folds diverge (in terms of backbone root mean square best fit) do not appear correspondingly high, even though blastp against the PDB database of determined structures gives only poor matches (low 20's in percent id) for either IDS or ARSK. The only mammalian (or even metazoan) sulfatases with crystallographic structures are ARSA, STS, and ARSB but these are too weak to allow accurate modeling of ARSK or IDS. | |||
Note the former sequences have low percent identity to each other -- low 30's -- yet have fairly similar folds. Somewhat better matches of ARSK occur with bacterial sulfatase, notably [http://www.rcsb.org/pdb/explore.do?structureId=3ED4 3ED4] of E. coli and [http://www.rcsb.org/pdb/explore.do?structureId=3B5Q 3B5Q] of Bacteroides thetaiotaomicron. | |||
It might be possible to model ARSK or IDS using these structures (or some consensus derived from them) but the outcome would not be nearly sufficiently accurate to depict substrate binding sites (ie, deduce the natural substrate for ARSK). There are substantial issues with deletions and insertions, though these presumably lie in loops outside the core beta sheet/alpha helix secondary structure. Conserved homologous disulfides could conceivably provide severe modeling restraints but these do not appear to exist. | |||
=== Conserved disulfides and glycosylation sites === | |||
Within vertebrates, ARSK has 3 cysteines conserved to some phylogenetic depth beyond the catalytic motif and 7 NxT/S potential glycosylation sites, one of which overlaps with a conserved cysteine. These number decrease if conservation throughout deuterostomes (or even to choanoflagellates) is demanded. Thus the proper way to summarize these sites is a table by phylogenetic depth; presumably those with greater antiquity have been conserved for more fundamental reasons. | |||
Two glycosylation sites and one cysteine are even conserved at homologous positions IDS (adjusting gap placement manually). This does not imply these sites have been conserved since the common ancestral protein. They could simply be favorable sites on the protein exterior where the same feature has been independently advantageous and so arisen multiple times. | |||
Just because cysteine pairs are conserved does not imply they form an disulfide; similarly NxT/S can also be conserved for other reasons than glycosylation. However assuming from the signal peptide a secreted location in an oxidizing environment, at least some of these will likely be realized. Conservation of unselected amino acids does not persist by accident over mammalian order time scales. | |||
It's been shown previously that the 17 human sulfatases do not conserve either type of site to any extent. This is surprising in the case of disulfides which are often fundamental to structural stability (eg trillions of years of branch length conservation in rhodpsins) but evidently have arisen multiple times de novo during the course of sulfatase evolution and acquired roles resulting in their subsequent conservation. | |||
Phylogenetic depth of potential glycosylation and disulfide sites | |||
ARSK IDS | |||
<font color=red>108 NYT vert</font><font color=red> 121 NFS</font> | |||
<font color=gray>166 NRT prim</font><font color=gray> 187 DVL</font> | |||
<font color=black>193 NYT terr</font><font color= gray > 215 SAS</font> | |||
<font color=red>262 NCT deut</font><font color=red> 280 NIS</font> | |||
<font color=black>263 C vert</font><font color= gray > 281 I</font> | |||
<font color= black >283 C vert</font><font color= gray > 304 V</font> | |||
<font color=gray>375 NLS terr</font><font color= gray > 390 LME</font> | |||
<font color=red>410 C deut</font><font color=red> 422 C</font> | |||
<font color=blue>413 NAS deut</font><font color= gray > 427 PSF</font> | |||
<font color=black>498 NYS vert</font><font color= gray > 499 YTV</font> | |||
[[Image:ArskGlyCys.png]] | |||
<br clear=all> | |||
=== Comparison of introns in ARSK and IDS === | |||
Given that the natural substrate of ARSK remains unknown, there is value in determining its nearest neighbor within the sulfatase gene family because some clues to the substrate might emerge. IDS emerges from alignment comparisons as the best candidate, but only with marginal support. Intron position and phase are often better conserved than amino acid identity. If introns between these two genes agree at least in part -- and other sulfatases fare far worse -- that would provide independent support. | |||
It is not easy to reliably compare [[Opsin_evolution:_ancestral_introns|intron position and phase]] in highly diverged homologs, especially if gaps are abundant as here. However the [[Opsin_evolution:_annotation_tricks|procedures]] worked out for opsins are applicable here: multiple orthologs of each gene are aligned together to improve gap placement and establish flanking invariant residues to limit uncertainty. This allows exon ends of the second gene to be painted on the first gene, with colors blue, red, and magenta encoding codon overhang (phase) of 0, 1 or 2 base pairs. The outcome of that is shown below for ARSK and IDS. | |||
Recall a gene of 500 amino acids has 1500 possible combinations of intron position and phase; accidental agreement (homoplasy) can occur but is [[Opsin_evolution:_ancestral_introns#Managing_homoplasy_in_intron_data|rare and manageable]]. Introns have been phenomenally conserved throughout deuterostome history, with gain and loss events approaching 1-2 per ten exon protein per ten billion years of branch length. (Here urochordates are an exception: Ciona introns turn over at a markedly higher rate; Drosophila is another species exhibiting intron churning.) | |||
None of the introns of IDS correspond to those of ARSK: | |||
>ARSK_homSap Homo sapiens (human) 544 aa 8 exons | |||
0 MLLLWVSVVAALALAVLAPGAGEQR<font color = red>RRAA</font>KAPNVVLVVSDSF 0 | |||
0 DGRLTFHPGSQVVKLPFINFMKTRGTSFLNA<font color = blue>YTNS</font>PICCPSRA 1 | |||
2 AMWSGLFTHLTESWNNFKGLDPNYTTWMDVMERHGYRTQKFGK<font color = red>LDYT</font>SGHHSIS 2 | |||
1 NRVEAWTRDVAFL<font color = red>LRQE</font>GRPMVNLIRNRTKVRVMERDWQNTDKAVNWLRKEAINYTEPFVIYLGLNLPHPYPS<font color = blue>PSSG</font>ENFGSSTFHTSLYWLEK 0 | |||
0 VSHDAIKIPKWSPLSEMHPVDYYSSYTKNCTGRF<font color = blue>TKKE</font>IKNIRAFYYAMCAETDAML 1 | |||
2 GEIILALHQLDLLQKTIVIY<font color = red>SSDH</font>GELAMEHRQFYKMSMYEASAHVPLLMMGPGIKAGLQVSNVVSLVDIYPTML 1 | |||
2 DIAGIPLPQ<font color = red>NLSG</font>YSLLPLSSETFKNEHKVKNLHPPWILSEFHGCNVNASTYMLRTNHWKYIAYSDGASILPQLF 1 | |||
2 DLSSDPDELTNVAVKFPEITYSLDQKLHSIINYPKVSASVHQYNKEQFIKWKQSIGQNYSNVIANLRWHQDWQKEPRKYENAIDQWLKTHMNPRAV* 0 | |||
>IDS_hsa 559 aa 9 exons | |||
0 MPPPRTGRGLLWLGLVLSSVCVALGSETQA<font color = red>NSTT 1</font> [can only be mapped approximately as amino terminal region is gappy] | |||
2 DALNVLLIIVDDLRPSLGCYGDKLVRSPNIDQLASHSLLFQN<font color = blue>AFAQ 0</font> | |||
0 QAVCAPSRVSFLTGRRPDTTRLYDFNSYWRVHAGNFSTIPQYFKENGYVTMSVGK<font color = red>VFHP 1</font> | |||
2 GISSNHTDDSPYSWSFPPYHPSSEKY<font color = blue>ENTK 0</font> | |||
0 TCRGPDGELHANLLCPVDVLDVPEGTLPDKQSTEQAIQLLEKMKTSASPFFLAVGYHKPHIPF<font color = blue>RYPK 0</font> | |||
0 EFQKLYPLENITLAPDPEVPDGLPPVAYNPWMDIRQREDVQALNISVPYGPIP<font color = blue>VDFQ 0</font> | |||
0 RKIRQSYFASVSYLDTQVGRLLSALDDLQLANSTIIAF<font color = red>TSDH 1</font> | |||
2 GWALGEHGEWAKYSNFDVATHVPLIFYVPGRTASLPEAGEKLFPYLDPFDSASQ<font color = red>LMEP 1</font> | |||
2 GRQSMDLVELVSLFPTLAGLAGLQVPPRCPVPSFHVELCREGKNLLKHFRFRDLEEDPYLPGNPRELIAYSQYPRPSDIPQ | |||
WNSDKPSLKDIKIMGYSIRTIDYRYTVWVGFNPDEFLANFSDIHAGELYFVDSDPLQDHNMYNDSQGGDLFQLLMP* 0 [cannot be mapped as carboxy terminus does not end with an intron] | |||
The image below shows a more ambitious comparison of introns in all 17 human sulfatases. | |||
[[Image:17sulfatasesComp.png]] | |||
<br clear=all> | |||
=== Comparison of indels in ARSK and IDS === | |||
An argument similar to introns applies to insertions and deletions (indels) which are also generally non-recurrent (homoplasy-free). Here if ARSK and IDS shared an ancestry to the exclusion of all other sulfatases, indels may have been fixed on their stem and so descended uniquely to both contemporary proteins. Here it can be seen from an alignment of just ARSK and IDS that each has certain deletions consistently in all species relative to the other protein. Here exact placement of gaps doesn't matter when framed by nearby conserved anchor residues on both sides, eg a gap of three amino acids can never be reconciled with a gap of two. | |||
The alignment below shows at least 8 indels that are strictly associated with either of ARSK or IDS. There could be a few more, depending if residual errors from certain sequences, especially Monosiga, can be removed. These errors arise both from original sequencing and assembly errors, the absence of validating transcripts, and the difficulty of locating splice junctions in weakly homologous genes. | |||
The 8 indel events cannot be assigned to a gene (ie resolved as insertion or deletion) without an outgroup. The best choice for that is the next closest blastp match, either GNS or one of the long sulfatases. However these have indels of their own. Previous alignments of all 17 human sulfatases have shown a puzzling number of indels for an enzyme. Some of these can be accounted for by acquisition of membrane-binding or other attributes, many lie in loops rather than secondary structure, but most remain unexplained. | |||
It should be recalled here that even the smallest mammalian sulfatase is "too big". That is, the sulfatase domain is perhaps the largest in all of PFAM despite the apparent simplicity of its hydrolysis reaction, lack of known binding partners, and minimal need for regulation of catabolism within lysozymes. If sulfatases are fusion proteins with multiple domains, that is not evident from the known xray structures. | |||
If some regions of the protein were functionally gratuitous, they would presumably have been eliminated by deletions eons ago. But even if the frequency of indel fixation is normalized for chain length and unselected loops, the rate still seems high. In part this may be attributed to very early duplication and divergence of sulfatases billions of years ago, in contrast to other vertebrate gene families which expanded far more recently. | |||
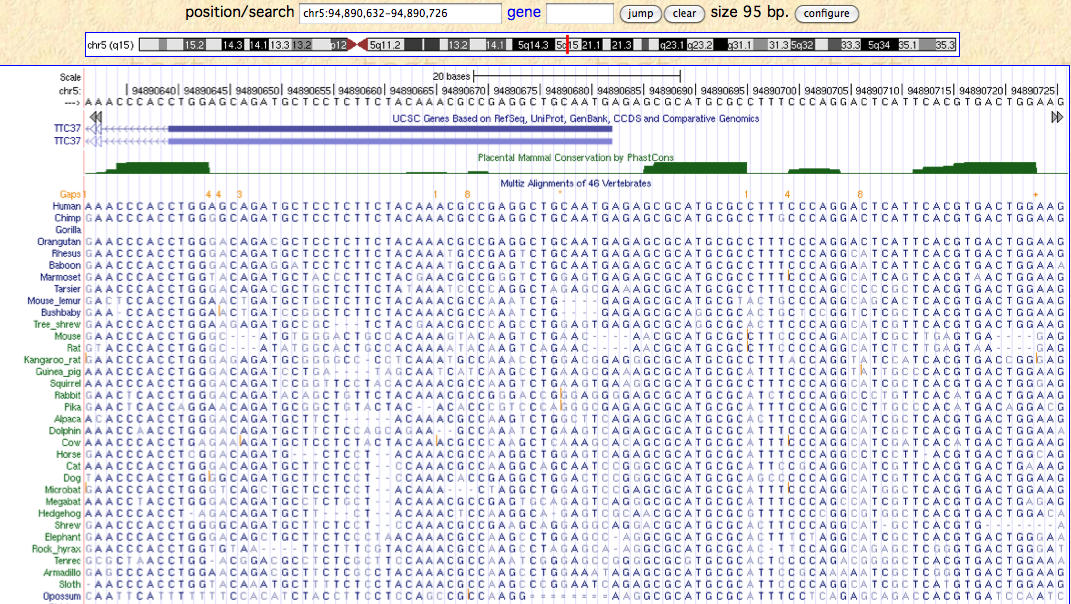
=== Overlap of ARSK transcription start with TTC37 === | === Overlap of ARSK transcription start with TTC37 === | ||
| Line 58: | Line 139: | ||
These motifs are fairly short, so their non-accidental occurence requires verification via conservation with comparative genomics as they cannot plausibly have arisen in human. Indeed these motifs are deeply conserved in vertebrates as is quickly seen from the [http://genome-test.cse.ucsc.edu/ 46-way alignment] of vertebrate genomes at UCSC. The [http://www.ncbi.nlm.nih.gov/pubmed/16024819 phastCons track] already identifies their conservation as a statistically significant occurence on a genomewide scale. | These motifs are fairly short, so their non-accidental occurence requires verification via conservation with comparative genomics as they cannot plausibly have arisen in human. Indeed these motifs are deeply conserved in vertebrates as is quickly seen from the [http://genome-test.cse.ucsc.edu/ 46-way alignment] of vertebrate genomes at UCSC. The [http://www.ncbi.nlm.nih.gov/pubmed/16024819 phastCons track] already identifies their conservation as a statistically significant occurence on a genomewide scale. | ||
Outside vertebrates, the nearest species with ARSK is Branchiostoma. The gene there is not longer syntenic with respect to TTC37. (This is also the case for Saccoglossus.) None of the motifs occurs exactly; one fragmentary match to the motif closest to the start codon is not statistically significant. The TFEB itself seems not to have a strict ortholog here either. Overall the prospects are not bioinformatically favorable for tracing the origin of these regulatory motifs. | |||
Note the motif CCCACCTGGA is not quite right as the last nucleotide does not belong in the motif and similarly for the other two -- it is unlikely that TFEB has changed its binding specificity only in humans or great apes. These motifs are better represented by profiles than by absolute sequence requirements. | Note the motif CCCACCTGGA is not quite right as the last nucleotide does not belong in the motif and similarly for the other two -- it is unlikely that TFEB has changed its binding specificity only in humans or great apes. These motifs are better represented by profiles than by absolute sequence requirements. | ||
| Line 75: | Line 158: | ||
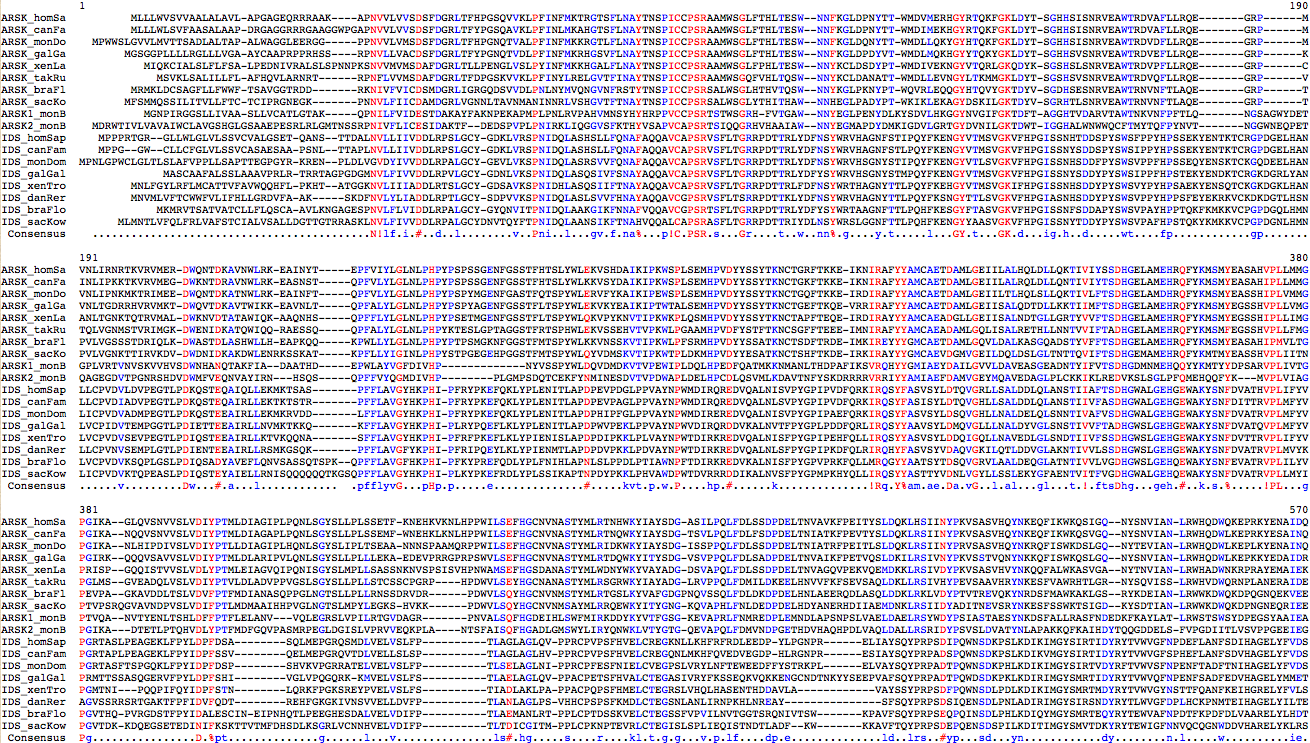
[[Image:ARSKalign.png|center]] | [[Image:ARSKalign.png|center]] | ||
=== ARSK reference sequences === | === ARSK, IDS, and choline sulfatase reference sequences === | ||
Only a sampler of vertebrate ARSK sequences are shown. The gene is present and well conserved in all 46 vertebrate genomes sequenced to date (reference sequences are pre-compiled at the proteinFasta link of the UCSC description page for ARSK). An ARSK ortholog is absent in all 30 sequenced non-deuterostome bilaterans as well as both cnidarian genomes, | Only a sampler of vertebrate ARSK sequences are shown. The gene is present and well conserved in all 46 vertebrate genomes sequenced to date (reference sequences are pre-compiled at the proteinFasta link of the UCSC description page for ARSK). An ARSK ortholog is absent in all 30 sequenced non-deuterostome bilaterans as well as both cnidarian genomes, trichoplax and sponge genomes, and numerous unicellular eukaryote genomes yet unmistakably present in two copies in choanoflagellate. Not all these genome assemblies have complete coverage but it is unlikely that all of a large 8 exon gene with conventional autosomal location (in vertebrates) would lack coverage so consistently. It will be possible to check a ctenophore genome shortly. | ||
>ARSK_homSap Homo sapiens (human) 544 aa 8 exons | >ARSK_homSap Homo sapiens (human) 544 aa 8 exons | ||
| Line 106: | Line 189: | ||
2 DLSSDPDELTNIATRFPEITLSLDQKLRSIINYPRVSASVHQYNKRQFISWKDSLGQNYTEVIANLRWHQDWLKEPLKYENAINQWLKTNTNM* 0 | 2 DLSSDPDELTNIATRFPEITLSLDQKLRSIINYPRVSASVHQYNKRQFISWKDSLGQNYTEVIANLRWHQDWLKEPLKYENAINQWLKTNTNM* 0 | ||
>ARSK_galGal (chicken) NM_001031415 | >ARSK_galGal Gallus gallus (chicken) NM_001031415 | ||
0 MGSGGPLLLLRGLLLVGAAYCAAPRPPRHSSRPNVLLVACDSF 0 | 0 MGSGGPLLLLRGLLLVGAAYCAAPRPPRHSSRPNVLLVACDSF 0 | ||
0 DGRLTFYPGNQTVDLPFINFMKRHGSVFLNAYTNSPI<font color =red>CCPSR</font>A 1 | 0 DGRLTFYPGNQTVDLPFINFMKRHGSVFLNAYTNSPI<font color =red>CCPSR</font>A 1 | ||
| Line 183: | Line 266: | ||
2 APAKKQFK 0 | 2 APAKKQFK 0 | ||
0 AIHDYTQQGDDELSFVPGDIITLVSVPPGEEIEGWLTGELNGRTGLFPDNFVEELPYVTCLAIFFSIPMFLFGCRHTSRPRTP* 0 | 0 AIHDYTQQGDDELSFVPGDIITLVSVPPGEEIEGWLTGELNGRTGLFPDNFVEELPYVTCLAIFFSIPMFLFGCRHTSRPRTP* 0 | ||
>IDS_homSap | |||
MPPPRTGRGLLWLGLVLSSVCVALGSETQANSTTDALNVLLIIVDDLRPSLGCYGDKLVRSPNIDQLASHSLLFQNAFAQQAV<font color =red>CAPSR</font>VSFLTGRRPDTT | |||
RLYDFNSYWRVHAGNFSTIPQYFKENGYVTMSVGKVFHPGISSNHTDDSPYSWSFPPYHPSSEKYENTKTCRGPDGELHANLLCPVDVLDVPEGTLPDKQ | |||
STEQAIQLLEKMKTSASPFFLAVGYHKPHIPFRYPKEFQKLYPLENITLAPDPEVPDGLPPVAYNPWMDIRQREDVQALNISVPYGPIPVDFQRKIRQSY | |||
FASVSYLDTQVGRLLSALDDLQLANSTIIAFTSDHGWALGEHGEWAKYSNFDVATHVPLIFYVPGRTASLPEAGEKLFPYLDPFDSASQLMEPGRQSMDL | |||
VELVSLFPTLAGLAGLQVPPRCPVPSFHVELCREGKNLLKHFRFRDLEEDPYLPGNPRELIAYSQYPRPSDIPQWNSDKPSLKDIKIMGYSIRTIDYRYT | |||
VWVGFNPDEFLANFSDIHAGELYFVDSDPLQDHNMYNDSQGGDLFQLLMP | |||
>IDS_canFam | |||
MPPGGWCLLCFGLVLSSVCASAESAAPSNLTTAPLNVLLIIVDDLRPSLGCYGDKLVRSPNIDQLASHSLLFQNAFAQQAV<font color =red>CAPSR</font>VSFLTGRRPDTTRL | |||
YDFNSYWRVHAGNFSTLPQYFKENGYVTMSVGKVFHPGISSNYSDDSPYSWSIPPYHPSSEKYENTKTCRGPDGELHANLLCPVDIADVPEGTLPDKQST | |||
EQAIRLLEKTKTSTRPFFLAVGYHKPHIPFRYPKEFQKLYPLENITLAPDPEVPAGLPPVAYNPWMDIRQREDVQALNLSVPYGPIPVDFQRKIRQSYFA | |||
SISYLDTQVGHLLSALDDLQLANSTIIVFASDHGWALGEHGEWAKYSNFDITTRVPLMFYVPGRTAPLPEAGEKLFPYIDPFSSVQELMEPGRQVTDLVE | |||
LLSLSPTLAGLAGLHVPPRCPVPSFHVELCREGQNLMKHFQVEDVEGDPHLRGNPRESIAYSQYPRPADSPQWNSDKPSLKDIKVMGYSIRTIDYRYTVW | |||
VGFSPHEFLANFSDVHAGELYFVDSDPLQDHNMYNDSQGKDLLRALMPF | |||
>IDS_monDom Monodelphis domestica XM_001376164 | |||
MPNLGPWCLGLTLSLAFVPPLLSAPTTEGPGYRKRENPLDLVGVDYIVVDDLRPALGCYGEVLVKSPNIDQLASRSVVFQNAFAQQAV<font color =red>CAPSR</font>VSFLTGR | |||
RPDTTRLYDFNSYWRVHSGNYSTIPQYFKENGYVTLSVGKVFHPGISSNHSDDFPYSWSVPPFHPSSEQYENSKTCKGQDEELHANLICPVDVADMPEGT | |||
LPDKQSTEEAIRLLEKMKRVDDLFFLAVGYHKPHIPFRYPKEFQKLYPLENITLAPDPHIPFGLPPVAYNPWMDIREREDVQALNISVPYGPIPAEFQRK | |||
IRQSYFASVSYLDSQVGHLLNALDELQLSNNTIVAFVSDHGWALGEHGEWAKYSNFDVATRVPLMFYVPGRTASFTSPGQKLFPYIDPFDSPSHVKVPGR | |||
RATELVELVSLFPTLSELAGLNIPPRCPFESFNIELCVEGPSLVRYLNFTEWEEDFFYSTRKPLELVAYSQYPRPADTPQWNSDKPHLKDIKIMGYSIRT | |||
VDYRFTVWVSFNPENFTADFTNIHAGELYFVDSDPLQDHNVYNQTVGIY | |||
>IDS_galGal | |||
MASCAAFALSSLAAAVPRLRTRRTAGPGDGMNVLFIVVDDLRPVLGCYGDNLVKSPNIDQLASQSIVFSNAYAQQAV<font color =red>CAPSR</font>VSFLTGRRPDTTRLYDFY | |||
SYWRVHSGNYSTMPQYFKENGYVTMSVGKVFHPGISSNYSDDYPYSWSIPPFHPSTEKYENDKTCRGKDGRLYANLVCPIDVTEMPGGTLPDIETTEEAI | |||
RLLNVMKTKKQKFFLAVGYHKPHIPLRYPQEFLKLYPLENITLAPDPWVPEKLPPVAYNPWVDIRQRDDVKALNVTFPYGPLPDDFQRLIRQSYYAAVSY | |||
LDMQVGLLLNALDYVGLSNSTIVVFTADHGWSLGEHGEWAKYSNFDVATQVPLMFYVPRMTTSSASQGERVFPYLDPFSHIVGLVPQGQRKKMVELVSLF | |||
STLAELAGLQVPPACPETSFHVALCTEGASIVRYFKSSEQKVQKKENGCNDTNKYYSEEPVAFSQYPRPADTPQWDSDKPKLKDIRIMGYSMRTIDYRYT | |||
VWVQFNPENFSADFEDVHAGELYMMETDPNQDNNIYNNTLHGHLFKKIVDFLKH | |||
>IDS_xenTro | |||
MNLFGYLRFLMCATTVFAVWQQHFLPKHTATGGKNVLIIIADDLRTSLGCYGDSAVKSPNIDHLASQSIIFTNAYAQQAV<font color =red>CAPSR</font>VSFLTGRRPDTTRLF | |||
DFNSYWRTHAGNYTTLPQYFKEHGYVTMSVGKIFHPGISSNHSDDYPYSWSVYPYHPSAEKYENSQTCKGKDGKLHANLVCPVDVSEVPEGTLPDIQSTE | |||
EAIRLLKTVKQQNASFFLAVGYHKPHIPFRFPKEFLKLYPIENISLAPDPDIPKKLPLVAYNPWTDIRKREDVQALNISFPYGPIPEHFQLLIRQSYYAS | |||
VSYLDDQIGQLLNAVEDLGLSNDTIIVFSSDHGWSLGEHGEWAKYSNFDVTTRVPLIFYVPGMTNIPQQPIFQYIDPFSTNLQRKFPGKSREYPVELVSL | |||
FSTIADLAKLPAPPACPQPSFHMELCTEGRSLVHQLHASENTHDDAVLAVAYSSYPRPSDFPQWNSDLPDLKDIKIMGYSMRTMDYRYTVWVGYNSTTFQ | |||
ANFKEIHGRELYFVLSDPNQDNNLYNQLLHLDIYKHFEFMNN | |||
>IDS_danRer6 551 chr14:22165989-22187666- | |||
MNVMLVFTCWWFVLIFHLLGRDVFAAKSKDFNVLYLIADDLRPTLGCYSDPVVKSPNIDQLASLSVVFHNAYAQQAV<font color =red>CGPSR</font>VSFLTSRRPDTTKLYDFN | |||
SYWRVHAGNYTTLPQYFKSNGYTTLSVGKVFHPGIASNHSDDYPYSWSVPPYHPPSFEYEKRKVCKDKDGTLHSNLLCPVNVSEMPLGTLPDIENTEEAI | |||
RLLRSMKGSQKPFFLAVGFYKPHIPFRIPQEYLKLYPIENMTLAPDPDVPKKLPDVAYNPWTDIRKREDVQALNLSFPYGPIPKDFQLRIRQHYFASVSY | |||
VDAQVGKILQTLDDVGLAKNTIVVLSSDHGWSLGEHGEWAKYSNFDVATRVPLMVYKAGVSSRRSRTGAKTFPFIDVFQDTREHFGKGKIVNSVVELLDV | |||
FPTLANLAGLPSVHHCPSPSFKMDLCTEGSNLANLIRNPKHLNREAYSFSQYPRPSDSIQENSDLPNLADIRIMGYSIRSNDYRYTLWVGFDPLHCKPNM | |||
TEIHAGELYILTEDPGQDNNLFDEFGHAALLNKFGTMPSWTESLKQHMMYFSSGLKSKGLS | |||
>IDS_braFlo Branchiostoma floridae (XM_002611665: flawed) BW796857 | |||
MKMRVTSATVATCLLFLQSCAAVLKNGAGESPNVLFLVIDDLRPALGCYGYQNVITPNIDQLAAKGIKFNNAFVQQAV<font color =red>CGPSR</font>TSFLTGRRPDTTRLYDF | |||
YSYWRTAAGNFTTLPQHFKESGYFTASVGKVFHPGGISSNFSDDAPYSWSVPAYHPPTQKFKMKKVCPGPDGQLHMNLVCPVDVKSQPLGSLPDIQSADY | |||
AVEFLQNVSASSQTSPKQPFFLAVGFHKPHIPFKYPREFQDLYPLFNIHLAPNLSLPPDLPTIAWNPFTDIRKREDVKALNISFPYGPVPRKFQLLMRQG | |||
YYAATSYTDSQVGRVLAALDEQGLATNTIVVLVGDHGWSLGEHQEWAKYSNFEVATRVPLILYVPGVTHQPVRGDSTFPYIDALESCINEIPNHQTLPEE | |||
GHESDALVELVDIFPTLAEMANLRTPPLCPTDSSKVELCTEGSSFVPVILNVTGGTSRQNIVTSWKPAVFSQYPRPSEQPQINSDLPHLKDIQYMGYSMR | |||
TEQYRYTEWVAFNPDTFKPDFDLVAARELYLHDTDELEDHNVAGKSEYRHLLTQLSQQLRKGWRNALPSQ | |||
>IDS_sacKow Saccoglossus kowalevskii XM_002733076 | |||
MLMNTLVFQLFRLVAFSTCIALVSALLDGTTGTRRASKLNVLFIVVDDLRPALGCYDNVTQYFTPNIDQLAANSIKFTNAHVQQAL<font color =red>CAPSR</font>ASFLTGRRP | |||
DTTRIYDLNSYWRSLGGNFTTLPQHFKENGYYAASVGKVFHPGISSNYTDDYPYSWSVPAFHPSTQKYKMKKVCPGPDGNLHMNLICPVDVKTQPEASLP | |||
DIQSTEYAIELLRNISQQQQQQTKGSQPFFLAVGYHKPHIPLKYPKEFRDLYPLSSIKAPTNPDYPKKLPHVAWDPWTDVRRRDDIKALNVSFPYGPMPK | |||
HYQLLIRQSYYASTTYVDNLVGYLLSSLEKYGFAENTVITFVGDHGWALGEHQEWAKYSNFDVATRVPLLMYIPGVTDKKDQEGSETEDINIFKSKTTVT | |||
MFDHSDLKSGRLVCNNHVELVDIFPTLTDICGITMPPLCPKNPTEVRLCTEGISLSPLIEQISTNDTLADFKWKKAVFTQYPRPSDEPQENSDSPILKDI | |||
TIMGYSMVTDKYRYTEWIGFNNVQCQGNWDDVHARELYKLRSDKMENNNVANDAQYKELTQKLANLLRKGWRHALP | |||
>IDS_monBre Monosiga brevicollis XM_001743372 | |||
mAFQSRGPNLLPDRAGEIIGVALSSLPLDGLNVLLIVVDDMRAELGTYGATHMITPHLDALAQDGMVFERAYVAISL<font color =red>CMPSR</font>TAFLTSRRPATTHNFVIA | |||
PNEQWRQTKGPNATTLPEFFKTVGGYRTYGMGKIFHGTTDEPYSWSAEMGDYYDWDNWTQYGNSMTYKCFDVPDNNLGDGIFADRAVNWINMFGADQANG | |||
SDTRPFFMGVGFHRPHIPYLVPKRYCDMYPPADEIPLAANPFKPEGMPDVAYSVSAGLRNFQDCAPLFENVSKCYDDPSWAFSNRVRRNYWAAISYIDAQ | |||
VGRIVQALKDNNLYDNTIVLFMGDHGVCTCTGRSTNFEHGTRIPLIIRDPSHTPARTAALVETVDIYPTLVDLAGLPSLETCAPGSMAALCTEGFSMRPL | |||
FTDPTRAWKSAAFSQYARPAPSPDNGFPADLFSPPLHVAGHREGVMGFTIRTNTYRYTNWVWFDPASATPHWNMSWGEELYNHTAQPVPDGLFNNENINL | |||
IDQPGLEPIIDKLRQALQAGWRAALPS | |||
>3ED4_escCol Escherichia coli | |||
MSLASLIGLAVCTGNAFSPALAAEAKQPNLVIIMADDLGYGDLATYGHQIVKTPNIDRLAQEGVKFTDYYAPAPL<font color =red>SSPSR</font>AGLLTGRMPFRTGIRSWIPS | |||
GKDVALGRNELTIANLLKAQGYDTAMMGKLHLNAGGDRTDQPQAQDMGFDYSLANTAGFVTDATLDNAKERPRYGMVYPTGWLRNGQPTPRADKMSGEYV | |||
SSEVVNWLDNKKDSKPFFLYVAFTEVHSPLASPKKYLDMYSQYMSAYQKQHPDLFYGDWADKPWRGVGEYYANISYLDAQVGKVLDKIKAMGEEDNTIVI | |||
FTSDNGPVTREARKVYELNLAGETDGLRGRKDNLWEGGIRVPAIIKYGKHLPQGMVSDTPVYGLDWMPTLAKMMNFKLPTDRTFDGESLVPVLEQKALKR | |||
EKPLIFGIDMPFQDDPTDEWAIRDGDWKMIIDRNNKPKYLYNLKSDRYETLNLIGKKPDIEKQMYGKFLKYKTDIDNDSLMKARGDKPEAVTWGEGHHHH | |||
>3B5Q_bacThe Bacteroides thetaiotaomicron 2.40A resolution | |||
GLALCGAAAQAQEKPNFLIIQCDHLTQRVVGAYGQTQGCTLPIDEVASRGVIFSNAYVGPL<font color =red>SQPSR</font>AALWSGHQTNVRSNSSEPVNTRLPENVPTL | |||
GSLFSESGYEAVHFGKTHDXGSLRGFKHKEPVAKPFTDPEFPVNNDSFLDVGTCEDAVAYLSNPPKEPFICIADFQNPHNICGFIGENAGVHTDRPISGP | |||
LPELPDNFDVEDWSNIPTPVQYICCSHRRXTQAAHWNEENYRHYIAAFQHYTKXVSKQVDSVLKALYSTPAGRNTIVVIXADHGDGXASHRXVTKHISFY | |||
DEXTNVPFIFAGPGIKQQKKPVDHLLTQPTLDLLPTLCDLAGIAVPAEKAGISLAPTLRGEKQKKSHPYVVSEWHSEYEYVTTPGRXVRGPRYKYTHYLE | |||
GNGEELYDXKKDPGERKNLAKDPKYSKILAEHRALLDDYITRSKDDYRSLKVDADPRCRNHTPGYPSHEGPGAREILKRK | |||
>CHOS_edwTar Edwardsiella tarda Choline-sulfatase DM42793 | |||
MSLSRREFLQRTAGGMAGVALGAPALAAGDAPAGTDTGAKMPPRNIVIITADQLARRGVGGYGNPQVNTPAIDSLIARGTRFEQAYCPYPL<font color =red>CAPSR</font>ACYW | |||
TGRLPHQTGVIANDSPNVPQDMVTLGELFSQAGYECRHFGKRHDYGALKGFTCADQVELPYDSPAAYPVDYDTREDVYCLQESLKYIDTLKGRDSDAPFM | |||
LAIEFNNPHNINGWTGAFAGPHGDIDGLGPLPPLLDNFDTSADLPNRPLAIQYACCTHNRVMQAANWNELNFRQYLKAYYHFTELADGFIGQVLSALRAS | |||
GHADDTLVVFFADHGDAMGAHRLVAKMNWFYEESTNVPLVFAGPGIRPQASSRHLTSLCDLLPTLCDYAGLTPPPGLYGRSLMPILRGEQPDGWRDEVIT | |||
QWNTDRNVDVQPARMLRTERYKYILYKENEEEELYDLQQDPGETRNLAHSPAHQAERQALRARFDEYVRNQVDPFYSQEAIIDRRWRSHLPGYHNHQGQT | |||
SIQVYQKEIRPLIMNKEFEKAREVRLALYRQARASYNGGV | |||
>CHOS_ruePom Ruegeria pomeroyi choline sulfatase YP_166053 | |||
MTNHPNLLVIVSDEHRKDAMGCAGHPIVKTPNLDALAARGTMFEAAYTPSPM<font color =red>CVPTR</font>AALATGDWIHRTGHWDSATPYAGQPRSWMHDLRDAGREVVSIG | |||
KLHFRATEDDNGFSQEILPMHVVGGIGWTVGLLRKNPPAYEAAAELAADVGVGASSYTDYDRAITAAAEAWLADPARQERPWAAFVSLVSPHYPLTCPEE | |||
WFALYDPDQMDLPVGYGQGLPDHAELRNIGGFFNYDAYFDAQKMREAKAAYYGLTSFMDDCVGRVLAALEAGGKADNTVVLYVSDHGDMMGDQGFWTKQV | |||
MYEASAGVPMIAAGPGIPAGHRVSTCTSLTDIAATARELCGLAAREDLPGLSLRSIATAPDDPDRAGFSEYHDGGSRTGTFMLRWGRWKYVHYVGEAPQL | |||
FDLERDPQELTDLAPRAAEDPDMRALLAEGEHRLRAICNPETVNARAFADQQRRIAELGGEEACRTGYSFNHTPVPQEGGAL | |||
[[Category:Comparative Genomics]] | |||
Latest revision as of 01:03, 4 December 2010
Introduction to sulfatases
Sulfatases are an old and deeply diverged family of hydratases that remove sulfate moieties from a variety of small and large molecules. Despite the apparent simplicity of this reaction, the sulfatase domain fold is perhaps the largest known for any enzyme and an unprecedented formyl glycine post-translational modification of encoded cysteine, serine or threonine is critical to activity. The fold is closely related to that of alkaline phosphatases, though primary sequence alignability has almost completely dissipated.
The 17 human paralogs reside either in lysozomes or endoplasmic reticulum. Mutations in these genes result in diseases that provide important clues as to natural substrates (which accumulate in lysosomal storage diseases). However only 8 of the 17 genes have an associated disease at OMIM as of Sept 2010. Functions of the remaining sulfatases have yet to be discovered, perhaps because the accumulating metabolite is not toxic or has an alternative catabolic pathway. Such diseases could be recessive and hence rare in the case of unassigned autosomal sulfatases.
ARSK is such a gene. First described in 2003 as SULFX, the substrate and function of ARSK remain unknown -- it has not yet been the focus of a single experimental paper. ARSK is a fairly typical sulfatase of 536 amino acids encoded by eight exons on human chr 5 with a conventional CPSRA formylglycine motif. It lacks overt membrane insertional regions and GPI terminal motif so is presumably soluble.
ARSK is however peculiar in several bioinformatic respects. Although clearly a full length duplicate of an ancestral sulfatase, its opaque evolutionary relationship to non-orthologous sulfatases makes it difficult to place in the sulfate gene tree. Its closest affinity (percent identity low 20's) is perhaps with IDS which removes the sulfate from iduronate, though the ARSK substrate may have drifted off to something else entirely during the 600+ million years since gene duplication.
A second unusual feature is that the 7 introns within the coding region of ARSK do not bear any relationship in position or phase to those of other human sulfatases. This suggests that the gene duplication event leading to ARSK and other sulfatases preceded the main era of gene intronation, ie sulfatses initially had no introns (as in bacterial genes) and were subsequently independently intronated in early eukaryotes. Once established, the introns of ARSK have been stable over billions of years of gene tree branch length.
The phylogenetic distribution of ARSK also raises many questions. Within deuterostomes, orthologs are readily located in representatives of all major subclades with the exception of echinoderms and tunicates. ARSK has evolved quite conservatively here, with the human protein still having 54% and 52% identity over 500 residues to Branchiostoma (amphioxus) and Saccoglossus (acornworm) respectively, despite divergences that preceded the Cambrian. Intron positions and phases are precisely preserved beyond two minor fission events, leaving no doubt of orthology within deuterostomes. However, ARSK is otherwise completely missing from other eumetazoans (ecdysozoa, lophotrochozoa, and cnidaria). Also missing from Trichoplax and sponge genomes, it makes its last eukaryotic appearance (two diverged paralogs) in Monosiga, a marine choanoflagellate, before fading into fungal and bacterial sequences of uncertain affinities.
A final oddity of ARSK observed early on is its close proximity to an apparently unrelated gene, TTC37 (twenty tetratricopeptide repeats 37): only 144 bp separate the two genes. These are transcribed divergently and could well share a bidirection promoter or overlap in 5' UTR. This relationship is by no means restricted to the human genes -- it is readily traced back throughout vertebrates. The putative chaperone function of TTC37 remains unspecified, though in June 2010 a disease has been assigned to it: trichohepatoenteric syndrome (THES) -- an "autosomal-recessive disorder characterized by life-threatening diarrhea in infancy, immunodeficiency, liver disease, trichorrhexis nodosa, facial dysmorphism, hypopigmentation, and cardiac defects". This does not immediately suggest why ARSK and TTC37 should be so closely linked.
ARSK phylogenetic distribution
As noted above, ARSK has evolved with above-average conservation in deuterostomes and especially vertebrates. Its apparent loss in echinoderms and tunicates -- which in itself is not unusual -- might be overturned if more genomes were sequenced from these clades. Its occurence in all earlier diverging lineages is restricted to Monosiga, which requires several independent losses as the species tree topology now stands. Monosiga thus illustrates the importance of thorough genomic sampling.
However even without the Monosiga sequences, it is certain that ARSK did not arise in deuterostomes either by horizontal gene transfer (from bacteria), nor from gene duplication and rapid divergence from a sulfatase exising in the bilateran ancestor, nor de nov from say junk dna. These options are all ruled out by its 7 immensely conserved GT-AG coding introns that do not resemble anything from these other sources. (A very high percentage of exons are precisely conserved between human and sponge implying the main era of intron creation occured much earlier.)
The Monosiga sequences below do have some problematic aspects. In part these arise from poor quality GenBank pipeline models of XM_001747506 and XM_001750805. The former even skips over small exon containing the catalytic site motif CPSRT as well as omitting a long distal exon; the latter model has a half dozen macro errors. Missing exons are however locatable using Blastx of the enveloping contig ABFJ01000822 against a small database of validated ARSK orthologs. However ambiguity remaining in earlier exons with weak homology that can only be resolved by transcript sequencing.
Despite these uncertainties, it is clear that intron positions and phases do not match those of deuterostomes very well. In particular, the latter all have a phase 1 intron starting one residue after the CCPSR motif. This motif and the following WSG pattern are easily recognized in Monosiga but there is no possibility of a GT-AG intron in the anticipated position:
agcccctgtatgctgtcccagccgaacttcgacttggtcgggccgtcacgt (The red cg in deuterostomes is splice donor GT.) A P V C C P S R T S T W S G R H
Despite unresolved intron issues, back-Blastp of Monosiga to the 17 sulfatases of human at GenBank gives ARSK_homSap as best match by a wide margin. When the target is all 'non-redundant' deuterostome sequences at GenBank, the best match is acornworm ARSK_sacKow followed by a long list of ARSK in other species. After these, much weaker matches to IDS sulfatases appear.
Curiously, when Blastp of Monosiga (or any bona fide ARSK) is restricted to sulfatase-rich bacteria, top matches are typically annotated as IDS-like or choline-sulfatases. This is consistent with a deep ancestral ARSK/IDS gene whose substrate was (and still is, in bacteria ) choline sulfate. That can be infered from the phylogenetic breadth of bacterial choline sulfatases.
After gene duplication and divergence, one copy may have changed its substrate over time in eukaryotes to iduronate (ie become IDS). The other copy became ARSK; its substrate today is unknown but might well still include choline sulfate. Later, this hypothesis goes, that molecule stopped being made in various lineages (eg arthropods) or was metabolized more effectively some other way, resulting in the subsequent loss of ARSK in those lineages (under the evolutionary principle of 'use it or lose it').
Comparison of ARSK and IDS evolutionary rates
IDS has a much more conventional phylogenetic distribution, with clear orthologs in many clades where ARSK is missing (eg tunicate, echinoderm, arthropods and cnidaria). Since choanoflagellates encode a distinct IDS with mediocre alignment with their ARSK, these genes diverged far earlier. However the rate of divergence of IDS (measured as divergence from human) is indistinguishable from that of ARSK over the 800 million years spanning the divergence of choanoflagellates from human.
The rates at which amino acid substitutions accrue in these two proteins are somewhat above average in the context of whole proteome comparisons of these species. Note divergence of sulfatases bottoms out at about 20% since a minimal core of residues is needed to define the fold and provide activity. Although subclades evolve at various rates, the divergence curve here has remained monotonic as plotted relative to the species divergence tree, ie the earlier a lineage diverged, the more diverged its protein comparison will be. Operationally, this simply means that species appear in correct phylogenetic order on Blastp output.
Blastp: score exp id score exp id ARSK_homSap Homo sapiens (human) 2871 1.2e-303 100% 2956 2.2e-312 100% IDS_homSap ARSK_canFam Canis familiarus (dog) 2555 3.6e-270 89% 2599 1.5e-274 87% IDS_canFam ARSK_monDom Monodelphis domestica (opossum) 2291 3.4e-242 79% 2238 2.7e-236 79% IDS_monDom ARSK_galGal Gallus gallus (chicken) 2118 7.3e-224 74% 1972 4.1e-208 69% IDS_galGal ARSK_xenLae Xenopus laevis (frog) 1864 6.0e-197 67% 1900 1.7e-200 67% IDS_xenLae ARSK_takRub Takifugu rubripes (fugu) 1747 1.5e-184 60% 1724 7.8e-182 62% IDS_takRub ARSK_braFlo Branchiostoma floridae (amphioxus) 1477 6.1e-156 54% 1704 7.5e-151 59% IDS_braFlo ARSK_sacKow Saccoglossus kowalevskii (acornworm) 1433 2.8e-151 52% 1488 7.9e-157 53% IDS_sacKow ARSK_monBre Monosiga brevicollis (choanoflagellate) 788 6.3e-83 33% 448 2.8e-061 36% IDS_monBre
The rates at which sulfatase folds diverge (in terms of backbone root mean square best fit) do not appear correspondingly high, even though blastp against the PDB database of determined structures gives only poor matches (low 20's in percent id) for either IDS or ARSK. The only mammalian (or even metazoan) sulfatases with crystallographic structures are ARSA, STS, and ARSB but these are too weak to allow accurate modeling of ARSK or IDS.
Note the former sequences have low percent identity to each other -- low 30's -- yet have fairly similar folds. Somewhat better matches of ARSK occur with bacterial sulfatase, notably 3ED4 of E. coli and 3B5Q of Bacteroides thetaiotaomicron.
It might be possible to model ARSK or IDS using these structures (or some consensus derived from them) but the outcome would not be nearly sufficiently accurate to depict substrate binding sites (ie, deduce the natural substrate for ARSK). There are substantial issues with deletions and insertions, though these presumably lie in loops outside the core beta sheet/alpha helix secondary structure. Conserved homologous disulfides could conceivably provide severe modeling restraints but these do not appear to exist.
Conserved disulfides and glycosylation sites
Within vertebrates, ARSK has 3 cysteines conserved to some phylogenetic depth beyond the catalytic motif and 7 NxT/S potential glycosylation sites, one of which overlaps with a conserved cysteine. These number decrease if conservation throughout deuterostomes (or even to choanoflagellates) is demanded. Thus the proper way to summarize these sites is a table by phylogenetic depth; presumably those with greater antiquity have been conserved for more fundamental reasons.
Two glycosylation sites and one cysteine are even conserved at homologous positions IDS (adjusting gap placement manually). This does not imply these sites have been conserved since the common ancestral protein. They could simply be favorable sites on the protein exterior where the same feature has been independently advantageous and so arisen multiple times.
Just because cysteine pairs are conserved does not imply they form an disulfide; similarly NxT/S can also be conserved for other reasons than glycosylation. However assuming from the signal peptide a secreted location in an oxidizing environment, at least some of these will likely be realized. Conservation of unselected amino acids does not persist by accident over mammalian order time scales.
It's been shown previously that the 17 human sulfatases do not conserve either type of site to any extent. This is surprising in the case of disulfides which are often fundamental to structural stability (eg trillions of years of branch length conservation in rhodpsins) but evidently have arisen multiple times de novo during the course of sulfatase evolution and acquired roles resulting in their subsequent conservation.
Phylogenetic depth of potential glycosylation and disulfide sites
ARSK IDS
108 NYT vert 121 NFS
166 NRT prim 187 DVL
193 NYT terr 215 SAS
262 NCT deut 280 NIS
263 C vert 281 I
283 C vert 304 V
375 NLS terr 390 LME
410 C deut 422 C
413 NAS deut 427 PSF
498 NYS vert 499 YTV
Comparison of introns in ARSK and IDS
Given that the natural substrate of ARSK remains unknown, there is value in determining its nearest neighbor within the sulfatase gene family because some clues to the substrate might emerge. IDS emerges from alignment comparisons as the best candidate, but only with marginal support. Intron position and phase are often better conserved than amino acid identity. If introns between these two genes agree at least in part -- and other sulfatases fare far worse -- that would provide independent support.
It is not easy to reliably compare intron position and phase in highly diverged homologs, especially if gaps are abundant as here. However the procedures worked out for opsins are applicable here: multiple orthologs of each gene are aligned together to improve gap placement and establish flanking invariant residues to limit uncertainty. This allows exon ends of the second gene to be painted on the first gene, with colors blue, red, and magenta encoding codon overhang (phase) of 0, 1 or 2 base pairs. The outcome of that is shown below for ARSK and IDS.
Recall a gene of 500 amino acids has 1500 possible combinations of intron position and phase; accidental agreement (homoplasy) can occur but is rare and manageable. Introns have been phenomenally conserved throughout deuterostome history, with gain and loss events approaching 1-2 per ten exon protein per ten billion years of branch length. (Here urochordates are an exception: Ciona introns turn over at a markedly higher rate; Drosophila is another species exhibiting intron churning.)
None of the introns of IDS correspond to those of ARSK: >ARSK_homSap Homo sapiens (human) 544 aa 8 exons 0 MLLLWVSVVAALALAVLAPGAGEQRRRAAKAPNVVLVVSDSF 0 0 DGRLTFHPGSQVVKLPFINFMKTRGTSFLNAYTNSPICCPSRA 1 2 AMWSGLFTHLTESWNNFKGLDPNYTTWMDVMERHGYRTQKFGKLDYTSGHHSIS 2 1 NRVEAWTRDVAFLLRQEGRPMVNLIRNRTKVRVMERDWQNTDKAVNWLRKEAINYTEPFVIYLGLNLPHPYPSPSSGENFGSSTFHTSLYWLEK 0 0 VSHDAIKIPKWSPLSEMHPVDYYSSYTKNCTGRFTKKEIKNIRAFYYAMCAETDAML 1 2 GEIILALHQLDLLQKTIVIYSSDHGELAMEHRQFYKMSMYEASAHVPLLMMGPGIKAGLQVSNVVSLVDIYPTML 1 2 DIAGIPLPQNLSGYSLLPLSSETFKNEHKVKNLHPPWILSEFHGCNVNASTYMLRTNHWKYIAYSDGASILPQLF 1 2 DLSSDPDELTNVAVKFPEITYSLDQKLHSIINYPKVSASVHQYNKEQFIKWKQSIGQNYSNVIANLRWHQDWQKEPRKYENAIDQWLKTHMNPRAV* 0 >IDS_hsa 559 aa 9 exons 0 MPPPRTGRGLLWLGLVLSSVCVALGSETQANSTT 1 [can only be mapped approximately as amino terminal region is gappy] 2 DALNVLLIIVDDLRPSLGCYGDKLVRSPNIDQLASHSLLFQNAFAQ 0 0 QAVCAPSRVSFLTGRRPDTTRLYDFNSYWRVHAGNFSTIPQYFKENGYVTMSVGKVFHP 1 2 GISSNHTDDSPYSWSFPPYHPSSEKYENTK 0 0 TCRGPDGELHANLLCPVDVLDVPEGTLPDKQSTEQAIQLLEKMKTSASPFFLAVGYHKPHIPFRYPK 0 0 EFQKLYPLENITLAPDPEVPDGLPPVAYNPWMDIRQREDVQALNISVPYGPIPVDFQ 0 0 RKIRQSYFASVSYLDTQVGRLLSALDDLQLANSTIIAFTSDH 1 2 GWALGEHGEWAKYSNFDVATHVPLIFYVPGRTASLPEAGEKLFPYLDPFDSASQLMEP 1 2 GRQSMDLVELVSLFPTLAGLAGLQVPPRCPVPSFHVELCREGKNLLKHFRFRDLEEDPYLPGNPRELIAYSQYPRPSDIPQ WNSDKPSLKDIKIMGYSIRTIDYRYTVWVGFNPDEFLANFSDIHAGELYFVDSDPLQDHNMYNDSQGGDLFQLLMP* 0 [cannot be mapped as carboxy terminus does not end with an intron]
The image below shows a more ambitious comparison of introns in all 17 human sulfatases.
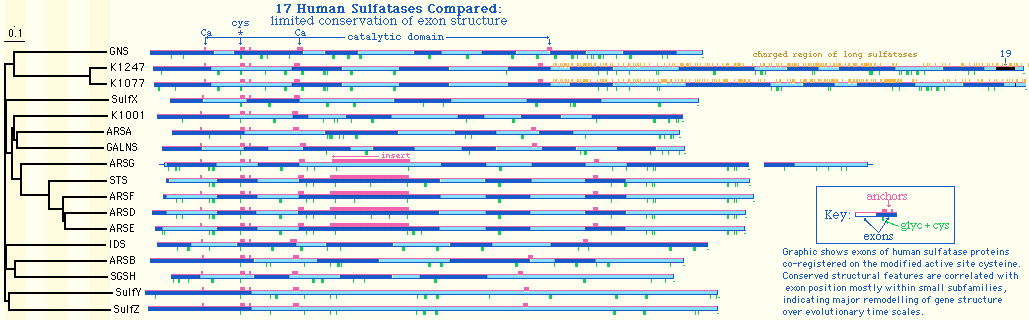
Comparison of indels in ARSK and IDS
An argument similar to introns applies to insertions and deletions (indels) which are also generally non-recurrent (homoplasy-free). Here if ARSK and IDS shared an ancestry to the exclusion of all other sulfatases, indels may have been fixed on their stem and so descended uniquely to both contemporary proteins. Here it can be seen from an alignment of just ARSK and IDS that each has certain deletions consistently in all species relative to the other protein. Here exact placement of gaps doesn't matter when framed by nearby conserved anchor residues on both sides, eg a gap of three amino acids can never be reconciled with a gap of two.
The alignment below shows at least 8 indels that are strictly associated with either of ARSK or IDS. There could be a few more, depending if residual errors from certain sequences, especially Monosiga, can be removed. These errors arise both from original sequencing and assembly errors, the absence of validating transcripts, and the difficulty of locating splice junctions in weakly homologous genes.
The 8 indel events cannot be assigned to a gene (ie resolved as insertion or deletion) without an outgroup. The best choice for that is the next closest blastp match, either GNS or one of the long sulfatases. However these have indels of their own. Previous alignments of all 17 human sulfatases have shown a puzzling number of indels for an enzyme. Some of these can be accounted for by acquisition of membrane-binding or other attributes, many lie in loops rather than secondary structure, but most remain unexplained.
It should be recalled here that even the smallest mammalian sulfatase is "too big". That is, the sulfatase domain is perhaps the largest in all of PFAM despite the apparent simplicity of its hydrolysis reaction, lack of known binding partners, and minimal need for regulation of catabolism within lysozymes. If sulfatases are fusion proteins with multiple domains, that is not evident from the known xray structures.
If some regions of the protein were functionally gratuitous, they would presumably have been eliminated by deletions eons ago. But even if the frequency of indel fixation is normalized for chain length and unselected loops, the rate still seems high. In part this may be attributed to very early duplication and divergence of sulfatases billions of years ago, in contrast to other vertebrate gene families which expanded far more recently.
Overlap of ARSK transcription start with TTC37
Recent papers establish that the transcription factor TFEB regulates the expression of lysosomal proteins (eg ARSK) via a CLEAR element in their promotor. These elements reportedly occur in the ARSK promotor-sequence at position -272 (TTCACGTGAC), -296 (CGCATGCGCC) and -348 (CCCACCTGGA).
These motifs are fairly short, so their non-accidental occurence requires verification via conservation with comparative genomics as they cannot plausibly have arisen in human. Indeed these motifs are deeply conserved in vertebrates as is quickly seen from the 46-way alignment of vertebrate genomes at UCSC. The phastCons track already identifies their conservation as a statistically significant occurence on a genomewide scale.
Outside vertebrates, the nearest species with ARSK is Branchiostoma. The gene there is not longer syntenic with respect to TTC37. (This is also the case for Saccoglossus.) None of the motifs occurs exactly; one fragmentary match to the motif closest to the start codon is not statistically significant. The TFEB itself seems not to have a strict ortholog here either. Overall the prospects are not bioinformatically favorable for tracing the origin of these regulatory motifs.
Note the motif CCCACCTGGA is not quite right as the last nucleotide does not belong in the motif and similarly for the other two -- it is unlikely that TFEB has changed its binding specificity only in humans or great apes. These motifs are better represented by profiles than by absolute sequence requirements.
The three motif sequences are non-palindromic so consequently are not applicable to TTC37 itself which lies on the opposite strand from ARSK -- even though one of the motifs lies within a 5'UTR exon of the TTC37 gene and another immediately precedes its transcription start. It is not known if TTC37 has upstream regulatory regions of its own and where these lie relative to the start of ARSK. Thus the two genes are not able to evolve completely independently but the constraints may not be too severe.
Metazoan genes are not organized into transcriptional unit operons as in bacteria. They may be coordinately regulated but this is rarely accomplished via physical proximity on a chromosome. Although normal function of TTC37 is not entirely clear from its disease phenotype and the natural substrate of ARSK sulfatase has not been established, it does not appear the two genes have coordinated expression or related functions. Their proximity and divergent transcription from the short shared intergenic region may simply have arisen from a chromosomal reorganization in early vertebrates. Now the genes are slightly intertwined meaning their separation from a further chromosomal rearrangement could be disadvantageous, causing the accidental adjacency to be conserved.
Alignment of diverse ARSK sequences
ARSK, IDS, and choline sulfatase reference sequences
Only a sampler of vertebrate ARSK sequences are shown. The gene is present and well conserved in all 46 vertebrate genomes sequenced to date (reference sequences are pre-compiled at the proteinFasta link of the UCSC description page for ARSK). An ARSK ortholog is absent in all 30 sequenced non-deuterostome bilaterans as well as both cnidarian genomes, trichoplax and sponge genomes, and numerous unicellular eukaryote genomes yet unmistakably present in two copies in choanoflagellate. Not all these genome assemblies have complete coverage but it is unlikely that all of a large 8 exon gene with conventional autosomal location (in vertebrates) would lack coverage so consistently. It will be possible to check a ctenophore genome shortly.
>ARSK_homSap Homo sapiens (human) 544 aa 8 exons 0 MLLLWVSVVAALALAVLAPGAGEQRRRAAKAPNVVLVVSDSF 0 0 DGRLTFHPGSQVVKLPFINFMKTRGTSFLNAYTNSPICCPSRA 1 2 AMWSGLFTHLTESWNNFKGLDPNYTTWMDVMERHGYRTQKFGKLDYTSGHHSIS 2 1 NRVEAWTRDVAFLLRQEGRPMVNLIRNRTKVRVMERDWQNTDKAVNWLRKEAINYTEPFVIYLGLNLPHPYPSPSSGENFGSSTFHTSLYWLEK 00 VSHDAIKIPKWSPLSEMHPVDYYSSYTKNCTGRFTKKEIKNIRAFYYAMCAETDAML 1 2 GEIILALHQLDLLQKTIVIYSSDHGELAMEHRQFYKMSMYEASAHVPLLMMGPGIKAGLQVSNVVSLVDIYPTML 1 2 DIAGIPLPQNLSGYSLLPLSSETFKNEHKVKNLHPPWILSEFHGCNVNASTYMLRTNHWKYIAYSDGASILPQLF 1 2 DLSSDPDELTNVAVKFPEITYSLDQKLHSIINYPKVSASVHQYNKEQFIKWKQSIGQNYSNVIANLRWHQDWQKEPRKYENAIDQWLKTHMNPRAV* 0 >ARSK_canFam Canis familiarus (dog) NM_001048117 0 MLLLWLSVFAASALAAPDRGAGGRRRGAAGGWPGAPNVVLVVSDSF 0 0 DGRLTFYPGSQAVKLPFINLMKAHGTSFLNAYTNSPICCPSRA 1 2 AMWSGLFTHLTESWNNFKGLDPNYTTWMDIMEKHGYRTQKFGKLDYTSGHHSIS 2 1 NRVEAWTRDVAFLLRQEGRPMINLIPKKTKVRVMEGDWKNTDRAVNWLRKEASNSTQPFVLYLGLNLPHPYPSPSSGENFGSSTFHTSLYWLKK 00 VSYDAIKIPKWSPLSEMHPVDYYSSYTKNCTGKFTKKEIKNIRAFYYAMCAETDAML 1 2 GEIILALRQLDLLQNTIVIYTSDHGELAMEHRQFYKMSMYEASAHIPLLMMGPGIKANQQVSNVVSLVDIYPTML 1 2 DIAGAPLPQNLSGYSLLPLSSEMFWNEHKLKNLHPPWILSEFHGCNVNASTYMLRTNQWKYIAYSDGTSVLPQLF 1 2 DLFSDPDELTNIATKFPEVTYSLDQKLRSIINYPKVSASVHQYNKEQFIKWKQSVGQNYSNVIANLRWHQDWLKEPRKYESAINQWLKTPH* 0 >ARSK_monDom Monodelphis domestica (opossum) XM_001364779 (wrong N-terminus) 0 MPWWSLGVVLMVTTSADLALTAPALWAGGLEERGGPPNVVLVMSDSF 0 0 DGRLTFHPGNQTVALPFINFMKKRGTLFLNAYTNSPICCPSRA 1 2 AMWSGLFTHLTESWNNFKGLDQNYTTWMDLLQKYGYHTQKFGKLDYTSGHHSIS 2 1 NRVEAWTRDVDFLLRQEGRPMVNLIPNKMKTRIMEEDWQNTDKATNWLRKEAINFTQPFVLYLGLNLPHPYPSPYMGENFGASTFQTSPYWLER 00 VFYKAIKIPEWSPLSEMHPVDYYSSYTKNCTGQFTKKEIRDIRAFYYAMCAETDAML 1 2 GEIILTLHQLSLLQKTIVLFTSDHGELAMDHRQFYKMSMYEASSHIPLVMMGPGIKANLHIPDIVSLVDIYPTLL 1 2 DIAGIPLHQNLSGYSLIPLTSEAANNNSPAAMQRPPWILSEFHGCNVNASTYMLRIDKWKYIAYSDGISSPPQLF 1 2 DLSSDPDELTNIATRFPEITLSLDQKLRSIINYPRVSASVHQYNKRQFISWKDSLGQNYTEVIANLRWHQDWLKEPLKYENAINQWLKTNTNM* 0 >ARSK_galGal Gallus gallus (chicken) NM_001031415 0 MGSGGPLLLLRGLLLVGAAYCAAPRPPRHSSRPNVLLVACDSF 0 0 DGRLTFYPGNQTVDLPFINFMKRHGSVFLNAYTNSPICCPSRA 1 2 AMWSGLFTHLTESWNNFKGLDPDYVTWMDLMQKHGYYTQKYGKLDYTSGHHSVS 2 1 NRVEAWTRDVEFLLRQEGRPKVNLTGDRRHVRVMKTDWQVTDKAVTWIKKEAVNLTQPFALYLGLNLPHPYPSPYAGENFGSSTFLTSPYWLEK 00 VKYEAIKIPTWTALSEMHPVDYYSSYTKNCTGEFTKQEVRRIRAFYYAMCAETDAML 1 2 GEIISALQDTDLLKKTIIMFTSDHGELAMEHRQFYKMSMYEGSSHVPLLVMGPGIRKQQQVSAVVSLVDIYPTML 1 2 DLARIPVLQNLSGYSLLPLLLEKAEDEVPRRGPRPSWVLSEFHGCNVNASTYMLRTDQWKYITYSDGVSVPPQLF 1 2 DLSADPDELTNVAIKFPETVQSLDKILRSIVNYPKVSSTVQNYNKKQFISWKQSLGQNYSNVIANLRWHQDWLKEPKKYEDAIDRWLSQREQRK* 0 >ARSK_xenLae Xenopus laevis (frog) 0 MIQKCIALSLFLFSALPEDNIVRALSLSPNNPKSNVVMVMSDAF 0 0 DGRLTLLPENGLVSLPYINFMKKHGALFLNAYTNSPICCPSRA 1 2 AMWSGLFPHLTESWNNYKCLDSDYPTWMDIVEKNGYVTQRLGKQDYKSGSHSLS 2 1 NRVEAWTRDVPFLLRQEGRPCANLTGNKTQTRVMALDWKNVDTATAWIQKAAQNHSQPFFLYLGLNLPHPYPSETMGENFGSSTFLTSPYWLQK 00 VPYKNVTIPKWKPLQSMHPVDYYSSYTKNCTAPFTEQEIRDIRAYYYAMCAEADGLL 1 2 GEIISALNDTGLLGRTYVVFTSDHGELAMEHRQFYKMSMYEGSSHIPLLIMGPRISPGQQISTVVSLVDLYPTML 1 2 EIAGVQIPQNISGYSLMPLLSASSNKNVSPSISVHPNWAMSEFHGSDANASTYMLWDNYWKYVAYADGDSVAPQLF 1 2 DLSSDPDELTNVAGQVPEKVQEMDKKLRSIVDYPKVSASVHVYNKQQFALWKASVGANYTNVIANLRWHADWNKRPRAYEMAIEKWIKSTRQH* 0 >ARSK_takRub Takifugu rubripes (fugu) 8 exons 504 aa single copy retained after whole genome duplication 0 MSVKLSALILLFLAFHQVLARNRTRPNFLVVMSDAF 0 0 DGRLTFDPGSKVVKLPFINYLRELGVTFINAYTNSPICCPSRA 1 2 AMWSGQFVHLTQSWNNYKCLDANATTWMDLLEVNGYLTKMMGKLDYTSGSHSvs 0 1 NRVEAWTRDVQFLLRQEGRPVTQLVGNMSTVRIMGKDWENIDKATQWIQQRAESSQQPFALYLGLNLPHPYKTESLGPTAGGSTFRTSPHWLEK 00 VSSEHVTVPKWLPGAAMHPVDFYSTFTKNCSGFFTEEEIMNIRAFYYAMCAEADAML 1 2 GQLISALRETHLLNNTVVIFTADHGELAMEHRQFYKMSMFEGSSHVPLLFMGPGLMSGVEADQLVSLVDIYPTVL 1 2 DLADVPPVGSLSGYSLLPLLSTCSSCPGRPHPDWVLSEYHGCNANASTYMLRSGRWKYIAYADGLRVPPQLF 1 2 DMILDKEELHNVVFKFSEVSAQLDKLLRSIVHYPEVSAAVHRYNKESFVAWRHTLGRNYSQVISSLRWHVDWQRNPLANERAIDEWLYGSF* 0 >ARSK_braFlo Branchiostoma floridae (amphioxus) XM_002594507 0 MRMKLDCSAGFLLFWWFTSAVGGTRDDRKNIVFVICDSM 0 0 DGRLIGRGQDSVVDLPNLNYMVQNGVNFRSTYTNSPICCPSRS 1 2 ALWSGLHTHVTQSWNNYKGLPKNYPTWQVRLEQQGYHTQVYGKTDYVSGDHSES 2 1 NRVEAWTRNVNFTLAQEGRPTPVLV 12 GSSSTDRIQLKDWASTDLASHWLLHEAPKQQKPWLLYLGLNLPHPYPTPSMGKNFGGSTFMTSPYWLKK VNSSKVTIPKWLPFSRMHPVDYYSSATKNCTSDFTRDEIMKIREYYYGMCAETDAML 1 2 GQVLDALKASGQADSTYVFFTSDHGELAMEHRQFYKMSMYEASAHIPMVLTGPEVPAGKAVDDLTSLVDVFPTFM 1 2 DIANASQPPGLNGTSLLPLLRNSSDRVDRPDWVLSQYHGCNVNMSTYMLRTGSLKYVAFGDGPNQVSSQLF 1 2 DLDKDPDELHNLAEERQDLASQLDDKLRKLVDYPTVTREVQKYNRDSFMAWKAKLGSRYKDEIANLRWWKDWQKDPQGNQEKVEEWLNNVVS* 0 >ARSK_sacKow Saccoglossus kowalevskii (acornworm) XM_002732823 0 MFSMMQSSILITVLLFTCTCIPRGNEGKPNNVLFIICDAM 0 0 DGRLVGNNLTAVNMANINNRLVSHGVTFTNAYTNSPICCPSRS 1 2 ALWSGLYTHITHAWNNHEGLPADYPTWKIKLEKAGYDSKILGKTDYVSGRHTLS 2 1 NRVEAWTRNVNFTLAQEGRPTPVLVGNKTTIRVKDVDWDNIDKAKDWLENRKSSKATKPFLLYIGINLPHPYSTPGEGEHPGGSTFMTSPYW LQYVDMSKVTIPKWTPLDKMHPVDYYESATKNCTSHFTKDEIRKIRAYYYGMCAEVDGMV 1 2 GEILDQLDSLGLTNTTQVIFTSDHGEMAMEHRQFYKMTMYEASSHVPLIITNPTVPSRQGVAVNDPVSLVDIFPTLM 1 2 DMAAIHHPVGLNGTSLMPYLEGKSHVKKPDWVLSQYHGCNVNMSAYMLRRQEWKYITYGNGKQVAPHLF 1 2 NLDEDPDELHDYANERHDIIAEMDNKLRSIIDYADITNEVSRYNKESFSSWKTSIGDKYSDTIANLRWWKDWQKDPNGNEQRIEEWLKSVE* 0 >ARSK1_monBre Monosiga brevicollis (choanoflagellate) ABFJ01000822 (XM_001747506: bad gene model) 0 MGNPIRGGSLLIVAASLLVCATLGTAKQPNILFVIDESTDAKAYFAKNPEKAPMPLPNLR 0 0 VPAHVMNSYHYHR 1 2 PPVCCPSRTSTWSGRH 0 0 FVTGAWNNYEGLPE 0 0 NYDLKYSDVLHKGGYNVGIFGKTDFTAGGHTVDARVTAWTNKVNFPFTLQNGSAGWYDETGPLVRTVNVSK 0 0 VVHVSDWNHANQTAKFIADAATHDEPWLAYVGFDIVHP 1 2 NYVSSPYWLDQVDMDKVTVPEWIPLDQLHPEDFQATMKKNMANLTHDPAFIKSVRQHYYGMIAE 2 1 YDAILGVVLDAVEASGEADNTY 0 0 IFVTSDHGDMNMEHQQYYK 0 0 MTYYDPSARVPLIVTGPTVQANVTYENLTSHLDFFPTFLELANV 1 2 VQLEGRSLVPILRTGVDAGRPNVALSQFHGDEIHLSWFMI 1 2 RKDDYKYVTFGSGKEVAPRLFNMREDPLEMNDLAPSNPSLVAELDAELRSYWDYPSIASTAESYNK 1 2 DSFALLRASFNDEDKFKAYLATLRWSTSWSYDPEGSYAAIEAWLKTPNSTFEWAFP* 0 >ARSK2_monBre Monosiga brevicollis (choanoflagellate) ABFJ01001665 (XM_001750805: horrible gene model) 0 MDRWTIVLVAVAIWCLAVGSHGLGSAAEPESRLRLGMTNSSRPNIVFLICESIDAKTFDEDSPVPLPNIRKLIQ 0 0 GGVSFKTHYVSAPVCAPSRTSIQQGRHVH 1 1 AAIAWNNYEGMAPDYDMKIGDVLGRTGYDVNILGKTDWT 1 IGGHALWNWWQCFTM 2 1 YTQFPYNVTNGGWNEQPETQAGE 1 2 GDVTPGNRSHDVDWMFVEQNVAYIRNHSQSQPFFVYQ 0 0 GMDIVHPP 0 0 LGMPSDQTCEKFYNMINESDVTVPDWAPLDELHPCDLQSVMLK 1 2 DNATAVTNFYSKDRRRRVRR 2 1 IYYAMIAEFDAMVGEYMQAVEDA 1 2 GLPLCKKIKLREDVKSLSGLPFQMEHQQFYKM 0 0 VPLVIAGPGIKADTETLPTQHVDLYPTFMDF 1 2 GQVPASMRPEGLDGISLVPRVVEQKPLANTSFAISQFHGADLGMSWYLIRYQ 0 0 NWKLVTYGTGQEVAPQLFDMVNDPGETHDVHAQHPDLVAQLDALLRSRIDYPSVSLDVATYNL 1 2 APAKKQFK 0 0 AIHDYTQQGDDELSFVPGDIITLVSVPPGEEIEGWLTGELNGRTGLFPDNFVEELPYVTCLAIFFSIPMFLFGCRHTSRPRTP* 0
>IDS_homSap MPPPRTGRGLLWLGLVLSSVCVALGSETQANSTTDALNVLLIIVDDLRPSLGCYGDKLVRSPNIDQLASHSLLFQNAFAQQAVCAPSRVSFLTGRRPDTT RLYDFNSYWRVHAGNFSTIPQYFKENGYVTMSVGKVFHPGISSNHTDDSPYSWSFPPYHPSSEKYENTKTCRGPDGELHANLLCPVDVLDVPEGTLPDKQ STEQAIQLLEKMKTSASPFFLAVGYHKPHIPFRYPKEFQKLYPLENITLAPDPEVPDGLPPVAYNPWMDIRQREDVQALNISVPYGPIPVDFQRKIRQSY FASVSYLDTQVGRLLSALDDLQLANSTIIAFTSDHGWALGEHGEWAKYSNFDVATHVPLIFYVPGRTASLPEAGEKLFPYLDPFDSASQLMEPGRQSMDL VELVSLFPTLAGLAGLQVPPRCPVPSFHVELCREGKNLLKHFRFRDLEEDPYLPGNPRELIAYSQYPRPSDIPQWNSDKPSLKDIKIMGYSIRTIDYRYT VWVGFNPDEFLANFSDIHAGELYFVDSDPLQDHNMYNDSQGGDLFQLLMP >IDS_canFam MPPGGWCLLCFGLVLSSVCASAESAAPSNLTTAPLNVLLIIVDDLRPSLGCYGDKLVRSPNIDQLASHSLLFQNAFAQQAVCAPSRVSFLTGRRPDTTRL YDFNSYWRVHAGNFSTLPQYFKENGYVTMSVGKVFHPGISSNYSDDSPYSWSIPPYHPSSEKYENTKTCRGPDGELHANLLCPVDIADVPEGTLPDKQST EQAIRLLEKTKTSTRPFFLAVGYHKPHIPFRYPKEFQKLYPLENITLAPDPEVPAGLPPVAYNPWMDIRQREDVQALNLSVPYGPIPVDFQRKIRQSYFA SISYLDTQVGHLLSALDDLQLANSTIIVFASDHGWALGEHGEWAKYSNFDITTRVPLMFYVPGRTAPLPEAGEKLFPYIDPFSSVQELMEPGRQVTDLVE LLSLSPTLAGLAGLHVPPRCPVPSFHVELCREGQNLMKHFQVEDVEGDPHLRGNPRESIAYSQYPRPADSPQWNSDKPSLKDIKVMGYSIRTIDYRYTVW VGFSPHEFLANFSDVHAGELYFVDSDPLQDHNMYNDSQGKDLLRALMPF >IDS_monDom Monodelphis domestica XM_001376164 MPNLGPWCLGLTLSLAFVPPLLSAPTTEGPGYRKRENPLDLVGVDYIVVDDLRPALGCYGEVLVKSPNIDQLASRSVVFQNAFAQQAVCAPSRVSFLTGR RPDTTRLYDFNSYWRVHSGNYSTIPQYFKENGYVTLSVGKVFHPGISSNHSDDFPYSWSVPPFHPSSEQYENSKTCKGQDEELHANLICPVDVADMPEGT LPDKQSTEEAIRLLEKMKRVDDLFFLAVGYHKPHIPFRYPKEFQKLYPLENITLAPDPHIPFGLPPVAYNPWMDIREREDVQALNISVPYGPIPAEFQRK IRQSYFASVSYLDSQVGHLLNALDELQLSNNTIVAFVSDHGWALGEHGEWAKYSNFDVATRVPLMFYVPGRTASFTSPGQKLFPYIDPFDSPSHVKVPGR RATELVELVSLFPTLSELAGLNIPPRCPFESFNIELCVEGPSLVRYLNFTEWEEDFFYSTRKPLELVAYSQYPRPADTPQWNSDKPHLKDIKIMGYSIRT VDYRFTVWVSFNPENFTADFTNIHAGELYFVDSDPLQDHNVYNQTVGIY >IDS_galGal MASCAAFALSSLAAAVPRLRTRRTAGPGDGMNVLFIVVDDLRPVLGCYGDNLVKSPNIDQLASQSIVFSNAYAQQAVCAPSRVSFLTGRRPDTTRLYDFY SYWRVHSGNYSTMPQYFKENGYVTMSVGKVFHPGISSNYSDDYPYSWSIPPFHPSTEKYENDKTCRGKDGRLYANLVCPIDVTEMPGGTLPDIETTEEAI RLLNVMKTKKQKFFLAVGYHKPHIPLRYPQEFLKLYPLENITLAPDPWVPEKLPPVAYNPWVDIRQRDDVKALNVTFPYGPLPDDFQRLIRQSYYAAVSY LDMQVGLLLNALDYVGLSNSTIVVFTADHGWSLGEHGEWAKYSNFDVATQVPLMFYVPRMTTSSASQGERVFPYLDPFSHIVGLVPQGQRKKMVELVSLF STLAELAGLQVPPACPETSFHVALCTEGASIVRYFKSSEQKVQKKENGCNDTNKYYSEEPVAFSQYPRPADTPQWDSDKPKLKDIRIMGYSMRTIDYRYT VWVQFNPENFSADFEDVHAGELYMMETDPNQDNNIYNNTLHGHLFKKIVDFLKH >IDS_xenTro MNLFGYLRFLMCATTVFAVWQQHFLPKHTATGGKNVLIIIADDLRTSLGCYGDSAVKSPNIDHLASQSIIFTNAYAQQAVCAPSRVSFLTGRRPDTTRLF DFNSYWRTHAGNYTTLPQYFKEHGYVTMSVGKIFHPGISSNHSDDYPYSWSVYPYHPSAEKYENSQTCKGKDGKLHANLVCPVDVSEVPEGTLPDIQSTE EAIRLLKTVKQQNASFFLAVGYHKPHIPFRFPKEFLKLYPIENISLAPDPDIPKKLPLVAYNPWTDIRKREDVQALNISFPYGPIPEHFQLLIRQSYYAS VSYLDDQIGQLLNAVEDLGLSNDTIIVFSSDHGWSLGEHGEWAKYSNFDVTTRVPLIFYVPGMTNIPQQPIFQYIDPFSTNLQRKFPGKSREYPVELVSL FSTIADLAKLPAPPACPQPSFHMELCTEGRSLVHQLHASENTHDDAVLAVAYSSYPRPSDFPQWNSDLPDLKDIKIMGYSMRTMDYRYTVWVGYNSTTFQ ANFKEIHGRELYFVLSDPNQDNNLYNQLLHLDIYKHFEFMNN >IDS_danRer6 551 chr14:22165989-22187666- MNVMLVFTCWWFVLIFHLLGRDVFAAKSKDFNVLYLIADDLRPTLGCYSDPVVKSPNIDQLASLSVVFHNAYAQQAVCGPSRVSFLTSRRPDTTKLYDFN SYWRVHAGNYTTLPQYFKSNGYTTLSVGKVFHPGIASNHSDDYPYSWSVPPYHPPSFEYEKRKVCKDKDGTLHSNLLCPVNVSEMPLGTLPDIENTEEAI RLLRSMKGSQKPFFLAVGFYKPHIPFRIPQEYLKLYPIENMTLAPDPDVPKKLPDVAYNPWTDIRKREDVQALNLSFPYGPIPKDFQLRIRQHYFASVSY VDAQVGKILQTLDDVGLAKNTIVVLSSDHGWSLGEHGEWAKYSNFDVATRVPLMVYKAGVSSRRSRTGAKTFPFIDVFQDTREHFGKGKIVNSVVELLDV FPTLANLAGLPSVHHCPSPSFKMDLCTEGSNLANLIRNPKHLNREAYSFSQYPRPSDSIQENSDLPNLADIRIMGYSIRSNDYRYTLWVGFDPLHCKPNM TEIHAGELYILTEDPGQDNNLFDEFGHAALLNKFGTMPSWTESLKQHMMYFSSGLKSKGLS >IDS_braFlo Branchiostoma floridae (XM_002611665: flawed) BW796857 MKMRVTSATVATCLLFLQSCAAVLKNGAGESPNVLFLVIDDLRPALGCYGYQNVITPNIDQLAAKGIKFNNAFVQQAVCGPSRTSFLTGRRPDTTRLYDF YSYWRTAAGNFTTLPQHFKESGYFTASVGKVFHPGGISSNFSDDAPYSWSVPAYHPPTQKFKMKKVCPGPDGQLHMNLVCPVDVKSQPLGSLPDIQSADY AVEFLQNVSASSQTSPKQPFFLAVGFHKPHIPFKYPREFQDLYPLFNIHLAPNLSLPPDLPTIAWNPFTDIRKREDVKALNISFPYGPVPRKFQLLMRQG YYAATSYTDSQVGRVLAALDEQGLATNTIVVLVGDHGWSLGEHQEWAKYSNFEVATRVPLILYVPGVTHQPVRGDSTFPYIDALESCINEIPNHQTLPEE GHESDALVELVDIFPTLAEMANLRTPPLCPTDSSKVELCTEGSSFVPVILNVTGGTSRQNIVTSWKPAVFSQYPRPSEQPQINSDLPHLKDIQYMGYSMR TEQYRYTEWVAFNPDTFKPDFDLVAARELYLHDTDELEDHNVAGKSEYRHLLTQLSQQLRKGWRNALPSQ >IDS_sacKow Saccoglossus kowalevskii XM_002733076 MLMNTLVFQLFRLVAFSTCIALVSALLDGTTGTRRASKLNVLFIVVDDLRPALGCYDNVTQYFTPNIDQLAANSIKFTNAHVQQALCAPSRASFLTGRRP DTTRIYDLNSYWRSLGGNFTTLPQHFKENGYYAASVGKVFHPGISSNYTDDYPYSWSVPAFHPSTQKYKMKKVCPGPDGNLHMNLICPVDVKTQPEASLP DIQSTEYAIELLRNISQQQQQQTKGSQPFFLAVGYHKPHIPLKYPKEFRDLYPLSSIKAPTNPDYPKKLPHVAWDPWTDVRRRDDIKALNVSFPYGPMPK HYQLLIRQSYYASTTYVDNLVGYLLSSLEKYGFAENTVITFVGDHGWALGEHQEWAKYSNFDVATRVPLLMYIPGVTDKKDQEGSETEDINIFKSKTTVT MFDHSDLKSGRLVCNNHVELVDIFPTLTDICGITMPPLCPKNPTEVRLCTEGISLSPLIEQISTNDTLADFKWKKAVFTQYPRPSDEPQENSDSPILKDI TIMGYSMVTDKYRYTEWIGFNNVQCQGNWDDVHARELYKLRSDKMENNNVANDAQYKELTQKLANLLRKGWRHALP >IDS_monBre Monosiga brevicollis XM_001743372 mAFQSRGPNLLPDRAGEIIGVALSSLPLDGLNVLLIVVDDMRAELGTYGATHMITPHLDALAQDGMVFERAYVAISLCMPSRTAFLTSRRPATTHNFVIA PNEQWRQTKGPNATTLPEFFKTVGGYRTYGMGKIFHGTTDEPYSWSAEMGDYYDWDNWTQYGNSMTYKCFDVPDNNLGDGIFADRAVNWINMFGADQANG SDTRPFFMGVGFHRPHIPYLVPKRYCDMYPPADEIPLAANPFKPEGMPDVAYSVSAGLRNFQDCAPLFENVSKCYDDPSWAFSNRVRRNYWAAISYIDAQ VGRIVQALKDNNLYDNTIVLFMGDHGVCTCTGRSTNFEHGTRIPLIIRDPSHTPARTAALVETVDIYPTLVDLAGLPSLETCAPGSMAALCTEGFSMRPL FTDPTRAWKSAAFSQYARPAPSPDNGFPADLFSPPLHVAGHREGVMGFTIRTNTYRYTNWVWFDPASATPHWNMSWGEELYNHTAQPVPDGLFNNENINL IDQPGLEPIIDKLRQALQAGWRAALPS
>3ED4_escCol Escherichia coli MSLASLIGLAVCTGNAFSPALAAEAKQPNLVIIMADDLGYGDLATYGHQIVKTPNIDRLAQEGVKFTDYYAPAPLSSPSRAGLLTGRMPFRTGIRSWIPS GKDVALGRNELTIANLLKAQGYDTAMMGKLHLNAGGDRTDQPQAQDMGFDYSLANTAGFVTDATLDNAKERPRYGMVYPTGWLRNGQPTPRADKMSGEYV SSEVVNWLDNKKDSKPFFLYVAFTEVHSPLASPKKYLDMYSQYMSAYQKQHPDLFYGDWADKPWRGVGEYYANISYLDAQVGKVLDKIKAMGEEDNTIVI FTSDNGPVTREARKVYELNLAGETDGLRGRKDNLWEGGIRVPAIIKYGKHLPQGMVSDTPVYGLDWMPTLAKMMNFKLPTDRTFDGESLVPVLEQKALKR EKPLIFGIDMPFQDDPTDEWAIRDGDWKMIIDRNNKPKYLYNLKSDRYETLNLIGKKPDIEKQMYGKFLKYKTDIDNDSLMKARGDKPEAVTWGEGHHHH >3B5Q_bacThe Bacteroides thetaiotaomicron 2.40A resolution GLALCGAAAQAQEKPNFLIIQCDHLTQRVVGAYGQTQGCTLPIDEVASRGVIFSNAYVGPLSQPSRAALWSGHQTNVRSNSSEPVNTRLPENVPTL GSLFSESGYEAVHFGKTHDXGSLRGFKHKEPVAKPFTDPEFPVNNDSFLDVGTCEDAVAYLSNPPKEPFICIADFQNPHNICGFIGENAGVHTDRPISGP LPELPDNFDVEDWSNIPTPVQYICCSHRRXTQAAHWNEENYRHYIAAFQHYTKXVSKQVDSVLKALYSTPAGRNTIVVIXADHGDGXASHRXVTKHISFY DEXTNVPFIFAGPGIKQQKKPVDHLLTQPTLDLLPTLCDLAGIAVPAEKAGISLAPTLRGEKQKKSHPYVVSEWHSEYEYVTTPGRXVRGPRYKYTHYLE GNGEELYDXKKDPGERKNLAKDPKYSKILAEHRALLDDYITRSKDDYRSLKVDADPRCRNHTPGYPSHEGPGAREILKRK >CHOS_edwTar Edwardsiella tarda Choline-sulfatase DM42793 MSLSRREFLQRTAGGMAGVALGAPALAAGDAPAGTDTGAKMPPRNIVIITADQLARRGVGGYGNPQVNTPAIDSLIARGTRFEQAYCPYPLCAPSRACYW TGRLPHQTGVIANDSPNVPQDMVTLGELFSQAGYECRHFGKRHDYGALKGFTCADQVELPYDSPAAYPVDYDTREDVYCLQESLKYIDTLKGRDSDAPFM LAIEFNNPHNINGWTGAFAGPHGDIDGLGPLPPLLDNFDTSADLPNRPLAIQYACCTHNRVMQAANWNELNFRQYLKAYYHFTELADGFIGQVLSALRAS GHADDTLVVFFADHGDAMGAHRLVAKMNWFYEESTNVPLVFAGPGIRPQASSRHLTSLCDLLPTLCDYAGLTPPPGLYGRSLMPILRGEQPDGWRDEVIT QWNTDRNVDVQPARMLRTERYKYILYKENEEEELYDLQQDPGETRNLAHSPAHQAERQALRARFDEYVRNQVDPFYSQEAIIDRRWRSHLPGYHNHQGQT SIQVYQKEIRPLIMNKEFEKAREVRLALYRQARASYNGGV >CHOS_ruePom Ruegeria pomeroyi choline sulfatase YP_166053 MTNHPNLLVIVSDEHRKDAMGCAGHPIVKTPNLDALAARGTMFEAAYTPSPMCVPTRAALATGDWIHRTGHWDSATPYAGQPRSWMHDLRDAGREVVSIG KLHFRATEDDNGFSQEILPMHVVGGIGWTVGLLRKNPPAYEAAAELAADVGVGASSYTDYDRAITAAAEAWLADPARQERPWAAFVSLVSPHYPLTCPEE WFALYDPDQMDLPVGYGQGLPDHAELRNIGGFFNYDAYFDAQKMREAKAAYYGLTSFMDDCVGRVLAALEAGGKADNTVVLYVSDHGDMMGDQGFWTKQV MYEASAGVPMIAAGPGIPAGHRVSTCTSLTDIAATARELCGLAAREDLPGLSLRSIATAPDDPDRAGFSEYHDGGSRTGTFMLRWGRWKYVHYVGEAPQL FDLERDPQELTDLAPRAAEDPDMRALLAEGEHRLRAICNPETVNARAFADQQRRIAELGGEEACRTGYSFNHTPVPQEGGAL